Abstract
Tryptophan synthase (TrpS) catalyzes the final steps in the biosynthesis of L-tryptophan from L-serine (Ser) and indole-3-glycerol phosphate (IGP). We report that native TrpS can also catalyze a productive reaction with L-threonine (Thr), leading to (2S,3S)-β-methyltryptophan. Surprisingly, β-substitution occurs in vitro with a 3.4-fold higher catalytic efficiency for Ser over Thr using saturating indole, despite >82,000-fold preference for Ser in direct competition using IGP. Structural data identify a novel product binding site and kinetic experiments clarify the atypical mechanism of specificity: Thr binds efficiently but decreases the affinity for indole and disrupts the allosteric signaling that regulates the catalytic cycle.
Graphical Abstract
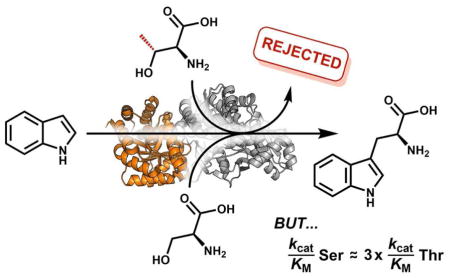
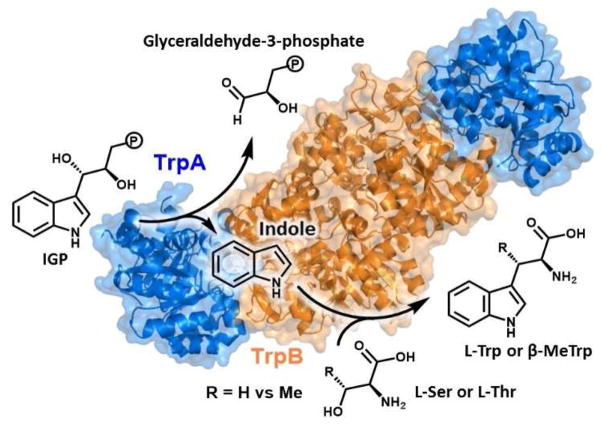
Tryptophan synthase (TrpS) is an αββα heteroenzyme complex present in all domains of life that catalyzes the formation of L-tryptophan (Trp) from 3-indole-D-glycerol-3′-phosphate (IGP) and L-serine (Ser; Figure 1).1–3 The α-subunit (TrpA) catalyzes the retro-aldol cleavage of IGP, releasing indole into an intramolecular tunnel that forms when each subunit is in a closed conformational state and extends ~20 Å to the β-subunit (TrpB) active site.4 TrpB utilizes a pyridoxal-phosphate (PLP) cofactor to effect the β-substitution of Ser with indole, yielding Trp. Recently, we used directed evolution to engineer TrpB from Pyrococcus furiosus (PfTrpB) to retain activity in the absence of its TrpA partner.5 We then further engineered this stand-alone enzyme to catalyze the efficient β-substitution of L-threonine (Thr), yielding (2S,3S)-β-methyltryptophan (β-MeTrp) in a single step.6 This non-canonical amino acid is an intermediate in the biosynthesis of maremycin and streptonigrin, and is natively produced from Trp through a three-enzyme, four-step pathway.7, 8 Given that Thr is a universal and abundant metabolite, we were curious whether the native PfTrpS enzyme can also synthesize β-MeTrp and, if so, how the enzyme discriminates between Ser and Thr to produce only Trp in vivo.
Figure 1.
Overview of the TrpS catalytic cycle. The α-subunit (blue) cleaves IGP in a retroaldol reaction. Indole is channeled into the β-subunit (orange), where it reacts with L-Ser or L-Thr to generate L-Trp or β-MeTrp, respectively.
PfTrpS was heterologously expressed and purified from Escherichia coli. Assaying for activity with Thr and IGP at 75 °C showed that the native enzyme can indeed synthesize β-MeTrp. To determine whether indole is efficiently tunneled through the enzyme complex or is instead released and re-enters during a separate catalytic cycle, we performed reactions in the presence of an equal volume of toluene, which can sequester indole. Under these conditions, Ser and IGP react with a coupling efficiency of >99% and only trace indole is released into solution, consistent with previous observations with TrpS from mesophilic organisms (Table 1).9 Under similar conditions with Thr, however, only 17% of the indole that is released from the α-subunit goes on to form β-MeTrp, demonstrating that the release of indole is decoupled from product formation. To verify that the enzyme is selective for Ser, we performed a direct competition experiment using equimolar Ser and Thr, and detected only Trp. This observation holds even when the reaction is conducted with a 1,000-fold molar excess of Thr over Ser: only Trp is observed, with no trace of β-MeTrp (Table S1).
Table 1.
Kinetic parameters of the Pyrococcus furiosus TrpSa
| Substrate | kcat substitution (s−1) | kcat deamination (s−1) | KM amino acid (mM) | KM indole (μM) | kcat/ KM amino acid (M−1s−1) | kcat/ KM indole (M−1s−1) | Coupling Efficiencyb (indole) | Coupling Efficiencyb (IGP) |
|---|---|---|---|---|---|---|---|---|
| L-Ser | 1.0 ± 0.1c | 0.055 ± 0.002 | 0.6 ± 0.1c | 20 ± 2c | 1,600 | 50,000 | >99% | >99% |
| L-Thr | 0.61 ± 0.04 | 0.47 ± 0.04 | 1.3 ± 0.2 | 1,400 ± 300d | 470 | 460 | 65 ± 10% | 17 ± 3% |
Errors were estimated from three independent experiments. All reactions conducted with 1–20 μM PfTrpS in potassium phosphate buffer pH 8.0.
Defined as the proportion of 1 equivalent of indole or IGP that reacts with a single equivalent of Ser or Thr to form product.
These values from Ref. 5.
This value was obtained by merging initial velocity data (shown in Figure S3) collected from the UV-vis and LC-MS.
To probe whether this reactivity profile is general to TrpS, we also assayed TrpS enzymes from Archaeoglobus fulgidus and Thermotoga maritima (67.1% and 55.9% sequence identity to PfTrpS, respectively).10 We found that each of these enzymes also performs a productive reaction with IGP and Thr. In the presence of Ser, however, and even with a 1,000-fold molar excess of Thr over Ser, there are at most trace amounts of β-MeTrp formed (Table S1). From these data, we calculate a specificity of ≥82,000 for Ser over Thr for each TrpS enzyme. Hence, there is a strong, evolutionarily conserved specificity for Ser within the TrpS family. Further study into the mechanism of discrimination using IGP as the indole source is convoluted, as product can form via two routes: either indole tunnels through the complex and immediately reacts in the β-subunit or, alternatively, indole exits the complex and re-enters to react during a separate iteration of the catalytic cycle. Therefore, we focused on identifying the mechanism of specificity in the β-site using exogenously added indole and Thr.
Previously, we observed that Thr binds non-covalently to the isolated β-subunit, indicating that the β-methyl group hinders entry into the catalytic cycle.6 We therefore performed a steady-state kinetic analysis of PfTrpS with Thr and indole, expecting that the enzyme would display a substantially lower catalytic efficiency with Thr. To our surprise, there is only a modest 1.6-fold decrease in kcat, and 2-fold increase in the KM for Thr (Table 1). Hence, the ratio of the catalytic efficiencies for each reactant, the specificity constant, is ~3.4 in favor of Ser. In contrast, direct competition using a 100-fold molar excess of Thr to Ser and indole shows that PfTrpS has a ~711-fold preference for Ser over Thr (Table S1). This apparent contradiction is reconciled when one considers that the specificity constant is defined for unimolecular reactions and that, while saturating indole makes the kinetics pseudo-first order in Ser or Thr, differences in reactivity with indole may still contribute to the overall specificity of the enzyme.11 Indeed, the KM for indole is 20 μM in the native reaction,5 but is 1,400 μM in the reaction with Thr (Table 1).
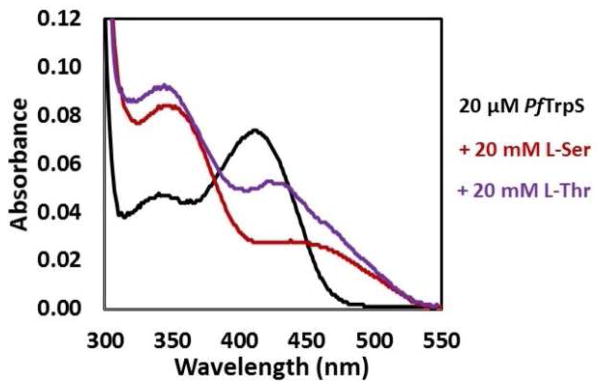
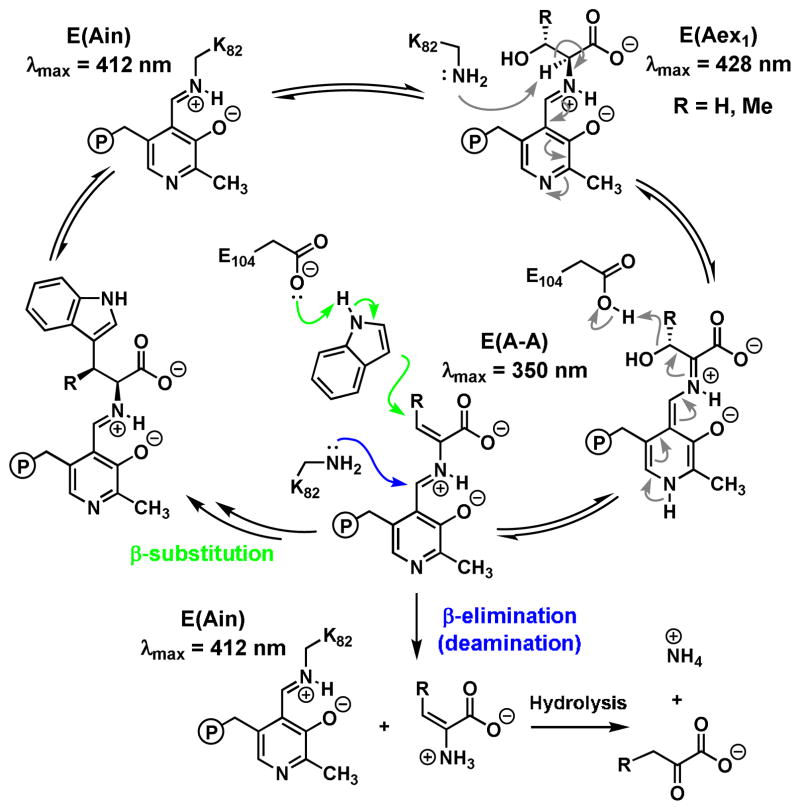
Comparison of the ratio of the third-order rate constants, kcat/KM(amino acid)/KM(indole) with units s−1M−2, has previously been used to compare the ‘overall catalytic efficiency’ of bimolecular reactions.12 However this parameter indicates a 250-fold preference for Ser, underestimating the observed ability of the enzyme to discriminate between Ser and Thr by almost 3-fold and indicating that additional factors are likely influencing catalysis. Whereas Thr binds the isolated PfTrpB non-covalently,6 the UV-vis spectrum of PfTrpS in the presence of Thr shows a prominent spectral shift indicative of covalent binding (Figure 2). The absorbance peak at 350 nm is consistent with accumulation of the E(A-A) intermediate (Scheme 1), and the peak at 425 nm is indicative of a minor population of E(Aex1). In addition to reaction with indole, the electrophilic E(A-A) species can undergo transimination with the active site lysine. This releases into solution dehydroalanine, whose subsequent hydrolysis yields ammonia and an α-keto acid.13 We observed that the rate of Thr deamination by PfTrpS is 8.5-fold faster than with Ser, competitive with the rate of β-substitution (Table 1). Further, we observed that Thr is deaminated even in the presence of indole (Figure S1). These experiments indicate the electrophilic intermediate with Thr, an aminocrotonate E(A–C), has a decreased lifetime that is subject to kinetic competition between β-substitution with indole and hydrolysis. The result is that when presented with a single equivalent of Thr and indole, the enzyme makes less than a full equivalent of product (Table 1). Hence, the deamination of Thr leads to a decreased coupling efficiency with indole, which effectively serves as a catalytic editing mechanism under these conditions.
Figure 2.
UV-vis absorbance spectra of PfTrpS. In the absence of substrate PfTrpS (20 μM) has an absorbance spectrum corresponding to E(Ain).3 Addition of 20 mM L-Ser yields a spectrum with λmax = 350 nm. A similar peak with λmax = 345 nm accumulates when L-Thr is added. This shift may reflect a mix of signals with α-ketobutyrate (λmax = 319 nm) formed from the fast deamination of L-Thr. The additional peak at ~425 nm corresponds to a small population of the E(Aex1) intermediate.
Scheme 1.
Catalytic cycle of tryptophan synthase for β-substitution and β-elimination
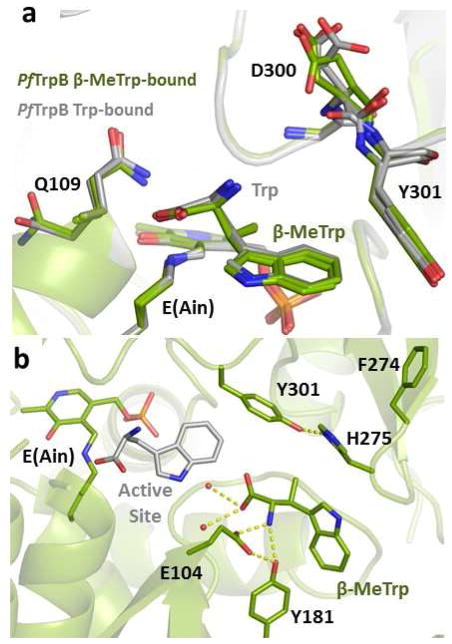
Previous structural and spectroscopic analysis of PfTrpB suggested that the β-methyl of Thr causes a steric clash that destabilizes the E(Aex1) intermediate.6 The spectral data above illustrate that this methyl group is moderately well-accommodated during the E(A–C) stage of the reaction, but the increased KM for indole with Thr suggests unfavorable interactions within the active site, likely steric clashes. To probe whether this additional methyl group may cause deleterious interactions during later stages of catalysis, we determined the X-ray crystal structure of β-MeTrp bound to PfTrpB at 1.8 Å resolution (Table S2). In contrast to the structures with Ser and Thr bound, the structure of β-MeTrp (Figure S2) is almost identical to the structure with Trp bound, consistent with a model in which the β-methyl is discriminated against early in the catalytic cycle (Figure 3a). Unexpectedly, we observed that β-MeTrp bound in a novel pose in one molecule of the asymmetric unit, with the indole sidechain protruding into the tunnel that extends from solution into the active site (Figure 3b, S2) or between the two subunits when in a fully closed state.4 The biological relevance of this pose is not immediately obvious; however the path between the active site and bulk solution is long and presumably some intermediate poses must be stabilized for facile entry and exit from the enzyme. Recent molecular dynamics simulations indicated that this exact region of the protein is de-wetted in solution and that it serves as a tunnel for the efficient diffusion of indole into the β-subunit active site.14 The new pose identified here constitutes the first direct structural evidence for substrate or product binding outside of the active site, which we hypothesize facilitates exit of the nascent Trp from the active site during the native catalytic cycle.
Figure 3.
β-MeTrp binding within the PfTrpB. (a) Two of the four PfTrpB subunits within the asymmetric unit show β-MeTrp bound in a pose superimposable with the Trp-bound conformation (from PDB ID: 5DW3). Minor structural changes are observed only in regions that display conformational heterogeneity between subunits of the same crystal, including Q109 and D300. (b) One of the four PfTrpB subunits shows β-MeTrp in a novel pose. Hydrogen bond interactions are shown in yellow dashes. For reference, a single molecule of Trp has been superimposed (gray) to indicate the new pose in relation to the active site.
Taken together with previous studies on the native TrpS mechanism, these data provide a view of a highly redundant mechanism for discriminating between Ser and Thr in PfTrpS. Both substrates bind in the β-site of the enzyme with nearly equal affinity, resulting in an unusual ‘specificity constant’ of ~3.4, despite a 711-fold greater preference for Ser when indole is the nucleophile. As the catalytic cycle is reversible up to the point of C–C bond formation, the majority of this discrimination occurs during this early phase. When Ser is bound, the coupling efficiency with indole exceeds 99%. In contrast, when Thr is bound, the KM for indole is higher, which decreases its population in the β-subunit active site. Additionally, the abortive deamination reaction with Thr is accelerated relative to Ser, which may decompose Thr before it reacts with indole. The biological relevance of this latter step, which would constitute a catalytic editing function, is difficult to assess from in vitro experiments using exogenously added indole. When IGP is used as substrate, we observed a substantial increase in the specificity of TrpS for Ser over Thr. Facile decomposition of Thr would be a metabolic burden in vivo and the aminocrotonate produced by decomposition is highly toxic, and cells possess enzymes to accelerate hydrolysis of this metabolite.15 Therefore, it seems unlikely that a significant proportion of TrpS specificity can be attributed to deamination of Thr. Instead, we hypothesize that allosteric effects are responsible for the increase in specificity when IGP is used instead of indole.
Studies of StTrpS have shown the deamination reaction is likely caused by premature opening of the communication (COMM) domain of TrpB that regulates the catalytic cycle.16 Hence, not only is the steady-state rate of catalysis lower with Thr, but the complex is likely in its closed state for less time. Notably, these open-close dynamics are an integral part of the TrpS allosteric signaling,17 and altered TrpB conformational dynamics with Thr may disrupt the intricate reciprocal signaling between subunits that coordinates the catalytic cycle. These allosteric properties are sensitive to the identity of the monovalent cation bound to TrpS, and the present study focusses on the potassium form of the enzyme, as this is predominant intracellular monovalent cation.3 Detailed analysis of allosteric events awaits pre-steady state analysis; however, some reasonable hypotheses can be made drawing from the detailed literature of StTrpS. In the native catalytic cycle, IGP binding in the α-site stimulates E(A-A) formation by stabilizing the closed conformation of the COMM domain, which in turn signals for retro-aldol cleavage of IGP to release indole.18, 19 This signaling system serves to release indole only when the tunnel is closed, thereby preventing its diffusion away from the complex.9 It is possible that IGP binding in the α-site has a greater effect stabilizing the electrophilic intermediate with Ser than with Thr. When the Thr E(A-C) is formed, it may still fail to efficiently signal for IGP cleavage and indole release.19, 20 Interestingly, we observed that the coupling efficiency between IGP and Thr, just 17%, is lower than when indole is added exogenously (Table 1). This decoupling convolutes experimental attempts to probe the catalytic cycle with Thr and IGP. Nonetheless, the decoupling indicates that when indole is released, one or both subunits re-opens prior to C–C bond formation, allowing indole to escape the complex. Hence, when Ser and Thr are in direct competition, there appear to be relatively few instances where the Thr E(A-C) and indole have the opportunity to react. These effects combine with the specificity inherent within the β-subunit to provide the >82,000-fold preference for Ser in PfTrpS.
Given that enzyme specificity can change during evolution and that Thr is an abundant metabolite,21 the question arises: why did nature evolve a three-enzyme pathway to synthesize β-MeTrp7, 8 when it already possessed the catalytic machinery to perform the reaction in the form of TrpS? Where specificity is driven by differences in the KM, mutations that alter binding affinity can readily swap the specificities of such enzymes.22 In contrast, TrpS has only a small preference for Ser over Thr and instead achieves specificity through catalytic and allosteric means, as described above. During directed evolution of PfTrpB for stand-alone activity with Thr, we found that the resultant enzyme was still highly specific for Ser,6 indicating that this catalytic mechanism of discrimination is robust to mutation. To a first approximation then, it appears that Nature does not use TrpS for β-MeTrp synthesis because the activity is completely suppressed under biological conditions and effectively hidden from metabolism. Additionally, this study reminds us that specificity parameters measured in the absence of competition in vitro may not be predictive of specificity that occurs when substrates are in direct competition in vivo.
Supplementary Material
Acknowledgments
Funding Sources
The Caltech Molecular Observatory is supported by the Gordon and Betty Moore Foundation, the Beckman Institute, and the Sanofi-Aventis Bioengineering Research Program at Caltech. A.R.B is supported by a Ruth Kirschstein NIH Postdoctoral Fellowship (F32G110851) and J.M.C. is supported by the Alfonso Martín Escudero Foundation.
We thank Dr. Sabine Brinkmann-Chen, Michael Herger, and Dr. David Romney for insightful discussions and experimental assistance. Crystallography experiments were supported by Dr. Jens Kaiser and the Caltech Molecular Observatory.
Footnotes
The authors declare no competing financial interests.
The coordinates for the PfTrpB structure with β-MeTrp bound are available from the RCSB Protein Data Bank at http://www.rcsb.org/pdb with PDB ID: 5T6M
The Supporting Information is available free of charge on the ACS Publications website.
Detailed Materials and Methods, tables of substrate specificities and crystallographic statistics. Figures of UV-spectra showing β-elimination and crystallographic maps demonstrating ligand binding.
References
- 1.Delmer DP, Mills SE. Biochim Biophys Acta. 1968;167(2):431–443. doi: 10.1016/0005-2744(68)90223-4. [DOI] [PubMed] [Google Scholar]
- 2.Miles EW, Bauerle R, Ahmed SA. Methods Enzymol. 1987;142:398–414. doi: 10.1016/s0076-6879(87)42051-x. [DOI] [PubMed] [Google Scholar]
- 3.Dunn MF. Arch Biochem Biophys. 2012;519(2):154–166. doi: 10.1016/j.abb.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hyde CC, Ahmed SA, Padlan EA, Miles EW, Davies DR. J Biol Chem. 1988;263(33):17857–17871. [PubMed] [Google Scholar]
- 5.Buller AR, Brinkmann-Chen S, Romney DK, Herger M, Murciano-Calles J, Arnold FH. Proc Natl Acad Sci USA. 2015;112(47):14599–14604. doi: 10.1073/pnas.1516401112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herger M, van Roye P, Romney DK, Brinkmann-Chen S, Buller AR, Arnold FH. J Am Chem Soc. 2016;138(27):8388–8391. doi: 10.1021/jacs.6b04836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zou Y, Fang Q, Yin H, Liang Z, Kong D, Bai L, Deng Z, Lin S. Angew Chem Int Ed. 2013;52(49):12951–12955. doi: 10.1002/anie.201306255. [DOI] [PubMed] [Google Scholar]
- 8.Kong D, Zou Y, Zhang Z, Xu F, Brock NL, Zhang L, Deng Z, Lin S. Sci Rep. 2016:6. doi: 10.1038/srep20273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson KS, Miles EW, Johnson KA. J Biol Chem. 1991;266(13):8020–8033. [PubMed] [Google Scholar]
- 10.Murciano-Calles J, Romney DK, Brinkmann-Chen S, Buller AR, Arnold FH. Angew Chem Int Ed. 2016;55(38):11577–11581. doi: 10.1002/anie.201606242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cornish-Bowden A. J Theor Biol. 1984;108(3):451–457. doi: 10.1016/s0022-5193(84)80045-4. [DOI] [PubMed] [Google Scholar]
- 12.Feeney R, Clarke AR, Holbrook JJ. Biochem Biophys Res Comm. 1990;166(2):667–672. doi: 10.1016/0006-291x(90)90861-g. [DOI] [PubMed] [Google Scholar]
- 13.Miles EW, McPhie P. J Biol Chem. 1974;249(9):2852–2857. [PubMed] [Google Scholar]
- 14.Hilario E, Caulkins BG, Huang YMM, You WL, Chang CEA, Mueller LJ, Dunn MF, Fan L. Biochim Biophys Acta-Prot Proteom. 2016;1864(3):268–279. doi: 10.1016/j.bbapap.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lambrecht JA, Schmitz GE, Downs DM. Mbio. 2013;4(1) doi: 10.1128/mBio.00033-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan YX, McPhie P, Miles EW. Biochemistry. 2000;39(16):4692–4703. doi: 10.1021/bi9921586. [DOI] [PubMed] [Google Scholar]
- 17.Niks D, Hilario E, Dierkers A, Ngo H, Borchardt D, Neubauer TJ, Fan L, Mueller LJ, Dunn MF. Biochemistry. 2013;52(37):6396–6411. doi: 10.1021/bi400795e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ngo H, Kimmich N, Harris R, Niks D, Blumenstein L, Kulik V, Barends TR, Schlichting I, Dunn MF. Biochemistry. 2007;46(26):7740–7753. doi: 10.1021/bi7003872. [DOI] [PubMed] [Google Scholar]
- 19.Brzovic PS, Ngo K, Dunn MF. Biochemistry. 1992;31(15):3831–3839. doi: 10.1021/bi00130a014. [DOI] [PubMed] [Google Scholar]
- 20.Kirschner K, Lane AN, Strasser AWM. Biochemistry. 1991;30(2):472–478. doi: 10.1021/bi00216a024. [DOI] [PubMed] [Google Scholar]
- 21.Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD. Nat Chem Biol. 2009;5(8):593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brinkmann-Chen S, Flock T, Cahn JKB, Snow CD, Brustad EM, McIntosh JA, Meinhold P, Zhang L, Arnold FH. Proc Natl Acad Sci USA. 2013;110(27):10946–10951. doi: 10.1073/pnas.1306073110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.