Summary
Plants are commonly exposed to abiotic and biotic stresses.
We used 350 Arabidopsis thaliana accessions grown under controlled conditions. We employed genome‐wide association analysis to investigate the genetic architecture and underlying loci involved in genetic variation in resistance to: two specialist insect herbivores, Pieris rapae and Plutella xylostella; and combinations of stresses, i.e. drought followed by P. rapae and infection by the fungal pathogen Botrytis cinerea followed by infestation by P. rapae.
We found that genetic variation in resistance to combined stresses by drought plus P. rapae was limited compared with B. cinerea plus P. rapae or P. rapae alone. Resistance to the two caterpillars is controlled by different genetic components. There is limited overlap in the quantitative trait loci (QTLs) underlying resistance to combined stresses by drought plus P. rapae or B. cinerea plus P. rapae and P. rapae alone. Finally, several candidate genes involved in the biosynthesis of aliphatic glucosinolates and proteinase inhibitors were identified to be involved in resistance to P. rapae and P. xylostella, respectively.
This study underlines the importance of investigating plant responses to combinations of stresses. The value of this approach for breeding plants for resistance to combinatorial stresses is discussed.
Keywords: abiotic stress, biotic stress, combined stresses, genome‐wide association, specialist herbivores
Introduction
During their life cycle, plants are exposed to diverse abiotic stresses, such as drought, flooding, heat, cold, nutrient deficiency or ozone (Shinozaki & Yamaguchi‐Shinozaki, 2007; Roy et al., 2011; Fahad et al., 2015; Mickelbart et al., 2015), and biotic stresses, such as attack by bacteria, fungi, viruses, insects or parasitic plants (Howe & Jander, 2008; Mithofer & Boland, 2012; Pieterse et al., 2012). Substantial progress has been made in the identification of genes that provide resistance to individual stresses (Smith & Clement, 2012). However, in natural ecosystems, plants suffer from combinations of stresses that occur simultaneously or sequentially. Recent studies have addressed this by investigating the phenotypic effect, transcriptomic changes and genetics underlying responses to combined stresses (De Vos et al., 2006; Atkinson et al., 2013; Rasmussen et al., 2013; Kissoudis et al., 2014). These studies have concluded that the effect of a combination of stresses can often not be predicted from the single stress effect at the phenotypic, transcriptomic or genetic level.
Herbivory by insects is one of the major stresses faced by plants: one‐quarter of all known eukaryotic species are insect herbivores (Futuyma & Agrawal, 2009). As a result of the strong selection pressure imposed on plants by insects, plants have evolved mechanisms to protect them from insects (Kessler & Baldwin, 2002; Schoonhoven et al., 2005). Plant traits that influence the degree of damage caused by insects can be classified into resistance (traits that limit the damage by the insect) and tolerance (traits that allow plants to compensate for insect damage) (Strauss & Agrawal, 1999; Stout, 2013). Furthermore, resistance and tolerance are mediated by distinct genetic mechanisms (Strauss & Agrawal, 1999; Carmona et al., 2011; Karinho‐Betancourt & Nunez‐Farfan, 2015). Resistance can be further divided into constitutive or induced defences (Schoonhoven et al., 2005; Mithofer & Boland, 2012; Stout, 2013). One of the best studied defence mechanisms of plants against insects is the myrosinase–glucosinolate system in the Brassicaceae family (Hopkins et al., 2009; Mithofer & Boland, 2012). Glucosinolates are hydrolysed by myrosinase enzymes on insect herbivory and their breakdown products are toxic to generalist insect herbivores (Fahey et al., 2001; Kliebenstein et al., 2005; Brachi et al., 2015). However, specialist insects, such as P. rapae and P. xylostella, have developed detoxification mechanisms and do not seem to be affected by the myrosinase–glucosinolate defence of brassicaceous plants (Wheat et al., 2007; De Vos et al., 2008; Müller et al., 2010). These two insect species are major pests in several crops from the Brassica genus (e.g. broccoli, cabbage, cauliflower) worldwide. For example, annual control costs of P. xylostella are estimated to be nearly US$ 4–5 billion (Zalucki et al., 2012). A good understanding of the genetic architecture of plant resistance against these insects and the identification of molecular mechanisms behind resistance would provide breeders with better tools to develop crops that are more resistant to these insect species.
In nature, insect herbivory commonly occurs simultaneously or sequentially with other abiotic and biotic stresses (Rizhsky et al., 2004; Mittler & Blumwald, 2010; Vile et al., 2012; Prasch & Sonnewald, 2013; Rasmussen et al., 2013; Kissoudis et al., 2014; Rivero et al., 2014; Sewelam et al., 2014; Stam et al., 2014; Suzuki et al., 2014). Several studies have shown that, when plants experience a certain stress, this may compromise the plant's ability to respond to subsequent stresses (Suzuki et al., 2014; Ramegowda & Senthil‐Kumar, 2015). Therefore, the study of single stresses does not provide good predictors of plant responses to multiple stresses. Plant hormones have emerged as major players in the control of the signal transduction pathways that regulate stress responses (Erb et al., 2012; Pieterse et al., 2012). Jasmonic acid (JA), salicylic acid (SA) and ethylene (ET) have emerged as important signalling molecules in plant defences against pathogens and insects, whereas abscisic acid (ABA) is important for resistance to abiotic stresses (Shinozaki & Yamaguchi‐Shinozaki, 2007; Pieterse et al., 2012). JA activates signalling pathways that mediate responses against chewing herbivores and necrotrophic fungi, whereas SA underlies responses to biotrophic pathogens (De Vos et al., 2005; Pieterse et al., 2012). Phytohormonal signalling pathways interact through ‘crosstalk’, which may allow plants to respond in a fast and cost‐effective manner to stresses (Vos et al., 2013). Therefore, a good understanding of phytohormonal networks is needed to understand how plants tailor their response to different stresses. Hence, the analysis of responses to combinatorial stresses may yield information on the signalling nodes that are involved in the tailoring of the plant's adaptive response to stress combinations. To this end, we decided to study combinations of stresses to which individual responses are highly divergent, but, at the same time, regulated by interacting plant hormones. Therefore, we studied the interaction between A. thaliana and the specialist insect P. rapae, which is a JA inducer, the necrotrophic fungus Botrytis cinerea, which is a JA and ET inducer, and drought, which is an ABA and JA inducer (Verhage et al., 2010; Pieterse et al., 2012; Vos et al., 2013).
The model plant of genetic studies, Arabidopsis thaliana, displays natural genetic variation in developmental and physiological traits, as well as in resistance to biotic and abiotic stresses (McKay et al., 2003; Alonso‐Blanco et al., 2009; Baxter et al., 2010; Juenger, 2013; Easlon et al., 2014). In addition, natural genetic variation for resistance to specialist and generalist insects has been reported (Jander et al., 2001; Kliebenstein et al., 2002; Pfalz et al., 2007). The causal genes for variation in resistance against generalist insects have been successfully identified (mostly glucosinolate biosynthesis‐related genes) (Kliebenstein et al., 2002; Zhang et al., 2006). Less information is available on genes underlying variation in resistance to specialist insects and combined stresses (Kliebenstein et al., 2002; Pfalz et al., 2007; Kliebenstein, 2014). Quantitative trait locus (QTL) mapping using bi‐parental or multi‐parental populations has been traditionally employed for the identification of genes responsible for natural genetic variation for a trait of interest (Alonso‐Blanco & Koornneef, 2000; Koornneef et al., 2004). However, QTL mapping has a low resolution and requires a lot of time and resources (Doerge, 2002; Koornneef et al., 2004; Kloth et al., 2012; Weigel, 2012). In recent years, large collections of A. thaliana natural accessions have been genotyped and re‐sequenced, enabling genome‐wide association (GWA) studies in this model plant (Atwell et al., 2010; Weigel, 2012). GWA makes use of linkage disequilibrium (LD), when two loci in the genome are statistically more or less often inherited together as a result of recombination history, to associate genotypes with phenotypes. GWA overcomes several of the drawbacks of QTL mapping: GWA offers higher resolution (in some cases, down to the causal gene), is less time consuming and requires fewer resources, and considers more allelic diversity (Nordborg & Weigel, 2008; Zhu et al., 2008; Korte & Farlow, 2013). However, association mapping has some limitations: it requires large population sizes; it can generate a large number of false positives as a result of population structure; it has low statistical power to identify rare alleles; and it has difficulties in dissecting complex traits (many rare variants of large effects or many common variants of small effects) (Nordborg & Weigel, 2008; Zhu et al., 2008; Korte et al., 2012). Therefore, both strategies complement each other, leading to a higher power of finding causal genetic variation (Zhu et al., 2008; Myles et al., 2009; Brachi et al., 2010; Kloth et al., 2012).
Here, we used a collection of 350 A. thaliana accessions to explore the natural variation to a range of combinations of abiotic and biotic stresses. We chose the following stresses: drought, herbivory by caterpillars of Pieris rapae and Plutella xylostella, and infection by the necrotrophic fungal pathogen Botrytis cinerea. Under controlled conditions, we investigated the natural genetic variation in: resistance to two specialist insects, i.e. P. rapae and P. xylostella; resistance to combined stresses imposed by drought plus P. rapae and the plant pathogen Botrytis cinerea plus P. rapae. Resistance was quantified as the reduction in plant biomass under stress compared with non‐stress conditions. Furthermore, we used GWA mapping to gain insights into the genetic architecture of these traits and to identify regions in the genome associated with variation in resistance.
Materials and Methods
Arabidopsis thaliana Hapmap population
We used a collection of 350 A. thaliana accessions from the Hapmap population (http://bergelson.uchicago.edu/wp-content/uploads/2015/04/Justins-360-lines.xls). This population was developed from a global collection of 5810 accessions with the purpose to minimize redundancy and relatedness, a common problem in GWA studies (Atwell et al., 2010; Platt et al., 2010; Chao et al., 2012). This population has been genotyped for 248 584 bi‐allelic single nucleotide polymorphisms (SNPs), as described in Atwell et al. (2010). After quality control and imputation, this set of SNPs was reduced to a set of 214 051 SNPs. For GWA analysis, we used only SNPs with a minor allele frequency (MAF) higher than 0.05, in order to prevent spurious associations, resulting in a total of 199 360 SNPs.
Plants, insects and pathogen
Plant growth conditions
Arabidopsis plants were grown under controlled conditions at 24 ± 1°C, 70 ± 10% relative humidity, 200 μmol m−2 s−1 photosynthetically active radiation and a diurnal cycle of 8 h : 16 h, light : dark. Seeds were vernalized at 4°C for 5 d in order to induce even germination. Plants were individually grown in 0.08‐l pots in a pasteurized (4 h, 80°C) commercial potting soil (Lentse potgrond, Lent, the Netherlands), which was mixed 1 : 1 (v/v) with autoclaved sand in Expt 1 and with pasteurized (4 h, 80°C) potting soil in Expt 2. Pots were accommodated in trays that were randomly distributed within a growth chamber. Plants were watered three times per week by adding water to the tray. Once per week, entomopathogenic nematodes were included (Entonem; http://www.koppert.nl/) to prevent infestation by fungus gnats.
Insect rearing
Pieris rapae L. (Small Cabbage White butterfly; Lepidoptera; Pieridae) was reared on Brussels sprouts plants (Brassica oleracea var. gemmifera cv Cyrus) in a growth chamber at 21 ± 1°C, 50–70% relative humidity and a diurnal cycle of 16 h : 8 h, light : dark.
Plutella xylostella L. (Diamondback moth; Lepidoptera; Plutellidae) was reared on Brussels sprouts plants (B. oleracea var. gemmifera cv Cyrus) in a growth chamber at 22 ± 1°C, 40–50% relative humidity and a diurnal cycle of 16 h : 8 h, light : dark.
Pathogen culture
The necrotrophic fungus B. cinerea, strain B0510 (Van der Ent et al., 2008), was grown on half‐strength potato dextrose agar (PDA) plates containing penicillin (100 μg ml−1) and streptomycin (200 μg ml−1) for 2 wk at room temperature. Spores were collected and re‐suspended in half‐strength potato dextrose broth (PDB; Difco Laboratories, Sparks, MD, USA) to a final density of 1.0 × 105 spores ml−1. After a 3‐h incubation period, the spores were used for inoculation (Thomma et al., 1998; Pre et al., 2008; Van der Ent et al., 2008).
Experimental design and treatments
The experimental design and treatments have been described in detail in Davila Olivas et al. (2016b). Briefly, two experiments were conducted. In Expt 1, we evaluated the growth of Arabidopsis plants after exposure to drought, herbivory by P. rapae, herbivory by P. rapae preceded by drought and herbivory by P. rapae preceded by B. cinerea infestation. The experiment was performed in 10 temporal blocks. Each block consisted of 37 randomly selected accessions plus three accessions that were present in all rounds (CS28780 (Tsu‐0), CS76113 (Col‐0) and CS76129 (Fei‐0)); the last block contained only 17 accessions. Within temporal blocks, plants were allocated in trays and the position of the tray in the rearing chamber was recorded as its position in one of the six racks, each with four shelves. The spatial location of each plant within a tray was recorded in terms of column C and row R. In each temporal block, accessions were exposed to the following five treatments: (1) no stress; (2) drought stress; (3) P. rapae herbivory; (4) drought and P. rapae; or (5) B. cinerea and P. rapae. Six replicates were included per accession and treatment combination; 11 400 plants were phenotyped in Expt 1: six replicates × 40 accessions (37 random accessions plus three accessions that were used in every temporal block) × nine temporal blocks × five treatments plus six replicates × (17 + 3 accessions) × five treatments for the last block. Plants were grown under similar conditions during the first 3 wk. Drought stress was imposed by withholding water for 7 d during the third week, whilst the rest of the plants were watered every 2 d with 1 l of water per tray. Withholding water for 7 d clearly resulted in water stress: the plants showed retarded growth and were smaller than well‐watered plants. Botrytis cinerea inoculation was carried out 24 h before P. rapae inoculation. Plants were inoculated with B. cinerea by pipetting 5 μl of spores suspended in half‐strength PDB (Difco Laboratories) at a concentration of 1 × 105 spores ml−1 on two leaves of the rosette. Plants were kept at 100% relative humidity for 24 h in order to ensure successful infection by B. cinerea. Four‐week‐old plants were exposed to herbivory by P. rapae as a single or combined stress. Plants were inoculated with two newly hatched first‐instar (L1) caterpillars that were allowed to feed for 5 d. At the time of inoculation, individual plants were placed on the inverted lid of a Petri dish and the trays were filled with water to prevent caterpillars from moving between plants. Whilst the plants were on the Petri dishes, they received the same watering regime as described above; however, watering was carried out by adding 20 ml of water to each Petri dish. Rosette fresh weight (FW) was quantified for all treatments (Supporting Information Fig. S1A).
In Expt 2, we evaluated the growth reduction in Arabidopsis after exposure to herbivory by P. xylostella. The experiment was performed in four temporal blocks. Within blocks, accessions were randomly distributed over 39 trays with nine accessions per tray plus one tray that contained eight accessions. In this experiment, accession Col‐0 was included to control for a positional effect within the chamber. Each tray contained both control and treatment for Col‐0 and for nine other accessions. Plants were randomized within the trays. In each block, all accessions were phenotyped; one replicate per accession was phenotyped at a time. We repeated this four times (temporal blocks), leading to four replicates per accession. Within blocks, accessions were exposed simultaneously to either (1) no stress or (2) herbivory by P. xylostella; 2800 plants were phenotyped in Expt 2: four replicates (temporal blocks) × 350 accessions × two treatments. Some of the accessions displayed germination problems and so we did not have sufficient replicates for the experiment; this reduced the dataset from 350 to 321 accessions. Plants were 4 wk old when they were inoculated with two L2 larvae. Larvae were allowed to feed for 5 d. At the time of inoculation, individual plants were placed on the inverted lid of a Petri dish and the trays were filled with water to prevent caterpillars from moving between plants. Whilst the plants were on the Petri dishes, they followed the same watering regime as described above; however, watering was carried out by adding 20 ml of water to each Petri dish. Rosette FW was quantified for all treatments (Fig. S1B).
Statistical analysis
Genotypic mean estimations
We obtained BLUEs (best linear unbiased estimators) for all genotype–treatment combinations as described in the literature (Jimenez‐Gomez et al., 2010; Filiault & Maloof, 2012; Riedelsheimer et al., 2012). BLUEs were estimated by a linear mixed model using the ASReml package in R (Butler et al., 2009).
where Y represents the rosette FW, GEN is the genotype (accession), TRT is the treatment factor, BLOCK represents the temporal block, RACK, SHELF, TRAY, C and R are factors that represent the spatial location of the plants within the chamber, and e is the residual error. GEN + TRT + GEN × TRT were fitted as a fixed effect, whereas all other variables were fitted as random effects (underlined).
Using BLUEs, for each stress, we estimated the percentage difference of rosette FW relative to control plants without stress. In the treatment in which plants were exposed to both drought and herbivory by P. rapae, the percentage difference in rosette FW was calculated relative to plants exposed to drought. Hereafter, we refer to the percentage of biomass reduction caused by drought, P. rapae herbivory, P. xylostella herbivory, drought plus P. rapae and B. cinerea plus P. rapae as ‘Drought’, ‘P. rapae’, ‘P. xylostella’, ‘Drought&Pieris’ and ‘Botrytis&Pieris’, respectively (Table S1).
Data inspection
We initially inspected the variation in response to each stress (Fig. 1). We observed that some accessions had larger biomass under treatment than under control conditions. We reasoned that these accessions displayed tolerance to the treatment. Because tolerance and resistance traits have a different genetic basis (Strauss & Agrawal, 1999; Carmona et al., 2011; Karinho‐Betancourt & Nunez‐Farfan, 2015), we only included data for accessions displaying a reduction in biomass under the treatment compared with control conditions (Table S1). This dataset was used for all downstream analyses.
Figure 1.
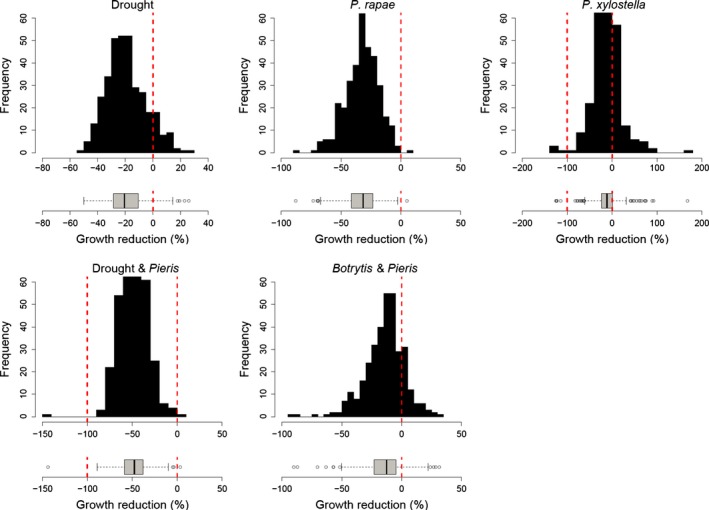
Variation in growth reduction in Arabidopsis plants exposed to drought, herbivory by Plutella xylostella, herbivory by Pieris rapae alone or preceded by drought or infection by the necrotrophic fungus Botrytis cinerea. Growth reduction was estimated from comparison with plants that had been grown without stress. For the Drought&Pieris treatment, growth reduction was estimated in comparison with plants exposed to drought only. Data subsets used for genome‐wide association analysis are indicated to the left of the zero and are delimited by either one or two red dashed lines. Box plots represent median value (thick line), the first plus third quartiles (box) and the lowest and highest values (whiskers); the circles represent outliers.
Phenotypic and genotypic correlations
Phenotypic correlations were estimated by Spearman correlation of the genotypic mean BLUEs for every possible combination of two traits. Spearman correlation analyses were implemented in the package Hmisc in R (Harrell, 2009). Genetic correlations reflect the overlap in polygenic effects amongst traits. A perfect genetic correlation (r g = 1) between two traits indicates that exactly the same loci control these traits, and a non‐perfect genetic correlation (r g < 1) reveals a mixture of unique and common genetic effects among traits. Genetic correlations were estimated according to the multi‐trait mixed model described in Korte et al. (2012).
Narrow‐sense heritability
Phenotypic variance can be decomposed into variance caused by genetic and environmental factors. Broad‐sense heritability (H 2) estimates the proportion of phenotypic variance that is caused by genetic factors. Genetic variance can be a result of additive, dominant or epistatic effects. Narrow‐sense heritability (h 2) captures the proportion of genetic variance that is caused by additive genetic effects. Narrow‐sense heritability is important because it is an indicator of how a population responds to artificial or natural selection (Wray & Visscher, 2008). Narrow‐sense heritability estimates for each response were estimated with the heritability package in R (Kruijer et al., 2015).
Genome‐wide association analysis
Variation in growth reduction under different stresses was linked to regions in the genome that explained the observed variation using a GWA analysis, carried out employing Fast‐LMM software, as described in Cao et al. (2011). Fast‐LMM assumes the following mixed model for each SNP:
where y is a vector of n phenotype values. X is a design matrix in which trait means are included with other fixed effects. In X, β is the effect of the Col‐0 allele. and are random effects. We tested the hypothesis β = 0 using generalized least squares (GLS), conditional on residual maximum likelihood (REML) estimates and for the genetic and environmental variance. The proportion of the genetic variance explained by each SNP was estimated using two methods: (1) the statistic proposed by Cox & Snell (1989), which is 1 − exp(−(2/n)(L 1 − L 0)); and (2) 2 × (β2 p(1 − p)/σ2), where β is the allele effect, p is the frequency and σ2 is the sample variance. Fast‐LMM corrects for population structure using a GRM (genetic relatedness matrix) instead of a kinship matrix as in Emmax software (Kang et al., 2010). Fast‐LMM is considered to be more powerful than Emmax because: (1) each SNP test is based on a local kinship matrix that consists of the GRM based on all markers, except those that are in a window of 20 kb on each side of the tested SNP; and (2) the genetic and residual variance components are estimated for each SNP, instead of assuming that these are constant across the genome. In order to reduce the amount of spurious associations caused by rare variants, a MAF of 5% was used.
Candidate gene selection
For the selection of candidate genes, an arbitrary threshold of −log10(P) ≥ 4 was considered. The same threshold has been used in other GWA studies on A. thaliana (Verslues et al., 2014; El‐Soda et al., 2015; Van Rooijen et al., 2015; Kooke et al., 2016). Regions containing SNPs with −log10(P) ≥ 4 were considered for further analysis as described by El‐Soda et al. (2015). A search window was defined by SNPs in LD (LD ≥ 0.5, if no SNPs were found at 0.5 the threshold was lowered to 0.4) in a window ± 20 kb with significant SNPs. SNPs in LD from the 250K array were enriched with SNPs in LD from 1001 genomes (http://1001genomes.org/), as described in Bac‐Molenaar et al. (2015). Thus, a search window was defined by the first and last SNP in LD. All genes within a search window were considered to be potential candidate genes. To narrow down the list of candidate genes, further analyses were performed. First, gene annotation from candidate genes was obtained from TAIR 10. Furthermore, candidate genes were enriched with gene expression data from different sources. Data from tissue exposed to the phytohormones JA, ABA or ET were obtained from a public database (http://bar.utoronto.ca/) (Toufighi et al., 2005). Expression data for A. thaliana plants infested with P. xylostella were obtained from Ehlting et al. (2008). RNA‐sequencing (RNA‐seq)‐based expression data for A. thaliana plants infested with P. rapae, drought and P. rapae, and B. cinerea and P. rapae were obtained from Davila Olivas et al. (2016a). RNA‐seq‐based expression data for drought responses were obtained from P. Huang et al. (unpublished). The data are summarized in Tables S2–S6.
Results
Variation within and between responses of A. thaliana to single or multiple stresses
We observed extensive variation among the accessions in the percentage of growth reduction for plants exposed to the different stresses addressed in this study (Fig. 1; Table 1). The largest variation was observed for the response to P. xylostella (CV = 78%), whereas the lowest variation was observed for the response to Drought&Pieris (CV = 31%) (Table 1). Narrow‐sense heritability estimates ranged from 0.17 to 0.52 (Table 1). No relationship between narrow‐sense heritability and variation in stress responses was observed (Table 1).
Table 1.
Summary of variation in the percentage of biomass reduction of 350 Arabidopsis thaliana accessions on exposure to drought, herbivory by Plutella xylostella and herbivory by Pieris rapae alone or preceded by drought or infection with the necrotrophic fungus Botrytis cinerea
| Trait | Min. | Mean | Max. | n | CV | h 2 | CI | va | ve |
|---|---|---|---|---|---|---|---|---|---|
| Botrytis&Pieris | 0.08 | 18.88 | 90.13 | 285 | 74 | 0.52 | 0.17–0.86 | 103.80 | 94.77 |
| P. rapae | 2.67 | 32.62 | 87.80 | 345 | 42 | 0.51 | 0.22–0.79 | 96.96 | 93.20 |
| P. xylostella | 0.03 | 21.66 | 82.22 | 234 | 78 | 0.42 | 0.13–0.78 | 121.71 | 166.41 |
| Drought | 0.29 | 22.67 | 50.07 | 307 | 48 | 0.42 | 0.15–0.76 | 49.99 | 68.16 |
| Drought&Pieris | 3.62 | 48.29 | 89.06 | 344 | 31 | 0.17 | 0.03–0.59 | 36.08 | 181.48 |
Traits are ordered by narrow‐sense heritability. Min., lowest value; Max., highest value; n, number of accessions analysed; CV, coefficient of variation (%); h 2, narrow‐sense heritability; CI, heritability 95% confidence intervals; va, additive genetic variance; ve, residual variance.
Genetic and phenotypic correlations among traits
To explore the relationships among different traits, we performed Spearman correlation analysis on the phenotypic values (Table 2). The response to drought displayed a negative correlation with the other traits. Furthermore, the largest phenotypic correlation was observed between the responses to Botrytis&Pieris and P. rapae (ρ = 0.52). A low phenotypic correlation was observed between the responses to P. xylostella and P. rapae (ρ = 0.15). Because phenotypic correlations may arise as a result of genetic and environmental factors, a better estimate of shared genetic basis between traits is genetic correlation. The largest genetic correlation was between the responses to Botrytis&Pieris and to P. rapae (r g = 0.98), followed by the responses to drought and to Botrytis&Pieris (r g = −0.81).
Table 2.
Phenotypic and genetic correlations among the percentages of biomass reduction in plants exposed to drought, herbivory by Plutella xylostella and herbivory by Pieris rapae alone or preceded by drought or infection with the necrotrophic fungus Botrytis cinerea
| Trait | Drought | P. rapae | Drought&Pieris | Botrytis&Pieris | P. xylostella |
|---|---|---|---|---|---|
| Drought | −0.65 | NC | −0.89 | −0.42 | |
| P. rapae | −0.25 | NC | 0.98 | 0.20 | |
| Drought&Pieris | −0.38 | 0.48 | NC | 0.64 | |
| Botrytis&Pieris | −0.29 | 0.53 | 0.40 | 0.33 | |
| P. xylostella | −0.12 | 0.15 | 0.16 | 0.14 |
NC, residual maximum likelihood did not converge. Phenotypic correlations (Spearman correlation coefficients) are indicated below the diagonal. Genetic correlations were estimated by residual maximum likelihood as in Korte et al. (2012). Genetic correlation estimates (r g) are indicated above the diagonal. Values below the diagonal that were not significant (P > 0.05 after Bonferroni correction) are indicated in bold.
Genetic architecture underlying variation in responses to single and multiple stresses
To obtain insight into the genetic architecture underlying the variation in responses to single stresses imposed by drought or P. rapae feeding and the two multiple stress situations, Drought&Pieris and Botrytis&Pieris, we performed a GWA analysis. We used a threshold of −log10(P) ≥ 4 to declare an SNP being associated with a trait. SNPs in LD were considered in a region of ± 20 kb from a significant SNP (Tables S2–S5). A summary of the GWA analysis for each trait is presented in Fig. 2 and Table 3. For the responses to single stresses, the numbers of significant SNPs amounted to a total of 20 (64 SNPs in LD) for the response to drought and 34 (78 SNPs in LD) for the response to P. rapae. For the responses to combined stresses, the numbers of significant SNPs were greater than in the responses to single stress situations: 38 (106 SNPs in LD) for the response to Drought&Pieris and 40 (106 SNPs in LD) for the response to Botrytis&Pieris. Effect sizes for the Col‐0 allele were estimated for each trait. For most of the traits, the significant SNPs displayed low effect sizes, except for the response to Botrytis&Pieris (Fig. 2b; Tables S2–S5). The response to drought displayed the lowest effect sizes, ranging from −4 (meaning that accessions with the Col‐0 allele have 4% less biomass reduction than accessions carrying the alternative allele) to 4. The response to Botrytis&Pieris displayed the highest effect sizes, ranging from −16 to 23. For most of the traits, the significant SNPs explained a low percentage of the genetic variance (Tables S2–S5). The maximum percentage of genetic variance explained by an SNP for the response to each stress was 7% for drought, 7% for P. rapae, 5% for Drought&Pieris and 12% for Botrytis&Pieris. Despite the moderate to high genetic correlations among traits, little overlap was observed between the significant SNPs, between the regions delimited by SNPs in LD (QTLs), and therefore also between the genes contained within QTLs (Fig. 2c).
Figure 2.
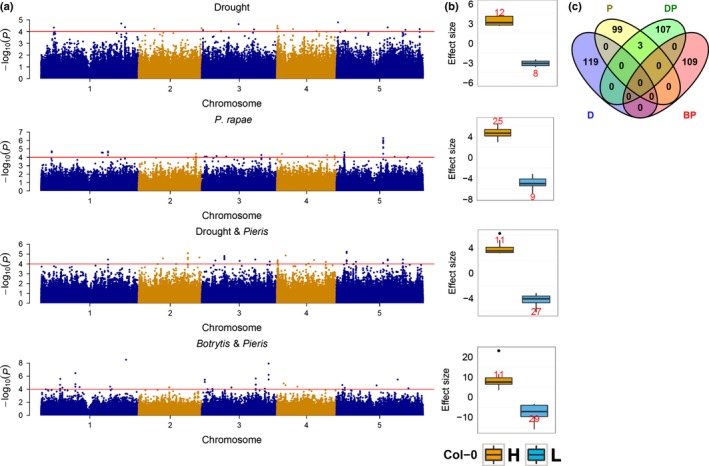
Genome‐wide association analysis of growth reduction in plants exposed to a single stress imposed by drought or herbivory by Pieris rapae or multiple stresses imposed by P. rapae preceded by drought or infection by the necrotrophic fungus Botrytis cinerea. (a) Manhattan plots. Red line indicates an arbitrary threshold set at −log10(P) ≥ 4, as described in the Materials and Methods section for the detection of significant associations. (b) Effect sizes for significant single nucleotide polymorphisms (SNPs). Effect sizes are indicated for the Col‐0 allele. Yellow and blue indicate a higher and lower reduction in biomass associated with the Col‐0 allele, respectively. The number of significant SNPs is indicated in red. Box plots represent the median value (thick line), the first plus third quartiles (box) and the lowest and highest values (whiskers); the circles represent outliers. (c) Candidate genes. Genes in a 20‐kb window of a significant SNP were considered as candidates. D, Drought; P, Pieris rapae; DP, Drought&Pieris; BP, Botrytis&Pieris.
Table 3.
Summary of genome‐wide association analysis per trait
| Trait | SNPs | SNPs in LDa | Strings | Singletons | QTLs | Genesb |
|---|---|---|---|---|---|---|
| Drought | 20 | 64 | 12 | 6 | 18 | 119 |
| P. rapae | 34 | 78 | 13 | 5 | 18 | 102 |
| Drought&Pieris | 38 | 106 | 13 | 6 | 19 | 110 |
| Botrytis&Pieris | 40 | 106 | 16 | 9 | 25 | 109 |
| P. xylostella | 57 | 238 | 22 | 10 | 32 | 141 |
Single nucleotide polymorphisms (SNPs) in linkage disequilibrium (LD) ≥ 0.5 were considered in a region of ± 20 kb from a significant SNP. Numbers of SNPs in LD are based on the 250K SNP array.
A search window was defined taking into consideration additional SNPs in LD from the 1001 genomes project (see the Materials and Methods section). All genes within a search window were considered as candidate genes.
QTL, quantitative trait locus.
Differences in genetic architecture underlying responses to two specialist insect herbivores
We also investigated the genetic architecture of A. thaliana's response to P. xylostella and compared it with the genetic architecture of the response to P. rapae. We identified a larger number of significant SNPs for the response to P. xylostella (57 SNPs plus 238 SNPs in LD) than for the response to P. rapae (34 SNPs plus 78 SNPs in LD) (Fig. 3; Table 3). Furthermore, the effect size of SNPs associated with the response to P. xylostella (from −20 to 22) was larger than that for the response to P. rapae (from −7 to 7) (Fig. 3b). The maximum percentage of genetic variance explained by the SNPs associated with the response to P. xylostella was 10%, whereas, for the response to P. rapae, it was 7% (Table S3, S6). No common significant SNPs, regions delimited by SNPs in LD (QTLs) and therefore also genes contained within QTLs were observed between P. xylostella and P. rapae (Fig. 3c).
Figure 3.
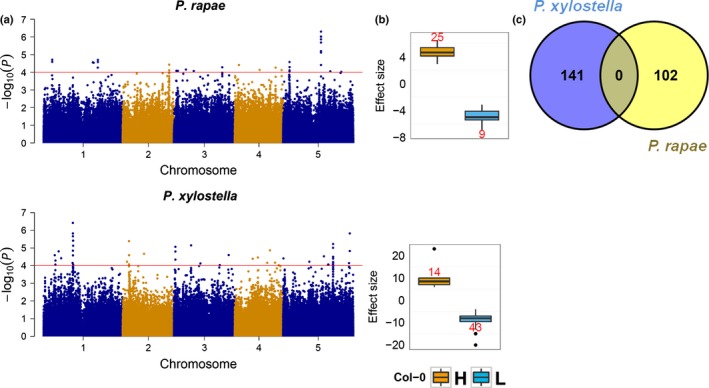
Genome‐wide association analysis of growth reduction in plants exposed to herbivory by the specialist insects Pieris rapae and Plutella xylostella. (a) Manhattan plots. Red line indicates an arbitrary threshold set at −log10(P) ≥ 4, as described in the Materials and Methods section for the detection of significant associations. (b) Effect sizes for significant single nucleotide polymorphisms (SNPs). Effect sizes are indicated for the Col‐0 allele. Yellow and blue indicate a higher and lower reduction in biomass associated with the Col‐0 allele, respectively. Number of significant SNPs is indicated in red. Box plots represent the median value (thick line), the first plus third quartiles (box) and the lowest and highest values (whiskers); the circles represent outliers. (c) Candidate genes. Genes in a 20‐kb window of a significant SNP were considered as candidates.
Candidate genes for drought resistance
Eighteen QTLs were identified for biomass reduction in response to drought. Within these regions, several genes that are known to play a role in drought acclimation were identified. For example, QTL 7 on chromosome 3 contained only one gene, AT3G17520, which encodes a late embryogenesis‐abundant protein (LEA protein). In general, LEA proteins have been suggested to play a protective role for other proteins under conditions of water stress in vegetative tissues (Battaglia et al., 2008). The closest significant SNP (Chr3: 5997119) explained 5% of the phenotypic variance; the Col‐0 allele was rare (86 accessions including Col‐0) and was associated with a greater reduction in A. thaliana FW (Table S2; Fig. S2). This gene was induced on drought stress and ABA application (Table S2). This suggests that this may be the causal gene for QTL 7.
Candidate genes involved in plant–insect interactions
We analysed the variation in growth reduction of Arabidopsis in response to two specialist insect herbivores, P. rapae and P. xylostella. GWA allowed the linking of this variation to several regions in the plant genome. We identified a total of 18 and 32 QTLs for the responses to P. rapae and P. xylostella, respectively (Table 3). Within these regions, several candidate genes with a known function in plant resistance against insect herbivores were identified.
For P. rapae, QTL 15 on chromosome 5 contained AT5G07690 (MYB29) and AT5G0700 (MYB76) (Table S3). The closest significant SNP (Chr5: 2454480) explained 4% of the phenotypic variance. The Col‐0 allele was rare (55 accessions including Col‐0) and was associated with a greater reduction in A. thaliana FW (Table S3; Fig. S3). Both genes were induced in response to P. rapae infestation. Furthermore, MYB76 was induced by JA and ET treatment (Table S3).
Another interesting QTL for the response to P. rapae was QTL 1 on chromosome 1, which contained AT1G10060 (BCAT‐1) and AT1G10070 (BCAT‐2) (Table S3). The closest significant SNP (Chr1: 3294935) explained 5% of the phenotypic variance; the Col‐0 allele was rare (89 accessions including Col‐0) and was associated with a greater reduction in A. thaliana FW (Table S3; Fig. S3). Furthermore, both genes were induced by P. rapae infestation and application of the phytohormones JA and ABA (Table S3).
For the response to P. xylostella, more QTLs were identified than for the response to P. rapae (Table 3). QTL 18 on chromosome 4 contained only two genes, AT4G11310 (CP1) and AT4G11320 (CP2). The closest significant SNP (Chr4: 3294935) explained 7% of the phenotypic variance; the Col‐0 allele was common (159 accessions including Col‐0) and was associated with a smaller reduction in A. thaliana FW (Table S6; Fig. S4). CP1 and CP2 were induced by both P. rapae and P. xylostella infestation. In addition, they were also induced by JA application (Table S6). Both CP1 and CP2 encode CYSTEINE PROTEASE enzymes (TAIR 10). CP2 has been implicated in increasing the resistance of cotton against Helicoverpa armigera (Mao et al., 2013).
Another example is QTL 32 on chromosome 5 which contains AT5G64080 (XYP1). The closest significant SNP (Chr5: 25640504) explained 9% of the phenotypic variance; the Col‐0 allele was common (178 accessions including Col‐0) and was associated with a smaller reduction in A. thaliana FW (Table S6; Fig. S4). XYP1 was induced by P. rapae and P. xylostella infestation (Table S6), and encodes a proteinase inhibitor/seed storage/lipid transfer protein. This type of protein has been implicated in anti‐nutritional defences against insect herbivores (Heidel‐Fischer et al., 2014).
Candidate genes for combined stresses
Nineteen and 25 QTLs were identified for the responses to the combined stresses Drought&Pieris and Botrytis&Pieris, respectively (Table 3). QTL 1 for Drought&Pieris and QTL 3 for P. rapae on chromosome 1 overlapped to some extent. The significant SNPs associated with each QTL were different, but the QTLs overlapped by SNPs in LD. The Col‐0 allele for significant SNPs was rare and was associated with a greater reduction in A. thaliana FW (Tables S3, S4; Figs S3, S5). AT1G55740 (SIP1) and AT1G55760 within this QTL displayed interesting expression patterns. SIP1 was induced by P. rapae infestation, drought and ABA application. AT1G55760 was induced by drought and ABA, but was repressed by JA application (Table S3, S4).
For the response to Drought&Pieris, QTL 10 on chromosome 4 and QTL 19 on chromosome 5 contained the bHLH transcription factors AT4G00480 (MYC1) and AT5G50915. The Col‐0 allele in both QTLs was rare and was associated with a smaller reduction in A. thaliana FW. Both genes were induced by P. rapae infestation and slightly induced by drought (Table S4). Natural variation in trichome density in A. thaliana has been associated with genetic variation in MYC1 (Symonds et al., 2011). Several other bHLH transcription factors (e.g. MYC2, MYC3, MYC4, MYC5) are well established in the literature as major regulators of JA‐ and ABA‐mediated responses, insect resistance and drought responses (Dombrecht et al., 2007; Shinozaki & Yamaguchi‐Shinozaki, 2007; Schweizer et al., 2013; Li et al., 2014; Qi et al., 2015). QTLs containing bHLH transcription factors were also identified for the responses to P. rapae (AT1G51140) and P. xylostella (AT1G12540) (Tables S3, S6).
For the response to Botrytis&Pieris, no bHLH transcription factors were identified. However, QTL 3 on chromosome 1 contained AT1G19210, an ERF/AP2 transcription factor (Table S5). The significant SNP with the highest effect within this QTL (Chr1: 6627245) explained 6% of the phenotypic variance; the Col‐0 allele was common (232 accessions including Col‐0) and was associated with a smaller reduction in A. thaliana FW (Table S5; Fig. S6). AT1G19210 was induced on P. rapae infection, drought, JA, ABA and ET application (Table S5). Several homologues of AT1G19210 (e.g. RAP2.1, RAP2.9, RAP2.10) have been implicated in tolerance to drought and freezing and resistance to necrotrophic fungi (Tsutsui et al., 2009; Dong & Liu, 2010).
Discussion
Genetic architecture of A. thaliana resistance to specialist insects
In this study, we have analysed the genetic architecture of A. thaliana responses to P. xylostella and P. rapae, two insect species specialized on the mustard family (Brassicaceae). We identified variation in resistance to both insect herbivores among A. thaliana accessions that is genetically determined, as indicated by the moderate narrow‐sense heritability estimates for the responses to both species (Table 1). Heritability estimates reported for resistance to generalist insects are higher than for specialist insects (Jander et al., 2001; Kliebenstein et al., 2002). For example, in the latter study, using two RIL populations, broad‐sense heritability estimates for resistance to the generalist Trichoplusia ni ranged from 0.26 to 0.31, whereas, for resistance to the specialist insect P. xylostella, it ranged from 0.12 to 0.18 (Kliebenstein et al., 2002). Although resistance to generalists is controlled by QTLs of large effect, resistance to specialists seems to be under the control of QTLs of small effect (Jander et al., 2001; Kliebenstein et al., 2002; Pfalz et al., 2007). Several studies have reported QTLs associated with insect resistance, but few have identified the causal loci (Jander et al., 2001; Pfalz et al., 2007; Ordas et al., 2009; Schranz et al., 2009; Prasad et al., 2012). The QTLs identified in this study for both insect species had small effects on plant phenotype (Tables S3, S6). However, none of the QTLs identified here were shared for resistance to the two specialist insect herbivores (Fig. 3; Table 3), suggesting that the resistance mechanisms are species specific. Similar results were obtained in a QTL study using P. brassicae and P. xylostella and A. thaliana, where no common QTLs were identified (Pfalz et al., 2007). Furthermore, microarray analyses have revealed that P. rapae and P. xylostella elicit different transcriptomic responses in A. thaliana, supporting the notion of species‐specific mechanisms of resistance (Ehlting et al., 2008; Bidart‐Bouzat & Kliebenstein, 2011).
QTL analyses in A. thaliana and other species in the Brassicaceae have identified several genes involved in the metabolism of glucosinolates as the source of resistance to generalist insects (Jander et al., 2001; Kliebenstein et al., 2002; Schranz et al., 2009). However, specialist insects, such as P. rapae and P. xylostella, have developed distinct detoxification mechanisms rendering glucosinolates ineffective (Schoonhoven et al., 2005; Wheat et al., 2007; De Vos et al., 2008; Müller et al., 2010; Agrawal et al., 2012). Interestingly, one of the QTLs identified here for resistance to P. rapae contained, as the most likely candidates, MYB29 and MYB76, encoding for two transcription factors involved in the induced production of aliphatic glucosinolates (Hirai et al., 2007). Indeed, the double mutant myb29myb28, which lacks aliphatic glucosinolates, is less preferred for feeding by P. rapae than is Col‐0 (Müller et al., 2010). Another QTL, identified for the response to P. rapae, contained, as most likely candidates, BCAT‐1 and BCAT‐2, which are enzymes involved in branched amino acid (leucine (Leu), valine (Val) and isoleucine (Ile)) (BCAA) metabolism (Diebold et al., 2002). Interestingly, homologues of these genes (BCAT‐3, BCAT‐4, BCAT‐6) have been implicated in the production of aliphatic glucosinolates (Schuster et al., 2006; Lachler et al., 2015). Furthermore, co‐expression networks have revealed that BCAT‐4 is co‐expressed with MYB29, MYB28 and several putative genes involved in Leu metabolism (Hirai et al., 2007). Interestingly, an evolutionary link has been suggested between aliphatic glucosinolates and BCAA metabolism (Schuster et al., 2006). In Boechera stricta, a species related to A. thaliana, QTL analysis identified a QTL that controls variation in allocation between methionine and BCAA‐derived glucosinolates and resistance to the generalist caterpillar T. ni (Schranz et al., 2009).
For P. xylostella, a small‐effect QTL near ERECTA on chromosome 2 has been reported in Arabidopsis and B. oleracea (Kliebenstein et al., 2002; Ramchiary et al., 2015). We identified two QTLs on chromosome 2. However, neither of these was in the vicinity of ERECTA.
Genetic architecture of resistance against multiple stresses
In complex environments, such as natural and agricultural ecosystems, plants experience several stresses that co‐occur (Mittler & Blumwald, 2010; Chan et al., 2011; Prasch & Sonnewald, 2013; Rasmussen et al., 2013; Kissoudis et al., 2014). Here, we compared the genetic architecture of the combined stresses imposed by drought plus P. rapae or B. cinerea plus P. rapae with the single stress imposed by P. rapae alone. We observed genetically determined variation for both combined stresses, as indicated by their narrow‐sense heritability values (Table 1). However, although the total phenotypic variance for resistance to drought plus P. rapae was larger than for the single stress situation, the proportion that was explained by genetic factors was dramatically lower (Table 1). This implies that there is little genetic variation for this trait, and this may have implications for the power of GWA analysis to identify true associations with this trait. Only a few studies have conducted QTL analysis on plant responses to combined stresses, and some have identified similar caveats. For example, in a study conducted in a maize population, a lower genetic variance was observed under a combination of drought plus heat than in the single stress situations (Cairns et al., 2013). Furthermore, in a tomato population exposed to a combination of salt and powdery mildew, a reduction in phenotypic variation in disease resistance was observed under combined stress in comparison with the single stress situation (Kissoudis et al., 2015). The low heritability and phenotypic variation under combined stresses may represent a pitfall for QTL identification and breeding for combined stresses.
However, for the combined stresses of B. cinerea plus P. rapae, no difference in narrow‐sense heritability was observed compared with the single stress imposed by P. rapae (Table 1). Furthermore, both traits displayed a high level of genetic correlation, suggesting that common genes influence both traits (Table 2). Despite the high genetic correlation between the response to B. cinerea plus P. rapae vs the response to the single stress P. rapae, no common QTLs were identified (Table 3; Fig. 2). It may be that the QTLs that underlie the similarity of both traits are QTLs of small effect that were not identified at the threshold used in this study. An alternative tool that may help to unravel the genetic commonality between these two traits is a multi‐trait GWA that allows for the identification of SNPs with common and opposite effects among highly correlated traits (Korte et al., 2012). This may increase the power of univariate GWAs for highly correlated traits (Korte et al., 2012).
Contrary to the limited overlap found between QTLs identified for combined stresses (Table 3; Fig. 2), other studies have identified a mixture of novel QTLs and QTLs that are present in the single stress case (Cairns et al., 2013; Makumburage et al., 2013). However, the effect of QTLs under stress combinations was never observed to be in the same direction as in the single stress case (Makumburage et al., 2013). Thus, the genetic architecture underlying single and combined stresses appears to be different. In addition to the few studies addressing QTL identification, several papers have addressed whole transcriptome changes in response to combinations of stresses (Atkinson et al., 2013; Prasch & Sonnewald, 2013; Rasmussen et al., 2013). These studies concluded that the transcriptional response to combined stresses was different from the single stress situation. Furthermore, up to 60% of the transcriptional changes in response to combined stresses could not be predicted from the response to each individual stress (Rasmussen et al., 2013).
Despite co‐occurrence being the rule rather than the exception under natural conditions, the importance of studying stress combinations has only just started to be acknowledged by the scientific community. It may be that the complexity of the experimental design, the number of possible stress combinations and the complex logistics have held back the adoption of this type of experiment. The present study, together with several studies on QTL mapping and transcriptomic changes under combinations of stresses, have concluded that responses to combined stresses cannot be predicted from the responses to individual stresses (Voelckel & Baldwin, 2004). This further underlines the complexity of the events that take place when plants are challenged by combinations of stresses, and highlights the importance of studying combinations of stresses in addition to studies of single stresses.
Finally, P. rapae and P. xylostella are major pests on Brassica crops, such as cabbage and broccoli (Agrawal & Kurashige, 2003; Zalucki et al., 2012). A good understanding of genetic architecture and the unequivocal identification of genes underlying variation in resistance will benefit the breeding process of cultivars that are more resistant to these insect pests.
In this study, we identified several candidate genes for resistance to two specialist insects (P. rapae and P. xylostella), an abiotic stress (drought) and two combined stresses (drought plus P. rapae; B. cinerea plus P. rapae). We have provided evidence using transcriptomic data from independent studies which show that these genes are differentially expressed when plants are exposed to the same stresses as addressed in this study. The genes identified here remain to be validated by, for example, allelic complementation or mutant experiments in future studies. However, it should be realized that a mutant effect is estimated in just one genetic background and tests all genetic (additive, dominance, epistatic) and genotype‐by‐environment effects for that position/gene simultaneously. A QTL allele effect in an association analysis represents the conditional difference between two groups of genotypes with alternative versions of an SNP, and so the additive effect in a more general sense than the additive effect in the mutant, where these groups are part of an association panel. Moreover, the data probably deal with quantitative phenotypic variation and small QTL effects; knockout mutants do not provide the best means to confirm such QTLs, as mutants are especially suitable for genes and QTLs with a qualitative effect. The present study, whilst revealing the genetic architecture underlying resistance to several environmental stresses, also highlights the complexity of studying combinations of stresses.
Author contributions
N.H.D.O., J.J.A.v.L., G.G. and M.D. planned and designed the research. N.H.D.O., C.L.W. and W.K. performed the experiments and/or analysed the data. N.H.D.O., J.J.A.v.L. and M.D. executed data interpretation. N.H.D.O., W.K., J.J.A.v.L. and M.D. wrote the manuscript.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Time scheme of treatments.
Fig. S2 Single nucleotide polymorphisms (SNPs) associated with reduction in Arabidopsis thaliana rosette fresh weight in response to drought.
Fig. S3 Single nucleotide polymorphisms (SNPs) associated with reduction in Arabidopsis thaliana rosette fresh weight in response to Pieris rapae.
Fig. S4 Single nucleotide polymorphisms (SNPs) associated with reduction in Arabidopsis thaliana rosette fresh weight in response to Plutella xylostella.
Fig. S5 Single nucleotide polymorphisms (SNPs) associated with reduction in Arabidopsis thaliana rosette fresh weight in response to drought plus Pieris rapae.
Fig. S6 Single nucleotide polymorphisms (SNPs) associated with reduction in Arabidopsis thaliana rosette fresh weight in response to Botrytis cinerea plus Pieris rapae.
Table S1 Percentage reduction in rosette fresh weight in Arabidopsis thaliana on stress exposure
Table S2 Candidate genes for reduction in Arabidopsis thaliana rosette fresh weight in response to drought
Table S3 Candidate genes for reduction in A. thaliana rosette fresh weight in response to Pieris rapae
Table S4 Candidate genes for reduction in Arabidopsis thaliana rosette fresh weight in response to drought plus Pieris rapae
Table S5 Candidate genes for reduction in Arabidopsis thaliana rosette fresh weight in response to Botrytis cinerea plus Pieris rapae
Table S6 Candidate genes for reduction in Arabidopsis thaliana rosette fresh weight in response to Plutella xylostella
Acknowledgements
We thank André Gidding, Frans van Aggelen and Léon Westerd for the rearing of insects. We are grateful to Fred van Eeuwijk for discussions and to Léon Westerd and Gerrie Wiegers for help with data collection. Furthermore, we thank two anonymous reviewers for their constructive comments on a previous version of the manuscript. This work was supported by The Netherlands Organization for Scientific Research (NWO) through the Technology Foundation, Perspective Programme ‘Learning from Nature’ (STW10988).
References
- Agrawal AA, Hastings AP, Johnson MTJ, Maron JL, Salminen JP. 2012. Insect herbivores drive real‐time ecological and evolutionary change in plant populations. Science 338: 113–116. [DOI] [PubMed] [Google Scholar]
- Agrawal AA, Kurashige NS. 2003. A role for isothiocyanates in plant resistance against the specialist herbivore Pieris rapae . Journal of Chemical Ecology 29: 1403–1415. [DOI] [PubMed] [Google Scholar]
- Alonso‐Blanco C, Aarts MGM, Bentsink L, Keurentjes JJB, Reymond M, Vreugdenhil D, Koornneef M. 2009. What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell 21: 1877–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso‐Blanco C, Koornneef M. 2000. Naturally occurring variation in Arabidopsis: an underexploited resource for plant genetics. Trends in Plant Science 5: 22–29. [DOI] [PubMed] [Google Scholar]
- Atkinson NJ, Lilley CJ, Urwin PE. 2013. Identification of genes involved in the response of Arabidopsis to simultaneous biotic and abiotic stresses. Plant Physiology 162: 2028–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwell S, Huang YS, Vilhjalmsson BJ, Willems G, Horton M, Li Y, Meng DZ, Platt A, Tarone AM, Hu TT et al 2010. Genome‐wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465: 627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bac‐Molenaar JA, Fradin EF, Becker FFM, Rienstra JA, van der Schoot J, Vreugdenhil D, Keurentjes JJB. 2015. Genome‐wide association mapping of fertility reduction upon heat stress reveals developmental stage‐specific QTLs in Arabidopsis thaliana . Plant Cell 27: 1857–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia M, Olvera‐Carrillo Y, Garciarrubio A, Campos F, Covarrubias AA. 2008. The enigmatic LEA proteins and other hydrophilins. Plant Physiology 148: 6–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter I, Brazelton JN, Yu DN, Huang YS, Lahner B, Yakubova E, Li Y, Bergelson J, Borevitz JO, Nordborg M et al 2010. A coastal cline in sodium accumulation in Arabidopsis thaliana is driven by natural variation of the sodium transporter AtHKT1;1. PLoS Genetics 6: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidart‐Bouzat MG, Kliebenstein D. 2011. An ecological genomic approach challenging the paradigm of differential plant responses to specialist versus generalist insect herbivores. Oecologia 167: 677–689. [DOI] [PubMed] [Google Scholar]
- Brachi B, Faure N, Horton M, Flahauw E, Vazquez A, Nordborg M, Bergelson J, Cuguen J, Roux F. 2010. Linkage and association mapping of Arabidopsis thaliana flowering time in nature. PLoS Genetics 6: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachi B, Meyer CG, Villoutreix R, Platt A, Morton TC, Roux F, Bergelson J. 2015. Co‐selected genes determine adaptive variation in herbivore resistance throughout the native range of Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 112: 4032–4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler DG, Cullis BR, Gilmour AR, Gogel BJ. 2009. ASReml‐R reference manual. Brisbane, Australia: Queensland Department of Primary Industries and Fisheries. [Google Scholar]
- Cairns JE, Crossa J, Zaidi PH, Grudloyma P, Sanchez C, Araus JL, Thaitad S, Makumbi D, Magorokosho C, Banziger M et al 2013. Identification of drought, heat, and combined drought and heat tolerant donors in maize. Crop Science 53: 1335–1346. [Google Scholar]
- Cao J, Schneeberger K, Ossowski S, Gunther T, Bender S, Fitz J, Koenig D, Lanz C, Stegle O, Lippert C et al 2011. Whole‐genome sequencing of multiple Arabidopsis thaliana populations. Nature Genetics 43: 956–963. [DOI] [PubMed] [Google Scholar]
- Carmona D, Lajeunesse MJ, Johnson MTJ. 2011. Plant traits that predict resistance to herbivores. Functional Ecology 25: 358–367. [Google Scholar]
- Chan EKF, Rowe HC, Corwin JA, Joseph B, Kliebenstein DJ. 2011. Combining genome‐wide association mapping and transcriptional networks to identify novel genes controlling glucosinolates in Arabidopsis thaliana . PLoS Biology 9: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao DY, Silva A, Baxter I, Huang YS, Nordborg M, Danku J, Lahner B, Yakubova E, Salt DE. 2012. Genome‐wide association studies identify heavy metal ATPase3 as the primary determinant of natural variation in leaf cadmium in Arabidopsis thaliana . PLoS Genetics 8: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DR, Snell EJ. 1989. Analysis of binary data. London, UK: Chapman and Hall. [Google Scholar]
- Davila Olivas NH, Coolen S, Huang P, Severing E, van Verk MC, Hickman R, Wittenberg AHJ, De Vos M, Prins M, van Loon JJA et al 2016a. Effect of prior drought and pathogen stress on Arabidopsis transcriptome changes to caterpillar herbivory. New Phytologist 210: 1344–1356. [DOI] [PubMed] [Google Scholar]
- Davila Olivas NH, Frago E, van Loon JJA, Thoen MPM, Kloth KJ, Becker F, van Heerwaarden J, Gort G, Keurentjes JJB, Dicke M. 2016b. Natural variation in life‐history strategy of Arabidopsis thaliana determines stress responses to drought and insects of different feeding guilds In: Davila Olivas NH, Ecogenomics of plant resistance to biotic and abiotic stresses, PhD thesis, Wageningen University, Wageningen, the Netherlands, 35–69. [Google Scholar]
- De Vos M, Kriksunov KL, Jander G. 2008. Indole‐3‐acetonitrile production from indole glucosinolates deters oviposition by Pieris rapae . Plant Physiology 146: 916–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos M, Van Oosten VR, Van Poecke RMP, Van Pelt JA, Pozo MJ, Mueller MJ, Buchala AJ, Metraux JP, Van Loon LC, Dicke M et al 2005. Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Molecular Plant–Microbe Interactions 18: 923–937. [DOI] [PubMed] [Google Scholar]
- De Vos M, Van Zaanen W, Koornneef A, Korzelius JP, Dicke M, Van Loon LC, Pieterse CMJ. 2006. Herbivore‐induced resistance against microbial pathogens in Arabidopsis . Plant Physiology 142: 352–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebold R, Schuster J, Daschner K, Binder S. 2002. The branched‐chain amino acid transaminase gene family in Arabidopsis encodes plastid and mitochondrial proteins. Plant Physiology 129: 540–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerge RW. 2002. Mapping and analysis of quantitative trait loci in experimental populations. Nature Reviews Genetics 3: 43–52. [DOI] [PubMed] [Google Scholar]
- Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, Fitt GP, Sewelam N, Schenk PM, Manners JM et al 2007. MYC2 differentially modulates diverse jasmonate‐dependent functions in Arabidopsis . Plant Cell 19: 2225–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong CJ, Liu JY. 2010. The Arabidopsis EAR‐motif‐containing protein RAP2.1 functions as an active transcriptional repressor to keep stress responses under tight control. BMC Plant Biology 10: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easlon HM, Nemali KS, Richards JH, Hanson DT, Juenger TE, McKay JK. 2014. The physiological basis for genetic variation in water use efficiency and carbon isotope composition in Arabidopsis thaliana . Photosynthesis Research 119: 119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlting J, Chowrira SG, Mattheus N, Aeschliman DS, Arimura GI, Bohlmann J. 2008. Comparative transcriptome analysis of Arabidopsis thaliana infested by diamond back moth (Plutella xylostella) larvae reveals signatures of stress response, secondary metabolism, and signalling. BMC Genomics 9: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El‐Soda M, Kruijer W, Malosetti M, Koornneef M, Aarts MGM. 2015. Quantitative trait loci and candidate genes underlying genotype by environment interaction in the response of Arabidopsis thaliana to drought. Plant, Cell & Environment 38: 585–599. [DOI] [PubMed] [Google Scholar]
- Erb M, Meldau S, Howe GA. 2012. Role of phytohormones in insect‐specific plant reactions. Trends in Plant Science 17: 250–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahad S, Hussain S, Matloob A, Khan FA, Khaliq A, Saud S, Hassan S, Shan D, Khan F, Ullah N et al 2015. Phytohormones and plant responses to salinity stress: a review. Plant Growth Regulation 75: 391–404. [Google Scholar]
- Fahey JW, Zalcmann AT, Talalay P. 2001. The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry 56: 5–51. [DOI] [PubMed] [Google Scholar]
- Filiault DL, Maloof JN. 2012. A genome‐wide association study identifies variants underlying the Arabidopsis thaliana shade avoidance response. PLoS Genetics 8: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futuyma DJ, Agrawal AA. 2009. Macroevolution and the biological diversity of plants and herbivores. Proceedings of the National Academy of Sciences, USA 106: 18054–18061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jr Harrell FE. 2009. Hmisc: Harrell miscellaneous. R package version 3.0. Vienna, Austria: R Foundation for Statistical Computing, http://cran.r-project.org/web/packages/Hmisc/index.html [accessed 26 September 2015]. [Google Scholar]
- Heidel‐Fischer HM, Musser RO, Vogel H. 2014. Plant transcriptomic responses to herbivory In: Voelckel C, Jander G, eds. Plant–insect interactions. Chichester, UK: John Wiley & Sons, 155–196. [Google Scholar]
- Hirai MY, Sugiyama K, Sawada Y, Tohge T, Obayashi T, Suzuki A, Araki R, Sakurai N, Suzuki H, Aoki K et al 2007. Omics‐based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proceedings of the National Academy of Sciences, USA 104: 6478–6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins RJ, van Dam NM, van Loon JJA. 2009. Role of glucosinolates in insect–plant relationships and multitrophic interactions. Annual Review of Entomology 54: 57–83. [DOI] [PubMed] [Google Scholar]
- Howe GA, Jander G. 2008. Plant immunity to insect herbivores. Annual Review of Plant Biology 59: 41–66. [DOI] [PubMed] [Google Scholar]
- Jander G, Cui JP, Nhan B, Pierce NE, Ausubel FM. 2001. The TASTY locus on chromosome 1 of Arabidopsis affects feeding of the insect herbivore Trichoplusia ni . Plant Physiology 126: 890–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez‐Gomez JM, Wallace AD, Maloof JN. 2010. Network analysis identifies ELF3 as a QTL for the shade avoidance response in Arabidopsis . PLoS Genetics 6: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juenger TE. 2013. Natural variation and genetic constraints on drought tolerance. Current Opinion in Plant Biology 16: 274–281. [DOI] [PubMed] [Google Scholar]
- Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, Sabatti C, Eskin E. 2010. Variance component model to account for sample structure in genome‐wide association studies. Nature Genetics 42: 348–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karinho‐Betancourt E, Nunez‐Farfan J. 2015. Evolution of resistance and tolerance to herbivores: testing the trade‐off hypothesis. PeerJ 3: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler A, Baldwin IT. 2002. Plant responses to insect herbivory: the emerging molecular analysis. Annual Review of Plant Biology 53: 299–328. [DOI] [PubMed] [Google Scholar]
- Kissoudis C, Chowdhury R, van Heusden S, van de Wiel C, Finkers R, Visser RGF, Bai YL, van der Linden G. 2015. Combined biotic and abiotic stress resistance in tomato. Euphytica 202: 317–332. [Google Scholar]
- Kissoudis C, van de Wiel C, Visser RGF, van der Linden G. 2014. Enhancing crop resilience to combined abiotic and biotic stress through the dissection of physiological and molecular crosstalk. Frontiers in Plant Science 5: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ. 2014. Quantitative genetics and genomics of plant resistance to insects In: Voelckel C, Jander G, eds. Plant–insect interactions. Chichester, UK: John Wiley & Sons, 235–262. [Google Scholar]
- Kliebenstein DJ, Kroymann J, Mitchell‐Olds T. 2005. The glucosinolate–myrosinase system in an ecological and evolutionary context. Current Opinion in Plant Biology 8: 264–271. [DOI] [PubMed] [Google Scholar]
- Kliebenstein D, Pedersen D, Barker B, Mitchell‐Olds T. 2002. Comparative analysis of quantitative trait loci controlling glucosinolates, myrosinase and insect resistance in Arabidopsis thaliana . Genetics 161: 325–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloth KJ, Thoen MPM, Bouwmeester HJ, Jongsma MA, Dicke M. 2012. Association mapping of plant resistance to insects. Trends in Plant Science 17: 311–319. [DOI] [PubMed] [Google Scholar]
- Kooke R, Kruijer W, Bours R, Becker F, Kuhn A, van de Geest H, Buntjer J, Doeswijk T, Guerra J, Bouwmeester H et al 2016. Genome‐wide association mapping and genomic prediction elucidate the genetic architecture of morphological traits in Arabidopsis . Plant Physiology 170: 2187–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Alonso‐Blanco C, Vreugdenhil D. 2004. Naturally occurring genetic variation in Arabidopsis thaliana . Annual Review of Plant Biology 55: 141–172. [DOI] [PubMed] [Google Scholar]
- Korte A, Farlow A. 2013. The advantages and limitations of trait analysis with GWAS: a review. Plant Methods 9: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korte A, Vilhjalmsson BJ, Segura V, Platt A, Long Q, Nordborg M. 2012. A mixed‐model approach for genome‐wide association studies of correlated traits in structured populations. Nature Genetics 44: 1066–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruijer W, Boer MP, Malosetti M, Flood PJ, Engel B, Kooke R, Keurentjes JJB, van Eeuwijk FA. 2015. Marker‐based estimation of heritability in immortal populations. Genetics 199: 379–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachler K, Imhof J, Reichelt M, Gershenzon J, Binder S. 2015. The cytosolic branched‐chain aminotransferases of Arabidopsis thaliana influence methionine supply, salvage and glucosinolate metabolism. Plant Molecular Biology 88: 119–131. [DOI] [PubMed] [Google Scholar]
- Li R, Weldegergis BT, Li J, Jung C, Qu J, Sun YW, Qian HM, Tee C, van Loon JJA, Dicke M et al 2014. Virulence factors of Geminivirus interact with MYC2 to subvert plant resistance and promote vector performance. Plant Cell 26: 4991–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makumburage GB, Richbourg HL, LaTorre KD, Capps A, Chen CX, Stapleton AE. 2013. Genotype to phenotype maps: multiple input abiotic signals combine to produce growth effects via attenuating signaling interactions in maize. G3‐Genes Genomes Genetics 3: 2195–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao YB, Xue XY, Tao XY, Yang CQ, Wang LJ, Chen XY. 2013. Cysteine protease enhances plant‐mediated bollworm RNA interference. Plant Molecular Biology 83: 119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay JK, Richards JH, Mitchell‐Olds T. 2003. Genetics of drought adaptation in Arabidopsis thaliana: I. Pleiotropy contributes to genetic correlations among ecological traits. Molecular Ecology 12: 1137–1151. [DOI] [PubMed] [Google Scholar]
- Mickelbart MV, Hasegawa PM, Bailey‐Serres J. 2015. Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nature Reviews Genetics 16: 237–251. [DOI] [PubMed] [Google Scholar]
- Mithofer A, Boland W. 2012. Plant defense against herbivores: chemical aspects. Annual Review of Plant Biology 63: 431–450. [DOI] [PubMed] [Google Scholar]
- Mittler R, Blumwald E. 2010. Genetic engineering for modern agriculture: challenges and perspectives. Annual Review of Plant Biology 61: 443–462. [DOI] [PubMed] [Google Scholar]
- Müller R, de Vos M, Sun JY, Sonderby IE, Halkier BA, Wittstock U, Jander G. 2010. Differential effects of indole and aliphatic glucosinolates on lepidopteran herbivores. Journal of Chemical Ecology 36: 905–913. [DOI] [PubMed] [Google Scholar]
- Myles S, Peiffer J, Brown PJ, Ersoz ES, Zhang ZW, Costich DE, Buckler ES. 2009. Association mapping: critical considerations shift from genotyping to experimental design. Plant Cell 21: 2194–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M, Weigel D. 2008. Next‐generation genetics in plants. Nature 456: 720–723. [DOI] [PubMed] [Google Scholar]
- Ordas B, Malvar RA, Santiago R, Sandoya G, Romay MC, Butron A. 2009. Mapping of QTL for resistance to the Mediterranean corn borer attack using the intermated B73 × Mo17 (IBM) population of maize. Theoretical and Applied Genetics 119: 1451–1459. [DOI] [PubMed] [Google Scholar]
- Pfalz M, Vogel H, Mitchell‐Olds T, Kroymann J. 2007. Mapping of QTL for resistance against the crucifer specialist herbivore Pieris brassicae in a new Arabidopsis inbred line population, Da(1)‐12xEi‐2. PLoS ONE 2: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CMJ, Van der Does D, Zamioudis C, Leon‐Reyes A, Van Wees SCM. 2012. Hormonal modulation of plant immunity. Annual Review of Cell and Developmental Biology 28: 489–521. [DOI] [PubMed] [Google Scholar]
- Platt A, Horton M, Huang YS, Li Y, Anastasio AE, Mulyati NW, Agren J, Bossdorf O, Byers D, Donohue K et al 2010. The scale of population structure in Arabidopsis thaliana . PLoS Genetics 6: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad K, Song BH, Olson‐Manning C, Anderson JT, Lee CR, Schranz ME, Windsor AJ, Clauss MJ, Manzaneda AJ, Naqvi I et al 2012. A gain‐of‐function polymorphism controlling complex traits and fitness in nature. Science 337: 1081–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasch CM, Sonnewald U. 2013. Simultaneous application of heat, drought, and virus to Arabidopsis plants reveals significant shifts in signaling networks. Plant Physiology 162: 1849–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pre M, Atallah M, Champion A, De Vos M, Pieterse CMJ, Memelink J. 2008. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiology 147: 1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi TC, Huang H, Song SS, Xie DX. 2015. Regulation of jasmonate‐mediated stamen development and seed production by a bHLH‐MYB complex in Arabidopsis . Plant Cell 27: 1620–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramchiary N, Pang W, Nguyen VD, Li X, Choi SR, Kumar A, Kwon M, Song HY, Begum S, Kehie M et al 2015. Quantitative trait loci mapping of partial resistance to diamondback moth in cabbage (Brassica oleracea L.). Theoretical and Applied Genetics 128: 1209–1218. [DOI] [PubMed] [Google Scholar]
- Ramegowda V, Senthil‐Kumar M. 2015. The interactive effects of simultaneous biotic and abiotic stresses on plants: mechanistic understanding from drought and pathogen combination. Journal of Plant Physiology 176: 47–54. [DOI] [PubMed] [Google Scholar]
- Rasmussen S, Barah P, Suarez‐Rodriguez MC, Bressendorff S, Friis P, Costantino P, Bones AM, Nielsen HB, Mundy J. 2013. Transcriptome responses to combinations of stresses in Arabidopsis . Plant Physiology 161: 1783–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedelsheimer C, Lisec J, Czedik‐Eysenberg A, Sulpice R, Flis A, Grieder C, Altmann T, Stitt M, Willmitzer L, Melchinger AE. 2012. Genome‐wide association mapping of leaf metabolic profiles for dissecting complex traits in maize. Proceedings of the National Academy of Sciences, USA 109: 8872–8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero RM, Mestre TC, Mittler R, Rubio F, Garcia‐Sanchez F, Martinez V. 2014. The combined effect of salinity and heat reveals a specific physiological, biochemical and molecular response in tomato plants. Plant, Cell and Environment 37: 1059–1073. [DOI] [PubMed] [Google Scholar]
- Rizhsky L, Liang HJ, Shuman J, Shulaev V, Davletova S, Mittler R. 2004. When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiology 134: 1683–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SJ, Tucker EJ, Tester M. 2011. Genetic analysis of abiotic stress tolerance in crops. Current Opinion in Plant Biology 14: 232–239. [DOI] [PubMed] [Google Scholar]
- Schoonhoven LM, Loon JJAv, Dicke M. 2005. Insect–plant biology. Oxford, UK: Oxford University Press. [Google Scholar]
- Schranz ME, Manzaneda AJ, Windsor AJ, Clauss MJ, Mitchell‐Olds T. 2009. Ecological genomics of Boechera stricta: identification of a QTL controlling the allocation of methionine‐ vs branched‐chain amino acid‐derived glucosinolates and levels of insect herbivory. Heredity 102: 465–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster J, Knill T, Reichelt M, Gershenzon J, Binder S. 2006. BRANCHED‐CHAIN AMINOTRANSFERASE4 is part of the chain elongation pathway in the biosynthesis of methionine‐derived glucosinolates in Arabidopsis . Plant Cell 18: 2664–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer F, Fernandez‐Calvo P, Zander M, Diez‐Diaz M, Fonseca S, Glauser G, Lewsey MG, Ecker JR, Solano R, Reymond P. 2013. Arabidopsis basic Helix‐Loop‐Helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell 25: 3117–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewelam N, Oshima Y, Mitsuda N, Ohme‐Takagi M. 2014. A step towards understanding plant responses to multiple environmental stresses: a genome‐wide study. Plant, Cell & Environment 37: 2024–2035. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi‐Shinozaki K. 2007. Gene networks involved in drought stress response and tolerance. Journal of Experimental Botany 58: 221–227. [DOI] [PubMed] [Google Scholar]
- Smith CM, Clement SL. 2012. Molecular bases of plant resistance to arthropods. Annual Review of Entomology 57: 309–328. [DOI] [PubMed] [Google Scholar]
- Stam JM, Kroes A, Li YH, Gols R, van Loon JJA, Poelman EH, Dicke M. 2014. Plant interactions with multiple insect herbivores: from community to genes. Annual Review of Plant Biology 65: 689–713. [DOI] [PubMed] [Google Scholar]
- Stout MJ. 2013. Reevaluating the conceptual framework for applied research on host plant resistance. Insect Science 20: 263–272. [DOI] [PubMed] [Google Scholar]
- Strauss SY, Agrawal AA. 1999. The ecology and evolution of plant tolerance to herbivory. Trends in Ecology & Evolution 14: 179–185. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Rivero RM, Shulaev V, Blumwald E, Mittler R. 2014. Abiotic and biotic stress combinations. New Phytologist 203: 32–43. [DOI] [PubMed] [Google Scholar]
- Symonds VV, Hatlestad G, Lloyd AM. 2011. Natural allelic variation defines a role for ATMYC1: trichome cell fate determination. PLoS Genetics 7: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma B, Eggermont K, Penninckx I, Mauch‐Mani B, Vogelsang R, Cammue BPA, Broekaert WF. 1998. Separate jasmonate‐dependent and salicylate‐dependent defense‐response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proceedings of the National Academy of Sciences, USA 95: 15107–15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ. 2005. The botany array resource: e‐northerns, expression angling, and promoter analyses. Plant Journal 43: 153–163. [DOI] [PubMed] [Google Scholar]
- Tsutsui T, Kato W, Asada Y, Sako K, Sato T, Sonoda Y, Kidokoro S, Yamaguchi‐Shinozaki K, Tamaoki M, Arakawa K et al 2009. DEAR1, a transcriptional repressor of DREB protein that mediates plant defense and freezing stress responses in Arabidopsis . Journal of Plant Research 122: 633–643. [DOI] [PubMed] [Google Scholar]
- Van der Ent S, Verhagen BWM, Van Doorn R, Bakker D, Verlaan MG, Pel MJC, Joosten RG, Proveniers MCG, Van Loon LC, Ton J et al 2008. MYB72 is required in early signaling steps of rhizobacteria‐induced systemic resistance in Arabidopsis . Plant Physiology 146: 1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rooijen R, Aarts MGM, Harbinson J. 2015. Natural genetic variation for acclimation of photosynthetic light use efficiency to growth irradiance in Arabidopsis . Plant Physiology 167: 1412–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhage A, van Wees SCM, Pieterse CMJ. 2010. Plant immunity: it's the hormones talking, but what do they say? Plant Physiology 154: 536–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues PE, Lasky JR, Juenger TE, Liu TW, Kumar MN. 2014. Genome‐wide association mapping combined with reverse genetics identifies new effectors of low water potential‐induced proline accumulation in Arabidopsis . Plant Physiology 164: 144–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vile D, Pervent M, Belluau M, Vasseur F, Bresson J, Muller B, Granier C, Simonneau T. 2012. Arabidopsis growth under prolonged high temperature and water deficit: independent or interactive effects? Plant, Cell & Environment 35: 702–718. [DOI] [PubMed] [Google Scholar]
- Voelckel C, Baldwin IT. 2004. Herbivore‐induced plant vaccination. Part II. Array‐studies reveal the transience of herbivore‐specific transcriptional imprints and a distinct imprint from stress combinations. Plant Journal 38: 650–663. [DOI] [PubMed] [Google Scholar]
- Vos IA, Pieterse CMJ, van Wees SCM. 2013. Costs and benefits of hormone‐regulated plant defences. Plant Pathology 62: 43–55. [Google Scholar]
- Weigel D. 2012. Natural variation in Arabidopsis: from molecular genetics to ecological genomics. Plant Physiology 158: 2–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheat CW, Vogel H, Wittstock U, Braby MF, Underwood D, Mitchell‐Olds T. 2007. The genetic basis of a plant–insect coevolutionary key innovation. Proceedings of the National Academy of Sciences, USA 104: 20427–20431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray R, Visscher P. 2008. Estimating trait heritability. Nature Education 1: 29. [Google Scholar]
- Zalucki MP, Shabbir A, Silva R, Adamson D, Liu SS, Furlong MJ. 2012. Estimating the economic cost of one of the world's major insect pests, Plutella xylostella (Lepidoptera: Plutellidae): just how long is a piece of string? Journal of Economic Entomology 105: 1115–1129. [DOI] [PubMed] [Google Scholar]
- Zhang ZY, Ober JA, Kliebenstein DJ. 2006. The gene controlling the quantitative trait locus EPITHIOSPECIFIER MODIFIER1 alters glucosinolate hydrolysis and insect resistance in Arabidopsis . Plant Cell 18: 1524–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu CS, Gore M, Buckler ES, Yu JM. 2008. Status and prospects of association mapping in plants. Plant Genome 1: 5–20. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Time scheme of treatments.
Fig. S2 Single nucleotide polymorphisms (SNPs) associated with reduction in Arabidopsis thaliana rosette fresh weight in response to drought.
Fig. S3 Single nucleotide polymorphisms (SNPs) associated with reduction in Arabidopsis thaliana rosette fresh weight in response to Pieris rapae.
Fig. S4 Single nucleotide polymorphisms (SNPs) associated with reduction in Arabidopsis thaliana rosette fresh weight in response to Plutella xylostella.
Fig. S5 Single nucleotide polymorphisms (SNPs) associated with reduction in Arabidopsis thaliana rosette fresh weight in response to drought plus Pieris rapae.
Fig. S6 Single nucleotide polymorphisms (SNPs) associated with reduction in Arabidopsis thaliana rosette fresh weight in response to Botrytis cinerea plus Pieris rapae.
Table S1 Percentage reduction in rosette fresh weight in Arabidopsis thaliana on stress exposure
Table S2 Candidate genes for reduction in Arabidopsis thaliana rosette fresh weight in response to drought
Table S3 Candidate genes for reduction in A. thaliana rosette fresh weight in response to Pieris rapae
Table S4 Candidate genes for reduction in Arabidopsis thaliana rosette fresh weight in response to drought plus Pieris rapae
Table S5 Candidate genes for reduction in Arabidopsis thaliana rosette fresh weight in response to Botrytis cinerea plus Pieris rapae
Table S6 Candidate genes for reduction in Arabidopsis thaliana rosette fresh weight in response to Plutella xylostella


