Abstract
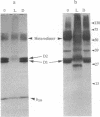
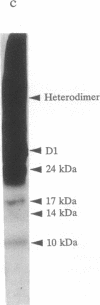
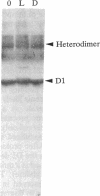
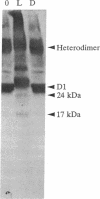
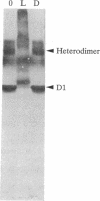
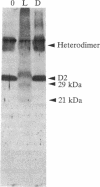
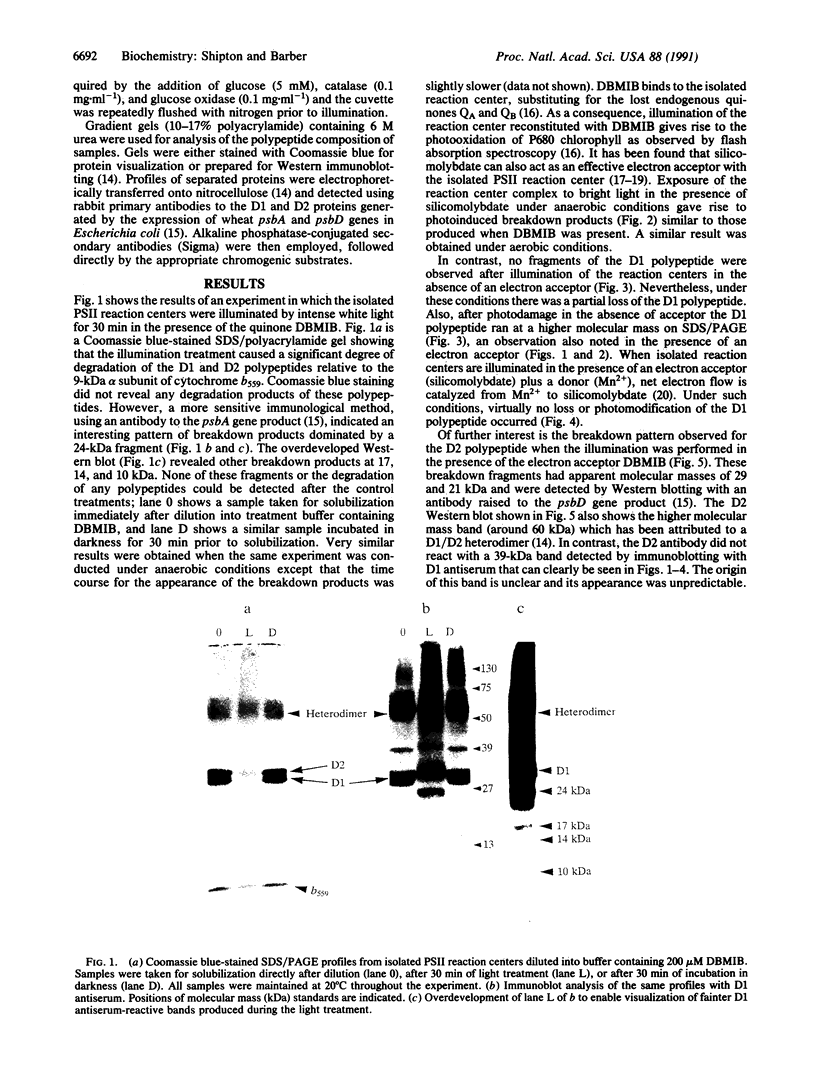
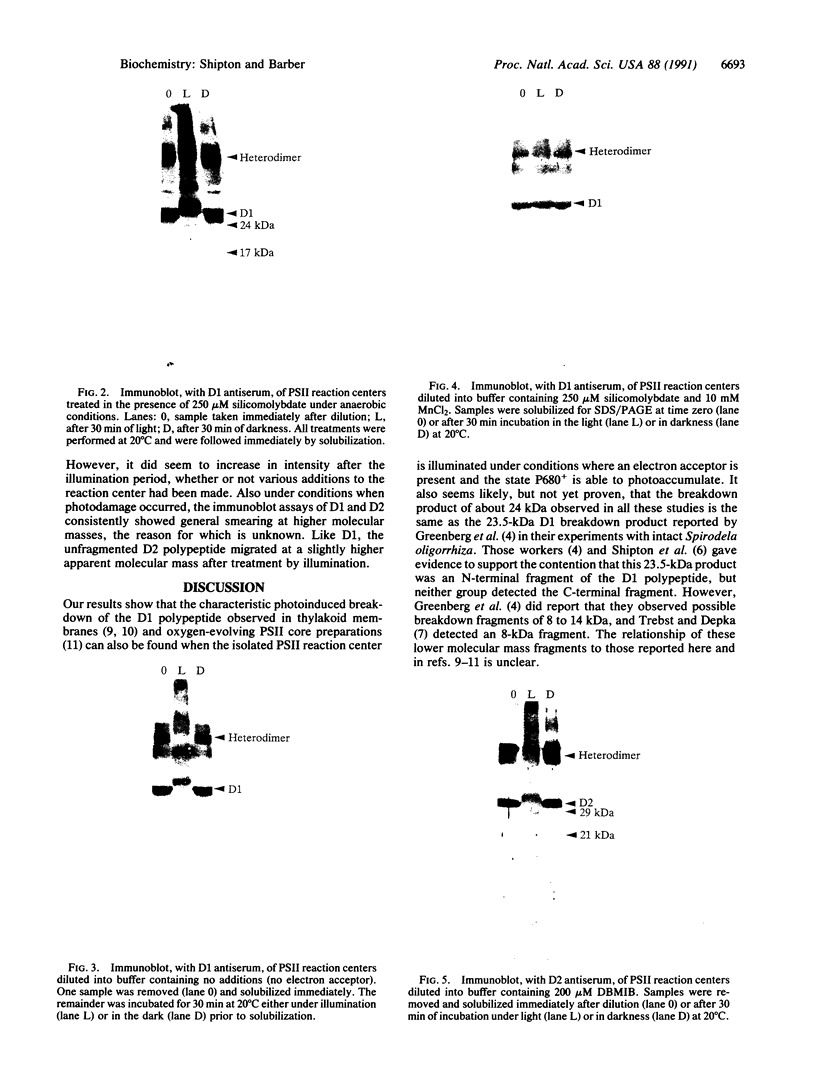
When the isolated D1/D2/cytochrome b559 complex was exposed to bright light, a distinctive pattern of D1 polypeptide fragments was observed under both aerobic and anaerobic conditions. The major degradation product had an apparent molecular mass of 24 kDa, while other fragments were detected at 17, 14, and 10 kDa by immunoblotting. This pattern was observed when the electron acceptors 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone or silicomolybdate were present during illumination. It is known that these conditions stabilize P680+ chlorophyll and bring about the photooxidation and destruction of pigments in the reaction center, particularly chlorophyll absorbing at 670 nm and beta-carotene. When P680+ was not allowed to accumulate, either by omission of an electron acceptor or by addition of both an electron donor (Mn2+) and an acceptor, no breakdown fragments were observed. In the former case, however, some degradation of the D1 and D2 polypeptides did occur. Under conditions that gave rise to the characteristic D1 breakdown pattern, the D2 polypeptide was also degraded to specific fragments detected at about 29 and 21 kDa by immunoblotting. The results indicate that the photoinduced degradation of D1 (and D2) does not involve exogenous proteases but is most likely an autoproteolytic process. Moreover, our data indicate that the photochemical damage giving rise to D1 and D2 degradation occurs on the oxidizing rather than the reducing side of photosystem II and involves photooxidation of the accessory pigments. The results are discussed in terms of D1 and D2 turnover and photoinhibition.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Greenberg B. M., Gaba V., Canaani O., Malkin S., Mattoo A. K., Edelman M. Separate photosensitizers mediate degradation of the 32-kDa photosystem II reaction center protein in the visible and UV spectral regions. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6617–6620. doi: 10.1073/pnas.86.17.6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg B. M., Gaba V., Mattoo A. K., Edelman M. Identification of a primary in vivo degradation product of the rapidly-turning-over 32 kd protein of photosystem II. EMBO J. 1987 Oct;6(10):2865–2869. doi: 10.1002/j.1460-2075.1987.tb02588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M., Inoue Y. A new photosystem II reaction center component (4.8 kDa protein) encoded by chloroplast genome. FEBS Lett. 1988 Dec 5;241(1-2):99–104. doi: 10.1016/0014-5793(88)81039-1. [DOI] [PubMed] [Google Scholar]
- Jegerschöld C., Styring S. Fast oxygen-independent degradation of the D1 reaction center protein in photosystem II. FEBS Lett. 1991 Mar 11;280(1):87–90. doi: 10.1016/0014-5793(91)80210-t. [DOI] [PubMed] [Google Scholar]
- Kyle D. J., Ohad I., Arntzen C. J. Membrane protein damage and repair: Selective loss of a quinone-protein function in chloroplast membranes. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4070–4074. doi: 10.1073/pnas.81.13.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanba O., Satoh K. Isolation of a photosystem II reaction center consisting of D-1 and D-2 polypeptides and cytochrome b-559. Proc Natl Acad Sci U S A. 1987 Jan;84(1):109–112. doi: 10.1073/pnas.84.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S., Wells R., Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986 Oct 17;234(4774):364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- Schuster G., Timberg R., Ohad I. Turnover of thylakoid photosystem II proteins during photoinhibition of Chlamydomonas reinhardtii. Eur J Biochem. 1988 Nov 1;177(2):403–410. doi: 10.1111/j.1432-1033.1988.tb14389.x. [DOI] [PubMed] [Google Scholar]
- Shipton C. A., Marder J. B., Barber J. Determination of catabolism of the photosystem II D 1 subunit by structural motifs in the polypeptide sequence. Z Naturforsch C. 1990 May;45(5):388–394. doi: 10.1515/znc-1990-0513. [DOI] [PubMed] [Google Scholar]
- Thompson L. K., Brudvig G. W. Cytochrome b-559 may function to protect photosystem II from photoinhibition. Biochemistry. 1988 Sep 6;27(18):6653–6658. doi: 10.1021/bi00418a002. [DOI] [PubMed] [Google Scholar]
- Virgin I., Ghanotakis D. F., Andersson B. Light-induced D1-protein degradation in isolated photosystem II core complexes. FEBS Lett. 1990 Aug 20;269(1):45–48. doi: 10.1016/0014-5793(90)81115-5. [DOI] [PubMed] [Google Scholar]