Summary
Gene regulatory networks are pivotal for many biological processes. In mouse embryonic stem cells (mESCs) the transcriptional network can be divided into three functionally distinct modules: Polycomb, Core and Myc. The Polycomb module represses developmental genes, while the Myc module is associated with proliferative functions and its mis-regulation is linked to cancer development. Here, we show that in mESCs the Polycomb Repressive Complex 2 (PRC2) associated protein EPOP (Elongin BC and Polycomb Repressive Complex 2-associated protein; a.k.a. C17orf96, esPRC2p48 and E130012A19Rik) co-localizes at chromatin with members of the Myc and Polycomb module. EPOP interacts with the transcription elongation factor Elongin BC and the H2B deubiquitinase USP7 to modulate transcriptional processes in mESCs similar to MYC. EPOP is commonly up-regulated in human cancer and its loss impairs the proliferation of several human cancer cell lines. Our findings establish EPOP as a transcriptional modulator, which impacts both Polycomb and active gene transcription in mammalian cells.
Graphical abstract
Introduction
To execute specific cellular function, each cell type establishes a unique gene regulatory network, which consists of cell-type specific DNA binding transcription factors and other associated regulatory proteins (Karlebach and Shamir, 2008). These networks remain robust and resist subtle perturbations, but are still flexible enough that upon external cues a deterministic transition to a distinct transcriptional program can take place. Embryonic stem cells (ESCs) are the prime example of both robustness and flexibility. They are characterized by the unique ability of unlimited self-renewal while retaining the capacity to differentiate into all three germ layers (Ng and Surani, 2011). In mouse ES cells (mESC) three distinguishable transcriptional modules have been identified: Polycomb, Core and Myc (Kim et al., 2010). The Polycomb module represses developmental genes due to the function of Polycomb group proteins (PcG), such as the Polycomb Repressive Complex 2 (PRC2) (Voigt et al., 2013). The pluripotency factors NANOG, SOX2 and POU5F1 are required to establish the Core pluripotency network in mESCs to support self-renewal and to prevent differentiation (Ng and Surani, 2011). The Myc module is typically associated with proliferative capacity, metabolism as well as self-renewal (Eilers and Eisenman, 2008). It has been proposed that erroneous activation of the Myc module in somatic cells contributes to tumorigenesis and accounts for an ES cell-like transcriptional signature in cancer cells (Kim et al., 2010). A further understanding of the components and mechanisms of the distinct transcriptional modules and their interplay is expected to provide the basis to better utilize stem cells for clinical purposes and to decipher the complexity of tumorigenesis.
EPOP (Elongin BC and Polycomb Repressive Complex 2-associated protein) (a.k.a. C17orf96, esPRC2p48 and E130012A19Rik) (GeneID: 103551 (mouse); 100170841 (human))(UniprotID: Q7TNS8 (mouse); A6NHQ4 (human)) has recently been identified as an interacting partner of the mammalian PRC2 (Alekseyenko et al., 2014; De Cegli et al., 2013; Smits et al., 2013; Zhang et al., 2011) but its role and mechanism of action are poorly understood. It is highly expressed in ES cells, during embryogenesis and in the adult brain (Liefke and Shi, 2015) and has been proposed to contribute to somatic cell reprogramming (Zhang et al., 2011) and neuronal differentiation (De Cegli et al., 2013). We have found that EPOP represses PRC2 function in vivo, possibly by interfering with PRC2 chromatin binding via the interaction of its C-terminal region with the VEFS box of SUZ12 (Liefke and Shi, 2015). EPOP also occupies actively transcribed genes and we observed a genome-wide alteration of H3K4me3 distribution upon EPOP knockdown, suggesting a global regulatory role (Liefke and Shi, 2015). Indeed, additional published work proposed EPOP as a pluripotency factor, which acts as a central regulatory hub in mESCs (De Cegli et al., 2013; Klein et al., 2015), implicating a more general regulatory role, beyond its function at PRC2 target genes. However, due to its unstructured and unique nature its precise role remains largely unknown (Liefke and Shi, 2015).
Here we show that in mESCs, in addition to the Polycomb Group protein (PcG)- occupied chromatin regions, EPOP also localizes to sites of actively transcribed genes that are typically targeted by members of the Myc module, including genes with a broad H3K4me3 domain, which is a newly identified functional element in cells (Benayoun et al., 2014; Chen et al., 2015). Upon EPOP depletion, we find genome-wide altered distributions of H3K4me3 and RNA polymerase II, with the strongest effects seen at the broad H3K4me3 domains. Via biochemical purification we identify the transcription elongation factor Elongin BC as the most abundant interacting partner of EPOP, in addition to PRC2. Further experiments suggest that EPOP and Elongin BC may cooperate with the deubiquitinase USP7 to affect H2B ubiquitination at promoters, which could indirectly affect H3K4me3 and RNA Polymerase II-mediated transcription. Although EPOP only modestly impacts gene expression in mESCs and mES cell biology, alteration of EPOP expression leads to pronounced changes of the genomic H3K4me3 pattern and altered gene expression in cancer cells. Furthermore, EPOP is up-regulated in many cancer types and proliferation of human cancer cells is positively correlated with the levels of EPOP. Together, this work establishes EPOP as an unusual chromatin regulator, which, in addition to its PRC2-related role, modulates chromatin landscape at active genes and their transcription in mammalian cells.
Results
EPOP is a putative regulator at active and repressed genes in mESCs
During embryogenesis, the rise of specific cell lineages is associated with the activation of PcG repressed developmental genes (Voigt et al., 2013), and the removal or de novo formation of highly transcribed broad H3K4me3 domains, which are linked to the cellular identity (Figure S1A-D, Table S1) (Benayoun et al., 2014; Chen et al., 2015; Kareta et al., 2015). The mechanisms by which those opposing gene groups are balanced and regulated are not well understood. To gain further insights and to identify regulators, we first performed a comprehensive analysis of published ChIP-Seq data from chromatin regulatory proteins in mESCs (Table S2) and investigated which of those factors are enriched at genes possessing a broad H3K4me3 domain, at PcG target genes, or both. As expected, the majority of these factors are enriched either at broad H3K4me3 domains or PcG target genes, demonstrating distinct regulatory mechanisms of these two gene groups (Figure 1A). However, some factors such as the ERK2 kinase or the PcG and Trithorax group (TrxG) related proteins KDM2B and KDM5B are enriched at both, PcG target genes and genes with broad H3K4me3 domains (Figure 1A,B). Those factors may therefore be involved in coordinating the expression of those gene groups during embryogenesis. Interestingly, the still yet poorly characterized PRC2-interacting protein EPOP is also enriched at both groups, prompting us to investigate this protein further in this context (Figure 1A,B) (Liefke and Shi, 2015).
Figure 1. EPOP co-occupies targets of the Myc and Polycomb module in mESCs.
A) Enrichment of chromatin regulators (Table S2) at genes with broad H3K4me3 domains and PRC2 target genes, relative to all genes. The color of the dot indicates enrichment at broad H3K4me3 domains (green), PRC2 target genes (red), or both (yellow). Trithorax (TrxG) and Polycomb group (PcG) associated proteins are underlined in green or red, respectively. B) Examples at the Esrrb gene locus, which possesses a PcG inhibited TSS (TSS 1, red box) and a broad H3K4me3 domain (TSS 2, green box). C) Heatmaps of ChIP-Seq data from EPOP and members of the Myc, Core and Polycomb module. Three independent antibodies (Ab1, −2, −3) for EPOP were used. CGIs = CpG islands. D) Browser view of ChIP-seq data of EPOP with two different antibodies (Ab1, Ab3) in control and Epop KO cells in comparison to RNAPII (8WG16) (Lin et al., 2011), H3K4me3 (from control cells) and input. Examples for EPOP enrichment at active genes, broad H3K4me3 domains (grey box) and PcG target genes are show. E) Venn diagram of the overlap significant peaks of EPOP (Ab1 and Ab3, red) with H3K4me3 (green) and RNA polymerase II (blue) or EZH2 (purple) and SUZ12 (orange). See also Figure S1 and S2.
EPOP occupies target sites of the Myc and Polycomb module in mESCs
Using CRISPR/Cas9, we generated Epop knockout (KO) mES cell lines (Figure S2A), which we found to have unaltered self-renewal capability and morphology (Figure S2B,C), and performed ChIP-Seq experiments using three independent EPOP antibodies (Ab1,-2,-3) in control versus the KO cells (Figure S2D-I). A comparison with publicly available data (Table S2, S3) showed that the EPOP binding pattern correlates with promoter-associated members of the Myc and Polycomb modules, but not with the enhancer-associated members of the Core module (Figure 1C) (Kim et al., 2010; Whyte et al., 2013), supporting a role for EPOP at active and repressed gene promoters. Consistently, EPOP peaks overlap with RNA polymerase II and the active histone mark H3K4me3 as well as with the PRC2 members, SUZ12 and EZH2 (Figure 1D,E). About sixty to eighty percent of EPOP peaks overlap with RNA polymerase II or H3K4me3 peaks (Figure 1E), demonstrating that the majority of EPOP binds to actively transcribed genes. A transcription-related role of EPOP is further supported by the finding that inhibition of transcription using DRB (5,6-dichloro-1-β-D-ribofuranosyl-1H-benzimidazole) or triptolide led to a reduced EPOP protein level at chromatin (Figure S3A,B), suggesting that the chromatin association of EPOP is partially dependent on active transcription. We also found enrichment of EPOP at broad H3K4me3 domains, which are commonly targeted by the Myc module and often found at pluripotency and house-keeping genes in mESCs (Figure 1D, S1A, E, F, S2G,H, Table S1). In addition, we confirmed the previously observed reduction of H3K4me3 at those domains upon EPOP depletion (Liefke and Shi, 2015) with two independent Epop CRISPR KO clones via ChIP-qPCR (Figure S3C).
To address whether EPOP affects the chromatin landscape during mESCs differentiation processes, we performed H3K4me3 ChIP-qPCR experiments upon retinoic acid (RA) treatment in control and Epop KO mESCs. In the absence of EPOP, the H3K4me3 removal at two investigated broad H3K4me3 domains occurred faster (Figure S3D), while the increase of H3K4me3 at some PRC2 target genes was delayed (Figure S3E). These results suggest that, consistent with its localization, EPOP is involved in regulating both active and repressed genes during differentiation and that the presence of EPOP favors the deposition of the active H3K4me3 mark. However, we found only minor differences in the levels of differentiation markers between wildtype and Epop KO cells (Figure S3F), suggesting that loss of EPOP is not sufficient to significantly impair the differentiation program of mESCs under this experimental condition.
EPOP influences the distributions of H3K4me3 and RNA polymerase II in mESCs
We investigated the effects of EPOP on chromatin modifications by Western blotting, and observed a global reduction of the active H3K4me3 histone mark (Figure 2A) in the Epop KO cells, consistent with a potential positive role for EPOP in transcriptional regulation. To investigate this possibility in more detail at a genome-wide level, we performed ChIP-Seq for H3K4me3 and the serine 5 phosphorylated RNA polymerase II in Epop KO versus control mESCs (Figure 2B) and found that Epop KO led to an increase of RNA polymerase II and H3K4me3 at the transcription start sites at genes with and without broad H3K4me3 domain (Figure 2C,D), supporting a regulatory role of EPOP at most actively transcribed gene promoters. In contrast, the levels of either H3K4me3 or RNA polymerase II were often reduced downstream from the TSS of genes with broad H3K4me3 domains (Figure 2B-D), suggesting that at these domains EPOP may play a role during transcription elongation. Consistently, calculation of RNA polymerase II travelling ratios, a measure for the balance between transcriptional pausing and elongation (Benayoun et al., 2014), showed elevated pausing upon Epop KO (Figure 2E). This effect is, however, less pronounced than what has been observed upon inhibition of MYC (Rahl et al., 2010) (Figure S3H).
Figure 2. EPOP genome-wide influences H3K4me3 and RNA polymerase II transcription.
A) Western blots of histone modifications in Epop KO cells in comparison to control cells. Actin or H3 is shown as loading control. B) Browser view of ChIP-Seq data for EPOP (from control cells, Ab1) as well as H3K4me3 and serine 5 phosphorylated RNA polymerase II in control and Epop KO cells at two genes with broad H3K4me3 domain (Tet1, Esrrb). The black/grey profiles show the subtraction (KO - control). The grey boxes indicate broad H3K4me3 domains. C) Heatmaps of the differences for H3K4me3 and serine 5 phosphorylated RNA polymerase II, at active and PRC2 target genes. At active genes the heatmaps are sorted after the H3K4me3 breadth. D) Profiles of H3K4me3 and serine 5 phosphorylated RNA polymerase II at active genes with (“broad”, n = 1000) and without broad H3K4me3 domain (“non-broad”, n = 20,256), as well PRC2 target genes (n = 2008) in control and Epop KO cells. The P values were calculated by ANOVA. E) Comparison of the travelling ratios for RNA polymerase II S5 in control and Epop KO cells. See also Figure S3H. The P values were calculated by a Kolmogorov–Smirnov test. See also Figure S3.
EPOP directly interacts with the transcription elongation factor SIII subunit Elongin BC and the deubiquitinase USP7
We next wished to gain further insights into mechanisms by which EPOP affects transcriptional processes and H3K4me3 and reasoned that EPOP might carry out its function at actively transcribed genes by interacting with additional proteins, other than PRC2. To address this possibility, we purified N-terminal Flag-HA tagged human EPOP, from human HeLa-S cells and mouse EPOP from mESCs using a tandem affinity purification approach (Figure 3A). Associated proteins were identified by mass spectrometry (MS) (Figure 3B). Consistent with previous reports (Alekseyenko et al., 2014), we identified all core members of PRC2 (SUZ12, EZH1, EZH2, EED), RBBP4/7 as well as all 3 PCL homologs (PHF1, MTF2, PHF19), but not JARID2, suggesting that EPOP may bind to a subset of PRC2 that lacks JARID2. We further detected several peptides for the deubiquitinase USP7, in line with the previously proposed association of EPOP with USP7 (Alekseyenko et al., 2014). In addition, we found high peptides numbers for TCEB1 (Elongin C) and TCEB2 (Elongin B)(Figure 3B), which form a heterodimer commonly known as Elongin BC, reported to be involved in transcriptional regulation via interaction with Elongin A (Aso et al., 1995). However, no peptides for Elongin A were detected in our purifications. Via co-immunoprecipitation, we confirmed the interaction of endogenous EPOP with endogenous Elongin B, SUZ12 and USP7 in mESCs (Figure 3C). Interestingly, Elongin B co-immunoprecipitated SUZ12, and vice versa, suggesting that EPOP may be able to bridge PRC2 with Elongin BC.
Figure 3. EPOP interacts with the transcription Elongation factor Elongin BC and USP7.
A) Silver staining of tandem affinity purification of EPOP from HeLa-S and mESCs. B) Mass spectrometry analysis of the purified complexes from HeLa-S and mESCs, as well as three samples (#4, #9, #12) corresponding to the HeLa-S glycerol gradient fractions shown in F (red). The numbers indicate unique and total peptide numbers, respectively. C) Endogenous co-immunoprecipitation of EPOP, SUZ12, Elongin B and USP7 in mESCs. The immunoprecipitated proteins were visualized by Western blotting. Two exposures are shown for EPOP. D) Semi-endogenous co-immunoprecipitation of wildtype and L40A mutant EPOP in HeLa-S. An extended blot is shown in Figure S4D. E) Co-immunoprecipitation experiment of wildtype and L40-mutated Flag-tagged EPOP versus HA-USP7 in HeLa-S cells. F) Silver staining of the glycerol gradient fractionation of the complex from HeLa-S cells. Asterisks indicate likely contaminants. Specific proteins in those fractions were visualized by Western blotting. Fraction 4, 9 and 12 were analyzed by MS (see B). The lower panel depicts three putative complexes formed by EPOP. The sequence of the N-terminal BC-Box is shown. See also Figure S4.
We next examined whether EPOP directly or indirectly interacts with Elongin BC and USP7. We found that bacterially purified EPOP interacted with the Elongin BC heterodimer in vitro (Figure S4A), suggesting a direct interaction, possibly via a BC-Box motif (P/S/T-L-x-x-x-C/S/A-x-x-x-L/I/A/V), which is the typical interaction motif for Elongin BC (Aso et al., 1996; Mahrour et al., 2008). We identified two putative BC-Boxes in EPOP (Figure S4B, C) and mutation of the critical leucine (L40A) in the N-terminal BC-Box strongly reduced the in vitro and in vivo interactions (Figure 3D, S4C, D). This demonstrates that EPOP possesses two separate interaction sites, a PRC2 interaction site at the C-terminus (Liefke and Shi, 2015) and an Elongin BC interaction site at the N-terminus. Consistently, Elongin BC is not required for EPOP to evict SUZ12 from chromatin, which depends on the C-terminal PRC2-interacting region of EPOP (Figure S4E-G and Figure 4A). Lastly, we also found a direct association of USP7 with EPOP and Elongin B and C in vitro (Figure S4H, I). Consistent with a potential cooperative interaction of EPOP/Elongin BC with USP7 we observed that the BC-box mutated EPOP appears to be compromised in its ability to interact with USP7 (Figure 3E).
Figure 4. EPOP recruits Elongin BC to chromatin.
A,B) Western blotting of proteins and histone modifications in control cells, Epop KO cells, and KO cells re-expressing either wildtype (WT) or L40A mutated EPOP. Actin or H3 is shown as loading control. C) Overlap of Elongin B ChIP-Seq peaks (brown) with RNA Polymerase II (blue) and EPOP (red) or SUZ12 (orange) and EZH2 (purple). D) Heatmaps of Elongin B ChIP-Seq in control, Epop KO cells and the subtraction (KO – control). The right panel shows the promoters profiles of Elongin B in control and KO cells (two replicates combined). The P values were calculated by ANOVA. See also Figure S5B C) Upper part: Genome browser view at Tet1 and Esrrb gene locus of ChIP-Seq data for EPOP, H3K4me3 and Elongin B in control and Epop KO cells. Lower part: ChIP-qPCR for Elongin B, EPOP and H3K4me3 at the Esrrb and Tet1 gene locus, in the four cell lines described in A). The primer positions are shown above. The values represent mean ± SD of two biological replicates. See also Figure S5.
EPOP forms distinctly composed complexes
To investigate whether EPOP forms common or distinct complexes with PRC2, Elongin BC and USP7 we subjected Flag-purified EPOP complexes to glycerol gradient fractionation, followed by Western blotting and MS (Figure 3B,F). Unexpectedly, the immuno-purified human EPOP complex from HeLa-S cells appeared to segregate into several distinctly composed sub-complexes. Specifically, we found that the high molecular weight fractions contained essentially the PRC2 complex and a small amount of Elongin BC and USP7 (MS for fraction 12), while the low molecular weight fractions contained a high amount of Elongin BC but no PRC2 members (MS for fraction 4). By Western blotting using antibodies for Elongin B and C as well as SUZ12 we confirmed a distinct distribution of Elongin BC and SUZ12 in those fractions. Interestingly, USP7 is mainly present in the intermediate molecular weight fractions (MS for fraction 9) and its distribution neither correlates with Elongin BC nor SUZ12, supporting the existence of several different EPOP complexes in HeLa-S cells (Figure 3F). A similar segregation was found when a purified mouse EPOP complex from mESCs was subjected to glycerol gradient analysis (Figure S4J), although we were unable to detect USP7 in those fractions.
Taken together, these results suggest that EPOP independently interacts with Elongin BC, USP7 and PRC2. It is thus possible that EPOP can assemble into multiple distinctly composed complexes that contain one or several of its interaction partners (Figure 3F).
EPOP recruits Elongin BC to chromatin in a BC-Box dependent manner
Since Elongin BC is a known transcription elongation factor (Aso et al., 1995), we speculated that the effects of EPOP on transcriptional processes may be dependent on its interaction with Elongin BC. To investigate the functional significance of this interaction, we re-introduced either wildtype (WT) or the L40A mutant EPOP into the Epop KO mESCs (Figure 4A). Analysis of the H3K4me3 histone mark by Western blotting demonstrated a reduced rescuing ability of the mutant compared with the wildtype, suggesting that the BC-box is important for this function of EPOP (Figure 4B). ChIP-Seq experiments using an Elongin B specific antibody in control or CRISPR/Cas9 mediated Tceb2 (Elongin B) KO cells (Figure S5A) showed a modest but significant enrichment of Elongin B at actively transcribed genes, including genes with broad H3K4me3 domain, as well as PRC2 target genes (Figure 4C,D, S5B). Consistent with a potentially EPOP-dependent recruitment of Elongin BC, we found in two independent biological replicates a reduced chromatin association of Elongin B in Epop KO cells (Figure 4D, S5B). This was further confirmed by ChIP-qPCR at two broad H3K4me3 domains where Elongin B recruitment could be rescued with the wildtype but not with the BC-box mutant of EPOP (Figure 4E). In contrast, Elongin BC is not required for EPOP’s chromatin association, since both the wildtype and the L40A mutant protein of EPOP bind to chromatin (Figure 4A,E). In line with a potential involvement of Elongin BC in the regulatory function of EPOP at active genes, rescue of the H3K4me3 levels at the investigated broad H3K4me3 domains with the BC-box mutant was compromised compared to wildtype EPOP (Figure 4E).
EPOP recruits USP7 to regulate H2B ubiquitination at promoter sites
Given that USP7 is a known H2B deubiquitinase (Maertens et al., 2010; Sarkari et al., 2009; van der Knaap et al., 2005) and Elongin BC has been proposed to be involved in regulating H2B mono-ubiquitination (H2Bub) (Hwang et al., 2011; Li et al., 2010) as well, we next investigated the role of EPOP for H2Bub, a histone modification commonly found at actively transcribed genes (Weake and Workman, 2008). We first examined whether EPOP may recruit USP7 to chromatin. Via ChIP-Seq we found an enrichment of USP7 downstream of the transcription sites (TSS) in control cells, which is significantly reduced in the Epop KO cells, suggesting that EPOP is important for USP7 association with chromatin (Figure 5A). To gain further insights into the role of EPOP, Elongin BC and USP7 in H2Bub regulation, we additionally generated Usp7 KO mESCs using CRISPR/Cas9 (Figure S5C) and performed H2Bub ChIP-Seq experiments in all three established KO mESCs cells versus control cells. Interestingly, we found that depletion of EPOP, Elongin B or USP7 induces a local increase of H2Bub directly downstream of the TSS (Figure 5B,C). This suggests that EPOP plays a role to remove H2Bub at promoter sites, possibly by enhancing the H2B deubiquitination through its associated protein USP7. Notably, the alteration of H2Bub resembles the changes of H3K4me3 and RNA polymerase II in the Epop KO cells (Figure 2C), supporting a previously proposed crosstalk between H2Bub, H3K4me3 and RNA polymerase II transcription (Batta et al., 2011; Henry et al., 2003; Kim et al., 2013; Pavri et al., 2006; Weake and Workman, 2008; Wu et al., 2013). Consistently, via ChIP-Seq we found a significant increase of H3K4me3 at the promoter regions in the Tceb2 and Usp7 KO cells (Figure S5D). However, only in the Tceb2 but not in the Usp7 KO cells we detected a reduction of H3K4me3 at the broad H3K4me3 domains, as observed before in the Epop KO cells (Figure 2C, S5D), suggesting that EPOP/Elongin BC may employ USP7-independent mechanisms at those domains. The decrease of H3K4me3 in the Tceb2 KO cells at the Tet1 and the Esrrb gene loci was confirmed by ChIP-qPCR with two independent KO clones (Figure S5D). Together, these findings raise the possibility that EPOP may cooperate with Elongin BC to influence USP7-mediated H2Bub deubiquitination at promoter sites, which may partially explain the effects of EPOP loss on H3K4me3 and RNA polymerase II transcription.
Figure 5. EPOP recruits USP7 to modulate H2B ubiquitination in mESCs.
A) Heatmaps (sorted as in Figure 2C) and profiles of USP7 ChIP-Seq in control and Epop KO cells, as well the subtraction (KO – control). B) Example browser view of USP7, Elongin B in control and Epop KO cells, as well as H2Bub in control versus Epop, Tceb2 or Usp7 KO cells. A local increase of H2Bub at promoter sites is indicated by the grey box. C) Heatmaps showing H2Bub in control and KO cells, and the subtraction. The profiles in A) and C) are shown for genes with and without broad H3K4me3 domain as well as PRC2 target genes. The P values were calculated via ANOVA. See also Figure S5.
EPOP has only minor impact on gene expression in mESCs
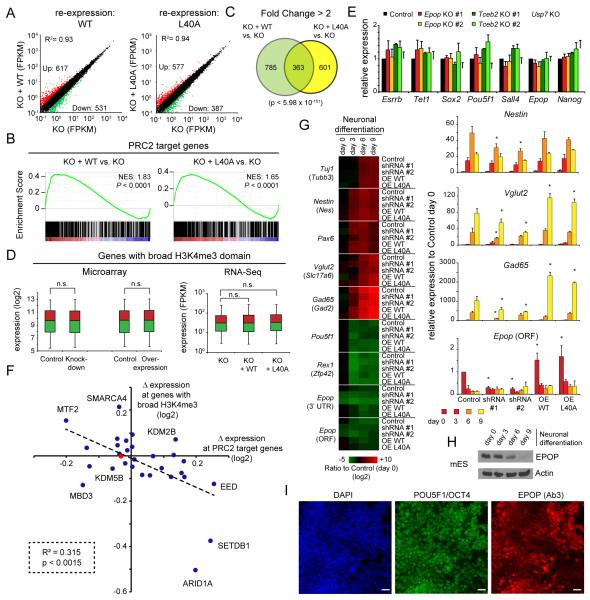
To determine the functional consequence of EPOP interaction with Elongin BC, we performed RNA-Seq after ectopic expression of either the wildtype (WT) or L40A mutant EPOP in the Epop KO cells (Figure 6A,B). We found 1148 genes whose expression levels were altered by 2-fold or more in KO cells expressing WT EPOP vs. KO cells. These genes are associated with transcription, development and axon guidance (Table S1), similar to what has been described before (De Cegli et al., 2013). Gene set enrichment analysis (GSEA) revealed that PRC2 targets were significantly de-repressed independently of the interaction with Elongin BC (Figure 6B), consistent with our finding that Elongin BC is not required for EPOP to inhibit PRC2, and explains the partial but significant overlap of affected genes in WT versus the mutant EPOP-expressing cells (Figure 6C). Surprisingly, neither our RNA-seq data, targeted RT-qPCR, nor previously published microarray data (De Cegli et al., 2013) found substantial changes of expression of active genes with broad H3K4me3 domains (Figure 6D,E). These results suggest that, despite the fact that EPOP localizes and regulates chromatin landscape at active genes in mESCs, especially those with broad H3K4me3 domains, the impact on gene expression is nonetheless minimal. This finding is reminiscent of what has been observed upon depletion of MYC and many other transcription factors in mESCs (Kim et al., 2010; Nishiyama et al., 2013). Analysis of publicly available microarray data (Table S4) also showed that depletion of many chromatin regulators in mESCs, including the broad H3K4me3 domain-affecting histone demethylase Kdm5b (Benayoun et al., 2014; Liefke and Shi, 2015), only subtly influences gene expression of genes with broad H3K4me3 domains (Figure 6F). This suggests extensive compensatory mechanisms, consistent with the finding that broad H3K4me3 domains in mESCs are governed by a large number of regulators (Figure 1A).
Figure 6. EPOP has low impact on gene expression in mESCs.
A) Comparison of RNA-Seq derived FPKM values of Epop KO and KO cells with re-expressed wildtype (WT) or L40A mutant EPOP. A two-fold cut-off was used for up-(red) and downregulated (green) genes. B) Overlap of affected genes in wildtype or mutant re-expressing cells. The P-value was calculated by a hypergeometric probability test. C) GSEA of PRC2 target genes using RNA-Seq data from wildtype and mutant re-expressing cells compared to KO cells. NES = Normalized Enrichment Score. D) Comparison of gene expression of genes with broad H3K4me3 domain using the RNA-Seq data or published Microarray data (De Cegli et al., 2013). n.s. = not significant. (Student’s t-test) E) RT-qPCR of gene expression of broad H3K4me3 domain possessing pluripotency genes in Epop, Tceb2 or Usp7 KO cells. The values show mean ± SD of duplicates. F) Average gene expression change at PRC2 target genes or genes with broad H3K4me3 domain upon depletion of specific chromatin regulators. Each dot represents an individual cell line depleted for a unique factor. Data are derived from expression profiles of each cell line compared to control cells (Table S4). The red dot marks EPOP. A significant negative correlation of expression changes of PRC2 target genes and genes with broad H3K4me3 domain can be observed. The p-value was calculated via ANOVA. G) Neuronal differentiation of mESC with depleted or over-expressed EPOP. The heatmap shows the expression relative to control cells at day 0, measured by RT-qPCR. The right panels show four examples genes, including the neuronal genes Vglut2 and Gad65. The values show mean ± SD of duplicates (* p < 0.05, Student’s t-test). H) Western blot for endogenous EPOP during neuronal differentiation. Actin is shown as loading control. I) Immunofluorescence of EPOP (using Ab3) and POU5F1/OCT4 in wildtype mESCs. scale bar = 20 μm
To gain additional insights into the biological role of EPOP in mESCs, we investigated neuronal differentiation of mESCs with reduced or elevated levels of EPOP (Figure 6G). Overexpression of EPOP enhanced the up-regulation of neuronal genes such as Vglut2 and Gad65, albeit modestly, while knockdown showed a modestly impaired up-regulation of those genes (Figure 6G), suggesting that EPOP may play a role in neuronal differentiation, consistent with a previous report (De Cegli et al., 2013). Since over-expression of the WT and the L40A mutant version of EPOP had similar consequences (Figure 6G), the observed expression effects might predominantly be due to the PRC2-related role of EPOP, in line with the larger influence on gene expression at PRC2 target genes in mESCs (Figure 6F).
Interestingly, we observed a down-regulation of EPOP expression during mESC differentiation (Figure 6G,H; S3F,G), which might be due to the fact that the mouse Epop gene (E130012A19Rik) is a direct target of several pluripotency factors, including NANOG, POU5F1, SOX2 and MYC (Figure S6A). Immunofluorescence experiments in wildtype mESCs further demonstrated that the levels of the EPOP protein vary among individual cells (Figure 6I) (more so than POU5F1/OCT4), similar to the reported heterogeneous (or “salt and pepper”) expression patterns described for critical pluripotency factors such as NANOG, ESRRB and KLF4 (Torres-Padilla and Chambers, 2014). Such heterogeneous expression patterns are believed to be associated with differential self-renewing and differentiation abilities of individual mESCs, suggesting that EPOP expression and function could be linked to the pluripotency network (Klein et al., 2015).
EPOP is a putative proto-oncogene
Given that highly expressed factors in ES cells are commonly activated during carcinogenesis (Ben-Porath et al., 2008; Kim et al., 2010) and EPOP influences transcriptional processes similar to the proto-oncogene MYC (Figure 2, S3H) (Rahl et al., 2010), we speculated that EPOP may be expressed in human cancer cells and could have an oncogenic role. Indeed, analysis of the TCGA (The Cancer Genome Atlas) data from human patients revealed that EPOP, as well as transcripts for Elongin B (TCEB2) and C (TCEB1), is often more highly expressed in cancer tissues compared with normal tissues (Figure 7A, Table S5). Moreover, in breast cancer patients, high levels of expression of EPOP, TCEB1 and TCEB2 are associated with poor prognosis (Figure 7B). In addition, we found that expression of EPOP positively correlates with a proliferation signature in many cancer tissues, such as breast and colon cancer (Figure 7C, Table S6), supporting the hypothesis that EPOP may positively influence cancer cell proliferation (Eilers and Eisenman, 2008; Hanahan and Weinberg, 2011). To investigate this further, we analyzed the consequence of EPOP knockdown with two independent shRNAs in several human cancer cell lines, which express low but detectable protein levels of EPOP (Figure 7D). Some cell lines (SH-SY5Y, MCF7, PC3, Colo205) showed reduced proliferation and slightly altered morphology upon depletion of EPOP (Figure 7E,F, Figure S6B), while other cancer cells (U2OS and HeLa-S) appeared unaffected (Figure 7F), suggesting that rapid proliferation of some cancer cells may require EPOP, although the underlying mechanisms are unclear at this time. The strongest proliferation defect upon EPOP knockdown was observed in the neuroblastoma-related SH-SY5Y cells (Figure 7F), which also showed the highest EPOP protein expression (Figure 7D), suggesting that proliferation of those cells may be particularly dependent on EPOP. This is further supported by a reduced proliferation of those cells upon infection with an EPOP CRISPR construct (Figure S6C). Similar results were also obtained upon depletion of Elongin B and C, supporting a potential cooperative function of EPOP and Elongin BC for the proliferation of those cells (Figure S6D, E). In support of this notion, we found that the reduced proliferation of SH-SY5Y cells upon EPOP knockdown could be rescued with the wildtype and with the PRC2-interacting domain deficient version (ΔCTR) of EPOP, but not with the L40A mutant version of EPOP (Figure S6G), suggesting that the interaction with Elongin BC is critical for EPOP to influence the proliferation of SH-SY5Y cells.
Figure 7. EPOP possesses oncogenic characteristics.
A) Average gene expression change of PRC2 and EPOP related proteins in cancer versus normal tissue using TCGA expression data (Table S5). The genes are sorted after the rank of the average. CCRCC = clear cell renal cell carcinoma, CCPRCC = clear cell papillary renal cell carcinoma, HaNC = head and neck cancer, LSCC = lung squamous cell carcinoma, LAC = lung adenocarcinoma, PRCC = papillary renal cell carcinoma. B) Kaplan-Meier plots (Gyorffy et al., 2010) for breast cancer patients expressing low or high levels of EPOP, TCEB1 and -2. C) Diagram showing the correlation coefficient R and its p-value of EPOP expression versus a proliferation index in cancer tissues. Each dot shows one cancer type (Table S6). The right panel shows two examples for breast and colon cancer. The p-values were calculated by ANOVA. D) Western blot of endogenous EPOP of investigated human cancer cell lines. Actin is shown a loading control. E) Knockdown of EPOP in SH-SY5Y cells analyzed by Western blotting. The asterisks in D) and E) indicate unspecific bands. F) Analysis of cancer cell proliferation upon EPOP knockdown with two independent shRNAs. G) Rescue experiment in SH-SY5Y cells with wildtype, C-terminal region deleted (ΔCTR) and the L40A mutant version of EPOP. The values in F and G show mean ± SD of triplicates. (* p < 0.05, Student’s t-test) H) H3K4me3 ChIP-Seq in SH-SY5Y cells upon knockdown or overexpression of EPOP. Genes with 25% (40% for strongly reduced) change of the H3K4me3 signal were considered as affected. Only H3K4me3 possessing promoters were included into the analysis (n = 17480). Specific groups were analzed by gene ontology (Table S1). Example genes are shown. See also Figure S6.
Since the human EPOP gene (C17orf96) is commonly occupied by MYC in human cancer cells (Figure S6F), we reasoned that the elevated EPOP expression in human cancer cells may be dependent on MYC. This hypothesis is further supported by a MYC-driven mouse lymphomagenesis model (Sabo et al., 2014), which shows about a six-fold up-regulation of the Epop transcript (Figure S6G). Furthermore, investigation of the TCGA expression data revealed a positive correlation of EPOP expression with that of MYC (Figure S6H). Interestingly, we found EPOP enriched at the MYC gene locus in SH-SY5Y cells (Figure S6I), suggesting that EPOP may also regulates MYC, revealing a potentially reciprocal regulation between MYC and EPOP. Indeed, RT-qPCR showed that knockdown of either EPOP or MYC in SH-SY5Y cells leads to the down-regulation of both EPOP and MYC (Figure S6J), raising the possibility that both proteins are involved in establishing a proliferation-promoting transcriptional network in these cells.
To address whether aberrant EPOP expression might contribute to an altered chromatin landscape in SH-SY5Y cells, we performed H3K4me3 ChIP-Seq after knockdown or overexpression of EPOP, respectively. Knockdown of EPOP led to a reduced H3K4me3 signal at cell cycle and housekeeping genes, including the MYC gene, while genes that showed constant or increased H3K4me3 levels are involved in neuronal differentiation processes, suggesting a possible shift of the transcriptional landscape that disfavors proliferation (Figure 7H, Table S1). Genes with decreased H3K4me3 are commonly occupied by MYC in human cancer cells (Figure S6K), further supporting the hypothesis that EPOP knockdown impairs the Myc regulatory network. When EPOP is over-expressed, the overall pattern remained largely unaffected, but we observed an increase of H3K4me3 at some genes involved in developmental processes, such as at the HoxA cluster, possibly due to inhibition of PRC2 (Figure 7H, Table S1). Consistent with the change of the H3K4me3 pattern, analysis of gene expression by RT-qPCR showed an altered transcription in EPOP knockdown and over-expressing cells (Figure S6L). Taken together, these results raise the possibility that anomalous expression of EPOP in human cancer cells may contribute to their proliferation (Hanahan and Weinberg, 2011).
Discussion
Our investigation establishes EPOP as an unusual chromatin regulator in mammalian cells. On one hand, it inhibits PRC2 chromatin binding (Liefke and Shi, 2015), while on the other hand it cooperates with Elongin BC and USP7 to modulate the chromatin landscape at actively transcribed genes (Figure 2 to 5), suggesting a versatile and complex function for EPOP in mammalian cells.
Despite its putative global gene regulatory role, we did not observe strong alterations of gene expression by manipulating EPOP protein level in mESCs (Figure 6). Only modest effects were seen at the lowly expressed PRC2 target genes, while highly expressed genes with broad H3K4me3 domains showed no significant changes. This is perhaps not unexpected as this has been seen for many other transcriptional (Nishiyama et al., 2013) and chromatin regulators (Figure 6F) (Clouaire et al., 2012; Jiang et al., 2011), highlighting the robustness of the transcriptional network in mESCs (Ng and Surani, 2011). Consistently, alteration of EPOP protein level has only subtle impact on mESCs differentiation processes (Figure 6G, S3F). The physiological relevance of EPOP may manifest upon long-term developmental processes, during which EPOP might cooperate with other chromatin regulators at active and repressed genes (Figure 1A) to contribute to a deterministic reprogramming of gene regulatory networks. For instance, it is possible that, besides its PRC2-related functions, EPOP may regulate the highly dynamic H2B ubiquitination during developmental processes (Fuchs et al., 2012). It would therefore be interesting to examine the consequences of loss of EPOP during murine embryogenesis in the future.
We identified Elongin BC as an interaction partner of EPOP and our work supports the hypothesis that EPOP cooperates with Elongin BC to modulate the local chromatin landscape (Figure 2 to 5). Elongin BC may influence EPOP’s association with other regulatory proteins, such as USP7. Alternatively, it could help supply Elongin BC and/or exclude other BC-box containing regulators. Possibly affected proteins include the TrxG proteins ARID1A/B (BAF250A/B) (Li et al., 2010), the Mediator complex subunit MED8 (Brower et al., 2002) and the transcription elongation factor Elongin A (Aso et al., 1995), but other BC-box containing proteins could be influenced as well (Mahrour et al., 2008). EPOP may modulate transcriptional processes by interacting with PRC2, Elongin BC, USP7 and possibly other regulators.
Many cancer types show increased transcript levels of EPOP compared to normal tissues and high EPOP expression in breast cancer correlates with poor prognosis (Figure 7A,B). Our work further demonstrated that reduction of EPOP leads to reduced proliferation of several human cancer cell lines from different origins, raising the possibility that EPOP may have an oncogenic role in multiple cancer types. Biochemical and genome-wide experiments suggest that EPOP generally functions to promote transcription, since it inhibits PRC2 in vivo (Liefke and Shi, 2015) and is involved in elevating RNA polymerase II pausing release and elongation (Figure 2). Many oncogenic proteins, such as MYC, have similar transcription-enhancing function (Lin et al., 2012; Nie et al., 2012; Rahl et al., 2010), which increases the proliferative capacity of the cell (Hanahan and Weinberg, 2011). Our finding of the interaction of EPOP with Elongin BC and USP7 to modulate the local chromatin landscape provides a possible molecular mechanism by which EPOP may enhance cancer cell proliferation. Future studies that validate and expand our initial investigation may lead to insights into the genesis of aberrant transcriptional networks in human cancer cells.
Experimental Procedures
Cell culture
E14 mouse ES cells (E14TG2a) were obtained from ATCC and cultured in DMEM, 15% FCS, 1 × L-Glutamine (Invitrogen), 1 × Non-essential amino acids (Invitrogen), 1 × Sodium pyruvate, 1 × Penicillin/Streptomycin (Invitrogen), 0.15% β-mercaptoethanol and 100 Units/ml of LIF (Millipore) on gelatin-coated plates. Human HeLa-S, U2OS, Colo205, PC3, MCF7, SH-SY5Y and 293T cells were cultured with DMEM, 10% FCS and 1 × Penicillin/Streptomycin (Invitrogen). Stable cell lines were obtained via infection with retroviral or lentiviral vectors harboring the appropriate construct and selected via puromycin or blasticidin.
ChIP, ChIP-Seq, RNA-Seq, qPCR
ChIP experiments were performed via cross-linking ChIP as described (Liefke and Borggrefe, 2014). For DNA shearing, bioruptor UCD-200 (diagenode) was used. For RNA-Seq whole RNA was prepared using trizol and purified using Magnetic beads mRNA Isolation Kit (BioLabs, #S1550S). After mRNA fragmentation by heating the sample for 6 min at 95 C, the mRNA was reverse-transcribed using SuperScript III (Invitrogen, 18080-044), followed by Second Strand Synthesis (Invitrogen, 10812-014). Libraries for ChIP and RNA-Seq were constructed of 10-50 ng DNA following Illumina’s® protocol and sequenced using an Illumina Genome Analyzer. Quantitative PCR (qPCR) were performed on a LightCycler 480 (Roche). Primers for ChIP-qPCR and RT-qPCR are presented in Table S7.
Immunofluorescence
Immunofluorescence experiments for comparing the three antibodies (Figure S2F) were performed as described (Liefke and Shi, 2015) with a final concentration of 1 ng/μL for each EPOP antibody. Immunofluorescence experiments comparing expression of EPOP with that of POU5F1/OCT4 in mESCs (Figure 6I) were performed as described (Lowell et al., 2006).
Supplementary Material
Highlights.
EPOP, a PRC2-associated protein, occupies both active and repressed genes in mESCs
EPOP interacts with Elongin BC and the H2B deubiquitinase USP7
It affects histone modifications, including H2Bub and H3K4me3, and RNA Polymerase II
EPOP is involved in cancer cell proliferation
Acknowledgments
We thank Ashwini Jambhekar for critical reading of the manuscript. We thank the members of the Shi lab for discussion, Andres Blanco and Andrew Elia for performing experiments that were not included in the final manuscript, Sebastian Dango and Benoît Laurent for experimental advice. We thank Luciano Di Croce and Malte Beringer for data and antibody sharing and discussion. This work is supported by the National Cancer Institute (R01 CA118487) to Y.S. and the German Research Foundation (DFG, LI 2057/1-1) to R.L.. Y.S. is an American Cancer Society Research Professor. Y.S. is a co-founder of Constellation Pharmaceuticals, Inc. and a member of its scientific advisory board.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
ChIP-Seq and RNA-Seq data are available under the GEO accession number GSE76536, GSE79318 and GSE85681.
Authors Contribution
R.L. conceived the study, performed most experiments, analyzed and interpreted the data, performed the bioinformatics analyses and wrote the paper. V.K-N. performed differentiation and immunofluorescence experiments in mESCs and helped writing the paper. Y.S. interpreted the data and co-wrote the paper.
References
- Alekseyenko AA, Gorchakov AA, Kharchenko PV, Kuroda MI. Reciprocal interactions of human C10orf12 and C17orf96 with PRC2 revealed by BioTAP-XL cross-linking and affinity purification. Proc Natl Acad Sci U S A. 2014;111:2488–2493. doi: 10.1073/pnas.1400648111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso T, Haque D, Barstead RJ, Conaway RC, Conaway JW. The inducible elongin A elongation activation domain: structure, function and interaction with the elongin BC complex. EMBO J. 1996;15:5557–5566. [PMC free article] [PubMed] [Google Scholar]
- Aso T, Lane WS, Conaway JW, Conaway RC. Elongin (SIII): a multisubunit regulator of elongation by RNA polymerase II. Science. 1995;269:1439–1443. doi: 10.1126/science.7660129. [DOI] [PubMed] [Google Scholar]
- Batta K, Zhang Z, Yen K, Goffman DB, Pugh BF. Genome-wide function of H2B ubiquitylation in promoter and genic regions. Genes Dev. 2011;25:2254–2265. doi: 10.1101/gad.177238.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benayoun BA, Pollina EA, Ucar D, Mahmoudi S, Karra K, Wong ED, Devarajan K, Daugherty AC, Kundaje AB, Mancini E, et al. H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell. 2014;158:673–688. doi: 10.1016/j.cell.2014.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower CS, Sato S, Tomomori-Sato C, Kamura T, Pause A, Stearman R, Klausner RD, Malik S, Lane WS, Sorokina I, et al. Mammalian mediator subunit mMED8 is an Elongin BC-interacting protein that can assemble with Cul2 and Rbx1 to reconstitute a ubiquitin ligase. Proc Natl Acad Sci U S A. 2002;99:10353–10358. doi: 10.1073/pnas.162424199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Chen Z, Wu D, Zhang L, Lin X, Su J, Rodriguez B, Xi Y, Xia Z, Chen X, et al. Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor-suppressor genes. Nat Genet. 2015;47:1149–1157. doi: 10.1038/ng.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouaire T, Webb S, Skene P, Illingworth R, Kerr A, Andrews R, Lee JH, Skalnik D, Bird A. Cfp1 integrates both CpG content and gene activity for accurate H3K4me3 deposition in embryonic stem cells. Genes Dev. 2012;26:1714–1728. doi: 10.1101/gad.194209.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cegli R, Iacobacci S, Flore G, Gambardella G, Mao L, Cutillo L, Lauria M, Klose J, Illingworth E, Banfi S, di Bernardo D. Reverse engineering a mouse embryonic stem cell-specific transcriptional network reveals a new modulator of neuronal differentiation. Nucleic Acids Res. 2013;41:711–726. doi: 10.1093/nar/gks1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M, Eisenman RN. Myc's broad reach. Genes Dev. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs G, Shema E, Vesterman R, Kotler E, Wolchinsky Z, Wilder S, Golomb L, Pribluda A, Zhang F, Haj-Yahya M, et al. RNF20 and USP44 regulate stem cell differentiation by modulating H2B monoubiquitylation. Mol Cell. 2012;46:662–673. doi: 10.1016/j.molcel.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J, Saffert RT, Kalejta RF. Elongin B-mediated epigenetic alteration of viral chromatin correlates with efficient human cytomegalovirus gene expression and replication. MBio. 2011;2:e00023–00011. doi: 10.1128/mBio.00023-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Shukla A, Wang X, Chen WY, Bernstein BE, Roeder RG. Role for Dpy-30 in ES cell-fate specification by regulation of H3K4 methylation within bivalent domains. Cell. 2011;144:513–525. doi: 10.1016/j.cell.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kareta MS, Gorges LL, Hafeez S, Benayoun BA, Marro S, Zmoos AF, Cecchini MJ, Spacek D, Batista LF, O'Brien M, et al. Inhibition of pluripotency networks by the Rb tumor suppressor restricts reprogramming and tumorigenesis. Cell Stem Cell. 2015;16:39–50. doi: 10.1016/j.stem.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlebach G, Shamir R. Modelling and analysis of gene regulatory networks. Nat Rev Mol Cell Biol. 2008;9:770–780. doi: 10.1038/nrm2503. [DOI] [PubMed] [Google Scholar]
- Kim J, Kim JA, McGinty RK, Nguyen UT, Muir TW, Allis CD, Roeder RG. The n-SET domain of Set1 regulates H2B ubiquitylation-dependent H3K4 methylation. Mol Cell. 2013;49:1121–1133. doi: 10.1016/j.molcel.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, Cantor AB, Orkin SH. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143:313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XS, Trojer P, Matsumura T, Treisman JE, Tanese N. Mammalian SWI/SNF--a subunit BAF250/ARID1 is an E3 ubiquitin ligase that targets histone H2B. Mol Cell Biol. 2010;30:1673–1688. doi: 10.1128/MCB.00540-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liefke R, Borggrefe T. Probing the epigenetic status at Notch target genes. Methods Mol Biol. 2014;1187:255–276. doi: 10.1007/978-1-4939-1139-4_20. [DOI] [PubMed] [Google Scholar]
- Liefke R, Shi Y. The PRC2-associated factor C17orf96 is a novel CpG island regulator in mouse ES cells. Cell Discov. 2015;1:15008. doi: 10.1038/celldisc.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Garrett AS, De Kumar B, Smith ER, Gogol M, Seidel C, Krumlauf R, Shilatifard A. Dynamic transcriptional events in embryonic stem cells mediated by the super elongation complex (SEC) Genes Dev. 2011;25:1486–1498. doi: 10.1101/gad.2059211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowell S, Benchoua A, Heavey B, Smith AG. Notch promotes neural lineage entry by pluripotent embryonic stem cells. PLoS Biol. 2006;4:e121. doi: 10.1371/journal.pbio.0040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens GN, El Messaoudi-Aubert S, Elderkin S, Hiom K, Peters G. Ubiquitin-specific proteases 7 and 11 modulate Polycomb regulation of the INK4a tumour suppressor. EMBO J. 2010;29:2553–2565. doi: 10.1038/emboj.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahrour N, Redwine WB, Florens L, Swanson SK, Martin-Brown S, Bradford WD, Staehling-Hampton K, Washburn MP, Conaway RC, Conaway JW. Characterization of Cullin-box sequences that direct recruitment of Cul2-Rbx1 and Cul5-Rbx2 modules to Elongin BC-based ubiquitin ligases. J Biol Chem. 2008;283:8005–8013. doi: 10.1074/jbc.M706987200. [DOI] [PubMed] [Google Scholar]
- Ng HH, Surani MA. The transcriptional and signalling networks of pluripotency. Nat Cell Biol. 2011;13:490–496. doi: 10.1038/ncb0511-490. [DOI] [PubMed] [Google Scholar]
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama A, Sharov AA, Piao Y, Amano M, Amano T, Hoang HG, Binder BY, Tapnio R, Bassey U, Malinou JN, et al. Systematic repression of transcription factors reveals limited patterns of gene expression changes in ES cells. Sci Rep. 2013;3:1390. doi: 10.1038/srep01390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabo A, Kress TR, Pelizzola M, de Pretis S, Gorski MM, Tesi A, Morelli MJ, Bora P, Doni M, Verrecchia A, et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature. 2014;511:488–492. doi: 10.1038/nature13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkari F, Sanchez-Alcaraz T, Wang S, Holowaty MN, Sheng Y, Frappier L. EBNA1-mediated recruitment of a histone H2B deubiquitylating complex to the Epstein-Barr virus latent origin of DNA replication. PLoS Pathog. 2009;5:e1000624. doi: 10.1371/journal.ppat.1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits AH, Jansen PW, Poser I, Hyman AA, Vermeulen M. Stoichiometry of chromatin-associated protein complexes revealed by label-free quantitative mass spectrometry-based proteomics. Nucleic Acids Res. 2013;41:e28. doi: 10.1093/nar/gks941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Padilla ME, Chambers I. Transcription factor heterogeneity in pluripotent stem cells: a stochastic advantage. Development. 2014;141:2173–2181. doi: 10.1242/dev.102624. [DOI] [PubMed] [Google Scholar]
- van der Knaap JA, Kumar BR, Moshkin YM, Langenberg K, Krijgsveld J, Heck AJ, Karch F, Verrijzer CP. GMP synthetase stimulates histone H2B deubiquitylation by the epigenetic silencer USP7. Mol Cell. 2005;17:695–707. doi: 10.1016/j.molcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Voigt P, Tee WW, Reinberg D. A double take on bivalent promoters. Genes Dev. 2013;27:1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Lee SY, Zhou B, Nguyen UT, Muir TW, Tan S, Dou Y. ASH2L regulates ubiquitylation signaling to MLL: trans-regulation of H3 K4 methylation in higher eukaryotes. Mol Cell. 2013;49:1108–1120. doi: 10.1016/j.molcel.2013.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Jones A, Sun CW, Li C, Chang CW, Joo HY, Dai Q, Mysliwiec MR, Wu LC, Guo Y, et al. PRC2 complexes with JARID2, MTF2, and esPRC2p48 in ES cells to modulate ES cell pluripotency and somatic cell reprogramming. Stem Cells. 2011;29:229–240. doi: 10.1002/stem.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.