Abstract
Plant-parasitic nematodes Meloidogyne spp induce an elaborate permanent feeding site characterized by the redifferentiation of root cells into multinucleate and hypertrophied giant cells. We have isolated by a promoter trap strategy an Arabidopsis thaliana formin gene, AtFH6, which is upregulated during giant cell formation. Formins are actin-nucleating proteins that stimulate de novo polymerization of actin filaments. We show here that three type-I formins were upregulated in giant cells and that the AtFH6 protein was anchored to the plasma membrane and uniformly distributed. Suppression of the budding defect of the Saccharomyces cerevisiae bni1Δ bnr1Δ mutant showed that AtFH6 regulates polarized growth by controlling the assembly of actin cables. Our results suggest that AtFH6 might be involved in the isotropic growth of hypertrophied feeding cells via the reorganization of the actin cytoskeleton. The actin cables would serve as tracks for vesicle trafficking needed for extensive plasma membrane and cell wall biogenesis. Therefore, determining how plant parasitic nematodes modify root cells into giant cells represents an attractive system to identify genes that regulate cell growth and morphogenesis.
INTRODUCTION
Reorganization of the cytoskeleton is essential for many celFlular processes, including cell morphogenesis and cell division, in animals and plants. Recently, new cytoskeletal components and regulatory proteins involved in plant cell growth and form have been identified (Wasteney and Galway, 2003). Among these genes, microtubule-associated proteins ZWI (Oppenheimer et al., 1997) and MOR1 (Whittington et al., 2001), profilin PFN (Ramachandran et al., 2000), actin ACT7 (Gilliland et al., 2003), and CDM family member SPIKE1 (Qiu et al., 2002) are required for cytoskeletal organization and normal cell growth and shape. Additionally in maize (Zea mays), the BRK1 gene encodes an 8-kD protein required for the formation of leaf epidermal cell lobes (Frank and Smith, 2002). Its mammalian homolog is found in a multiprotein complex implicated in the activation of Arp2,3-dependant actin polymerization.
The functions of microtubules in plant cell division and polarized growth are better understood than the role of actin microfilaments, which remains unclear (Banno and Chua, 2000). Actin-depolymerizing factor/cofilin and profilin are two well-studied plant actin-modulating proteins that act synergistically to regulate actin dynamics (Didry et al., 1998). Profilins in animal and fungal systems are known to interact with four major classes of poly-l-Pro–containing proteins belonging to the signal transduction cascade responsible for rearrangement of the actin cytoskeleton. Among them, formins are the first members of this group described in plants (Deeks et al., 2002). Formins, also known as formin homology (FH) proteins, are cytoskeleton-organizing proteins involved in cytokinesis, the establishment and maintenance of cell polarity (reviewed in Frazier and Field, 1997; Wasserman, 1998; Tanaka, 2000), vertebrate limb formation (Woychik et al., 1990), and the hearing process (Lynch et al., 1997). Several formins, such as the budding yeast proteins BNI1p and BNR1p (Kohno et al., 1996; Evangelista et al., 1997; Imamura et al., 1997) and the mammalian homolog of Drosophila melanogaster DIAPHANOUS (DIA) (Watanabe et al., 1997), are effectors of the Rho and Cdc-42 GTPases. Rho and Cdc-42 guanosine triphosphatases, which are two subgroups of the Rho family of Ras-related small GTP binding proteins, are signaling molecules that regulate several essential cellular processes, including actin dynamics. All FH proteins share two common structural features: a Pro-rich FH1 domain and a highly conserved FH2 domain (for review, see Frazier and Field, 1997). FH1 interacts with profilins and proteins containing SH3 and WWP/WW domains (Chan et al., 1996; Chang et al., 1997; Watanabe et al., 1997). The FH2 domain of BNI1p was recently shown to nucleate actin filaments and to associate with the barbed end of growing actin filaments (Pruyne et al., 2002; Sagot et al., 2002b). Although FH proteins are required for organization of the actin cytoskeleton, some formins also have been found to be implicated in microtubule cytoskeleton regulation (Emmons et al., 1995; Lee et al., 1999; Palazzo et al., 2001). Animal and fungal formins have been studied extensively, but little is known about the function of formins in plants. Banno and Chua (2000) reported the characterization of an FH protein, AFH1, in Arabidopsis thaliana. Overexpression of AFH1 in pollen tubes induced the formation of supernumerary actin cables, leading to tube broadening, growth depolarization, and growth arrest (Cheung and Wu, 2004). In silico analyses of the Arabidopsis genome have resulted in the identification of at least 21 genes predicted to encode FH proteins (Cvrckova, 2000; Deeks et al., 2002). However, except data obtained on AFH1, the localization of gene expression at the cellular and subcellular levels and function of the proteins encoded by these genes are still unknown.
In higher plants, various model systems, such as tip-growing cells (e.g., pollen tubes and root hairs) (Hepler et al., 2001), trichomes (Mathur et al., 1999), and morphogenetic mutants, mainly of Arabidopsis (Söllner et al., 2002), have been used to investigate the function of the cytoskeleton in cells during development. Host–pathogen interactions may also provide interesting model systems for the identification and analysis of the role of genes involved in plant development. In the case of the ontogenesis of nematode feeding sites induced by the plant-parasitic root-knot nematodes (Meloidogyne spp), parenchyma cells of the differentiating vascular cylinder are transformed into hypertrophied multinucleate giant cells from which the nematode feeds (Jones, 1981). These cells develop by repeated nuclear division without cytokinesis (Huang, 1985). The cell plate vesicles initially line up between the two daughter nuclei but are then dispersed, aborting the formation of a new cell plate (Jones and Payne, 1978). The fully differentiated giant cells are dramatically enlarged and may contain up to 150 polyploid nuclei that have also undergone extensive endoreduplication (Wiggers et al., 1990). The giant cell expands diffusely by isotropic growth to reach a final size ∼100 times that of root cortex cells. Mature giant cells function as transfer cells for the feeding nematode and are metabolically active, as shown by the presence of cell wall ingrowths adjacent to vascular tissue, breakdown of the large vacuole, and the dense granular cytoplasm with many organelles (Jones, 1981). Typical root-knots or galls are the primary visible symptom of infection and develop by hyperplasia and the division of cortical cells around giant cells. These complex morphological and physiological changes during establishment of the giant cells are reflected in altered gene expression (Gheysen and Fenoll, 2002). Molecular analysis of giant cell development has resulted in the identification of several plant genes that are upregulated during this process, including cell cycle markers, such as the mitotic cyclin gene CYCA2;1 (de Almeida-Engler et al., 1999), the APC activator gene CCS52a, encoding a protein involved in endoreduplication (Favery et al., 2002), and the actin genes ACT2 and ACT7 (de Almeida-Engler et al., 2004). The recent observation of the cytoskeleton architecture in giant cells revealed that major and essential rearrangements occur during the formation of nematode-induced feeding cells (de Almeida-Engler et al., 2004).
We investigated the molecular mechanisms underlying giant cell formation and tried to identify genes affecting cytoskeleton organization, cytokinesis, and polarized growth by means of a promoter trap strategy of genes expressed in giant cells in Arabidopsis. This biological screening method resulted in the identification of an Arabidopsis gene, AtFH6, that is upregulated at early stages of nematode feeding site formation and that encodes an FH protein. We report here the functional analysis of a plant FH protein, describing its spatial and temporal expression pattern during plant development and giant cell formation. In addition, immunolocalization and cell fractionation shows an unusual formin localization because AtFH6 is uniformly distributed and anchored to the plasma membrane. Finally, the suppression of the budding defect of the Saccharomyces cerevisiae bni1Δ bnr1Δ mutant shows that AtFH6 is a player in the regulation of polarized growth by controlling the assembly of actin cables. These results are consistent with AtFH6 being involved in actin cytoskeleton reorganization and its possible role in the control of plant cell growth.
RESULTS
The CSQ2-Tagged Line Displays β-Glucuronidase Activity at the Nematode Feeding Site
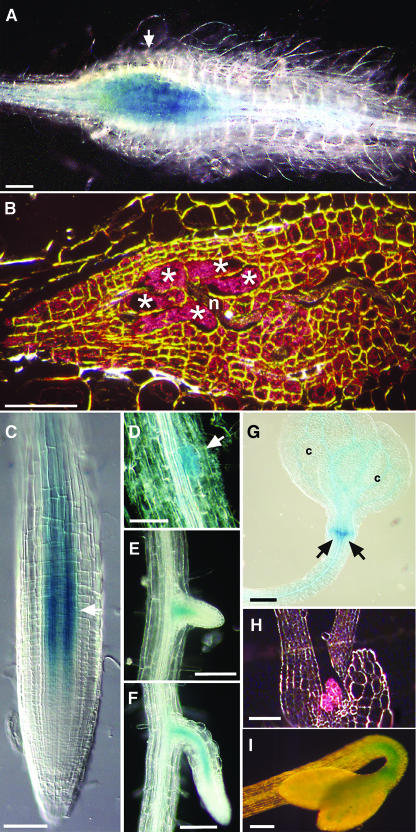
To isolate genes involved in the development of giant cells induced by Meloidogyne incognita, a promoter trap strategy was developed with a promoterless β-glucuronidase (GUS) construct being introduced randomly into the Arabidopsis genome via Agrobacterium tumefaciens T-DNA transformation (Favery et al., 1998). We screened 20,000 T-DNA–tagged Arabidopsis lines by GUS assay after root-knot nematode infection and identified 200 lines showing GUS induction in root galls. One of the lines, CSQ2, displayed early GUS activity in galls, which was detected <48 h after giant cell initiation (Figure 1A). GUS activity was detected over a 3-week period but was not detected thereafter in fully differentiated giant cells. Cross sections of 7-d-old galls clearly showed GUS staining in the giant cells and in the neighboring cells (Figure 1B). No GUS activity was detected in the cortical cells of the gall.
Figure 1.
The CSQ2 T-DNA–Tagged Arabidopsis Line Displays GUS Activity in Galls Induced by M. incognita and during Plant Development.
(A) and (B) GUS expression in galls induced by M. incognita.
(A) Localized GUS activity in a root gall 7 d post infection (dpi) (arrow).
(B) Sectioned gall shown in (A) seen by dark-field microscopy. GUS activity (seen as a pink precipitate) is observed in the giant cells and in the surrounding cells. Asterisks, giant cells; n, nematode.
(C) to (I) GUS expression during plant development.
(C) Root apex. Arrow shows region where GUS activity was stronger.
(D) to (F) Three stages during lateral root development (arrow).
(G) Young seedling 4 d after germination. c, cotyledon.
(H) Shoot apex of seedling 4 d after germination showing GUS-stained stipules examined by dark-field microscopy.
(I) Dark-grown seedling 4 d after germination showing GUS stain in the apical hook.
Bars in (A), (B), and (E) to (I) = 100 μm; bars in (C) and (D) = 50 μm.
During plant development, GUS expression was observed in differentiating cells of the vascular cylinder just above the root meristem (Figures 1C). GUS activity was also detected in the vascular tissue of the lateral root primordium and in the emerged lateral root (Figures 1D to 1F). GUS staining was observed neither in the root apical meristem nor in the differentiated root. In young seedlings, low levels of promoter activity were also detected in the vascular bundles of leaves and in the stipules (Figures 1G). In the shoots of older plants, GUS activity was restricted to the stipules (Figures 1H). In addition, in etiolated seedlings, GUS activity was detected only in the apical hook and not in the remaining elongated hypocotyl (Figure 1I).
Molecular Cloning of the CSQ2 Gene
The CSQ2 line carried a single T-DNA insert (data not shown). A 0.5-kb genomic DNA fragment adjacent to the right border of the T-DNA was isolated by kanamycin plasmid rescue (Bouchez et al., 1996) (Figure 2A). Sequence analysis, using The Arabidopsis Information Resource (TAIR), showed that the T-DNA had integrated into the predicted gene At5g67470 on chromosome 5. The corresponding CSQ2 cDNA was cloned by rapid amplification of cDNA ends (RACE) PCR. The CSQ2 cDNA is 3072 nucleotides long and contains an open reading frame of 899 amino acids (Figure 2A). Comparison of the genomic and cDNA sequences revealed that the CSQ2 gene contained four exons (Figure 2A). Integration of the T-DNA resulted in a 6-bp deletion and the insertion of a 23-bp filler sequence. The T-DNA had inserted into the first exon, 1444 bp downstream from the ATG, placing the ATG of the GUS gene in frame with the csq2 gene, resulting in a functional gene fusion.
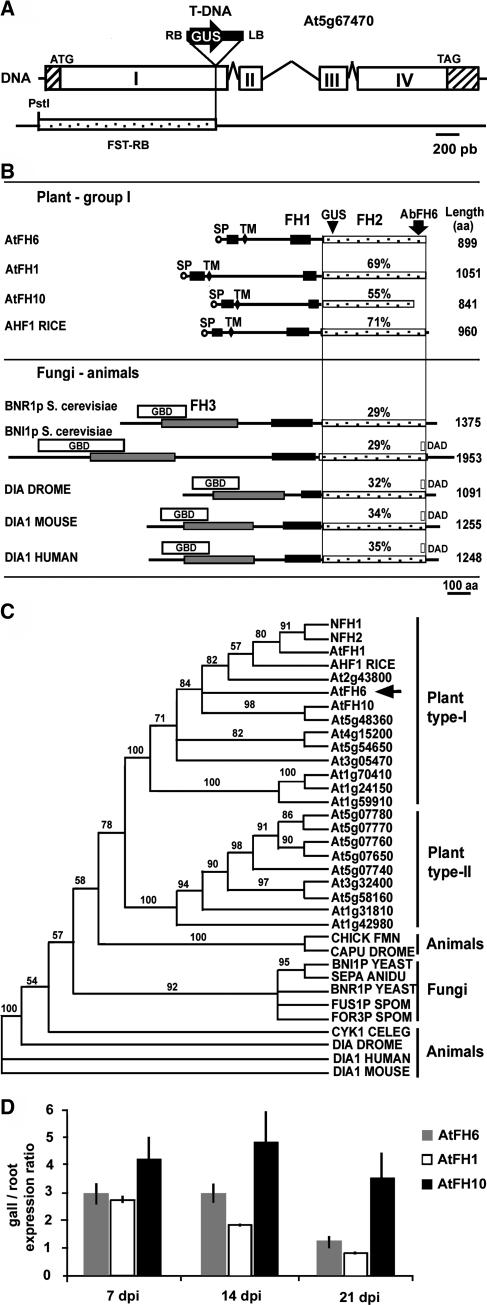
Figure 2.
The AtFH6 Gene Encodes an FH Protein.
(A) Organization of the AtFH6 gene and molecular analysis of the T-DNA insertion. Open boxes are the exons (I to IV, determined from cDNA), and the sequences between these boxes correspond to introns; hatched boxes indicate untranslated sequences. The T-DNA is inserted into the first exon. RB and LB correspond to the right and left T-DNA borders, respectively, and GUS the coding region of the β-glucuronidase gene. The initiation and stop codons are indicated. The RB flanking sequence (FST-RB) obtained by kanamycin rescue is shown.
(B) Schematic representation of AtFH6 and other FH-related proteins. All members share a central Pro-rich domain, FH1 (black boxes), and the FH2 domain (stippled boxes). The site of GUS fusion in the CSQ2 line is indicated. A second Pro-rich domain (black boxes) between a signal peptide (SP, circles) and a transmembrane segment (TM, lozenges) was observed only in plant type-I FH proteins. The GBD (white boxes), DAD (white boxes), and FH3 domains (gray boxes) are present in non-plant FH proteins. The percentage of similarity of sequences to the AtFH6 FH2 domain is indicated. Comparisons were made between three plant type-I FHs—Arabidopsis AtFH1 (Banno and Chua, 2000) and AtFH10 and rice AHF1—and five fungal and animal FH proteins—S. cerevisiae BNI1p and BNR1p (Evangelista et al., 1997; Imamura et al., 1997), Drosophila Dia (Dia Drome) (Castrillon and Wasserman, 1994), mouse DIA1 (Watanabe et al., 1997), and human DIA1 (Lynch et al., 1997).
(C) Neighbor-joining dendrogram of relationships among plant, yeast, and animal FH2 domains. The phylogenetic tree is based on the Clustal alignment of the FH2 domains of Arabidopsis FH proteins, rice AHF1, tobacco NFH1 and NFH2 (Banno and Chua, 2000)—and five fungal and six animal FH proteins—BNI1p, BNR1p, S. pombe FUS1p and FOR3p, Aspergillus nidulans SEPA, chicken FMN, Drosophila DIA and CAPPUCCINO (CAPU), mouse and human DIA1, and Caenorhabditis elegans CYK1. Bootstrap support (data resampled 100 times) for the apparent groupings is indicated.
(D) Quantitative RT-PCR analysis of the transcript abundance of AtFH6, AtFH1, and AtFH10 in galls 7, 14, and 21 dpi compared with noninfected root tissues.
CSQ2 Encodes an FH Protein
Analysis of the deduced amino acid sequence of the protein encoded by CSQ2 showed it to be a member of the FH protein family from Arabidopsis identified by in silico analysis (Deeks et al., 2002). It was therefore named A. thaliana Formin Homology protein 6 (AtFH6). The AtFH6 protein bears the two structural features common to FH proteins: a Pro-rich formin homology-1 (FH1; amino acids 317 to 398) domain in front of a conserved C-terminal FH2 domain (amino acids 452 to 899)—the hallmark of members of the formin family (Figure 2B). The FH1 domain of AtFH6 contains 43% Pro, with stretches of three to nine consecutive Pro residues, and two WW binding sequences (P/RPR as shown for the WW domains of the formin binding protein FBP30) (Chan et al., 1996) separated by a 26–amino acid spacer and several SH3 binding sequences (PxxP) (data not shown). The FH2 domain of AtFH6 was found to display 29 to 69% sequence similarity to the equivalent domains of the other 20 AtFH proteins. Similarity was greatest with AtFH1 (69%; Banno and Chua, 2000) and the rice (Oryza sativa) predicted protein AHF1 (71%; BAB86073) (Figure 2B). We investigated the possible relationships between plant FH sequences and between these sequences and those of known FHs from animals and fungi by protein sequence comparison algorithms, such as the neighbor-joining tree method (Figure 2C). The plant FH sequences clustered and formed a lineage distinct from that of fungi, insect, and vertebrate FH members. Plant FHs can be divided into two subfamilies—type I and type II—as previously described for AtFHs (Deeks et al., 2002). The AtFH6, AtFH1, and rice AHF1 proteins clearly belong to the type-I subclass (Figure 2C).
Unlike the FH proteins of animals and fungi, such as the budding yeast proteins BNI1p and BNR1p, Drosophila DIA, and mouse and human DIA, no N-terminal FH3 homology domain required for correct targeting (Petersen et al., 1998) or coiled-coil domain were identified in plant formins (Figure 2B). Moreover, the plant FHs characterized to date lack both the GTPase binding domain (GBD) (Watanabe et al., 1997) and the diaphanous autoregulatory domain (DAD) (Alberts, 2001). These two domains interact to inactivate the formin (Alberts, 2001; Sagot et al., 2002b). However, AtFH6 and the other type-I plant formins have an N-terminal domain with a putative signal peptide or membrane anchor (amino acids 1 to 24) and a transmembrane domain (107 to 129) adjacent to a second Pro-rich region, suggesting that this protein is targeted for secretion and integration into a membrane (Figure 2B). The region between the signal peptide and the transmembrane domain (amino acids 34 to 82) contains 43% Pro but is not rich in Ser and does not contain the SPPPP motif characteristic of plant cell wall extensin glycoproteins, as described for AtFH1 (Banno and Chua, 2000). Interestingly, this Pro-rich sequence contains a PLPPxxPxxPxxPxxPxxP sequence homologous to the WW binding sequence (PPLP as shown for the WW domains of FBP-11) and overlapping SH3 binding sequences, which mediate protein–protein interactions (Chan et al., 1996). Thus, AtFH6 may exert or regulate its biological functions by interacting with proteins containing SH3 and WW domains via this Pro-rich sequence and/or the FH1 domain.
Three AtFH Type-I Genes Are Expressed in Root Galls
We have characterized the role of AtFH6 in plant development and giant cell formation by isolating plants homozygous for the csq2 mutation. The csq2/csq2 plants were macroscopically not distinguishable from the wild type and developed normally. In addition, after nematode infection, csq2 homozygous mutant plants had a similar number of galls compared with wild-type plants. Giant cell structure and nematode development were also similar to what was observed in wild-type control plants. This lack of a visible phenotype may be because of (1) incomplete loss of protein function, as the N-terminal part of the protein in homozygous plants includes the FH1 domain fused to GUS, or more probably, (2) because of the presence of other AtFH genes with overlapping in their expression patterns, resulting in genetic redundancy.
Therefore, we have used gene-specific PCR primers to amplify each predicted transcript of the 21 AtFH genes. RT-PCR analysis showed that only two additional type-I AtFH genes, AtFH1 and AtFH10, were expressed in galls (data not shown). Quantitative RT-PCR showed that AtFH6, AtFH1, and AtFH10 were significantly upregulated in galls 7 and 14 dpi compared with uninfected nonmeristematic root fragments (Figure 2D). No upregulation of AtFH6 and AtFH1 was detected 21 dpi in fully differentiated giant cells. These results confirm the expression pattern of CSQ2 deduced from experiments with the GUS reporter gene and show that three AtFH genes are activated during gall formation induced by root-knot nematodes.
AtFH6 Encodes a Membrane Protein Located along the Plasma Membrane
The prediction of peptide domains and hydropathy profile determination suggested that AtFH6 is a membrane protein anchored to a plasma membrane or endomembrane. We have localized the AtFH6 protein using a polyclonal antibody (AbAtFH6) directed against part of the C-terminal sequence of the FH2 domain. In protein gel blots with an insoluble microsomal fraction prepared from young seedlings, we detected a single band at ∼99 kD (Figure 3A), consistent with the predicted molecular mass of AtFH6. No signal was detected in cytoplasmic fractions or in microsomal fractions obtained from the homozygous csq2/csq2 plants in which GUS replaced the C-terminal 418 amino acids of AtFH6 (Figure 3A). These results indicate that AbAtFH6 is specific for AtFH6 protein and does not cross-react with the other Arabidopsis FHs. In addition, protein gel blots performed with proteins from highly purified plasma membrane of Arabidopsis cells showed that AtFH6 protein is more abundant in this fraction compared with the endomembranes or the microsomal fraction (Figure 3B). No signal was detected in the cytosolic fraction.
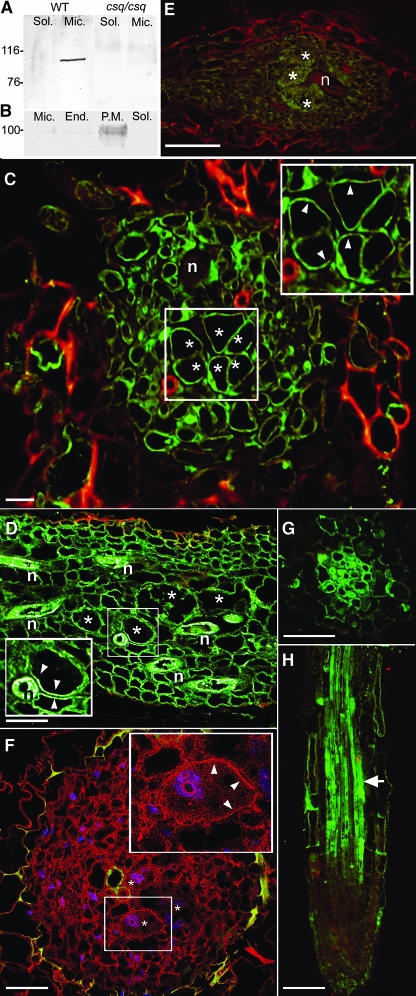
Figure 3.
Immunolocalization of AtFH6 and Actin in Galls and Root Apex.
(A) and (B) Cell fractionation and protein gel blot analyses of wild-type or homozygous csq2/csq2 plants.
(A) Mouse antibody AbAFH6 was used to detect the protein in soluble (Sol) and microsomal (Mic) fractions.
(B) The purified plasma membrane fraction (P.M.) of Arabidopsis wild-type cells contained a higher amount of AtFH6 than the microsomal fraction (Mic) or the endomembranes (End). No signal was detected in the cytosolic fraction (Sol).
(C) and (D) Immunolocalization of the AtFH6 protein in gall sections 5 (C) and 7 dpi (D) of wild-type plants. Fluorescent signal is detected in the plasma membrane (arrowheads) of giant cells (asterisks) and surrounding cells. n, nematodes.
(E) Immunolocalization of the AtFH6 protein in a gall section 7 dpi of csq2/csq2 plants showing no fluorescence.
(F) Immunofluorescence detection of actin in galls 5 dpi of wild-type plants. A detail of a giant cell (asterisks) showed actin localization in the cell cortex (arrowheads) and less within the cytoplasm. Actin is visualized in red and 4′,6-diamidino-2-phenylindole (DAPI)-stained nuclei in blue.
(G) and (H) Immunolocalization of the AtFH6 protein in a cross section and longitudinal section of root apex of wild-type plants. AtFH6 protein signal (arrow) is detected in the differentiation zone of the vascular cylinder.
Bars in (A) to (F) = 100 μm; bar in (G) = 50 μm.
Immunolocalization of the protein in galls, using AbAtFH6, confirmed the expression pattern observed with GUS staining (Figures 3C and 3D). The AtFH6 protein was detected only in giant cells and in neighboring cells. AtFH6 was detected only in the plasma membrane of giant cells, and no fluorescence was seen in the typically dense cytoplasm (Figures 3C and 3D). No signal was detected in galls of homozygous csq2/csq2 seedlings (Figure 3E). Immunostaining on sections of actin in giant cells revealed a higher density of actin label in the cell cortex than within the dense cytoplasm (Figure 3F). In root apex, AtFH6 staining was confined to differentiating cells within the vascular cylinder (Figures 3G and 3H), confirming the expression pattern observed with GUS staining. Cross sections showed that the AtFH6 protein was located along the plasma membrane as observed for gall cells.
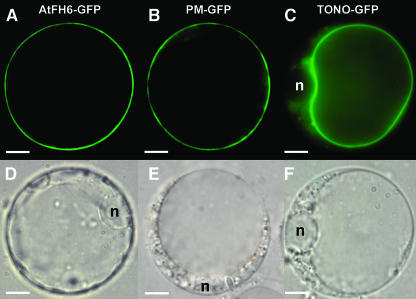
To confirm the AtFH6 subcellular localization, transient expression of fusion proteins with green fluorescent protein (GFP) was performed in protoplasts prepared from Arabidopsis cell suspensions. The green fluorescent signal clearly delineated the plasma membrane associated with the expression of the AtFH6:GFP fusion (Figures 4A and 4D) or the plasma membrane marker p31:GFP fusion (Figures 4B and 4E). GFP fusion with AtNRAMP3, a metal transporter targeted to the vacuolar membrane, led to a fluorescence (Figures 4C and 4F) confined to the tonoplast differently than observed for AtFH6.
Figure 4.
AtFH6 Is Targeted to the Plasma Membrane.
(A) to (F) Transient expression of GFP fusion proteins into Arabidopsis protoplasts.
(A) Fluorescence signal showing a plasma membrane targeting of AtFH6:GFP.
(B) Fluorescence signal showing expression of P31:GFP, a plasma membrane marker.
(C) Fluorescence signal showing expression of AtNRAMP3:GFP, a tonoplast marker.
(D) to (F) The transmission image of protoplasts shown in (A), (B), and (C), respectively. n, nucleus. Bars = 50 μm.
AtFH6 Suppress the Yeast bni1Δ bnr1Δ Mutant
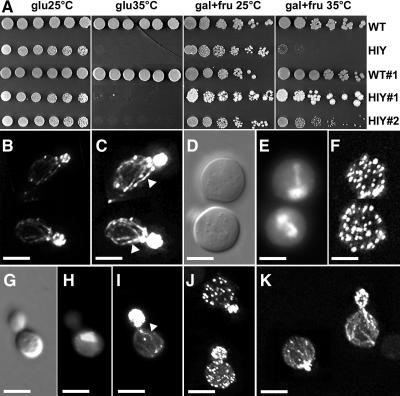
We investigated the role of AtFH6 in cytoskeleton organization by determining whether AtFH6 functionally suppressed bni1 and bnr1 deficiency in yeast. The FH proteins BNI1p and BNR1p of the budding yeast S. cerevisiae were recently shown to regulate polarized growth by controlling the assembly of actin cables (Evangelista et al., 2002; Sagot et al., 2002a). We introduced the full-length AtFH6 cDNA, under the control of a galactose-inducible promoter, into a yeast bni1Δ bnr1Δ mutant (HIY11). At the permissive temperature (25°C), HIY11 grew slower than the wild-type K699 strain, but nevertheless cells displayed the budding and wild-type phenotype (Figure 5A). At the restrictive temperature (35°C), all double mutant cells became enlarged (Figure 5D) and were unable to divide, as described by Imamura et al. (1997) (Figure 5A). DNA stained by DAPI also confirms that HIY11 cells are often multinucleate, indicating that synthesis and division of nuclear DNAs are not primarily affected (Figure 5E). The bni1Δ bnr1Δ cells containing pYES2, which carried AtFH6 (HIY11#1 and #2), grew at 35°C only in the presence of galactose, indicating suppression of the bni1Δ bnr1Δ phenotype (Figure 5A). Analysis of actin organization showed that, in wild-type cells, actin patches were polarized and mainly restricted to the bud (Figure 5B), whereas actin cables were oriented along the length of the cell (Figure 5C). By contrast, actin patches were randomly distributed in enlarged bni1Δ bnr1Δ cells, with no actin cables apparent (Figure 5F). In HIY#1 and HIY#2 cells, the budding defect and cell enlargement of the bni1Δ bnr1Δ mutant were rescued by suppression at the restrictive temperature (Figures 5G and 5H). In these budding cells, some actin cables were visible in the mother cell, and actin patches were polarized to the bud (Figure 5I). In some suppressed HIY cells, actin patches were still seen in the mother cells (Figure 5J). No growth defect or extra actin cables were detected in wild-type cells producing AtFH6 (WT#1) (Figure 5K), as previously reported after the overproduction of formin protein fragments (Evangelista et al., 2002; Sagot et al., 2002a). These experiments showed that AtFH6 is functionally similar to yeast FH proteins and that it plays a role in actin cytoskeleton reorganization.
Figure 5.
Suppression of the Yeast bniΔ bnr1Δ Mutant by AtFH6.
(A) AtFH6 expression suppress the temperature-sensitive defect of bniΔ bnr1Δ (HIY11) yeast. Serial dilutions of cultures of each transformant (bniΔ bnr1Δ mutant and wild-type K699) containing empty pYES2 (HIY11 and WT) or pYES2 AtFH6 (HIY#1, HIY#2, and WT#1) were spotted onto media lacking uracil but containing 1% glucose (glu) for repression or 1% galactose and 1% fructose (gal+fru) for induction and grown at permissive (25°C) or restrictive (35°C) temperature. HIY11 cells did not grow at 35°C, whereas HIY11#1 and #2 transformants grew at 35°C only in the presence of galactose.
(B), (C), (F), and (I) to (K) Projections of optical sections of yeast cells grown at 35°C in the presence of galactose and stained with fluorescent phalloidin, which binds to filamentous actin.
(B) Wild-type budding cells with polarized actin patches and cables.
(C) Image shown in (B) with contrast enhanced to make it easier to see actin cables (arrowheads).
(D) and (E) Large bniΔ bnr1Δ rounded cells visualized by differential interference contrast and fluorescence microscopy after DAPI staining of DNA, respectively.
(F) Enlarged bniΔ bnr1Δ mutants with randomly distributed actin patches.
(G) and (H) Suppressed HIY#1 budding cells expressing AtFH6 visualized by differential interference contrast and fluorescence microscopy after DAPI staining, respectively.
(I) Suppressed HIY#1 cells expressing AtFH6 with actin cables in the mother cell (arrowhead) and actin patches in the bud.
(J) In some suppressed HIY#1 budding cells, actin patches are still observed in the mother cells.
(K) WT#1 expressing AtFH6.
Bars = 5 μm.
DISCUSSION
The control of cell growth and polarity depends on a dynamic actin cytoskeleton that can reorganize in response to developmental and environmental stimuli. Among PLP profilin binding proteins, animal and fungal formins have been extensively studied, but their role in plants remains to be determined. Plant responses to pathogen attack involve dynamic reorganization of the cytoskeleton (reviewed in Kobayashi and Kobayashi, 2000) and therefore provide interesting model systems for the identification and analysis of the role of genes involved in plant development. Sedentary endoparasitic nematodes induce the differentiation of root cells into enlarged multinucleate feeding cells with a dense cytoplasm. It has been recently described that nematodes induce long-term rearrangements of the cytoskeleton during the infection process (de Almeida-Engler et al., 2004). Our goal is to understand how cytoskeleton organization is regulated during plant development and giant cell formation. We report here the functional analysis of a plant FH protein and suggest its role in plant cell growth.
AtFH6 Is a Member of the FH Family
The AtFH6 gene was identified by a promoter trap screen for genes upregulated in the hypertrophied multinucleate feeding cells induced by the sedentary plant-parasitic nematode M. incognita. The AtFH6 protein belongs to the formin protein family. The members of this family are involved in various aspects of morphogenesis, cell polarization, and cytokinesis in eukaryotes. From yeast to mammals, FH proteins fulfill these functions by controlling actin cytoskeleton rearrangements (reviewed in Frazier and Field, 1997; Wasserman, 1998; Tanaka, 2000). The founding members of this family—mouse formins encoded by the limb deformity (ld) locus—are required for limb and kidney development (Woychik et al., 1990). In Drosophila, the formin mutants dia and cappuccino display a loss of oocyte polarity and defects in cytokinesis in various tissues (Castrillon and Wasserman, 1994; Emmons et al., 1995). The budding yeast formins BNI1p and BNR1p are required for bud site selection and cytokinesis (Evangelista et al., 1997; Imamura et al., 1997) and regulate polarized growth by controlling the assembly of actin cables (Evangelista et al., 2002; Sagot et al., 2002a). Suppression of the bni1Δ bnr1Δ yeast mutant showed that plant AtFH6 fulfills a similar function to these two proteins in yeast and may therefore operate as part of the same or overlapping regulatory pathway. The rescued cells did not display the defects in bud emergence and subsequent depolarized growth observed in the double mutant (Imamura et al., 1997). Thus, the FH1 and FH2 domains of AtFH6 may be involved in controlling the organization of actin filaments and/or microtubules. The FH2 domain of BNI1p was recently shown to nucleate actin filaments and to associate with the barbed end of growing actin filaments (Pruyne et al., 2002; Sagot et al., 2002b; Pring et al., 2003).
AtFH6 Is a Type-I Plant FH Anchored to the Plasma Membrane
AtFH6 is a member of a multigene family of at least 21 FH2-containing proteins in Arabidopsis (Deeks et al., 2002). Plant FH genes have been classified into two subfamilies, based on their structures. AtFH6 is a type-I protein, defined on the basis of the presence of a specific N-terminal region containing a putative signal peptide, a Pro-rich domain, and a transmembrane domain. Immunolocalization, cell fractionation, and GFP fusion experiments demonstrated that AtFH6 is anchored to the plasma membrane. In animals and fungi, FH proteins are generally found in specific structures, such as the projection tip (FUS1p and Bni1p; Evangelista et al., 1997; Petersen et al., 1998), incipient bud site and neck (Bni1p and Bnr1p; Imamura et al., 1997), and the cell division ring (CDC12p; Chang et al., 1997). BNI1p is presumed to be linked to the plasma membrane through an anchoring protein system. Rho family members and SPA2p, a protein required for polarized growth, are candidates for anchoring proteins (Tanaka and Takai, 1998; Ozaki-Kuroda et al., 2001). In vertebrates, formins, unlike other FH proteins, are found in the nucleus (Trumpp et al., 1992).
Unlike fungal and animal FH proteins, plant type-I FH proteins lack the FH3 homology domain thought to be important for determining intracellular localization of formin family proteins (Petersen et al., 1998; Kitayama and Uyeda, 2003), the GBD (Watanabe et al., 1997), and the DAD (Alberts, 2001). The FH3 domain is involved in targeting to the projection tip of Schizosaccharomyces pombe (Petersen et al., 1998) and to the crowns of Dictyostelium discoideum (Kitayama and Uyeda, 2003). These very specific locations are quite peculiar; therefore, it is difficult to identify their possible counterparts in plant cells. GBD and DAD are involved in the inhibition of FH proteins, and binding of activated Rho GTPases to GBD releases this inhibition (Alberts, 2001; Sagot et al., 2002b). The absence of GBD and DAD domains in plant type-I FHs, the lack of interaction between AtFH1 and AtRAC1 (Banno and Chua, 2000), and the specific N-terminal region suggesting a direct plasma membrane anchorage indicate that FHs may be regulated by unique mechanisms in plants.
Function of AtFH6 in Plant Cells
During plant development, the spatial and temporal patterns of AtFH6 expression indicate that the protein may be involved in the early differentiation of vascular cylinder cells, especially in the root apex and during lateral root development. Root apex cells, where AtFH6 is detected, has been shown to exhibit a transient accelerating relative growth rate and cytoskeleton rearrangement (Sugimoto et al., 2000). Therefore, formin AtFH6 may play an adaptor role linking a morphogenetic signal to actin cytoskeleton remodeling important for growth and the first steps of differentiation, as suggested for ld gene products during chicken limb bud development (Trumpp et al., 1992). This hypothesis also has been suggested for the role of type-I formins, such as AtFH1, in mediating extracellular signals from female tissues to elicit the proper pollen tube growth response during pollination (Cheung and Wu, 2004).
AtFH6 expression is not observed in dividing cells of root apical meristems, suggesting that AtFH6 may not be required for cytokinesis. Furthermore, the suppression of Drosophila dia1 and dia2 mutants with AtFH6 failed to rescue the cytokinesis defect (data not shown). However, cytokinesis differs considerably in animals and plants (Assaad, 2001). It appears likely that at least some of the large number of AtFHs are required for cytokinesis, which may be considered to be a form of polarized secretion (Assaad, 2001).
Role of AtFH6 in Nematode Giant Cell Morphogenesis
We found that AtFH6 expression was induced in the early stages of giant cell formation and persisted until a final differentiated state. Giant cell formation involves complex cell shape changes and cell wall modifications arising from plant cell wall–modifying enzymes, such as endo-β-1,4-glucanases (Goellner et al., 2001). The recruitment of cytoskeleton organizing proteins is consistent with observations that these nematodes induce long-term rearrangements of the cytoskeleton during the infection process (de Almeida-Engler et al., 2004). Recently, it has been shown that giant cells at different stages of ontogeny showed randomly oriented actin cables going through the cytoplasm and cell cortex. Abnormal thick actin cables were longitudinally and transversely oriented mainly in the cell cortex in contrast with predominantly longitudinally oriented filaments observed in uninfected root tissue (de Almeida-Engler et al., 2004).
Formins are actin-nucleating proteins that stimulate de novo polymerization of actin filaments. We showed here that three type-I formins were upregulated in giant cells and that the AtFH6 protein was uniformly distributed throughout the plasma membrane. Therefore, we can speculate that AtFH6 distribution might be involved in the isotropic growth of these hypertrophied feeding cells via the reorganization of the actin cytoskeleton. The observed actin cables would serve as tracks for vesicle trafficking needed for extensive plasma membrane and cell wall biogenesis, as proposed in the polarized growth of budding yeast (Evangelista et al., 2002; Schott et al., 2002). Therefore, we believe that plant type-I formins may play a central role in organizing the actin cytoskeleton at the plasma membrane required for cell growth. Giant cells could be used in the future to explore potential formin interacting proteins and to identify regulatory mechanisms and signaling molecules responsible for actin cytoskeleton reorganization. Thus, determining how plant parasitic nematodes modify root cells into giant cells represents an attractive system to identify genes that regulate cell growth and morphogenesis.
METHODS
Plant Materials, Growth Conditions, and Nematode Infection
The T-DNA mutagenized Arabidopsis thaliana line collection (ecotype Wassilewskija [Ws]) was generated at Institut National de la Recherche Agronomique Versailles for promoter trap and gene tagging (Bechtold et al., 1993). The lines were screened individually for GUS expression after Meloidogyne incognita infection as previously described (Favery et al., 1998). For in vitro analyses, seeds were surface sterilized and grown on Gamborg B5 medium (Sigma, St. Louis, MO) containing 2% sucrose, 0.8% agar (plant cell culture tested; Sigma), and 50 μg/mL of kanamycin. Plates were inclined at an angle of 60° to allow the roots to grow along the surface. Kanamycin resistance was scored in 2-week-old seedlings. For nematode infection in vitro, 100 surface-sterilized freshly hatched J2 of M. incognita were added on each 2-week-old seedling as described previously (Sijmons et al., 1991). The plates were kept at 20°C with a 16-h photoperiod.
Histochemical Localization of GUS Activity and Microscopic Analyses
GUS activity was assayed histochemically with 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid as described by Favery et al. (1998). Galls, root apex, and shoot apical meristems were dissected from GUS-stained plants, fixed in 1% glutaraldehyde and 4% formaldehyde in 50 mM sodium phosphate buffer, pH 7.2, dehydrated, and embedded in Technovit 7100 (Heraeus Kulzer, Wehrheim, Germany) as described by the manufacturer. Sections (4 μm) were stained with 0.05% ruthenium red and mounted in DPX (BDH Laboratory Supplies, VWR International, Fontenay-sous-Bois, France). Sections were observed with a Zeiss Axioplan 2 microscope (Jena, Germany) using dark-field optics.
Isolation of T-DNA Flanking Sequences
Kanamycin plasmid rescue of the T-DNA right border flanking sequence was done as described by Bouchez et al. (1996). The genomic DNA from transgenic plants was isolated (Doyle and Doyle, 1990), digested by PstI (unique site at the beginning of the nptII gene), and ligated into pResc38 vector. Escherichia coli DH12S cells (Invitrogen, Carlsbad, CA) were transformed with the ligation mixture. Ampicillin and kanamycin selection provided clones containing the complementing region from the T-DNA plus a flanking genomic DNA region. PCR amplification of the T-DNA left border flanking sequences was done with T-DNA oligonucleotides T5 (5′-ctacaaattgccttttcttatcgac-3′) and the gene-specific primer AtFH6 3R (5′-aacgttcattcggtttcccg-3′). Amplification was performed with a cycle of 1 min denaturation at 94°C, 1 min annealing at 58°C, and 2 min extension at 72°C, repeated 35 times, with a final 10 min extension at 72°C using an MJ Research PTC-200 Peltier thermal cycler (Watertown, MA). The amplified fragment was sequenced by Genome Express (Grenoble, France).
Isolation of csq2/csq2 Plants
To isolate csq2/csq2 plants, we analyzed the segregation of the kanamycin marker carried by the T-DNA on progenies resulting from each of 20 selfed plants. Progenies of five plants segregated 100% kanamycin-resistant plants, indicating that they were homozygous for the csq2 allele. To confirm this results, PCR experiments were done with two CSQ2 primers, AtFH6 F3for (5′-agatcgagccactgtttggg-3′) and AtFH6 3R, which span the csq2 T-DNA insertion site, and a third primer (T5) specific for the sequence of T-DNA left border. When genomic DNA from heterozygous CSQ2 plants was used as a template, both a 1447-bp and a 1249-bp band were amplified, indicating the presence of both the mutant and wild-type alleles. By contrast, when DNA from csq2/csq2 plants was used as a template, only the 1447-bp product was obtained from amplifications with all three primers.
RNA, RACE, and RT-PCR Analysis
For RNA analysis on wild-type Arabidopsis, galls were excised 7, 14, and 21 dpi with M. incognita. For comparison, corresponding nonmeristematic root fragments were obtained from uninfected plants. Plant material was frozen in liquid nitrogen immediately after excision and stored at −80°C until use. Poly(A)+ RNA was isolated with a Quickprep Micro mRNA purification kit (Pharmacia P-L Biochemicals, Milwaukee, WI) according to the manufacturer's instructions. The 5′ and 3′ end of the CSQ2 cDNA was obtained by RACE PCR (Invitrogen) with mRNA from the wild-type Arabidopsis ecotype Ws. The primers 5.1R (5′-cagatgctccatcctatcct-3′) and nested 5.2R (5′-cacgggtcggttcaacagttccca-3′) were used for 5′RACE and primers 3.1 (5′-gtatttccacggaaacgctgcgag-3′) and nested 3.2 (5′-ctcggagttctcgataacgtttg-3′) for 3′RACE. PCR products were cloned in pGEM-T vector (Promega, Madison, WI) and sequenced. For RT-PCR analysis of the 20 additional Arabidopsis AtFH gene expression in galls, cDNAs were prepared from 500-ng mRNAs using the Superscript II reverse transcriptase (Invitrogen). PCR reactions were performed as described above using 10 pmole gene-specific primers deduced from the coding and EST sequences present in the TAIR database.
Real-time quantitative RT-PCR was performed using the ABI PRISM 7900HT sequence detection system (Applied Biosystems, Foster City, CA). Specific primers AtFH6Fw (5′-caagccggaggagatatcagac-3′), AtFH6Rev (5′-gccgtgttctccgactaaacc-3′), AtFH1Fw (5′-tgtttacttctagcctccgcatt-3′), AtFH1Rev (5′-ctaccctcctcctccggc-3′), AtFH10L (5′-tggtcttcaagccacttatatgttt-3′), AtFH10R (5′-tctggcttagatgtcgtgtca-3′), At5g10790L (5′-gccaaagctgtggagaaaag-3′), and At5g10790R (5′-tgtttaggcggaacggatac-3′) were designed from genes sequences using Primer3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). For each gene amplified, a standard curve was generated from duplicate series of five template dilutions to test PCR efficiencies. For quantification, the tested template was cDNAs from 7, 14, and 21 dpi galls. The reference template was cDNAs from uninfected nonmeristematic root fragments. PCR was conducted in duplicate in the presence of 1 ng of cDNA, 1.2 μL of each primer 2.5 μM, 5 μL of Sybr GREEN master mix, and distilled water to a final volume of 10 μL. PCR conditions were as described above with 10 min at 95°C and 45 cycles at 95°C for 10 s, 55°C for 10 s, and 72°C for 30 s. The results were standardized by comparing the data to reference the endogenous gene UBP22, which remain constant under the different treatment conditions. The quantification of gene expression was performed using the comparative CT method.
Sequence Analysis
The BLAST search program (Altschul et al., 1997) was used for sequence analysis and comparisons in the GenBank, EMBL, and SwissProt databases at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/cgi-bin/BLAST/) and in TAIR (http://www.arabidopsis.org/blast). Multiple sequence alignments and the unrooted neighbor-joining dendrogram were done with ClustalW (Thompson et al., 1994). The significance of the phylogenetic results was assessed by bootstrap analysis. For protein structure prediction, the following servers were used: ARAMEMNON (http://crombec.botanik.uni-koeln.de/aramemnon/) (Schwacke et al., 2003), SignalP version 1.1 (http://www.cbs.dtu.dk/services/SignalP) (Nielsen et al., 1997), and PSort (http://psort.nibb.ac.jp).
Cell Fractionation and Protein Gel Blot Analyses
For protein gel blot analysis, 7-d-old seedlings were homogenized in ice-cold extraction buffer consisting of 50 mM Tris-HCl, pH 7.2, 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, 1% Triton, and protease inhibitor cocktail (Roche Applied Science, Mannheim, Germany). The homogenate was then centrifuged at 2000g for 10 min after which the supernatant was collected into a chilled tube. The supernatant was centrifuged at 100,000g for 1 h to give a soluble fraction and a pellet, the microsomal fraction, dissolved in extraction buffer with Tris-HCl, pH 8.8. Fifty micrograms of protein were analyzed by SDS-PAGE. Extraction method and characterization of the highly purified plasma membrane (PM)–enriched fraction used is described by Marmagne et al. (2004). PM-enriched fraction was purified from microsomes by the two-phase partitioning between polyethylene glycol (upper phase containing plasma membrane vesicles) and dextran (lower phase, named endomembrane fraction, containing all the other membranes) (6.4% [w/w]). The purity of membrane preparations was estimated from enzymatic assays and completed by immunological tests using antibodies against antigens specifically associated with various membrane systems. The sensitivity of the Mg2+-ATPase activity to vanadate and KNO3 was used as a marker for the PM and the tonoplast, respectively. Cytochrome c oxidase activity was used as a marker for mitochondria. Immunological tests were performed with antibodies raised against (1) the plasma membrane H+-ATPase of Nicotiana plumbaginifolia, (2) the E37 protein from the inner envelope membrane of spinach (Spinacia oleracea) chloroplast, (3) a tobacco tonoplast protein, (4) the extrinsic protein Nad 9 of the wheat (Triticum aestivum) mitochondrial inner membrane, and (5) the outer membrane protein TOM40 of yeast mitochondria. Twenty micrograms of proteins were analyzed by SDS-PAGE.
The anti-AtFH6 mouse polyclonal antibody was produced (Agro-Bio, La Ferté St. Aubin, France) using a peptide corresponding to the last C-terminal 62 amino acids, expressed in E. coli with the pBAD TOPO TA expression kit (Invitrogen). Detections were performed using the ECL protein gel blotting system (Amersham Biosciences Europe, Saclay, France). Anti-AFH6 antibody and horseradish peroxidase–labeled anti-mouse IgG antibody were used at dilutions of 1:500 and 1:2000, respectively.
Immunolocalization in Arabidopsis
For immunolocalization, uninfected roots and galls of wild-type Ws and csq2/csq2 plants were fixed on 4% paraformaldehyde (Sigma) freshly prepared in Pipes buffer, pH 6.9. After dehydration and embedding in butyl-methylmethacrylate, immunolocalization was performed as described by de Almeida-Engler et al. (2004). After acetone treatment, sections were incubated in blocking solution (1% BSA in Pipes buffer) and then with the primary antibodies AbAtFH6 or the anti-ACT (ICN, Irvine, CA) diluted 500-fold in blocking solution. After washes, slides were incubated with secondary goat anti-mouse IgG fluorescein isothiocyanate conjugate antibodies (Sigma) diluted 200-fold in blocking solution. Mounted slides were then observed with a Zeiss Axioplan 2 microscope equipped for epifluorescence microscopy, and images were taken with a digital camera (AxioCam; Zeiss).
Transient Expression of Protein Fusions in Arabidopsis Protoplasts
The first exon sequence of AtFH6 was amplified by PCR using Gw5AtFH6-B (5′-aaaaagcaggcttcaccatgaaagctcttcaatccag-3′) and Gw3AtFH6-liter (5′-agaaagctgggtgccgtttctgaggaggtggagg-3′) primers and inserted into the pDONR207 donor vector using Gateway technology (Invitrogen). The C-terminal eGFP fusion was performed into the Gateway expression vector pK7FWG2 (Karimi et al., 2002). The tonoplast marker AtNRAMP3:GFP was kindly provided by S. Thomine. The fusion between the plasma membrane marker P31 and the GFP was described by Marmagne et al. (2004). The AtNRAMP3, the P31, and the AtFH6:GFP fusion constructs were transiently expressed in protoplasts from Arabidopsis cell suspensions by polyethylene glycol–mediated transformation as described by Thomine et al. (2003).
Yeast Transformation and Actin Staining
Thermosensitive strain bni1Δ bnr1Δ (HIY11; Mata, ura3, leu2, trp1, his3, ade2, bni1::HIS3, bnr1::TRP1) and wild-type strain K699 (Mata, ura3-52, leu2-3, -112, trp1-1, ade2-1, can1-100, his3-11, 15, ssd1-Δ2, GAL) were grown on YPD. AtFH6 cDNA was cloned behind the galactose-inductible promoter of the pYES2 plasmid (Invitrogen) using gap repair. The AtFH6 cDNA was amplified using primers GAP5′ (5′-gagaaaaaaccccggatcggactactagcagctgtaatacgactcactatagggaatattatggaagctcttcaatccag-3′) and GAP3′ (5′-gcggatgtggggggagggcgtgaatgtaagcgtgacataactaattacatgatgcggccctcacgtagaagagttgctgc-3′) containing 60 bp of sequence of pYES2 (underlined) for recombination in yeast. Yeast cells were transformed with 1 μg of HindIII/XbaI-linearized pYES2 and 1 μg of PCR-amplified AtFH6 cDNA, as described by Elble (1992), and plated on complete media lacking uracil and containing glucose (1%). For AtFH6 expression, transformants were transferred into the media containing 1% galactose and 1% fructose. The transformants were grown at 25°C and then spotted in 10-fold serial dilutions onto glucose and galactose-fructose media. Plates were then incubated for 5 d at the permissive (25°C) or restrictive (35°C) temperature for suppression analysis.
For actin staining, cells were grown overnight at 25°C in liquid medium containing 1% galactose/1% fructose, diluted to OD600 0.3, and transferred to 35°C for 3 h (sufficient for a doubling). Cells were fixed 1 h with 3.7% formaldehyde (Sigma) and washed twice with PBS. Cells were resuspended in 40 μL of 3.3 μM Alexa568-conjugated phalloidin (Molecular Probes, Eugene, OR) in PBS and incubated in the dark for 1 h. After two washes in PBS, actin cytoskeleton was visualized using a Deltavision (Applied Precision, Issaquah, WA) deconvolution microscopy system on an Olympus IX-70 microscope (Tokyo) with a 60× NA 1.4 objective. Optical Z-sections (0.1 μm) were taken through the cells. Images were deconvolved using softWoRX, and maximum intensity projections of Z-stacks were performed.
The GenBank accession numbers or TAIR locus identifiers for the sequences mentioned in this article are AY337456 (AtFH6), At3g25500 (AtFH1), At3g07540 (AtFH10), At5g62740 (P31), At5g10790 (UBP22), BAB86073 (rice AHF1), AF213695 (NFH1), AF213695 (NFH2), X62681 (chicken FMN), P48608 (Drosophila DIA), U34258 (CAPPUCCINO), P41832 (BNI1p), P40450 (BNR1p), L37838 (FUS1p), O94532 (FOR3p), P78621 (SEPA), AAM15566 (CYK1), XP109355 (mouse DIA1), and O60610 (human DIA1).
Acknowledgments
We thank Yoshimi Takai (Department of Molecular Biology and Biochemistry, Osaka University Graduate School/Faculty of Medicine, Japan) for the generous gift of bni1Δ bnr1Δ yeast mutant and Steven Wasserman (Department of Molecular Biology and Oncology, Dallas, TX) for the Drosophila dia1 and dia2 mutants. We would like to thank Frédéric Gaymard for providing us with Arabidopsis cells, Thomas Kroj and Dominique Thomas for helpful discussions, Xavier Sarda (Biogemma) for assistance with quantitative RT-PCR, and Philippe Castagnone for the phylogenetic analysis. We are grateful to Sébastien Thomine for the AtNRAMP3:GFP fusion plasmid and to Mansour Karimi (Plant Systems Biology, VIB University of Gent, Belgium) for the pK7FWG2 vector. We thank Institut National de la Recherche Agronomique Versailles for providing Arabidopsis T-DNA tagged lines. This work was supported by the French National Institute for Agronomic Research (Institut National de la Recherche Agronomique) and GENOPLANTE contracts AF1999046 and AF2001032. F.J. was supported by a fellowship of the Ministère de la Recherche et l'Enseignement Supérieure. R.A.A. was supported by La Ligue Contre Le Cancer. The deconvolution microscopy system was funded by the Fondation pour la Recherche Médicale.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.024372.
References
- Alberts, A.S. (2001). Identification of a carboxyl-terminal Diaphanous-related formin homology protein autoregulatory domain. J. Biol. Chem. 276, 2824–2830. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaad, F.F. (2001). Plant cytokinesis: Exploring the links. Plant Physiol. 126, 509–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banno, H., and Chua, N.-H. (2000). Characterization of the Arabidopsis Formin-like protein AFH1 and its interacting protein. Plant Cell Physiol. 41, 617–626. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., Elis, J., and Pelletier, G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana. C.R. Acad. Sci. Paris 316, 1194–1199. [Google Scholar]
- Bouchez, D., Vittorioso, P., Courtial, B., and Camilleri, C. (1996). Kanamycin rescue, a simple technique for the recovery of T-DNA flanking sequences. Plant Mol. Biol. Rep. 14, 115–123. [Google Scholar]
- Castrillon, D.H., and Wasserman, S.A. (1994). Diaphanous is required for cytokinesis in Drosophila and shares domains of similarity with the products of the limb deformity gene. Development 120, 3367–3377. [DOI] [PubMed] [Google Scholar]
- Chan, D.C., Bedford, M.T., and Leder, P. (1996). Formin binding proteins bear WWP/WW domains that bind proline-rich peptides and functionally resemble SH3 domains. EMBO J. 15, 1045–1054. [PMC free article] [PubMed] [Google Scholar]
- Chang, F., Drubin, D., and Nurse, P. (1997). Cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J. Cell Biol. 137, 169–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, A.Y., and Wu, H.-M. (2004). Overexpression of an Arabidopsis formin stimulates supernumerary actin cable formation from pollen tube cell membrane. Plant Cell 16, 257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvrckova, F. (2000). Are plant formins integral membrane proteins? Genome Biol. 1, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida-Engler, J., De Vleesschauwer, V., Burssens, S., Celenza, J.L., Inzé, D., Van Montagu, M., Engler, G., and Gheysen, G. (1999). Molecular markers and cell cycle inhibitors show the importance of the cell cycle progression in nematode-induced galls and syncytia. Plant Cell 11, 793–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida-Engler, J., Van Poucke, K., Karimi, M., De Groodt, R., Gheysen, G., Engler, G., and Gheysen, G. (2004). Dynamic cytoskeleton rearrangements in giant cells and syncytia of nematode-infected roots. Plant Cell 38, 12–26. [DOI] [PubMed] [Google Scholar]
- Deeks, M.J., Hussey, P.J., and Davies, B. (2002). Formins: Intermediates in signal-transduction cascades that affect cytoskeletal reorganization. Trends Plant Sci. 7, 492–498. [DOI] [PubMed] [Google Scholar]
- Didry, D., Carlier, M.F., and Pantaloni, D. (1998). Synergy between actin depolymerizing factor/cofilin and profilin in increasing actin filament turnover. J. Biol. Chem. 273, 25602–25611. [DOI] [PubMed] [Google Scholar]
- Doyle, J.J., and Doyle, D.J. (1990). Isolation of plant DNA from fresh tissue. Focus 12, 13–15. [Google Scholar]
- Elble, R. (1992). A simple and efficient procedure for transformation of yeasts. Biotechniques 13, 18–20. [PubMed] [Google Scholar]
- Emmons, S., Phan, H., Calley, J., Chen, W., James, B., and Mansea, L. (1995). cappuccino, a Drosophila maternal effect gene required for polarity of the egg and embryo, is related to the vertebrate limb deformity locus. Genes Dev. 9, 2482–2494. [DOI] [PubMed] [Google Scholar]
- Evangelista, M., Blundell, K., Longtime, M.S., Chow, C.J., Adames, N., Pringle, J.R., Peter, M., and Boone, C. (1997). Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science 276, 118–122. [DOI] [PubMed] [Google Scholar]
- Evangelista, M., Pruyne, D., Amberg, D.C., Boone, C., and Bretscher, A. (2002). Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat. Cell Biol. 4, 260–269. [DOI] [PubMed] [Google Scholar]
- Favery, B., Complainville, A., Vinardell, J.M., Lecomte, P., Vaubert, D., Mergaert, P., Kondorosi, A., Kondorosi, E., Crespi, M., and Abad, P. (2002). The endosymbiosis-induced genes ENOD40 and CCS52a are involved in endoparasitic-nematode interaction in Medicago truncatula. Mol. Plant-Microbe Interact. 15, 1008–1013. [DOI] [PubMed] [Google Scholar]
- Favery, B., Lecomte, P., Gil, N., Bechtold, N., Bouchez, D., Dalmasso, D., and Abad, P. (1998). RPE, a plant gene involved in early developmental steps of nematode feeding cells. EMBO J. 17, 6799–6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, M.J., and Smith, L.G. (2002). A small, novel protein highly conserved in plants and animals promotes the polarized growth and division of maize leaf epidermal cells. Curr. Biol. 12, 849–853. [DOI] [PubMed] [Google Scholar]
- Frazier, J.A., and Field, C.M. (1997). Actin cytoskeleton: Are FH proteins local organizers? Curr. Biol. 7, R414–R417. [DOI] [PubMed] [Google Scholar]
- Gheysen, G., and Fenoll, C. (2002). Gene expression in nematode feeding sites. Annu. Rev. Phytopathol. 40, 191–219. [DOI] [PubMed] [Google Scholar]
- Gilliland, L.U., Pawloski, L.C., Kandasamy, M.K., and Meagher, R.B. (2003). Arabidopsis actin gene ACT7 plays an essential role in germination and root growth. Plant J. 33, 319–328. [DOI] [PubMed] [Google Scholar]
- Goellner, M., Wang, X., and Davis, E.L. (2001). Endo-beta-1,4-glucanase expression in compatible plant-nematode interactions. Plant Cell 13, 2241–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler, P.K., Vidali, L., and Cheung, A.Y. (2001). Polarized cell growth in higher plants. Annu. Rev. Cell Dev. Biol. 17, 159–187. [DOI] [PubMed] [Google Scholar]
- Huang, C.S. (1985). Anatomy and physiology of giant cells induced by root-knot nematodes. In An Advanced Treatrise on Meloidogyne, J.N. Sasser and C.C. Carter, eds (Raleigh, NC: North Carolina State University Graphics), pp. 155–164.
- Imamura, H., Tanaka, K., Hihara, T., Umikawa, M., Kamei, T., Takahashi, K., Sasaki, T., and Takai, Y. (1997). Bni1p and Bnr1p: Downstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae. EMBO J. 16, 745–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, M.G.K. (1981). The development and function of plant cells modified by endoparasitic nematodes. In Plant Parasitic Nematodes, B.M. Zuckerman and R.A. Rhode, eds (New York: Academic Press), pp. 225–279.
- Jones, M.G.K., and Payne, H.L. (1978). Early stages of nematode-induced giant cell formation in roots of Impatiens balsamina. J. Nematol. 10, 70–84. [PMC free article] [PubMed] [Google Scholar]
- Karimi, M., Inze, D., and Depicker, A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195. [DOI] [PubMed] [Google Scholar]
- Kitayama, C., and Uyeda, T.Q.P. (2003). ForC, a novel type of formin family protein lacking an FH1 domain, is involved in multicellular development in Dictyostelium discoideum. J. Cell Sci. 116, 711–723. [DOI] [PubMed] [Google Scholar]
- Kobayashi, I., and Kobayashi, Y. (2000). Control of the response to biotic stresses. In Plant Microtubules: Potential for Biotechnology, P. Nick, ed (Berlin: Springer-Verlag), pp. 83–101.
- Kohno, H., Tanaka, K., Mino, A., Umikawa, M., Imamura, H., Fujiwara, T., Fujita, Y., Hotta, K., Qadota, H., Watanabe, G., Ohya, Y., and Takai, Y. (1996). Bni1p implicated in cytoskeletal control is a putative target of Rho1p small GFP binding protein in Saccharomyces cerevisiae. EMBO J. 15, 6060–6068. [PMC free article] [PubMed] [Google Scholar]
- Lee, L., Klee, S.K., Evangelista, M., Boone, C., and Pellman, D. (1999). Control of mitotic spindle position by the Saccharomyces cerevisiae formin Bni1p. J. Cell Biol. 144, 947–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, E.D., Lee, M.K., Morrow, J.E., Welcsh, P.L., Leon, P.E., and King, M.-C. (1997). Nonsyndromic deafness DFNA1 associated with mutation of a human homolog of the Drosophila gene diaphanous. Science 278, 1315–1318. [PubMed] [Google Scholar]
- Marmagne, A., Rouet, M.-A., Ferro, M., Rolland, N., Alcon, C., Joyard, J., Garin, J., Barbier-Brygoo, H., and Ephritikhine, G. (2004). Identification of new intrinsic proteins in Arabidopsis plasma membrane proteome. Mol. Cell. Proteomics 3, 675–691. [DOI] [PubMed] [Google Scholar]
- Mathur, J., Spielhofer, P., Kost, B., and Chua, N.-H. (1999). The actin cytoskeleton is required to elaborate and maintain spatial patterning during trichome cell morphogenesis in Arabidopsis thaliana. Development 126, 5559–5568. [DOI] [PubMed] [Google Scholar]
- Nielsen, H., Engelbrecht, J., Brunak, S., and Von Heijne, G. (1997). Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10, 1–6. [DOI] [PubMed] [Google Scholar]
- Oppenheimer, D.G., Pollock, M.A., Vacik, J., Szymanski, D.B., Ericson, B., Feldmann, K., and Marks, D. (1997). Essential role of a kinesin-like protein in Arabidopsis trichome morphogenesis. Proc. Natl. Acad. Sci. USA 94, 6261–6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki-Kuroda, K., Yamamoto, Y., Nohara, H., Kinoshita, M., Fujiwara, T., Irie, K., and Takai, Y. (2001). Dynamic localization and function of Bni1p at the sites of directed growth in Saccharomyces cerevisiae. Mol. Cell. Biol. 21, 827–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo, A.F., Cook, T.A., Alberts, A.S., and Gundersen, G.G. (2001). mDia mediates Rho-regulated formation and orientation of stable microtubules. Nat. Cell Biol. 3, 723–729. [DOI] [PubMed] [Google Scholar]
- Petersen, J., Nielsen, O., Egel, R., and Hagan, I.M. (1998). FH3, a domain found in formins, targets the fission yeast formin Fus1 to the projection tip during conjugation. J. Cell Biol. 141, 1217–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pring, M., Evangelista, M., Boone, C., Yang, C., and Zigmond, S.H. (2003). Mechanism of formin-induced nucleation of actin filaments. Biochemistry 42, 486–496. [DOI] [PubMed] [Google Scholar]
- Pruyne, D., Evangelista, M., Yang, C., Bi, E., Zigmond, S., Bretscher, A., and Boone, C. (2002). Role of formins in actin assembly: Nucleation and barbed end association. Science 297, 612–615. [DOI] [PubMed] [Google Scholar]
- Qiu, J.L., Jilk, R., Marks, M.D., and Szymanski, D.B. (2002). The Arabidopsis SPIKE1 gene is required for normal cell shape control and tissue development. Plant Cell 14, 101–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran, S., Christensen, H.E.M., Ishimaru, Y., Dong, C.-H., Chao-Ming, Y., Cleary, A.L., and Chua, N.-H. (2000). Profilin plays a role in cell elongation, cell shape, maintenance, and flowering in Arabidopsis. Plant Physiol. 124, 1637–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagot, I., Klee, S.K., and Pellman, D. (2002. a). Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nat. Cell Biol. 4, 42–50. [DOI] [PubMed] [Google Scholar]
- Sagot, I., Rodal, A.A., Moseley, J., Goode, B.L., and Pellman, D. (2002. b). An actin nucleation mechanism mediated by Bni1 and profilin. Nat. Cell Biol. 4, 626–631. [DOI] [PubMed] [Google Scholar]
- Schott, D., Huffaker, T., and Bretscher, A. (2002). Microfilaments and microtubules: The news from yeast. Curr. Opin. Microbiol. 5, 564–574. [DOI] [PubMed] [Google Scholar]
- Schwacke, R., Schneider, A., Van Der Graaff, E., Fischer, K., Catoni, E., Desimone, M., Frommer, W.B., Flugge, U.I., and Kunze, R. (2003). ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 131, 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijmons, P.C., Grundler, F.M.W., von Mende, N., Burrows, P.R., and Wyss, U. (1991). Arabidopsis thaliana as a new model host for plant-parasitic nematodes. Plant J. 1, 245–254. [Google Scholar]
- Söllner, R., Glässer, G., Wanner, G., Somerville, C.R., Jürgens, G., and Assaad, F.F. (2002). Cytokinesis-defective mutants of Arabidopsis. Plant Physiol. 129, 678–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto, K., Williamson, R.E., and Wasteney, G.O. (2000). New techniques enable comparative analysis of microtubule orientation, wall texture, and growth rate in intact roots of Arabidopsis. Plant Physiol. 124, 1493–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, K. (2000). Formin family proteins in cytoskeletal control. Biochem. Biophys. Res. Commun. 267, 479–481. [DOI] [PubMed] [Google Scholar]
- Tanaka, K., and Takai, Y. (1998). Control of reorganization of the actin cytoskeleton by Rho family small GTP-binding proteins in yeast. Curr. Opin. Cell Biol. 10, 112–116. [DOI] [PubMed] [Google Scholar]
- Thomine, S., Lelievre, F., Debarbieux, E., Schroeder, J.I., and Barbier-Brygoo, H. (2003). AtNRAMP3, a multispecific vacuolar metal transporter involved in plant responses to iron deficiency. Plant J. 34, 685–695. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumpp, A., Blundell, P.A., de la Pompa, J.L., and Zeller, R. (1992). The chicken limb deformity gene encodes nuclear proteins expressed in specific cell types during morphogenesis. Genes Dev. 6, 14–28. [DOI] [PubMed] [Google Scholar]
- Wasserman, S.A. (1998). FH proteins as cytoskeleton organizers. Trends Cell Biol. 8, 111–115. [DOI] [PubMed] [Google Scholar]
- Wasteney, G.O., and Galway, M.E. (2003). Remodeling the cytoskeleton for growth and form: An overview with some new views. Annu. Rev. Plant Biol. 54, 691–722. [DOI] [PubMed] [Google Scholar]
- Watanabe, N., Madaule, P., Reid, T., Ishizaki, T., Watanabe, G., Kakizuka, A., Saito, Y., Nakao, K., Jockusch, B.M., and Narumiya, S. (1997). p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J. 16, 3044–3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittington, A.T., Vugrek, O., Wei, K.J., Hasenbein, N.G., Sugimoto, K., Rashbrooke, M.C., and Wasteneys, G.O. (2001). MOR1 is essential for organizing cortical microtubules in plants. Nature 411, 610–613. [DOI] [PubMed] [Google Scholar]
- Wiggers, R.J., Starr, J.L., and Price, H.J. (1990). DNA content and variation in chromosome number in plant cells affected by Meloidogyne incognita and M. arenaria. Phytopathology 80, 1391–1395. [Google Scholar]
- Woychik, R.P., Maas, R.L., Zeller, R., Vogt, T.F., and Leder, P. (1990). ‘Formins’: Proteins deduced from the alternative transcripts of the limb deformity gene. Nature 346, 850–853. [DOI] [PubMed] [Google Scholar]