Abstract
Calpains (CAPN) are a family of Ca2+-dependent cysteine proteases that regulate various cellular functions by cleaving diverse substrates. Of the 15 mammalian calpains, CAPN8 and CAPN9 are two that are expressed predominantly in the gastrointestinal tract, where they interact to form a protease complex, termed G-calpain. However, because native G-calpain exhibits a highly restricted expression pattern, it has never been purified, and the interactions between CAPN8 and CAPN9 have not been characterized. Here, we clarified the molecular nature of G-calpain by using recombinant proteins and transgenic mice expressing FLAG-tagged CAPN8 (CAPN8-FLAG). Recombinant mouse CAPN8 and CAPN9 co-expressed in eukaryotic expression systems exhibited the same mobility as native mouse G-calpain in Blue Native-PAGE gels, and CAPN8-FLAG immunoprecipitation from stomach homogenates of the transgenic mice showed that CAPN9 was the only protein that associated with CAPN8-FLAG. These results indicated that G-calpain is a heterodimer of CAPN8 and CAPN9. In addition, active recombinant G-calpain was expressed and purified using an in vitro translation system, and the purified protease exhibited enzymatic properties that were comparable with that of calpain-2. We found that an active-site mutant of CAPN8, but not CAPN9, compromised G-calpain's substrate cleavage activity, and that the N-terminal helix region of CAPN8 and the C-terminal EF-hands of CAPN8 and CAPN9 were involved in CAPN8/9 dimerization. Furthermore, CAPN8 protein in Capn9−/− mice was almost completely lost, whereas CAPN9 was only partially lost in Capn8−/− mice. Collectively, these results demonstrated that CAPN8 and CAPN9 function as catalytic and chaperone-like subunits, respectively, in G-calpain.
Keywords: calcium, calpain, dimerization, protease, proteolysis, protease complex, dimerization, non-proteolytic function
Introduction
Calpains (Clan CA-C2, EC3.4.22.52–54) are a family of intracellular Ca2+-regulated cysteine proteases present in almost all eukaryotes and a few bacteria (1–4). Calpains modulate the structures and functions of diverse substrates, thereby regulating many biological processes, including apoptosis, cell cycling, myoblast fusion, and membrane repair; however, the underlying molecular mechanisms of these enzymes remain largely elusive (2, 5). The physiological importance of calpains has been demonstrated by reverse and forward genetics, and a number of pathological conditions including muscular dystrophies and gastropathy have been reported to be caused by aberrant calpain action (6–15).
The mammalian calpains are divided into two groups according to whether they exhibit ubiquitous or restricted expression patterns (3, 4). CAPN8 and CAPN9 belong to the latter group, and are predominantly expressed in the gastrointestinal mucous-secreting cells (i.e. pit cells in the stomach as well as goblet cells in intestines) (16–18). In contrast, the well characterized, conventional calpains, including calpain-1 and calpain-2 (also referred to as μ-calpain and m-calpain, respectively) are expressed in almost all cells. Calpain-1 and calpain-2 are heterodimers that consist of a distinct 80-kDa catalytic subunit (CAPN1 and CAPN2, respectively) and a common 28-kDa regulatory subunit (CAPNS1), which functions as a molecular chaperone for the catalytic subunits.
CAPN1 and CAPN2, as well as CAPN8 and CAPN9, consist of an N-terminal anchor helix (N), a highly conserved protease domain (CysPc,2 consisting of the protease core domains, PC1 and PC2), a calpain-type β-sandwich domain (CBSW), and a penta-EF-hand domain (PEF) (Fig. 1). In the absence of Ca2+, calpain-1 and calpain-2 are catalytically inactive, because PC1 and PC2 are far apart, preventing active-site formation (19, 20). The binding of Ca2+ to the PC1 and PC2 domains induces the active conformation (21–23). This activation process occurs concomitantly with the autolysis of the N-terminal anchor region and the Gly-rich domain (GR) of CAPNS1 (24, 25).
FIGURE 1.
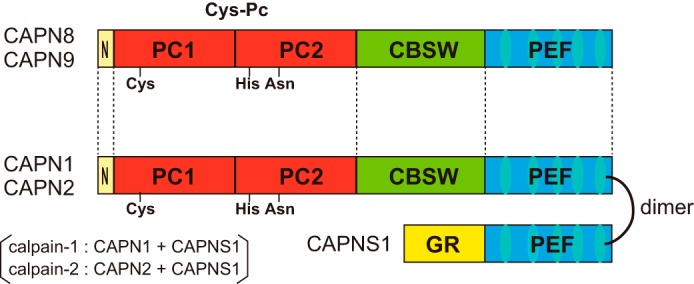
Schematic illustration of calpains. CAPN8 and CAPN9 have a domain structure typical of conventional calpains such as CAPN1 and CAPN2, consisting of an N-terminal anchor helix (N), a protease domain (CysPc) composed of protease core domains (PC1 and PC2), a calpain-type β-sandwich domain (CBSW), and a penta-EF-hand domain (PEF). CAPNS1 consists of a glycine-rich domain (GR) and a PEF domain.
It has long been believed that CAPN8 and CAPN9 are distinct proteases that are regulated independently, although they are both localized to the mucous-secreting cells in the gastrointestinal tract. Notably, recombinant CAPN8 is active when expressed alone, whereas recombinant CAPN9 requires the co-expression of CAPNS1 for its activity (26, 27).
CAPN8 and CAPN9 have been proposed to be involved in vesicle trafficking between the endoplasmic reticulum and Golgi apparatus, and in the suppression of tumorigenesis and gastric cancer, respectively (18, 28, 29). We recently demonstrated that CAPN8 and CAPN9 form an active protease complex, termed G-calpain, in which both are essential for its function, and that they play physiological roles in stress-induced gastric mucosal defense (15). By gel filtration analysis, native G-calpain was detected at a position corresponding to ∼180 kDa, suggesting that G-calpain contains one molecule each of CAPN8 and CAPN9 (79 kDa each), with possible additional small component(s) (15).
In this study, we sought to clarify the composition of G-calpain's subunits and the molecular mechanism underlying their interactions, which may shed light on how G-calpain is regulated for gastric mucosal defense. Our results showed that G-calpain is a heterodimer of CAPN8 and CAPN9 that requires the proteolytic activity of CAPN8 to cleave substrates.
Results
G-calpain Is a Heterodimer of CAPN8 and CAPN9
The purification of native G-calpain from rabbit, cow, and pig stomachs was not successful (data not shown), primarily due to its restricted expression, which is limited to the pit cells aligned on the thin surface of the gastric mucosa. In addition, antibodies recognizing CAPN8 and CAPN9 in these animals are not currently available. Thus, to determine whether G-calpain contains component(s) other than CAPN8 and CAPN9, in the 20-kDa size range, we compared the molecular weight of G-calpain in a mouse stomach homogenate to that of recombinant mouse CAPN8/9 co-expressed in COS7 cells or produced by a wheat germ in vitro translation system.
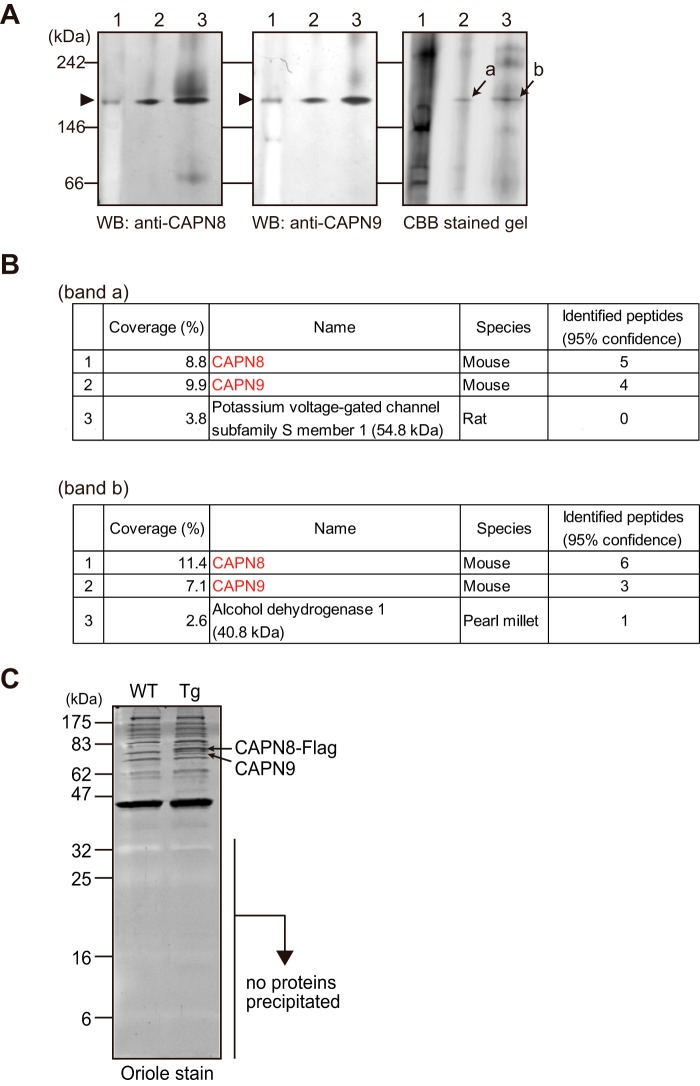
As shown in Fig. 2A, recombinant CAPN8/9 (lanes 2 and 3) was detected using Blue-Native (BN) PAGE at the same position as the native G-calpain in the stomach homogenate (lane 1) by both anti-CAPN8 and anti-CAPN9 antibodies. Coomassie Brilliant Blue staining also showed concentrated protein bands corresponding to those detected by the Western blotting analysis (arrows a and b). An in-gel tryptic digestion of these bands followed by mass spectrometry analysis revealed the presence of only two proteins, CAPN8 and CAPN9 (Fig. 2B).
FIGURE 2.
CAPN8 and CAPN9 form a heterodimer. A, native G-calpain in mouse stomach homogenate (lane 1), and recombinant mouse CAPN8 and CAPN9 co-expressed in COS7 cells (lane 2) or co-translated using a wheat germ in vitro translation system (lane 3) were detected by BN-PAGE followed by Western blotting (WB) with anti-CAPN8 (left panel) and anti-CAPN9 (middle panel) antibodies, and by CBB staining (right panel). Closed arrowheads in the left and middle panels indicate the G-calpain signal detected by anti-CAPN8 and anti-CAPN9 antibodies, respectively. Arrows a and b in the right panel indicate CBB-stained bands corresponding to the signals observed in lanes 2 and 3 of the left and middle panels, respectively. B, identification of the proteins in bands a and b from panel A by mass spectrometric analysis. Upper and lower panels show the raw data for bands a and b, respectively. C, anti-FLAG antibody immunoprecipitates of stomach homogenates prepared from WT and CAPN8-FLAG Tg mice (Tg). Immunoprecipitated proteins were subjected to SDS-PAGE followed by OrioleTM staining.
We also generated transgenic (Tg) mice that ubiquitously expressed C-terminally FLAG-tagged mouse CAPN8 (CAPN8-FLAG). In the stomach, expression level of total CAPN8 protein of the CAPN8-FLAG Tg mice was ∼2.5 times higher than that of wild-type mice (data not shown). Immunoprecipitation of the stomach homogenates prepared from the Tg mice with an anti-FLAG antibody revealed that CAPN9 co-precipitated with CAPN8-FLAG, but no additional co-precipitating proteins in the 5–30-kDa-size range were detected (Fig. 2C). Taken together, these findings indicated that G-calpain is a heterodimer of CAPN8 and CAPN9 and contains no other detectable subunits.
G-calpain Has Enzymatic Properties Similar to Those of Calpain-2
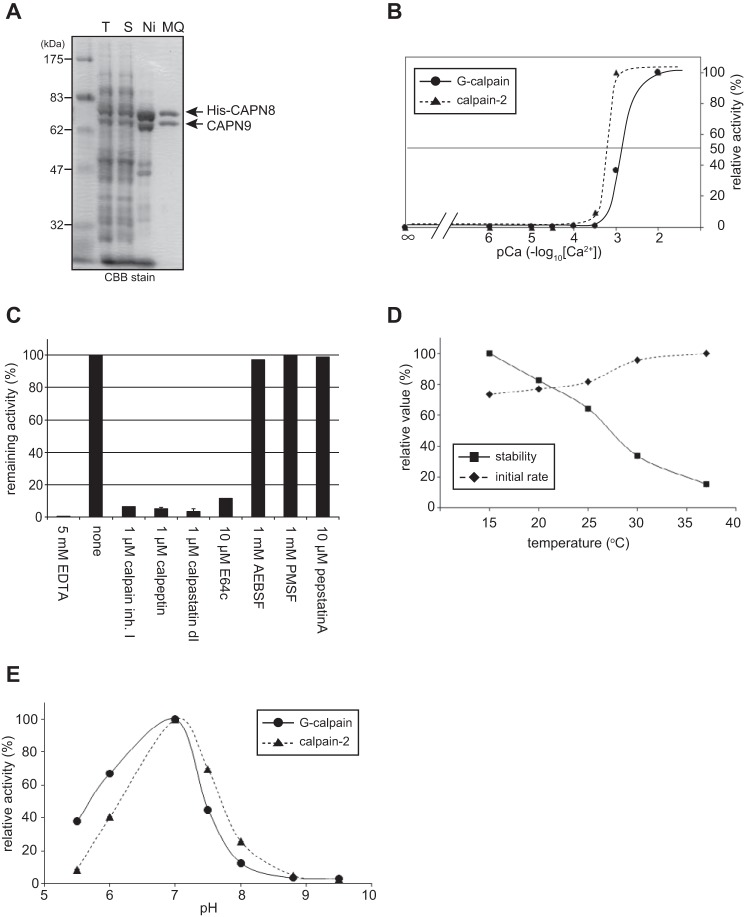
Because the co-expression of active mouse CAPN8 and CAPN9 in Escherichia coli was unsuccessful (data not shown), we used a large-scale version of the in vitro translation system described above to purify G-calpain. N-terminally His6-tagged mouse CAPN8 (His-CAPN8) and mouse CAPN9 were recovered primarily in the soluble fraction, co-eluted, and purified to homogeneity by sequential column chromatography (Fig. 3A). The purified enzyme retained more than 90% of the initial activity for at least 6 months at 4 °C.
FIGURE 3.
Purification and characterization of recombinant G-calpain. A, mouse CAPN8 and CAPN9 were co-translated using an in vitro translation system, and then purified by sequential column chromatography using Ni2+-affinity and MonoQ anion-exchange columns. Samples at each purification step were analyzed by SDS-PAGE. T, total translation reaction; S, supernatant after centrifugation of the reaction; Ni and NQ, peak fractions from the Ni2+-affinity and MonoQ columns, respectively. B, Ca2+ requirement of the Suc-LLVY-MCA hydrolyzing activity. G-calpain's activity was measured in the presence of various concentrations of Ca2+ and compared with that of calpain-2. The activities were standardized by defining the values at pCa = 2 as 100%. C, effect of inhibitors on G-calpain activity. The activities were standardized by defining the activity in the presence of 5 mm EDTA and in the absence of inhibitors as 0 and 100%, respectively. calpain inh. I, calpain inhibitor I; AEBSF, 4-(2-aminoethyl)benzenesulfonyl fluoride; calpastatin dI, recombinant human calpastatin domain 1 fragment. D, initial rate and stability of G-calpain activity at the indicated temperatures. For the initial activity rates, the activity was measured after the first minute of incubation, and then standardized by defining the value at 37 °C as 100%. To assess the activity stability, the activity was measured after a 60-min incubation, and then standardized by defining the value at 15 °C as 100%. E, pH dependence. The activities were standardized by defining the value at pH 7.0 as 100%.
Next, we compared the enzymatic properties of purified G-calpain with those of recombinant calpain-2 using a FRET-based fluorogenic substrate, succinyl-Leu-Leu-Val-Tyr-4-methyl-coumaryl-7-amide (Suc-LLVY-MCA). G-calpain exhibited a Ca2+-dependent proteolytic activity, which was effectively blocked by calpain inhibitors, such as calpastatin, calpeptin, calpain inhibitor I, and E64c, but not by inhibitors of Ser and Asp proteases (Fig. 3, B and C). These findings demonstrated that G-calpain possesses characteristics that are typical of conventional calpains.
The Ca2+ concentration required for half-maximal activity of G-calpain was ∼1 mm, which was higher than that of calpain-2 (∼0.4 mm) (Fig. 3B). G-calpain has more potential Ca2+-binding sites than calpain-2, because it contains two calpain isoforms. Although the 3D structure of G-calpain has not yet been solved, it is possible that G-calpain activation involves more dynamic movements between CAPN8 and CAPN9 that require the binding of more Ca2+ ions than are required for activating calpain-2.
The specific activity of G-calpain was ∼14% of that of calpain-2. However, the low specific activity was not the result of denaturation of the purified G-calpain, because the almost complete autolysis of G-calpain was observed (see Fig. 4B).
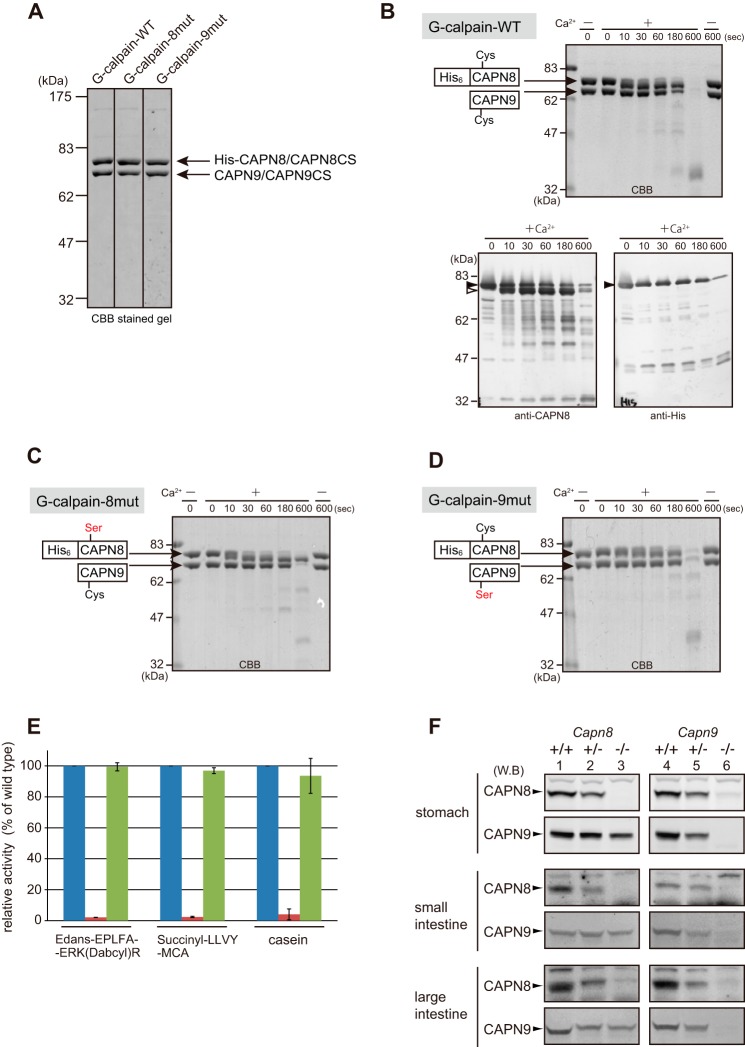
FIGURE 4.
CAPN8 and CAPN9 function as catalytic and regulatory subunits of G-calpain, respectively. A, purification of G-calpain mutants. The final fractions of G-calpain, G-calpain-8mut, and G-calpain-9mut eluted from the MonoQ column were analyzed by SDS-PAGE followed by CBB staining. B–D, SDS-PAGE analysis of the autolytic activity of G-calpain-WT (B, upper), G-calpain-8mut (C), and G-calpain-9mut (D), incubated with or without Ca2+ for 0, 10, 30, 60, and 180 s at 25 °C, and for 600 s at 37 °C. In B, SDS-PAGE was also followed by Western blotting with anti-CAPN8 (lower left panel) and anti-His antibodies (lower right panel). Closed and open arrowheads indicate full-length and N-terminally truncated CAPN8, respectively. E, the hydrolytic activities of G-calpain (blue bars), G-calpain-8mut (red bars), and G-calpain-9mut (green bars) for the indicated substrates. The activities were standardized by defining the activity of G-calpain-WT for each substrate as 100%. Error bars indicate means ± S.D. F, Western blotting (WB) analysis of stomach and intestinal tissue homogenates prepared from wild-type (lanes 1 and 4), Capn8+/− (lane 2), Capn8−/− (lane 3), Capn9+/− (lane 5), and Capn9−/− mice (lane 6) using anti-CAPN8 and anti-CAPN9 antibodies.
The initial activity (during the first 1 min of incubation) of G-calpain increased with rising temperature, whereas the activity after a 60-min incubation decreased (Fig. 3D). Although G-calpain exhibited slightly higher activity at acidic pH when compared with calpain-2, both calpains exhibited maximal activity at pH 7.0 (Fig. 3E). These results indicated that G-calpain and calpain-2 share similar enzymatic properties, regardless of their distinct subunit compositions.
CAPN8 and CAPN9 Play Distinct Roles in G-calpain
To analyze the proteolytic functions of G-calpain, two inactive mutants, G-calpain-8mut and G-calpain-9mut, in which the active-site Cys residue in His-CAPN8 and CAPN9, respectively, was mutated to Ser, were prepared in the same manner as WT G-calpain (G-calpain-WT). The yields and purity of the mutants were very similar to those of G-calpain-WT (Fig. 4A).
First, the autolytic process of G-calpain was analyzed using the mutants described above. The N terminus of CAPN8 underwent rapid proteolysis in the presence of Ca2+ (Fig. 4, B–D), similar to the activation process of conventional calpains (25). After a 10-min incubation, both subunits of G-calpain-WT and G-calpain-9mut, as well as CAPN9 in G-calpain-8mut, were almost completely degraded, whereas CAPN8 in G-calpain-8mut was only partially degraded. These results indicated that CAPN8 promotes autolysis and the proteolytic cleavage of CAPN9 to a similar extent, whereas CAPN9 exhibits autolytic activity to a greater extent than CAPN8-degrading activity.
Next, the proteolytic activity of G-calpain and its mutants was examined using Suc-LLVY-MCA, another FRET-based fluorogenic substrate, 5-[(2-aminoethyl) amino]naphthalene-1-sulfonic acid-Glu-Pro-Leu-Phe-Ala-Glu-Arg-Lys-4-((4-(diethylamino)phenyl)azo)benzoic acid-Arg (Edans-EPLFAERK-(Dabcyl)-R) (30), and casein as substrates. G-calpain-9mut exhibited almost identical levels of proteolytic activity against each of these substrates. In contrast, G-calpain-8mut exhibited barely detectable proteolytic activity against the same substrates (Fig. 4E). These data indicated that the substrate-proteolyzing activity of G-calpain is primarily due to the activity of CAPN8.
The results of our previous study suggested that CAPN8 and CAPN9 also function as chaperones for G-calpain (15). Thus, the stability of each of the G-calpain subunits was assessed in the absence, or in the presence of reduced levels, of the other subunit, using stomach and intestinal tissue homogenates prepared from WT, Capn8+/−, Capn8−/−, Capn9+/−, and Capn9−/− mice. As shown in Fig. 4F, CAPN8 was barely detectable in Capn9−/− mice (lane 6), as observed for CAPN2 in Capns1−/− mouse embryos (9). In contrast, CAPN9 down-regulation in Capn8−/− mice was moderate (compare lanes 3 and 6 in the CAPN9 rows). These observations suggested that the contribution of CAPN9 to G-calpain's stability is much greater than that of CAPN8.
The Interaction of CAPN8 and CAPN9 C-terminal EF5 Domains Is Essential for Dimerization
To investigate the mechanism underlying CAPN8/9 dimerization, several deletion mutants of each were prepared and analyzed for their binding abilities. To exclude possible proteolytic effects, protease-inactive CAPN8-C105S and CAPN9-C97S were used to construct the mutants (Fig. 5A). The series of deletion mutants was transiently expressed in COS7 cells in various combinations, and the soluble lysates were subjected to BN-PAGE and Western blotting analyses. As described above, the co-expression of CAPN8-FL and CAPN9-FL gave rise to a single 156-kDa band detected by both anti-CAPN8 and CAPN9 antibodies (Fig. 5B, lane 2 in the BN-PAGE panels).
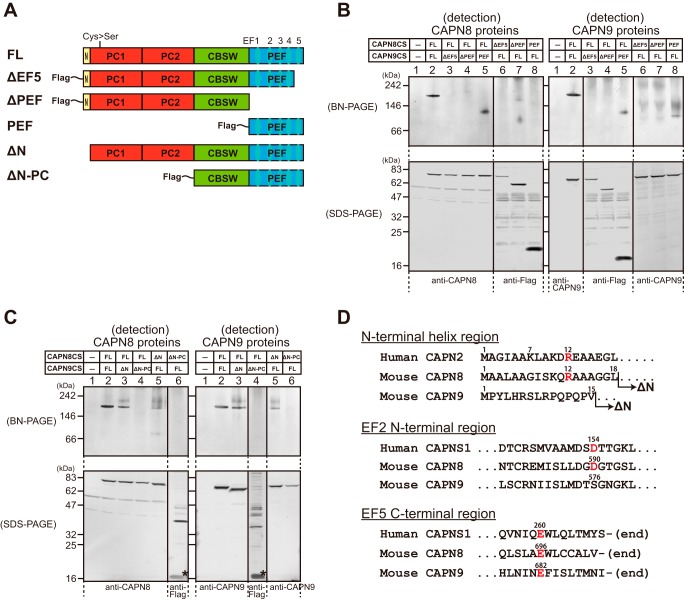
FIGURE 5.
Identification of domains involved in CAPN8/9 dimerization. A, schematic illustration of full-length CAPN8CS or CAPN9CS and their deletion mutants. ΔEF5, ΔPEF, PEF, ΔN, and ΔN-PC mutants lack EF5, PEF, all domains except for PEF, N, and N and PC domains, respectively. CBSW, calpain-type β-sandwich domain. B and C, the expression constructs shown in A were co-transfected into COS7 cells in the indicated combinations. The cell lysates (7 μg each) were subjected to BN-PAGE (upper panels) and SDS-PAGE (lower panels) followed by Western blotting with anti-CAPN8, anti-CAPN9, and anti-FLAG antibodies. Asterisks in C indicate degraded products of the ΔN-PC mutants. D, amino acid sequence alignments of the human CAPN2 and mouse CAPN8 and CAPN9 N-terminal anchor helix regions (upper), and of the human CAPNS1 and mouse CAPN8 and CAPN9 EF2 N-terminal regions (middle) and EF5 C-terminal regions (lower). Numbers above the sequences represent amino acid positions. Human CAPN2 Arg-12 and human CAPNS1 Asp-154 and Glu-260, and their corresponding residues in mouse CAPN8 and CAPN9, are shown in red.
As in the case of the conventional calpains (19, 20), deletion of the EF5 domain from either CAPN8 or CAPN9 (ΔEF5) resulted in the disappearance of the 156-kDa band (lanes 3 and 6). Conversely, the association of CAPN8-FL with CAPN9-PEF was observed (lane 5), whereas the association of CAPN9-FL with CAPN8-PEF was barely detected (lane 8). These findings indicated that the EF5 domains of CAPN8 and CAPN9 are necessary for subunit association like most of PEF family proteins (31, 32), and that although CAPN9-PEF can bind stably to CAPN8, the CAPN8-PEF is not sufficient to promote stable dimerization with CAPN9. Thus, the presence of additional interaction(s) between CAPN8 and CAPN9 may be necessary for dimerization.
The N Terminus of CAPN8 Plays an Important Role in Dimerization
The 3D structure of inactive calpain-2 shows that the N-terminal anchor helix of CAPN2 interacts with the PEF domain of CAPNS1, stabilizing the dimer formation (20, 25). Consistent with this finding, we found that the N-terminal deletion (ΔN) of CAPN8 prevented its dimerization with CAPN9-FL (Fig. 5C, lane 5). In contrast, the N-terminal deletion of CAPN9 had only a marginal effect on the interaction with CAPN8 (lane 3).
In addition, a larger deletion, encompassing both the N terminus and the CysPc domain (ΔN-PC), in either CAPN8 or CAPN9 not only completely abolished dimer formation (Fig. 5C, lanes 4 and 6), but also resulted in degradation of the truncated mutants (asterisks in lanes 6 and 4 of the left and right side in the SDS-PAGE panels, respectively). This finding suggested that the ΔN-PC mutants were unstable and were probably degraded by endogenous protease(s).
In calpain-2, Lys-7 and Arg-12 of CAPN2 make salt bridges with Asp-154 of EF2 and Glu-260 of EF5, respectively, in CAPNS1 (20, 25). Although the residues corresponding to Lys-7 and Asp-154 are not conserved in G-calpain, Arg-12 and Glu-260 correspond to Arg-12 of CAPN8 and Glu-682 of CAPN9, respectively (Fig. 5D). However, CAPN8-R12L and CAPN9-E682A were capable of forming WT-like dimers (data not shown). Therefore, although the N-terminal 18 residues of CAPN8 were found to be essential for its interaction with CAPN9, the specific residues required for dimerization were not identified.
Discussion
The current study is the first to our knowledge to describe the dimerization of distinct calpain catalytic isoforms. Using a combination of biochemical and genetic approaches, we have uncovered several mechanisms involved in the formation and stabilization of the G-calpain complex.
Unexpectedly, CAPN9 was found to play a role in CAPN8 stabilization with autolytic, but little proteolytic, activity. A previous analysis of the 3D structure of the CysPc domain of CAPN9 revealed that it is structurally less competent for catalysis than that of CAPN1 or CAPN2, despite the strong conservation of this domain in CAPN9 (33). In a separate study, recombinant CAPN9 was suggested to form an active heterodimer when co-expressed with CAPNS1 (26). The interaction between CAPN8 and CAPN9 may suppress the proteolytic activity of CAPN9 in G-calpain.
Non-proteolytic functions have been reported for CAPN3, which exhibits a proteolytic activity, and for CAPN6, which naturally lacks a proteolytic activity due to an amino acid substitution at the active-site Cys (to Lys) (34–37). Thus, CAPN9 is unique in that it exhibits little proteolytic activity despite the conserved active-site Cys, and functions to regulate the stability of another calpain isoform.
Notably, the activity of CAPN9 appears to be dispensable for G-calpain, because G-calpain-9mut exhibited a proteolytic activity that was comparable with that of G-calpain-WT (Fig. 4E). In contrast, our previous study showed that mice expressing active-site-inactive CAPN8 (corresponding to G-calpain-8mut) exhibited a stress-induced gastric injury that was as severe as the same injury observed in Capn8−/− and Capn9−/− mice (15), indicating the indispensability of the proteolytic activity of CAPN8. Further confirmation of the contrasting roles of CAPN8 and CAPN9 will require a phenotypic analysis of mice expressing catalytically inactive CAPN9.
Unlike calpain-2, G-calpain remained relatively active under acidic conditions. In mammalian cells, intracellular compartments such as the Golgi, lysosome, and endosome are acidic (38). We previously showed that recombinant CAPN8 expressed in cultured cells preferentially localizes to the Golgi and cleaves β-COP, a component of the COP-I vesicle involved in cargo transport between the Golgi and endoplasmic reticulum (18). G-calpain's proteolytic activity at lower pH is consistent with its localization and functions in acidic subcellular compartments.
The difference in specific activities between G-calpain and calpain-2 may reflect a difference in substrate preference/recognition. Indeed, CAPN3Δ (p94Δ), a CAPN3 splicing variant, exhibits a lower specific activity for such in vitro substrates with other unusual enzymatic characteristics, and limitedly cleaves calpastatin (39). The identification of substrates for G-calpain, including the candidate substrate β-COP, will contribute to our understanding of the physiological roles of G-calpain.
How do the N-terminal regions and EF5 domains of CAPN8 and CAPN9 contribute to their dimerization? Recent molecular modeling studies suggest that CAPN3, which is structurally similar to CAPN8 and CAPN9 except for three short insertions (NS, IS1, and IS2), has the potential to form a homodimer (40, 41). According to the model, the two CAPN3 molecules are aligned in opposite directions and dimerize through their PEF domains, and the N-terminal NS region is very close to the PEF domain. This model was based on the solved 3D structures of full-length CAPN2 and of the homodimeric CAPN3 PEF domains, and has the potential to be applied to all of the calpain catalytic isoforms that have the same domain structure.
The 3D structure of the full-length G-calpain is required to fully elucidate the interactions between CAPN8 and CAPN9. Our lab is currently preparing large amounts of recombinant G-calpain for use in solving its crystal structure.
Experimental Procedures
Antibodies and Protease Inhibitors
The anti-CAPN8 domain III polyclonal antibody (ab28215), anti-CAPN9 (V-18) polyclonal antibody, anti-FLAG monoclonal antibody (clone M2), and anti-His monoclonal antibody were purchased from Abcam, Santa Cruz Biotechnology, Sigma, and Novagen, respectively. The protease inhibitors calpeptin, calpain inhibitor I, the recombinant human calpastatin domain 1 fragment, E64c, PMSF, 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF), and pepstatin A, as well as general chemical reagents, were purchased from Merck Millipore, TaKaRa, Peptide Institute, Sigma, and Kanto Chemical.
Cloning and Construction of Expression Plasmids
Mouse Capn8 and Capn9 cDNAs corresponding to the full-length proteins (aa 1–703 and aa 1–690, respectively), as well as deletion mutants including ΔEF5 (aa 1–672 and aa 1–658), ΔPEF (aa 1–532 and aa 1–518), PEF (aa 533–703 and aa 519–690), ΔN (aa 19–703 and aa 16–690), and ΔN-PC (aa 356–703 and aa 519–690), were generated by PCR using Phusion High Fidelity DNA polymerase (New England Biolabs). For in vitro translation, the amplified cDNAs were ligated into pEU and pEU-His vectors (CellFree Sciences) to produce CAPN9 and N-terminally His-tagged CAPN8 proteins, respectively. For expression in COS7 cells, the amplified cDNAs were ligated into a modified pSRD vector (42) to produce CAPN8 and CAPN9 proteins with or without an N-terminal FLAG tag. Expression plasmids for CAPN8CS, CAPN9CS, CAPN8CS-R12L, and CAPN9CS-E682A were constructed by PCR-mediated site-directed mutagenesis as described previously (43).
Expression and Purification of G-calpain and Its Mutants, and of Calpain-2
For the expression and purification of mouse G-calpain and its mutants, the pEU and pEU-His constructs were co-transcribed and co-translated using the WEPRO7240 wheat germ cell-free expression kit (CellFree Sciences) according to the manufacturer's instructions. The purification was performed at 4 °C as follows. The translation mixture was ultracentrifuged at 50,000 × g for 20 min, and the supernatant was recovered and filtered through a 0.22-μm-pore filter. The supernatant was applied to a Ni2+-affinity column packed with 1 ml of cOmplete His tag purification resin (Roche Diagnostics) equilibrated with buffer A (20 mm Tris-HCl (pH 7.4), 300 mm NaCl, 1 mm DTT, 0.25 mm EGTA). The column was washed with 10 column volumes of buffer A containing 4 mm imidazole, and the proteins were eluted in a stepwise manner using 6 column volumes each of 40 mm imidazole in buffer A and 200 mm imidazole in buffer A, into collection tubes containing EDTA at a final concentration of 1 mm. The peak fractions were collected, dialyzed against TED buffer (20 mm Tris-HCl (pH 7.4), 1 mm EDTA, 1 mm DTT) overnight, and centrifuged at 20,000 × g for 20 min to remove debris. The dialyzed sample was applied to a MonoQ HR 10/10 anion-exchange column (GE Healthcare), equilibrated with TED buffer. The column was washed with 3 column volumes of TED buffer, and the G-calpain protein was eluted with a linear gradient of 0–0.5 mm NaCl in TED buffer, in 5 column volumes. The purity of the protein was verified by SDS-PAGE. Human calpain-2 was prepared as described previously (44). The peak fractions of both proteins were stored at 4 °C until use.
Protein Expression in COS7 Cells
The pSRD constructs were transfected into COS7 cells using Lipofectamine LTX (Life Technologies) according to the manufacturer's instructions. The cells were harvested 24 h after transfection, suspended in TED buffer containing 40 μm pepstatin A, 0.3 mm PMSF, and 50 μm leupeptin, sonicated by an ultrasonic disruptor (TOMY, UD-201), and centrifuged at 20,000 × g for 20 min. The supernatant was recovered, and then subjected to BN-PAGE or SDS-PAGE followed by Western blotting analysis.
Experimental Animals
All procedures using experimental animals were approved by the Animal Use and Care Committee of the Tokyo Metropolitan Institute of Medical Science, and the animals were treated according to the committee's guidelines. All mice were housed in specific pathogen-free facilities at our institute. CAPN8-FLAG Tg mice were generated as follows. The cDNA corresponding to mouse CAPN8 fused with a C-terminal FLAG tag was amplified by PCR using Phusion High-Fidelity DNA Polymerase, and then ligated into the pCALNL5 vector (45–47). The resultant plasmid, in which the Capn8-FLAG fragment was followed by a neomycin cassette (neor) flanked by loxP sequences at both ends, was cleaved with SalI and SfiI, releasing a 5.6-kbp fragment containing the CAG promoter-loxP-neor-loxP-Capn8-FLAG transgene fragment. The fragment was microinjected into 0.5-day-old C57BL/6 mouse embryos (ARK Resource) using a micromanipulator, and the embryos were transferred to the oviduct of pseudopregnant ICR females. The founder mice were mated with C57BL/6 mice to confirm germline transmission, and the germline transmitted mice were crossed with EIIa-Cre Tg mice, in which Cre recombinase is expressed from the zygote stage, to remove the neomycin cassette from the Capn8-FLAG transgene. The resulting CAPN8-FLAG Tg mice were inbred with C57BL/6 mice.
In-gel Digestion and Mass Spectrometric Analysis
The CBB-stained gel bands were excised, cut into pieces, and destained in 100 mm NH4HCO3 and 60% acetonitrile. The gel pieces were then dried in a vacuum centrifuge and treated with 10 μg/ml trypsin in 100 mm NH4HCO3 for 16 h at 37 °C. The resulting peptides were extracted from the gels in 60% acetonitrile, desalted using a ZipTip C18 column (Millipore), and analyzed with a 4800 MALDI TOF/TOFTM analyzer (SCIEX). Candidate proteins were identified by using ProteinPilot software (SCIEX).
Proteolytic Assays
Proteolytic activity was measured using the FRET-based fluorogenic substrates, Suc-LLVY-MCA and Edans-EPLFAERK-(Dabcyl)-R (30), or casein. To analyze the proteolysis of fluorogenic substrates, recombinant G-calpain or its mutants (2 μg each) or calpain-2 (0.3 μg) was incubated with 0.1 mm Suc-LLVY-MCA or 0.025 mm Edans-EPLFAERK-(Dabcyl)-R in reaction buffer (0.1 m Tris-HCl, 20 mm 2-mercaptoethanol, and 0.2% CHAPS) with varying Ca2+ concentrations, temperatures, or pH in a total volume of 40 μl. For pH values less than 7.0, Tris acetate buffer was used instead of Tris-HCl buffer. The reactions were stopped with 40 μl of 10% SDS and 1.2 ml of 0.1 m Tris-HCl (pH 9.5). Fluorescence generated by the release of MCA or Dabcyl was monitored by a spectrofluorophotometer (JASCO FP-8300), with excitation and emission wavelengths of 380 and 460 nm, respectively, for Suc-LLVY-MCA, and of 340 and 490 nm, respectively, for Edans-EPLFAERK-(Dabcyl)-R. To analyze the proteolysis of casein, recombinant G-calpain or its mutants (2 μg each) was incubated at 20 °C for 20 min, in reaction buffer (100 mm Tris-Cl (pH 7.5), 3 mg/ml casein, and 20 mm 2-mercaptoethanol) with 5 mm CaCl2 or 5 mm EDTA, in a total volume of 50 μl. After incubation, the reactions were stopped by adding 150 μl of 7% (v/v) trichloroacetic acid, incubated on ice for 30 min, and centrifuged at 15,000 rpm for 15 min at 4 °C. The A280 of the supernatant was measured using a spectrophotometer (SmartSpec-3000, Bio-Rad Laboratories). Calpain proteolytic activity was defined as a Ca2+-dependent increase in fluorescence or A280 values. To measure autolysis, 2 μg of recombinant G-calpain or its mutants was incubated in reaction buffer (20 mm Tris-HCl (pH 7.5), 1 mm DTT) with 5 mm EDTA or CaCl2 for 0, 10, 30, 60, or 180 s at room temperature (25 °C), and for 600 s at 37 °C.
Immunoprecipitations
Mouse gastric mucosa was scraped from the stomach of wild-type or CAPN8-FLAG Tg mice, and then homogenized in homogenization buffer (20 mm Tris-HCl (pH 7.5), 5 mm EDTA, 150 mm NaCl, 0.3 mm PMSF, 40 μm pepstatin A, and 50 μm leupeptin). Nonidet P-40 was added to 1.5 mg of the homogenate at a final concentration of 0.5%, and the samples were incubated with anti-FLAG M2-agarose (Sigma) for 2 h at 4 °C. After washing the anti-FLAG-agarose five times with 1 ml of homogenization buffer containing 0.5% Nonidet P-40, the bound proteins were eluted with 0.2 m glycine (pH 2.7), and the eluate was neutralized with Tris-HCl (pH 9.5), mixed with SDS-sample buffer, subjected to SDS-PAGE, and stained with OrioleTM fluorescent gel stain (Bio-Rad), a UV-based fluorescence imaging system that is as sensitive for protein visualization as silver staining.
Author Contributions
S. H. designed, performed, and analyzed the experiments, and wrote the paper. F. K., M. Y., H. Shitara, and M. M. provided technical assistance and contributed to the preparation of the figures. H. Sorimachi coordinated the study and wrote the paper. All of the authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank all of our laboratory members for valuable discussions, Dr. Choji Taya at the Laboratory of Transgenic Technology, Center for Basic Technology Research, Tokyo Metropolitan Institute of Medical Science for generating and maintaining CAPN8-FLAG Tg mice, and Dr. Leslie Miglietta and Dr. Karen Berg for critical reading of the manuscript.
This work was supported in part by Japan Society for the Promotion of Science (JSPS) KAKENHI Grants JP26450172 and JP23780152 (to S. H.) and JP15H02389 and JP23247021 (to H. Sorimachi), a Kato Memorial Bioscience Foundation research grant (to S. H.), a SUNTORY research grant (to S. H.), Open Partnership Joint Projects of the JSPS Bilateral Joint Research Projects (to H. Sorimachi), a Takeda Science Foundation research grant (to H. Sorimachi), the Council for Science, Technology and Innovation (CSTI), the Cross-ministerial Strategic Innovation Promotion Program (SIP), “Technologies for creating next-generation agriculture, forestry and fisheries” (funding agency: the Bio-oriented Technology Research Advancement Institution, National Agriculture and Food Research Organization (NARO)) (to H. Sorimachi), and a research grant of The Naito Foundation (to H. Sorimachi). The authors declare that they have no conflicts of interest with the contents of this article.
- CysPc
- calpain-type Cys protease conserved domain
- PC
- protease core domain
- PEF
- penta-EF-hand domain
- NS
- N-terminal sequence
- IS
- internal sequence
- BN-PAGE
- Blue-Native PAGE
- Tg
- transgenic
- CBB
- Coomassie Brilliant Blue
- Suc
- succinyl
- MCA
- 4-methyl-coumaryl-7-amide
- Edans
- 5-[(2-aminoethyl) amino]naphthalene-1-sulfonic acid
- (Dabcyl)-R
- 4-((4-(diethylamino)phenyl)azo)benzoic acid-Arg
- Cre
- causes recombination
- aa
- amino acids
- FL
- full-length.
References
- 1. Huang Y., and Wang K. K. (2001) The calpain family and human disease. Trends Mol. Med. 7, 355–362 [DOI] [PubMed] [Google Scholar]
- 2. Goll D. E., Thompson V. F., Li H., Wei W., and Cong J. (2003) The calpain system. Physiol. Rev. 83, 731–801 [DOI] [PubMed] [Google Scholar]
- 3. Sorimachi H., Hata S., and Ono Y. (2011) Impact of genetic insights into calpain biology. J. Biochem. 150, 23–37 [DOI] [PubMed] [Google Scholar]
- 4. Campbell R. L., and Davies P. L. (2012) Structure-function relationships in calpains. Biochem. J. 447, 335–351 [DOI] [PubMed] [Google Scholar]
- 5. Ono Y., Saido T. C., and Sorimachi H. (2016) Calpain research for drug discovery: challenges and potential. Nat. Rev. Drug Discov. 15, 854–876 [DOI] [PubMed] [Google Scholar]
- 6. Saito K., Elce J. S., Hamos J. E., and Nixon R. A. (1993) Widespread activation of calcium-activated neutral proteinase (calpain) in the brain in Alzheimer disease: a potential molecular basis for neuronal degeneration. Proc. Natl. Acad. Sci. U.S.A. 90, 2628–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Richard I., Broux O., Allamand V., Fougerousse F., Chiannilkulchai N., Bourg N., Brenguier L., Devaud C., Pasturaud P., Roudaut C., et al. (1995) Mutations in the proteolytic enzyme calpain 3 cause limb-girdle muscular dystrophy type 2A. Cell 81, 27–40 [DOI] [PubMed] [Google Scholar]
- 8. Horikawa Y., Oda N., Cox N. J., Li X., Orho-Melander M., Hara M., Hinokio Y., Lindner T. H., Mashima H., Schwarz P. E., del Bosque-Plata L., Horikawa Y., Oda Y., Yoshiuchi I., Colilla S., et al. (2000) Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat. Genet. 26, 163–175 [DOI] [PubMed] [Google Scholar]
- 9. Arthur J. S., Elce J. S., Hegadorn C., Williams K., and Greer P. A. (2000) Disruption of the murine calpain small subunit gene, Capn4: calpain is essential for embryonic development but not for cell growth and division. Mol. Cell Biol. 20, 4474–4481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Azam M., Andrabi S. S., Sahr K. E., Kamath L., Kuliopulos A., and Chishti A. H. (2001) Disruption of the mouse μ-calpain gene reveals an essential role in platelet function. Mol. Cell Biol. 21, 2213–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Syntichaki P., Xu K., Driscoll M., and Tavernarakis N. (2002) Specific aspartyl and calpain proteases are required for neurodegeneration in C. elegans. Nature 419, 939–944 [DOI] [PubMed] [Google Scholar]
- 12. Dutt P., Croall D. E., Arthur J. S., Veyra T. D., Williams K., Elce J. S., and Greer P. A. (2006) m-Calpain is required for preimplantation embryonic development in mice. BMC Dev. Biol. 6, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Russo I., Oksman A., Vaupel B., and Goldberg D. E. (2009) A calpain unique to alveolates is essential in Plasmodium falciparum and its knockdown reveals an involvement in pre-S-phase development. Proc. Natl. Acad. Sci. U.S.A. 106, 1554–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yamada M., Yoshida Y., Mori D., Takitoh T., Kengaku M., Umeshima H., Takao K., Miyakawa T., Sato M., Sorimachi H., Wynshaw-Boris A., and Hirotsune S. (2009) Inhibition of calpain increases LIS1 expression and partially rescues in vivo phenotypes in a mouse model of lissencephaly. Nat. Med. 15, 1202–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hata S., Abe M., Suzuki H., Kitamura F., Toyama-Sorimachi N., Abe K., Sakimura K., and Sorimachi H. (2010) Calpain 8/nCL-2 and calpain 9/nCL-4 constitute an active protease complex, G-calpain, involved in gastric mucosal defense. PLoS Genet. 6, e1001040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sorimachi H., Ishiura S., and Suzuki K. (1993) A novel tissue-specific calpain species expressed predominantly in the stomach comprises two alternative splicing products with and without Ca2+-binding domain. J. Biol. Chem. 268, 19476–19482 [PubMed] [Google Scholar]
- 17. Lee H. J., Sorimachi H., Jeong S. Y., Ishiura S., and Suzuki K. (1998) Molecular cloning and characterization of a novel tissue-specific calpain predominantly expressed in the digestive tract. Biol. Chem. 379, 175–183 [DOI] [PubMed] [Google Scholar]
- 18. Hata S., Koyama S., Kawahara H., Doi N., Maeda T., Toyama-Sorimachi N., Abe K., Suzuki K., and Sorimachi H. (2006) Stomach-specific calpain, nCL-2, localizes in mucus cells and proteolyzes the β-subunit of coatomer complex, β-COP. J. Biol. Chem. 281, 11214–11224 [DOI] [PubMed] [Google Scholar]
- 19. Hosfield C. M., Elce J. S., Davies P. L., and Jia Z. (1999) Crystal structure of calpain reveals the structural basis for Ca2+-dependent protease activity and a novel mode of enzyme activation. EMBO J. 18, 6880–6889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Strobl S., Fernandez-Catalan C., Braun M., Huber R., Masumoto H., Nakagawa K., Irie A., Sorimachi H., Bourenkow G., Bartunik H., Suzuki K., and Bode W. (2000) The crystal structure of calcium-free human m-calpain suggests an electrostatic switch mechanism for activation by calcium. Proc. Natl. Acad. Sci. U.S.A. 97, 588–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hata S., Sorimachi H., Nakagawa K., Maeda T., Abe K., and Suzuki K. (2001) Domain II of m-calpain is a Ca2+-dependent cysteine protease. FEBS Lett. 501, 111–114 [DOI] [PubMed] [Google Scholar]
- 22. Moldoveanu T., Hosfield C. M., Lim D., Elce J. S., Jia Z., and Davies P. L. (2002) A Ca2+ switch aligns the active site of calpain. Cell 108, 649–660 [DOI] [PubMed] [Google Scholar]
- 23. Hanna R. A., Campbell R. L., and Davies P. L. (2008) Calcium-bound structure of calpain and its mechanism of inhibition by calpastatin. Nature 456, 409–412 [DOI] [PubMed] [Google Scholar]
- 24. Chou J. S., Impens F., Gevaert K., and Davies P. L. (2011) m-Calpain activation in vitro does not require autolysis or subunit dissociation. Biochim. Biophys. Acta 1814, 864–872 [DOI] [PubMed] [Google Scholar]
- 25. Nakagawa K., Masumoto H., Sorimachi H., and Suzuki K. (2001) Dissociation of m-calpain subunits occurs after autolysis of the N-terminus of the catalytic subunit, and is not required for activation. J. Biochem. 130, 605–611 [DOI] [PubMed] [Google Scholar]
- 26. Lee H. J., Tomioka S., Kinbara K., Masumoto H., Jeong S. Y., Sorimachi H., Ishiura S., and Suzuki K. (1999) Characterization of a human digestive tract-specific calpain, nCL-4, expressed in the baculovirus system. Arch. Biochem. Biophys. 362, 22–31 [DOI] [PubMed] [Google Scholar]
- 27. Hata S., Doi N., Kitamura F., and Sorimachi H. (2007) Stomach-specific calpain, nCL-2/calpain 8, is active without calpain regulatory subunit and oligomerizes through C2-like domains. J. Biol. Chem. 282, 27847–27856 [DOI] [PubMed] [Google Scholar]
- 28. Liu K., Li L., and Cohen S. N. (2000) Antisense RNA-mediated deficiency of the calpain protease, nCL-4, in NIH3T3 cells is associated with neoplastic transformation and tumorigenesis. J. Biol. Chem. 275, 31093–31098 [DOI] [PubMed] [Google Scholar]
- 29. Yoshikawa Y., Mukai H., Hino F., Asada K., and Kato I. (2000) Isolation of two novel genes, down-regulated in gastric cancer. Jpn. J. Cancer Res. 91, 459–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cuerrier D., Moldoveanu T., and Davies P. L. (2005) Determination of peptide substrate specificity for μ-calpain by a peptide library-based approach: the importance of primed side interactions. J. Biol. Chem. 280, 40632–40641 [DOI] [PubMed] [Google Scholar]
- 31. Maki M., Kitaura Y., Satoh H., Ohkouchi S., and Shibata H. (2002) Structures, functions and molecular evolution of the penta-EF-hand Ca2+-binding proteins. Biochim. Biophys. Acta 1600, 51–60 [DOI] [PubMed] [Google Scholar]
- 32. Kitaura Y., Satoh H., Takahashi H., Shibata H., and Maki M. (2002) Both ALG-2 and peflin, penta-EF-hand (PEF) proteins, are stabilized by dimerization through their fifth EF-hand regions. Arch. Biochem. Biophys. 399, 12–18 [DOI] [PubMed] [Google Scholar]
- 33. Davis T. L., Walker J. R., Finerty P. J. Jr, Mackenzie F., Newman E. M., and Dhe-Paganon S. (2007) The crystal structures of human calpains 1 and 9 imply diverse mechanisms of action and auto-inhibition. J. Mol. Biol. 366, 216–229 [DOI] [PubMed] [Google Scholar]
- 34. Tonami K., Kurihara Y., Aburatani H., Uchijima Y., Asano T., and Kurihara H. (2007) Calpain 6 is involved in microtubule stabilization and cytoskeletal organization. Mol. Cell Biol. 27, 2548–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ojima K., Ono Y., Ottenheijm C., Hata S., Suzuki H., Granzier H., and Sorimachi H. (2011) Non-proteolytic functions of calpain-3 in sarcoplasmic reticulum in skeletal muscles. J. Mol. Biol. 407, 439–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tonami K., Kurihara Y., Arima S., Nishiyama K., Uchijima Y., Asano T., Sorimachi H., and Kurihara H. (2011) Calpain-6, a microtubule-stabilizing protein, regulates Rac1 activity and cell motility through interaction with GEF-H1. J. Cell Sci. 124, 1214–1223 [DOI] [PubMed] [Google Scholar]
- 37. Tonami K., Hata S., Ojima K., Ono Y., Kurihara Y., Amano T., Sato T., Kawamura Y., Kurihara H., and Sorimachi H. (2013) Calpain-6 deficiency promotes skeletal muscle development and regeneration. PLoS Genet. 9, e1003668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Weisz O. A. (2003) Acidification and protein traffic. Int. Rev. Cytol. 226, 259–319 [DOI] [PubMed] [Google Scholar]
- 39. Ono Y., Kakinuma K., Torii F., Irie A., Nakagawa K., Labeit S., Abe K., Suzuki K., and Sorimachi H. (2004) Possible regulation of the conventional calpain system by skeletal muscle-specific calpain, p94/calpain 3. J. Biol. Chem. 279, 2761–2771 [DOI] [PubMed] [Google Scholar]
- 40. Ravulapalli R., Campbell R. L., Gauthier S. Y., Dhe-Paganon S., and Davies P. L. (2009) Distinguishing between calpain heterodimerization and homodimerization. FEBS J. 276, 973–982 [DOI] [PubMed] [Google Scholar]
- 41. Partha S. K., Ravulapalli R., Allingham J. S., Campbell R. L., and Davies P. L. (2014) Crystal structure of calpain-3 penta-EF-hand (PEF) domain: a homodimerized PEF family member with calcium bound at the fifth EF-hand. FEBS J. 281, 3138–3149 [DOI] [PubMed] [Google Scholar]
- 42. Sorimachi H., Toyama-Sorimachi N., Saido T. C., Kawasaki H., Sugita H., Miyasaka M., Arahata K., Ishiura S., and Suzuki K. (1993) Muscle-specific calpain, p94, is degraded by autolysis immediately after translation, resulting in disappearance from muscle. J. Biol. Chem. 268, 10593–10605 [PubMed] [Google Scholar]
- 43. Ono Y., Shimada H., Sorimachi H., Richard I., Saido T. C., Beckmann J. S., Ishiura S., and Suzuki K. (1998) Functional defects of a muscle-specific calpain, p94, caused by mutations associated with limb-girdle muscular dystrophy type 2A. J. Biol. Chem. 273, 17073–17078 [DOI] [PubMed] [Google Scholar]
- 44. Hata S., Ueno M., Kitamura F., and Sorimachi H. (2012) Efficient expression and purification of recombinant human m-calpain using an Escherichia coli expression system at low temperature. J. Biochem. 151, 417–422 [DOI] [PubMed] [Google Scholar]
- 45. Niwa H., Yamamura K., and Miyazaki J. (1991) Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 108, 193–199 [DOI] [PubMed] [Google Scholar]
- 46. Kanegae Y., Lee G., Sato Y., Tanaka M., Nakai M., Sakaki T., Sugano S., and Saito I. (1995) Efficient gene activation in mammalian cells by using recombinant adenovirus expressing site-specific Cre recombinase. Nucleic Acids Res. 23, 3816–3821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kanegae Y., Takamori K., Sato Y., Lee G., Nakai M., and Saito I. (1996) Efficient gene activation system on mammalian cell chromosomes using recombinant adenovirus producing Cre recombinase. Gene. 181, 207–212 [DOI] [PubMed] [Google Scholar]