Abstract
Studies in model organisms have yielded considerable insights into the etiology of disease and our understanding of evolutionary processes. Caenorhabditis elegans is among the most powerful model organisms used to understand biology. However, C. elegans is not used as extensively as other model organisms to investigate how natural variation shapes traits, especially through the use of genome-wide association (GWA) analyses. Here, we introduce a new platform, the C. elegans Natural Diversity Resource (CeNDR) to enable statistical genetics and genomics studies of C. elegans and to connect the results to human disease. CeNDR provides the research community with wild strains, genome-wide sequence and variant data for every strain, and a GWA mapping portal for studying natural variation in C. elegans. Additionally, researchers outside of the C. elegans community can benefit from public mappings and integrated tools for comparative analyses. CeNDR uses several databases that are continually updated through the addition of new strains, sequencing data, and association mapping results. The CeNDR data are accessible through a freely available web portal located at http://www.elegansvariation.org or through an application programming interface.
INTRODUCTION
Model organisms are necessary to advance our understanding of the molecular underpinnings of biomedical traits and evolutionary processes. Caenorhabditis elegans is a small, free-living nematode found throughout the world. This nematode has several advantages that contribute to its power as an animal model. C. elegans is easily maintained in laboratory environments, has a 3 to 4 day generation time and produces ∼300 offspring per generation (1). The facile genetics and large experimental toolkit have made this organism a highly productive model in addressing biological questions. Furthermore, C. elegans has a transparent body that enables direct observation of developmental and physiological processes (2) and strains can be frozen in liquid nitrogen indefinitely creating a long-term resource for stable genetic stocks. The species also has a small genome that is comprehensively annotated (3). These experimental advances have yielded significant accomplishments, including mapping the cellular lineage of all 959 somatic cells in the hermaphrodite (4,5), a complete wiring diagram of the nervous system (6) and crucial insights into evolutionarily conserved RNA interference (7) and cell-signaling pathways (8).
Remarkably, the majority of discoveries facilitated by the study of C. elegans have come from the use of a single, laboratory-adapted strain from Bristol, England known as N2 (9). Because only one genetic background has been studied extensively, we have much more to learn by using the natural diversity present within this species (10,11). To address this significant gap in our experimental toolkit, a large global population of wild strains has been collected by the C. elegans community and citizen scientists (12,13). These strains serve as a reservoir of natural genetic variation that can be leveraged to understand the genetic drivers of evolutionary processes and the underlying causal variation for traits relevant to biomedicine using genome-wide association (GWA) mappings. These mappings correlate genotypic variation with phenotypic differences across a population to identify quantitative trait loci (QTL) (14).
Even though a few studies have shown the utility of GWA mappings to identify the genetic variation causing phenotypic differences across the C. elegans species (12,13,15–17), the technique has still not been widely adopted. One explanation for the lack of GWA studies in C. elegans is the diverse challenges associated with several necessary steps, each of which has corresponding difficulties. First, researchers require large collections of wild strains. To ensure the fidelity of these strains, care must be taken to avoid strain confusion (18). Second, researchers must genotype this large collection of wild strains to ascertain the genotypic variation for the population. The scale of this task is cost-prohibitive and organizationally difficult. Third, the large number of independent strains must be measured for a trait of interest. Finally, researchers must correlate genotypic variation with phenotypic differences using association mapping to identify QTL. This final task requires computational skills and knowledge of statistical genetics. Altogether, these tasks require considerable laboratory, bioinformatics, and statistical expertise often performed collaboratively.
One strategy used by several model organism communities to facilitate the study of natural variation is to develop centralized repositories of strains, genotype data, and analytical pipelines that perform GWA mappings, obviating the need for laboratories to develop all of these resources independently. For example, Drosophila strains can be obtained from the Drosophila Genetic Reference Panel, a collection of genotyped inbred lines from Raleigh, NC, USA (19). In turn, these lines can be measured for a trait of interest and submitted to a web portal that performs GWA mapping (20). Similar centralized repositories and association mapping portals exist for Arabidopsis thaliana (21–23), and Mus musculus (24,25).
Here, we introduce the C. elegans Natural Diversity Resource (CeNDR), a comprehensive database and set of tools for examining natural variation in C. elegans wild strains and performing GWA mappings. CeNDR organizes metadata on natural strains, provides tools to disseminate these strains to the community, offers whole-genome sequence and variant data for each strain, and enables users to perform GWA mappings and analyze the results. CeNDR also builds upon the ideas of existing resources with an application programming interface (API). CeNDR is freely accessible without registration at http://www.elegansvariation.org. Software used to run CeNDR is open source and is available at http://www.elegansvariation.org/Software. Below, we describe how CeNDR is implemented, relevant applications, the optimized toolkit, and future plans.
IMPLEMENTATION
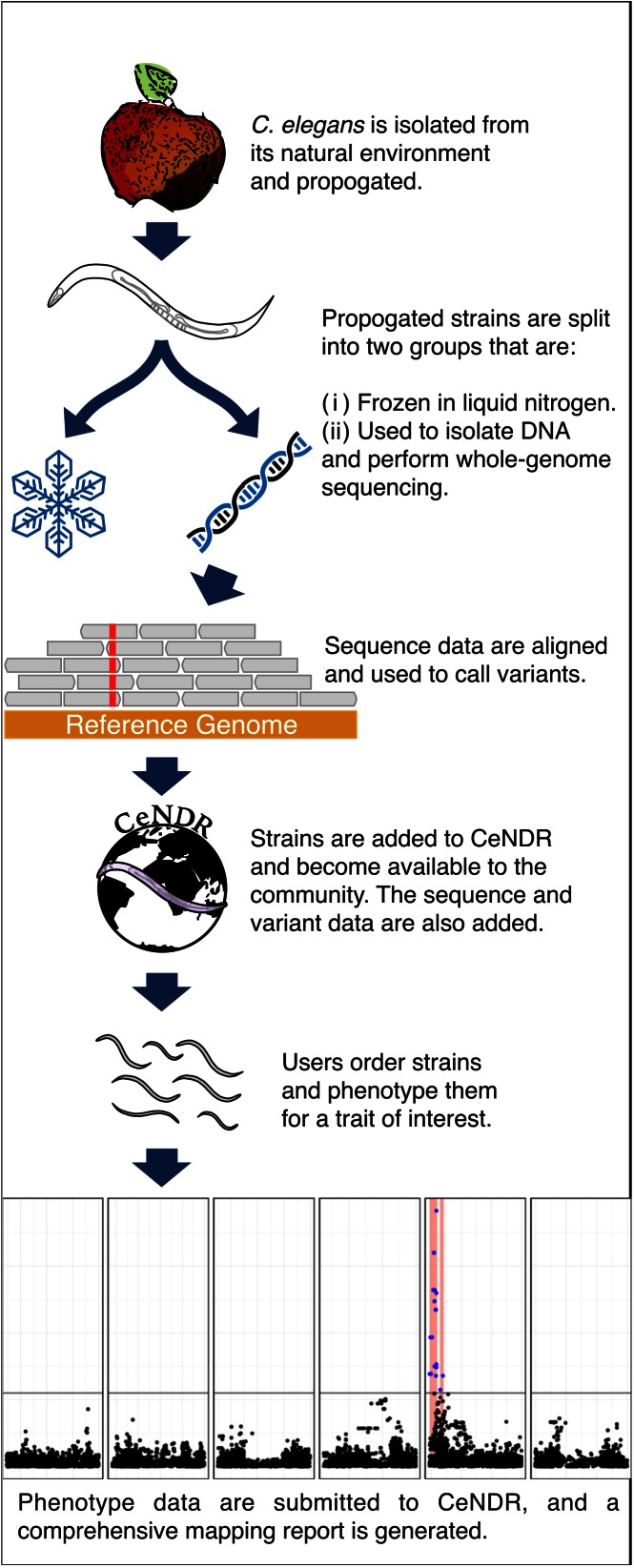
We have built CeNDR to facilitate the study of natural variation with three different areas of focus (Figure 1). First, CeNDR offers a platform for collecting, distributing and maintaining strains isolated from nature. Our laboratory amassed a large collection of wild strains from the C. elegans research community and has developed collection kits for isolating and processing additional strains. Following the receipt of new strains, a single hermaphrodite animal is propagated to ensure that the genotype is genetically distinct from a potentially heterogeneous wild population. We collect information on each strain such as its isolation location, date of collection, substrate where nematodes were found, elevation, etc. These data are integrated into the CeNDR database and can be browsed via a geographic interface on the website (Figure 2A). This dataset is also available for download or accessible through the API. After isolation and propagation of the strains, we split the population to freeze animals for long-term storage and to isolate DNA for whole-genome sequencing. This step ensures that the genotype information obtained from whole-genome sequencing can be connected directly back to a specific strain. Sample mix-ups and strain contamination (9,18) are possible when managing many strains and samples. However, our ability to retain frozen stocks allows us to verify the genetic identity of strains should the need arise and improves the data fidelity for downstream GWA mappings.
Figure 1.
Overview of the CeNDR focus areas.
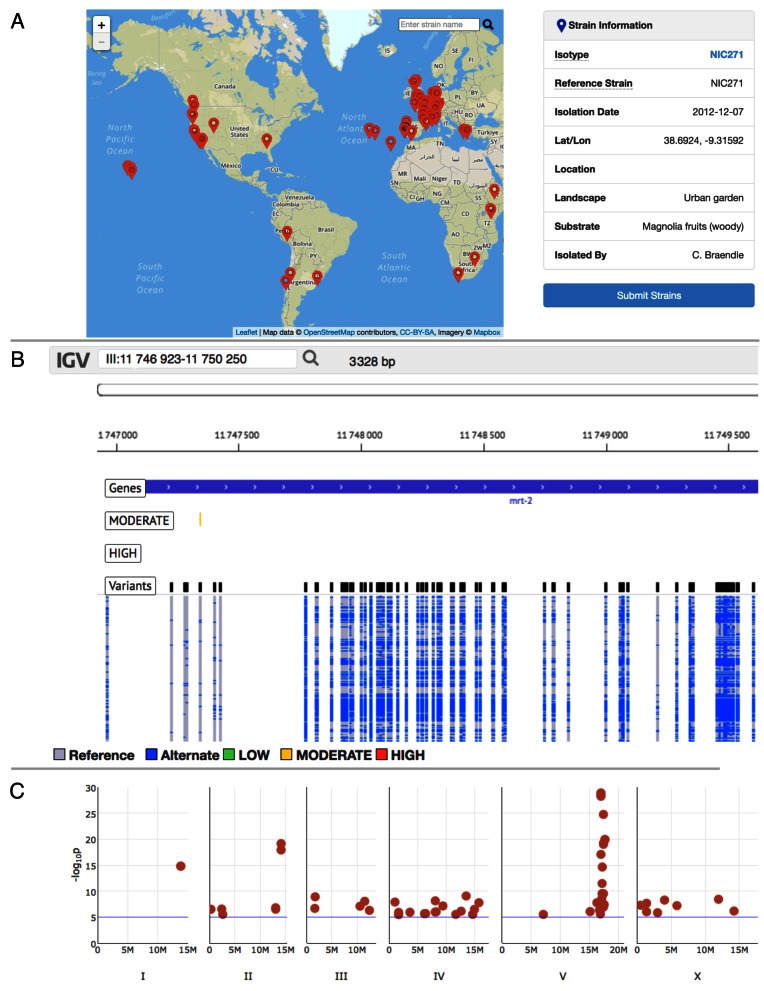
Figure 2.
Selected components of the CeNDR Resource. The following are screenshots of selected components of CeNDR. (A) A tool for interactive geographic exploration of wild isolates based on their isolation location (red markers). Additional information is displayed to the right of the map and is provided when hovering over isolation location. (B) A genome browser for examining genetic variation among wild isolates. Tracks for displaying genes, conservation, and the predicted effects of variants are also available. (C) The results from public statistically significant association mappings are added to a ‘cumulative’ Manhattan plot, which displays the positions of the most significant markers within a QTL confidence interval for each significant mapping.
Second, CeNDR offers whole-genome sequence and variant data of all archived wild isolates, along with metadata on gene conservation and functional studies. Most reproduction in C. elegans occurs through self-fertilization by hermaphrodites, resulting in the propagation of identical individuals near one another in nature. By contrast, distinct strains are sometimes found in the same isolation location. Therefore, we examine the concordance of genetic variation among strains and combine whole-genome sequence data for identical or nearly identical strains into isotypes, which represent genetically distinct genome-wide haplotypes from the same isolation location. The strain set for future GWA mapping experiments comprises a single representative strain from each isotype set. By combining sequence coverage of all strains within an isotype, we obtain high-coverage sequence data that are aligned and used to perform variant calling (see Software used for further details). All variant data are available through the API or can be downloaded in tab-delimited format or Variant Call Format files (26). Aligned sequence data is available in CRAM and BAM formats (27,28). Additionally, we have developed a genome browser for querying and visualizing genetic variation across the C. elegans species (Figure 2B). The genome browser allows users to toggle different tracks that detail genomic information. Available tracks include genes, conservation scores across nematode species (29) (e.g. phyloP (30) and phastCons (31)), single-nucleotide variants (SNV) identified within individual strains, and variant effects predicted with SnpEff (32).
Third, CeNDR combines whole-genome genotype data with measurements of quantitative traits to perform association mappings. The GWA mapping process is optimized for C. elegans, which has been used successfully in many applications (12,13,16). The GWA mapping portal is designed for non-experts and has several user-defined options along with drag-and-drop capabilities. Multiple traits can be submitted simultaneously and organized within a report, which can be kept private indefinitely, embargoed for one year, or made public. Public mapping reports that return significant QTL are added to an interactive graphic that shows all QTL identified to date (Figure 2C). CeNDR uses cloud-based virtual machines to perform GWA analyses. Results are stored in the CeNDR database, and the pipeline outputs a web-based report. Within these reports, we present users with figures, tables, interactive elements, and provide access to data in a tab-delimited format.
Additionally, we have incorporated several datasets from external sources designed to aid in comparative studies of genetic variation across diverse species and to facilitate the identification of candidate genes from GWA mappings. To query whether C. elegans natural variation affects genes conserved in other species, we integrated data from the Homologene database (33), associated human disease gene data listed in the Online Mendelian Inheritance in Man (OMIM) database (34), and a more nematode-focused collection of orthologs and paralogs available from WormBase (29). Once a QTL is identified, we created tools to browse the genes and potential functional connections underlying that genomic region. We integrated functional studies based on RNA interference (RNAi) screens and biochemical pathway predictions. Lastly, we developed features to enable CeNDR to interact with other services and allow access to the underlying databases through an API, which can be used to query, among other things, genetic variants, strain information, mapping report data, and C. elegans genes and homologs.
Software used
CeNDR website: the CeNDR website was developed using Flask (version 0.11.1). It is hosted using Google App Engine. MySQL (version 5.6.26) is used to store strain, variant, homology, and mapping data.
Sequence Analysis: raw FASTQ sequence data has been deposited under NCBI Bioproject accession PRJNA318647. Sequences were aligned to the WS245 reference genome using BWA (version 0.7.8-r455) (35). Optical/PCR duplicates were marked using PICARD (version 1.111). We used bcftools (version 1.3) to perform SNV calling (36), and SnpEff (version 4.1g) (32) to predict functional effects. Data were processed using additional scripts available at http://www.github.com/Andersenlab/vcf-kit.
Association Mapping: association mapping is performed on cloud-based virtual machines. Statistical analysis is performed using R (version 3.2.3) (37). Association mapping is performed within R using rrBLUP (version 4.4) (38). Graphics are generated using ggplot2 (version 2.0.0) (39). The CeNDR website and mapping pipelines are open source and are available on GitHub.com. See www.elegansvariation.org/software for details. We welcome community contributions.
Web-based visualization: the interactive genome browser is implemented using igv.js (version 1.0.0; github.com/igvteam/igv.js). d3.js (version 3; d3js.org), is used for certain interactive visualizations. Geographic visualizations are constructed using leaflet.js (version 0.7.7; leafletjs.com).
APPLICATIONS
Strain distribution and procurement
All wild C. elegans isolates in the CeNDR collection can be requested as individual strains or sets of strains. These sets are organized either into a small panel of 12 divergent strains to assess whether variation exists in a trait across the species or into several larger panels of 48 strains to measure quantitative traits for GWA mappings. Additionally, the data for each strain can be used to investigate ecological or environmental factors that influence C. elegans, including isolation location, substrate where the nematodes were found and the date of isolation. We also allow for anyone to submit C. elegans wild strains. Nematode collection kits are available from the Andersen research group and can be used to isolate new strains of C. elegans. As new strains are identified, they will be entered into CeNDR.
Functional studies of natural variation in C. elegans
Many C. elegans laboratories are interested in a single or small set of genes and the impacts of those genes on diverse traits. Traditional approaches used to study gene function involve the creation of loss-of-function alleles or overexpression of genes to assess phenotypic consequences. However, these methods may result in embryonic lethality or prohibit examination of more subtle aspects of gene function not observable under such extreme perturbations. For these reasons, we created tools to identify genetic variants and their predicted effects for any gene(s) of interest using a genome browser. In contrast to mutagenized strains, variants identified within wild isolates are less likely to be highly deleterious because those alleles would have been removed by natural selection if they negatively affect organismal fitness. Natural genetic variants can be integrated into a desirable genetic background, such as the laboratory-adapted strain N2 (9), using backcrossing or genome editing (40) to evaluate their effects on phenotype.
Comparative studies across Caenorhabditis nematodes and beyond
To investigate evolutionary processes that have occurred over longer time scales, comparative studies are often performed among different species. These studies, from Drosophila (41,42) to Arabidopsis (43), have taught us a great deal about the mechanisms of evolutionary change. Within the Caenorhabditis genus, comparisons of sex determination (44–48), mating behaviors (49) and gene expression regulation (50–52) are among many studies informing topics like the evolution of developmental mechanisms and behaviors. Within CeNDR, we built a homologous gene searching feature into the genome browser that can be used to identify C. elegans orthologs and examine genetic variation within these genes across nematodes and other species. Additionally, the genome browser includes tracks illustrating conservation using phyloP and phastCons scores across the Caenorhabditis genus and other nematode species. These tools allow investigators to rapidly assess whether a gene of interest has natural variation and whether that variation is in a gene region conserved across the genus. Additionally, we provide methods for researchers studying other organisms to identify homologs of their genes of interest in C. elegans and assess whether variation affects the functions of those genes. This tool gives non-C. elegans researchers an approach to test conserved gene functions in this highly tractable system.
Identifying genotype-phenotype correlations
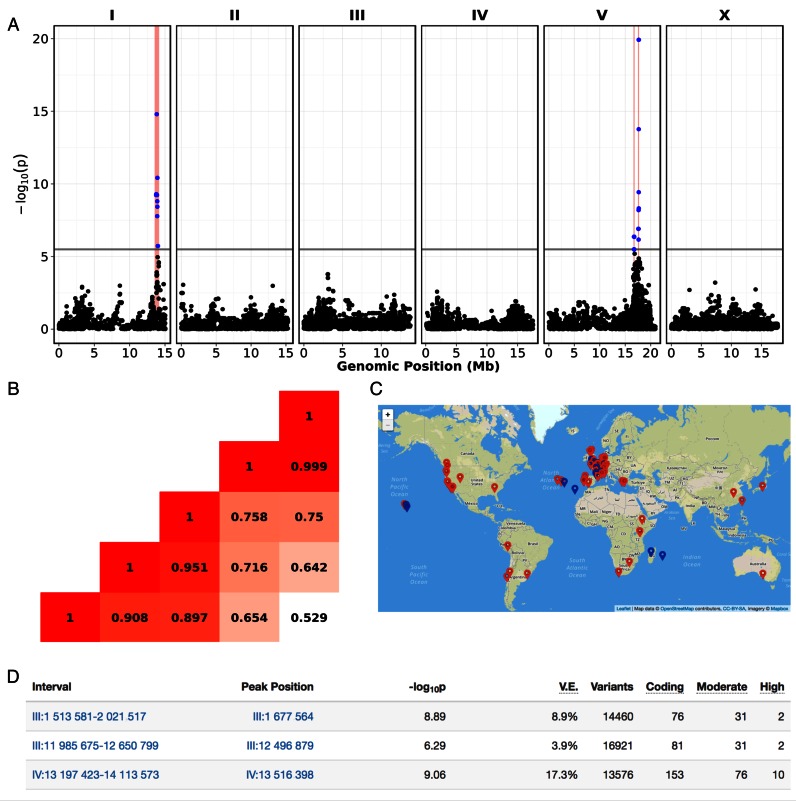
A central goal of GWA mapping is the identification of candidate genes and genetic variants responsible for phenotypic differences across a population. We provide a GWA mapping pipeline optimized for C. elegans (13). This pipeline produces an easy to understand report with figures, tables, descriptions and data aimed at helping users to narrow the list of genes and variants underlying significant GWA signals. Figures include Manhattan plots (Figure 3A) that provide visualization of significance values for all markers used in the statistical test of association and plots depicting the difference in phenotype with respect to genotype at the most significant marker within a QTL confidence interval. Because C. elegans has linkage disequilibrium even among chromosomes (53–55), the correlation of genotype and phenotype identified on multiple chromosomes could be caused by a single region alone. Figures illustrating the linkage disequilibrium among the most significant markers from each associated region are provided to help users interpret mapping results (Figure 3B). Mapping reports also provide two interactive visualizations. The first is a map of the geographic distribution of the most significant marker with the QTL confidence interval (Figure 3C). The second interactive visualization allows users to examine Tajima's D in associated regions, which can be used to suggest whether the genotype-phenotype correlation is caused by processes under neutral, directional, or balancing selection (56). Evidence of selection at a particular locus can indicate that the QTL could have a fitness consequence in nature. Also, a list of genes within the QTL confidence interval and the predicted effects of variants within those genes are provided (Figure 3D). To integrate results obtained from the study of natural variation with the extensive knowledgebase developed from experiments using the laboratory strain, we added tools to connect identified genetic correlations to external data about gene function, including RNAi phenotypes. The genes within a QTL confidence interval can also be connected to human disease genes through the OMIM database. These diverse connections could provide additional insights into the function of a particular gene and how natural variation might affect conserved processes.
Figure 3.
The GWA mapping reports within CeNDR. (A) Manhattan plots provide visualization of significance values for all markers used in the statistical test of association. The y-axis is the negative base 10 log of the P-value obtained from the statistical test of association. The x-axis is the genomic position in millions of base pairs. Markers with a -log10 P-value greater than the Bonferroni-corrected significance threshold (gray line) are considered to be significantly correlated with the phenotype, indicating that linked genetic variation could be causing the observed phenotypic variation. (B) Linkage disequilibrium among the most significant markers from each associated region is displayed. (C) An interactive plot of the geographic location of strains harboring either the reference or alternative marker at the most significant marker within the QTL confidence interval is shown. (D) A summary table of genes and other attributes within the QTL confidence interval is output. The number of protein-coding genes with variants, genes with moderate-impact variants, and genes with high-impact variants are provided.
DISCUSSION
The current version of the C. elegans Natural Diversity Resource (CeNDR, version 1.0.0; August 2016) provides a comprehensive set of tools for examining natural variation in C. elegans and supports a diverse array of applications spanning studies of evolutionary processes to traits conserved with humans. History has shown that centralized resources provide numerous benefits to research communities to address important scientific questions (23,29,33,34,57). CeNDR offers reduced redundancy of data collection (e.g. whole-genome sequencing) along with consistent data collection and organization as a centralized resource. Additionally, the unification of strain management facilitates studies of natural variation across the wide Caenorhabditis community and beyond. Because CeNDR is built as open-source software, it benefits from additional oversight and contributions from an active research community.
CeNDR builds upon the ideas of existing platforms designed to aid studies of natural variation in several ways. First, we uniquely provide access to strains, whole-genome sequence and variant data, and a GWA mapping pipeline within a singular resource. Second, CeNDR is highly extensible by enabling access to strain, variant and mapping report data through an API. Finally, we have developed tools to apply natural variation data beyond C. elegans, including tools for comparative analysis of genetic variation among species.
Future directions
CeNDR will continue to grow in three important areas. First, we will incorporate more wild C. elegans strains, sequence their genomes and identify natural variants. Each year, we will release a new validated set of strains to increase the statistical power of GWA mappings. Second, we will integrate additional classes of natural variants beyond SNVs, including transposon insertion, insertion-deletion, copy-number and genomic rearrangement variants. These additional classes of variation will better inform predictions of functional effects and improve our mapping resolution. Third, we will release new visualization and interactive tools to mine variation, quantitative phenotypes and conservation within and beyond Caenorhabditis.
Acknowledgments
We would like to thank members of the Andersen laboratory for critical comments on this manuscript. We also thank the CeNDR scientific advisory panel: Marie-Anne Félix, Matthew Rockman, Ann Rougvie, and Paul Sternberg.
FUNDING
National Institutes of Health (NIH) [R01GM107227]; American Cancer Society Research Scholar Award (to E.C.A.); Amazon Web Services Research Grant (to E.C.A.); Weinberg College of Arts and Sciences starter innovation award (to E.C.A.); National Science Foundation Graduate Research Fellowship [DGE-1324585 to D.E.C.]; Northwestern University Start-up Funds (to E.C.A.). Funding for open access charge: NIH [R01GM107227].
Conflict of interest statement. None declared.
REFERENCES
- 1.Wood W.B., Others . The Nematode Caenorhabditis Elegans. NY: Cold Spring Harbour Laboratory; 1987. [Google Scholar]
- 2.Corsi A.K., Wightman B., Chalfie M. A transparent window into biology: a primer on Caenorhabditis elegans. Genetics. 2015;200:387–407. doi: 10.1534/genetics.115.176099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 4.Sulston J.E., Schierenberg E., White J.G., Thomson J.N. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- 5.Kimble J., Hirsh D. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev. Biol. 1979;70:396–417. doi: 10.1016/0012-1606(79)90035-6. [DOI] [PubMed] [Google Scholar]
- 6.White J.G., Southgate E., Thomson J.N., Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 7.Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 8.Beitel G.J., Clark S.G., Horvitz H.R. Caenorhabditis elegans ras gene let-60 acts as a switch in the pathway of vulval induction. Nature. 1990;348:503–509. doi: 10.1038/348503a0. [DOI] [PubMed] [Google Scholar]
- 9.Sterken M.G., Snoek L.B., Kammenga J.E., Andersen E.C. The laboratory domestication of Caenorhabditis elegans. Trends Genet. 2015;31:224–231. doi: 10.1016/j.tig.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frézal L., Félix M.-A. C. elegans outside the Petri dish. Elife. 2015;4 doi: 10.7554/eLife.05849. doi:10.7554/eLife.05849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Félix M.-A., Braendle C. The natural history of Caenorhabditis elegans. Curr. Biol. 2010;20:R965–R969. doi: 10.1016/j.cub.2010.09.050. [DOI] [PubMed] [Google Scholar]
- 12.Andersen E.C., Gerke J.P., Shapiro J.A., Crissman J.R., Ghosh R., Bloom J.S., Félix M.-A., Kruglyak L. Chromosome-scale selective sweeps shape Caenorhabditis elegans genomic diversity. Nat. Genet. 2012;44:285–290. doi: 10.1038/ng.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cook D.C., Zdraljevic S., Tanny R.E., Seo B., Riccardi D.D., Noble L.M., Rockman M.V., Alkema M.J., Braendle C., Kammenga J.E., et al. The genetic basis of natural variation in Caenorhabditis elegans telomere length. Genetics. 2016;204:371–383. doi: 10.1534/genetics.116.191148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bush W.S., Moore J.H. Chapter 11: genome-wide association studies. PLoS Comput. Biol. 2012;8:e1002822. doi: 10.1371/journal.pcbi.1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rockman M.V., Kruglyak L. Recombinational landscape and population genomics of Caenorhabditis elegans. PLoS Genet. 2009;5:e1000419. doi: 10.1371/journal.pgen.1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghosh R., Andersen E.C., Shapiro J.A., Gerke J.P., Kruglyak L. Natural variation in a chloride channel subunit confers avermectin resistance in C. elegans. Science. 2012;335:574–578. doi: 10.1126/science.1214318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hodgkin J., Doniach T. Natural variation and copulatory plug formation in Caenorhabditis elegans. Genetics. 1997;146:149–164. doi: 10.1093/genetics/146.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGrath P.T., Rockman M.V., Zimmer M., Jang H., Macosko E.Z., Kruglyak L., Bargmann C.I. Quantitative mapping of a digenic behavioral trait implicates globin variation in C. elegans sensory behaviors. Neuron. 2009;61:692–699. doi: 10.1016/j.neuron.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mackay T.F.C., Richards S., Stone E.A., Barbadilla A., Ayroles J.F., Zhu D., Casillas S., Han Y., Magwire M.M., Cridland J.M., et al. The Drosophila melanogaster genetic reference panel. Nature. 2012;482:173–178. doi: 10.1038/nature10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang W., Massouras A., Inoue Y., Peiffer J., Ràmia M., Tarone A.M., Turlapati L., Zichner T., Zhu D., Lyman R.F., et al. Natural variation in genome architecture among 205 Drosophila melanogaster genetic reference panel lines. Genome Res. 2014;24:1193–1208. doi: 10.1101/gr.171546.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Childs L.H., Lisec J., Walther D. Matapax: an online high-throughput genome-wide association study pipeline. Plant Physiol. 2012;158:1534–1541. doi: 10.1104/pp.112.194027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seren Ü., Vilhjálmsson B.J., Horton M.W., Meng D., Forai P., Huang Y.S., Long Q., Segura V., Nordborg M. GWAPP: a web application for genome-wide association mapping in Arabidopsis. Plant Cell. 2012;24:4793–4805. doi: 10.1105/tpc.112.108068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamesch P., Berardini T.Z., Li D., Swarbreck D., Wilks C., Sasidharan R., Muller R., Dreher K., Alexander D.L., Garcia-Hernandez M., et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang H.M., Zaitlen N.A., Wade C.M., Kirby A., Heckerman D., Daly M.J., Eskin E. Efficient control of population structure in model organism association mapping. Genetics. 2008;178:1709–1723. doi: 10.1534/genetics.107.080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bennett B.J., Farber C.R., Orozco L., Kang H.M., Ghazalpour A., Siemers N., Neubauer M., Neuhaus I., Yordanova R., Guan B., et al. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res. 2010;20:281–290. doi: 10.1101/gr.099234.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonfield J.K., Mahoney M.V. Compression of FASTQ and SAM format sequencing data. PLoS One. 2013;8:e59190. doi: 10.1371/journal.pone.0059190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Howe K.L., Bolt B.J., Cain S., Chan J., Chen W.J., Davis P., Done J., Down T., Gao S., Grove C., et al. WormBase 2016: expanding to enable helminth genomic research. Nucleic Acids Res. 2016;44:D774–D780. doi: 10.1093/nar/gkv1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pollard K.S., Hubisz M.J., Rosenbloom K.R., Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cingolani P., Platts A., Wang L.L.L., Coon M., Nguyen T., Wang L.L.L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w 1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D., et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amberger J.S., Bocchini C.A., Schiettecatte F., Scott A.F., Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43:D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.R Core Team. R Core Team. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2013. [Google Scholar]
- 38.Endelman J.B. Ridge regression and other kernels for genomic selection with R Package rrBLUP. Plant Genome J. 2011;4:250–255. [Google Scholar]
- 39.Wickham H. ggplot2: Elegant Graphics for Data Analysis. NY: Springer; 2009. [Google Scholar]
- 40.Frøkjær-Jensen C. Exciting prospects for precise engineering of Caenorhabditis elegans genomes with CRISPR/Cas9. Genetics. 2013;195:635–642. doi: 10.1534/genetics.113.156521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drosophila 12 Genomes Consortium. Clark A.G., Eisen M.B., Smith D.R., Bergman C.M., Oliver B., Markow T.A., Kaufman T.C., Kellis M., Gelbart W., et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- 42.Ranz J.M., Castillo-Davis C.I., Meiklejohn C.D., Hartl D.L. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science. 2003;300:1742–1745. doi: 10.1126/science.1085881. [DOI] [PubMed] [Google Scholar]
- 43.Novikova P.Y., Hohmann N., Nizhynska V., Tsuchimatsu T., Ali J., Muir G., Guggisberg A., Paape T., Schmid K., Fedorenko O.M., et al. Sequencing of the genus Arabidopsis identifies a complex history of nonbifurcating speciation and abundant trans-specific polymorphism. Nat. Genet. 2016;48:1077–1082. doi: 10.1038/ng.3617. [DOI] [PubMed] [Google Scholar]
- 44.de Bono M., Hodgkin J. Evolution of sex determination in caenorhabditis: unusually high divergence of tra-1 and its functional consequences. Genetics. 1996;144:587–595. doi: 10.1093/genetics/144.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hill R.C., de Carvalho C.E., Salogiannis J., Schlager B., Pilgrim D., Haag E.S. Genetic flexibility in the convergent evolution of hermaphroditism in Caenorhabditis nematodes. Dev. Cell. 2006;10:531–538. doi: 10.1016/j.devcel.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 46.Woodruff G.C., Eke O., Baird S.E., Félix M.-A., Haag E.S. Insights into species divergence and the evolution of hermaphroditism from fertile interspecies hybrids of Caenorhabditis nematodes. Genetics. 2010;186:997–1012. doi: 10.1534/genetics.110.120550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haag E.S., Kimble J. Regulatory elements required for development of caenorhabditis elegans hermaphrodites are conserved in the tra-2 homologue of C. remanei, a male/female sister species. Genetics. 2000;155:105–116. doi: 10.1093/genetics/155.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo Y., Chen X., Ellis R.E. Evolutionary change within a bipotential switch shaped the sperm/oocyte decision in hermaphroditic nematodes. PLoS Genet. 2013;9:e1003850. doi: 10.1371/journal.pgen.1003850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rene Garcia L., LeBoeuf B., Koo P. Diversity in mating behavior of hermaphroditic and male–female Caenorhabditis nematodes. Genetics. 2007;175:1761–1771. doi: 10.1534/genetics.106.068304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tu S., Wu M.Z., Wang J., Cutter A.D., Weng Z., Claycomb J.M. Comparative functional characterization of the CSR-1 22G-RNA pathway in Caenorhabditis nematodes. Nucleic Acids Res. 2015;43:208–224. doi: 10.1093/nar/gku1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barrière A., Ruvinsky I. Pervasive divergence of transcriptional gene regulation in Caenorhabditis nematodes. PLoS Genet. 2014;10:e1004435. doi: 10.1371/journal.pgen.1004435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yanai I., Hunter C.P. Comparison of diverse developmental transcriptomes reveals that coexpression of gene neighbors is not evolutionarily conserved. Genome Res. 2009;19:2214–2220. doi: 10.1101/gr.093815.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cutter A.D. Nucleotide polymorphism and linkage disequilibrium in wild populations of the partial selfer Caenorhabditis elegans. Genetics. 2006;172:171–184. doi: 10.1534/genetics.105.048207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haber M., Schüngel M., Putz A., Müller S., Hasert B., Schulenburg H. Evolutionary history of Caenorhabditis elegans inferred from microsatellites: evidence for spatial and temporal genetic differentiation and the occurrence of outbreeding. Mol. Biol. Evol. 2005;22:160–173. doi: 10.1093/molbev/msh264. [DOI] [PubMed] [Google Scholar]
- 55.Barrière A., Félix M.-A. High local genetic diversity and low outcrossing rate in Caenorhabditis elegans natural populations. Curr. Biol. 2005;15:1176–1184. doi: 10.1016/j.cub.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 56.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Speir M.L., Zweig A.S., Rosenbloom K.R., Raney B.J., Paten B., Nejad P., Lee B.T., Learned K., Karolchik D., Hinrichs A.S., et al. The UCSC Genome Browser database: 2016 update. Nucleic Acids Res. 2016;44:D717–D725. doi: 10.1093/nar/gkv1275. [DOI] [PMC free article] [PubMed] [Google Scholar]