Abstract
Two geometric isomers related to pitiamide A, termed 1E-pitiamide B (1) and 1Z-pitiamide B (2), were isolated from a marine cyanobacterium collected from the shallow reef flat at Piti Bomb Holes, Guam, Mariana Islands. The structures of these analogues were elucidated using 1D and 2D NMR analysis. Pitiamide A, which had been previously described and had not been investigated in bioassays, was co-isolated. Pitiamides A and B were subjected to biological evaluation and they both showed antiproliferative effects on HCT116 cells with IC50 values of 1 – 5 μM. Pitiamide A was investigated individually and caused plasma membrane hyperpolarization and increase of intracellular calcium in HCT116 cells.
Keywords: Marine cyanobacteria, fatty acid amides, pitiamides, antiproliferative activity, membrane hyperpolarization
Introduction
As a rich source of secondary metabolites, marine cyanobacteria usually produce polyketides, polypeptides, hybrid polyketides and polypeptides or fatty acids derivatives with various biological activities, including anti-cancer, anti-microbial, protease inhibitory and neuromodulatory properties. One prevailing lipophilic class of compounds are fatty acid amides, which are featured by the presence of an amide bond in a fatty acid chain and in some cases with incorporated halogen atoms. A number of fatty acid amides have been isolated from marine cyanobacteria over the past two decades, including pitiamide A [1], malyngamides [2], jamaicamides [3], grenadamides [4, 5], hermitamides [6], semiplenamides [7], janthielamide A [8], kimbeamides [8], kalkitoxin [9], taveuniamides [10], besarhanamides [11] and credneramides [12]. Bioactivities associated with these fatty acid amides including cytotoxicity, voltage-gated sodium channel blockage or activation, cannabinoid receptors binding and others.
Pitiamide A was isolated from an extract of a mixed assemblage of Lyngbya majuscula and a Microcoleus sp. found growing on intact colonies of the hard coral Porites cylindrica on Guam [1]. This compound has not been linked to biological activity yet. An analogue of pitiamide A, named pitaimide B, was co-isolated with pitiamide A without full structure elucidation, but was hypothesized to be a mixture of E/Z isomers around the terminal double bond [1]. Here, we describe isolation and structure determination of these elusive isomers, 1E-pitiamide B (1) and 1Z-pitiamide B (2) using bioassay-guided fractionation along with the co-produced pitiamide A. We also describe the multifunctional bioactivities of pitiamides.
Results and Discussion
The cyanobacterium, a recollection of one that previously yielded pitiamide A [1], was collected on the shallow reef flat at Piti Bomb Holes, Guam, Mariana Islands. As previously noted, this iridescent cyanobacterium grows prominently on the tips of corals such as Porites cylindrica. Recently this cyanobacterium has been identified as Hydrocoleum majus [13]. Hydrocoleum is a polyphyletic group, and the taxonomy of marine cyanobacteria is under revision, so it is likely the taxonomic designation for this species will be revised. The nonpolar extract (MeOH-EtOAc, 1:1) was subjected to silica chromatography and various rounds of reversed-phase HPLC to yield pitiamide A, 1E-pitiamide B (1) and 1Z-pitiamide B (2) (Figure 1) using a HCT116 cancer cell growth-inhibition guided fractionation.
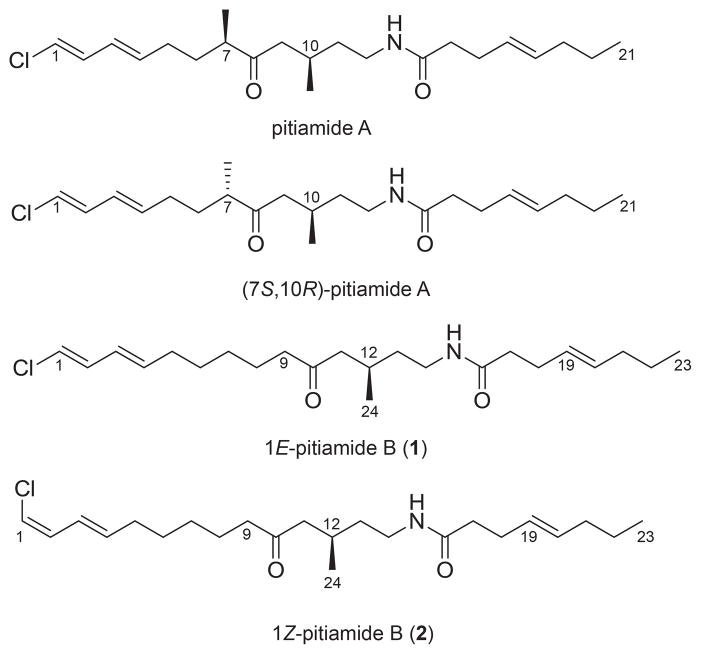
Figure 1.
Structures and absolute configuration of natural product pitiamide A, synthetic (7S,10R)-pitiamide A and 1E-pitiamide B (1) and 1Z-pitiamide B (2).
The HRESIMS spectrum of 1 showed a [M + NH4]+ peak at m/z 413.2926 and an isotopic peak of about one third intensity at m/z 415.2886, indicating the presence of one chlorine atom. The molecular formula of 1 was deduced as C23H38ClNO2, with 5 degrees of unsaturation. NMR analysis (Table 1) suggested its structural similarity to the co-isolated pitiamide A with the presence of a ketone (δC 211.2), an amide or ester linkage (δC 173.0), a terminally chlorinated conjugated diene (δC/δH 118.5/6.08, 133.7/6.40, 126.4/5.96, 135.8/5.68) and an isolated C-C double bond (δC/δH 128.6/5.40, 131.8/5.47).
Table 1.
NMR data for 1E-pitiamide B (1) and 1Z-pitiamide B (2) in CDCl3 (600 MHz)
| Position | 1E-pitiamide B (1) | 1Z-pitiamide B (2) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||
| δH mult. ( J in Hz) | δCa | COSY | HMBC | TOCSY | δH mult. ( J in Hz) | δCa | COSY | HMBC | TOCSY | |
| 1 | 6.08 d (13.1) | 118.5 | 2 | 2, 3 | 2, 3, 4, 5, 6, 7, 8, 9 | 5.88 d (7.0) | 116.22 | 2 | 2, 3, ( 5, 6) | 2, 3 |
| 2 | 6.4 dd (13.1, 10.8) | 133.7 | 1, 3 | 1, 3 | 1, 3, 4, 5, 6, 7 | 6.25 dd (10.3, 7.0) | 129.9 | 1, 3 | 1, 4 | 3, 4, 5, 6, 7, 8 |
| 3 | 5.96 dd (15.2, 10.8) | 126.4 | 2, 4, 5 | 1, 2, 4, 5, 6, 7, 8, 9 | 6.43 ddd (15.3, 10.3, 1.1) | 124.0 | 2, 4, 5 | 1, 2, 5 | 1, 2, 5, 6, 7, 8, 9 | |
| 4 | 5.68 dt (15.2, 7.1) | 135.8 | 3, 5 | 2, 6 | 1, 2, 3, 5, 6, 7, 8, 11a, 11b, 24 | 5.84 dt (15.3, 7.0) | 138.5 | 3, 5 | 2, 5, 6 | 2, 3, 5, 6, 7, 8, 9 |
| 5 | 2.07 m | 32.5 | 4, 6 | 3, 4, 6 | 1, 2, 3, 4, 6, 7, 8 | 2.14 dt (7.5, 7.5) | 32.8 | 3, 4, 6 | 3, 4, 6 | 2, 3, 4, 6, 7, 8, 9 |
| 6 | 1.39 m | 28.9 | 5, 7 | 4, 5 | 3, 4, 5, 7, 8, 9 | 1.42 m | 28.9 | 5, 7 | 4, 5, 8 | 2, 3, 4, 5, 7, 8, 9 |
| 7 | 1.29 m | 29 | 6, 8 | 8 | 3, 4, 5, 6, 8, 9 | 1.29 m | 29.0 | 6, 8 | 5, 8, 9 | 2, 3, 4, 5, 6, 8, 9 |
| 8 | 1.57 m | 23.7 | 7, 9 | 7, 9, 10 | 3, 4, 5, 6, 7, 9 | 1.57 m | 23.7 | 7, 9 | 7, 9, 10 | 2, 3, 4, 5, 6, 7, 9 |
| 9 | 2.38 t (7.4) | 43.8 | 8 | 7, 8, 10 | 6, 7, 8, 12 | 2.38 t (7.4) | 43.8 | 8 | 7, 8, 10 | 4, 6, 7, 8, 12 |
| 10 | 211.2 | 211.2 | ||||||||
| 11a | 2.39 dd (17.0, 7.0) | 49.7 | 11b, 12 | 10 | 11b, 12, 13a, 13b, 14a, 14b, 15, 24 | 2.39 dd (17.0, 7.0) | 49.7 | 11b, 12 | 10, 12, 13, 24 | 11b, 12, 13a, 13b, 14a, 14b, 15, 24 |
| 11b | 2.30 dd (17.0, 7.0) | 49.7 | 11a, 12, | 10 | 11a, 12, 13a, 13b, 14a, 14b, 15, 24 | 2.30 dd (17.0, 7.0) | 49.7 | 11a, 12 | 10, 12, 13, 24 | 11a, 12, 13a, 13b, 14a, 14b, 15, 24 |
| 12 | 2.06 m | 26.5 | 11a, 11b, 13a, 13b, 24 | 24 | 11a, 11b, 14a, 14b, 24 | 2.06 m | 26.5 | 11a, 11b,13a, 13b, 24 | 11, 13, 24 | 11a, 11b, 13a, 13b, 14a, 14b, 15, 24 |
| 13a | 1.43 m | 36.5 | 12, 13b, 14a, 14b | 24 | 13b, 14a, 14b, 24 | 1.43 m | 36.5 | 12, 13b, 14a, 14b | 11, 12, 14, 24 | 11b, 12, 13b, 14a, 14b, 15, 24 |
| 13b | 1.37 m | 36.5 | 12, 13a, 14a, 14b | 24 | 13a, 14a, 14b, 25 | 1.37 m | 36.5 | 13a, 14a, 14b | 11, 12, 14, 24 | 11b, 12, 13a, 14a, 14b, 15, 24 |
| 14a | 3.28 m | 37.4 | 13a, 13b, 14b, 15 | 11a, 11b, 12, 13a, 13b, 14b, 15, 24 | 3.28 m | 37.4 | 13a, 13b, 14b, 15 | 12, 13 | 11a, 11b, 12, 13a, 13b, 14b, 15, 24 | |
| 14b | 3.18 m | 37.4 | 13a, 13b, 14a, 15 | 11a, 11b, 12, 13a, 13b, 14a, 15, 24 | 3.18 m | 37.4 | 13a, 13b, 14a, 15 | 12 | 11a, 11b, 12, 13a, 13b, 14a, 15, 24 | |
| 15 | 5.66 | 14a, 14b | 14a, 14b | 5.68 br s | 14a, 14b | 12, 13a, 13b, 14a, 14b, 24 | ||||
| 16 | 173.0 | 173.0 | ||||||||
| 17 | 2.24 t (7.6) | 36.8 | 18 | 16, 18, 19 | 18, 19, 20, 21 | 2.24 t (7.6) | 36.8 | 18 | 18, 19, 20, 21 | |
| 18 | 2.32 m | 28.8 | 17, 19 | 16, 17, 20 | 17, 19, 20, 21 | 2.32 m | 28.8 | 17, 19 | 16, 17, 19, 20 | 17, 19, 20, 21 |
| 19 | 5.40 dt (15.2, 6.6) | 128.6 | 18, 20 | 17, 18, 20, 21, 22, 23 | 5.40 dt (15.2, 6.7) | 128.6 | 18, 20 | 18, 21 | 17, 18, 20, 21, 22, 23 | |
| 20 | 5.47 dt (15.2, 6.6) | 131.8 | 19, 21 | 17, 18, 19, 21, 22, 23 | 5.47 dt (15.2, 6.7) | 131.8 | 19, 21 | 18 | 17, 18, 19, 21, 22, 23 | |
| 21 | 1.95 dt (7.3, 7.2) | 34.8 | 20, 22 | 20, 19, 22, 23 | 17, 18, 19, 20, 22, 23 | 1.95 dt (7.3, 7.2) | 34.8 | 20, 22 | 19, 20, 22, 23 | 17, 18, 19, 20, 22, 23 |
| 22 | 1.34 m | 22.5 | 21, 23 | 20, 21, 23 | 19, 20, 21, 23 | 1.34 m | 22.5 | 21, 23 | 20, 21, 23 | 19, 20, 21, 23 |
| 23 | 0.87 t (7.3) | 13.8 | 22 | 21, 22 | 17, 18, 19, 20, 21, 22 | 0.87 t (7.3) | 13.8 | 22 | 21, 22 | 17, 18, 19, 20, 21, 22 |
| 24 | 0.92 d (6.7) | 20.2 | 12 | 11, 12, 13 | 11a, 11b, 12, 13a, 13b, 14a, 14b, 15 | 0.92 d (6.7) | 20.2 | 12 | 11, 13, 12 | 11a, 11b, 12, 13a, 13b, 14a, 14b, 15 |
Deduced from HSQC and HMBC experiments
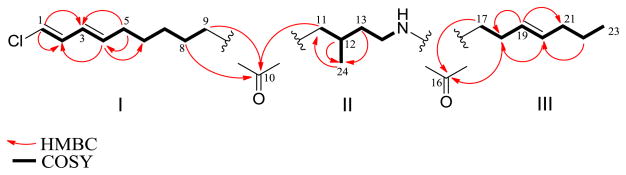
Compared to pitiamide A, 1 is a homologue of pitiamide A with a 14 amu higher molecular weight corresponding to additional methyl or methylene group. However, closer inspection of the 1H NMR revealed the presence of only one methyl doublet and one methyl triplet, while pitiamide A has three methyl groups including one terminal methyl group (two doublets and one triplet). The absence of one methyl branch in 1 suggested the presence of two additional methylenes, which are needed to account for MS-based molecular formula. The 3JH,H coupling constants for H-1/H-2 (13.1 Hz), H-2/H-3 (10.8 Hz), H-3/H-4 (15.2 Hz) suggested the same E,E configuration of the conjugated diene as pitiamide A. Interpretation of COSY data revealed partial structure I (Figure 2) which has five consecutive methylene groups (δH 2.07, 1.39, 1.29, 1.57, 2.38) adjacent to the terminally chlorinated conjugated diene. The connection of partial structure I was also confirmed by HMBC correlations. Partial structure II (Figure 2) was assigned based on COSY and HMBC correlations. The HMBC correlations between H-11, H-12, H-13 and C-24 supported the attachment of the methyl group (δC/δH 20.2/0.92) to the methine (δC/δH 26.5/2.06) in partial structure II. The 1H NMR and 13C NMR chemical shifts for C-14/H-14 (δC/δH 37.4/3.18, 3.28) indicated its direct attachment to a nitrogen atom. The 3JH,H coupling constants for H-19/H-20 (15.2 Hz) suggested an E configuration of the C-C double bond. COSY correlations combined with HMBC correlations established partial structure III (Figure 2). HMBC correlations between H-8, H-9, H-11 and C-10 (δC 211.2) constructed the linkage of partial structures I and II through a keto carbonyl group (δC 211.2). HMBC correlations between H-17, H-18 and C-16 linked partial structures II and III through an amide bond (δC 173.0).
Figure 2.
Partial structures of 1E-pitiamide B (1) with key COSY and HMBC correlations.
The same HRESIMS pattern was obtained for compound 2 with a [M + NH4]+ peak at m/z 413.2943 and an isotopic peak of about one third intensity at m/z 415.2924. A same molecular formula of 2 was deduced as C23H38ClNO2, with 5 degrees of unsaturation. Further interpretation of the COSY, HSQC, HMBC and TOSCY spectra indicated that 2 has an identical planar structure with 1. The 3JH,H coupling constants, however, revealed that the conjugated diene side chain of 2 has a different configuration from that of 1. The 3JH,H coupling constants for H-1/H-2 (7.0 Hz), H-2/H-3 (10.3 Hz), H-3/H-4 (15.3 Hz) suggested a Z,E configuration of the conjugated diene.
The absolute configuration was proposed as 12R for both 1 and 2 on the basis of the additivity rule of specific rotation [14]. Compounds 1 and 2 have very close structures with pitiamide A with one less stereocenter. The experimental specific rotations of (7S,10R)- and (7R,10R)- isomers of pitiamide A were reported as: [α]D +11.6° (c 0.49, CHCl3) and [α]D −30.3° (c 0.71, CHCl3) respectively [15] (Figure 1). In general, stereocenters more than three atoms removed from each other have negligible mixed-term contributions to the rotation of the plane of polarized light, as long as the rotation of the chain can occur freely [14]. The contribution of the α-ketone stereocenter at C-7 should therefore be about +20° for S and −20° for R configuration calculated on the basis of the additivity rule. These calculated results are also consistent with the observed specific rotation of other model compounds with an α-ketone stereocenter (Table 1S, Supporting Information). Therefore, the β-ketone stereocenter of pitiamide A at C-10 should have about −10° contribution for the R configuration. In contrast to pitamide A, 1 and 2 only have one β-ketone stereocenter at C-12. Compounds 1 and 2 have the same partial structure in three atom distance from the β-ketone stereocenter at C-12 as pitiamide A; we could therefore conclude that R configuration at C-12 of 1 and 2 should have contributed around −10° to their specific rotation, while S should have contributed around +10°. The experimental specific rotation measurement for 1 and 2 was [α]D20 −6.3° (c 0.011, CDCl3) and [α]D20 −8.8° (c 0.020, CDCl3) respectively, which is very close to the one we proposed for 12R-1 and 12R-2.
Pitiamide A was reported in 1997 and isolated with pitiamide B as a mixture of unstable isomers which differ in the configuration of the chlorinated olefin. Pitiamide B had the same molecular weight as 1and 2 evidenced by EIMS data: m/z 395, 6.7% rel. abundance [M]+; 397, 2.5% rel. abundance [M+2]+. Comparison of the NMR spectra of the original pitiamide B with spectra of 1 and 2 suggested that pitiamide B is the mixture of 1 and 2. Therefore, we refer to 1 and 2 as 1E-pitiamide B and 1Z-pitiamide B, respectively.
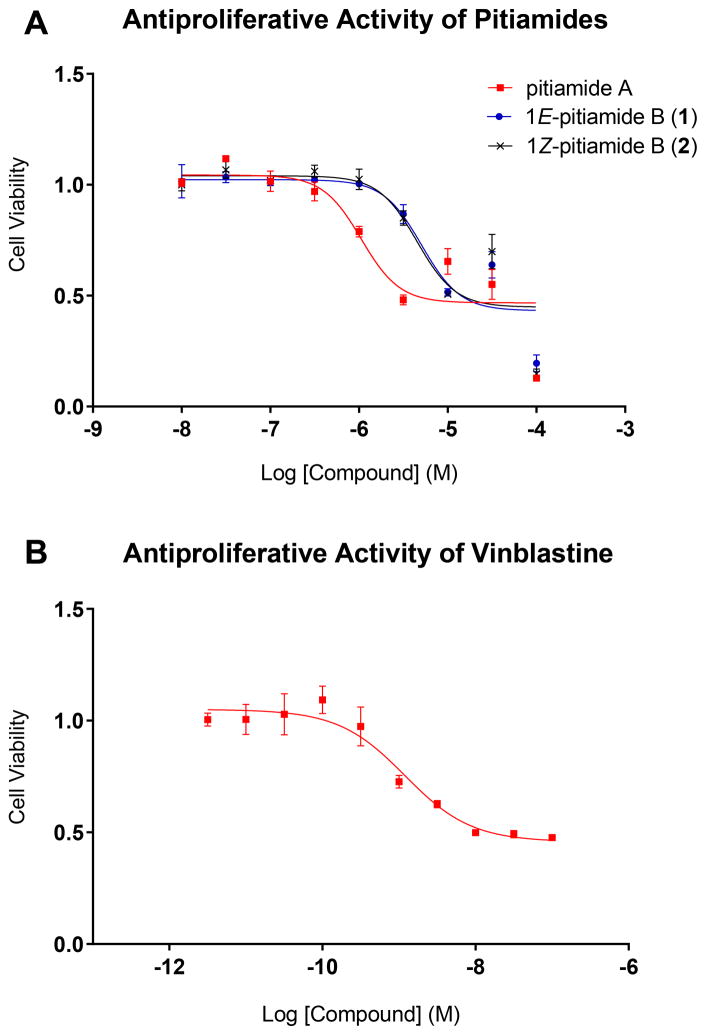
Pitiamide A, 1E-pitiamide B (1) and 1Z-pitiamide B (2) were evaluated for their antiproliferative effects towards HCT116 colorectal cancer cells, with vinblastine (IC50 = 1.2 nM) as a positive control. The IC50 values are 1.1 μM, 5.1 μM and 4.5 μM, respectively (Figure 3). Interestingly, the dose-response curves of pitiamides B were consistently rightward shifted by 4 to 5 fold compared with that of pitiamide A, suggesting a structure-activity relationship between the pitiamide compounds. Either the absence of α-ketone methyl or the increase in length of the methylene chain caused a shift of IC50 to a higher value.
Figure 3.
A) Antiproliferative activity of pitiamides on HCT116 cells. B) Antiproliferative activity of vinblastine on HCT116 cells. Vinblastine was used as a positive control.
Many fatty acid amides from marine cyanobacteria exhibit various neuromodulatory activities: voltage-gated sodium channel blocking activity (jamaicamides [3], janthielamide A, kimbeamides [8], kalkitoxin [9]), cannabinoid receptor binding activity (grenadamides [5], semiplenamides [7]), spontaneous calcium oscillations (credneramides [12]) and others, probably owing to their fatty acid structural features which may preferentially interact with cell membrane compartments and modulate ion channels or receptors on cell membranes.
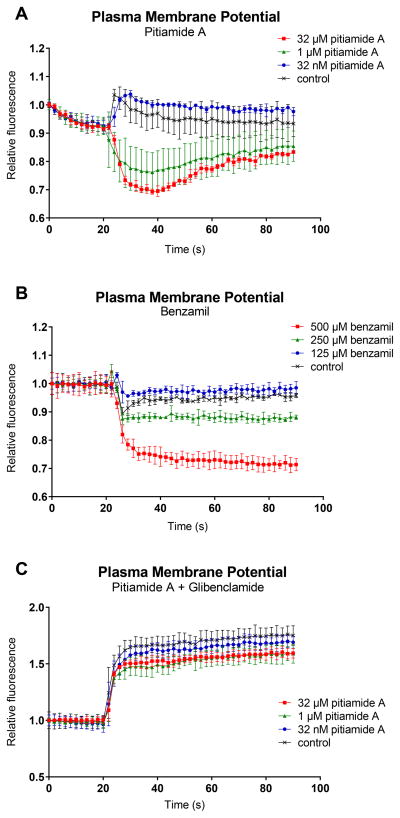
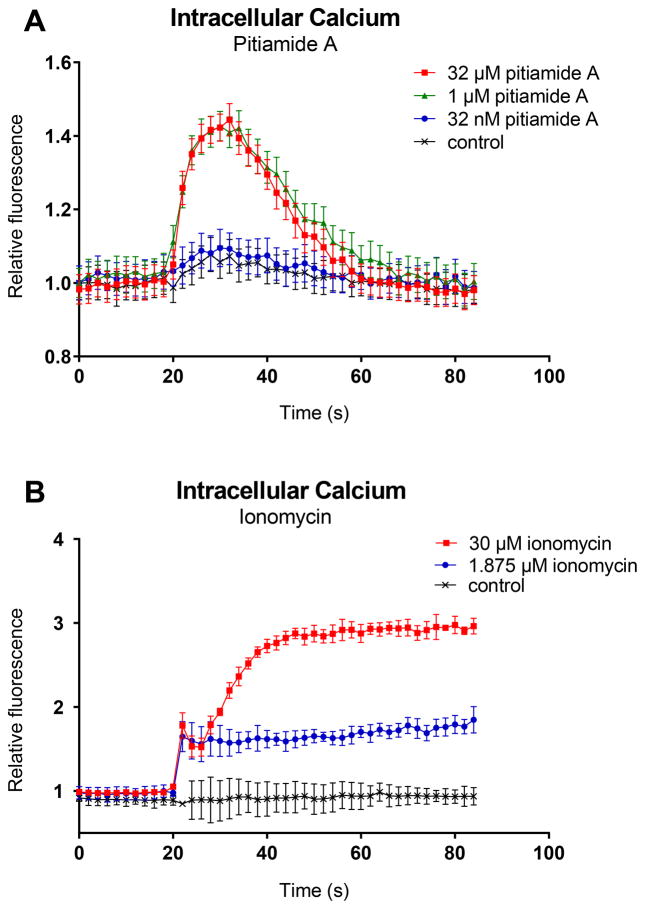
Consequently, pitiamide A was further evaluated for its effects on plasma membrane potential and intracellular calcium in HCT116 cells. We subjected pitiamide A to a fluorescence-based plasma membrane potential assay and intracellular calcium assay. The ability of acute exposure to pitiamide A to perturb plasma membrane potential of HCT116 cells was assessed using a fluorescent dye that was sensitive to plasma membrane potential. In this assay, hyperpolarization and depolarization of the plasma membrane are indicated by a decrease and increase in intracellular fluorescence, respectively. Acute pitiamide A exposure caused plasma membrane hyperpolarization at 32 μM and 1 μM, with no effect being observed at 32 nM (Figure 4A). Benzamil, an epithelial sodium channel blocker, was used as a positive control for plasma membrane hyperpolarizing effect (Figure 4B) [19]. Glibenclamide (Glyburide) is a potassium channel regulatory subunit inhibitor and causes membrane depolarization in a variety of cell types by preventing potassium influx [16, 17, 18]. In HCT116 cells, 12.5 μM glibenclamide induced depolarizing effect, while the pre-treatment of pitiamide A showed a dampened response by 10% (Figure 4C). The possible reason of this slight dampened effect caused by pitiamide A exposure could be that pitiamide A blocks the epithelial sodium channel and has negligible effect on the voltage-gated sodium channel. Consistent with the membrane potential changes, acute pitiamide A exposure also caused a rapid increase of intracellular calcium at 32 μM and 1 μM but not 32 nM (Figure 5A). We used the chelating calcium ionophore ionomycin as a positive control (Figure 5B). Ionomycin allows calcium to diffuse passively through cellular membranes [20].
Figure 4.
Measurement of plasma membrane potential in HCT116 cells after treatment of pitiamide A. Plasma membrane potential was monitored using FLIPR Membrane Potential Assay Kits (Molecular Devices). Hyperpolarization and depolarization of the plasma membrane are indicated by a decrease and increase in intracellular fluorescence respectively. A) Fluorescence traces were recorded in single wells using FlexStation3 (Molecular Devices) from 0 to 90 seconds in HCT116 cells, while pitiamide A at final concentrations of 32 μM, 1 μM, 32 nM or 1% EtOH was added at 20 s. B) Benzamil was used as a positive control for plasma membrane hyperpolarization effect in HCT116 cells. Benzamil at final concentrations of 500 μM, 250 μM, 125 μM or 1% EtOH was added to cells at 20 s. C) HCT116 cells were pre-treated with pitiamide A at final concentrations of 32 μM, 1 μM, 32 nM or 1% EtOH before glibenclamide (Glyburide) at final concentration of 12.5 μM was added at 20 s.
Figure 5.
Measurement of intracellular calcium in HCT116 cells after treatment of pitiamide A. Intracellular calcium was measured using FLIPR Calcium 6-QF Assay Kits (Molecular Devices). The increase of intracellular calcium was indicated by the increase of florescence intensity. A) Fluorescence traces were recorded using FlexStation3 (Molecular Devices) from 0 to 84 seconds, while pitiamide A at final concentrations of 32 μM, 1 μM, 32 nM or 1% EtOH was added at 20 s. B) Ionomycin was used as a positive control for intracellular calcium increasing effect. Ionomycin at final concentrations of 30 μM, 1.875 μM or 1% EtOH was added to cells at 20 s.
In conclusion, we identified two new pitiamide A analogues from marine cyanobacteria using bioassay-guided fractionation. Pitiamide A and its two analogues had antiproliferative effect on HCT116 cells. Pitiamide A was tested individually and caused membrane hyperpolarization and increase of intracellular calcium in HCT116. However, further studies on more relevant cell types are needed to elucidate pitiamide A’s effect on cell membrane compartments and ion channels. Also, further structure-activity relationship studies of pitiamides are warranted.
Materials and Methods
Chemicals
Benzamil (purity >98%) was purchased from Sigma Aldrich. Ionomycin (purity >97%), glibenclamide (purity >98%) and vinblastine (purity >98%) were purchased from Calbiochem.
General Experimental Procedures
Optical rotations were measured on a Perkin-Elmer 341 polarimeter. 1H and 2D NMR spectra were recorded in CDCl3 on Bruker Advance II 600 MHz spectrometer equipped with a 5 mm TXI cryogenic probe using residual solvent signals (δH 7.26; δC 77.0 ppm, CDCl3) as internal standards. HSQC and HMBC experiments were optimized for 1JCH = 145 and nJCH = 7 Hz, respectively. HRESIMS data were obtained using an Agilent LC-TOF mass spectrometer equipped with an APCI/ESI multimode ion source detector.
Extraction and Isolation
The sample VPG 14–38 was collected on May 31, 2014 in 1 m water on the shallow reef flat at Piti Bomb Holes, Guam, Mariana Islands. The sample was a recollection of one that previously yielded pitiamide A, and was previously described as predominately Lyngbya majuscula [1]. Recently this cyanobacterium has been identified as Hydrocoleum majus [13]. A voucher specimen of VPG 14–38 has been retained at the Smithsonian Marine Station. The freeze dried sample (10.26 g) was extracted with MeOH-EtOAc (1:1) (2 × 300 mL, each for 24 h) and the extract (2.4 g) was suspended in water (50 mL) and partitioned with EtOAc (3 × 50 mL). The EtOAc soluble part (246.6 mg) was then subjected to silica column (230–400 mesh, i.d. 2.5 × 40 cm, 50 g) using the gradient of hexane (300 mL), 30% EtOAc in hexane (300 mL), EtOAc (300 mL), 10% MeOH in EtOAc (300 mL) and EtOAc/MeOH 1:1 (300 mL) to elute five fractions. The EtOAc fraction (49.41 mg) was further subjected to semipreparative HPLC (Phenomenex Luna C18, 250 × 10 mm, 5μm, 2.0 mL/min; PDA detection) using an acetonitrile-H2O linear gradient (40–100% acetonitrile for 10 min and 100 % acetonitrile for 25 min). Fractions were pooled on the basis of retention times, 1H NMR analysis and low-resolution MS measurements to afford pitiamide A (tR 17.8 min, 0.92 mg) and a mixture of 1E-pitiamide B (1) and 1Z-pitiamide B (2) (tR 18.4–19.1 min, 0.87 mg). The mixture of pitiamide B was further purified by analytical HPLC (Phenomenex Kinetex 2.6 μm PFP, 150 × 4.60 mm, 1.0 mL/min; PDA detection) using an acetonitrile-H2O linear gradient (50% acetonitrile for 10 min, 50–100% acetonitrile for 20 min and 100% acetonitrile for 5 min) to yield 1E-pitiamide B (1) (tR 17.6 min, 0.12 mg) and 1Z-pitiamide B (2) (tR 16.9 min, 0.22 mg).
1E-pitiamide B (1): [α]D20 −6.3° (c 0.011, CDCl3); 1H NMR, COSY, TOCSY, HSQC and HMBC data, see Table 1; UV (MeOH) λmax (log ε) 235 (5.52); HRESIMS m/z [M + NH4]+ 413.2926 (calcd for C23H4235ClN2O2 413.2935), 415.2886 (calcd for C23H4237ClN2O2 415.2905) (3:1 [M + NH4]+ ion cluster)
1Z-pitiamide B (2): [α]D20 −8.8° (c 0.020, CDCl3); 1H NMR, COSY, TOCSY, HSQC and HMBC data, see Table 1; UV (MeOH) λmax (log ε) 235 (5.56); HRESIMS m/z [M + NH4]+ 413.2943 (calcd for C23H4235ClN2O2 413.2935), 415.2924 (calcd for C23H4237ClN2O2 415.2905) (3:1 [M + NH4]+ ion cluster)
Cell Viability Assay
HCT116 cells were cultured in Dulbecco’s modified Eagle medium (DMEM, Invitrogen) supplemented with 10% fetal bovine serum (FBS, Hyclone) under a humidified environment with 5% CO2 at 37 °C. HCT116 (10,000) cells were seeded in 96-well plates. Cells were treated with a series of concentrations of pitiamide A or its analogues dissolved in EtOH, 24 h postseeding. Cells were incubated for an additional 48 h before the addition of the MTT reagent. Cell viability was measured according to the manufacturer’s instructions (Promega, Madison, WI, USA). Treatments were done in triplicate. Nonlinear regression analysis was carried out using GraphPad Prism software for IC50 value calculations.
Membrane potential assay and intracellular calcium assay
HCT116 cells were maintained in DMEM medium supplemented with 10% FBS. On the day of the assay, cells were removed from plates with 0.05% trypsin, diluted in DMEM medium supplemented with 10% FBS before being seeded into a poly-D-lysine coated black clear bottom 96-well plate at a density of 2.5 × 104 cells per well and allowed to attach for 6 hours at 37 °C. Medium was removed and replaced with 45 μl of Hank’s balanced salt solution (HBSS) containing 20 mM HEPES (pH 7.4). The intracellular calcium assay and plasma membrane potential FLIPR reagents (Molecular Devices) were reconstituted according to the manufacturer’s instructions and 45 μl of each was added to separate wells. Prior to addition of the compounds by the FlexStation3, cells were incubated in the presence of the plasma membrane potential reagent for 1 h and the intracellular calcium assay reagent for 2 h at 37 °C.
Supplementary Material
Acknowledgments
This research was supported by the National Institutes of Health, NCI grant R01CA172310. We are grateful to L. Spiers for assistance with collection of the cyanobacterium and J. Biggs for general support on Guam.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Table S1, 1D NMR and 2D NMR spectra of compound 1, 2 and pitiamide A are available as Supporting Information.
References
- 1.Nagle DG, Park PU, Paul VJ. Pitiamide A, a new chlorinated lipid from a mixed marine cyanobacterial assemblage. Tetrahedron Lett. 1997;38:6969–6972. [Google Scholar]
- 2.Tan LT. Marine cyanobacteria: A treasure trove of bioactive secondary metabolites for drug discovery. In: Rahman A, editor. Studies in natural products chemistry. Elsevier; 2012. pp. 67–110. [Google Scholar]
- 3.Edwards DJ, Marquez BL, Nogle LM, McPhail K, Goeger DE, Roberts MA, Gerwick WH. Structure and biosynthesis of the jamaicamides, new mixed polyketide-peptide neurotoxins from the marine cyanobacterium Lyngbya majuscula. Chem Biol. 2004;11:817–833. doi: 10.1016/j.chembiol.2004.03.030. [DOI] [PubMed] [Google Scholar]
- 4.Jiménez JI, Vansach T, Yoshida WY, Sakamoto B, Pörzgen P, Horgen FD. Halogenated fatty acid amides and cyclic depsipeptides from an eastern caribbean collection of the cyanobacterium Lyngbya majuscula. J Nat Prod. 2009;72:1573–1578. doi: 10.1021/np900173d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sitachitta N, Gerwick WH. Grenadadiene and grenadamide, cyclopropyl-containing fatty acid metabolites from the marine cyanobacterium Lyngbya majuscula. J Nat Prod. 1998;61:681–684. doi: 10.1021/np970576a. [DOI] [PubMed] [Google Scholar]
- 6.Tan LT, Okino T, Gerwick WH. Hermitamides A and B, toxic malyngamide-type natural products from the marine cyanobacterium Lyngbya majuscula. J Nat Prod. 2000;63:952–955. doi: 10.1021/np000037x. [DOI] [PubMed] [Google Scholar]
- 7.Han B, McPhail KL, Ligresti A, Di Marzo V, Gerwick WH. Semiplenamides A-G, fatty acid amides from a Papua New Guinea collection of the marine cyanobacterium Lyngbya semiplena. J Nat Prod. 2003;66:1364–1368. doi: 10.1021/np030242n. [DOI] [PubMed] [Google Scholar]
- 8.Nunnery JK, Engene N, Byrum T, Cao Z, Jabba SV, Pereira AR, Matainaho T, Murray TF, Gerwick WH. Biosynthetically intriguing chlorinated lipophilic metabolites from geographically distant tropical marine cyanobacteria. J Nat Prod. 2012;77:4198–4208. doi: 10.1021/jo300160e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu M, Okino T, Nogle LM, Marquez BL, Williamson RT, Sitachitta N, Berman FW, Murray TF, McGough K, Jacobs R, Colsen K, Asano T, Gerwick WH. Structure, synthesis, and biological properties of Kalkitoxin, a novel neurotoxin from the marine cyanobacterium Lyngbya majuscula. J Am Chem Soc. 2000;122:12041–12042. [Google Scholar]
- 10.Williamson RT, Singh IP, Gerwick WH. Taveuniamides: new chlorinated toxins from a mixed assemblage of marine cyanobacteria. Tetrahedron. 2004;60:7025–7033. [Google Scholar]
- 11.Tan LT, Chang YY, Ashootosh T. Besarhanamides A and B from the marine cyanobacterium Lyngbya majuscula. Phytochemistry. 2008;69:2067–2069. doi: 10.1016/j.phytochem.2008.04.021. [DOI] [PubMed] [Google Scholar]
- 12.Malloy KL, Suyama TL, Engene N, Debonsi H, Cao Z, Matainaho T, Spadafora C, Murray TF, Gerwick WH. Credneramides A and B: neuromodulatory phenethylamine and isopentylamine derivatives of a vinyl chloride-containing fatty acid from cf. Trichodesmium sp. nov. J Nat Prod. 2011;75:60–66. doi: 10.1021/np200611f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palińska KA, Abed RM, Charpy L, Langlade MJ, Beltrán-Magos Y, Golubic S. Morphological, genetic and physiological characterization of Hydrocoleum, the most common benthic cyanobacterium in tropical oceans. Eur J Phycol. 2015;50(2):139–154. [Google Scholar]
- 14.Kondru RK, Lim S, Wipf P, Beratan DN. Synthetic and model computational studies of molar rotation additivity for interacting chiral centers: A reinvestigation of van’t Hoff’s principle. Chirality. 1997;9:469–477. doi: 10.1002/(SICI)1520-636X(1997)9:5/6<469::AID-CHIR13>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 15.Ribe S, Kondru RK, Beratan DN, Wipf P. Optical rotation computation, total synthesis, and stereochemistry assignment of the marine natural product pitiamide A. J Am Chem Soc. 2000;122:4608–4617. [Google Scholar]
- 16.Ashcroft FM, Rorsman P. Electrophysiology of the pancreatic β-cell. Prog Biophys Mol Bio. 1989;54:87–143. doi: 10.1016/0079-6107(89)90013-8. [DOI] [PubMed] [Google Scholar]
- 17.Babenko AP, Aguilar-Bryan L, Bryan J. A view of sur/KIR6, X, KATP channels. Annu Rev Physiol. 1998;60:667–687. doi: 10.1146/annurev.physiol.60.1.667. [DOI] [PubMed] [Google Scholar]
- 18.Reeve HL, Vaughan PF, Peers C. Glibenclamide inhibits a voltage-gated K+ current in the human neuroblastoma cell line SH-SY5Y. Neurosci Lett. 1992;135:37–40. doi: 10.1016/0304-3940(92)90130-y. [DOI] [PubMed] [Google Scholar]
- 19.Kleyman TR, Yulo T, Ashbaugh C, Landry D, Cragoe E, Karlin A, Al-Awqati Q. Photoaffinity labeling of the epithelial sodium channel. J Biol Chem. 1986;261:2839–2843. [PubMed] [Google Scholar]
- 20.Müller MS, Obel LF, Waagepetersen HS, Schousboe A, Bak LK. Complex actions of ionomycin in cultured cerebellar astrocytes affecting both calcium-induced calcium release and store-operated calcium entry. Neurochem Res. 2013;38:1260–1265. doi: 10.1007/s11064-013-1021-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.