Significance
Clustered genomic polymorphisms in DNA, such as microindels and stretches of nucleotide changes, play an important role in genome evolution. Here, we report a mutation mechanism responsible for such genomic polymorphisms where short, single-stranded DNA molecules invade double-stranded DNA and replace short genomic segments. We show, in a bacterial model organism, that the genomic replacements occur with very low levels of sequence identity (microhomologies). The invading DNA can be of intagenomic or foreign origin. Genotoxic stress, horizontally taken-up DNA, or lack of genome maintenance functions increase the mutation frequency up to 7,000-fold. Bioinformatic approaches suggest that this class of mutations is widespread in prokaryotes and eukaryotes and may have a role in tumorigenesis.
Keywords: illegitimate recombination, mutation, microindels
Abstract
In a screen for unexplained mutation events we identified a previously unrecognized mechanism generating clustered DNA polymorphisms such as microindels and cumulative SNPs. The mechanism, short-patch double illegitimate recombination (SPDIR), facilitates short single-stranded DNA molecules to invade and replace genomic DNA through two joint illegitimate recombination events. SPDIR is controlled by key components of the cellular genome maintenance machinery in the gram-negative bacterium Acinetobacter baylyi. The source DNA is primarily intragenomic but can also be acquired through horizontal gene transfer. The DNA replacements are nonreciprocal and locus independent. Bioinformatic approaches reveal occurrence of SPDIR events in the gram-positive human pathogen Streptococcus pneumoniae and in the human genome.
Short patches of clustered nucleotide variations are routinely observed in whole genome comparisons (1, 2). These sequence variations are substrates for natural selection, which shapes prokaryotic (3, 4) and eukaryotic (5, 6) genomes. Clustered nucleotide variations also play a role in oncogenesis where they add to the overall genomic instability (7, 8). Despite their significant biological role, the molecular mechanisms underlying formation of clustered nucleotide variations are not fully understood.
Known mechanisms responsible for clustered nucleotide variations include error-prone DNA polymerases (9) and conversions at imperfect palindromes through template-switching (10) (templated mutagenesis), which can generate tracts of single nucleotide changes, respectively. Down-regulation or loss of genes involved in mismatch repair can also lead to increased genome-wide point mutation frequencies that can result in random single-nucleotide variation (SNV) clusters. Moreover, cumulative SNVs have been described when genes for DNA-modifying enzymes were up-regulated (11). All these mechanisms typically result in tracts of single-nucleotide polymorphisms (SNPs).
More complex clustered genomic polymorphisms may also develop through point mutations accumulating in a small DNA tract over a short time or through independent insertion and deletion events (12). A number of RecA-independent mechanisms have been described and investigated in detail that lead to microdeletions without insertions, or to microinsertions without deletions, in both prokaryotic and eukaryotic organisms. Among these mechanisms are replication slippage (13) or copy number variations in microsatellite DNA (14), illegitimate recombination at microhomologies (15, 16), imprecise nonhomologous end joining (NHEJ) (17), DNA gyrase-mediated strand switching (18), and transposon scars. Two or more temporally independent deletion/insertion events at the same locus can result in clustered polymorphisms, although in retrospective studies, such sequential events are nearly impossible to verify.
The most diverse clusters of nucleotide variations are formed by microhomology-mediated end-joining (MMEJ). MMEJ has been observed in eukaryotes only and can repair DNA double-strand (ds) breaks in an error-prone way. During repair, MMEJ often generates short, direct, or inverted repeats (19) and occasionally incorporates ectopic DNA at the recombinant joints (20). MMEJ results in highly variable clustered polymorphisms at the recombinant joint and is now recognized as a driving force in rapidly evolving oncogenic cells (21). DNA polymerase theta (POLQ) has recently been identified as the key enzyme in MMEJ-directed error-prone repair, but many mechanistic details of its function remain elusive (22). To date, no POLQ-like genes have been identified in prokaryotes.
Due to the immense evolutionary and biomedical implications of how and why genetic diversity is generated in prokaryotic and eukaryotic organisms, the underlying mechanisms are intensively investigated. To study and quantify the formation of clustered polymorphisms, we developed a detection assay in the bacterium Acinetobacter baylyi. We demonstrate how regions of clustered, highly variable DNA sequence variations (ranging from 3 to 77 bp) can be formed by two coupled, microhomology-dependent illegitimate recombination (IR) events with free DNA single strands of intragenomic or external origin.
Results
Joined Double Illegitimate Recombinations Generate Clustered Polymorphisms.
To quantify and characterize clustered small indels and polymorphisms, we developed an in vivo detection construct (hisC::′ND5i′) (23) in the soil bacterium Acinetobacter baylyi ADP1. The construct is permissive for small IR events but largely refractory to single-nucleotide mutations. In this construct, two neighboring stop codons in a functionless 228-bp insert prevent expression of a histidine prototrophy marker gene (histidinol-phosphate aminotransferase; Fig. 1A). We found that spontaneous histidine-prototrophic (His+) mutants arose at low frequencies. Subsequent DNA sequencing analyses of individual His+ isolates revealed that the ′ND5i′ segment was frequently substituted with different heterologous segments of intragenomic origins. The substituting DNA segments were of similar or shorter length, eliminating or bypassing the stop codons (Fig. 1 B–E and Dataset S1), and their neighboring upstream and downstream nucleotide stretches were identical with DNA segments in otherwise fully heterologous DNA regions elsewhere in the genome (Fig. S1). Sequence analyses of these donor DNA fragments and the parental DNA sequences strongly suggested that integration occurred through hybridization at microhomologies (short identical DNA stretches) or at extended microhomologies (clusters of microhomologies interrupted by mismatches and gaps in heterologous DNA; Fig. 1 B–E and Supporting Information) followed by illegitimate recombinations. The recombinations occurred either at a single, contiguous microhomology (class 1 events; Fig. 1 B and C) or at two separate microhomologies on the same molecule (class 2 events; Fig. 1 D and E). The recombinations were nonreciprocal (Supporting Information) and independent of genomic locus and detection construct (Supporting Information). Together, these short-patch double illegitimate recombination (SPDIR) events led to highly variable polymorphisms at a single genetic locus and introduced multiple clustered nucleotide exchanges, DNA sequence replacements of variable length, or deletions accompanied by nucleotide changes at the deletion site (Dataset S1), resulting in highly diverse codon changes (Fig. S2). In all characterized SPDIR events, the source DNA of the acquired nucleotide polymorphisms was identified both for intragenomic and extragenomic (see below) origins (Dataset S1). Net nucleotide gains (maximum six base pairs) were observed in only a few cases. Although the SPDIR mechanism depends on microhomologies, the randomness of the genetic changes observed suggests a broad mutagenic potential.
Fig. 1.
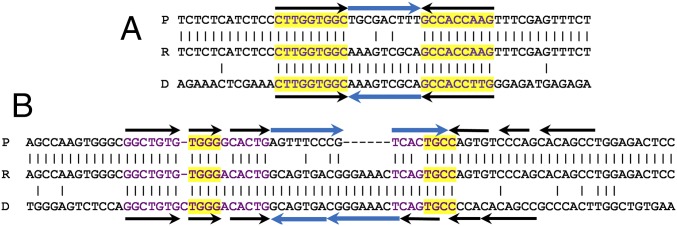
(A) Schematic illustration of the hisC::′ND5i′ detection construct for SPDIR. The genomic location and the sequence detail of the two stop codons are indicated (modified after ref. 23). The ′ND5i′ insert is shown in blue, and the translated codons are shown in black, with the two consecutive stop codons indicated in red. (B–E) Examples of clustered polymorphisms generated by SPDIR, shown as triple DNA alignments of the parental (His−; P), His+ recombinant (R), and donor (D) strands used for the double IR. Stop codons are indicated in red, and recombination sites are highlighted in yellow. Microhomologies (as approximated by ΔG0min) are in purple typeface. (B and C) Class 1 SPDIR events formed by two illegitimate joints at a single, contiguous extended microhomology. (D and E) Class 2 SPDIR events with illegitimate joints at separate (simple or extended) microhomologies, leading to complex replacements or deletions. The donor DNA originated from intragenomic loci [(B) Recurrent SPDIR mutation A26 (Dataset S1), putative ACIAD1938 gene; (C) SPDIR O106, putative ACIAD1581 gene; and (D) SPDIR R159, putative ACIAD2154 gene] except E. In E, the donor DNA was derived from Bacillus subtilis DNA (ipk gene) and acquired by A. baylyi through natural transformation. The complete set of experimentally found SPDIR sequences is listed in Dataset S1.
Fig. S1.
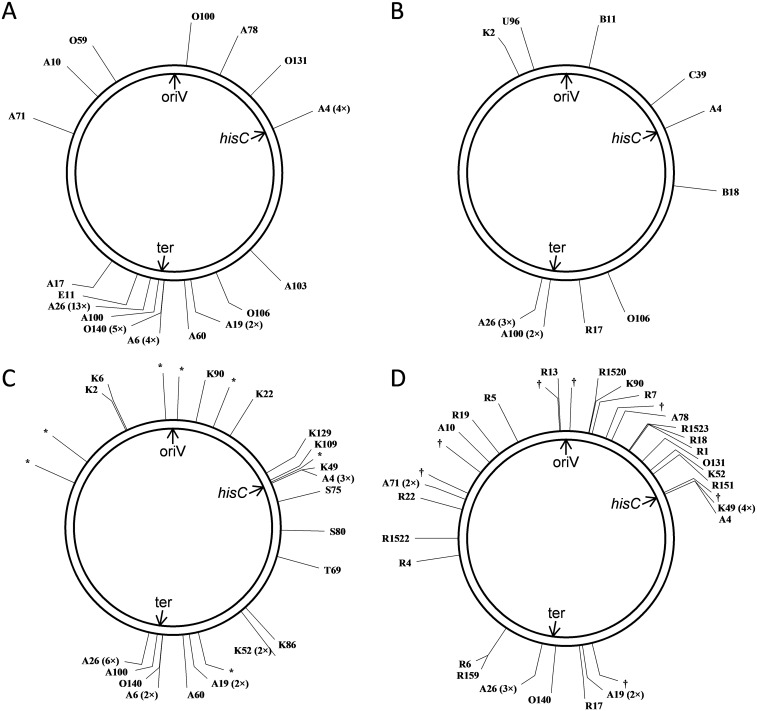
Schematic genomic positions of intragenomic donor DNA segments discovered in A. baylyi HisC+ SPDIR isolates. Numbers in brackets indicate the number of findings in independent experiments. The circles represent the A. baylyi chromosome (3.6 Mbp). oriV, origin of replication; ter, terminus of replication (approximate position); hisC, location of the hisC::′ND5i′ detection construct. All recombinant sequences are shown in Dataset S1. (A) Positions in recA+ strains (WT, ΔrecJ, ΔexoX, ΔrecJ ΔexoX, and ΔcomA ΔrecJ ΔexoX) without genotoxic stress or added DNA. (B) Positions in recA+ (WT and ΔrecJ ΔexoX) strains after exposure to ciprofloxacin or UV light. (C) Positions in recA+ strains without stress after addition of DNA (isolated from B. subtilis or salmon sperm or from isogenic A. baylyi hisC::′ND5i′ DNA). The donor DNA loci for K143 (Dataset S1) are identical with sequences from 23S rRNA genes of both B. subtilis (10 copies per genome) and A. baylyi (7 copies), and the A. baylyi positions are indicated with an asterisk. (D) Positions in ΔrecA strains (recJ+ exoX+ and ΔrecJ ΔexoX). The donor DNA for K1518 (Dataset S1) originates from one of the seven 16S rRNA genes (positions indicated by a dagger). Designed with pDRAW32 (www.acaclone.com).
Fig. S2.
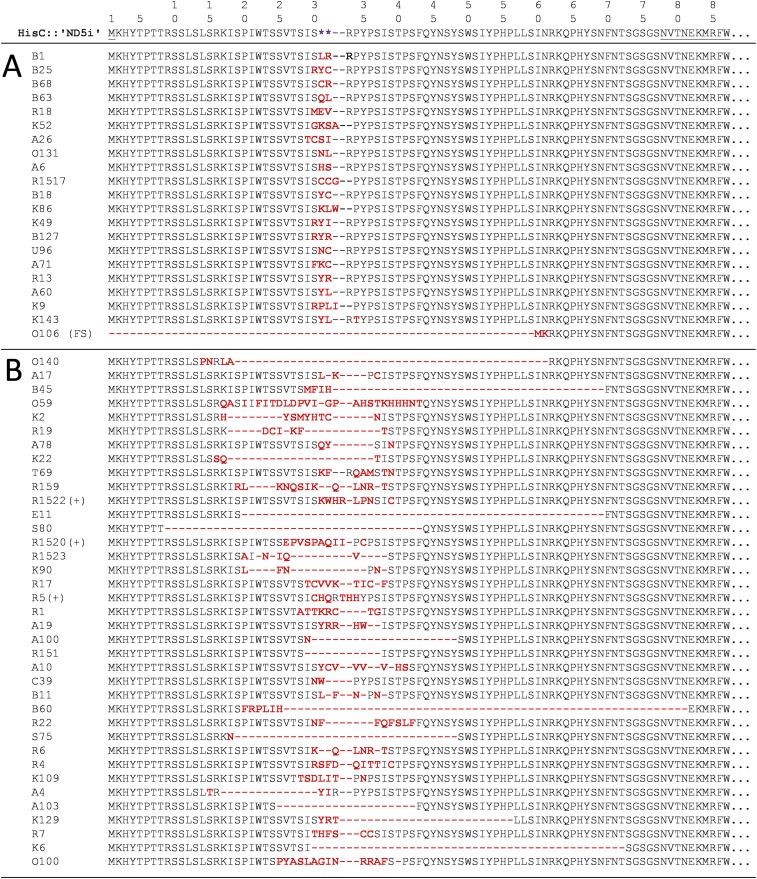
Deduced amino acid sequence alignments from the ′ND5i′ insert of the recombinant HisC from all experimentally obtained A. baylyi HisC+ revertant SPDIR isolates. The stop codons in the hisC::′ND5i′ detection allele are depicted as purple asterisks, and codon changes are indicated in red. The wt HisC codons flanking the ′ND5i′ insert are underlined in the top line. All corresponding DNA sequences are given in Dataset S1. (A) Class 1 SPDIR events formed at a single, contiguous microhomology typically contain two to four variable codon changes without change in protein length. In isolate O106 (Fig. 1C), the replacement introduced a frameshift (FS). (B) Illegitimate joints occurring at separate simple or extended microhomologies (class 2 SPDIR events) resulted in highly variable changes often associated with deletions. Events with total net amino acid gains are marked with (+).
Low Frequency of SPDIR Mutations in Wild-Type Cells.
We quantified occurrence of SPDIR experimentally in wild-type (WT) A. baylyi cells and found that His+ revertants were scarce (1.1 × 10−11; about 14-fold rarer than single point mutations; Table 1). The fraction of SPDIR mutation events among the His+ reversions was ∼5%, corresponding to a calculated SPDIR frequency of 5.6 × 10−13 (Table 1). This number is likely an underestimation due to limitations in the detection construct because SPDIR-generated substitutions that introduce stop codons or frameshifts or lead to improper protein folding remain undetected.
Table 1.
His+ and SPDIR frequencies in A. baylyi strains without and with genotoxic stress or addition of DNA
| A. baylyi ADP1 hisC::′ND5i′ relevant genotype | Amendment | Median His+ frequency | SPDIR fraction‡ | Calculated SPDIR frequency | n | |||
| CIP* (MIC) | UV, mJ260 nm | DNA† | Absolute | Relative | ||||
| Wild type | — | — | — | 1.1×10−11 | 5% (2/40) | 5.6×10−13 | =1 | 10 |
| ΔexoX | — | — | — | 3.1×10−11 | 8% (2/25) | 2.4×10−12 | 4.3 | 17 |
| ΔrecJ | — | — | — | 1.1×10−10 | 4% (1/25) | 4.3×10−12 | 7.7 | 9 |
| ΔrecJ ΔexoX | — | — | — | 4.6×10−11 | 34% (19/56) | 1.6×10−11 | 28 | 11 |
| ΔrecA | — | — | — | 4.4×10−11 | 8% (2/25) | 3.5×10−12 | 6.2 | 15 |
| ΔrecA ΔrecJ ΔexoX | — | — | — | 5.4×10−9 | 80% (32/40) | 4.3×10−9 | 7,722 | 14 |
| Wild type | 0.1 | — | — | 1.5×10−9 | 2% (1/50) | 3.0×10−11 | 53 | 11 |
| 0.25 | — | — | 7.1×10−9 | 5% (2/40) | 3.6×10−10 | 631 | 10 | |
| Wild type | — | 3.6 | — | 2.8×10−10 | 4% (2/46) | 1.2×10−11 | 21 | 13 |
| — | 10.8 | — | 8.2×10−9 | 0% (0/67) | <1.2×10−10 | <216 | 12 | |
| ΔrecJ ΔexoX | — | 3.6 | — | 1.3×10−9 | 25% (2/8) | 3.3×10−10 | 594 | 5 |
| — | 10.8 | — | 8.5×10−9 | 25% (3/12) | 2.1×10−9 | 3,782 | 5 | |
| Wild type | — | — | BS | 6.5×10−11 | 4% (1/25)§ | 2.6×10−12 | 4.6 | 10 |
| ΔrecJ ΔexoX | — | — | AB | 1.4×10−9 | 47% (8/17) | 6.6×10−10 | 1,173 | 10 |
| — | — | SS | 7.4×10−10 | 33% (7/21)§ | 2.5×10−10 | 439 | 5 | |
| — | — | BS | 5.5×10−10 | 51% (24/47)¶ | 2.8×10−10 | 500 | 9 | |
| ΔrecJ ΔexoX ΔcomA | — | — | — | 6.5×10−11 | 35% (8/23) | 2.3×10−11 | 40 | 10 |
| hisC+ trpE27 | — | — | — | 1.5×10-10# | n.a. | n.a. | n.a. | 11 |
n.a., not applicable.
CIP, ciprofloxacin supplemented at concentrations relative to the minimal inhibitory concentration (MIC) for A. baylyi wild type (62.5 ng⋅mL−1; modified Etest).
Supplemented with 300 ng⋅mL−1 genomic DNA from the following sources: BS, Bacillus subtilis 168; AB, A. baylyi hisC::′ND5i′; SS, salmon sperm DNA.
Identical genotypes were regarded as siblings originating from a single mutation event.
The SPDIR events formed with endogenous AB DNA.
Eight SPDIR events were formed with BS, and 15 events were formed with AB DNA. One donor DNA segment was present in both donor genomes.
Point mutation frequency, given as median Trp+ frequency.
The non-SPDIR His+ mutations were in most cases (>90% in WT) conferred by in frame deletions in ′ND5i′ [i.e., single illegitimate recombination (IR) events], both with and without microhomologies, and occasionally by different classes of mutations (Supporting Information). The fact that SPDIR occurred in the WT close to the detection limit in our specific experimental setup can explain lack of prior experimental discovery.
Single-Strand–Specific DNA Exonucleases Control SPDIR in Wild-Type Cells.
Microhomology-mediated IR events have been observed in prokaryotes and eukaryotes (15, 16) and are initiated by annealing of DNA single-strand ends. We hypothesized that SPDIR was initiated by hybridization of genomic dsDNA at exposed single-stranded (ss) gaps, loops, or replication forks, with ssDNA segments. In prokaryotes, free cytoplasmic DNA single strands are attacked by ss-specific DNA exonucleases (24) (ssExo), and in A. baylyi, these ssExo have been revealed as RecJ and ExoX (23). We therefore quantified SPDIR in ssExo-deficient mutants and found that the SPDIR frequency was elevated approximately sevenfold in ΔrecJ and fourfold in ΔexoX mutants (Table 1). The frequency was increased 28-fold in a ΔrecJ ΔexoX double mutant, which lacked all ssExo activity. In the ΔrecJ ΔexoX strain, SPDIR events produced about 34% of all His+ mutation events, whereas in WT and in the single mutants the proportion of SPDIR events was at least sixfold lower than in the ΔrecJ ΔexoX mutant (Table 1). These results confirmed that SPDIR is suppressed by ssExo in WT cells and indicate that SPDIR events depend on the presence of ssDNA in the cytoplasm.
SPDIR Is Inhibited by RecA Protein.
Cytoplasmic ssDNA is a cellular genome damage signal and can be bound by RecA protein to initiate recombinational repair and to trigger the SOS response (25). We deleted the recA gene of A. baylyi, and in the ΔrecA mutant we observed an about sixfold SPDIR frequency increase. Remarkably, in a ΔrecA ΔrecJ ΔexoX triple mutant, the SPDIR frequency was >7,700-fold higher than that of the WT, and SPDIR was the most common His+ mutation (80%; Table 1). The strong synergy effect suggests that SPDIR is controlled by factors beyond elimination of free cytoplasmic DNA. It is conceivable that binding of RecA protein to ssDNA efficiently prevents hybridization of ssDNA molecules, and molecules that escape RecA-binding frequently anneal at microhomologies. In WT cells, these microhomology-annealed molecules are attacked by ssExo and prevented from genomic integration, as observed in Escherichia coli (24) and A. baylyi (23). Alternatively, faithful recombinational DNA damage repair mediated by RecA together with ssExo prevents production of ssDNA remnants (26) (e.g., displaced strand fragments or flaps) that could act as donor molecules for SPDIR. These explanations are not mutually exclusive.
Exposure to Genotoxic Stress Increases SPDIR Frequencies.
IR frequencies are increased with accumulating genomic DNA damages, and the increase has been attributed to microhomology-mediated DNA end-joining events leading to deletions and other genomic rearrangements (27). We determined whether introduction of DNA strand breaks affected SPDIR frequency in A. baylyi. For this purpose, we treated growing cultures with subinhibitory concentrations of ciprofloxacin (a fluoroquinolone antibiotic interfering with DNA gyrase activity) (28), or with variable doses of UV (UV) light. Both agents result in replication blocks and lead to genome fragmentation (29, 30). We found that the His+ frequencies were increased up to at least 600-fold with increasing doses of ciprofloxacin or UV until viability was affected, and SPDIR events were detected at low proportions (2–5%) except after UV irradiation with 10.8 mJ (Table 1).
When we repeated the UV experiments with the ΔrecJ ΔexoX mutant, SPDIR events accounted for ∼25% of His+ events with both UV doses tested (Table 1). This ratio was lower than in untreated cells (34%), indicating that SPDIR is increased by two to three orders of magnitude with increasing DNA damage levels, which is in agreement with previous reports on IR (27). However, the increase of SPDIR events is lower than that of IR-mediated mutations such as deletions.
Natural Transformation Increases Frequency and Variability of SPDIR Events.
To explore the effect of exogenous DNA on SPDIR formation, we exploited the constitutive competence for natural transformation of WT A. baylyi cells (23). DNA molecules are taken up by the cells into the cytoplasm as single strands (31). We found that exposure to foreign DNA isolated from Bacillus subtilis resulted in a fourfold to fivefold elevated SPDIR frequency (Table 1). We repeated the experiments with the ΔrecJ ΔexoX mutant, using B. subtilis DNA, isogenic A. baylyi His− DNA, and DNA isolated from salmon sperm as donor DNA substrates. In the ΔrecJ ΔexoX strain, addition of the DNA substrates led to SPDIR frequencies about 15- to 40-fold higher than without added DNA (Table 1). Notably, when exposed to foreign DNA, about two thirds of the SPDIR mutations were formed with cognate DNA, and approximately one third were formed with taken-up DNA. This result is consistent with findings of previous reports showing that recombination attempts during natural transformation frequently result in DNA strand breaks and thus can damage genomic DNA (32). The DNA damages then lead to increased SPDIR frequencies, as observed in the experiments with ciprofloxacin and UV light. The RecA-independent recombination at the MH was strand orientation-specific (Supporting Information). In a transformation-deficient ΔcomA ΔrecJ ΔexoX triple mutant [lacking the ComA DNA uptake pore (23)], the SPDIR frequency was not different from that of the ΔrecJ ΔexoX mutant (Table 1).
These results confirm that SPDIR is primarily an intragenomic process and also demonstrate that natural transformation can be mutagenic through the SPDIR pathway. Consequently, clustered polymorphisms in the genome of some bacterial species can be the result of foreign DNA acquisition. However, in retrospective genome analyses it may often not be possible to identify the origin of donor DNA molecules due to the short length of the SPDIR-generated polymorphisms.
The Two IR Events of SPDIR Are Temporally Linked.
Three lines of evidence strongly suggest that SPDIR mutations form within a single generation before selection. First, we frequently found intragenomic donor DNA segments in SPDIR isolates with reverse complement orientation relative to the hisC::′ND5i′ allele (Table S1). In these cases, temporally independent IR events would result in lariat chromosome intermediates that cannot be replicated by the cell. Second, SPDIR events with foreign DNA (that is taken up by the cell as ssDNA fragments) require genomic integration through two IR events in a single generation to prevent potentially lethal dsDNA breaks. Third, many His+ colonies from the same primary cultures frequently carried unique, identical SPDIR mutations, both with intragenomic or exogenous donor DNA molecules. Such jackpot events strongly suggest that SPDIR mutations preexisted in the bulk culture (see Supporting Information and Table S1 for details). In our frequency calculations, identical mutations from the same assay were treated as single mutation events.
Table S1.
Distribution of His+ colony numbers in wild-type and mutant A. baylyi strains
| A. baylyi ADP1 hisC::′ND5i′ relevant genotype | Assay volume, mL | Mean total cell titer with SD, mL−1 | Mean His+ titer with SD, mL−1 | Number of His+ colonies per assay = X | Number of experiments | Ratio of variance (X) to mean (X) |
| Wild type | 20 | (1.3 ± 0.7) × 109 | (3.1 ± 6.0) × 10−2 | 0 (14×); 1 (3×); 2; 3; 4 | 20 | 2.2 |
| ΔexoX | 20 | (1.3 ± 0.4) × 109 | (5.7 ± 7.4) × 10−2 | 0 (16×); 1 (7×); 2 (6×); 3 (2×); 4 (2×); 6 | 34 | 1.9 |
| ΔrecJ | 20 | (6.7 ± 2.7) × 108 | (8.5 ± 8.2) × 10−2 | 0 (5×); 1 (6×); 2 (3×); 3 (3×); 5 (2×) | 19 | 1.5 |
| ΔrecJ ΔexoX | 20 | (1.1 ± 0,3) × 109 | (4.1 ± 15.5) × 10−1 | 0 (7×); 1 (7×); 2 (3×); 3; 7; 134 | 20 | 112.6 |
| ΔrecJ ΔexoX ΔcomA | 20 | (1.2 ± 0.4) × 109 | (8.3 ± 8.8) × 10−2 | 0 (6×); 1 (4×); 2 (7×); 3; 4; 7 | 20 | 1.8 |
| ΔrecA | 20 | (9.5 ± 2.8) × 108 | (5.7 ± 7.5) × 10−2 | 0 (13×); 1 (9×); 2 (4×); 3 (2×); 5; 6 | 30 | 2.0 |
| ΔrecJ ΔexoX ΔrecA | 20 | (4.6 ± 1.3) × 108 | (3.0 ± 1.4) × 100 | 17; 24; 27; 31 (3×); 35; 36; 37; 40 (2×); 41 (2×); 43; 45; 46; 47; 50; 56 (2×); 66; 67; 78; 102; 117; 134; 160; 172 | 28 | 27.6 |
A Model for SPDIR Caused by Cytoplasmic ssDNA Molecules.
We show that SPDIR events depend on the presence of ssDNA and are suppressed by key components of the genome maintenance machinery. A genomic integration model is depicted in Fig. 2. In that model, microhomologies are used by the cell to join unrelated DNA molecule ends, as has been demonstrated and quantified in previous studies for single (15) or multiple (24, 33) IR events. The model further builds on a proposed mechanism for strand orientation-specific, RecA-independent integration of short DNA molecules (23), in which we showed that fully homologous oligodeoxynucleotides (≥20 bp) could be chromosomally integrated in a single event during replication, acting as primers for Okazaki fragments (23, 24) (Supporting Information). In the present study, we demonstrate that microhomologies are sufficient for chromosomal integration at low but detectable frequencies during lagging strand DNA synthesis (Fig. 2, Fig. S3, and Supporting Information).
Fig. 2.
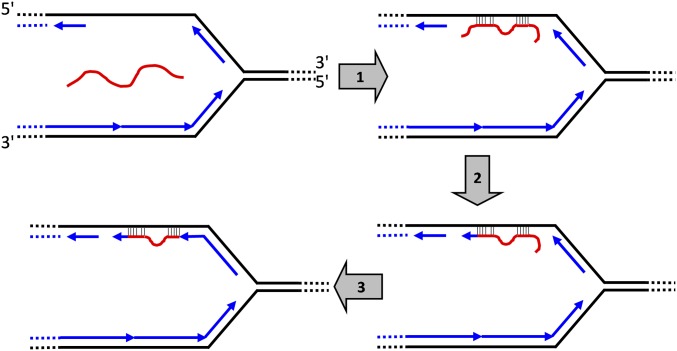
Model for SPDIR mechanism illustrated with a DNA replication fork (black indicates parental DNA strands; blue arrows indicate newly synthesized DNA strands). The proposed mechanism expands on a synthesis of several microhomology-dependent IR models (15, 23, 24, 33). In step 1, an ssDNA molecule (red) anneals at one or more microhomologous regions with exposed ssDNA segments at the discontinuously synthesized arm. In step 2, the potential 3′-extension is processed, and the hybridized molecule is extended by a DNA polymerase. In step 3, the potential 5′-overhang is removed, and the processed end is covalently joined with the newly synthesized 3′-end of the next Okazaki fragment.
Fig. S3.
(A) Sequence detail of the A. baylyi intragenomic donor DNA for the recurrent K52 SPDIR microindel mutation, located in the putative ACIAD0520 gene (Dataset S1) in opposite orientation compared with the hisC::′ND5i′ allele. (B) Sequence of the K52-tag heteroduplex dsDNA substrate used as donor DNA for natural transformation experiments, with the T/G mismatch indicated by the letter spacing and blue color. The mismatch was used as sequence tag to determine which strand was integrated in transformant SPDIR isolates and to distinguish transformation events from potential intragenomic SPDIR events. The mismatch position is a silent wobble position of an alanine codon. (C) Sequence detail of the hisC::′ND5i′ capture construct with details of microhomologies and illegitimate joints for comparison. The color codings are the same as in Fig. 1 B–E.
Bioinformatic Analyses Reveal Putative SPDIR Events in Streptococcus pneumoniae.
We hypothesized that SPDIR is a general genetic mechanism forming microindels and clustered polymorphisms with intragenomic DNA. To test this hypothesis, we searched for variations consistent with SPDIR in the gram-positive human pathogen Streptococcus pneumoniae and in human genomic DNA samples using bioinformatic approaches. We performed initial DNA sequence analyses on 203 pairwise genome alignments from the well-characterized S. pneumoniae PMEN1 lineage (34) collected over 30 y. We called clustered polymorphisms as a set of ≥3 cumulative single-nucleotide polymorphisms (SNPs) with no more than eight base pairs (bp) between each SNP (Supporting Information). We subsequently identified genomic DNA segments that could have served as potential donor molecules for SPDIR events.
For each microhomology, we calculated the minimal free energy of hybridization (35) (ΔG0min) as a proxy for the annealing stability properties of a microhomology. Conservatively, we only considered DNA segments that displayed a lower ΔG0min than the weakest microhomology found in the experimental studies with A. baylyi (Supporting Information and Dataset S1). Using these criteria, we obtained a set of eight putative SPDIR events that are in accordance with the thermodynamical requirements identified experimentally (Dataset S2).
Although identification of false-positive donor molecules cannot be excluded using this retrospective approach, the likelihood of random occurrence of identical DNA segments of typically 13 or more bp occurring in intragenomic DNA is low (Supporting Information). False-positives due to accumulated point mutations or alternative microindel-generating processes cannot be completely ruled out. On the basis of estimates of yearly point mutation rates in the PMEN1 lineage (35) (1.57 × 10−6) of ∼3.3 single-nucleotide changes per genome per year, the probability of multiple adjacent SNPs mimicking SPDIR events while the remainder of the genome remains unchanged is extremely low.
Bioinformatic Analyses Reveal Putative SPDIR Events in Human Genomes.
For humans, we isolated DNA from blood samples and colon cancer tissues from three individuals (36) and sequenced the DNA on an Illumina HiSeq 2000. We called clustered polymorphisms with donor molecules for SPDIR largely as described above (see Supporting Information for details). Altogether, we identified 94 putative SPDIR events (Table S2 and Dataset S3). Detailed analyses showed that more than half of these events were short clustered nucleotide variations present in various human sequence databases including alternative genome assemblies, suggesting that SPDIR contributes to the generation of human heterozygous alleles and that SPDIR is a mutation mechanism operative in humans.
Table S2.
Number of potential SPDIR events in human colon cancer tissue and peripheral blood control samples from three individuals
| Potential SPDIR events | Individual 1 | Individual 2 | Individual 3 | |||
| Cancer | Blood | Cancer | Blood | Cancer | Blood | |
| Inherited | 10 | 6 | 14 | 9 | 9 | 9 |
| Novel | 9 | 2 | 6 | 4 | 21 | 4 |
| Total | 19 | 8 | 20 | 13 | 30 | 13 |
The remaining insertion–deletion sequences were not found in available databases and were considered novel (Table S2). Seven novel putative SPDIR events were uniquely identified in DNA from blood, whereas a total of 33 putative novel events were identified in DNA from colon cancer tissue only (Table 2). Remarkably, 16 novel SPDIR events from cancer tissue formed at predicted hairpins and led to microinversions (Fig. 3) that in two cases were imprecise (Fig. 3B). These microinversions predictively formed through donor ssDNA molecules that originated from the same locus but reannealed with the DNA single strand as reverse complement. Donor DNA molecules from loci very close to the SPDIR site were also observed in the experimental studies (A. baylyi SPDIR isolates A4 and K49; Dataset S1), but the hisC::′ND5i′ detection construct did not contain stem–loop secondary structures. Close proximity between donor locus and recombinant microindel locus may increase the likelihood for a SPDIR event.
Table 2.
Combined numbers of novel SPDIR events from three human individuals
| Putative novel SPDIR events | Cancer | Cancer and blood | Blood |
| Total | 33 | 3 | 7 |
| Associated with genes | 20 | 1 | 4 |
| ORFs | 15 | 0 | 3 |
| Potential control regions | 5 | 1 | 1 |
| Tumorigenesis | 2 | 0 | 0 |
| Growth and proliferation, differentiation, apoptosis, DNA binding, and transcription | 8 | 1 | 2 |
| Other functions | 10 | 0 | 2 |
| Not associated with genes | 13 | 2 | 3 |
| Microinversions | 16 | 1 | 0 |
The potential SPDIR numbers for each human individual are listed in Table S2.
Fig. 3.
Examples of microinversions at predicted hairpins identified in cancer tissue from human individuals. Black arrows indicate the inverted repeats (IR), and blue arrows indicate the loop orientation. Other color codings are the same as in Fig. 1 B–E. All potential SPDIR events found in the human genomes are listed in Dataset S3. (A) The class 2 microinversion Z441, located in the protooncogene SASH1 of colon cancer tissue from individual 1. (B) Example of a microinversion that was fully annealed at the left IR but misannealed at the right IR (using an alternative microhomology for the right illegitimate joint), resulting in a net gain of six bp (Z2579; individual 3, colon cancer, intergenic region).
Three novel SPDIR events, including a single microinversion, were identified both from cancer tissue and from blood (Table 2), suggesting somatic mutations early in embryogenesis, spread of genetic material within the body, or previously unknown heterozygous alleles. The observed predominance of SPDIR in colon cancer tissue possibly reflects the reduced activity of genome maintenance functions generally observed in cancer cells (37, 38). This observation is consistent with the increased SPDIR frequencies of genome maintenance mutants in our experimental bacterial system (Table 1).
Discussion
In this study we identified a previously unrecognized mechanism, SPDIR, which generates clustered DNA polymorphisms. We show that SPDIR facilitates the formation of SNP clusters, microindels, and mosaic genes (experimentally observed substitutional insertion of up to 26 codons; Dataset S1). SPDIR occurs by ssDNA segments of intragenomic or extragenomic origins that invade and replace genomic DNA through two IR events.
Our genetic studies in A. baylyi with specific deletion mutants, together with the genotoxic stress and transformation experiments, clearly show that cytoplasmic ssDNA segments are responsible for SPDIR (Fig. 2). In wild-type cells, cytoplasmic ssDNA is a genomic damage signal, and the formation of ssDNA is tightly controlled (25). SPDIR can be classified both as a recombination and as a replication-associated mutation mechanism for clustered polymorphisms, with rare ssDNA segments acting as mutagens. Although oligonucleotides are known to recombine intracellularly or in the course of horizontal gene transfer (23, 24), and synthetic oligonucleotides are now widely used in targeted mutagenesis approaches, these events are based on DNA homology. SPDIR depends exclusively on microhomologies in otherwise heterologous DNA that can be as short as 12 bp and interrupted by mismatches and gaps.
SPDIR occurs rarely in A. baylyi wild-type cells. However, DNA damages increase the SPDIR frequency by orders of magnitude. Consequently, the cells turn into transient phenotypic mutators for microindels under genotoxic stress. The transient mutator phenotype does not require mutations in DNA repair genes, as frequently observed in mismatch repair-deficient mutators of prokaryotes and eukaryotes (39). It is conceivable that increased SPDIR frequencies can provide cells with a competitive advantage in fluctuating environments, as reported for genotypic mutators (40, 41). SPDIR can generate near-random genetic variations and alter entire protein domains in a single generation. It is thus tempting to speculate that SPDIR may be an important mechanism in protein evolution (42) following gene amplification and duplication events (43) (Supporting Information).
Our in silico identification of potential SPDIR events in both the gram-positive pathogen S. pneumoniae and in the human genome strongly suggests that SPDIR is a general mutation mechanism with relevance beyond our model organism A. baylyi. The identified microindel variants, together with the presence of intragenomic donor molecules, are consistent with the experimentally obtained SPDIR events and thus biologically plausible. Typical SPDIR-generated sequence changes are inaccessible by known point mutation or recombination processes, such as replication slippage, microhomology-dependent IR, NHEJ, DNA gyrase-mediated strand switching, or transpositions. However, sequence variations caused by SPDIR are comparable with those produced by MMEJ, a highly mutagenic DNA repair mechanism in eukaryotes (20). MMEJ is tightly down-regulated in healthy cells but often operative in tumor tissue. DNA double-strand breaks are repaired by MMEJ in an error-prone way, frequently leading to incorporation of ectopic DNA segments at the joints (20).
In our human tumor samples, we determined that 16 uniquely identified clustered polymorphisms were microinversions at predicted hairpins (Fig. 3 and Dataset S3). Microinversions at hairpins have been reported (44–46), but the mechanistic details of their formation remain elusive (45) and are considered unrelated to templated mutagenesis at imperfect hairpins (46). The formation of microinversions is also not consistent with our current understanding of the MMEJ or of other mutation mechanisms, and microinversions at hairpins have not been reported in MMEJ surveys (19, 20). However, microinversions can be explained most parsimoniously by SPDIR where the inverted repeats of the hairpins act as microhomologies and are used for the illegitimate joints (Fig. 3A), consistent with the model shown in Fig. 2. Our results indicate that SPDIR-caused mutations occur in colon cancer at elevated frequencies but not in the whole blood control. In many cancers, including those with up-regulated MMEJ, genome maintenance functions such as Rad51 (eukaryotic RecA homolog) and ssExo are down-regulated (47, 48). It is conceivable that SPDIR occurs at elevated frequencies in such tumor cells, as experimentally observed in the A. baylyi ΔrecA ΔrecJ ΔexoX triple mutant (Table 1). The role of SPDIR in cancer progression requires further exploration.
Materials and Methods
The A. baylyi mutant strains were constructed as described (23, 32, 49) with standard procedures (Supporting Information) and are listed in Table S3. The mutation experiments were conducted in liquid cultures that were inoculated with a single colony of a His− strain and aerated for 15 h at 30 °C in LB broth. The cells were washed, plated on M9 minimal medium with 10 mM succinate (M9S; His+ mutant titer) and in appropriate dilution on LB (total cell titer), and incubated at 30°. When applicable, ciprofloxacin or DNA was added before inoculation. When UV was used as DNA-damaging agent, the cells were grown for 11 h, washed in PBS, irradiated with a germicidal lamp, and then grown in LB for another 4 h. On the M9S selective plates, His− cells grow less than one generation.
Table S3.
Bacterial strains used in this study
| Strain | Genotype | Ref. |
| Acinetobacter baylyi | ||
| AL4 | ADP1 hisC::′ND5i′ rpoB1 alkM::(nptII' tg4) | This study |
| AL6 | AL4 ΔrecJ | This study |
| AL8 | AL4 ΔexoX | This study |
| KOM267 | AL4 ΔexoX | This study |
| KOM218 | AL4 ΔrecJ ΔexoX | 23 |
| KOM259 | AL4 ΔrecA::tetA | This study |
| KOM254 | AL4 ΔrecA::tetA ΔrecJ ΔexoX | This study |
| AL1 | AL4 ΔrecJ ΔexoX ΔcomA::(nptII sacB) | This study |
| KOM307 | ADP1 rpoB1 alkM::(nptII' tg4) hisA::NC2 ΔrecJ ΔexoX | This study |
| JV28 | ADP1 rpoB1 alkM::(nptII' tg4) trpE27 | 23 |
| Bacillus subtilis 168 | DSM23778 |
His+ colonies on M9S were picked after 40 h (recA+ strains) or 64 h (ΔrecA strains) and restreaked on M9S, and the recombinant hisC segment was amplified by PCR and Sanger-sequenced (Supporting Information). To identify ectopic inserts, the sequencing results were aligned with the A. baylyi genome and, when donor DNA for natural transformation was used, with donor DNA sequences, using BLAST (50).
The bioinformatic approaches are described in detail in Supporting Information. The R scripts are available from the authors upon request.
Two ethical boards reviewed the protocol for investigation of the human samples included in this study: the Regional Committee on Health Research Ethics (Case H-2–2012-FSP2) and the National Committee on Health Research Ethics (Case 1304226). Both review boards approved the human research and waivered the requirement for informed consent, in accordance with national legislation (Sundhedsloven) (36).
SI Materials and Methods
Bacterial Strains.
The A. baylyi ADP1 strains were derived from strain JV28 (51) using standard procedures (52) and are listed in Table S3. The hisC::′ND5i′, ΔrecJ, ΔexoX, ΔcomA::(nptII sacB), and ΔrecA::tetA alleles have been described elsewhere (23, 32, 49). The plasmid DNA constructs used for the strain manipulations are listed in Table S4. In short, strain AL4 was constructed by transformation of JV28 with trpE+ DNA and transformation of the resulting strain with linearized plasmid pKHhisC20 DNA. The resulting trpE+ hisC::(nptII sacB) mutant was subsequently transformed by linearized pKHhisC25, giving strain AL4 containing hisC::′ND5i′. From AL4, the recJ gene was deleted by transformation with linear pEK2 and subsequent transformation of the resulting strain by linear pEK3, giving strain AL6. AL4 ΔexoX strains were constructed by two different approaches: First, the exoX allele in AL4 was deleted using the linearized plasmids pSI2 and pSI3 in subsequent steps (giving strain AL8). Second, the recJ+ allele was crossed back into the ΔrecJ ΔexoX strain KOM218 (Table S3) with linearized pEK2 and subsequently with a PCR product covering the recJ+ ORF (giving strain KOM267). The recJ+ PCR product was amplified using primers recJ-up-f and recJ-down-r (Table S5) with Phusion DNA polymerase (New England Biolabs) and WT A. baylyi DNA as template. No differences in the His+ frequencies between AL8 and KOM267 were observed, and the data were pooled. The KOM218 strain was rendered nontransformable by deleting the comA gene through transformation with linearized pKHNH2 DNA (Table S4). The ΔrecA strains were constructed by transformation of the AL4 and KOM218 strains by purified DNA from strain JV33 that contained the ΔrecA::tetA allele (Table S3), giving strains KOM259 and KOM254, respectively. Cotransformation by unwanted DNA was ruled out with PCR and by phenotypic characterization.
Table S4.
Plasmids used in this study
Table S5.
Primers used in this study
| Name | DNA sequence (5′ to 3′) | Ref. |
| recJ-up-f | GTAAGCCTTCTGGACATATTTTGACC | 32 |
| recJ-down-r | AACTTGACTTACATTATTACAGTTCAGTTACG | 32 |
| hisC-ins-f | GACAAGCCATTTTTATTACACC | 23 |
| hisC-ins-r | CAATTACGACTACACGATCG | 23 |
| hisC-up-f | AATCTGGATGCACATGCACG | 23 |
| hisA-up-f | AAGCGACTCCTCTATTTCGG | This study |
| hisA-ins-r | GTCTGCTCGACATTTACACC | This study |
| A26-f | AGCGTCGCATAAGCGATTCC | This study |
| A26-r | ATTGGACTCGTACTGGCTGC | This study |
| A19-f | GAGGCAGTTGAAGCGAATGG | This study |
| A19-r | CCTCCTTAGGCATAAAGTCC | This study |
| O140-f | GCTTTAATTGCAAAGACACC | This study |
| O140-r | TTATCAGCGAATGAAATTCC | This study |
| recJ-ORF-f | ATGCGCACAATTCAAATCAAACAGC | 32 |
| recJ-ORF-r | TTATGTCTGATCTAAATGTAAGATGC | 32 |
| K52-f-tag | GATTTCTCATGACAACGACATCGGCAAATCAGCTCCCTACCCGTCGCGATCTTCTAAAAT | This study |
| K52-r-tag | ATTTTAGAAGATCGCGACGGGTAGGGGGCTGATTTGCCGATGTCGTTGTCATGAGAAATC | This study |
The hisA::NC2 allele was constructed as described for the hisC::′ND5i′ allele (23). Briefly, a 207 bp segment (named NC2) was genomically inserted between the start codon and the second codon of the hisA [N-(5′-phospho-l-ribosyl-formimino)-5-amino-1-(5′-phosphoribosyl)-4-imidazolecarboxamide isomerase] gene (ACIAD3398) of A. baylyi. The functionless NC2 insert consisted of 67 sense codons and two adjacent stop codons approximately in the center of the NC2 insert. The NC2 sequence was similar to the nucleotides 13280–13472 of woolly mammoth M20 mitochondrion (GenBank EU153450) DNA (nucleotides 1–193 of NC2), with the modification that the nucleotides 90 and 91 were changed from TA to GT, and the synthetic nucleotides CTGGTTCTGGCTCT (positions 194–207 of NC2). The A. baylyi ΔrecJ ΔexoX hisC+ strain carrying the hisA::NC2 allele was auxotrophic for histidine and was named KOM307. The hisC and hisA genes and the ′ND5i′ or NC2 inserts were present in the genome in single copies, and no DNA homologous to these segments was identified in the A. baylyi genome.
A. baylyi ΔrecJ ΔexoX mutants carrying either the hisC::NC2 or the hisA::′ND5i′ allele did not yield His+ colonies at detectable frequencies. Moreover, these mutants did not grow on minimal medium when the stop codons of the respective inserts were replaced with sense codons, suggesting that hisC::NC2 and hisA::′ND5i′ fusion constructs are nonfunctional.
Mutation Experiments.
Mutation assays were conducted as follows: Cultures were inoculated with a single, freshly grown auxotrophic A. baylyi colony in 20 mL liquid LB medium (in some experiments, 10 mL) and aerated at 30 °C for 15 h, with modifications for UV treatment described below. When applicable, ciprofloxacin [10-fold or fourfold below the minimal inhibitory concentration (MIC)] or DNA (300 ng⋅mL−1) were added before inoculation. Cells were harvested, washed twice, and resuspended in 0.1 volume PBS (52) and plated in appropriate dilution on LB (cell titer) and M9 medium (52) containing 10 mM succinate (M9S; His+ titer; maximum 2.5 × 109 cells distributed per plate). The plates were incubated at 30 °C for 16 (LB) and 40 (M9S) h (recA+ strains) or for 40 (LB) and 64 (M9S) h (ΔrecA strains). Colonies were counted, and His+ colonies were restreaked on M9S and incubated at 30 °C for 24 h.
The UV experiments were modified as follows: After 11 h, the cells were centrifuged for 10 min at 5,000 × g and 4 °C and washed and resuspended in PBS (4 °C) to a titer of (2–5) × 107 mL−1 (determined using a Neubauer hemocytometer), and 20 mL aliquots were transferred into a sterile Petri dish and irradiated with UV light (254 nm) with doses of 3.6 or 10.8 mJ using a germicidal lamp. The aliquots were repooled, centrifuged, and resuspended in prewarmed (30 °C) LB and grown for another 4 h in the dark before harvest. Omitting UV irradiation resulted in His+ frequencies indistinguishable from the unmodified experiments.
Approaches to identify mutator phenotypes were performed as described (23) with the following modifications: Briefly, cultures were inoculated in 7 mL LB and aerated at 30 °C for 15 h. The cultures were washed and resuspended in 7 mL PBS, and cells were plated on LB medium containing 100 mg⋅mL−1 ampicillin (1 mL; mutant titer) and in appropriate dilution on LB medium (cell titer). The plates were incubated at 30 °C for 16 (LB) and 72 h (LB with ampicillin), and the mutation frequency was calculated as ampicillin-resistant mutants per colony-forming unit.
Frequency Calculations.
His+ frequencies were determined as His+ titer divided by cell titer. Because the frequency distributions resembled Luria–Delbrück kinetics, we calculated the median His+ frequency. For the studies without genotoxic treatment or DNA addition, two independent experiments were combined and treated as one experiment before calculating the median. In some experiments, identical mutations (both SPDIR and non-SPDIR events such as deletions) were identified in more than one His+ isolate. Isolates with identical mutations from the same experiment were regarded as siblings and treated as a single mutation event. The SPDIR frequency was then calculated as number of SPDIR events per all events, multiplied with the median His+ frequency.
MIC Determinations.
The MIC of ciprofloxacin for A. baylyi ADP1 was determined with Etest reagent strips (BioMérieux) using the manufacturer’s instructions except for using solid LB medium.
PCR and DNA Sanger Sequencing.
PCR reactions were performed using Taq (Finnzymes) or Phusion (New England Biolabs) DNA polymerase according to the manufacturers’ recommendations. PCR products obtained were purified and sequenced with Sanger sequencing using the BigDye 3.1 cycle sequencing chemistry according to the manufacturer (Applied Biosystems) as follows: From randomly picked His+ isolates, the recombinant hisC segment was PCR-amplified using the primers hisC-ins-r and either hisC-up-f or hisC-ins-f (Table S5), and the PCR product was sequenced with hisC-up-f (in some cases: hisC-ins-f or hisC-ins-r) as sequencing primers. In experiments with the hisA::NC2 allele, the DNA sequences were characterized accordingly using the primers hisA-up-f and hisA-ins-r for PCR-amplification of the recombinant hisA segment and using hisA-up-f as sequencing primer. The sequences were aligned pairwise with the A. baylyi ADP1 hisC::′ND5i′ or hisA::NC2 sequence with BLAST (50) (megablast) and with the A. baylyi ADP1 genome (GenBank: NC_005966) (blastn with default settings or with parameters changed as follows: word size, 7; match/mismatch scores, 4 and −5) to determine the His+ mutation. In isolates obtained after amendment with B. subtilis DNA, the sequencing results were also aligned with the B. subtilis 168 genome sequence (NC_000964) using blastn as described above. Other chromosomal A. baylyi loci were PCR-amplified using primers A26-f and A26-r (part of ACIAD1938), A19-f and A19-r (parts of pcaD and pcaK), O140-f and O140-r (segment between ACIAD1855 and ACIAD1856), or recJ-ORF-f and recJ-ORF-r (recJ) and sequenced with A26-f, A19-r, O140-f, or recJ-ORF-f as sequencing primers (Table S5), and the sequencing results were aligned as described above.
Preparation of Heteroduplex DNA.
The K52-tag heteroduplex DNA substrate (Fig. S3) was generated by annealing the two near-complementary primers K52-f-tag and K52-r-tag as published (23).
Library Building and Illumina HiSeq 2000 DNA Sequencing.
Genomic DNA was extracted from biopsies according to instructions in QIAamp DNA Mini kit (Qiagen). Individually indexed DNA libraries were prepared from 0.5 to 1 μg of genomic DNA according to the Illumina Truseq DNA protocol (PE-940-2001) or an in-house protocol using the NEBnext E6070 (New England Biolabs) reagents (53). Pools of Illumina libraries were created using unique sequencing indices. Sequencing (100 bp paired-end) was performed on HiSeq 2000 instruments at BGI Europe.
ΔG0min Calculations.
The minimal free energy of hybridization for each simple or extended microhomology of illegitimate crossover joints was approximated for both complementary strand pairs using the nearest-neighbor values and mismatch and bulge penalties as published (35). ΔG0min were calculated as the mean of both forward and reverse complement strands for each microhomology.
Bioinformatic Identification of SPDIR Events in S. pneumoniae.
From 204 S. pneumoniae PMEN1 isolates (34), a maximum likelihood whole-genome phylogeny was reconstructed based on the core SNPs identified and aligned using Parsnp (54). A custom R script (www.r-project.org) was designed to identify potential SPDIR events in assembled genomes as follows: The DNA sequences of neighboring isolates in the tree were screened for microindels (clusters of greater than or equal to three nucleotide exchanges or gaps with no more than eight nucleotides between them) that were embedded in ≥50 bp of identical DNA sequences upstream and downstream with BLAST (50), yielding a total of 203 pairwise comparisons. Microindels were screened for potential intragenomic donor molecules that contained a microindel sequence, plus at least four identical nucleotides upstream and downstream. Pairwise BLAST alignments with pneumococcal genomes from the National Center for Biotechnology Information (NCBI) database were used to identify the ancestral and derived (microindel-containing) sequences. The custom R codes are available from the authors on request. From these results, events that could be explained by replication fork slippage or homology-driven recombination (microhomologies exceeding 35 bp; DNA sequence similarity neighboring the microindel 70% or higher) were excluded. The remaining results were evaluated for their thermodynamic properties, and all triple alignments that contained shorter or less stable microhomology than the shortest or thermodynamically weakest SPDIR events found in the experiments with A. baylyi (Dataset S1) were removed. In detail, the ΔG0min values for all microhomologies were calculated, and results that contained microhomologies above −4.46 kcal⋅mol−1 for SPDIR events at a single contiguous extended microhomology (class 1 events) or above −2.89 kcal⋅mol−1 for SPDIR events at two discrete microhomologies (class 2; cumulative: above −15.09 kcal⋅mol−1 for both microhomologies), as well as triple alignments with microhomologies (including mismatches and gaps) that were shorter than 12 bp (class 1) or 5 bp (class 2), were excluded.
Bioinformatic Identification of SPDIR Events in Human Samples.
Paired-end Illumina sequencing reads (100 nt) were mapped to the human genome (GRCh37/hg19; genome.ucsc.edu) using bwa (55), and read pairs where one pair could be mapped, whereas the other could not be mapped, were collected. The nonmapping reads were split in two terminal 30 nt sequences (flanking sequences) and a central 40 nt sequence, which were all separately mapped to hg19 again. In cases where two corresponding flanking sequences mapped in close proximity on the same strand (and in proximity to the sequence they were originally paired to, in opposite orientation), the genomic sequence covering the regions to which the flanking sequences mapped, as well as the region between these, was retrieved and aligned to the complete read sequence from which the flanking sequences were derived. The results were small clustered polymorphism patterns with fully homologous upstream and downstream sequences. From these alignment pairs, reads that contained single nucleotide variants more than 8 bp away from the polymorphism pattern, or mononucleotide repeats of at least 10 bp, were considered sequencing artifacts, and the reads were excluded. For identification of ancestral and derived (polymorphism-containing) sequences, the results were pairwise aligned with BLAST against the chimpanzee, bonobo, gorilla, and orangutan genomes. Triple alignments were built with the GRCh37 genome as reference according to the S. pneumoniae set and processed correspondingly. Heterozygous alleles were identified with BLAST using the Human G+T and nr/nt NCBI databases. Gene ontology terms and names were retrieved from the Uniprot website (www.uniprot.org/).
SI Results
The Templating DNA Segments for SPDIR Mutations Remain Unchanged.
The donor DNA segments of SPDIR mutants were located in both dispensable and essential genes (56) as well as in intergenic regions throughout the genome (Dataset S1). We investigated the sequence of the templating DNA source in 10 independent SPDIR-generated His+ A26 isolates (donor DNA patch located in the dispensable ACIAD1938 gene), 3 independent A19 isolates (pcaD; dispensable), 2 independent O140 isolates (intergenic region between nonessential ACIAD1855 and ACIAD1856 genes), and 1 U96 isolate (recJ, dispensable; Dataset S1) by DNA sequencing and found that in all cases the His+ isolates carried unmodified sequences at these loci. Although these results do not rule out a reciprocal exchange followed by a secondary event (restoration of the patch locus, or horizontal acquisition of the newly formed hisC+ allele by a sister cell), the results altogether confirm that SPDIR is a nonreciprocal recombination event. Consequently, SPDIR with cognate DNA segments lead to direct or inverted repeats of ≥12 bp that can increase genome instability.
SPDIR Occurs Preferentially at the Lagging Strand of DNA Replication.
If SPDIR was initiated by annealing of complementary single strands at MH, it can be hypothesized that the majority of initial hybridizations occurred at the lagging strand of replication because chromosomal DNA is exposed as ssDNA mainly during DNA replication, and the lagging strand remains as ssDNA in longer segments and for longer times, as demonstrated previously (23, 24). At the hisC::′ND5i′ allele, the replication fork always approaches from one direction (from the left in Fig. S3C). We tested this hypothesis with sequence-tagged heteroduplex DNA K52-tag DNA (Fig. S3B) that was added when inoculating cultures of the ΔrecJ ΔexoX strain (300 ng⋅mL−1). The K52-tag substrate carried a DNA sequence derived from the intragenomic donor DNA for the recurrent K52 SPDIR events identified in three independent experiments (Dataset S1) and was a 60-bp dsDNA substrate that carried the MH (24 bp) in the center. Compared with the genomic K52 DNA sequence, the K52-tag substrate was modified to carry two nucleotide changes to form a T/C mismatch that was used to determine by DNA sequencing which strand was integrated (Fig. S3). From 20 separate 20 mL assays, we recovered altogether eight individual SPDIR isolates that were formed with the K52-tag substrate. DNA sequencing revealed that all K52-tag-derived isolates were formed with the strand corresponding to the lagging strand of replication (bottom strand in Fig. S3B). This result confirms that SPDIR mutations are formed by annealing of single strands in the course of DNA replication (23, 24), presumably acting as primers for DNA extension (Fig. 2).
Donor DNA Molecules for SPDIR Are Distribution-Biased in Absence of Stress.
In the experiments with the wild-type and exonuclease-deficient recA+ strains, a distribution bias of the genomic donor DNA loci for the SPDIR events was observed: About 75% of the independent patch DNA source loci were located in a sector around the presumed terminus of replication (ter) site (Fig. S1A). It is assumed that ter is located at the GC skew (G − C/G + C) inversal site roughly opposite of the origin of replication (57). The distribution bias was less prominent after genotoxic stress, after addition of purified DNA, or in the absence of RecA protein (Fig. S1 B–D), i.e., when DNA strand breaks occurred or when free ssDNA was available. A. baylyi has no gene homologous to tus responsible for site-specific halting of replication forks in E. coli, and the mechanism of replication termination in A. baylyi is unknown. We speculate that termination of replication in A. baylyi is a DNA damage-inducing process and depends on DNA repair functions when two replication forks collide as reported for E. coli tus mutants (58) and that ssDNA molecules created in the course of such DNA repair events are the causing agents of SPDIR mutations in wild type.
SPDIR Is Locus and Context Independent.
To confirm that SPDIR occurred outside of the hisC locus and at sequences different from the recombinant ′ND5i′ capture segment, we constructed a new capture allele at a different genomic locus (hisA::NC2). The heterologous NC2 insert consisted of 67 sense codons and two stop joined codons approximately in the center of the insert, similar to the ′ND5i′ insert (Fig. 1A), but carried a fully heterologous DNA sequence compared with ′ND5i′. In the A. baylyi chromosome, the hisA gene is more than 1,000 kbp away from hisC. We replaced the hisA+ gene with the hisA::NC2 allele in an A. baylyi hisC+ ΔrecJ ΔexoX strain and confirmed that the resulting strain was auxotrophic for histidine due to the hisA mutation.
We determined the median His+ revertant frequency for the hisA::NC2 ΔrecJ ΔexoX mutant as described for the hisC::′ND5i′ strains as 1.7 × 10−11, which was about threefold lower than that of the corresponding hisC::′ND5i′ ΔrecJ ΔexoX mutant (Table 1). DNA sequencing revealed that about 25% of the hisA+ mutations were SPDIR events, corresponding to a calculated SPDIR frequency of 4.2 × 10−12. The SPDIR sequences and genomic and thermodynamic characterizations are given in Dataset S1. These results confirm that SPDIR mutations occur independently of the genomic locus and with any detection construct.
SPDIR Mutations Form in a Single Generation Before Selection.
It can be speculated that SPDIR events form through two temporally unlinked illegitimate recombination events. However, DNA is taken up by the cells in the course of natural transformation in the form of single-stranded, linear fragments, and these can be genomically integrated only by two IR events in a single generation [during replication (23)]. In addition, the intragenomic donor DNA segments for the SPDIR DNA replacements were located on the A. baylyi chromosome in the same orientation as the hisC::′ND5i′ allele or as reverse complement (Dataset S1). In the latter case, a single IR event at a MH would result in a lariat chromosome intermediate that cannot be replicated by the cell. Moreover, some donor DNA segments were located within genes essential for viability (56), and IR joints would disrupt these genes. For WT and mutant A. baylyi hisC::′ND5i′ strains, DNA sequencing revealed that the different SPDIR-generated His+ colonies from the same primary cultures were frequently caused by identical SPDIR events. We identified up to 24 identical SPDIR sequences from the same culture with a reverse complement-oriented donor DNA, and up to 10 identical SPDIR events from the same culture with a donor DNA from a gene essential for growth. Such events strongly support that His+ mutations, including SPDIR events, grow for several generations in the culture before selection. Comparisons of the mean His+ frequencies with their variance indicate that at high mutation frequencies, as observed for some mutants (ΔrecJ ΔexoX and ΔrecA ΔrecJ ΔexoX; Table S1) or under genotoxic stress, the mutations were distributed according to the Luria–Delbrück rather than Poisson kinetics (59). High variances were observed when many His+ mutations (jackpot events) occurred in the growth culture before selection. On the contrary, in wild-type cells and under conditions where the His+ frequencies were exceptionally low, the mutation frequency distribution resembled Poisson kinetics, although the variance was always greater than the mean (Table S1). This distribution can be attributed to the high number of experiments without colonies and the comparably low number of experiments which reduces the chance to encounter jackpot mutations (Table S1). Taken together, these results strongly suggest that the double illegitimate recombinations of SPDIR mutations typically occur in a single generation during culture growth in the absence of positive selection.
SPDIR Is Mechanistically Different from Double Illegitimate Recombination.
After UV irradiation, among all His+ revertants a unique double-IR insertion isolate was found (bottom entry in Dataset S1). This recombinant was clearly different from SPDIR isolates but resembled an A. baylyi natural transformant discovered previously that occurred by so-termed double illegitimate recombination (33) (DIR). Unlike in SPDIR isolates, here the two illegitimate joints did not occur at any microhomology but at 1-bp overlaps, and the insertion was a duplication of a nearby DNA segment (generating an interrupted direct repeat) and led to a net gain of 92 base pairs without loss. In contrast, SPDIR events usually led to substitutions or net deletions (total 3- to 168-bp loss), and only four SPDIR isolates were found that had obtained small net DNA insertions (1 to 6 bp; Dataset S1). The microhomology-independent DIR mechanism remains unknown. It is not known whether the lack of SPDIR events with large insertions is due to limitations of the detection constructs (tertiary or quarternary protein structure) or for other reasons.
Generation of His+ Phenotypes Through Different Mechanisms.
When not formed by SPDIR or DIR, His+ revertants typically occurred by deletions (i.e., single IR) in frame. These deletions occurred both at microhomologies and at sites with less than or equal to 2 or no nucleotide overlap, and the total amounts of DNA deleted ranged from 6 to 225 bp. In addition, one isolate with a single point mutation was identified (single insertion of a G residue, resulting in a novel start codon).
In repeatedly found cases, the His+ phenotype occurred by compensatory mutations elsewhere in the genome. We isolated genomic DNA from four of these isolates and used these DNA substrates to separately naturally transform the hisC::′ND5i′ strain toward His+ at high frequencies (about 10−4 to 10−3 transformants per recipient with 100 ng⋅mL−1 genomic DNA). In these isolates, the hisC::′ND5i′ allele was unchanged, and after substitution of the allele by hisC::(nptII sacB) through natural transformation these isolates remained prototrophic for histidine. In contrast, in SPDIR and deletion mutants the hisC::(nptII sacB) substitution resulted in His−. Moreover, these His+ hisC::′ND5i′ isolates frequently lost their His+ phenotype after repeated restreaking on LB medium (but remained His+ when repeatedly restreaked on M9S). In contrast, SPDIR isolates remained His+ after 10 or more restreakings without selection. The molecular bases for these suppressor phenotypes remain unclear.
SPDIR Events Can Mimic Point Mutation or Deletion Events.
Notably, in two cases, SPDIR isolates resembled double point mutants that had the two stop codons turned into sense codons (recombinants B18 and A60; Dataset S1). Although we cannot completely rule out the possibility of two individual nucleotide change events in one cell, several aspects strongly suggest SPDIR events in both cases: (i) The His+ isolates displayed no mutator phenotype (Table S6), and the expected calculated double point mutation frequency [(1.5 × 10−10 × 3)2 ≈ 2 × 10−19] is below detection limit (total number of cells plated in this study: ∼1013) and thus highly unlikely to occur; (ii) all nucleotide changes observed were transversions, whereas as replication errors, transitions occur more frequently than transversions; (iii) recombinant A60 (but no further putative point mutation combinations) was found repeatedly in independent experiments, whereas recombinant R18 was encountered only after addition of B. subtilis DNA; and (iv) in both recombinants, a microhomologous segment of 13 bp (R18: from B. subtilis) or 12 bp (A60: from A. baylyi) with sufficiently negative ΔG0min values was identified that could explain the nucleotide substitutions through two illegitimate crossovers. Similarly, several SPDIR mutants mimicking microhomology-independent deletion mutants were encountered (recombinants E11, S80, A100, R151, S75, A103, and K6; Dataset S1). For these isolates, matching donor DNA patches were identified that contained ≥14 identical consecutive bp, which is longer than expected in random DNA sequences (except for A103: 12 bp), and provided two sufficiently large microhomologies for IR (approximated by ΔG0min) at both ends that were not different from other SPDIR events.
Table S6.
Spontaneous mutation frequency of A. baylyi isolates
| Strain/isolate | hisC allele* | His phenotype | Spontaneous ApR mutation frequency† |
| AL4.H75 | A60 | His+ | (6.7 ± 0.9) × 10−7 |
| KOM218.H308 | A60 | His+ | (5.3 ± 5.2) × 10−7 |
| KOM218.H479 | B18 | His+ | (5.6 ± 2.6) × 10−7 |
| AL1 | hisC::′ND5i′ | His− | (6.4 ± 1.7) × 10−7 |
| KOM218 | hisC::′ND5i′ | His− | (6.7 ± 1.4) × 10−7 |
ApR, ampicillin resistance; means from three to five experiments with SDs.
Evolutionary Effects of SPDIR Mutations.
The experimentally observed SPDIR frequency in A. baylyi wild-type cells was 5.6 × 10−13 (Table 1) per marker, which is rare compared with other common mutation mechanisms such as SNVs, deletions, or transpositions. In our experimental system (hisC::′ND5i′), the selective marker was two adjacent stop codons, and the elimination of these codons through SPDIR was achieved by a set of highly diverse donor DNA molecules. The minimal number of changes experimentally observed in SPDIR isolates was two nucleotide substitutions, converting the two stop codons into sense codons, but often encompassed additional and more complex changes. The donor molecules were intragenomic but could also be received in the course of horizontal gene transfer, which broadens the potential for variability considerably. In this study we experimentally show that natural transformation increased the SPDIR frequency and led to genomic integration of taken-up foreign DNA molecules by SPDIR.
Compared with the SNV point mutation frequency in A. baylyi of about 1.5 × 10−10 [a specific A to G transition (23); Table 1], the SPDIR frequency is about 280 times lower. This consideration suggests that SPDIR-generated microindel mutations are much rarer than point mutations, which is consistent with the literature on mutation research. Nonetheless, reports repeatedly point out the role of microindels in evolution of both prokaryotes (2–4) and eukaryotes (1, 5, 6) and in tumorigenesis (7, 8).
Over time, mutations accumulate in lineages and lead to diversification. The selective effects, however, of different mutation types can be quite different. Although synonymous SNP changes that do not alter the sequence of the encoded protein are expected to have little if any selective effect, nonsynonymous changes that alter the protein sequence, at worst generating a premature stop codon, are expected to have large and predominantly negative effects on fitness. In contrast, SPDIR mutations are much more likely than SNPs to disrupt genes because they can substitute several adjacent codons and frequently result in frameshifts. The selective consequences of SPDIRs are hence expected to be graver than SNPs.
Beneficial SPDIR mutations are likely very rare but conceivable when two or more neighboring changes in a gene are necessary for a fitness increase. In the case of epistatic interactions in which single SNP intermediates reduce fitness relative to the ancestor, the probability that the first mutation becomes frequent enough to allow subsequent mutations is low. Two or more simultaneous SNP mutations are highly unlikely to occur (see above). SPDIR events provide a mechanism which, although rare, permits such leaps and may facilitate the exploration of fitness space.
Retrospective Identification of SPDIR Events in Prokaryotic Isolates.
Our results revealed that SPDIR events are primarily formed by nonreciprocal intragenomic recombination. In turn, SPDIR-facilitated microindels are in principle identifiable through the presence of the donor DNA patch segment, unless that segment has been lost or mutationally altered, in a bioinformatic approach. Pairwise comparisons using BLAST (50) revealed no SPDIR-generated clustered polymorphisms by pairwise comparisons of the two A. baylyi genomes available from GenBank, ADP1 (NC_005966), and DSM14961 (NZ_KB849623).
To investigate whether SPDIR events occurred in other prokaryotic species, we compared 204 closely related isolates of the gram-positive S. pneumoniae PMEN1 cluster (34) (main text). Clustered polymorphisms were called in 203 pairwise BLAST comparisons, and subsequently, in eight cases, potential donor DNA patch sequences, containing the polymorphism pattern and thermodynamically suitable microhomologous sequences for the two illegitimate crossovers, were identified, suggesting eight different SPDIR events (Dataset S2). The minimal length of the patch DNA segment identical at the putative SPDIR locus and in the donor segment was 13 bp, which was similar to the shortest length found in the A. baylyi experiments (12 bp). The calculated probability of two identical 13-mers is 4−13 × 2 ≈ 3.0 × 10−8, and the chance to occur in a PMEN1 genome (about 2.1 × 106 bp) and yield a false-positive is low. On the other hand, the obtained number of SPDIR events may be an underestimation due to genetic alteration or loss of donor DNA segments after the SPDIR event. In two out of the eight identified putative SPDIR cases, the same event was found multiple times (Dataset S2), implying horizontal spread of a positively selected mutation event.
Retrospective Identification of SPDIR Events in Human Blood and Tumor Tissue Samples.
We hypothesized that SPDIR was also a mechanism in eukaryotes and isolated and sequenced human DNA from colon cancer tissue samples, and peripheral blood control samples, of three different individuals, respectively (36) (main text). The number of Illumina paired-end sequence reads were comparable for all samples. The numbers (in million reads) were 78.5 (cancer) and 72.7 (blood) for individual 1, 96.5 (cancer) and 102.7 (blood) for individual 2, and 84.9 (cancer) and 100.7 (blood) for individual 3. Similar to the pneumococcal approach, potential SPDIR events were identified bioinformatically through aligning the short Illumina reads with a human reference genome (GRCh37/hg19). The results are listed in Table 2 and Table S2, and the sequence details and thermodynamic properties are given in detail in Dataset S3. The findings suggest that SPDIR-generated microindels occur among heterozygous variants in human populations as well as rare and possibly unique somatic mutations. The frequency of SPDIR-formed heterozygous alleles is similar in both tumor tissue and control blood sequences for all three individuals. The low number of such alleles found for each individual in both tumor and blood samples can be attributed to the short sequences of the paired-end reads, to the limitations of the identification method (Materials and Methods), and to the comparably low genome coverage and implies a considerable underestimation of the SPDIR count. On the other hand, false-positive identifications of donor DNA molecules can skew the results. For example, a random short identical segment of 16 bp is expected to occur about once in about 2.2 gigabase pairs (gbp), whereas the human haploid genome size is about 3.2 gbp. In our results, the majority of substituted segments including microhomologies for illegitimate crossovers at both sides is 19 bp or larger (Dataset S3).
Supplementary Material
Acknowledgments
We thank Sören Abel, Pia Abel zur Wiesch, Terje Johansen, Trond Lamark, Vidar Sørum, and Wilfried Wackernagel for helpful discussions and Jose Victor Moreno Mayar for assistance with bioinformatic approaches. We thank BGI Europe and the Danish National High Throughput Sequencing Centre for sequencing of the cancer samples and for technical assistance. The cancer work was supported by The Danish National Advanced Technology foundation (The GenomeDenmark platform, Grant 019-2011-2). This work was supported by The Arctic University of Norway and the Danish National Research Foundation (K.H.), the Academy of Finland Grant 251170 (to P.M.), the Finnish Centre of Excellence in Computational Inference Research Grant 259272 (to P.M.), and also by Norwegian Research Council Grant 204263/F20 (to P.J.J.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1615819114/-/DCSupplemental.
References
- 1.Cooper GM, et al. Characterization of evolutionary rates and constraints in three Mammalian genomes. Genome Res. 2004;14(4):539–548. doi: 10.1101/gr.2034704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mostowy R, et al. Heterogeneity in the frequency and characteristics of homologous recombination in pneumococcal evolution. PLoS Genet. 2014;10(5):e1004300. doi: 10.1371/journal.pgen.1004300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gibbons HS, et al. Comparative genomics of 2009 seasonal plague (Yersinia pestis) in New Mexico. PLoS One. 2012;7(2):e31604. doi: 10.1371/journal.pone.0031604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chewapreecha C, et al. Comprehensive identification of single nucleotide polymorphisms associated with beta-lactam resistance within pneumococcal mosaic genes. PLoS Genet. 2014;10(8):e1004547. doi: 10.1371/journal.pgen.1004547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mills RE, et al. Natural genetic variation caused by small insertions and deletions in the human genome. Genome Res. 2011;21(6):830–839. doi: 10.1101/gr.115907.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang S, Li J, Xu A, Huang G, You L. Small insertions are more deleterious than small deletions in human genomes. Hum Mutat. 2013;34(12):1642–1649. doi: 10.1002/humu.22435. [DOI] [PubMed] [Google Scholar]
- 7.Pu Y, et al. Association of an insertion/deletion polymorphism in IL1A 3′-UTR with risk for cervical carcinoma in Chinese Han Women. Hum Immunol. 2014;75(8):740–744. doi: 10.1016/j.humimm.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 8.Ahmad F, Lad P, Bhatia S, Das BR. Molecular spectrum of c-KIT and PDGFRA gene mutations in gastro intestinal stromal tumor: Determination of frequency, distribution pattern and identification of novel mutations in Indian patients. Med Oncol. 2015;32(1):424. doi: 10.1007/s12032-014-0424-7. [DOI] [PubMed] [Google Scholar]
- 9.Kunkel TA. DNA replication fidelity. J Biol Chem. 2004;279(17):16895–16898. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- 10.Viswanathan M, Lacirignola JJ, Hurley RL, Lovett ST. A novel mutational hotspot in a natural quasipalindrome in Escherichia coli. J Mol Biol. 2000;302(3):553–564. doi: 10.1006/jmbi.2000.4088. [DOI] [PubMed] [Google Scholar]
- 11.Nik-Zainal S, et al. Breast Cancer Working Group of the International Cancer Genome Consortium Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149(5):979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amos W. Even small SNP clusters are non-randomly distributed: Is this evidence of mutational non-independence? Proc Biol Sci. 2010;277(1686):1443–1449. doi: 10.1098/rspb.2009.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lovett ST, Drapkin PT, Sutera VA, Jr, Gluckman-Peskind TJ. A sister-strand exchange mechanism for recA-independent deletion of repeated DNA sequences in Escherichia coli. Genetics. 1993;135(3):631–642. doi: 10.1093/genetics/135.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bois P, Jeffreys AJ. Minisatellite instability and germline mutation. Cell Mol Life Sci. 1999;55(12):1636–1648. doi: 10.1007/s000180050402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ehrlich SD, et al. Mechanisms of illegitimate recombination. Gene. 1993;135(1-2):161–166. doi: 10.1016/0378-1119(93)90061-7. [DOI] [PubMed] [Google Scholar]
- 16.Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131(7):1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 17.Moore JK, Haber JE. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16(5):2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naito A, Naito S, Ikeda H. Homology is not required for recombination mediated by DNA gyrase of Escherichia coli. Mol Gen Genet. 1984;193(2):238–243. doi: 10.1007/BF00330674. [DOI] [PubMed] [Google Scholar]
- 19.Yu AM, McVey M. Synthesis-dependent microhomology-mediated end joining accounts for multiple types of repair junctions. Nucleic Acids Res. 2010;38(17):5706–5717. doi: 10.1093/nar/gkq379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mateos-Gomez PA, et al. Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination. Nature. 2015;518(7538):254–257. doi: 10.1038/nature14157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lorenz S, et al. Unscrambling the genomic chaos of osteosarcoma reveals extensive transcript fusion, recurrent rearrangements and frequent novel TP53 aberrations. Oncotarget. 2016;7(5):5273–5288. doi: 10.18632/oncotarget.6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kent T, Chandramouly G, McDevitt SM, Ozdemir AY, Pomerantz RT. Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase θ. Nat Struct Mol Biol. 2015;22(3):230–237. doi: 10.1038/nsmb.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Overballe-Petersen S, et al. Bacterial natural transformation by highly fragmented and damaged DNA. Proc Natl Acad Sci USA. 2013;110(49):19860–19865. doi: 10.1073/pnas.1315278110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dutra BE, Sutera VA, Jr, Lovett ST. RecA-independent recombination is efficient but limited by exonucleases. Proc Natl Acad Sci USA. 2007;104(1):216–221. doi: 10.1073/pnas.0608293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shinagawa H. SOS response as an adaptive response to DNA damage in prokaryotes. EXS. 1996;77:221–235. doi: 10.1007/978-3-0348-9088-5_14. [DOI] [PubMed] [Google Scholar]
- 26.Lyamichev V, Brow MA, Dahlberg JE. Structure-specific endonucleolytic cleavage of nucleic acids by eubacterial DNA polymerases. Science. 1993;260(5109):778–783. doi: 10.1126/science.7683443. [DOI] [PubMed] [Google Scholar]
- 27.Darmon E, Leach DR. Bacterial genome instability. Microbiol Mol Biol Rev. 2014;78(1):1–39. doi: 10.1128/MMBR.00035-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen CR, Malik M, Snyder M, Drlica K. DNA gyrase and topoisomerase IV on the bacterial chromosome: Quinolone-induced DNA cleavage. J Mol Biol. 1996;258(4):627–637. doi: 10.1006/jmbi.1996.0274. [DOI] [PubMed] [Google Scholar]
- 29.Malik M, Zhao X, Drlica K. Lethal fragmentation of bacterial chromosomes mediated by DNA gyrase and quinolones. Mol Microbiol. 2006;61(3):810–825. doi: 10.1111/j.1365-2958.2006.05275.x. [DOI] [PubMed] [Google Scholar]
- 30.Bonura T, Smith KC. Enzymatic production of deoxyribonucleic acid double-strand breaks after ultraviolet irradiation of Escherichia coli K-12. J Bacteriol. 1975;121(2):511–517. doi: 10.1128/jb.121.2.511-517.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith HO, Danner DB, Deich RA. Genetic transformation. Annu Rev Biochem. 1981;50:41–68. doi: 10.1146/annurev.bi.50.070181.000353. [DOI] [PubMed] [Google Scholar]
- 32.Kickstein E, Harms K, Wackernagel W. Deletions of recBCD or recD influence genetic transformation differently and are lethal together with a recJ deletion in Acinetobacter baylyi. Microbiology. 2007;153(Pt 7):2259–2270. doi: 10.1099/mic.0.2007/005256-0. [DOI] [PubMed] [Google Scholar]
- 33.Hülter N, Wackernagel W. Double illegitimate recombination events integrate DNA segments through two different mechanisms during natural transformation of Acinetobacter baylyi. Mol Microbiol. 2008;67(5):984–995. doi: 10.1111/j.1365-2958.2007.06096.x. [DOI] [PubMed] [Google Scholar]
- 34.Croucher NJ, et al. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331(6016):430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wetmur JG. Nucleic acid hybrids, formation and structure of. In: Meyers RA, editor. Encyclopedia of Molecular Biology and Molecular Medicine. Vol 4. VCH Press; New York: 1996. pp. 235–243. [Google Scholar]
- 36.Vinner L, et al. Investigation of human cancers for retrovirus by low-stringency target enrichment and high-throughput sequencing. Sci Rep. 2015;5:13201. doi: 10.1038/srep13201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoeijmakers JHJ. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411(6835):366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 38.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 39.Modrich P. Mechanisms and biological effects of mismatch repair. Annu Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- 40.Mao EF, Lane L, Lee J, Miller JH. Proliferation of mutators in A cell population. J Bacteriol. 1997;179(2):417–422. doi: 10.1128/jb.179.2.417-422.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taddei F, Vulić M, Radman M, Matić I. Genetic variability and adaptation to stress. EXS. 1997;83:271–290. doi: 10.1007/978-3-0348-8882-0_15. [DOI] [PubMed] [Google Scholar]
- 42.Alendé N, Nielsen JE, Shields DC, Khaldi N. Evolution of the isoelectric point of mammalian proteins as a consequence of indels and adaptive evolution. Proteins. 2011;79(5):1635–1648. doi: 10.1002/prot.22990. [DOI] [PubMed] [Google Scholar]
- 43.Sandegren L, Andersson DI. Bacterial gene amplification: Implications for the evolution of antibiotic resistance. Nat Rev Microbiol. 2009;7(8):578–588. doi: 10.1038/nrmicro2174. [DOI] [PubMed] [Google Scholar]
- 44.Kelchner SA, Wendel JF. Hairpins create minute inversions in non-coding regions of chloroplast DNA. Curr Genet. 1996;30(3):259–262. doi: 10.1007/s002940050130. [DOI] [PubMed] [Google Scholar]
- 45.Kim KJ, Lee HL. Widespread occurrence of small inversions in the chloroplast genomes of land plants. Mol Cells. 2005;19(1):104–113. [PubMed] [Google Scholar]
- 46.Schultz GE, Jr, Drake JW. Templated mutagenesis in bacteriophage T4 involving imperfect direct or indirect sequence repeats. Genetics. 2008;178(2):661–673. doi: 10.1534/genetics.107.083444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson LH, Schild D. Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat Res. 2001;477(1-2):131–153. doi: 10.1016/s0027-5107(01)00115-4. [DOI] [PubMed] [Google Scholar]
- 48.Chow TY, Choudhury SA. DNA repair protein: Endo-exonuclease as a new frontier in cancer therapy. Future Oncol. 2005;1(2):265–271. doi: 10.1517/14796694.1.2.265. [DOI] [PubMed] [Google Scholar]
- 49.Harms K, Schön V, Kickstein E, Wackernagel W. The RecJ DNase strongly suppresses genomic integration of short but not long foreign DNA fragments by homology-facilitated illegitimate recombination during transformation of Acinetobacter baylyi. Mol Microbiol. 2007;64(3):691–702. doi: 10.1111/j.1365-2958.2007.05692.x. [DOI] [PubMed] [Google Scholar]
- 50.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 51.de Vries J, Heine M, Harms K, Wackernagel W. Spread of recombinant DNA by roots and pollen of transgenic potato plants, identified by highly specific biomonitoring using natural transformation of an Acinetobacter sp. Appl Environ Microbiol. 2003;69(8):4455–4462. doi: 10.1128/AEM.69.8.4455-4462.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Lab Press; New York: 1989. [Google Scholar]
- 53.Seguin-Orlando A, et al. Ligation bias in illumina next-generation DNA libraries: Implications for sequencing ancient genomes. PLoS One. 2013;8(10):e78575. doi: 10.1371/journal.pone.0078575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Treangen TJ, Ondov BD, Koren S, Phillippy AM. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15(11):524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Berardinis V, et al. A complete collection of single-gene deletion mutants of Acinetobacter baylyi ADP1. Mol Syst Biol. 2008;4:174. doi: 10.1038/msb.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barbe V, et al. Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res. 2004;32(19):5766–5779. doi: 10.1093/nar/gkh910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sharma B, Hill TM. Insertion of inverted Ter sites into the terminus region of the Escherichia coli chromosome delays completion of DNA replication and disrupts the cell cycle. Mol Microbiol. 1995;18(1):45–61. doi: 10.1111/j.1365-2958.1995.mmi_18010045.x. [DOI] [PubMed] [Google Scholar]
- 59.Luria SE, Delbrück M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28(6):491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.