Abstract
Pollen-pistil interactions contribute to mate selection at the postmating, prezygotic level.
A PLACE FOR BOYS AND GIRLS TO MEET IN THE PLANT WORLD: THE PISTIL IS THE VENUE FOR MATE SELECTION
As a group, the flowering plants have remarkably diverse modes of reproduction that run the gamut from clonal propagation to obligate outcrossing. Articles in this volume focus on sexual reproduction, and there is great diversity even within this slice of the reproduction spectrum. Like all diverse systems, reproductive characters are the end results of molecular and genetic mechanisms that generate diversity. Selective pressures that act upon this diversity lead to the differentiation of lineages and the maintenance of their identity. The seemingly minuscule slice of the angiosperm life cycle represented in the postpollination to prezygotic phase, which is dominated by pollen-pistil interactions, plays a large role in these evolutionary processes.
The pistil is unique to angiosperms. It serves a protective role and functions as a conduit for pollen tubes to grow to the ovary, but it also provides a venue for pollen-pistil interactions that regulate pollen tube growth and, hence, fertilization. Although the earliest diverging angiosperm lineages do not possess the full set of canonical angiosperm pistil traits (Endress and Doyle, 2009; Endress, 2011, 2015; Lora et al., 2016), the core lineages (which display the most diversity) have a stigma specialized for the receipt of pollen and a style that pollen tubes must traverse before fertilization can occur. Some authors attribute angiosperm diversity to phenomena that depend on pollen-pistil interactions taking place in stigmas and styles, such as pollen competition and self-incompatibility (SI; Mulcahy and Mulcahy, 1988; Williams, 2012). Therefore, the evolved state of the pistil, as the organ that stands between pollen receipt and the ovule, allows it to assume a gatekeeper function, facilitating the growth of desirable pollen tubes and discouraging invaders and less desirable pollen (Heslop-Harrison, 2000).
Our focus is on pollen-pistil interactions that contribute to mate selection in the domains of SI and interspecific pollen rejection. Together, these processes define a window of genetic relationships for successful mating: SI prevents mating between very closely related individuals of the same species, and interspecific pollen rejection prevents crosses that are too distant. SI clearly contributes to a species’ long-term success, as SI lineages are genetically diverse and very persistent (Goldberg et al., 2010; Goldberg and Igić, 2012). However, transitions from SI to self-compatibility (SC) also are important. SC lineages emerge frequently and may make range expansion possible under conditions of mate limitation (Baker, 1955, 1967; Pannell et al., 2015) but SC lineages also may be vulnerable to extinction due to inbreeding depression. A species with any substantial level of outcrossing entertains the risk that the stigma will be challenged with pollen from other species. The ability to recognize and reject heterospecific pollen, therefore, is important to prevent wasted ovules as well as to maintain species integrity. Outcrossing species in complex plant communities often are challenged by heterospecific pollen (Arceo-Gómez and Ashman, 2014) and, thus, would be expected to maintain robust interspecific pollen recognition and rejection mechanisms.
A limited number of SI and interspecific pollen rejection mechanisms have been elucidated by genetic and biochemical analyses. We favor a synthetic view emphasizing the pistil’s overall mate selection function. This perspective provides a framework in which to view a range of pollen-pistil interactions that can function in both the generation and maintenance of species.
INTRASPECIFIC REPRODUCTIVE BARRIERS: SI
SI is a genetically controlled system for recognizing and rejecting self-pollen in plants with hermaphroditic flowers (de Nettancourt, 1997, 2001; Franklin-Tong, 2008). SI enforces outcrossing, but it is distinct from dioecy or phenological barriers to self-fertilization. The term is applied generally to genetically controlled systems that prevent selfing, and a variety of SI systems have evolved across the angiosperms. Because the simple genetics of SI provided a starting point for analyses, SI systems are the best understood pollen-pistil interactions at the molecular and genetic levels and, thus, provide paradigms for understanding interspecific pollen rejection.
Genetic control of SI usually derives from a single polymorphic locus called the S-locus. Although the S-locus has the same name in different families, the locus structure, the nature of the encoded genes, and the mechanisms of SI may be completely different (de Nettancourt, 2001). Heteromorphic SI, where alternate floral morphs are produced (e.g. long- and short-style Primula vulgaris; Darwin, 1877; Gilmartin and Li, 2010), is common, but outcrossing is achieved by structural barriers and not pollen-pistil interactions. In SI species with homomorphic flowers, the S-locus encodes pistil-expressed genes that provide for the recognition and rejection of self-pollen or the favoring of nonself pollen. Homomorphic SI is described as gametophytic if pollen compatibility is determined by its own S-haplotype and sporophytic if the compatibility of the haploid pollen is determined by the diploid sporophyte that produced it. In either case, the S-locus encodes genes controlling specificity on both the pollen and pistil sides. The genes and their functions have been studied extensively in three distinct SI systems (de Nettancourt, 1997, 2001; Franklin-Tong, 2008).
SI MECHANISMS: BRASSICACEAE
Sporophytic SI has been studied intensively in Brassicaceae. In simple cases where dominance does not play a role, sporophytic SI is quite selective, since a compatible pollination requires that there are no S-haplotypes in common between male and female partners (de Nettancourt, 1997, 2001; Franklin-Tong, 2008). The Brassicaceae pistil consists of a dry stigma with numerous papillar cells for pollen reception. Compatible pollen germinates on the papillar cell surface, and pollen tubes traverse a short transmitting tract before entering the ovary. Therefore, it is not surprising that SI pollen rejection is a rapid response occurring on the stigmatic papillar cells. The Brassicaceae SI system was one of the first to be successfully studied at the molecular level (Nasrallah and Wallace, 1967; Nasrallah et al., 1985). It is now established that each S-haplotype includes an allele of the S-locus receptor kinase (SRK) gene expressed in the stigma and a matching allele of an S-locus Cys-rich (SCR) gene expressed in the diploid tapetum of the anther, such that the encoded SCR protein is deposited in the exine of haploid pollen grains (Stein and Nasrallah, 1993; Schopfer et al., 1999; Takayama et al., 2000). SRK and SCR form a receptor-ligand pair that initiates self-pollen rejection. In an incompatible SI interaction in Brassica spp. and Arabidopsis, the SCR protein diffuses from the pollen exine and interacts with the SRK extracellular domain, which is located in the plasma membrane of the stigmatic papillar cells. SRK signals to intracellular proteins in the papillar cell, and downstream events, including protein phosphorylation and changes in intracellular calcium, lead to the rejection response (Takayama and Isogai, 2005; Nasrallah and Nasrallah, 2014; Iwano et al., 2015). In compatible pollinations, independent of SRK signaling, the papillar cells are stimulated to secrete material that presumably supports pollen tube growth (Safavian and Goring, 2013). In the incompatible response, a key secretory protein, exo70A1 (exocyst complex component 70A1), is destabilized, and secretory vesicles are targeted to a vacuole rather than secreted (Indriolo and Goring, 2014). Since diffusion of the pollen SCR polypeptide ligand to the SRK receptor induces the rejection response in the papillar cell, SC can be caused by a loss-of-function mutations in either pollen-expressed SCR or pistil-expressed SRK (Dwyer et al., 2013).
SI MECHANISMS: PAPAVER SPP.
Papaver rhoeas has a gametophytic SI system that also relies on receptor-ligand signaling and an induced rejection response. However, the S-specific polypeptide ligand is produced in the stigma and induces a rejection response in incompatible pollen tubes. Gametophytic SI is inherently less selective than sporophytic SI because it is the S-haplotype of the haploid male gametophyte that determines compatibility (de Nettancourt, 1997, 2001; Franklin-Tong, 2008). The pistil-side S-genes encode secreted Prs-S proteins, and the corresponding pollen-side genes encode Prp-S proteins that are located at the plasma membrane (Foote et al., 1994; Wheeler et al., 2010). Poppies have an unusual stigma with specialized spoke-like rays that receive pollen; although the anatomy differs from Brassicaceae in almost every detail, a common element is that pollen tubes only grow a short distance before entering the ovary. The SI response in Papaver spp. is likewise very rapid, and arrest of incompatible pollen tube growth occurs on the stigma.
Papaver SI is better understood at the physiological level than any other system, in part, because the response can be replicated faithfully in vitro (Franklin-Tong et al., 1988). In vitro systems have been used successfully to elucidate pollen tube cell biology and pollen tube attraction (Cheung et al., 1995; Palanivelu and Preuss, 2006; Okuda et al., 2009; Kanaoka and Higashiyama, 2015), but P. rhoeas is unique among pollen rejection systems because the SI response also can be reproduced faithfully. The addition of incompatible Prs-S protein to pollen tube growth medium rapidly initiates a sequence of events, including cessation of growth, changes in calcium currents and pH, and fragmentation of the actin cytoskeleton in pollen tubes (Franklin-Tong, 2008; Wilkins et al., 2014, 2015). Over longer time periods, a programmed cell death response is initiated, and pollen tube growth inhibition is irreversible (Bosch and Franklin-Tong, 2007, 2008). Strikingly, incompatible Prs-S protein alone is sufficient to induce this series of responses, and the Prp-S protein is sufficient to transduce the signal (Foote et al., 1994; de Graaf et al., 2012). While other SI systems require modifier genes (i.e. non-S-locus genes that are required for full function) on either the pollen or the pistil side, or both, Papaver SI appears to be simpler. The initial signaling to Prp-S in incompatible pollen tubes may directly affect intracellular calcium, leading to a complex but conserved series of events with cell death as the end point (Wilkins et al., 2014, 2015).
The proposition that Prs-S and Prp-S function together as a ligand-receptor signaling module that signals to a conserved cell death response has great potential significance. This hypothesis was tested recently by transferring the P. rhoeas SI system to Arabidopsis. Lin et al. (2015) showed that expressing Prs-S1 in Arabidopsis papillar cells and Prp-S1 in pollen (i.e. an incompatible Prs-S/Prp-S pair) results in SI (Lin et al., 2015). Given the phylogenetic distance between P. rhoeas and Arabidopsis, this exciting result suggests that the Papaver SI system may be used to manipulate compatibility in virtually any species, which has long been a goal of plant biotechnologists (see Advances).
S-RNASE-BASED SI
Species that display S-RNase-based SI have wet stigmas and long styles, and incompatible pollen tube rejection generally occurs in the style well after pollen tube germination (Fig. 1). S-RNases function as both pistil-side recognition proteins and as cytotoxins in SI, and their recognition function determines S-specificity on the pistil side. Allelic S-RNase sequences often show striking levels of divergence; for example, SA2- and SC10-RNase from Nicotiana alata show only 43% identity (McClure et al., 2000). S-specificity has been investigated by in vitro mutagenesis and domain-swap experiments (Kao and McCubbin, 1996; Matton et al., 1997; Zurek et al., 1997) with divergent results. Matton et al. (1997) converted S11-RNase into a chimeric sequence recognized as S13-RNase in Solanum chacoense by exchanging four residues between this pair of highly similar proteins. Intriguingly, exchanging just three residues results in a dual-specificity protein that caused the rejection of both S11 and S13 pollen (Matton et al., 1999). More recently, Soulard et al. (2013) generated another dual S11-/S13-RNase by incorporating an N-glycosylation site into a variable region. In contrast, 11 domain-swap experiments in Nicotiana and Petunia spp., where sequence segments ranging from 15 to 189 residues were exchanged, always resulted in chimeric S-RNases that failed to be recognized at all (Kao and McCubbin, 1996; Zurek et al., 1997). Thus, while S-RNases encode S-specificity information, it is not always localized to a specific region of the protein. As we discuss later, it is not completely clear how S-RNase specificity is perceived on the pollen side.
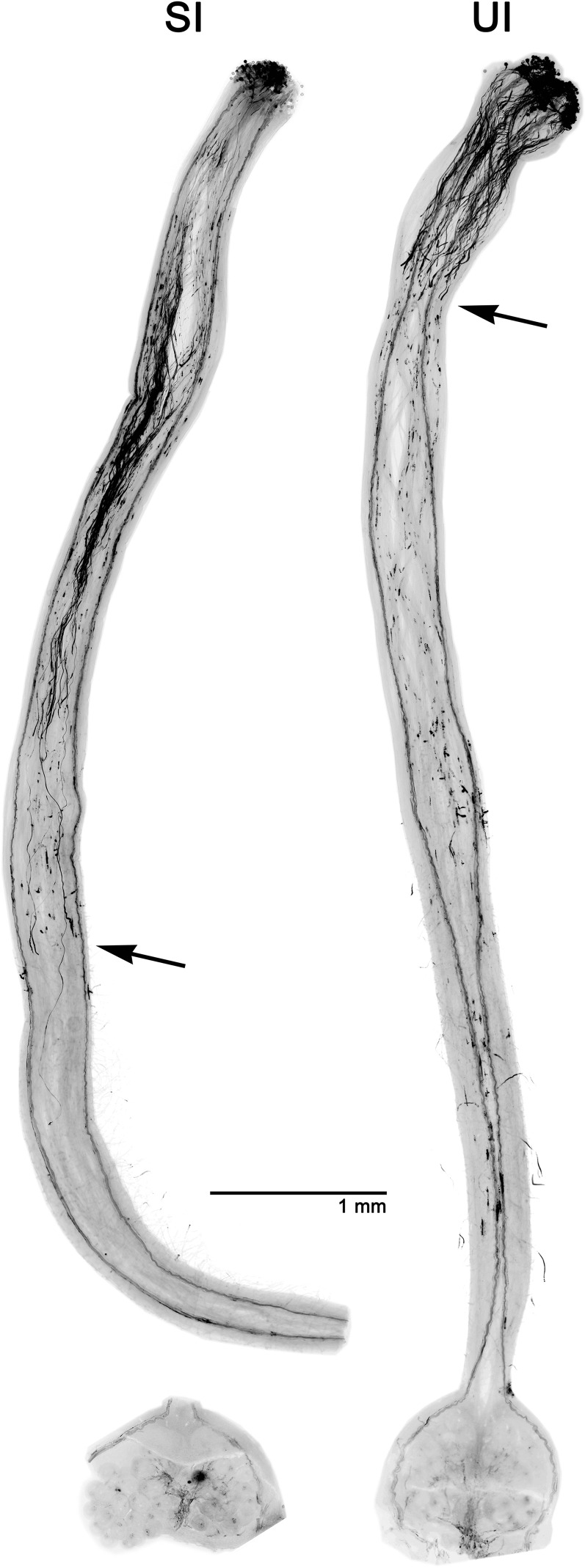
Figure 1.
Rapid interspecific unilateral incompatibility (UI) pollen rejection compared with SI. Pistils stained with Aniline Blue fluorochrome are shown. Left, Self-pollinated SI Solanum chilense LA2884. Right, SI S. chilense LA2884 pollinated by S. lycopersicum ‘VF36’. The UI rejection response in this example occurs closer to the stigma and is typical of crosses between SI green-fruited species and SC red-fruited species (Baek et al., 2015). Arrows indicate the position in the style where fewer than three pollen tubes pass at 48 h after pollination (compatible pollen tubes reach the ovary after 24 h). Images are by You Soon Baek and Suzanne Royer.
S-RNase conserved regions are crucial for their cytotoxic activity. Early studies identified five conserved regions, C1 to C5, that together account for about 30 of the approximately 200 residues in a typical S-RNase (Anderson et al., 1989; Ioerger et al., 1991). Conserved regions C2 and C3 each contain a His residue important for RNase activity (Kawata et al., 1988; McClure et al., 1989). The observation that pollen RNA is specifically degraded after self-pollination led to the suggestion that S-RNases function as S-specific cytotoxins in SI (McClure et al., 1989, 1990; Gray et al., 1991; Huang et al., 1994; Kowyama et al., 1994). Although there is considerable evidence that SI in Solanaceae is based on S-RNase cytotoxicity, recent evidence suggests that other effects also may be important. For example, Roldán et al. (2012) reported the disorganization of F-actin in incompatible N. alata pollen tubes, raising interesting parallels to Papaver SI. Moreover, there have been suggestions that S-RNase-based SI in Pyrus spp. (Rosaceae) involves a complex set of physiological responses, including a cell death response, reactive oxygen species, and the actin cytoskeleton (Liu et al., 2007; Wang et al., 2009, 2010). Although these Pyrus studies often rely on in vitro experiments that have not been thoroughly validated in vivo, there is good evidence that S-RNase-based SI in Prunus spp. differs fundamentally from that in Solanaceae (Tao and Iezzoni, 2010; Sassa, 2016). Thus, it is best to remain open to the possibility that S-RNase may have a series of effects on incompatible pollen tubes. At present, it is impossible to distinguish between direct effects of SI and indirect effects of reduced pollen tube growth or physiological decay in a dying pollen tube. Robust in vivo experiments are needed to determine the temporal series of events and distinguish between potential mechanisms in incompatible pollen tubes.
Although S-RNases are the crucial pistil-side determinants of S-specificity and also are involved directly in self-pollen rejection, they are not sufficient for pistil-side SI function. Three modifier genes have been identified that contribute to SI but that do not determine S-specificity (Fig. 2). Loss-of-function studies show that the Asn-rich HT proteins are required for SI in Nicotiana, Petunia, and Solanum spp. (McClure et al., 1999; O’Brien et al., 2002; Puerta et al., 2009). Most SI Solanum spp. express two HT genes designated HT-A and HT-B (Kondo et al., 2002; O’Brien et al., 2002). For example, with the exception of Solanum habrochaites, which expresses only an HT-A gene, the green-fruited species in the tomato clade express both HT-A and HT-B (Covey et al., 2010). These green-fruited species are predominantly SI or have recently undergone a shift in their mating system to SC. In contrast, cultivated tomato (Solanum lycopersicum) and its SC red- or orange-fruited relatives (Solanum galapagense, Solanum cheesmaniae, and Solanum pimpinellifolium; here referred to as red-fruited species) do not express HT proteins (Kondo et al., 2002). Interestingly, HT proteins are preferentially degraded after compatible pollination in Nicotiana spp. (Goldraij et al., 2006). The 120-kD glycoprotein (120K), another SI modifier, is an abundant S-RNase-binding protein located in the N. alata transmitting tract (Lind et al., 1994, 1996; Cruz-Garcia et al., 2005) that also is required for S-specific pollen rejection (Hancock et al., 2005). More recently, NaStEP (N. alta Stigma Expressed Protein) was identified as a proteinase inhibitor that is required for SI and also affects HT protein stability (Busot et al., 2008; Jiménez-Durán et al., 2013). Further insights into the functions of HT, 120K, and NaStEP are needed.
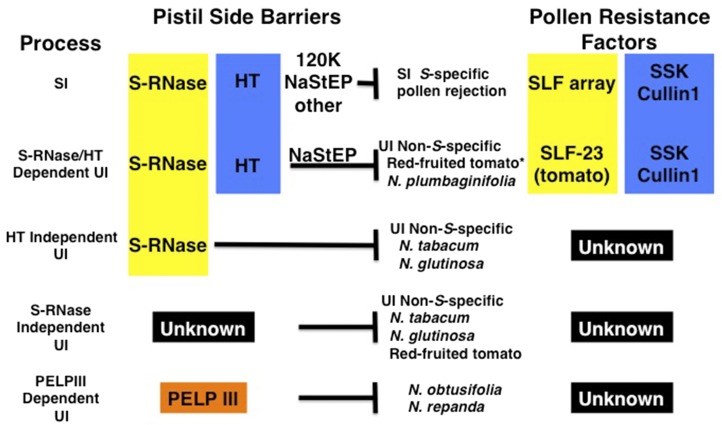
Figure 2.
Pollen rejection mechanisms in Solanaceae. At least five distinct processes can be recognized. The intraspecific S-RNase-dependent SI mechanism is the best characterized. SI genes are implicated in some interspecific UI mechanisms (yellow, SI genes that determine specificity; blue, SI modifier genes). It is especially noteworthy that three distinct processes are S-RNase dependent but use different modifier genes. Not all pollen and pistil factors have been identified in all species studied. Pollen-side UI factors have only been identified in tomato relatives. Only a single SLF gene, SLF-23, is known to function in UI. *, The requirement for a tomato equivalent of NaStEP is inferred from the similarity to N. plumbaginifolia pollen rejection but has not been confirmed experimentally. S-RNase-independent mechanisms function in several examples of interspecific UI pollen rejection. Several such mechanisms probably exist but are lumped together because they are undefined at present. One S-RNase-independent mechanism that has been characterized relies on PELPIII (Pistil Extensin-Like Protein III, orange).
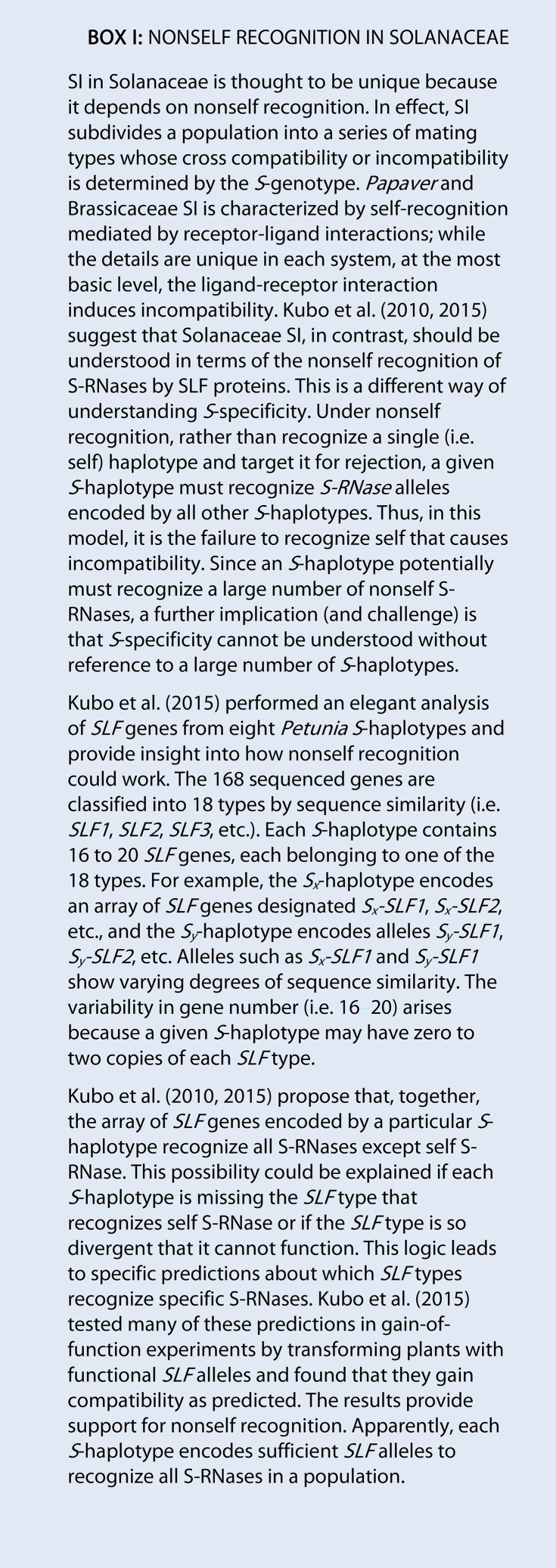
S-locus F-box (SLF) genes were first identified as pollen-expressed F-box protein-encoding genes linked to S-RNase in Antirrhinum spp. Because they showed low sequence polymorphism, SLF genes were not at first thought to function as pollen S-specificity determinants (Lai et al., 2002). However, Sijacic et al. (2004) confirmed an association with SI by a gain-of-function experiment in Petunia inflata. Recent transcriptome studies suggest that an array of 16 to 20 SLF genes function in SI in Petunia spp. (Box 2; Williams et al., 2014a; Kubo et al., 2015). Consistent with this finding, the genome sequence of SC Solanum pennellii LA0716 (a recently derived SC accession of an otherwise SI species) contains an array of 23 SLF genes, while the SC tomato genome shows, at most, four intact genes (Bolger et al., 2014; Li and Chetelat, 2015).
The SLF proteins are thought to function in SI as components of SCF complexes (SCFSLF), and the Petunia complexes have been the most thoroughly characterized. Li et al. (2014) recently expressed tagged SLF and recovered associated Rbx1-like, Skp1-like, and Cullin1-P (CUL1-P) proteins by coimmunoprecipitation. A reciprocal pull-down assay where the Skp1-like protein, PiSSK1 was tagged provided evidence for multiple SCFSLF complexes (Li et al., 2014). This is consistent with the suggestion that each member of the SLF protein array binds individually to SSK and CUL1-P to form a series of SCFSLF complexes that function together in SI (Kubo et al., 2010, 2015; Entani et al., 2014; Williams et al., 2014b). Several gain-of-function studies support the role of individual SLF genes in determining pollen-side S-specificity (Sijacic et al., 2004; Kubo et al., 2010, 2015). Therefore, the genes encoding the common SCF complex components that do not contribute to S-specificity (e.g. PiSSK and PiCUL1-P) are pollen-side modifier genes (Fig. 2). In the collaborative nonself recognition model (see Advances), a collection of SLF proteins brings all nonself S-RNases into the array of SCFSLF complexes for ubiquitylation and subsequent degradation (Qiao et al., 2004; Zhao et al., 2010; Entani et al., 2014; Williams et al., 2015).
The collaborative nonself recognition model is attractive, but other processes also may contribute to SI. Goldraij et al. (2006) showed that S-RNases are taken up by both compatible and incompatible pollen tubes and targeted to vacuoles. In incompatible pollen tubes, the endomembrane system eventually breaks down, and S-RNases are released into the pollen tube cytoplasm, while in compatible pollen tubes, S-RNases remain compartmentalized. Interestingly, S-RNase uptake and compartmentalization are not affected in HT-suppressed plants (Goldraij et al., 2006), so HT protein likely functions downstream of these processes. Either compartmentalization or degradation could be equally effective in preventing S-RNase cytotoxicity, but the relative contributions from these processes to SI are not clear. One suggestion is that the processes operate in parallel (McClure, 2009; Williams et al., 2015).
The hypothesis that pollen-side S-specificity is determined by nonself recognition through a collection of SLF proteins (rather than self-recognition by a single protein) requires a shift in thinking about specificity itself (Box 1). Under this model, each SLF protein is thought to recognize one to four nonself S-RNases, such that the collection of SLF proteins encoded by a given S-haplotype recognizes all S-RNases except the self protein (Kubo et al., 2010, 2015; Iwano and Takayama, 2012; Williams et al., 2015). The extensive transcriptome analysis and functional testing presented by Kubo et al. (2015) provide strong support for this model. However, it is worth revisiting results from S-RNase mutagenesis studies investigating pistil-side specificity. Under the nonself recognition model, a novel S-RNase that is not recognized by the SLF array in a given S-haplotype should cause pollen rejection (because no SLF protein is available to provide resistance). This is concordant with results from in vitro mutagenesis experiments in S. chacoense, where exchanging four residues between S11- and S13-RNase switched S-specificity (i.e. the S11/S13 chimera caused rejection of S13 pollen but not S11 pollen; Matton et al., 1999) but exchanging just three residues resulted in an S-RNase recognized in pollen as both S11 and S13 (i.e. caused the rejection of both S13 pollen and S11 pollen; Matton et al., 1999). This result can be explained if one or more SLF proteins in the S11 array recognize the former chimera but the latter is not recognized by any SLF in either the S11 or the S13 array. In contrast, the Petunia and Nicotiana S-RNase domain-swap experiments are not as easily accommodated (Kao and McCubbin, 1996; Zurek et al., 1997). It seems reasonable that exchanging 15 to 189 residues between allelic proteins should result, at least sometimes, in novel S-RNases that would not be recognized by the SLF array and, therefore, cause pollen rejection. However, none of the 11 chimeric S-RNases tested caused pollen rejection. Trivial explanations are unlikely because each of the nine Nicotiana spp. chimeras retain RNase activity (Zurek et al., 1997) and at least one functions in interspecific pollen rejection (Beecher et al., 2001). Under the collaborative nonself recognition model, the S-RNase domain-swap results could be explained if one or more SLF proteins recognized each of these 11 novel chimeric S-RNases. However, the functional testing reported so far suggests that most SLF proteins recognize a fixed set of S-RNases (Kubo et al., 2010, 2015). More information about how S-RNases are recognized by SLF proteins is needed to understand how the SLF array can recognize novel proteins.
INDUCED POLLEN REJECTION VERSUS CONSTITUTIVE BARRIER/RESISTANCE
Papaver and Brassicaceae SI are both systems characterized by a rapidly induced rejection response mediated by a ligand-receptor interaction. In Brassicaceae, signaling initiates the rejection response in the stigma papillar cell, while in Papaver, it is the pollen tube that responds to signaling. As the rejection response is induced in these systems, loss-of-function mutations in either the pollen- or pistil-side genes can cause SC, and, with some notable exceptions (Takada et al., 2013), the default for intraspecific pollination is compatibility. Iwano and Takayama (2012) refer to Papaver and Brassicaceae SI as self-recognition systems in the sense that the pollen- and pistil-expressed protein pair, encoded in a common (i.e. self) S-haplotype such as S-haplotype pairs of Prs-S/Prp-S and SCR/SRK proteins (i.e. self-recognition), interact to induce rejection.
SI in Solanaceae is fundamentally different from SI in either Brassicaceae or Papaver. In Solanaceae, pollen-side SI genes from a given S-haplotype recognize all pistil-side genes except the self gene (i.e. nonself recognition). SI in Solanaceae is not based on ligand-receptor signaling in the traditional sense; instead, pistil-side functions amount to a barrier to pollination, and pollen-side functions selectively provide resistance to nonself barriers. The key barrier proteins in SI Solanaceae spp. are pistil-expressed S-RNases (McClure et al., 1989), and the pollen-resistance factors are SLF proteins (Lai et al., 2002; Sijacic et al., 2004; Kubo et al., 2010). Thus, loss of the pistil-side barrier causes a mating system transition to SC, with potential consequences for the species’ evolutionary trajectory (Box 2). SI in Antirrhinum spp. is similar to the Solanaceae system (Xue et al., 1996; Lai et al., 2002; Huang et al., 2006; Chen et al., 2010). SI species in Rosaceae also rely on S-RNases and pollen-expressed F-box proteins; but curiously, species in the Pyreae tribe (e.g. apple [Malus domestica] and pear [Pyrus communis]) appear to have a nonself recognition system like Solanaceae, while those in Prunus (e.g. cherry [Prunus avium], apricot [Prunus armenaica], and almond [Prunus dulcis]) appear to have a self-recognition system (Sassa, 2016).
INTERSPECIFIC REPRODUCTIVE BARRIERS
Numerous adaptations and ecological factors, such as geographical distribution and pollinator specialization, help prevent hybridization between closely related species (Lowry et al., 2008; Widmer et al., 2009; Baack et al., 2015). However, the pistil also is a venue for mate selection at the interspecies level, since it is an arena for postmating, prezygotic barriers that rely on pollen-pistil interactions. This class of interspecific reproductive barriers includes conspecific pollen precedence, the preferential success of conspecific pollen compared with heterospecific pollen (Howard, 1999; Fishman et al., 2008; Lora et al., 2016), and pollen tube guidance and reception. Here, we focus on outright interspecific pollen tube rejection.
SI AND INTERSPECIFIC INCOMPATIBILITY: A COMMON TOOLKIT
Intraspecific and interspecific pollen rejection control mating at opposite ends of the genetic relationship continuum, yet these rejection responses often show similarities. Recent results show that some genes function at both levels and that some mechanisms are similar (see Advances). We view this as a manifestation of the overall function of the pistil as the organ for mate selection.
In a remarkably prescient article, Lewis and Crowe (1958) summarized crosses between many SI species and their SC relatives and emphasized a type of UI where “pollen of the self-compatible species was inhibited in the styles of the self-incompatible species, while no inhibitions occurred in the reciprocal cross.” They noted that this so-called SI × SC rule applies to many crosses in the Solanaceae and Scrophulariaceae (McGuire and Rick, 1954) and suggested that it was regulated by the S-locus (Lewis and Crowe, 1958). A recent comprehensive study of interspecific compatibility in the tomato clade demonstrated that the SI × SC rule is closely followed at the species level (Baek et al., 2015). In addition to the Solanaceae, UI consistent with the SI × SC rule has been recorded in Lilliaceae (Ascher and Peloquin, 1968), Poaceae (Heslop-Harrison, 1982), Brassicaceae (Sampson, 1962; Hiscock and Dickinson, 1993), and Orchidaceae (Pinheiro et al., 2015). Although the SI × SC rule applies broadly, differences between SI and UI, such as the timing of the response (Fig. 1; Ascher and Peloquin, 1968, Liedl et al., 1996) and the fact that exceptions to the rule can be identified easily, led some to challenge whether the S-locus functions at the interspecific level (Hogenboom, 1984; de Nettancourt, 1997, 2001). Recent studies clarify that UI is mechanistically linked to SI in some cases but not in others, and interpretations are complicated by genetic redundancy.
Genetic and molecular evidence shows that some forms of UI are related to SI. Quantitative trait locus (QTL) studies of tomato clade factors contributing to UI on the pistil side show linkage between the S-locus and a major UI QTL (Bernacchi and Tanksley, 1997) and a second that includes the Solanum HT-A and HT-B genes (Covey et al., 2010). Murfett et al. (1996) provided direct evidence that S-RNase functions in UI, but the results also reveal unexpected complexity. They found that expressing S-RNase from SI N. alata in SC Nicotiana plumbaginifolia pistils caused the rejection of pollen from Nicotiana tabacum but not pollen from N. plumbaginifolia. However, N. plumbaginifolia pollen was rejected if the S-RNase transgene was crossed into another background and expressed in conjunction with other pistil-side factors. Thus, S-RNase is implicated in rejecting pollen from both N. tabacum and N. plumbaginifolia, but the mechanisms are different because they show different dependencies on genetic background (Murfett et al., 1996). Other studies showed that HT protein is one of the pistil factors required for N. plumbaginifolia pollen rejection (Hancock et al., 2005). Thus, three S-RNase-dependent interspecific pollen rejection mechanisms are distinguished by reliance on modifier genes and pollen-side specificity (Fig. 2). The tomato clade offers additional insights, because its interspecific crossing relationships are better characterized than are Nicotiana’s. Expressing S-RNase and HT genes in SC S. lycopersicum, which normally lacks UI barriers, caused the rejection of pollen from all of the red-fruited SC tomato species but not from any green-fruited tomato species (Tovar-Méndez et al., 2014). Taken together, these studies clearly demonstrate that pistil-side SI and UI barriers share genetic factors. We suggest that they rely on a common genetic toolkit and that multiple UI mechanisms use distinct but overlapping sets of factors. This combinatorial aspect may allow the rejection of pollen from a large, variable, and unpredictable number of species with a limited set of genetic factors.
SI and UI also share genetic factors on the pollen side. Chetelat and Deverna (1991) identified three QTL from S. pennellii that are required for tomato pollen to overcome an S-RNase-dependent UI barrier. Fine-mapping the ui6.1 factor identified a CUL1 gene as a candidate UI factor (Li and Chetelat, 2010). A gain-of-function experiment showed that expressing the S. pennellii allele, SpCUL1, in conjunction with a chromosome 1 S. pennellii introgression, ui1.1, containing the S-locus was sufficient for pollen compatibility on an S-RNase-expressing pistil-side tester (Li and Chetelat, 2010). A loss-of-function study showed that CUL1 also functions in SI (Li and Chetelat, 2014). Recently, the ui1.1 factor was identified as a single SLF gene, SLF-23 (Li and Chetelat, 2015). Thus, SI and UI utilize shared factors for both the pollen- and pistil-side functions and are characterized by a pistil-barrier/pollen-resistance architecture.
DIFFERENCES BETWEEN SI AND UI: ADDITIONAL UI MECHANISMS
Although there is overlap between SI and UI, they are not identical. For instance, there are many exceptions to the SI × SC rule (e.g. SC × SC incompatibilities; Baek et al., 2015), and there are physiological and morphological differences between SI and UI. For example, pollen tube staining of pollinated pistils often shows that interspecific pollen tube rejection occurs closer to the stigma than SI pollen rejection (Fig. 1; Lewis and Crowe, 1958; Martin, 1964; Ascher and Peloquin, 1968; Liedl et al., 1996; Covey et al., 2010). In addition, ultrastructural studies comparing SI and UI in Solanum peruvianum self-pollinations versus cross-pollinations with S. lycopersicum show that the outer (noncallose, largely pectic) pollen tube cell wall appears to thicken during SI rejection and to be degraded during UI rejection (de Nettancourt et al., 1973, 1974).
Crucially, the level of specificity in SI and UI differs dramatically. SI is exquisitely specific and only causes the rejection of pollen with specific S-haplotypes, while UI interspecific pollen rejection causes the rejection of pollen from entire species or clades (Murfett et al., 1996; Beecher et al., 2001; Tovar-Méndez et al., 2014). Lewis and Crowe (1958) noted that pistils with any functional S-locus could inhibit interspecific SC pollen, and they referred to this as unitary action. Modern gain-of-function studies (Murfett et al., 1996; Beecher et al., 2001; Tovar-Méndez et al., 2014) confirm this large difference in specificity. The exact role (or roles) of S-RNase in UI could differ from its role in SI. In UI, for example, it is possible that S-RNases are not involved in pollen recognition per se but rather function downstream in signaling or inhibitory components of the pollen-rejection pathway. This type of system - separate recognition systems coupled to a common downstream pathway - has been proposed in Poaceae (Heslop-Harrison, 1982) and Brassicaceae (Kitashiba and Nasrallah, 2014). In fact, the rapid interspecific pollen tube rejection seen in UI (Fig. 1) may reflect a rapid deployment of S-RNase toxicity based on an alternative recognition system, or it is possible that S-RNase compartmentalization and degradation make different contributions to pollen resistance in SI and interspecific contexts.
Functional studies confirm that interspecific pollen rejection is more complex than SI in the sense that multiple UI mechanisms contribute to compatibility/incompatibility and, in addition, that redundancy is common. Redundancy is apparent in systems where gain-of-function studies show that pollen that is susceptible to S-RNase-dependent rejection (such as the Nicotiana and tomato studies described earlier; Murfett et al., 1996; Beecher et al., 2001; Tovar-Méndez et al., 2014) can also be rejected by SC accessions that entirely lack S-RNase (Martin, 1961; Murfett et al., 1996; Covey et al., 2010; Chalivendra et al., 2013; Tovar-Méndez et al., 2014; Baek et al., 2015; Broz et al., 2017). Redundancy complicates analysis, but the mechanisms can be elucidated through a combination of gain- and loss-of-function studies. It is difficult to understand the maintenance of multiple redundant pollen rejection systems. Laboratory experiments necessarily test a limited number of species on both the pistil and pollen sides. However, in natural contexts, outcrossing species’ pistils are challenged by pollen from many species (Ashman and Arceo-Gómez, 2013); therefore, they may deploy a variety of interspecific pollination barriers that appear to be redundant under laboratory conditions. We suggest that, overall, incompatibility between species can be understood as arising from the additive effects of the complement of pistil barriers and pollen resistance factors expressed in each species. In any case, elucidating these S-RNase-independent mechanisms for interspecific pollen rejection will provide new insights.
The nature and diversity of S-RNase-independent UI mechanisms are just beginning to be understood. Eberle et al. (2012, 2013) used an elegant semi in vivo assay to investigate SC × SC incompatibility between N. tabacum and relatives, such as Nicotiana repanda. They used genetic ablation to create hollow N. tabacum pistils with little or no transmitting tract and showed that the ability to reject pollen from N. repanda also was lost. However, when N. tabacum pistil extracts were introduced, pollen rejection was restored. Fractionation experiments suggested that the N. tabacum PELPIII (Pistil Extensin-Like Protein III) protein is sufficient to cause rejection in this assay (Fig. 2; Eberle et al., 2013). To our knowledge, this is the first example of a specific protein associated with S-RNase-independent pollen rejection.
There are also several examples of S-RNase-independent UI in the tomato clade. Loss of S-RNase expression or enzymatic activity has been documented in at least three SC accessions of otherwise SI species (Kondo et al., 2002; Covey et al., 2010) that nevertheless reject pollen from SC species, such as S. lycopersicum (Liedl et al., 1996; Baek et al., 2015). Chalivendra et al. (2013) showed that UI pollen rejection in one of these accessions (SC S. pennellii LA0716) is developmentally regulated in a manner similar to SI: interspecific pollen is accepted by very young pistils but rejected on mature pistils. This observation is important because it suggests that UI barriers are layered over an otherwise compatible state of the pistil and that UI genes and SI genes likely show similar developmental profiles. A recent study by Pease et al. (2016) exploited a transcriptomic approach to identify candidate UI genes. Among the top 20 genes showing high expression in UI-competent styles, cell wall/cell wall modification (five in the top 20) genes were prominent. One especially intriguing S. pennellii gene encodes a pectin methylesterase inhibitor (PMEI) that appears to be absent in the S. lycopersicum genome. The methylation state of pectin is critical to normal pollen tube growth; demethylated pectins form a rigid gel in the shaft of the pollen tube to allow the growth of pollen tubes through the transmitting tissue of the style, whereas PMEIs are localized specifically to the expandable and flexible tube apex, a structure that can respond to signaling and change directional growth (Bosch et al., 2005; Bosch and Hepler, 2005, 2006; Röckel et al., 2008; Hepler et al., 2013). Mutations in an Arabidopsis PMEI gene, VGD1, result in aberrant pollen tube growth and instability (Jiang et al., 2005), and exogenously adding maize (Zea mays) ZmPMEI1 protein causes the bursting of pollen tubes growing in vitro (Woriedh et al., 2013). Thus, it is plausible that the S. pennellii LA0716 PMEI gene could contribute to UI.
CONCLUSION
Genes that control mate selection control gene flow. The angiosperm pistil is the organ where many mate selection genes act. While it would be unwise to overemphasize one slice of the life cycle, these genes surely bear special importance in the generation of angiosperm diversity. The prevalence of SI systems and the persistence of SI lineages (Goldberg et al., 2010; Goldberg and Igić, 2012) are good evidence that genetic mechanisms controlling mating are adaptive.
Thirty years of research have identified genes that control the specificity of SI as well as some of the underlying physiological mechanisms. Yet to be determined are the biochemical interactions that define S-specificity and the roles of modifier genes that do not contribute to S-specificity per se (see Outstanding Questions). SI research has advanced, but it is only one example of a pollen-pistil interaction that controls mating. At present, we know that SI and interspecific pollination mechanisms share some genes and mechanisms, and it is now possible to clearly determine when they are similar to SI and when they are not. It also should be possible to better understand how the pollen-pistil genetic toolkit is applied to control mating in a broader sense.
Acknowledgments
We thank the IRB Tomato group (Roger Chetelat, Matt Hahn, and Leonie Moyle) for ongoing discussions and Melody Kroll for proofreading the article.
Glossary
- SI
self-incompatibility
- SC
self-compatibility
- UI
unilateral incompatibility
Footnotes
This work was supported by the U.S. National Science Foundation (grant no. MCB1127059).
Articles can be viewed without a subscription.
References
- Anderson MA, McFadden GI, Bernatzky R, Atkinson A, Orpin T, Dedman H, Tregear G, Fernley R, Clarke AE (1989) Sequence variability of three alleles of the self-incompatibility gene of Nicotiana alata. Plant Cell 1: 483–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arceo-Gómez G, Ashman TL (2014) Heterospecific pollen receipt affects self pollen more than outcross pollen: implications for mixed-mating plants. Ecology 95: 2964–2973 [Google Scholar]
- Ascher PD, Peloquin SJ (1968) Pollen tube growth and incompatibility following intra- and inter-specific pollinations in Lilium longiflorum. Am J Bot 55: 1230–1234 [Google Scholar]
- Ashman TL, Arceo-Gómez G (2013) Toward a predictive understanding of the fitness costs of heterospecific pollen receipt and its importance in co-flowering communities. Am J Bot 100: 1061–1070 [DOI] [PubMed] [Google Scholar]
- Baack E, Melo MC, Rieseberg LH, Ortiz-Barrientos D (2015) The origins of reproductive isolation in plants. New Phytol 207: 968–984 [DOI] [PubMed] [Google Scholar]
- Baek YS, Covey PA, Petersen JJ, Chetelat RT, McClure B, Bedinger PA (2015) Testing the SI × SC rule: pollen-pistil interactions in interspecific crosses between members of the tomato clade (Solanum section Lycopersicon, Solanaceae). Am J Bot 102: 302–311 [DOI] [PubMed] [Google Scholar]
- Baker HG. (1955) Self compatibility and establishment after long distance dispersal. Evolution 9: 347–349 [Google Scholar]
- Baker HG. (1967) Support for Baker’s law: as a rule. Evolution 21: 853. [DOI] [PubMed] [Google Scholar]
- Beecher B, Zurek D, McClure B (2001) Effects of RNases on rejection of pollen from Nicotiana tabacum and N. plumbaginifolia. Sex Plant Reprod 14: 69–76 [Google Scholar]
- Bernacchi D, Tanksley SD (1997) An interspecific backcross of Lycopersicon esculentum × L. hirsutum: linkage analysis and a QTL study of sexual compatibility factors and floral traits. Genetics 147: 861–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A, Scossa F, Bolger ME, Lanz C, Maumus F, Tohge T, Quesneville H, Alseekh S, Sørensen I, Lichtenstein G, et al. (2014) The genome of the stress-tolerant wild tomato species Solanum pennellii. Nat Genet 46: 1034–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M, Cheung AY, Hepler PK (2005) Pectin methylesterase, a regulator of pollen tube growth. Plant Physiol 138: 1334–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M, Franklin-Tong VE (2007) Temporal and spatial activation of caspase-like enzymes induced by self-incompatibility in Papaver pollen. Proc Natl Acad Sci USA 104: 18327–18332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M, Franklin-Tong VE (2008) Self-incompatibility in Papaver: signalling to trigger PCD in incompatible pollen. J Exp Bot 59: 481–490 [DOI] [PubMed] [Google Scholar]
- Bosch M, Hepler PK (2005) Pectin methylesterases and pectin dynamics in pollen tubes. Plant Cell 17: 3219–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M, Hepler PK (2006) Silencing of the tobacco pollen pectin methylesterase NtPPME1 results in retarded in vivo pollen tube growth. Planta 223: 736–745 [DOI] [PubMed] [Google Scholar]
- Busot GY, McClure B, Ibarra-Sánchez CP, Jiménez-Durán K, Vázquez-Santana S, Cruz-García F (2008) Pollination in Nicotiana alata stimulates synthesis and transfer to the stigmatic surface of NaStEP, a vacuolar Kunitz proteinase inhibitor homologue. J Exp Bot 59: 3187–3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz AK, Randle AR, Sianta SA, Tovar-Mendez A, McClure B (2017) Mating system transitions in Solanum habrochaites impact interactions between populations and species. New Phytol 213: 440–454 [DOI] [PubMed] [Google Scholar]
- Chalivendra SC, Lopez-Casado G, Kumar A, Kassenbrock AR, Royer S, Tovar-Mèndez A, Covey PA, Dempsey LA, Randle AM, Stack SM, et al. (2013) Developmental onset of reproductive barriers and associated proteome changes in stigma/styles of Solanum pennellii. J Exp Bot 64: 265–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Zhang B, Zhao Z, Sui Z, Zhang H, Xue Y (2010) ‘A life or death decision’ for pollen tubes in S-RNase-based self-incompatibility. J Exp Bot 61: 2027–2037 [DOI] [PubMed] [Google Scholar]
- Chetelat RT, Deverna JW (1991) Expression of unilateral incompatibility in pollen of Lycopersicon pennellii is determined by major loci on chromosomes 1, 6 and 10. Theor Appl Genet 82: 704–712 [DOI] [PubMed] [Google Scholar]
- Cheung AY, Wang H, Wu HM (1995) A floral transmitting tissue-specific glycoprotein attracts pollen tubes and stimulates their growth. Cell 82: 383–393 [DOI] [PubMed] [Google Scholar]
- Covey PA, Kondo K, Welch L, Frank E, Sianta S, Kumar A, Nuñez R, Lopez-Casado G, van der Knaap E, Rose JKC, et al. (2010) Multiple features that distinguish unilateral incongruity and self-incompatibility in the tomato clade. Plant J 64: 367–378 [DOI] [PubMed] [Google Scholar]
- Cruz-Garcia F, Hancock CN, Kim D, McClure B (2005) Stylar glycoproteins bind to S-RNase in vitro. Plant J 42: 295–304 [DOI] [PubMed] [Google Scholar]
- Darwin CR. (1877) The Different Forms of Flowers on Plants of the Same Species. John Murry, London [Google Scholar]
- de Graaf BHJ, Vatovec S, Juárez-Díaz JA, Chai L, Kooblall K, Wilkins KA, Zou H, Forbes T, Franklin FCH, Franklin-Tong VE (2012) The Papaver self-incompatibility pollen S-determinant, PrpS, functions in Arabidopsis thaliana. Curr Biol 22: 154–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nettancourt D. (1997) Incompatibility in angiosperms. Sex Plant Reprod 10: 185–199 [Google Scholar]
- de Nettancourt D. (2001) Incompatibility and Incongruity in Wild and Cultivated Plants, Ed 2. Springer-Verlag, Berlin [Google Scholar]
- de Nettancourt D, Devreux M, Bozzini A, Cresti M, Pacini E, Sarfatti G (1973) Ultrastructural aspects of the self-incompatibility mechanism in Lycopersicum peruvianum Mill. J Cell Sci 12: 403–419 [DOI] [PubMed] [Google Scholar]
- de Nettancourt D, Devreux M, Laneri U, Cresti M, Pacini E, Sarfatti G (1974) Genetical and ultrastructural aspects of self and cross incompatibility in interspecific hybrids between self-compatible Lycopersicum esculentum and self-incompatible L. peruvianum. Theor Appl Genet 44: 278–288 [DOI] [PubMed] [Google Scholar]
- Dwyer KG, Berger MT, Ahmed R, Hritzo MK, McCulloch AA, Price MJ, Serniak NJ, Walsh LT, Nasrallah JB, Nasrallah ME (2013) Molecular characterization and evolution of self-incompatibility genes in Arabidopsis thaliana: the case of the Sc haplotype. Genetics 193: 985–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle CA, Anderson NO, Clasen BM, Hegeman AD, Smith AG (2013) PELPIII: the class III pistil-specific extensin-like Nicotiana tabacum proteins are essential for interspecific incompatibility. Plant J 74: 805–814 [DOI] [PubMed] [Google Scholar]
- Eberle CA, Clasen BM, Anderson NO, Smith AG (2012) A novel pollen tube growth assay utilizing a transmitting tract-ablated Nicotiana tabacum style. Sex Plant Reprod 25: 27–37 [DOI] [PubMed] [Google Scholar]
- Endress PK. (2011) Evolutionary diversification of the flowers in angiosperms. Am J Bot 98: 370–396 [DOI] [PubMed] [Google Scholar]
- Endress PK. (2015) Patterns of angiospermy development before carpel sealing across living angiosperms: diversity, and morphological and systematic aspects. Bot J Linn Soc 178: 556–591 [Google Scholar]
- Endress PK, Doyle JA (2009) Reconstructing the ancestral angiosperm flower and its initial specializations. Am J Bot 96: 22–66 [DOI] [PubMed] [Google Scholar]
- Entani T, Kubo K, Isogai S, Fukao Y, Shirakawa M, Isogai A, Takayama S (2014) Ubiquitin-proteasome-mediated degradation of S-RNase in a Solanaceous cross-compatibility reaction. Plant J 78: 1014–1021 [DOI] [PubMed] [Google Scholar]
- Fishman L, Aagaard J, Tuthill JC (2008) Toward the evolutionary genomics of gametophytic divergence: patterns of transmission ratio distortion in monkeyflower (Mimulus) hybrids reveal a complex genetic basis for conspecific pollen precedence. Evolution 62: 2958–2970 [DOI] [PubMed] [Google Scholar]
- Foote HCC, Ride JP, Franklin-Tong VE, Walker EA, Lawrence MJ, Franklin FCH (1994) Cloning and expression of a distinctive class of self-incompatibility (S) gene from Papaver rhoeas L. Proc Natl Acad Sci USA 91: 2265–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin-Tong VE. (2008) Self-incompatibility in Papaver rhoeas: progress in understanding mechanisms involved in regulating self-incompatibility in Papaver. In VE Franklin-Tong, ed, Self-Incompatibility in Flowering Plants: Evolution, Diversity, and Mechanisms. Springer-Verlag, Berlin, pp 237–258 [Google Scholar]
- Franklin-Tong VE, Lawrence MJ, Franklin FCH (1988) An in vitro bioassay for the stigmatic product of the self-incompatibility gene in Papaver rhoeas L. New Phytol 110: 109–118 [Google Scholar]
- Gilmartin PM, Li J (2010) Homing in on heterostyly. Heredity (Edinb) 105: 161–162 [DOI] [PubMed] [Google Scholar]
- Goldberg EE, Igić B (2012) Tempo and mode in plant breeding system evolution. Evolution 66: 3701–3709 [DOI] [PubMed] [Google Scholar]
- Goldberg EE, Kohn JR, Lande R, Robertson KA, Smith SA, Igić B (2010) Species selection maintains self-incompatibility. Science 330: 493–495 [DOI] [PubMed] [Google Scholar]
- Goldraij A, Kondo K, Lee CB, Hancock CN, Sivaguru M, Vazquez-Santana S, Kim S, Phillips TE, Cruz-Garcia F, McClure B (2006) Compartmentalization of S-RNase and HT-B degradation in self-incompatible Nicotiana. Nature 439: 805–810 [DOI] [PubMed] [Google Scholar]
- Gray JE, McClure BA, Bönig I, Anderson MA, Clarke AE (1991) Action of the style product of the self-incompatibility gene of Nicotiana alata (S-RNase) on in vitro-grown pollen tubes. Plant Cell 3: 271–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock CN, Kent L, McClure BA (2005) The stylar 120 kDa glycoprotein is required for S-specific pollen rejection in Nicotiana. Plant J 43: 716–723 [DOI] [PubMed] [Google Scholar]
- Hepler PK, Rounds CM, Winship LJ (2013) Control of cell wall extensibility during pollen tube growth. Mol Plant 6: 998–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslop-Harrison J. (1982) Pollen-stigma interaction and cross-incompatibility in the grasses. Science 215: 1358–1364 [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison Y. (2000) Control gates and micro-ecology: the pollen-stigma interaction in perspective. Ann Bot (Lond) 85: 5–13 [Google Scholar]
- Hiscock SJ, Dickinson HG (1993) Unilateral incompatibility within the Brassicaceae: further evidence for the involvement of the self-incompatibility (S)-locus. Theor Appl Genet 86: 744–753 [DOI] [PubMed] [Google Scholar]
- Hogenboom NG. (1984) Incongruity: non-functioning of intercellular and intracellular partner relationships through non-matching information. In Linskens HF, Heslop-Harrison J, eds, Encyclopedia of Plant Physiology. Vol 17. Cellular Interactions. Springer-Verlag, Berlin, pp 640–654 [Google Scholar]
- Howard DJ. (1999) Conspecific sperm and pollen precedence and speciation. Annu Rev Ecol Syst 30: 109–132 [Google Scholar]
- Huang J, Zhao L, Yang Q, Xue Y (2006) AhSSK1, a novel SKP1-like protein that interacts with the S-locus F-box protein SLF. Plant J 46: 780–793 [DOI] [PubMed] [Google Scholar]
- Huang S, Lee HS, Karunanandaa B, Kao TH (1994) Ribonuclease activity of Petunia inflata S proteins is essential for rejection of self-pollen. Plant Cell 6: 1021–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indriolo E, Goring DR (2014) A conserved role for the ARC1 E3 ligase in Brassicaceae self-incompatibility. Front Plant Sci 5: 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioerger TR, Gohlke JR, Xu B, Kao T (1991) Primary structural features of the self-incompatibility protein in Solanaceae. Sex Plant Reprod 4: 81–87 [Google Scholar]
- Iwano M, Ito K, Fujii S, Kakita M, Asano-Shimosato H, Igarashi M, Kaothien-Nakayama P, Entani T, Kanatani A, Takehisa M, et al. (2015) Calcium signalling mediates self-incompatibility response in the Brassicaceae. Nat Plants 1: 15128. [DOI] [PubMed] [Google Scholar]
- Iwano M, Takayama S (2012) Self/non-self discrimination in angiosperm self-incompatibility. Curr Opin Plant Biol 15: 78–83 [DOI] [PubMed] [Google Scholar]
- Jiang L, Yang SL, Xie LF, Puah CS, Zhang XQ, Yang WC, Sundaresan V, Ye D (2005) VANGUARD1 encodes a pectin methylesterase that enhances pollen tube growth in the Arabidopsis style and transmitting tract. Plant Cell 17: 584–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Durán K, McClure B, García-Campusano F, Rodríguez-Sotres R, Cisneros J, Busot G, Cruz-García F (2013) NaStEP: a proteinase inhibitor essential to self-incompatibility and a positive regulator of HT-B stability in Nicotiana alata pollen tubes. Plant Physiol 161: 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaoka MM, Higashiyama T (2015) Peptide signaling in pollen tube guidance. Curr Opin Plant Biol 28: 127–136 [DOI] [PubMed] [Google Scholar]
- Kao TH, McCubbin AG (1996) How flowering plants discriminate between self and non-self pollen to prevent inbreeding. Proc Natl Acad Sci USA 93: 12059–12065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawata Y, Sakiyama F, Tamaoki H (1988) Amino-acid sequence of ribonuclease T2 from Aspergillus oryzae. Eur J Biochem 176: 683–697 [DOI] [PubMed] [Google Scholar]
- Kitashiba H, Nasrallah JB (2014) Self-incompatibility in Brassicaceae crops: lessons for interspecific incompatibility. Breed Sci 64: 23–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo K, Yamamoto M, Itahashi R, Sato T, Egashira H, Hattori T, Kowyama Y (2002) Insights into the evolution of self-compatibility in Lycopersicon from a study of stylar factors. Plant J 30: 143–153 [DOI] [PubMed] [Google Scholar]
- Kowyama Y, Kunz C, Lewis I, Newbigin E, Clarke AE, Anderson MA (1994) Self-compatibility in a Lycopersicon peruvianum variant (LA2157) is associated with a lack of style S-RNase activity. Theor Appl Genet 88: 859–864 [DOI] [PubMed] [Google Scholar]
- Kubo K, Entani T, Takara A, Wang N, Fields AM, Hua Z, Toyoda M, Kawashima S, Ando T, Isogai A, et al. (2010) Collaborative non-self recognition system in S-RNase-based self-incompatibility. Science 330: 796–799 [DOI] [PubMed] [Google Scholar]
- Kubo K, Paape T, Hatakeyama M, Entani T, Takara A, Kajihara K, Tsukahara M, Shimizu-Inatsugi R, Shimizu KK, Takayama S (2015) Gene duplication and genetic exchange drive the evolution of S-RNase-based self-incompatibility in Petunia. Nat Plants 1: 14005. [DOI] [PubMed] [Google Scholar]
- Lai Z, Ma W, Han B, Liang L, Zhang Y, Hong G, Xue Y (2002) An F-box gene linked to the self-incompatibility (S) locus of Antirrhinum is expressed specifically in pollen and tapetum. Plant Mol Biol 50: 29–42 [DOI] [PubMed] [Google Scholar]
- Lewis D, Crowe LK (1958) Unilateral interspecific incompatibility in flowering plants. Heredity 12: 233–256 [Google Scholar]
- Li S, Sun P, Williams JS, Kao TH (2014) Identification of the self-incompatibility locus F-box protein-containing complex in Petunia inflata. Plant Reprod 27: 31–45 [DOI] [PubMed] [Google Scholar]
- Li W, Chetelat RT (2010) A pollen factor linking inter- and intraspecific pollen rejection in tomato. Science 330: 1827–1830 [DOI] [PubMed] [Google Scholar]
- Li W, Chetelat RT (2014) The role of a pollen-expressed Cullin1 protein in gametophytic self-incompatibility in Solanum. Genetics 196: 439–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Chetelat RT (2015) Unilateral incompatibility gene ui1.1 encodes an S-locus F-box protein expressed in pollen of Solanum species. Proc Natl Acad Sci USA 112: 4417–4422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liedl BE, McCormick S, Mutschler MA (1996) Unilateral incongruity in crosses involving Lycopersicon pennellii and L. esculentum is distinct from self-incompatibility in expression, timing and location. Sex Plant Reprod 9: 299–308 [Google Scholar]
- Lin Z, Eaves DJ, Sanchez-Moran E, Franklin FCH, Franklin-Tong VE (2015) The Papaver rhoeas S determinants confer self-incompatibility to Arabidopsis thaliana in planta. Science 350: 684–687 [DOI] [PubMed] [Google Scholar]
- Lind JL, Bacic A, Clarke AE, Anderson MA (1994) A style-specific hydroxyproline-rich glycoprotein with properties of both extensins and arabinogalactan proteins. Plant J 6: 491–502 [DOI] [PubMed] [Google Scholar]
- Lind JL, Bönig I, Clarke AE, Anderson MA (1996) A style-specific 120 kDa glycoprotein enters pollen tubes of Nicotiana alata in vivo. Sex Plant Reprod 9: 75–86 [Google Scholar]
- Liu ZQ, Xu GH, Zhang SL (2007) Pyrus pyrifolia stylar S-RNase induces alterations in the actin cytoskeleton in self-pollen and tubes in vitro. Protoplasma 232: 61–67 [DOI] [PubMed] [Google Scholar]
- Lora J, Hormaza JI, Herrero M (2016) The diversity of the pollen tube pathway in plants: toward an increasing control by the sporophyte. Front Plant Sci 7: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry DB, Rockwood RC, Willis JH (2008) Ecological reproductive isolation of coast and inland races of Mimulus guttatus. Evolution 62: 2196–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin FW. (1961) The inheritance of self-incompatibility in hybrids of Lycopersicon esculentum Mill. × L. chilense Dun. Genetics 46: 1443–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin FW. (1964) The inheritance of unilateral incompatibility in Lycopersicum hirsutum. Genetics 50: 459–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matton DP, Luu DT, Xike Q, Laublin G, O’Brien M, Maes O, Morse D, Cappadocia M (1999) Production of an S RNase with dual specificity suggests a novel hypothesis for the generation of new S alleles. Plant Cell 11: 2087–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matton DP, Maes O, Laublin G, Xike Q, Bertrand C, Morse D, Cappadocia M (1997) Hypervariable domains of self-incompatibility RNases mediate allele-specific pollen recognition. Plant Cell 9: 1757–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure B. (2009) Darwin’s foundation for investigating self-incompatibility and the progress toward a physiological model for S-RNase-based SI. J Exp Bot 60: 1069–1081 [DOI] [PubMed] [Google Scholar]
- McClure B, Mou B, Canevascini S, Bernatzky R (1999) A small asparagine-rich protein required for S-allele-specific pollen rejection in Nicotiana. Proc Natl Acad Sci USA 96: 13548–13553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure BA, Cruz-Garcia F, Beecher BS, Sulaman W (2000) Factors affecting inter- and intra-specific pollen rejection in Nicotiana. Ann Bot 85: 113–123 [Google Scholar]
- McClure BA, Gray JE, Anderson MA, Clarke AE (1990) Self-incompatibility in Nicotiana alata involves degradation of pollen rRNA. Nature 347: 757–760 [Google Scholar]
- McClure BA, Haring V, Ebert PR, Anderson MA, Simpson RJ, Sakiyama F, Clarke AE (1989) Style self-incompatibility gene products of Nicotiana alata are ribonucleases. Nature 342: 955–957 [DOI] [PubMed] [Google Scholar]
- McGuire DC, Rick CM (1954) Self-incompatibility in species of Lycopersicon sect. Eriopersicon and hybrids with L. esculentum. Hilgardia 23: 101–124 [Google Scholar]
- Mulcahy DL, Mulcahy GB (1988) Test of the heterosis model. Sex Plant Reprod 1: 32–35 [Google Scholar]
- Murfett J, Strabala TJ, Zurek DM, Mou B, Beecher B, McClure BA (1996) S RNase and interspecific pollen rejection in the genus Nicotiana: multiple pollen rejection pathways contribute to unilateral incompatibility between self-incompatible and self-compatible species. Plant Cell 8: 943–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrallah JB, Kao TH, Goldberg ML, Nasrallah ME (1985) A cDNA clone encoding an S-locus-specific glycoprotein from Brassica oleracea. Nature 318: 263–267 [Google Scholar]
- Nasrallah JB, Nasrallah ME (2014) S-Locus receptor kinase signalling. Biochem Soc Trans 42: 313–319 [DOI] [PubMed] [Google Scholar]
- Nasrallah ME, Wallace DH (1967) Immunochemical detection of antigens in self-incompatibility genotypes of cabbage. Nature 213: 700–701 [Google Scholar]
- O’Brien M, Kapfer C, Major G, Laurin M, Bertrand C, Kondo K, Kowyama Y, Matton DP (2002) Molecular analysis of the stylar-expressed Solanum chacoense small asparagine-rich protein family related to the HT modifier of gametophytic self-incompatibility in Nicotiana. Plant J 32: 985–996 [DOI] [PubMed] [Google Scholar]
- Okuda S, Tsutsui H, Shiina K, Sprunck S, Takeuchi H, Yui R, Kasahara RD, Hamamura Y, Mizukami A, Susaki D, et al. (2009) Defensin-like polypeptide LUREs are pollen tube attractants secreted from synergid cells. Nature 458: 357–361 [DOI] [PubMed] [Google Scholar]
- Palanivelu R, Preuss D (2006) Distinct short-range ovule signals attract or repel Arabidopsis thaliana pollen tubes in vitro. BMC Plant Biol 6: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannell JR, Auld JR, Brandvain Y, Burd M, Busch JW, Cheptou PO, Conner JK, Goldberg EE, Grant AG, Grossenbacher DL, et al. (2015) The scope of Baker’s law. New Phytol 208: 656–667 [DOI] [PubMed] [Google Scholar]
- Pease JB, Guerrero RF, Sherman NA, Hahn MW, Moyle LC (2016) Molecular mechanisms of postmating prezygotic reproductive isolation uncovered by transcriptome analysis. Mol Ecol 25: 2592–2608 [DOI] [PubMed] [Google Scholar]
- Pinheiro F, Cafasso D, Cozzolino S, Scopece G (2015) Transitions between self-compatibility and self-incompatibility and the evolution of reproductive isolation in the large and diverse tropical genus Dendrobium (Orchidacae). Ann Bot (Lond) 116: 457–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puerta AR, Ushijima K, Koba T, Sassa H (2009) Identification and functional analysis of pistil self-incompatibility factor HT-B of Petunia. J Exp Bot 60: 1309–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H, Wang H, Zhao L, Zhou J, Huang J, Zhang Y, Xue Y (2004) The F-box protein AhSLF-S2 physically interacts with S-RNases that may be inhibited by the ubiquitin/26S proteasome pathway of protein degradation during compatible pollination in Antirrhinum. Plant Cell 16: 582–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röckel N, Wolf S, Kost B, Rausch T, Greiner S (2008) Elaborate spatial patterning of cell-wall PME and PMEI at the pollen tube tip involves PMEI endocytosis, and reflects the distribution of esterified and de-esterified pectins. Plant J 53: 133–143 [DOI] [PubMed] [Google Scholar]
- Roldán JA, Rojas HJ, Goldraij A (2012) Disorganization of F-actin cytoskeleton precedes vacuolar disruption in pollen tubes during the in vivo self-incompatibility response in Nicotiana alata. Ann Bot (Lond) 110: 787–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safavian D, Goring DR (2013) Secretory activity is rapidly induced in stigmatic papillae by compatible pollen, but inhibited for self-incompatible pollen in the Brassicaceae. PLoS ONE 8: e84286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson DR. (1962) Intergeneric pollen-stigma incompatibility in Cruciferae. Can J Genet Cytol 4: 38–49 [Google Scholar]
- Sassa H. (2016) Molecular mechanism of the S-RNase-based gametophytic self-incompatibility in fruit trees of Rosaceae. Breed Sci 66: 116–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopfer CR, Nasrallah ME, Nasrallah JB (1999) The male determinant of self-incompatibility in Brassica. Science 286: 1697–1700 [DOI] [PubMed] [Google Scholar]
- Sijacic P, Wang X, Skirpan AL, Wang Y, Dowd PE, McCubbin AG, Huang S, Kao TH (2004) Identification of the pollen determinant of S-RNase-mediated self-incompatibility. Nature 429: 302–305 [DOI] [PubMed] [Google Scholar]
- Soulard J, Qin X, Boivin N, Morse D, Cappadocia M (2013) A new dual-specific incompatibility allele revealed by absence of glycosylation in the conserved C2 site of a Solanum chacoense S-RNase. J Exp Bot 64: 1995–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein JC, Nasrallah JB (1993) A plant receptor-like gene, the S-locus receptor kinase of Brassica oleracea L. encodes a functional serine-threonine kinase. Plant Physiol 101: 1103–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada Y, Sato T, Suzuki G, Shiba H, Takayama S, Watanabe M (2013) Involvement of MLPK pathway in intraspecies unilateral incompatibility regulated by a single locus with stigma and pollen factors. G3 (Bethesda) 3: 719–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S, Isogai A (2005) Self-incompatibility in plants. Annu Rev Plant Biol 56: 467–489 [DOI] [PubMed] [Google Scholar]
- Takayama S, Shiba H, Iwano M, Shimosato H, Che FS, Kai N, Watanabe M, Suzuki G, Hinata K, Isogai A (2000) The pollen determinant of self-incompatibility in Brassica campestris. Proc Natl Acad Sci USA 97: 1920–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao R, Iezzoni AF (2010) The S-RNase-based gametophytic self-incompatibility system in Prunus exhibits distinct genetic and molecular features. Sci Hortic (Amsterdam) 124: 423–433 [Google Scholar]
- Tovar-Méndez A, Kumar A, Kondo K, Ashford A, Baek YS, Welch L, Bedinger PA, McClure BA (2014) Restoring pistil-side self-incompatibility factors recapitulates an interspecific reproductive barrier between tomato species. Plant J 77: 727–736 [DOI] [PubMed] [Google Scholar]
- Wang CL, Wu J, Xu GH, Gao YB, Chen G, Wu JY, Wu HQ, Zhang SL (2010) S-RNase disrupts tip-localized reactive oxygen species and induces nuclear DNA degradation in incompatible pollen tubes of Pyrus pyrifolia. J Cell Sci 123: 4301–4309 [DOI] [PubMed] [Google Scholar]
- Wang CL, Xu GH, Jiang XT, Chen G, Wu J, Wu HQ, Zhang SL (2009) S-RNase triggers mitochondrial alteration and DNA degradation in the incompatible pollen tube of Pyrus pyrifolia in vitro. Plant J 57: 220–229 [DOI] [PubMed] [Google Scholar]
- Wheeler MJ, Vatovec S, Franklin-Tong VE (2010) The pollen S-determinant in Papaver: comparisons with known plant receptors and protein ligand partners. J Exp Bot 61: 2015–2025 [DOI] [PubMed] [Google Scholar]
- Widmer A, Lexer C, Cozzolino S (2009) Evolution of reproductive isolation in plants. Heredity (Edinb) 102: 31–38 [DOI] [PubMed] [Google Scholar]
- Wilkins KA, Bosch M, Haque T, Teng N, Poulter NS, Franklin-Tong VE (2015) Self-incompatibility-induced programmed cell death in field poppy pollen involves dramatic acidification of the incompatible pollen tube cytosol. Plant Physiol 167: 766–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins KA, Poulter NS, Franklin-Tong VE (2014) Taking one for the team: self-recognition and cell suicide in pollen. J Exp Bot 65: 1331–1342 [DOI] [PubMed] [Google Scholar]
- Williams JH. (2012) Pollen tube growth rates and the diversification of flowering plant reproductive cycles. Int J Plant Sci 173: 649–661 [Google Scholar]
- Williams JS, Der JP, dePamphilis CW, Kao TH (2014a) Transcriptome analysis reveals the same 17 S-locus F-box genes in two haplotypes of the self-incompatibility locus of Petunia inflata. Plant Cell 26: 2873–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JS, Natale CA, Wang N, Li S, Brubaker TR, Sun P, Kao TH (2014b) Four previously identified Petunia inflata S-locus F-box genes are involved in pollen specificity in self-incompatibility. Mol Plant 7: 567–569 [DOI] [PubMed] [Google Scholar]
- Williams JS, Wu L, Li S, Sun P, Kao TH (2015) Insight into S-RNase-based self-incompatibility in Petunia: recent findings and future directions. Front Plant Sci 6: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woriedh M, Wolf S, Márton ML, Hinze A, Gahrtz M, Becker D, Dresselhaus T (2013) External application of gametophyte-specific ZmPMEI1 induces pollen tube burst in maize. Plant Reprod 26: 255–266 [DOI] [PubMed] [Google Scholar]
- Xue Y, Carpenter R, Dickinson HG, Coen ES (1996) Origin of allelic diversity in Antirrhinum S locus RNases. Plant Cell 8: 805–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Huang J, Zhao Z, Li Q, Sims TL, Xue Y (2010) The Skp1-like protein SSK1 is required for cross-pollen compatibility in S-RNase-based self-incompatibility. Plant J 62: 52–63 [DOI] [PubMed] [Google Scholar]
- Zurek DM, Mou B, Beecher B, McClure B (1997) Exchanging sequence domains between S-RNases from Nicotiana alata disrupts pollen recognition. Plant J 11: 797–808 [DOI] [PubMed] [Google Scholar]