Significance
The potential of proteasome inhibitors to prevent transplant rejection and to treat other immune disorders is hindered by mechanism-based toxicity from inhibition of constitutive proteasomes. Here, we demonstrate that briefly, reversibly, and selectively inhibiting the immunoproteasome prolonged the survival of transplanted hearts in mice and allowed long-term survival when combined with single-dose CTLA4-Ig. Immunoproteasome inhibition noncytotoxically reduced T-cell proliferation and the numbers of effector T cells in the allograft and draining nodes while increasing T-cell expression of exhaustion markers. The immunoproteasome thus appears to play a role in suppressing induction of T-cell exhaustion. Selective inhibition of the immunoproteasome may be a potential treatment option for the management of transplant rejection.
Keywords: allograft, effector T cells, immunoproteasome, memory T cells, T-cell exhaustion
Abstract
Constitutive proteasomes (c-20S) are ubiquitously expressed cellular proteases that degrade polyubiquitinated proteins and regulate cell functions. An isoform of proteasome, the immunoproteasome (i-20S), is highly expressed in human T cells, dendritic cells (DCs), and B cells, suggesting that it could be a potential target for inflammatory diseases, including those involving autoimmunity and alloimmunity. Here, we describe DPLG3, a rationally designed, noncovalent inhibitor of the immunoproteasome chymotryptic subunit β5i that has thousands-fold selectivity over constitutive β5c. DPLG3 suppressed cytokine release from blood mononuclear cells and the activation of DCs and T cells, diminished accumulation of effector T cells, promoted expression of exhaustion and coinhibitory markers on T cells, and synergized with CTLA4-Ig to promote long-term acceptance of cardiac allografts across a major histocompatibility barrier. These findings demonstrate the potential value of using brief posttransplant immunoproteasome inhibition to entrain a long-term response favorable to allograft survival as part of an immunomodulatory regimen that is neither broadly immunosuppressive nor toxic.
Allograft transplantation is an established, widespread intervention for organ failure. Unfortunately, complications often arise from prolonged administration of immunosuppressive agents to protect the graft from rejection. The drugs in current use cause broad immunosuppression. Some of them are toxic to cells outside the immune system, including to cells in the grafted organ. Because alloantibodies pose a great risk to allografts, one of the newest agents to be deployed in defense of the graft is bortezomib (1), a proteasome inhibitor that can kill plasma cells and was approved by the Food and Drug Administration (FDA) for the treatment of multiple myeloma (2). However, bortezomib inhibits the proteasomes in all cells, giving it a high potential for mechanism-based toxicity and requiring it to be used in transplantation medicine at subtherapeutic levels. Presumably, toxicity would be greatly diminished and efficacy improved if proteasomes could be inhibited selectively in immunocytes and especially in those that react to the alloantigens of the graft.
The possibility of inhibiting proteasomes selectively in certain types of cells arises from the existence of different isoforms of proteasome-associated proteases encoded by different genes whose expression is responsive both to cell lineage differentiation and to the cytokine milieu. Eukaryotic proteasomes have a barrel-shaped 20S core that contains two copies each of seven different α subunits and seven different β subunits arranged in four stacked rings in α1–7β1–7β1–7α1–7 fashion (3). Three β subunits are proteolytic: β1 has caspase-like activity, whereas β2 is tryptic and β5 is chymotryptic. The isoform that is constitutively expressed in all cells (c-20S) controls diverse functions ranging from signal transduction to cell cycle, allowing cells to adapt to circumstances quickly both pretranscriptionally and posttranscriptionally through proteolytic degradation of temporarily dispensable proteins (4). Moreover, products of the proteasome are the major source of antigenic oligopeptides for major histocompatibility complex (MHC) class I antigen presentation (5). However, mononuclear phagocytes, dendritic cells (DCs), and lymphocytes (6, 7), as well as cells at sites of inflammatory and immune reactions exposed to cytokines such as IFN-γ, express variable proportions of the immunoproteasome (i-20S), in which some of the β1c, β2c, and β5c catalytic subunits of c-20S are replaced by β1i, β2i, and β5i (also called LMP7), respectively (8, 9).
Bortezomib and the other FDA-approved proteasome inhibitors (carfilzomib and ixazomib) comparably target both c-20S and i-20S. One compound, which binds the proteasome covalently and irreversibly, has relatively greater activity on i-20S than on c-20S and has shown efficacy in mouse models of inflammatory bowel disease, arthritis, systemic lupus erythematosus, multiple sclerosis, and type I diabetes in association with modulation of the function of Th1 and Th17 cells (10–13). However, to our knowledge, selective immunoproteasome inhibitors have not been tested for their role in promoting allograft acceptance, nor have effects been described on T-cell exhaustion and coinhibitory markers on DCs.
We hypothesized, first, that immunoproteasome inhibitors might contribute to allograft acceptance in association with pleiotropic effects on DCs and T cells, and second, that their efficacy and mechanism-based safety would be proportional to their isoform selectivity for i-20S over c-20S, and specifically for β5i over β5c. With regard to the second hypothesis, we reasoned that whatever degree of isoform selectivity was demonstrable in short-term assays, as long as a covalently reacting and irreversible β5i inhibitor reacted to some degree with β5c, it would show diminishing isoform selectivity with time. In contrast, a non-covalently reacting, reversible immunoproteasome inhibitor should maintain its selectivity during prolonged incubation with proteasomes. The recent discovery of a class of highly selective, non-covalently reacting immunoproteasome inhibitors based on an N,C-capped dipeptidomimetic scaffold (14) encouraged us to test both sets of hypotheses.
We report here that members of a distinct but related class of immunoproteasome inhibitors—N,C-capped dipeptides—are orders of magnitude more isoform selective than an irreversible inhibitor and increasingly so the longer the exposure to the target. Such compounds suppressed cytokine release from blood mononuclear cells and the activation of DCs and T cells, diminished accumulation of effector T cells, promoted expression of exhaustion and coinhibitory markers on T cells, and, when given to mice for the first 14 d following transplant, synergized with a single dose of CTLA4-Ig to promote long-term acceptance of cardiac allografts across a major histocompatibility barrier.
Results
Characterization of Highly Selective, Noncovalent Immunoproteasome Inhibitors.
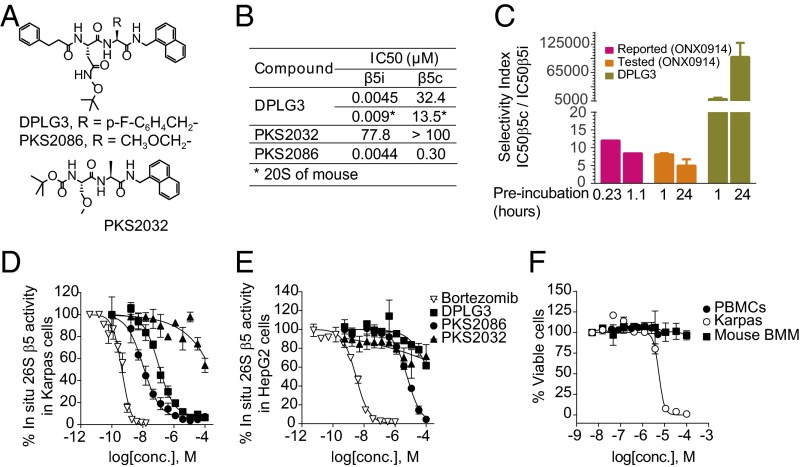
The immunoproteasome inhibitor whose favorable impact has been described in mouse models of autoimmune disease is a peptide epoxyketone that forms irreversible covalent bonds with the active site (10). Until recently, the only reported noncovalent inhibitor with relative selectivity for β5i was an N,C-capped dipeptide with selectivity of 20-fold (15). We recently described N,C-capped dipeptidomimetics with nanomolar IC50 values for β5i and >1,000-fold selectivity for β5i over β5c (14). For the present study, we designed and synthesized N,C-capped dipeptides, whose structure–activity relationship will be detailed elsewhere. Fig. 1A shows the structures of two active compounds, DPLG3 and PKS2086, and an inactive congener, PKS2032. DPLG3 inhibited β5i competitively with an IC50 of 4.5 nM (Fig. 1B) and PKS2086 did so with an IC50 of 4.4 nM (Fig. 1B), with <50% inhibition of β1i, β1c, β2i, or β2c at a concentration of 33.3 µM (SI Appendix, Table S1). PKS2032 was inactive against any subunit of c-20S or i-20S (Fig. 1B and SI Appendix, Table S1).
Fig. 1.
Design, rationale, and characterization of noncovalent β5i selective inhibitors. (A) Structures of DPLG3, PKS2086, and inactive congener PKS2032. (B) IC50 values of the compounds against β5i and β5c of human proteasomes and mouse proteasomes, the latter noted by an asterisk (*). Data are shown as mean ± SEM (n ≥ 3). (C) Selectivity indexes of ONX0914 and DPLG3 after 1- and 24-h preincubation with human c-20S and i-20S. “Reported” refers to data from ref. 10. Data are shown as mean ± SD (n = 2). (D) Inhibition of pan-β5 (β5i and β5c) activities in B lymphoma Karpas 1106P cells (β5i-dominant) and (E) hepatoma HepG2 cells (expressing β5c only) by DPLG3, PKS2086, and inactive congener PKS2032. Bortezomib was used as positive control. Pan-β5 activity was recorded with suc-LLVY-aminoluciferin, measuring luminescence of the hydrolysis product, aminoluciferin. (F) Assessment of DPLG3’s toxicity for PBMCs, mouse bone marrow macrophages (values of EC50 > 100 µM), and Karpas cells (EC50 = 5.53 µM). Data in D–F are means ± SEM (n ≥ 3).
The selectivity of DPLG3 for β5i over β5c was 7,200-fold without preincubation, 9,000-fold after 1 h of preincubation, and 99,000-fold with 24 h of preincubation (Fig. 1C). This may indicate a slow conformational change induced by binding of DPLG3. In contrast, the peptide epoxyketone inhibitor ONX0914 was reported to have an IC50 of 20 nM for β5i and 12-fold selectivity after 14 min of preincubation, with the selectivity falling to eightfold after 70 min (10). In our hands, ONX0914 showed eightfold selectivity after a 60-min preincubation, and this fell to fivefold after 24 h (Fig. 1C). Thus, by 24 h, the relative isoform selectivity of DPLG3 was four orders of magnitude greater than that of the irreversible inhibitor.
To understand the structural basis of selectivity, we built homology models of human i-20S and c-20S with the crystal structures of mouse i-20S and c-20S as templates (16). These models include only the domains surrounding the active site, that is, either β5i or β5c with β6. Docking studies were then carried out with DPLG3 (SI Appendix, Fig. S1). The enlarged S1 and smaller S3 pockets in β5i compared with β5c were previously identified as elements that contribute to the selectivity for ONX0914 (16). Consistent with this, the bulky, hydrophobic naphthyl group of DPLG3 is predicted to be accommodated better in the S1 of β5i than that of β5c. In addition, Ser27 in β5i is predicted to provide an H bond from its hydroxyl to the carbonyl of DPLG3, a bond that is not afforded by Ala27 in β5c.
At concentrations close to its action on the isolated immunoproteasome, DPLG3 inhibited β5i within Karpas lymphoma cells (Fig. 1D), which predominantly express the immunoproteasome (15), while having almost no effect on β5c within HepG2 human hepatoma cells (Fig. 1E) and no cytotoxic activity on human peripheral blood mononuclear cells (PBMCs) (17) or mouse bone marrow-derived macrophages (18) (Fig. 1F). In contrast, bortezomib inhibited proteasomes in both Karpas and HepG2 cells at roughly equivalent concentrations (0.5 and 3 nM, respectively) (Fig. 1 D and E) and was highly cytotoxic (EC50 of 1.7 nM against Karpas).
The Immunoproteasome Controls the Activation of Human Plasmacytoid DCs.
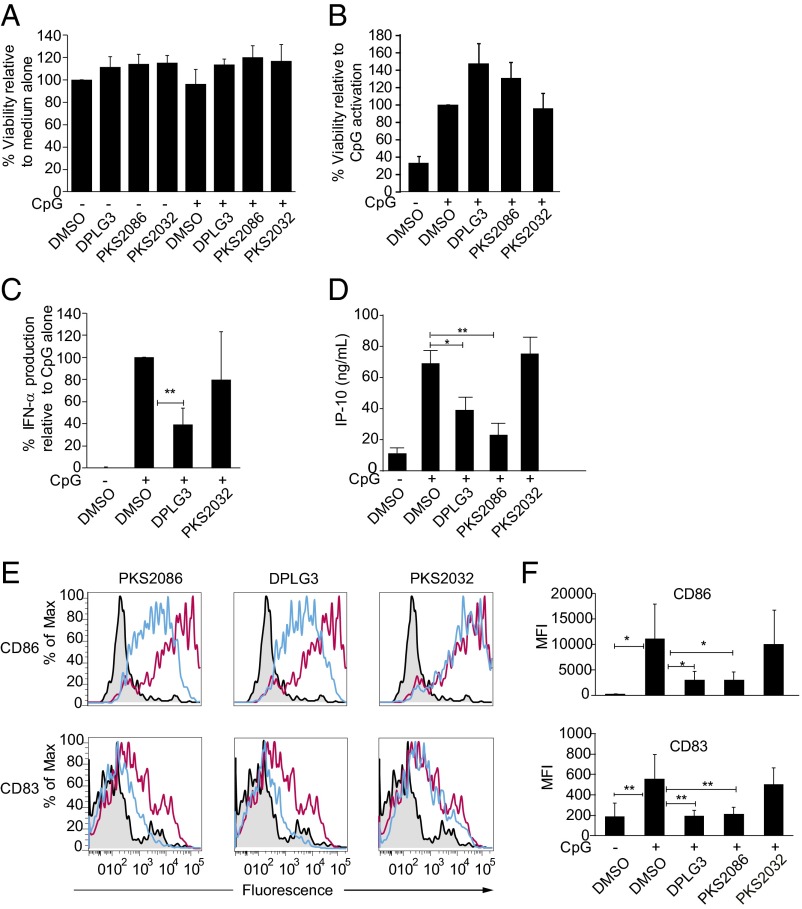
Although bortezomib has been shown to block cytokine production by human plasmacytoid DCs (PDCs), it has also been shown to induce apoptosis of these cells selectively among human PBMCs (19). Therefore, we compared effects of bortezomib and DPLG3 on PDCs, even though PDCs are not likely to be the major class of DC involved in presentation of allograft antigens. In contrast to what has been described with bortezomib, neither DLPG3 nor PKS2086 were toxic to PBMCs (Fig. 2A) or to PDCs purified from PBMCs (Fig. 2B). However, both compounds significantly inhibited the production of IFN-α and IP-10 by CpG-activated PBMCs, whereas the control compound, PKS2032, had no effect (Fig. 2 C and D). DPLG3 and PKS2086 but not PKS2032 also inhibited the CpG-induced increase in expression of the maturation markers CD83 and CD86 in purified PDCs (Fig. 2 E and F). These data demonstrate that the immunoproteasome controls both the production of cytokines and the induction of PDC maturation following TLR9 signaling, but its inhibition does not induce apoptosis of PDCs as observed with bortezomib (19).
Fig. 2.
Regulation of TLR9-mediated activation of human PBMCs and PDCs by the immunoproteasome. (A) Total PBMCs were prepared from the blood of four healthy volunteers and cultured for 24 h either alone, with inhibitors of the proteasome (DLPG3, PKS2086) or with a control compound (PKS2032) or in the presence of CpG-C (0.3 µM, TLR9 agonist) either alone or in the presence of DLPG3, PKS2086, or PKS2032 (1.5 µM) as indicated. The viability of the PBMCs was quantified at 24 h using a colorimetric (MTT) assay. (B) Purified PDCs were cultured for 24 h either alone, or in the presence of CpG-C (0.3 µM; TLR9 agonist) either alone or in the presence of DLPG3, PKS2086, or PKS2032 (1.5 µM) as indicated. The viability of PDCs was quantified compared with the CpG-activated cells by flow cytometry as described in Materials and Methods. (C and D) Total PBMCs from the blood of healthy volunteers (n = 6–8) were cultured either alone or in the presence of CpG-C (0.3 µM) either alone or with DLPG3, PKS2086, or PKS2032 (control) as indicated. IFN-α and IP-10 were quantified using an ELISA. (E and F) Purified PDCs from the blood of four healthy volunteers were cultured with CpG alone (red line) or with DLPG3, PKS2086, or PKS2032 (blue line), and the expression of CD83 and CD86 was measured by flow cytometry. One representative example (E) and averages of four donors (F) are shown. Data were analyzed using Mann–Whitney Student’s t test. All analyses were performed using Prism software, version 5 (GraphPad Software). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
Immunoproteasome Inhibitor-Induced Inhibition of T-Cell Proliferation in Vitro.
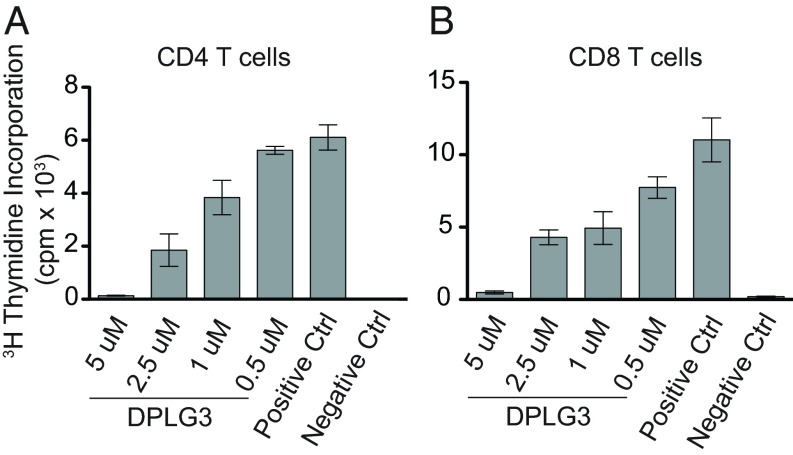
Prolongation of graft survival in the clinic requires suppression of T-cell responses. An immunoproteasome inhibitor might impair T-cell responses indirectly via its effects on DCs, such as those noted above, but could also exert direct effects on T cells. To test the latter possibility, we isolated CD4 and CD8 T cells from spleens of C57BL/6 mice and activated them with anti-CD3/CD28 in vitro. DPLG3 suppressed their proliferation in a concentration-dependent manner (Fig. 3 A and B). The concentrations of DPLG3 required to achieve the pharmacodynamic effect in T cells (Fig. 1) matched those yielding biologic effects, suggesting on-target action (20).
Fig. 3.
Suppression of T-cell proliferation by inhibition of β5i in vitro. (A) CD4+ T cells or (B) CD8+ T cells were stimulated with anti-CD3/CD28 in vitro in the presence of indicated concentrations of DPLG3 and their proliferation measured by thymidine incorporation. Data are representative of two separate experiments (n = 3–4 mice per group; *P < 0.05).
Immunoproteasome Inhibitor-Induced Decrease in Effector T Cells and Increase of T-Cell Exhaustion and Coinhibitory Markers in Vivo in Response to MHC-Disparate Tissue.
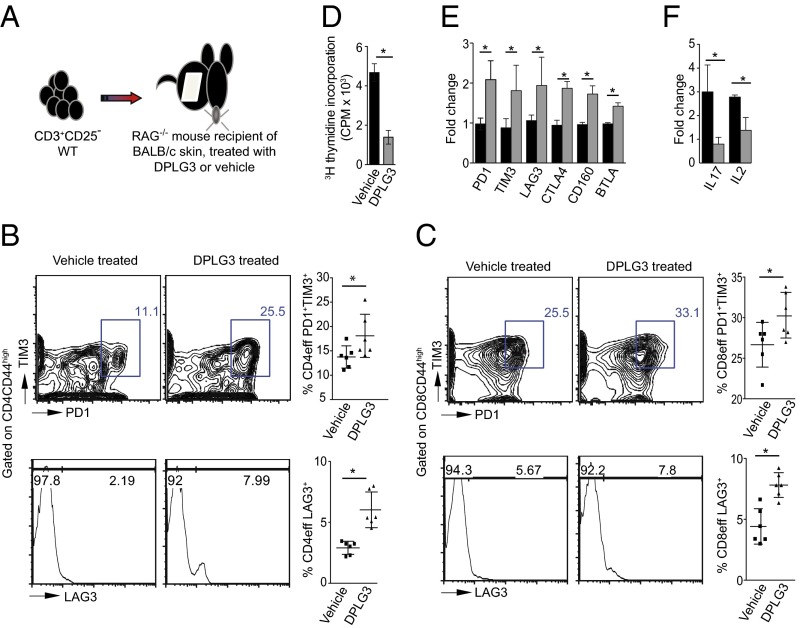
To see whether DPLG3’s modulation of in vitro T-cell responses would be recapitulated in vivo in T-cell responses to alloantigen, we adoptively transferred 6 × 106 CD3+CD25− T cells isolated from C57BL/6 FoxP3-GFP mice into RAG−/− C57BL/6 mice 1 d after a graft of BALB/c skin (Fig. 4A). The mice were then treated with DPLG3 or vehicle by i.p. injection once per day for 7 d. The dose of DPLG3 (25 mg/kg) was chosen arbitrarily after we tested tolerability at various doses up to 25 mg/kg and observed no adverse effects. Flow-cytometric analysis of cells isolated from the spleen 6 d post-adoptive transfer showed a decrease in CD4 effector T cells (GFP−CD4+CD44high) and CD8 effector T cells (GFP−CD8+CD44high) in DPLG3-treated mice compared with mice given vehicle (CD4: 41.60 ± 0.96 vs. 49.30 ± 2.90%, respectively; n = 6, P < 0.05; CD8: 35.82 ± 2.31 vs. 51.87 ± 1.90%, respectively; n = 6, P < 0.05). The exhaustion and coinhibitory markers PD1, TIM3, and LAG3 were expressed at higher levels on CD4 and CD8 effector T cells in DPLG3-treated mice than on cells from mice given vehicle (Fig. 4 B and C). To test the alloimmune memory response of DPLG3- and vehicle-treated recipients, we stimulated splenocytes isolated from recipient mice at day 7 post-adoptive transfer with irradiated BALB/c splenocytes. Splenocytes from DPLG3-treated recipients showed reduced proliferation to alloantigen (Fig. 4D; P < 0.05). Furthermore, flow-sorted CD4 effector T cells (GFP−CD4+CD44high) from spleens of DPLG3-treated mice showed a significant increase in gene expression of coinhibitory molecules such as PD1, TIM3, LAG3, CTLA4, CD160, and BTLA with a concomitant decrease of the inflammatory cytokines IL-17 and IL-2 compared with vehicle-treated mice (Fig. 4 E and F).
Fig. 4.
Up-regulation of T-cell exhaustion markers on CD4 and CD8 effector T cells in a skin transplant model by inhibition of β5i. (A) Cartoon depicting the methodology used in this experiment. C57BL/6 RAG−/− mice received a BALB/c skin allograft; on day 1 posttransplant, the mice were injected with 6 × 106 CD3+CD25− T cells isolated from splenocytes of C57BL/6 naive mice. (B) Representative flow cytometry analyses of splenocytes isolated from mice 7 d posttransplant. Data show increase in PD1, TIM3, and LAG3 expression on effector CD4CD44high T cells (n = 6; P < 0.05). (C) Representative figures of flow cytometry analysis of splenocytes isolated from mice 7 d posttransplant. Data show increase in PD1, TIM3, and LAG3 expression on effector CD8CD44high T cells (n = 6; P < 0.05). (D) Mixed-lymphocyte reaction to assess alloimmune responses by 3H-thymidine incorporation. Results show a marked reduction in T-cell proliferative responses in recipients treated with DPLG3 compared with those treated with vehicle (n = 4; P < 0.05). (E) Expression of coinhibitory molecules of flow-sorted CD4 effector T cells (GFP−CD4+CD44high) from spleens of DPLG3 and vehicle-treated mice. (F) Expression of IL-17 and IL-12 by flow-sorted CD4 effector T cells (GFP−CD4+CD44high) from spleens of DPLG3- and vehicle-treated mice (n = 3; *P < 0.05).
Alloimmunity-Induced Up-Regulation of β5i Expression in CD4 and CD8 Effector T Cells.
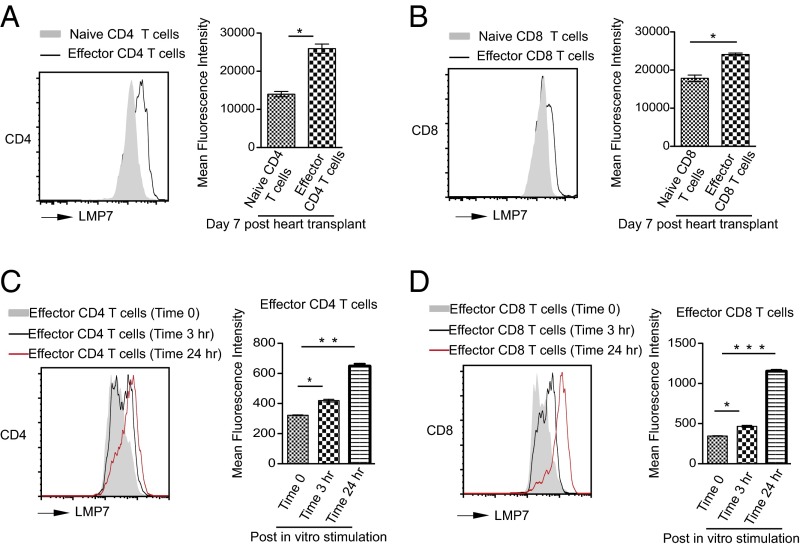
Before proceeding to test the impact of highly selective immunoproteasome inhibition on the host response to a vascularized organ allograft, we tested the impact of the allograft on expression of the inhibitor’s target, the β5i (LMP7) catalytic subunit, in naive and effector CD4 and CD8 T cells. We used a stringent, highly reproducible preclinical heart transplant mice model in which BALB/c hearts are transplanted into fully allogeneic C57BL/6 recipients. Splenocytes collected from C57BL/6 recipients of a BALB/c heart at day 7 posttransplantation, analyzed by flow cytometry, showed 2.0-fold and 1.3-fold increases in β5i (Fig. 5 A and B) in CD4 and CD8 effector T cells (CD44highCD62llow) compared with naive T cells (CD44lowCD62lhigh), respectively. To test whether this effect was cell autonomous, CD3 T cells were isolated from naive C57BL/6 mice and stimulated ex vivo with anti-CD3 and anti-CD28 antibodies. β5i expression in effector CD4 and CD8 T cells increased within 3 h poststimulation and peaked at 24 h (Fig. 5 C and D). Selective up-regulation of β5i in T cells reacting to alloantigen suggested that DPLG3 might not only be relatively selective for immunocytes over other cells but might have a relatively greater impact on antigen-activated effector T cells than on other T cells.
Fig. 5.
Up-regulation of β5i (LMP7) expression in CD4 and CD8 T cells upon alloactivation. Splenocytes of C57BL/6 mice recipients of BALB/c heart at day 7 posttransplantation were analyzed by flow cytometry. (A) Representative histogram showing a significant increase in β5i expression in CD4 effector T cells (CD4CD44highCD62llow) compared with naive T cells (CD4CD44lowCD62lhigh). (B) Representative histogram showing a significant increase in β5i expression in CD8 effector T cells (CD8CD44highCD62llow) compared with naive T cells (CD8CD44lowCD62lhigh). (C) Mean fluorescence intensity of LMP7 in naive and effector CD4 T cells (n = 3 mice; *P < 0.05). (D) Mean fluorescence intensity of β5i in naive and effector CD8 T cells (n = 3 mice; *P < 0.05).
Immunoproteasome Inhibitor-Induced Prolongation of Heart Allograft Survival and Reduction in Splenic Effector T Cells.
Increased β5i expression in alloimmune-activated T cells suggested that the immunoproteasome may play a critical role in T-cell function in alloimmunity. To test this, we transplanted BALB/c hearts into fully allogeneic C57BL/6 recipients and treated the transplanted mice with DPLG3 (25 mg/kg, i.p., per day) for 14 d beginning on day 0 posttransplant. Survival of the allograft was significantly prolonged compared with vehicle-treated recipients [median survival time (MST), 13 d vs. 7 d; *P < 0.01] (Fig. 6A). Accordingly, there was a marked reduction in the histologic signs of acute rejection in the allograft recovered at day 7 posttransplant from DPLG3-treated recipients (SI Appendix, Fig. S2A). Furthermore, we observed a decrease in the percentage of CD4 and CD8 effector memory T cells (CD44HighCD62LLow) in the spleen of DPLG3-treated recipients compared with vehicle-treated controls at day 7 posttransplant (CD4: 14.68 ± 1.08% vs. 17.72 ± 0.99%, respectively; n = 6, P < 0.05; CD8: 17.07 ± 2.54% vs. 29.08 ± 3.61%, respectively; n = 4, P < 0.05) (SI Appendix, Fig. S2B). The percentage of Tregs in the spleen of the DPLG3-treated recipients was not statistically significantly increased compared with wild type (WT) (11.63 ± 0.49% vs. 10.70 ± 0.32%, respectively; n = 6, P = 0.07). However, the ratio of Tregs to effector T cells was significantly up-regulated (SI Appendix, Fig. S2C). In contrast to the specific changes in effector T cells noted above, we saw no difference in the absolute count of total lymphocytes in the spleens of mice receiving a heart allograft and treated with DPLG3 or vehicle.
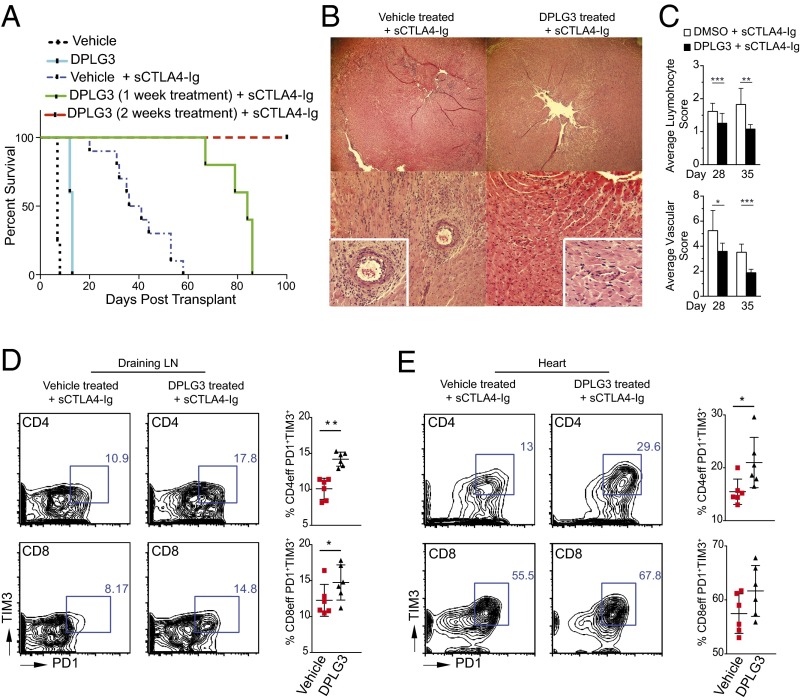
Fig. 6.
Contribution of β5i inhibition to up-regulation of exhaustion markers on effector T cells and prolonged survival of heart allografts when combined with a single low dose of CTLA4-Ig. (A) C57BL/6 recipients of BALB/c hearts treated with DPLG3 (25 mg/kg daily for 10 d) exhibited prolonged heart allograft survival compared with mice treated with vehicle (MST, 13 vs. 7 d, respectively; n = 4–5 mice/group; *P < 0.01). C57BL/6 recipients of BALB/c hearts treated with DPLG3 (25 mg/kg daily for 7 d) with a single low dose of CTLA4-Ig (sCTLA4-Ig) (250 µg on day 2) exhibited prolonged heart allograft survival compared with mice treated with vehicle and sCTLA4-Ig (MST, 84 and 38.5 d, respectively; n = 5–10 mice/group; P < 0.05). However, C57BL/6 recipients of BALB/c hearts treated both with DPLG3 (25 mg/kg daily for 14 d) and with sCTLA4-Ig had significantly longer allograft survival than mice treated with vehicle and sCTLA4-Ig (MST, >100 and 38.5 d, respectively; n = 5–10 mice/group; P < 0.05). (B) Representative examples of cardiac allograft histology at day 28 posttransplant show significantly lower grades of acute cellular rejection (H&E stain; original magnification, 10×) in mice treated with DPLG3 and sCTLA4-Ig compared with mice treated with vehicle and sCTLA4-Ig (n = 6 mice per group). (C) Cardiac allograft histology showed less lymphocyte infiltration (Top) and less vasculopathy (“vascular score”; Lower) at days 28 and 35 in recipients treated with DPLG3 plus sCTLA4-Ig compared with recipients treated with vehicle plus sCTLA4-Ig (Top). The vasculopathy score is a combination of vascular occlusion score and perivascular lymphocyte infiltration score for each of the coronary arteries (P < 0.05; n = 3–4 mice per group). (D) Representative contour plots of flow cytometry analysis of draining lymph nodes isolated from transplanted mice 28 d posttransplant after receiving 7-d treatment with DPLG3 or vehicle and sCTLA4-Ig. Data show increase in PD1 and TIM3 expression on effector CD4 CD44high and CD8 CD44high T cells (n = 6; P < 0.05). (E) Representative contour plots of flow cytometry analysis of lymphocytes isolated from heart allografts 28 d posttransplant. Data show increase in PD1 and TIM3 expression on effector CD4 CD44high T cells in mice treated with DPLG3 and sCTLA4-Ig compared with mice treated with vehicle and sCTLA4-Ig (n = 6; P < 0.05).
To test the alloimmune memory response of DPLG3- and vehicle-treated recipients, we stimulated splenocytes isolated from recipient mice at day 7 posttransplant with irradiated donor splenocytes ex vivo. Splenocytes from DPLG3-treated recipients showed reduced alloimmune responses as measured by 3H-thymidine incorporation (SI Appendix, Fig. S2D; P < 0.05).
Prolongation of Survival of a Fully Mismatched Heart Allograft by Brief Treatment with DPLG3 Together with a Single Dose of CTLA4-Ig.
Although single-dose CTLA4 Ig (sCTLA4-Ig) treatment (250 µg on day 2) of C57BL/6 recipients of BALB/c hearts resulted in moderate prolongation of allograft survival compared with untreated control (MST 38.5 vs. 7 d, respectively; P < 0.05), recipients treated with DPLG3 (25 mg/kg daily for 7 d) together with sCTLA4-Ig exhibited significant heart allograft survival prolongation compared with mice treated with vehicle and sCTLA4-Ig (MST: 84 and 38.5 d, respectively; n = 5–10 mice/group, P < 0.05) (Fig. 6A). Moreover, C57BL/6 recipients of BALB/c hearts treated with DPLG3 (25 mg/kg daily for 14 d) with sCTLA4-Ig exhibited much longer heart allograft survival than mice treated with vehicle and sCTLA4-Ig (MST: >100 and 38.5 d, respectively; n = 5–10 mice/group, P < 0.05) (Fig. 6A). All allografts that were not harvested for study sooner were monitored until day 130 posttransplant, at which time they were beating strongly. Histological examination of allografts harvested at day 28 posttransplant showed minimal signs of rejection in the group treated with DPLG3 plus sCTLA4-Ig compared with those treated with vehicle plus sCTLA4-Ig (Fig. 6B). At day 28 posttransplant, recipients treated with DPLG3 plus sCTLA4-Ig had fewer CD4+ and CD8+ effector T cells in the spleen (CD4: 13.83 ± 0.67 vs. 17.52 ± 0.43%, respectively; n = 6, P < 0.05; CD8: 3.51 ± 0.60 vs. 5.21 ± 0.51%, respectively; n = 6/group, P < 0.05) and draining lymph nodes (CD4: 14.37 ± 1.29 vs. 20.08 ± 1.20%, respectively; n = 6, P < 0.05; CD8: 2.85 ± 0.55 vs. 5.63 ± 0.23%, respectively; n = 6/group, P < 0.05) compared with vehicle plus sCTLA4-Ig–treated recipients. Cardiac allograft histology showed less lymphocyte infiltration at days 28 and 35 in recipients treated with DPLG3 plus sCTLA4-Ig compared with recipients treated with vehicle plus sCTLA4-Ig. Similarly, the vasculopathy score, a combination of vascular occlusion score and perivascular lymphocyte infiltration score for each of the coronary arteries, was lower in grafts of recipients treated with DPLG3 plus sCTLA4-Ig compared with recipients treated with vehicle plus sCTLA4-Ig (P < 0.05; n = 3–4 mice per group) (Fig. 6C).
Assessment of alloimmune responses in the periphery also showed a marked reduction in T-cell proliferative responses in recipients treated with DPLG3 plus sCTLA4-Ig compared with those treated with sCTLA4-Ig alone, as judged by 3H-thymidine incorporation (3,916 ± 1,561 dpm vs. 7,794 ± 1,467 dpm, respectively; n = 6, P < 0.05). Moreover, expression of PD1 and TIM3 increased on effector CD4 CD44high and CD8 CD44high T cells in the draining lymph nodes of mice given DPLG3 plus sCTLA4-Ig compared with those given sCTLA4-Ig alone (Fig. 6D; n = 6, P < 0.05). A similar pattern was observed in CD4 CD44high and CD8 CD44high T cells infiltrating the allografts of mice treated with DPLG3 plus sCTLA4-Ig compared with those given sCTLA4-Ig alone (Fig. 6E; n = 6, P = 0.05).
Discussion
Here, we describe a noncovalent N,C-capped dipeptide inhibitor of immunoproteasome subunit β5i (LMP7) with nanomolar potency whose selectivity for β5i over β5c rose from 7,200-fold to 99,000-fold as preincubation was extended from 0 to 24 h. The increase in selectivity with time reflected a time-dependent fall in IC50 for β5i. We speculate that this may result from slow conformational changes favorable to binding of the inhibitor in the active site of β5i in response to noninhibitory occupancy of other β subunits. In any event, after 24 h of preincubation, the isoform selectivity of DPLG3 was four orders of magnitude greater than that of the peptide epoxyketone β5i inhibitor, ONX0914 (10), tested under the same conditions. A recent study reported a variant of ONX0914 with up to 600-fold isoform selectivity (21). Nonetheless, as another irreversibly acting peptide epoxyketone, the new agent is expected to inhibit β5c to a greater extent the longer that exposure continues, as well as to present a risk for inhibiting some unrelated targets (22).
The exceptionally high, time-enhanced isoform selectivity of the reversibly acting N,C-capped dipeptide provided a biochemical rationale for expecting its mechanism-based effects to be limited to cells engaged in immunologic responses, as opposed to acting on nonimmunocytes. Moreover, increased expression of the target, β5i, in effector T cells reacting to antigen provided a biologic rationale for expecting the impact on immunocytes to be relatively restricted to those engaged in response to antigen at the time of treatment. Finally, the impact of an N,C-capped dipeptide immunoproteasome inhibitor on at least two classes of immunocytes—DCs and T cells—may contribute to the profound and prolonged impact on graft survival that resulted from transient treatment with the immunoproteasome inhibitor in combination with a single dose of CTLA4-Ig. Given the importance of alloantibody in chronic graft rejection, the impact of immunoproteasome inhibition on B cells and plasma cells warrants analysis as well, and will be the subject of future studies.
The pleiotropic effects of selective immunoproteasome inhibition reflect both the protean biologic roles of the ubiquitin–proteasome system in all cells, and the variable degree to which c-20S subunits are replaced by i-20S subunits in some cells. Here, we show a selective up-regulation of β5i in T cells reacting to alloantigens, which suggests a specific role for this subunit in antigen-reactive effector T cells compared with other T cells. We are particularly intrigued that selective immunoproteasome inhibition led to the induction of exhaustion and coinhibitory markers on, and reduction in the proportions of, effector T cells in the allografts and recipients’ spleens. This suggests that a previously unappreciated biologic function of the immunoproteasome is to protect antigen-reacting effector T cells from exhaustion or inhibition. T-cell exhaustion is a state characterized by a progressive loss of production of certain cytokines such as IL-2 followed by a proliferation defect and apoptosis (23). Exhausted T cells express a variety of coinhibitory receptors, including PD1, TIM3, LAG3, CTLA4, CD160, and BLTA, that modulate the intracellular signaling responsible for this state (24). Although reversal of T-cell exhaustion may be useful in the treatment of chronic infections and cancers, induction of T-cell exhaustion may promote tolerance to autoantigens and alloantigens (24, 25). T-cell exhaustion has recently been shown to play a central role in determining clinical outcome in multiple autoimmune diseases (26), and it has been suggested that targeted induction of exhaustion could benefit patients with an aggressive course of disease. Nonselective proteasome inhibitors have shown benefit in small trials in patients with lupus nephritis, autoimmune hemolytic anemia, and autoantibody-associated mesangioproliferative glomerulonephritis (27), but it is unknown whether the benefit was related to induction of T-cell exhaustion, reduction in autoantibodies, and/or other mechanisms.
Highly selective immunoproteasome β5i inhibitors such as DPLG3 represent powerful tools to dissect the role of the β5i subunit in various disease models and the impact of inhibition at different stages of the pathogenic process. Mice lacking β5i have limitations in this regard, both because absence of β5i precedes disease onset and because there is extensive compensation by β5c.
Further study is required to identify the signaling, transcriptional, and posttranscriptional mechanisms by which immunoproteasome inhibition leads to changes in immunocyte activation, proliferation, surface marker expression, and secretion of cytokines. The present results call attention to the potential value of using brief posttransplant immunoproteasome inhibition to entrain a long-term response favorable to allograft survival as part of an immunomodulatory regimen that is neither broadly immunosuppressive nor cytotoxic.
Materials and Methods
Synthesis and Characterization of β5i Inhibitors.
The synthetic route and methods of compound characterization were similar to those reported for N,C-capped dipeptidomimetics (15, 28), with modifications as given in the supplemental online material. All compounds were >95% pure. ONX0914 was obtained from BPBio. Human immunoproteasomes were purified from PBMCs and constitutive proteasomes from red blood cells. Suc-LLVY-AMC and Ac-ANW-AMC were purchased from Boston Biochem. Enzymatic assays were monitored on a Molecular Devices SpectraMax M5 plate reader. Cell viability was determined with ATP-lite assay kit (PerkinElmer). Cell-based Proteasome-Glo (G8660; Promega) was used to measure 20S chymotryptic-like β5 activity and its inhibition in cells.
Biochemical Characterization of β5i Inhibitors.
Assays with isolated proteasomes and in intact cells were performed as reported (29). Briefly, Karpas 1106P cells (80,000/well) were incubated with compound at indicated concentrations for 1 h at 37 °C. The plate was then spun at 188 × g for 1 min and the supernatant was removed. The overall β5 activity including β5i and β5c in each well was measured in situ with the Proteasome-Glo assay kit according to the manufacturer’s instructions. Luminescence was recorded on a SpectraMax M5 plate reader. Relative percentage of relative light units was used to calculate IC50 values.
Cell Cultures.
We cultured Karpas1106P B lymphoma cell line (catalog no. 06072607; Aldrich) (15), mouse bone marrow-derived macrophages (18), PBMCs (17, 30), and HepG2 human hepatoma cells (31). Karpas cells were cultured in complete medium and the other cells in complete medium with 10% (vol/vol) FBS instead of 20% (vol/vol), all at 37 °C in a humidified air/5% (vol/vol) CO2. Cells in a 96-well plate were treated with compounds at indicated concentrations for 72 h at 37 °C in with 5% (vol/vol) CO2. Viable cells were counted using CellTiter-Glo assay kit. EC50 values were calculated using PRISM.
Human PBMC and PDC Assays.
Buffy coats were obtained from the New York Blood Center (Long Island City, NY) with informed consent and used under a protocol approved by the institutional review board of the Hospital for Special Surgery. Total PBMCs were prepared using standard protocol (17) and cells (3 × 105 cells/well) were cultured for 24 h in 96-well plate. The viability of PBMCs was quantified using a colorimetric (MTT) assay (Millipore) according to manufacturer’s instructions. PDCs were isolated using positive selection using BDCA-4–conjugated beads (Miltenyi Biotec) as described (32). PDCs were 94–99% as determined by flow cytometry (BDCA2+ CD123+) and cells (2–5 × 104 cells per well) were cultured in U-bottom 96-well plate. Cells were activated with a phosphorothioate CpG-B or -C ODN at 0.3–1 μM (TLR9 agonist) as described (32), and their viability was assessed by flow cytometry using propidium iodide. Cells were cultured for 24 h, and human IFN-α and IP-10 production were assayed by ELISA with reagents from PBL Biomedical Laboratories and from Thermo Fisher, respectively. Monoclonal antibodies used for flow cytometry included the following: anti-CD3, anti-CD14, anti-CD19, anti-HLA-DR anti-CD83, anti-CD86 (BD Biosciences), anti-CD123, and anti-BDCA-2 (Miltenyi Biotec).
Mice.
Female C57BL/6 (H-2b), BALB/c (H-2d), FoxP3-GFP knock-in mice on a C57BL/6 background and Rag−/− mice on a BALB/c background were obtained from The Jackson Laboratory. Mice were 6–10 wk of age (20–25 g) at the start of the experiments. They were housed in accordance with institutional and NIH guidelines. The Harvard Medical School Animal Management Committee approved all animal experiments.
Skin Transplantation.
Full-thickness trunk skin grafts (1 cm2) harvested from BALB/c donors were transplanted onto the flank of Rag−/− C57BL/6 recipient mice, sutured with 6.0 silk, and secured with dry gauze and a bandage for 7 d.
Cardiac Transplantation.
Vascularized intraabdominal heterotopic transplantation of cardiac allografts was performed using microsurgical techniques (33). The survival of cardiac allografts was assessed by daily palpation. Rejection was defined as complete cessation of cardiac contractility as determined by direct visualization and confirmed by histology.
Histological and Immunohistochemical Assessment.
Five-micrometer-thick formalin-fixed paraffin-embedded sections were stained with standard hematoxylin and eosin (H&E) stain.
Isolation of Lymphocytes from Hearts.
Cardiac allografts were removed, perfused with PBS, minced finely with a razor blade, and digested at 37 °C with 1 mg/mL collagenase in 1 mL of complete RPMI medium 1640 for 1 h. Cells in the supernatant were washed twice, centrifuged at 620 × g using Percoll solutions at 33% (vol/vol) (cell suspension) and 66% (vol/vol). Lymphocytes were aspirated at the interface.
Histology Score.
Four sections stained with H&E were inspected from the allografts of each of three mice per group. To score lymphocyte infiltration, each section was divided into six sectors and lymphocyte infiltration in each sector was graded using a scale modified from the International Society for Heart and Lung Transplantation (0, no lymphocyte infiltration; 1, less than 25% lymphocyte infiltration; 2, 25–50% lymphocyte infiltration; 3, 50–75% lymphocyte infiltration; 4, more than 75% lymphocyte infiltration with hemorrhage and/or necrosis). The score for each sector was averaged, and the average from each of the four sections was used as the lymphocyte infiltration score for each heart. The severity of vasculopathy was determined by a combination of vascular occlusion score and perivascular lymphocyte infiltration score for each coronary vessel. Vascular occlusion was scored from grade 0–3 as follows: grade 0 (no or minimal, <10%), grade 1 (10–50% occlusion), grade 2 (50–75% occlusion), and grade 3 (more than 75% occlusion). The perivascular lymphocyte infiltration was scored as grade 0 (no lymphocyte infiltration), grade 1 (mild lymphocyte infiltration), 2 (moderate lymphocyte infiltration), and 3 (severe lymphocyte infiltration), and then added to the vascular occlusion score, for a final score that could range from 0 to 6.
CD3/CD28 T-Cell Stimulation Assay and Mixed-Lymphocyte Reaction.
Anti-CD3 Ab (100 μL) and soluble anti-CD28 Ab (1 μg/mL; BD Biosciences) were dispensed to wells of a 96-well flat-bottom plate containing C57BL/6 CD4 or CD8 T cells and the indicated concentrations of DPLG3 or the DMSO vehicle (final concentration of 1% DMSO). For the mixed-lymphocyte reaction, irradiated WT BALB/c splenocyte stimulators and C57BL/6 splenocyte responders from DPLG3-treated or vehicle-treated mice were added to each well in a 96-well round-bottom plate, then pulsed with 1 μCi of tritiated thymidine 9 h before the end of the 72-h assay. Incorporation efficiency was determined by scintillation counting.
Flow Cytometry.
Anti-mouse Abs against CD62L, CD44, CD4, CD25, CD8, FoxP3, GrB, PD1, TIM3, LAG3, LMP7, annexin, 7-amino-actinomycin D (7-AAD), and Ki67 were purchased from BD Biosciences. Cells recovered from spleens and peripheral lymphoid tissues were analyzed on a FACSCanto II flow cytometer (BD Biosciences) with FlowJo software, version 9.3.2 (Tree Star).
RNA Sequencing.
Total RNA was isolated from flow-sorted CD4 effector T cells (GFP−CD4+CD44high) and from spleens of DPLG3-treated mice using RNeasy Micro kit (QIAGEN) and subjected to library preparation using NEBNext Ultra RNA Library prep for Illumina kit (New England Biolabs). Library quality was evaluated by Bioanalyzer with High Sensitivity chips (Agilent Technologies). Sequencing was performed on a HiSeq 2000 (Illumina) by 2 × 50-bp paired-end reads at the Biopolymers Facility of Harvard Medical School. We used Bcbio_nextgen (https://github.com/chapmanb/bcbio-nextgen/) to process the RNA-seq data. Briefly, cutadapt (https://github.com/marcelm/cutadapt/) was used to trim adapters; trimmed reads were aligned to human reference genome (GRCh37) with tophat2; and read count for each gene was calculated by HT-seq. Genes with low expression (fragments per kilobase of transcript per million mapped reads < 1 across all samples) were filtered out.
Statistical Analyses.
Kaplan–Meier survival graphs were constructed and a log-rank comparison of the groups was used to calculate P values. The unpaired t test was used for comparison of experimental groups examined by Luminex assay, flow cytometry, and mixed-lymphocyte reactions. Differences were considered to be significant for P ≤ 0.05. Prism software was used for data analysis and graphing (GraphPad Software). Data represent means ± SEM.
Supplementary Material
Acknowledgments
We thank Dr. J. David Warren at The Abby and Howard P. Milstein Synthetic Chemistry Core Facility at Weill Cornell Medicine for assistance and Drs. Jane Salmon and Xiaoping Qing at Hospital for Special Surgery for discussions. This work was supported by an American Heart Association grant (to J.A.), by the Milstein Program in Translational Medicine and Chemical Biology (C.F.N.), by the Alliance for Lupus Research (G.L.), and by the Daedalus Fund for Innovation at Weill Cornell Medicine (G.L.). The Department of Microbiology and Immunology at Weill Cornell Medicine is supported by the William Randolph Hearst Trust.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1618548114/-/DCSupplemental.
References
- 1.Neubert K, et al. The proteasome inhibitor bortezomib depletes plasma cells and protects mice with lupus-like disease from nephritis. Nat Med. 2008;14(7):748–755. doi: 10.1038/nm1763. [DOI] [PubMed] [Google Scholar]
- 2.Altun M, et al. Effects of PS-341 on the activity and composition of proteasomes in multiple myeloma cells. Cancer Res. 2005;65(17):7896–7901. doi: 10.1158/0008-5472.CAN-05-0506. [DOI] [PubMed] [Google Scholar]
- 3.Baumeister W, Walz J, Zühl F, Seemüller E. The proteasome: Paradigm of a self-compartmentalizing protease. Cell. 1998;92(3):367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 4.Goldberg AL. Functions of the proteasome: From protein degradation and immune surveillance to cancer therapy. Biochem Soc Trans. 2007;35(Pt 1):12–17. doi: 10.1042/BST0350012. [DOI] [PubMed] [Google Scholar]
- 5.Rock KL, et al. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78(5):761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 6.Kim MS, et al. A draft map of the human proteome. Nature. 2014;509(7502):575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilhelm M, et al. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509(7502):582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- 8.Foss GS, Prydz H. Interferon regulatory factor 1 mediates the interferon-gamma induction of the human immunoproteasome subunit multicatalytic endopeptidase complex-like 1. J Biol Chem. 1999;274(49):35196–35202. doi: 10.1074/jbc.274.49.35196. [DOI] [PubMed] [Google Scholar]
- 9.Griffin TA, et al. Immunoproteasome assembly: Cooperative incorporation of interferon gamma (IFN-gamma)-inducible subunits. J Exp Med. 1998;187(1):97–104. doi: 10.1084/jem.187.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muchamuel T, et al. A selective inhibitor of the immunoproteasome subunit LMP7 blocks cytokine production and attenuates progression of experimental arthritis. Nat Med. 2009;15(7):781–787. doi: 10.1038/nm.1978. [DOI] [PubMed] [Google Scholar]
- 11.Ichikawa HT, et al. Beneficial effect of novel proteasome inhibitors in murine lupus via dual inhibition of type I interferon and autoantibody-secreting cells. Arthritis Rheum. 2012;64(2):493–503. doi: 10.1002/art.33333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Basler M, et al. Inhibition of the immunoproteasome ameliorates experimental autoimmune encephalomyelitis. EMBO Mol Med. 2014;6(2):226–238. doi: 10.1002/emmm.201303543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Basler M, Dajee M, Moll C, Groettrup M, Kirk CJ. Prevention of experimental colitis by a selective inhibitor of the immunoproteasome. J Immunol. 2010;185(1):634–641. doi: 10.4049/jimmunol.0903182. [DOI] [PubMed] [Google Scholar]
- 14.Singh PK, et al. Immunoproteasome β5i-selective dipeptidomimetic inhibitors. ChemMedChem. 2016;11(19):2127–2131. doi: 10.1002/cmdc.201600384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blackburn C, et al. Characterization of a new series of non-covalent proteasome inhibitors with exquisite potency and selectivity for the 20S beta5-subunit. Biochem J. 2010;430(3):461–476. doi: 10.1042/BJ20100383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huber EM, et al. Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity. Cell. 2012;148(4):727–738. doi: 10.1016/j.cell.2011.12.030. [DOI] [PubMed] [Google Scholar]
- 17.Vogt G, Nathan C. In vitro differentiation of human macrophages with enhanced antimycobacterial activity. J Clin Invest. 2011;121(10):3889–3901. doi: 10.1172/JCI57235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bryk R, et al. Selective killing of nonreplicating mycobacteria. Cell Host Microbe. 2008;3(3):137–145. doi: 10.1016/j.chom.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirai M, et al. Bortezomib suppresses function and survival of plasmacytoid dendritic cells by targeting intracellular trafficking of Toll-like receptors and endoplasmic reticulum homeostasis. Blood. 2011;117(2):500–509. doi: 10.1182/blood-2010-05-284737. [DOI] [PubMed] [Google Scholar]
- 20.Lightcap ES, et al. Proteasome inhibition measurements: Clinical application. Clin Chem. 2000;46(5):673–683. [PubMed] [Google Scholar]
- 21.Kasam V, Lee NR, Kim KB, Zhan CG. Selective immunoproteasome inhibitors with non-peptide scaffolds identified from structure-based virtual screening. Bioorg Med Chem Lett. 2014;24(15):3614–3617. doi: 10.1016/j.bmcl.2014.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huber EM, et al. Systematic analyses of substrate preferences of 20S proteasomes using peptidic epoxyketone inhibitors. J Am Chem Soc. 2015;137(24):7835–7842. doi: 10.1021/jacs.5b03688. [DOI] [PubMed] [Google Scholar]
- 23.Wherry EJ. T cell exhaustion. Nat Immunol. 2011;12(6):492–499. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- 24.Kuchroo VK, Anderson AC, Petrovas C. Coinhibitory receptors and CD8 T cell exhaustion in chronic infections. Curr Opin HIV AIDS. 2014;9(5):439–445. doi: 10.1097/COH.0000000000000088. [DOI] [PubMed] [Google Scholar]
- 25.Sarraj B, et al. Impaired selectin-dependent leukocyte recruitment induces T-cell exhaustion and prevents chronic allograft vasculopathy and rejection. Proc Natl Acad Sci USA. 2014;111(33):12145–12150. doi: 10.1073/pnas.1303676111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKinney EF, Lee JC, Jayne DR, Lyons PA, Smith KG. T-cell exhaustion, co-stimulation and clinical outcome in autoimmunity and infection. Nature. 2015;523(7562):612–616. doi: 10.1038/nature14468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feist E, Burmester GR, Krüger E. 2016. The proteasome—victim or culprit in autoimmunity. Clin Immunol S1521-6616(16)30212-1.
- 28.Lin G, et al. N,C-capped dipeptides with selectivity for mycobacterial proteasome over human proteasomes: Role of S3 and S1 binding pockets. J Am Chem Soc. 2013;135(27):9968–9971. doi: 10.1021/ja400021x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan H, Angelo NG, Warren JD, Nathan CF, Lin G. Oxathiazolones selectively inhibit the human immunoproteasome over the constitutive proteasome. ACS Med Chem Lett. 2014;5(4):405–410. doi: 10.1021/ml400531d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin G, et al. Inhibitors selective for mycobacterial versus human proteasomes. Nature. 2009;461(7264):621–626. doi: 10.1038/nature08357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gold B, et al. Nonsteroidal anti-inflammatory drug sensitizes Mycobacterium tuberculosis to endogenous and exogenous antimicrobials. Proc Natl Acad Sci USA. 2012;109(40):16004–16011. doi: 10.1073/pnas.1214188109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guiducci C, et al. Properties regulating the nature of the plasmacytoid dendritic cell response to Toll-like receptor 9 activation. J Exp Med. 2006;203(8):1999–2008. doi: 10.1084/jem.20060401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Corry RJ, Winn HJ, Russell PS. Heart transplantation in congenic strains of mice. Transplant Proc. 1973;5(1):733–735. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.