TATA-box insertion in the upstream untranslated sequences of genes enhances adaptation to selective pressure.
Abstract
Intraspecific genetic variation is essential for the responses and adaption of plants to evolutionary challenges, such as changing environmental conditions. The development of the Earth’s aerobic atmosphere has increased the demand for iron (Fe) in organisms, and Fe deficiency has become a limiting environmental factor for plant growth. Here, we demonstrate that genus Malus adapt to Fe deficiency through modification of the Iron-Regulated Transporter1 (IRT1) promoter. Specifically, an IRT1 mutant allele with a TATA-box insertion in the promoter region upstream of the coding region exhibited increased IRT1 expression. The altered IRT1 promoter is responsible for enhancing Fe uptake. Increasing the number of synthetic repeat TATA-boxes correlates with increased promoter activity. Furthermore, we demonstrate that the insertion of the TATA-box correlates with an increase in transcriptional activation via specific binding of the transcription factor IID (MDP0000939369). Taken together, these results indicate that an allelic insertion of a TATA-box in a gene promoter has allowed apple to adapt to the selective pressure posed by Fe deficiency. More broadly, this study reveals a new mechanism for enhancing gene expression to help plants adapt to different environments, providing new insights into molecular genetic divergence in plants.
A diverse portfolio of crops that can adapt to potentially adverse environmental conditions is essential to meet the demands of increasingly intensive agricultural production (Huang and Han, 2014), particularly in the context of climate change and global population increases. Genetic variants of cultivated species derived from wild progenitors of plants under both natural and human selection provide vital sources of this type of genetic variation. Therefore, there is considerable interest in identifying allelic variants in natural populations and determining whether they have a selective advantage under specific conditions. Some variants account for all of the heritability of gene expression attributable to cis-regulatory elements. Variations in the expression of genes associated with agriculturally important traits due to transposon insertion or differential expansion of microsatellite repeat sequences have been identified (Sureshkumar and Lee, 2009; Wang et al., 2012).
A critical factor that can limit crop production is the availability of the micronutrient Fe, which plays important roles in a wide range of cellular processes, including photosynthesis and respiration (Bonner and Varner, 1976; Briat et al., 1995; Le, 2002). During the early period of the Earth’s history, the atmosphere was anoxic, and water-soluble ferrous Fe was the form of Fe used by early organisms. However, the development of an oxygen-rich atmosphere has resulted in the loss of the bioavailable form of Fe, Fe (II), and the accumulation of insoluble Fe (III; Crichton and Pierre, 2001; Darbani et al., 2013). Therefore, Fe is increasingly becoming a limiting factor for plant growth and development (Abadía et al., 2011). To adapt to these changes, plants have developed two Fe uptake strategies (I and II; Marschner et al., 1986a; Marschner et al., 1986b; Briat et al., 1995), which are exhibited in nongraminaceous and graminaceous plants, respectively. The Strategy I response, also known as the Reduction Strategy, involves the reduction of ferric chelates at the root surface and absorption of the generated ferrous ions across the root plasma membrane. Strategy II relies on the biosynthesis and secretion of Fe(III)-solubilizing molecules, termed mugineic acids, which chelate Fe; therefore, this strategy is also known as the Chelation Strategy (Marschner et al., 1986a; Marschner et al., 1986b; Briat et al., 1995). In Strategy I plants, the transporter Iron-Regulated Transporter1 (IRT1) plays a major role in regulating Fe homeostasis, and its expression is induced by Fe (Henriques et al., 2002; Vert et al., 2002). Some species, such as tomato (Solanum lycopersicum), lettuce (Lactuca sativa), and peanut (Arachis hypogaea), show different levels of resistance to Fe deficiency, depending on the genotype (Li and Wang, 1995; Chen et al., 2001; Xu et al., 2012; Xia et al., 2013). QTL studies investigating the genetic basis of mineral tolerance in crops have focused on mineral concentrations in seeds or leaves, and most have targeted Strategy II plants, such as rice (Oryza sativa), wheat (Triticum aestivum), and barley (Hordeum vulgare; Alonso-Blanco et al., 2009). Despite these efforts, the molecular basis of the natural variation in Fe uptake systems and the mechanisms determining Fe uptake affinity remain largely undefined.
In this study, we provide evidence that a TATA-box insertion in the IRT1 promoter in the genus Malus increases the expression of IRT1, which accounts for the increase in Fe uptake in various Malus species. Furthermore, our findings suggest that the binding of transcription factor IID (TFIID) to the IRT1 promoter is enhanced by the presence of a TATA-box insertion, leading to increased IRT1 transcript levels and enhanced Fe uptake. This represents a novel mechanism for genetic divergence in which a TATA-box insertion promotes environmental adaption.
RESULTS
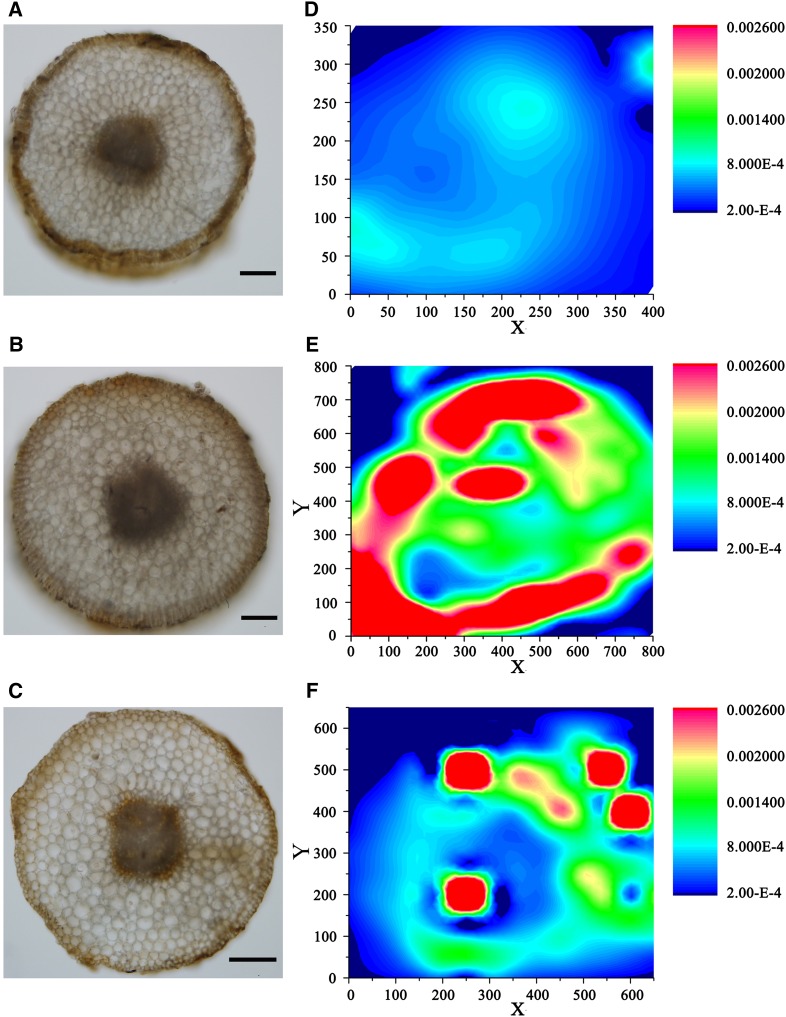
Microtomography Analysis of Fe Distribution in Roots Reveals Variants with Fe-Deficiency Resistance
Among members of the genus Malus, Malus xiaojinensis and Malus baccata have well-developed root systems and are valued in China as native apple rootstocks with good grafting compatibility for the development of new apple cultivars (Han et al., 1994, 1998). However, while M. xiaojinensis is an Fe-efficient species, M. baccata has much lower Fe uptake efficiency. We confirmed this difference using microtomography analysis of Fe distribution in the roots of plants of both species. Figure 1 shows x-ray fluorescence (XRF) maps of Fe distribution patterns in cross-sections of roots under normal (40 μm Fe2+) and low (4 μm Fe2+) Fe conditions. Under both conditions, the Fe intensity in cross-sections of M. xiaojinensis roots was high, whereas reduced levels or the complete absence of Fe was detected in M. baccata (Fig. 1, D and E). Fe, the ferrous-iron (Fe2+) form termed “active Fe,” which can be extracted with weak acids and some chelating agents, is closely associated with Fe chlorosis (Köseoğlu and Açikgöz, 1995; Zha et al., 2014). Furthermore, we measured the active Fe contents in M. xiaojinensis and M. baccata supplied with 0 μm Fe2+ or 40 μm Fe2+ for 0, 3, and 6 d. Under Fe-sufficient conditions, the active Fe contents were significantly lower in the roots of M. baccata than in those of M. xiaojinensis. Similarly, continuous Fe deficiency induced a more rapid decrease in Fe levels in the roots of M. baccata versus M. xiaojinensis. Therefore, the different Fe levels are consistent with different levels of Fe uptake efficiency between M. xiaojinensis and M. baccata (Fig. 2B).
Figure 1.
XRF microtomography showing that the Fe content is higher in M. xiaojinensis roots than in M. baccata roots. Qualitative spatial distribution and concentration gradients of Fe in latitudinal sections of (A and D) M. baccata (sufficient Fe supply, 40 μm Fe2+), (B and E) M. xiaojinensis (sufficient Fe supply, 40 μm Fe2+), and (C and F) M. Xiaojinensis roots (deficient Fe supply, 0 μm Fe2+). Bar color (blue to red) reflects Fe contents (low to high). The SR-μXRF signals for the map were collected in 50-μm steps. The areas mapped for A and D, B and E, and C and F were 350 μm × 400 μm, 800 μm × 800 μm, and 650 μm × 650 μm, respectively. Scale bars in A, B, and C indicate 50 µm, 100 µm, and 100 µm, respectively.
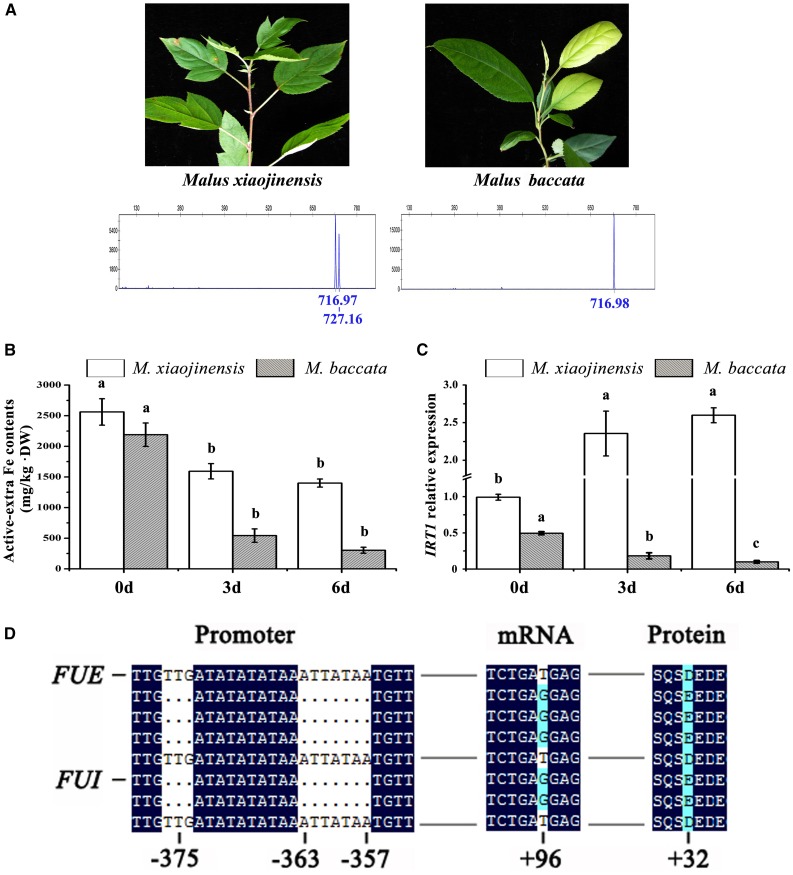
Figure 2.
Fe-deficiency tolerance, root Fe content, and IRT1 expression in M. xiaojinensis and M. baccata. A, Fe-deficiency symptoms in M. xiaojinensis and M. baccata grown under Fe-deficient conditions and short tandem repeats analysis of the IRT1 promoter. The peak at 716 corresponds to the FUE promoter sequence with the TATA-box, and the peak at 727 corresponds to the FUI promoter sequence without the TATA-box. B, Root Fe content in M. xiaojinensis and M. baccata supplied with 0 μm Fe2+ or 40 μm Fe2+ for 0, 3, and 6 d. C, Relative expression of IRT1 in M. xiaojinensis and M. baccata supplied with 0 μm Fe2+ or 40 μm Fe2+ for 0, 3, and 6 d. D. Variations in the IRT1 promoter fragment between the FUE and FUI alleles. (See full sequences in Supplemental Fig. S1.)
Identification of an IRT1 Mutant Allele with a TATA-Box Insertion
We previously reported that M. xiaojinensis has higher IRT1 mRNA levels than M. baccata (Zha et al., 2014) and that Fe deficiency induces a substantial increase in IRT1 expression in M. xiaojinensis, but not in M. baccata (Fig. 2C), suggesting that a modified allele of IRT1 with enhanced expression might be responsible for the enhanced Fe efficiency of this species. To elucidate the genetic mechanism underlying this difference, we analyzed the gene sequences of IRT1 from M. xiaojinensis and M. baccata. As shown in Figure 2. xiaojinensis has a TTG…ATTATAA insertion in the promoter of the IRT1 coding region that is absent in the equivalent sequence of M. baccata. The insertion, −363 to −357 bp from the ATG start site, is predicted to be a core promoter element TATA-box. Alleles containing or lacking the TATA-box insertion were designated FUE (Fe uptake efficiency) and FUI (Fe uptake inefficiency), respectively (accession no. KX549782, KX549781; Fig. 2D; Supplemental Fig. S1).
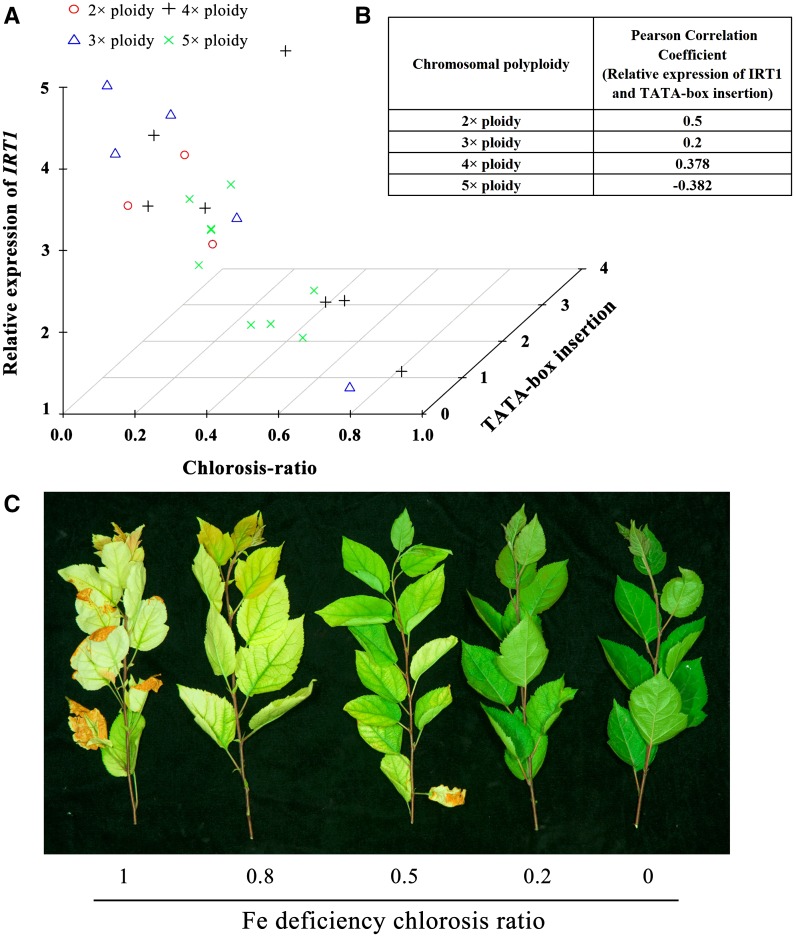
We confirmed the association between the FUE allele and Fe uptake efficiency by analyzing segregating progeny from a cross between M. xiaojinensis (FUE/FUI) and M. baccata (FUI/FUI). We observed a strong negative correlation (R2 = 0.7536, P < 0.0001) between the appearance of leaf chlorosis, a notable phenotypic symptom of Fe deficiency (Zhu et al., 2015), and the expression of IRT1 in roots from the progeny of both M. xiaojinensis and M. baccata, highlighting the importance of IRT1 in the regulation of Fe uptake (Supplemental Fig. S2). The progeny of M. xiaojinensis × M. baccata were hybrids showing 2×, 3×, 4×, or 5× ploidy. Since gene copy number can significantly contribute to phenotype, we reasoned that differences in ploidy might directly result in differences in uptake efficiency among progeny. To eliminate the possibility that absolute or relative gene dosage effects contribute to these differences, we individually analyzed the association between the FUE allele and IRT1 expression in plants of the same ploidy level and found that the presence of a high copy number of the TATA-box insertion helped prevent Fe deficiency-induced chlorosis in 2×, 3×, and 4× plants (Fig. 3). Furthermore, our results show a positive correlation between the frequency of TATA-box insertion and IRT1 expression in 2×, 3×, and 4× plants. However, in 5× plants, no correlation between chlorosis and TATA-box insertion was observed that could be explained by gene dosage effects (Fig. 3). Therefore, the results of genetic analysis indicate that the TATA-box insertion conferred Fe-deficiency resistance to most of the progeny.
Figure 3.
Analysis of segregating progeny from a cross between M. xiaojinensis and M. baccata. A, Correlation analysis between relative expression levels of IRT1 in the progeny and the TATA-box insertion percentage. B, Fe-deficient chlorosis index in young leaves of the hybrid combinations (M. xiaojinensis and M. baccata) under Fe-deficient treatment. C, Varied phenotypic symptoms of Fe deficiency in the segregating progeny.
A TATA-Box Insertion in the Promoter of IRT1 Alters Its Expression
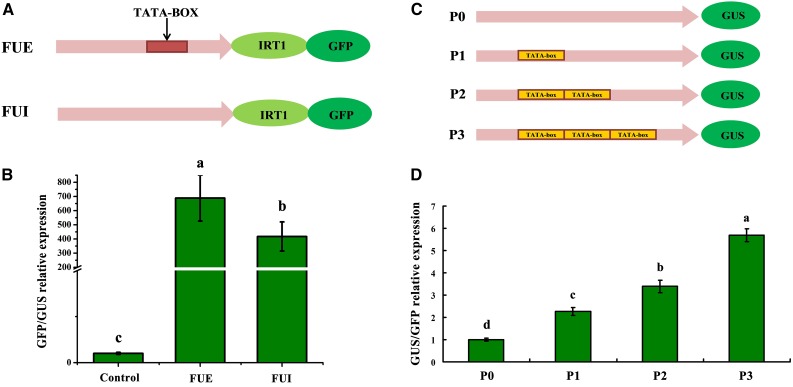
Since the TATA-box can play a role in the activation of eukaryotic genes transcribed by RNA polymerase II (Burley and Roeder, 1996), a possible consequence of the TATA-box insertion within the IRT1 promoter is an increase in the basal activity of the promoter. We investigated this possibility using transient expression assays in Arabidopsis (Arabidopsis thaliana) mesophyll protoplasts. We fused the FUE and FUI promoter sequences to the green fluorescence protein (GFP) coding sequence, expressed the fusion construct in protoplasts, and measured GFP expression relative to that of coexpressed GUS driven by the constitutive 35S promoter. The GFP reporter exhibited 1.58-fold greater expression when driven by the FUE promoter compared to the FUI promoter (Fig. 4). We also assessed the activities of the promoters via transient expression in tobacco (Nicotiana benthamiana) leaves harboring each promoter fused to the GUS coding sequence. Again, the FUE promoter had higher activity than the FUI promoter (Supplemental Fig. S3). Taken together, these results suggest that the TATA-box insertion enhances the basal activity of the IRT1 promoter.
Figure 4.
Functional analysis of TATA-box insertion in Arabidopsis protoplasts. A, Diagrams of the reporter constructs used for transient expression analysis. B, Transient expression analysis of FUE and FUI in Arabidopsis protoplasts. Arabidopsis protoplasts were transfected with FUE and FUI promoter-GFP fusions, and GFP expression was measured relative to that of GUS driven by the constitutive 35S promoter. Error bars in B indicate sd of three biological replicates. C, Schematic of promoters with different numbers of TATA-boxes ranging from zero (P0) to three (P3). D, Transient expression analysis of P0 to P3 in Arabidopsis protoplasts. Arabidopsis protoplasts were transfected with P0-GUS, P1-GUS, P2-GUS, and P3-GUS fusions, and GUS expression was measured relative to that of GFP driven by the constitutive 35S promoter. Error bars in D indicate sd of six biological replicates.
To further investigate the effects of the TATA-box insertion on IRT1 promoter activity, we produced a series of synthetic constructs to test the effects of the number of TATA-box repeat units in the upstream region of IRT1 on its transcriptional activity. These constructs, which were based on the native promoter sequences, but with the addition of zero to three TATA-boxes fused to the GUS reporter (Fig. 4C; Supplemental Data S1), were assayed as above. The results show a strong positive correlation between the number of repeat TATA-box insertions and promoter activity (Fig. 4D).
We also found that the TATA-box insertion is tightly linked to a base mutation in the coding sequence at +96 bp (G/T), which results in a missense mutation and an amino acid change from E to D (Fig. 2D). To assess the effects of this mutation of FUE on Fe uptake, we performed functional complementation of yeast (Saccharomyces cerevisiae) mutant DEY1453 (fet3fet4). The coding sequence of both FUE and FUI successfully complemented the high-affinity Fe uptake defect of the mutant. We therefore conclude that the TATA-box-linked base mutation does not affect the function of IRT1 (Supplemental Fig. S4).
FUE:IRT1-Overexpressing Plants Exhibit Increased Fe Storage
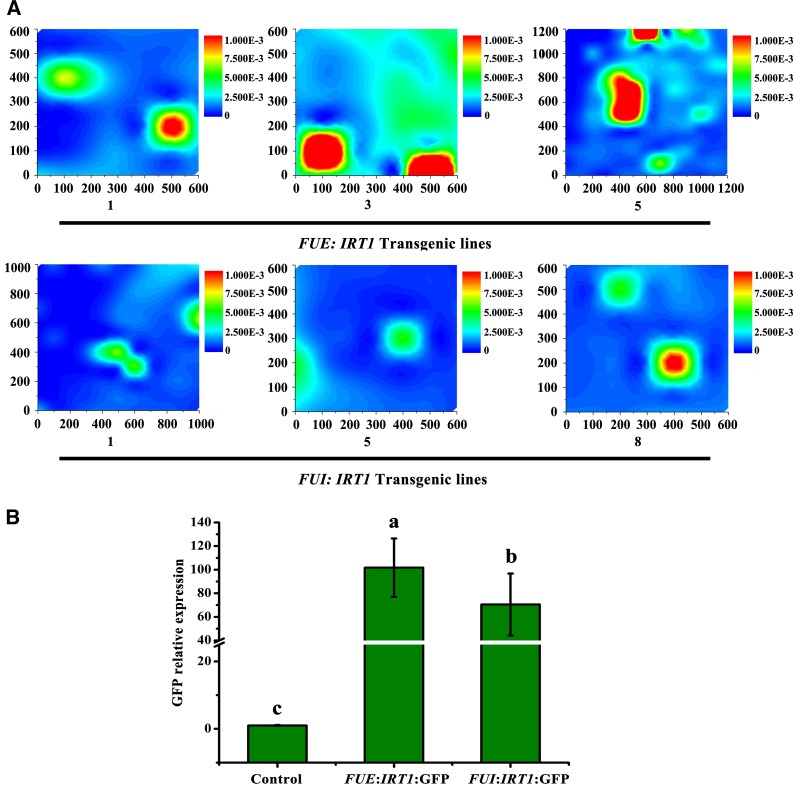
To further investigate the effects of the TATA-box insertion on Fe storage, we introduced IRT1 driven by either the FUE or FUI promoter into tobacco. XRF analyses showed that more Fe accumulated in transgenic FUE:IRT1 versus FUI:IRT1 plants (Fig. 5A). Moreover, IRT1 transcript levels were significantly higher in FUE:IRT1 roots than in FUI:IRT1 roots. We also found that the FUE:IRT1 promoter increased the expression of the GFP reporter 1.5-fold in roots relative to the FUI:IRT1 promoter (Fig. 5B).
Figure 5.
The TATA-box insertion in FUE increases Fe accumulation. A, XRF microtomography of tobacco leaves transformed with FUE:IRT1:GFP and FUI:IRT1:GFP. Bar color (blue to red) reflects the Fe contents (low to high). The SR-μXRF signals for the map were collected in 100-μm steps. The areas mapped for FUE:IRT transgenic lines 1, 3, and 5 were 600 μm × 600 μm, 600 μm × 600 μm, and 1,200 μm × 1,200 μm, respectively. The areas mapped for FUI:IRT transgenic lines 1, 5, and 8 were 1,000 μm × 1,000 μm, 600 μm × 600 μm, and 600 μm × 600 μm, respectively. B, Relative expression of GFP in transgenic tobacco. Error bars in B indicate sd of three biological replicates.
The TATA-Box Insertion Enhances IRT1 Expression via the Specific Binding of TFIID
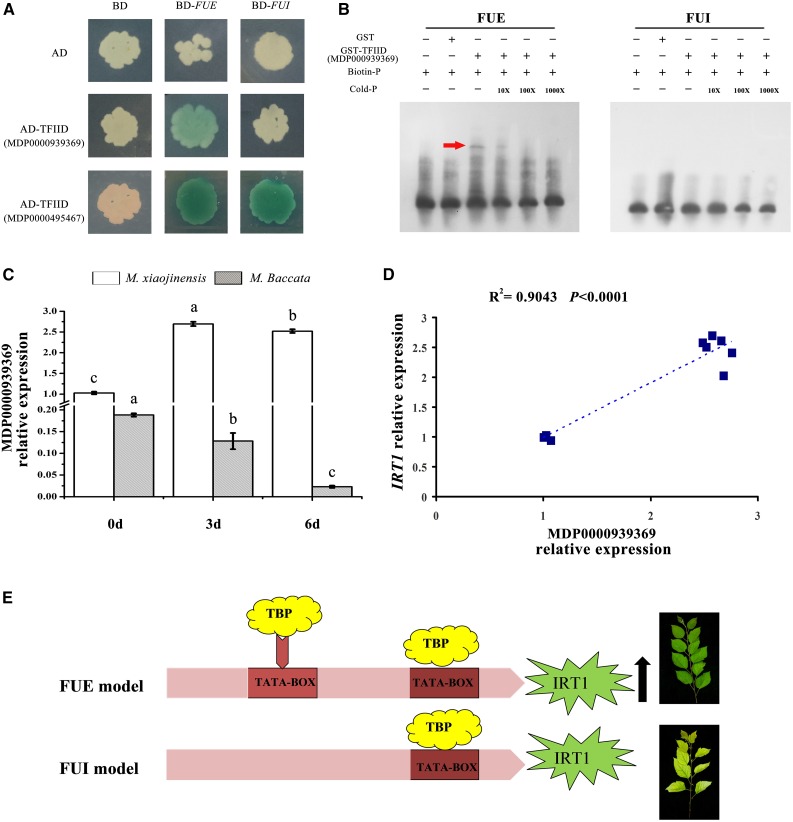
RNA polymerase II-mediated transcription by the preinitiation complex begins with the binding of the TFIID to the TATA-box. We therefore investigated the binding of TFIID to the TATA-box identified in this study using yeast one-hybrid analysis. Phylogenetic analysis identified 29 genes that are most closely related to TFIID in the apple genome (Supplemental Fig. S5), 11 of which are expressed in the roots of M. xiaojinensis. We therefore investigated the ability of the corresponding proteins to bind to the TATA-box (Supplemental Table S1). One TFIID protein (MDP0000495467) bound to both the FUE and FUI promoters, with no difference in binding activity detected between the two. However, another TFIID protein (MDP0000939369) bound to the FUE promoter, but not to the FUI promoter (Fig. 6A). To confirm this finding, we analyzed in vitro binding using electrophoretic mobility shift assays (EMSAs). Recombinant glutathione S-transferase-tagged TFIID protein (MDP0000939369) was purified from Escherichia coli and incubated with DNA probes representing the FUE or FUI promoter (Supplemental Table S2). MDP0000939369 specifically bound only to biotin-labeled FUE (Fig. 6B). Finally, we measured TATA box binding protein (TBP; MDP0000939369) expression in M. xiaojinensis and M. baccata supplied with 0 μm Fe2+ or 40 μm Fe2+ for 0, 3, or 6 d, finding that Fe deficiency induced a substantial increase in TBP expression in M. xiaojinensis, but not in M. baccata (Fig. 6C). In addition, TBP expression was positively correlated with IRT1 expression (R2 = 0.9043, P < 0.001; Fig. 6D).
Figure 6.
Presence of the TATA-box correlates with an increase in transcriptional activation by specific TFIID protein binding. A, Yeast one-hybrid assay showing the activity of LacZ reporters driven by the FUE and FUI promoters and activated by the activation domain (AD) fused with MDP0000495467 or MDP0000939369. Empty vectors pJG4-5 and pLacZi were used as negative controls. B, Gel-shift analysis of MDP0000939369 binding to the promoters of FUE. EMSAs of GST-labeled probe fused with TFIID protein (MDP0000939369); DNA probes represent the FUE or FUI (probe sequences are shown in Supplemental Table S2). Protein/DNA complexes are indicated by arrows. C, Relative expression of MDP0000939369 in M. xiaojinensis and M. baccata supplied with 0 μm Fe2+ or 40 μm Fe2+ for 0, 3, and 6 d. D, Correlation analysis between relative expression levels of IRT1 and MDP0000939369. E, Schematic model of the regulation of IRT1 expression by the TATA-box insertion regulatory module.
Based on the data presented in this study, we propose a model for the regulation of IRT1 expression by the TATA-box insertion regulatory module. The model suggests that the specific binding of TFIID protein (MDP0000939369) to FUE leads to an increase in IRT1 transcript levels and, consequently, Fe uptake (Fig. 6E).
Evolutionary Analysis of the TATA-Box Insertion
To confirm the role of the FUE allele in the evolution of Malus species, we allelotyped the FUE/FUI loci of 52 accessions belonging to four different Malus species (Phipps et al., 1990; Yao et al., 2015; Supplemental Table S1): the 52 accessions represent the diversity of the four Malus species. We found that the nucleotide diversity (Θ and π values) of the IRT1 allele was significantly lower in Malus domestica than in the other species (Table I). Tajima’s D tests (Tajima, 1989) detected departure from neutrality for the IRT1 genes of Malus sieversii and M. domestica, indicating that the IRT1 allele is under selection.
Table I. Nucleotide polymorphisms in IRT1 in various Malus species.
| m | Ps | Θ | π | D | Neutral Mutation Range (Tajima’s D Value) | |
|---|---|---|---|---|---|---|
| M. toringoides | 5 | 0.065693 | 0.031533 | 0.032263 | 0.174412 | −1.255–1.737 |
| M. baccata | 21 | 0.030797 | 0.00856 | 0.00842 | −0.06069 | −1.585–1.709 |
| M. sieversii | 11 | 0.030581 | 0.010441 | 0.005783 | −2.02136 | −1.572–1.710 |
| M. domestica | 15 | 0.016514 | 0.005079 | 0.002412 | −1.96923 | −1.584–1.780 |
DISCUSSION
Our data suggest that a TATA-box insertion in the upstream regulatory region of IRT1 functions in a novel regulatory mechanism. Specifically, we present evidence that the TATA-box insertion enhances IRT1 expression to control Fe uptake efficiency as a result of specific binding of TFIID to the TATA box.
Plants exhibit substantial natural variation in mineral use efficiency, root uptake, translocation from roots to shoots, and accumulation (Alonso-Blanco et al., 2009). QTL studies in various crop species have focused on seed or leaf mineral concentrations, primarily in rice, wheat, and barley, which use the Chelation Strategy (Strategy II) to acquire Fe. These studies have resulted in the identification of several natural alleles differing in missense mutations in conserved motifs exhibiting reduced P-Type ATPase activity and copper (Cu) translocation to the shoot in Arabidopsis (Kobayashi et al., 2008) as well as a 1-kb insertion in the promoter that increases the expression of citrate transporter gene HvAACT1 in several Al-tolerant barley cultivars (Fujii et al., 2012). In addition, natural variation in the rice Nramp aluminum transporter gene results in significant differences in Al tolerance (Li et al., 2014). Finally, a deletion in the promoter of a mitochondrial molybdenum transporter gene in Arabidopsis is associated with reduced gene expression and low molybdenum (Mo) levels in the shoot, suggesting that this regulatory mutation is the causal nucleotide polymorphism (Baxter et al., 2008). QTL studies of the Strategy I plant Arabidopsis have focused on the accumulation of several minerals, including nitrogen (Loudet et al., 2003; Harada et al., 2004), potassium (Harada and Leigh, 2006), Cu (Sasaki et al., 2008), Mo (Baxter et al., 2008), and sodium (Rus et al., 2006). Compared to Arabidopsis, a relatively high level of genetic variation has been maintained between or among domesticated crops and their close wild relatives (Huang and Han, 2014). Self-incompatible species, such as apple, have particularly high levels of genetic variation; the use of highly heterozygous Malus genotypes has allowed us to demonstrate that a TATA-box insertion in the promoter of a key gene involved in the regulation of Fe homeostasis contributes to Fe-uptake efficiency. Our results provide compelling evidence that natural allelic differences in promoter sequences alter Fe uptake efficiency in this Strategy I plant.
The best-characterized core-promoter elements, which can function independently or synergistically, are TATA elements, located 25 bp upstream of the transcription start site (Burley and Roeder, 1996). The TBP was first identified as a component of TFIID, a universal transcription initiation factor (Nikolov and Burley, 1994). Thus, in addition to identifying a polymorphic gene, our study lays the foundation for elucidating the detailed regulatory mechanism associated with natural variation. Based on the newly identified role of the FUE allele in Fe uptake, we identified a specific TFIID protein (MDP0000939369) that binds to the TATA box. Our model provides a new framework for future investigations of mechanisms driving molecular genetic divergence in plants. Moreover, our results establish that a synthetic repeat TATA-box insertion in the promoter can enhance gene expression, which might represent a general tool for precisely regulating gene expression.
Polyploidy has a major influence on gene flow in species. Multiple ploidy levels exist within many species, and gene flow between ploidy levels is restricted by the low fitness of the progeny produced by crosses between ploidy levels (Martinez-Reyna and Vogel, 2002; Henry and Crawford, 2005). In addition, the physiological and genetic effects of polyploidy may directly result in adaptive differentiation within a species (Otto and Whitton, 2000). However, the role of polyploidy in shaping intraspecific diversity is poorly understood (Levin, 1983; Ramsey and Schemske, 2002). Cultivated apple is an example of a plant with secondary polyploidy (Velasco et al., 2010), as the wild Malus species, M. xiaojinensis, is tetraploid (Wang et al., 2012). The progeny of M. xiaojinensis × M. baccata comprise hybrids of 2×, 3×, 4×, or 5× ploidy (Wang et al., 2012). The physiological and genetic effects of polyploidy may directly result in adaptive differentiation within a species. Since the complexity of ploidy differences complicates efforts to quantify the contribution of the TATA-box insertion to iron deficiency tolerance in the progeny, we performed separate Pearson correlation analyses with plants of the same ploidy level to better understand the relationship between the TATA-box insertion and the Fe deficiency tolerance phenotype. Taken together, the results of relative genetic analysis between the FUE allele and phenotype suggest that the TATA-box insertion contributes to the Fe deficiency-resistant trait, but this contribution varies with ploidy level.
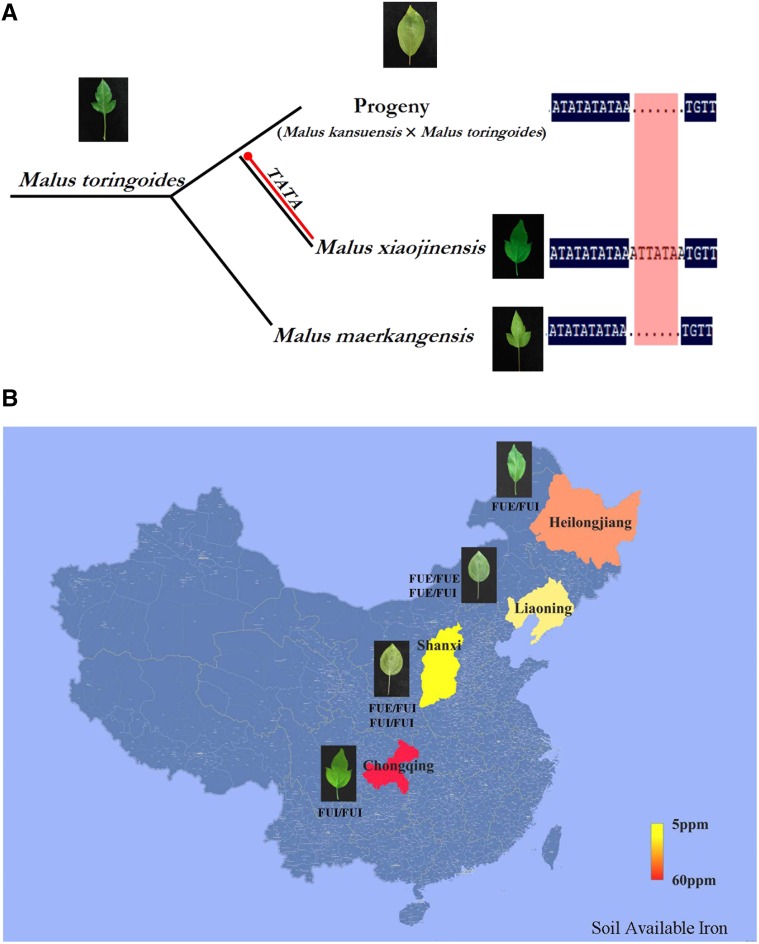
Phylogenetic analysis has suggested that IRT genes from dicotyledonous species Arabidopsis and M. xiaojinensis are closely related and belong to a single clade (Victoria et al., 2012). Additionally, the close relationship between dicotyledon and monocotyledon IRT genes suggests incipient divergence in these two groups (Guerinot, 2000; Mäser, 2001; Vert et al., 2002; Gross et al., 2003), which we found evidence for in our analysis of IRT1. Malus toringoides species, comprising the hybridization complex M. toringoides, Malus kansuensis, and Malus transitoria, produce highly phenotypically variable progenies due to their self-incompatibility or long juvenile phase. M. xiaojinensis is a product of the complicated evolutionary history of M. toringoides (Chen et al., 2000), which originated in (and is distributed throughout) Sichuan province. The soil type in this region is brown acidic, with a pH value slightly <7 and high levels of available Fe. Therefore, there is no anticipated selective pressure for Fe homeostasis-related genes in plants in this region. We found that wild relatives of M. xiaojinensis growing in similar soils are homozygous for FUI and therefore lack a TATA-box insertion in IRT1 (Fig. 7A). Consistently, in our evolutionary analysis of TATA-box insertions, we did not detect significant departure from neutrality in M. toringoides species. Although the ecological effects of the genetic insertion, which occurred in a geographic region with high levels of available Fe, have not yet been assessed, our findings suggest that the TATA-box insertion may have promoted the migration of M. xiaojinensis from the acidic soils of southwest China to the alkaline soils in the north.
Figure 7.
Evolution of the TATA-box in the IRT1 promoter in the genus Malus. A, Phylogenetic analysis of the TATA-box in the IRT1 promoters of M. toringoides, M. toringoides × M. kansuensis progeny, M. maerkangensis, and M. xiaojinensis. B, TATA-box insertion in the IRT1 promoter was induced by geographic ecological selective pressure. The data about available Fe in soil were obtained from published sources (Chen et al., 2000; Cai et al., 2002; Li et al., 2009).
Geographic analyses of genetic variation in several plant species have revealed clear genetic signals of local adaptation caused by differences in selective regimes among locations (Linhart and Grant, 1996). At the beginning of our study, we confirmed that M. baccata is homozygous for a lack of the TATA-box insertion. However, we subsequently found M. baccata plants of different ecotypes containing a TATA insertion from regions with alkaline soil (Fig. 7B). M. baccata and M. xiaojinensis do not share a common ancestor (Chen et al., 2001), suggesting that the TATA insertion in Fe uptake-efficient genus Malus occurred independently. Our results provide evidence that the acquisition of tolerance to Fe deficiency is an adaptive trait of the genus Malus. This adaptation likely facilitates the expansion of apple production areas using genotypes tolerant to low Fe availability.
Previous studies have suggested that natural variation in cis elements can underlie phenotypic variation (Sureshkumar and Lee, 2009). We found that this variation can occur in the form of a TATA-box insertion in a gene promoter. This model provides an important framework for future investigations of the role of molecular genetic divergence in environmental adaption.
MATERIALS AND METHODS
Materials and Treatments
For XRF analysis, Malus xiaojinensis and Malus baccata seedlings were propagated on Murashige and Skoog (MS) medium containing 0.5 mg/L 6-benzylaminopurine and 0.5 mg/L indole-3-butytric acid for 1 mo. The seedlings were transferred to MS medium with 1.0 mg/L indole-3-butytric acid for rooting. After an additional 1.5 mo, the rooted seedlings were transferred to Hoagland’s nutrient solution, pH 6.0 (Han et al., 1994), and cultured at 24°C, 60% to 70% relative humidity with a photoperiod of 16 h light/8 h dark under a light intensity of 250 mmol quanta m−2s−1. The nutrient solution was replaced weekly. After 3 weeks, the plants were placed in Hoagland nutrient solution containing 0 μm (Fe deficient) or 40 μm (Fe sufficient) FeNaEDTA and grown for 6 d.
To measure IRT1 expression, M. xiaojinensis, M. baccata, and M. xiaojinensis × M. baccata progeny seedlings were grown as above under Fe-sufficient conditions. Roots were collected for RNA isolation and gene expression analysis.
To analyze active Fe levels and the expression of IRT1 and the selected transcription factor genes, M. xiaojinensis and M. baccata seedlings were grown in Hoagland nutrient solution under Fe-deficient conditions for 0, 3, and 6 d and their roots analyzed.
To sequence the IRT1 coding region and upstream region, leaves were collected from M. xiaojinensis, M. baccata, and M. xiaojinensis × M. baccata progeny plants at the Institute of Forestry and Pomology, Beijing Academy of Agricultural and Forestry Sciences, the Institute of Botany, Chinese Academy of Sciences, and the horticulture research garden in Changping district, Beijing, respectively.
To evaluate leaf chlorosis, the in vitro-rooted progeny of M. xiaojinensis × M. baccata were transplanted to the field in May 2013 and cultured in fine quartz sand in a 90 × 30 × 30 cm (L × W × H) plastic container. The plants were drip-irrigated with Hoagland nutrient solution containing 4 μm Fe (for Fe-deficient treatment) and 40 μm Fe (for Fe-sufficient treatment).
To analyze the evolution of the TATA-box insertion in the IRT1 promoter, the leaves of 67 Malus species were collected from different regions of China: Malus toringoides, Southwest University in Chongqing; M. baccata, Shanxi, Liaoning, Heilongjiang; Malus domestica and Malus sieversii, the horticulture research garden, Changping district, Beijing. The leaves were collected for DNA isolation and gene expression detection.
Synchrotron Radiation Microscopic XRF Analysis
The micro-XRF microspectroscopy experiment was performed at the 4W1B endstation at the Beijing Synchrotron Radiation Facility, which runs a 2.5-GeV electron beam with a current of 150 mA to 250 mA. The incident x-ray energy was monochromatized with a W/B4C Double-Multilayer-Monochromator at 15 keV and focused to a 50-μm-diameter beam using a polycapillary lens. M. xiaojinensis and M. baccata roots was sliced transversely with a scalpel. To obtain a flat surface, a Cryotome was used to produce approximately 200-μm-thick latitudinal root sections. The samples were placed on Kapton tape and freeze-dried in a vacuum freeze dryer (LGJ-10B, Beijing Four-Ring Science Instrument Factory). Two-dimensional mapping was performed in step-mode: the sample was held on a precision motor-driven stage, scanning 50 μm stepwise. A Si (Li) solid-state detector was used to detect XRF emission lines with a live time of 60 s. Data reduction and processing were performed using PyMCA (Kim et al., 2006; Solé et al., 2007).
Cloning and Sequencing of the IRT1 Genome Fragment
Genomic DNA was extracted from leaves of Malus species and their progeny using the CTAB method (Gasic et al., 2004). The PCR primers listed below were used to amplify the different MdIRT1 DNA fragments with KOD FX DNA polymerase (Toyobo). The amplified product was purified, ligated into the pEASY-T1 vector (TransGen Biotechnology Co., Ltd., Beijing, China), cloned into Escherichia coli strain DH5α, and sequenced. The PCR primers aIRT251-F: 5′-GCAGCTGCCAACCTCAATCC-3′ and aIRT251-R: 5′-CGACCGGCTTGACACCAAAA-3′ were used to amplify the region 5′ 578 bp + IRT1 (3 exons + 2 introns) 1,637 bp + 87 bp 3′. The primers IRT721-F: 5′-AAGCCACCGGAGGACATGG-3′ and IRT721-R: 5′-GGGCTATGATTTTGAG GGGCA-3′ were used to amplify the region 5′ 621 bp + IRT1 (100 bp cDNA) to detect the sequence mutation. Short tandem repeats analysis was performed using fluorescently labeled primers IRT721-F: 5′-AAGCCACCGGAGGACATGG-3′ and IRT721-R: 5′-GGGCTATGATTTTGAGGGGCA-3′ (Beijing Microread Genetics Co., Ltd.).
Total RNA Isolation and Gene Expression Analysis
Total RNA was extracted from root tissue using a modified CTAB method (Gasic et al., 2004), followed by DNase I digestion to remove any DNA contamination (TaKaRa Biotechnology Co., Ltd.). To generate cDNA, the RNA samples were reverse-transcribed using oligo(dT) primer and reverse transcriptase (TaKaRa Biotechnology Co., Ltd.) according to the manufacturer’s instructions.
The relative expression levels of IRT1 were measured using an Applied Biosystems 7500 real-time PCR system and SYBR Green fluorescent dye (Takara Biotechnology Co., Ltd.). The primers MdIRT1-F: 5′-TGACAAGGGAGAAAACGGAGAC-3′ and MdIRT1-R: 5′-AACAACTGAATGGACAATGATACCC-3′ were designed using Primer Premier 5 (Zhang and Gao, 2004), and the efficiency of the primers was confirmed by semi-quantitative RT-PCR (reverse transcription-PCR). β-actin was used as the reference gene. The primers were actin-F: 5′-TGGCATATACTCTGGAGGCT-3′ and actin-R: 5′TGGTGAGGCTCTATTCCAAC-3′. MdIRT1 relative expression levels were calculated according to the 2−ΔΔCT method (Livak and Schmittgen, 2001). Each reaction was performed with three biological replicates.
Measuring Chlorosis in the Leaves in M. xiaojinensis × M. baccata Progeny
Leaf chlorosis was evaluated as previously described (Xu et al., 2012; Zhu et al., 2015).
Separation Ratio of TATA-box Insertions or Deletions in the IRT1 Promoters of M. xiaojinensis × M. baccata Progeny
To investigate the influence of the TATA-box on iron deficiency resistance in the progeny of M. xiaojinensis × M. baccata, progeny with 2×, 3×, 4×, and 5× ploidy were used to measure the separation ratio of TATA-box insertion or deletion. DNA fragments (721 bp) were PCR amplified using primers IRT721-F (5′-AAGCCACCGGAGGACATGG-3′) and IRT721-R (5′-GGGCTATGATTTTGAGGGGCA-3′), ligated into the pEASY-T1 vector, and cloned into E. coli strain DH5α. The sequences of 20 monoclonal colonies per M. xiaojinensis × M. baccata progeny (grown in ampicillin LB medium) were used as the basis for estimating the proportion of TATA-box insertions.
Analysis of Active Fe
Active Fe was evaluated by polarized Zeeman atomic absorption spectrophotometry (Z-5000, Hitachi) as previously described (Zha et al., 2014).
Construction of Recombinant Plasmids
Fusion products of the promoters with or without the TATA-box and the two versions of the full-length IRT1 open reading frame were generated using overlap PCR technology (Lee et al., 2010). The promoters were amplified with the primers a-IRT600-F (5′-AAGCCACCGGAGGACATGG-3′) and a-IRT600-R (5′-GTGGCAGCCATTGACCCTGAT-3′). The full-length IRT1 open reading frame was amplified with the primers b-IRT1100-F (5′-ATCAGGGTCAATGGCTGCCAC-3′) and b-IRT1100-R (5′-AGCCCAC TTTGCCAATA-3′). The recombinant fusion products were renamed FUE and FUI and cloned into the pGWEB404 vector with GFP as a tag using the Gateway method (Curtis and Grossniklaus, 2003).
Arabidopsis Protoplast Isolation and Transformation
Protoplasts were isolated from fully expanded healthy leaves of 3- to 4-week-old Arabidopsis (Arabidopsis thaliana) using a modified version of a published protocol (Sheen, 2001). Protoplasts were transformed with polyethylene glycol essentially as described (He et al., 2007; Yoo et al., 2007) using recombinant plasmid DNA including FUE or FUI, with empty vector used as the negative control. The detail protocol is presented in Supplemental Method S2. After 16 h, the protoplasts were harvested and stored at −80°C for RNA extraction and GFP expression analysis.
Investigating Promoter Activity in IRT1 Promoters with Different Numbers of TATA-Box Insertions
To investigate the effects of different numbers of TATA-box insertions on IRT1 promoter activity, IRT promoters with 0, 1, 2, or 3 copies of the TATA-box were generated by artificial gene synthesis (Beijing Kwinbon Biotechnology Co., Ltd. China) and renamed as P0, P1 (−358), P2 (−410, −358), and P3 (−415, −410, −358), respectively. The promoters were inserted into the vector pCAMBIA1301 (containing the GUS reporter gene) between the NcoI and KpnI sites. The recombinant plasmids were transformed into Arabidopsis protoplast as described above, with empty vector containing the 35S promoter used as the control. Protoplasts were harvested and stored at −80°C for RNA extraction and GUS expression analysis.
Generation of Transgenic Tobacco
The constructed plasmids containing FUE or FUI were introduced into Agrobacterium tumefaciens (strain EHA105) cells, which were used to transform tobacco (Nicotiana benthamiana) using the leaf disc method (Horsch et al., 1989), with empty vector used as the negative control. Transgenic plants were selected based on kanamycin resistance and transferred to MS liquid medium after rooting. After 2 weeks, transgenic plants were used to detect the expression of GFP in roots, while the leaves were used for synchrotron radiation microscopic XRF analysis SR-μXRF analysis of Fe content.
Identifying TATA-Box-Binding Proteins via Yeast One-Hybrid Analysis
The TBP domains were identified using the search term “TATA-box binding protein (TBP)” in the Arabidopsis Information Resource Web site (http://www.arabidopsis.org/). The domains in the Genome Database for Rosaceae Web site (https://www.rosaceae.org/) were used queries to search for homologous transcription factors in the apple genome, revealing 29 genes. A phylogenetic analysis of these genes (generated using MEGA5.2) is shown in Supplemental Figure S3 (Tamura et al., 2011). Gene-specific PCR primers were used to amplify the sequences (Supplemental Table S1). Total RNA was extracted from the roots of M. xiaojinensis plants grown under normal and Fe-deficient conditions using the modified CTAB method described above and treated with DNase I (TaKaRa Biotechnology Co., Ltd.); cDNA generation and quantitative RT-PCR analysis were performed as described above. According to the quantitative RT-PCR results, only 11 of the 29 transcription factors were expressed under Fe-deficient conditions; the corresponding PCR products were purified and cloned into the pEASY Blunt Simple T1 vector (TransGen) according to the manufacturer’s instructions, followed by sequencing (GENEWIZ).
Yeast One-Hybrid Analysis
Yeast one-hybrid analysis was performed to investigate the interactions of the 11 transcription factors with the two IRT1 promoters (with and without the TATA-box). Using EcoRI and XhoI digestion and T4 ligation reactions, the transcription factor genes were inserted into the AD vector (pJG4-5; Clontech Matchmaker One-Hybrid System User Manual), and the two promoters (with or without the TATA-box) were inserted into the BD vector (pLacZi). Vector construction and screening were performed according to the manufacturer’s instructions, with minor modifications (Supplemental Method S2).
IRT1 Nucleotide Polymorphism Analyses in Various Malus Genotypes
To investigate the DNA sequence diversity in the IRT1 promoter, DNA fragments (721 bp) were PCR amplified using primers IRT721-F (5′-AAGCCACCGGAGGACATGG-3′) and IRT721-R (5′-GGGCTATGATTTTGAGGGGCA-3′). Leaves from 67 Malus species from different regions of China were collected and used for DNA amplification. TransTaq DNA Polymerase High Fidelity (TransGen) was used for PCR to minimize DNA synthesis errors. The PCR products were purified and sequenced (GENEWIZ), and the sequences were analyzed using MEGA5.2 (Tamura et al., 2011).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers FUE (KX549782), FUI (KX549781).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Sequences of the 1,500-bp FUE and FUI promoters.
Supplemental Figure S2. Correlation analysis between chlorosis ratios and relative expression levels of IRT1 in the progeny of M. xiaojinensis and M. baccata under Fe-deficiency treatment.
Supplemental Figure S3. Transient GUS expression under the control of the FUE and FUI promoters in tobacco.
Supplemental Figure S4. Complementation test of IRT1 Fe2+ uptake in the fet3fet4 yeast mutant grown on Fe-deficient medium supplemented with 55 μm bathophenanthrolinedisulfonic acid disodium salt, pH 5.8.
Supplemental Figure S5. Phylogenetic analysis of homologous transcription factors in the apple and Arabidopsis genomes.
Supplemental Table S1. Primers for the transcription factor genes used in the yeast one-hybrid assay.
Supplemental Table S2. Probe sequences used for EMSA.
Supplemental Data S1. Sequences of IRT1 promoters with different numbers of the TATA-box insertion.
Supplemental Methods S1. Arabidopsis protoplast isolation and polyethylene glycol-mediated gene transformation.
Supplemental Methods S2. Modified yeast one-hybrid analysis protocol.
Supplemental Methods S3. Functional complementation in yeast.
Supplementary Material
Acknowledgments
The micro-XRF beam time was granted by the 4W1B endstation of the Beijing Synchrotron Radiation Facility, Institute of High Energy Physics, Chinese Academy of Sciences. The staff members of 4W1B are acknowledged for their help with measurements and data analysis. Dr. Ronald F. Korcak is acknowledged for his comments on the experiments and technical editing of the manuscript.
Glossary
- EMSA
electrophoretic mobility shift assay
- MS
Murashige and Skoog
- TFIID
transcription factor IID
- XRF
x-ray fluorescence
Footnotes
This work was financially supported by the National Natural Science Foundation of China (nos. 31272139 and 31401840), Beijing Natural Science Foundation (no. 6154028), China Postdoctoral Science Foundation (no. 2016M590157), Special Fund for Agro-scientific Research in the Public Interest (no. 201203075 and No.201303093), Beijing Municipal Education Commission (CEFF-PXM2016_014207_000038), and Key Labs Nutrition and Physiology for Horticultural Crops, and Physiology and Molecular Biology of Tree Fruit. J.K.C.R. is supported by an affiliation with the Atkinson Center for a Sustainable Future.
Articles can be viewed without a subscription.
References
- Abadía J, Vázquez S, Rellán-Álvarez R, El-Jendoubi H, Abadía A, Alvarez-Fernández A, López-Millán AF (2011) Towards a knowledge-based correction of iron chlorosis. Plant Physiol Biochem 49: 471–482 [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C, Aarts MG, Bentsink L, Keurentjes JJ, Reymond M, Vreugdenhil D, Koornneef M (2009) What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell 21: 1877–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter I, Muthukumar B, Park HC, Buchner P, Lahner B, Danku J, Zhao K, Lee J, Hawkesford MJ, Guerinot ML, et al. (2008) Variation in molybdenum content across broadly distributed populations of Arabidopsis thaliana is controlled by a mitochondrial molybdenum transporter (MOT1). PLoS Genet 4: e1000004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner J, Varner F, editors (1976) Plant Biochemistry. Academic, New York. [Google Scholar]
- Briat JO, Fobis-Loisy I, Grignon N, Lobraux SP, Pascal N, Savino G, Thoiron SV, Wir N, Wuytswinkel O (1995) Cellular and molecular aspects of iron metabolism in plants. Biol Cell 84: 69–81 [Google Scholar]
- Burley SK, Roeder RG (1996) Biochemistry and structural biology of transcription factor IID (TFIID). Annu Rev Biochem 65: 769–799 [DOI] [PubMed] [Google Scholar]
- Cai J, Cai SL, Lu JL (2002) Vertical variation of element contents in the main soil types of Heilongjiang province. Word Geology 21: 364–367 (Chinese) [Google Scholar]
- Chen GL, Li SJ, Zhang FS, Li CJ (2001) The adaptive mechanism of Fe deficiency to susceptibility of Fe stress of lettuce cultivars. Plant Nutr and Fert Sci 7: 117–120 [Google Scholar]
- Chen MH, Li XL, Zhang YG (2000) Malus xiaojinensis cheng et jiang-a promising stock for apple trees. J Southwest Agr Univ 22: 383–386 (Chinese) [Google Scholar]
- Crichton RR, Pierre JL (2001) Old iron, young copper: from Mars to Venus. Biometals 14: 99–112 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darbani B, Briat JF, Holm PB, Husted S, Noeparvar S, Borg S (2013) Dissecting plant iron homeostasis under short and long-term iron fluctuations. Biotechnol Adv 31: 1292–1307 [DOI] [PubMed] [Google Scholar]
- Fujii M, Yokosho K, Yamaji N, Saisho D, Yamane M, Takahashi H, Sato K, Nakazono M, Ma JF (2012) Acquisition of aluminium tolerance by modification of a single gene in barley. Nat Commun 3: 713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasic K, Hernandez A, Korban SS (2004) RNA extraction from different apple tissues rich in polyphenols and polysaccharides for cDNA library construction. Plant Mol Biol Report 22: 437–438 [Google Scholar]
- Gross BL, Schwarzbach AE, Rieseberg LH (2003) Origin(s) of the diploid hybrid species Helianthus deserticola (Asteraceae). Am J Bot 90: 1708–1719 [DOI] [PubMed] [Google Scholar]
- Guerinot ML. (2000) The ZIP family of metal transporters. Biochimica et Biophysica Acta-Biomembranes 1465: 190–198 [DOI] [PubMed] [Google Scholar]
- Han ZH, Wang Q, Shen T (1994) Comparison of some physiological and biochemical characteristics between iron-efficient and iron-inefficient species in the genus. J Plant Nutr 17: 1257–1264 [Google Scholar]
- Han ZH, Shen T, Korcak RF, Baligar VC (1998) Iron absorption by iron-efficient and -inefficient species of apples. J Plant Nutr 21: 181–190 [Google Scholar]
- Harada H, Kuromori T, Hirayama T, Shinozaki K, Leigh RA (2004) Quantitative trait loci analysis of nitrate storage in Arabidopsis leading to an investigation of the contribution of the anion channel gene, AtCLC-c, to variation in nitrate levels. J Exp Bot 55: 2005–2014 [DOI] [PubMed] [Google Scholar]
- Harada H, Leigh RA (2006) Genetic mapping of natural variation in potassium concentrations in shoots of Arabidopsis thaliana. J Exp Bot 57: 953–960 [DOI] [PubMed] [Google Scholar]
- He P, Shan L, Sheen J (2007) The use of protoplasts to study innate immune responses. In Ronald PC, ed, Plant-Pathogen Interactions: Methods and Protocols, Humana Press, Totowa, New Jersey, pp 1–9 [DOI] [PubMed] [Google Scholar]
- Henriques R, Jásik J, Klein M, Martinoia E, Feller U, Schell J, Pais MS, Koncz C (2002) Knock-out of Arabidopsis metal transporter gene IRT1 results in iron deficiency accompanied by cell differentiation defects. Plant Mol Biol 50: 587–597 [DOI] [PubMed] [Google Scholar]
- Henry JD, Crawford JR (2005) The short-form version of the Depression Anxiety Stress Scales (DASS-21): construct validity and normative data in a large non-clinical sample. Br J Clin Psychol 44: 227–239 [DOI] [PubMed] [Google Scholar]
- Horsch RB, Fry J, Hoffmann N, Neidermeyer J, Rogers SG, Fraley RT (1989) Leaf Disc Transformation. Plant molecular biology manual. Springer, the Netherlands, pp 63–71 [Google Scholar]
- Huang X, Han B (2014) Natural variations and genome-wide association studies in crop plants. Annu Rev Plant Biol 65: 531–551 [DOI] [PubMed] [Google Scholar]
- Kim SA, Punshon T, Lanzirotti A, Li L, Alonso JM, Ecker JR, Kaplan J, Guerinot ML (2006) Localization of iron in Arabidopsis seed requires the vacuolar membrane transporter VIT1. Science 314: 1295–1298 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Kuroda K, Kimura K, Southron-Francis JL, Furuzawa A, Kimura K, Iuchi S, Kobayashi M, Taylor GJ, Koyama H (2008) Amino acid polymorphisms in strictly conserved domains of a P-type ATPase HMA5 are involved in the mechanism of copper tolerance variation in Arabidopsis. Plant Physiol 148: 969–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köseoğlu AT, Açikgöz V (1995) Determination of iron chlorosis with extractable iron analysis in peach leaves. J Plant Nutr 18: 153–161 [Google Scholar]
- Lee J, Shin MK, Ryu DK, Kim S, Ryu WS (2010) Insertion and deletion mutagenesis by overlap extension pcr. Methods Mol Biol 634: 137–46 [DOI] [PubMed] [Google Scholar]
- Le N. (2002) The role of iron in cell cycle progression and the proliferation of neoplastic cells. Biochimica et Biophysica Acta Rev Can 1603: 31–46 [DOI] [PubMed] [Google Scholar]
- Levin DA. (1983) Polyploidy and novelty in flowering plants. Am Nat 122: 1–25 [Google Scholar]
- Li SF, Chen Y, Wang J, Lv LT, Li G, Hao XL (2009) Study on abundance evaluation of available microelements in soil in Liaoning province and its grading criteria. Liaoning Agr Sci 1: 31–38 (Chinese) [Google Scholar]
- Li SJ, Wang LP (1995) Screening of hydroponically grown lecttuce cultivars for tolerating iron deficiency stress and their phyfioligical characteristic. Yuan Yi Xue Bao 22: 147–152 [Google Scholar]
- Li JY, Liu J, Dong D, Jia X, McCouch SR, Kochian LV (2014) Natural variation underlies alterations in Nramp aluminum transporter (NRAT1) expression and function that play a key role in rice aluminum tolerance. PNAS 111: 6503–6508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhart YB, Grant MC (1996) Evolutionary significance of local genetic differentiation in plants. Annu Rev Ecol Syst 27: 237–277 [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Merigout P, Talbotec J, Daniel-Vedele F (2003) Quantitative trait loci analysis of nitrogen use efficiency in Arabidopsis. Plant Physiol 131: 345–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner H, Romheld V, Kissel M (1986a) Different strategies in higher plants in mobilization and uptake of iron. J Plant Nutr 9: 695–713 [Google Scholar]
- Marschner H, Römheld V, Horst WJ, Martin P (1986b) Root-induced changes in the rhizosphere: importance for the mineral nutrition of plants. Z Pflanzenernährung Bodenkunde 149: 441–456 [Google Scholar]
- Martinez-Reyna JM, Vogel KP (2002) Incompatibility systems in switchgrass. Crop Sci 42: 1800–1805 [Google Scholar]
- Mäser P, Thomine S, Schroeder JI, Ward JM, Hirschi K, Sze H, Talke IN, Amtmann A, Maathuis FJ, Sanders D, et al. (2001) Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol 126: 1646–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolov DB, Burley SK (1994) 2.1 A resolution refined structure of a TATA box-binding protein (TBP). Nat Struct Biol 1: 621–637 [DOI] [PubMed] [Google Scholar]
- Otto SP, Whitton J (2000) Polyploid incidence and evolution. Annu Rev Genet 34: 401–437 [DOI] [PubMed] [Google Scholar]
- Phipps JB, Robertson KR, Smith PG, Rohrer JR (1990) A checklist of the subfamily Maloideae (Rosaceae). Can J Bot 68: 2209–2269 [Google Scholar]
- Ramsey J, Schemske DW (2002) Neopolyploidy in flowering plants. Annu Rev Ecol Syst 33: 589–639 [Google Scholar]
- Rus A, Baxter I, Muthukumar B, Gustin J, Lahner B, Yakubova E, Salt DE (2006) Natural variants of AtHKT1 enhance Na+ accumulation in two wild populations of Arabidopsis. PLoS Genet 2: e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Yoneyama N, Nakamura Y, Kobayashi N, Ikemoto Y, Moriwaki T, Kimura H (2008) Optical probe of carrier doping by X-ray irradiation in the organic dimer Mott insulator κ-(BEDT-TTF)_2Cu[N(CN)_2]Cl. Phys Rev Lett 101: 206403. [DOI] [PubMed] [Google Scholar]
- Sheen J. (2001) Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol 127: 1466–1475 [PMC free article] [PubMed] [Google Scholar]
- Solé VA, Papillon E, Cotte M, Walter P, Susini J (2007) A multiplatform code for the analysis of energy-dispersive X-ray fluorescence spectra. Spectrochim Acta B At Spectrosc 62: 63–68 [Google Scholar]
- Sureshkumar M, Lee C (2009) Biocatalytic reactions in hydrophobic ionic liquids. J Mol Catal, B Enzym 60: 1–12 [Google Scholar]
- Tajima F. (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D, et al. (2010) The genome of the domesticated apple (Malus × domestica Borkh.). Nat Genet 42: 833–839 [DOI] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dédaldéchamp F, Gaymard F, Guerinot ML, Briat JF, Curie C (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victoria FdeC, Bervald CMP, da Maia LC, de Sousa RO, Panaud O, de Oliveira AC, Gustafson P (2012) Phylogenetic relationships and selective pressure on gene families related to iron homeostasis in land plants. Genome 55: 883–900 [DOI] [PubMed] [Google Scholar]
- Wang L, Han D, Gao C, Wang Y, Zhang X, Xu X, Han Z (2012) Paternity and ploidy segregation of progenies derived from tetraploid Malus xiaojinensis. Tree Genet Genomes 8: 1469–1476 [Google Scholar]
- Xia YL, Liao BS, Mao JX, Zeng Y, Qi Y, Liao JH, Jing YL, Cui FH, Ren ZL, et al. (2013) Evaluation of lime induced iron-deficiency chlorosis tolerance of peanut on calcareous and purplish soil in Sichuan. J Oil Crop Sci 35: 326–330 [Google Scholar]
- Xu LS, Wang L, Wang R, Wang Y, Zhang XZ, Xu XF, Han ZH (2012) Segregation of tolerence to iron deficiency in apomictic and hybrid progeny of Malus xiaojinensis. Guoshu Xuebao 29: 770–775 [Google Scholar]
- Yao Y, Cao Q, Vasilakos AV (2015) EDAL: an energy-efficient, delay-aware, and lifetime-balancing data collection protocol for heterogeneous wireless sensor networks. IEEE ACM Trans Netw 23: 810–823 [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=17585298&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- Zha Q, Wang Y, Zhang XZ, Han ZH (2014) Both immanently high active iron contents and increased root ferrous uptake in response to low iron stress contribute to the iron deficiency tolerance in Malus xiaojinensis. Plant Sci 214: 47–56 [DOI] [PubMed] [Google Scholar]
- Zhang XY, Gao YN (2004) To design PCR primers with Oligo 6 and Primer Premier 5. Bioinformatiocs 4: 3 [Google Scholar]
- Zhu M, Wang R, Kong P, Zhang X, Wang Y, Wu T, Jia W, Han Z (2015) Development of a dot blot macroarray and its use in gene expression marker-assisted selection for iron deficiency tolerant apple rootstocks. Euphytica 202: 469–477 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.