Abstract
X-ray analysis of enzyme–DNA interactions is very informative in revealing molecular contacts, but provides neither quantitative estimates of the relative importance of these contacts nor information on the relative contributions of specific and nonspecific interactions to the total affinity of enzymes for specific DNA. A stepwise increase in the ligand complexity approach is used to estimate the relative contributions of virtually every nucleotide unit of synthetic DNA containing abasic sites to its affinity for apurinic/apyrimidinic endonuclease (APE1) from human placenta. It was found that APE1 interacts with 9–10 nt units or base pairs of single-stranded and double-stranded ribooligonucleotides and deoxyribooligonucleotides of different lengths and sequences, mainly through weak additive contacts with internucleotide phosphate groups. Such nonspecific interactions of APE1 with nearly every nucleotide within its DNA-binding cleft provides up to seven orders of magnitude (ΔG° ∼ −8.7 to −9.0 kcal/mol) of the enzyme affinity for any DNA substrate. In contrast, interactions with the abasic site together with other specific APE1–DNA interactions provide only one order of magnitude (ΔG° ∼ −1.1 to −1.5 kcal/mol) of the total affinity of APE1 for specific DNA. We conclude that the enzyme's specificity for abasic sites in DNA is mostly due to a great increase (six to seven orders of magnitude) in the reaction rate with specific DNA, with formation of the Michaelis complex contributing to the substrate preference only marginally.
INTRODUCTION
Apurinic/apyrimidinic (AP) or abasic sites are formed in DNA by spontaneous base loss and as a result of treatment with certain chemical (acids, alkylating agents, etc.) or physical mutagens (UV or ionizing radiation) (1). Excision of damaged DNA bases by DNA glycosylases also creates AP site repair intermediates (1). The steady-state level of AP sites in living cells is estimated at 5–20 lesions per 106 bases and the rate of their formation at 10–30 lesions per 106 bases per hour (2). Thus, most AP sites are removed from DNA soon after formation.
Normally, AP sites are processed by AP endonucleases (1), which recognize AP sites and cleave the DNA phosphodiester backbone 5′ to the lesion to create a free 3′-OH terminus suitable for priming DNA polymerases (1). The major human AP endonuclease, APE1, is homologous to the major Escherichia coli AP endonuclease Xth, both sharing a common structural fold with DNase I (3–5). Both APE1 and Xth are constitutively expressed (1). In other eukaryotes the primary constitutive AP endonuclease (e.g. budding yeast's Apn1p) belongs to a family of which the second, inducible E.coli AP endonuclease Nfo is a prototypic member (1,3–5). AP endonucleases of the APE1/Xth family are small (30–40 kDa), monomeric, divalent metal cation-dependent enzymes (1,3).
In the past decade, significant progress has been made in the detailed analysis of specific protein–DNA interactions by X-ray crystallography, one of the most informative methods for analysis of biomolecules [for recent reviews, see (6–13) and references therein]. Crystal structures of uncomplexed APE1 and wild-type and mutant APE1 bound to AP site-containing DNA in the presence of different divalent cations offer many details of possible mechanisms of AP site recognition and DNA hydrolysis by this enzyme (14–16). Other AP endonucleases have also been subject to crystallographic study (17,18). However, a static structure provides neither quantitative estimates of the relative importance of individual contacts nor the relative contributions of strong and weak specific and nonspecific contacts to the total affinity of an enzyme for DNA (6,7). Only a handful of DNA-dependent enzymes have been analyzed with respect to the relative contributions of thermodynamic (complex formation) and kinetic (reaction rate constant) steps of catalysis to their affinity for DNA or substrate specificity, e.g. EcoRI, EcoRV and BamHI restriction endonucleases (19–21). It has been proposed that the specific protein–DNA recognition complex closely resembles the transition state complex, such that very tight binding to the recognition site in DNA does not inhibit catalysis but instead provides energy efficiently utilized along the path to the transition state (22). A recently developed mathematical model of competing specific and nonspecific binding sites provides an elegant way to estimate the contribution of nonspecific contacts to specific binding through titration with nonspecific DNA (23).
Studies of a number of DNA-dependent enzymes [reviewed in (6,7)] have shown that complex formation, including formation of contacts between an enzyme and specific sequences, cannot provide for either high enzyme affinity for DNA or substrate specificity. Virtually all nucleotide units within the DNA-binding cleft interact with these enzymes, and high affinity (five to eight orders of magnitude) is mainly provided by numerous weak interactions between the enzyme and various structural elements of many nucleotide units. Transition from nonspecific to specific DNA is accompanied by the strengthening of some contacts existing for nonspecific DNA and by the formation of new contacts (6,7). However, specific interactions between enzymes and cognate DNA are usually also weak, and the relative contribution of specific interactions to the enzyme's total affinity for DNA is rather small and does not exceed one to two orders of magnitude (6,7). On the other hand, after binding to the enzyme, DNA undergoes multiple conformational changes to reach the catalytically proficient structure; as a result, the reaction rate is highly accelerated for specific DNA. Enzyme specificity is thus provided at the stages of the enzyme-dependent adjustment of DNA conformation and directly by chemical steps of catalysis.
It is clear that specific protein–DNA complexes vary greatly in their structural properties and in the thermodynamic strategy they use to traverse energy barriers along the reaction coordinate. The relative importance of the structures of proteins and DNA, their conformational changes, conformational dynamics and additional interactions within protein and DNA molecules is probably individual for each enzyme, demanding case-by-case analysis. To evaluate the relative contributions of individual DNA elements to the enzyme affinity for long DNA, a new approach, stepwise increase in ligand complexity, or SILC [reviewed in (6,7)], has been used for a number of DNA-dependent enzymes (24–34) to yield thermodynamic models, in some cases, related to their established three-dimensional structures. In this study, we report a quantitative characterization of the structural determinants of substrate specificity of human APE1. The SILC approach is used to probe for interactions of the enzyme with a series of model ligands and substrates [single-stranded (ss) and double stranded (ds) specific and nonspecific oligodeoxynucleotides (ODNs)], and the results are analyzed using a thermodynamic model of specific DNA recognition.
MATERIALS AND METHODS
Enzymes
Electrophoretically homogeneous APE1 (∼37 kDa; 3.1 × 103 U/mg) was purified from human placenta by ammonium sulfate fractionation and sequential chromatography on hydroxyapatite (Pharmacia), Fractogel Toyopearl HW-55 (TOSOH, Japan) and CM-Trisacryl M (Pharmacia), and analyzed by SDS–PAGE as described previously (35). Escherichia coli Ung and bacteriophage T4 polynucleotide kinase were purchased from SibEnzyme (Novosibirsk, Russia).
Oligonucleotides
All unmodified ODNs and oligoribonucleotides (ORNs) were synthesized using standard phosphoramidite methods. ODNs containing a tetrahydrofuran AP site analogue [(3-hydroxytetrahydrofuran-2-yl)methyl phosphate; F] were synthesized as described previously (36). ODNs with an aldehydic AP site (2,3-dihydroxy-5-oxopentyl phosphate; R) were prepared from ODNs containing uracil at the appropriate position by Ung treatment. ODNs with a reduced AP site (2,3,5-trihydroxypentyl phosphate) were prepared from the respective oligonucleotides containing an aldehydic AP site by treatment with NaBH4 as described previously (37). ODNs containing any type of AP site or unmodified adenine in any position are further coded as NXM, where N is the length of the ODN, X is the type of base or AP site (A, F or R), and M is the position of the abasic site from the 5′-terminus. Thus, 14F8 stands for (pT)7pF(pT)6 and 24R8 for d(CTAGTCARCACTGTCTGTGGATACC). Concentrations of ODNs were determined using calculated extinction coefficients (38). If ds ODNs were required, ODNs containing an AP site were annealed to complementary ODNs, which are coded as N(Y)M, where N is the length of the ODN, (Y) is the base opposite to the AP site (A, C, G or T) and M is the distance from the 5′-terminus to the AP site in the complementary AP site-containing ODN. ODNs were radioactively labelled at the 5′-terminus using [γ-32P]ATP and polynucleotide kinase according to the manufacturer's instructions.
Enzyme activity assay
One unit of APE1 is defined as the amount of the enzyme that hydrolyses 1 nmol of phosphodiester bonds in AP DNA in 1 min at 37°C (35). The reaction mixtures (30–60 μl) contained 10 mM Tris-HCl (pH 7.6), 50 mM KCl, 2 mM MgCl2 and various concentrations of AP ODN(s). Reactions were initiated by adding 2–3 U of APE1 and stopped after incubation for 2–30 min at 37°C by addition of an equal volume of formamide gel-loading buffer (80% formamide, 15% glycerol, 10 mM EDTA). Reaction products were separated by 20% PAGE in the presence of 7 M urea. Gels were autoradiographed and the pieces corresponding to the bands were cut out and measured by Cerenkov counting. In inhibition experiments, calf thymus DNA was used as a substrate after partial nicking by DNase I, labeling with the Klenow fragment and [3H]TTP, and acidic depurination (35,39). Cleavage of such [3H]DNA by APE1 releases short acid-soluble fragments, while acid-insoluble radioactivity decreases to a background value.
To introduce one to two AP sites per molecule of pUC18 plasmid DNA, the plasmid was incubated in 100 mM sodium citrate (pH 5.0) at 70°C for 5 min and precipitated with ethanol (35).
Reaction mixtures (50 μl) contained 7 A260 U/ml of polymeric [3H]AP DNA or AP pUC18 plasmid DNA, the buffer described above, 160 mM KCl (the concentration optimal for polymeric DNA substrates) and various concentrations of inhibitor ODNs. Reactions were initiated by adding 6–8 U of APE1. Aliquots of 5–7 μl were withdrawn every 2–5 min and transferred onto 2.5 mm Whatman 3MM disks presoaked in 5% trichloroacetic acid. The disks were washed eight times in 5% trichloroacetic acid for 5 min at 4°C and then once in ice-cold acetone, dried and monitored for radioactivity in a Minibeta counter (LKB). All measurements (initial rates) were taken within the linear regions of the time courses and APE1 concentration curves.
APE1-dependent hydrolysis of AP pUC18 plasmid DNA was analyzed using electrophoresis in 1% agarose gels (35). The gels were stained with ethidium bromide, photographed and the images were scanned. The enzyme activity was estimated from a decrease in the intensity of the bands corresponding to supercoiled and circular DNA.
Kinetic parameters
The KM and Vmax values were calculated by least-squares nonlinear regression fitting using Microcal Origin v5.0 software. KI values were determined using different concentrations of inhibitors by least-squares nonlinear regression fitting (40,41). Values for IC50 were determined for varying concentrations of the inhibitor (0.1–10 IC50) at the [3H]AP DNA concentration equaling 2KM (7 A260 U/ml). Errors in IC50 were within 10–20%. From the equation for competitive inhibition (40,41), IC50 = 3KI at [S] = 2KM; errors in KI were within 10–30%.
RESULTS AND DISCUSSION
Competitive inhibition of APE1 by ODNs
We have determined the KM and Vmax (kcat) values for some substrates used in the reaction catalyzed by APE1. The values of observed kcat depended on the structure of the ODN substrate; they were found to be 0.27 s−1 for a 14mer ds 5′-[32P][(pT)7(pR)(pT)6] and 6.7 s−1 for a 24mer ds d(GTACGTARCCACAGACAGTGATGA). Since molecules of long [3H]AP DNA from calf thymus are heterogeneous in length, a true kcat value for [3H]AP DNA could not be determined by the method of acid-soluble products used for this substrate. Therefore, we have estimated the kcat value (2.0 s−1) for high molecular weight AP DNA using a pUC18 plasmid containing one to two AP sites per molecule. The observed values of kcat in the case of good substrates (2–7 s−1) were in good agreement with previously published kcat values for ODN substrates and preparations of recombinant APE1: 1.8 (15) and 10 s−1 (42).
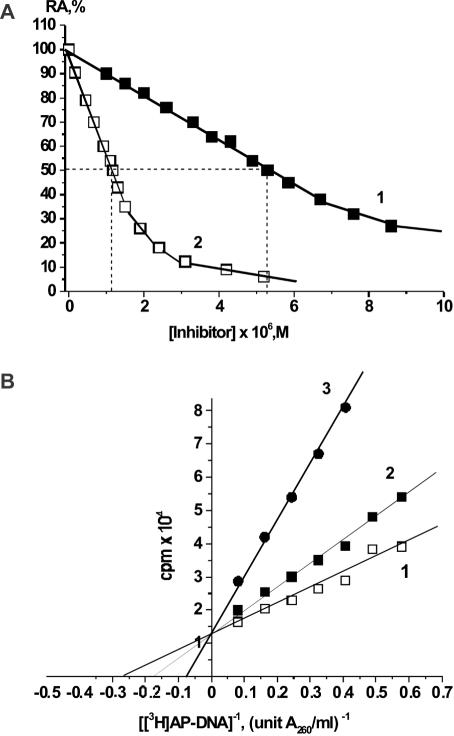
We have found that APE1 can bind different short specific and nonspecific ss and ds ODNs and that this binding inhibits the APE1 reaction (35) (Figure 1a). The inhibition was competitive for both [3H]AP DNA and ds 5′-[32P][(pT)7(pR)(pT)6] as substrates (Figure 1b). Therefore, KI provides an estimate of the affinity (Kd ∼ KI) of the APE1 DNA-binding site for ODNs (Figure 1b). Since most of the short ODNs had relatively low affinities for APE1, their KI values were calculated from the respective IC50 values. For competitive inhibition, IC50 = KI([S]/KM + 1) and under the conditions used ([S0] = 2KM), IC50 = 3KI; KI values calculated in this way were in excellent agreement with those determined experimentally for all tested ss and ds ODNs (see Tables 1–3). It should be mentioned that, despite the optimal concentration of KCl in the case of [3H]AP DNA (160 mM) being higher than that for the oligonucleotide substrate (<120 mM, 50 mM was used), the KI values for noncleaved ODNs determined using [3H]AP DNA and ds 5′-[32P][(pT)7(pR)(pT)6] as substrates, demonstrating different kcat and optimal concentrations of KCl, were comparable within the errors of the experimental methods. Higher concentration of KCl in the case of AP DNA is probably necessary to neutralize the negative charges of the polymeric substrates, which may be important for reaching the optimal conformation of these DNA in the complex with APE1 (see below).
Figure 1.
(a) Dependencies of the initial rate of APE1-catalyzed cleavage of [3H]AP DNA (3.5 U A260/ml) on the concentration of ss d(pA)10 (1) and ss d[(pT)7pR(pT)6] (2). The activity of APE1 in the absence of inhibitors was taken as 100%. (b) Lineweaver–Burk plot of the dependence of the initial rate of APE1-catalyzed accumulation of the acid-soluble short [3H]DNA product on the concentration of [3H]AP DNA at different concentrations of d(pT)10: 0 μM (1), 1.6 μM (2) and 7.8 μM (3) d(pT)10.
Table 1. Affinity of APN for minimal ligands, their derivatives, ss and ds homo-deoxy ODNsa.
| Ligand | IC50 (μM) | KI (μM)b | Ligand | IC50 (μM) | KI (μM)b | Ligand | IC50 (μM) | KI (μM)b |
|---|---|---|---|---|---|---|---|---|
| NaH2PO4 | 1080 | 360 | D-ribose | >0.5 M | >0.17 M | d(pR) | 75 | 25 |
| dAMP | 495 | 165 | dTMP | 490 | 163.3 | dCMP | 490 | 163.3 |
| Interlink phosphate (calculated) | N/A | 100 or 264c | d(pF) | 177 | 59.0 | dGMP | 500 | 166.6 |
| ss ODNs | ||||||||
| d(pA)2 | 150 | 50 | d(pT)2 | 350 | 116.6 | d(pC)2 | 420 | 140 |
| d(pT)3 | 200 | 66.7 | d(pC)3 | 180 | 60 | |||
| d(pA)4 | 100 | 33.3 | d(pT)4 | 135 | 45 | |||
| d(pC)5 | 65 | 21.7 | ||||||
| d(pA)6 | 51.6 | 17.2 | d(pT)6 | 74 | 24.6 | |||
| d(pC)7 | 30 | 10 | ||||||
| d(pA)8 | 7.5 | 2.5 | d(pT)8 | 25 | 8.3 | |||
| d(pC)9 | 14 | 4.7 | ||||||
| d(pA)10 | 5.0 | 1.66 | d(pT)10 | 7.5 | 2.5 | d(pC)10 | 10 | 3.33 |
| d(pT)11 | 7.5 | 2.5 | d(pC)11 | 10 | 3.33 | |||
| d(pA)12 | 5.4 | 1.7 | d(pT)12 | 7.5 | 2.5 | |||
| d(pC)13 | 10 | 3.33 | ||||||
| d(pA)14 | 5.3 | 1.7 | d(pT)14 | 7.5 | 2.5 | |||
| d(pT)15 | 7.7 | 2.57 | ||||||
| d(pA)16 | 5.1 | 1.66 | ||||||
| d[(pF)3pT] | 34 | 11.7 | d(pG)2 | 306 | 102 | |||
| d[(pF)5pT] | 15.6 | 5.2 | d(pG)4 | 115 | 38.3 | |||
| d[(pF)7pT] | 6.9 | 2.3 | d(pG)6 | 43 | 14.4 | |||
| d[(pF)9pT] | 3.0 | 1.0 | d(pG)8 | 16.3 | 5.4 | |||
| ds ODNs | ||||||||
| d(pA)2·d(pT)2 | 110 | 36.6 | d(pA)8·d(pT)8 | 28.0 | 9.3 | d(pA)14·d(pT)14 | 1.0 | 0.33 |
| d(pA)4·d(pT)4 | 80 | 26.6 | d(pA)10·d(pT)10 | 1.0 | 0.33 | d(pA)16·d(pT)16 | 1.0 | 0.33 |
| d(pA)6·d(pT)6 | 35.5 | 11.8 | d(pA)12·d(pT)12 | 1.1 | 0.36 | d(pA)20·d(pT)20 | 1.0 | 0.33 |
N/A, not applicable.
aStandard error in experimentally determined KI and IC50 values was 10–30%; mean of three to four measurements are given.
bKI values shown in bold were determined directly, others were calculated from the respective IC50 values.
cKI values for internucleotide phosphates were calculated from the log-dependencies for d(pN)n (264 mM) and for d[(pF)npT] (100 mM) by extrapolation of the lines to n = 0 (Figure 1).
Table 3. Affinities of APE1 for ORNs and their duplexesa.
| Ligand | IC50 (μM)a | KI (μM) | Ligand | IC50 (μM)b | KI (μM) | Ligand | IC50 (μM)b | KI (μM) |
|---|---|---|---|---|---|---|---|---|
| ss ORNs | ||||||||
| AMP | 1120 | 373 | UMP | 5620 | 1873 | CMP | 1340 | 447 |
| (pA)2 | 789 | 263 | (pC)2 | 949 | 316 | |||
| (pA)3 | 597 | 199 | ||||||
| (pA)4 | 550 | 183 | (pU)4 | 2100 | 700 | (pC)4 | 641 | 214 |
| (pA)6 | 300 | 100 | (pU)6 | 1129 | 339 | (pC)6 | 361 | 120 |
| (pA)8 | 110 | 36.7 | (pU)8 | 534 | 178 | (pC)8 | 212 | 70.8 |
| (pA)9 | 80.7 | 26.9 | (pU)9 | 475 | 158 | |||
| (pA)10 | 80 | 26.7 | (pU)10 | 471 | 157 | (pC)10 | 134 | 44.7 |
| (pA)12 | 90 | 30 | (pU)11 | 477 | 159 | (pC)14 | 114 | 38 |
| (pA)16 | 90 | 30 | (pU)16 | 455 | 158 | |||
| ds ORNs | ||||||||
| (pU)4·(pA)4 | 405 | 135 | (pA)4·d(pT)4 | 22.7 | 7.59 | |||
| (pU)6·(pA)6 | 49.8 | 16.6 | (pA)6·d(pT)6 | 64.1 | 21.4 | |||
| (pU)9·(pA)9 | 50 | 16.7 | (pA)8·d(pT)8 | 18.93 | 6.3 | |||
| (pU)10·(pA)10 | 30 | 10 | (pA)10·d(pT)10 | 7.5 | 2.5 | |||
| (pA)11·d(pT)11 | 7.5 | 2.5 | ||||||
| (pU)16·(pA)16 | 30 | 10 | (pA)16·d(pT)16 | 7.5 | 2.5 |
aError in directly determined KI (shown in bold) and IC50 values was 10–30%; means of three to four measurements are given.
bKI values not in bold were calculated from the respective IC50 values.
Additive interaction of APE1 with nucleotide units of deoxyribooligonucleotides
The formation of the primary complex between APE1 and DNA was analyzed using the SILC approach (6,7). The Gibbs free energy characterizing complex formation can be expressed as a sum of the ΔG° values for individual contacts (40,41): ΔG° = ΔG°1 + ΔG°2 + ··· + ΔG°n, where ΔG°i = −RT ln Kdi, Kdi indicating the contribution of an individual contact. Hence, the overall Kd value characterizing the complex formation is the product of the Kd values for all individual contacts:
![]()
Interactions between APE1 and DNA were found to be additive not only at the level of individual strands of the duplex but also at the level of individual units of a long DNA. Table 1 shows that the minimal ligands of APE1 are orthophosphate (Pi; KI = 360 μM) and deoxynucleotide monophosphates (dNMPs) (KI ∼165 μM). Thus, dNMPs interact with the active center of APE1 recognizing free nucleotides through both nucleoside and phosphate groups, with the latter making the major contribution.
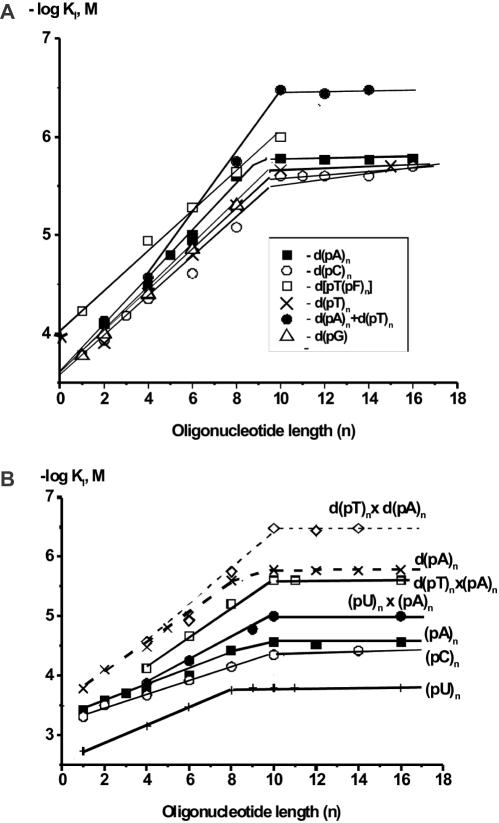
To assess the additivity of APE1 interactions with ODNs, the data in Table 1 were analyzed as logarithmic dependencies of KI (or Kd = KI) for d(pN)n versus the number of mononucleotide units n (Figure 2A). The linear log-dependencies for ss d(pN)n (1 ≤ n ≤ 10) provide evidence of the additivity of ΔG° for the interaction of 9–10 individual d(pN)n units with APE1. This number of nucleotide units of ss d(pN)n interacting within the globule of APE1 agrees well with previously published results that are characteristic of APE1 (43,44) and other DNA-binding enzymes of 30–40 kDa (6,7,31).
Figure 2.
Dependencies of −logKI on the length of inhibitor (n) for ss and ds deoxyribooligonucleotides (a) and ribooligonucleotides (b). (a) d(pT)n (crosses), d(pA)n (filled squares), d(pC)n (open circles), d(pG)n (triangles), d[(pF)npT] (open squares), d(pT)n·d(pA)n (filled circles). (b) (pU)n (crosses), (pC)n (open circles), (pA)n (filled squares), (pU)n·(pA)n (closed circles); the curves for d(pA)n (crosses) and d(pT)n·d(pA)n (diamonds) are given for comparison.
Values of f, the increase in APE1 affinity for various d(pN)n for a unit increase in their length, were evaluated from the slopes of the linear parts of these curves [Figure 2A; f(d(pC)n) = 1.53, f(d(pT)n) = 1.58, f(d(pG)n) = 1.62, f(d(pA)n) = 1.66]. The monotonic increase in Kd, reflecting interactions between the enzyme and one unit of ss DNA, equals the reciprocal of the f factor (KI = 1/f = 0.60–65 M). Thus, the KI values characterizing the affinity of the APE1 active center for nonspecific dNMPs (165 μM) are 3.6–3.9 × 103-fold lower than the KI (0.60–0.65 M) characterizing the enzyme interaction with any of the additional 8–9 nt of an extended ODN. The interaction of APE1 with all units of homo-d(pN)n is additive and the Kd (or KI) values for any ODN can be obtained by multiplying Kd for the minimal ligand (dNMP) with Kd = 1/f for each of the mononucleotide units:
![]()
Uracil DNA glycosylase (UDG) (28), Topo I (32,33) and the template-binding sites of many DNA polymerases (24,25,45) interact with DNA not only through weak electrostatic interactions with internucleotide phosphates but also through weak hydrophobic or van der Waals interactions with nucleobases. However, the affinities of other enzymes, such as Fpg (30,31) or EcoRI (27), for any d(pN)n does not depend on the relative hydrophobicity of the nucleobases, indicating that these enzymes essentially do not contact the DNA bases but mainly interact with the sugar–phosphate backbone. In the case of APE1, the increase in affinity for d(pN)n proceeds along with the increase in the relative hydrophobicity of the bases (C < T < G < A, cf. the respective f values above).
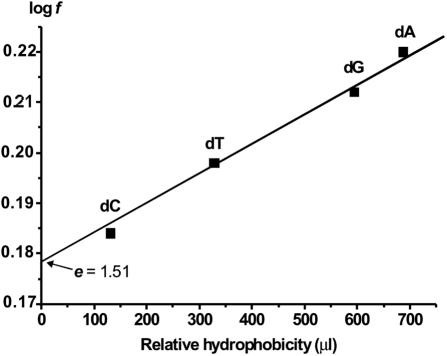
Extrapolation of the dependencies of log f versus the relative hydrophobicity of the bases to zero hydrophobicity gives an estimate of the increase in affinity of enzymes for DNA due to electrostatic interactions with a single internucleotide phosphate group. Consequently, it is termed the electrostatic (e) factor (6,7). For APE1 interacting with ss DNA, we found that e = 1.51 (Figure 3). As a measure of base hydrophobicity, the retention time of respective nucleosides during the isocratic elution from a reverse-phase column was used, as described previously (46). Given the additive character of interactions of the structural elements of d(pF) and the bases of d(pN)n to the DNA affinity of APE1, a factor of increase in affinity due to hydrophobic interactions of the enzyme with a single base (h factor) can be estimated as h = f/e. For APE1, h = 1.01, 1.05, 1.07 and 1.1 for d(pC)n, d(pT)n, d(pG)n and d(pA)n, respectively. Thus, the interaction of APE1 with each nucleotide unit of ss ODNs is a superposition of weak electrostatic and hydrophobic or van der Waals interactions with the individual structural elements and can be described as
Figure 3.
Logarithmic dependencies of factor f for APE1 on the relative hydrophobicity of nucleotide bases of homo-d(pN)n estimated from isocratic reverse phase chromatography of different nucleosides according to ref. (46). Extrapolation of the curve to zero hydrophobicity corresponding to orthophosphate gives an electrostatic factor e = 1.51.
![]()
where Kd[Pi] is the Kd value for the minimal orthophosphate ligand, and the numbers of C, T, G and A bases in d(pN)n are c, t, g and a, respectively. Experimentally measured KI values can be compared with the values calculated using Equation 1 for several hetero-d(pN)n (Table 2), and it was found that experimental and calculated KI values coincide within experimental error.
Table 2. Affinity of APE1 for hetero-deoxy ODNs and their duplexesa.
| Code | Sequence | KI(exp.) (μM)b | KI(calc.) (μM)c | KI(ss):KI(ds) | KI(specific):KI(nonspecific) |
|---|---|---|---|---|---|
| Nonspecific ss heterooligonucleotides | |||||
| ss dp(CTCCCTTCCT) | 3.1 ± 0.3 | 3.3 ± 0.3 | N/A | N/A | |
| ss dp(CTCACACACT) | 2.6 ± 0.3 | 2.9 ± 0.3 | N/A | N/A | |
| ss dp(GAAGAGAAGA) | 2.2 ± 0.4 | 1.9 ± 0.2 | N/A | N/A | |
| ss dp(CTAGTCA A CA)d | 2.6 ± 0.3 | 2.4 ± 0.3 | N/A | N/A | |
| Nonspecific and specific ss and ds homo-ODNs containing one irregular nucleotide | |||||
| 14C8 | ss d[(pT)7pC(pT)6] | 2.5 ± 0.3 | 2.6 ± 0.3 | 3.0 | |
| ds d[(pT)7pC(pT)6]·d(pA)14 | 0.83 ± 0.1 | N/D | |||
| 14G8 | ss d[(pT)7pG(pT)6] | 3.0 ± 0.3 | 2.6 ± 0.3 | 2.0 | ss/ss = 6.4–7.7 |
| ds d[(pT)7pG(pT)6]·d(pA)14 | 1.5 ± 0.2 | N/D | ds/ds = 6.4–11.5 | ||
| 14R8 | ss d[(pT)7pR(pT)6] | 0.39 ± 0.2 | 0.38 ± 0.4 | 3.0 | |
| ds d[(pT)7pR(pT)6]·d(pA)14 | 0.13 ± 0.15 | N/D | ss/ss = 5.0–6.0 | ||
| 14F8 | ss d[(pT)7pF(pT)6] | 0.5 ± 0.1 | N/D | 3.0 | ds/ds = 2.0 |
| ds d[(pT)7pF(pT)6]·d(pA)14 | 0.16 ± 0.03 | N/D | |||
| Nonspecific and specific ss and ds heterooligonucleotides | |||||
| 24A8 | ss dp(CTAGTCA A CACTGTCTGTGGATAC) | 2.1 ± 0.3 | 2.5 ± 0.3e | 4.2 | |
| ds dp(CTAGTCA A CACTGTCTGTGGATAC) | 0.5 ± 0.1 | 0.8 ± 0.2f | ss/ss = 6.0 | ||
| 24R8 | ss dp(CTAGTCA R CACTGTCTGTGGATAC) | 0.35 ± 0.3 | 0.39 ± 0.3e | 2.7 | ds/ds = 3.8 |
| ds dp(CTAGTCA R CACTGTCTGTGGATAC) | 0.13 ± 0.05 | 0.12 ± 0.2f | |||
N/A, not applicable, N/D, not determined.
aMean ± SE of three measurements are given.
bKI values shown in bold were determined directly, others were calculated from the respective IC50 values.
cCalculated KI values were estimated from Equation 1 using the following factors: Kd[Pi] = 264 μM, e = 1.51, hC = 1.01, hT = 1.05, hG = 1.07, hA = 1.10.
dThe italicized sequence of the decanucleotide corresponds to the 5′-sequence within long oligonucleotides 24A8 and 24R8 (the variable nucleotide in the eighth position is shown in bold).
eSince the DNA-binding site of APE1 contains 10 nucleotide-binding subsites, KI values for ss 24A8 and 24R8 were calculated for the decamers GTCAACACTG and GTCARCACTG, respectively.
fThe KI values for ds 24A8 and ds 24R8 were calculated by simple division of the calculated KI for ss 24A8 and ss 24R8 by an average value of a ratio of KI(ss)/KI(ds) = 3.0, calculated using six values from this table.
Interestingly, this expression describes the interaction of ss (or ds) DNA with any of the sequence-independent enzymes investigated so far, as well as the interaction of nonspecific DNA with most sequence-dependent enzymes (6,7). Different enzymes differ only in the values of e and hN factors. For example, e factors for DNA polymerases and UDG are 1.52 and 1.35, respectively, whereas for Fpg and EcoRI they are equal to 1 (hN = 1) (24,27,30). APE1 mostly interacts with the sugar–phosphate backbone of DNA (e = 1.51) and its hydrophobic or van der Waals interactions with nucleobases are less significant (h = 1.01–1.1).
Interaction of APE1 with the sugar–phosphate backbone of oligonucleotides
To confirm the predominant interactions of APE1 with the DNA backbone, and to estimate the contribution of the backbone structural units to the formation of weak additive contacts between APE1 and ODNs, we synthesized abasic oligomers, d[(pF)npT], where F is a tetrahydrofuran analogue of deoxyribose (Table 1). Since a d(pF) monomer has higher affinity for the active center of APE1 (KI = 59 μM) than dNMPs (165 μM), the log-dependence for d[(pF)npT] is shifted upward from nonspecific d(pN)n, but the slope of this line was slightly lower that that for d(pC)n (Figure 2); the factor f = 1.50 differs very little from the electrostatic factor e = 1.51 found for different homo-d(pN)n as described above. Thus, 9 out of 10 links of d[(pF)9pT] interact with APE1 with virtually the same efficiency as the deoxyribose phosphate structural elements of the backbone of nonspecific d(pN)n.
A 1.53–1.66-fold change in affinity on d(pN)n elongation by one nucleotide unit (corresponding to a change in ΔG° of −0.26 to −0.31 kcal/mol) is lower than would be expected for strong electrostatic contacts (up to −1.0 kcal/mol) or hydrogen bonds (−2 to −6 kcal/mol) (41), but is comparable with the values for weak hydrophobic, ion–dipole and dipole–dipole interactions (41). The crystal structure of human APE1 reveals that the enzyme possesses a preexisting positively charged surface for DNA binding and inserts loops into both grooves of DNA (14,15). This strip of positive potential probably underlies APE1 interactions with internucleotide phosphate groups of specific and nonspecific DNA. Negatively charged internucleotide groups of nonspecific ODNs could interact with the DNA-binding groove of APE1 through dipolar electrostatic forces rather than electrostatic interactions between point charges. Thus, the interaction between APE1 and DNA may resemble interaction between surfaces of opposite charge (6,7). It seems reasonable that APE1 could use a specific distribution of charged and neutral amino acid residues in the DNA-binding site for interactions with internucleotide phosphates and nucleobases, respectively. Therefore, in addition to weak hydrophobic or van der Waals interactions between DNA bases and amino acids of inserted protein loops, transition of DNA bases from water to an even slightly more hydrophobic environment of DNA-binding subsites can also lead to a favorable gain in energy during complex formation. The increase in APE1 affinity for DNA per base by a factor of 1.01–1.1 (ΔG° = −0.26 to −0.31 kcal/mol) is comparable with a gain in energy upon transfer of nucleobases from water to 1–3 M aqueous methanol (6,7).
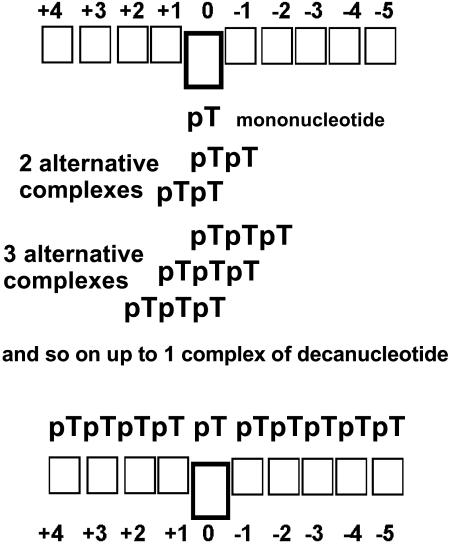
From the data discussed it can be concluded that the sugar–phosphate moiety of dNMPs [or each nucleotide of d(pN)n] interacts with the active center of APE1 through relatively strong nonspecific contacts with their phosphate groups and significantly weaker contacts with bases. ODNs containing two or more nucleotides can form several thermodynamically comparable microscopic complexes with APE1; the number of such complexes increases with increasing ODN length when n ≤ 5 and decreases when n ≥ 6, until d(pN)10, which can form only one complex with the enzyme (Figure 4). All interactions of APE1 with the nucleotide units of ODNs, except one unit that presumably fits directly into the active center, are weak and additive.
Figure 4.
Schematic structure of DNA binding site of APE1. The DNA-binding site of the enzyme consists of two sets of ten subsites each, but only one set of subsites interacting with the cleaved strand, shown in the figure, contains a specific subsite (‘0’ subsite) with increased affinity for one specific or nonspecific nucleotide unit of DNA. Lengthening of nonspecific d(pN)n (1 ≤ n ≤ 10) leads to the formation of several alternative thermodynamically comparable complexes of these ODNs with different subsites on the enzyme.
Additive interaction of APE1 with nucleotide units of ribooligonucleotides
According to structural data, APE1 introduces a kink into the helix of specific DNA (15). Structural characteristics of RNA and DNA differ markedly in solution: ds RNA usually exists in the A form and ds DNA in the B form, while ss r(pN)n adopt much more rigid nonflexible structures as compared with d(pN)n (47). Therefore, it was interesting to compare APE1 interaction with d(pN)n and r(pN)n (Tables 1 and 3). The affinity of the APE1 active center for AMP (373 μM) and CMP (447 μM) was 2–2.7-fold lower than that for dNMPs (163–165 μM), while the affinity for UMP (1873 μM) was 11.4-fold lower. The log-dependencies for (pA)n and (pU)n were linear for 1 ≤ n ≤ 8–9, and only for (pC)n was the curve linear up to n = 10 (Figure 2B). The values of f factors for d(pC)n (1.53), d(pT)n (1.58) and d(pA)n (1.66) are slightly higher compared with those for the respective ribooligonucleotides (pC)n (1.29), (pU)n (1.38) and (pA)n (1.40). Thus, not only can the active site of APE1 distinguish between conformationally different ribonucleotides and deoxyribonucleotides, but other subsites of the enzyme can interact with 9 out of 10 nucleotides less efficiently as well. At the level of decanucleotides, the difference between interaction of APE1 with 9 nt units of d(pN)10 and r(pN)10 was estimated as factors of 7.1, 5.3 and 4.9 for dA/rA, dT/rU and dC/rC, respectively. Overall, d(pN)10 and r(pN)10 interact with APE1 due to superposition of the same nonspecific interactions with internucleotide phosphates and bases. Most probably, (pN)n cannot be kinked by APE1 in the same way as d(pN)n, resulting in their lower affinity. Interestingly, the log-dependencies are linear up to 10 residues only for (pC)n, which, of all r(pN)n, possesses the highest conformational flexibility (47), while the affinity of (pA)n and (pU)n increases only up to n = 8 (Figure 2B). One possible explanation is that the two terminal nucleotides of (pA)n and (pU)n accommodated in the DNA-binding cleft of APE1 could lie far away from its positively charged region. Thus, the increased affinity for d(pN)n could stem from bringing the oppositely charged surfaces of APE1 and ODNs closer together as a result of easier conformational changes in the DNA and APE1 structures.
Affinity of APE1 for nonspecific DNA duplexes
Some enzymes, such as UDG, partially melt ds d(pN)10 and contact both strands of this relatively short ODN almost independently (28). In contrast, DNA polymerases and Topo I interact with both base-paired DNA strands (24–26,32,33). However, the contribution of the second strand to the affinity of any enzyme for ds DNA is usually much smaller than that of the first strand. A remarkable feature of the behavior of Topo I and DNA polymerases is the ‘assembly’ and subsequent stabilization of correct duplexes for which the melting temperature (Tm) in solution is substantially lower than the reaction temperature (6,7,24–26,32,33).
Figure 2 and Tables 1 and 3 show that the minimal ligand exhibiting duplex properties toward APE1 is d(pT)6–8·d(pA)6–8, and for an octamer duplex the Tm in solution [21°C, calculated according to ref. (48)] is lower than the reaction temperature (37°C). Shorter duplexes with Tm significantly lower than the reaction temperature behave as ss ODNs and not as duplexes under the present reaction conditions (Tables 1 and 3). Thus, short duplexes are weakly stabilized by their interaction with APE1. Similar to ss ODNs, a linear increase in log KI for duplexes was found up to n = 10. The affinity of APE1 for d(pT)n·d(pA)n is ∼5-fold higher than that for ss d(pA)n. The change in APE1 affinity for d(pT)n·d(pA)n (n ≥ 6) is described formally by the same equation as for ss ODN (see above), but the factor f increases from 1.58 [for d(pT)n] or 1.66 [for d(pA)n] to 2.44. The ratio of these f factors (characterizing an increase in affinity due to the addition of a single unit of the second strand) is 1.47–1.54 (ΔG° = −0.23 to −0.26 kcal/mol). Note that the formation of a single A:T or G:C pair in solution is characterized by ΔG° values of −1.2 to −1.9 kcal/mol and −2.0 to −2.8 kcal/mol, respectively (6,7). Interestingly, the affinity of an ORN duplex (pA)10·(pU)10 is only 2.7-fold higher than that for ss (pA)10 (Table 3), whereas the ratio of KI values for d(pA)10 and d(pA)10·d(pT)10 is 5 (Table 1). The addition of d(pT)n to a complementary (pA)n strand does not lead to an increase in the affinity of the mixed d(pT)10·(pA)10 duplex (KI = 2.5 μM) compared with that for d(pT)10 (KI = 2.5 μM) (Tables 1 and 3; Figure 2B). Thus, the contribution of the second strand is much lower than that of the first strand. In addition, APE1 seems to be unable to distort the solution structure of the RNA–RNA and RNA–DNA duplexes.
Thermodynamic model of APE1 interaction with nonspecific DNA
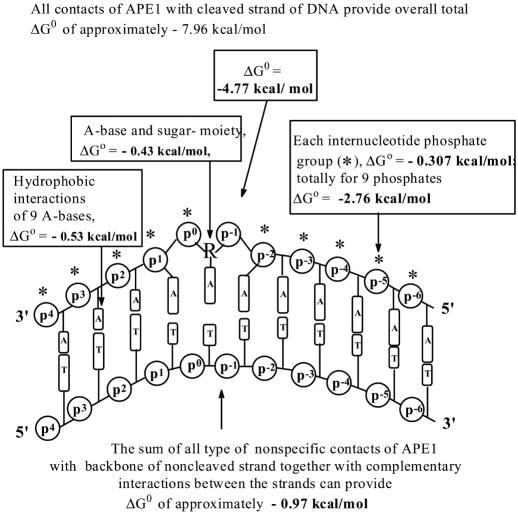
The contribution of interactions of any unit of nonspecific d(pN)10 (KI = 163–167 μM, ΔG° = −5.2 kcal/mol) with APE1 does not depend on the particular base (Table 1). The relative contribution of a phosphate group can be approximately estimated (ΔG° = −4.77 kcal/mol) from the KI value for orthophosphate (360 μM). Thus, the contributions of the nucleoside moiety of any dNMP unit of an ODN can be estimated from the difference in ΔG° for the nucleotides and orthophosphate as −0.43 kcal/mol. Since nine d(pA) nucleotide units of one strand of ds d(pA)10 interact with APE1 through weak additive contacts (f = 1.66; Kd = 0.6 M; ΔG° = −0.31 kcal/mol), the net relative contribution of these nine nucleotides may be estimated as ΔG° = −2.76 kcal/mol, ΔG° of the nine internucleotide phosphates as −2.23 kcal/mol and that of the nine bases as −0.53 kcal/mol. Thus, all contacts of APE1 with the poly(dA) strand interacting with the enzyme's DNA binding groove provide ΔG° of −7.96 kcal/mol. From the ratio of Kd values (equal to ∼5, or Kd = 0.2 M), characterizing the increased affinity for ds d(pA)10–16·d(pT)10–16 compared with d(pA)10–16, the contribution of the 10 nt units of the second strand to the affinity of ds DNA may be estimated as ΔG° = −0.97 kcal/mol. Extrapolation of structural data to APE1 complexed with undamaged DNA (3,15) suggests that, in order to search for lesions, the enzyme severely distorts and possibly melts DNA locally. Taking this into account, all interactions of APE1 with nonspecific DNA can be summarized using the thermodynamic model shown in Figure 5.
Figure 5.
Thermodynamic model of APE1 interactions with nonspecific DNA. For the enzyme subsites interacting with the cleaved strand, the ΔG° values characterizing their contacts with the d(pA)n chain of d(pA)n·d(pT)n are given; for the subsites interacting with the noncleaved strand, the ΔG° values refer to their contacts with the d(pT)n chain. All types of nonspecific additive interactions of APE1 with the d(pA)n·d(pT)n duplex provide ΔG° ∼ −7.96 kcal/mol.
Contribution of a specific AP site in DNA to its affinity for APE1
The relative contributions of an AP site to the total affinity of APE1 can be estimated for specific DNA. The KI values for duplexes corresponding to specific 14X8 and 24X8 ODNs (Table 2) were determined by using them as inhibitors of the APE1 cleavage of apurinized ds polymeric [3H]DNA. The increase in affinity on transition from nonspecific ss d(pT)14 (Table 1) and ss 14X8 (14C8 and 14G8) to specific ss 14X8 and ss 24X8 varied from 6.4 to 8.6 depending on the ODN sequence and length. The ratio of KI values for ss nonspecific 24A8 and specific 24R8 was equal to 6.0 (Table 2).
Transition from nonspecific ds d(pT)14 (Table 1) and ds 14N8 (14C8 and 14G8) to specific ds 14R8 and ds 24R8 (Table 2) led to a decrease in KI by a factor of 6.4–11.5. The affinity for specific ds 24R8 is only 3.8-fold higher than that for nonspecific 24A8 duplexes (Table 2). Interestingly, APE1 bound free deoxyribose-5′-phosphate 6.6-fold more effectively than various dNMPs. Thus, a relative contribution of specific interactions of APE1 with the natural AP site is comparable at the level of minimal ligands (6.6-fold), ss d(pN)n and ds ODNs (3.8–11.5-fold) (Table 2).
The affinity of APE1 for the 14F8:d(pA)14 duplex (0.16 μM; Table 2) was 2.1-fold higher than that for d(pT)14:d(pA)14 (0.33 μM) (Table 1). A very similar 2.5-fold difference in the affinity was observed for the phosphorylated tetrahydrofuran analogue d(pF) and various dNMPs. Thus, the contribution of specific and nonspecific interactions of different nucleotides of specific DNA to its total affinity for APE1 is nearly additive. The same situation occurs for two other repair enzymes, UDG and Fpg (6,7,28,30).
Thermodynamic model of APE1 interaction with specific DNA
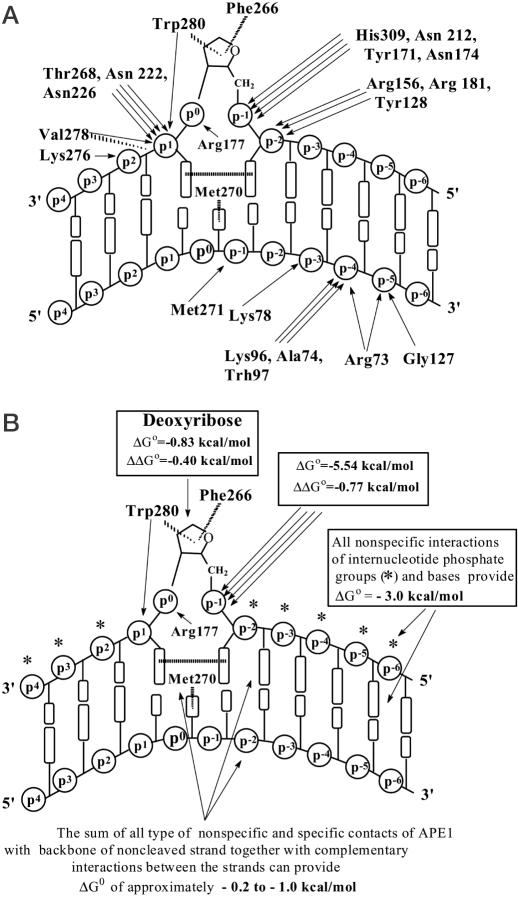
According to X-ray crystallographic data, APE1 electrostatically orients a rigid, preformed DNA-binding face and inserts loops into the DNA helix through both the major and the minor groove, stabilizing the target AP site in an extrahelical conformation. APE1-bound DNA is severely distorted, with the DNA bent at about 35° and the helical axis kinked by ∼5 Å. Figure 6 presents a summary of APE1 contacts with specific DNA (3,15). Immediately 3′ to the AP site, APE1 forms several bonds with two phosphates (p2 and p3) and braces the AP DNA backbone for the double loop insertion. At a position opposite to the everted AP site, Met-270 is inserted through the minor groove to pack against the orphaned base partner of the abasic site and occupy the space where it would be found in regular B-DNA. Above the abasic site, Arg-177 is inserted through the major groove and provides a hydrogen bond to the AP site 3′ phosphate (p1). Interactions in the major groove are unusual for base excision repair enzymes and, as the sequence and conformation of the Arg-177 loop is unique to APE1, it probably reflects specific APE1 functions. On the 5′ side of the lesion, the side chains of several amino acids residues contact the p-1 and p-2 phosphates of the damaged strand and the p-1, p-3, p-4 and p-5 phosphates of the undamaged strand, which results in a widening of the minor groove by ∼2 Å (3,15). These 5′ contacts may anchor the DNA for the kinking caused by the loop insertion at a position 3′ of the extrahelical abasic site.
Figure 6.
(A) Schematic representation of contacts between APE1 and specific ds DNA revealed by X-ray crystallography (3,15). Arrows indicate interactions between the various amino acid residues and structural elements of DNA, assisting the sharp DNA kinking (see text for details). (B) Thermodynamic model of APE1 interactions with specific DNA, displaying ΔG° values characterizing different contacts and strengthening of some contacts in comparison with nonspecific DNA (see Figure 5). The total ΔΔG° value characterizing a change in all types of interactions upon transition from nonspecific to specific DNA can be estimated at −1.1 to −1.5 kcal/mol.
Specific binding of extrahelical AP sites occurs in a hydrophobic pocket bordered by Phe-266, Trp-280 and Leu-282, which pack against the hydrophobic face and edge of the abasic deoxyribose. All listed interactions between APE1 and AP DNA stabilize the extrahelical AP site conformation and effectively lock APE1 onto the AP DNA.
As discussed above, the active site of APE1 can efficiently interact with different nucleotide units of DNA. At the same time, tight packing of the abasic deoxyribose against Phe-266, Trp-280 and Leu-282 should prevent productive binding of normal deoxynucleotides (3,15). We have shown previously that some sequence-specific enzymes have increased affinity for free deoxynucleotides compared with the same deoxynucleotide units within DNA (6,7,30,31). This may result from the absence of steric hindrance to free nucleotide binding, or from the restrictions imposed in a longer d(pN)n on nucleotide eversion or on a particular conformational change necessary for productive interaction of a unit of d(pN)n with the catalytic center of the enzyme. As free dNMPs, deoxyribose-5′-phosphate, deoxyribose and orthophosphate are the least restrained in their search for optimal binding interactions, their Kd values may put an upper estimate on the affinity of the respective elements of long DNA for the active site of APE1.
The affinity of APE1 for deoxyribose-5′-phosphate [d(pR); KI = 25 μM] and its tetrahydrofuran analogue [d(pF), KI = 59 μM] is 6.6- and 2.8-fold higher than that for nonspecific dNMPs (KI = 163–167 μM; Table 1). This increase in the affinity of APE1 for d(pR) in comparison with dNMPs can be a result of better interaction of the enzyme with the sugar moiety of d(pR). On the other hand, removal of the base from dNMPs could also lead to a remarkable strengthening of the enzyme's contacts with both sugar and phosphate groups. KI for the internucleotide phosphates in abasic DNA may be estimated at ∼100 μM by extrapolation of the line for d[(pF)npT] to n = 0 (Figure 1). Thus, a difference in the affinity for dNMPs and d(pF) (2.8-fold) is comparable with the ratio of KI values for the internucleotide phosphates of d(pN)n and d[(pF)npT] (3.0-fold). Therefore, the transition from d(pN)n to d[(pF)npT] leads mainly to strengthening of the enzyme active center contacts with only one internucleotide phosphate, while the contribution of the tetrahydrofuran moiety to the affinity for ODNs is very low. A similar situation probably occurs at the AP site unit in the AP DNA; detectable inhibition of APE1 by free deoxyribose was observed only at very high concentrations of this ligand (IC50 ≥ 0.5 M; KI ≥ 0.17 M). Assuming comparable contributions of the phosphate groups of d(pF) and d(pR) to their affinity for APE1 (KI = 100 μM; ΔG° = −5.54 kcal/mol), the contribution of the deoxyribose moiety of d(pR) can be estimated at KI = 0.25 M (ΔG° = −0.83 kcal/mol), a value that is in agreement with inhibition by free deoxyribose. Thus, the difference in APE1 affinity for deoxyribose itself and deoxyribose plus the adenine base within dAMP can be estimated at ΔΔG° = −0.4 kcal/mol (Figure 6B).
A significant overall 6.6-fold increase in the d(pR) affinity as compared with that for dNMP was observed, which is in good agreement with the steric restrictions imposed by Phe-266, Trp-280 and Leu-282 discussed above. However, the efficiency of APE1 interactions with AP sites in DNA is likely to depend strongly on the everted conformation of this nucleotide, which allows its 5′-phosphate to form stronger contacts with the enzyme (Figure 6). This strengthening may result from DNA backbone compression (3,15) and from displacement of the p-1 to a position where it can form more efficient contacts with four amino acid residues of the enzyme (Figure 6).
Depending on the sequence of ODNs used, a transition from nonspecific ss to different specific ss AP ODNs led to an increase in their affinity by a factor of 6.0–7.7 (Table 2). This difference remained nearly the same (3.8–11.5-fold) for different ds nonspecific versus specific AP ODNs (Table 2), and all these increase factors were comparable with the ratio of the KI values for dNMPs and d(pR) (6.6). Thus, the affinity improvement for different ss and ds AP sites containing specific compared with the respective nonspecific substrate (Table 2) is 6.0–11.5-fold (Kd = 0.087–0.26 M, ΔG° = −0.81 to −1.47 kcal/mol). The increase in affinity of APE1 for specific ds ODNs compared with that for specific ss ODNs is 2.7–4.2 (Table 2), which is comparable with the 2–5-fold difference between KI values for ss and ds nonspecific ODNs (Tables 1 and 2).
It is quite possible that some of the nonspecific contacts between APE1 and the internucleotide phosphate groups or nucleobases of the cleavable strand of specific ds ODNs are different from the contacts arising in nonspecific d(pN)n duplexes. Given the drastic APE1-dependent changes in the structure of specific DNA (Figure 6A), there could be a weakening of some contacts and strengthening of others formed by enzymes at the stage of primary complex formation. However, our data suggest that overall there is no remarkable thermodynamic difference between the majority of these contacts in specific and nonspecific ODNs. Taking into account a comparable difference in the affinity of APE1 for specific and nonspecific ligands at the level of a single dNMP DNA element, ss and ds DNAs, the contribution of all nonspecific contacts can be approximately put at ΔG° ∼ −3.3 kcal/mol (Figure 6B).
Transition from nonspecific to specific ODNs is probably also accompanied by some reorganization of contacts between APE1 and the second strand as well as between both DNA strands. Nevertheless, the average additional increase in the affinity for nonspecific (ΔG° = −0.97 kcal/mol) and specific DNAs due to the presence of the complementary strand may be characterized by similar values of Kd = 0.2–0.5 M and ΔG° = −0.2 to −1.0 kcal/mol. The contributions of the AP site and the second strand of ds DNA to the affinity depend to some extent on the DNA sequence. However, at the level of d(pR), ss and ds AP ODNs, the affinity of APE1 for specific ligands in comparison with nonspecific ones usually increases by a factor of 3.8–6.6 (ΔG° = −0.8 to −1.1 kcal/mol), most probably reflecting the contribution of the d(pR) unit to the affinity of these ligands for the active center of APE1. The recognition of specific AP DNA by APE1 can be generally described using the thermodynamic model shown in Figure 6B.
The efficiency of specific contact formation by APE1, as in the case of all studied DNA-dependent enzymes (6,7,31), does not exceed one to two orders of affinity, while the relative contribution of nonspecific interactions to the total affinity is four to five orders of magnitude greater (6,7). Formation of the enzyme–DNA complex cannot alone explain the observed specificity of enzyme catalysis. All the enzymes investigated to date, including APE1, interact with noncognate RNA–RNA and RNA–DNA duplexes with affinities comparable with those for DNA–DNA duplexes, and the affinity for such complexes is still only one to two orders of magnitude lower than that for specific DNA–DNA duplexes (6,7). However, the enzymes do not catalyze conversion of noncognate duplexes even at their saturating concentrations. The specificity of DNA-dependent enzymes lies in the kcat term; the rate is usually elevated by four to eight orders of magnitude upon transition from nonspecific to specific DNAs (6,7).
Kinetic factors: reaction rate and the specificity of APE1 action
Previous studies showed that APE1 could in principle cleave DNA at nonmodified nucleotides but only at high enzyme concentrations and longer incubation times (104–107-fold) compared with AP DNA (35,39). APE1 cannot hydrolyze nonspecific DNA with noticeable efficiency: the rate of nonspecific enzyme action decreases by six to eight orders of magnitude (35,39). The catalytic stage appears significantly more sensitive to the DNA structure than the stage of the enzyme–DNA complex formation. The rate of APE1-dependent hydrolysis of ODNs notably depends on the AP site structure; the rate for ds 14R8 decreased 10–14-fold when the natural aldehydic AP site was replaced with an AP site bearing a hydroxy group (NaBH4-reduced deoxyribose). This situation is similar to human UDG, where even minimal modifications of uracil, deoxyribose or internucleotide phosphate (e.g. introduction of a fluorine atom at certain positions) of a dUMP unit of specific ODNs often do not change the affinity for this ligand but result in a decrease in kcat that is less than three to four orders of magnitude, sometimes abolishing uracil excision altogether (49). However, the gross DNA structure does not seem to influence kcat, since its value was similar for high-molecular-weight plasmid DNA and the oligonucleotide substrates, both measured in this study and reported in the literature (42). The independence of kcat on DNA length was also observed for DNA repair glycosylases such as Fpg (50).
From the structure of the specific APE1–DNA complex, it is evident that enzyme-dependent DNA conformation adjustment involves pronounced kinking of both strands (15). It is known that the AP site significantly increases the ability of DNA to be kinked (51,52). However, ss d[(pT)7pR(pT)6] was a relatively poor substrate for APE1 and was not significantly hydrolyzed after 1 h of incubation, whereas ss hetero-ODNs of the same length containing AP sites were effectively cleaved after 10 min (35). The duplex of d[(pT)7pRd(pT)6] with d(pA)14 was cleaved 7-fold better (35). At the same time, the Vmax values of hetero ss AP ODNs were 10–15-fold higher than that for ds homo AP ODNs, while the Vmax values of hetero ss and ds AP ODNs differed by only 2–3-fold depending on the sequence (35). These data indicate that APE1 can distort ss as well as ds DNA, but the efficiency of DNA adjustment to the conformation optimal for catalysis depends on the DNA sequence.
Examples of many DNA-dependent enzymes show that the adjustment of DNA structure to the optimal conformation depends both on the initial structure in solution and on its flexibility in the enzyme-driven direction (6,7). The ability of different ds ODNs to be kinked and partially melted, necessary for DNA distortion by APE1, depends on several structural characteristics of DNA (33). DNA kinking and bending is notably facilitated in pyrimidine–purine sequences, which favor bending towards the major groove, and in regions with sterically unfavorable minor groove interactions between N3 and NH2 of guanine and N3 of adenine (33). Hetero-ODNs with the AP site incorporated in the context of a more flexible and easily kinked trinucleotide ARC demonstrated the highest affinity and Vmax values. Thus, in contrast to a quite rigid ss d(pT)n, the structure of ss specific hetero-ODNs can probably be changed by the enzyme much more easily. As shown above, addition of a complementary strand even to intrinsically rigid homo ss AP ODNs can convert such a ligand into a good APE1 substrate (35). This result agrees well with the important role of the second DNA strand in productive DNA distortion by APE1, as evident from the structural data (3,15). Similar results have been observed for human UDG (28) and for Fpg (53). All these data suggest that the second strand can be actively involved in attaining the optimal DNA conformation in complexes with repair enzymes. The capability of both strands of specific ds DNA to be distorted by APE1 may be very important for more productive formation of all APE1–DNA contacts revealed by X-ray crystallography (3,15). Introduction of an additional AP site into the second strand of a 24mer hetero-ODN leads to an 8–10-fold increase in affinity over two alternative duplexes containing a single AP site in either of the strands (35). Such an increase in the affinity, however, did not lead to a significant increase in the cleavage rate. Therefore, it cannot be excluded that the formation of a limited number of strong contacts between APE1 and the two strands of AP DNA is not obligatory for the productive eversion of the AP site from DNA.
Comparison of APE1 with other DNA-dependent enzymes
APE1, like many other DNA-depending enzymes (UDG, Fpg, Topo I, EcoRI, HIV integrase), interacts efficiently with both specific and nonspecific ss and ds ODNs (27,28,30–34) through contacts with the internucleotide phosphate groups and bases of DNA. The factor e (1.51) for APE1 (reflecting its interaction with one internucleotide phosphate of nonspecific DNA) is comparable with e factors for other enzymes: UDG (1.35), DNA polymerases (1.52), Fpg (1.54), RNA helicase (1.61), Topo I (1.67), EcoRI (2.0) and DNA ligase (2.14) (6,7,24–28,30–34). The factor h (1.01–1.10) for APE1 is remarkably lower than that for other enzymes interacting with bases of ss or ds DNA: DNA polymerases (1.03–1.32), Topo I (1.04–1.4), UDG (1.04–1.41), RNA helicase (1.05–1.59) and DNA ligase (1.1–1.62) (6,7,24–28,30–34). The relative contribution of the second strand to the affinity of APE1 for ds DNA is much lower than that for the first strand, again similar to other enzymes analyzed. Recognition of nonspecific DNA by sequence- and structure-specific DNA-dependent enzymes may be considered a first stage of specific DNA recognition. This stage of the primary complex formation due to nonspecific interactions between DNA and enzymes provides high affinity of any DNA-dependent enzyme for any DNA (6,7). High affinity of the enzymes to nonspecific DNA allows their ‘sliding’ along DNA to the site containing a specific sequence, lesion or structural element (6,7). The positively charged DNA-binding grooves of the enzymes and the negatively charged DNA sugar–phosphate backbone can interact during primary complex formation through many weak additive contacts. Since all these contacts are thermodynamically nearly equal, the enzymes can easily slide along DNA in search of specific elements, which are then recognized in unique enzyme-specific ways (6,7).
APE1 binds the DNA minor groove via a conserved minor-groove widening loop (3,15), suggesting that the enzyme could search for AP sites by using this loop to slightly distort DNA. Minor-groove widening is probably a conserved function of the four-layered α,β-sandwich fold, as similar interactions are also seen in bovine DNase I (54,55) and in E.coli Xth (17). The penetration of the DNA minor groove anchors one half of APE1 to the DNA, while the electrostatic attraction between the positive APE1 DNA-binding groove and the negative DNA phosphodiester backbone ensures that the entire enzyme molecule remains properly oriented. In this half-bound configuration APE1 can slide progressively along DNA, scanning for regions that can accommodate the kinking induced by the enzyme (3). Only abasic DNA can be deformed in this manner within the constraints of the APE1 abasic nucleotide-binding pocket (Figure 6) (3,15).
APE1 belongs to a group of highly specific DNA-dependent enzymes that catalyze the conversion of specific DNA four to eight orders of magnitude more effectively than that for nonspecific DNA ([(6,7) and references therein]. The increase in affinity of such enzymes for specific ds ODNs compared with nonspecific ones was estimated at 7–10-fold (UDG, Fpg), 50–70-fold (HIV integrase), 50–100-fold (EcoRI), 200–250-fold (Topo I) (6,7) (27–34) and 6–11-fold for APE1 (this study). Thus, the efficiency of specific contact formation between such enzymes and DNA does not exceed one to two orders of affinity and the relative contribution of nonspecific interactions to the total affinity is four to six orders of magnitude greater than that of specific interactions. Although these enzymes do not act on nonspecific DNA, the formation of a primary complex cannot alone explain their specificity. At the same time, the low affinity of enzymes for specific parts of their substrates can be of biological significance. An increase in the affinity for specific sequences limited to one to two orders of magnitude ensures a relatively short lifetime for a specific complex. The specificity of enzyme action can thus be provided by the impossibility of productive enzyme-dependent deformation of nonspecific DNA during the short existence time of the complex. For several DNA-dependent enzymes (including APE1), the conformational adjustment step of the reaction, in contrast to DNA binding, is extremely sensitive for specific DNA elements, and it is this step that determines the reaction rates for different DNAs (6,7,24–34). According to structural data, APE1 cannot promote productive eversion of a normal nucleotide into the enzyme active site pocket (3,15) and therefore a satisfactory orbital overlap and high reaction rate cannot be achieved. The formation of specific bonds between the extrahelical abasic site and amino acid residues in the active site (Figure 6) is most probably one of the final stages in the selection of specific DNA by APE1. After formation of such contacts, the reaction can be accelerated by six to seven orders of magnitude. Specific contacts between APE1 and the sugar moiety of d(pR) can provide, at most, a 6.6-fold increase in the affinity for specific DNA. Experimentally determined increase in affinity for AP DNA, compared with nonspecific DNA, does not exceed 3.8–11-fold. Moreover, this small increase arises not only from APE1-specific interaction with the sugar moiety of the AP site but also from strengthening of the enzyme contacts with other parts of the cleaved and noncleaved strands of AP DNA (Figure 6). Thus, the actual thermodynamic contribution of APE1-specific interaction with the extrahelical AP site is remarkably low. In general, recognition of small ligands by enzymes is based on the formation of several strong contacts (hydrogen bonds, electrostatic contacts, stacking interactions, etc.) with specific structural elements. Interestingly, during formation of a specific complex of ds DNA with EcoRI, 12 hydrogen bonds are formed, providing in total only about two orders of affinity (27). This means that the energy of each of these 12 bonds is rather low (ΔG° ∼ −0.23 kcal/mol) and comparable with the energy of weak additive nonspecific interactions (6,7,27). Only one order of affinity (ΔG° ∼ −0.28 to −0.36 kcal/mol) is accounted for by five pseudo-Watson–Crick hydrogen bonds formed by a uracil residue with UDG (28). Similar weak specific contacts with nucleotides of DNA were observed for all other investigated enzymes (6,7), indicating that formation of specific contacts between enzymes and DNA is not very important at the stage of protein–DNA complexation. This hydrogen bond energetic summary does not take into account the solvation reorganization energies (enthalpy and entropy) of hydrogen-bond networks such as these and must thus be a lower limit for the isolated hydrogen bond contributions in these cases. On the contrary, such contacts are extremely important at the stage of adjustment of DNA and enzyme conformations, and only in the case of specific DNA do specific contacts provide a very precise alignment of electronic orbitals of the reacting atoms.
Acknowledgments
ACKNOWLEDGEMENTS
This research was made possible in part by grants from the Wellcome Trust UK (070244/Z/03/Z), Presidium of the Russian Academy of Sciences (Physicochemical Biology Program 10.5), Russian Foundation for Basic Research (01-04-48892, 02-04-49605), Russian Ministry of Education (PD02-1.4-469), Award no. NO-008-X1 of the U.S. Civilian Research & Development Foundation for the Independent States of the Former Soviet Union (CRDF), Russian Science Support Foundation (to D.O.Z.) and funds from the Siberian Division of the Russian Academy of Sciences.
REFERENCES
- 1.Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. ASM Press, Washington, DC. [Google Scholar]
- 2.Atamna H., Cheung,I. and Ames,B.N. (2000) A method for detecting abasic sites in living cells: age-dependent changes in base excision repair. Proc. Natl Acad. Sci. USA, 97, 686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mol C.D., Hosfield,D.J. and Tainer,J.A. (2000) Abasic site recognition by two apurinic/apyrimidinic endonuclease families in DNA base excision repair: the 3′ ends justify the means. Mutat. Res., 460, 211–229. [DOI] [PubMed] [Google Scholar]
- 4.Eisen J.A. and Hanawalt,P.C. (1999) A phylogenomic study of DNA repair genes, proteins, and processes. Mutat. Res., 435, 171–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aravind L., Walker,D.R. and Koonin,E.V. (1999) Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res., 27, 1223–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nevinsky G.A. (1995) Important role of weak interactions in long DNA and RNA molecule recognition by enzymes. Mol. Biol. (Moscow), 29, 6–19. [PubMed] [Google Scholar]
- 7.Bugreev D.V. and Nevinsky,G.A. (1999) Possibilities of the method of step-by-step complication of ligand structure in studies of protein–nucleic acid interactions: mechanisms of functioning of some replication, repair, topoisomerization, and restriction enzymes. Biochemistry (Moscow), 64, 237–249. [PubMed] [Google Scholar]
- 8.Phillips S.E.V. and Moras,D. (1999) Protein–nucleic acid interactions. Curr. Opin. Struct. Biol., 9, 11–13. [DOI] [PubMed] [Google Scholar]
- 9.Tainer J.A. and Friedberg,E.C. (2000) Dancing with the elephants: envisioning the structural biology of DNA repair pathways. Mutat. Res., 460, 139–141. [DOI] [PubMed] [Google Scholar]
- 10.Luscombe N.M., Austin,S.E., Berman,H.M. and Thornton,J.M. (2000) An overview of the structures of protein-DNA complexes. Genome Biol., 1, 1–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pingoud A. and Jeltsch,A. (2001) Structure and function of type II restriction endonucleases. Nucleic Acids Res., 29, 3705–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rice P.A. and Baker,T.A. (2001) Comparative architecture of transposase and integrase complexes. Nature Struct. Biol., 8, 302–307. [DOI] [PubMed] [Google Scholar]
- 13.Aggarwal A.K. and Doudna,J.A. (2003) Protein–nucleic acid interactions. Curr. Opin. Struct. Biol., 13, 3–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorman M.A., Morera,S., Rothwell,D.G., de La Fortelle,E., Mol,C.D., Tainer,J.A., Hickson,I.D. and Freemont,P.S. (1997) The crystal structure of the human DNA repair endonuclease HAP1 suggests the recognition of extra-helical deoxyribose at DNA abasic sites. EMBO J., 16, 6548–6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mol C.D., Izumi,T., Mitra,S. and Tainer,J.A. (2000) DNA-bound structures and mutants reveal abasic DNA binding by APE1 DNA repair and coordination. Nature, 403, 451–456. [DOI] [PubMed] [Google Scholar]
- 16.Beernink P.T., Segelke,B.W., Hadi,M.Z., Erzberger,J.P., Wilson,D.M.,III and Rupp,B. (2001) Two divalent metal ions in the active site of a new crystal form of human apurinic/apyrimidinic endonuclease, Ape1: implications for the catalytic mechanism. J. Mol. Biol., 307, 1023–1034. [DOI] [PubMed] [Google Scholar]
- 17.Mol C.D., Kuo,C.F., Thayer,M.M., Cunningham,R.P. and Tainer,J.A. (1995) Structure and function of the multifunctional DNA-repair enzyme exonuclease III. Nature, 374, 381–386. [DOI] [PubMed] [Google Scholar]
- 18.Hosfield D.J., Guan,Y., Haas,B.J., Cunningham,R.P. and Tainer,J.A. (1999) Structure of the DNA repair enzyme endonuclease IV and its DNA complex: double-nucleotide flipping at abasic sites and three-metal-ion catalysis. Cell, 98, 397–408. [DOI] [PubMed] [Google Scholar]
- 19.Lesser D.R., Kurpiewski,M.R. and Jen-Jacobson,L. (1990) The energetic basis of specificity in the EcoRI endonuclease-DNA interaction. Science, 250, 776–786. [DOI] [PubMed] [Google Scholar]
- 20.Engler L.E., Welch,K.K. and Jen-Jacobson,L. (1997) Specific binding by EcoRV endonuclease to its DNA recognition site GATATC. J. Mol. Biol., 269, 82–101. [DOI] [PubMed] [Google Scholar]
- 21.Engler L.E., Sapienza,P., Dorner,L.F., Kucera,R., Schildkraut,I. and Jen-Jacobson,L. (2001) The energetics of the interaction of BamHI endonuclease with its recognition site GGATCC. J. Mol. Biol., 307, 619–636. [DOI] [PubMed] [Google Scholar]
- 22.Jen-Jacobson L. (1997) Protein-DNA recognition complexes: conservation of structure and binding energy in the transition state. Biopolymers, 44, 153–180. [DOI] [PubMed] [Google Scholar]
- 23.Tsodikov O.V., Holbrook,J.A., Shkel,I.A. and Record,M.T.,Jr (2001) Analytic binding isotherms describing competitive interactions of a protein ligand with specific and nonspecific sites on the same DNA oligomer. Biophys. J., 81, 1960–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolocheva T.I., Nevinsky,G.A., Levina,A.S., Khomov,V.V. and Lavrik,O.I. (1991) The mechanism of recognition of templates by DNA polymerases from pro- and eukaryotes as revealed by affinity modification data. J. Biomol. Struct. Dyn., 9, 169–186. [DOI] [PubMed] [Google Scholar]
- 25.Ljach M.V., Kolocheva,T.I., Gorn,V.V., Levina,A.S. and Nevinsky,G.A. (1992) The affinity of the Klenow fragment of E. coli DNA-polymerase 1 to primers containing bases noncomplementary to the template and hairpin-like elements. FEBS Lett., 300, 18–20. [DOI] [PubMed] [Google Scholar]
- 26.Kolocheva T.I., Maksakova,G.A., Zakharova,O.D. and Nevinsky,G.A. (1996) The algorithm of estimation of the Km values for primers in DNA synthesis catalyzed by human DNA polymerase α. FEBS Lett., 399, 113–116. [DOI] [PubMed] [Google Scholar]
- 27.Kolocheva T.I., Maksakova,G.A., Bugreev,D.V. and Nevinsky,G.A. (2001) Interaction of endonuclease EcoRI with short specific and nonspecific oligonucleotides. IUBMB Life, 51, 189–195. [DOI] [PubMed] [Google Scholar]
- 28.Vinogradova N.L., Bulychev,N.V., Maksakova,G.A., Johnson,F. and Nevinskii,G.A. (1998) Uracil DNA glycosylase: interpretation of X-ray data in the light of kinetic and thermodynamic studies. Mol. Biol. (Moscow), 32, 400–409. [PubMed] [Google Scholar]
- 29.Ishchenko A.A., Koval,V.V., Fedorova,O.S., Douglas,K.T. and Nevinsky,G.A. (1999) Structural requirements of double and single stranded DNA substrates and inhibitors, including a photoaffinity label, of Fpg protein from Escherichia coli. J. Biomol. Struct. Dyn., 17, 301–310. [DOI] [PubMed] [Google Scholar]
- 30.Ishchenko A.A., Vasilenko,N.L., Sinitsina,O.I., Yamkovoy,V.I., Fedorova,O.S., Douglas,K.T. and Nevinsky,G.A. (2002) Thermodynamic, kinetic, and structural basis for recognition and repair of 8-oxoguanine in DNA by Fpg protein from Escherichia coli. Biochemistry, 41, 7540–7548. [DOI] [PubMed] [Google Scholar]
- 31.Zharkov D.O., Ishchenko,A.A., Douglas,K.T. and Nevinsky,G.A. (2003) Recognition of damaged DNA by Escherichia coli Fpg protein: insights from structural and kinetic data. Mutat. Res., 531, 141–156. [DOI] [PubMed] [Google Scholar]
- 32.Bugreev D.V., Buneva,V.N., Sinitsina,O.I. and Nevinskii,G.A. (2003) The mechanism of the supercoiled DNA recognition by eukaryotic type I topoisomerases. I. The enzyme interaction with nonspecific oligonucleotides [in Russian]. Bioorg. Khim., 29, 163–174. [DOI] [PubMed] [Google Scholar]
- 33.Bugreev D.V., Sinitsina,O.I., Buneva,V.N. and Nevinskii,G.A. (2003) The mechanism of supercoiled DNA recognition by eukaryotic type I topoisomerases. II. A comparison of the enzyme interaction with specific and nonspecific oligonucleotides [in Russian]. Bioorg. Khim., 29, 277–289. [DOI] [PubMed] [Google Scholar]
- 34.Bugreev D.V., Baranova,S., Zakharova,O.D., Parissi,V., Desjobert,C., Sottofattori,E., Balbi,A., Litvak,S., Tarrago-Litvak,L. and Nevinsky,G.A. (2003) Dynamic, thermodynamic, and kinetic basis for recognition and transformation of DNA by human immunodeficiency virus type 1 integrase. Biochemistry, 42, 9235–9247. [DOI] [PubMed] [Google Scholar]
- 35.Beloglazova N.G., Petruseva,I.O., Bulychev,N.V., Maksakova,G.A., Johnson,F. and Nevinskii,G.A. (1997) Isolation and substrate specificity of apurine/apyrimidine endonuclease from human placenta. Mol. Biol. (Moscow), 31, 1104–1111. [PubMed] [Google Scholar]
- 36.Takeshita M., Chang,C.-N., Johnson,F., Will,S. and Grollman,A.P. (1987) Oligodeoxynucleotides containing synthetic abasic sites. Model substrates for DNA polymerases and apurinic/apyrimidinic endonucleases. J. Biol. Chem., 262, 10171–10179. [PubMed] [Google Scholar]
- 37.Castaing B., Boiteux,S. and Zelwer,C. (1992) DNA containing a chemically reduced apurinic site is a high affinity ligand for the E. coli formamidopyrimidine-DNA glycosylase. Nucleic Acids Res., 20, 389–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cantor C.R., Warshaw,M.M. and Shapiro,H. (1970) Oligonucleotide interactions. 3. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers, 9, 1059–1077. [DOI] [PubMed] [Google Scholar]
- 39.Beloglazova N.G., Lokhova,I.A., Maksakova,G.A., Tsvetkov,I.V. and Nevinskii,G.A. (1996) Apurine/apyrimidine endonuclease from human placenta. Recognition of apurinized DNA by the enzyme. Mol. Biol. (Moscow), 30, 220–230. [PubMed] [Google Scholar]
- 40.Cornish-Bowden A. (1976) Principles of Enzyme Kinetics. Butterworths, London. [Google Scholar]
- 41.Fersht A. (1985) Enzyme Structure and Mechanism. 2nd edn. W. H. Freeman & Co., New York. [Google Scholar]
- 42.Lucas J.A., Masuda,Y., Bennett,R.A.O., Strauss,N.S. and Strauss,P.R. (1999) Single-turnover analysis of mutant human apurinic/apyrimidinic endonuclease. Biochemistry, 38, 4958–4964. [DOI] [PubMed] [Google Scholar]
- 43.Wilson D.M. III, Takeshita,M. and Demple,B. (1997) Abasic site binding by the human apurinic endonuclease, Ape, and determination of the DNA contact sites. Nucleic Acids Res., 25, 933–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nguyen L.H., Barsky,D., Erzberger,J.P. and Wilson,D.M.,III (2000) Mapping the protein-DNA interface and the metal-binding site of the major human apurinic/apyrimidinic endonuclease. J. Mol. Biol., 298, 447–459. [DOI] [PubMed] [Google Scholar]
- 45.Kolocheva T.I., Levina,A.S. and Nevinsky,G.A. (1996) Recognition of the primers containing different modified nucleotide units by the Klenow fragment of DNA polymerase I from E. coli. Biochimie, 78, 201–203. [DOI] [PubMed] [Google Scholar]
- 46.Doronin S.V., Lavrik,O.I., Nevinsky,G.A. and Podust,V.N. (1987) The efficiency of dNTP complex formation with human placenta DNA polymerase α as demonstrated by affinity modification. FEBS Lett., 216, 221–224. [DOI] [PubMed] [Google Scholar]
- 47.Saenger W. (1984) Principles of Nucleic Acid Structure. Springer-Verlag, New York. [Google Scholar]
- 48.Breslauer K.J., Frank,R., Blocker,H. and Marky,L.A. (1986) Predicting DNA duplex stability from the base sequence. Proc. Natl Acad. Sci. USA, 83, 3746–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kubareva E.A., Volkov,E.M., Vinogradova,N.L., Kanevsky,I.A., Oretskaya,T.S., Kuznetsova,S.A., Brevnov,M.G., Gromova,E.S., Nevinsky,G.A. and Shabarova,Z.A. (1995) Modified substrates as probes for studying uracil-DNA glycosylase. Gene, 157, 167–171. [DOI] [PubMed] [Google Scholar]
- 50.Zaika E.I., Perlow,R.A., Matz,E., Broyde,S., Gilboa,R., Grollman,A.P. and Zharkov,D.O. (2004) Substrate discrimination by formamidopyrimidine-DNA glycosylase: a mutational analysis. J. Biol. Chem., 279, 4849–4861. [DOI] [PubMed] [Google Scholar]
- 51.Ayadi L., Coulombeau,C. and Lavery,R. (2000) The impact of abasic sites on DNA flexibility. J. Biomol. Struct. Dyn., 17, 645–653. [DOI] [PubMed] [Google Scholar]
- 52.Barsky D., Foloppe,N., Ahmadia,S., Wilson,D.M.,III and MacKerell,A.D.,Jr (2000) New insights into the structure of abasic DNA from molecular dynamics simulations. Nucleic Acids Res., 28, 2613–2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ishchenko A.A., Bulychev,N.V., Maksakova,G.A., Johnson,F. and Nevinsky,G.A. (1997) Recognition and conversion of single- and double-stranded oligonucleotide substrates by 8-oxoguanine-DNA glycosylase from Escherichia coli. Biochemistry (Moscow), 62, 204–211. [PubMed] [Google Scholar]
- 54.Lahm A. and Suck,D. (1991) DNase I-induced DNA conformation. 2 Å structure of a DNase I-octamer complex. J. Mol. Biol., 222, 645–667. [DOI] [PubMed] [Google Scholar]
- 55.Weston S.A., Lahm,A. and Suck,D. (1992) X-ray structure of the DNase I-d(GGTATACC)2 complex at 2.3 Å resolution. J. Mol. Biol., 226, 1237–1256. [DOI] [PubMed] [Google Scholar]