Abstract
Gene regulation at the post-transcriptional level is frequently based on cis- and trans-acting factors on target mRNAs. We found a C-rich element (CRE) in mu-opioid receptor (MOR) 3′-untranslated region (UTR) to which poly (rC) binding protein 1 (PCBP1) binds, resulting in MOR mRNA stabilization. RNA immunoprecipitation and RNA EMSA revealed the formation of PCBP1-RNA complexes at the element. Knockdown of PCBP1 decreased MOR mRNA half-life and protein expression. Stimulation by forskolin increased cytoplasmic localization of PCBP1 and PCBP1/MOR 3′-UTR interactions via increased serine phosphorylation that was blocked by protein kinase A (PKA) or (phosphatidyl inositol-3) PI3-kinase inhibitors. The forskolin treatment also enhanced serine- and tyrosine-phosphorylation of AU-rich element binding protein (AUF1), concurrent with its increased binding to the CRE, and led to an increased interaction of poly A binding protein (PABP) with the CRE and poly(A) sites. AUF1 phosphorylation also led to an increased interaction with PCBP1. These findings suggest that a single co-regulator, PCBP1, plays a crucial role in stabilizing MOR mRNA, and is induced by PKA signaling by conforming to AUF1 and PABP.
Keywords: Mu opioid receptor, RNA stability, 3′-Untranslated region, RNA binding protein, Protein kinase A signaling
1. Introduction
In eukaryotes, the 3′-untranslated region (3′-UTR) of mRNA regulates many post-transcriptional regulatory pathways, such as mRNA localization, stability, and translation efficiency, via either its length or specific sequence elements for 3′-UTR-binding proteins and microRNAs (Pesole et al., 2001). The average length of the 3′-UTR increases strikingly from 200 nucleotides (nt) in plants and fungi (Tanaka et al., 2011) to > 1000 nt in humans and other vertebrates (Pesole et al., 2001), suggesting that long 3′-UTRs play a role in the regulation of the more complicated gene expression in higher vertebrates. Several target genes known to be involved in human diseases are depended on cis- and/or trans-factors acting at the 3′-UTR of mRNA (Conne et al., 2000; Chatterjee and Pal, 2009; Han et al., 2010; Chaudhury et al., 2011). A major class of cis-acting elements that regulates mRNA stability includes AU-rich elements (AREs) and is often found in the 3′-UTR of short-lived mRNA (Caput et al., 1986; Shaw and Kamen, 1986; Zou et al., 2010). AREs mainly recruit mRNA degradation proteins (i.e., AUF1, also called hnRNPD) (Pan et al., 2005; Barker et al., 2012); however, mRNA stabilizing proteins, such as HuR and pp32, have also been reported to bind to and be recruited to AREs (Peng et al., 1998; Brennan et al., 2000). Reports have shown the importance of 3′-UTR length and ARE location within the 3′-UTR as key determinants of RNA/protein interaction and translational control of beta2-adrenergic receptor mRNA (Subramaniam et al., 2004; Subramaniam et al., 2011).
C-rich element (CRE) has also been reported to be involved in the regulation of mRNA stability. In erythroid cells, α-globin mRNA stability is regulated by a sequence-specific RNA-protein complex at a CRE region in the 3′-UTR of α-globin mRNA (Weiss and Liebhaber, 1994). This α-complex is a multisubunit structure and may be a general determinant of mRNA stability (Chkheidze et al., 1999). Sustained stability of α-globin mRNA is important during all stages of erythropoiesis in order to have steady expression of α-globin mRNA and its protein (Waggoner and Liebhaber, 2003). PCBP1 as well as several other RNA binding proteins, such as PCBP2, AUF1, hnRNPK, and poly(A)-binding protein (PABP), are constituents of the α-complex that binds to the CRE motif to prevent mRNA degradation (Kiledjian et al., 1997; Chkheidze et al., 1999; Wang et al., 1999; Chaudhury et al., 2010). PCBP1 has been shown to regulate stability of many other mRNAs, such as androgen receptor (Yeap et al., 2002), β-globin (Yu and Russell, 2001), collagens I and III (Thiele et al., 2004), disabled-2 (Dab2) and interleukin-like EMT inducer (ILEI) (Chaudhury et al., 2011), endothelial nitric oxide synthase (Ho et al., 2013), erythropoietin (Czyzyk-Krzeska and Bendixen, 1999), folate receptor (Tang et al., 2011), neurofilament-M (Thyagarajan and Szaro, 2008), renin (Skalweit et al., 2003), and tyrosine hydroxylase (Czyzyk-Krzeska and Beresh, 1996).
Recent reports indicate that microRNAs regulate the MOR gene by binding to target sites in the 3′-UTR of MOR (Hwang et al., 2012). Opioids increased the expression of three microRNAs, miR-23b (Wu et al., 2009), let-7 (He et al., 2010), and miR-339-3p (Wu et al., 2013), resulting in downregulation of the MOR gene at the post-transcriptional level. Inhibition of the expression of the microRNAs in vivo by microRNA inhibitors resulted in upregulation of the MOR gene (Wu et al., 2009, 2013) and partially attenuated opioid tolerance in the case of let-7 (He et al., 2010). These studies suggest that microRNAs may play a causal role, at least partially, during development of opioid tolerance by increasing MOR expression. Besides microRNAs, several other factors also regulate gene expression by binding to the non-coding regions of the RNA. For example, a report suggested that short tandem repeats (STRs) in MOR 3′-UTR, especially T and TA repeats, may contribute to mRNA stability by affecting the binding of mRNA degradation or stabilizing proteins, which consequently leads to the high morphine sensitivity observed in mouse strains with long T and TA STRs (Shigeta et al., 2008). Several ARE motifs have been found at positions 4–5 and 8–9 kb (mouse) and 11–12 kb (human) downstream from the stop codon in the 3′-UTR (Wu et al., 2005; Kasai et al., 2006). The current search for conserved motifs in the MOR 3′-UTR reveals several other conserved motifs known to function as cis-acting elements for up- or downregulation of genes. However, until now the roles of these cis-acting elements have not been fully studied.
In this study, we found that PCBP1 binds to a novel target site (CRE) of MOR 3′-UTR and enhances MOR mRNA stability by binding to several other factors. We studied the effect and regulation of PCBP1 binding on the MOR gene and identified novel roles for PCBP1 interaction with different RNA-binding factors. We have attempted to demonstrate for the first time that a single regulator can function as a major factor in the formation of an RNA-binding protein complex that stabilizes the target gene MOR.
2. Materials and methods
2.1. Materials
Forskolin (344270), JNK inhibitor II, Ly294002, NF-κB inhibitor [6-amino-4-(4-phenoxyphenylethylamino)quinazoline], PD98059, PP2 [4-amino-5-(4-chlorophenyl)-7-(t-butyl)pyrazolo(3,4-d)pyrimidine], and PP3 [4-amino-7-phenylpyrazolo(3,4-d)pyrimidine] were purchased from EMD Biosciences. SB203580 and U0126 were purchased from Cell Signaling. Actinomycin-D, cycloheximide, H89 (H-89 dihydrochloride hydrate), and leptomycin B were purchased from Sigma. Phorbol-12-myristate-13-acetate (PMA) was purchased from Fisher Scientific. siRNAs of hnRNPK (s67648), PCBP1 (s76740), and negative control (#1, 4390843) were purchased from Invitrogen. We obtained control shRNA and AUF1 shRNA (pSilencer-AUF1) (Chang et al., 2010) through the kindness of Dr. Wengong Wang (Peking University, China). PCBP1 shRNA (sc-38269-SH) was purchased from Santa Cruz.
2.2. Cell culture and plasmid construction
Cell culture and plasmid transfection in neuroblastoma NMB cells were performed as described previously (Hwang et al., 2004). For drug treatment of NMB cells, cells were plated to a density of 2 × 105 cells/well in 12-well culture plates 15 h before the drug treatment; 48 h after transfection, cells were harvested and used for isolation of total RNA or luciferase assay. Cultures of P19 cells and the procedures to differentiate P19 cells (AP4d) have been described previously (Hwang et al., 2007).
pMUTR and pSVUTR were described previously (Wu et al., 2008). PCBP1 expression plasmid (myc-tagged pcDNA4-PCBP1, also called pcDNA4-αCP1) was described previously (Choi et al., 2008). Mutations of PCBP1 for phosphorylation, NES, and NLS sites in pcDNA4-PCBP1 were generated by PCR site-directed mutagenesis using high-fidelity Pfu DNA polymerase according to the manufacturer's protocol (Quikchange, Stratagene) with the respective PCR primer sets (Table 1). To generate plasmid 5′-UTR, uAUG(+) plasmid (Song et al., 2007) was digested with HindIII and NcoI, and a 301-bp DNA fragment of the MOR 5′-UTR was isolated. This fragment was inserted into the HindIII/NcoI sites of pSVPA (Wu et al., 2008) and was designated 5′-UTR plasmid. DNA fragment A, containing + 1 ~ + 197-bp of the 3′-UTR fragment downstream of the MOR stop codon, was obtained by PCR amplification of the rpL32 region in pMUTR with primers Xba-S and RPfse-AS (Table 1). Fragment A was digested with XbaI and FseI and inserted into the same sites of 5′-UTR plasmid to create 5′-UTR-Ag plasmid (Fig. 1B). Similarly, DNA fragment B (441-bp in size) was generated by PCR amplification of the AP1/Brd-containing region of pMUTR with primers XbaA-S and RPfseA-AS and inserted into the 5′-UTR plasmid, creating 5′-UTR-Bg plasmid. DNA fragment B contains two AP-1 sites (Activator Protein-1, 5′TGAGTCA-3′) (Halazonetis et al., 1988) in the region from + 2582 to + 2972 (downstream of the MOR stop codon) of MOR 3′-UTR and one Brd-box (Brd, 5′AGCTTTA-3′) at + 2675 of the MOR 3′-UTR (Lai et al., 1998; Lai, 2002). DNA fragment C (172-bp in size) was generated by PCR amplification of the k-boxcontaining of pMUTR with primers XbaB-S and RPfseB-AS and inserted into the 5′-UTR plasmid to create 5′-UTR-Cg plasmid. DNA fragment C contains three conserved motifs, ADH-DRE (in reverse direction, 5′ AAGGCTGA-3′) (Parsch et al., 1999), k-box (k-box2 in the MOR 3′-UTR, 5′TGTGAT-3′) (Wu et al., 2008), and IGHA1 (immunoglobulin heavy constant alpha 1, 5′ATTTTCAT-3′) (Maeda et al., 1987). All vectors were confirmed by DNA sequencing and restriction enzyme analyses. RegRNA (Huang et al., 2006) was used to search for the regulatory RNA motifs in MOR 3′-UTR.
Table 1.
List of primers used in this study.
| Namea | Primer (5′→3′) | Location | Notes |
|---|---|---|---|
| Xba-S | TAATTCTAGAGTCGGGGCGGACG | +1–+13b | Xba I site underlined |
| RPfse-AS | AGTGGCCGGCCTGACGTGTAGCGTGAATAGCC | +161–+141b | Fse I site underlined |
| XbaA-S | GACTCTAGAAGAAATTGAGTCATGCTT | +2576–+2593b | Xba I site underlined |
| RPfseA-AS | AGTGGCCGGCCGAGAGGAACAGGAGTGAATCC | +3002–+2982b | Fse I site underlined |
| XbaB-S | CACTCTAGAGTCTGAACTCACA | +8306–+8318b | Xba I site underlined |
| RPfseB-AS | AGTGGCCGGCCCTAACTCAGTTCCGAGTTAAG | +8457–+8437b | Fse I site underlined |
| LUC-S1 | CTGGAAGATGGAACCGCTGGAG | 150-bp LUC in PCR size | |
| LUC-AS1 | AGCTTCTGCCAACCGAACGGAC | ||
| Gal-S1 | CCGGTGGGTGAAGACCAGAAAC | 151-bp β-gal in PCR size | |
| Gal-AS1 | GTGCTGCAAGGCGATTAAGTTG | ||
| Rch-S1 | CAGACCCTCGCTAAACTTAG | +18–+37b | 230-bp in PCR size, RPL32 site |
| Rch-AS1 | CCATTGGATGTTTCTGTACATG | +247–+226b | RPL32 site |
| Rch-S5 | GATGCGCCTCCGTGTACTTCTAAG | −237–−214c | 240-bp in PCR size, 5′-UTR region |
| Rch-AS5 | CATGGTTCTGAATGCTTGCTG | −18–+3c | 5′-UTR region |
| Rch-S8 | CGTATGAATTAAATGATGTGTGGC | +1903–+1926b | 181-bp in PCR size, 3′-UTR repeat |
| Rch-AS8 | AATGGACCGATTCAAGAACAAG | +2083–+2062b | 3′-UTR repeat |
| Rch-S11 | AAACTGTGTGAGATCCATAGGC | +9423–+9444b | 250bp in PCR size, CA repeat 2 |
| Rch-AS11 | ACAACAAACCACATACTCACGTC | +9672–+9650b | CA repeat 2 |
| Rch-S2 | CTTGCAGAGAAGACTAAGCTATC | +9807–+9829b | 303-bp in PCR size, 3′-UTR end |
| Rch-AS2 | GCAAGAAATCACAATACAGCC | +10109–+10089b | 3′-UTR end |
| mMOR-S7 | CATGGCCCTCTATTCTATCGTGT | +213–+235c | 151bp in PCR size, exon 1 |
| mMOR-AS7 | CAGCGTGCTAGTGGCTAAGG | +363–+344c | exon 2 |
| Rch-S6 | GCCGGTCCTCATCATCACTGTG | +729–+750c | 235-bp in PCR size, exon 3 |
| Rch-AS6 | GCAGAAGTGCCAGGAAACAGTC | +963–+942c | exon 3 |
| Rch-S4 | CTGCAGAACCAAGAGGTTACC | +8134–+8154b | 320-bp in PCR size, AR site |
| Rch-AS4 | CTCAGTTCCGAGTTAAGCTAATG | +8453–+8431b | AR site |
| S43A-Sd | GAGGATCCGCGAGGAGgcCGGCGCGCGGATC | eSer→Ala | |
| S43A-ASd | GATCCGCGCGCCGgcCTCCTCGCGGATCCTC | ||
| Y200A-Sd | GCAGCGACGCGGCGGGCgcaCCCCACGCCAC | eTyr→Ala | |
| Y200A-ASd | GTGGCGTGGGGtgcGCCCGCCGCGTCGCTGC | ||
| S246A-Sd | GGTGGCAAGACAACAGgCTCACTTTGCCATG | eSer→Ala | |
| S246A-ASd | CATGGCAAAGTGAGcCTGTTGTCTTGCCACC | ||
| S309A-Sd | GAGATCCGCCAGATGgCCGGGGCCCAGATC | eSer→Ala | |
| S309A-ASd | GATCTGGGCCCCGGcCATCTGGCGGATCTC | ||
| S348_349A-S | CTAATCAATGCCAGGCTTgCCgCTGAGAAGGG | eSerSer→AlaAla | |
| S348_349A-ASd | CCCTTCTCAGcGGcAAGCCTGGCATTGATTAG | ||
| NLS1-Sf | CAGCTCTCCAGgGaTccAAGGCTATTGG | eGluValLys→GlyIleGln | |
| NLS1-ASf | CCAATAGCCTTggAtCcCTGGAGAGCTG | ||
| NLS2-Sf | GGCTCTTCTGGAAttCAGGTCACcATggCTGGCTCGGC | eArg→Ile, eThrIleThr→ThrMetAla | |
| NLS2-ASf | GCCGAGCCAGccATgGTGACCTGaaTTCCAGAAGAGCC | ||
| NES1-Sf | CGTGACTCgagCCATTCccaTGgTGATGCAC | eLeuThr→ArgAla, eArgLeuLeu→ProMetVal | |
| NES1-ASf | GTGCATCAcCAtggGAATGGctcGAGTCACG | ||
| NES2-Sf | CTGCCTGGTCAccaTGGAGACGCTCTC | eMetLeu→ThrMet | |
| NES2-ASd | GAGAGCGTCTCCAtggTGACCAGGCAG | ||
| RPLflank-S | UCCUGGAAAAGCAUCUGAUCCUGCAUCAUUCAAAGUCA | +93–+131b | RNA oligonucleotide |
S and AS in the primer name indicate sense and antisense primers, respectively.
Numbers are PCR primer sites downstream (+ number) from MOR stop codon excluding the introduced restriction enzymes sites.
Numbers are PCR primer sites downstream or upstream from MOR start codon.
Bold lower cases indicate mutated nucleotides. Bold/underlined sequence represents mutated regions to change amino acids.
Mutated amino acids sequences caused by underlined nucleotides sequence changes.
Bold sequence represent mutated regions to change amino acids and also created newly restriction enzyme site for cloning purpose. Bold lower cases indicate mutated nucleotides.
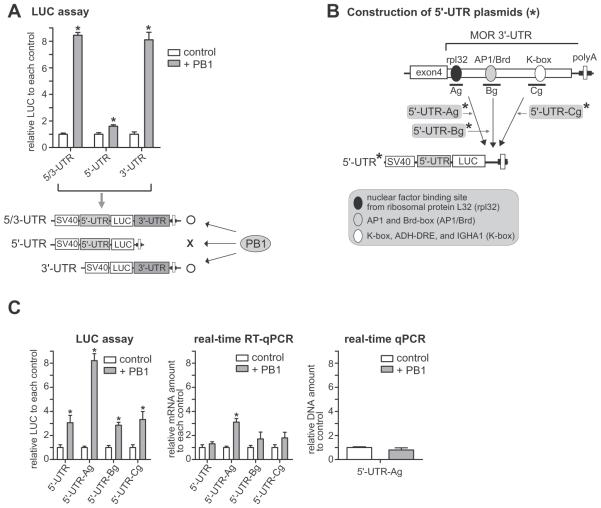
Fig. 1.
PCBP1 (PB1), an RNA-binding protein, upregulates luciferase (LUC) reporter activity through MOR 3′-UTR, primarily post-transcriptionally. A) Co-expression of PCBP1 enhances luciferase reporter activity (LUC) by both constructs MOR 5/3-UTR and MOR 3′-UTR, but not by MOR 5′-UTR. Three different LUC constructs (5′-UTR, 3′-UTR, and 5/3-UTR) were cotransfected with vector (control) or with PCBP1 and transfected into MOR-positive NMB cells. The results are given as luciferase activity normalized against β-galactosidase activity from cotransfected pCH110. The drawing under the graph shows the structures of the plasmids and a summary of results, “O” and “X” indicate an increase and little or no change of reporter activities, respectively. B) Schematic diagram of the cloning of conserved sites of the MOR 3′-UTR into the 5′-UTR plasmid. Asterisks indicate the names of plasmid constructs. The major conserved motifs of MOR 3′-UTR are indicated by ovals and described in the box. C) The three plasmids (5′-UTR-Ag, 5′-UTR-Bg, and 5′-UTR-Cg) as well as 5′-UTR were separately transfected with or without PCBP1 into NMB cells and analyzed for LUC activity (left graph), real-time RT-qPCR (LUC transcript level, middle graph), and real-time qPCR (LUC DNA amount, right graph). The results are given as LUC transcript and DNA amounts (primer set LUC-S1 and LUC-AS1) normalized against β-galactosidase transcript and DNA amounts (primer set Gal-S1 and Gal-AS1) from cotransfected plasmid. All data shown are the mean of three independent experiments with at least two different plasmid preparations. The error bars indicate the range of standard error. Asterisks indicate statistically significant findings. *P < 0.05 vs. control.
2.3. Transfection and luciferase reporter assay
Transfection of plasmid DNAs was performed with Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions. Single (Luciferase Assay System, Promega) and dual luciferase assays (Promega) were performed as described (Wu et al., 2013). For transfection in neuronal differentiated P19 cells, we used cells grown in suspension for transfection with plasmids or shRNAs using Lipofectamine 2000 carried out as described (Jalali et al., 2011; Wu et al., 2013). 48 h after transfection, cells were plated at a density of 6 × 106 cells per 10 cm tissue culture dish; 4 days after plating, the cells were harvested and processed for flow cytometry.
To assay RNA stability in vitro, NMB cells were plated at a density of 7× 105 cells per well in 6-well plates and transfected with the indicated DNAs (Fig. 3B), followed by treatment with Act-D (5 μg/ml) for 1 to 24 h. Analysis of mRNA decay profiles (with half-life) was performed as described previously (Wang et al., 2002; Wu et al., 2013). Relative mRNA levels of LUC and β-gal were measured by RT-PCR and real-time RT-quantitative PCR (RT-qPCR) with LUC and β-gal primers (Table 1). For the in vivo mRNA stability assay, P19 cells were plated at a density of 5× 105 cells per well in 12-well plates and transfected with the indicated DNAs (Fig. 3E), followed by treatment with TSA (10 nM) for 6 h for MOR stimulation as described previously (Hwang et al., 2007). After the MOR stimulation, cells were treated with Act-D (5 μg/ml) for 1 to 24 h. Non-treated cells were included as a control. Experiments were done in triplicate.
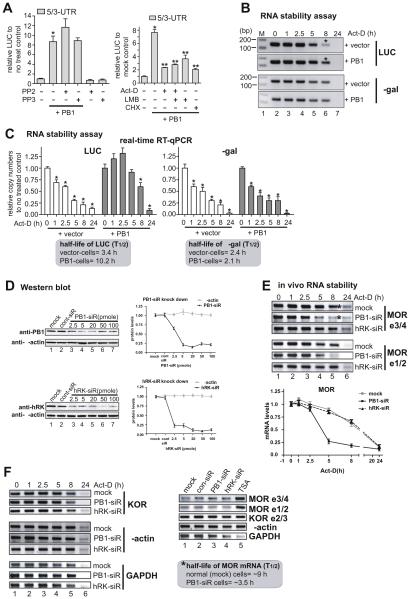
Fig. 3.
Induction of MOR mRNA stability by PCBP1. A) PCBP1-mediated upregulation was examined in MOR UTR constructs by LUC assay with an inhibitor of c-Src family kinases, PP2, along with its inactive control PP3 (left graph), and inhibitors of transcription, actinomycin-D (Act-D), translation cycloheximide (CHX), and nuclear export leptomycin-B (LMB) (right graph). *P < 0.05 vs. control; **P < 0.05 vs. non-treated PB1-transfectant. B) mRNA stability of LUC analyzed by RT-PCR. C) mRNA stability of LUC analyzed by real-time RT-qPCR. Primer sets LUC-S1 and LUC-AS1 were used for luciferase transcript and set Gal-S1 and Gal-AS1 for β-galactosidase transcript from the cotransfected internal control. Asterisks in B indicate the major bands that differ between vector control and PB1-transfectant. *P < 0.05 vs. control (C). D) Protein levels of PCBP1 (PB1, upper gel image) and hnRNPK (hRK, lower gel image). Indicated amounts of siRNAs of PB1 and hRK were transfected into P19 cells, and the protein levels of the cells were analyzed in Western blots using the respective antibodies. Anti-β-actin was used as a gelloading control. Quantitative analyses of the Western blot results are shown on the right side. E) In vivo mRNA stability of MOR is decreased in PCBP1-knockdown cells. In vivo mRNA stability of MOR was examined by RT-PCR with two sets of MOR-specific PCR primers: MOR e3/4 primers [mMOR_E3-S and mMOR_E4-AS, (Hwang et al., 2007)] covering MOR exons 3 and 4, and MOR e1/2 primers (mMOR-S7 and mMOR-AS7, Table 1) covering exon 1 and 2. Asterisks indicate the major band that differs between control siRNA- and PB1 siRNA-transfectant (PB1-siR) cells. hnRNPK siRNA (hRK-siR) was used as a control for the subfamily member of PCBP1. The lower panel shows the quantitative difference in the bands after averaging results obtained from RT-PCRs with two MOR primer sets (MOR e3/4 and MOR e1/2). F) Other genes were examined in the knockdown cells by RT-PCR. PCR primers for KOR (Kim et al., 2011), β-actin (Hwang et al., 2007), and GAPDH (Smirnov et al., 2001) were used as described previously. In panels E and F, MOR in P19 cells was stimulated by TSA as described previously (Hwang et al., 2007) before the RNA stability assay was done. siRNA did not change the expression levels of genes tested in the non-TSA-stimulated cells except for the GAPDH gene. The half-life (T½) of the MOR transcript is shown in the box in F. These results are representative of three different experiments.
2.4. RNA electrophoretic mobility shift assay (RNA EMSA)
RNA-protein interactions were performed as described previously with minor modifications (Bagga et al., 1998). For the RNA-protein interactions, 8 μg of cytoplasmic extract (CE) was incubated with 1 μl of 1 mM spermidine, 1 μl of heparin (50 mg/ml), 1 μl of 32P-labeled RNA probe (~ 200 kcpm), 0.5 μl of 25 mM EDTA, and 4 μl of Buffer D (20 mM Hepes-KOH, 20% glycerol, 100 mM KCl, 0.2 mM EDTA, 1 mM DTT) in a 10 μl reaction at 25 °C for 30 min. For antibody supershift experiments, antibodies and competitor RNAs were pre-incubated with the protein mixture before the addition of RNA probe. Gels were visualized on a phosphor screen with a Phosphor Imager (Storm 840; Molecular Dynamics).
In vitro translation was performed using the TnT T7 Quick Coupled Transcription/Translation System (Promega, #L1170) with 1 μg plasmid DNA, according to the manufacturer's instructions. For the protein kinase A reaction (PKA, NEB #P6000S), in vitro translated PCBP1 (PB1-IVT) was incubated with 25 U of PKA in PKA buffer at 30 °C for 1 h. The untreated reaction was performed under the same buffer conditions with no PKA. Cytoplasmic extracts were obtained as described (Dignam, 1990).
2.5. Real-time quantitative PCR (qPCR) and qRT-PCR
Total RNA was isolated with TRI reagent (Molecular Research Center) and treated with Turbo DNase I (2 U/μg RNA, Applied Biosystems). One-step RT-PCR was performed by using a OneStep RT-PCR Kit (Qiagen) with the primers described in Table 1 as well as β-actin (Hwang et al., 2007) and GAPDH (Smirnov et al., 2001) primers. PCR conditions were 32 cycles (for MOR) or 20 cycles (for β-actin and GAPDH) of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min followed by 72 °C for 10 min. PCR bands were quantified by ImageQuant 5.2 (Amersham) and also verified by DNA sequence analysis.
Real-time qPCR and qRT-PCR was performed as described previously (Hwang et al., 2007; Wu et al., 2013) with a Quantitect SYBR Green RT-PCR Kit (Qiagen) and DNA isolated with TRI reagent. For real-time RT-qPCR, total RNA was treated with DNase I as above. The relative expression levels were analyzed as described previously (Hwang et al., 2007; Wu et al., 2013). The RT-PCR and real-time RT-qPCR experiments were repeated at least three times for statistical significance.
2.6. Co-immunoprecipitation and RNA immunoprecipitation (RIP)
The procedure for co-immunoprecipitation was performed as previously described with some modifications (Hwang et al., 2009). Whole cell lysates were isolated from NMB cells with or without forskolin treatment and subjected to immunoprecipitation and Western blot analysis with the following antibodies: anti-PCBP1 (T-18, sc-16504), anti-HuR (3A2, sc-5261), and anti-PABP (F-2, sc-166027) from Santa Cruz; antiAUF1 (GTX101814) from GeneTex; anti-pan-phosphorylated protein (61-8300) from Invitrogen; and anti-phospho-serine (p-Ser, 05-1000) and anti-phospho-tyrosine (p-Tyr, 06-427) from Millipore.
RNA immunoprecipitation (RIP) was performed as described previously (Gilbert and Svejstrup, 2006; Hwang et al., 2011) with some modifications. To have enough endogenous mouse MOR expression, differentiated neuronal mouse P19 cells were used. The neuronal differentiation of the cells was performed as described previously (Hwang et al., 2007). On the 4th day after plating (AP4d) when MOR expression level reaches a plateau, cells were treated with forskolin or inhibitors; the crosslinking RNA-protein complexes were then formed with 1% formaldehyde at room temperature for 20 min. The cross-linking reaction was quenched by the addition of 0.2 M glycine. After the cell lysates were sonicated for 5 cycles of 10 s each at 40% amplitude with a sonicator (XL-2020, Heat Systems), 60 U of RNase-free DNAase I (Roche) was used to remove DNA. Routine ChIP procedures were performed as described (Gilbert and Svejstrup, 2006; Hwang et al., 2011) with the following antibodies: anti-PCBP1, anti-PABP, anti-AUF1, hnRNPK (D-6, sc-28380, Santa Cruz), non-specific antibody anti-gal4 (sc-577, Santa Cruz), and normal rabbit serum (PI) as a control. The immunoprecipitated RNA was analyzed by RT-PCR and real-time RT-qPCR with the respective PCR primer sets (Table 1, Figs. 2AB, 4C, 6B, and 7C).
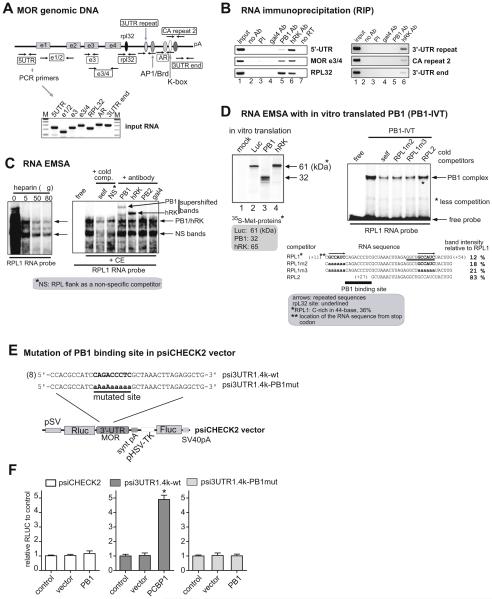
Fig. 2.
PCBP1 specifically binds to a novel site in MOR 3′-UTR. A) PCR primer locations in MOR genomic DNA used for RNA immunoprecipitation (RIP). Primer set names and locations are shown in rectangles. Sequences of primers used in RIP are listed in Table 1. The gel image shows that PCR products from input RNA isolated from mouse MOR-positive cells AP4d (Hwang et al., 2007) were the expected sizes. B) Data from RIP show the interaction of PCBP1 on the rpL32 site of MOR 3′-UTR. Details of the relative RNA-binding affinities of PCBP1 and hnRNPK (hRK) are presented in Table 2. PI is pre-immune serum. C) Specific activity of the binding of PCBP1 to the C-rich element (CRE) of the 44-base RPL1 RNA segment. The left panel shows RNA EMSA performed with radiolabeled RPL1 RNA probe in the presence of P19 cytoplasmic extracts (CE, 8 μg) and the indicated amounts of heparin. The sequence-specific complexes are indicated by the arrows. In the right panel, a supershift EMSA assay using antibodies indicates that PCBP1 binds to the RPL1 RNA probe. The sequence of RPLflank-S, used as a nonspecific RNA competitor (Table 1), abolished the lower complex on the image indicated as NS (non-specific) bands. These results are representative of three different experiments. D) The left panel shows that radioisotope-labeled proteins were produced by in vitro translation in the presence of 35S-methionine. Proteins were analyzed on a 12% SDS-PAGE gel. Protein size in kiloDalton (kDa) is indicated by the arrow. The right panel shows RNA EMSA performed with in vitro translated PCBP1 (PB1-IVT). RNA sequences of the RNA probe and competitors are shown below the gel image. The identified PCBP1 binding motif is indicated by a solid bar at the region of + 17 ~ + 26. E) Construction of a mutated binding site for PCBP1. A 1.4-kb 3′-UTR fragment was inserted into psiCHECK2 vector (Promega), and the resulting plasmid was designated psi3UTR1.4k-wt. The bold sequence in psi3UTR1.4k-wt was mutated as indicated in the underlined sequence, and the resulting plasmid was designated as psi3UTR1.4k-PB1mut. The number in parentheses indicates the distance from the stop codon; the insert begins at +1. F) The three plasmids were cotransfected with PCBP1, vector, and control into NMB cells, and the cells analyzed for their relative firefly luciferase (Fluc) activity normalized against Renilla luciferase (Rluc) activity. *P < 0.05 vs. control.
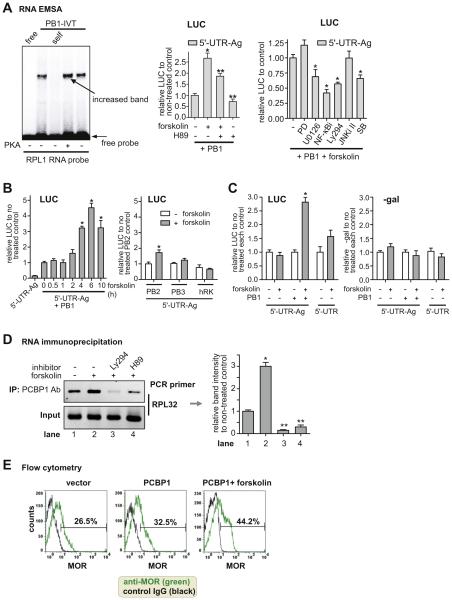
Fig. 4.
Forskolin, an activator of protein kinase A (PKA), enhances PCBP1-mediated MOR upregulation. A) RNA EMSA shows that PKA enhances the interaction of PCBP1 with the RPL1 RNA probe. PB1-IVT was treated with PKA (25 U) as indicated. Free probe (free) without the protein and self competitor (self, 10-fold excess) were included as controls. The middle graph of LUC reporter analysis shows that forskolin stimulates PCBP1-mediated upregulation of the reporter-containing RPL site of MOR 3′-UTR (5′-UTR-Ag). Cells were treated with forskolin (10 μM) for 4 h and were pretreated with PKA inhibitor H89 as indicated for 30 min prior to forskolin treatment. In the right graph, other inhibitors were used for co-treatment: PD98059 (PD, MEK1 inhibitor), U0126 (MEK1/2 inhibitor,) NF-κBi (NF-κB inhibitor), Ly294002 (Ly294, PI3-kinase inhibitor), JNKi II (SP600125, JNK inhibitor II), and SB203580 (SB, MAPK kinase p38 inhibitor). *P < 0.05 vs. control; **P < 0.05 vs. forskolin-treated PB1-transfectant. B) Luciferase reporter analysis. In the left graph, plasmid DNA 5′-UTR-Ag was transfected with and without PCBP1 into NMB cells. The time of forskolin treatment is indicated in hours (h). Other RNA-binding factors, PCBP2 (PB2), PCBP3 (PB3), and hnRNPK (hRK), were cotransfected with 5′-UTR-Ag as shown in the right graph. LUC reporter activities are relative to that of PB2-non-treated control. C) In the left graph, 5′-UTR-Ag or 5′-UTR was cotransfected with PB1 (+) or vector (−) as indicated. Forskolin treatment and no treatment are indicated (+ and −). As a control experiment, pCH110 was cotransfected with all samples, and β-galactosidase (β-gal) activity was measured as shown in the right graph. *P < 0.05 vs. non-treated control. D) RNA immunoprecipitation was performed as in Fig. 2B. Cells were pretreated with inhibitors Ly294 and H89 as indicated for 30 min prior to forskolin treatment. RIP PCR primers (Rch-S1 and Rch-AS1) were used to detect the rpL32 site. The right graph shows the band intensities relative to non-treated control (lane 1) from the gel images on the left. *P < 0.05 vs. control; **P < 0.05 vs. forskolin-treated sample (lane 2). E) Flow cytometric data shows that MOR expression was increased by overexpression of PCBP1 (from 26.5% to 32.5%) and further enhanced by forskolin treatment (from 32.5% to 44.2%). The percentage of MOR expression (green) was relative to the base level obtained from control IgG (black). The result is representative of three different experiments.
Fig. 6.
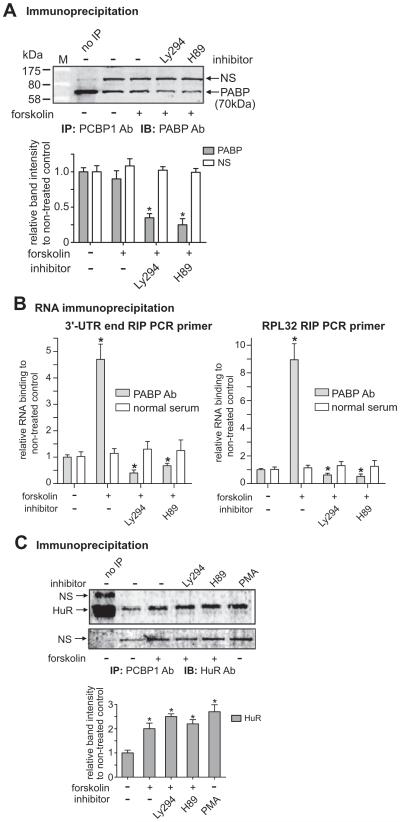
Constitutive interaction of PCBP1 with poly(A)-binding protein (PABP) was decreased by co-treatments with H89 as well as Ly294. A) Antibodies of PCBP1 and PABP were used for IP and IB, respectively. The graph shows the band intensities of PABP relative to the NS band. The same sized band was observed in IPs with other antibodies, AUF, PCBP1, and HuR, which suggests that the band represents ribonucleoproteins (RNPs) complex. The steady interaction of PCBP1 with PABP is reduced by co-treatments with Ly294 and H89. M is the Protein MW marker (Bio-Rad, P7708). B) The RIP was performed as in Fig. 2B except with PABP Ab. Two RIP PCR primer sets, 3′-UTR end (left graph) and RPL32 (right graph), were used for the RIPs as indicated. Normal goat serum (normal serum) was used for each control RIP. *P < 0.05 vs. non-treated control. C) The interaction of PCBP1 with HuR, known to be an mRNA stabilizing factor, is increased by forskolin treatment, and the interaction is not changed by co-treatments with Ly294 and H89. Antibodies of PCBP1 and HuR were used with protein extracts of NMB cells for IP and IB, respectively. The graph shows the relative band intensities of HuR compared to that of the non-treated control after normalizing to the respective NS bands (shown in the lower gel image). Cells were co-treated with forskolin and Ly294 or H89 as indicated. *P < 0.05 vs. non-treated control.
Fig. 7.
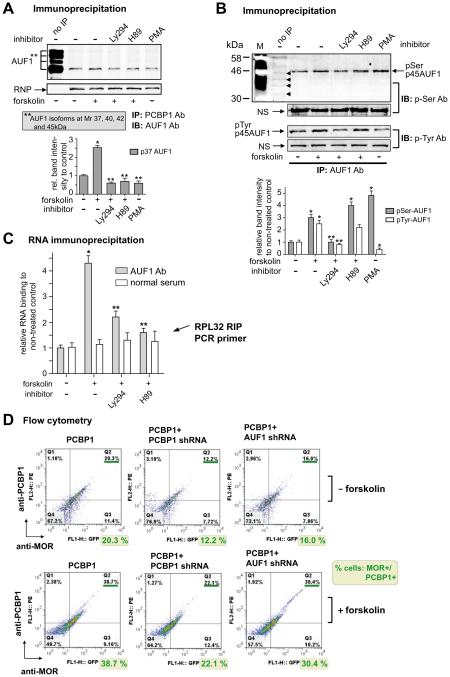
AUF1 is involved in PCBP1-mediated MOR induced by forskolin. A) Interaction of PCBP1 with AUF1, known as mRNA destabilizing factor, is increased by forskolin as analyzed by IP. Antibodies to PCBP1 and AUF1 were used with protein extracts of NMB cells for IP and IB, respectively. The graph shows the band intensities of the AUF1-p37 isoform from the treated and non-treated control after normalizing to their NS bands. Ly294 and H89 were co-treated with forskolin as indicated. *P < 0.05 vs. non-treated control; **P < 0.05 vs. forskolin-treated sample. B) AUF1 antibody was used for IP and antibodies to phospho-serine (p-Ser Ab, upper two gel images) and phospho-tyrosine (p-Tyr Ab, lower two images) for IB. Four arrowheads in the upper image indicate the basal level of serine phosphorylation of the potential AUF1 isoforms (p45, p42, p40, and p37). The white bands in lane M were caused by pencil marks on the PVDF membrane. M is the protein MW marker (Bio-Rad). C) The RIP was performed as in Fig. 6B except with AUF1 Ab. RIP PCR primer set RPL32 was used for the RIPs as indicated. *P < 0.05 vs. non-treated control; **P < 0.05 vs. forskolin-treated sample. D) Flow cytometric data plots displayed in four quadrants to show cell populations with or without expressions of PCBP1 and MOR proteins. The transfected plasmid DNAs are indicated above each plots. The cells in the lower three plots were treated with forskolin while those in the upper three plots were not. Percentages of PCBP1 + and MOR+ cells (from upper right quadrant) were rewritten below each plot. The result is representative of three different experiments.
2.7. Immunocytochemistry and flow cytometry
To detect changes in the subcellular location of PCBP1, we performed immunocytochemistry as described with minor modifications (Wu et al., 2013). Cells were plated at a density of 1 × 105 cells per well in 4-well chamber slides (Nalge Nunc) and 24 h later were transfected with PCBP1 and its mutants using Effectene (Qiagen). One day later, cells were treated with forskolin (10 μM) for 6 h, fixed with 4% paraformaldehyde in PBS for 10 min at room temperature, and permeabilized with 0.1% Triton X-100 for 10 min at room temperature. PCBP1 was visualized using mouse anti-myc antibody (2276, Cell Signaling), followed by incubation with secondary goat anti-mouse antibody coupled to Alexa-488 (Molecular Probes). Propidium iodide (PI, Sigma P4170) was used to stain nuclei. Cell images were analyzed on image analysis software MetaMorph® (Molecular Devices).
Flow cytometry was performed as described previously (Wu et al., 2013) to determine changes in MOR protein level as a result of PCBP1 and forskolin treatment of MOR-positive cells (neuronal differentiated/transfected P19 cells). Cells were harvested and approximately 5 × 105 cells were used for flow cytometric analyses. Flow cytometric analyses were performed on a FACScalibur (BD Biosciences) equipped with two lasers for excitation at 488 nm and 635 nm. At least 10,000 cells were analyzed in each sample. Data were collected and analyzed with FlowJo software (Tree Star).
3. Results
3.1. Upregulation of the MOR gene by RNA-binding protein PCBP1 at the post-transcriptional level
In an effort to identify a factor(s) that increases MOR expression at the post-transcriptional level, we studied DNA sequences near or downstream of the MOR stop codon, where, in general, significant RNA elements are located and involved in post-transcriptional regulation. We found a C-rich region of the 3′-UTR (a 44-base sequence) downstream (+ 11 ~ + 54 bases) of the MOR stop codon, which may bind to poly(C) binding factors (also called α-CPs), such as hnRNPK and PCBP1 ~ 4 (Kong et al., 2003). We cotransfected these factors with luciferase(LUC) reporter plasmids containing MOR UTRs and analyzed the cells with LUC reporter assays. Among the factors studied, we found that PCBP1 specifically enhanced LUC reporter activity through the MOR 3′-UTR (Fig. 1A), suggesting that the 3′-UTR of the MOR gene is associated with its own regulation at the post-transcriptional level. The results in Fig. 1A show that overexpression of PCBP1 increased the reporter activities in the 5/3-UTR and 3′-UTR constructs, but not in the 5′-UTR construct, indicating the effect of PCBP1 depends on the presence or absence of the 5′- and 3′-UTR regions of the MOR gene.
To determine if the C-rich region or other sequence of the 3′-UTR region is responsible for PCBP1-mediated upregulation, we constructed plasmid 5′-UTR-Ag containing the C-rich region (rpL32 site) as well as two additional plasmids 5′-UTR-Bg and 5′-UTR-Cg from the 5′-UTR as shown in Fig. 1B. Plasmid 5′-UTR-Ag contains the 44-base C-rich sequence, which includes the sequence 5′GCUGCCAUC-3′ (underlined sequence in Fig. 2D) that is a binding motif for a nuclear factor for the rpL32 (ribosomal protein L32) gene (Atchison et al., 1989; Moura-Neto et al., 1989), as analyzed by a web-based software, RegRNA (http://regrna.mbc.nctu.edu.tw) (Huang et al., 2006), and designated the RNA region to rpL32. Ribosomal protein L32, a component of the large 60S subunit in eukaryotic ribosome (80S), is localized in the cytoplasm and plays an important role in protein synthesis (Dudov and Perry, 1984). The exact function and identity of the nuclear factor for rpL32 is still not known. Other conserved motifs are listed in the box in Fig. 1B.
In the LUC reporter assays shown in Fig. 1C left graph, overexpression of PCBP1 increased the LUC reporter activity of 5′-UTR-Ag approximately 8-fold as compared to that of the control; whereas overexpression of PCBP1 in the other constructs (5′-UTR, 5′-UTR-Bg, and 5′-UTR-Cg) increased the reporter activities in the range of 3-fold. This indicates that the increased basal reporter activity by PCBP1 may be due to a 5′-UTR sequence that contains an internal ribosome entry segment (IRES) site. The IRES in another gene (Bag-1) has been reported to bind to PCBP1, resulting in an increase in the activity of the gene in vitro and in vivo (Pickering et al., 2003; Sawicka et al., 2008).
To determine whether PCBP1-mediated upregulation occurs at the transcriptional or post-transcriptional level, two real-time PCR reactions (RT-qPCR and qPCR) were performed as shown in Fig. 1C middle and right graphs, respectively. The middle graph shows that increased transcription (approximately 3-fold) by PCBP1 was observed only with 5′-UTR-Ag but not with the other plasmids, suggesting a possible interaction between PCBP1 and an RNA sequence in 5′-UTR-Ag. The right graph indicates that similar amounts of DNA were transfected (for simplicity we show data from only two samples).
3.2. Identification of the binding site of PCBP1 in the 3′-UTR of MOR mRNA
Although the approximate location of the binding site of PCBP1 on the 3′-UTR was determined from the LUC reporter assays shown in Fig. 1, we performed RNA immunoprecipitation (RIP) to further define the site. RIP is able to detect protein binding to RNA in vivo as well as indicate the approximate location of the binding site. Fig. 2A shows the locations and expected sizes for PCR primer sets for the immunoprecipitated RNA. As shown in Fig. 2B, PCBP1 binding to MOR mRNA appears localized specifically to the rpL32 site of the 3′-UTR; whereas hnRNPK appears to bind non-specifically to different extents to several regions, such as 5′-UTR, MOR e3/4 (exon 3 and 4 region), rpL32 site, and 3′-UTR repeat sites (Table 2). Since hnRNPK belongs to the same subfamily as poly(C)-binding proteins, such as PCBP1, it was used as a non-specific positive control for this and the experiments described below.
Table 2.
Relative RNA-binding affinity by RNA-binding factors in RNA immunoprecipitation.
| Factor | Region in MOR mRNA | Relative RNA-binding affinitya |
|---|---|---|
| PCBP1 (PB1) | 5'-UTR | − |
| E3/4 | ++ | |
| RPL32 | +++ | |
| 3′-UTR repeat | − | |
| CA repeat 2 | − | |
| 3′-UTR end | − | |
| hnRNPK (hRK) | 5'-UTR | ++++ |
| E3/4 | ++ | |
| RPL32 | +++ | |
| 3′-UTR repeat | ++ | |
| CA repeat 2 | + | |
| 3′-UTR end | +/− |
Relative binding affinity was measured by setting the highest (++++) level with hnRNPK's binding in 5′-UTR region and the lowest level (−) with gal4's binding in 5′-UTR region.
To further define the binding site of PCBP1, RNA EMSA was performed with cytoplasmic extracts (CE) of P19 cells, which express endogenous PCBP1 at a high level. In the first RNA EMSA, the addition of CE resulted in the appearance of multiple retarded bands, which most likely correspond to multiple protein molecules binding nonspecifically to one molecule of RNA (Fig. 2C left panel, left lane). This reduced the clarity of the assay. Similar observations were made with several of the other probes (data not shown). Therefore, we tried adding heparin to the RNA EMSA to reduce this effect. Heparin, a highly sulfated glycosaminoglycan that mimics the high negative charge of nucleic acids, has been used as a DNA and RNA nonspecific competitor (Lozinski et al., 2009). As shown in Fig. 2C, the addition of increasing amounts (5–80 μg/reaction) of heparin to EMSA with RPL1 RNA as a probe reduced the background signal on the gel. This was presumably due to a decrease in the non-specific binding of protein and resulted in the appearance of two major, more highly focused bands (arrows) with improved intensity. As a result, we used 50 μg of heparin per reaction in further experiments.
To define exactly which RNA sequences of the 44-base RPL1 are essential for PCBP1 binding, three additional RNA oligonucleotides were synthesized as indicated in Fig. 2D: RPL1m2, RPL1m3, and RPL2. The RPL1m2 RNA fragment contains a mutation in the first of the two repeated RNA sequences (arrows above sequences) where RNA-binding proteins are frequently shown to bind. The RPL1m3 RNA fragment has mutations in both of the repeats, the second of which overlaps the rpL32 site (underlined sequence). The RPL2 RNA fragment has a deletion from + 11 to + 27 where more C bases are located among 44-base sequences of the RPL1.
These RNA oligonucleotides were then tested for their ability to compete with the RPL1 element in the formation of RNA-protein complexes. In vitro translated PCBP1 (PB1-IVT) was used as a source of protein in RNA EMSA to avoid possible interference from other subfamily members of the poly(C)-binding proteins, such as hnRNPK (hRK) and PCBP2, 3, and 4. The RNA-protein complexes were effectively abolished by a 2.5-fold excess of competitors RPL1m2 and RPL1m3 (see also their relative band intensities compared to RPL1 as shown below the gel image), but not by RPL2 (Fig. 2D). These data suggest that the single-stranded RNA sequence shared by RPL1, RPL1m2, and RPL1m3, but not RPL2 is probably important for the binding of PCBP1 to the RPL1 cis-acting element. Examination of this sequence revealed a putative PCBP1-binding site, 5′CAGACCCUC-3′, at the region of + 17 ~ + 26, marked by the solid bar in Fig. 2D. Furthermore, the sequence of the RPL1 probe contains a high percentage (36%) of cytosines and thus may be considered a new type of CRE. Fig. 2D shows in vitro translation products (35S-Met labeled) of expected sizes on an SDS-PAGE gel.
To confirm the importance of this PCBP binding motif in the regulation of MOR 3′-UTR reporter activity, two plasmids were constructed in psiCHECK2 vector, psi3UTR1.4k-wt and psi3UTR1.4k-PB1mut. Construct psi3UTR1.4k-wt contains a 1.4-kb fragment of the 3′-UTR (from the stop codon to 1.4-kb downstream). In construct psi3UTR1.4k-PB1mut, the PCBP1 binding motif in the wild-type construct psi3UTR1.4k-wt is mutated. As shown in Fig. 2E and F, when compared with the wild-type construct and control vector (psiCHECK2), the mutation of the PCBP1 binding motif (psi3UTR1.4k-PB1mut) resulted in the loss of the enhancement in luciferase activity seen with PCBP1. This suggests that the PCBP1 binding motif we identified is important for the regulation of MOR 3′-UTR reporter activity.
3.3. PCBP1 increases mRNA stability of MOR
It has been reported that PCBP1 (as well as hnRNPK) binds to the differentiation-control element (DICE) of the 3′-UTR of reticulocyte-15-lipoxygenase mRNA (r15-LOX) and silences the translation of LOX mRNA by destabilizing its mRNA (Reimann et al., 2002; Ostareck-Lederer and Ostareck, 2004; Naarmann et al., 2008). We found four putative DICE sites in the MOR 3′-UTR (+ 3251, + 3473, + 6755, and + 9623) and attempted to determine the effect of PCBP1 on these sites by overexpressing PCBP1 with MOR UTR constructs followed by analysis in a luciferase assay, as shown in Fig. 3A. Since the interaction of PCBP1 (as well as hnRNPK) with the LOX DICE is regulated by c-Src kinase (Naarmann et al., 2008), we used the c-Src kinase inhibitor PP2. The upregulation by PCBP1 was not blocked by PP2; in fact, it increased slightly although not significantly. PP3, the inactive control of the inhibitor PP2, did not affect the upregulation (Fig. 3A), suggesting no involvement of a DICE-mediated mechanism or c-Src kinase.
We then performed a luciferase reporter assay with inhibitors of transcription or translation to determine which step was involved in the PCBP1-mediated upregulation (Fig. 3A, right graph). Both inhibitors, Act-D (inhibitor of transcription) and CHX (inhibitor of translation), efficiently abolished the PCBP1-mediated upregulation seen in the PCBP1-transfected control. Interestingly, an inhibitor of nuclear export, leptomycin B (LMB), also decreased the upregulation. LMB is known to inhibit export of many RNAs, including c-Fos and Cox-2 mRNAs, by inhibiting the export of ribonucleoproteins (RNPs, a group that includes PCBP1), resulting in translational inhibition in cytoplasm (Brennan et al., 2000; Jang et al., 2003). The results of our assays suggest that PCBP1 regulates MOR expression by regulating MOR mRNA transport from nucleus to cytoplasm at the post-transcriptional level.
Therefore, we decided to analyze mRNA stability that occurs in cytoplasm, as shown in Fig. 3B and C. MOR-positive NMB cells were cotransfected with the MOR UTR construct (5/3-UTR) and either vector or PCBP1 plasmid. As an internal control, β-gal expression plasmid was also included in the transfection. The cells were used for both RT-PCR and real-time PCR. As shown in Fig. 3B (RT-PCR) and 3C (real-time PCR), reporter LUC mRNA stability was enhanced by overexpression of PCBP1 compared to that of the control vector transfection. The mRNA stability of the control β-gal was unchanged.
To examine mRNA stability in vivo, we performed siRNA-mediated knockdown of genes in P19 cells as shown in Fig. 3D–F. Since our target MOR gene is from mouse, mouse P19 cells were used. Fig. 3D shows the optimization of the concentrations of each siRNA required to effectively silence the target genes (PCBP1 and hnRNPK). We used a concentration of 20 pmol of siRNA for experiments with the PCBP1 gene. In the in vivo mRNA stability assay, the stability of MOR mRNA was drastically reduced in PCBP1-knockdown cells while control and hnRNPK-knockdown cells showed no change in mRNA stability (Fig. 3E, asterisks used for comparison). The half-life of MOR mRNA under normal conditions (control) is approximately 9 h; the half-life was reduced to approximately 3.5 h in the absence of PCBP1. Fig. 3F shows no change in other genes (KOR, β-actin, and GAPDH) in siRNA-knockdown cells, and Fig. 3F shows that siRNAs did not change the expression levels of the genes tested in un-stimulated cells except for changes in GAPDH mRNA level by siRNAs. These results suggest that PCBP1 upregulates the MOR gene via stabilization of target MOR mRNA.
3.4. Forskolin enhances PCBP1 activity on MOR expression
We screened several enzymes for their effect on PCBP1-RNA binding and found that treatment of the in vitro translated PCBP1 protein with protein kinase A (PKA) led to increased binding to the RPL1 RNA probe, shown in Fig. 4A. Forskolin has been used as an activator of PKA in many studies (Chen et al., 1998). To further confirm this enhancement of PCBP1 activity on MOR, transient transfection experiments with PCBP1 were performed with or without forskolin treatment and signaling modulators (several inhibitors), as shown in Fig. 4B. The data from the in vitro experiment (Fig. 4B, left panel) show that LUC reporter activity was increased (~2.6-fold) by forskolin. This increase was reduced to approximately 1.8-fold by PKA inhibitor H89, indicating involvement of PKA in the PCBP1-mediated enhancement. As shown in Fig. 4B right panel, co-treatment of forskolin and PI3-kinase inhibitor Ly294002 (Ly294) led to a decrease in the reporter activity to ~ 60% compared to that of the forskolin-treated control, suggesting an association between PI3-kinase pathway and PCBP1-mediated enhancement. Co-treatment with NF-κB inhibitor (NF-κBi) also reduced the activity to ~ 40%; other inhibitors showing some degree of reduction were U0126 and SB203580 (SB). The p42/44 MAPK inhibitor PD98059 (PD) and JNK inhibitor II (JNKi II) did not affect the enhancement. Fig. 4B (left graph) shows optimization of the time of treatment with forskolin (10 μM); we treated for 4 h in further experiments. Forskolin treatment with PCBP2, PCBP3, and hnRNPK showed little or no change in LUC reporter activity (Fig. 4B, right graph). The data from the LUC assay in Fig. 4C show that LUC reporter activity was increased by forskolin in the presence of PCBP1 and 5′-UTR-Ag, whereas β-gal activity was not affected.
We used RNA immunoprecipitation to determine if forskolin treatment and/or inhibitors can affect the endogenous binding of PCBP1 with MOR 3′-UTR. As shown in Fig. 4C, when MOR-positive cells (neuronal differentiated P19 cells) were treated with forskolin, PCBP1 binding on the rpL32 site was increased approximately 3-fold (lane 2) as compared to non-treated control (lane 1). The increased PCBP1 binding was decreased to 30% with H89 pretreatment (lane 4) and 10% with Ly294 pretreatment (lane 3), thus confirming the increase of endogenous PCBP1 binding to MOR 3′-UTR induced by forskolin and the involvement of the PI3-kinase pathway and PKA. In addition, in vitro RNA EMSA showed that pretreatment with Ly294 and/or CHX led to a decrease in the binding of the cytoplasmic protein to the rpL32 site compared to non-treated control (data not shown). This indicates the involvement of both the PI3-kinase pathway and protein synthesis in the binding of PCBP1 with RNA.
3.5. Endogenous MOR expression is increased by PCBP1 and further enhanced by forskolin co-treatment
To examine the effect of the enhanced binding of PCBP1 to MOR 3′-UTR, we attempted to determine the direct effect of PCBP1 on endogenous MOR. From flow cytometry with MOR antibody, as shown in Fig. 4D, MOR expression was shown to increase in PCBP1-transfected cells (32.5%, middle plot) compared to vector-transfected cells (26.5%, left plot). The PCBP1-mediated enhancement of MOR expression was further increased by forskolin co-treatment (44.2%, right plot). The percentage of MOR expression in each plot was calculated based on the base level of the control IgG (black).
3.6. Forskolin induces phosphorylation of PCBP1 at the serine residues
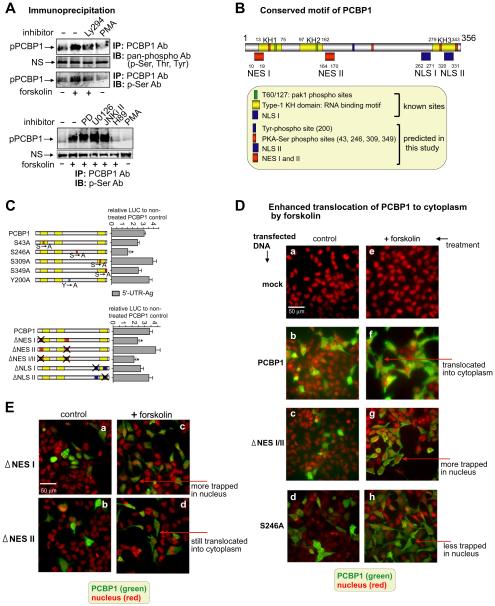
To determine which amino residues of PCBP1 are phosphorylated by forskolin, we performed immunoprecipitation using phospho-specific antibodies (Fig. 5A). Pan-phospho antibody was used as an immunoblotting antibody (IB Ab) to detect phosphorylation in the sample that was immunoprecipitated by PCBP1 antibody (IP Ab). The phosphorylated band (pPCBP1) increased in samples with forskolin treatment but was reduced in samples co-treated with Ly294 (Fig. 5A top gel). The third gel of Fig. 5A shows that use of phospho-serine antibody (p-Ser Ab) resulted in a similar pattern, indicating that serine residue(s) of PCBP1 can be phosphorylated by forskolin. Antibodies to p-Tyr or p-Thr showed no changes in the phosphorylation of PCBP1 (data not shown). PMA, an activator of protein kinase C (PKC), was used as a control since it has been reported that an increase in PKC activity by PMA resulted in a decrease in the MOR mRNA level (Gies et al., 1997). In experiments with both pan-phospho and p-Ser antibodies, PMA treatment caused some decrease in the basal phosphorylation of PCBP1 compared to that of the non-treated control, suggesting that a further decrease in basal phosphorylation of PCBP1 may be associated with the reduction of MOR mRNA level. This awaits further investigation. The fourth gel image in Fig. 5A shows that co-treatment with forskolin and H89, a PKA inhibitor, also caused a decrease in Ser phosphorylation of PCBP1.
Fig. 5.
Forskolin treatment enhances serine phosphorylation of PCBP1 as analyzed by immunoprecipitation. A) The upper set of gel images shows the enhanced phosphorylation of PCBP1 with forskolin treatment by immunoprecipitation (IP) with antibody to PCBP1 and immunoblotting (IB) with antibody to pan-phospho. The second gel image shows that the nonspecific (NS) band used as the control is unchanged. The third gel image shows the increase in serine phosphorylation of PCBP1 with forskolin treatment detected with p-Ser antibody. Ly294 blocks the phosphorylation, and the basal level of phosphorylation was slightly decreased by treatment with only phorbol-12-myristate-13-acetate (PMA), the activator of PKC. The lower set of gel images show that an inhibitor of PKA, H89, blocks the serine phosphorylation of PCBP1 induced by forskolin although other inhibitors do not block the phosphorylation. PMA alone, used as a control, did not show any change in PCBP1 phosphorylation. B) Schematic representation of PCBP1. Numbers near each motif's symbol indicate the locations of conserved motifs, details of which are described in the box. C) Analysis of mutations of PCBP1 in 5′-UTR-Ag containing the rpL32 site. As seen in the upper panel, potential phosphorylation sites of PCBP1 were mutated as indicated and analyzed for their LUC reporter activities compared to wild-type PCBP1-transfectant. “S → A” and “Y → A” on the phosphorylation sites indicate the locations of amino acids mutated to alanine residues. In the lower drawing, either nuclear export signals (NES) or nuclear localization signals (NLS) sites were mutated as indicated and their relative reporter activities were analyzed as above. All samples were treated with forskolin. X indicates mutated sites. D) Cytoplasmic translocation of PCBP1 was enhanced by forskolin. Plasmid DNA from PCBP1, ΔNES I/II, and S246A was transfected into NMB cells and treated with forskolin. Myc antibody was used to detect the subcellular location of overexpressed myc-tagged PCBP1 and its mutants in green. Propidium iodide (PI) was used to stain the nuclei in red. In fluorescent image a, the scale bar represents 50 μm. E) Fluorescent images were obtained as described in the above Fig. 5D, except using two PCBP1 mutant plasmids (ΔNES I and ΔNES II).
It has been reported that mitogen-stimulated phosphorylation of threonines (Thr) 60 and 127 of PCBP1 in a Pak1 (p21-activated kinase 1)-sensitive manner reduced its binding, resulting in an increase in its translational inhibition through a DICE of 3′-UTR (Meng et al., 2007). From our data of Fig. 5A, it seems that forskolin did not phosphorylate Thr residues of PCBP1; instead it phosphorylated Ser residues and functioned through its unique target site (rpL32) rather than through DICE of 3′-UTR. Other co-treatments with forskolin and PD, U0126, and JNKi II, further increased the serine phosphorylation of PCBP1 seen with forskolin treatment alone, which provides more complicated data for interpreting the role of the enhanced serine phosphorylation when taking the functional results in Fig. 4B into account.
3.7. Finding a novel PCBP1 site phosphorylated by forskolin
To define the target site(s) in the amino acids sequence of PCBP1 phosphorylation by forskolin, we first analyzed the conserved motifs in PCBP1 by using web-based software: PROSITE (Sigrist et al., 2002; Sigrist et al., 2013), KinasePhos (Huang et al., 2005), and GPS (Xue et al., 2005). For subcellular localization, NetNES 1.1 (la Cour et al., 2004) and WoLF PSORT (Horton et al., 2007) were used to predict the putative NES motif and NLS, respectively. Fig. 5B isa schematic drawing of the conserved sites in PCBP1 as analyzed by these programs. Conserved sites published previously, Thr60/127 (Meng et al., 2007), type-1 KH domain (Makeyev and Liebhaber, 2002), and NLS I (Chaudhury et al., 2010), are shown as known sites in the box under the schematic drawing. Conserved sites predicted from this study are also drawn.
After determining the potential phosphorylated sites in PCBP1, we mutated serine residues as well as the tyrosine residue at amino acid 200 in the conserved PKA-phosphorylated sites and analyzed for activity in the LUC reporter assay (Fig. 5C). Forskolin-mediated enhancement of PCBP1 on the rpL32 site (5′-UTR-Ag) was reduced in S246A-mutated PCBP1 by 50% compared to that of the control (wild-type PCBP1 transfectant treated with forskolin). Transfections of S43A- and S349A-mutated PCBP1s showed minor reductions in assay activity to 76% and 77% of the control, respectively; and the two other constructs, S309A and Y200A, showed some increases, 124% and 116%, respectively. These functional analyses suggest that serine 246 residue in PCBP1 plays an important role in the forskolin-mediated enhancement, possibly through phosphorylation of the serine residue by PKA.
3.8. Cytoplasmic translocation of PCBP1 by forskolin
We have shown (Fig. 3A) that LMB, an inhibitor of nuclear export, causes a decrease in PCBP1-mediated upregulation of the MOR reporter construct. Therefore, we attempted to determine if forskolin treatment leads to a change in the subcellular location of PCBP1 from the nucleus where it is mainly expressed. Immunocytochemistry data show that in NMB cells transfected with PCBP1 (Fig. 5D, panel b) the majority of PCBP1 is located in the nucleus with a minor amount in cytoplasm. Panel f indicates that in the same PCBP1 transfectant treated with forskolin the PCBP1 is translocated to the cytoplasmic regions. The translocation of PCBP1 on treatment by forskolin was abolished when the NES I/II sites were mutated (panels c and g). In the functional assay shown in Fig. 5C bottom graph, mutations in NES I and NES I/II led to a decrease in the forskolin-mediated enhancement of PCBP1 by 70.6% and 63%, respectively, whereas a mutation in NES II alone led to only a minor change. This is consistent with our observation shown in Fig. 5D that when the NES I mutant is treated with forskolin (panel c) more PCBP1 is trapped in the nucleus, whereas, the NES II mutant still shows a translocation of PCBP1 to cytoplasm with only a minor nuclear amount remaining (panel d). The NLS I and NLS II mutants did not display any major changes in the reporter assay. Serine residue 246 does not seem necessary for the translocation of PCBP1 since forskolin treatment of the S246A mutant (Fig. 5D, panel h) shows less PCBP1 trapped in the nucleus as compared to the NES I/II mutant (panel g). However, that serine residue is still functionally important because, as noted above, a mutation at that site led to decreased enhancement of PCBP1-mediated luciferase activity (Fig. 5C). All these results suggest that forskolin induces translocation of PCBP1 to cytoplasm through the NES I motif of PCBP1.
3.9. Involvement of other RNA-binding factors on PCBP1 function
It is known that PCBP1 binds to poly(A)-binding protein PABP, a protein that binds to the poly(A) tail of mRNA to prevent endoribonuclease (ErEN) cleavage by binding to the CRE in the α-globin 3′-UTR for mRNA stabilization (Wang and Kiledjian, 2000). PABP was shown to enhance the efficiency of the binding of PCBPs to α-globin 3′-UTR, which in turn protected the ErEN-target sequence. Additionally, the binding of PABP to the poly(A) tail was augmented by PCBPs, implying that a stable higher order structural network is involved in stabilization of the α-globin mRNA.
From the immunoprecipitation data in Fig. 6A, it can be seen that PCBP1 interacted constitutively with PABP and the interaction was reduced to 35% and 25% after co-treatment with Ly294 and H89, respectively. Forskolin treatment did not lead to increased interaction, suggesting that PABP plays a role in the prevention of mRNA destabilization by PCBP1. To test whether forskolin directly regulates PABP binding to MOR 3′-UTR, RNA immunoprecipitation (RIP) was performed with PABP antibody (Fig. 6B). Indeed, forskolin treatment did lead to increased PABP binding to MOR RNA, the 3′-UTR end (left graph) and rpL32 (right graph). The increased binding observed with forskolin treatment was dramatically reduced by treatment with Ly294 and H89, suggesting the involvement of PABP in PCBP1-mediated enhancement through PKA and PI3-kinase pathways.
3.10. Interaction of PCBP1 with an isoform of AUF1 is enhanced by forskolin
Two of the most extensively studied RNA binding proteins are the AU-rich element RNA-binding protein 1 (AUF1, also called hnRNPD) and HuR. AUF1 consists of four isoforms of 37, 40, 42, and 45 kDa as a result of alternative transcript splicing. The AUF1 isoforms possess distinct RNA-binding affinities, abilities to destabilize mRNA (through ARE-mediated mRNA decay, AMD), and even subcellular locations either in the nucleus or cytoplasm (Gratacós and Brewer, 2010). HuR and AUF1, have been shown to exert opposing effects on the stability and translation of several target mRNAs. HuR binding generally leads to mRNA stabilization resulting in increased protein production. Conversely, binding of AUF1 isoforms generally leads to rapid degradation of mRNA resulting in reduced protein production (Lal et al., 2004; Hinman and Lou, 2008), although, in some instances, AUF1 binding enhances mRNA stability and translation (Sela-Brown et al., 2000; Bell et al., 2005; Liao et al., 2007). It was previously reported that all isoforms of AUF1 interact with PCBP1 in vitro. On the other hand, PCBP2 and hnRNPK have lesser or no ability to bind with AUF1, respectively (Kiledjian et al., 1997).
Therefore, we examined the interaction of PCBP1 with AUF1 in our cell system (MOR + NMB cells) by immunoprecipitation (Fig. 7A). Surprisingly, PCBP1 interacted with only p37AUF1, unlike a previous observation that all four AUF1 isoforms interact with GST-fused PCBP1 in vitro (Kiledjian et al., 1997). The interaction of p37AUF1 and PCBP1 was increased about 2.5-fold by treatment with forskolin as compared to the non-treated control, while the interaction was reduced by cotreatments with Ly294 or H89. With PMA treatment alone, the interaction level remained the same as that of the non-treated control. The discrepancy between the observation by Kiledjian et al. and ours on the interaction of PCBP1 with AUF1 isoforms might be due to the use of different cell systems and needs further study.
3.11. Enhanced serine/tyrosine phosphorylation of AUF1 by forskolin treatment
It is known that the phosphorylation state of AUF1 affects AMD efficiency (Wilson et al., 2003a,b). One isoform of AUF1 (p40AUF1) can be phosphorylated at Ser83 and Ser87, and its phosphorylation states regulate p40AUF1 binding to ARE resulting in changes in mRNA stability (Wilson et al., 2003a; Gratacós and Brewer, 2010). Global proteomic profiling showed a potential phosphorylation site at Thr91 (Molina et al., 2007), but to our knowledge no investigation on the role of Thr91 phosphorylation in AUF1 has been done thus far.
We examined this by performing phosphorylation assays in the forskolin-treatment system using AUF1 and phospho-specific antibodies for immunoprecipitation analyses (Fig. 7B). In the upper gel of the IP after serine phosphorylation of AUF1, it appears that surprisingly the p45 isoform, but not the p40 or p37 isoforms, was enhanced approximately 3-fold by forskolin treatment as compared to the non-treated control. The enhanced serine phosphorylation was blocked by cotreatment with Ly294, whereas co-treatment with H89 did not inhibit it. Treatment with PMA alone led to an approximately 4.5-fold increase in serine phosphorylation compared to the level of the non-treated control. In a previous report, phosphorylated Ser83 and Ser87 of p40AUF1 were shown to be dephosphorylated by PMA treatment in THP-1 monocytic leukemia cells, concomitantly resulting in stabilization of the target mRNAs, IL1β, and tumor necrosis factor (TNF)-α (Wilson et al., 2003a). There was no indication in our current study on how serine phosphorylation of p45AUF1 but not p40AUF1 was increased by PMA contrary to the work previously reported (Wilson et al., 2003a).
We also examined changes in the tyrosine phosphorylation state with forskolin and PMA treatment (lower two gel images in Fig. 7B). Tyrosine phosphorylation of p45AUF1 was increased approximately 2.5-fold by forskolin, and this increase was blocked by co-treatment with Ly294 but not H89. The level of tyrosine phosphorylation was decreased by approximately 40% upon PMA treatment. This evidence of tyrosine phosphorylation of p45AUF1 might agree with previously published data that demonstrated that an oncogenic tyrosine kinase (known as nucleophosmin-anaplastic lymphoma kinase, NPM-ALK) interacted with p45AUF1 following tyrosine phosphorylation, resulting in stabilization of the target ARE-mRNAs (Fawal et al., 2006).
Since AUF1 is one of the RNPs that also include HuR, PABP, and PCBPs, we performed RIP (Fig. 7C) to study the interaction of AUF1 and the MOR 3′-UTR, where we had previously located direct RNA interaction with PCBP1 and possibly indirect interactions with PABP. AUF1 binding to the rpL32 site was increased about 4.2-fold with forskolin treatment as compared to the non-treated control, and the increased binding was efficiently blocked by treatment with H89 as well as to a lesser extent by treatment with Ly294 (Fig. 7C). This suggests that the interaction of AUF1 with the rpL32 site, presumably indirectly through PCBP1, is regulated by the PKA and PI3-kinase pathways.
To determine the requirement for AUF1 in the PCBP1-mediated upregulation of MOR, we performed flow cytometry with antibodies to PCBP1 and MOR (Fig. 7D). MOR-positive cells (AP4d, neuronally differentiated cells) were transfected with PCBP1 along with shRNAs of PCBP1 or AUF1. With no treatment (− forskolin), cells with MOR +/ PCBP1 + (upper right quadrant) were decreased in knockdown cells of PCBP1 (PCBP1 + PCBP1 shRNA) by 12.3% and of AUF1 (PCBP1 + AUF1 shRNA) by 16%, compared to control cells (PCBP1, 20.3%). In addition, cells with MOR+/PCBP1 − (lower right quadrant) were decreased in the knockdown cells relative to the control cells (11.4%) to 7.72% (PCBP1 + PCBP1 shRNA) and 7.86% (PCBP1 + AUF1 shRNA). In forskolin-treated cells (+ forskolin), MOR +/PCBP1 + cells were also decreased in both knockdown cells, 22.1% and 30.4%, compared to control cells (PCBP1, 38.7%). Cells with MOR +/PCBP1 − (lower right quadrant) were increased slightly, but not significantly, in the knockdown cells, suggesting a requirement for both factors for forskolin-mediated upregulation of MOR.
All these results (see the summary in Fig. 8) suggest that forskolin increases the phosphorylation of PCBP1 and AUF1 via the PKA and PI3-kinase pathways and translocates the PCBP1 to cytoplasm, where it forms a complex with PABP, resulting in stabilizing its target, MOR mRNA.
Fig. 8.
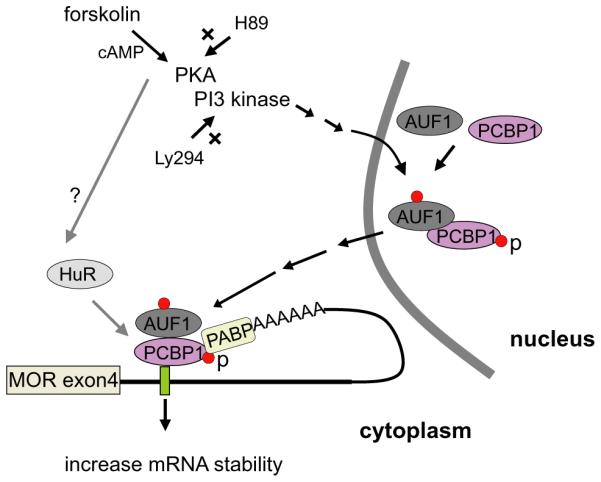
A summary of the proposed mechanism for the PCBP1-mediated stabilization of MOR mRNA is shown. Red dots indicate phosphorylation of residues in proteins (p) and X indicates involvement of inhibitor. The question mark (?) indicates a pathway unidentified in this study.
4. Discussion
We have previously identified trans-acting factors, such as miR-23b and miR-339, that suppress translation of MOR mRNA through its own 3′-UTR (Wu et al., 2008, 2013; Hwang et al., 2012). Others have also found a trans-acting miRNA, let-7, that interacts with its binding site in the MOR 3′-UTR and suppresses MOR translation (He et al., 2010). Despite these reports of negative regulators of MOR expression, much is still unknown about how this gene is expressed or enhanced in MOR positive (+) cells and tissue, such as brain, at the post-transcriptional level. In an attempt to find a positive regulator, we took a close look at the DNA sequences downstream of the stop codon (+ 1) and found a region that is C-rich in a 44-base sequence from + 11 to + 54. A similar C-rich element (CRE) is known to be a general binding site for poly(C) binding factors, such as hnRNPK and PCBP1 ~ 4 (Lee et al., 2007). Therefore, we overexpressed poly(C) binding factor PCBP1 and its family member hnRNPK with MOR 3′-UTR constructs in MOR + cells and studied their functions as positive regulators. PCBP1 was bound specifically to the CRE, whereas hnRNPK was bound ubiquitously throughout MOR mRNA (Fig. 2B and Table 2). PCBP1, assisted by RNA binding factors AUF1 and PABP, enhanced MOR expression through mRNA stabilization. Serine phosphorylation of PCBP1 by forskolin enhanced MOR upregulation as well as increased translocation of PCBP1 to cytoplasm, which is a novel finding.
Among RNA-binding proteins, HuR (Hu-antigen R) and its neuronal homologues, HuB, HuC, and HuD, have been associated with post-transcriptional stabilization of mRNA for cyclin, vascular endothelial growth factor, TNF-α, c-Myc, interleukins, and cell-cycle modulators p21, p27, and p53 (Brennan and Steitz, 2001). HuR is also known to share its binding site with PCBP1 through conserved regions, such as the UC-rich region of the androgen receptor 3′-UTR (Yeap et al., 2002) and the UVC-responsive region of p21(WAF1) 3′-UTR (Giles et al., 2003). The two factors may function cooperatively for mRNA stabilization. In the case of the renin gene, both PCBP1 and HuR stabilize renin mRNA but bind to different regions of the 3′-UTR: CRE for PCBP1 and ARE for HuR (Morris et al., 2004). Forskolin, which augments renin mRNA stability in Calu-6 cells, increases the binding of several proteins, including HuR and PCBP1, to the renin 3′-UTR, which in turn affects renin mRNA stability and thus overall expression of the protein, suggesting that cyclic AMP (cAMP) is a crucial regulator of the post-transcriptional control of renin expression (Adams et al., 2003). It was not known previously if PCBP1 interacts directly with HuR in the ribonucleoprotein (RNP) complex they form. In Fig. 6C, we show that they do interact with each other and this interaction is increased by forskolin treatment. However, co-treatment with both Ly294 and H89 did not affect the increased interaction, thus indicating no involvement of PI3-kinase and PKA in the forskolin-enhanced interaction of PCBP1 with HuR. The involvement of HuR in the function of PCBP1 remains to be investigated further.
As mentioned, mitogenic stimulation of human cells results in phosphorylation of Thr60/127 in a Pak1-sensitive manner. This Pak1-dependent phosphorylation of PCBP1 reduces binding and increases translational inhibition through a DICE of the 3′-UTR (Meng et al., 2007). In MOR 3′-UTR, there are four putative DICE sites. As shown in Fig. 3A, c-Src kinase inhibitor (PP2) along with its inactive control (PP3) did not affect the PCBP1-mediated MOR upregulation, suggesting no involvement of a DICE-mediated mechanism. Instead, we found that additional novel phosphorylation of PCBP1, especially on serine residues, was induced by PKA activation and stimulated by forskolin. Serine phosphorylation of PCBP1 was involved in further upregulation of the target gene, MOR, via increased interaction with AUF1. The phosphorylation of AUF1 serine and tyrosine residues was also induced by PKA activation. It is generally known that AUF1 and a number of other RNA-binding proteins, such as BRF1, KSRP, TTP, and other members of the hnRNP family, destabilize target gene mRNAs by binding to ARE of target 3′-UTRs (Gratacós and Brewer, 2010); however, this has also been reported to increase the stability and translation of some target transcripts (Kiledjian et al., 1997; Raineri et al., 2004; Zucconi and Wilson, 2011). In studies on human α-globin mRNA, stability of the mRNA is mediated by a specific RNP complex at the 3′-UTR. This RNP complex, termed an α-complex, contains PCBP1, PCBP2 and AUF1, and was shown to stabilize the half-life of α-globin mRNA in vitro and in vivo. This raises the possibility that the interaction of AUF1 and various co-factors may dictate whether there is a positive or negative effect on the stability of mRNAs.
Interestingly, in vitro the four isoforms (p37, p40, p42, and p45) of AUF1 differ considerably in their ARE binding affinities. The two isoforms with the highest binding affinities, p37 and p42, exert the most profound effect on ARE-mRNA stability (Gratacós and Brewer, 2010). When we looked for direct interaction of PCBP1 with AUF1, we observed, as shown in Fig. 7A, that only the p37 isoform interacted with PCBP1. The interaction was increased on forskolin treatment, whereas it was decreased on co-treatment with either Ly274 or H89. AUF1 is present predominantly in the nucleus, and all isoforms shuttle between the nucleus and cytoplasm (Barreau et al., 2005). Both p37AUF1 and p40AUF1 contain a nuclear import signal (NIS) at their C-terminal domains; the insertion of a nuclear export signal (NES) in p42AUF1 and p45AUF1 disrupts the NIS to promote cytoplasmic localization (Sarkar et al., 2003). Deletion of the C-terminal domain of p37AUF1 suggested a mechanism by which p37 enters the nucleus, binds to target mRNAs, and brings them to the cytoplasm where the effects are shown (Chen et al., 2004). Other studies (He and Schneider, 2006) identified 14-3-3 as an AUF1-interacting protein that interacts strongly with p37AUF1 and to a lesser extent with p40AUF1, but not with p42AUF1 and p45AUF1. The interaction between 14-3-3 and p37AUF1 results in retention of AUF1 in the cytoplasm (He and Schneider, 2006). This explains that the location of AUF1 protein is tightly controlled and its isoforms are shuttled to different locations to facilitate their activities.
Therefore, we were interested in determining if p37AUF1 interacts with PCBP1 along with MOR mRNA exclusively in the nucleus to shuttle the complex to the cytoplasm for mRNA stabilization, as with 14-3-3 and p37AUF1 (He and Schneider, 2006). Although the IP data in Fig. 7A demonstrate the interaction of PCBP1 with p37AUF1, surprisingly, the IP results in Fig. 7B indicate that forskolin treatment leads to increased serine phosphorylation of only p45AUF1, indicating a distinct signaling pathway different from previous findings. The Ser phosphorylation of AUF1 by forskolin was reduced by Ly294 treatment but was unchanged by H89, suggesting the involvement of the PI3-kinase pathway in the Ser phosphorylation and not the PKA pathway. A previous report showed that AUF1 serine residues 87 and 83 are phosphorylated by PKA and glycogen synthase kinase 3-beta (GSK), respectively (Gratacós and Brewer, 2010). It is an interesting coincidence that GSK-3 is a critical downstream element in the PI3-kinase pathway and that the serine phosphorylation by forskolin seems to occur at Ser83 of AUF1 because Ly294 blocked the phosphorylation (Fig. 7B). The phosphorylation state of AUF1 is known to affect mRNA decay kinetics (Gratacós and Brewer, 2010), possibly through changes in ARE-binding affinity, protein oligomerization potential, mRNP conformation, and interaction with other proteins (Wilson et al., 2003b; Zucconi et al., 2010).
Tyrosine phosphorylation of AUF1 was also increased by forskolin treatment, and the phosphorylation was reduced by Ly294 treatment but not by H89, similar to the effects seen with serine phosphorylation (Fig. 7B). PMA reduced the basal tyrosine phosphorylation level, which was the opposite of what was seen with serine phosphorylation after PMA treatment. Tyrosine phosphorylation of AUF1 has not been reported previously, so further investigation of tyrosine phosphorylation may provide insights in understanding the mechanisms of AUF1/ PCBP1-mediated mRNA stabilization.
Forskolin activates adenylate cyclase, which leads to increased levels of cAMP resulting in PKA activation. This cAMP-dependent PKA is crucial for several opioid-related effects, including opioid withdrawal (Rothman et al., 2009), opioid receptor desensitization, opioid intracellular signaling, development of hyperalgesia, and molecular adaptation by chronic opioids (Law et al., 2004). Following chronic morphine administration, the addition of an antagonist, such as naloxone, results in markedly increased levels of cAMP (termed a cAMP “super-activation”) that is a signal of a severe side effect in the treatment of opioid addiction (Law et al., 2004). In the present study, we found that cAMP-dependent PKA plays an important role in regulating the expression of MOR, which is a core factor in several opioid-related effects. Therefore, it would be interesting to define the relationship between the PKA pathway and opioid effects, especially through the upregulation of MOR expression by forskolin.
Acknowledgements
We thank Geneva Davis for editorial assistance with the manuscript. This work was supported by the National Institutes of Health National Institutes of Drug Abuse [Grants DA000564, DA001583, DA011806, DA011190, DA013926]; and by the A & F Stark Fund of the Minnesota Medical Foundation.
Abbreviations
- PCBP1
poly(rC) binding protein 1
- MOR
mu opioid receptor
- AUF1
AU-rich element RNA-binding protein 1
- PABP
poly (A)-binding protein
- 3′-UTR
3′-untranslated region
- PKA
protein kinase A
- PKC
protein kinase C
- CRE
C-rich element
- RNA-EMSA
RNA electrophoretic mobility shift assay
- ARE
AU-rich elements
- hnRNPs
heterogeneous nuclear ribonucleoproteins
- HuR
human antigen R
- RNPs
ribonucleoproteins
- ILEI
interleukin-like EMT inducer
- Dab2
disabled-2
- STR
short tandem repeats
- RIP
RNA immunoprecipitation
- nt
nucleotide
- rpL32
ribosomal protein L32
- IRES
internal ribosome entry segment
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- DICE
differentiation-control element
- LOX
lipoxygenase
- LMB
leptomycin B
- LUC
luciferase
- PMA
phorbol 12-myristate 13-acetate
Footnotes
The authors declare no conflict of interest.
References
- Adams DJ, Beveridge DJ, van der Weyden L, Mangs H, Leedman PJ, Morris BJ. HADHB, HuR, and CP1 bind to the distal 3′-untranslated region of human renin mRNA and differentially modulate renin expression. J. Biol. Chem. 2003;278:44894–44903. doi: 10.1074/jbc.M307782200. [DOI] [PubMed] [Google Scholar]
- Atchison ML, Meyuhas O, Perry RP. Localization of transcriptional regulatory elements and nuclear factor binding sites in mouse ribosomal protein gene rpL32. Mol. Cell. Biol. 1989;9:2067–2074. doi: 10.1128/mcb.9.5.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga PS, Arhin GK, Wilusz J. DSEF-1 is a member of the hnRNP H family of RNA-binding proteins and stimulates pre-mRNA cleavage and polyadenylation in vitro. Nucleic Acids Res. 1998;26:5343–5350. doi: 10.1093/nar/26.23.5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker A, Epis MR, Porter CJ, Hopkins BR, Wilce MC, Wilce JA, Giles KM, Leedman PJ. Sequence requirements for RNA binding by HuR and AUF1. J. Biochem. 2012;151:423–437. doi: 10.1093/jb/mvs010. [DOI] [PubMed] [Google Scholar]
- Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell O, Silver J, Naveh-Many T. Identification and characterization of cis-acting elements in the human and bovine PTH mRNA 3′-untranslated region. J. Bone Miner. Res. 2005;20:858–866. doi: 10.1359/JBMR.041227. [DOI] [PubMed] [Google Scholar]
- Brennan CM, Steitz JA. HuR and mRNA stability. Cell. Mol. Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan CM, Gallouzi IE, Steitz JA. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J. Cell Biol. 2000;151:1–14. doi: 10.1083/jcb.151.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. U S A]–>Proc. Natl. Acad. Sci. U. S. A. 1986;83:1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang N, Yi J, Guo G, Liu X, Shang Y, Tong T, Cui Q, Zhan M, Gorospe M, Wang W. HuR uses AUF1 as a cofactor to promote p16INK4 mRNA decay. Mol. Cell. Biol. 2010;30:3875–3886. doi: 10.1128/MCB.00169-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Pal JK. Role of 5′- and 3′-untranslated regions of mRNAs in human diseases. Biol. Cell. 2009;101:251–262. doi: 10.1042/BC20080104. [DOI] [PubMed] [Google Scholar]
- Chaudhury A, Chander P, Howe PH. Heterogeneous nuclear ribonucleoproteins (hnRNPs) in cellular processes: focus on hnRNP E1's multifunctional regulatory roles. RNA. 2010;16:1449–1462. doi: 10.1261/rna.2254110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury A, Hussey GS, Howe PH. 3′-UTR-mediated post-transcriptional regulation of cancer metastasis: beginning at the end. RNA Biol. 2011;8:595–599. doi: 10.4161/rna.8.4.16018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TC, Hinton DR, Zidovetzki R, Hofman FM. Up-regulation of the cAMP/ PKA pathway inhibits proliferation, induces differentiation, and leads to apoptosis in malignant gliomas. Lab. Investig. 1998;78:165–174. [PubMed] [Google Scholar]
- Chen CY, Xu N, Zhu W, Shyu AB. Functional dissection of hnRNP D suggests that nuclear import is required before hnRNP D can modulate mRNA turnover in the cytoplasm. RNA. 2004;10:669–680. doi: 10.1261/rna.5269304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chkheidze AN, Lyakhov DL, Makeyev AV, Morales J, Kong J, Liebhaber SA. Assembly of the alpha-globin mRNA stability complex reflects binary interaction between the pyrimidine-rich 3′ untranslated region determinant and poly(C) binding protein alphaCP. Mol. Cell. Biol. 1999;19:4572–4581. doi: 10.1128/mcb.19.7.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HS, Song KY, Hwang CK, Kim CS, Law PY, Wei LN, Loh HH. A proteomics approach for identification of single strand DNA-binding proteins involved in transcriptional regulation of mouse micro opioid receptor gene. Mol. Cell. Proteomics. 2008;7:1517–1529. doi: 10.1074/mcp.M800052-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conne B, Stutz A, Vassalli JD. The 3′ untranslated region of messenger RNA: a molecular ‘hotspot’ for pathology? Nat. Med. 2000;6:637–641. doi: 10.1038/76211. [DOI] [PubMed] [Google Scholar]
- Czyzyk-Krzeska MF, Bendixen AC. Identification of the poly(C) binding protein in the complex associated with the 3′ untranslated region of erythropoietin messenger RNA. Blood. 1999;93:2111–2120. [PubMed] [Google Scholar]
- Czyzyk-Krzeska MF, Beresh JE. Characterization of the hypoxia-inducible protein binding site within the pyrimidine-rich tract in the 3′-untranslated region of the tyrosine hydroxylase mRNA. J. Biol. Chem. 1996;271:3293–3299. doi: 10.1074/jbc.271.6.3293. [DOI] [PubMed] [Google Scholar]
- Dignam JD. Preparation of extracts from higher eukaryotes. Methods Enzymol. 1990;182:194–203. doi: 10.1016/0076-6879(90)82017-v. [DOI] [PubMed] [Google Scholar]
- Dudov KP, Perry RP. The gene family encoding the mouse ribosomal protein L32 contains a uniquely expressed intron-containing gene and an unmutated processed gene. Cell. 1984;37:457–468. doi: 10.1016/0092-8674(84)90376-3. [DOI] [PubMed] [Google Scholar]
- Fawal M, Armstrong F, Ollier S, Dupont H, Touriol C, Monsarrat B, Delsol G, Payrastre B, Morello D. A “liaison dangereuse” between AUF1/hnRNPD and the oncogenic tyrosine kinase NPM-ALK. Blood. 2006;108:2780–2788. doi: 10.1182/blood-2006-04-014902. [DOI] [PubMed] [Google Scholar]
- Gies EK, Peters DM, Gelb CR, Knag KM, Peterfreund RA. Regulation of mu opioid receptor mRNA levels by activation of protein kinase C in human SH-SY5Y neuroblastoma cells. Anesthesiology. 1997;87:1127–1138. doi: 10.1097/00000542-199711000-00017. [DOI] [PubMed] [Google Scholar]
- Gilbert C, Svejstrup JQ. RNA immunoprecipitation for determining RNA-protein associations in vivo. Curr. Protoc. Mol. Biol. 2006 doi: 10.1002/0471142727.mb2704s75. (p. Unit 27.4., Chapter 27) [DOI] [PubMed] [Google Scholar]
- Giles KM, Daly JM, Beveridge DJ, Thomson AM, Voon DC, Furneaux HM, Jazayeri JA, Leedman PJ. The 3′-untranslated region of p21WAF1 mRNA is a composite cis-acting sequence bound by RNA-binding proteins from breast cancer cells, including HuR and poly(C)-binding protein. J. Biol. Chem. 2003;278:2937–2946. doi: 10.1074/jbc.M208439200. [DOI] [PubMed] [Google Scholar]
- Gratacós FM, Brewer G. The role of AUF1 in regulated mRNA decay. Wiley Interdiscip. Rev. RNA. 2010;1:457–473. doi: 10.1002/wrna.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halazonetis TD, Georgopoulos K, Greenberg ME, Leder P. c-Jun dimerizes with itself and with c-Fos, forming complexes of different DNA binding affinities. Cell. 1988;55:917–924. doi: 10.1016/0092-8674(88)90147-x. [DOI] [PubMed] [Google Scholar]
- Han SP, Tang YH, Smith R. Functional diversity of the hnRNPs: past, present and perspectives. Biochem. J. 2010;430:379–392. doi: 10.1042/BJ20100396. [DOI] [PubMed] [Google Scholar]
- He C, Schneider R. 14-3-3sigma is a p37 AUF1-binding protein that facilitates AUF1 transport and AU-rich mRNA decay. EMBO J. 2006;25:3823–3831. doi: 10.1038/sj.emboj.7601264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Yang C, Kirkmire CM, Wang ZJ. Regulation of opioid tolerance by let-7 family microRNA targeting the mu opioid receptor. J. Neurosci. 2010;30:10251–10258. doi: 10.1523/JNEUROSCI.2419-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell. Mol. Life Sci. 2008;65:3168–3181. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho JJ, Robb GB, Tai SC, Turgeon PJ, Mawji IA, Man HS, Marsden PA. Active stabilization of the human eNOS mRNA by hnRNP E1 protects against antisense RNA and microRNAs. Mol. Cell. Biol. 2013;33:2029–2046. doi: 10.1128/MCB.01257-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HD, Lee TY, Tzeng SW, Horng JT. KinasePhos: a web tool for identifying protein kinase-specific phosphorylation sites. Nucleic Acids Res. 2005;33:W226–W229. doi: 10.1093/nar/gki471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HY, Chien CH, Jen KH, Huang HD. RegRNA: an integrated web server for identifying regulatory RNA motifs and elements. Nucleic Acids Res. 2006;34:W429–W434. doi: 10.1093/nar/gkl333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang CK, Kim CS, Choi HS, McKercher SR, Loh HH. Transcriptional regulation of mouse mu opioid receptor gene by PU.1. J. Biol. Chem. 2004;279:19764–19774. doi: 10.1074/jbc.M400755200. [DOI] [PubMed] [Google Scholar]
- Hwang CK, Song KY, Kim CS, Choi HS, Guo XH, Law PY, Wei LN, Loh HH. Evidence of endogenous mu opioid receptor regulation by epigenetic control of the promoters. Mol. Cell. Biol. 2007;27:4720–4736. doi: 10.1128/MCB.00073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang CK, Song KY, Kim CS, Choi HS, Guo XH, Law PY, Wei LN, Loh HH. Epigenetic programming of mu opioid receptor gene in mouse brain is regulated by MeCP2 and Brg1 chromatin remodeling factor. J. Cell. Mol. Med. 2009;13:3591–3615. doi: 10.1111/j.1582-4934.2008.00535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang CK, Law P-Y, Wei L-N, Loh HH. Regulation of the Transcription of G Protein-coupled Receptor Genes. In: Stevens CW, editor. Methods for the Discovery and Characterization of G Protein-coupled Receptors. Humana Press; Totowa, NJ: 2011a. pp. 49–69. (Stevens, C.W.)Stevens, C.W.s) [Google Scholar]
- Hwang CK, Wagley Y, Law PY, Wei LN, Loh HH. MicroRNAs in opioid pharmacology. J. NeuroImmune Pharmacol. 2012;7:808–819. doi: 10.1007/s11481-011-9323-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalali A, Bassuk AG, Kan L, Israsena N, Mukhopadhyay A, McGuire T, Kessler JA. HeyL promotes neuronal differentiation of neural progenitor cells. J. Neurosci. Res. 2011;89:299–309. doi: 10.1002/jnr.22562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang BC, Munoz-Najar U, Paik JH, Claffey K, Yoshida M, Hla T. Leptomycin B, an inhibitor of the nuclear export receptor CRM1, inhibits COX-2 expression. J. Biol. Chem. 2003;278:2773–2776. doi: 10.1074/jbc.C200620200. [DOI] [PubMed] [Google Scholar]
- Kasai S, Han W, Ide S, Hata H, Takamatsu Y, Yamamoto H, Uhl GR, Sora I, Ikeda K. Involvement of the 3′ non-coding region of the mu opioid receptor gene in morphine-induced analgesia. Psychiatry Clin. Neurosci. 2006;60:S11–S17. [Google Scholar]
- Kiledjian M, DeMaria CT, Brewer G, Novick K. Identification of AUF1 (heterogeneous nuclear ribonucleoprotein D) as a component of the alpha-globin mRNA stability complex. Mol. Cell. Biol. 1997;17:4870–4876. doi: 10.1128/mcb.17.8.4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DK, Hwang CK, Wagley Y, Law PY, Wei LN, Loh HH. p38 mitogenactivated protein kinase and PI3-kinase are involved in up-regulation of mu opioid receptor transcription induced by cycloheximide. J. Neurochem. 2011b;116:1077–1087. doi: 10.1111/j.1471-4159.2010.07163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong J, Ji X, Liebhaber SA. The KH-domain protein alpha CP has a direct role in mRNA stabilization independent of its cognate binding site. Mol. Cell. Biol. 2003;23:1125–1134. doi: 10.1128/MCB.23.4.1125-1134.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- la Cour T, Kiemer L, Molgaard A, Gupta R, Skriver K, Brunak S. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 2004;17:527–536. doi: 10.1093/protein/gzh062. [DOI] [PubMed] [Google Scholar]
- Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- Lai EC, Burks C, Posakony JW. The K box, a conserved 3′ UTR sequence motif, negatively regulates accumulation of enhancer of split complex transcripts. Development. 1998;125:4077–4088. doi: 10.1242/dev.125.20.4077. [DOI] [PubMed] [Google Scholar]
- Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004;23:3092–3102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law PY, Loh HH, Wei LN. Insights into the receptor transcription and signaling: implications in opioid tolerance and dependence. Neuropharmacology. 2004;47(Suppl. 1):300–311. doi: 10.1016/j.neuropharm.2004.07.013. [DOI] [PubMed] [Google Scholar]
- Lee PT, Liao PC, Chang WC, Tseng JT. Epidermal growth factor increases the interaction between nucleolin and heterogeneous nuclear ribonucleoprotein K/ poly(C) binding protein 1 complex to regulate the gastrin mRNA turnover. Mol. Biol. Cell. 2007;18:5004–5013. doi: 10.1091/mbc.E07-04-0384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao B, Hu Y, Brewer G. Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat. Struct. Mol. Biol. 2007;14:511–518. doi: 10.1038/nsmb1249. [DOI] [PubMed] [Google Scholar]
- Lozinski T, Bolewska K, Wierzchowski KL. Equivalence of Mg2+ and Na+ ions in salt dependence of the equilibrium binding and dissociation rate constants of Escherichia coli RNA polymerase open complex. Biophys. Chem. 2009;142:65–75. doi: 10.1016/j.bpc.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Maeda H, Araki K, Kitamura D, Wang J, Watanabe T. Nuclear factors binding to the human immunoglobulin heavy-chain gene enhancer. Nucleic Acids Res. 1987;15:2851–2869. doi: 10.1093/nar/15.7.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev AV, Liebhaber SA. The poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms. RNA. 2002;8:265–278. doi: 10.1017/s1355838202024627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Q, Rayala SK, Gururaj AE, Talukder AH, O'Malley BW, Kumar R. Signaling-dependent and coordinated regulation of transcription, splicing, and translation resides in a single coregulator, PCBP1. U S A]–>Proc. Natl. Acad. Sci. U. S. A. 2007;104:5866–5871. doi: 10.1073/pnas.0701065104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina H, Horn DM, Tang N, Mathivanan S, Pandey A. Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. U S A]–>Proc. Natl. Acad. Sci. U. S. A. 2007;104:2199–2204. doi: 10.1073/pnas.0611217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris BJ, Adams DJ, Beveridge DJ, van der Weyden L, Mangs H, Leedman PJ. cAMP controls human renin mRNA stability via specific RNA-binding proteins. Acta Physiol. Scand. 2004;181:369–373. doi: 10.1111/j.1365-201X.2004.01307.x. [DOI] [PubMed] [Google Scholar]
- Moura-Neto R, Dudov KP, Perry RP. An element downstream of the cap site is required for transcription of the gene encoding mouse ribosomal protein L32. U S A]–>Proc. Natl. Acad. Sci. U. S. A. 1989;86:3997–4001. doi: 10.1073/pnas.86.11.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naarmann IS, Harnisch C, Flach N, Kremmer E, Kuhn H, Ostareck DH, OstareckLederer A. mRNA silencing in human erythroid cell maturation: heterogeneous nuclear ribonucleoprotein K controls the expression of its regulator c-Src. J. Biol. Chem. 2008;283:18461–18472. doi: 10.1074/jbc.M710328200. [DOI] [PubMed] [Google Scholar]
- Ostareck-Lederer A, Ostareck DH. Control of mRNA translation and stability in haematopoietic cells: the function of hnRNPs K and E1/E2. Biol. Cell. 2004;96:407–411. doi: 10.1016/j.biolcel.2004.03.010. [DOI] [PubMed] [Google Scholar]
- Pan YX, Chen H, Kilberg MS. Interaction of RNA-binding proteins HuR and AUF1 with the human ATF3 mRNA 3′-untranslated region regulates its amino acid limitation-induced stabilization. J. Biol. Chem. 2005;280:34609–34616. doi: 10.1074/jbc.M507802200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsch J, Stephan W, Tanda S. A highly conserved sequence in the 3′-untranslated region of the Drosophila Adh gene plays a functional role in Adh expression. Genetics. 1999;151:667–674. doi: 10.1093/genetics/151.2.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng SS, Chen CY, Xu N, Shyu AB. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesole G, Mignone F, Gissi C, Grillo G, Licciulli F, Liuni S. Structural and functional features of eukaryotic mRNA untranslated regions. Gene. 2001;276:73–81. doi: 10.1016/s0378-1119(01)00674-6. [DOI] [PubMed] [Google Scholar]
- Pickering BM, Mitchell SA, Evans JR, Willis AE. Polypyrimidine tract binding protein and poly r(C) binding protein 1 interact with the BAG-1 IRES and stimulate its activity in vitro and in vivo. Nucleic Acids Res. 2003;31:639–646. doi: 10.1093/nar/gkg146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raineri I, Wegmueller D, Gross B, Certa U, Moroni C. Roles of AUF1 isoforms, HuR and BRF1 in ARE-dependent mRNA turnover studied by RNA interference. Nucleic Acids Res. 2004;32:1279–1288. doi: 10.1093/nar/gkh282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimann I, Huth A, Thiele H, Thiele BJ. Suppression of 15-lipoxygenase synthesis by hnRNP E1 is dependent on repetitive nature of LOX mRNA 3′-UTR control element DICE. J. Mol. Biol. 2002;315:965–974. doi: 10.1006/jmbi.2001.5315. [DOI] [PubMed] [Google Scholar]
- Rothman RB, Ananthan S, Bilsky EJ. Novel Opioid Antagonists With Mixed/dual Selectivity. In: Dean RL, Bilsky EJ, Negus SS, editors. Opiate Receptors and Antagonists: From Bench to Clinic. Humana Press; Totowa, NJ: 2009. pp. 137–151. (Dean, R.L., Bilsky, E.J. and Negus, S.S.(Dean, R.L., Bilsky, E.J. and Negus, S.S.s) [Google Scholar]
- Sarkar B, Lu JY, Schneider RJ. Nuclear import and export functions in the different isoforms of the AUF1/heterogeneous nuclear ribonucleoprotein protein family. J. Biol. Chem. 2003;278:20700–20707. doi: 10.1074/jbc.M301176200. [DOI] [PubMed] [Google Scholar]
- Sawicka K, Bushell M, Spriggs KA, Willis AE. Polypyrimidine-tract-binding protein: a multifunctional RNA-binding protein. Biochem. Soc. Trans. 2008;36:641–647. doi: 10.1042/BST0360641. [DOI] [PubMed] [Google Scholar]
- Sela-Brown A, Silver J, Brewer G, Naveh-Many T. Identification of AUF1 as a parathyroid hormone mRNA 3′-untranslated region-binding protein that determines parathyroid hormone mRNA stability. J. Biol. Chem. 2000;275:7424–7429. doi: 10.1074/jbc.275.10.7424. [DOI] [PubMed] [Google Scholar]
- Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- Shigeta Y, Kasai S, Han W, Hata H, Nishi A, Takamatsu Y, Hagino Y, Yamamoto H, Koide T, Shiroishi T, Kasai K, Tsunashima K, Kato N, Ikeda K. Association of morphine-induced antinociception with variations in the 5′ flanking and 3′ untranslated regions of the mu opioid receptor gene in 10 inbred mouse strains. Pharmacogenet. Genomics. 2008;18:927–936. doi: 10.1097/FPC.0b013e32830d0b9e. [DOI] [PubMed] [Google Scholar]
- Sigrist CJ, Cerutti L, Hulo N, Gattiker A, Falquet L, Pagni M, Bairoch A, Bucher P. PROSITE: a documented database using patterns and profiles as motif descriptors. Brief. Bioinform. 2002;3:265–274. doi: 10.1093/bib/3.3.265. [DOI] [PubMed] [Google Scholar]
- Sigrist CJ, de Castro E, Cerutti L, Cuche BA, Hulo N, Bridge A, Bougueleret L, Xenarios I. New and continuing developments at PROSITE. Nucleic Acids Res. 2013;41:D344–D347. doi: 10.1093/nar/gks1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalweit A, Doller A, Huth A, Kahne T, Persson PB, Thiele BJ. Post-transcriptional control of renin synthesis: identification of proteins interacting with renin mRNA 3′-untranslated region. Circ. Res. 2003;92:419–427. doi: 10.1161/01.RES.0000059300.67152.4E. [DOI] [PubMed] [Google Scholar]
- Smirnov D, Im HJ, Loh HH. Delta-opioid receptor gene: effect of Sp1 factor on transcriptional regulation in vivo. Mol. Pharmacol. 2001;60:331–340. doi: 10.1124/mol.60.2.331. [DOI] [PubMed] [Google Scholar]
- Song KY, Hwang CK, Kim CS, Choi HS, Law PY, Wei LN, Loh HH. Translational repression of mouse mu opioid receptor expression via leaky scanning. Nucleic Acids Res. 2007;35:1501–1513. doi: 10.1093/nar/gkm034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam K, Chen K, Joseph K, Raymond JR, Tholanikunnel BG. The 3′-untranslated region of the beta2-adrenergic receptor mRNA regulates receptor synthesis. J. Biol. Chem. 2004;279:27108–27115. doi: 10.1074/jbc.M401352200. [DOI] [PubMed] [Google Scholar]
- Subramaniam K, Kandasamy K, Joseph K, Spicer EK, Tholanikunnel BG. The 3′-untranslated region length and AU-rich RNA location modulate RNA-protein interaction and translational control of beta(2)-adrenergic receptor mRNA. Mol. Cell. Biochem. 2011;352:125–141. doi: 10.1007/s11010-011-0747-z. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Sakai Y, Yamada O, Shintani T, Gomi K. In silico analysis of 3′-endprocessing signals in Aspergillus oryzae using expressed sequence tags and genomic sequencing data. DNA Res. 2011;18:189–200. doi: 10.1093/dnares/dsr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YS, Khan RA, Zhang Y, Xiao S, Wang M, Hansen DK, Jayaram HN, Antony A. Incrimination of heterogeneous nuclear ribonucleoprotein E1 (hnRNP-E1) as a candidate sensor of physiological folate deficiency. J. Biol. Chem. 2011;286:39100–39115. doi: 10.1074/jbc.M111.230938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele BJ, Doller A, Kahne T, Pregla R, Hetzer R, Regitz-Zagrosek V. RNA-binding proteins heterogeneous nuclear ribonucleoprotein A1, E1, and K are involved in post-transcriptional control of collagen I and III synthesis. Circ. Res. 2004;95:1058–1066. doi: 10.1161/01.RES.0000149166.33833.08. [DOI] [PubMed] [Google Scholar]
- Thyagarajan A, Szaro BG. Dynamic endogenous association of neurofilament mRNAs with K-homology domain ribonucleoproteins in developing cerebral cortex. Brain Res. 2008;1189:33–42. doi: 10.1016/j.brainres.2007.11.012. [DOI] [PubMed] [Google Scholar]
- Waggoner SA, Liebhaber SA. Regulation of alpha-globin mRNA stability. Exp. Biol. Med. (Maywood) 2003;228:387–395. doi: 10.1177/153537020322800409. [DOI] [PubMed] [Google Scholar]
- Wang Z, Kiledjian M. The poly(A)-binding protein and an mRNA stability protein jointly regulate an endoribonuclease activity. Mol. Cell. Biol. 2000;20:6334–6341. doi: 10.1128/mcb.20.17.6334-6341.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Day N, Trifillis P, Kiledjian M. An mRNA stability complex functions with poly(A)-binding protein to stabilize mRNA in vitro. Mol. Cell. Biol. 1999;19:4552–4560. doi: 10.1128/mcb.19.7.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Liu CL, Storey JD, Tibshirani RJ, Herschlag D, Brown PO. Precision and functional specificity in mRNA decay. U S A]–>Proc. Natl. Acad. Sci. U. S. A. 2002;99:5860–5865. doi: 10.1073/pnas.092538799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss IM, Liebhaber SA. Erythroid cell-specific determinants of alpha-globin mRNA stability. Mol. Cell. Biol. 1994;14:8123–8132. doi: 10.1128/mcb.14.12.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson GM, Lu J, Sutphen K, Suarez Y, Sinha S, Brewer B, Villanueva-Feliciano EC, Ysla RM, Charles S, Brewer G. Phosphorylation of p40AUF1 regulates binding to A+U-rich mRNA-destabilizing elements and protein-induced changes in ribonucleoprotein structure. J. Biol. Chem. 2003a;278:33039–33048. doi: 10.1074/jbc.M305775200. [DOI] [PubMed] [Google Scholar]
- Wilson GM, Lu J, Sutphen K, Sun Y, Huynh Y, Brewer G. Regulation of A+-rich element-directed mRNA turnover involving reversible phosphorylation of AUF1. J. Biol. Chem. 2003b;278:33029–33038. doi: 10.1074/jbc.M305772200. [DOI] [PubMed] [Google Scholar]
- Wu Q, Hwang CK, Yao S, Law PY, Loh HH, Wei LN. A major species of mouse mu-opioid receptor mRNA and its promoter-dependent functional polyadenylation signal. Mol. Pharmacol. 2005;68:279–285. doi: 10.1124/mol.105.012567. [DOI] [PubMed] [Google Scholar]
- Wu Q, Law PY, Wei LN, Loh HH. Post-transcriptional regulation of mouse mu opioid receptor (MOR1) via its 3′ untranslated region: a role for microRNA23b. FASEB J. 2008;22:4085–4095. doi: 10.1096/fj.08-108175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Zhang L, Law PY, Wei LN, Loh HH. Long-term morphine treatment decreases the association of mu-opioid receptor (MOR1) mRNA with polysomes through miRNA23b. Mol. Pharmacol. 2009;75:744–750. doi: 10.1124/mol.108.053462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Hwang CK, Zheng H, Wagley Y, Lin HY, Kim DK, Law PY, Loh HH, Wei LN. MicroRNA 339 down-regulates mu-opioid receptor at the post-transcriptional level in response to opioid treatment. FASEB J. 2013;27:522–535. doi: 10.1096/fj.12-213439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Zhou F, Zhu M, Ahmed K, Chen G, Yao X. GPS: a comprehensive www server for phosphorylation sites prediction. Nucleic Acids Res. 2005;33:W184–W187. doi: 10.1093/nar/gki393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeap BB, Voon DC, Vivian JP, McCulloch RK, Thomson AM, Giles KM, Czyzyk-Krzeska MF, Furneaux H, Wilce MC, Wilce JA, Leedman PJ. Novel binding of HuR and poly(C)-binding protein to a conserved UC-rich motif within the 3′-untranslated region of the androgen receptor messenger RNA. J. Biol. Chem. 2002;277:27183–27192. doi: 10.1074/jbc.M202883200. [DOI] [PubMed] [Google Scholar]
- Yu J, Russell JE. Structural and functional analysis of an mRNP complex that mediates the high stability of human beta-globin mRNA. Mol. Cell. Biol. 2001;21:5879–5888. doi: 10.1128/MCB.21.17.5879-5888.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou T, Rao JN, Liu L, Xiao L, Yu TX, Jiang P, Gorospe M, Wang JY. Polyamines regulate the stability of JunD mRNA by modulating the competitive binding of its 3′ untranslated region to HuR and AUF1. Mol. Cell. Biol. 2010;30:5021–5032. doi: 10.1128/MCB.00807-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucconi BE, Wilson GM. Modulation of neoplastic gene regulatory pathways by the RNA-binding factor AUF1. Front. Biosci. 2011;16:2307–2325. doi: 10.2741/3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucconi BE, Ballin JD, Brewer BY, Ross CR, Huang J, Toth EA, Wilson GM. Alternatively expressed domains of AU-rich element RNA-binding protein 1 (AUF1) regulate RNA-binding affinity, RNA-induced protein oligomerization, and the local conformation of bound RNA ligands. J. Biol. Chem. 2010;285:39127–39139. doi: 10.1074/jbc.M110.180182. [DOI] [PMC free article] [PubMed] [Google Scholar]