Abstract
Encapsulating bacteria within constrained microenvironments can promote the manifestation of specialized behaviors. Using double-emulsion droplet-generating microfluidic synthesis, live Bacillus subtilis were encapsulated in a semi-permeable membrane comprised of poly(ethylene glycol)-b-poly(D,L-lactic acid). This mPEG-PDLLA membrane was sufficiently permeable to permit exponential bacterial growth, metabolite-induced gene expression, and rapid biofilm growth. The biodegradable microparticles retained structural integrity for several days, and could be successfully degraded with time or sustained bacterial activity. Microencapsulated B. subtilis successfully captured and contained sodium selenite added outside the polymersomes, converting the selenite into elemental selenium nanoparticles that were selectively retained inside the polymer membrane. This remediation of selenium using polymersomes has high potential for reducing the toxicity of environmental selenium contamination, as well as allowing selenium to be harvested from areas not amenable to conventional waste or water treatment.
Keywords: microparticles, microfluidics, double-emulsion, bacteria, biofilm, selenite, nanoparticles
Introduction
The research and development of polymeric particles has surged in recent decades, particularly for the encapsulation of therapeutic molecules. Poly(lactide-co-glycolide), or PLGA, has been studied extensively as a delivery vehicle for chemotherapeutics, proteins, nucleic acids, and other macromolecules (Danhier et al. 2012). Among FDA-approved biodegradable polymers, PLGA is one of the most popular, owing to its long clinical history, favorable degradation characteristics, and tunable drug release profile (Jain 2000; Makadia and Siegel 2011). Block copolymer blends that combine PLGA with a hydrophilic polymer, such as polyethylene glycol (PEG), produce bilayer vesicles when self-assembled in aqueous solutions (Chen et al. 2011). These synthetic polymersomes have proven promising for the immunoisolation of transplanted mammalian cells across a range of applications including hormone replacement, glucose management, macular degeneration treatment, neurodegenerative disease management, and cancer treatment (Olabisi 2015). Injectable PLGA microspheres containing a glucagon-like peptide-1 agonist have been used for sustained glucose management in diabetic patients (DeYoung et al. 2011) and are also being investigated for the transplantation of pancreatic islet cells (Borg and Bonifacio 2011).
Bacteria microencapsulation has been investigated for a variety of applications, with the most advanced application being oral probiotics. Here, microparticles are used to protect probiotic bacteria from the harsh conditions of gastric passage (Kailasapathy 2002). Because probiotics are taken orally, naturally occurring polysaccharide polymers, such as alginate, chitosan, and gellan, have historically been considered the microencapsulation material of choice (Prakash et al. 2011). Unfortunately, the use of these natural polymers is limited by several physical constraints that are not readily overcome, and thus these materials are not well suited for applications requiring tunable diffusivity or degradation. The wide variety of synthetic polymers available, combined with advances in more physiological assembly techniques, now allows for the design of more advanced bacterial encapsulation systems. For example, the combination of PLGA-encapsulated prebiotics with probiotic bacteria in an alginate matrix was found to enhance the survival of probiotic bacteria under acidic conditions (Cook et al. 2014).
Microfluidic technology is ideally suited for the assembly of synthetic polymersomes. Unlike bulk synthesis methods, microfluidic devices are able to handle small liquid volumes, tune mixing conditions, and allow precise control over the reaction environment to produce microparticles with a narrower size distribution and improved loading efficiency. Of the different microfluidic techniques developed to date, the flow-focusing technique pioneered by Weitz et al. (Utada et al. 2005; Ho et al. 2008) allows synthetic polymersomes to be continuously synthesized inside glass capillaries compatible with a wide range of polymers and solvents. This technique produces water/oil/water (w/o/w) double-emulsion droplets that form solid-shell particles following solvent evaporation from the oil phase. Optimization of the capillary diameter, reagents, and flow rates readily produces highly reproducible batches of microparticles (Utada et al. 2005; Ho et al. 2008). Scale-up is performed by lengthening production runs or operating multiple devices in parallel. By enabling reproducibility, versatility, and tight control over the particle production rate, this microfluidic assembly technique has introduced a new era of encapsulation technologies ranging from academic to industrial. Double-emulsion microfluidics have successfully been used to microencapsulate Bacillus subtilis (B. subtilis) in silicon oil for templated biofilm growth (Chang et al. 2015) and Escherichia coli (E. coli) in fluoropolyether-polyethylene glycol for the study of quorum-quenching (Zhang et al. 2013).
Constraining bacteria within synthetic polymersomes opens the opportunity for performing nanoliter-scale chemical reactions. Specialized bacterial functions, such as bioremediation of heavy metals from soil and water, would no longer be limited to industrial scale bioreactors and could instead be performed directly in the field. One such potential application is the selective capture and containment of selenium. Selenium is a trace mineral essential for human health but toxic when consumed in excess. Selenium has also been reported to have important anti-bacterial (Wang and Webster 2013; Tran and Webster 2013; Chudobova et al. 2014; Estevam et al. 2015; Wang et al. 2015) and anti-cancer properties (Wallenberg et al. 2014; Fernandes and Gandin 2015). Of all the essential elements, however, selenium has one of the narrowest ranges between dietary deficiency (40 µg/day) and toxic levels (400 µg/day) (World Health Organization 1996). The rate at which selenium compounds are released into the environment by industrial emissions greatly exceeds natural recycling (Fordyce 2013), and thus represents a major environmental risk (U.S. Environmental Protection Agency 1991). A variety of bacteria have the capacity to reduce selenite into elemental selenium nanoparticles (Lapage and Bascomb 1968; Oremland et al. 2004), and this bioconversion renders the selenium less toxic to organisms (Agency for Toxic Substances and Disease Registry 2003). Extracting such nanoparticles from the environment, however, continues to be a major challenge outside dedicated waste and water treatment plants (Dungan and Frankenberger 1999).
In this work, we describe the microfluidic synthesis of biodegradable synthetic polymer microspheres containing B. subtilis for selenium remediation. Live bacteria were fully encapsulated within semi-permeable membranes comprised of poly(ethylene glycol)-b-poly(D,L-lactic acid) (mPEG-PDLLA) using a glass capillary-based double-emulsion microfluidic device. The physical and mechanical properties of the mPEG-PDLLA membrane were characterized following different incubation temperatures and mechanical disruption techniques. Cell viability, as well as the rate of cell proliferation, was assessed by microscopy after placing the polymersomes under optimal bacterial growth conditions. Using B. subtilis, a gram-positive soil bacterium carrying an inducible reporter gene, encapsulated bacteria of both motile and biofilm-forming phenotypes were demonstrated to be capable of metabolizing a lactose mimic added outside the polymersomes. Widefield and transmission electron microscopy confirmed that encapsulated B. subtilis retained the capacity to reduce sodium selenite and demonstrated that the resulting nanoparticles were selectively contained inside the polymersome. These findings open new opportunities for extracting selenium from the environment, as well as serving as a model for the encapsulation of bacteria in semi-permeable membranes for a variety of applications.
Materials and methods
Materials and device fabrication
Microfluidic devices were fabricated following procedures outlined by Weitz et al. (Ho et al. 2008) Briefly, two glass capillaries (O.D. 1 mm) were tapered to diameters of approximately 20 and 200 µm using a Flaming/Browning micropipette puller (Sutter P-97). The tip of the smaller (injection) capillary was coated with n-octadecyltrimethoxysilane (Sigma-Aldrich) while the tip of the larger (collection) capillary was coated with 2-[methoxy(polyethyleneoxy)propyl]trimethoxysilane (Gelest, Inc.). The capillaries were coaxially aligned within a square glass capillary tube (O.D. 1.05 mm) with an intercapillary distance of 100 µm. Three 20-gauge syringe tips were epoxied over the square capillary termini to allow injection of each liquid phase with syringe pumps via polyethylene tubing (O.D. 1.32 mm). For polymersome synthesis, three liquid phases were prepared: An aqueous solution comprised of Lysogeny broth (LB, Becton-Dickson), an organic polymer solution containing 2.5 mg/mL of diblock copolymer poly(ethylene glycol)-b-poly(D,L-lactic acid) (mPEG-PDLLA, 5000 MW mPEG and 50,000 MW PDLLA, Polyscitech) and 1.0 mg/mL of poly(lactic acid) (PLA, 15,000 MW, Polysciences) dissolved in a 2:1 ratio of toluene : chloroform, and an aqueous solution comprised of 10 wt% poly(vinyl alcohol) (PVA, 13,000–23,000 MW, 98% hydrolyzed, Sigma-Aldrich) dissolved by heating in deionized water.
Microparticle assembly
Polymersomes were synthesized at room temperature with a continuous flow-focusing microfluidic device to produce water/oil/water (w/o/w) double-emulsion droplets. The inner aqueous phase was injected into the organic phase, which flowed around the injection capillary. The resulting single emulsion was pinched off in the collection capillary by the continuous PVA phase, which opposed the flow of the single emulsion. The double-emulsion droplets subsequently flowed through the collection capillary and exited the device. The individual liquid phases were held at constant flow rates between 1000–1800 µL/hr for the inner aqueous phase, 2000–3500 µL/hr for the organic phase, and 8,000–12,000 µL/hr for the continuous PVA phase. Droplet generation was monitored visually using an optical microscope coupled to a 1000 fps digital camera (Casio EX-FH20), and individual pump flow rates were adjusted as needed for each device to maximize double-emulsion droplet formation. Droplets were produced at a maximum rate of 700 droplets per second. The suspended droplets were flowed into a glass scintillation vial containing LB to a final ratio of 1:1 by volume. To remove excess PVA, the double-emulsion droplets were allowed to settle to the bottom for 30 seconds, after which the supernatant was aspirated and replaced with an equivalent volume of LB. Solvent removal was performed in a two-stage evaporation process under a chemical fume hood. In the first step, polymersomes were placed uncovered in a 40-mm glass Petri dish for 3.5 hours to allow the volatile chloroform to evaporate and promote polymer migration to the solvent-LB interface. Subsequently, the Petri dish was covered overnight to allow the immiscible toluene to phase separate from the polymer.
Bacteria microencapsulation
Bacillus subtilis (B. subtilis) is a gram-positive, mesophilic soil-dwelling bacterium capable of growing at a range of temperatures. Under laboratory conditions, B. subtilis is able to sustain growth in temperatures spanning from 11°C (Nichols et al. 1995) to 52°C (Holtmann & Bremer, 2004). A sudden drop from the standard growth temperature (from 37 °C down to 15°C) is generally used in the laboratory to elicit the cold-shock response, after which the cells still maintain their viability but with altered gene expression to promote stable growth at the lower temperature (Budde et al. 2006). B. subtilis bacteria (strain NCIB3610) were transformed with an IPTG-inducible GFP as an inducible marker of cell viability (Sambrook and Russell 2001). B. subtilis (YCN041: amyE::Physpank-GFP::spec) were cultured in LB liquid medium (Lysogenic broth, consisting of 10 g tryptone, 5 g yeast extract, and 10 g NaCl per liter) at 37°C on a shaker at 225 RPM for 12–18 hr before particle synthesis. B. subtilis were resuspended in fresh LB at a concentration of 5 × 107 to 5 × 108 cells/mL (based on calibrated measurements of optical density) immediately prior to encapsulation.
Particle characterization
Particle size, concentration, and mechanical stability were assessed by optical microscopy. Replicates of five 10 µL droplets of polymersome suspension were placed in single depression concave slides (AmScope), imaged using Leica MSV269 dissecting microscope equipped with a 6 × objective (~100 particles/field-of-view), and analyzed using ImageJ software v1.49. Particle integrity was determined by counting the number of intact particles following mechanical disruption. The particles were subjected to 10 minutes of centrifugation at 18,400g, 30 seconds of vortexing at 3,000 RPM, or 3 seconds of sonication, and were subsequently imaged and quantified. Particle stability with time was determined using particles containing bacteria (or LB alone) under different static storage conditions, namely 4°C, 23°C, and 37°C. To promote bacterial growth, polymersomes held at 37°C were also subjected to shaking culture using an incubator at 225 RPM (New Brunswick Scientific). To identify the mechanism of bacteria-mediated polymersome degradation, empty polymersomes (LB only) were co-incubated with fresh bacteria supernatant and free bacteria in shaking culture at 37°C. Groups were compared as pairs using a student’s t-test, with a p-value ≤ 0.05 considered significant.
Bacterial growth and cross-membrane transport
Growth of encapsulated bacteria was induced by diluting the polymersome suspension 1:1 in fresh LB and placing the mixture on a shaker (225 RPM) at 37°C for up to 96 hours. To determine whether encapsulated bacteria were metabolically active, transformed B. subtilis (YCN041: amyE::Physpank-GFP, spec in NCIB 3610) were induced to express green fluorescent protein (GFP) through the addition of 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG), a molecular mimic of a lactose metabolite that triggers transcription of genes controlled under the lac operon. IPTG was added directly to the supernatant before shaking to determine whether the polymersome membrane is permeable to small molecule metabolites. Widefield fluorescent and phase-contrast images of polymersomes were acquired on a Leica AF6000 microscope at 40 × magnification. Confocal laser scanning microscope images were acquired on an Olympus Fluoview FV1000 microscope equipped with a 100mW Argon laser at 40 × magnification. All image analysis was performed using ImageJ software v1.49. Representative widefield images collected during the low-density exponential growth phase (3, 3.5, and 4 hours) were utilized to count the number of bacteria per polymersome to measure the population doubling time. Representative confocal images collected at 3µm slice intervals over a 40 – 60 µm sample thickness were utilized to generate z-projections and 3D reconstructions.
Selenium remediation
Encapsulated bacteria were grown at 37°C in the presence of 3 mM sodium selenite (Na2SeO3) added to the supernatant. Polymersomes were placed on a shaker (225 RPM) for up to 8 hours. The presence of selenium nanoparticles inside polymersomes was visualized using wide-field microscopy following 8 hours co-incubation. B. subtilis co-incubated with sodium selenite for 8 hours were dried on 300-mesh copper-coated carbon grids, and negatively stained with a 1.5% uranyl acetate solution, and analyzed using a JEM-1010 (JEOL) transmission electron microscope (TEM). Selenium nanoparticle size was measured from TEM images using ImageJ software v1.49. The measured and expected number of Se atoms per nanoparticle, as well as per polymersome, were estimated based on measurements of nanoparticle diameter, nanoparticle number, and polymersome diameter.
Results
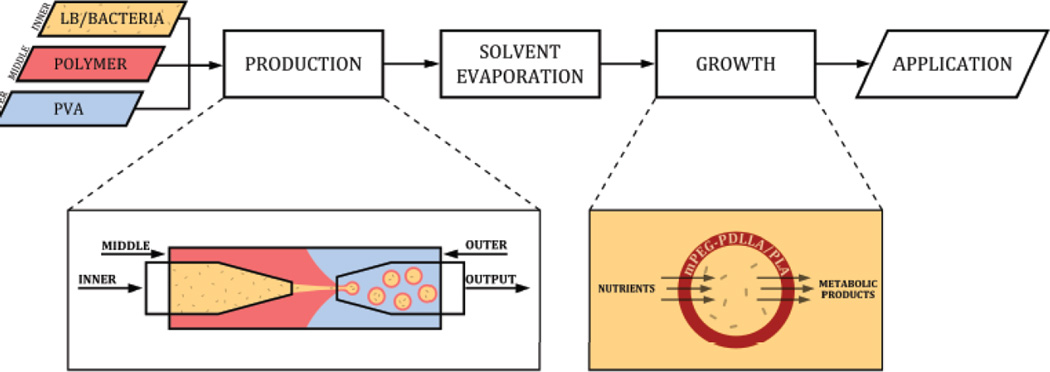
We optimized a flow-focusing microfluidic device to generate semi-permeable polymeric microparticles containing live B. subtilis bacteria (Fig. 1). During the production stage, an aqueous phase comprised of bacteria in LB growth media (inner layer) was flowed into an organic polymer solution containing the diblock copolymer mPEG-PDLLA (middle layer) and was subsequently pinched off by an aqueous solution of PVA. The resulting water/oil/water (w/o/w) double-emulsion droplets contained bacteria encapsulated in a layer of biodegradable polymer. Droplet production was verified by video capture at 1000 fps (Video 1). Intact polymersomes were collected in LB and solvent evaporation was performed to produce a semipermeable polymersome membrane for nutrient and metabolite exchange.
Fig. 1.
Overview of polymersome production. Three individual liquid phases, including bacteria suspended in Lysogeny broth (LB) growth media (inner), mPEG-PDLLA polymer in organic solvent (middle), and PVA in water (outer) are continuously injected into a flow-focusing microfluidic device to create water/oil/water double-emulsion droplets. The double-emulsion droplets are washed with LB and then subjected to overnight solvent evaporation to promote polymer migration and phase separation. Bacteria growth and metabolism is observed following polymersome incubation at 37°C. The polymersome allows the diffusion of nutrients and waste products across its semi-permeable membrane while preventing the release of bacteria. Abbreviations: poly(ethylene glycol)-b-poly(D,L-lactic acid) (mPEG-PDLLA); poly(vinyl alcohol) (PVA).
mPEG-PDLLA polymersome stability and degradation properties
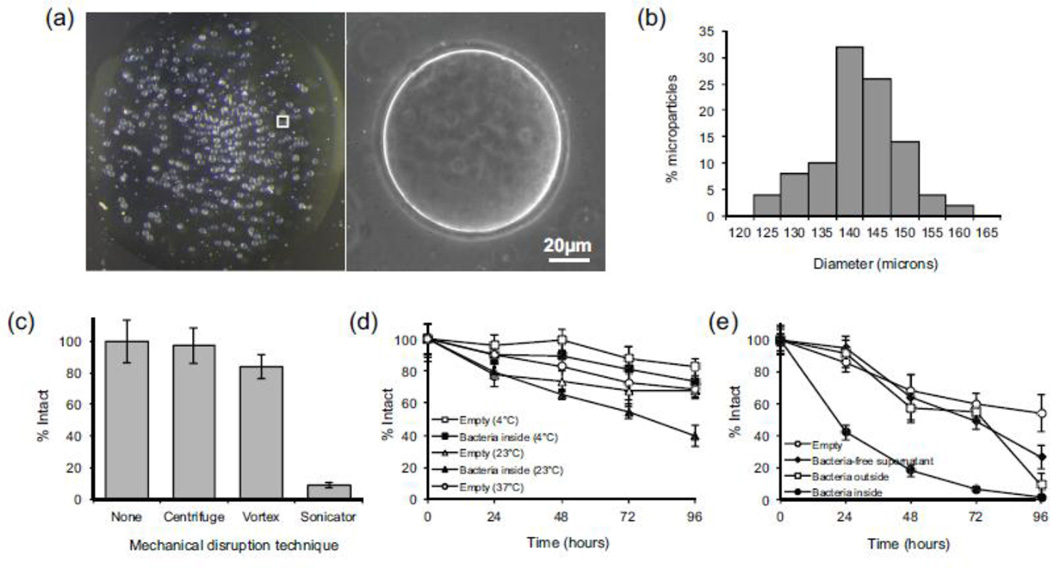
Monodisperse spherical polymersomes of uniform diameter were reproducibly synthesized at a maximum rate of ~42,000 particles per minute (Fig. 2). The mean polymersome size was dependent on the diameter of the tapered upstream capillary, as well as the device flow rates. Following solvent evaporation, particles within a single batch varied from 125 to 165 µm, with a mean diameter of 145 ± 7 µm (Fig. 2B). The overall batch-to-batch variation in mean diameter was less than 5%, a level of reproducibility similar to that reported for microfluidic synthesis of other microparticles (Ho et al. 2008). The integrity of the polymersomes when subjected to standard separation and resuspension processes, such as centrifugation, vortexing, and sonication, was evaluated. By quantifying the polymersome concentration before and after each process, it was determined that repeat centrifugation and vortexing had little effect on polymersome integrity (Fig. 2C), whereas sonication resulted in complete disruption of the polymersome membrane.
Fig. 2.
Physical properties of mPEG-PDLLA polymersomes. (a) Widefield and phase contrast images of a 10µL droplet of polymersomes following solvent evaporation, acquired at 6 × and 20 × magnification respectively. (b) Polymersome size distribution following solvent evaporation. (c) Integrity of polymersomes subjected to mechanical disruption by centrifuge (18,400 g for 10 min), vortex (3000 RPM for 30 s), and probe sonicator (3 s). * Statistically significant (p<0.01). (d) Polymersome degradation following storage under static conditions at 4°C, 23°C, and 37°C, plotted as the percentage of intact polymersomes as a function of time. (e) Polymersome degradation under shaking conditions at 37°C.
Polymersome stability was evaluated under storage and growth conditions. All polymersomes containing bacteria, as well as those containing LB alone (empty particles), displayed time-dependent degradation. Empty particles stored under static conditions showed the least amount of degradation, with no significant differences observed between empty polymersomes stored at 4°C, 23°C, and 37°C for 96 hours (Fig. 2D). Polymersomes containing B. subtilis stored at 23°C showed decreased stability compared to empty particles. To stimulate bacterial growth, polymersomes incubated at 37°C were subjected to continuous shaking (Fig. 2E). Empty polymersomes that were shaken showed a small but statistically significant (p=0.03) decrease in particle count compared to those in static culture, possibly a result of non-specific adhesion of polymersomes to the test tube wall. Polymersomes containing bacteria displayed an increased tendency to rupture, with ~45% of polymersomes remaining after 24 hours compared to 85% remaining without bacteria. By 96 hours, over 90% of the polymersomes that contained bacteria were ruptured.
Encapsulated bacteria demonstrate proliferation and biofilm formation capacity
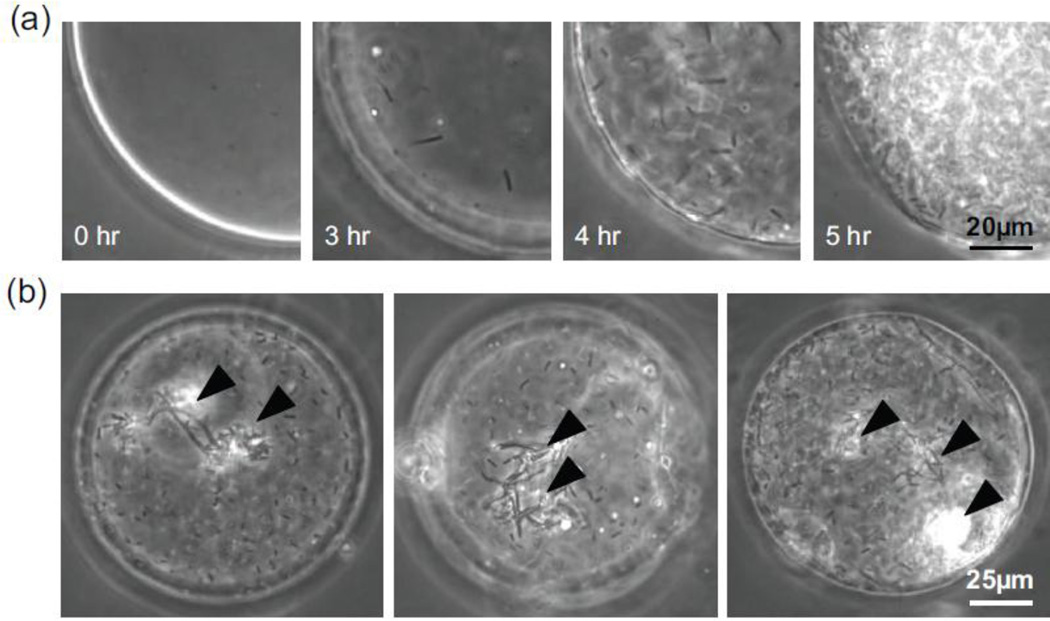
Bacterial viability following microencapsulation was assessed by imaging polymersomes after incubation in shaking culture conditions at 37°C (Fig. 3). We found that up to 70% of the polymersomes contained one or more cells capable of proliferating within 3 hours. We found that encapsulated bacteria could be stored at 4°C for at least one week then grown at 37°C (data not shown). Images of encapsulated bacteria collected at half-hour intervals revealed an initial lag phase followed by exponential growth (Fig. 3A). While many of the bacteria inside the polymersome were highly motile, cell proliferation was also accompanied by the formation of multicellular structures that floated within the polymersome (Videos 2, 3) and grew with time (Fig. 3B). Using ImageJ to count individual cells during the early growth phase (hours 3 – 4), we observed exponential growth with an estimated doubling time of approximately 25 minutes.
Fig. 3.
Growth of bacteria encapsulated in semi-permeable mPEG-PDLLA. (a) Phase-contrast images of B. subtilis encapsulated at a starting concentration of 2 × 108 cells/ml. The bacteria displayed exponential growth following incubation at 37°C. (b) Bacteria transitioned from single planktonic cells into ordered multicellular structures (arrows) as early as 4 hr following incubation.
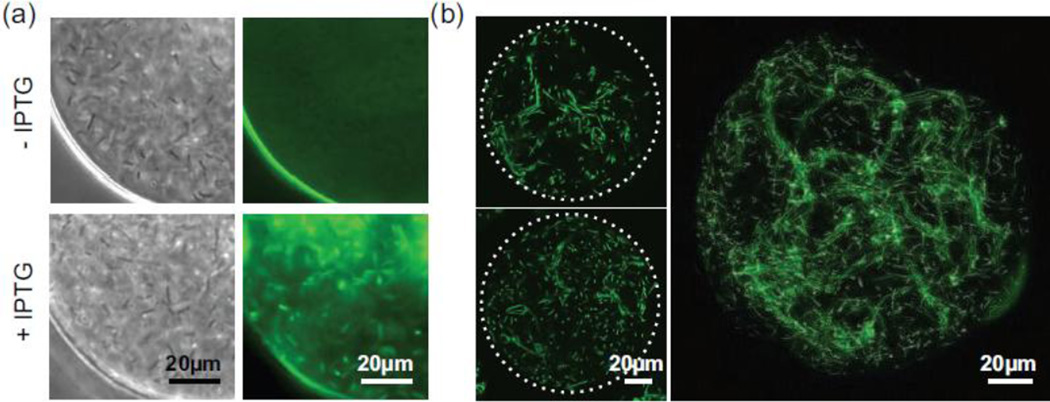
To demonstrate that the polymer membrane was permeable to small molecules, intact polymersomes were incubated at 37°C in LB containing isopropyl β-D-1-thiogalactopyranoside (IPTG, 238 MW, 1 mM), a lactose homolog that promotes GFP expression in genetically modified B. subtilis (Fig. 4). GFP expression was subsequently induced in the bacteria within the polymersomes, indicating that IPTG crossed the polymer membrane (Fig. 4A). Highly uniform GFP expression was observed as early as 30 minutes after IPTG addition. Importantly, this GFP expression was limited to polymersomes treated with IPTG. Using laser scanning confocal microscopy, dense, rope-like, multicellular structures indicative of a biofilm were observed throughout highly populated polymersomes. Representative images are shown in Fig. 4B and Videos 4, 5.
Fig. 4.
GFP expression by encapsulated bacteria following isopropyl β-D-1-thiogalactopyranoside (IPTG) transport across the polymersome membrane. (a) Widefield brightfield and fluorescence images of encapsulated bacteria in the presence and absence of IPTG. The addition of IPTG to the supernatant induces GFP expression (green) by B. subtilis. (b) Representative confocal microscopy images of B. subtilis in IPTG-treated polymersomes incubated at 37°C for 4.5 hr. Three-dimensional biofilm formation is characterized by the formation of dense, multicellular rope-like structures that can be visualized in a single image plane (3µm, left) as well as a maximum intensity projection sampled over a larger thickness (40µm, right).
Encapsulated bacteria convert selenite into selenium nanoparticles
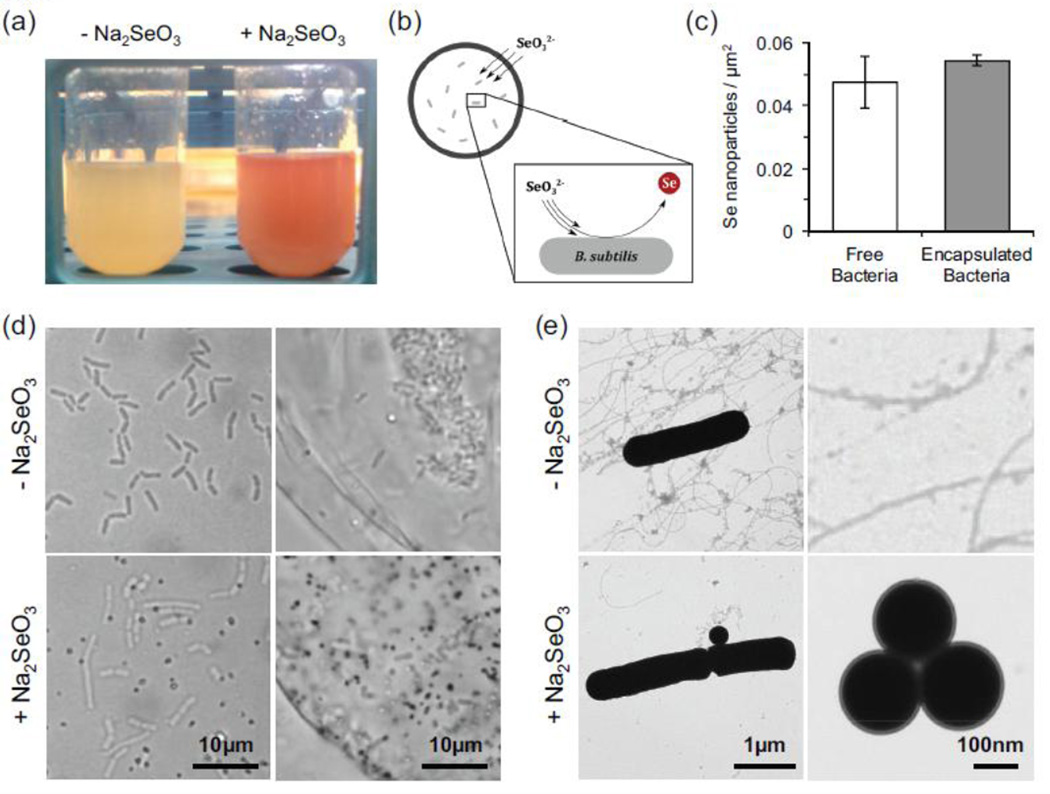
In a proof-of-concept exercise, we utilized B. subtilis to selectively convert sodium selenite (Na2SeO3, 3mM) into elemental selenium (Fig. 5). We found that a culture of B. subtilis produced a distinct red color following 8 hours of co-incubation with sodium selenite (Fig. 5A), indicative of selenite reduction into elemental selenium. Wide-field microscopy revealed that the elemental selenium takes the form of nanoparticles. When selenite was added to the polymersome supernatant, selenium nanoparticles were formed inside the polymersomes (Fig. 5B). The quantity of selenium nanoparticles produced by free and encapsulated bacteria was quantified using wide-field microscopy after 8hr (Fig. 5C). No statistically significant differences were observed, indicating that the polymersome membrane does not present a significant barrier to selenite diffusion. The selective accumulation of selenium nanoparticles inside the polymersomes suggests that the selenite diffused across the polymer membrane and became trapped inside upon conversion to selenium (Fig. 5D). Transmission electron microscopy revealed individual and clustered nanoparticles of ~200–350nm diameter following 8 hours of co-incubation at 37°C (Fig. 5E). We found that polymersome-mediated selenite reduction generated nanoparticles containing as much as 8 × 108 atoms of selenium per nanoparticle within 8 hours. The total amount of selenium accumulated per polymersome was over 100 times higher than expected for an equivalent volume supernatant containing only selenite (~2.8 × 1011 vs 2.2 × 109 Se atoms), indicating that selenite reduction produces a positive diffusion gradient for continued selenite entry into polymersomes.
Fig. 5.
Conversion of soluble selenite (Na2SeO3) to elemental selenium (Se) by free and encapsulated bacteria. (a) Eight hours after sodium selenite addition, B. subtilis solutions turn visibly red, indicative elemental selenium nanoparticle formation. (b) Proposed model of selenium remediation. (c) Quantification of selenium nanoparticle synthesis by free and encapsulated bacteria, as measured by optical microscopy. (d) Widefield images of encapsulated bacteria in the presence and absence of Na2SeO3. The addition of Na2SeO3 to the supernatant results in formation of selenium nanoparticles (dark dots) by free and encapsulated B. subtilis. (e) Transmission electron microscopy images of nanoparticle formation by B. subtilis.
Discussion
Several methodologies were employed to maintain bacterial viability throughout the microencapsulation process. B. subtilis was selected as a model organism due to its ability to survive adverse environmental conditions and its prevalence in environments where excessive soluble selenium compounds may be found. Bacteria were suspended in LB growth media to provide sufficient nutrients for survival during solvent evaporation. The diblock copolymer membrane was selected to contain a mixture of poly(D)- and (L)-lactic acid enantiomers in order to increase membrane permeability (Jain 2000), as well as to permit rapid microparticle degradation for environmental applications. The concentration of the PLA homopolymer was optimized to increase membrane stability while still allowing for cross-membrane transport of nutrients and waste (Martino et al. 2012). The double-emulsion droplets were collected into a vial filled with LB media, and additional washing of the droplets with LB helped to equalize the osmotic pressure across the polymer phase prior to solvent evaporation. Finally, the two-step overnight evaporation stabilized the double emulsions by transitioning the oil layer into a thin shell bilayer.
Following production, polymersome stability was evaluated through mechanical and incubation studies. We found that the polymersomes were able to withstand standard laboratory techniques such as centrifugation and vortexing. However, sonication caused complete disruption and could be used as a method for selectively releasing the contents of the polymersomes. Furthermore, incubation studies indicated that degradation did not appear to be solely caused by mechanical means, such as internal bacterial overgrowth, since empty polymersomes cultured with bacteria on the outside also displayed a significant decrease (p<0.01) in stability at 96 hours compared to empty polymersomes in LB. Interestingly, a decrease in stability was also measured when empty polymersomes were incubated with cell-free supernatant from an overnight bacterial culture, suggesting that the mechanism for polymersome degradation involves digestion by one or more secreted products. It is well known that a variety of microorganisms secrete poly(lactic acid) depolymerases, including B. subtilis (Mayumi et al. 2008). This may be one reason for accelerated degradation when bacteria are present in solution.
We found that up to 70% of the polymersomes contained proliferating bacteria, in contrast to previous reports of viability at 5% or less when encapsulated in PLGA using bulk synthesis techniques (Della Porta et al. 2012). Given that the typical time for heat-activated germination of B. subtilis spores into metabolically active cells exceeds 4 hours (Setlow 2013), it is unlikely that the proliferating bacteria originated from spores at the start of incubation. B. subtilis have previously been reported to double once every 12 hours when encapsulated inside microdroplets comprised of silicon oil (Chang et al. 2015). In contrast, we observed that B. subtilis encapsulated in mPEG-PDLLA are capable of proliferating in an exponential fashion with an estimated doubling time of approximately 25 minutes. This is in close agreement with the minimum doubling time of 21 minutes reported for B. subtilis under optimal growth conditions (Yano et al. 2013).
Wild-type B. subtilis encapsulated in droplets of silicone oil have been reported to form a thin shell of biofilm that adhered at the water-oil interface (Chang et al. 2015). Because silicone oil is non-permeable to nutrients, the rate of biofilm formation in such a system is limited by nutrient availability as well as number of bacteria encapsulated inside the droplet. In contrast, biofilm was formed rapidly inside the nutrient-permeable microparticles, with multicellular structures visible as early as 4 hours after incubation at 37°C. Additionally, the biofilm formed inside polymersomes did not appear to require a substrate for adhesion. Considering that biofilm formation is largely regulated by bacterial density (Lopez et al. 2009), we believe that the ability of B. subtilis to reach high density in the physically constrained, nutrient-rich microenvironment of the polymersome is key to rapid biofilm formation. Considering that biofilm formation in conventional cell culture can take multiple days, the polymersome microencapsulation strategy offers an attractive way to quickly form biofilms for the study of biofilm growth and treatment.
A major advantage of microencapsulation is the ability to selectively harvest bacteria and their products from the supernatant. In particular, we would expect cellular products to which the polymer membrane is impermeable to remain inside the polymersome. A variety of bacteria have been reported to reduce selenite, including B. subtilis (Garbisu et al. 1996). In our studies, selenium nanoparticles were frequently associated with bacteria, suggesting that early nanoparticle growth might occur on the cell wall, thereby preventing the nanoparticles from escaping through the polymersome membrane while still small enough to cross. This bioconversion of selenite into selenium using polymersomes is attractive for reducing selenium toxicity, as well as allowing selenium to be readily harvested from environments not amenable to conventional waste or water treatment. Given the biodegradable nature of the mPEG-PDLLA, it is not necessary for the harvesting process be 100% efficient, since any unharvested microparticles will naturally and rapidly degrade, releasing selenium in a less toxic form. Potential applications for such a technology include remediation of agricultural irrigation runoff into the Kesterson Reservoir in California (Wu 2004), mining water runoff into the Blackfoot Watershed in Idaho (Myers 2013) and other projects where concentrations of selenium compounds in soils, surface waters, and crops are sufficiently high to be of concern.
In conclusion, we have identified a method to encapsulate and grow bacteria in polymer membranes using a double-emulsion droplet-generating microfluidic device. A major advantage of the device is its versatility: the reagents and assembly conditions can be interchanged with ease to adapt polymersome size, permeability, and stability for specific applications. We found that B. subtilis encapsulated in mPEG-PDLLA membranes remain viable throughout polymersome synthesis and display the capacity to proliferate when raising the temperature to 37°C. Bacteria growth is exponential and is accompanied by rapid biofilm formation, demonstrating that the polymersome membrane is sufficiently permeable to permit optimal bacteria growth. The microencapsulated bacteria are capable of converting soluble selenite to selenium nanoparticles that are retained inside the polymersome, opening opportunities for rapid selenium remediation from polluted environments. A further study of semi-permeable bacterial encapsulation, including expansion to new bacteria species and polymer formulations, is likely to generate microenvironments conducive to the sustained capture (or release) of bacteria products, with applications ranging from the prevention of biofouling to the enhancement of bioreactor efficiency.
Supplementary Material
Acknowledgments
Special thanks to Alireza Abbaspourrad and David A.Weitz for the demonstration of double-emulsion microfluidic device assembly techniques.
Funding
This work was supported in part by: National Science Foundation (NSF) DGE-0965843, Department of Defense (DOD) W81XWH-09-2-0001, National Cancer Institute (NCI) 1R25CA174650-01A, a startup grant from Northeastern University, and the Electronics Materials Research Institute at Northeastern University. Undergraduates J.B., K.G., and C.K. were supported by the Northeastern University Provost Undergraduate Research Award, Honors Early Research Grant, and Advanced Research/Creative Endeavor Award.
Footnotes
Compliance with Ethical Standards
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of Interest
Author 1 declares that he has no conflict of interest.
Author 2 declares that he has no conflict of interest.
Author 3 declares that he has no conflict of interest.
Author 4 declares that he has no conflict of interest.
Author 5 declares that he has no conflict of interest.
Author 6 declares that he has no conflict of interest.
Author 7 declares that he has no conflict of interest.
Author 8 declares that she has no conflict of interest.
References
- Agency for Toxic Substances and Disease Registry. Toxicological Profile for Selenium (Update) Atlanta: 2003. [PubMed] [Google Scholar]
- Borg DJ, Bonifacio E. The Use of Biomaterials in Islet Transplantation. Curr. Diab. Rep. 2011;11:434–444. doi: 10.1007/s11892-011-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde I, Steil L, Scharf C, Volker U, Bremer E. Adaptation of Bacillus subtilis to growth at low temperature: a combined transcriptomic and proteomic appraisal. Microbiol. 2006;152:831–853. doi: 10.1099/mic.0.28530-0. [DOI] [PubMed] [Google Scholar]
- Chang CB, Wilking JN, Kim S-H, Shum HC, Weitz DA. Monodisperse Emulsion Drop Microenvironments for Bacterial Biofilm Growth. Small. 2015;11:3954–3961. doi: 10.1002/smll.201403125. [DOI] [PubMed] [Google Scholar]
- Chen S, Cheng S-X, Zhuo R-X. Self-Assembly strategy for the preparation of polymer-based nanoparticles for drug and gene delivery. Macromol Biosci. 2011;11:576–589. doi: 10.1002/mabi.201000427. [DOI] [PubMed] [Google Scholar]
- Chudobova D, Cihalova K, Dostalova S, Ruttkay-Nedecky B, Rodrigo MAM, Tmejova K, Kopel P, Nejdl L, Kudr J, Gumulec J, Krizkova S, Kynicky J, Kizek R, Adam V. Comparison of the effects of silver phosphate and selenium nanoparticles on Staphylococcus aureus growth reveals potential for selenium particles to prevent infection. FEMS Microbiol Lett. 2014;351:195–201. doi: 10.1111/1574-6968.12353. [DOI] [PubMed] [Google Scholar]
- Cook MT, Tzortzis G, Charalampopoulos D, Khutoryanskiy VV. Microencapsulation of a synbiotic into PLGA/alginate multiparticulate gels. Int J Pharm. 2014;466:400–408. doi: 10.1016/j.ijpharm.2014.03.034. [DOI] [PubMed] [Google Scholar]
- Danhier F, Ansorena E, Silva JM, Coco R, Le Breton A, Préat V. PLGA-based nanoparticles: an overview of biomedical applications. J Control Release. 2012;161:505–522. doi: 10.1016/j.jconrel.2012.01.043. [DOI] [PubMed] [Google Scholar]
- Della Porta G, Castaldo F, Scognamiglio M, Paciello L, Parascandola P, Reverchon E. Bacteria microencapsulation in PLGA microdevices by supercritical emulsion extraction. J Supercrit Fluids. 2012;63:1–7. [Google Scholar]
- DeYoung MB, MacConell L, Sarin V, Trautmann M, Herbert P. Encapsulation of Exenatide in Poly-(d,l-Lactide-Co-Glycolide) Microspheres Produced an Investigational Long-Acting Once-Weekly Formulation for Type 2 Diabetes. Diabetes Technol. Ther. 2011;13:1145–1154. doi: 10.1089/dia.2011.0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dungan RS, Frankenberger WT. Microbial transformations of selenium and the bioremediation of seleniferous environments. Bioremediat J. 1999;3:171–188. [Google Scholar]
- Estevam EC, Witek K, Faulstich L, Nasim MJ, Latacz G, Dominguez-Alvarez E, Kiec-Kononowicz K, Demasi M, Handzlik J, Jacob C. Aspects of a distinct cytotoxicity of selenium salts and organic selenides in living cells with possible implications for drug design. Molecules. 2015;20:13894–13912. doi: 10.3390/molecules200813894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes AP, Gandin V. Selenium compounds as therapeutic agents in cancer. Biochim Biophys Acta. 2015;1850:1642–1660. doi: 10.1016/j.bbagen.2014.10.008. [DOI] [PubMed] [Google Scholar]
- Fordyce F. Selenium Deficiency and Toxicity in the Environment. In: Selinus O, editor. Essentials of Medical Geology. 2nd. Springer; 2013. pp. 375–416. [Google Scholar]
- Garbisu C, Ishii T, Leighton T, Buchanan BB. Bacterial reduction of selenite to elemental selenium. Chem Geol. 1996;132:199–204. [Google Scholar]
- Ho CS, Kim JW, Weitz DA. Microfluidic fabrication of monodisperse biocompatible and biodegradable polymersomes with controlled permeability. J Am Chem Soc. 2008;130:9543–9549. doi: 10.1021/ja802157y. [DOI] [PubMed] [Google Scholar]
- Holtmann G, Bremer E. Thermoprotection of Bacillus subtilis by exogenously provided glycine betaine and structurally related compatible solutes: involvement of the Opu transporters. J Bacteriol. 2004;186:1683–1693. doi: 10.1128/JB.186.6.1683-1693.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain RA. The manufacturing techniques of various drug loaded biodegradable poly(lactide-co-glycolide) (PLGA) devices. Biomaterials. 2000;21:2475–2490. doi: 10.1016/s0142-9612(00)00115-0. [DOI] [PubMed] [Google Scholar]
- Kailasapathy K. Microencapsulation of probiotic bacteria: technology and potential applications. Curr Issues Intest Microbiol. 2002;3:39–48. [PubMed] [Google Scholar]
- Lapage SP, Bascomb S. Use of selenite reduction in bacterial classification. J Appl Bacteriol. 1968;31:568–580. doi: 10.1111/j.1365-2672.1968.tb00407.x. [DOI] [PubMed] [Google Scholar]
- Lopez D, Vlamakis H, Losick R, Kolter R. Paracrine signaling in a bacterium. Genes Dev. 2009;23:1631–1638. doi: 10.1101/gad.1813709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makadia HK, Siegel SJ. Poly Lactic-co-Glycolic Acid (PLGA) as biodegradable controlled drug delivery carrier. Polymers (Basel) 2011;3:1377–1397. doi: 10.3390/polym3031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino C, Kim SH, Horsfall L, Abbaspourrad A, Rosser SJ, Cooper J, Weitz DA. Protein expression, aggregation, and triggered release from polymersomes as artificial cell-like structures. Angew Chemie - Int Ed. 2012;51:6416–6420. doi: 10.1002/anie.201201443. [DOI] [PubMed] [Google Scholar]
- Mayumi D, Akutsu-Shigeno Y, Uchiyama H, Nomura N, Nakajima-Kambe T. Identification and characterization of novel poly(DL-lactic acid) depolymerases from metagenome. Appl Microbiol Biotechnol. 2008;79:743–750. doi: 10.1007/s00253-008-1477-3. [DOI] [PubMed] [Google Scholar]
- Myers T. Remediation scenarios for selenium contamination, Blackfoot watershed, southeast Idaho, USA. Hydrogeol J. 2013;21:655–671. [Google Scholar]
- Nichols DS, Nichols PD, McMeekin TA. Ecology and physiology of psychrophilic bacteria from Antarctic saline lakes and sea-ice. Sci Prog. 1995;78:311–348. [Google Scholar]
- Olabisi RM. Cell microencapsulation with synthetic polymers. J Biomed Mater Res A. 2015;103:846–859. doi: 10.1002/jbm.a.35205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oremland RS, Herbel MJ, Blum JS, Langley S, Beveridge TJ, Ajayan PM, Sutto T, Ellis AV, Curran S. Structural and spectral features of selenium nanospheres produced by se-respiring bacteria. Appl Environ Microbiol. 2004;70:52–60. doi: 10.1128/AEM.70.1.52-60.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S, Tomaro-Duchesneau C, Saha S, Cantor A. The gut microbiota and human health with an emphasis on the use of microencapsulated bacterial cells. J Biomed Biotechnol. 2011;2011:981214. doi: 10.1155/2011/981214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell D. Molecular Cloning: A Laboratory Manual. 3rd. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Setlow P. Summer meeting 2013 – when the sleepers wake: the germination of spores of Bacillus species. J Appl Microbiol. 2013;115:1251–1268. doi: 10.1111/jam.12343. [DOI] [PubMed] [Google Scholar]
- Tran PA, Webster TJ. Antimicrobial selenium nanoparticle coatings on polymeric medical devices. Nanotechnology. 2013;24:155101. doi: 10.1088/0957-4484/24/15/155101. [DOI] [PubMed] [Google Scholar]
- U.S. Environmental Protection Agency. Integrated Risk Information System (IRIS): Selenium and Compounds. Washington, DC: 1991. [Google Scholar]
- Utada AS, Lorenceau E, Link DR, Kaplan PD, Stone HA, Weitz DA. Monodisperse double emulsions generated from a microcapillary device. Science. 2005;308:537–541. doi: 10.1126/science.1109164. [DOI] [PubMed] [Google Scholar]
- Wallenberg M, Misra S, Bjornstedt M. Selenium cytotoxicity in cancer. Basic Clin Pharmacol Toxicol. 2014;114:377–386. doi: 10.1111/bcpt.12207. [DOI] [PubMed] [Google Scholar]
- Wang Q, Larese-Casanova P, Webster TJ. Inhibition of various gram-positive and gram-negative bacteria growth on selenium nanoparticle coated paper towels. Int J Nanomedicine. 2015;10:2885–2894. doi: 10.2147/IJN.S78466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Webster TJ. Biomimetics. John Wiley & Sons, Inc; 2013. Nanostructured selenium – a novel biologically-inspired material for antibacterial medical device applications; pp. 203–220. [Google Scholar]
- World Health Organization. Trace elements in human nutrition and health. 1996
- Wu L. Review of 15 years of research on ecotoxicology and remediation of land contaminated by agricultural drainage sediment rich in selenium. Ecotoxicol Environ Saf. 2004;57:257–269. doi: 10.1016/S0147-6513(03)00064-2. [DOI] [PubMed] [Google Scholar]
- Yano K, Wada T, Suzuki S, Tagami K, Matsumoto T, Shiwa Y, Ishige T, Kawaguchi Y, Masuda K, Akanuma G, Nanamiya H, Niki H, Yoshikawa H, Kawamura F. Multiple rRNA operons are essential for efficient cell growth and sporulation as well as outgrowth in Bacillus subtilis. Microbiology. 2013;159:2225–2236. doi: 10.1099/mic.0.067025-0. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ho Y-P, Chiu Y-L, Chan HF, Chlebina B, Schuhmann T, You L, Leong KW. A programmable microenvironment for cellular studies via microfluidics-generated double emulsions. Biomaterials. 2013;34:4564–4572. doi: 10.1016/j.biomaterials.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.