Abstract
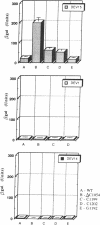
Site-directed mutagenesis was performed on a sequence motif within the 3' major domain of Escherichia coli 16S rRNA shown previously to be important for peptide chain termination. Analysis of stop codon suppression by the various mutants showed an exclusive response to UGA stop signals, which was correlated directly with the continuity of one or the other of two tandem complementary UCA sequences (bases 1199-1204). Since no other structural features of the mutated ribosomes were hampered and the translation initiation and elongation events functioned properly, we propose that a direct interaction occurs between the UGA stop codon on the mRNA and the 16S rRNA UCA motif as one of the initial events of UGA-dependent peptide chain termination. These results provide evidence that base pairing between rRNA and mRNA plays a direct role in termination, as it has already been shown to do for initiation and elongation.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen P. N., Noller H. F. Mutations in ribosomal proteins S4 and S12 influence the higher order structure of 16 S ribosomal RNA. J Mol Biol. 1989 Aug 5;208(3):457–468. doi: 10.1016/0022-2836(89)90509-3. [DOI] [PubMed] [Google Scholar]
- Barta A., Steiner G., Brosius J., Noller H. F., Kuechler E. Identification of a site on 23S ribosomal RNA located at the peptidyl transferase center. Proc Natl Acad Sci U S A. 1984 Jun;81(12):3607–3611. doi: 10.1073/pnas.81.12.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin F., Ehresmann C., Romby P., Mougel M., Colin J., Lempereur L., Bachellerie J. P., Ebel J. P., Ehresmann B. Higher-order structure of domain III in Escherichia coli 16S ribosomal RNA, 30S subunit and 70S ribosome. Biochimie. 1987 Oct;69(10):1081–1096. doi: 10.1016/0300-9084(87)90008-3. [DOI] [PubMed] [Google Scholar]
- Baudin F., Mougel M., Romby P., Eyermann F., Ebel J. P., Ehresmann B., Ehresmann C. Probing the phosphates of the Escherichia coli ribosomal 16S RNA in its naked form, in the 30S subunit, and in the 70S ribosome. Biochemistry. 1989 Jul 11;28(14):5847–5855. doi: 10.1021/bi00440a022. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Backman K. Plasmids of Escherichia coli as cloning vectors. Methods Enzymol. 1979;68:245–267. doi: 10.1016/0076-6879(79)68018-7. [DOI] [PubMed] [Google Scholar]
- Bonham-Smith P. C., Bourque D. P. Translation of chloroplast-encoded mRNA: potential initiation and termination signals. Nucleic Acids Res. 1989 Mar 11;17(5):2057–2080. doi: 10.1093/nar/17.5.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J., Ullrich A., Raker M. A., Gray A., Dull T. J., Gutell R. R., Noller H. F. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of E. coli. Plasmid. 1981 Jul;6(1):112–118. doi: 10.1016/0147-619x(81)90058-5. [DOI] [PubMed] [Google Scholar]
- Cech T. R. The chemistry of self-splicing RNA and RNA enzymes. Science. 1987 Jun 19;236(4808):1532–1539. doi: 10.1126/science.2438771. [DOI] [PubMed] [Google Scholar]
- Dahlberg A. E. The functional role of ribosomal RNA in protein synthesis. Cell. 1989 May 19;57(4):525–529. doi: 10.1016/0092-8674(89)90122-0. [DOI] [PubMed] [Google Scholar]
- Dams E., Hendriks L., Van de Peer Y., Neefs J. M., Smits G., Vandenbempt I., De Wachter R. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 1988;16 (Suppl):r87–173. doi: 10.1093/nar/16.suppl.r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper D. E., Deckman I. C., Vartikar J. V. Physical studies of ribosomal protein-RNA interactions. Methods Enzymol. 1988;164:203–220. doi: 10.1016/s0076-6879(88)64044-4. [DOI] [PubMed] [Google Scholar]
- Gourse R. L., Takebe Y., Sharrock R. A., Nomura M. Feedback regulation of rRNA and tRNA synthesis and accumulation of free ribosomes after conditional expression of rRNA genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1069–1073. doi: 10.1073/pnas.82.4.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983 Dec;35(3 Pt 2):849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Gutell R. R., Weiser B., Woese C. R., Noller H. F. Comparative anatomy of 16-S-like ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1985;32:155–216. doi: 10.1016/s0079-6603(08)60348-7. [DOI] [PubMed] [Google Scholar]
- Hui A. S., Eaton D. H., de Boer H. A. Mutagenesis at the mRNA decoding site in the 16S ribosomal RNA using the specialized ribosome system in Escherichia coli. EMBO J. 1988 Dec 20;7(13):4383–4388. doi: 10.1002/j.1460-2075.1988.tb03337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänfler A., Kleuvers B., Göringer H. U. The involvement of base 1054 in 16S rRNA for UGA stop codon dependent translational termination. Nucleic Acids Res. 1990 Oct 11;18(19):5625–5632. doi: 10.1093/nar/18.19.5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob W. F., Santer M., Dahlberg A. E. A single base change in the Shine-Dalgarno region of 16S rRNA of Escherichia coli affects translation of many proteins. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4757–4761. doi: 10.1073/pnas.84.14.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lang A., Friemert C., Gassen H. G. On the role of the termination factor RF-2 and the 16S RNA in protein synthesis. Eur J Biochem. 1989 Apr 1;180(3):547–554. doi: 10.1111/j.1432-1033.1989.tb14680.x. [DOI] [PubMed] [Google Scholar]
- Makosky P. C., Dahlberg A. E. Spectinomycin resistance at site 1192 in 16S ribosomal RNA of E. coli: an analysis of three mutants. Biochimie. 1987 Aug;69(8):885–889. doi: 10.1016/0300-9084(87)90216-1. [DOI] [PubMed] [Google Scholar]
- Maly P., Brimacombe R. Refined secondary structure models for the 16S and 23S ribosomal RNA of Escherichia coli. Nucleic Acids Res. 1983 Nov 11;11(21):7263–7286. doi: 10.1093/nar/11.21.7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell. 1989 May 19;57(4):585–597. doi: 10.1016/0092-8674(89)90128-1. [DOI] [PubMed] [Google Scholar]
- Moazed D., Stern S., Noller H. F. Rapid chemical probing of conformation in 16 S ribosomal RNA and 30 S ribosomal subunits using primer extension. J Mol Biol. 1986 Feb 5;187(3):399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- Murgola E. J., Hijazi K. A., Göringer H. U., Dahlberg A. E. Mutant 16S ribosomal RNA: a codon-specific translational suppressor. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4162–4165. doi: 10.1073/pnas.85.12.4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrullo L. A., Gallagher P. J., Elseviers D. The role of 2-methylthio-N6-isopentenyladenosine in readthrough and suppression of nonsense codons in Escherichia coli. Mol Gen Genet. 1983;190(2):289–294. doi: 10.1007/BF00330653. [DOI] [PubMed] [Google Scholar]
- Prescott C. D., Göringer H. U. A single mutation in 16S rRNA that affects mRNA binding and translation-termination. Nucleic Acids Res. 1990 Sep 25;18(18):5381–5386. doi: 10.1093/nar/18.18.5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottmann N., Kleuvers B., Atmadja J., Wagner R. Mutants with base changes at the 3'-end of the 16S RNA from Escherichia coli. Construction, expression and functional analysis. Eur J Biochem. 1988 Oct 15;177(1):81–90. doi: 10.1111/j.1432-1033.1988.tb14347.x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Schüler D., Brimacombe R. The Escherichia coli 30S ribosomal subunit; an optimized three-dimensional fit between the ribosomal proteins and the 16S RNA. EMBO J. 1988 May;7(5):1509–1513. doi: 10.1002/j.1460-2075.1988.tb02970.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z. H., Fox T. D. Substitution of an invariant nucleotide at the base of the highly conserved '530-loop' of 15S rRNA causes suppression of yeast mitochondrial ochre mutations. Nucleic Acids Res. 1989 Jun 26;17(12):4535–4539. doi: 10.1093/nar/17.12.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern S., Powers T., Changchien L. M., Noller H. F. RNA-protein interactions in 30S ribosomal subunits: folding and function of 16S rRNA. Science. 1989 May 19;244(4906):783–790. doi: 10.1126/science.2658053. [DOI] [PubMed] [Google Scholar]
- Stöffler G., Tate W. P., Caskey C. T. Ribosomal proteins cross-linked to peptide chain termination release factor 2. J Biol Chem. 1982 Apr 25;257(8):4203–4206. [PubMed] [Google Scholar]
- Thompson J., Cundliffe E., Dahlberg A. E. Site-directed mutagenesis of Escherichia coli 23 S ribosomal RNA at position 1067 within the GTP hydrolysis centre. J Mol Biol. 1988 Sep 20;203(2):457–465. doi: 10.1016/0022-2836(88)90012-5. [DOI] [PubMed] [Google Scholar]
- Valle R. P., Morch M. D. Stop making sense: or Regulation at the level of termination in eukaryotic protein synthesis. FEBS Lett. 1988 Aug 1;235(1-2):1–15. doi: 10.1016/0014-5793(88)81225-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartikar J. V., Draper D. E. S4-16 S ribosomal RNA complex. Binding constant measurements and specific recognition of a 460-nucleotide region. J Mol Biol. 1989 Sep 20;209(2):221–234. doi: 10.1016/0022-2836(89)90274-x. [DOI] [PubMed] [Google Scholar]
- Weiss R. B., Dunn D. M., Dahlberg A. E., Atkins J. F., Gesteland R. F. Reading frame switch caused by base-pair formation between the 3' end of 16S rRNA and the mRNA during elongation of protein synthesis in Escherichia coli. EMBO J. 1988 May;7(5):1503–1507. doi: 10.1002/j.1460-2075.1988.tb02969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamao F., Muto A., Kawauchi Y., Iwami M., Iwagami S., Azumi Y., Osawa S. UGA is read as tryptophan in Mycoplasma capricolum. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2306–2309. doi: 10.1073/pnas.82.8.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano R., Yura T. Suppression of the Escherichia coli rpoH opal mutation by ribosomes lacking S15 protein. J Bacteriol. 1989 Mar;171(3):1712–1717. doi: 10.1128/jb.171.3.1712-1717.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaug A. J., Cech T. R. The intervening sequence RNA of Tetrahymena is an enzyme. Science. 1986 Jan 31;231(4737):470–475. doi: 10.1126/science.3941911. [DOI] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis of DNA fragments cloned into M13 vectors. Methods Enzymol. 1983;100:468–500. doi: 10.1016/0076-6879(83)00074-9. [DOI] [PubMed] [Google Scholar]