Abstract
The mushroom bodies are bilaterally arranged structures in the protocerebrum of Drosophila and most other insect species. Mutants with altered mushroom body structure have been instrumental not only in establishing their role in distinct behavioral functions but also in identifying the molecular pathways that control mushroom body development. The mushroom body miniature1 (mbm1) mutation results in grossly reduced mushroom bodies and odor learning deficits in females. With a survey of genomic rescue constructs, we have pinpointed mbm1 to a single transcription unit and identified a single nucleotide exchange in the 5′ untranslated region of the corresponding transcript resulting in a reduced expression of the protein. The most obvious feature of the Mbm protein is a pair of C2HC zinc fingers, implicating a function of the protein in binding nucleic acids. Immunohistochemical analysis shows that expression of the Mbm protein is not restricted to the mushroom bodies. BrdUrd labeling experiments indicate a function of Mbm in neuronal precursor cell proliferation.
Adaptive behavior of animals and humans requires functional neuronal circuits in the brain. The genetic programs that control the generation of these circuits by providing an adequate number of neurons, establishing neuronal connectivity, and remodeling them during development and in response to external stimuli during adulthood are just beginning to emerge. The mushroom bodies (MBs), a prominent neuropil structure of the insect brain (1), have become an attractive model system to study many aspects of this intricate network. Functional studies have established a role of the MBs in olfactory learning and memory, controlling locomotor activity, performing visual context generalization, and decision making (2, 3). On the other hand, the structural organization and the development of the MBs have been described in great detail in refs. 4–10. In the adult fly Drosophila melanogaster, ≈2,500 intrinsic neurons (Kenyon cells) build up one MB. The Kenyon cell bodies are located in the dorsal cortex and extend their dendritic branches into the calyx, where prominent inputs from other brain regions are received. The Kenyon cell axons fasciculate in the peduncle and extend rostroventral, where most of them bifurcate to form a system of medially and dorsally projecting lobes. Each MB arises from a group of four apparently equipotent neuronal stem cells (neuroblasts), each of which generates in a sequential manner several types of Kenyon cells during larval and pupal stages. MB γ neurons are born before the mid-third-larval instar, then α′/β′ neurons are born, and finally the α/β neurons are added at pupal stages (6, 8). The nomenclature of the Kenyon cells refers to the corresponding dorsally and medially projecting MB lobes in the adult fly (see Fig. 2). More recently, immunohistochemical studies have identified additional subtypes of Kenyon cells (9). Yet, the anatomical description disregards the structural plasticity of the adult MBs as a consequence of changes in living conditions and experience (11). Furthermore, Kenyon cell axons and dendrites undergo massive remodeling during metamorphosis to establish adult-specific branching patterns. The axons of the γ neurons, which bifurcate in a dorsal and a medial branch in the larvae, degenerate and regrow only in the medial direction (8, 10, 12–14).
Fig. 2.
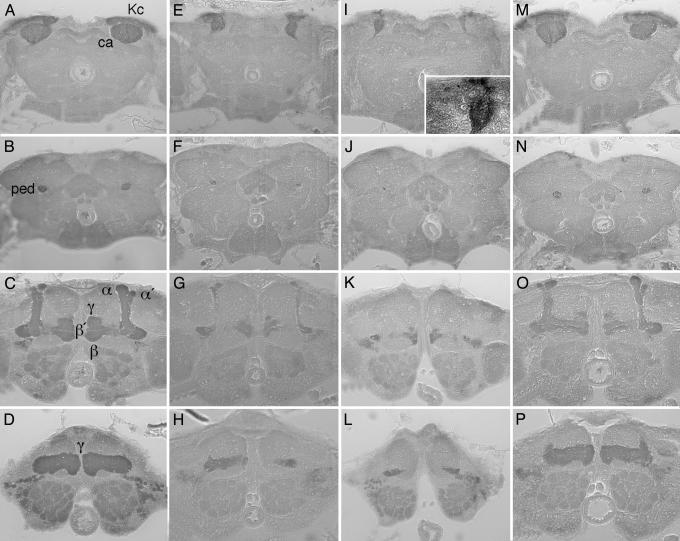
Analysis of the adult mbm1 phenotype. Frontal sections of paraffin-embedded heads of WT females (A–D), mbm1 females with a moderate (E–H)ora strong (I–L) phenotype, and Df(2L)A1/mbm1 females carrying a single copy the rescue construct P[TW115] (M–P). Photographs were taken after staining with the anti-Leonardo antibody (33) at the level of the Kenyon cell (Kc) bodies/calyx (A, E, I, and M), the peduncle (B, F, J, and N), and the α/β (C, G, K, and O), and the γ (D, H, L, and P) lobe system. In the WT, the Kenyon cell bodies are located in the dorsal-posterior cortex. Kenyon cell dendrites and extrinsic input fibers form the calyx (ca). The Kenyon cell fibers form the peduncle (ped) extending anterior-ventrally where they divide to form the dorsally and medially projecting lobe system. Two major dorsally projecting lobes (α and α′) and three medially projecting lobes (β, β′, and γ) are visible; the additional subdivisions described by Strausfeld et al. (9) are not discernible at this resolution. Despite the severe reduction in the number of Leonardo-positive cells (E and I), the structural subdivision of the MBs is maintained in mbm1 (F–H). Even in the most severe cases (J–L), a rudimentary lobe system can be detected. Note also the many unstained cell bodies surrounding the Leonardo-positive Kenyon cells in mbm1 (I Inset). The MB defect of Df(2L)A1/mbm1 females is rescued to nearly WT appearance by the genomic construct P[TW115] (M–P).
What are the genetic programs controlling MB development? The first relevant mutants were identified nearly 25 years ago in screens for altered MB structure (15). More recently, mosaic techniques and gain-of-function systems have allowed searching for genes that control MB development among other (vital) processes outside of the MBs (e.g., refs. 16 and 17). Some of the genes identified so far control the number or proliferation pattern of MB neuroblasts (18–20); others regulate Kenyon cell axon growth and guidance (7, 21) or are required for remodeling of Kenyon cell axons during metamorphosis (22–24).
In this article, we describe the identification of the gene mushroom body miniature (mbm). Mutant mbm1 females have grossly reduced MBs, which correlates with odor-learning deficits (25, 26). The most prominent structural feature of the Mbm protein is a pair of zinc fingers of the C2HC type, which indicates a function of the protein in binding nucleic acids. Expression of the Mbm protein is not restricted to the MBs, suggesting a function of Mbm in other aspects of brain development. In particular, when the hypomorphic mbm1 allele was placed over a noncomplementing deficiency, the number of proliferating cells in larval brains is greatly reduced.
Materials and Methods
Genetics and Transgenic Lines. Flies were raised at 25°C on standard cornmeal food. mbm1 (formerly mbmN337) was identified in a mass histology screen of ethyl methanesulfonate-treated cn, bw, sp flies (25). The strain was outcrossed with WT Berlin flies to remove the genetic markers. The breakpoints of Df(2L)net-PMF and Df(2L)al are described in Alcedo et al. (27). To generate deletions that remove the genomic sequences between the proximal breakpoint of Df(2L)net-PMF and the distal break-point of Df(2L)al, a P element (P[lacW]319) located in the PlC21C gene close to the distal breakpoint of Df(2L)al was mobilized. One strain (P[lacW]185/319) carrying the original P insertion plus an additional P insertion ≈18 kb more distally was used for a jump-out mutagenesis to isolate strains that lack both P insertions. These strains were characterized genetically by complementation tests and by Southern blot analysis (T.T., unpublished data).
Genomic fragments used for rescue experiments were derived from a genomic walk in the 21B region (27) and were cloned into the pW8 vector (28). Transgenic lines were generated by injecting Qiagen-purified plasmid DNA into w1118 embryos. The transgenic line P[AatII] was provided by Markus Noll (University of Zürich, Zürich). For rescue experiments, transgenic flies carrying the indicated genomic rescue construct on the third chromosome were crossed to Df(2L)A1 flies. Males derived from this cross were mated with mbm1 females. The female progeny of the genotype mbm1/Df(2L)A1; P[gen-rescue]/+ was analyzed by mass histology for rescue of the MB phenotype. For each genomic rescue construct, at least two independent insertion lines were tested. Expression of the Mbm protein under Gal4/UAS control was achieved by cloning the Berkeley Drosophila Genome Project cDNA LD12178 (29) into the pUAST transformation vector (30).
Immunohistochemistry and Western Blot Analysis. A peptide (KFRDPQQELDNHQPN) corresponding to amino acids 67–81 of the Mbm protein (Fig. 6, which is published as supporting information on the PNAS web site) was synthesized and used to immunize rabbits by a commercial supplier. Fixation and immunostainings of third-instar larval brains were performed as described in ref. 31 by using the affinity-purified anti-Mbm, mouse anti-fasciclin II (Developmental Studies Hybridoma Bank, Iowa City, IA), and mouse anti-phospho-histone H3 (Cell Signaling Technology, Beverly, MA) antibodies. The corresponding Alexa488- and Cy3-conjugated secondary antibodies were purchased from Molecular Probes and Dianova (Hamburg, Germany). Optical sections of brains were recorded with a step size of 0.8 μm with a Leica TCS (Wetzlar, Germany) laser confocal microscope. Images were processed with amira software (TGS, San Diego). For BrdUrd labeling experiments, dissected brains from third-instar larvae were incubated for 1.5 h in Schneider's medium containing 150 μg/ml BrdUrd. After fixation in 2% paraformaldehyde for 15 min, the brains were stained with the anti-D-Mef2 serum (kindly provided by H. Nguyen, Albert Einstein College of Medicine, New York) in combination with a horseradish peroxidase-conjugated secondary antibody and the Vector Laboratories SG detection kit. BrdUrd incorporation was detected after denaturation of the DNA with 2 M HCl (30 min) by using the BrdUrd in situ detection kit (Pharmingen). For immunostainings of paraffin-embedded adult brains (32), heads were cut in 7-μm serial sections. After deparaffinization in xylene and rehydration, sections were incubated with the anti-Leonardo antibody (33) and further processed with the ABC kit (Vector Laboratories). For Western blot analysis, protein lysates from 10 heads of male or female adult flies or 15 brain/eye-antennal imaginal disk complexes dissected from male or female third-instar larvae of the indicated genotype were separated by SDS/PAGE. After transfer on nitrocellulose, the blot was probed with the anti-Mbm antibody (1:200).
Results and Discussion
The Anatomy of the MBs in mbm1 Animals. Mutant mbm1 flies display a sexually dimorphic phenotype. The MBs of mbm1 females develop normally until the beginning of the third larval instar, when the axons of the Kenyon cells start to degenerate inappropriately. Kenyon cell perikarya apparently survive, but no regeneration of the axons is seen during metamorphosis, leading to a grossly reduced MB structure in the adult (25, 26). In the WT, degeneration of the γ-neuron axons becomes evident no earlier than the early pupal stage (Fig. 1 A and C). Antifasciclin II staining revealed the appearance of hole-like structures in the lobe system, which become infiltrated by glia cell processes to engulf degenerating axons (13, 14). In mbm1, hole-like structures are already seen at late third-larval instar, indicating a premature degeneration of axons (Fig. 1B). At early pupal stage, only residual anti-fasciclin II staining can be detected in mbm1 (Fig. 1D). In addition, a general reduction in the size of the lobe system was observed. To determine whether the reduced MB neuropil size seen in adult mbm1 females results from a selective loss of γ-neuron axons or whether other Kenyon cell subtypes are also affected by the mutation, frontal sections from heads of WT and mbm1 flies were stained with an antibody against the Drosophila 14-3-3 homologue Leonardo (33). Leonardo protein can be detected in the perikarya, dendrites, and axonal projections of the α/β, α′/β′, and γ neurons of WT flies (Fig. 2 A–D). The MB phenotype of mbm1 is variable. In mbm1 females displaying a moderate phenotype, the structural subdivision of the MBs is maintained (Fig. 2 E–H). In particular, the α/α′, β/β′, and γ-lobes can be distinguished, but there is an overall reduction in size. Even in animals with a strong mbm1 phenotype (Fig. 2 I–L), rudimentary calyx, peduncle, and lobe structures are formed. Thus, mbm1 affects all Kenyon cell subtypes in the adult fly. The anti-Leonardo stainings also revealed that the reduction in MB neuropil size always correlates with a reduction in the number of Leonardo-positive Kenyon cell bodies (Fig. 2 E and I). This finding appears to contradict the previous notion that the Kenyon cell bodies in mbm1 might survive into adulthood despite deprivation of dendritic and axonal branches at late larval development (25, 26). However, the remaining Leonardo-positive Kenyon cell bodies are surrounded by a large number of densely packed, unstained cells (Fig. 2I Inset). These cells might represent former Kenyon cells, which have lost their projections and concomitantly no longer express Kenyon-cell-specific proteins. Alternatively, but not mutually exclusive to the former explanation, mutations in mbm might also interfere with the generation of Kenyon cells. The expression pattern of the Mbm protein and the defects observed in cell proliferation assays indicate such a function (see Localization of Mbm in the Developing Nervous System).
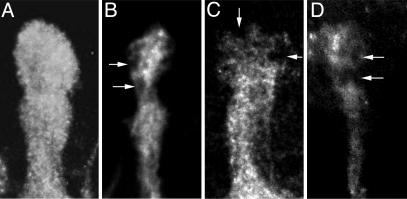
Fig. 1.
Premature degeneration of MB axons in mbm1. Shown are the dorsal lobes of WT (A and C) and mbm1 (B and D) MBs. (A) In late third-instar larvae, axon branches of the γ-neurons that form the dorsal lobe are uniformly labeled with the anti-fasciclin II antibody. Six hours after puparium formation (APF), degeneration of γ-neuron axons in WT animals becomes evident by the appearance of large unstained holes (arrows in C). The mbm1 mutation is characterized by a general size reduction of the lobe system and the appearance of unstained holes already at third larval instar (arrows in B). (D) Six hours APF, only residual anti-fasciclin II staining can be detected in mbm1.
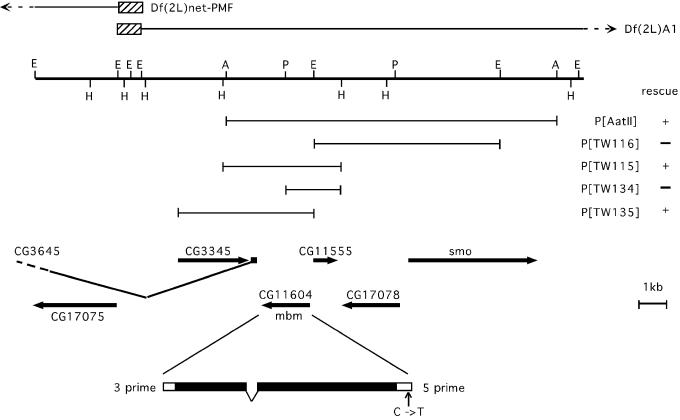
Identification of the mbm Gene Locus. The mbm1 mutation was initially mapped by complementation analysis with the deficiencies Df(2L)net-PMF and Df(2L)al to the distal region of chromosome 2L. We noticed that the MB phenotype of transheterozygous Df(2L)al/mbm1, and in particular of Df(2L)net-PMF/mbm1 flies, was even more variable than in homozygous mbm1 flies. Mapping of the breakpoints of these deficiencies by Southern blotting (ref. 27 and T.T., unpublished data) showed that localization of the mbm gene was not straightforward. This conclusion is because Df(2L)net-PMF and Df(2L)al proved to be nonoverlapping deficiencies that are separated by more than 30 kb, a stretch of DNA that encompasses 12 known or predicted transcription units. The complementation and mapping results could be explained if mbm mapped under one deficiency and the other contained an additional mutation. Alternatively, the mbm gene might reside within the 30-kb genomic interval with both deficiencies exerting a silencing effect on the expression of the mbm gene. In the case of the terminal deficiency Df(2L)net-PMF, genes near the breakpoint come in close vicinity to telomeric heterochromatin, which could lead to silencing of gene expression (telomeric position effect, ref. 34). Although Df(2L)al is not adjacent to a telomere, based on the variability of the MB phenotype seen in Df(2L)al/mbm1 animals, we suspect that it too depresses mbm expression through an indirect mechanism. To test the hypothesis that the mbm gene is deleted by neither deficiency but is localized to the 30-kb interval, we generated a set of overlapping deficiencies by jump-out mutagenesis of a P-element strain (P[lacW]185/319), which carries two P-element insertions in the vicinity of the distal breakpoint of Df(2L)al. Classification of these deficiencies by complementation analysis verified that only deficiencies that remove genomic material distal to the smoothened (smo) gene did not complement the mbm1 mutation (Fig. 3). Indeed, the mbm1 phenotype became more pronounced in these transheterozygous animals. Only very rarely, females were recovered and, in contrast to the original mbm1 mutation, also the surviving males had reduced MBs. Mapping of the distal breakpoint of one deficiency, Df(2L)A1, narrowed the mbm gene to a genomic interval of 10 kb distal to smo (Fig. 3). To identify the mbm transcription unit within this genomic segment, a set of overlapping genomic rescue constructs was generated. Only constructs encompassing the predicted CG11604 transcription unit (Fig. 3) were able to rescue the semilethality and the MB phenotype of mbm1/Df(2L)A1 animals (Fig. 2 M–P). As an independent verification that CG11604 indeed corresponds to the mbm gene, we sequenced genomic DNA isolated from mbm1 flies and the parental cn, bw, sp strain, which was used for mutagenesis. One C to T nucleotide exchange was detected in the 5′ untranslated region of the CG11604 transcript, thereby introducing an additional translational start codon (Fig. 3). This ATG codon is followed after 33 nucleotides by an in-frame TAA stop codon, thus creating a short ORF just in front of the predicted ORF of the CG11604 transcription unit. Because of the disassembly of the eukaryotic ribosomal complex after translation of an ORF, it can be assumed that this short additional ORF blocks at least to some degree the translation of the CG11604 ORF in mbm1 flies.
Fig. 3.
Genomic organization and identification of the mbm gene. The thick horizontal line represents a physical map of the genomic scaffold sequence AE003590 (position 263820–283640) (36). Restriction sites for AatII (A), EcoRI (E), HindIII (H), and HpaI (P) are indicated. The lines above the genomic sequence indicate the extension of the deficiencies Df(2L)net-PMF in the distal direction and Df(2L)A1 in the proximal direction. The proximal breakpoint of Df(2L)net-PMF and the distal breakpoint of Df(2L)A1 are located in the hatched boxes. Df(2L)al is located outside the map in proximal direction. Below the genomic map, the rescue constructs used in this study are shown. Their ability to rescue the mbm1/Df(2L)A1 phenotype is indicated with + or –. Predicted or known genes in this region are shown by arrows. The exon–intron structure of the mbm transcription unit CG11604 is diagrammed at the bottom. Filled boxes represent the ORF. In mbm1, aCtoT transition is found in the 5′ untranslated region.
To validate the expression of the Mbm protein in flies, an antiserum against the Mbm protein was generated and used in Western blot analysis. The CG11604 transcription unit is predicted to encode a polypeptide of 539 aa with a calculated molecular mass of 62 kDa (Fig. 6). On Western blots of brain homogenates of male and female third-instar larvae, the anti-serum recognizes a major protein band of ≈83 kDa and a slightly faster migrating protein isoform (Fig. 7, which is published as supporting information on the PNAS web site). Only very low levels of Mbm protein are detected in adult flies. Despite the sexually dimorphic MB phenotype of mbm1 flies, the expression level of the Mbm protein is equally reduced in male and female larvae (Fig. 7). These results show that mbm1 is a hypomorphic allele. The discrepancy between the predicted molecular mass of 62 kDa of the Mbm protein and the observed doublet of protein bands at 83 kDa on Western blots prompted us to verify the specificity of the antiserum. Transgenic flies, which allowed expression of a cDNA corresponding to the mbm gene (29) under Gal4/UAS control, were generated. High levels of Mbm protein accumulate in adult flies upon heat shock-induced expression of the mbm transgene. Again, the same doublet of protein bands was detected by the anti-Mbm antiserum (Fig. 7). The appearance of two protein isoforms, which are derived from a single cDNA, also suggests that the Mbm protein is subject to posttranslational modification.
In summary, we have provided genetic and molecular evidence that the CG11604 transcription unit corresponds to the mbm gene. Furthermore, we could show that a single point mutation in the 5′ untranslated region of the mbm transcript leads to reduction in Mbm protein expression.
Mbm Is a Zinc-Finger Protein. The Mbm protein does not display a significant overall homology to other proteins. Characteristic features of the protein are several domains that are highly enriched in certain amino acids. At the N terminus, an arginine/glycine-rich region is found, followed by a proline-rich region and several clusters of acidic or basic amino acids (Fig. 6). The most prominent structural feature of Mbm is a pair of zinc fingers of the Cys-Xaa2-Cys-Xaa4-His-Xaa4-Cys type located in the C-terminal half of the protein (amino acid positions 354–367 and 371–386). This type of zinc finger (sometimes referred to as zinc knuckle because of the short intervening sequences between the ion-contacting cysteine and histidine residues) was originally identified in the nucleocapsid proteins of retroviruses but has now been recognized as a general structural motif present in proteins of many eukaryotes (for a comprehensive list with >4,000 entries, see the Pfam protein families database at www.sanger.ac.uk/Software/Pfam). In general, it is thought that the Cys-Xaa2-Cys-Xaa4-His-Xaa4-Cys motif mediates binding to RNA or DNA. Binding studies with the two Cys-Xaa2-Cys-Xaa4-His-Xaa4-Cys fingers present in the nucleocapsid protein p7 of HIV have shown that they function concomitantly in binding RNA. Positively charged residues present within the zinc-finger structure and in the flanking sequences contribute to establish contacts with RNA (35). Mbm has a similar tandem arrangement of two Cys-Xaa2-Cys-Xaa4-His-Xaa4-Cys fingers, which are separated by only three amino acids (Fig. 6). Several basic amino acids are present N-terminal to and within both zinc-finger structures.
Besides mbm, few other genes in the Drosophila genome encode proteins with two or more consecutive Cys-Xaa2-Cys-Xaa4-His-Xaa4-Cys fingers (36). A protein with two zinc fingers but unknown biological function is encoded by CG9715. The Drosophila homologue of the cellular nucleic acid binding protein (CNBP) contains six zinc-finger domains. Vertebrate members of the CNBP family have been shown to promote expression of the c-myc and the colony stimulating factor-1 genes (37, 38). On the other hand, CNBP has been reported to bind to the 5′ untranslated sequences of ribosomal mRNAs (39). The first evidence that Mbm is able to bind to nucleic acids comes from an in vitro assay with a recombinant Mbm protein, which binds in a zinc-finger-dependent manner to DNA-coated cellulose beads (A.E. and T.R., unpublished data).
Localization of Mbm in the Developing Nervous System. By using the anti-Mbm antiserum, the expression pattern of the Mbm protein in the developing brain was determined. The finding that CNBP is able to bind to both RNA and DNA and can be found in the nucleus or the cytoplasm (40, 41) also raises the question of the subcellular localization of the Mbm protein.
Mbm shows a widespread expression in third-instar larval brains with no apparent difference between males and females (Fig. 4A). It can be detected at low levels in the MB neuropil. Most prominently, Mbm protein is found in the nuclei of many cells outside of the MBs. In mbm1, a global reduction in staining is seen in the brains of male and female larvae (data not shown). This result corresponds to the Western blot analysis (Fig. 7), but cannot explain the sexually dimorphic phenotype of mbm1. In a group of large cells, which, based on their large size, are neuroblasts, Mbm can be found either in the cell nucleus or in the cytoplasm (Fig. 4B). The change in subcellular localization correlates with the cell cycle. This result becomes evident by costaining with an antibody directed against phosphorylated histone H3, which marks chromosomes from late G2 phase throughout mitosis. In Fig. 4B, a group of three neuroblasts, which are closely associated with the Kenyon cell body layer and might therefore represent MB neuroblasts, is outlined. In one cell, Mbm is found in the nucleus; in the two phospho-histone H3-positive cells, Mbm localizes to the cytoplasm. We also noticed that cells in anaphase are devoid of detectable levels of Mbm protein (Fig. 4B, arrowhead). Direct evidence for a possible function of Mbm in cell proliferation was provided by BrdUrd pulse-labeling experiments. Dissected third-instar larval brains from WT and mbm1/Df(2L)A1 females were analyzed for incorporation of BrdUrd into newly synthesized DNA within a time window of 90 min. To identify the region in the dorsal brain where the MB neuroblasts are localized, we costained with an antiserum against D-Mef2, which selectively labels Kenyon cells (42). As shown in Fig. 5A, two to four BrdUrd-labeled cells are derived from a single, BrdUrd-positive neuroblast in WT brains. Beneath several neuroblasts, large clusters of D-Mef2-stained Kenyon cells can be recognized. In mbm1/Df(2L)A1 animals, there is a dramatic reduction in the number of BrdUrd- and D-Mef2-positive cells (Fig. 5B). In many cases, only single cells or two-cell clusters are labeled with BrdUrd, indicating a general defect in the proliferation pattern of central brain neuroblasts.
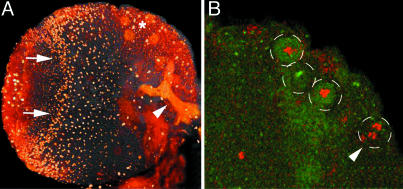
Fig. 4.
Staining of brains from third-instar larvae with the anti-Mbm anti-serum. (A) The projection view of one brain hemisphere shows the nuclear localization of the Mbm protein in many cells. The arrows point to the anlagen of the optic lobes. In addition, the antibody labels the neuropil of the larval MBs (arrowhead). In a group of three large cells (*), the Mbm protein localizes to the cytoplasm. (B) Costaining of brains with the anti-Mbm antiserum (green) and an antibody against phospho-histone H3 (red). A single confocal section (0.8 μm) at the level of the Kenyon cell body layer is shown. The Kenyon cell bodies are barely visible below the three outlined neuroblasts. The arrowhead points to a dividing cell at anaphase.
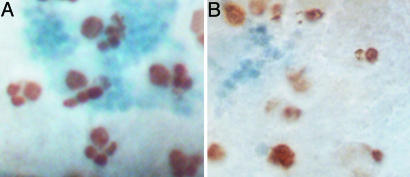
Fig. 5.
Altered cell proliferation in brains of mbm1/Df(2L)A1 larvae. Dorsal views of brains of late third-instar larvae labeled with BrdUrd (brown) and the D-Mef2 antibody (blue). In the central brain of the WT larvae (A), the large neuroblasts are faced at one side by two to four BrdUrd-positive ganglion mother cells or neurons. The Kenyon cells (blue) are localized beneath the MB neuroblasts and, therefore, most of them are out of focus. Only scattered BrdUrd-positive cells and small D-Mef2 cell clusters are seen in mbm1/Df(2L)A1 brains (B).
Again, these results parallel findings with the CNBP protein. Knockout experiments in mice have shown that CNBP is essential for forebrain formation (43). In conjunction with other studies, which have demonstrated an increase of c-myc promotor activity and cell proliferation upon overexpression of CNBP (44), it has been proposed that CNBP regulates forebrain formation through induction of c-Myc expression, which in turn stimulates cell proliferation and differentiation (43).
Conclusions
The mbm gene codes for a protein with two consecutive zinc-finger domains and nucleic acid binding properties. The Mbm protein cycles in larval neuroblasts in amount and subcellular localization. The protein is also found in low concentration in the larval MB neuropil. Mutational analysis shows that it is an essential gene involved in CNS development. In the original mutant mbm1 in which the developmental defects are largely confined to the female MBs, less Mbm protein is synthesized in both sexes. The molecular and systemic functions of Mbm are still poorly understood. The described binding of zinc-finger proteins, such as CNBP to both RNA and DNA, together with the finding that Mbm and CNBP can be localized to the cytoplasm and the nucleus, allow for a function in transcriptional and translational control. Mbm appears to regulate proliferation of neuronal precursor cells but also structural plasticity of Kenyon cells in the third-instar larva and during metamorphosis. Verification of Mbm as a DNA- or RNA-binding protein, together with the identification of the nucleotide sequences recognized by Mbm, should help to identify putative target genes and to understand the biological functions of Mbm in more detail.
Supplementary Material
Acknowledgments
We thank Markus Noll for several fly strains and the genomic walk of the 21B/C region; Susan Albert for the initial characterization of the 21B genomic region; Heike Wecklein for technical assistance; Efthimios Skoulakis (Texas A&M University, College Station) for the anti-Leonardo antibody; and Hanh Nguyen for the anti-D-Mef2 serum. This work was supported by grants from the Deutsche Forschungsgemeinschaft (to M.H. and T.R.), the Fonds der Chemischen Industrie (to M.H.), and the Human Frontier Science Program (to M.H.).
Abbreviations: CNBP, cellular nucleic acid binding protein; MB, mushroom body.
Data deposition: The sequence reported in this paper has been deposited in the TrEMBL database (accession no. Q9VPM5).
References
- 1.Strausfeld, N. J., Hansen, L., Li, Y., Gomez, R. S. & Ito, K. (1998) Learn. Mem. 5, 11–37. [PMC free article] [PubMed] [Google Scholar]
- 2.Heisenberg, M. (2003) Nat. Rev. Neurosci. 4, 266–275. [DOI] [PubMed] [Google Scholar]
- 3.Zars, T., Fischer, M., Schulz, R. & Heisenberg, M. (2000) Science 288, 672–675. [DOI] [PubMed] [Google Scholar]
- 4.Crittenden, J. R., Skoulakis, E. M., Han, K. A., Kalderon, D. & Davis, R. L. (1998) Learn. Mem. 5, 38–51. [PMC free article] [PubMed] [Google Scholar]
- 5.Heisenberg, M. (1998) Learn. Mem. 5, 1–10. [PMC free article] [PubMed] [Google Scholar]
- 6.Ito, K., Awano, W., Suzuki, K., Hiromi, Y. & Yamamoto, D. (1997) Development (Cambridge, U.K.) 124, 761–771. [DOI] [PubMed] [Google Scholar]
- 7.Kurusu, M., Awasaki, T., Masuda-Nakagawa, L. M., Kawauchi, H., Ito, K. & Furukubo-Tokunaga, K. (2002) Development (Cambridge, U.K.) 129, 409–419. [DOI] [PubMed] [Google Scholar]
- 8.Lee, T., Lee, A. & Luo, L. (1999) Development (Cambridge, U.K.) 126, 4065–4076. [DOI] [PubMed] [Google Scholar]
- 9.Strausfeld, N. J., Sinakevitch, I. & Vilinsky, I. (2003) Microsc. Res. Tech. 62, 151–169. [DOI] [PubMed] [Google Scholar]
- 10.Zhu, S., Chiang, A. S. & Lee, T. (2003) Development (Cambridge, U.K.) 130, 2603–2610. [DOI] [PubMed] [Google Scholar]
- 11.Heisenberg, M., Heusipp, M. & Wanke, C. (1995) J. Neurosci. 15, 1951–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Technau, G. & Heisenberg, M. (1982) Nature 295, 405–407. [DOI] [PubMed] [Google Scholar]
- 13.Awasaki, T. & Ito, K. (2004) Curr. Biol. 14, 668–677. [DOI] [PubMed] [Google Scholar]
- 14.Watts, R. J., Schuldiner, O., Perrino, J., Larsen, C. & Luo, L. (2004) Curr. Biol. 14, 678–684. [DOI] [PubMed] [Google Scholar]
- 15.Heisenberg, M. (1980) Basic Life Sci. 16, 373–390. [DOI] [PubMed] [Google Scholar]
- 16.Nicolai, M., Lasbleiz, C. & Dura, J. M. (2003) J. Neurobiol. 57, 291–302. [DOI] [PubMed] [Google Scholar]
- 17.Reuter, J. E., Nardine, T. M., Penton, A., Billuart, P., Scott, E. K., Usui, T., Uemura, T. & Luo, L. (2003) Development (Cambridge, U.K.) 130, 1203–1213. [DOI] [PubMed] [Google Scholar]
- 18.Guan, Z., Prado, A., Melzig, J., Heisenberg, M., Nash, H. A. & Raabe, T. (2000) Proc. Natl. Acad. Sci. USA 97, 8122–8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott, E. K., Lee, T. & Luo, L. (2001) Curr. Biol. 11, 99–104. [DOI] [PubMed] [Google Scholar]
- 20.Lee, T., Winter, C., Marticke, S. S., Lee, A. & Luo, L. (2000) Neuron 25, 307–316. [DOI] [PubMed] [Google Scholar]
- 21.Ng, J., Nardine, T., Harms, M., Tzu, J., Goldstein, A., Sun, Y., Dietzl, G., Dickson, B. J. & Luo, L. (2002) Nature 416, 442–447. [DOI] [PubMed] [Google Scholar]
- 22.Lee, T., Marticke, S., Sung, C., Robinow, S. & Luo, L. (2000) Neuron 28, 807–818. [DOI] [PubMed] [Google Scholar]
- 23.Zheng, X., Wang, J., Haerry, T. E., Wu, A. Y., Martin, J., O'Connor, M. B., Lee, C. H. & Lee, T. (2003) Cell 112, 303–315. [DOI] [PubMed] [Google Scholar]
- 24.Watts, R. J., Hoopfer, E. D. & Luo, L. (2003) Neuron 38, 871–885. [DOI] [PubMed] [Google Scholar]
- 25.Heisenberg, M., Borst, A., Wagner, S. & Byers, D. (1985) J. Neurogenet. 2, 1–30. [DOI] [PubMed] [Google Scholar]
- 26.de Belle, J. S. & Heisenberg, M. (1996) Proc. Natl. Acad. Sci. USA 93, 9875–9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alcedo, J., Ayzenzon, M., Von Ohlen, T., Noll, M. & Hooper, J. E. (1996) Cell 86, 221–232. [DOI] [PubMed] [Google Scholar]
- 28.Klemenz, R., Weber, U. & Gehring, W. J. (1987) Nucleic Acids Res. 15, 3947–3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stapleton, M., Liao, G., Brokstein, P., Hong, L., Carninci, P., Shiraki, T., Hayashizaki, Y., Champe, M., Pacleb, J., Wan, K., et al. (2002) Genome Res. 12, 1294–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brand, A. H. & Perrimon, N. (1993) Development (Cambridge, U.K.) 118, 401–415. [DOI] [PubMed] [Google Scholar]
- 31.Gaul, U., Mardon, G. & Rubin, G. M. (1992) Cell 68, 1007–1019. [DOI] [PubMed] [Google Scholar]
- 32.Heisenberg, M. & Bohl, K. (1979) Z. Naturforsch. 34, 143–147. [Google Scholar]
- 33.Skoulakis, E. M. & Davis, R. L. (1996) Neuron 17, 931–944. [DOI] [PubMed] [Google Scholar]
- 34.Boivin, A., Gally, C., Netter, S., Anxolabehere, D. & Ronsseray, S. (2003) Genetics 164, 195–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Urbaneja, M. A., Kane, B. P., Johnson, D. G., Gorelick, R. J., Henderson, L. E. & Casas-Finet, J. R. (1999) J. Mol. Biol. 287, 59–75. [DOI] [PubMed] [Google Scholar]
- 36.Celniker, S. E., Wheeler, D. A., Kronmiller, B., Carlson, J. W., Halpern, A., Patel, S., Adams, M., Champe, M., Dugan, S. P., Frise, E., et al. (2002) Genome Biol. 3, RESEARCH0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michelotti, E. F., Tomonaga, T., Krutzsch, H. & Levens, D. (1995) J. Biol. Chem. 270, 9494–9499. [DOI] [PubMed] [Google Scholar]
- 38.Konicek, B. W., Xia, X., Rajavashisth, T. & Harrington, M. A. (1998) DNA Cell Biol. 17, 799–809. [DOI] [PubMed] [Google Scholar]
- 39.Pellizzoni, L., Lotti, F., Maras, B. & Pierandrei-Amaldi, P. (1997) J. Mol. Biol. 267, 264–275. [DOI] [PubMed] [Google Scholar]
- 40.Armas, P., Cabada, M. O. & Calcaterra, N. B. (2001) Dev. Growth Differ. 43, 13–23. [DOI] [PubMed] [Google Scholar]
- 41.Calcaterra, N. B., Palatnik, J. F., Bustos, D. M., Arranz, S. E. & Cabada, M. O. (1999) Dev. Growth Differ. 41, 183–191. [DOI] [PubMed] [Google Scholar]
- 42.Schulz, R. A., Chromey, C., Lu, M. F., Zhao, B. & Olson, E. N. (1996) Oncogene 12, 1827–1831. [PubMed] [Google Scholar]
- 43.Chen, W., Liang, Y., Deng, W., Shimizu, K., Ashique, A. M., Li, E. & Li, Y. P. (2003) Development (Cambridge, U.K.) 130, 1367–1379. [DOI] [PubMed] [Google Scholar]
- 44.Shimizu, K., Chen, W., Ashique, A. M., Moroi, R. & Li, Y. P. (2003) Gene 307, 51–62. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.