ABSTRACT
The UL16 tegument protein of herpes simplex virus 1 (HSV-1) is conserved among all herpesviruses and plays many roles during replication. This protein has an N-terminal domain (NTD) that has been shown to bind to several viral proteins, including UL11, VP22, and glycoprotein E, and these interactions are negatively regulated by a C-terminal domain (CTD). Thus, in pairwise transfections, UL16 binding is enabled only when the CTD is absent or altered. Based on these results, we hypothesized that direct interactions occur between the NTD and the CTD. Here we report that the separated and coexpressed functional domains of UL16 are mutually responsive to each other in transfected cells and form complexes that are stable enough to be captured in coimmunoprecipitation assays. Moreover, we found that the CTD can associate with itself. To our surprise, the CTD was also found to contain a novel and intrinsic ability to localize to specific spots on mitochondria in transfected cells. Subsequent analyses of HSV-infected cells by immunogold electron microscopy and live-cell confocal imaging revealed a population of UL16 that does not merely accumulate on mitochondria but in fact makes dynamic contacts with these organelles in a time-dependent manner. These findings suggest that the domain interactions of UL16 serve to regulate not just the interaction of this tegument protein with its viral binding partners but also its interactions with mitochondria. The purpose of this novel interaction remains to be determined.
IMPORTANCE The HSV-1-encoded tegument protein UL16 is involved in multiple events of the virus replication cycle, ranging from virus assembly to cell-cell spread of the virus, and hence it can serve as an important drug target. Unfortunately, a lack of both structural and functional information limits our understanding of this protein. The discovery of domain interactions within UL16 and the novel ability of UL16 to interact with mitochondria in HSV-infected cells lays a foundational framework for future investigations aimed at deciphering the structure and function of not just UL16 of HSV-1 but also its homologs in other herpesviruses.
KEYWORDS: domains, HSV-1, herpes simplex virus, mitochondria, tegument, UL16
INTRODUCTION
Herpes simplex virus 1 (HSV-1) contains a 373-amino-acid tegument protein, UL16 (Fig. 1), which is conserved among all families of herpesviruses (1–15). Encoded by the late-expressing 16th open reading frame in the unique long segment of the viral genome, UL16 initially localizes to both the nucleus and the cytoplasm of the infected cell but later accumulates in the cytoplasm (4, 16). There, a population of UL16 is found to be stably associated with an unknown partner on cytoplasmic capsids (16, 17), where it plays a role in envelopment by providing bridging functions via its interaction with at least three viral membrane proteins: UL11, glycoprotein E (gE), and VP22 (12, 18–43). Thus, UL16-null mutants produce only 1/10 the number of wild-type virions (1, 12), and electron microscopy of cells infected with such a mutant revealed an accumulation of partially wrapped capsids in the cytoplasm (12). However, UL16 also participates in other parts of the replication cycle, including cell-to-cell spread, syncytium formation, rearrangements of the tegument upon binding of the virus to the host cell, and unknown events in the nucleus (26, 44).
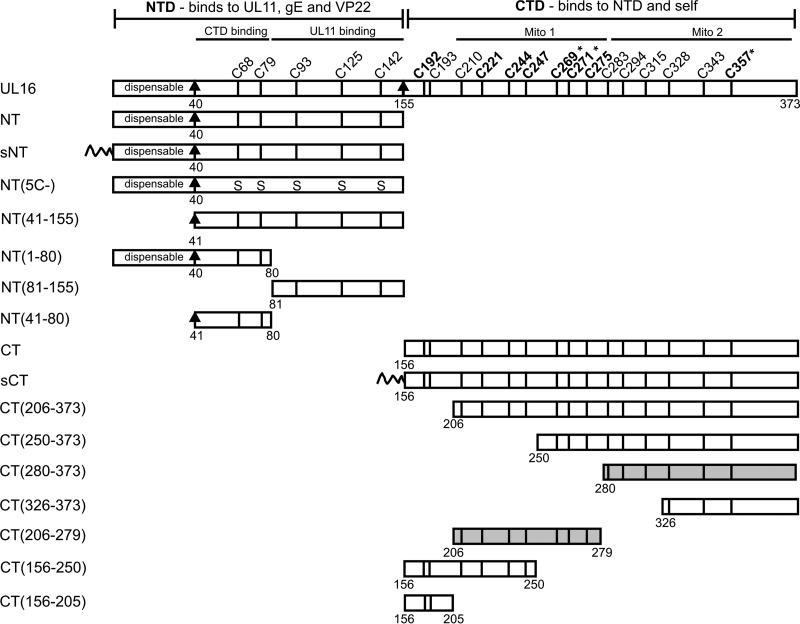
FIG 1.
UL16 and the deletion mutants used in this study. The N- and C-terminal domains of UL16 are shown, and mutants expressing each of these are referred to by “NT” and “CT” names. The first 40, nonconserved residues are dispensable for binding to UL11, gE, and VP22. Positions and residue numbers of the 20 cysteines of UL16 are indicated. The eight conserved cysteines reside in the regulatory CTD and are shown in bold. The five nonconserved cysteines in the NTD were replaced with serines to generate the NT(5C−) mutant. The first 10 amino acids of the Src oncoprotein, which act as a membrane-targeting sequence (shown as a wavy line), were added to the N-terminal ends of NT and CT to generate sCT and sNT. Asterisks indicate the three cysteines that each activate binding of full-length UL16 to UL11 when changed to serine. Two subregions of the CTD that are sufficient for mitochondrial targeting (Mito 1 and Mito 2) are shaded.
Binding of UL16 to its three membrane partners occurs via an N-terminal domain (NTD) (Fig. 1), which maps to the first 155 amino acids (36); however, each of these interactions is inefficient when full-length UL16 is used in pairwise transfections. Deletion of amino acids 156 to 373 dramatically enhances each of the three interactions, suggesting the presence of a C-terminal domain (CTD) (Fig. 1) that negatively regulates binding (12, 36, 37). Consistent with this, single-amino-acid changes within the CTD of full-length UL16 also activate binding to UL11, although they do not stimulate binding to gE and VP22 (12, 36, 37). Moreover, UL16 binds directly to UL21, and although the sites of contact are unknown, this interaction also activates binding to UL11 but, again, not to gE (26, 45–50). Thus, it seems that UL16 has at least two functional domains: an NTD that mediates binding to partners on the membrane and a CTD that regulates those interactions in a complicated manner (36).
The initial purpose of the experiments described here was to test the hypothesis that the NTD and CTD of UL16 interact directly with one another. The results strongly support this and also revealed the ability of the CTD to interact with itself. In the course of these experiments, we unexpectedly discovered that the CTD has an intrinsic ability to accumulate on mitochondria in transfected cells, while the full-length, wild-type protein transiently associates with mitochondria in HSV-1-infected cells. Thus, the domain interactions of UL16 serve to regulate not just the interaction of this tegument protein with its viral binding partners but also its interactions with mitochondria.
RESULTS
Colocalization of independently expressed N- and C-terminal domains of UL16.
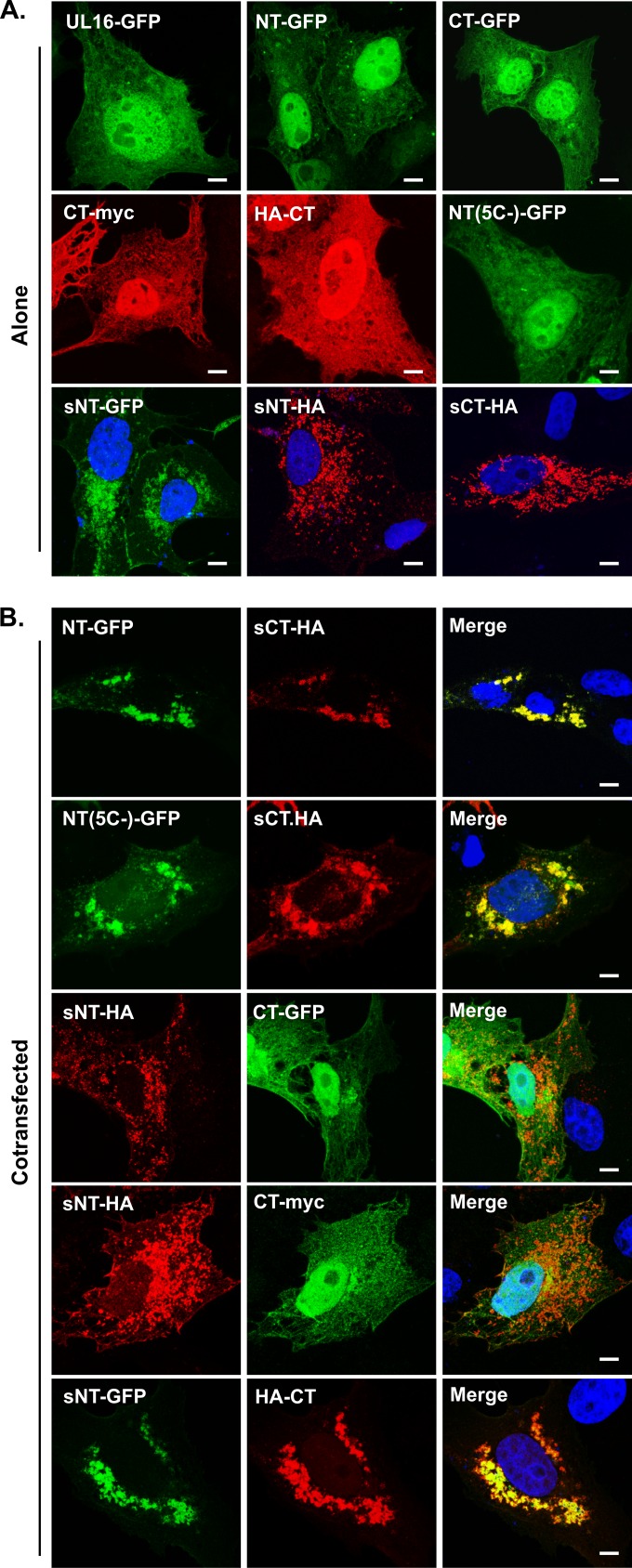
To test for interactions between the N- and C-terminal domains of UL16 (Fig. 1), plasmids encoding tagged versions (NT and CT constructs) were transfected either alone or together into Vero cells. Whether they were expressed individually or coexpressed, the NT-GFP and CT constructs (tagged with a hemagglutinin [HA] or myc epitope) were distributed throughout the cytoplasm and nuclei of transfected cells (data not shown), similarly to full-length UL16 (Fig. 2A). The identical localization patterns of the two domains obscured the visualization of any interactions; to circumvent this problem, the first 10 amino acids of the Src oncoprotein, which provide a membrane-targeting motif, were attached to the N terminus of CT-HA to generate sCT-HA. The Src peptide is well known to relocalize a variety of proteins to membranes (28, 36, 51–55). Accordingly, sCT-HA was not present in the nucleus but accumulated massively on punctate-looking cytoplasmic membranes (Fig. 2A, bottom right panel) and to some extent on the plasma membrane (not shown). In support of domain interactions, a dramatic relocalization of NT-GFP to sCT-HA was observed in cells expressing both constructs (Fig. 2B, top row). In particular, NT-GFP lost its diffuse cytoplasmic and nuclear localization in these cells. Moreover, in the presence of NT-GFP, the localization pattern of sCT-HA also changed from scattered punctate structures in the cytoplasm to more compact and slightly globule-appearing structures in the juxtanuclear region, suggesting that both domains are mutually responsive to each other. Immunostaining with antibodies against the aggresome marker vimentin revealed that these globular structures where NT and CT colocalize are not uninteresting protein aggregates (data not shown). To ascertain whether the green fluorescent protein (GFP) tag itself provided the interaction with the C-terminal domain, a plasmid encoding only this tag was used, and no relocalization of GFP to sCT-HA was observed (not shown), suggesting that the relocalization of NT and sCT-HA is indeed specific. Similarly, coexpression with UL16-GFP resulted in no change in sCT-HA localization (not shown). Together, these results confirmed that the relocalization of NT-GFP and sCT-HA to globular structures in the juxtanuclear region is not merely due to overexpression of two different proteins in the same cell.
FIG 2.
Colocalization of the N- and C-terminal domains of UL16 in mammalian cells. Vero cells were transfected to express the indicated domains of UL16 either alone (A) or in combinations (B). At 16 to 20 h posttransfection, the cells were fixed, and the expressed fusion proteins were revealed either by GFP fluorescence (green) or by staining with antibodies specific for the myc or HA tag (red). Nuclei were stained with DAPI (blue). The images were obtained by confocal microscopy. See Fig. 1 for details of the UL16 constructs. Bars, 3.8 μm.
The presence of multiple cysteines in UL16 (5 in NT and 15 in CT) (Fig. 1) raised the possibility of a disulfide-linked interaction between the NT and CT domains. To address this, an NT-GFP chimera lacking all five cysteines, named NT(5C−) (36), was coexpressed with sCT-HA. Confocal microscopy revealed a massive relocalization of NT(5C−)-GFP to intracellular sites where sCT-HA accumulated (Fig. 2B, row 2). The diffuse cytoplasmic and nuclear localization pattern of NT(5C−)-GFP seen when it was expressed individually (Fig. 2A) was completely lost in the presence of CT. These results show that interdomain interactions do not involve disulfide bonds or strictly require the presence of the five nonconserved cysteines in the N-terminal domain.
We also tried adding the Src membrane-binding peptide to the N-terminal domain, thereby creating sNT-HA. When expressed by itself, this derivative was found to be absent from the nucleus and accumulated on cytoplasmic and plasma membranes (Fig. 2A). However, the C-terminal constructs CT-GFP and CT-myc could not be relocalized to this Src chimera (Fig. 2B, rows 3 and 4). When the GFP tag was moved to the N-terminal domain, the resulting construct, sNT-GFP, was found to cause substantial relocalization of HA-CT (Fig. 2B, row 5) or CT-myc (not shown) in ∼60% of the cotransfected cells; neither CT derivative was found in the nucleus. From these data, it appears that the presence of GFP on the N-terminal domain either stabilizes the interaction with CT or helps to expose the region of NT that binds to CT. Further studies are needed to distinguish between these possibilities. In any case, it is clear from these results that the N- and C-terminal domains of UL16 can respond to each other when coexpressed in the same cell.
Coimmunoprecipitation of the N- and C-terminal domains.
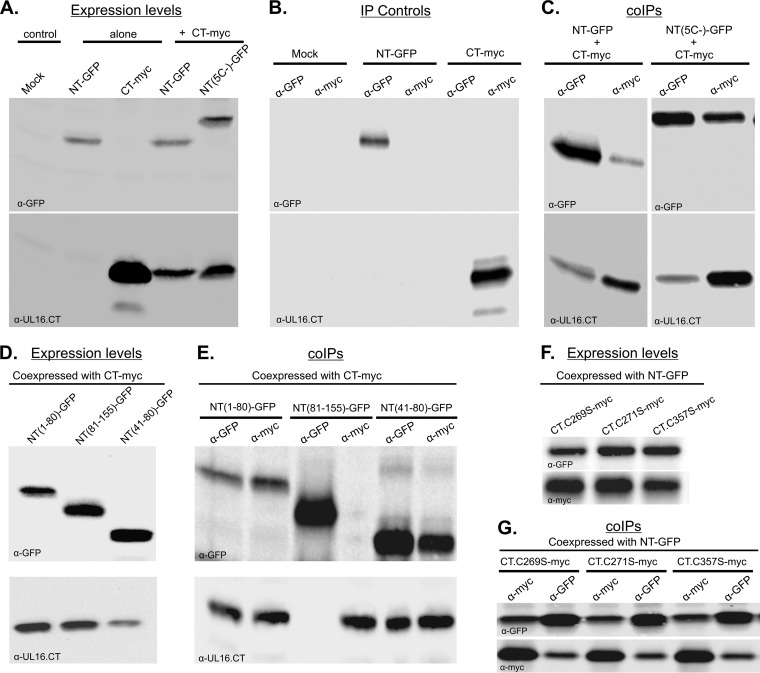
To test for domain interactions in a different assay that does not depend on the Src membrane-binding peptide, NT-GFP and CT-myc were expressed either individually or together in Vero cells, and coimmunoprecipitations (co-IPs) were performed. Immunoblotting of the cell lysates showed that all the constructs were expressed at detectable levels (Fig. 3A). Immunoprecipitations performed with lysates of mock-transfected or singly transfected cells demonstrated the specificities of the antibodies for the GFP and myc tags (Fig. 3B). With these controls working as expected, it is clear that coexpressed NT-GFP and CT-myc were readily coimmunoprecipitated with either antibody, indicating the formation of NT-CT complexes (Fig. 3C, left panel). To corroborate the nonrequirement of disulfide bond formation for NT-CT complex formation, the NT-GFP chimera lacking all five cysteines [NT(5C−)] was coexpressed with CT-myc (Fig. 3A), and both anti-GFP and anti-myc sera yielded efficient precipitation of both proteins (Fig. 3C, right panel). Importantly, NT-CT complexes were not detected when the two domains were expressed separately and their cell lysates mixed prior to immunoprecipitations (not shown). Thus, the complexes were made only by coexpression of the domains in the same cell, and interactions were not the result of irrelevant, spontaneous disulfide bonds forming when cells were broken open.
FIG 3.
Coimmunoprecipitation of independently expressed N- and C-terminal domains of UL16. Plasmids encoding NT or CT (or their derivatives) were transfected into Vero cells either alone or together, as indicated. At 16 to 20 h posttransfection, cell lysates were prepared, and these were either analyzed by immunoblotting to measure the input expression levels (A, D, and F) or incubated with the antibodies shown at the tops of the lanes to collect interacting complexes, which were then analyzed by immunoblotting (B, C, E, and G). The antibody used for immunoblotting is indicated at the bottom left of each panel.
To define subregions required for the interaction, truncation mutants of NT-GFP (Fig. 1) were coexpressed with CT-myc at readily detected levels (Fig. 3D). Immunoprecipitations with anti-GFP or anti-myc serum revealed that NT(1–80aa)-GFP and CT-myc formed a complex (Fig. 3E, lanes 1 and 2). In contrast, no interaction was found when a fragment containing amino acid residues 81 to 155 was used (Fig. 3E, lanes 3 and 4). This fragment has been shown to be sufficient for interaction with the tegument protein UL11 (36); hence, the amino acid sequence required for domain interactions does not overlap that binding site (see Discussion). Further analyses revealed that residues 41 to 80 are sufficient for NT-CT complex formation (Fig. 3E, lanes 5 and 6) in this assay. Consistent with this, a chimera expressing NT(41–155aa)-GFP was found to be competent for binding to CT-myc (not shown), suggesting that the first 40 residues of UL16, which are dispensable for interactions with UL11 and UL21 (40, 47), are not critical for NT-CT complex formation either. Attempts were made to find the minimal portion of the CT that is sufficient to interact with NT; however, those experiments failed because the truncated derivatives were found to be associated with mitochondria and pelleted out of solution while cell lysates were being prepared for immunoprecipitation (see below).
Activating mutations in CT do not disrupt domain interactions.
Previous studies have shown that single-amino-acid substitutions at three conserved cysteines (C269S, C271S, and C357S, within the CT) of full-length UL16 will activate it for efficient binding to UL11 (36). Based on this, we hypothesized that these substitutions would disrupt the interaction between the NT and CT constructs to reveal the UL11-binding site located in the NTD. To address this, NT-GFP was coexpressed individually with CT.C269S-myc, CT.C271S-myc, or CT.357S-myc and subjected to co-IP assays (Fig. 3F). Surprisingly, none of the cysteine-to-serine substitutions had any effect on NT-CT complex formation (Fig. 3G). In further support of these findings, distinct relocalization of NT-GFP and the CT mutants (C269S, C271S, and C357S) to punctate structures was observed (data not shown; also see the mitochondrial experiments, below). Consistent with the interaction seen in the co-IP assay, a massive relocalization of NT-GFP to the perinuclear region was also observed for cotransfections with Src-tagged CT derivatives with the C269S, C271S, or C357S substitution (not shown). Thus, it appears that in full-length UL16, each activating substitution must alter at least one interaction between the N- and C-terminal domains, but others that are not altered presumably must exist (see Discussion).
Evidence of CT-CT interactions.
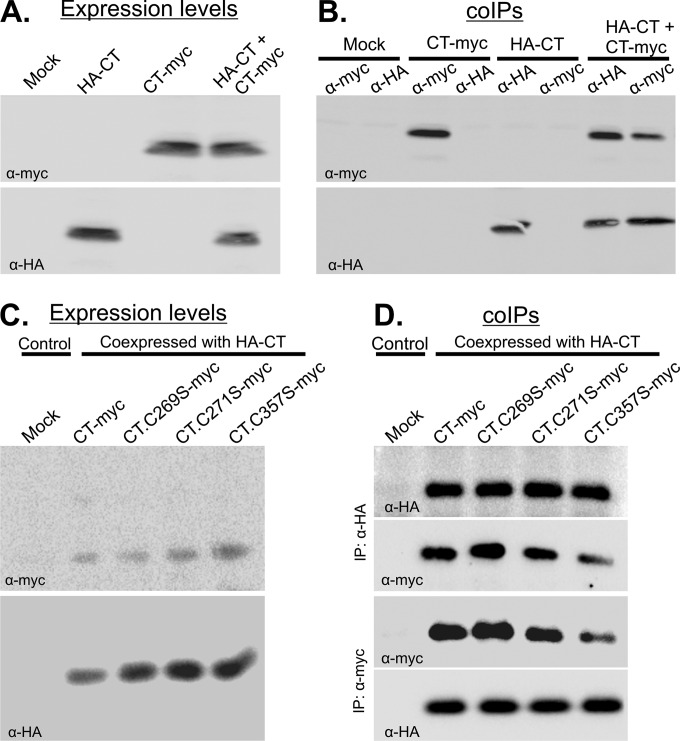
Having found the interaction between the NT and CT domains, we considered the possibility of self-interactions for each domain. For the C-terminal domain, tagged derivatives of CT (HA-CT and CT-myc) were expressed singly or together and analyzed by co-IPs (Fig. 4A). IPs from lysates prepared from the single transfections showed the specific pulldown of each protein with its respective antibody (Fig. 4B, lanes 1 to 6). However, when the two were coexpressed, immunoblotting of the precipitated protein complexes with either antibody provided unequivocal evidence of CT-CT complex formation (Fig. 4B, lanes 7 and 8). Controls in which protein A or G beads were added in the absence of antibodies showed that nonspecific binding did not occur (not shown). Mixing of lysates from cells individually expressing HA-CT or CT-myc did not enable coprecipitation of the proteins (not shown), suggesting that CT-CT complexes are formed only within intact cells and are not the result of spontaneous, irrelevant disulfide bonding.
FIG 4.
Self-interaction of the CTD even when activating substitutions are present. CT-myc and its derivatives were expressed either alone or with HA-CT in Vero cells for 16 to 20 h, and cell lysates were prepared. These were either analyzed by immunoblotting to measure the input expression levels (A and C) or incubated with the indicated antibodies specific for the HA or myc tag to collect complexes, which were then analyzed by immunoblotting (B and D). The antibody used for each immunoblot is indicated at the bottom left of each panel.
To ascertain whether the Cys-to-Ser substitutions in the C-terminal domain interfere with CT-CT interactions, HA-CT was coexpressed with myc-tagged variants that contained the C269S, C271S, or C357S mutation (Fig. 4C). Co-IPs revealed that these substitutions did not disrupt the CT-CT interaction (Fig. 4D). Similarly, for cotransfections in which both HA- and myc-tagged versions of the C-terminal domain had the Cys-to-Ser substitutions, complexes were still formed with the same efficiency (not shown). This implies that the cysteine substitution mutants that enable UL11 binding to full-length UL16 in cells do not interfere with CT-CT interactions, at least when the CT is expressed separately in cells.
Attempts to identify the sequences involved in CT-CT interaction were discouraging due to the tendency of the CT truncation mutants (Fig. 1) to pellet in the insoluble fractions during the process of lysate preparation (not shown; see the mitochondrial experiments, below, for further explanation). Also, in vitro binding assays to confirm CT-CT (or NT-CT) interactions were not possible because of difficulties in purifying the cysteine-rich C-terminal domain from Escherichia coli lysates.
Attempts to find self-association of the N-terminal domain were also made. For this purpose, sNT-HA was coexpressed with NT-GFP in Vero cells. Confocal microscopy revealed that only a meager population of NT-GFP responded to the presence of sNT-HA (not shown). Experiments with swapped tags on these proteins were discouraging because NT-HA was found to be expressed very poorly, even in cotransfections. Thus, we found no evidence for self-interaction of the N-terminal domain.
Addition of myristate causes CT to accumulate on mitochondria.
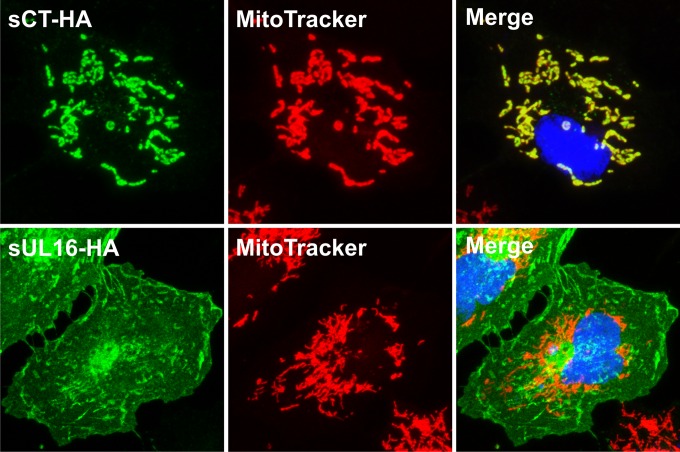
Although the evidence for interactions between the N- and C-terminal domains of UL16 was clear, we were surprised to find that the first 10 residues of Src, which include a myristoylation signal and three basic (lysine) residues for membrane binding, directed CT to subcellular locations different from those observed for full-length UL16 (Fig. 5). All the cells expressing full-length UL16 had signals near the cell periphery, but this was found in only ∼20% of cells expressing the sCT-HA chimera (not shown).
FIG 5.
Addition of myristate directs CT but not full-length UL16 to mitochondria. Plasmids encoding the indicated constructs were transfected into Vero cells, and at 16 to 20 h posttransfection, the cells were stained with MitoTracker Red. The cells were then fixed and costained with a mouse anti-HA monoclonal antibody followed by incubation with an Alexa Fluor 488-conjugated secondary antibody (green). Nuclei were revealed by staining with DAPI (blue). Images were collected with a confocal microscope, and representative stacks are shown.
To gain insight into the nature of the cellular compartments where the majority of sCT-HA accumulated in punctate structures, a variety of organellar markers were utilized (not shown). To our surprise, the results revealed that sCT-HA colocalized completely with staining for the mt-hsp70 antibody, a mitochondrial marker, and not with Golgi apparatus and endoplasmic reticulum (ER) markers (not shown). Costaining of sCT-HA with MitoTracker Red (a mitochondrion-staining dye) confirmed the overlap of sCT-HA signals and mitochondrial signals (Fig. 5). However, the full-length Src-tagged version of UL16 (sUL16-HA) not only failed to colocalize with both mitochondrial markers (Fig. 5 and data not shown) but also lacked any overlap with Golgi and ER compartments (not shown). The punctate pattern of sNT-HA (Fig. 2B) also colocalized with mitochondria, but only in about 40% of transfected cells (not shown). Taken together, these results suggest that the Src peptide in some way helps the N- and C-terminal domains of UL16 to accumulate on mitochondria, unlike the similarly sized HA epitope tag (Fig. 2A). However, UL16 must also contribute, because the Src peptide alone does not have this ability. For example, we found that our previously described Src-UL11 construct (28, 36) does not accumulate on mitochondria (data not shown).
Activating mutations enable full-length UL16 to localize to mitochondria.
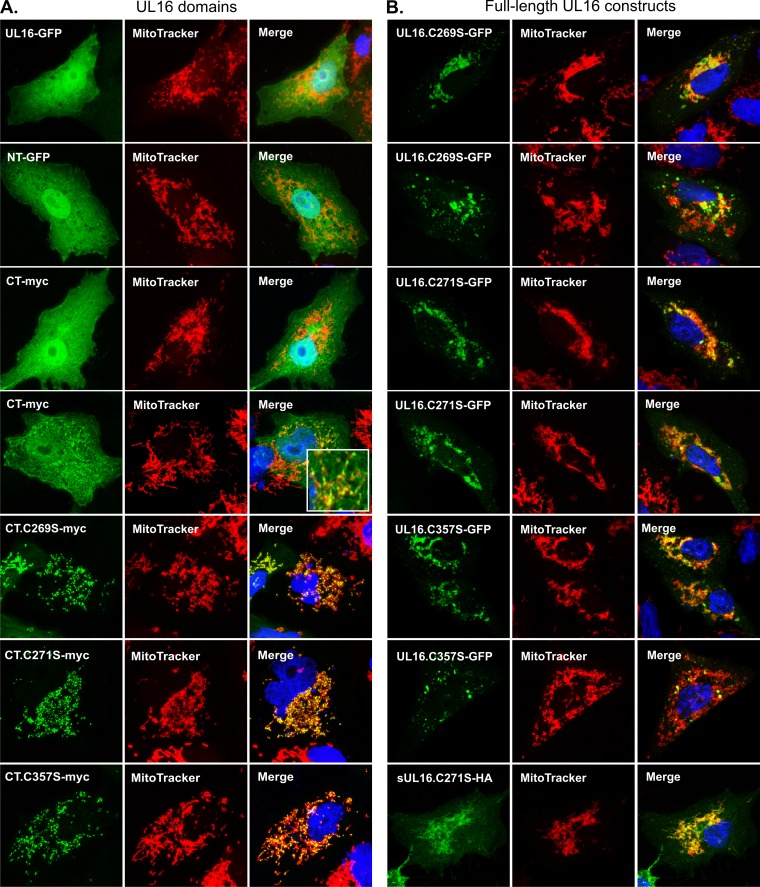
Further evidence that UL16 contains intrinsic mitochondrial targeting information was found in studies performed in the absence of the Src peptide. Although confocal microscopy revealed no mitochondrial localization for full-length UL16-GFP or NT-GFP (Fig. 6A, rows 1 and 2), approximately 10% of the cells transfected with CT-myc contained a population that did colocalize with MitoTracker Red (Fig. 6A, rows 3 and 4) and mt-Hsp70 (not shown). Attempts to increase this percentage were made by using a variety of conditions, including heat shock, apoptosis induction, hydrogen peroxide treatment, and glucose and ATP deprivation. None were able to translocate full-length UL16 or its N-and C-terminal domains to mitochondria (data not shown). Coexpression of binding partners (UL11, glycoprotein E, and UL21) also failed to redirect these constructs to mitochondria (not shown). In contrast, when the activating Cys-to-Ser substitutions (C269S, C271S, and C357S) (36) were built into CT-myc, all the mutants showed dramatic colocalization with mitochondria in ∼70% of transfected cells, and unlike wild-type CT-myc, these mutants were completely absent from the nuclei (Fig. 6, rows 5 to 7). In ∼30% of the cells, these CT-myc mutants behaved like their wild-type counterpart and were present in both the cytoplasm and the nucleus (not shown). Similar mitochondrial localization of CT mutants was observed in several human cell lines (HeLa, 293T, and HFF-1), indicating that the ability of CT mutants to accumulate at mitochondria is not cell type specific.
FIG 6.
Mitochondrial localization of UL16 is triggered by substitutions in its C-terminal domain. The indicated derivatives of full-length UL16 and its N- and C-terminal domains were expressed individually in Vero cells and detected by GFP fluorescence or by staining with antibodies specific for the myc or HA tag (green). Mitochondria were stained with MitoTracker Red. Confocal images of the various full-length constructs are shown in row 1 of panel A (wild type) and all rows of panel B (two different cells for each mutant). All the remaining images are of an NT or CT construct of UL16. Two different cells expressing wild-type CT are shown in rows 3 and 4 of panel A, with the inset showing an enlarged region to highlight its occasional mitochondrial localization. Mutants of CT are shown in the bottom three rows of panel A.
To test whether the Cys-to-Ser substitutions could confer mitochondrial targeting to full-length UL16, we reexamined three constructs that we had previously described as forming aggregates and punctate structures of an unknown nature (36). Staining with MitoTracker Red revealed that the aggregates were closely associated with mitochondria and occasionally intertwined with mitochondria (Fig. 6B). In fact, in cells where the proteins were slightly overexpressed, the mitochondrial network appeared to collapse to a juxtanuclear position (Fig. 6B, rows 1 and 3). Furthermore, when the substitutions were introduced into the Src-tagged version of full-length UL16 (sUL16-HA), the constructs (sUL16-C269S-HA, sUL16-271S-HA, and sUL16-C357S-HA) were found to be dramatically relocalized to mitochondria (Fig. 6B, bottom row, and data not shown), unlike the parental construct, sUL16-HA (Fig. 5, row 2). These results suggest that full-length UL16 has the ability to interact with mitochondria, perhaps in a regulated manner (also see the results of studies of infected cells as described in the final section, below).
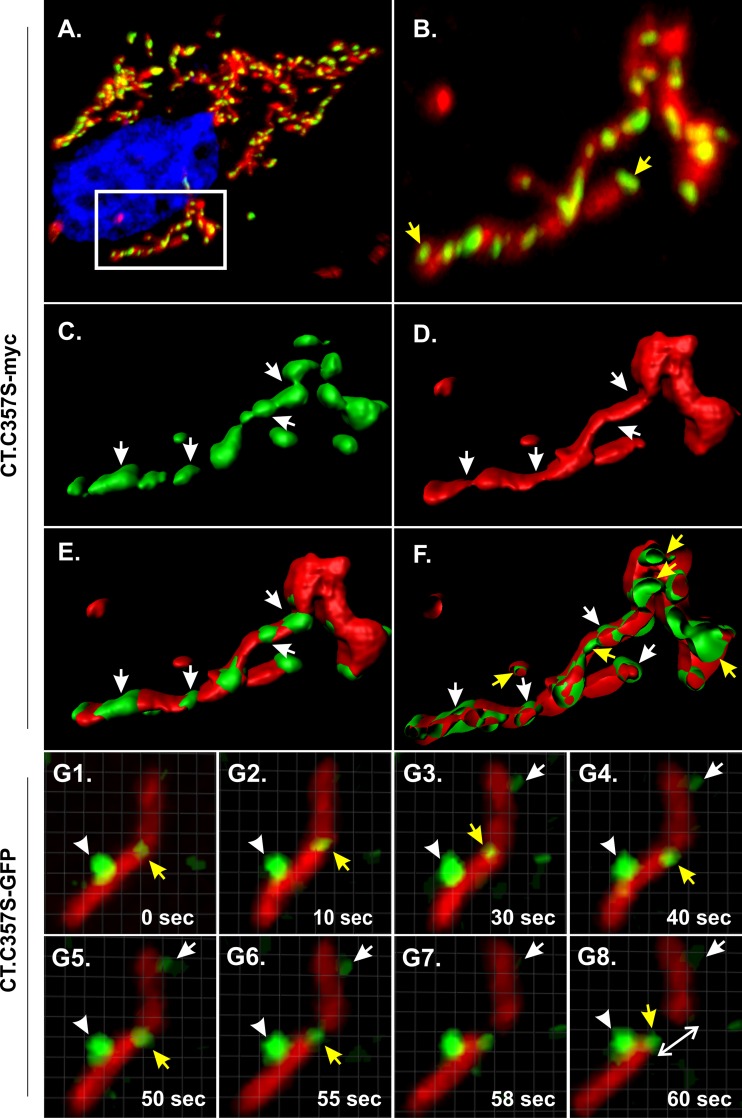
CT mutants overlap mitochondrial fission sites.
A close examination of the imaging data collected for the CT-myc C269S, C271S, and C357S mutants revealed that they were not present homogenously on the mitochondria but in fact were arranged as distinct puncta on the mitochondrial tubule, giving a “beads-on-a-string” appearance (Fig. 7A and B and data not shown). Three-dimensional (3D) reconstructions of the stacked images confirmed that these puncta were indeed present at discrete locations, and these often appeared narrow and constricted (Fig. 7C to E). Rotation of the reconstructed images at different angles revealed that these punctate structures were in fact present on both sides of the mitochondrial tubules and appeared to be wrapping or holding them (see Movie S1 in the supplemental material). However, when the surface-rendered images were clipped in the x-y plane, the green signal could be seen outside, wrapping the mitochondria (white arrows in Fig. 7F), as well as inside the mitochondrial tubule (yellow arrows in Fig. 7F). Similar punctate staining patterns were observed when cysteines were replaced with alanines instead of serines or when the cysteine-to-serine substitutions were built into a CT chimera with an HA tag at the C terminus (not shown).
FIG 7.
Mutants of CT are found in puncta that form discrete foci on mitochondrial tubules. CT derivatives containing an activating cysteine substitution (C357S) and fused to either a myc tag (A to F) or GFP (G1 to G8) were expressed in transfected Vero cells and examined at a high magnification to investigate their association with mitochondria. (A) Representative confocal stack obtained from imaging of fixed cells. The mutant was stained with rabbit polyclonal anti-myc and an Alexa Fluor 488-conjugated secondary antibody (green), while mitochondria were stained with MitoTracker Red and nuclei were stained with DAPI (blue). (B) Zoomed view of the boxed area in panel A. Yellow arrows indicate puncta where the mutant accumulated at the tips or ends of mitochondrial tubules. (C to E) 3D reconstructions of the images from panel B (also see Movie S1 in the supplemental material). White arrows indicate sites where green puncta overlap the narrow or constricted regions of mitochondrial tubules. (F) xy-plane-clipped view of the image reconstructed from panel B. White and yellow arrows indicate the presence of green signals outside and inside mitochondria, respectively. (G1 to G8) Live-cell imaging of Vero cells expressing CT.C357S-GFP and stained with MitoTracker Red. Images were captured at a rate of 2 frames/s in a single plane. Representative time-lapse images from Movie S2 are displayed. Yellow arrows indicate the localization of green puncta at sites of mitochondrial fission, whereas the white arrows and arrowheads indicate discrete puncta that remained associated with the mitochondrial tubule during the whole event. The white double-headed arrow indicates the separation of a newly formed mitochondrion from the parent tubule.
To better understand the nature of the green puncta and their interaction with mitochondria, live-cell imaging was performed with a GFP-tagged derivative, CT.C357S-GFP. Unexpectedly, mitochondrial localization appeared to be hampered by the GFP tag, and most of the expressed protein was found diffusely in the cytoplasm and the nucleus of the cell (not shown). However, we did observe green puncta on mitochondria (the “beads-on-a-string” phenotype seen with the myc-tagged derivative) in ∼20% of the cells. Thus, only cells with weak cytoplasmic staining and bright puncta were selected for live-cell imaging. In these cells, the puncta could be seen to overlap mitochondrial fission sites often (Movie S2 and still frames from the recording, which are displayed in Fig. 7, panels G1 to G8), and in most cases, they remained attached to the ends of the newly formed tubules. Identical results were obtained for recordings of 10 different cells expressing CT.C357S-GFP, with ∼30 different mitochondrial tubules analyzed in each (not shown). Although the green puncta were present at multiple sites along the mitochondrial tubules and had nearly equal spacing, only some of these sites gave rise to divisions during the period of observation. We also observed many examples of puncta coming together and fusing as would be expected for vesicular structures (see Discussion).
Mapping of mitochondrial targeting sequences.
In an attempt to identify residues needed for mitochondrial targeting, 36 single-amino-acid substitutions were inserted into the parental CT-myc construct or its variant containing the C271S substitution (referred to as UL16CT-C271S), which is already active for mitochondrial localization. Charged, polar, and hydrophobic residues were selected and replaced individually with alanines. Analysis of these CT-myc derivatives revealed 11 newly described sites that can be changed to strongly activate mitochondrial localization (resembling the C269S, C271S, and C357S mutants), 13 changes that cause partial localization, and 12 changes that do not induce localization (Table 1). Unfortunately, none of the 36 changes interfered with the ability of CT.C271S to accumulate on mitochondria (not shown), and thus they failed to reveal residues critical for targeting.
TABLE 1.
Classification of UL16 CT mutants in transfected Vero cells
| Group | Localization phenotype | Mutants |
|---|---|---|
| 1 | Diffuse in cytoplasm and nucleus (rarely on mitochondria) | CT-myc, CT(156–250aa)-myc, CT(156–205aa)-myc, T162A, Q165A, E174A, D176A, L217A, W232A, D254A, N257A, W260A, R312A, Q321A, D331A |
| 2 | Diffuse in cytoplasm and nucleus (70% of cells), mitochondrial/punctate (30% of cells) | E168A, D214A, L230A, E245A, H252A, T266A, W277A, Q282A, T284A, Q288A, D290A, Q309A, R327A |
| 3 | Diffuse in cytoplasm and nucleus (30% of cells), mitochondrial/punctate (70% of cells) | V181A, E188A, W200A, Y209A, W211A, C269S/A, C271S/A, F298A, D303A, D318A, E322A, W340A, F345A, C357S/A |
| 4 | Exclusively mitochondrial | CT(206–373aa)-myc, CT(250–373aa)-myc, CT(280–373aa)-myc, CT(206–279aa)-myc |
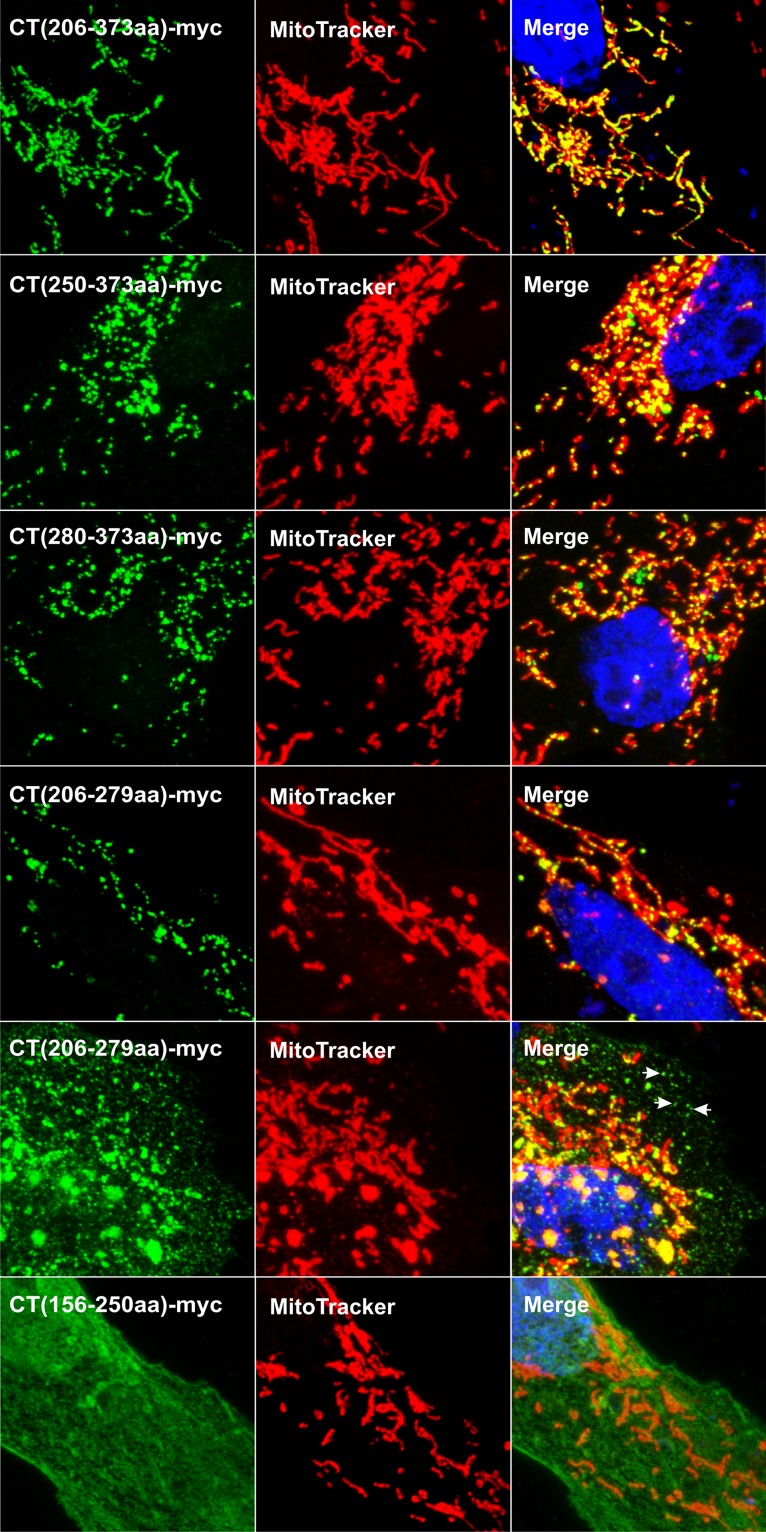
As an alternative for mapping the mitochondrial targeting information, several deletion mutants of CT-myc were analyzed (Fig. 1). Confocal microscopy revealed that the CT(206–373aa), CT(250–373aa), CT(280–373aa), and CT(206–279aa) mutants have robust targeting properties that produce discrete puncta on mitochondria (Fig. 8, rows 1 to 4). Of these, the CT(206–279aa) mutant was found to also accumulate in puncta separate from mitochondria in ∼50% of the transfected cells (Fig. 8, row 5, white arrows). The CT(326–373aa) fragment failed to be expressed at detectable levels (not shown). In contrast, the CT(156–250aa) (Fig. 8, row 6) and CT(156–205aa) (not shown) fragments were expressed well, but neither localized to mitochondria. These data (summarized in Table 1) suggest the presence of two, nonoverlapping targeting sequences in the C-terminal domain of UL16 (shaded fragments in Fig. 1). This redundancy likely explains why none of the single-amino-acid substitutions were able to prevent mitochondrial localization of CT.C271S.
FIG 8.
Presence of two independent mitochondrial targeting regions in CT. Vero cells were transfected with the indicated myc-tagged deletion mutants (see Fig. 1) and analyzed for mitochondrial localization ∼20 h later. The expressed fragments of CT were visualized by staining with anti-myc and an Alexa Fluor 488-conjugated secondary antibody (green), and mitochondria were labeled with MitoTracker Red. Representative confocal stacks are shown.
Analysis of UL16-mitochondrion interactions in HSV-1-infected cells.
To ascertain whether UL16 associates with mitochondria in HSV-1-infected cells, recombinant viruses encoding either UL16-HA or UL16-GFP were generated. Tags were placed at the C-terminal end of UL16 to avoid interfering with the many binding activities that are located within the N-terminal domain. At a high multiplicity of infection (MOI = 5), HSV.UL16-HA propagated like the wild-type virus, whereas a small difference in the yield of infectious virus (less than a half-log reduction) was observed for HSV.UL16-GFP (Fig. 9A). When fewer cells were initially infected (MOI = 1), lower viral yields were obtained with both tagged viruses (data not shown), which together with reductions in plaque size (Fig. 9B) suggests a defect in virus spreading. However, neither tag interfered with other known properties of UL16, including its level of expression (not shown), its packaging into virions (not shown), and its ability to interact with viral binding partners (26, 36, 38). Hence, these two viruses were used to examine the interaction of UL16 with mitochondria in infected cells.
FIG 9.
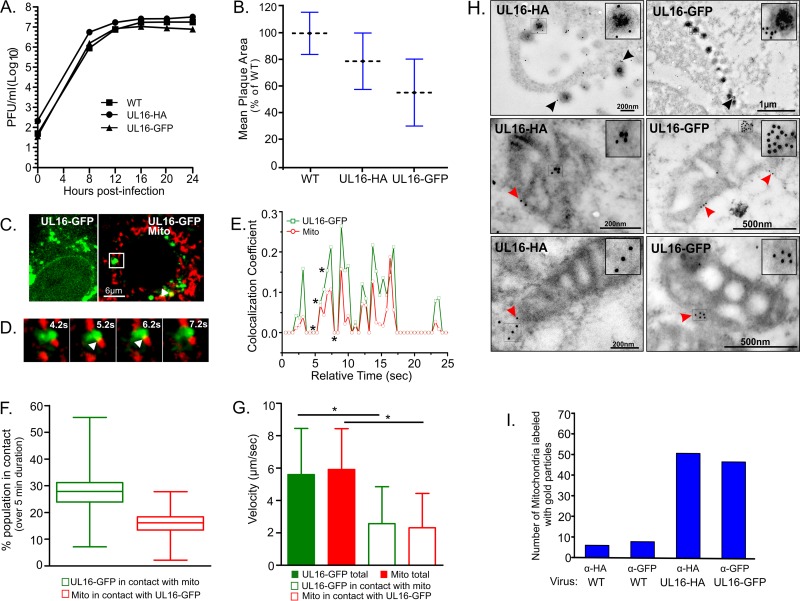
UL16-mitochondrion interactions in HSV-1-infected cells. (A) Replication kinetics of UL16-tagged viruses. Vero cells were infected with the specified viruses at an MOI of 5 for 1 h at 37°C, and virions remaining on the cell surface were inactivated with a brief acidic wash prior to incubation in growth medium. The cultures (cells together with medium) were harvested at the indicated time points, and plaque assays were done to measure the total amount of infectious virus present. WT, wild type. (B) Comparison of plaque sizes. For each of the viruses, two plates of Vero cells were infected and overlaid with methylcellulose. At 4 days postinfection, cells were fixed and stained, and plaque sizes were measured (n = 60 per virus). The average plaque sizes (dashed lines) were plotted relative to that of wild-type virus. (C) Localization of UL16-GFP in live-cell recordings of HSV.UL16-GFP-infected cells. The images are from two different cells, one showing a typical distribution of UL16-GFP puncta and nuclear rim staining (left) and the other stained to visualize mitochondria (right). The UL16-GFP punctum in the white rectangle was analyzed at a higher magnification by live-cell recording at a rate of 2 frames/s for 5 min (Movie S3), and this image is a still frame from time zero. (D) Additional still frames from Movie S3, showing four different time points. White arrowheads indicate the points of overlap of the UL16-GFP (green) and mitochondrial (red) signals. (E) Changes in Coste's overlap coefficient values were calculated every 0.5 s for both the green and red channels during the first 25 s of the recording. Asterisks mark the time points for which images are displayed in panel D. (F) Box-and-whisker plots were created to display the changes in percentage of the UL16-GFP and mitochondrial populations in contact with each other, calculated by an object-based quantitation method. Whiskers, maximum to minimum; middle line, median; box, interquartile range (75th to 25th percentiles). (G) Comparison of particle velocity values for the UL16-GFP and mitochondrial puncta. Error bars show standard errors of the means. *, P < 0.0001. (H) Electron micrographs of immunogold-stained Vero cells infected with either HSV.UL16-HA (left panels) or HSV.UL16-GFP (right panels) at an MOI of 5 and analyzed at 12 to 14 h postinfection. Black arrowheads point to gold-labeled extracellular virions. Red arrowheads point to gold particles that are on or near mitochondria. (I) Quantitation of gold-labeled mitochondria. Primary antibodies used for immunogold labeling are indicated.
Live-cell confocal microscopy was used to investigate HSV.UL16-GFP-infected cells during the late phase of the replication cycle (8 to 24 h postinfection). At these time points, UL16-GFP was found to light up the nucleus and the nuclear rim, and it exhibited diffuse localization in the cytoplasm, along with many distinct punctate structures (Fig. 9C, left panel). To address whether these puncta associated with mitochondria, the cells were stained with MitoTracker Red and monitored for short durations. This revealed that a few of the puncta were in contact with mitochondria (Fig. 9C, right panel); however, these interactions were transient and lasted only a few seconds (Fig. 9D and Movie S3). Time-dependent, concordant changes in the colocalization coefficients for the UL16-GFP puncta and mitochondria further strengthened the evidence of transient and repeated contact formation between the two (Fig. 9E). Also, an object-based colocalization method was used to specifically measure the fraction of each population that was in contact over time (Fig. 9F). This method is independent of the fluorophore ratiometric numbers and calculates the number of green (UL16-GFP) or red (mitochondria) objects that overlap between two channels (green and red) to indicate their colocalization. The percentages of UL16-GFP puncta in contact with mitochondria ranged from 7 to 55% during a 5-min interval (n = 600 time points measured every 0.5 s over 300 s), with a mean value ± standard error of the mean (SEM) of 27.02% ± 0.259%, while the mitochondrial population in contact with UL16-GFP varied from 2% to 27%, with a mean ± SEM of 15.3% ± 0.178%. To ascertain whether contacts between the UL16-GFP puncta and mitochondria affected the movements of either, the velocities of all the members of each population were measured every 0.5 s for 300 s. In analyzing the total populations, only a minor difference in velocity was observed (Fig. 9G, solid bars), but this was not surprising because the punctum-mitochondrion interactions were dynamic, with the majority of each population not in contact at any given time. In contrast, the contacting members of each population had strikingly reduced velocities (Fig. 9G, open bars). Thus, it seems quite clear that the UL16-GFP puncta were not just coming close to mitochondria but were directly interacting with them in some way.
Based on their large sizes and movements, the UL16-GFP puncta seem to be membrane vesicles of some sort. Of course, it is well established that a population of UL16 associates with cytoplasmic capsids to help drive their envelopment via interactions with the viral membrane proteins UL11 and gE (16, 17, 36–38, 40). This population of UL16 is packaged into nascent virions, which reside in cytoplasmic vesicles awaiting transport to the plasma membrane. If the UL16-GFP puncta correspond to this population of vesicles, then they should be less evident when the production of capsids is blocked. To test this possibility, one of the genes essential for capsid assembly, UL18, was deleted to make HSV.ΔUL18/UL16-GFP. This mutant was propagated in G5 complementing cells (12, 56), where it produced close-to-wild-type amounts of virions (not shown). In noncomplementing cells, UL16-GFP puncta were still observed, and these again exhibited time-dependent colocalization with mitochondria (not shown), with the contacts between the two populations being highly dynamic (Movie S4) and causing reductions in mean particle velocities that were similar to what was seen with the parental virus (not shown). Thus, the formation of UL16-GFP puncta is not a capsid-driven process, and further experiments will be needed to ascertain their identity.
For yet another method of detecting UL16 in association with mitochondria, electron microscopy was used to examine immunogold-labeled sections. Cells infected with either HSV.UL16-HA or HSV.UL16-GFP were fixed, embedded, sectioned, exposed to mouse monoclonal antibodies specific for the appropriate tag, and then stained with a gold-conjugated anti-mouse antibody. None of the antibodies bind to the Fc receptor of gE/gI (57–59), and to confirm this, wild-type HSV-infected cells were used as a negative control (not shown). As expected, gold particles were evident in the extracellular virions released from HSV.UL16-HA- or HSV.UL16-GFP-infected cells (Fig. 9H, top panels) but not in those from wild-type-infected cells (not shown).
In the cytoplasm, gold particles were frequently observed on or in the vicinity of mitochondria in cells infected with the tagged viruses (Fig. 9H, middle and lower panels), but these were very infrequent in the cells infected with the wild type (not shown). Immunogold staining methods have inherent difficulties, and the one utilized here suffers from poor preservation of membrane structures, which made it difficult to find intact mitochondria in thin sections. Another method that does not have this problem (60) exhibited high background immunogold staining and could not be used (not shown). In an attempt to quantify the data, many sections were examined to find 70 unambiguous mitochondria for each sample, and those with associated gold particles were counted. The resulting numbers were much higher for the cells infected with the tagged viruses than for those infected with the wild type (Fig. 9I). Although the number of mitochondria that did not have immunogold particles is unknown, this experiment provides further evidence that UL16 is associated with some mitochondria at any given time in the infection.
To gain further insight, recombinant viruses harboring activating cysteine substitutions within UL16 (C269S, C271S, and C357S) were made, but these did not provide information regarding the mitochondrial interaction (data not shown). UL16 was poorly expressed by the virus with the C269S substitution, and thus it replicated similarly to a UL16-null mutant (12). The viruses with the C271S or C357S mutation expressed normal amounts of mutant UL16 and replicated similarly to the wild type; however, they exhibited 40 to 50% reductions in plaque size (data not shown). Given that UL16 is involved in different steps of the virus replication cycle, for example, virus assembly and cell spread (12, 26), the defects seen in these viruses are difficult to interpret and to assign to one specific function. Further studies will be required to find mutants that affect the mitochondrial function without affecting the other roles of UL16.
DISCUSSION
Although UL16 has many viral binding partners and participates in several different parts of the HSV-1 replication cycle, little is known about what it actually does (beyond binding) and how it works. Previous studies suggested that this conserved tegument protein has two distinct functional domains: an NTD that binds certain viral proteins on the membrane and a CTD that somehow regulates the binding domain. The initial goal of this study was to learn whether the domains of UL16 physically interact, and the results show that they do. However, in the course of this investigation, mutants were discovered that have a striking ability to accumulate on mitochondria. Subsequent analyses of HSV-1-infected cells revealed that UL16 does indeed interact with mitochondria, but in a transient manner. The significance of all these findings is discussed below.
Domain interactions in UL16.
The colocalization and coimmunoprecipitation experiments presented here clearly demonstrate that the NTD and CTD of UL16 can interact in a robust manner when coexpressed separately. However, the interaction is much more complex than anticipated. For example, in the context of full-length UL16, activating substitutions in the CTD change the NTD so that it can interact with UL11, and if there is only one interacting surface between the two domains, then they should no longer interact when coexpressed separately, but they did. We speculate that there are multiple binding surfaces between the domains. This is based on two observations. First, the binding sites for UL11, VP22, and gE seem to be distinct, although their precise locations within the NTD are unknown. For example, there is a binding site within residues 81 to 155 that recognizes the acidic cluster of UL11 (36, 38, 40), but the acidic cluster in the cytoplasmic tail of gE is not required for UL16 binding (37), suggesting the use of a distinct site for this interaction. Second, the cysteine substitutions that induce full-length UL16 to bind efficiently to UL11 do not enable binding to gE and VP22 (12, 36). Likewise, interaction of UL21 with UL16 only exposes the UL11-binding site, but removal of the entire CTD enables the NTD to interact efficiently with all three partners (12, 36, 37). Thus, we imagine that each of the distinct binding sites of the NTD is draped in some manner over the surface of the CTD, remaining sequestered until the proper signals come along to reveal the sites to their binding partners.
Although the mapping data show that residues 41 to 80 are sufficient for binding to the CTD, this result does not rule out other NTD-CTD interactions. Indeed, the binding sites for two UL16 partners (UL11 and gE) are known to become exposed upon cell disruption, whereas the interdomain interaction provided by this small region obviously occurs in the oxidizing environment of cell lysates (but does not involve disulfide bonds). For example, it has long been known that full-length UL16 in cell lysates will efficiently bind to GST-UL11 despite the presence of the C-terminal regulatory domain (36, 38, 40). Perhaps oxidation of cysteines within the CTD alters interactions with the NTD to release the UL11-binding site (located within residues 81 to 155). The gE-UL16 interaction is similar in that it occurs efficiently in cell lysates but not in cotransfected cells (37); however, the VP22-UL16 interaction is different in that it occurs only in intact cells (12) and cannot be demonstrated with assays that require the disruption of cells (e.g., glutathione S-transferase [GST] pulldown and coimmunoprecipitation assays) (our unpublished data). It is possible that residues 41 to 80 contain the VP22-binding site, which is not exposed in cell lysates, but it may be that this segment serves only to link the NTD to the CTD. Collectively, these observations offer further evidence that distinct sites are used for the binding partners of UL16 and are consistent with the hypothesis of multiple NTD-CTD interactions.
It seems unlikely that intradomain interactions within the NTD are involved in regulating the binding of UL16 to its partners. After all, complete removal of the CTD enables efficient interactions of the NTD with each of its three membrane-binding partners (12, 36, 37), and this would not be expected if one binding site in the NTD interacts with another to regulate it. Consistent with this, we were unable to find evidence of NTD-NTD interactions; however, our experiments required the use of foreign tags, which might have had an adverse effect on the assay.
In contrast to the NTD, the CTD of UL16 was found to self-associate with a high efficiency. This was not due to the tagged derivatives binding independently to mitochondria, because the method used for the coimmunoprecipitation assay included a high-speed centrifugation step to remove these organelles prior to collection of complexes with the antisera. Moreover, the interaction was not the result of disulfide bonds forming between CTD molecules when the cells were disrupted, because colysis of cells that separately expressed the tagged derivatives did not yield interactions. We attempted to find evidence of CTD-CTD interactions with a colocalization assay, but those experiments required the use of the Src membrane-binding peptide, and the results were modest (data not shown). Self-interactions may be inhibited when the CTD is bound directly to the membrane. Attempts to map sites within the CTD that mediate the NTD-CTD and CTD-CTD interactions were not possible because of the propensity of these fragments to accumulate on mitochondria. Perhaps future studies of purified fragments would enable more to be learned; however, such studies will be complicated by the 20 cysteines in UL16.
Since the CTD can self-interact, it seemed likely that full-length UL16 would be able to do this as well. Indeed, studies of the UL16 homolog from a gammaherpesvirus (Epstein-Barr virus) have shown that it can multimerize in transfected cells (61). We used coimmunoprecipitation and colocalization assays to look for interactions among tagged derivatives of full-length UL16, but these experiments did not yield convincing results (not shown). There are several possible explanations for this, as follows. (i) Perhaps UL16 molecules do not have the capacity to self-associate, and the interactions observed between CTDs are an artifact of expressing this fragment. (ii) Perhaps UL16 molecules interact with a high efficiency as they come off the ribosome, which would preclude two differently tagged derivatives from coming together. (iii) Perhaps the NTD-CTD interaction hides the site in the CTD used for self-association and it is revealed when UL16 binds to one of its partners. For example, binding of UL21 might reveal the CTD self-association site while also exposing the UL11-binding site. (iv) Perhaps the poorly understood interaction of UL16 with the capsid (16, 17, 44) is needed to promote CTD-CTD interactions. Further studies are needed to ascertain which of these possibilities is correct.
Interaction of UL16 with mitochondria.
The ability of UL16 to localize to mitochondria was an unexpected discovery, and one that has not been made for any of the homologs in other herpesviruses. Overall, our data show that the wild-type protein does not accumulate on mitochondria when expressed alone but that a wide variety of mutants acquire this ability, with some accumulating almost exclusively on this organelle. In retrospect, it is now clear that cytoplasmic puncta that we previously described as being aggregates of mutant forms of UL16 (36) were actually mitochondrion-associated proteins. However, this property is not limited to mutants, as wild-type UL16 (tagged with GFP or an HA epitope) was found to interact with mitochondria in a transient manner in HSV-1-infected cells.
How does UL16 reach mitochondria? We initially favored a direct targeting mechanism, but a variety of software tools that are commonly used to predict the subcellular localization of eukaryotic proteins did not provide a convincing argument for mitochondrial targeting (data not shown). As our studies progressed, it became evident that the targeting information is contained in the CTD of UL16, and although this domain had only weak activity when expressed alone, three simple modifications were found to induce its robust accumulation on mitochondria. One was the attachment of the first 10 amino acids of the Src oncoprotein, which contains an N-terminal myristoylation motif. This peptide is well known for its ability to target a variety of different proteins to the cytoplasmic face of the plasma membrane (28, 36, 51–55), but in this case it also enabled the CTD to reach mitochondria. A second method was to insert single-amino-acid substitutions in the CTD that are known to activate full-length UL16 so that it can bind to UL11 (36). These substitutions have been shown to endow membrane-binding activity on both the CTD and the full-length protein (36). The third way to induce robust accumulation of the CTD on mitochondria was to delete any portion of it except for the regions encompassing amino acids 206 to 279 and 280 to 373. While we do not understand how the subdomains of the CTD interact with mitochondria, these results collectively suggest that membrane interactions are a prerequisite for mitochondrial targeting.
If membrane interactions are required for mitochondrial targeting, this explains why the wild-type UL16 protein does not have activity when expressed alone. In transfected cells, this protein appears to be in a closed conformation that is unable to associate with membranes, as shown by its presence in the soluble fraction in membrane flotation assays and its diffuse distribution throughout the cytoplasm and the nucleus (26, 36, 37). In contrast, in infected cells, wild-type UL16 can interact with several viral membrane proteins (i.e., UL11, VP22, and gE).
What might be the role of membranes in the targeting mechanism? Perhaps the most insightful observation was provided by live-cell imaging experiments, which showed UL16-GFP trafficking to mitochondria on the cytoplasmic surfaces of vesicular structures. These fluorescent puncta had the ability to merge and divide independently of mitochondria. In HSV-1-infected cells, the puncta were observed to transiently come in close contact with the surfaces of mitochondria, which resulted in slower movements for each until the two separated again. Although the origin of these vesicles is unclear, it is likely that UL16 is bound to them via its interaction with UL11, VP22, or gE (12, 36–38, 40), although it is possible that additional viral binding partners have yet to be discovered. However, vesicular structures were also observed with some UL16 mutants, even in the absence of other viral proteins in transfected cells. Thus, UL16 may have the ability to interact with as yet unidentified host membrane proteins or have a hidden membrane-binding sequence that can be activated by mutations. In this regard, it is interesting to compare UL16 to the cellular Bax protein, which is normally distributed in a diffuse pattern in the cytoplasm but translocates to the outer membrane of mitochondria during apoptosis, where it oligomerizes to induce pore formation and triggers release of cytochrome c into the cytoplasm (62–67). The mitochondrion-binding peptide of Bax is C-terminally located and is normally sequestered within the much larger, N-terminal regulatory domain (65, 67–69). Interactions with activator proteins, such as Bim and tBid, induce a conformational change in Bax to reveal the binding peptide (62, 70), and as with UL16, single-amino-acid changes have been found that induce binding to mitochondrial membranes (69). However, there is no sequence similarity between the mitochondrion-binding sequences of UL16 and Bax (not shown).
Since membrane binding seems to be correlated with UL16-mitochondrion interactions, the most likely route to mitochondria in infected cells may be through mitochondrion-associated membranes (MAMs), which are well-known sites of ER-mitochondrion contacts (71–73). MAMs are hubs for lipid synthesis and transfer, signaling, metabolic flow, and apoptosis (71, 74–83). A precedent for this route is provided by the pUL37x1 protein of human cytomegalovirus, which has been shown to traffic to mitochondria by passing through MAMs (84–86). Perhaps UL16 utilizes its interaction with gE, which is made in the ER, to gain access to mitochondria via MAMs. Consistent with MAM-dependent routing, the experiments described here revealed UL16 mutants that localized at or adjacent to mitochondrial fission sites, which have been shown to reside within MAMs (87).
What might be the purpose of the UL16-mitochondrion interaction? Perhaps UL16 plays a role in regulating apoptosis on the outer membrane of mitochondria. Perhaps it is targeted to MAMs to alter the various cellular processes that take place there. Perhaps it goes to MAMs to assemble with other viral proteins, such as gE, for subsequent purposes related to virion assembly. In any case, the answer to this question warrants further investigation of UL16 and its homologs in other herpesviruses.
MATERIALS AND METHODS
Cells and antibodies.
Vero and G5 (a gift from Prashant Desai, John Hopkins University) cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco) essentially as described before (12, 36). Rabbit polyclonal antibodies against GFP and full-length UL16 have been described previously (19, 38). Peptide antibodies used in immunoprecipitation assays specifically recognize a C-terminal region (amino acids 357 to 371) in UL16 and were produced in rabbits. The following antibodies were obtained from Sigma-Aldrich: mouse monoclonal anti-HA and anti-GFP, rabbit polyclonal anti-HA, rabbit polyclonal anti-myc, and mouse monoclonal anti-Golgi 58K. Mouse monoclonal antibodies recognizing heat shock protein 70 (Hsp70) were purchased from Enzo Life Sciences.
Mammalian expression plasmids.
Expression vectors encoding UL16-GFP and its derivatives, namely, NT, NT(5C−), CT, NT(41–80aa), NT(1–80aa), NT(41–155aa), NT(81–155aa), UL16.C269S/A, UL16.C271S/A, UL16.C357S/A, CT.C269S/A, CT.C271S/A, and CT.C357S/A, have been described previously (36, 38, 40). CT-myc, NT-HA, and HA-CT were generated in the pEGFP-N2 vector by use of an In-Fusion Ecodry cloning kit (Clontech). The CT-myc and HA-CT constructs were used as parent plasmids to individually mutate the cysteines at positions 269, 271, and 357 to either serine or alanine. QuikChange mutagenesis was used to incorporate the following single-amino-acid substitutions into the pCT-myc and pCT.C271S backgrounds: T162A, Q165A, E168A, E174A, D176A, V181A, E188A, W200A, Y209A, W211A, D214A, L217A, L230A, W232A, E245A, H252A, D254A, N257A, W260A, T266A, W277A, Q282A, T284A, Q288A, D290A, F298A, D303A, Q309A, R312A, D318A, Q321A, E322A, R327A, D331A, W340A, and F345A. pCT-myc was used as a template for QuikChange deletion mutagenesis to generate the following myc-epitope-tagged constructs: pCT(156–205aa), pCT(156–250aa), pCT(206–373aa), pCT(206–279aa), pCT(250–373aa), pCT(280–373aa), and pCT(326–373aa).
The sUL16-HA, sNT-HA, sNT-GFP, and sCT-HA constructs were cloned into the pEGFP-N2 vector by use of an In-Fusion EcoDry cloning kit (Clontech). The cysteines at positions 269, 271, and 357 were individually replaced with serines in the sUL16-HA and sCT-HA backgrounds to generate the following chimeras: sUL16.C269S-HA, sUL16.C271S-HA, sUL16.C357S-HA, sCT.C269S-HA, sCT.C271S-HA, and sCT.C357S-HA. In all these plasmids, the UL16 derivatives were tagged with an N-terminal amino acid sequence (GSSKSKPKDL) from the v-Src oncogene (28, 36, 51–55, 87). The expression vector sUL11-HA, encoding a Src-tagged UL11 protein, has been described previously (28, 36). The plasmids mCherry-KDEL, encoding an ER marker (88), and mito-BFP, encoding a mitochondrial marker (87), were kind gifts from Gia Voeltz (University of Colorado).
Immunoprecipitation assays.
Vero cells grown to 70% confluence in 60- or 100-mm plates were transfected singly or in combination with plasmids encoding the UL16 N- or C-terminal domain (or derivatives) via Lipofectamine 2000 (Invitrogen). At 16 to 20 h posttransfection, the cells were washed twice with phosphate-buffered saline (PBS), harvested in PBS, pelleted, lysed with 0.5% NP-40 lysis buffer, and subjected to immunoprecipitation to capture antibody-protein complexes essentially as described before (37). The captured proteins were resolved by SDS-PAGE, transferred to nitrocellulose membranes, incubated with primary antibodies specific against UL16, the UL16 C terminus, GFP, myc, or HA, and treated with True-Blot horseradish peroxidase (HRP)-conjugated rabbit or mouse IgG secondary antibodies (eBioscience). The blots were then developed by use of enhanced chemiluminescence (ECL) reagents (Pierce).
Viruses.
Wild-type HSV-1 was derived from a bacterial artificial chromosome (BAC) containing the KOS strain genome (89). Recombinant viruses HSV.UL16-HA and HSV.UL16-GFP were constructed via homologous recombination in Escherichia coli by the GalK selection method, as described previously (90). To generate the HSV.UL16-GFP/ΔUL18 virus, the UL18 gene was deleted from the genome of HSV.UL16-GFP by using the GalK selection method as mentioned above. Clones were screened by PCR, HindIII restriction digestion analyses, and sequencing of the corresponding regions in the BAC DNA. The positive BAC plasmids were purified by use of a NucleoBond 100 BAC kit (Macherey-Nagel) and transfected into Vero or G5 cells by use of Lipofectamine 2000 (Invitrogen). After 3 to 4 days, when the transfected monolayers showed cytopathic effects, the cells and medium were harvested to generate the transfection and amplified viral stocks.
Virus growth kinetics and plaque assays.
Vero or G5 cells were infected with the specified viruses at a multiplicity of infection (MOI) of either 1 or 5. After 1 h of incubation at 37°C, the cells were first washed with acid buffer (135 mM NaCl, 10 mM KCl, 40 mM citric acid, pH 3.0), followed by one rinse with DMEM. The cells were then maintained in DMEM containing 2% fetal bovine serum (FBS). At the indicated time points postinfection, infected cells and medium were harvested separately and processed as follows. The medium was cleared of cells by centrifugation at 3,000 rpm for 5 min prior to freezing. Infected cells were scraped into PBS, washed three times with PBS, and freeze-thawed three times to release cell-associated viruses. Samples were titrated on either Vero or G5 cells by using a standard plaque assay. For plaque size analyses, plaque areas were calculated using ImageJ software (NIH), and the mean plaque area relative to that of wild-type virus was plotted.
Viral protein expression and incorporation assay.
Vero cells were infected with wild-type HSV-1 or the recombinant virus HSV.UL16-HA or HSV.UL16-GFP at an MOI of 5. At 16 to 24 h postinfection, the medium and infected cells were harvested. Cells were pelleted by centrifugation at 3,000 rpm for 5 to 10 min. Extracellular virions were pelleted from the medium by centrifugation through a 30% sucrose cushion at 4°C for 1 h at 26,000 rpm in a Beckman SW41 rotor. Both cell pellets and virion pellets were resuspended in sample buffer and heat denatured, and equal quantities were loaded into SDS-PAGE gels for Western blot analyses to visualize virion incorporation and intracellular protein expression of UL16 proteins.
Immunofluorescence assay.
Cells grown on coverslips were transfected with 0.5 to 2.5 μg of plasmid DNA encoding UL16 or its derivatives. At 16 to 20 h posttransfection, the cells were fixed with 3.7% paraformaldehyde (PFA) for 10 min, permeabilized, and subsequently blocked for 30 min or overnight in blocking buffer as described previously (36). The samples were incubated with primary antibodies for 1 h at room temperature, rinsed with PBS, and then stained for 45 min with Alexa Fluor 488- or Alexa Fluor 568-conjugated goat anti-mouse or rabbit IgG F(ab′)2 fragment, or a combination thereof. Nuclear DNA was stained with DAPI (4′,6-diamidino-2-phenylindole; Molecular Probes) for 5 min; the samples were then mounted in Aqua-Polymount medium (Polysciences Inc.) and examined by confocal microscopy.
Mitochondrial staining in immunofluorescence assays.
Live transfected cells were incubated with DMEM containing a 100 nM to 150 mM concentration of the mitochondrion-staining dye MitoTracker Red CMXRos (Molecular Probes) for 20 min in a CO2 incubator set at 37°C. Excess dye was removed by washing cells twice with fresh DMEM for 5 min each in a CO2 incubator. Stained cells were quickly washed two more times with PBS at room temperature, fixed in 3.7% PFA, and immunostained as described above. To confirm the results of MitoTracker Red staining, a plasmid encoding a mitochondrion-targeting peptide tagged with a blue fluorescent protein (mito-BFP) (87) was transfected with the UL16 constructs to substitute for MitoTracker Red staining.
Confocal microscopy of fixed-cell samples.
A confocal microscope (Leica AOBS SP8; Leica Microsystems, Heidelberg, Germany) was used for all cell imaging experiments. Three-dimensional (xyz) imaging was performed on fixed-cell samples, and optical slices were obtained every 0.33 μm over the entire cell volume. Final 3D projections of all optical slices, including surface rendering, were generated using either ImageJ (NIH), IMARIS (Bitplane, Switzerland), or VOLOCITY (PerkinElmer, United Kingdom).
Live-cell imaging.
For live-cell experiments, a heated live-cell stage equipped with a humidified 5% CO2 perfusion system (Tokai Hit) was attached to the confocal microscope. Vero cells grown on glass-bottomed dishes (MatTek) were either transfected with GFP-tagged UL16 constructs or infected with HSV.UL16-GFP at an MOI of 1 or 3. At 16 to 20 h posttransfection and at different time points postinfection, the medium in the dishes was replaced with a DMEM solution containing 15 to 30 nM MitoTracker Red, and stained cells were subjected to time-lapse two-dimensional (2D) imaging (xyt) in a single plane. Time-dependent colocalization analyses of UL16-GFP-positive puncta and mitochondria were performed using VOLOCITY to calculate the thresholded Coste's colocalization coefficients (M1 and M2) (91). For live-cell tracking of objects, namely, UL16-GFP-positive puncta and mitochondria, or these objects in contact with each other, the measurement module of the VOLOCITY software program was utilized. The 2D time series images (xyt) were background corrected, and the objects, either mitochondria or UL16-GFP-positive punctate structures, were then identified as accurately as possible for each time point by the thresholding procedure. A tracking algorithm based on the centroid measurement was applied to determine the movement of the objects over time. Parameters such as the velocities of the objects (either UL16-GFP-positive puncta or mitochondria) and the fractions of objects in contact were then extracted from the tracking data sets. The data were compiled, plotted, and subjected to statistical analysis by use of GraphPad Prism v6 or Origin Lab v9.1.
Immunogold electron microscopy.
Vero cells grown on Thermanox coverslips (Electron Microscopy Sciences) were infected with wild-type HSV-1 or its recombinants at an MOI of 5. At 12 to 14 h postinfection, the cells were rinsed three times with 0.1 M phosphate buffer and fixed with ice-cold fixative (4% PFA and 0.2% glutaraldehyde in 0.1 M phosphate buffer, pH 7.2) for 1 h at 4°C. Cells were then washed three times for 5 min each with 0.1 M phosphate buffer, pH 7.2, followed by 15 to 20 min of incubation at room temperature in an aldehyde-quenching buffer (0.05 M glycine, 0.1 M phosphate buffer, pH 7.2). Cells were dehydrated, infiltrated with LR White resin (Electron Microscopy Sciences), and then allowed to polymerize in a 50°C incubator for 2 to 3 days prior to sectioning. Grids containing sections were washed with 0.05 M glycine in PBS for 10 min at room temperature, transferred to blocking buffer (5% bovine serum albumin [BSA], 5% goat serum, 0.1% cold-water fish skin [CWFS] gelatin, 10% porcine serum, 15 mM sodium azide in PBS) overnight at 4°C, and incubated with primary antibodies (monoclonal anti-HA and anti-GFP antibodies) for 90 min at room temperature. After 5 washes (5 min each), grids were incubated for 1 h with 10-nm-colloidal-gold-conjugated secondary anti-mouse IgG antibody (Sigma), washed with PBS, and postfixed in a 2% glutaraldehyde-PBS solution. Sections were stained with 1% alkaline lead citrate and 2% uranyl acetate for 2 min and then photographed with a JEOL JEM-1400 digital capture transmission electron microscope (Microscopy Imaging Core, Penn State University College of Medicine). Electron micrographs of infected cells were used to quantify staining of UL16 on mitochondrial tubules with gold-conjugated antibodies. At least 30 individual micrographs from each experiment (n = 2) were used, and a total of 70 clearly defined, intact-looking mitochondria were analyzed for labeling with gold particles. Only mitochondria labeled with at least 2 gold particles were considered positive, and those stained with single or no gold particles were considered negative for the UL16 signal.
Supplementary Material
ACKNOWLEDGMENTS
We extend thanks to Rebecca Craven for a critical reading of the manuscript and to Carol B. Wilson for excellent technical support. We also thank the Penn State Hershey College of Medicine Microscopy Imaging Facility for use of a Leica SP8 confocal microscope (supported by NIH grant 1S10OD010756-01A1).
This work and authors P.C., A.S., and J.C. were supported by an NIH grant (grant R01AI071286) to J.W.W. J.H. and D.Z. were supported by NIH training grants K99AI110648 and T32CA060395, respectively. T.A. was supported by NIH grants 1S10OD010756-01A1 and 1S10OD018124-01A1.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01995-16.
REFERENCES
- 1.Baines JD, Roizman B. 1991. The open reading frames UL3, UL4, UL10, and UL16 are dispensable for the replication of herpes simplex virus 1 in cell culture. J Virol 65:938–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo H, Wang L, Peng L, Zhou ZH, Deng H. 2009. ORF33 of a gammaherpesvirus encodes a tegument protein essential for virion morphogenesis and egress. J Virol 83:10582–10595. doi: 10.1128/JVI.00497-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maninger S, Bosse JB, Lemnitzer F, Pogoda M, Mohr CA, von Einem J, Walther P, Koszinowski UH, Ruzsics Z. 2011. M94 is essential for the secondary envelopment of murine cytomegalovirus. J Virol 85:9254–9267. doi: 10.1128/JVI.00443-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nalwanga D, Rempel S, Roizman B, Baines JD. 1996. The UL16 gene product of herpes simplex virus 1 is a virion protein that colocalizes with intranuclear capsid proteins. Virology 226:236–242. doi: 10.1006/viro.1996.0651. [DOI] [PubMed] [Google Scholar]
- 5.Oshima S, Daikoku T, Shibata S, Yamada H, Goshima F, Nishiyama Y. 1998. Characterization of the UL16 gene product of herpes simplex virus type 2. Arch Virol 143:863–880. doi: 10.1007/s007050050338. [DOI] [PubMed] [Google Scholar]
- 6.Wing BA, Lee GC, Huang ES. 1996. The human cytomegalovirus UL94 open reading frame encodes a conserved herpesvirus capsid/tegument-associated virion protein that is expressed with true late kinetics. J Virol 70:3339–3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johannsen E, Luftig M, Chase MR, Weicksel S, Cahir-McFarland E, Illanes D, Sarracino D, Kieff E. 2004. Proteins of purified Epstein-Barr virus. Proc Natl Acad Sci U S A 101:16286–16291. doi: 10.1073/pnas.0407320101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kattenhorn LM, Mills R, Wagner M, Lomsadze A, Makeev V, Borodovsky M, Ploegh HL, Kessler BM. 2004. Identification of proteins associated with murine cytomegalovirus virions. J Virol 78:11187–11197. doi: 10.1128/JVI.78.20.11187-11197.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klupp BG, Hengartner CJ, Mettenleiter TC, Enquist LW. 2004. Complete, annotated sequence of the pseudorabies virus genome. J Virol 78:424–440. doi: 10.1128/JVI.78.1.424-440.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunn W, Chou C, Li H, Hai R, Patterson D, Stolc V, Zhu H, Liu F. 2003. Functional profiling of a human cytomegalovirus genome. Proc Natl Acad Sci U S A 100:14223–14228. doi: 10.1073/pnas.2334032100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen MR, Hsu TY, Lin SW, Chen JY, Yang CS. 1991. Cloning and characterization of cDNA clones corresponding to transcripts from the BamHI G region of the Epstein-Barr virus genome and expression of BGLF2. J Gen Virol 72:3047–3055. doi: 10.1099/0022-1317-72-12-3047. [DOI] [PubMed] [Google Scholar]
- 12.Starkey JL, Han J, Chadha P, Marsh JA, Wills JW. 2014. Elucidation of the block to herpes simplex virus egress in the absence of tegument protein UL16 reveals a novel interaction with VP22. J Virol 88:110–119. doi: 10.1128/JVI.02555-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phillips SL, Bresnahan WA. 2012. The human cytomegalovirus (HCMV) tegument protein UL94 is essential for secondary envelopment of HCMV virions. J Virol 86:2523–2532. doi: 10.1128/JVI.06548-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das S, Ortiz DA, Gurczynski SJ, Khan F, Pellett PE. 2014. Identification of human cytomegalovirus genes important for biogenesis of the cytoplasmic virion assembly complex. J Virol 88:9086–9099. doi: 10.1128/JVI.01141-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu JJ, Avey D, Li W, Gillen J, Fu B, Miley W, Whitby D, Zhu F. 2015. ORF33 and ORF38 of Kaposi's sarcoma-associated herpesvirus interact and are required for optimal production of infectious progeny viruses. J Virol 90:1741–1756. doi: 10.1128/JVI.02738-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meckes DG Jr, Wills JW. 2007. Dynamic interactions of the UL16 tegument protein with the capsid of herpes simplex virus. J Virol 81:13028–13036. doi: 10.1128/JVI.01306-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meckes DG Jr, Marsh JA, Wills JW. 2010. Complex mechanisms for the packaging of the UL16 tegument protein into herpes simplex virus. Virology 398:208–213. doi: 10.1016/j.virol.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Britt WJ, Jarvis M, Seo JY, Drummond D, Nelson J. 2004. Rapid genetic engineering of human cytomegalovirus by using a lambda phage linear recombination system: demonstration that pp28 (UL99) is essential for production of infectious virus. J Virol 78:539–543. doi: 10.1128/JVI.78.1.539-543.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baird NL, Yeh PC, Courtney RJ, Wills JW. 2008. Sequences in the UL11 tegument protein of herpes simplex virus that control association with detergent-resistant membranes. Virology 374:315–321. doi: 10.1016/j.virol.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiu YF, Sugden B, Chang PJ, Chen LW, Lin YJ, Lan YC, Lai CH, Liou JY, Liu ST, Hung CH. 2012. Characterization and intracellular trafficking of Epstein-Barr virus BBLF1, a protein involved in virion maturation. J Virol 86:9647–9655. doi: 10.1128/JVI.01126-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dingwell KS, Brunetti CR, Hendricks RL, Tang Q, Tang M, Rainbow AJ, Johnson DC. 1994. Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. J Virol 68:834–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dingwell KS, Doering LC, Johnson DC. 1995. Glycoproteins E and I facilitate neuron-to-neuron spread of herpes simplex virus. J Virol 69:7087–7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dingwell KS, Johnson DC. 1998. The herpes simplex virus gE-gI complex facilitates cell-to-cell spread and binds to components of cell junctions. J Virol 72:8933–8942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuchs W, Granzow H, Veits J, Mettenleiter TC. 2012. Identification and functional analysis of the small membrane-associated protein pUL11 of avian infectious laryngotracheitis virus. Virus Res 163:599–608. doi: 10.1016/j.virusres.2011.12.014. [DOI] [PubMed] [Google Scholar]
- 25.Fulmer PA, Melancon JM, Baines JD, Kousoulas KG. 2007. UL20 protein functions precede and are required for the UL11 functions of herpes simplex virus type 1 cytoplasmic virion envelopment. J Virol 81:3097–3108. doi: 10.1128/JVI.02201-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han J, Chadha P, Starkey JL, Wills JW. 2012. Function of glycoprotein E of herpes simplex virus requires coordinated assembly of three tegument proteins on its cytoplasmic tail. Proc Natl Acad Sci U S A 109:19798–19803. doi: 10.1073/pnas.1212900109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kopp M, Granzow H, Fuchs W, Klupp BG, Mundt E, Karger A, Mettenleiter TC. 2003. The pseudorabies virus UL11 protein is a virion component involved in secondary envelopment in the cytoplasm. J Virol 77:5339–5351. doi: 10.1128/JVI.77.9.5339-5351.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loomis JS, Bowzard JB, Courtney RJ, Wills JW. 2001. Intracellular trafficking of the UL11 tegument protein of herpes simplex virus type 1. J Virol 75:12209–12219. doi: 10.1128/JVI.75.24.12209-12219.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacLean CA, Clark B, McGeoch DJ. 1989. Gene UL11 of herpes simplex virus type 1 encodes a virion protein which is myristylated. J Gen Virol 70:3147–3157. doi: 10.1099/0022-1317-70-12-3147. [DOI] [PubMed] [Google Scholar]
- 30.MacLean CA, Dolan A, Jamieson FE, McGeoch DJ. 1992. The myristylated virion proteins of herpes simplex virus type 1: investigation of their role in the virus life cycle. J Gen Virol 73:539–547. doi: 10.1099/0022-1317-73-3-539. [DOI] [PubMed] [Google Scholar]
- 31.Sadaoka T, Serada S, Kato J, Hayashi M, Gomi Y, Naka T, Yamanishi K, Mori Y. 2014. Varicella-zoster virus ORF49 functions in the efficient production of progeny virus through its interaction with essential tegument protein ORF44. J Virol 88:188–201. doi: 10.1128/JVI.02245-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saldanha CE, Lubinski J, Martin C, Nagashunmugam T, Wang L, Van Der Keyl H, Tal-Singer R, Friedman HM. 2000. Herpes simplex virus type 1 glycoprotein E domains involved in virus spread and disease. J Virol 74:6712–6719. doi: 10.1128/JVI.74.15.6712-6719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schimmer C, Neubauer A. 2003. The equine herpesvirus 1 UL11 gene product localizes to the trans-Golgi network and is involved in cell-to-cell spread. Virology 308:23–36. doi: 10.1016/S0042-6822(02)00060-0. [DOI] [PubMed] [Google Scholar]
- 34.Silva MC, Yu QC, Enquist L, Shenk T. 2003. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J Virol 77:10594–10605. doi: 10.1128/JVI.77.19.10594-10605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baines JD, Roizman B. 1992. The UL11 gene of herpes simplex virus 1 encodes a function that facilitates nucleocapsid envelopment and egress from cells. J Virol 66:5168–5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chadha P, Han J, Starkey JL, Wills JW. 2012. Regulated interaction of tegument proteins UL16 and UL11 from herpes simplex virus. J Virol 86:11886–11898. doi: 10.1128/JVI.01879-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yeh PC, Han J, Chadha P, Meckes DG Jr, Ward MD, Semmes OJ, Wills JW. 2011. Direct and specific binding of the UL16 tegument protein of herpes simplex virus to the cytoplasmic tail of glycoprotein E. J Virol 85:9425–9436. doi: 10.1128/JVI.05178-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loomis JS, Courtney RJ, Wills JW. 2003. Binding partners for the UL11 tegument protein of herpes simplex virus type 1. J Virol 77:11417–11424. doi: 10.1128/JVI.77.21.11417-11424.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han J, Chadha P, Meckes DG Jr, Baird NL, Wills JW. 2011. Interaction and interdependent packaging of tegument protein UL11 and glycoprotein E of herpes simplex virus. J Virol 85:9437–9446. doi: 10.1128/JVI.05207-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yeh PC, Meckes DG Jr, Wills JW. 2008. Analysis of the interaction between the UL11 and UL16 tegument proteins of herpes simplex virus. J Virol 82:10693–10700. doi: 10.1128/JVI.01230-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maringer K, Stylianou J, Elliott G. 2012. A network of protein interactions around the herpes simplex virus tegument protein VP22. J Virol 86:12971–12982. doi: 10.1128/JVI.01913-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mbong EF, Woodley L, Dunkerley E, Schrimpf JE, Morrison LA, Duffy C. 2012. Deletion of the herpes simplex virus 1 UL49 gene results in mRNA and protein translation defects that are complemented by secondary mutations in UL41. J Virol 86:12351–12361. doi: 10.1128/JVI.01975-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Y, Cui Z, Zhang Z, Wei H, Zhou Y, Wang M, Zhang XE. 2009. The tegument protein UL94 of human cytomegalovirus as a binding partner for tegument protein pp28 identified by intracellular imaging. Virology 388:68–77. doi: 10.1016/j.virol.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 44.Meckes DG Jr, Wills JW. 2008. Structural rearrangement within an enveloped virus upon binding to the host cell. J Virol 82:10429–10435. doi: 10.1128/JVI.01223-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baines JD, Koyama AH, Huang T, Roizman B. 1994. The UL21 gene products of herpes simplex virus 1 are dispensable for growth in cultured cells. J Virol 68:2929–2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Wind N, Wagenaar F, Pol J, Kimman T, Berns A. 1992. The pseudorabies virus homology of the herpes simplex virus UL21 gene product is a capsid protein which is involved in capsid maturation. J Virol 66:7096–7103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harper AL, Meckes DG Jr, Marsh JA, Ward MD, Yeh PC, Baird NL, Wilson CB, Semmes OJ, Wills JW. 2010. Interaction domains of the UL16 and UL21 tegument proteins of herpes simplex virus. J Virol 84:2963–2971. doi: 10.1128/JVI.02015-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klupp BG, Bottcher S, Granzow H, Kopp M, Mettenleiter TC. 2005. Complex formation between the UL16 and UL21 tegument proteins of pseudorabies virus. J Virol 79:1510–1522. doi: 10.1128/JVI.79.3.1510-1522.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mbong EF, Woodley L, Frost E, Baines JD, Duffy C. 2012. Deletion of UL21 causes a delay in the early stages of the herpes simplex virus 1 replication cycle. J Virol 86:7003–7007. doi: 10.1128/JVI.00411-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Metrick CM, Chadha P, Heldwein EE. 2015. The unusual fold of herpes simplex virus 1 UL21, a multifunctional tegument protein. J Virol 89:2979–2984. doi: 10.1128/JVI.03516-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pellman D, Garber EA, Cross FR, Hanafusa H. 1985. An N-terminal peptide from p60src can direct myristylation and plasma membrane localization when fused to heterologous proteins. Nature 314:374–377. doi: 10.1038/314374a0. [DOI] [PubMed] [Google Scholar]
- 52.Sigal CT, Zhou W, Buser CA, McLaughlin S, Resh MD. 1994. Amino-terminal basic residues of Src mediate membrane binding through electrostatic interaction with acidic phospholipids. Proc Natl Acad Sci U S A 91:12253–12257. doi: 10.1073/pnas.91.25.12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wills JW, Craven RC, Achacoso JA. 1989. Creation and expression of myristylated forms of Rous sarcoma virus Gag protein in mammalian cells. J Virol 63:4331–4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wills JW, Craven RC, Weldon RA Jr, Nelle TD, Erdie CR. 1991. Suppression of retroviral MA deletions by the amino-terminal membrane-binding domain of p60src. J Virol 65:3804–3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaplan JM, Mardon G, Bishop JM, Varmus HE. 1988. The first seven amino acids encoded by the v-src oncogene act as a myristylation signal: lysine 7 is a critical determinant. Mol Cell Biol 8:2435–2441. doi: 10.1128/MCB.8.6.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Person S, Desai P. 1998. Capsids are formed in a mutant virus blocked at the maturation site of the UL26 and UL26.5 open reading frames of herpes simplex virus type 1 but are not formed in a null mutant of UL38 (VP19C). Virology 242:193–203. doi: 10.1006/viro.1997.9005. [DOI] [PubMed] [Google Scholar]
- 57.Johansson PJ, Nardella FA, Sjöquist J, Schröder AK, Christensen P. 1989. Herpes simplex type 1-induced Fc receptor binds to the Cgamma2-Cgamma3 interface region of IgG in the area that binds staphylococcal protein A. Immunology 66:8–13. [PMC free article] [PubMed] [Google Scholar]
- 58.Sprague ER, Martin WL, Bjorkman PJ. 2004. pH dependence and stoichiometry of binding to the Fc region of IgG by the herpes simplex virus Fc receptor gE-gI. J Biol Chem 279:14184–14193. doi: 10.1074/jbc.M313281200. [DOI] [PubMed] [Google Scholar]
- 59.Sprague ER, Wang C, Baker D, Bjorkman PJ. 2006. Crystal structure of the HSV-1 Fc receptor bound to Fc reveals a mechanism for antibody bipolar bridging. PLoS Biol 4:e148. doi: 10.1371/journal.pbio.0040148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamamoto A, Masaki R. 2010. Pre-embedding nanogold silver and gold intensification. Methods Mol Biol 657:225–235. doi: 10.1007/978-1-60761-783-9_18. [DOI] [PubMed] [Google Scholar]
- 61.Liu X, Cohen JI. 2015. Epstein-Barr virus (EBV) tegument protein BGLF2 promotes EBV reactivation through activation of the p38 mitogen-activated protein kinase. J Virol 90:1129–1138. doi: 10.1128/JVI.01410-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Desagher S, Osen-Sand A, Nichols A, Eskes R, Montessuit S, Lauper S, Maundrell K, Antonsson B, Martinou JC. 1999. Bid-induced conformational change of Bax is responsible for mitochondrial cytochrome c release during apoptosis. J Cell Biol 144:891–901. doi: 10.1083/jcb.144.5.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eskes R, Desagher S, Antonsson B, Martinou JC. 2000. Bid induces the oligomerization and insertion of Bax into the outer mitochondrial membrane. Mol Cell Biol 20:929–935. doi: 10.1128/MCB.20.3.929-935.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.George NM, Evans JJ, Luo X. 2007. A three-helix homo-oligomerization domain containing BH3 and BH1 is responsible for the apoptotic activity of Bax. Genes Dev 21:1937–1948. doi: 10.1101/gad.1553607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gross A, Jockel J, Wei MC, Korsmeyer SJ. 1998. Enforced dimerization of BAX results in its translocation, mitochondrial dysfunction and apoptosis. EMBO J 17:3878–3885. doi: 10.1093/emboj/17.14.3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hsu YT, Wolter KG, Youle RJ. 1997. Cytosol-to-membrane redistribution of Bax and Bcl-X(L) during apoptosis. Proc Natl Acad Sci U S A 94:3668–3672. doi: 10.1073/pnas.94.8.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wolter KG, Hsu YT, Smith CL, Nechushtan A, Xi XG, Youle RJ. 1997. Movement of Bax from the cytosol to mitochondria during apoptosis. J Cell Biol 139:1281–1292. doi: 10.1083/jcb.139.5.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goping IS, Gross A, Lavoie JN, Nguyen M, Jemmerson R, Roth K, Korsmeyer SJ, Shore GC. 1998. Regulated targeting of BAX to mitochondria. J Cell Biol 143:207–215. doi: 10.1083/jcb.143.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nechushtan A, Smith CL, Hsu YT, Youle RJ. 1999. Conformation of the Bax C-terminus regulates subcellular location and cell death. EMBO J 18:2330–2341. doi: 10.1093/emboj/18.9.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim H, Tu HC, Ren D, Takeuchi O, Jeffers JR, Zambetti GP, Hsieh JJ, Cheng EH. 2009. Stepwise activation of BAX and BAK by tBID, BIM, and PUMA initiates mitochondrial apoptosis. Mol Cell 36:487–499. doi: 10.1016/j.molcel.2009.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Giorgi C, De Stefani D, Bononi A, Rizzuto R, Pinton P. 2009. Structural and functional link between the mitochondrial network and the endoplasmic reticulum. Int J Biochem Cell Biol 41:1817–1827. doi: 10.1016/j.biocel.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hayashi T, Rizzuto R, Hajnoczky G, Su TP. 2009. MAM: more than just a housekeeper. Trends Cell Biol 19:81–88. doi: 10.1016/j.tcb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Raturi A, Simmen T. 2013. Where the endoplasmic reticulum and the mitochondrion tie the knot: the mitochondria-associated membrane (MAM). Biochim Biophys Acta 1833:213–224. doi: 10.1016/j.bbamcr.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 74.Ardail D, Privat JP, Egret-Charlier M, Levrat C, Lerme F, Louisot P. 1990. Mitochondrial contact sites. Lipid composition and dynamics. J Biol Chem 265:18797–18802. [PubMed] [Google Scholar]
- 75.Ardail D, Gasnier F, Lerme F, Simonot C, Louisot P, Gateau-Roesch O. 1993. Involvement of mitochondrial contact sites in the subcellular compartmentalization of phospholipid biosynthetic enzymes. J Biol Chem 268:25985–25992. [PubMed] [Google Scholar]
- 76.Rusinol AE, Cui Z, Chen MH, Vance JE. 1994. A unique mitochondria-associated membrane fraction from rat liver has a high capacity for lipid synthesis and contains pre-Golgi secretory proteins including nascent lipoproteins. J Biol Chem 269:27494–27502. [PubMed] [Google Scholar]
- 77.Vance JE. 1990. Phospholipid synthesis in a membrane fraction associated with mitochondria. J Biol Chem 265:7248–7256. [PubMed] [Google Scholar]
- 78.Vance JE. 2015. Phospholipid synthesis and transport in mammalian cells. Traffic 16:1–18. doi: 10.1111/tra.12230. [DOI] [PubMed] [Google Scholar]
- 79.Hayashi T, Su TP. 2007. Sigma-1 receptor chaperones at the ER-mitochondrion interface regulate Ca(2+) signaling and cell survival. Cell 131:596–610. doi: 10.1016/j.cell.2007.08.036. [DOI] [PubMed] [Google Scholar]
- 80.Mendes CC, Gomes DA, Thompson M, Souto NC, Goes TS, Goes AM, Rodrigues MA, Gomez MV, Nathanson MH, Leite MF. 2005. The type III inositol 1,4,5-trisphosphate receptor preferentially transmits apoptotic Ca2+ signals into mitochondria. J Biol Chem 280:40892–40900. doi: 10.1074/jbc.M506623200. [DOI] [PubMed] [Google Scholar]
- 81.Pinton P, Giorgi C, Siviero R, Zecchini E, Rizzuto R. 2008. Calcium and apoptosis: ER-mitochondria Ca2+ transfer in the control of apoptosis. Oncogene 27:6407–6418. doi: 10.1038/onc.2008.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rizzuto R, Brini M, Murgia M, Pozzan T. 1993. Microdomains with high Ca2+ close to IP3-sensitive channels that are sensed by neighboring mitochondria. Science 262:744–747. doi: 10.1126/science.8235595. [DOI] [PubMed] [Google Scholar]
- 83.Rizzuto R, Pinton P, Carrington W, Fay FS, Fogarty KE, Lifshitz LM, Tuft RA, Pozzan T. 1998. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science 280:1763–1766. doi: 10.1126/science.280.5370.1763. [DOI] [PubMed] [Google Scholar]
- 84.Bozidis P, Williamson CD, Wong DS, Colberg-Poley AM. 2010. Trafficking of UL37 proteins into mitochondrion-associated membranes during permissive human cytomegalovirus infection. J Virol 84:7898–7903. doi: 10.1128/JVI.00885-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Williamson CD, Colberg-Poley AM. 2009. Access of viral proteins to mitochondria via mitochondria-associated membranes. Rev Med Virol 19:147–164. doi: 10.1002/rmv.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang A, Hildreth RL, Colberg-Poley AM. 2013. Human cytomegalovirus inhibits apoptosis by proteasome-mediated degradation of Bax at endoplasmic reticulum-mitochondrion contacts. J Virol 87:5657–5668. doi: 10.1128/JVI.00145-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Friedman JR, Lackner LL, West M, Dibenedetto JR, Nunnari J, Voeltz GK. 2011. ER tubules mark sites of mitochondrial division. Science 334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zurek N, Sparks L, Voeltz G. 2011. Reticulon short hairpin transmembrane domains are used to shape ER tubules. Traffic 12:28–41. doi: 10.1111/j.1600-0854.2010.01134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gierasch WW, Zimmerman DL, Ward SL, Vanheyningen TK, Romine JD, Leib DA. 2006. Construction and characterization of bacterial artificial chromosomes containing HSV-1 strains 17 and KOS. J Virol Methods 135:197–206. doi: 10.1016/j.jviromet.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 90.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res 33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Costes SV, Daelemans D, Cho EH, Dobbin Z, Pavlakis G, Lockett S. 2004. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys J 86:3993–4003. doi: 10.1529/biophysj.103.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.