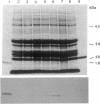
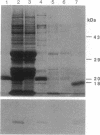
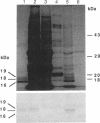
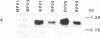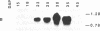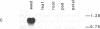Abstract
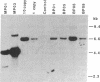
Oleosins are small hydrophobic abundant proteins localized in the oil bodies of plant seeds. An oleosin gene from the monocotyledonous maize (Zea mays L.) was transferred into the dicotyledonous Brassica napus L. using Agrobacterium-mediated transformation. The maize oleosin gene was placed under the control of either its own promoter/terminator or the promoter/terminator of a Brassica seed storage protein (napin) gene. Southern blot analyses of individual transformed plants suggested that the oleosin gene from either construct was incorporated into the Brassica chromosomes without appreciable structural alterations. The amount of construct incorporated was from 1 to >10 copies per haploid genome, depending on the individual transformant. Maize oleosin mRNA and protein were detected only in the transformants containing the napin gene promoter/terminator constructs; these transformants were studied further. Northern blot analyses of RNA isolated from different tissues and seeds of different developmental stages indicated that the maize oleosin mRNA was present only in the maturing seed. Approximately 1% of the total protein in mature seed was represented by maize oleosin. Subcellular fractionation of the mature seed revealed that 90% or more of the maize oleosin, as well as the Brassica oleosin, was localized in the oil bodies. The results show that a monocotyledonous oleosin possesses sufficient targeting information for its proper intracellular transport in a dicotyledon and also suggest that the napin gene promoter/terminator of Brassica, or equivalent seed storage protein regulatory elements of other plant species, may be used to express genes for the genetic engineering of seed oils.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adelman J. P., Hayflick J. S., Vasser M., Seeburg P. H. In vitro deletional mutagenesis for bacterial production of the 20,000-dalton form of human pituitary growth hormone. DNA. 1983;2(3):183–193. doi: 10.1089/dna.1983.2.183. [DOI] [PubMed] [Google Scholar]
- Altenbach S. B., Pearson K. W., Meeker G., Staraci L. C., Sun S. M. Enhancement of the methionine content of seed proteins by the expression of a chimeric gene encoding a methionine-rich protein in transgenic plants. Plant Mol Biol. 1989 Nov;13(5):513–522. doi: 10.1007/BF00027311. [DOI] [PubMed] [Google Scholar]
- Beachy R. N., Chen Z. L., Horsch R. B., Rogers S. G., Hoffmann N. J., Fraley R. T. Accumulation and assembly of soybean beta-conglycinin in seeds of transformed petunia plants. EMBO J. 1985 Dec 1;4(12):3047–3053. doi: 10.1002/j.1460-2075.1985.tb04044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Hatzopoulos P., Franz G., Choy L., Sung R. Z. Interaction of nuclear factors with upstream sequences of a lipid body membrane protein gene from carrot. Plant Cell. 1990 May;2(5):457–467. doi: 10.1105/tpc.2.5.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazawa S., Osumi T., Hashimoto T., Ohno K., Miura S., Fujiki Y. Peroxisome targeting signal of rat liver acyl-coenzyme A oxidase resides at the carboxy terminus. Mol Cell Biol. 1989 Jan;9(1):83–91. doi: 10.1128/mcb.9.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy D. J., Cummins I., Kang A. S. Synthesis of the major oil-body membrane protein in developing rapeseed (Brassica napus) embryos. Integration with storage-lipid and storage-protein synthesis and implications for the mechanism of oil-body formation. Biochem J. 1989 Feb 15;258(1):285–293. doi: 10.1042/bj2580285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu R. D., Huang A. H. Oleosin KD 18 on the surface of oil bodies in maize. Genomic and cDNA sequences and the deduced protein structure. J Biol Chem. 1990 Feb 5;265(4):2238–2243. [PubMed] [Google Scholar]
- Qu R., Wang S. M., Lin Y. H., Vance V. B., Huang A. H. Characteristics and biosynthesis of membrane proteins of lipid bodies in the scutella of maize (Zea mays L.). Biochem J. 1986 Apr 1;235(1):57–65. doi: 10.1042/bj2350057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinkels B. W., Evers R., Borst P. The topogenic signal of the glycosomal (microbody) phosphoglycerate kinase of Crithidia fasciculata resides in a carboxy-terminal extension. EMBO J. 1988 Apr;7(4):1159–1165. doi: 10.1002/j.1460-2075.1988.tb02926.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzen J. T., Lai Y. K., Chan K. L., Huang A. H. Oleosin isoforms of high and low molecular weights are present in the oil bodies of diverse seed species. Plant Physiol. 1990 Nov;94(3):1282–1289. doi: 10.1104/pp.94.3.1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance V. B., Huang A. H. The major protein from lipid bodies of maize. Characterization and structure based on cDNA cloning. J Biol Chem. 1987 Aug 15;262(23):11275–11279. [PubMed] [Google Scholar]
- Voelker T. A., Herman E. M., Chrispeels M. J. In vitro mutated phytohemagglutinin genes expressed in tobacco seeds: role of glycans in protein targeting and stability. Plant Cell. 1989 Jan;1(1):95–104. doi: 10.1105/tpc.1.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsu L. Y., Jacks T. J. Spherosome membranes: half unit-membranes. Plant Physiol. 1972 Jun;49(6):937–943. doi: 10.1104/pp.49.6.937. [DOI] [PMC free article] [PubMed] [Google Scholar]