Abstract
Upon encountering nutrient stress conditions, plant cells undergo extensive metabolic changes and induce nutrient recycling pathways for their continued survival. The role of nutrient mobilization in the response of Arabidopsis suspension cells to Suc starvation was examined. Vacuolar autophagy was induced within 24 h of starvation, with increased expression of vacuolar proteases that are likely to be required for degradation of cytoplasmic components delivered to the vacuole, and thus for nutrient recycling. After 48 h of starvation, culture viability began to decrease, and substantial cell death was evident by 72 h. To provide further insight into the pathways required for survival during Suc deficit, transcriptional profiling during Suc starvation was performed using the ATH1 GeneChip array containing 22,810 probe sets. A significant increase in transcript levels was observed for 343 genes within 48 h of starvation, indicating a response to nutrient stress that utilizes the recycling of cellular components and nutrient scavenging for maintaining cell function, the protection of the cell from death through activation of various defense and stress response pathways, and regulation of these processes by specific protein kinases and transcription factors. These physiological and molecular data support a model in which plant cells initiate a coordinated response of nutrient mobilization at the onset of Suc depletion that is able to maintain cell viability for up to 48 h. After this point, genes potentially involved in cell death increase in expression, whereas those functioning in translation and replication decrease, leading to a decrease in culture viability and activation of cell death programs.
All organisms are dependent on nutrients from the environment for their continued viability and growth. In plants, the availability of nitrogen in the soil and the presence of adequate light are necessary for proper synthesis of proteins, lipids, and polysaccharides. Plants have evolved both general and specific systems for survival during periods of nutrient stress that may be brought on by extended darkness. These systems utilize stored polysaccharides and recycled cellular components to replace missing nutrients in order to prevent severe decreases in the amount of respiratory substrates and maintain important biochemical pathways during Suc starvation (Aubert et al., 1996).
Major responses to carbon limitation include directed release of stored nutrients and degradation of proteins, starch, and fatty acids (Journet et al., 1986; Koch, 1996). Journet et al. performed a comprehensive study of the biochemical changes occurring in sycamore cells during starvation and observed a decrease in the amount of Suc, starch, protein, and phospho- and galactolipids, suggesting an induction of degradative pathways for these components. This large-scale degradation was accompanied by decreases in the activity of glycolytic enzymes and a loss in respiratory capacity. Vacuole-specific proteolysis during carbon starvation was induced by extended periods of darkness in maize (Zea mays) roots and young leaves (Brouquisse et al., 1991, 1998; James et al., 1993), with increased expression of a specific vacuolar Ser endopeptidase (RSIP), leading to an increase in free amino acids. Fatty acid catabolism was also found to increase during Glc starvation in maize root tips (Dieuaide et al., 1992, 1993). Through mobilization of the breakdown products of these metabolic processes, the cell is able to maintain those pathways and systems most essential for its survival for an extended period of time.
One of the key degradative processes for survival of eukaryotic cells during periods of nutrient starvation is vacuolar autophagy. During autophagy, portions of cytoplasm are transported to the vacuole or lysosome for degradation (for review, see Huang and Klionsky, 2002), and this process is initiated by nutrient starvation in fungi, plants, and animals. Carbon starvation, and autophagy induced by carbon starvation, has been characterized in maize root tips (Brouquisse et al., 1991; James et al., 1993) and whole plants (Brouquisse et al., 1998), as well as rice (Oryza sativa; Chen et al., 1994), sycamore (Acer pseudoplatanus; Aubert et al., 1996), tobacco (Nicotiana tabacum; Moriyasu and Ohsumi, 1996), and Arabidopsis (Bassham, 2002) suspension cell cultures during Suc starvation. Changes observed in the starving cells include an increase in the size of the vacuole, a decrease in the amount of cytoplasm, and an increase in the activity of vacuolar enzymes (Moriyasu and Ohsumi, 1996). Large portions of cytoplasm, including organelles, are enveloped by membrane-bound vesicles to form autophagosomes and transported to the vacuole for degradation, releasing nutrients for use in other pathways. The loss of oxidation potential seen in starving plant cells may also stem from the breakdown of mitochondria during this process (Journet et al., 1986). Studies in sycamore cells, using electron microscopy and the accumulation of phosphorylcholine as a marker for an increase in autophagy, suggest that a decrease in the supply of respiratory substrates is responsible for the onset of autophagy (Aubert et al., 1996).
Genetic studies of autophagy in yeast (Saccharomyces cerevisiae) have revealed a group of mutants that are sensitive to nitrogen starvation. Autophagy in yeast requires a unique conjugation system, involving a number of proteins, which drives the initiation of macroautophagy, the formation of autophagosomes, and the control of autophagosome size (Mizushima et al., 1998a, 1998b; Klionsky and Ohsumi, 1999; Ichimura et al., 2000). In Arabidopsis, genes encoding potential homologs for many of these yeast autophagy proteins have been found. Arabidopsis knockout mutants of the homologs of the yeast autophagy genes APG9, APG7, and VTI1 showed increased sensitivity to nitrogen deficiency and an early senescence phenotype (Doelling et al., 2002; Hanaoka et al., 2002; Surpin et al., 2003).
Starvation has been shown to induce the expression of a number of genes in various plant systems (Koch, 1996; Yu, 1999), including genes involved in transport and degradation of nutrients. Suc starvation in cucumber suspension cells increased the expression of glyoxylate cycle genes encoding malate synthase and isocitrate lyase (Graham et al., 1994), and carbohydrate depletion of the roots of a Citrus reticulata hybrid led to the induction of genes involved in Suc transport and carbohydrate metabolism (Li et al., 2003). Genes encoding various vacuolar enzymes are specifically up-regulated by Suc starvation in suspension cells (Bassham, 2002), including invertase (Tymowska-Lalanne and Kreis, 1998), vacuolar processing enzyme-γ (VPE-γ Kinoshita et al., 1999), and aleurain (Ahmed et al., 2000), emphasizing the importance of this organelle in the degradation of macromolecules. One of the best-studied examples of Suc starvation-regulated gene expression is the increase of α-amylases during Suc starvation of cultured rice cells (Chen et al., 1994), concomitant with the degradation of amyloplasts, suggesting an increase in starch breakdown. A TATCCA element present in the rice α-amylase promoters is recognized by three novel MYB transcription factors that are also up-regulated by starvation (Lu et al., 2002). The promoters of the dark-inducible branched-chain-α-keto acid dehydrogenase subunits are activated during sugar starvation (Fujiki et al., 2000); however, they do not contain the TATCCA cis-element, suggesting that other cis-acting transcriptional elements are also involved in gene expression changes during Suc starvation in plants.
While a significant amount of information is now available on the biochemical responses of plants to Suc depletion, in many cases the genes responsible for these responses and their regulation remain unknown. Here, we characterize the physiological and morphological changes that occur in Arabidopsis suspension cells during Suc starvation and use Affymetrix GeneChip analysis to determine the changes in gene expression that may be responsible for these characteristic effects. We have identified transcripts encoding degradative enzymes and putative membrane transporters whose levels increase dramatically during Suc starvation, and therefore are likely to function in starvation responses. mRNA levels for a number of genes predicted to encode transcription factors and signal transduction components also increased, and these genes are potentially involved in regulation of the observed responses. Genes known to be up-regulated during other stress conditions were also identified, suggesting that, in addition to specific responses, general stress response pathways are induced during starvation. In contrast, genes encoding components of the translational apparatus and proteins functioning in cell division decreased in expression during starvation, suggesting that these processes are specifically down-regulated during Suc starvation.
RESULTS
Changes in Cellular Morphology and Growth during Suc Starvation of Arabidopsis Suspension Cell Cultures
An Arabidopsis ecotype Columbia suspension cell culture was chosen for initial studies on Suc starvation because the cells lack mature plastids and are incapable of photosynthesis. The cells are therefore dependent on the Suc in the growth medium for their sole source of carbon. Suspension cell cultures also allow for homogeneity in the type of tissue used for the experiment, thus synchronizing the onset of starvation responses. To analyze the effect of Suc depletion on cell morphology and growth, suspension cultures were grown for 3 d in the presence or absence of Suc. Samples were taken at 0, 6, 24, 48, and 72 h and used for microscopy and RNA extraction.
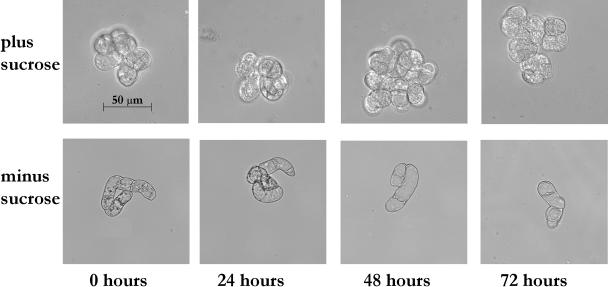
Light microscopy of the cells at various time points during starvation revealed similar morphology to that seen in previous studies in tobacco (Moriyasu and Ohsumi, 1996) and Arabidopsis (Bassham, 2002). The size of the vacuole began to increase and the cytoplasmic volume was reduced after 24 h of Suc starvation (Fig. 1). After 48 h of starvation, much of the cytoplasm, including its organelles, was gone, and the vacuolar compartment took up most of the cellular volume, whereas after 72 h, the incidence of cell death increased. Autophagosome formation was observed in the cells after 24 and 48 h of starvation (A.L. Contento, Y. Xiong, and D.C. Bassham, unpublished data), and an increase in expression of the autophagy marker APG8 occurred at these time points (see Fig. 7). These cytological changes indicate that autophagy is induced by 24 h after transfer to Suc-free medium, and continues for at least another 24 h.
Figure 1.
Cellular morphology changes during Suc starvation. Arabidopsis suspension cell cultures were transferred to Suc-free medium and subcellular structure analyzed after 0, 24, 48, and 72 h, compared with control cultures. Scale bar is equal to 50 μm.
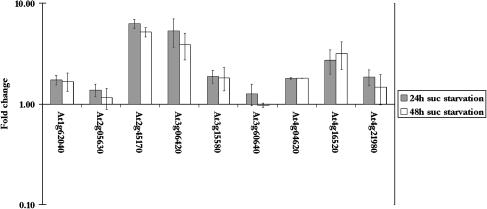
Figure 7.
Changes in transcript levels of putative APG8/ATG8 genes in Arabidopsis suspension cells during Suc starvation. The fold change in transcript level after 24 h (gray bars) and 48 h (white bars) was calculated from the mean intensities of two sets of biological replicates. Error bars indicate se. The scale is logarithmic.
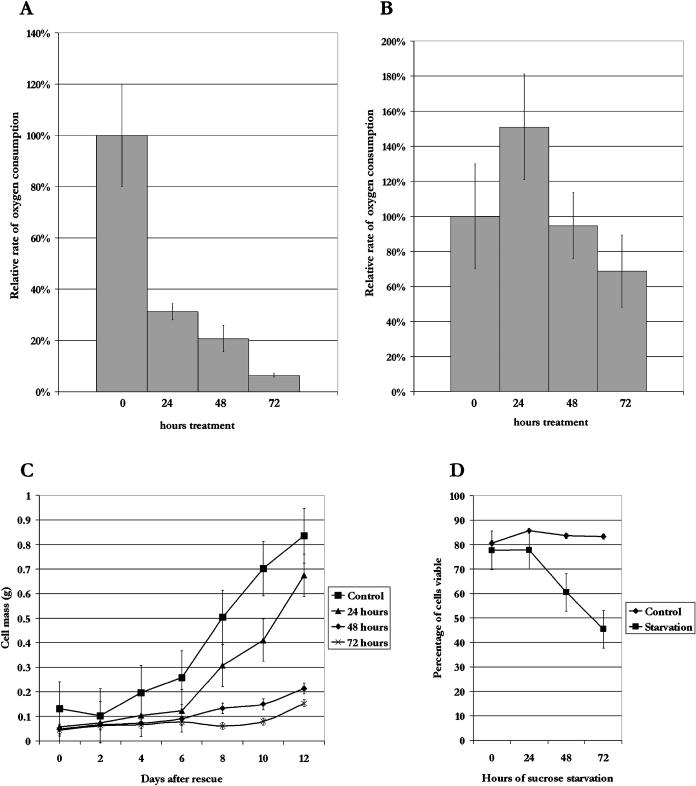
To correlate these morphological changes with physiological activity of the cultures, respiration rate was measured during starvation. Compared with the 0-h time point, the rate of respiration was reduced by approximately 70% after 24 h of starvation, and still further after 48 h of starvation to approximately 20% of the control (Fig. 2A). By 72 h, respiration decreased almost to zero, potentially reflecting a loss in culture viability and cell death. In contrast, a small increase was seen after 24 h in control (plus Suc) time courses (Fig. 2B), possibly due to the transfer to fresh medium. Even after 72 h, only a 30% decrease in respiration rate was observed, indicating that control cultures are active throughout the study. In addition, the total RNA level in the cells remained constant for the first 48 h of starvation, whereas after 72 h, total RNA levels decreased dramatically (data not shown), also suggesting that cell death is occurring by this time point.
Figure 2.
Analysis of oxygen consumption and culture viability during Suc starvation. Suspension cultures were grown with or without Suc for 0, 24, 48, or 72 h. The rate of respiration per gram of cells was determined for starved cells (A) and for control cells (B). Oxygen consumption rate is represented as a percentage of the 0-h control sample. C, Suspension cultures were starved of Suc for 0, 24, 48, or 72 h, after which they were rescued by replacing the starvation medium with Suc-containing medium. Samples (5 mL) were taken every 48 h after rescue for 12 d and the fresh weight of cells was measured for each sample. D, Suspension cell cultures were starved for 3 d, with 5-mL samples taken every 24 h. The cells were stained with fluorescein diacetate and viewed under a UV fluorescence microscope to determine the percentage of viable cells. Error bars indicate se.
To determine the viability of the culture during starvation, suspension cells were grown in the absence of Suc for up to 3 d, after which the medium was replaced with medium containing Suc, and growth continued. The fresh weight of the cells was measured during the rescue period to evaluate recovery of the cultures (Fig. 2C). Cultures starved for 24 h were able to recover in a manner similar to nonstarved cultures, but by 48 h, a decrease in the viability of the culture was observed. The cultures starved for 48 and 72 h never resumed growth, even after several weeks, indicating that starvation is irreversible by 48 h. The small increase in fresh weight of 48- and 72-h cultures during the time course in Figure 2C may be due to an increase in water content, rather than growth of the cells. A similar result was observed in starved maize root tips, where water content increased steadily during starvation, leading to an increase in fresh weight, even though dry weight decreased (Brouquisse et al., 1991). To determine the percentage of living cells, cells were stained with fluorescein diacetate at each time point after initiation of starvation, and the percentage of viable cells was calculated (Fig. 2D). Control samples, washed with Suc-containing medium, consistently demonstrated approximately 80% living cells. Viability was maintained after 24 h of starvation, while the number of living cells dropped sharply afterward, to less than 50% after 72 h of starvation. When the vital staining data are taken together with the cell rescue data, it is evident that while more than half of the cells are still living by 48 h of starvation, and respiration is maintained at similar rates to cultures starved for 24 h, the culture is not able to recover. This suggests that a change in the physiology of the cell culture is taking place between 24 and 48 h of starvation, leading to a loss of reversibility of starvation.
Increase in Expression of Genes Encoding Vacuolar Proteases during Starvation
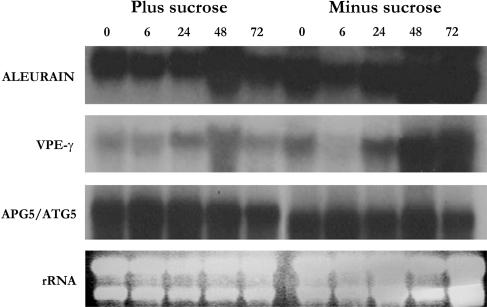
A number of sugar-responsive genes have previously been identified (e.g. Sheen, 1990; Koch 1996; Fujiki et al., 2000), and typically increase in expression rapidly after removal of Suc. However, our focus is on nutrient mobilization processes during starvation, rather than direct sugar regulation, and these processes occur at much later time points after sugar removal. Expression of genes encoding the vacuolar proteases aleurain-like protein (AALP; Ahmed et al., 2000) and VPE-γ (Kinoshita et al., 1999) are known to increase under conditions of stress, including Suc starvation, potentially to break down and recycle damaged and nonessential proteins (Bassham, 2002). The increase in the expression of these vacuolar genes was therefore used as a marker to determine when nutrient remobilization occurred, using the stably expressed autophagy 5 gene (APG5/ATG5) as a control (Fig. 3). AALP and VPE-γ transcript levels were found to increase as the starvation time course progressed, with maximum transcript levels observed after 48 h of starvation, whereas no increase in expression was seen in nonstarved control cultures. These data suggest that an increase in vacuolar function and autophagy occurs within 48 h of starvation in Arabidopsis suspension cultures as a component of the nutrient stress response.
Figure 3.
Transcript levels of vacuolar enzymes increase during Suc starvation. Total RNA was extracted from suspension cells after 0, 6, 24, 48, or 72 h of Suc starvation, or control cells grown in the presence of Suc. RNA gel blots were probed with labeled cDNAs for the vacuole-specific proteases aleurain (AALP; At5g60360) and VPE-γ (At4g32940). An APG5/ATG5 (At5g17290) probe and ethidium bromide-stained rRNA were used as controls for equal loading.
Gene Expression Profiles in Response to Suc Starvation
From the above data, most of the morphological and molecular responses of Arabidopsis suspension cells to starvation occur by 48 h in Suc-free medium. We therefore examined the changes in gene expression that occur between 0 and 48 h of Suc starvation. RNA was extracted from cultures grown for 0, 24, or 48 h after transfer to starvation medium, or 48 h in Suc-containing medium as a control. As an additional control for osmotic differences in the growth media, in one sample Suc was replaced with polyethylene glycol (PEG) to maintain iso-osmotic conditions. These total RNA samples were used as templates for labeled cRNA synthesis and hybridized to Arabidopsis ATH1 Genome GeneChip microarrays (Affymetrix, Santa Clara, CA), which contain probe sets representing 22,810 unique genes. Microarray Suite 5.0 (Affymetrix) and GeneSpring (Silicon Graphics, Redwood, CA) were used to normalize the data from each experiment, and probe sets showing changes in transcript levels in the starved cells, as compared to the nonstarvation controls, were identified.
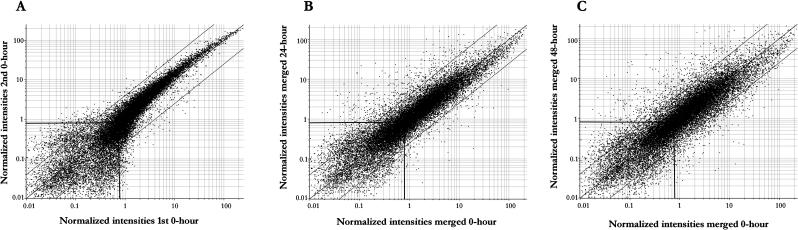
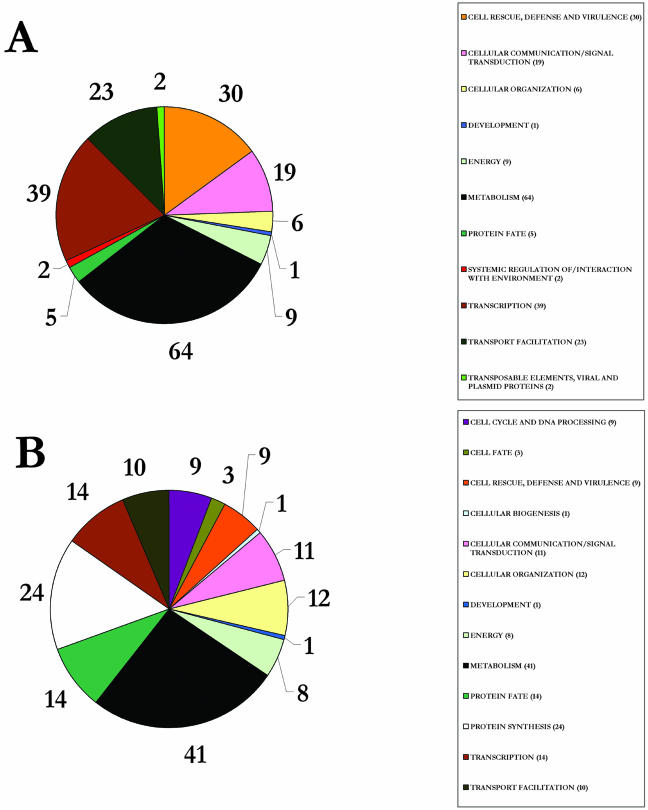
Scatter plots comparing expression data between biological replicates of control samples (0 h; Fig. 4A) demonstrated that the majority (99%) of the genes varied in expression by less than fourfold. It was therefore decided that a fourfold cutoff would be used to identify expression changes between samples, as this would allow identification of differentially expressed genes with a high confidence. Genes that fell outside of this cutoff when comparing replicate samples were excluded from further analysis. Probe sets showing normalized signal intensities of less than 0.8 in all samples were considered to be below the limit of detection and were not analyzed further. Figure 4, B and C, show scatter plots of signal intensities from 24- and 48-h starved samples against the control 0-h sample. RNA levels corresponding to many different genes can be seen to increase or decrease dramatically during Suc starvation. Genes showing a fourfold increase or decrease in transcript levels in at least one of the starvation samples, compared to the 0- and 48-h control samples, were identified as being significantly induced. In addition, any transcripts whose expression did not change in the PEG osmotic control, as compared with nonstarved controls, were discarded. Using these criteria, a total of 343 genes was found to have increased mRNA levels when subjected to Suc starvation, whereas the mRNA level of 263 genes decreased (see supplemental data, available at www.plantphysiol.org). The genes were assigned a functional classification based on the Munich Information Center for Protein Sequencing (MIPS) database (Fig. 5; Schoof et al., 2002). Approximately 40% of the genes identified are not yet classified based on known function and/or sequence or structure homology.
Figure 4.
Scatter plots of Arabidopsis ATH1 GeneChip data. Normalized signal intensities are plotted, with guide lines on each graph representing a fourfold change, increasing or decreasing in signal, and indicating a signal intensity of 0.8, used as the lower limit of detection. A, Two biological replicates are compared for the 0-h time point. B, Comparison of 0- and 24-h starvation samples. C, Comparison of 0- and 48-h starvation samples. For B and C, signal intensities are the average of two biological replicates.
Figure 5.
Functional categories of genes. Each gene was assigned a functional category based on the known or putative function of its protein, according to the Munich Information Center for Protein Sequencing Functional Category database. Pie charts show the number of probe sets identified in each category that show at least a 4-fold increase (A) or decrease (B) in transcript level during starvation. Those categorized as unclassified, 143 probe sets for A and 104 probe sets for B, are excluded from the pie charts for clarity.
Genes Up-Regulated during Starvation
Nine of the genes that increase in expression during starvation are classified as being involved in energy production, including two light-harvesting chlorophyll a/b-binding proteins and five PSI and PSII component proteins. A number of photosynthetic genes have been found to be induced by sugar starvation in maize (Sheen, 1990), and the increase in transcript levels of these genes in our samples could be considered a response designed to increase the level of photosynthesis, even though the suspension culture used is nonphotosynthetic.
Over 15% of the identified genes are involved in metabolic processes including carbohydrate metabolism, Tyr, Ile, and Val amino acid metabolism, protein and lipid degradation, and trehalose metabolism. Numerous members of various glycosyl hydrolase families are represented in the data and may be involved in carbohydrate degradation. Two forms of branched-chain-α-keto acid dehydrogenase may be involved in amino acid metabolism and have been documented as expressed during darkness-induced starvation (Fujiki et al., 2000, 2002). A group of putative lipases may be involved in lipid degradation during starvation. Twenty-four of the genes identified encode proteins potentially involved in transport facilitation, including a number of carbohydrate and ion transporters, presumably for the scavenging of nutrients. These metabolic and transport proteins are most likely involved in a cellular response to compensate for the Suc starvation by seeking out new sources of nutrients and initiating the use of other metabolic substrate pathways.
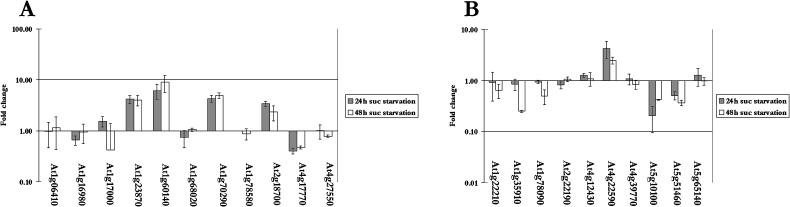
Three of the 11 trehalose-6-phosphate synthases (TPS; Fig. 6A) and one of the 10 trehalose 6-phosphate phosphatases (TPP; Fig. 6B) found in Arabidopsis are significantly up-regulated upon Suc starvation. Trehalose 6-phosphate (T-6-P) levels have been shown to regulate carbohydrate utilization and growth through regulation of glycolysis (Schluepmann et al., 2003). Schluepmann et al. have shown in Arabidopsis that supplied sugar leads to a reduction in T-6-P levels, which leads to decreased carbohydrate metabolism and growth. An increase in T-6-P levels, via increased levels of TPS, should cause an increase in carbohydrate utilization. A knockout mutant in the TPS1 gene is embryo lethal, indicating that this gene is required for embryo development (Eastmond et al., 2002). Interestingly, the mutant embryos arrest at the time when deposition of storage reserves is initiated, and a dramatic increase in the sugar supply to the embryo occurs in wild-type seeds, again indicating a correlation between sugar metabolism and T-6-P level. The physiological function of the remaining TPS genes is as yet unknown, but the increase in expression of three of these genes during Suc starvation may also lead to an increase in T-6-P and therefore to enhanced carbohydrate metabolism.
Figure 6.
Genes encoding trehalose metabolic enzymes change in expression during starvation. The fold change in transcript level of TPS (A) and TPP (B) genes after 24 h (gray bars) and 48 h (white bars) was calculated from the mean intensities of two sets of biological replicates. Error bars indicate se. The scale is logarithmic.
Two TPP genes also change in expression during starvation, one increasing and the other decreasing in mRNA level (Fig. 6B). TPP breaks down T-6-P to trehalose, thus having an opposing effect on T-6-P levels when compared with TPS. When TPS and TPP were expressed in rice as a gene fusion, an increase in trehalose levels was observed, and the transgenic plants showed an increased tolerance to various abiotic stresses (Garg et al., 2002). We also observed that two of the TPP genes were responsive to changes in osmotic potential (At5g10100 and At5g51460 in Fig. 6B). Clearly, much additional work is needed before the precise function of T-6-P and trehalose in starvation and other stress conditions, and the role of each of the TPS and TPP isoforms, can be determined.
Morphological analysis of suspension cells during Suc starvation (Fig. 1) indicated that vacuolar autophagy occurred by 24 h of starvation. In agreement with this, a transcript encoding a homolog of the yeast APG8/ATG8 protein displayed increased transcript levels under these conditions (Fig. 7). APG8/ATG8 in yeast is thought to be required for lipid recruitment and regulation of the size of autophagosomes during autophagy (Kirisako et al., 1999, 2000; Ichimura et al., 2000). An increase in the amount of Arabidopsis APG8/ATG8 therefore suggests an increase in autophagy. There are nine homologs of the yeast APG8/ATG8 gene in Arabidopsis, but only one of them (At2g45170) increased significantly in expression due to starvation of the suspension cells (Fig. 7), possibly indicating a functional specialization of members of the gene family. A second APG8/ATG8 gene (At3g06420) increased during starvation only under osmotic stress conditions, suggesting that members of this family may be involved in response to diverse environmental stresses.
Thirty-nine genes with increased expression during starvation are predicted to be involved in transcription, including a group of 4 basic region/Leu zipper (bZIP) motif transcription factors, 6 MYB transcription factors, 2 WRKY transcription factors, and a RAV1 DNA-binding element (Table I). The bZIP transcription factor family has 75 members that have been shown to regulate processes including pathogen defense, light and stress signaling, seed maturation, and flower development (for review, see Jakoby et al., 2002). The RAV1 DNA-binding protein has been cloned from Arabidopsis and contains amino acid sequence domains that are unique to plants (Kagaya et al., 1999). No specific physiological function or target genes are currently known for these proteins, but the promoters of a 2-Cys peroxiredoxin gene and an auxin-binding protein gene have been shown to contain binding regions for RAV1. However, neither of these genes shows increased expression in our experiment. Three MYB proteins have recently been found to mediate α-amylase expression during periods of sugar starvation in rice (Lu et al., 2002). It is possible that the six MYB family proteins found during this study could have similar regulatory control in Arabidopsis during nutrient stress. The WRKY transcription factor family has 74 members in Arabidopsis and they have been suggested to be involved in responses to biotic and abiotic stresses, defense responses, and regulation of senescence (for review, see Eulgem et al., 2000).
Table I.
Transcription-related genes that show a significant increase in transcript level in Arabidopsis suspension cells during Suc starvation
| Probe Seta | AGIb | 24c | 48d | Description |
|---|---|---|---|---|
| 260999_at | At1g26580 | 2.15 | 3.68 | Hypothetical protein; similar to putative MYB family transcription factor |
| 260887_at | At1g29160 | 2.31 | 3.54 | Dof zinc finger protein; similar to ascorbate oxidase promoter-binding protein |
| 260882_at | At1g29280 | 4.92 | 6.55 | WRKY family transcription factor |
| 262552_at | At1g31350 | 3.65 | 4.36 | F-box protein family |
| 260632_at | At1g62360 | 5.89 | 3.79 | Homeobox protein, putative |
| 264942_at | At1g67340 | 3.89 | 4.36 | F-box protein family |
| 260220_at | At1g74650 | 1.91 | 3.68 | MYB family transcription factor |
| 259982_at | At1g76410 | 30.63 | 29.54 | Putative RING zinc finger protein |
| 261395_at | At1g79700 | 25.56 | 31.83 | Ovule development protein, putative; similar to AINTEGUMENTA |
| 265321_at | At2g18280 | 3.93 | 8.09 | F-box containing tubby family protein |
| 266695_at | At2g19810 | 3.96 | 4.54 | Putative CCCH-type zinc finger protein |
| 245078_at | At2g23340 | 16.13 | 25.39 | Putative AP2 domain transcription factor |
| 265989_at | At2g24260 | 2.35 | 4.09 | bHLH protein |
| 266656_at | At2g25900 | 7.85 | 9.86 | Putative CCCH-type zinc finger protein |
| 258975_at | At3g01970 | 3.30 | 4.18 | WRKY family transcription factor |
| 258795_at | At3g04570 | 2.48 | 4.05 | Expressed protein; similar to putative DNA-binding proteins |
| 258890_at | At3g05690 | 6.32 | 4.08 | Putative transcription factor |
| 256255_at | At3g11280 | 10.14 | 9.51 | MYB family transcription factor, putative |
| 257265_at | At3g14980 | 6.53 | 8.35 | PHD-finger protein, putative |
| 256914_at | At3g23880 | 8.02 | 13.46 | F-box protein family |
| 252751_at | At3g43430 | 5.50 | 7.12 | Putative protein; RING-H2 zinc finger protein ATL4 |
| 252340_at | At3g48920 | 47.63 | 52.86 | MYB family transcription factor |
| 252081_at | At3g51910 | 3.98 | 5.94 | Heat shock transcription factor HSF30 |
| 254919_at | At4g11360 | 3.76 | 3.22 | RING-H2 finger protein RHA1b |
| 245331_at | At4g14410 | 4.06 | 4.28 | bHLH protein |
| 254681_at | At4g18140 | 2.80 | 3.86 | Putative protein; HYA22 protein |
| 253872_at | At4g27410 | 4.68 | 9.62 | Putative protein |
| 253061_at | At4g37610 | 9.86 | 10.66 | Putative protein; SPOP |
| 251066_at | At5g01880 | 31.26 | 34.96 | Putative protein; RING-H2 finger protein RHA3a |
| 250463_at | At5g10030 | 5.55 | 3.40 | bZIP transcription factor, OBF4 |
| 246432_at | At5g17490 | 5.21 | 5.20 | RGA-like protein; putative member of the VHIID domain transcription factor family RGAL |
| 246755_at | At5g27920 | 3.48 | 5.16 | F-box protein family |
| 245925_at | At5g28770 | 8.42 | 9.33 | bZIP family transcription factor; similar to seed storage protein opaque-2(bZIP family) |
| 248606_at | At5g49450 | 13.82 | 12.58 | bZIP family transcription factor |
| 248618_at | At5g49620 | 2.61 | 3.74 | MYB family transcription factor |
| 247868_at | At5g57620 | 3.80 | 5.96 | MYB family transcription factor |
| 247696_at | At5g59780 | 3.19 | 4.72 | MYB family transcription factor |
| 247199_at | At5g65210 | 3.97 | 4.36 | bZIP transcription factor, TGA1 |
| 247052_at | At5g66700 | 3.76 | 2.32 | Homeodomain transcription factor-like |
The fold change values reported were calculated using the mean intensities of two sets of biological replicates.
Affymetrix probe set.
Arabidopsis Genome Initiative number.
Fold change between the normalized intensities of 0-h Suc control sample and 24-h Suc starvation sample.
Fold change between the normalized intensities of 0-h Suc control sample and 48-h Suc starvation sample.
Thirty of the genes are known or predicted to function in cell rescue, defense, and virulence. These genes included reactive oxygen-scavenging enzymes and several disease resistance proteins, and are most likely involved in the nonspecific response to the stress of Suc starvation in Arabidopsis cells. Catalase-3 was found to have a large increase in expression. Of the three Arabidopsis catalase proteins, catalase-3, a class III catalase, is commonly found in glyoxysomes and may be involved in the removal of the H2O2 created during β-oxidation (Willekens et al., 1995; Dat et al., 2000). Three genes for DnaJ proteins also show an increase in transcript levels. The J-domain family of proteins has 89 members in Arabidopsis and DnaJ-like proteins can act independently or in conjugation with DnaK and GrpE as molecular chaperones during environmental stresses, such as heat shock (Miernyk, 2001). Interestingly, the three genes showing an increase in transcript levels during Suc starvation all lie within the same subgroup of J-domain proteins and have all been predicted to be localized within the plastid. It is possible that these proteins could be involved in proper chloroplast assembly during periods of nutrient stress. Five potential disease resistance Toll/interleukin-1 receptor class proteins (R genes) were also found during this study. Many R genes are involved in the local hypersensitive defense response to pathogen attack (Meyers et al., 2002), although the specific function of the genes that we identified is not yet known.
Nineteen of the genes are predicted to function in cellular communication and signal transduction (Table II), including seven protein kinases. The transcript levels of three of these genes, an ethylene-responsive element-binding factor, a putative phosphatidylinositol-4-phosphate-5-kinase, and an auxin-regulated protein, are all found to increase during phosphate starvation as well (Wu et al., 2003). It is possible that these genes may be involved in a general starvation response, although, in contrast to Suc starvation, phosphate starvation involves a decrease in photosynthesis and carbon metabolism. However, it may be suggested that a lack of different nutrients induces the expression of similar genes and thus activates the same signal transduction pathway.
Table II.
Cellular communication and signal transduction-related genes that show a significant increase in transcript level in Arabidopsis suspension cells during Suc starvation
| Probe Seta | AGIb | 24c | 48d | Description |
|---|---|---|---|---|
| 263657_at | At1g04440 | 4.89 | 6.48 | Putative casein kinase I |
| 262531_at | At1g17230 | 2.42 | 7.05 | Putative Leu-rich receptor protein kinase |
| 260855_at | At1g21920 | 3.15 | 4.71 | Phosphatidylinositol-4-phosphate-5-kinase, putative |
| 260974_at | At1g53440 | 3.57 | 4.56 | Receptor-like Ser/Thr kinase, putative |
| 260774_at | At1g78290 | 10.00 | 12.97 | Ser/Thr protein kinase, putative |
| 267477_at | At2g02710 | 6.21 | 7.64 | Putative receptor-like protein kinase |
| 266019_at | At2g18750 | 3.24 | 4.93 | Calmodulin-binding protein |
| 267254_at | At2g23030 | 7.74 | 13.16 | Putative protein kinase |
| 266324_at | At2g46710 | 4.89 | 4.09 | Putative rac GTPase-activating protein |
| 257892_at | At3g17020 | 2.52 | 4.10 | Expressed protein; similar to ER6 protein |
| 251665_at | At3g57040 | 3.53 | 5.37 | Response reactor 4 |
| 254901_at | At4g11530 | 6.30 | 4.79 | Ser/Thr kinase-like protein (fragment); receptor-like protein kinase RLK3 |
| 250981_at | At5g03140 | 4.32 | 7.35 | Receptor like protein kinase |
| 250697_at | At5g06800 | 1.87 | 3.55 | Putative protein; contains similarity to transfactor |
| 246028_at | At5g21170 | 6.50 | 5.47 | AKIN beta1 |
| 249550_at | At5g38210 | 3.22 | 3.67 | Wall-associated kinase 4 (wak4) |
| 249361_at | At5g40540 | 2.70 | 3.63 | Protein kinase ATN1 |
| 247790_at | At5g58720 | 2.71 | 3.80 | Putative PRL1-associated protein |
| 247540_at | At5g61590 | 15.19 | 17.63 | Ethylene-responsive element-binding factor-like ERF5_ARATH |
The fold change values reported were calculated using the mean intensities of two sets of biological replicates.
Affymetrix probe set.
Arabidopsis Genome Initiative number.
Fold change between the normalized intensities of 0-h Suc control sample and 24-h Suc starvation sample.
Fold change between the normalized intensities of 0-h Suc control sample and 48-h Suc starvation sample.
Genes Down-Regulated during Starvation
Our main focus is on genes that increase in expression during starvation, as these are likely to function in the stress response. However, an examination of genes that decrease in mRNA level during Suc starvation reveals a striking number of genes involved in translation (Fig. 5B). These include 19 ribosomal proteins and two translation initiation factors, eIF4A and eIF2γ (Table III). This indicates that a general decrease in protein synthesis may occur, particularly at later time points during starvation, as the mRNA levels continue to decrease throughout the time course. It is known that, in yeast and animal cells, translational activity is tightly linked to nutritional status and is controlled by the target of rapamycin (TOR) kinase pathway (Barbet et al., 1996; Proud, 2004). A TOR homolog is present in Arabidopsis (Menand et al., 2002), and it is likely that translation is also regulated by TOR during starvation in plant cells.
Table III.
Genes involved in protein synthesis showing a significant decrease in transcript level in Arabidopsis suspension cells during Suc starvation
| Probe Seta | AGIb | 24c | 48d | Description |
|---|---|---|---|---|
| 256065_at | At1g07070 | −2.48 | −4.99 | Ribosomal protein, putative; similar to ribosomal protein L35a GI:57118 (Rattus norvegicus); supported by full-length cDNA: Ceres: 2778. |
| 263691_at | At1g26880 | −2.57 | −4.86 | 60s ribosomal protein L34; identical to GB:Q42351, location of EST 105E2T7, gb|T22624; supported by full-length cDNA: Ceres:7182. |
| 265147_at | At1g51380 | −2.08 | −5.76 | Eukaryotic translation initiation factor 4A (eIF-4A), putative; contains DEAD/DEAH box helicase; similar to PIR:S71280 from (Arabidopsis) |
| 259678_at | At1g77750 | −2.64 | −3.80 | Putative 30S ribosomal protein S13; similar to putative 30S ribosomal protein S13, chloroplast precursor GB:P42732 (Arabidopsis) |
| 266061_at | At2g18720 | −1.29 | −4.98 | Translation initiation factor eIF2γ subunit, putative |
| 266980_at | At2g39390 | −2.26 | −4.43 | 60S ribosomal protein L35; supported by full-length cDNA: Ceres:11583. |
| 266822_at | At2g44860 | −2.17 | −3.59 | 60S ribosomal protein L30; supported by full-length cDNA: Ceres:34564. |
| 259130_at | At3g02190 | −2.28 | −3.61 | Putative ribosomal protein L39; similar to ribosomal protein L39 GB:P51424 (Arabidopsis); supported by full-length cDNA: Ceres:946. |
| 258521_at | At3g06680 | −2.83 | −6.50 | Ribosomal protein L29, putative; similar to 60S ribosomal protein L29 GB:P25886 from (Rattus norvegicus) |
| 258532_at | At3g06700 | −2.72 | −5.88 | Ribosomal protein L29, putative; similar to ribosomal protein L29 GI:7959366 (Panax ginseng); supported by full-length cDNA: Ceres:315. |
| 258296_at | At3g23390 | −2.10 | −3.89 | Putative ribosomal protein; similar to ribosomal protein L41 GB:AAA34366 (Candida maltosa); supported by full-length cDNA: Ceres: 13557. |
| 252643_at | At3g44590 | −2.94 | −5.13 | Acidic ribosomal protein P2-like; acidic ribosomal protein P2, maize, PIR:S54179 |
| 252287_at | At3g49080 | −2.29 | −4.38 | 30S ribosomal protein S9-like; similar to 30S ribosomal proteins |
| 251538_at | At3g58660 | −2.91 | −5.45 | Putative protein; PBK1 protein, Homo sapiens, EMBL:HSA7398; supported by cDNA: gi_17979348 |
| 255657_at | At4g00810 | −2.67 | −4.32 | Acidic ribosomal protein p1; similar to acidic ribosomal protein p1; supported by full-length cDNA: Ceres:26442. |
| 255455_at | At4g02930 | −2.09 | −3.89 | Mitochondrial elongation factor Tu |
| 254981_at | At4g10480 | −2.61 | −4.77 | Putative alpha NAC; stung similarity to nascent polypeptide associated complex α-chain-human, PIR2:S49326; supported by cDNA: gi_15027908_gb_AY045811.1_ |
| 254763_at | At4g13170 | −2.76 | −5.52 | Ribosomal protein L13a-like protein; ribosomal protein L13a, Lupinus luteus, PID:e1237871; supported by cDNA: gi_15529277_gb_AY052263.1_ |
| 245311_at | At4g14320 | −1.85 | −3.59 | Ribosomal protein; supported by full-length cDNA: Ceres: 18153. |
| 254355_at | At4g22380 | −2.29 | −5.76 | Ribosomal protein L7Ae-like; NHP2/RS6 FAMILY PROTEIN, Homo sapiens, PID:g4826860; supported by cDNA: 2673. |
| 253598_at | At4g30800 | −3.79 | −7.85 | Ribosomal protein S11-like; ribosomal protein S11, Arabidopsis, PIR2:C35542; supported by cDNA: gi_15028244_gb_AY046037.1_ |
| 247900_at | At5g57290 | −1.91 | −3.55 | 60S acidic ribosomal protein P3; supported by full-length cDNA: Ceres: 8695. |
| 247739_at | At5g59240 | −3.49 | −7.28 | 40S ribosomal protein S8-like; 40S ribosomal protein S8, Prunus armeniaca, EMBL:AF071889 |
| 247566_at | At5g61170 | −2.33 | −3.93 | 40S ribosomal protein S19-like; 40S ribosomal protein S19, Oryza sativa, SWISSPROT:RS19_ORYSA |
The fold change values reported were calculated using the mean intensities of two sets of biological replicates.
Affymetrix probe set.
Arabidopsis Genome Initiative number.
Fold change between the normalized intensities of 0-h Suc control sample and 24-h Suc starvation sample.
Fold change between the normalized intensities of 0-h Suc control sample and 48-h Suc starvation sample.
Genes involved in cell division also decrease in expression during Suc starvation (Fig. 5B), as might be expected with growth arrest of the culture. These include the Arabidopsis homologs of the DNA replication factors RPA1 (Van der Knaap et al., 1997) and MCM3 (Stevens et al., 2002), two B-type cyclins (Ito et al., 1998), and the cytokinesis-specific syntaxin KNOLLE (Lukowitz et al., 1996; Lauber et al., 1997). Each of these genes is known to be under cell cycle control, either in Arabidopsis or in other species, and the decrease in expression during starvation may therefore be due to a cessation in cell division. In addition, seven genes encoding putative histone proteins were identified. As histones are synthesized preferentially during the S-phase (Callard and Mazzolini, 1997), although lower amounts are synthesized throughout the cell cycle, the reduced expression during starvation could again be linked to cell division arrest.
Analyses of Expression Profiles of Up-Regulated Genes
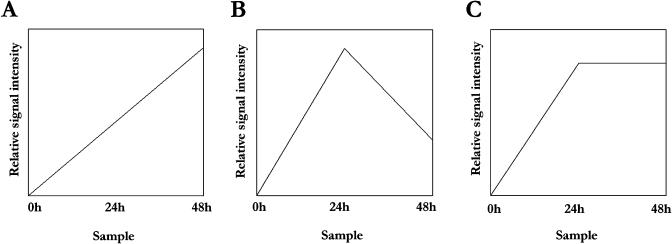
The expression patterns of the 343 starvation-induced transcripts were classified into three groups, using k-means cluster analysis and Pearson correlations corresponding to (1) an increase in RNA level from 24 to 48 h; (2) a decrease from 24 to 48 h; and (3) no change between 24 and 48 h of starvation (Fig. 8). As might be expected, genes involved in metabolism and transcription, the two categories containing the highest number of genes, were spread throughout the three groups. In contrast, most of the genes classified as functioning in membrane transport and those in stress responses and defense were found in groups 1 and 3, with very few genes showing a decrease in expression at the later time points. These gene products, therefore, appear to be required throughout the time course, indicating that nutrient scavenging and general stress responses continue even during extreme starvation conditions. Strikingly, a large number of the genes predicted to function in disease resistance or pathogen response increased in expression throughout the time course. A characteristic feature of plant-pathogen responses is cell death, and one possibility is that the increase in expression of these genes plays a role in the loss of culture viability seen after 48 h of starvation. Several potential plasma membrane aquaporins also increase at 48 h, and it is possible that these proteins cause changes in membrane permeability leading to cell death. Brouquisse et al. (1991) measured the intracellular osmolarity in excised maize root tips during Suc starvation and saw a sharp decrease in osmolarity concomitant with loss of reversibility of the starvation responses. They proposed that this is likely to be due to a rapid increase in permeability of the plasma membrane and tonoplast, eventually causing cell death, and a similar process may be occurring in our suspension cultures. Genes involved in signal transduction showed perhaps the most interesting trend, with subsets of transcripts increasing or decreasing between 24 and 48 h of starvation, but very few remaining constant in expression between these times. This suggests that distinct signaling pathways are activated at different starvation time points, and the kinetics of activation of these pathways may hint as to their role in the nutrient stress response.
Figure 8.
Expression patterns observed over a 48-h Suc starvation time course. Three distinct patterns of gene expression were identified in the subset of genes that were up-regulated by Suc starvation, illustrated by plotting relative signal intensity for 0-h control and 24- and 48-h Suc starvation samples. A, A significant increase in signal intensity from 24 to 48 h is observed. This pattern correlates with k-means cluster group 1. B, A decrease in relative signal intensity is seen between the 24- and 48-h starvation samples. This pattern correlates with k-means cluster group 2. C, No significant change in signal intensities is seen between the 24- and 48-h starvation samples. This pattern correlates with k-means cluster group 3.
The promoter regions (1,000 nucleotides upstream of the start codon) of genes in each of the three groups were examined for the presence of common regulatory elements using the AlignAce program (Hughes et al., 2000). However, no clear candidates for cis-acting motifs were obtained. In addition, promoters were analyzed for known motifs in the PLACE database (Higo et al., 1999) using the Signal Scan program (Prestridge, 1991). Several of the genes in group 1 had a large number (up to 11) of WRKY transcription factor binding motifs in their upstream regions, and two WRKY transcription factors are also in this group, suggesting that they may be involved in the regulation of these genes. However, these motifs are commonly found within the genome, and the significance of this finding remains to be determined experimentally.
Verification of GeneChip Expression Data
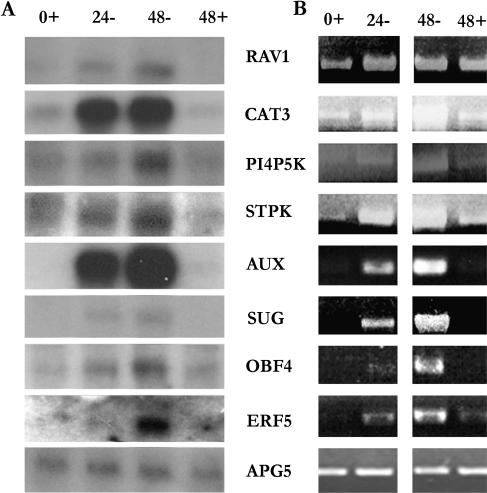
Northern-blot analysis was used to verify the increased transcript levels of eight genes identified by the GeneChip analysis: RAV1 DNA-binding protein (RAV1), catalase-3 (CAT3), a putative phosphatidylinositol-4-phosphate-5-kinase-like protein (PI4P-5K), a Ser/Thr protein kinase (STPK), a potential auxin-regulated protein kinase (AUX), a sugar transporter (SUG), a bZIP family transcription factor (OBF4), and an ethylene-responsive element-binding factor (ERF5). These genes were chosen to represent diverse functional categories and various overall transcript levels. In each case, the trend in transcript level was the same for the northern-blot hybridizations and GeneChip analysis (Fig. 9A).
Figure 9.
Verification of GeneChip expression data by RNA blot analysis and RT-PCR. Total RNA from Arabidopsis suspension cell cultures after 24- and 48-h Suc starvation (24 and 48), and 0- and 48-h nonstarved controls (0+ and 48+) was used for RNA blot hybridizations (A) and for RT-PCR reactions (B) for eight genes identified by the GeneChip analysis as induced during starvation. The genes chosen were: At1g13260 (RAV1), a DNA-binding protein; At1g20620 (CAT3), catalase-3; At1g21920 (PI4P5K), a phosphoinositol-4-phosphate-5-kinase; At1g78290 (STPK), a Ser/Thr kinase; At2g33830 (AUX), a putative auxin-regulated protein kinase; At4g36670 (SUG), a sugar transporter; At5g10030 (OBF4), a bZIP transcription factor; and At5g61590 (ERF5), an ethylene-responsive element-binding factor. The Arabidopsis APG5/ATG5 gene (At5g17290) was used as a loading control in both experiments.
As several of these genes are members of gene families that could potentially cross-hybridize with the above probes, the results were confirmed by reverse transcription (RT)-PCR using transcript-specific primers (Fig. 9B). The identity of the RT-PCR products was confirmed by DNA sequencing. In each case, a good correlation was observed between the RNA blots, RT-PCR, and GeneChip analysis (Fig. 9B), confirming that the GeneChip analysis has reliably identified specific genes whose transcript levels increase during carbon starvation.
DISCUSSION
Suc Starvation in Arabidopsis Suspension Cells Induces Increases in the Transcript Levels of 343 Genes
In this study, we report increases in the transcript levels of 343 distinct genes that we hypothesize are involved in the response to Suc starvation in Arabidopsis suspension cells. Zero-, 24-, and 48-h starvation time points were chosen for GeneChip analysis, based on the observation that most of the morphological changes of the suspension cultures occurred between 24 and 48 h after transfer to Suc-free medium, and that the increase in expression of vacuolar enzymes showed similar kinetics. Cluster analysis revealed that the genes showing an increase in mRNA level during starvation showed three possible expression patterns. Twenty-two genes showed a significant decrease in transcript levels between 24 and 48 h of starvation, and 84 genes showed a significant increase. The transcripts showing a decrease in expression after 24 h may potentially be involved with the initiation and regulation of the nutrient mobilization and recycling responses that occur at this time. These transcripts encode proteins with a wide variety of potential functions, including the SUC1 Suc transporter, a bZIP transcription factor, a homeodomain protein, and TPP. After 24 h of Suc starvation, they may be down-regulated during a shift to a response for the increasing severity of the nutrient stress. Processes that respond to these increasing levels of nutrient stress or that are involved in cell death may regulate the genes that showed an increase in expression after 24 h. Examples of these genes include three of the cytochrome p450 family genes, a putative heat shock transcription factor, and RAV1. These proteins could be required for a rising metabolic response to nutrient starvation or the activation of proteins designed to deal with the increasing level of stress. Surprisingly, expression of most of the genes did not change between 24 and 48 h, suggesting that many of the processes involved in the plant response to Suc starvation are maintained between 24 and 48 h.
One point that needs to be considered is the transition, occurring between 24 and 48 h of starvation, from a survival response to cell death, evident in the loss of culture and cell viability (Fig. 2). Autophagy is induced in the starved cultures by 24 h (Figs. 1 and 7), presumably to recycle nutrients for cell survival, and continues throughout the time course. In mammalian cells, autophagic or type II programmed cell death has been characterized as showing the morphological hallmarks of autophagy (Bursch et al., 2000), although the relationship between the mechanism of autophagy as a survival response and autophagic cell death has been the subject of some debate. Recently, two proteins involved in autophagy, ATG7 and Beclin1, identified originally in yeast as being required for survival during starvation (Kametaka et al., 1998; Mizushima et al., 1998b), have also been shown to function during autophagic cell death in mammals (Yu et al., 2004). This indicates that starvation-induced autophagy and autophagic cell death share at least some of the same machinery. In plants, cell death with morphological characteristics of autophagic cell death has been observed (Lam, 2004), and it is possible that a similar mechanism is responsible for cell death at later time points during Suc starvation.
Suc Starvation May Induce Changes to Carbohydrate Metabolism and Degradation of Proteins and Fatty Acids as Alternate Sources of Respiratory Substrates
With Suc as the sole carbon source and an inability to perform photosynthesis, the cells used in this study were forced to seek alternative sources of metabolic substrates to maintain cellular respiration. α- and β-galactosidases, as well as a number of other glycosyl hydrolases, showed increased transcript levels. This may suggest an attempt to switch to other types of carbohydrate metabolism, as an alternative source of metabolic substrates, by the suspension cells. Transcripts encoding enzymes involved in Tyr (homogentisate 1,2-dioxygenase), Ile, Leu, Val, (α-keto acid dehydrogenase), Lys (Lys-ketoglutarate reductase), and Arg catabolism (Orn aminotransferase) also increase during starvation. Increases in proteolysis have already been witnessed in starving maize root tips (James et al., 1993). This, coupled with the observed increase in the transcript levels of carboxypeptidases, suggests an increase in proteolysis and possible utilization of amino acids from unnecessary proteins as another source of carbon and/or energy to maintain vital functions in the starving cells. This protein degradation is likely to occur at least in part in the vacuole, as vacuolar autophagy increases during starvation by both morphological criteria (Fig. 1) and increase in expression of the autophagy-specific gene APG8/ATG8 (Fig. 7).
Increases in the transcript levels of a phospholipase, a triacylglycerol lipase, a putative lipase, and transcripts for genes involved in fatty acid oxidation suggest a possible increase in the breakdown of fatty acids as well. It has been reported that β-oxidation increases in plants during carbon starvation (Dieuaide et al., 1992, 1993), and transcript levels for a peroxisomal acyl-CoA synthase and a peroxisomal acyl-CoA oxidase, shown to be unique to plants and involved in short-chain acyl-CoA oxidation (Hayashi et al., 1999), increase under our experimental conditions. Acyl-CoA oxidase is a hydrogen peroxide-generating enzyme, and the increase in transcript for the peroxisomal catalase-3 may be in response to excess hydrogen peroxide produced by an increase in β-oxidation. A potential increase in Ile, Leu, and Val degradation is suggested by induction of two mitochondrial α-keto acid dehydrogenase (BCKDH) transcripts. Products from this degradation must feed into β-oxidation in order to be completely catabolized (Graham and Eastmond, 2002). An increase in transcript levels of an α-dioxygenase-peroxidase suggests that α-oxidation also increases. Hamburg et al. (1999) have suggested that α-oxidation is involved in the induction of the plant response to pathogens, and the products of α-oxidation may also act as signals for the response to nutrient stress. Overall, these results indicate an apparent switch toward the degradation and oxidation of fatty acids in carbon-starved cells.
Like Suc starvation responses, senescence also involves the breakdown of macromolecules and mobilization of nutrients, in this case to different parts of the plant. Parallels may therefore be drawn between these two processes and it has been suggested that senescence may be induced by sugar depletion in some species (Yoshida, 2003). Four senescence-associated genes showed altered expression levels during starvation, two increasing and two decreasing (see supplemental tables; Oh et al., 1996; Panavas et al., 1999; Quirino et al., 1999), suggesting that these genes may in fact be responding to changes in Suc concentration. A comparison of gene expression profiles during starvation with expression profiles during senescence of leaves or flowers reveals some similarities, such as an increase in catalase-3 (Swidzinski et al., 2002), several transcription factors (At2g23340, At5g10030, At5g65210; Chen et al., 2002), and genes potentially involved in lipid metabolism and protein degradation (Bhalerao et al., 2003; van Doorn et al., 2003). However, the majority of genes studied do not show similar trends during senescence and starvation, indicating that while senescence and starvation responses share some common pathways, these processes also have unique features and requirements, as might be expected due to their distinct physiological roles in stress and development.
Increased Levels of Transcription Factors and Proteins Involved in Signal Transduction May Activate and Regulate the Responses to Suc Starvation in Arabidopsis Suspension Cells
A recent microarray study of Arabidopsis transcription factors has determined that many of the factors found in this species are multifunctional for responses to environmental stresses and hormones (Chen et al., 2002) and, in concurrence with these results, 39 potential transcription factor genes displayed increased transcript levels in our study. These include several genes of the bZIP family, members of which have been shown to be involved in light and stress signaling and seed maturation (for review, see Jakoby et al., 2002). One of the bZIP factors is from a group of proteins that may regulate seed storage protein production and responses to environmental or pathogen stresses. Another three of the bZIP proteins are from a group linked to defense against pathogens and development. Two WRKY transcription factors were also identified. WRKY proteins bind to the (T)(T)TGAC(C/T) W-box motif (de Pater et al., 1996), and members of the protein family have been shown to be expressed during plant wounding (Hara et al., 2000), and as early regulators of plant defense (Eulgem et al., 1999). Of the two genes identified, WRKY45 is also up-regulated in response to pathogen infection and salicylic acid (Dong et al., 2003), implicating this factor in regulation of diverse stress responses, whereas the expression pattern of WRKY65 is unknown. Considering that the WRKY family has 74 members in Arabidopsis, it is likely that these two WRKY proteins are involved in the regulation of gene expression during carbon nutrient stresses. These transcription factors may be involved in the regulation of the many metabolic and stress-related proteins that were induced during starvation.
One well-characterized system of transcriptional regulation during starvation is the expression of amylase genes in rice. The amylase3 promoter contains a TATCCA element that has been shown to serve as an enhancer for sugar starvation-induced expression (Lu et al., 2002). Variants of the TATCCA element are found upstream of 18 amylase genes isolated from various plant species (Yu, 1999) and other sugar starvation- induced genes, such as the cucumber malate synthase and isocitrate lyase genes (Graham et al., 1994) and the maize Suc synthase gene (Koch, 1996). These findings suggest that the TATCCA element could be a common cis-regulatory element used during sugar starvation. Three novel rice MYB transcription factors were shown to bind to the TATCCA region and mediate α-amylase expression (Lu et al., 2002). None of the starvation-induced genes above increased substantially in expression under our experimental conditions; however, several potential MYB transcription factors showed an increase, and may function in the up-regulation of a subset of the identified starvation-induced genes. Future study of these transcription factors should lead to a better understanding of their targets and the pathways that they regulate.
Regulation of starvation responses is likely to involve signaling cascades, typically involving a series of protein kinases. While these pathways are often not transcriptionally controlled, a number of genes predicted to function in cellular communication and signal transduction increase in expression during Suc starvation. These genes include a large group of protein kinases, a phosphatidylinositol-4-phosphate-5-kinase-like protein, and several proteins known to play a role in hormone-signaling pathways. The precise function of these proteins, and whether some of the observed starvation responses are hormonally controlled, remains to be determined.
Genes Functioning in Translation, Metabolism, and Cell Division Are Down-Regulated during Starvation
A number of ribosomal protein genes show significant decreases in mRNA level during starvation. In addition, two putative translation initiation factors also decrease in expression, together suggesting that translation activity declines over the starvation time course. In all eukaryotes, the TOR kinase is thought to act as a master regulator of multiple nutrient starvation responses, including autophagy, protein synthesis, ribosome biosynthesis, and some transcriptional responses (Raught et al., 2001). During starvation, TOR is inactivated, leading to inhibition of translation both by direct inactivation of translation initiation factors and by the inhibition of transcription of ribosomal protein and translation initiation factor genes (Cardenas et al., 1999; Powers and Walter, 1999). TOR is present in Arabidopsis; however, a T-DNA knockout mutation in the TOR gene is embryo lethal (Menand et al., 2002), and its function in nutrient stress responses is therefore difficult to assess. We hypothesize that the observed decrease in expression of a large number of ribosomal protein genes, concomitant with the induction of autophagy, may be due to the action of the Arabidopsis TOR protein, as seen in yeast and animal cells.
A second notable group of down-regulated genes is those that are known or expected to be cell cycle-regulated. Two mitotic B-type cyclins that probably function in cell cycle progression (John et al., 2001) are included, as is the syntaxin homolog KNOLLE (Lukowitz et al., 1996), required for the fusion of transport vesicles to form the cell plate during cytokinesis. Genes encoding two proteins involved in DNA replication also decrease in expression. RPA1 is required for DNA repair as well as replication in yeast (Umezu et al., 1998) and thus plays a role in both stress responses and cell division. A rice homolog has been implicated in DNA replication and is highly expressed in proliferating cells (van der Knaap et al., 1997), suggesting a similar function in plants. The MCM3 protein functions in yeast at the G1/S transition in the initiation of DNA replication (Tye, 1999), and Arabidopsis MCM3 is transcriptionally regulated in a cell cycle-dependent manner (Stevens et al., 2002). In addition, several histone proteins show a decrease in mRNA level after Suc starvation; histones are known to be more highly expressed during DNA replication (Callard and Mazzolini, 1997). It is likely that the change in expression for these genes is due to the inhibition of growth of the cell cultures in the absence of Suc (Fig. 2).
As might be expected during a period of Suc limitation, genes involved in some aspects of metabolism also decrease in expression, including those potentially involved in glycolysis, the pentose phosphate pathway, and nucleotide, amino acid, and fatty acid biosynthesis. This suggests that certain metabolic pathways are suppressed to conserve resources during nutrient starvation. This resembles the suppression of metabolism observed in situations of dormancy, where unnecessary metabolic pathways are shut down to allow survival until environmental conditions are encountered that are favorable for resumption of growth (Bewley, 1997; Shimizu and Mori, 1998; Pnueli et al., 2002). The extent of this similarity at the level of gene expression remains to be determined.
It should be noted that, while a number of genes that have previously been reported to be regulated by Suc starvation were also identified in our analysis (e.g. several din genes [Fujiki et al., 2000] and glycosyl hydrolases [Lee et al., 2004]), others did not show a significant change in our experiments. This may be due partially to differences in experimental design; our aim was to identify genes involved in starvation responses and, in particular, nutrient mobilization, rather than sugar-regulated genes, and thus an extended starvation period was used. In addition, differences between plant species are evident; for example, the glyoxylate cycle genes, malate synthase and isocitrate lyase, are up-regulated during starvation of cucumber cultured cells (Graham et al., 1994), but show no increase during starvation of Arabidopsis. Finally, we excluded genes that appeared to be regulated by osmotic conditions from our analysis. Several of the genes identified by Lee et al. (2004) were excluded in this manner and may, in fact, respond to osmotic changes or, most likely from our data, a combination of osmotic and starvation stresses.
In conclusion, we have identified a group of genes showing significant changes in transcript level during a 48-h period of Suc starvation in Arabidopsis suspension cells. Many of the genes that increase in expression appear to be involved in nutrient mobilization and scavenging, responses apparently intended to allow survival under nutrient-limiting conditions. Some of the predicted transcription factors and signaling molecules identified are expected to function in the regulation of these responses. In addition, a number of genes previously shown to be regulated by biotic or abiotic stress conditions were up-regulated, suggesting that general stress response pathways are induced as well as those specific to Suc starvation. In contrast, genes that function in translation and replication decreased in expression during starvation. The amount and activities of the encoded proteins will now have to be determined to confirm the significance of the transcriptional regulation. However, in yeast, polysome microarray analysis of TOR-regulated responses has indicated a surprisingly strong correlation between increases in gene transcription and translation, providing an amplification of responses, termed potentiation (Preiss et al., 2003), and validating the use of transcriptional analysis to identify pathways induced under specific conditions. The challenge now will be to determine the physiological and biochemical functions of the identified genes and pathways, both at the cellular level and in the context of the whole plant. The characterization of the phenotypes of knockout mutants in genes up-regulated by starvation, both in terms of the whole-plant response when grown under nutrient stress conditions and the biochemical and molecular changes that result from the mutation, should allow the elucidation of the role of the gene in the starvation response. In the case of putative transcription factors, identification of the subset of genes that are regulated by each factor, either by transcriptional profiling of knockout mutants or transgenic plants overexpressing the factor, transient expression analysis, or in vitro DNA-binding analysis, will be critical in determining its function in survival during starvation. Analysis of the contribution of the induced pathways to the tolerance of nutrient stress will lead to a clearer understanding of the global response to starvation in plants.
MATERIALS AND METHODS
Growth of Arabidopsis Suspension Cell Cultures
An Arabidopsis Columbia-0 suspension cell culture was obtained from Dr. S.B. Gelvin and maintained by subculturing weekly into 50-mL of Murashige and Skoog Minimal Organics medium (Gibco-BRL, Gaithersburg, MD), 2% (w/v) Suc, 1 μg mL−1 naphthalene acetic acid (Sigma-Aldrich, St. Louis), and 50 ng mL−1 kinetin (Sigma-Aldrich). Cultures were grown in Erlenmeyer flasks at room temperature, under ambient light, with constant shaking (115-rpm rotation).
Suc Starvation Treatment of Suspension Cell Cultures
All starvation time courses were begun using suspension cells 3 d after subculturing, at an approximate cell density of 2 × 105 cells mL−1. Cultures were washed three times with either Suc-containing medium for control samples, medium lacking Suc for starvation samples, or medium containing 0.058 m polyethylene glycol (PEG 4000) for osmoticum-replaced samples. After the third wash, 50 mL of the appropriate medium were added and the cells were grown for up to 72 h on a rotational shaker using the conditions described above. Samples (2 mL) were removed for morphological analysis and RNA extraction after 0, 6, 24, 48, and 72 h.
Measurement of Respiration Rate
One-milliliter samples from suspension cultures starved for 0, 24, 48, and 72 h, and control cultures grown in Suc-containing medium, were pelleted and the fresh weight determined. Cells were then resuspended in the appropriate fresh medium (1 mL). Oxygen consumption was measured using an O2 electrode (Rank Brothers, Bottisham, Cambridge, UK) at 25°C and recorded by a Houston Instrument Chart Recorder (GTCO CalComp, Columbia, MD). Data were converted into nm O2 consumed per min and standardized by total fresh weight. Each experiment was repeated three times.
Fluorescein Cell Viability Assay
For fluorescein staining (Chaves et al., 2002), 5-mL samples of Suc-starvation and control cultures were taken after 0, 24, 48, and 72 h. The medium was removed from each sample and replaced with 5 mL of phosphate-buffered saline. Fluorescein diacetate in ethanol was added to each sample to a concentration of 2.5 μg mL−1, followed by incubation at room temperature for 3 min. The samples were then placed on ice and 300 cells from each sample were analyzed using UV fluorescence microscopy. Fluorescing cells were counted as living.
Culture Viability Assay
After Suc starvation of suspension cultures for 0, 24, 48, and 72 h, as described above, the starvation medium was replaced with Suc-containing medium, and growth continued. Five-milliliter samples were taken every 48 h for 12 d, and cell volume and fresh-cell mass were measured for each time point.
Total RNA Isolation and Northern-Blot Analyses
Suspension cell samples were collected, the medium was removed, and the cells were stored at −80°C until RNA extractions were performed. Total RNA for northern-blot analysis, RT-PCR, and GeneChip microarray analysis was isolated using a TRIzol extraction method (http://www.science.siu.edu/plantbiology/PLB420/DNA.Techniques/TRIzol.method.html), followed by the RNeasy Clean-Up protocol (Qiagen, Valencia, CA), as recommended by the University of Iowa DNA Core Facility, to obtain the best results for use with Affymetrix's GeneChip Expression Analysis system (Affymetrix, Santa Clara, CA). Northern-blot analyses were performed using probes consisting of radiolabeled cDNA fragments corresponding to At5g60360 (AALP; Ahmed et al., 2000), At4g32940 (VPE-γ; Kinoshita et al., 1999), At3g30775 (PRODH; Nakashima et al., 1998), and At5g17290 (APG5/ATG5). All primers used in this study are listed in Table IV. Hybridization was performed using the manufacturer's protocol for UltraHyb solution (Ambion, Austin, TX).
Table IV.
Primers used
| AGIa | Product | Sequence |
|---|---|---|
| At5g60360 | AALP | FOR 5′-ACCATCAGGCATTTGGAAAA-3′ |
| REV 5′-TCTTCCCCATCTCCATCTTG-3′ | ||
| At4g32940 | VPE-γ | FOR 5′-CGGATCTAGCGGATATTGGA-3′ |
| REV 5′-TCAGGAAGAAGCCCTTCAAA-3′ | ||
| At3g30774 | PRODH | FOR 5′-ATGGCAACCCGTCTTCTCCGAACAA-3′ |
| REV 5′-TTACGCAATCCCGGCGATTAATCTC-3′ | ||
| At5g17290 | APG5/ATG5 | FOR 5′-CGAAGGAAGCGGTCAAGTAT-3′ |
| REV 5′-ATCACCGTTCATGACAGAGG-3′ | ||
| At1g13260 | RAV1 | FOR 5′-GTCGACATGGAATCGAGTAGCGTTGAT-3′ |
| REV 5′-CCTAGGTTACGAGGCGTCAAAGATGCG-3′ | ||
| At1g20620 | CAT3 | FOR 5′-GTCGACATGGATCCTTACAAGTATCGTCCTTCAAGC-3′ |
| REV 5′-GCGGCCGCCTAGATGCTTGGCCTCACGTTCAGACGGCT-3′ | ||
| At1g21920 | PI4P-5K | FOR 5′-GTCGACATGGAAAAACAGGCGAAGCTA-3′ |
| REV 5′-CCTAGGTCAACTTTGACAAAATTTACC-3′ | ||
| At1g78290 | STPK | FOR 5′-GTCGACATGGAGAGGTACGAAATAGTC-3′ |
| REV 5′-CCTAGGTCACAAAGGGGAAAGGAGATC-3′ | ||
| At2g33830 | AUX | FOR 5′-GTCGACATGTGGGATGAAACTGTAGCC-3′ |
| REV 5′-CCTAGGTCAACGGTGCTTGCTCCTAGT-3′ | ||
| At4g36670 | SUG | FOR 5′-GTCGACATGGCCGATCAAATCTCCGGC-3′ |
| REV 5′-CCTAGGCTAAGCTGCACCGTTTTCGCC-3′ | ||
| At5g10030 | OBF4 | FOR 5′-GTCGACATGAATACAACCTCGACACAT-3′ |
| REV 5′-CCTAGGTTACGTTGGTTCACGTTGCCT-3′ | ||
| At5g61590 | ERF5 | FOR 5′-GTCGACATGGAGACTTTTGAGGAAACG-3′ |
| REV 5′-TCTAGATTAGTTTGATGACGATGATGA-3′ |
Arabidopsis Genome Initiative number.
GeneChip Analysis and Expression Data Processing
RNA was isolated from suspension cells starved for 0, 24, or 48 h, from cells grown for 48 h in the presence of Suc, and from 48-h starved cultures containing 0.058 m PEG 4000 as an osmotic control. In each case, RNA was pooled from three independent experiments and used to synthesize cRNA, which was hybridized to an ATH1 Arabidopsis GeneChip microarray (Affymetrix) containing 22,810 probe sets. Two independent biological replicate microarray hybridizations were performed for all samples, except for the 48-h no-Suc plus PEG 4000 sample. Each replicate was from an independent starvation time course, started from different subcultures, and performed at different times. Synthesis of cRNA, hybridization to the ATH1 GeneChips, chip scanning, and data accumulation were performed at the University of Iowa DNA Core Facility, using the Affymetrix-recommended protocol. Data were accumulated using Affymetrix's MicroArray Suite version 5.0. Relative expression intensities were generated in the form of average difference values. GeneSpring (Silicon Genetics, Redwood, CA) was used for data normalization, data visualization, and cluster analysis. Using GeneSpring, the median expression level of each chip was normalized to a value of 1 by dividing each measurement for each probe set by the 50th percentile of all measurements on that chip. MicroArray Suite 5.0 calculated the 50th percentile using the average difference of all probe sets labeled present in at least one sample. Normalized values less than 0 were set to 0. Fold change was calculated by dividing the average difference for each experimental starvation sample by the average difference for the untreated 0-h sample. Probe sets with a fourfold (rounded up) or greater change in expression levels in either of the Suc starvation samples compared with the 0- and 48-h controls were identified. A normalized intensity value of 0.8 was used as a cutoff for reliable detection, and also removed a negative bias in lower intensity values when comparing scatter plots of replicate samples from different hybridizations. Probe sets that did not also show a change in transcript level of at least 2-fold (rounded up) in the starvation samples containing PEG 4000, as compared to the 0- and 48-h controls, were discarded as potentially being due to osmotic differences. Using these criteria, transcript levels for 343 unique genes were found to increase during starvation, whereas 263 genes showed a decrease in transcript level. The probe sets for the up-regulated genes were used to perform k-means cluster analysis, using five clusters, with 100 iterations, comparing similarity by Pearson correlation. Individual Pearson correlations were also performed using a 0.95 minimum correlation based on the characteristic expression patterns of the following probe sets: At3g48360 (set1), At1g66280 (set 2), and At1g20620 (set 3). Four of the five clusters were combined into two separate clusters, based on the similarity in their expression patterns and comparison with the Pearson correlations for the individual probe sets.
Verification of GeneChip Data by Northern-Blot Hybridization and RT-PCR
Oligonucleotide primers were designed for eight gene transcripts (At1g13260, At1g20620, At1g21920, At1g78290, At2g33830, At4g36670, At5g10030, and At5g61590) that showed an increase in expression in the starvation samples, according to the GeneChip data. First-strand cDNA was synthesized from 2 μg of DNase-treated total RNA from suspension cells using Superscript II Reverse Transcriptase (Invitrogen, Carlsbad, CA), followed by PCR amplification. The products were ligated into the pGEM-Teasy vector system (Promega, Madison, WI) and gene identity was verified by DNA sequencing performed at the Iowa State University DNA Sequencing and Synthesis Facility. RT-PCR samples were used to semiquantitatively determine the relative amount of product by visualization using DNA agarose electrophoresis. The products were also used as probes for northern-blot hybridization analysis, as described above.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Supplementary Material
Acknowledgments
We thank Drs. Steven Whitham and Ron Mittler for critical reading of the manuscript, Dr. Carol Foster for helpful suggestions on data analysis, and Dr. Dan Voytas for providing the Arabidopsis suspension culture.
This work was supported by the Plant Responses to the Environment Program of the National Research Initiative Competitive Grants Program, U.S. Department of Agriculture (grant no. 2002–35100–12034 to D.C.B.), and by the Iowa State University Plant Sciences Institute.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.044362.
References
- Ahmed SU, Rojo E, Kovaleva V, Venkataraman S, Dombrowski JE, Matsuoka K, Raikhel NV (2000) The plant vacuolar sorting receptor AtELP is involved in transport of NH2-terminal propeptide-containing vacuolar proteins in Arabidopsis thaliana. J Cell Biol 149: 1335–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert S, Gout E, Bligny R, Marty-Mazars D, Barrieu F, Alabouvette J, Marty F, Douce R (1996) Ultrastructural and biochemical characterization of autophagy in higher plant cells subjected to carbon deprivation: control by the supply of mitochondria with respiratory substrates. J Cell Biol 133: 1251–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbet NC, Schneider U, Helliwell SB, Stansfield I, Tuite MF, Hall MN (1996) TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell 7: 25–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham DC (2002) Golgi-independent trafficking of macromolecules to the plant vacuole. Adv Bot Res 38: 65–92 [Google Scholar]
- Bewley JD (1997) Seed germination and dormancy. Plant Cell 9: 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalerao R, Keskitalo J, Sterky F, Erlandsson R, Bjorkbacka H, Birve SJ, Karlsson J, Gardestrom P, Gustafsson P, Lundeberg J, et al (2003) Gene expression in autumn leaves. Plant Physiol 131: 430–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouquisse R, Gaudillere JP, Raymond P (1998) Induction of a carbon-starvation-related proteolysis in whole maize plants submitted to light/dark cycles and to extended darkness. Plant Physiol 117: 1281–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouquisse R, James F, Raymond P, Pradet A (1991) Study of glucose starvation in excised maize root tips. Plant Physiol 96: 619–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursch W, Ellinger A, Gerner CH, Frohwein U, Schulte-Hermann R (2000) Programmed cell death (PCD). Apoptosis, autophagic PCD, or others? Ann N Y Acad Sci 926: 1–12 [DOI] [PubMed] [Google Scholar]
- Callard D, Mazzolini L (1997) Identification of proliferation-induced genes in Arabidopsis thaliana. Characterization of a new member of the highly evolutionarily conserved histone H2A.F/Z variant subfamily. Plant Physiol 115: 1385–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas ME, Cutler NS, Lorenz MC, Di Como CJ, Heitman J (1999) The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev 13: 3271–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves I, Regalado AP, Chen M, Ricardo CP, Showalter AM (2002) Programmed cell death induced by (β-D-galactosyl)3 Yariv reagent in Nicotiana tabacum BY-2 suspension-cultured cells. Physiol Plant 116: 548–553 [Google Scholar]
- Chen M-H, Liu L-F, Chen Y-R, Wu HK, Yu S-M (1994) Expression of α-amylases, carbohydrate metabolism, and autophagy in cultured rise cells is coordinately regulated by sugar nutrient. Plant J 6: 625–636 [DOI] [PubMed] [Google Scholar]
- Chen W, Provart NJ, Glazebrook J, Katagiri F, Chang HS, Eulgem T, Mauch F, Luan S, Zou G, Whitham SA, et al (2002) Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14: 559–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dat J, Vandenabeele S, Vranova E, Van Montagu M, Inze D, Van Breusegem F (2000) Dual action of the active oxygen species during plant stress responses. Cell Mol Life Sci 57: 779–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pater S, Greco V, Pham K, Memelink J, Kijne J (1996) Characterization of a zinc-dependent transcriptional activator from Arabidopsis. Nucleic Acids Res 24: 4624–4631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieuaide M, Brouquisse R, Pradet A, Raymond P (1992) Increased fatty acid β-oxidation after glucose starvation in maize root tips. Plant Physiol 99: 595–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieuaide M, Couee I, Pradet A, Raymond P (1993) Effects of glucose starvation on the oxidation of fatty acids by maize root tip mitochondria and peroxisomes: evidence for fatty acid β-oxidation and acyl-coA dehydrogenase activity in higher plants. Biochem J 296: 199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling JH, Walker JM, Friedman EM, Thompson AR, Vierstra RD (2002) The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem 277: 33105–33114 [DOI] [PubMed] [Google Scholar]
- Dong J, Chen C, Chen Z (2003) Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol Biol 51: 21–37 [DOI] [PubMed] [Google Scholar]
- Eastmond PJ, van Dijken AJ, Spielman M, Kerr A, Tissier AF, Dickinson HG, Jones JD, Smeekens SC, Graham IA (2002) Trehalose-6-phosphate synthase 1, which catalyses the first step in trehalose synthesis, is essential for Arabidopsis embryo maturation. Plant J 29: 225–235 [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Sommssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5: 199–206 [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Schmelzer E, Hahlbrock K, Somssich IE (1999) Early nuclear events in plant defense signaling: rapid gene activation by WRKY transcription factors. EMBO J 18: 4689–4699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y, Ito M, Nishida I, Watanabe A (2000) Multiple signaling pathways in gene expression during sugar starvation. Pharmacological analysis of din gene expression in suspension-cultured cells of Arabidopsis. Plant Physiol 124: 1139–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y, Masaki I, Takashi I, Ikou N, Watanabe A (2002) Activation of the promoters of Arabidopsis genes for the branched-chain-α-keto acid dehydrogenase complex in transgenic tobacco BY-2 cells under sucrose starvation. Plant Cell Physiol 43: 275–280 [DOI] [PubMed] [Google Scholar]
- Garg AK, Kim JK, Owens TG, Ranwala AP, Choi YD, Kochian LV, Wu RJ (2002) Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc Natl Acad Sci USA 99: 15898–15903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham IA, Denby KJ, Leaver CJ (1994) Carbon catabolite repression regulates glyoxylate cycle gene expression in cucumber. Plant Cell 6: 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham IA, Eastmond PJ (2002) Pathways of straight and branched chain fatty acid catabolism in higher plants. Prog Lipid Res 41: 156–181 [DOI] [PubMed] [Google Scholar]
- Hamburg M, Sanz A, Castresana C (1999) α-Oxidation of fatty acids in higher plants. Identification of a pathogen-inducible oxygenase (piox) as an α-dioxygenase and biosynthesis of 2-hydroperoxylinolenic acid. J Biol Chem 274: 24503–24513 [DOI] [PubMed] [Google Scholar]
- Hanaoka H, Noda T, Shirano Y, Kato T, Hayashi H, Shibata D, Tabata S, Ohsumi Y (2002) Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol 129: 1181–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Yagi M, Kusano T, Sano H (2000) Rapid systemic accumulation of transcripts encoding a tobacco WRKY transcription factor upon wounding. Mol Gen Genet 263: 30–37 [DOI] [PubMed] [Google Scholar]
- Hayashi H, De Bellis L, Ciurli A, Kondo M, Hayashi M, Nishimura M (1999) A novel acyl-CoA oxidase that can oxidize short-chain acyl-CoA in plant peroxisomes. J Biol Chem 273: 8301–8307 [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang WP, Klionsky DJ (2002) Autophagy in yeast: a review of the molecular machinery. Cell Struct Funct 27: 409–420 [DOI] [PubMed] [Google Scholar]
- Hughes JD, Estep PW, Tavazoie S, Church GM (2000) Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J Mol Biol 296: 1205–1214 [DOI] [PubMed] [Google Scholar]
- Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, et al (2000) A ubiquitin-like system mediates protein lipidation. Nature 408: 488–492 [DOI] [PubMed] [Google Scholar]
- Ito M, Iwase M, Kodama H, Lavisse P, Komamine A, Nishihama R, Machida Y, Watanabe A (1998) A novel cis-acting element in promoters of plant B-type cyclin genes activates M phase-specific transcription. Plant Cell 10: 331–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Droge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7: 106–111 [DOI] [PubMed] [Google Scholar]
- James F, Brouquisse R, Pradet A, Raymond P (1993) Changes in proteolytic activities in glucose-starved maize root tips: regulation by sugars. Plant Physiol Biochem 31: 845–856 [Google Scholar]
- John PC, Mews M, Moore R (2001) Cyclin/Cdk complexes: their involvement in cell cycle progression and mitotic division. Protoplasma 216: 119–142 [DOI] [PubMed] [Google Scholar]
- Journet EP, Bligny R, Douce R (1986) Biochemical changes during sucrose deprivation in higher plant cells. J Biol Chem 261: 3193–3199 [PubMed] [Google Scholar]
- Kagaya Y, Ohmiya K, Hattori T (1999) RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in plants. Nucleic Acids Res 27: 470–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kametaka S, Okano T, Ohsumi M, Ohsumi Y (1998) Apg14p and Apg6/Vps30p form a protein complex essential for autophagy in the yeast, Saccharomyces cerevisiae. J Biol Chem 273: 22284–22291 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Yamada K, Hiraiwa N, Kondo M, Nishimura M, Hara-Nishimura I (1999) Vacuolar processing enzyme is up-regulated in the lytic vacuoles of vegetable tissues during senescence and various stressed conditions. Plant J 19: 43–53 [DOI] [PubMed] [Google Scholar]
- Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y (1999) Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol 147: 435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y (2000) The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol 151: 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Ohsumi Y (1999) Vacuolar import of proteins and organelles from the cytoplasm. Annu Rev Cell Dev Biol 15: 1–32 [DOI] [PubMed] [Google Scholar]
- Koch KE (1996) Carbohydrate modulated gene expression in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 509–540 [DOI] [PubMed] [Google Scholar]
- Lam E (2004) Controlled cell death, plant survival and development. Nat Rev Mol Cell Biol 5: 305–315 [DOI] [PubMed] [Google Scholar]
- Lauber MH, Waizenegger I, Steinmann T, Schwarz H, Mayer U, Hwang I, Lukowitz W, Jurgens G (1997) The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J Cell Biol 139: 1485–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EJ, Koizumi N, Sano H (2004) Identification of genes that are up-regulated in concert during sugar depletion in Arabidopsis. Plant Cell Environ 27: 337–345 [Google Scholar]
- Li CY, Weiss D, Goldschmidt EE (2003) Effects of carbohydrate starvation on gene expression in citrus root. Planta 217: 11–20 [DOI] [PubMed] [Google Scholar]
- Lu C-A, Ho T-H, Ho S-L, Yu S-M (2002) Three novel MYB proteins with one DNA binding repeat mediate sugar and hormone regulation of α-amylase gene expression. Plant Cell 14: 1963–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W, Mayer U, Jurgens G (1996) Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell 84: 61–71 [DOI] [PubMed] [Google Scholar]
- Menand B, Desnos T, Nussaume L, Berger F, Bouchez D, Meyer C, Robaglia C (2002) Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc Natl Acad Sci USA 99: 6422–6427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Morgante M, Michelmore RW (2002) TIR-X and TIR-NBS proteins: two new families related to disease resistance TIR-NBS-LRR proteins encoded in Arabidopsis and other plant genomes. Plant J 32: 77–92 [DOI] [PubMed] [Google Scholar]
- Miernyk JA (2001) The J-domain proteins of Arabidopsis thaliana: an unexpectedly large and diverse family of chaperones. Cell Stress Chaperones 6: 209–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Sugita H, Yoshimori T, Ohsumi Y (1998. a) A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J Biol Chem 273: 33889–33892 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Sugita H, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y (1998. b) A protein conjugation system essential for autophagy. Nature 395: 395–398 [DOI] [PubMed] [Google Scholar]
- Moriyasu Y, Ohsumi Y (1996) Autophagy in tobacco suspension-cultured cell in response to sucrose starvation. Plant Physiol 111: 1233–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Satoh R, Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K (1998) A gene encoding proline dehydrogenase is not only induced by proline and hypoosmolarity, but is also developmentally regulated in the reproductive organs of Arabidopsis. Plant Physiol 118: 1233–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SA, Lee SY, Chung LK, Lee CH, Nam HG (1996) A senescence-associated gene of Arabidopsis thaliana is distinctively regulated during natural and artificially induced leaf senescence. Plant Mol Biol 30: 739–754 [DOI] [PubMed] [Google Scholar]
- Panavas T, Pikula A, Reid PD, Rubinstein B, Walker EL (1999) Identification of senescence-associated genes from daylily petals. Plant Mol Biol 40: 237–248 [DOI] [PubMed] [Google Scholar]
- Pnueli L, Hallak-Herr E, Rozenberg M, Cohen M, Goloubinoff P, Kaplan A, Mittler R (2002) Molecular and biochemical mechanisms associated with dormancy and drought tolerance in the desert legume Retama raetam. Plant J 31: 319–330 [DOI] [PubMed] [Google Scholar]
- Powers T, Walter P (1999) Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol Biol Cell 10: 987–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss T, Baron-Benhamou J, Ansorge W, Hentze MW (2003) Homodirectional changes in transcriptome composition and mRNA translation induced by rapamycin and heat shock. Nat Struct Biol 10: 1039–1047 [DOI] [PubMed] [Google Scholar]
- Prestridge DS (1991) SIGNAL SCAN: a computer program that scans DNA sequences for eukaryotic transcriptional elements. Comput Appl Biosci 7: 203–206 [DOI] [PubMed] [Google Scholar]
- Proud CG (2004) Role of mTOR signaling in the control of translation initiation and elongation by nutrients. Curr Top Microbiol Immunol 279: 215–244 [DOI] [PubMed] [Google Scholar]
- Quirino BF, Normanly J, Amasino RM (1999) Diverse range of gene activity during Arabidopsis thaliana leaf senescence includes pathogen-independent induction of defense-related genes. Plant Mol Biol 40: 267–278 [DOI] [PubMed] [Google Scholar]
- Raught B, Gingras AC, Sonenberg N (2001) The target of rapamycin (TOR) proteins. Proc Natl Acad Sci USA 98: 7037–7044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluepmann H, Pellny T, van Dijken A, Smeekens S, Paul M (2003) Trehalose 6-phosphate is indispensable for carbohydrate utilization and growth in Arabidopsis thaliana. Proc Natl Acad Sci USA 100: 6849–6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H, Zaccaria P, Gundlach H, Lemcke K, Rudd S, Kolesov G, Arnold R, Mewes HW, Mayer KF (2002) MIPS Arabidopsis thaliana Database (MAtDB): an integrated biological knowledge resource based on the first complete plant genome. Nucleic Acids Res 30: 91–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J (1990) Metabolic repression of transcription in higher plants. Plant Cell 2: 1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S, Mori H (1998) Analysis of cycles of dormancy and growth in pea axillary buds based on mRNA accumulation patterns of cell cycle-related genes. Plant Cell Physiol 39: 255–262 [DOI] [PubMed] [Google Scholar]
- Stevens R, Mariconti L, Rossignol P, Perennes C, Cella R, Bergounioux C (2002) Two E2F sites in the Arabidopsis MCM3 promoter have different roles in cell cycle activation and meristematic expression. J Biol Chem 277: 32978–32984 [DOI] [PubMed] [Google Scholar]
- Surpin M, Zheng H, Morita MT, Saito C, Avila E, Blakeslee JJ, Bandyopadhyay A, Kovaleva V, Carter D, Murphy A, et al (2003) The VTI family of SNARE proteins is necessary for plant viability and mediates different protein transport pathways. Plant Cell 15: 2885–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swidzinski JA, Sweetlove LJ, Leaver CJ (2002) A custom microarray analysis of gene expression during programmed cell death in Arabidopsis thaliana. Plant J 30: 431–446 [DOI] [PubMed] [Google Scholar]
- Tye BK (1999) MCM proteins in DNA replication. Annu Rev Biochem 68: 649–686 [DOI] [PubMed] [Google Scholar]
- Tymowska-Lalanne Z, Kreis M (1998) Expression of the Arabidopsis thaliana invertase gene family. Planta 207: 259–265 [DOI] [PubMed] [Google Scholar]
- Umezu K, Sugawara N, Chen C, Haber JE, Kolodner RD (1998) Genetic analysis of yeast RPA1 reveals its multiple functions in DNA metabolism. Genetics 148: 989–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Knaap E, Jagoueix S, Kende H (1997) Expression of an ortholog of replication protein A1 (RPA1) is induced by gibberellin in deepwater rice. Proc Natl Acad Sci USA 94: 9979–9983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doorn WG, Balk PA, van Houwelingen AM, Hoeberichts FA, Hall RD, Vorst O, van der Schoot C, van Wordragen MF (2003) Gene expression during anthesis and senescence in Iris flowers. Plant Mol Biol 53: 845–863 [DOI] [PubMed] [Google Scholar]
- Willekens H, Inze D, Van Montagu M, Van Camp W (1995) Catalases in plants. Mol Breed 1: 207–228 [Google Scholar]
- Wu P, Ma L, Hou X, Wang M, Wu Y, Liu F, Deng XW (2003) Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiol 132: 1260–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S (2003) Molecular regulation of leaf senescence. Curr Opin Plant Biol 6: 79–84 [DOI] [PubMed] [Google Scholar]
- Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S, Baehrecke EH, Lenardo MJ (2004) Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science 304: 1500–1502 [DOI] [PubMed] [Google Scholar]
- Yu S-M (1999) Cellular and genetic responses of plants to sugar starvation. Plant Physiol 121: 687–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.