Abstract
LINE-1 (L1) retrotransposons are dynamic elements. They have the potential to cause great genomic change because of their ability to ‘jump’ around the genome and amplify themselves, resulting in the duplication and rearrangement of regulatory DNA. Active L1, in particular, are often thought of as tightly constrained, homologous and ubiquitous elements with well-characterized domain organization. For the past 30 years, model organisms have been used to define L1s as 6–8 kb sequences containing a 5′-UTR, two open reading frames working harmoniously in cis, and a 3′-UTR with a polyA tail. In this study, we demonstrate the remarkable and overlooked diversity of L1s via a comprehensive phylogenetic analysis of elements from over 500 species from widely divergent branches of the tree of life. The rapid and recent growth of L1 elements in mammalian species is juxtaposed against the diverse lineages found in other metazoans and plants. In fact, some of these previously unexplored mammalian species (e.g. snub-nosed monkey, minke whale) exhibit L1 retrotranspositional ‘hyperactivity’ far surpassing that of human or mouse. In contrast, non-mammalian L1s have become so varied that the current classification system seems to inadequately capture their structural characteristics. Our findings illustrate how both long-term inherited evolutionary patterns and random bursts of activity in individual species can significantly alter genomes, highlighting the importance of L1 dynamics in eukaryotes.
Keywords: transposable element, retrotransposon, LINE, eukaryotes, evolution
Introduction
Transposable elements (TEs) are repetitive DNA sequences found in genomes scattered across the tree of life, and are often called ‘jumping genes’ because of their ability to replicate and move to new genomic locations. As such, they provide an important source of genome variation at both the species and individual level (Lynch 2006). Eukaryotic TEs are categorized based on their mechanism of retrotransposition. Class I retrotransposons use a copy-and-paste mechanism via an RNA intermediate, allowing massive amplification of copy number, which has the potential to cause substantial genomic change. Class II DNA transposons are more restricted because of their cut-and-paste mechanism. Retrotransposons are further divided into elements with (LTR) and without (non-LTR) long terminal repeats. Non-LTR elements comprise long interspersed elements (LINEs) and short interspersed elements (SINEs). LINEs are autonomous because they encode their own proteins for retrotransposition, whereas SINEs are non-autonomous and depend (in trans) on LINE-expressed proteins.
Long interspersed element 1 (LINE-1 or L1) is a well-known group of non-LTR retrotransposons found primarily in mammals (Kazazian 2000). Given their presence in both plant and animal species, L1s are very ancient elements; and it is assumed that they are ubiquitous across eukaryotes. More importantly, they are one of the most active autonomous elements in mammals, covering as much as 18% of the human genome (Furano 2000; Lander et al. 2001) and accountable for about 30% through amplification of processed pseudogenes and Alu SINEs (Esnault et al. 2000; Dewannieux et al. 2003; Graham and Boissinot 2006). This means that L1s are major drivers of evolution, capable of wreaking havoc on the genome through gene disruption (Kazazian 1998), alternative splicing (Kondo-Iida et al. 1999) and overexpression leading to cancer development and progression (Chen et al. 2005; Kaer and Speek 2013).
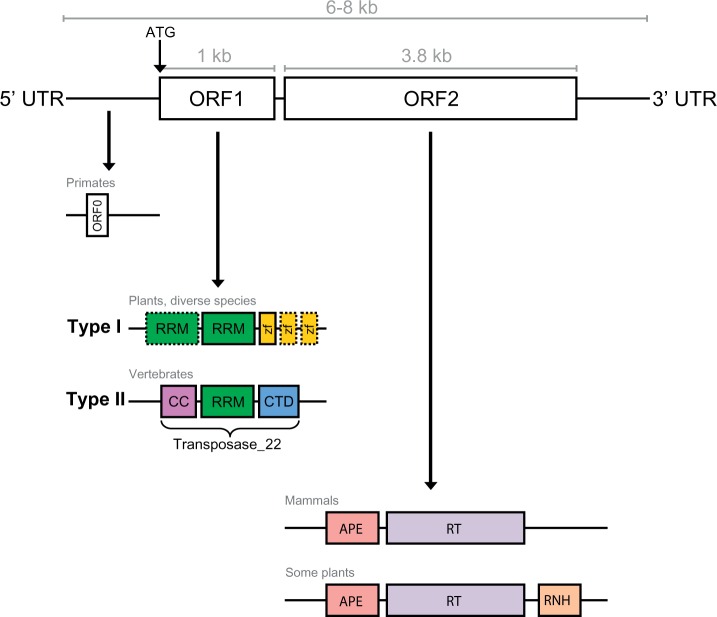
In the literature, active L1s are defined as 6–8 kb elements containing a 5′-untranslated region (5′-UTR) with an internal promoter; two open reading frames (ORF1 and ORF2) separated by an intergenic region; and a 3′ UTR containing a polyA tail (Furano 2000) (see fig. 1). ORF2 is around 3.8 kb in length, translating to a 150-kDa protein (ORF2p) which encodes an apurinic endonuclease and reverse transcriptase (RT) necessary for retrotransposition. ORF1 is much smaller (1 kb nucleotide sequence; ORF1p is only 40 kDa) and thought to have RNA-binding functionality (Furano 2000; Cost et al. 2002). This widely accepted structure has been used for over 30 years to identify putatively active elements in mammalian genomes (Scott et al. 1987). More recently, however, L1s with significant structural variations have been discovered – to the extent that the current terminology on what constitutes an L1 seems inadequate and limiting.
Fig. 1.—
Conventional L1 structure and known variants. A functional L1 retrotransposon is 6–8 kb in length and contains two ORFs, both of which encode proteins for retrotransposition. ORF0 has recently been discovered in primates and is thought to facilitate retrotransposition. L1 ORF1 sequences are divided into two types: Type II is widespread throughout vertebrates, while Type I has only been found in diverse plants and non-mammalian animals such as amphibians and fish. Likewise, domain variants of ORF2 with an additional ribonuclease domain have been found in some plant species (described in the main text). UTR, untranslated region; ORF, open reading frame; RRM, RNA recognition motif; zf, gag-like Cys2HisCys zinc knuckle; CC, coiled-coil; CTD, C-terminal domain; APE, apurinic endonuclease; RT, reverse transcriptase; RNH, ribonuclease H domain.
For example, some plant species have been shown to contain an additional ribonuclease H domain (RNH) in ORF2p downstream of the RT domain, possibly acquired from domain shuffling between plants, bacteria, and Archaea (Smyshlyaev et al. 2013). The domains located within ORF1p can also vary drastically. Khazina and Weichenrieder (2009) classified retrotransposon ORF1 proteins into five types based on the presence and grouping of different domains, and indicated in which species/transposons each type was most commonly found. Type I ORF1p contains at least one RNA recognition motif (RRM) with a Cys2HisCys (CCHC) zinc knuckle, and is found in some plant L1s. Type II is the typical mammalian L1 ORF1p ‘Transposase 22’ (Finn et al. 2010), consisting of a coiled-coil (CC), single RRM and C-terminal domain. Type III and IV ORF1s are supposedly restricted to archaic elements such as CR1s (Chicken repeat 1) (Kapitonov and Jurka 2003) and L2s (Nakamura et al. 2012) and Type V are unclassified. However, even these classifications are insufficient. Metcalfe and Casane (2014) found that Jockey superfamily elements (especially CR1s and L2s) contain every possible type described by Khazina and Weichenrieder (2009), as well as further subtypes. This raises the question of whether L1s are also diverse in their structure, rather than being confined to Type II or I.
Some L1s do not appear to have an ORF1 region (Odon et al. 2013). For a long time, it was thought that co-expression of both ORF1p and ORF2p in cis was necessary for retrotransposition (Moran et al. 1996). However, L1 copies containing a disrupted ORF1p but intact ORF2p retain the ability to mobilise SINEs within the genome, as shown by Dewannieux et al. (2003) with a defective ORF1p mutant. Perhaps most intriguingly of all, recent evidence suggests the possibility of a third ORF in L1 elements: ORF0, an antisense open reading frame upstream of ORF1 (Denli et al. 2015). This ORF0 is very short, encoding a 71 amino acid peptide, and is thought to be primate-specific. Overexpression of ORF0p leads to a significant increase in L1 mobility, which may help explain the high retrotransposition activity of L1 in some primates (e.g. humans).
Growing evidence (Kordis et al. 2006; Waters et al. 2007; Blass et al. 2012; Tollis and Boissinot 2013; Heitkam et al. 2014) suggests that the current model of L1 activity is insufficient. The idea that ORF1p + ORF2p in cis = retrotransposition fails to capture variation between different organisms, particularly beyond the mammalian lineage. In this study, we provide a definitive and comprehensive phylogenetic analysis of L1 content and activity in over 500 species from widely divergent branches of the tree of life. The genomes selected include plants, arthropods, sauropsids, mammals, and other, more primitive eukaryotic species. We also include several cases of closely related organisms (within the same genus or species) to look for L1 differences between individuals, and the effects of different genome assembly methods. For each genome, we searched for the presence of L1 elements; and if found, characterized the elements as active or inactive and identified the domains in each of the ORF proteins. Our findings effectively illustrate the overlap between inherited evolutionary patterns and random individual bursts of activity, allowing a much broader understanding of TE dynamics in eukaryotes.
Materials and Methods
Extraction and Characterization of L1 Repeats from Taxa with Full Genome Data
Almost all of the genomes used in this study (499 out of 503) are publicly available from the National Center for Biotechnology Information (NCBI) (Sayers et al. 2012) or UCSC Genome Browser (Kent et al. 2002). Supplementary table S1, Supplementary Material online lists the systematic name, common name, version, source and submitter of each genome assembly, and marks which genomes were privately acquired. If there was both a GenBank and RefSeq version for the genome, the GenBank version was used by default. Supplementary table S2, Supplementary Material online shows the total genome sequence length and scaffold/contig N50 values, giving an approximation of the assembly quality. Supplementary table S3, Supplementary Material online compares the different sequencing technologies and methods. A phylogenetic representation of the genomic dataset was inferred using Archaeopteryx (Zmasek 2015) to download the Tree of Life (Maddison and Schulz 2007) topology for all Eukaryota (node identifier 3, ∼76,000 species). The tree was extended (e.g. descendants added where necessary) to include all of the 503 genomes, and species not included in this study were removed. Out-dated branches were changed using OrthoDB (Kriventseva et al. 2015), OrthoMaM (Douzery et al. 2014), NCBI Taxonomy (Sayers et al. 2012) and recent publications (Murphy et al. 2001; Beck et al. 2006; Janecka et al. 2007) as references (see supplementary fig. S1, Supplementary Material online).
L1 hits were initially identified in each genome using an iterative query-driven method based on sequence similarity, as seen in Walsh et al. (2013). The original query L1 sequences were obtained from Repbase (Jurka et al. 2005) by searching for anything listed as ‘L1’ or ‘Tx1’ (subgroup of the L1 clade) for all taxa. Cow and horse L1s were also obtained from past analyses (Adelson et al. 2009, 2010). All of the accumulated query sequences were concatenated into one file, which was used as the input query to run LASTZ v1.02.00 (Harris 2007) with at least 80% length coverage. BEDTools v2.17.0 (Quinlan and Hall 2010) was used to merge overlapping hit intervals from different queries and extract a non-redundant set of L1 sequences in FASTA format. For each genome, the output hits were globally aligned with MUSCLE v3.8.31 (Edgar 2004) to produce a species consensus with Geneious v7.0.6 (Kearse et al. 2012). Genomes with a substantial number of hits required clustering with UCLUST v7.0.959_i86linux32 (Edgar 2010) before aligning. The species consensus sequences were then added to the query file (see supplementary fig. S2, Supplementary Material online). This process was repeated three times, to accommodate inclusion of new genomes at various stages in the pipeline and to include diverse L1s to the set of queries.
To control for difference in genome assembly quality, we also used the TBLASTN program (Altschul et al. 1990) to search the non-redundant NCBI nucleotide database (NR) and high throughput genomic sequences (HTGS) (Sayers et al. 2012). TBLASTN search parameters were default except the e-value was changed to 1e−5. Input was the concatenated ORF1p and ORF2p from 13 full-length L1-clade elements from Repbase (Jurka et al. 2005), spanning each order/clade (where available), and consisting of mammalian L1/diverse L1/diverse Tx1 elements (see supplementary table S4, Supplementary Material online for exact queries and TBLASTN results). To determine the reliability of low-scoring hits, each hit was extracted as a nucleotide sequence and screened with CENSOR (Kohany et al. 2006) against the entire Repbase library of known repeats. This provided a ‘reciprocal best-hit’ check: Hits were kept if the best hit from CENSOR was an L1, and discarded if the best hit was another repetitive sequence (e.g. retrotransposons BovB or CR1).
Confirmed L1 sequences from the TBLASTN approach were used as species-specific queries to re-run LASTZ on each genome. Then, the sequences from each species were concatenated into a final query file (>3 million L1s, both fragment and full-length copies) for the last round of LASTZ extraction. The Repbase library (with CENSOR) was again used to verify L1s with a reciprocal best-hit check. Supplementary table S5, Supplementary Material online shows the results from the final LASTZ extraction, with notes comparing the number of L1s found to previous studies. Sample code for each step is available online (https://github.com/AdelaideBioinfo/L1-dynamics).
Both the LASTZ and TBLASTN approaches are limited by the quality and quantity of available nucleotide data whether it is from the genome assembly or nucleotide databases (NR/HTGS). As such, the L1 status of each species (e.g. L1 presence versus absence) was determined based on the union of the two methods (see Supplementary table S7, Supplementary Material online).
Identification of Intact Open Reading Frames
BEDTools (Quinlan and Hall 2010) was used to extend each L1 hit by 1kb either side before the ORF analysis, to overcome incomplete 5′ and 3′ ends that may be missing crucial start/stop codons. Geneious (Kearse et al. 2012) was then used to scan for open reading frames that were at least 80% of the expected length (≥ 800 bp for ORF1 and ≥3 kb for ORF2 – see supplementary fig. S4, Supplementary Material online). ORF sequences which satisfied the length requirements were subjected to a series of tests to confirm their functionality: Each ORF had to be complete with a start codon, stop codon and no debilitating mutations in between (such as premature stop codons or too many ambiguous nucleotides). For ORF1, the start codon had to be a methionine (ATG) (Penzkofer et al. 2005) and ORF2p sequences had to have a confirmed RT domain. After translation, both ORF1p and ORF2p candidates were checked for similarity to known domains using HMM–HMM comparison (Finn et al. 2011) against the Pfam 28.0 database (Finn et al. 2010) as at May 2015 (includes 16,230 families).
ORF1p sequences were initially screened for known L1 ORF1p domains (e.g. Transposase_22, RRM, zf-CCHC). Sequences containing at least one of these domains were kept as ‘confirmed’ ORF1p. Confirmed ORF1p sequences often contained other, associated domains: ‘probable’ ORF1p domains, such as DUF4283 in plants. A library was generated containing probable ORF1p-associated domains and used to re-screen the unconfirmed ORF1p candidates. Matching sequences were categorized as ‘probable ORF1p’ (see supplementary fig. S7, Supplementary Material online ). This resulted in three categories of L1 ORF proteins: Confirmed ORF2p, confirmed ORF1p, and probable ORF1p. Nucleotide L1 sequences were given label prefixes according to their ORF composition: ORF1_ (confirmed ORF1p), ORF2_ (confirmed ORF2p), probORF1_ (probable ORF1p), ORF1_ORF2_ (both ORF proteins confirmed), or probORF1_ORF2_ (confirmed ORF2p, probable ORF1p). Supplementary table S6, Supplementary Material online summarizes the ORF content in each genome. Only ORF sequences that passed all the tests were included in subsequent analyses.
Classification of Potentially Active L1 Elements
An L1 was defined as a potentially active candidate if it contained an intact ORF2 (regardless of the state of ORF1), as this means that it is either fully capable of retrotransposing itself (Moran et al. 1996; Heras et al. 2006) or it can cause activity in the genome by mobilizing SINEs (Dewannieux et al. 2003). The ORF2 sequence had to satisfy the criteria listed above (≥ 3kb nucleotide sequence, complete with start and stop codons and no inactivating mutations, and confirmed RT domain). L1 elements containing intact ORF2, and thus potentially active, were typically full-length or near full-length (e.g. >4.5 kb). Genomes with low copy number were further checked for contamination: For example, the potentially active L1s were not considered valid if they came from short, isolated scaffolds or showed suspiciously high similarity to another (divergent) species.
Dendrogram Construction from Nucleotide L1 Sequences
Full-length L1 sequences (or near full-length, as long as they included an intact ORF2) were globally aligned using MUSCLE (Edgar 2004). Mammalian species required iterative clustering with UCLUST (Edgar 2010) before aligning, due to the huge number of hits. Clustering identities ranged from 70 to 95%. Alignments were trimmed with Gblocks (Castresana 2000) to remove large gaps (default parameters, allowed gap positions: with half). The dominant active clusters for each species were represented as dendrograms, or unrooted tree diagrams, using FastTree v2.1.8, double-precision version (i.e. compiled with –DUSE_DOUBLE) (Price et al. 2010). Archaeopteryx v0.9901 beta (Zmasek 2015) was used to visualise and annotate each tree based on the ORF labels.
Phylogenetic Analysis of Conserved L1 Amino Acid Residues
Two methods were tested to depict the evolutionary dynamics of potentially active L1 elements. First, we inferred an ORF2p consensus tree: All confirmed ORF2 sequences in each species were extracted, translated and globally aligned with MUSCLE (Edgar 2004). The consensus for each species was generated in Geneious (Kearse et al. 2012) using majority rule (most common bases, fewest ambiguities) and a base was regarded ambiguous if coverage at that position was < 3 sequences (unless the alignment had ≤ 3 sequences, in which case this was changed to < 2 sequences). This produced a single L1 ORF2p consensus for each species. These consensus sequences were globally aligned using MUSCLE (Edgar 2004) and a phylogeny was inferred with maximum likelihood using FastTree, double precision compilation (Price et al. 2010).
Another phylogeny was inferred using just the RT domains within ORF2p. For each confirmed ORF2p sequence, the RT domain was extracted using the envelope coordinates from the HMMer domain hits table (--domtblout) (Finn et al. 2011), with minimum length 200 amino acid residues. RT domains from all species were collated into one file (37,994 sequences total), which was then clustered with USEARCH (Edgar 2010) at 90% identity. Each cluster was defined as a L1 RT-family (3508 families total). Only RT-families containing more than five members were included in the phylogenetic analysis. Two RT domains from Repbase (Jurka et al. 2005) were also included: A CR1 element from Anopheles gambiae (Ag-CR1-22), to act as the outgroup, and Zepp from Chlorella vulgaris, as a sister element to the L1s found in Coccomyxa subellipsoidea. As before, alignments were performed using MUSCLE, Geneious was used to extract a consensus for each family, and FastTree was used to infer a maximum likelihood phylogeny. A second tree was built using the neighbor-joining method and tested with bootstrapping (1,000 replicates).
Clustering Analysis of L1 ORF1 Proteins
A reliable phylogeny could not be inferred from ORF1p sequences because of the high variation in non-mammalian species. Instead, ORF1p sequences were clustered using an all-against-all BLAST (Altschul et al. 1990) approach. The BLAST was performed using BLAST v2.2.24 and NCBI-BLAST v2.2.27+ (Altschul et al. 1990) with the following parameters: -p blastp, -e 1e−10, -m 8 (for tabular output). Based on the BLAST results, the ORFs were then clustered using SiLiX software (Miele et al. 2011) with default parameters and –net to create a net file which contains all the pairs taken into account after filtering.
Results
Ubiquity of L1 across Plants and Animals
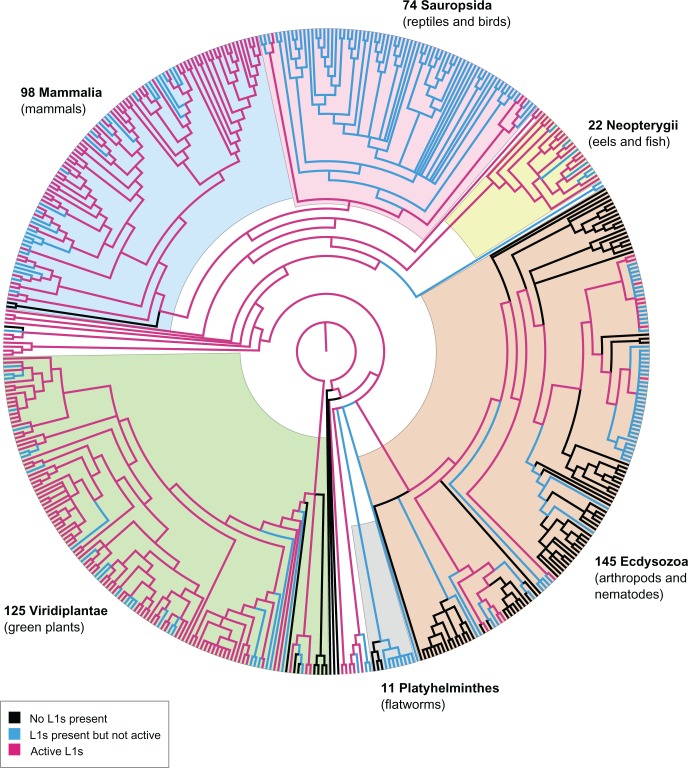
To simplify discussion of the results, we define three different states that a genome can be in, in terms of L1 content: Absent (L1−), meaning that no L1s were detected in the genome; present (L1+), meaning that L1s were found in partial or full-length form; and potentially active (L1*), meaning that at least one putatively active L1 was found in the genome (using either the TBLASTN or LASTZ method). L1− and L1+ are mutually exclusive (a genome cannot have both presence and absence of L1s), whereas L1* is the potentially active subset of L1+. Using this ternary system, we screened 503 eukaryotic species representing key clades of the tree of life (125 plants, 145 protostomes, 98 mammals, 74 sauropsids, 22 neopterygians, 11 flatworms, and 28 other species) (fig. 2; see supplementary fig. S1, Supplementary Material online). Of these, 407 species were found to be L1+. L1 copy number was highest in mammals, with thousands of full-length L1 sequences found in almost every mammalian species analysed (with the exception of monotremes, which are L1−).
Fig. 2.—
Phylogenetic representation of genomic dataset. Species relationships between the 503 representative genomes used in this study were depicted using Archaeopteryx to download the Tree of Life topology for all Eukaryota (node id 3) and extract the 503 species of interest. Out-dated branches were updated using OrthoDB, OrthoMaM, NCBI Taxonomy and recent publications as references. Labels indicate the major groups present in this dataset. Branches are colored to indicate the L1 state of each genome, as shown in the legend.
L1s also appeared frequently in plants (118/125 L1+ plant species), but colonized far less of each genome (e.g. typical copy number between 10 and 1,000 L1s). Fish, non-avian reptiles and amphibians showed consistent presence but similarly low copy numbers compared with mammals. Birds had an exceptionally low (yet consistent) L1 copy number: Only one full-length L1 element was found in most of the bird species analysed (and multiple fragments), yet this element was conserved through enough species that it is likely an ancient remnant of L1 from a common ancestor.
In the protostomes, L1 presence was verified in all mosquito and fly species, but appeared sporadically elsewhere. Fragments were found in all Schistosoma flatworms, as well as Clonorchis sinensis. The remaining ‘primitive’ orders contained multiple full-length L1 families, with the exception of Tentaculata (Mnemiopsis leidyi), Placozoa (Trichoplax adhaerens), and Porifera (Amphimedon queenslandica). Supplementary table S5, Supplementary Material online contains a summary of the L1 sequences found in each genome and the length distribution of the hits.
Dead or Alive – How Many L1s Have Retained Their Activity?
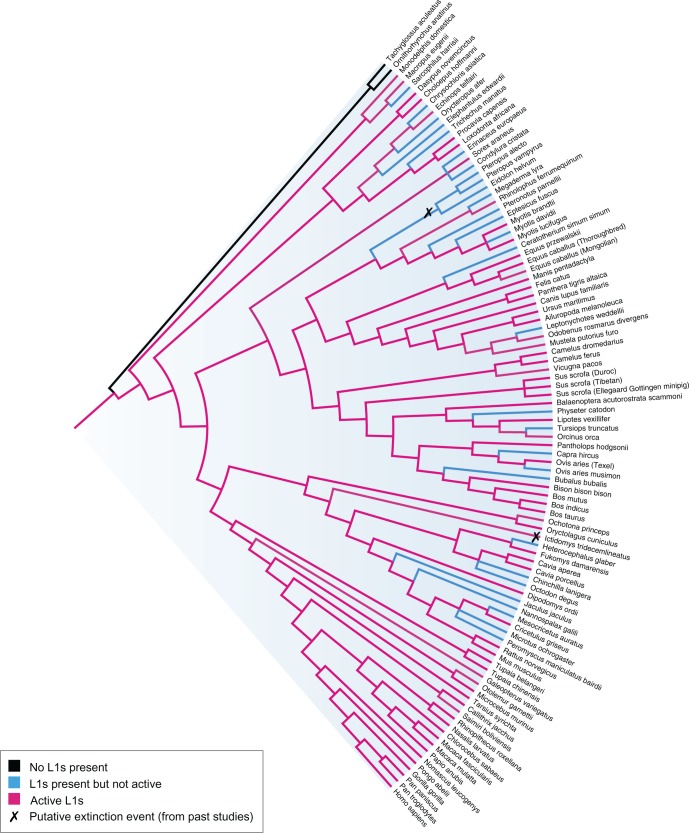
Of the 407 L1+ eukaryotes, 206 species were further determined to be L1*: 92 plants, 67 mammals, and 47 non-mammalian animal species. This is illustrated in fig. 2 (full tree, no node labels – see supplementary fig. S5, Supplementary Material online), fig. 3 (mammals) and fig. 4 (plants). Although all coloured branches indicate presence (L1+), the potentially active subset (L1*) is coloured magenta, so in this case the blue branches (L1+ −L1*) indicate species that only contain ‘extinct’ L1s (i.e. present but inactive). Because the L1 state of each genome is only observable at the tree tips, the phylogeny was annotated based on the notion that the most parsimonious explanation is a loss of activity, not a gain (hence ancestral branches are coloured ‘active’ if any of the descendants display activity). Noticeably, despite the ubiquitous presence of L1 across the mammalian lineage, L1 in quite a few mammalian species or subgroups (e.g. megabats, some rodents, and Afrotherian mammals) appear extinct. In contrast, other mammals seem to be bursting with L1 activity: Including several species (e.g. minke whale, antelope, snub-nosed monkey, panda, baiji) which have not been studied before in the context of L1 retrotransposition.
Fig. 3.—
Mammalian phylogeny reveals ubiquitous L1 presence (except for monotremes) and possible extinction events. Genomes are classified as L1 absent (L1−) (black), L1 present but inactive (L1+–L1*) (blue) or L1 active (L1*) (red). Putative extinction events from past studies are marked.
Fig. 4.—
Plant phylogeny showing the sporadic distribution of active L1 and the L1 state of each genome (colored branches). Brassicales and Poales stand out as the dominant L1* families. Orders containing more than three representative genomes are named.
Previously, the human genome has been used as a model for high retrotranspositional activity. Numerous studies have found that L1 retrotransposition rates differ substantially between primate lineages, for example, human versus chimpanzee (Gregory et al. 2002; Mathews et al. 2003; Lee et al. 2007). That is particularly evident with this new comparison of human versus snub-nosed monkey. For example: In the human genome, we identified 266 potentially active, both-ORF-intact L1s, and other studies have quoted similar numbers [e.g. Penzkofer et al. (2005) estimate ∼150 on L1 Base]. Of such L1* candidates, <50% are active in cell culture: Brouha et al. (2003) predict that there are only about 80–100 active L1s in the average human, although this varies between individuals (Seleme et al. 2006; Beck et al. 2010). The snub-nosed monkey genome, on the other hand, contains 2549 both-ORF-intact L1* candidates. More than 95% of these would have to be determined inactive upon experimental analysis to obtain a comparable number to human; so the retrotransposition potential of snub-nosed monkey is substantially higher than that of human or any other primate.
L1 activity persists beyond the mammalian lineage as well. Almost every order that exhibits L1 presence contains L1* species (the two exceptions being Platyhelminthes and Chondrichthyes, where the presence is solely due to L1 fragments). Birds similarly contain L1 fragments or low copy number full-length elements, yet the ORF2 region is heavily degraded and mutated.
In plants, the L1 state of species seems to mirror mammalian genomes. Brassicales and Poales stand out as the most dominant orders, with each member bearing a significant number of active L1s. Another notable L1* species is Coccomyxa subellipsoidea, which only contains 15 L1 elements but every single one of these elements is putatively active and almost identical, suggesting recent retrotransposition. This genome also appears as a discrepancy in our tree; it is one of the only instances where a L1* species is phylogenetically placed next to a L1− species (fig. 4). However, given that our dataset does not contain all species, this could be a result of incomplete sampling and hence incorrect placement of the species. The ancestral branch was coloured red (L1*) despite the absence of L1s in several descendent species, because another study shows that Chlorella vulgaris (sister to Chlorella variabilis, which is marked L1+) contains active L1-like Zepp elements 98% identical to Coccomyxa subellipsoidea (Higashiyama et al. 1997).
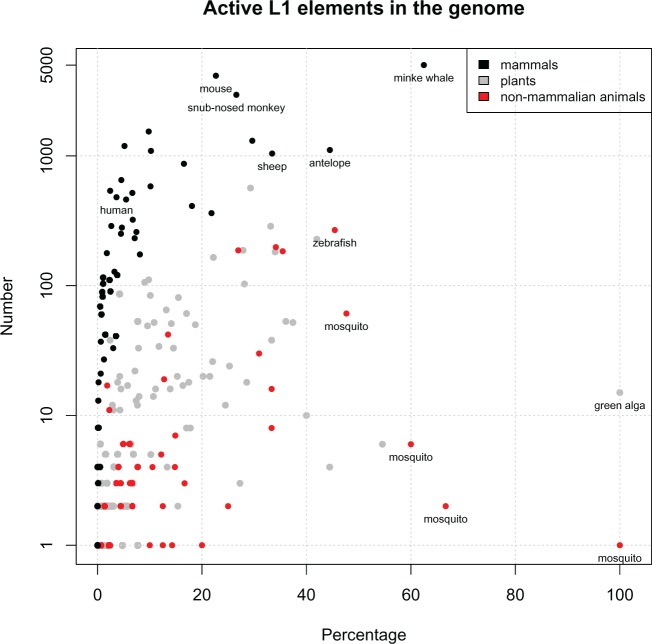
Finally, the number of potentially active L1s found in each genome was compared with the total number of near full-length L1s in that genome, to get a percentage estimate of L1 activity per species (fig. 5; see supplementary table S8, Supplementary Material online). We found that mammalian species often contain a large number of inactive elements, so the percentage of active L1s is relatively low (e.g. <20%). In contrast, non-mammalian species (animals and plants) seem to have a higher proportion of active L1s in the genome despite the lower copy number; so the centroid of the graph is shifted to the right.
Fig. 5.—
Distribution of active L1 elements reveals several ‘hyperactive’ mammalian species. The y-axis shows the number of active L1 in the genome; the x-axis shows the percentage of active L1s in the genome (i.e. # active L1/# near full-length L1 × 100, as described in supplementary table S8, Supplementary Material online). Non-mammalian animal species (red) and plants (gray) appear to have high retrotranspositional potential but low observable L1 activity in the genome. In contrast, mammals (black) typically have a very high L1 copy number, but the majority of these are inactive. The labelled mammalian species stand out as L1 ‘hyperactive’ species because they are the most likely to be currently replicating and expanding within the genome.
Mammalian Species Typically Have a Dominant Active Cluster
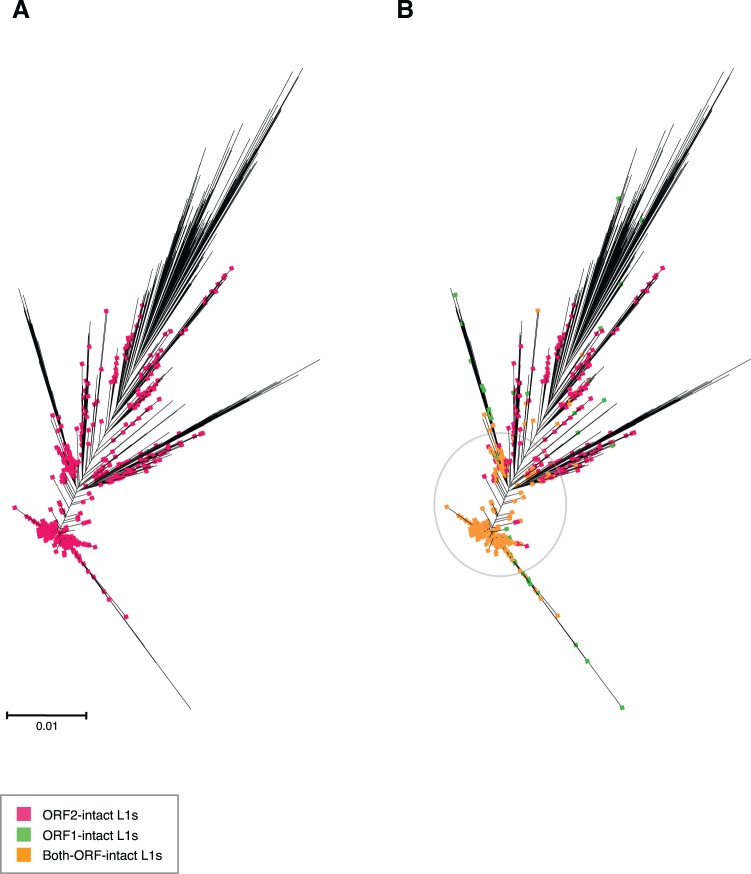
The clustering and dendrogram construction of L1 nucleotide sequences revealed that most mammals contain one large, dominant active cluster of closely related elements. As mentioned before, snub-nosed monkey is a remarkably active species in a comparatively inactive subgroup (i.e. primates). The cluster depicted in figure 6 contains 1742 full-length L1 (1337 both-ORF-intact and another 195 ORF2-intact) with 95.2% pairwise identity, which was used to construct an unrooted maximum likelihood tree highlighting elements with ORF1 intact, ORF2 intact, or both ORFs intact. Almost all of the L1s in this cluster have both ORFs intact and are clustered on the shorter branches, indicating very recent activity.
Fig. 6.—
Master lineage model predominant in most mammalian species, including snub-nosed monkey Rhinopithecus roxellana. (a) Maximum likelihood dendrogram inferred using FastTree double precision version, from full-length L1 nucleotide sequences extracted from genomic data. Sequences were clustered with UCLUST and globally aligned with MUSCLE. Species with a clearly dominant L1* cluster were classified as master lineage models, as shown in Supplementary table 9. Sequences in the alignment were tagged to indicate which ORFs were intact and visualized using Archaeopteryx. This figure highlights the ORF2-intact L1s. (b) Same as (a), but here the highlighting also shows ORF1-intact L1s and both-ORF-intact L1s. Both-ORF-intact L1s are tightly clustered on the short branches in the middle.
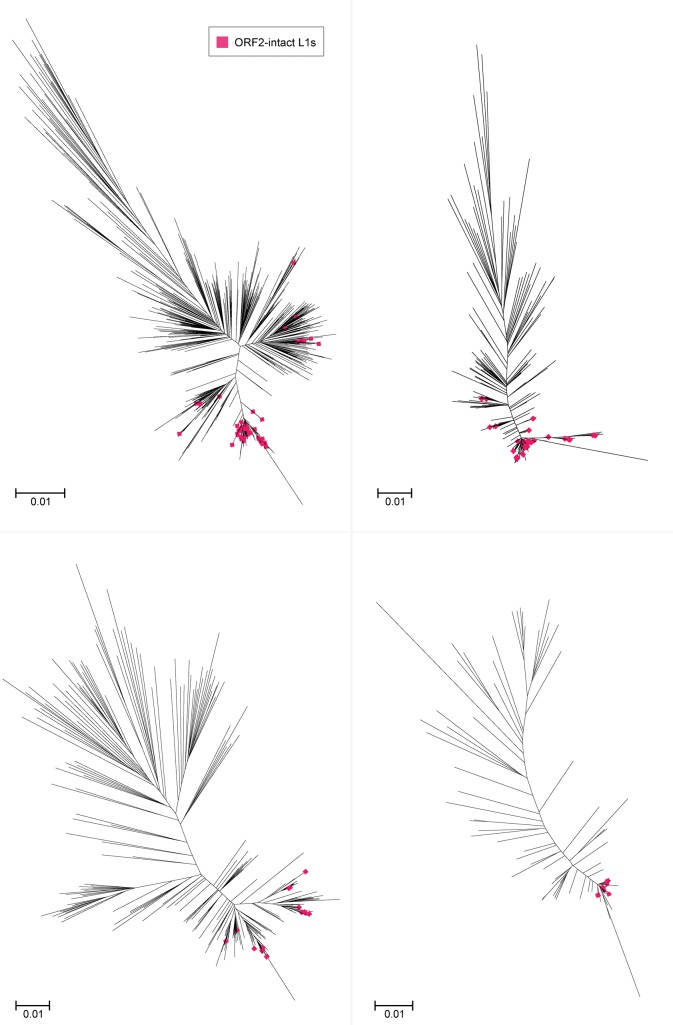
However, in some species it is obvious that there is more than one significant active cluster. Horse (Equus caballus) is a well-known example of a species with five L1 (equine) subfamilies, two of which contain active elements (Adelson et al. 2010). Megabats are also known to have harboured multiple contemporaneous L1 lineages, although those lineages are now extinct (Yang et al. 2014). Nonetheless, this multiple lineage phenomenon seems to extend to the microbat subgroup as well: figure 7 depicts the clustering and dendrogram construction for Myotis lucifugus, where there is no discernible dominant cluster. The elements in each cluster are >70% similar to each other, but the clusters themselves are distinct at this level (see supplementary table S9, Supplementary Material online). Once again, we see a tendency for active L1s to converge on the short branches.
Fig. 7.—
Multiple L1 lineages present in the Myotis lucifugus genome. Maximum likelihood dendrogram inferred using FastTree from full-length L1 nucleotide sequences extracted from full genome species data. As in Fig. 6, sequences were clustered with UCLUST, aligned with MUSCLE, annotated with Geneious and visualized with Archaeopteryx. Only ORF2-intact L1s are highlighted.
RT Domain Reveals Distinct L1 Groups
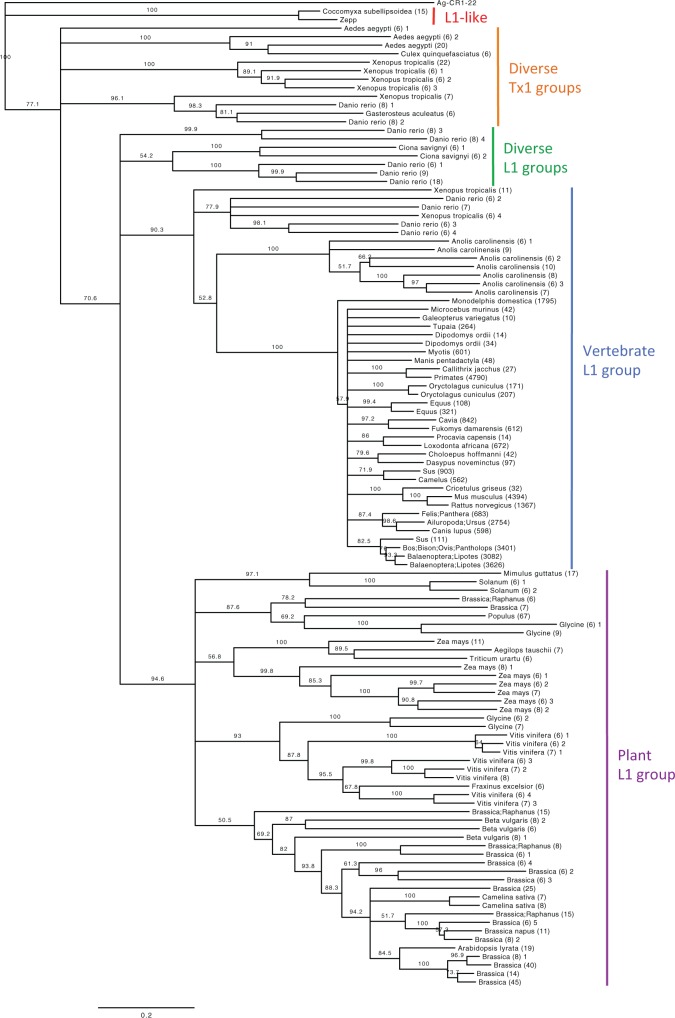
The phylogenetic analysis of RT families (fig. 8) clearly illustrated differences between L1 groups. Two L1 clades are immediately obvious: Vertebrate L1s, with the shortest observed branch lengths, and plant L1s, displaying significantly longer branches and lower support values. The rest of the phylogeny is made up of diverse L1 and Tx1 groups from combinations of fish, amphibians, mosquitos, sea squirts, and green algae.
Fig. 8.—
Phylogenetic analysis of RT families shows the overall hierarchy of L1/Tx1 groups. Rooted Neighbor-Joining tree based on amino acid RT domains. This tree represents the bootstrap consensus after 1,000 replicates, with nodes that have confidence values over 50% labelled. CR1 from Anopheles gambiae (outgroup) and Zepp from Chlorella vulgaris (98% identical to Coccomyxa subellipsoidea L1s) were obtained from Repbase. Only RT-families with >5 members at > 90% identity are shown in this tree. Nodes are labelled as follows: By species name if there is only one species in the family (e.g. Loxodonta africana); by genus name if there are multiple species of the same genus (e.g. Sus); by multiple genus names if there are multiple genera in the family (e.g. Ailuropoda; Ursus); and by clade name if there are more than five genera (e.g. Primates). The number in parentheses after the node name indicates the number of elements in the family.
Mammalian species form a hard polytomy, vaguely reflecting expected species relationships but without accurate subclass structure. This is most likely due to the sporadic sampling of species (based on data availability). In addition, the mammalian RT-families all have a large number of shared amino acids, making it difficult to reliably distinguish subfamilies. This is especially true for primates, which all grouped together as a single RT-family (4790 members with >90% identity) except for the strepsirrhine primate Microcebus murinus. The striking lack of diversity supports the idea of a rapid L1 explosion in the mammalian lineage following a severe population bottleneck (Kordis et al. 2006).
In contrast, non-mammalian animals contain multiple distinct L1 lineages and are not restricted to a single group or clade. This phenomenon has been explored in depth for fish (Duvernell et al. 2004; Furano et al. 2004; Blass et al. 2012), Anole lizard (Novick et al. 2009; Tollis and Boissinot 2013), Xenopus frogs (Kojima and Fujiwara 2004; Kordis et al. 2006) and African mosquitos (Biedler and Tu 2003). Fish and amphibians are the only known species to contain both mammalian-like vertebrate L1s, and diverse L1/Tx1 families (representatives Danio rerio and Xenopus tropicalis shown in fig. 8). Note that figure 8 only shows RT families within confirmed ORF2p, ≥200 amino acids in length, and containing >5 members at 90% identity, to reduce the dataset to a manageable number for visualization.
The plant L1 group (excluding Coccomyxa subellipsoidea) is divided into five subclades: The largest of which is made up of Brassicales species plus Beta vulgaris (Caryophyllales) (fig. 8). Brassicales is one of the most L1-active orders (fig. 4; see supplementary table S5, Supplementary Material online) and contains multiple L1 lineages. This is evident by the ORF2p analysis: Excluding Carica papaya (L1−), all Brassicales species contain both the typical RT (RVT_1), as well as diverse RT and ribonuclease combinations (e.g. RVT_1 + RVT_3/RNH, see supplementary table S10, Supplementary Material online). The ORF1p analysis similarly revealed novel L1 lineages within Brassicales species Camelina sativa, Aethionema arabicum, and Arabis alpina, characterized by the presence of N-terminal RRMs (see supplementary table S11, Supplementary Material online). Beta vulgaris contains these same RRM-ORF1p, known as the BNR lineage (Heitkam and Schmidt 2009) – which is probably why Beta vulgaris is the only non-Brassicales species to appear in this L1 subgroup (fig. 8). Heitkam et al. (2014) suggested that the RRM domain substitutes the RNA-binding function of the zinc finger. A number of other plant species were found to include RRM-ORF1p (see supplementary table S11, Supplementary Material online), supporting the idea that L1s can recruit functional domains from their host to contribute to retrotransposition (Heitkam et al. 2014).
Variation of ORF1 Proteins across Species
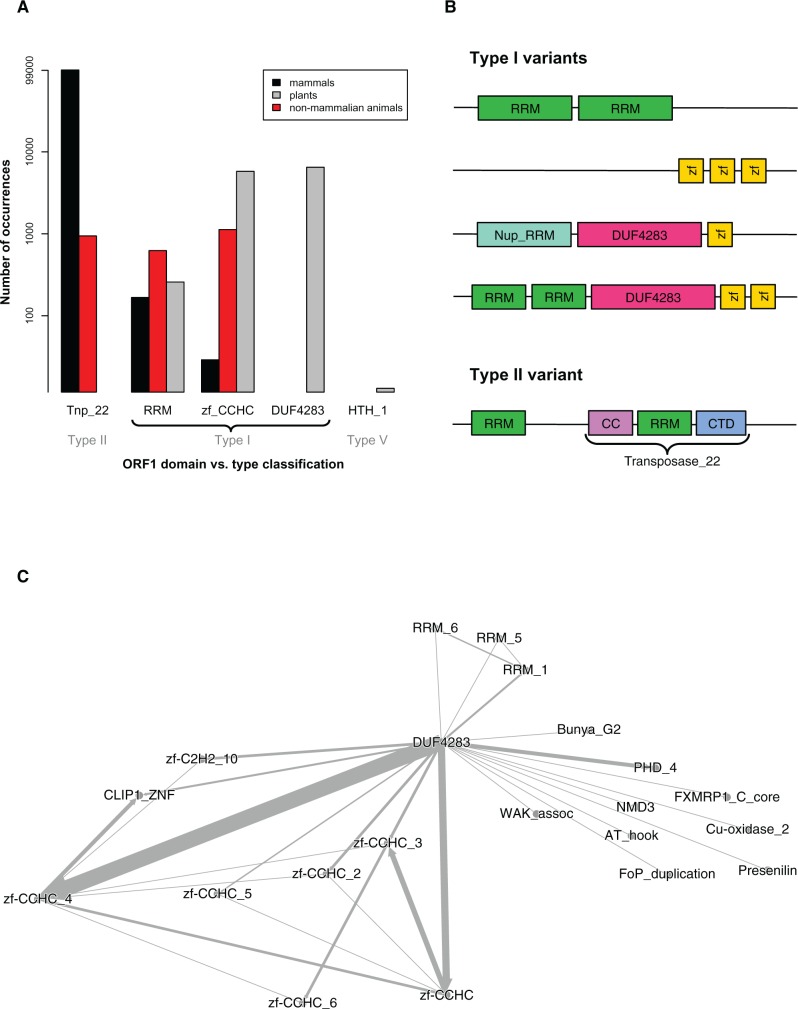
The variability found in ORF1 sequences, from both plants and animals, is staggering. Khazina and Weichenrieder (2009) defined Type II ORF1p as the Transposase_22 domain, and Type I ORF1p as a combination of RRM and zf-CCHC domains (fig. 1). Mammalian species are dominated by Transposase_22 ORF1 proteins (fig. 9a); as expected from the Type II classification. However, some mammalian species also contain ORF1 proteins with RRM or zf-CCHC domains – which are more characteristic of Type I, and are likely very ancient. There was even a Type II variant found: Several ORF1p in Myotis lucifugus display an RRM domain before the expected Transposase_22 (fig. 9b), which has not been previously documented.
Fig. 9.—
ORF1p clustering and domain identification analysis. (a) ORF1p domain summary from HMM–HMM comparison. Transposase_22 (Tnp_22), RNA recognition motifs (RRM), and zinc fingers (zf-CCHC) are known ORF1p domains. The y-axis shows the number of times these appeared in each group of species (mammals, non-mammalian animals, plants), on a log scale. Several unknown domains also appeared frequently; for example, DUF4283 was found in every plant species except Coccomyxa subellipsoidea, which harboured HTH_1 ORF1 proteins instead. (b) Variants of Type I ORF1 proteins. Type I ORF1p typically has at least 1 RRM and 1 zf-CCHC; Type II ORF1p is characterized as the Transposase_22 domain. This figure highlights type variants found in the analyzed species: for example, lack of zf-CCHC motifs, seen in mosquitos; lack of RRM domains, seen in sea squirts; Nup_RRM instead of RRM, seen in some plants; over-representation of unknown DUF4283 domain in almost all plants; and an additional RRM before the Transposase_22 in some mammals, for example, bat Myotis lucifigus. Supplementary table S11, Supplementary Material online shows the ORF1p domains in each species. (c) Directed network graph of Type I ORF1 protein domains found in plants. Each ORF1p in each L1 (in each plant species) was screened using HMMer against the Pfam database. The highest-scoring domain hit was ranked first; other domains also found within that ORF1p sequence were listed next, by decreasing score. This was used to construct a network graph of the associated domains. DUF4283 was the most frequently seen, highest scoring domain – it is the centroid of the graph. RRM and zf-CCHC domains are associated with this domain (especially zf-CCHC_4), but it is the unknown domain that acts as the vital ORF1p identifier in plants.
Non-mammalian animals contain the typical Type II ORF1p, Type I ORF1p, and assorted combinations of RRM/zf-CCHC domains. These appear as variants of Type I ORF1p (fig. 9b) but are consistent with the Tx1 clade of retrotransposons and RT-based phylogeny (fig. 8). There are numerous studies that describe these domains in depth, for example, Kojima and Fujiwara (2004) and Kordis et al. (2006).
In plants there were many ORF1p with RRM or zf-CCHC domains, indicative of Type I proteins. As mentioned above, several species harboured novel Nup_RRM or RRM domains. However, the overwhelmingly dominant plant ORF1p domain was DUF4283: An uncharacterized domain of unknown function (Finn et al. 2010). Figure 9c shows a directed network graph of the most frequently seen ORF1p domains across Viridiplantae. For all other species, this graph is centred around Transposase_22, RRM or zf-CCHC domains (see supplementary fig. S7a–f, Supplementary Material online). In plants, DUF4283 appears to act as the primary ORF1p classifier, strongly associated with zf-CCHC_4 (fig. 9c).
Coccomyxa subellipsoidea does not contain any of these domains – instead, the entire ORF1p region is enveloped by HTH_1 (fig. 9a): A bacterial regulatory helix-turn-helix protein of the LysR family (Finn et al. 2010). Coccomyxa subellipsoidea L1s are 98% identical to Zepp (fig. 8), a LINE-like retrotransposon found in Chlorella vulgaris (Higashiyama et al. 1997). Chlorella vulgaris was not included in this study as the assembly is only available in contig form. However, another Chlorella species (C. variabilis) was included and showed minimal, fragmented L1 presence (fig. 4). Given that Coccomyxa subellipsoidea and C. vulgaris share such high L1 identity, yet this is missing from the closely related C. variabilis species, it is possible that a horizontal transfer event occurred between the first two species. Alternatively, TEs have a tendency to take necessary proteins directly from their host (Abrusan et al. 2013; Heitkam et al. 2014); this may also explain the newly acquired HTH_1 proteins.
Antisense Characteristics of Active L1s
The analysis of ORF1 and ORF2 sequences across genomes led to the discovery of an antisense open reading frame overlapping ORF1. This novel ORF was initially noticed in the panda genome (Ailuropoda melanoleuca), where it is present in almost every L1 element that has both ORFs intact (1157/1200). As a result, we screened each genome for strictly active L1s (i.e. both ORF1 and ORF2 intact) to determine whether other species contained similar antisense ORFs (i.e. overlapping ORF1 in the reverse direction and about 1 kb in length). Apart from panda, only eight other mammalian species contained anything remotely similar (fig. 10a), albeit at lower copy number. No such reverse ORFs were found in any of the non-mammalian animal or plant species. Interestingly, these ORFs only appeared in mammalian species with a substantial number of active L1s (e.g. minke whale, baiji, dog, rat), suggesting that they might somehow contribute to L1 retrotransposition; yet they are noticeably absent from all of the primates, including snub-nosed monkey. They are also clearly distinct from the primate-specific antisense ORF0 (Denli et al. 2015), which is much shorter and upstream of ORF1.
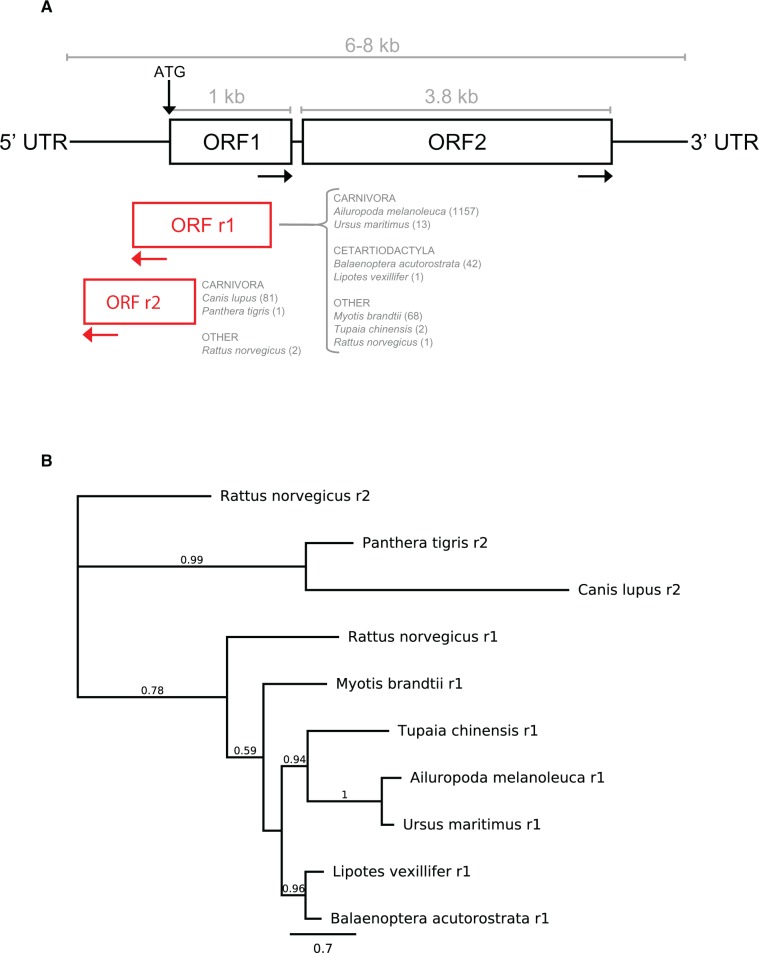
Fig. 10.—
Novel antisense open reading frames found in some mammals. (a) Characteristics and distribution of the antisense ORFs. The position and approximate size of the novel antisense ORFs, as well as the order/species they are found in and the number of L1s that contain this ORF (in brackets). These ORFs have no known functional domains. (b) Antisense ORFp species consensus tree. Maximum likelihood phylogeny inferred using FastTree from extracted and aligned L1 reverse ORFp consensus sequences. Expected species relationships appear preserved within the r1 and r2 clades.
Using the same procedure as previously described for ORF2p, we extracted and aligned the reverse ORF proteins in each species to generate a representative consensus sequence, then aligned the consensus sequences and inferred maximum likelihood and Neighbor-Joining phylogenies (fig. 10b shows the maximum likelihood tree). The only difference between the trees was the position of Myotis brandtii (outgroup to minke whale/baiji on NJ tree, with low support). The reverse ORF proteins found in dog Canis lupus and Siberian tiger Panthera tigris appear to be a distinct type of reverse ORFp, denoted r2. Both r1 and r2 ORFs were found in the rat genome (Rattus norvegicus). All reverse ORF proteins were checked for similarity to known domains using HMMer (Finn et al. 2011). The most significant hits came from Myotis brandtii (r1 ORF, only 19/68 non-redundant sequences), which showed homology to the Pico_P1A picornavirus coat protein; and Canis lupus (r2 ORF, all 81/81 non-redundant sequences), which showed a range of hits from various transporter and initiation molecules (e.g. ZIP: Zinc transporter, Rrn6: RNA polymerase I-specific transcription–initiation factor, Afi1: Docking domain of Afi1 for Arf3 in vesicle trafficking).
Discussion
Extinction of L1s in Mammalian Taxa – Known Versus New Events
An L1 element is called ‘extinct’ if it completely loses its ability to retrotranspose. If there is very low (but still extant) activity in the genome, this has been referred to as ‘quiescence’ rather than extinction (Yang et al. 2014). Figure 3 shows all of the known cases of L1 extinction (not quiescence) out of the 98 mammalian species analysed in this study: Three pteropodid bats (Cantrell et al. 2008; Yang et al. 2014) and the thirteen-lined ground squirrel Ictidomys tridecemlineatus (Platt and Ray 2012). Interestingly, the TBLASTN analysis found intact ORF2 in nucleotide sequences from squirrel – so in figure 3, this species is annotated L1-active. It is possible that squirrel is a case of quiescence rather than extinction, or the ORF2 regions are structurally conserved rather than functional. Other confirmed cases of L1 extinction include the spider monkey (Boissinot et al. 2004) and all studied Sigmondontinae rodents except for the Sigmodontini tribe (Casavant et al. 2000; Grahn et al. 2005), which were not included in this study because there are no public genome assemblies available.
Novel L1 extinction species candidates found in this study include eight rodents, five cetartiodactyls, one carnivore, one perissodactyl, four bats, two Insectivora, four Afrotherian mammals and one marsupial (fig. 3). Gallus et al. (2015) recently investigated L1 dynamics in Tasmanian devil – their results also suggest that this marsupial has lost L1 functionality. To our knowledge, the remaining species have not been previously studied as L1 extinction candidates, although some closely related species have been, for example, Peromyscus californicus (Casavant et al. 1998).
Evidence of a retro-element extinction event is often difficult to confirm, because we cannot determine whether it occurred in the individual genome or at the species level. The easiest extinction event to observe is one that is ancestral, such that a large monophyletic group of species all lack evidence of recent L1 activity (Grahn et al. 2005). For example, Cantrell et al. (2008) confirmed L1 extinction of the Pteropodidae megabat family by showing that the event had been inherited in 11 sampled genera. There are no other monophyletic extinction events shown in the mammalian phylogeny (fig. 3). Instead, all of the new L1 extinction candidate species appear paraphyletic or polyphyletic.
There are several possible explanations for these occurrences. First, these may be individual organism-specific changes – as with the putative extinction of L1s in the ground squirrel, which corresponded to a steady decline of all TE classes in that genome (Platt and Ray 2012), or the similar scenario seen in Tasmanian devil (Gallus et al. 2015). Second, the re-emergence or persistence of L1 activity in closely related species suggests that these are examples of quiescence rather than extinction. This may especially be true for rodents, where we already know of several extinct/quiescent species (Casavant et al. 1998, 2000). Such a scenario suggests that there is a fine line between calling an L1 active or extinct, and a lot of these rodents may have only recently become inactive. The fact that numerous rodent species (eight in fig. 3 alone, not including previous studies) have no intact ORF2 argues that the entire group may be headed towards L1 extinction (disregarding mouse and rat, which are extraordinarily L1-active). The naked mole rat (Heterocephalus glaber) and blind mole rat (Nannospalax galili) are among these putatively ‘L1-extinct’ species: Two species renowned for their cancer resistance. Given the deleterious effects that L1 activity can cause, if these rodents are truly L1-extinct, it would likely be a consequent of robust host suppression mechanisms (Deininger et al. 2003; Han and Boeke 2005).
Lastly, it is possible that these supposedly extinct species appear so because of the draft quality of the genome assemblies used. There are several cases (e.g. wallaby Macropus eugenii) where intact ORF2 could only be found in the NR/HTGS NCBI databases, not in the genome assembly. Indeed, many of the species colored in blue (e.g. Leptonychotes weddellii, Bubalus bubalis) have short Illumina read assemblies with low contig N50 values – making it virtually impossible to find perfectly intact ORF2 sequences. Gallus et al. (2015) experienced the same problem when mining the Tasmanian devil genome for intact L1s. More reliable analyses such as long read Sanger sequencing or in situ hybridization would be needed to confirm complete loss or presence of L1 activity (Grahn et al. 2005; Cantrell et al. 2008).
The Difference between Retrotransposition Potential and Activity
The majority of this study focuses on identifying L1 elements that have retrotransposition potential, and therefore may be active within the genome and causing change. But what does it mean for an L1 to be active? We can label an element as having the potential to be active by looking for intact open reading frames, or calculating the proportion of intact full-length L1s in the genome. But to be truly active, the element must provide evidence that it is doing something in the genome, not just that it has the potential to. So for L1 elements, effective activity should be confirmable by substantial replication and propagation of the element throughout the genome.
The distribution of L1* proportions shown in figure 5 clearly illustrates this concept. There are three things that are immediately obvious in this figure: (1) non-mammalian animal species (shown in red) and plant species (e.g. green alga) have a surprisingly high proportion of potentially active elements but low copy number; (2) the majority of mammals have a huge number of potentially active L1s, but a consistently low (<20%) proportion; (3) several mammalian species (e.g. minke whale, antelope, snub-nosed monkey, mouse, sheep) stand out because they have a high L1* proportion, unlike the other mammals. The variation between species illustrates those that are potentially active versus those that are truly active. However, we cannot establish a population variance because for almost all cases there is only one individual per species, due to the available data.
Addressing the first of these observations – non-mammalian species (plants and animals) all seem to have a relatively low L1 copy number. This is not unexpected in itself; many of these elements are divergent and have accumulated mutations, suggesting that they are older than their mammalian counterparts (as shown by the longer branch lengths in fig. 8). What is surprising is that, based on the identification of intact ORFs, a large proportion of L1s in these genomes seem putatively active. For instance, green alga (Coccomyxa subellipsoidea) only has 15 full-length L1s, yet all 15 of them are apparently active. But are these L1s really active? Such low copy number would suggest that there is high retrotransposition potential, but low effectiveness or a high turnover rate.
In contrast, we know that mammalian species typically have a high L1 copy number (Lander et al. 2001; Mouse Sequencing Consortium et al. 2002). We also know that L1 retrotransposition is extremely inefficient because the vast majority of new insertions are 5′ truncated and thus inactive (Sassaman et al. 1997; Boissinot et al. 2000). This seems to be the case for most of the mammals analyzed in this study: Although they have a high number of active L1s, the number of inactive L1s is much greater (∼80%); hence they have a low level of observable activity within the genome.
However, there are a few mammals that have both a high L1 copy number and a high active percentage in the genome. Indeed, the most significantly ‘hyperactive’ species (minke whale) has never been mentioned before in the context of L1 activity, yet it contains 5006 active L1s that make up more than 62% of the total full-length L1 content in the genome – far surpassing the retrotranspositional activity of mouse. This directly contradicts the belief that most full-length L1s are inactive or truncated during replication. As such, it is a good indication that these species are truly active, not just potentially active. These L1s are dynamically replicating and expanding within the genome, resulting in a large copy number of elements that share high pairwise identity with each other. Therefore, out of the 206 putatively active species found in this analysis, these five genomes would be the best model organisms for studying genomic change due to L1 retrotransposition.
The Master Lineage Paradigm
The master lineage model is an evolutionary scenario where the active elements in a genome give rise to a single active lineage that dominates long-term retrotransposition (Clough et al. 1996). Phylogenetic analyses such as dendrogram constructions are often used to give an indication of existent lineages (Grahn et al. 2005; Adelson et al. 2009), under the rationale that longer branch lengths represent accumulated mutations (including insertions and deletions) due to age, whereas shorter branch lengths signify younger, closely related elements with little nucleotide divergence from the master template. If all of the active elements form polytomies with very short branch lengths, as opposed to multiple divergent clusters, then this would be an example of a strict master lineage model.
It is hypothesized that there is selective pressure for the master LINE (and/or SINE) lineage to monopolise active retrotransposition in mammalian model organisms (Platt and Ray 2012). Our data supports this – all of the ‘hyperactive’ species and many of the potentially active ones contain a single active L1 family/cluster, as shown in figure 6 with the snub-nosed monkey example. This seems somewhat counterintuitive; given the vast number of active elements, it should be feasible for numerous independent lineages to amplify, over time. A possible explanation is that the single lineage we observe is due to a master element that was particularly effective at evading host suppression mechanisms, and thus initiated widespread retrotransposition throughout the genome.
In some species with relatively low active copy number, such as Myotis lucifugus (fig. 7), there appear to be multiple simultaneously active lineages. Myotis lucifigus also contains some L1 elements with a peculiar Type II ORF1p variant (fig. 9b), and some ORF1p with the traditional Transposase_22 domain, supporting the theory of different L1 lineages. A similar situation was observed in the (now extinct) megabat L1s (Yang et al. 2014) and two putatively active L1 lineages in rodent Peromyscus californicus (Casavant et al. 1998). There are various theories as to how multiple lineages may arise; for example, after a period of low activity, multiple ‘stealth driver’ (Cordaux and Batzer 2009) elements may be driven to retrotranspose at the same time; or horizontal acquisition of a retroelement from a different species can produce a foreign active lineage alongside the native lineage. Nonetheless, not much is known about how both lineages can be maintained, if there really is selective pressure to adhere to a master model. Yang et al. (2014) speculate that if the lineages are specialized in different tissue types (e.g. male germ line vs. female germ line), they can co-exist without competition – however, this is countered by the observation that in mouse, most L1 retrotransposition events seem to occur in the early embryo rather than in germ cells (Kano et al. 2009). Furthermore, the fact that we do not observe any high copy number species harboring more than one lineage suggests that multiple lineages are inhibitory to retrotransposition: Either through competition, or because it increases the chance that both lineages will be detected and suppressed by regulatory mechanisms, so neither lineage can effectively proliferate within the genome.
Discordance between ORF Nomenclature and Domain Classification
A predictable side effect of having access to more data and discovering new domains is that the existing nomenclature may need revision to reflect this new information. Based on the existing Type system for ORF1p elements (Khazina and Weichenrieder 2009), mammals typically have Type II; non-mammalian animals have both Types I and II; plants have variants of Type I; and the single remaining plant species (Coccomyxa subellipsoidea) belongs to Type V: Unclassified ORF proteins (fig. 9a and b). Such a categorization can be misleading because it implies that Type I sequences are alike and share high amino acid similarity – and even the HTH_1 domain in C. subellipsoidea cannot be that distantly related, by virtue of it being an ‘ORF1p’. But at what point does a domain variant become too different to be an ORF1p? A phylogeny of ORF1p could not be reliably inferred because of the extreme variation found within these sequences, and the all-against-all clustering analysis showed that there are multiple independent ORF1p clusters within each species - despite using the default settings where two proteins in a pair are included in the same family if the homologous segment pairs have at least 35% similarity over 80% coverage (Penel et al. 2009). The protein domain network diagrams (e.g. fig. 9c) further show that the ‘known’ ORF1 domains are not always the key identifiers, and there are numerous strongly associated domains that are often overlooked.
Accordingly, we propose a more informative revision to the nomenclature to refer to ORF proteins by the dominant functional domain(s); for example, ORF2p = RVT_1-ORFp for mammals, or (RVT_1 + RVT_3)-ORFp for most plants (see supplementary table S10 and fig. S6 a–g, Supplementary Material online). Likewise, ORF1p = HTH_1-ORFp for C. subellipsoidea. This allows us to forego predetermined Type or ORF# labels, especially for unusual cases. The discovery of additional ORF proteins such as the primate-specific ORF0 (Denli et al. 2015) or the reverse ORF proteins found in this study (fig. 10) makes a compelling argument for re-naming.
Confounding Bias Due to Genome Assembly Quality
Advances in technology mean that genomes are now being sequenced at alarmingly fast rates. However, once sequenced, many genomes tend to remain in their error riddled, scaffolded state. The majority of genomes used in this study are draft assemblies, so it is important to check that the quality of the assembly is not affecting the results (either by restricting the ability to detect repetitive 6kb elements, or by creating false positive hits from misread errors). Accordingly, we analysed independently-assembled closely related species (within the same genus or species) and used multiple searching strategies (e.g. LASTZ with genomic data versus TBLASTN with nucleotide databases). Consider the three horse genomes included in this study: Equus przewalski (submitted by IMAU, contig N50 of 57,610, SOAPdenovo assembly method used), Equus caballus Thoroughbred (submitted by GAT, contig N50 of 112,381, ARACHNE2.0 assembly method used) and Equus caballus Mongolian (submitted by IMAU, contig N50 of 40,738, SOAPdenovo assembly method used) (see supplementary tables S1–S3, Supplementary Material online). Based on the submitter, contig N50 and assembly method, Equus przewalski and the Mongolian Equus caballus would be expected to be the most similar. Based on species relationships, one would expect the two Equus caballus horses to be more similar. However, the actual findings show that while all three horses are marked L1*, only Equus przewalski and Equus caballus (Thoroughbred) have intact ORF2 in the genome. Equus caballus (Mongolian) was determined L1-active solely based on the TBLASTN results. This is a known problem with using draft assemblies – and it has been detailed previously with the Tasmanian Devil genome (Gallus et al. 2015), as well as the wallaby and cat genomes (Pontius et al. 2007; Renfree et al. 2011). It is likely that as genome assemblies improve, it will become possible to detect more ORF2-intact, active L1 (although the overall L1-status is unlikely to change).
As a contrasting example, the three Arabidopsis species that were submitted independently (A. halleri: TokyoTech, A. lyrata: JGI, A. thaliana: Arabidopsis Information Resource), have very different contig N50 values (A. halleri: 2864, A. lyrata: 227,391, A. thaliana: 11,194,537) and used different sequencing strategies (A. halleri: Illumina, A. lyrata: Sanger, A. thaliana: BAC physical map then Sanger sequencing of BACs) have very similar results in terms of L1 presence, activity and open reading frame structure. In fact, Illumina seems to be the most widely used sequencing technology across all the genomes (mammalian, non-mammalian, and plant) but it does not appear to introduce platform specific artifacts. This is encouraging because it demonstrates that draft genomes can be used to study repetitive sequences such as L1s, as long as suitable quality controls are taken into account.
The assembly level does not seem to hinder the ability to detect highly L1-active species (more so the ability to confirm L1 extinction). Out of the five so-called ‘hyperactive’ mammalian species labelled in figure 5, three (minke whale, snub-nosed monkey, antelope) are scaffold-level assemblies, whereas two (mouse and sheep) are chromosome-level with noticeably higher N50 values. One might argue that this just shows that draft assemblies are more likely to have duplication or misread errors, leading to greater L1 copy number. However, a de-duplication test of these genomes found very few identical hits (e.g. minke whale contains 13,681 L1s over 3 kb: The largest cluster of duplicates had 47 elements, and only two L1s shared the same 1 kb flanking region). This suggests that the majority of identical hits are likely to be true duplicates rather than assembly errors.
Implications for Our Perception of Genome Evolution
This study complements those of Kordis et al. (2006) (deuterostomes), Khan et al. (2006) (primates), Sookdeo et al. (2013) (mouse), Yang et al. (2014) (megabats), Metcalfe and Casane (2014) (Jockey non-LTR elements), and Heitkam et al. (2014) (plants) in demonstrating the diversity of TE evolutionary patterns across species. We have identified over 10 million L1 sequences from 503 different genomes, including ORF1 and ORF2 proteins with novel domain variations that strain the current L1 classification system. While most animals and plants still exhibit some form of L1 activity, the discovery of new extinction candidates leaves us better equipped to identify common factors in the genomic landscape that contribute to TE suppression (particularly in species with desirable characteristics, such as cancer resistance). Conversely, investigation into ‘hyperactive’ species such as minke whale and snub-nosed monkey, whose retrotranspositional activity seems to far surpass that of human, rat and mouse, could be used to study the extent to which L1s cause genomic change. Perhaps the presence of reverse ORFs helps the L1 in these species to attain hyperactivity. Multiple lines of evidence suggest that L1s can form an ‘ORF-anage’ by recruiting functional domains from the host, thus propagating their activity in the genome. As always, it is likely that our findings here are only the very tip of the iceberg. We present this data with the hope that it will provide a definitive reference for future studies, aiding our understanding of eukaryotic evolution.
Supplementary Material
Supplementary figures S1–S7 and Supplementary tables S1–S11 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We would like to thank our collaborators (Broad Institute of MIT and Harvard for the chromosome-level elephant genome, and Frank Grutzner from the University of Adelaide for the echidna assembly) for making private genome assemblies available to us. We would also like to thank Iain Searle for taking the time to read the manuscript in full and offer helpful comments, and the anonymous reviewers for their clear, detailed feedback. Finally, this paper would not be possible without the invaluable insights of Lu Zeng, R language mastery of Reuben Buckley, HMMer knowledge of Zhipeng Qu, and extraordinary IT support from Matt Westlake.
Literature Cited
- Abrusan G, Szilagyi A, Zhang Y, Papp B. 2013. Turning gold into ‘junk’: transposable elements utilize central proteins of cellular networks. Nucleic Acids Res. 41:3190–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelson DL, Raison JM, Edgar RC. 2009. Characterization and distribution of retrotransposons and simple sequence repeats in the bovine genome. Proc Natl Acad Sci U S A. 106:12855–12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelson DL, Raison JM, Garber M, Edgar RC. 2010. Interspersed repeats in the horse (Equus caballus); spatial correlations highlight conserved chromosomal domains. Anim Genet. 41 (Suppl 2):91–99. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Beck CR, et al. 2010. LINE-1 retrotransposition activity in human genomes. Cell 141:1159–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck RM, Bininda-Emonds OR, Cardillo M, Liu FG, Purvis A. 2006. A higher-level MRP supertree of placental mammals. BMC Evol Biol. 6:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedler J, Tu Z. 2003. Non-LTR retrotransposons in the African malaria mosquito, Anopheles gambiae: unprecedented diversity and evidence of recent activity. Mol Biol Evol. 20:1811–1825. [DOI] [PubMed] [Google Scholar]
- Blass E, Bell M, Boissinot S. 2012. Accumulation and rapid decay of non-LTR retrotransposons in the genome of the three-spine stickleback. Genome Biol Evol. 4:687–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissinot S, Chevret P, Furano AV. 2000. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol Biol Evol. 17:915–928. [DOI] [PubMed] [Google Scholar]
- Boissinot S, Roos C, Furano AV. 2004. Different rates of LINE-1 (L1) retrotransposon amplification and evolution in New World monkeys. J Mol Evol. 58:122–130. [DOI] [PubMed] [Google Scholar]
- Brouha B, et al. 2003. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci U S A. 100:5280–5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantrell MA, Scott L, Brown CJ, Martinez AR, Wichman HA. 2008. Loss of LINE-1 activity in the megabats. Genetics 178:393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casavant NC, Lee RN, Sherman AN, Wichman HA. 1998. Molecular evolution of two lineages of L1 (LINE-1) retrotransposons in the california mouse, Peromyscus californicus . Genetics 150:345–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casavant NC, et al. 2000. The end of the LINE? Lack of recent L1 activity in a group of South American rodents. Genetics 154:1809–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552. [DOI] [PubMed] [Google Scholar]
- Chen JM, Stenson PD, Cooper DN, Ferec C. 2005. A systematic analysis of LINE-1 endonuclease-dependent retrotranspositional events causing human genetic disease. Hum Genet. 117:411–427. [DOI] [PubMed] [Google Scholar]
- Clough JE, Foster JA, Barnett M, Wichman HA. 1996. Computer simulation of transposable element evolution: random template and strict master models. J Mol Evol. 42:52–58. [DOI] [PubMed] [Google Scholar]
- Cordaux R, Batzer MA. 2009. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 10:691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cost GJ, Feng Q, Jacquier A, Boeke JD. 2002. Human L1 element target-primed reverse transcription in vitro. EMBO J. 21:5899–5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger PL, Moran JV, Batzer MA, Kazazian HH., Jr. 2003. Mobile elements and mammalian genome evolution. Curr Opin Genet Dev. 13:651–658. [DOI] [PubMed] [Google Scholar]
- Denli AM, et al. 2015. Primate-Specific ORF0 Contributes to Retrotransposon-Mediated Diversity. Cell 163:583–593. [DOI] [PubMed] [Google Scholar]
- Dewannieux M, Esnault C, Heidmann T. 2003. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 35:41–48. [DOI] [PubMed] [Google Scholar]
- Douzery EJ, et al. 2014. OrthoMaM v8: a database of orthologous exons and coding sequences for comparative genomics in mammals. Mol Biol Evol. 31:1923–1928. [DOI] [PubMed] [Google Scholar]
- Duvernell DD, Pryor SR, Adams SM. 2004. Teleost fish genomes contain a diverse array of L1 retrotransposon lineages that exhibit a low copy number and high rate of turnover. J Mol Evol. 59:298–308. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. [DOI] [PubMed] [Google Scholar]
- Esnault C, Maestre J, Heidmann T. 2000. Human LINE retrotransposons generate processed pseudogenes. Nat Genet. 24:363–367. [DOI] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR. 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39:W29–W37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38:D211–D222. 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furano AV. 2000. The biological properties and evolutionary dynamics of mammalian LINE-1 retrotransposons. Prog Nucleic Acid Res Mol Biol. 64:255–294. [DOI] [PubMed] [Google Scholar]
- Furano AV, Duvernell DD, Boissinot S. 2004. L1 (LINE-1) retrotransposon diversity differs dramatically between mammals and fish. Trends Genet. 20:9–14. [DOI] [PubMed] [Google Scholar]
- Gallus S, et al. 2015. Evolutionary histories of transposable elements in the genome of the largest living marsupial carnivore, the Tasmanian devil. Mol Biol Evol. 32:1268–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham T, Boissinot S. 2006. The genomic distribution of L1 elements: the role of insertion bias and natural selection. J Biomed Biotechnol. 2006:75327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grahn RA, Rinehart TA, Cantrell MA, Wichman HA. 2005. Extinction of LINE-1 activity coincident with a major mammalian radiation in rodents. Cytogenet Genome Res. 110:407–415. [DOI] [PubMed] [Google Scholar]
- Gregory SG, et al. 2002. A physical map of the mouse genome. Nature 418:743–750. [DOI] [PubMed] [Google Scholar]
- Han JS, Boeke JD. 2005. LINE-1 retrotransposons: modulators of quantity and quality of mammalian gene expression?. Bioessays 27:775–784. [DOI] [PubMed] [Google Scholar]
- Harris RS. 2007. Improved Pairwise Alignment of Genomic DNA. Ph.D. Thesis, The Pennsylvania State University.
- Heitkam T, et al. 2014. Profiling of extensively diversified plant LINEs reveals distinct plant-specific subclades. Plant J. 79:385–397. [DOI] [PubMed] [Google Scholar]
- Heitkam T, Schmidt T. 2009. BNR - a LINE family from Beta vulgaris - contains a RRM domain in open reading frame 1 and defines a L1 sub-clade present in diverse plant genomes. Plant J. 59:872–882. [DOI] [PubMed] [Google Scholar]
- Heras SR, et al. 2006. L1Tc non-LTR retrotransposons from Trypanosoma cruzi contain a functional viral-like self-cleaving 2A sequence in frame with the active proteins they encode. Cell Mol Life Sci. 63:1449–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashiyama T, Noutoshi Y, Fujie M, Yamada T. 1997. Zepp, a LINE-like retrotransposon accumulated in the Chlorella telomeric region. EMBO J. 16:3715–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janecka JE, et al. 2007. Molecular and genomic data identify the closest living relative of primates. Science 318:792–794. [DOI] [PubMed] [Google Scholar]
- Jurka J, et al. 2005. Repbase update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110:462–467. [DOI] [PubMed] [Google Scholar]
- Kaer K, Speek M. 2013. Retroelements in human disease. Gene 518:231–241. [DOI] [PubMed] [Google Scholar]
- Kano H, et al. 2009. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 23:1303–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. 2003. The esterase and PHD domains in CR1-like non-LTR retrotransposons. Mol Biol Evol. 20:38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH., Jr. 1998. Mobile elements and disease. Curr Opin Genet Dev. 8:343–350. [DOI] [PubMed] [Google Scholar]
- Kazazian HH., Jr. 2000. Genetics. L1 retrotransposons shape the mammalian genome. Science 289:1152–1153. [DOI] [PubMed] [Google Scholar]
- Kearse M, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, et al. 2002. The human genome browser at UCSC. Genome Res. 12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan H, Smit A, Boissinot S. 2006. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res. 16:78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khazina E, Weichenrieder O. 2009. Non-LTR retrotransposons encode noncanonical RRM domains in their first open reading frame. Proc Natl Acad Sci U S A. 106:731–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohany O, Gentles AJ, Hankus L, Jurka J. 2006. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics 7:474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima KK, Fujiwara H. 2004. Cross-genome screening of novel sequence-specific non-LTR retrotransposons: various multicopy RNA genes and microsatellites are selected as targets. Mol Biol Evol. 21:207–217. [DOI] [PubMed] [Google Scholar]
- Kondo-Iida E, et al. 1999. Novel mutations and genotype-phenotype relationships in 107 families with Fukuyama-type congenital muscular dystrophy (FCMD). Hum Mol Genet. 8:2303–2309. [DOI] [PubMed] [Google Scholar]
- Kordis D, Lovsin N, Gubensek F. 2006. Phylogenomic analysis of the L1 retrotransposons in Deuterostomia. Syst Biol. 55:886–901. [DOI] [PubMed] [Google Scholar]
- Kriventseva EV, et al. 2015. OrthoDB v8: update of the hierarchical catalog of orthologs and the underlying free software. Nucleic Acids Res. 43:D250–D256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860–921. [DOI] [PubMed] [Google Scholar]
- Lee J, et al. 2007. Different evolutionary fates of recently integrated human and chimpanzee LINE-1 retrotransposons. Gene 390:18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. 2006. The origins of eukaryotic gene structure. Mol Biol Evol. 23:450–468. [DOI] [PubMed] [Google Scholar]
- Maddison DR, Schulz KS. The Tree of Life Web Project. Available at: http://tolweb.org. (2007).
- Mathews LM, Chi SY, Greenberg N, Ovchinnikov I, Swergold GD. 2003. Large differences between LINE-1 amplification rates in the human and chimpanzee lineages. Am J Hum Genet. 72:739–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalfe CJ, Casane D. 2014. Modular organization and reticulate evolution of the ORF1 of Jockey superfamily transposable elements. Mob DNA 5:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele V, Penel S, Duret L. 2011. Ultra-fast sequence clustering from similarity networks with SiLiX. BMC Bioinformatics 12:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran JV, et al. 1996. High frequency retrotransposition in cultured mammalian cells. Cell 87:917–927. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520–562. [DOI] [PubMed] [Google Scholar]
- Murphy WJ, et al. 2001. Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science 294:2348–2351. [DOI] [PubMed] [Google Scholar]
- Nakamura M, Okada N, Kajikawa M. 2012. Self-interaction, nucleic acid binding, and nucleic acid chaperone activities are unexpectedly retained in the unique ORF1p of zebrafish LINE. Mol Cell Biol. 32:458–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick PA, Basta H, Floumanhaft M, McClure MA, Boissinot S. 2009. The evolutionary dynamics of autonomous non-LTR retrotransposons in the lizard Anolis carolinensis shows more similarity to fish than mammals. Mol Biol Evol. 26:1811–1822. [DOI] [PubMed] [Google Scholar]
- Odon V, et al. 2013. APE-type non-LTR retrotransposons of multicellular organisms encode virus-like 2A oligopeptide sequences, which mediate translational recoding during protein synthesis. Mol Biol Evol. 30:1955–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penel S, et al. 2009. Databases of homologous gene families for comparative genomics. BMC Bioinformatics 10 (Suppl 6):S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzkofer T, Dandekar T, Zemojtel T. 2005. L1Base: from functional annotation to prediction of active LINE-1 elements. Nucleic Acids Res. 33:D498–D500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt RN, 2nd, Ray DA. 2012. A non-LTR retroelement extinction in Spermophilus tridecemlineatus. Gene 500:47–53. [DOI] [PubMed] [Google Scholar]
- Pontius JU, et al. 2007. Initial sequence and comparative analysis of the cat genome. Genome Res. 17:1675–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. 2010. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renfree MB, et al. 2011. Genome sequence of an Australian kangaroo, Macropus eugenii, provides insight into the evolution of mammalian reproduction and development. Genome Biol. 12:R81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassaman DM, et al. 1997. Many human L1 elements are capable of retrotransposition. Nat Genet. 16:37–43. [DOI] [PubMed] [Google Scholar]
- Sayers EW, et al. 2012. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 40:D13–D25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott AF, et al. 1987. Origin of the human L1 elements: proposed progenitor genes deduced from a consensus DNA sequence. Genomics 1:113–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seleme MC, et al. 2006. Extensive individual variation in L1 retrotransposition capability contributes to human genetic diversity. Proc Natl Acad Sci U S A. 103:6611–6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyshlyaev G, Voigt F, Blinov A, Barabas O, Novikova O. 2013. Acquisition of an Archaea-like ribonuclease H domain by plant L1 retrotransposons supports modular evolution. Proc Natl Acad Sci U S A. 110:20140–20145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sookdeo A, Hepp CM, McClure MA, Boissinot S. 2013. Revisiting the evolution of mouse LINE-1 in the genomic era. Mob DNA 4:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollis M, Boissinot S. 2013. Lizards and LINEs: selection and demography affect the fate of L1 retrotransposons in the genome of the green anole (Anolis carolinensis). Genome Biol Evol. 5:1754–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh AM, Kortschak RD, Gardner MG, Bertozzi T, Adelson DL. 2013. Widespread horizontal transfer of retrotransposons. Proc Natl Acad Sci U S A. 110:1012–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters PD, Dobigny G, Waddell PJ, Robinson TJ. 2007. Evolutionary history of LINE-1 in the major clades of placental mammals. PLoS One 2:e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Brunsfeld J, Scott L, Wichman H. 2014. Reviving the dead: history and reactivation of an extinct l1. PLoS Genet. 10:e1004395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmasek C. 2015. Archaeopteryx: visualization, analysis, and editing of phylogenetic trees. Available at: https://sites.google.com/site/cmzmasek/home/software/archaeopteryx.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.