Abstract
Cereal starch production forms the basis of subsistence for much of the world's human and domesticated animal populations. Starch concentration and composition in the maize (Zea mays ssp mays) kernel are complex traits controlled by many genes. In this study, an association approach was used to evaluate six maize candidate genes involved in kernel starch biosynthesis: amylose extender1 (ae1), brittle endosperm2 (bt2), shrunken1 (sh1), sh2, sugary1, and waxy1. Major kernel composition traits, such as protein, oil, and starch concentration, were assessed as well as important starch composition quality traits, including pasting properties and amylose levels. Overall, bt2, sh1, and sh2 showed significant associations for kernel composition traits, whereas ae1 and sh2 showed significant associations for starch pasting properties. ae1 and sh1 both associated with amylose levels. Additionally, haplotype analysis of sh2 suggested this gene is involved in starch viscosity properties and amylose content. Despite starch concentration being only moderately heritable for this particular panel of diverse maize inbreds, high resolution was achieved when evaluating these starch candidate genes, and diverse alleles for breeding and further molecular analysis were identified.
INTRODUCTION
As a result of increased demands on food production from escalating population growth and environmental degradation, interest in improved breeding strategies for agricultural crops is growing. Progress in cereal starch production is especially important because these starches comprise 55 to 75% of daily human food intake and are the main source of food for domestic animals (Pan, 2000). In addition to being the largest production crop in the world (http://apps.fao.org), maize (Zea mays ssp mays) has numerous starch mutants that provide a unique source of specialty starches, including amylose-free waxy, important for many industrial applications (Lambert, 2001), and the sugary mutants, responsible for the production of popular sweet maize varieties (Pan, 2000). Interest is also growing in the use of maize as fuel, marked by recent breeding attempts to enhance efficiency in fermentation to ethanol (Dien et al., 2002). Thus, identifying those genes and alleles in maize that control traits such as grain yield, starch concentration, and starch quality is an important step in meeting the future goals of both agriculture and industry.
Research on well-known mutants of maize has helped elucidate key genes involved in the starch pathway. Sucrose transported into the maize kernel is converted to UDP-glucose and fructose by the major isoform of sucrose synthase, encoded by the shrunken1 (sh1) gene (Chourey and Nelson, 1976). sh2 and brittle endosperm2 (bt2) encode the large and small subunits, respectively, of ADP-glucose pyrophosphorylase (AGPase), which converts ADP-glucose into glucose-1-phosphate, the substrate for starch synthases (Tsai and Nelson, 1966; Bae et al., 1990; Bhave et al., 1990). Generally regarded as the rate-limiting step in starch biosynthesis, AGPase is allosterically regulated by 3-phosphoglycerate and Pi and thus is a target for controlling starch yield through the modification of its allosteric effector sites (Stark et al., 1992).
Starch synthases then sequentially add glucose-1-phosphate molecules onto the nonreducing ends of a growing starch chain. Granule-bound starch synthase, encoded by the waxy1 (wx1) locus in maize, is solely responsible for amylose production (Nelson and Rines, 1962; Shure et al., 1983). Amylose-free starches caused by the wx1 mutant in maize have long been of commercial interest (Deatherage et al., 1954). Mutants of a second gene, amylose extender1 (ae1), result in maize kernels with higher amounts of amylose than nonmutant kernels (Fisher et al., 1996; Kim et al., 1998). The ae1 gene codes for the starch branching enzyme IIb isoform, which hydrolyzes α(1→4) linkages and reattaches these chains with α(1→6) branch points found in amylopectin. sugary1 (su1) encodes a debranching enzyme of the isoamylase type. Mutant su1 kernels contain the highly branched, water-soluble phytoglycogen and constituted the original sweet corns (James et al., 1995). To obtain the semicrystalline formation of amylopectin, it may be that the correct ratio of starch branching to debranching enzymes is important (Ball et al., 1996), but the role of isoamylase in conjunction with branching enzymes has not yet been resolved, and differing models have been proposed (reviewed in Smith, 2001).
Although scientists understand the basic structure of the molecule itself, there is still much to be learned about starch metabolism and the organization of the starch granule. Maize starch is composed of 21% amylose, a mostly linear chain of α(1→4) linked glucose molecules, and 79% amylopectin, a more highly branched molecule of α(1→4) linkages with α(1→6) branch points. Such high amylopectin content accommodates the long-term storage of starch in the kernel endosperm, where its semicrystalline nature allows for efficient packaging into granules. Attempts at producing amylopectin in vitro, however, have been unsuccessful, producing instead an animal-like glycogen product (Guan et al., 1995). In addition to the exact nature of amylopectin formation, several enzymes involved in starch biosynthesis are also poorly understood because many of these enzymes have multiple isoforms (Fisher et al., 1996; Gao et al., 1996; Huang and Wang, 1998; Sidebottom et al., 1998; Beckles et al., 2001).
Although geneticists and biochemists have identified many relevant genes, the ability to modify starch products for future gain will depend on an increased level of understanding regarding specific alleles that modify starch concentration and composition. Linkage mapping has identified several regions in the maize genome that have an effect on kernel starch concentration, with some of these quantitative trait loci (QTL) corresponding to starch biosynthesis genes. One of the most documented genes, sh2, colocalizes with a QTL effect on protein and starch levels (Goldman et al., 1993) and correlates with amylose levels (Prioul et al., 1999; Séne et al., 2000). Another positional candidate gene, sh1, has been linked to starch and protein concentration (Berke and Rocheford, 1995). Based on mutational studies, bt2, ae1, su1, sh1, sh2, and wx1 are functional candidate genes thought to affect either starch concentration or starch composition quality. Mutant kernels for all six genes (individually) contain severely reduced starch concentrations or altered amylose/amylopectin ratios. As such, all six genes provide good starting points in the quest for functional polymorphisms that directly affect starch metabolism in maize.
Although the aforementioned QTL studies are initially suggestive, their resolution is often on the order of 10 centimorgans (cM) or more. Because this distance can correspond to millions of bases in maize, real evaluation of individual genes has not occurred. By contrast, association mapping can provide a high-resolution alternative for the evaluation of these candidate genes and has the potential to evaluate a wide range of alleles (Buckler and Thornsberry, 2002; Flint-Garcia et al., 2003). Though common in human genetics (Lander and Schork, 1994; Risch and Merikangas, 1996), association approaches have only recently been applied to plant populations (Flint-Garcia et al., 2003). The comparatively high resolution provided by association mapping is dependent upon the amount of linkage disequilibrium (LD), or the nonrandom association of alleles, present in a species. Two LD studies in maize for both diverse inbreds as well as traditional landraces suggest that in most cases LD decays rapidly within genes, usually within 2000 bp (Remington et al., 2001; Tenaillon et al., 2001). Therefore, high-resolution association mapping is possible in maize. This resolution can be reduced when genes have been recent targets of selection, as has been the case for several genes in the starch pathway (Whitt et al., 2002) and kernel carotenoids (Palaisa et al., 2003).
One difficulty, however, in applying association methods is that LD can be present as a result of genetic drift, selection, or population admixture. Thus, as sometimes seen in human populations, LD can contain the confounding effect of population substructure, resulting in a high frequency of false positive associations (Lander and Schork, 1994). To control for this type of structure, Pritchard et al. (2000a) developed a statistical approach that assigns membership to various subpopulations by determining the amount of genotypic correlation based on unlinked, random markers. Thornsberry et al. (2001) adapted Pritchard's approach for use with quantitative variation and then successfully applied it to the evaluation of maize flowering time. By including estimates of population structure in this analysis, the risk of obtaining false positive associations was reduced (Thornsberry et al., 2001).
Interestingly, these population structure estimation procedures—initially designed for outbred populations—are useful for evaluating these diverse maize lines for several reasons (Remington et al., 2001; Thornsberry et al., 2001; Liu et al., 2003). First, most of these inbreds are unrelated to one another because they are essentially derived from extremely outbred landraces or from synthetic populations. Closely related pairs of lines were also eliminated when first choosing a diverse maize panel for this study. Second, many of the breeding crosses were essentially random. Finally, the actual inbreeding process is not modeled or relevant to these population structure estimates because the genotypes are treated as haplotypes for analysis (Falush et al., 2003).
In this study, six maize candidate genes involved in kernel starch biosynthesis (ae1, bt2, sh1, sh2, su1, and wx1) were tested for associations with starch concentration and starch composition quality using the structured association method of Thornsberry et al. (2001). Each gene was sequenced in a diverse set of maize inbreds, a germplasm that captured 80 to 90% of the microsatellite diversity found in maize landraces (Liu et al., 2003). The use of these maize inbreds reduced the analysis to essentially one haplotype per line, allowing for examination of additive effects only. By locating those allelic regions associated with either starch concentration or composition, polymorphisms identified in this survey can be used in future genetic and breeding studies to manipulate these important agronomic traits.
RESULTS
Principal Component Analysis
Phenotypic trait values for kernel composition (near infrared [NIR]) and starch pasting traits from summer (Clayton, NC 2001; summerC) and winter (Homestead, FL 1998; winterH) replications were analyzed separately using principal component analysis (PCA) on the covariance matrix of traits. PCA takes complex correlated data arranged in multidimensional space and reduces the high dimensionality of the data into more simple, linearized axes while retaining as much of the original variation as possible. All correlated components of sample data will form a correlation matrix, where the variances of the transformed, standardized data along an axis (eigenvectors) are the principal components. Such axes correspond to the largest eigenvalues in the direction of the largest variation of the data. PCA was used in this study to reduce multiple testing in the association analyses by summarizing the phenotypes over the various replications and by combining correlated traits into single PCA indexes. PCA is appropriate for these kernel data, where protein, oil, and starch compositions in cereals are correlated traits (Dudley and Lambert, 1992, 2004).
PCA results for kernel composition along with subjective interpretations of eigenvectors for each factor are included in Table 1. Cumulatively, three factors explained 55% of the variation in phenotypes, where factor one alone explained 34%. Table 1 also shows the results of PCA for starch pasting values of the winterH and summerC field seasons. Cumulatively, three factors explained 91 and 94% of the variance in pasting and viscosity traits seen in maize kernels from the winterH and summerC environments, respectively.
Table 1.
Results of Kernel Composition and Each Starch Pasting PCA
| Principal Component Factor | Proportion of Variance | Cumulative Variance | Interpretationa |
|---|---|---|---|
| NIR-1 | 0.34 | 0.34 | Oil and protein accumulation versus starch |
| NIR-2 | 0.13 | 0.47 | Oil production versus protein production |
| NIR-3 | 0.08 | 0.55 | Genotype × environment effects |
| Starch Pasting-1b | 0.63/0.61 | 0.63/0.61 | All viscosity traits and consistencyc |
| Starch Pasting-2 | 0.20/0.23 | 0.83/0.84 | Peak temperatured and peak timee |
| Starch Pasting-3 | 0.08/0.10 | 0.91/0.94 | Pasting temperature |
Interpretation of the trait(s) driving each principal component were determined by summing the absolute values of PCA eigenvectors (standardized values of the PCA covariance matrix) over all environments for each phenotypic trait. The greater the absolute value for a trait, the more weight that trait was given in the amount of its contribution to driving the variance seen for a particular PCA factor.
All starch pasting variance values are reported as winterH/summerC.
Gel consistency: cool paste viscosity minus hot paste viscosity (Pa·s).
Peak time: time required to reach peak (s).
Peak temperature: temperature of peak viscosity (°C).
Extent of LD in Starch Genes
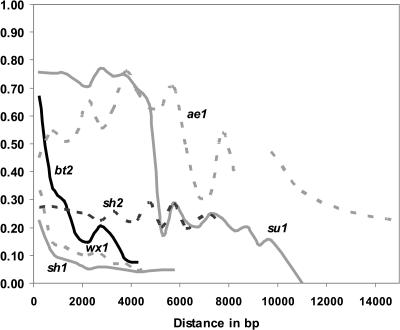
Levels of LD decay varied across the six starch genes, as also seen in the Remington et al. (2001) study. Whereas LD declines rapidly for many genes in this diverse inbred panel (<2000 bp), three of the six starch genes studied here (ae1, sh2, and su1) had substantial LD present for 6000 to 20,000 bp (Figure 1). Thus, in these three cases, the resolution of association mapping is more likely to approach the gene level, rather than subgene resolution. All three genes with extensive LD have been recent targets of selection (Whitt et al., 2002).
Figure 1.
LD Plots of Squared Correlations of Allele Frequencies (r2) against Distance between Polymorphic Sites for the Six Candidate Genes.
The lines indicate average r2 for 500-bp windows for polymorphisms with a minimum frequency of 0.10. The ae1 graph is based on 1000-bp windows because there were so few polymorphisms per window.
Associations with Kernel Composition and Starch Pasting Properties
Of the six starch genes sampled, four showed significant associations (P ≤ 0.05) for one or more traits (Table 2). Overall, ae1 associated with pasting temperature (summerC starch pasting factor three) and amylose content. bt2 associated with oil versus protein production (kernel composition factor two). sh1 showed an association with a general genotype × environment (G × E) effect (kernel composition factor three) and with amylose content in the summerC replication. Lastly, sh2 associated with a general G × E effect (kernel composition factor three) and with starch viscosity characteristics (summerC starch pasting factor one). Additionally, haplotype analysis of sh2 indicated an effect on amylose content in the winterH environment.
Table 2.
Results for Overall Gene Association Analyses Using Logistic Regression
| Kernel Composition Factor
|
Starch Pasting Factor, WinterH
|
Starch Pasting Factor, SummerC
|
Amylose Content
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 | WinterH | SummerC |
| ae1 | * | ** | * | ||||||||
| bt2 | ** | ||||||||||
| sh1 | * | ** | * | ** | |||||||
| sh2 | ** | ** | * | ||||||||
| su1 | |||||||||||
| wx1 | |||||||||||
*, ** = P ≤ 0.10 and 0.05, respectively. These are gene-wise P-values that account for multiple testing among sites. Exact P-values can be found in the text. Blank cells indicate P > 0.10.
Associations that were less significant (P ≤ 0.10) were also identified for those genes with high diversity (e.g., sh1, sh2, and wx1) and correspondingly low statistical power, but these will not be examined in detail (Table 2). The false discovery rate was estimated to be ∼50% for associations where P ≤ 0.10. When considering only the better replicated NIR data, then the estimated false discovery rate decreases to 21%. Overall, for associations where P < 0.05, only one or two false positives may exist.
We also empirically tested for false positives by associating the kernel composition NIR data with 10 genes in unrelated pathways (d8, d3, zflA, zhd1, zmLD, fae2, ra1, tb1, zb7, and zb12) using the three principal components for 30 tests. Only one test was significant at the 0.05 level (d8 with PC2), which is slightly less than what is expected from chance. Even in this case, it is likely that the effects of d8 on flowering translate into environmentally induced differences in grain filling. Such differences are a consequence of evaluating such a diverse panel of maize inbreds in any one environment, as these lines vary considerably in terms of photoperiod adaptations because of their native habitats. Thus, if these adaptive differences are taken into account, this empirical test resulted in no false positives, suggesting that most, if not all, of the starch associations identified in this study are a result of true linkage.
In the following detailed gene analyses, specific site(s) with the most significant results from overall gene testing are discussed; however, because of LD, associations with traits should be viewed as associations by haplotype, and not necessarily a specific polymorphism.
ae1
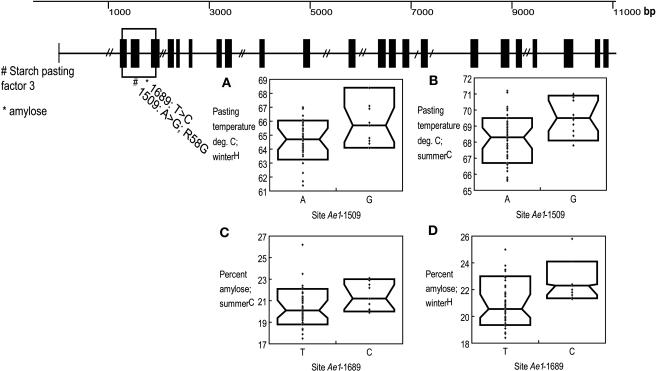
Overall, ae1 significantly associated with pasting temperature (summerC starch pasting factor three; Table 2) (logistic regression; P = 0.02). The most significant site was 1509, an adenosine to guanine transition located in exon two, hereafter referred to as Ae1-1509(G↔A). The Ae1-1509(G) allele and associated haplotype were found in 13 lines out of the total 102, including both nonstiff stalk and semitropical lines (Table 3). Single nucleotide polymorphism (SNP) Ae1-1509(G) caused a nonsynonymous change in the predicted AE1 protein sequence, converting Arg to Gly at amino acid 58 (R58G). The ae1 orthologs in rice (Oryza sativa), wheat (Triticum aestivum), barley (Hordeum vulgare), and potato (Solanum tuberosum) all contain a Gly in the predicted amino acid sequences of the starch branching enzyme, whereas the majority of maize lines sampled in this study contained an Arg residue. The overall effect of ae1 on pasting temperature (starch pasting factor three) was also seen in the winterH replication, although at a marginal level of significance (Table 2). Taken alone, SNP Ae1-1509(G) showed a significant effect on starch pasting temperature in both summer and winter field replications (general linear model [GLM]; P = 0.0002 and 0.03, respectively) (Figure 2). In both replications, the Ae1-1509(G) allele associated with 2.0 and 1.6% higher average pasting temperatures for summerC and winterH, respectively.
Table 3.
Maize Inbreds Surveyed and a Listing of Significant Alleles in the Association Tests
| Maize Inbred | Subpopulation | Allele | Maize Inbred | Subpopulation | Allele |
|---|---|---|---|---|---|
| 38-11 | NSS | K55 | NSS | C | |
| A6a | ST | B | Ki3 | ST | B |
| A272a | ST | B | Ki9a | ST | B |
| A441-5 | NSS | B, F, H | Ki11 | ST | |
| A554 | NSS | Ki21a | ST | D, E | |
| A619 | NSS | C, D, E | Ki43 | ST | B |
| A632 | SS | C | Ki44 | NSS | F, H |
| B14Aa | SS | C | Ky21a | NSS | C |
| B37a | NSS | C | M37W | ST | |
| B68 | SS | M162W | NSS | B | |
| B73a | SS | Mo17a | NSS | B, D | |
| B84 | SS | C, D | Mo24W | NSS | D |
| B97a | NSS | C | MS153 | NSS | |
| B103a | Mixed | C | N192 | SS | C |
| B104 | SS | C | N28Hta | SS | C |
| C103 | NSS | B, F, H | NC250 | NSS | C |
| CI187-2a | NSS | C, D | NC258 | NSS | B |
| CM7 | NSS | B | NC260a | NSS | C |
| CM105 | SS | C | NC296 | ST | B |
| CM174 | SS | C | NC298 | ST | B |
| CML5 | ST | NC300 | ST | B | |
| CML10 | ST | F, H | NC304 | ST | B |
| CML61 | ST | F, H | NC320 | ST | B, C |
| CML91 | ST | C | NC338 | ST | B |
| CML247 | ST | C, E | NC348a | ST | B |
| CML254a | ST | E | NC350 | ST | B |
| CML258a | ST | B | NC352 | ST | B |
| CML261 | ST | C | NC354 | ST | B |
| CML277 | ST | ND246 | NSS | C, E, F, H | |
| CML281 | ST | B | Oh7B | NSS | |
| CML287 | ST | B, C, E | Oh43a | NSS | C, E, G |
| CML333a | ST | C, F, H | P39a | NSS | D |
| CMV3 | NSS | B | Pa91a | NSS | |
| D940Ya | NSS | B | Q6199 | ST | B, E |
| EP1a | NSS | B, E, F, H | SA24 | NSS | B, D |
| F2a | NSS | E | SC55 | ST | B, G |
| F7 | NSS | C, E | SC213 | Mixed | B, C |
| F44 | NSS | E | Sg18 | NSS | |
| F2834T | NSS | C, F, H | T8 | ST | F, G, H |
| GT112 | NSS | C, F, H | T232a | NSS | |
| H95 | NSS | G | Tx601a | ST | C |
| H99 | NSS | C | Tzi8 | NSS | |
| HP301 | NSS | C | Tzi10 | ST | B, E, F, H |
| I-29a | Mixed | B, E, G | Tzi18 | ST | B, E |
| I137TN | NSS | H | U267Y | NSS | B, C |
| I205a | NSS | C | Va26 | NSS | E |
| Ia2132a | NSS | F, H | W64A | NSS | C |
| IDS28a | NSS | D | W117Ht | NSS | D, E |
| IL14H | NSS | C | W153Ra | NSS | E |
| IL101a | NSS | C | W182B | NSS | C, E |
| IL1677a | NSS | C | Wf9 | NSS |
The subpopulation classification is based on Remington et al. (2001). Subpopulations are denoted by the following: NSS, nonstiff stalk; SS, stiff stalk; ST, subtropical/tropical. Allele designations are as follows: B, line containing polymorphism Sh1-1210G; C, line containing polymorphism Sh1-775C; D, line containing polymorphism Sh2-3674-1; E, line containing polymorphism Bt2-925T; F, line containing polymorphism Ae1-1509G; G, line containing polymorphism Sh2-3842G; H, line containing polymorphism Ae1-1689C.
Member of the 32-line subset for which all six genes (except ae1) were sequenced over their entirety.
Figure 2.
Genetic Structure of ae1 Sequenced from 32 Maize Taxa.
The gene area outlined with a box denotes the region sampled in the full set of 102 taxa. Sites significant for starch pasting factor three (#) and amylose (*) are highlighted. Allele Ae1-1509G associated with higher pasting temperatures in both the summerC and winterH replications ([A] and [B]). Allele Ae1-1689 associated with higher amylose in both summerC and winterH replications. Insets show the distribution of the data: the median for the data points is marked by the middle horizontal line; the upper and lower horizontal lines highlight the 10th and 90th percentiles. Figures for genes bt2, sh1, and sh2 are set up in the same fashion. Note that the diagonal lines (//) in the gene picture are intronic areas and exon 15 not sequenced in the initial 32 lines.
When summerC and winterH replications were analyzed simultaneously, ae1 also showed an overall association with amylose content (logistic regression; P = 0.03). The most significant site was 1689, a SNP located in intron two containing a T↔C transition. The polymorphic allele Ae1-1689(C) was in very strong LD with allele Ae1-1509(G) (LD; r2 = 0.9) discussed above. In both summer and winter replications, lines containing the Ae1-1689C allele had 5.1 and 6.5% higher amylose content relative to those lines with the more common allele, Ae1-1689(T) (GLM; P = 0.02 and 0.004, respectively) (Figure 1). Overall, this polymorphism explained 7 and 14% of the total variation in the two seasons, respectively.
bt2
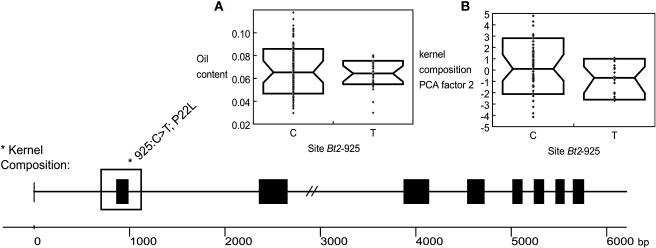
Overall, bt2 associated significantly with kernel composition factor two, which affected oil and protein levels (Table 2; logistic regression; P = 0.05). The most significant site was 925 located in exon one, in which a C↔T transition has occurred (Figure 2). Within the ∼400 bp sampled in the 102 lines, eight other SNPs and one insertion/deletion (indel) occurred in significant LD with site 925, revealing a haplotype. When considering the entire bt2 gene alignment of 32 taxa, this distinct haplotype encompassed ∼1000 bp at the 5′ end of the gene. The SNP at site 925, hereafter referred to as Bt2-925(T), caused a nonsynonymous change in the N-terminal region of the BT2 protein, converting Pro to Leu at amino acid 22 (P22L). Whereas the mean oil content between lines varying for the Bt2-925(T) allele was not significantly different, the variance in oil content in lines with the Bt2-925(T) allele was significantly lower than lines with the more common allele (F test; P < 0.002) (Figure 3). Nineteen lines out of 102 contained polymorphism Bt2-925(T) and were of nonstiff stalk or semitropical origin (Table 3).
Figure 3.
Genetic Structure of bt2 with the Significant Region Associated with Kernel Composition Factor Two for Oil.
The last exon of bt2 (exon nine) was not sequenced. Diagonal lines near position 3000 (//) denote an area not sequenced because of a highly repetitive 250-bp stretch.
sh1
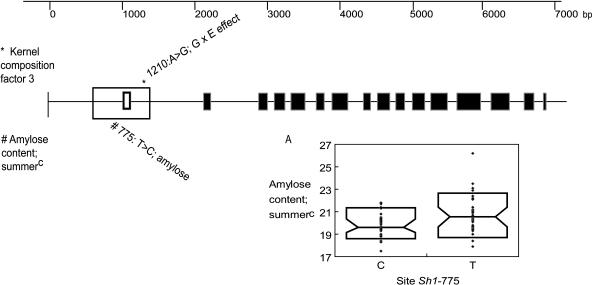
Overall, sh1 associated with a general G × E effect (kernel composition factor three; Table 2) (logistic regression; P = 0.02). PCA weighted all kernel traits similarly for factor three but in opposite directionality across replications, suggesting the G × E effect. The most significant site was SNP Sh1-1210(A↔G) located in intron one (Figure 4). The Sh1-1210(G) allele occurred in 35 out of the 102 lines surveyed, the majority being of tropical origin (Table 3).
Figure 4.
Genetic Structure of sh1 and the Region Significantly Associated with Kernel Composition Factor Three for G × E Effects and for Amylose Content.
The black, unfilled exon denotes noncoding exon one of sh1.
sh1 also associated with amylose content in the summerC replication (logistic regression; P = 0.002) (Table 2). The most significant polymorphism was SNP Sh1-775(T↔C), which is located within the promoter region (Figure 4). Lines with the Sh1-775(C) allele contain 4.6% less amylose relative to the more common allele, Sh1-775(T) (GLM; P = 0.0001). Forty-one lines out of 102 contained the Sh1-775(C) allele, and it explained ∼12% of the total variation. All three subpopulations have representative lines containing allele Sh1-775(C) (Table 3); however, the stiff stalks had the highest frequency of Sh1-775(C) at 75%. The amount of LD between Sh1-775(T↔C) and other polymorphic sites in sh1 was low (LD; r2 < 0.30).
sh2
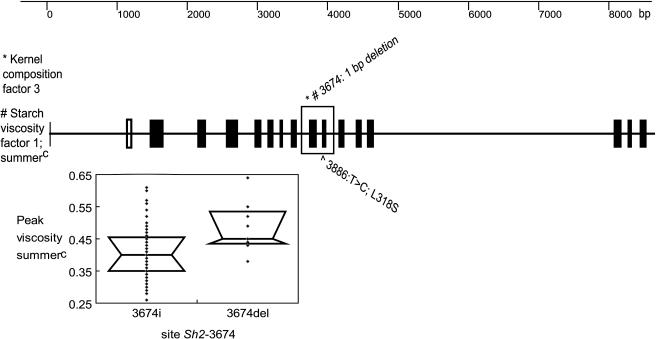
Overall gene analysis for sh2 indicated a significant association with a general G × E effect (kernel composition factor three; Table 2) (logistic regression; P = 0.035). The most significant polymorphism included a 1-bp deletion in intron eight at site 3674, Sh2-3674(INDEL1) (Figure 5). Examination of the sequence alignment for the entire gene from all 32 lines revealed that allele Sh2-3674(del1) is in LD with a suite of other polymorphisms, including multiple SNPs, an 8-bp insertion in the promoter (581 bp away from the noncoding exon one), an 11-bp deletion located in intron 10, and a 67-bp deletion in intron 13 (site 4640), indicating an obvious haplotype. Allele Sh2-3674(del1) occurred in 10 out of the 102 lines surveyed, mainly in nonstiff stalks (Table 3). This same polymorphic site in sh2 also showed an overall association with starch viscosity characteristics (summerC starch pasting factor one; Table 2) (logistic regression; P = 0.05).
Figure 5.
Genetic Structure of sh2 and the Region Significantly Associated with Kernel Composition Factor Three for G × E Effects and with Starch Pasting Factor One for Multiple Viscosity Traits.
Only the trait peak viscosity is shown for space conservation for the allelic distribution of the data.
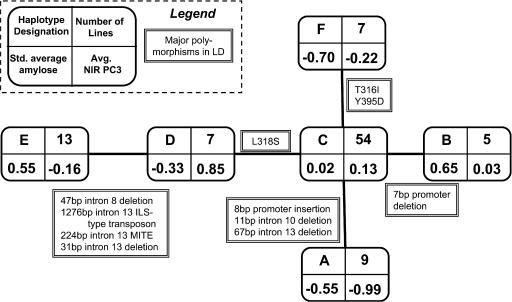
Because sh2 contained several clearly defined polymorphic alleles, haplotype analysis was performed to increase power to detect associations. Six haplotypes based on six polymorphic sites were tested for associations with seven viscosity traits (breakdown, consistency, cool paste viscosity, hot paste viscosity, peak viscosity, setback, and trough viscosity) and with amylose content, using GLM with population structure (Figure 6). sh2 haplotypes showed significance mainly in the summerC replication, where associations were seen with consistency, cool paste viscosity, hot paste viscosity, peak viscosity, and trough viscosity (Table 4). Significant associations were seen with amylose content and starch breakdown in the winterH environment. Furthermore, a comparison of sh2 haplotypes was performed between winterH and summerC replications for mean amylose content and kernel composition factors one through three (Figure 6). Haplotype A containing the Sh2-3674(del1) polymorphism (which associated significantly with both kernel composition and starch pasting traits) had the largest negative weighting with kernel composition factor three (the G × E factor) and one of the largest negative effects on amylose content. A different sh2 haplotype, B, had the greatest (positive) least squares mean in amylose content compared with the other five haplotypes. This B haplotype contains the most significant polymorphism in the overall gene analysis, polymorphism Sh2-3842(G), which was marginally significant for amylose content in winterH (logistic regression; P = 0.095) (Table 2). Significance increased using haplotype analysis, where the strength of an association with amylose content was P = 0.01 (Table 4).
Figure 6.
sh2 Haplotype Network.
Sequencing of a 500-bp region of sh2 revealed six major haplotypes, with haplotype C being the most common with 54 member lines. The least squares mean for amylose content (both winterH and summerC replications) and kernel composition factor three (the G × E factor) are reported for each haplotype, signifying the haplotype's average effect on these traits. Major polymorphisms of interest and in LD occurring outside the analyzed region of sh2 are indicated along the branches where the mutations likely occurred.
Table 4.
Results of sh2 Haplotype Analysis
| Field Replication P-Values
|
||
|---|---|---|
| Trait | WinterH | SummerC |
| Amylose | 0.010 | 0.287 |
| Breakdowna | 0.006 | 0.367 |
| Gel consistencyb | 0.524 | 0.054 |
| Cool paste viscosityc | 0.573 | 0.014 |
| Hot paste viscosityd | 0.480 | 0.008 |
| Peak viscositye | 0.174 | 0.041 |
| Setbackf | 0.556 | 0.065 |
| Trough viscosityg | 0.541 | 0.004 |
| Pasting PC1 | 0.415 | 0.025 |
| Pasting PC2 | 0.185 | 0.680 |
| Pasting PC3 | 0.961 | 0.801 |
Highly weighted pasting traits in PC factor one were tested individually for associations. Significant test results are in boldface.
Breakdown: peak viscosity minus trough viscosity, indication of breakdown in viscosity of paste during 95°C holding period (Pa·s).
Gel consistency: cool paste viscosity minus hot paste viscosity (Pa·s).
Cool paste viscosity: viscosity of paste cooled to 50°C (Pa·s).
Hot paste viscosity: final viscosity after cooking at 95°C (Pa·s).
Peak viscosity: maximum viscosity recorded during heating and holding cycles (Pa·s).
Setback: cool paste viscosity minus trough viscosity, indication of retrogradation of cooked rice during cooling (Pa·s).
Trough viscosity: minimum viscosity after peak (Pa·s).
The Séne et al. (2000) QTL study found a significant effect on amylose content that colocalized with sh2. Their study compared genotypes F2 and Iodent (I205-like). Genotype I205 contains the most common haplotype in the sample, C, whereas genotype F2 contains haplotype D. Although these two haplotypes were significantly different for kernel composition PC3 (P = 0.03), they were not significant for amylose content in summerC. One particular polymorphism of interest found within genotype F2, but not in the I205 haplotype, is a Leu to Ser amino acid substitution at residue 318. Polymorphism L318S is also found in a second haplotype, E, where member lines contained a high percentage of amylose. When haplotypes D and E containing L318S were compared against the Iodent haplotype C, a significant difference in kernel composition PC3 was seen.
DISCUSSION
Six major genes controlling starch content in maize were analyzed using an association approach to identify those haplotypes affecting starch content or quality, which could be used in maize improvement. Mutational studies have already shown that ae1, bt2, sh1, sh2, su1, and wx1 have major effects on either the amount of starch produced or the type, as indicated by different amylose or amylopectin levels.
Phenotypic traits were organized into three major groupings for this analysis: kernel composition, starch pasting properties, and amylose content. Significant kernel composition associations were found in the three starch production genes sh1, sh2, and bt2. Tests for association with either starch pasting characteristics and/or amylose content were significant in sh1, sh2, and ae1.
Resolution and Power Implications for Detecting Associations
Because association approaches rely on the variation produced by evolution for mapping, the evolutionary history of a particular locus affects both genetic resolution and statistical power. The genetic resolution of association approaches is directly dependent upon the structure of LD, and generally the rapid decay in association populations provides high resolution. Prior work using the same diverse maize panel at Dwarf8 revealed rapid LD decay, and associations were not seen in genes within 1 cM (Thornsberry et al., 2001). By contrast, LD at the Y1 locus—with its near perfect penetrance and the target of very strong selection in this last century—can be more extensive (Palaisa et al., 2003). Among the six genes studied here, three exhibit LD decay within 2000 bp, whereas the remainder had slower LD decay (although still within a gene's length). This lack of decay is almost certainly because of diversity reductions caused by selection (Whitt et al., 2002). Ancestral recombination can still be retained after some selection bottlenecks, however, as evidenced by the rapid LD decay of bt2 despite its being a target of selection. Although LD decay varied among the six genes studied here, the extent of LD decay and, thus, the resolution of this association approach still exceeded that of linkage mapping by several orders of magnitude.
In terms of statistical power, association approaches can be limited by several factors. (1) Allele class frequency directly affects the statistical power of all mapping approaches. In a standard linkage population with two inbred founders, alleles have a frequency of 50% (excluding segregation distortion), whereas in an association study, allele frequency can be highly variable. As such, there will always be less statistical power to evaluate rare alleles in an association study. Our findings revealed few associations for those genes with low diversity (ae1, bt2, and su1), which may result from a lack of power to test rare alleles. However, because selection events can skew allele frequencies (with deleterious alleles becoming very rare or extinct), perhaps all functional variation was eliminated from these loci as a result of selection—leaving no associations to detect.
(2) The number of alleles per locus must also be considered when detecting associations because more alleles require more statistical tests per locus. In this study, we performed gene-wise P-value corrections, resulting in a higher stringency per SNP for more diverse loci. Most of the robust associations, however, occurred in sh1 and sh2 (logistic regression; P = 0.002 and 0.02 overall, respectively)—two of the three most diverse genes in this survey. Although greater numbers of polymorphic sites will decrease statistical power, it is encouraging to note the strength of the associations identified here for several diverse genes for traits with modest heritability.
(3) The interactions between diverse alleles across the genome may have limited the statistical power needed to evaluate some functional polymorphisms. Although the extremely diverse nature of the germplasm used in this study no doubt ensured that diverse alleles with large effects were present in the population, differences in days to flower make it likely that only those alleles with consistent effects across the season were detected. Although such diversity in grain-filling times does not cause false positives (as evidenced by our empirical test using genes in unrelated pathways), it may reduce the power needed to detect associations.
To address these issues, statistical power can be improved be increasing sample size. This not only increases the possibility of capturing more instances of less common alleles but also provides avenues for dealing with the confounding effects of epistasis. In this study, if the number of genotypes was increased from 100 to 300 or 400, not only could many rare or weaker alleles be evaluated (Long and Langley, 1999) but lower Type I error rates could also be set, resulting in lower false discovery rates. Increasing the sample size would also permit the inclusion of interaction terms in the models, separate statistical tests for each population, or use of specific polymorphisms with known effects as cofactors. Although increasing sample size is statistically straightforward, empirical researchers must overcome the limitations posed by phenotyping many genotypes in replicated environments.
In addition to increased sample size, testcross evaluations (cross all the diverse lines to a tester line and then evaluate the F1 offspring) may also provide an efficient means of reducing genetic background interactions among the lines. Even with the dilution of additive effects that is expected for such testcrosses, the reduction in epistasis that is also achieved may improve the evaluation of additive effects.
Evaluating QTL with Linkage and Association Analysis
QTL mapping studies for starch traits using recombinant inbred line (RIL) or F2 populations have traditionally identified regions that span 10 to 20 cM, corresponding to as many as 20 million bases. By contrast, this study used association analyses to identify a suite of polymorphisms within a few thousand bases, resulting in a substantial increase in resolution. This high resolution raises the question of whether candidate gene association analysis should replace mapping with RIL or F2 populations. A comparison of this association study with RIL mapping suggests that a combination of both these methods provides the most powerful experimental approach for the following three reasons.
First, there is an important tradeoff between statistical power and resolution for all mapping approaches. In standard linkage populations, the equivalent of 50 independent regions are surveyed, whereas in a high-resolution association panel (as in this study) >50,000 such independent regions must be surveyed. Although this may become feasible on a molecular level in the near future, it would still require the phenotyping of a vast number of lines to scan the genome. Genome scan power estimates for alleles with modest effects may require thousands to tens of thousands of individuals (McGinnis et al., 2002). Phenotyping of distinct genotypes would be limiting.
Second, candidate genes with an established position under QTL peaks exhibited more associations than did those outside such peaks. For example, 7 of 11 associations were found in those two genes (sh1 and sh2) that colocalized with QTL for kernel traits, a result that is significantly more than expected (χ2 test; P = 0.03). This suggests that RIL mapping is an efficient way to eliminate the first 90% of the genome.
Third, although the use of RIL and F2 populations in QTL studies tests a maximum of only two alleles, RIL and F2 populations do have substantial statistical power to contrast these alleles. To illustrate this point, associations found in sh2 in this study were compared with a QTL study done by Séne et al. (2000) that found an effect on amylose content near sh2. Although our study was able to confirm that these two alleles have an environment-dependent effect on kernel quality, we were unable to confirm the specific amylose effects, partially because of genotype by environment interactions. We identified a candidate polymorphism, L318S, as a possible basis for the results of both this and the Séne study. Larger populations and, therefore, more statistical power are probably needed to effectively apply association approaches for those genes with numerous alleles, as seen in wx1.
Overall, RIL populations can powerfully contrast pairs of alleles with low resolution, whereas association analysis provides a high-resolution evaluation of numerous alleles with uneven statistical power. Exploiting the complementary strengths and weaknesses of both approaches should allow efficient QTL evaluation of the genome.
Potential Functional Polymorphisms
Our association analysis was successful in locating regions and haplotypes that affect maize kernel composition. This has suggested several specific biochemical hypotheses that should be tested in the future: (1) a sh1 polymorphism located in intron one associated with a G × E effect—this intron has been related to gene activity (Vasil et al., 1989; Clancy et al., 1994; Clancy and Hannah, 2002). (2) Allele Bt2-925(T), located in bt2's exon one and causing a P22L polymorphism, associated with a decrease in variance in oil content. Genotypes with the Bt2-925(T) allele resulted in reduced variability by cutting out the high and low extremes in oil production. (3) sh2 is the other AGPase gene with significant associations—out of the six sh2 haplotypes examined (Figure 6), two in particular showed interesting effects on pasting traits. Haplotype A, which contains deletion polymorphism Sh2-3674(del1), not only seems to have different effects on kernel composition traits under different environmental conditions, but also may have a negative effect on amylose. Furthermore, the A haplotype also displays the highest means over the other five haplotypes for significant viscosity traits in summerC: consistency, cool paste viscosity, hot paste viscosity, peak viscosity, and setback (data not shown). (4) Variations in ae1, a branching enzyme gene, are likely to have an effect on amylose content and/or pasting properties. One particular polymorphism in exon two, Ae1-1509(G), caused the nonsynonymous change R58G in the predicted protein sequence. The phenotypic effect seen with this mutation was higher pasting temperatures. The R58G mutation occurred within the identified transit peptide and is located near the cleavage site in front of the N terminus of the mature BEII protein (Fisher et al., 1993). Although an increase in amylose content was seen in lines with the Ae1-1689(C) polymorphism, in LD with site 1509, it is unclear what causes an increase in pasting temperature, an important indicator of stability in food processing of starches. Amylose content and pasting temperature did not significantly correlate (data not shown); however, these traits could be related to changes through physicochemical properties of the amylopectin branch lengths, thereby contributing to the stability of the starch under stress.
It should be noted that although the above polymorphisms were the most suggestive functionally, there were in almost all cases other polymorphisms with equal statistical support. Additionally, although large portions of the genes and promoter regions were sequenced for the test associations, we cannot rule out closely linked regions based on the structure of LD at a given locus.
Genotype by Environment Interactions
The multiple occurrences of starch genes that associate with genotype by environment interactions in kernel composition are hardly surprising for a moderately heritable pathway, diverse germplasm, and diverse environments. Of the four field locations where kernels were phenotyped for kernel composition traits, West Lafayette, IN (2000) displayed poorer grain quality because of a stressed environment in comparison with the other environments. G × E associations with kernel composition were seen in both sh1 and sh2.
sh1 associated significantly with a G × E effect at an intronic polymorphism, Sh1-1210 (see above), whereas sh2 associated significantly with a G × E effect at a 1-bp deletion at position 3674; however, this particular polymorphism was in significant LD with a suite of polymorphisms located throughout the entire gene to form a limited number of haplotypes, thus limiting resolution. Therefore, the causative polymorphism may not even be located within the sh2 gene, but reside instead in closely associated flanking regions. This latter association may involve the SH2 subunit of the AGPase enzyme. Evidence for AGPase suggests that alternate alleles produce enzymes that perform differently in a stressed environment, affecting factors such as heat lability or altering SH2:BT2 interactions. Mutations in the SH2 subunit have been shown to increase its stability, thereby increasing SH2:BT2 interactions (Greene and Hannah, 1998).
The G × E nature of sh2 may also be reflected in the results of viscosity associations (Table 4). Haplotype analysis of sh2 allowed for a more powerful examination of the pasting traits driving PC1. In the winterH replication, haplotypes differed significantly in amylose content and starch breakdown, but not in the summerC environment. Conversely, in the summerC environment, almost all remaining viscosity traits thought to drive PC1 (consistency, cool paste viscosity, hot paste viscosity, peak viscosity, and trough viscosity) were significant by sh2 haplotype but were not significant in the winterH replication. Therefore, sh2 haplotypes may be showing G × E effects in viscosity traits as well, but more than two replications are needed to obtain adequate statistical power to detect this. Differing associations between summerC and winterH environments on downstream traits, such as starch pasting, may be influenced by the results we obtained for the G × E nature of sh2 on general kernel composition traits.
One commonality this study shared with the Séne study (Séne et al., 2000) is that the effect of sh2 on amylose was not seen in all environments. Because sh2 showed a significant G × E effect on kernel composition, we propose the hypothesis that with higher ambient temperatures, the heat labile nature of AGPase (Greene and Hannah, 1998), in which sh2 encodes a subunit, varies between sh2 haplotypes and has an epistatic effect on amylose production. When the two haplotypes of F2 and the Iodent I205 were compared, significance with the G × E effect on kernel composition was also seen, further explaining the inconsistent results for amylose between environments.
Population Structure and Association Analysis
Although sh2 has a significant effect on the amylose/amylopectin ratio in certain environments, a previously reported association with overall starch may be a false positive result. Prioul et al. (1999) found an association with a SacI restriction site within the sh2 gene in a sample of 46 unrelated maize inbreds but did not control for population structure. Our analysis also finds a significant association if population structure is ignored because tropical, stiff stalk, and nonstiff stalk germplasm all have different mean starch levels. Although the Prioul association may reflect the G × E nature of sh2, another possibility is that this association with starch is purely a result of population structure. Population structure was significantly related to basic kernel composition (PC1) (GLM; P < 0.02) and accounted for 10% of the variation. Because kernel quality is correlated with population structure, associations performed without a population structure correction need to be reevaluated. The particular story at sh2 may also be complicated by a tightly linked locus to sh2 with effects on starch, as suggested by a recent QTL study based on crosses of the Illinois high and low protein maize strains that not only found a QTL at sh2, but also more strongly at a linked marker (Dudley et al., 2004).
To effectively use association approaches in the study of plant genetics, population structure must be considered to prevent false positive results (Knowler et al., 1988; Pritchard et al., 2000a, 2000b). By controlling for population structure, Thornsberry et al. (2001) were able to locate polymorphisms within the Dwarf8 gene that associated with flowering time variation in maize. In this study, estimates of population structure for the diverse set of inbred lines, as determined in Remington et al. (2001), were incorporated into all analyses. This allowed for the detection of significant associations by increasing power through the use of an unlinked set of markers. In some instances, however, correcting for population structure actually caused several genes to lose significance (data not shown). This loss of significance can have two causes: (1) this was a nonfunctional polymorphism and the association was caused by population structure, or (2) the polymorphism is functionally related but the polymorphism distribution coincides with population structure. The second case results in a functionally false negative result. If this is the case, then the polymorphism needs to be reevaluated in alternative population structures.
Implications
To date, only a handful of QTL have been dissected to the gene level in plants. By building on previous linkage mapping populations, this study used association approaches to identify at least three additional genes with QTL effects. This study has supplied breeders with a set of high-resolution markers for a set of six starch genes. Ultimately, these markers can be used to meet specific starch or yield goals by incorporating desirable alleles into maize germplasm. This study has also provided a wealth of candidate polymorphisms for future analysis by molecular biologists and biochemists to further elucidate this critical plant pathway. As previously acknowledged, the modest sample size perhaps limited power to detect all associations in such a diverse germplasm. By reevaluating these candidate polymorphisms in larger association populations and in testcrosses, additional functional polymorphisms may yet be discovered. As such, this study represents a promising beginning to evaluating functional nucleotide diversity in the maize starch pathway.
METHODS
Plant Materials
The 102 maize inbreds (Zea mays ssp mays) used in this study represent most of the diversity available to breeding programs around the world, retaining at least 80% of the microsatellite diversity found in maize landraces (Liu et al., 2003). These inbreds were divided into three subpopulations based on simple sequence repeat data described by Remington et al. (2001): the stiff stalks, the nonstiff stalks, and the subtropical/tropicals (Table 3). Note that some lines traditionally known as stiff stalk lines (B37, for example) are grouped with the nonstiff stalk lines, and the one white popcorn in the study (I29) is grouped with the tropicals here. The inbreds were grown in one-row plots in a randomized complete block design at four different field sites in the U.S., with a total of six replications: Homestead, FL (winter 1998 to 1999); West Lafayette, IN (summer 2000); Clayton, NC (summer 2001) in two replications; Urbana, IL (summer 2001) in two replications. Ten to fifteen plants were self-pollinated by hand in each row; ears were then harvested at maturity and dried and shelled.
Starch Isolation and Phenotyping
Approximately 10 grams of seed from each inbred was pooled from several ears and ground using an M-2 Stein Mill for 90 s. Kernel starch, oil, protein, and moisture percentage were measured from ∼600 mg of ground sample using a Dickey-john GAS III NIR light reflectance machine (Hymowitz et al., 1974). All six replications were phenotyped by NIR.
Maize starches were isolated by salt steeping (Rani and Bhattacharya, 1995) and freeze dried. Starch true amylose content was determined in triplicate with an amylose/amylopectin assay kit (K-AMYL; Megazyme International, Wicklow, Ireland) following a simplified concanavalin A procedure (Gibson et al., 1997).
Starch pasting properties of maize starches were determined using a controlled stress rheometer (AR 1000-N, Rheolyst; TA Instruments, Dover, DE) at a constant shear rate of 200 s−1. The rheometer was fitted with a polysulfone cone, which had a diameter of 4 cm and an angle of 4°. A microviscoamylographic method, which only requires 100 mg of sample, was used. The temperature program used consisted of four segments: (1) 45 to 95°C in 3.5 min, (2) holding at 95°C for 2.3 min, (3) 95 to 50°C in 3.5 min, and (4) holding at 50°C for 1.25 min. Triplicate aqueous maize starch suspensions (8% w/w) were prepared using deionized water. Suspensions were degassed while stirring under vacuum for 15 min (Ibáñez, 2002). The definitions for measured properties include the following: pasting temperature, temperature of initial viscosity increase; peak time, time required to reach peak; peak temperature, temperature of peak viscosity; peak viscosity, maximum viscosity recorded during heating and holding cycles; trough, minimum viscosity after peak; hot paste viscosity, final viscosity after cooking at 95°C; cool paste viscosity, viscosity of paste cooled to 50°C; breakdown, difference (−) between peak viscosity and trough, indicating breakdown in viscosity of paste during 95°C holding period; setback, difference (−) between cool paste viscosity and trough; gel consistency, difference (−) between cool paste viscosity and hot paste. The controlled stress rheometer was operated using TA navigator software (version 4.0; TA Instruments) for a personal computer. For true amylose content and starch pasting properties, kernels from the environments Homestead, FL (1998) and Clayton, NC (2001; one replication) were phenotyped. Sixty-seven nonsweet lines were scored from Homestead, and 90 lines were scored from Clayton; discrepancy in number of samples phenotyped by NIR analysis and starch pasting assays were a result of limited samples available for the starch pasting assays after NIR measurements.
Amplification and Sequencing
The six candidate genes and promoter regions were amplified and direct sequenced in 32 lines as described previously by Whitt et al. (2002). Candidate genomic regions either from the coding region and/or promoter regions were chosen based on the position of amino acid changes and substantial indel polymorphisms. Selected regions were sampled in a set of 102 maize inbreds as follows: for ae1, exon one through exon three, exon 12 through exon 14, and exon 16 through exon 18; for bt2, the promoter through intron one; for sh1, the promoter through the noncoding exon one and a portion of intron one; for sh2, intron eight through intron 10; for su1, the promoter through exon one and exon 13 through intron 14; for wx1, exon one through exon two and exon eight through exon nine. SNP/indel positions referred to in the text correspond to alignment positions from sequences submitted by Whitt et al. (2002). Major alleles are defined in Table 5.
Table 5.
Sequence Context of the Major Polymorphisms Identified through the Association Analysis
| Polymorphism Name | Sequence (5′/3′) |
|---|---|
| Ae1-1509A>G | GCGCGGCGGCCGCGGCC[A/G]GGAAGGCGGTCATGGTTCCT |
| Ae1-1689T>C | CCTCTTT[T/C]TGGATGCTATTTGAGAACAA |
| Bt2-925T>C | CGCCGAGCAGC[T/C]AATTCCAAAGCGTGACAAAGCCGCTG |
| Sh1-775T>Ca | GGTCTGAAC[T/C]TT[T/C]CCGAAACCAGCCAGCCATTGGTCT |
| Sh1-1210A>Gb | ATCTGCTGG[T/C]C[C/G]CGGTAGAAAAGA[T/C]C[A/G]TGTCCC |
| Sh2-3674-1c | {[T/-]ACTGAT}GTTGCAGAGAGTTGAGACCAACTTCCTGAGCT |
| Sh2-3842T>G | TCTATCCAACTCCTAGT[G/T]TACCTTCTAACAGTGT |
The first polymorphic site, [T/C], is the significantly associated site.
The significantly associated site is the last polymorphic site, [A/G].
The significant polymorphism is the 1-bp deletion indicated by [T/-]. A third allele included lines that had a 47-bp deletion that encompassed this area. The sequence in braces is the last part of this deletion and therefore is missing in these lines.
Statistics
To summarize the data over multiple replications, PCA of the correlation matrix was performed on both the NIR and the starch pasting data using SAS software (SAS, 1999) for 97 nonsweet taxa from 102 maize inbreds. Although sequence alignments for the six candidate loci and phenotypic data from five sampled sweet maize lines were made and are available (Ia2132, Il14H, Il101, Il677a, and P39), these low-starch mutants were removed from the association analyses to avoid false results produced by these outliers (the sample number of sweet lines was too low to form its own subpopulation). Three separate PCAs were performed: one for total NIR data and one each for starch pasting data from the winter (Homestead, FL 1998) and from the summer (Clayton, NC 2001) replications, referred to hereafter as winterH and summerC, respectively. To handle missing NIR data for the PCA, missing data were imputed using the KNN impute program (Troyanskaya et al., 2001) with K set to five nearest neighbors. The major principal components (PC) found for NIR and for each starch pasting analysis were then used as traits in association tests for the 97 taxa. Amylose content for both summerC and winterH replications, not included in the PCAs, was used directly to test for associations. Starch pasting and amylose data were not pooled from the two environments, as this would have required excessive imputation.
Tests for association (Thornsberry et al., 2001) and LD (Hill and Robertson, 1968) were performed using the software package TASSEL, available at http://www.maizegenetics.net. SNPs or indels at a site frequency of 0.05 or greater among the 97 inbreds were evaluated using TASSEL. All association tests were run with population structure included, using logistic regression as described by Thornsberry et al. (2001). One thousand permutations of the data were run to account for multiple tests within a gene, and a significant association was called if the P-value of the most significant polymorphism in a region was seen in <5% of the permutations.
To examine possible allelic effects of significant polymorphisms, post hoc statistical tests (GLM) were used to further dissect PC-associated effects in those genes with significantly associated PC traits. These tests were used to determine whether the sample means of actual trait values were significantly different between lines with the best site polymorphism in an association and those lines without the polymorphism. GLM models in SAS (SAS, 1999) included estimates of population structure, and reported P-values are from the Type III sum of squares (i.e., the effect after the population structure has been removed).
Sequence data for the six genes for the 102 genotypes included in this article have been deposited with the EMBL/GenBank data libraries under the following accession numbers: ae1 (AY290043 to AY290190 and AY290192 to AY290304), bt2 (AY290600 to AY290700), sh1 (AY290403 to AY290503), sh2 (AY324882 to AY324981), su1 (AY290305 to AY290599), and wx1 (AF544068 to AF544099).
Acknowledgments
We would like to express our appreciation to Lauren McIntyre at Purdue University for evaluation of the West Lafayette, IN replication. We also thank Natalie Stevens, Sherry Flint-Garcia, and the anonymous reviewers for their helpful comments on the manuscript and Jason Dinges and Martha James for help on su1. Sequencing was done at the North Carolina State University Genome Research Laboratory. This work was supported by a grant from the National Science Foundation (DBI-9872631 and DBI-0321467) and by the USDA's Agricultural Research Service.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Edward S. Buckler (esb33@cornell.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.025700.
References
- Bae, J.M., Giroux, M.J., and Hannah, L.C. (1990). Cloning and characterization of the Brittle-2 gene of maize. Maydica 35, 317–322. [Google Scholar]
- Ball, S., Guan, H.P., James, M.G., Myers, A.M., Keeling, P.L., Mouille, G., Buleon, A., Colonna, P., and Preiss, J. (1996). From glycogen to amylopectin: A model for the biogenesis of the plant starch granule. Cell 86, 349–352. [DOI] [PubMed] [Google Scholar]
- Beckles, D.M., Smith, A.M., and ap Rees, T. (2001). A cytosolic ADP-glucose pyrophosphorylase is a feature of graminaceous endosperms, but not of other starch-storing organs. Plant Physiology 125, 818–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berke, T.G., and Rocheford, T. (1995). Quantitative trait loci for flowering, plant and ear height, and kernel traits in maize. Crop Sci. 35, 1542–1549. [Google Scholar]
- Bhave, M.R., Lawrence, S., Barton, C., and Hannah, L.C. (1990). Identification and molecular characterization of Shrunken-2 cDNA clones of maize. Plant Cell 2, 581–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler, E.S., and Thornsberry, J.M. (2002). Plant molecular diversity and applications to genomics. Curr. Opin. Plant Biol. 5, 107–111. [DOI] [PubMed] [Google Scholar]
- Chourey, P.S., and Nelson, O.E. (1976). Enzymatic deficiency conditioned by shrunken 1 mutations in maize. Biochem. Genet. 14, 1041–1055. [DOI] [PubMed] [Google Scholar]
- Clancy, M., and Hannah, L.C. (2002). Splicing of the maize sh1 first intron is essential for enhancement of gene expression, and a T-rich motif increases expression without affecting splicing. Plant Physiol. 130, 918–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy, M., Vasil, V., Hannah, L.C., and Vasil, I.K. (1994). Maize shrunken-1 intron and exon regions increase gene expression in maize protoplasts. Plant Sci. 98, 151–161. [Google Scholar]
- Deatherage, W.L., Macmasters, M.M., Vineyard, M.L., and Bear, R.P. (1954). A note on starch of high amylose content from corn with high starch content. Cereal Chem. 31, 50–53. [Google Scholar]
- Dien, B., Bothast, R., Nichols, N., and Cotta, M. (2002). The U.S. corn ethanol industry: An overview of current technology and future prospects. Int. Sugar J. 104, 7. [Google Scholar]
- Dudley, J.W., Dijkhuizen, A., Paul, C., Coates, S.T., and Rocheford, T.R. (2004). Effects of random mating on marker-QTL associations in the cross of the Illinois high protein x Illinois low protein maize strains. Crop Sci. 44, 1419–1428. [Google Scholar]
- Dudley, J.W., and Lambert, R.J. (1992). 90 generations of selection for oil and protein in maize. Maydica 37, 81–87. [Google Scholar]
- Dudley, J.W., and Lambert, R.J. (2004). 100 generations of selection for oil and protein in corn. Plant Breed. Rev. 24, 79–110. [Google Scholar]
- Falush, D., Stephens, M., and Pritchard, J.K. (2003). Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 164, 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, D.K., Boyer, C.D., and Hannah, L.C. (1993). Starch branching enzyme II from maize endosperm. Plant Physiol. 102, 1045–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, D.K., Gao, M., Kim, K.N., Boyer, C.D., and Guiltinan, M.J. (1996). Allelic analysis of the maize amylose-extender locus suggests that independent genes encode starch-branching enzymes LLa and LLb. Plant Physiol. 110, 611–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint-Garcia, S.A., Thornsberry, J.M., and Buckler, E.S. (2003). Structure of linkage disequilibrium in plants. Annu. Rev. Plant Biol. 54, 357–374. [DOI] [PubMed] [Google Scholar]
- Gao, M., Fisher, D.K., Kim, K.N., Shannon, J.C., and Guiltinan, M.J. (1996). Evolutionary conservation and expression patterns of maize starch branching enzyme I and IIb genes suggests isoform specialization. Plant Mol. Biol. 30, 1223–1232. [DOI] [PubMed] [Google Scholar]
- Gibson, T.S., Solah, V.A., and McCleary, B.V. (1997). A procedure to measure amylose in cereal starches and flours with concanavalin A. J. Cereal Sci. 25, 111–119. [Google Scholar]
- Goldman, I.L., Rocheford, T., and Dudley, J.W. (1993). Quantitative trait loci influencing protein and starch concentration in the Illinois long term selection maize strains. Theor. Appl. Genet. 87, 217–224. [DOI] [PubMed] [Google Scholar]
- Greene, T.W., and Hannah, L.C. (1998). Enhanced stability of maize endosperm ADP-glucose pyrophosphorylase is gained through mutants that alter subunit interactions. Proc. Natl. Acad. Sci. USA 95, 13342–13347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, H.P., Kuriki, T., Sivak, M., and Preiss, J. (1995). Maize branching enzyme catalyzes synthesis of glycogen-like polysaccharide in glgB-deficient Escherichia coli. Proc. Natl. Acad. Sci. USA 92, 964–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, W.G., and Robertson, A. (1968). Linkage disequilibrium in finite populations. Theor. Appl. Genet. 38, 226–231. [DOI] [PubMed] [Google Scholar]
- Huang, D.Y., and Wang, A.Y. (1998). Purification and characterization of sucrose synthase isozymes from etiolated rice seedlings. Biochem. Mol. Biol. Int. 46, 107–113. [DOI] [PubMed] [Google Scholar]
- Hymowitz, T., Dudley, J.W., Collins, F.I., and Brown, C.M. (1974). Estimations of protein and oil concentration in corn, soybean, and oat seed by near-Infrared light reflectance. Crop Sci. 14, 713–715. [Google Scholar]
- Ibáñez, A.M. (2002). A Study of Rice Pasting Properties of Rice Flour and Starch as Affected by Rice Variety and Physicochemical Properties. PhD dissertation (Davis, CA: University of California).
- James, M.G., Robertson, D.S., and Myers, A.M. (1995). Characterization of the maize gene Sugary1, a determinant of starch composition in kernels. Plant Cell 7, 417–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K.N., Fisher, D.K., Gao, M., and Guiltinan, M.J. (1998). Molecular cloning and characterization of the amylose-extender gene encoding starch branching enzyme IIB in maize. Plant Mol. Biol. 38, 945–956. [DOI] [PubMed] [Google Scholar]
- Knowler, W.C., Williams, R.C., Pettitt, D.J., and Steinberg, A.G. (1988). Gm3;5,13,14 and Type 2 diabetes mellitus: An association in American Indians with genetic admixture. Am. J. Hum. Genet. 43, 520–526. [PMC free article] [PubMed] [Google Scholar]
- Lambert, R.J. (2001). High-oil corn hybrids. In Specialty Corns, A.R. Hallauer, ed (Boca Raton, FL: CRC Press), pp. 131–154.
- Lander, E.S., and Schork, N.J. (1994). Genetic dissection of complex traits. Science 265, 2037–2048. [DOI] [PubMed] [Google Scholar]
- Liu, K., Goodman, M., Muse, S., Smith, J.S., Buckler, E., and Doebley, J. (2003). Genetic structure and diversity among maize inbred lines as inferred from DNA microsatellites. Genetics 165, 2117–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, A.D., and Langley, C.H. (1999). The power of association studies to detect the contribution of candidate genetic loci to variation in complex traits. Genome Res. 9, 720–731. [PMC free article] [PubMed] [Google Scholar]
- McGinnis, R., Shifman, S., and Darvasi, A. (2002). Power and efficiency of the TDT and case-control design for association scans. Behav. Genet. 32, 135–144. [DOI] [PubMed] [Google Scholar]
- Nelson, O.E., and Rines, H.W. (1962). The enzymatic deficiency in waxy mutant of maize. Biochem. Biophys. Res. Commun. 9, 297–300. [DOI] [PubMed] [Google Scholar]
- Palaisa, K.A., Morgante, M., Williams, M., and Rafalski, A. (2003). Contrasting effects of selection on sequence diversity and linkage disequilibrium at two phytoene synthase loci. Plant Cell 15, 1795–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, D. (2000). Starch synthesis in maize. In Carbohydrate Reserves in Plants: Synthesis and Regulation, A.K. Gupta and N. Kaur, eds (Amsterdam: Elsevier), pp. 125–146.
- Prioul, J.L., Pelleschi, S., Sene, M., Thevenot, C., Causse, M., de Vienne, D., and Leonardi, A. (1999). From QTLs for enzyme activity to candidate genes in maize. J. Exp. Bot. 50, 1281–1288. [Google Scholar]
- Pritchard, J.K., Stephens, M., and Donnelly, P. (2000. a). Inference of population structure using multilocus genotype data. Genetics 155, 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard, J.K., Stephens, M., Rosenberg, N.A., and Donnelly, P. (2000. b). Association mapping in structured populations. Am. J. Hum. Genet. 67, 170–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani, M.R.S., and Bhattacharya, K.R. (1995). Microscopy of rice starch granules during cooking. Starch/Stärke 47, 334–337. [Google Scholar]
- Remington, D.L., Thornsberry, J.M., Matsuoka, Y., Wilson, L.M., Whitt, S.R., Doebley, J., Kresovich, S., Goodman, M.M., and Buckler, E.S. (2001). Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc. Natl. Acad. Sci. USA 98, 11479–11484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch, N., and Merikangas, K. (1996). The future of genetic studies of complex human diseases. Science 273, 1516–1517. [DOI] [PubMed] [Google Scholar]
- SAS (1999). Statistical Analysis Systems. (Cary, NC: SAS Institute).
- Séne, M., Causse, M., Damerval, C., Thevenot, C., and Prioul, J.L. (2000). Quantitative trait loci affecting amylose, amylopectin and starch content in maize recombinant inbred lines. Plant Physiol. Biochem. 38, 459–472. [Google Scholar]
- Shure, M., Wessler, S., and Fedoroff, N. (1983). Molecular identification and isolation of the Waxy locus in maize. Cell 35, 225–233. [DOI] [PubMed] [Google Scholar]
- Sidebottom, C., Kirkland, M., Strongitharm, B., and Jeffcoat, R. (1998). Characterization of the difference of starch branching enzyme activities in normal and low-amylopectin maize during kernel development. J. Cereal Sci. 27, 279–287. [Google Scholar]
- Smith, A.M. (2001). The biosynthesis of starch granules. Biomacromolecules 2, 335–341. [DOI] [PubMed] [Google Scholar]
- Stark, D.M., Timmerman, K.P., Barry, G.F., Preiss, J., and Kishore, G.M. (1992). Regulation of the amount of starch in plant tissues by ADP-glucose pyrophosphorylase. Science 258, 287–292. [DOI] [PubMed] [Google Scholar]
- Tenaillon, M.I., Sawkins, M.C., Long, A.D., Gaut, R.L., Doebley, J.F., and Gaut, B.S. (2001). Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp. mays L.). Proc. Natl. Acad. Sci. USA 98, 9161–9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornsberry, J.M., Goodman, M.M., Doebley, J., Kresovich, S., Nielsen, D., and Buckler IV, E.S. (2001). Dwarf8 polymorphisms associate with variation in flowering time. Nat. Genet. 28, 286–289. [DOI] [PubMed] [Google Scholar]
- Troyanskaya, O., Cantor, M., Sherlock, G., Brown, P., Hastie, T., Tibshirani, R., Botstein, D., and Altman, R.B. (2001). Missing value estimation methods for DNA microarrays. Bioinformatics 17, 520–525. [DOI] [PubMed] [Google Scholar]
- Tsai, C.Y., and Nelson, O. (1966). Starch-deficient maize mutant lacking adenosine diphosphate glucose pyrophosphorylase activity. Science 151, 341–343. [DOI] [PubMed] [Google Scholar]
- Vasil, V., Clancy, M., Ferl, R.J., Vasil, I.K., and Hannah, L.C. (1989). Increased gene expression by the 1st intron of maize Shrunken-1 locus in grass species. Plant Physiol. 91, 1575–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitt, S.R., Wilson, L.M., Tenaillon, M.I., Gaut, B.S., and Buckler, E.S. (2002). Genetic diversity and selection in the maize starch pathway. Proc. Natl. Acad. Sci. USA 99, 12959–12962. [DOI] [PMC free article] [PubMed] [Google Scholar]