Abstract
Lignin, a cross-linked macromolecule of hydrophobic aromatic structure, provides additional rigidity to a plant cell wall. Although it is an integral part of the plant cell, presence of lignin considerably reduces the quality of the fiber of fiber-yielding plants. Decreasing lignin in such plants holds significant commercial and environmental potential. This study aimed at reducing the lignin content in jute-a fiber crop, by introducing hpRNA-based vectors for downregulation of two monolignoid biosynthetic genes- cinnamate 4-hydroxylase (C4H) and caffeic acid O-methyltransferase (COMT). Transgenic generations, analyzed through Southern, RT-PCR and northern assays showed downregulation of the selected genes. Transgenic lines exhibited reduced level of gene expression with ~ 16–25% reduction in acid insoluble lignin for the whole stem and ~13–14% reduction in fiber lignin content compared to the control lines. Among the two transgenic plant types one exhibited an increase in cellulose content and concomitant improvement of glucose release. Composition of the lignin building blocks was found to alter and this alteration resulted in a pattern, different from other plants where the same genes were manipulated. It is expected that successful COMT-hpRNA and C4H-hpRNA transgenesis in jute will have far-reaching commercial implications leading to product diversification and value addition.
Growing environmental awareness together with a gradual curb of fossil fuels has led to a paradigm shift in the search for new fuel reserve and has recognized the potential of living materials as a source of renewable energy. Bioethanol, produced from biomass, is considered a promising transportation fuel, for which the major plant types used are sugarcane and corn1. However, bioethanol production from these crops has its limitations since they provide little or no net life cycle benefit in terms of carbon dioxide and other greenhouse gas (GHG) emission reductions2 and also because of their recognizable incorporation in our food supply chain3. On the other hand, lignocellulose biomass obtained from agricultural remains, forest residues (hardwood and softwood), herbaceous and woody energy crops- is renewable, cheap, adaptable to less cultivable land and readily obtainable with 10–50 billion tons produced per year at the global level4. It has therefore been considered an attractive replacement of food-crop based source of biofuels2. Lignocellulosic biomass is also a potential source of raw materials for the paper and pulp industry5.
Nevertheless, the popularity of lignocellulose as an alternative source of biofuel, paper and pulping industry is severely hindered by its structural features. Crystalline cellulose embedded in a hemicellulose and lignin matrix, renders enzymatic, microbial or chemical conversion of cellulose to glucose difficult6. Existing biomass fermentation procedures for fuels and chemicals are generally expensive principally because of this recalcitrance, which in turn lead to limited commercialization of bioethanol7. Recent plant genetic engineering studies have aimed to decrease the cost by lowering physical hindrance of plant cell wall and to increase the cellulose and/or overall crop biomass yield8. Accessibility of plant cell wall polysaccharides to digestion is limited by many factors, including the presence of lignin, a phenylpropanoid polymer in vascular tissues and fibers9. Attempts have been made to lower the lignin content and/or change the composition by reducing the need for expensive and harsh pretreatments required to disintegrate the same and to allow access to cellulose10. There has also been an intense focus of research to alter lignocellulosic materials currently used in paper as well as pulp industries in order to improve pulp production, reduce the amount of energy required and the environmental pollution associated with biomass processing11.
Jute (Corchorus sp.), a lignocellulosic annual plant grown abundantly in Bangladesh, India, China and Thailand, is known for its fiber quality and is widely used for the manufacture of flexible packing fabrics, carpet backings, decorative fabrics, automotive headlining and other applications12. Agro-based nature, annually renewable, biodegradable and obtainable at a low cost put jute at an advantage13. Biodegradable nature of jute fiber and the potential high yield of cellulose biomass per acre have increased the global interest in jute. Textile and paper industries are also interested in its potential as an important ingredient for producing paper and fine textiles14. As with other lignocellulosic plants the drawback of jute when used as a source for such purposes is the abundance of lignin polymer that renders the plant material almost inaccessible for downstream processes. The high amount of lignin in jute makes it coarse and impedes use of the same as a textile fiber.
In this context, the current study aimed at modifying the lignin biosynthetic pathway of jute (Corchorus olitorius var. O-9897) for induced altered lignin composition, increase in cellulose content and enzymatic digestibility. This was attempted by downregulating two major genes, cinnamate 4-hydroxylase (C4H) and caffeic acid 3-O methyltransferase (COMT) of the lignin biosynthetic pathway by using the technique of RNAi15. Ten different enzymes catalyze a series of hydroxylation, methylation and side chain reduction reactions of monolignol precursors16. C4H is involved near the beginning of the lignin pathway which hydroxylates cinnamic acid to form p-coumaric acid17. COMT functions late in the monolignol biosynthetic pathway and, despite its name, methylates 5-hydroxyconiferyl aldehyde and 5-hydroxyconiferyl alcohol to form S unit precursors, sinapyl aldehyde and sinapyl alcohol, respectively18 These two genes, COMT and C4H do not appear to overlap with the plant defense machinery19. Transgenic jute lines with reduced expression of COMT and C4H genes were found to have lowered lignin content, altered lignin composition, increase in total cellulose content and improvement, albeit small in enzymatic saccharification. Moreover, none of these alterations led to any growth compensation. The results bolster the potential of lignin modification as a tool for improving lignocellulosic crops like jute so that it may be used more efficiently as a source of biofuel, paper pulp and textile fiber.
Results
Gene identification, construction of RNAi vector and validation
Full length sequences of each selected gene namely, caffeic acid O-methyltransferase (COMT) and cinnamate 4-hydroxylase (C4H) in jute were retrieved by using traditional gene walking method with degenerate primers designed from plants with sequence similarity to jute like Populus, Vitis, Gossypium etc20. This was followed by cloning and sequencing of the same.
Successful COMT-hpRNA and C4H-hpRNA constructs were designed from the conserved region of the respective genes (details given in methods). A set of construct specific PCRs revealed an inverted orientation of the two copies of COMT-hpRNA and C4H-hpRNA sequences. Determination of full-length sequence of the recombinant destination vectors (see methods) further validated the same (Supplementary Fig. 1.1, 1.2).
Transformation of jute plants
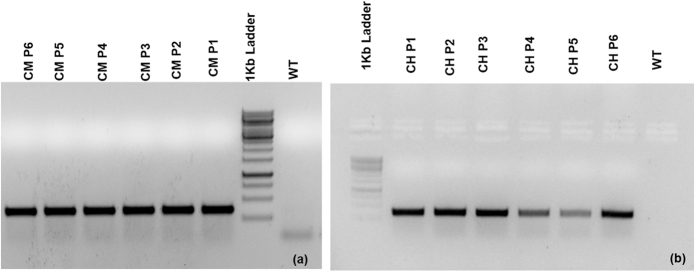
An in planta protocol for jute transformation developed by Sajib et al.21 was used to introduce the COMT-hpRNA and C4H-hpRNA based constructs into C. olitorius var 0–9897 independently to give rise to two different groups of transgenic lines (COMT-hpRNA and C4H-hpRNA lines). Initial confirmation of transgenesis was done by amplification of a region of NPT(II) (neomycin phosphotransferase II, determinant of kanamycin resistance) reporter gene using genomic DNA isolated from the leaves of randomly selected transgenic plants from each group and wild type control. PCR results showed prominent presence of the reporter gene in transgenic plants. No signal was obtained for the wild type control. Among the positive transgenic plants, six chosen arbitrarily for further analysis were designated CM P1 -P 6 for COMT-hpRNA and CH P1 - P6 for C4H-hpRNA plant lines (Fig. 1a,b).
Figure 1. Confirmation of initial transgenesis.
Genomic DNA isolated from randomly selected transformed plants was PCR amplified with NPT(II) primer for COMT-hpRNAi (a) and C4H-hpRNAi (b) lines to confirm the initial transgenesis.
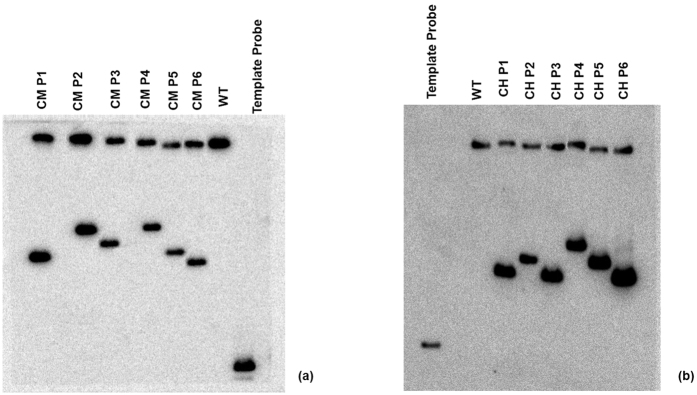
Successive T1 generations of each line mentioned above were screened by their growth on media containing 400 mM kanamycin, the resistance gene for which, is present in the RNAi vector. For both the genes ~75% of the assumed transformants were able to grow in the antibiotic media, indicating efficient transformation. Seedlings tested positive were used for subsequent molecular studies. Southern blot analysis was performed for T1 plants of the six lines of each transgenic type to further confirm the incorporation of hpRNA for both genes in the jute genome. Single conspicuous bands were detected in the blotting experiments when hpRNA precursor specific probes were used (Fig. 2a,b).
Figure 2. Southern analysis of primary hpRNAi transformants of jute.
Blots were probed with [α32P] labeled hpRNAi construct specific region of different (a) COMT-hpRNA and (b) C4H-hpRNA transgenic plants.
Downregulation of COMT and C4H genes in jute
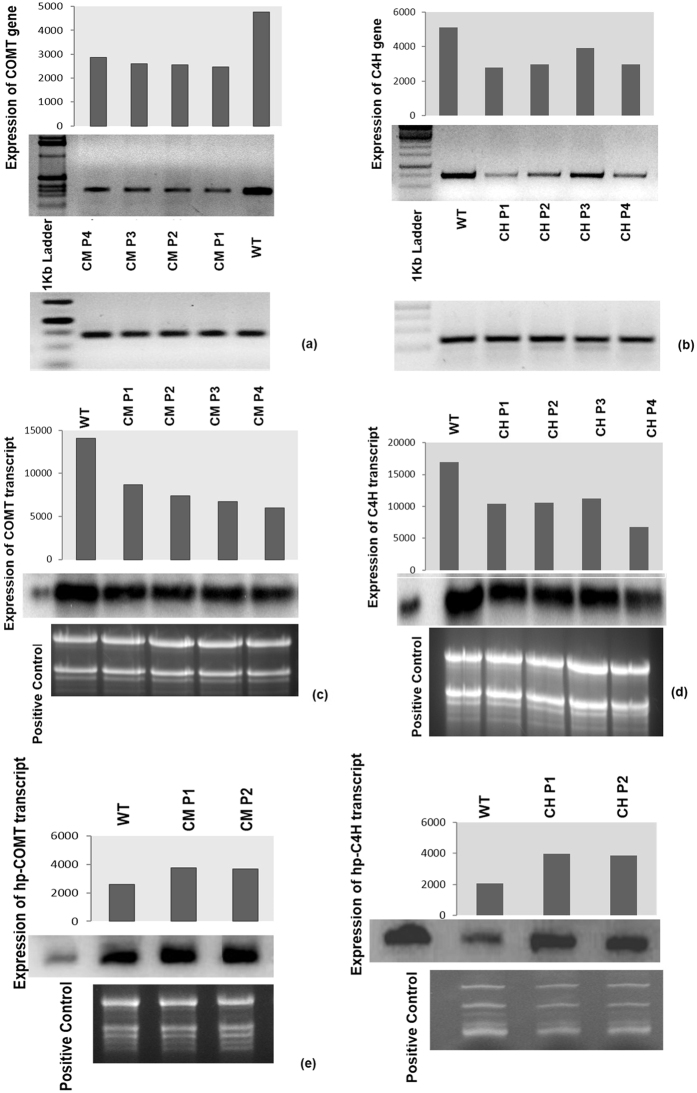
Among these six lines of each transgenic type found positive by Southern analysis, four (CM P1-P4 and CH P1-P4) from each group were then considered for assessing the functionality of the hpRNA constructs. The hpRNA for COMT and C4H appeared to be effective in activating the RNAi pathway leading to depleted gene expression. Expression was found to drop in comparison to the wildtype for both genes when monitored by semi-quantitative RT-PCR (Fig. 3a,b). Also, in the northern blots, intensities of the bands for expressed genes were found to be lower in comparison to non-transgenic plants for both lines, representing suppression of the respective genes and hence validating the functionality of the hpRNA constructs (Fig. 3c,d). Further experiments were conducted for T1, T2 and T3 generations of CM P1-P2 as well as CH P1-P2 plants. Each experiment (measurement of lignin and cellulose content and the amount of glucose released) was conducted taking into account biological duplicates with three technical replicates. Representative lines were also subjected to hpRNA northern blot analysis (Fig. 3e,f) to check their expression levels in transgenic plants. The blot showed an opposite trend of expression confirming successful and functional hpRNA transgenesis for both the genes. Signals were also observed in wild type samples because the hpRNAs were designed from endogenous COMT and C4H genes of C. olitorius. However, the intensities were much higher in transgenic lines in comparison to the wild type.
Figure 3. Expression analysis of downregulated genes as a proof of functionality of the RNAi construct.
RT-PCR of COMT (a) and C4H (b) gene along with actin gene as loading control. Northern blot with [α32P] labeled cDNA probe of COMT (c) and C4H gene (d). For hp-northerns, the probes were hpRNAi amplicon specific sequences of COMT (e) and C4H (f) genes respectively. The figures show intensities of bands normalized with respect to 28S rRNA.
Phenotypical and histochemical analysis
Morphological assessments in terms of plant height, width, pod number and average pod length of all plants grown under field conditions were compared (Table 1) in order to determine if there were any differences in the vegetative and reproductive stages of the transformed jute generations. Plants were randomly chosen from the selected lines of each group. No distinct morphological variations with respect to wild type plants were observed. Plant height, width and pod length were considerably same in control and each type of transgenic plant generations (p = 0.3, 0.037 and 0.058 respectively). Random variations in the number of pods (ranging from 58–63) were seen in both control and different transgenic plant types. This therefore cannot be attributed to any effect of gene downregulation.
Table 1. Measurement of various growth and yield parameters of wild type and transgenic lines.
| Parameters | wild type |
COMT-hpRNA line |
C4H-hpRNA line |
||||
|---|---|---|---|---|---|---|---|
| T1 | T2 | T3 | T1 | T2 | T3 | ||
| Plant Height (cm) | 335.24 ± 0.25a | 334.84 ± 0.32a | 334.7 ± 0.49a | 335.52 ± 0.32a | 334.8 ± 0.31a | 335.5 ± 0.32a | 335.4 ± 0.4a |
| Plant width (cm) | 1.188 ± 0.02a | 1.18 ± 0.003a | 1.18 ± 0.0003a | 1.19 ± 0.004a | 1.22 ± 0.003a | 1.19 ± 0.008a | 1.20 ± 0.005a |
| Pod Number | 61.8 ± 1.06ab | 58 ± 0.44c*** | 60 ± 0.44abc | 59 ± 0.31bc | 62 ± 0.31a | 60 ± 0.31abc | 58 ± 0.31c*** |
| Pod Length (cm) | 5.00 ± 0.007a | 4.9 ± 0.005a | 5.00 ± 0.003a | 4.9 ± 0.003a | 4.9 ± 0.005a | 4.9 ± 0.005a | 5.00 ± 0.004a |
Results are given as means ± standard error. Sample size = 5. Statistical analyses were done using one way ANOVA and Tukey’s test with P value < 0.001 considered as highly significant. (P value is given in Supplementary Table 2). ‘***’denotes P < 0.001, ‘**’denotes P < 0.01. For each individual parameter, means that do not share a letter are significantly different. (cm = centimeter).
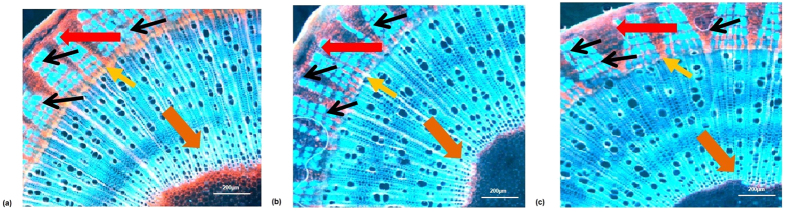
Histochemical analysis of stem cross sections of wild type and T3 generations of the selected plant lines was performed using phloroglucinol (a dye which specifically stains native lignin due to the formation of a purple to reddish chromophore between lignin hydroxycinnamaldehyde end-groups and phloroglucinol), showed significantly lower deposition of lignin in both transgenic type. This is indicated by a reduction in the staining intensity when compared to control plants (Fig. 4). Variation in epidermal lignin content was not much for the different plant types but decreased lignin accumulation was observed in the phloem fiber (the region from where jute fiber is obtained) and the cortex of the transgenic lines compared to the wild type. The vascular cambium ring of each transgenic type was almost devoid of lignin. Within these plants the parenchymal tissue of the pith region, showed significant reduction of lignin deposition in comparison to the wild type. The overall lignin content appeared to be lower for C4H-hpRNA than that of COMT-hpRNA lines which supported the value obtained for Klason lignin (Table 2).
Figure 4. Histochemical assay of lignin.
Lignin deposition observed in wildtype plants (a) and transgenic lines COMT-hpRNA (b) and C4H-hpRNA (c). The arrows indicate difference in lignin deposition in transgenic and wild type jute plants (red: epidermis; orange: pith; yellow: vascular cambium). The area between the epidermis and vascular cambium (indicated by black arrows) is the bast region from where the jute fiber is obtained. This area as observed is significantly less lignified in transgenic plants than the wild type.
Table 2. A summary of lignin content and composition, cellulose content, enzymatic glucose release and a comparative analyses between wild type and transgenic jute plants.
| Parameters | Wildtype |
COMT-hpRNA lines |
C4H-hpRNA lines |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 |
P2 |
P1 |
P2 |
||||||||||
| T1 | T2 | T3 | T1 | T2 | T3 | T1 | T2 | T3 | T1 | T2 | T3 | ||
| % Klason lignin (whole stem) | 29.50 ± 0.04a | 25.61 ± 0.10b*** | 24.22 ± 0.21c*** | 23.81 ± 0.13c*** | 25.35 ± 0.16b*** | 23.97 ± 0.16c*** | 23.95 ± 0.14c*** | 22.90 ± 0.12d*** | 22.85 ± 0.15d*** | 22.96 ± 0.07d*** | 22.85 ± 0.14d*** | 22.56 ± 0.07d*** | 22.79 ± 0.16d*** |
| % Lignin reduction (whole stem) | 13.18 | 17.90 | 19.28 | 14.08 | 18.76 | 18.82 | 22.36 | 22.56 | 22.17 | 22.54 | 23.54 | 22.73 | |
| % Klason lignin (fiber) | 13.46 ± 0.02a | 11.61 ± 0.12bcd*** | 11.68 ± 0.14bcd*** | 11.54 ± 0.17bcd*** | 11.72 ± 0.16bc*** | 11.58 ± 0.11bcd*** | 11.86 ± 0.06b*** | 11.05 ± 0.18d*** | 11.74 ± 0.18bc*** | 11.97 ± 0.09b*** | 11.71 ± 0.13bc*** | 11.16 ± 0.07 cd*** | 11.41 ± 0.13bcd*** |
| % Lignin reduction (fiber) | 13.72 | 13.24 | 14.25 | 12.91 | 13.97 | 11.86 | 17.89 | 12.82 | 11.07 | 13.03 | 17.07 | 15.23 | |
| % Cellulose (whole stem) | 30.37 ± 0.13d | 31.55 ± 0.14abc*** | 31.55 ± 0.05abc*** | 31.49 ± 0.11abc*** | 31.79 ± 0.13a*** | 31.87 ± 0.02a*** | 31.70 ± 0.07ab*** | 31.18 ± 0.16c*** | 31.23 ± 0.06bc*** | 31.58 ± 0.15abc*** | 31.45 ± 0.05abc*** | 31.42 ± 0.07abc*** | 31.64 ± 0.05abc*** |
| % Increase in cellulose content (whole stem) | 3.88 | 3.89 | 3.69 | 4.68 | 4.95 | 4.39 | 2.67 | 2.84 | 3.99 | 3.56 | 3.45 | 4.19 | |
| Ratio of cellulose to lignin | 1.00 f | 1.23e*** | 1.30d*** | 1.32bcd*** | 1.25e*** | 1.33bcd*** | 1.36ab*** | 1.37a*** | 1.37a*** | 1.38a*** | 1.38a*** | 1.39a*** | 1.39a*** |
| Amount of glucose released (mg/g of sample) | 76.87 ± 0.31b | 76.01 ± 0.63b | 76.11 ± 0.55b | 76.01 ± 0.63b | 76.04 ± 0.61b | 76.06 ± 0.51b | 76.06 ± 0.47b | 78.46 ± 0.23a*** | 78.18 ± 0.16ab*** | 78.36 ± 0.18a*** | 78.38 ± 0.22a*** | 78.03 ± 0.28ab*** | 78.37 ± 0.17a*** |
| H (%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| S (%) | 52 ± 0.57e | 47.33 ± 0.67 f*** | 46.67 ± 0.33 f*** | 51.67 ± 0.88e | 46.67 ± 0.33 f*** | 47.33 ± 0.33 f*** | 50.67 ± 0.88e | 61.67 ± 1.20a*** | 55.33 ± 0.67 cd*** | 53.33 ± 0.33d*** | 59.00 ± 0.58ab*** | 57.00 ± 0.58bc*** | 52.67 ± 0.33d*** |
| G (%) | 48.00 ± 0.58b | 52.67 ± 0.67a*** | 53.33 ± 0.33a*** | 48.33 ± 0.88b | 53.33 ± 0.33a*** | 52.67 ± 0.33a*** | 49.33 ± 0.88b | 38.33 ± 1.20 f*** | 44.67 ± 0.67 cd*** | 46.67 ± 0.33bc*** | 41.00 ± 0.58ef*** | 43.00 ± 0.58de*** | 47.33 ± 0.33bc*** |
| S/G= | 1.1e | 0.9e | 0.9e | 1.1e | 0.9e | 0.9e | 1.1e | 1.8a*** | 1.2 cd*** | 1.2 cd*** | 1.5ab*** | 1.4bc*** | 1.1e |
| P coumarate (%)* | 11.33 ± 1.02c | 9.33 ± 1.76d** | 4.66 ± 0.57 g** | 9 ± 0.57de** | 8.95 ± 0.81e** | 5.78 ± 0.95 g** | 9.21 ± 1.05d** | 15.7 ± 2.19a** | 11.67 ± 0.88c | 8.33 ± 2.19ef** | 14.78 ± 1.56b** | 11.12 ± 1.72c | 9.26 ± 2.06d** |
The sample size was six for lignin measurement, cellulose content and glucose release estimations including two biological replicates and technical triplicates. Three replicates were used for analyses of lignin composition. Results are given as means ± standard error. Statistical analyses were done using one way ANOVA and Tukey’s test with P value < 0.001 considered as highly significant (P value is given in Supplementary Table 2). ‘***’denotes P < 0.001, ‘**’denotes P < 0.01. For each individual parameter, means that do not share a letter are significantly different. P coumarate content expressed as a fraction of the total lignin aromatic units (H + G + S).
Lignin content and composition
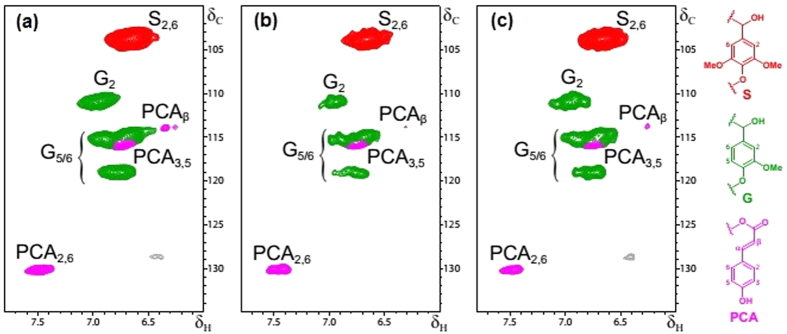
To estimate if the lignin content was reduced as an effect of downregulation of genes under consideration after incorporation of hpRNA, amount of lignin was estimated by the Klason lignin method. For measuring lignin reduction, three generations of the two lines (CM P1-P2 and CH P1-P2) of each transgenic plant type were taken into consideration and compared to the control. Samples used were randomly selected ensuring two biological replicates with technical triplicates of T1 - T3 generations. Different constituents of lignin were determined by 2D NMR spectroscopy (Fig. 5). Substantial decrease in the lignin content was found for both transgenic type in comparison to the control. On an average, 16% reduction in the lignin content was found for the whole stem of the COMT -hpRNA- line and about 13.5% for the fiber alone when compared with wild type data. These COMT-hpRNA lines showed almost similar S/G ratio to that of the wild type. As for C4H -hpRNA- line, lignin reduction was calculated to be about 22% for the whole stem and an average of 14% for the fiber. The S/G ratio was found to increase in these lines in comparison to the wild type. Over all, C4H-hpRNA lines showed more decrease in lignin content than the COMT-hpRNA lines. S/G ratios were considerably different, up to the T2 generations for both transgenic plant types. However, T3 generations of these plants had an S/G ratio similar to that of the wild type.
Figure 5.
2D-HSQC-NMR spectra of the aromatic/unsaturated regions from whole jute stems representative of (a) wild type plants, (b) C4H-hpRNA lines, and (c) COMT-hpRNA lines. Main cross-signals in the aromatic/unsaturated region (δC/δH 100–135/6.0–7.8) of the HSQC spectra corresponded to the aromatic rings and unsaturated side-chains of the different guaiacyl (G) and syringyl (S) lignin units, and to p-coumarates (PCA) that are associated to the lignin. Main lignin units detected in the aromatic region of the 2D HSQC NMR spectra of jute plants are displayed in the right side: G, guaiacyl units; S, syringyl units; PCA, p-coumarates.
Effect on cellulose content
In our study, both types of transgenic lines showed a considerable increase in the cellulose content in all three generations. This increase was up to 4% for COMT- hpRNA lines compared to the wild type with a concomitant increase in cellulose-to-lignin ratio (Table 2). In C4H- hpRNA lines, cellulose content on an average increased 3.5%, together with an increase in cellulose/lignin ratio. Modification induced by reduced expression of COMT- led to substantially more accumulation of cellulose than lines with reduced C4H expression. However, cellulose-to-lignin ratio was better in C4H- hpRNA, because of the lower lignin content than that of COMT- hpRNA lines.
Reduced recalcitrance in transgenic plants
Efficiency of glucose release (as a parameter of reduced recalcitrance) was measured for the transgenic lines. With reduced C4H transcript level, considerably more glucose was released (an increase of about 2%) from these transgenic lines than the wild type (Table 2). However, for COMT-hpRNA lines the amount of glucose release was almost the same as the control.
Discussion
The current trend of altering lignin biosynthetic pathway with a view to reducing the amount of lignin is largely effective in enhancing the profitable usability of lignified plant-based resources. Jute, a ligno-cellulose-rich fiber crop, delineates a new sphere with respect to lignin manipulation. In the light of several literature reports5,22,23, it can be assumed that lignin engineering can reinforce the use of jute as a sustainable source of bio-based materials for commercial purposes (viz. textile, paper and pulping, biofuel). This study emphasized on reducing the lignin content in jute, using the RNAi technique (siRNA) for fine-tuned alteration of the lignin biosynthetic pathway.
RNA interference (RNAi) is based on sequence-specific RNA degradation that follows formation of double-stranded RNA (dsRNA) homologous in sequence to the target gene24. RNAi vector mediated gene silencing has increased the efficiency of genetic manipulation in plants because of its ease of application and possibilities for genome-wide reverse genetics25. With the advent of gene silencing techniques, many studies have shown that efficient repression of target genes can be induced by expressing self-complimentary hpRNA constructs, known as hpRNAi26. Gene constructs encoding intron spliced RNA with a self-complementary hairpin (hp) structure are known to induce post-transcriptional gene silencing with almost 100% efficiency when directed against viruses or endogenous genes and transgenes27. Because the method of RNAi is particularly sequence specific, elucidation of the very sequence of jute genes was imperative. Due to unavailability of jute genome sequence in public database, first challenge of this study was to retrieve the full length sequence of the selected genes. Traditional method of gene walking was found to be effective because of sequence similarity of these genes among different species.
Lignin content and composition are two aspects that contribute to recalcitrance of a cell wall28. Among plants the lignin content can vary from 15% to 40%29. Deposition of lignin in cell walls is a crucial step in the adaptation of plants to a land habitat, as such, plants tolerate up to 40% reduction in lignin without major adverse effects on normal plant growth and development in greenhouse conditions30. Lignin is measured by the Klason method which determines the total residue remaining after removal of cell wall polysaccharides by sulfuric acid31. We observed significant reduction in lignin content in both types of transgenic plants (C4H-hpRNA and COMT-hpRNA lines) (Table 2). Effect of down regulating COMT expression has previously been reported for transgenic lines of tobacco23, maize32, alfalfa33, switchgrass34 etc. in which down regulation of COMT expression was found to accompany a reduction in the lignin amount, consistent with our findings. Decreasing C4H gene expression in tobacco35, alfalfa6 and eucalyptus36 showed a similar reduction in the lignin content.
Lignin is composed of a combination of sinapyl (S), coniferyl (G), and 4-coumaryl (H) alcohol subunits which polymerize with each other10. Composition of lignin varies widely based on plant species and subunit availability. Relative levels of S, G, and H lignin subunits are expressed as the S/G/H ratio. Since, H subunits are in insignificant amount in woody species37 S/G ratio is therefore the general expression of lignin subunits. In addition to the three main monolignols, lignin contains small amount of units from incomplete monolignol biosynthesis and integrates various other phenylpropanoid units, such as hydroxycinnamyl aldehydes, acetates, p-coumarates, p-hydroxybenzoates, and tyramine ferulates38. Since the polymerization of monolignols is random, lignin composition is highly plastic and amenable to manipulation39. Lignins rich in G units has relatively more carbon-carbon bonds than lignin rich in S lignin, as a consequence, lignins essentially made of G units are less susceptible to delignification than lignins made of S units40. Changes in lignin monomer composition are most readily visualized from the aromatic regions of two-dimensional NMR (HSQC) 1H–13C correlation signals41. Increasing the S-to-G ratio by altering lignin biosynthesis related gene expression can make a cell wall somewhat easier to degrade42.
The S/G ratio in this study was found to increase in C4H-hpRNA lines as opposed to other experiments of downregulation of C4H gene in tobacco35, alfalfa6 and eucalyptus36, in which C4H downregulation showed a drop in S/G ratio. The discrepancy between C4H downregulation and corresponding change in S/G ratio has been enigmatic to researchers. Because of its relative position in the pathway, the S/G ratio is expected to rise when C4H expression is lowered. In contrast to other reports artificial downregulation of C4H in jute supported the expected rise in the S/G ratio. The increase in S/G ratio in the transgenic lines of C4H-downregulated plants is due to an increase in the S lignin level (Table 2). Differences in lignin composition in C4H-hpRNA transgenic lines in jute and other plants cannot be linearly described because of the metabolic complexity of lignin biosynthetic pathway. Although this pathway has been studied for more than a century now, there are still labyrinths that are not fully understood9. Variation in S/G lignin ratio as an effect of downregulation of C4H in jute and other plants may not be purely the result of a change in the overall fluctuation of the pathway determined by the extent of downregulation of the individual enzymes. Metabolic channeling may function at different stages of the phenylpropanoid biosynthetic pathway43, although there is no direct confirmation yet of the presence of separate metabolic channels leading to G and S lignin44. It is also possible that there are isoforms of C4H present in jute as reported for oilseed rape45, orange46, poplar47 and white popinac48 which influence the metabolic steps of the pathway. Recently a predictive kinetic metabolic-flux (PKMF) model has been proposed and validated in poplar which shows that two paralogs of C4H are kinetically distinct with different Km and Kcat values and contribute differentially to the metabolic-flux converting cinnamic acid to 4-coumaric acid; this model with experimental validations showed that a reduction in C4H expression, resulted in low lignin and a high S/G ratio49. An increase in p-coumarate in transgenic generations of C4H-hpRNA line (Table 2) suggests that although C4H, responsible for biosynthesis of p-coumarate is down-regulated, a second pathway responsible for altering lignin composition could possibly be active.
An increase in the amount of cellulose and a doubled cellulose to lignin ratio have been reported for most severely lignin-reduced transgenic trees of aspen50 and poplar51. In our study, the transgenic lines showed an increased level of cellulose content (Table 2). This might be due to relative locations of the enzymes in the lignin biosynthetic pathway which influence the associated surge in cellulose content because it is assumed that, cellulose synthesis is normally substrate limited and reducing the flow at different stages of the lignin pathway increases the availability of carbon for cellulose deposition50.
Reduced recalcitrance may be referred to as an escalation in sugar release after enzymatic saccharification relative to the control samples under a defined pretreatment condition. In other words, it is an increase in the digestibility of polysaccharide components to liberate sugar22. There are many reports indicating that biomass digestibility is negatively correlated with lignin concentration. Genetic and transgenic approaches to manipulate these parameters have resulted in varying degrees of success with regard to improving cell wall digestibility6,52. Caffeic acid 3-O-methyltransferase (COMT) downregulation has led to increased saccharification in tobacco53, sorghum54, alfalfa33 and switchgrass34. Positive effect of C4H downregulation in saccharification and glucose release has also been reported in eucalyptus. For jute in our case, the amount of glucose released from the different transgenic plants (C4H-hpRNA and COMT-hpRNA lines) may be attributed to S/G ratios of the same. S/G ratio influences biomass digestibility with S lignin being favorable for hydrolysis because of less crosslinking compared to G lignin53. Between the two, C4H-hpRNA lines showed a decrease in lignin content, increase in S/G ratio and consequently better sugar release compared to COMT-hpRNA lines which showed a drop in the S lignin content. However, improvement in digestibility was not as large as has been seen for other plants. Our findings are in line with a report which documents that reduced lignin does not substantially improve the saccharification potential of transgenic poplars55, ascribed to complex biological aspect of plants which includes increase in phenolics as a consequence of perturbation of the phenylpropanoid pathway.
One remarkable observation of this study is that, different generations of both transgenic plant types showed no vital change in their phenotypic features (Table 1). This finding is of much significance, because the popularity of lignin engineering is frequently questioned due to observed growth retardation of transgenic plants with reduction in the percentage of their biomass56. For example in Arabidopsis57 C4H downregulation had resulted in dwarfism. In this respect, jute has been found to be more robust to lignin perturbation, emphasizing its potential industrial applications.
Conclusion
In an earlier study, we had used artificial miRNA for manipulating the lignin pathway in order to reduce lignin content in jute by targeting two genes (coumarate 3- hydroxylase, C3H and ferulate 5-hydroxylase, F5H) different from the ones used in this study58. In the present study we have independently downregulated two other genes of the lignin biosynthetic pathway (COMT and C4H) by RNAi. Results show significant downregulation of transcripts as well as lignin content in the transgenic lines with an associated increase in cellulose and slight improvement in digestibility. UV fluorescence microscopy of stem cross sections of transgenic lines corroborate the biochemical estimation of lignin content. Even though digestibility testing does not show much improvement in release of sugars as compared to other transgenic plants, it supports the conventional theory of S/G ratio with respect to effective enzymatic saccharification. It is also possible that plants with different genetic make-ups have different responses to the same manipulation22. These data suggest that downregulation of lignin biosynthetic genes may lower the lignin content but the consequences of decreased lignin may not be the same in all modified plants. Discrepancies in the significances of such modifications appear to be rife.
Methods
Plant material
Seeds of Corchorus olitorius (var. 0–9897), one of the two cultivated jute species, were collected from the Physiology Department, Bangladesh Jute Research Institute (BJRI), Dhaka.
Gene identification approach
Full length sequences of the two genes (C4H and COMT) used in the study were retrieved by using PCR based walking method. At first, parts of the two genes were identified using degenerate primers designed from conserved domains of the same genes from different plants followed by PCR amplifications, TA cloning (Invitrogen, USA) of the amplicons according to the manufacturer’s instruction, and sequencing. On the basis of the first sequence data, jute specific primers for particular genes were designed and gene walking approach was then followed from subsequent sequence data to extract the whole sequence of the two genes.
hpRNAi constructs
Once the sequence was known, RNAi primers were designed from the conserved coding region to amplify the products. COMT-RNAi and C4H-RNAi products were amplified (primer sequences can be found in Supplementary Table-1) followed by TA vector mediated cloning to determine the RNAi amplicons (Supplementary Fig. 2). Then these amplicons were incorporated into pENTER11 plasmids and the constructs were confirmed by colony PCR and restriction digestion with enzymes (BamHI/XhoI), the restriction sites for which are present in the constructs. Finally, the recombinant GATEWAY™ pENTER11 plasmids (COMT-pENTER11 and C4H-pENTER11) were mobilized into GATEWAY™ pK7GWIWG (II)59 destination vector using LR reaction (Supplementary Fig. 3.1, 3.2) in order to introduce the hairpin construct into the same.
Agrobacterium mediated in-planta transformation
The hairpin constructs were then introduced into C. olitorius var 0–9897 by using a tissue culture independent Agrobacterium tumefaciens mediated gene transformation method21. Shoot tips of young jute plants (15–20 cm in height) were pricked with a fine needle. After an hour, the injured region was infected with a few drops of A. tumefaciens suspension, having an O.D. of 0.80 at 600 nm in YMB (Yeast Mannitol Broth) medium, containing the respective constructs. This was followed by a second infection in the same region an hour later. The infected plants were then incubated in dark for 3 days at 28 °C. After that the plants were grown under normal conditions of light.
Genomic DNA PCR
The presence of the transgenes in the putative transformants (COMT -hpRNA lines and C4H –hpRNA lines) was assessed using genomic DNA from leaves of transformed (kanamycin-resistant) and non-transformed jute plants isolated by using the CTAB (N-Cetyl-N, N, N-trimethylammonium bromide) method. The presence of the reporter gene NPT(II), was identified by performing PCR using the primers, NPT (II) forward (5′-CCGTAAAGCACGAGGAAGTC-3′) and NPT (II) reverse (5′-ATGGGGATT GAACAAGATGG-3′). The PCR reaction included initial denaturation at 95 °C for 5 min followed by 30 cycles at 94 °C for 1 min, annealing at 56 °C for 30 s and extension at 72 °C for 30 s. The program ended with a final extension step for 7 min at 72 °C. Amplification products of approximately 350 bp were analyzed on a 0.8% agarose gel.
Southern blot analysis
20 μg of genomic DNA from PCR-positive hpRNAi lines for COMT and C4H together with the wild type control were digested with BamHI/HindIII, electrophoresed and blotted on to Hybond N + membranes (GE Healthcare Life Sciences, UK) by overnight capillary transfer.
A 250 bp hpRNAi construct was used as a probe and purified in a G25 column (GE Healthcare Life Sciences, UK) according to the supplier’s protocol. The probes were labeled with [α32P] dCTP (PerkinElmer Life Sciences, USA). After hybridization for 20 h at 68 °C, the membrane was washed once with 2X saline-sodium citrate (SSC) containing 0.1% sodium dodecyl sulfate (SDS) at 60 °C for 20 min, then washed with 0.5X SSC containing 0.1% SDS at 60 °C for 30 min and finally washed with 2X SSC at room temperature and stored until exposure. The membranes were subjected to autoradiography using a TYPHOON phosphor imager (GE Healthcare Life Sciences, UK).
Northern blot analysis
Total RNA was extracted from wild type and kanamycin screened 0–9897 jute seedlings of hpRNA transgenic lines (COMT & C4H) using the guanidium thiocynate extraction method. For northern blot of the transcripts 30 μg of total RNA from each plant sample was resolved on a 1.2% formaldehyde-agarose gel and transferred on to Hybond N + membranes (GE Healthcare Life Sciences, UK). The probes that spanned the entire coding sequence of the corresponding genes were prepared using [α32P] dCTP labelled cDNA of COMT and C4H genes individually.
For northern blot of hpRNA, total RNA was isolated from non-transgenic and transgenic hpRNA (COMT and C4H) lines using TRIZOL reagent and following the manufacturer’s protocol (Invitrogen). 20 μg of each sample was resolved on a 12% denaturing urea-PAGE gel. The RNA was then blotted onto a Hybond-N + membrane (GE Healthcare Life Sciences, UK) by Semi-Dry Transfer Cell (Bio-Rad Laboratories, Inc, CA). Probes were prepared using [α32P] dCTP labelled cDNAs that enclosed the respective hpRNAi amplicon specific sequences of COMT and C4H genes. Probes were purified in a G25 column (GE Healthcare Life Sciences, UK) according to the supplier’s protocol. Hybridization was carried out at 42 °C using a standard protocol. The membranes were subjected to autoradiography using a TYPHOON phosphor imager (GE Healthcare Life Sciences, UK).
Reverse transcriptase polymerase chain reaction (RT-PCR) analysis
Approximately 2 μg of total RNA from non-transgenic and transgenic jute seedlings was used for first-strand cDNA synthesis, followed by gene specific PCR. cDNA was prepared in a 20 μL reaction volume, which contained 2 μL of reverse transcriptase (200 U/μL), using SuperScriptTM II reverse transcriptase kit (Invitrogen, USA) according to the manufacturer’s manual. The reaction was carried out with an initial incubation at 50 °C for 50 min, then 5 min at 85 °C to inactivate the enzyme. Next RNAse H was added followed by an incubation at 37 °C for 20 min to remove the cDNA-RNA hybrid. 1 μL of the cDNA sample was used as a template for PCR amplification with COMT GSP forward/COMT GSP reverse and C4H GSP forward /C4H GSP reverse primers (Supplementary Table 1) to amplify the respective samples. PCR of the same cDNA samples was carried out using gene-specific primers for actin to be used as a loading control in gel electrophoresis. All PCRs included 30 cycles of denaturation at 95 °C for 30 s, 40 s at the optimal temperature for each gene specific primer set (Supplementary Table 1) for annealing and 40 s at 72 °C for extension. RT-PCR band intensities were quantified using the ImageJ software (http://rsbweb.nih.gov/ij/index.html).
Lignin content
Lignin content of jute whole stem and fiber was measured to determine the amount of acid insoluble lignin in jute for both transgenic and non-transgenic plants. To get the average lignin value of a plant, only the mid-section (~16 cm) of the stem was used in this study. A modified method of ribbon retting was followed for isolating the jute fibers60. Briefly, green ribbons (outer skin) of jute were stripped manually from barks of mature plants and submerged for 35 days in a tank in 1:10 ribbon to water ratio using water from the city’s regular supply. Next the fibers were washed and dried for downstream chemical measurements. A modified Klason lignin estimation method was used to estimate acid insoluble lignin or AIL as described by Tanmoy et al.61.
Lignin composition
Lignin composition was determined from integration of the 13C-1H correlation signals in the 2D-NMR Heteronuclear Single Quantum Coherence (HSQC) of whole stems. Around 50 mg of finely divided (ball-milled) jute whole stem was swollen in 0.75 mL of Dimethyl sulfoxide-d6 (DMSO-d6) according to the method previously described62. 2D HSQC spectra were recorded at 25 °C on a Bruker AVANCE III 500 MHz instrument equipped with a cryogenically-cooled 5 mm TCI gradient probe with inverse geometry. 2D 13C-1H correlation spectra were carried out using an adiabatic HSQC pulse program (Bruker standard pulse sequence ‘hsqcetgpsisp2.2′) and the following parameters: spectra acquired from 10 to 0 ppm in F2 (1H) using 1000 data points for an acquisition time of 140 ms, an interscan delay of 1 s, and from 165 to 0 ppm in F1 (13C) using 256 increments of 32 scan, for a total acquisition time of 2 h 40 min. The 1JCH used was 145 Hz. Processing used typical matched Gaussian apodization in 1H and a squared cosine bell in 13C. The central solvent peak was used as an internal reference (δC/δH 39.5/2.49). 2D-NMR cross-signals were assigned by literature comparison. A semi quantitative analysis of the volume integrals of the HSQC correlation peaks was performed using Bruker’s Topspin 3.2 processing software. In the aromatic/unsaturated region, C2/H2 correlations from H, G, and S lignin units and from p-coumarates were used to estimate their relative abundances.
Histochemistry and fluorescence microscopy
Lignified tissues were identified in the cross sections through histochemistry and fluorescence microscopy. Cross sectioning of jute stem samples was done using a microtome (Thermo Scientific™ HM 325) and sections were stained with phloroglucinol and observed under a light microscope (Nikon ECLIPES 50i) with a magnification of 100X. Presence of lignin was considered when the tissues were stained red. The fluorescence images were captured with a CCD (charged-couple device) camera.
Cellulose content
Cellulose content of both wildtype and transgenic plants was determined using cold anthrone reagent63. Samples were taken from the whole stem. They were first digested by an 80% acetic acid: 100% nitric acid (8:1) solution and the precipitates were digested with 72% sulphuric acid. Addition of anthrone resulted in a green colored solution which gave an absorbance at 620 nm.
Enzymatic saccharification
For enzymatic saccharification, first the neutral detergent fiber (NDF) was prepared64 and was then incubated for 24 h at 37 °C using commercial cellulase (Sigma, Source : Aspergillus niger) in 0.1 N sodium acetate buffer, pH 4.8, at 80 FPU/g NDF and measurement of reducing sugars was made by using dinitrosalicylic acid at 540 nm65.
Statistical analysis
Five randomly selected plants of each generation of the selected lines were considered for phenotypical analyses. For lignin and cellulose content and enzymatic saccharification, two randomly selected biological replicates and technical triplicates of all three generations of each plant line for both transgenic plant types (CM P1-P2 and CH P1-P2) were taken into account. For lignin composition study, technical triplicates were taken from each plant line. Standard errors were calculated by using Microsoft excel tool pack 2013. For each parameter, the means, one way ANOVA test and Tukey’s test (for evaluating differences in each plant type) were performed using the R program. The alpha value was set at 0.001 (99.99% confidence). P value < 0.001 was considered as significant.
Additional Information
How to cite this article: Shafrin, F. et al. Modification of Monolignol Biosynthetic Pathway in Jute: Different Gene, Different Consequence. Sci. Rep. 7, 39984; doi: 10.1038/srep39984 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
Farhana Shafrin gratefully acknowledges the Bangladesh Academy of Sciences (BAS) for the fellowship she received as a Ph.D Research Fellow. The authors would like to thank Prof. Sudhir K Sopory and V S Chauhan of ICGEB, New Delhi for providing Farhana Shafrin the facilities required for her stay and work at the ICGEB. The authors also thank BJRI (Bangladesh Jute Research Institute) for providing the jute seeds. Authors acknowledge the contribution of Nadim Ashraf in designing the experiment and Aubhishek Zaman for his help in in planta transformation. The research was supported by a financial grant received from the BAS-USDA-PALS Program.
Footnotes
Author Contributions F.S. and H.K. designed the experiment. F.S. performed the experimental research, data analysis and manuscript writing. ASF performed the manuscript writing with F.S. H.K., J.C. del R and N Sanan-Mishra performed the manuscript editing. R.A. worked with F.S. for construct validation. R.A. and S.K.S. contributed in in planta transformation experiments. K.H. performed Klason lignin estimation. N Sanan-Mishra helped with expression scrutiny, northern and Southern blot analysis of samples. J.R., A.G. and J.C. del R determined the lignin composition. A.S.F., A.A., M.S. and F.S. performed the cellulose and enzymatic saccharification assay and statistical analysis. M.S. helped F.S. with the histochemical assay. All authors have approved the final manuscript for publication.
References
- Kim S. & Dale B. E. Life cycle assessment of various cropping systems utilized for producing biofuels: Bioethanol and biodiesel. Biomass Bioenergy 29, 426–439 (2005). [Google Scholar]
- Prasad A., Sotenko M., Blenkinsopp T. & Coles S. R. Life cycle assessment of lignocellulosic biomass pretreatment methods in biofuel production. Int. J. Life Cycle Assess. 21, 44–50 (2016). [Google Scholar]
- Taherzadeh M. J. & Karimi K. Acid-based hydrolysis processes for ethanol from lignocellulosic materials: a review. BioResources 2, 472–499 (2007). [Google Scholar]
- Greene N. In Growing Energy: How Biofuels Can Help End America’s Oil Dependence (Natural Resources Defence Council, 2004). [Google Scholar]
- Kuhad R. C. & Singh A. Lignocellulose biotechnology: current and future prospects. Crit. Rev. Biotechnol. 13, 151–172 (1993). [Google Scholar]
- Chen F. & Dixon R. A. Lignin modification improves fermentable sugar yields for biofuel production. Nat. Biotechnol. 25, 759–761 (2007). [DOI] [PubMed] [Google Scholar]
- Aden A. & Foust T. Technoeconomic analysis of the dilute sulfuric acid and enzymatic hydrolysis process for the conversion of corn stover to ethanol. Cellulose 16, 535–545 (2009). [Google Scholar]
- Ragauskas A. J. et al. The path forward for biofuels and biomaterials. Science 311, 484–489 (2006). [DOI] [PubMed] [Google Scholar]
- Boerjan W., Ralph J. & Baucher M. Lignin biosynthesis. Annu. Rev. Plant Biol. 54, 519–546 (2003). [DOI] [PubMed] [Google Scholar]
- Baucher M., Monties B., Montagu M. V. & Boerjan W. Biosynthesis and genetic engineering of lignin. Crit. Rev. Plant Sci. 17, 125–197 (1998). [Google Scholar]
- Chabannes M. et al. Strong decrease in lignin content without significant alteration of plant development is induced by simultaneous down‐regulation of cinnamoyl CoA reductase (CCR) and cinnamyl alcohol dehydrogenase (CAD) in tobacco plants. Plant J. 28, 257–270 (2001). [DOI] [PubMed] [Google Scholar]
- Samantab A. Effect of selective chemical treatments of jute fibre on textile-related properties and processibility. Indian J. Fibre Text. 29, 85–99 (2004). [Google Scholar]
- Uddin M., Ullah S., Shahinur S. & Kaysar M. A. Study on the chemical modification process optimization and spinning properties of jute fibre. Indian J. of Nat. Fibers 2, 155–164 (2015). [Google Scholar]
- Wazni M. W., Islam A. S., Taliaferro J. M., Anwar N. & Sathasivan K. Novel ESTs from a jute (Corchorus olitorius L.) cDNA library. Plant Tissue Cult. Biotechnol 17, 173–182 (2007). [Google Scholar]
- Wesley S. V. et al. Construct design for efficient, effective and high‐throughput gene silencing in plants. Plant J. 27, 581–590 (2001). [DOI] [PubMed] [Google Scholar]
- Bonawitz N. D. & Chapple C. The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu. Rev. Genet. 44, 337–363 (2010). [DOI] [PubMed] [Google Scholar]
- Humphreys J. M. & Chapple C. Rewriting the lignin roadmap. Curr. Opin. Plant Biol. 5, 224–229 (2002). [DOI] [PubMed] [Google Scholar]
- Bout S. & Vermerris W. A candidate-gene approach to clone the sorghum Brown midrib gene encoding caffeic acid O-methyltransferase. Mol. Genet. Genomics 269, 205–214 (2003). [DOI] [PubMed] [Google Scholar]
- Chapple C., Ladisch M. & Meilan R. Loosening lignin’s grip on biofuel production. Nat. Biotechnol. 25, 746–748 (2007). [DOI] [PubMed] [Google Scholar]
- Samira R., Moosa M. M., Alam M. M., Keka S. I. & Khan H. ‘In silico’ analysis of jute SSR library and experimental verification of assembly. Plant Omics 3(2), 57–65 (2010). [Google Scholar]
- Sajib A. A. et al. Tissue culture independent transformation for Corchorus olitorius. Plant Cell Tiss. Org. Cult. 95, 333–340 (2008). [Google Scholar]
- Li X., Weng J. K. & Chapple C. Improvement of biomass through lignin modification. Plant J. 54, 569–581 (2008). [DOI] [PubMed] [Google Scholar]
- Sewalt V. J., Ni W., Jung H. G. & Dixon R. A. Lignin impact on fiber degradation: increased enzymatic digestibility of genetically engineered tobacco (Nicotiana tabacum) stems reduced in lignin content. J. Agric. Food Chem. 45, 1977–1983 (1997). [Google Scholar]
- Baulcombe D. RNA silencing in plants. Nature 431, 356–363 (2004). [DOI] [PubMed] [Google Scholar]
- Travella S., Klimm T. E. & Keller B. RNA interference-based gene silencing as an efficient tool for functional genomics in hexaploid bread wheat. Plant Physiol. 142, 6–20 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson J. M., Fusaro A. F., Wang M. & Waterhouse P. M. RNA silencing platforms in plants. FEBS Letters 579, 5982–5987 (2005). [DOI] [PubMed] [Google Scholar]
- Smith N. A. et al. Gene expression: total silencing by intron-spliced hairpin RNAs. Nature 407, 319–320 (2000). [DOI] [PubMed] [Google Scholar]
- Vanholme R. et al. Metabolic engineering of novel lignin in biomass crops. New Phytol. 196, 978–1000 (2012). [DOI] [PubMed] [Google Scholar]
- Sarkanen K. V. & Ludwig C. H. In Lignins: Occurrence, Formation, Structure And Reactions. (John Wiley and Sons, New York, 1971). [Google Scholar]
- Zhong R., Morrison W. H., Himmelsbach D. S., Poole F. L. & Ye Z.-H. Essential role of caffeoyl coenzyme A O-methyltransferase in lignin biosynthesis in woody poplar plants. Plant Physiol. 124, 563–578 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean J. F. Lignin analysis in Methods In Plant Biochemistry And Molecular Biology, 199–215 (1997). [Google Scholar]
- Piquemal J. et al. Downregulation of caffeic acid O-methyltransferase in maize revisited using a transgenic approach. Plant Physiol. 130, 1675–1685 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D., Chen F., Inoue K., Blount J. W. & Dixon R. A. Downregulation of caffeic acid 3-O-methyltransferase and caffeoyl CoA 3-O-methyltransferase in transgenic alfalfa: impacts on lignin structure and implications for the biosynthesis of G and S lignin. The Plant Cell 13, 73–88 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C. et al. Genetic manipulation of lignin reduces recalcitrance and improves ethanol production from switchgrass. Proc Natl Acad Sci USA 108, 3803–3808 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewalt V. J. et al. Reduced lignin content and altered lignin composition in transgenic tobacco down-regulated in expression of L-phenylalanine ammonia-lyase or cinnamate 4-hydroxylase. Plant Physiol. 115, 41–50 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes R. W. et al. Downregulation of p-coumaroyl quinate/shikimate 3′-hydroxylase (C3′ H) and cinnamate 4-hydroxylase (C4H) genes in the lignin biosynthetic pathway of Eucalyptus urophylla × E. grandis leads to improved sugar release. Biotechnol. Biofuels 8, 1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R., Demedts B., Morreel K., Ralph J. & Boerjan W. Lignin biosynthesis and structure. Plant Physiol. 153, 895–905 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sederoff R. R., MacKay J. J., Ralph J. & Hatfield R. D. Unexpected variation in lignin. Curr. Opin. Plant Biol. 2, 145–152 (1999). [DOI] [PubMed] [Google Scholar]
- Loqué D., Scheller H. V. & Pauly M. Engineering of plant cell walls for enhanced biofuel production. Curr. Opin. Plant Biol. 25, 151–161 (2015). [DOI] [PubMed] [Google Scholar]
- Lapierre C. et al. Structural alterations of lignins in transgenic poplars with depressed cinnamyl alcohol dehydrogenase or caffeic acid O-methyltransferase activity have an opposite impact on the efficiency of industrial kraft pulping. Plant Physiol. 119, 153–164 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson N. A. et al. Manipulation of guaiacyl and syringyl monomer biosynthesis in an Arabidopsis cinnamyl alcohol dehydrogenase mutant results in atypical lignin biosynthesis and modified cell wall structure. The Plant Cell 27, 2195–2209 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart J. Altering Lignin Composition to Improve Biofuel Production. The Plant Cell 27, 2082–2082 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel B. S. Metabolic channeling in plants. Annu. Rev. Plant Biol. 55, 85–107 (2004). [DOI] [PubMed] [Google Scholar]
- Chen F. et al. Multi‐site genetic modulation of monolignol biosynthesis suggests new routes for formation of syringyl lignin and wall‐bound ferulic acid in alfalfa (Medicago sativa L.). Plant J. 48, 113–124 (2006). [DOI] [PubMed] [Google Scholar]
- Chen A.-H., Chai Y.-R., Li J.-N. & Chen L. Molecular cloning of two genes encoding cinnamate 4-hydroxylase (C4H) from oilseed rape (Brassica napus). BMB Reports 40, 247–260 (2007). [DOI] [PubMed]
- Betz C., McCollum T. G. & Mayer R. T. Differential expression of two cinnamate 4-hydroxylase genes in ‘Valencia’ orange (Citrus sinensis Osbeck). Plant Mol. Biol. 46, 741–748 (2001). [DOI] [PubMed] [Google Scholar]
- Ro D. K., Mah N., Ellis B. E. & Douglas C. J. Functional characterization and subcellular localization of poplar (Populus trichocarpa × Populus deltoides) cinnamate 4-hydroxylase. Plant Physiol. 126, 317–329 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Omer S., Patel K. & Khan B. M. Cinnamate 4-Hydroxylase (C4H) genes from Leucaena leucocephala: a pulp yielding leguminous tree. Mol. Biol. Rep. 40, 1265–1274 (2013). [DOI] [PubMed] [Google Scholar]
- Wang J. P. et al. Complete proteomic-based enzyme reaction and inhibition kinetics reveal how monolignol biosynthetic enzyme families affect metabolic flux and lignin in Populus trichocarpa. The Plant Cell 26, 894–914 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W.-J. et al. Repression of lignin biosynthesis promotes cellulose accumulation and growth in transgenic trees. Nat. Biotechnol. 17, 808–812 (1999). [DOI] [PubMed] [Google Scholar]
- Wang H., Xue Y., Chen Y., Li R. & Wei J. Lignin modification improves the biofuel production potential in transgenic Populus tomentosa. Ind. Crops Prod. 37, 170–177 (2012). [Google Scholar]
- Van Acker R. et al. Lignin biosynthesis perturbations affect secondary cell wall composition and saccharification yield in Arabidopsis thaliana. Biotechnol Biofuels 6, 46 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vailhé M. A. B. et al. Effect of modification of the O‐methyltransferase activity on cell wall composition, ultrastructure and degradability of transgenic tobacco. J. Sci. Food Agric. 72, 385–391 (1996). [Google Scholar]
- Sattler S. E. et al. Identification and characterization of four missense mutations in brown midrib 12 (Bmr12), the caffeic O-methyltranferase (COMT) of sorghum. BioEnergy Research 5, 855–865 (2012). [Google Scholar]
- Voelker S. L. et al. Antisense downregulation of 4CL expression alters lignification, tree growth, and saccharification potential of field-grown poplar. Plant Physiol. 154, 874–886 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonawitz N. D. & Chapple C. Can genetic engineering of lignin deposition be accomplished without an unacceptable yield penalty? Curr. Opin. Biotechnol. 24(2), 336–343 (2013). [DOI] [PubMed] [Google Scholar]
- Schilmiller A. L., Stout J. et al. Mutations in the cinnamate 4‐hydroxylase gene impact metabolism, growth and development in Arabidopsis. Plant J 60(5), 771–782 (2009). [DOI] [PubMed] [Google Scholar]
- Shafrin F., Das S. S., Sanan-Mishra N. & Khan H. Artificial miRNA-mediated downregulation of two monolignoid biosynthetic genes (C3H and F5H) cause reduction in lignin content in jute. Plant Mol. Biol. 89, 511–527 (2015). [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D. & Depicker A. GATEWAY™ vectors for Agrobacterium - mediated plant transformation. Trends Plant Sci. 7(5), 193–195 (2002). [DOI] [PubMed] [Google Scholar]
- Banik S. et al. Ribbon retting of jute-a prospective and eco-friendly method for improvemfent of fibre quality. Ind Crops Prod. 17(3), 183–190 (2003). [Google Scholar]
- Tanmoy A., Alum M., Islam M., Farzana T. & Khan H. Jute (Corchorus olitorius var. O-72) stem lignin: variation in content with age. Bangladesh J. Botany 43, 309–314 (2015). [Google Scholar]
- Rencoret J. et al. HSQC-NMR analysis of lignin in woody (Eucalyptus globulus and Picea abies) and non-woody (Agave sisalana) ball-milled plant materials at the gel state 10th EWLP, Stockholm, Sweden, August 25–28, 2008. Holzforschung, 63(6), 691–698 (2009). [Google Scholar]
- Updegraff D. M. Semimicro determination of cellulose inbiological materials. Anal. Biochem. 32, 420–424 (1969). [DOI] [PubMed] [Google Scholar]
- Van Soest P. V., Robertson J. & Lewis B. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 74, 3583–3597 (1991). [DOI] [PubMed] [Google Scholar]
- Miller G. L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31, 426–428 (1959). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.