Abstract
The circadian clock and light signaling regulate CONSTANS function through intricate mechanisms that reside in phloem companion cells of leaves for controlling photoperiodic flowering in Arabidopsis.
Plants sense changes in day length (= photoperiod) as a reliable seasonal cue to regulate important developmental transitions such as flowering. Integration of various external light information into the circadian clock-controlled mechanisms enables plants to precisely measure photoperiod changes in the surrounding environment. The core mechanism of photoperiodic measurement is regulation of CONSTANS (CO) activity, which takes place in phloem companion cells in leaves. Until recently, it remained unclear whether plants possess specific variations of the clock for this regulation. Now it is known that a specific circadian timing mechanism in the vascular tissue is essential for photoperiodic flowering. In addition to spatial tissue-specific regulation, temporal regulation of CO activity is also important. The identification and characterization of multiple regulators that physically interact with CO and influence its function in the morning in long days are two recent advances in photoperiodic regulation of flowering time. It seems that CO acts as a network hub to integrate various external and internal signals into the photoperiodic flowering pathway. CO regulates the amount of FLOWERING LOCUS T (FT) transcripts and FT protein moves from companion cells of leaf phloem to the shoot apical meristem. The protein that helps long-distance transport of FT protein was also identified recently. Here, we introduce recent advances in tissue-specific variations of the circadian clock and the emerging picture of the intricate connections of transcriptional regulators through CO in the morning, which all facilitate plants knowing when to flower.
Properly timing the floral transition is crucial for reproductive success. It can also influence the early development of offspring. To optimize this timing, plants constantly monitor changes in the surrounding environment. Among various environmental factors, observing changes in day length (= photoperiod) is the most reliable way for many organisms to know the specific time of year. Therefore, photoperiodic regulation is one of the major flowering time mechanisms and it has been studied since it was first reported in 1920 (Song et al., 2015). Arabidopsis (Arabidopsis thaliana), as it flowers mainly in spring, can sense lengthening days to induce flowering and became a suitable model to study photoperiodic flowering regulation. (Andrés and Coupland, 2012; Song et al., 2013; Pajoro et al., 2014; Shim and Imaizumi, 2015).
In principle, photoperiodic flowering mechanisms can be divided into three parts: light input, circadian clock, and output. Light information is integrated into innate photoperiodic timing mechanisms governed by the circadian clock to induce genes that trigger flowering. In flowering in Arabidopsis, light is perceived by various photoreceptors, such as the red/far-red light photoreceptor phytochrome (phy), and two classes of blue-light photoreceptors, cryptochrome (cry) and the ZEITLUPE (ZTL)/FLAVIN BINDING, KELCH REPEAT, F-BOX1 (FKF1)/LOV KELCH PROTEIN2 (LKP2) family of proteins. Photoperiodic information is ultimately converted into transcript levels of the FT gene (Song et al., 2015). FT is the florigen, as it is synthesized in leaves in long days and transported to the shoot apical meristem to start orchestrating expression of multiple floral identify genes, such as APETALA1 and LEAFY (Abe et al., 2005; Golembeski and Imaizumi, 2015). FT is mainly induced by the transcriptional activator CO in long days (Song et al., 2015). Therefore, the clock-controlled mechanism by which photoperiodic information regulates CO function is the key mechanism in Arabidopsis.
In this review, we first discuss our current view of circadian clock architecture and recent advances in tissue specificity in the plant clock. Next, we introduce recent updates on photoperiodic regulation of flowering, focusing on the complex regulation of CO function.
CIRCADIAN CLOCK ARCHITECTURE AND TISSUE SPECIFICITY
A Molecular Framework of the Circadian Oscillator
Recent genomic, biochemical, and computational approaches have greatly advanced our understanding of molecular architecture of the circadian clock in Arabidopsis (Hsu and Harmer, 2014; Shim and Imaizumi, 2015; Millar, 2016). Most clock components possess transcriptional activity, and are interconnected by time-resolved multiple feedback regulation to form this oscillator. Throughout the day, each clock gene also coordinates the expression of numerous output genes to regulate the timing of various physiological responses, such as growth and underlying metabolic regulation, hormone and stress responses, and so on (Farré and Weise, 2012; Shim and Imaizumi, 2015; Atamian and Harmer, 2016). Therefore, defects in the circadian clock have adverse effects on plant fitness (Green et al., 2002; Dodd et al., 2005).
In the current model of the Arabidopsis circadian clock, most components function as repressors (Fig. 1). At dawn, two MYB transcription factors, CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), repress evening-phased genes (Nagel et al., 2015; Kamioka et al., 2016). This repression is partly dependent on the function of the CONSTITUTIVE PHOTOMORPHOGENIC10 (COP10)-DE-ETIOLATED1-DAMAGED DNA BINDING1 complex, a negative regulator for photomorphogenesis (Lau et al., 2011). To repress transcription, CCA1 and LHY bind to related cis-elements called Evening Element and the CCA1 Binding Site (Harmer and Kay, 2005). Chromatin immunoprecipitation sequencing analyses brought us, to our knowledge, a new insight regarding potential variation in CCA1 binding sites. In addition to Evening Element and the CCA1 Binding Site, DNA motifs that contain G-box or CT repeats are significantly enriched in the CCA1 binding regions (Nagel et al., 2015; Kamioka et al., 2016), indicating that CCA1 may form complexes with different transcription factors that alter the DNA binding preferences of complexes that contain CCA1. Previously, genetic results implied that CCA1 and LHY directly act as transcriptional activators for the morning-phased genes PSEUDO RESPONSE REGULATOR9 (PRR9) and PRR7 (Farré et al., 2005). However, recent transient induction analysis demonstrated that CCA1 can directly repress the expression of both PRR9 and PRR7 (Kamioka et al., 2016), suggesting that the previous result can be caused by indirect effects of cca1 and lhy mutations.
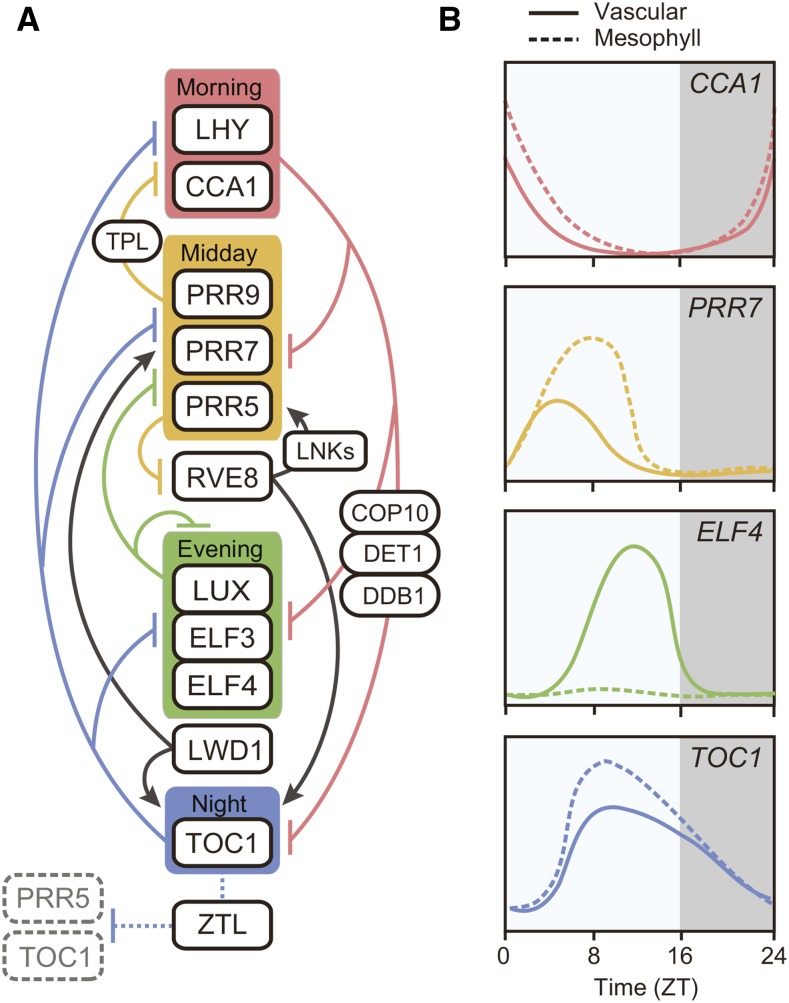
Figure 1.
A simplified model of the circadian clock architecture and tissue-specific expression profiles of core clock genes in Arabidopsis. A, Intricate transcriptional repression mechanisms interlocked with core clock components comprise the Arabidopsis circadian clock. In the morning, CCA1 and LHY directly bind to the promoters and repress the transcription of the clock genes expressed from midday to evening: PRR9, PRR7, PRR5, TOC1, LUX, ELF3, and ELF4. The CCA1/LHY-dependent repression is partly mediated by the interaction with COP10-DE-ETIOLATED1 (DET1)-DAMAGED DNA BINDING1 (DDB1) complex. During the day, PRR9, PRR7, and PRR5, all accompanied with TPL general repressor machinery, directly repress the expression of CCA1 and LHY. In the evening, ELF3, ELF4, and LUX form a complex referred to as the Evening Complex, and repress PRR9 and LUX transcription. LWD1, which binds to ELF3, induces PRR9, PRR5, and TOC1 expression. During the night, TOC1 reciprocally represses the core clock genes expressed from morning to evening (CCA1, LHY, PRR9, PRR7, TOC1, ELF4, and LUX) by directly binding to their promoters. As a transcriptional activator, RVE8, which peaks during the day, directly induces the expression of PRR5, TOC1, and most likely other evening-expressed genes that possess Evening Elements at their promoters. PRR5 in turn represses RVE8 transcription. LNK1 and LNK2 act as coactivators for RVE8 to induce PRR5 and TOC1. PRR5 and TOC1 proteins are subjected to 26S proteasome-mediated degradation at night by clock-associated F-box protein ZTL. Solid lines indicate transcriptional regulation, while dotted lines indicate posttranscriptional regulation. B, Daily expression profiles of clock genes show tissue specificity in long days. Solid lines represent expression patterns in vascular tissues, and dotted lines represent those in mesophyll cells. The expression profiles are drawn based on the results presented in Endo et al. (2014).
From early in the morning to the first-half part of the night, PRR9, PRR7, and PRR5 redundantly repress the transcription of CCA1 and LHY via G-box-like cis-elements (Nakamichi et al., 2010, 2012; Liu et al., 2016; Fig. 1). This depression of CCA1 and LHY levels during the day allows the induction of evening-phased genes, such as those encoding EARLY FLOWERING3 (ELF3), ELF4 proteins, and LUX ARRYTHMO (LUX), a GARP-type MYB transcription factor. LUX, ELF3, and ELF4 form a protein complex referred to as the Evening Complex that represses PRR9 and LUX expression through binding to LUX binding sites in the promoters (Dixon et al., 2011; Helfer et al., 2011; Nusinow et al., 2011; Herrero et al., 2012). Affinity purification-coupled mass spectrometry analysis revealed that Evening Complex interacts not only with other clock components [TIMING OF CAB EXPRESSION1 (TOC1), and LIGHT REGULATED WD1 (LWD1), etc.] but also with red-light signaling components (all five phytochromes, and COP1, etc.) in vivo (Huang et al., 2016). Interestingly, ELF3 loses its interaction with all light-signaling components in the phytochrome B (phyB) mutant background, suggesting that the phyB-ELF3 complex is one of the signaling hubs that connects red light signaling with the circadian clock. (Huang et al., 2016). This type of biochemical approach is powerful for deciphering the complex network architecture of the clock protein interactome, as well as for discovering new regulators overlooked by genetic screening.
At night, a pseudo-response regulator, TOC1 (also known as PRR1) protein becomes abundant and contributes to the repression of CCA1 and LHY transcription through direct binding to G-box related sequences (Gendron et al., 2012; Huang et al., 2012b; Fig. 1). In addition, TOC1 interacts with PHYTOCHROME INTERACTING FACTOR3, which binds to G-box, to repress transcription of their co-target genes (Soy et al., 2016). TOC1-dependent repression is gradually removed toward the end of the night by TOC1 protein degradation controlled by ZTL E3 ubiquitin ligase and its homologs, FKF1 and LKP2 (Más et al., 2003; Baudry et al., 2010; Fig. 1). The ZTL family of proteins also target PRR5 for degradation at night (Kiba et al., 2007; Baudry et al., 2010).
In addition to these repressors, a few activators are now known to exist in the core loops of the circadian clock. LWD1 and LWD2, two related WD repeat proteins, directly act as activators of PRR9, PRR5, and TOC1 (Wu et al., 2008; Wang et al., 2011; Fig. 1). The lwd1 lwd2 double mutant showed light dosage-dependent shorter period phenotypes, suggesting that LWDs also function in the light input pathway to the clock (Wang et al., 2011).
Another activator that has been identified is REVEILLE8 (RVE8; Fig. 1). In the early afternoon, RVE8 directly activates transcription of PRR5, TOC1, and likely other evening phased genes (Farinas and Más, 2011; Rawat et al., 2011; Nakamichi et al., 2012; Hsu et al., 2013). RVEs are close homologs of CCA1/LHY and share Evening Element binding sites (Hsu et al., 2013). RVE8 physically interacts with NIGHT LIGHT-INDUCIBLE AND CLOCK-REGULATED GENES1 (LNK1) and LNK2 during the day in a circadian manner (Xie et al., 2014). Its function is partially dependent on LNKs (Xie et al., 2014); however, it remains unknown how LNKs act as co-activators due to the lack of functional domains. Intriguingly, LNKs can also bind to CCA1 and LHY (Xie et al., 2014), and LNKs act as repressors for anthocyanin biosynthesis genes and work antagonistically with RVE8 (Pérez-García et al., 2015). Given that LNKs act downstream of phyB signaling (Rugnone et al., 2013), LNKs also have roles in the light signaling pathway.
Recent mathematical modeling revealed that adding the RVE8 activation loop to the previously published repressilator model did not drastically change the parameters used in the repressilator model (Fogelmark and Troein, 2014). Although genes involved in light input or nontranscriptional regulation still need to be incorporated in future models to capture all aspects of the molecular clock, this implies that the activation loop is not essential for oscillator function. Rather, it can confer robustness of the clock to a wide range of environmental conditions (Fogelmark and Troein, 2014). Given that the circadian clock regulates many physiological responses in various cell types, it could be advantageous to have a more robust and adjustable oscillator. In the next section, we introduce this view of tissue specificity in the molecular clock.
Tissue Specificity in Clock Function
As the circadian clock can regulate many aspects of plant physiology and that most clock components are widely expressed in different tissues, it was assumed that each cell had a cell-autonomous clock. Consistent with this idea, different phases of rhythmicity can be maintained within the same plant (Thain et al., 2000). Previous reports showed that a local cell-to-cell rhythm coupling mechanism exists, and it creates spatiotemporal waves of clock gene expression especially under constant light conditions (Fukuda et al., 2007; Wenden et al., 2012). Local coupling of rhythms among neighboring cells is also observed in duckweed cells, but light signal overwrites local coupling effect and masks heterogeneity of individual cell rhythmicity (Muranaka and Oyama, 2016). The Arabidopsis clock in the shoot apex can be distinguished from other tissues in the sense that individual cells are tightly coupled to synchronize rhythmicity (Takahashi et al., 2015). In addition, micrografting experiments demonstrated that the clock in the shoot apex can affect circadian rhythms in root tissues, although the actual nature of the systemic signaling component for rhythm coupling remains unknown (Takahashi et al., 2015). It is noteworthy that mathematical modeling suggests that the difference in clock properties between the shoot and root can be explained by light sensitivity of clock entrainment in each tissue (Bordage et al., 2016). Supporting this idea, tissue-specific clock properties, such as phase variation in clock genes, or different sensitivity to temperature signals, has been reported in intact Arabidopsis plants (Thain et al., 2002; Michael et al., 2003; Yakir et al., 2011). Recent tissue-specific microarray time-course analysis revealed that the clock genes in mesophyll cells and vascular cells show distinct expression profiles from each other (Fig. 1; Endo et al., 2014). By disrupting clock functions in specific tissues, it can be shown that the vascular clock also affects rhythmic expression of core clock genes in mesophyll cells (Endo et al., 2014). In addition, vascular-specific expression of clock genes is essential for photoperiodic flowering regulation, while the circadian clock in the epidermis plays a crucial role in temperature-mediated hypocotyl elongation (Shimizu et al., 2015). These results suggest that the circadian clock in each tissue contributes differently to regulating specific physiological responses. It seems that the vascular tissue possesses a more complicated clock that may provide more accurate timing information for seasonal flowering.
PHOTOPERIODIC REGULATION OF FLOWERING TIME
The photoperiod-sensing mechanisms described below reside in phloem companion cells in leaves (Golembeski and Imaizumi, 2015). As discussed above, the circadian clock that exists in the vascular tissues is essential for proper photoperiodic flowering. This is because a key transcriptional activator of the photoperiodic pathway, CO, is regulated by the circadian clock and light signaling pathways (Putterill et al., 1995; Samach et al., 2000; Suárez-López et al., 2001). Complex multiple layers of regulation ensure CO only induces FT under preferable environmental conditions. In this section, we summarize recent progress in the molecular mechanisms of photoperiodic flowering, featuring regulation related to CO abundance and activity.
Temporal Transcriptional Regulation of CO
Transcriptional and posttranslational regulation of CO is important for incorporating day-length information into the flowering mechanism. In the morning, CO transcription is repressed by the CYCLING DOF FACTOR (CDF) family of proteins that bind to DOF binding sites in the CO promoter (Imaizumi et al., 2005; Fornara et al., 2009). There is natural variation in the number of tandem repeats (two to four) of the DOF binding sites in the CO promoter among Arabidopsis wild-type accessions. When the number of DOF binding sites is higher, the daytime suppression of CO is greater and flowering time is consequently delayed more (Rosas et al., 2014). Temporal expression profiles of CDFs are directly controlled by the circadian clock (Nakamichi et al., 2007, 2012; Ito et al., 2008; Fornara et al., 2009). Transcription of CDFs is induced in the morning by CCA1 and LHY (Niwa et al., 2007), and repressed by PRR9, PRR7, and PRR5 in the afternoon (Nakamichi et al., 2012). The blue-light photoreceptor E3 ubiquitin ligase FKF1 removes CDF-dependent repression of CO transcription in the long-day afternoon (Sawa et al., 2007; Fornara et al., 2009). Once FKF1 observes blue light through its LOV domain, it forms a complex with GIGANTEA (GI). The FKF1-GI complex recognizes CDF1 (and CDF2, and likely other CDFs as well) as substrates for degradation. This temporal degradation of CDF proteins in long days is achieved by the coincidence of the circadian clock-controlled timing of FKF1 and GI expression with perception of light by FKF1 (Sawa et al., 2007). In short days, the contribution of FKF1 to CDF degradation is negligible, as FKF1 is mostly expressed in the dark. Once CDF proteins are removed by the FKF1-GI complex from the CO promoter, the basic Helix-Loop-Helix transcriptional activators FLOWERING BHLH1 (FBH1), FBH2, FBH3, and FBH4 bind to E-box cis-elements in the CO promoter to activate its transcription (Ito et al., 2012). Even though FBHs are strong activators of CO, CO expression in fbh quadruple mutants suggests that there may be other unknown positive regulators of CO transcription.
Posttranslational Regulation of CO Protein
Similar to transcriptional regulation, posttranslational regulation of CO protein is also tightly controlled by intricate mechanisms (Andrés and Coupland, 2012; Shim and Imaizumi, 2015; Song et al., 2015; Fig. 2). CO transcript is highly expressed from the late afternoon to the dawn, but CO protein only accumulates in the late afternoon in long days. To set a narrow time window for CO protein stabilization, plants utilize multiple photoreceptors and E3 ubiquitin ligases.
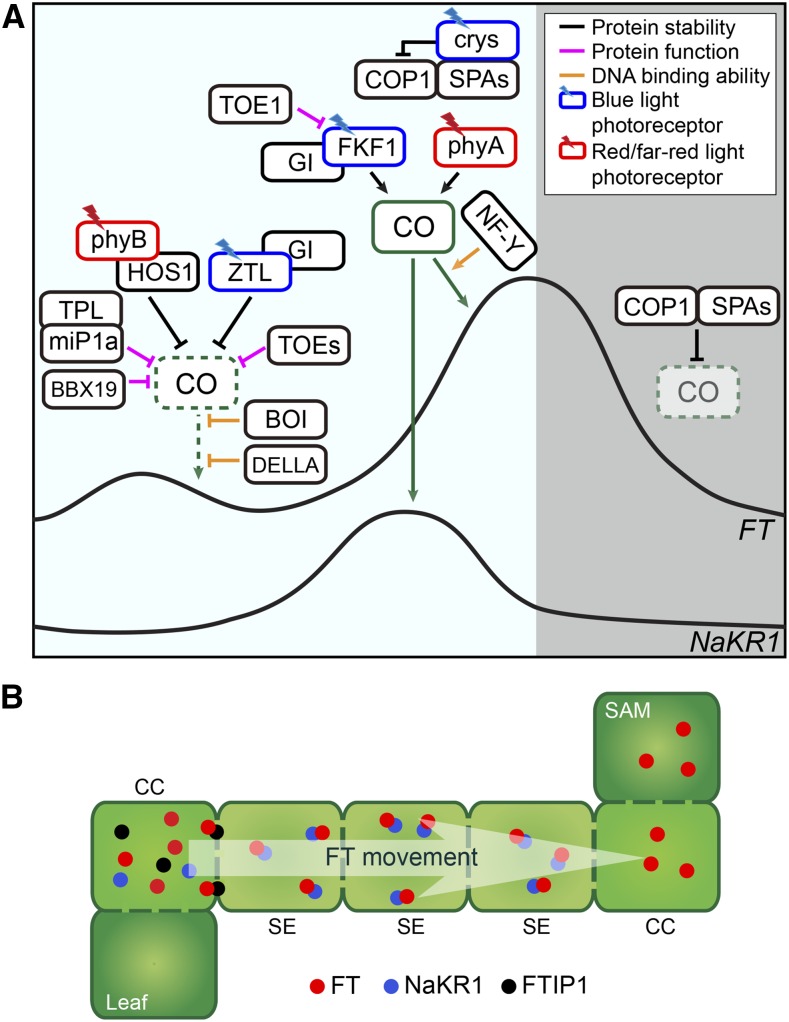
Figure 2.
Photoperiodic regulation of FT expression by CO, and FT protein movement. A, Arabidopsis plants possess multiple regulatory components to specifically induce FT expression in the afternoon in long days. During the morning, the abundance of CO protein and its activity are decreased through interactions with various proteins. These mechanisms inhibit CO-dependent activation of FT in the morning. CO protein is destabilized through phyB, using two different E3 ubiquitin ligases, HOS1 and ZTL. Red light absorbed by phyB induces formation of the phyB-HOS1 complex that degrades CO. Together with GI protein, ZTL also destabilizes CO. In addition to protein stability regulation, transcriptional activity of CO is also directly suppressed by two B-BOX-containing proteins, BBX19 and miP1a. Both proteins are highly expressed in the morning to reduce the amount of active CO protein. MiP1a-dependent suppression of CO activity is attained by direct recruitment of TPL cotranscriptional repressor. Two AP2-domain transcription factors, TOE1 and TOE2, can also form a complex with CO to suppress CO activity in the morning. DELLA protein, the key repressor of the GA signaling pathway, interacts with CO to prevent its function potentially by interrupting its interaction with NF-Y. The RING domain protein, BOI, inhibits DNA binding activity of CO protein to the FT promoter. In the afternoon, CO protein is stabilized by two classes of blue-light photoreceptors: crys and FKF1 protein. Crys negatively regulate the function of the COP1-SPA complexes that degrade CO. The FKF1-GI complex binds to CO to stabilize it through unknown mechanisms. TOE1 may counteract the FKF1-dependent stabilization of CO by competing with CO for the same interacting domain of FKF1. Far-red light absorbed by phyA also contributes to stabilization of CO in the afternoon. Once CO is stabilized in the afternoon, CO activates transcription of FT. The NF-Y complex enhances the binding of CO protein to the FT promoter. CO also activates transcription of the FT transporter gene, NaKR1, in long-day afternoons. During the night, the COP1-SPA complexes actively degrade CO protein. Therefore, FT levels decline during the night. CO symbols drawn with the dotted line indicate a lower abundance of CO protein. B, Long-distance transport of FT protein is mediated by its interacting partners. FT is transported from companion cells of leaf phloem tissues (where it is synthesized) to the shoot apical meristem. Two transporter proteins are important for FT movement. FTIP1 is localized in the plasmodesmata that connects companion cells to the sieve elements, and is required for transport of FT from companion cells to sieve elements, whereas NaKR1 is responsible for long-distance movement of FT through sieve elements to the SAM. CCs, companion cells; SAM, shoot apical meristem; SEs, sieve elements.
Once there is light, various photoreceptors participate in posttranslational regulation of CO protein to orchestrate CO protein accumulation. Blue-light photoreceptors, cry1 and cry2, stabilize CO in a light-dependent manner by attenuating COP1-SUPPRESSOR OF PHYA-105 1 (SPA1) activity throughout the day (Liu et al., 2008b, 2011; Zuo et al., 2011). Cry1 physically interacts with SPA1 to interrupt the formation of the COP1-SPA1 complex. Cry2 also binds to SPA1 in response to blue light, and the light-dependent cry2-SPA1 interaction enhances the cry2-COP1 interaction to suppress the function of COP1. Two phytochromes, phyA and phyB, act antagonistically on CO protein stability regulation. CO protein is destabilized by red light through phyB, but stabilized by far-red light through phyA (Valverde et al., 2004). PhyB-dependent regulation of CO stability can be explained by two distinct mechanisms [i.e. HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1 (HOS1) and PHYTOCHROME-DEPENDENT LATE-FLOWERING (PHL)] (Endo et al., 2013; Lazaro et al., 2015). In the morning, HOS1 E3 ubiquitin ligase restricts accumulation of CO protein by degrading it (Lazaro et al., 2012, 2015). Because HOS1 and phyB have a similar effect on CO protein stability in the morning, it is conceivable that they could be in the same pathway. Consistent with this idea, it has been reported that HOS1 is genetically in the same pathway for FT induction with phyB in long days (Lazaro et al., 2015). Similar to phyB, HOS1 is also required for red light-mediated degradation of CO. In addition, HOS1 physically interacts with phyB and CO. PhyB-dependent CO destabilization can therefore operate through HOS1 in the morning. PhyB-dependent destabilization of CO can be relieved in the afternoon by PHL (Endo et al., 2013). In the afternoon, PHL functions as a positive regulator of flowering by countering the inhibitory effect of phyB on CO stability. PHL interacts with phyB and CO under red light conditions. Therefore, PHL interferes with phyB-dependent destabilization of CO in the afternoon, and this regulation could help plants to secure the amount of CO protein required for further stabilization by blue light until late afternoon. It is unclear how phyA stabilizes CO in the afternoon, but it could be the mechanism by which phyA disrupts the function of the COP1-SPA complex (Sheerin et al., 2015). Light-activated phyA (and also phyB) competes with COP1 for binding to SPA1 and SPA2, leading to inactivation of the COP1-SPA complex. (Hajdu et al., 2015).
In addition to these photoreceptor-mediated mechanisms, there is the ZTL/FKF1-dependant mechanism that directly regulates CO protein stability throughout the day. In the morning, ZTL, likely together with GI, negatively controls CO stability (Song et al., 2014), while in the afternoon FKF1 stabilizes CO protein in a blue-light-dependent manner (Song et al., 2012). As GI also interacts with CO directly, FKF1, GI, and CO potentially form a trimeric complex and stabilize CO (Song et al., 2014). Once plants are in the dark, the COP1-SPA complex degrades CO protein (Jang et al., 2008; Liu et al., 2008b; Sarid-Krebs et al., 2015).
Apart from CO stability regulation, CO activity is also regulated by physical interactions with other B-BOX domain proteins (BBX): BBX19, microProtein 1a (miP1a)/BBX30, and miP1b/BBX31 (Wang et al., 2014; Graeff et al., 2016; Fig. 2). BBX19 possesses two B-BOX domains at the N terminus but lacks the CCT domain. BBX19 is highly expressed in the morning and BBX19 protein binds to CO to suppress its function; therefore, suppression of BBX19 only affects FT expression in the morning. Two small B-BOX proteins, MiP1a and MiP1b (previously known as BBX30 and BBX31), also form a complex with CO to prevent its function (Khanna et al., 2009; Graeff et al., 2016). Similar to BBX19, MiP1a is highly expressed in the morning in long days. MiP1a suppresses CO-dependent activation of FT through recruiting the TOPLESS (TPL) transcriptional repressor on the FT promoter. Together with BBX19, they form an additional regulatory loop of CO for FT activation in the morning.
AP2 type transcription factors, TARGET OF EAT1 (TOE1) and TOE2, repress FT expression. These AP2 type proteins are age-dependent regulators of FT, as they are posttranscriptionally regulated by miR172 (Aukerman and Sakai, 2003; Chen, 2004; Song et al., 2013). TOE1 also represses FT expression by physically suppressing the function of CO on the FT promoter (Zhang et al., 2015). TOE1-dependent suppression of FT requires its EAR-like motif, a binding site of TPL. Therefore, TOE1 likely represses CO-dependent FT activation through recruiting the TPL repressor complex. TOE1 also interacts with FKF1 and may interfere with the FKF1-CO interaction, resulting in destabilization of CO protein in the afternoon.
Transcriptional Regulation of FT
In addition to CO, there are multiple components that affect the transcription of FT (please see details in Andrés and Coupland, 2012; Shim and Imaizumi, 2015; Song et al., 2015). In this section, we focus on introducing recent updates related to the functions of two transcriptional activators of FT, CO, and CRYPTOCHROME-INTERACTING BASIC HELIX-LOOP-HELIX (CIB).
Once CO is stabilized in the late afternoon in long days, CO binds to the cis-element referred to as CO Responsive Element (CORE) in the FT promoter (Tiwari et al., 2010). Even though two CORE motifs (that are located within approximately 250 bps upstream from the transcriptional start site of the FT promoter) are crucial for FT induction, the 5.3 kb upstream region of the FT promoter that contains the CCAAT-binding motif is also required for its full induction (Adrian et al., 2010). CO and CO-like proteins physically interact with NUCLEAR FACTOR-Y (NF-Y) transcription factors, and genetic results support the functional interdependency of CO and NF-Y (Ben-Naim et al., 2006; Wenkel et al., 2006). However, it was difficult to explain how their interaction occurs in vivo, because the physical distance between the upstream NF-Y binding site and the COREs is more than 5 kb apart. The question regarding this physical limitation was recently answered. The chromatin looping of the FT locus brings the distal NF-Y binding site in close proximity to the CORE sequences near the transcriptional start site (Cao et al., 2014). The degree of looping becomes greater in the late afternoon when FT is induced in long days. Interestingly, the chromatin looping in the FT locus is not solely dependent on the presence of either CO or NF-Y (Cao et al., 2014).
Even though the contribution of gibberellic acid (GA) to flowering time is more pronounced in short days, GA is also required for proper induction of flowering through FT in long days (Galvão et al., 2012; Porri et al., 2012). Mutations in GA signaling or the depletion of active GA pools cause late flowering with lower expression of FT in long days. It was recently demonstrated that GA signaling can be integrated into the photoperiodic pathway partly through physical interaction of DELLA with CO (Xu et al., 2016; Yu et al., 2016; Fig. 2). DELLA proteins act as negative regulators of FT expression in vascular tissues (Galvão et al., 2012). Co-expression of DELLA protein attenuates CO-dependent activation of the FT promoter in Arabidopsis protoplasts (Yu et al., 2016). This suppression could be attained by dosage-dependent inhibition of DELLA proteins on CO-NF-Y complex formation, as an increasing amount of DELLA proteins results in a decreased amount of CO and NF-Y interaction (Xu et al., 2016). The expression profile of REPRESSOR OF GA1-3 protein, a member of the DELLA protein family, diurnally oscillates with a trough in the afternoon when CO induces FT expression. GA also regulates CO function through BOTRYTIS SUSCEPTIBLE1 INTERACTOR (BOI; Nguyen et al., 2015). BOI interacts with CO but it does not affect CO protein stability; instead, it influences the DNA binding ability of CO to the FT promoter. BOI also physically associates with the FT promoter to negatively regulate its expression, and GA reduces the binding of BOI on the FT promoter. Therefore, BOI can compete with CO on the FT promoter to restrict CO function.
Other transcription factors, CIBs (CIB1, CIB2, CIB4, and CIB5), redundantly activate FT expression (Liu et al., 2008a, 2013c). CIBs form various combinations of heterodimers that possess higher affinities to E-box sequences (CANNTG). CIB1 was originally characterized as the G-box (CACGTG) binding factor in vitro, which binds to the E-box in vivo (Liu et al., 2008a). Heterodimer formation seems to be important for shifting the preference of binding sites from G-box to E-box. As FT promoter only possesses the E-box, CIBs must exist as heterodimers on the promoter. The presence of blue light is important for CIB function, as its transcriptional activity is enhanced by cry2 and the protein is stabilized by ZTL and LKP2 in a light-dependent manner (Liu et al., 2008a, 2013a). In contrast to CO, when CIB1 is constitutively overexpressed in plants, it can increase FT levels only in the afternoon when FT is usually expressed in long days (Liu et al., 2008a). This implies that CIBs may require other photoperiodic regulators to induce FT expression.
FT Protein Movement
Once FT protein is synthesized in companion cells of leaf phloem in long days, it travels through the vascular system to the shoot apical meristem and triggers the phase transition from vegetative to reproductive growth (Liu et al., 2013b; Putterill and Varkonyi-Gasic, 2016; Fig. 2). Two membrane-associated proteins, FT-INTERACTING PROTEIN1 (FTIP1) and SODIUM POTASSIUM ROOT DEFECTIVE1 (NaKR1), are identified as molecular transporters of FT movement (Liu et al., 2012; Zhu et al., 2016; Fig. 2). FTIP1, which is localized in the ER in companion cells, and in the plasmodesmata between companion cells and sieve elements, interacts with FT protein in plasmodesmata, and it is required for FT export from companion cells to the sieve elements (Liu et al., 2012). Although FTIP1 expression is not affected by photoperiod (Liu et al., 2012), the phloem-expressed MYB transcription factor FE/ALTERED PHLOEM DEVELOPMENT, which contributes to FT expression in long days, is required for the expression of FTIP1 (Abe et al., 2015), indicating the connection of FT induction and FT movement. There is another example for this connection. NaKR1 is required for long distance movement of FT protein through sieve elements (Zhu et al., 2016). NaKR1 expression is highly induced in the afternoon in long days but not in short days, and CO is crucial for long day-specific induction of NaKR1 (Fig. 2). CO physically associates with CORE-like sequences in the NaKR1 promoter to induce its expression (Zhu et al., 2016). The movement of FT from companion cells to the distal shoot apical meristem is regulated by at least two distinct mechanisms (Fig. 2). Therefore, photoperiodic flowering mechanisms that exist in companion cells also facilitate FT movement to the short apical meristem.
FUTURE PERSPECTIVES
It has been more than a decade since we learned the tissue-specific existence of the photoperiodic time measurement mechanism (because CO and FT expression is restricted to phloem companion cells; Takada and Goto, 2003). Until recently, it remained unknown whether the upstream circadian clock structure and functions also had tissue-specific variations. As discussed in this article, recent work demonstrated that the expression patterns of the clock genes show tissue-specific variations in Arabidopsis, and that the clock in each specific tissue differently affects each specific output, as it was shown that the functional vascular clock is essential for controlling photoperiodic flowering (Endo et al., 2014; Shimizu et al., 2015). As was also shown recently, the plant clock in each cell can synchronize within and across the tissues (Wenden et al., 2012; Takahashi et al., 2015; Muranaka and Oyama, 2016), although they can independently sustain circadian rhythmicity in clock gene transcription. Similar to the synchronization of the clock-regulated genes observed, do the phloem companion cells that express FT also communicate with each other to coordinate the timing of expression of FT?
Unlike cry2 and phyA, at least, phyB expressed in mesophyll cells (but not in vascular tissues) regulates photoperiodic flowering (Endo et al., 2005, 2007; Kirchenbauer et al., 2016), indicating the presence of intertissue communication. To investigate inter- and intratissue cell-to-cell communication in the photoperiodic pathway, we first need to understand which gene is expressed exactly where. To obtain transcriptome data from specific tissue types and/or a single cell, the laser capture microdissection and/or isolation-of-nuclei-tagged-in-specific-cell-types (INTACT) method (Deal and Henikoff, 2011) could be useful for preparing samples. Obtaining results from these types of analyses is necessary to begin investigating potential cell-to-cell communication for flowering time regulation.
Within the last several years, our understanding of the molecular mechanisms of photoperiodic flowering time regulation has advanced due to the identification of new regulators that affect CO transcriptional activity, the finding of chromatin structure changes in the FT locus, and the characterization of additional components that regulate CO protein stability. As CO-dependent regulation of FT is well conserved in other plant species (Shrestha et al., 2014; Song et al., 2015), regulation of CO activity is the important mechanism for controlling flowering time. For instance, in the short-day plant, rice (Oryza sativa), the CO ortholog, Heading date1 (Hd1), functions as both activator and repressor of rice FT, Heading date 3a (Hd3a). This functional conversion was regulated by light signaling perceived by phytochrome, but the mechanism of the conversion remained unknown. Similar to the regulation of CO function described in this article, recent work demonstrated that the physical interactions of Hd1 with other regulators likely control this functional conversion. Hd1 interacts with the protein that contains the CCT domain, Grain number, plant height, and heading date7, which is a negative regulator of flowering in long days. This interaction makes Hd1 the repressor of Hd3a (Nemoto et al., 2016). This finding is important for understanding how Hd1 suppresses rice flowering in noninductive conditions. A mechanism similar to the CO-BBX functional interaction may also exist in rice. The B-box protein, OsBBX14, which has two B-BOX domains, counteracts Hd1 function by suppressing expression of Hd3a (Bai et al., 2016). Including OsBBX14, other OsBBXs show diurnal oscillation of expression with peaks in the morning or night (Huang et al., 2012a). Based on findings in Arabidopsis, Hd1 and OsBBX14 may form a complex to regulate flowering time. These examples from rice clearly signify the importance of studies in various plant species to learn the conserved as well as unique flowering mechanisms, both of which must be important for the adaptation of each species to the environment. Further studies are awaited to investigate the presence of similar mechanisms in other crop species, such as long-day plants wheat (Triticum aestivum) and barley (Hordeum vulgare).
As discussed above, multiple repressors interact with CO to repress its activity in the morning, and some of the mechanisms are likely conserved in other plants. In addition, often these mechanisms are long-day specific. Why do Arabidopsis plants possess multiple repressive mechanisms of CO in the morning? Does it mean plants can fine-tune the activity of CO depending on various external and internal conditions in the morning? For instance, in addition to age-dependent regulation (Aukerman and Sakai, 2003; Song et al., 2013), low temperature in the morning can also strengthen the function of TOEs to inhibit flowering by down-regulating miRNA172s (Lee et al., 2010). BBX19 can link retrograde signals derived from chloroplasts with flowering time regulation (Wang and Dehesh, 2015), potentially through the function of CO. In addition, integration of major GA signaling regulators, DELLA proteins, into the regulation of CO function also made our view of flowering pathways more complex. Expression of DELLA is controlled by the circadian clock with peak expression in the morning (Arana et al., 2011). In addition to DELLA-CO regulation, expression of the GA biosynthesis genes is negatively regulated by floral repressors, such as SHORT VEGETATIVE PHASE and TEMPRANILLO1 (Osnato et al., 2012; Andrés et al., 2014). It will be of great interest to learn how and when signal integrations of internal or external cues occur to regulate CO function to optimize flowering time to the everchanging environment.
Acknowledgments
We apologize to researchers whose relevant studies were not cited in this review due to page limitations.
Footnotes
This work was supported by NIH grant (GM079712) and Next-Generation BioGreen 21 Program (SSAC, PJ011175, Rural Development Administration, Republic of Korea) to T.I. A.K. is supported by JSPS Postdoctoral Fellowships for Research Abroad.
Articles can be viewed without a subscription.
References
- Abe M, Kaya H, Watanabe-Taneda A, Shibuta M, Yamaguchi A, Sakamoto T, Kurata T, Ausín I, Araki T, Alonso-Blanco C (2015) FE, a phloem-specific Myb-related protein, promotes flowering through transcriptional activation of FLOWERING LOCUS T and FLOWERING LOCUS T INTERACTING PROTEIN 1. Plant J 83: 1059–1068 [DOI] [PubMed] [Google Scholar]
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 [DOI] [PubMed] [Google Scholar]
- Adrian J, Farrona S, Reimer JJ, Albani MC, Coupland G, Turck F (2010) cis-Regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell 22: 1425–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés F, Coupland G (2012) The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13: 627–639 [DOI] [PubMed] [Google Scholar]
- Andrés F, Porri A, Torti S, Mateos J, Romera-Branchat M, García-Martínez JL, Fornara F, Gregis V, Kater MM, Coupland G (2014) SHORT VEGETATIVE PHASE reduces gibberellin biosynthesis at the Arabidopsis shoot apex to regulate the floral transition. Proc Natl Acad Sci USA 111: E2760–E2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arana MV, Marín-de la Rosa N, Maloof JN, Blázquez MA, Alabadí D (2011) Circadian oscillation of gibberellin signaling in Arabidopsis. Proc Natl Acad Sci USA 108: 9292–9297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atamian HS, Harmer SL (2016) Circadian regulation of hormone signaling and plant physiology. Plant Mol Biol 91: 691–702 [DOI] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H (2003) Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai B, Zhao J, Li Y, Zhang F, Zhou J, Chen F, Xie X (2016) OsBBX14 delays heading date by repressing florigen gene expression under long and short-day conditions in rice. Plant Sci 247: 25–34 [DOI] [PubMed] [Google Scholar]
- Baudry A, Ito S, Song YH, Strait AA, Kiba T, Lu S, Henriques R, Pruneda-Paz JL, Chua N-H, Tobin EM, Kay SA, Imaizumi T (2010) F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell 22: 606–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Naim O, Eshed R, Parnis A, Teper-Bamnolker P, Shalit A, Coupland G, Samach A, Lifschitz E (2006) The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant J 46: 462–476 [DOI] [PubMed] [Google Scholar]
- Bordage S, Sullivan S, Laird J, Millar AJ, Nimmo HG (2016) Organ specificity in the plant circadian system is explained by different light inputs to the shoot and root clocks. New Phytol 212: 136–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S, Kumimoto RW, Gnesutta N, Calogero AM, Mantovani R, Holt BF III (2014) A distal CCAAT/NUCLEAR FACTOR Y complex promotes chromatin looping at the FLOWERING LOCUS T promoter and regulates the timing of flowering in Arabidopsis. Plant Cell 26: 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2004) A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303: 2022–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Henikoff S (2011) The INTACT method for cell type-specific gene expression and chromatin profiling in Arabidopsis thaliana. Nat Protoc 6: 56–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon LE, Knox K, Kozma-Bognar L, Southern MM, Pokhilko A, Millar AJ (2011) Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis. Curr Biol 21: 120–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd AN, Salathia N, Hall A, Kévei E, Tóth R, Nagy F, Hibberd JM, Millar AJ, Webb AA (2005) Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309: 630–633 [DOI] [PubMed] [Google Scholar]
- Endo M, Mochizuki N, Suzuki T, Nagatani A (2007) CRYPTOCHROME2 in vascular bundles regulates flowering in Arabidopsis. Plant Cell 19: 84–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Nakamura S, Araki T, Mochizuki N, Nagatani A (2005) Phytochrome B in the mesophyll delays flowering by suppressing FLOWERING LOCUS T expression in Arabidopsis vascular bundles. Plant Cell 17: 1941–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Shimizu H, Nohales MA, Araki T, Kay SA (2014) Tissue-specific clocks in Arabidopsis show asymmetric coupling. Nature 515: 419–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Tanigawa Y, Murakami T, Araki T, Nagatani A (2013) PHYTOCHROME-DEPENDENT LATE-FLOWERING accelerates flowering through physical interactions with phytochrome B and CONSTANS. Proc Natl Acad Sci USA 110: 18017–18022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinas B, Más P (2011) Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. Plant J 66: 318–329 [DOI] [PubMed] [Google Scholar]
- Farré EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA (2005) Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol 15: 47–54 [DOI] [PubMed] [Google Scholar]
- Farré EM, Weise SE (2012) The interactions between the circadian clock and primary metabolism. Curr Opin Plant Biol 15: 293–300 [DOI] [PubMed] [Google Scholar]
- Fogelmark K, Troein C (2014) Rethinking transcriptional activation in the Arabidopsis circadian clock. PLOS Comput Biol 10: e1003705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F, Panigrahi KCS, Gissot L, Sauerbrunn N, Rühl M, Jarillo JA, Coupland G (2009) Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell 17: 75–86 [DOI] [PubMed] [Google Scholar]
- Fukuda H, Nakamichi N, Hisatsune M, Murase H, Mizuno T (2007) Synchronization of plant circadian oscillators with a phase delay effect of the vein network. Phys Rev Lett 99: 098102. [DOI] [PubMed] [Google Scholar]
- Galvão VC, Horrer D, Küttner F, Schmid M (2012) Spatial control of flowering by DELLA proteins in Arabidopsis thaliana. Development 139: 4072–4082 [DOI] [PubMed] [Google Scholar]
- Gendron JM, Pruneda-Paz JL, Doherty CJ, Gross AM, Kang SE, Kay SA (2012) Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc Natl Acad Sci USA 109: 3167–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golembeski GS, Imaizumi T (2015) Photoperiodic regulation of florigen function in Arabidopsis thaliana. Arabidopsis Book 13: e0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeff M, Straub D, Eguen T, Dolde U, Rodrigues V, Brandt R, Wenkel S (2016) MicroProtein-mediated recruitment of CONSTANS into a TOPLESS trimeric complex represses flowering in Arabidopsis. PLoS Genet 12: e1005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RM, Tingay S, Wang ZY, Tobin EM (2002) Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol 129: 576–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdu A, Ádám É, Sheerin DJ, Dobos O, Bernula P, Hiltbrunner A, Kozma-Bognár L, Nagy F (2015) High-level expression and phosphorylation of phytochrome B modulates flowering time in Arabidopsis. Plant J 83: 794–805 [DOI] [PubMed] [Google Scholar]
- Harmer SL, Kay SA (2005) Positive and negative factors confer phase-specific circadian regulation of transcription in Arabidopsis. Plant Cell 17: 1926–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfer A, Nusinow DA, Chow BY, Gehrke AR, Bulyk ML, Kay SA (2011) LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr Biol 21: 126–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero E, Kolmos E, Bujdoso N, Yuan Y, Wang M, Berns MC, Uhlworm H, Coupland G, Saini R, Jaskolski M, Webb A, Gonçalves J, et al. (2012) EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 24: 428–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PY, Devisetty UK, Harmer SL (2013) Accurate timekeeping is controlled by a cycling activator in Arabidopsis. eLife 2: e00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PY, Harmer SL (2014) Wheels within wheels: the plant circadian system. Trends Plant Sci 19: 240–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Yoo CY, Bindbeutel R, Goldsworthy J, Tielking A, Alvarez S, Naldrett MJ, Evans BS, Chen M, Nusinow DA (2016) PCH1 integrates circadian and light-signaling pathways to control photoperiod-responsive growth in Arabidopsis. eLife 5: e13292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Zhao X, Weng X, Wang L, Xie W (2012a) The rice B-box zinc finger gene family: genomic identification, characterization, expression profiling and diurnal analysis. PLoS One 7: e48242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Pérez-García P, Pokhilko A, Millar AJ, Antoshechkin I, Riechmann JL, Más P (2012b) Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336: 75–79 [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA (2005) FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309: 293–297 [DOI] [PubMed] [Google Scholar]
- Ito S, Niwa Y, Nakamichi N, Kawamura H, Yamashino T, Mizuno T (2008) Insight into missing genetic links between two evening-expressed pseudo-response regulator genes TOC1 and PRR5 in the circadian clock-controlled circuitry in Arabidopsis thaliana. Plant Cell Physiol 49: 201–213 [DOI] [PubMed] [Google Scholar]
- Ito S, Song YH, Josephson-Day AR, Miller RJ, Breton G, Olmstead RG, Imaizumi T (2012) FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proc Natl Acad Sci USA 109: 3582–3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S, Marchal V, Panigrahi KCS, Wenkel S, Soppe W, Deng X-W, Valverde F, Coupland G (2008) Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J 27: 1277–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamioka M, Takao S, Suzuki T, Taki K, Higashiyama T, Kinoshita T, Nakamichi N (2016) Direct repression of evening genes by CIRCADIAN CLOCK-ASSOCIATED1 in the Arabidopsis circadian clock. Plant Cell 28: 696–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R, Kronmiller B, Maszle DR, Coupland G, Holm M, Mizuno T, Wu S-H (2009) The Arabidopsis B-box zinc finger family. Plant Cell 21: 3416–3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T, Henriques R, Sakakibara H, Chua N-H (2007) Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by an SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell 19: 2516–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchenbauer D, Viczián A, Ádám É, Hegedűs Z, Klose C, Leppert M, Hiltbrunner A, Kircher S, Schäfer E, Nagy F (2016) Characterization of photomorphogenic responses and signaling cascades controlled by phytochrome-A expressed in different tissues. New Phytol 211: 584–598 [DOI] [PubMed] [Google Scholar]
- Lau OS, Huang X, Charron JB, Lee JH, Li G, Deng XW (2011) Interaction of Arabidopsis DET1 with CCA1 and LHY in mediating transcriptional repression in the plant circadian clock. Mol Cell 43: 703–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaro A, Mouriz A, Piñeiro M, Jarillo JA (2015) Red light-mediated degradation of CONSTANS by the E3 ubiquitin ligase HOS1 regulates photoperiodic flowering in Arabidopsis. Plant Cell 27: 2437–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaro A, Valverde F, Piñeiro M, Jarillo JA (2012) The Arabidopsis E3 ubiquitin ligase HOS1 negatively regulates CONSTANS abundance in the photoperiodic control of flowering. Plant Cell 24: 982–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Yoo SJ, Lee JH, Kim W, Yoo SK, Fitzgerald H, Carrington JC, Ahn JH (2010) Genetic framework for flowering-time regulation by ambient temperature-responsive miRNAs in Arabidopsis. Nucleic Acids Res 38: 3081–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Zuo Z, Liu H, Liu X, Lin C (2011) Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev 25: 1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang Q, Liu Y, Zhao X, Imaizumi T, Somers DE, Tobin EM, Lin C (2013a) Arabidopsis CRY2 and ZTL mediate blue-light regulation of the transcription factor CIB1 by distinct mechanisms. Proc Natl Acad Sci USA 110: 17582–17587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, Lin C (2008a) Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322: 1535–1539 [DOI] [PubMed] [Google Scholar]
- Liu L, Liu C, Hou X, Xi W, Shen L, Tao Z, Wang Y, Yu H (2012) FTIP1 is an essential regulator required for florigen transport. PLoS Biol 10: e1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Zhu Y, Shen L, Yu H (2013b) Emerging insights into florigen transport. Curr Opin Plant Biol 16: 607–613 [DOI] [PubMed] [Google Scholar]
- Liu LJ, Zhang YC, Li QH, Sang Y, Mao J, Lian HL, Wang L, Yang HQ (2008b) COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20: 292–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TL, Newton L, Liu MJ, Shiu SH, Farré EM (2016) A G-box-like motif is necessary for transcriptional regulation by circadian pseudo-response regulators in Arabidopsis. Plant Physiol 170: 528–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Li X, Li K, Liu H, Lin C (2013c) Multiple bHLH proteins form heterodimers to mediate CRY2-dependent regulation of flowering-time in Arabidopsis. PLoS Genet 9: e1003861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Más P, Kim WY, Somers DE, Kay SA (2003) Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426: 567–570 [DOI] [PubMed] [Google Scholar]
- Michael TP, Salome PA, McClung CR (2003) Two Arabidopsis circadian oscillators can be distinguished by differential temperature sensitivity. Proc Natl Acad Sci USA 100: 6878–6883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ. (2016) The intracellular dynamics of circadian clocks reach for the light of ecology and evolution. Annu Rev Plant Biol 67: 595–618 [DOI] [PubMed] [Google Scholar]
- Muranaka T, Oyama T (2016) Heterogeneity of cellular circadian clocks in intact plants and its correction under light-dark cycles. Sci Adv 2: e1600500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel DH, Doherty CJ, Pruneda-Paz JL, Schmitz RJ, Ecker JR, Kay SA (2015) Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc Natl Acad Sci USA 112: E4802–E4810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua NH, Sakakibara H (2010) PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22: 594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kiba T, Kamioka M, Suzuki T, Yamashino T, Higashiyama T, Sakakibara H, Mizuno T (2012) Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc Natl Acad Sci USA 109: 17123–17128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kita M, Niinuma K, Ito S, Yamashino T, Mizoguchi T, Mizuno T (2007) Arabidopsis clock-associated pseudo-response regulators PRR9, PRR7 and PRR5 coordinately and positively regulate flowering time through the canonical CONSTANS-dependent photoperiodic pathway. Plant Cell Physiol 48: 822–832 [DOI] [PubMed] [Google Scholar]
- Nemoto Y, Nonoue Y, Yano M, Izawa T (2016) Hd1, a CONSTANS ortholog in rice, functions as an Ehd1 repressor through interaction with monocot-specific CCT-domain protein Ghd7. Plant J 86: 221–233 [DOI] [PubMed] [Google Scholar]
- Nguyen KT, Park J, Park E, Lee I, Choi G (2015) The Arabidopsis RING domain protein BOI inhibits flowering via CO-dependent and CO-independent mechanisms. Mol Plant 8: 1725–1736 [DOI] [PubMed] [Google Scholar]
- Niwa Y, Ito S, Nakamichi N, Mizoguchi T, Niinuma K, Yamashino T, Mizuno T (2007) Genetic linkages of the circadian clock-associated genes, TOC1, CCA1 and LHY, in the photoperiodic control of flowering time in Arabidopsis thaliana. Plant Cell Physiol 48: 925–937 [DOI] [PubMed] [Google Scholar]
- Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farré EM, Kay SA (2011) The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osnato M, Castillejo C, Matías-Hernández L, Pelaz S (2012) TEMPRANILLO genes link photoperiod and gibberellin pathways to control flowering in Arabidopsis. Nat Commun 3: 808. [DOI] [PubMed] [Google Scholar]
- Pajoro A, Biewers S, Dougali E, Leal Valentim F, Mendes MA, Porri A, Coupland G, Van de Peer Y, van Dijk AD, Colombo L, Davies B, Angenent GC (2014) The (r)evolution of gene regulatory networks controlling Arabidopsis plant reproduction: a two-decade history. J Exp Bot 65: 4731–4745 [DOI] [PubMed] [Google Scholar]
- Pérez-García P, Ma Y, Yanovsky MJ, Más P (2015) Time-dependent sequestration of RVE8 by LNK proteins shapes the diurnal oscillation of anthocyanin biosynthesis. Proc Natl Acad Sci USA 112: 5249–5253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porri A, Torti S, Romera-Branchat M, Coupland G (2012) Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development 139: 2198–2209 [DOI] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857 [DOI] [PubMed] [Google Scholar]
- Putterill J, Varkonyi-Gasic E (2016) FT and florigen long-distance flowering control in plants. Curr Opin Plant Biol 33: 77–82 [DOI] [PubMed] [Google Scholar]
- Rawat R, Takahashi N, Hsu PY, Jones MA, Schwartz J, Salemi MR, Phinney BS, Harmer SL (2011) REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genet 7: e1001350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas U, Mei Y, Xie Q, Banta JA, Zhou RW, Seufferheld G, Gerard S, Chou L, Bhambhra N, Parks JD, Flowers JM, McClung CR, et al. (2014) Variation in Arabidopsis flowering time associated with cis-regulatory variation in CONSTANS. Nat Commun 5: 3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugnone ML, Faigón Soverna A, Sanchez SE, Schlaen RG, Hernando CE, Seymour DK, Mancini E, Chernomoretz A, Weigel D, Más P, Yanovsky MJ (2013) LNK genes integrate light and clock signaling networks at the core of the Arabidopsis oscillator. Proc Natl Acad Sci USA 110: 12120–12125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Sarid-Krebs L, Panigrahi KC, Fornara F, Takahashi Y, Hayama R, Jang S, Tilmes V, Valverde F, Coupland G (2015) Phosphorylation of CONSTANS and its COP1-dependent degradation during photoperiodic flowering of Arabidopsis. Plant J 84: 451–463 [DOI] [PubMed] [Google Scholar]
- Sawa M, Nusinow DA, Kay SA, Imaizumi T (2007) FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318: 261–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheerin DJ, Menon C, zur Oven-Krockhaus S, Enderle B, Zhu L, Johnen P, Schleifenbaum F, Stierhof YD, Huq E, Hiltbrunner A (2015) Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27: 189–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim JS, Imaizumi T (2015) Circadian clock and photoperiodic response in Arabidopsis: from seasonal flowering to redox homeostasis. Biochemistry 54: 157–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu H, Katayama K, Koto T, Torii K, Araki T, Endo M (2015) Decentralized circadian clocks process thermal and photoperiodic cues in specific tissues. Nat Plants 1: 15163. [DOI] [PubMed] [Google Scholar]
- Shrestha R, Gómez-Ariza J, Brambilla V, Fornara F (2014) Molecular control of seasonal flowering in rice, Arabidopsis and temperate cereals. Ann Bot (Lond) 114: 1445–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Estrada DA, Johnson RS, Kim SK, Lee SY, MacCoss MJ, Imaizumi T (2014) Distinct roles of FKF1, GIGANTEA, and ZEITLUPE proteins in the regulation of CONSTANS stability in Arabidopsis photoperiodic flowering. Proc Natl Acad Sci USA 111: 17672–17677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Ito S, Imaizumi T (2013) Flowering time regulation: photoperiod- and temperature-sensing in leaves. Trends Plant Sci 18: 575–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Shim JS, Kinmonth-Schultz HA, Imaizumi T (2015) Photoperiodic flowering: time measurement mechanisms in leaves. Annu Rev Plant Biol 66: 441–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Smith RW, To BJ, Millar AJ, Imaizumi T (2012) FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336: 1045–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soy J, Leivar P, González-Schain N, Martín G, Diaz C, Sentandreu M, Al-Sady B, Quail PH, Monte E (2016) Molecular convergence of clock and photosensory pathways through PIF3-TOC1 interaction and co-occupancy of target promoters. Proc Natl Acad Sci USA 113: 4870–4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120 [DOI] [PubMed] [Google Scholar]
- Takada S, Goto K (2003) Terminal flower2, an Arabidopsis homolog of heterochromatin protein1, counteracts the activation of flowering locus T by CONSTANS in the vascular tissues of leaves to regulate flowering time. Plant Cell 15: 2856–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Hirata Y, Aihara K, Más P (2015) A hierarchical multi-oscillator network orchestrates the Arabidopsis circadian system. Cell 163: 148–159 [DOI] [PubMed] [Google Scholar]
- Thain SC, Hall A, Millar AJ (2000) Functional independence of circadian clocks that regulate plant gene expression. Curr Biol 10: 951–956 [DOI] [PubMed] [Google Scholar]
- Thain SC, Murtas G, Lynn JR, McGrath RB, Millar AJ (2002) The circadian clock that controls gene expression in Arabidopsis is tissue specific. Plant Physiol 130: 102–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Shen Y, Chang HC, Hou Y, Harris A, Ma SF, McPartland M, Hymus GJ, Adam L, Marion C, Belachew A, Repetti PP, et al. (2010) The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol 187: 57–66 [DOI] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006 [DOI] [PubMed] [Google Scholar]
- Wang C, Dehesh K (2015) From retrograde signaling to flowering time. Plant Signal Behav 10: e1022012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CQ, Guthrie C, Sarmast MK, Dehesh K (2014) BBX19 interacts with CONSTANS to repress FLOWERING LOCUS T transcription, defining a flowering time checkpoint in Arabidopsis. Plant Cell 26: 3589–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wu JF, Nakamichi N, Sakakibara H, Nam HG, Wu SH (2011) LIGHT-REGULATED WD1 and PSEUDO-RESPONSE REGULATOR9 form a positive feedback regulatory loop in the Arabidopsis circadian clock. Plant Cell 23: 486–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenden B, Toner DL, Hodge SK, Grima R, Millar AJ (2012) Spontaneous spatiotemporal waves of gene expression from biological clocks in the leaf. Proc Natl Acad Sci USA 109: 6757–6762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, Samach A, Coupland G (2006) CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18: 2971–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JF, Wang Y, Wu SH (2008) Two new clock proteins, LWD1 and LWD2, regulate Arabidopsis photoperiodic flowering. Plant Physiol 148: 948–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Wang P, Liu X, Yuan L, Wang L, Zhang C, Li Y, Xing H, Zhi L, Yue Z, Zhao C, McClung CR, et al. (2014) LNK1 and LNK2 are transcriptional coactivators in the Arabidopsis circadian oscillator. Plant Cell 26: 2843–2857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Li T, Xu PB, Li L, Du SS, Lian HL, Yang HQ (2016) DELLA proteins physically interact with CONSTANS to regulate flowering under long days in Arabidopsis. FEBS Lett 590: 541–549 [DOI] [PubMed] [Google Scholar]
- Yakir E, Hassidim M, Melamed-Book N, Hilman D, Kron I, Green RM (2011) Cell autonomous and cell-type specific circadian rhythms in Arabidopsis. Plant J 68: 520–531 [DOI] [PubMed] [Google Scholar]
- Yu D, Hu Y, Wang H, Pan J, Li Y, Lou D (2016) The DELLA-CONSTANS transcription factor cascade integrates gibberellic acid and photoperiod signaling to regulate flowering. Plant Physiol 172: 479–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Wang L, Zeng L, Zhang C, Ma H (2015) Arabidopsis TOE proteins convey a photoperiodic signal to antagonize CONSTANS and regulate flowering time. Genes Dev 29: 975–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Liu L, Shen L, Yu H (2016) NaKR1 regulates long-distance movement of FLOWERING LOCUS T in Arabidopsis. Nat Plants 2: 16075. [DOI] [PubMed] [Google Scholar]
- Zuo Z, Liu H, Liu B, Liu X, Lin C (2011) Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr Biol 21: 841–847 [DOI] [PMC free article] [PubMed] [Google Scholar]