Abstract
Aplastic anemia is an acquired bone marrow failure characterized by marrow hypoplasia, a paucity of hematopoietic stem and progenitor cells, and pancytopenia of the peripheral blood, due to immune attack on the bone marrow. In aplastic anemia, a major challenge is to develop immune biomarkers to monitor the disease. We measured circulating microRNAs in plasma samples of aplastic anemia patients in order to identify disease-specific microRNAs. A total of 179 microRNAs were analyzed in 35 plasma samples from 13 aplastic anemia patients, 11 myelodysplastic syndrome patients, and 11 healthy controls using the Serum/Plasma Focus microRNA Polymerase Chain Reaction Panel. Subsequently, 19 microRNAs from the discovery set were investigated in the 108 plasma samples from 41 aplastic anemia patients, 24 myelodysplastic syndrome patients, and 43 healthy controls for validation, confirming that 3 microRNAs could be validated as dysregulated (>1.5-fold change) in aplastic anemia, compared to healthy controls. MiR-150-5p (induction of T-cell differentiation) and miR-146b-5p (involvement in the feedback regulation of innate immune response) were elevated in aplastic anemia plasma, whereas miR-1 was decreased in aplastic anemia. By receiver operating characteristic curve analysis, we developed a logistic model with these 3 microRNAs that enabled us to predict the probability of a diagnosis of aplastic anemia with an area under the curve of 0.86. Dysregulated expression levels of the microRNAs became normal after immunosuppressive therapy at 6 months. Specifically, miR-150-5p expression was significantly reduced after successful immunosuppressive therapy, but did not change in non-responders. We propose 3 novel plasma biomarkers in aplastic anemia, in which miR-150-5p, miR-146b-5p, and miR-1 can serve for diagnosis and miR-150-5p for disease monitoring. Clinicaltrials.gov identifiers:00260689, 00217594, 00961064.
Introduction
The disease, aplastic anemia (AA) is caused in most patients by immune-mediated destruction of hematopoietic stem and progenitor cells (HSPCs), resulting in trilineage marrow hypoplasia and pancytopenia of the peripheral blood. The responsiveness of a significant proportion of AA patients to immunosuppressive therapies (IST) is the best evidence of an underlying immune pathophysiology: the majority of patients show hematologic improvement after only transient T-cell depletion by antithymocyte globulins (ATGs).1,2 Although the immune pathophysiology of AA is well characterized,3–5 there are no biomarkers that would allow a better understanding of the immunological status of an individual AA patient, including disease severity and response to therapy.6 Reliable biomarkers that correlated with disease severity or response would be useful for individual treatment decisions and in clinical trials.
MicroRNAs (miRNAs) are a group of small, conserved, non-coding RNA molecules that primarily modulate gene expression, post-transcriptionally by hybridization to complementary sequences in the 3′ untranslated region of corresponding messenger RNAs (mRNAs).7 MiRNAs contribute to the pathophysiology of important human diseases.8,9 Emerging evidence supports the fact that miRNAs have important roles in controlling and modulating immunity.10 Dysregulation of miRNAs can lead to autoimmune diseases, such as rheumatoid arthritis (RA) and multiple sclerosis (MS).11,12 Although miRNA regulation of each target results in small changes in gene expression, the network activity of miRNAs affecting hundreds of genes simultaneously can produce dramatic changes in cell behavior.13 We have recently reported that downregulation of miR-126-3p and miR-145-5p promotes CD4+ and CD8+ T-cell activation by increasing MYC and PIK3R2 expression levels in AA patients.14
MiRNAs can also be detected outside the cell. Extracellular miRNAs are cell-free circulating molecules residing in microvesicles, exosomes, and microparticles.15 These circulating miRNAs can be detected and quantified in biofluids, such as serum, plasma, urine, and saliva.16 Circulating miRNAs mirror physiological and pathophysiological conditions and have high stability in stored patient samples, allowing them to serve as biomarkers for various diseases.17 In particular, the detection of miRNA levels in blood plasma and serum has the potential for early cancer diagnosis and to predict prognosis and response to therapy.18,19 Recent studies have also identified circulating miRNAs as biomarkers to monitor the disease state in T cell-mediated autoimmune diseases, such as MS and myasthenia gravis (MG).20,21 However, miRNAs have yet to be explored in the serum or plasma of AA. The purpose of our study was to analyze the circulating miRNA profile in the plasma of AA patients and to assess whether specific miRNAs could serve as new biomarkers for AA.
Methods
Patients and treatment
The study population was 183 human subjects who were enrolled on clinical protocols between 2006 and 2015 at the NIH’s National Heart, Lung, and Blood Institute (NHLBI) (Bethesda, MD, USA). Samples were collected after informed consent was obtained in accordance with the Declaration of Helsinki. All human subjects were enrolled on clinical protocols approved by the NHLBI Institutional Review Board.
Ethylenediaminetetraacetic acid (EDTA) anticoagulated plasma samples were obtained from patients and age-matched healthy blood donors. Standard criteria were used for the diagnosis of AA and the assessment of disease severity.22 All AA patients were diagnosed as severe AA and none had received IST at the time of sampling. All AA patients received IST [horse-ATG + cyclosporine (CsA) or rabbit-ATG + CsA] on a clinical research protocol (clinicaltrials.gov identifier:00260689).2,23 EDTA plasma samples from myelodysplastic syndrome (MDS) were used for comparison (clinicaltrials.gov identifier:00217594 or clinicaltrials.gov identifier:00961064).24 Healthy controls (HC) were recruited from donors of the National Institutes of Health Clinical Center Department of Transfusion Medicine.
A discovery set (n=35) included 13 AA patients without IST at the time of sampling, 11 MDS patients, and 11 blood donors as HC. A validation set (n=108) consisted of 41 treatment-naïve AA patients, 24 MDS patients, and 43 HC. Demographics of the discovery and validation sets are shown in the Online Supplementary Table S1. To assess the effect of IST, 40 out of 41 AA patients with plasma samples available both before and after 6 months of IST were analyzed further.
RNA isolation and cDNA synthesis
Blood samples collected in EDTA tubes were centrifuged and stored at -80°C until use. RNA isolation from 200 ml EDTA plasma was performed using the miRCURY RNA Isolation Kit - Biofluids (Exiqon, Vedbaek, Denmark), according to the manufacturer’s instructions. RNA isolation efficiency was monitored with three synthetic RNA spike-ins at different concentrations (UniSp2, UniSp4, and UniSp5). Isolated RNA samples were employed for cDNA synthesis with the Universal cDNA Synthesis Kit II (Exiqon). Two synthetic RNA spike-ins in different concentrations (UniSp6 and cel-miR-39-3p) were used to check for reverse transcription reactions and polymerase chain reaction (PCR) inhibitors. Prepared cDNAs were stored at -20°C until use.
MiRNA profiling
Initial miRNA detection screening with the discovery set (n=35) was performed using the Serum/Plasma Focus microRNA PCR Panel, 384 well (V4.M) and the ExiLENT SYBR Green Master Mix (Exiqon). This panel allows for the analysis of 179 human miRNAs and was used to profile the discovery set of 13 AA, 11 MDS, and 11 HC (Online Supplementary Table S2). The information for validation of the miRNA profiling by a custom PCR panel is shown in the Online Supplementary Table S3 and Online Supplementary Experimental Methods.
Statistics
Statistical analysis was performed using the GenEx6 software (Exiqon) and SPSS 23.0 software, and graphs were generated using GraphPad PRISM version 6.0 (GraphPad Software, Inc., La Jolla, CA, USA).
More detailed information is provided in the Online Supplementary Experimental Methods.
Results
A distinct circulating miRNA profile in AA, compared to MDS and HC
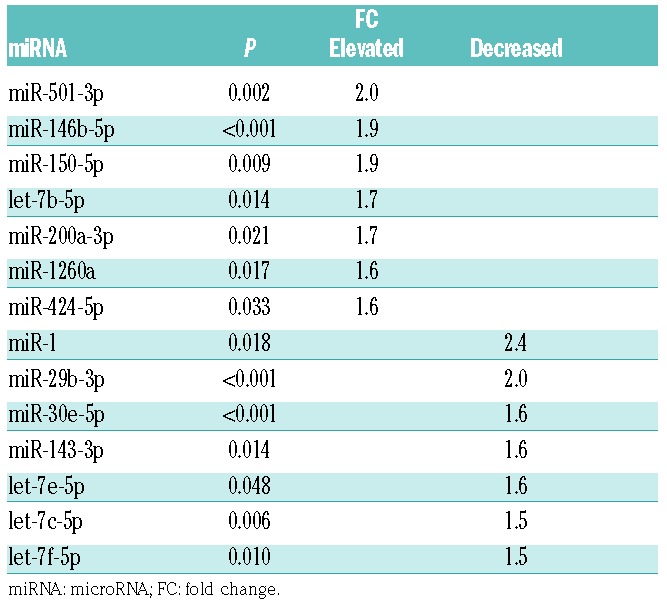
To examine whether there were unique AA-associated miRNAs, we first analyzed 35 discovery set plasma samples (13 untreated AA, 11 MDS, and 11 HC) without hemolysis (ΔCT<7) using the Serum/Plasma Focus microRNA PCR Panel (179 miRNAs). Of 179 miRNAs, 178 miRNAs showed amplification in more than 60% of the samples. When compared between AA and HC, 14 miRNAs displayed more than a 1.5-fold change (FC) (P<0.05 by t-test; Figure 1A and Table 1): 7 miRNAs were significantly upregulated (miR-501-3p, miR-146b-5p, miR-150-5p, let-7b-5p, miR-200a-3p, miR-1260a, and miR-424-5p) and 7 miRNAs were significantly downregulated (miR-1, miR-29b-3p, miR-30e-5p, miR-143-3p, let-7e-5p, let-7c-5p, and let-7f-5p) in AA, compared to HC. Hierarchical clustering showed that the plasma miRNA signature distinguished AA from HC (Figure 1B). The comparison of different groups by one-way ANOVA revealed multiple differentially expressed circulating miRNAs (P<0.05): 12, 47, and 36 miRNAs distinguished AA from HC, MDS from HC, and AA from MDS, respectively (Online Supplementary Table S4).
Figure 1.
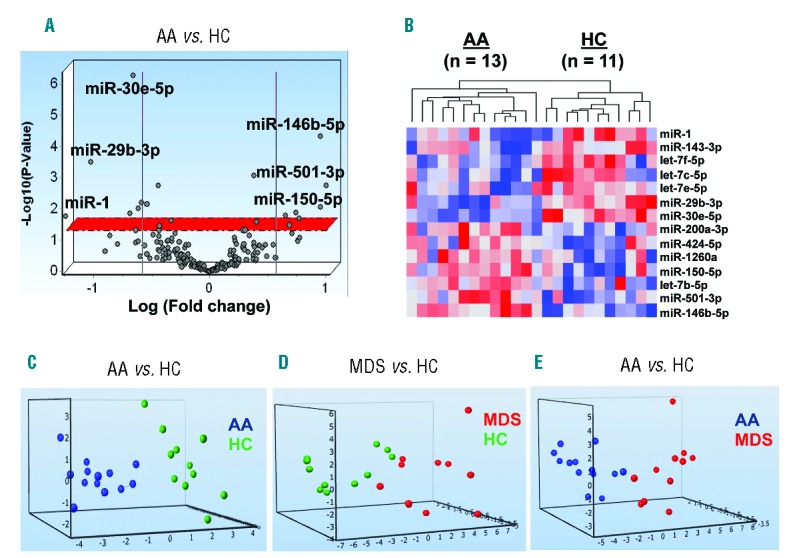
Distinct circulating microRNA (miRNA) expression profiles of AA patients compared to MDS and HC in the discovery set. (A) A volcano plot of 178 miRNA expression levels in the plasma of AA patients (n=13) and HC (n=11) in the discovery set. The x-axis displays the estimated expression difference measured in log2. Vertical lines refer to a 1.5-fold expression difference between two groups, showing that miRNAs highly expressed in AA or HC are on the right or the left, respectively, in which 6 miRNAs with higher fold changes are depicted. The y-axis shows the significance of the expression difference measured in −log10 of the P-value. The horizontal red line represents our cut-off for the significance at P<0.05. (B) A heatmap analysis visualizes hierarchical clustering of 14 miRNAs in the plasma from 13 AA patients and 11 HC. A red-blue color scale indicates normalized miRNA expression levels (red: high, blue: low). (C) Principal component analysis (PCA) plots of significantly (P<0.05) and differentially expressed miRNAs from the discovery set. Blue circles = AA, red circles = MDS, and green circles = HC. AA: aplastic anemia; HC: healthy control; MDS: myelodysplastic syndrome.
Table 1.
Differentially expressed miRNAs (>1.5 FC) in the aplastic anemia (AA) discovery set.
To further address differences of plasma miRNA levels between individual groups, a principal component analysis (PCA) of differentially expressed miRNAs was performed, resulting in distinct profiles between AA and MDS vs. HC, and between AA vs. MDS (Figure 1C–E). The PCA plots clearly visualized potential distinct grouping of the compared disease populations and controls, providing a basis to validate results of the discovery set in a separate cohort. The Online Supplementary Figure S1 summarizes all the analysis steps for the candidate miRNAs.
Validation of miRNA expression profiles
To validate findings in the discovery set, 19 miRNAs, including 17 candidate miRNAs from the discovery set (Online Supplementary Table S4), were selected to investigate in a separate cohort of AA, MDS, and HC (n=108; Online Supplementary Table S1) using the custom PCR array plate (Online Supplementary Table S3): 12 miRNAs (let-7f-5p, miR-1, miR-1260a, miR-143-3p, miR-146b-5p, miR-150-5p, miR-20a-5p, miR-21-5p, miR-26b-5p, miR-29b-3p, miR-30e-5p, and miR-501-3p) differentially expressed between AA and HC (P<0.05 by one-way ANOVA); 5 miRNAs (let-7g-5p, miR-181a-5p, miR-22-3p, miR-29c-3p, and miR-424-5p) significantly different by 3 group comparison (AA vs. MDS vs. HC); and additional 2 miRNAs (let-7a-5p and miR-144-5p) that have been reported to distinguish MDS from HC in previous reports.19,25 Box plots of 19 miRNA expression levels in the discovery set are shown in the Online Supplementary Figure S2. As a hemolysis marker (miR-23a-3p-miR-451a) was >7 in 3 HC and 3 patient plasma samples (1 AA and 2 MDS); these 6 samples were excluded from further analysis. All of the selected 19 miRNAs were amplified by quantitative real time PCR (qRT-PCR) in >60% of the samples, then subjected to further analysis. Of the 5 reference genes, miR-106a-5p and miR-320a were selected to calculate ΔCT and relative expression, as these 2 miRNAs displayed stable amplification in all of the individual samples with good expression (CT≤30, Online Supplementary Figure S3). Group comparison revealed distinct expression profiles at statistical significance (>1.5-FC, P<0.05): upregulated miR-150-5p and miR-146b-5p and downregulated miR-1 in AA, compared to HC (Figure 2A,D); upregulated miR-146b-5p and miR-22-3p in MDS, compared to HC (Figure 2B,D); and downregulated miR-1, miR-22-3p, and miR-424-5p in AA, compared to MDS (Figure 2C,D). Summary data (AA and MDS vs. HC, and AA vs. MDS) obtained from the validation set are shown in Table 2. When AA was compared to HC, 6 miRNAs showed significant association with AA: miR-1 (P=0.004), let-7g-5p (P=0.04), miR-146b-5p (P=0.0001), miR-26b-5p (P=0.049), miR-150-5p (P=0.015), and miR-181a-5p (P=0.029). Additionally, these 6 miRNAs were subjected to multivariate analysis to determine which miRNA associated with AA: miR-1 [odds ratio (OR)=0.34, adjusted P=0.005], miR-146b-5p (OR=3.82, adjusted P=0.03), and miR-150-5p (OR=2.19, adjusted P=0.04), that were differentially expressed in AA patients in the discovery set (Online Supplementary Table S4), significantly and independently associated with the AA diagnosis (Table 3).
Figure 2.
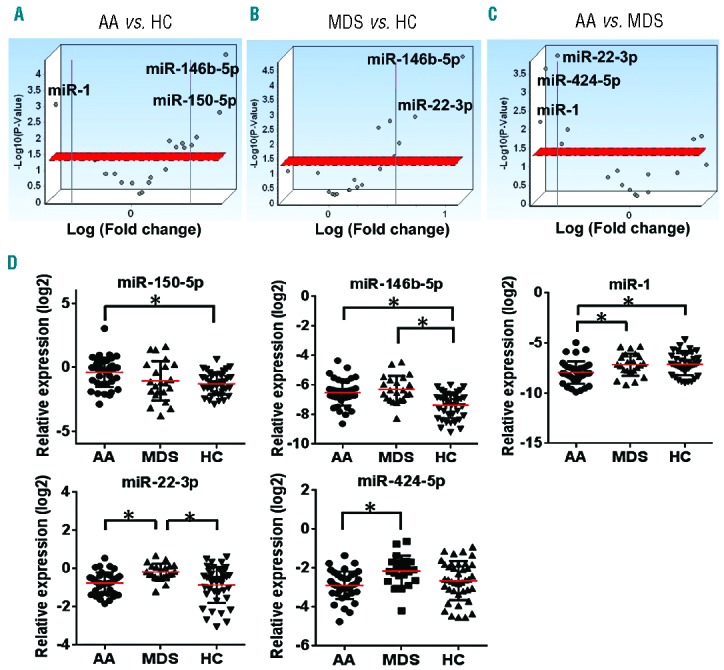
Validation of the circulating microRNA (miRNA) expression profiles in the validation set. (A-B) Volcano plots of 19 miRNA expression levels in the plasma of AA (n=41), MDS (n=24), and HC (n=43) in the validation set. The x-axis is the estimated difference in expression measured in log2; vertical lines refer to a 1.5-fold difference in expression between the two groups. MiRNAs highly expressed in AA (MDS) or HC are on the right or the left, respectively. The y-axis is the significance of the difference measured in −log10 of the P-value; the horizontal red line represents our cut-off for significance at P<0.05. (C) Volcano plots of 19 miRNA expression levels in the plasma of AA (n=41) and MDS patients (n=24) in the validation set. MiRNAs highly expressed in AA or MDS are on the right or the left, respectively. (D) miR-150-5p, miR-146b-5p, miR-1, miR-22-3p, and miR-424-5p expression in the plasma of AA (n=41), MDS (n=24), and HC (n=43). *P<0.05 (one-way ANOVA). AA: aplastic anemia; HC: healthy control; MDS: myelodysplastic syndrome.
Table 2.
Association between miRNAs and groups in validation set.
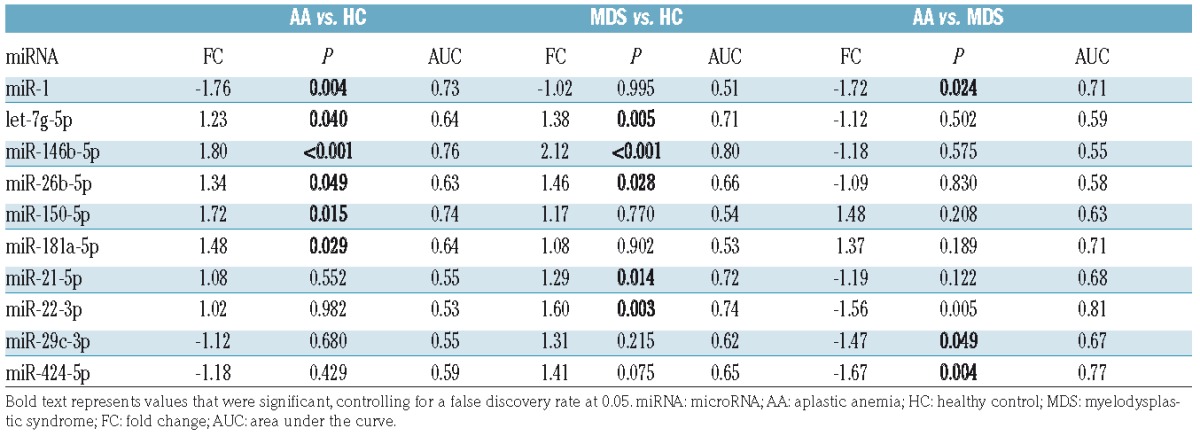
Table 3.
Multivariate logistic regression model in the validation set.
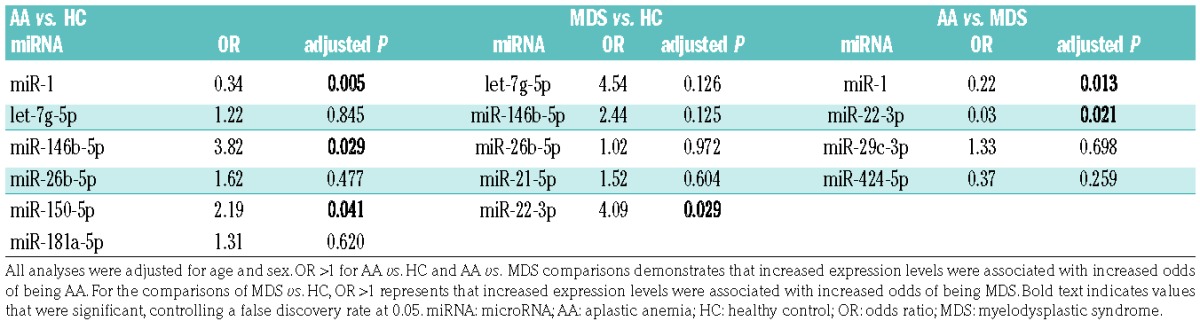
The comparison between MDS and HC revealed 5 miRNAs that exhibited significant association with MDS: let-7g-5p (P=0.005), miR-146b-5p (P=0.0003), miR-26b-5p (P=0.028), miR-21-5p (P=0.014), and miR-22-3p (P=0.003). Subsequent multivariate analysis of these 5 miRNAs showed that only miR-22-3p, which was differentially expressed in the discovery set (Online Supplementary Table S4), significantly associated with MDS (OR=4.09, adjusted P=0.03; Table 3); 4 other miRNAs were not significantly associated with MDS, in part due to the high correlations of the 4 miRNAs with each other. When AA was compared to MDS, 4 miRNAs displayed significant association with AA: miR-1 (P=0.024), miR-22-3p (P=0.005), miR-29c-3p (P=0.049), and miR-424-5p (P=0.004). In multivariate analysis of these miRNAs, significant, independent association with AA was observed only for miR-1 (OR=0.22, adjusted P=0.01) and miR-22-3p (OR=0.03, adjusted P=0.02; Table 3).
Development of a diagnostic panel for AA composed of three plasma miRNAs
Next, the sensitivity and specificity of the miRNAs for the diagnosis of AA were evaluated by receiver operating characteristic (ROC) curve analysis using the validation set: there was strong association of miR-146b-5p [95% confidence interval (CI), 0.65–0.86, P=0.0001], miR-150-5p (95% CI, 0.63–0.85, P=0.0002), and miR-1 (95% CI, 0.62–0.85, P=0.0004) with AA [area under the curve (AUCs) of 0.76, 0.74, and 0.73, respectively; Figure 3A]. Logistic regression was employed to determine the best combination of miRNAs to diagnose AA, demonstrating that a linear combination of expression levels of miR-150-5p, miR-146b-5p, and miR-1 produced the best model. A miRNA biomarker panel composed of miR-150-5p, miR-146b-5p, and miR-1 provided significantly increased AUC of 0.86 (95% CI, 0.78–0.94, P<0.00001; Figure 3A), compared with the use of each miRNA alone. Cut-off values of the diagnostic performances of the model were determined based on the maximum of Youden’s Index of the ROC curve (cut-off value >0.44 for AA vs. HC). In the 3 miRNA-combined panel, a sensitivity of 82% and a specificity of 80% were obtained for the diagnosis of AA, compared with HC.
Figure 3.
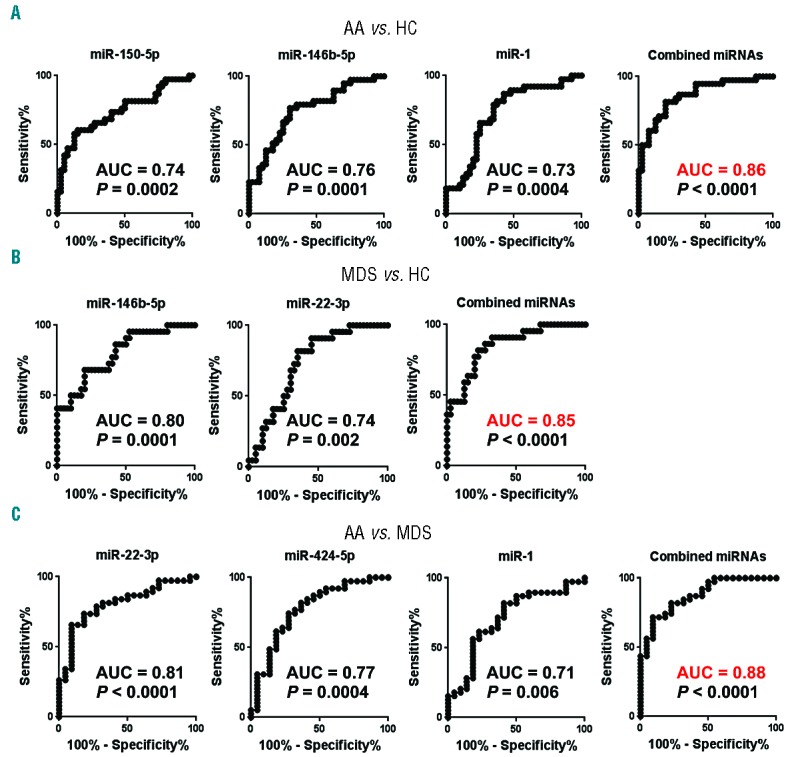
Receiver operating characteristic (ROC) curves of dysregulated miRNAs in the validation set. ROC curves for individual miRNAs in AA vs. HC (A), MDS vs. HC (B), and AA vs. MDS (C). Logistic regression demonstrated a linear combination of values of miRNAs for the compared groups: miR-150-5p, miR-146b-5p, and miR-1 produced the model for AA diagnosis (the equation of the Combined miRNA panel = 4.728 + 0.446 × miR-150-5p + 1.725 × miR-146b-5p − 1.022 × miR-1); miR-146b-5p and miR-22-3p produced the model for MDS diagnosis (the equation of Combined miRNA panel = 9.547 + 1.416 × miR-146b-5p + 1.089 × miR-22-3p), and miR-22-3p, miR-424-5p, and miR-1 produced the model for AA diagnosis compared to MDS (the equation of Combined miRNA panel = −13.117 − 2.046 × miR-22-3p − 1.384 × miR-424-5p − 1.221 × miR-1). The ROC curve of the miRNA panel was generated based on the predicted probability for each patient. Predicted probability = Exponential function (Exp) (Combined miRNA panel) / [1+ Exp (Combined miRNA panel)]. AA: aplastic anemia; HC: healthy control; MDS: myelodysplastic syndrome; AUC: area under the curve; miRNA: microRNA.
By comparing MDS to HC in the validation set, a strong association was observed between miR-146b-5p and MDS, with AUC of 0.80 (95% CI, 0.68–0.91, P=0.0001), and miR-22-3p, with AUC of 0.74 (95% CI, 0.61–0.86) (Figure 3B). In the 2 miRNA-combined panel (miR-146b-5p and miR-22-3p), AUC was increased to 0.85 (95% CI, 0.76–0.95, P<0.00001; Figure 3B). The comparison between AA and MDS revealed strong association of miR-22-3p with AA, having an AUC of 0.81 (95% CI, 0.70–0.92, P<0.0001; Figure 3C), miR-424-5p, with AUC of 0.77 (95% CI, 0.65–0.90), and miR-1 with AUC of 0.71 (95% CI, 0.57–0.85) (Figure 3C). Increased AUC (0.88) was achieved by using a panel composed of these 3 miRNAs (95% CI, 0.79–0.96, P<0.00001; Figure 3C).
Correlations of miRNAs with clinical parameters in AA
In addition to group comparisons, correlation analysis between differentially expressed miRNAs and clinical parameters [complete blood count (CBCs) (absolute neutrophil count (ANC), absolute reticulocyte count (ARC), and platelet count) and age] was performed using the validation set to assess their correlation with disease severity. At the onset of AA, miR-150-5p and miR-146b-5p showed modest but significant negative correlations with platelet counts (r=-0.33, P=0.025) and ARC (r=-0.34, P=0.020), respectively (Online Supplementary Table S5 and Online Supplementary Figure S4). MiR-1 positively correlated with ANC (r=0.30, P=0.038; Online Supplementary Table S5 and Online Supplementary Figure S4). Thus, expression levels of the 3 miRNAs (elevated miR-150-5p and miR-146b-5p and reduced miR-1) were correlated to clinical parameters in AA. There was no correlation between age and the differentially expressed miRNAs (data not shown).
Effects of IST on miRNA expression in AA patients
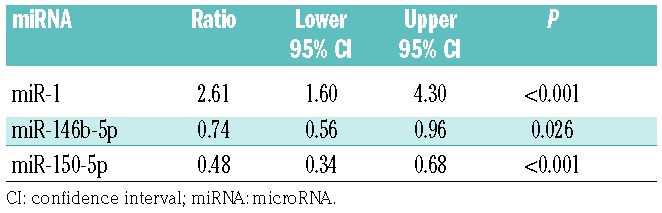
To address whether IST affected the circulating miRNA profile of AA, 40 AA patients with plasma samples before commencing IST and 6 months after IST (Online Supplementary Table S6) were analyzed. Statistically significant changes after IST were detected in all 3 miRNA (miR-150-5p, miR-146b-5p, and miR-1) levels, suggesting the restoration of dysregulated miRNA expressions after therapy (P=0.0001 for miR-150-5p, P=0.0263 for miR-146b-5p, and P=0.0003 for miR-1; Figure 4A and Table 4). In particular, a sharp miR-150-5p decline was seen after IST (48%, 95% CI: 34–68%), compared to before IST (Figure 4A). The use of horse-ATG (n=23) or rabbit-ATG (n=17) did not significantly affect the miRNA expression changes after IST: statistically significant changes after IST were detected for all 3 miRNA levels in both groups.
Figure 4.
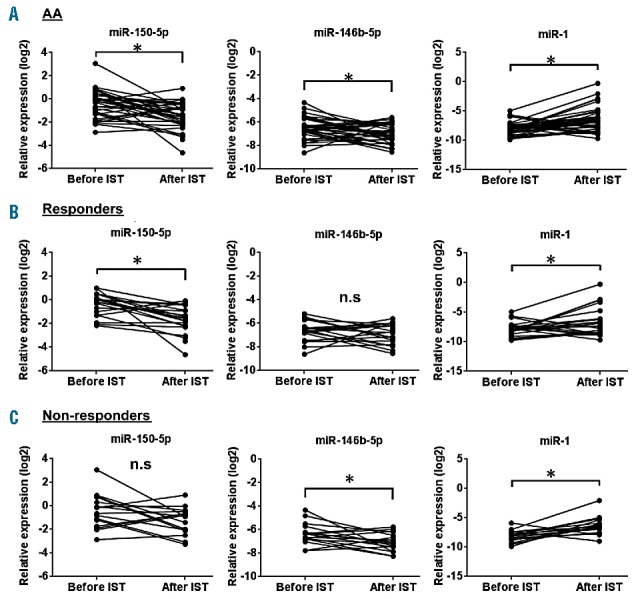
MicroRNA (miRNA) expression changes after IST. (A) MiR-150-5p, miR-146b-5p, and miR-1 expression in the plasma of AA patients at onset (n=40) and after IST at 6 months (n=40). (B) MiR-150-5p, miR-146-5p, and miR-1 expression in the plasma of AA patients (responders) at onset (n=23) and after IST at 6 months (n=23). (C) MiR-150-5p, miR-146-5p, and miR-1 expression in the plasma of AA patients (non-responders) at onset (n=17) and after IST at 6 months (n=17). *P<0.05 (paired two-tailed t-test). AA: aplastic anemia; IST: immunosuppressive therapy; N.S: not significant.
Table 4.
Changes in miRNA expression after immumosuppresive therapies (IST).
Of 40 AA patients, 23 patients (58%) achieved partial responses (PR) or complete responses (CR) while the remaining 17 patients (42%) were non-responders 6 months after IST. When we analyzed the effects of IST in responders (either PR or CR) and non-responders, a reduced miR-150-5p level was observed in responders (P=0.0005; Figure 4B), but there were no statistically significant changes of miR-150-5p expression in non-responders (Figure 4C), indicating that the restoration of miR-150-5p levels after IST was associated with successful treatment. Restoration of miR-1 levels was also observed both in responders and non-responders after IST (Figure 4B,C). Further, miRNA expression levels were compared between responders and non-responders before IST to assess whether miRNA signatures were useful for predicting response to IST. When expression levels of 19 miRNAs at onset were compared between responders (n=23) and non-responders (n=17), none of them was significantly different (Online Supplementary Figure S5).
Discussion
In recent publications, circulating miRNAs have been described as noninvasive biomarkers for many different diseases.17–21 Challenges in the analysis of circulating miRNAs include pre-analytical decisions (such as sample storage and processing) and post-analytical assessments (data normalization).17 Detailed analyses of miRNA spectra in serum and plasma recommend plasma over serum to reduce sample-to-sample variations induced by serum coagulation,26 and we used EDTA anticoagulated plasma in this study to avoid such variations. To characterize miRNA signatures in the plasma of AA, the Serum/Plasma Focus microRNA PCR Panel was used, as it is considered optimal in terms of reproducibility, sensitivity, accuracy, and specificity.27 We found that circulating miRNAs were differentially expressed in AA and MDS vs. HC and in AA vs. MDS, which was validated in a larger separate cohort using a different normalization method. By multivariate analysis, we identified the 5 miRNAs (miR-150-5p, miR-146-5p, miR-1, miR-22-3p, and miR-424-5p) that were significantly associated with AA or MDS, and these miRNA expression patterns were similar between the 2 cohorts. Our study showed the possible utility of “liquid biopsy” to distinguish AA and MDS.
Starting with a total of 179 miRNAs, 19 from the discovery set were investigated and 3 miRNAs were validated as dysregulated in AA. Two miRNAs (miR-150-5p and miR-146b-5p) and miR-1 were significantly elevated and decreased in the plasma of AA patients, respectively. Pathway analysis revealed that these 3 miRNAs targeted important immune-related functions (Figure 5). One of miR-150-5p targets is NOTCH3: the regulation of the Notch pathway through miR-150-5p may impact T-cell development.28 Another target of miR-150-5p is the transcription factor C-MYB which plays an essential role in T-cell differentiation.29 MiR-150-5p is an immuno-miRNA considered to be a crucial regulator of T-cell processes.30 Since AA is a T cell mediated disease, it is interesting that miR-150-5p is selectively expressed in immature resting T cells and then strongly upregulated with their maturation and differentiation of the T-cell progresses.28 MiR-150-5p is a circulating biomarker in female MG patients without immunosuppressive drug treatment, as the miR-150-5p level is reduced with clinical improvement after thymectomy.21 Why miRNA expression changes is unknown. MiRNAs are released into the circulation as a result of apoptotic and necrotic cell death.31 Active secretion is also a potential source of cell-free miRNAs,32 and miR-150-5p is transferred to plasma with CD8+ T cell-derived exosomes.33 We detected increased expression of miR-150-5p in plasma, which might be due to the secretion from active T cells in AA. Of interest, we also observed a negative correlation between miR-150-5p levels and platelet counts. MiR-150-5p is preferentially expressed in megakaryocytic lineage cells and it drives megakaryocyte-erythrocyte progenitor (MEP) differentiation toward megakaryocytes.34 A mechanistic link between thrombopoietin (TPO)-induced miR-150-5p upregulation and megakaryopoiesis has been described.35 Circulating TPO levels are very high in AA.36 Therefore, considering our data demonstrating high miR-150-5p expression in AA plasma, and previous reports,34–36 it is plausible that miR-150-5p promotes megakaryopoiesis in AA. The miR-146b-5p plasma level was able to differentiate both AA and MDS from HC. In addition, pathway analysis identified innate immune pathways related to Toll-like receptors that might be important both in AA and MDS (Figure 5).5,37 MiR-146b-5p decreases TNFa expression in THP-1 monocytes by targeted repression of IRAK1 and TRAF6,38 and miR-146b-5p is involved in feedback regulation of the innate immune response.39 Previous studies have also described alterations of miR-146a-5p and/or miR-146b-5p levels to be associated with inflammatory diseases.11 In a recent study, the regulatory roles of miR-146b-5p in erythropoiesis and megakaryopoiesis have been reported.40 MiR-1 has BCL2 as its target,41 and circulating miR-1 is a potential novel biomarker in acute myocardial infarction.42 MiR-1 exhibits anti-inflammatory roles in asthma mouse models and in inflammatory myopathies.43,44 Decreased levels of miR-1 might reflect an active inflammatory status in AA patients. The recovery of miR-1 levels after IST might be mediated by the immunological effects of ATG + CsA. Collectively, our data suggest that the miRNA signature in AA plasma samples may reflect the aberrant immune response and dysregulated hematopoiesis in AA.
Figure 5.
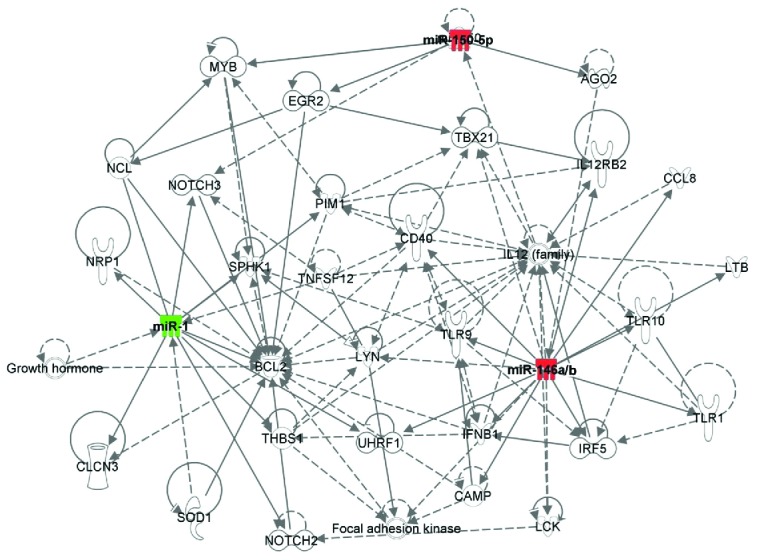
Ingenuity Pathway Analysis (IPA) to identify immune targets of the selected three microRNAs (miRNAs). Shown are network genes of the dysregulated 3 miRNAs (miR-1, miR-146b-5p, and miR-150-5p) in aplastic anemia (AA) plasma, compared to healthy control (HC). Color intensity indicates upregulation (red) and downregulation (green). Solid and dotted lines represent direct and indirect relationships between genes.
We included MDS patients in our study to compare AA to another bone marrow failure syndrome. Others have described an association of miRNA expression with MDS subtypes and disease outcome.45 For example, reduced expression levels of 5 miRNAs (miR-378a-3p, miR-143-3p, miR-143-5p, miR-145-5p, and miR-146a-5p) have been reported in MDS with del(5q),46,47 but these reports focused on miRNA expression in HSPCs. Circulating let-7a and miR-16 from MDS predicted progression-free survival and overall survival,19 and a 7-miRNA signature (let-7a-5p, miR-144-5p, miR-16-5p, miR-25-3p, miR-451a, miR-651, and miR-655) was an independent predictor of survival in MDS.25 Based on the literature, we included 2 miRNAs (let-7a-5p and miR-144-5p) into the custom PCR panel for validation, but we did not observe association of let-7a-5p and miR-144-5p with MDS, probably due to the heterogeneity of the disease, or variations between the sample processing and detection protocols, or our particular MDS patient cohort. In our study, MDS patients were relatively young, compared to the median age of >70 years for typical MDS. Patients enrolled on our research protocols might be more likely to have an immune-mediated pathology compared to typical MDS, with a higher response rate to alemtuzumab.24 Thus, patient selection might lead to the underestimation of our results when comparing AA and MDS. Distinguishing between AA and MDS is often difficult.48 We found that the miR-146b-5p expression level was significantly elevated both in AA and MDS, and miR-1 and miR-22-3p could distinguish AA and MDS, perhaps reflecting commonalities and differences between both AA and MDS. MiR-22-3p, which could distinguish MDS with AA and MDS with HC in our cohort, is an oncogenic miRNA and is upregulated in MDS.49 Regarding autoimmune cytopenias, 7 plasma miRNAs (miR-302c-3p, miR-483-5p, miR-410, miR-544a, miR-302a-3p, miR-223-3p, and miR-597) are differentially expressed in immune thrombocytopenia (ITP).50 These results also suggest the miRNA signatures in AA are disease-related and not simply a reflection of blood counts.
The analysis of serial plasma samples in 40 AA patients suggest potential utility of 3 dysregulated miRNAs (miR-150-5p, miR-146b-5p, and miR-1) as disease biomarkers for diagnosis, as their expression levels were significantly different from those of healthy donors, and levels tended to normalize after IST. Specifically, miR-150-5p may represent a response marker in AA, as its plasma concentration expression was significantly decreased in responders but not in non-responders after IST. Similarly, in cancer patients, not all dysregulated miRNAs at diagnosis have clinical relevance for monitoring, and only some miRNAs normalize after successful treatment.18
Our study showed rather modest miRNA expression changes in AA plasma, but miRNAs may affect biologic functions even with subtle alterations in expression levels.51 ROC curve analysis revealed the potential clinical utility of combined miRNA panels for future applications. Our current study had other limitations, such as the relatively small number of patients. Nonetheless, our data strongly suggest that aberrant miRNA expression was disease-related, especially as the restoration of miRNA levels was observed after IST. In our recent work, we identified concurrent downregulation of 4 miRNAs (miR-126-3p, miR-145-5p, miR-199a-5p, and miR-223-3p) in both CD4+ and CD8+ T cells from AA patients,14 but dysregulation of the 4 miRNAs was not observed in AA plasma samples in the study herein. Previous reports in autoimmune diseases12,20,52–59 and cancers,60–63 have also demonstrated discrepancies of expression profiles between cellular and circulating miRNAs. Systemic lupus erythematosus (SLE) patients exhibit distinct miRNA expression profiles in T cells52–55 and plasma,56 and miRNA profiles in T cells12,57,58 and plasma20,59 are inconsistent in MS. Although circulating miRNAs originate from cells, not all cellular miRNAs can be identified as circulating miRNAs in biofluids. Indeed, miRNA expression profiles are cell-type specific. Of note, regardless of discrepancies between cellular and circulating miRNA profiles, we could observe some common miRNA signatures in both AA and MG, such as decreased miR-145-5p in T cells14,64 and increased miR-150-5p in plasma specimens.21
In conclusion, we demonstrate that expression levels of 3 dysregulated miRNAs in AA plasma associate with clinical parameters and normalize after IST, suggesting their use as potential biomarkers in AA. Importantly, we developed a diagnostic logistic model using combined miRNA panels for diagnosis. Additional studies in larger patient cohorts are required to validate miRNAs as disease biomarkers for diagnostic and therapeutic purposes in bone marrow failure.
Supplementary Material
Acknowledgments
The authors would like to thank Kinneret Broder for assistance in obtaining healthy volunteer samples and Camilo Canel and Barbara R. Gould for designing the custom plate.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/102/1/69
Funding
This research was supported by the Intramural Research Program of the NIH, National Heart, Lung, and Blood Institute, USA.
References
- 1.Young NS, Calado RT, Scheinberg P. Current concepts in the pathophysiology and treatment of aplastic anemia. Blood. 2006;108(8):2509–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scheinberg P, Nunez O, Weinstein B, Biancotto A, Wu CO, Young NS. Horse versus rabbit antithymocyte globulin in acquired aplastic anemia. N Engl J Med. 2011;365(5):430–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Risitano AM, Kook H, Zeng W, Chen G, Young NS, Maciejewski JP. Oligoclonal and polyclonal CD4 and CD8 lymphocytes in aplastic anemia and paroxysmal nocturnal hemoglobinuria measured by V beta CDR3 spectratyping and flow cytometry. Blood. 2002;100(1):178–183. [DOI] [PubMed] [Google Scholar]
- 4.Sloand E, Kim S, Maciejewski JP, Tisdale J, Follmann D, Young NS. Intracellular interferon-gamma in circulating and marrow T cells detected by flow cytometry and the response to immunosuppressive therapy in patients with aplastic anemia. Blood. 2002; 100(4):1185–1191. [DOI] [PubMed] [Google Scholar]
- 5.Zeng W, Kajigaya S, Chen G, Risitano AM, Nunez O, Young NS. Transcript profile of CD4+ and CD8+ T cells from the bone marrow of acquired aplastic anemia patients. Exp Hematol. 2004;32(9):806–814. [DOI] [PubMed] [Google Scholar]
- 6.Hosokawa K, Muranski P, Feng X, et al. Memory stem T cells in autoimmune disease: high frequency of circulating CD8+ memory stem cells in acquired aplastic anemia. J Immunol. 2016;196(4):1568–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132(1):9–14. [DOI] [PubMed] [Google Scholar]
- 8.Quiat D, Olson EN. MicroRNAs in cardiovascular disease: from pathogenesis to prevention and treatment. J Clin Invest. 2013; 123(1):11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marcucci G, Mrozek K, Radmacher MD, Garzon R, Bloomfield CD. The prognostic and functional role of microRNAs in acute myeloid leukemia. Blood. 2011; 117(4): 1121–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136(1):26–36. [DOI] [PubMed] [Google Scholar]
- 11.Nakasa T, Miyaki S, Okubo A, et al. Expression of microRNA-146 in rheumatoid arthritis synovial tissue. Arthritis Rheum. 2008;58(5):1284–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du C, Liu C, Kang J, et al. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat immunol. 2009;10(12):1252–1259. [DOI] [PubMed] [Google Scholar]
- 13.Simpson LJ, Ansel KM. MicroRNA regulation of lymphocyte tolerance and autoimmunity. J Clin Invest. 2015;125(6):2242–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hosokawa K, Muranski P, Feng X, et al. Identification of novel microRNA signatures linked to acquired aplastic anemia. Haematologica. 2015;100(12):1534–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Creemers EE, Tijsen AJ, Pinto YM. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res. 2012;110(3): 483–495. [DOI] [PubMed] [Google Scholar]
- 16.Etheridge A, Lee I, Hood L, Galas D, Wang K. Extracellular microRNA: a new source of biomarkers. Mutat Res. 2011;717(1–2):85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grasedieck S, Sorrentino A, Langer C, et al. Circulating microRNAs in hematological diseases: principles, challenges, and perspectives. Blood. 2013;121(25):4977–4984. [DOI] [PubMed] [Google Scholar]
- 18.Schwarzenbach H, Nishida N, Calin GA, Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol. 2014;11(3):145–156. [DOI] [PubMed] [Google Scholar]
- 19.Zuo Z, Calin GA, de Paula HM, et al. Circulating microRNAs let-7a and miR-16 predict progression-free survival and overall survival in patients with myelodysplastic syndrome. Blood. 2011;118(2):413–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gandhi R, Healy B, Gholipour T, et al. Circulating microRNAs as biomarkers for disease staging in multiple sclerosis. Ann Neurol. 2013;73(6):729–740. [DOI] [PubMed] [Google Scholar]
- 21.Punga T, Le Panse R, Andersson M, Truffault F, Berrih-Aknin S, Punga AR. Circulating miRNAs in myasthenia gravis: miR-150-5p as a new potential biomarker. Ann Clin Transl Neurol. 2014;1(1):49–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Camitta BM, Thomas ED, Nathan DG, et al. Severe aplastic anemia: a prospective study of the effect of early marrow transplantation on acute mortality. Blood. 1976;48(1):63–70. [PubMed] [Google Scholar]
- 23.Scheinberg P, Nunez O, Weinstein B, Wu CO, Young NS. Activity of alemtuzumab monotherapy in treatment-naive, relapsed, and refractory severe acquired aplastic anemia. Blood. 2012;119(2):345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sloand EM, Olnes MJ, Shenoy A, et al. Alemtuzumab treatment of intermediate-1 myelodysplasia patients is associated with sustained improvement in blood counts and cytogenetic remissions. J clin oncol. 2010;28(35):5166–5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuo Z, Maiti S, Hu S, et al. Plasma circulating-microRNA profiles are useful for assessing prognosis in patients with cytogenetically normal myelodysplastic syndromes. Mod Pathol. 2015;28(3):373–382. [DOI] [PubMed] [Google Scholar]
- 26.Wang K, Yuan Y, Cho JH, McClarty S, Baxter D, Galas DJ. Comparing the MicroRNA spectrum between serum and plasma. PLoS One. 2012;7(7):e41561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mestdagh P, Hartmann N, Baeriswyl L, et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods. 2014; 11(8):809–815. [DOI] [PubMed] [Google Scholar]
- 28.Ghisi M, Corradin A, Basso K, et al. Modulation of microRNA expression in human T-cell development: targeting of NOTCH3 by miR-150. Blood. 2011;117(26): 7053–7062. [DOI] [PubMed] [Google Scholar]
- 29.Lieu YK, Kumar A, Pajerowski AG, Rogers TJ, Reddy EP. Requirement of c-myb in T cell development and in mature T cell function. Proc Natl Acad Sci USA. 2004; 101(41):14853–14858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kroesen BJ, Teteloshvili N, Smigielska-Czepiel K, et al. Immuno-miRs: critical regulators of T-cell development, function and ageing. Immunology. 2015;144(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids–the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8(8):467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat cell biol. 2007;9(6):654–659. [DOI] [PubMed] [Google Scholar]
- 33.Bryniarski K, Ptak W, Martin E, et al. Free extracellular miRNA functionally targets cells by transfecting exosomes from their companion cells. PLoS One. 2015; 10(4):e0122991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu J, Guo S, Ebert BL, et al. MicroRNA-mediated control of cell fate in megakaryocyte-erythrocyte progenitors. Dev Cell. 2008;14(6):843–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barroga CF, Pham H, Kaushansky K. Thrombopoietin regulates c-Myb expression by modulating micro RNA 150 expression. Exp Hematol. 2008;36(12):1585–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng X, Scheinberg P, Wu CO, et al. Cytokine signature profiles in acquired aplastic anemia and myelodysplastic syndromes. Haematologica. 2011;96(4):602–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei Y, Dimicoli S, Bueso-Ramos C, et al. Toll-like receptor alterations in myelodysplastic syndrome. Leukemia. 2013;27(9): 1832–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103(33):12481–12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Neill LA, Sheedy FJ, McCoy CE. MicroRNAs: the fine-tuners of Toll-like receptor signalling. Nat Rev Immunol. 2011;11(3):163–175. [DOI] [PubMed] [Google Scholar]
- 40.Zhai PF, Wang F, Su R, et al. The regulatory roles of microRNA-146b-5p and its target platelet-derived growth factor receptor alpha (PDGFRA) in erythropoiesis and megakaryocytopoiesis. J Biol Chem. 2014;289(33):22600–22613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang Y, Zheng J, Sun Y, Wu Z, Liu Z, Huang G. MicroRNA-1 regulates cardiomyocyte apoptosis by targeting Bcl-2. Int Heart J. 2009;50(3):377–387. [DOI] [PubMed] [Google Scholar]
- 42.Ai J, Zhang R, Li Y, et al. Circulating microRNA-1 as a potential novel biomarker for acute myocardial infarction. Biochem Biophys Res Commun. 2010;391(1):73–77. [DOI] [PubMed] [Google Scholar]
- 43.Georgantas RW, Streicher K, Greenberg SA, et al. Inhibition of myogenic microRNAs 1, 133, and 206 by inflammatory cytokines links inflammation and muscle degeneration in adult inflammatory myopathies. Arthritis Rheumatol. 2014;66(4):1022–1033. [DOI] [PubMed] [Google Scholar]
- 44.Takyar S, Vasavada H, Zhang JG, et al. VEGF controls lung Th2 inflammation via the miR-1-Mpl (myeloproliferative leukemia virus oncogene)-P-selectin axis. J Exp Med. 2013;210(10):1993–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rhyasen GW, Starczynowski DT. Deregulation of microRNAs in myelodysplastic syndrome. Leukemia. 2012;26(1):13–22. [DOI] [PubMed] [Google Scholar]
- 46.Starczynowski DT, Kuchenbauer F, Argiropoulos B, et al. Identification of miR-145 and miR-146a as mediators of the 5q-syndrome phenotype. Nat med. 2010;16(1):49–58. [DOI] [PubMed] [Google Scholar]
- 47.Votavova H, Grmanova M, Dostalova Merkerova M, et al. Differential expression of microRNAs in CD34+ cells of 5q- syndrome. J hematol oncol. 2011;4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Afable MG, 2nd, Wlodarski M, Makishima H, et al. SNP array-based karyotyping: differences and similarities between aplastic anemia and hypocellular myelodysplastic syndromes. Blood. 2011;117(25):6876–6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song SJ, Ito K, Ala U, et al. The oncogenic microRNA miR-22 targets the TET2 tumor suppressor to promote hematopoietic stem cell self-renewal and transformation. Cell Stem Cell. 2013;13(1):87–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bay A, Coskun E, Oztuzcu S, Ergun S, Yilmaz F, Aktekin E. Plasma microRNA profiling of pediatric patients with immune thrombocytopenic purpura. Blood Coagul Fibrinolysis. 2014;25(4):379–383. [DOI] [PubMed] [Google Scholar]
- 51.Mestdagh P, Van Vlierberghe P, De Weer A, et al. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10(6):R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ding S, Liang Y, Zhao M, et al. Decreased microRNA-142-3p/5p expression causes CD4+ T cell activation and B cell hyperstimulation in systemic lupus erythematosus. Arthritis Rheum. 2012;64(9):2953–2963. [DOI] [PubMed] [Google Scholar]
- 53.Lu MC, Lai NS, Chen HC, et al. Decreased microRNA(miR)-145 and increased miR-224 expression in T cells from patients with systemic lupus erythematosus involved in lupus immunopathogenesis. Clin Exp Immunol. 2013;171(1):91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stagakis E, Bertsias G, Verginis P, et al. Identification of novel microRNA signatures linked to human lupus disease activity and pathogenesis: miR-21 regulates aberrant T cell responses through regulation of PDCD4 expression. Ann Rheum Dis. 2011;70(8): 1496–1506. [DOI] [PubMed] [Google Scholar]
- 55.Zhao S, Wang Y, Liang Y, et al. MicroRNA-126 regulates DNA methylation in CD4+ T cells and contributes to systemic lupus erythematosus by targeting DNA methyltransferase 1. Arthritis Rheum. 2011;63(5):1376–1386. [DOI] [PubMed] [Google Scholar]
- 56.Carlsen AL, Schetter AJ, Nielsen CT, et al. Circulating microRNA expression profiles associated with systemic lupus erythematosus. Arthritis Rheum. 2013;65(5):1324–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jernas M, Malmestrom C, Axelsson M, et al. MicroRNA regulate immune pathways in T-cells in multiple sclerosis (MS). BMC Immunol. 2013;14:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lindberg RL, Hoffmann F, Mehling M, Kuhle J, Kappos L. Altered expression of miR-17-5p in CD4+ lymphocytes of relapsing-remitting multiple sclerosis patients. Eur J Immunol. 2010;40(3):888–898. [DOI] [PubMed] [Google Scholar]
- 59.Kacperska MJ, Jastrzebski K, Tomasik B, Walenczak J, Konarska-Krol M, Glabinski A. Selected extracellular microRNA as potential biomarkers of multiple sclerosis activity–preliminary study. J Mol Neurosci. 2015;56(1):154–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tanaka M, Oikawa K, Takanashi M, et al. Down-regulation of miR-92 in human plasma is a novel marker for acute leukemia patients. PLoS One. 2009;4(5):e5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhi F, Cao X, Xie X, et al. Identification of circulating microRNAs as potential biomarkers for detecting acute myeloid leukemia. PLoS One. 2013;8(2):e56718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jongen-Lavrencic M, Sun SM, Dijkstra MK, Valk PJ, Lowenberg B. MicroRNA expression profiling in relation to the genetic heterogeneity of acute myeloid leukemia. Blood. 2008;111(10):5078–5085. [DOI] [PubMed] [Google Scholar]
- 63.Garzon R, Volinia S, Liu CG, et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111(6):3183–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang J, Zheng S, Xin N, et al. Identification of novel MicroRNA signatures linked to experimental autoimmune myasthenia gravis pathogenesis: down-regulated miR-145 promotes pathogenetic Th17 cell response. J Neuroimmune Pharmacol. 2013;8(5):1287–1302. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


