Abstract
Complementing genome sequence with deep transcriptome and proteome data could enable more accurate assembly and annotation of newly sequenced genomes. Here, we provide a proof-of-concept of an integrated approach for analysis of the genome and proteome of Anopheles stephensi, which is one of the most important vectors of the malaria parasite. To achieve broad coverage of genes, we carried out transcriptome sequencing and deep proteome profiling of multiple anatomically distinct sites. Based on transcriptomic data alone, we identified and corrected 535 events of incomplete genome assembly involving 1196 scaffolds and 868 protein-coding gene models. This proteogenomic approach enabled us to add 365 genes that were missed during genome annotation and identify 917 gene correction events through discovery of 151 novel exons, 297 protein extensions, 231 exon extensions, 192 novel protein start sites, 19 novel translational frames, 28 events of joining of exons, and 76 events of joining of adjacent genes as a single gene. Incorporation of proteomic evidence allowed us to change the designation of more than 87 predicted “noncoding RNAs” to conventional mRNAs coded by protein-coding genes. Importantly, extension of the newly corrected genome assemblies and gene models to 15 other newly assembled Anopheline genomes led to the discovery of a large number of apparent discrepancies in assembly and annotation of these genomes. Our data provide a framework for how future genome sequencing efforts should incorporate transcriptomic and proteomic analysis in combination with simultaneous manual curation to achieve near complete assembly and accurate annotation of genomes.
Genome assembly and subsequent genome annotation are largely driven by computational pipelines, which ultimately provide a set of predicted protein-coding and noncoding genes. In recent years, next-generation sequencing technologies have been used to achieve high coverage of large genomes in a cost-effective fashion (Schatz et al. 2010; Salzberg et al. 2012). However, there are inherent challenges in genome assembly because of repetitive sequences, high GC content, and chimeric reads (Schatz et al. 2010). Indeed, substantial differences in genome assemblies have been reported when the same data sets were analyzed by different methodologies (Earl et al. 2011; Salzberg et al. 2012) demonstrating a critical need for developing protocols for accurate assembly of genomic sequences.
Genome annotation, especially identification of protein-coding genes, is of high priority once the genome is sequenced. Annotation of protein-coding genes is currently based on gene prediction algorithms (Renuse et al. 2011; Megy et al. 2012; Jiang et al. 2014). The annotated genes remain as hypothetical sequences until validated through experiments. The experimental data might include transcriptomic data, which can improve the predicted gene models (Denoeud et al. 2008; Gerstein et al. 2010; Guo et al. 2014; Kelkar et al. 2014; Woo et al. 2014; Wu et al. 2014; Yu et al. 2014; Linde et al. 2015). More recently, proteomic data from mass spectrometry experiments have been used for validating protein-coding genes (Brunner et al. 2007; Gupta et al. 2008; Merrihew et al. 2008; Chaerkady et al. 2011; Kelkar et al. 2011; Bock et al. 2014; Castellana et al. 2014; Kim et al. 2014; Trapp et al. 2014; Wilhelm et al. 2014). Though transcriptomic and proteomic data have been successfully used in correction of genome annotation errors in prokaryotes and lower eukaryotes, the approach is yet to be used efficiently for complex eukaryotic genomes (Armengaud 2009). Annotation of genes in eukaryotic genome is more complex and error prone owing to the presence of long introns, repetitive sequences, alternative splicing, noncoding RNAs, and large genome size.
Transcriptomic and proteomic data have been used for improving annotation of genes (Bock et al. 2014; Kelkar et al. 2014; Woo et al. 2014; Wu et al. 2014). However, the potential application of these data sets in the correction of incomplete genome assemblies has not been realized thus far, and only few studies have demonstrated utility of RNA-seq data in improving incomplete genome assemblies (Mortazavi et al. 2010; Xue et al. 2013). Therefore, as a proof of concept, we used an integrated approach to improve genome assembly and annotation of newly assembled genomes of Anopheles stephensi and 15 other related genomes (Fig. 1). Anopheles stephensi is one of the major vectors of malaria in Asia. Genomes of two strains of Anopheles stephensi (Indian and SDA-500 strains, with corresponding genome assemblies referred to as AsteI2 and AsteS1, respectively) have recently been sequenced and are available through VectorBase (Jiang et al. 2014; Neafsey et al. 2015). A total of 12,350 genes are annotated in AsteI2, whereas the AsteS1 assembly has 13,652 annotated genes. We carried out transcriptomic and proteomic analysis of four and 15 organs of Anopheles stephensi, respectively. We used a combination of computational workflows and expert manual curation to identify and correct errors in genome assembly and annotation of genes. We also used this unique opportunity to test whether the novel findings from the Anopheles stephensi genome could be used to improve the accuracy of assembly and annotation of 15 genomes of other Anopheles species whose genomes have also been recently sequenced (Neafsey et al. 2015).
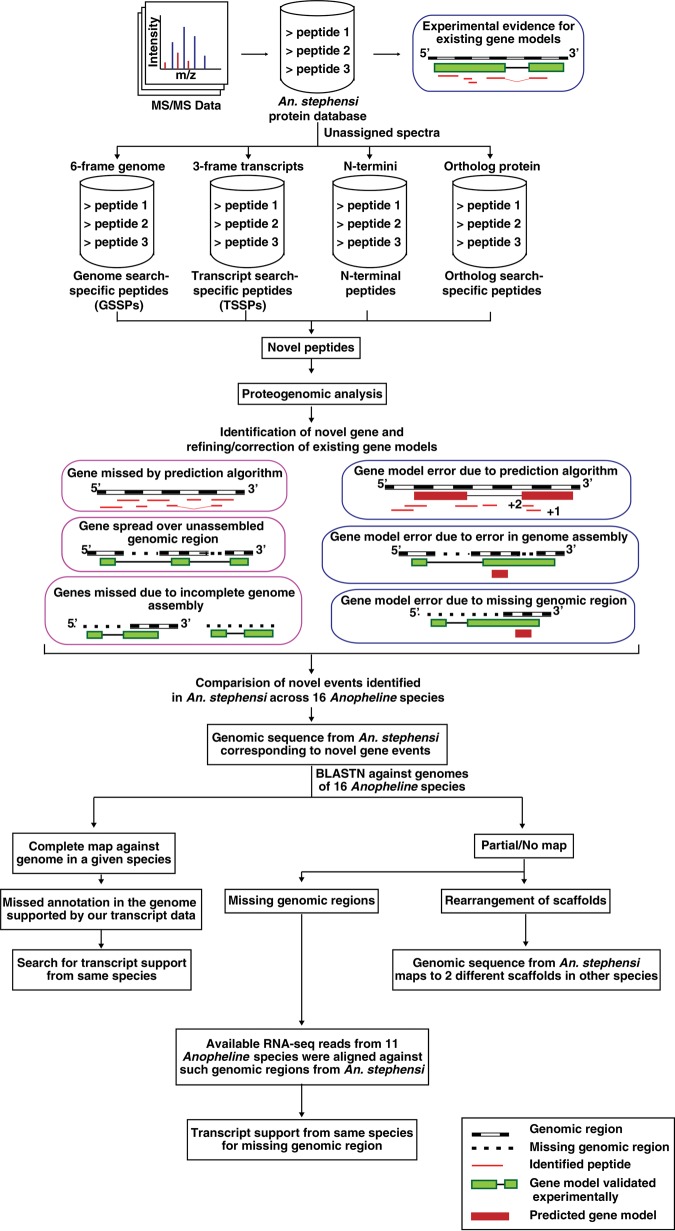
Figure 1.
Overview of pipeline used for correction of genome annotation and genome assembly using transcriptomic and proteomic data. MS/MS spectra, which did not assign to the known protein database, were further searched against six-frame translated genome, three-frame translated transcripts, and Anopheles gambiae protein database. Further analysis of these peptides resulted in identification of novel protein-coding genes and revised gene annotations, which were compared against 15 other Anopheline species.
Results
Transcriptomic and proteomic landscape of Anopheles stephensi
We carried out transcriptomic and proteomic analyses of multiple tissues from the Indian strain of An. stephensi to obtain comprehensive coverage (Fig. 2A). Because only limited RNA-seq data were available for individual organs in VectorBase, we selected four adult tissues for deep transcriptomic analysis. When we mapped the RNA-seq data from these four tissues—Malpighian tubules, fat body, midgut, and ovary—to the two available genome assemblies for An. stephensi, we observed 3673 intergenic transcripts in the case of AsteI2 and 1920 intergenic transcripts in the case of AsteS1. Figure 2B represents one such example in which one of the existing assemblies (AsteI2) did not have any predicted gene at all. The other assembly (AsteS1) had a predicted gene, but our transcriptomic data provided evidence for extension of the predicted gene model as well as the presence of additional unannotated exons. The novel transcripts along with the proteomic data were used for improving the genome assembly and annotation errors using a novel integrative proteogenomic analysis pipeline (Fig. 1). The relative utility of transcriptomic and proteomic data has been described in greater detail in the Discussion. Supplemental Table S5.11 provides the number of novel annotations or corrections that resulted from proteomic evidence alone or from both RNA-seq and proteomic evidence.
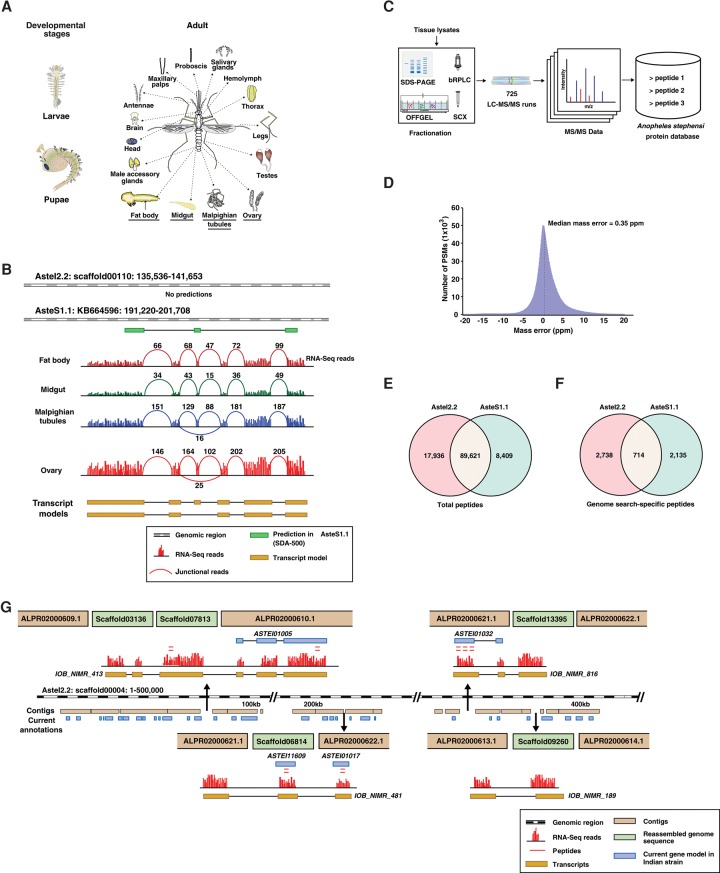
Figure 2.
Schematic representation of the workflow and summary of proteomic data. (A) Adult tissues and developmental stages of the Indian strain of An. stephensi that were dissected and processed for transcriptomic or proteomic analysis. (B) Revised annotation of An. stephensi genome based on RNA-seq evidence. The numbers represent the junctional reads identified in each tissue, and the two transcript models shown are splice variants identified based on RNA-seq data. (C) Broad overview of mass spectrometry–based proteomic analysis of multiple tissues. (D) Median mass error of the peptide spectral matches identified in the study. (E) Total number of peptides identified against AsteI2 and AsteS1 assembly. (F) Total number of genome search-specific peptides identified against the two assemblies. (G) Insertion of five small scaffolds in genome gap regions of scaffold00004.
We also undertook tandem mass spectrometry–based proteomic profiling of An. stephensi. To obtain broad coverage of An. stephensi proteome, we extracted proteins from larvae, pupae, and 15 tissues from adult mosquitoes and subjected them to multiple fractionation strategies (Fig. 2C). The peptide fractions were analyzed on Fourier transform mass spectrometers, and both precursor and fragment ions were measured in the Orbitrap in high resolution mode. In all, analysis of 725 fractions by LC-MS/MS generated more than 5 million tandem mass spectra that were searched against predicted proteins from the AsteI2 assembly. About 2.4 million peptide spectrum matches were identified with a median mass error of 350 parts per billion (Fig. 2D), corresponding to 92,628 peptides. False discovery rate (FDR) for such large data sets does not scale as expected, and therefore protein level FDR could be erroneous in such studies (Wilhelm et al. 2014; Savitski et al. 2015). Therefore, in our study, we used 1% peptide level FDR for each tissue separately and rejected the peptides that did not qualify for this threshold. In addition, we confirmed MS/MS spectra for a subset of peptides by analyzing synthetic peptides as discussed in greater detail below (Validation of MS/MS spectra for novel peptides). A similar approach was followed in two recent studies published on the human proteome, which involved such large data sets (Kim et al. 2014; Wilhelm et al. 2014).
The high-quality proteomic data allowed us to identify 8303 genes, of which 8041 were already annotated, thereby providing experimental evidence for predicted annotations. To compare the annotations of AsteI2 and AsteS1 assemblies, we searched the proteomic data against 13,251 predicted proteins from the SDA-500 strain. Both peptide matches to annotated proteins (Fig. 2E) as well as genome search-specific peptides for each assembly (Fig. 2F) revealed that although the large majority of peptide matches were shared, there were several that were unique to each assembly (Supplemental Tables S1, S2).
Improving genome assembly through an integrative analysis pipeline
The AsteI2 and AsteS1 genome assemblies are organized into 23,371 and 1100 discrete scaffolds, respectively. The high number of scaffolds in AsteI2 reflects a decision to include short sequence fragments that contain repetitive sequences (Jiang et al. 2014). Annotation of protein-coding regions can be missed by computational pipelines owing to gaps in genomic regions. BAC-end sequences and PacBio reads were used for better scaffolding and gap filling in the AsteI2 genome assembly (Jiang et al. 2014). After quality filtering, only 46 such scaffold links were retained, which connected 22 scaffolds. In contrast, we used our RNA-seq data to identify and improve incomplete genome assemblies. We mapped and assembled our RNA-seq data against the AsteI2 and AsteS1 genome assemblies. Transcripts uniquely assembled against AsteS1 were mapped against the AsteI2 using BLAST to identify incomplete genome assemblies in AsteI2 and vice versa. Using another approach, we also aligned the RNA-seq data to the reference genome with a maximum of four mismatches using TopHat2 (Kim et al. 2013). TopHat-Fusion (Kim and Salzberg 2011), an attribute of TopHat2, was implemented with the “fusion-search” option to identify transcripts spanning multiple scaffolds in each genome assembly (Supplemental Fig. S1). Thus, transcripts were obtained based on the mapping of reads across multiple scaffolds, which permitted sequential alignment of these scaffolds and allowed us to insert short scaffolds within genomic gaps.
In all, we identified 1084 transcripts that spanned across 1046 discrete scaffolds in the Indian strain (Supplemental Table S3). Of all scaffolds, we observed an unusually high frequency of gaps in scaffold00004 in the AsteI2 assembly and were able to introduce 44 short scaffolds in 28 genomic gaps using transcript data. Figure 2G illustrates one such example in which we used transcript evidence for inserting five genomic regions into gaps contained within scaffold00004. Altogether, such events led to the revision of 794 known annotations in AsteI2, of which 54 were also supported by peptide evidence. In addition, seven novel protein-coding regions were identified that were previously missed because of incomplete genome assembly. On the other hand, only 22 such scaffolds could be linked using BAC-end sequences (Jiang et al. 2014). The additional improvements of scaffold assembly and gap closing demonstrated in our study provide a measure of advantage of using RNA-seq data in addition to the DNA-based scaffolding. Similar analysis of AsteS1 led to identification of 150 transcripts spanning across 166 discrete scaffolds. Supplemental Figure S2 illustrates how insertion of scaffold KB665166 in a gap within the scaffold KB664481 revised the annotation of the ASTE007054 gene.
Identification of unannotated mitochondrial genes
To identify mitochondrial genes, we aligned the RNA-seq data against the An. stephensi mitochondrial genome. We obtained evidence of transcription for all 13 protein-coding mitochondrial genes in An. stephensi (Fig. 3A; Supplemental Table S4.1). Surprisingly, we observed that the coding regions for three mitochondrial genes in AsteI2 were interrupted, although they were correctly annotated as single exon genes in the genome of a related Anopheline genome, Anopheles gambiae. Upon examination, we observed that the RNA-seq data provided definitive evidence that some nucleotides were missed in the corresponding genomic loci, most likely owing to sequencing errors. After accounting for these nucleotides, a continuous coding region for all the three genes was easily established (Fig. 3B; Supplemental Table S4.2). In fact, in a previous study, transcripts for these 13 genes have already been predicted in An. stephensi based on alignment of RNA-seq from An. stephensi data against the mitochondrial genome of An. gambiae (Hittinger et al. 2010). In order to detect proteins encoded by the mitochondrial genes, we also searched MS/MS data against three-frame translated transcripts and six-frame translated mitochondrial genome and obtained peptide level evidence for seven protein-coding mitochondrial genes.
Figure 3.
Reannotation of the An. stephensi genome based on transcriptomic and proteomic evidence. (A) Alignment of RNA-seq data against the 15.4-kb-long mitochondrial genome reveals transcript evidence (colored arrows) for 13 mitochondrial genes, of which seven were also identified at the protein level (red lines). (B) Insertion of a nucleotide base in mitochondrial genome based on RNA-seq evidence. (C) Identification of a novel gene in the AsteI2 and AsteS1 assemblies. (D) Alternate protein start site based on the presence of an upstream N-terminally acetylated peptide.
Improving the accuracy of predictions from genome annotation
To identify genes that might have escaped prediction by computational pipelines, we searched unmatched mass spectra against custom databases containing a six-frame translation of the genome and a three-frame translation of the transcriptome from the Indian strain of An. stephensi (Fig. 1). In addition, we searched MS/MS data against the protein database of An. gambiae to identify peptides for which the corresponding genomic region might be missing in An. stephensi. Using these databases, we identified 1429 potentially novel peptides through searches against translated transcripts of An. stephensi, 1434 novel peptides through An. gambiae protein database and 5082 novel peptides through translated genome of An. stephensi. The mean Sequest and Mascot score for known peptides (peptides corresponding to annotated proteins) were 3.5 and 55.3, respectively. On the other hand, the mean Sequest and Mascot score for novel peptides (identified through the proteogenomic searches) were 3.7 and 57.1, respectively (Supplemental Information Fig. A). We manually evaluated mass spectra of these peptides and investigated them further for corresponding transcripts or orthologous sequences from An. gambiae. In 12 cases, orthologous proteins in An. gambiae were identified by multiple peptides, but the corresponding genomic regions were missing in the AsteI2 assembly. By mapping these orthologous protein sequences against the genomic data itself, we were able to identify the missing genomic regions corresponding to these peptides.
In all, the novel peptides identified above resulted in the identification of 365 protein-coding genes, which were missed in both genome assemblies (Supplemental Table S5.1). Evolutionary conservation analysis of these novel genes was performed across Anopheline species, using OrthoDB (Kriventseva et al. 2015). The level of conservation was similar for both the known and novel protein coding regions of the genome (Supplemental Information Fig. B). Figure 3C shows identification of one such novel protein-coding gene, which showed the presence of dUTPase domain. Although the majority of the novel protein-coding regions were expressed in multiple tissues, a subset of them was tissue-restricted, e.g., 62 unannotated regions were exclusively expressed in testes. Similarly, of the four novel genes identified uniquely in chemosensory appendages, two were putative odorant receptors. Identification of missed features in current genome annotations also led to identification of 917 gene correction events in An. stephensi involving 151 examples of novel exons, 231 exon extensions, 297 protein extensions, 192 alternate protein start sites (including 77 cases confirmed by acetylated peptides and 115 cases of N-terminal protein extension identified by nonacetylated peptides), 76 events of joining of genes, 28 events of joining of exons, and 19 cases of translation of a reading frame that was different from the annotated frame (Supplemental Tables S5.2–S5.7). In most of these cases, novel peptides resulted in revision of existing annotations of both strains (Supplemental Fig. S3A). In 76 cases, our analysis led to merging of two adjacent independent gene models into a single gene (Supplemental Table S5.6). For example, we identified a junctional peptide, which suggested that the genes annotated as ASTEI00796 and ASTEI00797 in AsteI2 were in fact derived from a single gene. This single gene model was also supported by transcript evidence (Supplemental Fig. S3B). In contrast, Supplemental Figure S3C illustrates a case in which our data indicate that an existing gene annotation in both strains should be altered to indicate the existence of two separate genes (Supplemental Table S5.7). These two genes exhibit differential expression at the transcript level, and one of them was identified on the basis of an N-terminally acetylated peptide, again confirming that the second transcript indeed encoded a different protein (Supplemental Table S5.8). In some cases, peptides mapping to known protein-coding regions provided evidence for alternate frame of translation. In one such example, we identified multiple peptides that mapped to a second exon of ASTEI05717 gene but in a different frame of translation. This revised frame of translation suggested the presence of chaperonin subunit 10 domain in the gene (Supplemental Fig. S3D).
As most mature proteins are known to be acetylated in their N termini, N-terminally acetylated peptides discovered by mass spectrometry can assist in precise identification of protein start sites (Kim et al. 2014). Using this strategy, we confirmed translational start sites of 1507 proteins and found 77 cases of alternate start sites. One such example where an N-terminally acetylated peptide indicated an upstream start site (ASTEI00264) is shown in Figure 3D. In four cases, N-terminally acetylated peptides mapping to a downstream region of a gene indicated splitting of the gene into two independent genes with independent translational start sites. In each case, the peptides mapped to existing downstream exons in an alternate frame of translation, suggesting a novel protein sequence for the split gene (Supplemental Fig. S3C). This was further supported by RNA-seq data, which showed differential expression for the two independent genes. In the absence of such complementary experimental data sets and integrative analysis, two distinct genes can be misannotated as a single gene by gene prediction algorithms. Similar analysis with previous genome assemblies and protein databases of An. stephensi in VectorBase, i.e., AsteI1, resulted in identification of more than 320 novel gene models. We performed RT-PCR and sequenced the amplicons for 50 such novel events (48 novel genes and two gene model corrections). The cDNA sequences were submitted to dbEST (http://www.ncbi.nlm.nih.gov/nucest?LinkName=biosample_nucest&from_uid=1837904). Forty-nine of these 50 novel events were incorporated in the revised assembly, AsteI2 (Jiang et al. 2014).
Revising genome assemblies and annotations across 15 other Anopheles species
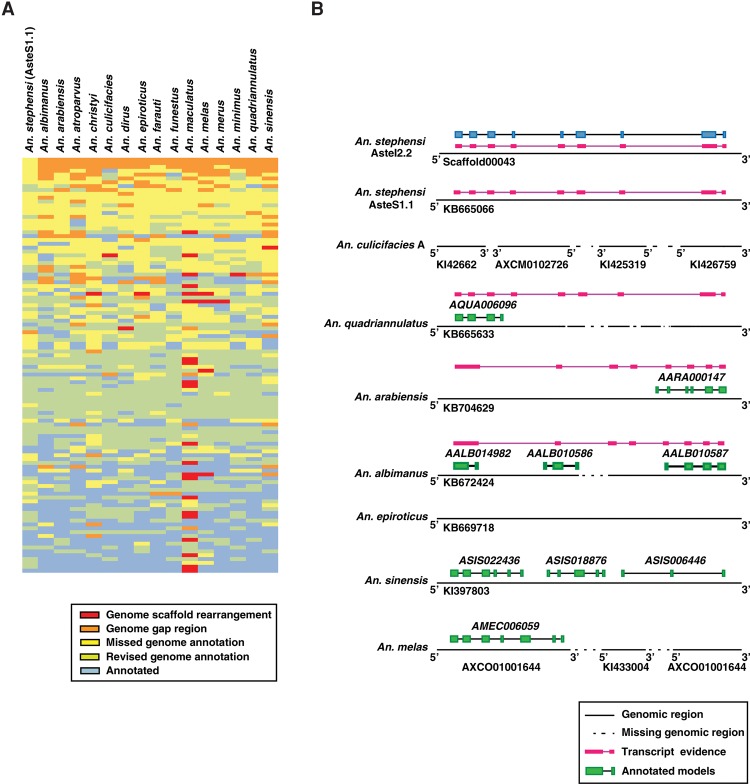
We took the unannotated/novel protein-coding regions from AsteI2 and mapped them against 15 other Anopheline genomes using BLASTN to check whether the novel genes identified in An. stephensi were annotated in other Anopheline genomes or were missed during their annotation (Supplemental Table S6). If the corresponding genomic sequence was present, we looked for the presence of any annotated gene model within the region. In the absence of any annotated gene, we proposed a novel protein-coding region and designated it as a “missed genome annotation.” In other cases, we proposed revision of an existing gene model and termed it a “revised genome annotation.” In the large majority of cases, these claims were supported by transcript evidence from the respective species. Absence of all or part of the corresponding genomic region could possibly be due to inter-species differences or incomplete genome sequence. We referred to such events as a “genome gap region.” If the novel transcripts identified in An. stephensi spanned ends of two scaffolds in a given species, it was considered a possible genome scaffold rearrangement event. Figure 4A represents a heat map of such events across 16 Anopheline genomes. Figure 4B represents one such example in which a novel gene identified in An. stephensi Indian strain was mapped against the other Anopheline genomes and either led to the identification of a novel gene, revision of an existing gene, or joining of adjacent genes in respective genome assemblies. In the case of An. maculatus, the annotation of this gene model was missed because the corresponding genomic region spanned five independent short scaffolds (Supplemental Fig. S4A). In all, using this strategy, we identified 102 examples in An. maculatus in which genes could not be predicted because of incomplete assembly of scaffolds in those regions of the genome. These observations are in agreement with the fact that the genome of An. maculatus is comprised of scaffolds that are greater in number and smaller in size than scaffolds from other Anopheline genomes. In most cases, the missed genome annotations identified across the 15 genomes were supported by RNA-seq evidence from the same species (Supplemental Fig. S4B). Overall, we revised annotations of more than 5800 genomic loci across the 16 Anopheline genomes.
Figure 4.
Comparison of annotations across 16 Anopheline genomes. (A) A schematic representation comparing 100 novel representative annotations identified in An. stephensi with 15 other Anopheline species. The 100 novel events identified in An. stephensi were selected at random and mapped across the 15 Anopheline genomes to check for orthologous sequences in these species. (B) Comparison of one such gene (annotation shown in blue in An. stephensi) provided evidence for revised genome annotation in An. quadriannulatus, An. Arabiensis, and An. albimanus.
Intersection of genome, transcriptome, and proteome data for annotation of noncoding RNAs
Identification of noncoding RNAs is based on computational predictions. Recently, some annotated “noncoding RNAs” in the human genome have been shown to be translated (Bánfai et al. 2012; Chocu et al. 2014; Kim et al. 2014; Wilhelm et al. 2014). To determine the protein-coding potential of intergenic transcripts in An. stephensi, we first analyzed them with the protein coding potential assessment tool (CPAT) (Wang et al. 2013). Of a total of 3423 transcripts, corresponding to 3375 intergenic loci obtained from four tissues, 3110 transcripts (corresponding to 3088 intergenic loci) were predicted to be noncoding RNAs, which included 2436 single exon and 674 multiexon transcripts (Supplemental Table S7). The median ORF length of predicted noncoding RNA was comparatively smaller, i.e., 153 base pairs. On the other hand, the median ORF length of protein coding annotated genes was 1240 base pairs (Supplemental Information Fig. C). However, certain genes can be misannotated as noncoding RNA and in reality get translated in spite of having a short ORF. We used our in-depth proteomic data to further evaluate the coding potential of these predicted noncoding RNAs. In all, 1% (23 transcripts) of all predicted single exon noncoding RNAs and 12% of all predicted multiexon noncoding RNAs (77 transcripts belonging to 64 gene loci) were determined to be protein coding based on peptide evidence. These transcripts would have been incorrectly annotated as noncoding RNAs in the absence of proteomic data. Figure 5A represents a multiexon transcript, which was predicted as noncoding RNA but had a number of peptide hits. Further domain analysis of the translated protein showed presence of ATPase domain, confirming it to be a bona fide protein that would otherwise have been misannotated. Most of such computationally predicted noncoding RNAs had translational evidence from multiple tissues, whereas a subset was expressed at the protein level only in specific tissues (Fig. 5B). The sequence coverage and spectral counts for peptides encoded by tissue-restricted and predicted noncoding RNAs were lower than those that were ubiquitously expressed. Ubiquitously expressed proteins, in general, were also more abundant. When we carried out a similar analysis on known proteins, a similar trend was observed between the tissue-restricted and ubiquitously expressed known proteins (Supplemental Information Fig. D).
Figure 5.

Translational evidence for predicted noncoding RNAs. (A) Multiple peptides from various tissues were identified that corresponded to a computationally predicted noncoding RNA. (B) Relative expression of proteins encoded by computationally predicted noncoding RNAs that were detected across tissues.
Validation of MS/MS spectra for novel peptides
The novel peptides reported in this study were based on matching of tandem mass spectra to translated transcriptome, genome, or orthologous proteins. We manually evaluated the MS/MS spectral quality of the novel peptides to remove false discoveries. In addition, to experimentally validate their fragmentation patterns, we synthesized 175 novel peptides selected at random across the various proteogenomic categories. These peptides were then analyzed using the same instrument parameters on the mass spectrometer that was used for proteomic analysis of the tissues from An. stephensi. MS/MS fragmentation patterns of these synthetic peptides were then manually compared with the MS/MS spectra generated from the proteomic analysis of the tissues (see “Experimentally validated MS/MS spectra” in Supplemental Information). Notably, all 175 peptide identifications were validated upon comparison of the MS/MS fragmentation patterns to that of synthetic peptides. A similar approach has been undertaken in recent studies describing identification of novel peptides (Kim et al. 2014; Wilhelm et al. 2014; Yagoub et al. 2015).
Discussion
We describe a systematic approach for an integrated transcriptomic and proteomic data–based reanalysis of genome assembly and annotation using An. stephensi genome as a proof of principle. We demonstrated both the need and the utility for simultaneous large-scale transcriptomic and proteomic analysis as an integral part of whole-genome sequencing projects by using recently generated whole-genome sequences of 16 Anopheline species. To the best of our knowledge, this is the first such effort in which transcriptomic and proteomic data were used to identify and correct a large number of incomplete genome assemblies in an insect. A recent study on proteomics informed by transcriptomics (PIT) demonstrated integration of transcriptomic and proteomic data to identify known and novel ORFs in the transcripts (Fan et al. 2015). The tool enables searching of the proteomic data against a known protein database and translated transcripts. However, the study does not represent comprehensive annotation of the genome by using in-depth transcriptomic and proteomic data and is limited to visualization of these experimental data against the genome using a genome browser. Also, it does not demonstrate the utility of RNA-seq data in complementing genome assembly process. Peppy, a tool designed for proteogenomic analysis, generates a peptide database from a genome and matches these in silico generated peptides against MS/MS spectra provided as input (Risk et al. 2013). In this study, three novel peptides were identified by searching against the human genome using proteomic data, but there was no integration of RNA-seq data. Another proteogenomic integration tool, QUILTS (Quantitative Integrated Library of Translated SNPs/Splicing), allows protein variant discovery using whole-genome, transcriptome, and proteomic data sets (Ruggles et al. 2016). The utility of this tool was demonstrated by the identification of tumor-specific novel variant peptides and junctional peptides. However, it lacks identification of diverse examples of genome assembly and genome annotation events, which we have demonstrated in our study. For example, identification of novel exons, alternate start sites, exon extension, joining of genes, and translational evidence for noncoding RNAs have not been studied with this tool. None of the aforementioned studies describe annotation of any genome by using in-depth transcriptomic and proteomic data or demonstrate the utility of RNA-seq data in complementing the genome assembly process. Further, these tools are primarily tested on the well-annotated human genome, and hence, they do not define the limitations of existing genome annotation pipelines for newly sequenced genomes of eukaryotic organisms.
As demonstrated by our methods, the use of transcriptomic and proteomic data in the genome assembly and annotation could complement the process of genome annotation. The overall cost of our extensive proteomic analysis presented here was almost six times higher than transcriptomic analysis alone. However, considering the time and cost involved in the annotation of a newly sequenced genome, these costs are minimal. Although transcriptomic data are more economical than proteomic analysis, a large subset of additional information provided by proteomic data could not have been inferred from transcript data alone. For instance, the protein-coding potential of the computationally designated noncoding RNAs could not be confirmed without proteomics data. The noncoding status of this subset of genes would further reduce the likelihood of them being considered for any further studies focusing on the protein-based functions. In addition, translational evidence for intergenic transcripts with predicted protein-coding potential indicated that these were likely novel protein-coding genes. Proteomic data acquired from multiple tissues and developmental stages provided translational evidence for 365 novel genes identified in this study, of which 45 were identified by proteomic data alone. Supplemental Table S5.11 summarizes the number of novel events confirmed by RNA-seq and proteomic data. Similarly, identification of alternate translational start sites and events of an alternate frame of translation was aided by proteomic data that could not have been inferred from RNA-seq data.
We believe that demonstration of the utility of proteogenomic approaches in the correction of incomplete genome assemblies in An. stephensi by this study will provide a platform for genomic and bioinformatics researchers to develop newer tools to carry out similar analysis across recently annotated eukaryotic genomes. The pipeline adopted in this study for mapping transcriptomic and proteomic data onto genome sequences is a cost-effective strategy that can provide a framework for developing more automated workflows, including manual curation for genome annotation. Accurate genome annotation along with proteomic and transcriptomic landscape will provide a better interpretation of a sequenced genome and aid in further investigation related to the biology of the sequenced organisms.
Methods
Mosquito rearing and sample collection
Anopheles stephensi mosquitoes were grown in the insectary of the National Institute of Malaria Research, Field Station, Goa, under ambient conditions (humidity 70 ± 5%, temperature 27 ± 2°C, and a photoperiod:scotoperiod of 12:12 h). Adult mosquitoes were grown on 10% glucose soaked in a cotton pad. Glucose fed, 2–4 d emerged adult An. stephensi females were dissected to obtain antennae, proboscis, maxillary palps, brain, head, salivary glands, thorax, midgut, Malpighian tubules, fat body, and ovaries. Hemolymph was collected by applying a slight pressure on the thorax of cold anesthetized female mosquitoes after cutting the proboscis. Male adult mosquitoes were dissected for midgut and male reproductive organs. All the organs were dissected in 0.65% normal saline under wild stereomicroscope. In addition to dissected organs, third instar larvae and pupae were also collected and preserved at −80°C until use. The tissues dissected for RNA-seq analysis were stored in RNAlater until RNA extraction.
RNA isolation and RNA-seq analysis
Four tissues (midgut, Malpighian tubule, ovary, and fat body) of female An. stephensi mosquitoes, grown in the insectary of National Institute of Malaria Research, Field Station, Goa, were homogenized using a MINILYS benchtop homogenizer and Precelly Lysis Kit (PEQLAB). Total RNA was extracted using miRNeasy kit (Qiagen) according to the manufacturer's protocol and RNA with a RIN value ranging between 9 and 10 was used for library preparation. The RNA-seq libraries were constructed for each tissue using the Illumina TruSeq RNA Sample Preparation Kit v3 as described previously with some minor modification (for details, see Supplemental Methods; Kelkar et al. 2014). The clusters generated from the final library were sequenced on an Illumina HiScanSQ system to obtain a total of about 223 million paired-end reads of 101 bp in length. Reads from the two technical replicates (from two different lanes) of the same RNA-seq library were combined together to represent sequencing readouts for samples from each tissue. The reads were aligned using the Bowtie 2 (Version 2.1.0) (Langmead and Salzberg 2012) against the genome of Anopheles stephensi (Indian strain), downloaded from “VectorBase” (http://www.vectorbase.org/). The aligned reads were assembled using the TopHat (Version 2.0.10) (Kim et al. 2013) and Cufflinks (Version 2.1.1) (Trapnell et al. 2010; Roberts et al. 2011) pipeline. Details regarding the data analysis pipeline and the parameters used are provided in the Supplemental Methods.
Protein-coding potential analysis of transcripts
Protein-coding potential of intergenic transcripts were analyzed using a coding potential assessment tool based on the probability score (Wang et al. 2013). Default settings with the fly model as reference and coding potential cutoff of >0.39 was used. Transcripts with a coding potential score less than 0.39 were considered to be noncoding RNA.
Proteomic analysis
Various mosquito organs (salivary glands, brain, midgut, fat body, Malpighian tubules, ovary, testes, and male accessory glands) were lysed by homogenization followed by sonication in lysis buffer. The extracted proteins were processed using the Filter Aided Sample Preparation (FASP) method of sample preparation as described earlier for LC-MS/MS analysis (Kim et al. 2014). In total, we performed 725 LC-MS/MS runs for samples fractionated using basic pH Reversed Phase Liquid Chromatography (bRPLC), SDS-PAGE, offgel, and strong cation exchange (SCX) methods on the latest hybrid Orbitrap mass spectrometers (for details, see Supplemental Methods).
Mass spectrometry analysis
In this study, we performed a total of 725 LC-MS/MS analyses, of which 120 bRPLC fractions (from salivary glands, midgut, Malpighian tubules, fat body, and testes) were performed on a LTQ-Orbitrap Elite (Thermo Scientific) mass spectrometer interfaced with Easy-nanoLC II nano flow liquid chromatography system (Thermo Scientific), 12 fractions of brain bRPLC were analyzed on a Q-Exactive hybrid Orbitrap, and the remaining 593 fractions (including bRPLC, in-gel, and offgel fractions) were analyzed on a LTQ-Orbitrap Velos mass spectrometer interfaced with Proxeon Easy nLC system (Thermo Scientific). Further details regarding data acquisition settings are provided in the Supplemental Methods.
Database searches
The raw data obtained were processed using a unique workflow consisting of multiple search nodes on Proteome Discoverer (Version 1.4.1.14) software (Thermo Fisher Scientific). The data were searched against An. stephensi protein databases (Astel2.1 – Indian strain and AsteS1.0 – SDA500 strain) from VectorBase using Sequest and Mascot search algorithms. The searches were performed sequentially against An. stephensi protein database, three-frame translated RNA-seq-based transcript database, An. gambiae protein database, and six-frame translated An. stephensi genome databases, respectively (as shown in Fig. 1). The search parameters are described in detail in the Supplemental Methods. A FDR cutoff of 1% at the peptide level was used for identification. We carried out combined Mascot and Sequest search for each tissue. We retained all of the peptide assignments that were identified by Mascot and Sequest along with the corresponding scores for each PSM in separate columns. In all, 90% of PSMs were identified by both search engines, ∼5% by Sequest alone and 5% by Mascot alone. However, if a given scan ID or spectrum was assigned to different peptides by Mascot and Sequest, they were discarded to maintain a stringency. We achieved this using an in-house program, which identified the individual spectral assignment based on the combination of scan ID, fraction number, and file names. Quantitation of the proteins identified across the tissues was performed by counting the total number of PSMs for all the peptides corresponding to a protein (for details, see Supplemental Methods).
Proteogenomic analysis of An. stephensi
To enable identification of novel protein-coding regions in the An. stephensi genome, we searched proteomic data against three-frame translated RNA-seq transcripts, six-frame translated An. stephensi genome, and An. gambiae protein databases using a unique search workflow on Proteome Discoverer (Version 1.4.1.14) software (Thermo Fisher Scientific). These peptides were manually analyzed using the Proteogenomics workflow (as described in Fig. 1) to identify novel genes missed in the annotation pipeline along with those missed due to gaps in the genome assembly. The peptides identified in these searches also provided evidence for novel coding regions contributing to the revision of annotated gene models due to the inherent limitations of the existing annotation pipeline or due to the incomplete genome assembly, as described previously (Kelkar et al. 2014; Kim et al. 2014; Supplemental Methods). Details of the identification and revision of genome sequencing and assembly errors in An. stephensi are provided in the Supplemental Methods.
Validation of novel identifications through an LC-MS/MS analysis of synthetic peptides
A total of 175 identified peptides were selected at random from various categories of proteogenomics analysis and synthesized as four pools of synthetic peptides (JPT Peptide Technologies). Two pools contained 50 synthetic peptides each, which were derived from identifications based on LTQ-Orbitrap Velos, and the other two pools contained 35 and 40 peptides, respectively, that were derived from identifications based on the LTQ-Orbitrap Elite mass spectrometer. Each pool was dried and diluted using 0.1% formic acid. Peptides from each pool were subjected to LC-MS/MS analysis on the LTQ-Orbitrap Velos and LTQ-Orbitrap Elite mass spectrometers. Fragmentation patterns of these 175 synthetic peptides were then manually compared and validated with that of peptides identified in proteogenomic analysis (Supplemental Information).
Genome annotation and genome assembly improvements across 16 Anopheline species
Novel events thus identified in An. stephensi were cross-checked in the other 16 Anopheline species (An. stephensi SDA500, An. arabiensis, An. quadriannulatus, An. merus, An. melas, An. christyi, An. epiroticus, An. maculatus (sp. B), An. funestus, An. minimus s.s. (sp. A), An. culicifacies A, An. farauti, An. dirus s.s. (sp. A), An. atroparvus, An. sinensis, and An. albimanus), recently sequenced by Broad Institute and available on VectorBase (as described in Fig. 1).
RT-PCR analysis
Total RNA was isolated from midgut, ovary, salivary gland, testes, and whole female mosquitoes. RT-PCR validation was carried out for 48 novel genes and two gene correction events. Primers were designed in the exonic regions of the alternate gene models, and specific amplicons were purified and sequenced. Primer sequences and associated details are provided in Supplemental Table S5.10.
Data access
The mass spectrometry–based proteomics data from this study have been submitted to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository under the data set identifier PXD001128. The RNA-seq-based transcriptomic data from this study have been submitted to the NCBI Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra) and the Transcriptome Shotgun Assembly (TSA; https://www.ncbi.nlm.nih.gov/genbank/tsa/) database under accession numbers SRP043489 and GBVY00000000.1, respectively. cDNA sequences of 50 novel genes identified against the previous An. stephensi genome assembly annotations (ASTEI1) from VectorBase have been submitted to dbEST (https://www.ncbi.nlm.nih.gov/nucest) under the GenBank accession numbers JZ152704.1–JZ152780.1.
Supplementary Material
Acknowledgments
This paper is funded by the joint research project to NIMR and IOB entitled “Characterization of Malaria vector Anopheles stephensi Proteome and Transcriptome” (EMR/2014/000444) from the Science and Engineering Research Board (SERB), Department of Science and Technology, Government of India. T.S.K.P. is also supported by the DBT Program Support grant on “Development of infrastructure and a computational framework for analysis of proteomic data” (BT/01/COE/08/05). We also thank Infosys Foundation for financial support to IOB. A.P. and P.S. were funded by a pilot grant from the Johns Hopkins Malaria Research Institute. This paper bears the NIMR publication screening committee approval No. 009/2015. H.G. is a Wellcome Trust-DBT India Alliance Early Career Fellow. We thank the Council of Scientific and Industrial Research, Department of Biotechnology, University Grants Commission, Indian Council of Medical Research and Department of Science and Technology, Government of India for research fellowships to M.K., S.K.S., G.D., R.S.N., S.M.P., A.K.M. (IOB), S.S.M., M.K.G., S.B.D., D.S.K., P.R., N.S., S.D.Y., K.K.D., R.R., A.A.K., A.R., G.J.S., S.C., and R.V. M.D. is funded by the Faculty Improvement Program of Siddaganga Institute of Technology, Tumkur.
Author contributions: A.P., T.S.K.P., H.G., and A.K. designed the study and experimental workflow. T.S.K.P., A.K.M., M.K., S.K.S., and G.D. contributed equally to this manuscript. T.S.K.P., A.K.M., M.K., S.K.S., G.D., M.K.G., D.S.K., and S.B.D. dissected mosquitoes and collected organs, carried out sample preparation for mass spectrometry, and data analysis. C.W., S.K.S., and G.D. prepared RNA samples for RNA-seq analysis. A.K. and A.K.M. reared and provided larvae, pupae, and adult mosquitoes for this study. M.K., R.S.N., S.M.P., S.C., G.J.S., and C.H.N. performed LC-MS/MS; C.W. constructed RNA-seq libraries and generated transcriptomic (RNA-seq) data; A.K.M. (IOB), A.H.P., J.A., C.J.M., and S.S.M. prepared databases and processed the transcriptomic and proteomic data using Perl and Python scripts; I.V.S., Z.T., B.H., and X.J. provided genomic and transcriptomic (RNA-seq) data of the An. stephensi Indian strain; M.K., S.K.S., G.D., M.K.G., S.B.D., P.R., D.S.K., T.S.K.P., H.G., and A.P. performed proteogenomic analysis; N.K. and P.S. assisted with analysis and interpretation of data; N.S., S.D.Y., K.K.D., A.A.K., H.S.S., R.R., A.P.J., M.D., S.J., A.R., G.J.S., S.C., K.M.P., and R.V. assisted with manual validation of novel and revised protein-coding genes; G.D., R.S.N., M.K., and S.K.S. illustrated figures and prepared Supplemental Tables with the help of other authors; and A.P., T.S.K.P., H.G., A.K., M.K., S.K.S., G.D., and A.K.M. wrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.201368.115.
References
- Armengaud J. 2009. A perfect genome annotation is within reach with the proteomics and genomics alliance. Curr Opin Microbiol 12: 292–300. [DOI] [PubMed] [Google Scholar]
- Bánfai B, Jia H, Khatun J, Wood E, Risk B, Gundling WE Jr, Kundaje A, Gunawardena HP, Yu Y, Xie L, et al. 2012. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res 22: 1646–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock T, Chen WH, Ori A, Malik N, Silva-Martin N, Huerta-Cepas J, Powell ST, Kastritis PL, Smyshlyaev G, Vonkova I, et al. 2014. An integrated approach for genome annotation of the eukaryotic thermophile Chaetomium thermophilum. Nucleic Acids Res 42: 13525–13533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner E, Ahrens CH, Mohanty S, Baetschmann H, Loevenich S, Potthast F, Deutsch EW, Panse C, de Lichtenberg U, Rinner O, et al. 2007. A high-quality catalog of the Drosophila melanogaster proteome. Nat Biotechnol 25: 576–583. [DOI] [PubMed] [Google Scholar]
- Castellana NE, Shen Z, He Y, Walley JW, Cassidy CJ, Briggs SP, Bafna V. 2014. An automated proteogenomic method uses mass spectrometry to reveal novel genes in Zea mays. Mol Cell Proteomics 13: 157–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaerkady R, Kelkar DS, Muthusamy B, Kandasamy K, Dwivedi SB, Sahasrabuddhe NA, Kim MS, Renuse S, Pinto SM, Sharma R, et al. 2011. A proteogenomic analysis of Anopheles gambiae using high-resolution Fourier transform mass spectrometry. Genome Res 21: 1872–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chocu S, Evrard B, Lavigne R, Rolland AD, Aubry F, Jégou B, Chalmel F, Pineau C. 2014. Forty-four novel protein-coding loci discovered using a proteomics informed by transcriptomics (PIT) approach in rat male germ cells. Biol Reprod 91: 123. [DOI] [PubMed] [Google Scholar]
- Denoeud F, Aury JM, Da Silva C, Noel B, Rogier O, Delledonne M, Morgante M, Valle G, Wincker P, Scarpelli C, et al. 2008. Annotating genomes with massive-scale RNA sequencing. Genome Biol 9: R175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl D, Bradnam K, St John J, Darling A, Lin D, Fass J, Yu HO, Buffalo V, Zerbino DR, Diekhans M, et al. 2011. Assemblathon 1: a competitive assessment of de novo short read assembly methods. Genome Res 21: 2224–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Saha S, Barker G, Heesom KJ, Ghali F, Jones AR, Matthews DA, Bessant C. 2015. Galaxy integrated omics: web-based standards-compliant workflows for proteomics informed by transcriptomics. Mol Cell Proteomics 14: 3087–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. 2010. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330: 1775–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Bird DM, Nielsen DM. 2014. Improved structural annotation of protein-coding genes in the Meloidogyne hapla genome using RNA-Seq. Worm 3: e29158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N, Benhamida J, Bhargava V, Goodman D, Kain E, Kerman I, Nguyen N, Ollikainen N, Rodriguez J, Wang J, et al. 2008. Comparative proteogenomics: combining mass spectrometry and comparative genomics to analyze multiple genomes. Genome Res 18: 1133–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hittinger CT, Johnston M, Tossberg JT, Rokas A. 2010. Leveraging skewed transcript abundance by RNA-Seq to increase the genomic depth of the tree of life. Proc Natl Acad Sci 107: 1476–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Peery A, Hall A, Sharma A, Chen XG, Waterhouse RM, Komissarov A, Riehl MM, Shouche Y, Sharakhova MV, et al. 2014. Genome analysis of a major urban malaria vector mosquito, Anopheles stephensi. Genome Biol 15: 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelkar DS, Kumar D, Kumar P, Balakrishnan L, Muthusamy B, Yadav AK, Shrivastava P, Marimuthu A, Anand S, Sundaram H, et al. 2011. Proteogenomic analysis of Mycobacterium tuberculosis by high resolution mass spectrometry. Mol Cell Proteomics 10: M111.011627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelkar DS, Provost E, Chaerkady R, Muthusamy B, Manda SS, Subbannayya T, Selvan LD, Wang CH, Datta KK, Woo S, et al. 2014. Annotation of the zebrafish genome through an integrated transcriptomic and proteomic analysis. Mol Cell Proteomics 13: 3184–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Salzberg SL. 2011. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol 12: R72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Madugundu AK, Kelkar DS, Isserlin R, Jain S, et al. 2014. A draft map of the human proteome. Nature 509: 575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriventseva EV, Tegenfeldt F, Petty TJ, Waterhouse RM, Simao FA, Pozdnyakov IA, Ioannidis P, Zdobnov EM. 2015. OrthoDB v8: update of the hierarchical catalog of orthologs and the underlying free software. Nucleic Acids Res 43: D250–D256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linde J, Duggan S, Weber M, Horn F, Sieber P, Hellwig D, Riege K, Marz M, Martin R, Guthke R, et al. 2015. Defining the transcriptomic landscape of Candida glabrata by RNA-Seq. Nucleic Acids Res 43: 1392–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megy K, Emrich SJ, Lawson D, Campbell D, Dialynas E, Hughes DS, Koscielny G, Louis C, Maccallum RM, Redmond SN, et al. 2012. VectorBase: improvements to a bioinformatics resource for invertebrate vector genomics. Nucleic Acids Res 40: D729–D734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrihew GE, Davis C, Ewing B, Williams G, Kall L, Frewen BE, Noble WS, Green P, Thomas JH, MacCoss MJ. 2008. Use of shotgun proteomics for the identification, confirmation, and correction of C. elegans gene annotations. Genome Res 18: 1660–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Schwarz EM, Williams B, Schaeffer L, Antoshechkin I, Wold BJ, Sternberg PW. 2010. Scaffolding a Caenorhabditis nematode genome with RNA-seq. Genome Res 20: 1740–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neafsey DE, Waterhouse RM, Abai MR, Aganezov SS, Alekseyev MA, Allen JE, Amon J, Arcà B, Arensburger P, Artemov G, et al. 2015. Mosquito genomics. Highly evolvable malaria vectors: the genomes of 16 Anopheles mosquitoes. Science 347: 1258522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renuse S, Chaerkady R, Pandey A. 2011. Proteogenomics. Proteomics 11: 620–630. [DOI] [PubMed] [Google Scholar]
- Risk BA, Spitzer WJ, Giddings MC. 2013. Peppy: proteogenomic search software. J Proteome Res 12: 3019–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Pimentel H, Trapnell C, Pachter L. 2011. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 27: 2325–2329. [DOI] [PubMed] [Google Scholar]
- Ruggles KV, Tang Z, Wang X, Grover H, Askenazi M, Teubl J, Cao S, McLellan MD, Clauser KR, Tabb DL, et al. 2016. An analysis of the sensitivity of proteogenomic mapping of somatic mutations and novel splicing events in cancer. Mol Cell Proteomics 15: 1060–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg SL, Phillippy AM, Zimin A, Puiu D, Magoc T, Koren S, Treangen TJ, Schatz MC, Delcher AL, Roberts M, et al. 2012. GAGE: a critical evaluation of genome assemblies and assembly algorithms. Genome Res 22: 557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitski MM, Wilhelm M, Hahne H, Kuster B, Bantscheff M. 2015. A scalable approach for protein false discovery rate estimation in large proteomic data sets. Mol Cell Proteomics 14: 2394–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz MC, Delcher AL, Salzberg SL. 2010. Assembly of large genomes using second-generation sequencing. Genome Res 20: 1165–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapp J, Geffard O, Imbert G, Gaillard JC, Davin AH, Chaumot A, Armengaud J. 2014. Proteogenomics of Gammarus fossarum to document the reproductive system of amphipods. Mol Cell Proteomics 13: 3612–3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Park HJ, Dasari S, Wang S, Kocher JP, Li W. 2013. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res 41: e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm M, Schlegl J, Hahne H, Moghaddas Gholami A, Lieberenz M, Savitski MM, Ziegler E, Butzmann L, Gessulat S, Marx H, et al. 2014. Mass-spectrometry-based draft of the human proteome. Nature 509: 582–587. [DOI] [PubMed] [Google Scholar]
- Woo S, Cha SW, Merrihew G, He Y, Castellana N, Guest C, MacCoss M, Bafna V. 2014. Proteogenomic database construction driven from large scale RNA-seq data. J Proteome Res 13: 21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Zhang H, Lin W, Hao Y, Ren L, Zhang C, Li N, Wei H, Jiang Y, He F. 2014. Discovery of novel genes and gene isoforms by integrating transcriptomic and proteomic profiling from mouse liver. J Proteome Res 13: 2409–2419. [DOI] [PubMed] [Google Scholar]
- Xue W, Li JT, Zhu YP, Hou GY, Kong XF, Kuang YY, Sun XW. 2013. L_RNA_scaffolder: scaffolding genomes with transcripts. BMC Genomics 14: 604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagoub D, Tay AP, Chen Z, Hamey JJ, Cai C, Chia SZ, Hart-Smith G, Wilkins MR. 2015. Proteogenomic discovery of a small, novel protein in yeast reveals a strategy for the detection of unannotated short open reading frames. J Proteome Res 14: 5038–5047. [DOI] [PubMed] [Google Scholar]
- Yu Y, Fuscoe JC, Zhao C, Guo C, Jia M, Qing T, Bannon DI, Lancashire L, Bao W, Du T, et al. 2014. A rat RNA-Seq transcriptomic BodyMap across 11 organs and 4 developmental stages. Nat Commun 5: 3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.