Abstract
MicroRNAs (miRNAs) regulate gene output by targeting degenerate elements in mRNAs and have undergone drastic expansions in higher metazoan genomes. The evolutionary advantage of maintaining copies of highly similar miRNAs is not well understood, nor is it clear what unique functions, if any, miRNA family members possess. Here, we study evolutionary patterns of metazoan miRNAs, focusing on the targeting preferences of the let-7 and miR-10 families. These studies reveal hotspots for sequence evolution with implications for targeting and secondary structure. High-throughput screening for functional targets reveals that each miRNA represses sites with distinct features and regulates a large number of genes with cooperative function in regulatory networks. Unexpectedly, given the high degree of similarity, single-nucleotide changes grant miRNA family members with distinct targeting preferences. Together, our data suggest complex functional relationships among miRNA duplications, novel expression patterns, sequence change, and the acquisition of new targets.
MicroRNAs (miRNAs) are ∼22-nucleotide (nt) noncoding RNAs that regulate a multitude of biological processes in metazoans, and have been implicated in various diseases. MiRNAs recognize target elements primarily in 3′ untranslated regions (3′ UTRs) of messenger RNA (mRNA) by Watson-Crick and G:U wobble pairing. These elements are short and degenerate and are driven largely by perfect complementarity in nucleotides 2–7 of the miRNA, which is termed the “seed” region (Lewis et al. 2005). However, the presence of a perfect seed does not always confer regulation by complementary miRNAs (Grimson et al. 2007; Wolter et al. 2015b), and there are several alternative modes of miRNA target recognition that do not utilize canonical seed pairing (Ha et al. 1996; Moss et al. 1997; Reinhart et al. 2000; Johnston and Hobert 2003; Lal et al. 2009; Shin et al. 2010; Gabriely et al. 2011; Chi et al. 2012; Wolter et al. 2015b). The result of miRNA targeting is typically a modest reduction in protein output (Baek et al. 2008) by deadenylation and subsequent mRNA degradation (Wu et al. 2006). However, translation inhibition without a reduction of mRNA levels is frequently observed (Guo et al. 2010; Wu et al. 2010).
The modest effect of miRNA targeting on individual protein levels suggests that miRNAs may productively impact cell fate by targeting multiple components of regulatory networks. This has been demonstrated with multiple approaches: experimentally, by measuring mRNA abundance following miRNA perturbations (Linsley et al. 2007); bioinformatically, using multiple features of target sites and pathway annotations (Tsang et al. 2010); and by high-throughput screening for direct miRNA targets (Wolter et al. 2015b). However, the complexity of targeting principles, mechanisms of translational repression, and high frequency of noncanonical targets complicates the prediction and detection of cooperative networks of bona fide miRNA targets using traditional approaches. Reporter assays overcome these caveats by measuring RNA interactions that result in functional repression at the protein level. However, these assays are costly and difficult to perform at a scale conducive to the discovery of miRNA regulated gene networks.
miRNAs have undergone several pronounced expansions in metazoan genomes (Hertel et al. 2006; Berezikov 2011). These expansions coincide with evolving organismal complexity yet seldom correspond to increases in the number of protein coding genes (Berezikov 2011). While expansions are driven in part by the de novo evolution of novel miRNAs, the majority arise from duplications of existing miRNAs. These miRNAs are rarely lost following duplication events, suggesting selective pressure to acquire and maintain new miRNAs (Wheeler et al. 2009). Following duplication, the primary sequence of miRNAs frequently evolves at positions outside the seed region, creating families of miRNAs that have distinct sequence variations. The advantage of the expanded miRNA repertoire of higher metazoans is unclear, but it has been suggested that copies of highly similar miRNAs provide robustness to gene regulatory networks (Ebert and Sharp 2012). This has led to the assumption that miRNA family members target largely, if not completely, overlapping gene sets. However, the extent to which nucleotide changes impart novel targeting specificity to miRNA family members has not been systematically studied, and it is unclear what unique functions individual miRNA family members possess. Here we report our efforts to characterize evolutionary trends in metazoan miRNA families, quantify targeting specificity between miRNA family members, and assess how these families evolve to regulate gene networks.
Results
miRNA families exhibit position-specific functional signatures
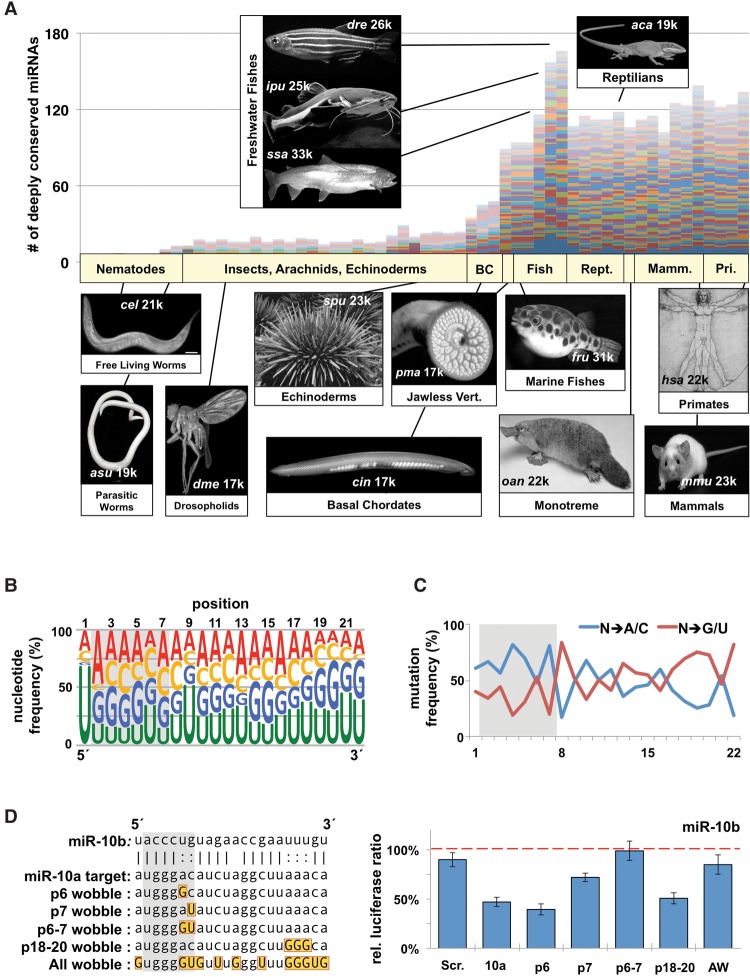
We hypothesized that evolutionary patterns may reveal information about the function of miRNA family members in higher metazoans. To test this idea, we analyzed the sequences of the most deeply conserved metazoan miRNA families (n = 3265 distinct miRNAs, 62 families), with the sole criteria being that each family must possess at least one homolog in the human genome (Supplemental Table S1; Kozomara and Griffiths-Jones 2014). The raw number of miRNAs showed that these families have undergone a drastic expansion at the base of the vertebrate lineage, and outside of freshwater, teleosts have not changed dramatically in vertebrates (Fig. 1A). When correcting for the number of miRNAs per species, we observed that the number of miRNAs that exist in multicopy families has increased about fourfold in vertebrates, with 85% of deeply conserved human miRNAs belonging to families that contain at least two members (Supplemental Fig. S1A).
Figure 1.
The evolution of highly conserved miRNA families reveals functional signatures. (A) We downloaded all metazoan pre-miRNAs and mature miRNAs from miRBase and identified 62 miRNA families that contain the most members in metazoans. Plotted are the raw numbers of pre-miRNAs in each species, with each colored bar representing a single miRNA family. Images represent the prominent model organism in each major phylogenetic grouping. Numbers on each image represent approximate number of protein coding genes. (B) Nucleotide content by position of the mature miRNA strand for all conserved miRNA families within metazoans. Data represent 3685 miRNAs from 62 different families in all metazoans. Relative position is based on the most frequent 5′ nucleotide from sequencing data. Seed region is marked with a gray box. (C) We measured the specific mutation frequency of specific nucleotides away from the conserved base in the family, toward either A/C or G/U. (D) Various permutations of the miR-10a complementary sequence were cloned into the SV40 3′ UTR (left). Gold boxes mark wobble nucleotides in targets, each of which was tested for repression by miR-10b in dual luciferase assays in HEK293T cells (right). Repression was measured as the change in relative luciferase ratio in response to miRNA cotransfection. Values were normalized to a negative miRNA control (dashed line) (n = 3). All error bars are SEM unless otherwise noted. (Scr.) Scrambled sequence; (10a) miR-10a; (AW) all wobble.
These miRNA families showed no bias toward either the 5p or 3p arms (Supplemental Fig. S1B), and nucleotide content confirmed the expected complementarity in total nucleotide content between the miRNA and miRNA* strands (Supplemental Fig. S1C). When we looked at individual positions within the miRNA, we observed an enrichment of U residues in the first position in the miRNA strand, which plays a role in Argonaute incorporation (Fig. 1B; Wang et al. 2012). There is a preference for A residues in the second position and U residues in the ninth position, as well as an overall enrichment of G/U nucleotides in the 3′ end. Conversely, the miRNA* strand showed no positional nucleotide bias beyond a depletion for U residues in the first position (Supplemental Fig. S1D).
Next, we explored if each position favored specific point mutations, reasoning that this measure would independently assess preferences in each position regardless of the identity of the conserved nucleotide. When we grouped nucleotide changes based on ability to form wobble base pairs, we noticed that G/U mutations were generally depleted in the seed region, neutral in the central region, and enriched in the 3′ end (Fig. 1C). To assess the tolerance of wobbles during miRNA repression, we tested the ability of human miR-10b to repress an artificial target site (Fig. 1D). In agreement with the evolutionary trends (Fig. 1C), G:U wobble pairing had no impact at position 6 (p6), or in p18–20, but was not well tolerated in p7 (Fig. 1D). Previous experiments found that wobble base-pairing in the seed negatively impacts the affinity and translational repression between a miRNA and target mRNA; however, wobble pairing in p6 was not tested in these experiments (Brennecke et al. 2005; Wee et al. 2012). Our results are consistent with the idea that specificity in the majority of the seed region via Watson-Crick base-pairing is a conserved feature of miRNA:mRNA interactions, but expands upon the details of positional specificity. Furthermore, wobble pairing in the 3′ end of the miRNA may be advantageous by increasing the frequency of base-pairing in canonical miRNA targets.
miRNA families evolve at specific hotspots
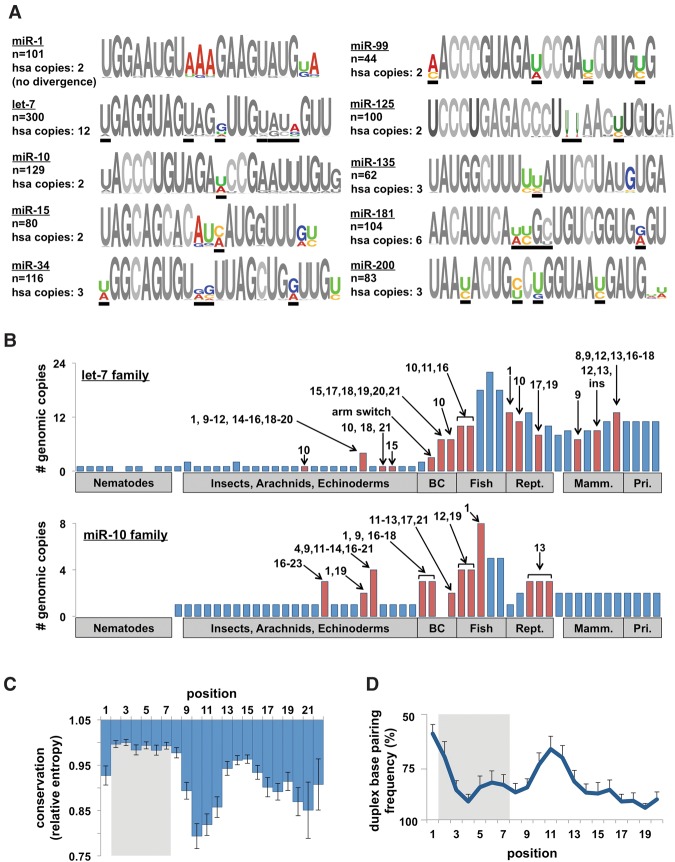
Next, we investigated patterns of sequence variation within miRNA families. Alignments of families revealed conserved nucleotide changes at specific positions, which we refer to as evolutionary “hotspots” (Fig. 2A). We observed that the majority of deeply conserved miRNAs belonging to multimember families have experienced at least one nucleotide change, which are frequently conserved in humans (Fig. 2A, black bars). When we investigated where in the metazoan phylogeny these hotspots first appear, we find that in the absence of duplication events there is rarely sequence evolution (Fig. 2B; Supplemental Fig. S2). Conversely, duplications are almost invariably followed by the emergence of sequence changes.
Figure 2.
Evolutionary patterns of highly conserved miRNA families. (A) Sequence logos for 10 representative miRNA families from all metazoan species. Colored nucleotides represent highly variable positions across species (hotspots). Number of miRNAs in each family and the number of copies in the human genome are noted. Black bars represent the positions that are variable within the human families. (B) The number of family members of let-7 and miR-10b in metazoan species. Arrows and numbers specify the position(s) of nucleotide changes within the mature miRNA relative to the nearest species. The numbers represent changes unique to that species, not cumulative changes. Red bars denote species with sequence changes. (C) We calculated the conservation score (entropy) of each position in all 62 families and combined them to reveal general trends in sequence evolution. Conservation score normalized to most conserved position (p3). (D) Frequency of base-pairing between miRNA/miRNA* strands in the miRNA duplex. Base-pairing information was extracted from alignments on miRBase. This figure is representative of 62 families in eight species.
An analysis of the positional conservation scores in all 62 miRNA families revealed that evolutionary hotspots center on nucleotides 9–12 of the mature miRNA (Fig. 2C). Outside the seed, conservation is highest at positions 13–15 and decreases until the 3′ end of the miRNA (Fig. 2C). Conversely, the miRNA* strand is uniformly conserved at positions 1–8 and 13–18 and is poorly conserved in positions 9–13 (Supplemental Fig. S3A). The mirrored conservation between the miRNA and miRNA* strands suggested that the secondary structure of the miRNA duplex may influence sequence evolution. To investigate this, we measured the frequency of base-pairing at each position in the miRNA duplex in eight model species (Supplemental Fig. S3B) and observed a relatively low frequency of base-pairing at the 5′ end of the miRNA, as well as a high frequency at the 3′ end (Fig. 2D). We also observed a bias in the position of single-nucleotide bulges in the 5′ end of the miRNA strand and the 3′ end of the miRNA* strand (Supplemental Fig. S3C). These results highlight the asymmetry of the miRNA duplex, which plays a role in strand selection during Argonaute incorporation (Schwarz et al. 2003). Surprisingly, we also observed that p10–12 are unpaired as frequently as p1–2, suggesting that relaxed base-pairing constraints may contribute to the high frequency of nucleotide changes observed in the central region of these miRNA families.
Different miRNAs employ distinct targeting footprints
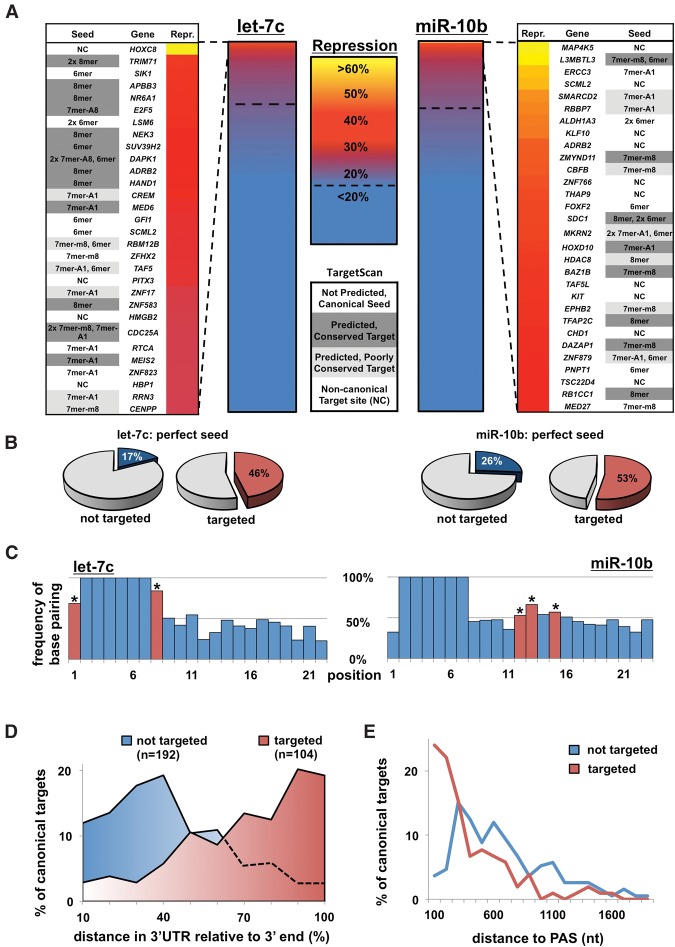
Evidence from RNA-seq and in vitro binding affinity experiments suggests that the central region of miRNAs may not substantially contribute to target recognition (Grimson et al. 2007; Wee et al. 2012). However, as mRNA abundance and binding strength may not be directly indicative of the miRNA's effect on protein levels (Guo et al. 2010; Wu et al. 2010), we wondered if similar patterns would be obtained when measuring protein output directly. To test this, we employed a high-throughput luciferase-based screen called Luminescent Identification of Functional Elements in 3′ UTRs (3′ LIFE) (Wolter et al. 2015a, b). We tested two deeply conserved miRNAs (let-7c and miR-10b) against a target library composed of 1407 full-length human 3′ UTRs, corresponding to proteins with regulatory functions including kinases, RNA-binding proteins, and transcription factors (h3′ UTRome v1) (Kotagama et al. 2015). We identified 334 putative targets (>20% repression, P < 0.05) (Fig. 3A,B; Supplemental Table S2), which were enriched for canonical target sites (Fig. 3B). When we looked for the presence of targets with a p6 wobble, we did not observe enrichment within the let-7c or miR-10b targets (Supplemental Fig. S4A), suggesting that while these miRNAs may be capable of utilizing this motif, it is not preferred.
Figure 3.
3′ LIFE assay identifies 3′ UTRs repressed by let-7c and miR-10b. (A) Summary of 3′ LIFE screens. We tested 1407 3′ UTRs in triplicate for targeting by each miRNA in cotransfection experiments in HEK293T cells (∼13,000 dual luciferase reactions). A 3′ UTR was considered a putative target if it exhibited >20% repression and P < 0.05. The top 30 repressed genes for each miRNA are listed adjacent to each heatmap, with the type(s) of miRNA target site(s). Predicted targets, both deeply and poorly conserved, were identified using TargetScan 7.0. (B) Characteristics of miRNA target sites in 3′ UTR library separated by target score measured by the 3′ LIFE assay. (C) Alignments of target sites from the top 50 hits with a canonical seed for let-7c (left) and miR-10b (right). Asterisks represent statistically significant nucleotides (P < 0.05, χ2). (D,E) Distance of putative miRNA targets to the mRNA end. 3′ UTRs in the library with one canonical miRNA target site and one canonical polyadenylation sequence (AAUAAA) were analyzed for the relative distance between the poly(A) site and the miRNA target site. Genes were separated based on targeting status.
In order to identify which positions cooperate with canonical seeds in granting functional repression, we analyzed the sequence downstream from seed sites in the top 50 canonical target sites for each miRNA (Fig. 3C). Interestingly, the targeting footprints of let-7c and miR-10b exhibit distinct patterns. Consistent with the classic model of miRNA targeting (Grimson et al. 2007), let-7c strongly favors 7mers and 8mers (Fig. 3C). The position with the least complementarity, p12, is the site with the most heterogeneity in the human let-7 miRNAs (Fig. 2A), suggesting that other family members may utilize this position in these targets. In contrast, miR-10b relies more heavily on positions 12, 13, and 15 (Fig. 3C). To further characterize miRNA binding preferences, we performed a targeted screen for two additional miRNAs, miR-34a and miR-125b (Supplemental Fig. S4B–D), and found that each miRNA exhibits distinct targeting footprints. Together, this suggests that individual miRNAs may utilize distinct features of target sites for the recognition and repression of canonical targets.
Of the 773 genes in the 3′ LIFE screen that contain a canonical target site, only 163 (21%) show any measure of repression (Fig. 3B). To test how the position of miRNA targets impacts the strength of repression, we measured the relative distance between the miRNA target site and the 3′ UTR end (Fig. 3D,E). This revealed that functional target sites tend to cluster toward the 3′ end of the mRNA (Fig. 3D). When looking at absolute distance to the poly(A) tail, we find that when a canonical target site is located in the last 200 nt of an mRNA, it will lead to functional repression the majority of the time (Fig. 3E). However, beyond this distance, additional factors may influence the ability of a miRNA to functionally repress a target mRNA.
let-7c and miR-10b target multiple genes in regulatory networks
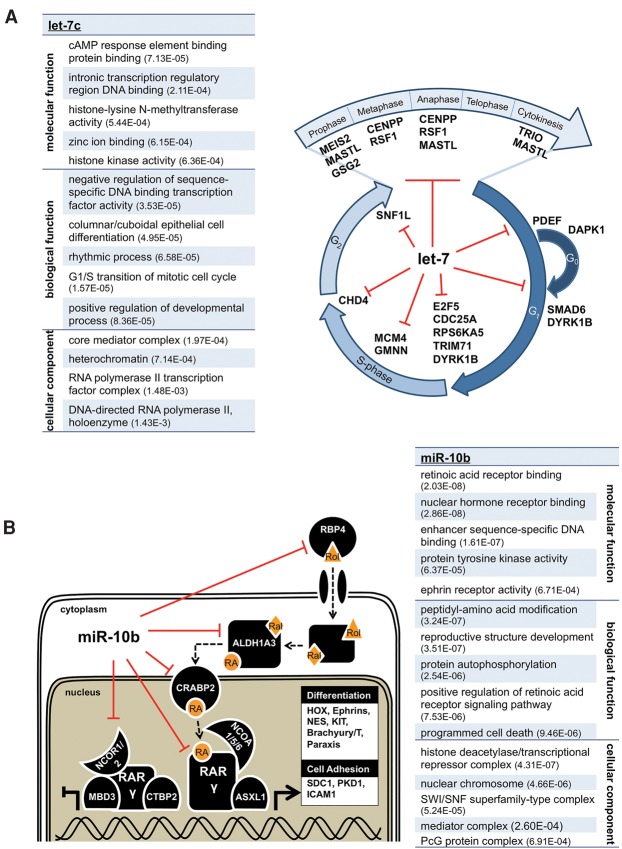
To identify networks targeted by these miRNAs, we performed a gene ontology (GO) analysis of the hits obtained in our screen and found that each miRNA repressed genes with cooperative function in specific biological pathways (Fig. 4). Let-7c targeted multiple members of the general transcription machinery (splicing, mediator complex, and general transcription factors), as well as many key proteins involved in cell cycle checkpoints, DNA replication, and mitosis (Fig. 4A; Johnson et al. 2007). MiR-10b targeted multiple ephrin receptors, mediator subunits, and genes implicated in apoptosis. We also found that miR-10b targeted genes at nearly all levels of the retinoic acid (RA) signaling pathway (Foley et al. 2011; Gabriely et al. 2011), including a nuclear RA receptor (RARG), RA-metabolite transport proteins (RBP4, CRABP2), a rate-limiting enzyme in RA synthesis (ALDH1A3), several RA receptor cofactors, and downstream effectors of the RA response (Fig. 4B). To validate endogenous targeting of these genes, we depleted miR-10 in HEK293T cells using a doxycycline inducible miRNA sponge (Supplemental Fig. S5A,B; Szulc et al. 2006; Ma et al. 2010) and observed that as levels of miR-10 family members decrease, mRNA levels of target genes increase (Supplemental Fig. S5C,D). One of these genes, RARG, possesses two unpredicted canonical target sites, but deletion analysis revealed that only the most distal site was functional (Supplemental Fig. S5E,F). Combined with the modest repression we observe for the majority of targets, the striking convergence of targets into cooperative networks provides evidence that miRNAs effectively impact cell fate by targeting multiple components of regulatory networks.
Figure 4.
let-7c and miR-10b target multiple genes in regulatory networks. (A) GO term analysis of genes repressed by let-7c; P-value noted in parentheses. let-7c targets multiple genes associated with cell cycle checkpoint progression, DNA synthesis, and several steps of mitosis. All listed genes were identified as targets by the 3′ LIFE screen (repression >20%, P < 0.05). (B) GO term analysis of miR-10b targets, highlighting that miR-10b targets multiple genes in the retinoic acid signaling pathway. Genes with experimental evidence for physical association are adjacent to each other. miR-10b regulates several key components of the pathway, including extracellular transport and delivery of the retinoic acid precursor retinol (Rol), the conversion of retinaldehyde (Ral) to retinoic acid (RA), cytoplasm to nucleus transport of RA to nuclear hormone receptors, one of the RA receptors (RAR gamma), and multiple effectors of RA activation.
Nucleotide changes at evolutionary hotspots in the human let-7 and miR-10 families modulate targeting preferences
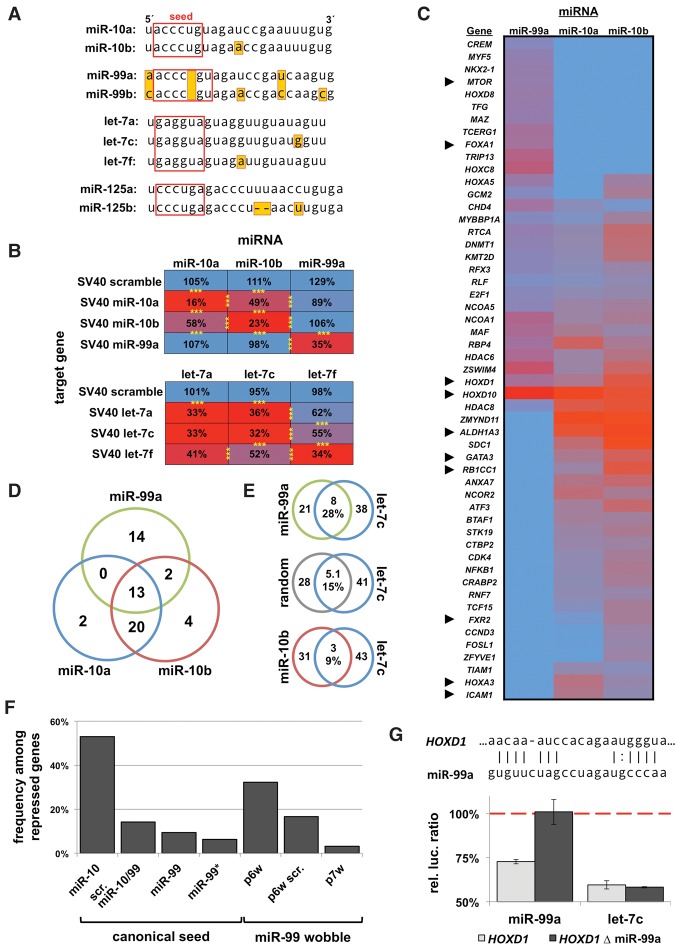
In order to test if the hotspots within miRNA families impact targeting specificity, we studied the targeting preferences of three intertwined human miRNA families (let-7, miR-10, and miR-125). The miR-10 family is composed of five miRNAs (Fig. 5A). MiR-10a and miR-10b differ by a single nucleotide at p12, which occurred at the base of the vertebrate lineage and is maintained in all subsequent species (Fig. 2B). This family also includes miR-99a/b and miR-100, which differ from miR-10a/b primarily by a single-nucleotide deletion in the seed (Fig. 5A). In contrast, miR-125a/b differs by a 2-nt insertion that arose at the base of the vertebrate lineage (Fig. 2A; Supplemental Fig. S2). MiR-99a/b, miR-100, and miR-125a/b are located within miRNA clusters alongside let-7c/e/a.
Figure 5.
miRNA family members have distinct target specificity. (A) miRNA sequences and their family members used in this study. (B) Targeting specificity of miR-10 and let-7 family members for perfectly complementary targets in the SV40 3′ UTR. Results are represented as percentage of luciferase signal normalized to a negative miRNA control. n = 4; (***) P < 0.001. (C) Results of a 3′ LIFE assay testing 375 3′ UTRs for targeting by miR-99a, miR-10a, and miR-10b (n = 5, total ∼8000 dual luciferase reactions). Heatmap of 3′ UTRs with repression (rep. <0.8, P < 0.05) by any one of the three miRNAs. Arrowheads denote genes further discussed in this study. (D) Venn diagram of genes exhibiting any degree of repression from screen in C. (E) Commonly regulated genes between genomically clustered miRNAs (miR-99a/let-7c) share more common targets than nonclustered miRNAs (miR-10/let-7c). (F) Occurrence of miR-99 seed in repressed genes. (Column 1) Frequency of miR-10b seed among genes repressed by miR-10b; (column 2) scrambled permutation of the miR-10/miR-99 seed among miR-99 hits; (column 3) canonical miR-99; (column 4) miR-99* seed; (columns 5–7) miR-99 seeds with wobbles allowed in indicated positions. (G) Deletion analysis of the p6 wobble seed motif targeted by miR-99a in the 3′ UTR of HOXD1 (nucleotides 577–599 from stop codon) rescues repression by miR-99a, but not let-7c, which also targets this 3′ UTR (n = 4).
To test the specificity of miRNA family members, we cloned perfectly complementary target sites for miR-10 and let-7 family members into a minimal 3′ UTR and tested the ability of these miRNAs to repress targets using luciferase assays (Fig. 5B). While miR-10a and miR-10b were capable of cross-repression, the single-nucleotide mismatch at p12 disrupted ∼50% of the repression by each miRNA (Fig. 5B). Let-7a and let-7c (difference at p19) equally repressed each other's targets, while let-7f (p12), also demonstrated target specificity. This confirms that in cases of perfect complementarity (slicing) (Nakanishi et al. 2012), there are positional differences in the effect of mismatched base-pairing on miRNA targeting (Fig. 5B).
Because endogenous miRNA target sites rarely utilize perfect complementarity, we next characterized the extent to which miR-10a/b/99a target a panel of 374 3′ UTRs (Fig. 5C). When comparing the identity of target genes without considering strength of repression, we found that the majority were commonly regulated by both miR-10a and miR-10b and uniquely regulated by miR-99a (Fig. 5C,D). We also found that miR-99a and let-7c commonly repress 28% of target genes, suggesting cooperativity between these clustered miRNAs (Fig. 5E).
We observed a significant overlap between the miR-10a/b and miR-99a target genes (Fig. 5D), which was surprising given that target predictions for these miRNAs are completely nonoverlapping (Supplemental Fig. S6A). We were unable to identify a direct target site for miR-99 in the HOXD10 3′ UTR by deletion analysis (data not shown), but deletion of the target site for miR-10 suggests that these miRNAs target different sites (Supplemental Fig. S6B). We also found only 20% of miR-99 predicted targets were repressed in our screen, and >90% of targets were not predicted (Supplemental Fig. S6C). When we looked for canonical miR-99 target sites in repressed 3′ UTRs, we found a depletion of seed sites (9%) (Fig. 5F), which is consistent with an overall depletion of this seed sequence in the human 3′ UTRome (data not shown). However, we did find enrichment for target sites with wobble base-pairing in p6 (Fig. 5F) and validated the direct targeting of one of these sites in HOXD1 (Fig. 5G). Together, these data suggest that miR-99a may prefer noncanonical target sites.
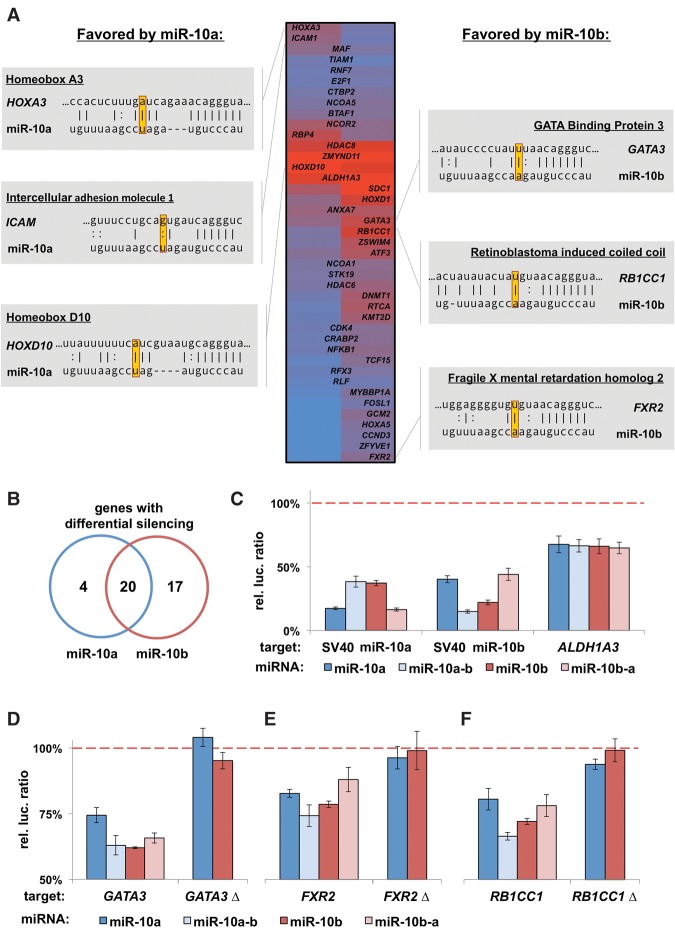
We next focused on targeting differences between miR-10a and miR-10b, finding that these miRNAs differentially repressed 51% of genes (>8% repression difference) (Fig. 6A,B). Importantly, alignments for miR-10a or miR-10b targets with a canonical seed revealed that all the targets favored by miR-10a and the majority of miR-10b targets possess a complementary nucleotide at p12 (Fig. 6A). Because the miR-10 family differs at a position that is sensitive to slicing (p12), we also tested two families that differ at other positions (let-7, miR-125) (Fig. 5A). By using small libraries enriched for target 3′ UTRs, we found that each miRNA preferentially repressed a large portion of these genes (Supplemental Fig. S7A,B). Strikingly, alignments for the let-7 targets revealed that all of the target sites favored by one family member contain the complementary nucleotide to the preferred miRNA (Supplemental Fig. S7C). Together, these data suggest that evolutionary hotspots provide distinct target preferences, regardless of where in the miRNA the hotspot occurs.
Figure 6.
The evolutionary hotspot in the miR-10 family grants target specificity. (A) Heatmap of miR-10a and miR-10b targets showing hits that exhibit repression more than two times the standard error of the 3′ LIFE assay. Genes targeted more strongly by miR-10a or miR-10b are listed on the left or right, respectively. Alignment of top genes with canonical target sites shows that nucleotides in position 12 favor the respective miRNA. (B) Count of genes with differential silencing activity between family members. (C) We reciprocally mutated p12 of each miRNA and tested for the target preference of specific genes with luciferase assays in HEK293T cells (n = 4). Repression of the perfectly complementary sequence in each SV40 3′ UTR shows switching of target preference. ALDH1A3, which contains multiple miR-10 target sites, was not affected by these mutations. Results normalized to a negative control miRNA (dashed line). (D–F) Luciferase assay showing genes favored by miR-10b switch target preference by mutant miRNAs (n = 4). (Δ) Deletion of the miR-10 target site in each 3′ UTR listed in A.
We then sought to determine experimentally if the single-nucleotide change between miR-10a/b affects target preference. We reciprocally mutated p12 of each miRNA and tested for targeting specificity of the wild-type and mutant miRNAs. Both miR-10a and miR-10b switched targeting preferences for a fully complementary target (Fig. 6C). Conversely, ALDH1A3, which was equally repressed by both miRNAs, was not affected by these mutations (Fig. 6C). Because miR-10b preferentially targeted more genes than miR-10a, we focused on three targets favored by miR-10b and found that the point mutation reversed the targeting preferences of these genes (Fig. 6D–F).
MiRNA family members exhibit tissue-specific expression
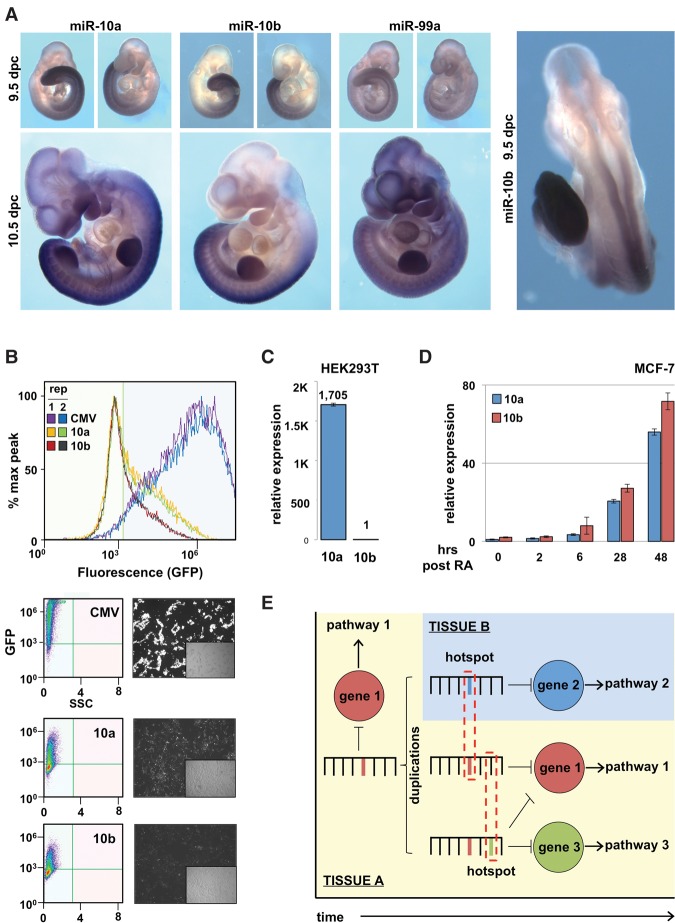
We hypothesized that the appearance of hotspots may accompany novel expression patterns, allowing the enhanced repression of target genes expressed in novel contexts. To address this, we studied the expression patterns of the miR-10 family during embryogenesis in 9.5- and 10.5-d-old mouse embryos (Fig. 7A; Supplemental Table S3). miR-99a is ubiquitously expressed at both stages, while miR-10a and miR-10b share largely overlapping expression patterns along the dorsal aspect of the embryos (neural tube, paraxial mesoderm, and lateral plate mesoderm), extending from the midthoracic region to the tailbud, where RA signaling is repressed (Cunningham and Duester 2015). However, miR-10a is more broadly expressed anteriorly, including in the branchial arches and developing brain (Fig. 7A; Supplemental Table S3). These divergentexpression patterns are also in agreement with miR-10a/b expression in adult mouse tissues (Stadthagen et al. 2013). To test if these differences are due to changes in the composition of the miRNA promoters, we isolated ∼2000 base pairs (bp) upstream of each miRNA and tested their ability to drive GFP expression in HEK293T cells. We found the MIR10A promoter is significantly more active than the MIR10B promoter in these cells (Fig. 7B), which was mirrored by expression of the mature miRNAs (Fig. 7C). We then studied the expression dynamics of these two miRNAs in response to RA in a breast ductal adenocarcinoma cell line (MCF-7) (Fig. 7D). While the expression of both mature miRNAs increases dramatically after treatment, their expression levels are not identical, suggesting differences in expression dynamics.
Figure 7.
miR-10 family members have divergent expression patterns. (A) Whole-mount in situ hybridizations of mouse embryos at 9.5 and 10.5 d post-coitus (dpc), using LNA-based probes to detect the expression pattern of mature miR-10a, miR-10b, and miR-99a. Data are representative of three embryos. (B) We cloned the promoter regions (2000 bp) of MIR10A (10a) and MIR10B (10b) and tested their ability to drive GFP expression by transient transfection in HEK293T cells. CMV promoter was used as a positive control to indicate transfection efficiency. Fluorescence quantified by flow cytometry (n = 2) and representative fluorescent images of cells. (C) qRT-PCR of mature miR-10a (10a) and miR-10b (10b) levels in HEK293T cells (n = 3). (D) Expression dynamics of mature miR-10a (10a) and miR-10b (10b) following 10 μM retinoic acid in MCF-7 cells (n = 3). (E) Model: Over evolutionary time, miRNAs evolve to regulate specific mRNAs within pathways. Following a miRNA duplication event, there is frequent sequence divergence between paralogous miRNAs, along with the emergence of novel miRNA expression patterns. The sequence of target genes expressed in these novel contexts may influence specific nucleotide changes to facilitate more efficient repression by the duplicated miRNA.
To expand on these observations, we explored the expression patterns for the let-7, miR-10, and miR-99 families in normal human tissues using miRGator, a curated miRNA expression database that compiles TCGA, GEO, and SRA data sets (Cho et al. 2013). The expression patterns of individual family members were surprisingly distinct, with many tissues favoring expression of only one family member (Supplemental Figs. S8, S9). Interestingly, the family members that have undergone duplications without sequence divergence (let-7a, let-7f) are more similar to each other than other family members. Together, the difference in target preference and expression patterns between family members suggests a connection between the functional divergence of miRNA family members with the coexpression of novel target genes.
Discussion
miRNA families evolve at specific hotspots
Our study found that miRNA families evolve at specific hotspots, which occur most frequently between p10–12. Poor conservation typically implies relaxed selective pressure, yet there are conflicting reports as to whether an miRNA's central region plays a role in miRNA target identification. A past study using miRNA overexpression found that p9–11 are the most mismatched nucleotides in canonical targets that lead to decreases in mRNA abundance (Grimson et al. 2007). In vitro biochemical approaches converge on similar findings, demonstrating p10–11 plays a relatively minimal role in stabilizing the interaction between miRNA and mRNA (Wee et al. 2012). In contrast, complementarity at p10–11 is critical for the slicing activity of deeply conserved Argonaute proteins (Elbashir et al. 2001; Meister et al. 2004; Nakanishi et al. 2012), and a recent study analyzing miRNA:mRNA chimeras found targeting motif preferences spanning the entirety of the miRNA (Moore et al. 2015). In agreement with a functional role for the central region, our study finds that miRNAs preferentially utilize nucleotides along the length of the miRNA, including the central region. A previous study also found that the 3′ UTR nucleotides that align to the middle portion of miRNAs are poorly conserved (Grimson et al. 2007), concluding that poor conservation implies a lack of function. Given our findings that the central region of the miRNA duplex is under relaxed secondary structure constraints, we speculate that poor conservation in this region may be mirrored in target sites and may not be an indication of a lack of function per se. However, the biological relevance of interactions detected with our approach should be validated in secondary experiments due to the use of exogenous constructs.
The majority of genes regulated by members of the same miRNA family vary in the strength of repression
The expansion of miRNA families in higher metazoans has been suggested to provide robustness to gene regulatory networks, and it is commonly assumed family members target completely overlapping sets of genes. However, our study found a strong connection between duplications and sequence divergence. In line with this, our results confirm the overlap between targets of miR-10a/b and let-7a/c, but we also find that the majority of the genes exhibit different degrees of repression. Given that a single-nucleotide change in the miR-10 family has the ability to modulate target specificity, we speculate that multiple changes, such as in the let-7 family, may exaggerate the targeting differences between family members. In tissues where multiple miRNA family members are coexpressed, miRNA sequence changes that enhance the repression of specific genes may allow a given miRNA family to fortify its biological function via regulation of a broader set of target genes. The emergence of novel expression patterns may also shape miRNA sequence evolution through coexpression with novel targets. In support of this hypothesis, we observe clear differences in expression patterns of family members. However, it is difficult to infer individual relationships from this anecdotal evidence because bona fide miRNA targets have been observed in both cases of coexpression and mutually exclusive expression (Ebert and Sharp 2012).
Conclusion
Here we find that the ability of a miRNA to regulate a large set of genes is enhanced by membership in a larger miRNA family (Fig. 7E). The pronounced expansion of miRNA families at the base of the vertebrate lineage suggests this may be especially relevant during the development of higher metazoans. Nucleotide changes at specific hotspots may allow each family member to specialize and preferentially regulate a specific set of genes, which may be driven by novel expression patterns of duplicated miRNAs. Furthermore, our data suggest that miRNA family members may not equally repress target genes, which is especially relevant for the development of synthetic antisense oligonucleotide therapeutics based on existing miRNAs (Kole et al. 2012).
Methods
MiRNA conservation analysis
Pre-miRNA and mature miRNA sequences were downloaded from miRBase release 21 (Kozomara and Griffiths-Jones 2014). Conserved metazoan miRNAs were defined as those that have at least 20 annotated homologs across all species and at least one copy in Homo sapiens. Family alignments were performed using ClustalO (McWilliam et al. 2013), and output files were used in Weblogo (Crooks et al. 2004) to calculate positional nucleotide contents and entropy values. Internal and external insertions and deletions were omitted from analysis if they were present in less than 10% of miRNAs.
MiRNA and 3′ UTR cloning
3′ UTRs and miRNAs were cloned as previously described (Kotagama et al. 2015; Wolter et al. 2015b). In brief, ∼200 nt upstream of and downstream from the pre-miRNA were cloned into an intron in pLIFE:miRNA by Gateway cloning. All mutagenesis experiments were performed using site directed mutagenesis (Stratagene).
3′ LIFE assay
The 3′ LIFE assay for was performed as previously described (Wolter et al. 2015a,b). In brief, each miRNA was tested with either five (miR-10, let-7, and miR-125 family screens) or three replicates (miR-10b and let-7c screens) in HEK293T cells. For the analysis in Figure 3, we used the complete human 3′ UTRome v1, consisting of 1407 3′ UTRs not enriched for targets of any specific miRNA (Kotagama et al. 2015). For the analysis in Figure 5, we used 375 3′ UTRs enriched for canonical seed sequences for miR-10 and miR-99, and for the analysis in Supplemental Figures S4–S7, we used 189 3′ UTRs enriched with genes containing canonical seed sequences for let-7, mir-34, and miR-125. Repression of a 3′ UTR by a miRNA was measured by calculating the Firefly/Renilla ratios compared to a negative control and normalized to the mean of each 96-well plate. Putative hits were called as those that had a repression index less than 0.80 and a P < 0.05. Differences in repression between miRNA family members were calculated by determining the standard error the 3′ LIFE assay (∼0.04), and differences between miRNAs greater than 2× the SE were considered divergent.
Target site distance analysis
We identified all the 3′ UTRs in the 3′ LIFE library that contain only one canonical miRNA target site, and filtered to keep 3′ UTRs with one annotated 3′ UTR based on RefSeq annotation. We then calculated the distance to the end of the transcript and the distance to the miRNA target site.
GO term analysis
Analysis of GO terms was performed with ToppGene software (Chen et al. 2009). We removed common results between both miRNAs, because the 3′ LIFE library was created using predefined criteria.
MiRNA sponge experiments
The TetR::RFP gene and tetracycline responsive GFP::miR-10 sponge construct were integrated into the genome of HEK293T cells via sequential lentivirus infection. Cells were treated with 5 µM doxycycline for 72 h with three replicates for each condition. For plasmid construction, see the Supplemental Material.
In situ hybridizations
In situ hybridizations were performed on CD-1 mouse embryos with miRCURY LNA probes (Exiqon). For detailed protocol, see the Supplemental Material.
Retinoic acid treatment and qPCR from cell lines
MCF-7 cells were treated with 10 µM ATRA (Sigma). Total RNA was extracted from cell lines using TRIzol (Invitrogen). To quantify miRNA expression, we used Taqman probes for miR-10a and miR-10b (Invitrogen). For mRNAs, we performed cDNA synthesis using SuperScriptIII (Invitrogen) and polydT primer and quantified using SYBR Green MM (Invitrogen) (ΔΔCt, normalization to ACTB).
Promoter cloning and transfection
The MIR10A/B promoters were amplified from genomic DNA from the ENCODE Project Consortium Tier 1 cell line GM12878. Promoters were recombined with the reading frame from eGFP and the SV40 3′ UTR using LR Clonase 2+ (Invitrogen). Cells line were transfected with Lipofectamine 3000 (ThermoFisher) per the manufacturer's protocol. Cells were counted 48 h post-transfection, capturing 10,000 events, using the Attune flow cytometer (Applied Biosystems).
Supplementary Material
Acknowledgments
We thank Stephen Blazie, Heather Geissel, Karen Anderson, and Josh LaBaer for advice and discussion. We also thank the Anderson laboratory, Mitch McGee, and the LaBaer laboratory for sharing reagents and instrumentation; Ian Shoemaker and Merica Vorachitti for technical advice; and Shwetal Mehta for cell lines. J.M.W. is the Maher Alumni Scholar and would like to thank the Maher family for their generous support of this research. This work is supported by funds from the College of Liberal Arts and Science, the Biodesign Institute at Arizona State University, the National Cancer Institute (1R21CA179144–01A1), and the National Institute of General Medical Sciences (1R01GM118796-01).
Author contributions: M.M. and J.M.W. conceived of this project and prepared the manuscript. H.H.T.L. performed the literature review for the network analysis, the bioinformatic miRNA expression analysis, and the miRNA target distance analysis. A. Linse participated in the bioinformatic analysis of miRNA sequences. T.-D.N. and K.K. participated in the qPCR experiments. V.A.G. participated in the miR-10b and let-7c 3′ LIFE screen and the miRNA promoter experiments. A. Lynch and A.R. performed the in situ hybridizations. J.M.W. participated in all experiments.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.209361.116.
Freely available online through the Genome Research Open Access option.
References
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. 2008. The impact of microRNAs on protein output. Nature 455: 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E. 2011. Evolution of microRNA diversity and regulation in animals. Nat Rev Genet 12: 846–860. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM. 2005. Principles of microRNA-target recognition. PLoS Biol 3: e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Bardes EE, Aronow BJ, Jegga AG. 2009. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 37: W305–W311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Hannon GJ, Darnell RB. 2012. An alternative mode of microRNA target recognition. Nat Struct Mol Biol 19: 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S, Jang I, Jun Y, Yoon S, Ko M, Kwon Y, Choi I, Chang H, Ryu D, Lee B, et al. 2013. MiRGator v3.0: a microRNA portal for deep sequencing, expression profiling and mRNA targeting. Nucleic Acids Res 41: D252–D257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14: 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham TJ, Duester G. 2015. Mechanisms of retinoic acid signalling and its roles in organ and limb development. Nat Rev Mol Cell Biol 16: 110–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA. 2012. Roles for microRNAs in conferring robustness to biological processes. Cell 149: 515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T. 2001. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev 15: 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley NH, Bray I, Watters KM, Das S, Bryan K, Bernas T, Prehn JH, Stallings RL. 2011. MicroRNAs 10a and 10b are potent inducers of neuroblastoma cell differentiation through targeting of nuclear receptor corepressor 2. Cell Death Differ 18: 1089–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriely G, Yi M, Narayan RS, Niers JM, Wurdinger T, Imitola J, Ligon KL, Kesari S, Esau C, Stephens RM, et al. 2011. Human glioma growth is controlled by microRNA-10b. Cancer Res 71: 3563–3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. 2007. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 27: 91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha I, Wightman B, Ruvkun G. 1996. A bulged lin-4/lin-14 RNA duplex is sufficient for Caenorhabditis elegans lin-14 temporal gradient formation. Genes Dev 10: 3041–3050. [DOI] [PubMed] [Google Scholar]
- Hertel J, Lindemeyer M, Missal K, Fried C, Tanzer A, Flamm C, Hofacker IL, Stadler PF, Students of Bioinformatics Computer Labs. 2006. The expansion of the metazoan microRNA repertoire. BMC Genomics 7: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, et al. 2007. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res 67: 7713–7722. [DOI] [PubMed] [Google Scholar]
- Johnston RJ, Hobert O. 2003. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 426: 845–849. [DOI] [PubMed] [Google Scholar]
- Kole R, Krainer AR, Altman S. 2012. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat Rev Drug Discov 11: 125–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotagama K, Babb CS, Wolter JM, Murphy RP, Mangone M. 2015. A human 3′ UTR clone collection to study post-transcriptional gene regulation. BMC Genomics 16: 1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. 2014. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42: D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O'Day E, Chowdhury D, Dykxhoorn DM, Tsai P, Hofmann O, et al. 2009. miR-24 inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3′ UTR microRNA recognition elements. Mol Cell 35: 610–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20. [DOI] [PubMed] [Google Scholar]
- Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, Johnson JM, Cummins JM, Raymond CK, Dai H, et al. 2007. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol 27: 2240–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Reinhardt F, Pan E, Soutschek J, Bhat B, Marcusson EG, Teruya-Feldstein J, Bell GW, Weinberg RA. 2010. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol 28: 341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliam H, Li W, Uludag M, Squizzato S, Park YM, Buso N, Cowley AP, Lopez R. 2013. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res 41: W597–W600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 15: 185–197. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Scheel TK, Luna JM, Park CY, Fak JJ, Nishiuchi E, Rice CM, Darnell RB. 2015. miRNA-target chimeras reveal miRNA 3′-end pairing as a major determinant of Argonaute target specificity. Nat Commun 6: 8864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss EG, Lee RC, Ambros V. 1997. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 88: 637–646. [DOI] [PubMed] [Google Scholar]
- Nakanishi K, Weinberg DE, Bartel DP, Patel DJ. 2012. Structure of yeast Argonaute with guide RNA. Nature 486: 368–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906. [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115: 199–208. [DOI] [PubMed] [Google Scholar]
- Shin C, Nam JW, Farh KK, Chiang HR, Shkumatava A, Bartel DP. 2010. Expanding the microRNA targeting code: functional sites with centered pairing. Mol Cell 38: 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadthagen G, Tehler D, Hoyland-Kroghsbo NM, Wen J, Krogh A, Jensen KT, Santoni-Rugiu E, Engelholm LH, Lund AH. 2013. Loss of miR-10a activates Lpo and collaborates with activated Wnt signaling in inducing intestinal neoplasia in female mice. PLoS Genet 9: e1003913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulc J, Wiznerowicz M, Sauvain MO, Trono D, Aebischer P. 2006. A versatile tool for conditional gene expression and knockdown. Nat Methods 3: 109–116. [DOI] [PubMed] [Google Scholar]
- Tsang JS, Ebert MS, van Oudenaarden A. 2010. Genome-wide dissection of microRNA functions and cotargeting networks using gene set signatures. Mol Cell 38: 140–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Zhang Z, O'Loughlin E, Lee T, Houel S, O'Carroll D, Tarakhovsky A, Ahn NG, Yi R. 2012. Quantitative functions of Argonaute proteins in mammalian development. Genes Dev 26: 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee LM, Flores-Jasso CF, Salomon WE, Zamore PD. 2012. Argonaute divides its RNA guide into domains with distinct functions and RNA-binding properties. Cell 151: 1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler BM, Heimberg AM, Moy VN, Sperling EA, Holstein TW, Heber S, Peterson KJ. 2009. The deep evolution of metazoan microRNAs. Evol Dev 11: 50–68. [DOI] [PubMed] [Google Scholar]
- Wolter JM, Kotagama K, Babb CS, Mangone M. 2015a. Detection of miRNA targets in high-throughput using the 3′LIFE assay. J Vis Exp 99: e52647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolter JM, Kotagama K, Pierre-Bez AC, Firago M, Mangone M. 2015b. 3′LIFE: a functional assay to detect miRNA targets in high-throughput. Nucleic Acids Res 42: e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Fan J, Belasco JG. 2006. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci 103: 4034–4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu E, Thivierge C, Flamand M, Mathonnet G, Vashisht AA, Wohlschlegel J, Fabian MR, Sonenberg N, Duchaine TF. 2010. Pervasive and cooperative deadenylation of 3′UTRs by embryonic microRNA families. Mol Cell 40: 558–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.