Abstract
Cytokines of the transforming growth factor β (TGF-β) family, including TGF-βs, bone morphogenic proteins (BMPs), activins, and Nodal, play crucial roles in embryonic development and adult tissue homeostasis by regulating cell proliferation, survival, and differentiation, as well as stem-cell self-renewal and lineage-specific differentiation. Smad proteins are critical downstream mediators of these signaling activities. In addition to regulating the transcription of direct target genes of TGF-β, BMP, activin, or Nodal, Smad proteins also participate in extensive cross talk with other signaling pathways, often in a cell-type- or developmental stage-specific manner. These combinatorial signals often produce context-, time-, and location-dependent biological outcomes that are critical for development. This review discusses recent progress in our understanding of the cross talk between Smad proteins and signaling pathways of Wnt, Notch, Hippo, Hedgehog (Hh), mitogen-activated protein (MAP), kinase, phosphoinositide 3-kinase (PI3K)-Akt, nuclear factor κB (NF-κB), and Janus kinase/signal transducers and activators of transcription (JAK/STAT) pathways.
Smad proteins transduce signals directly from TGF-β ligands to the nucleus. In addition, Smads participate in cross talk with other signaling pathways (e.g., Wnt), often in a cell type- or developmental stage-specific manner.
The transforming growth factor β (TGF-β) family of cytokines, including TGF-βs, bone morphogenic proteins (BMPs), and activins, regulates a wide array of biological activities in various cell types and at different developmental stages. Smad proteins are critical mediators of TGF-β, BMP, and activin signaling (Feng and Derynck 2005; Heldin and Moustakas 2011; Massagué 2012). On phosphorylation by the activated type-I receptor kinase, the receptor-associated R-Smads form a heteromeric complex with the co-Smad and translocate into the nucleus, where they interact with sequence-specific DNA-binding cofactors and transcriptional coactivators or corepressors to regulate the transcription of target genes. Additionally, the activity of this Smad pathway can be regulated by positive and negative modulators, including the inhibitory Smads, Smad6 and Smad7, the corepressors Ski and SnoN, and the Smurf family of E3 ubiquitin ligases.
The Smad pathway is integrated into the intracellular signaling network through cross talk with other signaling pathways, and these cross talk activities play important roles in the regulation of various biological responses. The cross talk can occur at multiple levels: by altering the expression and activities of ligands, antagonists, receptors, and signaling components; by incorporating into transcription complexes and/or inducing changes in chromatin modification complexes that globally impact gene expression; and by direct interactions between Smads and other intracellular signaling components. This review discusses the cross talk of Smads with Wnt, Notch, Hippo, Hedgehog (Hh), mitogen-activated protein (MAP) kinase, phosphoinositide 3-kinase (PI3K)-Akt, nuclear factor κB (NF-κB), and JAK-STAT signaling pathways, with a focus on the direct interactions among key signaling components. This review does not discuss the cross talk between TGF-β-activated non-Smad signaling pathways and other signaling pathways.
CROSS TALK WITH Wnt SIGNALING
The Wnt signaling pathways regulate many aspects of vertebrate development and play important roles in cell-fate determination, self-renewal, and maintenance of stem and early progenitor cells. Deregulation of Wnt signaling is associated with various types of human cancer, including colorectal cancer and leukemia. The canonical Wnt signaling pathway is initiated on binding of a Wnt ligand to its cognate receptor Frizzled and the transmembrane protein Lrp5 or Lrp6, and is primarily mediated by β-catenin (Nusse 2012). In the absence of a Wnt ligand, the newly synthesized β-catenin is found in the destruction complex with the adenomatous polyposis coli (APC) tumor suppressor and scaffolding protein Axin, where it is phosphorylated by casein kinase I (CKI) and glycogen synthase kinase-3β (GSK-3β) and targeted for degradation. On ligand binding, Lrp5 or Lrp6 binds to Axin in a Wnt- and phosphorylation-dependent manner, leading to the formation of the complex containing Dishevelled (Dvl), Axin, and GSK-3β. As a consequence, the kinase activity of GSK-3β is inhibited, resulting in stabilization of β-catenin. β-catenin then translocates into the nucleus and binds to the closely related T-cell factor (TCF) or lymphoid enhancer–binding factor (LEF) transcription factors. With the help of additional nuclear components, including BCL9, Pygopos, and cAMP-response element-binding (CREB)-binding protein (CBP), this binding converts TCF or LEF from transcriptional repressors into activators. Wnt signaling also regulates planar cell polarity through the noncanonical pathway, by activating Rho and Rac signaling, and modulates calcium release through G-protein-dependent activation of the phospholipase C (PLC) pathway (Krausova and Korinek 2014).
Combinatorial TGF-β and Wnt Signaling Is Essential for Early Development and Tissue Homeostasis
Wnt signaling benefits from extensive cross talk with other signaling pathways, particularly TGF-β and BMP signaling, and the combinatorial signaling often occurs in early embryos to allow overlapping signaling pathways to specify different territories and cell fates. In early embryos, extensive mutual regulation and cross talk between Wnt and Nodal/activin/BMP pathways and later between Wnt and BMP signaling exist at multiple levels, and these interactions are essential for embryonic patterning and development of multiple lineages. For example, in Drosophila, the BMP ligand Decapentaplegic (Dpp) and Wnt ligand Wingless (Wg) cooperate to pattern the wings, legs, imaginal discs, brain and midgut (Attisano and Labbé 2004). In Xenopus, signals from both pathways are critical for the establishment of Spemann’s organizer and activation of many organizer-specific genes, including those encoding Twin, Goosecoid, chordin, and Cerberus, as well as dorsal fate specification in mesoderm and endoderm (Cui et al. 1996; Crease et al. 1998; Zorn et al. 1999; Labbé et al. 2000; Nishita et al. 2000; Schohl and Fagotto 2002; Xanthos et al. 2002). In zebrafish, the two pathways together regulate posterior mesoderm formation by synergistically activating the expression of posterior mesoderm genes such as tbx6 (Szeto and Kimelman 2004). In mouse embryos, Wnt signaling modulates the expression of the BMP target gene Msx2, either directly or through induction of expression of BMP ligands, thereby influencing cell fates in the ectoderm and the neural crest (Hussein et al. 2003). In the dorsal telencephalon, Wnt and BMP signaling regulate graded emx2 expression in a cooperative manner (Theil et al. 2002).
In adult tissues, Wnt and BMP signaling often interact to ensure proper tissue homeostasis by regulating the expression of common key target genes, and aberrant signaling in either pathway often contributes to carcinogenesis and diseases. Compound heterozygote mice lacking both Smad4 and APC develop more intestinal or pancreatic tumors than deletion of APC alone, and deletion of Smad2 accelerates colon cancer progression in APC-deficient mice (Takaku et al. 1998; Cullingworth et al. 2002; Hamamoto et al. 2002). However, a separate study reported that compound Smad2/Apc heterozygotes are indistinguishable from Apc-null mice in intestinal tumor progression (Takaku et al. 2002), and argued that Smad4 plays a more prominent role in coordinating with Wnt signaling in the intestine. In support of these observations, TGF-β and Wnt were shown to synergize in the transcription activation of the Wnt target gene encoding gastrin, a promoter of gastrointestinal cancer, indicating that TGF-β and Wnt signaling can cooperate to promote tumorigenesis (Lei et al. 2004).
Mechanistically, the TGF-β/BMP and Wnt pathways coordinate to regulate development and homeostasis, likely by controlling the self-renewal and differentiation of stem cells. In mouse embryonic stem (ES) cells (mESCs), BMP, acting together with leukemia inhibitory factor (LIF), maintains pluripotency and is essential for self-renewal (Ying et al. 2003). In the presence of both TGF-β and Wnt signaling, however, BMP induces a posterior primitive-streak (PS)-like fate and promotes differentiation of PS-like cells into Flk1-expressing hematopoietic mesoderm (Nostro et al. 2008). In the Flk1-expressing hematopoietic mesoderm, BMP activates Wnt signaling, and the two signals then act together to activate the Cdx-Hox pathway, leading to blood cell–fate commitment (Lengerke et al. 2008). The presence of TGF-β and Wnt signaling is required for the initial inductive activity of BMP, because inhibition of either of these signals abolishes the inductive activity. Similarly, in human ES cells (hESCs), BMP induces mesendoderm differentiation together with fibroblast growth factor 2 (FGF2), and this activity requires TGF-β or Wnt signaling (Yu et al. 2011). In early neural crest stem cells, Wnt promotes sensory neurogenesis, whereas BMP antagonizes Wnt signaling to suppress differentiation and neurogenesis (Kleber et al. 2005). BMP also suppresses Wnt signaling to maintain a proper balance in self-renewal of intestinal stem cells in a phosphatase and tensin homolog (PTEN)-Akt pathway-dependent manner. BMP enhances the activity of PTEN, leading to inactivation of Akt and inhibition of the nuclear accumulation, and transcription activity of β-catenin (He et al. 2004), resulting in inhibition of Wnt signaling. Finally, in transformed mammary epithelial cells, TGF-β and Wnt signaling synergize to induce activation of the epithelial–mesenchymal transition (EMT) program, and function in an autocrine fashion to maintain the resulting stem-cell state (Scheel et al. 2011). Thus, a common theme that emerges from these observations is that the outcome of signaling cross talk is determined by the context of the signaling environment and that multiple signal inputs, rather than BMP or Wnt alone, are needed to allow stem-cell fate determination (Kimelman and Griffin 2000; Loose and Patient 2004). This theme is frequently repeated in cross talk among other pathways as well.
Cross Talk between TGF-β Family and Wnt Signaling Occurs at Multiple Levels
On receptor activation, cross talk between TGF-β family and Wnt signaling can occur at multiple levels (Fig. 1).
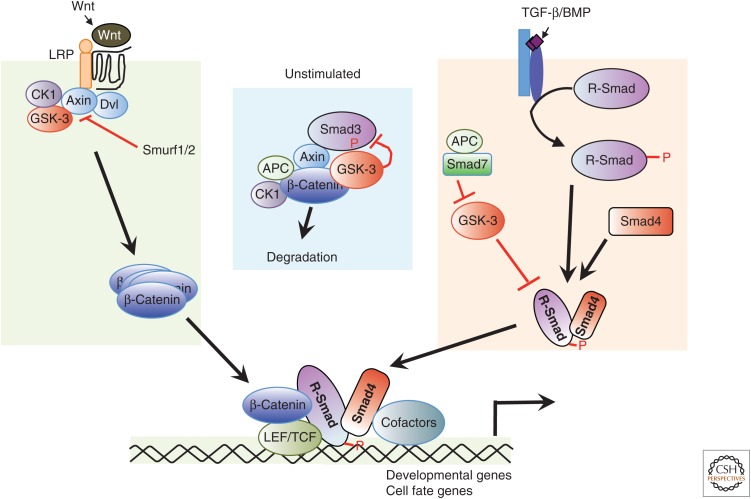
Figure 1.
Cross talk between the transforming growth factor β (TGF-β) family and Wnt signaling at multiple points. In the absence of TGF-β stimulation (middle), Smad3 can form a complex with Axin and glycogen synthase kinase (GSK)-3β, where it is phosphorylated by GSK-3β, leading to its degradation. In the presence of TGF-β or bone morphogenic proteins (BMPs) stimulation (right), GSK-3β also phosphorylates the activated R-Smads (Smad1 or Smad3) in the linker region to inhibit their activity and promote degradation. Wnt signaling inhibits GSK-3β and stabilizes the Smad proteins. Other components of the TGF-β pathway, including Smurf1, Smurf2, and Smad7, also modulate Wnt signaling. In response to stimulation by Wnt, the canonical Wnt pathway and the Smad pathway can synergize to activate transcription of target genes. Smad3 facilitates β-catenin nuclear translocation and coordinates with the complex of β-catenin and T-cell factor (TCF) or lymphoid enhancer–binding factor 1 (LEF1) at regulatory promoter sequences of target genes that contain TCF- or LEF1-binding sites and/or Smad-binding sequences to regulate gene expression.
Reciprocal Regulation of the Expression of Pathway Ligands and Antagonists
Wnt signaling modulates the expression of BMP or Nodal ligands, coreceptor or BMP antagonists in embryos, adult stem cells, and cancer cells (Guo and Wang 2009), whereas BMP-2 and BMP-4 regulate the expression of Wnt-8 in Xenopus (Hoppler and Moon 1998) or Wnt-7c in chicken embryonic mesenchymal cells (Jin et al. 2006). These regulations are likely to be critical for establishing proper morphogen gradients during cell-fate determination.
Direct Physical Interaction between and Modification of Key Components of the Two Pathways in the Cytoplasm and/or Nucleus
A well-documented mechanism of Smad regulation by Wnt signaling is through phosphorylation of Smad proteins in the linker region by GSK-3β (Fuentealba et al. 2007; Millet et al. 2009; Aragon et al. 2011). In mammalian cells and Xenopus embryos, in the absence of Wnt, GSK-3β phosphorylates the linker region of Smad1, resulting in its polyubiquitylation and degradation. Wnt signaling inhibits GSK-3β activity and prevents Smad1 linker phosphorylation, leading to Smad1 stabilization (Fuentealba et al. 2007; Aragon et al. 2011). Similarly, GSK-3β phosphorylates Smad3 in the linker region on Ser204, and this phosphorylation appears to inhibit the transcription activity of Smad3. Mutation of Ser204 to alanine strengthens the interaction of Smad3 with transcription coactivators, and promotes its ability to activate target genes and its ability to induce cell-cycle arrest (Millet et al. 2009; Wang et al. 2009a). In the absence of TGF-β, Axin and GSK-3β can bind to Smad3 to promote its degradation. GSK-3β phosphorylates Smad3 at Thr66, leading to its ubiquitylation and degradation, and this phosphorylation is further enhanced in the presence of Axin. Through this linker phosphorylation, Wnt signaling can control the basal level of Smad3 activity in cells (Guo et al. 2008).
GSK-3β phosphorylation of Smad1 or Smad3 appears to be a critical step in the sequential regulation of Smad activation and subsequent destruction in response to BMP or TGF-β and Wnt signals. Smad proteins are first activated by BMP or TGF-β signaling through phosphorylation at two carboxy-terminal serines. This activation is followed by a series of phosphorylation events at the linker region that is mediated by extracellular signal-regulated kinase (Erk) or p38 MAP kinases, or cyclin-dependent kinase (CDK)8 or CDK9, which prime the Smad proteins for binding to and phosphorylation by GSK-3β (Fuentealba et al. 2007; Aragon et al. 2011). The regulation of Smad proteins by GSK-3β in the presence of BMP or TGF-β signals not only serves to inactivate Smad signaling, but also provides a path for the Wnt ligand to directly regulate Smad activity. In vivo epistatic experiments in Xenopus embryos indicate that Smad1 phosphorylation by GSK-3β plays a key role in mediating the effects of Wnt signaling on neural development at the gastrula stage and in ectodermal cells. Furthermore, overexpression of Wnt-8 induced epidermal differentiation dependent on activation of Smad1, 5, and/or 8 by BMP (Fuentealba et al. 2007).
Negative regulation of Smad activity through linker phosphorylation by GSK-3β has also been observed in Drosophila (Eivers et al. 2009, 2011; Quijano et al. 2011). In Drosophila, Mad is capable of signaling in both the Dpp (BMP subfamily) and Wingless (Wnt family) pathways, and the pathway choice depends on the phosphorylation state of Mad. Signaling downstream of Dpp requires the carboxy-terminal phosphorylation of Mad, whereas unphosphorylated Mad participates in canonical Wingless signaling to restrict self-renewing mitosis by interacting with the transcription factors Armadillo and Pangolin (homologs of β-catenin and TCF, respectively). Both Wingless and Dpp-induced functions of Mad are terminated by GSK-3β-dependent linker phosphorylation. Thus, Drosophila Mad can exist in three functional states depending on the phosphorylation status. Given the conservation of Zw3/GSK-3β phosphorylation sites in vertebrate Smad1, 5, and 8, it is possible that this triphasic response to Wingless- and TGF-β family- or BMP-dependent Smad phosphorylation may also be conserved during vertebrate embryonic development (Shimmi and Newfeld 2013).
Smad proteins and Wnt pathway components can also physically interact to regulate the activity of each other (Fig. 1). Smad3 has been found in the same complex as Axin and CKIɛ, and GSK-3β in transfected cells as well as human mesenchymal stem cells (MSCs), in the absence of TGF-β stimulation in which Smad3 can be phosphorylated and inhibited by CKIɛ or GSK-3β (Furuhashi et al. 2001; Waddell et al. 2004; Jian et al. 2006). The interaction of Axin and Smad3 appears to facilitate the phosphorylation of Smad3 by the active TGF-β type I receptor (TβRI) kinase, resulting in enhanced transcriptional activation of reporter constructs (Furuhashi et al. 2001). Smad3 also plays an essential role in shuttling β-catenin into the nucleus, likely through TGF-β-induced phosphorylation of Smad3 and the subsequent reduction in the interaction of Smad3 with GSK-3β (Jian et al. 2006). Dissociation of this protein complex allows cotranslocation of β-catenin and Smad3 into the nucleus, with Smad3 acting as a chaperone, and this regulation is required for the stimulation of MSC proliferation and inhibition of MSC osteogenic differentiation by TGF-β1.
Other positive and negative regulators of the Smad pathway can also mediate cross talk with the canonical Wnt pathway. For example, Smurf1 and Smurf2 have been shown to inhibit Wnt signaling by targeting Axin for ubiquitylation, but using distinct mechanisms and with different consequences. Smurf2 induces polyubiquitylation of Axin at Lys505, leading to its degradation (Kim and Jho 2010). Reducing endogenous Smurf2 levels results in accumulation of Axin and a subsequent decrease in β-catenin signaling. Smurf1, on the other hand, ubiquitylates Axin at Lys789 and Lys821 mainly through the Lys29 ubiquitin linkage, which disrupts the association of Axin with Lrp5 or Lrp6, leading to attenuation of Wnt signaling (Fei et al. 2013). In addition to the Smurf proteins, Smad7 and p38 MAP kinase (MAPK) together regulate the expression of APC and cell migration in prostate cancer cells in response to TGF-β (Ekman et al. 2012). Smad7 forms a complex with APC and acts as an adaptor protein for the p38 MAPK and GSK-3β kinases to facilitate TGF-β- and p38 MAPK–dependent inactivation of GSK-3β, leading to accumulation of β-catenin and recruitment of APC to the microtubule plus end in the leading edge of migrating prostate cancer cells. The Smad7–APC complex also links TβRI to the microtubule system to regulate TGF-β-dependent cell migration.
Finally, cross talk between dSno, the fly homolog of SnoN, and Wnt signaling in Drosophila wing development has also been reported (Quijano et al. 2010). Analysis of the loss of function mutant of dSno reveals the presence of ectopic margin bristles and campaniform sensilla in the anterior wing blade, whereas the gain of function of dSno mutation results in a loss of bristles and sensilla, features usually controlled by Wingless (Wg) signaling. These phenotypes are consistent with a role of dSno in the antagonism of Wg signaling. The biochemical mechanism by which dSno cross talks to Wg signaling has not been defined, and such a cross talk has not been reported in vertebrate systems.
Convergence at Transcription Complexes Assembled at Target Gene Regulatory Sequences
The transcription complex containing β-catenin and TCF or LEF1 often functions as the signal coordinator that interacts with the Smad proteins to mediate Wnt-TGF-β family cross talk (Labbé et al. 2000; Nishita et al. 2000; Hussein et al. 2003; Szeto and Kimelman 2004). In response to Wnt signaling and BMP or TGF-β stimulation, R-Smads, including Smad1, Smad2, and Smad3 as well as Smad4, directly associate with TCF or LEF1 to form a transcriptional activation complex on the promoter DNA. The promoter regions of many Wnt- and BMP- or TGF-β-responsive genes, such as Xtwin, tbx6, Msx2, and gastrin, often contain Smad-binding element (SBE) and TCF- or LEF1-binding sites in juxtaposition, such that the Smad proteins and TCF or LEF1, present in the same transcription complex, can simultaneously bind to their own recognition sequences and synergize to activate transcription (Labbé et al. 2000; Nishita et al. 2000; Hussein et al. 2003; Szeto and Kimelman 2004). Optimal activation of these genes under physiological concentrations usually requires the synergy of the two pathways. Genome-wide chromatin immunoprecipitation-sequencing (ChIP-Seq) mapping studies reveal that the binding sites for Smad1, 5, and/or 8 often overlap with those for the key pluripotency transcription factors Oct4, Sox2, and Nanog, as well as STAT3 (downstream of LIF) in mESCs (Chen et al. 2008). Smad1 and TCF7l1/TCF3 have been found to co-occupy target sites together with the Oct4/Nanog/Sox2 complex in the pluripotency target genes in the ES cells (Chen et al. 2008; Cole et al. 2008). In addition, both Smad1 and TCF7L2 co-occupy sites with master regulators adjacent to hematopoietic genes to regulate hematopoietic stem-cell fate (Trompouki et al. 2011). These data suggest that TGF-β family and Wnt signaling extensively cross talk at many levels, and that multiple signaling inputs are integrated into the core transcription factor network to regulate target gene expression in a cooperative manner. Together, they regulate embryonic development, tissue homeostasis, and carcinogenesis, and modulate the self-renewal and differentiation of embryonic and adult stem cells.
CROSS TALK WITH NOTCH SIGNALING
Notch signaling is triggered by the binding of the cell-surface Notch receptor to its ligands Delta, Serrate, or Lag-2 (DSL family ligands), located at the surface of neighboring cells on cell–cell contact. This binding results in two proteolytic cleavage events, first at the extracellular domain by the membrane-associated metalloprotease tumor necrosis factor α–converting enzyme (TACE), also known as ADAM17 to shed the extracellular domain, and second within the transmembrane domain by the γ-secretase activity of a multiprotein complex containing presenilin, APH1, nicastrin, and PEN2, leading to the release of the signaling Notch intracellular domain (NICD) from the cell membrane. The NICD then translocates into the nucleus and binds to DNA-binding proteins of the CBF1/RBPjk/Su(H)/Lag1 (CSL) family (typified by hairless, RBP-Jκ, and CBF1). Binding of NICD to the DNA-bound RBP-Jκ then displaces the RBP-Jκ-associated histone deacetylase corepressor complex and recruits the coactivator P/CAF, converting RBP-Jκ from a transcription repressor to an activator. The NICD/RBP-Jκ complex activates the primary Notch target genes, including members of the Hairy/Enhancer of split (HES) and HES-related repressor protein (HERP) families of basic/helix–loop–helix transcription repressors (Nowell and Radtke 2013). The HES and HERP proteins subsequently regulate the expression of downstream tissue-specific transcription factors.
Notch signaling is an evolutionarily conserved pathway that regulates stem-cell-fate determination and differentiation during embryonic development, tissue homeostasis, and carcinogenesis. Many developmental processes that are regulated by Notch signaling are also controlled by TGF-β family ligands including BMPs, thus setting the stage for frequently occurring cross talk between the two pathways. Several studies of the cross talk between BMP and Notch pathways were performed in cell-line-based differentiation models, such as myogenic or osteoblast differentiation of C2C12 myoblasts and MC3T3 pre-osteoblasts. In such cell-culture systems, BMPs can synergize with Notch signaling by enhancing transcription activation of Notch target genes, such as Hes5, Hey1, Herp2, Hes1, and Hesr1, to inhibit myogenic differentiation of C2C12 myoblasts and to suppress differentiation of neuroepithelial precursor cells (Dahlqvist et al. 2003; Takizawa et al. 2003; de Jong et al. 2004; Itoh et al. 2004; Zamurovic et al. 2004; Nobta et al. 2005). Notch ligands also enhance BMP-induced osteoblast differentiation of C2C12 myoblasts and MC3T3 pre-osteoblasts (Tezuka et al. 2002; Nobta et al. 2005). Dominant negative inhibition of Notch signaling by expressing an extracellular domain of Notch, or downregulation of Notch1 expression using siRNA, impairs BMP-induced osteoblast differentiation (Nobta et al. 2005). In addition, Notch signaling may lead to negative feedback regulation of BMP-induced osteoblast differentiation, because excess BMP signaling or prolonged overexpression of NICD induces Hey1 expression, which then interacts with and inhibits Runx2, thus inhibiting osteoblast differentiation (Zamurovic et al. 2004).
Similar to BMPs, TGF-β can also cooperate with Notch to induce Hes1, Hey1, and Jag1 expression in a Smad3-dependent manner through a Smad3–NICD interaction (Blokzijl et al. 2003; Zavadil et al. 2004). In keratinocytes, NMuMG mammary epithelial cells and primary kidney tubular epithelial cells, Notch signaling is required for TGF-β-induced EMT and cell differentiation (Zavadil et al. 2004), as well as TGF-β-induced cytostasis and expression of TGF-β target genes, including the gene encoding the CDK inhibitor p21CIP1 (Niimi et al. 2007). Similarly, in epithelial ovarian cancer cells, the Notch and TGF-β pathways form a reciprocal regulatory loop that enhances the expression and activities of each other to promote EMT (Zhou et al. 2016). In other cases, however, Notch signaling was found to antagonize TGF-β-induced growth arrest and transcription, and reducing Notch1 expression using siRNA or inhibition of Notch4 signaling using a γ-secretase inhibitor restored TGF-β-induced cytostatic responses (Rao and Kadesch 2003; Masuda et al. 2005; Sun et al. 2005). In one study, NICD blocks TGF-β signaling through sequestration of p300 or CBP away from Smad3 (Masuda et al. 2005). These diverse and sometimes conflicting results may be because of the different cellular contexts in the cell culture model systems used. They may also reflect the complexity of signaling networks that may produce different signaling outcomes depending on other signaling pathways. This complexity highlights the importance of evaluating the signaling outcome in the context of the entire signaling network, as well as the necessity of understanding the mechanistic details of cross talk among different pathways.
Mechanistically, the cross talk between Notch and TGF-β/BMP signaling can occur at multiple levels. TGF-β and Nodal affect the expression of the Notch ligands Delta2 or Jagged1 and the Notch target gene Hey1 in a variety of cell types (Zavadil et al. 2004; Hudson and Yasuo 2006; Hudson et al. 2007). Several Smad proteins, including Smad3 and Smad1 or Smad5 have been shown to directly associate with NICD (Blokzijl et al. 2003; Zavadil et al. 2004) and, through this interaction, Smads are recruited to the regulatory sequences of key Notch target genes to enhance their expression in conjunction with NICD/RBP-Jκ. In some cases, the Smad3–NICD interaction enables synergistic activation of Notch target genes (Blokzijl et al. 2003; Zavadil et al. 2004; Niimi et al. 2007), whereas, in others, Smad3 and NICD antagonize each other, through either sequestration of p300 or CBP away from Smad3 by NICD (Masuda et al. 2005) or direct binding of Notch4 NICD to Smad3 to inhibit its activity (Rao and Kadesch 2003; Masuda et al. 2005; Sun et al. 2005).
Other studies document cross talk between Notch and TGF-β pathways in vivo, during development and tissue regeneration, and reveal further layers of complexity in the signaling modulation. In muscle stem cells (satellite cells), TGF-β and Notch signals antagonize each other to control the regenerative competence of these stem cells. Although TGF-β inhibits satellite cell proliferation and differentiation, through activation of Smad3, Notch signaling enhances regeneration, partially by blocking the binding of Smad3 to its target promoters (Carlson et al. 2008). In embryonic endothelial cells, activin receptor-like kinase 1 (ALK-1) signaling, activated by its high-affinity ligands BMP-9 or BMP-10 and mediated by Smad1, 5, and/or 8, cooperates with Notch signaling to inhibit angiogenesis (Larrivée et al. 2012). Although, in this case, the detailed mechanism has yet to be defined, the BMP-activated Smads could directly bind to regulatory sequences of key Notch target genes, such as Hey1 and Hey2, to activate their expression in a manner independent of canonical Notch activation.
Another mechanism of cross talk between TGF-β and Notch signaling has also been reported, in which the TβRI receptor and Notch signaling may cooperate to promote prostate cancer invasion through a common γ-secretase subunit that can cleave both receptors at the transmembrane domains (Gudey et al. 2014). In prostate cancer cells, TGF-β increases the abundance and activity of presenilin 1 (PS1), a catalytic core component of the γ-secretase complex, through tumor necrosis factor (TNF) receptor-associated factor 6 (TRAF6)-mediated ubiquitylation and activation of PS1. This results in cleavage of TβRI in the transmembrane domain to generate its intracellular domain (ICD), which then translocates into the nucleus and enhances prostate cancer cell invasion. This ICD can interact and colocalize with NICD, and their association promotes cell-invasive behavior.
Taken together, Notch and TGF-β/BMP signaling show frequent cross talk in a variety of cell types and tissues. However, the outcomes and mechanisms of these cross-talk activities vary depending on the cellular context and possibly the activity of other signaling pathways, such as Wnt and Hippo pathways, which are also involved in the regulation of similar physiological and pathological processes.
CROSS TALK WITH HIPPO SIGNALING
The Hippo pathway is evolutionarily conserved from Drosophila to mammals and plays important roles in the regulation of organ size, embryonic development, tumorigenesis, and stem-cell self-renewal (Yu and Guan 2013). The canonical core Hippo kinase complex in mammals comprises two kinases, Mst1 or Mst2 and Lats1 or Lats2. The Mst kinase forms a complex with the Sav1 adaptor protein to phosphorylate and activate the Lats kinase. The activated Lats kinase, in association with the tumor-suppressor Mob, then phosphorylates and inhibits the transcription coactivators TAZ and/or YAP (Dong et al. 2007; Zhao et al. 2007; Hao et al. 2008; Lei et al. 2008; Oka et al. 2008). TAZ and YAP do not bind to DNA directly, but can be recruited to specific target promoter sequences through binding to the TEAD transcription factors (Wu et al. 2008; Zhang et al. 2008, 2009), and regulate the expression of genes that are essential for proliferation, apoptosis, EMT, and various other developmental processes (Ota and Sasaki 2008; Zhao et al. 2008, 2009; Zhang et al. 2009; Lian et al. 2010). TAZ and YAP can be phosphorylated at multiple sites and inhibited by the Lats kinases (Wang et al. 2009b). In particular, phosphorylation of TAZ at Ser89 (equivalent to YAP Ser127) allows its binding to 14-3-3, leading to cytoplasmic sequestration (Dong et al. 2007; Hao et al. 2008; Lei et al. 2008; Zhao et al. 2008), and phosphorylation at Ser311 primes it to be further phosphorylated by CK1ɛ at Ser314, which mediates binding to the F-box containing E3 ubiquitin ligase β-TrCP, leading to subsequent ubiquitylation and degradation of TAZ (Liu et al. 2010). The activities of YAP and TAZ can be regulated by extracellular diffusible signals and growth factors, as well as signals generated by cell–cell junctions, cell density and polarity, tissue architecture, and mechanotransduction (Dupont et al. 2011; Aragona et al. 2013; Park and Guan 2013). These signals include ligands that bind to G-protein-coupled receptors, such as those for lysophosphatidic acid, thrombin, and epinephrine, which have been shown to promote or inhibit nuclear translocation of YAP and TAZ, growth factors such as epidermal growth factor (EGF), insulin-like growth factor (IGF), or Wnt, and signals sensing cell–cell adhesion, junctional structures, polarity, and matrix stiffness. Various polarity complexes, including Crumbs, Scribble, and cadherin complexes, have been shown to regulate Hippo signaling and the localization and stability of YAP and TAZ (Piccolo et al. 2014).
The first reported cross talk between TGF-β and Hippo signaling involves binding of YAP to Smad7, leading to enhanced inhibition of TGF-β signaling (Ferrigno et al. 2002). YAP and TAZ also bind to other Smad proteins and participate in the regulation of BMP or TGF-β signaling through distinct mechanisms. In Drosophila, the YAP homolog Yorkie binds to Mad, the homolog of BMP-activated Smads, and promotes Mad-dependent transcription (Alarcón et al. 2009; Oh and Irvine 2011). In mammalian cells, YAP can bind effectively to the PPxY motif in Smad1 through its two WW domains, and this binding is further strengthened by the phosphorylation of the Smad1 linker region by CDK9 (Alarcón et al. 2009). The YAP–Smad1 binding supports Smad1-dependent transcription, and is required for the suppression of neural differentiation of mESCs by BMP. In contrast, TAZ does not bind well to Smad1, possibly because of the presence of only one WW domain in TAZ, and may only affect TGF-β but not BMP signaling. In response to TGF-β signaling, TAZ and YAP were shown to associate with heteromeric Smad2/3/4 complexes and dictate their intracellular localization (Varelas et al. 2008). By binding to the Crumbs polarity complex, which promotes the phosphorylation, cytoplasmic localization, and inhibition of YAP and TAZ, TAZ and YAP also block nuclear localization of Smad2 and Smad3, and control the cell density inhibition of TGF-β/Smad signaling in murine EpH4 mammary epithelial cells (Varelas et al. 2008). However, another study indicates that most cell types show functional TGF-β signaling under both high and low cell-density culture conditions, and that Smad nuclear localization in response to TGF-β occurs independent of the YAP or TAZ levels (Nallet-Staub et al. 2015). These data suggest that the inhibition of TGF-β signaling by cell density is limited to polarized epithelial cells and largely reflects the polarized distribution of the TGF-β receptors and not the levels of YAP or TAZ (Nallet-Staub et al. 2015).
YAP or TAZ were shown to participate with Smads in the same transcription complexes at promoters of target genes. In hESCs, a regulatory transcription complex consisting of TAZ or YAP, TEADs, and Smad2 or Smad3, as well as the pluripotent factor Oct4 was identified, and may function in a switch-like manner to regulate the maintenance of pluripotency and cell-fate specification in conjunction with other transcription factors (Beyer et al. 2013). TGF-β and Hippo signaling also converge at the level of transcription regulation of common target genes, such as the gene-encoding connective tissue growth factor (CTGF). In mesothelioma cells with mutations in the Hippo pathway, a YAP-TEAD4-Smad3-p300 complex forms at the CTGF promoter to activate its expression, leading to malignant progression of mesothelioma (Fujii et al. 2012). Such cooperation between YAP or TAZ and Smads in transcription complexes also extends to Drosophila. In Drosophila, the BMP ligand Dpp and the Fat-Hippo pathway synergize to promote growth, and this cross talk is mediated by a direct interaction between Mad and Yorkie, which forms a transcription activating complex at the promoter of the gene for bantam microRNA to promote its expression (Oh and Irvine 2011). YAP and Smad2 or Smad3 also antagonize each other’s activities during endodermal differentiation of hESCs. Although YAP suppresses the transcription elongation of mesendodermal lineage genes by promoting the binding of negative transcription elongation factors to the regulatory region of these genes, activin, acting through Smad2 and/or Smad3, acts in concert with Wnt-3a-β-catenin signaling to counteract YAP and promotes the expression of these genes (Estaras et al. 2015).
In addition to the direct cross talk between YAP or TAZ and Smads, the negative regulators of TGF-β/Smad signaling, Ski and SnoN, also affect the stability and transcription activity of YAP and TAZ by directly binding to components of the Hippo core kinase complex, and modifying the kinase activity of Lats2 and the phosphorylation of YAP and TAZ. In particular, Ski binds to Lats2, Sav, NF2, and Mob, and increases the affinity of the Lats2–Sav interaction to enhance the kinase activity of Lats2, leading to increased phosphorylation of TAZ and YAP, and their cytoplasmic accumulation and degradation (Rashidian et al. 2015). In addition, Ski also induces TAZ degradation and suppresses its biological activity in a Lats2-independent manner. Consistent with these findings, Ski inhibits TAZ-induced transformation and EMT of human breast cancer cells in vitro and metastasis in xenograft mouse models in vivo (Rashidian et al. 2015). Interestingly, the ability of Ski to block TAZ and YAP signaling is independent of its ability to antagonize the Smad proteins (Rashidian et al. 2015).
SnoN also interacts with the Hippo kinase complex but, different from Ski, this interaction involves different components of the Hippo complex and, more importantly, results in different outcomes (Zhu et al. 2016). SnoN binds strongly to Lats2 and Sav, weakly to Mst2, but not to Mob or TAZ, and these interactions prevent the binding of Lats2 to TAZ and the phosphorylation of TAZ, leading to TAZ stabilization. Consistent with this, SnoN enhances the transcriptional and oncogenic activities of TAZ, and reducing SnoN decreases TAZ expression as well as malignant progression of breast cancer cells. The intracellular localization and expression levels of SnoN itself are sensitive to cell density and are regulated by the cell polarity complex–associated Hippo kinases. SnoN is localized to the basolateral domain in polarized epithelia and forms a complex with the Scribble polarity protein and its associated Lats2 kinase. The Lats2 kinase that is activated by the Scribble complex can induce downregulation of SnoN and TAZ expression, thereby suppressing the proliferative potential of epithelial cells (Zhu et al. 2016). Thus, SnoN is a critical component of the Hippo regulatory network that receives signals from the tissue architecture and polarity to coordinate the activity of intracellular signaling pathways. As our understanding of the Hippo pathway deepens and new components are identified, more modes of cross talk between TGF-β family signaling and Hippo signaling will be revealed to coordinate various biological processes.
CROSS TALK WITH HEDGEHOG SIGNALING
The Hh signaling pathway is evolutionarily conserved and is required for embryonic patterning, tissue repair, and regeneration. It also plays an important role in tumorigenesis, as mutations in Hh pathway components that cause constitutive activation of the pathway have been identified in several types of human cancer, such as basal cell carcinomas (BCCs) and medulloblastoma. Hh signaling is controlled by two cell-surface transmembrane proteins, the Patched receptor (PTCH1 or PTCH2) and the 7-membrane-spanning receptor-like protein Smoothened (SMO), and is intracellularly mediated by Gli (glioma-associated oncogene homolog) proteins of the Krüppel family of zinc finger transcription factors. In the absence of ligand, PTCH1 and PTCH2 repress the activity of SMO. This results in the phosphorylation of Gli by several protein kinases, including protein kinase A (PKA), GSK-3β, and CK1, and subsequent proteasome-mediated cleavage of Gli into amino-terminal truncated forms that act as repressors of Hh target genes (Hui and Angers 2011). Binding of Hh ligand abolishes the inhibition of SMO by PTCH, leading to the activation and translocation of Gli proteins into the nucleus to control the expression of Hh target genes.
During embryonic development and oncogenesis, TGF-β/BMP signaling often regulates the expression of Hh ligands and pathway components, and Hh/Gli can also induce the expression of TGF-β or BMP proteins, sometimes in a tissue- or cell-type-specific manner to regulate lineage-specific development (Perrot et al. 2013). More often, TGF-β can directly regulate the expression of Gli proteins, and Gli may mediate some TGF-β responses independent of Hh signaling. In the developing cerebellum, BMP-2 and BMP-4 antagonize the proliferative function of Sonic Hedgehog (Shh) by downregulating SMO and Gli1 expression (Rios et al. 2004). TGF-β has been shown to inhibit PKA activity, while concomitantly inducing Gli2 and Gli1 expression (Perrot et al. 2013). The Gli2 gene is a direct transcription target of the TGF-β/Smad pathway in a variety of cell types, including keratinocytes and fibroblasts, and a number of cancer cells such as melanoma. In several mouse breast cancer metastasis models, including an intracardiac tumor inoculation bone metastasis model and in progression from ductal carcinoma in situ (DCIS) to invasive carcinoma, TGF-β induces the expression of Gli2 and, subsequently, Gli1, independent of Hh signaling, to promote bone metastasis (Hui and Angers 2011; Johnson et al. 2011). Activation of Gli2 transcription by TGF-β involves the actions of both Smad3 and β-catenin (Dennler et al. 2009). In response to TGF-β, Smad3 and β-catenin are recruited to distinct elements in the Gli2 regulatory gene sequences to induce its expression. This activation of Gli2 expression is not blocked by cyclopamine, an inhibitor of SMO, suggesting that TGF-β-induced Gli2 expression occurs independent of the SMO/Hh pathway (Dennler et al. 2007, 2009). Consistent with this ability of TGF-β/Smad signaling to directly activate Gli2 expression, high Gli2 levels are detected in many malignant tumor cells, including melanoma, breast cancer, glioblastoma, and ovarian cancer, that also express high levels of TGF-β (Edson et al. 2010; Steg et al. 2012). High Gli2 expression is associated with loss of E-cadherin expression and increased tumor cell invasion, suggesting that high Gli2 and Gli1 levels in melanoma and breast cancer cells may mediate TGF-β-induced EMT and tumor progression. Silencing Gli2 expression or pharmacological inhibition of the TβRI kinase both result in inhibition of bone metastasis and downregulation of prometastatic genes, encoding PTHrP, interleukin-11, CXCR4, and osteopontin (Javelaud et al. 2011, 2012; Mohammad et al. 2011).
A more direct mode of cross talk is mediated by functional interaction between Gli proteins and Smads at common target promoter sequences. In zebrafish embryos, the eng2a promoter integrates repressive signals from BMPs and activating signals from Hh, and the cross talk between the two pathways defines the spatial pattern of eng2a gene expression. In this case, Gli2 and Smad1 both bind to eng2a regulatory sequences to modulate its expression (Maurya et al. 2011). Gli1 can also function as an effector of TGF-β signaling in pancreatic cancer cell lines to promote cell survival. In the presence of TGF-β, Gli1 forms a complex with Smad2 and Smad4 at BCL2 promoter sequences to stimulate its expression, leading to cell survival (Nye et al. 2014).
Thus, in malignant human cancer cells, expression of Gli proteins is often induced by TGF-β signaling, and they in turn mediate the tumor-promoting activity of TGF-β by forming transcription complexes with the Smad proteins. Whether this regulatory cooperation model also operates in untransformed cells or during normal tissue development and homeostasis is yet to be determined. More investigation in this area is clearly needed.
CROSS TALK WITH MAP KINASE PATHWAYS
The TGF-β/Smad and MAP kinase pathways are functional in most if not all cell types at all stages of development and during disease development and progression. TGF-β and BMPs can directly activate the Erk, c-Jun amino-terminal kinase (JNK), and p38 MAP kinase pathways independent of Smad proteins to regulate cell motility, EMT, cell differentiation, and survival. TGF-β has also been shown to indirectly upregulate the Erk and p38 MAP kinase activity by inducing the expression of ligands or receptors that activate these pathways (Vinals and Pouysségur 2001; Takekawa et al. 2002). This section focuses on the cross talk and mutual regulation between the Smad pathway and the Erk, JNK, and p38 MAPK pathways.
Cooperative Interactions between Erk MAPK and Smad Signaling
In mammals, the Smad and MAPK pathways are often critical components of the same signaling network that is essential for most cellular processes (Guo and Wang 2009). In mESCs, BMPs cooperate with LIF, which signals through the Erk MAPK pathway to maintain the pluripotency state. When both BMP and LIF signaling are activated, BMP suppresses neuroectoderm differentiation, whereas LIF signaling inhibits the differentiation to mesoderm and endoderm lineages (Ying et al. 2003). Erk MAPK activity is critical in mouse ES cell-fate determination—low Erk activity is required for ES cell self-renewal (Burdon et al. 1999; Kunath et al. 2007; Stavridis et al. 2007) and high Erk activity induces differentiation (Yoshida-Koide et al. 2004). Although LIF signaling can directly activate the Erk activity through the gp130 subunit of the LIF receptor (Fukada et al. 1996; Burdon et al. 1999), BMP-4 can attenuate Erk activation by upregulating the expression of an Erk phosphatase, Dusp9, thereby maintaining a properly balanced Erk activity to ensure self-renewal of mESCs (Li et al. 2012). In hESCs, BMPs act together with FGF2, which signals through the Erk MAPK pathway (Yu et al. 2011) to drive mesendoderm differentiation. Thus, multiple signaling inputs, often involving Smad signaling and the Erk MAPK pathway, regulate stem-cell fate.
Mechanistically, the two pathways often directly interact and mutually regulate the activities or expression of each other. In cancer cells, activation of TGF-β signaling and the HER2-Ras-Erk MAPK pathway often leads to the production and/or secretion of additional growth factors and cytokines. TGF-β can induce the expression of platelet-derived growth factor, which activates the Erk MAPK pathway, whereas Erk MAPK can promote the expression of TGF-β1 or Smad3, resulting in enhanced activation of both pathways. At the intracellular signaling and transcription level, the Erk MAPK and Smad pathways often associate with each other, and these interactions can result in either elevation or inhibition of Smad activity by Erk MAPK, depending on the specific target genes and cell types. A well-defined cooperative cross talk occurs at regulatory DNA sequences of TGF-β target genes, many of which contain tissue plasminogen activator (TPA)-responsive elements (TREs) that can be bound by AP-1 transcription factors or bipartite TRE-SBE (Smad-binding element) sequences. Smad3/4 complexes, either by themselves or in conjunction with AP-1, can bind to some of these TREs to mediate TGF-β responses (Yingling et al. 1997; Attisano and Wrana 2000). On activation, Erk MAPK can phosphorylate AP-1, which then binds to TRE sequences or physically interacts with Smads to mediate synergistic activation of TGF-β-responsive promoters with bipartite TRE-SBE sequences (Zhang et al. 1998; Liberati et al. 1999; Wong et al. 1999). Furthermore, activation of MEK1 also induces Smad3 transcription, thereby enhancing Smad3 signaling in epithelial and smooth muscle cells (Ross et al. 2007). The cooperation between the Erk MAPK and Smad pathways has been well documented in many physiological and pathological processes, including tooth and palate development (Xu et al. 2008), TGF-β-induced autophagy (Kiyono et al. 2009) and progression of aortic diseases (Holm et al. 2011).
Downregulation of TGF-β Signaling by Erk MAPK Pathway
In addition to cooperative interactions between Erk MAPK signaling and Smads at the level of target gene transcription, the two pathways also directly modify the activities of each other in the cytoplasm (Fig. 2). In human cancer cells, constitutively active Ras-Erk MAPK can antagonize TGF-β-induced apoptosis and cell-cycle arrest to promote proliferation, while allowing for promigratory and proinvasive functions of TGF-β. The inhibition of TGF-β/Smad signaling by Ras-Erk MAPK signaling can occur through several mechanisms. Erk MAPK signaling can downregulate TGF-β signaling by inducing cleavage of cell-surface TβRI (Liu et al. 2009). This shedding of ectodomain of TβRI is mediated by TACE/ADAM17, which is activated by Erk MAPK. This regulation occurs in both untransformed cells and in cancer cells, and functions to attenuate all TβRI-dependent cellular processes. In addition, Erk MAPK can directly phosphorylate the linker region of various Smad proteins to alter their subcellular localization and inhibit their transcription activity. The Smad linker region is highly flexible in structure and contains many serines and threonines in the context of proline residues, favoring phosphorylation by MAPKs and GSK-3β. In human cancer cells, Erk MAPK activated by oncogenic Ras phosphorylates Smad3 on at least three residues, Thr178, Ser203, and Ser207, both in vitro and in vivo. Erk-mediated phosphorylation of these sites inhibits Smad3 transcriptional activity (Matsuura et al. 2005) and nuclear localization (Kretzschmar et al. 1997).
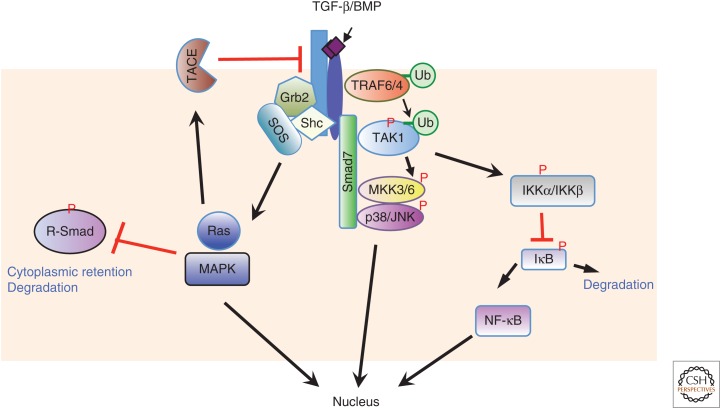
Figure 2.
Transforming growth factor β (TGF-β) cross talks to NF-κB and mitogen-activated protein (MAP) kinase pathways. TGF-β receptors activate p38 and c-Jun amino-terminal kinase (JNK) MAPK pathways and NF-κB pathways through receptor-associated factor (TRAF4) or TRAF6 and TGF-β-activated kinase 1 (TAK1), and Ras-Erk MAPK through Grb2 and Shc. The activated Ras-Erk MAPK and p38 pathways can trigger activation of TACE, leading to cleavage of TβRI, and phosphorylation of Smads in the linker region, sequestering it in the cytoplasm and promoting its degradation.
Erk MAPKs also antagonize BMP function by phosphorylating the linker regions of Smad1 at multiple Ser or Thr residues, and this phosphorylation primes them for further phosphorylation by GSK-3β. These sequential phosphorylation events create a docking site for the Smurf1 E3 ubiquitin ligase that targets Smad1 for polyubiquitylation and degradation. The binding of Smurf1 also blocks the interaction of Smad1 with the nuclear pore complex (Sapkota et al. 2007). As a result of this reduced Smad1 nuclear translocation and expression, BMP signaling can be effectively suppressed by growth factors that activate Erk MAPK signaling, including EGF, FGF, and IGF (Kretzschmar et al. 1997; Sapkota et al. 2007; Eivers et al. 2009). Indeed, FGF-induced Erk MAPK activation relieves BMP repression to induce neural differentiation of Xenopus embryonic cells and rat neural precursor cells (Kuroda et al. 2005; Bilican et al. 2008).
In addition to the linker regions of R-Smads, MAPKs also phosphorylate and regulate the expression levels of Smad4 and Smad7. MEK-Erk MAPK signaling activated by oncogenic Ras results in phosphorylation of Smad4 and decreases its protein stability (Saha et al. 2001). JNK and p38 MAPK have been found to preferentially phosphorylate tumor-derived mutant Smad4 to promote its degradation (Liang et al. 2004). Finally, Erk, JNK, and p38 MAPK have all been implicated in activating the expression of the Smad7 gene (Brodin et al. 2000; Uchida et al. 2001; Dowdy et al. 2003).
Activation of MAPK Signaling by TGF-β
TGF-β is a potent activator of the Erk MAPK pathway through Smad-independent mechanisms. First, the TGF-β receptors can be phosphorylated on tyrosine residues. The TβRII cytoplasmic domain is autophosphorylated on three tyrosines (Lawler et al. 1997). In a manner analogous to receptor tyrosine kinase activation, these phosphorylated tyrosine residues in TβRII create docking sites for the recruitment of SH2 domain proteins. Src-mediated phosphorylation of TβRII on Tyr284 results in the recruitment of Grb2 and Shc, leading to p38 MAPK activation (Galliher and Schiemann 2007). Similarly, activated TβRI contains an intrinsic tyrosine kinase activity, in addition to the well-characterized Ser-Thr kinase activity, and can phosphorylate Shc directly on tyrosine and serine residues (Lee et al. 2007). Phosphorylated Shc associates with TβRI and recruits Grb2 and SOS, leading to activation of Ras-Erk MAPK signaling (Fig. 2).
Through its Smad-independent signaling, TβRI activates TAK1 (Sorrentino et al. 2008; Yamashita et al. 2008; Mu et al. 2012), a MAPK kinase kinase (MAPKKK) family member known to be an important activator of the p38 MAPK pathway (Fig. 2) (Yamaguchi et al. 1995). This activation is mediated by TRAF6, which was initially identified as an adaptor protein that activates NF-κB signaling in response to interleukin-1 (Cao et al. 1996; Ishida et al. 1996) and TRAF4, which is differentially expressed in metastatic breast cancer (Regnier et al. 1995). Both TRAF6 and TRAF4 contain RING-domain E3 ubiquitin ligase activity. On TGF-β stimulation, TRAF6 associates with TβRI at a conserved consensus motif (basic residue-X-P-X-E-X-X aromatic/acidic residue), leading to autoubiquitylation of TRAF6 and subsequent Lys63-linked polyubiquitylation of TAK1 (Sorrentino et al. 2008). TRAF4 also associates with the activated TGF-β receptor complex and stabilizes TβRI by antagonizing Smurf2-mediated TβRI degradation. Similar to TRAF6, this association of TRAF4 with TβRI also promotes Lys63-linked autoubiquitylation of TRAF4 as well as polyubiquitylation of TAK1 (Zhang et al. 2013). The Lys63-linked TAK1 polyubiquitylation causes its activation through either a conformational change or recruitment of the TAK1-binding proteins 2 and 3 (TAB2 and TAB3) (Xia et al. 2009). Once activated, TAK1 functions as a MAPKKK to stimulate activation of MKK3 and/or MKK6, leading to p38 MAPK activation (Sorrentino et al. 2008). TAK1 also phosphorylates IκB-kinase α (IKKα) to activate NF-κB signaling (Wang et al. 2001). Through these pathways, TGF-β-induced activation of TAK1 and the p38 MAPK and JNK MAPK pathways has been implicated in the regulation of apoptosis, cell migration, and EMT (Adhikari et al. 2007; Sorrentino et al. 2008; Yamashita et al. 2008; Heldin et al. 2009; Landström 2010; Zhang et al. 2013).
These noncanonical TGF-β-induced TAK1-p38 MAPK or JNK pathways can additionally be regulated in positive or negative manners by the inhibitory Smads (Fig. 2). Smad7 associates with TAK1, MKK3, and p38 MAPK to facilitate activation of the TAK1-p38 MAPK pathway in human prostate cancer cells, leading to apoptosis (Edlund et al. 2003). In contrast, Smad6 inhibits TGF-β1-induced activation of TRAF6-TAK1-p38 MAPK and/or -JNK signaling by recruiting the A20 deubiquitylase to abolish Lys63-linked polyubiquitination of TRAF6 (Jung et al. 2013).
Given the complexity and multiple levels of the mutual regulation, the net outcome of the Erk MAPK pathway cross talk with TGF-β signaling is highly complex depending on the cellular context and influences of other signaling inputs.
CROSS TALK BETWEEN Smad PROTEINS AND THE PI3K-Akt PATHWAY
The PI3K-Akt pathway regulates diverse cellular responses, including glucose homeostasis, cell proliferation and growth, motility, and survival. On activation by a variety of extracellular stimuli, PI3K generates 3′-phosphoinositides (PI(3,4)P2 and PI(3,4,5)P3) that recruit target proteins with lipid-binding domains to the plasma membrane. The Ser-Thr kinase Akt/protein kinase B (PKB) is an important downstream effector of PI3K and initiates a kinase cascade that plays a critical role in the regulation of cell survival (Downward 2004). Akt contains a pleckstrin homology (PH) domain at its amino terminus that mediates interaction with the 3′-phosphoinositides, leading to its translocation to the cell membrane where it is subsequently phosphorylated at two key residues, Thr308 and Ser473. Plasma membrane localization and phosphorylation are both required for optimal activation of Akt. Activated Akt has been shown to phosphorylate important proteins in the apoptotic machinery, including Forkhead box O (FOXO) transcription factors, Bax and Bad, as well as IKK and Mdm2, to modulate cell proliferation and survival. This pathway is negatively regulated by the lipid phosphatases PTEN and SH2-containing inositol 5′-phosphatase (SHIP). PTEN and SHIP dephosphorylate PI(3,4,5)P3 and reverse the action of PI3K (Rohrschneider et al. 2000).
Several targets of Akt play important roles in the regulation of cellular metabolism and protein synthesis, including mammalian target of rapamycin (mTOR) and GSK-3β. mTOR is a large Ser-Thr kinase that can be found in two complexes, mTOR complex 1 (mTORC1) and mTORC2. mTORC1 consists of mTOR, Raptor, mLST8, and PRAS40 and, in response to activation by Akt, phosphorylates S6 kinase 1 (S6K1) and eukaryotic initiation factor 4E-binding protein 1 (4E-BP1) to increase protein translation and synthesis. mTORC2 is composed of mTOR, Rictor, mSin1, and mLST8 and can phosphorylate Akt on Ser473, an event required for full activation of Akt (Bozulic and Hemmings 2009; Zoncu et al. 2011). The biological functions of mTORC2 are less defined.
Extensive cross talk between TGF-β and PI3K pathways has been reported for various cell types including stem cells and cancer cells. The cross talk activities are often complex and can result in mutual activation or inhibition dependent on the cellular context and biological processes involved. In hESCs, activin-induced Smad2 and/or Smad3 signaling can modulate cell-fate decisions depending on the status of PI3K activation. In the presence of robust PI3K signals, Smad2 and Smad3 activate the expression of the pluripotency gene Nanog to maintain self-renewal. However, low PI3K activity switches Smad2/3 signaling to direct mesendoderm differentiation (Singh et al. 2012). The mechanism underlying this switch appears to involve Erk MAPK and Wnt signaling. Activation of mesendoderm gene expression requires the activities of Smad2 and Smad3 as well as β-catenin. When PI3K activity is low, β-catenin can bind to regulatory DNA sequences of lineage-specific genes and, together with Smad2 and/or Smad3, activates their expression to induce differentiation. High PI3K activity inhibits Erk MAPK signaling to promote GSK-3β activation, leading to inhibition of β-catenin. Under this condition, Smad2 and Smad3 signaling activates Nanog expression, but is not sufficient to activate mesendoderm gene expression, thereby promoting self-renewal.
TGF-β/Smad signaling inhibits cell proliferation in epithelial and lymphoid cells and can induce apoptosis in resting B cells and hepatocytes. PI3K-Akt signaling has been shown to antagonize the proapoptotic and cytostatic activity of TGF-β/Smad signaling to promote survival through both Akt kinase-dependent and -independent mechanisms. Akt was shown to directly bind and sequester Smad3 in the cytosol, and thus prevent Smad3-dependent growth inhibition and apoptosis in hepatocytes (Conery et al. 2004; Remy et al. 2004). Additionally, Akt phosphorylates FOXO and prevents its nuclear localization, and formation of the FOXO–Smad complex required for expression of p15INK4B and p21CIP1, effectively blocking the cytostatic responses of TGF-β (Seoane et al. 2004). This ability of the PI3K-Akt pathway to inhibit the cytostatic activity of Smad signaling may play an important role in the switch of TGF-β signaling from a tumor-suppressor pathway to a tumor-promoting activity at late stages of tumorigenesis.
In cancer cells, the PI3K-Akt pathway cooperates with TGF-β or BMP to regulate EMT, cell migration, tumor metastasis, and cell differentiation. In a number of cell types, including fibroblasts, keratinocytes, and hepatic stellate cells, the PI3K-Akt pathway is an important mediator of TGF-β-induced activation of various EMT responses (Asano et al. 2004; Jeong and Kim 2004; Lechuga et al. 2004), and inhibition of PI3K or Akt by pharmacological inhibitors or dominant negative mutants block TGF-β-induced transcription of target promoters, EMT, and cell migration as well as BMP-induced osteoblast differentiation (Ghosh-Choudhury et al. 2002). TGF-β signaling can activate the PI3K-Akt pathway either directly or indirectly. In keratinocytes and mammary epithelial cells, stimulation with TGF-β results in phosphorylation of Akt at Ser473 and activation of its kinase activity. This activation appears to be Smad-independent and may be mediated by a RhoA-dependent mechanism (Bakin et al. 2000). The integrin-linked kinase (ILK) has also been reported to be involved in Akt activation by TGF-β (Lee et al. 2004). Activated Akt enhances Smad3 transcriptional activity to induce collagen I expression in human mesangial cells (Runyan et al. 2004). In this case, activation of the PI3K-Akt pathway alone is not sufficient to increase gene expression, and its ability to phosphorylate Smad3 on residues outside the carboxy-terminal region is necessary for optimal activation of Smad3. A similar enhancement of Smad1- and/or Smad5-mediated transcription activation by PI3K-Akt signaling has also been reported for BMP-induced colony-stimulating factor 1 (CSF-1) expression during osteoclast differentiation (Mandal et al. 2009).
In neurons and fibroblasts, TGF-β can activate PI3K indirectly, for example, by inducing the expression of secreted growth factors (Vinals and Pouysségur 2001; Horowitz et al. 2004) to promote cell proliferation and survival (Zhu et al. 2001, 2004; Horowitz et al. 2004; Wilkes et al. 2005). TGF-β can also indirectly activate PI3K by inducing the expression of several microRNAs. In hepatoma cells and glomerular mesangial cells, TGF-β can induce the expression of miR-216a/217 and miR-21, leading to enhanced EMT, expanded stem-cell population, and metastasis of hepatoma (Kato et al. 2009; Xia et al. 2013). miR-216a/217 is a negative regulator of Smad7 and PTEN. By inhibiting Smad7 and PTEN expression, high levels of miR-216a/217 can promote TGF-β signaling and PI3K-Akt pathway activation (Kato et al. 2009; Xia et al. 2013). miR-21 also targets PTEN to promote mesangial cell hypertrophy and matrix protein synthesis through an Akt -mTORC1 pathway (Dey et al. 2012). Finally, both TGF-β and BMP signaling have been reported to regulate the transcription or protein levels of PTEN in a number of cancer or cell types. None of these modes of regulation appear to be direct and, therefore, are likely to involve signaling pathways other than Smads (Guo and Wang 2009).
During EMT, TGF-β induces increased cell size and protein content (Lamouille and Derynck 2007; Lamouille et al. 2012). This process is mediated by mTORC1, which activates translation initiation to increase protein content. The completion of TGF-β-induced EMT also requires activation of mTORC2, which promotes cell migration and invasion. Furthermore, activation of mTORC1 and mTORC2 in response to TGF-β is mediated by the PI3K-Akt pathway. The TGF-β receptors can form an indirect complex with the p85 regulatory subunit of PI3K, resulting in its activation. This, in turn, leads to activation of Akt and formation and activation of mTORC1 and mTORC2.
The PI3K-Akt pathway can also directly enhance the stability of TβRI, by regulating the activity of the deubiquitylating enzyme, ubiquitin-specific protease 4 (USP4) (Zhang et al. 2012). USP4 is activated by Akt phosphorylation at a conserved Ser445, resulting in its translocation from the nucleus to the plasma membrane, where USP4, together with USP11 or USP15, binds directly to TβRI, leading to its deubiquitylation and stabilization at the plasma membrane. Thus, by activating USP4, the PI3K-Akt pathway enhances TGF-β signaling to promote EMT in breast cancer cells. Additionally, activation of Akt leads to inhibition of GSK-3β, which promotes Smad3 polyubiquitylation and degradation (Lim et al. 2012). Inactivation of GSK-3β leads to Smad3 stabilization and enhanced TGF-β signaling. The PI3K-Akt pathway could also induce phosphorylation of Smad3 at residues preceding the carboxy-terminal region, leading to increased transcription activity of Smad3, thereby enhancing TGF-β signaling (Runyan et al. 2004).
CROSS TALK BETWEEN THE SMADs AND THE JAK-STAT PATHWAYS
The JAK-STAT pathways are activated by cytokines and growth factors to regulate cell growth, differentiation, and survival. Without stimulation, latent STAT proteins exist as monomers or nonphosphorylated N-domain-mediated dimers and shuttle between the cytoplasm and nucleus. On stimulation by ligands, the cytoplasmic JAK kinases are activated by tyrosine phosphorylation and dimerization. The activated JAK kinases subsequently phosphorylate STAT proteins on tyrosine residues, allowing the formation of active SH2-mediated dimers. These phosphorylated STAT dimers are retained in the nucleus and bind regulatory gene sequences containing an interferon-γ-activated sequence (GAS) consensus recognition motif to activate transcription. In mammals, the STAT family consists of seven members (STAT1, 2, 3, 4, 5a, 5b, and 6) that mediate signaling in response to a diverse array of extracellular ligands (Li 2008).
Both Smads and STATs are intimately involved in pluripotency and differentiation transcription programs, and often combine in the same transcription complexes (Chen et al. 2008). A direct cross talk between STAT3 and Smad1 is required for BMP-2- and LIF-induced differentiation of primary fetal neural progenitor cells into astrocytes (Nakashima et al. 1999). STAT3, activated by JAK kinase in response to LIF, binds to the amino-terminal region of the transcription coactivators p300, whereas Smad1, activated by BMP-2, interacts with the carboxy-terminal region of p300. This complex of Smad1 and STAT3 bridged by p300 at the promoter is required for the astrocyte-specific activation of glial fibrillary acidic protein (GFAP) expression. During tumorigenesis, STAT3 was shown to directly bind Smad3 and to block its ability to bind to DNA and form a complex with Smad4, attenuating the activity of TGF-β in inducing cell-cycle arrest and promoting EMT (Wang et al. 2015).
Additional studies show that TGF-β can regulate JAK-STAT signaling either in a positive or negative manner, depending on the cell type. In T lymphocytes, TGF-β inhibits interleukin-12-induced activation of JAK2 and subsequent phosphorylation and activation of STAT3 and STAT4 (Bright and Sriram 1998; Pardoux et al. 1999). In the liver, the activated hepatic stellate cells produce CTGF in response to TGF-β to promote liver fibrosis, and this process can be mediated by STAT3 (Liu et al. 2013). STAT3 activation in response to TGF-β requires the TβRI receptor, but is independent of the Smad proteins. Instead, this activation depends on the PI3K and MAPK pathways, and is mediated by the JAK1 kinase that induces STAT3 phosphorylation and activation. Thus, STAT3 could act as a downstream effector of TGF-β signaling in hepatic stellate cells. The JAK-STAT pathway can also indirectly regulate Smad3 activity by enhancing the expression of the inhibitory Smad7. In a human fibrosarcoma-derived cell line, interferon-γ, acting through JAK1 and STAT1, induces expression of Smad7, which then inhibits phosphorylation and activation of Smad3 (Ulloa et al. 1999).
In the mammary gland, prolactin, signaling through JAK2 and STAT5, enables alveologenesis and lactation. TGF-β inhibits prolactin signaling to block mammary epithelial proliferation and differentiation. In response to TGF-β, the Smad2/3/4 complex inhibits the transcription activity of STAT5 by blocking its interaction with the transcription coactivator CBP, leading to inhibition of mammary gland differentiation and lactation (Cocolakis et al. 2008). This may be one of the mechanisms by which TGF-β suppresses STAT5 activity in the mammary gland. Interestingly, TGF-β expression is known to peak in mid-pregnancy, but the inhibitory activity of TGF-β must be suppressed to allow alveologenesis and lactation. This suppression is achieved by SnoN, a potent negative regulator of the Smad proteins (Jahchan et al. 2012). SnoN expression is transiently and sharply elevated at the end of pregnancy, and elevated SnoN promotes STAT5 signaling by enhancing its stability, thereby sharply increasing the activity of prolactin signaling at the onset of lactation. SnoN−/– mice display severe defects in alveologenesis and lactogenesis, and mammary epithelial cells from these mice fail to undergo proper morphogenesis. These defects can be rescued by an active STAT5. Thus, SnoN enables cross talk to coordinate TGF-β and prolactin signaling to regulate alveologenesis and lactogenesis.
CROSS TALK WITH NF-κB/IKK SIGNALING
The NF-κB/Rel family comprises NF-κB1 (p50/p105), NF-κB2 (p52/p100), RelA (p65), c-Rel, and RelB, which function as dimeric transcription factors. NF-κB was originally identified as an important transcription factor that mediates various immune and inflammatory responses. Subsequently, NF-κB signaling was found to contribute to a broad range of biological processes, including cell adhesion, differentiation, proliferation, autophagy, senescence, and cell survival. Deregulated NF-κB activity is apparent in a number of diseases, including cancer, arthritis, chronic inflammation, asthma, neurodegenerative diseases, and heart disease (Hinz and Scheidereit 2013). In the absence of stimulating signals, NF-κB dimers are sequestered in the cytoplasm by binding to the inhibitory IκB proteins. In the canonical pathway, proinflammatory cytokines, growth factors, and antigen receptors activate an IKK complex consisting of IKKα, IKKβ, and NF-κB essential modulator (NEMO), and the activated IKK complex phosphorylates IκB at critical serine residues, leading to its ubiquitylation by the E3 ubiquitin ligase SCFβTrCP and proteasomal degradation. The freed NF-κB/Rel complexes are further activated by phosphorylation and translocate into the nucleus where they induce target gene expression. This canonical signaling pathway strictly depends on NEMO, whereas the two catalytic subunits (IKKα, IKKβ) may be more redundant (Hinz and Scheidereit 2013).
The noncanonical pathway is activated by a specific group of receptors, such as the receptors for the TNF family members lymphotoxin-α/β or CD40L, and induces stabilization and activation of NF-κB interacting kinase (NIK). NIK then phosphorylates IKKα, which, in turn, phosphorylates the carboxy-terminal residues in NF-κB2 p100, leading to its proteasomal processing to generate the transcriptionally competent NF-κB or p52/RelB. NF-κB or p52/RelB then translocates to the nucleus and induces target gene expression. The noncanonical pathway shows a slower kinetics, is independent of IKKβ and NEMO, and plays a critical role in the development of lymphoid organs (Hinz and Scheidereit 2013).
TGF-β can synergize with TNF-α or interleukin-1 to activate type VII collagen gene expression through both the NF-κB-binding site and SBE sites in regulatory gene sequences (Kon et al. 1999), suggesting a convergence of the two pathways at common target genes. NF-κB can be activated by TGF-β and mediate transcription activation of TGF-β target genes in a variety of cell types (Lopez-Rovira et al. 2000; Ogawa et al. 2004; Yeh et al. 2008). For example, TGF-β increases the migration and cell-surface αvβ3 integrin expression in chondrosarcoma cells (Yeh et al. 2008), promotes EMT in pancreatic cancer cells (Brandl et al. 2010), survival of osteoclasts (Gingery et al. 2008), proliferation and differentiation of keratinocytes (Descargues et al. 2008), and activates transcription of NF-κB target genes (Freudlsperger et al. 2013), all in an NF-κB-dependent manner. Activation of the NF-κB by TGF-β can be mediated by both Smad-dependent and Smad-independent pathways. The Smad-dependent mechanism often involves a physical interaction between Smad3 and NF-κB or its activator IKKα (Lopez-Rovira et al. 2000; Descargues et al. 2008; Brandl et al. 2010; Hogan et al. 2013). Smad3 was shown to physically interact with NF-κB or p52/RelB to activate JunB expression (Lopez-Rovira et al. 2000). IKKα can associate with Smad3 in response to TGF-β and undergo nuclear translocation. This is necessary for TGF-β-induced downregulation of E-cadherin expression and transcription activation of the genes encoding Slug and Snail in pancreatic cancer cells (Brandl et al. 2010). In mouse keratinocytes, this interaction results in the recruitment of Smad3 to regulatory DNA sequences of several genes encoding the Myc antagonists, Mad1, Mad2, and Ovol1, to inhibit cell-cycle progression and promote differentiation of keratinocytes in a Smad4-independent manner (Descargues et al. 2008). In addition to activating NF-κB on IKK-mediated phosphorylation, TGF-β also induces acetylation of p65/RelA, dependent on Smad3 and Smad4, PKA, and the coactivator p300. This acetylation of p65/RelA at Lys221 is required for the synergy of TGF-β in enhancing the activation of the DNA-binding and transcription activity of NF-κB in response to bacteria (Ishinaga et al. 2007). Thus, NF-κB and IKKα can function both as signaling components for NF-κB activation and as an important interface for cross talk between NF-κB and TGF-β pathways.
TGF-β can also activate NF-κB in a Smad-independent manner. In multiple cell types, including osteoclasts, head and neck squamous cell carcinoma (HNSCC) cells, murine B cells, and hepatocytes, TGF-β induces NF-κB activation by TAK1 (Arsura et al. 2003; Gingery et al. 2008; Mao et al. 2011; Freudlsperger et al. 2013; Zhang et al. 2013). This activation requires Lys158 of TAK1, and is mediated by TRAF6- or TRAF4-dependent polyubiquitylation of TAK1 at Lys158 (Wang et al. 2001; Sorrentino et al. 2008; Xia et al. 2009; Mao et al. 2011; Zhang et al. 2013). Once activated, TAK1 proceeds to phosphorylate and activate IKKα, leading to NF-κB signaling (Wang et al. 2001; Xia et al. 2009). Mutation of Lys158 abolishes TGF-β-induced TAK1 activation and subsequent IKK, JNK, and p38 activation by TAK1 (Mao et al. 2011). RhoA-Rho-associated kinase (ROCK) that is activated by TAK1 in response to TGF-β can also phosphorylate and activate IKKβ, leading to NF-κB activation (Kim et al. 2014). In addition to TAK1, TGF-β also acts through the PI3K-Akt pathway to increase phosphorylation of IKKα/β and subsequent phosphorylation of IκB and NF-κB, leading to increased integrin expression and cell migration (Yeh et al. 2008).
Although most reports suggest that TGF-β signaling activates NF-κB, TGF-β1 has also been found to repress NF-κB signaling in human intestinal lamina propria mononuclear cells (LPMCs). In these cells, TGF-β1 suppresses TNF-α-induced activation of NF-κB p65 by increasing IκB transcription (Monteleone et al. 2004). This inhibition of NF-κB by TGF-β could be caused by a negative feedback loop. Indeed, in murine B cells and hepatocytes, TGF-β could induce an initial activation of NF-κB, which then leads to increased transcription of IκB, resulting eventually in the inhibition of NF-κB signaling (Arsura et al. 2003).
NF-κB/RelA can also inhibit TGF-β/Smad signaling by inducing Smad7 expression. In fibroblasts stimulated with lipopolysaccharide (LPS) or proinflammatory cytokines, activated NF-κB/RelA can induce transcription of the Smad7 gene (Bitzer et al. 2000). Similarly, in HNSCC cells, NF-κB activated by TAK1 in response to TGF-β can also increase Smad7 expression, which then suppresses TGF-β/Smad signaling (Freudlsperger et al. 2013). Through this feedback cross talk, NF-κB may contribute to the attenuation of cytostatic responses of TGF-β during malignant progression in human cancer cells.
CROSS TALK WITH PLURIPOTENCY- AND LINEAGE-SPECIFIC TRANSCRIPTION FACTORS
In the cells, multiple inputs from various pathways inevitably converge on transcription complexes at regulatory DNA sequences of target genes. Smad proteins, acting as transcription factors, are important components of these transcription complexes or chromosome-modification complexes, and physical and functional interactions between Smads and other high-affinity DNA-binding proteins or lineage-specific transcription factors are essential for signal integration and cooperation. Thus, cross talk with various transcription factors at regulatory gene sequences is an inherent feature of Smad function. Consistent with this, recent genome-wide profiling analyses using ChIP-Seq in mouse and human ES cells have revealed that Smad proteins exist in transcription complexes together with pluripotency transcription factors or chromosome modifiers to regulate stem-cell-fate decision. ChIP-Seq profiling of the binding sites of 13 transcription factors and two transcription regulators in mESCs showed that an ES-specific Smad-binding pattern often contains Smad1 in a complex with Sox2 and Oct4 (Chen et al. 2008). In hESCs, Smad2 and Smad3 acting downstream of Nodal and activin signaling physically associates with Oct4, Sox2, and Nanog at target DNA sequences as part of the transcription program to maintain pluripotency (Teo et al. 2011). During differentiation toward mesendoderm, Nanog induces the initial expression of the endoderm-specific transcription factor Eomes. Smad2 and/or Smad3 then exchange their transcriptional partners from the pluripotency factors (Oct4, Sox2, and Nanog) to lineage-specification factors such as Eomes or FOXH1 to enable a differentiation program (Teo et al. 2011). Similarly, Smad3 was found to co-occupy the genome with various master transcription factors in a lineage-specific manner (Mullen et al. 2011). One of the mechanisms by which Smad2 or Smad3 may activate the expression of lineage-specific genes is by recruiting the histone demethylase JMJD3 to Nodal target promoters (Dahle et al. 2010), thereby directly acting on the repressive chromatin state to induce their activation. Indeed, using ChIP-Seq analysis in hESCs, Smad2 or Smad3 was found to accumulate at regulatory promoter sequences of endoderm genes, and this coincided with the recruitment of histone demethylase JMJD3 and an increase in histone acetylation (Kim et al. 2011).
CONCLUDING REMARKS
The past decade has seen major progress in the area of stem-cell biology, and the availability of genome-wide research tools further facilitates the investigation on the dynamic changes in global signaling networks and transcription complexes. Mirroring their importance in embryonic development, TGF-β family proteins and activated Smad signaling are found to be essential players in stem-cell self-renewal and differentiation. Through extensive interactions with other signaling pathways and pluripotency or lineage-specific transcription factors, these TGF-β family pathways are effectively integrated into the cellular signaling network, which, depending on the dosage, timing, and location of various ligand inputs, interprets the combinatorial signals and produces an integrated output that specifies cell-fate decisions within the physiological context. Although the roles of the TGF-β family in various biological processes may appear complex and sometimes even confusing, the interpretation of the outcomes of TGF-β signaling has to be placed under the specific physiological context and take into consideration the presence of other signaling pathways. A future challenge is to accurately predict biological outcomes from these combinatorial signaling activities. The exciting progress in genome-wide mapping technologies may reveal a full signaling network at mechanistic levels in cells under varying physiological contexts, and mathematical modeling combined with biochemical analyses of global signaling networks may yield important information in this area.
ACKNOWLEDGMENTS
I apologize to the researchers whose work was not included in this review because of space limitations. K.L. is supported by the National Institutes of Health (NIH) R21 CA187632.
Footnotes
Editors: Rik Derynck and Kohei Miyazono
Additional Perspectives on The Biology of the TGF-β Family available at www.cshperspectives.org
REFERENCES
- Adhikari A, Xu M, Chen ZJ. 2007. Ubiquitin-mediated activation of TAK1 and IKK. Oncogene 26: 3214–3226. [DOI] [PubMed] [Google Scholar]
- Alarcón C, Zaromytidou AI, Xi Q, Gao S, Yu J, Fujisawa S, Barlas A, Miller AN, Manova-Todorova K, Macias MJ, et al. 2009. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-β pathways. Cell 139: 757–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragon E, Goerner N, Zaromytidou AI, Xi Q, Escobedo A, Massagué J, Macias MJ. 2011. A Smad action turnover switch operated by WW domain readers of a phosphoserine code. Genes Dev 25: 1275–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragona M, Panciera T, Manfrin A, Giulitti S, Michielin F, Elvassore N, Dupont S, Piccolo S. 2013. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell 154: 1047–1059. [DOI] [PubMed] [Google Scholar]
- Arsura M, Panta GR, Bilyeu JD, Cavin LG, Sovak MA, Oliver AA, Factor V, Heuchel R, Mercurio F, Thorgeirsson SS, et al. 2003. Transient activation of NF-κB through a TAK1/IKK kinase pathway by TGF-β1 inhibits AP-1/SMAD signaling and apoptosis: Implications in liver tumor formation. Oncogene 22: 412–425. [DOI] [PubMed] [Google Scholar]
- Asano Y, Ihn H, Yamane K, Jinnin M, Mimura Y, Tamaki K. 2004. Phosphatidylinositol 3-kinase is involved in α2(I) collagen gene expression in normal and scleroderma fibroblasts. J Immunol 172: 7123–7135. [DOI] [PubMed] [Google Scholar]
- Attisano L, Labbé E. 2004. TGFβ and Wnt pathway cross-talk. Cancer Metastasis Rev 23: 53–61. [DOI] [PubMed] [Google Scholar]
- Attisano L, Wrana JL. 2000. Smads as transcriptional comodulators. Curr Opin Cell Biol 12: 235–243. [DOI] [PubMed] [Google Scholar]
- Bakin AV, Tomlinson AK, Bhowmick NA, Moses HL, Arteaga CL. 2000. Phosphatidylinositol 3-kinase function is required for transforming growth factor β-mediated epithelial to mesenchymal transition and cell migration. J Biol Chem 275: 36803–36810. [DOI] [PubMed] [Google Scholar]
- Beyer TA, Weiss A, Khomchuk Y, Huang K, Ogunjimi AA, Varelas X, Wrana JL. 2013. Switch enhancers interpret TGF-β and Hippo signaling to control cell fate in human embryonic stem cells. Cell Rep 5: 1611–1624. [DOI] [PubMed] [Google Scholar]
- Bilican B, Fiore-Heriche C, Compston A, Allen ND, Chandran S. 2008. Induction of Olig2 precursors by FGF involves BMP signalling blockade at the Smad level. PLoS ONE 3: e2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitzer M, von Gersdorff G, Liang D, Dominguez-Rosales A, Beg AA, Rojkind M, Böttinger EP. 2000. A mechanism of suppression of TGF-β/SMAD signaling by NF-κB/RelA. Genes Dev 14: 187–197. [PMC free article] [PubMed] [Google Scholar]
- Blokzijl A, Dahlqvist C, Reissmann E, Falk A, Moliner A, Lendahl U, Ibanez CF. 2003. Cross-talk between the Notch and TGF-β signaling pathways mediated by interaction of the Notch intracellular domain with Smad3. J Cell Biol 163: 723–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozulic L, Hemmings BA. 2009. PIKKing on PKB: Regulation of PKB activity by phosphorylation. Curr Opin Cell Biol 21: 256–261. [DOI] [PubMed] [Google Scholar]
- Brandl M, Seidler B, Haller F, Adamski J, Schmid RM, Saur D, Schneider G. 2010. IKKα controls canonical TGFβ-SMAD signaling to regulate genes expressing SNAIL and SLUG during EMT in panc1 cells. J Cell Sci 123: 4231–4239. [DOI] [PubMed] [Google Scholar]
- Bright JJ, Sriram S. 1998. TGF-β inhibits IL-12-induced activation of Jak-STAT pathway in T lymphocytes. J Immunol 161: 1772–1777. [PubMed] [Google Scholar]
- Brodin G, Ahgren A, ten Dijke P, Heldin CH, Heuchel R. 2000. Efficient TGF-β induction of the Smad7 gene requires cooperation between AP-1, Sp1, and Smad proteins on the mouse Smad7 promoter. J Biol Chem 275: 29023–29030. [DOI] [PubMed] [Google Scholar]
- Burdon T, Stracey C, Chambers I, Nichols J, Smith A. 1999. Suppression of SHP-2 and ERK signalling promotes self-renewal of mouse embryonic stem cells. Dev Biol 210: 30–43. [DOI] [PubMed] [Google Scholar]
- Cao Z, Xiong J, Takeuchi M, Kurama T, Goeddel DV. 1996. TRAF6 is a signal transducer for interleukin-1. Nature 383: 443–446. [DOI] [PubMed] [Google Scholar]
- Carlson ME, Hsu M, Conboy IM. 2008. Imbalance between pSmad3 and Notch induces CDK inhibitors in old muscle stem cells. Nature 454: 528–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. 2008. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117. [DOI] [PubMed] [Google Scholar]
- Cocolakis E, Dai M, Drevet L, Ho J, Haines E, Ali S, Lebrun JJ. 2008. Smad signaling antagonizes STAT5-mediated gene transcription and mammary epithelial cell differentiation. J Biol Chem 283: 1293–1307. [DOI] [PubMed] [Google Scholar]
- Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA. 2008. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev 22: 746–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conery AR, Cao Y, Thompson EA, Townsend CM Jr, Ko TC, Luo K. 2004. Akt interacts directly with Smad3 to regulate the sensitivity to TGF-β-induced apoptosis. Nat Cell Biol 6: 366–372. [DOI] [PubMed] [Google Scholar]
- Crease DJ, Dyson S, Gurdon JB. 1998. Cooperation between the activin and Wnt pathways in the spatial control of organizer gene expression. Proc Natl Acad Sci 95: 4398–4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Tian Q, Christian JL. 1996. Synergistic effects of Vg1 and Wnt signals in the specification of dorsal mesoderm and endoderm. Dev Biol 180: 22–34. [DOI] [PubMed] [Google Scholar]
- Cullingworth J, Hooper ML, Harrison DJ, Mason JO, Sirard C, Patek CE, Clarke AR. 2002. Carcinogen-induced pancreatic lesions in the mouse: Effect of Smad4 and Apc genotypes. Oncogene 21: 4696–4701. [DOI] [PubMed] [Google Scholar]
- Dahle O, Kumar A, Kuehn MR. 2010. Nodal signaling recruits the histone demethylase Jmjd3 to counteract polycomb-mediated repression at target genes. Sci Signal 3: ra48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlqvist C, Blokzijl A, Chapman G, Falk A, Dannaeus K, Ibanez CF, Lendahl U. 2003. Functional Notch signaling is required for BMP4-induced inhibition of myogenic differentiation. Development 130: 6089–6099. [DOI] [PubMed] [Google Scholar]
- de Jong DS, Steegenga WT, Hendriks JM, van Zoelen EJ, Olijve W, Dechering KJ. 2004. Regulation of Notch signaling genes during BMP2-induced differentiation of osteoblast precursor cells. Biochem Biophys Res Commun 320: 100–107. [DOI] [PubMed] [Google Scholar]
- Dennler S, André J, Alexaki I, Li A, Magnaldo T, ten Dijke P, Wang XJ, Verrecchia F, Mauviel A. 2007. Induction of sonic hedgehog mediators by transforming growth factor-β: Smad3-dependent activation of Gli2 and Gli1 expression in vitro and in vivo. Cancer Res 67: 6981–6986. [DOI] [PubMed] [Google Scholar]
- Dennler S, André J, Verrecchia F, Mauviel A. 2009. Cloning of the human GLI2 promoter: Transcriptional activation by transforming growth factor-β via SMAD3/β-catenin cooperation. J Biol Chem 284: 31523–31531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descargues P, Sil AK, Sano Y, Korchynskyi O, Han G, Owens P, Wang XJ, Karin M. 2008. IKKα is a critical coregulator of a Smad4-independent TGFβ-Smad2/3 signaling pathway that controls keratinocyte differentiation. Proc Natl Acad Sci 105: 2487–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey N, Ghosh-Choudhury N, Kasinath BS, Choudhury GG. 2012. TGFβ-stimulated microRNA-21 utilizes PTEN to orchestrate AKT/mTORC1 signaling for mesangial cell hypertrophy and matrix expansion. PLoS ONE 7: e42316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. 2007. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130: 1120–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowdy SC, Mariani A, Janknecht R. 2003. HER2/Neu- and TAK1-mediated up-regulation of the transforming growth factor β inhibitor Smad7 via the ETS protein ER81. J Biol Chem 278: 44377–44384. [DOI] [PubMed] [Google Scholar]
- Downward J. 2004. PI 3-kinase, Akt and cell survival. Semin Cell Dev Biol 15: 177–182. [DOI] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, et al. 2011. Role of YAP/TAZ in mechanotransduction. Nature 474: 179–183. [DOI] [PubMed] [Google Scholar]
- Edlund S, Bu S, Schuster N, Aspenström P, Heuchel R, Heldin NE, ten Dijke P, Heldin CH, Landström M. 2003. Transforming growth factor-β1 (TGF-β)-induced apoptosis of prostate cancer cells involves Smad7-dependent activation of p38 by TGF-β-activated kinase 1 and mitogen-activated protein kinase kinase 3. Mol Biol Cell 14: 529–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edson MA, Nalam RL, Clementi C, Franco HL, Demayo FJ, Lyons KM, Pangas SA, Matzuk MM. 2010. Granulosa cell-expressed BMPR1A and BMPR1B have unique functions in regulating fertility but act redundantly to suppress ovarian tumor development. Mol Endocrinol 24: 1251–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eivers E, Fuentealba LC, Sander V, Clemens JC, Hartnett L, De Robertis EM. 2009. Mad is required for wingless signaling in wing development and segment patterning in Drosophila. PLoS ONE 4: e6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eivers E, Demagny H, Choi RH, De Robertis EM. 2011. Phosphorylation of Mad controls competition between wingless and BMP signaling. Sci Signal 4: ra68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekman M, Mu Y, Lee SY, Edlund S, Kozakai T, Thakur N, Tran H, Qian J, Groeden J, Heldin CH, et al. 2012. APC and Smad7 link TGFβ type I receptors to the microtubule system to promote cell migration. Mol Biol Cell 23: 2109–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estaras C, Benner C, Jones KA. 2015. SMADs and YAP compete to control elongation of β-catenin: LEF-1-recruited RNAPII during hESC differentiation. Mol Cell 58: 780–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei C, Li Z, Li C, Chen Y, Chen Z, He X, Mao L, Wang X, Zeng R, Li L. 2013. Smurf1-mediated Lys29-linked nonproteolytic polyubiquitination of axin negatively regulates Wnt/β-catenin signaling. Mol Cell Biol 33: 4095–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng XH, Derynck R. 2005. Specificity and versatility in TGF-β signaling through Smads. Annu Rev Cell Dev Biol 21: 659–693. [DOI] [PubMed] [Google Scholar]
- Ferrigno O, Lallemand F, Verrecchia F, L’Hoste S, Camonis J, Atfi A, Mauviel A. 2002. Yes-associated protein (YAP65) interacts with Smad7 and potentiates its inhibitory activity against TGF-β/Smad signaling. Oncogene 21: 4879–4884. [DOI] [PubMed] [Google Scholar]
- Freudlsperger C, Bian Y, Contag Wise S, Burnett J, Coupar J, Yang X, Chen Z, Van Waes C. 2013. TGF-β and NF-κB signal pathway cross-talk is mediated through TAK1 and SMAD7 in a subset of head and neck cancers. Oncogene 32: 1549–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentealba LC, Eivers E, Ikeda A, Hurtado C, Kuroda H, Pera EM, De Robertis EM. 2007. Integrating patterning signals: Wnt/GSK3 regulates the duration of the BMP/Smad1 signal. Cell 131: 980–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii M, Toyoda T, Nakanishi H, Yatabe Y, Sato A, Matsudaira Y, Ito H, Murakami H, Kondo Y, Kondo E, et al. 2012. TGF-β synergizes with defects in the Hippo pathway to stimulate human malignant mesothelioma growth. J Exp Med 209: 479–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukada T, Hibi M, Yamanaka Y, Takahashi-Tezuka M, Fujitani Y, Yamaguchi T, Nakajima K, Hirano T. 1996. Two signals are necessary for cell proliferation induced by a cytokine receptor gp130: Involvement of STAT3 in anti-apoptosis. Immunity 5: 449–460. [DOI] [PubMed] [Google Scholar]
- Furuhashi M, Yagi K, Yamamoto H, Furukawa Y, Shimada S, Nakamura Y, Kikuchi A, Miyazono K, Kato M. 2001. Axin facilitates Smad3 activation in the transforming growth factor β signaling pathway. Mol Cell Biol 21: 5132–5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galliher AJ, Schiemann WP. 2007. Src phosphorylates Tyr284 in TGF-β type II receptor and regulates TGF-β stimulation of p38 MAPK during breast cancer cell proliferation and invasion. Cancer Res 67: 3752–3758. [DOI] [PubMed] [Google Scholar]
- Ghosh-Choudhury N, Abboud SL, Nishimura R, Celeste A, Mahimainathan L, Choudhury GG. 2002. Requirement of BMP-2-induced phosphatidylinositol 3-kinase and Akt serine/threonine kinase in osteoblast differentiation and Smad-dependent BMP-2 gene transcription. J Biol Chem 277: 33361–3368. [DOI] [PubMed] [Google Scholar]
- Gingery A, Bradley EW, Pederson L, Ruan M, Horwood NJ, Oursler MJ. 2008. TGF-β coordinately activates TAK1/MEK/AKT/NFκB and SMAD pathways to promote osteoclast survival. Exp Cell Res 314: 2725–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudey SK, Sundar R, Mu Y, Wallenius A, Zang G, Bergh A, Heldin CH, Landström M. 2014. TRAF6 stimulates the tumor-promoting effects of TGFβ type I receptor through polyubiquitination and activation of presenilin 1. Sci Signal 7: ra2. [DOI] [PubMed] [Google Scholar]
- Guo X, Wang XF. 2009. Signaling cross-talk between TGF-β/BMP and other pathways. Cell Res 19: 71–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Ramirez A, Waddell DS, Li Z, Liu X, Wang XF. 2008. Axin and GSK3- control Smad3 protein stability and modulate TGF-β signaling. Genes Dev 22: 106–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamoto T, Beppu H, Okada H, Kawabata M, Kitamura T, Miyazono K, Kato M. 2002. Compound disruption of Smad2 accelerates malignant progression of intestinal tumors in Apc knockout mice. Cancer Res 62: 5955–5961. [PubMed] [Google Scholar]
- Hao Y, Chun A, Cheung K, Rashidi B, Yang X. 2008. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J Biol Chem 283: 5496–5509. [DOI] [PubMed] [Google Scholar]
- He XC, Zhang J, Tong WG, Tawfik O, Ross J, Scoville DH, Tian Q, Zeng X, He X, Wiedemann LM, et al. 2004. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-β-catenin signaling. Nat Genet 36: 1117–1121. [DOI] [PubMed] [Google Scholar]
- Heldin CH, Moustakas A. 2011. Role of Smads in TGFβ signaling. Cell Tissue Res 347: 21–36. [DOI] [PubMed] [Google Scholar]
- Heldin CH, Landstrom M, Moustakas A. 2009. Mechanism of TGF-β signaling to growth arrest, apoptosis, and epithelial-mesenchymal transition. Curr Opin Cell Biol 21: 166–176. [DOI] [PubMed] [Google Scholar]
- Hinz M, Scheidereit C. 2013. The IκB kinase complex in NF-κB regulation and beyond. EMBO Rep 15: 46–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan KA, Ravindran A, Podolsky MA, Glick AB. 2013. The TGFβ1 pathway is required for NFκB-dependent gene expression in mouse keratinocytes. Cytokine 64: 652–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm TM, Habashi JP, Doyle JJ, Bedja D, Chen Y, van Erp C, Lindsay ME, Kim D, Schoenhoff F, Cohn RD, et al. 2011. Noncanonical TGFβ signaling contributes to aortic aneurysm progression in Marfan syndrome mice. Science 332: 358–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppler S, Moon RT. 1998. BMP-2/-4 and Wnt-8 cooperatively pattern the Xenopus mesoderm. Mech Dev 71: 119–129. [DOI] [PubMed] [Google Scholar]
- Horowitz JC, Lee DY, Waghray M, Keshamouni VG, Thomas PE, Zhang H, Cui Z, Thannickal VJ. 2004. Activation of the pro-survival phosphatidylinositol 3-kinase/AKT pathway by transforming growth factor-β1 in mesenchymal cells is mediated by p38 MAPK-dependent induction of an autocrine growth factor. J Biol Chem 279: 1359–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson C, Yasuo H. 2006. A signalling relay involving Nodal and Delta ligands acts during secondary notochord induction in Ciona embryos. Development 133: 2855–2864. [DOI] [PubMed] [Google Scholar]
- Hudson C, Lotito S, Yasuo H. 2007. Sequential and combinatorial inputs from Nodal, Delta2/Notch and FGF/MEK/ERK signalling pathways establish a grid-like organisation of distinct cell identities in the ascidian neural plate. Development 134: 3527–3537. [DOI] [PubMed] [Google Scholar]
- Hui CC, Angers S. 2011. Gli proteins in development and disease. Annu Rev Cell Dev Biol 27: 513–537. [DOI] [PubMed] [Google Scholar]
- Hussein SM, Duff EK, Sirard C. 2003. Smad4 and β-catenin co-activators functionally interact with lymphoid-enhancing factor to regulate graded expression of Msx2. J Biol Chem 278: 48805–48814. [DOI] [PubMed] [Google Scholar]
- Ishida T, Mizushima S, Azuma S, Kobayashi N, Tojo T, Suzuki K, Aizawa S, Watanabe T, Mosialos G, Kieff E, et al. 1996. Identification of TRAF6, a novel tumor necrosis factor receptor-associated factor protein that mediates signaling from an amino-terminal domain of the CD40 cytoplasmic region. J Biol Chem 271: 28745–28748. [DOI] [PubMed] [Google Scholar]
- Ishinaga H, Jono H, Lim JH, Kweon SM, Xu H, Ha UH, Koga T, Yan C, Feng XH, Chen LF, et al. 2007. TGF-β induces p65 acetylation to enhance bacteria-induced NF-κB activation. EMBO J 26: 1150–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh F, Itoh S, Goumans MJ, Valdimarsdottir G, Iso T, Dotto GP, Hamamori Y, Kedes L, Kato M, ten Dijke P. 2004. Synergy and antagonism between Notch and BMP receptor signaling pathways in endothelial cells. EMBO J 23: 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahchan NS, Wang D, Bissell MJ, Luo K. 2012. SnoN regulates mammary gland alveologenesis and onset of lactation by promoting prolactin/Stat5 signaling. Development 139: 3147–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javelaud D, Alexaki VI, Dennler S, Mohammad KS, Guise TA, Mauviel A. 2011. TGF-β/SMAD/GLI2 signaling axis in cancer progression and metastasis. Cancer Res 71: 5606–5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javelaud D, Pierrat MJ, Mauviel A. 2012. Crosstalk between TGF-β and hedgehog signaling in cancer. FEBS Lett 586: 2016–2025. [DOI] [PubMed] [Google Scholar]
- Jeong HW, Kim IS. 2004. TGF-β1 enhances βig-h3-mediated keratinocyte cell migration through the α3β1 integrin and PI3K. J Cell Biochem 92: 770–780. [DOI] [PubMed] [Google Scholar]
- Jian H, Shen X, Liu I, Semenov M, He X, Wang XF. 2006. Smad3-dependent nuclear translocation of β-catenin is required for TGF-β1-induced proliferation of bone marrow-derived adult human mesenchymal stem cells. Genes Dev 20: 666–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin EJ, Lee SY, Choi YA, Jung JC, Bang OS, Kang SS. 2006. BMP-2-enhanced chondrogenesis involves p38 MAPK-mediated down-regulation of Wnt-7a pathway. Mol Cells 22: 353–359. [PubMed] [Google Scholar]
- Johnson RW, Nguyen MP, Padalecki SS, Grubbs BG, Merkel AR, Oyajobi BO, Matrisian LM, Mundy GR, Sterling JA. 2011. TGF-β promotion of Gli2-induced expression of parathyroid hormone-related protein, an important osteolytic factor in bone metastasis, is independent of canonical Hedgehog signaling. Cancer Res 71: 822–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung SM, Lee JH, Park J, Oh YS, Lee SK, Park JS, Lee YS, Kim JH, Lee JY, Bae YS, et al. 2013. Smad6 inhibits non-canonical TGF-β1 signalling by recruiting the deubiquitinase A20 to TRAF6. Nat Commun 4: 2562. [DOI] [PubMed] [Google Scholar]
- Kato M, Putta S, Wang M, Yuan H, Lanting L, Nair I, Gunn A, Nakagawa Y, Shimano H, Todorov I, et al. 2009. TGF-β activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat Cell Biol 11: 881–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Jho EH. 2010. The protein stability of Axin, a negative regulator of Wnt signaling, is regulated by Smad ubiquitination regulatory factor 2 (Smurf2). J Biol Chem 285: 36420–36426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SW, Yoon SJ, Chuong E, Oyolu C, Wills AE, Gupta R, Baker J. 2011. Chromatin and transcriptional signatures for Nodal signaling during endoderm formation in hESCs. Dev Biol 357: 492–504. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Kim JG, Moon MY, Park SH, Park JB. 2014. IκB kinase γ/nuclear factor-κB-essential modulator (IKKγ/NEMO) facilitates RhoA GTPase activation, which, in turn, activates Rho-associated KINASE (ROCK) to phosphorylate IKKβ in response to transforming growth factor (TGF)-β1. J Biol Chem 289: 1429–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimelman D, Griffin KJ. 2000. Vertebrate mesendoderm induction and patterning. Curr Opin Genet Dev 10: 350–356. [DOI] [PubMed] [Google Scholar]
- Kiyono K, Suzuki HI, Matsuyama H, Morishita Y, Komuro A, Kano MR, Sugimoto K, Miyazono K. 2009. Autophagy is activated by TGF-β and potentiates TGF-β-mediated growth inhibition in human hepatocellular carcinoma cells. Cancer Res 69: 8844–8852. [DOI] [PubMed] [Google Scholar]
- Kleber M, Lee HY, Wurdak H, Buchstaller J, Riccomagno MM, Ittner LM, Suter U, Epstein DJ, Sommer L. 2005. Neural crest stem cell maintenance by combinatorial Wnt and BMP signaling. J Cell Biol 169: 309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon A, Vindevoghel L, Kouba DJ, Fujimura Y, Uitto J, Mauviel A. 1999. Cooperation between SMAD and NF-κB in growth factor regulated type VII collagen gene expression. Oncogene 18: 1837–1844. [DOI] [PubMed] [Google Scholar]
- Krausova M, Korinek V. 2014. Wnt signaling in adult intestinal stem cells and cancer. Cell Signal 26: 570–579. [DOI] [PubMed] [Google Scholar]
- Kretzschmar M, Doody J, Massagué J. 1997. Opposing BMP and EGF signalling pathways converge on the TGF-β family mediator Smad1. Nature 389: 618–622. [DOI] [PubMed] [Google Scholar]
- Kunath T, Saba-El-Leil MK, Almousailleakh M, Wray J, Meloche S, Smith A. 2007. FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development 134: 2895–2902. [DOI] [PubMed] [Google Scholar]
- Kuroda H, Fuentealba L, Ikeda A, Reversade B, De Robertis EM. 2005. Default neural induction: Neuralization of dissociated Xenopus cells is mediated by Ras/MAPK activation. Genes Dev 19: 1022–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé E, Letamendia A, Attisano L. 2000. Association of Smads with lymphoid enhancer binding factor 1/T cell-specific factor mediates cooperative signaling by the transforming growth factor-β and Wnt pathways. Proc Natl Acad Sci 97: 8358–8363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamouille S, Derynck R. 2007. Cell size and invasion in TGF-β-induced epithelial to mesenchymal transition is regulated by activation of the mTOR pathway. J Cell Biol 178: 437–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamouille S, Connolly E, Smyth JW, Akhurst RJ, Derynck R. 2012. TGF-β-induced activation of mTOR complex 2 drives epithelial-mesenchymal transition and cell invasion. J Cell Sci 125: 1259–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landström M. 2010. The TAK1-TRAF6 signalling pathway. Int J Biochem Cell Biol 42: 585–589. [DOI] [PubMed] [Google Scholar]
- Larrivée B, Prahst C, Gordon E, del Toro R, Mathivet T, Duarte A, Simons M, Eichmann A. 2012. ALK1 signaling inhibits angiogenesis by cooperating with the Notch pathway. Dev Cell 22: 489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawler S, Feng XH, Chen RH, Maruoka EM, Turck CW, Griswold-Prenner I, Derynck R. 1997. The type II transforming growth factor-β receptor autophosphorylates not only on serine and threonine but also on tyrosine residues. J Biol Chem 272: 14850–14859. [DOI] [PubMed] [Google Scholar]
- Lechuga CG, Hernandez-Nazara ZH, Dominguez Rosales JA, Morris ER, Rincon AR, Rivas-Estilla AM, Esteban-Gamboa A, Rojkind M. 2004. TGF-β1 modulates matrix metalloproteinase-13 expression in hepatic stellate cells by complex mechanisms involving p38MAPK, PI3-kinase, AKT, and p70S6K. Am J Physiol Gastrointest Liver Physiol 287: G974–G987. [DOI] [PubMed] [Google Scholar]
- Lee YI, Kwon YJ, Joo CK. 2004. Integrin-linked kinase function is required for transforming growth factor β-mediated epithelial to mesenchymal transition. Biochem Biophys Res Commun 316: 997–1001. [DOI] [PubMed] [Google Scholar]
- Lee MK, Pardoux C, Hall MC, Lee PS, Warburton D, Qing J, Smith SM, Derynck R. 2007. TGF-β activates Erk MAP kinase signalling through direct phosphorylation of ShcA. EMBO J 26: 3957–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei S, Dubeykovskiy A, Chakladar A, Wojtukiewicz L, Wang TC. 2004. The murine gastrin promoter is synergistically activated by transforming growth factor-β/Smad and Wnt signaling pathways. J Biol Chem 279: 42492–42502. [DOI] [PubMed] [Google Scholar]
- Lei QY, Zhang H, Zhao B, Zha ZY, Bai F, Pei XH, Zhao S, Xiong Y, Guan KL. 2008. TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol Cell Biol 28: 2426–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengerke C, Schmitt S, Bowman TV, Jang IH, Maouche-Chrétien L, McKinney-Freeman S, Davidson AJ, Hammerschmidt M, Rentzsch F, Green JB, et al. 2008. BMP and Wnt specify hematopoietic fate by activation of the Cdx-Hox pathway. Cell Stem Cell 2: 72–82. [DOI] [PubMed] [Google Scholar]
- Li WX. 2008. Canonical and non-canonical JAK-STAT signaling. Trends Cell Biol 18: 545–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Fei T, Zhang J, Zhu G, Wang L, Lu D, Chi X, Teng Y, Hou N, Yang X, et al. 2012. BMP4 Signaling Acts via dual-specificity phosphatase 9 to control ERK activity in mouse embryonic stem cells. Cell Stem Cell 10: 171–182. [DOI] [PubMed] [Google Scholar]
- Lian I, Kim J, Okazawa H, Zhao J, Zhao B, Yu J, Chinnaiyan A, Israel MA, Goldstein LS, Abujarour R, et al. 2010. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev 24: 1106–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M, Liang YY, Wrighton K, Ungermannova D, Wang XP, Brunicardi FC, Liu X, Feng XH, Lin X. 2004. Ubiquitination and proteolysis of cancer-derived Smad4 mutants by SCFSkp2. Mol Cell Biol 24: 7524–7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberati NT, Datto MB, Frederick JP, Shen X, Wong C, Rougier-Chapman EM, Wang XF. 1999. Smads bind directly to the Jun family of AP-1 transcription factors. Proc Natl Acad Sci 96: 4844–4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JH, Jono H, Komatsu K, Woo CH, Lee J, Miyata M, Matsuno T, Xu X, Huang Y, Zhang W, et al. 2012. CYLD negatively regulates transforming growth factor-β-signalling via deubiquitinating Akt. Nat Commun 3: 771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Xu P, Lamouille S, Xu J, Derynck R. 2009. TACE-mediated ectodomain shedding of the type I TGF-β receptor downregulates TGF-β signaling. Mol Cell 35: 26–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CY, Zha ZY, Zhou X, Zhang H, Huang W, Zhao D, Li T, Chan SW, Lim CJ, Hong W, et al. 2010. The hippo tumor pathway promotes TAZ degradation by phosphorylating a phosphodegron and recruiting the SCFβ-TrCP E3 ligase. J Biol Chem 285: 37159–37169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Liu H, Meyer C, Li J, Nadalin S, Konigsrainer A, Weng H, Dooley S, ten Dijke P. 2013. Transforming growth factor-β (TGF-β)-mediated connective tissue growth factor (CTGF) expression in hepatic stellate cells requires Stat3 signaling activation. J Biol Chem 288: 30708–30719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loose M, Patient R. 2004. A genetic regulatory network for Xenopus mesendoderm formation. Dev Biol 271: 467–478. [DOI] [PubMed] [Google Scholar]
- Lopez-Rovira T, Chalaux E, Rosa JL, Bartrons R, Ventura F. 2000. Interaction and functional cooperation of NF-κB with Smads. Transcriptional regulation of the JunB promoter. J Biol Chem 275: 28937–28946. [DOI] [PubMed] [Google Scholar]
- Mandal CC, Ghosh Choudhury G, Ghosh-Choudhury N. 2009. Phosphatidylinositol 3 kinase/Akt signal relay cooperates with Smad in bone morphogenetic protein-2-induced colony stimulating factor-1 (CSF-1) expression and osteoclast differentiation. Endocrinology 150: 4989–4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao R, Fan Y, Mou Y, Zhang H, Fu S, Yang J. 2011. TAK1 lysine 158 is required for TGF-β-induced TRAF6-mediated Smad-independent IKK/NF-κB and JNK/AP-1 activation. Cell Signal 23: 222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J. 2012. TGFβ signalling in context. Nat Rev Mol Cell Biol 13: 616–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda S, Kumano K, Shimizu K, Imai Y, Kurokawa M, Ogawa S, Miyagishi M, Taira K, Hirai H, Chiba S. 2005. Notch1 oncoprotein antagonizes TGF-β/Smad-mediated cell growth suppression via sequestration of coactivator p300. Cancer Sci 96: 274–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura I, Wang G, He D, Liu F. 2005. Identification and characterization of ERK MAP kinase phosphorylation sites in Smad3. Biochemistry 44: 12546–12553. [DOI] [PubMed] [Google Scholar]
- Maurya AK, Tan H, Souren M, Wang X, Wittbrodt J, Ingham PW. 2011. Integration of Hedgehog and BMP signalling by the engrailed2a gene in the zebrafish myotome. Development 138: 755–765. [DOI] [PubMed] [Google Scholar]
- Millet C, Yamashita M, Heller M, Yu LR, Veenstra TD, Zhang YE. 2009. A negative feedback control of transforming growth factor-β signaling by glycogen synthase kinase 3-mediated Smad3 linker phosphorylation at Ser-204. J Biol Chem 284: 19808–19816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad KS, Javelaud D, Fournier PG, Niewolna M, McKenna CR, Peng XH, Duong V, Dunn LK, Mauviel A, Guise TA. 2011. TGF-β-RI kinase inhibitor SD-208 reduces the development and progression of melanoma bone metastases. Cancer Res 71: 175–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteleone G, Mann J, Monteleone I, Vavassori P, Bremner R, Fantini M, Del Vecchio Blanco G, Tersigni R, Alessandroni L, Mann D, et al. 2004. A failure of transforming growth factor-β1 negative regulation maintains sustained NF-κB activation in gut inflammation. J Biol Chem 279: 3925–3932. [DOI] [PubMed] [Google Scholar]
- Mu Y, Gudey SK, Landström M. 2012. Non-Smad signaling pathways. Cell Tissue Res 347: 11–20. [DOI] [PubMed] [Google Scholar]
- Mullen AC, Orlando DA, Newman JJ, Loven J, Kumar RM, Bilodeau S, Reddy J, Guenther MG, DeKoter RP, Young RA. 2011. Master transcription factors determine cell-type-specific responses to TGF-β signaling. Cell 147: 565–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Yanagisawa M, Arakawa H, Kimura N, Hisatsune T, Kawabata M, Miyazono K, Taga T. 1999. Synergistic signaling in fetal brain by STAT3-Smad1 complex bridged by p300. Science 284: 479–482. [DOI] [PubMed] [Google Scholar]
- Nallet-Staub F, Yin X, Gilbert C, Marsaud V, Ben Mimoun S, Javelaud D, Leof EB, Mauviel A. 2015. Cell density sensing alters TGF-β signaling in a cell-type-specific manner, independent from Hippo pathway activation. Dev Cell 32: 640–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimi H, Pardali K, Heldin CH, Moustakas A. 2007. Notch signaling is necessary for epithelial growth arrest by TGF-β. J Cell Biol 176: 695–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishita M, Hashimoto MK, Ogata S, Laurent MN, Ueno N, Shibuya H, Cho KW. 2000. Interaction between Wnt and TGF-β signalling pathways during formation of Spemann’s organizer. Nature 403: 781–785. [DOI] [PubMed] [Google Scholar]
- Nobta M, Tsukazaki T, Shibata Y, Xin C, Moriishi T, Sakano S, Shindo H, Yamaguchi A. 2005. Critical regulation of bone morphogenetic protein-induced osteoblastic differentiation by Delta1/Jagged1-activated Notch1 signaling. J Biol Chem 280: 15842–15848. [DOI] [PubMed] [Google Scholar]
- Nostro MC, Cheng X, Keller GM, Gadue P. 2008. Wnt, activin, and BMP signaling regulate distinct stages in the developmental pathway from embryonic stem cells to blood. Cell Stem Cell 2: 60–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowell C, Radtke F. 2013. Cutaneous Notch signaling in health and disease. Cold Spring Harb Perspect Med 3: a017772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusse R. 2012. Wnt signaling. Cold Spring Harb Perspect Biol 4: a011163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nye MD, Almada LL, Fernandez-Barrena MG, Marks DL, Elsawa SF, Vrabel A, Tolosa EJ, Ellenrieder V, Fernandez-Zapico ME. 2014. The transcription factor GLI1 interacts with SMAD proteins to modulate transforming growth factor β-induced gene expression in a p300/CREB-binding protein-associated factor (PCAF)-dependent manner. J Biol Chem 289: 15495–15506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa K, Chen F, Kuang C, Chen Y. 2004. Suppression of matrix metalloproteinase-9 transcription by transforming growth factor-β is mediated by a nuclear factor-κB site. Biochem J 381: 413–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Irvine KD. 2011. Cooperative regulation of growth by Yorkie and Mad through Bantam. Dev Cell 20: 109–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T, Mazack V, Sudol M. 2008. Mst2 and Lats kinases regulate apoptotic function of Yes kinase-associated protein (YAP). J Biol Chem 283: 27534–27546. [DOI] [PubMed] [Google Scholar]
- Ota M, Sasaki H. 2008. Mammalian TEAD proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development 135: 4059–4069. [DOI] [PubMed] [Google Scholar]
- Pardoux C, Ma X, Gobert S, Pellegrini S, Mayeux P, Gay F, Trinchieri G, Chouaib S. 1999. Downregulation of interleukin-12 (IL-12) responsiveness in human T cells by transforming growth factor-β: Relationship with IL-12 signaling. Blood 93: 1448–1455. [PubMed] [Google Scholar]
- Park HW, Guan KL. 2013. Regulation of the Hippo pathway and implications for anticancer drug development. Trends Pharmacol Sci 34: 581–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrot CY, Javelaud D, Mauviel A. 2013. Overlapping activities of TGF-β and Hedgehog signaling in cancer: Therapeutic targets for cancer treatment. Pharmacol Ther 137: 183–199. [DOI] [PubMed] [Google Scholar]
- Piccolo S, Dupont S, Cordenonsi M. 2014. The biology of YAP/TAZ: Hippo signaling and beyond. Physiol Rev 94: 1287–1312. [DOI] [PubMed] [Google Scholar]
- Quijano JC, Stinchfield MJ, Zerlanko B, Gibbens YY, Takaesu NT, Hyman-Walsh C, Wotton D, Newfeld SJ. 2010. The Sno oncogene antagonizes Wingless signaling during wing development in Drosophila. PLoS ONE 5: e11619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quijano JC, Stinchfield MJ, Newfeld SJ. 2011. Wg signaling via Zw3 and Mad restricts self-renewal of sensory organ precursor cells in Drosophila. Genetics 189: 809–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao P, Kadesch T. 2003. The intracellular form of Notch blocks transforming growth factor β-mediated growth arrest in Mv1Lu epithelial cells. Mol Cell Biol 23: 6694–6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashidian J, Le Scolan E, Ji X, Zhu Q, Mulvihill MM, Nomura D, Luo K. 2015. Ski regulates Hippo and TAZ signaling to suppress breast cancer progression. Sci Signal 8: ra14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regnier CH, Tomasetto C, Moog-Lutz C, Chenard MP, Wendling C, Basset P, Rio MC. 1995. Presence of a new conserved domain in CART1, a novel member of the tumor necrosis factor receptor-associated protein family, which is expressed in breast carcinoma. J Biol Chem 270: 25715–25721. [DOI] [PubMed] [Google Scholar]
- Remy I, Montmarquette A, Michnick SW. 2004. PKB/Akt modulates TGF-β signalling through a direct interaction with Smad3. Nat Cell Biol 6: 358–365. [DOI] [PubMed] [Google Scholar]
- Rios I, Alvarez-Rodriguez R, Marti E, Pons S. 2004. BMP2 antagonizes sonic hedgehog-mediated proliferation of cerebellar granule neurones through Smad5 signalling. Development 131: 3159–3168. [DOI] [PubMed] [Google Scholar]
- Rohrschneider LR, Fuller JF, Wolf I, Liu Y, Lucas DM. 2000. Structure, function, and biology of SHIP proteins. Genes Dev 14: 505–520. [PubMed] [Google Scholar]
- Ross KR, Corey DA, Dunn JM, Kelley TJ. 2007. SMAD3 expression is regulated by mitogen-activated protein kinase kinase-1 in epithelial and smooth muscle cells. Cell Signal 19: 923–931. [DOI] [PubMed] [Google Scholar]
- Runyan CE, Schnaper HW, Poncelet AC. 2004. The phosphatidylinositol 3-kinase/Akt pathway enhances Smad3-stimulated mesangial cell collagen I expression in response to transforming growth factor-β1. J Biol Chem 279: 2632–2639. [DOI] [PubMed] [Google Scholar]
- Saha D, Datta PK, Beauchamp RD. 2001. Oncogenic ras represses transforming growth factor-β/Smad signaling by degrading tumor suppressor Smad4. J Biol Chem 276: 29531–29537. [DOI] [PubMed] [Google Scholar]
- Sapkota G, Alarcon C, Spagnoli FM, Brivanlou AH, Massagué J. 2007. Balancing BMP signaling through integrated inputs into the Smad1 linker. Mol Cell 25: 441–454. [DOI] [PubMed] [Google Scholar]
- Scheel C, Eaton EN, Li SH, Chaffer CL, Reinhardt F, Kah KJ, Bell G, Guo W, Rubin J, Richardson AL, et al. 2011. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell 145: 926–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schohl A, Fagotto F. 2002. β-catenin, MAPK and Smad signaling during early Xenopus development. Development 129: 37–52. [DOI] [PubMed] [Google Scholar]
- Seoane J, Le HV, Shen L, Anderson SA, Massagué J. 2004. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell 117: 211–223. [DOI] [PubMed] [Google Scholar]
- Shimmi O, Newfeld SJ. 2013. New insights into extracellular and post-translational regulation of TGF-β family signalling pathways. J Biochem 154: 11–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AM, Reynolds D, Cliff T, Ohtsuka S, Mattheyses AL, Sun Y, Menendez L, Kulik M, Dalton S. 2012. Signaling network crosstalk in human pluripotent cells: A Smad2/3-regulated switch that controls the balance between self-renewal and differentiation. Cell Stem Cell 10: 312–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrentino A, Thakur N, Grimsby S, Marcusson A, von Bulow V, Schuster N, Zhang S, Heldin CH, Landstrom M. 2008. The type I TGF-β receptor engages TRAF6 to activate TAK1 in a receptor kinase-independent manner. Nat Cell Biol 10: 1199–1207. [DOI] [PubMed] [Google Scholar]
- Stavridis MP, Lunn JS, Collins BJ, Storey KG. 2007. A discrete period of FGF-induced Erk1/2 signalling is required for vertebrate neural specification. Development 134: 2889–2894. [DOI] [PubMed] [Google Scholar]
- Steg AD, Katre AA, Bevis KS, Ziebarth A, Dobbin ZC, Shah MM, Alvarez RD, Landen CN. 2012. Smoothened antagonists reverse taxane resistance in ovarian cancer. Mol Cancer Ther 11: 1587–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Lowther W, Kato K, Bianco C, Kenney N, Strizzi L, Raafat D, Hirota M, Khan NI, Bargo S, et al. 2005. Notch4 intracellular domain binding to Smad3 and inhibition of the TGF-β signaling. Oncogene 24: 5365–5374. [DOI] [PubMed] [Google Scholar]
- Szeto DP, Kimelman D. 2004. Combinatorial gene regulation by BMP and Wnt in zebrafish posterior mesoderm formation. Development 131: 3751–3760. [DOI] [PubMed] [Google Scholar]
- Takaku K, Oshima M, Miyoshi H, Matsui M, Seldin MF, Taketo MM. 1998. Intestinal tumorigenesis in compound mutant mice of both Dpc4 (Smad4) and Apc genes. Cell 92: 645–656. [DOI] [PubMed] [Google Scholar]
- Takaku K, Wrana JL, Robertson EJ, Taketo MM. 2002. No effects of Smad2 (Madh2) null mutation on malignant progression of intestinal polyps in ApcΔ716 knockout mice. Cancer Res 62: 4558–4561. [PubMed] [Google Scholar]
- Takekawa M, Tatebayashi K, Itoh F, Adachi M, Imai K, Saito H. 2002. Smad-dependent GADD45β expression mediates delayed activation of p38 MAP kinase by TGF-β. EMBO J 21: 6473–6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa T, Ochiai W, Nakashima K, Taga T. 2003. Enhanced gene activation by Notch and BMP signaling cross-talk. Nucleic Acids Res 31: 5723–5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo AK, Arnold SJ, Trotter MW, Brown S, Ang LT, Chng Z, Robertson EJ, Dunn NR, Vallier L. 2011. Pluripotency factors regulate definitive endoderm specification through eomesodermin. Genes Dev 25: 238–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezuka K, Yasuda M, Watanabe N, Morimura N, Kuroda K, Miyatani S, Hozumi N. 2002. Stimulation of osteoblastic cell differentiation by Notch. J Bone Miner Res 17: 231–239. [DOI] [PubMed] [Google Scholar]
- Theil T, Aydin S, Koch S, Grotewold L, Ruther U. 2002. Wnt and BMP signalling cooperatively regulate graded Emx2 expression in the dorsal telencephalon. Development 129: 3045–3054. [DOI] [PubMed] [Google Scholar]
- Trompouki E, Bowman TV, Lawton LN, Fan ZP, Wu DC, DiBiase A, Martin CS, Cech JN, Sessa AK, Leblanc JL, et al. 2011. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell 147: 577–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida K, Suzuki H, Ohashi T, Nitta K, Yumura W, Nihei H. 2001. Involvement of MAP kinase cascades in Smad7 transcriptional regulation. Biochem Biophys Res Commun 289: 376–381. [DOI] [PubMed] [Google Scholar]
- Ulloa L, Doody J, Massagué J. 1999. Inhibition of transforming growth factor-β/SMAD signalling by the interferon-γ/STAT pathway. Nature 397: 710–713. [DOI] [PubMed] [Google Scholar]
- Varelas X, Sakuma R, Samavarchi-Tehrani P, Peerani R, Rao BM, Dembowy J, Yaffe MB, Zandstra PW, Wrana JL. 2008. TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat Cell Biol 10: 837–848. [DOI] [PubMed] [Google Scholar]
- Vinals F, Pouysségur J. 2001. Transforming growth factor β1 (TGF-β1) promotes endothelial cell survival during in vitro angiogenesis via an autocrine mechanism implicating TGF-α signaling. Mol Cell Biol 21: 7218–7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell DS, Liberati NT, Guo X, Frederick JP, Wang XF. 2004. Casein kinase Iɛ plays a functional role in the transforming growth factor-β signaling pathway. J Biol Chem 279: 29236–29246. [DOI] [PubMed] [Google Scholar]
- Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. 2001. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412: 346–351. [DOI] [PubMed] [Google Scholar]
- Wang G, Matsuura I, He D, Liu F. 2009a. Transforming growth factor-β-inducible phosphorylation of Smad3. J Biol Chem 284: 9663–9673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Degerny C, Xu M, Yang XJ. 2009b. YAP, TAZ, and Yorkie: A conserved family of signal-responsive transcriptional coregulators in animal development and human disease. Biochem Cell Biol 87: 77–91 [DOI] [PubMed] [Google Scholar]
- Wang G, Yu Y, Sun C, Liu T, Liang T, Zhan L, Lin X, Feng XH. 2015. STAT3 selectively interacts with Smad3 to antagonize TGF-β. Oncogene 10.1038/onc.2015.446. [DOI] [Google Scholar]
- Wilkes MC, Mitchell H, Penheiter SG, Dore JJ, Suzuki K, Edens M, Sharma DK, Pagano RE, Leof EB. 2005. Transforming growth factor-β activation of phosphatidylinositol 3-kinase is independent of Smad2 and Smad3 and regulates fibroblast responses via p21-activated kinase-2. Cancer Res 65: 10431–10440. [DOI] [PubMed] [Google Scholar]
- Wong C, Rougier-Chapman EM, Frederick JP, Datto MB, Liberati NT, Li JM, Wang XF. 1999. Smad3-Smad4 and AP-1 complexes synergize in transcriptional activation of the c-Jun promoter by transforming growth factor β. Mol Cell Biol 19: 1821–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Liu Y, Zheng Y, Dong J, Pan D. 2008. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev Cell 14: 388–398. [DOI] [PubMed] [Google Scholar]
- Xanthos JB, Kofron M, Tao Q, Schaible K, Wylie C, Heasman J. 2002. The roles of three signaling pathways in the formation and function of the Spemann organizer. Development 129: 4027–4043. [DOI] [PubMed] [Google Scholar]
- Xia ZP, Sun L, Chen X, Pineda G, Jiang X, Adhikari A, Zeng W, Chen ZJ. 2009. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature 461: 114–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H, Ooi LL, Hui KM. 2013. MicroRNA-216a/217-induced epithelial-mesenchymal transition targets PTEN and SMAD7 to promote drug resistance and recurrence of liver cancer. Hepatology 58: 629–641. [DOI] [PubMed] [Google Scholar]
- Xu X, Han J, Ito Y, Bringas P Jr, Deng C, Chai Y. 2008. Ectodermal Smad4 and p38 MAPK are functionally redundant in mediating TGF-β/BMP signaling during tooth and palate development. Dev Cell 15: 322–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K, Shirakabe K, Shibuya H, Irie K, Oishi I, Ueno N, Taniguchi T, Nishida E, Matsumoto K. 1995. Identification of a member of the MAPKKK family as a potential mediator of TGF-β signal transduction. Science 270: 2008–2011. [DOI] [PubMed] [Google Scholar]
- Yamashita M, Fatyol K, Jin C, Wang X, Liu Z, Zhang YE. 2008. TRAF6 mediates Smad-independent activation of JNK and p38 by TGF-β. Mol Cell 31: 918–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh YY, Chiao CC, Kuo WY, Hsiao YC, Chen YJ, Wei YY, Lai TH, Fong YC, Tang CH. 2008. TGF-β1 increases motility and αvβ3 integrin up-regulation via PI3K, Akt and NF-κB-dependent pathway in human chondrosarcoma cells. Biochem Pharmacol 75: 1292–1301. [DOI] [PubMed] [Google Scholar]
- Ying QL, Nichols J, Chambers I, Smith A. 2003. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 115: 281–292. [DOI] [PubMed] [Google Scholar]
- Yingling JM, Datto MB, Wong C, Frederick JP, Liberati NT, Wang XF. 1997. Tumor suppressor Smad4 is a transforming growth factor β-inducible DNA binding protein. Mol Cell Biol 17: 7019–7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida-Koide U, Matsuda T, Saikawa K, Nakanuma Y, Yokota T, Asashima M, Koide H. 2004. Involvement of Ras in extraembryonic endoderm differentiation of embryonic stem cells. Biochem Biophys Res Commun 313: 475–481. [DOI] [PubMed] [Google Scholar]
- Yu FX, Guan KL. 2013. The Hippo pathway: Regulators and regulations. Genes Dev 27: 355–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P, Pan G, Yu J, Thomson JA. 2011. FGF2 sustains NANOG and switches the outcome of BMP4-induced human embryonic stem cell differentiation. Cell Stem Cell 8: 326–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamurovic N, Cappellen D, Rohner D, Susa M. 2004. Coordinated activation of notch, Wnt, and transforming growth factor-β signaling pathways in bone morphogenic protein 2-induced osteogenesis. Notch target gene Hey1 inhibits mineralization and Runx2 transcriptional activity. J Biol Chem 279: 37704–37715. [DOI] [PubMed] [Google Scholar]
- Zavadil J, Cermak L, Soto-Nieves N, Bottinger EP. 2004. Integration of TGF-β/Smad and Jagged1/Notch signalling in epithelial-to-mesenchymal transition. EMBO J 23: 1155–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Feng XH, Derynck R. 1998. Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-β-induced transcription. Nature 394: 909–913. [DOI] [PubMed] [Google Scholar]
- Zhang L, Ren F, Zhang Q, Chen Y, Wang B, Jiang J. 2008. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev Cell 14: 377–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Liu CY, Zha ZY, Zhao B, Yao J, Zhao S, Xiong Y, Lei QY, Guan KL. 2009. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. J Biol Chem 284: 13355–13362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhou F, Drabsch Y, Gao R, Snaar-Jagalska BE, Mickanin C, Huang H, Sheppard KA, Porter JA, Lu CX, et al. 2012. USP4 is regulated by AKT phosphorylation and directly deubiquitylates TGF-β type I receptor. Nat Cell Biol 14: 717–726. [DOI] [PubMed] [Google Scholar]
- Zhang L, Zhou F, Garcia de Vinuesa A, de Kruijf EM, Mesker WE, Hui L, Drabsch Y, Li Y, Bauer A, Rousseau A, et al. 2013. TRAF4 promotes TGF-β receptor signaling and drives breast cancer metastasis. Mol Cell 51: 559–572. [DOI] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. 2007. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev 21: 2747–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Lin JD, Wang CY, Chinnaiyan AM, Lai ZC, et al. 2008. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev 22: 1962–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Kim J, Ye X, Lai ZC, Guan KL. 2009. Both TEAD-binding and WW domains are required for the growth stimulation and oncogenic transformation activity of yes-associated protein. Cancer Res 69: 1089–1098. [DOI] [PubMed] [Google Scholar]
- Zhou J, Jain S, Azad AK, Xu X, Yu HC, Xu Z, Godbout R, Fu Y. 2016. Notch and TGFβ form a positive regulatory loop and regulate EMT in epithelial ovarian cancer cells. Cell Signal 28: 838–849. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Ahlemeyer B, Bauerbach E, Krieglstein J. 2001. TGF-β1 inhibits caspase-3 activation and neuronal apoptosis in rat hippocampal cultures. Neurochem Int 38: 227–235. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Culmsee C, Klumpp S, Krieglstein J. 2004. Neuroprotection by transforming growth factor-β1 involves activation of nuclear factor-κB through phosphatidylinositol-3-OH kinase/Akt and mitogen-activated protein kinase-extracellular-signal regulated kinase1,2 signaling pathways. Neuroscience 123: 897–906. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Le Scolan E, Jahchan N, Ji X, Xu A, Luo K. 2016. SnoN antagonizes the Hippo kinase complex to promote TAZ signaling during breast carcinogenesis. Dev Cell 37: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. 2011. mTOR: From growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 12: 21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn AM, Butler K, Gurdon JB. 1999. Anterior endomesoderm specification in Xenopus by Wnt/β-catenin and TGF-β signalling pathways. Dev Biol 209: 282–297. [DOI] [PubMed] [Google Scholar]