Abstract
Although Rho GTPases are essential molecular switches involved in many cellular processes, an unbiased experimental comparison of their interaction partners was not yet performed. Here, we develop quantitative GTPase affinity purification (qGAP) to systematically identify interaction partners of six Rho GTPases (Cdc42, Rac1, RhoA, RhoB, RhoC, and RhoD), depending on their nucleotide loading state. The method works with cell line or tissue-derived protein lysates in combination with SILAC-based or label-free quantification, respectively. We demonstrate that qGAP identifies known and novel binding partners that can be validated in an independent assay. Our interaction network for six Rho GTPases contains many novel binding partners, reveals highly promiscuous interaction of several effectors, and mirrors evolutionary relationships among Rho GTPases.
The Ras superfamily of small guanosine triphosphatases (Ras GTPases) consists of more than 150 members in mammals and conserved orthologs in all eukaryotes (1). As molecular switches, they cycle between an active GTP- and an inactive GDP-bound state. GTPase-activating proteins (GAPs)1 stimulate the slow intrinsic GTPase activity, which inactivates the GTPase. Conversely, guanine nucleotide exchange factors (GEFs) promote release of GDP, which is replaced by GTP, thereby transforming the GTPase into the active state. The GTP-bound form binds to downstream effector proteins to initiate their specific cellular function. In this manner, GTPases control numerous biological processes, including cytoskeletal rearrangements, membrane dynamics, and gene expression. Rho GTPases form a subfamily of the Ras superfamily (2). Of its 22 mammalian members, RhoA, Rac1, and Cdc42 have been studied most intensively and are best known for their role in regulating the actin cytoskeleton (3).
Identifying effector proteins is key to understanding Rho GTPase function. More than 70 effector proteins have already been identified for each of the three prototypical family members, RhoA, Rac1, and Cdc42 (4). However, new effector proteins are still being discovered, and little is known about effectors of less well studied family members. Systematic screens for GTPase effectors employed the yeast two-hybrid approach (5) or immobilized GTPases for affinity purification (6, 7). However, these approaches are semiquantitative at best, which makes it difficult to distinguish loading-state-specific binders from constitutive interactors and nonspecific contaminants. Due to these challenges, an unbiased interactor screen for multiple Rho GTPases has not yet been reported.
We sought to systematically identify proteins that interact with Rho GTPases in a loading-state-specific manner. Quantitative affinity purification combined with mass spectrometry is a powerful technology that can be used to identify protein–protein interactions (PPIs) in an unbiased way (8–11). Here, we develop quantitative GTPase affinity purification (qGAP) as a novel variant of quantitative affinity purification combined with mass spectrometry to systematically identify Rho GTPase interaction partners. We then employ qGAP to screen for interaction partners of the three prototypical Rho GTPases (Cdc42, Rac1, RhoA) and three additional family members (RhoB, RhoC, RhoD). We employ qGAP in a SILAC-dependent way with lysates of cultured cells (SILAC-qGAP) and a label-free manner (LF-qGAP) with tissue samples. We show that qGAP identifies many well-known effectors and dozens of potential new interaction partners. Importantly, new interaction partners can be validated with a high success rate in an independent assay. Our interaction network reflects evolutionary relationships among Rho GTPases and reveals promiscuous binding of several known effectors.
RESULTS
Establishing SILAC-qGAP
Quantitative affinity purification combined with mass spectrometry can be used to study cellular dynamics of PPIs (8, 9, 11–13). The principle is to compare the abundance of proteins copurifying with the bait to an internal control. The approach can thus distinguish specific interaction partners from nonspecific contaminants: While true interaction partners are more abundant in the bait sample, nonspecific contaminants show a 1:1 ratio. This strategy has been successfully employed to identify both constitutive PPIs and dynamic PPIs that are regulated by cellular signaling events and/or posttranslational modifications (14–17).
We reasoned that quantitative affinity purification combined with mass spectrometry should also allow us to identify GTPase effector proteins in an unbiased manner (Fig. 1 A). To establish the method, we recombinantly expressed Rho GTPases as GST-fusion proteins in Escherichia coli and purified them to homogeneity. In order to test whether or not purified recombinant GTPases were enzymatically active, we initially monitored the intrinsic GTPase activities of RhoA, Cdc42, and Rac1 by an HPLC-based method. All three Rho GTPases showed GTP-hydrolysis activity (Fig. S1) with rate constants similar to previously published data (18–21). To further evaluate their functionality, we added HEK cell lysates to the reactions overexpressing citrine-labeled GAPs specific for each GTPase (22–24). Lysates of citrine-expressing control cells increased GTP hydrolysis, presumably due to the presence of endogenously expressed GAPs in the lysate (Fig. S1). The presence of citrine-labeled GAPs dramatically increased GTP hydrolysis (Fig. S1). These data clearly show that the purified recombinant GTPases used in our experiments were fully active and capable of interacting with known interaction partners.
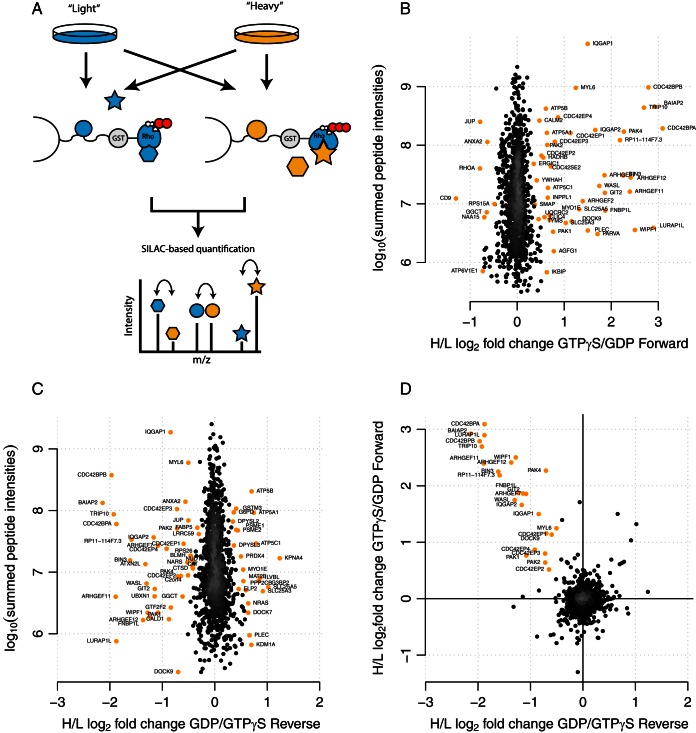
Fig. 1.
Identification of Cdc42 interaction partners with SILAC-qGAP. (A) Rho GTPases were expressed, purified, loaded with either GDP or GTPγS, and bound to a Sepharose matrix. Protein samples were obtained from SILAC-labeled cells. Proteins from H- and l-labeled cells were pulled down with Cdc42GTPγS and Cdc42GDP in a forward (arrows) and reverse experiment. Bound proteins were eluted and analyzed via mass spectrometry. Significant outliers were calculated (labeled in orange) for the forward (B) and reverse (C) experiment. Proteins that are significant outliers in the forward and reverse experiment and that were specific to one nucleotide form in both experiments are considered specific interaction partners of Cdc42 (D, labeled in orange).
In order to test whether or not we can identify activation state-specific binders, we loaded recombinant Cdc42 with GDP or GTPγS, a GTP-analog that is inefficiently hydrolyzed (25). Differentially loaded fusion proteins were then covalently coupled to N-hydroxysuccinimide-activated Sepharose beads. These beads were used to pull-down interacting proteins from differentially SILAC-labeled HeLa cell lysates. After washing, proteins were combined, eluted, and analyzed by high resolution shotgun proteomics and MaxQuant (26). In total, we identified over 1,000 proteins (Fig. 1B). As expected, the vast majority of these proteins had SILAC log2 fold changes around 0, indicating that they do not interact with Cdc42 in a loading-state-specific way. We then selected significant outliers in the distribution of log2 fold changes as putative specific interaction partners of the GTP- and GDP-loaded forms (calculation with the software package Perseus, “significance B,” Benjamini Hochberg false discovery rate < 5% (26)). Most of these putative binders showed preferential interaction with the GTPγS-loaded form. This is consistent with the idea that the GTP-bound form mediates most downstream functions (27, 28). As an additional control, we also performed a label swap experiment (Fig. 1C). We then plotted the log2 fold changes from both experiments against each other (Fig. 1D). Specific binders were required to be significant outliers in both experiments and to show inverted ratios. Note that the same heavy and light lysates were used for both the forward and reverse experiment. Therefore, requiring an inverted ratio in the label swap eliminates proteins that deviate from the 1:1 ratio already in the lysate. According to these criteria, 25 proteins were defined as loading-state-specific binders. All of them showed preferential binding to the GTPγS-loaded form. The list contains several well-known Cdc42 interactors like Cdc42 binding proteins (CDCBPA and CDC42BPB), Cdc42 effector proteins (CDC42EP1, 2, 3, and 4) as well as BAIAP2. Collectively, these data indicate that our assay can reliably identify Cdc42 effectors.
Establishing LF-qGAP
One limitation of the experiments described above is that they are based on lysate derived from HeLa cells. Hence, proteins that are not expressed in this cell line cannot be detected. We therefore sought to employ lysate derived from tissues as broader source of proteins for our qGAP assay (Fig. 2A). We chose mouse brain lysate since five of the six Rho GTPases studied here are expressed in this tissue (only RhoC seems to be expressed in macrophages or glandular cell types (29)). In addition, different cell types are present in brain, which should lead to a large number of potential binding proteins being available in the lysate. For quantification, we chose a label-free approach that was used previously to study interaction partners of GFP-fusion proteins (30, 31).
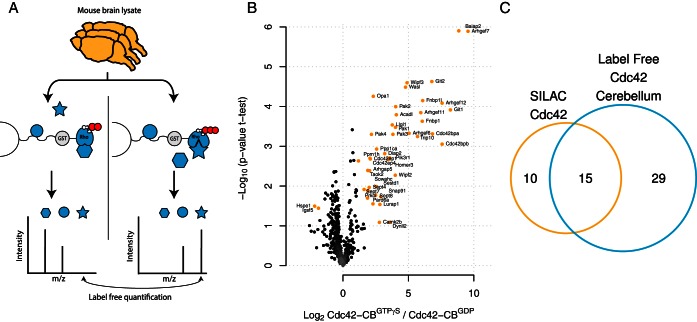
Fig. 2.
Identification of Cdc42 interaction partners with LF-qGAP from mouse cerebellum. (A) Protein lysates from mouse cerebellum (n = 3 independent samples) were incubated with Cdc42GTPγS or Cdc42GDP coupled to beads. Bound proteins were eluted and analyzed via mass spectrometry in triplicates. Differences in intensities between runs (Log2 Cdc42GTPγS/Cdc42GDP) and statistical significance (-Log10(p value t test)) were calculated. (B) Specific interactors are distinguished from background proteins based on a combination of the log2 fold change and the t test p value (significantly different proteins in orange), Cdc42GTPγS (right) or Cdc42GDP (left) (colored in red). (C) We identified more interaction partners for LF-qGAP (44 proteins) than for SILAC-qGAP (25) with a significant overlap (15).
To test the performance of (LF-qGAP, we repeated pull-downs with Cdc42 using protein lysate prepared from mouse cerebellum (Figs. 2A and 2B). In total, we performed six pull-downs—three with the GDP-loaded form and three with the GTPγS-loaded form. After proteomic analysis of all six samples, we used label-free quantification by MaxQuant (32) and displayed the data as a volcano plot (Fig. 2B). We accepted proteins as loading-state-specific binders based on a combination of the t test p value and the fold change at a permutation-based false discovery rate of 0.05 (30). In total, these experiments identified 42 and two potential interactors of the GTPγS- and GDP-loaded form, respectively. The increased number of identified interaction partners compared with the SILAC data (Fig. 2C) is probably due to a combination of technical (experimental setup, data analysis) and biological factors (different protein levels, cofactors, and posttranslational modifications). For example, we observed that interaction partners that we exclusively found in brain tissue pull-downs tend to be more abundant in brain tissue than in HeLa cell lysate (data not shown). Thus, LF-qGAP is not necessarily superior to the SILAC-qGAP. The choice of the lysate should generally be guided by the biological question.
LF-qGAP for Six Rho GTPases
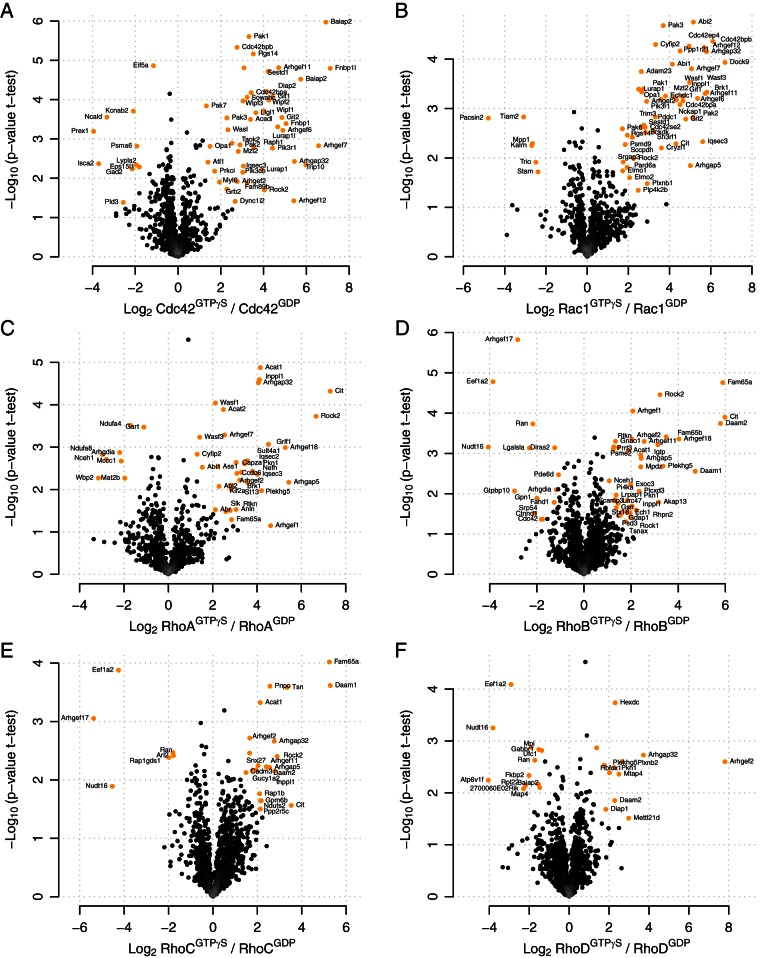
Encouraged by these results, we used LF-qGAP to screen for interaction partners of Rac1, Cdc42, RhoA, RhoB, RhoC, and RhoD (Fig. 3 and Fig. S2). For Rac1, Cdc42, and RhoA, we also used lysates derived from different brain regions (cerebellum, cerebrum, and hippocampus) while whole brain lysates were used for RhoB, RhoC, and RhoD. Every experiment was performed in biological triplicates. We performed ten experiments (six Rho GTPases and varying brain tissues) with six pull-downs for each experiment (three replicates for GDP and GTP each, i.e. a total number of 60 individual pull-downs). From these, we generated an unbiased map of Rho GTPase interaction partners. For each GTPase, we identified between 24 and 73 potential interactors. In total, this amounted to 293 interactions (Supplemental Table 1). Most of them were specific for the GTPγS-loaded form, consistent with the idea that the GTP-loaded form mediates most downstream functions. Many of the proteins we identified as GTPγS-specific binders are well-known effectors (like N-WASP for Cdc42, Rhotekin for RhoA and RhoB) while others are novel (such as mitotic spindle organizing protein 2 for Cdc42). Similarly, some of the GDP-specific interactions are known (e.g. Tiam2-Rac1) and some are new (e.g. Traf7-Rac1).
Fig. 3.
LF-qGAP from brain lysates for six Rho GTPases. Cerebrum pull-downs for (A) Cdc42, (B) Rac1, (C) RhoA. Pull-downs from whole brain lysates for (D) RhoB, (E) RhoC, (F) RhoD. As before, specific interactors (colored in orange) are distinguished from background proteins based on a combination of the log2 fold change and the t test p value. GTPγS-specific interactors are expected in the upper right corner, interactors of the GDP-form in the upper left corner.
The analysis above is based on the direct comparison of the GTPγS and the GDP-loaded form of an individual Rho GTPase. While this is the most relevant control strategy, we would like to point out that our data can be analyzed in different ways. For example, differences in interactors between GTP-loaded forms of two different GTPases could be identified by directly comparing them to each other (e.g. Rac1- GTPγS with Cdc42-GTPγS).
Comparison to Published Data
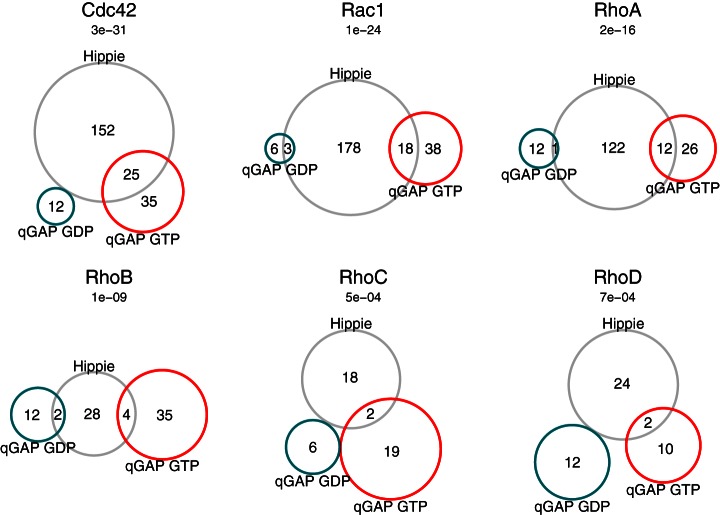
To systematically compare our data with known interactors, we used an integrated protein interaction database, HIPPIE, as a reference (33). We found that known targets are highly significantly overrepresented in our dataset (Fig. 4, p values between 7 × 10−4 and 3 × 10−31, p(X ≥ x), hypergeometric test). Since several literature-described Rho GTPase interaction partners are not in the reference database (e.g. Plekhg5 for RhoA (34) and Arhgap5 for Rac1 (35)) but were identified in our screen, we expect the number of true positive identifications of qGAP to be even higher. In addition, our data also contain proteins that are probably indirect binders. For example, we found association of lethal giant larvae homolog 1 with Cdc42, which is probably mediated via the complex of PAR-6A and atypical protein kinase C (aPKC-lambda/iota) that were also both identified. Whether or not detection of indirect interactors is a strength or weakness is a matter of perspective. In any case, our data show that qGAP hits are highly enriched for proteins involved in Rho GTPase biology.
Fig. 4.
Overlap with published interactions. Interactors identified by qGAP (GDP specific interactors in turquoise; GTPγS specific interactors in red) with interactors listed in the HIPPIE protein-protein interaction Database (gray circle). p values indicate significance of the enrichment of overlapping interactors between qGAP and HIPPIE and are given above each Venn diagram.
Validation
For each Rho GTPase, we identified dozens of novel interaction partners (see also colored lines in Fig. 6A). Thus, our dataset provides a valuable resource for exploring Rho GTPase biology. However, since qGAP is an in vitro assay, it is not clear if identified potential interactors also associate with their respective Rho GTPase in living cells. We therefore sought to validate qGAP hits using an independent method. Ideally, this method should (i) assess interactions of endogenous untagged proteins in vivo, (ii) provide an unbiased quantitative read out, and (iii) differentiate between the GTP- and the GDP-loaded form of the Rho GTPase. The proximity ligation assay (PLA) uses oligonucleotides attached to antibodies against two target proteins that guide the formation of circular DNA strands when bound in close proximity (36). The DNA can be amplified and visualized, which enables unbiased quantification of endogenous protein complexes at single-molecule resolution. The GTP-loading state of endogenous Rho GTPases can be manipulated with the bacterial toxins CNFY and CNF1 (37, 38). These toxins deamidate a single glutamine residue that is required for GTPase activity. Thus, toxin treatment locks the endogenous Rho GTPases in the GTP-bound state. CNF1 targets RhoA at Gln63 and Rac1 and Cdc42 at Gln61 while CNFY is specific for RhoA (39–41).
Fig. 6.
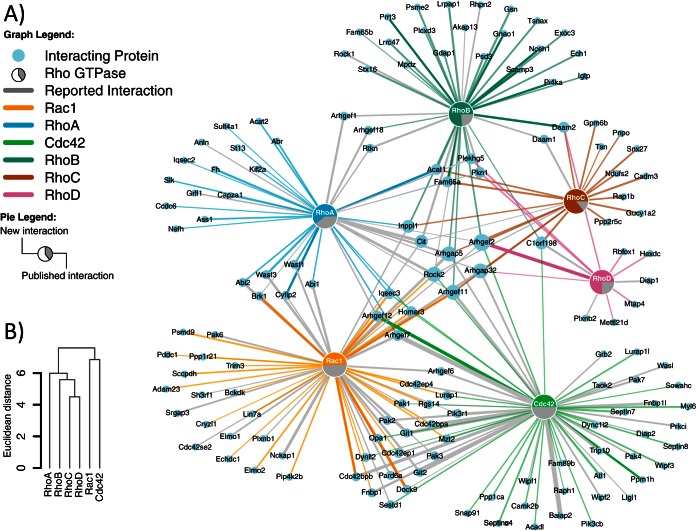
(A) Interaction network between Rho GTPases and identified binding partners of the GTPγS-loaded form. Known (gray) and new (colored) interactions are visualized by line colors and the pie charts for every Rho GTPase. The width of edges corresponds to relative intensity ratio between GDP- and GTPγS-bound Rho GTPase. (B) Clustering Rho GTPases based on shared binding partners mirrors their phylogenetic relationships.
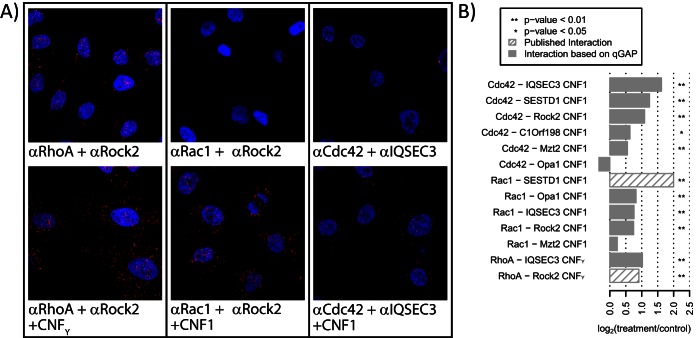
We reasoned that combining PLA with CNF1 or CNFY treatment should allow us to validate qGAP interaction partners in situ. HeLa cells were serum starved, treated with the toxin or vehicle control for 1 h, and fixed. We then used PLA to assess the proximity of Rho GTPases with identified interaction partners using specific antibodies. The PLA signal between RhoA and the known effector Rho-associated protein kinase 2 (Rock2) increased upon toxin treatment, indicating that Rock2 associates with activated RhoA (Fig. 5A, left). Rock2 was originally described to be specific for RhoA (42). Interestingly, qGAP also detected association of Rock2 with Rac1 and Cdc42, and this was validated by our PLA-based approach (Figs. 5A center and 5B). PLA also validated interaction of Cdc42 with IQSEC3 (Fig. 5A, right). In total, our PLA-based method validated 9/11 tested novel interactions (Fig. 5B and Fig. S3). The data suggest that most of the novel qGAP interactors also associate with their cognate Rho GTPase in a loading-state-specific manner in situ.
Fig. 5.
Validation of loading-state-dependent interactions in situ with the proximity ligation assay (PLA). (A) Cells were treated with CNFY (for RhoA activation) or with CNF1 (Cdc42 and Rac1 activation), fixed, costained with oligonucleotide-coupled antibodies against the Rho GTPase and interactors. The basal PLA signal (red) increased upon toxin treatment for RhoA-Rock2 (known interaction), Rac1-Rock2 and Cdc42-IQSEC3 (both novel). (B) Automated quantification shows that toxin treatment significantly increased PLA signals for 9/11 tested novel interactions.
A Protein–Protein Interaction Map for Rho GTPases
To provide a global overview, we assembled our data into the first protein–protein interaction network for multiple Rho GTPases (Fig. 6A). Many proteins were found to interact only with a single Rho GTPase. However, several proteins such as Rock2, Arhgef2, and Arhgap5 were surprisingly promiscuous. For Rock2, promiscuous association with Rac1 and Cdc42 was also validated with the proximity ligation assay (Fig. 5). The number of shared effectors is probably even higher since proteins may have escaped detection in individual experiments. Thus, our data indicate extensive crosstalk between Rho GTPases. Our network also shows that Rac1 and Cdc42 share more interaction partners with each other than with RhoA, B, C, and D. This is consistent with the higher level of sequence homology between Rac1 and Cdc42. Intriguingly, hierarchical clustering of the six Rho GTPases based on our interaction network results in a dendrogram that resembles their phylogenetic relationships (Fig. 6B). Hence, closely related family members share more interaction partners than distantly related ones. It is tempting to speculate that orthologs of some of the shared targets already interacted with their common ancestor.
DISCUSSION
Rho proteins are key regulators of the actin cytoskeleton and involved in a plethora of biological processes. However, a quantitative screen for interaction partners of multiple Rho GTPases has not yet been published. This is probably mainly due to the technical challenges involved. For example, while the yeast two-hybrid approach was used to screen for interaction partners of Cdc42 and Rac1 (5), this method cannot distinguish between the GTP- and the GDP-loaded forms of the GTPase. Moreover, previous attempts to identify interactors in pull-down assays required huge amounts of input material (e.g. 14 bovine brains for one experiment) and were not quantitative (6, 7). Here, we combined the pull-down assay with SILAC-based or label-free quantification to establish qGAP as a novel means to identify Rho GTPase interaction partners. We expect that this method is also applicable to the other ∼150 proteins of the Ras superfamily. Using qGAP, we generate the first extensive Rho interaction network including the representative family members RhoA, Rac1, Cdc42, RhoB, RhoC, and RhoD. Our network reveals that several effectors such as Rock2 bind highly promiscuously. The network also reflects phylogenetic relationships between Rho family members. Hence, the specificity of individual effectors appears to mirror the evolution of Rho GTPase signaling. Finally, we report many novel Rho GTPase interaction partners and thus provide a useful resource for the community.
Despite the advantages of the qGAP method, it is important to also keep limitations in mind. First, the method is based on an in vitro pull-down. It is therefore not clear if identified interactions also occur in living cells. While 9 out of 11 tested interactions were validated using the proximity ligation assay and bacterial toxins (Fig. 5), we cannot provide independent evidence for all novel interactions. A second limitation is that we can only detect interactions of proteins that are present in the lysate used for the pull-down assay. The comparison of interaction partners identified in HeLa cell lysate and brain lysate shows that tissue choice is an important factor (Fig. 2). It should also be noted that differences in experimental design for some bait proteins might influence our interaction network (Fig. 6). Finally, qGAP detects both direct and indirect interactions. This can be seen as an advantage or a disadvantage of the method. In any case, it is important to keep this point in mind when interpreting the data.
It is also important to note that experiments with HeLa and brain lysate differ in several ways (choice of tissue, SILAC versus LFQ, statistical analysis, mass spectrometers used). This paper does not intend to compare both approaches. The choice of the biological system and therefore also the method for quantification will typically depend on the biological question of interest: Cell-culture-based systems lend themselves to SILAC-based quantification while studying in vivo systems is usually easier with label-free methods. Our results show that qGAP works with both.
Following up on the biological significance of novel interactors is beyond the scope of this manuscript. However, we would like to point at a number of interesting observations that could be addressed in future studies. One remarkable finding is that ARHGEF 1, 2, 6, 7, 11, 12, and 18 consistently associated with the GTPγS-bound form of the respective Rho protein. It is well known that catalytic domains of GEFs preferentially bind to the nucleotide-free form of small GTPases (43). However, several GEF proteins have been previously shown to bind to the GTP-bound form of their GTPase via adjacent domains, like PH (Pleckstrin homolgy domain) domains (44). In fact, all of the above-mentioned GEFs have been reported to associate with GTP-loaded GTPases (44, 45). Hence, our data corroborate the previously postulated noncanonical binding mode.
Our data for RhoB, RhoC, and RhoD are particularly informative since these GTPases are less well characterized. RhoB and RhoC are closely related to RhoA, yet they have different effects on cell shape and migration (46). Our interaction data may therefore help to understand their nonredundant function. For example, we detect Rhophilin-2 only in the RhoB-GTPγS pull-downs, and this protein was previously shown to be specific for RhoB (47). Similarly, we also see AKAP13 (the lymphoid blast crisis oncogene) only in RhoB pull-downs. This is intriguing since both AKAP13 and RhoB are specifically involved in apoptosis induction by farnesyl transferase inhibitors (48, 49).
In summary, we develop qGAP, to identify interaction partners of Rho GTPases dependent on the loading state. We present the first extensive screen for Rho GTPase interaction partners with many new potential binding partners that can be validated with a high success rate by an orthogonal method. Our first systematic comparison of Rho GTPase effectors provides a rich resource for the community that has so far been lacking.
EXPERIMENTAL PROCEDURES
Experimental Design and Statistical Rationale
For SILAC experiments, protein samples were obtained from adherent HeLa cell culture. Pull-downs were performed with lysate from one dish (15 cm), and beads from one heavy and one light experiment were mixed before elution (see Fig. 1). A label swap experiment was carried out. Label-free experiments were performed as biological triplicates for each nucleotide form of a Rho GTPase. This allowed us to do a t test on the obtained data. Additionally, several tissues were examined (hippocampus, cerebellum, cerebrum, whole brain, see Fig. 2).
Protein Expression and Purification
N-terminal GST-fusion proteins of RhoA, RhoB, RhoC, RhoD, Rac1, and Cdc42 were expressed in E. coli BL21 (DE3) Rosetta. N-terminal GST-tagged CNF1 and CNFY were expressed in E. coli Tuner pLysS. Bacteria were grown at 37 °C in TB medium supplemented with 100 μg/ml ampicillin and 34 μg/ml chloramphenicol until an OD600 of 0.4. Subsequently, protein expression was induced by addition of 40 μm isopropyl β-d-1-thiogalactopyranoside and bacteria grown at 18 °C and 180 rpm shaking for at least 15 h. Bacteria were lysed by either passing at least twice through a microfluidizer or by 6 × 10 s of sonication (UP200S, 0.85 amplitude, 0.5 s cycle time, sonotrode DRH-S2) with cooling steps in between. The lysate was cleared from debris by centrifugation (50,000 g, 4 °C, 45 min, JA-12 rotor) and filtered through a 0.2 μm pore size filter.
Affinity purification was performed with GSH Sepharose columns at a flow rate of 1 ml/min. The column was equilibrated with 6 CV (column volumes) equilibration buffer (20 mm HEPES, pH 7.5, 150 mm NaCl, 5 mm MgCl2, 2.5 mm DTT). The filtered bacteria lysate was applied, the column was washed with 30 CV washing buffer (20 mm HEPES, pH 7.5, 300 mm NaCl, 5 mm MgCl2, 2.5 mm DTT) followed by 5 CV equilibration buffer and bound proteins were finally eluted with 5 CV elution buffer (20 mm HEPES, pH 7.5, 150 mm NaCl, 5 mm MgCl2, 2.5 mm DTT, 20 mm glutathione). Size exclusion chromatography was performed for RhoA, Rac1, and Cdc42 with concentrated protein solutions with a Superdex 200 16/60 column equilibrated with 2 CV equilibration buffer. The peak fractions containing the protein of interest were pooled and concentrated.
Nucleotide exchange was performed by EDTA-driven Mg2+-depletion as described previously (50). 200 μm of the respective Rho GTPase were incubated with 15 mm EDTA, 150 mm NH4SO4, and 10 mm of GDP or 2 mm GTPγS (in 1 m HEPES, pH 7.5) overnight at 4 °C. The exchange reaction was stopped by addition of 30 mm MgCl2. Excess nucleotide was removed by Amicon concentrator or buffer exchange via FPLC. Determination of bound nucleotide was followed standard HPLC protocols (51). GST-CNF1 and CNFY were stored at −20 °C in 50 mm Tris, pH 7.5, 50% glycerol.
GTPase Activity Assays
GTPase activity of RhoA, Cdc42, and Rac1 was determined by monitoring GTP to GDP ratios over time by HPLC measurements (52, 53). GTPases were loaded with GTP prior to measurements. Nucleotide exchange of RhoA was performed by EDTA-driven Mg2+-depletion (50). Due to the fast intrinsic GTP hydrolysis rates of Cdc42 and Rac1, nucleotide-free forms of GTPase were generated prior to GTP loading as described before (53, 54).
For monitoring RhoA activity 50–100 μm GTP loaded GTPase was quickly thawed and diluted in assay buffer (20 mm HEPES, 150 mm NaCl, 5 mm MgCl2, 2.5 mm DTT, pH 7.5) or prepared cell lysates in a 1:5 ratio. Hydrolysis reaction was assayed at room temperature and stopped by adding trichloroacetic acid (final concentration of 20%). Samples were centrifuged and neutralized. HPLC measurements were carried out using a reversed-phase C18 column (Reversed-phase ODS-2 Hypersil HPLC column, Thermo Scientific) and a precolumn (Hypersil ODS guard column, Agilent Technologies) for adsorbing residual denatured protein. Measurements were carried out in HPLC-buffer containing 10 mm tetrabutylammoniumbromide and 100 mm potassium phosphate (pH 6,5) with 7.5% acetonitrile. Nucleotide peaks were detected by measuring adsorption at 254 nm and quantified by integration.
GTPase activity of Cdc42 and Rac1 was monitored as described (18). Briefly, 50–100 μm nucleotide-free GTPase was quickly mixed with 50 μm GTP in GAP buffer (30 mm Tris, 10 mm MgCl2, 3 mm DTT, 10 mm K2HPO4/KH2PO4, pH 7.5). Subsequently, assay buffer or prepared cell lysate was added in a 1:5 ratio. Samples were collected as described for RhoA and subjected to HPLC analysis.
Preparation of Cell Lysates for GTPase Activity Assays
HEK 293T cells were cultivated at 37 °C with 5% CO2 in DMEM culture media supplemented with 5% fetal calf serum (FCS) and 1% penicillin/streptomycin. For GTPase activity assays lysates of cells transfected with GAP proteins with a known activity toward the respective GTPase (RhoA: STARD13, Cdc42: CDGAP; Rac1: ChimerinH206K; all encoded as mCitrine fusion proteins on pLP-mCitrine C1 SP vectors) were used. Transfection was mediated using the polyethylenimine-mediated DNA transfer method. Cells were lysed in Nonidet P-40 lysis buffer (1% Nonidet P-40, 10% glycerol, 20 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1 mm EGTA, 5 mm NaF, Pefabloc, Benzonase), centrifuged, and the supernatant was washed three times in a VWR 15 ml concentrator (10 kDa cut-off) with assay buffer. The total protein concentration of the lysate was adjusted to 10 μg/μl. Lysates were immediately used in a GAP assay.
Preparation of Mouse Brain Lysate
Mouse brains were dissected from female BL/6 mice at an age of 12 weeks (kind gift of Dr. Ibanez-Tallon, MDC Berlin). Brains were either used as a whole (RhoB, RhoC, RhoD) or separated into a hippocampal (Cdc42), cerebellar (Cdc42, Rac1, RhoA), and remaining cerebrum fraction (Cdc42, Rac1, RhoA). The tissue samples were lysed in lysis buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Triton X-100, 1 mm EDTA, 1 mm EGTA, and protease inhibitors (Roche)) by application of 50–100 strokes in a dounce homogenizer on ice. The lysate was cleared from debris by two subsequent centrifugation steps at 20,000 g (4 °C, 20 min) and directly used for experiments. The input for a single pull-down was 500 μg (SILAC), 90 μg (hippocampus-LFQ), 700 μg (cerebellum-LFQ), 1400 μg cerebrum-LFQ), and 1500 μg (whole-brain-LFQ).
Pull-Down Assays
For pull-down assays cell lysates were freshly prepared from mouse brains. Experiments were conducted as triplicates, resulting in six single pull-downs per Rho GTPase (three for the GDP and three for the GTPγS-loaded protein). Each pull-down was analyzed separately by mass spectrometry and quantified using label-free quantification (see below). For SILAC experiments, lysate was prepared from labeled HeLa cells in adherent culture. The same lysis buffer as for mice brains was used. Sepharose beads with an active N-hydroxysuccinimide group were used for covalent coupling of recombinant proteins (GE Healthcare). 150 μl of bead slurry were used per single pull-down. Storage solution was removed from bead slurry by centrifugation (1000 g, 2 min). Beads were washed in ice-cold equilibration (1 mm HCl) buffer and incubated with 500 μg of the recombinant protein for at least 2 h at room temperature. The beads were subsequently washed in buffer A (0.5 m ethanolamine, pH 8.3, 0.5 m NaCl), buffer B (0.1 m sodium acetate, pH 4.0, 0.5 m NaCl) and incubated in buffer A for 30 min. After wash steps with buffers B, A, and again B, the cell lysate was added to the beads and incubation was performed for 30 min at 4 °C. The supernatant was removed, the beads washed in pull-down wash buffer (50 mm Tris, pH 7.4, 300 mm NaCl, 5 mm MgCl2) twice, and bound proteins were eluted with 200 μl U/T buffer (10 mm HEPES, pH 8.0, 6 m urea, 2 m thiourea) by shaking at 1400 rpm on an Thermo shaker (Eppendorf) for 15 min. Eluted proteins were precipitated in 70% ethanol.
Proximity Ligation Assay
HeLa cells were seeded on poly-l-lysine coated 18-well μ-slides (ibidi, Martinsried, Germany) for 1 day in standard cell culture medium (450 cells/well). After 24 h, the medium was replaced by starvation medium (0.2% FCS) and cells were cultivated for another 24 h. Purified cytotoxic necrotizing factors (0.4 g/l CNF1-GST or CNFY-GST in 50 mm Tris, pH 7.5, 50% glycerol) were dissolved in prewarmed PBS. Cells were then incubated for 1 h with either toxin at a final concentration of 0.4 μg/ml or the glycerol/PBS vehicle control. Cells were briefly washed in PBS, fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100 in PBS. Cells were washed in PBS, and unspecific binding of antibodies was blocked by incubation in PBS containing 1% BSA, 0.05% Tween20 for 20 min. Reagents for PLA were obtained from Olink Bioscience (Duolink® in situ orange starter kit). The primary antibody solution was applied for 1 h at 37 °C in a humidity chamber. Antibodies were combinations of one from mouse and one from rabbit donors. The two PLA probes were mixed, diluted 1:5 in antibody diluent buffer, and left for 20 min. The primary antibody was removed from the chamber slide. The slide was washed once; the PLA probe solution was added and incubated for 1 h at 37 °C. The probes were removed, the slide was washed twice for 5 min under gentle agitation, and the ligation–ligase solution was added to each sample and incubated in a preheated humidity chamber for 30 min at 37 °C. For amplification, the slide was washed twice for 2 min and the amplification–polymerase solution was added and incubated for 100 min in a preheated humidity chamber for 80 min at 37 °C. Finally, the slides were washed, dried, and In Situ Mounting Medium, including DAPI was added. Nonspecific signals were assessed by single primary antibody staining. Information about the antibodies used and their dilution can be found in Supplemental Table 2.
Microscopy and Processing of Images
Images were taken with a Leica TCS SP5 confocal microscope (Leica Microsystems, Wetzlar, Germany) using a 63x objective and Leica LAS AF software. Standard parameters were: pinhole of 2.5 AE, resolution of 2048 × 2048 pixels, line average of 4. Pictures were processed with ImageJ (version 1.44, Bethesda, MD) using the LOCI plugin. Pictures were imported without autoscale, threshold was set with a minimum of 30 and a maximum of 255, and “analyze particle” function was used for counting of spots.
Sample Preparation and Mass Spectrometry
Protein pellets were redissolved in U/T buffer and subsequently reduced with dithiothreitol and alkylated with iodoacetamide. Proteins were digested with LysC and trypsin. The peptides were desalted off-line and analyzed by online LC-MS on an EASY-nLC system (Thermo Scientific) coupled to a Q-Exactive-Orbitrap (Thermo Fisher Scientific, Waltham MA) for cerebrum and whole-brain samples or an LTQ-Orbitrap-Velos (Thermo Fisher Scientific) for hippocampus and cerebellum samples. SILAC samples were analyzed on an LTQ-Orbitrap XL (Thermo Fisher Scientific) system. 5 μl peptide samples were loaded onto a fritless microcolumn (75 μm inner diameter packed in-house with ReproSil-Pur C18-AQ 3-μm resin, Dr. Maisch GmbH). Peptides were eluted with an 8–60% acetonitrile gradient and 0.5% formic acid. Runs were performed as 4 h gradients at a flow rate of 200 nl/min. Peptides were ionized at currents of 2.2 kV (Q-Exactive and Orbitrap XL) and 2.3 kV (Orbitrap Velos). The Q-Exactive Orbitrap device was operated in data-dependent mode with a TOP10 method as previously described (55). One full scan (m/z range = 300 - 1650, r = 70,000, target value: 106 ions, maximum injection time = 20 ms) was used to detect precursor ions. The 10 most intense ions with a charge state greater than1 were selected for fragmentation (r = 17,500, target value = 106 ions, isolation window = 3 m/z, maximum injection time = 60 ms). Dynamic exclusion time for fragmented precursor ions was set to 30 s. The Velos Orbitrap was operated in the data-dependent mode with a standard TOP20 method. One full scan (m/z range = 300 - 1700, r = 60,000, target value = 106 ions) was used to detect precursor ions. The 20 most intense ions with a charge state greater than 1 were selected for fragmentation (target value 3000 ions, isolation window = 2 m/z). Dynamic exclusion time for fragmented precursor ions was set to 60 s.
The Orbitrap XL operated in data-dependent mode with one full scan in the Orbitrap analyzer (m/z = 300–1700; resolution = 60,000; target value = 106 ions). The five most intense ions with a charge state greater than 1 were selected (target value = 5,000) and fragmented in the linear trap quadrupole using CID (collision induced dissociation) (35% normalized collision energy and wideband activation enabled). Dynamic exclusion for selected precursor ions was 60 s.
Data Analysis and Label-Free Quantification with MaxQuant and Perseus
MS raw data files were analyzed with the MaxQuant software package (version 1.3.0.5) that includes the search engine Andromeda, using default settings (26). Carbamidomethylation on cysteines was used as fixed modification and oxidation on methionine and acetylated N terminus as variable modifications. Trypsin was used as protease, including cleavage between arginine or lysine and proline. Missed cleavages were set to two. Peptides were identified with 6 ppm precursor mass deviation and 20 ppm fragment mass deviation for Orbitrap Q Exactive devices. Samples from the LTQ-Orbitrap XL and Velos Orbitrap were allowed to have up to 6 ppm precursor mass deviation and 0.5 Da fragment mass deviation. Proteins were searched against the Uniprot mouse database (version June 2012, 59,377 entries) for LF-qGAP and Uniprot human database (version June 2012, 86,898 entries) for SILAC-qGAP. Identifications were filtered to a false discovery rate of 0.01 at the PSM (peptide spectrum match) and protein level using the target–decoy approach. The available Requantify option in MaxQuant was enabled. For label-free quantification, we directly compared peptide peak intensities between corresponding runs (match between runs enabled, LFQ min ratio count = 2) using the label-free quantification algorithm (LFQ) implemented in MaxQuant. Files produced by MaxQuant were further processed using the Perseus software package (version 1.3.0.4). The “LFQ Intensity” for each experiment was selected as expression value. The matrix was filtered for columns “Only identified by site,” “Contaminant,” and “Reverse hits.” For each set of experiments, we only accepted proteins that were quantified in all three replicates of either the GTPγS- or GDP-bound form. The logarithm to the base of 10 was taken from LFQ intensities and missing values imputed by sampling from a normal distribution (width 0.3, downshift 1.8). Assuming that mainly low abundant peaks are the cause for missing data in shotgun proteomics, the imputation settings aim to simulate the distribution of low abundant peaks. Missing data were imputed in each experiment individually (column per column). The Gaussian distribution used for imputation was modified by using 0.3 width (in standard deviation of the real data) and a 1.8 fold downshift (in standard deviation units of the valid data).
The significance of the difference in protein abundance between the GTPγS and GDP pull-downs was calculated with a two-sided t test. Specific interactors were then selected based on the combination of the t test p values and the log2 fold differences as previously described (false discovery rate 5%, s0 = 0.5) (56–58). Most interaction partners had at least two unique peptides. For five potential interactors that were identified by a single unique peptide, we provide annotated spectra in Fig. S4 (Cit, Fnbp1l, Pak3, Rps27, Syp). Arhgef7 is listed with zero unique peptides due to the presence of several splice variants in the Uniprot database. The interaction network was plotted using the igraph R-package (59). Enrichment of known Rho GTPase interactions in identified binding partners was tested against HIPPIE Database (33) using the hypergeometric test functions phyper and dhyper from R (version 3.2.2).
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD004190.
Supplementary Material
Acknowledgments
We thank Martha Hergeselle, Christian Sommer, and Sabine Werner for excellent technical assistance. Anne Ridley (Cancer Research UK), Gudula Schmidt (Freiburg University), and Alfred Wittinghofer (MPI of Molecular Physiology) kindly provided constructs.
Footnotes
Author contributions: O.R., O.D., and M.S. designed the research; F.E.P. and L.V. performed research; O.R. and O.D. contributed new reagents or analytic tools; F.E.P. and H.Z. analyzed data; and F.E.P., H.Z., and M.S. wrote the paper.
* This project was supported by the German Research Foundation (SFB958, Scaffolding of Membranes) and the Helmholtz Association.
 This article contains supplemental material.
This article contains supplemental material.
The authors declare that they have no conflict of interest.
1 The abbreviations used are:
- GAP
- GTPase-activating protein
- CRIB
- Cdc42/Rac interactive binding
- GEF
- GTPase exchange factor
- GDP
- guanosine diphosphate
- GTP
- guanosine triphosphate
- GTPase
- guanosine triphosphatases
- HIPPIE
- human integrated protein–protein interaction reference
- LFQ
- label-free quantification
- PLA
- proximity ligation assay
- PPI
- protein–protein interaction
- qGAP
- quantitative GTPase affinity purification
- Ras
- rat sarcoma
- Rho
- Ras homolog
- SILAC
- stable isotope labeling by/with amino acids in cell culture
- CNF
- cytotoxic necrotizing factor.
REFERENCES
- 1. Wennerberg K., Rossman K. L., and Der C. J. (2005) The Ras superfamily at a glance. J. Cell Sci. 118, 843–846 [DOI] [PubMed] [Google Scholar]
- 2. Jaffe A. B., and Hall A. (2005) Rho GTPases: Biochemistry and biology. Annu. Rev. Cell Dev. Biol. 21, 247–269 [DOI] [PubMed] [Google Scholar]
- 3. Ridley A. J. (2006) Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 16, 522–529 [DOI] [PubMed] [Google Scholar]
- 4. Bustelo X. R., Sauzeau V., and Berenjeno I. M. (2007) GTP-binding proteins of the Rho/Rac family: Regulation, effectors and functions in vivo. Bioessays 29, 356–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aspenström P., Lindberg U., and Hall A. (1996) Two GTPases, Cdc42 and Rac, bind directly to a protein implicated in the immunodeficiency disorder Wiskott–Aldrich syndrome. Curr. Biol. 6, 70–75 [DOI] [PubMed] [Google Scholar]
- 6. Kato K., Yazawa T., Taki K., Mori K., Wang S., Nishioka T., Hamaguchi T., Itoh T., Takenawa T., Kataoka C., Matsuura Y., Amano M., Murohara T., and Kaibuchi K. (2012) The inositol 5-phosphatase SHIP2 is an effector of RhoA and is involved in cell polarity and migration. Mol. Biol. Cell. 23, 2593–2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Christoforidis S., and Zerial M. (2000) Purification and identification of novel Rab effectors using affinity chromatography. Methods 20, 403–410 [DOI] [PubMed] [Google Scholar]
- 8. Paul F. E., Hosp F., and Selbach M. (2011) Analyzing protein–protein interactions by quantitative mass spectrometry. Methods 54, 387–395 [DOI] [PubMed] [Google Scholar]
- 9. Vermeulen M., Hubner N. C., and Mann M. (2008) High confidence determination of specific protein–protein interactions using quantitative mass spectrometry. Curr. Opin. Biotechnol. 19, 331–337 [DOI] [PubMed] [Google Scholar]
- 10. Meyer K., and Selbach M. (2015) Quantitative affinity purification mass spectrometry: a versatile technology to study protein–protein interactions. Front. Genet. 6, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gingras A. C., Gstaiger M., Raught B., and Aebersold R. (2007) Analysis of protein complexes using mass spectrometry. Nat. Rev. Mol. Cell Biol. 8, 645–654 [DOI] [PubMed] [Google Scholar]
- 12. Gavin A. C., Maeda K., and Kühner S. (2011) Recent advances in charting protein–protein interaction: Mass spectrometry-based approaches. Curr. Opin. Biotechnol. 22, 42–49 [DOI] [PubMed] [Google Scholar]
- 13. Zinn N., Hopf C., Drewes G., and Bantscheff M. (2012) Mass spectrometry approaches to monitor protein–drug interactions. Methods 57, 430–440 [DOI] [PubMed] [Google Scholar]
- 14. Tinti M., Kiemer L., Costa S., Miller M. L., Sacco F., Olsen J. V., Carducci M., Paoluzi S., Langone F., Workman C. T., Blom N., Machida K., Thompson C. M., Schutkowski M., Brunak S., Mann M., Mayer B. J., Castagnoli L., and Cesareni G. (2013) The SH2 domain interaction landscape. Cell Rep. 3, 1293–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Selbach M., and Mann M. (2006) Protein interaction screening by quantitative immunoprecipitation combined with knockdown (QUICK). Nat. Methods 3, 981–983 [DOI] [PubMed] [Google Scholar]
- 16. Vermeulen M., Eberl H. C., Matarese F., Marks H., Denissov S., Butter F., Lee K. K., Olsen J. V., Hyman A. A., Stunnenberg H. G., and Mann M. (2010) Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 142, 967–980 [DOI] [PubMed] [Google Scholar]
- 17. Selbach M., Paul F. E, Brandt S. Guye P., Daumke O., Backert S., Dehio C., and Mann M. (2009) Host cell interactome of tyrosine-phosphorylated bacterial proteins. Cell Host Microbe 5, 397–403 [DOI] [PubMed] [Google Scholar]
- 18. Jaiswal M., Fansa E. K., Dvorsky R., and Ahmadian M. R. (2013) New insight into the molecular switch mechanism of human Rho family proteins: Shifting a paradigm. Biol. Chem. 394, 89–95 [DOI] [PubMed] [Google Scholar]
- 19. Eberth A., Dvorsky R., Becker C. F., Beste A., Goody R. S., and Ahmadian M. R. (2005) Monitoring the real-time kinetics of the hydrolysis reaction of guanine nucleotide-binding proteins. Biol. Chem. 386, 1105–1114 [DOI] [PubMed] [Google Scholar]
- 20. Mazhab-Jafari M. T., et al. (2010) Real-time NMR study of three small GTPases reveals that fluorescent 2′(3′)-O-(N-methylanthraniloyl)-tagged nucleotides alter hydrolysis and exchange kinetics. J. Biol. Chem. 285, 5132–5136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang B., Wang Z. X., and Zheng Y. (1997) Characterization of the interactions between the small GTPase Cdc42 and its GTPase-activating proteins and putative effectors. Comparison of kinetic properties of Cdc42 binding to the Cdc42-interactive domains. J. Biol. Chem. 272, 21999–22007 [DOI] [PubMed] [Google Scholar]
- 22. Ching Y. P., Wong C. M., Chan S. F., Leung T. H., Ng D. C., Jin D. Y., and Ng I. O. (2003) Deleted in liver cancer (DLC) 2 encodes a RhoGAP protein with growth suppressor function and is underexpressed in hepatocellular carcinoma. J. Biol. Chem. 278, 10824–10830 [DOI] [PubMed] [Google Scholar]
- 23. Lamarche-Vane N., and Hall A. (1998) CdGAP, a novel proline-rich GTPase-activating protein for Cdc42 and Rac. J. Biol. Chem. 273, 29172–29177 [DOI] [PubMed] [Google Scholar]
- 24. Wegmeyer H., Egea J., Rabe N., Gezelius H., Filosa A., Enjin A., Varoqueaux F., Deininger K., Schnütgen F., Brose N., Klein R., Kullander K., and Betz A. (2007) EphA4-dependent axon guidance is mediated by the RacGAP alpha2-chimaerin. Neuron 55, 756–767 [DOI] [PubMed] [Google Scholar]
- 25. Cherfils J., Ménétrey J., Le Bras G., Janoueix-Lerosey I., de Gunzburg J., Garel J. R., and Auzat I. (1997) Crystal structures of the small G protein Rap2A in complex with its substrate GTP, with GDP and with GTPgammaS. EMBO J. 16, 5582–5591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p. p. b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 27. Etienne-Manneville S., and Hall A. (2002) Rho GTPases in cell biology. Nature 420, 629–635 [DOI] [PubMed] [Google Scholar]
- 28. Ridley A. J. (2001) Rho GTPases and cell migration. J. Cell Sci. 114, 2713–2722 [DOI] [PubMed] [Google Scholar]
- 29. Uhlen M., Oksvold P., Fagerberg L., Lundberg E., Jonasson K., Forsberg M., Zwahlen M., Kampf C., Wester K., Hober S., Wernerus H., Björling L., and Ponten F. (2010) Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 28, 1248–1250 [DOI] [PubMed] [Google Scholar]
- 30. Hubner N. C., Bird A. W., Cox J., Splettstoesser B., Bandilla P., Poser I., Hyman A., and Mann M. (2009) Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J. Cell Biol. 189, 739–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hein M. Y., Hubner N. C., Poser I., Cox J., Nagaraj N., Toyoda Y., Gak I. A., Weisswange I., Mansfeld J., Buchholz F., Hyman A. A., and Mann M. (2015) A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 163, 712–723 [DOI] [PubMed] [Google Scholar]
- 32. Cox J., Matic I., Hilger M., Nagaraj N., Selbach M., Olsen J. V., and Mann M. (2009) A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 4, 698–705 [DOI] [PubMed] [Google Scholar]
- 33. Schaefer M. H., Fontaine J. F., Vinayagam A., Porras P., Wanker E. E., and Andrade-Navarro M. A. (2012) HIPPIE: Integrating protein interaction networks with experiment based quality scores. PLoS ONE 7, e31826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. De Toledo M., Coulon V., Schmidt S., Fort P., and Blangy A. (2001) The gene for a new brain specific RhoA exchange factor maps to the highly unstable chromosomal region 1p36.2–1p36.3. Oncogene 20, 7307–7317 [DOI] [PubMed] [Google Scholar]
- 35. Burbelo P. D., Miyamoto S., Utani A., Brill S., Yamada K. M., Hall A., and Yamada Y. (1995) p190-B, a new member of the Rho GAP family, and Rho are induced to cluster after integrin cross-linking. J. Biol. Chem. 270, 30919–30926 [DOI] [PubMed] [Google Scholar]
- 36. Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K. J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L. G., and Landegren U. (2006) Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000 [DOI] [PubMed] [Google Scholar]
- 37. Aktories K., and Baribieri J. T. (2005) Bacterial cytotoxins: Targeting eukaryotic switches. Nat. Rev. Microbiol. 3, 397–410 [DOI] [PubMed] [Google Scholar]
- 38. Munro P., and Lemichez E. (2005) Bacterial toxins activating Rho GTPases. Curr. Top. Microbiol. Immunol. 291, 177–190 [DOI] [PubMed] [Google Scholar]
- 39. Schmidt G., Sehr P., Wilm M., Selzer J., Mann M., and Aktories K. (1997) Gln 63 of Rho is deamidated by Escherichia coli cytotoxic necrotizing factor-1. Nature 387, 725–729 [DOI] [PubMed] [Google Scholar]
- 40. Lerm M., Selzer J., Hoffmeyer A., Rapp U. R., Aktories K., and Schmidt G. (1999) Deamidation of Cdc42 and Rac by Escherichia coli cytotoxic necrotizing factor 1: Activation of c-Jun N-terminal kinase in HeLa cells. Infect. Immun. 67, 496–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hoffmann C., Pop M., Leemhuis J. Schirmer J., Aktories K., and Schmidt G. (2004) The Yersinia pseudotuberculosis cytotoxic necrotizing factor (CNFY) selectively activates RhoA. J. Biol. Chem. 279, 16026–16032 [DOI] [PubMed] [Google Scholar]
- 42. Leung T., Manser E., Tan L., and Lim L. (1995) A novel serine/threonine kinase binding the Ras-related RhoA GTPase which translocates the kinase to peripheral membranes. J. Biol. Chem. 270, 29051–29054 [DOI] [PubMed] [Google Scholar]
- 43. Bos J. L., Rehmann H., and Wittinghofer A. (2007) GEFs and GAPs: Critical elements in the control of small G proteins. Cell 129, 865–877 [DOI] [PubMed] [Google Scholar]
- 44. Medina F., Carter A. M., Dada O., Gutowski S., Hadas J., Chen Z., and Sternweis P. C. (2013) Activated RhoA is a positive feedback regulator of the Lbc family of Rho guanine nucleotide exchange factor proteins. J. Biol. Chem. 288, 11325–11333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Baird D., Feng Q., and Cerione R. A. (2005) The Cool-2/alpha-Pix protein mediates a Cdc42-Rac signaling cascade. Curr. Biol. 15, 1–10 [DOI] [PubMed] [Google Scholar]
- 46. Ridley A. J. (2013) RhoA, RhoB and RhoC have different roles in cancer cell migration. J. Microsc. 251, 242–249 [DOI] [PubMed] [Google Scholar]
- 47. Mircescu H., Steuve S., Savonet V., Degraef C., Mellor H., Dumont J. E., Maenhaut C., and Pirson I. (2002) Identification and characterization of a novel activated RhoB binding protein containing a PDZ domain whose expression is specifically modulated in thyroid cells by cAMP. Eur. J. Biochem. 269, 6241–6249 [DOI] [PubMed] [Google Scholar]
- 48. Prendergast G. C. (2001) Actin' up: RhoB in cancer and apoptosis. Nat. Rev. Cancer 1, 162–168 [DOI] [PubMed] [Google Scholar]
- 49. Raponi M., Harousseau J. L., Lancet J. E., Löwenberg B., Stone R., Zhang Y., Rackoff W., Wang Y., and Atkins D. (2007) Identification of molecular predictors of response in a study of tipifarnib treatment in relapsed and refractory acute myelogenous leukemia. Clin. Cancer Res. 13, 2254–2260 [DOI] [PubMed] [Google Scholar]
- 50. Tucker J., Sczakiel G., Feuerstein J., John J., Goody R. S., and Wittinghofer A. (1986) Expression of p21 proteins in Escherichia coli and stereochemistry of the nucleotide-binding site. EMBO J. 5, 1351–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lenzen C., Cool R. H., and Wittinghofer A. (1995) Analysis of intrinsic and CDC25-stimulated guanine nucleotide exchange of p21ras-nucleotide complexes by fluorescence measurements. Meth. Enzymol. 255, 95–109 [DOI] [PubMed] [Google Scholar]
- 52. Eberth A., and Ahmadian M. R. (2009) In vitro GEF and GAP assays. Curr. Protoc. Cell Biol. 43, 14.9.1–14.9.25 [DOI] [PubMed] [Google Scholar]
- 53. Jaiswal M., Dubey B. N., Koessmeier K. T., Gremer L., and Ahmadian M. R. (2012) Biochemical assays to characterize Rho GTPases. Meth. Mol. Biol. 827, 37–58 [DOI] [PubMed] [Google Scholar]
- 54. John J., Sohmen R., Feuerstein J., Linke R., Wittinghofer A., and Goody R. S. (1990) Kinetics of interaction of nucleotides with nucleotide-free H-ras p21. Biochemistry 29, 6058–6065 [DOI] [PubMed] [Google Scholar]
- 55. Kelstrup C. D., Young C., Lavallee R., Nielsen M. L., and Olsen J. V. (2012) Optimized fast and sensitive acquisition methods for shotgun proteomics on a quadrupole Orbitrap mass spectrometer. J. Proteome Res. [DOI] [PubMed] [Google Scholar]
- 56. Tusher V. G., Tibshirani R., and Chu G. (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U.S.A. 98, 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cox J., Hein M. Y., Luber C. A., Paron I., Nagaraj N., and Mann M. (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M. Y., Geiger T., Mann M., and Cox J. (2016) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 [DOI] [PubMed] [Google Scholar]
- 59. Csardi G., and Nepusz T. (2006) The igraph Software Package for Complex Network Research. InterJournal Complex Systems 1695, 1–9 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.