Abstract
Topoisomerases are unique enzymes that regulate torsional stress in DNA to enable critical genome functions including DNA replication and transcription. While all cells in an organism require topoisomerases, the nervous system in particular shows a critical need for these enzymes to maintain normal function. A spectrum of inherited human neurologic syndromes including neurodegeneration, schizophrenia and intellectual impairment are associated with aberrant topoisomerase function. Much remains unknown regarding tissue-specific neural topoisomerase function or the connections between these enzymes and disease aetiology. Precisely how topoisomerases regulate genome dynamics within the nervous system is a critical research question.
Introduction
The unwinding of DNA strands is essential for template access by replication and transcription machinery. However, these vital activities invariably generate torsional stress (a force generated by twisting) and topology issues in DNA. For example, the unwinding of DNA to form a negatively supercoiled structure incurs concomitant over-winding (positive supercoiling) elsewhere in DNA, which can profoundly disrupt transcription1,2. During DNA replication there is also a risk of tangled or knotted DNA, which in turn can result in detrimental damage or defective chromosomal segregation3-6. Thus, the regulation of frequent torsional stress or entanglement during DNA replication and transcription is essential for cellular homeostasis, and is achieved by the concerted action of multiple topoisomerases. In this progress article, I discuss recent studies that broaden our understanding of how topoisomerase function in the nervous system integrates neural homeostasis. These new findings underscore how these enzymes coordinate many facets of brain health.
1. Basic topoisomerase biology
Topoisomerases are remarkable enzymatic machines that exert exquisite control over DNA topology to regulate essential genomic transactions5-9. Topoisomerases introduce transient DNA breaks that relax tightly wound DNA to facilitate replication and transcription5,6. Topoisomerases also help to protect the cell against a spectrum of potential DNA lesions that can impact cellular function, such as replication-associated damage or R-loop formation during transcription (an RNA-DNA hybrid associated with an unannealed DNA single strand)5,6,8-11. These critical roles suggest that abnormal topoisomerase activity could be a potential catastrophe for cellular homeostasis. Indeed, aberrant topoisomerase activity is linked to multiple human syndromes12-16.
1.1 Mammalian topoisomerases
Mammalian topoisomerases comprise seven distinct enzymes, Top1, Top1mt, Top2α and 2β, Top3α and 3β and Spo11 (Fig. 1). These are categorized as type I [Top1 and Top3] or type II [Top2 and Spo11] depending upon whether a transient single strand or a double strand break respectively is introduced into DNA5,9,11. All topoisomerases are critical during organismal development and maintenance: Top1, Top2α and Top3α are essential for DNA replication6,17-19, while Top1, Top2β and Top3β are required for transcriptional modulation in differentiated cells15,20-22. Spo11 is a germ cell-specific topoisomerase involved in meiosis23, while Top1mt (a mitochondria specific Top1) functions during mitochondrial DNA replication to ensure organelle homeostasis24,25.
Figure 1. Mammalian Topoisomerases.
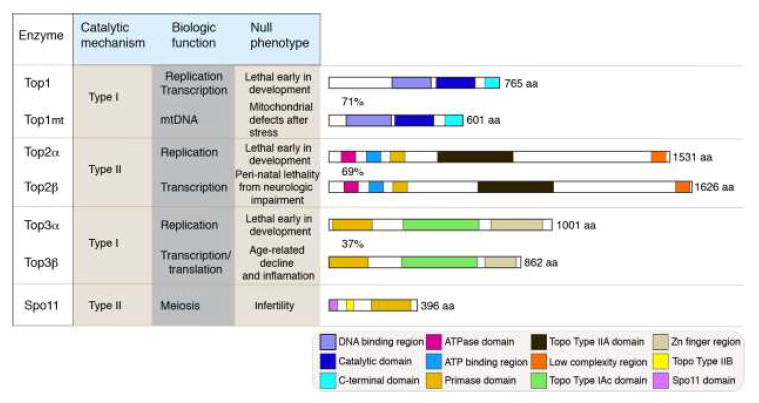
There are seven mammalian topoisomerases indicated with individual biochemical characteristics. Top1 and Top3 are Type 1 topoisomerases, which cleave a single DNA strand, while Top2 and Spo11 are Type II topoisomerases and create a double strand break. SPO11 is specific for meiosis and is only expressed in germ cells. Each topoisomerase has unique biologic functions, reflected by the diversity of phenotypes that occur when a particular enzyme is inactivated in the mouse. Topoisomerase primary amino acid (aa) sequence highlighting enzyme-specific motifs are depicted to scale. Similar motifs are present in related topoisomerases; motif identities are listed in the lower boxed panel. Motifs were determined based on a BLAST search at the NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi; zinc finger domains in Top3β were identified via other analysis16). Despite possessing a similar primary structure, Top3 isoforms are only 37% identical at the amino acid level, compared with 71% and 69% for Top1 and Top2 respectively; aa is amino acids.
Although there are many topoisomerases, there is a common theme in the way they function, as all contain an active site tyrosine that covalently attaches to DNA and in doing so creates a strand break. This occurs via a nucleophilic attack by the active site tyrosine of the topoisomerase towards the phosphodiester bond of DNA, generating a strand scission and a reversible enzyme-DNA covalent linkage5. This results in a 3’phosphate-linked product in the case of Top1, but a 5’phosphate-linkage for topoisomerase 2 (Fig. 2). Features other than the catalytic domain of the protein regulate enzyme activity. For instance, topoisomerase 2α and 2β, exhibit structural similarity around the active site, but each contains a distinct C-terminal region, which is important for controlling differences in topoisomerase function in replicating vs. non-replicating cells26.
Figure 2. Topoisomerase function.
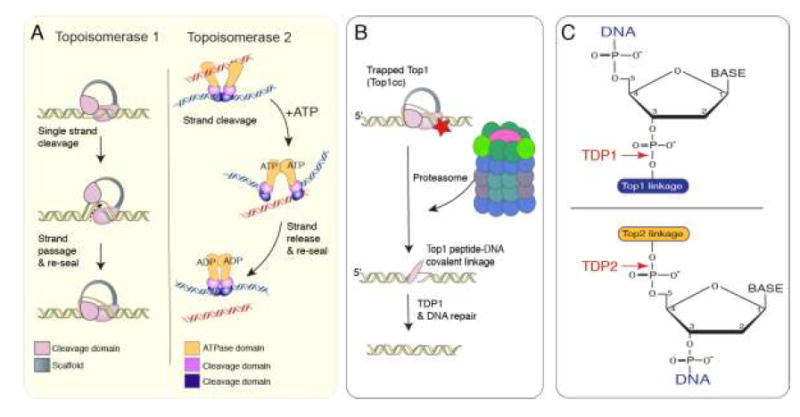
A. Topoisomerases generate a transient single- or double-strand break in DNA to relive torsional stress. Top1 creates a single-strand break in one strand of DNA and passes the other strand through the nick, before resealing the break. Top2 creates a double strand break and passes another duplex through the break, releasing the strand from the enzyme, prior to resealing. In contrast to Top1, Top2 utilizes ATP for the strand cleavage reaction5,8. B. Topoisomerase activities are transient and are normally not detected as DNA damage. If a topoisomerase encounters DNA damage (red star) this can cause abortive activity resulting in a Topoisomerase cleavage complex (Top1cc) covalently attached to DNA, which is detected as DNA damage. To remove this complex the proteasome degrades the topoisomerase leaving a peptide fragment, which is a substrate for TDP1 (in the case of topoisomerase 1). After TDP1 processing, the DNA single strand break is repaired via base excision repair. C. Specific tyrosyl DNA phosphodiesterases (TDP1 and TDP2) are utilized to cleave the Top1 or Top2 DNA adducts that reside at either the 3’ (Top1) or 5’ (Top2) position. For TDP1 this occurs via a nucleophilic attack by an active site histidine, and in the case of TDP2, hydrolytic cleavage of the phosphodiester bond occurs by an asparagine residue33,35.
While torsional stress regulation is key during DNA replication, DNA catenation (intertwining of daughter strands) will also occur and must be prevented to avoid chromosomal damage upon cell division5. Similarly, positive supercoiling forms during transcription as the DNA unwinds, which blocks RNA polymerase movement and must be relieved to allow polymerase passage for successful gene transcription27-29. However, intrinsic topoisomerase function poses a risk for the cell, as any perturbations during DNA strand cleavage can result in an abortive reaction, whereby the topoisomerase becomes covalently trapped on the DNA, which is potentially catastrophic for a cell9. In this scenario, the DNA break becomes persistent, rather than transient, and is signalled as DNA damage. As is the case for other types of DNA damage, this initiates an elaborate DNA damage-signalling response13,30. In order to alleviate DNA damage generated by abortive topoisomerase activity, specialized repair enzymes exist that release the remaining covalent topoisomerase peptide from the DNA (after proteasome-mediated degradation of the trapped topoisomerase31,32) before the breaks are repaired by the general DNA repair machinery13,33-35. Two tyrosyl-DNA phosphodiesterases (TDP1 and TDP2) release DNA from a trapped topoisomerase after abortive topoisomerase 1 or 2 activity respectively (Fig.2)33,35-39. Failure of TDP1 or TDP2 activity will lead to DNA damage accumulation that can impact cellular function, which results in neurologic disease12-14,30,40.
Recently Top3β was unexpectedly shown to have RNA topoisomerase activity16. Top3β binds mRNA and importantly, can directly catalyse topoisomerase reactions on RNA substrates. Thus, a need for torsional stress control during RNA functions seems likely. Mitochondria contain a 16 kb genome for which topoisomerases are also important during replication and transcription; mammalian mitochondria contain Top1mt25, Top2β41 and Top3α42.
1.2 Topoisomerase inactivation in mice
Mouse models have shown that topoisomerases have broad biologic roles. Disruption of topoisomerase 1, 2α or 3α results in lethality very early during embryonic development, underscoring their important roles in DNA replication17-19. Top2β inactivation results in perinatal death due to respiratory distress from diaphragm innervation defects43, while Top3β-null mice develop to maturity but show widespread inflammation and have a shortened lifespan44. This phenotypic spectrum suggests specific physiologic roles for the respective topoisomerases; Top1, 2α and 3α during replication, but specialized roles for Top2β and 3β after tissue formation. Inactivation of the mitochondrial specific form of Top1 is compatible with viability, and mice are relatively normal unless challenged with stress that requires enhanced mitochondrial functions45. Consistent with its specific role in meiosis, Spo11 inactivation result in a viable, but infertile mouse23.
2. Neural topoisomerase function
While the basic enzymatic function of topoisomerases in neural cells is likely similar to that in non-neural cells, there are particular aspects of the nervous system that require special consideration. For instance, some unique properties pertinent to genome integrity that set neural cells apart are the higher metabolic demands and oxygen consumption in this tissue and also the dependence on neuronal synaptic function13. The specific requirements for topoisomerases are different during neurogenesis, where proliferation regulation is critical, from those in the mature brain, where transcription regulation is paramount. The shift from neurogenesis to differentiation and maturation can coincide with changes in relative expression of topoisomerase isoforms. For instance, in the rat cerebellum a defined transition from Top2α to Top2β expression is observed as granule neuron precursors differentiate in this tissue46.
An initial indication of the critical importance of topoisomerase function in the murine nervous system was the observation that germline inactivation of Top2β led to perinatal death43. This resulted from an inability of the newborn mouse to breathe due to diaphragm innervation defects, although widespread defects throughout the nervous system were also observed43,47. Further to these studies, a conditional Top2β allele with gene deletion driven by Foxgl-cre, which primarily targets cortical structures, showed lamination defects and broad disruption of neurogenesis47. Similar to germline Top2β disruption, Top2βFoxg1-cre mice were also perinatal lethal, indicating that demise of the Top2β-/- mice is due to specific disruption of Top2β function in the nervous system.
2.1 Top2β regulates neural gene expression
Studies to address the impact of Top2β loss on transcription showed that in Top2β-/- embryos at mid-late gestation, only a small fraction of genes (<3%) were altered in mutant embryos21. However, while only a modest reduction in overall gene expression occurred, nearly a third of developmentally relevant transcripts were altered, likely contributing to the neural abnormalities in the Top2β-/- mice.
Analysis of Top2β-/- embryonic stem cells showed normal growth/proliferation and differentiation into neural progenitors, although at terminal differentiation neurons underwent apoptosis48. This also occurred in differentiating cortical neurons in the Top2β-/- embryonic nervous system, and was associated with decreased expression of genes important for neurogenesis48. Thus, Top2β is largely dispensable during cellular proliferation, but important after differentiation. Top2β binding to gene promoters was associated with histone H3 dimethylation at lysine 4, which influenced the expression of genes including the neurotrophin P75 receptor, whose deregulation potentially contributed to apoptosis of Top2β-/- cortical neurons48.
In Top2β-/- embryos, normal retinal development around mid-gestation was observed, indicating that loss of Top2β didn’t strongly impact early retinogenesis. To determine Top2β function at later stages of retinogenesis (and to bypass Top2β-/- lethality), Top2β deletion was directed to retinal progenitors via Dkk2-cre (which deletes Top2β in the retina commencing at E10). Top2βDkk2-cre mice showed pronounced lamination defects and neurodegeneration in the retina as differentiation commenced postnatally49. This was despite all retinal cell types (including photoreceptors, horizontal, amacrine and bipolar cells) being formed initially prior to degeneration, indicating that Top2β plays an essential role in the survival and maintenance of differentiated retinal cells.
Recently, Top2β has been linked to activity-dependent gene expression in the brain via the generation of DNA double strand breaks that activate the DNA damage-signalling pathway22,50,51. In this scenario, Top2β-induced expression of immediate early genes was associated with the formation of double strand breaks, which activate histone H2AX phosphorylation at serine 139, forming γH2AX (a characteristic event reflecting DNA damage). As topoisomerases typically create transient breaks that are not signalled as DNA damage (i.e. are not associated with γH2AX formation), this particular role during neural gene activation implies a non-canonical mode of topoisomerase activity. An earlier study also reported DNA double strand breaks as a result of Top2β activity involving components of the DNA damage pathway in MCF7 cells associated with ligand-dependent gene expression51. Further, DNA nicking by Top1 has shown to activate gene expression by relieving torsional stress at enhancers and activating eRNA synthesis (enhancer RNA; non-coding transcripts from gene enhancers that correlate with gene activity52), via a mechanism also involving the DNA damage response53. Additional analysis of Top2β function in other neural populations and compartments are warranted to more fully delineate Top2β function.
2.2 Top1 and neural gene expression
Top1 is required in post-mitotic neurons20,54,55 and in replicating cells19; its inactivation in the germline is lethal very early during development. Top1 is widely expressed throughout the nervous system, with high relative activity in various cortical area and in the cerebellum, and immunolocalization indicate higher protein levels in some inhibitory neurons56. It was also reported that Top1 activity varied across different brain regions between genders, as male mice typically showed an age-dependent decline in brain Top1 activity56. Topoisomerase activity has also been linked to neurotransmitter signalling as glutamate or GABA can modulate Top1 activity in the mouse cerebellum57. Recent findings have indicated a particular need for Top1 activity in the normal transcription of long genes (those >100 kb) in the nervous system20,54,55. Specific inhibitors of Top1 such as topotecan were shown to selectively reduce expression of long genes, amongst which were many associated with synaptic function and linked to autism20,58. Notably, Top2 inhibitors such as etoposide were also found to reduce long gene expression, indicating that multiple topoisomerases influence this class of gene expression20. For instance, in Top2β-/- embryonic cortex the Cajal-Retzius protein Reelin, which is also encoded by a large gene (~500 kb), was markedly decreased accounting for migration defects in the mutant embryonic brain47. These data suggest a specific requirement for topoisomerases in the regulation of long neural gene expression, especially those involved in synaptic function. More broadly, agents that inhibit topoisomerase function, or that cause DNA damage, which can trap topoisomerase on DNA, may influence pathology in various neurologic syndromes.
Because topoisomerases are critical for long neural gene expression, this feature might potentially be leveraged to ameliorate human neurologic conditions. MeCP2 (methyl CpG binding protein 2) is a methyl-binding transcriptional regulator responsible for preventing Rett Syndrome. Loss of MECP2 results in an inappropriate increase in long neural gene expression, potentially disrupting normal synaptic function59,60. However, Top1 inhibition returned expression to normal levels in neurons, demonstrating that at least in principle, this strategy could be beneficial in Rett Syndrome59.
In other scenarios, topoisomerase inhibition also modulates important neurocognitive functions. Angelman syndrome is a severe neurodevelopmental disease that results from disruption of the maternal allele of the ubiquitin ligase E3A (UBE3A). Topoisomerase inhibitors were identified in a screen for compounds that could activate the paternal UBE3A allele (which is epigenetically silenced in neurons), to compensate for the defective maternal allele61. Top1 inhibition activated UBE3A expression that was correlated with R-loop formation (normally repressed by topoisomerases) within the paternal SNORD116 locus, which controls expression of an antisense UBE3A transcript62. Thus, in principal, therapeutic manipulation of topoisomerase activity is a potential approach to treatment of these seemingly intractable neurologic syndromes. Of course, the collateral impact of topoisomerase inhibition on neural function will require evaluation before these approaches can be considered.
3. Neurologic disease and topoisomerases
A clear demonstration of the need for topoisomerase activity in the brain has come from inherited human syndromes (Table 1) resulting from mutations in genes related to topoisomerase function12-15. Somewhat unexpectedly, although these syndromes result from germline mutations, they appear to exclusively impact the nervous system, a scenario akin to other inherited genome instability syndromes13. In the case of Topoisomerases 1 and 2, neurologic diseases arise when enzymes required for resolution of aberrant topoisomerase activity are defective (Fig. 2), rather than direct mutations of the topoisomerases themselves12,14. However, direct Top3β mutation have been identified in individuals with schizophrenia and cognitive impairment15.
Table 1.
| Disease | Gene | Molecular defect | Clinical presentation |
|---|---|---|---|
| SCAN1 | TDP1 [Tyrosyl DNA phosphodiesterases-1] | Disables TDP1 function and cannot repair 3’Top1-DNA covalent linkage after abortive Top1 activity. [Refs 12, 38] | Spinocerebellar ataxia with axonal neuropathy. White matter/myelin defects |
| Seizures, Intellectual impairment and ataxia. | TDP2 [Tyrosyl DNA phosphodiesterases-2] | Inactivating TDP2 mutations. Cannot repair 5’Top2-DNA covalent linkage after abortive Top2 activity. [Refs 14, 39] | Initial presentation with seizures and intellectual impairment. Later development of ataxia. |
| Schizophrenia | Top3β [Topoisomerase 3β] | Homozygous TOP3β deletion. [Ref 15] | Psychotic disorder with marked alterations in sensory stimuli response. |
| Learning disability/cognition impairment | Top3β | Mutations and loss encompassing TOP3Blocus is at-risk for neurodevelopmental disorders. [Refs 15, 16] | Lower IQ, defective cognitive and learning functions. |
| Autism | Top1 [Topoisomerase 1] | Defective Top1 activity may lead to decreased neural long gene expression involving genes linked to autism and cognition. [Ref 20] | Inability to engage in normal social interaction. |
The first identification of a need for careful regulation of neural topoisomerase-1 activity was the finding that TDP1 mutations cause spinocerebellar ataxia with axonal neuropathy 1 (SCAN1) (REF 10). TDP1 cleaves the peptide-DNA linkage at the site of a stalled Top1, which allows for DNA repair and continuation of transcription33. As TDP1 is a repair factor for multiple types of DNA 3’ end damage34,38, it is formally possible that DNA damage other than trapped Top1 might also contribute to the spectrum of SCAN1 neuropathology. However, an inherited inactivating mutation in TDP2, an enzyme whose only known function is cleaving trapped Top2 from DNA37,39 was also found to result in a neurologic disease characterized by seizures and intellectual disability14. Thus, aberrant Top2 activity resulting in transcription disruption appears to be an underlying cause of this neuropathology, suggesting a similar scenario for abortive Top1 activity arising from defective TDP1 in SCAN1.
Topoisomerases have also been identified as potential etiologic agents in other neurodegenerative syndromes including ataxia telangiectasia63, a disease resulting from mutation of the DNA damage response kinase, ataxia telangiectasia, mutated (ATM). An increase in trapped Top1 resulting in elevated DNA damage was identified in brain tissue lacking ATM and shown to result from defective ATM-dependent Top1 turnover64. This was also the situation in brain tissue from mice harbouring mutations in different components of base excision repair64. DNA lesions resulting from repair defects in this pathway caused the trapping of topoisomerases, a known outcome of topoisomerase encounters with damaged DNA9,64. Thus, general genome damage may indirectly perturb topoisomerase function.
3.1 Top3β loss is linked to schizophrenia
While diseases associated with Top1 and Top2 occur via mutation of enzymes required to correct aberrant activity, direct inactivation of Top3β occurs in cases of familial schizophrenia15. Using genetic homogeneity present in Finish populations, it was found that some individuals with an increased risk of schizophrenia and intellectual disability had lost a single copy of Top3β15. Additionally, four individuals with intellectual impairment were identified that showed a homozygous loss of Top3β, and two of these were also diagnosed with schizophrenia.
Top3β is a type I DNA topoisomerase, but was recently shown to also possess RNA topoisomerase activity15,16. Top3β was found to bind RNA, to interact with RNA binding proteins and to associate with polyribosomes and RNA stress granules15,16. Critically, Top3β (but not Top3α) was also shown to directly catalyse topoisomerase activity on RNA and a point mutation that inactivates DNA topoisomerase activity also inactivated this activity towards RNA16. Top3β also bound multiple mRNAs that are encoded by genes linked to schizophrenia and autism15. While Top3β is clearly important in the nervous system, the aspects of RNA function that require its activity remain uncertain. Recently, many novel facets of RNA biology have been uncovered and multiple new forms of functionally important RNAs have been identified65-67. Amongst these, some forms may require topological modulation, such as higher order structures in mRNA or the case of circular RNA, torsional constraints may occur.
Biochemically, Top3β interacts with the N-terminal region of the tudor domain containing protein 3 (TDRD3), a transcriptional co-activator and the fragile X mental retardation protein (FMRP) can also bind TDRD3 at the C-terminus16,68. Fragile X is the most common inherited cause of intellectual disability and autism, and usually results from loss (via trinucleotide CGG repeat expansion) of the mRNA-binding protein, FMRP that functions at many synapses to inhibit translation stimulated by metabotropic glutamate receptors69. Recruitment of Top3β to transcribed genes occurs by TDRD3 binding methylated arginine residues in proteins such as histones68,70. Thus, TDRD3 acts as a scaffold that functionally integrates Top3β and FMRP. This complex also localizes to polyribosomes and stress granules, suggesting a role alongside FMRP for Top3β in protein translation16. Importantly, the endogenous Top3β-TDRD3-FMRP complex has been identified in the mouse brain16. As mutations in FMRP are found in individuals with fragile X syndrome and disease-causing FRMP mutations disrupt the interaction with Top3β/TDRD3, this complex may ensure correct mRNA translation. Further, in drosophila, Top3β mutations led to abnormal neuromuscular junctions or alterations of the rough eye phenotype, similar to those observed when FRMP was mutated or over expressed16. Therefore, Top3β potentially modulates RNA processes at both the transcriptional and translational level.
Given the striking link between specific loss of Top3β in some individuals with schizophrenia, and its interactions with FRMP and RNA, this topoisomerase may be a potent modulator of translational control in the brain, and therefore an important determinant for neurocognitive homeostasis. However, Top3β is a DNA topoisomerase71 (although substantial amounts of Top3β are localized to the cytoplasm115,16), so it remains speculative if the phenotypes associated with Top3β loss are solely from perturbation of RNA function. If an RNA substrate is the key determinant of neuropathology after Top3β disruption, then what is the role for Top3β in DNA transactions? Nonetheless, these provocative new findings for Top3β broaden the functional repertoire for topoisomerases, and point to an increasing range of general neural processes requiring these enzymes.
Conclusions and perspective
Topoisomerases are critical for fundamental aspects of neural function. Their primary functions, to cleave DNA strands to provide torsional stress relief or to untangle replicating DNA, provides essential cellular controls during replication and transcription. In the case of Top3β, topoisomerase activity involving RNA may be a key function in the nervous system. The connections between Top3β loss in schizophrenia15 and the role for Top1 in regulating long genes linked to autism spectrum disorder20 underscores a dynamic interplay between these enzymes in the maintenance of synaptic function.
Disease-associated defects in topoisomerase activity mostly impact the nervous system, raising the question of what particular lesion is responsible for cellular demise in the absence of normal topoisomerase function? Faulty topoisomerase function will likely have a different impact during neurogenesis than in the mature brain. As DNA replication is a main driving force during neurogenesis, replication fork-associated DNA strand breaks will be most detrimental to the developing nervous system13. In comparison, the high transcriptional activity of the mature brain coupled with high oxidative metabolism and associated free radicals that are generated causes frequent DNA strand breaks13,34, which in turn can trap topoisomerase on DNA, resulting in DNA damage accumulation9. During transcription, topoisomerases prevent genome instability by restraining R-Loop formation, which can lead to genomic damage and cell death68,72-74. In the case of RNA topoisomerase activity, Top3β might regulate circular RNAs. This class of RNA has been suggested to act as a sink for microRNA, which could in turn regulate mRNA translation66. Recently, Top1 was identified as a key component of the host response against pathogens, which was responsible for inflammatory gene expression75, suggesting that topoisomerases might be important components of neuroinflammation.
Illuminating additional functional requirements for topoisomerase in the nervous system will expand our knowledge of what is an already considerable functional diversity of these enzymes. Importantly, chemical manipulation of topoisomerases offers potential as a therapeutic strategy to aid in the treatments of cognitive and neurodegenerative disease.
Acknowledgments
I thank the NIH (NS-37956, CA-21765), the CCSG (P30 CA21765) and the American Lebanese and Syrian Associated Charities of St. Jude Children’s Research Hospital for financial support. Space constraints limited the number of primary research papers cited.
References
- 1.Gartenberg MR, Wang JC. Positive supercoiling of DNA greatly diminishes mRNA synthesis in yeast. Proc Natl Acad Sci U S A. 1992;89:11461–11465. doi: 10.1073/pnas.89.23.11461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Joshi RS, Pina B, Roca J. Positional dependence of transcriptional inhibition by DNA torsional stress in yeast chromosomes. The EMBO journal. 2010;29:740–748. doi: 10.1038/emboj.2009.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bermejo R, Lai MS, Foiani M. Preventing replication stress to maintain genome stability: resolving conflicts between replication and transcription. Mol Cell. 2012;45:710–718. doi: 10.1016/j.molcel.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 4.McClendon AK, Rodriguez AC, Osheroff N. Human topoisomerase IIalpha rapidly relaxes positively supercoiled DNA: implications for enzyme action ahead of replication forks. J Biol Chem. 2005;280:39337–39345. doi: 10.1074/jbc.M503320200. [DOI] [PubMed] [Google Scholar]
- 5.Vos SM, Tretter EM, Schmidt BH, Berger JM. All tangled up: how cells direct, manage and exploit topoisomerase function. Nature reviews. Molecular cell biology. 2011;12:827–841. doi: 10.1038/nrm3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nature reviews. Molecular cell biology. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 7.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 8.Nitiss JL. DNA topoisomerase II and its growing repertoire of biological functions. Nature reviews Cancer. 2009;9:327–337. doi: 10.1038/nrc2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pommier Y, et al. Repair of topoisomerase I-mediated DNA damage. Prog Nucleic Acid Res Mol Biol. 2006;81:179–229. doi: 10.1016/S0079-6603(06)81005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andersen SL, Sloan RS, Petes TD, Jinks-Robertson S. Genome-destabilizing effects associated with top1 loss or accumulation of top1 cleavage complexes in yeast. PLoS Genet. 2015;11:e1005098. doi: 10.1371/journal.pgen.1005098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen SH, Chan NL, Hsieh TS. New mechanistic and functional insights into DNA topoisomerases. Annu Rev Biochem. 2013;82:139–170. doi: 10.1146/annurev-biochem-061809-100002. [DOI] [PubMed] [Google Scholar]
- 12.Takashima H, et al. Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat Genet. 2002;32:267–272. doi: 10.1038/ng987. [DOI] [PubMed] [Google Scholar]
- 13.McKinnon PJ. Maintaining genome stability in the nervous system. Nature neuroscience. 2013;16:1523–1529. doi: 10.1038/nn.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomez-Herreros F, et al. TDP2 protects transcription from abortive topoisomerase activity and is required for normal neural function. Nat Genet. 2014;46:516–521. doi: 10.1038/ng.2929. [DOI] [PubMed] [Google Scholar]
- 15.Stoll G, et al. Deletion of TOP3beta, a component of FMRP-containing mRNPs, contributes to neurodevelopmental disorders. Nature neuroscience. 2013;16:1228–1237. doi: 10.1038/nn.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu D, et al. Top3beta is an RNA topoisomerase that works with fragile X syndrome protein to promote synapse formation. Nature neuroscience. 2013;16:1238–1247. doi: 10.1038/nn.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akimitsu N, et al. Enforced cytokinesis without complete nuclear division in embryonic cells depleting the activity of DNA topoisomerase IIalpha. Genes Cells. 2003;8:393–402. doi: 10.1046/j.1365-2443.2003.00643.x. [DOI] [PubMed] [Google Scholar]
- 18.Li W, Wang JC. Mammalian DNA topoisomerase IIIalpha is essential in early embryogenesis. Proc Natl Acad Sci U S A. 1998;95:1010–1013. doi: 10.1073/pnas.95.3.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morham SG, Kluckman KD, Voulomanos N, Smithies O. Targeted disruption of the mouse topoisomerase I gene by camptothecin selection. Mol Cell Biol. 1996;16:6804–6809. doi: 10.1128/mcb.16.12.6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King IF, et al. Topoisomerases facilitate transcription of long genes linked to autism. Nature. 2013;501:58–62. doi: 10.1038/nature12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lyu YL, et al. Role of topoisomerase IIbeta in the expression of developmentally regulated genes. Mol Cell Biol. 2006;26:7929–7941. doi: 10.1128/MCB.00617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madabhushi R, et al. Activity-Induced DNA Breaks Govern the Expression of Neuronal Early-Response Genes. Cell. 2015;161:1592–1605. doi: 10.1016/j.cell.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keeney S. Spo11 and the Formation of DNA Double-Strand Breaks in Meiosis. Genome Dyn Stab. 2008;2:81–123. doi: 10.1007/7050_2007_026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Douarre C, et al. Mitochondrial topoisomerase I is critical for mitochondrial integrity and cellular energy metabolism. PLoS One. 2012;7:e41094. doi: 10.1371/journal.pone.0041094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H, et al. Human mitochondrial topoisomerase I. Proc Natl Acad Sci U S A. 2001;98:10608–10613. doi: 10.1073/pnas.191321998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Linka RM, et al. C-terminal regions of topoisomerase IIalpha and IIbeta determine isoform-specific functioning of the enzymes in vivo. Nucleic Acids Res. 2007;35:3810–3822. doi: 10.1093/nar/gkm102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mondal N, Parvin JD. DNA topoisomerase IIalpha is required for RNA polymerase II transcription on chromatin templates. Nature. 2001;413:435–438. doi: 10.1038/35096590. [DOI] [PubMed] [Google Scholar]
- 28.Pedersen JM, et al. DNA Topoisomerases maintain promoters in a state competent for transcriptional activation in Saccharomyces cerevisiae. PLoS Genet. 2012;8:e1003128. doi: 10.1371/journal.pgen.1003128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, Wang JC, Liu LF. Involvement of DNA topoisomerase I in transcription of human ribosomal RNA genes. Proc Natl Acad Sci U S A. 1988;85:1060–1064. doi: 10.1073/pnas.85.4.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin CP, Ban Y, Lyu YL, Desai SD, Liu LF. A ubiquitin-proteasome pathway for the repair of topoisomerase I-DNA covalent complexes. The Journal of biological chemistry. 2008;283:21074–21083. doi: 10.1074/jbc.M803493200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mao Y, Sun M, Desai SD, Liu LF. SUMO-1 conjugation to topoisomerase I: A possible repair response to topoisomerase-mediated DNA damage. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:4046–4051. doi: 10.1073/pnas.080536597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ashour ME, Atteya R, El-Khamisy SF. Topoisomerase-mediated chromosomal break repair: an emerging player in many games. Nature reviews. Cancer. 2015;15:137–151. doi: 10.1038/nrc3892. [DOI] [PubMed] [Google Scholar]
- 34.Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 35.Pommier Y, et al. Tyrosyl-DNA-phosphodiesterases (TDP1 and TDP2) DNA Repair (Amst) 2014;19:114–129. doi: 10.1016/j.dnarep.2014.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gomez-Herreros F, et al. TDP2-dependent non-homologous end-joining protects against topoisomerase II-induced DNA breaks and genome instability in cells and in vivo. PLoS Genet. 2013;9:e1003226. doi: 10.1371/journal.pgen.1003226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nitiss JL, Nitiss KC. Tdp2: a means to fixing the ends. PLoS Genet. 2013;9:e1003370. doi: 10.1371/journal.pgen.1003370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.El-Khamisy SF, et al. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature. 2005;434:108–113. doi: 10.1038/nature03314. [DOI] [PubMed] [Google Scholar]
- 39.Cortes Ledesma F, El Khamisy SF, Zuma MC, Osborn K, Caldecott KW. A human 5’-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature. 2009;461:674–678. doi: 10.1038/nature08444. [DOI] [PubMed] [Google Scholar]
- 40.McKinnon PJ. DNA repair deficiency and neurological disease. Nat Rev Neurosci. 2009;10:100–112. doi: 10.1038/nrn2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang H, et al. Increased negative supercoiling of mtDNA in TOP1mt knockout mice and presence of topoisomerases IIalpha and IIbeta in vertebrate mitochondria. Nucleic Acids Res. 2014;42:7259–7267. doi: 10.1093/nar/gku384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y, Lyu YL, Wang JC. Dual localization of human DNA topoisomerase IIIalpha to mitochondria and nucleus. Proc Natl Acad Sci U S A. 2002;99:12114–12119. doi: 10.1073/pnas.192449499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang X, Li W, Prescott ED, Burden SJ, Wang JC. DNA topoisomerase IIbeta and neural development. Science. 2000;287:131–134. doi: 10.1126/science.287.5450.131. [DOI] [PubMed] [Google Scholar]
- 44.Kwan KY, Wang JC. Mice lacking DNA topoisomerase IIIbeta develop to maturity but show a reduced mean lifespan. Proc Natl Acad Sci U S A. 2001;98:5717–5721. doi: 10.1073/pnas.101132498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khiati S, et al. Lack of mitochondrial topoisomerase I (TOP1mt) impairs liver regeneration. Proc Natl Acad Sci USA. 2015;112:11282–11287. doi: 10.1073/pnas.1511016112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsutsui K, Tsutsui K, Sano K, Kikuchi A, Tokunaga A. Involvement of DNA topoisomerase IIbeta in neuronal differentiation. J Biol Chem. 2001;276:5769–5778. doi: 10.1074/jbc.M008517200. [DOI] [PubMed] [Google Scholar]
- 47.Lyu YL, Wang JC. Aberrant lamination in the cerebral cortex of mouse embryos lacking DNA topoisomerase IIbeta. Proc Natl Acad Sci USA. 2003;100:7123–7128. doi: 10.1073/pnas.1232376100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tiwari VK, et al. Target genes of Topoisomerase IIbeta regulate neuronal survival and are defined by their chromatin state. Proc Natl Acad Sci U S A. 2012;109:E934–943. doi: 10.1073/pnas.1119798109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Y, et al. Topoisomerase IIbeta is required for proper retinal development and survival of postmitotic cells. Biol Open. 2014;3:172–184. doi: 10.1242/bio.20146767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bunch H, et al. Transcriptional elongation requires DNA break-induced signalling. Nat Commun. 2015;6:10191. doi: 10.1038/ncomms10191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ju BG, et al. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 52.Heinz S, Romanoski CE, Benner C, Glass CK. The selection and function of cell type-specific enhancers. Nature reviews Molecular cell biology. 2015;16:144–154. doi: 10.1038/nrm3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Puc J, et al. Ligand-dependent enhancer activation regulated by topoisomerase-I activity. Cell. 2015;160:367–380. doi: 10.1016/j.cell.2014.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mabb AM, et al. Topoisomerase 1 Regulates Gene Expression in Neurons through Cleavage Complex-Dependent and -Independent Mechanisms. PLoS One. 2016;11:e0156439. doi: 10.1371/journal.pone.0156439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zylka MJ, Simon JM, Philpot BD. Gene length matters in neurons. Neuron. 2015;86:353–355. doi: 10.1016/j.neuron.2015.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Plaschkes I, Silverman FW, Priel E. DNA topoisomerase I in the mouse central nervous system: Age and sex dependence. J Comp Neurol. 2005;493:357–369. doi: 10.1002/cne.20793. [DOI] [PubMed] [Google Scholar]
- 57.Zehorai E, Eitan E, Hershfinkel M, Sekler I, Priel E. Glutamate regulates the activity of topoisomerase I in mouse cerebellum. Mol Neurobiol. 2008;38:242–252. doi: 10.1007/s12035-008-8044-x. [DOI] [PubMed] [Google Scholar]
- 58.Mabb AM, et al. Topoisomerase 1 inhibition reversibly impairs synaptic function. Proc Natl Acad Sci USA. 2014;111:17290–17295. doi: 10.1073/pnas.1413204111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gabel HW, et al. Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature. 2015;522:89–93. doi: 10.1038/nature14319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sugino K, et al. Cell-type-specific repression by methyl-CpG-binding protein 2 is biased toward long genes. J Neurosci. 2014;34:12877–12883. doi: 10.1523/JNEUR0SCI.2674-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang HS, et al. Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons. Nature. 2012;481:185–189. doi: 10.1038/nature10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Powell WT, et al. R-loop formation at Snord116 mediates topotecan inhibition of Ube3a-antisense and allele-specific chromatin decondensation. Proc Natl Acad Sci USA. 2013;110:13938–13943. doi: 10.1073/pnas.1305426110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McKinnon PJ. ATM and the molecular pathogenesis of ataxia telangiectasia. Annu Rev Pathol. 2012;7:303–321. doi: 10.1146/annurev-pathol-011811-132509. [DOI] [PubMed] [Google Scholar]
- 64.Katyal S, et al. Aberrant topoisomerase-1 DNA lesions are pathogenic in neurodegenerative genome instability syndromes. Nature neuroscience. 2014;17:813–821. doi: 10.1038/nn.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hansen TB, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 66.Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20:1829–1842. doi: 10.1261/rna.047126.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Memczak S, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 68.Yang Y, et al. Arginine methylation facilitates the recruitment of TOP3B to chromatin to prevent R loop accumulation. Mol Cell. 2014;53:484–497. doi: 10.1016/j.molcel.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bhakar AL, Dolen G, Bear MF. The pathophysiology of fragile X (and what it teaches us about synapses) Annu Rev Neurosci. 2012;35:417–443. doi: 10.1146/annurev-neuro-060909-153138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang Y, et al. TDRD3 is an effector molecule for arginine-methylated histone marks. Mol Cell. 2010;40:1016–1023. doi: 10.1016/j.molcel.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seki T, Seki M, Onodera R, Katada T, Enomoto T. Cloning of cDNA encoding a novel mouse DNA topoisomerase III (Topo IIIbeta) possessing negatively supercoiled DNA relaxing activity, whose message is highly expressed in the testis. J Biol Chem. 1998;273:28553–28556. doi: 10.1074/jbc.273.44.28553. [DOI] [PubMed] [Google Scholar]
- 72.Aguilera A, Garcia-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol Cell. 2012;46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 73.Skourti-Stathaki K, Proudfoot NJ. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 2014;28:1384–1396. doi: 10.1101/gad.242990.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tuduri S, et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol. 2009;11:1315–1324. doi: 10.1038/ncb1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rialdi A, et al. Topoisomerase 1 inhibition suppresses inflammatory genes and protects from death by inflammation. Science. 2016;352:aad7993. doi: 10.1126/science.aad7993. [DOI] [PMC free article] [PubMed] [Google Scholar]


