Abstract
Plant mitochondria contain three rRNA genes, rrn26, rrn18 and rrn5, the latter two being co-transcribed. We have recently identified a polynucleotide phosphorylase-like protein (AtmtPNPase) in Arabidopsis mitochondria. Plants downregulated for AtmtPNPase expression (PNP− plants) accumulate 18S rRNA species polyadenylated at internal sites, indicating that AtmtPNPase is involved in 18S rRNA degradation. In addition, AtmtPNPase is required to degrade the leader sequence of 18S rRNA, a maturation by-product excised by an endonucleolytic cut 5′ to the 18S rRNA. PNP− plants also accumulate 18S rRNA precursors correctly processed at their 5′ end but containing the intergenic sequence (ITS) between the 18S and 5S rRNA. Interestingly, these precursors may be polyadenylated. Taken together, these results suggest that AtmtPNPase initiates the degradation of the ITS from 18S precursors following polyadenylation. To test this, we overexpressed in planta a second mitochondrial exoribonuclease, AtmtRNaseII, that degrades efficiently unstructured RNA including poly(A) tails. This resulted also in the detection of 18S rRNA precursors showing that AtmtRNaseII is not able to degrade the ITS but can impede the action of AtmtPNPase in initiating the degradation of the ITS. These results show that AtmtPNPase is essential for several aspects of 18S rRNA metabolism in Arabidopsis mitochondria.
INTRODUCTION
The genome of higher plant mitochondria contains three ribosomal RNA (rRNA) genes, rrn26, rrn18 and rrn5, encoding the 26S, 18S and 5S rRNAs, respectively. A common organization of rRNA genes is observed in higher plants, i.e. the proximity of rrn18 and rrn5 genes that are co-transcribed and a more distal rrn26 gene. The observation of multiple transcripts containing rRNAs in various plant species indicates that the mature rRNAs are generated by processing from longer precursor molecules (1,2). Indeed, there is apparently no efficient transcription termination downstream of the rRNA genes in plants. For instance in maize, transcription proceeds both in vivo and in organello several kilobases beyond the 3′ extremities of rrn26 and rrn5 genes. However, accumulation of stable transcripts corresponding to these regions has not been observed (3). In some species such as wheat and rye, tRNAs or t-elements, sequences that can fold into tRNA-like structures, are located in the close vicinity of rRNA genes (4,5). Excision of the tRNA or t-element sequences by the tRNA processing machinery might be involved in rRNA processing in these species (4,5). However, these features are not conserved among different plant species and the general molecular mechanisms and ribonucleases involved in rRNA maturation remain to be identified in plant mitochondria. In contrast, rRNA maturation processes are well characterized in Saccharomyces cerevisiae and Escherichia coli (6–8). In these organisms, rRNA maturation involves the combined action of endonucleases and exonucleases. In E.coli, in the absence of RNases T, PH, D and BN, precursors of all stable RNAs except for the 16S rRNA harbor 3′ extensions of a few genomically encoded nucleotides. Interestingly, these extensions are followed by one to seven adenosine residues (9). The adenylation of the unprocessed RNAs might serve as a means to degrade defective RNAs (9). Indeed, polyadenylation triggers degradation of RNA not only in E.coli but also in chloroplasts and in plant and trypanosome mitochondria (10–13). In Chlamydomonas reinhardtii chloroplasts, 5S rRNAs may also be polyadenylated, indicating that polyadenylation-mediated decay is involved in the degradation of rRNAs in these organelles (14). More recently, degradation of polyadenylated rRNA has also been reported in yeast nuclei (15).
Recently, we identified two exoribonucleases, AtmtPNPase and AtmtRNaseII, in Arabidopsis thaliana mitochondria (16). These proteins belong to the polynucleotide phosphorylase (PNPase) and RNaseII protein families, respectively. AtmtPNPase is essential for at least some stages of development but we can downregulate its expression in seedlings by overexpressing a tagged version of AtmtPNPase (PNPase-tag), which triggers the phenomenon of co-suppression (16). These plants, silenced for the expression of the AtmtPNPase gene, were named PNP− plants. A PNPase-like protein is also present in the chloroplasts of A.thaliana where it is not only involved in the degradation of mRNAs but also plays a role in both mRNA and 23S rRNA maturation (17–20). AtmtRNaseII expression is not essential, as viable mutants homozygous for three distinct T-DNA insertions in AtmtRNaseII gene have been identified and named rnaII-1 to 3 mutants (16). Both AtmtPNPase and AtmtRNaseII are involved in the maturation of 3′ extremities of atp9 mRNAs in Arabidopsis mitochondria (16). To determine the possible roles of AtmtPNPase and AtmtRNaseII in 18S rRNA metabolism, we analyzed both maturation intermediates and polyadenylation status of this rRNA in PNP− and rnaII mutant plants. We show here that AtmtPNPase plays multiple roles in 18S rRNA maturation and degradation, whereas 18S rRNA metabolism is not impaired in rnaII mutants. First, the leader sequence corresponding to the sequence between the promoter and the 5′ extremity of the 18S rRNA accumulates in PNP−plants when compared with wild-type plants. Thus, AtmtPNPase is required to degrade this maturation by-product. Second, we observed an increase of polyadenylated degradation intermediates of 18S rRNA in PNP− plants, suggesting an involvement of AtmtPNPase in 18S rRNA degradation. Third, in PNP− plants we could detect 18S rRNA precursors harboring mature 5′ ends but most of the intergenic sequence (ITS) present between the 18S and 5S rRNAs. In addition, these precursors may be polyadenylated. These results suggest that AtmtPNPase initiates the degradation of the ITS from polyadenylated 18S rRNA precursors. To confirm this hypothesis, we overexpressed in planta a second mitochondrial exoribonuclease, AtmtRNaseII, which is able to efficiently degrade poly(A) tails as well as other unstructured RNA. Upon overexpression of AtmtRNaseII, we could detect 18S rRNA precursors that are correctly processed at their 5′ extremity but not at their 3′ extremity, showing that AtmtRNaseII cannot degrade the ITS and impedes the action of AtmtPNPase, possibly by degrading poly(A) tails. These results demonstrate that AtmtPNPase is required for several aspects of 18S rRNA metabolism.
MATERIALS AND METHODS
Plant material
All A.thaliana plants used in this study are of columbia ecotype. The transgenic plants, PNP+ and R+ overexpress AtmtPNPase (CAB43865) or AtmtRNaseII (AAQ62877), respectively, fused at their C-terminus to the FLAG peptide and the calmodulin binding peptide. Downregulation of the expression of AtmtPNPase gene (PNP− plants) was obtained by co-suppression in lines that initially overexpressed the tagged version of AtmtPNPase. A detailed description of these plants has been published previously (16).
Primer sequences
All numbers referring to primers given here are as in NC_001284, the accession number of the complete mitochondrial genome sequence of Arabidopsis. The reverse primers P1, P2, P4, P6 and P8 correspond to nucleotides 362891-909, 362980-998, 361331-348, 363284-301 and 361284-304, respectively. The forward primers P3, P5 and P7 correspond to nucleotides 361460-482, 361319-337 and 363332-352, respectively.
Miscellaneous techniques
Total RNA was extracted using the TRI-reagent (Molecular Research Center) according to the manufacturer's instructions. Northern blot experiments were performed using 1.5% (w/v) agarose/formaldehyde or 12% (w/v) acrylamide–bisacrylamide (19:1)/7 M urea gels according to standard procedures. After fractionation, RNA was transferred to Hybond-N+ membrane (Amersham). Prehybridization and hybridization were performed in 7% (w/v) SDS, 0.5 M NaPi, pH 7.2 at 45°C. All probes consisted of the indicated primers labeled with [γ−32P] ATP and polynucleotide kinase (Fermentas). Three washings were conducted at 45°C in 2× SSC, 0.1% (w/v) SDS for 20 min. Western blot analysis was conducted according to standard protocols using anti-AtmtRNaseII antibodies raised in rabbits against a peptide corresponding to the 16 C-terminal amino acids.
PCR methods and cloning
Mapping of poly(A) sites by RT–PCR was carried out as in (21) except that the sequence of the adapter primer was 5′-GAATTCCATGTCGACGGTCTCA-3′. Circular RT–PCR (cRT–PCR) was used to determine both 5′ and 3′ extremities of 18S rRNA and its precursors. Five micrograms of total RNA were incubated with 40 U of T4 RNA ligase (New England Biolabs) in the supplied buffer supplemented with 2 U of RNase inhibitor and 1 U of RQ1 DNase (Promega) in a total volume of 25 μl. After phenol–chloroform extraction and ethanol precipitation, cDNAs were synthesized using Superscript™ II RNaseH− reverse transcriptase (Invitrogen) and the reverse primer P1 (Figure 1A). The region containing the junction of 5′ and 3′ extremities was then amplified by RT–PCR using forward and reverse primers as indicated. PCR amplification consisted of 30 cycles of 30 s duration at 94°C, 30 s at 50°C, 1 min at 72°C. All cRT–PCR products were cloned using the TOPO cloning kit (Invitrogen).
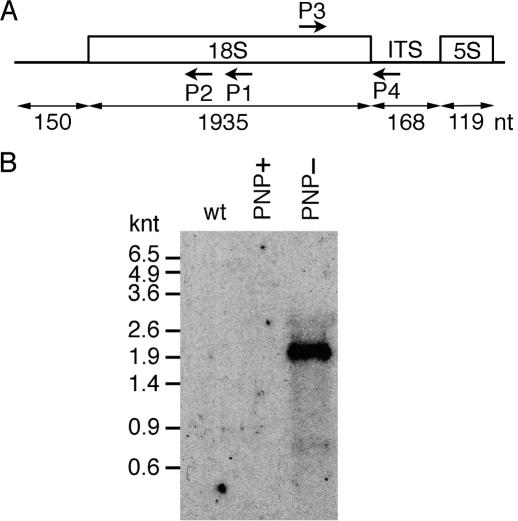
Figure 1.
18S precursors accumulate in PNP− plants. (A) Location of primers P1–P4 on a scheme of 18S–5S co-transcripts. Arrows indicate the sense or reverse orientation of the primers. Sizes of the different part of the 18S locus are indicated in nucleotides. (B) Northern analysis of 18S precursors. Total RNA (5 μg) from wild-type plants (wt), plants overexpressing PNPase-tag (PNP+) or plants silenced for the expression of AtmtPNPase gene (PNP−) were separated on a 1.5% agarose/formaldehyde gel, transferred to a nylon membrane and hybridized with the 5′ [32P]-labeled primer P4. Autoradiography was performed for 24 h at –80°C using one intensifying screen.
Purification of PNPase-tag and RNaseII-tag and RNase activity test
Both purification and activity tests were performed as described previously (16). RNA substrates consisted of the last 136 nt of potato atp9 mRNAs either with or without a poly(A) tail of 19 adenosines (22).
RNase H treatment
Three micrograms of total RNA were incubated with 100 pmol of the reverse primer P4 in the presence of 2.5 U of RNase H (USB) as recommended by the manufacturer. After phenol extraction and ethanol precipitation, RNA samples were treated for northern blot analysis as detailed above.
RESULTS
18S rRNA precursors accumulate in PNP− plants
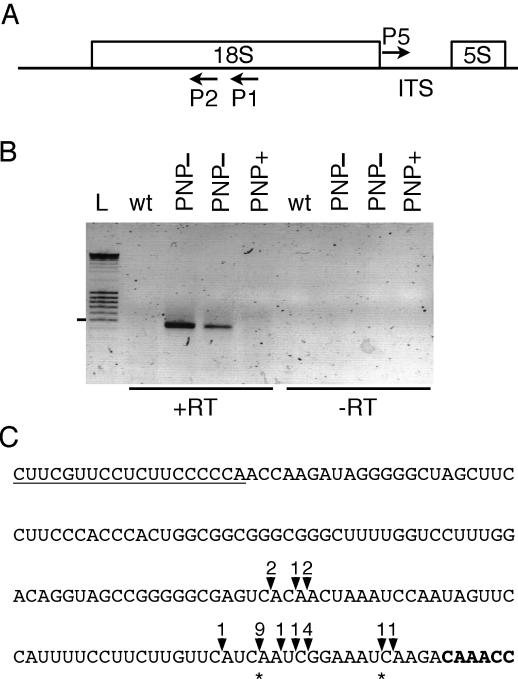
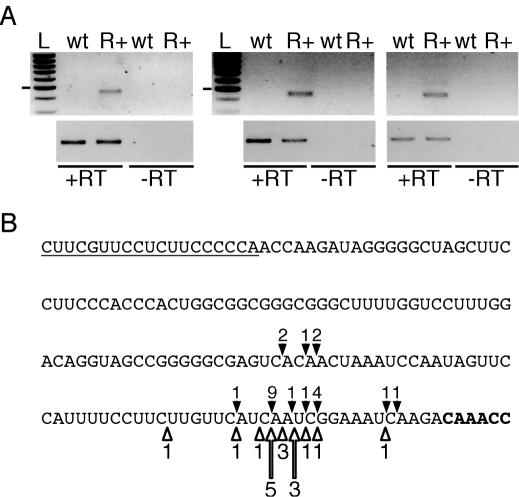
The 5′ and 3′ extremities of mitochondrial 18S rRNA were mapped in wild-type Arabidopsis plants. We used the cRT–PCR method in which RNAs were circularized by T4 RNA ligase prior to cDNA synthesis and PCR amplification of the region encompassing the ligated 5′ and 3′ ends (23). The location of primer P1, used to initiate cDNA synthesis, as well as the location of primers P2 and P3, used for PCR amplification, are shown in Figure 1A. The 5′ and 3′ extremities of mature 18S rRNA in wild-type plants were mapped at nucleotides 363282 and 361349 in NC_001284, the accession number of the Arabidopsis complete mitochondrial genome sequence. To detect 3′ unprocessed 18S precursors in wild-type, rnaII, PNP+ and PNP− plants by northern blot analysis, we used the antisense primer, P4 located immediately downstream of the mature 3′ end of 18S rRNA (Figure 1A). No signals were observed for rnaII plants (data not shown), wild-type and PNP+ plants (Figure 1B). In contrast, a distinct band of ∼2000 nt was detected in PNP− plants (Figure 1B). To confirm that this RNA species corresponds to 18S precursors, we performed cRT–PCR experiments using primer P1 for cDNA synthesis and primers P2 and P5 for PCR amplification. Primer P5 corresponds to a sense primer located 11 nt downstream of the mature 18S rRNA 3′ extremity (Figure 2A). A band of 450 bp was obtained for PNP− samples but not for wild-type or PNP+ samples (Figure 2B). Cloning and sequencing of 23 PCR products revealed that they correspond to 18S precursors harboring 5′ extremities identical to the mature 18S rRNA but with unprocessed 3′ ends (Figure 2C). Most of the 3′ extremities of 18S precursors (78%) were mapped in a small window of 15 nt located 4 nt upstream of the 5S sequence (Figure 2C).
Figure 2.
Characterization of 18S precursors in PNP− plants. (A) Location of primers P1, P2 and P5 used to map the 5′ and 3′ extremities of 18S precursors in PNP– plants by cRT–PCR. (B) Negative image of an ethidium bromide-stained agarose gel of cRT–PCR products amplified using primers P2 and P5. L, DNA ladder consisting of bands with 100 bp increments. The position of the 500 bp band is marked on the left. Lane labeling is as in Figure 1A. The presence or absence of reverse transcriptase during the cDNA synthesis step is indicated by +RT or −RT, respectively. (C) Positions of 3′ extremities of 18S precursors in PNP− plants are indicated by solid arrowheads on the sequence corresponding to the reverse sequence from nucleotides 361174–361337 in NC_001284, the accession number of the Arabidopsis mitochondrial genome sequence. The 5′ sequence of the 5S rRNA is shown in bold. Contrary to the database annotation, the 5′ extremity of the 5S rRNA in Arabidopsis mitochondria is located at nucleotide 361179 and not at 361178 in NC_001284. The 5′ extremity of the 5S rRNA was mapped by cRT–PCR (data not shown). The sequence of the primer P5 is underlined. The number of clones obtained are indicated above each arrowhead. Asterisks indicate the position of short oligo(A) tails of 4–6 nt observed for two clones.
Thus, 18S rRNA precursors accumulate in PNP− plants in contrast to wild-type plants, as shown by northern blot analysis (Figure 1B). These precursors are correctly processed at their 5′ extremity but not at their 3′, extremity, as they contain most of the ITS (Figure 2C).
Wild-type mitochondria contain polyadenylated 18S rRNA species
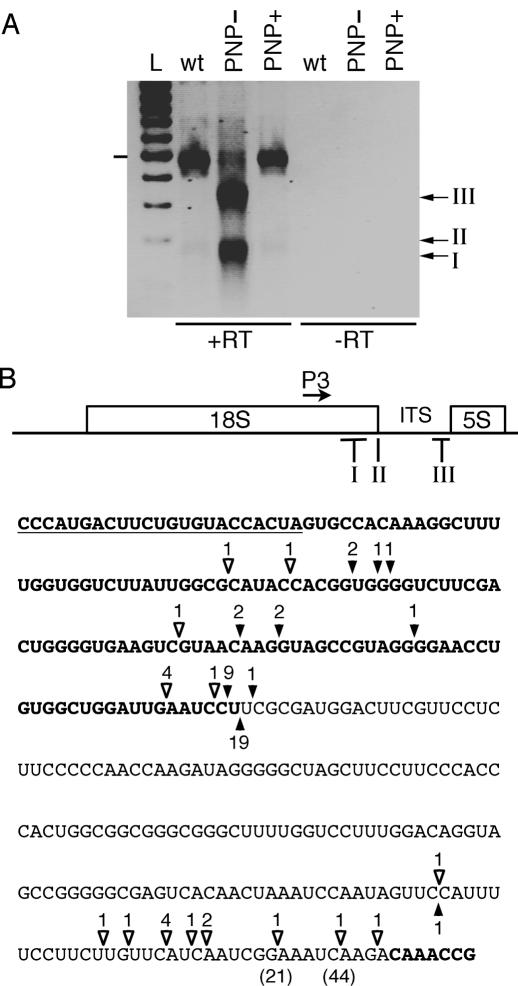
Interestingly, two of the cRT–PCR clones described above contained short oligo(A) tails of 4–6 adenosines (Figure 2C). We thus performed RT–PCR experiments to detect possible poly(A) tails at the 3′ ends of 18S rRNA or its precursors in wild-type, PNP+ and PNP− plants. cDNA synthesis was initiated using an oligo(dT)12-adapter primer. The resulting cDNAs were amplified using a forward primer (P3), ending 111 nt upstream to the mature 3′ end of the 18S rRNA, in combination with the adapter primer. Identical RT–PCR patterns were observed for both wild-type plants and PNP+ plants (Figure 3A). Sequence analysis of PCR products from wild-type samples revealed that the major band resulted from the annealing of the oligo(dT)12-adapter primer to a adenosine-rich region located 3′ to the 5S rRNA (nucleotides 361013-361060 in NC_001284). These PCR products were therefore unlikely to represent true polyadenylated RNA species but were amplified from 18S–5S co-transcripts. However, we also detected clones corresponding to truly polyadenylated RNAs. Such clones contained poly(A) tails that were longer than the oligo(dT) and/or localized at sites lacking adenosine residues in the genomic sequence (solid arrowheads in Figure 3B). These clones were identified by sequencing only those inserts whose size did not correspond to the major band amplified from 18S–5S co-transcripts. The clones representing truly polyadenylated RNAs were divided into three classes (I to III) (solid arrowheads in Figure 3B). Class I consists of clones (9/39) with poly(A) sites localized upstream of the 3′ extremities of mature 18S rRNA. As these poly(A) sites are internal to the 18S sequence and as we and others have shown the existence of polyadenylation-mediated decay in plant mitochondria (21,22,24,25), these RNA species are likely to represent 18S degradation intermediates. Most clones analyzed (29/39) belong to class II and are polyadenylated at or very close to the mature 3′ extremity, similar to what we and others have described for several mitochondrial mRNAs (21,22,24,25). Only 1 out of 39 clones belongs to class III. This clone corresponds to a 18S rRNA precursor polyadenylated at a site localized in the ITS, 35 nt upstream of the 5S rRNA (Figure 3B).
Figure 3.
Characterization of polyadenylated 18S rRNA species. (A) Negative image of an ethidium bromide-stained gel of RT–PCR products amplified from oligo(dT)12-adapter primed cDNAs using the adapter primer in combination with the forward primer P3 ending 111 nt upstream of the mature 3′ ends of 18S rRNA. Migration of PCR products corresponding to classes I–III of poly(A) sites are indicated on the right. All labeling are as in Figure 2B. (B) Positions of polyadenylation sites of 18S rRNA species in wild-type (solid arrowheads) and PNP− (open arrowheads) plants. The position of the forward primer P3 used in RT–PCR experiments is shown by an arrow on the scheme of the 18S–5S co-transcript and its sequence is underlined in the sequence given below the scheme. Locations of the classes I, II and III of poly(A) sites are indicated on the scheme. The sequence shown corresponds to the reverse sequence from nucleotides 361192–361462 with respect to the numbering in NC_001284. The 3′ and 5′ sequences of the 18S and 5S rRNAs, respectively, are shown in bold. The number of clones obtained is indicated above each arrowhead. Sizes of two poly(A) tails situated in adenosine-rich regions are indicated for two clones in parentheses. These sizes include the 12 adenosines due to the oligo(dT)12.
These results demonstrate that degradation intermediates and mature 18S rRNAs as well as also 18S rRNA precursors may be polyadenylated in wild-type plant mitochondria, albeit to low levels.
Polyadenylated 18S rRNA degradation intermediates and precursors accumulate in PNP− plants
The same type of RT–PCR experiments revealed a different polyadenylation pattern in PNP− plants (Figure 3A). PCR products corresponding to classes I and III were clearly more abundant in PNP− plants than in wild-type plants (see discussion). The position of the polyadenylation sites detected in PNP− plants are indicated by open arrowheads in Figure 3B. One poly(A) site was mapped only 2 nt upstream of the mature 3′ end of 18S rRNA. It is thus unknown whether this clone corresponds to a polyadenylated mature 18S rRNA (class II) or to a defective, degradation intermediate (class I) (Figure 3B). Nevertheless, these results show that class II clones are less abundant or absent in PNP− plants, whereas they are the most abundant type of clones in wild-type plants. In PNP− plants, the 3′ ends of most 18S precursors (12 out of 13 clones of class III) map in a region of 24 nt, upstream of the 5S sequence. This region is similar to the region where we mapped the 3′ ends of 18S precursors by cRT–PCR. However, at the nucleotide level, only a few of the 3′ extremities mapped by cRT–PCR are common to the polyadenylation sites mapped here (compare Figures 2C and 3B).
Two poly(A) sites of 18S precursors located a few nucleotides upstream of the 5S are in regions containing 3 consecutive adenosines (Figure 3B). However, an artifactual priming of the oligo(dT)12 primer was not considered as the poly(A) tails of 21 and 44 nt detected for these clones exceeded the size of the oligo(dT)12 primer. In addition, we detected one clone corresponding to polyadenylated 18S rRNA precursors ending at the 5′ end of the 5S rRNA. This indicates that an endoribonucleolytic cut could occur precisely at the 5′ end of the 5S rRNA or at one nucleotide upstream of the 5S rRNA. Since this nucleotide is an adenosine, we could not distinguish whether it is genomically encoded or added post-transcriptionally by polyadenylation. The ragged 3′ ends of other 18S precursors could be explained either by additional endoribonucleolytic cuts or, more likely, by the presence of AtmtRNAseII that could trim these precursors but could not progress much further than a few nucleotides (see below).
These data demonstrate that the absence of AtmtPNPase results in a qualitative modification of the polyadenylation pattern of 18S rRNA. In addition, the presence of polyadenylated 18S rRNA species in PNP− plants and the fact that these tails are almost exclusively composed of adenosine (data not shown) strongly suggest that poly(A) tails are synthesized by a true poly(A) polymerase and not by AtmtPNPase in plant mitochondria.
AtmtRNaseII can degrade poly(A) tails
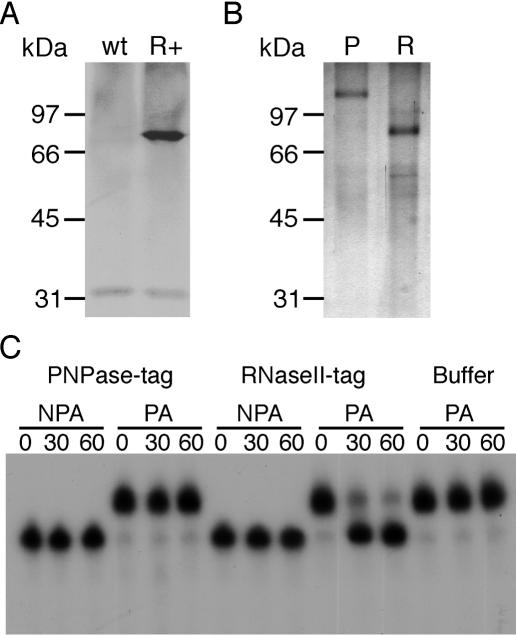
Polyadenylation of 18S rRNA precursors raised the possibility that polyadenylation is involved in triggering the degradation of the ITS or of the entire precursors by AtmtPNPase. To investigate this hypothesis, we looked for a way to alter the polyadenylation of these precursors in planta to determine whether this would compromise the action of AtmtPNPase. In E.coli, RNaseII degrades poly(A) tails efficiently and its overexpression can stabilize transcripts that are otherwise degraded by PNPase (26). We had previously detected a poly(A) degradation activity in mitochondrial protein extracts (22) and the recently identified AtmtRNaseII is a potential candidate for such an activity. Therefore, we overexpressed a tagged version of this protein (RNaseII-tag) in Arabidopsis plants (Figure 4A). RNaseII-tag consists of full-length AtmtRNaseII fused at its C-terminus to the FLAG peptide and the calmodulin binding peptide (16). These affinity tags were used to purify RNaseII-tag (Figure 4B). We also purified an inactive, tagged version of AtmtPNPase (PNPase-tag) from transgenic plants (Figure 4B). PNPase-tag was used as a negative control for the activity test to ensure that no contaminating RNase activities were present in our protein fractions. Purified PNPase-tag and RNaseII-tag were incubated with polyadenylated or non-polyadenylated RNA substrates. These RNA substrates correspond to the last 136 nt of potato atp9 mRNAs either with or without a poly(A) tail of 19 adenosines that we previously used to detect a poly(A) degradation activity (22). RNaseII-tag was not able to degrade the non-polyadenylated RNA substrate (Figure 4C). However, it removed the poly(A) tail from the polyadenylated RNA substrate very efficiently (Figure 4C). This ability to degrade poly(A) tails was further confirmed using various polyadenylated and non-polyadenylated RNA substrates (data not shown).
Figure 4.
AtmtRNaseII can degrade poly(A) tails. (A) Western blot analysis of wild-type plants (wt) and plants overexpressing RNaseII-tag (R+). (B) Silver-stained SDS–PAGE of purified PNPase-tag (P) and RNaseII-tag (R) proteins. (C) Activity of RNaseII-tag. Uniformly [α-32P] UTP labeled non-polyadenylated (NPA) or polyadenylated (PA) RNA substrates consisted of the last 136 nt of potato atp9 mRNAs either with or without a poly(A) tail of 19 adenosines (22). Incubation times are shown in minutes above each lane. Aliquots of degradation reactions were fractionated on 7 M urea/6% (w/v) acrylamide gels before autoradiography.
These results show that RNaseII-tag efficiently removes poly(A) tails from RNA substrates. However, RNaseII-tag is not a poly(A) specific enzyme but rather catalyses the degradation of any unstructured RNA [(16), data not shown].
Overexpression of RNaseII-tag mimics the downregulation of AtmtPNPase
We then analyzed by cRT–PCR whether 18S rRNA precursors could be detected upon overexpression of RNaseII-tag in Arabidopsis plants. As shown above, 18S rRNA precursors were not amplified from wild-type samples probably due to their lower abundance. In contrast, we detected 18S rRNA precursors in plants overexpressing RNaseII-tag (Figure 5A). Sequence analysis of 17 PCR products revealed that they indeed correspond to 18S rRNA with mature 5′ ends but with unprocessed 3′ extremities. Again, these precursor 3′ extremities are localized in the restricted region situated upstream of the 5S rRNA sequence, as has been observed in PNP− plants (Figure 5B). In addition, most 3′ extremities of 18S precursors were mapped at identical nucleotides in plants overexpressing the RNaseII-tag and in PNP− plants (Figure 5B).
Figure 5.
Detection of 18S rRNA precursors in plants overexpressing RNaseII-tag. (A) Negative images of ethidium bromide-stained gels of cRT–PCR products obtained as described in Figure 2. L, DNA ladder consisting of bands with 100 bp increments. The position of the 500 bp band is marked on the left. wt, wild-type plant. R+, plants overexpressing RNaseII-tag. The results presented for R+ plants have been obtained in three independent experiments. To ensure that similar amounts of cDNAs were used, primers allowing for cDNA synthesis and PCR amplification of a 500 bp fragment of the cox2 intron were included. These amplifications are shown on the lower panels. The presence or absence of reverse transcriptase during the cDNA synthesis step is indicated by +RT or −RT, respectively. (B) Position of 3′ extremities of 18S rRNA precursors in plants overexpressing RNaseII-tag (open arrowheads) as compared with PNP− plants (solid arrowheads). All features are as in Figure 2C.
These results show that overexpression of AtmtRNaseII, an exoribonuclease that degrades unstructured RNA including poly(A) tails, mimics the downregulation of AtmtPNPase, i.e. in both cases similar 18S rRNA precursors accumulate. In addition, upon overexpression of AtmtRNaseII in vivo, the action of AtmtPNPase is impeded as 18S rRNA precursors are stabilized.
The 5′ end of 18S rRNA is generated by an endonucleolytic cut and AtmtPNPase is responsible for the degradation of an 18S maturation by-product
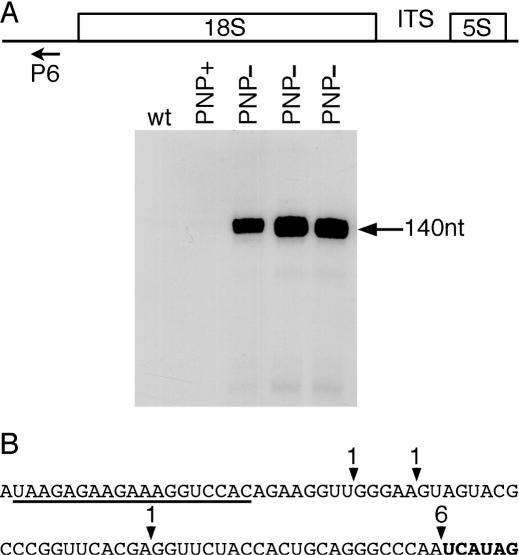
The 18S promoter has been previously mapped upstream of the 5′ end of the 18S rRNA in Arabidopsis mitochondria (27). However, it is not known whether the 5′ extremity of the 18S is generated by an endonucleolytic cut and/or involves a putative 5′ to 3′ exoribonuclease. To discriminate between these possibilities, we performed a northern blot analysis on wild-type and PNP− plants using an oligonucleotide (P6) complementary to a region located between the 18S promoter and the mature 5′ end of 18S rRNA. A signal corresponding to an abundant RNA of ∼140 nt was observed in PNP− plants, whereas no signals were detected in wild-type plants (Figure 6A). To determine whether these RNAs could be polyadenylated, we performed RT–PCR experiments. Oligo(dT)12-adapter primed cDNAs were subjected to PCR amplification using a forward primer (P7) located downstream of the 18S promoter in combination with the adapter primer. Cloning and sequencing of the PCR products revealed the existence of polyadenylated RNA species (6 out of 9 clones) ending at either the 5′ extremity of the 18S rRNA or two nucleotides upstream of this extremity (Figure 6B). Again, this imprecision is due to the fact that the two nucleotides upstream to the 5′ end of the 18S sequence are adenosines. The remaining three clones exhibited a poly(A) tail located upstream of the 5′ end of 18S rRNA. These alternative poly(A) sites could be generated either by an alternative endonucleolytic cut or by the stalling of a second exoribonuclease activity, such as AtmtRNaseII.
Figure 6.
Accumulation of 18S 5′ maturation by-product in PNP− plants. (A) Detection of the 18S leader sequence by northern blot analysis. Total RNA (5μg) from wild-type plants (wt), plants overexpressing PNPase-tag (PNP+) or plants silenced for the expression of AtmtPNPase gene (PNP−) were fractionated on a 12% acrylamide/7 M urea gel, transferred to a nylon membrane and hybridized with the 5′ [32P]-labeled primer P6 complementary to a region located between the 18S promoter and the mature 5′ end of 18S rRNA. Autoradiography was performed for 24 h at −80°C using one intensifying screen. (B) Polyadenylation sites in the leader sequence. The sequence shown corresponds to the reverse sequence from nucleotides 363275–363313 with respect to the numbering in NC_001284. The sequence of the forward primer P7 used in RT–PCR experiments is underlined. The 5′ sequence of the 18S is shown in bold. Positions of polyadenylation sites are indicated by solid arrowheads. The number of clones obtained are shown above each arrowhead.
Thus, PNP− plants accumulate an RNA species corresponding to the region downstream of the 18S promoter and the 5′ end of mature 18S rRNA. These data imply that the 5′ extremity of 18S is generated by an endonucleolytic cut and that the maturation by-product (i.e. the 5′ leader) is degraded by AtmtPNPase as it accumulates in PNP− plants.
The ITS is not excised by an endonucleolytic cut 3′ to the 18S rRNA and 18S rRNA precursors accumulate to levels comparable with that of the leader sequence in PNP− plants
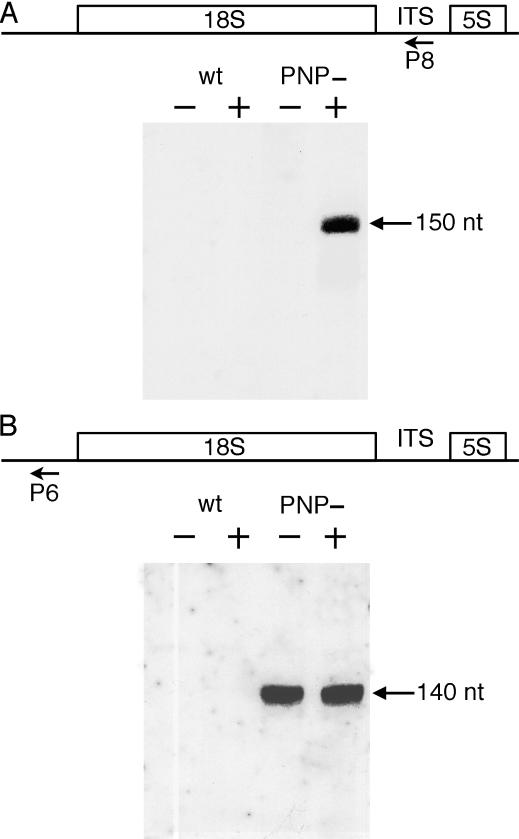
Our results have shown that two endoribonucleolytic cuts occur 5′ to the 18S and 5S rRNA resulting in the accumulation of 18S rRNA precursors in PNP− plants. We then determined whether an additional cleavage could also occur 3′ to the 18S rRNA. This would result in excising the whole ITS from 18S precursors. We thus compared the respective levels of 18S precursors and of the putative free ITS by northern blot analysis. However, these RNA species exhibit rather different sizes, from 168 to >2000 nt. To obtain comparable sizes of both RNA species, total RNA from PNP− plants was hybridized with experimentally determined saturating amounts of the antisense primer P4, located just dowstream of the mature 18S 3′ ends and then treated with RNase H. This RNase H treatment cleaves off the 18S rRNA sequence from the 18S precursors and the released ITS was detected by northern blot using a labeled primer (P8) complementary to a region within the ITS (see Figure 7A). Under these experimental conditions, 18S precursors could no longer be observed by northern blot using agarose gels (data not shown). However, an RNA species of ∼150 nt corresponding to the ITS was detected by northern blot using 12% acrylamide gels (Figure 7A). In contrast, the ITS was not detected in samples treated with RNase H but lacking antisense primer P4. These results show that the ITS is associated with the 18S sequence and is not excised by an endonucleolytic cleavage 3′ to the 18S rRNA in PNP− plants (Figure 7A).
Figure 7.
Respective levels of 18S leader and precursors in PNP− plants. The position and orientation of primers P6 and P8 are indicated on the schemes. Three micrograms of total RNA from wild-type (wt) or PNP− plants were treated with RNase H in the absence or presence of primer P4, as indicated above each lane by minus and plus signs, respectively. RNA were then fractionated on a 12% acrylamide/7 M urea gel, transferred to a nylon membrane, hybridized with the 5′ [32P]-labeled primer P8 (A) or P6 (B) and exposed to X-ray film for 24 h at −80°C using one intensifying screen. P6 and P8 primers have identical theoretical melting temperatures.
The same blot was then hybridized with the antisense primer P6 to detect the leader sequence (Figure 7B). The accumulation of the leader sequence can be used as a marker for the amount of primary 18S–5S transcripts that have been produced and processed in cells where AtmtPNPase was no longer expressed. Comparable signal intensities were observed in both experiments (compare Figures 7A and B). Identical results were observed by reversing the sequential order of P6 and P8 hybridizations or by simultaneous hybridization (data not shown). The use of two different labeled primers precludes an absolute quantification of different RNA species. However, these results show that the majority of 18S–5S co-transcripts are converted to 18S precursors in PNP− plants.
DISCUSSION
Our results show that downregulation of AtmtPNPase affects several aspects of the 18S rRNA metabolism. The polyadenylation pattern of 18S RNA is modified and a maturation by-product and 18S RNA precursors accumulate in PNP− plants.
We show by RT–PCR experiments that 18S rRNA may be polyadenylated at internal sites (class I clones), at their mature 3′ extremities (class II clones) and also at the 3′ ends of 18S rRNA precursors (class III clones). In these RT–PCR experiments, the priming of the oligo(dT)12 primer to the adenosine-rich regions 3′ to the 5S rRNA may be used as an internal control assuming that 18S–5S co-transcripts are not preferentially destabilized in PNP− plants. PCR products originating from the priming of the oligo(dT)12 in the adenosine-rich regions do not reflect truly polyadenylated RNA but rather indicate the level of 18S–5S co-transcripts. These PCR products are by far the most abundant products for wild-type and PNP+ samples. For wild-type plants, clones corresponding to classes I–III could be characterized only by selecting the inserts whose size would not correspond to that of 18S–5S co-transcripts. On the contrary, for PNP− samples, PCR products originating from the priming of the oligo(dT)12 in the adenosine-rich regions were hardly obtained. Most clones corresponded to classes I and III. Thus, in PNP− plants, polyadenylated degradation intermediates (class I clones) and 18S precursors (class III clones) are more abundant when compared with the 18S–5S co-transcripts than in wild-type plants. The accumulation of 18S rRNA polyadenylated at internal sites in PNP− plants indicates the involvement of AtmtPNPase and polyadenylation in the degradation of rRNA, similar to the situation described for E.coli (11). Although rRNA degradation may involve common mechanisms in bacteria and plant mitochondria, rRNA maturation appears to be quite different between these organisms. In E.coli, rRNA precursors are matured by endonucleases such as RNase E, RNase G and RNase III that ultimately cut relatively close to what will become the 3′ mature ends. RNase T is involved in the maturation process of 23S and 5S rRNAs but its role is restricted to the trimming of a few nucleotides (28–30). In plant mitochondria, the situation is strikingly different. It has been shown in maize, both in organello and in vivo, that transcription proceeds well beyond the rrn genes (3). Indeed, up to 21kb flanking the rrn genes are actively transcribed although no stable transcripts corresponding to these sequences have been detected. This implies that large regions of these pre-transcripts have to be removed post-transcriptionally. This phenomenon is more restricted for the 18S rRNA in Arabidopsis and probably in other species since the proximity of the 18S and 5S sequences is conserved in higher plants. We have shown here that an endonucleolytic cut at or near the 5′ end of the 5S rRNA releases 18S precursors, but still 168 nt corresponding to the ITS have to be removed.
The accumulation of 18S precursors in PNP− plants raises two hypotheses that are not mutually exclusive. Either AtmtPNPase is involved in degrading excess of 18S precursors or, alternatively, AtmtPNPase is required in one step of the 18S rRNA 3′ processing, namely the initiation of the degradation of the ITS. Proper biogenesis of mitoribosomes involves the coordinated assembly of nuclear-encoded proteins with mitochondrially encoded proteins and three rRNAs. If the production of the mitochondrial rRNA is not the limiting factor in the biogenesis of mitoribosomes, the excess of 18S RNA precursors would be degraded by AtmtPNPase. Degradation of unprocessed and defective rRNA has been reported in E.coli (9,31). However, we also favor the direct involvement of AtmtPNPase in the 3′ processing of 18S rRNA. This hypothesis is supported by the detection in PNP− plants of a single RNA species corresponding to the entire 18S rRNA plus the ITS by northern analysis. In contrast, we could not detect an RNA species corresponding to the sole ITS (i.e. cleaved at the 3′ and 5′ of the 18S and 5S, respectively). Thus, this processing step is probably not performed by an endoribonucleolytic cut at or near the 3′ extremities of the 18S rRNA. Alternatively, if this hypothetical endoribonucleolytic cut exits, it would require the action of AtmtPNPase as an absolute prerequisite. In addition, most 3′ ends of 18S precursors mapped by cRT–PCR are located in a restricted area 5′ to the 5S. Thus, these precursors contain most of the ITS. We did not observe 18S precursors harboring only a few nucleotides downstream of the 18S rRNA sequence. Finally, analysis of polyadenylated 18S rRNA species by RT–PCR using a forward primer within the 18S revealed several poly(A) sites localized within the 18S (i.e. degradation intermediates) or in the small region upstream of the 5S rRNA. Thus, in planta, no additional enzymes besides AtmtPNPase are able to initiate the degradation of the ITS, once the 18S–5S co-transcript has been cleaved 5′ to the 5S. In particular, the presence of 18S precursors in plants overexpressing AtmtRNaseII indicate that AtmtRNaseII cannot perform this maturation step and thus cannot compensate for the lack of AtmtPNPase in PNP− plants. Although polyadenylated 18S precursors containing the entire ITS have been detected by RT–PCR (Figure 3B), most precursors that accumulate in PNP− plants and plants overexpressing AtmtRNaseII lack 11–19 nt of the ITS. Interestingly, these few nucleotides cannot be predicted to form double-stranded structures by the MFold program. It is thus possible that this region remains unstructured and therefore could be degraded by AtmtRNaseII. Overexpressed AtmtRNaseII could either compete with AtmtPNPase in a stochastic manner or impede the action of AtmtPNPase by degrading poly(A) tails of 18S precursors. In the latter case, this would imply that polyadenylation is involved in the 18S rRNA maturation process by providing a recognition site for AtmtPNPase. To date, polyadenylation was reported to be mainly involved in targeting RNA species for degradation in E.coli. However, both bacterial poly(A) polymerase I and PNPase are involved in the 3′ maturation of CI RNA, the P4 phage immunity factor (32,33). CI RNA is a short, stable RNA that can induce premature transcription termination in the P4 early operon, which results in establishing prophage immunity by inhibiting the expression of lytic genes. It is processed from larger precursors using endonucleases such as RNase P and RNase E and also by polyadenylation and PNPase activity (33). Although this pathway of 3′ maturation may not be used for endogenous E.coli RNAs, the data presented here suggest that, in plant mitochondria, polyadenylation could be involved in the 3′ maturation process of the 18S rRNA by AtmtPNPase. However, AtmtPNPase might not be sufficient and so additional ribonucleases or other trans-factors could be involved to establish 18S rRNA mature 3′ ends. Further work is needed to fully understand the molecular mechanism of 18S rRNA 3′ maturation. Nevertheless, our present results show that AtmtPNPase plays multiple roles in the metabolism of 18S rRNA that cannot be compensated by another exoribonuclease in Arabidopsis mitochondria.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by the Centre National de la Recherche Scientifique (CNRS, France) and by the French Ministry of Research through a PhD fellowship to R.P. and a Action Concertée Incitative Jeunes Chercheurs to D.G.
REFERENCES
- 1.Mulligan R.M., Maloney,A.P. and Walbot,V. (1988) RNA processing and multiple transcription initiation sites result in transcript size heterogeneity in maize mitochondria. Mol. Gen. Genet., 211, 373–380. [DOI] [PubMed] [Google Scholar]
- 2.Maloney A.P., Traynor,P.L., Levings,C.S., III and Walbot,V. (1989) Identification in maize mitochondrial 26S rRNA of a short 5′-end sequence possibly involved in transcription initiation and processing. Curr. Genet., 15, 207–212. [DOI] [PubMed] [Google Scholar]
- 3.Finnegan P.M. and Brown,G.G. (1990) Transcriptional and post-transcriptional regulation of RNA levels in maize mitochondria. Plant Cell, 2, 71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanic-Joyce P.J., Spencer,D.F. and Gray,M.W. (1990) In vitro processing of transcripts containing novel tRNA-like sequences (‘t-elements’) encoded by wheat mitochondrial DNA. Plant Mol. Biol., 15, 551–559. [DOI] [PubMed] [Google Scholar]
- 5.Coulthart M.B., Spencer,D.F. and Gray,M.W. (1993) Comparative analysis of a recombining-repeat-sequence family in the mitochondrial genomes of wheat (Triticum aestivum L.) and rye (Secale cereale L.). Curr. Genet., 23, 255–264. [DOI] [PubMed] [Google Scholar]
- 6.Srivastava A.K. and Schlessinger,D. (1990) Mechanism and regulation of bacterial ribosomal RNA processing. Annu. Rev. Microbiol., 44, 105–129. [DOI] [PubMed] [Google Scholar]
- 7.Butler J.S. (2002) The yin and yang of the exosome. Trends Cell. Biol., 12, 90–96. [DOI] [PubMed] [Google Scholar]
- 8.Granneman S. and Baserga,S.J. (2004) Ribosome biogenesis: of knobs and RNA processing. Exp. Cell. Res., 296, 43–50. [DOI] [PubMed] [Google Scholar]
- 9.Li Z., Pandit,S. and Deutscher,M.P. (1998) Polyadenylation of stable RNA precursors in vivo. Proc. Natl Acad. Sci. USA, 95, 12158–12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schuster G., Lisitsky,I. and Klaff,P. (1999) Polyadenylation and degradation of mRNA in the chloroplast. Plant Physiol., 120, 937–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dreyfus M. and Regnier,P. (2002) The poly(A) tail of mRNAs: bodyguard in eukaryotes, scavenger in bacteria. Cell, 111, 611–613. [DOI] [PubMed] [Google Scholar]
- 12.Ryan C.M., Militello,K.T. and Read,L.K. (2003) Polyadenylation regulates the stability of Trypanosoma brucei mitochondrial RNAs. J. Biol. Chem., 278, 32753–32762. [DOI] [PubMed] [Google Scholar]
- 13.Gagliardi D., Stepien,P.P., Temperley,R.J., Lightowlers,R.N. and Chrzanowska-Lightowlers,Z.M. (2004) Messenger RNA stability in mitochondria: different means to an end. Trends Genet., 20, 260–267. [DOI] [PubMed] [Google Scholar]
- 14.Komine Y., Kwong,L., Anguera,M.C., Schuster,G. and Stern,D.B. (2000) Polyadenylation of three classes of chloroplast RNA in Chlamydomonas reinhadtii. RNA, 6, 598–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuai L., Fang,F., Butler,J.S. and Sherman,F. (2004) Polyadenylation of rRNA in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 101, 8581–8586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perrin R., Meyer,E.H., Zaepfel,M., Kim,Y.J., Mache,R., Grienenberger,J.M., Gualberto,J.M. and Gagliardi,D. (2004) Two exoribonucleases act sequentially to process mature 3′-ends of atp9 mRNAs in Arabidopsis mitochondria. J. Biol. Chem., 279, 25440–25446. [DOI] [PubMed] [Google Scholar]
- 17.Hayes R., Kudla,J., Schuster,G., Gabay,L., Maliga,P. and Gruissem,W. (1996) Chloroplast mRNA 3′-end processing by a high molecular weight protein complex is regulated by nuclear encoded RNA binding proteins. EMBO J., 15, 1132–1141. [PMC free article] [PubMed] [Google Scholar]
- 18.Lisitsky I., Kotler,A. and Schuster,G. (1997) The mechanism of preferential degradation of polyadenylated RNA in the chloroplast. The exoribonuclease 100RNP/polynucleotide phosphorylase displays high binding affinity for poly(A) sequence. J. Biol. Chem., 272, 17648–17653. [DOI] [PubMed] [Google Scholar]
- 19.Lisitsky I. and Schuster,G. (1999) Preferential degradation of polyadenylated and polyuridinylated RNAs by the bacterial exoribonuclease polynucleotide phosphorylase. Eur. J. Biochem., 261, 468–474. [DOI] [PubMed] [Google Scholar]
- 20.Walter M., Kilian,J. and Kudla,J. (2002) PNPase activity determines the efficiency of mRNA 3′-end processing, the degradation of tRNA and the extent of polyadenylation in chloroplasts. EMBO J., 21, 6905–6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gagliardi D. and Leaver,C.J. (1999) Polyadenylation accelerates the degradation of the mitochondrial mRNA associated with cytoplasmic male sterility in sunflower. EMBO J., 18, 3757–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gagliardi D., Perrin,R., Marechal-Drouard,L., Grienenberger,J.M. and Leaver,C.J. (2001) Plant mitochondrial polyadenylated mRNAs are degraded by a 3′- to 5′-exoribonuclease activity, which proceeds unimpeded by stable secondary structures. J. Biol. Chem., 276, 43541–43547. [DOI] [PubMed] [Google Scholar]
- 23.Kuhn J. and Binder,S. (2002) RT-PCR analysis of 5′ to 3′-end-ligated mRNAs identifies the extremities of cox2 transcripts in pea mitochondria. Nucleic Acids Res., 30, 439–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lupold D.S., Caoile,A.G. and Stern,D.B. (1999) Polyadenylation occurs at multiple sites in maize mitochondrial cox2 mRNA and is independent of editing status. Plant Cell, 11, 1565–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuhn J., Tengler,U. and Binder,S. (2001) Transcript lifetime is balanced between stabilizing stem-loop structures and degradation-promoting polyadenylation in plant mitochondria. Mol. Cell. Biol., 21, 731–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marujo P.E., Hajnsdorf,E., Le Derout,J., Andrade,R., Arraiano,C.M. and Regnier,P. (2000) RNase II removes the oligo(A) tails that destabilize the rpsO mRNA of Escherichia coli. RNA, 6, 1185–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giese A., Thalheim,C., Brennicke,A. and Binder,S. (1996) Correlation of nonanucleotide motifs with transcript initiation of 18S rRNA genes in mitochondria of pea, potato and Arabidopsis. Mol. Gen. Genet., 252, 429–436. [DOI] [PubMed] [Google Scholar]
- 28.Li Z. and Deutscher,M.P. (1995) The tRNA processing enzyme RNase T is essential for maturation of 5S RNA. Proc. Natl Acad. Sci. USA, 92, 6883–6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Z., Pandit,S. and Deutscher,M.P. (1998) 3′ exoribonucleolytic trimming is a common feature of the maturation of small, stable RNAs in Escherichia coli. Proc. Natl Acad. Sci. USA, 95, 2856–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Z., Pandit,S. and Deutscher,M.P. (1999) Maturation of 23S ribosomal RNA requires the exoribonuclease RNase T. RNA, 5, 139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng Z.F. and Deutscher,M.P. (2003) Quality control of ribosomal RNA mediated by polynucleotide phosphorylase and RNase R. Proc. Natl Acad. Sci. USA, 100, 6388–6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piazza F., Zappone,M., Sana,M., Briani,F. and Deho,G. (1996) Polynucleotide phosphorylase of Escherichia coli is required for the establishment of bacteriophage P4 immunity. J. Bacteriol., 178, 5513–5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Briani F., Del Vecchio,E., Migliorini,D., Hajnsdorf,E., Regnier,P., Ghisotti,D. and Deho,G. (2002) RNase E and polyadenyl polymerase I are involved in maturation of CI RNA, the P4 phage immunity factor. J. Mol. Biol., 318, 321–331. [DOI] [PubMed] [Google Scholar]