Communication between astrocytes and neurons has been difficult to study because cultured astrocytes do not resemble those in vivo. Wolfes et al. develop a stellate astrocyte monoculture with physiological characteristics and find that VAMP2 and SYT7 mark distinct vesicle populations in astrocytes.
Abstract
Interactions between astrocytes and neurons rely on the release and uptake of glial and neuronal molecules. But whether astrocytic vesicles exist and exocytose in a regulated or constitutive fashion is under debate. The majority of studies have relied on indirect methods or on astrocyte cultures that do not resemble stellate astrocytes found in vivo. Here, to investigate vesicle-associated proteins and exocytosis in stellate astrocytes specifically, we developed a simple, fast, and economical method for growing stellate astrocyte monocultures. This method is superior to other monocultures in terms of astrocyte morphology, mRNA expression profile, protein expression of cell maturity markers, and Ca2+ fluctuations: In astrocytes transduced with GFAP promoter–driven Lck-GCaMP3, spontaneous Ca2+ events in distinct domains (somata, branchlets, and microdomains) are similar to those in astrocytes co-cultured with other glia and neurons but unlike Ca2+ events in astrocytes prepared using the McCarthy and de Vellis (MD) method and immunopanned (IP) astrocytes. We identify two distinct populations of constitutively recycling vesicles (harboring either VAMP2 or SYT7) specifically in branchlets of cultured stellate astrocytes. SYT7 is developmentally regulated in these astrocytes, and we observe significantly fewer synapses in wild-type mouse neurons grown on Syt7−/− astrocytes. SYT7 may thus be involved in trafficking or releasing synaptogenic factors. In summary, our novel method yields stellate astrocyte monocultures that can be used to study Ca2+ signaling and vesicle recycling and dynamics in astrocytic processes.
Introduction
Astrocytes fine-tune synaptic transmission and plasticity through Ca2+ signaling and releasing molecules, e.g., via vesicular exocytosis. To mediate vesicular release, astrocytes express vesicle-associated proteins similar to those in neurons (Hamilton and Attwell, 2010; Schubert et al., 2011). These proteins can be targeted in astrocytes alone by using astrocyte monocultures, a simple approach that is easier to manipulate and visualize than astrocytes in brain slices or in vivo and facilitates studying cells from neonatal lethal mutants. The approaches currently used include growing MD astrocytes (named after the protocol’s authors McCarthy and de Vellis [1980]), immunopanned (IP) astrocytes (Foo et al., 2011), and induced pluripotent stem cell (iPSC)–derived astrocytes (Table 1; Krencik and Zhang, 2011). However, these protocols either yield polygonal astrocytes (e.g., MD astrocytes) unlike those in vivo or are lengthy and require many ingredients (e.g., IP and iPSC-derived astrocytes).
Table 1. Common and recently developed astrocyte monoculture protocols.
| Culture method | Cell source | Advantages | Potential disadvantages |
|---|---|---|---|
| MD: McCarthy and de Vellis (1980) | perinatal (often P0–2) | simple, quick, cheap, single optical plane (useful for studying exo-/endocytosis) | polygonal morphology, protein expression profile unlike freshly isolated astrocytes (Foo et al., 2011), uses serum |
| IP: Foo et al. (2011) | P0–14 | stellate morphology, gene expression like freshly isolated astrocytes, serum free | lengthy protocol, requires many reagents, uses one antibody to isolate astrocytes (may only target a subpopulation) |
| iPSC derived: Krencik and Zhang (2011) | iPSCs | human astrocytes, can use cells from disease-specific tissue, maintain for months, serum free | takes >3 mo to prepare (and longer to mature) |
| 3-D matrix: Puschmann et al. (2013) and Placone et al. (2015)* | P1–3 | stellate, 3-D morphology, serum free*, murine/human | requires 3-D matrices |
Comparison of three recently published protocols for growing astrocyte monocultures and the commonly used MD protocol, including advantages and potential disadvantages of each protocol, the cell source, and original references. In light of techniques that require single optical planes, we considered 3-D morphology a disadvantage.
A faster way to culture stellate astrocytes is using 3-D matrices (Puschmann et al., 2013; Placone et al., 2015). Yet, the medium must be carefully chosen, as the high concentration of heparin-binding epidermal growth factor (HBEGF) used by Puschmann et al. (2014) was later shown to de-differentiate astrocytes. Moreover, some approaches require 2-D culture systems, e.g., when imaging Ca2+ events within single cells or analyzing vesicle fusion at the cell surface using pHluorin reporters.
Prior to 3-D matrix studies, the MD protocol was criticized not only for generating polygonal astrocytes, but also for the use of serum, which has high batch variability and is not well defined. Thus, Morita et al. (2003) first used serum-free media for growing astrocytes with thin processes and glutamate-inducible, but no spontaneous Ca2+ fluctuations—although spontaneous Ca2+ fluctuations occur in brain slices and in vivo (Nett et al., 2002; Shigetomi et al., 2013a; Srinivasan et al., 2015). Moreover, some of the added growth factors (i.e., EGF and TGFα) can induce reactive astrocytes (Tsugane et al., 2007), so that a more defined medium with fewer growth factors (as described below) may be more applicable.
To analyze vesicle-associated proteins and their recycling in astrocyte monocultures (that have a more stellate in vivo–like morphology and allow imaging of single optical planes), we developed a simple protocol with a defined medium composition. This novel protocol (termed AWESAM: a low-cost easy stellate astrocyte method) was superior to others tested in terms of ease of preparation, in vivo–like morphology, and spontaneous Ca2+ signaling profiles.
Considering the thin astrocytic processes that contact neurons in vivo, we hypothesized that the thin processes of stellate astrocytes in vitro would similarly represent sites of distinct Ca2+ signaling and vesicle turnover. Using our novel method, we found two distinct constitutively recycling vesicle populations in astrocytes marked by VAMP2 (a regulator of vesicle exocytosis) or the Ca2+ sensor synaptotagmin 7 (SYT7), where SYT7 expression in astrocytes was necessary for normal synapse number in developing neural networks.
Materials and methods
Ethical approval
All animal experiments were performed in accordance with the guidelines for German animal welfare. All animals were euthanized by CO2. Chemicals were from Carl Roth (Germany) unless otherwise indicated.
Cell culture
Because neurons co-cultured with glia (referred to as co-cultures) are best prepared from embryonic day (E) 18–19 tissue, we chose to prepare the majority of astrocytic monocultures from E19 animals rather than postnatal day (P) 0–4 tissue (Martineau et al., 2008; Malarkey and Parpura, 2011) to allow direct comparison with co-cultures. We prepared IP astrocytes from P8 animals as suggested by the authors of the immunopanning protocol (Foo et al., 2011; Foo, 2013) but also compared P8, P4, and P0 preparations.
Co-cultures of neurons and glia, MD, and AWESAM astrocyte monocultures were prepared from E19 Wistar rat pups by standard techniques (Dichter, 1978; McCarthy and de Vellis, 1980) with minor modifications. Pregnant rats were euthanized by CO2, pups were removed from the uterus and decapitated, and cortices were dissected in 4°C dissection medium (10 mM HEPES [cat. no. 15630080; Gibco] in Hank’s balanced salt solution [cat. no. 14170112; Gibco]). Cortices were cut into small pieces and incubated in 0.05% trypsin-EDTA (cat. no. 25200056; Gibco) in a 37°C water bath. After 20 min, the tissue was washed in 4°C dissection medium three times, after which the tissue was triturated in 1 ml NB+ (2% B27 supplement [cat. no. 17504044; Gibco], 2 mM GlutaMAX [cat. no. 35050061; Gibco], and 5,000 U/ml penicillin and 5,000 µg/ml streptomycin [cat. no. 15070063; Gibco] in neurobasal medium [cat. no. 21103049; Gibco]). The cell suspension was then filtered through a 100-µm nylon cell strainer (cat. no. 352360; BD), prewet with 4.5 ml NB+. Next, another 4.5 ml NB+ was added to wash out cells stuck to the cell strainer. Using the Trypan blue (cat. no. T8154; Sigma-Aldrich) exclusion technique, cells were counted and plated on 0.04% polyethylenimine (PEI)–coated cell culture dishes preequilibrated with medium in a Hera Cell 240i cell culture incubator (Thermo Fisher Scientific) at 37°C and 5% CO2. Co-cultures were plated at 50,000 cells/well on 12-mm glass coverslips preequilibrated with NB+ in the incubator (or 2 × 106 cells/dish for generating immunoblot samples).
For MD and AWESAM astrocyte monocultures, cells were plated in preequilibrated DMEM+ (10% FCS [cat. no. 10437028; Gibco] and 5,000 U/ml penicillin and 5,000 µg/ml streptomycin [cat. no. 15070063; Gibco] in DMEM [cat. no. 41966029; Gibco]) at 500,000 cells/10 cm dish. On day in vitro (DIV) 7, the dishes were placed on a shaker (Heidolph Rotamax 120) in the incubator and shaken at 110 rpm for 6 h. After shaking, the medium (including loosened neurons and nonastrocytic glia) was exchanged with preheated 1× PBS. PBS was then replaced by 37°C 0.25% trypsin-EDTA (except for samples for Western blots, for which fresh preequilibrated medium was added instead of trypsin-EDTA; such cultures were then maintained in the incubator). After incubating the cultures with trypsin-EDTA in the incubator for 5 min, 5 ml DMEM+ was added to inactivate the trypsin, and cells were loosened by forceful pipetting, and, in a fresh tube, centrifuged in an Eppendorf centrifuge 5810 R at 3,220 g for 4 min at 20°C. The supernatant was then removed, and the pellet was resuspended in 1 ml DMEM+ for MD astrocytes or NB+ containing 5 ng/ml HBEGF (cat. no. 4643; Sigma-Aldrich) for AWESAM astrocytes. Cells were plated at 10,000 (MD) or 5,000 (AWESAM) cells/well on 0.04% PEI–coated, 12-mm glass coverslips. Cultures were then maintained in the incubator, and half the medium was exchanged once a week.
Astrocyte monocultures were also generated by immunopanning (Foo et al., 2011), according to the recently published protocol (Foo, 2013) with minor modifications: All 4°C steps described here were performed at room temperature in the original protocol (Foo, 2013), but we found that cooling cells (and some solutions) improved cell viability. The following solutions were prepared before immunopanning.
The following compounds were from Sigma-Aldrich: BSA (cat. no. A8806), 10× EBSS (cat. no. E7510), N-acetyl cysteine (cat. no. A8199), progesterone (cat. no. P8783), putrescine (cat. no. P5780), sodium selenite (cat. no. S5261), and transferrin (cat. no. T-1147). Trypsin inhibitor was from Worthington Biochemical Corporation (cat. no. LS003086).
Enzyme stock solution (200 ml) contained the following: 20 ml of 10× EBSS (final concentration 1×), 2.4 ml of 30% D(+)-glucose (final concentration 0.46%), 5.2 ml of 1 M NaHCO3 (final concentration 26 mM), 2 ml of 50 mM EDTA (final concentration 0.5 mM), and 170.4 ml ddH2O; brought to 200 ml with ddH2O and filtered through 0.22-µm filter and stored at 4°C.
Inhibitor stock solution (500 ml) contained the following: 50 ml of 10× EBSS (final concentration 1×), 6 ml of 30% D(+)-glucose (final concentration 0.46%), 13 ml of 1 M NaHCO3 (final concentration 26 mM), and 431 ml ddH2O; brought to 500 ml with ddH2O and filtered through 0.22-µm filter and kept at 4°C.
10× low ovomucoid (200 ml) contained the following: 150 ml dPBS, 3 g BSA, and 3 g trypsin inhibitor, pH 7.4; brought to 200 ml with dPBS and filtered; aliquots were stored at −20°C.
10× high ovomucoid (200 ml) contained the following: 150 ml dPBS, 6 g BSA, and 6 g trypsin inhibitor, pH 7.4; brought to 200 ml with dPBS and filtered; aliquots were stored at −20°C.
100× Sato (50 ml) contained the following: 50 ml neurobasal, 500 mg transferrin (final concentration 100 µg/ml), 500 mg BSA (final concentration 100 µg/ml), 80 mg putrescine (final concentration 16 µg/ml), progesterone 12.5 µl of 2.5 mg/100 µl EtOH stock (final concentration 60 ng/ml; 0.2 µM), and sodium selenite 500 µl of 4 mg + 10 µl 1 N NaOH in 10 ml neurobasal (final concentration 40 ng/ml); filtered through pre-rinsed 0.22-µm filters; aliquots were stored at −20°C.
IP astrocyte base medium (300 ml) contained the following: 145.5 ml of 50% neurobasal, 145.5 ml of 50% DMEM, 6 ml P/S stock of 100 U penicillin and 100 µg/ml streptomycin, 1 mM sodium pyruvate (already included in medium), 292 µg/ml l-glutamine (already included in medium), 3 ml of 100× stock of 1× Sato, and 1.5 mg of 5 µg/ml N-acetyl cysteine.
30% FCS in 50/50 mixture of DMEM and neurobasal contained the following: 30% FCS in 50% DMEM/50% neurobasal; filtered through 0.22-µm filter.
Immunopanning dishes were prepared by coating each of six 15-cm Petri dishes with 25 ml of 50 mM Tris-HCl, pH 9.5 (sterilized), per dish and the following secondary antibodies at 4°C overnight. 1× secondary only: 60 µl goat anti–mouse IgG + IgM (H+L; 115-005-044; Jackson ImmunoResearch Laboratories, Inc.); 1× BSL-1: 20 µl of 20 mg/ml BSL-1 (cat. no. L-1100; Vector Laboratories); 1× CD45: 60 µl goat anti–rat IgG H+L (112-005-167; Jackson ImmunoResearch Laboratories, Inc.); 2× O4: 60 µl goat anti–mouse IgM µ-chain specific (115-005-020; Jackson ImmunoResearch Laboratories, Inc.); and 1× ITGB5: 60 µl of 60 µl goat anti–mouse IgG + IgM (H+L).
The next day, 22 ml enzyme stock solution was bubbled with CO2 until the solution turned from red to orange; the solution was then kept in a 34°C water bath. Similarly, 2 × 21 ml and 1 × 10 ml inhibitor stock solutions were bubbled but then kept at room temperature. 1.5 ml of 10× low Ovo and 100 µl of 0.4% DNase (cat. no. LS002007; Worthington Biochemical Corporation) were added to each of the two 21-ml aliquots of bubbled inhibitor stock (low Ovo), whereas 2 ml of 10× high Ovo and 20 µl of 0.4% DNase were added to the 10-ml aliquot of bubbled inhibitor stock solution (high Ovo). 70 ml of 0.2% and 40 ml of 0.02% BSA were prepared in 1× PBS and kept at 4°C. Except for the “BSL-1” dish, all 15-cm dishes were washed with 1× PBS three times, after which PBS was replaced with 12 ml of 0.2% BSA for the “secondary only” dish, 12 ml of 0.2% BSA including 20 µl anti–mouse CD45 (550539; BD) for the “CD45” dish, 12 ml of 0.2% BSA including 20 µl anti-O4 (MAB345; EMD Millipore) for each “O4” dish, and 12 ml of 0.2% BSA including 20 µl anti–human integrin β 5 (14-0497-82; eBioscience) for the “ITGB5” dish. The dishes were then incubated at room temperature for ≥2 h.
P7 rat pups were decapitated, and cortices were dissected in 4°C 1× PBS under a light microscope (Olympus) in a semi-sterile dissection hood. 20 min before the end of dissection, 100 U papain (cat. no. LS 03126; Worthington Biochemical Corporation) and 3.6–4.2 mg l-cysteine hydrochloride monohydrate (cat. no. C7880; Sigma-Aldrich) were mixed in 34°C enzyme stock solution. Cortices were transferred into small drops of 4°C 1× PBS and cut into ≈1-mm3 pieces with a no. 10 scalpel blade.
For cell dissociation, the enzyme solution (including papain and l-cysteine) was filtered through a 0.22-µm filter (cat. no. SLGV013SL; EMD Millipore) and added to the brain pieces alongside 100 µl of 0.4% DNase. Brain pieces in enzyme solution were bubbled with CO2 on a 34°C heat block (and shaken every 10 min). After 40 min, digested brain pieces were collected in a tube and washed five times with 4.5 ml low Ovo solution. After the last wash, 4 ml low Ovo solution was added, and the brain pieces were triturated by pipetting. Single cells at the top of the suspension (brain pieces sank to the bottom) were collected in a separate tube with 4 ml low Ovo solution using a 1 ml pipette. Trituration steps were repeated (adding 4.5 ml low Ovo solution every time) until the majority of brain pieces were gone. Next, 12 ml high Ovo solution was carefully layered beneath the single cell suspension, after which the tube with the cell suspension/high Ovo biphasic mix was centrifuged at 110 g at 4°C for 5 min. The supernatant was then discarded, and the cell pellet was resuspended in 9 ml of 4°C 0.02% BSA. This cell suspension was then filtered into a 50-ml tube through an autoclaved 30-µm Nitex mesh, after which the filter was washed with 3 ml of 4°C 0.02% BSA. The tube with the cells was then left in a 37°C water bath for 45 min.
After the dissociation, immunopanning steps were performed at room temperature. The 15-cm dishes prepared earlier were washed with 1× PBS three times, after which the cell suspension was passed through the following series of immunopanning steps, for which the dishes were shaken at the half-way point of each incubation: (a) “secondary only” dish for 10 min, (b) “BSL-1” dish for 10 min, (c) “CD45” dish for 20 min, (d) one “O4” dish for 15 min, (e) the second “O4” dish for 15 min, and (f) “ITGB5” dish for 40 min. The “ITGB5” dish was subsequently washed with 1× PBS five times. 200 U trypsin (cat. no. T9935; Sigma-Aldrich) was added to 8 ml of preequilibrated 1× EBSS and pipetted onto the “ITGB5” dish, which was then incubated at 37°C for 3 min. The side of the dish was tapped to dislodge and resuspend cells in 20 ml preequilibrated 30% FCS (in 1:1 neurobasal/DMEM). The cells were then added to a fresh tube with 200 µl of 0.4% DNase and centrifuged at 170 g at 4°C for 11 min. The supernatant was discarded, and the cell pellet was resuspended in 4°C 0.02% BSA. Cells were counted using the Trypan blue exclusion method and plated at 30,000 cells/well on 12-mm 0.04% PEI–coated coverslips or (for Western blot samples) at 2 × 106 cells/10-cm dish in preequilibrated IP astrocyte medium (IPm: 5 ng/ml HBEGF in 1:1 neurobasal/DMEM). IP astrocyte cultures were maintained in the incubator, and half the medium was exchanged once a week.
Immunocytochemistry
Cultures were fixed in 4% paraformaldehyde in 0.1 M phosphate buffer for 30 min on DIV14, 16, or 21 and then washed in 1× PBS 5 × 3 min. Samples were then incubated in buffer D (2% donkey serum, 0.1% Triton X-100, and 0.05% NaN3 in 2× PBS) for 30 min and then treated with primary antibodies diluted in buffer D at 4°C overnight. The next day, samples were washed in 1× PBS for 5 × 3 min and subsequently treated with secondary antibodies diluted in buffer D at room temperature in the dark for 2 h. During secondary antibody incubation, phalloidin Att390 (cat. no. 50556; Sigma-Aldrich) or phalloidin Alexa Fluor 647 (cat. no. A-12379; Invitrogen) was applied to some samples according to the manufacturer’s suggestions to label F-actin. Samples were washed in 1× PBS 5 × 3 min, and coverslips were mounted on glass slides via Fluoromount (cat. no. K048; Diagnostic BioSystems). In some cases, 10 µM DAPI (cat. no. D1306; Invitrogen) was added to the mounting reagent.
The following primary antibodies were used. From Abcam: mouse anti-CD11B (ab8879) diluted 1:200, mouse anti-GFP (ab1218) diluted 1:2,000, rabbit anti-GFP (ab290) diluted 1:2,000, chick anti-GFP (ab13970) diluted 1:2,000, chick anti–myelin basic protein (MBP; ab106583) diluted 1:2,000, rabbit anti-RAB7 (ab74906) diluted 1:500, mouse anti-S100β (ab4066) diluted 1:200, rabbit anti-SYT17 (ab76274) diluted 1:1,000, and mouse anti–βIII-tubulin (ab78078) diluted 1:100. From Biosensis: chick anti-MAP2 (C-1382-50) diluted 1:5,000. From Enzo Life Sciences: mouse anti-GAPDH (11101127) diluted 1:1,000 and mouse anti-LAMP1 (ADI-VAM-EN-001_D) diluted 1:100. From EMD Millipore: mouse anti-synaptophysin (SYP; mab368) diluted 1:2,000. From NeuroMab: mouse anti-ALDH1L1 (75164 and 75140) diluted 1:500. From Synaptic Systems: rabbit anti-EEA1 (237002) diluted 1:500, guinea pig anti-GFAP (173005) diluted 1:1,000, mouse anti-nestin (312011) diluted 1:1,000, mouse anti-RAB3A (107111) diluted 1:2,000, mouse anti-SNAP25 (111011) diluted 1:1,000, guinea pig anti-SYP (101004) diluted 1:1,000, mouse anti-SYT1 (105011) diluted 1:1,000, guinea pig anti-SYT1 (105105) diluted 1:500 for antibody internalization assay, rabbit anti-SYT3 (105033) diluted 1:1,000, rabbit anti-SYT4 (105143) diluted 1:1,000, rabbit anti-SYT5 (105053) diluted 1:1,000, rabbit anti-SYT7 (105173) diluted 1:200, rabbit anti-SYT11 (270003) diluted 1:1,000, mouse anti-syntaxin1 (110011) diluted 1:2,000, mouse anti-VAMP2 (104211) diluted 1:4,000, rabbit anti-VAMP2 (104202) diluted 1:1,000, guinea pig anti-VAMP2 (104204) diluted 1:500, rabbit anti-VAMP4 (136002) diluted 1:100, rabbit anti–vesicular glutamate transporter (VGLUT) isoform 1 (VGLUT1; 135303) diluted 1:1,000, guinea pig anti-VGLUT2 (135404) diluted 1:1,000, rabbit anti-VGLUT3 (135203) diluted 1:2,000, and rabbit anti-vimentin (172002) diluted 1:1,000.
The following secondary antibodies were used. From Abcam (all HRP coupled): goat anti–guinea pig IgG (ab97155) diluted 1:5,000 and rabbit anti–chicken IgY (ab97140) diluted 1:2,000. From Bio-Rad Laboratories (all HRP coupled): goat anti–mouse IgG (1706516) diluted 1:2,000 and goat anti–rabbit IgG (1706515) diluted 1:2,000. From Invitrogen: donkey anti–mouse Alexa Fluor 488 (A-21202) diluted 1:1,000, goat anti–rabbit Alexa Fluor 488 (A-11008) diluted 1:1,000, donkey anti–mouse Alexa Fluor 546 (A-10036) diluted 1:2,000, goat anti–rabbit Alexa Fluor 546 (A-11010) diluted 1:2,000, goat anti–guinea pig Alexa Fluor 546 (A-11074) diluted 1:2,000, goat anti–chick Alexa Fluor 546 (A-11040) diluted 1:2,000, donkey anti–mouse Alexa Fluor 647 (A-31571) diluted 1:1,000, goat anti–guinea pig Alexa Fluor 647 (A-21450) diluted 1:1,000, and goat anti–chick Alexa Fluor 647 (A-21449) diluted 1:1,000.
RNA extraction, deep sequencing, and RNA-Seq data processing
RNA-Seq experiments were performed as previously described (Centeno et al., 2016). In brief, RNA was isolated from DIV7 cultures versus DIV14 MD and AWESAM cultures in triplicates using Tri reagent (Sigma-Aldrich) according to the manufacturer’s protocol. RNA-Seq libraries were prepared using the TruSeq RNA Sample Preparation v2 kit (Illumina). The library quality was checked using an Agilent 2100 Bioanalyzer (Agilent Technologies). The sample concentration was measured by a Qubit dsDNA HS Assay kit and adjusted to 2 nM before sequencing (50 bp) on a HiSeq 2000 sequencer (Illumina) using TruSeq SR Cluster kit v3-cBot-HS and TruSeq SBS kit v3-HS according to the manufacturer’s instructions.
Counts were obtained as described previously (Centeno et al., 2016). In brief, RNA-Seq reads were aligned to the Rn4 genome using bowtie2 v2.0.2 and then subsequently converted to .sam and .bam files via SAMtools v0.1.18 with the Nov. 2004 (Baylor 3.4/rn4) UCSC annotation. Gene counts were generated using featureCounts v1.4.6. For further analysis, counts of biological replicate samples were averaged and rounded. Cell type–specific gene expression information was obtained from the Brain RNA-Seq Database (Zhang et al., 2014), filtering for genes that are preferentially expressed in astrocytes, neurons, oligodendrocytes, microglia, and endothelial cells. Specifically, to construct a list of cell type–enriched genes, we filtered for the top 300 most expressed genes that are differentially expressed between each cell type of interest and all other cell types (e.g., astrocytic gene expression vs. gene expression in all other cell types combined). We next converted the cell type–specific gene lists from mouse gene IDs to rat identifiers using Rnor_6.0 from Ensembl-BioMart’s mart-view (Yates et al., 2016). The cell type–specific gene expression data were measured in FPKM values.
Fluorescein in situ apoptosis detection
For the apoptosis test comparing IP and NB+H astrocytes grown in HBEGF-containing with HBEGF-free media, the ApopTag Fluorescein In Situ Apoptosis Detection kit (cat. no. S7110; EMD Millipore) was used according to the manufacturer’s recommendations.
Immunoblotting
For Western blots, cell culture samples were harvested on DIV9, 14, and 21 by washing cultures with 1× PBS, replacing PBS with 500 µl of fresh 1× PBS and then detaching cells using a cell scraper (cat. no. CC76000220; CytoOne). This cell suspension was then triturated five times with a 27-gauge needle (cat. no. 4710004020; HSW Fine-ject) and centrifuged at 400 g at 4°C for 10 min. The resulting supernatant was centrifuged as before in a fresh tube, and the protein concentration of the final supernatant was determined via the BCA protein assay kit (cat. no. 712853; EMD Millipore).
Brain lysate was prepared from adult C57B/6J mice. Isolated brains were dissected in homogenization buffer (320 mM sucrose, 4 mM HEPES-KOH, pH 7.4) at 4°C and homogenized in 500 µl homogenization buffer in 1.5-ml tubes on ice at 900 rpm in a laboratory stirrer (VWR VOS 14 S40). The homogenized brain lysate was centrifuged at 1,000 g at 4°C for 10 min, and the supernatant was snap-frozen in liquid nitrogen for storage at −80°C.
For gel electrophoresis, 1.5-mm-thick 10% acrylamide running gels (including 5% stacking gels) were run in anode buffer (1 M Tris in dH2O, pH 8.9) in the outer chamber and cathode buffer (1 M Tris, 1 M Tricin, and 1% SDS in dH2O) in the inner cassette. 5 µg protein/lane was loaded, as well as the PageRuler molecular weight marker (cat. no. SM0671; Fermentas), and the gel was run at 60 V, switching to 100 V after 15 min. After 1 h, the gel, fiber pads, nitrocellulose membranes, and filter papers were equilibrated in blotting buffer (200 mM glycine, 25 mM Tris, 0.04% SDS, and 20% methanol) for 10 min. The transfer was performed at 100 mA for 1 h, after which the membranes were blocked in 5% milk in PBS-T (0.05% Tween 20 in 1× PBS) for 1 h. Membranes were then incubated in primary antibodies in 5% milk in PBS-T at 4°C overnight. The next day, membranes were washed in 1× PBS 5 × 5 min and incubated in secondary antibodies in 5% milk in PBS-T at 4°C overnight. The next day, membranes were washed in 1× PBS 5 × 5 min and then incubated in ECL solution (1.28% [wt/vol] Tris-HCl and 0.23% [wt/vol; Luminol Na+ salt [cat. no. A4685-1g; Sigma-Aldrich] in dH2O, pH 8.6, 0.01% p-coumaric acid [cat. no. 9008-5g; Sigma-Aldrich] in DMSO, and 0.008% H2O2) for 1 min. Proteins were detected via the Fujifilm LAS-3000 imaging system (R&D Systems). For reprobing, membranes were washed in 1× PBS for 10 min and blocked in 5% milk in PBS-T for 1 h before primary antibody incubation.
Transfection and antibody internalization assays
For antibody internalization assays of transfected pHluorin-SYT7, 24-well plate cell cultures were transfected in 37°C serum-free medium. For each well, 0.7 µl Lipofectamine 2000 transfection reagent (cat. no. 11668027; Invitrogen) was prepared in 50 µl medium in one tube, and 0.7 µg plasmid DNA was prepared in 50 µl medium in a second tube and incubated at room temperature. For tetanus toxin (TeNT) experiments, 800 nM purified TeNT light chain peptides were also introduced to cells by transfection, which had previously been successful (Kuo et al., 2010); TeNT peptides were a gift from Y. Park and R. Jahn (Max Planck Institute for Biophysical Chemistry, Göttingen, Germany). After 5 min, both solutions were mixed, and the Lipofectamine/DNA mix was incubated at room temperature for 30 min.
Medium was removed (and saved for co-cultures only) before adding 400 µl of fresh preheated neurobasal medium per well. 100 µl of the Lipofectamine/DNA mix was added per well and incubated at 37°C and 5% CO2 for 2 h, after which medium was removed and cells were washed once in fresh preheated medium. Subsequently, saved medium was added to co-cultures, whereas fresh preheated NB+H was added to AWESAM cultures. Transfection efficiency was ≈10% in co-cultures and ≈20% in astrocyte monocultures.
For the antibody internalization assay, cells that had been transfected 2 d before were moved to new wells containing anti-GFP antibodies (1:500) in preequilibrated astrocyte-stimulating solution: 50 µM glutamate, 10 mM KCl, 135 mM NaCl, 2 mM CaCl2, 5.5 mM glucose, 2 mM MgCl2, and 20 mM HEPES; or neuron-stimulating solution: 50 mM KCl, 95 mM NaCl, 2 mM CaCl2, 5.5 mM glucose, 2 mM MgCl2, and 20 mM HEPES (for Fig. 7 B [control] and Fig. 8 C); or nonstimulating solution: 2 mM KCl, 143 mM NaCl, 2 mM CaCl2, 5.5 mM glucose, 2 mM MgCl2, and 20 mM HEPES. Cultures were kept in either solution in the incubator for 15 min and then fixed as described for immunostaining. The same assay was also performed on untransfected co-cultures, to which anti-SYT1 antibodies (diluted 1:500) were added in preheated stimulating solution for neurons.
Figure 7.
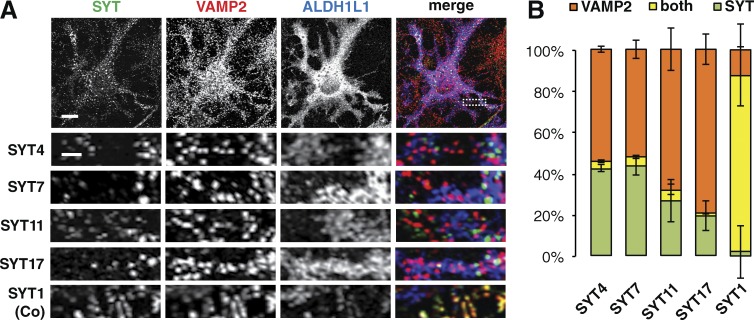
SYT isoforms are not located on VAMP2-harboring vesicles in astrocytes. (A) Immunocytochemistry showing that SYT isoforms present in astrocytes, SYT4, 7, 11, and 17, do not colocalize with VAMP2 in DIV16 AWESAM astrocytes (marked by ALDH1L1), whereas SYT1 colocalizes with VAMP2 in neurons of DIV14 co-cultures (Co), as a positive control. Higher-magnification images of the boxed area (outlined by a dashed line) for SYT4 are shown below each panel. Images per experiment: n = 7 (SYT4), 8 (SYT7), 6 (SYT11), 7 (SYT17), and 4 (SYT1). Bars: (top panels) 10 µm; (zoomed-in bottom panels) 2 µm. (B) Bar graphs showing relative amounts of SYT4, 7, 11, or 17 and VAMP2 puncta, and puncta positive for both VAMP2 and the indicated SYT isoform. Error bars indicate SEM.
Figure 8.
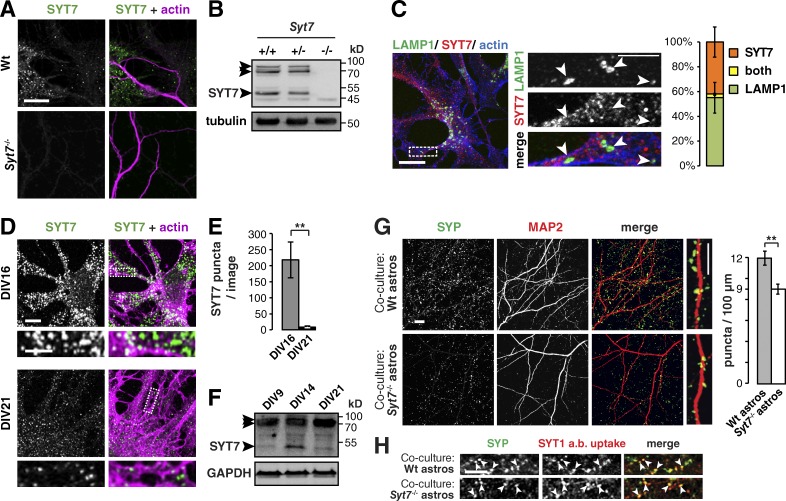
When astrocytes lack SYT7, neurons have fewer synapses. (A) Immunocytochemistry of DIV14 wild-type (Wt) and Syt7−/− co-cultures stained for actin and SYT7, where SYT7 puncta appear only in wild-type samples. Images per condition: n = 12. (B) Immunoblot showing three SYT7 splice isoforms at 47, 70, and ≈80 kD (arrowheads) in Syt7+/+ and Syt7+/− but not Syt7−/− adult brain lysate. Tubulin served as a loading control. (C) Immunocytochemistry and bar graph showing that the lysosomal protein LAMP1 (arrowheads) and SYT7 do not colocalize in DIV16 AWESAM astrocytes. The bar graph shows relative amounts of LAMP1 and SYT7 puncta and puncta positive for both (n = 6 images). (D) Immunocytochemistry comparing SYT7 expression levels in DIV16 and DIV21 AWESAM astrocytes (n = 5 images for DIV16, n = 3 images for DIV21). (C and D) Higher-magnification images of the boxed area (outlined by a dashed line) are shown in right (C) and bottom (D) panels. (E) Bar graph showing that astrocytes contain significantly more SYT7 puncta on DIV16 than DIV21. (F) Immunoblot showing that SYT7 expression is up-regulated in DIV14 AWESAM astrocytes versus DIV9 and DIV21. GAPDH served as a loading control. (G) Immunocytochemistry showing SYP puncta on dendrites (MAP2) of wild-type neurons grown on either wild-type astrocytes or Syt7−/− astrocytes. The bar graph shows that wild-type neurons have significantly fewer SYP puncta per 100-µm dendrite when grown on Syt7−/− astrocytes instead of wild-type astrocytes (n = 12 images for wild type and 14 images for Syt7−/−). (H) Antibody internalization assay showing that anti-SYT1 antibodies taken up during stimulation with 50 mM KCl solution colocalize with SYP (arrowheads) in DIV14 neurons grown on wild-type or Syt7−/− astrocytes (n = 6). Error bars indicate SEM, where **, P < 0.01 by unpaired two-tailed Student’s t test. Bars: (large view panels) 10 µm; (zoomed-in panels) 5 µm.
Generation of Lck-GCaMP3 adeno-associated virus (AAV) and viral transduction
Recombinant chimeric AAV1/2 particles were generated by transfecting HEK293 cells with a 1:1:2:4 ratio of plasmids for AAV1 (pH21) and AAV2 (pRV1) cap and rep sequences, the adenovirus helper-plasmid (pFΔ6), and the vector plasmid pZac2.1 gfaABC1D-Lck-GCaMP3 (a gift from B. Khakh, University of California, Los Angeles, Los Angeles, CA; Addgene plasmid #44330) using calcium phosphate transfection. 48–72 h after transfection, cells were harvested in 1× PBS and lysed by three times repeated freezing (−80°C) and thawing (37°C). Cell debris was removed by centrifuging at 10,000 g for 5 min. Crude AAV supernatant was transferred to a 1.5-ml tube and stored at 4°C. Before transduction, half the culture medium was exchanged with fresh preheated medium. Cultures were subsequently transduced with 0.3 µl AAV particles/well of a 24-well plate on DIV7–9, and half of the medium was exchanged 7 d later. Infected cultures were used for Ca2+ imaging 10 d after transduction. Cell health after transduction was monitored by assessing process integrity by brightfield imaging before Ca2+ imaging with fluorescence. IP astrocytes appeared less robust than other culture types and exhibited deteriorating health in 50% of the cases.
Microscopy
Fixed samples and GCaMP3-infected, live cell cultures were imaged using a LSM710 confocal microscope (ZEISS), equipped with 405-, 488-, 561-, and 633-nm lasers. For Ca2+ imaging, coverslips were placed in a low profile open bath chamber (cat. no. RC-40LP; Warner Instruments), and immediately placed on an LSM710 confocal microscope for imaging, before which the imaging chamber had been filled with preheated nonstimulating solution (as used in the antibody internalization assay). Spontaneous Ca2+ signaling (as indicated by GCaMP3 fluorescence increases) was monitored for 200 s at room temperature, and cells were only imaged for a 30-min period after removal from the incubator to ensure cell health (some cells became unhealthy, as evident from inclusion bodies that appeared after >45-min periods). Images were obtained using Zen Black software. Within the same experiment, image acquisition, light intensity, electron multiplication (EM gain), exposure time, and acquisition rate (1 Hz) were kept the same. Plan-Apochromat 20×/0.8, 40×/1.3 oil DIC, and 63×/1.4 oil DIC objectives (ZEISS) were used for fixed imaging. For live cell Ca2+ imaging, a 20×/1.0 DIC objective (WPlan-Apochromat; ZEISS) was used.
Colocalization and Ca2+ imaging analysis
Colocalization analysis of fixed samples was achieved using MetaMorph Microscopy Automation and Image Analysis software. SYP-positive puncta per µm dendrite were counted manually in a blinded fashion using ImageJ/Fiji software (National Institutes of Health).
Ca2+ events of GCaMP3 signal in astrocytes were analyzed using the ImageJ/Fiji plugin GECIquant (Srinivasan et al., 2015) to define soma, branchlet, and microdomain regions, within which Ca2+ fluctuations were quantified. A custom-written MATLAB code was used to exclude all regions of interest with >10% total frames of 0 fluorescence (in at least two consecutive frames) and to plot Ca2+ fluctuations within each region from the absolute fluorescence values output by GECIquant, as dF/F. For somatic events in MD and IP astrocytes (which were slow rising and infrequent), the baseline fluorescence was calculated as the mean of 15 frames before and 15 frames after the minimum fluorescence value (i.e., mean of 30 frames total). An event was then defined as having at least 30 frames above 3.5× RMS (root mean square) noise. The event amplitude was calculated as the mean of 30 frames around the maximum value. For somatic events in co-culture and AWESAM astrocytes, the mean of the 75th percentile (mean of 75% of the total frames with the lowest fluorescence values) was used to calculate the baseline; an event was defined as fluorescence values above a threshold of 3.5× RMS noise, where single points above the threshold or the maximum value of a cluster of points above the threshold was used to define amplitude and half-width. Branchlet events were defined as for the soma in each condition, except in co-culture and AWESAM cultures, where events were defined as being above the 92nd percentile of all events (to exclude small “events” that resembled noise upon visual inspection). The baseline for microdomain events was defined as the mean of all fluorescence values in each trace (because microdomain events were shorter lived and lower in amplitude than somatic and branchlet events, the mean of all fluorescence values best approximated baseline). Microdomain events were then defined as above for branchlet events. Peak half-width was defined as the duration of peak width at half-maximal amplitude.
Results
A specific medium composition leads to stellate astrocyte morphology
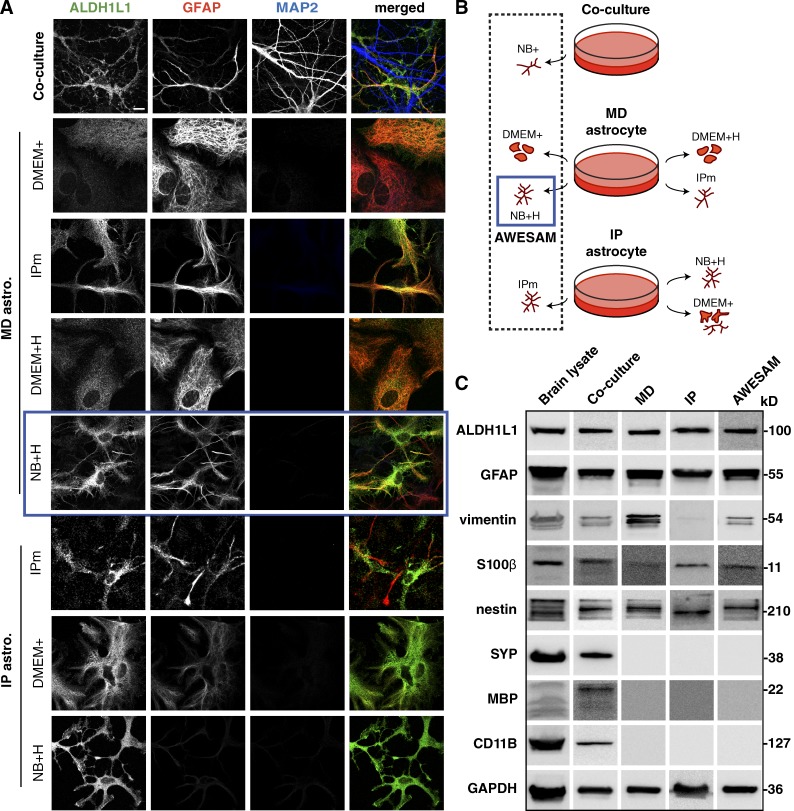
To analyze the role of specific vesicle-associated proteins in astrocytes, we first established a protocol for growing stellate astrocyte monocultures in a defined medium in the absence of neurons. First, we assessed astrocyte morphology in three culture types: co-cultures, MD, and IP astrocytes (Fig. 1 A), where we tested different media compositions (Fig. 1 B). Defined medium containing HBEGF was previously shown to promote astrocytic Ca2+ oscillations (Morita et al., 2003) and a stellate morphology in astrocytes obtained by immunopanning (Foo et al., 2011). We therefore tested whether HBEGF alone might promote a stellate morphology in astrocytes. Using the astrocyte-specific markers ALDH1L1 and GFAP (expressed in the majority of astrocytes), we found that astrocytes grown in DMEM with serum (DMEM+) adopted a polygonal morphology regardless of the astrocyte isolation method used. Contrary to our hypothesis that HBEGF alone induces a stellate morphology, MD astrocytes grown in DMEM with HBEGF (DMEM+H) remained polygonal. Reasoning that astrocytes may adopt a more in vivo–like stellate morphology when cultured in media that more closely mimics surrounding neurons, we applied neurobasal medium with and without HBEGF. We found that monocultures grown in neurobasal medium with HBEGF (NB+H) and in 1:1 DMEM/neurobasal medium with HBEGF (IPm) featured stellate astrocytes, similar to astrocytes co-cultured with neurons, irrespective of the astrocyte isolation method. Astrocytes grown in neurobasal medium without HBEGF did not survive. Thus, the simple mix of HBEGF in neurobasal medium was the only medium composition that produced stellate astrocytes (even in astrocytes prepared using shaking/the MD method). We termed this novel preparation AWESAM (Fig. 1, blue squares).
Figure 1.
Different media yield morphologically different astrocytes. (A) Immunocytochemistry of DIV14 co-cultures and DIV16 astrocyte monocultures treated with different media. ALDH1L1 and GFAP label astrocytes; MAP2 labels neuronal dendrites. MD and IP astrocyte cultures were grown in different media as explained in B: Co-cultures were directly plated in NB+, whereas MD astrocyte monocultures growing in DMEM with 10% FCS (DMEM+) were shaken on DIV7 and then grown in DMEM+ for classical MD astrocytes, NB+ with HBEGF (NB+H), DMEM+ with HBEGF (DMEM+H), or the medium used in the Foo et al. (2011) IP astrocyte protocol (IPm), which is a 1:1 mix of DMEM and NB+H. IP astrocytes were generated as described by Foo (2013) and grown in IPm, NB+H, or DMEM+. In A and B, the blue square marks MD cultures grown in NB+H (AWESAM astrocytes), which we selected for further analysis because of the stellate morphology and ease of production. Images per experiment in A, from top to bottom: n = 6, 4, 8, 4, 8, 5, 4, and 6. Bar, 10 µm. (C) Immunoblots of adult rat brain lysate and DIV14 co-culture, MD, IP, or AWESAM astrocyte lysate. Antibodies targeted general markers of astrocytes (ALDH1L1 and GFAP), immature or reactive astrocytes (vimentin and GFAP), mature astrocytes (S100β), neurons (SYP), oligodendrocytes (MBP), and microglia (CD11B); GAPDH served as a loading control.
All astrocyte monocultures tested were free of neurons, oligodendrocytes, and microglia, marked by SYP, MBP, and CD11B, respectively (Fig. 1 C). MD, IP, and AWESAM astrocytes expressed comparable amounts of ALDH1L1 and GFAP (Fig. 1 C). AWESAM astrocytes expressed less vimentin (which is up-regulated in reactive astrocytes [Schiffer et al., 1986]) than MD astrocytes, and IP astrocytes expressed the least vimentin. Although all monocultures showed similar nestin expression levels, S100β levels were slightly higher in AWESAM astrocytes than in MD astrocytes, suggesting that AWESAM astrocytes are more mature than MD astrocytes because nestin and S100β are expressed by immature and mature astrocytes, respectively (Messam et al., 2000; Raponi et al., 2007).
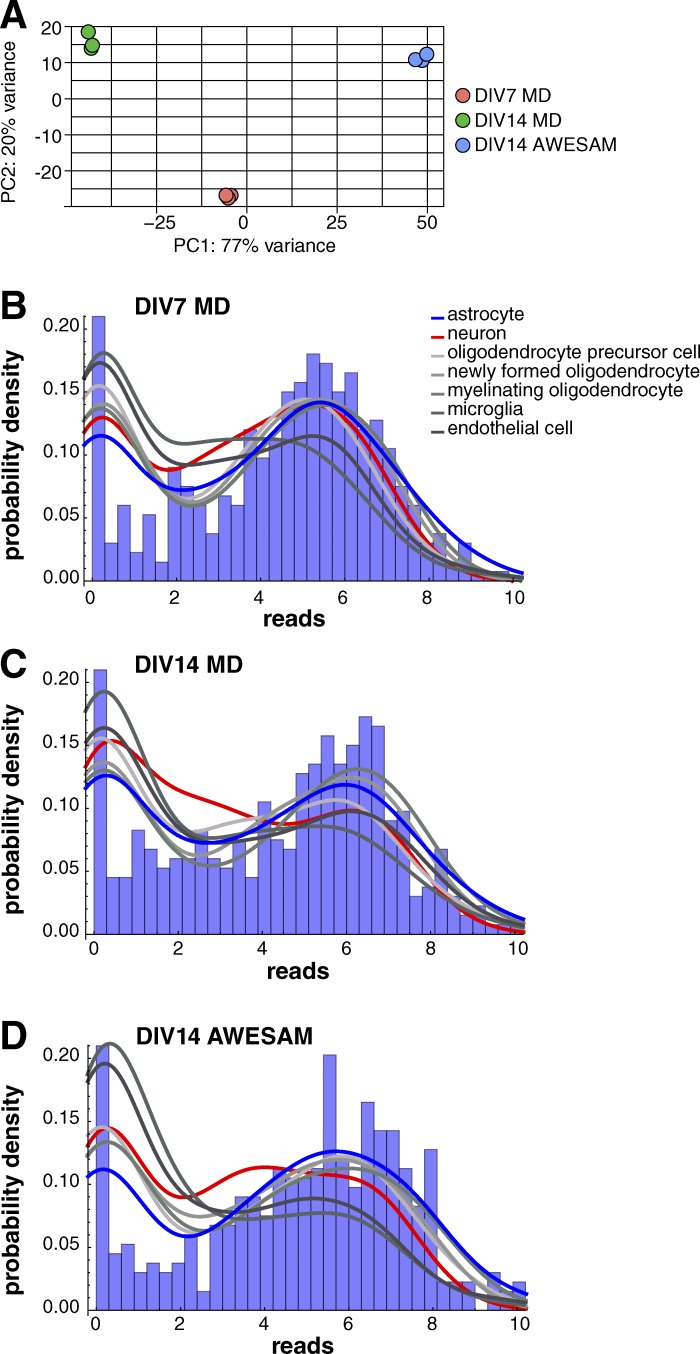
To further assess whether AWESAM cells have astrocytic properties, we conducted gene expression profiling. We compared our RNA-Seq data from three different culture conditions (DMEM+ until DIV7, subsequent culture in NB+H until DIV14 to generate AWESAM astrocytes, or continued culture in fresh DMEM+ until DIV14 to generate MD astrocytes) with published in vivo–derived cell type–specific gene expression (Zhang et al., 2014). Overall, our RNA-Seq data were of high quality, with 14.8 million mapped reads per sample, where cultured cell replicates showed distinct clustering by principal component analysis (Fig. 2 A). For each culture condition, we plotted probability density functions of reads versus number of 300 astrocyte, neuron, oligodendrocyte, microglia, or endothelial cell type–specific genes identified by Zhang et al. (2014) (Fig. 2, B–D). Cultured cells that are more astrocytic are expected to have more astrocytic genes with higher reads and less with lower reads and, conversely, more nonastrocyte-specific genes with lower reads and less with higher reads; i.e., the blue line should be lower for astrocytes than for all other cell types on the left and higher than for all other cell types on the rightmost peak of the plot. For AWESAM astrocytes, the blue line was indeed lowest on the left and highest on the right, indicating greater expression of astrocyte-specific genes and less expression of nonastrocyte-specific genes, compared with DIV7 and DIV14 MD astrocytes (Fig. 2, B–D). Thus, our culture method of switching the cells to NB+H medium after DIV7 until DIV14 (AWESAM astrocytes) increased their astrocytic specificity.
Figure 2.
AWESAM astrocytes show astrocyte-specific gene expression. (A) Cultured cell replicates show distinct clustering. The first two principal components (PC) for the regularized log transformation of the original counts for all DIV7 MD (red), DIV14 MD (green), and DIV14 AWESAM (blue) culture triplicates allow clear segmentation of conditions. (B–D) Gene expression values of 300 astrocyte (blue), neuron (red), oligodendrocyte, microglia, and endothelial (shades of gray) cell type–specific genes for each cell type (i.e., 2,100 genes total) plotted as probability density for DIV7 MD (B), DIV14 MD (C), and DIV14 AWESAM (D) cells, with the corresponding astrocyte-specific gene expression histogram underneath. AWESAM astrocytes show increased astrocyte-specific gene expression (and less nonastrocyte-specific gene expression) compared with DIV7 and DIV14 MD astrocytes.
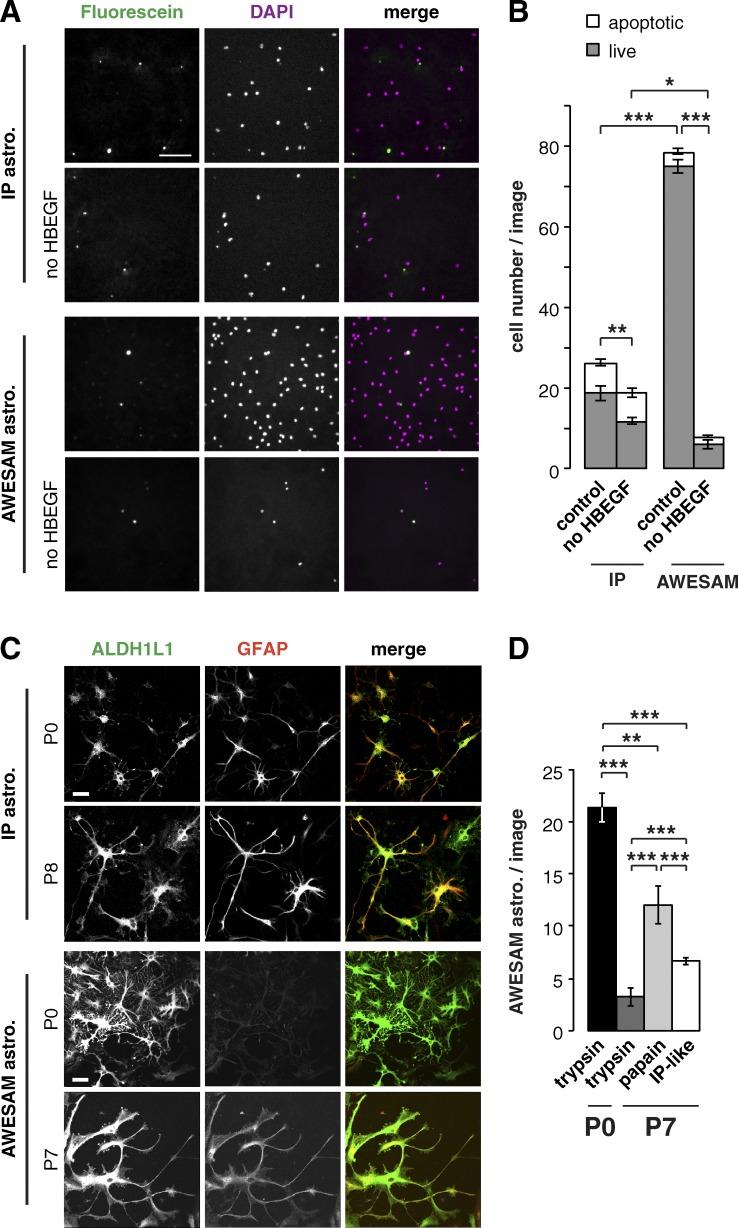
We found that astrocyte proliferation and survival depended on both HBEGF and the initial plating density (Fig. 3, A and B). Astrogliogenesis peaks shortly after birth (Sauvageot and Stiles, 2002; Tien et al., 2012), offering the highest yield of astrocytes, but older tissue contains more mature cells. Yet, the older the tissue source, the more difficult it becomes to dissect cortical tissue and generate healthy cell cultures. Because we prepared AWESAM and IP astrocytes from different developmental stages (E19 and P8, respectively), we also compared the morphology of AWESAM and IP astrocytes from P0 and P7/8 rats (Fig. 3, C and D). Both AWESAM and IP astrocytes derived from either age were stellate and expressed the astrocyte markers ALDH1L1 and GFAP. Postnatally derived AWESAM astrocytes (Fig. 3 C) reflected that cortical astrocytes may express only little or no GFAP (Cahoy et al., 2008; Zamanian et al., 2012) compared with other brain areas. We also tested the method of cell dissociation: To generate AWESAM astrocyte cultures from P7 rats, we digested dissected tissue with either trypsin or papain or as in the IP protocol (“IP-like,” where three to four times more tissue is used and digested in papain bubbled with carbogen for 40 min, then repeatedly triturated in 1.5% BSA solution, and centrifuged through 3% BSA solution). All three methods of tissue digestion produced stellate AWESAM astrocytes similar to P0 astrocytes digested with trypsin, where P0 tissue yielded significantly more astrocytes than older tissue (Fig. 3 D). To generate more astrocytes from P7 rats, papain digestion resulted in the highest cell yield. Thus, tissue age and method of enzymatic digestion determined cell number, where the AWESAM method could be applied to older tissue, too.
Figure 3.
HBEGF promotes astrocyte proliferation, and older tissue yields fewer but viable astrocytes. (A) Immunocytochemistry of DIV14 IP astrocytes plated in IPm at 10,000 cells/12-mm coverslip and AWESAM astrocytes plated in NB+H at 5,000 cells/12-mm coverslip. Fluorescein marks apoptotic cells; DAPI marks cell nuclei. (B) Quantification of apoptotic versus live cell number in media with or without HBEGF in absolute numbers (n = 12 images from 4 cultures). (C) Immunocytochemistry of DIV14 IP and AWESAM astrocytes derived from P0 versus P7/P8 rats, where ALDH1L1 and GFAP depict astrocyte morphology (for IP astrocytes, n = 12 [P0] and 11 [P7]; for AWESAM astrocytes, n = 6). Bars: (A) 100; (C) 25 µm. (D) Quantification of AWESAM astrocytes counted per image when dissociated with trypsin or papain or as described by Foo (2013) but without immunopanning before plating (“IP-like”; n = 8 [trypsin], n = 6 [papain], and n = 16 [IP-like]). Error bars indicate SEM, where *, P < 0.05; **, P < 0.01; ***, P < 0.001 by unpaired two-tailed Student’s t test.
Astrocyte Ca2+ signaling depends on the medium
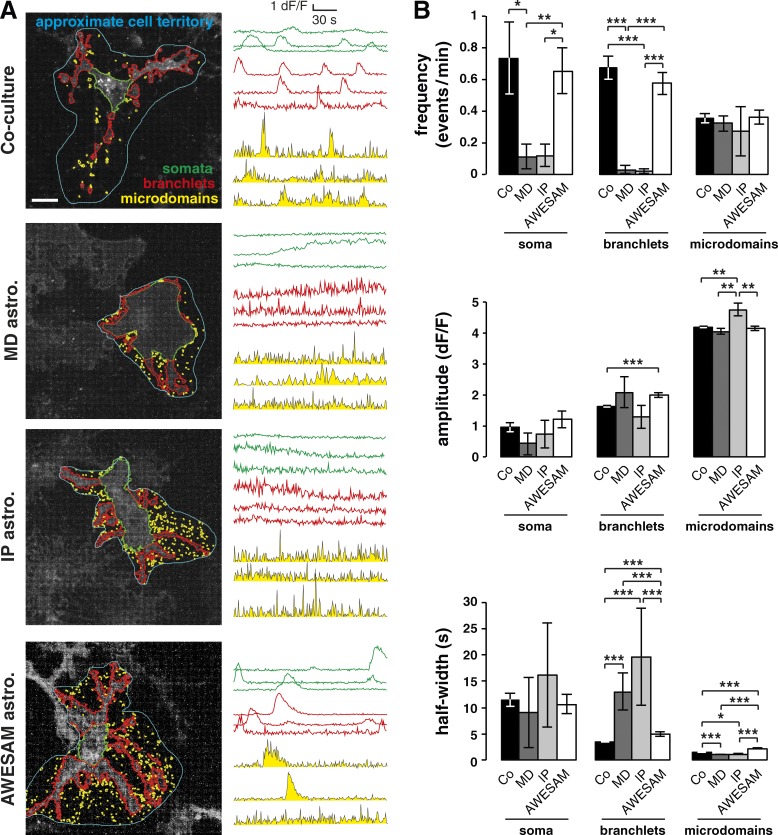
Astrocytes communicate with other glia and neurons through Ca2+ signaling, so intracellular [Ca2+] changes are often used to visualize an astrocyte’s response to stimuli (Cornell-Bell et al., 1990; Dani et al., 1992; Dallwig and Deitmer, 2002; Agulhon et al., 2008). To characterize spontaneous AWESAM astrocyte Ca2+ fluctuations and compare them with Ca2+ fluctuations in co-cultured, MD, and IP astrocytes, we transduced cultures with AAV particles of the 1/2 serotype encoding gfaABC1D-Lck-GCaMP3, which expresses membrane-targeted GCaMP3 via the astrocyte-specific GFAP promoter (Shigetomi et al., 2013a). We categorized Ca2+ signals in three domains: somata, branchlets, and microdomains (Fig. 4 A), as previously defined by Tong et al. (2013) using GECIquant software. Spontaneous Ca2+ fluctuations identified by fluorescence intensity changes in GCaMP3 signal occurred in astrocytes of all culture types and in all domains.
Figure 4.
AWESAM astrocytes show Ca2+ signaling as in astrocytes co-cultured with other glia and neurons. Spontaneous Ca2+ signaling in DIV16 co-cultures, and MD, IP, and AWESAM astrocyte monocultures, measured by Gfap promoter–driven Lck-GCaMP3 fluorescence changes in astrocytes. (A) Representative images of transduced astrocytes, where estimated territories (marked in blue) were analyzed for Ca2+ fluctuations in somata (green), branchlets (red), and microdomains (yellow). Approximate cell territories were not analyzed but used to define subdomains; example dF/F traces represent Ca2+ fluctuations from three regions of interest (ROIs) for each subdomain, as detected and quantified using the semiautomated software GECIquant (Srinivasan et al., 2015). Bar, 20 µm. (B) Quantitation of Ca2+ event frequency, amplitude, and half-width for each subdomain type in different culture types. For each of the four cultures types, one cell (one soma) in each of 9 (co-culture), 8 (MD), 5 (IP), or 11 (AWESAM) different videos was analyzed; branchlet ROIs: n = 431 (co-culture), n = 18 (MD), n = 7 (IP), and n = 177 (AWESAM); microdomain ROIs: n = 1,356 (co-culture), n = 414 (MD), n = 68 (IP), and n = 617 (AWESAM). Error bars indicate SEM, where *, P < 0.05; **, P < 0.01; ***, P < 0.001 by unpaired two-tailed Student’s t test.
As illustrated in example traces (Fig. 4 A), astrocytes from co-cultures and AWESAM cultures had similar Ca2+ event properties, which were distinct from those in MD and IP cultures: Co-cultured and AWESAM astrocytes showed numerous sharply rising and decaying, short Ca2+ events, but MD and IP astrocytes showed fewer, predominantly slowly rising and decaying Ca2+ changes that often lasted longer than 2 min (Fig. 4 A, MD culture, middle somatic trace).
We quantified astrocytic Ca2+ event frequency, amplitude, and duration (half-width) across all cultures (Fig. 4 B). In somata and branchlets, event frequency was similarly high in co-cultured and AWESAM astrocytes, whereas event frequency was lower in MD and IP astrocytes. In contrast, microdomain Ca2+ event frequency was similar across all cultures.
Ca2+ event amplitude in microdomains of co-cultured, MD, and AWESAM astrocytes was similar, whereas IP astrocytes featured slightly (but significantly) higher amplitude compared with other cultures. In somata and branchlets, Ca2+ event amplitude was similar in all cultures (although co-cultured astrocytes showed slightly lower Ca2+ event amplitude in branchlets than AWESAM astrocytes).
Ca2+ event duration, defined as event half-width, in somata and microdomains was similar across all culture types. In branchlets, MD and IP astrocytes exhibited significantly longer-lasting Ca2+ events than astrocytes in co-cultures or AWESAM cultures. Compared with somata and branchlets, microdomain Ca2+ event duration was significantly shorter in all culture types tested, although MD and IP astrocytes featured slightly (but significantly) shorter Ca2+ events than co-cultured and AWESAM astrocytes.
Astrocytes in all culture types shared two features: (1) higher Ca2+ event amplitude in microdomains than in branchlets or somata, and (2) significantly shorter Ca2+ event duration in microdomains than in branchlets or somata; i.e., microdomains in all cultures showed short, high-amplitude Ca2+ fluctuations.
In co-cultured and AWESAM astrocytes, Ca2+ event amplitude was significantly higher the smaller the domain (somata < branchlet < microdomain; P < 0.01 for soma vs. branchlets of AWESAM astrocytes, P < 0.001 for all other conditions). Conversely, in co-cultured and AWESAM astrocytes, Ca2+ event duration was significantly shorter the smaller the domain (somata > branchlet > microdomain; P < 0.001). In contrast, MD and IP astrocytes showed no significant differences between somata and branchlets (except for Ca2+ event amplitude in MD somata vs. branchlets; P < 0.05). Thus, only co-culture and AWESAM astrocytes showed distinct Ca2+ event amplitude and duration characteristics, i.e., a functional difference, between the three domains.
In summary, in terms of Ca2+ signaling, co-culture and AWESAM astrocytes showed similar Ca2+ signaling characteristics, which were distinct from MD and IP astrocytes: (a) Co-cultured and AWESAM astrocytes had higher Ca2+ event frequencies in somata and branchlets than MD or IP astrocytes; (b) co-cultured and AWESAM astrocytes featured long Ca2+ events in branchlets compared with MD and IP astrocytes; and (c) Ca2+ event amplitude and duration only varied significantly across all three domains in co-cultured and AWESAM astrocytes.
Astrocytes express several vesicle-associated proteins
Similar to neurons, astrocytes express several vesicle-associated proteins required for loading cargo into vesicles, vesicle fusion, and vesicle recycling (Parpura et al., 1995; Hepp et al., 1999; Mittelsteadt et al., 2009; Schubert et al., 2011). Through such vesicular release of gliotransmitters, astrocytes influence synaptic signaling and plasticity (Jourdain et al., 2007; Perea et al., 2009; Sasaki et al., 2011; Carlsen and Perrier, 2014). Therefore, we also tested whether astrocytes from AWESAM, MD, IP, and co-cultures express known vesicle-associated proteins by Western blot analysis.
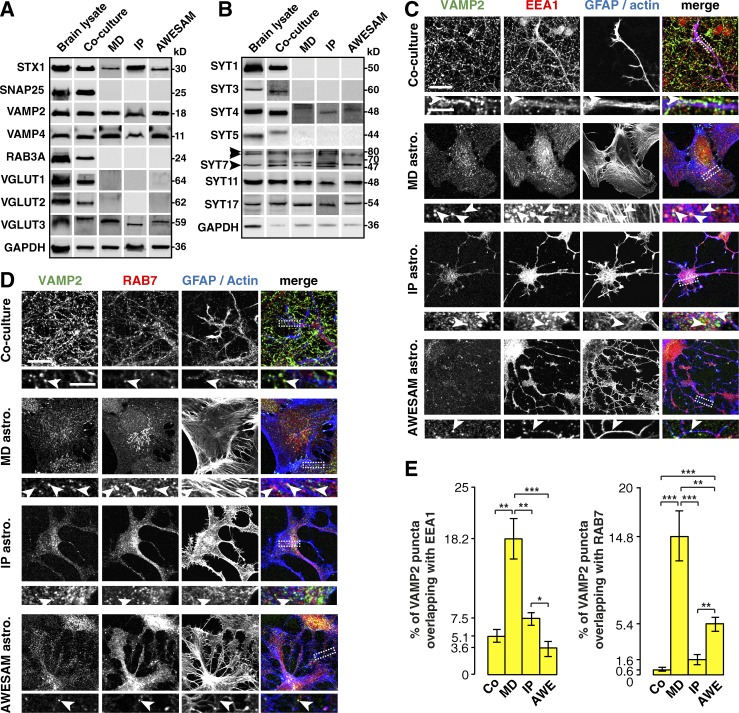
All cultures expressed syntaxin1, VAMP2, and VAMP4—proteins implicated in exocytotic vesicle release—as well as VGLUT3 (Fig. 5 A). In contrast, only cells from brain lysate and co-cultures expressed SNAP25, RAB3A, and VGLUT isoforms 1 and 2, suggesting that these proteins are present exclusively in neurons and not in astrocytes.
Figure 5.
Different astrocyte culture types express the same vesicle-associated proteins. (A) Immunoblots of adult rat brain, DIV14 co-culture, and MD, IP, and AWESAM astrocyte monoculture lysates. Proteins that form the SNARE complex in neuronal synaptic vesicle fusion (syntaxin1 [STX1] and VAMP2) appear in all samples, whereas SNAP25 is absent from astrocyte monocultures. VAMP4 and VGLUT3 are expressed in all samples, whereas RAB3A and VGLUT1 and 2 are absent from astrocyte monocultures. GAPDH served as a loading control (note that the same GAPDH blot appears in Fig. 1, where samples were processed together). (B) Immunoblots of the same samples as in A. Although brain and co-culture lysates contain all SYT isoforms tested, astrocyte monocultures express only SYT4, 7, 11, and 17. Three SYT7 splice isoforms appear at 47, 70, and ≈80 kD (arrowheads), as reported by Fukuda et al. (2002). GAPDH served as a loading control. (C and D) Immunocytochemistry of DIV14 co-cultures and DIV16 MD, IP, and AWESAM astrocytes, where antibodies label putative recycling vesicles (VAMP2), early endosomes (EEA1), and late endosomes (RAB7), GFAP labels astrocytes in co-cultures, and actin labels all cells in astrocyte monocultures. Higher-magnification images of the boxed area (outlined by a dashed line) are shown below each panel. VAMP2 colocalizes with endosomal proteins (arrowheads) in several areas of MD astrocytes but only rarely in co-culture, IP, or AWESAM astrocytes. Images per experiment in C: n = 6 (co-culture), n = 10 (MD and IP), n = 11 (AWESAM); in D: n = 6 per condition. Bars: (top panels) 10 µm; (zoomed-in bottom panels) 5 µm. (E) Bar graphs showing the percentage of VAMP2 puncta that colocalize with EEA1 or RAB7 puncta in astrocytes of co-cultures and MD, IP, and AWESAM cultures in C and D. Error bars indicate SEM, where *, P < 0.05; **, P < 0.01; ***, P < 0.001 by unpaired two-tailed Student’s t test.
SYT isoforms are good candidates for regulated fusion in astrocytes because many isoforms act as Ca2+ sensors (Bhalla et al., 2005; Hui et al., 2005; Chapman, 2008), where SYT1 is essential for controlled neurotransmitter release (Geppert et al., 1994; Nishiki and Augustine, 2004). Astrocytes express SYT4, 5, 7, and 11 mRNA in vivo (Cahoy et al., 2008; Mittelsteadt et al., 2009), and SYT4 and SYT11 protein in MD astrocyte cultures has been reported (Zhang et al., 2004; Sreetama et al., 2016). To test whether AWESAM and IP astrocytes also express SYT proteins, we compared different cultures via Western blot analysis (Fig. 5 B). In concordance with mRNA studies (Zhang et al., 2004; Mittelsteadt et al., 2009), all astrocyte cultures expressed SYT4, 7, and 11. We also found SYT17 protein in astrocytes, which has not been previously reported. Brain lysate and co-cultures contained SYT1, 3, and 5, suggesting that these isoforms are present in neurons and not in astrocytes.
Several groups have found that astrocytes express VAMP2 on vesicular structures that undergo fusion (Coco et al., 2003; Crippa et al., 2006; Malarkey and Parpura, 2011; Singh et al., 2014). However, reports of vesicular exocytosis in astrocytes have largely relied on polygonal MD astrocytes. We therefore tested whether AWESAM astrocytes can be used for studying gliotransmitter release in astrocytes with a more in vivo–like morphology. We first tested whether VAMP2 colocalized with proteins of early (EEA1 positive) or late (RAB7 positive) endosomes in astrocytes. Interestingly, VAMP2 colocalized with both EEA1 and RAB7 in MD astrocytes but rarely in co-culture, IP, or AWESAM astrocytes (Fig. 5, C–E). Thus, MD astrocytes may incorporate VAMP2 into endosomes, whereas VAMP2 is distinct from endosomal markers in stellate astrocytes of co-cultures and IP and AWESAM cultures.
VAMP2- and SYT7-harboring vesicles recycle constitutively in astrocytic processes
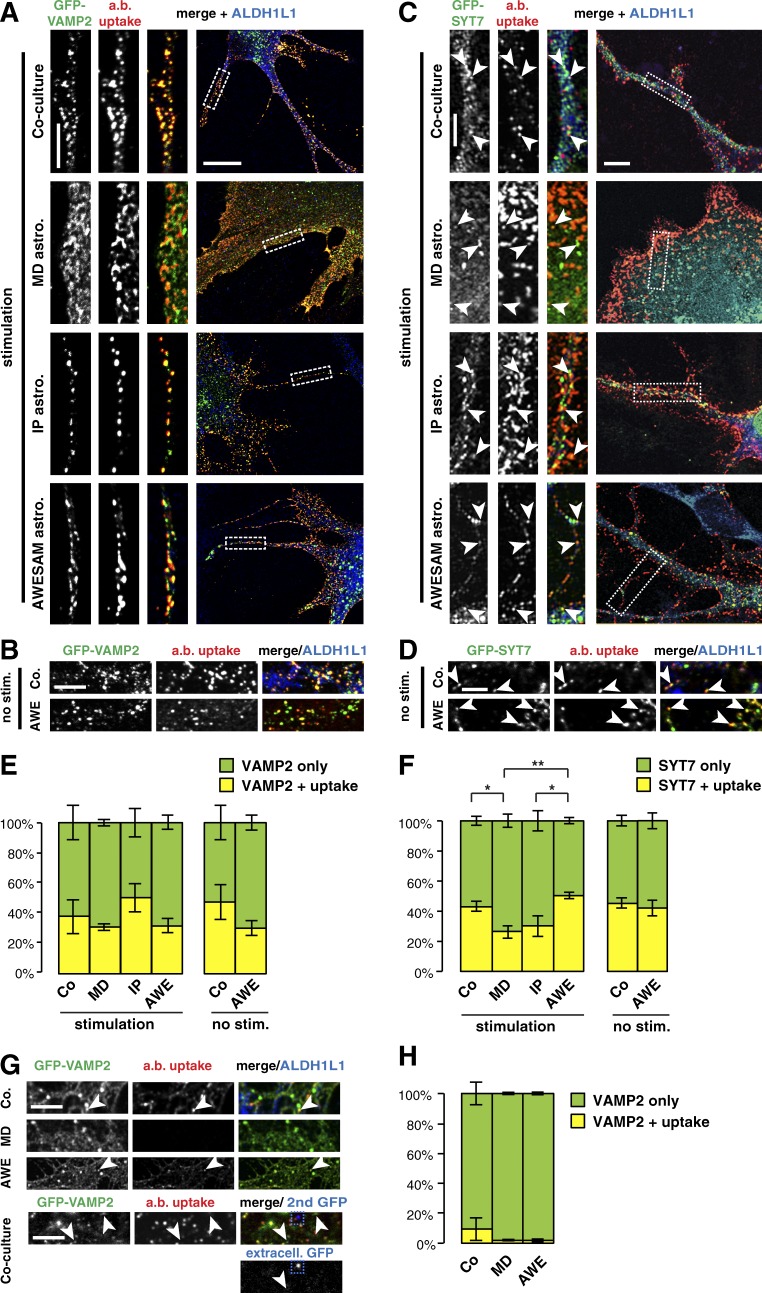
To investigate whether VAMP2 is present on recycling vesicles, we transfected co-cultured and MD, IP, and AWESAM astrocytes with GFP-VAMP2 for an antibody internalization assay. We stimulated live astrocytes with 50 µM glutamate solution that included anti-GFP antibodies, which only label recycling VAMP2-harboring vesicles in which the lumenal domain GFP tag is exposed to the extracellular solution. After fixation and permeabilization, secondary antibodies against GFP were applied to label recycling vesicles with a red fluorophore, compared with total GFP labeled with a green fluorophore to amplify the signal of transfected GFP-VAMP2. Strikingly, in stellate-shaped astrocytes, most VAMP2-harboring vesicles recycled in thin astrocytic processes (Fig. 6 A). MD astrocytes featured only thick processes (or no processes), and the GFP signal was not as clearly punctate as in other culture types. We then tested whether VAMP2-harboring vesicle recycling occurred in the absence of stimulation (Fig. 6 B). Interestingly, VAMP2-harboring vesicles also recycled in astrocytes that were not stimulated, suggesting that these vesicles undergo constitutive recycling.
Figure 6.
Astrocytes constitutively recycle VAMP2- and SYT7-harboring vesicles. (A) Immunocytochemistry of DIV14 co-cultures and DIV16 MD, IP, and AWESAM astrocytes (labeled by ALDH1L1) shows that anti-GFP antibodies taken up by recycling GFP-VAMP2 vesicles are often distinct in the soma but colocalize in astrocytic processes. Images per condition: n = 6. (B) Immunocytochemistry of DIV14 co-cultures and DIV16 AWESAM astrocytes also shows recycling GFP-VAMP2 vesicles in nonstimulating conditions. Images per condition: n = 6. (C) Immunocytochemistry of DIV14 co-cultures and DIV16 MD, IP, and AWESAM astrocytes shows that GFP-SYT7 puncta and anti-GFP antibody uptake colocalize (indicated by arrowheads) as in A when astrocytes were stimulated with 50 µM glutamate. Images per condition: n = 6 for co-cultures, MD, and AWESAM astrocytes; n = 4 for IP astrocytes. (A and C) Higher-magnification images of the boxed area (outlined by a dashed line) are shown in the left panels. (D) Immunocytochemistry of DIV14 co-cultures and DIV16 AWESAM astrocytes also shows recycling GFP-SYT7 vesicles (indicated by arrowheads) in nonstimulating conditions. Images per condition: n = 6. (E) Bar graphs showing similar levels of antibody uptake in GFP-VAMP2–transfected astrocytes of all cultures shown in A and B. (F) Bar graphs showing similar levels of antibody uptake in GFP-SYT7–transfected astrocytes of co-cultures compared with AWESAM and IP cultures. (G) Immunocytochemistry of anti-GFP antibody uptake in DIV14 co-cultures and DIV16 MD, and AWESAM astrocytes cotransfected with GFP-VAMP2 and TeNT, where VAMP2 function is impaired while GFP tags remain in the vesicle lumen. IP astrocytes did not survive cotransfections. Bottom panels show the signal of an anti-GFP antibody added to fixed GFP-VAMP2–transfected DIV14 co-cultures without permeabilizing the membrane. Only little GFP-VAMP2 was found at the membrane (blue dashed square), and only few puncta stained positive for GFP-VAMP2 but negative for anti-GFP antibody uptake (arrowheads). Images per condition: n = 6. (H) Bar graph showing antibody uptake in astrocytes cotransfected with GFP-VAMP2 and TeNT in the cultures shown in G. Error bars indicate SEM, where *, P < 0.05; **, P < 0.01 by unpaired two-tailed Student’s t test. Bars: (large view panels) 10 µm; (zoomed-in panels) 5 µm.
In neurons, VAMP2 and SYT1 are key mediators of vesicle fusion (Littleton et al., 1993; Geppert et al., 1994; Schoch et al., 2001). Of the four SYT isoforms we found in astrocytes, SYT4 and 11 are negative regulators of vesicle fusion (Bhalla et al., 2008; Dean et al., 2009), and SYT17 is predicted to lack a transmembrane domain (Fukuda and Mikoshiba, 2001). Thus, we next focused on SYT7 as a candidate protein for regulated exocytosis of astrocytic vesicles. To investigate whether SYT7 localizes to any recycling vesicles, we also transfected co-cultured and MD, IP, and AWESAM astrocytes with GFP-SYT7 for the antibody internalization assay. As for VAMP2, GFP-SYT7–harboring vesicles recycled with, but also without, glutamate stimulation (Fig. 6, C and D).
When quantifying antibody uptake, we found that astrocytes from all four culture types recycled similar amounts of VAMP2-harboring vesicles (Fig. 6 E). However, for SYT7-harboring vesicles, MD astrocytes showed significantly less vesicle recycling compared with co-cultured and AWESAM (but not IP) astrocytes (Fig. 6 F).
To investigate whether the constitutive recycling we observed in GFP-VAMP2–transfected astrocytes was indeed VAMP2 dependent, we cotransfected cultures with GFP-VAMP2 and TeNT, which cleaves VAMP2 (rendering VAMP2 nonfunctional but leaving the lumenal GFP tag). IP astrocytes did not survive this treatment, but co-cultured, MD, and AWESAM astrocytes showed greatly reduced antibody uptake after glutamate stimulation, confirming that the vesicle recycling we observed required functional VAMP2 (Fig. 6, G and H). To validate that GFP puncta represent internalized proteins (and not proteins on the cell surface), we also stained with an anti-GFP antibody after fixing but before permeabilizing. This revealed only little GFP on the membrane, suggesting that the majority of labeled GFP-VAMP2 was internalized (Fig. 6 G).
VAMP2 and SYT isoforms do not colocalize in astrocytes
Synaptic vesicles in neurons harbor SNARE proteins including VAMP2, which is required for vesicle fusion, and SYT, which promotes regulated fast exocytosis of these vesicles (Geppert et al., 1994). Several studies hint at regulated vesicle fusion in astrocytes (Araque et al., 1998; Calegari et al., 1999; Bezzi et al., 2004; Vardjan et al., 2014), but we found that both VAMP2- and SYT7-harboring vesicles recycled constitutively. VAMP2 and SYT7 might therefore be on either the same or distinct vesicle types in astrocytes. Alternatively, other astrocytic SYTs may localize to VAMP2-harboring vesicles. To test this, we co-immunostained astrocytes for VAMP2 and SYT4, 7, 11, or 17. Although both VAMP2 and SYT stainings appeared punctate (Fig. 7 A), we detected no colocalization of these puncta in astrocytic processes compared with VAMP2 and SYT1 in neuronal processes as a positive control (Fig. 7, A and B).
Astrocytic SYT7 affects synapse number
SYT7 antibody immunostaining revealed a punctate pattern present only in wild-type (but not Syt7−/−) co-cultures (Fig. 8 A). In Syt7+/+ and Syt7+/− but not Syt7−/− brain lysate, all three SYT7 splice isoforms appeared (Fig. 8 B), where the lowest molecular weight splice isoform relates to SYT7 function (Fukuda et al., 2002). Although SYT7 is not localized to lysosomes in MD astrocytes (Sreetama et al., 2016), SYT7 was found on lysosomes in other cell types (Martinez et al., 2000; Arantes and Andrews, 2006; Flannery et al., 2010). We therefore tested SYT7 localization to lysosomes marked by LAMP1. We found that SYT7 did not colocalize with the lysosomal protein LAMP1 (Fig. 8 C), indicating its absence on lysosomes in stellate-shaped AWESAM astrocytes.
Astrocytes expressed significantly more SYT7 puncta on DIV16 than DIV21 (Fig. 8, D and E; P = 0.0015), and corresponding Western blot analysis showed that astrocytes up-regulate the lowest molecular weight SYT7 splice isoform between DIV9 and 14 and down-regulate it after DIV14 (Fig. 8 F), at which point synaptogenesis is completed in vitro (Goslin et al., 1990; Ohno and Sakurai, 2005). Because astrocytic SYT7 was developmentally regulated, we next asked whether astrocytic SYT7 expression was relevant for neuronal synapse number. Indeed, wild-type neurons had 25% fewer synapses when grown on Syt7−/− astrocytes versus wild-type astrocytes (Fig. 8 G; P = 0.0016). Using a SYT1 lumenal antibody internalization assay, we found that SYT1-harboring synaptic vesicles recycled normally (Fig. 8 H), indicating that astrocytic SYT7 affected synapse number but not synaptic vesicle recycling.
Discussion
We developed a new protocol to easily generate stellate astrocyte monocultures, which revealed that Ca2+ signaling in AWESAM astrocytes is more similar to that of astrocytes co-cultured with neurons than MD astrocytes (which are traditionally used to examine regulated exocytosis in astrocytes) or IP astrocytes. Although we found that vesicle-associated regulatory proteins are present in all culture types, the distribution and subcellular localization differed: In MD but not other astrocyte cultures, VAMP2 was localized to endosomes, and SYT7-harboring vesicles recycle less compared with co-culture and AWESAM astrocytes. Our findings show that VAMP2- and SYT7-harboring vesicles recycle constitutively in thin processes, which MD astrocytes lack.
A central component of the AWESAM protocol is HBEGF, where a concentration of 5 ng/ml promotes IP astrocyte survival via EGF receptor signaling (Foo et al., 2011). HBEGF not only promotes astrocyte migration in simulated injury cultures (Faber-Elman et al., 1996), but also enhances astrocyte proliferation (Kornblum et al., 1999). In vivo, brain cells increasingly express HBEGF from around E14 onwards (and at high levels during astrogliogenesis). Thus, HBEGF may aid astrocyte survival, proliferation, or differentiation (Puschmann et al., 2013) and thereby influence astrocyte morphology and function. Apart from supporting in vivo–like morphology, HBEGF batch quality is controlled (unlike for serum), and HBEGF enhances astrocyte proliferation as much as serum in astrocyte monocultures in 3-D matrices (Puschmann et al., 2014). However, HBEGF can also induce de-differentiation in zebrafish retinal Müller glia (Wan et al., 2012) and at 10 ng/ml, in vitro (Puschmann et al., 2014). We used HBEGF at 5 ng/ml as suggested by Foo et al. (2011) and saw a mostly stellate morphology and did not observe a bipolar morphology that Puschmann et al. (2014) reported as indicative of de-differentiation. Overall, our comparison of different medium compositions with method of preparation emphasizes the importance of the extracellular environment on astrocytes in monoculture. Our gene expression profiling further suggests that switching cells from DMEM + 10% FCS to 5 ng/ml HBEGF in NB medium after 7 d in culture to generate AWESAM astrocytes may increase the expression of in vivo astrocyte-specific genes.
Our data show spatially segregated, spontaneous Ca2+ fluctuations in cultured astrocytes, where AWESAM astrocytes are most similar to astrocytes co-cultured with other glia and neurons: Compared with MD and IP astrocytes, co-cultured and AWESAM astrocytes featured higher Ca2+ event frequencies in somata and branchlets and shorter Ca2+ events in branchlets (where MD and IP astrocyte Ca2+ events were at least twice as long).
AWESAM astrocyte Ca2+ event frequencies (which were higher than those in MD and IP astrocyte somata and branchlets) and amplitudes were comparable with those in astrocytes in brain slices (Shigetomi et al., 2013a) and in vivo (Srinivasan et al., 2015). In contrast, astrocytes from MD and IP cultures had no or only few Ca2+ events in somata and branchlets, matching previous data from Ca2+ indicator dyes in IP astrocytes (Foo et al., 2011; Zhang et al., 2015) and unlike in brain slices and in vivo (Shigetomi et al., 2013a; Srinivasan et al., 2015). However, Ca2+ event duration was longer in all cultured astrocyte types we tested than in brain slices and in vivo, which may be because of the stringent criteria we used to define Ca2+ events (where we applied high amplitude thresholds to avoid detecting false positive events but may have excluded some smaller events closer to the noise). Although our findings in cultured astrocytes cannot be directly compared with brain slice or in vivo data because of methodological and analysis differences, the resemblance of Ca2+ event frequency and amplitude suggests that co-cultured and AWESAM (but not MD or IP) astrocytes are useful models for studying Ca2+ fluctuations in cultures.
Because AWESAM astrocytes were so similar to co-cultured astrocytes, in terms of Ca2+ fluctuations, morphology, and expression of markers of cell maturity, we conclude that our novel AWESAM protocol generates cultures superior to other monocultures.
Astrocytes use both internal and extracellular Ca2+ sources (Deitmer et al., 2009). Inositol triphosphate type 2 receptor (IP3R2)–mediated Ca2+ release from internal stores (Petravicz et al., 2008; Agulhon et al., 2010) largely supports somatic Ca2+ fluctuations but does not account for all Ca2+ signaling in astrocytes because astrocytic Ca2+ fluctuations persist in Ip3r2−/− mice (Srinivasan et al., 2015). It is therefore likely that branchlet and microdomain Ca2+ fluctuations also result from Ca2+ influx via TRPA1 channels or Na+/Ca2+ exchangers (Kirischuk et al., 1997; Minelli et al., 2007; Shigetomi et al., 2013b). It would be interesting to further dissect which domains in astrocytes use which Ca2+ source or sources and how similar Ca2+ events are in astrocytes in culture versus in vivo.
Interestingly, organic Ca2+ indicator dyes revealed that extracellular Ca2+ underlies event frequency of large events that occur simultaneously in larger domains of adjacent cells but not microdomains (Di Castro et al., 2011). However, the same study reports much lower branchlet Ca2+ event frequency than Lck-GCaMP–based in vivo experiments, emphasizing that membrane-tethered, genetically encoded Ca2+ indicators more accurately report microdomain Ca2+ signals in astrocytes than organic dyes (Shigetomi et al., 2010a; Tong et al., 2013). Future experiments using Lck-GCaMPs may reveal specific Ca2+ sources for signaling within astrocytic branchlets and microdomains in stellate astrocytes in culture, e.g., by depleting internal Ca2+ stores or removing extracellular Ca2+.
The AWESAM culture system allows spatially resolving Ca2+ fluctuations within different astrocytic domains of stellate astrocytes. Our study is the first to analyze Ca2+ fluctuations in IP astrocytes transduced with the Ca2+ indicator Lck-GCaMP3 and the first to compare Ca2+ fluctuations in somata, branchlets, and microdomains of astrocytes from different culture protocols. Previously, IP astrocyte Ca2+ signaling was only studied using Fluo-4 (Foo et al., 2011), whereas the membrane-tethered Lck-GCaMP3 used in our study is superior to Ca2+ indicator dyes as it reports Ca2+ events even in fine processes (Shigetomi et al., 2010b). We further illustrate that the semiautomated software GECIquant (Srinivasan et al., 2015) is applicable to cultured astrocytes.
Apart from Ca2+ signaling, MD astrocytes also differed from other cultured astrocytes in terms of the subcellular localization of VAMP2 to endosomes, where VAMP2 was distinct from endosomes in AWESAM, IP, and co-cultured astrocytes. VAMP2 was previously found to undergo regulated exocytosis in astrocytes, but only in MD cultures, which do not form thin processes like AWESAM astrocytes (Crippa et al., 2006; Malarkey and Parpura, 2011; Singh et al., 2014). Our results indicate that VAMP2 recycles in astrocytic process and does so constitutively. Small synaptic vesicle–like vesicles have been isolated from MD astrocytes via VAMP2 (Crippa et al., 2006; Martineau et al., 2013) to study astrocytic vesicle characteristics, but as we found another, distinct subpopulation of recycling astrocytic vesicles harboring SYT7, it is likely that only a subpopulation of vesicles was assessed in these studies.
We found two distinct populations of vesicles that undergo constitutive recycling in astrocytes: VAMP2 and SYT7. In cultures in which astrocytes lacked SYT7, wild-type neurons had significantly fewer synapses. Thus, SYT7 promotes synapse number, possibly through trafficking or releasing synaptogenic factors. We further found that astrocytes developmentally up-regulate the main SYT7 splice isoform when synapses are forming in the brain, which may therefore act specifically during synaptogenesis. Because VAMP2- and SYT7-harboring vesicles both recycled constitutively in astrocytes but did not colocalize, astrocytic SYT7-harboring vesicles may represent a distinct vesicle population that affects synapse number.
Although SYT7 localizes to lysosomes in cultured neurons, fibroblasts, and macrophages (Martinez et al., 2000; Arantes and Andrews, 2006; Flannery et al., 2010), we found SYT7 in nonlysosomal compartments in AWESAM astrocytes, which is also the case in MD astrocytes (Sreetama et al., 2016). Instead, Sreetama et al. (2016) suggest that SYT11 mediates lysosomal exocytosis for repair in injured MD astrocytes. We found SYT11, as well SYT4, 7, and 17 protein, in stellate AWESAM astrocytes. Different SYT isoforms may thus perform different functions in astrocytes, where SYT7 is involved in constitutive release, e.g., during early postnatal development.
The AWESAM method is a simple, fast, and economical way of generating stellate astrocyte monocultures with a more in vivo–like morphology and gene expression than MD astrocytes and Ca2+ signaling more similar to astrocytes co-cultured with other glia and neurons. Thus, AWESAM cultures allow visualization of localized vesicle recycling and Ca2+ signaling in astrocytic processes, branchlets, and microdomains—important sites of astrocytic function in vivo.
Acknowledgments
We thank Prof. Khakh and Dr. Srinivasan (University of California, Los Angeles, Los Angeles, CA) for introducing us to GECIquant and technical support with this plugin. We are grateful to Prof. Ahmed Mansouri and Dr. Zeeshan Ahmad for help with the apoptosis assay. We thank Prof. Hülsmann (University of Göttingen, Göttingen, Germany) for helpful discussion of the manuscript.
This work was supported by the European Research Council (FP7/260916), Alexander von Humboldt-Stiftung (Sofja Kovalevskaja), and Deutsche Forschungsgemeinschaft (CNMPB, DE1951, and SFB889).
The authors declare no competing financial interests.
Sharona E. Gordon served as editor.
Footnotes
Abbreviations used:
- AAV
- adeno-associated virus
- AWESAM
- a low-cost easy stellate astrocyte method
- DIV
- day in vitro
- HBEGF
- heparin-binding epidermal growth factor
- IP
- immunopanned
- iPSC
- induced pluripotent stem cell
- MBP
- myelin basic protein
- PEI
- polyethylenimine
- SYP
- synaptophysin
- SYT
- synaptotagmin
- TeNT
- tetanus toxin
- VGLUT
- vesicular glutamate transporter
References
- Agulhon C., Petravicz J., McMullen A.B., Sweger E.J., Minton S.K., Taves S.R., Casper K.B., Fiacco T.A., and McCarthy K.D.. 2008. What is the role of astrocyte calcium in neurophysiology? Neuron. 59:932–946. 10.1016/j.neuron.2008.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agulhon C., Fiacco T.A., and McCarthy K.D.. 2010. Hippocampal short- and long-term plasticity are not modulated by astrocyte Ca2+ signaling. Science. 327:1250–1254. 10.1126/science.1184821 [DOI] [PubMed] [Google Scholar]
- Arantes R.M.E., and Andrews N.W.. 2006. A role for synaptotagmin VII-regulated exocytosis of lysosomes in neurite outgrowth from primary sympathetic neurons. J. Neurosci. 26:4630–4637. 10.1523/JNEUROSCI.0009-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araque A., Parpura V., Sanzgiri R.P., and Haydon P.G.. 1998. Glutamate-dependent astrocyte modulation of synaptic transmission between cultured hippocampal neurons. Eur. J. Neurosci. 10:2129–2142. 10.1046/j.1460-9568.1998.00221.x [DOI] [PubMed] [Google Scholar]
- Bezzi P., Gundersen V., Galbete J.L., Seifert G., Steinhäuser C., Pilati E., and Volterra A.. 2004. Astrocytes contain a vesicular compartment that is competent for regulated exocytosis of glutamate. Nat. Neurosci. 7:613–620. 10.1038/nn1246 [DOI] [PubMed] [Google Scholar]
- Bhalla A., Tucker W.C., and Chapman E.R.. 2005. Synaptotagmin isoforms couple distinct ranges of Ca2+, Ba2+, and Sr2+ concentration to SNARE-mediated membrane fusion. Mol. Biol. Cell. 16:4755–4764. 10.1091/mbc.E05-04-0277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla A., Chicka M.C., and Chapman E.R.. 2008. Analysis of the synaptotagmin family during reconstituted membrane fusion. Uncovering a class of inhibitory isoforms. J. Biol. Chem. 283:21799–21807. 10.1074/jbc.M709628200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy J.D., Emery B., Kaushal A., Foo L.C., Zamanian J.L., Christopherson K.S., Xing Y., Lubischer J.L., Krieg P.A., Krupenko S.A., et al. 2008. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci. 28:264–278. 10.1523/JNEUROSCI.4178-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calegari F., Coco S., Taverna E., Bassetti M., Verderio C., Corradi N., Matteoli M., and Rosa P.. 1999. A regulated secretory pathway in cultured hippocampal astrocytes. J. Biol. Chem. 274:22539–22547. 10.1074/jbc.274.32.22539 [DOI] [PubMed] [Google Scholar]
- Carlsen E.M., and Perrier J.-F.. 2014. Purines released from astrocytes inhibit excitatory synaptic transmission in the ventral horn of the spinal cord. Front. Neural Circuits. 8:60 10.3389/fncir.2014.00060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centeno T.P., Shomroni O., Hennion M., Halder R., Vidal R., Rahman R.-U., and Bonn S.. 2016. Genome-wide chromatin and gene expression profiling during memory formation and maintenance in adult mice. Sci. Data. 3:160090 10.1038/sdata.2016.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman E.R. 2008. How does synaptotagmin trigger neurotransmitter release? Annu. Rev. Biochem. 77:615–641. 10.1146/annurev.biochem.77.062005.101135 [DOI] [PubMed] [Google Scholar]
- Coco S., Calegari F., Pravettoni E., Pozzi D., Taverna E., Rosa P., Matteoli M., and Verderio C.. 2003. Storage and release of ATP from astrocytes in culture. J. Biol. Chem. 278:1354–1362. 10.1074/jbc.M209454200 [DOI] [PubMed] [Google Scholar]
- Cornell-Bell A.H., Finkbeiner S.M., Cooper M.S., and Smith S.J.. 1990. Glutamate induces calcium waves in cultured astrocytes: long-range glial signaling. Science. 247:470–473. 10.1126/science.1967852 [DOI] [PubMed] [Google Scholar]
- Crippa D., Schenk U., Francolini M., Rosa P., Verderio C., Zonta M., Pozzan T., Matteoli M., and Carmignoto G.. 2006. Synaptobrevin2-expressing vesicles in rat astrocytes: insights into molecular characterization, dynamics and exocytosis. J. Physiol. 570:567–582. 10.1113/jphysiol.2005.094052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallwig R., and Deitmer J.W.. 2002. Cell-type specific calcium responses in acute rat hippocampal slices. J. Neurosci. Methods. 116:77–87. 10.1016/S0165-0270(02)00030-4 [DOI] [PubMed] [Google Scholar]
- Dani J.W., Chernjavsky A., and Smith S.J.. 1992. Neuronal activity triggers calcium waves in hippocampal astrocyte networks. Neuron. 8:429–440. 10.1016/0896-6273(92)90271-E [DOI] [PubMed] [Google Scholar]
- Dean C., Liu H., Dunning F.M., Chang P.Y., Jackson M.B., and Chapman E.R.. 2009. Synaptotagmin-IV modulates synaptic function and long-term potentiation by regulating BDNF release. Nat. Neurosci. 12:767–776. 10.1038/nn.2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitmer J.W., Singaravelu K., and Lohr C.. 2009. Calcium ion signaling in astrocytes. In Astrocytes in (patho)physiology of the nervous system. Haydon P.G., and Parpura V., editors. Springer US, New York, NY: 201–224. 10.1007/978-0-387-79492-1_8 [DOI] [Google Scholar]
- Di Castro M.A., Chuquet J., Liaudet N., Bhaukaurally K., Santello M., Bouvier D., Tiret P., and Volterra A.. 2011. Local Ca2+ detection and modulation of synaptic release by astrocytes. Nat. Neurosci. 14:1276–1284. 10.1038/nn.2929 [DOI] [PubMed] [Google Scholar]
- Dichter M.A. 1978. Rat cortical neurons in cell culture: culture methods, cell morphology, electrophysiology, and synapse formation. Brain Res. 149:279–293. 10.1016/0006-8993(78)90476-6 [DOI] [PubMed] [Google Scholar]
- Faber-Elman A., Solomon A., Abraham J.A., Marikovsky M., and Schwartz M.. 1996. Involvement of wound-associated factors in rat brain astrocyte migratory response to axonal injury: in vitro simulation. J. Clin. Invest. 97:162–171. 10.1172/JCI118385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannery A.R., Czibener C., and Andrews N.W.. 2010. Palmitoylation-dependent association with CD63 targets the Ca2+ sensor synaptotagmin VII to lysosomes. J. Cell Biol. 191:599–613. 10.1083/jcb.201003021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo L.C. 2013. Purification of rat and mouse astrocytes by immunopanning. Cold Spring Harb. Protoc. 2013:421–432. 10.1101/pdb.prot074211 [DOI] [PubMed] [Google Scholar]
- Foo L.C., Allen N.J., Bushong E.A., Ventura P.B., Chung W.-S., Zhou L., Cahoy J.D., Daneman R., Zong H., Ellisman M.H., and Barres B.A.. 2011. Development of a method for the purification and culture of rodent astrocytes. Neuron. 71:799–811. 10.1016/j.neuron.2011.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M., and Mikoshiba K.. 2001. The N-terminal cysteine cluster is essential for membrane targeting of B/K protein. Biochem. J. 360:441–448. 10.1042/bj3600441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M., Ogata Y., Saegusa C., Kanno E., and Mikoshiba K.. 2002. Alternative splicing isoforms of synaptotagmin VII in the mouse, rat and human. Biochem. J. 365:173–180. 10.1042/bj20011877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geppert M., Goda Y., Hammer R.E., Li C., Rosahl T.W., Stevens C.F., and Südhof T.C.. 1994. Synaptotagmin I: a major Ca2+ sensor for transmitter release at a central synapse. Cell. 79:717–727. 10.1016/0092-8674(94)90556-8 [DOI] [PubMed] [Google Scholar]
- Goslin K., Schreyer D.J., Skene J.H., and Banker G.. 1990. Changes in the distribution of GAP-43 during the development of neuronal polarity. J. Neurosci. 10:588–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton N.B., and Attwell D.. 2010. Do astrocytes really exocytose neurotransmitters? Nat. Rev. Neurosci. 11:227–238. 10.1038/nrn2803 [DOI] [PubMed] [Google Scholar]
- Hepp R., Perraut M., Chasserot-Golaz S., Galli T., Aunis D., Langley K., and Grant N.J.. 1999. Cultured glial cells express the SNAP-25 analogue SNAP-23. Glia. 27:181–187. [DOI] [PubMed] [Google Scholar]
- Hui E., Bai J., Wang P., Sugimori M., Llinas R.R., and Chapman E.R.. 2005. Three distinct kinetic groupings of the synaptotagmin family: candidate sensors for rapid and delayed exocytosis. Proc. Natl. Acad. Sci. USA. 102:5210–5214. 10.1073/pnas.0500941102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdain P., Bergersen L.H., Bhaukaurally K., Bezzi P., Santello M., Domercq M., Matute C., Tonello F., Gundersen V., and Volterra A.. 2007. Glutamate exocytosis from astrocytes controls synaptic strength. Nat. Neurosci. 10:331–339. 10.1038/nn1849 [DOI] [PubMed] [Google Scholar]
- Kirischuk S., Kettenmann H., and Verkhratsky A.. 1997. Na+/Ca2+ exchanger modulates kainate-triggered Ca2+ signaling in Bergmann glial cells in situ. FASEB J. 11:566–572. [DOI] [PubMed] [Google Scholar]
- Kornblum H.I., Zurcher S.D., Werb Z., Derynck R., and Seroogy K.B.. 1999. Multiple trophic actions of heparin-binding epidermal growth factor (HB-EGF) in the central nervous system. Eur. J. Neurosci. 11:3236–3246. 10.1046/j.1460-9568.1999.00744.x [DOI] [PubMed] [Google Scholar]
- Krencik R., and Zhang S.-C.. 2011. Directed differentiation of functional astroglial subtypes from human pluripotent stem cells. Nat. Protoc. 6:1710–1717. 10.1038/nprot.2011.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C.-L., Oyler G., and Shoemaker C.B.. 2010. Lipid and cationic polymer based transduction of botulinum holotoxin, or toxin protease alone, extends the target cell range and improves the efficiency of intoxication. Toxicon. 55:619–629. 10.1016/j.toxicon.2009.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littleton J.T., Stern M., Schulze K., Perin M., and Bellen H.J.. 1993. Mutational analysis of Drosophila synaptotagmin demonstrates its essential role in Ca2+-activated neurotransmitter release. Cell. 74:1125–1134. 10.1016/0092-8674(93)90733-7 [DOI] [PubMed] [Google Scholar]
- Malarkey E.B., and Parpura V.. 2011. Temporal characteristics of vesicular fusion in astrocytes: examination of synaptobrevin 2-laden vesicles at single vesicle resolution. J. Physiol. 589:4271–4300. 10.1113/jphysiol.2011.210435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martineau M., Galli T., Baux G., and Mothet J.-P.. 2008. Confocal imaging and tracking of the exocytotic routes for D-serine-mediated gliotransmission. Glia. 56:1271–1284. 10.1002/glia.20696 [DOI] [PubMed] [Google Scholar]
- Martineau M., Shi T., Puyal J., Knolhoff A.M., Dulong J., Gasnier B., Klingauf J., Sweedler J.V., Jahn R., and Mothet J.-P.. 2013. Storage and uptake of D-serine into astrocytic synaptic-like vesicles specify gliotransmission. J. Neurosci. 33:3413–3423. 10.1523/JNEUROSCI.3497-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez I., Chakrabarti S., Hellevik T., Morehead J., Fowler K., and Andrews N.W.. 2000. Synaptotagmin VII regulates Ca2+-dependent exocytosis of lysosomes in fibroblasts. J. Cell Biol. 148:1141–1150. 10.1083/jcb.148.6.1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy K.D., and de Vellis J.. 1980. Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J. Cell Biol. 85:890–902. 10.1083/jcb.85.3.890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messam C.A., Hou J., and Major E.O.. 2000. Coexpression of nestin in neural and glial cells in the developing human CNS defined by a human-specific anti-nestin antibody. Exp. Neurol. 161:585–596. 10.1006/exnr.1999.7319 [DOI] [PubMed] [Google Scholar]
- Minelli A., Castaldo P., Gobbi P., Salucci S., Magi S., and Amoroso S.. 2007. Cellular and subcellular localization of Na+–Ca2+ exchanger protein isoforms, NCX1, NCX2, and NCX3 in cerebral cortex and hippocampus of adult rat. Cell Calcium. 41:221–234. 10.1016/j.ceca.2006.06.004 [DOI] [PubMed] [Google Scholar]
- Mittelsteadt T., Seifert G., Alvárez-Barón E., Steinhäuser C., Becker A.J., and Schoch S.. 2009. Differential mRNA expression patterns of the synaptotagmin gene family in the rodent brain. J. Comp. Neurol. 512:514–528. 10.1002/cne.21908 [DOI] [PubMed] [Google Scholar]
- Morita M., Higuchi C., Moto T., Kozuka N., Susuki J., Itofusa R., Yamashita J., and Kudo Y.. 2003. Dual regulation of calcium oscillation in astrocytes by growth factors and pro-inflammatory cytokines via the mitogen-activated protein kinase cascade. J. Neurosci. 23:10944–10952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett W.J., Oloff S.H., and McCarthy K.D.. 2002. Hippocampal astrocytes in situ exhibit calcium oscillations that occur independent of neuronal activity. J. Neurophysiol. 87:528–537. [DOI] [PubMed] [Google Scholar]
- Nishiki T., and Augustine G.J.. 2004. Dual roles of the C2B domain of synaptotagmin I in synchronizing Ca2+-dependent neurotransmitter release. J. Neurosci. 24:8542–8550. 10.1523/JNEUROSCI.2545-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno T., and Sakurai M.. 2005. Critical period for activity-dependent elimination of corticospinal synapses in vitro. Neuroscience. 132:917–922. 10.1016/j.neuroscience.2005.01.056 [DOI] [PubMed] [Google Scholar]
- Parpura V., Fang Y., Basarsky T., Jahn R., and Haydon P.G.. 1995. Expression of synaptobrevin II, cellubrevin and syntaxin but not SNAP-25 in cultured astrocytes. FEBS Lett. 377:489–492. 10.1016/0014-5793(95)01401-2 [DOI] [PubMed] [Google Scholar]
- Perea G., Navarrete M., and Araque A.. 2009. Tripartite synapses: astrocytes process and control synaptic information. Trends Neurosci. 32:421–431. 10.1016/j.tins.2009.05.001 [DOI] [PubMed] [Google Scholar]
- Petravicz J., Fiacco T.A., and McCarthy K.D.. 2008. Loss of IP3 receptor-dependent Ca2+ increases in hippocampal astrocytes does not affect baseline CA1 pyramidal neuron synaptic activity. J. Neurosci. 28:4967–4973. 10.1523/JNEUROSCI.5572-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Placone A.L., McGuiggan P.M., Bergles D.E., Guerrero-Cazares H., Quiñones-Hinojosa A., and Searson P.C.. 2015. Human astrocytes develop physiological morphology and remain quiescent in a novel 3D matrix. Biomaterials. 42:134–143. 10.1016/j.biomaterials.2014.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puschmann T.B., Zandén C., De Pablo Y., Kirchhoff F., Pekna M., Liu J., and Pekny M.. 2013. Bioactive 3D cell culture system minimizes cellular stress and maintains the in vivo-like morphological complexity of astroglial cells. Glia. 61:432–440. 10.1002/glia.22446 [DOI] [PubMed] [Google Scholar]
- Puschmann T.B., Zandén C., Lebkuechner I., Philippot C., de Pablo Y., Liu J., and Pekny M.. 2014. HB-EGF affects astrocyte morphology, proliferation, differentiation, and the expression of intermediate filament proteins. J. Neurochem. 128:878–889. 10.1111/jnc.12519 [DOI] [PubMed] [Google Scholar]
- Raponi E., Agenes F., Delphin C., Assard N., Baudier J., Legraverend C., and Deloulme J.-C.. 2007. S100B expression defines a state in which GFAP-expressing cells lose their neural stem cell potential and acquire a more mature developmental stage. Glia. 55:165–177. 10.1002/glia.20445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T., Kuga N., Namiki S., Matsuki N., and Ikegaya Y.. 2011. Locally synchronized astrocytes. Cerebral Cortex. 21:1889–1900. 10.1093/cercor/bhq256 [DOI] [PubMed] [Google Scholar]
- Sauvageot C.M., and Stiles C.D.. 2002. Molecular mechanisms controlling cortical gliogenesis. Curr. Opin. Neurobiol. 12:244–249. 10.1016/S0959-4388(02)00322-7 [DOI] [PubMed] [Google Scholar]
- Schiffer D., Giordana M.T., Migheli A., Giaccone G., Pezzotta S., and Mauro A.. 1986. Glial fibrillary acidic protein and vimentin in the experimental glial reaction of the rat brain. Brain Res. 374:110–118. 10.1016/0006-8993(86)90399-9 [DOI] [PubMed] [Google Scholar]
- Schoch S., Deák F., Königstorfer A., Mozhayeva M., Sara Y., Südhof T.C., and Kavalali E.T.. 2001. SNARE function analyzed in synaptobrevin/VAMP knockout mice. Science. 294:1117–1122. 10.1126/science.1064335 [DOI] [PubMed] [Google Scholar]
- Schubert V., Bouvier D., and Volterra A.. 2011. SNARE protein expression in synaptic terminals and astrocytes in the adult hippocampus: a comparative analysis. Glia. 59:1472–1488. 10.1002/glia.21190 [DOI] [PubMed] [Google Scholar]
- Shigetomi E., Kracun S., and Khakh B.S.. 2010a Monitoring astrocyte calcium microdomains with improved membrane targeted GCaMP reporters. Neuron Glia Biol. 6:183–191. 10.1017/S1740925X10000219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigetomi E., Kracun S., Sofroniew M.V., and Khakh B.S.. 2010b A genetically targeted optical sensor to monitor calcium signals in astrocyte processes. Nat. Neurosci. 13:759–766. 10.1038/nn.2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigetomi E., Bushong E.A., Haustein M.D., Tong X., Jackson-Weaver O., Kracun S., Xu J., Sofroniew M.V., Ellisman M.H., and Khakh B.S.. 2013a Imaging calcium microdomains within entire astrocyte territories and endfeet with GCaMPs expressed using adeno-associated viruses. J. Gen. Physiol. 141:633–647. 10.1085/jgp.201210949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigetomi E., Jackson-Weaver O., Huckstepp R.T., O’Dell T.J., and Khakh B.S.. 2013b TRPA1 channels are regulators of astrocyte basal calcium levels and long-term potentiation via constitutive d-serine release. J. Neurosci. 33:10143–10153. 10.1523/JNEUROSCI.5779-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P., Jorgačevski J., Kreft M., Grubišić V., Stout R.F. Jr., Potokar M., Parpura V., and Zorec R.. 2014. Single-vesicle architecture of synaptobrevin2 in astrocytes. Nat. Commun. 5:3780 10.1038/ncomms4780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreetama S.C., Takano T., Nedergaard M., Simon S.M., and Jaiswal J.K.. 2016. Injured astrocytes are repaired by Synaptotagmin XI-regulated lysosome exocytosis. Cell Death Differ. 23:596–607. 10.1038/cdd.2015.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan R., Huang B.S., Venugopal S., Johnston A.D., Chai H., Zeng H., Golshani P., and Khakh B.S.. 2015. Ca2+ signaling in astrocytes from Ip3r2−/− mice in brain slices and during startle responses in vivo. Nat. Neurosci. 18:708–717. 10.1038/nn.4001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien A.-C., Tsai H.-H., Molofsky A.V., McMahon M., Foo L.C., Kaul A., Dougherty J.D., Heintz N., Gutmann D.H., Barres B.A., and Rowitch D.H.. 2012. Regulated temporal-spatial astrocyte precursor cell proliferation involves BRAF signalling in mammalian spinal cord. Development. 139:2477–2487. 10.1242/dev.077214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong X., Shigetomi E., Looger L.L., and Khakh B.S.. 2013. Genetically encoded calcium indicators and astrocyte calcium microdomains. Neuroscientist. 19:274–291. 10.1177/1073858412468794 [DOI] [PubMed] [Google Scholar]
- Tsugane M., Nagai Y., Kimura Y., Oka J., and Kimura H.. 2007. Differentiated astrocytes acquire sensitivity to hydrogen sulfide that is diminished by the transformation into reactive astrocytes. Antioxid. Redox Signal. 9:257–269. 10.1089/ars.2007.9.257 [DOI] [PubMed] [Google Scholar]
- Vardjan N., Kreft M., and Zorec R.. 2014. Regulated exocytosis in astrocytes is as slow as the metabolic availability of gliotransmitters: Focus on glutamate and ATP. Adv. Neurobiol. 11:81–101. 10.1007/978-3-319-08894-5_5 [DOI] [PubMed] [Google Scholar]
- Wan J., Ramachandran R., and Goldman D.. 2012. HB-EGF is necessary and sufficient for Müller glia dedifferentiation and retina regeneration. Dev. Cell. 22:334–347. 10.1016/j.devcel.2011.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates A., Akanni W., Amode M.R., Barrell D., Billis K., Carvalho-Silva D., Cummins C., Clapham P., Fitzgerald S., Gil L., et al. 2016. Ensembl 2016. Nucleic Acids Res. 44:D710–D716. 10.1093/nar/gkv1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamanian J.L., Xu L., Foo L.C., Nouri N., Zhou L., Giffard R.G., and Barres B.A.. 2012. Genomic analysis of reactive astrogliosis. J. Neurosci. 32:6391–6410. 10.1523/JNEUROSCI.6221-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Fukuda M., Van Bockstaele E., Pascual O., and Haydon P.G.. 2004. Synaptotagmin IV regulates glial glutamate release. Proc. Natl. Acad. Sci. USA. 101:9441–9446. 10.1073/pnas.0401960101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen K., Sloan S.A., Bennett M.L., Scholze A.R., O’Keeffe S., Phatnani H.P., Guarnieri P., Caneda C., Ruderisch N., et al. 2014. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34:11929–11947. 10.1523/JNEUROSCI.1860-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Sloan S.A., Clarke L.E., Caneda C., Plaza C.A., Blumenthal P.D., Vogel H., Steinberg G.K., Edwards M.S.B., Li G., et al. 2015. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron. 89:37–53. 10.1016/j.neuron.2015.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]