Abstract
Pleuromutilins are antibiotics that selectively inhibit bacterial translation and are semisynthetic derivatives of the naturally occurring tricyclic diterpenoid pleuromutilin, which received its name from the pleuromutilin-producing fungus Pleurotus mutilus. Tiamulin and valnemulin are two established derivatives in veterinary medicine for oral and intramuscular administration. As these early pleuromutilin drugs were developed at a time when companies focused on major antibacterial classes, such as the β-lactams, and resistance was not regarded as an issue, interest in antibiotic research including pleuromutilins was limited. Over the last decade or so, there has been a resurgence in interest to develop this class for human use. This has resulted in a topical derivative, retapamulin, and additional derivatives in clinical development. The most advanced compound is lefamulin, which is in late-stage development for the intravenous and oral treatment of community-acquired bacterial pneumonia and acute bacterial skin infections. Overall, pleuromutilins and, in particular, lefamulin are characterized by potent activity against Gram-positive and fastidious Gram-negative pathogens as well as against mycoplasmas and intracellular organisms, such as Chlamydia spp. and Legionella pneumophila. Pleuromutilins are unaffected by resistance to other major antibiotic classes, such as macrolides, fluoroquinolones, tetracyclines, β-lactam antibiotics, and others. Furthermore, pleuromutilins display very low spontaneous mutation frequencies and slow, stepwise resistance development at sub-MIC in vitro. The potential for resistance development in clinic is predicted to be slow as confirmed by extremely low resistance rates to this class despite the use of pleuromutilins in veterinary medicine for >30 years. Although rare, resistant strains have been identified in human- and livestock-associated environments and as with any antibiotic class, require close monitoring as well as prudent use in veterinary medicine. This review focuses on the structural characteristics, mode of action, antibacterial activity, and resistance development of this potent and novel antibacterial class for systemic use in humans.
Pleuromutilins are potent antibiotics that have been used in veterinary medicine for several decades. Recent efforts to develop derivatives for human use (e.g., topical retapamulin and systemical lefamulin) have been successful.
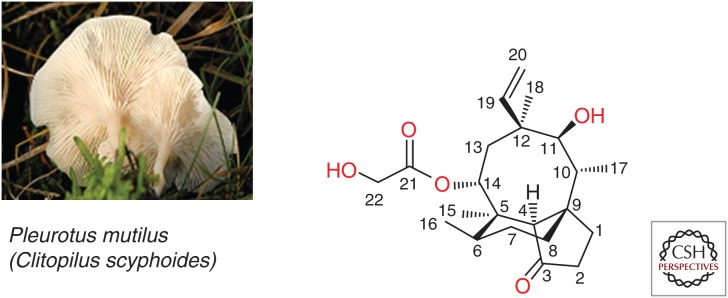
Pleuromutilins are a well-known class of antibiotics discovered in the 1950s by the isolation of the naturally occurring pleuromutilin from Pleurotus mutilus (now renamed Clitophilus scyphoides), an edible mushroom (Fig. 1) (Kavanagh et al. 1951). Semisynthetic derivatizations have led to tiamulin and valnemulin, which were introduced to veterinary medicine in 1979 and 1999, respectively, for the treatment of pulmonary and intestinal infections caused by Mycoplasma spp., Brachyspira spp., and Lawsonia intracellularis in pigs, poultry, and, to some extent, in rabbits. Despite the use of pleuromutilins for treatment in veterinary medicine for more than three decades, resistance development has been uncommon. This can likely be attributed to several factors including the unique and highly specific mode of action of the pleuromutilins. Further, this class has not been used for enhancement of food-producing animal production (e.g., as growth promoters or for enhancement of feed efficiency) unlike the tetracyclines, penicillins, or sulfonamides (EMA 2014a,b). Even though oral valnemulin has been reported to be efficacious in the treatment of persistent or life-threatening mycoplasma infection in humans (Heilmann et al. 2001), no pleuromutilin had received marketing authorization by the end of the last century.
Figure 1.
Structure of pleuromutilin with numbering system (Arigoni 1968). (Left panel from Lindsey 2006.)
In the new millennium, interest in the pleuromutilin class significantly increased as evidenced by the development of new derivatives for human use. Retapamulin, a topical agent, was the first to be approved for the treatment of impetigo and infected small lacerations, abrasion or sutured wounds caused by Staphylococcus aureus and Streptococcus pyogenes (FDA 2007; EMA 2008). More recently, lefamulin, the first pleuromutilin for intravenous and oral administration, has entered into late-stage clinical development for the treatment of community-acquired bacterial pneumonia (CABP) and acute bacterial skin and skin structure infections (ABSSSIs). BC-7013 and BC-3205, which have a similar antibacterial profile but differ in ADMET properties, are in early-stage clinical development for topical and oral administration, respectively. In addition, recent research has been directed at further extending the antibacterial spectrum to include the ESKAPE pathogens (Boucher et al. 2009; Paukner et al. 2014a,b, 2015a,b,c; Strickmann et al. 2014; Wicha and Ivezic-Schoenfeld 2014; Wicha et al. 2015b).
To the question “Are pleuromutilins finally fit for human use?” (Novak 2011), which has been raised because of unjustified anecdotal concerns regarding metabolic stability, gastrointestinal side effects, cardiac safety, or intravenous tolerability, a clear response can be given: Yes. In a phase 2 study, lefamulin was well-tolerated and showed comparable efficacy to IV vancomycin in patients with ABSSSI (Prince et al. 2013). Despite challenging medicinal chemistry, a number of compounds are in the pipeline and further developments in this antibiotic class are anticipated.
PLEUROMUTILINS—MODE OF ACTION, ACTIVITY, AND RESISTANCE
Structure
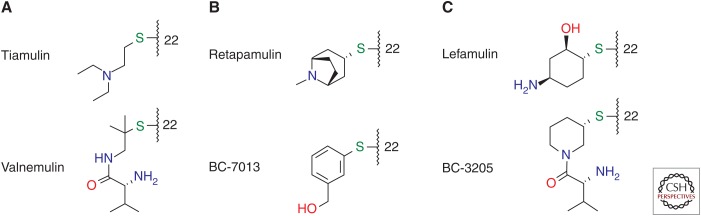
The diterpenoid pleuromutilin comprises a tricyclic scaffold with unique annelation of a five-, six-, and eight-membered ring and eight stable chiral centers, as well as a glycolic ester moiety forming the side chain also regarded as an extension at position C14 (Fig. 1) (Anchel 1952; Arigoni 1962, 1968; Birch et al. 1963, 1966). Remarkable efforts have been made to achieve chemical modifications at several positions of the tricyclic core (Naegeli 1961; Berner et al. 1980, 1981, 1983, 1987; Brooks and Hunt 2000; Bacqué et al. 2002; Springer et al. 2003, 2008; Takadoi et al. 2007; Wang et al. 2009; Paukner et al. 2015b), as well as biotransformation (Hanson et al. 2002), and the total synthesis of this unique scaffold (Gibbons 1982; Paquette and Bullman-Page 1985; Paquette and Wiedemann 1985; Bacqué et al. 2003; Liu et al. 2011; Lotesta et al. 2011; Ruscoe et al. 2015). Most modifications, however, are primarily performed at the glycolic side chain of pleuromutilin with replacement of the terminal hydroxyl group or the entire side chain resulting in semisynthetic pleuromutilin analogs of two main types: (1) the flexible sulfanylacetyl, and (2) the rigid acylcarbamate linker type. Despite significant efforts in the field of acylcarbamate pleuromutilins by the GlaxoSmithKline group (Hunt 2000; Brooks et al. 2001; Andemichael et al. 2009), only sulfanylacetyl derivatives, mostly with one basic function at the side chain, have progressed beyond phase 1 clinical studies. The early work of the Sandoz group resulted in lipophilic orally available veterinary products, tiamulin (Egger and Reinshagen 1978) and valnemulin (Fig. 2A) (Berner and Vyplel 1987), whereas work from GlaxoSmithKline and Sandoz/Nabriva led to the lipophilic topical products retapamulin (Berry et al. 1999) and BC-7013 (Fig. 2B) (Thirring et al. 2007). Extensive modification of the C14 side chain culminated in lefamulin (Fig. 2C) (Mang et al. 2008), the first pleuromutilin with optimized physicochemical characteristics, including exceptional solubility, potent antimicrobial activity, and excellent ADMET properties including metabolic stability enabling administration by both the intravenous and oral routes.
Figure 2.
Structures of side chains at C22. Side chains of various pleuromutilin derivatives: (A) veterinary, (B) topical human, and (C) systemical human.
Mode of Action
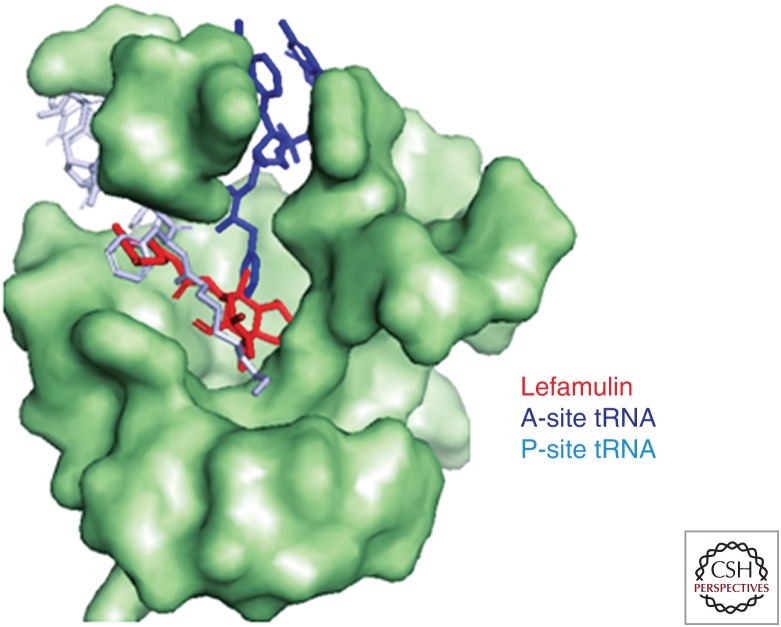
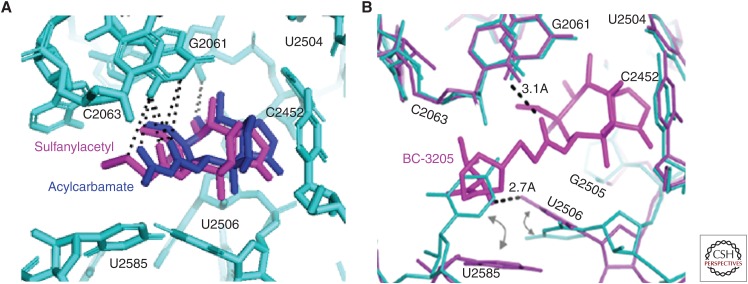
Pleuromutilins inhibit bacterial protein synthesis by binding to the central part of domain V of the 50S ribosomal subunit at the peptidyl transferase center (PTC), which prevents the correct positioning of the CCA ends of tRNAs for peptide transfer in the A- and P-site, thereby inhibiting peptide bond formation (Hogenauer 1975; Hogenauer and Ruf 1981; Hogenauer et al. 1981; Poulsen et al. 2001; Schlunzen et al. 2004; Long et al. 2006; Davidovich et al. 2007). Figure 3 shows the positioning of lefamulin in the PTC of the bacterial ribosome in relation to the positions of A- and P-site tRNA. Positioning is similar for various pleuromutilin derivatives in that the tricyclic core is located in a pocket close to the A-site, whereas the C14 side chain extends toward the P-site hindering the 3′-end tRNA A- to P-site rotary motion, as shown by various footprinting and crystallographic studies for tiamulin, valnemulin (Poulsen et al. 2001; Schlunzen et al. 2004; Long et al. 2006; Davidovich et al. 2007), retapamulin (Yan et al. 2006), lefamulin (Nabriva, unpubl.), and BC-3205 (Eyal et al. 2015). Crystallography data using ribosomal preparations from Deinococcus radiodurans and S. aureus show that the tricyclic pleuromutilin core interacts with the ribosomal nucleotides mainly through hydrophobic interactions, van der Waal forces, and hydrogen bonds with nucleotides of domain V of 23S rRNA, namely, A2503, U2504, G2505, U2506, C2452, and U2585 (Fig. 4). For the tricyclic pleuromutilin core, specific hydrogen bonds have been reported for the hydroxyl group at C11 with nucleotides G2505 or A2503 (Davidovich et al. 2007) and for the hydroxyl group at C2 (present in only few selected derivatives) with G2505. Further, hydrogen bonds have been reported for the C21 carboxyl group and the sulfur in sulfanylacetyl or acyl in acylcarbamate of the C14 extension (linker) with the nucleotide G2061, which are similar for both linker types (Schlunzen et al. 2004; Davidovich et al. 2007). In previous studies using D. radiodurans ribosomes, it was concluded that the rest of the C14 extension is involved only in minor hydrophobic interactions (Davidovich et al. 2007). Recent studies using S. aureus ribosomes, however, clearly showed additional hydrogen bonds of the C14 extensions, specifically the amino groups of BC-3205 and lefamulin, with the nucleotides U2506 (Eyal et al. 2015) and A2062 (A Yonath, Z Eyal, E Zimmerman, et al., unpubl.), respectively. Most notably, the C14 extensions of all pleuromutilins sterically interfere with the highly flexible nucleotides U2585 and U2506 causing rotational movements of these nucleotides, which consequently interact with each other by the formation of one or more additional hydrogen bonds or at least van der Waal or similar interactions. This closing of the binding pocket around pleuromutilins, also regarded as the induced-fit mechanism, tightens the binding of pleuromutilins to the ribosome (Davidovich et al. 2007; Eyal et al. 2015) and leads to the protection of these nucleotides in footprinting experiments (Schlunzen et al. 2004). Interestingly, the amino group of the C14 extension of BC-3205 forms a hydrogen bond with U2506 in S. aureus causing a larger shift of U2506, consequently further stabilizing BC-3205 in the binding pocket and indicating a better fit of this molecule in the pocket (Eyal et al. 2015).
Figure 3.
Lefamulin positioning in the peptidyl transferase center (PTC). PTC of the bacterial ribosome in relation to the positions of A- and P-site tRNA. Red, Lefamulin; blue, A-site tRNA; teal, P-site tRNA.
Figure 4.
Interaction network. (A) The tricyclic pleuromutilin core and sulfanylacetyl as well as acylcarbamate linker (side chains omitted for clarity), and (B) the C14 side chain extension with nucleotides of the peptidyl transferase center (PTC). Hydrogen bonds are shown as dotted lines (Schlunzen et al. 2004; Davidovich et al. 2007; Eyal et al. 2015).
It has been further hypothesized that pleuromutilins might also interfere with translation initiation or at an early point of the elongation cycle with particular sensitivity of the first peptide bond formation (Hunt 2000; Novak 2011). This is based on the fact that (1) radiolabeled tiamulin did not bind to the 50S subunit of the ribosome once elongation has begun, (2) addition of tiamulin to intact cells led to the formation of defective initiation complexes reflected by the depletion of the polysome pool through blockage of reinitiation (Dornhelm and Hogenauer 1978), and (3) retapamulin partially inhibited binding of fMet-tRNA during initiation complex formation (Yan et al. 2006).
Coupled in vitro transcription/translation (TT) assays with bacterial ribosomes have shown high specificity of pleuromutilins for the inhibition of bacterial protein translation, whereas no effect on eukaryotic nonorganelle protein synthesis was observed for tiamulin and retapamulin (Yan et al. 2006). The specificity for bacterial protein synthesis was also confirmed for lefamulin with IC50 values of 0.51 µM and 0.31 µM in Escherichia coli and S. aureus TT-assays, respectively, whereas the IC50 in the eukaryotic TT-assay was with 952 µM >2000-fold higher than the IC50 in the bacterial system. Cycloheximide, a TT inhibitor of eukaryotic protein synthesis, and puromycin, a nonspecific inhibitor of bacterial and eukaryotic TT, were used as controls. Comparison of IC50 values of various pleuromutilin derivatives showed a slight trend of higher IC50 for E. coli than for S. aureus (Table 1; Nabriva, unpubl.). It further showed that the IC50 does not necessarily correlate with high antibacterial activity; for example, BC-7013, which has the highest IC50, was one of the most active compounds in vitro (Biedenbach et al. 2009). Additional important factors other than translation inhibition, such as intracellular concentration, uptake, or efflux, contribute to in vitro antimicrobial activity as well (Paukner et al. 2014b).
Table 1.
Inhibition of bacterial and eukaryotic in vitro transcription–translation by various pleuromutilin derivatives
| IC50 (CI95) (µM) | |||
|---|---|---|---|
| Compound | Escherichia coli | Staphylococcus aureus | Eukaryotic (reticulocyte lysate system) |
| Pleuromutilin | 0.76 (0.63–0.92) | 1.73 (1.22–2.44) | ND |
| Tiamulin | 0.50 (0.44–0.57) | 0.36 (0.32–0.42) | ND |
| Valnemulin | 0.59 (0.54–0.66) | 0.38 (0.35-0.41) | ND |
| Retapamulin | 0.69 (0.64–0.76) | 0.35 (0.32–0.39) | 850 (562–1287) |
| Lefamulin | 0.51 (0.45–0.57) | 0.31 (0.29–0.33) | 952 (732–1238) |
| BC-3205 | 0.62 (0.56–0.68) | 0.49 (0.44–0.54) | >100 |
| BC-7013 | 0.74 (0.65–0.83) | 0.64 (0.59–0.69) | >100 |
| Cycloheximide | >100 | >100 | 0.44 (0.29–0.68) |
| Puromycin | 0.39 (0.34–0.46) | 0.19 (0.16–0.23) | 0.31 (0.27–0.36) |
Unpublished data (Nabriva).
ND, not determined.
Antibacterial Activity
The antibacterial spectrum of the pleuromutilins is characterized by potent activity against Gram-positive organisms including staphylococcal species (e.g., community-acquired methicillin-resistant S. aureus [CA-MRSA], hospital-acquired MRSA [HA-MRSA], vancomycin-resistant S. aureus [VRSA], vancomycin-intermediate S. aureus [VISA], heteroresistant VISA [hVISA]), streptococcal species (e.g., penicillin-resistant Streptococcus pneumoniae [PRSP], multidrug-resistant S. pneumoniae [MDR-SP]), and Enterococcus faecium (particularly vancomycin-resistant strains [VRE]), as well as activity against fastidious Gram-negatives, including Haemophilus spp., Moraxella catarrhalis, Neisseria spp., and Legionella pneumophila (Rittenhouse et al. 2006; Sader et al. 2012a,b; Paukner et al. 2013c). Pleuromutilins also display potent activity against mycoplasmas, ureaplasmas, chlamydia (Hannan et al. 1997), and Brachispira hyodysenteriae (Karlsson et al. 2001). Activity against anaerobic organisms has been seen for retapamulin, lefamulin, and BC-7013, including Propionibacterium acnes (Goldstein et al. 2006), Peptostreptococcus spp., Prevotella spp., Porphyromonas spp., Fusobacterium spp., and Clostridium perfringens, whereas pleuromutilins generally show weak activity against strains from the Bacteroides fragilis group (Odou et al. 2007; Paukner et al. 2013a). Pleuromutilin activity against Clostridium difficile varies and is dependent on the C14 side chain; retapamulin and lefamulin possess no relevant to weak activity, whereas BC-7013 has potent activity with 78% of isolates inhibited at concentrations of ≤1 µg/mL (Nabriva, unpubl.). The lack of lefamulin activity against B. fragilis group and Enterobacteriaceae is anticipated to result in limited disruption to the normal gastrointestinal microbiome and potentially a lower propensity to be associated with C. difficile infection (CDI). Further studies are warranted to assess the effect of the pleuromutilins on the gut microbiome and its impact on CDI.
No relevant activity was observed against Enterococcus faecalis, Enterobacteriaceae, and nonfermenting Gram-negatives, such as Acinetobacter baumannii and Pseudomonas aeruginosa, although coupled in vitro transcription–translation assay results showed inhibition of the bacterial translation in these organisms and would suggest also antibacterial in vitro activity (Nabriva, unpubl.). Research at Nabriva revealed that the intrinsic resistance of Enterobacteriaceae is caused by the efflux of pleuromutilins mediated by the AcrAB-TolC efflux pump. This is supported by the fact that AcrAB-TolC deficient E. coli strains were susceptible to pleuromutilins (Paukner et al. 2014b) and that the minimum inhibitory concentration (MIC) values against Enterobacteriaceae were significantly reduced by the addition of efflux pump inhibitors (e.g., PAβN). Furthermore, recent new pleuromutilin derivatives partially overcome efflux and show increased activity against Enterobacteriaceae, including carbapenem-resistant isolates. These so-called “extended spectrum pleuromutilins” (ESPs) are characterized by the modification of the tricyclic pleuromutilin core at C12 (Paukner et al. 2014a,b, 2015a,b,c; Strickmann et al. 2014; Wicha and Ivezic-Schoenfeld 2014; Wicha et al. 2015b). Further investigations are needed to identify the mechanism(s) responsible for decreased susceptibility in nonfermenting organisms and E. faecalis.
Given that lefamulin is the first in-class IV and oral pleuromutilin antibiotic to advance into late-stage clinical development, additional information is provided on its activity. The antibacterial spectrum of lefamulin against a recent strain collection is well matched to the profile required for the empiric treatment of patients with CABP and ABSSSI (Table 2). Notably, lefamulin was equipotent against S. aureus strains that were community-acquired or healthcare-associated and its activity was not negatively influenced by the presence of Panton-Valentine leukocidin (PVL). Compared with macrolides, lincosamides, fluoroquinoloes, tetracyclines, β-lactams, linezolid, and vancomycin, lefamulin was among the most active compounds in vitro and the lefamulin activity was unaffected by resistance or multidrug resistance to these antibiotic classes (Paukner et al. 2013c). Lefamulin’s activity is not adversely affected by presence of serum (≤95% v/v) or lung surfactant (≤1 mg/mL Survanta; 4% v/v). Lefamulin has shown high intracellular concentration in macrophages (Paukner et al. 2013b), achieves excellent penetration into human tissues including epithelial lining fluid of the lung (Zeitlinger et al. 2016), has oral bioavailability, potent in vivo efficacy in skin and pulmonary infection mouse models (Wicha et al. 2010, 2013, 2015a), and possesses a low potential for resistance development. Moreover, lefamulin has shown efficacy in patients with ABSSSI infection caused primarily by MRSA (including PVL-producing CA-MRSA) comparable to that of vancomycin (Prince et al. 2013). To date, lefamulin has been well tolerated in phase 1 and 2 clinical studies involving exposure of more than 400 subjects. Lefamulin also possesses potent activity (MIC90 values ≤2 µg/ml) against organisms causing sexually transmitted infections (STIs), including resistant Neisseria gonorrhoeae, Mycoplasma genitalium, Chlamydia trachomatis, or Haemophilus ducreyi, warranting further evaluation of this drug for treatment of STIs (Paukner et al. 2013a).
Table 2.
Antibacterial activity of lefamulin and comparators
| MIC90 (μg/mL) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | n | LMU | ERY | CLI | DOX | TGC | LZD | VAN | DAP | LEV | PEN |
| Organisms causing predominantly SSSI and bacteremia | |||||||||||
| Staphylococcus aureus | 5527 | 0.12 | >4 | >2 | 0.25 | 0.25 | 1 | 1 | 0.5 | >4 | – |
| MSSA | 3157 | 0.12 | >4 | ≤0.25 | 0.25 | 0.25 | 2 | 1 | 0.5 | 1 | – |
| MRSA | 2370 | 0.25 | >4 | >2 | 1 | 0.25 | 1 | 1 | 0.5 | >4 | – |
| CoNS | 878 | 0.12 | >4 | >2 | 2 | 0.25 | 1 | 2 | 0.5 | >4 | – |
| Enterococcus faecium | 536 | 4 | >4 | – | >8 | 0.25 | 1 | >16 | 2 | >4 | – |
| Vancomycin-nonsusceptible | 304 | 0.25 | >4 | – | >8 | 0.25 | 1 | >16 | 2 | >4 | – |
| β-Haemolytic Streptococcus spp. | 763 | 0.03 | >4 | >2 | 8 | 0.06 | 1 | 0.5 | 0.25 | 1 | 0.06 |
| Viridans group Streptococcus spp. | 245 | 0.5 | >4 | ≤0.25 | >8 | 0.06 | 1 | 0.5 | 0.5 | 2 | 0.5 |
| n | LMU | ERY | AZI | DOX | TGC | LEV | SXT | CXM | IMI | PEN | |
| Organisms causing predominantly RTI | |||||||||||
| Streptococcus pneumoniaea | 1473 | 0.25 | >4 | >4 | 8 | 0.06 | 1 | >4 | 8 | 0.5 | 4 |
| Haemophilus influenzaeb | 360 | 2 | 8 | 2 | 0.5 | 0.25 | – | >4 | 2 | 1 | – |
| Moraxella catarrhalis | 253 | 0.25 | 0.25 | ≤0.25 | 0.25 | 0.25 | – | ≤0.5 | 2 | ≤0.12 | >4 |
| Legionella pneumophilac | 30 | 0.5 | 0.25 | 0.12 | – | – | 0.12 | – | – | – | – |
| Mycoplasma pneumoniae | 50 (4)d | 0.006 | 0.0025–0.005f | ≤0.0003f | 0.04–0.04f | – | – | – | – | – | – |
| Chlamydia pneumoniae | 50 (2)e | 0.04 | 0.04–0.16f | 0.08–0.16f | 0.04–0.08f | – | – | – | – | – | – |
Data adapted from Paukner et al. (2013c) and Sader (2012b).
MRSA, Methicillin-resistant S. aureus; MSSA, methicillin-susceptible S. aureus; CoNS, coagulase-negative Staphylococcus spp.; AZI, azithromycin; CLI, clindamycin; CXM, cefuroxime; DAP, daptomycin; DOX, doxycycline; ERY, erythromycin; IMI, imipenem; LEV, levofloxacin; LMU, lefamulin; LZD, linezolid; PEN, penicillin; SXT, trimethoprim-sulfamethoxazole; TGC, tigecycline; VAN, vancomycin.
a61.3% of S. pneumoniae isolates were penicillin-susceptible.
b23.6% of H. influenzae isolates were β-lactamase-positive.
cMICs determined by agar dilution using charcoal-supplemented BCYEα medium.
dLMU was tested against n = 50 isolates, whereas comparators were tested against n = 4 isolates.
eLMU was tested against n = 50 isolates, whereas comparators were tested against n = 2 isolates.
fRange of MICs for isolates tested.
Resistance and Cross-Resistance
The unique mode of action of pleuromutilins and the binding to highly conserved ribosomal targets implies a low probability of resistance development and lack of cross-resistance with other antibiotic classes including protein synthesis inhibitors, such as macrolides, ketolides, or fusidic acid (Yan et al. 2006). The binding sites and mode of action of pleuromutilins can be clearly differentiated from those of oxazolidinones, lincosamides, phenicols, and streptogramins; however, pleuromutilins also have partly overlapping interaction sites with these antibacterials (Schlunzen et al. 2004). Consequently, resistance mechanisms exist that can mediate cross-resistance with these antibacterials, albeit with an exceedingly low incidence.
Pleuromutilins have shown a low potential for resistance development in vitro as shown in various studies for tiamulin and valnemulin in Brachyspira spp. (Karlsson et al. 2001; Pringle et al. 2004), Mycoplasma spp. (Long et al. 2009; Li et al. 2011), S. aureus and E. coli (Miller et al. 2008), for retapamulin in S. aureus and S. pyogenes (Kosowska-Shick et al. 2006; Gentry et al. 2007), and for lefamulin in S. aureus, S. pneumoniae, and S. pyogenes (Paukner et al. 2012). Generally, the spontaneous mutation frequencies are low (≤10−9) with no stable resistant mutants selected at four- to eightfold MIC. In multipassage experiments, resistance developed in a slow and step-wise manner with multiple mutations required to cause high-level resistance. Mutations in 23S rRNA, rplC, and rplD genes encoding the large ribosomal proteins L3 and L4, have been identified as the primary resistance mechanism in vitro. In clinical isolates, two additional resistance mechanisms have been identified: the acquisition of vga(A) encoded or related ATP-binding cassette (ABC)-F transporters and the acquisition of cfr encoding the Cfr methyltransferase. The common denominator is the alteration of the pleuromutilin target site.
Mutations in the 23S RNA gene (rrn) at positions G2032A, C2055A, A2058, A2058G, A2059G, G2061U, G2447A/U, C2499A, A2503U, U2504A/G, and A2572U were primarily observed in laboratory-selected Brachyspira spp., Mycoplasma spp., and in clinical isolates. Mutations in 23S rRNA have been described to confer resistance only in Mycoplasma spp. and Brachyspira spp., which only have a single copy of 23S rRNA, whereas staphylococcal and streptococcal species have multiple copies (Pringle et al. 2004; Miller et al. 2008; Long et al. 2009; Li et al. 2010; Hillen et al. 2014). Experiments with single-copy rrn knockout strains of E. coli illustrated that “the copy number of 23S rRNA is the limiting factor in the selection of 23S rRNA mutants” (Miller et al. 2008). In Brachyspira, resistance is often associated with additional mutations in rplC (Pringle et al. 2004; Long et al. 2009; Li et al. 2010, 2011; Hidalgo et al. 2011).
Mutations and deletions in the rplC and rplD genes, although L3 and L4 do not directly interact with the pleuromutilins, can cause conformational changes in the PTC and hinder correct positioning of the pleuromutilins in the pocket formed between the nucleotides G2576 with U2506 and G2505 (Eyal et al. 2015). Mutations in rplC and rplD have been described for Staphylococcus spp. (Pringle et al. 2004; Kosowska-Shick et al. 2006; Gentry et al. 2007; Miller et al. 2008; Paukner et al. 2012) and together with mutations in 23S rRNA for B. hyodysenteriae (Hillen et al. 2014). Notably, mutations in rplC have also been associated with considerable loss of fitness (Gentry et al. 2007). Pleuromutilin resistance by mutational changes in rplC and 23S rRNA develops gradually and in a stepwise manner both in vitro and in vivo, suggesting that multiple mutations are needed to achieve high-level resistance. (Karlsson et al. 2001; Gentry et al. 2007; Miller et al. 2008; Hidalgo et al. 2011; Paukner et al. 2012).
ABC-F transporters encoded by vga(A) and its variants vga(A)v, vga(A)LC, vga(B), vga(C), vga(D), vga(E), and lsa(E) have been described to confer resistance to pleuromutilins, streptogramin A, and lincosamides in Staphylococcus spp., E. faecium, and Erysypelothrix rhusiopathiae. Isolates were collected almost exclusively from animal species, predominantly swine (Kadlec and Schwarz 2009; Kadlec et al. 2010; Overesch et al. 2011; Schwendener and Perreten 2011; Li et al. 2013, 2014a,b; Zhang et al. 2015). MRSA isolates collected from humans appeared to be related to animal-associated lineages of S. aureus such as ST398 (Lozano et al. 2012). Recent studies concluded that ABC-F transporters, which lack a transmembrane domain, likely mediate resistance by the interference of translation at the PTC and by the action as efflux transporter. This is based on the homology of vga(A) and variants with the ABC-F transporter EttA, which is a translation factor binding to the tRNA exit site (E-site) (Lenart et al. 2015).
Last, the rarely encountered methyltransferase Cfr, methylating the nucleotide A2503 of 23S rRNA, can confer resistance. Because of steric hindrance, binding of phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramins (PhLOPS antibiotics) is prohibited, which results in the PhLOPS-resistance phenotype. The cfr gene was originally identified in coagulase-negative staphylococci from animals and has been detected mostly in livestock-associated staphylococci (Kehrenberg et al. 2005, 2009; Alba et al. 2015; Feltrin et al. 2015; Moon et al. 2015) but, more recently, has also been found in a limited number of staphylococcal isolates from humans including one outbreak of a cfr-positive MRSA in a Spanish hospital, which was terminated by reduction of linezolid use and infection-control measures (Sanchez et al. 2010; Shore et al. 2010, 2016). Cfr was also found in nonstaphylococcal species collected, with the exception of E. faecalis, exclusively from livestock animals and related farm environments: one out of 1230 E. coli isolates collected from pigs, ducks, and chickens in China, in one Proteus vulgaris out of 557 nasal swabs of Chinese swine and a porcine Bacillus spp., as well as a Macrococcus caseolyticus and Jeotgalicoccus pinnipedialis isolate (Wang et al. 2011, 2012a,b,c,d). Cfr has also been detected in an E. faecalis isolate collected from a Chinese animal as well as in an animal-associated isolate from a patient in Thailand. Cfr has been located on the chromosome and on various plasmids or transferrable elements indicating the ability to spread (Locke et al. 2012; Shen et al. 2013; Li et al. 2015; Shore et al. 2016). In vitro, the cfr-carrying plasmid isolated from human E. faecalis was only transferrable by conjugation to another E. faecalis laboratory strain, whereas it was not transferrable to S. aureus or E. faecium (Diaz et al. 2012). Recently, the transferability of cfr-carrying plasmids from S. epidermidis to MRSA by conjugation or transduction was shown, indicating a role of S. epidermidis as a potential reservoir for cfr spread (Cafini et al. 2016).
Most importantly, it should be noted that despite the characterization of isolates resistant to pleuromutilins and the descriptions of mechanisms conferring resistance, the rate of resistance to pleuromutilins remains low. In the SENTRY surveillance program conducted with lefamulin in 2010, the total incidence of pleuromutilin resistance was 0.18% for S. aureus and 3.4% for coagulase-negative Staphylococcus spp. Among the S. aureus isolates, 0.018% harbored the cfr gene, 0.11% harbored the vga(A) gene, and 0.05% had mutations in rplC. Among coagulase-negative Staphylococcus spp., the incidences for cfr, vga(A), and rplD alterations were 0.11%, 2.5%, and 0.34%, respectively (Paukner et al. 2013c). The low prevalence of cfr is consistent with data collected in the course of linezolid-resistance monitoring (Table 3).
Table 3.
Cfr-positive isolates collected from human in the course of global surveillance studies
| Investigation period | Number of resistant Staphylococcus aureus isolates per total screened isolates (%) | Number of resistant CoNS isolates per total screened isolates (%) | Country, surveillance program | References |
|---|---|---|---|---|
| 2015 | 0/2434 (0%) | 1/465 (0.22%) | Europe, ZAAPS | Flamm et al. 2016 |
| 2014 | 0/3560 (0%) | 0/956 (0%) | Worldwide, ZAAPS | Mendes et al. 2016 |
| 2012 | 1/4077 (0.02%) | 3/905 (0.33%) | Worldwide, ZAAPS | Mendes et al. 2014 |
| 2011 | 0/3884 (0%) | 3/928 (0.32%) | Worldwide, ZAAPS | Flamm et al. 2013 |
| 2010 | 1/5527 (0.018%) | 2/823 (0.24%) | Worldwide, lefamulin, SENTRY | Paukner et al. 2013c |
| 2010 | 0/2875 (0%) | 2/855 (0.23%) | Worldwide, ZAAPS | Flamm et al. 2012 |
| 2002–2009 | 0/5952 (0%) | 2/2132 (0.09%) | Worldwide, ZAAPS | Ross et al. 2011 |
| 2007 | 0/3000 (0%) | 0/716 | Worldwide, ZAAPS | Jones et al. 2009 |
| 1999–2010 | 1/2215 (0.045%) | ND | Spain | Sierra et al. 2013 |
Note: Annual Appraisal of Potency and Spectrum (ZAAPS) Program and SENTRY Surveillance Program.
CoNS, coagulase-negative Staphylococcus spp.
CONCLUDING REMARKS
In summary, pleuromutilins display potent antibacterial activity against a variety of Gram-positive, fastidious Gram-negative and atypical respiratory bacterial pathogens—a profile well suited to treat human infections, including CABP, ABSSSI, and STI. Chemical modifications have led to derivatives with optimized physicochemical properties and improved ADME properties, allowing for intravenous and oral dosing in humans; of these analogs, lefamulin is the most advanced in clinical development. The incidence of pleuromutilin-resistant bacterial isolates is low despite the use of tiamulin and valnemulin in veterinary medicine for more than 30 years. The availability of topical retapamulin in human medicine since 2007 and selection pressure for cfr by the use of linezolid over the past two decades does not appear to have had a major effect on the incidence of pleuromutilin-resistant bacterial isolates among organisms causing infections in humans. Nevertheless, close monitoring of resistance development to pleuromutilins is warranted, along with prudent use of oxazolidinones and veterinary pleuromutilins to maintain low resistance rates and retain the potent activity of this novel antibacterial class against pathogens that have acquired resistance to other established antibiotic classes.
ACKNOWLEDGMENTS
We thank the Nabriva team and particularly W. W. Wicha for kind support for compilation of this review.
Footnotes
Editors: Lynn L. Silver and Karen Bush
Additional Perspectives on Antibiotics and Antibiotic Resistance available at www.perspectivesinmedicine.org
REFERENCES
- Alba P, Feltrin F, Cordaro G, Porrero MC, Kraushaar B, Argudin MA, Nykasenoja S, Monaco M, Stegger M, Aarestrup FM, et al. 2015. Livestock-associated methicillin resistant and methicillin susceptible Staphylococcus aureus sequence type (CC)1 in European farmed animals: High genetic relatedness of isolates from Italian cattle herds and humans. PLoS ONE 10: e0137143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anchel M. 1952. Chemical studies with pleuromutilin. J Biol Chem 199: 133–139. [PubMed] [Google Scholar]
- Andemichael YW, Chen J, Clawson JS, Dai W, Diederich A, Downing SV, Freyer AJ, Liu P, Oh LM, Patience DB, et al. 2009. Process development for a novel pleuromutilin-derived antibiotic. Org Process Res Dev 13: 729–738. [Google Scholar]
- Arigoni D. 1962. The structure of a new class of terpene. Gazz Chim Ital 92: 884–901. [Google Scholar]
- Arigoni D. 1968. Some studies in the biosynthesis of terpenes and related compounds. Pure Appl Chem 17: 331–348. [DOI] [PubMed] [Google Scholar]
- Bacqué E, Pautrat F, Zard SZ. 2002. A flexible strategy for the divergent modification of pleuromutilin. Chem Commun 20: 2312–2313. [DOI] [PubMed] [Google Scholar]
- Bacqué E, Pautrat F, Zard SZ. 2003. A concise synthesis of the tricyclic skeleton of pleuromtuilin and a new approach to cycloheptenes. Org Lett 5: 325–328. [DOI] [PubMed] [Google Scholar]
- Berner H, Vyplel H. 1987. Pleuromutilin derivatives process for their use and their preparation. U.S. Patent 4,675,330. [Google Scholar]
- Berner H, Schulz G, Schneider H. 1980. Synthesis of an AB-trans-annulated derivative of the tricyclic diterpene pleuromutilin via an intramolecular 1,5-hydride shift. Tetrahedron 36: 1807–1811. [Google Scholar]
- Berner H, Schulz G, Schneider H. 1981. Chemistry of pleuromutilins. II: Synthesis of 12-desvinylpleuromutilin. Tetrahedron 37: 915–919. [Google Scholar]
- Berner H, Vyplel H, Schulz G. 1983. Chemistry of pleuromutilins. VII: Base-induced transannular 1,4-hydride shift in 8-substituted pleuromutilin derivatives. Monatshefte für Chemie 114: 501–507. [Google Scholar]
- Berner H, Vyplel H, Schulz G, Stuchlik P. 1984. Chemistry of pleuromutilins. VIII: Functionalization at C-13 by intramolecular nitrene insertion. Tetrahedron 40: 919–923. [Google Scholar]
- Berner H, Vyplel H, Schulz G, Stuchlik P. 1986. Chemistry of pleuromutilins. XI: Inversion of configuration of the vinyl group at carbon 12 by reversible retro-en-cleavage. Monatshefte für Chemie 117: 1073–1080. [Google Scholar]
- Berner H, Vyplel H, Schulz G. 1987. Chemistry of pleuromutilins. XII: A cyclopropyl conjugated system within the tricyclic skeleton of the diterpene pleuromutilin: Formation and synthetic use. Tetrahedron 43: 765–770. [Google Scholar]
- Berry V, Dabbs S, Frydrych CH, Hunt E, Woodnutt G, Sanderson FD. 1999. Pleuromutilin derivatives as antimicrobials. Patent WO199921855. [Google Scholar]
- Biedenbach DJ, Jones RN, Ivezic-Schoenfeld Z, Paukner S, Novak R. 2009. In vitro antibacterial spectrum of BC-7013, a novel pleuromutilin derivative for topical use in humans. 49th Interscience Conference on Antimicrobial Agents and Chemotherapy (ICAAC) San Francisco, CA, September 12–15. [Google Scholar]
- Birch AJ, Cameron DW, Holzapfel CW, Richards RW. 1963. The diterpenoid nature of pleuromutilin. Chem Ind 5: 374–375. [Google Scholar]
- Birch AJ, Holzapfel CW, Richards RW. 1966. The structure and some aspects of the biosynthesis of pleuromutilin. Tetrahedron 8: 359–387. [Google Scholar]
- Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. 2009. Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 48: 1–12. [DOI] [PubMed] [Google Scholar]
- Brooks G, Hunt E. 2000. Mutilin 14-ester derivatives having antibacterial activity. Patent WO2000037074. [Google Scholar]
- Brooks G, Burgess W, Colthurst D, Hinks JD, Hunt E, Pearson MJ, Shea B, Takle AK, Wilson JM, Woodnutt G. 2001. Pleuromutilins. Part 1: The identification of novel mutilin 14-carbamates. Bioorg Med Chem 9: 1221–1231. [DOI] [PubMed] [Google Scholar]
- Cafini F, Nguyen le TT, Higashide M, Roman F, Prieto J, Morikawa K. 2016. Horizontal gene transmission of the cfr gene to MRSA and Enterococcus: Role of Staphylococcus epidermidis as a reservoir and alternative pathway for the spread of linezolid resistance. J Antimicrob Chemother 71: 587–592. [DOI] [PubMed] [Google Scholar]
- Davidovich C, Bashan A, Auerbach-Nevo T, Yaggie RD, Gontarek RR, Yonath A. 2007. Induced-fit tightens pleuromutilins binding to ribosomes and remote interactions enable their selectivity. Proc Natl Acad Sci 104: 4291–4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz L, Kiratisin P, Mendes RE, Panesso D, Singh KV, Arias CA. 2012. Transferable plasmid-mediated resistance to linezolid due to cfr in a human clinical isolate of Enterococcus faecalis. Antimicrob Agents Chemother 56: 3917–3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornhelm P, Hogenauer G. 1978. The effects of tiamulin, a semisynthetic pleuromutilin derivative, on bacterial polypeptide chain initiation. Eur J Biochem 91: 465–473. [DOI] [PubMed] [Google Scholar]
- Egger H, Reinshagen H. 1978. Pleuromutilin esters. U.S. Patent 4,208,326. [Google Scholar]
- EMA. 2008. EPAR for Altargo (retapamulin). European Medicines Agency, London. [Google Scholar]
- EMA. 2014a. Reflection paper on use of pleuromutilins in food-producing animals in the European Union: Development of resistance and impact on human and animal health. European Medicines Agency, London. [DOI] [PubMed] [Google Scholar]
- EMA. 2014b. Sales of veterinary antimicrobial agents in 26 EU/EEA countries in 2012. European Medicines Agency, London. [Google Scholar]
- Eyal Z, Matzov D, Krupkin M, Wekselman I, Paukner S, Zimmerman E, Rozenberg H, Bashan A, Yonath A. 2015. Structural insights into species-specific features of the ribosome from the pathogen Staphylococcus aureus. Proc Natl Acad Sci 112: E5805–E5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA. 2007. Retapamulin. U.S. Food and Drug Administration, Silver Spring, MD. [Google Scholar]
- Feltrin F, Alba P, Kraushaar B, Ianzano A, Argudin MA, Di Matteo P, Porrero MC, Aarestrup FM, Butaye P, Franco A, et al. 2015. A livestock-associated, multidrug-resistant, methicillin-resistant Staphylococcus aureus clonal complex 97 lineage spreading in dairy cattle and pigs in Italy. Appl Environ Microbiol 82: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flamm RK, Farrell DJ, Mendes RE, Ross JE, Sader HS, Jones RN. 2012. ZAAPS Program results for 2010: An activity and spectrum analysis of linezolid using clinical isolates from 75 medical centres in 24 countries. J Chemother 24: 328–337. [DOI] [PubMed] [Google Scholar]
- Flamm RK, Mendes RE, Ross JE, Sader HS, Jones RN. 2013. An international activity and spectrum analysis of linezolid: ZAAPS Program results for 2011. Diagn Microbiol Infect Dis 76: 206–213. [DOI] [PubMed] [Google Scholar]
- Flamm RK, Mendes RE, Streit JM, Sader HS, Hogan PA, Jones RN. 2016. Activity of linezoli when tested against contemporary European bacterial clinical isolates (2015). 26th European Congress of Clinical Microbiology and Infectious Diseases, ESCMID Amsterdam, April 9–12. [Google Scholar]
- Gentry DR, Rittenhouse SF, McCloskey L, Holmes DJ. 2007. Stepwise exposure of Staphylococcus aureus to pleuromutilins is associated with stepwise acquisition of mutations in rplC and minimally affects susceptibility to retapamulin. Antimicrob Agents Chemother 51: 2048–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons EG. 1982. Total synthesis of (+)-pleuromutilin. J Am Chem Soc 104: 1767–1769. [Google Scholar]
- Goldstein EJ, Citron DM, Merriam CV, Warren YA, Tyrrell KL, Fernandez HT. 2006. Comparative in vitro activities of retapamulin (SB-275833) against 141 clinical isolates of Propionibacterium spp., including 117 P. acnes isolates. Antimicrob Agents Chemother 50: 379–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan PC, Windsor HM, Ripley PH. 1997. In vitro susceptibilities of recent field isolates of Mycoplasma hyopneumoniae and Mycoplasma hyosynoviae to valnemulin (Econor), tiamulin and enrofloxacin and the in vitro development of resistance to certain antimicrobial agents in Mycoplasma hyopneumoniae. Res Vet Sci 63: 157–160. [DOI] [PubMed] [Google Scholar]
- Hanson RL, Matson JA, Brzozowski DB, LaPorte TL, Springer DM, Patel RN. 2002. Hydroxylation of mutilin by Streptomyces griseus and Cunninghamella echinulata. Org Process Res Dev 6: 482–487. [Google Scholar]
- Heilmann C, Jensen L, Jensen JS, Lundstrom K, Windsor D, Windsor H, Webster D. 2001. Treatment of resistant mycoplasma infection in immunocompromised patients with a new pleuromutilin antibiotic. J Infect 43: 234–238. [DOI] [PubMed] [Google Scholar]
- Hidalgo A, Carvajal A, Vester B, Pringle M, Naharro G, Rubio P. 2011. Trends towards lower antimicrobial susceptibility and characterization of acquired resistance among clinical isolates of Brachyspira hyodysenteriae in Spain. Antimicrob Agents Chemother 55: 3330–3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillen S, Willems H, Herbst W, Rohde J, Reiner G. 2014. Mutations in the 50S ribosomal subunit of Brachyspira hyodysenteriae associated with altered minimum inhibitory concentrations of pleuromutilins. Vet Microbiol 172: 223–229. [DOI] [PubMed] [Google Scholar]
- Hogenauer G. 1975. The mode of action of pleuromutilin derivatives. Location and properties of the pleuromutilin binding site on Escherichia coli ribosomes. Eur J Biochem 52: 93–98. [DOI] [PubMed] [Google Scholar]
- Hogenauer G, Ruf C. 1981. Ribosomal binding region for the antibiotic tiamulin: Stoichiometry, subunit location, and affinity for various analogs. Antimicrob Agents Chemother 19: 260–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogenauer G, Egger H, Ruf C, Stumper B. 1981. Affinity labeling of Escherichia coli ribosomes with a covalently binding derivative of the antibiotic pleuromutilin. Biochemistry 20: 546–552. [DOI] [PubMed] [Google Scholar]
- Hunt E. 2000. Pleuromutlin antibiotics. Drugs Future 25: 1163–1168. [Google Scholar]
- Jones RN, Kohno S, Ono Y, Ross JE, Yanagihara K. 2009. ZAAPS International Surveillance Program (2007) for linezolid resistance: Results from 5591 Gram-positive clinical isolates in 23 countries. Diagn Microbiol Infect Dis 64: 191–201. [DOI] [PubMed] [Google Scholar]
- Kadlec K, Schwarz S. 2009. Novel ABC transporter gene, vga(C), located on a multiresistance plasmid from a porcine methicillin-resistant Staphylococcus aureus ST398 strain. Antimicrob Agents Chemother 53: 3589–3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadlec K, Pomba CF, Couto N, Schwarz S. 2010. Small plasmids carrying vga(A) or vga(C) genes mediate resistance to lincosamides, pleuromutilins and streptogramin A antibiotics in methicillin-resistant Staphylococcus aureus ST398 from swine. J Antimicrob Chemother 65: 2692–2693. [DOI] [PubMed] [Google Scholar]
- Karlsson M, Gunnarsson A, Franklin A. 2001. Susceptibility to pleuromutilins in Brachyspira (Serpulina) hyodysenteriae. Anim Health Res Rev 2: 59–65. [PubMed] [Google Scholar]
- Kavanagh F, Hervery A, Robbins WJ. 1951. Antibiotic substances from basidiomycetes. VIII: Pleurotus mutilus (Fr.) Sacc. and Pleurotus Passeckerianus Pilat. Proc Natl Acad Sci 37: 570–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrenberg C, Schwarz S, Jacobsen L, Hansen LH, Vester B. 2005. A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: Methylation of 23S ribosomal RNA at A2503. Mol Microbiol 57: 1064–1073. [DOI] [PubMed] [Google Scholar]
- Kehrenberg C, Cuny C, Strommenger B, Schwarz S, Witte W. 2009. Methicillin-resistant and -susceptible Staphylococcus aureus strains of clonal lineages ST398 and ST9 from swine carry the multidrug resistance gene cfr. Antimicrob Agents Chemother 53: 779–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosowska-Shick K, Clark C, Credito K, McGhee P, Dewasse B, Bogdanovich T, Appelbaum PC. 2006. Single- and multistep resistance selection studies on the activity of retapamulin compared to other agents against Staphylococcus aureus and Streptococcus pyogenes. Antimicrob Agents Chemother 50: 765–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenart J, Vimberg V, Vesela L, Janata J, Balikova Novotna G. 2015. Detailed mutational analysis of Vga(A) interdomain linker: Implication for antibiotic resistance specificity and mechanism. Antimicrob Agents Chemother 59: 1360–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li BB, Shen JZ, Cao XY, Wang Y, Dai L, Huang SY, Wu CM. 2010. Mutations in 23S rRNA gene associated with decreased susceptibility to tiamulin and valnemulin in Mycoplasma gallisepticum. FEMS Microbiol Lett 308: 144–149. [DOI] [PubMed] [Google Scholar]
- Li BB, Wu CM, Wang Y, Shen JZ. 2011. Single and dual mutations at positions 2058, 2503 and 2504 of 23S rRNA and their relationship to resistance to antibiotics that target the large ribosomal subunit. J Antimicrob Chemother 66: 1983–1986. [DOI] [PubMed] [Google Scholar]
- Li B, Wendlandt S, Yao J, Liu Y, Zhang Q, Shi Z, Wei J, Shao D, Schwarz S, Wang S, et al. 2013. Detection and new genetic environment of the pleuromutilin-lincosamide-streptogramin A resistance gene lsa(E) in methicillin-resistant Staphylococcus aureus of swine origin. J Antimicrob Chemother 68: 1251–1255. [DOI] [PubMed] [Google Scholar]
- Li J, Li B, Wendlandt S, Schwarz S, Wang Y, Wu C, Ma Z, Shen J. 2014a. Identification of a novel vga(E) gene variant that confers resistance to pleuromutilins, lincosamides and streptogramin A antibiotics in Staphylococci of porcine origin. J Antimicrob Chemother 69: 919–923. [DOI] [PubMed] [Google Scholar]
- Li XS, Dong WC, Wang XM, Hu GZ, Wang YB, Cai BY, Wu CM, Wang Y, Du XD. 2014b. Presence and genetic environment of pleuromutilin-lincosamide-streptogramin A resistance gene lsa(E) in Enterococci of human and swine origin. J Antimicrob Chemother 69: 1424–1426. [DOI] [PubMed] [Google Scholar]
- Li D, Wu C, Wang Y, Fan R, Schwarz S, Zhang S. 2015. Identification of multiresistance gene cfr in methicillin-resistant Staphylococcus aureus from pigs: Plasmid location and integration into a staphylococcal cassette chromosome mec complex. Antimicrob Agents Chemother 59: 3641–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey J. 2006. Ecology of Commanster, www.commanster.eu/commanster.html.
- Liu J, Lotesta SD, Sorensen EJ. 2011. A concise synthesis of the molecular framework of pleuromutilin. Chem Commun 47: 1500–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke JB, Rahawi S, Lamarre J, Mankin AS, Shaw KJ. 2012. Genetic environment and stability of cfr in methicillin-resistant Staphylococcus aureus CM05. Antimicrob Agents Chemother 56: 332–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long KS, Hansen LH, Jakobsen L, Vester B. 2006. Interaction of pleuromutilin derivatives with the ribosomal peptidyl transferase center. Antimicrob Agents Chemother 50: 1458–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long KS, Poehlsgaard J, Hansen LH, Hobbie SN, Bottger EC, Vester B. 2009. Single 23S rRNA mutations at the ribosomal peptidyl transferase centre confer resistance to valnemulin and other antibiotics in Mycobacterium smegmatis by perturbation of the drug binding pocket. Mol Microbiol 71: 1218–1227. [DOI] [PubMed] [Google Scholar]
- Lotesta SD, Liu J, Yates EV, Krieger I, Sacchettini JC, Freundlich JS, Sorensen EJ. 2011. Expanding the pleuromutilin class of antibiotics by de novo chemical synthesis. Chem Sci 2: 1258–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano C, Aspiroz C, Rezusta A, Gomez-Sanz E, Simon C, Gomez P, Ortega C, Revillo MJ, Zarazaga M, Torres C. 2012. Identification of novel vga(A)-carrying plasmids and a Tn5406-like transposon in methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis of human and animal origin. Int J Antimicrob Agents 40: 306–312. [DOI] [PubMed] [Google Scholar]
- Mang R, Heilmayer W, Badegruber R, Strickmann D, Novak R, Ferencic M, Bulusu ARCM. 2008. Pleuromutilin derivatives for the treatment of diseases mediated by microbes. Patent WO2008113089. [Google Scholar]
- Mendes RE, Hogan PA, Streit JM, Jones RN, Flamm RK. 2014. Zyvox Annual Appraisal of Potency and Spectrum (ZAAPS) program: Report of linezolid activity over 9 years (2004–12). J Antimicrob Chemother 69: 1582–1588. [DOI] [PubMed] [Google Scholar]
- Mendes RE, Hogan PA, Jones RN, Sader HS, Flamm RK. 2016. Surveillance for linezolid resistance via the Zyvox Annual Appraisal of Potency and Spectrum (ZAAPS) programme (2014): Evolving resistance mechanisms with stable susceptibility rates. J Antimicrob Chemother 10.1093/jac/dkw052. [DOI] [PubMed] [Google Scholar]
- Miller K, Dunsmore CJ, Fishwick CW, Chopra I. 2008. Linezolid and tiamulin cross-resistance in Staphylococcus aureus mediated by point mutations in the peptidyl transferase center. Antimicrob Agents Chemother 52: 1737–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon DC, Tamang MD, Nam HM, Jeong JH, Jang GC, Jung SC, Park YH, Lim SK. 2015. Identification of livestock-associated methicillin-resistant Staphylococcus aureus isolates in Korea and molecular comparison between isolates from animal carcasses and slaughterhouse workers. Foodborne Pathog Dis 12: 327–334. [DOI] [PubMed] [Google Scholar]
- Naegeli P. 1961. “Zur Kenntnis des Pleuromutilins” [Notice about pleuromutilin]. PhD thesis, ETH, Zurich. [Google Scholar]
- Novak R. 2011. Are pleuromutilin antibiotics finally fit for human use? Ann NY Acad Sci 1241: 71–81. [DOI] [PubMed] [Google Scholar]
- Odou MF, Muller C, Calvet L, Dubreuil L. 2007. In vitro activity against anaerobes of retapamulin, a new topical antibiotic for treatment of skin infections. J Antimicrob Chemother 59: 646–651. [DOI] [PubMed] [Google Scholar]
- Overesch G, Buttner S, Rossano A, Perreten V. 2011. The increase of methicillin-resistant Staphylococcus aureus (MRSA) and the presence of an unusual sequence type ST49 in slaughter pigs in Switzerland. BMC Vet Res 7: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette LA, Bullman-Page PC. 1985. A relay approach to (+)-pleuromutilin. II: Preparation of an advanced optically pure intermediate. Tetrahedron Lett 26: 1607–1611. [Google Scholar]
- Paquette LA, Wiedemann PE. 1985. A relay to (+)-pleuromutilin. I: De novo synthesis of a levorotatory tricyclic lactone subunit. Tetrahedron Lett 26: 1603–1606. [Google Scholar]
- Paukner S, Clark C, Ivezic-Schoenfeld Z, Kosowska-Shick K. 2012. Single- and multistep resistance selection with the pleuromutilin antibiotic BC-3781. 52nd Interscience Conference on Antimicrobial Agents and Chemotherapy (ICAAC) San Francisco, September 9–12. [Google Scholar]
- Paukner S, Gruss A, Fritsche TR, Ivezic-Schoenfeld Z, Jones RN. 2013a. In vitro activity of the novel pleuromutilin BC-3781 tested against bacterial pathogens causing sexually transmitted diseases (STD). Abstracts of the Fifty-Third Interscience Conference on Antimicrobial Agents and Chemotherapy Denver, September 10–13. [Google Scholar]
- Paukner S, Krause K, Gruss A, Keepers T, Gomez M, Bischinger A, Strickmann DB, Ivezic-Schoenfeld Z. 2013b. Accumulation of the pleuromutilin antibiotic BC-3781 in murine macrophages and effect of lung surfactant on the BC-3781 in vitro activity. 53rd Interscience Conference on Antimicrobial Agents and Chemotherapy (ICAAC) Denver, CO, September 10–13. [Google Scholar]
- Paukner S, Sader HS, Ivezic-Schoenfeld Z, Jones RN. 2013c. Antimicrobial activity of the pleuromutilin antibiotic BC-3781 against bacterial pathogens isolated in the SENTRY antimicrobial surveillance program in 2010. Antimicrob Agents Chemother 57: 4489–4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paukner S, Kollmann H, Thirring K, Heilmayer W, Ivezic-Schoenfeld Z. 2014a. Antibacterial in vitro activity of novel extended spectrum pleuromutilins against Gram-positive and -negative bacterial pathogens. 24th European Congress of Clinical Microbiology and Infectious Diseases (ECCMID) Barcelona, Spain, May 10–13. [Google Scholar]
- Paukner S, Strickmann D, Ivezic-Schoenfeld Z. 2014b. Extended spectrum pleuromutilins: Mode-of-action studies. 24th European Congress of Clinical Microbiology and Infectious Diseases (ECCMID) Barcelona, Spain, May 10–13. [Google Scholar]
- Paukner S, Kollmann H, Riedl R, Ivezic-Schoenfeld Z. 2015a. Kill curves of the novel extended-spectrum pleuromutilin antibiotic BC-9529. 25th European Congress of Clinical Microbiology and Infectious Diseases (ECCMID) Copenhagen, Denmark, April 25–28. [Google Scholar]
- Paukner S, Wicha WW, Heilmayer W, Thirring K, Riedl R. 2015b. Extended spectrum pleuromutilins: potent translation inhibitors with broad-spectrum antibacterial activity in vitro and in vivo. ICAAC/ICC 2016 San Diego, CA, September 17–21. [Google Scholar]
- Paukner S, Wicha WW, Thirring K, Kollmann H, Ivezic-Schoenfeld Z. 2015c. In vitro and in vivo efficay of novel extended spectrum pleuromutilins against S. aureus and S. pneumoniae. 25th European Congress of Clinical Microbiology and Infectious Diseases (ECCMID) Copenhagen, Denmark, April 25–28. [Google Scholar]
- Poulsen SM, Karlsson M, Johansson LB, Vester B. 2001. The pleuromutilin drugs tiamulin and valnemulin bind to the RNA at the peptidyl transferase centre on the ribosome. Mol Microbiol 41: 1091–1099. [DOI] [PubMed] [Google Scholar]
- Prince WT, Ivezic-Schoenfeld Z, Lell C, Tack KJ, Novak R, Obermayr F, Talbot GH. 2013. Phase II clinical study of BC-3781, a pleuromutilin antibiotic, in treatment of patients with acute bacterial skin and skin structure infections. Antimicrob Agents Chemother 57: 2087–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle M, Poehlsgaard J, Vester B, Long KS. 2004. Mutations in ribosomal protein L3 and 23S ribosomal RNA at the peptidyl transferase centre are associated with reduced susceptibility to tiamulin in Brachyspira spp. isolates. Mol Microbiol 54: 1295–1306. [DOI] [PubMed] [Google Scholar]
- Rittenhouse S, Biswas S, Broskey J, McCloskey L, Moore T, Vasey S, West J, Zalacain M, Zonis R, Payne D. 2006. Selection of retapamulin, a novel pleuromutilin for topical use. Antimicrob Agents Chemother 50: 3882–3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JE, Farrell DJ, Mendes RE, Sader HS, Jones RN. 2011. Eight-year (2002–2009) summary of the linezolid (Zyvox Annual Appraisal of Potency and Spectrum; ZAAPS) program in European countries. J Chemother 23: 71–76. [DOI] [PubMed] [Google Scholar]
- Ruscoe RE, Fazakerley NJ, Huang H, Flitsch S, Procter DJ. 2015. Copper-catalyzed double additions and radical cyclization cascades in the re-engineering of the antibacterial pleuromutilin. Chem Eur J 21: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sader HS, Biedenbach DJ, Paukner S, Ivezic-Schoenfeld Z, Jones RN. 2012a. Antimicrobial activity of the investigational pleuromutilin compound BC-3781 tested against Gram-positive organisms commonly associated with acute bacterial skin and skin structure infections. Antimicrob Agents Chemother 56: 1619–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sader HS, Paukner S, Ivezic-Schoenfeld Z, Biedenbach DJ, Schmitz FJ, Jones RN. 2012b. Antimicrobial activity of the novel pleuromutilin antibiotic BC-3781 against organisms responsible for community-acquired respiratory tract infections (CARTIs). J Antimicrob Chemother 67: 1170–1175. [DOI] [PubMed] [Google Scholar]
- Sanchez GM, De la Torre MA, Morales G, Pelaez B, Tolon MJ, Domingo S, Candel FJ, Andrade R, Arribi A, Garcia N, et al. 2010. Clinical outbreak of linezolid-resistant Staphylococcus aureus in an intensive care unit. JAMA 303: 2260–2264. [DOI] [PubMed] [Google Scholar]
- Schlunzen F, Pyetan E, Fucini P, Yonath A, Harms JM. 2004. Inhibition of peptide bond formation by pleuromutilins: The structure of the 50S ribosomal subunit from Deinococcus radiodurans in complex with tiamulin. Mol Microbiol 54: 1287–1294. [DOI] [PubMed] [Google Scholar]
- Schwendener S, Perreten V. 2011. New transposon Tn6133 in MRSA ST398 contains vga(E), a novel streptogramin A-, pleuromutilin-, and lincosamide-resistance gene. Antimicrob Agents Chemother 55: 4900–4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Wang Y, Schwarz S. 2013. Presence and dissemination of the multiresistance gene cfr in Gram-positive and Gram-negative bacteria. J Antimicrob Chemother 68: 1697–1706. [DOI] [PubMed] [Google Scholar]
- Shore AC, Brennan OM, Ehricht R, Monecke S, Schwarz S, Slickers P, Coleman DC. 2010. Identification and characterization of the multidrug resistance gene cfr in a Panton-Valentine leukocidin-positive sequence type 8 methicillin-resistant Staphylococcus aureus IVa (USA300) isolate. Antimicrob Agents Chemother 54: 4978–4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore AC, Lazaris A, Kinnevey PM, Brennan OM, Brennan GI, OC B, Fessler AT, Schwarz S, Coleman DC. 2016. First report of cfr-encoding plasmids in the pandemic sequence type 22 methicillin-resistant Staphylococcus aureus Staphylococcal cassette chromosome mec type-IV clone. Antimicrob Agents Chemother 22: 3007–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra JM, Camoez M, Tubau F, Gasch O, Pujol M, Martin R, Dominguez MA. 2013. Low prevalence of Cfr-mediated linezolid resistance among methicillin-resistant Staphylococcus aureus in a Spanish hospital: Case report on linezolid resistance acquired during linezolid therapy. PLoS ONE 8: e59215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer DM, Sorenson ME, Huang S, Connolly TP, Bronson JJ, Matson JA, Hanson RL, Brzozowski DB, LaPorte TL, Patel RN. 2003. Synthesis and activity of a C-8 keto pleuromutilin derivative. Bioorg Med Chem Lett 13: 1751–1753. [DOI] [PubMed] [Google Scholar]
- Springer DM, Goodrich JT, Luh B-Y, Bronson JJ, Gao Q, Huang S, DenBleyker K, Dougherty TJ, Fung-Tomc J. 2008. Antibacterial pleuromutilin derivatives based on alternate core structures: Arigoni and Birch chemistry revisited. Lett Drug Des Discov 5: 327–331. [Google Scholar]
- Strickmann D, Wicha WW, Kollmann H, Ivezic-Schoenfeld Z. 2014. In vitro metabolism and in vivo pharmacokinetics of novel extended spectrum pleuromutilin antibiotics. 24th European Congress of Clinical Microbiology and Infectious Diseases (ECCMID) Barcelona, Spain, May 10–13. [Google Scholar]
- Takadoi M, Sato T, Fukuda Y. 2007. Synthesis and antibacterial activity of novel C-12 substituted mutilins. 47th Interscience Conference on Antimicrobial Agents and Chemotherapy (ICAAC) Chicago, IL, September 17–20. [Google Scholar]
- Thirring K, Ascher G, Paukner S, Heilmayer W, Novak R. 2007. Mutilin derivatives and their use as pharmaceutical. Patent WO2007079515. [Google Scholar]
- Wang H, Andemichael YW, Vogt FG. 2009. A scalable synthesis of 2S-hydroxymutilin via modified rubottom oxidation. J Organ Chem 74: 478–481. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang Y, Wu CM, Schwarz S, Shen Z, Zhang W, Zhang Q, Shen JZ. 2011. Detection of the staphylococcal multiresistance gene cfr in Proteus vulgaris of food animal origin. J Antimicrob Chemother 66: 2521–2526. [DOI] [PubMed] [Google Scholar]
- Wang Y, He T, Schwarz S, Zhou D, Shen Z, Wu C, Wang Y, Ma L, Zhang Q, Shen J. 2012a. Detection of the staphylococcal multiresistance gene cfr in Escherichia coli of domestic-animal origin. J Antimicrob Chemother 67: 1094–1098. [DOI] [PubMed] [Google Scholar]
- Wang Y, Schwarz S, Shen Z, Zhang W, Qi J, Liu Y, He T, Shen J, Wu C. 2012b. Co-location of the multiresistance gene cfr and the novel streptomycin resistance gene aadY on a small plasmid in a porcine Bacillus strain. J Antimicrob Chemother 67: 1547–1549. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang Y, Schwarz S, Shen Z, Zhou N, Lin J, Wu C, Shen J. 2012c. Detection of the staphylococcal multiresistance gene cfr in Macrococcus caseolyticus and Jeotgalicoccus pinnipedialis. J Antimicrob Chemother 67: 1824–1827. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang W, Wang J, Wu C, Shen Z, Fu X, Yan Y, Zhang Q, Schwarz S, Shen J. 2012d. Distribution of the multidrug resistance gene cfr in Staphylococcus species isolates from swine farms in China. Antimicrob Agents Chemother 56: 1485–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicha WW, Ivezic-Schoenfeld Z. 2014. In vivo activity of extended spectrum pleuromutilins in murine sepsis model. 24th European Congress of Clinical Microbiology and Infectious Diseases (ECCMID) Barcelona, Spain, May 10–13. [Google Scholar]
- Wicha WW, Ivezic-Schoenfeld Z, Novak R. 2010. Pharmacokinetics, mass balance and tissue distribution of [14C]-BC-3781 in non-pigmented rats. 20th European Congress of Clinical Microbiology and Infectious Diseases (ECCMID) Vienna, April 10–13. [Google Scholar]
- Wicha WW, Fischer E, Kappes BC, Ivezic-Schoenfeld Z. 2013. Comparative pharmacodynamics of BC-3781 in murine Streptococcus pneumoniae—Thigh and lung infection models. 53rd Interscience Conference on Antimicrobial Agents and Chemotherapy (ICAAC) Denver, May 18–21. [Google Scholar]
- Wicha WW, Paukner S, Strickmann D, Bhavnani SM, Ambrose PG. 2015a. Pharmacokinetics-pharmacodynamics of lefamulin in a neutropenic murine lung infection model. ICAAC/ICC 2015 San Diego, September 17–21. [Google Scholar]
- Wicha WW, Paukner S, Strickmann D, Thirring K, Kollmann H, Heilmayer W, Ivezic-Schoenfeld Z. 2015b. Efficacy of novel extended spectrum pleuromutilins against E. coli in vitro and in vivo. 25th European Congress of Clinical Microbiology and Infectious Diseases (ESCMID) Copenhagen, April 25–28. [Google Scholar]
- Yan K, Madden L, Choudhry AE, Voigt CS, Copeland RA, Gontarek RR. 2006. Biochemical characterization of the interactions of the novel pleuromutilin derivative retapamulin with bacterial ribosomes. Antimicrob Agents Chemother 50: 3875–3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger M, Schwameis R, Burian A, Burian B, Matzneller P, Muller M, Wicha WW, Strickmann DB, Prince W. 2016. Simultaneous assessment of the pharmacokinetics of a pleuromutilin, lefamulin, in plasma, soft tissues and pulmonary epithelial lining fluid. J Antimicrob Chemother 71: 1022–1026. [DOI] [PubMed] [Google Scholar]
- Zhang A, Xu C, Wang H, Lei C, Liu B, Guan Z, Yang C, Yang Y, Peng L. 2015. Presence and new genetic environment of pleuromutilin-lincosamide-streptogramin A resistance gene lsa(E) in Erysipelothrix rhusiopathiae of swine origin. Vet Microbiol 177: 162–167. [DOI] [PubMed] [Google Scholar]