Abstract
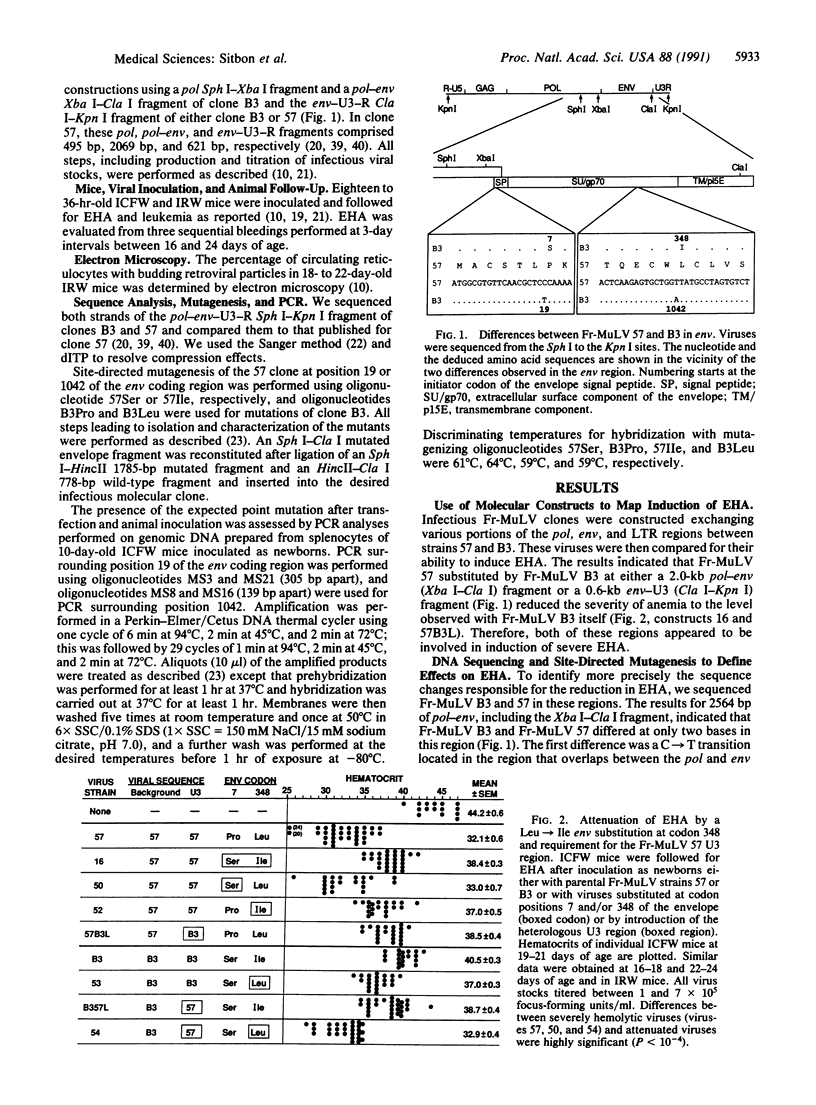
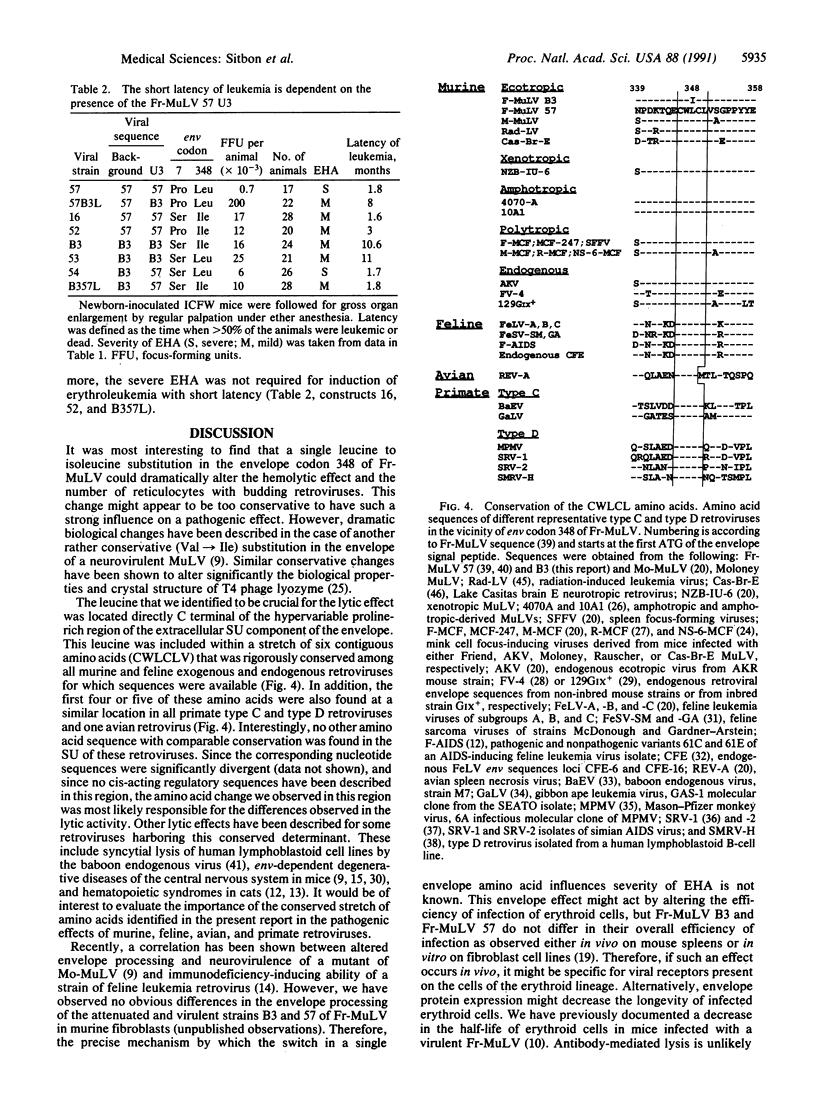
Friend murine leukemia virus is a replication-competent retrovirus that contains no oncogene and that exerts lytic and leukemogenic properties. Thus, newborn mice inoculated with Friend murine leukemia virus develop severe early hemolytic anemia before appearance of erythroleukemia. To identify the retroviral determinants regulating these effects, we used chimeric infectious constructions and site-directed point mutations between a virulent Friend murine leukemia virus strain and a naturally occurring variant attenuated in lytic and leukemogenic effects. We found that severe hemolytic anemia was always associated with higher numbers of blood reticulocytes with budding retroviral particles. Furthermore, a remarkably conservative leucine to isoleucine change in the extracellular SU component of the retroviral envelope was sufficient to attenuate this lytic effect. Also, this leucine at position 348 of the envelope precursor protein was located within the only stretch of five amino acids that is conserved in the extracellular SU component of all murine, feline, and primate type C and type D retroviral envelopes. This observation suggested an important structural function for this yet undescribed conserved sequence of the envelope. Lastly, we observed that lytic and leukemogenic effects were attenuated by a deletion of a second repeat in the transcriptional enhancer region of the viral long terminal repeats of the variant strain.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alber T., Sun D. P., Wilson K., Wozniak J. A., Cook S. P., Matthews B. W. Contributions of hydrogen bonds of Thr 157 to the thermodynamic stability of phage T4 lysozyme. Nature. 1987 Nov 5;330(6143):41–46. doi: 10.1038/330041a0. [DOI] [PubMed] [Google Scholar]
- Chatis P. A., Holland C. A., Hartley J. W., Rowe W. P., Hopkins N. Role for the 3' end of the genome in determining disease specificity of Friend and Moloney murine leukemia viruses. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4408–4411. doi: 10.1073/pnas.80.14.4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatis P. A., Holland C. A., Silver J. E., Frederickson T. N., Hopkins N., Hartley J. W. A 3' end fragment encompassing the transcriptional enhancers of nondefective Friend virus confers erythroleukemogenicity on Moloney leukemia virus. J Virol. 1984 Oct;52(1):248–254. doi: 10.1128/jvi.52.1.248-254.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S. K., Baroudy B. M., Holmes K. L., Fredrickson T. N., Lander M. R., Morse H. C., 3rd, Hartley J. W. Biologic and molecular genetic characteristics of a unique MCF virus that is highly leukemogenic in ecotropic virus-negative mice. Virology. 1989 Jan;168(1):90–100. doi: 10.1016/0042-6822(89)90407-8. [DOI] [PubMed] [Google Scholar]
- Clark S. P., Kaufhold R., Chan A., Mak T. W. Comparison of the transcriptional properties of the Friend and Moloney retrovirus long terminal repeats: importance of tandem duplications and of the core enhancer sequence. Virology. 1985 Jul 30;144(2):481–494. doi: 10.1016/0042-6822(85)90288-0. [DOI] [PubMed] [Google Scholar]
- Colleaux L., d'Auriol L., Betermier M., Cottarel G., Jacquier A., Galibert F., Dujon B. Universal code equivalent of a yeast mitochondrial intron reading frame is expressed into E. coli as a specific double strand endonuclease. Cell. 1986 Feb 28;44(4):521–533. doi: 10.1016/0092-8674(86)90262-x. [DOI] [PubMed] [Google Scholar]
- Delassus S., Sonigo P., Wain-Hobson S. Genetic organization of gibbon ape leukemia virus. Virology. 1989 Nov;173(1):205–213. doi: 10.1016/0042-6822(89)90236-5. [DOI] [PubMed] [Google Scholar]
- DesGroseillers L., Jolicoeur P. Mapping the viral sequences conferring leukemogenicity and disease specificity in Moloney and amphotropic murine leukemia viruses. J Virol. 1984 Nov;52(2):448–456. doi: 10.1128/jvi.52.2.448-456.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golemis E. A., Speck N. A., Hopkins N. Alignment of U3 region sequences of mammalian type C viruses: identification of highly conserved motifs and implications for enhancer design. J Virol. 1990 Feb;64(2):534–542. doi: 10.1128/jvi.64.2.534-542.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilhot S., Hampe A., D'Auriol L., Galibert F. Nucleotide sequence analysis of the LTRs and env genes of SM-FeSV and GA-FeSV. Virology. 1987 Nov;161(1):252–258. doi: 10.1016/0042-6822(87)90194-2. [DOI] [PubMed] [Google Scholar]
- Holland C. A., Thomas C. Y., Chattopadhyay S. K., Koehne C., O'Donnell P. V. Influence of enhancer sequences on thymotropism and leukemogenicity of mink cell focus-forming viruses. J Virol. 1989 Mar;63(3):1284–1292. doi: 10.1128/jvi.63.3.1284-1292.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch W., Hunsmann G., Friedrich R. Nucleotide sequence of the envelope gene of Friend murine leukemia virus. J Virol. 1983 Jan;45(1):1–9. doi: 10.1128/jvi.45.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch W., Zimmermann W., Oliff A., Friedrich R. Molecular analysis of the envelope gene and long terminal repeat of Friend mink cell focus-inducing virus: implications for the functions of these sequences. J Virol. 1984 Mar;49(3):828–840. doi: 10.1128/jvi.49.3.828-840.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D. V., Berry B. T., Roy-Burman P. Nucleotide sequence and distinctive characteristics of the env gene of endogenous feline leukemia provirus. J Virol. 1989 May;63(5):2379–2384. doi: 10.1128/jvi.63.5.2379-2384.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz J., Celander D., Crowther R. L., Patarca R., Perkins D. W., Haseltine W. A. Determination of the leukaemogenicity of a murine retrovirus by sequences within the long terminal repeat. 1984 Mar 29-Apr 4Nature. 308(5958):467–470. doi: 10.1038/308467a0. [DOI] [PubMed] [Google Scholar]
- Levy D. E., Lerner R. A., Wilson M. C. Normal expression of polymorphic endogenous retroviral RNA containing segments identical to mink cell focus-forming virus. J Virol. 1985 Dec;56(3):691–700. doi: 10.1128/jvi.56.3.691-700.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. P., D'Andrea A. D., Lodish H. F., Baltimore D. Activation of cell growth by binding of Friend spleen focus-forming virus gp55 glycoprotein to the erythropoietin receptor. Nature. 1990 Feb 22;343(6260):762–764. doi: 10.1038/343762a0. [DOI] [PubMed] [Google Scholar]
- Li Y., Golemis E., Hartley J. W., Hopkins N. Disease specificity of nondefective Friend and Moloney murine leukemia viruses is controlled by a small number of nucleotides. J Virol. 1987 Mar;61(3):693–700. doi: 10.1128/jvi.61.3.693-700.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linemeyer D. L., Ruscetti S. K., Menke J. G., Scolnick E. M. Recovery of biologically active spleen focus-forming virus from molecularly cloned spleen focus-forming virus-pBR322 circular DNA by cotransfection with infectious type C retroviral DNA. J Virol. 1980 Sep;35(3):710–721. doi: 10.1128/jvi.35.3.710-721.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda M., Yoshikura H. Construction and characterization of the recombinant Moloney murine leukemia viruses bearing the mouse Fv-4 env gene. J Virol. 1990 Mar;64(3):1033–1043. doi: 10.1128/jvi.64.3.1033-1043.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merregaert J., Nuyten J. M., Janowski M. Nucleotide sequence of the envelope gene of radiation leukemia virus. Virology. 1985 Jul 30;144(2):457–467. doi: 10.1016/0042-6822(85)90286-7. [DOI] [PubMed] [Google Scholar]
- Oda T., Ikeda S., Watanabe S., Hatsushika M., Akiyama K., Mitsunobu F. Molecular cloning, complete nucleotide sequence, and gene structure of the provirus genome of a retrovirus produced in a human lymphoblastoid cell line. Virology. 1988 Dec;167(2):468–476. [PubMed] [Google Scholar]
- Oliff A. I., Hager G. L., Chang E. H., Scolnick E. M., Chan H. W., Lowy D. R. Transfection of molecularly cloned Friend murine leukemia virus DNA yields a highly leukemogenic helper-independent type C virus. J Virol. 1980 Jan;33(1):475–486. doi: 10.1128/jvi.33.1.475-486.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott D., Friedrich R., Rein A. Sequence analysis of amphotropic and 10A1 murine leukemia viruses: close relationship to mink cell focus-inducing viruses. J Virol. 1990 Feb;64(2):757–766. doi: 10.1128/jvi.64.2.757-766.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbaugh J., Donahue P. R., Quackenbush S. L., Hoover E. A., Mullins J. I. Molecular cloning of a feline leukemia virus that induces fatal immunodeficiency disease in cats. Science. 1988 Feb 19;239(4842):906–910. doi: 10.1126/science.2893454. [DOI] [PubMed] [Google Scholar]
- Paquette Y., Hanna Z., Savard P., Brousseau R., Robitaille Y., Jolicoeur P. Retrovirus-induced murine motor neuron disease: mapping the determinant of spongiform degeneration within the envelope gene. Proc Natl Acad Sci U S A. 1989 May;86(10):3896–3900. doi: 10.1073/pnas.86.10.3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss M. L., Mullins J. I., Hoover E. A. Posttranslational modifications distinguish the envelope glycoprotein of the immunodeficiency disease-inducing feline leukemia virus retrovirus. J Virol. 1989 Jan;63(1):189–195. doi: 10.1128/jvi.63.1.189-195.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power M. D., Marx P. A., Bryant M. L., Gardner M. B., Barr P. J., Luciw P. A. Nucleotide sequence of SRV-1, a type D simian acquired immune deficiency syndrome retrovirus. Science. 1986 Mar 28;231(4745):1567–1572. doi: 10.1126/science.3006247. [DOI] [PubMed] [Google Scholar]
- Rassart E., Nelbach L., Jolicoeur P. Cas-Br-E murine leukemia virus: sequencing of the paralytogenic region of its genome and derivation of specific probes to study its origin and the structure of its recombinant genomes in leukemic tissues. J Virol. 1986 Dec;60(3):910–919. doi: 10.1128/jvi.60.3.910-919.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick-Roguel N., Burstein H., Hamburger J., Panet A., Eldor A., Vlodavsky I., Kotler M. Cytocidal effect caused by the envelope glycoprotein of a newly isolated avian hemangioma-inducing retrovirus. J Virol. 1989 Oct;63(10):4325–4330. doi: 10.1128/jvi.63.10.4325-4330.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel N., Hoover E. A., Dornsife R. E., Mullins J. I. Pathogenic and host range determinants of the feline aplastic anemia retrovirus. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2758–2762. doi: 10.1073/pnas.85.8.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saal F., Cavalieri F., Sitbon M., Périés J. Baboon Endogenous C type Virus induces syncytium formation in Epstein barr virus (EBV) carrying lymphoblastoid human cell lines. Arch Virol. 1983;75(1-2):151–155. doi: 10.1007/BF01314137. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe A. H., Hunter J. J., Chassler P., Jaenisch R. Role of abortive retroviral infection of neurons in spongiform CNS degeneration. Nature. 1990 Jul 12;346(6280):181–183. doi: 10.1038/346181a0. [DOI] [PubMed] [Google Scholar]
- Sitbon M., Ellerbrok H., Pozo F., Nishio J., Hayes S. F., Evans L. H., Chesebro B. Sequences in the U5-gag-pol region influence early and late pathogenic effects of Friend and Moloney murine leukemia viruses. J Virol. 1990 May;64(5):2135–2140. doi: 10.1128/jvi.64.5.2135-2140.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitbon M., Evans L., Nishio J., Wehrly K., Chesebro B. Analysis of two strains of Friend murine leukemia viruses differing in ability to induce early splenomegaly: lack of relationship with generation of recombinant mink cell focus-forming viruses. J Virol. 1986 Jan;57(1):389–393. doi: 10.1128/jvi.57.1.389-393.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitbon M., Sola B., Evans L., Nishio J., Hayes S. F., Nathanson K., Garon C. F., Chesebro B. Hemolytic anemia and erythroleukemia, two distinct pathogenic effects of Friend MuLV: mapping of the effects to different regions of the viral genome. Cell. 1986 Dec 26;47(6):851–859. doi: 10.1016/0092-8674(86)90800-7. [DOI] [PubMed] [Google Scholar]
- Sodroski J., Goh W. C., Rosen C., Campbell K., Haseltine W. A. Role of the HTLV-III/LAV envelope in syncytium formation and cytopathicity. 1986 Jul 31-Aug 6Nature. 322(6078):470–474. doi: 10.1038/322470a0. [DOI] [PubMed] [Google Scholar]
- Sonigo P., Barker C., Hunter E., Wain-Hobson S. Nucleotide sequence of Mason-Pfizer monkey virus: an immunosuppressive D-type retrovirus. Cell. 1986 May 9;45(3):375–385. doi: 10.1016/0092-8674(86)90323-5. [DOI] [PubMed] [Google Scholar]
- Speck N. A., Renjifo B., Golemis E., Fredrickson T. N., Hartley J. W., Hopkins N. Mutation of the core or adjacent LVb elements of the Moloney murine leukemia virus enhancer alters disease specificity. Genes Dev. 1990 Feb;4(2):233–242. doi: 10.1101/gad.4.2.233. [DOI] [PubMed] [Google Scholar]
- Szurek P. F., Yuen P. H., Ball J. K., Wong P. K. A Val-25-to-Ile substitution in the envelope precursor polyprotein, gPr80env, is responsible for the temperature sensitivity, inefficient processing of gPr80env, and neurovirulence of ts1, a mutant of Moloney murine leukemia virus TB. J Virol. 1990 Feb;64(2):467–475. doi: 10.1128/jvi.64.2.467-475.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer R. M., Power M. D., Bryant M. L., Gardner M. B., Barr P. J., Luciw P. A. Sequence relationships of type D retroviruses which cause simian acquired immunodeficiency syndrome. Virology. 1987 Apr;157(2):317–329. doi: 10.1016/0042-6822(87)90274-1. [DOI] [PubMed] [Google Scholar]
- Vogt M., Haggblom C., Swift S., Haas M. Envelope gene and long terminal repeat determine the different biological properties of Rauscher, Friend, and Moloney mink cell focus-inducing viruses. J Virol. 1985 Jul;55(1):184–192. doi: 10.1128/jvi.55.1.184-192.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen P. H., Malehorn D., Knupp C., Wong P. K. A 1.6-kilobase-pair fragment in the genome of the ts1 mutant of Moloney murine leukemia virus TB that is associated with temperature sensitivity, nonprocessing of Pr80env, and paralytogenesis. J Virol. 1985 May;54(2):364–373. doi: 10.1128/jvi.54.2.364-373.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]


