The Leu-rich repeat receptor-like kinases SERK1 and SERK2 affect anther cell differentiation as a partners of EMS1 in Arabidopsis.
Abstract
Cell signaling pathways mediated by leucine-rich repeat receptor-like kinases (LRR-RLKs) are essential for plant growth, development, and defense. The EMS1 (EXCESS MICROSPOROCYTES1) LRR-RLK and its small protein ligand TPD1 (TAPETUM DETERMINANT1) play a fundamental role in somatic and reproductive cell differentiation during early anther development in Arabidopsis (Arabidopsis thaliana). However, it is unclear whether other cell surface molecules serve as coregulators of EMS1. Here, we show that SERK1 (SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1) and SERK2 LRR-RLKs act redundantly as coregulatory and physical partners of EMS1. The SERK1/2 genes function in the same genetic pathway as EMS1 in anther development. Bimolecular fluorescence complementation, Förster resonance energy transfer, and coimmunoprecipitation approaches revealed that SERK1 interacted biochemically with EMS1. Transphosphorylation of EMS1 by SERK1 enhances EMS1 kinase activity. Among 12 in vitro autophosphorylation and transphosphorylation sites identified by tandem mass spectrometry, seven of them were found to be critical for EMS1 autophosphorylation activity. Furthermore, complementation test results suggest that phosphorylation of EMS1 is required for its function in anther development. Collectively, these data provide genetic and biochemical evidence of the interaction and phosphorylation between SERK1/2 and EMS1 in anther development.
Plants and animals share similar mechanisms for cell-cell and cell-environment communications. Signaling in multicellular organisms relies predominantly on receptor kinases, and this is particularly true for plants. In Arabidopsis (Arabidopsis thaliana), more than 600 genes encode receptor-like kinases (RLKs), with 223 Leu-rich repeat (LRR) RLKs forming the largest family of RLKs (McCarty and Chory, 2000; Shiu et al., 2004; Torii, 2004; Zhao, 2009). Extensive research has shown that LRR-RLKs are involved in a wide range of plant growth, developmental, and physiological processes as well as defense responses, including the regulation of shoot and root meristem sizes, cell fate determination and patterning, steroid hormone signaling, vascular patterning, organ size and shape regulation, organ abscission, pollen tube reception, defense responses, plant transpiration, nodulation, and nitrogen acquisition (Torii, 2004; Zhao, 2009; Gursanscky et al., 2016; Ma et al., 2016; Shinohara et al., 2016; Takeuchi and Higashiyama, 2016; Wang et al., 2016). So far, only a few coreceptors have been identified for the many functionally characterized LRR-RLKs.
In flowering plants, successful sexual reproduction depends on the specification of distinct types of somatic and reproductive cells that yield male and female gametophytes. The anther, where male gametophytes (pollen) are produced, typically has four lobes (microsporangia; Goldberg et al., 1993; Scott et al., 2004; Zhao, 2009; Feng et al., 2013; Walbot and Egger, 2016). Within each of these lobes in a mature anther, the central reproductive microsporocytes (or pollen mother cells) are surrounded by four concentrically organized somatic cell layers: epidermis, endothecium, middle layer, and tapetum (Zhao et al., 2002; Zhao, 2009). Microsporocytes give rise to pollen via meiosis, while somatic cells, particularly the tapetum, are required for the normal development and release of pollen. Our previous studies demonstrate that the EMS1 (EXCESS MICROSPOROCYTES1) LRR-RLK-linked signal transduction pathway plays a fundamental role in somatic and reproductive cell differentiation during early anther development. In Arabidopsis, anthers in the ems1 (also known as extra sporogenous cells) mutant have no tapetal cells; instead, they produce excess microsporocytes (Canales et al., 2002; Zhao et al., 2002). Furthermore, we found that TPD1 (TAPETUM DETERMINANT1) is a small secreted Cys-rich protein ligand of EMS1 (Yang et al., 2003; Jia et al., 2008; Huang et al., 2016c). Our recent results show that TPD1 is secreted from precursors of microsporocytes/microsporocytes (Huang et al., 2016c). TPD1 interacts with EMS1 that is localized at the plasma membrane of tapetal precursor cells/tapetal cells. The activated TPD1-EMS1 signaling initially promotes parietal cell division to form tapetal precursor cells and then determines the fate of functional tapetal cells. Disruption of MSP1 (MULTIPLE SPOROCYTE 1) and TDL1A (also known as MIL2 [MICROSPORELESS2]) genes in rice (Oryza sativa) as well as the MAC1 (MULTIPLE ARCHESPORIAL CELLS1) gene in maize (Zea mays) causes similar anther phenotypes to that of ems1 and tpd1 mutants (Nonomura et al., 2003; Zhao et al., 2008; Hong et al., 2012; Wang et al., 2012). MSP1 encodes an EMS1-like protein, while MIL2/TDL1A and MAC1 genes encode a TPD1-like protein, suggesting that the function of TPD1-EMS1 signaling is conserved in anther cell differentiation. However, it is not known whether EMS1 functions together with a coreceptor to determine somatic cell fate during anther development.
In Arabidopsis, SERK (SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE) LRR-RLKs are involved in multiple signaling pathways by partnering with other LRR-RLKs, such as BRI1 (BRASSINOSTEROID INSENSITIVE1), ER/ERL (ERECTA/ERECTA-LIKE), HAE/HSL2 (HAESA/HAESA-LIKE2), PSKR1 (PHYTOSULFOKINE [PSK] RECEPTOR1), FLS2 (FLAGELLIN-SENSING2), and EFR (ELONGATION FACTOR-TU [EF-Tu] RECEPTOR; Ma et al., 2016). SERK1 and SERK2 might act redundantly as coreceptors of EMS1, because the serk1 serk2 double mutant phenocopies ems1, although neither the serk1 nor serk2 single mutant shows detectable anther defects (Zhao et al., 2002; Albrecht et al., 2005; Colcombet et al., 2005). In this study, we report that SERK1/2 serves as an EMS1 partner to control anther cell differentiation. We show that SERK1/2 genetically interacted with EMS1 in anther development. Additionally, bimolecular fluorescence complementation (BiFC), Förster resonance energy transfer (FRET), and coimmunoprecipitation (co-IP) assays demonstrated that SERK1 biochemically interacted with EMS1. Transphosphorylation of EMS1 by SERK1 is essential for enhancing EMS1 kinase activity. Furthermore, tandem mass spectrometry analysis identified a total of 12 in vitro autophosphorylation and transphosphorylation sites of EMS1, and seven of them were found to be important for EMS1 autophosphorylation activity. Our results also suggest that phosphorylation of EMS1 is required for its function in planta. Collectively, our data support that SERK1/2 functions as partners of EMS1 to control anther cell fate determination in Arabidopsis.
RESULTS
SERK1/2 and EMS1 Function in the Same Genetic Pathway for Anther Development
Neither the serk1-1 nor serk2-1 single mutant has a detectable phenotype, but anthers in the serk1 serk2 double mutant show an identical phenotype to the ems1 anther (Zhao et al., 2002; Albrecht et al., 2005; Colcombet et al., 2005). Therefore, SERK1 and SERK2 might function redundantly as coreceptors of EMS1 during anther development. To investigate the genetic interaction between SERK1/2 and EMS1 genes, we identified a novel weak allele of EMS1 (SALK_051989) with the T-DNA insertion located 497 bp upstream of the ATG start codon (Supplemental Fig. S1A). We named this new allele ems1-2, and the previously identified null EMS1 mutant ems1 was renamed as ems1-1 (Zhao et al., 2002). Reverse transcription-PCR and quantitative reverse transcription-PCR results showed that the expression of EMS1 was reduced in ems1-2 anthers compared with those of the wild type (Supplemental Fig. S1, B and C). There was no difference in growth between wild type and ems1-2 plants; however, the fertility of the ems1-2 mutant was reduced slightly, as indicated by the fact that a few early siliques were completely or partially sterile (Supplemental Fig. S2, A and E). Pollen staining examination revealed a small reduction of viable pollen grains in ems1-2 partially sterile anthers compared with those of the wild type (Supplemental Fig. S2, B and F).
We then performed genetic analysis between EMS1 and SERK1/2. Examination of silique growth and pollen viability showed that serk1-1 (Supplemental Fig. S2, G and H) and serk2-1 (Supplemental Fig. S2, I and J) single null mutants had normal male fertility. Conversely, similar to the ems1-1 strong mutant (Supplemental Fig. S2, C and D) and the serk1-1 serk2-1 double mutant (Supplemental Fig. S2, K and L), the ems1-2 serk1-1 double mutant was sterile and did not produce pollen grains in the anther (Supplemental Fig. S2, M and N), although the fertility reduction of ems1-2 serk2-1 was not as severe as that of ems1-2 serk1-1 (Supplemental Fig. S2, O and P). Moreover, the ems1-1 serk1-1 serk2-1 triple mutant was completely sterile (Supplemental Fig. S2, Q and R).
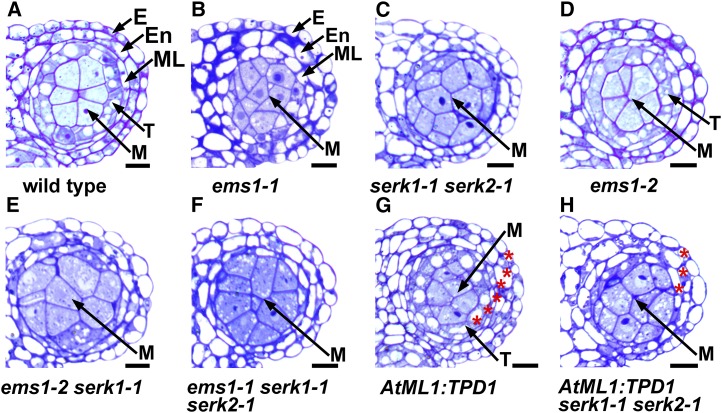
To further examine anther cell differentiation defects in those mutants, we conducted anther semithin sectioning. In the wild-type anther, the reproductive microsporocytes are surrounded by a layer of somatic cells called tapetum (Fig. 1A). In contrast, anthers from the ems1-1 single mutant (Fig. 1B) and the serk1-1 serk2-1 double mutant (Fig. 1C) completely lacked the tapetum but produced excess microsporocytes, as reported previously (Zhao et al., 2002; Albrecht et al., 2005; Colcombet et al., 2005). Similar to serk1-1 and serk2-1 single mutants (Albrecht et al., 2005; Colcombet et al., 2005), the ems1-2 anther formed normal tapetum and microsporocytes (Fig. 1D). Conversely, the ems1-2 serk1-1 anther (Fig. 1E) resembled the phenotype of ems1-1 (Fig. 1B) and serk1-1 serk2-1 (Fig. 1C), and so did the ems1-1 serk1-1 serk2-1 triple mutant (Fig. 1F).
Figure 1.
SERK1/2 genetically interacts with EMS1 in anther development. Semithin sections show one anther lobe at stage 5. A, A mature wild-type anther lobe showing epidermis (E), endothecium (En), middle layer (ML), tapetum (T), and microsporocytes (M). B and C, Anther lobes from the ems1-1 strong mutant (B) and the serk1-1 serk2-1 double mutant (C) showing no tapetum but excess microsporocytes. D, The ems1-2 weak mutant anther lobe showing normal tapetum and microspocytes, which is similar to the wild type (A). E and F, Anther lobes from ems1-2 serk1-1 double (E) and ems1-1 serk1-1 serk2-1 triple (F) mutants showing no tapetum but excess microsporocytes, which resemble the ems1-1 single mutant (B) and the serk1-1 serk2-1 double mutant (C). G, The AtML1:TPD1 anther lobe exhibiting six layers of anther wall cells (indicated by red asterisks), which is two layers more than that of the wild type (A). H, The AtML1:TPD1 serk1-1 serk2-1 anther lobe showing a similar structure to that of serk1-1 serk2-1 (C). Bars = 10 µm.
Ectopic expression of the EMS1 ligand TPD1 in anther epidermis using the AtML1 promoter (AtML1:TPD1) caused the formation of six somatic cell layers (Fig. 1G). However, this phenotype was absent in the serk1 serk2 anther (Fig. 1H), suggesting that SERK1/2 is required for the EMS1 signaling. Both SERK1:SERK1 and SERK2:SERK2 were capable of rescuing the fertility of ems1-2 serk1-1 plants, whereas SERK1:SERK1 had a slightly higher complementation efficiency than SERK2:SERK2 (Supplemental Fig. S3, A and B). Thus, our data agree with previous findings that SERK1 and SERK2 function redundantly in anther development (Albrecht et al., 2005; Colcombet et al., 2005).
Taken together, our genetic results support that SERK1/2 and EMS1 function in the same genetic pathway, suggesting that SERK1 and SERK2 act redundantly as partners of EMS1.
SERK1 Interacts with EMS1
The primary amino acid sequences of SERK1 and SERK2 share 90% identity (Albrecht et al., 2005) and are functionally redundant. Therefore, we took three approaches to examine a potential biochemical interaction between SERK1 and EMS1.
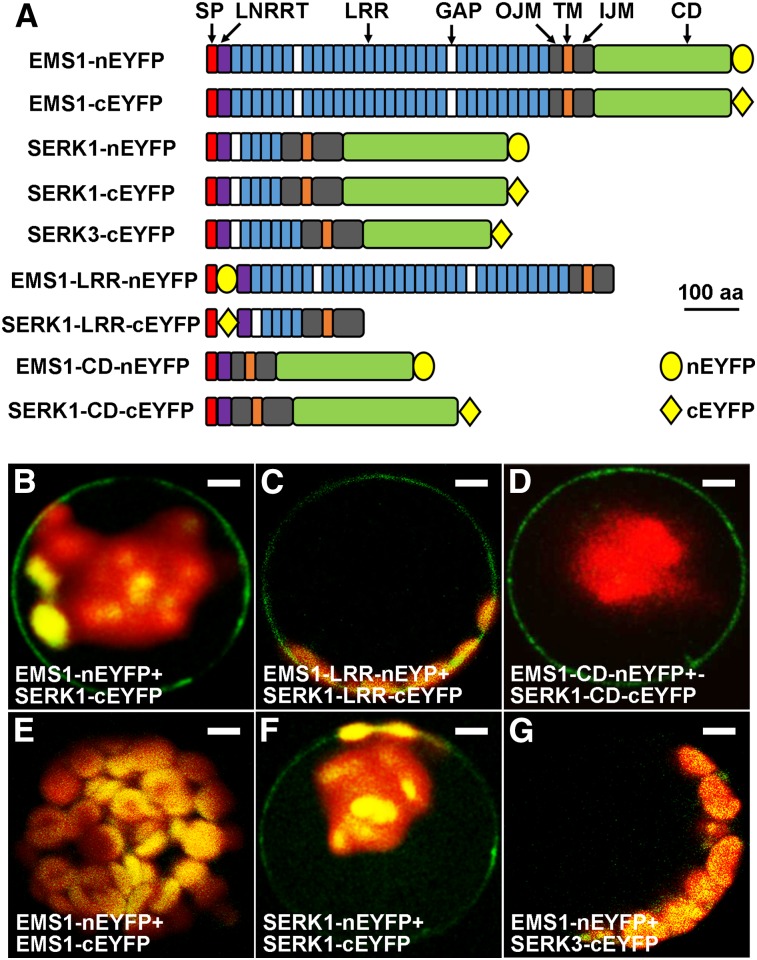
First, we performed BiFC assays in Arabidopsis protoplasts. Our results showed that the full-length EMS1 interacted with the full-length SERK1 at the plasma membrane (Fig. 2, A and B). We further found that SERK1-EMS1 interaction occurred in their LRR (Fig. 2, A and C) and cytoplasmic kinase domain (CD; Fig. 2, A and D), indicating that both the LRR extracellular domain and the CD are responsible for the SERK1-EMS1 interaction. Moreover, no EMS1-EMS1 interaction was detected (Fig. 2, A and E), while SERK1 interacted with itself at the plasma membrane (Fig. 2, A and F). In addition, our control results showed that EMS1 did not interact with SERK3 (BAK1) (Fig. 2, A and G), suggesting that SERK1 interacts specifically with EMS1.
Figure 2.
BiFC results showing that SERK1 interacts with EMS1 via LRR and kinase domains. A, Schematic diagrams showing EMS1, SERK1, and SERK3 (control) primary structures as well as truncated versions for BiFC assays. SP, Signal peptide; LRRNT, LRR-containing N terminus; GAP, non-LRR gap region between two LRRs; OJM, outer juxtamembrane domain; TM, transmembrane domain; IJM, inner juxtamembrane domain. All constructs contain SP, OJM, TM, and IJM domains. Yellow ovals represent the N-terminal EYFP (nEYFP), and yellow diamonds represent the C-terminal EYFP (cEYFP). The size bar (aa = amino acids) only indicates sizes of EMS1, SERK1, and SERK3 but not nEYFP or cEYFP. B, Confocal image showing that the full-length EMS1 interacts with the full-length SERK1 at the plasma membrane. C and D, Confocal images showing that EMS1 interacts with SERK1 at the plasma membrane via their LRR domains (C) and kinase domains (D). E, Confocal image showing no EMS1-EMS1 interaction. F, Confocal image showing that SERK1 interacts with itself at the plasma membrane. G, No interaction between EMS1 and SERK3 (control). Bars = 10 µm.
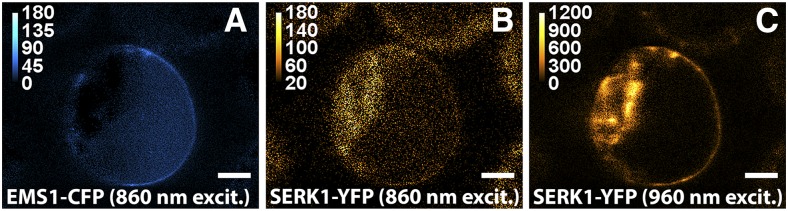
Second, we carried out FRET spectrometry experiments to quantify the SERK1-EMS1 interaction via a two-photon microspectroscope (Raicu et al., 2009; Biener et al., 2013). FRET efficiencies were calculated using images generated by unmixing pixel-level spectra of donors upon excitation at the 860-nm wavelength (EMS1-cyan fluorescent protein [CFP]; Fig. 3A) and acceptor (SERK1-yellow fluorescent protein [YFP]) upon excitation at the 860-nm (Fig. 3B) and 960-nm (Fig. 3C) wavelengths. A FRET efficiency of 24% ± 5% was obtained from 48 regions of interest at the plasma membrane from cells coexpressing EMS1-CFP and SERK1-YFP (Supplemental Table S1). Our FRET results indicated a strong interaction between SERK1 and EMS1.
Figure 3.
Typical fluorescence images used to probe interaction between SERK1 and EMS1 using FRET in vivo. FRET assays show fluorescence images of the donor (EMS1-CFP) excited only at the 860-nm wavelength (A) and the acceptor (SERK1-YFP) following excitation at the 860-nm (B) and the 960-nm (C) wavelengths. Signals were obtained from spectral unmixing of images in cells coexpressing EMS1-CFP and SERK1-YFP. Bars = 10 µm.
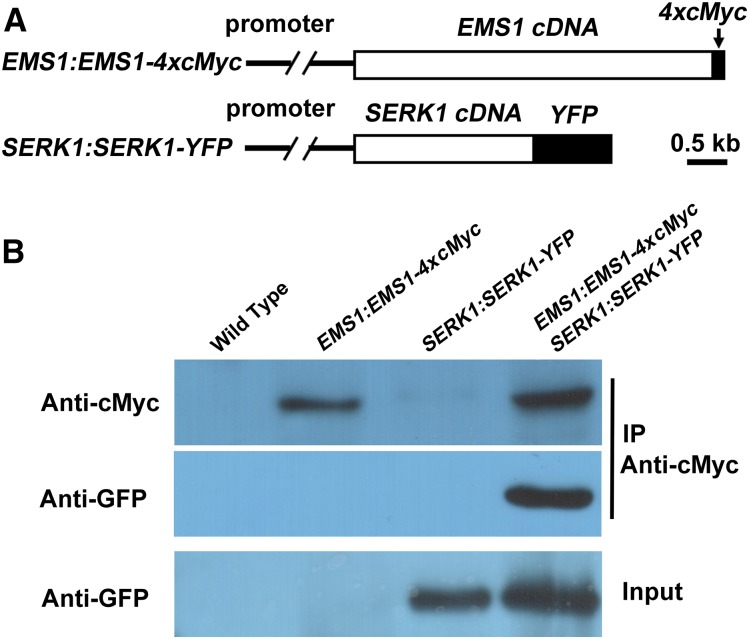
Third, we performed co-IP to investigate whether SERK1 interacts with EMS1 in planta. We generated EMS1:EMS1-4xcMyc SERK1:SERK1-YFP double transgenic plants that express the full-length EMS1 fused with four cMyc tags (EMS1-4xcMyc) and the full-length SERK1 fused with YFP (SERK1-YFP) using their native promoters (Fig. 4A). Membrane proteins extracted from young buds of double transgenic and control plants (wild-type and single transgenic plants) were immunoprecipitated with an anti-cMyc antibody. The precipitated proteins were then analyzed by western blot with anti-cMyc and anti-GFP antibodies. Our result showed that the SERK1-YFP protein was coimmunoprecipitated by EMS1-4xcMyc (Fig. 4B), suggesting that SERK1 interacts with EMS1 in planta.
Figure 4.
co-IP results showing that SERK1 interacts with EMS1 in planta. A, Schematic diagrams showing construct structures of EMS1:EMS1-4xcMyc and SERK1:SERK1-YFP. B, SERK1 interacts with EMS1 in planta in a co-IP assay. Compared with wild-type, EMS1:EMS1-4xcMyc, and SERK1:SERK1-YFP single transgenic plants, both EMS1-4xcMyc and SERK1-YFP were detected using proteins from young buds of EMS1:EMS1-4xcMyc SERK1:SERK1-YFP double transgenic plants when immunoprecipitated (IP) by the anti-cMyc antibody.
Collectively, our results suggest that SERK1 interacts biochemically with EMS1.
Transphosphorylation of EMS1 by SERK1 Is Essential for Enhancing EMS1 Kinase Activity
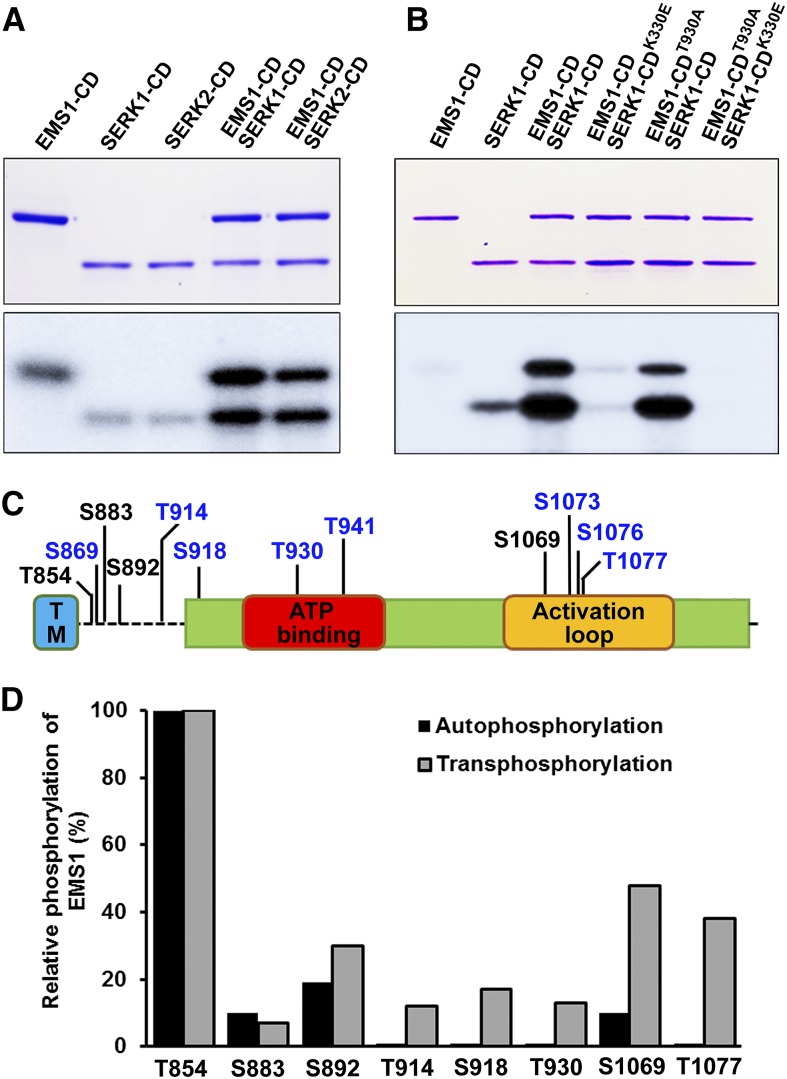
Previous studies have demonstrated that EMS1 and SERK1/2 encode active protein kinases with autophosphorylation activity (Shah et al., 2001b; Zhao et al., 2002; Karlova et al., 2009). Our results showed that autophosphorylation activities were detected in the EMS1-CD, SERK1-CD, and SERK2-CD (Fig. 5A). Phosphorylation levels of EMS1-CD, SERK1-CD, and SERK2-CD were enhanced significantly when EMS1-CD was incubated with SERK1-CD or SERK2-CD (Fig. 5A), suggesting that SERK1/2 and EMS1 strongly transphosphorylate each other.
Figure 5.
In vitro transphosphorylation activities between EMS1 and SERK1/2. A and B, In vitro kinase assays were performed using EMS1-CD, SERK1-CD, and SERK2-CD in the presence of [γ-32P]ATP. Top gels, Input proteins stained with Coomassie Brilliant Blue. Bottom gels, Phosphorylation changes analyzed by autoradiography. EMS1-CDT930A and SERK1-CDK330E are inactive forms of EMS1 and SERK1 kinases, respectively. Consistent results were obtained from three independent repeats. C, Identified in vitro autophosphorylation (in black) and transphosphorylation (in blue) sites in the EMS1-CD via mass spectrometry. S, Ser; T, Thr. D, Relative phosphorylation level changes of specific residues in autophosphorylated and transphosphorylated EMS1-CD. Relative phosphorylation was calculated based on the ratio of spectral counts for total versus phosphorylated peptides identified by mass spectrometry analysis.
A previous study found that SERK1-CDK330E has no kinase activity (Shah et al., 2001b). When the active form of EMS1-CD was incubated with SERK1-CDK330E, almost no phosphorylation was detected in either EMS1-CD or SERK1-CDK330E (Fig. 5B). We further generated a dead EMS1-CD version, EMS1-CDT930A (details shown in Fig. 6). Interestingly, after coincubation, the phosphorylation levels were still enhanced in both EMS1-CDT930A and SERK1-CD, while, as expected, no phosphorylation was found in either EMS1-CDT930A or SERK1-CDK330E when EMS1-CDT930A and SERK1-CDK330E were coincubated (Fig. 5B). Thus, these results suggest that the SERK1-EMS1 interaction leads to transphosphorylation between SERK1/2 and EMS1. SERK1 is essential for enhancing EMS1 kinase phosphorylation activity. The autophosphorylation activity of EMS1 is not important for EMS1-SERK1 transphosphorylation in vitro.
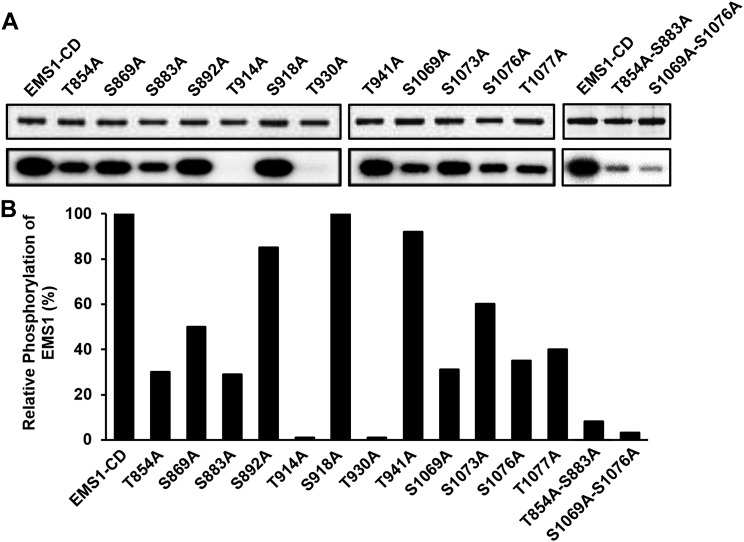
Figure 6.
Effects of mutations in identified Ser and Thr residues on EMS1 autophosphorylation activities. A, Top gels, Coomassie Brilliant Blue staining showing equal amounts of the examined proteins. Bottom gels, Autoradiography showing EMS1 autophosphorylation activities. Consistent results were obtained from three independent repeats. B, Level changes of mutated EMS1 autophosphorylation activities. Relative phosphorylation activity for each mutated EMS1-CD was quantified based on comparison with the native form of EMS1-CD, set as 100%.
EMS1 Is Transphosphorylated by SERK1 on Multiple Ser and Thr Residues
To demonstrate how SERK1 transphosphorylates EMS1, we identified in vitro autophosphorylation and transphosphorylation sites in EMS1-CD by tandem mass spectrometry. A total of four autophosphorylation sites (Thr-854, Ser-883, Ser-892, and Ser-1069) were identified from the autophosphorylated EMS1-CD (Fig. 5C; Supplemental Fig. S4; Supplemental Table S2). Besides these four autophosphorylation sites, we also identified eight transphosphorylated sites (Thr-914, Thr-930, Thr-941, and Thr-1077 as well as Ser-869, Ser-918, Ser-1073, and Ser-1076) from the SERK1-CD phosphorylated EMS1-CD (Fig. 5C; Supplemental Fig. S4; Supplemental Table S2). Among the 12 identified in vitro phosphorylation sites, five of them (Thr-854, Ser-869, Ser-883, Ser-892, and Thr-914) are located in the EMS1 inner juxtamembrane domain (Fig. 5C). Seven phosphorylation sites (Ser-918, Thr-930, Thr-941, Ser-1069, Ser-1073, Ser-1076, and Thr-1077) were found in the kinase domain, wherein Thr-930 and Thr-941 are in the ATP-binding domain and Ser-1069, Ser-1073, Ser-1076, and Thr-1077 are in the activation loop region (Fig. 5C).
Among four autophosphorylation sites, quantitative mass spectrometry analysis revealed that relative phosphorylation rates of Ser-892 and Ser-1069 were increased by SERK1-CD transphosphorylation, while phosphorylation rates of Thr-854 and Ser-883 remained unchanged (Fig. 5D). Together, our results suggest that transphosphorylation and the enhancement of autophosphorylated sites by SERK1 cause the increase of EMS1 phosphorylation level and possibly EMS1 kinase activity.
Mutations of Identified Phosphorylation Sites Affect EMS1 Kinase Activity
To test whether the identified Ser and Thr phosphorylation sites affect EMS1 kinase activity, we performed site-directed mutagenesis of these Ser or Thr residues to Ala and then assessed their autophosphorylation changes in vitro. Our results showed that EMS1-CDT914A and EMS1-CDT930A lost kinase activity, whereas kinase activities of EMS1-CDT854A, EMS1-CDS883A, EMS1-CDS1069A, EMS1-CDS1076A, and EMS1-CDT1077A were decreased significantly (Fig. 6). EMS1-CDT854A-S883A and EMS1-CDS1069A-S1076A, which carry double mutations, had further decreased kinase activities compared with their corresponding single mutants (Fig. 6).
Both Thr-854 and Ser-883 sites are located in the inner juxtamembrane domain, suggesting that phosphorylation of these two residues in the inner juxtamembrane domain is important for maintaining kinase activity in vitro. In addition, substitutions of Ser-1069, Ser-1076, or Thr-1077 with Ala in the activation loop domain resulted in a great decrease in autophosphorylation level compared with the wild type. Moreover, S1069A and S1076A double mutations nearly abolished EMS1 kinase activity, providing further evidence to support that autophosphorylation in the activation loop is required for general kinase activity.
Phosphorylation Is Required for EMS1 Biological Function
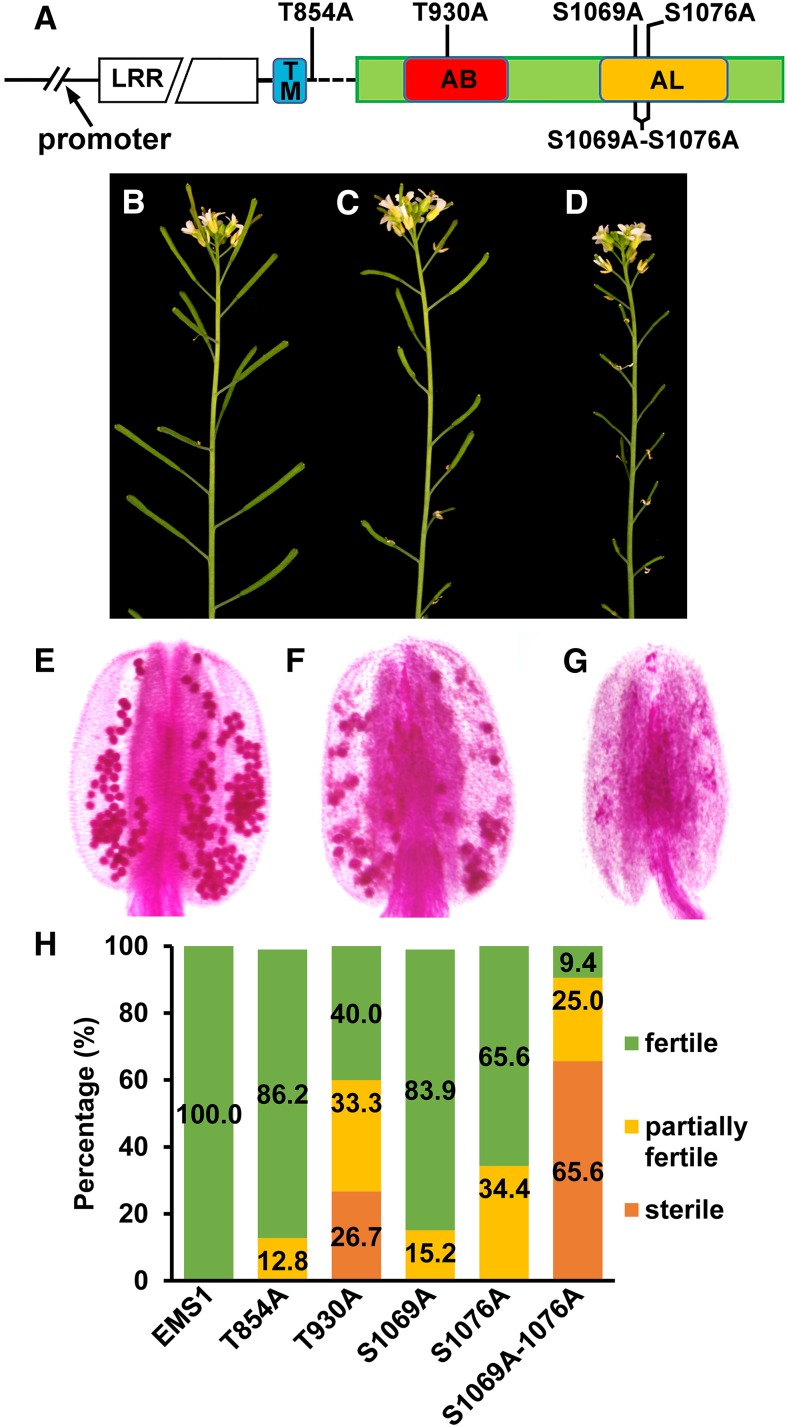
Phosphorylation of specific Ser and Thr residues in LRR-RLK is essential for kinase activation and the recognition of downstream signaling components. To assess the functional significance of identified in vitro phosphorylation sites in EMS1, we representatively chose four phosphorylation residues (T854A, in the inner juxtamembrane domain; T930A, in the ATP-binding domain; and S1069A and S1076A, in the activation loop region) and then tested the ability of five versions of EMS1 mutations to rescue the ems1-1 fertility (Fig. 7A). In each complementation assay, 40 independent transgenic lines were analyzed (Supplemental Fig. S6). The complementation experiments resulted in three kinds of phenotypes in T2 plants: wild-type-like plants with fully recovered fertility (Fig. 7B), partially fertile plants with some short siliques (Fig. 7C), and completely sterile plants indicated by the production of all short siliques (Fig. 7D). Examination of the pollen viability showed viable pollen grains in anthers from wild-type-like plants (Fig. 7E). Reduced pollen grains were found in anthers from partially sterile plants (Fig. 7F), while no pollen was observed in anther from sterile plants (Fig. 7G). Our results showed that EMS1:EMS1T854A and EMS1:EMS1S1069A, which harbor a single mutation in the juxtamembrane region and the activation loop, respectively, almost completely rescued the ems1-1 phenotype (Fig. 7H). Although EMS1:EMS1T930A and EMS1:EMS1S1076A partially complemented the ems1-1 phenotype, the complementation rate of EMS1:EMS1T930A was lower than that of EMS1:EMS1S1076A. Thr-930 is in the ATP-binding domain and Ser-1076 is in the activation loop (Fig. 5C). The T930A mutation abolished, while the S1076A mutation decreased, EMS1 kinase in vitro autophosphorylation activity, which might explain the functional difference between Thr-930 and Ser-1076. Moreover, EMS1:EMS1S1069A- S1076A failed to rescue the ems1-1 phenotype, further supporting that the normal phosphorylation of EMS1 is critical for its biological function in anther development.
Figure 7.
Functional analysis of EMS1 in vitro phosphorylation sites in planta. A, Schematic diagram showing the EMS1 gene structure and locations of mutated in vitro phosphorylation residues. TM, Transmembrane domain; AB, ATP-binding domain; AL, activation loop. B to D, Representative phenotypes found in T2 transgenic plants in the ems1-1 mutant background: fully fertile wild-type-like plants (B), partially sterile plants (C), and completely sterile plants (D). E to G, Pollen staining results showing viable pollen grains from wild-type-like plants (E), reduced viable pollen grains from partially sterile plants (F), and no pollen from completely sterile plants (G). H, Complementation effects of five mutated EMS1 versions in the ems1-1 mutant background. Values are means of 40 independent lines grown under the same condition.
DISCUSSION
In this study, we provide several lines of evidence to support that SERK1/2 serves as a partner of EMS1 for anther development. In Arabidopsis, SERKs with five members in the LRR-RLK II subfamily play vital roles in growth, development, and defense via acting as coreceptors/partners of other LRR-RLKs (Shiu et al., 2004; Ma et al., 2016). The BRI1 receptor and its redundant coreceptors SERK1, SEKR3 (BAK1), and SERK4 control brassinosteroid signaling (Li et al., 2002; Nam and Li, 2002; Hothorn et al., 2011; She et al., 2011; Gou et al., 2012; Santiago et al., 2013). Both ER/ERLs and SERK1/SERK2/SERK3/SERK4 are required for stomatal patterning (Shpak et al., 2005; Meng et al., 2015). EPF (EPIDERMAL PATTERNING FACTOR) small proteins trigger the interaction of ER/ERL with SERK1 to SERK4, while the TOO MANY MOUTHS-SERK association is independent on EPFs (Lee et al., 2015; Meng et al., 2015). Loss-of-function mutants of SERK1 to SERK4 genes exhibit the same phenotype in floral organ abscission as that of hae hsl2 and ida (inflorescence deficient in abscission) mutants (Butenko et al., 2003; Cho et al., 2008; Meng et al., 2016). IDA is perceived by the HAE ectodomain, which may trigger the heterodimerization between HAE and SERKs (Meng et al., 2016; Santiago et al., 2016). PSK binding prompts the heterodimerization between PSKR1 and SERK1/SERK2/SERK3 (Wang et al., 2015). In the innate immune response, SERKs also play a role as coreceptors of LRR-RLKs involved in pattern recognition receptor signaling. For example, the 22-residue bacterial flagellin oligopeptide (flg22) elicits assembly of the active FLS2-BAK1 (receptor-coreceptor) signaling complex to mediate plant defense (Chinchilla et al., 2007; Heese et al., 2007; Roux et al., 2011; Sun et al., 2013). In addition, EF-Tu (elf18) elicits the formation of EFR-BAK1 heterodimer (Roux et al., 2011). Our results suggest that SERK1 and SERK2 may function redundantly as coreceptors of EMS1 during anther development in Arabidopsis.
Studies on crystal structures have revealed the molecular mechanisms underlying the assembly of LRR-RLK receptor-coreceptor complexes in the presence of their ligands. The binding of brassinolide (the most active brassinosteroid) to the BRI1 island domain induces BRI1-SERK1 interface formation, which enables a tight BRI1-SERK1 interaction and, consequently, activation of the BRI1-SERK1 complex (Hothorn et al., 2011; She et al., 2011; Santiago et al., 2013). The heterodimerization between FLS2 and BAK1 is triggered by flg22 (Sun et al., 2013). flg22 binds to the concave surface of the FLS2 LRR domain across 14 LRRs (LRR3–LRR16) via both C- and N-terminal segments of flg22. Two interfaces are formed between FLS2 and BAK1 LRRs. The direct interface may explain the flg22-independent formation of the FLS2-BAK1 complex, while the indirect interface that is induced by flg22 binding may contribute to the full activation of the FLS2-BAK1 complex.
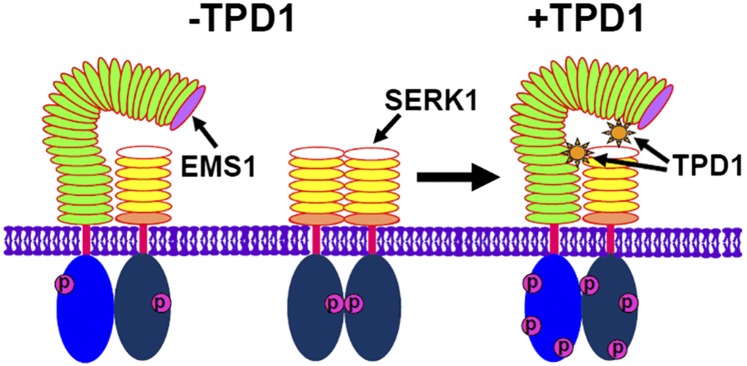
Both EMS1 and BRI1 are in the same subfamily of LRR, Xb (Shiu et al., 2004). Using the RaptorX program (http://raptorx.uchicago.edu/; Källberg et al., 2014), we simulated the EMS1 LRR three-dimensional structure based on the BRI1 crystal structure. The EMS1 LRR structure exhibits a twisted right-handed superhelix, where LRR2 creates the small loop and LRR23 to LRR25 generate the large loop (Supplemental Fig. S5). Our previous studies show that TPD1 is a small Cys-rich protein ligand of EMS1 (Jia et al., 2008; Huang et al., 2016b, 2016c). We identified two TPD1-binding sites in the EMS1 LRR ectodomain. TPD1 binds to the first three EMS1 LRRs (Huang et al., 2016c). The K(104)N mutation in LRR2 impairs the biological function of EMS1 (Canales et al., 2002) and the TPD1-EMS1 interaction. Moreover, TPD1 may interact with EMS1 in the vicinity of the large loop (Huang et al., 2016c). Our BiFC results demonstrate that EMS1 can interact with SERK1 at the plasma membrane in the absence of TPD1, suggesting that the TPD1 ligand is not required for the preassembly of the EMS1-SERK1 heterodimer. Formation of the FLS2-BAK1 complex is flg22 independent (Sun et al., 2013), whereas the association of BRI1 and BAK1 requires brassinolide (Hothorn et al., 2011; She et al., 2011; Santiago et al., 2013). No EMS1-EMS1 interaction was detected in this study (Fig. 2, A and E); however, previous studies (Shah et al., 2001a; Russinova et al., 2004; Hink et al., 2008) and our results (Fig. 2, A and F) show that SERK1 forms a homodimer at the plasma membrane. Therefore, we hypothesize that, without TPD1, EMS1 and SERK1 remain as a relaxed EMS1-SERK1 heterodimer and a SERK1-SERK1 homodimer at the plasma membrane, respectively (Fig. 8). In the presence of TPD1, consecutive binding of two TPD1 molecules induces the assembly of an active and stable SERK1-EMS1 heterodimer. More specifically, TPD1 first binds to the EMS1 LRR2-constituted domain, which causes the conformational rearrangement of the interface between EMS1 and SERK1. EMS1 then becomes more competent for an additional binding of TPD1, which reinforces/activates the EMS1-SERK1 complex. The EMS1 kinase activity was greatly enhanced by SERK1 via transphosphorylation. Transphosphorylation still occurs between SERK1-CD and EMS1-CDT930A, which has no detectable autophosphorylation. In contrast, SERK1-CDK330E, whose autophosphorylation activity is abolished (Karlova et al., 2009), was unable to transphosphorylate EMS1-CD, indicating that the transphosphorylation of EMS1 by its partner SERK1 plays a vital role in activating the EMS1-SERK1 signaling complex.
Figure 8.
Hypothetical model for the assembly of the active EMS1-SERK1/2 complex at the plasma membrane. Without the ligand TPD1, EMS1 and SERK1 remain as relaxed EMS1-SERK1 heterodimers and SERK1-SERK1 homodimers at the plasma membrane. Two TPD1 molecules consecutively bind to EMS1, inducing the assembly of an active SERK1-EMS1 heterodimer.
Studies on BRI1 and SERK3 (BAK1) LRR-RLKs showed that the phosphorylation sites identified by in vitro analyses have an excellent correlation to the in vivo phosphorylation sites (Wang et al., 2005, 2008). Furthermore, among 24 in vitro-identified phosphorylation sites in SERK1, only three sites are due to bacterial transphosphorylation (Karlova et al., 2009). A recent study reported the identification of in vitro autophosphorylation sites in 73 LRR-RLKs, including SERK1 but not EMS1 (Mitra et al., 2015). In this study, we identified five in vitro Ser and Thr phosphorylation sites in the EMS1 inner juxtamembrane domain and six in vitro Ser and Thr phosphorylation sites in the EMS1 ATP-binding domain and activation loop. Phosphorylation occurs in both conserved and nonconserved residues in LRR-RLKs (Mitra et al., 2015). Similarly, alignment of the EMS1-CD with its most related LRR-RLKs (BRI1, BRL1, BRL2, and BRL3) shows that only the Ser-892, Thr-914, and Ser-1076 phosphorylation sites are conserved, but nine other phosphorylation sites are diverse (Supplemental Fig. S7; Wang et al., 2005, 2008; Mitra et al., 2015). Our studies showed that the S1076A mutation reduced 65% of EMS1 autophosphorylation activity. Although EMS1S1076A was capable of partially rescuing the ems1-1 fertility, EMS1S1069A- S1076A, which carries mutations of two phosphorylation residues in the activation loop, almost failed to complement the ems1-1 phenotype. The mutations of BRI1 S858A, T880A, and S1042A, which are equivalent to EMS1 S892A, T914A, and S1076A, do not affect BRI1 autophosphorylation activity, but their abilities to phosphorylate the peptide substrate were decreased. BRI1S858A and BRI1T880A are able to rescue the bri1-5 mutant phenotype. Consistent with our findings, BRI1S1042A just partially complements the bri1-5 mutant phenotype, which might be due to the S1042A mutation having a strong effect on substrate phosphorylation. In addition, the mutation S1044/5A in the BRI1 activation loop results in loss of function in BRI1 (Wang et al., 2005), as does the mutation T455A in the BAK1 (SERK3) activation loop (Wang et al., 2008). Furthermore, HAES856A (in the activation loop), which has an abolished autophosphorylation activity, is capable of partially rescuing the hae hsl2 abscission-deficient mutant (Taylor et al., 2016). Although the phosphorylation mimic HAES856D-S861D maintains some autophosphorylation activity, it fails to complement the hae hsl2 phenotype. HAES856D-S861D has an enhanced Tyr autophosphorylation activity, which might affect the HAE biological function. BRI1 Thr-1049 in the activation loop is required for BRI1 autophosphorylation and transphosphorylation activities as well as its in planta function (Wang et al., 2005). Thr-1049 is conserved; however, we did not find that the EMS1 equivalent residue Thr-1082 could be phosphorylated. It would be interesting to characterize the effect of the T1082A mutation on EMS1 kinase activity and in vivo function. Although Thr-705 in the rice XA21 LRR-RLK juxtamembrane domain is required for XA21 autophosphorylation and XA21-mediated innate immunity, mutation of its equivalent residue Thr-880 in BRI1 has no effect on BRI1 autophosphorylation and function (Wang et al., 2005; Chen et al., 2010). Collectively, our results support previous findings that conserved phosphorylation residues in the juxtamembrane domain and activation loop are important for kinase activity and function.
The nonconserved phosphorylation residues also are essential for kinase activity and function. The single mutations T854A, S869A, and S883A in the EMS1 juxtamembrane domain decreased significantly, while the T854A-S883A double mutation further reduced EMS1 kinase activity. Conversely, The T872A mutation in the BRI1 juxtamembrane domain increases BRI1 kinase activity (Wang et al., 2005). BRI1S981A (in the ATP-binding domain), which has a normal kinase activity, functions as the wild-type BRI1 (Wang et al., 2005; Oh et al., 2012). However, the phosphorylation-mimic mutation in Ser-891 deactivates BRI1 phosphorylation and impairs BRI1 biological function (Oh et al., 2012). The S938A mutation in FLS2 abolishes immune responses (Cao et al., 2013). The phosphorylation-mimic mutation S938D or S938E has no effect on the immune responses, but it does not lead to constitutive activation of FLS2 signaling without the flg22 ligand either. Functional studies showed that EMS1T930A partially lost its function in complementing the ems1-1 mutant phenotype. This is consistent with our in vitro data that, although EMS1-CDT930A completely lost its in vitro autophosphorylation activity, it still could be transphosphorylated by SERK1. Thus, our results further suggest that the transphosphorylation of EMS1 by SERK1/2 is critical for transmitting downstream signaling. Collectively, we identified EMS1 conserved and diverse in vitro autophosphorylation and transphosphorylation sites, which paves the way for studying the functions of individual phosphorylation residues in activating the EMS1-SERK1 signaling complex and in establishing the substrate specificity in the future.
Cell signaling has been studied extensively in the context of many physiological and developmental processes, but little is known about how signal transduction pathways regulate somatic and reproductive cell differentiation in plants (Zhao et al., 2002; Jia et al., 2008; Zhao, 2009; Feng et al., 2013; Huang et al., 2016c; Walbot and Egger, 2016). So far, most identified ligands for functionally studied LRR-RLKs are small molecules and oligopeptides (Fletcher et al., 1999; Wang et al., 2001; Matsubayashi et al., 2002; Tang et al., 2002; Caño-Delgado et al., 2004; Kinoshita et al., 2005; Ito et al., 2006; Kondo et al., 2006; Stenvik et al., 2008; Shimada et al., 2011; Lee et al., 2012; Torii, 2012; Butenko et al., 2014; Huang et al., 2014; Matsubayashi, 2014). Although using similar coreceptors/partners, BRI1-SERK1, FLS2-BAK1, and EMS1-SERK1 are activated by different types of ligands. Future studies on the EMS1-TPD1-SERK1 structure will not only provide insight into the molecular mechanisms underlying the assembly and activation of LRR-RLK signaling complexes induced by a small protein ligand but also will fundamentally advance our understanding of somatic and reproductive cell fate determination during plant sexual reproduction.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
All Arabidopsis (Arabidopsis thaliana) plants were grown at 22°C under a photoperiod of 16 h of light/8 h of dark. The wild-type plants used are Arabidopsis ecotypes Columbia and Landsberg erecta. All T-DNA insertion lines, including SALK_044330 (serk1-1), SALK_058020 (serk2-1), and SALK_051989 (ems1-2), were obtained from the Arabidopsis Biological Resource Center and genotyped by PCR. The previously identified strong ems1 mutant was renamed as ems1-1 (Zhao et al., 2002). Primers for genotyping are listed in Supplemental Table S3.
Pollen Staining and Anther Semithin Sectioning
Alexander pollen staining and anther semithin sectioning were conducted as described in our previous studies (Huang et al., 2016a, 2016c).
BiFC Assay
The EMS1-nEYFP construct was generated by inserting the N-terminal enhanced YFP (nEYFP) after the full-length EMS1 cDNA without the stop codon in pSAT1-nEYFP-C1-B (Tzfira et al., 2005; Huang et al., 2016c). The SERK1-cEYFP construct was made by introducing the C-terminal EYFP (cEYFP) after the full-length SERK1 cDNA without the stop codon in pSAT1-cEYFP-C1-B. Constructs EMS1-cEYFP, SERK1-nEYFP, SERK3-cEYFP, EMS1-CD-nEYFP, and SERK1-CD-cEYFP were created similarly. To generate EMS1-LRR-nEYFP and SERK1-LRR-cEYFP constructs, nEYFP and cEYFP were inserted after EMS1 and SERK1 signal peptides followed by their LRR, outer juxtamembrane, transmembrane, and inner juxtamembrane domains. All the constructs contain signal peptide, outer juxtamembrane, transmembrane, and inner juxtamembrane domains. Primers for generating all constructs and more details about constructs are included in Supplemental Tables S3 and S4.
For high-quality plasmid preparation, the PureYield Plasmid Midiprep System (Promega) was used to isolate plasmids. A pair of tested constructs was used to cotransfect Arabidopsis protoplasts that were prepared from 4-week-old leaves (Yoo et al., 2007). Constructs with half EYFP and SERK3 were used as negative controls. Samples were observed after 16 h with a Leica TCS SP2 laser scanning confocal microscope using a 63×/1.4 water-immersion objective lens.
FRET Assay
To generate EMS1-CFP and SERK1-YFP constructs, full-length EMS1 and SERK1 cDNAs without stop codons were fused with CFP and YFP, respectively (Supplemental Tables S3 and S4). The resulting EMS1-CFP and SERK1-YFP plasmids were cotransfected into Arabidopsis mesophyll protoplasts. Protoplasts transfected by single plasmids were used as controls. Acquisition of spectrally resolved fluorescence images, spectral unmixing of donor, acceptor, and autofluorescence signals, as well as determination of FRET efficiency were performed as described previously (Raicu and Singh, 2013; King et al., 2016). Briefly, cells coexpressing EMS1-CFP and SERK1-YFP (controls: cells expressing only EMS1-CFP or SERK1-YFP) were imaged using an optical microspectroscope (OptiMiS) with the line-scan excitation powered by a femtosecond laser emitting near-infrared light pulses with tunable emission wavelength between 780 and 1,040 nm (Raicu et al., 2009; Biener et al., 2013). The spectrally resolved images were then unmixed to generate two-dimensional fluorescence maps of donor and acceptor (Raicu and Singh, 2013). The elementary donor (CFP) and acceptor (YFP) spectra as well as the autofluorescence spectra were obtained from measurements of cells that expressed only EMS1-CFP, only SERK1-YFP, and that were devoid of any fluorescent protein, respectively. The unmixing protocol has been optimized for cells presenting substantial autofluorescence emission with the final goal to determine donor and acceptor emission images free from any autofluorescence (Mannan et al., 2013). Since the laser wavelength of 860 nm (selected as optimal for donor excitation) also caused slight excitation of the acceptor, we did not attempt to compute the FRET efficiency for every image pixel from the donor and acceptor fluorescence maps (Raicu and Singh, 2013), or else the FRET efficiency map would have been contaminated by such spurious excitation. Instead, we used a method introduced recently, which employed two excitation wavelengths (860 and 960 nm) to account for the direct excitation of the acceptor and compute the average FRET efficiency for regions of interest (at the plasma membrane; Stoneman et al., 2016). In order to reduce noise and avoid singularities at pixels with zero intensity values, a signal-to-noise threshold of 1.5 sd was applied to FRET efficiency calculations.
co-IP Assay
To generate EMS1:EMS1-4xcMyc SERK1:SERK1-YFP double transgenic plants, plants harboring EMS1:EMS1-4xcMyc were crossed with SERK1:SERK1-YFP plants. SERK1:SERK1-YFP and EMS1:EMS1-4xcMyc complement the serk1-1 serk2-1 and ems1-1 mutant phenotypes, respectively (Albrecht et al., 2005; Jia et al., 2008). Young inflorescences were harvested from wild-type, single transgenic, and double transgenic plants. Membrane protein preparation and co-IP were performed as described in our previous study (Jia et al., 2008). Briefly, about 0.5 g of young buds was ground in liquid nitrogen and extracted with extracting buffer (50 mm Tris, pH 8.8, 10 mm EDTA, 150 mm NaCl, 10% glycerol [v/v], 20 mm NaF, 1 mm Na2MoO4, 0.1 mm Na3VO4, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitor cocktail [Roche]). Crude proteins were centrifuged two times at 5,000g for 5 min at 4°C. Then, the supernatants were centrifuged at 90,000 rpm for 40 min at 4°C. The pellets were suspended in membrane solubilization buffer (100 mm Tris-HCl, pH 7.3, 150 mm NaCl, 1 mm EDTA, 10% glycerol, 20 mm NaF, 1% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitor cocktail) with sonication. After further centrifugation at 90,000 rpm for 20 min at 4°C, the protein extracts in supernatant were then incubated with 30 μL of Myc-Trap beads (ChromoTek) at 4°C for 2 h. Finally, after washing five times with membrane solubilization buffer, membrane proteins were eluted with 2× SDS-PAGE loading buffer and analyzed by western blotting using anti-GFP (GeneScript) and anti-cMyc (Roche) antibodies.
In Vitro Kinase Assay
To express proteins for in vitro kinase assay, the cytoplasmic domain of EMS1 (amino acids 849–1,192) was amplified by PCR and inserted in frame with a glutathione S-transferase (GST) tag into pGEX-4T-2 (Promega). Site-direct mutated versions of EMS1 and SERK1 were generated via overlapping PCR. The cytoplasmic domains of SERK1 (amino acids 270–625) and SERK2 (amino acids 281–628) were PCR amplified and inserted in frame with a His tag into the vector pET-28a (GE Healthcare). Primers for generating all constructs and more details about constructs are included in Supplemental Tables S3 and S4.
The resulting vectors were transformed into Escherichia coli BL21 Star (DE3) cells (Invitrogen). Recombinant proteins were induced with 1 mm Isopropyl-β-D-thiogalactopyranoside for GST fusions and 0.5 mm Isopropyl-β-D-thiogalactopyranoside for His fusions at 16°C for 16 h. GST fusion proteins were purified by Glutathione Sepharose 4B (GE Healthcare), while recombinant His-tagged fusion proteins were purified by His Mag Sepharose Ni (GE Healthcare).
In vitro kinase assays were performed as described in our previous studies with slight modifications (Zhao et al., 2002). Briefly, 1 μg of purified proteins was incubated at 25°C for 30 min in the kinase buffer that contained 50 mm HEPES (pH 7.5), 10 mm MgCl2, 10 mm MnCl2, 1 mm dithiothreitol, 100 μm ATP, and 0.5 μL of [γ-32P]ATP (Perkin-Elmer Life Sciences) in a total volume of 20 μL. Reactions were terminated by adding 2× SDS-PAGE loading buffer and boiling for 10 min at 95°C. For transphosphorylation assay, MnCl2 was reduced to 2 mm in the kinase buffer. Autophosphorylation and transphosphorylation reactions were separated on a 12% SDS-PAGE gel, and the gel was stained with Coomassie Brilliant Blue. The gel was finally scanned on the Amersham Storm 860 imaging system (Amersham Biosciences) for filmless autoradiography imaging capture. The relative phosphorylation level was calculated by ImageJ (https://imagej.nih.gov/ij/).
Identification of in Vitro Phosphorylation Sites in EMS1-CD by Mass Spectrometry
For tandem mass spectrometer analysis, the purified recombinant EMS1-CD protein was autophosphorylated or transphosphorylated by SERK1-CD in vitro in the presence of ATP. After SDS-PAGE, protein bands were excised from PAGE gels. In-gel digestion was then performed using trypsin. As described in our previous studies (Ahsan et al., 2013), samples were analyzed in data-dependent tandem mass spectrometry mode on an Easy-nanoLC-1000 device coupled with Orbitrap Elite and LTQ Orbitrap XL ETD mass spectrometers (Thermo Scientific).
Complementation Analysis
To study the functional redundancy of SERK1 and SEKR2, 2-kb promoters and full-length cDNAs of SERK1 and SERK2 were PCR amplified (Supplemental Tables S3 and S4) and cloned into the pENTR-TOPO vector, respectively (Invitrogen). The resulting constructs SERK1:SERK1 and SERK2:SERK2 were subcloned into the pEarleyGate-301 vector via recombination reaction (Invitrogen). After confirmation by restriction digestion and sequencing, two plasmids were introduced into the Agrobacterium tumefaciens strain GV3101. Strains harboring SERK1:SERK1 and SERK2:SERK2 were used to transform ems1-2 serk1-1+/− plants using the floral dip method (Clough and Bent, 1998). The transformants were screened with 1% Basta (PlantMedia).
To test the functional significance of identified phosphorylation sites in EMS1, site mutations (T854A, T930A, S1069A, S1076A, and S1069A-S1076A) were generated by a series of overlapping PCR amplifications (Supplemental Tables S3 and S4) in the EMS1 gene that contains its 1,740-bp promoter and full-length coding region. After sequencing verification, all constructs (EMS1:EMS1T854A, EMS1:EMS1T930A, EMS1:EMS1S1069A, EMS1:EMS S1076A, and EMS1:EMS1S1069-1076A) and the control construct EMS1:EMS1 were subcloned into the binary vector pGWB1, and the resulting plasmids were introduced into the A. tumefaciens strain GV3101. The ems1-1+/− plants were transformed using the floral dip method (Clough and Bent, 1998).
T1 seedlings were screened on one-half-strength Murashige and Skoog medium with 50 mg mL−1 kanamycin and 50 mg mL−1 hygromycin. In the T1 generation, transgenic plants in the wild-type background were removed and transgenic plants in the ems1-1+/− background were kept after PCR genotyping (Zhao et al., 2002; Supplemental Fig. S6; Supplemental Table S3). In the T2 generation, the first PCR was performed to detect the Ds insertion with primers OMCP489 and OMC500, the second PCR was carried out with primers OMCP499 and OMC500 to test whether plants were in the ems1-1−/− background (homozygous for Ds transposable element), and the third PCR was performed with primers ZP1034, ZP1040, and ZP1606 to further verify whether plants contain the transgene. Only transgenic plants in the ems1-1 mutant background were chosen for fertility and anther staining analysis.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Identification of the ems1-2 weak allele.
Supplemental Figure S2. Fertilities of wild-type, ems1-1, ems1-2, serk1-1, serk2-1, serk1-1 serk2-1, ems1-2 serk1-1, ems1-2 serk2-1, and ems1-1 serk1-1 serk2-1 plants.
Supplemental Figure S3. Functional redundancy of SERK1 and SERK2 in rescuing the ems1-2 serk1-1 male-sterile phenotype.
Supplemental Figure S4. Representative tandem mass spectrometry spectra for identifying in vitro autophosphorylation and transphosphorylation sites in the EMS1 kinase domain.
Supplemental Figure S5. Simulated structure of the EMS1 LRR domain.
Supplemental Figure S6. Genotyping of EMS1:EMS1 transgenic plants in the ems1-1 mutant background.
Supplemental Figure S7. Alignment of the EMS1 cytoplasmic domain with its four most closely related LRR-RLKs (BRI1, BRL1, BRL2, and BRL3) as well as FLS2 and HAE.
Supplemental Table S1. Calculation of FRET efficiency between SERK1 and EMS1 using a two-excitation-wavelength scheme.
Supplemental Table S2. Summary of in vitro autophosphorylation and transphosphorylation sites in the EMS1 kinase domain identified by mass spectrometry.
Supplemental Table S3. Primers used in this study.
Supplemental Table S4. Constructs generated in this study.
Supplementary Material
Acknowledgments
We thank X. Wang and S. Clouse for advice on mass spectrometry analysis; S. Clouse for critical reading of the article; T. Schuck, J. Gonnering, and P. Engevold for plant care; P. He, and J. Sheen for the protoplast transfection; C. Albrecht for the SERK1:SERK1-YFP construct and seeds; R. Wang for contributions to the identification of the ems1-2 mutant; X. Peng for use of the Amersham Storm 860 imaging system; the Arabidopsis Biological Resource Center for TPD1 cDNA, TPD1-, EMS1-, SERK1-, SERK2-, and SERK3-containing BAC clones, as well as SALK_044330 (serk1-1), SALK_058020 (serk2-1), and SALK_051989 (ems1-2) lines; T. Nakagawa for pGBW vectors; C. Pikaard for pEARLEY vectors; and S. Gelvin for pSAT vectors.
Glossary
- BiFC
bimolecular fluorescence complementation
- FRET
Förster resonance energy transfer
- co-IP
coimmunoprecipitation
Footnotes
This work was supported by the National Science Foundation (grant nos. IOS–0721192 and IOS–1322796 to D.Z. and grant nos. PHY-1126386 and PHY-1058470 to V.R.), the Research Growth Initiative at the University of Wisconsin-Milwaukee (to D.Z. and V.R.), the University of Wisconsin-Madison/University of Wisconsin-Milwaukee Intercampus Research Incentive Grants Program (to D.Z.), the Greater Milwaukee Foundation (Shaw Scientist Award to D.Z.), and the University of Wisconsin-Milwaukee Research Foundation (Bradley Catalyst Award to D.Z.).
Articles can be viewed without a subscription.
References
- Ahsan N, Huang Y, Tovar-Mendez A, Swatek KN, Zhang J, Miernyk JA, Xu D, Thelen JJ (2013) A versatile mass spectrometry-based method to both identify kinase client-relationships and characterize signaling network topology. J Proteome Res 12: 937–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht C, Russinova E, Hecht V, Baaijens E, de Vries S (2005) The Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES1 and 2 control male sporogenesis. Plant Cell 17: 3337–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biener G, Stoneman MR, Acbas G, Holz JD, Orlova M, Komarova L, Kuchin S, Raicu V (2013) Development and experimental testing of an optical micro-spectroscopic technique incorporating true line-scan excitation. Int J Mol Sci 15: 261–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butenko MA, Patterson SE, Grini PE, Stenvik GE, Amundsen SS, Mandal A, Aalen RB (2003) Inflorescence deficient in abscission controls floral organ abscission in Arabidopsis and identifies a novel family of putative ligands in plants. Plant Cell 15: 2296–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butenko MA, Wildhagen M, Albert M, Jehle A, Kalbacher H, Aalen RB, Felix G (2014) Tools and strategies to match peptide-ligand receptor pairs. Plant Cell 26: 1838–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canales C, Bhatt AM, Scott R, Dickinson H (2002) EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis. Curr Biol 12: 1718–1727 [DOI] [PubMed] [Google Scholar]
- Caño-Delgado A, Yin Y, Yu C, Vafeados D, Mora-García S, Cheng JC, Nam KH, Li J, Chory J (2004) BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 131: 5341–5351 [DOI] [PubMed] [Google Scholar]
- Cao Y, Aceti DJ, Sabat G, Song J, Makino S, Fox BG, Bent AF (2013) Mutations in FLS2 Ser-938 dissect signaling activation in FLS2-mediated Arabidopsis immunity. PLoS Pathog 9: e1003313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Chern M, Canlas PE, Jiang C, Ruan D, Cao P, Ronald PC (2010) A conserved threonine residue in the juxtamembrane domain of the XA21 pattern recognition receptor is critical for kinase autophosphorylation and XA21-mediated immunity. J Biol Chem 285: 10454–10463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nürnberger T, Jones JD, Felix G, Boller T (2007) A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500 [DOI] [PubMed] [Google Scholar]
- Cho SK, Larue CT, Chevalier D, Wang H, Jinn TL, Zhang S, Walker JC (2008) Regulation of floral organ abscission in Arabidopsis thaliana. Proc Natl Acad Sci USA 105: 15629–15634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colcombet J, Boisson-Dernier A, Ros-Palau R, Vera CE, Schroeder JI (2005) Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASES1 and 2 are essential for tapetum development and microspore maturation. Plant Cell 17: 3350–3361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Zilberman D, Dickinson H (2013) A conversation across generations: soma-germ cell crosstalk in plants. Dev Cell 24: 215–225 [DOI] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM (1999) Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283: 1911–1914 [DOI] [PubMed] [Google Scholar]
- Goldberg RB, Beals TP, Sanders PM (1993) Anther development: basic principles and practical applications. Plant Cell 5: 1217–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou X, Yin H, He K, Du J, Yi J, Xu S, Lin H, Clouse SD, Li J (2012) Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLoS Genet 8: e1002452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gursanscky NR, Jouannet V, Grünwald K, Sanchez P, Laaber-Schwarz M, Greb T (2016) MOL1 is required for cambium homeostasis in Arabidopsis. Plant J 86: 210–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese A, Hann DR, Gimenez-Ibanez S, Jones AM, He K, Li J, Schroeder JI, Peck SC, Rathjen JP (2007) The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA 104: 12217–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hink MA, Shah K, Russinova E, de Vries SC, Visser AJ (2008) Fluorescence fluctuation analysis of Arabidopsis thaliana somatic embryogenesis receptor-like kinase and brassinosteroid insensitive 1 receptor oligomerization. Biophys J 94: 1052–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong L, Tang D, Shen Y, Hu Q, Wang K, Li M, Lu T, Cheng Z (2012) MIL2 (MICROSPORELESS2) regulates early cell differentiation in the rice anther. New Phytol 196: 402–413 [DOI] [PubMed] [Google Scholar]
- Hothorn M, Belkhadir Y, Dreux M, Dabi T, Noel JP, Wilson IA, Chory J (2011) Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 474: 467–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Smith AR, Zhang T, Zhao D (2016a) Creating completely both male and female sterile plants by specifically ablating microspore and megaspore mother cells. Front Plant Sci 7: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wijeratne AJ, Tang C, Zhang T, Fenelon RE, Owen HA, Zhao D (2016b) Ectopic expression of TAPETUM DETERMINANT1 affects ovule development in Arabidopsis. J Exp Bot 67: 1311–1326 [DOI] [PubMed] [Google Scholar]
- Huang J, Zhang T, Linstroth L, Tillman Z, Otegui MS, Owen HA, Zhao D (2016c) Control of anther cell differentiation by the small protein ligand TPD1 and its receptor EMS1 in Arabidopsis. PLoS Genet 12: e1006147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang WJ, Liu HK, McCormick S, Tang WH (2014) Tomato pistil factor STIG1 promotes in vivo pollen tube growth by binding to phosphatidylinositol 3-phosphate and the extracellular domain of the pollen receptor kinase LePRK2. Plant Cell 26: 2505–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Nakanomyo I, Motose H, Iwamoto K, Sawa S, Dohmae N, Fukuda H (2006) Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 313: 842–845 [DOI] [PubMed] [Google Scholar]
- Jia G, Liu X, Owen HA, Zhao D (2008) Signaling of cell fate determination by the TPD1 small protein and EMS1 receptor kinase. Proc Natl Acad Sci USA 105: 2220–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Källberg M, Margaryan G, Wang S, Ma J, Xu J (2014) RaptorX server: a resource for template-based protein structure modeling. Methods Mol Biol 1137: 17–27 [DOI] [PubMed] [Google Scholar]
- Karlova R, Boeren S, van Dongen W, Kwaaitaal M, Aker J, Vervoort J, de Vries S (2009) Identification of in vitro phosphorylation sites in the Arabidopsis thaliana somatic embryogenesis receptor-like kinases. Proteomics 9: 368–379 [DOI] [PubMed] [Google Scholar]
- King C, Stoneman M, Raicu V, Hristova K (2016) Fully quantified spectral imaging reveals in vivo membrane protein interactions. Integr Biol 8: 216–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Caño-Delgado A, Seto H, Hiranuma S, Fujioka S, Yoshida S, Chory J (2005) Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433: 167–171 [DOI] [PubMed] [Google Scholar]
- Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, Fukuda H, Sakagami Y (2006) A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313: 845–848 [DOI] [PubMed] [Google Scholar]
- Lee JS, Hnilova M, Maes M, Lin YC, Putarjunan A, Han SK, Avila J, Torii KU (2015) Competitive binding of antagonistic peptides fine-tunes stomatal patterning. Nature 522: 439–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Kuroha T, Hnilova M, Khatayevich D, Kanaoka MM, McAbee JM, Sarikaya M, Tamerler C, Torii KU (2012) Direct interaction of ligand-receptor pairs specifying stomatal patterning. Genes Dev 26: 126–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222 [DOI] [PubMed] [Google Scholar]
- Ma X, Xu G, He P, Shan L (2016) SERKing coreceptors for receptors. Trends Plant Sci 21: 1017–1033 [DOI] [PubMed] [Google Scholar]
- Mannan MA, Shadrick WR, Biener G, Shin BS, Anshu A, Raicu V, Frick DN, Dey M (2013) An ire1-phk1 chimera reveals a dispensable role of autokinase activity in endoplasmic reticulum stress response. J Mol Biol 425: 2083–2099 [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y. (2014) Posttranslationally modified small-peptide signals in plants. Annu Rev Plant Biol 65: 385–413 [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y, Ogawa M, Morita A, Sakagami Y (2002) An LRR receptor kinase involved in perception of a peptide plant hormone, phytosulfokine. Science 296: 1470–1472 [DOI] [PubMed] [Google Scholar]
- McCarty DR, Chory J (2000) Conservation and innovation in plant signaling pathways. Cell 103: 201–209 [DOI] [PubMed] [Google Scholar]
- Meng X, Chen X, Mang H, Liu C, Yu X, Gao X, Torii KU, He P, Shan L (2015) Differential function of Arabidopsis SERK family receptor-like kinases in stomatal patterning. Curr Biol 25: 2361–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Zhou J, Tang J, Li B, de Oliveira MV, Chai J, He P, Shan L (2016) Ligand-induced receptor-like kinase complex regulates floral organ abscission in Arabidopsis. Cell Rep 14: 1330–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra SK, Chen R, Dhandaydham M, Wang X, Blackburn RK, Kota U, Goshe MB, Schwartz D, Huber SC, Clouse SD (2015) An autophosphorylation site database for leucine-rich repeat receptor-like kinases in Arabidopsis thaliana. Plant J 82: 1042–1060 [DOI] [PubMed] [Google Scholar]
- Nam KH, Li J (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110: 203–212 [DOI] [PubMed] [Google Scholar]
- Nonomura K, Miyoshi K, Eiguchi M, Suzuki T, Miyao A, Hirochika H, Kurata N (2003) The MSP1 gene is necessary to restrict the number of cells entering into male and female sporogenesis and to initiate anther wall formation in rice. Plant Cell 15: 1728–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh MH, Wang X, Clouse SD, Huber SC (2012) Deactivation of the Arabidopsis BRASSINOSTEROID INSENSITIVE 1 (BRI1) receptor kinase by autophosphorylation within the glycine-rich loop. Proc Natl Acad Sci USA 109: 327–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raicu V, Singh DR (2013) FRET spectrometry: a new tool for the determination of protein quaternary structure in living cells. Biophys J 105: 1937–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raicu V, Stoneman MR, Fung R, Melnichuk M, Jansma DB, Rath S, Pisteri LF, Fox M, Wells JW, Saldin DK (2009) Determination of supramolecular structure and spatial distribution of protein complexes in living cells. Nat Photonics 3: 107–113 [Google Scholar]
- Roux M, Schwessinger B, Albrecht C, Chinchilla D, Jones A, Holton N, Malinovsky FG, Tör M, de Vries S, Zipfel C (2011) The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23: 2440–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russinova E, Borst JW, Kwaaitaal M, Caño-Delgado A, Yin Y, Chory J, de Vries SC (2004) Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 16: 3216–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J, Brandt B, Wildhagen M, Hohmann U, Hothorn LA, Butenko MA, Hothorn M (2016) Mechanistic insight into a peptide hormone signaling complex mediating floral organ abscission. eLife 5: e15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J, Henzler C, Hothorn M (2013) Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases. Science 341: 889–892 [DOI] [PubMed] [Google Scholar]
- Scott RJ, Spielman M, Dickinson HG (2004) Stamen structure and function. Plant Cell (Suppl) 16: S46–S60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah K, Gadella TW Jr, van Erp H, Hecht V, de Vries SC (2001a) Subcellular localization and oligomerization of the Arabidopsis thaliana somatic embryogenesis receptor kinase 1 protein. J Mol Biol 309: 641–655 [DOI] [PubMed] [Google Scholar]
- Shah K, Vervoort J, de Vries SC (2001b) Role of threonines in the Arabidopsis thaliana somatic embryogenesis receptor kinase 1 activation loop in phosphorylation. J Biol Chem 276: 41263–41269 [DOI] [PubMed] [Google Scholar]
- She J, Han Z, Kim TW, Wang J, Cheng W, Chang J, Shi S, Wang J, Yang M, Wang ZY, et al. (2011) Structural insight into brassinosteroid perception by BRI1. Nature 474: 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Sugano SS, Hara-Nishimura I (2011) Positive and negative peptide signals control stomatal density. Cell Mol Life Sci 68: 2081–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara H, Mori A, Yasue N, Sumida K, Matsubayashi Y (2016) Identification of three LRR-RKs involved in perception of root meristem growth factor in Arabidopsis. Proc Natl Acad Sci USA 113: 3897–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Karlowski WM, Pan R, Tzeng YH, Mayer KF, Li WH (2004) Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16: 1220–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak ED, McAbee JM, Pillitteri LJ, Torii KU (2005) Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309: 290–293 [DOI] [PubMed] [Google Scholar]
- Stenvik GE, Tandstad NM, Guo Y, Shi CL, Kristiansen W, Holmgren A, Clark SE, Aalen RB, Butenko MA (2008) The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in Arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell 20: 1805–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneman MR, Paprocki JD, Biener G, Yokoi K, Shevade A, Kuchin S, Raicu V (2016) Quaternary structure of the yeast pheromone receptor Ste2 in living cells. BBA Biomembranes (in press) [DOI] [PubMed] [Google Scholar]
- Sun Y, Li L, Macho AP, Han Z, Hu Z, Zipfel C, Zhou JM, Chai J (2013) Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 342: 624–628 [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Higashiyama T (2016) Tip-localized receptors control pollen tube growth and LURE sensing in Arabidopsis. Nature 531: 245–248 [DOI] [PubMed] [Google Scholar]
- Tang W, Ezcurra I, Muschietti J, McCormick S (2002) A cysteine-rich extracellular protein, LAT52, interacts with the extracellular domain of the pollen receptor kinase LePRK2. Plant Cell 14: 2277–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor I, Wang Y, Seitz K, Baer J, Bennewitz S, Mooney BP, Walker JC (2016) Analysis of phosphorylation of the receptor-like protein kinase HAESA during Arabidopsis floral abscission. PLoS ONE 11: e0147203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii KU. (2004) Leucine-rich repeat receptor kinases in plants: structure, function, and signal transduction pathways. Int Rev Cytol 234: 1–46 [DOI] [PubMed] [Google Scholar]
- Torii KU. (2012) Mix-and-match: ligand-receptor pairs in stomatal development and beyond. Trends Plant Sci 17: 711–719 [DOI] [PubMed] [Google Scholar]
- Tzfira T, Tian GW, Lacroix B, Vyas S, Li J, Leitner-Dagan Y, Krichevsky A, Taylor T, Vainstein A, Citovsky V (2005) pSAT vectors: a modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol Biol 57: 503–516 [DOI] [PubMed] [Google Scholar]
- Walbot V, Egger RL (2016) Pre-meiotic anther development: cell fate specification and differentiation. Annu Rev Plant Biol 67: 365–395 [DOI] [PubMed] [Google Scholar]
- Wang CJ, Nan GL, Kelliher T, Timofejeva L, Vernoud V, Golubovskaya IN, Harper L, Egger R, Walbot V, Cande WZ (2012) Maize multiple archesporial cells 1 (mac1), an ortholog of rice TDL1A, modulates cell proliferation and identity in early anther development. Development 139: 2594–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Li H, Han Z, Zhang H, Wang T, Lin G, Chang J, Yang W, Chai J (2015) Allosteric receptor activation by the plant peptide hormone phytosulfokine. Nature 525: 265–268 [DOI] [PubMed] [Google Scholar]
- Wang T, Liang L, Xue Y, Jia PF, Chen W, Zhang MX, Wang YC, Li HJ, Yang WC (2016) A receptor heteromer mediates the male perception of female attractants in plants. Nature 531: 241–244 [DOI] [PubMed] [Google Scholar]
- Wang X, Goshe MB, Soderblom EJ, Phinney BS, Kuchar JA, Li J, Asami T, Yoshida S, Huber SC, Clouse SD (2005) Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell 17: 1685–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kota U, He K, Blackburn K, Li J, Goshe MB, Huber SC, Clouse SD (2008) Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell 15: 220–235 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J (2001) BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410: 380–383 [DOI] [PubMed] [Google Scholar]
- Yang SL, Xie LF, Mao HZ, Puah CS, Yang WC, Jiang L, Sundaresan V, Ye D (2003) Tapetum determinant1 is required for cell specialization in the Arabidopsis anther. Plant Cell 15: 2792–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhao D. (2009) Control of anther cell differentiation: a teamwork of receptor-like kinases. Sex Plant Reprod 22: 221–228 [DOI] [PubMed] [Google Scholar]
- Zhao DZ, Wang GF, Speal B, Ma H (2002) The excess microsporocytes1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev 16: 2021–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, de Palma J, Oane R, Gamuyao R, Luo M, Chaudhury A, Hervé P, Xue Q, Bennett J (2008) OsTDL1A binds to the LRR domain of rice receptor kinase MSP1, and is required to limit sporocyte numbers. Plant J 54: 375–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.