Abstract
Chromatin associated proteins are key regulators of many important processes in the cell. Trypanosoma cruzi, a protozoa flagellate that causes Chagas disease, alternates between replicative and nonreplicative forms accompanied by a shift on global transcription levels and by changes in its chromatin architecture. Here, we investigated the T. cruzi chromatin proteome using three different protocols and compared it between replicative (epimastigote) and nonreplicative (trypomastigote) forms by high-resolution mass spectrometry. More than 2000 proteins were identified and quantified both in chromatin and nonchromatin extracts. Besides histones and other known nuclear proteins, trypanosomes chromatin also contains metabolic (mainly from carbohydrate pathway), cytoskeleton and many other proteins with unknown functions. Strikingly, the two parasite forms differ greatly regarding their chromatin-associated factors composition and amount. Although the nucleosome content is the same for both life forms (as seen by MNase digestion), the remaining proteins were much less detected in nonreplicative forms, suggesting that they have a naked chromatin. Proteins associated to DNA proliferation, such as PCNA, RPA, and DNA topoisomerases were exclusively found in the chromatin of replicative stages. On the other hand, the nonreplicative stages have an enrichment of a histone H2B variant. Furthermore, almost 20% of replicative stages chromatin-associated proteins are expressed in nonreplicative forms, but located at nonchromatin space. We identified different classes of proteins including phosphatases and a Ran-binding protein, that may shuttle between chromatin and nonchromatin space during differentiation. Seven proteins, including those with unknown functions, were selected for further validation. We confirmed their location in chromatin and their differential expression, using Western blotting assays and chromatin immunoprecipitation (ChIP). Our results indicate that the replicative state in trypanosomes involves an increase of chromatin associated proteins content. We discuss in details, the qualitative and quantitative implication of this chromatin set in trypanosome chromatin biology. Because trypanosomes are early-branching organisms, this data can boost our understanding of chromatin-associated processes in other cell types.
Chromatin is formed by DNA complexed with proteins and RNAs. Approximately 146 bp of DNA wraps around the histone octamer (composed by two copies of each of the canonical histones H2A, H2B, H3, and H4) forming the nucleosomes, the basic structural unit of chromatin. A fifth histone (H1) associates with the nucleosomes and seals the DNA turns (1). Chromatin is the substrate of many important processes, like DNA repair and replication, gene regulation and transcriptional control. Classically, chromatin can be divided as euchromatin, composed of less condensed and actively transcribed regions, and heterochromatin, comprising more compacted and silent regions. However, this binary classification is becoming obsolete as meta-analysis of the interaction of proteins with DNA, and histone post-translational modifications (PTMs)1, indicates a more complex pattern with at least five different chromatin subtypes (2).
Over the last years, many tools have been used to analyze chromatin proteome. Histones PTMs and chromatin binding proteins have been detected by specific antibodies followed by mass spectrometry (MS) and chromatin immunoprecipitation (ChIP) analyses (3–5). MS became a powerful technique for epigenetics research. For example, many studies based on MS made advances in characterizing human mitotic chromosomes (6–10). The first studies detected less than 80 proteins, whereas in 2010, a proteomic study together with machine learning techniques identified ∼4000 proteins, including many uncharacterized proteins associated with the chromatin (11). More recently, more than 3900 phosphorylation sites were identified in proteins interacting with the chromatin (12).
T. cruzi is the etiological agent of Chagas disease, a parasitic disease that affects millions of people, mainly in Latin America. During its life cycle, T. cruzi has distinct cellular forms that are either able to divide (epimastigote and amastigote) or to actively infect cells (trypomastigote) (13). As other eukaryotes, T. cruzi has its chromatin organized into nucleosome filaments forming 10 nm fibers. However, neither 30 nm fibers, nor condensed chromosomes are observed at mitosis (14). The T. cruzi nucleosome is composed of canonical histones that are very divergent from fungi and other metazoan (15, 16). Histone H1 is also found in these parasites, but it is formed only by regions similar to the C-terminal domain of higher eukaryotes. It is believed that the lack of the histone H1 globular domain may be associated with a more relaxed chromatin structure observed in trypanosomes.
During life cycle, trypanosomes change their nuclear structure. The epimastigote form presents a round nucleus, a defined nucleolus and relatively small amounts of peripheral heterochromatin. In contrast, the trypomastigote form exhibits an elongated nucleus, no identifiable nucleolus and a more abundant and dispersed heterochromatin. These changes are accompanied by a decrease in transcription rates when the replicative forms transform into nonreplicative ones (17). It is unknown, however, how these changes in the nuclear structure are achieved during the differentiation and what characterizes the chromatin in these different stages.
Here we analyzed the chromatin content of epimastigotes and trypomastigotes by high resolution mass spectrometry to identify and quantify proteins in chromatin (C) as well as in nonchromatin (NC) fractions. Different classes of well-known chromatin proteins and proteins with apparent nonchromatin related function were detected. Surprisingly, the nonreplicative stages have a very poor protein content but an enrichment of a histone H2B variant when compared with replicative stages. Comparing proteins that may shuttle between chromatin and nonchromatin space upon differentiation, we found two interesting groups with different biological functions as well as putative chromatin proteins with different location life form-dependent. Additionally, we confirmed the association of some proteins (with unknown functions) to chromatin and discussed in details their possible roles regarding different regulatory aspects of trypanosomes chromatin biology.
EXPERIMENTAL PROCEDURES
1. Parasite Cultures
T. cruzi (Y strain) epimastigotes were cultured in liver infusion tryptose (LIT) medium supplemented with 10% fetal bovine serum (FBS) at 28 °C (18). Trypomastigotes forms (Y strain) were obtained from the supernatant of infected monolayers of LLC-MK2 cells in culture medium DMEM with 10% FBS at 37 °C, 5% CO2, as described previously (19).
2. Chromatin Extraction
Chromatin was extracted from 5 × 108 epimastigotes and trypomastigotes forms of T. cruzi using either protocol 1, 2 or 3. Protocol 1 was based on (20) whereas protocol 2 was based on (21). Both are described in details at (22). Protocol 3 was described in (23), with a few modifications. 5 × 108 epimastigotes and trypomastigotes were resuspended in buffer A (10 mm HEPES pH 7.9, 10 mm KCl, 1.5 mm de MgCl2, 340 mm sucrose, 10% glycerol, 1 mm DTT, 10 mm sodium butyrate, 0.1% Triton X-100, supplemented with phosphatase and protease inhibitors, 50 mm NaF, 1 mm Na3VO4, 1 mm de PMSF and cOmpleteTM EDTA-free protease inhibitor mixture - Roche) and incubated on ice for 8 min and centrifuged at 1300 × g for 5 min at 4 °C. The supernatant was saved (nonchromatin fraction) and the pellet was washed once in buffer A. The pellet was then resuspended in buffer B (3 mm EDTA, 0.2 mm EGTA, 1 mm DTT, 10 mm sodium butyrate, 50 mm NaF, 1 mm Na3VO4,1 mm PMSF and cOmpleteTM EDTA free protease inhibitor mixture -Roche) and incubated on ice for 30 min. Samples were centrifuged at 1700 × g for 5 min at 4 °C and the pellet was resuspended in buffer B with 250 U of benzonase and incubated for 30 min at 37 °C under agitation (1,400 rpm). A buffer containing 8 m urea, 75 mm NaCl, 50 mm Tris-HCl pH 8 was added to the samples, and the tubes were sonicated for 3 cycles of 30 s/45 s rest on a sonicator bath. Samples were centrifuged at 21,000 × g for 10 min at 4 °C and the final supernatant was kept for analysis (chromatin fraction). The nonchromatin fraction from protocol 1 and 2 correspond to the supernatant of the first lysis round.
3. Protein Digestion and Stage Tip Fractionation (SCX)
In-gel tryptic protein digestion was performed as described in (24). In-solution digestion was described in details at (22). Briefly, after TCA precipitation, 150 μg of protein extracts were reduced with 5 mm of DTT for 30 min at room temperature, alkylated with 14 mm of iodoacetamide in the dark for 30 min and digested with 0.5 μg of Lys-C (Promega, Madison, Wisconsin) for 4 h at 37 °C, under agitation (900 rpm). Sequentially, samples were digested with 0.75 μg of trypsin (Sigma) in the presence of 10 mm of Tris-HCl pH 8 and 2 mm CaCl2 overnight at 37 °C, under agitation (900 rpm). The reactions were stopped with 5% formic acid and vacuum dried. After protein digestion, the peptides were cleaned up for detergent removal by hydrophilic interaction chromatography-HILIC- (The Nest Group, Inc., Southborough, Massachusetts), according to instructions of the manufacturer. Samples were redissolved in 400 μl of 0.1% TFA and desalinated using the Sep-pak Light tC18 column (Waters, Milford, Massachusetts). After desalination, samples were fractionated using strong cation exchange (SCX) offline chromatography as described in (25), with some modifications (22). In short, peptides were eluted in six fractions based on ammonium acetate concentration (50, 100, 200, and 500 mm) in 0.3% TFA/20% acetonitrile, followed by 5 and 15% of ammonium hydroxide in 80% acetonitrile. Samples were dried and analyzed by mass spectrometry.
4. LC-MS/MS Analysis
Peptides were resuspended in 0.1% formic acid and injected in an in-house made 5 cm reversed phase pre-column (inner diameter 100 μm, filled with a 10 μm C18 Jupiter resins -Phenomenex, Torrance, California) coupled to a nano HPLC (NanoLC-1DPlus, Proxeon, Thermo Fischer Scientific, Waltham, Massachusetts). The peptide fractionation was carried on an in-house 10 cm reversed phase capillary emitter column (inner diameter 75 μm, filled with 5 μm C18 Aqua resins-Phenomenex) with a gradient of 2–35% of acetonitrile in 0.1% formic acid for 52 min followed by a gradient of 35–95% for 5 min at a flow rate of 300 nl/min. The eluted peptides were directly analyzed in LTQ-OrbitrapVelos (Thermo Scientific). The source voltage and the capillary temperature were set at 1.9 kV and 200 °C, respectively. The mass spectrometer was operated in a data-dependent acquisition mode to automatically switch between one Orbitrap full-scan and ten ion trap tandem mass spectra. The FT scans were acquired from m/z 200 to 2000 with mass resolution of 30,000. MS/MS spectra were acquired at normalized collision energy of 35%. Singly charged and charge-unassigned precursor ions were excluded. The dynamic exclusion was set as: 45 s for exclusion duration; 500 for exclusion list size and 30 s for repeat duration.
5. Data Analysis
The raw data were processed in software environment MaxQuant (26) version 1.3.0.5 and Andromeda Search engine (27). Proteins were identified by searching against the complete database sequence of Trypanosoma cruzi-Cl Brener (downloaded at TriTryp DB, release 4.2 - 23,311 sequences) together with a set of commonly observed contaminants. Carbamidomethylation (C) was set as fixed modification whereas oxidation (M) and acetylation (N-terminal) as variable modifications; maximal number of modification per peptide of 5; maximal missed cleavages of 2; MS1 tolerance of 6 ppm; MS2 of 0.5 Da; maximum false peptide and protein discovery rates of 0.01. For matching between runs, the time window was 2 min. The bioinformatics analysis was performed using Perseus software (http://www.perseus-framework.org/). Protein matching to the reverse (or contaminants) database, or identified only by modified peptides, were filtered out. Proteins were identified by at least one unique peptide. Relative protein quantification was performed using the LFQ algorithm of MaxQuant (28) using minimum ratio count of two. Protein quantification was based on LFQ values of “razor and unique peptides” of unmodified peptides, using the Proteingroups.txt file. Proteins with to 2 or more valid LFQ values (not a NaN) in at least one study group were considered for analysis, the remaining proteins were filtered out. When indicated, LFQ values were transformed in log scale. Differentially expressed proteins were statistically analyzed using t test considering FDR = 0.05, s0 = 0.2 and 250 number of randomization using Perseus platform. For shuttle analysis, proteins that were expressed (at least two valid LFQ values) only in C or only in NC extracts were excluded from this analysis. An average of LFQ values were obtained from chromatin proteins and divided by the corresponding LFQ values from NC after imputation of missing values by normal distribution (considering width = 0.3 and down shift = 1.8.) according to Perseus. Euclidean distances were used for hierarchical clustering of ratios of C/NC using Perseus. Gene Ontology (GO) term analysis was performed using the “Gene Ontology enrichment tool” from http://tritrypdb.org/ (release 27). In order to compare the p values among protocols and life forms, p value cutoff was set as 1.0; however, term enrichments were only considered significant when p value was < 0.05. Clustering analysis of supplemental Fig. S7 was carried out using an in-house program coded in Python programming language together with the SciPy scientific library. Firstly, missing values were imputed drawing from a normal distribution whose parameters were estimated from their own data. After that, for each protein, a z-normalization among different replicates were performed. Finally, hierarchical clustering for both proteins (rows) and replicates (columns) were produced; for each clustering, we adopted an average linkage method, and also the Euclidean distance to compute pairwise distances. Raw data used in data analysis is available at ftp://MSV000080261@massive.ucsd.edu.
6. Experimental Design and Statistical Rationale
For MS analysis, chromatin extracts of epimastigotes and trypomastigotes have been obtained in three biological replicates. Proteins were considered present only if they had a valid LFQ value (not NaN) in at least 2 replicates of each group. An average of LFQ and iBAQ values were obtained for each group (epimastigotes and trypomastigotes) as well as ratios of C/NC. We concluded that three biological replicates were enough to reach a stationary number of identified proteins, because a good agreement among biological replicates was observed. For example, proteins exclusively found in replicates 1, 2, and 3 of epimastigotes were only 4.5, 3.5, and 5.4%, respectively, of total number of proteins identified/quantified. We noted that a second replicate increases protein number by 7.5%, whereas the addition of a third replicate leads to an addition of only 3.5%. A similar conclusion was obtained for quantitative data (LFQ and iBAQ values). We observed that pairwise analysis of biological replicates shows, on average, r = 0.957 (supplemental Fig. S6). Pearson correlation and volcano plots were done in Perseus. Coomassie-stained gels, Western blotting and electropherograms results show a representative image out of (at least) 3 replicates obtained from independent biological replicates. MNase digestion was also performed using T.cruzi life forms obtained from three biological replicates.
7. Plasmid Constructs, Protein Expression, and Purification
The genes of the proteins selected for target validation were amplified from T. cruzi genomic DNA using the following primers (5′-3′): TcCLB.506779.150, GAATTCCCGCCAACAAAAGGAGGG (forward) and AAGCTTCTACACCACCTT CGCGTAGG (reverse); TcCLB.509747.90, GAATTCTCCACTCCGTTGTGCACG (forward) andAAGCTTTCACGCCAAGTACCCCATC (reverse); TcCLB.510513.40, GAATTCGAGCTACATGTATTTGATTTTGATGG (forward) and CTCGAGCTA TTTCCCTCGATACCGAG (reverse); TcCLB.511439.40, GAATTCCTGTTTTCA TGCGTGGAG and AAGCTTTCAGTACTTGCCGCCAA (reverse); TcCLB.508177.70, GAATTCAGTGTATTTATCATTCCCCATTCCGG (forward) and AAGCTTTCAAAATGAGCCCAAACCGAGTCG (reverse); TcCLB.504001.20, GAA TTCTCTTGCGATGCTGAAGAACC (forward) and AAGCTTTTACTGACCTTTTTC AGCACT (reverse). All genes were amplified in full length, except TcCLB.508177.70, in which primers were designed to amplify a specific and smaller fragment to facilitate its subsequent expression. The purified PCR was cloned into pET28a (+) in E. coli Bl21 DE3, and cells were grown in LB broth with kanamycin (50 μg/ml). Expression of the recombinant was induced when cultured reached A600 = 0.6 with 1 mm of isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h at 37 °C. After induction, the cells were centrifuged at 6000 g for 15 min at 4 °C and ressuspended in 50 mm Tris-HCl pH 7.4 or pH 6.8 (for TcCLB.510513.40), 300 mm NaCl, 1% sarkosyl, 1 mm PMSF, 5 mm DTT and cOmpleteTM EDTA free protease inhibitor mixture (Roche). Cells were lysed on ice using an ultrasonic disruptor UD-201 (Tomy Seiko) for 6 cycles of sonication for 30 s and cooling on ice for 1 min. The lysate was centrifuged at 18,000 × g for 30 min. For soluble proteins (TcCLB.506779.150 and TcCLB.509747.90), the supernatant obtained was used directly for purification by affinity chromatography on nickel charge resin Ni-NTA agarose (Qiagen). For insoluble proteins (TcCLB.510513.40, TcCLB.511439.40, TcCLB.508177.70, and TcCLB.504001.20) the pellet was resuspended in 50 mm Tris-HCl pH 7.4 or pH 6.8, 7.6 m urea, then the sample was centrifuged at 10,000 × g for 30 min and the supernatant was applied to the resin. For purification, the resin was washed with 5 column volumes of MilliQ water and pre-equilibrated with 5 column volumes of 50 mm Tris-HCl pH 7.4, 300 mm NaCl (soluble proteins) or 50 mm Tris-HCl pH 7.4 or pH 6.8, 7.6 m urea (insoluble proteins). The sample was loaded three times through the column, washed with 10 column volumes of 50 mm Tris-HCl pH 7.4, 300 mm NaCl (soluble proteins) or 50 mm Tris-HCl pH 7.4 or pH 6.8, 7.6 m urea, 5 mm imidazole (insoluble proteins). For protein elution, an imidazole gradient from 50 mm to 500 mm (with two column volumes) was made. The recombinant proteins were visualized by SDS-PAGE, cut from the gel and used for polyclonal antibody production by Proteimax (www.proteimaxnet.com.br). The molecular size of TcCLB.508177.70 is 94 kDa but only a fragment of 16.7 kDa was expressed. The proteins TcCLB.506779.150 (15.6 kDa), TcCLB.509747.90 (47 kDa), TcCLB.510513.40 (34 kDa), TcCLB.511439.40 (52 kDa), (16.7 kDa), and TcCLB.504001.20 (13 kDa) were expressed in their full length.
8. Western Blotting
Samples were separated by 15% or 10% SDS-PAGE and transferred to 0.45 μm nitrocellulose membrane for 1 h at 25 V, using a semidry transfer apparatus in transfer buffer (25 mm Tris, 192 mm glycine, pH 8.3). The membrane was blocked in phosphate-buffered saline (PBS), 0.1% Tween 20 containing dry milk (5% w/v) for 1 h, then incubated with primary antibody (anti-eIF5A 1:10000 (29), anti-histone H3 1:3000 (abcam), anti-BIP 1:10000 (30), anti-HSP70 1:10000 (31), anti-methyl glutaconyl-CoA (MgCoA) 1:10000 (32), anti-TcOrc1/Cdc6 1:1000 (21), anti-TcCLB.506779.150 1:2000, anti-TcCLB.509747.90 1:1000, anti-TcCLB.510513.40 1:3000, anti-TcCLB.511439.40 1:2000, anti-TcCLB.508177.70 1:1000, anti-TcCLB.504001.20 1:1000 (the last six antibodies were produced by Proteimax) in blocking solution at 4 °C overnight. The membrane was washed three times in PBS-0.1% Tween 20 incubated with secondary antibody anti-rabbit or mouse (for anti-TcOrc1/Cdc6) conjugated with HRP 1:5000 in blocking solution, for 1 h, and washed three times in PBS-0.1% Tween 20. Antibody reactive bands were visualized using the ECL detection kit (GE Healthcare Life Sciences) in Western blot chemiluminescence imaging platform Alliance 4.7 (UVITEC Cambridge).
9. Chromatin Immunoprecipitation (ChIP)
ChIP assays of 8 × 108 epimastigotes and trypomastigotes forms were performed as described in (33). Samples were incubated with 3 μg of anti-TcCLB.506779.150, anti-TcCLB.511439.40, and anti-TcCLB.508177.70 and with the same amount of the respective pre-immune serum of each protein. Samples were also incubated with anti-histone H3 (abcam) and without an antibody as a control. Chromatin was eluted with 250 μl of elution buffer (50 mm Tris-HCl pH 8.0, 10 mm EDTA and 1% SDS) and cross-links were reversed by incubation at 65 °C overnight. DNA was purified by phenol-chloroform extraction and quantified using NanoDrop 2000 spectrophotometer.
10. Micrococal Nuclease Digestion
T. cruzi epimastigotes or trypomastigotes (108 parasites) were washed in lysis buffer (1 mm potassium l-glutamate, 250 mm Sucrose, 2.5 mm CaCl2, 1 mm PMSF) and after centrifuged (at 1700 × g for 5 min) pellets were lysed with lysis buffer containing 0.1% Triton X-100. The supernatants were discarded and the pellets were washed two times with lysis buffer without detergent. Samples were incubated with 1500 U of Micrococcal nuclease (Thermo Scientific) for about 30 min at 37 °C then they were supplemented with 10 μl of proteinase K (20 mg/ml) and incubated at 56 °C for 3 h. The DNA was extracted by phenol-chloroform method, analyzed at 2100 Bioanalyzer (Agilent) and quantified using a NanoDrop 2000 spectrophotometer.
RESULTS
Proteomics Analysis of Chromatin and Nonchromatin Associated Proteins
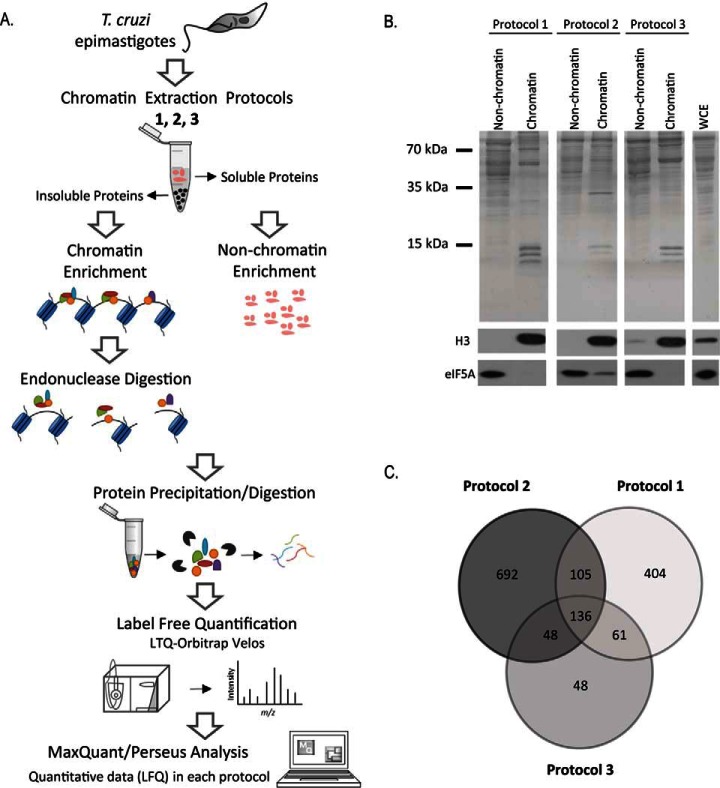
Chromatin is a very dynamic structure in which proteins can be linked to DNA with different strengths and affinities. To better obtain a set of chromatin-associated proteins, we extracted chromatin from T. cruzi by using three different protocols namely protocol 1, 2, and 3 as described by (20, 21, 23), as well as their nonchromatin fractions for comparison. The utilization of these three protocols could circumvent the fact that some chromatin proteins are difficult to extract. Chromatin proteins tightly associated with DNA were separated from nonchromatin fractions by centrifugation, submitted to endonuclease digestion, protein precipitation and MS analysis (Fig. 1A).
Fig. 1.
A, Outline of the workflow to identify chromatin-associated proteins in T. cruzi. Epimastigotes were harvested and chromatin and nonchromatin enriched extracts were obtained by three different extraction protocols (1, 2 and 3) described at Materials and Methods. After protein precipitation and digestion, peptides were analyzed by LC-MS/MS. Raw data were analyzed by MaxQuant/Perseus software. B, Protein fractionation after chromatin extraction of epimastigotes using three different protocols. Upper panel: SDS-PAGE of nonchromatin and chromatin fractions obtained by protocols 1, 2 and 3. Molecular marker and whole cell extract (WCE) are indicated. Lower panels: Western blotting using antibodies against eIF5A (a nonchromatin protein) and histone H3 (chromatin protein). C, Venn diagram showing the number of proteins found by MS in each chromatin-enrichment protocol.
To evaluate the presence of chromatin and nonchromatin proteins, the fractions obtained by the three protocols were submitted to Western blotting using antibodies to histone H3, a known chromatin protein, and the eukaryotic translation initiation factor 5A (eIF5A) a predominant cytosolic protein (29) (Fig. 1B). This latter can also be found in the nuclear space in some contexts (34). In addition, chromatin extracts were also probed against a mitochondrial (methyl glutaconyl-CoA- MgCoA), an endoplasmic reticulum (BiP) (30) and other cytoplasmic (Hsp70) (31) marker (supplemental Fig. S1). Protocol 3 seems to have more contaminants mainly from cytoplasm and mitochondria. The enrichment of the histone and the lower levels of other cytoplasmic markers in the chromatin fraction show the effectiveness of the three protocols. Importantly, protocols 1 and 2 were previously used to obtain histones and to analyze pre-replication factors in parasites, respectively (21, 35) whereas protocol 3 was used to obtain chromatin associated proteins from mammals (23).
High resolution proteomics analysis was performed after enzymatic digestion and strong cation exchange (SCX)-stage tip decomplexation. Altogether, 2254 proteins were identified and quantified based on extracted ion chromatogram (XIC) by label free quantification. Combined analysis of all samples, identified 19674 peptides from which 15488 were unique peptides (supplemental Table S1). For the chromatin-enriched fractions, 706, 981, and 293 proteins were identified in protocol 1, 2, and 3, respectively, totalizing 1494 proteins (Fig. 1C). For nonchromatin fractions 1088, 872, and 1143 proteins were identified in protocol 1, 2, and 3, respectively (supplemental Fig. S2). Surprisingly, only 136 proteins were common to all chromatin fractions, showing that each protocol identified a different subset of proteins, which highlights the difficulty of establishing a set of chromatin proteins based on just one purification procedure. In contrast, the nonchromatin extracts contain 700 proteins that were detected in all three protocols (supplemental Fig. S2).
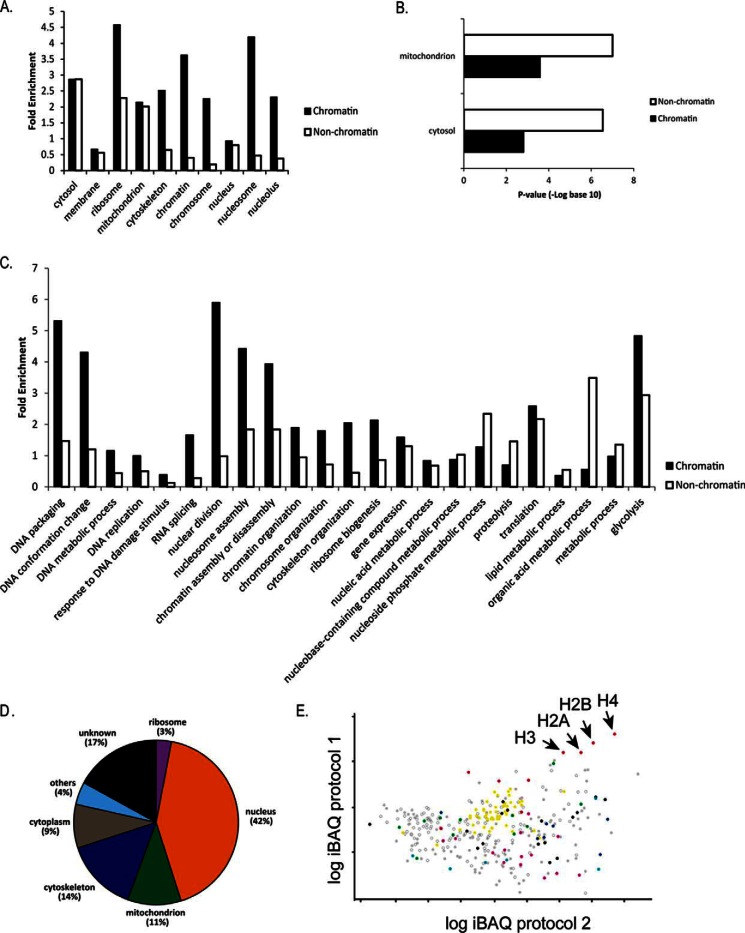
To obtain a set of more stringent chromatin-associated proteins, we selected proteins that were identified in at least two different extraction protocols. This approach retrieved 349 proteins (supplemental Table S2). Gene ontology (GO) (cellular component) terms such as “nucleus,” “chromatin,” “chromosome,” “nucleolus,” and “nucleosome” were enriched in proteins present in this subset, in comparison to a similar subset composed of proteins from nonchromatin extracts (Fig. 2A). Although the fold enrichment for “cytosol” and “mitochondrion” were the same for proteins obtained from nonchromatin and chromatin extracts, -log p values for this latter were much lower (Fig. 2B). Moreover, GO terms associated with biological processes located preferentially at nuclear spaces (“DNA packaging,” “nuclear division,” “nucleosome assembly,” “RNA splicing”), are enriched in chromatin-extracts, whereas processes mainly located primarily at cytoplasm (“nucleoside phosphate metabolic process” (36), “vesicle mediated transport,” “metabolic process”) were more enriched at nonchromatin extracts (Fig. 2C).
Fig. 2.
Classification of chromatin and nonchromatin extracts identified in at least two enrichment protocols. A, Fold enrichment related to the whole proteome according to the predicted localization based on GO search for cellular component. B, p value (log base 10) from Fischer's exact test for proteins predicated to be located at mitochondria and cytosol. C, Predicted function based on GO search for biological process. Fold enrichment is shown as in B. D, Distribution of iBAQ values of proteins identified in at least two protocols according to GO cellular component. E, Scatter plot of iBAQ values of chromatin-associated proteins from protocols 1 and 2. Categories from GO predicted biological function were highlighted: translation (yellow), nucleosome assembly (red), microtubule-based moviment (green), DNA and RNA binding/regulation of replication/transcription/mitosis (black), oxidation-reduction process (light blue), protein folding (dark blue), metabolic process/carbohydrate (pink), hypotheticals (open circle).
Interestingly, we found enriched proteins involved in the “glycolysis” (p value: 0.0038) and the “glucose metabolic process” (p value: 0.0048) as well as “generation of precursor metabolites and energy” (p value: 0.0042) in chromatin-extracts. Four proteins from the glycolytic pathway were found in the chromatin associated-extracts including fructose-bisphosphate aldolase (TcCLB.504163.50), glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (TcCLB.506943.50), hexokinase (TcCLB.508951.20), and pyruvate dehydrogenase E1 component alpha subunit (TcCLB.507831.70).
The absolute abundance of proteins was estimated using the iBAQ (Intensity- Based Absolute Quantification) (37) tool from MaxQuant. From the 349 putative chromatin associated proteins, 42% abundance was detected from proteins associated with the term “nucleus,” whereas 9% came from proteins associated with “cytoplasm,” showing that our preparations are enriched with proteins located at the nucleus (Fig. 2D). It is important to note that, 17% of proteins are classified as “hypothetical proteins” with unknown location and function.
As expected, histones were present within the highest abundant group for all protocols. RNA binding proteins and high mobility group proteins were detected as highly abundant, as well. However, some proteins with unrelated chromatin functions were also identified with high abundance, such as kinetoplastid membrane protein KMP-11, paraflagellar rod proteins, and cytoskeleton components (α e β tubulin, dynein) (supplemental Table S2, Fig. 2E and supplemental Fig. S3). Further experimental validation is necessary to confirm whether these proteins represent contaminants in our preparations, or if they are proteins with an undescribed chromatin function.
It is worth to emphasize that we likewise detected highly abundant hypothetical proteins. From the 50-most abundant proteins in each chromatin extraction protocol, 11 were hypothetical, including seven (TcCLB.507105.50, TcCLB.511733.90, TcCLB.507165.30, TcCLB.504153.280, TcCLB.508719.30, TcCLB.507625.120, and TcCLB.505807.60) that were detected in at least two different protocols. Interestingly, TcCLB.504153.280 has a domain of SMC (structural maintenance of chromosomes), which is involved in chromosome segregation (38).
Epimastigotes Chromatin Contains More Protein Diversity Than Trypomastigotes
Chromatin is a very dynamic and organized structure that can be changed by alterations in the surrounding environment. Therefore, we aimed to identify and compare the chromatin-associated proteins in epimastigotes and trypomastigotes. Both live in different environmental conditions. The latter is the nonproliferative/infective form of T. cruzi and presents some important differences regarding chromatin structure and transcriptional levels when compared with the first.
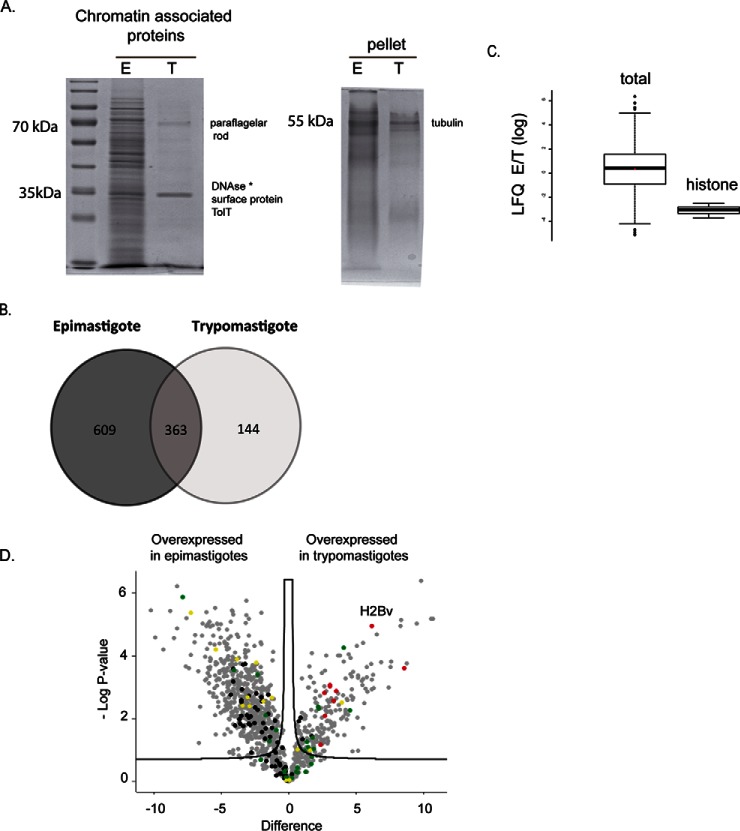
Whole cell and chromatin extracts of proliferative forms yields 4.3 and 7.6 (an average of the three protocols) times more micrograms of proteins per cell when compared with the nonproliferative forms (supplemental Fig. S4C). These differences were visible in proteins fractionated by SDS-PAGE (Fig. 3A and supplemental Fig. S4A). It is striking that trypomastigote chromatin extracts show a modest protein diversity and quantity when compared with epimastigotes regardless of the extraction protocol. It is important to note that the lack of proteins in the chromatin extracts of trypomastigotes was not because of a tight association of proteins with DNA as observed by a low amount of proteins remaining in the pellet.
Fig. 3.
A, SDS-PAGE analysis of chromatin-extracts (protocol 2) from epimastigotes and trypomastigotes normalized by number of cells. At left, proteins obtained from supernatant of DNase treatment, at right, the remaining pellet that were solubilized with Coomassie sample buffer. B, Venn diagram showing a comparison of proteins identified exclusively in epimastigotes (609) and trypomastigotes (144) forms and those proteins found in both forms (363) after chromatin extraction and MS analysis. C, Comparison of LFQ ratios (in log scale) of epimastigotes and trypomastigotes from all chromatin associated proteins (left) and from histones (right). * Deoxyribonuclease-1 from Bos taurus. D, Volcano plot of chromatin-associated proteins from epimastigotes and trypomastigotes. Differentially expressed proteins were obtained considering p value <0.05 and s0 = 0.2. Missing data were imputed by normal distribution. Ribosomal (black), histone (red) and cytoskeleton proteins (green) and RNA binding proteins (yellow).
Nonreplicative Forms Contain the Majority of Chromatin Associated Proteins in Lower Levels Than Epimastigotes
To evaluate chromatin of replicative and nonreplicative forms in more details, chromatin extracts from protocol 2 (in biological triplicates) were analyzed by high-resolution mass spectrometry. One thousand one hundred and sixteen proteins (1116) were identified/quantified in chromatin extracts (Fig. 3B). supplemental Fig. S6 shows that the biological replicates were in a good correlation. Considering proteins that were identified in at least two replicates from epimastigotes or trypomastigotes, we could classify proteins expressed predominantly in epimastigotes (609 proteins) and trypomastigote (144 proteins), as well as 363 proteins that are common to both forms of the parasite (Fig. 3B, supplemental Table S3). These results confirm that epimastigotes contain more protein diversity than trypomastigotes, indicating that trypomastigotes have fewer chromatin-associated proteins.
Accordingly, in the group of proteins commonly expressed in both forms, all five histones were identified. Among the proteins present only in epimastigotes, we found several nuclear proteins involved in processes such as transcription/splicing: RNA-binding proteins, small nuclear ribonucleoproteins, and replication: replication factor A 28 kDa subunit- (TcCLB.510821.50) and 51 kDa subunit (TcCLB.510901.60), DNA topoisomerase IA (TcCLB.510121.160), proliferative cell nuclear antigen (TcCLB.508277.150), and a nucleosome assembly protein-like protein (TcCLB.505983.20). Among the proteins preferentially expressed in trypomastigotes, we found ATP-dependent DEAD/H RNA helicase (TcCLB.508973.50), heat shock protein-like protein DnaJ homolog (TcCLB.509157.80), RNA-binding protein 6 (TcCLB.508153.680), transcription elongation factor 1 homolog (ELOF1-TcCLB.504247.30), and a histone H2B variant (TcCLB.506779.150).
Quantitative proteomics analysis indicates that globally, proteins are equally expressed in both life forms (LFQ E/T ratio ∼ 0, Fig. 3C) however, histones are overrepresented in our trypomastigotes extracts. In fact, 29% of trypomastigote's chromatin content is composed of histones, comparing to 5% of epimastigotes chromatin (Fig. 4B). However, both life forms contain the same number of nucleosomes, as seen by microccocal nuclease digestion (supplemental Fig. S5). The nucleosomal DNA obtained from both forms was the same (supplemental Fig. S5B) indicating that epimastigotes and trypomastigotes contain the same number of nucleosomes per cell.
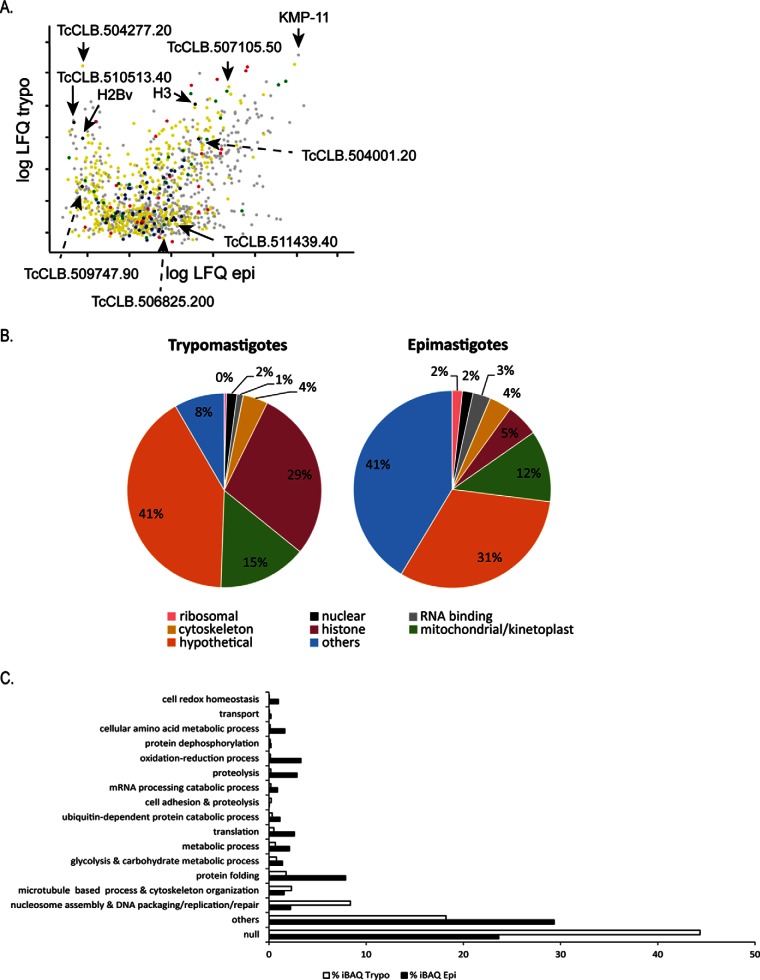
Fig. 4.
Absolute abundance evaluation using iBAQ. A, Scatter plot of iBAQ values of chromatin-associated proteins from epimastigotes and trypomastigotes. Protein extracts were obtained by using protocol 2. Hypotheticals (yellow), ribosomal (black), histone (red) and cytoskeleton proteins (green) are shown. B, Relative percentage of iBAQ values according to the indicated categories. Note that 29% of trypomastigotes extracts correspond to histone proteins. C, GO process terms and their corresponding percentage of iBAQ values in epimastigotes and trypomastigotes.
Hierarchical clustering confirms the good agreement between biological replicates (supplemental Fig. S7). In addition, it shows a clear distinction of expression pattern between chromatin-associated proteins. The great majority of proteins are more expressed in epimastigotes whereas a small subset is more represented in trypomastigotes. Statistical analysis (p value < = 0.05, s0 = 0.2) from the 1116 analyzed proteins indicates that 840 (75%) proteins are differentially expressed during life forms and, 215 proteins are more expressed in trypomastigotes chromatin (Fig. 3D). This latter set, however, contains proteins that are equally expressed (per cell) per life form. This result should be analyzed with caution, as MS analysis were performed from equal mass of protein samples. Taken together, these data suggest that besides having less protein diversity, trypomastigotes contain lower amounts of proteins associated with their chromatin, which further corroborates that they have a naked chromatin.
One of the most abundant proteins found in trypomastigotes chromatin extracts is TcCLB.504277.20, a hypothetical protein with sequence homology to TolT (Fig. 4A). Another high abundant and still uncharacterized protein is TcCLB507105.50 (Fig. 4A), which is present in both life forms. It corresponds to a 17 kDa protein with isoelectric point of 11.63. These biochemical characteristics resemble proteins that are in close proximity to DNA, such as histones and high mobility proteins. It is possible that this hypothetical protein may represent an undescribed protein that interacts with DNA.
In agreement to high DNA and RNA-metabolic activity, ribosomal proteins and RNA binding proteins are detected in higher amounts in epimastigotes chromatin extracts when compared with trypomastigotes chromatin extracts (Fig. 4B and supplemental Table S3).
In order to investigate if any important biological function could be distinct in chromatin from these two life forms, we searched at GO biological function. In accordance to Fig. 4C, all analyzed terms, except two (“microtubule-based process and cytoskeleton organization” and “nucleosome assembly and DNA packaging/replication/repair”) were more abundant in epimastigotes (Fig. 4C).
The term “protein folding” comprises almost 8% of protein abundance of epimastigotes chromatin but only 1.2% of trypomastigotes (Fig. 4C). This term includes mainly chaperone proteins that are involved in the proper folding of nascent proteins. Two prefoldins were found only in epimastigote chromatin extracts (TcCLB.506859.90 and TcCLB.510629.430). Prefoldins are cochaperones that work on folding of actin and tubulin, however, they are also found in nucleus and associated with gene transcription (39). Terms associated with “oxidation and reduction” and “cell redox homeostasis”, are also more abundant in epimastigotes (3.3% and 0.14%) when compared with trypomastigotes (1% and 0.04%) chromatin extracts (Fig. 4C).
Trypomastigotes also contains proteins associated with metabolism. However, these proteins are three times more abundant in epimastigotes than in trypomastigotes chromatin. Specifically, proteins from glycolysis pathway are two times more abundant in epimastigotes chromatin (Fig. 4C and supplemental Table S3). As it will be discussed below, GAPDH, a glycolytic enzyme, has been previously shown to be associated with T.cruzi chromatin (33).
Chromatin Extraction and ChIP Confirms Chromatin Association of Selected Candidates
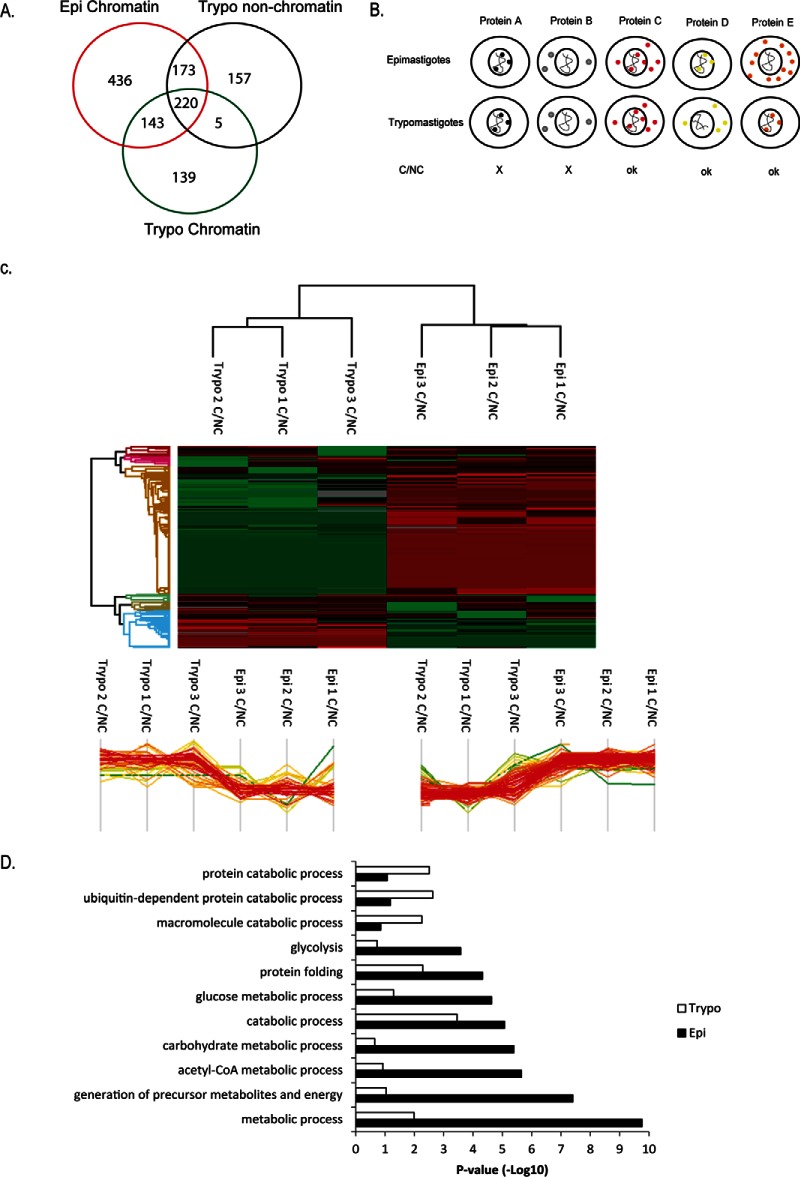
Because of the large number of putative chromatin-associated proteins classified as hypothetical proteins, we aimed to confirm their localization by chromatin fractionation and chromatin immunoprecipitation (ChIP). The chosen proteins were preferentially hypothetical proteins expressed mainly in epimastigotes (TcCLB.511439.40 and TcCLB.508177.70), or in trypomastigotes (TcCLB.506779.150 -histone H2B variant, TcCLB.509747.90 and TcCLB.510513.40), or common to both forms (TcCLB.509471.59-histone H3 and TcCLB.504001.20). This latter protein was classified as a shuttle protein as described in the next section.
After a search on blastp (http://blast.ncbi.nlm.nih.gov/Blast.cgi), using the hypothetical proteins as queries, we observed that the TcCLB.510513.40 protein, despite not having a known chromatin related domain, is similar to a protein involved in nucleic acid binding (OB protein containing fold) of Dictyostelium discoideum, and to a protein related to nucleolar rRNA processing (GAR1 protein) of Fusarium fujikuroi. The protein TcCLB.504001.20 has an Alba superfamily domain. Alba protein is a chromosomal protein that coats archaeal DNA but does not compact it (40), and may play a role in maintenance of chromatin architecture and, thereby, in transcription repression (41).
From this selection, the corresponding genes were cloned, and the recombinant proteins were expressed in E. coli and affinity purified (supplemental Fig. S8) for polyclonal antibody production. The resulting immune sera were used to check for the presence of the target proteins in the chromatin after protein extraction in epimastigotes and trypomastigotes forms (Fig. 5A). As expected, the proteins that were preferentially expressed in trypomastigotes (TcCLB.506779.150, TcCLB.509747.90, and TcCLB.510513.40) are mainly present in the chromatin fraction of these parasites forms rather than epimastigotes. The TcCLB.511439.40 and TcCLB.508177.70 proteins were found in the chromatin fraction of epimastigotes but not of trypomastigotes. Antibodies against proteins commonly expressed in the two forms (TcCLB.504001.20 and histone H3) also confirmed the presence of these proteins in the chromatin of both forms. eIF5A and TcOrc1/Cdc6, a protein involved in DNA replication, were used as a cytosolic and chromatin controls, respectively. We confirmed that this latter is associated with epimastigote chromatin, but not to trypomastigote chromatin (21, 42). Unfortunately, none of the antibodies raised against selected candidates worked for immunofluorescence assays.
Fig. 5.
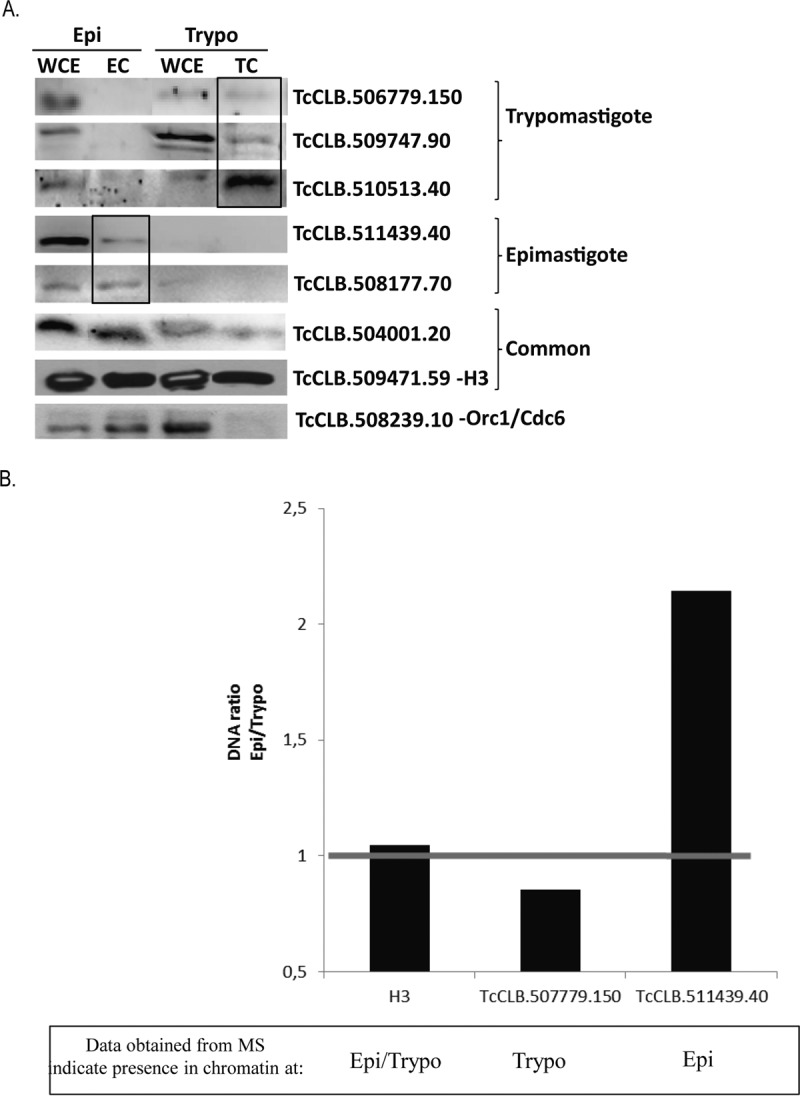
A, Western blot of whole cell extracts (WCE) and chromatin extracts (EC or TC) of epimastigotes (Epi) and trypomastigotes (Trypo) forms using antibodies against the indicated proteins. Proteins expressed preferably in trypomastigotes, epimastigotes and expressed in both forms (common) are indicated. Antibodies against the Orc1/Cdc6 present mainly in the chromatin of epimastigotes were used as a control. B, T. cruzi epimastigote or trypomastigote cells were submitted to a chromatin immunoprecipitation assay using antibodies or pre-immune serum as indicated. After immunoprecipitation, cross-links were reversed, and DNA was extracted and quantified. Percentage of the immunoprecipitated DNA related to input after subtraction of percentage of DNA immunoprecipitated by preimmune serum was calculated. Graph indicates the ratio of these values between epimastigote and trypomastigote cells.
Chromatin immunoprecipitation (ChIP) assays were performed to confirm if specific life stage proteins were associated with chromatin (Fig. 5B). Because of the high amount of parasites necessary for this experiment, we chose three proteins for validation: TcCLB.506779.150, expressed mainly in trypomastigotes; TcCLB.511439.40, expressed mainly in epimastigotes, and TcCLB.509471.59, expressed in both forms. ChIP was performed for each antibody as well as its corresponding preimmune serum in both parasite forms. The percentage of DNA immunoprecipitated related to the input is shown in supplemental Fig. S9, where a higher percentage of DNA is obtained in comparison with the preimmune serum, indicating antibody specificity to a DNA binding protein. As expected, for TcCLB.506779.150, we detected more immunoprecipitated DNA from trypomastigotes than from epimastigotes. For TcCLB.511439.40, we observed the opposite. In Fig. 5B, it is showed ratios of epimastigote and trypomastigote DNA immunoprecipitated by the indicated antibody, confirming their preferential location at DNA from trypomastigotes (TcCLB.506779.150), epimastigotes (TcCLB.511439.40), and from both forms (TcCLB.509471.59).
Different Classes of Proteins Shuttle Between Chromatin and Nonchromatin Territories in T. cruzi Life Forms
As fewer proteins were found in nonreplicative chromatin, we asked if any of chromatin-associated protein from replicative stage would be found in the nonchromatin (NC) space of nonreplicative stage. Thus, we analyzed by quantitative MS the NC content of both life forms looking for proteins that may shuttle between chromatin and NC space during T. cruzi differentiation. Using a very simple criterion (presence or absence based on quantitative data - LFQ values), we detected 173 putative chromatin-associated proteins from epimastigotes that were located at NC extracts of trypomastigotes, indicating that they are indeed expressed, however they have a different location in the nonreplicative forms (Fig. 6A).
Fig. 6.
A, Venn diagram comparing proteins detected by MS at chromatin extracts of epimastigotes and trypomastigotes and nonchromatin (NC) extracts of trypomastigotes. B, Outline of proteins used in analysis of chromatin-nonchromatin shuttle during life cycle. Proteins expressed either on chromatin (protein A) or cytoplasm (protein B) were not included in these analysis. C, Hierarchical clustering of quantitative proteomics data from ratios of epimastigotes and trypomastigotes chromatin (C) (3 biological replicates) and NC extracts obtained by protocol 2. Below, two main clusters are represented. At left (Cluster 420), proteins whose C/NC ratio in trypomastigotes are higher than in epimastigotes and at right (cluster 418), proteins whose C/NC ratio are higher in epimastigotes. D, p values of GO process enriched at cluster 420 (trypomastigotes) and 418 (epimastigotes).
In order to have more insights into proteins that may shuttle between chromatin and NC during differentiation, we compared quantitative MS data from all proteins except those that were expressed only in chromatin or only in NC extracts as depicted at Fig. 6B. We calculated the ratios of quantitative data (LFQ values) of a given protein obtained from chromatin and NC extracts. Hierarchical clustering analysis (Fig. 6C) shows two interesting clusters whose ratios C/NC are life form dependent, highlighting proteins that are predominantly presented bounded or not to chromatin depending of life stage. Two hundred and seventy-one (271) proteins have C/NC higher in epimastigotes than trypomastigotes, whereas the opposite is true for 79 proteins.
GO analysis indicated that these groups differ greatly among their biological function. Interestingly, terms associated with metabolic process GO:0008152 (epi p value = 1.72E-10; trypo p value = 0.01031); generation of precursor metabolites and energy GO:0006091 (epi p value = 3.93E-08; trypo p value = 0.09122); acetyl-CoA metabolic process GO:0006084 (epi p value = 2.23E-06; trypo p value = 0.11814), glucose metabolic process GO:0006006 (epi p value = 2.31E-05; trypo p value = 0.0512) are enriched in epimastigotes C/NC. However, in trypomastigotes C/NC, we found an enrichment of GO terms associated mainly ubiquitin-dependent protein catabolic process GO:0006511 (trypo p value = 0.00235, epi p value = 0.06550), macromolecule catabolic process GO:0009057 (trypo p value = 0.00552, epi p value = 0.13929), and protein catabolic process (trypo p value = 0.00310, epi p value = 0.08425) (Fig. 6D).
Many subunits of proteasome were found enriched for trypomastigotes C/NC compared with epimastigotes (TcCLB.503613.20, TcCLB.504069.10, TcCLB.504213.120, TcCLB.506885.350) although proteasome regulatory ATPase subunits (TcCLB.504147.200, TcCLB.506857.90, TcCLB.506859.20) and proteasome regulatory nonATPase subunit (TcCLB.508741.300) were found enriched for C/NC in epimastigotes (supplemental Table S4, clusters 418 and 420). Protozoan proteasomes contain multiple α and β subunits, as other eukaryotes, and inhibition of proteasomal function suspends cell cycle progression and morphological differentiation in Trypanosoma (43, 44). The proteasome components found here may play a role in differentiation or cell cycle of trypanosomes regulating important chromatin components.
Different classes of RNA binding proteins are enriched either in epimastigotes or trypomastigotes C/NC, namely:TcCLB.511727.270, TcCLB.511517.70, TcCLB.506755.260, TcCLB.510859.10, TcCLB.511621.50, and TcCLB.508413.50. In addition, two different phosphatases (TcCLB.506925.150 and TcCLB.511491.100) were found enriched in C/NC extracts of epimastigotes compared with trypomastigotes and vice-versa, respectively. Protein phosphorylation/dephosphorylation is important for regulation of many signaling pathways and has been shown to be important in T. cruzi differentiation (45, 46). It is tempting to propose that these enzymes may represent important regulators of transformation by controlling dephosphorylation steps life-form and location specific.
Intriguing, ratios C/NC of a Ran-binding protein 1 (BP1) (TcCLB.507099.30) are thousand times higher in trypomastigotes than in epimastigotes. This protein associates with a Ran (Ras-related nuclear) small GTPase protein, which inhibits the Ran exchange of GDP for GTP catalyzed by the chromatin-bound protein RCC1 (Regulator of Chromosome Condensation 1). Once bounded to GTP, Ran is responsible (together with exportins) for protein nuclear exportation (47, 48). Once high ratios of Ran-BP1 is found on trypomastigotes chromatin, our data suggest that protein nuclear exportation is impaired in trypomastigotes.
DISCUSSION
T. cruzi is a good model for chromatin study as it alternates between replicative and nonreplicative forms accompanied by a shift on their global transcription levels and changes in their chromatin architecture. In addition, in comparison to higher eukaryotes, their histones differ greatly regarding primary structure, however they are also subjected to modifications that are differentially expressed upon cell differentiation (22). Here, one of the most surprising findings is that the nonreplicative form not only has few proteins in its chromatin but also a less diverse protein repertoire. This observation raised important questions: How is the DNA metabolism affected by the lack of some proteins in chromatin? Would it be related to the replication arrest and low global transcription levels observed in trypomastigotes? What are the essential chromatin proteins? Would chromatin of other nondividing cells also contain low levels and little diversity of proteins? To our knowledge, there are no chromatin proteome analyses of such cell for comparison. These are important questions that need be addressed in the chromatin field and the growing robustness of proteomics analysis may be able to start answering them.
Here, we identified many known chromatin associated proteins, such as histones, PCNAs, transcription factors, HMG proteins, and components of spliceosome, among others. From ten small nuclear ribonucleoprotein proteins found in T. cruzi genome (Non-Esmeraldo-like), we were able to find five of them: TcCLB.508257.150- SmD3; TcCLB.510531.54; TcCLB.507007.74- Sm-F; TcCLB.511499.59- Sm-E; TcCLB.511725.174- Sm-G. The last three were found mainly in replicative forms. All trypanosome mRNAs are transcribed as long polycistronic units that are cotranscriptionally processed to monocistronic mRNAs by trans-splicing (49). Thus, in contrast to mammalians, the maturation of mRNA occurs mainly at nuclear space in close association with chromatin (50). Thereby, it was not surprise that we detect many components of spliceosome in our chromatin extracts.
We also detected some proteins with no apparent chromatin-related functions, such as cytoskeleton proteins, ribosomal proteins, mitochondrial proteins, and proteins associated with metabolic process. Whether they represent proteins with unknown chromatin functions or if they are mere contaminants of our preparations need to be further verified. In the following, we discuss their possible roles in chromatin emphasizing some evidence of them. Nevertheless, it is important to stress that trypanosomes have a closed mitosis (51), which may contribute to lower contamination levels from cytoplasmic compartments.
Concerning cytoskeleton proteins, there is evidence that actin and vimentin are involved in chromatin remodeling and DNA binding, respectively (52–54). In addition, there is growing evidence that chromatin associated proteins act as moonlight proteins, interacting with microtubules via their chromatin-binding domains and nuclear localization signal (55). In this regard, one of the most abundant detected protein was a kinetoplastid membrane protein 11 (KMP-11), that is a microtubule-bound protein localized at the basal body of Trypanosoma brucei (56). The basal body is essentially a centriole that controls the growth of microtubules. Whether it can participate on mitotic spindle formation (justifying its identification) or not is an open question.
Regarding proteins associated with metabolic process, here we found mainly those related to carbohydrate metabolism (fructose-bisphosphate aldolase (TcCLB.504163.50), glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (TcCLB.506943.50); hexokinase (TcCLB.508951.20), pyruvate dehydrogenase E1 component alpha subunit (TcCLB.507831.70)). It was previously shown that GAPDH associates with T. cruzi telomeric DNA during the replicative phase of life cycle (epimastigote) (33), where authors showed that the GAPDH-telomere association and NAD+/NADH balance changed throughout the T. cruzi life cycle. The presence of metabolic proteins at T. cruzi chromatin, corroborates the growing evidences that associate chromatin and metabolism. It has been suggested that chromatin may function as a sensor to cell metabolism therefore transforming intermediate metabolites into epigenetic changes (57).
In addition, kinetoplast-associated proteins were found in our preparations. Kinetoplast is a structure found in trypanosomatids that is composed of a single mitochondrion (58). It is possible that the protocols used here also extracted mitochondrial DNA and, in turn, proteins associated with it, as it is very abundant in trypanosomes. However, only traces of MgCoA (a mitochondrial protein) were detected at chromatin from protocol 2 (supplemental Fig. S1).
T. cruzi alters between the nonproliferative/infective form (trypomastigote) and proliferative/noninfective form (epimastigote) that, as discussed above, has peculiar differences regarding morphology and chromatin structure. Here, we found that epimastigotes chromatin has much more protein diversity than trypomastigotes. A model highlighting important differences observed between life forms is shown at Fig. 7. Ribosomal proteins and RNA binding proteins are enriched at epimastigotes chromatin, which may be a consequence of more DNA and RNA metabolic activity found in this life form. Nevertheless, it is striking how trypomastigotes chromatin is naked: histones represent almost one-third of their total chromatin content. In contrast, only 5% of epimastigotes chromatin is composed of histones. Besides that, we observed that chromatin associated proteins are less expressed in nonreplicative forms. Important proteins associated with DNA replication, such as PCNA, RPA, and DNA topoisomerases were found in replicative stages but not in nonproliferative stages. As trypanosomes regulate their genes mainly post transcriptionally (59), we speculate that an important regulation through life stage transformation may modulate transcripts/protein stabilization/degradation in order to arrest proliferation through DNA replication blockage.
Fig. 7.
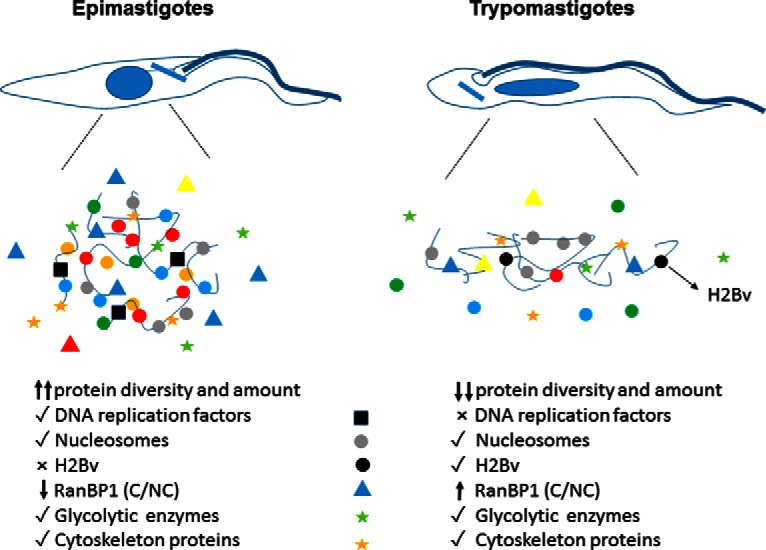
Model of chromatin content of replicative and nonreplicative stages of T. cruzi.
It was previously shown that trypomastigotes contains a poorly transcribed and highly condensed chromatin (17). To date, no protein was assigned to be associated with this compaction. Curiously, this life form contains the majority of histone H1 in a phosphorylated form that is more weakly associated with the chromatin (60). In Drosophila, it was shown that the repressive chromatin could be classified in at least three types (namely, black, green, and blue) according to a unique combination of a subset of proteins and/or histone PTMs. For example, proteins associated to the known pathways of gene repression, including the Polycomb and HP1 pathways, are found in blue and green chromatin (2). In T.cruzi, no gene for HP-1 or polycomb has been identified so far. However, they contain a NUP-1, a nucleoskeleton protein similar to lamins, that is involved with the organization of heterochromatin and epigenetic control (61). Here, we found NUP-1 both in epimastigotes and trypomastigotes chromatin-enriched extracts, although they are up-regulated in trypomastigotes extracts.
We envisage that a protein mainly expressed in trypomastigote chromatin extracts could play a role on the differential chromatin compaction found between both life forms. In this regard, it is interesting that we found a histone variant H2B, (H2Bv- TcCLB.506779.150) expressed mainly in trypomastigote chromatin. The replacement of canonical histones by histones variants (62), causes profound effects on chromatin structure. In T. brucei, H2B variant dimerizes with the histone variant H2AZ, and both are absent from transcription sites (63, 64). These data are important given that we found H2Bv mainly expressed in trypomastigotes forms, which present low levels of transcription. In addition, we detected an acetylation at H2Bv N terminus, possibly enhancing the regulation complexity (data not shown). Whether this histone is associated with heterochromatin formation/maintenance needs further investigation.
We have validated the presence and differential expression in chromatin extracts of seven proteins. Because of the difficulty to obtain antibodies for all of them, we chose representants from hypothetical proteins (as well as histones) that were expressed in one or both life forms. The confirmation of their association with chromatin gave us confidence that our data set could help on the holistic understanding of chromatin function and structure in parasites. The high number of hypothetical proteins identified here as putative chromatin-associated protein is challenging. Some of them have conserved regions that might suggest their function, whereas for others, no available clue is present. Their characterization may reveal important aspects of chromatin biology considering that T. cruzi is an early-branching organism.
Finally, we looked for proteins that may shuttle between chromatin and nonchromatin spaces during differentiation. Almost 20% of epimastigote's chromatin-associated proteins were found in trypomastigotes nonchromatin extracts, indicating they are expressed in the nonproliferative forms but with a different localization. Changing protein location may be one of the strategies used by trypanosomes to regulate protein function. How this mislocalization is achieved is an open question, however we have found a very interesting protein involved in nucleocytoplasmic transport that may play a role on that. High ratios of C/NC of the Ran-binding protein 1 (BP1) (TcCLB.507099.30) is found at trypomastigotes. This protein inhibits the exchange of GDP for GTP into Ran proteins that, in turn, interferes with protein nuclear exportation (47, 48). The fact that high ratios of this protein is found at trypomastigotes chromatin and the fact that this life form exhibits a poor chromatin content, indicates that important differences into nucleocytoplasmic transport exist in life forms that, in turn, could be related to the different chromatin profile and amount found in different T.cruzi life forms.
Supplementary Material
Acknowledgments
We thank Ismael Feitosa Lima, Ivan Novaski Avino, Mariana Morone for technical assistance and Dr. Sergio Schenkman for eIF5A, HSP70, MgCoA and BiP antibodies. We thank Dr. Solange Serrano, Dr. Leo Iwai and Dr. Sergio Schenkman for reading this manuscript and providing critical comments.
Footnotes
Author contributions: T.C.d., M.E., and J.P.d. designed research; T.C.d., S.G.C., F.d.V., R.P.L., M.d.L., C.B.d., M.d.R., and J.P.d. performed research; O.H.T. contributed new reagents or analytic tools; T.C.d., S.G.C., M.d.R., M.E., and J.P.d. analyzed data; T.C.d., M.E., and J.P.d. wrote the paper.
* This work was supported by fellowships from CNPq and by grants (#11/22619-7, and #13/07467-1, São Paulo Research Foundation (FAPESP)). T.C.L.J., M.C.L., and C.B.A. were supported by a fellowship from FAPESP.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- PTM
- post-translational modifications
- C
- chromatin-associated protein
- NC
- nonchromatin associated protein
- ChIP
- chromatin immunoprecipitation
- SCX
- stage tip fractionation
- LFQ
- label free quantification
- GO
- gene ontology
- MNase
- micrococcal nuclease
- iBAQ
- intensity based absolute quantitation
- GAPDH
- glyceraldehyde 3-phosphate dehydrogenase.
REFERENCES
- 1. Harvey A. C., and Downs J. A. (2004) What functions do linker histones provide? Mol. Microbiol. 53, 771–775 [DOI] [PubMed] [Google Scholar]
- 2. Filion G. J., van Bemmel J. G., Braunschweig U., Talhout W., Kind J., Ward L. D., Brugman W., de Castro I. J., Kerkhoven R. M., Bussemaker H. J., and van Steensel B. (2010) Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143, 212–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Imhof A., and Bonaldi T. (2005) “Chromatomics” the analysis of the chromatome. Mol. bioSystems 1, 112–116 [DOI] [PubMed] [Google Scholar]
- 4. Kustatscher G., Wills K. L., Furlan C., and Rappsilber J. (2014) Chromatin enrichment for proteomics. Nat. Protocols 9, 2090–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Johnson D. S., Mortazavi A., Myers R. M., and Wold B. (2007) Genome-wide mapping of in vivo protein-DNA interactions. Science 316, 1497–1502 [DOI] [PubMed] [Google Scholar]
- 6. Morrison C., Henzing A. J., Jensen O. N., Osheroff N., Dodson H., Kandels-Lewis S. E., Adams R. R., and Earnshaw W. C. (2002) Proteomic analysis of human metaphase chromosomes reveals topoisomerase II alpha as an Aurora B substrate. Nucleic Acids Res. 30, 5318–5327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gassmann R., Henzing A. J., and Earnshaw W. C. (2005) Novel components of human mitotic chromosomes identified by proteomic analysis of the chromosome scaffold fraction. Chromosoma 113, 385–397 [DOI] [PubMed] [Google Scholar]
- 8. Uchiyama S., Kobayashi S., Takata H., Ishihara T., Hori N., Higashi T., Hayashihara K., Sone T., Higo D., Nirasawa T., Takao T., Matsunaga S., and Fukui K. (2005) Proteome analysis of human metaphase chromosomes. J. Biol. Chem. 280, 16994–17004 [DOI] [PubMed] [Google Scholar]
- 9. Takata H., Uchiyama S., Nakamura N., Nakashima S., Kobayashi S., Sone T., Kimura S., Lahmers S., Granzier H., Labeit S., Matsunaga S., and Fukui K. (2007) A comparative proteome analysis of human metaphase chromosomes isolated from two different cell lines reveals a set of conserved chromosome-associated proteins. Genes Cells 12, 269–284 [DOI] [PubMed] [Google Scholar]
- 10. Ohta S., Bukowski-Wills J. C., Sanchez-Pulido L., Alves Fde L., Wood L., Chen Z. A., Platani M., Fischer L., Hudson D. F., Ponting C. P., Fukagawa T., Earnshaw W. C., and Rappsilber J. (2010) The protein composition of mitotic chromosomes determined using multiclassifier combinatorial proteomics. Cell 142, 810–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kustatscher G., Hegarat N., Wills K. L., Furlan C., Bukowski-Wills J. C., Hochegger H., and Rappsilber J. (2014) Proteomics of a fuzzy organelle: interphase chromatin. EMBO J. 33, 648–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maiolica A., de Medina-Redondo M., Schoof E. M., Chaikuad A., Villa F., Gatti M., Jeganathan S., Lou H. J., Novy K., Hauri S., Toprak U. H., Herzog F., Meraldi P., Penengo L., Turk B. E., Knapp S., Linding R., and Aebersold R. (2014) Modulation of the chromatin phosphoproteome by the Haspin protein kinase. Mol. Cell. Proteomics 13, 1724–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tyler K. M., and Engman D. M. (2001) The life cycle of Trypanosoma cruzi revisited. Int. J. Parasitol. 31, 472–481 [DOI] [PubMed] [Google Scholar]
- 14. Hecker H., Betschart B., Bender K., Burri M., and Schlimme W. (1994) The chromatin of trypanosomes. Int. J. Parasitol. 24, 809–819 [DOI] [PubMed] [Google Scholar]
- 15. Alsford S., and Horn D. (2004) Trypanosomatid histones. Mol. Microbiol. 53, 365–372 [DOI] [PubMed] [Google Scholar]
- 16. Elias M. C., Nardelli S. C., and Schenkman S. (2009) Chromatin and nuclear organization in Trypanosoma cruzi. Future Microbiol. 4, 1065–1074 [DOI] [PubMed] [Google Scholar]
- 17. Elias M. C., Marques-Porto R., Freymuller E., and Schenkman S. (2001) Transcription rate modulation through the Trypanosoma cruzi life cycle occurs in parallel with changes in nuclear organisation. Mol. Biochem. Parasitol. 112, 79–90 [DOI] [PubMed] [Google Scholar]
- 18. Camargo E. P. (1964) Growth and differentiation in Trypanosoma cruzi. I. Origin of metacyclic trypanosomes in liquid media. Revista do Instituto de Medicina Tropical de Sao Paulo 6, 93–100 [PubMed] [Google Scholar]
- 19. Nogueira N., and Cohn Z. (1976) Trypanosoma cruzi: mechanism of entry and intracellular fate in mammalian cells. J. Exp. Med. 143, 1402–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Toro G. C., and Galanti N. (1990) Trypanosoma cruzi histones. Further characterization and comparison with higher eukaryotes. Biochem. Int. 21, 481–490 [PubMed] [Google Scholar]
- 21. Godoy P. D., Nogueira-Junior L. A., Paes L. S., Cornejo A., Martins R. M., Silber A. M., Schenkman S., and Elias M. C. (2009) Trypanosome prereplication machinery contains a single functional orc1/cdc6 protein, which is typical of archaea. Eukaryotic Cell 8, 1592–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. de Jesus T. C., Nunes V. S., Lopes M. C., Martil D. E., Iwai L. K., Moretti N. S., Machado F. C., de Lima-Stein M. L., Thiemann O. H., Elias M. C., Janzen C., Schenkman S., and da Cunha J. P. (2016) Chromatin Proteomics Reveals Variable Histone Modifications during the Life Cycle of Trypanosoma cruzi. J. Proteome Res. 15, 2039–2051 [DOI] [PubMed] [Google Scholar]
- 23. Torrente M. P., Zee B. M., Young N. L., Baliban R. C., LeRoy G., Floudas C. A., Hake S. B., and Garcia B. A. (2011) Proteomic interrogation of human chromatin. PloS One 6, e24747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hanna S. L., Sherman N. E., Kinter M. T., and Goldberg J. B. (2000) Comparison of proteins expressed by Pseudomonas aeruginosa strains representing initial and chronic isolates from a cystic fibrosis patient: an analysis by 2-D gel electrophoresis and capillary column liquid chromatography-tandem mass spectrometry. Microbiology 146, 2495–2508 [DOI] [PubMed] [Google Scholar]
- 25. Rappsilber J., Mann M., and Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nature Protocols 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 26. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 27. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 28. Cox J., Hein M. Y., Luber C. A., Paron I., Nagaraj N., and Mann M. (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chung J., Rocha A. A., Tonelli R. R., Castilho B. A., and Schenkman S. (2013) Eukaryotic initiation factor 5A dephosphorylation is required for translational arrest in stationary phase cells. Biochemical J. 451, 257–267 [DOI] [PubMed] [Google Scholar]
- 30. Bangs J. D., Uyetake L., Brickman M. J., Balber A. E., and Boothroyd J. C. (1993) Molecular cloning and cellular localization of a BiP homologue in Trypanosoma brucei. Divergent ER retention signals in a lower eukaryote. J. Cell Sci. 105, 1101–1113 [DOI] [PubMed] [Google Scholar]
- 31. McDowell M. A., Ransom D. M., and Bangs J. D. (1998) Glycosylphosphatidylinositol-dependent secretory transport in Trypanosoma brucei. Biochem. J. 335, 681–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Moretti N. S., da Silva Augusto L., Clemente T. M., Antunes R. P., Yoshida N., Torrecilhas A. C., Cano M. I., and Schenkman S. (2015) Characterization of Trypanosoma cruzi Sirtuins as possible drug targets for Chagas disease. Antimicrobial Agents Chemother. 59, 4669–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pariona-Llanos R., Pavani R. S., Reis M., Noel V., Silber A. M., Armelin H. A., Cano M. I., and Elias M. C. (2015) Glyceraldehyde 3-phosphate dehydrogenase-telomere association correlates with redox status in Trypanosoma cruzi. PloS One 10, e0120896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lebska M., Ciesielski A., Szymona L., Godecka L., Lewandowska-Gnatowska E., Szczegielniak J., and Muszynska G. (2010) Phosphorylation of maize eukaryotic translation initiation factor 5A (eIF5A) by casein kinase 2: identification of phosphorylated residue and influence on intracellular localization of eIF5A. J. Biol. Chem. 285, 6217–6226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. da Cunha J. P., Nakayasu E. S., de Almeida I. C., and Schenkman S. (2006) Post-translational modifications of Trypanosoma cruzi histone H4. Mol. Biochem. Parasitol. 150, 268–277 [DOI] [PubMed] [Google Scholar]
- 36. Van Rompay A. R., Johansson M., and Karlsson A. (2000) Phosphorylation of nucleosides and nucleoside analogs by mammalian nucleoside monophosphate kinases. Pharmacol. Therap. 87, 189–198 [DOI] [PubMed] [Google Scholar]
- 37. Schwanhausser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., and Selbach M. (2011) Global quantification of mammalian gene expression control. Nature 473, 337–342 [DOI] [PubMed] [Google Scholar]
- 38. Cobbe N., and Heck M. M. (2004) The evolution of SMC proteins: phylogenetic analysis and structural implications. Mol. Biol. Evol. 21, 332–347 [DOI] [PubMed] [Google Scholar]
- 39. Millan-Zambrano G., and Chavez S. (2014) Nuclear functions of prefoldin. Open Biol. 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Aravind L., Iyer L. M., and Anantharaman V. (2003) The two faces of Alba: the evolutionary connection between proteins participating in chromatin structure and RNA metabolism. Genome Biol. 4, R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sandman K., and Reeve J. N. (2005) Archaeal chromatin proteins: different structures but common function? Curr. Opinion Microbiol. 8, 656–661 [DOI] [PubMed] [Google Scholar]
- 42. Calderano S., Godoy P., Soares D., Sant'Anna O. A., Schenkman S., and Elias M. C. (2014) ORC1/CDC6 and MCM7 distinct associate with chromatin through Trypanosoma cruzi life cycle. Mol. Biochem. Parasitol. 193, 110–113 [DOI] [PubMed] [Google Scholar]
- 43. da Cunha J. P., Nakayasu E. S., Elias M. C., Pimenta D. C., Tellez-Inon M. T., Rojas F., Munoz M. J., Manuel M., Almeida I. C., and Schenkman S. (2005) Trypanosoma cruzi histone H1 is phosphorylated in a typical cyclin dependent kinase site accordingly to the cell cycle. Mol. Biochem. Parasitol. 140, 75–86 [DOI] [PubMed] [Google Scholar]
- 44. Mutomba M. C., and Wang C. C. (1998) The role of proteolysis during differentiation of Trypanosoma brucei from the bloodstream to the procyclic form. Mol. Biochem. Parasitol. 93, 11–22 [DOI] [PubMed] [Google Scholar]
- 45. Gonzalez J., Cornejo A., Santos M. R., Cordero E. M., Gutierrez B., Porcile P., Mortara R. A., Sagua H., Da Silveira J. F., and Araya J. E. (2003) A novel protein phosphatase 2A (PP2A) is involved in the transformation of human protozoan parasite Trypanosoma cruzi. Biochem. J. 374, 647–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Grellier P., Blum J., Santana J., Bylen E., Mouray E., Sinou V., Teixeira A. R., and Schrevel J. (1999) Involvement of calyculin A-sensitive phosphatase(s) in the differentiation of Trypanosoma cruzi trypomastigotes to amastigotes. Mol. Biochem. Parasitol. 98, 239–252 [DOI] [PubMed] [Google Scholar]
- 47. Lonhienne T. G., Forwood J. K., Marfori M., Robin G., Kobe B., and Carroll B. J. (2009) Importin-beta is a GDP-to-GTP exchange factor of Ran: implications for the mechanism of nuclear import. J. Biol. Chem. 284, 22549–22558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sazer S., and Dasso M. (2000) The ran decathlon: multiple roles of Ran. J. Cell Sci. 113, 1111–1118 [DOI] [PubMed] [Google Scholar]
- 49. Agabian N. (1990) Trans splicing of nuclear pre-mRNAs. Cell 61, 1157–1160 [DOI] [PubMed] [Google Scholar]
- 50. Gunzl A. (2010) The pre-mRNA splicing machinery of trypanosomes: complex or simplified? Eukaryotic Cell 9, 1159–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhou Q., Hu H., and Li Z. (2014) New insights into the molecular mechanisms of mitosis and cytokinesis in trypanosomes. Int. Rev. Cell Mol. Biol. 308, 127–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Oma Y., and Harata M. (2011) Actin-related proteins localized in the nucleus: from discovery to novel roles in nuclear organization. Nucleus 2, 38–46, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Olave I. A., Reck-Peterson S. L., and Crabtree G. R. (2002) Nuclear actin and actin-related proteins in chromatin remodeling. Ann. Rev. Biochem. 71, 755–781 [DOI] [PubMed] [Google Scholar]
- 54. Ivaska J., Pallari H. M., Nevo J., and Eriksson J. E. (2007) Novel functions of vimentin in cell adhesion, migration, and signaling. Exp. Cell Res. 313, 2050–2062 [DOI] [PubMed] [Google Scholar]
- 55. Yokoyama H. (2016) Chromatin-Binding Proteins Moonlight as Mitotic Microtubule Regulators. Trends Cell Biol. 26, 161–164 [DOI] [PubMed] [Google Scholar]
- 56. Li Z., and Wang C. C. (2008) KMP-11, a basal body and flagellar protein, is required for cell division in Trypanosoma brucei. Eukaryotic Cell 7, 1941–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Janke R., Dodson A. E., and Rine J. (2015) Metabolism and epigenetics. Ann. Rev. Cell Develop. Biol. 31, 473–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Povelones M. L. (2014) Beyond replication: division and segregation of mitochondrial DNA in kinetoplastids. Mol. Biochem. Parasitol. 196, 53–60 [DOI] [PubMed] [Google Scholar]
- 59. Vanhamme L., and Pays E. (1995) Control of gene expression in trypanosomes. Microbiol. Rev. 59, 223–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Marques Porto R., Amino R., Elias M. C., Faria M., and Schenkman S. (2002) Histone H1 is phosphorylated in non-replicating and infective forms of Trypanosoma cruzi. Mol. Biochem. Parasitol. 119, 265–271 [DOI] [PubMed] [Google Scholar]
- 61. DuBois K. N., Alsford S., Holden J. M., Buisson J., Swiderski M., Bart J. M., Ratushny A. V., Wan Y., Bastin P., Barry J. D., Navarro M., Horn D., Aitchison J. D., Rout M. P., and Field M. C. (2012) NUP-1 Is a large coiled-coil nucleoskeletal protein in trypanosomes with lamin-like functions. PLoS Biol. 10, e1001287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bernstein E., and Hake S. B. (2006) The nucleosome: a little variation goes a long way. Biochem. Cell Biol. 84, 505–517 [DOI] [PubMed] [Google Scholar]
- 63. Mandava V., Janzen C. J., and Cross G. A. (2008) Trypanosome H2Bv replaces H2B in nucleosomes enriched for H3 K4 and K76 trimethylation. Biochem. Biophys. Res. Commun. 368, 846–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lowell J. E., Kaiser F., Janzen C. J., and Cross G. A. (2005) Histone H2AZ dimerizes with a novel variant H2B and is enriched at repetitive DNA in Trypanosoma brucei. J. Cell Sci. 118, 5721–5730 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.