Abstract
Dynamic reprogramming of transcriptional networks enables cells to adapt to a changing environment. Thus, it is crucial not only to understand what gene targets are regulated by a transcription factor (TF) but also when. This review explores the way TFs function with respect to time, paying particular attention to discoveries made in plants - where coordinated, genome-wide responses to environmental change is crucial to the survival of these sessile organisms. We investigate the molecular mechanisms that mediate transient TF-DNA binding, and assess how these rapid and dynamic interactions translate to long-term temporal regulation of genomes. We also discuss how current molecular techniques can catch, and sometimes miss, transient TF-target interactions that underlie dynamic cellular responses.
Keywords: Transcription factor, transcription dynamics, transient DNA binding, plant genomics, epigenetics
Graphical abstract
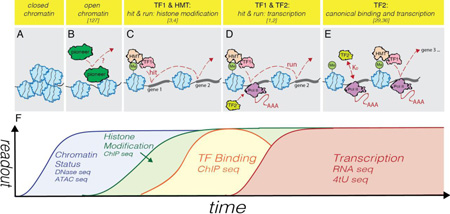
Tracking genome-wide dynamic transcription over time. (A–B) Closed chromatin can be rendered accessible through pioneer TF binding. (C) Post translational modification of histones via histone methyltransferases (HMTs) can earmark loci for transcriptional activation. In the hit-and-run transcriptional model, a TF can first assist with localizing HMTs to target sites, and as well as (D) activate transcription. Once a loci has been 'hit', the hit-and-run complex can move on to activate other sites (E) while transcriptional output is maintained by a secondary TF (TF2). (F) Complex molecular dynamics can be monitored genome-wide over time by combining different genomic techniques.
Introduction
Over the last few decades, discovering the gene targets of transcription factors (TFs) has been a central aim of molecular biology. Understanding which TFs regulate what gene targets have helped explain how fundamental biological processes are mediated, such as how a cell chooses its lineage, how it responds to stress, and how it maintains homeostasis. While mapping the complete set of interactions between TFs and gene targets genome-wide can provide a holistic view of gene regulation within an organism [5, 6], characterizing all possible regulatory interactions within a genome is proving an arduous task. Higher eukaryotes typically code hundreds to thousands of TFs [7, 8], and since each TF frequently regulates hundreds to thousands of gene targets, genomes can potentially encode well over 1 × 105 unique regulatory interactions. Consequently, the topology of gene regulatory networks in eukaryotes is highly integrated, complex, and by no means as yet delineated in any species.
If untangling this web of gene regulatory interactions did not seem challenging enough, gene regulatory networks are not static. Organisms create dynamic gene expression patterns by temporally regulating their TFs both transcriptionally and post-translationally [6, 9–11]. Consequently, gene regulatory networks are inherently plastic, where external signals and regulatory interactions between TFs and their targets cause gene expression patterns to be in constant flux. This fluid property of regulatory networks makes them, in a sense, 'moving targets' - their structure cannot be captured at a single time-point or within a single measurement. Only through observing how TFs function over time, can transient gene regulatory events be detected, and the full array of TF-target interactions within a genome captured.
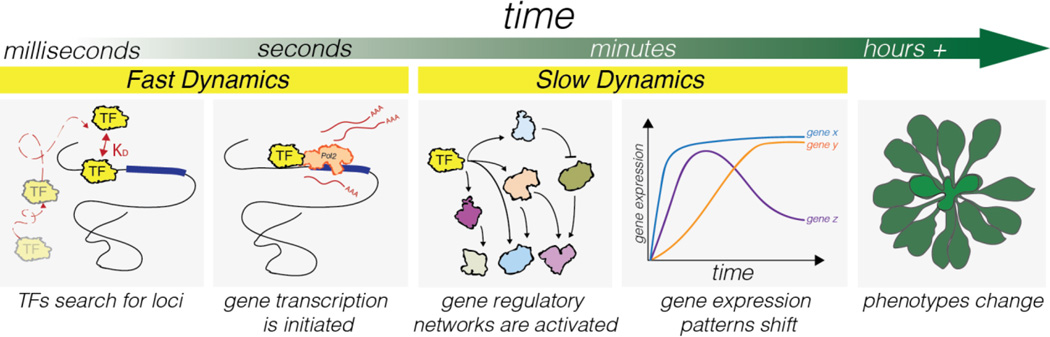
By what means do TFs temporally regulate the expression of target genes within a genome? Examining TF behavior over time reveals that they are dynamic in two discrete ways, as outlined in Figure 1. (i) One way is observed at the molecular level, where monitoring TF molecules has indicated that a TF's interaction at a target locus is highly transient, lasting only seconds (dubbed 'fast dynamics', as illustrated in Figure 1) [12–16]. When this fleeting TF-target interaction is the rate-limiting step for transcription initiation, it results in stochastic gene expression [17, 18]. (ii) Changing the frequency at which TF-target binding events occur is another dynamic property of TFs (dubbed 'slow dynamics'). Through altering TF-DNA binding equilibriums or chromatin structure, the probability of TF binding events occurring can be changed, and thus lead to changes gene differential expression patterns. [19–21]. Understanding how rapid and transient binding of TFs at individual loci at the molecular level (i) translates to temporal gene regulation at the genomic level (ii), is the goal of this review.
Figure 1. TFs display dynamic properties over several orders of time.
Within seconds, TFs can locate target loci within a genome and initiate transcription (a 'fast dynamic'). Such binding events can activate gene regulatory network cascades, creating gene regulation patterns that persist from minutes to hours (a 'slow dynamic'). By changing expression patterns of gene regulatory networks, phenotypes can change over time.
While drawing from insights made across Eukarya, this article will play particular attention to plants. Plant gene regulatory networks are probably larger than those found in other kingdoms, since size and complexity of TF families in plants is highest among eukaryotes [8]. Plant's larger network size may stem from their sessile nature; they must be highly adaptive to their immediate environment in order to survive, and thus may require larger gene regulatory spaces to generate suitable adaptive phenotypes. Delineating the topologies of plant gene networks is a promising approach for crop scientists to generate novel plant phenotypes, since rewiring of gene regulatory networks through mutations in TF coding and regulatory regions has been found a principle driver of crop domestication phenotypes [22, 23].
A Chance Encounter
What are the molecular mechanisms that govern how long a TF resides at a gene target, and how often that target is bound? In this section, we summarize the TF-DNA binding dynamics displayed at the molecular level. While these highly transient interactions have been studied across a range of eukaryotes at the single molecule level, they have not yet been observed directly in plants. Consequently for the time being, plant molecular biologists must assume that TF-DNA interaction dynamics detected in other eukaryotes also apply to plants.
Measuring binding-length times of single TF molecules within isolated cells has been accomplished through a variety of methods, such as single molecule tracking, fluorescence recovery after photobleaching, and fluorescence correlation spectroscopy (a review of these approaches can be found [12]). Together, these techniques have shown that the time a TF spends residing at its canonical binding sequence lasts for only seconds. For example, the binding time for the human TF STAT1 is 0.5 s [13], the tumor suppressor TF p53 binds for ~3.6 s [14, 15], the glucocorticoid receptor binds for ~6.5 s [15, 24], and Sox2 in embryonic stem cells binds its targets for 12 s [16].
As yet, it is unclear if the differences between TF residence times are related to the strength of interaction between TF and its binding sequence (as captured by dissociation rates measured by in vitro methods) whether binding length time is also regulated via interactions with other trans-acting factors in vivo [25–27], or even if the difference simply stems from technical variability within the techniques themselves [28]. However, evidence suggests that longer residence times of the TF on the target promoter do indeed increase mRNA output. For example, in S. cerevisiae, time spent by the TF Rap1 at a locus was positively correlated with RNA Polymerase (Pol II) recruitment and amount of mRNA molecules produced. Conversely, genes bound by Rap1 for shorter periods of time had lower transcriptional output (known as 'treadmilling') [29]. However, weak, treadmilling interactions may not always be biologically insignificant. Modeling evidence performed in E. coli suggest that weak binding by TF repressors can actually be favorable for reducing noise within gene regulatory systems, while at the same time allowing target gene expression to remain high [30].
After dissociating from target loci, TFs are free to rebind elsewhere in the genome. Even though it is in close proximity, the chances of a TF re-associating with the same locus diminishes once it has moved further away than its own diameter [31]. When unbound, TFs rely on passive diffusion to forage for their respective binding sequence within the nucleus (regarded as 3D movement) [31, 32]. TFs can also interact non-specifically with accessible DNA and scan along the double helix for a short period of time (milliseconds) over short distances of 45 – 100 bp [32, 33]. Regarded as 1D movement, this scanning ability greatly reduces locus search times [32–34]. Reports vary on the amount of time spent between 1D and 3D search modes; while some estimate approximately 75% of TFs may be unbound and diffuse within the nucleus, other reports suggest that up to 90% may be actively scanning DNA through 1D interactions [28, 31, 32, 34].
The amount of time it takes a TF to find a gene target varies depending on genome size and number of targets present. It is estimated to take up to 5 hours for one molecule of a TF (Mbp1p) to find a single target in the yeast genome [35]. Conversely, the lac repressor in E. coli requires only 270 s [34]. At these rates, having only TF per nucleus will not propagate gene regulatory signals effectively. Thus, having a population of TFs searching simultaneously is critical. In yeast, the number of TF molecules per cell can range from 4 × 103 – 2.5 × 106 - where increasing concentration reduces the effective search time [15, 35–38]. However, even at high concentrations not all gene targets will be saturated by TF-binding; there will still be many instances when target loci remain vacant [15, 36].
Since TF diffusion within the nucleus is random, the chance of a TF binding to a particular cis-element at any particular time is stochastic. Thus, when TF-target binding is the rate-limiting step, TF search times result in stochastic gene expression [32, 35], where increasing TF concentration improves both the probability of transcription occurring as well as the consistency of gene expression. Visualization of mRNA molecules within single cells through in situ hybridization of antisense mRNA probes and live mRNA imaging have captured stochastic expression events caused by TF binding [17, 39]. Transcript production from a single locus shows that transcription occurs intermittently in transcriptional bursts, where increasing TF concentration within the cell increases the size of burst events, and thus mRNA output [17]. This effect is consistent with the observation that TFs do not stably bind loci, and supports the hypothesis that transcription is reinitiated each time a TF associates with a target.
Stochastic binding of TFs can cause to stochastic gene expression, but does this noise filter through to effect protein abundance? For a particular locus, stochastic mRNA production can cause protein abundance to be stochastic if mRNA or protein degradation rates are rapid. Conversely, if the transcripts or proteins produced are stable, then rapid fluctuations in mRNA abundance will be buffered [18]. Noise in protein translation may be additionally buffered by the heat shock protein HSP90, a chaperone that mediates many protein regulatory interactions [40]. This has evidence in Arabidopsis, where inhibition of HSP90 produced a range of diverse phenotypes from genetically identical plants. Since this phenotypic variation could not be genetic, it is possible that losing HSP90 buffering capacity allows for stochastic fluctuations in protein translation to leak through and effect phenotype [18, 41]. Highly Occupied Target (HOT) regions - regions of DNA that contain a high proportion of non-canonical TF binding [42] - may function like HSP90 by reducing the noise caused by translational bursts in TF production. By sequestering TFs away from gene targets, off-target binding to 'decoy' sites may buffer fluctuations in TF concentration, thereby maintaining a mean number of active TFs within the cell over time [43, 44].
Another way genomes may reduce the effect of TF search times and transient binding causing stochastic fluctuations in gene expression is by regulating loci with multiple TFs. Evidence indicates that gene expression amplitude increases with number of TF binding sites at promoters [27]. Also, yeast one-hybrid assays have revealed that 52 different TFs bind to modulate expression of the Arabidopsis master clock gene CCA1 [45]. Similarly the Arabidopsis nitrate transporter NRT2.1 has been found regulated by at least 5 TFs in vivo [1, 19, 46–48]. By not depending on one rate-limiting TF to activate gene expression, mRNA production may be more consistent over time. Indeed, binding of multiple TFs to a promoter has been associated with broad, high gene expression patterns in Arabidopsis [49] and other eukaryotes [50], where individual TFs may only have a small impact expression levels. Similarly, recruitment of general transcriptional co-factors or histone modifiers that are prerequisites for transcription initiation may weaken the effect of TF search times causing stochastic gene expression [51].
Improving The Odds
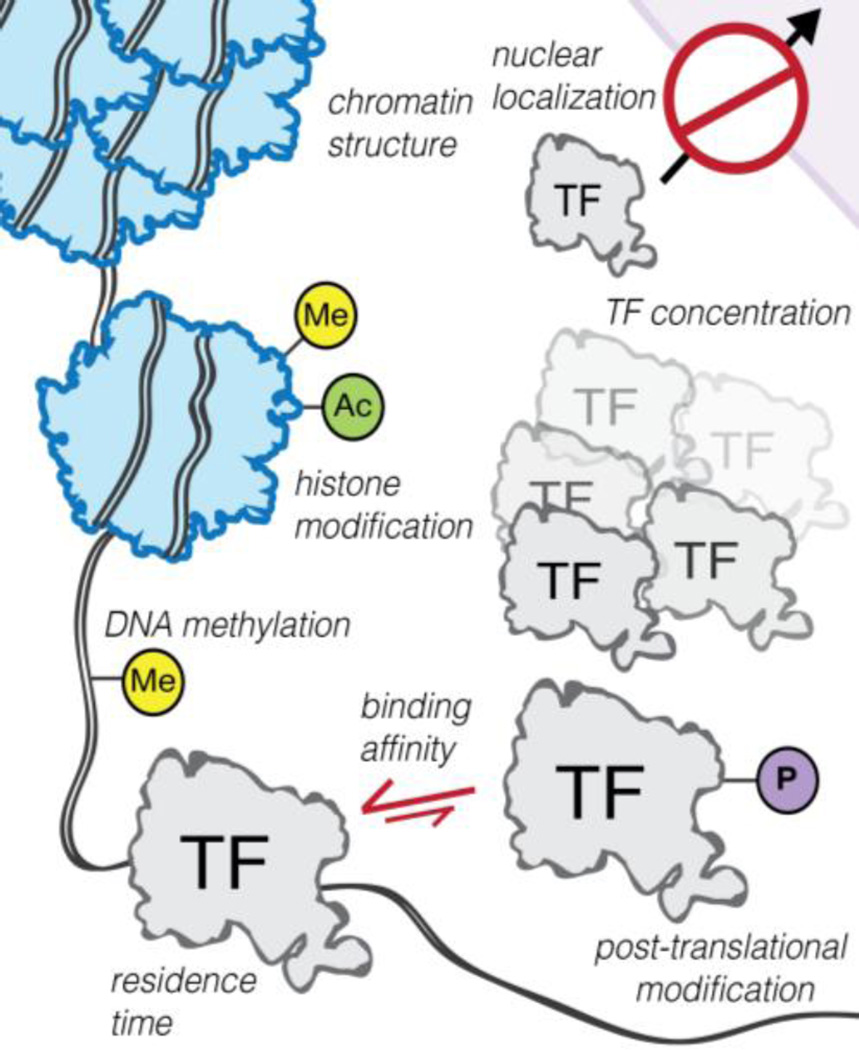
Passive movement and transient binding of TFs determine the timing and consistency of gene expression. When gene regulation patterns need to change, TF signal transduction can be altered by changing the abundance of active TF molecules within the cell, as discussed in the previous section. In this section, we discuss the additional molecular strategies that are employed by plants to alter TF binding dynamics (as shown in Figure 2), as well as how rapidly these strategies can be implemented by the cell.
Figure 2. Modes of Changing TF-DNA Binding Dynamics.
Many molecular factors drive TF-DNA associations within the cell. For example, localizing TFs within the nucleus can be a rapid means to promote TF-DNA interactions in response to a specific signaling cue or stress. Altering the epigenome through histone or chromatin modification can globally transform the TF-DNA binding landscape to drive environmental responses or developmental programs.
Post-translational regulation can be employed to change the TF equilibrium between nucleus and cytoplasm. Expressing TFs and then concentrating them within the nucleus when they are needed is a more rapid means of signal transduction than requiring TFs to be first transcribed. For example, the TF NLP7 is expressed constitutively within the Arabidopsis root, and its nuclear export signal allows it to freely diffuse across the nuclear membrane [19, 52]. However upon exposing the root to nitrate, NLP7 becomes retained within the nucleus, possibly through a post-translational modification that inhibits its nuclear export signal. In this way exposure to nitrate effectively concentrates the TF NLP7 within the nucleus in under 3 minutes, and consequently increases NLP7-target interactions, including those involved in nitrogen assimilation [19].
By the same means, the plant pathogen response, which also benefit from an immediate gene regulatory response, is in part mediated by TF nuclear localization dynamics, as demonstrated for TFs bZIP10 and EDS1 [53–56]. Interestingly, just like NLP7, these TFs were found within the nucleus at basal levels under normal conditions; a change in their relative concentration within the nucleus allowed the cell to launch pathogen-related gene regulatory responses. This suggests that a nominal level of TF-DNA interactions occur before nuclear localization is signaled, and thus that gene regulatory pathways are not always simply 'on-off' systems, but rather exist on a quantitative continua [36].
Altering the abundance of active TFs within nuclei is not the only means by which cells can affect temporal TF-DNA binding. Genomes can also alter TF-DNA binding dynamics through epigenetic regulation. One way to change TF binding ability epigenetically is via post translational modification of histones. Histone acetylation or methylation patterns can promote or occlude TF binding at loci [57–60], though the specific molecular interactions responsible are not yet clear. Histone modifications may cause nucleosomes to weaken their interaction with DNA and thus make TF binding more favorable, or may directly mediate TF binding events by regulating TF dissociation rates [26, 61, 62]. Histone modifications are regulated by the dynamic binding of chromatin modifying proteins such as histone acetyltransferases (HATs), which, like TFs, also associate with their target for the order of seconds [63]. However, unlike TFs, HATs need only associate once with a locus to have a lasting effect on transcription, since the acetyl post-translational modification remains after HAT dissociation [64]. In this way, HATs are able to rapidly acetylate a large number of target loci genome-wide (the same case can for transient DNA binding can be made for histone methyltransferases (HMTs) [64]). Histone marks have been found dynamically regulated to synchronize gene activation over time. For example, H3K4me3 and H3K56ac marks oscillate with loci associated with the circadian clock [65, 66]. Post translational modification of histones has also proven dynamic in response to environmental stress. In Arabidopsis drought stress induces H3K4me3 marks at drought responsive genes to serve as a form of transcriptional memory; earmarking sites for rapid re-activation under recurring drought stress [67, 68]. Indeed, in rice [69, 70] and maize [71] histone modifications have also proven dynamic in response to abiotic stress and have an effect on transcriptional output.
Like histone modification, the DNA methylation landscape is dynamic over time, and is another means for genomes to control TF-DNA interactions. For example, DNA methylation patterns have been shown to be dynamic in Arabidopsis in response to pathogen infection or salicylic acid treatment - where these changes correlate with changes gene expression [72]. However, unlike histone modifications, which can be either permissive or restrictive to transcription [57, 73], the addition of methyl groups of promoter regions inhibits gene expression in most cases, possibly by inhibiting TF binding or transcriptional machinery recruitment [59, 74, 75]. Indeed, via an ex vivo TF binding assay, it was found that 76% of TF-DNA interactions could be inhibited when DNA methylation occurred within the binding motif [76].
Changing chromatin accessibility through dynamically decompressing heterochromatic regions is another means of mediating TF-DNA associations. While TFs are often predicted to bind many thousands of sites within a genome, closed chromatin occludes most of these associations - typically, predicted sites are found bound only in areas of open chromatin [36, 49, 74, 77–80]. In this way, compacting DNA into chromatin probably reduces the time TFs require to discover their targets, by allowing the DNA that remains open to act as a sink for TF binding. However, changes in accessible chromatin is perhaps reserved for changing global gene regulatory patterns within a cell rather than as a means to rapidly promote specific TF-DNA interactions in response to a stimuli. This is suggested through chromatin structure proving dynamic during cell development and differentiation periods [81, 82], where altering accessibly is a way to canalize distinct gene regulatory profiles and promote cell identity. Indeed, this role of maintaining cell identity is indicated through open chromatin regions varying between different tissue types and developmental stages in Arabidopsis [79, 83, 84] and rice [78].
Detecting TF-Target Interactions Genome-Wide
In the previous sections, we discussed the molecular mechanisms that govern transient TF-DNA interactions. Measuring such TF-DNA dynamics has largely drawn from single locus or single molecule approaches, which can accurately quantify transient interactions down to the millisecond. However, such approaches are unable identify the genome-wide targets TFs transiently bind to and regulate. In contrast, techniques such as ChIP-seq and ChIP-chip (referred to here collectively as ChIP) or RNA-seq and gene expression microarrays (referred to here collectively as transcriptomics) can capture a TF's regulatory role across the entire genome. However, while these genomic approaches are powerful tools to assess TF function, they are unable to detect TF-DNA dynamics that occur in the order of seconds. In the following section we discuss how this trade-off can impact genomic analysis of dynamic TF activity.
ChIP remains the prevailing technique to learn TF binding sites genome-wide in vivo. Briefly, this technique relies upon immobilizing TFs bound at their regulatory site by covalently cross-linking the TF to DNA by treating with an aldehyde. In this way, ChIP 'freezes time' and stabilized TF-DNA interactions can be captured with an antibody selective to the TF. The resulting TF-bound precipitated DNA can be quantified to reveal the binding events of a TF within a genome. The time it takes to infiltrate plant tissues with an aldehyde is on the order of minutes, suggesting that this stress is unlikely to cause a large transcriptional response before cell death and thus distort ChIP readout (however, it may be enough time for post-translational regulation of TFs to occur, which may indeed bias ChIP outcome). To obtain enough DNA sequence to reliably identify TF binding sites, a population of cells is required, where increasing population size often provides better resolution of binding events. In this way, ChIP provides a global snapshot of the collective distribution of TF binding events across a cell population at a particular point in time. Typically, entire plants tissues (such as the shoot or root tissue) are utilized as starting material, whereby different cell layers are not discriminated. Consequently, if TF binding activity is specific to a specific cell layer (for example TFs TMO5 [85] or SHR [86] in the Arabidopsis root), such interactions will be diluted across the cell population.
While single molecule studies demonstrate that not all target loci will be bound by a TF in every cell at the time of cross-linking, by working across a population, ChIP finds sites that are bound in a statistically significant subset of cells. TF-DNA interactions that are found highly enriched indicate a larger proportion of cells have the TF bound at that locus at the time of aldehyde treatment. Conversely, weaker ChIP enrichments are TF interactions that occur less frequently, or more transiently, within the population. Consequently, while the degree of ChIP enrichment is often thought to report the 'strong' or 'weak' binding properties of a TF, strictly speaking, ChIP peaks actually reflect the proportion of cells within the population that have that TF bound. In this way, ChIP does not provide a binary read-out; it is a quantitative measurement that reports the degree of binding at a given time-point. This presents a conundrum - since TF binding at any one particular time will never be saturated at a locus across a cell population, as indicated by single molecule studies, how many times does a TF need to be found bound at a particular locus within the population for that interaction to be considered biologically meaningful?
A common way to discriminate if a TF-DNA binding event detected by ChIP has an effect on transcription, and thus is biologically meaningful, is to alter the expression of the TF itself. This is routinely achieved by detecting gene misregulation events within over-expression or knock-out mutant lines. Genes that are bound by a TF as well as differentially expressed when TF expression itself is perturbed are considered bona fide targets of that TF [87]. Overlapping ChIP and transcriptomic gene sets in this way is considered the gold standard of determining which gene targets a TF both binds and regulates.
However, the overlap of TF-bound and TF-regulated genes is often poor. While this intersection is never expected to be complete, reports from both plant studies and across Eukarya shows that this approach show that only between 1 to 40 % of TF-bound targets lead to target gene regulation, as indicated in Figure 3 [19, 29, 88–96]. How can data that falls outside this intersection be interpreted? Below, we discuss how the insensitivity of these genomic techniques to tissue-specific or transient TF-DNA interactions may cause this overlap to be smaller than it should.
Figure 3. A large fraction of TF-regulated genes are not stably bound to a TF.
Overlapping TF binding data and genes found differentially expressed due to TF perturbation (in gene knock out or over expression lines) can reveal genes that a TF both binds and regulates. However, overlapping datasets in this way, as shown for 11 TF studies performed across Eukarya, typically validates less than a quarter of the gene targets bound by ChIP [15,25, 71–79]. This finding suggests that a large portion of TF-regulated genes may be only transient bound to TFs at time-points not captured in these static studies.
I. Transcriptomics can fails to report differential gene expression
Often more than three quarters of all binding events found by ChIP (Figure 3) are found to have no consequence on target gene expression. Does this mean that the remaining binding interactions detected by ChIP do not lead to transcription?
Evidence suggests that ChIP enrichment that is highly significant stem from binding events that occurs in the vast majority of the cell population, and thus are more likely to be found differentially expressed through TF perturbation studies. In contrast, less significant ChIP enrichment, is thought less likely to indicate a regulatory event [36, 97, 98]. Undoubtedly, some degree of error is introduced by the technical limitations of ChIP itself [99], which may contribute to false positives. Also, TF binding is not always gene proximal, it can occur non-specifically open chromatin regions [36, 49, 74, 77–80], and regulation may require additional TF partners [85, 86]. But it is unlikely that these limitations explain all those TF binding events detected by ChIP that are not found to have an effect on transcription.
However, these lowly enriched TF-target interactions may not be contributing the poor overlap between ChIP and transcriptome analyses, as shown in Figure 3. Another possibility is that transcriptomes generated from TF perturbation studies may fail to report gene expression changes caused by low frequency or highly transient TF-target binding events. After all, when ChIP detects TF binding at gene proximal loci at these lower frequencies, they could initiate or repress transcription within the cells where these events occur. However these rarer transcription events may be too sensitive to be captured when transcriptomics is performed using a population of cells.
This concurs with single-cell RNA-seq analyses, which indicates transcriptomes vary dramatically between single cells within a population [100, 101]. While individual cells are known to hold distinct transcriptome profiles, governed in part by transient TF regulatory interactions, this transcriptome heterogeneity is rendered invisible when cells are sequenced as a population [102]. In contrast, the heterogeneity of TF binding that contributes to these distinct profiles between individual cells is revealed through differences in ChIP enrichment. Consequently, although less frequent binding events are often considered false positive interactions, they may be biologically meaningful within the subset of cells where they occur.
II. ChIP can fail to report binding events
Another reason for the poor overlap between ChIP and transcriptomic data sets (Figure 3) is that while transcriptomics generated from TF perturbation studies may report true positive TF-DNA regulatory interactions, ChIP may fail to detect these binding events. It is well established that ChIP suffers from a high false-negative rate - where TF targets are found differentially regulated in transcriptomic studies, however binding enrichment determined by ChIP fails to pass significance tests [99]. Higher false negative rates in ChIP may be expected for TFs whose residence times are highly transient (as seen for STAT1 at 0.5 s compared to Sox2 at 12 s) [13, 16] for TFs that are very dilute within the nucleus, or TFs which exhibit weak target binding affinities [30]. Under such conditions, ChIP enrichment at target loci would be expected to be lower, as less common targets within the cell population would be bound at any particular time. Following from this, the reproducibility among ChIP replicates may also be diminished, since the TF is too dilute to occupy the majority of targets consistently.
Another reason why ChIP may fail to report binding events is that those binding events are dynamic with respect to the TF's own active concentration within the nucleus. Consequently, TF occupancy is dependent on the time ChIP is performed. This can only be rectified by conducting ChIP over a time series, as discussed in the next section.
Let Some Time Pass
Another limitation of overlapping TF binding and data from transcriptome perturbation studies, as shown in Figure 3, is that this approach keeps time static. Since binding profiles change over time with a TFs active concentration or with DNA accessibility, intersecting genomic datasets in this way fails to capture how TFs dynamically regulate their targets over time. Similarly, validating TF bound targets through perturbing a TF's own expression provides no context to how those TF-target interactions play out over time.
In contrast, measuring both TF binding and transcriptomes over time can catch time-dependent interactions that are not visible at a single time point. Performing ChIP over time can reveal both the location and duration of binding events, and complimenting this approach with transcriptomics can indicate if those bound loci are differentially expressed (see Figure 4). Performing genomic techniques over time in this way is not sensitive enough to detect transient TF-DNA interactions or transcriptional bursting that occur within seconds, as seen in single molecule studies. However these approaches will reveal the consequence of transient TF-target interactions within a genome that occur in the order of minutes, thus indicating how these temporal interactions result in changes in the regulation of gene networks in the whole genome. Furthermore, this approach enables detecting both binding and transcriptional output without needing to perturb TF expression itself.
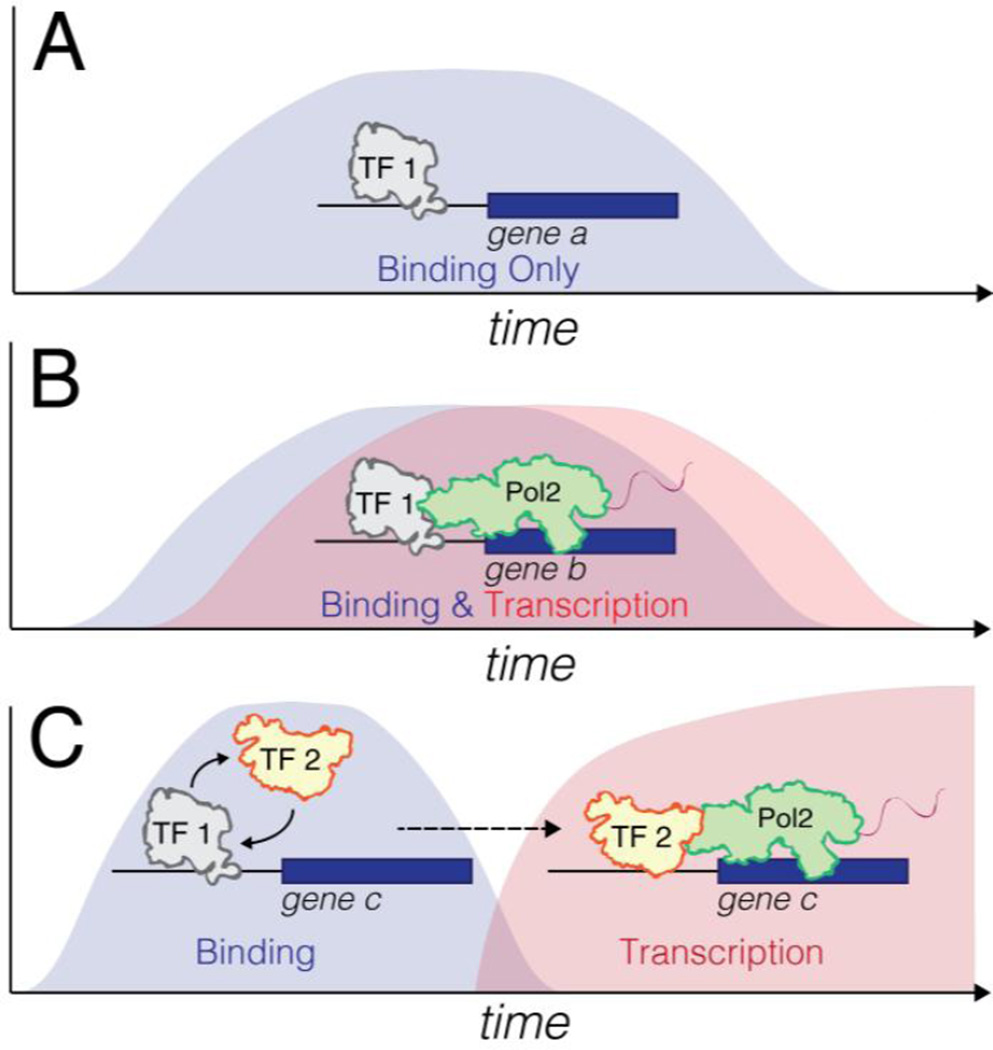
Figure 4. Tracking TF behavior over time uncovers new modes of transient TF action.
Performing ChIP and transcriptomics over time can reveal dynamic binding of a TF (A) and if binding leads to transcription (B). (C) Binding and transcription events can also be decoupled. In the 'hit-and-run' molecular phenotype [1–3] transient binding of TF 1 allows recruitment of secondary TFs (TF 2), where secondary TFs can continue initiating transcription without the hit-and-run TF1 [4].
One instance where TF binding and expression were monitored temporally genome-wide explored the role of the TF EIN3 in Arabidopsis ethylene signaling over a 24 hour period [103]. EIN3 expression was induced by treating plants with the plant hormone ethylene, and EIN3 was found to bind dynamically to its gene targets, where binding peaked 4 hours after treatment [103]. Performing transcriptomics over time revealed a subset of these EIN3-bound targets whose differential expression in response to ethylene treatment correlated with TF binding. Indeed, by integrating time-series ChIP-seq and RNA-seq data, 4 waves of temporally distinct transcriptional responses mediated by EIN3 binding over time were revealed [103]. Importantly EIN3 binding was depleted after 24 hours, indicating that ethylene induced EIN3 gene regulation had returned to its previous steady state. In this way, an entire array of gene targets that EIN3 acts upon over time within the context of ethylene signaling had been elucidated [103].
In a similar manner, monitoring transcriptomes over time in plants has revealed temporal expression of genes involved in plant immunity [104] as well as seed development [105]. Observing TF binding patterns or transcriptomics over time has also been successful in deciphering how the temporal expression of TF subnetworks governing multipotent differentiation evolve [106], and how D. melanogaster developmental programs are driven by temporal TF-DNA interactions [107–109]. In these instances, the dynamic binding behavior TFs, and the tempo of TF signal propagation, would not have been revealed had only one time point been observed.
Time-series transcriptome experiments have been valuable for deriving regulatory edges between not just a single TF and its targets (like for EIN3 [103]), but also for mapping gene regulatory networks that link multiple TFs with their targets. For example, through modeling ordinary differential equations that employ transcriptomic time series data, the Arabidopsis circadian clock [110, 111] and gene networks that governs flowering time [112] have been delineated.
An emerging approach to model dynamic gene regulatory from transcriptomic time-series datasets is through state-space modeling - a machine learning approach that can predict gene regulatory interactions from time-series expression profiles of TFs and their putative targets [113]. State-space modeling synthesizes Bayesian and Markovian approaches to learn from time series data, and estimates the quantitative influence of expressed TFs on each potential gene target [113, 114]. This maps regulatory edges between TFs and their targets, and consequently produces a gene regulatory network predictive of TF-target interactions. Such networks are able to resolve time-dependent interactions between TFs and their targets, such as feed-forward loops. Indeed, state-space modeling has been used in this way to understand which TFs drive genes responsible for leaf senescence in Arabidopsis [115], and has also been able to delineate the regulatory interactions between TFs and the genes responsible for nitrogen transport and assimilation Arabidopsis roots [116]. Crucially, since such regulatory networks are generated from temporal data, they can be used to extrapolate how transcriptomes might look in future untested time-points - thus achieving the ultimate goal of systems biology.
Short-Term TF Binding, Long-Term Expression
While computationally modeling how gene regulatory dynamics unfold over hours can reveal the topology of plant gene regulatory networks, performing very fine scaled time-series experimental analyses of TF-target interactions over the order of minutes have also proven fruitful. Observing gene regulatory responses that occur within smaller time-frames have been able to capture TF-DNA dynamics that would have otherwise been missed in over longer periods. Indeed, such approaches have uncovered new transient transcriptional mechanisms, as reviewed below.
I. Hit-and-run transcription
One example is the activity of 'hit-and-run' TFs, where transient binding of a TF has an enduring effect on transcriptional output that persists after the TF has dissociated from its target. The hit-and-run model is illustrated in Figure 4C, as well within the graphical abstract. This model proposes that TF binding (the 'hit') can organize a transcriptional complex, including recruitment of other TFs, so that transcription can continue after the initiating TF is no longer bound (the 'run'). By this means, TF signaling can speed up, since the hit-and-run TF need only bind once to a locus to initiate a lasting transcriptional response. The hit-and-run model was first proposed in the 1980s [117] and validated on a single gene basis in animals [118, 119]. To date, there are three published examples of this hit-and-run mechanism in plants [1–3].
Genome-wide evidence for hit-and-run transcription in plants was first discovered for bZIP1, a TF responsible for nutrient signaling in Arabidopsis, using a cell-based assay called TARGET ('Transient Assay Reporting Genome-wide Effects of Transcription factors') [1, 120]. In this assay, bZIP1 was transiently overexpressed in isolated Arabidopsis root cells as a TF-GR (Glucocorticoid Receptor domain) fusion protein. In this way, the TF-GR was sequestered in the cytoplasm through HSP90 heat shock protein interacting with GR. Treatment with dexamethasone, which displaces HSP90, enabled bZIP1 to be translocated to the nucleus and bind to its gene targets. This ability to control TF nuclear import using the TF-GR system has previously been used in planta (for review see [121]). Using the TF-GR system to control TF nuclear import in isolated cells allowed ChIP to be performed 1 minute after nuclear entry of bZIP1 - a time scale not feasible in whole plant tissues due to time it takes to infiltrate aldehydes [1, 120]. Using this approach, bZIP1 was discovered to transiently bind its targets over time; binding of some loci occurred as early as 1 and 5 minute after nuclear entry, but were not found bound later at 30 and 60 min [1].
While early TF-target binding of bZIP1 was transient, expression levels of these targets remained high as long as 5 hours after the initial binding event was detected [1]. To investigate whether mRNA was still being actively transcribed from these loci at later time points when TF-binding was not detected, and thus provide evidence for hit-and-run transcription, cells were treated with 4tU – a uracil analog that is incorporated into actively transcribed RNA [122–124]. By selecting for and quantifying 4tU labeled mRNA, a snapshot of actively transcribed genes was obtained. Gene targets of bZIP1 that were bound at earlier time points - and no longer bound at later time points - were still being actively transcribed after bZIP1 binding at these loci was depleted [2]. This indicates that bZIP1 binding, while transient, allows for transcription to continue long after its departure.
It has been proposed that hit-and-run transcription may result from the ability of a TF to modify histones and subsequently promote future regulatory interactions, where evidence for this molecular phenotype exists across eukaryotes [1, 25, 125, 126]. Additional genome-wide evidence for hit-and-run transcription in Arabidopsis that supports this mode of action has been obtained for the heat shock response factor TF HSFA2. HSFA2 is induced by heat stress, and was found to bind to its targets 30 min after heat treatment. However, TF binding was transient; consistent with the hit-and-run molecular phenotype, TF-target binding events were depleted after 28 hours [3]. During target binding HSFA2 was found to introduce, either directly or indirectly, H3K4me2 and H3K4me3 methylation marks at some of its gene targets. Introducing histone marks allowed these targets to be continuously transcribed, even after HSFA2 expression was itself depleted. Introducing these marks also allowed some of the targets transcribed at higher rates upon second exposure to heat treatment [3]. The ability of bZIP1 and HSFA2 to bind allows a transient interaction to have a lasting effect on transcription. Thus these hit-and-run TFs share a similar property to histone modifying proteins (such as HATs, as described earlier), which also need bind only once have an enduring effect on a locus.
The hypothesis that hit-and-run transcription involves chromatin modification is reminiscent of the activity of pioneer TFs in animals. However, the hit-and-run transcriptional model differs from that described of pioneer TFs. Although they share similar nucleosome remodeling phenotypes, pioneer TFs in animal development are thought to remain stably bound to enable binding of secondary TF partners [127–131]. Also pioneer TFs appear to trigger binary developmental switches in animals [132], whereas hit-and-run TFs may be considered 'transiently acting pioneer TFs', what are responsible for a more fluid response to environmental change.
II. Polymerase Pausing
Other examples of where TF binding and transcription events do not occur simultaneously over time have begun to emerge. Evidence across Eukarya suggest that many genes having undergone transcription initiation have Pol II complexes paused downstream within the promoter proximal region. Only when pause release factor pTEFb is recruited can transcription elongation continue [133]. Pausing of Pol II after transcription initiation probably constitutes an extra gene regulatory step, where a single rate limiting interaction allows transcriptional elongation to proceed rapidly. Indeed, the TF c-myc was found to perform such a rate-limiting function by recruiting pTEFb [134]. Pausing of Pol II implies that TF binding events that initiate transcription could occur at a much earlier time than transcript output.
Widespread Pol II pausing has been detected in H. sapiens, where pausing has been detected at 28% of all genes [135]. Currently, there is conflicting evidence on whether Pol II pausing exists as a general gene regulatory mechanism in planta. Genomic evidence in maize indicates that this form of regulation is missing [136]. The contrary was found in Arabidopsis, where Pol II pausing was enriched at drought-responsive genes after plants had been exposed to drought stress, suggesting pausing was being employed to prime rapid reactivation of these loci [68]. And unlike in metazoan, Pol II was found paused along the length of the gene, rather than being found promoter proximal [68]. Future research efforts should focus on confirming whether indeed pausing is present in plants, and whether pausing allows TF binding events to be decoupled from transcriptional output.
Moving Forward
Single molecule tracking and high-throughput sequencing techniques have each revealed one half of the transient properties of TF-target binding and its consequence on gene expression over time. Single molecule studies have indicated that TF binding is rapid, and results in stochastic mRNA production. This stochasticity most likely contributes to heterogeneity within transcriptomes among clonal cell populations. However, observing TF binding activity at the single molecule level cannot reveal the targets they act upon genome-wide. Conversely, transcriptome profiling and ChIP approaches have reported gene targets of TFs genome-wide, as well as detected how changing the equilibrium of active TFs allow for dynamic gene regulation. However, genomic techniques require populations of cells as starting material. Pooling cells together in this way renders an 'averaged' global profile of TF status, where disparate TF binding profiles or transcriptomes present across individuals can only be indirectly observed through comparing variances in gene expression between cell populations.
The advent of single-cell sequencing technologies promises to reveal both the stochastic TF binding events seen in single molecule studies, as well what effect these events have on a cell's transcriptome. By performing ChIP-seq at the single cell level, it will become possible to observe how transient TF binding properties lead to unique TF binding profiles within cell populations. Single cell ChIP-seq has already been able to detect subtle but distinct variations in histone landscapes between ES cells, suggesting that this technique will prove successful in detecting TF binding patterns in the near future [137]. Through single-cell RNA-seq, it has already become possible to see how such transient TF binding drives stochastic transcription events at the genome scale.
Single cell RNA-seq has revealed that transcriptome profiles vary dramatically between cells within a cell populations [100–102, 138]. For example, single-cell sequencing of the Arabidopsis quiescent center (QC), a stem cell niche in roots, has revealed unique transcriptomes between individual cells. Among the 30 QC cells sequenced, around 10,000 genes were expressed - however only 9.4 % were found expressed in all cells. This indicates that expression profiles between individual cells differ widely, especially for genes expressed at low levels. This kind of resolution was lost when QC cells were pooled and sequenced, as pooling renders an averaged transcriptome profile representative of a population [139, 140]. These differences of expression states within homogenous cell populations are probably driven in part by that transient TF binding events generating unsynchronized gene regulatory networks. The next step then is to understand how gene regulatory networks at the single cell level function over time. Work towards this has also been achieved in the Arabidopsis root. Through employing single cell transcriptome sequencing to monitor how excised root tips regenerate over a 46 hour period, it was found that individual stele cells can switch gene regulatory patterns and adopted the transcriptomic fingerprint of cell types that had been removed, such as QC cells [141].
Concluding Remarks
While extraordinary progress has been made in mapping the targets of TFs across many model organisms, these interactions are often rendered as a static web of interactions. While the temporal properties of TF activity is often overlooked, understanding how TFs function with respect to time can provide crucial context to how genomes drive gene regulatory changes.
This review has assessed how TFs work over time, focusing specifically to those discoveries made in plants. It has discussed how rapid and transient TF binding at loci cause stochastic gene expression, how altering TF equilibriums and DNA accessibility changes the probability of transcriptional events occurring, and consequently how genomes redirect their gene expression programs. This review has also detailed how genomic techniques can sometimes miss TF-DNA interactions, and how single cell genomic techniques promise to capture the effects of transient TF activity on stochastic gene expression, which are obscured when analyzing whole cell populations.
Ultimately, by considering time as a key variable when exploring TF function, it becomes possible to not only detect what regulatory connections a TF mediates within the genome space - but also how these interactions change with time. Capturing this 'space-time' feature of TF activity can provide greater insight into how genomes dynamically regulate themselves.
Highlights.
Transient TF-DNA interactions dynamically regulate gene networks.
Plant epigenomes can regulate transient TF-DNA interactions.
Single time-point genomic analyses can miss transient or localized TF-DNA interactions.
Detecting 'hit-and-run' transcriptional regulation genome-wide in planta reveals new insights into dynamic gene regulation.
Acknowledgments
The authors would like to thank Ying Li for her assistance with meta-data analysis.
Work on dynamic transcription in plants has been supported in our lab by NIH GM032877 and NSF MCB-1412232; and by fellowships NIH NIGMS Fellowship 1F32GM116347; EMBO Long-term Fellowship ALTF 1449-2015.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Para A, et al. Hit-and-run transcriptional control by bZIP1 mediates rapid nutrient signaling in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(28):10371–10376. doi: 10.1073/pnas.1404657111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doidy J, et al. "Hit-and-Run" transcription: de novo transcription initiated by a transient bZIP1 "hit" persists after the "run". BMC Genomics. 2016;17(1):92. doi: 10.1186/s12864-016-2410-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lamke J, et al. A hit-and-run heat shock factor governs sustained histone methylation and transcriptional stress memory. EMBO J. 2016;35(2):162–175. doi: 10.15252/embj.201592593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varala K, et al. "Hit-and-Run" leaves its mark: Catalyst transcription factors and chromatin modification. Bioessays. 2015;37(8):851–856. doi: 10.1002/bies.201400205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee TI, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298(5594):799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 6.Alon U. Network motifs: theory and experimental approaches. Nature Reviews Genetics. 2007;8(6):450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 7.Vaquerizas JM, et al. A census of human transcription factors: function, expression and evolution. Nature Reviews Genetics. 2009;10(4):252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- 8.Shiu SH, Shih MC, Li WH. Transcription factor families have much higher expansion rates in plants than in animals. Plant Physiology. 2005;139(1):18–26. doi: 10.1104/pp.105.065110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luscombe NM, et al. Genomic analysis of regulatory network dynamics reveals large topological changes. Nature. 2004;431(7006):308–312. doi: 10.1038/nature02782. [DOI] [PubMed] [Google Scholar]
- 10.Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(21):11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simon I, et al. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell. 2001;106(6):697–708. doi: 10.1016/s0092-8674(01)00494-9. [DOI] [PubMed] [Google Scholar]
- 12.Mueller F, et al. Quantifying transcription factor kinetics: At work or at play? Critical Reviews in Biochemistry and Molecular Biology. 2013;48(5):492–514. doi: 10.3109/10409238.2013.833891. [DOI] [PubMed] [Google Scholar]
- 13.Speil J, et al. Activated STAT1 Transcription Factors Conduct Distinct Saltatory Movements in the Cell Nucleus. Biophysical Journal. 2011;101(11):2592–2600. doi: 10.1016/j.bpj.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mazza D, et al. A benchmark for chromatin binding measurements in live cells. Nucleic Acids Research. 2012;40(15) doi: 10.1093/nar/gks701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morisaki T, et al. Single-molecule analysis of transcription factor binding at transcription sites in live cells. Nature Communications. 2014;5 doi: 10.1038/ncomms5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen JJ, et al. Single-Molecule Dynamics of Enhanceosome Assembly in Embryonic Stem Cells. Cell. 2014;156(6):1274–1285. doi: 10.1016/j.cell.2014.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raj A, et al. Stochastic mRNA synthesis in mammalian cells. Plos Biology. 2006;4(10):1707–1719. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raj A, van Oudenaarden A. Nature, Nurture, or Chance: Stochastic Gene Expression and Its Consequences. Cell. 2008;135(2):216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marchive C, et al. Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nature Communications. 2013;4 doi: 10.1038/ncomms2650. [DOI] [PubMed] [Google Scholar]
- 20.Rey G, et al. Genome-Wide and Phase-Specific DNA-Binding Rhythms of BMAL1 Control Circadian Output Functions in Mouse Liver. Plos Biology. 2011;9(2) doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ni L, et al. Dynamic and complex transcription factor binding during an inducible response in yeast. Genes & Development. 2009;23(11):1351–1363. doi: 10.1101/gad.1781909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Century K, Reuber TL, Ratcliffe OJ. Regulating the regulators: The future prospects for transcription-factor-based agricultural biotechnology products. Plant Physiology. 2008;147(1):20–29. doi: 10.1104/pp.108.117887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doebley JF, Gaut BS, Smith BD. The molecular genetics of crop domestication. Cell. 2006;127(7):1309–1321. doi: 10.1016/j.cell.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 24.Becker M, et al. Dynamic behavior of transcription factors on a natural promoter in living cells. Embo Reports. 2002;3(12):1188–1194. doi: 10.1093/embo-reports/kvf244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voss TC, et al. Dynamic Exchange at Regulatory Elements during Chromatin Remodeling Underlies Assisted Loading Mechanism. Cell. 2011;146(4):544–554. doi: 10.1016/j.cell.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo Y, et al. Nucleosomes accelerate transcription factor dissociation. Nucleic Acids Research. 2014;42(5):3017–3027. doi: 10.1093/nar/gkt1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hansen AS, O'shea EK. Promoter decoding of transcription factor dynamics involves a trade-off between noise and control of gene expression. Molecular Systems Biology. 2013;9 doi: 10.1038/msb.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mueller F, Wach P, McNally JG. Evidence for a common mode of transcription factor interaction with chromatin as revealed by improved quantitative fluorescence recovery after photobleaching. Biophysical Journal. 2008;94(8):3323–3339. doi: 10.1529/biophysj.107.123182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lickwar CR, et al. Genome-wide protein-DNA binding dynamics suggest a molecular clutch for transcription factor function. Nature. 2012;484(7393):251-U141. doi: 10.1038/nature10985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gronlund A, Lotstedt P, Elf J. Transcription factor binding kinetics constrain noise suppression via negative feedback. Nature Communications. 2013;4 doi: 10.1038/ncomms2867. [DOI] [PubMed] [Google Scholar]
- 31.Loverdo C, et al. Quantifying Hopping and Jumping in Facilitated Diffusion of DNA-Binding Proteins. Physical Review Letters. 2009;102(18) doi: 10.1103/PhysRevLett.102.188101. [DOI] [PubMed] [Google Scholar]
- 32.Li GW, Xie XS. Central dogma at the single-molecule level in living cells. Nature. 2011;475(7356):308–315. doi: 10.1038/nature10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hammar P, et al. The lac Repressor Displays Facilitated Diffusion in Living Cells. Science. 2012;336(6088):1595–1598. doi: 10.1126/science.1221648. [DOI] [PubMed] [Google Scholar]
- 34.Elf J, Li GW, Xie XS. Probing transcription factor dynamics at the single-molecule level in a living cell. Science. 2007;316(5828):1191–1194. doi: 10.1126/science.1141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Larson DR, et al. Real-Time Observation of Transcription Initiation and Elongation on an Endogenous Yeast Gene. Science. 2011;332(6028):475–478. doi: 10.1126/science.1202142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Biggin MD. Animal Transcription Networks as Highly Connected, Quantitative Continua. Developmental Cell. 2011;21(4):611–626. doi: 10.1016/j.devcel.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 37.Bradley MN, Zhou LA, Smale ST. C/EBP beta regulation in lipopolysaccharide-stimulated macrophages. Molecular and Cellular Biology. 2003;23(14):4841–4858. doi: 10.1128/MCB.23.14.4841-4858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oppenhei Jh, et al. Limited Binding-Capacity Sites for L-Triiodothyronine in Rat-Liver Nuclei - Nuclear-Cytoplasmic Interrelation, Binding Constants, and Cross-Reactivity with L-Thyroxine. Journal of Clinical Investigation. 1974;53(3):768–777. doi: 10.1172/JCI107615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chubb JR, et al. Transcriptional pulsing of a developmental gene. Current Biology. 2006;16(10):1018–1025. doi: 10.1016/j.cub.2006.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pearl LH, Prodromou C. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annual Review of Biochemistry. 2006;75:271–294. doi: 10.1146/annurev.biochem.75.103004.142738. [DOI] [PubMed] [Google Scholar]
- 41.Queitsch C, Sangster TA, Lindquist S. Hsp90 as a capacitor of phenotypic variation. Nature. 2002;417(6889):618–624. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- 42.Yip KY, et al. Classification of human genomic regions based on experimentally determined binding sites of more than 100 transcription-related factors. Genome Biology. 2012;13(9) doi: 10.1186/gb-2012-13-9-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soltani M, et al. Nonspecific transcription factor binding can reduce noise in the expression of downstream proteins. Physical Biology. 2015;12(5) doi: 10.1088/1478-3975/12/5/055002. [DOI] [PubMed] [Google Scholar]
- 44.Burger A, Walczak AM, Wolynes PG. Abduction and asylum in the lives of transcription factors. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(9):4016–4021. doi: 10.1073/pnas.0915138107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pruneda-Paz JL, et al. A Genome-Scale Resource for the Functional Characterization of Arabidopsis Transcription Factors. Cell Reports. 2014;8(2):621–631. doi: 10.1016/j.celrep.2014.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alvarez JM, et al. Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. Plant Journal. 2014;80(1):1–13. doi: 10.1111/tpj.12618. [DOI] [PubMed] [Google Scholar]
- 47.Guan PZ, et al. Nitrate foraging by Arabidopsis roots is mediated by the transcription factor TCP20 through the systemic signaling pathway. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(42):15267–15272. doi: 10.1073/pnas.1411375111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Widiez T, et al. High Nitrogen Insensitive 9 (Hni9)-Mediated Systemic Repression of Root No3- Uptake Is Associated with Changes in Histone Methylation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(32):13329–13334. doi: 10.1073/pnas.1017863108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heyndrickx KS, et al. A Functional and Evolutionary Perspective on Transcription Factor Binding in Arabidopsis thaliana. Plant Cell. 2014;26(10):3894–3910. doi: 10.1105/tpc.114.130591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ouyang ZQ, Zhou Q, Wong WH. ChIP-Seq of transcription factors predicts absolute and differential gene expression in embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(51):21521–21526. doi: 10.1073/pnas.0904863106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Metivier R, et al. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115(6):751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 52.Li Y, Varala K, Coruzzi GM. From milliseconds to lifetimes: tracking the dynamic behavior of transcription factors in gene networks. Trends in Genetics. 2015;31(9):509–515. doi: 10.1016/j.tig.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garcia AV, et al. Balanced Nuclear and Cytoplasmic Activities of EDS1 Are Required for a Complete Plant Innate Immune Response. Plos Pathogens. 2010;6(7) doi: 10.1371/journal.ppat.1000970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaminaka H, et al. bZIP10-LSD1 antagonism modulates basal defense and cell death in Arabidopsis following infection. Embo Journal. 2006;25(18):4400–4411. doi: 10.1038/sj.emboj.7601312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garcia AV, Parker JE. Heaven's Gate: nuclear accessibility and activities of plant immune regulators. Trends in Plant Science. 2009;14(9):479–487. doi: 10.1016/j.tplants.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 56.Moore JW, Loake GJ, Spoel SH. Transcription Dynamics in Plant Immunity. Plant Cell. 2011;23(8):2809–2820. doi: 10.1105/tpc.111.087346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang XY, et al. Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biology. 2009;10(6) doi: 10.1186/gb-2009-10-6-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wapinski OL, et al. Hierarchical Mechanisms for Direct Reprogramming of Fibroblasts to Neurons. Cell. 2013;155(3):621–635. doi: 10.1016/j.cell.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rothbart SB, Strahl BD. Interpreting the language of histone and DNA modifications. Biochimica Et Biophysica Acta-Gene Regulatory Mechanisms. 2014;1839(8):627–643. doi: 10.1016/j.bbagrm.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoffman BG, et al. Locus co-occupancy, nucleosome positioning, and H3K4me1 regulate the functionality of FOXA2-, HNF4A-, and PDX1-bound loci in islets and liver. Genome Research. 2010;20(8):1037–1051. doi: 10.1101/gr.104356.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ernst J, Kellis M. Interplay between chromatin state, regulator binding, and regulatory motifs in six human cell types. Genome Research. 2013;23(7):1142–1154. doi: 10.1101/gr.144840.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Slattery M, et al. Absence of a simple code: how transcription factors read the genome. Trends in Biochemical Sciences. 2014;39(9):381–399. doi: 10.1016/j.tibs.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Phair RD, et al. Global nature of dynamic protein-chromatin interactions in vivo: Three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Molecular and Cellular Biology. 2004;24(14):6393–6402. doi: 10.1128/MCB.24.14.6393-6402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Venkatesh S, Workman JL. Histone exchange, chromatin structure and the regulation of transcription. Nature Reviews Molecular Cell Biology. 2015;16(3):178–189. doi: 10.1038/nrm3941. [DOI] [PubMed] [Google Scholar]
- 65.Song HR, Noh YS. Rhythmic oscillation of histone acetylation and methylation at the Arabidopsis central clock loci. Molecules and Cells. 2012;34(3):279–287. doi: 10.1007/s10059-012-0103-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Malapeira J, Khaitova LC, Mas P. Ordered changes in histone modifications at the core of the Arabidopsis circadian clock. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(52):21540–21545. doi: 10.1073/pnas.1217022110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim JM, et al. Transition of Chromatin Status During the Process of Recovery from Drought Stress in Arabidopsis thaliana. Plant and Cell Physiology. 2012;53(5):847–856. doi: 10.1093/pcp/pcs053. [DOI] [PubMed] [Google Scholar]
- 68.Ding Y, Fromm M, Avramova Z. Multiple exposures to drought 'train' transcriptional responses in Arabidopsis. Nature Communications. 2012;3 doi: 10.1038/ncomms1732. [DOI] [PubMed] [Google Scholar]
- 69.Mukhopadhyay P, et al. Stress-Mediated Alterations in Chromatin Architecture Correlate with Down-Regulation of a Gene Encoding 60S rpL32 in Rice. Plant and Cell Physiology. 2013;54(4):528–540. doi: 10.1093/pcp/pct012. [DOI] [PubMed] [Google Scholar]
- 70.Tsuji H, et al. Dynamic and reversible changes in histone H3-Lys4 methylation and H3 acetylation occurring at submergence-inducible genes in rice. Plant and Cell Physiology. 2006;47(7):995–1003. doi: 10.1093/pcp/pcj072. [DOI] [PubMed] [Google Scholar]
- 71.Casati P, et al. Histone acetylation and chromatin remodeling are required for UV-B-dependent transcriptional activation of regulated genes in maize. Plant Cell. 2008;20(4):827–842. doi: 10.1105/tpc.107.056457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dowen RH, et al. Widespread dynamic DNA methylation in response to biotic stress. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(32):E2183–E2191. doi: 10.1073/pnas.1209329109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang XY, et al. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. Plos Biology. 2007;5(5):1026–1035. doi: 10.1371/journal.pbio.0050129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489(7414):75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang XY, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell. 2006;126(6):1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 76.O'Malley RC, et al. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell. 2016;165(5):1280–1292. doi: 10.1016/j.cell.2016.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Franco-Zorrilla JM, et al. DNA-binding specificities of plant transcription factors and their potential to define target genes. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(6):2367–2372. doi: 10.1073/pnas.1316278111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang WL, et al. High-resolution mapping of open chromatin in the rice genome. Genome Research. 2012;22(1):151–162. doi: 10.1101/gr.131342.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang WL, et al. Genome-Wide Identification of Regulatory DNA Elements and Protein-Binding Footprints Using Signatures of Open Chromatin in Arabidopsis. Plant Cell. 2012;24(7):2719–2731. doi: 10.1105/tpc.112.098061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.John S, et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nature Genetics. 2011;43(3):264-U116. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sachs M, et al. Bivalent Chromatin Marks Developmental Regulatory Genes in the Mouse Embryonic Germline In Vivo. Cell Reports. 2013;3(6):1777–1784. doi: 10.1016/j.celrep.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Winter DR, Amit I. The role of chromatin dynamics in immune cell development. Immunological Reviews. 2014;261(1):9–22. doi: 10.1111/imr.12200. [DOI] [PubMed] [Google Scholar]
- 83.Pajoro A, et al. Dynamics of chromatin accessibility and gene regulation by MADS-domain transcription factors in flower development. Genome Biology. 2014;15(3) doi: 10.1186/gb-2014-15-3-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sullivan AM, et al. Mapping and Dynamics of Regulatory DNA and Transcription Factor Networks in A-thaliana. Cell Reports. 2014;8(6):2015–2030. doi: 10.1016/j.celrep.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 85.De Rybel B, et al. A bHLH Complex Controls Embryonic Vascular Tissue Establishment and Indeterminate Growth in Arabidopsis. Developmental Cell. 2013;24(4):426–437. doi: 10.1016/j.devcel.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 86.Clark NM, et al. Tracking transcription factor mobility and interaction in Arabidopsis roots with fluorescence correlation spectroscopy. Elife. 2016;5 doi: 10.7554/eLife.14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Walhout AJM. What does biologically meaningful mean? A perspective on gene regulatory network validation. Genome Biology. 2011;12(4) doi: 10.1186/gb-2011-12-4-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Monke G, et al. Toward the identification and regulation of the Arabidopsis thaliana ABI3 regulon. Nucleic Acids Research. 2012;40(17):8240–8254. doi: 10.1093/nar/gks594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hull RP, et al. Combined ChIP-Seq and transcriptome analysis identifies AP-1/JunD as a primary regulator of oxidative stress and IL-1 beta synthesis in macrophages. Bmc Genomics. 2013;14 doi: 10.1186/1471-2164-14-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Arenhart RA, et al. New Insights into Aluminum Tolerance in Rice: The ASR5 Protein Binds the STAR1 Promoter and Other Aluminum-Responsive Genes. Molecular Plant. 2014;7(4):709–721. doi: 10.1093/mp/sst160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bianco S, et al. LRH-1 Governs Vital Transcriptional Programs in Endocrine-Sensitive and -Resistant Breast Cancer Cells. Cancer Research. 2014;74(7):2015–2025. doi: 10.1158/0008-5472.CAN-13-2351. [DOI] [PubMed] [Google Scholar]
- 92.Lee J, et al. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell. 2007;19(3):731–749. doi: 10.1105/tpc.106.047688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li CB, et al. Genome-Wide Characterization of cis-Acting DNA Targets Reveals the Transcriptional Regulatory Framework of Opaque2 in Maize. Plant Cell. 2015;27(3):532–545. doi: 10.1105/tpc.114.134858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang HY, et al. Genome-wide mapping of the HY5-mediated genenetworks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant Journal. 2011;65(3):346–358. doi: 10.1111/j.1365-313X.2010.04426.x. [DOI] [PubMed] [Google Scholar]
- 95.Morohashi K, Grotewold E. A Systems Approach Reveals Regulatory Circuitry for Arabidopsis Trichome Initiation by the GL3 and GL1 Selectors. Plos Genetics. 2009;5(2) doi: 10.1371/journal.pgen.1000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gorski JJ, et al. Profiling of the BRCA1 transcriptome through microarray and ChIP-chip analysis. Nucleic Acids Research. 2011;39(22):9536–9548. doi: 10.1093/nar/gkr679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fisher WW, et al. DNA regions bound at low occupancy by transcription factors do not drive patterned reporter gene expression in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(52):21330–21335. doi: 10.1073/pnas.1209589110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li XY, et al. The role of chromatin accessibility in directing the widespread, overlapping patterns of Drosophila transcription factor binding. Genome Biology. 2011;12(4) doi: 10.1186/gb-2011-12-4-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen YW, et al. Systematic evaluation of factors influencing ChIP-seq fidelity. Nature Methods. 2012;9(6):609-+. doi: 10.1038/nmeth.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jaitin DA, et al. Massively Parallel Single-Cell RNA-Seq for Marker-Free Decomposition of Tissues into Cell Types. Science. 2014;343(6172):776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zeisel A, et al. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347(6226):1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 102.Luo YP, et al. Single-Cell Transcriptome Analyses Reveal Signals to Activate Dormant Neural Stem Cells. Cell. 2015;161(5):1175–1186. doi: 10.1016/j.cell.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chang KN, et al. Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis. Elife. 2013;2 doi: 10.7554/eLife.00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Maekawa T, et al. Conservation of NLR-triggered immunity across plant lineages. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(49):20119–20123. doi: 10.1073/pnas.1218059109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li GS, et al. Temporal patterns of gene expression in developing maize endosperm identified through transcriptome sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(21):7582–7587. doi: 10.1073/pnas.1406383111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.May G, et al. Dynamic Analysis of Gene Expression and Genome-wide Transcription Factor Binding during Lineage Specification of Multipotent Progenitors. Cell Stem Cell. 2013;13(6):754–768. doi: 10.1016/j.stem.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jakobsen JS, et al. Temporal ChIP-on-chip reveals Biniou as a universal regulator of the visceral muscle transcriptional network. Genes & Development. 2007;21(19):2448–2460. doi: 10.1101/gad.437607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sandmann T, et al. A temporal map of transcription factor activity: Mef2 directly regulates at all stages of muscle target genes development. Developmental Cell. 2006;10(6):797–807. doi: 10.1016/j.devcel.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 109.Sandmann T, et al. A core transcriptional network for early mesoderm development in Drosophila melanogaster. Genes & Development. 2007;21(4):436–449. doi: 10.1101/gad.1509007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bujdoso N, Davis SJ. Mathematical modeling of an oscillating gene circuit to unravel the circadian clock network of Arabidopsis thaliana. Frontiers in Plant Science. 2013;4 doi: 10.3389/fpls.2013.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pokhilko A, et al. The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Molecular Systems Biology. 2012;8 doi: 10.1038/msb.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Valentim FL, et al. A Quantitative and Dynamic Model of the Arabidopsis Flowering Time Gene Regulatory Network. Plos One. 2015;10(2) doi: 10.1371/journal.pone.0116973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Krouk G, et al. Gene regulatory networks in plants: learning causality from time and perturbation. Genome Biology. 2013;14(6) doi: 10.1186/gb-2013-14-6-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mirowski P, et al. Classification of patterns of EEG synchronization for seizure prediction. Clinical Neurophysiology. 2009;120(11):1927–1940. doi: 10.1016/j.clinph.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 115.Breeze E, et al. High-Resolution Temporal Profiling of Transcripts during Arabidopsis Leaf Senescence Reveals a Distinct Chronology of Processes and Regulation. Plant Cell. 2011;23(3):873–894. doi: 10.1105/tpc.111.083345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Krouk G, et al. Predictive network modeling of the high-resolution dynamic plant transcriptome in response to nitrate. Genome Biol. 2010;11(12):R123. doi: 10.1186/gb-2010-11-12-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schaffner W. Gene-Regulation - a Hit-and-Run Mechanism for Transcriptional Activation. Nature. 1988;336(6198):427–428. doi: 10.1038/336427a0. [DOI] [PubMed] [Google Scholar]
- 118.Eadara JK, Hadlock KG, Lutter LC. Chromatin structure and factor site occupancies in an in vivo-assembled transcription elongation complex. Nucleic Acids Research. 1996;24(20):3887–3895. doi: 10.1093/nar/24.20.3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.McNally JG, et al. The glucocorticoid receptor: Rapid exchange with regulatory sites in living cells. Science. 2000;287(5456):1262–1265. doi: 10.1126/science.287.5456.1262. [DOI] [PubMed] [Google Scholar]
- 120.Bargmann BOR, et al. TARGET: A Transient Transformation System for Genome-Wide Transcription Factor Target Discovery. Molecular Plant. 2013;6(3):978–980. doi: 10.1093/mp/sst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yamaguchi N, et al. Identification of Direct Targets of Plant Transcription Factors Using the GR Fusion Technique. Plant Functional Genomics: Methods and Protocols, 2nd Edition. 2015;1284:123–138. doi: 10.1007/978-1-4939-2444-8_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sidaway-Lee K, et al. Direct measurement of transcription rates reveals multiple mechanisms for configuration of the Arabidopsis ambient temperature response. Genome Biology. 2014;15(3) doi: 10.1186/gb-2014-15-3-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cleary MD, et al. Biosynthetic labeling of RNA with uracil phosphoribosyltransferase allows cell-specific microarray analysis of mRNA synthesis and decay. Nature Biotechnology. 2005;23(2):232–237. doi: 10.1038/nbt1061. [DOI] [PubMed] [Google Scholar]
- 124.Duffy EE, et al. Tracking Distinct RNA Populations Using Efficient and Reversible Covalent Chemistry. Molecular Cell. 2015;59(5):858–866. doi: 10.1016/j.molcel.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lefevre P, et al. Developmentally regulated recruitment of transcription factors and chromatin modification activities to chicken lysozyme cis-regulatory elements in vivo. Molecular and Cellular Biology. 2003;23(12):4386–4400. doi: 10.1128/MCB.23.12.4386-4400.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hoogenkamp M, et al. Early chromatin unfolding by RUNX1: a molecular explanation for differential requirements during specification versus maintenance of the hematopoietic gene expression program. Blood. 2009;114(2):299–309. doi: 10.1182/blood-2008-11-191890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes & Development. 2011;25(21):2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sherwood RI, et al. Discovery of directional and nondirectional pioneer transcription factors by modeling DNase profile magnitude and shape. Nature Biotechnology. 2014;32(2):171-+. doi: 10.1038/nbt.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sun YJ, et al. Zelda overcomes the high intrinsic nucleosome barrier at enhancers during Drosophila zygotic genome activation. Genome Research. 2015;25(11):1703–1714. doi: 10.1101/gr.192542.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Foo SM, et al. Zelda Potentiates Morphogen Activity by Increasing Chromatin Accessibility. Current Biology. 2014;24(12):1341–1346. doi: 10.1016/j.cub.2014.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yu HB, et al. Coassembly of REST and its cofactors at sites of gene repression in embryonic stem cells. Genome Research. 2011;21(8):1284–1293. doi: 10.1101/gr.114488.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Iwafuchi-Doi M, Zaret KS. Pioneer transcription factors in cell reprogramming. Genes & Development. 2014;28(24):2679–2692. doi: 10.1101/gad.253443.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Nechaev S, Adelman K. Pol II waiting in the starting gates: Regulating the transition from transcription initiation into productive elongation. Biochimica Et Biophysica Acta-Gene Regulatory Mechanisms. 2011;1809(1):34–45. doi: 10.1016/j.bbagrm.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rahl PB, et al. c-Myc Regulates Transcriptional Pause Release. Cell. 2010;141(3):432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Core LJ, Waterfall JJ, Lis JT. Nascent RNA Sequencing Reveals Widespread Pausing and Divergent Initiation at Human Promoters. Science. 2008;322(5909):1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Erhard Karl F, Jr, Joy-El RB, Talbot NCD, McClish Allison E, Hollick Jay B. Nascent Transcription Affected by RNA Polymerase IV in Zea mays. Genetics. 199:1107–1125. doi: 10.1534/genetics.115.174714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rotem A, et al. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nature Biotechnology. 2015;33(11):1165-U91. doi: 10.1038/nbt.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Macosko EZ, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161(5):1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Nawy T, et al. Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell. 2005;17(7):1908–1925. doi: 10.1105/tpc.105.031724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Efroni I, et al. Quantification of cell identity from single-cell gene expression profiles. Genome Biology. 2015;16 doi: 10.1186/s13059-015-0580-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Efroni I, et al. Root Regeneration Triggers an Embryo-like Sequence Guided by Hormonal Interactions. Cell. 2016;165(7):1721–1733. doi: 10.1016/j.cell.2016.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]