A metabolic network-guided genome-wide association study of seed free amino acids facilitates the identification of a histidine-specific transporter in Arabidopsis.
Abstract
Amino acids are essential for proper growth and development in plants. Amino acids serve as building blocks for proteins but also are important for responses to stress and the biosynthesis of numerous essential compounds. In seed, the pool of free amino acids (FAAs) also contributes to alternative energy, desiccation, and seed vigor; thus, manipulating FAA levels can significantly impact a seed’s nutritional qualities. While genome-wide association studies (GWAS) on branched-chain amino acids have identified some regulatory genes controlling seed FAAs, the genetic regulation of FAA levels, composition, and homeostasis in seeds remains mostly unresolved. Hence, we performed GWAS on 18 FAAs from a 313-ecotype Arabidopsis (Arabidopsis thaliana) association panel. Specifically, GWAS was performed on 98 traits derived from known amino acid metabolic pathways (approach 1) and then on 92 traits generated from an unbiased correlation-based metabolic network analysis (approach 2), and the results were compared. The latter approach facilitated the discovery of additional novel metabolic interactions and single-nucleotide polymorphism-trait associations not identified by the former approach. The most prominent network-guided GWAS signal was for a histidine (His)-related trait in a region containing two genes: a cationic amino acid transporter (CAT4) and a polynucleotide phosphorylase resistant to inhibition with fosmidomycin. A reverse genetics approach confirmed CAT4 to be responsible for the natural variation of His-related traits across the association panel. Given that His is a semiessential amino acid and a potent metal chelator, CAT4 orthologs could be considered as candidate genes for seed quality biofortification in crop plants.
Free amino acids (FAAs) play a pivotal role in the central metabolism of plants. FAAs serve as building blocks for protein synthesis and also are precursors for osmolytes, alternative energy, hormones, and key secondary metabolites (Rai, 2002; Araújo et al., 2010; Tzin and Galili, 2010; Angelovici et al., 2011). Studies of both developing and germinating seeds also have implicated FAAs in proper seed development and germination (Angelovici et al., 2010; Galili and Amir, 2013). Still, the genetic control of FAA metabolism in seed remains poorly understood. One reason is that FAA metabolism is tightly intertwined with essential cellular processes in plants, and manipulating FAA levels can have strong deleterious, pleiotropic effects on the entire system (Guyer et al., 1995; Galili, 2011; Ingle, 2011; Pratelli and Pilot, 2014). For example, increasing Lys and Thr inhibits the activity of Asp kinase via a feedback inhibition loop (Clark and Lu, 2015). At high concentrations, this inhibition can lead to starvation of a downstream metabolic product, Met, which inhibits plant growth (Bright et al., 1982; Rognes et al., 1983; Heremans and Jacobs, 1995). In addition, several amino acid catabolic products, such as those emanating from branched-chain amino acids (BCAAs) or Lys degradation pathways, can be channeled toward cellular energy production under both standard and stress growth conditions (Angelovici et al., 2009, 2011; Araújo et al., 2010; Peng et al., 2015).
Reconstructions of seed metabolic networks in several model species and tissues have offered important insights into these underlying metabolic interactions and regulation (Lu et al., 2008; Toubiana et al., 2013, 2015). For example, a correlation-based network metabolic reconstruction of FAA levels revealed that individual FAAs are strongly correlated during seed development compared with leaf or fruit development (Toubiana et al., 2012), which suggests a much tighter interaction of this metabolic network in seeds. Similarly, two correlation-based network analyses of more than 50 primary metabolite profiles spanning late seed maturation and desiccation demonstrated that FAAs form a highly interconnected metabolic cluster with only positive correlations (Angelovici et al., 2009; Toubiana et al., 2012). Interestingly, studies also suggest that the latent dynamics of the metabolic network are strongly connected to the underlying reaction pathway structure (Steuer et al., 2003; Weckwerth, 2003). Weckwerth et al. (2004) demonstrated that the correlation network properties of metabolites better capture the systemic responses to genetic alterations than do changes to metabolic levels (Weckwerth et al., 2004). In addition, Gu et al. (2010) found that knocking out mitochondrial isovaleryl-CoA dehydrogenase, an enzyme in the Leu degradation pathway, results in a significant increase in 12 FAAs in a seed-specific manner (Gu et al., 2010). Taken together, these findings suggest that the inherent coordination of seed FAAs could be used as a tool to further unravel the genetic basis of quantitative traits, such as amino acids.
Forward genetic approaches, such as linkage analysis, have proven to be powerful for identifying quantitative trait loci (QTLs) that control phenotypic variation for phenological and metabolic traits, such as flowering time and levels of carotenoids, tocochromanols, and amino acids in seed (Kowalski et al., 1994; Alonso-Blanco et al., 1998; Yan et al., 2000; Wong et al., 2004; Wentzell et al., 2007; Chander et al., 2008; Balasubramanian et al., 2009; Vallabhaneni and Wurtzel, 2009; Gutiérrez-Rojas et al., 2010; Maloney et al., 2010; Kochevenko and Fernie, 2011). Nevertheless, traditional linkage analysis to identify QTLs in biparental recombinant inbred line (RIL) populations has two major weaknesses: it captures narrow levels of allelic diversity as a result of only two parental lines, and it provides low mapping resolution due to the overall limited number of recombination events that occur while constructing the RIL population (Yu and Buckler, 2006; Korte and Farlow, 2013). While both the extent of allelic diversity and the density of recombination break points can be enhanced through the construction of multiparent advanced generation intercross populations (Balasubramanian et al., 2009; Kover et al., 2009), the resulting mapping populations from such a time-consuming effort would not reflect the frequencies and combinations of alleles found in the natural population (Weigel, 2012). In recent years, a number of genome-wide association studies (GWAS) have exploited historical recombination events in large association panels of unrelated individuals assembled to capture the phenotypic variability of a wide range of complex traits, allowing it to offer higher mapping resolution of causal loci (Atwell et al., 2010; Huang et al., 2011; Li et al., 2012; Ramstein et al., 2015; Scossa et al., 2016). Although it has its own set of limitations, such as possible spurious associations due to cryptic population structure and multiple levels of relatedness, GWAS often overcomes the two major weaknesses inherent to QTL detection in RIL populations (Pritchard and Donnelly, 2001; Yu and Buckler, 2006; Platt et al., 2010a, 2010b; Trontin et al., 2011; Korte and Farlow, 2013). As such, GWAS has been employed successfully to better resolve the genetic basis of many primary and secondary metabolites (e.g. carotenoids, tocochromanols, glucosinolates, and organic acids) in several model systems, including rice (Oryza sativa), maize (Zea mays), and Arabidopsis (Arabidopsis thaliana; Chan et al., 2011; Riedelsheimer et al., 2012; Gonzalez-Jorge et al., 2013, 2016; Lipka et al., 2013; Chen et al., 2014; Owens et al., 2014; Verslues et al., 2014). In several cases, GWAS has confirmed genes and their orthologs identified via mutant screens, metabolic QTLs, and other methods (Clarke et al., 1995; Wong et al., 2004; Vallabhaneni and Wurtzel, 2009; Yan et al., 2010; Wurtzel et al., 2012). Recently, several GWAS have successfully identified genes involved in the regulation of the absolute levels of BCAAs, Pro, Lys, and Tyr, in both maize and Arabidopsis (Riedelsheimer et al., 2012; Angelovici et al., 2013; Verslues et al., 2014). Although effective at detecting large-effect genes, GWAS have been less successful at identifying rare variants with large effects or many common variants with small effects (Trontin et al., 2011; Korte and Farlow, 2013). The latter is often due to the low statistical power of most currently available association panels (Aranzana et al., 2005; Nordborg et al., 2005). Several approaches have been developed to enhance the detection of small-effect genes, including one that uses coexpression networks to prioritize multiple GWAS candidates (Chan et al., 2011).
It was demonstrated previously that derived traits generated from absolute levels of metabolites can provide unique insights into a metabolic network. These traits include, for example, the sum of related metabolites or the ratio of two related metabolites, such as precursors or products (Sauer et al., 1999; Weckwerth et al., 2004; Wentzell et al., 2007). With both linkage analysis and GWAS, derived traits that represent ratios based on metabolic pathways or known interactions have generated more significant associations compared with associations from absolute levels of metabolites (Wentzell et al., 2007; Vallabhaneni and Wurtzel, 2009; Wurtzel et al., 2012; Angelovici et al., 2013; Gonzalez-Jorge et al., 2013; Lipka et al., 2013; Owens et al., 2014). One explanation for this phenomenon could be the higher heritability of metabolic ratios compared with content traits (Wentzell et al., 2007). A recent GWAS of Arabidopsis seed BCAA-related traits found that the ratio of Ile versus the BCAA family (i.e., Ile, Leu, and Val) or even Ile versus total amino acids has a stronger association by several orders of magnitude compared with absolute levels of Ile (Angelovici et al., 2013). GWAS of the ratio of δ-tocotrienol to the sum of γ- and α-tocotrienols in maize grain also found a more highly significant association with a known tocochromanol biosynthesis gene than any of the absolute tocotrienol levels (Lipka et al., 2013). Lastly, a joint-linkage-assisted GWAS of the ratio of two homoterpenes in the maize nested association mapping panel facilitated the identification of a new cytochrome 450 gene involved in the pathway (Richter et al., 2016).
Here, we report and compare findings from GWAS performed on a broad range of biochemically based and network-derived FAA ratios calculated from a previous quantification of 18 FAAs in an Arabidopsis association panel (Angelovici et al., 2013). These metabolic ratios integrate both the extensive biochemical knowledge of amino acid pathways and the inherent coordinated behaviors of FAAs in seeds as deduced from a correlation-based network topology. This integrated approach uncovered novel metabolic clusters and the genes that regulate them. An in-depth molecular analysis of a significantly associated genomic region, pinpointed using this network-guided GWAS, facilitated the identification of a seed-specific His regulator not detected by GWAS either with the absolute levels of FAA in dry seeds or with calculated metabolic ratios based on biochemical affiliation. This study supports the potential of the network-guided GWAS approach to elucidate the genetic basis of a complex coordinated metabolic network, such as the FAA network.
RESULTS
Characterization of the Natural Variation of Free Amino Acids and Their Relationships in Dry Seeds
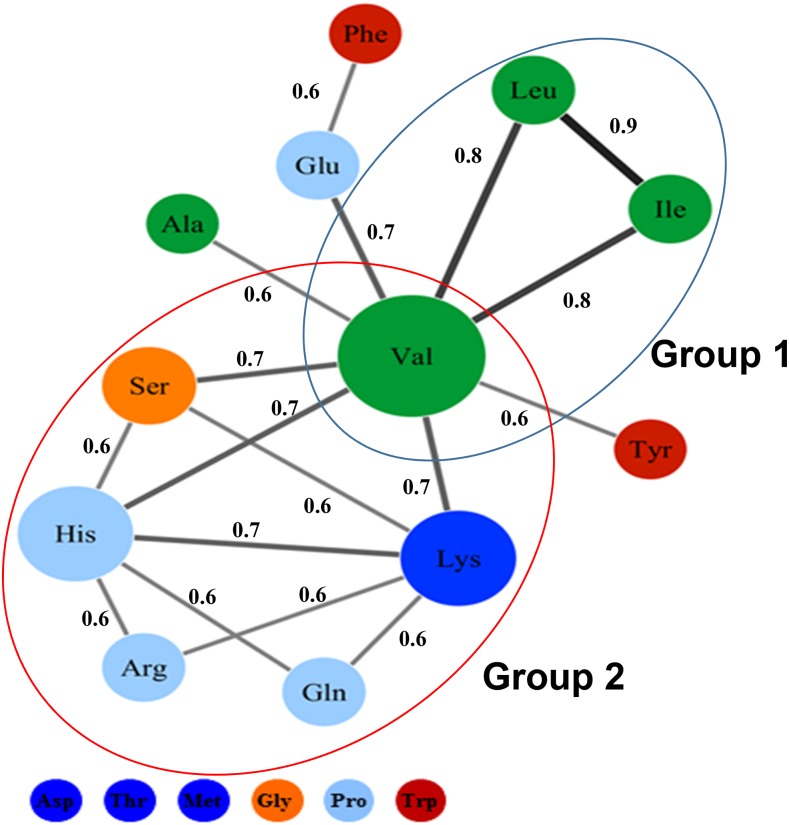
We previously performed a comprehensive analysis of 18 FAA levels quantified from three biological replicates of a 313-accession Arabidopsis diversity panel (Angelovici et al., 2013; Supplemental Data Set S1). Across the diversity panel, Glu and Asp are the most abundant amino acids, and Met and His are the least abundant, with average relative compositions of 37%, 20%, 0.63%, and 0.96%, respectively (Table I). Met and Ile vary the most across the panel (range greater than 4-fold), and Thr, Trp, Lys, and Glu vary the least (range less than 2-fold; Table I). Broad-sense heritabilities of the absolute levels of FAAs are 0.5 to 0.8 for most amino acids except Thr and Trp, which are 0.11 and 0.24, respectively (Table I). We performed a correlation-based network analysis to evaluate the relationships among the absolute levels of the 18 FAAs. Pairwise correlation coefficients among the 18 FAAs’ back-transformed best linear unbiased predictors (BLUPs) were calculated using the Spearman’s rank correlation method, and the results were visualized as a network of nodes and edges (Fig. 1; Supplemental Table S1). Each node in the network represents an amino acid, and each edge/line represents a correlation-based significant relationship between a pair of nodes (P < 0.001, rsp ≥ 0.6). The network consists of 12 connected amino acids, and all correlation coefficients are positive. Interestingly, the connectivity of Glu is relatively low despite its abundance. Ala, Phe, and Tyr also have low connectivity. Asp, Thr, Met, Gly, Pro, and Trp are disconnected from the network. The network topology also indicates two highly interconnected groups. Group 1 includes the BCAA family (i.e. Ile, Leu, and Val; Fig. 1), which is consistent with previous studies (Binder, 2010; Angelovici et al., 2013). Group 2 is novel and includes six amino acids from four amino acid families: Ser, Gln, Arg and His, Val, and Lys (Fig. 1). Val has the highest number of connections (nodal degree) and the highest hub score, followed by His, Ser, and Lys, indicating its centrality to the network (Supplemental Table S1B).
Table I. Mean, relative composition, mean range, and broad sense heritability of 18 FAA absolute levels in dry seeds.
| Trait | Back-Transformed BLUPs |
Broad Sense Heritability | se | |||||
|---|---|---|---|---|---|---|---|---|
| Mean | se | Relative Composition | se | Mean Range |
||||
| Minimum | Maximum | |||||||
| nmol mg seed−1 | % of total | nmol mg seed−1 | ||||||
| Met | 0.08 | 0.001 | 0.63 | 0.01 | 0.04 | 0.18 | 0.80 | 0.02 |
| His | 0.13 | 0.001 | 0.96 | 0.01 | 0.08 | 0.26 | 0.60 | 0.04 |
| Ile | 0.15 | 0.002 | 1.17 | 0.01 | 0.07 | 0.33 | 0.76 | 0.02 |
| Leu | 0.16 | 0.002 | 1.23 | 0.01 | 0.10 | 0.27 | 0.66 | 0.03 |
| Gln | 0.18 | 0.002 | 1.41 | 0.01 | 0.12 | 0.31 | 0.49 | 0.05 |
| Tyr | 0.19 | 0.001 | 1.43 | 0.01 | 0.13 | 0.28 | 0.53 | 0.04 |
| Pro | 0.22 | 0.002 | 1.66 | 0.02 | 0.13 | 0.41 | 0.54 | 0.04 |
| Lys | 0.24 | 0.001 | 1.81 | 0.01 | 0.18 | 0.34 | 0.56 | 0.04 |
| Val | 0.37 | 0.003 | 2.83 | 0.02 | 0.21 | 0.63 | 0.71 | 0.03 |
| Phe | 0.39 | 0.004 | 2.98 | 0.02 | 0.24 | 0.64 | 0.71 | 0.03 |
| Gly | 0.43 | 0.004 | 3.31 | 0.02 | 0.26 | 0.71 | 0.43 | 0.05 |
| Thr | 0.51 | 0.001 | 3.85 | 0.03 | 0.43 | 0.58 | 0.11 | 0.09 |
| Arg | 0.57 | 0.005 | 4.38 | 0.03 | 0.40 | 1.02 | 0.53 | 0.05 |
| Ala | 0.62 | 0.006 | 4.78 | 0.04 | 0.43 | 1.13 | 0.60 | 0.04 |
| Ser | 0.71 | 0.006 | 5.44 | 0.04 | 0.47 | 1.10 | 0.64 | 0.03 |
| Trp | 0.72 | 0.003 | 5.58 | 0.04 | 0.56 | 0.89 | 0.24 | 0.06 |
| Asp | 2.61 | 0.029 | 19.93 | 0.18 | 1.58 | 5.16 | 0.77 | 0.02 |
| Glu | 4.76 | 0.030 | 36.63 | 0.13 | 3.51 | 6.55 | 0.64 | 0.03 |
Figure 1.
Correlation-based network of seed FAA relationships across the Arabidopsis diversity panel. The Spearman’s rank correlation matrix was calculated from the back-transformed BLUPs of the absolute levels. The nodes represent FAA levels, and the edges represent correlations between the different FAA levels. Network edge parameters are rsp ≥ 0.6 and P ≤ 0.001. The node color denotes the amino acid family (red, shikimic acid; orange, Ser; green, pyruvate and BCAA; blue, Glu; dark blue, Asp); node size reflects nodal degree; and edge width reflects the strength of the correlation. The Spearman’s rank correlation coefficient is represented near each edge. The two highly connected groups are circled in blue and red. Free His is included within the red-circled group together with Ser, Gln, Lys, Arg, and Val.
Approach 1: GWAS of Absolute and Relative FAA Levels and Ratios Based on a Priori Biochemical Knowledge
To uncover potential regulators of FAAs in seeds, we calculated 98 traits (Supplemental Table S2) using known metabolic pathways and biochemical interactions among the 18 FAAs. The traits fall into four categories: (1) the absolute levels of the quantified FAAs (in nmol mg−1 dry seeds); (2) the sum of amino acids that constitute a biochemical family (e.g. the sum of all FAAs that belong to the Asp family: Asp, Thr, Ile, and Met); (3) the relative composition (i.e. each amino acid as a percentage of the total FAA quantified; e.g. Ile/total); and (4) the ratios based on the amino acid biosynthetic and degradative pathways and known interactions among their enzymes, such as competition between pathways and feedback loops. Category 4 ratios include a single FAA divided by the sum of its biochemical family (e.g. Lys/Asp family), ratios of FAAs from competing metabolic branches (e.g. Thr/Met; Galili, 1995), and ratios of FAAs that can serve as precursors to downstream products (e.g. Glu/Gln). The full list of these biochemically based traits is presented in Supplemental Table S2. Hereafter, we use the one-letter code annotation of the amino acids to describe the FAA traits; a string of one-letter codes describes the sum of FAAs (e.g. ILV is the sum of Ile, Leu, and Val).
We conducted a GWAS on all 98 traits using the methodology described by Angelovici et al. (2013). Consistent with this previous study, we found several BCAA-related traits to have significant associations, including with BCAT2 (At1G10070; Supplemental Data Set S2). However, beyond those already reported (Angelovici et al., 2013), no major, novel GWAS signals were discovered from this elaborated analysis of all BCAA-related traits (21 traits in total, including all traits with BCAAs in the denominator; Supplemental Table S2). Therefore, these BCAA-related traits will not be addressed further here and instead will be referenced in subsequent sections for comparative purposes only. GWAS on the remaining 77 non-BCAA-related FAA traits produced only one significant single-nucleotide polymorphism (SNP)-trait association at the 5% false discovery rate (FDR) level. We found SNP184159 to be significantly associated with the sum of Gly and Ser absolute levels (GS), which, notably, are two of the three members of the Ser family (Ser, Cys, and Gly; Table II, region 1; Supplemental Data Set S2; Supplemental Fig. S1). SNP184159 is located on chromosome 5 at position 12,282,814 bp within At5G32623, which is annotated as a pseudogene/transposon. An estimation of pairwise linkage disequilibrium (LD) between SNP184159 and SNPs located in the surrounding ±20-kb genomic region found moderately strong LD (r2 = 0.518) with an SNP at position 12,277,752 bp (SNP184146). This nearly 5-kb upstream SNP is located in a gene annotated as DEFENSIN-LIKE (DEFL family protein; At5G32619), the closest functional gene (located 4,723 bp upstream) to SNP184159 (Supplemental Fig. S1).
Table II. Summary of GWAS results for a genomic region that contains significant associations at the 5% FDR level identified by approach 1 and three genomic regions that contain significant associations at the 5% FDR level identified by approach 2.
NA, Not applicable.
| Region Identifier | Maximal Chromosomal Range for Locus | Trait | No. of Significant SNPs in the Region | Most Significant SNP | P Value of the Most Significant SNP after FDR Correction | Gene | Gene Annotation |
|---|---|---|---|---|---|---|---|
| Summary of significant associations determined/calculated by approach 1 (not including BCAA traits) | |||||||
| 1 | Chromosome 5, 12,282,814 | GS | 1 | 184,159 | 4.32E-02 | At5G32619 | Pseudogene |
| Summary of significant associations determined/calculated by approach 2 (not including BCAA traits) | |||||||
| 2 | Chromosome 1, 19,661,134 | H/LIVKHSQR | 1 | 32,041 | 4.97E-02 | At1G52780 | Protein of unknown function (DUF2921) |
| 3 | Chromosome 3, 912,617–941,537 | H/KHSQR | 18 | 82,058 | 9.24E-04 | At3G03710 | Polyribonucleotide nucleotidyltransferase, putative |
| H/KHSQRV | 13 | 82,067 | 1.26E-03 | At3G03710 | Polyribonucleotide nucleotidyltransferase, putative | ||
| H/LIVKHSQR | 12 | 82,067 | 3.47E-03 | At3G03710 | Polyribonucleotide nucleotidyltransferase, putative | ||
| H/KHR | 10 | 82,066 | 2.35E-02 | NA | |||
| 4 | Chromosome 5, 26,246,837–26,289,644 | S/LIVKHSQR | 4 | 212,610 | 2.39E-02 | NA | |
Approach 2: Network-Guided GWAS of Seed FAAs
The assessment of metabolic ratios based only on current biochemical knowledge yielded few new insights and is likely constrained by our limited understanding of FAA pathways and their interactions. Because the GWAS of the 77 non-BCAA-related FAA traits yielded only one new association (for the GS trait), we evaluated the use of an unbiased approach based on a correlation network topology for trait determination. The method is based on the assumption that FAA correlations across the association panel are driven, in part, by genetics. Hence, while conceptually similar to approach 1, the metabolic ratios generated via a network topology are based on metabolic cluster affiliation rather than on biochemical family affiliation. We calculated traits as a ratio of connected metabolic pairs or as a ratio of a single amino acid to its fully or partially connected metabolic group. Thirteen such groups were so defined (Supplemental Table S3), with most being partial versions of the two highly correlated groups (group 1 or group 2) or the entire network (Fig. 1; Supplemental Table S3). A total of 92 traits were derived using this approach (Supplemental Table S3), several of which overlap with the traits determined using approach 1 (marked with asterisks in Supplemental Table S3). As with approach 1, no major novel genomic regions were detected among the 31 BCAA-related network-derived traits, and these traits will be addressed separately in a comparison of the two approaches. GWAS performed on the remaining 61 network-derived traits produced three unique genomic regions with significant associations at the 5% FDR level for five FAA-related ratios (Table II; Supplemental Data Set S3): four ratios are His-related traits, and one ratio is a Ser-related trait (Table II). Two of the three identified genomic regions have highly significant GWAS signals. A genomic region on chromosome 5 is strongly associated with the S/LIVKHSQR trait (Table II), while a second region on chromosome 3 is strongly associated with four His-related traits (Table II). The latter region has significant SNP-trait associations spanning 29 kb (positions 912,617–941,537 bp; Supplemental Data Set S3).
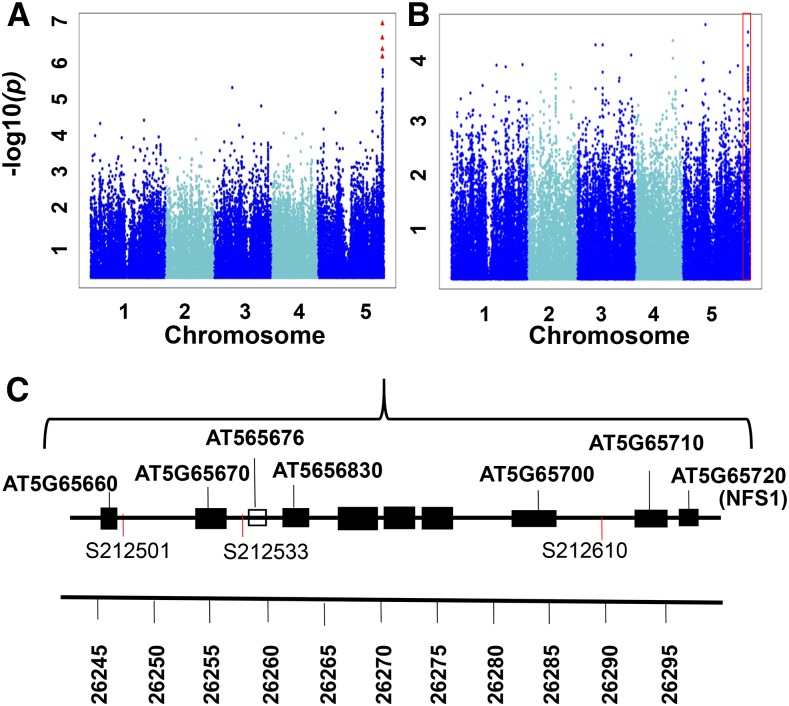
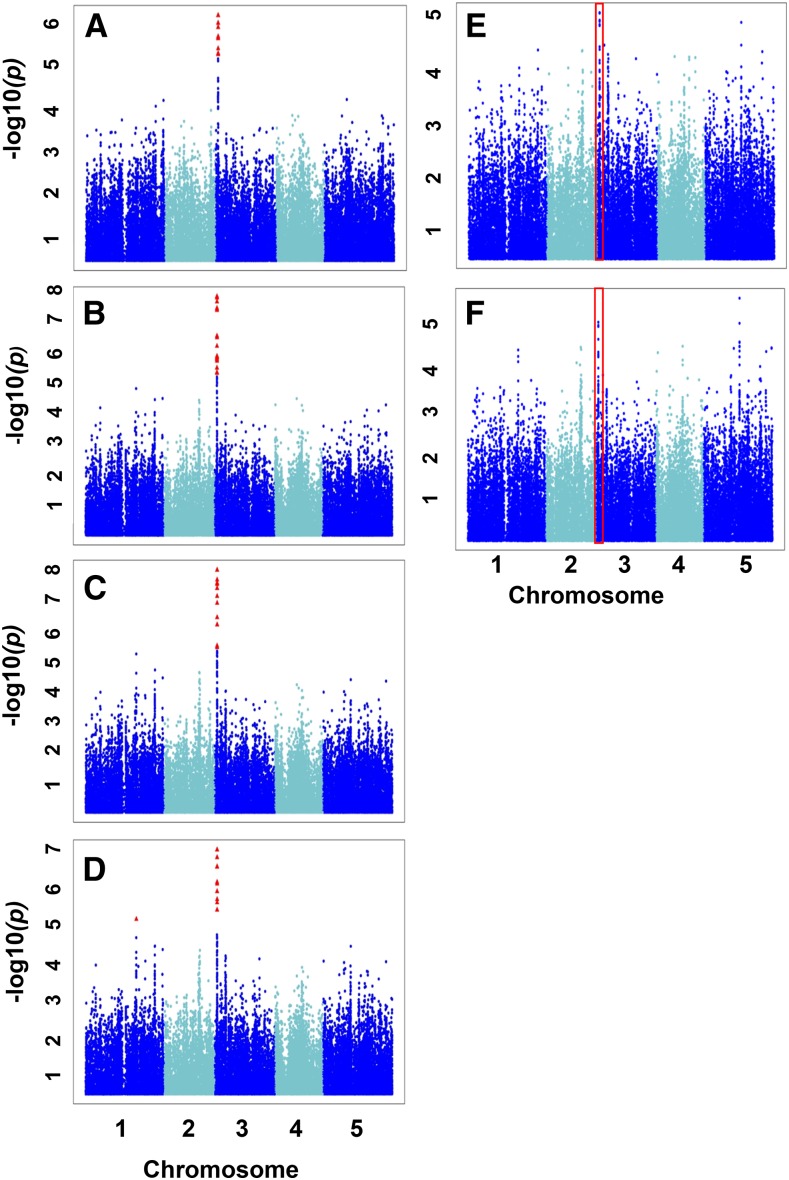
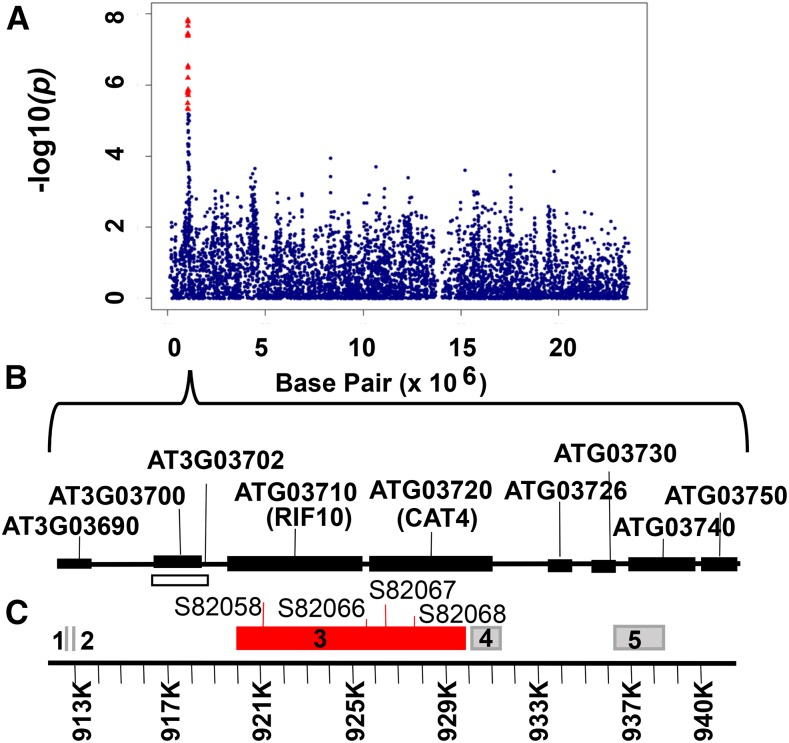
Four SNPs on chromosome 5 are significantly associated with the S/LIVKHSQR trait (Table II; Fig. 2). Three of these SNPs (i.e. SNP212610, SNP212533, and SNP212501) have similar raw P values and FDR-corrected P values of 2.43E-02 to 3.92E-02 (Fig. 2C; Supplemental Data Set S3) and explain 9.4%, 8.7%, and 8.3%, respectively, of the total phenotypic variation. The three SNPs are located relatively far from each other (at positions 26,289,644, 26,258,036, and 26,246,837 bp, respectively) and show weak LD with each other (r2 < 0.26; Supplemental Fig. S2). All three SNPs also demonstrate weak LD with SNPs within any of their surrounding genes in a 200-kb region (r2 < 0.23; Supplemental Fig. S2). However, no SNPs were called within a gene located 6,609 bp upstream of SNP212610 and that is annotated as a Cys desulfurase (At5G65720; NITROGEN FIXATION S-LIKE 1 [NFS1]). This is an amino acid biosynthetic gene responsible for the conversion of Cys to Ala. Because Ser is the precursor of Cys, this gene would seem a leading candidate as a regulator, except that, as with many amino acid biosynthetic genes that are not redundant, its knockout is lethal (Frazzon et al., 2007). We also detected two of the three most significant SNPs for the S/LIVKQHSR trait (i.e. SNP212501 and SNP212610) among the top 20 SNPs of the S/total GWAS (SNPs 2 and 16, respectively; Supplemental Table S4), suggesting that the network-guided approach increased our ability to identify significant candidate genes. The four His-related traits are H/KHR, H/KHSQR, H/KHSQRV, and H/LIVKHSQR, and the top three associated SNPs for these traits are SNP82066, SNP82058, and SNP82067, respectively, on chromosome 3 (Figs. 3 and 4; Table II; Supplemental Data Set S3). These three SNPs explain 9% to 11% of the total phenotypic variation for the four His-related traits (Supplemental Data Set S3). SNP82058 is located in a gene annotated as a polyribonucleotide nucleotidyltransferase (RESISTANT TO INHIBITION WITH FSM [RIF10]; At3G03710; Fig. 4B). SNP82066 is located in an intragenic region upstream of RIF10 and downstream of a gene annotated as CATIONIC AMINO ACID TRANSPORTER4 (CAT4; At3G03720; Fig. 4B), and SNP82067 is located within CAT4. In total, all the significant SNPs across all four traits span a 28,920-bp region that contains nine open reading frames (Fig. 4B). Pairwise LD estimates for the ±100-kb region centered on the most significant SNP (i.e. SNP82058) indicate strong LD among all these SNPs and patterns of long-range LD in the region, which may account for the multiple significant associations (Supplemental Fig. S3). Interestingly, SNP82066, SNP82058, and SNP82067 are significantly associated with the four His network-derived traits but not with any His-related traits determined by approach 1, such as H/total or H/EHPRG (Glu family). Nevertheless, these SNPs are among the top three SNPs with the lowest P values for H/EHPRG and among the fifth and sixth lowest P values for H/total (Fig. 3, E and F; Supplemental Table S4). Beyond chromosome 3, H/LIVKHSQR is the only trait with an additional significant SNP-trait association, which occurs for SNP32041 at position 19,661,134 bp on chromosome 1 (Fig. 3D; Table II; Supplemental Data Set S3). This SNP is located in a gene with unknown function (At1G52780), although, the PANTHER classification system (http://www.pantherdb.org/panther/family.do?clsAccession=PTHR19241) records this gene as an ATP-binding cassette transporter.
Figure 2.
GWAS summary of S/LIVKHSQR. A and B, Scatterplots of the association results from a unified mixed model analysis of S/LIVKHSQR (A) and S/total (B) traits across the five Arabidopsis chromosomes. The negative log10-transformed P values from the GWAS analysis are plotted against the genomic physical positions. P values for SNPs that are statistically significant for a trait at 5% FDR are in red. The red box represents the corresponding chromosomal region of this significant signal in the S/total Manhattan plot. C, Graphical representation of the genes within the vicinity of SNP212501, SNP212533, and SNP212610. Genes are represented by black boxes, and a pseudogene is represented by the white box. At5G65720 encodes a Cys desulfurase (NFS1) that is the leading candidate gene for S/LIVKHSQR.
Figure 3.
GWAS for the His-related traits from the two approaches. Scatterplots show the association results from a unified mixed model analysis for H/KHR (A), H/KHSQR (B), H/KHSQRV (C), H/LIVKHQSR (D), H/EHPRG (Glu family; E), and H/total (F) traits across the five Arabidopsis chromosomes. The negative log10-transformed P values from the GWAS analysis are plotted against the genomic physical position. P values for SNPs that are statistically significant for a trait at 5% FDR are in red. Traits A through D share a similar significant GWAS signal on chromosome 3. Red boxes represent the corresponding chromosomal region of this signal for traits E and F.
Figure 4.
GWAS of H/KHSQR. A, Scatterplot of the association results from a unified mixed model analysis of H/KHSQR on chromosome 3 only. The negative log10-transformed P values from the GWAS analysis are plotted against the genomic physical position. P values for SNPs that are statistically significant for a trait at 5% FDR are in red. B, Graphical representation of genes and haploblocks within the genomic region spanning SNPs that have significant associations for H/KHSQR. Genes are represented by black boxes, and a pseudogene is represented by the white box. C, Haploblocks are represented by gray boxes. Haploblock 3 (shown in red) contains the four most significant SNPs for the GWAS-assisted His-related traits.
Comparison between GWAS of BCAA-Related Traits Derived from Approaches 1 and 2
GWAS of BCAA traits had previously characterized BCAT2 as an important gene for determining BCAA-related traits in seeds. Our GWAS results from the BCAA-related traits derived using both approaches 1 and 2 are consistent with this previous study (Supplemental Data Sets S2 and S3; Supplemental Table S5). Nevertheless, among all the BCAA-related traits analyzed, the network-derived trait I/LIVKHSQR has a slightly elevated significant association (P value of 4.02E-08) with the previously characterized tagging SNP5373 on chromosome 1, whose strongest association from approach 1 is 5.27E-07 for the I/total trait. SNP5373 explains 22% of the phenotypic variation of the I/LIVKHSQR trait compared with 19% of the I/total trait (Supplemental Data Sets S2 and S3). More pronounced observations were obtained using linkage analysis of dry seed FAAs from a Bayreuth-0 (Bay) × Shahdara (Sha) RIL population (Loudet et al., 2002; Angelovici et al., 2013). This mapping population contains significantly different haplotype pairs for the significantly associated genomic region identified in GWAS of BCAA on chromosome 1 (Angelovici et al., 2013) and was used previously to independently confirm a GWAS association. A linkage analysis of the same data using the network-derived BCAA traits (I, L, or V)/LIVKHSQR and (I, L, or V)/LIVKHSQRYEFA was performed and compared with the previous linkage analysis (Table III). We again found that the large-effect QTL on chromosome 1 detected using approach 1 shows a substantial increase in significance for the I/LIVKHSQR and I/LIVKHSQRYEFA network-derived traits (18.46 and 22.57 log of odds [LOD] scores, respectively) and explains more of the total phenotypic variation (r2 = 41.8% and 48.4%, respectively). The strongest effect QTL from the previous BCAA-related trait linkage analysis yielded LOD scores that range from 6.06 to 9.3 and explain 16.7% to 25.55% of the total phenotypic variation (Table III; Angelovici et al., 2013).
Table III. QTL analysis of the network-guided BCAA traits from the Bay × Sha mapping population.
Only QTLs that overlap the approximately 6- to 12-centimorgan (cM) chromosome 1 interval previously detected by GWAS are shown. Parameters were obtained from the Plant Breeding and Biology Quantitative Trait Loci (PLABQTL) software program using phenotypic and genotypic data from 158 RILs. Asterisks represent the most significant QTLs described by Angelovici et al. (2013); all traits are represented in one letter code.
| Trait | Chromosome | Position | Supporting Interval | LOD | High Parent | r2 | Allelic Effect Estimates | |
|---|---|---|---|---|---|---|---|---|
| cM | % | |||||||
| I/LIVKHSQR | 1 | 8 | 6 | 12 | 18.46 | Sha | 41.8 | −0.52 |
| I/LIVKHSQRYEFA | 1 | 10 | 6 | 12 | 22.57 | Sha | 48.4 | −0.186 |
| L/LIVKHSQR | 1 | 8 | 4 | 12 | 6.06 | Sha | 16.4 | −0.321 |
| L/ LIVKHSQRYEFA | 1 | 8 | 6 | 12 | 10.89 | Sha | 27.8 | −0.16 |
| I/IVL* | 1 | 8 | 6 | 12 | 9.3 | Sha | 23.9 | −0.011 |
| L/total* | 1 | 8 | 6 | 14 | 9.91 | Sha | 25.5 | −0.119 |
| I | 1 | 8 | 4 | 14 | 6.06 | Sha | 16.7 | −0.021 |
Multiple-Locus Mixed Model and Haplotype Analysis of the Significant His-Related SNP-Trait Associations
We chose the strong GWAS signal from the His-related traits for further dissection and validation. To resolve the complex signal on chromosome 3, we performed a multiple-locus mixed model (MLMM) approach that uses stepwise selection (Segura et al., 2012) in the vicinity of the most significant SNPs for each trait. All SNPs with a minor allele frequency greater than or equal to 0.05 in a ±100-kb region were considered for inclusion in the final model (Supplemental Table S5). The optimal models contain only one SNP for each trait tested: SNP82058 for the H/KHSQR trait, SNP82066 for the H/KHR trait, and SNP82067 for the H/LIVKHSQR and H/KHSQRV traits (Fig. 4; Supplemental Table S6). To validate the results, we reran the GWAS using a unified mixed model that included the respective optimal SNP as a covariate for each trait. No significant associations were detected for any traits with either SNP (Supplemental Fig. S4), including the additional SNP-trait association detected on chromosome 1 for the H/LIVKHSQR trait (Fig. 3D; Table II). A haplotype analysis of the region spanning all the significant SNPs on chromosome 3 (i.e. 912,617–941,537 bp; Table II; Supplemental Data Set S3) identified five haploblocks (Supplemental Table S7), and the four most significant SNPs trace back to haploblock 3 (chromosome 3, 919,927–929,830 bp). Haploblock 3 (9.903 kb; Fig. 4B) spans a large portion of both the RIF10 (919,542–925,338 bp) and CAT4 (925,612–930,974 bp) genes and contains 16 SNPs: nine in RIF10, six in CAT4, and one between the two genes. When unified mixed linear models were fitted, the most significant contrasting haplotype pair found for haploblock 3 was ACAAGTCACGATGTTA versus ATTCATCTAAGTGCAT, with frequencies of 0.262 and 0.099, respectively (Supplemental Fig. S5A). These haplotypes represent the low and high levels, respectively, of the His-derived traits (Supplemental Fig. S5B). Although the differences in the phenotype averages between the ecotype groups harboring the two haplotypes are small (7%–9%), they are highly significant (P values of 1.2E-05–2.38E-07; Supplemental Fig. S5B).
Functional Characterization of CAT4 and RIF10
To further test whether CAT4 or RIF10 is a regulator/effector of the His-related traits, we used quantitative reverse transcription (RT)-PCR to determine the transcription levels of each gene at four different stages of seed development in the Columbia-0 (Col-0) background: early maturation (12 d after flowering [DAF]), midmaturation (15 DAF), late maturation (18 DAF), and complete desiccation (dry seeds; Supplemental Fig. S6A). We found that CAT4 transcription levels increase toward midmaturation and decrease moderately during desiccation and that RIF10 transcript levels are low and constant throughout maturation and desiccation. According to the Arabidopsis eFP Browser (http://bbc.botany.utoronto.ca/efp/cgi-bin/efpWeb.cgibin/efpWeb.cgi; Winter et al., 2007), CAT4 expression levels are relatively high in dry seeds compared with vegetative tissues, and RIF10 is highly expressed in the vegetative tissue and decreases greatly during seed development (Supplemental Fig. S6, B and C).
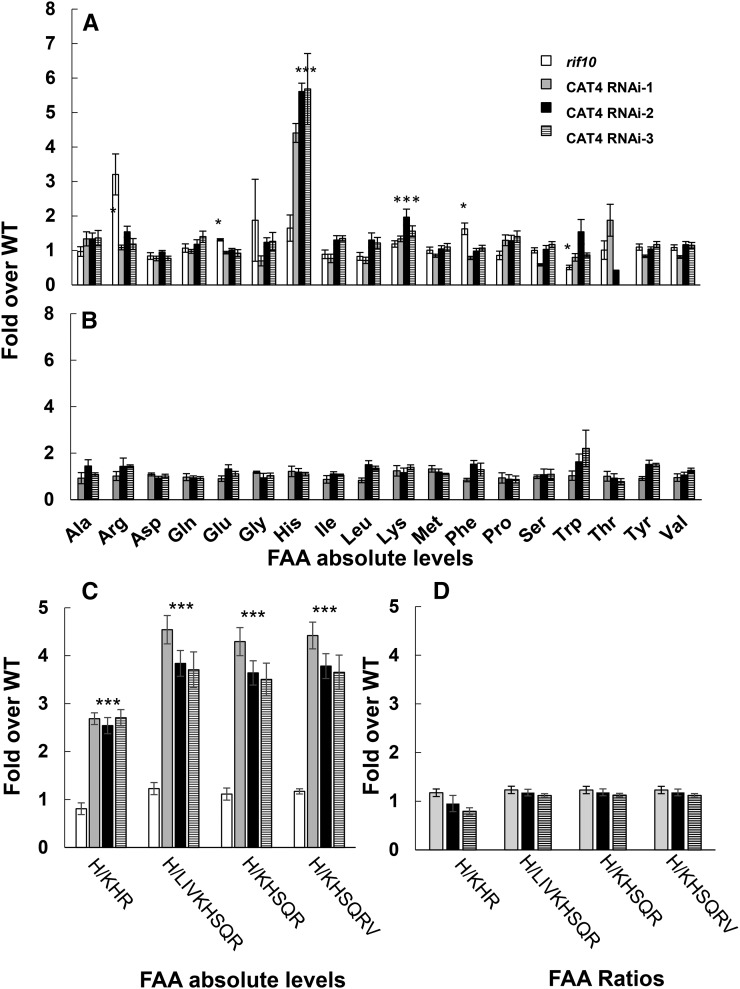
Based on results from the expression analysis and on its annotation as a cationic amino acid transporter, CAT4 seemed the most likely candidate gene responsible for the natural variation in the His-related traits. Hence, we employed a transgenic strategy to test the effect of the null or knockdown mutants of both CAT4 and RIF10 on the FAA content of dry seeds. Since T-DNA insertion lines for CAT4 were not available, we used an RNA interference (RNAi) approach to knock down the gene. We transformed the Agrikola construct CATMAa02630 into the Col-0 background and used an empty vector with similar antibiotic resistance as a control. We recovered three homozygous lines with reductions of more than 70% at the mRNA level (Supplemental Fig. S7A) and then performed dry seed and leaf FAA quantification analyses on these lines (Fig. 5). The results show a seed-specific significant increase in absolute levels of His in all three independent RNAi lines (Fig. 5; Supplemental Data Set S4); the seed-specific increases in His absolute levels ranged from 4- to 6-fold (Fig. 5A), and a 2.5- to 4.5-fold increase in the His-relative ratios is significantly associated with this region (Fig. 5C). Interestingly, Lys levels have small but significantly increased levels in all three RNAi lines (fold increase, 1.4–2; P value, 0.03–0.00015; Fig. 5). We isolated and characterized the rif10 alleles of the homozygous T-DNA insertion lines from the SALK T-DNA collection (SALK_013306 and SALK_037353); one allele (SALK_013306) has a severe growth phenotype and therefore was excluded from the analysis. RT-PCR using primers that flank the T-DNA insertion of rif10 was used to confirm that the line lacks the transcripts (Supplemental Fig. S7B). An FAA quantification analysis of the dry seeds from rif10 showed small but significant effects on the absolute levels of Gln, Asp, Phe, and Trp but not on His or any His-related ratios (Fig. 5, A and C). Taken together, these results confirm that CAT4 significantly affects the absolute and relative levels of His and is the gene responsible for the natural variation.
Figure 5.
Seed and leaf FAA-related traits measured from three cat4 RNAi lines and rif10. Absolute FAA levels were measured from dry seeds (A) and leaves (B) harvested from 2-week-old plants. Significant ratios were calculated from both dry seeds (C) and leaves (D). Values represent fold changes in FAA for the three CAT4 RNAi lines (gray, black, and striped bars) and rif10 (white bars; seeds only) relative to their respective controls (wild type [WT] with an empty vector). Averages and se values were calculated from four measurements. *, FAA traits that have a significant difference (P < 0.05, Student’s t test) between the wild type and rif10; ***, FAA traits that have a significant difference (P < 0.05, Student’s t test) between the wild type and all three CAT4 RNAi lines. For the FAA content, see Supplemental Data Set S4.
DISCUSSION
Previous metabolic QTL mapping and GWAS on both primary and secondary metabolite profiles have demonstrated that analyses of metabolic ratios based on known biochemical interactions yield stronger associations than GWAS performed on the corresponding metabolites’ absolute levels (Wentzell et al., 2007; Vallabhaneni and Wurtzel, 2009; Wurtzel et al., 2012; Angelovici et al., 2013; Gonzalez-Jorge et al., 2013; Lipka et al., 2013; Owens et al., 2014). Our study demonstrates that a network-guided metabolite-based-GWAS performed on unbiased ratios derived from a correlation-based network topology improves the selection of traits for downstream GWAS and can facilitate further identification of significant associations among genomic regions that affect the natural variation of a tight, interconnected FAA metabolic network, such as amino acids (Table II; Supplemental Table S5). Our approach utilizes the metabomelic network analysis differently than previous network-assisted GWAS. For example, previous studies mainly used gene coexpression networks to either prioritize candidate genes retrieved from GWAS performed on absolute metabolite levels (Chan et al., 2011; Lee et al., 2011; Greene et al., 2015; Matsuda et al., 2015) or to identify unknown metabolites from combined metabolites and gene coexpression networks (Krumsiek et al., 2012). To date, interconnected metabolic traits have been analyzed using multivariate GWAS, factor analysis, or principal component analysis-based GWAS (Shen et al., 2013; Brachi et al., 2015). Our approach, which relies instead on single amino acids to a single or a group of amino acids derived from the network topology, may yield results whose biological relevance is comparatively clearer. The correlation-based network of the seed FAAs facilitated the visualization of both characterized and uncharacterized metabolic interactions (Fig. 1). The genetics that underlie the strong correlation of group 1 (composed of all the BCAAs) is most likely due to the four shared biosynthetic enzymes and one shared catabolic gene, BCAT2, that is associated with the natural variation of BCAA levels in seeds (Binder et al., 2007; Binder, 2010; Angelovici et al., 2013). A second metabolic cluster (group 2; Fig. 1), consisting of FAAs from four amino acid families, is an uncharacterized metabolic module. Interestingly, Val is the central hub that connects the two parts of the network (Fig. 1).
This network is consistent with previous studies showing that perturbations in BCAA pathway genes induce multiple changes of amino acid levels in a seed-specific manner (Gu et al., 2010; Angelovici et al., 2013). The network-guided metabolite-based-GWAS we performed on the FAAs of dry Arabidopsis seed identified two significantly associated genomic regions (Table II, regions 3 and 4) that could not have been detected otherwise. One region is significantly associated with a Ser-related trait (S/LIVKHSQR), while the other region is significantly associated with four His-related traits (H/KHR, KHSQR, KHSQRV, and LIVKHSQR; Table II; Figs. 2–4; Supplemental Data Sets S2 and S3). Interestingly, the most significant SNPs (P < 0.05 after FDR correction) detected for these five traits are among the top 20 SNPs with the lowest P value for the corresponding traits generated using approach 1: that is, S/total, H/total, and H/EHPRG (representing the H/Glu family; Table II; Supplemental Table S4). Nevertheless, the P values (after FDR correction) for these traits are not significant and range from 0.28 to 0.97. These results suggest that the network-guided GWAS increased the power of our analysis. It is arguable that an SNP ranking approach may have extracted the same candidate genes; however, when testing nearly 100 traits (as with approach 1), that approach could yield hundreds of candidate genes and, therefore, might not be as useful at prioritizing candidate genes for validation. The linkage analysis of the unique network of BCAA-related traits performed on dry seed FAAs measured from the Bay × Sha mapping population (Loudet et al., 2002; Angelovici et al., 2013) further supports the increase in our analysis power. A previous linkage analysis of the free BCAA-related traits in this population identified a large-effect QTL on chromosome 1 (containing the quantitative trait gene BCAT2) for L/total, I/IVL, and I with LOD scores of 9.91, 9.3, and 6.06, respectively, that explained 25.5%, 23.9%, and 16.7% of the total phenotypic variation, respectively (Table III; Angelovici et al., 2013). In contrast, linkage analysis of this FAA data set with the unique network-guided BCAA-related traits found that the same QTLs for the I/LIVKHSQRYEFA and I/LIVKHSQR traits have LOD scores of 22 and 19, respectively, and explained 48% and 42% of the total phenotypic variation (Table III). Notably, the increase in significance is specific to the Ile-related traits and not the Leu-related ratios, which remained comparable throughout the analyses (Tables II and III; Supplemental Table S5). These results suggest that network-guided metabolic ratios are genetically less complex and, therefore, are more likely to be detected in a GWAS on a relatively small population size (Korte and Farlow, 2013). It is also possible that a metabolic ratio approach reduces the inclusion of confounding effects, such as seed size, which repeatedly colocalizes with several amino acid levels (Joosen et al., 2013).
These results are consistent with observations of linkage analysis of glucosinolate metabolic ratios, which also have been shown to be highly useful for detecting metabolic QTLs, perhaps due to their high heritability (Wentzell et al., 2007). In addition, the metabolic network was reconstructed from the relationship of amino acids across a natural population of dry seeds; hence, it is possible that these metabolic ratios represent a unique interaction with seeds compared with a general metabolic ratio, which may or may not be represented in the specific tissue of interest. This possibility is of special importance for metabolites, such as amino acids, that are known to be differentially regulated in leaves and seeds (Toubiana et al., 2012). However, the latter hypothesis, although consistent with our findings, should be regarded with caution because the network is constructed from a metabolic data set derived from an association panel and, therefore, captures correlative patterns influenced by the metabolic network and genetic relatedness between accessions. This helps zoom in on the genetics but may overestimate the physiological metabolic relationships.
Interestingly, all the network-guided traits with significant SNP-trait associations belong to the five FAAs traits with the highest network hub scores (Table II; Supplemental Table S1B), which indicates the relevance of the network properties for network-guided metabolome-GWAS. This observation is of importance to cases of metabolic networks that have many possible metabolic ratio traits; instead of analyzing all possible ratios, one could start by analyzing the ratios of metabolites with the highest hub scores. It is also consistent with the observation that network dynamics is strongly connected to the underlying reaction pathway structure (Steuer et al., 2003; Weckwerth, 2003) and is more sensitive to genetic permutation than the metabolic steady-state levels. For example, a correlation-based network analysis of potato (Solanum tuberosum) leaves with a Suc synthase antisense construct and the wild type found more significant changes in correlations and ratios of metabolites compared with metabolite absolute levels, which had mostly small, nonsignificant differences (Weckwerth et al., 2004). Similarly, a transgenic Arabidopsis plant with induced high Lys levels in seeds (Angelovici et al., 2009) demonstrated wider and more significant effects on the correlation network properties compared with the absolute levels of the metabolites.
The strongest GWAS signal from among all the network-guided metabolic ratios was for four His-related traits on chromosome 3 (Table II; Figs. 3 and 4). His plays an essential role in plant reproduction, growth, chelation, and transport of metal ions. Blocking His biosynthesis in Arabidopsis leaves, for example, causes increased expression of several genes involved in other amino acid synthesis pathways and affects the levels of several FAAs (Guyer et al., 1995; Stepansky and Leustek, 2006; Ingle, 2011). Nevertheless, the regulation of His metabolism as well as its entire catabolic pathway remain unclear (Stepansky and Leustek, 2006; Ingle, 2011). Our results shed light on the genetic regulation of free His in seeds and also provide a clue to the localization of its catabolism. The GWAS signal for the four His-related traits spans 18 significant SNPs that comprise nine open reading frames on chromosome 3 (Fig. 4). Haploblock and MLMM analyses (Segura et al., 2012) of the region indicate that the four most significant SNPs for all four traits are located in haploblock 3 (Fig. 4; Supplemental Tables S6 and S7). The average difference for all four traits between the two accession groups containing the most significantly different haplotype pairs for haploblock 3 is highly significant (Supplemental Fig. S5B). Together, these findings suggest that the variation in all four His-related traits is genetically driven by a polymorphism in this haploblock, which spans only two genes, RIF10 and CAT4. The functional annotations and transcription analyses of both RIF10 and CAT4 strongly indicate that CAT4 is the causal gene (Supplemental Fig. S6). CAT4 transcription levels are induced during seed maturation, while RIF10 transcription is maintained at very low absolute and relative levels (Supplemental Fig. S7, A–C). Moreover, results from an amino acid analysis demonstrated that only cat4 RNAi lines have a significant effect on the absolute and relative levels of His, thus confirming that CAT4 is the causal gene (Fig. 5). According to the FAA analysis, CAT4 is functional in seed but not in leaves (Fig. 5, B and C), an observation that is consistent with a previous study (Yang et al., 2014b). CAT4 is localized primarily to the tonoplast, but it has been detected in the endoplasmic reticulum. It belongs to a gene family composed of nine genes that are defined by their similarity to the mammalian CAT gene (Su et al., 2004). While CAT8 and CAT9 are localized partially in the tonoplast, only CAT2 and CAT4 are localized primarily in the tonoplast (Carter et al., 2004; Su et al., 2004; Yang et al., 2014a, 2015). Like CAT4, CAT2 and CAT9 are involved in amino acid metabolism, but these genes have a broad-spectrum effect on FAA homeostasis in Arabidopsis leaves (Yang et al., 2014a, 2015). Studies of other amino acid transporters located on the plasma membrane (e.g. Arabidopsis AMINO ACID PERMEASE1 [AAP1]) have demonstrated the importance of these proteins for uptake by the embryo; for example, the aap1 null mutant shows a reduction in storage proteins and seed yield (Sanders et al., 2009; Tegeder, 2014). Nevertheless, no transporter, to date, displays such a specific effect on any single amino acid in seed, let alone His. In general, the functional role of FAAs’ preferential sequestering to the vacuole and its regulation is not clear; vacuoles store a large pool of FAAs, although at a much lower concentration than the cytosol. For example, around 50% of the FAA pool was found in illuminated barley (Hordeum vulgare) protoplast vacuoles and shown to be diurnally regulated, with increased loading of amino acids into the vacuole at night (Dietz et al., 1990; Winter et al., 1994). Moreover, a metabolic analysis of these protoplasts suggests that some amino acids (e.g. His, Ala, Trp, Met, and Ser) accumulate in the vacuole while all others are preferentially found outside the vacuole (Tohge et al., 2011).
Our data demonstrate that, in vivo, CAT4 is a His transporter, despite previous, unsuccessful attempts to identify the activity of CAT4 (import/export) by heterologous expression in yeast and oocytes (Su et al., 2004). In light of the accumulation of His only in the seeds of the cat4 mutants, we hypothesize that CAT4 is involved in His export from the vacuole to the cytosol for downstream utilization in this tissue. However, in contrast to leaf vacuoles, which are involved in multiple functions (e.g. turgor maintenance, protoplasmic homeostasis, lysis, and metabolite sequestering and storage), seed vacuoles mainly specialize in storage of proteins (Höfte and Chrispeels, 1992; Paris et al., 1996; Herman and Larkins, 1999; Marty, 1999). The directionality of the transporter (export or import from the vacuoles) and its functional relevance in the unique context of metabolic regulation during seed development and filling and desiccation (Baud et al., 2002, 2008; Fait et al., 2006) should be investigated further. Interestingly, a GWAS conducted on free metabolite levels in maize leaves revealed a significant association between free Tyr and Lys levels and a cationic amino acid transporter homolog (Riedelsheimer et al., 2012). This finding implies an important role for CAT transporter family members in the regulation of FAA levels in crops. Hence, it is worthwhile to consider CAT transporters as potentially beneficial for biofortification (since His is a semiessential amino acid) as well as useful in heavy metal resistance in crops. Several studies have demonstrated that increasing His by manipulating its biosynthetic pathway leads to severe growth defects (Stepansky and Leustek, 2006), likely a consequence of the interconnectedness of its metabolic pathway with several essential processes and its high energetic cost (Ingle et al., 2005). Our results suggest that engineering CAT homologs in crops may provide a method to increase His levels while circumventing these pleiotropic effects in crops.
MATERIALS AND METHODS
Plant Growth and Seed Collection
All Arabidopsis (Arabidopsis thaliana) genotypes were grown at 18°C to 21°C (night/day) under long-day conditions (16 h of light/8 h of dark). Plant growth of the Arabidopsis diversity panel (Nordborg et al., 2005; Platt et al., 2010a; Horton et al., 2012) is described by Angelovici et al. (2013). For the developing seed analysis, flowers of Col-0 were marked at specific times (12 ± 1, 15 ± 1, 18 ± 1, and 20 ± 1 DAF), and seeds were collected and stored at −80°C. For leaf analysis, 2-week-old plants were collected and stored at −80°C.
Isolation of T-DNA Insertion Mutants and RNAi Lines
The T-DNA SALK_037353c (rif10-1) and SALK_013306 (rif10-2) insertion lines were obtained from the Salk collection. Homozygous plants were isolated by genomic PCR using gene-specific primers in combination with the T-DNA left border primer. The lack of transcripts from RIF10 was validated by RT-PCR using RNA isolated from seeds of the respective mutants and primers of the coding region [At3G03710 cDNA LP, 5′-ATGTTGACGAGTCCCAGTAAC-3′; At3G03710 Ins RP1, 5′-GCCATTTGTTTATCACCAAGCGT-3′; CAT4 (1) F, 5′-GTGCGAGTTTGTTGGGTTCC-3′; CAT4 (1) R, 5′-CCATGTCCCAGCTCCAATGT-3′; PM ACT2 F, 5′-CAGCATCATCACAAGCATCC-3′; and PM ACT2 R, 5′-CCGTTGTCCTGAGGTTCTGT-3′]. The RNAi construct was designed and obtained from the Agrikola gene-specific tag collection (CATMA3a02630; Hilson et al., 2004) using Gateway technology. The pDonor (PN253177) containing the CAT4 identified gene-specific tag was transferred into the pHellsgate vector and then transformed into Arabidopsis by Agrobacterium tumefaciens-mediated gene transfer using the floral dip method (Clough and Bent, 1998). Homozygous lines with a single insertion and lines segregating for multiple insertions were isolated by selection with the appropriate antibiotic. Knockdown lines were defined by a 70% reduction in CAT4 transcription.
RIF10 and CAT4 Transcript Analyses
Total RNA was isolated from dry seeds using the hot borate method (Birtić and Kranner, 2006) followed by DNase treatment using TURBO DNA-free DNase (Ambion). First-strand cDNA was synthesized from 1 μg of total RNA with SuperScript II H2 reverse transcriptase (Invitrogen) and an oligo(dT) primer. Transcript levels were determined by quantitative real-time PCR using SYBR Green Master Mix (Applied Biosystems) with ACTIN2 (At3G18780) mRNA as an internal control. Primers were as follows: qPCR CAT4 (3) F, 5′-GACACAAAGGAGGGTTTCTCTG-3′; qPCR CAT4 (3) R, 5′-AGATCATGCTTCCAATAAGTAGCC-3′; ACTIN-RT-F, 5′-CAGCATCATCACAAGCATCC-3′; ACTIN-RT-R, 5′-CCGTTGTCCTGAGGTTCTGT-3′; qPCR RIF10(3) F, 5′-AGGGCGAAAGCGATTATTAGT-3′; and qPCR RIF10(3) R, 5′-CTCCTACTTTATAGGCATCCTCTG-3′.
Plant Extraction and Analysis of Seed Amino Acids Using Liquid Chromatography-Tandem Mass Spectrometry
FAA extraction for the liquid chromatography-tandem mass spectrometry analysis was performed using a previously described method (Angelovici et al., 2013). The method was modified from Gu et al. (2007) to include selected ion pairs for 11 additional heavy amino acid standards. Under these conditions, Cys and Asn do not have reliable selected ion monitoring pairs and, therefore, were excluded from the analyses.
Data Source and Analysis
Network Analysis
Seed FAA data from the diversity panel were obtained from Angelovici et al. (2013). Briefly, 18 FAAs were quantified from three independent outgrowths (biological replicates) of the Arabidopsis diversity panel (Atwell et al., 2010; Platt et al., 2010a). Replicates were integrated into a single value using the BLUP model and then reverse transformed. Spearman’s rank correlation was then used to produce a correlation matrix. The threshold of the Spearman’s rank correlation coefficient and the significance of correlation were used to describe different network properties (i.e. average node degree, network density, and diameter; Toubiana et al., 2013). Thus, a list of correlation matrices was created using different r value thresholds from 0.1 to 0.9 with steps equal to 0.1. The threshold of r ≥ 0.6 was chosen based on the stabilization of average node degree and the network diameter calculated at each step. The P value that reflects significance of correlation did not strongly affect network density and was selected on a level equal to 0.001. The significant correlation matrix was created using the freely distributed R software (version 3.0.1; www.r-project.org), and the network visualization and analysis was applied using Cytoscape, version 3.0 (Shannon et al., 2003), using a previously described method (Batushansky et al., 2016). Each node represents a specific amino acid; the node properties (color and size) reflect attributes of biochemical pathways and nodal degree, respectively. Each edge represents a correlation between adjacent nodes, and the edge width reflects the strength of correlation. The Cytoscape organic layout was used for network graphical output. Additional network properties (i.e. diameter and transitivity or clustering coefficient) were calculated using the igraph R package (version 3.0.1; http://igraph.org/r/).
GWAS, LD, MLMM, and Haplotype Analyses
All traits including ratios were treated independently. Metabolic ratios were derived prior to the calculation of BLUPs to minimize noise. For each trait, the outlier removal, optimal transformation, and BLUP calculation were performed as described previously (Angelovici et al., 2013; Gonzalez-Jorge et al., 2013). The BLUPs were used as phenotypic data for the GWAS and the haplotype analyses. Variance component estimates from each fitted model were used to estimate the broad-sense heritability of each trait (Holland et al., 2010; Hung et al., 2012), and se values for the heritability estimates were approximated using the delta method (Holland et al., 2010). GWAS and MLMM analyses were conducted as described previously (Angelovici et al., 2013). Haplotypes were created using the confidence interval method in Haploview version 4.2 (Barrett et al., 2005).
Linkage Analysis
The network-guided ratios were used as quantitative values in the identification of QTLs. The PLABQTL software package was used for composite interval mapping (Utz and Melchinger, 1996). A permutation analysis (1,000) was performed to calculate the critical LOD score (a = 0.05) of 3.11. Genotypic data used for the analysis were obtained from Loudet et al. (2002; http://dbsgap.versailles.inra.fr/vnat/Documentation/33/DOC.html). Cofactors used for calculations were automatically chosen by PLABQTL.
Accession Numbers
Sequence data can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At3G03720, CAT4; At3G03710, RIF10; At2G26190, calmodulin-binding family protein; At5G2623, pseudogene; At5G32619, encodes a DEFL family protein; At3G28970, AAR3; At3G28960, transmembrane amino acid transporter family protein; At5G65700, BARELY ANY MERISTEM 1 (BAM1); At5G65710, HAESA-LIKE2; At5G65720, ATNFS1; At5G65683, Waive phenotype 3 (WAV3) HOMOLOG2; At5G65670, INDOLE-3-ACETIC ACID INDUCIBLE 9 (IAA9); At5G65660, Hyp-rich glycoprotein family protein; At1G52780, DUF2921; At1G10090, ERD4; and At1G10070, BCAT2.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. GWAS summary of the Gly and Ser absolute sum.
Supplemental Figure S2. GWAS summary of S/LIVKHSQR.
Supplemental Figure S3. GWAS of H/KHSQR.
Supplemental Figure S4. Conditional GWAS of the four most significant His-related traits.
Supplemental Figure S5. Haplotype analysis of haploblock 3.
Supplemental Figure S6. Expression levels of CAT4 and RIF10.
Supplemental Figure S7. CAT4 and RIF10 transcript levels in the mutant lines.
Supplemental Table S1. Comparative analysis of the transitivity (clustering coefficient) of the Arabidopsis accessions’ metabolic networks and equalized random networks and the weighted hub score of the network nodes.
Supplemental Table S2. List of seed FAA traits calculated from the quantification of 18 FAAs and known biochemical interactions.
Supplemental Table S3. List of seed FAA traits determined from the network topology (approach 2).
Supplemental Table S4. GWAS results of the top 20 SNPs from the H/total, H/EHPRG (Glu family), and S/total GWAS.
Supplemental Table S5. Summary of GWAS results on the BCAA-related traits.
Supplemental Table S6. Summary of the MLMM analysis of the four His-related traits with a GWAS signal on chromosome 3, using all SNPs within ±100 kb of the most significant SNP in the region.
Supplemental Table S7. Haplotype analysis was performed using the Haploview software program on the genomic region chromosome 3, 912,617 to 941,537, which had significant SNP associations with the His-related traits.
Supplemental Data Set S1. The 313 accessions used in this study and the back-transformed BLUPs of the FAA absolute levels quantified for each.
Supplemental Data Set S2. Summary of GWAS results of all SNPs with significant associations at 5% FDR identified by approach 1, not including BCAA-related traits and only BCAA-related traits.
Supplemental Data Set S3. Summary of GWAS results of all SNPs with significant associations at 5% FDR for all the network-assisted derived GWAS traits, not including BCAA-related traits and only BCAA-related traits.
Supplemental Data Set S4. Seed free amino acid profiles of rif10, CAT4 RNAI-1, RNAI-2, and RNAI-3 and leaf free amino acid profiles of CAT4 RNAI-1, RNAI-2, and RNAI-3 along with their wild-type (Col-0) control.
Supplementary Material
Acknowledgments
We thank Dr. David Toubiana for suggestions on the network analysis and Melody Kroll for helping with the editing of the article.
Glossary
- FAA
free amino acid
- BCAA
branched-chain amino acid
- QTL
quantitative trait locus
- RIL
recombinant inbred line
- GWAS
genome-wide association studies
- BLUP
best linear unbiased predictor
- SNP
single-nucleotide polymorphism
- FDR
false discovery rate
- LD
linkage disequilibrium
- Bay
Bayreuth-0
- Sha
Shahdara
- LOD
log of the odds
- MLMM
multiple-locus mixed model
- RT
reverse transcription
- Col-0
Columbia-0
- DAF
days after flowering
- RNAi
RNA interference
- PLABQTL
Plant Breeding and Biology Quantitative Trait Loci
Footnotes
This work was supported by the U.S.-Israel Binational Agricultural Research and Development Fund (postdoctoral award no. ALTF 29–2011 to R.A.) and the National Science Foundation (grant no. 0922493 to D.D.).
Articles can be viewed without a subscription.
References
- Alonso-Blanco C, El-Assal SE, Coupland G, Koornneef M (1998) Analysis of natural allelic variation at flowering time loci in the Landsberg erecta and Cape Verde Islands ecotypes of Arabidopsis thaliana. Genetics 149: 749–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelovici R, Fait A, Fernie AR, Galili G (2011) A seed high-lysine trait is negatively associated with the TCA cycle and slows down Arabidopsis seed germination. New Phytol 189: 148–159 [DOI] [PubMed] [Google Scholar]
- Angelovici R, Fait A, Zhu X, Szymanski J, Feldmesser E, Fernie AR, Galili G (2009) Deciphering transcriptional and metabolic networks associated with lysine metabolism during Arabidopsis seed development. Plant Physiol 151: 2058–2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelovici R, Galili G, Fernie AR, Fait A (2010) Seed desiccation: a bridge between maturation and germination. Trends Plant Sci 15: 211–218 [DOI] [PubMed] [Google Scholar]
- Angelovici R, Lipka AE, Deason N, Gonzalez-Jorge S, Lin H, Cepela J, Buell R, Gore MA, Dellapenna D (2013) Genome-wide analysis of branched-chain amino acid levels in Arabidopsis seeds. Plant Cell 25: 4827–4843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranzana MJ, Kim S, Zhao K, Bakker E, Horton M, Jakob K, Lister C, Molitor J, Shindo C, Tang C, et al. (2005) Genome-wide association mapping in Arabidopsis identifies previously known flowering time and pathogen resistance genes. PLoS Genet 1: e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo WL, Ishizaki K, Nunes-Nesi A, Larson TR, Tohge T, Krahnert I, Witt S, Obata T, Schauer N, Graham IA, et al. (2010) Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell 22: 1549–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwell S, Huang YS, Vilhjálmsson BJ, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone AM, Hu TT, et al. (2010) Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465: 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S, Schwartz C, Singh A, Warthmann N, Kim MC, Maloof JN, Loudet O, Trainer GT, Dabi T, Borevitz JO, et al. (2009) QTL mapping in new Arabidopsis thaliana advanced intercross-recombinant inbred lines. PLoS ONE 4: e4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265 [DOI] [PubMed] [Google Scholar]
- Batushansky A, Toubiana D, Fait A (2016) Correlation-based network generation, visualization, and analysis as a powerful tool in biological studies: a case study in cancer cell metabolism. BioMed Res Int 2016: 8313272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Boutin JP, Miquel M, Lepiniec L, Rochat C (2002) An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol Biochem 40: 151–160 [Google Scholar]
- Baud S, Dubreucq B, Miquel M, Rochat C, Lepiniec L (2008) Storage reserve accumulation in Arabidopsis: metabolic and developmental control of seed filling. The Arabidopsis Book 6: e0113, doi/10.1199/tab.0113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder S. (2010) Branched-chain amino acid metabolism in Arabidopsis thaliana. The Arabidopsis Book 8: e0137, doi/10.1199/tab.0137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder S, Knill T, Schuster J (2007) Branched-chain amino acid metabolism in higher plants. Physiol Plant 129: 68–78 [Google Scholar]
- Birtić S, Kranner I (2006) Isolation of high-quality RNA from polyphenol-, polysaccharide- and lipid-rich seeds. Phytochem Anal 17: 144–148 [DOI] [PubMed] [Google Scholar]
- Brachi B, Meyer CG, Villoutreix R, Platt A, Morton TC, Roux F, Bergelson J (2015) Coselected genes determine adaptive variation in herbivore resistance throughout the native range of Arabidopsis thaliana. Proc Natl Acad Sci USA 112: 4032–4037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright SWJ, Miflin BJ, Rognes SE (1982) Threonine accumulation in the seeds of a barley mutant with an altered aspartate kinase. Biochem Genet 20: 229–243 [DOI] [PubMed] [Google Scholar]
- Carter C, Pan S, Zouhar J, Avila EL, Girke T, Raikhel NV (2004) The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell 16: 3285–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EKF, Rowe HC, Corwin JA, Joseph B, Kliebenstein DJ (2011) Combining genome-wide association mapping and transcriptional networks to identify novel genes controlling glucosinolates in Arabidopsis thaliana. PLoS Biol 9: e1001125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chander S, Guo YQ, Yang XH, Zhang J, Lu XQ, Yan JB, Song TM, Rocheford TR, Li JS (2008) Using molecular markers to identify two major loci controlling carotenoid contents in maize grain. Theor Appl Genet 116: 223–233 [DOI] [PubMed] [Google Scholar]
- Chen W, Gao Y, Xie W, Gong L, Lu K, Wang W, Li Y, Liu X, Zhang H, Dong H, et al. (2014) Genome-wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism. Nat Genet 46: 714–721 [DOI] [PubMed] [Google Scholar]
- Clark TJ, Lu Y (2015) Analysis of loss-of-function mutants in aspartate kinase and homoserine dehydrogenase genes points to complexity in the regulation of aspartate-derived amino acid contents. Plant Physiol 168: 1512–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke JH, Mithen R, Brown JK, Dean C (1995) QTL analysis of flowering time in Arabidopsis thaliana. Mol Gen Genet 248: 278–286 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dietz KJ, Jäger R, Kaiser G, Martinoia E (1990) Amino acid transport across the tonoplast of vacuoles isolated from barley mesophyll protoplasts: uptake of alanine, leucine, and glutamine. Plant Physiol 92: 123–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fait A, Angelovici R, Less H, Ohad I, Urbanczyk-Wochniak E, Fernie AR, Galili G (2006) Arabidopsis seed development and germination is associated with temporally distinct metabolic switches. Plant Physiol 142: 839–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazzon AP, Ramirez MV, Warek U, Balk J, Frazzon J, Dean DR, Winkel BS (2007) Functional analysis of Arabidopsis genes involved in mitochondrial iron-sulfur cluster assembly. Plant Mol Biol 64: 225–240 [DOI] [PubMed] [Google Scholar]
- Galili G. (1995) Regulation of lysine and threonine synthesis. Plant Cell 7: 899–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili G. (2011) The aspartate-family pathway of plants: linking production of essential amino acids with energy and stress regulation. Plant Signal Behav 6: 192–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili G, Amir R (2013) Fortifying plants with the essential amino acids lysine and methionine to improve nutritional quality. Plant Biotechnol J 11: 211–222 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Jorge S, Ha SH, Magallanes-Lundback M, Gilliland LU, Zhou A, Lipka AE, Nguyen YN, Angelovici R, Lin H, Cepela J, et al. (2013) Carotenoid cleavage dioxygenase4 is a negative regulator of β-carotene content in Arabidopsis seeds. Plant Cell 25: 4812–4826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Jorge S, Mehrshahi P, Magallanes-Lundback M, Lipka AE, Angelovici R, Gore MA, DellaPenna D (2016) ZEAXANTHIN EPOXIDASE activity potentiates carotenoid degradation in maturing seed. Plant Physiol 171: 1837–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene CS, Krishnan A, Wong AK, Ricciotti E, Zelaya RA, Himmelstein DS, Zhang R, Hartmann BM, Zaslavsky E, Sealfon SC, et al. (2015) Understanding multicellular function and disease with human tissue-specific networks. Nat Genet 47: 569–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L, Jones AD, Last RL (2007) LC-MS/MS assay for protein amino acids and metabolically related compounds for large-scale screening of metabolic phenotypes. Anal Chem 79: 8067–8075 [DOI] [PubMed] [Google Scholar]
- Gu L, Jones AD, Last RL (2010) Broad connections in the Arabidopsis seed metabolic network revealed by metabolite profiling of an amino acid catabolism mutant. Plant J 61: 579–590 [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Rojas A, Betrán J, Scott MP, Atta H, Menz M (2010) Quantitative trait loci for endosperm modification and amino acid contents in quality protein maize. Crop Sci 50: 870–879 [Google Scholar]
- Guyer D, Patton D, Ward E (1995) Evidence for cross-pathway regulation of metabolic gene expression in plants. Proc Natl Acad Sci USA 92: 4997–5000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heremans B, Jacobs M (1995) Threonine accumulation in a mutant of Arabidopsis thaliana (L.) Heynh. with an altered aspartate kinase. J Plant Physiol 146: 249–257 [Google Scholar]
- Herman EM, Larkins BA (1999) Protein storage bodies and vacuoles. Plant Cell 11: 601–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilson P, Allemeersch J, Altmann T, Aubourg S, Avon A, Beynon J, Bhalerao RP, Bitton F, Caboche M, Cannoot B, et al. (2004) Versatile gene-specific sequence tags for Arabidopsis functional genomics: transcript profiling and reverse genetics applications. Genome Res. 14: 2176–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfte H, Chrispeels MJ (1992) Protein sorting to the vacuolar membrane. Plant Cell 4: 995–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland JB, Nyquist WE, Cervantes-Martínez CT (2010) Estimating and interpreting heritability for plant breeding: an update. In Janick J, ed, Plant Breeding Reviews, Vol 22 John Wiley & Sons, Oxford, doi/10.1002/9780470650202.ch2 [Google Scholar]
- Horton MW, Hancock AM, Huang YS, Toomajian C, Atwell S, Auton A, Muliyati NW, Platt A, Sperone FG, Vilhjálmsson BJ, et al. (2012) Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat Genet 44: 212–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Zhao Y, Wei X, Li C, Wang A, Zhao Q, Li W, Guo Y, Deng L, Zhu C, et al. (2011) Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat Genet 44: 32–39 [DOI] [PubMed] [Google Scholar]
- Hung HY, Browne C, Guill K, Coles N, Eller M, Garcia A, Lepak N, Melia-Hancock S, Oropeza-Rosas M, Salvo S, et al. (2012) The relationship between parental genetic or phenotypic divergence and progeny variation in the maize nested association mapping population. Heredity (Edinb) 108: 490–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingle RA. (2011) Histidine biosynthesis. The Arabidopsis Book 9: e0141, doi/10.1199/tab.0141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingle RA, Mugford ST, Rees JD, Campbell MM, Smith JAC (2005) Constitutively high expression of the histidine biosynthetic pathway contributes to nickel tolerance in hyperaccumulator plants. Plant Cell 17: 2089–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosen RVL, Arends D, Li Y, Willems LAJ, Keurentjes JJB, Ligterink W, Jansen RC, Hilhorst HWM (2013) Identifying genotype-by-environment interactions in the metabolism of germinating Arabidopsis seeds using generalized genetical genomics. Plant Physiol 162: 553–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochevenko A, Fernie AR (2011) The genetic architecture of branched-chain amino acid accumulation in tomato fruits. J Exp Bot 62: 3895–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korte A, Farlow A (2013) The advantages and limitations of trait analysis with GWAS: a review. Plant Methods 9: doi/10.1186/1746-4811-9-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kover PX, Valdar W, Trakalo J, Scarcelli N, Ehrenreich IM, Purugganan MD, Durrant C, Mott R (2009) A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet 5: e1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski SP, Lan TH, Feldmann KA, Paterson AH (1994) QTL mapping of naturally-occurring variation in flowering time of Arabidopsis thaliana. Mol Gen Genet 245: 548–555 [DOI] [PubMed] [Google Scholar]
- Krumsiek J, Suhre K, Evans AM, Mitchell MW, Mohney RP, Milburn MV, Wägele B, Römisch-Margl W, Illig T, Adamski J, et al. (2012) Mining the unknown: a systems approach to metabolite identification combining genetic and metabolic information. PLoS Genet 8: e1003005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Blom UM, Wang PI, Shim JE, Marcotte EM (2011) Prioritizing candidate disease genes by network-based boosting of genome-wide association data. Genome Res 21: 1109–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Yang X, Xu S, Cai Y, Zhang D, Han Y, Li L, Zhang Z, Gao S, Li J, et al. (2012) Genome-wide association studies identified three independent polymorphisms associated with α-tocopherol content in maize kernels. PLoS ONE 7: e36807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka AE, Gore MA, Magallanes-Lundback M, Mesberg A, Lin H, Tiede T, Chen C, Buell CR, Buckler ES, Rocheford T (2013) Genome-wide association study and pathway-level analysis of tocochromanol levels in maize grain. G3 (Bethesda) 3: 1287–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Camilleri C, Bouchez D, Daniel-Vedele F (2002) Bay-0 × Shahdara recombinant inbred line population: a powerful tool for the genetic dissection of complex traits in Arabidopsis. Theor Appl Genet 104: 1173–1184 [DOI] [PubMed] [Google Scholar]
- Lu Y, Savage LJ, Ajjawi I, Imre KM, Yoder DW, Benning C, Dellapenna D, Ohlrogge JB, Osteryoung KW, Weber AP, et al. (2008) New connections across pathways and cellular processes: industrialized mutant screening reveals novel associations between diverse phenotypes in Arabidopsis. Plant Physiol 146: 1482–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney GS, Kochevenko A, Tieman DM, Tohge T, Krieger U, Zamir D, Taylor MG, Fernie AR, Klee HJ (2010) Characterization of the branched-chain amino acid aminotransferase enzyme family in tomato. Plant Physiol 153: 925–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty F. (1999) Plant vacuoles. Plant Cell 11: 587–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda F, Nakabayashi R, Yang Z, Okazaki Y, Yonemaru J, Ebana K, Yano M, Saito K (2015) Metabolome-genome-wide association study dissects genetic architecture for generating natural variation in rice secondary metabolism. Plant J 81: 13–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M, Hu TT, Ishino Y, Jhaveri J, Toomajian C, Zheng H, Bakker E, Calabrese P, Gladstone J, Goyal R, et al. (2005) The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol 3: e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens BF, Lipka AE, Magallanes-Lundback M, Tiede T, Diepenbrock CH, Kandianis CB, Kim E, Cepela J, Mateos-Hernandez M, Buell CR, et al. (2014) A foundation for provitamin A biofortification of maize: genome-wide association and genomic prediction models of carotenoid levels. Genetics 198: 1699–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris N, Stanley CM, Jones RL, Rogers JC (1996) Plant cells contain two functionally distinct vacuolar compartments. Cell 85: 563–572 [DOI] [PubMed] [Google Scholar]
- Peng C, Uygun S, Shiu SH, Last RL (2015) The impact of the branched-chain ketoacid dehydrogenase complex on amino acid homeostasis in Arabidopsis. Plant Physiol 169: 1807–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt A, Horton M, Huang YS, Li Y, Anastasio AE, Mulyati NW, Agren J, Bossdorf O, Byers D, Donohue K, et al. (2010a) The scale of population structure in Arabidopsis thaliana. PLoS Genet 6: e1000843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt A, Vilhjálmsson BJ, Nordborg M (2010b) Conditions under which genome-wide association studies will be positively misleading. Genetics 186: 1045–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli R, Pilot G (2014) Regulation of amino acid metabolic enzymes and transporters in plants. J Exp Bot 65: 5535–5556 [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Donnelly P (2001) Case-control studies of association in structured or admixed populations. Theor Popul Biol 60: 227–237 [DOI] [PubMed] [Google Scholar]
- Rai VK. (2002) Role of amino acids in plant responses to stresses. Biol Plant 45: 481–487 [Google Scholar]
- Ramstein GP, Lipka AE, Lu F, Costich DE, Cherney JH, Buckler ES, Casler MD (2015) Genome-wide association study based on multiple imputation with low-depth sequencing data: application to biofuel traits in reed canarygrass. G3 (Bethesda) 5: 891–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter A, Schaff C, Zhang Z, Lipka AE, Tian F, Köllner TG, Schnee C, Preiß S, Irmisch S, Jander G, et al. (2016) Characterization of biosynthetic pathways for the production of the volatile homoterpenes DMNT and TMTT in Zea mays. Plant Cell 28: 2651–2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedelsheimer C, Lisec J, Czedik-Eysenberg A, Sulpice R, Flis A, Grieder C, Altmann T, Stitt M, Willmitzer L, Melchinger AE (2012) Genome-wide association mapping of leaf metabolic profiles for dissecting complex traits in maize. Proc Natl Acad Sci USA 109: 8872–8877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rognes SE, Bright SW, Miflin BJ (1983) Feedback-insensitive aspartate kinase isoenzymes in barley mutants resistant to lysine plus threonine. Planta 157: 32–38 [DOI] [PubMed] [Google Scholar]
- Sanders A, Collier R, Trethewy A, Gould G, Sieker R, Tegeder M (2009) AAP1 regulates import of amino acids into developing Arabidopsis embryos. Plant J 59: 540–552 [DOI] [PubMed] [Google Scholar]
- Sauer U, Lasko DR, Fiaux J, Hochuli M, Glaser R, Szyperski T, Wüthrich K, Bailey JE (1999) Metabolic flux ratio analysis of genetic and environmental modulations of Escherichia coli central carbon metabolism. J Bacteriol 181: 6679–6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scossa F, Brotman Y, De Abreu e Lima F, Willmitzer L, Nikoloski Z, Tohge T, Fernie AR (2016) Genomics-based strategies for the use of natural variation in the improvement of crop metabolism. Plant Sci 242: 47–67 [DOI] [PubMed] [Google Scholar]
- Segura V, Vilhjálmsson BJ, Platt A, Korte A, Seren Ü, Long Q, Nordborg M (2012) An efficient multi-locus mixed-model approach for genome-wide association studies in structured populations. Nat Genet 44: 825–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Alam M, Fikse F, Rönnegård L (2013) A novel generalized ridge regression method for quantitative genetics. Genetics 193: 1255–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepansky A, Leustek T (2006) Histidine biosynthesis in plants. Amino Acids 30: 127–142 [DOI] [PubMed] [Google Scholar]
- Steuer R, Kurths J, Fiehn O, Weckwerth W (2003) Interpreting correlations in metabolomic networks. Biochem Soc Trans 31: 1476–1478 [DOI] [PubMed] [Google Scholar]
- Su YH, Frommer WB, Ludewig U (2004) Molecular and functional characterization of a family of amino acid transporters from Arabidopsis. Plant Physiol 136: 3104–3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegeder M. (2014) Transporters involved in source to sink partitioning of amino acids and ureides: opportunities for crop improvement. J Exp Bot 65: 1865–1878 [DOI] [PubMed] [Google Scholar]
- Tohge T, Ramos MS, Nunes-Nesi A, Mutwil M, Giavalisco P, Steinhauser D, Schellenberg M, Willmitzer L, Persson S, Martinoia E, et al. (2011) Toward the storage metabolome: profiling the barley vacuole. Plant Physiol 157: 1469–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toubiana D, Batushansky A, Tzfadia O, Scossa F, Khan A, Barak S, Zamir D, Fernie AR, Nikoloski Z, Fait A (2015) Combined correlation-based network and mQTL analyses efficiently identified loci for branched-chain amino acid, serine to threonine, and proline metabolism in tomato seeds. Plant J 81: 121–133 [DOI] [PubMed] [Google Scholar]
- Toubiana D, Fernie AR, Nikoloski Z, Fait A (2013) Network analysis: tackling complex data to study plant metabolism. Trends Biotechnol 31: 29–36 [DOI] [PubMed] [Google Scholar]
- Toubiana D, Semel Y, Tohge T, Beleggia R, Cattivelli L, Rosental L, Nikoloski Z, Zamir D, Fernie AR, Fait A (2012) Metabolic profiling of a mapping population exposes new insights in the regulation of seed metabolism and seed, fruit, and plant relations. PLoS Genet 8: e1002612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trontin C, Tisné S, Bach L, Loudet O (2011) What does Arabidopsis natural variation teach us (and does not teach us) about adaptation in plants? Curr Opin Plant Biol 14: 225–231 [DOI] [PubMed] [Google Scholar]
- Tzin V, Galili G (2010) The biosynthetic pathways for shikimate and aromatic amino acids in Arabidopsis thaliana. The Arabidopsis Book 8: e0132, doi/10.1199/tab.0132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utz HF, Melchinger AE (1996) PLABQTL: a program for composite interval mapping of QTL. Journal of Agricultural Genomics 2: 1–6 [Google Scholar]
- Vallabhaneni R, Wurtzel ET (2009) Timing and biosynthetic potential for carotenoid accumulation in genetically diverse germplasm of maize. Plant Physiol 150: 562–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues PE, Lasky JR, Juenger TE, Liu TW, Kumar MN (2014) Genome-wide association mapping combined with reverse genetics identifies new effectors of low water potential-induced proline accumulation in Arabidopsis. Plant Physiol 164: 144–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weckwerth W. (2003) Metabolomics in systems biology. Annu Rev Plant Biol 54: 669–689 [DOI] [PubMed] [Google Scholar]
- Weckwerth W, Loureiro ME, Wenzel K, Fiehn O (2004) Differential metabolic networks unravel the effects of silent plant phenotypes. Proc Natl Acad Sci USA 101: 7809–7814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D. (2012) Natural variation in Arabidopsis: from molecular genetics to ecological genomics. Plant Physiol 158: 2–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentzell AM, Rowe HC, Hansen BG, Ticconi C, Halkier BA, Kliebenstein DJ (2007) Linking metabolic QTLs with network and cis-eQTLs controlling biosynthetic pathways. PLoS Genet 3: 1687–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter H, Robinson DG, Heldt HW (1994) Subcellular volumes and metabolite concentrations in spinach leaves. Planta 193: 530–535 [Google Scholar]
- Wong HK, Chan HK, Coruzzi GM, Lam HM (2004) Correlation of ASN2 gene expression with ammonium metabolism in Arabidopsis. Plant Physiol 134: 332–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzel ET, Cuttriss A, Vallabhaneni R (2012) Maize provitamin A carotenoids, current resources, and future metabolic engineering challenges. Front Plant Sci 3: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Kandianis CB, Harjes CE, Bai L, Kim EH, Yang X, Skinner DJ, Fu Z, Mitchell S, Li Q, et al. (2010) Rare genetic variation at Zea mays crtRB1 increases beta-carotene in maize grain. Nat Genet 42: 322–327 [DOI] [PubMed] [Google Scholar]
- Yan N, Doelling JH, Falbel TG, Durski AM, Vierstra RD (2000) The ubiquitin-specific protease family from Arabidopsis: AtUBP1 and 2 are required for the resistance to the amino acid analog canavanine. Plant Physiol 124: 1828–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Krebs M, Stierhof YD, Ludewig U (2014a) Characterization of the putative amino acid transporter genes AtCAT2, 3 & 4: the tonoplast localized AtCAT2 regulates soluble leaf amino acids. J Plant Physiol 171: 594–601 [DOI] [PubMed] [Google Scholar]
- Yang H, Postel S, Kemmerling B, Ludewig U (2014b) Altered growth and improved resistance of Arabidopsis against Pseudomonas syringae by overexpression of the basic amino acid transporter AtCAT1. Plant Cell Environ 37: 1404–1414 [DOI] [PubMed] [Google Scholar]
- Yang H, Stierhof YD, Ludewig U (2015) The putative Cationic Amino Acid Transporter 9 is targeted to vesicles and may be involved in plant amino acid homeostasis. Front Plant Sci 6: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Buckler ES (2006) Genetic association mapping and genome organization of maize. Curr Opin Biotechnol 17: 155–160 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.