ABSTRACT
The survival of all organisms is dependent on complex, coordinated responses to environmental cues. Non-coding RNAs have been identified as major players in regulation of gene expression, with recent evidence supporting roles for long non-coding (lnc)RNAs in both transcriptional and post-transcriptional control. Evidence from our laboratory shows that lncRNAs have the ability to form hybridized structures called R-loops with specific DNA target sequences in S. cerevisiae, thereby modulating gene expression. In this Point of View, we provide an overview of the nature of lncRNA-mediated control of gene expression in the context of our studies using the GAL gene cluster as a model for controlling the timing of transcription.
KEYWORDS: Environment, gene expression, helicase, long non-coding, metabolism, ncRNA, R-loops
Abbreviations
- DRIP
DNA/RNA immunoprecipitation
- lncRNA
long non-coding RNA
- mRNA
mRNA
- ncRNA
non-coding RNA
- lncRNA
long non-coding RNA
- RNP
ribonucleoprotein complex
Non-coding RNAs (ncRNAs) are a family of RNAs that display a wide range of biochemically distinct roles.1 Over the past 50 years, many different classes of ncRNAs have been described, including microRNAs (miRNAs),2,3 small nucleolar RNAs (snoRNAs),4,5 small nuclear RNAs (snRNAs),6 small Cajal body-specific RNAs (scaRNAs),7 enhancer RNAs (eRNAs),8 and long non-coding RNAs (lncRNAs).9 LncRNAs are a diverse class of ncRNAs found in all eukaryotes from mammals10 to unicellular organisms, such as Trichomonas vaginalis,11 Plasmodium falciparum,12 and the budding yeast Saccharomyces cerevisiae.13 The generally accepted definition of a lncRNA is a transcript that lacks a long or conserved open reading frame and is greater than 200 nucleotides in length.1,14 Initially theorized to be the result of spontaneous, aberrant transcription initiation, lncRNAs were first dismissed as spurious “sloppy” transcription.15 However, the characterization of gene regulatory functions by mammalian lncRNAs, such as XIST16 and HOTAIR,17 made it clear that many of these transcripts are much more than extraneous transcriptional products.
Upwards of 30,000 lncRNAs have been identified in mammalian cells to date.18 Thus far, many lncRNAs have been shown to control the expression of protein-coding genes by recruiting chromatin remodeling factors to specific gene loci.19 Histone modifying enzymes and nucleosome remodeling proteins that interact with lncRNAs include G9a,20 LSD1,21 PRC2,21,22 SWI/SNF,23 and MLL,24 indicating a diversity of interactions. LncRNAs have also been described as miRNA sponges,25 facilitators of cytoplasmic mRNA decay,26 and remodelers of the 3D chromatin architecture,27 indicating a broad array of activities. In addition to biochemical diversity, a common theme for lncRNA function is temporal control of gene expression, regulating cellular programs that require highly specific and well timed responses to extracellular stimuli.28–30 Unsurprisingly, misregulation of lncRNAs has been linked to the development of a multitude of human diseases including cancer31 and heart failure.32
The budding yeast Saccharomyces cerevisiae has been a robust system for genetic and molecular investigation of gene expression steps since its introduction as a model organism almost 30 y ago.33 Thus far, ∼2,000 lncRNAs have been identified in budding yeast, which vary both in presence and abundance depending on growth conditions.34–36 To survive in the wild, yeasts must rapidly adapt to changing environmental conditions, such as heat and osmotic stress and nutrient availability. The natural habitat for S. cerevisiae is fresh and decaying fruit where carbon sources are abundant and diverse.37,38 These yeast preferentially use glucose as a carbon source, however, they have the ability to switch their metabolic profile to use alternative sugars, a response which is essential to environmental adaptation.39 This switch involves reprogramming of upwards of 40% of the yeast transcriptome as a result of derepression and transcriptional activation of genes necessary for metabolism of sugars other than glucose.40,41
A key component of the glucose to galactose metabolic switch is the GAL gene cluster consisting of GAL1, GAL10, and GAL7. These genes exist in 3 distinct transcriptional states depending on the carbon source in the media. In the presence of glucose, the GAL cluster is repressed via association of glucose-dependent transcription factors, Mig1 and Nrg1, and the associated Tup1/Cyc8 (Tup1/Ssn6) co-repressor complex.42,43 In the presence of galactose, the Gal4 transcriptional activator associates with GAL genes and facilitates transcriptional activation by promoting recruitment of co-activators and RNA polymerase II (RNA P II).44 The third transcriptional state, called the “derepressed” or “non-induced” state, occurs when yeast are grown in the presence of raffinose. In this condition, the GAL genes are neither actively repressed nor are they induced.45 This ability to control the transcriptional states by manipulation of carbon sources has made the GAL cluster an exceptional model gene locus for studying transcription initiation, termination, and chromatin remodeling events for decades.44,46
In addition to regulatory proteins, the GAL cluster also contains 2 lncRNAs, the GAL10 and GAL10s lncRNAs, which originate from a bidirectional promoter within the 3′ end of the GAL10 open reading frame.47 The GAL10 lncRNA is transcribed in an antisense orientation with respect to the GAL10 protein-coding gene and overlaps both GAL10 and GAL1, whereas the GAL10s lncRNA is expressed in the opposite orientation and runs through the downstream GAL7 promoter region.34,47 Initial studies of the GAL10 antisense lncRNA showed that expression of this non-coding molecule correlates with repression of GAL10 and GAL1 transcription when low levels of glucose are available in a complex sugar mixture, similar to conditions in the wild.47 This repressive role was later supported by single transcript microscopy studies which demonstrated suppression of leaky transcription of the GAL cluster genes in the absence of GAL10 antisense lncRNA expression.48 Repression occurs in cis47 consistent with a transcriptional interference mode of action, whereby the process of transcription, rather than the lncRNA, regulates expression of an overlapped, protein-coding gene. This type of lncRNA-dependent regulation has been demonstrated for other genes in budding yeast as well as mammalian cells.28,49
The first discovery of transcriptional interference by a ncRNA was regulation of the SER3 gene, which encodes 3-phosphoglycerate dehydrogenase, an enzyme integral to glycolysis and serine biosynthesis. In 2004, it was reported that serine-dependent transcription of the SRG1 lncRNA disrupts binding of general transcription factors to the downstream SER3 gene promoter, thereby regulating transcription of this protein-coding gene.28 Interestingly, SRG1 transcription is activated by the presence of serine in the media, suggesting direct regulation of an lncRNA by nutrient availability. Repression of SER3 is mediated by changes in nucleosome occupancy within the SER3 promoter that occur during transcription of SRG1.50 When cells become serine-deprived, SRG1 is no longer transcribed and SER3 repression is released.51 These studies demonstrated that lncRNAs can regulate gene expression via multiple modalities in response to extracellular nutrient status.
Interestingly, the same lncRNA can also have more than one biochemical function in vivo. In addition to transcriptional repression, our group discovered that the GAL10 and GAL10s lncRNAs also function in transcriptional activation from transcriptionally repressive (+glucose) to activating (−galactose) conditions.52 During this switch, the GAL lncRNAs enhance induction of the GAL protein coding genes from a repressed state by interfering with binding of the Tup1/Cyc8-corepressor complex. This occurs, presumably, by physical occlusion of the complex and results in derepression of the glucose-repressed GAL genes and allows for faster transcriptional activation in the presence of galactose (Fig. 1). Consistently, the GAL lncRNAs have no effect on induction from the derepressed (+raffinose) state, but rather confer a specific fitness advantage to yeast cells over those lacking the GAL lncRNAs during a glucose to galactose switch.48,53 GAL lncRNA-dependent induction occurs when the GAL lncRNAs are encoded in trans, indicating that lncRNA-dependent transcriptional induction and repression are mechanistically distinct. This result is reminiscent of Air (Airn), which also functions in mechanistically distinct cis and trans roles to regulate gene expression.54,55
Figure 1.
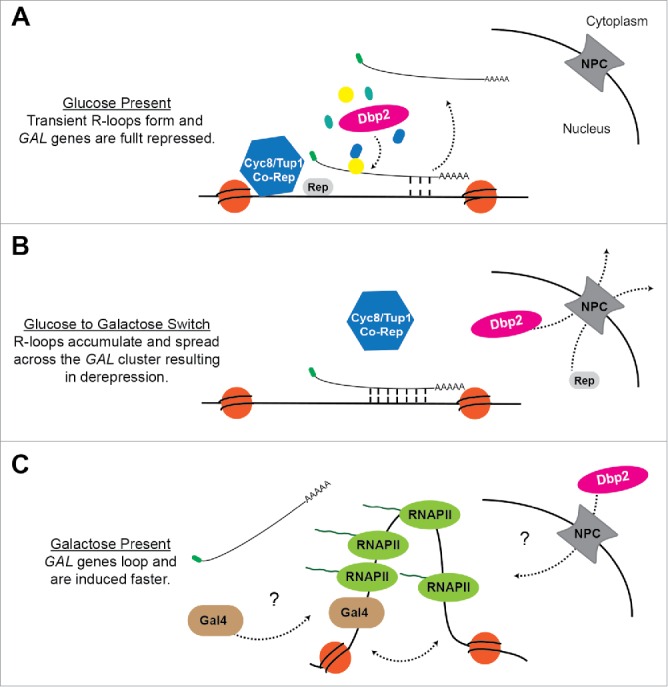
A model for regulation of the GAL cluster by GAL lncRNA R-loops and Dbp2. (A) Glucose Repression When glucose is available, Dbp2 is localized in the nucleus74 and prevents the accumulation of R-loops at the GAL gene cluster53 via the assembly of Dbp2-dependent lncRNA-protein complexes.56 This allows the successful docking of the Cyc8/Tup1 co-repressor complex and subsequent repression. (B) Carbon Source Switch During a switch from glucose to galactose, Dbp2 is actively relocalized to the cytoplasm via export through the nuclear pore complex (NPC).74 This may alter lncRNP assembly and cause R-loop formation and spreading across the GAL cluster. R-loop formation likely interferes with binding of transcriptional repressors and the Cyc8/Tup1 corepressor complex, causing derepression of the GAL genes. (C) Galactose Induction In the presence of galactose, the Gal4 transcriptional activator is released from the Gal80 inhibitor (not pictured),96 enabling recruitment of co-activators and RNA P II.97 This results in altered chromatin structure, as evidenced by looping of the GAL10 promoter and terminator,53 which may enhance transcriptional induction. It is currently unknown how the GAL lncRNAs are cleared from chromatin or how Dbp2 is re-imported following long-term growth in galactose.
Dbp2 is a co-transcriptional RNA chaperone and bona fide RNA helicase whose activity is necessary for assembly of RNA-binding proteins, Yra1, Mex67, and Nab2 onto poly(A)+ RNAs.56,57 During transcription, both messenger and long non-coding RNAs are co-transcriptionally bound with a set of RNA-binding proteins to form a ribonucleoprotein complex (RNP), which then undergoes a variety of processing steps, including capping, splicing, termination, and polyadenylation. Loss of DBP2 also results in over-accumulation and 3′ extension of the GAL10s lncRNA,58 suggesting a connection between Dbp2 and proper biogenesis of the GAL lncRNAs. Interestingly, we found that cells lacking DBP2 display a much more rapid lncRNA-dependent transcriptional induction of the GAL genes, as well as reduced association of the Cyc8 corepressor.52 This suggested that Dbp2 and/or its role in RNP assembly, antagonizes the transcriptionally activating role of the GAL lncRNAs and that dbp2Δ cells could be used as a tool to decipher the mechanism of transcriptional induction by these non-coding molecules.
Seminal studies from the Aguilera lab demonstrated that mutations in genes needed for mRNA export, transcription elongation, and termination result in formation of R-loops.59,60 R-loops are structures that form when an RNA base pairs with one strand of a DNA double helix, resulting in formation of an RNA:DNA hybrid and a displaced stretch of single-stranded DNA. R-loops can be both biologically beneficial as well as etiological agents of DNA damage. Aberrant formation and/or stabilization of R-loops results in genomic instability as a result of prolonged exposure of a single-stranded DNA to the environment and/or by interfering with DNA replication machinery.61,62 In contrast, R-loops play beneficial roles by mediating CRISPR interference,63,64 promoting IgG class switching,65,66 and by regulating transcription.67,68 These structures can form in cis60 or trans69 in vivo, either by “threading back” of nascent RNAs during transcription by RNA Pol II, through the action of the homologous recombination machinery, or associated RNA-binding proteins as in the case of CRISPR.60,70,71
Given that RNP assembly defects cause R-loops,60,70 we speculated that loss of DBP2 might promote formation of these structures between the GAL lncRNAs and GAL gene promoters. To test this, we asked if ectopic expression of human RNAse H1 in dbp2Δ cells would prevent rapid induction of the GAL genes by the GAL lncRNAs. Strikingly, this was the case, suggesting that RNA-DNA hybrids were involved in lncRNA-dependent induction of the GAL genes in dbp2Δ cells.72 To determine if GAL lncRNA R-loops formed at the GAL cluster, we conducted DNA:RNA immunoprecipitation (DRIP) using the S9.6 RNA:DNA hybrid antibody. DRIP revealed that loss of DBP2 results in accumulation of lncRNA-dependent R-loops across all 3 GAL cluster genes, consistent with the length of the GAL10 antisense lncRNA and 3′ extended GAL10s lncRNA.58 Interestingly, the nuclear RNA decay enzyme RRP6 also controls R-loop formation in mammalian cells between eRNAs and enhancers,73 indicating that RNP assembly and/or processing may dictate the function of numerous lncRNAs. Since simultaneous loss of DBP2 and RRP6 is lethal in yeast, indicating functional overlap,58 it will be interesting to determine if orthologs of Dbp2 (DDX5 in mammals) also controls R-loop formation in multicellular eukaryotes.
While investigating the role of the GAL lncRNAs, we serendipitously discovered that Dbp2 is transported to the cytoplasm in response to glucose deprivation or a glucose to galactose shift, suggesting that wild type yeast actively export Dbp2 from the nucleus to alter gene expression.74 Strikingly, nuclear depletion of Dbp2 using the “anchor away” strategy75 resulted in time-dependent accumulation of R-loops beginning at the 5′ end of GAL1 and spreading across GAL1, GAL10 and through the 3′ end of GAL7.53 Removal of the lncRNAs through genomic deletion reduced R-loops at the 5′ end of GAL1 and abolished detection of these structures across the rest of the GAL cluster locus. This suggested that Dbp2 regulates formation of lncRNA R-loops at the GAL genes (Fig. 1A-B), thereby controlling lncRNA-dependent transcriptional regulation in response to nutrient availability. Moreover, this suggests that GAL lncRNA R-loops promote induction of the GAL genes from transcriptionally repressive conditions. In support of a role for R-loops in depression of the GAL cluster, we found that ectopic expression of RNase H1 reduced induction of the GAL genes to wild type levels in dbp2Δ cells.53 The fact that INO1, an inducible gene without an overlapping, annotated lncRNA, induced at the same rate regardless of the presence of RNAse H1 argues against a general reduction in transcription.53
Because we observed R-loop accumulation across the entire GAL cluster, this leads one to wonder how these structures could specifically prevent Cyc8 association without impacting subsequent steps of transcriptional activation by Gal4, coactivators, and RNAPII (Fig. 1C). One possibility is that these R-loops are transient, allowing recruitment of transcriptional activators. Resolution of R-loops in vivo would be accomplished by the activity of endogenous RNase H enzymes as well as RNA-DNA helicases such as Sen1/Senataxin.70,76,77 It is also likely that these R-loops are cleared by transcription of another lncRNA molecule. The latter is supported by the fact that high levels of transcription correlate with the presence of R-loops in wild type cells,78 which would be predicted to interfere with transcription if these structures were static. If this is the case, one would predict that most chromatin-bound factors would be displaced by these R-loops, irrespective of their role in transcriptional regulation. Cyc8 would be impacted specifically, not necessarily because of the R-loops, but because of the combined action of these structures with regulated export of glucose-dependent transcription factors to the cytoplasm.79,80 However, very little is known about the dynamics of R-loops in vivo.
Another possibility is that the GAL lncRNA R-loops are discontinuous, forming along regions that impact Cyc8 but not Gal4 and RNA P II. It should be noted that DRIP assays depend on chromatin shearing to an average DNA size and is, thus, not at nucleotide resolution. Two new reports have attempted to address this concern by either the addition of S1 nuclease to prevent RNA:DNA hybrid loss or by sequencing the isolated RNA following immunoprecipitation.81,82 However, a weakness of these studies and of our own investigations of the GAL cluster is the reliance on the S9.6 antibody as the sole reagent available for detection of RNA-DNA hybrids. Because this antibody shows both length and sequence bias,83 new methods will need to be developed to precisely determine the sites of R-loop formation in vivo.
The GAL lncRNAs offer a striking example of how non-coding RNA molecules can promote adaptation. While we do not know if the GAL lncRNAs serve as a paradigm for other lncRNAs, because very few have been functionally characterized, the majority of those that have been studied thus far are linked to environmental sensing and stress response (Table 1). This role would be consistent with the emerging theme for mammalian lncRNAs cellular differentiation programs and human disease states. Additionally, 43% of single-nucleotide polymorphisms associated with human disorders are found outside of protein-coding regions [7], suggesting that mutations in the non-coding genome have been underappreciated. S. cerevisiae likely offer the perfect model system for mechanistic investigation of lncRNA activities, given recent evolutionary studies showing an increased reliance on antisense RNAs for fine tuning gene expression following loss of the microRNA pathway in this species.84
Table 1.
Individually characterized lncRNAs of Saccharomyces cerevisiae. 65% of functionally characterized lncRNAs in S. cerevisiae have gene targets that function in pathways associated with metabolism or nutrient sensing/transport. AS = antisense. Us = upstream.
| Functionally characterized lncRNAs in S. cerevisiae. | ||||
|---|---|---|---|---|
| Name | Function | Target Gene | Target gene function | Metabolism/Nutrition related |
| MMF1 AS RNA29 | Promotes induction | MMF1 | Mitochondrial protein | YES |
| ASP3 lncRNA98 | Upregulation | ASP3 | Asparagine catabolism | YES |
| PHO84 AS RNA99-101 | Repression | PHO84 | Phosphate metabolism | YES |
| SRG128,50,51 | Repression | SER3 | Serine / glycine biogenesis | YES |
| PHO5 AS RNA102 | Promotes induction | PHO5 | Phosphate metabolism | YES |
| usURA2.103 | Repression | URA2 | Pyrimidine biogenesis | YES |
| usDCI1.104 | Repression | DCI1 | Fatty acid metabolism | YES |
| GAL10 lncRNA47,48,105 | Repression | GAL genes | Galactose metabolism | YES |
| GAL10/GAL10s lncRNAs.52,53 | Promotes induction | GAL genes | Galactose metabolism | YES |
| KCS1 lncRNAs106 | Translational interference | KCS1 | Inositol hexa/hepta bisphosphate kinase | YES |
| SUT71930,35 | Repression | SUR7 | Plasma membrane protein | YES |
| CDC28 AS RNA29 | Promotes induction | CDC28 | Cell cycle regulator | NO |
| RTL.107 | Repression | Ty1 | Retrotransposon | NO |
| ICR1.108 | Repression | FLO11 | Cell surface glycoprotein | NO |
| PWR1.108 | Upregulation | FLO11 | Cell surface glycoprotein | NO |
| IME4 AS RNA109,110 | Repression | IME4 | Meiosis regulator | NO |
| IRT1.109 | Repression | IME1 | Meiosis regulator | NO |
Moving forward, there are many aspects of lncRNA-dependent gene regulation that remain to be addressed. First, it is currently unknown how many lncRNAs utilize R-loops as a mechanism for gene regulation. It has been speculated for some time that direct hybridization of lncRNAs with genomic DNA could be a mechanism for locus-specific targeting, however, with the exception of RNA:DNA:DNA triplexes,85–87 this has yet to be broadly demonstrated. Another question is how an RNA invades a DNA duplex, an activity that is thermodynamically unfavorable. Interestingly, evidence from the Koshland laboratory points to direct roles for Rad51 and Rad52, components of the homologous recombination machinery, for promoting R-loop formation in trans.69 However, it is unknown what features of the RNA and/or DNA locus are recognized to mediate RNA:DNA hybridization and how the RNA helicase Dbp2 antagonizes this process. One possibility is that the lncRNA-protein composition and structure dictates the ability of this molecule to form R-loops.
Another question is how many other lncRNAs, in fungi or other species, promote the timing of gene expression. It is likely that many lncRNAs detected to date are spurious, non-functional products of transcription. However, future studies may be informed by analyses of transcription induction or repression kinetics, rather than reliance on lncRNA-dependent changes in steady state transcript levels. Finally, as more efforts are put forth to decipher the role of the non-coding genome, it must be emphasized that some of these long non-coding RNAs have been detected with translating ribosomes.88 Although no examples of lncRNAs that produce functional peptides have been documented to date, we should continue to view the term “non-coding” with caution until a much larger subset of these molecules have been functionally validated.
Aberrant expression of lncRNAs is associated with various diseases such as prostate cancer,89 breast cancer,90 HIV,91 Type-2 diabetes,92,93 and obesity,93,94 underscoring a need for understanding the precise roles of lncRNAs. It should be emphasized that, although pioneering discoveries regarding lncRNA scaffolding of chromatin remodeling factors21 and remodeling of the 3D genome95 have relied on mammalian systems, transcriptional interference28 and lncRNA R-loops were initially discovered in budding yeast.53 As we continue to define biological roles for individual lncRNAs, it is essential to complement our studies of multicellular eukaryotes with simple model organisms, which have provided and continue to provide mechanistic paradigms for gene regulation.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank all members of the Tran laboratory for insightful discussions, especially Sara Cloutier, for critical reading of this manuscript.
Funding
This work was supported by NIH GM097332 to E.J.T.
References
- 1.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012; 81:145-66; PMID:22663078; http://dx.doi.org/ 10.1146/annurev-biochem-051410-092902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambros V. microRNAs: Tiny regulators with great potential. Cell 2001; 107:823-6; PMID:11779458; http://dx.doi.org/ 10.1016/S0092-8674(01)00616-X [DOI] [PubMed] [Google Scholar]
- 3.Lagos-quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science 2001; 294:853-8; PMID:11679670; http://dx.doi.org/ 10.1126/science.1064921 [DOI] [PubMed] [Google Scholar]
- 4.Muramatsu M, Hodnett JL, Busch H. Base composition of fractions of nuclear and nucleolar ribonucleic acid obtained by sedimentation and chromatography. J Biol Chem 1966; 241:1544-50; PMID:5946613 [PubMed] [Google Scholar]
- 5.Penman S, Smith I, Holtzman E. Ribosomal RNA synthesis and processing in a particulate site in the HeLa cell nucleus. Science 1966; 154:786-9; PMID:5919449; http://dx.doi.org/ 10.1126/science.154.3750.786 [DOI] [PubMed] [Google Scholar]
- 6.Jarmolowski A, Zagorski J, Li H V, Fournier MJ. Identification of essential elements in U14 RNA of Saccharomyces cerevisiae. EMBO J 1990; 9:4503-9; PMID:2265615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darzacq X, Jády BE, Verheggen C, Kiss AM, Bertrand E, Kiss T. Cajal body-specific small nuclear RNAs: A novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J 2002; 21:2746-56; PMID:12032087; http://dx.doi.org/ 10.1093/emboj/21.11.2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim T, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-haley K, Kuersten S, et al.. Widespread transcription at neuronal activity-regulated enhancers. 2010; 465:182-7; PMID:20393465; http://dx.doi.org/ 10.1038/nature09033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagano T, Fraser P. Emerging similarities in epigenetic gene silencing by long noncoding RNAs. Mamm Genome 2009; 20:557-62; PMID:19727951; http://dx.doi.org/ 10.1007/s00335-009-9218-1 [DOI] [PubMed] [Google Scholar]
- 10.Chowdhury D, Choi YE, Brault ME. Charity begins at home: non-coding RNA functions in DNA repair. Nat Rev Mol Cell Biol 2013; 14:181-9; PMID:23385724; http://dx.doi.org/ 10.1038/nrm3523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woehle C, Kusdian G, Radine C, Graur D, Landan G, Gould SB. The parasite Trichomonas vaginalis expresses thousands of pseudogenes and long non-coding RNAs independently from functional neighbouring genes. BMC Genomics 2014; 15:1-12; PMID:24382143; http://dx.doi.org/ 10.1186/1471-2164-15-906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Broadbent KM, Broadbent JC, Ribacke U, Wirth D, Rinn JL, Sabeti PC. Strand-specific RNA sequencing in Plasmodium falciparum malaria identifies developmentally regulated long non-coding RNA and circular RNA. BMC Genomics 2015; 16:454; PMID:26070627; http://dx.doi.org/ 10.1186/s12864-015-1603-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamashita A, Shichino Y, Yamamoto M. The long non-coding RNA world in yeasts. Biochim Biophys Acta 2015; 1859:147-54; PMID:26642900; http://dx.doi.org/ 10.1016/j.bbagrm.2015.08.003 [DOI] [PubMed] [Google Scholar]
- 14.Ruiz-Orera J, Messeguer X, Subirana JA, Alba MM. Long non-coding RNAs as a source of new peptides. Elife 2014; 3:e03523; PMID:25233276; http://dx.doi.org/ 10.7554/eLife.03523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol 2007; 14:103-5; PMID:17277804; http://dx.doi.org/ 10.1038/nsmb0207-103 [DOI] [PubMed] [Google Scholar]
- 16.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS ZY. Role of histone H3 Lysine 27 Methylation in X inactivation. Science 2002; 298:1039-43; PMID:12351676; http://dx.doi.org/ 10.1126/science.1076997 [DOI] [PubMed] [Google Scholar]
- 17.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al.. Functional demarcation of active and silent chromatin domains in human HOX Loci by noncoding RNAs. Cell 2007; 129:1311-23; PMID:17604720; http://dx.doi.org/ 10.1016/j.cell.2007.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fantom Consortium T The transcriptional landscape of the mammalian genome. Science 2006; 309:1559-63; PMID:16141072; http://dx.doi.org/18951091 10.1126/science.1112014 [DOI] [PubMed] [Google Scholar]
- 19.Han P, Chang CP. Long non-coding RNA and chromatin remodeling. RNA Biol 2016; 12:1094-98; PMID:26177256; http://dx.doi.org/18951091 10.1080/15476286.2015.1063770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-DiNardo D, Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell 2008; 32:232-46; PMID:18951091; http://dx.doi.org/ 10.1016/j.molcel.2008.08.022 [DOI] [PubMed] [Google Scholar]
- 21.Tsai M, Manor O, Wan Y, Mosammaparast N, Wang JK, Shi Y, Segal E, Chang HY. Long Noncoding RNA as modular scaffold of histone modification complexes. Science 2010; 329:689-93; PMID:20616235; http://dx.doi.org/ 10.1126/science.1192002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park M, Salgado JM, Ostroff L, Helton TD, Camenzind G, Harris KM, Ehlers MD, Augusta G. Functional demarcation of active and silent chromatin domains in human HOX Loci by Non-Coding RNAs. Cell 2007; 52:817-30; PMID:17604720; http://dx.doi.org/25831520 10.1016/j.cell.2007.05.022 [DOI] [Google Scholar]
- 23.Kawaguchi T, Tanigawa A, Naganuma T, Ohkawa Y, Souquere S, Pierron G, Hirose T. SWI/SNF chromatin-remodeling complexes function in noncoding RNA-dependent assembly of nuclear bodies. Proc Natl Acad Sci U S A 2015; 112:4304-9; PMID:25831520; http://dx.doi.org/ 10.1073/pnas.1423819112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al.. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011; 472:120-4; PMID:21423168; http://dx.doi.org/ 10.1038/nature09819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paci P, Colombo T, Farina L. Computational analysis identifies a sponge interaction network between long non-coding RNAs and messenger RNAs in human breast cancer. BMC Syst Biol 2014; 8:83; PMID:25033876; http://dx.doi.org/ 10.1186/1752-0509-8-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gong C, Maquat LE. lncRNAs transactivate Staufen1-mediated mRNA decay by duplexing with 3′UTRs via Alu elements. Nature 2011; 470:284-8; PMID:21307942; http://dx.doi.org/ 10.1038/nature09701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melé M, Rinn JL “Cat's Cradling” the 3D Genome by the Act of LncRNA Transcription. Mol Cell 2016; 62:657-64; PMID:27259198; http://dx.doi.org/15175754 10.1016/j.molcel.2016.05.011 [DOI] [PubMed] [Google Scholar]
- 28.Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 2004; 429:571-4; PMID:15175754; http://dx.doi.org/ 10.1038/nature02538 [DOI] [PubMed] [Google Scholar]
- 29.Nadal-Ribelles M, Solé C, Xu Z, Steinmetz LM, de Nadal E, Posas F. Control of Cdc28 CDK1 by a Stress-Induced lncRNA. Mol Cell 2014; 53:549-61; PMID:24508389; http://dx.doi.org/ 10.1016/j.molcel.2014.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu Z, Wei W, Gagneur J, Clauder-Munster S, Smolik M, Huber W, Steinmetz LM. Antisense expression increases gene expression variability and locus interdependency. Mol Syst Biol 2011; 7:468; PMID:21326235; http://dx.doi.org/ 10.1038/msb.2011.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fatima R, Akhade VS, Pal D, Rao SM. Long noncoding RNAs in development and cancer: potential biomarkers and therapeutic targets. Mol Cell Ther 2015; 3:5; PMID:26082843; http://dx.doi.org/ 10.1186/s40591-015-0042-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greco S, Zaccagnini G, Perfetti A, Fuschi P, Valaperta R, Voellenkle C, Castelvecchio S, Gaetano C, Finato N, Beltrami AP, et al.. Long noncoding RNA dysregulation in ischemic heart failure. J Transl Med 2016; 14:183; PMID:27317124; http://dx.doi.org/ 10.1186/s12967-016-0926-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Botstein D, Fink GR. Yeast: An experimental organism for 21st century biology. Genetics 2011; 189:695-704; PMID:22084421; http://dx.doi.org/ 10.1534/genetics.111.130765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Dijk EL, Chen CL, D'Aubenton-Carafa Y, Gourvennec S, Kwapisz M, Roche V, Bertrand C, Silvain M, Legoix-Né P, Loeillet S, et al.. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature 2011; 475:114-7; PMID:21697827; http://dx.doi.org/ 10.1038/nature10118 [DOI] [PubMed] [Google Scholar]
- 35.Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Münster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. Bidirectional promoters generate pervasive transcription in yeast. Nature 2009; 457:1033-7; PMID:19169243; http://dx.doi.org/ 10.1038/nature07728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neil H, Malabat C, d'Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A, Aubenton-carafa Y, Xu Z, Steinmetz LM, Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature 2009; 457:1038-42; PMID:19169244; http://dx.doi.org/ 10.1038/nature07747 [DOI] [PubMed] [Google Scholar]
- 37.Vadkertiová R, Molnárová J, Vránová D, Sláviková E. Yeasts and yeast-like organisms associated with fruits and blossoms of different fruit trees. Can J Microbiol 2012; 58:1344-52; PMID:23210991; http://dx.doi.org/12589024 10.1139/cjm-2012-0468 [DOI] [PubMed] [Google Scholar]
- 38.Wheeler RT, Kupiec M, Magnelli P, Abeijon C, Fink GR. A Saccharomyces cerevisiae mutant with increased virulence. Proc Natl Acad Sci U S A 2003; 100:2766-70; PMID:12589024; http://dx.doi.org/ 10.1073/pnas.0437995100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Demir O, Aksan Kurnaz I. An integrated model of glucose and galactose metabolism regulated by the GAL genetic switch. Comput Biol Chem 2006; 30:179-92; PMID:16679066; http://dx.doi.org/ 10.1016/j.compbiolchem.2006.02.004 [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, Pierce M, Schneper L, Güldal CG, Zhang X, Tavazoie S, Broach JR. Ras and Gpa2 mediate one branch of a redundant glucose signaling pathway in yeast. PLoS Biol 2004; 2:610-22; PMID:15138498; http://dx/doi.org/18303986 10.1371/journal.pbio.0020128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zaman S, Lippman SI, Zhao X, Broach JR. How Saccharomyces responds to nutrients. Annu Rev Genet 2008; 42:27-81; PMID:18303986; http://dx.doi.org/ 10.1146/annurev.genet.41.110306.130206 [DOI] [PubMed] [Google Scholar]
- 42.Williams FE, Varanasi U, Trumbly RJ. The CYC8 and TUP1 proteins involved in glucose repression in Saccharomyces cerevisiae are associated in a protein complex. Mol Cell Biol 1991; 11:3307-16; PMID:2038333; http://dx.doi.org/ 10.1128/MCB.11.6.3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou H, Winston F. NRG1 is required for glucose repression of the SUC2 and GAL genes of Saccharomyces cerevisiae. BMC Genet 2001; 2:5; PMID:11281938; http://dx.doi.org/ 10.1186/1471-2156-2-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Traven A, Jelicic B, Sopta M. Yeast Gal4: a transcriptional paradigm revisited. EMBO Rep 2006; 7:496-9; PMID:16670683; http://dx.doi.org/ 10.1038/sj.embor.7400679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnston M, Flick JS, Pexton T. Multiple mechanisms provide rapid and stringent glucose repression of GAL gene expression in Saccharomyces cerevisiae. Mol Cell Biol 1994; 14:3834-41; PMID:8196626; http://dx.doi.org/ 10.1128/MCB.14.6.3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flick JS, Johnston M. GRR1 of Saccharomyces cerevisiae is required for glucose repression and encodes a protein with leucine-rich repeats. Mol Cell Biol 1991; 11:5101-12; PMID:1922034; http://dx.doi.org/ 10.1128/MCB.11.10.5101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Houseley J, Rubbi L, Grunstein M, Tollervey D, Vogelauer M. A ncRNA Modulates Histone Modification and mRNA Induction in the Yeast GAL Gene Cluster. Mol Cell 2008; 32:685-95; PMID:19061643; http://dx.doi.org/ 10.1016/j.molcel.2008.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lenstra TL, Coulon A, Chow CC, Larson DR, Lenstra TL, Coulon A, Chow CC, Larson DR. Single-molecule imaging reveals a switch between spurious and functional ncRNA transcription article single-molecule imaging reveals a switch between spurious and functional ncRNA transcription. Mol Cell 2015; 60:597-610; PMID:26549684; http://dx.doi.org/ 10.1016/j.molcel.2015.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 2002; 415:810-3; PMID:11845212; http://dx.doi.org/ 10.1038/415810a [DOI] [PubMed] [Google Scholar]
- 50.Hainer SJ, Pruneski JA, Mitchell RD, Monteverde RM, Martens JA. Intergenic transcription causes repression by directing nucleosome assembly. Genes Dev 2011; 25:29-40; PMID:21156811; http://dx.doi.org/ 10.1101/gad.1975011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martens JA, Wu PYJ, Winston F. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev 2005; 19:2695-704; PMID:16291644; http://dx.doi.org/ 10.1101/gad.1367605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cloutier SC, Wang S, Ma WK, Petell CJ, Tran EJ. Long noncoding RNAs promote transcriptional poising of inducible genes. PLoS Biol 2013; 11:32-4; PMID:24260025; http://dx.doi.org/26833086 10.1371/journal.pbio.1001715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cloutier SC, Wang S, Ma WK, Al Husini N, Dhoondia Z, Ansari A, Pascuzzi PE, Tran EJ. Regulated formation of lncRNA-DNA hybrids enables faster transcriptional induction and environmental adaptation. Mol Cell 2016; 61:393-404; PMID:26833086; http://dx.doi.org/ 10.1016/j.molcel.2015.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 2008; 322:1717-21; PMID:18988810; http://dx.doi.org/ 10.1126/science.1163802 [DOI] [PubMed] [Google Scholar]
- 55.Latos PA, Pauler FM, Koerner MV, Şenergin HB, Hudson QJ, Stocsits RR, Allhoff W, Stricker SH, Klement RM, Warczok KE, et al.. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 2012; 338:1469-72; PMID:23239737; http://dx.doi.org/ 10.1126/science.1228110 [DOI] [PubMed] [Google Scholar]
- 56.Ma WK, Cloutier SC, Tran EJ. The DEAD-box protein Dbp2 functions with the RNA-binding protein Yra1 to promote mRNP assembly. J Mol Biol 2013; 425:3824-38; PMID:23721653; http://dx.doi.org/ 10.1016/j.jmb.2013.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ma WK, Paudel BP, Xing Z, Sabath IG, Rueda D, Tran EJ. Recruitment, duplex unwinding and protein-mediated inhibition of the dead-box RNA helicase Dbp2 at actively transcribed chromatin. J Mol Biol 2016; 428:1091-106; PMID:26876600; http://dx.doi.org/ 10.1016/j.jmb.2016.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cloutier SC, Ma WK, Nguyen LT, Tran EJ. The DEAD-box RNA helicase Dbp2 connects RNA quality control with repression of aberrant transcription. J Biol Chem 2012; 287:26155-66; PMID:22679025; http://dx.doi.org/ 10.1074/jbc.M112.383075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Santos-Pereira JM, Aguilera A. R loops: new modulators of genome dynamics and function. Nat Rev Genet 2015; 16:583-97; PMID:26370899; http://dx.doi.org/ 10.1038/nrg3961 [DOI] [PubMed] [Google Scholar]
- 60.Aguilera A, García-Muse T. R Loops: From transcription byproducts to threats to genome stability. Mol Cell 2012; 46:115-24; PMID:22541554; http://dx.doi.org/ 10.1016/j.molcel.2012.04.009 [DOI] [PubMed] [Google Scholar]
- 61.Gan W, Guan Z, Liu J, Gui T, Shen K, Manley JL, Li X. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev 2011; 25:2041-56; PMID:21979917; http://dx.doi.org/ 10.1101/gad.17010011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hatchi E, Skourti-Stathaki K, Ventz S, Pinello L, Yen A, Kamieniarz-Gdula K, Dimitrov S, Pathania S, McKinney KM, Eaton ML, et al.. BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol Cell 2015; 57:636-47; PMID:25699710; http://dx.doi.org/ 10.1016/j.molcel.2015.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rutkauskas M, Sinkunas T, Siksnys V, Seidel R, Rutkauskas M, Sinkunas T, Songailiene I, Tikhomirova MS, Siksnys V, Seidel R. Directional R-Loop formation by the CRISPR-Cas surveillance complex cascade provides efficient article directional R-Loop formation by the CRISPR-Cas surveillance complex cascade provides efficient off-target site rejection. CellReports 2015; 10:1534-43; PMID:25753419; http://dx.doi.org/24912165 10.1016/j.celrep.2015.01.067 [DOI] [PubMed] [Google Scholar]
- 64.Szczelkun MD, Tikhomirova MS, Sinkunas T, Gasiunas G, Karvelis T, Pschera P, Siksnys V, Seidel R. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc Natl Acad Sci U S A 2014; 111:9798-803; PMID:24912165; http://dx.doi.org/ 10.1073/pnas.1402597111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vaidyanathan B, Chaudhuri J. Epigenetic codes programing class switch recombination. Front Immunol 2015; 6:405; PMID:26441954; http://dx.doi.org/ 10.3389/fimmu.2015.00405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu K, Chedin F, Hsieh C-L, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol 2003; 4:442-51; PMID:12679812; http://dx.doi.org/ 10.1038/ni919 [DOI] [PubMed] [Google Scholar]
- 67.Ginno PA, Lott PL, Christensen HC, Korf I, Chédin F. R-Loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell 2012; 45:814-25; PMID:22387027; http://dx.doi.org/ 10.1016/j.molcel.2012.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sun Q, Csorba T, Skourti-Stathaki K, Proudfoot NJ, Dean C. R-Loop stabilization represses antisense transcription at the arabidopsis FLC Locus. Science 2013; 340:619-21; PMID:23641115; http://dx.doi.org/ 10.1126/science.1234848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wahba L, Gore SK, Koshland D. The homologous recombination machinery modulates the formation of RNA-DNA hybrids and associated chromosome instability. Elife 2013; 2013:1-20; PMID:23795288; http://dx.doi.org/24990962 10.7554/eLife.00505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Skourti-Stathaki K, Proudfoot NJ. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev 2014; 28:1384-96; PMID:24990962; http://dx.doi.org/ 10.1101/gad.242990.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jiang F, Taylor DW, Chen JS, Kornfeld JE, Zhou K, Thompson AJ, Nogales E, Doudna JA. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science 2016; 351:867-71; PMID:26841432; http://dx.doi.org/ 10.1126/science.aad8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wahba L, Amon JD, Koshland D, Vuica-Ross M. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol Cell 2011; 44:978-88; PMID:22195970; http://dx.doi.org/ 10.1016/j.molcel.2011.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pefanis E, Wang J, Rothschild G, Lim J, Kazadi D, Sun J, Federation A, Chao J, Elliott O, Liu Z, et al.. RNA exosome regulated long non-coding RNA transcription controls super-enhancer activity. Cell 2015; 21:193-201; PMID:25957685; http://dx.doi.org/25164881 10.1016/j.cell.2015.04.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Beck ZT, Cloutier SC, Schipma MJ, Petell CJ, Ma WK, Tran EJ. Regulation of glucose-dependent gene expression by the RNA helicase Dbp2 in Saccharomyces cerevisiae. Genetics 2014; 198:1001-14; PMID:25164881; http://dx.doi.org/ 10.1534/genetics.114.170019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Haruki H, Nishikawa J, Laemmli UK. The anchor-away technique: Rapid, conditional establishment of yeast mutant phenotypes. Mol Cell 2008; 31:925-32; PMID:18922474; http://dx.doi.org/ 10.1016/j.molcel.2008.07.020 [DOI] [PubMed] [Google Scholar]
- 76.Skourti-Stathaki K, Proudfoot NJ, Gromak N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell 2011; 42:794-805; PMID:21700224; http://dx.doi.org/ 10.1016/j.molcel.2011.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hausen P, Stein H. Ribonuclease H. An enzyme degrading the RNA moiety of DNA-RNA hybrids. Eur J Biochem 1970; 14:278-83; PMID:5506170; http://dx.doi.org/ 10.1111/j.1432-1033.1970.tb00287.x [DOI] [PubMed] [Google Scholar]
- 78.Chan YA, Aristizabal MJ, Lu PYT, Luo Z, Hamza A, Kobor MS, Stirling PC, Hieter P. Genome-wide profiling of yeast DNA:RNA hybrid prone sites with DRIP-chip. PLoS Genet 2014; 10:e1004288; PMID:24743342; http://dx.doi.org/10556086 10.1371/journal.pgen.1004288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.DeVit MJ, Johnston M. The nuclear exportin Msn5 is required for nuclear export of the Mig1 glucose repressor of Saccharomyces cerevisiae. Curr Biol 1999; 9:1231-41; PMID:10556086; http://dx.doi.org/ 10.1016/S0960-9822(99)80503-X [DOI] [PubMed] [Google Scholar]
- 80.Papamichos-Chronakis M, Gligoris T, Tzamarias D. The Snf1 kinase controls glucose repression in yeast by modulating interactions between the Mig1 repressor and the Cyc8-Tup1 co-repressor. EMBO Rep 2004; 5:368-72; PMID:15031717; http://dx.doi.org/ 10.1038/sj.embor.7400120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wahba L, Costantino L, Tan FJ, Zimmer A, Koshland D. S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev 2016; 30:1327-38; PMID:27298336; http://dx.doi.org/ 10.1101/gad.280834.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nadel J, Athanasiadou R, Lemetre C, Wijetunga NA, Ó Broin P, Sato H, Zhang Z, Jeddeloh J, Montagna C, Golden A, et al.. RNA:DNA hybrids in the human genome have distinctive nucleotide characteristics, chromatin composition, and transcriptional relationships. Epigenetics Chromatin 2015; 8:46; PMID:26579211; http://dx.doi.org/ 10.1186/s13072-015-0040-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Boguslawski SJ, Smith DE, Michalak MA, Mickelson KE, Yehle CO, Patterson WL, Carrico RJ. Characterization of monoclonal antibody to DNA•RNA and its application to immunodetection of hybrids. J Immunol Methods 1986; 89:123-30; PMID:2422282; http://dx.doi.org/ 10.1016/0022-1759(86)90040-2 [DOI] [PubMed] [Google Scholar]
- 84.Alcid EA, Tsukiyama T. Expansion of antisense lncRNA transcriptomes in budding yeast species since the loss of RNAi. Nat Struct Mol Biol 2016; 23:450-5; PMID:27018804; http://dx.doi.org/ 10.1038/nsmb.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 2007; 445:666-70; PMID:17237763; http://dx.doi.org/ 10.1038/nature05519 [DOI] [PubMed] [Google Scholar]
- 86.Schmitz KM, Mayer C, Postepska A, Grummt I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev 2010; 24:2264-9; PMID:20952535; http://dx.doi.org/ 10.1101/gad.590910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Postepska-Igielska A, Giwojna A, Gasri-Plotnitsky L, Schmitt N, Dold A, Ginsberg D, Grummt I. LncRNA Khps1 regulates expression of the Proto-oncogene SPHK1 via Triplex-mediated changes in chromatin structure. Mol Cell 2015; 60:626-36; PMID:26590717; http://dx.doi.org/ 10.1016/j.molcel.2015.10.001 [DOI] [PubMed] [Google Scholar]
- 88.Smith JE, Alvarez-Dominguez JR, Kline N, Huynh NJ, Geisler S, Hu W, Coller J, Baker KE. Translation of small open reading frames within unannotated RNA transcripts in Saccharomyces cerevisiae. Cell Rep 2014; 7:1858-66; PMID:24931603; http://dx.doi.org/ 10.1016/j.celrep.2014.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang A, Zhang J, Kaipainen A, Lucas JM, Yang H. Long non-coding RNA, a newly deciphered “code” in prostate cancer. Cancer Lett 2016; 375(2):323-30; PMID:26965999; http://dx.doi.org/18836484 10.1016/j.canlet.2016.03.003 [DOI] [PubMed] [Google Scholar]
- 90.Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene 2009; 28:195-208; PMID:18836484; http://dx.doi.org/ 10.1038/onc.2008.373 [DOI] [PubMed] [Google Scholar]
- 91.Rice AP. Roles of microRNAs and long-noncoding RNAs in human immunodeficiency virus replication. Wiley Interdiscip Rev RNA 2015; 6:661-70; PMID:26394053; http://dx.doi.org/ 10.1002/wrna.1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Arnes L, Akerman I, Balderes DA, Ferrer J, Sussel L. βlinc1 encodes a long noncoding RNA that regulates islet β-cell formation and function. Genes Dev 2016; 30:502-7; PMID:26944677; http://dx.doi.org/ 10.1101/gad.273821.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kim J, Kim KM, Noh JH, Yoon JH, Abdelmohsen K, Gorospe M. Long noncoding RNAs in diseases of aging. Biochim Biophys Acta - Gene Regul Mech 2016; 1859:209-21; PMID:26141605; http://dx.doi.org/24490137 10.1016/j.bbagrm.2015.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Szeto CYY, Lin CH, Choi SC, Yip TTC, Ngan RKC, Tsao GSW, Li Lung M. Integrated mRNA and microRNA transcriptome sequencing characterizes sequence variants and mRNA-microRNA regulatory network in nasopharyngeal carcinoma model systems. FEBS Open Bio 2014; 4:128-40; PMID:24490137; http://dx.doi.org/ 10.1016/j.fob.2014.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Minajigi A, Froberg JE, Wei C, Sunwoo H, Kesner B, Colognori D, Lessing D, Payer B, Boukhali M, Haas W, et al.. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science 2015; PMID:26089354; http://dx.doi.org/ 10.1126/science.aab2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jiang F, Frey BR, Evans ML, Friel JC, Hopper JE. Gene activation by dissociation of an inhibitor from a transcriptional activation domain. Mol Cell Biol 2009; 29:5604-10; PMID:19651897; http://dx.doi.org/ 10.1128/MCB.00632-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bryant GO, Ptashne M. Independent recruitment in vivo by Gal4 of two complexes required for transcription. Mol Cell 2003; 11:1301-9; PMID:12769853; http://dx.doi.org/ 10.1016/S1097-2765(03)00144-8 [DOI] [PubMed] [Google Scholar]
- 98.Huang YC, Chen HT, Teng SC. Intragenic transcription of a noncoding RNA modulates expression of ASP3 in budding yeast. RNA 2010; 16:2085-93; PMID:20817754; http://dx.doi.org/ 10.1261/rna.2177410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Castelnuovo M, Rahman S, Guffanti E, Infantino V, Stutz F, Zenklusen D. Bimodal expression of PHO84 is modulated by early termination of antisense transcription. Nat Struct {&} Mol Biol 2013; 20:851-8; http://dx.doi.org/ 10.1038/nsmb.2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell 2007; 131:706-17; PMID:18022365; http://dx.doi.org/ 10.1016/j.cell.2007.09.014 [DOI] [PubMed] [Google Scholar]
- 101.Camblong J, Beyrouthy N, Guffanti E, Schlaepfer G, Steinmetz LM, Stutz F. Trans-acting antisense RNAs mediate transcriptional gene cosuppression in S. cerevisiae. Genes Dev 2009; 23:1534-45; PMID:19571181; http://dx.doi.org/ 10.1101/gad.522509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Uhler JP, Hertel C, Svejstrup JQ. A role for noncoding transcription in activation of the yeast PHO5 gene. Proc Natl Acad Sci U S A 2007; 104:8011-6; PMID:17470801; http://dx.doi.org/ 10.1073/pnas.0702431104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Thiebaut M, Colin J, Neil H, Jacquier A, Séraphin B, Lacroute F, Libri D. Futile cycle of transcription initiation and termination modulates the response to nucleotide shortage in S. cerevisiae. Mol Cell 2008; 31:671-82; PMID:18775327; http://dx.doi.org/ 10.1016/j.molcel.2008.08.010 [DOI] [PubMed] [Google Scholar]
- 104.Kim T, Xu Z, Clauder-Munster S, Steinmetz LM, Buratowski S. Set3 HDAC mediates effects of overlapping non-coding transcription on gene induction kinetics. Cell 2012; 150:1158-69; PMID:22959268; http://dx.doi.org/ 10.1016/j.cell.2012.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pinskaya M, Gourvennec S, Morillon A. H3 lysine 4 di- and tri-methylation deposited by cryptic transcription attenuates promoter activation. EMBO J 2009; 28:1697-707; PMID:19407817; http://dx.doi.org/ 10.1038/emboj.2009.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nishizawa M, Komai T, Katou Y, Shirahige K, Ito T, Toh-E A. Nutrient-regulated antisense and intragenic RNAs modulate a signal transduction pathway in yeast. PLoS Biol 2008; 6:2817-30; PMID:19108609; http://dx.doi.org/ 10.1371/journal.pbio.0060326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Berretta J, Pinskaya M, Morillon A. A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S. cerevisiae. Genes Dev 2008; 22:615-26; PMID:18316478; http://dx.doi.org/ 10.1101/gad.458008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bumgarner SL, Dowell RD, Grisafi P, Gifford DK, Fink GR. Toggle involving cis-interfering noncoding RNAs controls variegated gene expression in yeast. TL - 106. Proc Natl Acad Sci U S A 2009; 106:18321-6; PMID:19805129; http://dx.doi.org/ 10.1073/pnas.0909641106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Van Werven FJ, Neuert G, Hendrick N, Lardenois A, Buratowski S, Van Oudenaarden A, Primig M, Amon A. Transcription of two long noncoding RNAs mediates mating-type control of gametogenesis in budding yeast. Cell 2012; 150:1170-81; PMID:22959267; http://dx.doi.org/ 10.1016/j.cell.2012.06.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hongay CF, Grisafi PL, Galitski T, Fink GR. Antisense transcription controls cell fate in saccharomyces cerevisiae. Cell 2006; 127:735-45; PMID:17110333; http://dx.doi.org/ 10.1016/j.cell.2006.09.038 [DOI] [PubMed] [Google Scholar]


