Abstract
Isolates of the rice blast fungus Magnaporthe grisea that carry the gene encoding Avirulence Conferring Enzyme1 (ACE1) are specifically recognized by rice (Oryza sativa) cultivars carrying the resistance gene Pi33. This recognition enables resistant plants to activate a defense response. ACE1 was isolated by map-based cloning and encodes a putative hybrid between a polyketide synthase and a nonribosomal peptide synthetase, enzymes involved in microbial secondary metabolism. ACE1 is expressed exclusively during fungal penetration of host leaves, the time point at which plant defense reactions are triggered. Ace1 appears to be localized in the cytoplasm of the appressorium. Mutation of the putative catalytic site of the β-ketoacyl synthase domain of Ace1 abolishes recognition of the fungus by resistant rice. This suggests that Ace1 biosynthetic activity is required for avirulence. Our results are consistent with the hypothesis that the fungal signal recognized by resistant rice plants is the secondary metabolite whose synthesis depends on Ace1.
INTRODUCTION
In the interaction between plants and their microbial pathogens, naturally occurring disease resistance often depends on specific interactions governed by pathogen avirulence (AVR) and plant disease resistance (R) genes (Flor, 1971). In these interactions, the pathogen produces a signal under the control of the AVR gene, whose recognition by the plant requires R gene activity. R genes encode related proteins that are classified based on the presence of leucine-rich repeats, nucleotide binding site, leucine zipper, Toll/Interleukin-1 receptor, and transmembrane domains (Martin et al., 2003). The predominant class of R proteins belongs to the nucleotide binding site–leucine-rich repeat type and appears to be directly involved in translating the presence of the pathogen-derived AVR signal into a plant response leading to resistance. It was originally proposed that the R protein acts as receptor for the AVR signal. However, after a series of unsuccessful attempts to demonstrate direct interaction between R protein and the corresponding AVR factor, this situation is now considered to occur rarely (Hammond-Kosack and Parker, 2003). Accumulating evidence supports a model termed the guard hypothesis that proposes that the R protein recognizes a complex containing the AVR signal and other plant proteins (van der Biezen and Jones, 1998).
Understanding the molecular basis of disease resistance requires not only knowledge about the mechanisms underlying perception of AVR signals by the host but also about the biological processes involved in avirulence signaling and their role in pathogenicity. Most of our current knowledge about AVR genes was gained from studies on plant pathogenic bacteria. More than 40 bacterial AVR genes were characterized to date, mostly in the genera Pseudomonas and Xanthomonas (Vivian and Arnold, 2000). Most bacterial AVR genes encode proteins without known functions that share a common active delivery system into the host cell (type III secretion). Bacterial AVR genes with known functions belong to the AvrBs3 family that displays typical features of eukaryotic transcription factors and modulate host gene expression (Marois et al., 2002) and to the AvrPtoB family that suppresses plant defense by inhibition of host programmed cell death (Abramovitch et al., 2003). The two unrelated Pseudomonas AVR factors AvrRpm1 and AvrB interact with RIN4, a protein that is involved in regulating basal defense responses in Arabidopsis thaliana. These interactions support the guard hypothesis because RIN4 appears to be guarded by the Arabidopsis R protein RPM1 (Mackey et al., 2002).
Despite the fact that fungi represent some of the most damaging crop pathogens, our knowledge about fungal AVR genes is still limited. Twelve fungal AVR genes have been characterized to date (Farman et al., 2002; Knogge, 2002; Luderer et al., 2002a, 2002b; Dodds et al., 2004) as well as one oomycete AVR gene (Shan et al., 2004). They encode small proteins or peptides without homology to each other that are secreted into host plants during infection. As for the bacterial AVR factors, predicting function for the proteins encoded by fungal AVR genes has proven difficult. NIP1 from the barley (Hordeum vulgare) pathogen Rhynchosporium secalis encodes a peptidic toxin that contributes to pathogenicity by inducing necrotic lesions (Rohe et al., 1995). ECP2 from the tomato (Lycopersicon esculentum) pathogen Cladosporium fulvum encodes a protein without known biochemical function that acts as pathogenicity factor (Laugé et al., 1998), and AVR4 from C. fulvum encodes a secreted protein that protects chitin from fungal cell walls (van den Burg et al., 2004). Finally, AVR-Pita from the rice (Oryza sativa) pathogen Magnaporthe grisea encodes a putative zinc-dependent metalloprotease that is dispensable for pathogenicity (Orbach et al., 2000). The interaction between AVR-Pita and the corresponding rice R protein Pi-ta is one of the few examples of direct interaction between R and AVR proteins (Jia et al., 2000). Clearly, more fungal AVR genes need to be characterized to investigate the spectrum of signals recognized by resistant host plants. In this context, the M. grisea–rice pathosystem is an attractive model for studying the interaction between an avirulent fungal plant pathogen and its resistant host. M. grisea is responsible for the most devastating disease of cultivated rice, called blast. Disease control relies mainly on the use of resistant rice cultivars, and several AVR genes have been identified in this fungus using classical genetics (Notteghem et al., 1994). Besides the above mentioned AVR-Pita, only one other AVR gene, AVR-CO39, and a set of host specificity genes, the PWL1-4 genes, have been characterized so far (Kang et al., 1995; Sweigard et al., 1995; Farman et al., 2002).
The AVR gene Avirulence Conferring Enzyme1 (ACE1) from M. grisea, previously named AVR-Irat7, was identified in a cross between Guy11, a hermaphroditic isolate from South America, and ML25, a male isolate from Africa (Silué et al., 1992). Guy11 is unable to infect resistant rice cultivars Irat7, DJ8-341, and Carreon, whereas the full-sib progeny 2/0/3 is virulent on these cultivars (Dioh et al., 2000). The rice resistance gene Pi33 is responsible for the recognition of isolates carrying ACE1 (Berruyer et al., 2003). Pi33 was identified in different semidwarf indica rice cultivars, including the high yielding rice cultivar IR64 extensively used for rice breeding at the International Rice Reseach Institute. Pi33 mapped on rice chromosome 8 and was distinguished from known neighboring resistance genes using allelism tests (Berruyer et al., 2003). ACE1 segregates as a single Mendelian locus in the backcross between Guy11 and 2/0/3. Using progeny from this backcross, a genetic map was constructed based on restriction fragment length polymorphism (RFLP) and random amplified polymorphic DNA (RAPD) markers. ACE1 was found to cosegregate with RAPD marker OPE-Y13, which maps on chromosome 1 at 20 centimorgans from AVR1-CO39 (Farman and Leong, 1998; Dioh et al., 2000).
Here, we describe the cloning and characterization of the AVR gene ACE1 from M. grisea. ACE1 encodes a putative hybrid between a polyketide synthase (PKS) and a nonribosomal peptide synthetase (NRPS). PKS and NRPS are two distinct classes of enzymes involved in the production of secondary metabolites. ACE1 is expressed specifically during penetration, and its translation product appears to be localized in the cytoplasm of appressoria. A single amino acid exchange in the β-ketoacyl synthase catalytic site of Ace1 abolishes recognition of the fungus by resistant cultivars. This suggests that Ace1 biosynthetic activity is required for avirulence and that the detection of the invading fungal pathogen involves recognition of a secondary metabolite whose biosynthesis depends on Ace1.
RESULTS
Cloning of M. grisea Avirulence Gene ACE1
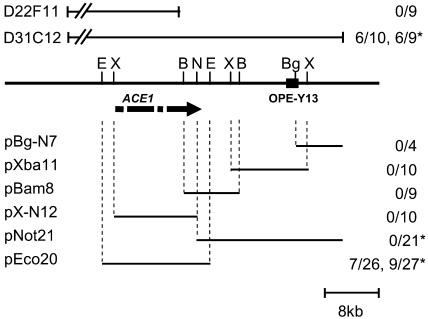
The single copy RAPD marker OPE-Y13 that cosegregates with the avirulence gene ACE1 (Dioh et al., 2000) was used to screen a genomic cosmid library from avirulent progeny 96/0/76 (Dioh et al., 1997). The cosmid clone D31C12 was found to carry OPE-Y13. Using cosmid ends as probes, we isolated a single overlapping cosmid, D22F11. Together, both cosmids span a genomic region of 55 kb (Figure 1). To test for presence of ACE1, circular cosmids D31C12, D22F11, and the empty cosmid vector pMOcosX were introduced by transformation into virulent parent 2/0/3. Three pMOcosX transformants and nine D22F11 transformants were inoculated on susceptible rice cultivar Maratelli and resistant rice cultivar Irat7 carrying Pi33. These transformants were virulent on both cultivars (Figure 1), indicating that cosmid D22F11 does not carry ACE1. By contrast, six of ten transformants obtained with D31C12 were unable to infect resistant cultivar Irat7 (Figure 1), although they retained full pathogenicity toward susceptible cultivar Maratelli (data not shown), suggesting that D31C12 carries ACE1.
Figure 1.
Isolation of ACE1 by Complementation.
Schematic representation of the ACE1 locus in Guy11, cosmids, and subclones tested for complementation to avirulence. The genomic locus is represented as a solid line, and the positions of RAPD marker OPE-Y13 and relevant restriction sites are indicated as follows: E, EcoRI; X, XbaI; B, BamHI; N, NotI; Bg, BglII. The ACE1 ORF is indicated as an arrow, and interruptions indicate positions of the three introns. Above the genomic locus map, the relative positions of the noncomplementing cosmid D22F11 and the complementing cosmid D31C12 are shown. Below the genomic locus map, the relative positions of D31C12 subclones used for complementation are shown. Fractions given to the right of each construct indicate the number of avirulent transformants identified over the number of transformants tested. Numbers without asterisks represent results obtained with virulent parent 2/0/3, numbers with asterisks represent results obtained with virulent field isolate PH14. Cosmid D31C12 and plasmid pEco20 complement both virulent strains to avirulence, demonstrating that they carry ACE1.
To locate ACE1 within D31C12, five overlapping subclones (pBg-N7, pXba11, pBam8, pX-N12, and pEco20; Figure 1) were constructed in the vector pCB1004 and introduced by transformation into virulent parent 2/0/3. The resulting transformants and two control 2/0/3 transformants obtained with cloning vector pCB1004 were inoculated on cultivars Irat7 and Maratelli. The 33 transformants obtained with the subclones pBg-N7, pXba11, pBam8, and pX-N12 and the two 2/0/3-pCB1004 transformants were virulent toward both resistant and susceptible rice cultivars (Table 1, Figure 1). By contrast, subclone pEco20 yielded seven avirulent transformants toward resistant cultivar Irat7 among 26 tested (Figure 1, transformant HBE20 in Figure 2B). These avirulent transformants were fully pathogenic toward susceptible cultivar Maratelli, suggesting that pEco20 carries ACE1. To confirm their specific interaction with Pi33, these avirulent transformants were inoculated on rice cultivars IR1529, C101lac, and Bala that carry Pi33 but are of different genetic origin than Irat7 (Berruyer et al., 2003). Rice cultivar Sariceltik was included as additional susceptible control. As expected, transformants that were avirulent toward Irat7 were also avirulent toward IR1529, C101lac, and Bala (Table 1), confirming that pEco20 carries the dominant avirulence gene ACE1 that interacts specifically with Pi33.
Table 1.
Pathogenicity Assays of 2/0/3-ACE1 Transformants and Guy11-ace1 Deletion Mutants
| Cultivar | Guy11 | 2/0/3 | 2/0/3-31C12 #5 | 2/0/3-pMOcosX#1 | 2/0/3-pEco20 (HBE20) | 2/0/3-pCB1004#48 | Guy11-HB26 | Guy11-HB92 | Guy11-HB24 (Ectopic) | Guy11-HB16 (Ectopic) | Guy11-HB33 (Ectopic) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Irat7 (R) | AVR | vir | AVR | vir | AVR | vir | vir | vir | AVR | AVR | AVR |
| IR1529 (R) | AVR | vir | AVR | vir | AVR | vir | vir | vir | AVR | AVR | AVR |
| C101lac (R) | AVR | vir | AVR | vir | AVR | vir | vir | vir | AVR | AVR | AVR |
| Bala (R) | AVR | vir | AVR | vir | AVR | vir | vir | vir | AVR | AVR | AVR |
| LS1 (s) | vir | vir | – | – | vir | vir | vir | vir | vir | vir | – |
| LTH (s) | vir | vir | – | – | vir | vir | vir | vir | vir | vir | – |
| Maratelli (s) | vir | vir | vir | vir | vir | vir | vir | vir | vir | vir | vir |
| Sariceltik (s) | vir | vir | vir | vir | vir | vir | vir | vir | vir | vir | vir |
R, resistant (carrying Pi33); s, susceptible; AVR, avirulent; vir, virulent; –, not tested.
Figure 2.
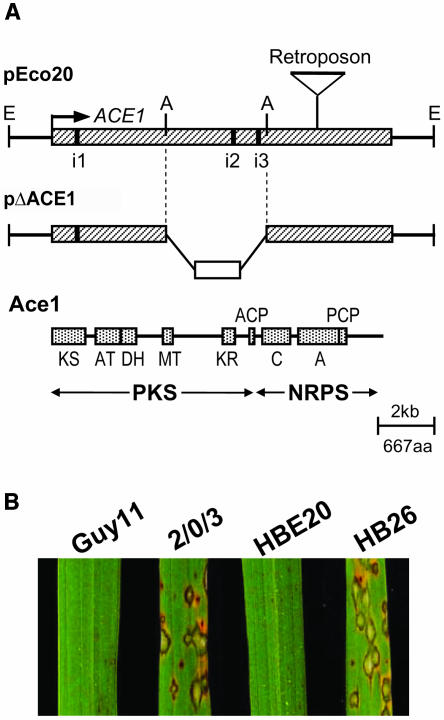
Organization of Avirulence Gene ACE1 and Analysis of Its Partial Deletion Mutants.
(A) Physical map of the complementing plasmid pEco20, insertion site of retroposon in 2/0/3 ace1 virulent allele, the gene replacement vector pΔACE1, and Ace1 protein encoded by ACE1. The ACE1 ORF is represented as a hatched box, the three introns (i1 to i3) as black bars, and restriction sites as E for EcoRI and A for AgeI. In the ace1 allele from virulent parent 2/0/3, a 2-kb sequence is inserted at 2.6 kb before the ACE1 stop codon indicated by a retroposon sign on the pEco20 physical map. In the gene replacement vector pΔACE1, the hph resistance gene (open box) replaces an internal 3.6-kb AgeI fragment. In the ACE1 translation product (Ace1), stippled boxes represent enzymatic domains (drawn to scale): KS, β-ketoacyl synthase; AT, acyltransferase; DH, dehydratase; MT, methyl-transferase; KR, β-ketoreductase; ACP, acyl carrier protein; C, condensation domain; A, AMP binding domain; PCP, peptidyl carrier protein. PKS designates the polyketide synthase part of Ace1 and NRPS the nonribosomal peptide synthetase module of Ace1. aa, amino acids.
(B) Infection assays on resistant rice cultivar C101lac. Seedlings were spray inoculated with avirulent parent Guy11, virulent parent 2/0/3, 2/0/3 transformant HBE20 carrying pEco20, and Guy11 ace1 deletion mutant HB26. Typical symptoms on rice leaves were obtained 7 d after inoculation. Guy11 is unable to infect the Pi33-carrying rice cultivar C101lac. Isolate 2/0/3 is virulent on C101lac and induces typical susceptible lesions. The 2/0/3 transformant HBE20 is avirulent on C101lac as a result of the introduction of ACE1 carried by pEco20. HB26 is virulent on C101lac as a result of the partial deletion of ACE1 by gene replacement using the pΔACE1 construct.
To corroborate the results of the complementation assays performed with virulent parent 2/0/3, we tested some selected D31C12 subclones for their ability to confer avirulence to the virulent field isolate PH14. Cosmid D31C12 and the subclones pEco20 and pNot21 that together span the entire genomic fragment carried by D31C12, were introduced by transformation into PH14. Six of nine transformants obtained with D31C12 and nine of 27 obtained with pEco20 were unable to infect the Pi33 rice cultivars Irat7 and IR1529 but were fully virulent toward Maratelli (Figure 1). By contrast, the 21 PH14 transformants carrying pNot21 or the empty vector pCB1004 were as virulent on resistant rice cultivars as the recipient strain PH14. These results indicate that cosmid D31C12 and its subclone pEco20 have the ability to confer avirulence toward rice cultivars carrying Pi33 independently of the virulent strain used as a recipient in transformation.
Subclone pEco20 carries a genomic EcoRI fragment of 15.4 kb. Sequence analysis of this fragment revealed a large open reading frame (ORF) of 12.5 kb interrupted by a few stop codons that correspond to three putative introns of 87, 91, and 70 bp, respectively (Figure 2A). The positions of these introns were confirmed experimentally by RT-PCR using total RNA extracted from barley leaves infected with avirulent isolate Guy11 17 h after inoculation and sequencing of the amplification products. Removal of the introns from the genomic sequence revealed an ORF of 12.1 kb. Transcription initiation and polyadenylation sites of ACE1 were determined by rapid amplification of both cDNA ends (5′ and 3′ RACE). The 5′ and 3′ untranslated regions (UTRs) of ACE1 were found to be 198 and 144 bp in length, respectively. The complete ACE1 transcript is therefore expected to be 12,450-bp long. Besides the complete ACE1 ORF, the complementing subclone pEco20 carries 1390 bp of upstream sequence (promoter region) and 1297 bp of downstream sequence (terminator region).
To prove that this ORF is the ACE1 gene responsible for the avirulence of Guy11 toward rice cultivars carrying Pi33, we constructed mutants of Guy11 carrying a partial deletion of this ORF (ace1Δ:hph). To this end, we constructed a gene replacement vector pΔACE1 to replace an internal ACE1 3.6-kb AgeI fragment by a hygromycin B resistance cassette (Figure 2A). After the introduction of pΔACE1 into Guy11 by transformation, we identified two transformants (HB26 and HB92) amongst 100 tested that had replaced their ACE1 allele by the ace1Δ:hph allele. The two ace1 deletion mutants HB26 and HB92 were inoculated on Pi33 rice cultivars Irat7, IR1529, C101lac, and Bala and on the susceptible cultivars Maratelli and Sariceltik. The avirulent parental strain Guy11, the virulent strain 2/0/3, and three transformants (HB16, HB24, and HB33) that resulted from the ectopic integration of pΔACE1 served as controls. As expected, the three ectopic transformants HB16, HB24, and HB33 and the avirulent isolate Guy11 were unable to infect resistant cultivars but were fully pathogenic on Maratelli and Sariceltik (Table 1). By contrast, both ace1 deletion mutants HB26 and HB92 were as pathogenic as the virulent parent 2/0/3 on the susceptible rice cultivars and on rice cultivars Irat7, IR1529, C101lac, and Bala, which contain Pi33 (Table 1, HB26 in Figure 2B). These results demonstrate that the 12.5-kb ORF carried by pEco20 is the ACE1 avirulence gene that interacts with the resistance gene Pi33.
Because the virulent ace1 deletion mutants HB26 and HB92 are still pathogenic on rice, ACE1 does not appear to be required for pathogenicity of M. grisea under our standard infection conditions. To test whether ACE1 contributes to the aggressiveness of this fungus, we performed a quantitative pathogenicity assay with the virulent ace1 deletion mutants HB26 and HB92, avirulent parent Guy11, and the two avirulent ectopic transformants HB16 and HB24. Additionally, we included avirulent transformant HBE20, control transformant 2/0/3-pCB1004, and virulent parent 2/0/3 (Table 1). Inoculations were performed on the following cultivars that do not carry Pi33: Sariceltik (highly susceptible japonica cultivar), Lijiangxintuanheigu (LTH; susceptible japonica cultivar), Maratelli (susceptible japonica cultivar), and Lung Sheng 1 (LS1; partially resistant japonica cultivar). These cultivars were inoculated with M. grisea conidial suspensions using two different methods, injection and spray inoculation. In these assays, virulent ace1 deletion mutants HB26 and HB92 induce the same number of susceptible lesions on rice leaves (data not shown) as Guy11 and avirulent isogenic strains (HB16 and HB24). Furthermore, these lesions have the same size as those induced by avirulent strains (data not shown). These results suggest that ACE1 does not contribute to lesion formation or invasive growth under our infection conditions. However, we cannot exclude that ACE1 contributes to the aggressiveness of this fungus on other rice cultivars/host plants or under different infection conditions.
Loss of Avirulence in Virulent Parent 2/0/3
A survey of rice M. grisea isolates collected in rice fields at various worldwide locations revealed that virulence toward Pi33 is less frequent in nature than avirulence (Berruyer et al., 2003). To understand the molecular events responsible for the loss of avirulence in the virulent parent 2/0/3, we compared the structure of the ACE1 locus in 2/0/3 and avirulent parent Guy11. Genomic DNA gel blot hybridization revealed that ACE1 is present in the genome of 2/0/3. RFLP analysis revealed a single difference in hybridization patterns between both strains. An internal 1.9-kb XhoI fragment of the ACE1 allele from Guy11 appeared to be increased in size by 2 kb in 2/0/3 (see Supplemental Figure 1 online). The corresponding genomic region from 2/0/3 was amplified by PCR and sequenced. This revealed a 2-kb insertion in the last exon of ACE1 (Figure 2A) that contains a fragment of the MGL Line1-like retroposon (GenBank accession number AF018033). Because no further RFLP was detected between Guy11 and 2/0/3, this insertion is likely to be responsible for the loss of avirulence.
ACE1 Encodes a Putative PKS/NRPS
The ACE1 gene encodes a predicted 4035–amino acid polypeptide with a molecular mass of 438 kD. The deduced Ace1 polypeptide exhibits a high degree of similarity to type I fungal PKS at the N terminus and to bacterial NRPS at the C terminus. The PKS lovastatin nonaketide synthase (LNKS) from Aspergillus terreus, a key enzyme involved in the biosynthesis of the antihypercholesterolemic lovastatin, shows a high degree of similarity to Ace1 (37% identity and 53% similarity). Fungal type I PKS are composed of a set of enzymatic domains that are used iteratively to catalyze the sequential condensation and modification of acyl-CoA precursors (Hutchinson et al., 2000). Ace1 is predicted to contain six enzymatic domains characteristic of fungal PKS (Figure 2A): β-ketoacyl synthase (KS), acyltransferase (AT), dehydratase (DH), methyltransferase (MT), β-ketoreductase (KR), and acyl carrier protein (ACP). The conserved active site motifs of KS, AT, DH, MT, and ACP domains (Hopwood and Sherman, 1990; Aparicio et al., 1996; Hopwood, 1997) were identified in the Ace1 protein sequence. The sequence of the putative NADPH binding site of the Ace1 KR domain (GLTGGLA) does not comply with the conserved consensus sequence described for PKS and fatty acid synthase (FAS) (GGxGxLG) and is therefore likely to be nonfunctional (Mathur and Kolattukudy, 1992). However, the KR domain is not essential for polyketide backbone biosynthesis but is involved in its modification by β-ketoreduction. Therefore, the PKS part of Ace1 is likely to be functional.
The C-terminal 1435–amino acid sequence of Ace1 is highly similar to MxaA from the myxobacterium Stigmatella auranthiaca (30% identity and 45% similarity). NRPS are composed of repeated, coordinated groups of enzymatic domains called modules. Each module consists of a condensation domain (C-domain), an adenylation domain (A-domain), and a peptidyl carrier protein (PCP) and is responsible for activation and covalent attachment of an amino acid (Marahiel et al., 1997). The C terminus of Ace1 represents a single complete NRPS module composed of a C-domain, an A-domain, and a PCP domain (Figure 2A). Because all core sequences described for bacterial NRPS (Marahiel et al., 1997) were identified in Ace1 protein sequence (shown for core motif C3 in Table 2), the NRPS module of Ace1 is likely to be functional. The structural basis of substrate recognition and activation by bacterial amino acid activating A-domains has been determined (Conti et al., 1997), allowing the deduction of the specificity-conferring code of bacterial adenylation domains. This code is contained in the signature sequence that is composed of the amino acid residues that line the amino acid binding pocket of the A-domain (Stachelhaus et al., 1999; Challis et al., 2000). Following the example of bacterial A-domains, we tried to predict which amino acid is activated by the Ace1 adenylation domain by comparing its signature sequence to the 31 bacterial NRPS signature sequences described by Stachelhaus and colleagues. Ace1 A-domain displayed closest similarity (70% identity) to Orn activating domains (DMAQLGGINK for Ace1 and DMENLGLINK for Orn1). However, because these signatures were defined for A-domains from bacterial NRPS, their specificity may be different from eukaryotic NRPS. In summary, our analyses suggest that Ace1 is a functional enzyme corresponding to a hybrid between a PKS and a NRPS. Previously, functional PKS/NRPS hybrids have been described exclusively in prokaryotes (Silakowski et al., 1999).
Table 2.
Amino Acid Sequences of Core Motif C3 from the NRPS Condensation Domaina
| Enzyme | Sequence |
|---|---|
| LNKS | HRLVGDG |
| Ace1 | HHINMDG |
| Conserved motif | HHxxxDG |
The core motif C3 is essential for NRPS catalytic activity (Marahiel et al., 1997).
Ace1 Belongs to a Family of Fungal PKS/NRPS
Among fungal PKS with known biochemical activities, LNKS shows the highest degree of sequence identity to Ace1 (37%). LNKS carries the same PKS domains as Ace1 (KS, AT, DH, MT, KR, and ACP) and displays at its C terminus a sequence with similarity to NRPS condensation domains (C-domains; Hendrickson et al., 1999). In contrast with Ace1, the KR domain from LNKS is functional (Hutchinson et al., 2000), but the C-domain from the LNKS NRPS module lacks the second essential His of the core 3 motif HHxxxDG (Table 2; Marahiel et al., 1997) and is therefore unlikely to be functional. Additionally, LNKS does not carry the adenylation and PCP domains required for a functional NRPS module. The MlcA gene from Penicillium citrinum (Abe et al., 2002) encodes an enzyme with a biosynthetic activity similar to LNKS that carries a C-domain at its C terminus but has no adenylation and PCP domains.
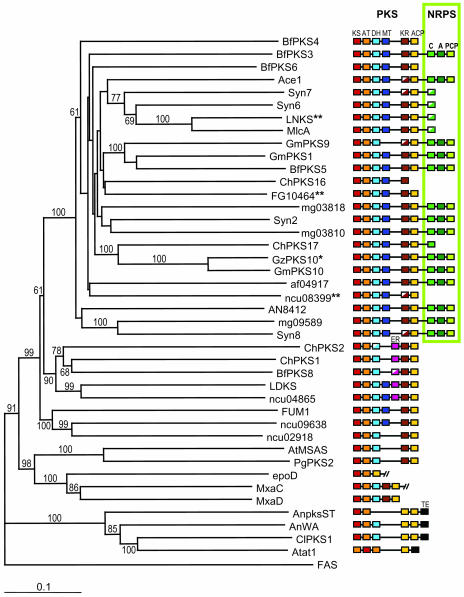
The search for sequences homologous to Ace1 in fungal genome databases from the Broad Institute (http://www.broad.mit.edu/annotation/fungi/fgi/) and the Institute for Genomic Research (http://www.tigr.org) revealed 10 putative PKS/NRPS (Figure 3). In the M. grisea genome sequence, we identified apart from Ace1, five PKS/NRPS with a complete NRPS module (mg03810, mg03818, mg09589, Syn2, and Syn8) and two LNKS-like PKS/NRPS (Syn6 and Syn7) with a truncated NRPS module. Three of these PKS/NRPS are encoded by hypothetical proteins deduced from M. grisea ORFs (mg03810, mg03818, and mg09589) and four by novel genes we identified in the M. grisea genome (Syn2, Syn6, Syn7, and Syn8). Putative PKS/NRPS were also identified as hypothetical proteins deduced from ORFs of Aspergillus nidulans (AN8412), A. fumigatus (af04917), and Fusarium graminearum (FG10464) genomes. Independently, a phylogenomic analysis of fungal type I PKS reported the presence of seven other putative PKS/NRPS in the genomes of the ascomycetes Botryotinia fuckeliana, Cochliobolus heterostrophus, Gibberella moniliformis, and G. zeae (Kroken et al., 2003). A phylogenetic analysis of these putative PKS/NRPS was performed with the protein sequences of their KS and AT domains (see Supplemental Figure 2 online) using the neighbor-joining method. We included fungal PKS representatives of the monophyletic groups described by Kroken and coauthors, bacterial PKS (epoD, MxaC, and MxaD) related to fungal PKS, and the FAS from the silk moth Bombyx mori as outgroup. This phylogenetic analysis showed that PKS/NRPS clustered as a single group distinct from other fungal PKS because the node supporting this cluster has a bootstrap value of 100 (Figure 3). Interestingly, five PKS (BfPKS4, BfPKS6, ChPKS16, FG10464, and ncu08399) clustered with PKS/NRPS, suggesting they derive from PKS/NRPS by the complete loss of their NRPS module. Whereas all genomes from filamentous ascomycetes analyzed appeared to contain PKS/NRPS encoding genes, we could not identify such genes in the genomes of the hemi-ascomycetous yeast Saccharomyces cerevisae, the archeo-ascomycete Schizosaccharomyces pombe, and the basidiomycetes Ustilago maydis, Coprinus cinereus, and Cryptococcus neoformans. Overall, we did not identify PKS/NRPS orthologous to Ace1 in other fungal genomes, although we clearly showed that Ace1 belongs to a novel family of fungal PKS/NRPS.
Figure 3.
Phylogeny of Fungal PKS Based on Protein Sequence of KS and AT Domains.
The consensus phylogenetic tree was obtained using the neighbor-joining method with distances. Bootstrap values are indicated above or below nodes of the tree. Colored boxes represent PKS and NRPS enzymatic domains (see Figure 2 for definitions). Sequences from the following organisms were included in this analysis (see Methods for accession numbers): M. grisea (Ace1, Syn2, Syn6,7,8, mg03810, mg03818, and mg09589); A. fumigatus (af04917); A. nidulans (AN8412, AnpksST, and AnWA); F. graminearum (FG10464); N. crassa (ncu08399, ncu04865, ncu09638, and ncu02918); A. terreus (LNKS, LDKS, AtMSAS, and Atat1); P. citrinum (MlcA); P. griseofulvum (PgPKS2), C. heterostrophus (ChPKS1); G. moniliformis (FUM1), and C. lagenarium (ClPKS1). We added fungal PKS and PKS/NRPS sequences described by Kroken et al. (2003) from C. heterostrophus (ChPKS2,16,17), B. fuckeliana (BfPKS3,4,5,6,8), G. moniliformis (GmPKS1,9,10), and G. zeae (GzPKS10) and bacterial PKS sequences (epoD, MxaC, and MxaD) as references. FAS from the silk moth Bombyx mori was included as an outgroup to root the tree and PKS/NRPS cluster as a monophyletic group. *, Although GzPKS10 is only described as a partial PKS sequence, its relatedness to GmPKS10 (90% identity) suggests that it has a full NRPS module as GmPKS10. **, FG10464 is almost identical (>91% identity) to GzPKS9 from Kroken et al. (2003). ncu08399 is identical to NcPKS4 from Kroken et al. (2003). LNKS is identical to lovB from Kroken et al. (2003).
ACE1 Is Expressed Exclusively during Penetration of M. grisea into the Leaf
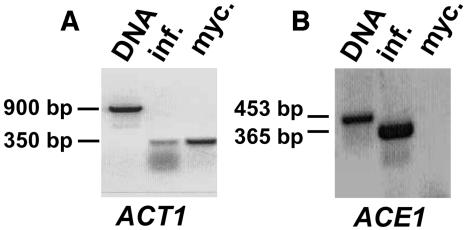
The expression of ACE1 in the avirulent M. grisea isolate Guy11 during mycelial growth and barley infection was analyzed by RT-PCR. RNA was isolated from mycelium grown in axenic culture and from infected barley leaves at the onset of penetration 17 h after inoculation. Primers were designed to amplify an ACE1 genomic fragment of 453 bp carrying the first intron and corresponding to a 365-bp cDNA. The actin-encoding gene ACT1 served as constitutive control. RT-PCR products were cloned and sequenced to confirm the specificity of amplification. An RT-PCR product corresponding to the ACT1 transcript was obtained from both RNA samples (Figure 4A). By contrast, the ACE1 transcript was detected only in RNA isolated from infected leaves but not in the mycelial RNA sample (Figure 4B). This suggested that ACE1 expression is infection specific.
Figure 4.
Expression Pattern of ACE1 in Avirulent Isolate Guy11.
(A) PCR and RT-PCR amplification of the constitutively expressed ACT1 gene as control of RNA integrity. The region spanning the first three introns of ACT1 was amplified. Lane 1, ACT1 PCR product obtained from genomic DNA (size, 900 bp). Lane 2, ACT1 RT-PCR product obtained from RNA isolated from barley leaves infected with Guy11 17 h after infection (size, 350 bp). Lane 3, ACT1 RT-PCR product obtained from RNA isolated from Guy11 mycelial culture. The amplification products were separated by agarose gel electrophoresis and stained with ethidium bromide.
(B) PCR and RT-PCR amplification of ACE1 from avirulent parent Guy11. The region spanning the first intron of ACE1 was amplified. Lane 1, ACE1 PCR product obtained from genomic DNA (size, 453 bp). Lane 2, ACE1 RT-PCR product obtained from RNA isolated from barley leaves infected with Guy11 17 h after inoculation (size, 365 bp). Lane 3, No ACE1 RT-PCR product was obtained with RNA isolated from Guy11 mycelial culture, suggesting that ACE1 is not expressed in mycelium.
To follow the expression of ACE1 during infection, we fused the green fluorescent protein (GFP) reporter gene to the ACE1 promoter and terminator sequences. The resulting reporter construct, pProACE1:GFP, was introduced by transformation into avirulent isolate Guy11. Microscopic analysis of transformants carrying the ProACE1:GFP reporter construct confirmed that ACE1 is not expressed at detectable levels in mycelium during axenic growth of the fungus in culture (data not shown). Transformants carrying the ProACE1:GFP reporter construct were inoculated on rice and barley leaves. They did not display GFP fluorescence in spores and germ tubes differentiated on the surface of rice and barley leaves. The characteristic green fluorescence of GFP was first detected in mature appressoria 15 h after inoculation of barley or rice leaves (data not shown). GFP fluorescence was strongest during penetration of the leaf surface ∼24 h after inoculation (data not shown). After penetration into the epidermal cell (∼36 h), GFP fluorescence was also detected in infectious hyphae (Figure 5A), illustrating cytoplasmic continuity between both structures (Bourett and Howard, 1990; Koga, 1994). The GFP fluorescence rapidly disappeared once secondary infectious hyphae began to spread within leaf tissues (∼48 h; data not shown).
Figure 5.
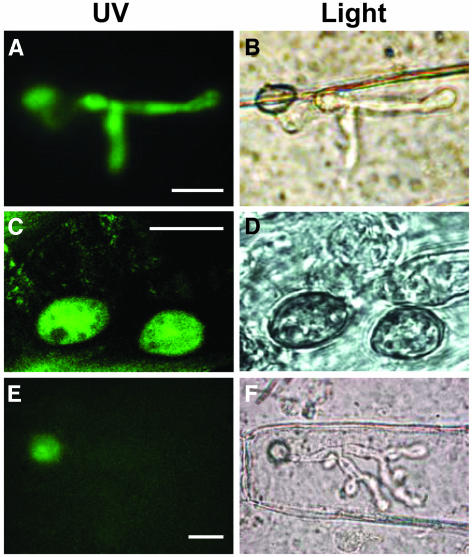
Localization of Ace1-GFP Fusion Protein in M. grisea Appressoria.
(A) GFP fluorescence of appressoria and infectious hypha after penetration of barley leaves. Conidia of transformant IF22 expressing GFP under the control of the ACE1 promoter were inoculated on detached barley leaves and incubated at 26°C. At 36 h after infection, epidermal strips were removed from the infected leaf and observed under a microscope. Under blue light, GFP fluorescence is detected both in appressorium and in infectious hyphae. Bar = 10 μm.
(B) Same view as (A) under bright field. An appressorium and its infectious hypha located in the underlying epidermal cell are visible.
(C) Ace1-GFP fluorescence of appressoria on the surface of barley leaves. Conidia of transformant HB41 expressing the Ace1-GFP fusion protein under the control of ACE1 promoter were inoculated on detached barley leaves incubated for 24 h at 26°C and observed with confocal laser scanning microscopy. Under blue light, fluorescence of the Ace1-GFP fusion protein is detected exclusively in the cytoplasm of the appressoria. Bar = 10 μm.
(D) Same view as (C) under bright field. Two germinated spores and appressoria are visible on the leaf surface.
(E) Ace1-GFP fluorescence of appressoria after penetration of barley leaves. Conidia of transformant HB44 expressing the Ace1-GFP fusion protein were inoculated on detached barley leaves and incubated at 26°C. Epidermal strips were removed from the infected leaf 40 h after inoculation and observed under a microscope. Under blue light, GFP fluorescence is detected in the appressorium but not in infectious hypha. Bar = 10 μm.
(F) Same view as (E) under bright field. An appressorium and its infectious hypha located in the underlying epidermal cell are visible.
A similar appressorium-specific expression pattern was described for the M. grisea genes GAS1 (MAS3), GAS2 (MAS1), CBP1, and AI68463 (Banno et al., 2003; Takano et al., 2003). We searched for common transcription factor binding sites in the promoter sequences of ACE1, GAS1 (AF363065), GAS2 (AF264035), CBP1 (BG809764), and AI068463. As control, we performed the same analysis on promoters from the constitutively expressed M. grisea genes encoding γ-actin (MG03982.4) and α-tubulin (MG06650.4). This revealed potential binding sites similar to the yeast stress response element (Transfac ID: F$STRE) in the promoters of GAS1, GAS2, CBP1, and AI068463, but not ACE1. This motif is located at similar distances from the transcription start site (TSS) of GAS1, GAS2, CBP1, and AI068463 (position −307 to −504, see Methods) and was not detected in the promoters of ACT1 and TUB1. Thus, stress-related signaling may be involved in the regulation of expression of these four genes but is less likely to play a role in the regulation of ACE1 expression. The promoters of all five appressorium-specific genes, including ACE1, carry potential GAL4 binding sites (Transfac ID: F$GAL4) that were not detected for the constitutive genes. However, the positions of these motifs vary considerably between promoters (position −136 to −685). It remains to be determined experimentally whether these putative binding sites play a role in the regulation of ACE1 and other appressorium-specific genes.
Ace1-GFP Fusion Protein Is Localized in the Cytoplasm of the Appressorium
Fungal AVR genes characterized to date encode proteins that are either known or suspected to be secreted during infection (Knogge, 2002). By contrast, PKS and NRPS are typically cytoplasmic enzymes. Ace1 does not display a signal peptide sequence at its N terminus and was predicted to be cytoplasmic by PSORT II software (psort.nibb.ac.jp). To determine the cellular localization of Ace1, we constructed the vector pACE1:GFP, which encodes a C-terminal fusion between Ace1 and GFP, expressed under the control of the ACE1 promoter and terminator sequences. We tested the effect of C terminus GFP fusion on Ace1 biological activity by introducing pACE1:GFP into the virulent parent 2/0/3. Nine of the eleven transformants obtained displayed a strong GFP fluorescence in their appressoria. These nine transformants were avirulent toward rice cultivars Bala and C101Lac carrying Pi33, whereas they were fully virulent toward susceptible cultivar Maratelli. The two transformants that did not display GFP fluorescent appressoria remained virulent toward Bala and C101Lac. These results demonstrated that the Ace1-GFP fusion protein is functional as it conferred avirulence when introduced in a virulent strain. Microscopic observations of the early infection process of these avirulent transformants (Figures 5C and 5E) showed that the kinetics of Ace1-GFP expression is similar to the kinetics observed with transformants expressing the ProACE1:GFP reporter construct (Figure 5A). Fluorescence of the Ace1-GFP protein was detected exclusively in the cytoplasm of appressoria (Figures 5C and 5E) during penetration of the fungus into barley or rice leaves, with the same expression pattern on susceptible and resistant rice cultivars (data not shown). Interestingly, the fluorescence of the fusion protein remained in the cytoplasm of the appressorium even after emergence of the infectious hyphae (Figure 5E). This is in contrast with the situation observed for transformants expressing GFP under the control of the ACE1 promoter (Figure 5A) that displayed primary infectious hyphae with a strong GFP fluorescence. Fluorescence of the Ace1-GFP fusion protein disappeared gradually after completion of penetration and was not detected 48 h after inoculation (data not shown). Because we never detected Ace1-GFP fusion protein outside the appressorium, our observations suggest that Ace1 is not secreted into infected plant tissues. This raised the possibility that the host resistance response is not triggered by Ace1 protein but may depend on the biosynthesis of a metabolite by Ace1.
Site-Directed Mutagenesis of the Putative KS Domain of Ace1 Suggests That Its Enzymatic Activity Is Required for Avirulence
To test the hypothesis that Ace1 is involved in the biosynthesis of a secondary metabolite recognized by resistant rice plants, we decided to modify a domain of Ace1 that should be essential for its proposed enzymatic activity. We chose to modify the putative KS domain of Ace1 because it is known to be essential for the biosynthetic activity of PKS and mammalian FAS. Furthermore, the catalytic site motif of KS domains of bacterial PKS, fungal PKS, and mammalian FAS are highly similar in protein sequence and structure, and it has been well established that in these enzymes, a universally conserved Cys residue of the KS active site is essential for its catalytic activity (Keating and Walsh, 1999). Using site-directed mutagenesis, we replaced this Cys by an Ala (ace1C183A allele) because this mutation is known to abolish the enzymatic activity of bacterial PKS and mammalian FAS without altering their folding and dimerization (Kao et al., 1996; Rangan et al., 1998). The ace1C183A allele was fused to GFP to follow the expression of the mutant protein. The resulting construct, pace1C183A:GFP was introduced by transformation into virulent parent 2/0/3. Among 44 transformants tested, 23 displayed a GFP fluorescence localized in the cytoplasm of appressoria, similar to those of transformants expressing a functional Ace1-GFP protein (Figure 6A). Whereas the 2/0/3 Ace1-GFP transformants were avirulent, transformants expressing the Ace1C183A-GFP protein were as virulent on cultivars Bala and C101lac that carry Pi33 as the virulent recipient strain 2/0/3 (Figure 6B). These results demonstrate that the ace1C183A:GFP allele is unable to confer avirulence. Because the Ace1C183A mutant is likely to have lost its ability to synthesize a polyketide, the observed loss of avirulence-conferring activity of the mutant protein is consistent with the hypothesis that the enzymatic activity of Ace1 synthase is required for avirulence.
Figure 6.
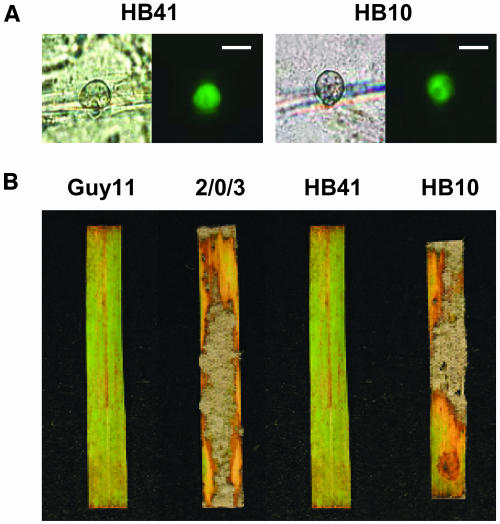
Lack of Avirulence Conferring Activity of the ace1C183A Allele.
(A) Conidia of the transformants HB41 and HB10 were inoculated on detached barley leaves and incubated at 26°C. Epidermal strips were removed from the infected leaf 20 h after infection and observed under a microscope. Appressoria were observed under bright-field illumination (left) or UV light (right). The GFP fluorescence of Ace1-GFP and Ace1C183A-GFP provides a control of the expression of the transgenes in appressoria and the correct localization of their respective translation product. Expression and localization of Ace1-GFP (HB41) and Ace1C183-GFP (HB10) were similar, indicating that the Ace1C183-GFP protein is expressed and localized as wild-type Ace1 protein. Bars = 8 μm.
(B) Avirulent isolate Guy11, virulent parent 2/0/3, and transformants of 2/0/3 expressing the ACE1:GFP fusion (HB41) or the mutant ace1C183A:GFP allele (HB10) were inoculated on resistant rice cultivar C101lac. Guy11 and HB41 carrying ACE1:GFP are avirulent. Transformant HB10 and 2/0/3 carrying the mutant ace1C183A:GFP allele are virulent.
DISCUSSION
Ace1-Mediated Avirulence Signaling
Fungal avirulence genes isolated to date encode small proteins that are known or supposed to be secreted during infection and are recognized by resistant plants (Knogge, 2002). The plant–pathogen interaction controlled by the avirulence gene ACE1 from M. grisea and the rice resistance gene Pi33 appears to be different because ACE1 encodes a large cytoplasmic enzyme of 4035 amino acids that is unlikely to be secreted. Based on sequence homology, Ace1 protein appears to be a fungal PKS fused at its C terminus to a single complete NRPS module. Ace1 contains all sequence motifs known to be essential for the enzymatic activity of PKS and NRPS (Aparicio et al., 1996; Hopwood, 1997; Marahiel et al., 1997). These results suggest that Ace1 is an active enzyme that is likely to be involved in the biosynthesis of a secondary metabolite. Fungal PKS and NRPS are known to be involved in the production of mycotoxins and host-specific toxins (HSTs) (Sweeney and Dobson, 1999; Wolpert et al., 2002). Wolpert and colleagues already pointed out that important parallels exist between the molecular and biochemical responses of host plants to HSTs and host plant defense reactions triggered by avirulence signals. The characterization of the PKS/NRPS Ace1 and its role in avirulence signaling may provide an example of the overlap between the concepts of HTS action and avirulence. Given the proposed biochemical function of Ace1, it is tempting to speculate that the metabolite produced by Ace1 could act as toxin that interferes with vital cellular functions or defense responses of the host plant, as illustrated by the Cochliobolus carbonum–maize (Zea mays) interaction (Walton, 1996). C. carbonum race 1 produces the NRPS-derived cyclic peptide HC-toxin that acts as HST. Specific maize cultivars are resistant to C. carbonum race 1, and this resistance depends on the presence of the dominant HM1 gene in maize. The HM1 resistance gene encodes a reductase that detoxifies HC-toxin. Lack of HC-toxin production results in complete loss of pathogenicity of C. carbonum on maize. By contrast, we did not observe a significant reduction in pathogenicity of M. grisea ace1 deletion mutants. This suggests that Ace1 is not involved in the production of a toxin that acts as major pathogenicity determinant. Still, it remains possible that the proposed metabolite contributes to the aggressiveness of this fungus. Further experiments are needed to address this question.
To investigate the mechanisms involved in the recognition of the fungus triggered by ACE1, we addressed the following question. Is the Ace1 protein recognized by resistant rice cultivars? First, we determined the localization of Ace1 in the fungal cell to assess if this protein is secreted into infected host tissues where it could directly trigger resistance. For this study, we used a vector expressing an ACE1:GFP translational fusion under the control of ACE1 promoter that confers avirulence. Ace1-GFP fusion protein was detected exclusively in the cytoplasm of penetrating appressoria. We were unable to detect Ace1-GFP in the surrounding areas of appressoria. After penetration, the Ace1-GFP remained detectable in appressoria but not in infectious hyphae differentiated inside epidermal cells. This is in contrast with transformants that express GFP under the control of the ACE1 promoter. In these strains, the cytoplasmic GFP is detectable both in appressoria and primary infection hyphae. Because there is a cytoplasmic continuity between appressoria and infectious hyphae, our observations suggest that Ace1-GFP fusion protein is retained in the appressorium. Similar observations have been described for the appressorium-specific proteins Gas1 and Gas2 from M. grisea (Xue et al., 2002), suggesting that retention of specific proteins in the appressorium during penetration is a common phenomenon. Localization of Ace1-GFP in the cytoplasm of the appressorium argues against its secretion into the underlying epidermal cell. However, it remains possible that small amounts of Ace1-GFP fusion protein below our detection limit are expressed in the infection hypha, secreted and delivered to plant cells. Additionally, Ace1 degradation products may be secreted and delivered to plant cells, triggering host resistance. Taking into consideration these possibilities, we decided to create a mutant protein that differs from the wild type only by the loss of its enzymatic function. The mutant Ace1C183A protein fulfills these requirements as it carries a single amino acid modification in the KS domain catalytic site. The same amino acid exchange has been introduced in the KS domains of a bacterial PKS and mammalian FAS, where it has been shown to abolish enzymatic activity without interfering with the folding and dimerization of the mutant protein (Kao et al., 1996; Rangan et al., 1998). Thus, if direct recognition of Ace1 protein or its degradation products was involved in avirulence signaling, it should occur in the same way in transformants expressing Ace1C183A. However, we found that Ace1C183A was not able to confer avirulence to virulent strain 2/0/3, despite its correct expression and localization in appressoria. The most likely explanation for this lack of recognition is the loss of KS enzymatic activity of Ace1C183A and the consequent loss of secondary metabolite production. To validate this hypothesis, we tried to identify the ACE1 metabolite by liquid chromatography tandem mass spectrometry analysis of extracts from infected barley leaves 24 h after inoculation with either an avirulent or a virulent M. grisea isolate. These attempts were unsuccessful (data not shown), possibly because of the low amount of fungal material present in infected leaf tissue combined with the appressorium-specific expression of ACE1. Because appressoria formed on the artificial surfaces Teflon and Mylar do not appear to express ACE1 (data not shown), we could not use these supports to generate large amounts of appressoria for the detection of the metabolite. Although we were not able to detect the ACE1 metabolite, our results support the following hypothesis: Ace1 enzymatic activity is required for avirulence and is involved in the synthesis of a metabolite that triggers the recognition of the avirulent fungus by rice plants that carry the resistance gene Pi33.
To some extent, the requirement of a functional Ace1 enzyme for avirulence parallels results described for the bacterial type III–secreted Cys proteases AvrBsT and AvrPphB. For these three AVR factors, it was demonstrated that protease enzymatic activity is required for avirulence (Orth et al., 2000; Shao et al., 2003). Shao and coworkers postulated that these AVR proteases act on host plant target proteins and that the corresponding degradation products are the signals recognized by R proteins. The indirect signaling proposed for ACE1 is different because this fungal protein is not expected to act through an enzymatic modification of host proteins, but through the biosynthesis of a secondary metabolite that may serve as avirulence signal. To our knowledge, there is currently only one precedent for metabolite-mediated avirulence signaling, described in the interaction between Pseudomonas syringae and soybean (Glycine max). In this case, the avirulence gene AvrD directs the production of a group of small glycolipid molecules called syringolides (Kobayashi et al., 1990). These amphipathic molecules induce the hypersensitive response only in soybean cultivars carrying the Rpg4 resistance gene, possibly through interference with a photorespiration enzyme of the host (Atkinson et al., 1996; Okinaka et al., 2002). However, the role of syringolide production in the biology of Pseudomonas remains somewhat enigmatic. Apart from two sequences with similarity to AvrD that have been discovered in the genomes of Ralstonia solanacearum and Streptomyces coelicolor A3(2), AvrD genes were found exclusively in P. syringae isolates (Bentley et al., 2002; Salanoubat et al., 2002). Therefore, syringolides do not appear to be a major class of metabolites from plant pathogenic bacteria. By contrast, many microorganisms produce secondary metabolites using biosynthetic pathways that involve PKS or NRPS. In particular, plant-pathogenic microbes are notable producers of polyketides or nonribosomal peptides (Lugtenberg et al., 2002; Wolpert et al., 2002). Our analysis of ACE1-mediated avirulence signaling implies that the spectrum of molecular structures that can be recognized by resistant host plants is broad and that it could include PKS/NRPS-derived metabolites. Recognition of this class of metabolites by plants may provide disease resistance against a large variety of microbial pathogens through the perception of metabolites they produce during infection.
Ace1 Belongs to a Novel Family of Fungal PKS/NRPS
Ace1 belongs to a novel family of fungal PKS/NRPS that gathers at least 17 hypothetical proteins and two enzymes. Two fungal PKS/NRPS have known biochemical functions, the lovastatin biosynthetic enzyme LNKS from A. terreus (Kennedy et al., 1999) and the related enzyme MlcA from P. citrinum (Abe et al., 2002). LNKS and MlcA lack the adenylation and PCP domains required for a functional NRPS module. Therefore, their metabolic products may not be representative of the compounds produced by PKS/NRPS with a complete NRPS module. Two PKS/NRPS with a structure similar to LNKS were identified in the genome of M. grisea (Syn6 and Syn7). We identified eight PKS/NRPS with a complete NRPS module in the genomes of the ascomycetes M. grisea, A. nidulans, and A. fumigatus (see Supplemental Figure 3 online). Independently, a phylogenomic analysis of fungal type I PKS genes reported the presence of six novel putative PKS/NRPS with a full NRPS module in the genomes of the ascomycetes B. fuckeliana, C. heterostrophus, and G. moniliformis (Kroken et al., 2003). This study is complementary to ours in terms of the genomes analyzed and reaches similar conclusions. Fungal PKS/NRPS belong to a monophyletic group (Figure 3) and are likely to derive from an ancestral enzyme corresponding to a type I fungal PKS fused to a complete NRPS module. This family may have evolved through duplications and independent truncations of the C-terminal NRPS module. Partial truncation may have led to PKS/NRPS related to LNKS (Syn6 and Syn7) or to ChPKS17. Independent loss of the entire NRPS module may have led to putative PKS, such as BfPKS4, BfPKS6, ChPKS16, FG10464, and ncu08399.
The genome of M. grisea appears to be relatively rich in PKS/NRPS with either a full or a truncated NRPS module (eight PKS/NRPS) compared with other fungi, such as B. fuckeliana (three PKS/NRPS), C. heterostrophus (one PKS/NRPS), and G. moniliformis (three PKS/NRPS) (Kroken et al., 2003). This may indicate a recent evolution of this gene family by gene duplication in M. grisea. By contrast, the closely related pyrenomycete Neurospora crassa has only one sequence related to fungal PKS/NRPS that may have lost its NRPS module. M. griseae and N. crassa are phylogenetically related species that differ most importantly in their respective life styles because the former is a pathogen of cereals, whereas the latter is a tropical soil/bark saprophyte. After the recent completion of the genome sequence of both organisms, one hope is that the direct comparison of their genomes may provide some insight into what it takes to make a pathogen. In this context, it is tempting to speculate that the observed richness of the Magnaporthe genome in PKS/NRPS may be part of the important differences between the pathogen and the saprophyte.
METHODS
Fungal Strains, Growth Conditions, and Transformation
The M. grisea strains used in this study are listed in Table 3. Guy11 is a fertile rice field isolate pathogenic on rice (Oryza sativa) and barley (Hordeum vulgare) (Notteghem et al., 1994), and 2/0/3 is a sib-progeny from the cross (Guy11 × ML25) (Silué et al., 1992). As all rice pathogenic Magnaporthe grisea isolates, Guy11 and 2/0/3 are also pathogenic on barley. Fungal strains were grown and stored as described (Dioh et al., 2000). M. grisea strains were transformed as described (Sweigard et al., 1995; Clergeot et al., 2001), with one modification for transformation of isolate Guy11 to hygromycin resistance: after incubation of the transformation plates at 30°C for 20 h, 5 mL of overlay agar (1% granulated agar [Difco, Becton Dickinson, Franklin Lakes, NY] and 300 mg/L of hygromycin) were added to each plate to reduce the occurrence of background growth from nontransformed colonies.
Table 3.
M. grisea Strains Used in This Study
| Strain | Description/Reference |
|---|---|
| Guy11 | ACE1+ field isolate (Silué et al., 1992) |
| 2/0/3 | ace1− progeny of a cross between Guy11 and ML25 (Silué et al., 1992) |
| PH14 | ace1− field isolate (Berruyer et al., 2003) |
| HBE20 | ACE1+ transformant of 2/0/3 carrying pACE1 (this study) |
| HB26 | ace1− deletion mutant of Guy11 obtained with pΔACE1 (this study) |
| HB92 | ace1− deletion mutant of Guy11 obtained with pΔACE1 (this study) |
| HB16 | ACE1+ ectopic transformant of Guy11 carrying pΔACE1 (this study) |
| HB24 | ACE1+ ectopic transformant of Guy11 carrying pΔACE1 (this study) |
| HB33 | ACE1+ ectopic transformant of Guy11 carrying pΔACE1 (this study) |
| IF22 | ACE1+ transformant of Guy11 carrying pProACE1:GFP (this study) |
| HB41 | ACE1+ transformant of 2/0/3 carrying pACE1:GFP (this study) |
| HB44 | ACE1+ transformant of 2/0/3 carrying pACE1:GFP (this study) |
| HB10 | ace1− transformant of 2/0/3 carrying pace1C183A:GFP (this study) |
Phenotypic Analysis and Cytology
Rice cultivars Maratelli, Sariceltik, LTH, and LS1 were used as susceptible controls because they do not carry Pi33. Rice cultivars Irat7, IR1529, C101lac, and Bala were used as resistant controls because they carry the resistance gene Pi33. These rice cultivars were grown in the greenhouse as described (Dioh et al., 2000). Spray infections and detached leaf assays were performed as described (Silué et al., 1992; Clergeot et al., 2001). Fluorescence of the Ace1-GFP fusion protein was observed using a Nikon Optiphot-2 epi-fluorescence microscope equipped with a GFP-specific filter (excitation, 475/20; emission, 510/23; ref, XF100-2-NS4A; Nikon, Tokyo). Laser scanning electron confocal microscopy and image analyses were performed at the Centre de Quantimétrie (Lyon, France).
Nucleic Acid Extraction Analysis and Cloning Procedures
Genomic DNA was isolated from M. grisea following the miniprep procedure (Sweigard et al., 1997) with the following modifications: for protoplasting, Glucanex (Novo Nordisc Ferment, Dittingen, Switzerland) was used at 30 mg/mL in 0.7 M NaCl, pH adjusted to 6; protoplasts were lysed by vigorous vortexing with 0.3 g glass beads (425 to 600 μm, acid washed) for 45 s. Total RNA was extracted from M. grisea liquid cultures and infected barley leaves using the hot acid phenol protocol (Ausubel et al., 1994). For RFLP analysis, 5 μg of each genomic DNA were digested with XhoI, separated by gel electrophoresis on a 1% agarose gel, and transferred by vacuum blot to positively charged nylon membrane (Hybond-N+; Amersham Biosciences). Hybridization was performed with a probe composed of three individually labeled genomic restriction fragments spanning the ACE1 ORF from 123 to 11,860 bp: a 2.8-kb EcoRV fragment, a 4.4-kb EcoRV fragment, and a 4.5-kb EcoRV-NotI fragment. Labeling was performed using the Megaprime DNA labeling system (Amersham Biosciences, Piscataway, NJ). Restriction enzyme digestion, gel electrophoresis, and hybridization were performed using standard procedures (Sambrook et al., 1989). For cloning purposes, the Escherichia coli strains DH5α (Bethesda Research Laboratories, Gaithersburg, MD) and JM110 (Stratagene, La Jolla, CA) were used. Molecular methods followed described protocols (Sambrook et al., 1989).
RT-PCR
RT-PCR was performed with 6 μg of total RNA using ReadyToGo You-Prime first-strand beads (Amersham Biosciences) according to the manufacturer's protocol. Positions of introns in the ACE1 sequence were identified using the following strategy. Putative exon junction regions were amplified by RT-PCR using total RNA extracted from infected barley leaves 17 h after inoculation and the following primer pairs: intron 1, H-OL1 (for primer sequences, see Table 4) and H-OL2; intron 2, H-OL3 and H-OL4; intron 3, H-OL5 and H-OL6. PCR products were cloned in pGEM T-easy (Promega, Madison, WI) according to the manufacturer's protocol and sequenced. Transcription initiation and polyadenylation site of ACE1 were determined by 5′ and 3′ RACE using GENE RACER kit (Invitrogen, Carlsbad, CA) and sequence analysis of amplification products. The 5′ UTR was amplified with gene-specific primer H-OL7, and the 3′ UTR was amplified using gene-specific primer H-OL8. The actin encoding gene ACT1 was used as control in RT-PCR experiments. An EST corresponding to ACT1 was isolated from a M. grisea cDNA library prepared from mycelial RNA (Clergeot et al., 2001) and used to identify the corresponding cosmid from a 96/0/76 genomic library (Dioh et al., 1997). Comparison of cDNA and genomic sequence revealed the presence of four introns. In the RT-PCR experiments, the region carrying the first three introns was amplified using primers H-OL9 and H-OL10.
Table 4.
Primers Used in This Study
| Primer | Sequence (5′/3′) | Purpose |
|---|---|---|
| H-OL1 | TTGCCTCCATTGTGCTGAAG | RT-PCR (amplification of ACE1 intron 1) |
| H-OL2 | GCCTTCTGTGTGCCCAAT | RT-PCR (amplification of ACE1 intron 1) |
| H-OL3 | CCGCCGTCGTCACTCCCACC | RT-PCR (amplification of ACE1 intron 2) |
| H-OL4 | TGACAGAGGACAGGAAGACG | RT-PCR (amplification of ACE1 intron 2) |
| H-OL5 | CTCTTCTCGGTCGGCAACAC | RT-PCR (amplification of ACE1 intron 3) |
| H-OL6 | CGACGGCGACCAGTGAATCC | RT-PCR (amplification of ACE1 intron 3) |
| H-OL7 | ACATTGCTGGTTCCGTGGTGTTTATGA | 5′ RACE of ACE1 |
| H-OL8 | CATCACGGGCGAGGGCACCAAT | 3′ RACE of ACE1 |
| H-OL9 | TTGCGATTAGCGTCCATTGT | RT-PCR (amplification of ACT1 introns 1 to 3) |
| H-OL10 | TTCATCAGGTAGTCGGTCAA | RT-PCR (amplification of ACT1 introns 1 to 3) |
| H-OL11 | CGGAAATCCGCATCTCTAGCTTGGCGC | Construction of pProACE1:GFP |
| H-OL12 | GTCACCCATGGTTGGTGATTTATTTGTTT | Construction of pProACE1:GFP |
| H-OL13 | GGCCGGC | Construction of pProACE1:GFP |
| H-OL14 | GCCACTCGGAATAAACAGAGGGTGGTC | Construction of pProACE1:GFP |
| H-OL15 | GCTGAAGGGCGGCCGCTGATTCATGAACTTTGGAATACTGTAACTAG | Construction of pACE1:GFP |
| H-OL16 | CAGTATTCCAAAGTTCATGACCATGGTC | Construction of pACE1:GFP |
| H-OL17 | CCGTCCTTCAGC | Construction of pACE1:GFP |
| H-OL18 | GCTGAAGGACGGGACCATGGTCATGAA | Construction of pACE1:GFP |
| H-OL19 | CTTTGGAATACTG | Construction of pace1C183A:GFP |
| H-OL20 | GCTGAAGGGCGGCCGCTGATTCATGAACTTTGGAATACTGTAACTAG | Construction of pace1C183A:GFP |
| GCCACTCGGAATAAACAGAGGGTGGTC | ||
| CGGTCACGGTCGATACCGCAGCTAGCTCCAGCTTGGTGGCCG | ||
| CGGCCACCAAGCTGGAGCTAGCTGCGGTATCGACCGTGACCG |
Sequence Analysis
Sequencing was performed by Genome Express (Meylan, France). Sequence analysis was performed using DNA Strider 1.2 (Marck, 1988), BLAST (Altschul et al., 1997; http://www.ncbi.nlm.gov/BLAST/), PSORT (Nakai, 2000; http://psort.nibb.ac.jp/), and pfam 6.5 (Bateman et al., 2002; http://pfam.wustl.edu/).
Promoter Analysis
For each gene, the TSS was determined by comparing EST sequences to the corresponding genomic sequence. The 1 kb of DNA sequence upstream from TSS was examined using the following software: TRES (http://bioportal.bic.nus.edu sg/tres/), MatInspector (http://www.genomatix.de), TF Scan (http://bioweb.pasteur.fr), and OTFBS (http://www.bioinfo.tsinghua.edu.cn/∼zhengjsh/OTFBS/). Putative binding sites for the yeast stress response element (Transfac ID: F$STRE) were identified in the promoters of GAS1 (positions −504 and −558), GAS2 (position −416), CBP1 (position −307), and AI068463 (position −331). Furthermore, putative GAL4 binding motifs were detected in the promoters of ACE1 (positions −136 and −610), GAS1 (position −387), GAS2 (position −670), CBP1 (overlapping motifs on the plus and minus strand, positions −158 and −160 and positions −668 and −685).
Identification of Ace1 Enzymatic Domains
The following domains were identified using the Pfam 6.5 database: KS, N-terminal and C-terminal domains (PF00109 and PF02801), AT (PF00698), condensation (C-) domain (PF00668), AMP binding enzyme (A-domain) (PF00501), and phosphopantheteine attachment site (ppt-domain) (PF00550). The remaining domains (DH, MT, KR, and ACP) were identified by direct comparison of the Ace1 sequence with the sequences of LNKS from Aspergillus terreus (AF151722), Pks1 from Cochliobolus heterostrophus (U68040), and Fum5 from Gibberella moniliformis (AF155773).
Phylogeny of Fungal PKS and PKS/NRPS
The Ace1 protein sequence was used as a query to search the nonredundant protein database at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/). Additional fungal PKS/NRPS were identified using the TBLASTN program and Ace1 protein sequence as a query in the following databases. The fungal genomes database Web site (http://www.broad.mit.edu/annotation/fungi/fgi) was used for M. grisea, Neurospora crassa, Aspergillus nidulans, Fusarium graminearum, Ustilago maydis, Coprinus cinereus, and Cryptococcus neoformans genome sequences. The Institute for Genomic Research Web site (http://www.tigr.org) was used for the preliminary genomic sequences of Aspergillus fumigatus. Annotated ORFs with significant similarity to Ace1 were used directly for the analysis (mg03810, mg03818, mg09589, ncu08399, AN8412, af04917, and FG10464). Sequences that displayed high degrees of similarity but were not annotated (Syn2, Syn6, Syn7, and Syn8) were processed as follows. The nucleotide sequence spanning the region of similarity to ACE1 was recovered and compared with ACE1 at the protein level using BLASTX. Putative introns were detected as gaps in the collinearity of the aligned protein sequences, followed by manual search for motifs characteristic of fungal intron splice donor and acceptor sites (Edelmann and Staben, 1994). The protein sequences were obtained from the translation of ORF sequences resulting from the removal of the putative introns. Protein sequences from the KS and AT domains of each PKS and PKS/NRPS were aligned using the CLUSTALW program (http://www.ch.embnet.org/software/ClustalW.html). Ace1, fungal PKS, and PKS/NRPS sequences were analyzed with PAUP 4.0b10 (Swofford, 2002) using the neighbor-joining distance method. The following enzymes were included in the analysis: AnpksST (AAA81586) and AnWA (CAA46695) from A. nidulans; LNKS (AF151722), LDKS (AF141925), AtMSAS (AAC49814), and Atat1 (BAB88688) from Aspergillus terreus; ClPKS1 (BAA18956) from Colletotrichum lagenarium; ChPks1 (AAB08104) from Cochliobolus heterostrophus; Fum1 (AAD43562) from Gibberella moniliformis; mLcA (BAC20564) from Penicillium citrinum; PgPKS2 (AAB49684) from Penicillium griseofulvum. The following hypothetical proteins deduced from ORFs identified in genomic sequences were included in the analysis: mg03810 (EEA50051), mg03818 (EEA50059), mg09589 (EEA48052) from M. grisea, ncu02918.1 (EEA36364), ncu04865.1 (EEA29886), ncu08399.1 (EEA34002), and ncu09638.1 (EAA28933) from N. crassa, AN8412 (EAA67034) from A. nidulans, af04917 from A. fumigatus, and FG10464.1 (EAA68296) from F. graminearum. Syn2, Syn6, Syn7, and Syn8 were identified in the M. grisea genome during this study (SYN2 as AJ704623, SYN6 as BN000505, SYN7 as BN000506, and SYN8 as BN000507). We added fungal PKS and PKS/NRPS sequences described by Kroken et al. (2003) from C. heterostrophus (ChPKS2,16,17), B. fuckeliana (BfPKS3,4,5,6,8), G. moniliformis (GmPKS1,9,10), and G. zeae (GzPKS10, partial sequence). GzPKS10 is highly related to GmPKS10 (90% identity) and is likely to have a full NRPS module as GmPKS10. The following three PKS/NRPS (GzPKS9, BfPKS7, and GmPKS8) identified by Kroken et al. were not added to this analysis because they lack some sequences of their KS or AT domains or have only KS domains (GzPKS9). GzPKS9 is almost identical (>91% identity) to FG10464. We also included in the analysis bacterial PKS epoD (AAF26922), MxaC (AAK57187), and MxaD (AAK57188) related to fungal PKS. FAS from Bombyx mori (U67866) served as outgroup to root the tree.
Plasmid Constructions
The cosmid library is described by Dioh et al. (1997). For cosmid D31C12 subclones, cosmid DNA was digested with each of the following enzymes: BamHI, EcoRI, NotI, and XbaI and in double digests with BglII plus NotI and XbaI plus NotI, respectively. The following genomic fragments were recovered and cloned into pCB1004 (Sweigard et al., 1997): for pBam8, an 8.3-kb BamHI fagment; for pEco20, a 15.4-kb EcoRI fragment; for pNot21, a 21.0-kb NotI fragment (one NotI site is derived from the cosmid polylinker); for Xba11, an 11.1-kb XbaI fragment; for pBg-N7, a 6.7-kb BglII-NotI fragment (NotI site derived from polylinker); for pX-N12, a 12.1-kb XbaI-NotI fragment.
For pΔACE1, the 15.4-kb EcoRI fragment carrying ACE1 was cloned into pBluescript II KS−. The resulting vector was digested with AgeI, and the internal 3.5-kb AgeI fragment was replaced by the 1.4-kb hygromycin resistance cassette from pCB1004 (Sweigard et al., 1997).
For pProACE1:GFP, the 1.6-kb ACE1 promoter region was amplified using primers H-OL11 and H-OL12, introducing an NcoI site at the start codon of ACE1; 1.5-kb ACE1 terminator region were amplified using primers H-OL13 and H-OL14, introducing a NotI site at the stop codon of ACE1. The resulting PCR products were cloned in pGEMT-easy (Promega) and sequenced. Then, the ACE1 promoter was excised as an EcoRI-NcoI fragment and the terminator as NotI-EcoRI fragment, and both were fused to the NcoI-NotI fragment from pEGFP (Clontech, Palo Alto, CA) carrying the EGFP reporter gene (Lorang et al., 2001). The ProACE1:GFP transcriptional fusion was introduced into a pBluescript II KS− (Stratagene) carrying at its EcoRV site the hygromycine resistance cassette from pCB1003 .
For pACE1:GFP, an NcoI site was introduced at the ACE1 stop codon by PCR using primers H-OL15 and H-OL16. The ACE1 terminator region was amplified with primers H-OL17 and H-OL18, introducing a NotI site immediately downstream of the stop codon of ACE1. The NcoI-NotI GFP cassette of pEGFP (Clontech) was cloned between the 3′ end of the ACE1 ORF and the ACE1 terminator. Expression of the tagged allele is under the control of the native promoter.
For pace1C183A:GFP, the KS domain coding sequence of ACE1 was subjected to site-directed mutagenesis using primers H-OL19 and H-OL20 and the QuikChange site-directed mutagenesis kit (Stratagene). This leads to replacement of catalytic residue Cys183 with Ala, introducing a diagnostic NheI restriction site. The resulting PCR product was sequenced, and the 470-bp BspEI-FseI fragment carrying the introduced mutation was used to replace the native 470-bp BspEI-FseI fragment in pACE1:GFP.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under the following accession numbers: ACE1 as AJ704622, SYN2 as AJ704623, SYN6 as BN000505, SYN7 as BN000506, and SYN8 as BN000507.
Supplementary Material
Acknowledgments
We thank T. Fonseca and J. Staunton (Chemistry Department, Cambridge University, UK) for attempts to detect the Ace1 metabolite by liquid chromatography tandem mass spectrometry analysis, K. Percet for assistance in subcloning, and J. Collemare for helpful discussion of the manuscript. The Fungal Genome Initiative sequencing has been supported by the National Human Genome Research Institute, the National Science Foundation, and the USDA and has included collaborations with several academic and commercial groups. Sequencing of A. fumigatus was funded by the National Institute of Allergy and Infectious Diseases U01 AI 48830 to D. Denning and W. Nierman, the Wellcome Trust, and Fondo de Investicagiones Sanitarias. This work was supported by European Union Grants ERBFMRX-CT-98-0241 (Training and Mobility of Researchers Programme project CEREPAT), ICA4-CT-2000-30021 (International Cooperation Development Programme project RESIDIV), Centre National de la Recherche Scientifique, and Bayer CropScience.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instruction for Authors (www.plantcell.org) is: Marc-Henri Lebrun (marc-henri.lebrun@bayercropscience.com).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.022715.
References
- Abe, Y., Suzuki, T., Ono, C., Iwamoto, K., Hosobuchi, M., and Yoshikawa, H. (2002). Molecular cloning and characterization of an ML-236B (compactin) biosynthetic gene cluster in Penicillium citrinum. Mol. Genet. Genomics 267, 636–646. [DOI] [PubMed] [Google Scholar]
- Abramovitch, R.B., Kim, Y.J., Chen, S., Dickman, M.B., and Martin, G.B. (2003). Pseudomonas type III effector AvrPtoB induces plant disease susceptibility by inhibition of host programmed cell death. EMBO J. 22, 60–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio, J.F., Molnar, I., Schwecke, T., Konig, A., Haydock, S.F., Khaw, L.E., Staunton, J., and Leadlay, P.F. (1996). Organization of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: Analysis of the enzymatic domains in the modular polyketide synthase. Gene 169, 9–16. [DOI] [PubMed] [Google Scholar]
- Atkinson, M.M., Midland, S.L., Sims, J.J., and Keen, N.T. (1996). Syringolide 1 triggers Ca2+ influx, K+ efflux, and extracellular slkalization in soybean cells carrying the disease-resistance gene Rpg4. Plant Physiol. 112, 297–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1994). Current Protocols in Molecular Biology. (New York: Wiley).
- Banno, S., Kimura, M., Tokai, T., Kasahara, S., Higa-Nishiyama, A., Takahashi-Ando, N., Hamamoto, H., Fujimura, M., Staskawicz, B.J., and Yamaguchi, I. (2003). Cloning and characterization of genes specifically expressed during infection stages in the rice blast fungus. FEMS Microbiol. Lett. 222, 221–227. [DOI] [PubMed] [Google Scholar]
- Bateman, A., Birney, E., Cerruti, L., Durbin, R., Etwiller, L., Eddy, S.R., Griffiths-Jones, S., Howe, K.L., Marshall, M., and Sonnhammer, E.L. (2002). The Pfam protein families database. Nucleic Acids Res. 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley, S.D., et al. (2002). Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417, 141–147. [DOI] [PubMed] [Google Scholar]
- Berruyer, R., Adreit, H., Milazzo, J., Gaillard, S., Berger, A., Dioh, W., Lebrun, M.H., and Tharreau, D. (2003). Identification and fine mapping of Pi33, the rice resistance gene corresponding to the Magnaporthe grisea avirulence gene ACE1. Theor. Appl. Genet. 107, 1139–1147. [DOI] [PubMed] [Google Scholar]
- Bourett, T.M., and Howard, R.J. (1990). In vitro development of penetration structures is the rice blast fungus Magnaporthe grisea. Can. J. Bot. 68, 329–342. [Google Scholar]
- Challis, G.L., Ravel, J., and Townsend, C.A. (2000). Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem. Biol. 7, 211–224. [DOI] [PubMed] [Google Scholar]
- Clergeot, P.H., Gourgues, M., Cots, J., Laurans, F., Latorse, M.P., Pepin, R., Tharreau, D., Notteghem, J.L., and Lebrun, M.H. (2001). PLS1, a gene encoding a tetraspanin-like protein, is required for penetration of rice leaf by the fungal pathogen Magnaporthe grisea. Proc. Natl. Acad. Sci. USA 98, 6963–6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti, E., Stachelhaus, T., Marahiel, M.A., and Brick, P. (1997). Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidin S. EMBO J. 16, 4174–4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dioh, W., Tharreau, D., and Lebrun, M.H. (1997). RAPD-based screening of genomic libraries for positional cloning. Nucleic Acids Res. 25, 5130–5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dioh, W., Tharreau, D., Notteghem, J.L., Orbach, M., and Lebrun, M.H. (2000). Mapping of avirulence genes in the rice blast fungus, Magnaporthe grisea, with RFLP and RAPD markers. Mol. Plant-Microbe Interact. 13, 217–227. [DOI] [PubMed] [Google Scholar]
- Dodds, P.N., Lawrence, G.J., Catanzariti, A.M., Ayliffe, M.A., and Ellis, J.G. (2004). The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell 16, 755–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann, S.E., and Staben, C. (1993). A statistical analysis of sequence features within genes from Neurospora crassa. Exp. Mycol. 18, 70–81. [Google Scholar]
- Farman, M.L., Eto, Y., Nakao, T., Tosa, Y., Nakayashiki, H., Mayama, S., and Leong, S.A. (2002). Analysis of the structure of the AVR1-CO39 avirulence locus in virulent rice-infecting isolates of Magnaporthe grisea. Mol. Plant-Microbe Interact. 15, 6–16. [DOI] [PubMed] [Google Scholar]
- Farman, M.L., and Leong, S.A. (1998). Chromosome walking to the AVR1-CO39 avirulence gene of Magnaporthe grisea: Discrepancy between the physical and genetic maps. Genetics 150, 1049–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor, H.H. (1971). Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 9, 275–296. [Google Scholar]
- Hammond-Kosack, K.E., and Parker, J.E. (2003). Deciphering plant-pathogen communication: Fresh perspectives for molecular resistance breeding. Curr. Opin. Biotechnol. 14, 177–193. [DOI] [PubMed] [Google Scholar]
- Hendrickson, L., Davis, C.R., Roach, C., Nguyen, D.K., Aldrich, T., McAda, P.C., and Reeves, C.D. (1999). Lovastatin biosynthesis in Aspergillus terreus: Characterization of blocked mutants, enzyme activities and a multifunctional polyketide synthase gene. Chem. Biol. 6, 429–439. [DOI] [PubMed] [Google Scholar]
- Hopwood, D.A. (1997). Genetic contributions to understanding polyketide synthases. Chem. Rev. 97, 2465–2498. [DOI] [PubMed] [Google Scholar]
- Hopwood, D.A., and Sherman, D.H. (1990). Molecular genetics of polyketides and its comparison to fatty acid biosynthesis. Annu. Rev. Genet. 24, 37–66. [DOI] [PubMed] [Google Scholar]
- Hutchinson, C.R., Kennedy, J., Park, C., Kendrew, S., Auclair, K., and Vederas, J. (2000). Aspects of the biosynthesis of non-aromatic fungal polyketides by iterative polyketide synthases. Antonie Van Leeuwenhoek 78, 287–295. [DOI] [PubMed] [Google Scholar]
- Jia, Y., McAdams, S.A., Bryan, G.T., Hershey, H.P., and Valent, B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19, 4004–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, S., Sweigard, J.A., and Valent, B. (1995). The PWL host specificity gene family in the blast fungus Magnaporthe grisea. Mol. Plant-Microbe Interact. 8, 939–948. [DOI] [PubMed] [Google Scholar]
- Kao, C.M., Pieper, R., Cane, D.E., and Khosla, C. (1996). Evidence for two catalytically independent clusters of active sites in a functional modular polyketide synthase. Biochemistry 35, 12363–12368. [DOI] [PubMed] [Google Scholar]
- Keating, T.A., and Walsh, C.T. (1999). Initiation, elongation, and termination strategies in polyketide and polypeptide antibiotic biosynthesis. Curr. Opin. Chem. Biol. 3, 598–606. [DOI] [PubMed] [Google Scholar]
- Kennedy, J., Auclair, K., Kendrew, S.G., Park, C., Vederas, J.C., and Hutchinson, C.R. (1999). Modulation of polyketide synthase activity by accessory proteins during lovastatin biosynthesis. Science 284, 1368–1372. [DOI] [PubMed] [Google Scholar]
- Knogge, W. (2002). Avirulence determinants and elicitors. In The Mycota XI, F. Kempken, ed (Berlin, Heidelberg: Springler-Verlag), pp. 289–310.
- Kobayashi, D.Y., Tamaki, S.J., and Keen, N.T. (1990). Molecular characterization of avirulence gene D from Pseudomonas syringae pv. tomato. Mol. Plant-Microbe Interact. 3, 94–102. [DOI] [PubMed] [Google Scholar]
- Koga, H. (1994). Hypersensitive death, autofluorescence, and ultrastructural changes in cells of leaf sheaths of susceptible and resistant near-isogenic lines of rice (Pi-zt) in relation to penetration and growth of Pyricularia oryzae. Can. J. Bot. 72, 1463–1477. [Google Scholar]
- Kroken, S., Glass, N.L., Taylor, J.W., Yoder, O.C., and Turgeon, B.G. (2003). Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc. Natl. Acad. Sci. USA 100, 15670–15675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugé, R., Joosten, M.H., Haanstra, J.P., Goodwin, P.H., Lindhout, P., and De Wit, P.J. (1998). Successful search for a resistance gene in tomato targeted against a virulence factor of a fungal pathogen. Proc. Natl. Acad. Sci. USA 95, 9014–9018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorang, J.M., Tuori, R.P., Martinez, J.P., Sawyer, T.L., Redman, R.S., Rollins, J.A., Wolpert, T.J., Johnson, K.B., Rodriguez, R.J., Dickmann, M.B., and Ciufetti, L.M. (2001). Green fluorescent protein is lighting up fungal biology. Appl. Environ. Microbiol. 67, 1987–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luderer, R., de Kock, M.J.D., Robert, H.L.D., de Wit, P.J., and Joosten, M.H. (2002. b). Functional analysis of cysteine residues of ECP elicitor proteins of the fungal tomato pathogen Cladosporium fulvum. Mol. Plant Pathol. 3, 91–95. [DOI] [PubMed] [Google Scholar]
- Luderer, R., Takken, F.L., de Wit, P.J., and Joosten, M.H. (2002. a). Cladosporium fulvum overcomes Cf-2-mediated resistance by producing truncated AVR2 elicitor proteins. Mol. Microbiol. 45, 875–884. [DOI] [PubMed] [Google Scholar]
- Lugtenberg, B.J., Chin, A.W.T.F., and Bloemberg, G.V. (2002). Microbe-plant interactions: Principles and mechanisms. Antonie Van Leeuwenhoek 81, 373–383. [DOI] [PubMed] [Google Scholar]
- Mackey, D., Holt, B.F., Wiig, A., and Dangl, J.L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108, 743–754. [DOI] [PubMed] [Google Scholar]
- Marahiel, M.A., Stachelhaus, T., and Mootz, H.D. (1997). Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 97, 2651–2674. [DOI] [PubMed] [Google Scholar]
- Marck, C. (1988). ‘DNA Strider’: A ‘C’ program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 16, 1829–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marois, E., Van den Ackerveken, G., and Bonas, U. (2002). The Xanthomonas type III effector protein AvrBs3 modulates plant gene expression and induces cell hypertrophy in the susceptible host. Mol. Plant-Microbe Interact. 15, 637–646. [DOI] [PubMed] [Google Scholar]
- Martin, G.B., Bogdanove, A.J., and Sessa, G. (2003). Understanding the functions of plant disease resistance proteins. Annu. Rev. Plant Physiol. Plant Mol. Biol. 54, 23–61. [DOI] [PubMed] [Google Scholar]
- Mathur, M., and Kolattukudy, P.E. (1992). Molecular cloning and sequencing of the gene for mycocerosic acid synthase, a novel fatty acid elongating multifunctional enzyme, from Mycobacterium tuberculosis var. bovis Bacillus Calmette-Guerin. J. Biol. Chem. 267, 19388–95. [PubMed] [Google Scholar]
- Nakai, K. (2000). Protein sorting signals and prediction of subcellular localization. Adv. Protein Chem. 54, 277–344. [DOI] [PubMed] [Google Scholar]
- Notteghem, J.L., Tharreau, D., Silue, D., and Roumen, E. (1994). Present knowledge on rice resistance genetics and strategies for analysis of M. grisea pathogenicity and avirulence genes. In Rice Blast Disease, R. Zeigler, S.A. Leong, and P.S. Teng, eds (Wallingford, UK: Commonwealth Agricultural Bureau International), pp. 155–166.
- Okinaka, Y., Yang, C.H., Herman, E., Kinney, A., and Keen, N.T. (2002). The P34 syringolide elicitor receptor interacts with a soybean photorespiration enzyme, NADH-dependent hydroxypyruvate reductase. Mol. Plant-Microbe Interact. 15, 1213–1218. [DOI] [PubMed] [Google Scholar]
- Orbach, M.J., Farrall, L., Sweigard, J.A., Chumley, F.G., and Valent, B. (2000). A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi-ta. Plant Cell 12, 2019–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth, K., Xu, Z., Mudgett, M.B., Bao, Z.Q., Palmer, L.E., Bliska, J.B., Mangel, W.F., Staskawicz, B., and Dixon, J.E. (2000). Disruption of signaling by Yersinia effector YopJ, a ubiquitin-like protein protease. Science 290, 1594–1597. [DOI] [PubMed] [Google Scholar]
- Rangan, V.S., Joshi, A.K., and Smith, S. (1998). Fatty acid synthase dimers containing catalytically active beta-ketoacyl synthase or malonyl/acetyltransferase domains in only one subunit can support fatty acid synthesis at the acyl carrier protein domains of both subunits. J. Biol. Chem. 273, 34949–34953. [DOI] [PubMed] [Google Scholar]
- Rohe, M., Gierlich, A., Hermann, H., Hahn, M., Schmidt, B., Rosahl, S., and Knogge, W. (1995). The race-specific elicitor, NIP1, from the barley pathogen, Rhynchosporium secalis, determines avirulence on host plants of the Rrs1 resistance genotype. EMBO J. 14, 4168–4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salanoubat, M., et al. (2002). Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415, 497–502. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Shan, W., Cao, M., Leung, D., and Tyler, B.M. (2004). The Avr1b locus of Phytophthora sojae encodes an elicitor and a regulator required for avirulence on soybean plants carrying resistance gene Rps1b. Mol. Plant-Microbe Interact. 17, 394–403. [DOI] [PubMed] [Google Scholar]
- Shao, F., Golstein, C., Ade, J., Stoutemyer, M., Dixon, J.E., and Innes, R.W. (2003). Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 301, 1230–1233. [DOI] [PubMed] [Google Scholar]
- Silakowski, B., Schairer, H.U., Ehret, H., Kunze, B., Weinig, S., Nordsiek, G., Brandt, P., Blocker, H., Hofle, G., Beyer, S., and Muller, R. (1999). New lessons for combinatorial biosynthesis from myxobacteria. The myxothiazol biosynthetic gene cluster of Stigmatella aurantiaca DW4/3-1. J. Biol. Chem. 274, 37391–37399. [DOI] [PubMed] [Google Scholar]
- Silué, D., Tharreau, D., and Notteghem, J.L. (1992). Identification of Magnaporthe grisea avirulence genes to seven rice cultivars. Phytopathology 82, 1462–1467. [Google Scholar]
- Stachelhaus, T., Mootz, H.D., and Marahiel, M.A. (1999). The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6, 493–505. [DOI] [PubMed] [Google Scholar]
- Sweeney, M.J., and Dobson, A.D. (1999). Molecular biology of mycotoxin biosynthesis. FEMS Microbiol. Lett. 175, 149–163. [DOI] [PubMed] [Google Scholar]
- Sweigard, J.A., Carroll, A.M., Kang, S., Farrall, L., Chumley, F.G., and Valent, B. (1995). Identification, cloning, and characterization of PWL2, a gene for host species specificity in the rice blast fungus. Plant Cell 7, 1221–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweigard, J.A., Chumley, F., Carroll, A.M., Farrall, L., and Valent, B. (1997). A series of vectors for fungal transformation. Fungal Genet. Newsl. 44, 52–53. [Google Scholar]
- Swofford, D.L. (2002). PAUP*: Phylogenetic Analysis Using Parsimony. (Sunderland, MA: Sinauer Associates).
- Takano, Y., Choi, W., Mitchell, T.K., Okuno, T., and Dean, R.A. (2003). Large scale parallel analysis of gene expression during infection-related morphogenesis of Magnaporthe grisea. Mol. Plant Pathol. 4, 337–346. [DOI] [PubMed] [Google Scholar]
- van den Burg, H.A., Spronk, C.A., Boeren, S., Kennedy, M.A., Vissers, J.P., Vuister, G.W., de Wit, P.J., and Vervoort, J. (2004). Binding of the AVR4 elicitor of Cladosporium fulvum to chitotriose units is facilitated by positive allosteric protein-protein interactions: The chitin-binding site of AVR4 represents a novel binding site on the folding scaffold shared between the invertebrate and the plant chitin-binding domain. J. Biol. Chem. 279, 16786–16796. [DOI] [PubMed] [Google Scholar]
- van der Biezen, E.A., and Jones, J.D. (1998). Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem. Sci. 23, 454–456. [DOI] [PubMed] [Google Scholar]
- Vivian, A., and Arnold, D.L. (2000). Bacterial effector genes and their role in host-pathogen interactions. J. Plant Pathol. 82, 163–178. [Google Scholar]
- Walton, J.D. (1996). Host-selective toxins: Agents of compatibility. Plant Cell 8, 1723–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolpert, T.J., Dunkle, L.D., and Ciuffetti, L.M. (2002). Host-selective toxins and avirulence determinants: What's in a name? Annu. Rev. Phytopathol. 40, 251–285. [DOI] [PubMed] [Google Scholar]
- Xue, C., Park, G., Choi, W., Zheng, L., Dean, R.A., and Xu, J.R. (2002). Two novel fungal virulence genes specifically expressed in appressoria of the rice blast fungus. Plant Cell 14, 2107–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.