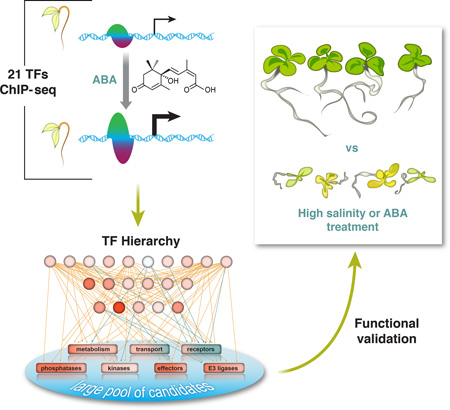
Abstract
Environmental stresses are universally encountered by microbes, plants and animals. Yet systematic studies of stress-responsive transcription factor (TF) networks in multi-cellular organisms have been limited. The phytohormone abscisic acid (ABA) influences the expression of thousands of genes, allowing us to characterize complex stress-responsive regulatory networks. Using chromatin immunoprecipitation sequencing, we identified genome-wide targets of 21 ABA-related TFs to construct a comprehensive regulatory network in Arabidopsis thaliana. Determinants of dynamic TF binding and a hierarchy among TFs were defined, illuminating the relationship between differential gene expression patterns and ABA pathway feedback regulation. By extrapolating regulatory characteristics of observed canonical ABA pathway components, we identified a new family of transcriptional regulators modulating ABA and salt responsiveness and demonstrated their utility to modulate plant resilience to osmotic stress.
Graphical Abstract
Transcription is a key step in gene expression. There have been concerted efforts to map functional elements in human, fly and worm (1–3), including a large number of cis-regulatory elements identified by profiling transcription factor (TF) binding using chromatin immunoprecipitation sequencing (ChIP-seq). One area that remains largely unexplored is how stimulus modulates TF binding and subsequent transcriptome changes. Furthermore, compared to studies in animals, very few comprehensive in vivo TF binding datasets are available for the Plantae kingdom. To begin to address this knowledge gap, we generated more than 100 ChIP-seq and time-series RNA-seq datasets to characterize a stimulus-influenced transcriptional network and map functional cis-regulatory elements in the reference plant Arabidopsis thaliana, focused on the phytohormone abscisic acid (ABA). The response to ABA provides an excellent model for the examination of stimulus-influenced transcriptional regulation. ABA triggers differential expression (DE) of thousands of genes including many TFs, providing a robust response that enables modeling of complex gene regulatory networks. Moreover, ABA’s role in a variety of plant processes is of significant importance to both fundamental biology and agriculture (4, 5).
ABA plays a pivotal role in optimizing water use in plants and is required for both seed development and responses to multiple environmental stresses such as drought and high salinity. In Arabidopsis thaliana, ABA is recognized by the PYR/PYL/RCAR receptor proteins (6–8). Binding of ABA triggers the interaction of PYR/PYL/RCARs with group A PP2C protein phosphatases and de-represses the SnRK2 protein kinases (7, 9). SnRK2s subsequently activate substrates such as transcription factors (TFs) and elicit ABA responses (10, 11). While many TFs are known to contribute to the ABA responses (8), little is known about their target genes and the way these targets are combinatorially regulated. In vitro approaches including the recently described Arabidopsis cistrome dataset have enabled identification of DNA motifs and inference of the associated TFs (13–17). However, accurate predictions are still challenging due to many large, multi-member TF families. Furthermore, it is difficult to establish a direct link between TF binding and transcriptome changes or to address the dynamics of TF regulation through in vitro assays. Therefore, we applied ChIP-seq to unambiguously discover TF targets, examine stimulus-influenced TF binding dynamics and link them to subsequent transcriptome changes.
ChIP-seq analyses of ABA responses
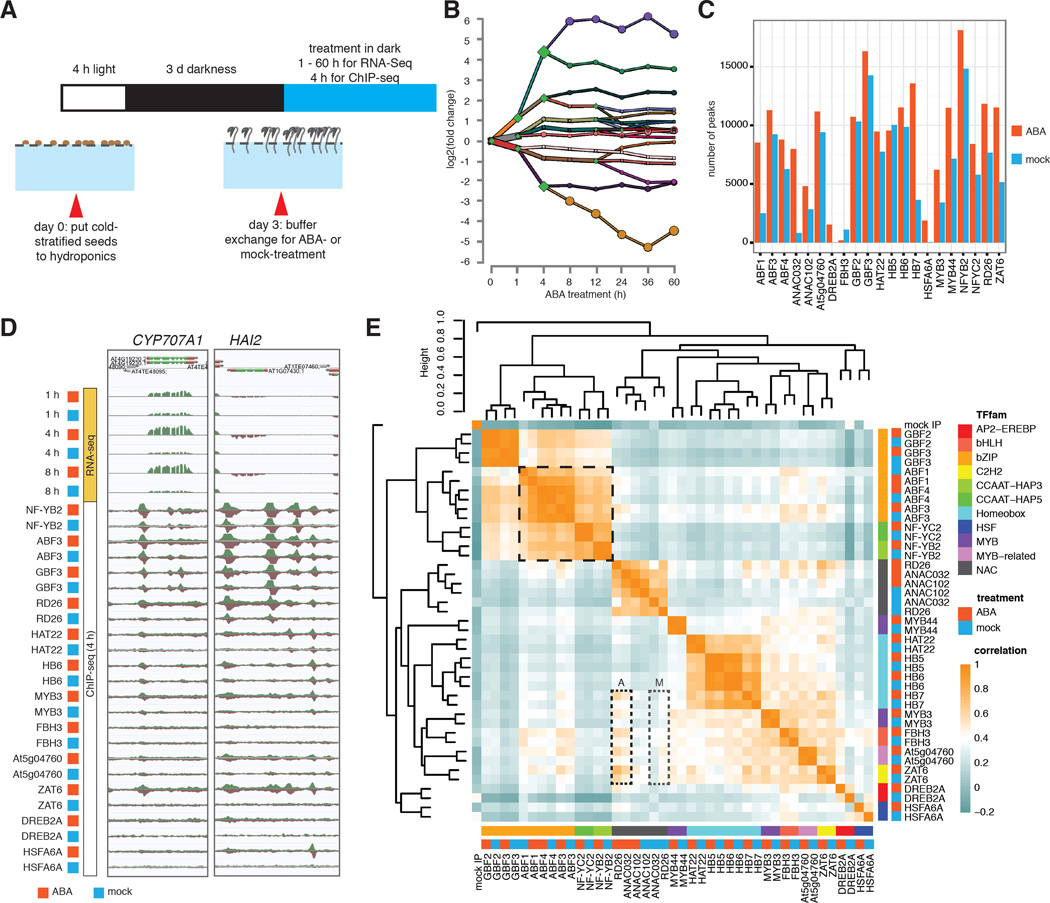
We profiled the genome-wide binding dynamics of a diverse collection of TFs using ChIP-seq to develop an in planta ABA transcriptional regulatory network. We first surveyed ABA-responsive transcripts in Arabidopsis thaliana, by generating strand-specific RNA-seq libraries from 3-day-old etiolated whole seedlings treated with either 10 uM (+/−)-ABA or an ethanol-containing mock for 1, 4, 8, 12, 24, 36 and 60 hours (Fig. 1A). Among 18,310 expressed genes, 3,061 are DE (FDR < 0.01, Table S1) (18) for at least one time point. One hour of ABA treatment leads to moderate DE of many genes, and most transcriptional responses plateau after 8 hours (Figs. 1B, S1). On the basis of gene expression data, we performed ChIP-seq experiments at four hours post ABA dose. We selected TFs based on responsiveness to ABA and published evidence, aiming to provide a good representation of TF families (Fig. S2; Tables S2, S3). In general, highly expressed and responsive TFs were chosen in each representative TF family because in the context of an in planta experimental framework, the impact of these TFs on gene expression can be more effectively investigated compared with their weakly expressed homologs. All TF genes were epitope tagged by a recombineering-based approach (19), mostly with large DNA transformable artificial chromosomes, allowing the TFs to be expressed under their native promoters and genomic context (Table S3). The final dataset consisted of one hundred and twenty-two ChIP-seq experiments of 21 TFs from 11 families, including mock- and ABA-treated conditions.
Figure 1. TF identity and hormone treatment determine genome-wide binding profiles.
(A) Growing Arabidopsis thaliana in hydroponics allows convenient buffer exchange for hormone treatment. (B) DREM reconstructed RNA expression paths 60 hours post ABA exposure. Each path corresponds to a set of genes that are co-expressed. Split nodes (green diamonds) represent a temporal event where a group of genes co-expressed up to that point diverge in expression, most likely due to regulatory events. Most splits are observed up to and including the 4h time point, indicating that the majority of regulatory events occur at the first 4 hours. (C) The number of ChIP-seq peaks varies greatly between TFs and treatment conditions. (D) ABA mediated differential gene expression and altered dynamics of TF binding as exemplified by CYP707A1 and HAI2 genes. (E) Comparison of binding correlations based on normalized ChIP-seq read counts near binding sites shows that TFs from same family often have similar binding profiles. TF-TF interaction (bZIPs and NF-Y, black dashed box) and hormone treatment (RD26 and ANAC032, dotted boxes A and M for ABA- and mock-treatment) also contribute to binding profile similarities between TFs.
Overall, the number of binding sites (termed “peaks”) varies greatly across TFs and between treatments (Fig. 1C). Most TFs gain bindings sites across the genome after ABA treatment (Fig. 1C), consistent with the fact that these TFs are induced by ABA at both the transcript and protein level (Figs. 1D, S2–3). As exemplified by CYP707A1 and HAI2 (20, 21), two important genes regulating ABA catabolism and signaling, respectively, the dynamic binding of TFs elicited by ABA is often accompanied by altered transcript abundance of the target genes (Fig. 1D). Comparing the genome wide binding profiles of these TFs, taking into account the binding location and strength, revealed that the TFs are generally grouped by family and known physical interactions (Fig. 1E) (22). Interestingly, the binding profiles between NAC and other TF families become more similar after ABA treatment (Fig. 1E, box A vs. M), suggesting ABA prompts coordinated regulation of target genes by these TFs.
Hormonal effects on TF binding and expression of target genes
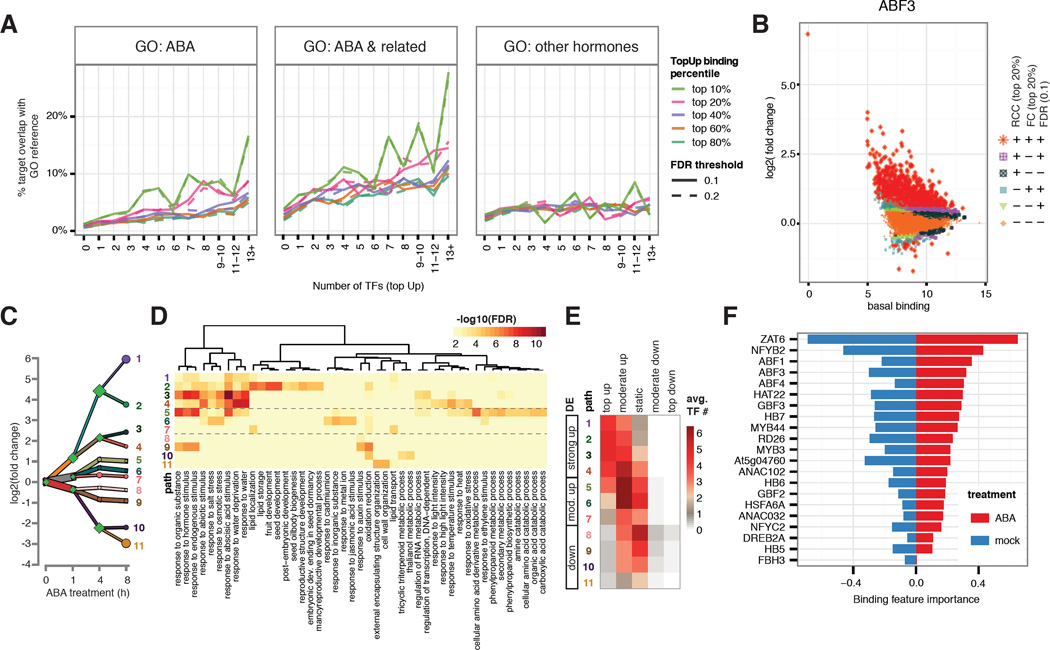
We observed dramatic changes TF binding at promoter regions of several known components of ABA signaling pathway (Fig. 1D), so we investigated whether dynamic binding may predict genes function in the ABA pathway. To quantify hormone-dependent, locus-specific change of TF binding, we compared ChIP-seq peaks of each TF between ABA- and mock-treated conditions by performing differential binding analysis of the sequencing reads under the peaks (23). Three measures of differential binding were calculated for each peak: 1) normalized read count change (RCC) that measures absolute changes of binding, 2) fold change (FC) that measures relative changes of binding, and 3) statistically significant differential binding (FDR). Since there are limited down-regulated binding events in our dataset, we focused on up-regulated binding to determine the optimal cutoff of RCC, FC and FDR to define the relationship between dynamic binding targets and genes involved in ABA response. We extracted three groups of Arabidopsis thaliana genes (Table S4) based on gene ontology (GO) annotation (24). Group 1 contains 493 genes involved in ABA response, Group 2 contains 1452 genes involved in responses to either ABA or other related processes such as water deprivation, osmotic stress, salt stress, cold, seed development and stomatal movement. Group 3 contains 999 genes involved in responses to other hormones after excluding genes shared with Group 2. Three observations emerged from comparing these lists to the TF target gene lists defined by various thresholds on RCC, FC and FDR. First, when Group 1 and 2 genes were used as a reference set, the percentage of TF targets overlapping with the set increases with the number of bound TFs (Fig. 2A, panels 1–2). By contrast, there is very little, if any, increase when Group 3 (other hormone genes) was used as the reference (Fig. 2A, panel 3). Second, an increase of RCC and FC threshold beyond top 20% boosted the percentage of target genes involved in ABA-related responses but not genes-related to other hormones (Fig 2A, panels 3). This improvement is even more obvious for genes targeted by multiple TFs. Lastly, FDR thresholds of 0.1 and 0.2 show little differences across all analyses. These results support the premise that dynamic binding by multiple TFs is an important feature to specifically recover genes involved in ABA-related responses. We thus chose top 20% RCC and FC and FDR 0.1 as the cutoff for follow-up analyses. As shown in Figs. 2B and S4, peaks passing all three thresholds were designated as top-ranked up- (“top up”) or down-regulated (“top down”) whereas those failing all thresholds were designated as static; all remaining peaks were classified as moderately up- (“moderate up”) or down-regulated (“moderate down”). For all tested TFs except FBH3 and HB5, peaks tend to gain binding instead of maintaining or losing binding after ABA treatment.
Figure 2. Dynamic TF binding triggered by ABA treatment correlates with gene function and expression.
(A) Genes targeted by higher number of TFs with dynamic binding events (x-axis) have higher percentage overlap (y-axis) with genes annotated with ABA and ABA-related GO terms, but not with GO terms specific to other hormones. This positive correlation is stronger for target genes associated with stronger dynamics (different color lines). (B) Hormone-dependent, locus-specific TF binding dynamics vary greatly across the genome. Log2 (fold change) of TF binding upon ABA treatment (y-axis) was plotted against basal binding measured as log2 (normalized read counts) under mock treatment (x-axis). Peaks were classified by three criteria: read count change (RCC, within top 20%), fold change (FC, within top 20%), and DiffBind FDR (less than 0.1). Peaks satisfying all three criteria were designated as top dynamic (+++) and those failing all three were designated as static (---). The remaining peaks were designated as moderately dynamic. (C) DREM analysis shows 11 paths of DE genes after 8 hours of ABA treatment. (D) Each DREM path is enriched for specific GO terms. (E) Level of DE is correlated with multi-TF dynamic binding. (F) Ridge regression model for differential expression at 4 hour using binding strength in both ABA- and mock-treated condition includes contribution from multiple TF in both conditions. Regression coefficients are plotted as relative importance of the binding features.
We then explored the relationship between dynamic TF binding triggered by ABA treatment and gene expression. The Dynamic Regulatory Event Miner (DREM) (25) reports 11 paths of DE genes for the first 8 hours of ABA treatment (Fig. 2C). As shown in Fig. S5, combining DREM with DNA motifs from PBM, AGRIS and DAP-seq databases recovered few TFs in our dataset, likely due to a low overlap of these TFs with the databases (13, 15, 26). The DREM model identified TFs from all ChIPped families except for CCAAT-HAP3 and CCAAT-HAP5, which do not bind DNA in in vitro assays as a monomer (27). In addition, although TF binding was examined at the single time point, we observed a positive correlation between the number of dynamically bound TF and the magnitude of DE across all time points (Fig. S6), suggesting TF binding data at 4 hour post ABA dose can explain a broad temporal span of gene expression. ABA-related GO terms such as seed development and response to salt / osmotic stress / water deprivation were enriched in up-regulated genes, whereas a few growth-related terms such as response to auxin stimulus and cell wall organization were enriched in down-regulated genes (Fig. 2D). We observed a distinct distribution of dynamic binding category across DREM paths (Figs. 2E, S1, Table S5). The extent of multi-TF dynamic binding is associated with the magnitude of differential gene expression. For example, highly up-regulated genes are often targeted by multiple TFs through “top up” binding. Moderately up-regulated genes are more commonly targeted by multiple TFs through “moderate up” binding. Down-regulated genes are rarely associated with “top up” binding. Instead, these genes are predominantly associated with either static binding by multiple TFs or down-regulated TF binding. These data suggest that DE at the whole seedling level is often subject to a combinatorial regulation by multiple TFs. As an independent validation, we built a regression model of differential expression using peak signals in ABA and mock-treated conditions as features without hard thresholds for the level of dynamic binding. The resulting model reveals that multiple TF binding features such as ZAT6, NF-YB2 and ABF factors in both ABA- and mock-treated conditions contribute to differential expression of target genes (Figs. 2F, S7).
Determinants of differential transcription factor binding
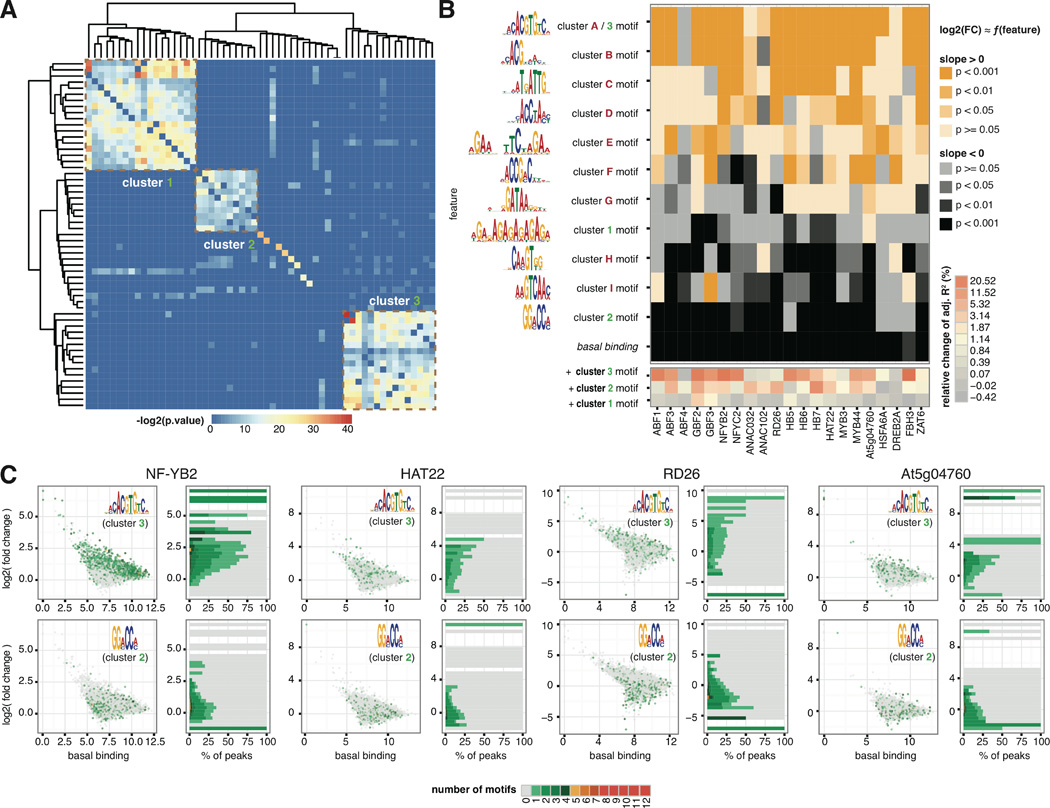
With the discovery of tens of thousands of differential binding events, we were interested in whether we could identify features that may predict binding dynamics. First we performed motif discovery by MEME-ChIP (28) to identify enriched motifs of all 21 ChIPped TFs from the strongest 600 peaks after either ABA or mock hormone treatment. A complete collection of the motifs is available at www.ABAtf.net. To investigate whether there are additional motifs that correlated with TF binding dynamics, we also performed motif discovery on both dynamic and static peaks for a handful of TFs. These factors, NF-YB2, ABF1, ABF4, FBH3, MYB3, RD26, ZAT6 and HB7, were selected to represent a variety of TF families. Pairwise comparison of primary and secondary motifs discovered from dynamic and static peaks across the selected TFs revealed three major clusters (Fig. 3A). Cluster 1 motifs are composed of (AG)n repeats. Cluster 2 motifs contain a (A/G)G(A/C)CC(A/C) consensus sequence, whereas Cluster 3 comprises G-box motifs (Fig. 3B). To examine the contributors to binding dynamics, we used linear regression to model the fold change of binding as a function of variables including basal binding of the TF (under mock treatment) and the number of occurrences (counts) of a set of non-redundant sequence features that capture the diversity of motifs. These sequence features were selected from major clusters of all the motifs found in the strongest 600 peaks (Cluster A-I motifs) and the dynamic and static peaks (Cluster 1 to 3 motifs) (Fig. S8). Examining the p-values of the regression coefficients (Fig. 3B) suggests that the primary motifs of ABF (which also represent cluster 3 motifs) and ANAC TFs are associated with enhanced dynamic binding, whereas basal binding and Cluster 2 motifs are associated with a negative impact on binding dynamics for a broad range of TFs (Fig. 3B, Table S6). Including Cluster 3 or Cluster 1 motifs in the regression results increases the explained variability by up to 20% (Fig. 3B). To visualize the impact of Cluster 3 G-box motif and Cluster 2 motif at the resolution of individual peaks, we plotted basal binding of TFs quantified by normalized read count against log2 fold change of binding after ABA treatment and assigned a color to individual binding events on the basis of the count of motifs in the same peak (Fig. 3C). The proportion of peaks containing Cluster 3 motif increases along with log2 fold change of binding, whereas the proportion of peaks containing Cluster 2 common motif are negatively correlated with log2 fold change of binding. These data suggest that the binding of a TF to Cluster 3 motif (likely the ABFs) and the binding of an unknown family of TFs to Cluster 2 motif, positively and negatively regulate the binding dynamics of neighboring TFs.
Figure 3. Determinants of TF binding dynamics.
(A) Hierarchical clustering of motifs enriched in dynamic and static peaks revealed three clusters. Each entry in the distance matrix is –log2(p-value) of motif similarity reported by Tomtom (44). (B) Linear regression of differential binding using basal binding and non-redundant sequence features identified positive and negative determinants of dynamic TF binding. Heatmap colors map to two-tailed t-test p-values on the regression coefficients for the null hypothesis that the coefficient is zero. The sequence features were selected from motifs enriched in the strongest peaks in ABA- and mock-treated conditions as well as dynamic and static peaks (Fig. S8). (C) Scatter plots on the left: basal binding of TFs quantified by normalized read count in the peak (x-axis) against log2(fold change) of TF binding after ABA treatment (y-axis), with color of each dot mapped to the number of indicated motifs in the same peak. The occurrence of Cluster 3 and Cluster 2 motifs over the distributions of log2 (fold change) of binding are shown in histograms on the right, with the same color code as the scatter plot. Proportion of peaks containing Cluster 3 motif increases along with log2(fold change) of TF binding for the indicated TFs, whereas proportion of peaks containing Cluster 2 motif are negatively correlated with log2(fold change) of TF binding.
Construction of an ABA response network
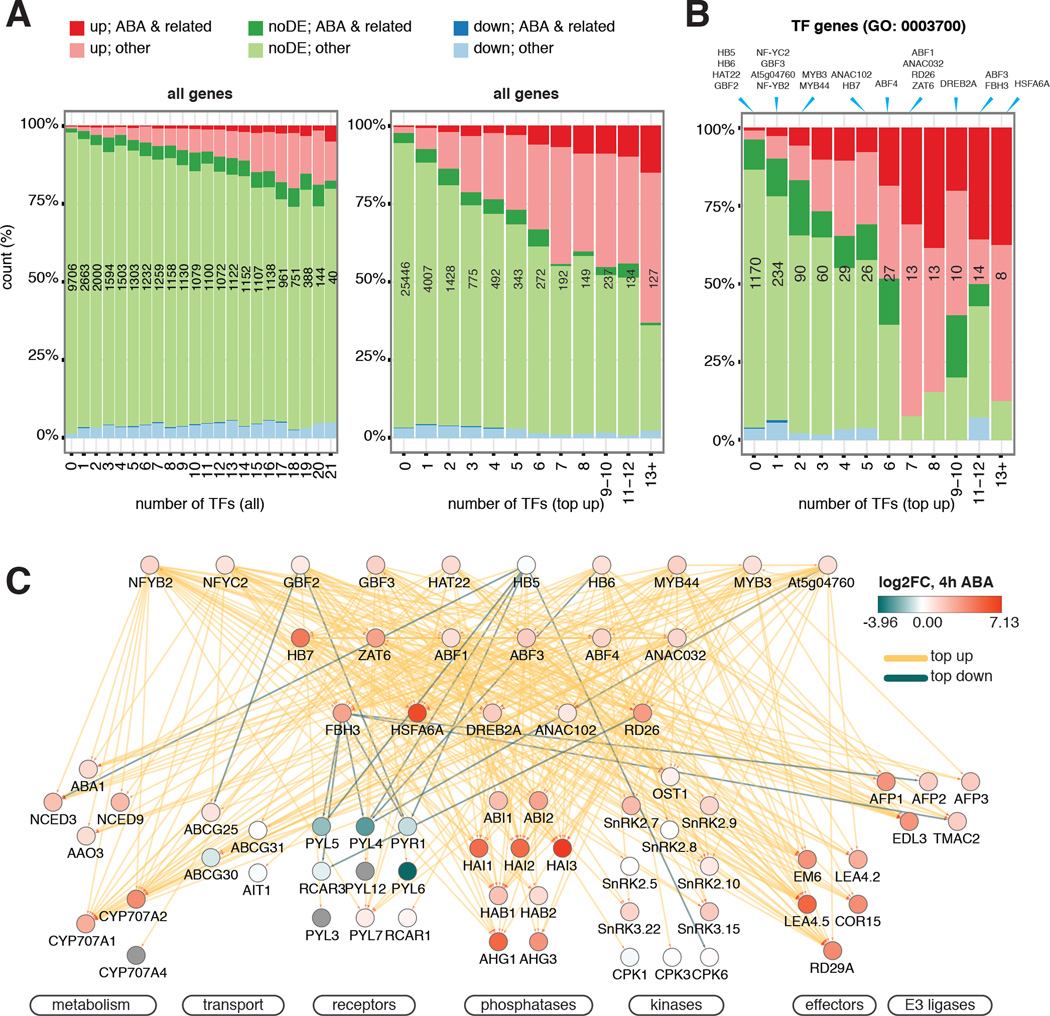
To confirm that dynamic binding is more robust than total binding in predicting gene expression and genes involved in ABA and related responses, we compared the expression and functional composition of genes grouped by the number of targeting TFs through either any type of binding or “top up” binding (Fig. 4A). Representation of both genes associated with ABA-related GO terms and ABA up-regulated genes increase more rapidly with the increase number of TFs that have “top up” binding. Therefore, we decided to use top-ranked dynamic TF binding triggered by ABA treatment to demonstrate the wiring of this ABA network using the core ABA metabolic and signaling genes and to calculate the hierarchical height of TFs in the network (Figs. 4B–C, S9A). TFs in the network are organized into three tiers by their hierarchical height (Figs. 4C, S9A). The level of DE of lower tier TFs is often amplified compared to upper tier TFs, which results in greater changes in binding dynamics likely as a result of greater protein accumulation (Figs. S3, 4, 9A). Negative regulators of ABA response, including genes encoding ABA catabolic enzymes, protein phosphatase 2Cs and E3 ligases, are often induced by ABA and are heavily targeted by multiple TFs through highly up-regulated TF binding (Fig. 4C). By contrast, positive regulators of ABA response can either be up-regulated due to increased TF binding, or down-regulated due to reduced TF binding (Fig. 4C). These results point to a transcriptional feedback strategy in ABA response, presumably to allow rapid restoration of normal growth once stress is lifted. Because some transcriptional responses triggered by ABA are similar to those triggered by natural stresses (Figs. S9A–B) such as high saline conditions, we also expect to see a similar organization of regulatory networks for other osmotic-related stresses.
Figure 4. TF network integrates expression and connectivity features of genes in ABA response.
(A–B) Expression and functional composition of all genes (A) and TF genes (B) are grouped by the number of targeting TFs through either any kind of binding or “top up” binding. “Top up” binding is a better predictor for both ABA-related BP functions and DE than “all” binding. The number of genes in each bin is shown in black. The bins to which of the TFs included in this study belong are indicated at the top of (B). (C) ABA pathway genes are subject to extensive feedback regulations and multi-TF dynamic binding. ChIPped TFs are arranged in three tiers by normalized hierarchy height. Target genes are grouped by function. Node color depicts changes of transcript abundance after 4 hours of ABA treatment. Edge color corresponds to TF binding dynamic categories.
Extensive targeting by ABA-responsive TFs appears to be specific to the ABA pathway, as the core ABA genes are targeted by significantly more TFs through “top up” binding than genes from other plant hormones (Fig. S10, Table S7). However, instances of hormone crosstalk can be observed in dynamically targeted DE genes. For example, both RGA-like 3 (RGL3), a master regulator of gibberellin response, and ACC synthase 2 (ACS2), an ethylene biosynthesis gene, were reported to be ABA-responsive (29, 30). We observed that dynamic binding is mainly contributed by the bZIP and the NF-Y factors to the promoter of RGL3, and by a diverse family of TFs to the gene body of ACS2 (Fig. S11). These results demonstrate the utility of these data to pinpoint regulatory regions that might modulate the expression of genes in one hormone response pathway by another.
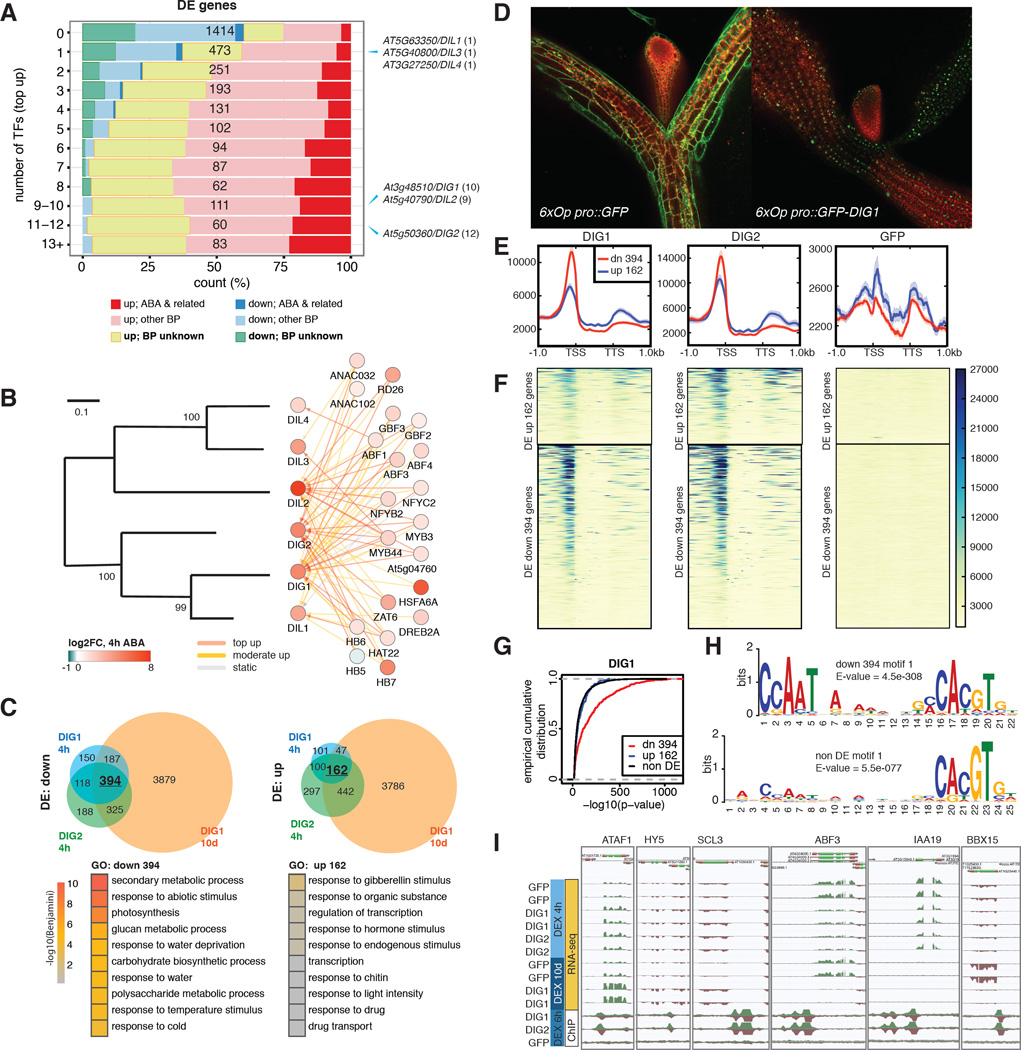
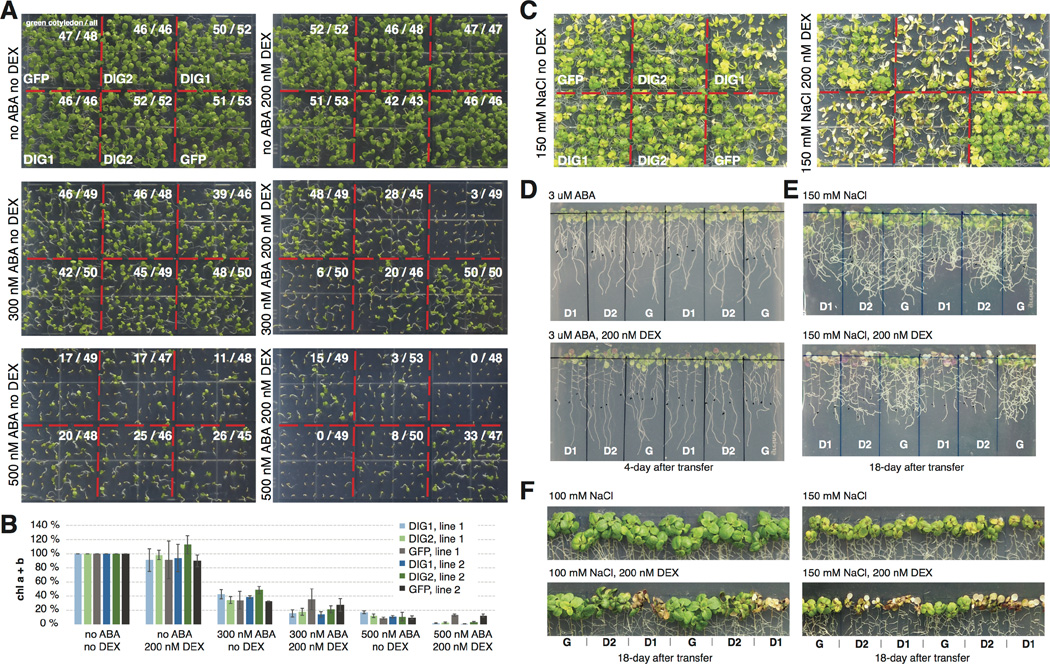
No GO term besides the ABA-related ones was enriched in DE genes heavily targeted by the 21 TFs through “top-up” binding. This is partially because more than one third (12136 / 33601) of the genes in the genome of Arabidopsis thaliana still have no information regarding their biological processes (BP) (Fig. 5A, Table S4). On the basis of a “guilt-by-association” paradigm (31), we speculate that many BP-unknown genes in Fig. 5A are also involved in ABA responses (Table S8). As a proof of principle, we functionally characterized a family in which all the members are BP-unknown and DE in response to ABA. In particular, three members in this family, At3g48510, At5g50360 and At5g40790 are heavily targeted by TFs through “top up” binding (Fig. 5A–B). Little is known about this family except that the proper expression of At3g48510 relies on core ABA signaling (32). In addition, predicted proteins of this family contain no known domains. We generated dexamethasone (DEX)-inducible lines expressing GFP fusion of the two most heavily targeted genes, At3g48510 and At5g50360. Analysis by RNA-seq showed that a few hundred DE genes were consistently identified from both short-term (4h) and long-term (10d) DEX induction of the two genes (Fig. 5C, Table S8). To reflect their regulation and function, these genes were named Dynamic Influencer of Gene expression 1 (DIG1) and DIG2 and their homologs were named DIG-likes (DILs). ABA-related GO terms such as response to water deprivation were enriched in DIG down-regulated genes (Fig. 5C). Confocal imaging further showed DIGs were localized to the nucleus (Fig. 5D). We then tested whether the DIGs are transcriptional regulators. ChIP-seq of DEX inducible GFP-DIGs showed that the DIGs bind chromatin. Moreover, stronger binding was observed in the promoter of DIG down-regulated genes than up-regulated ones or non-DE genes (Fig. 5E–F). De novo motif discovery identified a CCAAT(n)8 ABRE motif strongly enriched near the DIG1 binding sites within 1 kb of DIG down-regulated genes. By contrast, either a weaker motif or no similar motif was enriched near DIG binding sites in the corresponding regions of non-DE genes or DIG up-regulated genes (Fig. 5G–H). Several ABA-responsive or developmental TFs are targeted by DIGs and differentially expressed upon the induction of DIGs (Table S9). Among these, ATAF1, HY5 and ABF3 have been linked to ABA sensitivity (33–35), whereas HY5, SCL3 and perhaps IAA19 have developmental roles (34, 36, 37). Sequence analysis revealed that DIGs are conserved between monocots and dicots (Fig. S11). A remotely-related clade of DIG contains a gene Sdr4, which regulates seed dormancy in rice (38) (Fig. S12). The Sdr4 paralog in Arabidopsis is also dynamically targeted by multiple ABA-responsive TFs and differentially expressed in response to ABA (Table S1). However, the functionally important amino acid residues of Sdr4 are not conserved in the DIGs and their homologs (Fig. S13) (38). Therefore, genes in the DIG and Sdr4 clades may exert ABA-related functions through distinct mechanisms. Finally, inducible expression of DIGs enhances ABA sensitivity as assayed by cotyledon greening (Figs. 6A–B) and lateral root growth (Fig. 6D). Similarly, enhanced growth inhibition of DIG lines can also be observed after prolonged growth under high NaCl conditions (Figs. 6C, 6E–F). Combined, our results suggest that DIGs are a family of transcriptional regulators with broad roles that include regulating gene expression affecting ABA sensitivity and salt stress responses.
Figure 5. Network analysis identifies new transcriptional regulators of ABA response.
(A) Expression and functional composition of DE grouped by the number of targeting TFs through “top up” binding. Number of genes in each bin is shown in black. The bins to which the DIG/DIL genes belong are indicated on the right, with number of targeting TFs shown in parentheses. (B) DIG/DILs are regulated by multiple ABA-responsive TFs. Left panel: a phylogram of Arabidopsis DIG/DIL proteins. Right Panel: TFs targeting the DIG/DIL genes. (C) DEX-induction of DIGs results in DE of stress- and water-related genes. Upper panels: DE genes by DIGs after indicated period of DEX treatment. Lower panels: top GO terms enriched in DIG DE genes. (D) Confocal imaging of 9-day-old DEX-treated transgenic seedlings shows DIG1 is nuclear localized. (E–F) Metagene profiles (E) and heatmaps (F) of normalized ChIP-seq read counts surrounding DIG DE genes. Down-regulated genes are often associated with strong DIG binding in their promoters. (G) Empirical cumulative distributions of -log10(p-value) of ChIP-seq peaks of DIG1 show it bound more strongly to DIG down-regulated genes than to up-regulated or non-DE genes. (H) A CCAAT(n)8 ABRE motif is strongly enriched near DIG1 binding sites residing within 1 kb of DIG down-regulated genes. Either a weaker motif or no similar motif is enriched in the corresponding regions of non-DE genes or DIG up-regulated genes. (I) Induction of DIGs results in DE of ABA- and developmental-related TFs.
Figure 6. DIG inducible lines exhibit enhanced sensitivity to ABA and salt.
(A–B) ABA-dependent delay of cotyledon greening in 8-day-old seedlings were further amplified upon DEX-mediated induction of DIG1 and DIG2 compared to GFP control quantified by count of green cotyledons (A) and measurement of relative chlorophyll content (B). Error bar reflects the 95% confidence interval around the mean estimate calculated from 3 biological replicates of ca. 50 eight-day-old seedlings each. (C) NaCl-dependent bleaching was observed in 4-week-old plants upon DEX-mediated induction of DIG1 and DIG2. (D-E) DEX-mediated induction of DIG1 (D1) and DIG2 (D2) resulted in more severe inhibition of lateral root growth than GFP (G) control plants on ABA (D) and NaCl (E) plates. (F) DEX-mediated induction of DIG1 and DIG2 led to over-accumulation of pigments in leaves. In D-F, seedlings were transferred to the indicated plates after grown on LS plates for seven days.
Conclusions
We performed a systematic study of a transcriptional network by combining dynamic binding data of 21 TFs and time series RNA-seq data in response to a stimulus by the plant hormone ABA. We found that dynamic TF binding measured at a single time point correlated with the transcriptome changes over a prolonged span of time. Consistent with yeast and animals (2, 3, 39, 40), transcription of genes in Arabidopsis are often subject to a complex regulation of multiple TFs. We further demonstrated that dynamic binding, especially by multiple TFs, is more functionally relevant than static TF binding in correlation with differential gene expression. We speculate that this is because an expression scheme with coordinated changes in the binding dynamics of multiple TFs would ensure robust responsiveness of target genes to a stimulus. This observation would have a broad application to both plants and other species, including prioritizing studies of 1) TF binding events and cis-regulatory elements, 2) functionally unknown genes in a pathway.
In plants, studies of transcriptional regulation are often focused on master regulators (33, 41, 42). Our data confirmed the importance of master regulators in plants. For instance, we showed that ABFs and a physical interactor NF-YB2 ranked as top contributors to explain gene expression. In addition, the primary binding motif of ABFs also enhances the binding dynamics of many other ABA-responsive TFs. However, more than just the master regulators are required to attain complex transcriptome changes to a stimulus, as many ABA-responsive genes are dynamically targeted by multiple TFs. Therefore, ABA response can be viewed as orchestrated by a handful of master regulators and facilitated by other TFs, where coordinated signaling and regulatory response lead to a rapid elicitation of transcriptome changes.
As indicated by GO annotation, network analyses of this study recovered genes affecting all aspects of ABA-related processes such as seed development and response to osmotic stresses. In planta ectopic expression of of several members of a newly discovered family of transcriptional regulators also exhibited altered response to both ABA and high salinity. Therefore, although we carried out the experiments using seedlings, the discoveries may be directly applicable to a broader range of development stages and stress scenarios. Emerging technologies to optimize plant water use have been developed based on the in-depth characterization of ABA perception (43). Knowledge derived from further studies of the genes uncovered in this study may also prove valuable to global agriculture, possibly enabling new strategies for plants to respond to the challenges of ongoing drought and groundwater depletion in changing environments.
Materials and Methods
Plant materials
Recombineering lines for the ChIP-seq experiments were generated as previously described (19) with minor modifications. A YPet-6xHis-3xFLAG tag and a 3xFLAG-YPet tag were designed for C-terminus and N-terminus fusion to the TFs of interest (Table S3, Fig. S14). To abolish weak dimerization of YPet (45), an A206K point mutation was introduced by primers 5’- ATCCTTGAAGAGCTTAGACTGGTAAGA -3’ and 5’- TCTTACCAGTCTAAGCTCTTCAAGGAT -3’. After floral dip of wild-type Col-0 plants, T1 seeds were pooled and transgenic plants were selected on plates containing 1x Linsmaier and Skoog (LS) pH buffered basal salts, pH 5.7 (Caisson laboratories, UT, USA, cat. # LSP03-1LT) with 0.7% agar and 15 µg/ml glufosinate ammonium (Fisher Scientific, NH, USA, cat. # N-12111-250MG). Single-insertional transgenic lines were selected by Chi-square test from T2 plants on 1x LS plates containing 15 µg/ml glufosinate ammonium. The expression of the tagged TFs was confirmed by western blotting. Homozygous transgenic lines were selected from the subsequent generation for bulking seeds. DEX-inducible lines for the functional characterization of DIGs were generated by cloning the coding sequence of DIGs into p35S::LhGR-p6xOP::mGFP-attL1-ccdB-attR1 cassette by LR combination. After floral dip of wild-type Col-0 plants, T1 seeds were pooled and transgenic plants were selected by hygromycin.
ChIP-seq experiments and analysis
0.4 g seeds were surface sterilized by 50% bleach + 0.05% Triton-X100 for 10 minutes. After 4 days of stratification at 4 °C, seeds were spread on nylon mesh (Component Supply, FL, USA, cat. # U-CMN-215) in 6 hydroponics (Sigma-Aldrich, MO, USA, cat. # P1552) containing 1x pH buffered LS basal salts. After exposure under light for 4 hours to enhance germination, seeds were grown in dark at 22 °C for 3 days. Etiolated seedlings were then switched to 1x LS buffer containing either (+/−)-ABA (MP biomedicals LLC, CA, USA, cat. # 190673) dissolved in 100% ethanol at a final concentration of 10 µM or ethanol alone as mock and treated for 4 hours in dark before ChIP as previously described (46). Briefly, harvested seedlings were cross-linked by 1% formaldehyde solution (Sigma-Aldrich, cat. # F8775) under vacuum for 20 minutes. After nuclei isolation, chromatin was sonicated to 100–400 bp fragments. Tagged TFs in the transgenic lines were immunoprecipitated by a rabbit polyclonal anti-GFP antibody (Thermo Fisher Scientific, MA, USA, cat. # A11122). After elution, reverse crosslinking and DNA purification, Illumina TruSeq libraries were constructed according to manufacturer’s protocols. All ChIP-seq experiments in both ABA and ethanol mock-treatment conditions were done with biological replicates. Uniquely mapped sequencing reads to the TAIR10 genome assembly (Bowtie v0.12.7) (47, 48) were used to call peaks by the IDR pipeline utilizing MACS2 (49) with mock IP of wild-type Col-0 ChIPped by the anti-GFP antibody as a control. Peaks with a p-value <= 1e-16 were kept and differential binding of TFs were analyzed by DiffBind (v1.10.1 with edgeR 3.0.8) (23). To calculate TF binding similarity in Figure 1E, the center of peaks (termed “summits”) of all ChIPped TFs were pooled together to create a union list. Sequencing coverage within 50 base pairs of summits in the union list was counted and normalized by deepTools (v1.5.8) (50). Pairwise Pearson correlation between samples was used as entries in the distance matrix to plot the heat map in Fig. 1E. Hierarchy height of ChIPped TFs was calculated as previously described (40):
where O and I are out-degree and in-degree of examined TF through top-ranked dynamic binding. Peaks in each dynamic binding categories were associated to TAIR10 annotated genes within 1000 bp from the summit of the peaks, using the R BioConductor package ChIPpeakAnno (v2.12.1) (51).
Motif discovery and modeling the contribution of individual motif to TF binding dynamics
De novo motif discovery was carried out by meme-chip (meme 4.9.1) using a background file calculated from TAIR10 intergenic sequences (47). Top five enriched motifs identified within 50 base pairs of the summits were filtered at e-value cutoff of 1e-05. To model the contribution of individual features, a set of non-redundant sequence features were selected to represent the overall motif diversity. To do this we first assembled a set of 135 motifs in our dataset, consisted of the two most enriched motifs in the top 600 peaks in ABA- and mock-treated conditions for each TF, as well as top five motifs enriched in dynamic and static peaks for all TFs. The motifs were clustered by applying hierarchical clustering using motif distances calculated by Pearson Correlated Coefficients as column comparison metric and Ungapped Smith-Waterman alignment method (52, 53). Dynamic tree cut of the clustering dendrogram (54) identified 19 major clusters (color of dendrogram branch and the left of the annotation tracks in Fig. S8). As several of the clusters contain similar motifs (for example, the G-boxes and the AG-rich motifs are split into multiple clusters), we selected 11 sequence features to capture the diversity in this set of motifs indicated by dark red color of motif name and dark red color in the right annotation track in Fig. S8. Basal binding was measured as log2(normalized read counts) under mock treatment and occurrences of motifs were assessed by FIMO (55) at the p-value cutoff of 0.0004. These features were used to fit log2(fold change) of the binding of indicated TFs in Fig. 2D between ABA- and mock treatment. Relative changes of explained variability was calculated as:
where R2′ and R2 are the adjusted R2 from lm() output that includes and excludes Cluster II or Cluster III motif as a feature, respectively.
RNA-seq experiments and analysis
For ABA time series experiments, two biological replicates of 3-day-old etiolated, hydroponic-grown wild-type Col-0 seedlings were treated either by 10 µM (+/−)-ABA (MP biomedicals LLC, cat. # 190673) dissolved in ethanol or ethanol-only mock control for 1, 4, 8, 12, 24, 36, and 60 hours. For DEX treatment, short term experiment was carried out by treating 3-day-old etiolated, DEX-inducible GFP-DIG1, GFP-DIG2 or GFP lines with 10 µM DEX (Sigma Aldrich, cat. # D9184); long term experiment was carried out by growing the same lines of plants containing 500 nM DEX for 10 days. Total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen, CA, USA, Cat. # 74903), and cDNA libraries were constructed using the TruSeq Stranded Total RNA LT Sample Prep Kit (Illumina, CA, USA, Cat. # 15032611) according to manufacturers' instructions. Single-end reads were generated by the HiSeq 2500 Sequencing System (Illumina) and mapped to TAIR10 genome assembly using TopHat 2 (v2.0.8) (56). Mapped reads with mapping score equal to or larger than 10 were counted by HTSeq (v0.5.4) (57) and analyzed by edgeR (v3.6.2) (18) to identify differentially expressed genes using contrasts between ABA- and mock-treated samples at each time point and false discovery rate 0.01 or 0.05 as thresholds.
DREM
The Dynamic Regulatory Events Miner (DREM) (25, 58), integrates TF-gene interactions from ChIP-seq experiments with time series gene expression data to identify patterns of temporal gene expression, the associated regulators and the dynamics of the interactions. Splits in the reconstructed network (green nodes in Figs. 1B, 2C, S1, S5) represent divergence of genes that are co-regulated up to that point and can be annotated by DREM with the TFs that are predicted to regulate them, allowing us to associate the temporal information (the timing of the splits) with the interaction information either directly measured by ChIP-seq (Fig. S1) or inferred from the AGRIS database (26), PBM (13) and DAP-seq (15) data (Fig. S5). The analysis performed here used the log fold change of 3061 DE genes (Table S1) identified in the ABA time series RNA-seq data. DREM paths were created using all DE genes without further filtering.
For GO enrichment in DE genes targeted by categories of dynamic binding peaks (Figure 2D), we defined the genes in distinct DREM paths as foreground, and all expressed genes as background, and retrieved the Functional Annotation Chart with EASE score (modified p-value) threshold of 0.1 and count threshold of 2, using functionalities provided by the R BioConductor package RDAVIDWebService (59) to query the DAVID web service (60). The GO terms in GO_TERM_BP_FAT with FDR<=1% from all target gene sets are combined, and the enrichment p-values of these terms are retrieved for each gene set to create the heatmap in Figure 2D. If a term is not reported to be significant for a target set, its p-value is set to 0.1 (the p-value threshold).
Modeling the contribution of individual TF to gene expression
We adopted an approach similar to previous regression-based models that relate gene expression to TF binding (61, 62). We first defined TF affinity score (TFAS), Aij, for TF j on gene i, using the peak closest to the TSS of the gene:
where g is the log2 TMM normalized read counts of the peak, d is the distance of the peak summit to TSS. d0 is set to 1000. For N genes and M TF, we constructed one N × M TFAS matrix for the ABA treatment and one for the ethanol mock treatment, and concatenated these two matrices horizontally to create a final N × 2M matrix A. We centered and scaled each column of A and fit a log-linear model:
where Yi is the fold change in expression of gene i at 4h ABA treatment compared to mock. We limited the model training and testing to genes that are differentially expressed at 4h with FDR<=0.01 and those are not differentially expressed at all time points (FDR>0.7). A glmnet regression model (63) was trained on 75% of the genes by 5 repeats of 10-fold cross-validation using the caret package in R (64) with tuning metric set to RMSE and the elastic net mixing parameter α = 0 to allow selection correlated TFAS features. The “best” rule was used to choose a value for the tuning parameter (in this case, the regularization parameter λ), i.e., a value that minimized the average RMSE of the regression on the 50 resampling of the training set. The glmnet model was then fitted using the chosen λ value to arrive at the regression coefficients in the final model. The unscaled coefficients of the TFAS features are plotted as binding feature importance in Fig. 2F.
Confocal imaging
Nine-day-old DEX inducible GFP and GFP-DIG1 seedlings grown on 1x LS plates containing 200 nM DEX and 300 nM ABA were imaged by Zeiss 710 confocal microscope under an Argon laser at 488 nm. GFP signal was captured within the 493 – 548 nm emission window and was pseudo-colored in green. Auto-fluorescence from chloroplasts was captured within the 569–695 nm emission window and was pseudo-colored in red.
Sequence analysis of DIGs and their homologs
Protein sequence of DIG1 was used as a query to search for homologous protein sequences in Arabidopsis thaliana by the BLASTP search tool on EnsemblPlants (65). The resulting six protein sequences (Q9FK36, Q9SMP6, Q9FGW7, Q9LK28, Q9FKS6, Q9FKS7) were used to query Arabidopsis thaliana, Glycine max, Solanum lycopersicum, Oryza sativa japonica and Zea mays by BLASP, resulting in 21 homologous sequences. These sequences were aligned by MEGA6 (66) using distance-based maximum likelihood method, and bootstrap values were generated from 1000 replications.
Chlorophyll measurement
Chlorophyll content was determined as previously described (67). Briefly, each sample consisting of ca. 50 seeds were germinated and grown on LS plates supplemented with or without ABA and DEX for eight days. The seedlings were collected and ground in liquid nitrogen. Chlorophyll were extracted by 80% acetone until pellets were almost white. Absorbance was measured at 647 and 664 nm in a DU-730 spectrophotometer (Beckman Coulter, CA, USA). Chlorophyll content was determined as
Chlorophyll content of each transgenic line was then normalized by the corresponding seedlings grown on LS plates containing no DEX or ABA. The 95% confidence interval around the mean estimate was calculated from 3 biological replicates.
Supplementary Material
Acknowledgments
We thank F. Turck and K.N.Chang for discussions of the ChIP procedures; J. Alonso for discussions of the recombineering procedures; J. Chory, N. Fedoroff and M. Zander for comments; C. Serrano, J. P. Saldierna and A. Nasamran for cloning and plant maintenance; U. Pedmale and M. Xie for sharing plasmids; Jamie Simon for for assistance with graphics. L.S. and S.C.H. were supported by Salk Pioneer Postdoctoral Fellowships. This work was supported by grants from the Gordon and Betty Moore Foundation (GBMF 3034 to J.R.E), NSF (MCB-1024999 to J.R.E. and DBI- 1356505 to Z.B.-J.) and NIH (U01 HL122626-01 to Z.B.-J.). J.R.E. is an investigator of the Howard Hughes Medical Institute. All of the reported RNA-Seq and ChiP-seq data has been deposited at NCBI (GEO accession no. GSE80568). Genome brower tracks and motifs of our dataset can be viewed at www.ABAtf.net.
References and Notes
- 1.Consortium EP, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerstein M. Integrative Analysis of the Caenorhabditis elegans Genome by the modENCODE Project. Science (80-.) 2010;47:7–10. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roy S, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- 5.Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. 2010;61 doi: 10.1146/annurev-arplant-042809-112122. http://www.ncbi.nlm.nih.gov/pubmed/20192755. [DOI] [PubMed] [Google Scholar]
- 6.Park S-Y, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma Y, et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 8.Santiago J, et al. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature. 2009;462:665–668. doi: 10.1038/nature08591. [DOI] [PubMed] [Google Scholar]
- 9.Santiago J, et al. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 2009;60:575–588. doi: 10.1111/j.1365-313X.2009.03981.x. [DOI] [PubMed] [Google Scholar]
- 10.Furihata T, et al. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. U. S. A. 2006;103:1988–1993. doi: 10.1073/pnas.0505667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujii H, et al. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462:660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujita Y, Fujita M, Shinozaki K, Yamaguchi-Shinozaki K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 2011;124:509–525. doi: 10.1007/s10265-011-0412-3. [DOI] [PubMed] [Google Scholar]
- 13.Weirauch MT, et al. Determination and Inference of Eukaryotic Transcription Factor Sequence Specificity. Cell. 2014;158:1431–1443. doi: 10.1016/j.cell.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jolma A, et al. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature. 2015;527:384–388. doi: 10.1038/nature15518. [DOI] [PubMed] [Google Scholar]
- 15.O’Malley RC, et al. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell. 2016;165:1280–1292. doi: 10.1016/j.cell.2016.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cumbie JS, Filichkin SA, Megraw M. Improved DNase-seq protocol facilitates high resolution mapping of DNase I hypersensitive sites in roots in Arabidopsis thaliana. Plant Methods. 2015;11:42. doi: 10.1186/s13007-015-0087-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 2015;2015:21.29.1–21.29.9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alonso JM, Stepanova AN. Bacterial artificial chromosomes. 2015;1227 [Google Scholar]
- 20.Lim CW, Kim JH, Baek W, Kim BS, Lee SC. Functional roles of the protein phosphatase 2C, AtAIP1, in abscisic acid signaling and sugar tolerance in Arabidopsis. Plant Sci. 2012;187:83–88. doi: 10.1016/j.plantsci.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 21.Okamoto M, et al. CYP707A1 and CYP707A2, which encode abscisic acid 8’-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol. 2006;141:97–107. doi: 10.1104/pp.106.079475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumimoto RW, et al. NUCLEAR FACTOR Y Transcription Factors Have Both Opposing and Additive Roles in ABA-Mediated Seed Germination. PLoS One. 2013;8:e59481. doi: 10.1371/journal.pone.0059481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stark R. DiffBind : Differential binding analysis of ChIP-Seq peak data. 2015:1–31. [Google Scholar]
- 24.Gene Ontology Consortium, Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schulz MH, et al. DREM 2.0: Improved reconstruction of dynamic regulatory networks from time-series expression data. BMC Syst. Biol. 2012;6:104. doi: 10.1186/1752-0509-6-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Celli F, et al. AGRIS: providing access to agricultural research data exploiting open data on the web. F1000Research. 2015;4:110. doi: 10.12688/f1000research.6354.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oldfield A, et al. Histone-Fold Domain Protein NF-Y Promotes Chromatin Accessibility for Cell Type-Specific Master Transcription Factors. Mol. Cell. 2014;55:708–722. doi: 10.1016/j.molcel.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bailey TL, Johnson J, Grant CE, Noble WS. The MEME Suite. Nucleic Acids Res. 2015;43:W39–W49. doi: 10.1093/nar/gkv416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piskurewicz U, Lopez-Molina L. The GA-signaling repressor RGL3 represses testa rupture in response to changes in GA and ABA levels. Plant Signal.Behav. 2009;4:63–65. doi: 10.4161/psb.4.1.7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee IC, et al. Age-dependent action of an ABA-inducible receptor kinase, RPK1, as a positive regulator of senescence in arabidopsis leaves. Plant Cell Physiol. 2011;52:651–662. doi: 10.1093/pcp/pcr026. [DOI] [PubMed] [Google Scholar]
- 31.Lee I, Blom UM, Wang PI, Shim JE, Marcotte EM. Prioritizing candidate disease genes by network-based boosting of genome-wide association data. Genome Res. 2011;21:1109–1121. doi: 10.1101/gr.118992.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lumba S, et al. A mesoscale abscisic acid hormone interactome reveals a dynamic signaling landscape in arabidopsis. Dev. Cell. 2014;29:360–372. doi: 10.1016/j.devcel.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 33.Yoshida T, et al. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010;61:672–685. doi: 10.1111/j.1365-313X.2009.04092.x. [DOI] [PubMed] [Google Scholar]
- 34.Lee J, et al. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell. 2007;19:731–749. doi: 10.1105/tpc.106.047688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jensen MK, et al. ATAF1 transcription factor directly regulates abscisic acid biosynthetic gene NCED3 in Arabidopsis thaliana. FEBS Open Bio. 2013;3:321–327. doi: 10.1016/j.fob.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Z-L, et al. Scarecrow-like 3 promotes gibberellin signaling by antagonizing master growth repressor DELLA in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 2011;108:2160–2165. doi: 10.1073/pnas.1012232108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kohno M, Takato H, Horiuchi H, Fujita K, Suzuki S. Auxin-nonresponsive grape Aux/IAA19 is a positive regulator of plant growth. Mol. Biol. Rep. 2012;39:911–917. doi: 10.1007/s11033-011-0816-0. [DOI] [PubMed] [Google Scholar]
- 38.Sugimoto K, et al. Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice. Proc. Natl. Acad. Sci. U. S. A. 2010;107:5792–5797. doi: 10.1073/pnas.0911965107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu H, Gerstein M. Genomic analysis of the hierarchical structure of regulatory networks. Proc. Natl. Acad. Sci. U. S. A. 2006;103:14724–14731. doi: 10.1073/pnas.0508637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gerstein MB, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smaczniak C, et al. Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc. Natl. Acad. Sci. U. S. A. 2012;109:1560–1565. doi: 10.1073/pnas.1112871109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moreno-Risueno MA, et al. Transcriptional control of tissue formation throughout root development. Science (80-.) 2015;6:1–20. doi: 10.1126/science.aad1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park S-Y, et al. Agrochemical control of plant water use using engineered abscisic acid receptors. Nature. 2015;520:545–548. doi: 10.1038/nature14123. [DOI] [PubMed] [Google Scholar]
- 44.Bailey TL, Johnson J, Grant CE, Noble WS. The MEME Suite. Nucleic Acids Res. 2015;43:W39–W49. doi: 10.1093/nar/gkv416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat. Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 46.Song L, Koga Y, Ecker J. Profiling of Transcription Factor Binding Events by Chromatin Immunoprecipitation Sequencing (ChIP-seq) Curr. Protoc. plant Biol. 2016:293–306. doi: 10.1002/cppb.20014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lamesch P, et al. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012;40 doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Langmead B. Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinforma. 2010 doi: 10.1002/0471250953.bi1107s32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Landt SG, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42:W187–W191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu LJ, et al. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics. 2010;11:237. doi: 10.1186/1471-2105-11-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mahony S, Auron PE, Benos PV. DNA familial binding profiles made easy: Comparison of various motif alignment and clustering strategies. PLoS Comput. Biol. 2007;3:0578–0591. doi: 10.1371/journal.pcbi.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mercier E, Gottardo R. Motif Identification and Validation MotIV. 2014:1–15. [Google Scholar]
- 54.Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: The Dynamic Tree Cut package for R. Bioinformatics. 2008;24:719–720. doi: 10.1093/bioinformatics/btm563. [DOI] [PubMed] [Google Scholar]
- 55.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anders S, Pyl PT, Huber W. HTSeq A Python framework to work with high-throughput sequencing data. 2014 doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wise A, Bar-Joseph Z. cDREM: Inferring Dynamic Combinatorial Gene Regulation. J. Comput. Biol. 2015;22:324–333. doi: 10.1089/cmb.2015.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fresno C, Fernández Ea. RDAVIDWebService: A versatile R interface to DAVID. Bioinformatics. 2013;29:2810–2811. doi: 10.1093/bioinformatics/btt487. [DOI] [PubMed] [Google Scholar]
- 60.Jiao X, et al. DAVID-WS: A stateful web service to facilitate gene/protein list analysis. Bioinformatics. 2012;28:1805–1806. doi: 10.1093/bioinformatics/bts251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ouyang Z, Zhou Q, Wong WH. ChIP-Seq of transcription factors predicts absolute and differential gene expression in embryonic stem cells. Proc. Natl. Acad. Sci. U. S. A. 2009;106:21521–21526. doi: 10.1073/pnas.0904863106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McLeay RC, Lesluyes T, Cuellar Partida G, Bailey TL. Genome-wide in silico prediction of gene expression. Bioinformatics. 2012;28:2789–2796. doi: 10.1093/bioinformatics/bts529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 64.Kuhn M. Building Predictive Models in R Using the caret Package. J. Stat. Softw. 2008;28:1–26. [Google Scholar]
- 65.Kersey PJ, et al. Ensembl Genomes 2013: scaling up access to genome-wide data. Nucleic Acids Res. 2014;42:D546–D552. doi: 10.1093/nar/gkt979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Petrillo E, et al. A Chloroplast Retrograde Signal Regulates Nuclear Alternative Splicing. Science (80-.) 2014;344:427–430. doi: 10.1126/science.1250322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 69.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fujita M, et al. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 2004;39:863–876. doi: 10.1111/j.1365-313X.2004.02171.x. [DOI] [PubMed] [Google Scholar]
- 71.Tran L-SP, et al. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell. 2004;16:2481–2498. doi: 10.1105/tpc.104.022699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jensen MK, et al. The Arabidopsis thaliana NAC transcription factor family: structure-function relationships and determinants of ANAC019 stress signalling. Biochem. J. 2010;426:183–196. doi: 10.1042/BJ20091234. [DOI] [PubMed] [Google Scholar]
- 73.Liu Q, et al. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Narusaka Y, et al. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 2003;34:137–148. doi: 10.1046/j.1365-313x.2003.01708.x. [DOI] [PubMed] [Google Scholar]
- 75.Lee S-J, et al. DREB2C interacts with ABF2, a bZIP protein regulating abscisic acid-responsive gene expression, and its overexpression affects abscisic acid sensitivity. Plant Physiol. 2010;153:716–727. doi: 10.1104/pp.110.154617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang P, et al. Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc. Natl. Acad. Sci. U. S. A. 2013;110:11205–11210. doi: 10.1073/pnas.1308974110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Takahashi Y, et al. bHLH transcription factors that facilitate K+ uptake during stomatal opening are repressed by abscisic acid through phosphorylation. Sci. Signal. 2013;6:ra48. doi: 10.1126/scisignal.2003760. [DOI] [PubMed] [Google Scholar]
- 78.Liu W, et al. bHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism. New Phytol. 2014;201:1192–1204. doi: 10.1111/nph.12607. [DOI] [PubMed] [Google Scholar]
- 79.Chen YT, Liu H, Stone S, Callis J. ABA and the ubiquitin E3 ligase KEEP on GOING affect proteolysis of the Arabidopsis thaliana transcription factors ABF1 and ABF3. Plant J. 2013;75:965–976. doi: 10.1111/tpj.12259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Finkelstein R, Gampala SSL, Lynch TJ, Thomas TL, Rock CD. Redundant and distinct functions of the ABA response loci ABA-insensitive(ABI)5 and ABRE-binding factor (ABF)3. Plant Mol. Biol. 2005;59:253–267. doi: 10.1007/s11103-005-8767-2. [DOI] [PubMed] [Google Scholar]
- 81.Kang J, Choi H, Im M, Kim SY. Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell. 2002;14:343–357. doi: 10.1105/tpc.010362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lu G, Paul aL, McCarty DR, Ferl RJ. Transcription factor veracity: is GBF3 responsible for ABA-regulated expression of Arabidopsis Adh? Plant Cell. 1996;8:847–857. doi: 10.1105/tpc.8.5.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ciftci-Yilmaz S, Mittler R. The zinc finger network of plants. Cell. Mol. Life Sci. 2008;65:1150–1160. doi: 10.1007/s00018-007-7473-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu XM, et al. Phosphorylation of the zinc finger transcriptional regulator ZAT6 by MPK6 regulates Arabidopsis seed germination under salt and osmotic stress. Biochem. Biophys. Res. Commun. 2013;430:1054–1059. doi: 10.1016/j.bbrc.2012.12.039. [DOI] [PubMed] [Google Scholar]
- 85.Mittler R, et al. Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett. 2006;580:6537–6542. doi: 10.1016/j.febslet.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Conditions HS, et al. Arabidopsis Cys2 / His2-Type Zinc-Finger Proteins Function as Transcription Repressors under Drought. Society. 2004;136:2734–2746. doi: 10.1104/pp.104.046599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Causier B, Ashworth M, Guo W, Davies B. The TOPLESS Interactome: A Framework for Gene Repression in Arabidopsis. Plant Physiol. 2012;158:423–438. doi: 10.1104/pp.111.186999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kagale S, Links MG, Rozwadowski K. Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol. 2010;152:1109–1134. doi: 10.1104/pp.109.151704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ariel FD, Manavella PA, Dezar CA, Chan RL. The true story of the HD-Zip family. Trends Plant Sci. 2007;12:419–426. doi: 10.1016/j.tplants.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 90.Johannesson H, Wang Y, Engström P. DNA-binding and dimerization preferences of Arabidopsis homeodomain-leucine zipper transcription factors in vitro. Plant Mol. Biol. 2001;45:63–73. doi: 10.1023/a:1006423324025. [DOI] [PubMed] [Google Scholar]
- 91.Himmelbach A, Hoffmann T, Leube M, Höhener B, Grill E. Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J. 2002;21:3029–3038. doi: 10.1093/emboj/cdf316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Köllmer I, Werner T, Schmülling T. Ectopic expression of different cytokinin-regulated transcription factor genes of Arabidopsis thaliana alters plant growth and development. J. Plant Physiol. 2011;168:1320–1327. doi: 10.1016/j.jplph.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 93.Rivero RM, et al. Delayed leaf senescence induces extreme drought tolerance in a flowering plant. Proc. Natl. Acad. Sci. U. S. A. 2007;104:19631–19636. doi: 10.1073/pnas.0709453104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hwang SM, et al. Functional characterization of Arabidopsis HsfA6a as a heat-shock transcription factor under high salinity and dehydration conditions. Plant. Cell Environ. 2014;37:1202–1222. doi: 10.1111/pce.12228. [DOI] [PubMed] [Google Scholar]
- 95.Chen NZ, et al. AtHAP3b Plays a Crucial Role in the Regulation of Flowering Time in Arabidopsis during Osmotic Stress. J Biochem Mol Biol. 2007;40:1083–1089. doi: 10.5483/bmbrep.2007.40.6.1083. [DOI] [PubMed] [Google Scholar]
- 96.Jung C, et al. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 2008;146:623–635. doi: 10.1104/pp.107.110981. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.