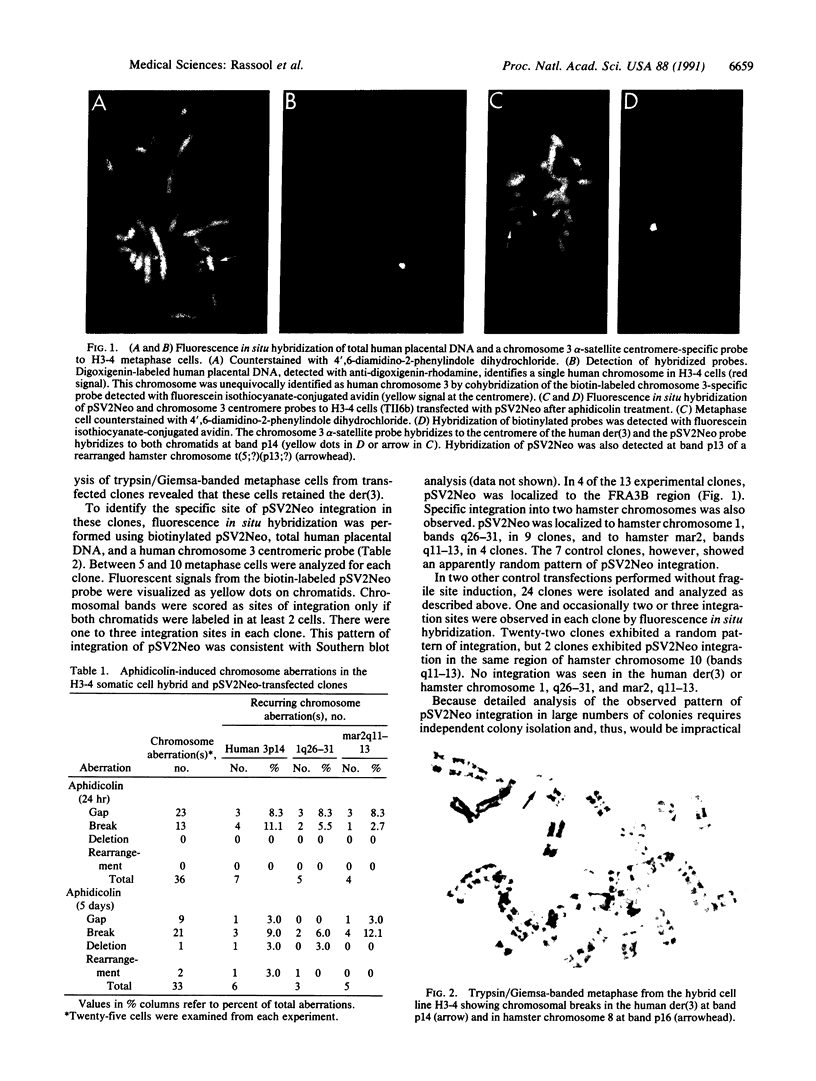
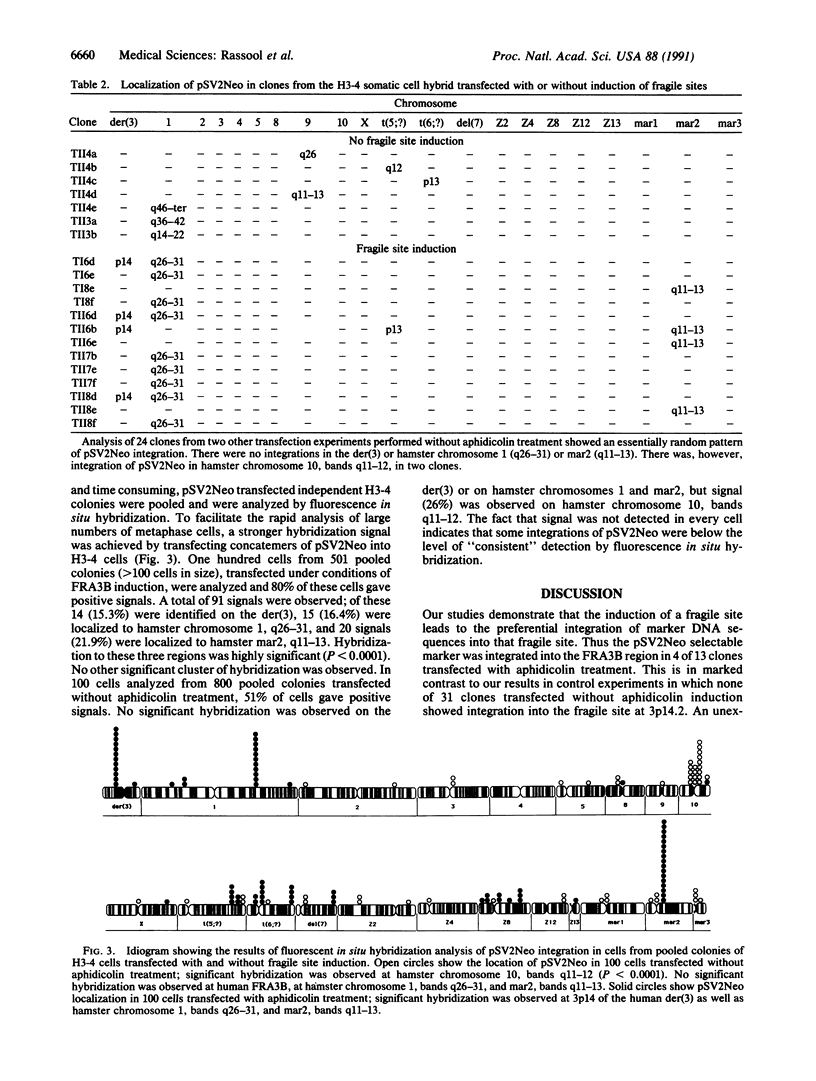
Abstract
Fragile sites are specific regions of chromosomes that are prone to breakage. In cells cultured under conditions that induce fragile site expression, high levels of inter- and intrachromosomal recombination have been observed involving chromosomal bands containing fragile sites. To determine whether expression of specific fragile sites would facilitate preferential integration of exogenous DNA at these recombination hot spots, the vector pSV2Neo was transfected into a Chinese hamster-human somatic cell hybrid containing a derivative chromosome 3 as its only human component. Chromosome 3 contains a common fragile site at band 3p14.2 (FRA3B) that is induced by aphidicolin. Both cells induced to express FRA3B and the uninduced control cells were transfected with the pSV2Neo selectable plasmid. In situ hybridization of a biotin-labeled pSV2Neo probe to metaphase chromosomes revealed one to three integration sites in each stably transfected clone. Four of 13 clones transfected under conditions of FRA3B induction showed integration of pSV2Neo at 3p14; these clones also showed specific integration into hamster chromosome 1 and a rearranged chromosome characteristic of CHO cells (mar2). The 7 control clones, however, showed an apparently random pattern of pSV2Neo integration. Significant hybridization of pSV2Neo to both FRA3B and Chinese hamster chromosomes 1 and mar2 was seen in 100 cells from pooled colonies transfected after treatment with aphidicolin. These results suggest that preferential integration of marker DNA into human and Chinese hamster fragile sites occurs with exposure to aphidicolin. The nature of the DNA sequences at fragile sites is unknown and, despite a number of approaches, these sequences have not yet been isolated; our procedure may represent an approach to the cloning of fragile sites.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chen C., Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987 Aug;7(8):2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaven L. L., Petersen D. F. The chromosomes of CHO, an aneuploid Chinese hamster cell line: G-band, C-band, and autoradiographic analyses. Chromosoma. 1973;41(2):129–144. doi: 10.1007/BF00319690. [DOI] [PubMed] [Google Scholar]
- Djalali M., Adolph S., Steinbach P., Winking H., Hameister H. A comparative mapping study of fragile sites in the human and murine genomes. Hum Genet. 1987 Oct;77(2):157–162. doi: 10.1007/BF00272384. [DOI] [PubMed] [Google Scholar]
- Glover T. W., Stein C. K. Chromosome breakage and recombination at fragile sites. Am J Hum Genet. 1988 Sep;43(3):265–273. [PMC free article] [PubMed] [Google Scholar]
- Hastie N. D., Allshire R. C. Human telomeres: fusion and interstitial sites. Trends Genet. 1989 Oct;5(10):326–331. doi: 10.1016/0168-9525(89)90137-6. [DOI] [PubMed] [Google Scholar]
- Heitz D., Rousseau F., Devys D., Saccone S., Abderrahim H., Le Paslier D., Cohen D., Vincent A., Toniolo D., Della Valle G. Isolation of sequences that span the fragile X and identification of a fragile X-related CpG island. Science. 1991 Mar 8;251(4998):1236–1239. doi: 10.1126/science.2006411. [DOI] [PubMed] [Google Scholar]
- LeBeau M. M., Rowley J. D. Heritable fragile sites in cancer. Nature. 1984 Apr 12;308(5960):607–608. doi: 10.1038/308607a0. [DOI] [PubMed] [Google Scholar]
- Lichter P., Tang C. J., Call K., Hermanson G., Evans G. A., Housman D., Ward D. C. High-resolution mapping of human chromosome 11 by in situ hybridization with cosmid clones. Science. 1990 Jan 5;247(4938):64–69. doi: 10.1126/science.2294592. [DOI] [PubMed] [Google Scholar]
- McKeithan T. W., Shima E. A., Le Beau M. M., Minowada J., Rowley J. D., Diaz M. O. Molecular cloning of the breakpoint junction of a human chromosomal 8;14 translocation involving the T-cell receptor alpha-chain gene and sequences on the 3' side of MYC. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6636–6640. doi: 10.1073/pnas.83.17.6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore E. E., Jones C., Kao F. T., Oates D. C. Synteny between glycinamide ribonucleotide synthetase and superoxide dismutase (soluble). Am J Hum Genet. 1977 Jul;29(4):389–396. [PMC free article] [PubMed] [Google Scholar]
- Nussbaum R. L., Airhart S. D., Ledbetter D. H. Recombination and amplification of pyrimidine-rich sequences may be responsible for initiation and progression of the Xq27 fragile site: an hypothesis. Am J Med Genet. 1986 Jan-Feb;23(1-2):715–721. doi: 10.1002/ajmg.1320230162. [DOI] [PubMed] [Google Scholar]
- Pellicer A., Wigler M., Axel R., Silverstein S. The transfer and stable integration of the HSV thymidine kinase gene into mouse cells. Cell. 1978 May;14(1):133–141. doi: 10.1016/0092-8674(78)90308-2. [DOI] [PubMed] [Google Scholar]
- Rowley J. D., Diaz M. O., Espinosa R., 3rd, Patel Y. D., van Melle E., Ziemin S., Taillon-Miller P., Lichter P., Evans G. A., Kersey J. H. Mapping chromosome band 11q23 in human acute leukemia with biotinylated probes: identification of 11q23 translocation breakpoints with a yeast artificial chromosome. Proc Natl Acad Sci U S A. 1990 Dec;87(23):9358–9362. doi: 10.1073/pnas.87.23.9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Feichtinger W., Jessberger A., Köhler J., Lange R. The fragile site (16) (q22). I. Induction by AT-specific DNA-ligands and population frequency. Hum Genet. 1986 Sep;74(1):67–73. doi: 10.1007/BF00278788. [DOI] [PubMed] [Google Scholar]
- Southern P. J., Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J Mol Appl Genet. 1982;1(4):327–341. [PubMed] [Google Scholar]
- Sutherland G. R. Heritable fragile sites on human chromosomes I. Factors affecting expression in lymphocyte culture. Am J Hum Genet. 1979 Mar;31(2):125–135. [PMC free article] [PubMed] [Google Scholar]
- Sutherland G. R., Ledbetter D. H. Report of the committee on cytogenetic markers. Cytogenet Cell Genet. 1989;51(1-4):452–458. doi: 10.1159/000132804. [DOI] [PubMed] [Google Scholar]
- Tommerup N., Poulsen H., Brøndum-Nielsen K. 5-Fluoro-2'-deoxyuridine induction of the fragile site on Xq28 associated with X linked mental retardation. J Med Genet. 1981 Oct;18(5):374–376. doi: 10.1136/jmg.18.5.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkerk A. J., Pieretti M., Sutcliffe J. S., Fu Y. H., Kuhl D. P., Pizzuti A., Reiner O., Richards S., Victoria M. F., Zhang F. P. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991 May 31;65(5):905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- Warren S. T., Zhang F., Licameli G. R., Peters J. F. The fragile X site in somatic cell hybrids: an approach for molecular cloning of fragile sites. Science. 1987 Jul 24;237(4813):420–423. doi: 10.1126/science.3603029. [DOI] [PubMed] [Google Scholar]
- Yunis J. J., Soreng A. L., Bowe A. E. Fragile sites are targets of diverse mutagens and carcinogens. Oncogene. 1987 Mar;1(1):59–69. [PubMed] [Google Scholar]
- Yunis J. J., Soreng A. L. Constitutive fragile sites and cancer. Science. 1984 Dec 7;226(4679):1199–1204. doi: 10.1126/science.6239375. [DOI] [PubMed] [Google Scholar]