The global proteome and ubiquitylome were negatively correlated, and ubiquitination could be involved in the degradation of proteins during ethylene-mediated corolla senescence in petunias.
Abstract
Petal senescence is a complex programmed process. It has been demonstrated previously that treatment with ethylene, a plant hormone involved in senescence, can extensively alter transcriptome and proteome profiles in plants. However, little is known regarding the impact of ethylene on posttranslational modification (PTM) or the association between PTM and the proteome. Protein degradation is one of the hallmarks of senescence, and ubiquitination, a major PTM in eukaryotes, plays important roles in protein degradation. In this study, we first obtained reference petunia (Petunia hybrida) transcriptome data via RNA sequencing. Next, we quantitatively investigated the petunia proteome and ubiquitylome and the association between them in petunia corollas following ethylene treatment. In total, 51,799 unigenes, 3,606 proteins, and 2,270 ubiquitination sites were quantified 16 h after ethylene treatment. Treatment with ethylene resulted in 14,448 down-regulated and 6,303 up-regulated unigenes (absolute log2 fold change > 1 and false discovery rate < 0.001), 284 down-regulated and 233 up-regulated proteins, and 320 up-regulated and 127 down-regulated ubiquitination sites using a 1.5-fold threshold (P < 0.05), indicating that global ubiquitination levels increase during ethylene-mediated corolla senescence in petunia. Several putative ubiquitin ligases were up-regulated at the protein and transcription levels. Our results showed that the global proteome and ubiquitylome were negatively correlated and that ubiquitination could be involved in the degradation of proteins during ethylene-mediated corolla senescence in petunia. Ethylene regulates hormone signaling transduction pathways at both the protein and ubiquitination levels in petunia corollas. In addition, our results revealed that ethylene increases the ubiquitination levels of proteins involved in endoplasmic reticulum-associated degradation.
Flowers have limited life spans and are irreversibly programmed to undergo senescence; therefore, they represent an excellent model system in which to study senescence (Jones et al., 2005). Postharvest longevity is an important characteristic of cut flowers. Studying petal senescence may provide insight into the mechanisms of plant senescence in general and provide a means to improve the vase lives of cut flowers (Borochoy et al., 1997).
Senescence is regulated at several levels, including mRNA, protein, and posttranslational modification (PTM; van Doorn and Woltering, 2008; Woo et al., 2013). The gaseous plant hormone ethylene exerts significant effects on flower senescence (Abeles, 1992; Ecker, 1995; Douglas, 2014). Many flowers are classified as ethylene sensitive, including petunia (Petunia hybrida) and carnation (Dianthus caryophyllus; Woltering and Van Doorn, 1988). In these flowers, ethylene production peaks close to senescence. The application of exogenous ethylene enhances this process, whereas inhibition of ethylene synthesis or activity slows senescence (Reid and Wu, 1992). Previous studies have demonstrated that ethylene treatment can extensively alter transcriptome and proteome profiles in plants (Prayitno et al., 2006; Mayuoni et al., 2011; Slade et al., 2012; Cheng et al., 2013).
Protein degradation is one of the hallmarks of senescence (Shahri and Tahir, 2014). Ubiquitination, a well-known PTM, plays important roles in protein degradation (Wilkinson, 2000). Ubiquitin is a highly conserved 76-amino acid polypeptide that is found throughout the eukaryotic kingdom. In vivo, polyubiquitin chains are most frequently linked through Lys-48, and the canonical ubiquitin signal is recognized by the 26S proteasome and thereby targets tagged proteins for degradation (Peng et al., 2003). Among six other Lys residues of ubiquitin, at least four (Lys-6, Lys-11, Lys-29, and Lys-63) can function as a linkage for polyubiquitin chains (Arnason and Ellison, 1994; Peng et al., 2003). Lys-11- and Lys-29-linked polyubiquitin chains may target proteins to the proteasome (Johnson et al., 1995; Baboshina and Haas, 1996). Conjugation of monoubiquitylation is a regulatory modification involved in diverse processes, including transcription, histone function, endocytosis, DNA repair, viral budding, and membrane trafficking (Schnell and Hicke, 2003; Passmore and Barford, 2004). The attachment of the ubiquitins to proteins involves three classes of enzyme: ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin ligases (E3; Hochstrasser, 1995). Ubiquitinated substrates may be degraded to peptides by the multisubunit 26S protease. However, no attempts have been made to perform PTM analysis to characterize the ubiquitination of the proteome or the association between modifications and the proteome during flower senescence in response to ethylene.
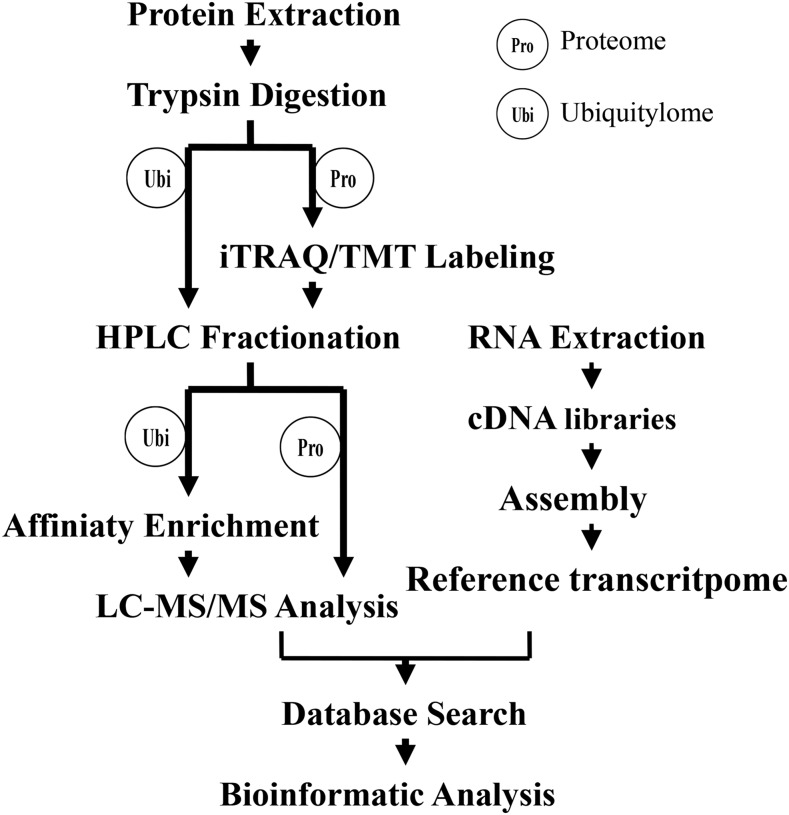
Petunia has served as a model plant for the molecular and biochemical analysis of flower senescence (Gerats and Vandenbussche, 2005). In this study, a reference transcriptome data set from petunia was first obtained via RNA sequencing. Then, using iTRAQ and a label-free quantitative strategy involving antibody-based affinity enrichment and high-resolution liquid chromatography-tandem mass spectrometry analysis, we generated proteome and ubiquitylome analyses of petunia corollas with and without ethylene treatment (Fig. 1). In total, 51,799 unigenes, 3,606 proteins, and 2,270 ubiquitination sites were quantified in response to 16 h of ethylene treatment. Ethylene treatment altered the proteome and ubiquitylome profiles of petunia corollas. The correlation between the proteome and ubiquitylome also is described. Finally, the function of ubiquitination in protein degradation during ethylene-mediated corolla senescence in petunia and the effects of ethylene on proteins involved in hormone biosynthesis, signaling transduction, amino acid biosynthesis, endoplasmic reticulum-associated degradation (ERAD), and other processes are discussed.
Figure 1.
Systematic workflow for quantitative profiling of the global proteome and ubiquitylome in petunia corollas upon ethylene treatment. LC-MS/MS, Liquid chromatography-tandem mass spectrometry.
RESULTS AND DISCUSSION
Ethylene Treatment Accelerates Corolla Wilting and Decreases Fresh Weight and Total Protein Content
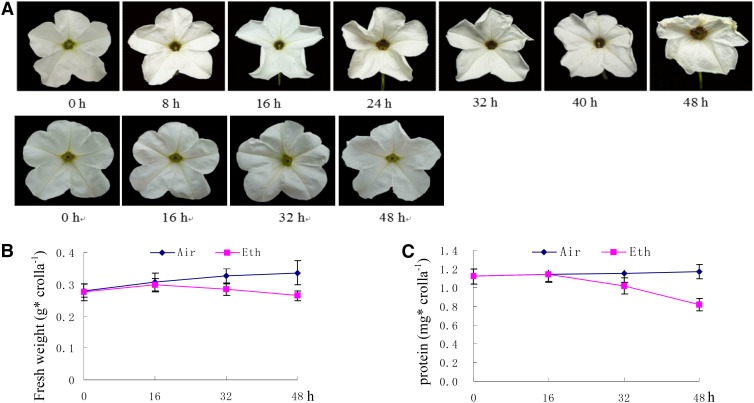
The petunia evaluated (cv Mitchell) exhibited the first visible symptom of senescence, the wilting of the corolla, at approximately 16 h after 2 µL L−1 ethylene treatment. The margins of the corollas began to involute, and a few translucent dots appeared in the corollas (Fig. 2A); however, the corolla fresh weight and protein content remained constant. At 32 h after ethylene treatment, the petunias exhibited obvious symptoms of senescence (Fig. 2, B and C), and the corolla fresh weight and protein content decreased to approximately 87% and 88%, respectively, compared with air-treated, control corollas. These decreases coincided with corolla wilting. Air-treated petunia corollas were fully turgid 0 to 48 h after flower opening, exhibited no symptoms of senescence, and visually were indistinguishable from flowers at anthesis (Fig. 2A). We selected a 2 µL L−1, 16-h ethylene treatment and a 16-h air treatment to perform transcriptome, proteome, and ubiquitylome analyses.
Figure 2.
Effect of ethylene on flowers of cv Mitchell. A, Flower profiles with ethylene treatment (top) or without treatment (bottom). B, Fresh weights of corollas with or without ethylene treatment. C, Protein contents of corollas with or without ethylene treatment. Corollas were collected from at least five flowers on various days after flower opening. Total protein was determined using the Bradford assay. Data represent means of three replicates ± se. Experiments were conducted at least twice with similar results.
Ethylene Treatment Increases Ubiquitin in Petunia Corollas at the Protein Level
To examine the effects of ethylene on the ubiquitin protein, western blotting was performed to examine the expression patterns of ubiquitin in petunia corollas in response to ethylene treatment. As shown in Supplemental Figure S1, ethylene treatment significantly increased the expression of ubiquitin at the protein level in petunia corollas. These results implied that the ubiquitin-proteasome system may play a role during ethylene-mediated corolla senescence.
RNA Sequencing and Assembly
To comprehensively construct the complete transcriptome of cv Mitchell, eight tissues, the roots, stems, leaves, buds (0.4 cm), buds (0.8 cm), corollas (8 h post ethylene treatment), corollas (16 h post ethylene treatment), and corollas (16 h post air treatment), were harvested for RNA isolation. Shotgun libraries were constructed and sequenced on an Illumina High-Seq 2000 platform according to the manufacturer’s instructions. In total, approximately 247.25 million paired-end reads with read lengths of 100 bp were generated (Supplemental Table S1). After quality checks, adapter trimming, and size selection, de novo assembly was performed using Trinity. A final high-quality data set of 72,249 unigenes longer than 200 bp with an average length of 820 bp and an N50 of 1,379 bp was obtained (Supplemental Table S2; Sequence Read Archive accession no. SRP077541).
To perform functional annotation of the petunia transcriptome, the unigene sequences were BLAST searched against the National Center for Biotechnology Information nonredundant protein database and the SwissProt, Cluster of Orthologous Groups, and Kyoto Encyclopedia of Genes and Genomes (KEGG) protein databases with a cutoff E value of 10−5. A total of 41,035 unigenes (56.8% of the total assembled unigenes) were aligned to the four protein databases (Supplemental Table S3; Supplemental Fig. S2). The 40,341 predicted amino acid sequences of the unigenes are shown in Supplemental File Exc S1. Tandem mass spectra were searched against these sequences to analyze the proteome and ubiquitylome, the analysis of which we focused on in this study.
Ethylene Treatment Alters the Transcriptome in Petunia Corollas
To quantify the expression levels of the transcripts of 16-h ethylene and air treatment corollas, HTseq was used to count the read numbers mapped to each gene based on the 72,249 genes in the petunia reference transcriptome. These data were then normalized to reads in a given unigene per million mapped reads. A total of 51,799 unigenes available for both ethylene and air treatment were analyzed. This analysis indicated that 20,751 unigenes were differentially expressed (absolute log2 fold change > 1 and false discovery rate < 0.001), including 14,448 (69.6%) down-regulated and 6,303 (30.4%) up-regulated unigenes, whereas 31,048 unigenes were not differentially expressed. Of the 20,751 differentially expressed unigenes (DEGs), 15,472 were annotated, including 10,753 down-regulated and 4,719 up-regulated unigenes after ethylene treatment (Supplemental File Exc S2). Previous studies showed that ethylene treatment resulted in 935 down-regulated and 1,666 up-regulated genes in the auxiliary bud tissue of soybean (Glycine max; Prayitno et al., 2006), and ethylene treatment resulted in 331 (50%) down-regulated and 330 (50%) up-regulated genes in Citrus reticulata fruits (Mayuoni et al., 2011), which suggested a differential impact of ethylene on different species and tissues or differences attributable to ethylene treatment time or concentration.
To investigate the influence of the DEGs on pathways, a statistical pathway enrichment analysis of ethylene and air treatment corollas was performed based on the KEGG database using fold change and false discovery rate. The DEGs from 16-h ethylene and air treatment corollas were enriched in 22 KEGG metabolic pathways (Supplemental File Exc S2). The top 10 P < 0.05 metabolic pathways of the DEGs in ethylene and air treatment corollas were as follows: plant hormone signal transduction, photosynthesis, carotenoid biosynthesis, inositol phosphate metabolism, photosynthesis-antenna proteins, homologous recombination, ubiquinone and other terpenoid-quinone biosynthesis, flavonoid biosynthesis, Phe, Tyr, and Trp biosynthesis, and porphyrin and chlorophyll metabolism.
Significant pathway enrichment analysis showed that plant hormone signal transduction was the most important pathway in the ethylene-versus-air comparison, and plant hormone signal transduction was the key biological event. Plant hormone signal transduction is very important for hormone-induced biochemical changes during plant growth, development, and environmental information-processing pathways. Previous studies showed that ethylene interacts with plant hormones at different levels to form a network of signaling pathways connected by antagonistic and synergistic interactions (Sun et al., 2006; Stepanova et al., 2007). Our evidence indicated that the genes involved in plant hormone signal transduction play important roles in ethylene-induced senescence in petunia corolla.
Confirmation of DEG Data by Quantitative Real-Time PCR
To confirm the results of the gene expression analysis obtained using DEG data, transcriptional regulation revealed by RNA sequencing was assessed in a biologically independent experiment using quantitative real-time (qRT) PCR. We randomly selected 20 genes as candidate genes. The results for the 20 candidate genes are shown in Supplemental Figure S3. Overall, the qRT PCR data were in agreement (pairwise correlation coefficient of 0.87, P = 5.1092E-7) with the DEG results. Thus, our data showed that the DEG technique for counting transcripts reflects transcript abundance and can be used for gene expression analysis in an organism lacking genome information.
Ethylene Treatment Changes the Proteome Profile in Petunia Corollas
To examine the whole proteome in corollas in response to ethylene, three biological replicates were analyzed for each treatment. In total, 5,189 protein groups were identified from petunia, among which 3,606 proteins were quantified. A total of 233 proteins were up-regulated and 284 proteins were down-regulated (with a threshold of 1.5-fold) in response to ethylene (P < 0.05) with a high degree of repeatability (Supplemental File Exc S3).
To elucidate the functional differences between the down-regulated and up-regulated proteins, the quantified proteins were analyzed for Gene Ontology (GO) enrichment based on clustering analysis (Supplemental Fig. S4; Supplemental File Exc S4). In the cellular component category, many of the down-regulated proteins were enriched in the ribosome and ribosomal subunit category, whereas the up-regulated proteins were not enriched in any cellular component category. In the genus Iris, one of the earliest ultrastructural senescence symptoms is the loss of the majority of ribosomes (Van Doorn et al., 2003). In harvest-induced senescence in detached Arabidopsis (Arabidopsis thaliana) plants, genes involved in ribosome biogenesis and assembly are down-regulated (Chang et al., 2015). These results suggest that protein processing might be suppressed during senescence in plants.
In terms of biological processes, a large portion of the up-regulated proteins were highly enriched in the heterocycle catabolic process, cellular nitrogen compound catabolic process, aromatic compound catabolic process, disaccharide metabolic process, organic cyclic compound catabolic process, Suc metabolic process, and others. In petunia, it has been found that elements such as carbon, nitrogen, phosphorus, potassium, and some metal ions are reduced in corollas during pollination-induced senescence (Paul and Frigerio, 2007). These results suggest that a different nutrient remobilization program operates during pollination- or ethylene-induced senescence. Moreover, it has been shown that carbohydrates are transported primarily in the phloem during petal senescence (van Doorn and Woltering, 2008). In our results, down-regulated proteins were enriched in the organ nitrogen compound biosynthetic process, aromatic amino acid family metabolic process, aromatic amino acid family biosynthetic process, cellular amino acid biosynthetic process, small molecule biosynthetic process, organic acid biosynthetic process, carboxylic acid biosynthetic process, aromatic compound biosynthetic process, and others. These results suggest that ethylene treatment likely promotes many catabolic processes while inhibiting certain biosynthetic processes, suggesting an intrinsic role for ethylene as a senescence enhancer.
The analysis of molecular functions showed that many of the up-regulated proteins were highly enriched for the following: oxidoreductase activity, acting on paired donors, iron ion binding, transferase activity, hexosyl groups, transition metal ion binding, Cys-type peptidase activity, UDP-glucosyltransferase activity, Suc synthase activity, heme binding, transferase activity, transferring glycosyl groups, tetrapyrrole binding, glucosyltransferase activity, and UDP-glycosyltransferase activity. The down-regulated proteins were enriched in transferase activity, transferring alkyl or aryl groups, structural constituent of ribosome, Met adenosyltransferase activity, and 3-deoxy-7-phosphoheptulonate synthase activity. The term transferase activity was observed to occur among both up-regulated and down-regulated proteins in the ontology of molecular functions, suggesting the impact of ethylene on protein modification and the important role of protein modification during corolla senescence in petunia.
Comparative Analysis of Proteome and Transcriptome Data
To compare the proteome with the transcriptome, all significantly differentially expressed mRNAs were first matched with quantifiable proteins (Supplemental File Exc S5), and then the proteins were compared with their cognate mRNAs by sorting the proteins according to their ethylene-air ratio. A positive correlation of r = 0.39 was observed when all significantly changed mRNAs with a cognate protein were considered, regardless of the direction of the change (Supplemental Fig. S5, A and F). Restricting the analysis to pairs in which the mRNA was up-regulated markedly increased the correlation (r = 0.49; Supplemental Fig. S5, B and F), while no correlation (r = 0.08) between transcript and protein abundance was observed for transcripts with significantly decreased abundance upon ethylene treatment (Supplemental Fig. S5C). This indicates that, contrary to expectations, the vast majority of the down-regulated mRNAs were not associated with lower abundance proteins. For protein/mRNA pairs in which the protein was significantly up-regulated, the highest positive correlation (r = 0.53) between the two levels was calculated (Supplemental Fig. S5, D and F). A weak positive correlation was observed between protein and mRNA for significantly down-regulated proteins (r = 0.21; Supplemental Fig. S5, E and F).
Numerous reports have suggested that RNA transcript accumulation is not always conveyed to the final product protein (Shemesh-Mayer et al., 2015). For example, a negative correlation between mRNA and protein accumulation patterns was found in Arabidopsis in response to cold treatment (Nakaminami et al., 2014). The lack of correlation between mRNA and protein levels has been attributed to differences in translational efficiency, codon usage/bias, mRNA versus protein stability, posttranslational modifications, sequencing depth, and proteomic approach (Alberch, 1991; Gygi et al., 1999; Pigliucci, 2010; Ghazalpour et al., 2011; Rodrigues et al., 2012). In this study, the number of mRNA copies in the sample and the subcellular localization of the protein restricted the number of identified proteins relative to the detection of their cognate transcripts (Supplemental Fig. S6). Comparing the number of reads recorded for transcripts corresponding to identified and not identified proteins, a transition is reached at around 20 reads, under which the products of the majority of transcripts were not detected (Supplemental Fig S6A). In addition, proteins tightly associated with membranes are underrepresented in the pool of identified proteins relative to the predicted proteome (Supplemental Fig. S6B).
Ethylene Treatment Changes the Ubiquitylome Profile in Petunia Corollas
Ubiquitination is a posttranslational mechanism that is important for protein quality control, DNA repair, cell survival, and cell death in eukaryotes (Kerscher et al., 2006). Ethylene is an important senescence hormone and has been observed to induce a drop in protein content. In previous studies, ubiquitin E3 ligase was found to be closely related to ethylene in plants (Potuschak et al., 2003; Xu et al., 2007; Qiao et al., 2009); therefore, the effects of ethylene treatment on the protein ubiquitylome were investigated in this work.
Proteome-wide enrichment of ubiquitination is based on its distinct di-Gly remnant (K-ε-GG). In this work, we combined label-free immunoaffinity enrichment using a high-quality anti-K-ε-GG antibody (PTM Biolabs) and high-resolution mass spectrometry to quantify protein ubiquitination in petunia corollas with and without ethylene treatment. In total, after obtaining three replicates for each treatment, 3,263 Lys ubiquitination (Kub) sites in 1,611 protein groups were identified, among which 2,270 sites in 1,221 proteins were accurately quantified, possessing consistent quantification ratios in at least two of the three liquid chromatography-tandem mass spectrometry analyses. From these, 127 (28.4%) sites in 118 proteins were quantified as down-regulated targets and 320 (71.6%) sites in 246 proteins were quantified as up-regulated targets at a threshold of 1.5 (P < 0.05; Supplemental File Exc S6). These results suggested that ethylene treatment greatly increased the level of ubiquitination in petunia corollas.
To elucidate the functions of the proteins that underwent ubiquitination, KEGG pathway analysis was performed. A number of vital pathways, including those related to the spliceosome, RNA transport, mRNA surveillance pathway, endocytosis, and ATP-binding cassette (ABC) transporters, were enriched among proteins with Kub sites (Supplemental File Exc S7). These results suggested that ubiquitination might be highly associated with RNA metabolism, endocytosis, and ABC transporters. Alternative pre-mRNA splicing is thought to provide a mechanism to increase the complexity of the proteome and introduce additional layers to regulate gene expression in different cell types and during development (Zhou and Fu, 2013). A previous study showed that the ubiquitination of histone H2B modulates spliceosome assembly and function in budding yeast (Saccharomyces cerevisiae; Zhou and Fu, 2013). The ubiquitination of proteins associated with the spliceosome may change the alternative pre-mRNA splicing that takes place during corolla senescence.
To elucidate the functional differences between proteins with up-regulated and down-regulated ubiquitination, enrichment-based clustering analyses were performed (Fig. 3; Supplemental File Exc S7). In the cellular component analysis, we found that proteins associated with vesicles were highly enriched among proteins with down-regulated Kub sites. Coated vesicles represent vital transport intermediates in all eukaryotic cells (Paul and Frigerio, 2007). The down-regulated ubiquitination of proteins associated with vesicles may play important roles in cell death or senescence. Conversely, proteins with up-regulated Kub sites were observed in the nucleosome, DNA-binding complex, DNA-packaging complex, and protein-DNA complex. The degradation of nucleic acids by specific nucleases during flower senescence has been observed in various flower systems, and a range of transcription factors have been found to be differentially regulated during development and senescence in various flower systems (Shahri and Tahir, 2014). These results suggest that ubiquitination might play an important role in the nucleus, including in transcription regulation and DNA repair, during ethylene-mediated senescence in petunia.
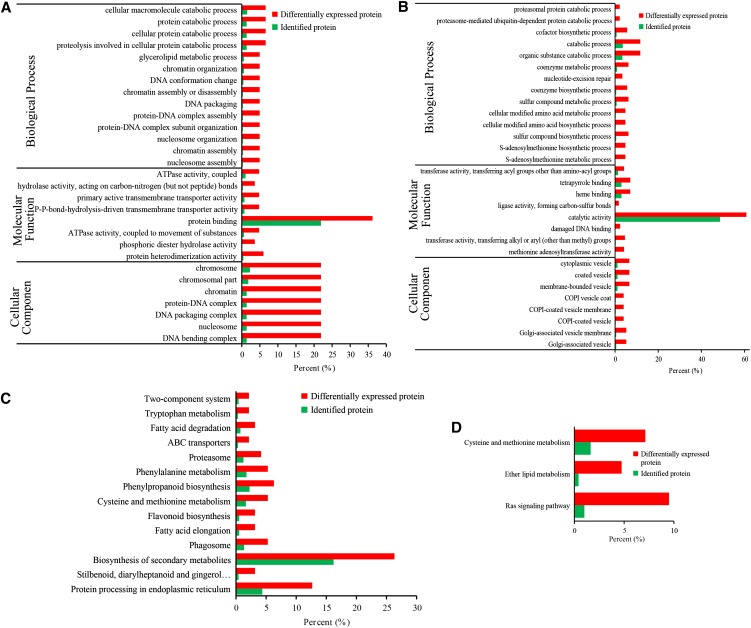
Figure 3.
Functional enrichment analysis of proteins with up-regulated and down-regulated Kub sites. A and B, GO-based enrichment analysis of proteins with up-regulated (A) and down-regulated (B) Kub sites. C and D, KEGG pathway-based enrichment analysis of proteins with up-regulated (C) and down-regulated (D) Kub sites. The percentage of differentially expressed proteins indicates the ratio of the mapping proteins to all mapping proteins. The percentage of identified proteins indicates the ratio of the background proteins to all background proteins. The significance level was set at P < 0.05 (Fisher’s exact test). These data come from Supplemental File Exc S7.
In the biological process analysis of ubiquitination, up- and down-regulated Kub proteins were enriched in 28 processes, including proteasome-mediated ubiquitin-dependent protein catabolic process, proteasomal protein catabolic process, and others, implying that ubiquitinated proteins may be involved in a wide range of biological processes in plants (Fig. 3, A and B).
In the molecular function analysis, proteins with binding activity, catalytic activity, and transporter activity were enriched among proteins containing both up-regulated and down-regulated Kub sites. Previous studies have shown that ions and amino acids are transferred to vegetative organs during senescence in unpollinated petunia petals (Shibuya et al., 2013). These results suggested that proteins demonstrating changes in ethylene-mediated ubiquitination are connected to protein interactions, DNA transcription, and ion and protein transport.
KEGG pathway analysis of proteins whose ubiquitination quantitatively changed revealed a number of vital pathways. The protein-processing pathways in the endoplasmic reticulum, stilbenoid and diarylheptanoid biosynthesis, phagosome, fatty acid elongation, flavonoid biosynthesis, Cys metabolism, Met metabolism, phenylpropanoid biosynthesis, Phe metabolism, proteasome, ABC transporters, and others were enriched among proteins with up-regulated Kub sites. Proteins with down-regulated Kub sites were enriched in pathways involving Ras signaling, ether lipid metabolism, Cys metabolism, Met metabolism, and others (Fig. 3, C and D). These results indicate that ubiquitination was associated with protein processing, protein degradation, and secondary metabolites.
From protein domain analysis, we observed that protein domains associated with S-adenosyl-Met synthetase, ubiquitin-like, NmrA, small GTP binding, and others were enriched in proteins with up-regulated Kub sites, whereas histone core and histone fold, ubiquitin-like, zinc finger, and other protein domains were enriched in down-regulated quantiles (Supplemental File Exc S7). We also identified 27 Kub sites in 14 histones, including in H1D, H1.2, H2B, H2A, H3, H4, and various histone isoforms, in this study, among which 16 sites in 10 histones were quantified (Supplemental File Exc S7). The ubiquitination levels of six Kub sites in five histones decreased. Five Kub sites were even down-regulated by over 10-fold, whereas no up-regulated Kub sites were identified, suggesting that ethylene negatively regulates the ubiquitination of histones and may play critical roles in regulating many processes within the nucleus, including transcription initiation and elongation, silencing, and DNA repair, by decreasing the ubiquitination levels of histones in petunia corollas. In Drosophila, Tetrahymena, and mammalian cells, the ubiquitylated forms of histones H2A and H2B were associated specifically with actively transcribed genes, making histone ubiquitination one of the first markers of transcriptionally active chromatin to be recognized (Muratani and Tansey, 2003).
Sequence Properties of Ubiquitinated Proteins
To understand the properties of the identified Kub sites in petunia, we used the Motif-X program to compare the position-specific frequencies of the amino acid residues surrounding all ubiquitinated Lys residues.
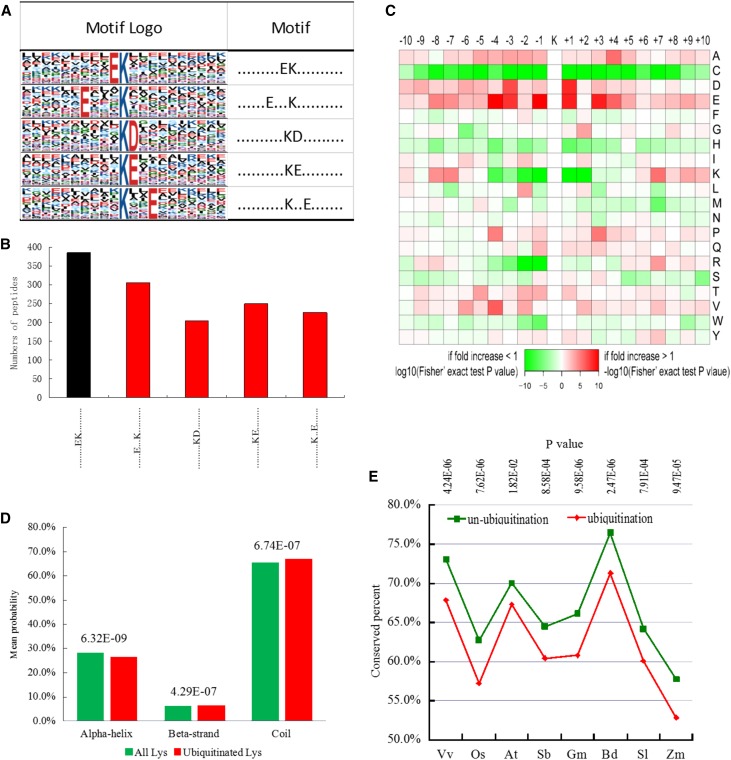
Of the 3,265 Kub peptides, we identified a total of five conserved motifs for 1,373 unique sites, which accounted for approximately 42% of the sites identified (Supplemental File Exc S8). The five unique sites were designated .........EK.........., ......E...K.........., ..........KD........., ..........KE........., and ..........K..E......., and they exhibited different abundances (. indicates any amino acid; Fig. 4A). Among them, .........EK.......... has been reported previously (Xie et al., 2015), while the other four motifs are novel (Fig. 4B, red columns), which may provide insight into ethylene signaling in petunia as well as in plants in general. A survey of these motifs revealed that only two distinct residues are found upstream or downstream of the ubiquitinated Lys (Fig. 4A), including acidic Asp (D) and Glu (E), whereas in rice (Oryza sativa), only neutral Ala (A) and acidic Glu (E) were observed surrounding ubiquitinated Lys residues (Xie et al., 2015). These results show the differences in ubiquitinated Lys motifs between the dicotyledon petunia and the monocotyledon rice.
Figure 4.
Motif analysis of all the identified Kub sites in petunia. A, Ubiquitination motifs and the conservation of Kub sites. The height of each letter corresponds to the frequency of that amino acid residue in that position. The central K refers to the ubiquitinated Lys. B, Number of identified peptides containing ubiquitinated Lys in each motif. The red columns represent novel motifs. C, Amino acid sequence properties of ubiquitylation sites. The heat map shows significant position-specific underrepresentation or overrepresentation of amino acids flanking the modification sites. D, Predicted protein secondary structures near Kub sites. Probabilities for different secondary structures (coil, α-helix, and β-strand) of modified Lys residues were compared with the secondary structure probabilities of all Lys residues or all Ser/Thr/Tyr in all proteins identified in this study. E, Evolutionary conservation of ubiquitylated and nonubiquitylated Lys residues on protein orthologs in selected eukaryotic species: Vv, Vitis vinifera; Os, Oryza sativa japonica; At, Arabidopsis thaliana; Sb, Sorghum bicolor; Gm, Glycine max; Bd, Brachypodium distachyon; Sl, Solanum lycopersicum; Zm, Zea mays.
To further examine the properties of amino acids surrounding ubiquitination sites, the frequencies of neighboring amino acid residues were analyzed for ubiquitinated Lys residues using iceLogo (Colaert et al., 2009). We observed a significant preference for hydrophilic residues such as Glu and Asp at positions adjacent to ubiquitinated Lys residues (+1, +3, −1, and −3; Fig. 4C). In mammals, a significant preference for hydrophobic residues, such as Phe, Tyr, Trp, Leu, Ile, and Val, adjacent to ubiquitinated Lys residues has been observed (Wagner et al., 2011). These results indicate the different properties of amino acids surrounding ubiquitination sites when comparing plants and mammals.
In addition to primary sequences around Kub sites, protein secondary structure has been found to be informative in Kub site prediction (Gnad et al., 2011). Therefore, we integrated protein secondary structure features using NetSurfP software (Muller et al., 2010). The probabilities of different secondary structures (coil, α-helix, and β-strand) near ubiquitinated Lys sites were compared with the secondary structure probabilities of all Lys sites on proteins identified in this study. Ubiquitinated Lys sites occurred significantly more frequently in unstructured regions of proteins (P = 6.74E-07 for coil) and less frequently in structured regions (P = 6.32E-09 for α-helix and P = 4.29E-07 for β-strand; Fig. 4D). However, in mammals, ubiquitinated Lys residues are marginally, yet significantly, more frequently present in structured regions of proteins than in unstructured regions (Wagner et al., 2011), indicating a difference in ubiquitinated Lys sites between plants and mammals.
In mammals, ubiquitinated Lys is significantly more conserved than nonubiquitinated Lys (Wagner et al., 2011). To study the evolutionary conservation of ubiquitinated Lys and nonubiquitinated Lys in plants, we aligned petunia proteins with their respective orthologs from eight other plant species. The results unexpectedly showed that ubiquitinated Lys residues are significantly less conserved than nonubiquitinated Lys residues, suggesting that ubiquitinated Lys residues do not maintain a stronger selective pressure compared with nonubiquitinated Lys residues in plants (Fig. 4E). It appears that ubiquitination occurs primarily in nonconserved Lys positions in petunia corollas, and further experiments are required to validate this possible evolutionary mechanism.
The Correlation between the Global Proteome and Ubiquitylome
Ubiquitination is well known for its role in proteasome-mediated protein degradation. The expression of proteins in corollas also may be regulated by ubiquitination. In this work, among the 5,189 proteins identified, 1,161 were ubiquitinated (Supplemental Fig. S8). The quantitative proteome and ubiquitylome of ethylene-treated corollas were both obtained to study the interaction between the proteome and ubiquitylome.
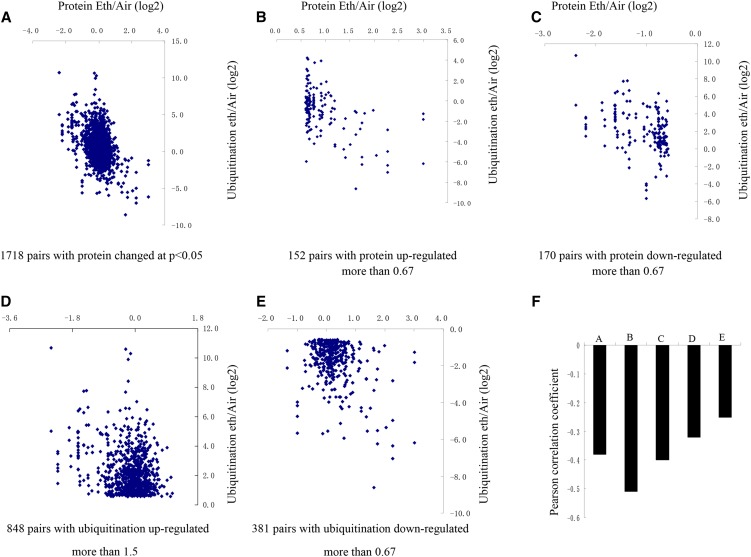
The correlation between the whole proteome and ubiquitylome during senescence in corollas was analyzed based on the quantitative results obtained in this study. There were 985 quantified proteins that were also found to undergo ubiquitination, and 2,270 Kub sites in 1,221 proteins were quantified. Of the 985 quantified proteins, 66 proteins were down-regulated and 96 were up-regulated. Quantitative ratios from the proteome and ubiquitylome were compared upon ethylene treatment, as shown in Figure 4. Pearson’s correlation coefficient, a statistical measure of the strength of a linear relationship between paired data, is denoted by r and is, by design, constrained between −1 and 1. Positive values denote positive linear correlation, negative values denote negative linear correlation, and a value of 0 denotes no linear correlation. The closer the value is to 1 or −1, the stronger the linear correlation. The Pearson’s correlation coefficient was calculated as −0.38 when all significantly altered proteins were considered in terms of their ubiquitination, regardless of the direction of the change (Fig. 5, A and F). In addition, the overlap between differentially expressed proteins and ubiquitination is shown in Figure 4B and Supplemental File Exc S9. A total of 67 proteins exhibited opposing changes in protein and ubiquitination levels, whereas only 10 proteins demonstrated consistent changes. Therefore, the global proteome and ubiquitylome were negatively correlated, which implies that, to a certain extent, the changing pattern of the proteome was opposite that of the ubiquitylome following ethylene treatment. Restricting the analysis to pairs of up-regulated proteins and pairs of down-regulated proteins increased the correlation (r = −0.51 and −0.4, respectively; Fig. 5, B, C, and F). For ubiquitination/protein pairs with significantly up-regulated and significantly down-regulated ubiquitination, two weak negative correlations were observed (r = −0.32 and −0.25, respectively; Fig. 5, D–F). These results suggested that proteome expression levels were negatively regulated by ubiquitination.
Figure 5.
Concordance between changes in proteins and their ubiquitination. A to E, Correlation between protein and ubiquitination fold changes upon ethylene treatment for all ubiquitination-protein pairs (A), significantly up-regulated proteins (B), significantly down-regulated proteins (C), significantly up-regulated ubiquitination (D), and significantly down-regulated ubiquitination (E). F, Pearson correlations of the comparisons shown in A to E.
It should be noted that the ubiquitylome reveals the status of proteins that are ubiquitinated but not those already subjected to 26S proteasome degradation, because these degraded proteins will not be detectable in the ubiquitylome. Thus, the ubiquitylome does not truly reflect the status of protein degradation. If ones takes into account these proteins already subjected to 26S proteasome degradation, the ubiquitylome value is higher than the present total value; however, this does not change the conclusion regarding the negative correlation between the global proteome and ubiquitylome but, rather, supports this conclusion. In addition, aside from proteasome-mediated degradation, ubiquitination has many other roles in protein modification, such as altering biochemical properties and subcellular protein localization (Shabek and Zheng, 2014); this partially explains why the negative correlation observed between the proteome and the ubiquitylome was not very strong.
Several spectra corresponding to sites from proteins that undergo ubiquitination are presented in Supplemental Figure S9.
Involvement of Ubiquitination in the Degradation of Proteins during Ethylene-Mediated Corolla Senescence in Petunias
The degradation of proteins in developing tissues is a notable process during senescence (Shahri and Tahir, 2014). In the transcriptome obtained in this study, 144 unigenes encoding putative ubiquitin-protein ligases (35 E3 ubiquitin-protein ligases, 72 F-box proteins, and 37 U-box proteins), six unigenes encoding ubiquitin proteins, and seven unigenes encoding 26S proteasome subunits up-regulated by ethylene were identified (Supplemental File Exc S10). In the proteome, ethylene treatment resulted in 284 down-regulated and 233 up-regulated proteins, and among them, four putative ubiquitin ligases were up-regulated (Supplemental File Exc S11). Moreover, 246 quantified proteins also underwent ubiquitination, and their up-regulated Kub sites were identified; among them, 44 proteins were down-regulated and only eight proteins were up-regulated with respect to protein concentration. In addition, 118 quantified proteins underwent ubiquitination, and their down-regulated Kub sites were identified in this study; among these, 23 proteins were up-regulated and only two proteins were down-regulated with respect to protein concentration following ethylene treatment (Supplemental File Exc S9). Of the 18 ubiquitinated proteins identified only in the control, 17 were up-regulated and only one was down-regulated by ethylene at the protein level, while of the 11 ubiquitinated proteins identified only in corollas following ethylene treatment, nine were down-regulated and only two were up-regulated by ethylene at the protein level (Supplemental File Exc S9). Silencing the expression of a gene homolog to MjXB3 in petunia resulted in an extension in flower life (Xu et al., 2007). Proteomic analysis of pollination-induced corolla senescence in petunia identified a ubiquitin-conjugating enzyme (E2) that was up-regulated by pollination, accelerating flower senescence (Bai et al., 2010). These results indicate the involvement of ubiquitination in protein degradation during ethylene-mediated corolla senescence in petunia. In addition, the proteasome system was apparently up-regulated during petal senescence in daylily (Hemerocallis spp.; Courtney et al., 1994; Müller et al., 2004) and daffodil (Narcissus pseudonarcissus; Hunter et al., 2002). In carnation, several transcripts homologous to genes encoding various components of the 26S proteasome machinery, including RPT6, RPN2, a RING finger protein, and a U-box-containing protein, all were induced during carnation petal senescence (Hoeberichts et al., 2007). Feeding isolated Iris spp. petals with Z-Leu-Leu-Nva-H, an inhibitor of proteasome activity, led to a significant delay in the time to visible senescence (Pak and van Doorn, 2005), indicating that proteasome action limits senescence. In addition, Arabidopsis UPL5, a HECT E3 ubiquitin ligase, negatively regulates leaf senescence through the degradation of WRKY53 and ensures that senescence is executed in the correct time frame (Miao and Zentgraf, 2010).
To elucidate the function of proteins with opposite trends in protein and ubiquitination levels, KEGG pathway enrichment-based clustering analyses were performed (Supplemental Fig. S7). The protein-processing pathways for flavonoid biosynthesis, Phe metabolism, phenylpropanoid and secondary metabolite biosynthesis, and others were enriched among proteins with up-regulated Kub sites and down-regulated protein levels. Previous studies suggested that ethylene treatment reduced the biosynthesis of phenylpropanoid and secondary metabolites in petunia (Negre et al., 2003; Underwood et al., 2005; Schuurink et al., 2006). It is possible that ubiquitination could be involved in the degradation of the proteins in these pathways during ethylene-mediated flower senescence. Proteins with down-regulated Kub sites and up-regulated protein levels were enriched in pathways involving SNARE interaction in vesicular transport and Gal metabolism.
The canonical view of protein ubiquitination posits that the entire pool of a targeted protein becomes ubiquitinated and is degraded subsequently. However, Kim et al. (2011) and Swaney et al. (2013) showed that most cases of increased ubiquitination were not accompanied by corresponding reductions in protein abundance. Similarly, in this study, 221 and 96 proteins demonstrating increased and decreased ubiquitination, respectively, were not accompanied by corresponding reductions and increases in protein abundance. One reasonable explanation is that complex signaling may be at play, in which specific Kub sites are utilized as degradation markers and others serve to modulate protein function.
The regulatory pathways in flower senescence were divided into three phases: the signaling phase, the regulatory phase, and the execution phase (Tripathi and Tuteja, 2007). Protein degradation, as well as the hydrolysis of nucleic acids, lipids, and carbohydrates, take place in the execution phase (Tripathi and Tuteja, 2007). Our results suggested the involvement of ubiquitination in the degradation of proteins during ethylene-mediated corolla senescence in petunias. Taken together, the large amounts of protein ubiquitination underlie corolla senescence. Moreover, PhXB3 silencing delayed flower senescence in petunia (Xu et al., 2007).
Involvement of Nonproteasomal Proteases in the Degradation of Proteins during Ethylene-Mediated Corolla Senescence in Petunias
The activity of nonproteasomal protease has been found to increase prior to visible senescence (Stephenson and Rubinstein, 1998; Pak and van Doorn, 2005). Of these proteases, Cys proteases have been reported exclusively to be involved in and thought to mediate the remobilization of essential nutrients from senescing floral tissues. In this study, in the transcriptome, 37 nonproteasomal proteases, including six Cys proteases, three metalloproteases, two Ser proteases, three subtilisin proteases, and nine Asp proteases, were up-regulated by ethylene in petunia corollas (Supplemental File Exc S12). Proteomic analysis showed that three Cys proteases, two metalloproteases, and one Asp protease were up-regulated by ethylene in this study (Supplemental File Exc S11). Cys protease genes have been reported to be up-regulated during senescence in petunia (Jones et al., 2005). These results implied that nonproteasomal proteases, including Cys proteases, metalloproteases, and Asp proteases, are likely also involved in the degradation of proteins during ethylene-mediated corolla senescence in petunias.
Changes of the Autophagy Proteins after Ethylene Treatment
Autophagy is one of the main mechanisms of degradation and remobilization of macromolecules (Shahri and Tahir, 2011). Shibuya et al. (2013) suggested that ethylene is a key regulator of autophagy in petal senescence of petunia. Ethylene inhibitor treatment in pollinated flowers delayed the induction of homologs of an autophagy-related gene (PhATG8), and ethylene treatment rapidly up-regulated PhATG8 homologs in petunia petals. Arabidopsis AtATG8 mRNA levels increased in senescing leaves (Doelling et al., 2002). In Arabidopsis, a number of autophagy genes (ATG) had been knocked out, which resulted in hastened leaf yellowing (Hanaoka et al., 2002; Yoshimoto et al., 2004). In this study, PhATG8b (Unigene0018716) and PhATG11 (Unigene0069693) protein levels were increased after ethylene treatment. In addition, PhATG18H (Unigene0007523), PhATG3 (Unigene0031140), and PhATG2 (Unigene0011829) were identified. No down-regulated autophagy-related protein was identified (Supplemental File Exc S13). These results suggested that autophagy occurs during the senescence of corollas, is promoted by ethylene, and plays an important role in petal senescence.
In mammals and yeast, two ubiquitin-like systems, the autophagy-defective12 (Apg12) system and the Apg8 system, are required for autophagy (Ohsumi, 2001). Phosphorylation and ubiquitination were crucial for autophagy induction, regulation, and fine-tuning and were influenced by a variety of stimuli (McEwan and Dikic, 2011). In this study, to our knowledge for the first time, the ubiquitination of ATG8b (Lys-11), a ubiquitin-like protein, was up-regulated by 3.486-fold by ethylene, suggesting that ubiquitination could be involved in ethylene-induced autophagy in a plant.
Effects of Ethylene Treatment on Hormone Biosynthesis and Signaling Transduction Pathways
S-Adenosyl-Met, a precursor for ethylene biosynthesis and polyamine synthesis, is the methyl group donor for many cellular molecules, including nucleic acids, proteins, and lipids (Yang and Hoffman, 1984; Schuurink et al., 2006). The formation of S-adenosyl-Met is catalyzed by S-adenosyl-Met synthetase (SAMS). In this study, we found 11 Kub sites in five SAMSs (PhSAMS1a, Unigene0023828, Lys-169, Lys-175, Lys-226, and Lys-340; PhSAMS3a, Unigene0028250, Lys-78; PhSAMS3b, Unigene0028252, Lys-67 and Lys-364; PhSAMS1b, Unigene0023825, Lys-94; and PhSAMS1c, Unigene0023827, Lys-67, Lys-71, and Lys-120) that were significantly up-regulated by ethylene. Among them, eight Kub sites were up-regulated by more than 10-fold (Unigene0023828, Lys-226, Lys-169, and Lys-175; Unigene0028250, Lys-78; Unigene0028252, Lys-67 and Lys-364; Unigene0023825, Lys-94; and Unigene0023827, Lys-120). Accordingly, in the proteome, the abundance of five SAMSs (Unigene0023828, Unigene0028250, Unigene0028252, Unigene0023825, and Unigene0023827) decreased following ethylene treatment (Fig. 6; Supplemental File Exc S13), suggesting that ethylene negatively regulates S-adenosyl-Met abundance. However, ethylene treatment did not result in a general decrease in ethylene biosynthesis. It is possible that the S-adenosyl-Met cycle and polyamine biosynthesis are negatively regulated by ethylene.
Figure 6.
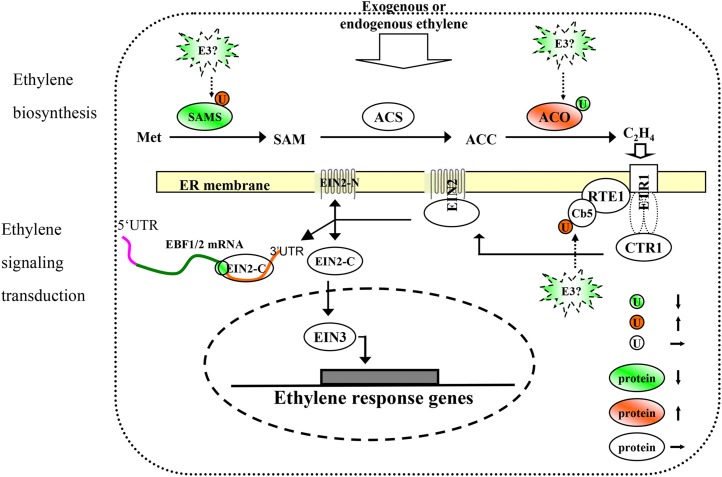
Effects of ethylene on the proteins engaged in ethylene biosynthesis and signaling transduction pathways in petunia. Differentially expressed proteins based on statistical significance in this study are framed in ovals, and differentially ubiquitinated and phosphorylated proteins are framed in circles. Red shapes indicate up-regulation; green shapes indicate down-regulation; and white shapes indicate no significant changes upon ethylene treatment. ACC, 1-Aminocyclopropane-1-carboxylic acid; ACO, ACC oxidase; ACS, ACC synthase; Cb5, cytochrome b5; CTR1, CONSTITUTIVE TRIPLE RESPONSE1; EIN, ETHYLENE INSENSITIVE; EIN2-C, EIN2 C-terminal end; EIN2-N, EIN2 N-terminal end; ETR1, ETHYLENE RESPONSE1; RTE1, REVERSION TO ETHYLENE SENSITIVITY1; SAM, S-adenosyl-Met; SAMS, S-adenosyl-Met synthetase; U, ubiquitination; UTR, untranslated region.
1-Aminocyclopropane-1-carboxylic acid synthase (ACS) is the rate-limiting enzyme of ethylene synthesis. Previous research has suggested that ACS family proteins are up-regulated by ethylene and that ETO1/EOL, calcium-dependent protein kinase (CDPK), 14-3-3, and mitogen-activated protein kinase (MAPK) interact with ACS family proteins, modulating their stability in plants (Xu and Zhang, 2014). However, in this study, in both protein and ubiquitination analyses, ACS family proteins were not identified.
The discovery of two plant MAPK substrates, ACS2 and ACS6, which are type I ACS isoforms, revealed ACS phosphorylation regulation by AtMPK3 and AMPK6, two functionally redundant stress/pathogen-responsive MAPKs in Arabidopsis. In this study, two Kub sites in PhMAPK6 (Unigene0025211, Lys-57 and Lys-95), a homolog of AtMAK6, were identified. The ubiquitination levels of 14-3-3 (Unigene0024326, Lys-48) and PhCDPK30 (Unigene0029654, Lys-389; greater than 4-fold) increased after ethylene treatment, which may maintain protein abundance and promote the activity of ACS to alter their biochemical properties.
1-Aminocyclopropane-1-carboxylic acid oxidase (ACO) is another key enzyme in ethylene biosynthesis, and antisense ACO RNA delayed flower senescence in transgenic carnation (Savin et al., 1995). In this study, to our knowledge for the first time, the ubiquitination of PhACO3 (Unigene0022854, Lys-41) was identified and was found to be down-regulated more than 15-fold by ethylene treatment. Accordingly, PhACO3 (Unigene0022854) protein levels were up-regulated following ethylene treatment, suggesting that ubiquitination could be involved in PhACO3 degradation and in ethylene biosynthesis. Consistent with these results, ethylene production increases in corollas during flower senescence in petunia (Liu et al., 2011).
Ethylene receptors are encoded by a multigene family that can be divided into subfamilies 1 and 2. Kevany et al. (2007) suggested that the receptors LeETR4 and LeETR6 were degraded rapidly in the presence of ethylene and that degradation likely occurs through the 26S proteasome-dependent pathway in tomato (Solanum lycopersicum) plants. In Arabidopsis, the ethylene-induced decrease in ETR2 levels is not affected by cycloheximide, an inhibitor of protein biosynthesis, but is affected by proteasome inhibitors, indicating a role for the proteasome in ETR2 degradation (Chen et al., 2007). However, those authors did not provide direct evidence of the ubiquitination of ethylene receptors. In our study, a Kub site on PhETR2 (Unigene0010512, Lys-359) was identified. These results suggested the involvement of ubiquitination in ethylene receptor degradation and ethylene signaling.
EIN2 acts downstream of ethylene receptors and upstream of EIN3/EIL and is involved in the regulation of flower senescence. Qiao et al. (2009) reported that the stability of EIN2 is modulated by the two F-box proteins, ETP1/2, via ubiquitination, but the ubiquitination of PhEIN2 was not observed in this study. In addition, it was proposed that EIN3 is targeted by the F-box proteins EBF1/2 in Arabidopsis (Potuschak et al., 2003). However, PhEILs, PhEBF1, and PhEBF2 were not identified in this study at either the protein or ubiquitination level.
A recent study showed that Arabidopsis Cb5 proteins are involved in ethylene signaling, and RTE1 interacts physically with AtCb5-B, AtCb5-C, AtCb5-D, and AtCb5-E (Chang et al., 2014). The Kub sites of two Cb5s (PhCb5B, Unigene0023698, Lys-35; and PhCb5E, Unigene0016038, Lys-51) were up-regulated more than 4-fold by ethylene in this study, which further supported the involvement of ubiquitination in ethylene signaling in petunia.
Ethylene is an important regulator of flower senescence. The results mentioned above illustrated protein and ubiquitination levels in ethylene biosynthesis and demonstrated that signaling pathways can be regulated by ethylene. These findings, including the ubiquitination of PhACO3, PhETR2, PhCb5B, and PhCb5E, significantly advance our understanding of the mechanisms underlying ethylene biosynthesis and signaling transduction (Fig. 6).
Ethylene appears to be a negative regulator of abscisic acid (ABA) action during germination, although it was confirmed to exert a positive synergistic effect on ABA action by modulating the overall carbon status in Arabidopsis roots (Ghassemian et al., 2000; Gazzarrini and McCourt, 2001; Cheng et al., 2009). In carnation, ABA has been found to accelerate flower senescence (Ronen and Mayak, 1981). A large increase in ABA levels was observed in the gynoecium prior to or concomitant with the upsurge in ethylene (Onoue et al., 2000). In this study, the enzymes related to ABA biosynthesis, PhDXS (Unigene0009358), PhPDS3 (Unigene0017870), PhNCED4 (Unigene0037462), and PhSDR (Unigene0012764), were down-regulated between 1.5- and 3-fold at the protein level by ethylene (Supplemental Fig. S10A; Supplemental File Exc S13). Additionally, the ABA signaling component PP2C, a major negative regulator of ABA signaling, inhibits SnRK2, a positive regulator of ABA signaling, thus inhibiting the activation of the ABA pathway (Umezawa et al., 2010). In this study, PP2C (PhPP2C, Unigene0006325; and PhPP2C58, Unigene0014490) and SnRK2A (Unigene0014500) increased at the protein level after ethylene treatment. These results hinted that ethylene likely negatively regulates ABA biosynthesis and signaling transduction in petunia corollas. In rose (Rosa spp.) petals, the external application of ethylene accelerated senescence and induced a rise in endogenous ABA-like activity (Mayak and Halevy, 1972). In petunia, ethylene might directly affect senescence in petals without requiring the involvement of the ABA pathway.
Many components of the auxin efflux (but not influx) system have been shown to be activated by PTM (Delbarre et al., 1998; Zourelidou et al., 2014). In this study, ethylene did not change the abundance of proteins involved in auxin signaling or that of efflux or influx transporters. However, two Kub sites on indole-3-acetic acid (IAA)/auxin (AUX) repressors (PhIAA14, Unigene0023390, Lys-26 and Lys-106) were up-regulated more than 20- and 7-fold by ethylene, respectively (Supplemental Fig. S10B; Supplemental File Exc S13). Leitner et al. (2012) showed that ubiquitination of the PIN2 auxin carrier protein governs the hormonally controlled adaptation of Arabidopsis root growth. Ethylene treatment significantly increased the ubiquitination level of PhPIN4 (Unigene0020360, Lys-331 and Lys-438). It is noteworthy that the auxin influx transport proteins AUX1/LAX (Unigene0019926 and Unigene0070491) were ubiquitinated, and ethylene treatment significantly increased the ubiquitination of PhAUX1 (Unigene0019926, Lys-5; greater than 5-fold). To the best of our knowledge, the ubiquitination of AUX1 has not been reported previously. In addition, a third class of auxin transporters includes phosphoglycoproteins that belong to the ABCB subgroup of the ABC transporter superfamily. ABCB1 and ABCB19 have been shown to play direct roles in the cellular efflux of auxin (Titapiwatanakun and Murphy, 2009). In this study, the ubiquitination level of PhABPB2 (Unigene0047722, Lys-882) increased, whereas the ubiquitination level of another site in PhABPB2 (Lys-315) decreased, after ethylene treatment. These results suggested that, in petunia corollas, ethylene might play an important role in auxin transport, including both influx and efflux. It is possible that the inhibition of auxin transport, a process that inhibits senescence (Teale et al., 2006), accelerated corolla senescence.
In summary, during ethylene-mediated corolla senescence, ethylene appeared to affect the biosynthesis and signal transduction pathways of plant hormones such as ABA, auxin, and ethylene itself at the transcript, protein, and ubiquitination levels in this study. In addition, it should be noted that the omics changes in this study may be caused directly or indirectly by ethylene treatment.
Changes of Proteins Involved in Suc Biosynthesis and Transport after Ethylene Treatment
During petal senescence in the genera Alstroemeria (Breeze et al., 2004) and Iris (Van Doorn et al., 2003), the transcript abundance of a gene encoding a triose phosphate isomerase and those of genes encoding Suc synthase increased. In Alstroemeria spp., the transcripts of a gene encoding a cell wall invertase also became more abundant (van Doorn and Woltering, 2008). In this study, three Suc synthases (PhSS7, Unigene0008278; PhSS6, Unigene0012766; and PhSS1, Unigene0025892) were increased in protein level after ethylene treatment. Two Kub sites in Suc synthases (PhSS1, Lys-190; and PhSS2, Unigene0011388, Lys-65) were down-regulated by ethylene (Supplemental File Exc S13), which may alter the activity of Suc synthase. These data suggested an increase in Suc synthesis in corollas after ethylene treatment.
Petal senescence was accompanied by a high sugar concentration in the phloem (van Doorn and Woltering, 2008). In order to reach the phloem, the sugars must be transferred, at some point, through a membrane. Several genes encoding sugar transporters were up-regulated during Alstroemeria spp. and carnation petal senescence (Breeze et al., 2004; Hoeberichts et al., 2007). In this study, five Kub sites in three sugar transporters (PhERD6, Unigene0030195, Lys-277; PhSWEET10a, Unigene0064435, Lys-28, Lys-44K, and Lys-22; and PhSWEET10b, Unigene0027205, Lys-225) were down-regulated by ethylene. PhSWEET10a and PhSWEET11 (Unigene0027207) were increased in protein level after ethylene treatment (Supplemental File Exc S13). These data suggested that ethylene-mediated petal senescence was probably accompanied by a high sugar concentration and that the sugar was transported to the developing tissues in petunia.
Changes of Proteins Involved in the Biosynthesis of Volatile Organic Compounds after Ethylene Treatment
Petunia has become a model in which to study the biosynthesis and regulation of floral volatile benzenoids and phenylpropanoids, which are produced from shikimate-derived l-Phe (Boatright et al., 2004). Several genes encoding shikimate enzymes (Colquhoun et al., 2010; Maeda et al., 2010) and subsequent branched pathways have been identified and characterized in petunia. Underwood et al. (2005) demonstrated that multiple components of the emission of volatile benzenoids and phenylpropanoids and the transcripts of genes involved in benzenoid and phenylpropanoid biosynthesis are negatively regulated by ethylene in cv Mitchell. In this study, seven of the eight enzymes related to Phe biosynthesis decreased at the protein level in the presence of ethylene, including 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase (Unigene0014414), 3-dehydroquinate synthase (Unigene0006116), 5-enolpyruvate shikimate-3-phosphate (PhEPSPS, Unigene0021752), 3-dehydroquinate synthase (Unigene0006116), and chorismate synthase (PhCS, Unigene0026072). In the phenylpropanoid pathway, Phe ammonia lyase (Unigene0017590 and Unigene0035641; greater than 3-fold), 4-coumarate:CoA ligase (Ph4CL1, Unigene0030548), phenylacetaldehyde synthase (Unigene0024129), acyl-activating enzyme (PhAAE11, Unigene0028342), and two caffeoyl-CoA O-methyltransferases (PhCCOMT1, Unigene0026144; and PhCCOMT2, Unigene002614) also were down-regulated at the protein level by ethylene (Supplemental Fig. S11; Supplemental File Exc S13). These results suggested that ethylene negatively regulates the biosynthesis of Phe, benzenoids, and phenylpropanoids, which is consistent with a previous report (Underwood et al., 2005).
To confirm the reduction of these proteins by ethylene treatment, specific antibodies against PhCS, PhPAL1, Ph4CL1, PhAAE11, and PhEPSPS proteins were prepared and western blotting was performed. The results showed that all eight proteins were reduced by ethylene treatment (Supplemental Fig. S14A), which is consistent with the iTRAQ results.
In the ubiquitylome, the ubiquitination levels of shikimate 5-dehydrogenase (Unigene0001508, Lys-114 and Lys-504; greater than 15-fold), cinnamate-4-hydroxylase (Unigene0023326, Lys-268), coniferyl alcohol acetyltransferase (Unigene0011295, Lys-176; greater than 11-fold), isoeugenol synthase (Unigene0003787, Lys-39; and Unigene0015809, Lys-47), eugenol synthase (Unigene0016673, Lys-85), benzoic acid/salicylic acid carboxyl methyltransferase (Unigene0029058, Lys-274 and Lys-188; greater than 10-fold), PhCCOMT1 (Unigene0026144, Lys-159; greater than 35-fold), and cinnamyl alcohol dehydrogenase (Unigene0026909, Lys-354; greater than 35-fold) increased after ethylene treatment (Supplemental Fig. S11). These results implied that, aside from alterations at the mRNA level, ethylene regulated the abundance of proteins associated with floral scent biosynthesis at the ubiquitination level in petunia and that ubiquitination might play an important role in floral scent biosynthesis.
Ethylene Treatment Decreases the Abundance of Proteins Involved in Amino Acid Biosynthesis
In addition to the enzymes in the Phe biosynthesis pathway mentioned above, ethylene treatment significantly decreased the protein abundance of enzymes related to the biosynthesis of other amino acids, including His, Tyr, Met, Ser, and Lys biosynthesis (Supplemental Fig. S12; Supplemental File Exc S13). In contrast, previous studies have revealed considerable synthesis of specific amino acids in cells undergoing senescence in Sandersonia aurantiaca and carnation as well as the accumulation of these amino acids in the phloem (van Doorn and Woltering, 2008). These results illustrate the different levels of amino acid synthesis that occur in different species undergoing senescence.
Ethylene Treatment Increases the Ubiquitination Levels of Proteins Involved in ERAD
In yeast, mammalian, and plant cells, unfolded or misfolded proteins generated in the rough endoplasmic reticulum (ER) are degraded predominantly by ERAD, which involves ubiquitination, retrotranslocation, and degradation by the cytosolic proteasome (Smith et al., 2011). In ERAD, the family of ER-localized HSP70 proteins (known as BiPs) recognizes and binds to exposed hydrophobic patches of incompletely folded or misfolded proteins in an ATP-dependent manner (Buck et al., 2007). Arabidopsis BiPs were thought to contribute to the ER retention of two mutant BR receptors (Hong et al., 2008). BiPs and their associated factor, ERdj3B (an Arabidopsis ER-localized DnaJ homolog), also were involved in the biogenesis and folding control of EFR (Nekrasov et al., 2009). In this study, ethylene treatment increased the ubiquitination levels of PhHSP70 (Unigene0027213, Lys-560 and Lys-91) and a DnaJ homolog subfamily A member (PhDnaJ2, Unigene0027373, Lys-66; greater than 10-fold; Supplemental Fig. S13; Supplemental File Exc S13).
In ERAD, processed substrates are delivered to the cytosolic proteasome by CDC48 in association with RAD23 and DSK2, two ubiquitin receptors (Raasi and Wolf, 2007). UBX-containing proteins likely recruit AtCDC48A to the ER membrane (Rancour et al., 2004). In Arabidopsis, RAD23 proteins also play an important role in the cell cycle, morphology, and fertility of plants through their delivery of substrates to the 26S proteasome (Farmer et al., 2010). In this study, ethylene treatment increased the ubiquitination levels of PhCDC48C/P19 (Unigene0026112, Lys-280) and three PhRAD23d proteins (Unigene0018393, Lys-51 [greater than 10-fold]; Unigene0018392, Lys-18, Lys-28, Lys-62, and Lys-9; and Unigene0020741, Lys-18).
In Arabidopsis, ERAD substrates may be processed through antagonistic interactions between UFD2 and UFD3, along with unknown enzymes and the deubiquitinating enzyme OTU1, and/or through deglycosylation by the cytoplasmic peptide N-glycanase (PNGase) PNG1 (Raasi and Wolf, 2007). AtPNG1 may contain suspected PNGase activity and could stimulate the degradation of two mutant variants of RTA in an N-glycan-dependent manner in yeast cells (Diepold et al., 2007; Masahara-Negishi et al., 2012). Here, ethylene treatment increased the ubiquitination levels of PhPNG1P (Unigene0025382, Lys-104) and PhOUT2 (Unigene0047836, Lys-57 and Lys-161). In addition, ethylene treatment altered the ubiquitination abundance of S-PHASE KINASE-ASSOCIATED PROTEIN1 (Unigene0020623, Lys-79 and Lys-51), the molecular chaperone HSP90 (PhHSP90a, Unigene0029683, Lys-212 and Lys-277; and PhHSP90b, Unigene0029681, Lys-376), and B-CELL RECEPTOR-ASSOCIATED PROTEIN31 (Unigene0007191, Lys-84; and Unigene0003563, Lys-419).
The ER is a well-controlled microenvironment that facilitates proper protein synthesis and folding and is highly susceptible to stress conditions (Liu and Howell, 2010). The accumulation of unfolded or misfolded proteins activates the unfolded protein response pathway and, if unsuccessful, leads to cell death (Deng et al., 2013). The above results implied the important role of ethylene in the regulation of ERAD in plants. To our knowledge, this is the first report of a relationship between ethylene and ERAD in plants, particularly in the context of ubiquitination regulation. Further exploration of these Kub protein targets may provide insight into previously unknown effectors of the ethylene signaling pathway. In addition, ERAD might be associated with corolla senescence in petunia, as the ubiquitination abundance of several proteins involved in ERAD was changed significantly during ethylene-mediated senescence.
Confirmation of the Ubiquitination of Certain Proteins by Western Blotting
To confirm the ubiquitination of proteins utilizing the K-ε-GG antibody, we performed western blotting. Proteins whose ubiquitination was not reported previously were selected as candidates. More evidence has indicated that ER-associated degradation plays important roles in plant development, including senescence (Guerra and Callis, 2012). We selected three proteins, PhCDC48C/P19 (Unigene0026112), PhRAD23d (Unigene0018393), and PhPNG1P (Unigene0025382), which were involved in ERAD, to further examine their ubiquitination by western blotting. Two additional proteins, PhACO3 (Unigene0022854) and PhAUX1 (Unigene0019926), also were selected. Synthetic peptide versions of these proteins were used as immunogens to immunize rabbits for antibody production. Total proteins were extracted from corollas treated with air, ethylene, and both ethylene and MG132. Western blotting using the antibodies raised against these proteins showed that protein abundance was higher in plants treated with both ethylene and MG132 compared with plants treated only with ethylene (Supplemental Fig. S14B), which further confirmed the ubiquitination of these proteins (Kevany et al., 2007).
CONCLUSION
This study provides a global and comparative analysis of transcriptome, proteome, and ubiquitylome regulation by ethylene and offers further insights into the dynamics of individual Kub sites. Our results revealed Kub site motifs not observed previously in plants, and these novel plant Kub site motifs could lead to future discoveries of novel ubiquitin ligase-substrate interactions. We also revealed that the global proteome and ubiquitylome were negatively correlated because of the important function of ubiquitination in protein degradation (Wilkinson, 2000). Several putative ubiquitin ligases were up-regulated by ethylene at the protein and transcription levels. These results demonstrated the important roles of ubiquitination in the degradation of proteins during ethylene-mediated corolla senescence in petunias. We analyzed the effects of ethylene on several aspects of flower senescence. To our knowledge for the first time, our results revealed the effects of ethylene on proteins involved in ERAD and identified many novel ubiquitination sites in several proteins, including PhETR2, PhACO, PhCb5s, and PhAUX1. The provided data set may serve as an important resource for the functional analysis of Kub in petunia and facilitate the elucidation of the senescence process in this model petunia. In addition, it should be pointed out that some changes unveiled by omics in this study could be the outcomes of senescence, and the ubiquitination type (polyubiquitination or monoubiquitination) of the specific Kub sites of proteins in this study needs further study.
MATERIALS AND METHODS
Plant Material
Petunia (Petunia hybrida ‘Mitchell’) plants were grown under greenhouse conditions (22°C–25°C) and a natural photoperiod (Liu et al., 2011). Flowers were emasculated 1 d before they were fully open to prevent self-pollination. Eight to 10 petunia flowers were harvested at the anthesis stage (corollas 90° reflexed) and then placed immediately in tap water. Buds were collected at about 3 weeks after the first flower opened. Stems, leaves, and roots were collected from plants at the vegetative stage, when the plants were approximately 10 cm in height. All tissues were frozen in liquid nitrogen and stored at –80°C until used for RNA extraction. Fresh weights were measured immediately before freezing (Liu et al., 2011). All experiments were conducted at least three times with independently collected and extracted tissues unless noted otherwise.
Ethylene Treatment
Petunia flowers were treated with ethylene according to previously described protocols (Tan et al., 2014). Petunia flowers were harvested at anthesis, and their stems were recut to 5 cm, placed in flasks with distilled water, and subsequently treated with 2 µL L–1 ethylene for 0, 8, 16, 24, 32, 40, and 48 h. Corollas from eight to 10 flowers were collected at each time point, immediately frozen in liquid nitrogen, and stored at –80°C for subsequent RNA or protein extraction.
Proteasome inhibitor studies were performed by spraying corollas with an 80 μm MG132 solution (8% dimethyl sulfoxide) 4 h prior to 16 h of ethylene treatment. Control corollas were sprayed with an 8% dimethyl sulfoxide solution (Kevany et al., 2007).
RNA Extraction, Library Construction, and Sequencing
The total RNA of each sample listed above was isolated using the Trizol kit (Promega) following the manufacturer’s instructions. Then, total RNA was treated with RNase-free DNase I (Takara Bio) for 30 min at 37°C to remove residual DNA. RNA quality was verified using a 2100 Bio-analyzer (Agilent Technologies) and also was checked by RNase-free agarose gel electrophoresis. Next, poly(A) mRNA was isolated using oligo(dT) beads (Qiagen). All mRNA was broken into short fragments by adding fragmentation buffer. First-strand cDNA was generated using random hexamer-primed reverse transcription followed by the synthesis of the second-strand cDNA using RNase H and DNA polymerase I. The cDNA fragments were purified using the QIA Quick PCR extraction kit. These purified fragments were then washed with elution buffer for end-reparation poly(A) addition and ligated to sequencing adapters. Following agarose gel electrophoresis and extraction of cDNA from gels, the cDNA fragments were purified and enriched by PCR to construct the final cDNA library. The cDNA library was sequenced on the Illumina sequencing platform (Illumina HiSeq 2000) using paired-end technology by Gene Denovo. A Perl program was written to select clean reads by removing low-quality sequences (there were more than 50% bases with quality lower than 20 in one sequence), reads with more than 5% N bases (bases unknown), and reads containing adaptor sequences.
Read Alignment and Normalization of Gene Expression Levels
Sequencing reads were mapped to a reference sequence by SOAPaligner/soap2 (Li et al., 2009), a tool designed for short sequence alignment. The coverage of reads in one gene was used to calculate the expression level of that gene. Using this method, we obtained the expression levels of all genes detected.
Reads that could be mapped uniquely to a gene were used to calculate the expression level. The gene expression level was measured by the number of uniquely mapped reads per kilobase of exon region per million mapable reads (RPKM). The formula was defined as below:
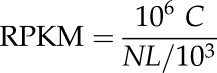 |
in which C was the number of reads uniquely mapped to the given gene, N was the number of reads uniquely mapped to all genes, and L was the total length of exons from the given gene. For genes with more than one alternative transcript, the longest transcript was selected to calculate the RPKM. The RPKM method eliminates the influence of different gene lengths and sequencing discrepancies on the gene expression calculation. Therefore, the RPKM value can be used directly to compare the differences in gene expression among samples. All expression data statistical analysis and visualization were conducted with the R package (http://www.r-project.org/).
qRT PCR Assays
qRT PCR assays were performed according to the methods of Liu et al. (2011). Total RNA was extracted from the samples of corollas and digested with RNase-free DNase I followed by reverse transcription according to the manufacturer’s instruction. PCR analysis was performed with the cDNA extracted from different samples as a template.
Quantitative PCR was performed on the LightCycler 480 Real-Time PCR system (Roche). Samples were subjected to thermal cycling conditions of DNA polymerase activation at 95°C for 4 min; 40 cycles of 45 s at 95°C, 45 s at 52°C or 55°C, 45 s at 72°C, and 45 s at 80°C; and a final elongation step of 7 min at 72°C was performed. The amplicon was analyzed by electrophoresis and sequenced once for identity confirmation. The sequences of all primers used for real-time PCR analysis are described in Supplemental Table S4. Petunia ACTIN was used as the housekeeping gene to quantify cDNA abundance. Primer specificity was determined by melting curve analysis; a single, sharp peak in the melting curve ensured that a single, specific DNA species had been amplified. Quantification was based on analysis of the threshold cycle value as described by Pfaffl (2001).
Protein Extraction
Petunia corollas were ground in liquid nitrogen, then the cell powder was transferred to a 5-mL centrifuge tube and sonicated three times on ice using a high-intensity ultrasonic processor (Scientz) in lysis buffer (8 m urea, 1% Triton X-100, 65 mm dithiothreitol and 0.1% protease inhibitor cocktail). The remaining debris was removed by centrifugation at 20,000g at 4°C for 10 min. Finally, the protein was precipitated with cold 15% TCA for 2 h at −20°C. After centrifugation at 4°C for 10 min, the supernatant was discarded. The remaining precipitate was washed with cold acetone three times. The protein was redissolved in buffer (8 m urea and 100 mm tetraethylammonium bromide [TEAB], pH 8), and the protein concentration was determined with the 2-D Quant kit (GE Healthcare) according to the manufacturer’s instructions. Three biological replicates were performed.
Preparation of Specific Antibodies against Proteins and Western-Blot Analysis
The synthetic peptides of proteins were used as an antigen for antibody production in rabbit from Abmart (www.ab-mart.com.cn/). These antibodies were used for blotting analysis.
Western-blot analyses were performed according to the methods of Tatsuki and Mori (2001). Proteins were separated using SDS-PAGE (10% acrylamide gels) and blotted onto nitrocellulose membranes (BA-S 85; Schleicher & Schuell). The membrane was blocked with 5% skim milk and 0.05% Tween 20 in Tris-buffered saline (50 mm Tris-HCl, pH 8, and 150 mm NaCl). Purified ubiquitin antibody or anti-glyceraldehyde-3-phosphate dehydrogenase antibody (internal reference) was used at a concentration of 50 mg mL−1. The membrane was washed with 0.05% Tween 20 in Tris-buffered saline and then reacted with horseradish peroxidase-conjugated goat anti-rabbit IgG (Pierce) at a dilution of 1:20,000. Detection was achieved using Super Signal West Femto (Pierce). Three biological replicates were performed.
Trypsin Digestion
For digestion, the protein solution was reduced with 10 mm dithiothreitol for 1 h at 37°C and alkylated with 20 mm IAA for 45 min at room temperature in darkness. For trypsin digestion, the protein sample was diluted by adding 100 mm TEAB to a urea concentration of less than 2 m. Finally, trypsin was added at a 1:50 trypsin:protein mass ratio for the first digestion overnight and a 1:100 trypsin:protein mass ratio for a second 4-h digestion. Approximately 100 μg of protein for each sample was digested with trypsin for the following experiments.
Tandem Mass Tag Labeling
After trypsin digestion, peptide was desalted with a Strata X C18 SPE column (Phenomenex) and vacuum dried. Peptide was reconstituted in 0.5 m TEAB and processed according to the manufacturer’s protocol for the six-plex Tandem Mass Tag (TMT) kit. Briefly, 1 unit of TMT reagent (defined as the amount of reagent required to label 100 μg of protein) was thawed and reconstituted in 24 μL of acetonitrile. The peptide mixtures were then incubated for 2 h at room temperature and pooled, desalted, and dried by vacuum centrifugation.
HPLC Fractionation
The sample was then fractionated into fractions by high-pH reverse-phase HPLC using an Agilent 300 Extend C18 column (5-μm particles, 4.6 mm i.d., 250 mm length). Briefly, peptides were first separated with a gradient of 2% to 60% acetonitrile in 10 mm ammonium bicarbonate, pH 10, over 80 min into 80 fractions, Then, the peptides were combined into 18 fractions and dried by vacuum centrifugation.
Affinity Enrichment
To enrich Kub peptides, tryptic peptides dissolved in NETN buffer (100 mm NaCl, 1 mm EDTA, 50 mm Tris-HCl, and 0.5% Nonidet P-40, pH 8) were incubated with prewashed antibody beads (PTM Biolabs) at 4°C overnight with gentle shaking. The beads were washed four times with NETN buffer and twice with distilled, deionized water. The bound peptides were eluted from the beads with 0.1% trifluoroacetic acid. The eluted fractions were combined and vacuum dried. The resulting peptides were cleaned with C18 ZipTips (Millipore) according to the manufacturer’s instructions, followed by LC-MS/MS analysis.
LC-MS/MS Analysis
Three parallel analyses for each fraction were performed. LC-MS/MS analysis was performed according to previously described protocols (Wu et al., 2015). Peptides were dissolved in 0.1% fatty acids and loaded directly onto a reverse-phase precolumn (Acclaim PepMap 100; Thermo Scientific). Peptide separation was performed using a reverse-phase analytical column (Acclaim PepMap RSLC; Thermo Scientific). The gradient was composed of an increase from 8% to 25% solvent B (0.1% fatty acids in 98% acetonitrile) over 26 min, 25% to 38% in 8 min, climbing to 85% in 4 min, and then holding at 85% for the last 4 min, all at a constant flow rate of 280 nL min−1 on an EASY-nLC 1000 ultra-performance liquid chromatography system. The resulting peptides were analyzed with the Q Exactive Plus Hybrid Quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific).
The peptides were subjected to a nanospray ionization source followed by tandem mass spectrometry (MS/MS) in Q Exactive Plus (Thermo) coupled online to the ultra-performance liquid chromatograph. Intact peptides were detected in the Orbitrap at a resolution of 70,000. Peptides were selected for MS/MS using a normalized collision energy setting of 30; ion fragments were detected in the Orbitrap at a resolution of 17,500. A data-dependent procedure that alternated between one mass spectrometry scan followed by 20 MS/MS scans was applied for the top 20 precursor ions above a threshold ion count of 1.5E4 in the mass spectrometry survey scan with 30-s dynamic exclusion. The electrospray voltage applied was 2 kV. Automatic gain control was used to prevent overfilling of the ion trap; 5E4 ions were accumulated for the generation of MS/MS spectra. For mass spectrometry scans, the mass-to-charge ratio scan range was 350 to 1,800. The fixed first mass was set as 100 mass-to-charge ratio.
Database Search
The resulting MS/MS data were processed using MaxQuant with an integrated Andromeda search engine (version 1.4.1.2). Tandem mass spectra were searched against a database (40,341 sequences) made from RNA sequencing of petunias in this study.
For proteomic peptides, Trypsin/P was used as a cleavage enzyme, allowing up to two missed cleavages. Mass error was set to 10 ppm for precursor ions and to 0.02 D for fragment ions. Carbamidomethyl on Cys, TMT-6plex (N-term), and TMT-6plex (K) were specified as fixed modifications, and oxidation on Met was specified as a variable modification. The false discovery rate was adjusted to less than 1%, and peptide ion score was set to greater than 20.
For Kub peptides, Trypsin/P was specified as a cleavage enzyme, allowing up to three missed cleavages. First, the search range was set to 5 ppm for precursor ions, and the main search range was set to 5 ppm and 0.02 D for fragment ions. Carbamidomethyl on Cys was specified as a fixed modification, and GlyGly on Lys and oxidation on Met were specified as variable modifications. The label-free quantification method was label-free quantification, false discovery rate was adjusted to less than 1%, while the minimum score for modified peptides was set to greater than 40.
Bioinformatic Analysis
Bioinformatic analysis was performed according to previously described protocols (Wu et al., 2015; Xie et al., 2015). GO term association and enrichment analysis were performed using the Database for Annotation, Visualization, and Integrated Discovery. The KEGG database was used to annotate protein pathways (Kanehisa and Goto, 2000). The KEGG online service tool KAAS was used to annotate the proteins’ KEGG database descriptions. The annotation results were mapped on the KEGG pathway database using the KEGG online service tool KEGG Mapper. The domain annotation was performed with InterProScan on the InterPro domain database via Web-based interfaces and services. WoLF PSORT was used to predict subcellular localization (Horton et al., 2007). The CORUM database was used to annotate protein complexes. Motif-X software was used to analyze the models of the sequences with amino acids in specific positions of ubiquityl-21-mers (10 amino acids upstream and downstream of the Kub site) in all of the protein sequences. In addition, the IPI Arabidopsis (Arabidopsis thaliana) proteome was used as the background database, and the other parameters were set to default values. The setting parameters for searching motifs using Motif-X software were occurrences 20 and the Bonferroni corrected P = 0.005. Protein-protein interaction networks were analyzed with the IntAct database (http://www.ebi.ac.uk/intact/). The protein-protein interaction network map was generated with Cytoscape software (Shannon et al., 2003).
Protein Quantitative Ratio Analysis
Protein quantitative ratio was calculated as the median of all unique peptide ratios. Student’s t test was performed to investigate differentially expressed protein effects. In order to meet the condition of Student’s t test, logarithmic transformation was performed to achieve the ratio of all peptides. Then, Student’s t test was explored to calculate the P value.
Ubiquitinated Protein Secondary Structure Analysis
In this study, we show the distribution of ubiquitinated and nonubiquitinated amino acids in protein secondary structures. The probabilities for different secondary structures (α-helix, β-strand, and coil) of ubiquitinated Lys were compared with the secondary structure probabilities of all Lys in all identified proteins. We further investigated the local secondary structures of proteins using NetSurfP, a software that predicts the surface accessibility and secondary structure of amino acids in an amino acid sequence.
Conservation Analysis of Ubiquitinated Proteins
To analyze the conservation of ubiquitinated proteins, first, proteins homologous to the ubiquitinated proteins identified in this study were obtained by BLASTing in the UniProtKB database against eight species: rice (Oryza sativa japonica), Brachypodium distachyon, Sorghum bicolor, Zea mays, Arabidopsis, soybean (Glycine max), tomato (Solanum lycopersicum), and Vitis vinifera. All of the protein sequences of these eight species were downloaded from the UniProtKB database. The sequence alignment software BLASTp was used to obtain the homologous proteins of the ubiquitinated proteins in this study. To find conserved sites, the multiple sequence alignment software MUSCLE was used to align homologs.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number FN014209 (petunia ACTIN). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (Vizcaino et al., 2010) via the Proteomics Identification Database partner repository with the dataset identifiers PXD005470 and PXD005457.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Effects of ethylene on the expression of ubiquitin in protein level.
Supplemental Figure S2. Venn diagram of annotation results against four protein databases.
Supplemental Figure S3. Confirmation of digital gene expression data by qRT-PCR.
Supplemental Figure S4. Functional enrichment analysis of differently expressed proteins.
Supplemental Figure S5. Concordance between changes in the abundance of mRNA and its encoded protein.
Supplemental Figure S6. Detection of mRNAs and their cognate proteins.
Supplemental Figure S7. KEGG pathway enrichment heat map of proteins with opposite trends in protein and ubiquitination levels.
Supplemental Figure S8. Venn diagram of proteomics and ubiquitinomic identification.
Supplemental Figure S9. MS/MS spectra of several ubiquitinated proteins.
Supplemental Figure S10. Effects of ethylene on the proteins engaged in the ABA and auxin signaling transduction pathway.
Supplemental Figure S11. Effects of ethylene on floral scent biosynthesis in petunia.
Supplemental Figure S12. Effects of ethylene on the amino acid biosynthesis pathway in petunia.
Supplemental Figure S13. Effects of ethylene on ERAD in petunia.
Supplemental Figure S14. Confirmation of proteome and ubiquitylome data.
Supplemental Table S1. Summary of Illumina paired-end sequencing and assembly.
Supplemental Table S2. Statistics of RNA sequencing results.
Supplemental Table S3. Statistics of the annotation of unigenes in four databases.
Supplemental Table S4. Primer sequences of 20 random genes used in quantitative PCR.
Supplemental File Exc S1. Predicted amino acid sequences of the coding sequence unigenes of the transcriptome.
Supplemental File Exc S2. Differently expressed genes from ethylene and air treatment of corollas.
Supplemental File Exc S3. Ethylene treatment changes the proteome profile in petunia corollas.
Supplemental File Exc S4. Enrichment of proteins up/down-regulated by ethylene.
Supplemental File Exc S5. Comparative analysis of proteome and transcriptome data.
Supplemental File Exc S6. Ethylene treatment changes the ubiquitylome profile in corollas in petunia.
Supplemental File Exc S7. Enrichment of proteins with Kub sites up/down-regulated.
Supplemental File Exc S8. Ubiquitination sites of proteins in petunia.
Supplemental File Exc S9. Correlation between the global proteome and ubiquitylome.
Supplemental File Exc S10. Ubiquitin proteins up/down-regulated by ethylene treatment.
Supplemental File Exc S11. Proteasome and nonproteasome proteases involved in the degradation of proteins.
Supplemental File Exc S12. Nonproteasomal proteases up-regulated by ethylene treatment.
Supplemental File Exc S13. Changes of the senescence-related proteins after ethylene treatment.
Supplementary Material
Acknowledgments
We thank Guangzhou Gene Denovo Biotechnology and Jingjie PTM Biolabs for providing the methods for partial data analysis.
Glossary
- PTM
posttranslational modification
- ERAD
endoplasmic reticulum-associated degradation
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- DEG
differentially expressed unigene
- qRT
quantitative real-time
- GO
Gene Ontology
- Kub
lysine ubiquitination
- ABA
abscisic acid
- ER
endoplasmic reticulum
- RPKM
reads per kilobase of exon region per million mapable reads
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- MS/MS
tandem mass spectrometry
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31270736, 31661143047, and 31470700).
References
- Abeles FB. (1992) Ethylene in Plant Biology. Academic Press, San Diego [Google Scholar]
- Alberch P. (1991) From genes to phenotype: dynamical systems and evolvability. Genetica 84: 5–11 [DOI] [PubMed] [Google Scholar]
- Arnason T, Ellison MJ (1994) Stress resistance in Saccharomyces cerevisiae is strongly correlated with assembly of a novel type of multiubiquitin chain. Mol Cell Biol 14: 7876–7883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baboshina OV, Haas AL (1996) Novel multiubiquitin chain linkages catalyzed by the conjugating enzymes E2EPF and RAD6 are recognized by 26 S proteasome subunit 5. J Biol Chem 271: 2823–2831 [DOI] [PubMed] [Google Scholar]
- Bai S, Willard B, Chapin LJ, Kinter MT, Francis DM, Stead AD, Jones ML (2010) Proteomic analysis of pollination-induced corolla senescence in petunia. J Exp Bot 61: 1089–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boatright J, Negre F, Chen X, Kish CM, Wood B, Peel G, Orlova I, Gang D, Rhodes D, Dudareva N (2004) Understanding in vivo benzenoid metabolism in petunia petal tissue. Plant Physiol 135: 1993–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borochoy A, Spiegelstein H, Philosoph-Hadas S (1997) Ethylene and flower petal senescence: interrelationship with membrane lipid catabolism. Physiol Plant 100: 606–612 [Google Scholar]
- Breeze E, Wagstaff C, Harrison E, Bramke I, Rogers H, Stead A, Thomas B, Buchanan-Wollaston V (2004) Gene expression patterns to define stages of post-harvest senescence in Alstroemeria petals. Plant Biotechnol J 2: 155–168 [DOI] [PubMed] [Google Scholar]
- Buck TM, Wright CM, Brodsky JL (2007) The activities and function of molecular chaperones in the endoplasmic reticulum. Semin Cell Dev Biol 18: 751–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Clay JM, Chang C (2014) Association of cytochrome b5 with ETR1 ethylene receptor signaling through RTE1 in Arabidopsis. Plant J 77: 558–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Zhang L, Jia Z, Gu H, Hong K, Gong D (2015) Early differential gene expression profiling of harvest-induced senescence in detached Arabidopsis plants. Acta Physiol Plant 37: 1–10 [Google Scholar]
- Chen YF, Shakeel SN, Bowers J, Zhao XC, Etheridge N, Schaller GE (2007) Ligand-induced degradation of the ethylene receptor ETR2 through a proteasome-dependent pathway in Arabidopsis. J Biol Chem 282: 24752–24758 [DOI] [PubMed] [Google Scholar]
- Cheng WH, Chiang MH, Hwang SG, Lin PC (2009) Antagonism between abscisic acid and ethylene in Arabidopsis acts in parallel with the reciprocal regulation of their metabolism and signaling pathways. Plant Mol Biol 71: 61–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Liu J, Yang X, Ma R, Liu Q, Liu C (2013) Construction of ethylene regulatory network based on the phytohormones related gene transcriptome profiling and prediction of transcription factor activities in soybean. Acta Physiol Plant 35: 1303–1317 [Google Scholar]
- Colaert N, Helsens K, Martens L, Vandekerckhove J, Gevaert K (2009) Improved visualization of protein consensus sequences by iceLogo. Nat Methods 6: 786–787 [DOI] [PubMed] [Google Scholar]
- Colquhoun TA, Schimmel BC, Kim JY, Reinhardt D, Cline K, Clark DG (2010) A petunia chorismate mutase specialized for the production of floral volatiles. Plant J 61: 145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney SE, Rider CC, Stead AD (1994) Changes in protein ubiquitination and the expression of ubiquitin-encoding transcripts in daylily petals during floral development and senescence. Physiol Plant 91: 196–204 [Google Scholar]
- Delbarre A, Muller P, Guern J (1998) Short-lived and phosphorylated proteins contribute to carrier-mediated efflux, but not to influx, of auxin in suspension-cultured tobacco cells. Plant Physiol 116: 833–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Srivastava R, Howell SH (2013) Endoplasmic reticulum (ER) stress response and its physiological roles in plants. Int J Mol Sci 14: 8188–8212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diepold A, Li G, Lennarz WJ, Nürnberger T, Brunner F (2007) The Arabidopsis AtPNG1 gene encodes a peptide: N-glycanase. Plant J 52: 94–104 [DOI] [PubMed] [Google Scholar]
- Doelling JH, Walker JM, Friedman EM, Thompson AR, Vierstra RD (2002) The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem 277: 33105–33114 [DOI] [PubMed] [Google Scholar]
- Douglas CC. (2014) An open framework for dynamic big-data-driven application systems (DBDDAS) development. Procedia Comput Sci 29: 1246–1255 [Google Scholar]
- Ecker JR. (1995) The ethylene signal transduction pathway in plants. Science 268: 667–675 [DOI] [PubMed] [Google Scholar]
- Farmer LM, Book AJ, Lee KH, Lin YL, Fu H, Vierstra RD (2010) The RAD23 family provides an essential connection between the 26S proteasome and ubiquitylated proteins in Arabidopsis. Plant Cell 22: 124–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzarrini S, McCourt P (2001) Genetic interactions between ABA, ethylene and sugar signaling pathways. Curr Opin Plant Biol 4: 387–391 [DOI] [PubMed] [Google Scholar]
- Gerats T, Vandenbussche M (2005) A model system for comparative research: Petunia. Trends Plant Sci 10: 251–256 [DOI] [PubMed] [Google Scholar]
- Ghassemian M, Nambara E, Cutler S, Kawaide H, Kamiya Y, McCourt P (2000) Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell 12: 1117–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazalpour A, Bennett B, Petyuk VA, Orozco L, Hagopian R, Mungrue IN, Farber CR, Sinsheimer J, Kang HM, Furlotte N, et al. (2011) Comparative analysis of proteome and transcriptome variation in mouse. PLoS Genet 7: e1001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnad F, Gunawardena J, Mann M (2011) PHOSIDA 2011: the posttranslational modification database. Nucleic Acids Res 39: D253–D260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra DD, Callis J (2012) Ubiquitin on the move: the ubiquitin modification system plays diverse roles in the regulation of endoplasmic reticulum- and plasma membrane-localized proteins. Plant Physiol 160: 56–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gygi SP, Rochon Y, Franza BR, Aebersold R (1999) Correlation between protein and mRNA abundance in yeast. Mol Cell Biol 19: 1720–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka H, Noda T, Shirano Y, Kato T, Hayashi H, Shibata D, Tabata S, Ohsumi Y (2002) Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol 129: 1181–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M. (1995) Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr Opin Cell Biol 7: 215–223 [DOI] [PubMed] [Google Scholar]
- Hoeberichts FA, van Doorn WG, Vorst O, Hall RD, van Wordragen MF (2007) Sucrose prevents up-regulation of senescence-associated genes in carnation petals. J Exp Bot 58: 2873–2885 [DOI] [PubMed] [Google Scholar]
- Hong Z, Jin H, Tzfira T, Li J (2008) Multiple mechanism-mediated retention of a defective brassinosteroid receptor in the endoplasmic reticulum of Arabidopsis. Plant Cell 20: 3418–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35: W585–W587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter DA, Steele BC, Reid MS (2002) Identification of genes associated with perianth senescence in daffodil (Narcissus pseudonarcissus L. ‘Dutch Master’). Plant Sci 163: 13–21 [Google Scholar]
- Johnson ES, Ma PC, Ota IM, Varshavsky A (1995) A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem 270: 17442–17456 [DOI] [PubMed] [Google Scholar]
- Jones ML, Chaffin GS, Eason JR, Clark DG (2005) Ethylene-sensitivity regulates proteolytic activity and cysteine protease gene expression in petunia corollas. J Exp Bot 56: 2733–2744 [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28: 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M (2006) Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol 22: 159–180 [DOI] [PubMed] [Google Scholar]
- Kevany BM, Tieman DM, Taylor MG, Cin VD, Klee HJ (2007) Ethylene receptor degradation controls the timing of ripening in tomato fruit. Plant J 51: 458–467 [DOI] [PubMed] [Google Scholar]
- Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, et al. (2011) Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell 44: 325–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner J, Petrášek J, Tomanov K, Retzer K, Pařezová M, Korbei B, Bachmair A, Zažímalová E, Luschnig C (2012) Lysine63-linked ubiquitylation of PIN2 auxin carrier protein governs hormonally controlled adaptation of Arabidopsis root growth. Proc Natl Acad Sci USA 109: 8322–8327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25: 1966–1967 [DOI] [PubMed] [Google Scholar]
- Liu J, Li J, Wang H, Fu Z, Liu J, Yu Y (2011) Identification and expression analysis of ERF transcription factor genes in petunia during flower senescence and in response to hormone treatments. J Exp Bot 62: 825–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Howell SH (2010) Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. Plant Cell 22: 2930–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H, Shasany AK, Schnepp J, Orlova I, Taguchi G, Cooper BR, Rhodes D, Pichersky E, Dudareva N (2010) RNAi suppression of Arogenate Dehydratase1 reveals that phenylalanine is synthesized predominantly via the arogenate pathway in petunia petals. Plant Cell 22: 832–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masahara-Negishi Y, Hosomi A, Della Mea M, Serafini-Fracassini D, Suzuki T (2012) A plant peptide: N-glycanase orthologue facilitates glycoprotein ER-associated degradation in yeast. Biochim Biophys Acta 1820: 1457–1462 [DOI] [PubMed] [Google Scholar]
- Mayak S, Halevy AH (1972) Interrelationships of ethylene and abscisic acid in the control of rose petal senescence. Plant Physiol 50: 341–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayuoni L, Sharabi-Schwager M, Feldmesser E, Porat R (2011) Effects of ethylene degreening on the transcriptome of mandarin flesh. Postharvest Biol Technol 60: 75–82 [Google Scholar]
- McEwan DG, Dikic I (2011) The three musketeers of autophagy: phosphorylation, ubiquitylation and acetylation. Trends Cell Biol 21: 195–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Zentgraf U (2010) A HECT E3 ubiquitin ligase negatively regulates Arabidopsis leaf senescence through degradation of the transcription factor WRKY53. Plant J 63: 179–188 [DOI] [PubMed] [Google Scholar]
- Müller F, Adori C, Sass M (2004) Autophagic and apoptotic features during programmed cell death in the fat body of the tobacco hornworm (Manduca sexta). Eur J Cell Biol 83: 67–78 [DOI] [PubMed] [Google Scholar]
- Muller J, Szklarczyk D, Julien P, Letunic I, Roth A, Kuhn M, Powell S, von Mering C, Doerks T, Jensen LJ, et al. (2010) eggNOG v2.0: extending the evolutionary genealogy of genes with enhanced non-supervised orthologous groups, species and functional annotations. Nucleic Acids Res 38: D190–D195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratani M, Tansey WP (2003) How the ubiquitin-proteasome system controls transcription. Nat Rev Mol Cell Biol 4: 192–201 [DOI] [PubMed] [Google Scholar]
- Nakaminami K, Matsui A, Nakagami H, Minami A, Nomura Y, Tanaka M, Morosawa T, Ishida J, Takahashi S, Uemura M, et al. (2014) Analysis of differential expression patterns of mRNA and protein during cold-acclimation and de-acclimation in Arabidopsis. Mol Cell Proteomics 13: 3602–3611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre F, Kish CM, Boatright J, Underwood B, Shibuya K, Wagner C, Clark DG, Dudareva N (2003) Regulation of methylbenzoate emission after pollination in snapdragon and petunia flowers. Plant Cell 15: 2992–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov V, Li J, Batoux M, Roux M, Chu ZH, Lacombe S, Rougon A, Bittel P, Kiss-Papp M, Chinchilla D, et al. (2009) Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. EMBO J 28: 3428–3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi Y. (2001) Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol 2: 211–216 [DOI] [PubMed] [Google Scholar]
- Onoue T, Mikami M, Yoshioka T, Hashiba T, Satoh S (2000) Characteristics of the inhibitory action of 1,1-dimethyl-4-(phenylsulfonyl) semicarbazide (DPSS) on ethylene production in carnation (Dianthus caryophyllus L.) flowers. Plant Growth Regul 30: 201–207 [Google Scholar]
- Pak C, van Doorn WG (2005) Delay of Iris flower senescence by protease inhibitors. New Phytol 165: 473–480 [DOI] [PubMed] [Google Scholar]
- Passmore LA, Barford D (2004) Getting into position: the catalytic mechanisms of protein ubiquitylation. Biochem J 379: 513–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul MJ, Frigerio L (2007) Coated vesicles in plant cells. Semin Cell Dev Biol 18: 471–478 [DOI] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP (2003) A proteomics approach to understanding protein ubiquitination. Nat Biotechnol 21: 921–926 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigliucci M. (2010) Genotype-phenotype mapping and the end of the ‘genes as blueprint’ metaphor. Philos Trans R Soc Lond B Biol Sci 365: 557–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potuschak T, Lechner E, Parmentier Y, Yanagisawa S, Grava S, Koncz C, Genschik P (2003) EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115: 679–689 [DOI] [PubMed] [Google Scholar]
- Prayitno J, Imin N, Rolfe BG, Mathesius U (2006) Identification of ethylene-mediated protein changes during nodulation in Medicago truncatula using proteome analysis. J Proteome Res 5: 3084–3095 [DOI] [PubMed] [Google Scholar]
- Qiao H, Chang KN, Yazaki J, Ecker JR (2009) Interplay between ethylene, ETP1/ETP2 F-box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes Dev 23: 512–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raasi S, Wolf DH (2007) Ubiquitin receptors and ERAD: a network of pathways to the proteasome. Semin Cell Dev Biol 18: 780–791 [DOI] [PubMed] [Google Scholar]
- Rancour DM, Park S, Knight SD, Bednarek SY (2004) Plant UBX domain-containing protein 1, PUX1, regulates the oligomeric structure and activity of Arabidopsis CDC48. J Biol Chem 279: 54264–54274 [DOI] [PubMed] [Google Scholar]
- Reid MS, Wu M (1992) Ethylene and flower senescence. Plant Growth Regul 11: 37–43 [Google Scholar]
- Rodrigues RS, Boldrini-França J, Fonseca FP, de la Torre P, Henrique-Silva F, Sanz L, Calvete JJ, Rodrigues VM (2012) Combined snake venomics and venom gland transcriptomic analysis of Bothropoides pauloensis. J Proteomics 75: 2707–2720 [DOI] [PubMed] [Google Scholar]
- Ronen M, Mayak S (1981) Interrelationship between abscisic acid and ethylene in the control of senescence processes in carnation flowers. J Exp Bot 32: 759–765 [Google Scholar]
- Savin KW, Baudinette SC, Graham MW (1995) Antisense ACC oxidase RNA delays carnation petal senescence. HortScience 30: 970–972 [Google Scholar]
- Schnell JD, Hicke L (2003) Non-traditional functions of ubiquitin and ubiquitin-binding proteins. J Biol Chem 278: 35857–35860 [DOI] [PubMed] [Google Scholar]
- Schuurink RC, Haring MA, Clark DG (2006) Regulation of volatile benzenoid biosynthesis in petunia flowers. Trends Plant Sci 11: 20–25 [DOI] [PubMed] [Google Scholar]
- Shabek N, Zheng N (2014) Plant ubiquitin ligases as signaling hubs. Nat Struct Mol Biol 21: 293–296 [DOI] [PubMed] [Google Scholar]
- Shahri W, Tahir I (2011) Flower senescence: strategies and some associated events. Bot Rev 77: 152–184 [Google Scholar]
- Shahri W, Tahir I (2014) Flower senescence: some molecular aspects. Planta 239: 277–297 [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemesh-Mayer E, Ben-Michael T, Rotem N, Rabinowitch HD, Doron-Faigenboim A, Kosmala A, Perlikowski D, Sherman A, Kamenetsky R (2015) Garlic (Allium sativum L.) fertility: transcriptome and proteome analyses provide insight into flower and pollen development. Front Plant Sci 6: 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya K, Niki T, Ichimura K (2013) Pollination induces autophagy in petunia petals via ethylene. J Exp Bot 64: 1111–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slade WO, Ray WK, Williams PM, Winkel BS, Helm RF (2012) Effects of exogenous auxin and ethylene on the Arabidopsis root proteome. Phytochemistry 84: 18–23 [DOI] [PubMed] [Google Scholar]
- Smith MH, Ploegh HL, Weissman JS (2011) Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science 334: 1086–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Yun J, Likhacheva AV, Alonso JM (2007) Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 19: 2169–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson P, Rubinstein B (1998) Characterization of proteolytic activity during senescence in daylilies. Physiol Plant 104: 463–473 [Google Scholar]
- Sun J, Cardoza V, Mitchell DM, Bright L, Oldroyd G, Harris JM (2006) Crosstalk between jasmonic acid, ethylene and Nod factor signaling allows integration of diverse inputs for regulation of nodulation. Plant J 46: 961–970 [DOI] [PubMed] [Google Scholar]
- Swaney DL, Beltrao P, Starita L, Guo A, Rush J, Fields S, Krogan NJ, Villén J (2013) Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat Methods 10: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y, Liu J, Huang F, Guan J, Zhong S, Tang N, Zhao J, Yang W, Yu Y (2014) PhGRL2 protein, interacting with PhACO1, is involved in flower senescence in the petunia. Mol Plant 7: 1384–1387 [DOI] [PubMed] [Google Scholar]
- Tatsuki M, Mori H (2001) Phosphorylation of tomato 1-aminocyclopropane-1-carboxylic acid synthase, LE-ACS2, at the C-terminal region. J Biol Chem 276: 28051–28057 [DOI] [PubMed] [Google Scholar]
- Teale WD, Paponov IA, Palme K (2006) Auxin in action: signalling, transport and the control of plant growth and development. Nat Rev Mol Cell Biol 7: 847–859 [DOI] [PubMed] [Google Scholar]
- Titapiwatanakun B, Murphy AS (2009) Post-transcriptional regulation of auxin transport proteins: cellular trafficking, protein phosphorylation, protein maturation, ubiquitination, and membrane composition. J Exp Bot 60: 1093–1107 [DOI] [PubMed] [Google Scholar]
- Tripathi SK, Tuteja N (2007) Integrated signaling in flower senescence: an overview. Plant Signal Behav 2: 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Nakashima K, Miyakawa T, Kuromori T, Tanokura M, Shinozaki K, Yamaguchi-Shinozaki K (2010) Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol 51: 1821–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood BA, Tieman DM, Shibuya K, Dexter RJ, Loucas HM, Simkin AJ, Sims CA, Schmelz EA, Klee HJ, Clark DG (2005) Ethylene-regulated floral volatile synthesis in petunia corollas. Plant Physiol 138: 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn WG, Balk PA, van Houwelingen AM, Hoeberichts FA, Hall RD, Vorst O, van der Schoot C, van Wordragen MF (2003) Gene expression during anthesis and senescence in Iris flowers. Plant Mol Biol 53: 845–863 [DOI] [PubMed] [Google Scholar]
- van Doorn WG, Woltering EJ (2008) Physiology and molecular biology of petal senescence. J Exp Bot 59: 453–480 [DOI] [PubMed] [Google Scholar]
- Vizcaino JA, Cote R, Reisinger F, Barsnes H, Foster JM, Rameseder J, Hermjakob H, Martens L (2010) The Proteomics Identifications database: 2010 update. Nucleic Acids Res 38: D736–D742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner SA, Beli P, Weinert BT, Nielsen ML, Cox J, Mann M, Choudhary C (2011) A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol Cell Proteomics 10: 013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KD. (2000) Ubiquitination and deubiquitination: targeting of proteins for degradation by the proteasome. Semin Cell Dev Biol 11: 141–148 [DOI] [PubMed] [Google Scholar]
- Woltering EJ, Van Doorn WG (1988) Role of ethylene in senescence of petals: morphological and taxonomical relationships. J Exp Bot 39: 1605–1616 [Google Scholar]
- Woo HR, Kim HJ, Nam HG, Lim PO (2013) Plant leaf senescence and death: regulation by multiple layers of control and implications for aging in general. J Cell Sci 126: 4823–4833 [DOI] [PubMed] [Google Scholar]
- Wu Q, Cheng Z, Zhu J, Xu W, Peng X, Chen C, Li W, Wang F, Cao L, Yi X, et al. (2015) Suberoylanilide hydroxamic acid treatment reveals crosstalks among proteome, ubiquitylome and acetylome in non-small cell lung cancer A549 cell line. Sci Rep 5: 9520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Kang H, Liu W, Wang GL (2015) Comprehensive profiling of the rice ubiquitome reveals the significance of lysine ubiquitination in young leaves. J Proteome Res 14: 2017–2025 [DOI] [PubMed] [Google Scholar]
- Xu J, Zhang S (2014) Regulation of ethylene biosynthesis and signaling by protein kinases and phosphatases. Mol Plant 7: 939–942 [DOI] [PubMed] [Google Scholar]
- Xu X, Jiang CZ, Donnelly L, Reid MS (2007) Functional analysis of a RING domain ankyrin repeat protein that is highly expressed during flower senescence. J Exp Bot 58: 3623–3630 [DOI] [PubMed] [Google Scholar]
- Yang SF, Hoffman NE (1984) Ethylene biosynthesis and its regulation in higher plants. Annu Rev Plant Physiol 35: 155–189 [Google Scholar]
- Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y (2004) Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16: 2967–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Fu XD (2013) Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma 122: 191–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zourelidou M, Absmanner B, Weller B, Barbosa IC, Willige BC, Fastner A, Streit V, Port SA, Colcombet J, de la Fuente van Bentem S, et al. (2014) Auxin efflux by PIN-FORMED proteins is activated by two different protein kinases, D6 PROTEIN KINASE and PINOID. eLife 3: e2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.