SUMMARY
Beneficial microorganisms hold promise for the treatment of numerous gastrointestinal diseases. The transfer of whole microbiota via fecal transplantation has already been shown to ameliorate the severity of diseases such as Clostridium difficile infection, inflammatory bowel disease, and others. However, the exact mechanisms of fecal microbiota transplant efficacy and the particular strains conferring this benefit are still unclear. Rationally designed combinations of microbial preparations may enable more efficient and effective treatment approaches tailored to particular diseases. Here we use an infectious disease, C. difficile infection, and an inflammatory disorder, the inflammatory bowel disease ulcerative colitis, as examples to facilitate the discussion of how microbial therapy might be rationally designed for specific gastrointestinal diseases. Fecal microbiota transplantation has already shown some efficacy in the treatment of both these disorders; detailed comparisons of studies evaluating commensal and probiotic organisms in the context of these disparate gastrointestinal diseases may shed light on potential protective mechanisms and elucidate how future microbial therapies can be tailored to particular diseases.
KEYWORDS: Clostridium difficile, fecal microbiota transplantation, microbiota, probiotics, ulcerative colitis
INTRODUCTION
Fecal microbiota transplantation (FMT) is an effective and promising therapy for a number of gastrointestinal (GI) diseases, including Clostridium difficile infection (CDI) and inflammatory bowel disease (IBD). The simultaneous administration of a community of microorganisms in FMT is thought to exert therapeutic effects by restoring functions to the diseased intestine normally conferred by the native microbiota. The particular beneficial strains in FMT are currently incompletely defined, but an improved understanding of the therapeutic benefits conferred by individual microbial strains could enable tailored applications of microbial therapy that circumvent the logistical and ethical issues currently surrounding FMT.
Techniques of FMT administration vary, with fecal preparations being given via oral capsules, nasogastric tubes, nasoduodenal tubes, colonoscopy, or enema (1, 2). Both related and unrelated donors have been used. Donor screening in either case is necessary to reduce the risk of the spread of infectious diseases or other health conditions. Studies originally focused on the use of fresh feces, but frozen fecal preparations have also been shown to be effective (3), and this finding has facilitated the setup of stool banks as a source of preparations (4).
A recent review of case series studies found no serious adverse events attributable to FMT (5), but this procedure is not entirely without risks. There have been two reported cases of patients contracting norovirus following FMT, although transmission was not linked to the donor in these cases (6). There is also concern that FMT in immunocompromised patients could lead to the acquisition of opportunistic infections, but the available data suggest that this is not a common problem for this patient population (7, 8). However, there is some evidence that FMT can lead to the development of noninfectious diseases. FMT for CDI has been linked to relapses in IBD (7, 9) and to the development of peripheral neuropathy, Sjögren's disease, idiopathic thrombocytopenic purpura, and rheumatoid arthritis (10). There has also been a reported case of the development of obesity following FMT from an overweight donor (11), but further study is needed to understand the impacts of FMT beyond the GI tract. Furthermore, while donor screening is necessary to reduce the risk of the spread of disease, recruitment and screening of donors are difficult processes with low rates of success (12). FMT is currently designated a biological agent by the FDA, and physicians must submit an investigational new drug application to administer FMT for any indication other than recurrent CDI (13). Uncertainty about the potential long-term effects of FMT and how to appropriately regulate this treatment has limited its use (14). In contrast, development of treatments containing only the effective components of FMT by using combinations of specific microbial strains would alleviate many of these drawbacks that result largely from the undefined nature of fecal preparations.
The focus of this review is to highlight the mechanisms of action by which specific strains of microorganisms known as probiotics exert beneficial effects on the intestinal environment. This information could be used to refine FMT into rationally designed combination microbial therapies that will provide specific benefits of FMT without the potential risks associated with unknown components. Probiotics, defined as live microorganisms that confer health benefits when consumed, are usually administered as individual strains or small cocktails of strains and have been shown to reduce the severity of several infectious and inflammatory diseases of the GI tract (15–18). There are several suggested mechanisms by which these diverse microorganisms may confer protection (19–21), including effects on the composition of the resident microbiota, the GI epithelial barrier, and host immune responses. However, in the context of particular diseases, certain functions may confer a greater degree of benefit. Infectious diseases, for example, may require reinforcement of the GI barrier, maintenance or restoration of a normal microbiota, and perhaps direct antipathogen effects. In contrast, diseases with an autoimmune component may be mitigated by probiotic strains that decrease inflammatory responses of the mucosal immune system. Given that the potency of each of these potential mechanisms differs on a strain-specific level, informed selection of probiotic strains to be administered therapeutically in place of FMT is essential.
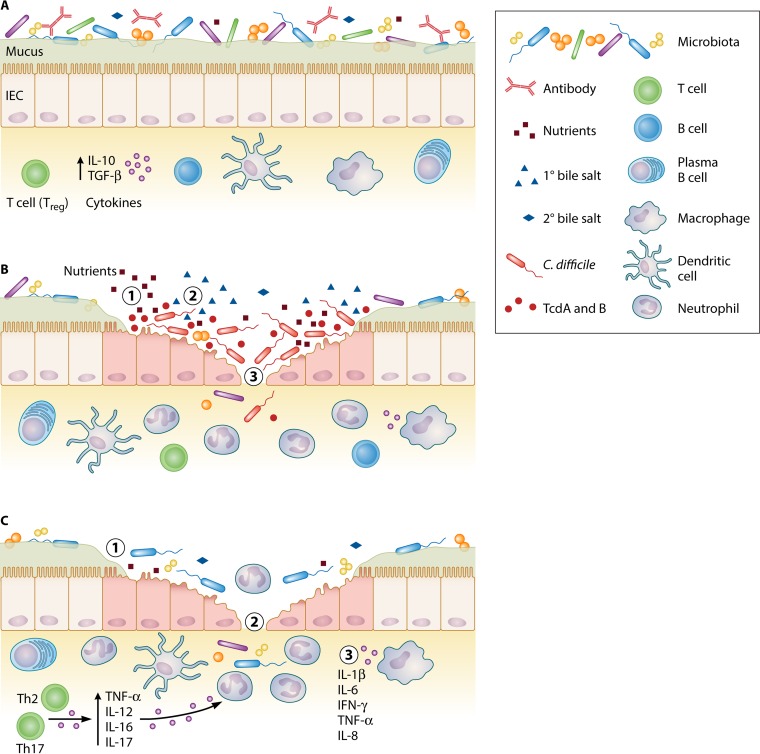
In this review, we use CDI and the inflammatory bowel disease ulcerative colitis (UC) as illustrative cases to explore how microbial therapy might be tailored to either infectious or autoimmune diseases. Both CDI and UC are serious GI diseases that are increasing in prevalence (22, 23). Numerous trials have demonstrated the effectiveness of FMT for CDI, especially for recurrent infections, and recent smaller-scale trials have suggested that UC may also be treated with microbial therapy (22–24). In the case of CDI, pseudomembranous colitis arises from colonization with pathogenic C. difficile and direct toxin-mediated damage of the host GI epithelium (Fig. 1A and B). In contrast, UC develops when genetically susceptible individuals exhibit a breakdown of the GI barrier due to aberrant inflammatory immune responses to microbial antigens (22) (Fig. 1A and C). Comparison of these two diseases with disparate pathogenic mechanisms allows consideration of how particular probiotic strains may be more appropriate in certain disease contexts. We may thus gain insight into which particular organisms could best be applied to the treatment of these and other infectious and inflammatory GI diseases.
FIG 1.
The gastrointestinal mucosa in health, CDI, and UC. (A) The healthy mucosa is characterized by a diverse microbiota that confers colonization resistance and proper immunomodulation; few freely available nutrients; low levels of primary bile salts relative to secondary bile salts; secretory antibody capable of sequestering commensals, pathogens, and other antigens; an intact barrier with healthy epithelial cells and thick layers of mucus containing antimicrobial peptides; few immune cells; and a cytokine milieu dominated by anti-inflammatory cytokines such as IL-10 and TGF-β. (B) Disruption of the microbiota results in increased nutrients permissive for C. difficile growth (1) and high concentrations of primary bile salts relative to secondary bile salts (2). These changes promote C. difficile spore germination and growth to high concentrations within the intestine. C. difficile toxins damage epithelial cytoskeletal components, leading to cell death and ulcerations (3). Probiotics may promote colonization resistance through multiple mechanisms, including competition for nutrients and the generation of secondary bile salts that prevent C. difficile germination. Probiotics may also directly inhibit the growth of C. difficile by producing bacteriocins or other inhibitory compounds. Some probiotics produce antitoxin proteases and may stimulate antibody production to sequester C. difficile and toxin. Reinforcing epithelial barriers and modulating inflammation may also promote healing and limit injurious host responses to infection. (C) Ulcerative colitis is characterized by an altered microbiota of decreased diversity (1), damage to the gastrointestinal epithelium (2), as well as aberrant, overly inflammatory host immune responses (3). By helping to maintain a normal microbiota and reinforce the barrier function of the epithelium, probiotics may limit exposure to inflammatory signals. Modulation of the mucosal immune system, including the cytokine milieu, neutrophil infiltration and function, and T cell differentiation, may also help redress aberrant responses to luminal antigens and prevent host-mediated damage to the mucosa. Abbreviations: IEC, intestinal epithelial cell; IFN, interferon; IL, interleukin; TcdA and TcdB, C. difficile toxins A and B, respectively; TGF, transforming growth factor; TNF, tumor necrosis factor.
To permit discussion of potential microbial therapeutics for infectious diseases as exemplified by CDI and for inflammatory diseases as exemplified by UC, this review is divided into two major sections. We begin each section with an overview of disease pathophysiology followed by a discussion of applicable therapeutic traits identified for particular probiotic and commensal organisms. Emphasis is placed on probiotic strains for which clinical trials have been conducted for the diseases of interest, although additional commensal strains shown to have potential benefits in experimental systems are also considered. By identifying specific organisms with particular mechanisms of action, we can inform studies and trials of rationally combined microbial therapeutics tailored to individual infectious or inflammatory GI diseases.
C. DIFFICILE INFECTION
CDI is an increasing health problem, leading to nearly 500,000 diagnoses and approximately 30,000 deaths annually in the United States alone (25). C. difficile is an obligate anaerobe but can survive for months in the external environment as a dormant spore (26, 27). Spores are highly resistant to many environmental stresses, including ethanol-based disinfectants (28). In susceptible hosts, ingested spores germinate in response to bile salts and amino acids found in the intestine (29). Some individuals develop asymptomatic colonization with C. difficile, while others develop pathogenic CDI. Symptoms of CDI range from mild diarrhea to severe pseudomembranous colitis and death (30). Both asymptomatic and diseased individuals shed infectious spores in their feces that can then spread and infect new hosts (31).
CDI is a toxin-mediated disease, and it has been suggested that patients asymptomatically colonized by C. difficile may have more robust neutralizing immune responses against C. difficile toxins than patients who develop symptoms (32). Most C. difficile strains encode two toxins, TcdA and TcdB, but strains that produce only TcdB or no toxins have also been isolated; only strains without toxins are considered to be avirulent (33–36). TcdA and TcdB bind to any of a number of host cell receptors (37–40) and, once inside host cells, act as monoglucosyltransferases to inactivate Rho family GTPases (41, 42). This inactivation leads to rounding and death of GI epithelial cells, disrupting the epithelial barrier (33, 43, 44). Some strains of C. difficile also encode a binary toxin, C. difficile transferase (CDT) (45), which ADP-ribosylates actin and leads to actin depolymerization and rearrangement of microtubules (45, 46).
In addition to effects on cell death and proliferation, C. difficile toxins perturb the intestinal epithelial barrier by affecting cytoskeletal components and junctional complexes (47). Both TcdA and TcdB mediate the dissociation of the proteins zonula occludins 1 (ZO-1) and ZO-2 in epithelial tight junctions, leading to the separation of F-actin (48) and modulating the integrity of the epithelial barrier (49). The influx of luminal compounds across the intestinal barrier exposes immune cells to bacterial components as well as numerous inflammatory damage-associated molecular patterns (DAMPs) from necrotic epithelial cells. TcdA also disrupts epithelial cell polarization, thus affecting the distribution of Toll-like receptors (TLRs) and the nature and magnitude of immune responses to DAMPs (49). Maintaining the integrity of the junctional complexes between epithelial cells and reinforcing the integrity of the epithelial barrier may thus help to limit damage induced by C. difficile toxins and host inflammatory responses (Fig. 1B).
Risk Factors for Developing CDI
A healthy and intact gut microbiota decreases susceptibility to CDI, a phenomenon known as colonization resistance (50). Indeed, recent studies of CDI in humans have found that decreased microbial diversity is associated with severe and recurrent CDI (51) and have also identified patterns of microbiota change associated with recovery from CDI (52). Antibiotic exposure is the primary risk factor for the development of symptomatic CDI because this treatment perturbs the gut microbiota and reduces colonization resistance (50). Broad-spectrum antibiotics are of the greatest concern for the development of CDI; clindamycin, cephalosporins, aminopenicillins, and fluoroquinolones are all particularly associated with an increased risk of CDI (53–55). Antibiotic treatment depletes members of the two dominant bacterial phyla in the gut, the Bacteroidetes and Firmicutes (56, 57). Antibiotics also lead to increases in the numbers of Proteobacteria, which are associated with susceptibility to CDI in humans (56–59). Studies in both humans and animals have indicated that changes in the microbiota brought on by antibiotic treatment can be long lasting, although this depends on the antibiotic used (56, 57, 60, 61). These changes in the gut microbiota facilitate the development of CDI following antibiotic therapy.
In addition to antibiotic use, other factors that influence susceptibility to CDI include age, exposure to health care environments, the use of proton pump inhibitors for conditions such as peptic ulcers, and the production of antitoxin antibodies. Asymptomatic colonization with C. difficile is common in infants; in fact, it is estimated that up to 21 to 48% of infants are asymptomatically colonized with C. difficile (62). Although it is not known why colonized infants generally do not develop disease, it has been suggested that they could be protected by a lack of functional toxin receptors or by antibodies in breast milk (63). Asymptomatic colonization can also occur in adults (62), but old age is a risk factor for the development of symptomatic CDI (64–66). The elderly are thought to be more susceptible because of changes in their gut microbiomes, immunosenescence, increased exposure to health care environments, antibiotic use, and other comorbidities (64, 67). Hospitalization is a major risk factor for both the asymptomatic carriage of C. difficile and the acquisition of pathogenic CDI (26). Proton pump inhibitors are thought to increase the risk of CDI by altering the composition of the gut microbiota (68).
Natural anti-C. difficile TcdA and TcdB antibodies in the general population have been proposed to be protective factors against disease development (69, 70). Toxin-reactive IgG and IgA can be detected in the intestine and serum and have the potential to block toxin binding to epithelial receptors and promote toxin clearance from the intestine (71). The presence of antibodies that are reactive to C. difficile TcdA has been positively correlated with asymptomatic carriage of C. difficile (32), although there are conflicting reports regarding whether naturally occurring antitoxin antibodies and intravenous immunoglobulin therapy (IVIG) affect the disease course (32, 69, 70, 72–83). Questions thus remain as to the extent to which antibody levels may confer protection against CDI.
Treatment of CDI and Disease Recurrence
Treatment of CDI generally involves prescription of either metronidazole or vancomycin. Metronidazole is the preferred treatment for mild disease due to its cost-effectiveness, but it is associated with rates of treatment failure higher than those for vancomycin in severe and complicated cases of CDI (84). Severe and recurrent cases may be treated with combination therapy of intravenous metronidazole with oral vancomycin or with a vancomycin taper. A more recently developed antibiotic, fidaxomicin, has a cure rate similar to that of vancomycin (85) but is currently recommended only for recurrent CDI due to its expense (84). In particularly complicated cases, surgical intervention may be required to remove the infected colon (84).
Recurrence of CDI following completion of treatment is common, occurring in up to 20 to 40% of cases after one episode and in up to 60% of cases after a first reoccurrence (86). This can occur via a relapse of the initial infection or from reinfection with spores from the environment (87, 88). The risk of recurrence is high because current therapies are limited to antibiotics that kill much of the gut microbiota along with C. difficile. This wholesale killing results in decreased colonization resistance due to the suppression of levels of Bacteroidetes and Firmicutes (89–91). Vancomycin in particular has dramatic and long-lasting effects on the composition of the microbiota (61). Fidaxomicin, in contrast, has the least profound effect on the gut microbiota (90) and is associated with lower rates of CDI recurrence than vancomycin (85, 90). The serious problem of recurrence has led to interest in nonantibiotic therapies to treat CDI, including microbial-based therapies such as FMT and probiotics.
Fecal Microbiota Transplantation and CDI
FMT seeks to reconstitute a healthy gut microbiota and colonization resistance against CDI through the administration of fecal preparations from a healthy donor. Numerous studies have shown that FMT restores microbial diversity in recipients (92–95). A recent study by van Nood et al. demonstrated that FMT dramatically increased cure rates among patients receiving vancomycin therapy for recurrent CDI (92). A recent review of case series studies demonstrated that 85% of 480 patients with recurrent CDI were successfully treated by using FMT (5), illustrating the potential of the use of microbes as a therapy to restore colonization resistance against CDI. Although whole fecal samples are most often administered in FMT, a few studies have demonstrated the use of defined bacterial consortia to cure CDI in mice (96) and humans (97–99). The RePOOPulate study, for example, recently utilized a defined mixture of 33 fecal bacterial strains to treat recurrent CDI (97). Trials testing the ability of the feces-derived bacterial spore preparation SER-109 to prevent CDI recurrence have unfortunately shown conflicting results (100, 101). Animal studies comparing FMT and defined bacterial consortia found that both approaches speed the restoration of the intestinal microbiota after antibiotic administration and promote the recovery of host secretory IgA (sIgA), intestinal epithelial MUC2, and defensin levels (102). Still, although FMT and more specific formulations are thought to restore colonization resistance by increasing gut microbial diversity, the exact mechanisms involved and the specific microbial species responsible for inhibiting C. difficile growth are not well described. Understanding the mechanisms underlying the efficacy of FMT for the treatment of CDI could lead to the development of more defined probiotic therapeutics to reestablish colonization resistance and ameliorate disease.
Clinical Trials Evaluating Probiotic Efficacy against CDI
Numerous clinical trials over the past few decades have evaluated the efficacy of probiotics against C. difficile (Table 1), identifying some individual strains and cocktails of beneficial microbes that may be candidates for further use in rationally designed combined microbial therapies. These trials tested primarily lactic acid-producing bacteria, including Lactobacillus, Streptococcus, and Bifidobacterium species, and the probiotic yeast Saccharomyces boulardii. Most studies have evaluated the ability of probiotics to prevent primary CDI in patients receiving antibiotic therapy, although a few have specifically considered the prevention of recurrent CDI. The majority of trials to date have been unable to determine a statistically significant benefit of probiotic administration for the prevention of CDI, although a recent meta-analysis found benefits of some probiotic formulations (103). Many studies are limited by a number of biases, including a lack of appropriate randomization, poorly defined outcome measures, and reliance on post hoc analyses. Critically, most studies are small in scale and underpowered (104). Particularly in studies considering antibiotic-associated diarrhea as a primary outcome and C. difficile infection as a secondary outcome, low incidences of CDI in small study populations limit evidence of efficacy (105). Results of individual trials are also difficult to compare, as the selection and preparation of probiotic agents, treatment lengths, study methods, patient populations, and means of identifying CDI cases all vary.
TABLE 1.
Clinical trials evaluating probiotic efficacy in preventing primary and recurrent CDI
| Study type and reference | Yr | Species (daily dose[s]) | Endpoint | Patient population | Conclusion(s) |
|---|---|---|---|---|---|
| Bacterial trials showing benefit | |||||
| 334 | 2007 | Lactobacillus casei, Lactobacillus bulgaricus, Streptococcus thermophilus (4.2 × 1010 CFU) | Primary CDI | 112 adults | Decreased incidence of primary CDI in patients receiving antibiotics when given probiotic bacteria |
| 335 | 2010 | Lactobacillus acidophilus CL1285, L. casei LBC80R (5 × 1010 CFU or 1011 CFU) | Primary CDI | 255 adult inpatients | Low- and high-dose probiotic mixtures confer protection against acquisition of primary CDI in adult patients |
| Bacterial trials showing no benefit | |||||
| 336 | 2001 | LGG (2 × 1010 CFU) | Primary CDI | 267 adults | No statistically significant difference in primary CDI in adults with probiotic administration |
| 337 | 2004 | L. acidophilus, Bifidobacterium bifidum (2 × 1010 CFU) | Primary CDI | 138 adults >65 yr old | No statistically significant difference in primary CDI in elderly patients with probiotic administration |
| 338 | 2005 | LGG (80 mg lyophilized LGG given with 640 mg inulin) | Recurrent CDI | 15 adults with recurrent CDI | No significant difference in recurrent CDI detected |
| 339 | 2007 | L. acidophilus, B. bifidum, L. bulgaricus, S. thermophilus (1.5 × 109 CFU of each) | Primary CDI | 42 adults | No statistically significant difference in primary CDI in adults with probiotic administration |
| 340 | 2007 | L. acidophilus CL1285, L. casei LBC80R (5 × 1010 CFU) | Primary CDI | 89 adult inpatients | No statistically significant difference in primary CDI with probiotic administration |
| 341 | 2008 | L. acidophilus (Florajen) (6 × 1010 CFU) | Primary CDI | 40 adult inpatients | No statistically significant difference |
| 342 | 2009 | L. plantarum 299v (5 × 1010 CFU) | Recurrent CDI | 20 adults with at least 1 CDI episode in previous 2 mo | No statistically significant difference |
| 343 | 2010 | BIO-K+ CL128 (L. acidophilus CL1285 and L. casei) (49 g and then 98 g) | Primary CDI | 437 adult inpatients | No statistically significant difference |
| 106 | 2013 | L. acidophilus CUL60 and CUL21, B. bifidum CUL20 and CUL34 (6 × 1010 CFU) | Primary CDI | 2,941 adult inpatients >65 yr old | No statistically significant difference in primary CDI with probiotic administration |
| S. boulardii trials showing benefit | |||||
| 110 | 1994 | S. boulardii (1 g; 3 × 1010 CFU) | Recurrent CDI | 124 adults with initial and recurrent CDI | Combination of antibiotic and S. boulardii therapy decreases CDI recurrence relative to antibiotics alone |
| 344 | 2000 | S. boulardii (1 g) | Recurrent CDI | 32 adults with CDI | Statistically significant decrease in CDI recurrence with S. boulardii administration in combination with high-dose vancomycin but not metronidazole or low-dose vancomycin |
| 345 | 2005 | S. boulardii (500 mg) | Primary CDI | 246 children treated for otitis media or respiratory infections | S. boulardii decreased the risk of CDI in children receiving antibiotics although with a borderline level of significance |
| S. boulardii trials showing no benefit | |||||
| 346 | 1989 | S. boulardii (1 g) | Primary CDI | 180 adult patients | No statistically significant decrease in CDI |
| 347 | 1989 | S. boulardii (1 g) | Recurrent CDI | 13 patients | Non-statistically significant decrease in CDI diarrhea with S. boulardii administration |
| 348 | 1995 | S. boulardii (1 g; 3 × 1010 CFU) | Primary CDI | 193 adult patients receiving antibiotics | No significant difference in incidence of primary CDI between groups |
| 349 | 1998 | S. boulardii (226 mg) | Primary CDI | 69 patients >65 yr old receiving antibiotics | No statistically significant difference in incidence of CDI |
| 350 | 2006 | S. boulardii (1 × 1010 CFU) | Primary CDI | 151 adults receiving antibiotics | No statistically significant difference in incidence of CDI |
A recent large clinical trial tested the use of Lactobacillus acidophilus (CUL60 and CUL21) and Bifidobacterium bifidum (CUL20 and CUL34) in older patients receiving antibiotic therapy and found no benefit in terms of diarrhea severity or abdominal symptoms (106). Nevertheless, a few earlier trials and meta-analyses found benefits of other probiotic strains for the treatment of CDI. One meta-analysis found beneficial effects of using Saccharomyces boulardii, Lactobacillus rhamnosus GG (LGG), and certain probiotic mixtures to reduce the risk of antibiotic-associated diarrhea and of using S. boulardii to reduce the risk of CDI (15), although there has been some criticism of the trials included in this study (107, 108). Of the four trials able to meet the stringent criteria of a Cochrane review in 2008 (109), only one showed a significant benefit of probiotics (S. boulardii) for preventing CDI recurrence (110). Thus, although transfer of the whole microbiota through FMT can be effective in treating CDI, the administration of currently available individual probiotic strains and some cocktails does not appear to reliably confer protection.
Rational design of probiotic cocktails that provide the protective effects associated with FMT while avoiding the transfer of potentially deleterious strains would provide a much needed therapy for CDI. Below, we discuss individual strains associated with colonization resistance and inhibition of the deleterious effects of C. difficile. Further study of these strains could lead to the development of effective probiotic therapies for CDI.
Microbial Taxa Associated with Colonization Resistance against CDI
Recent studies have attempted to identify individual commensal microbes associated with colonization resistance or susceptibility to CDI in both humans and animal models. This work has the potential to uncover taxa that are responsible for the efficacy of FMT against CDI and that could be incorporated into future defined therapeutic cocktails. Several studies have identified bacterial taxa associated with colonization resistance versus the development of CDI in antibiotic-treated mice challenged with C. difficile. In general, mice that remain healthy after challenge with C. difficile exhibit increased levels of Firmicutes relative to mice that develop CDI (111). The families Porphyromonadaceae and Lachnospiraceae (111, 112) and the genera Lactobacillus, Alistipes, and Turicibacter (112) are also associated with colonization resistance against CDI in mice. In contrast, the Escherichia and Streptococcus genera (112) and the Enterobacteriaceae family (111) are correlated with increased susceptibility to CDI. A recent analysis identified individual bacterial species associated with colonization resistance in antibiotic-treated mice (113). This study identified Clostridium scindens, Clostridium saccharolyticum, Moryella indoligenes, Pseudoflavonifractor capillosus, Porphyromonas catoniae, Barnesiella intestinihominis, Clostridium populeti, Blautia hansenii, and Eubacterium eligens as being protective against CDI (113). The majority of these species belong to Clostridia cluster XIVa (phylum Firmicutes) (113). It has been suggested that members of Clostridia cluster XIVa may protect against C. difficile colonization through their ability to metabolize bile salts, as discussed below.
Human studies have also implicated particular microbial taxa in modulating susceptibility to CDI. In general, high levels of members of the phylum Bacteroidetes, consisting of strictly Gram-negative anaerobes, are thought to be protective against CDI, whereas increased numbers of Proteobacteria are thought to increase susceptibility (58, 59, 99, 114, 115). These correlations are also consistent with observations that FMT recipients have increased levels of Bacteroidetes and decreased levels of Proteobacteria following recovery from CDI (92, 93, 95, 116). More specifically, the family Ruminococcaceae and the genus Blautia are also associated with colonization resistance to CDI, while multiple groups have found that the family Peptostreptococcaceae and the genera Enterococcus and Lactobacillus are associated with susceptibility (58, 94, 113, 115, 117, 118).
Not all bacteria within the same group confer equivalent benefits of colonization resistance in humans. Some taxa of the family Lachnospiraceae (belonging to Clostridia cluster XIVa) are associated with protection in humans and mice (58, 115, 118), and FMT has also been shown to increase levels of Lachnospiraceae (116, 117, 119). However, some taxa within this family are actually associated with increased susceptibility to CDI (115). There are also conflicting reports regarding the role of some bacteria. For example, some studies associate streptococci with colonization resistance (118), and others associate them with susceptibility (58, 113). Such examples highlight the need for both experimental reproducibility and species- and strain-level specificity when determining probiotic potential. In order to develop targeted probiotic therapies, more studies will be needed to determine which bacterial strains are able to confer colonization resistance.
Mechanisms of Colonization Resistance against CDI
The mechanisms by which commensal bacteria mediate colonization resistance against C. difficile are incompletely understood; however, several possible mechanisms of colonization resistance are discussed below. It is likely that successful probiotic therapeutics for CDI would restore colonization resistance by one or more of these mechanisms.
Nutrient availability and competition for resources.
Commensal bacteria are thought to provide colonization resistance by occupying nutrient niches that could be exploited by C. difficile (120). Levels of nutrients and metabolites in the mouse gut are substantially altered by antibiotic treatment (121, 122), presumably due to the elimination of bacteria with specific metabolic functions. This change in nutrient availability in turn favors C. difficile growth. Antibiotic-treated, CDI-susceptible mice exhibit elevated intestinal levels of carbohydrates (121) and sialic acid (123), which enhance C. difficile growth. C. difficile has also been shown to consume succinate in vitro, which is present at higher concentrations in mice following antibiotic treatment (124). It is likely that restoration of the gut metabolome to a preantibiotic state through FMT is a factor in restoring colonization resistance to CDI.
The concept of niche exclusion in the gut environment has led to interest in the use of nontoxigenic C. difficile (NTCD) as a probiotic to prevent recurrent infections by toxigenic strains. This strategy is based on observations that people asymptomatically colonized with C. difficile are less likely than uncolonized individuals to develop symptomatic CDI when hospitalized (125). Administration of NTCD following clindamycin treatment in the hamster model protects most animals from death due to challenge with toxigenic C. difficile (126, 127). Human studies have also shown some promise for this strategy: phase 1 clinical trials indicated that oral ingestion of NTCD strain VP20621 is safe in healthy humans (128), and phase 2 trials showed that 11% of patients who received VP20621 developed recurrent CDI, in contrast to 30% of patients who received placebo (129). Although the mechanism by which NTCD is able to prevent colonization by toxigenic C. difficile has not been thoroughly investigated, it is hypothesized that prior NTCD colonization allows NTCD to outcompete newly introduced toxigenic strains (129). NTCD is thus an intriguing illustration of how certain bacterial species may occupy particular niches within the gut and provide colonization resistance against toxigenic C. difficile. However, it should be noted that toxigenic strain 630Δerm can share toxin genes with NTCD strains via horizontal gene transfer in vitro (130). Whether this transfer would be a potential danger in vivo by converting NTCD to a toxigenic form remains to be seen.
Bile salt metabolism and colonization resistance.
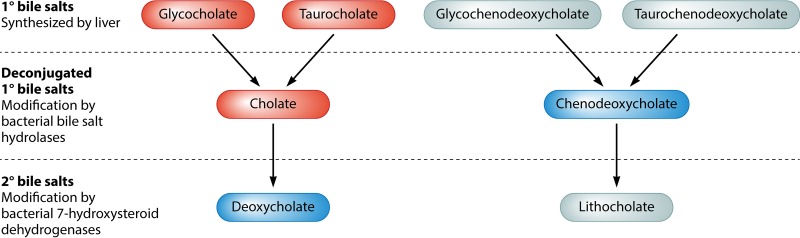
Levels of different bile salts in the gut are thought to affect C. difficile colonization by directly modulating its germination and growth. The primary bile salts glycocholate (GCA), glycochenodeoxycholate, taurocholate (TA), and taurochenodeoxycholate are synthesized by the liver to aid in the breakdown, digestion, and absorption of lipids in the small intestine (Fig. 2) (131). Although most bile salts are reabsorbed in the ileum and recycled by the liver, about 5% of bile salts pass into the large intestine, where they act as substrates for bacterial modification (131). Primary bile salts are deconjugated from their amino acid groups by bacterial bile salt hydrolases to make cholate (CA) and chenodeoxycholate (CDCA) (131). These bile salts can be further modified by bacterial 7-hydroxysteroid dehydrogenases to form the secondary bile salts deoxycholate (DCA) and lithocholate (LCA) (131). Although a wide variety of bacteria are capable of carrying out bile salt deconjugation, only a few intestinal bacteria can synthesize secondary bile salts (131). CDCA inhibits the germination of C. difficile, while TA, GCA, and CA all enhance C. difficile germination (29, 132). DCA is also capable of enhancing the germination of C. difficile spores but inhibits the growth of vegetative cells (29, 132). These findings suggest that levels of different bile salts in the intestine exert fine control over C. difficile germination and outgrowth.
FIG 2.
Summary of bile salt metabolism. Primary bile salts (1°) produced by the host liver are modified and deconjugated by intestinal bacteria to form secondary bile salts (2°). Bile salts that stimulate the germination of C. difficile spores and thus increase susceptibility to CDI are shown in red. Bile salts that are known to inhibit the sporulation or outgrowth of C. difficile and therefore contribute to colonization resistance are shown in blue. Probiotics with 7-hydroxysteroid dehydrogenase activity may enhance colonization resistance by decreasing the intestinal levels of glycocholate, taurocholate, and cholate and by increasing the levels of deoxycholate.
Antibiotic treatment results in alterations in bile salt levels that favor the germination and growth of C. difficile. Intestinal extracts from antibiotic-treated mice contain higher levels of primary bile salts than do extracts from untreated mice (121, 133, 134). Spores incubated with intestinal extracts from antibiotic-treated mice also germinate better than do spores incubated with untreated mouse extracts (133). The addition of the bile salt chelator cholestyramine to these intestinal extracts eliminated C. difficile germination, showing that germination occurs in response to bile salts in the mouse intestine (133). Furthermore, patients with CDI have higher levels of primary bile salts and lower levels of secondary bile salts than do healthy controls (135), and FMT has been shown to restore bile salt levels to those observed in healthy individuals (136). FMT efficacy thus appears to be mediated at least in part by restoring normal bile salt metabolism in CDI patients.
The role of secondary bile salts in protecting against C. difficile colonization suggests that bacteria with 7-hydroxysteroid dehydrogenase activity could be used as probiotics against CDI. C. scindens, a Clostridia cluster XIVa bacterium that produces a 7α-hydroxysteroid dehydrogenase, has been associated with colonization resistance against CDI in both mice and humans (113). The administration of C. scindens to antibiotic-treated mice restored DCA and LCA concentrations to preantibiotic levels, and intestinal contents from these mice were shown to inhibit the growth of vegetative C. difficile (113). Furthermore, feeding antibiotic-treated mice C. scindens prior to challenge with C. difficile significantly improved survival (113). C. scindens may thus be an attractive candidate for inclusion in novel probiotic formulations.
Production of anti-C. difficile compounds.
The production of molecules by the gut microbiota that have direct antibacterial activity may also contribute to colonization resistance against C. difficile. Organic acids produced by bacteria have been proposed to inhibit the in vitro growth of C. difficile, with culture supernatants from strains of Lactobacillus, Lactococcus, and Bifidobacterium species demonstrating pH-dependent anti-C. difficile activity (137–139). Growth of C. difficile is also inhibited by supernatants from Bacillus amyloliquefaciens cultures (140). Although the exact inhibitory molecule(s) within these culture supernatants remains to be identified, antibiotic-treated mice given B. amyloliquefaciens prior to challenge with C. difficile exhibit decreased disease severity (140), suggesting that these molecules are also active in vivo. Both lacticin 3147, produced by Lactococcus lactis strain DP3147 (141, 142), and thuricin CD, produced by Bacillus thuringiensis DPC 6431 (143), are bacteriocins that are inhibitory to C. difficile. Thuricin CD has potent activity against C. difficile without any apparent significant effects on other gut commensals (91, 143); however, mouse studies suggest that B. thuringiensis DPC 6431 spores pass through the mouse GI tract without germinating, limiting the probiotic potential of this strain (144). In contrast, lacticin 3147 is inhibitory toward other gut commensals (141), likely limiting its potential for restoring colonization resistance. More research is thus needed to identify strains that can both produce compounds specific for C. difficile and remain in the C. difficile-infected GI tract long enough to exert an effect.
Other Mechanisms of Action of Beneficial Microbes and Probiotics against CDI
Inactivation of C. difficile toxins.
Factors that directly target C. difficile toxins also have the potential to ameliorate disease by limiting the damaging effects of CDI on the GI epithelium. S. boulardii has been shown to secrete a 54-kDa protease capable of degrading TcdA, TcdB, and their brush border membrane receptors in vitro (145, 146). Blocking this protease abrogated protective effects of S. boulardii against C. difficile toxin-mediated epithelial cell damage in vitro (146), suggesting that S. boulardii protects against CDI pathogenesis at least in part via toxin degradation. However, this stands in contrast to data from another study that found no increase in survival of mice administered toxins preincubated with S. boulardii (147). The role of S. boulardii in direct toxin inactivation thus remains incompletely understood.
Probiotics also have the potential to inactivate C. difficile toxins indirectly by increasing the production of antitoxin neutralizing antibodies. TcdA-reactive IgM and IgA antibodies are induced by the administration of S. boulardii in vivo (148). One hypothesis is that such antibodies could prevent the binding of TcdA to its receptors on epithelial cells, thus limiting histological damage. This hypothesis is supported by data from a study in which a cocktail of monoclonal antibodies directed against TcdA and TcdB was administered intraperitoneally to hamsters prior to C. difficile challenge. This approach was found to protect against GI damage and death from CDI (149, 150). The administration of the TcdB-reactive antibody bezlotoxumab in combination with either metronidazole or vancomycin has also been shown to decrease rates of CDI recurrence in humans (151). It is unclear whether organisms other than S. boulardii can also induce antibodies with neutralization activity against C. difficile toxins. More studies are needed to determine the degree of protection conferred by C. difficile toxin-specific antibodies and to identify probiotic strains capable of stimulating such responses.
Antibody-mediated control of C. difficile bacteria.
Several studies have shown that the administration of probiotic organisms can increase total secretory IgA levels in rodents (148, 152–154), which may contribute to the control of C. difficile bacteria (72, 77–81). S. boulardii, for example, increases total secretory IgA levels in conventional rats and mice as well as in germfree mice colonized with S. boulardii (148, 152–155). Studies of Bifidobacterium animalis subsp. lactis BB-12, Escherichia coli EMO, and Lactobacillus casei and L. rhamnosus strains showed effects on total secretory IgA levels in rodent models (153, 156–158). However, the mechanisms by which some probiotics increase secretory IgA levels are not well understood, and more studies are needed to determine whether such changes in antibody production could protect against CDI.
Inhibition of mucus layer disruption.
Mucus forms a semipermeable barrier between the GI epithelium and the lumen. It consists of mucin glycoproteins, which are produced by goblet cells within the epithelium (159). The secreted glycoprotein MUC2 and the membrane-bound mucins MUC1, MUC3, and MUC17 form a dense meshwork to which numerous bioactive molecules, including trefoil factor peptides, resistin-like molecule β (RELMβ), Fc-γ binding protein, and antimicrobial peptides, as well as commensal bacteria are able to bind (160, 161). This mucus barrier normally prevents the direct contact of bacteria with the epithelium.
CDI is associated with changes in mucus thickness and composition (162) that promote the binding of C. difficile to mucus and increase the risk of epithelial cell damage from C. difficile toxins (163–166). Intestinal biopsy specimens from CDI patients show decreased MUC2 expression levels relative to those in healthy patients (162). C. difficile and CDI stool samples decrease MUC2 levels and alter mucus oligosaccharide composition in cultured human intestinal epithelial cells (162). Incubation with TcdA also decreases mucin exocytosis in the HT29-Cl.16E human colonic goblet cell line (167). As such, a key mechanism of FMT and probiotics in protecting against CDI may be to restore mucus composition in order to maintain an effective barrier.
A limited number of probiotics have been well studied with regard to the modulation of mucin production. Intestinal epithelial cells exposed to Lactobacillus plantarum 299v or LGG have been shown to upregulate MUC2 (168) and MUC3 (169) expression, respectively. In the case of LGG, this upregulation is mediated via the secreted soluble protein p40, which activates the epidermal growth factor receptor and induces mucin expression from GI epithelial cells (170). Preincubation of epithelial cells in vitro with L. rhamnosus ATCC 7469 has been shown to maintain mucin expression upon incubation with enterotoxigenic E. coli (ETEC) (171). Interestingly, an increase in mucus layer thickness via the addition of exogenous mucus increased the ability of L. rhamnosus to prevent the adherence and pathogenic effects of ETEC, suggesting that an intact mucus layer may support the protective effects of probiotics. The induction of increased mucus and mucin expression has also been noted for the probiotic bacterial cocktail VSL#3 (L. casei, L. plantarum, L. acidophilus, L. delbrueckii subsp. bulgaricus, B. longum, B. breve, B. infantis, and Streptococcus salivarius subsp. thermophilus) incubated with HT29 cells in vitro (172) as well as in vivo when fed to laboratory rats (173). A probiotic yeast strain, Saccharomyces cerevisiae CNCM I-3856, also upregulates MUC1 mRNA expression in epithelial cells in vitro (174), possibly via the induction of butyrate (175, 176). However, some probiotic strains, such as E. coli Nissle 1917, have minimal effects on mucus (172). In addition to species differences, the in vivo abilities of particular probiotics to affect the mucus layer may furthermore differ depending on the age (177) and overall GI microbiota composition (178) of patients. Thus, currently, only some probiotic strains are clearly capable of influencing mucus production, and more research is needed to evaluate their effects on restoring mucus specifically in the context of CDI.
Maintenance of the intestinal epithelial cell barrier and tight junction expression.
Microbes may promote the maintenance of the epithelial barrier between luminal contents and host cells through the modulation of mucus production (as discussed above) or by influencing regulatory factors, such as cytokines, that affect intestinal permeability (see the discussion below on the cytokine milieu). However, many probiotics have also been shown to influence the barrier function of epithelial cells by modulating the expression of junctional complexes (Table 2). These complexes normally seal together adjacent epithelial cells and prevent the indiscriminate translocation of particles from the gut lumen into host tissues. Although the role of junctional complexes in the pathogenesis of CDI is not well studied, it is possible that reinforcing the GI epithelial barrier via modulation of junctional complexes may help to reduce the leakiness associated with CDI-induced inflammation and possibly help repair damage induced by C. difficile toxins.
TABLE 2.
Effects of probiotics on the gastrointestinal epitheliuma
| Organism and genus | Species | Strain(s), company, or trade name | Effect(s) on epithelial barrier | Model system(s) | Reference |
|---|---|---|---|---|---|
| Gram-positive bacteria | |||||
| Lactobacillus | L. rhamnosus | GG (ATCC 53103) | TER ↑ | Alcoholic liver disease in male Sprague-Dawley rats | 351 |
| L. rhamnosus | GG (ATCC 53103) | Prevented ↓ in ZO-1, claudin-1, symplekin, p130, and fordin | Chronic alcohol feeding in mice | 293 | |
| L. rhamnosus | GG (ATCC 53103) | Occludin, claudin-1, ZO-1 ↑ when given with gliadin | Caco-2 cells | 352 | |
| L. rhamnosus | GG (ATCC 53103) p40 and p75 proteins | PKCε and PKCβI membrane translocalization; prevents occludin, ZO-1, E-cadherin, and B-catenin redistribution in ERK1/2- and PKC-dependent manners | Caco-2 cells exposed to H2O2 | 353 | |
| L. rhamnosus | GG (ATCC 53103) | TER, claudin-1, ZO-1, and occludin ↑ | In vitro human epidermal keratinocytes | 185 | |
| L. rhamnosus | ATCC 7469 | ZO-1, TLR2, and TLR4 ↑; PKCα unchanged; prevents mucus disruption | ETEC-infected IPEC-J2 cells | 171 | |
| L. acidophilus | ATCC 4356 | TER ↑, ↑ occludin and ZO-1 phosphorylation | Control and EIEC-infected Caco-2 cells | 354 | |
| L. plantarum | ATCC 10241 | Transient TER ↑ | In vitro human epidermal keratinocytes | 185 | |
| L. plantarum | CGMCC 1258 | Prevented ↓ in occludin | ETEC-infected piglets | 355 | |
| L. plantarum | 299v | No change in bacterial translocation to cervical and mesenteric lymph nodes | 5-FU-treated rats | 356 | |
| Streptococcus | S. thermophilus | ATCC 19258 | TER ↑, ↑ occludin and ZO-1 phosphorylation | Control and EIEC-infected Caco-2 cells | 354 |
| Bifidobacterium | B. bifidum | WU12 | ↑ occludin mRNA in Caco-2 cells after TNF-α exposure | Caco-2 cells | 192 |
| B. longum | ATCC 51870 | TER (TLR2 dependent), claudin-1 and -4, ZO-1, and occludin ↑ | In vitro human epidermal keratinocytes | 185 | |
| B. infantis | Isolated from VSL#3 | Prevented TNF-α- and IFN-γ-induced TER ↓, ↑ claudin-3 and -4, occludin, and ZO-1; prevented redistribution of claudin-1 and occludin in vivo | T84 cells, IL-10-deficient mice | 357 | |
| Gram-negative bacteria | |||||
| Escherichia | E. coli | Nissle 1917 | ↑ ZO-1 in the absence of inflammation; ↑ ZO-1 and ZO-2 in DSS colitis; ↓ recruitment of inflammatory leukocytes to colon | Monoassociated mice and DSS colitis | 358 |
| Probiotic cocktails | L. rhamnosus and L. helveticus | R0011 and R0052 (Lacidofil) | Intestinal permeability ↓, ↓ bacterial adherence to epithelium | Chronic stress in rats | 359 |
| S. thermophilus and L. acidophilus | ATCC 19258 and ATCC 4356 | ↑ TER, ↑ phosphorylation of occludin and ZO-1 | Caco-2 cells, EIEC-infected Caco-2 cells | 354 | |
| Yeast | |||||
| Saccharomyces | S. boulardii | Biocodex | No change in TER in control or infected T84 cells; partial protection from ↑ HRP flux in Shigella flexneri coinfection; restoration or preservation of claudin-1 and ZO-2 expression at later time points | Control and Shigella flexneri-infected T84 cells | 190 |
| S. boulardii | Biocodex | Prevents EPEC-induced activation of the ERK1/2 MAP kinase pathway; preservation of ZO-1 distribution | EPEC-stimulated T84 cells | 186 | |
| S. boulardii | Biocodex | Prevented EHEC-induced MLC phosphorylation linked to ↓ TER | EHEC-infected T84 cells | 360 | |
| S. boulardii | Biocodex | Inhibited IL-1β- and TcdA-induced ↑ in IL-8 expression, ERK1/2 and JNK/SAPK but not p38 activation; ↓ ERK1/2 activation in TcdA-treated ileal loop | NCM460 human colonocytes; mouse ileal loop | 361 | |
| S. boulardii | Perenterol | ↑ brush border enzyme activity | Duodenal biopsy specimens from S. boulardii-treated healthy human volunteers | 362 | |
| S. cerevisiae | CNCM I-3856 | No effect on barrier function | IPEC-1 cells with ETEC exposure | 174 |
DSS, dextran sodium sulfate; EHEC, enterohemorrhagic E. coli; EIEC, enteroinvasive E. coli; EPEC, enteropathogenic E. coli; ERK1/2, extracellular signal-regulated kinase 1/2; HRP, horseradish peroxidase; IPEC-1, newborn piglet intestinal epithelial cell line; JNK/SAPK, c-Jun N-terminal kinase/stress-activated protein kinase; MAP, mitogen-activated protein; MLC, myosin light chain; PKC, protein kinase C; TER, transepithelial resistance; 5-FU, 5-fluorouracil; ↑, increase; ↓, decrease.
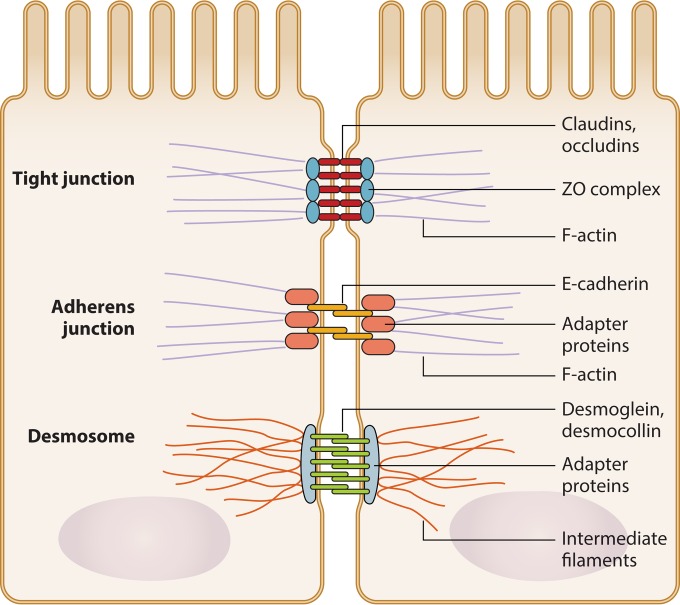
Junctional complexes are composed of tight junctions, adherens junctions, gap junctions, and desmosomes (179) (Fig. 3). Most work on junctional complexes and probiotic organisms has centered on tight junctions, whose transmembrane components include claudins, occludins, and junction-associated molecule (JAM) family proteins. These transmembrane components interact with plaque proteins, including zonula occludens (ZO) family members (180), in order to mediate intracellular signaling and cytoskeletal reorganization (181, 182). The expression of tight junction molecules in the healthy gut is modulated by numerous environmental signals, including metabolic compounds such as acetate and short-chain fatty acids (SCFAs) (183), although the exact mechanisms by which this occurs are still unclear. Some studies suggest that butyrate, an SCFA whose level increases with probiotic administration, decreases intestinal permeability through the induction of AMP-activated protein kinase activity and the increased assembly of tight junctions in Caco-2 monolayers (184). Junctional complex expression is also influenced by innate immune functions of epithelial cells, such as by TLR recognition of microbial ligands (185).
FIG 3.
Epithelial cell junctional complex. Junctional complexes hold together adjacent epithelial cells. Tight junctions at the apical end of junctional complexes are composed of occludin and claudin proteins that span the intercellular space and bind intracellular adapter proteins, such as zonula occludens (ZO) complex proteins. Adherens junctions are composed of E-cadherins and adapter proteins. Desmosomes are formed of desmoglein and desmocollin proteins that bind internal adapter proteins. Adapter proteins associated with tight junctions, adherens junctions, and desmosomes in turn bind components of the cytoskeleton, including F-actin or intermediate filaments.
Several probiotic organisms are capable of modulating junctional complexes to restore or maintain the intestinal epithelial barrier. The probiotic yeast S. boulardii increases the expression of ZO-1 in T84 cells (186) and has been associated with decreased intestinal permeability in numerous studies (155, 187–190). Similarly, both Bifidobacterium longum and LGG have been shown to induce the upregulation of claudin-1, ZO-1, and occludin protein levels in keratinocytes (185). Intriguingly, the in vitro increase in keratinocyte transepithelial electrical resistance (TER) induced by a B. longum lysate, but not by an L. rhamnosus GG lysate, was abrogated in the presence of a TLR2-neutralizing antibody, suggesting that these bacteria act on different pathways to influence tight junction molecule expression (185). An in vivo infectious model recently demonstrated that a defined mixture of 33 probiotic bacterial strains prevented the Salmonella enterica serovar Typhimurium-induced disruption of ZO-1 and claudin-1 in mice and ameliorated disease severity (191).
There is evidence that the effects of probiotics on epithelial cell junctional complexes are highly strain specific. One study using Caco-2 cells exposed to probiotics found that while all tested Bifidobacterium strains increased TER, a measure of barrier integrity, only 6 of 15 tested Lactobacillus strains showed a similar increase (192). Even fewer strains were able to prevent the tumor necrosis factor alpha (TNF-α)-induced decrease in TER (192). Furthermore, the effect of the most protective strain of B. bifidum (WU12) on TER was strikingly attenuated when it was heat killed, suggesting that metabolic or secreted factors produced by B. bifidum mediate beneficial effects (192). Another study found that L. plantarum L2 was able to reduce TNF-α levels, intestinal epithelial cell apoptosis, and ileal mucosa erosion in an ischemia reperfusion injury model (193). By reinforcing the GI epithelial barrier, probiotic organisms may help to repair or prevent damage induced by C. difficile toxins or host inflammatory immune cells.
Summary of Potential Mechanisms of Action of FMT against CDI and Implications for Probiotics
C. difficile infection is a toxin-mediated disease that leads to severe damage of the GI mucosa (Fig. 1B). Numerous factors may help to prevent initial colonization with C. difficile or to maintain an asymptomatic infection and limit damage after sporulation in susceptible individuals.
The phenomenon of colonization resistance in preventing CDI is particularly well studied and presents one major mechanism through which beneficial microbes may help to ameliorate disease pathogenesis and symptoms. Strains that are able to alter bile salt concentrations or limit the availability of other resources may discourage growth of and colonization by C. difficile. Delivery of NTCD is also a promising novel therapy due to its potential competition with toxigenic C. difficile for an intestinal niche (126, 127). Future work administering C. scindens and NTCD (128, 129) holds promise for the use of these strains as preventative therapies in antibiotic-treated patients or as treatments for CDI. Further studies on colonization resistance will help identify additional microbes that could be beneficial in treating CDI.
Direct targeting of C. difficile or its toxins is another way in which probiotics may protect against CDI even after pathogen colonization and sporulation. Indeed, the probiotic yeast S. boulardii has been found to secrete a protease capable of degrading C. difficile toxin A (145, 146). It is interesting to note that this is the only probiotic strain for which such direct anti-C. difficile toxin activity has been identified and one of the few strains shown to have efficacy against CDI in clinical trials (109, 110). The identification of other yeast or bacterial strains with antitoxin activity may provide further potential therapeutic strains.
Other probiotic strains may help to ameliorate disease symptoms and limit damage by promoting the reinforcement and repair of the epithelial barrier. Such reinforcement may help protect the host from increased exposure to C. difficile toxins. In vitro studies also suggest that the effects of probiotics may be greater with an intact mucus layer, suggesting that probiotics may be more beneficial as prophylactic agents. However, further studies are needed to determine whether the effects of these probiotic strains seen in vitro confer protection in the context of CDI.
Finally, the administration of probiotic organisms may be beneficial by harnessing the host immune response to alleviate CDI disease progression and symptoms. For example, increasing the production of secretory IgA may promote the sequestration of toxins within the intestinal lumen (72, 77–81, 148). Stimulation of pattern recognition receptors (PRRs) such as TLRs has also been found to limit CDI severity (194). However, such a strategy must be pursued with care: it has also been hypothesized that some degree of damage in CDI may be immune mediated, with decreased toxin-associated damage being seen in mice deficient in neutrophils, mast cells, or the inflammatory cytokine gamma interferon (IFN-γ) (194). Although live probiotics may have adverse effects in severely immunocompromised individuals (195, 196), probiotic strains that are able to attenuate inflammatory responses in immunocompetent hosts may thus limit host-induced histological damage and improve disease symptoms. In order to identify optimal probiotics for the treatment of CDI, it will be crucial to identify those strains that are able to alleviate symptoms associated with deleterious inflammatory responses without undermining the ability to control C. difficile infection. Current knowledge of immunomodulatory effects of probiotics and implications for their use in GI diseases are discussed in further detail below in the context of UC.
ULCERATIVE COLITIS
Ulcerative colitis is a serious GI disorder currently affecting an estimated 1 million to 1.3 million people in the United States (197, 198). UC is more common in developed countries and in urban areas. The incidence and prevalence of UC and Crohn's disease (CD), another common form of IBD, are both highest in northern Europe and North America; however, incidence is also increasing in other regions of the world, including South America and Africa (197). Although often grouped together with CD, UC has an etiology distinct from that of CD, with different associated genes, inciting factors, responses to therapies, and affected bowel regions (22, 199).
UC pathology is characterized by diffuse mucosal inflammation and histological alterations limited to the mucosal layer of the colon (200). The inflammation seen in UC is chronic but waxes and wanes in intensity. Various degrees of immune cell infiltration may be observed in the mucosa depending on whether the individual is experiencing active disease or remission (22). In active disease, lymphocytes, plasma cells, and granulocytes may all be seen within the mucosa (201). Ulcerations, goblet cell depletion, and fewer crypts are also observed. In advanced disease, epithelial cells may undergo dysplasia and increase the risk of epithelial cancer (202, 203). Symptoms of mild to moderate disease may include rectal bleeding, diarrhea, and abdominal cramping, while more severe cases may present with fever, weight loss, anemia, and severe abdominal pain (22). UC may also cause extra-abdominal symptoms affecting the eyes, kidneys, and joints (204).
Risk Factors for Developing Ulcerative Colitis
The development of UC is thought to be a multihit process, with genetic predispositions leading to disease only upon exposure to as-yet-poorly understood environmental triggers. Several genetic correlations have been identified, with a recent meta-analysis identifying 47 loci associated with IBD, 19 of which were specific for susceptibility to UC rather than CD (199). Still, twin studies have shown that the overall genetic concordance for UC is low relative to those for CD and other genetic diseases (22). Environmental exposures related to a Western diet and lifestyle have also been linked to the development of UC (205, 206). Other known epidemiological risk factors include appendectomy (207) and smoking (208), both of which reduce disease risk.
Ulcerative Colitis Pathophysiology
Although the exact mechanisms of UC pathogenesis are still incompletely understood, disease is generally believed to stem from inflammatory immune responses to the microbiota in genetically susceptible individuals (209). The major factors contributing to active disease are thought to include impaired barrier integrity of the GI epithelium, an altered microbiota, and aberrant immune responses to GI antigens and microbes; these factors are discussed in more detail below (Fig. 1C). Other factors that may also play a role, such as adiposity, regulatory RNA, angiogenesis, and the inflammasome, have been reviewed elsewhere (210) and are not discussed here.
Intestinal permeability.
Intestinal permeability is a major component of UC pathology and may serve as a potential novel therapeutic target (211). Breakdown of the epithelial barrier may lead to increased and prolonged exposure to bacterial antigens or other insults that in turn may compound inflammatory responses and intestinal damage. Whether intestinal permeability is a cause or a consequence of disease is still a question of debate. However, several genome-wide association studies (GWASs) have identified numerous UC susceptibility loci that contain genes involved in intestinal permeability and pathogen recognition, suggesting a causative effect (199, 212, 213). Many of these genes are known to be expressed by epithelial cells, including GNA12, which is associated with tight junction assembly (199); CDH1, encoding the adherens protein E-cadherin (199, 212); and LAMB1, encoding the laminin beta 1 subunit expressed by the intestinal basement membrane. Some studies have also found UC susceptibility to be associated with polymorphisms in the multidrug resistance 1 gene (MDR1) (also known as ABCB1) encoding P-glycoprotein, a protein responsible for pumping substances out of epithelial cells to help maintain barrier function (213, 214).
The mucus layer that forms an additional barrier between epithelial cells and the GI lumen is dysregulated and thinned in individuals with UC (215, 216). This is proposed to be the result of defects in mucin production as well as increased numbers of mucus-degrading (mucolytic) bacteria in individuals with UC (217). Indeed, MUC2-deficient mice spontaneously develop colitis, demonstrating the need for this factor for the maintenance of gut homeostasis (215). Nod-like receptor pyrin domain-containing protein 6 (NLRP6), which is known to be important for mucin exocytosis from epithelial cells, has also been linked to colitis susceptibility in mouse models (218, 219). UC patients have significantly reduced numbers of mucin-containing goblet cells in uninflamed ileal biopsy specimens relative to controls (220), suggesting that dysregulation of mucus production occurs even in the absence of host inflammatory cell responses. Decreased mucus layer thickness allows increased contact between the microbiota and the epithelium in UC patients (221) and may exacerbate immunostimulation and inflammation.
The microbiota and dysbiosis.
The microbiota of UC patients is vastly different from those of healthy controls, although it is unclear whether this is a cause or a consequence of the chronic inflammation associated with UC. Dysbiosis may be influenced by genetic risk factors leading to impaired intestinal epithelial barrier integrity as well as dietary factors such as high intake of fat, refined sugar, iron, and aluminum (222).
There are alterations in several bacterial groups within the microbiota of UC patients relative to healthy individuals. Like those suffering from CDI, UC patients have decreased prevalences of Bacteroidetes and Firmicutes and increased prevalences of Actinobacteria and Proteobacteria, especially Enterobacteriaceae (223, 224). UC patients were also specifically found to have increased prevalences of Porphyromonadaceae and enteroadherent E. coli in addition to decreased prevalences of Prevotella, Catenibacterium, Streptococcus, and Asteroleplasma species relative to healthy patients (224, 225). Patients with active UC disease have also been reported to have a decreased prevalence of Lactobacillus species relative to patients in remission (226).
The mechanisms by which dysbiosis influences the development of UC are currently unclear; however, it is possible that dysbiosis early in life may predispose individuals to UC by negatively affecting the maturation of the immune system (227). GI immune tissues such as Peyer's patches, isolated lymphoid follicles, and mesenteric lymph nodes are all underdeveloped in the absence of microbial stimulation (228). Indeed, models known to develop spontaneous colitis, including interleukin 10 (IL-10)- and T cell receptor-deficient mice, do not develop colitis if they are raised under germfree conditions (228–230), indicating that aberrant immune responses to a deregulated microbiota play a role in inciting colitis. Furthermore, cohousing of wild-type mice with colitis-prone Tbx21−/− Rag−/− mice induces the development of colitis in wild-type mice (228). Although the exact signaling pathways through which this susceptibility is conferred are still unclear, these data suggest that exposure to certain colitogenic strains of bacteria within a dysbiotic microbiota can be sufficient to induce colitis. Together, these studies demonstrate that dysbiosis is both a consequence of immune deregulation and a factor that affects disease susceptibility and progression.
Aberrant immune responses.
UC is characterized by the infiltration and activation of many immune cells in the mucosa, including neutrophils, macrophages (231), and T cells (232). These inflammatory cells are recruited and activated by the production of numerous chemokines and cytokines that are upregulated in the mucosa of UC patients, further promoting inflammation and damage in active disease (233). Serum levels of chemokines that attract monocytes, dendritic cells, T cells, and neutrophils, including CXCL5 and CCL23, are elevated in UC patients compared to healthy controls (234). Levels of macrophage migration inhibitory factor (MIF), macrophage inflammatory protein 3 (MIP3) (CCL23), monocyte chemoattractant protein 1 (MCP-1) (CCL2), MIP3β (CCL21), and granulocyte chemotactic protein 2 (CXCL6) are also elevated in the periphery of UC patients (234). CCL25-CCR9 interactions, which regulate leukocyte recruitment to the intestine, also play a role in mediating colitis (235). Novel antibodies such as vedolizumab and PF-00547659, which prevent homing of leukocytes to the gut, have been found to ameliorate symptoms of active UC in clinical trials (22). The ability to modulate immune cell recruitment and the level of inflammatory cytokines may thus confer protection against increased disease severity.
TNF-α.
In addition to their role in immune cell recruitment, inflammatory cytokines can be directly pathogenic. The best example of this is TNF-α, which promotes fibroblast proliferation, increased adhesion molecule expression, neutrophil activation, disruption of junctional complexes, and the production of proinflammatory cytokines such as IFN-γ (236). UC patients have increased levels of TNF-α relative to healthy controls (237). The administration of infliximab, a monoclonal antibody against TNF-α, has shown some success in the treatment of steroid-refractory UC (238), highlighting the critical role of this cytokine in mediating disease pathogenesis.
Th2 cells.
Despite the abundance of pro- and anti-inflammatory cytokines, such as IL-12, TNF-α, IL-1β, IL-16, and transforming growth factor β (TGF-β), that can be found in UC patients (234, 237, 239–241), UC has traditionally been considered a CD4+ T helper cell type 2 (Th2) disease (242, 243). This view stemmed from the observation that increased levels of Th2-associated cytokines, including IL-5 and IL-13, can be measured in UC patients and experimental colitis models (239, 243, 244). Th2-associated cytokines have been shown in some studies to induce damaging effects at the mucosa. IL-13, for example, is thought to mediate epithelial cell cytotoxicity, apoptosis, and barrier dysfunction in some situations (239, 245). However, the importance of Th2 cytokines in UC pathogenesis relative to other cytokine pathways is currently unclear.
Th17 cells.
Recent evidence also suggests an important role for Th17 cells, a subset of CD4+ T cells that secrete primarily IL-17 (242, 243), in UC pathogenesis. Th17 cells and their associated cytokines increase neutrophil recruitment to areas of inflammation, as discussed below; however, the extent to which Th17-associated cytokines such as IL-17A are pathogenic versus protective is controversial (246). A recent GWAS identified numerous Th17-related genes associated with UC susceptibility (199). Multiple genes in the IL-23 pathway that induces Th17 cell differentiation, including IL23R, JAK2, STAT3, and IL12B, were also associated with susceptibility to both UC and CD (199). Both rodents with colitis and patients with active UC disease have increased amounts of IL-17 and Th17 cells in the mucosa relative to controls (247–249). Although some studies have shown that antibody depletion of IL-17 increases the severity of acute colitis in mice (250), other mouse experiments conversely demonstrated that IL-17 receptor (IL-17R) deficiency reduces colitis severity (251). Novel drugs that block IL-17 activity have also been shown to confer protection in models of chronic colitis (252, 253). Thus, there is currently much evidence to suggest a critical role for Th17 cells and their associated cytokines in UC pathogenesis.
Neutrophils.
The recruitment and activation of neutrophils at the intestinal mucosa are striking features of UC pathophysiology (254, 255). Neutrophils are innate immune cells that normally protect the host against microbial pathogens and dying cells through pathogen phagocytosis and the production of reactive oxygen species, antimicrobial peptides, and proteases such as elastase that are exuded from specialized granules. Numbers of neutrophils are increased in both the periphery (256) and colons (257) of UC patients. Neutrophils secrete both proinflammatory factors such as IL-17 (258), leukotrienes, and CXCL8 (257) as well as anti-inflammatory cytokines such as IL-10 (259). Matrix metalloproteases, which are involved in the activation of chemokines such as CXCL5 and CXCL8, are also secreted by neutrophils to facilitate the recruitment of additional immune cells.
The exact role played by neutrophil expansion and activation in UC pathogenesis has been the subject of much debate, with different experimental colitis models suggesting different effects of neutrophils on disease severity. Neutrophils are important in wound healing and the maintenance of homeostatic processes through their phagocytosis of damaging cellular debris as well as through the secretion of growth-promoting factors such as vascular endothelial growth factor (VEGF), lipoxins, and protectins (257). Some studies have reported that depletion of Gr1+ CD11b+ cells, including neutrophils, exacerbates mouse models of colitis, suggesting a protective role for neutrophils (260, 261). However, other studies have demonstrated the opposite effect (262), perhaps due to differences in neutrophil depletion methods.
Although neutrophils are normally short-lived cells, a buildup of neutrophils in chronic UC inflammation can overwhelm the ability of resident macrophages to clear this cell population, leading to neutrophil necrosis and the release of damaging granule contents (257). Thus, the ability of certain factors to either inhibit (IL-8, IL-1, IFN-γ, granulocyte-macrophage colony-stimulating factor [GM-CSF], and C5a) (257, 263) or promote (IL-10 and TNF-α) (264, 265) neutrophil apoptosis can influence the degree of tissue damage. The massive transmigration of neutrophils through the epithelium and the release of elastase have also been associated with decreased expression levels of tight junction and adherens junction proteins (266). Elevated levels of fecal elastase have been found to correlate with disease severity in UC patients (267). It thus appears that neutrophils may contribute to both disease pathogenesis and recovery in UC.
Treatment of Ulcerative Colitis
Unfortunately, current treatment options for UC are limited and unable to induce remission in all patients. Given the inflammatory nature of this disease, most treatments entail immunosuppression. First-line treatments, typically sulfasalazine and 5-aminosalicylates, including mesalamine, olsalazine, and balsalazide, induce remission in about 50% of patients (22, 268). If 5-aminosalicylate therapy fails, patients with milder UC may be prescribed oral glucocorticoids or immunosuppressives (269). Azathioprine (270), 6-mercaptopurine (271), and monoclonal antibody inhibitors of TNF-α, including infliximab (272) and adalimumab (273, 274), have all shown efficacy as immunosuppressives for UC. In more severe cases, patients may receive intravenous glucocorticoids or cyclosporine to attempt to induce remission (269, 275). Maintenance therapy during remission may include oral or rectal 5-aminosalicylates or thiopurines, azathioprine, or 6-mercaptopurine.
Side effects of these treatments can be serious, including acute pancreatitis and bone marrow suppression (276). Patients who are unable to tolerate treatment or whose disease does not respond to treatment may develop serious complications such as toxic megacolon, bowel perforation, uncontrolled bleeding, and carcinoma or high-grade dysplasia, each of which is an indication for colectomy (277). Unlike CD, colectomy is often curative for UC. However, as many as 40 to 50% of patients develop pouchitis, whereby the artificial rectum surgically created from ileal tissue after colectomy becomes inflamed (278). Pouch failure is estimated to occur in 4 to 10% of patients (22, 279). This inflammatory condition is thought to result from changes in the microbiota within the ileal pouch, but the disease mechanism is still unclear (279). These side effects and the often limited effectiveness of current treatments mean that novel treatments for UC are needed.
Ulcerative Colitis and Fecal Microbiota Transplantation
Given that UC is thought to stem from dysbiosis and aberrant immune responses to the microbiota, there was early interest in the use of probiotics and FMT to treat UC. However, the mechanisms by which FMT may ameliorate UC are unknown, and the use of FMT as adjunctive therapy remains controversial (280–282). Following FMT, IBD patients exhibit microbiome compositions that resemble those of their donors (24, 283–285). A recent randomized clinical trial comparing the efficacy of FMT to that of a water enema control found a significant difference in levels of remission between the two groups, with 24% of FMT-treated patients achieving clinical remission (24). However, another randomized clinical trial in the same year reported no statistically significant difference in remission rates between patients who received FMT from a healthy donor (41% remission) and control patients who received FMT using their own feces (25% remission) (285). A recent meta-analysis of case series studies of FMT for the treatment of IBD showed that 45% of patients achieved clinical remission following treatment, with higher rates of remission being observed for CD patients than for UC patients (286). More research is needed to determine why some IBD patients receiving FMT experience remission (283) while others have no change in symptoms despite alterations in their gut microbiomes (284). The identification of microbial taxa that are associated with remission in patients who respond to FMT treatment could result in the development of more targeted probiotic therapeutics with greater efficacy.
Clinical Trials of Probiotics and Ulcerative Colitis
Although this field is still in its infancy, recent clinical trials and meta-analyses suggest that probiotics may be a viable option for adjuvant therapy in some UC patients (Table 3). A recent systematic review of clinical trials evaluating probiotics for the treatment of IBD concluded that although there was no evidence to suggest benefit in CD treatment, probiotics and prebiotics were useful in helping to induce and maintain remission of UC (16). Twenty-one trials using probiotics for UC treatment were identified in this review, with most trials considering either E. coli Nissle 1917 or the probiotic cocktail VSL#3. One double-blind double-dummy study showed that E. coli Nissle 1917 therapy was as effective as mesalamine in maintaining remission (17). Another study showed significantly greater induction of remission among pediatric UC patients treated with VSL#3 than among placebo-treated controls (18). A few smaller-scale studies showed that other probiotics, including Bio-Three (Enterococcus faecalis, Clostridium butyricum, and Bacillus mesentericus) (287) and Bifidobacterium breve (288), also reduced disease activity. Thus, although there is still a paucity of well-designed, large-scale, randomized, controlled trials, there is the potential that microbial therapy could serve as a viable alternative to pharmacological therapy for UC.
TABLE 3.
Clinical trials evaluating probiotic efficacy in maintenance or induction of UC remissiona
| Study type and reference | Yr | Species (daily dose) | Primary outcome(s) | Patient population | Conclusion(s) |
|---|---|---|---|---|---|
| Probiotic trials showing benefit | |||||
| 363 | 2006 | LGG (1.8 × 1010 CFU with or without mesalamine) | Relapse (UC symptoms requiring treatment or increase in CAI to >4) | 187 patients with UC in remission for <12 mo | Increased relapse-free time with probiotics relative to mesalamine, no difference in relapse rate at 6 or 12 mo |
| 364 | 2005 | Bifidobacterium longum (2 × 1011 CFU) plus Synergy (6 g fructooligosaccharide/inulin) twice daily | CAI, bowel habit index, sigmoidoscopy score, histology score, and immune parameters (colonic TNF-α, IL-1α, serum C-reactive protein, human beta defensins) | 16 patients with active UC | CAI significantly reduced in the probiotic group, TNF-α and IL-1α levels lower in the probiotic group than in the placebo group after 4 wk (P = 0.0177 and P = 0.0051, respectively), defensin levels not different |
| 365 | 1997 | E. coli Nissle 1917 (2.5 × 1010 viable CFU daily for 4 days and twice daily for the remainder of the study) | Time to relapse (CAI of ≥4) | 120 patients with chronic UC in remission | E. coli (16% relapse rate) as effective as mesalamine treatment (11.3% relapse rate) in maintaining remission |
| 326 | 1999 | E. coli Nissle 1917 (2 capsules with 2.5 × 1010 CFU viable bacteria twice a day) | Time to remission, rate of relapse after induction of remission | 116 patients with clinically active UC | E. coli Nissle 1917 plus steroids was similar to mesalamine plus steroids in inducing remission (OR, 1.35; 95% CI, 0.6 to 3.04), relapse rate was lower in the probiotic group (67% vs 73% in controls; P < 0.05), no difference in duration or mean time to remission |
| 17 | 2004 | E. coli Nissle 1917 (2.5 × 109–25 × 109 viable CFU once daily for 4 days and twice daily for the remainder of the study) | Time to relapse (CAI of >6 or increase of 3 points and CAI of >4; endoscopic index of >4 and histological signs of acute inflammation) | 327 patients with UC in remission | E. coli Nissle 1917 (36.4% relapse rate) as effective as mesalamine (33.9% relapse rate) in maintaining remission |
| 325 | 2010 | E. coli Nissle 1917 (daily 40-, 20-, or 10-ml enemas containing 108 CFU/ml) | Clinical remission (DAI of ≤2) | 90 patients with moderate distal activity in UC | Dose-dependent increase in remission with E. coli therapy (by per-protocol but not intention-to-treat analysis), time to remission was shortest with the highest dose |
| 366 | 2005 | VSL#3 (1.8 × 1012 CFU twice daily) | Remission (UCDAI of ≤2) or response (UCDAI decrease of ≥3 points) | Adult patients with ≥2-wk history of active UC not responsive to mesalamine | VSL#3 promotes remission |
| 18 | 2009 | VSL#3 (wt-based dose between 4.5 × 1012 and 18 × 1012 CFU) | Remission rate and time to relapse | 29 pediatric patients with newly diagnosed UC | VSL#3-treated patients were more likely to achieve remission and had fewer relapses and lower endoscopic and histological scores at 6 and 12 mo or point of relapse |
| 367 | 2010 | VSL#3 (3.6 × 1012 viable lyophilized bacteria) | Decrease in UCDAI of ≥50% | 144 patients with mild to moderate relapsing UC | VSL#3 reduced UCDAI scores (3 points or more) and rectal bleeding but not endoscopic scores or physician's rate of disease activity; trend toward increased remission in the VSL#3 treatment group (P = 0.069) |
| 368 | 2009 | VSL#3 (3.6 × 1012 lyophilized bacteria) | 50% reduction in UCDAI score | 147 adult patients with mild to moderate UC | VSL#3 was significantly better than placebo in inducing remission and improving UCDAI scores (P < 0.001) |
| 369 | 1999 | VSL#3 (3 g twice daily for 12 mo) | Remission maintenance | 20 patients with UC intolerant to 5-ASA in remission | 15/20 VSL#3-treated patients (75%) were still in remission at 12 mo |
| 370 | 2012 | Bifid Triple Viable (6 capsules of Bacillus acidophilus, B. bifidum, and streptococci) | Clinic symptom score, colon mucosa inflammation score, and immune indices | 82 adult patients with active UC | Decreased clinical symptoms and mucosal inflammation scores in the probiotic group |
| 287 | 2007 | Bio-Three (2 mg Enterococcus faecalis T-110, 10 mg Clostridium butyricum TO-A, and 10 mg Bacillus mesentericus TO-A) | Improved UCDAI scores | 20 patients with mild to moderate UC | Remission (UCDAI score of <2) in 45% of patients (9/20) and response (decrease in UCDAI of >3 points) in 10% of patients (2/20) |
| 288 | 2003 | Yakult (B. breve, B. bifidum, L. acidophilus YIT 0168 in 100 ml with at least 1010 viable bacteria) | Exacerbation of clinical symptoms (increased frequency of bowel movements or abdominal pain or appearance or increased frequency of blood or mucus in movements) | 21 adult UC patients | Some protection in preventing exacerbation of UC symptoms (3/11 probiotic-treated vs 9/10 control patients developed exacerbated symptoms) |
| 371 | 2010 | S. boulardii (250 mg 3 times daily for 4 wk) | Improved clinical score | 25 patients with mild to moderate UC | Reduced CAI scores with S. boulardii |
| Probiotic trials showing no benefit | |||||
| 372 | 2015 | B. longum 536 (2 doses of 3 × 1011 freeze-dried viable CFU 3 times daily) | Remission (UCDAI of ≤2) | 56 patients with mild to moderate UC | No difference in remission or UCDAI scores in probiotic vs placebo (containing dextrin) groups; rectal bleeding was reduced with probiotics |
| 373 | 2004 | VSL#3 (9 × 1011 lyophilized bacteria plus either balsalazide or mesalamine) | Remission (based on clinical evaluation and diary card) | 90 patients with mild to moderate ulcerative colitis | Remission was similar with VSL#3 plus balsalazide and placebo plus balsalazide, but time to remission was shorter in the probiotic group (4 vs 7 avg days in probiotic and placebo groups, respectively; P < 0.01) |
| 374 | 2010 | VSL#3 (3.6 × 1012 bacteria) | Clinical response and remission as defined by UCDAI | 28 patients with mild to moderate UC | 10/14 VSL#3-treated patients showed a clinical response relative to 5/14 control patients (P = 0.064) |
| 375 | 2004 | B. breve, B. bifidum, and L. acidophilus (1 × 1010 CFU in fermented milk plus sulfasalazine or 5-ASA) | Remission rate and CAI | 20 patients with moderate to severe UC | No improvement in CAI scores over placebo (OR, 0.64; 95% CI, 0.10 to 4.10), improved endoscopic activity index score (P < 0.01) and histological scores (P < 0.01) in the probiotic group vs no improvement in the placebo group |
| 376 | 2011 | Probio-Tec AB-25 (1.25 × 1010 CFU of both L. acidophilus LA-5 and B. animalis subsp. lactis BB-12) | Time to relapse (SCCAI of >4 or endoscopic changes) | 32 patients in remission ≥4 wk | No significant clinical benefit for maintaining remission |
CAI, clinical activity index; DAI, disease activity index; UCDAI, ulcerative colitis daily activity index; SCCAI, simple clinical colitis activity index; 5-ASA, 5-aminosalicylic acid; CI, confidence interval; OR, odds ratio; wt, wild type. Scoring criteria for CAI, DAI, UCDAI, and SCCAI differ between studies.
Protective Mechanisms of Probiotics against Ulcerative Colitis
As discussed above, UC is a multifactorial disease characterized by complex genetics, dysbiosis, and aberrant activation of the inflammatory immune response. Epithelial barrier breakdown and subsequent increased exposure to microbial products further stimulate immune responses and host immune-induced epithelium damage. The ability of probiotics to promote epithelial barrier integrity, either directly by influencing junctional complexes or indirectly by affecting the cytokine milieu and immune cell activation, will thus likely have profound effects on UC disease severity by limiting exposure to inflammatory signals and repairing host-induced epithelium damage. Probiotics that are able to influence antigen-presenting cell (APC) activation, as well as the downstream recruitment of effector immune cells, may also modulate immune cell responses to inflammatory signals. There are thus multiple mechanisms through which probiotics may limit the inflammation and barrier disruption observed in individuals with UC. Some of these immunomodulatory mechanisms discovered for specific probiotics that will be of potential interest in the design of probiotic cocktail therapy for UC are discussed below.
Maintenance of the microbiota.
The ability of probiotic strains to restore or maintain the composition of the microbiota may help prevent inflammatory immune signaling induced by dysbiosis. Probiotic administration has been shown to limit dysbiosis in many disease states, including colitis models, and to restore a normal microbiota composition more quickly than with placebo (289–291). Patients with minimal hepatic encephalopathy secondary to cirrhosis who were treated with LGG showed significantly increased prevalences of Lachnospiraceae and Clostridia cluster XIV and decreased prevalences of Enterobacteriaceae and Porphyromonadaceae relative to placebo-treated controls (292). LGG has also been shown to prevent the increased prevalences of Alcaligenes and Corynebacterium species observed with chronic alcohol feeding in mice (293). Similarly, S. boulardii treatment in a diabetic mouse model led to increased prevalences of Bacteroidetes and decreased prevalences of Firmicutes closer to levels observed in normal mice (294). Administration of S. boulardii to antibiotic-treated mice also resulted in a faster return to preantibiotic levels of specific bacterial strains, such as increased levels of Clostridium coccoides-Eubacterium rectale group members, including butyrate producers, and decreased levels of Enterobacteriaceae and Bacteroides species (291). Restoration of butyrate producers may be especially helpful in the context of UC, which is associated with decreased levels of butyrate-producing bacteria (228).
Probiotics can also modulate the metabolic profile of the microbiota, suggesting a means by which these organisms may help prevent alterations in the microbiota and limit dysbiosis. For example, LGG administration to healthy 65- to 80-year-old individuals induced no change in overall microbiota composition, as determined by 16S rRNA sequencing, with a few exceptions, such as increased levels of the butyrate producers Roseburia and Eubacterium (289). However, the expression levels of genes involved in bacterial motility and chemotaxis were increased in certain commensal species, including Bifidobacterium, leading to the suggestion that LGG can promote interactions between certain microbes and the host epithelium (289). More research is needed to determine if the effects of probiotic organisms on microbiota composition and metabolic activity would confer protection in the context of human UC.
Maintenance of intestinal epithelium integrity and barrier function.
Reinforcing the damaged GI epithelial barrier is a further potential avenue by which probiotics may limit inflammatory responses in UC patients. As discussed above for CDI, specific probiotics can directly influence the expression level, composition, and organization of the mucus layer and junctional complex components. However, a key feature of barrier dysfunction in UC is immune-mediated dysregulation of epithelial junctions via inflammatory cytokines, providing another avenue by which probiotic organisms may limit damage associated with UC (Table 2).
Inflammatory cytokines such as TNF-α, IFN-γ, and IL-23 are known to increase epithelial barrier breakdown and can be modulated by probiotic strains (295, 296). Several probiotic strains have been reported to downregulate TNF-α and IFN-γ production in mouse models of colitis, including Lactobacillus brevis SBC 8803, Lactobacillus fermentum, Lactobacillus salivarius subsp. salivarius, Bifidobacterium lactis (243), and mixtures of Lactobacillus and Bifidobacterium species (243, 297). S. boulardii also decreases TNF-α expression in mice (188). Infectious models also demonstrate the ability of probiotics to limit GI inflammatory cytokine secretion, with LGG, for example, partially preventing the ETEC-induced increase in TNF-α expression in IPEC-J2 cells (171). Although the mechanisms underlying these probiotic-mediated decreases in inflammatory cytokine levels and the associated barrier disruption are not well described, it is possible that probiotics act at least in part by modulating the overall cytokine milieu and inducing the production of anti-inflammatory cytokines.
Dampening inflammation through modulating the cytokine milieu.
The ability of probiotics to influence the cytokine milieu can have profound effects on disease severity by modulating the level of harmful host inflammatory immune responses. The anti-inflammatory cytokines IL-10 and TGF-β decrease the production of inflammatory cytokines, including IL-12p70, TNF-α, IL-1β, and IFN-γ (236). In this manner, IL-10 and TGF-β are able to dampen host immune responses and limit the inflammation-mediated deregulation of barrier integrity (295, 298). Indeed, the absence of IL-10 in mouse models significantly increases susceptibility to colitis, and IL-10 supplementation can ameliorate the severity of chemically induced colitis in mice (299, 300). Importantly, multiple probiotic species capable of ameliorating disease severity in colitis models, including Lactobacillus species, Bifidobacterium species, and E. coli, increase the production of the anti-inflammatory cytokine IL-10 (301–306) and decrease the expression levels of the inflammatory cytokines TNF-α (243) and IFN-γ (243, 297).
Given the key role of APCs in directing the balance of the cytokine milieu, numerous studies have screened probiotics for potential effectiveness in the treatment of colitis based on the ratio of inflammatory to anti-inflammatory cytokines that they induce from APCs (Table 4). Indeed, the levels of APC activation and cytokine production induced by different probiotic species vary significantly (307), and there are reports that some probiotic strains actually inhibit the effects of more stimulatory strains (307, 308). For example, the addition of the weak inflammatory cytokine inducer Lactobacillus reuteri with L. casei prevented the previously noted high levels of activation markers (major histocompatibility complex class II [MHC-II] and CD86) and inflammatory cytokines (IL-12, IL-6, and TNF-α) induced by L. casei, although IL-10 levels were not affected (307). Thus, it is possible that the addition of particular strains may diminish potential beneficial effects of other strains in probiotic cocktails. Some probiotics have also been reported to induce the expression of APC activation markers while simultaneously limiting cell activation in response to inflammatory stimuli. For example, while one study reported strong CD80, CD86, and CCR7 upregulation in human APCs exposed to S. boulardii (309), previous studies found that S. boulardii inhibited the lipopolysaccharide-induced upregulation of CD40, CD80, and CCR7 expression in human myeloid cells in vitro (310). It will be important to assess specific inhibitory properties of individual probiotic organisms on mucosal APCs, which are known to have vastly different phenotypic profiles than in vitro bone marrow- or monocyte-derived phagocytic cells, in order to more accurately assess whether these probiotics may have beneficial effects in the context of UC.
TABLE 4.
Immunological effects of probiotic strainsa
| Organism and genus | Species | Strain(s), company, or trade name | Effect(s) on immune system | Model(s) | Reference |
|---|---|---|---|---|---|
| Gram-positive bacteria | |||||
| Lactobacillus | L. acidophilus | NCFMTM | Induced IL-12p70 and IL-10 in a dose-dependent manner; ↑ IL-23, IL-6, IL-12p40, and CXCL1 expression | Human monocyte-derived DCs | 303 |
| L. acidophilus | X37 | Strong ↑ IL-12 and TNF-α; ↑ activation markers CD40, CD83, CD86, and HLA-DR | Human monocyte-derived DCs | 308 | |
| L. fermentum | CECT 5716 | ↓ IL-6 at wk 2 in the therapeutic group relative to TNBS-only controls; no effect on MPO levels | TNBS colitis | 321 | |
| ↓ colonic MPO levels relative to TNBS controls, ↓ TNF-α relative to controls | TNBS colitis | 377 | |||
| L. fermentum | Lf1 | ↑ SOD1 expression | DSS colitis | 378 | |
| L. paracasei | Z11 | Strong ↑ IL-12 and TNF-α; ↑ activation markers CD40, CD83, CD86, and HLA-DR | Human monocyte-derived DCs | 308 | |
| L. plantarum | HY115 | ↓ IL-1β, IFN-γ, and TNF-α expression compared to DSS controls | DSS colitis | 323 | |
| ↑ cytoplasmic IκBα and decreased nuclear NF-κB compared to DSS controls | |||||
| L. brevis | HY7401 | ↓ IL-1β, IFN-γ, and TNF-α expression compared to DSS controls; decreased intestinal epithelial MPO activity compared to DSS-treated controls; ↑ cytoplasmic IκBα and ↓ nuclear NF-κB compared to DSS controls | DSS colitis | 323 | |
| L. reuteri | ATCC 55730 | ↓ TNF-α relative to TNBS controls | TNBS colitis | 377 | |
| L. reuteri | R2LC, JCM 5869, ATC PTA 4659, ATCC 55730 | Prevented ↑ in colonic P-selectin, ↓ numbers of rolling leukocytes in submucosal and mucosal vessels | DSS colitis | 379 | |
| L. reuteri | DSM 12246:12002 | Strong ↑ IL-10 | Human monocyte-derived DCs | 308 | |
| L. salivarius | Ls-33 | ↑ IL-10 secretion, IL-12p40, IL-23, IL-6, and CXCL1 expression | Human monocyte-derived DCs | 303 | |
| L. rhamnosus | GG (ATCC 53103) | ↓ IL-23, IL-17, and CD40 expression | LPS-stimulated T84 and HT29 cultures | 317 | |
| L. rhamnosus | GG (ATCC 531030) | ↓ hepatic MPO levels (neutrophil infiltration) | Alcohol-fed rats | 351 | |
| L. rhamnosus | GG (ATCC 53103) | Prevented ↑ in hepatic TNF-α levels | Chronic alcohol feeding in mice | 293 | |
| L. rhamnosus | ATCC 7469 | ↑ TLR2 and TLR4 expression; ↓ TNF-α with pretreatment | ETEC-infected IPEC-J2 cells | 171 | |
| Bifidobacterium | B. infantis | 35624 | ↑ IL-10 secretion; ↓ IL-12p70 secretion and ↑ IL-10 secretion in LPS-, IFN-γ-, and TNF-α-stimulated cells; ↑ IL-23, IL-12p40, IL-6, and CXCL1 expression | Human monocyte-derived DCs | 303 |
| B. infantis | Guangzhou Baoxing Biotechnology Company | ↑ IL-2, IL-12p40, RORγT, IL-23, and IL-17A expression in MLNs relative to untreated TNBS controls | TNBS colitis | 380 | |
| B. infantis | Riken Lab | ↓ IL-17 production in ex vivo-stimulated colonocytes | DSS-stimulated mouse colonocytes | 304 | |
| B. infantis | JCM 1222 | ↓ IL-17A and IFN-γ and ↑ IL-10 expression in T cells; ↓ CD40 and CD80 expression on IECs | T cells stimulated ex vivo with IECs from DSS-treated mice; DSS-treated mice | 318 | |
| B. infantis | Isolated from VSL#3 | ↓ IFN-γ secretion, ↑ TGF-β | IL-10-deficient mice | 357 | |
| B. animalis subsp. lactis | Bb12 | Strong ↑ IL-10 | Human monocyte-derived DCs | 308 | |
| B. bifidum | Riken Lab | ↓ IL-17 production in ex vivo-stimulated colonocytes | DSS-stimulated mouse colonocytes | 304 | |
| B. bifidum | S131, Z9 | Strong ↑ IL-10 | Human monocyte-derived DCs | 308 | |
| B. catenulatum | Riken Lab | ↓ IL-17 production in ex vivo-stimulated colonocytes | DSS-stimulated mouse colonocytes | 304 | |
| B. breve | DSMZ 20213 | ↓ IL-23, IL-17, and CD40 expression | LPS-stimulated T84 and HT29 cultures | 317 | |
| B. longum | Q45, Q46 | Strong ↑ IL-10 | Human monocyte-derived DCs | 308 | |
| Gram-negative bacteria | |||||
| Escherichia | E. coli | Nissle 1917 | ↑ IL-12p70 and IL-10 in a dose-dependent manner; ↓ IL-12p70 secretion and ↑ IL-10 secretion in LPS-, IFN-γ-, and TNF-α-stimulated moDCs; ↑ IL-23 and IL-6 expression; no change in IL-17 | Human monocyte-derived DCs | 303 |
| E. coli | Nissle 1917 O6:K5:H1, F18 OR:K1:H5, BJ4 OR:K:H2, MG1655 OR:K:H48, UTI | Strong ↑ IL-10; weak ↑ TNF-α and IL-12p70 | Human monocyte-derived DCs | 308 | |
| Probiotic cocktails | |||||
| Duolac Gold (mixture of 6 probiotics) | B. lactis, B. longum, B. bifidum, L. acidophilus, L. rhamnosus, Streptococcus thermophilus | KCTC 11904BP, 12200BP, 12199BP; KCTC 11906BP, 12202BP; KCTC 11870BP | ↓ IL-6 in serum and colon relative to DSS control mice | DSS colitis | 322 |
| VSL#3 | L. casei, L. plantarum, L. acidophilus, L. delbrueckii subsp. bulgaricus, B. longum, B. breve, B. infantis, and Streptococcus salivarius subsp. thermophilus | VSL Pharmaceuticals | ↑ primarily IL-12p70 over IL-10; ↑ IL-23, IL-12p40, IL-6, and CXCL1 expression | Human monocyte-derived DCs | 303 |
| VSL#3 | ↑ IL-10 and ↓ IL-12 in colonic lamina propria DCs; no change in IL-6 or IL-13 | Patients with mild to moderate UC | 374 | ||
| Mix 1 | Lactobacillus acidophilus Bar 13 and Bifidobacterium longum Bar 33 (1:1) | Barilla G&R F.lli SPA (Parma, Italy) | ↓ serum IL-12, TNF-α, MCP-1, and IFN-γ and ↑ IL-10 compared to TNBS-treated controls; ↓ total CD4+ cells and ↓ γδ+ lamina propria T cells, ↑ Tregs relative to TNBS-treated controls | TNBS colitis in mice | 297 |
| Mix 2 | L. plantarum Bar 10, Streptococcus thermophilus Bar 20, and B. animalis subsp. lactis Bar 30 (1:1:1) | Barilla G&R F.lli SPA | ↓ serum IL-12, TNF-α, MCP-1, and IFN-γ and ↑ IL-10 compared to TNBS-treated controls | TNBS colitis in mice | 297 |
| Yeast | |||||
| Saccharomyces | S. boulardii | Biocodex | ↓ plasma IL-6, IL-4, IL-1β, and TNF-α compared to control mice | Db/db mice | 294 |
| S. boulardii | Biocodex | Prevented IL-8 ↑ (viable but not heat-killed S. boulardii); viable yeast prevented EHEC-induced NF-κB DNA binding and MAP kinase activation | EHEC-infected T84 cells | 360 | |
| S. boulardii | Biocodex | S. boulardii supernatant ↓ IL-8 mRNA and protein production, prevented IκBα degradation, and reduced NF-κB DNA binding | IL-1β- and TNF-α-stimulated HT29 cells | 314 | |
| S. boulardii | Biocodex | ↓ ERK, JNK, and NF-κB activation and IL-8 secretion | S. flexneri-stimulated T84 cells | 190 | |
| S. boulardii | Floratil | ↑ serum IL-10 (viable or heat-killed S. boulardii) | Murine intestinal obstruction model | 155 | |
| S. boulardii | Bioflor | ↑ PPARγ expression; ↓ IL-8 secretion from unstimulated and stimulated HT29 cells in a PPARγ-dependent manner; ↑ PPARγ, ↓ IL-8, IL-1β, IL-6, IL-8R, TNF-α, and iNOS expression in healthy and TNBS-treated colons | TNF-α- and IL-1β-stimulated HT29 cells; TNBS colitis in rats | 381 | |
| S. boulardii | Merck SA | ↑ Kupffer cells; earlier and higher-level TNF-α, IL-12, and IFN-γ responses to E. coli B41 infection | Monoassociated Swiss/NIH mice | 154 | |
| S. boulardii | Biocodex | ↓ T cell infiltrate and NF-κB activation in colon; no ↑ in IL-10 or TGF-β in MLNs or colon; no ↑ in CXCL9, CXCL10, CXCL11, CCL4, or CCL5 in MLNs; ↑ P-selectin interaction with activated T cells and ↑ T cell accumulation in MLNs | Lymphocyte transfer SCID mouse model of colitis | 382 | |
| S. boulardii | Biocodex | Preincubation prevents TNF-α expression and caspase 8 and 9 activation, ↓ IL-8, IL-6, IL-1β, TNF-α, and IFN-γ | T84 human colonic cell line | 383 | |
| S. boulardii | Perenterol | No effect on sIgA; ↑ CD25 expression by peripheral CD4+ T cells; no change in peripheral B or T cell numbers | PBMCs of S. boulardii-treated healthy human volunteers | 362 | |
| S. boulardii | Ultra-Levure, Biocodex | Reduced iNOS levels in macrophages; high dose of S. boulardii reduced colon citrulline in diarrhea | IFN-γ-stimulated mouse macrophage RAW 264-7 cells; castor oil-induced diarrhea in male Wistar rats | 384 | |
| S. boulardii | Biocodex | ↓ ERK, JNK, and NF-κB activation; IL-8 secretion from control and infected cells; ↓ PMN transmigration across infected T84 cells or recruitment to human fetal colonic xenografts | Shigella flexneri-infected T84 cells and human fetal colonic xenografts | 190 | |
| S. boulardii | Biocodex | ↓ CD40, CD80, CCR7, TNF-α, and IL-6 expression; S. boulardii ↑ IL-10 expression | LPS-stimulated human myeloid CD1c+ CD11c+ CD123− DCs | 310 | |
| S. boulardii | Reflor, Biocodex | ↑ serum NO compared to TNBS-treated controls | TNBS colitis in rat | 385 | |
| S. boulardii | Ardeypharm | Small ↑ TNF-α and CXCL1 expression; little effect on IL-10 and IL-12p70 | Human monocyte-derived DCs | 303 | |
| S. boulardii | Biocodex | ↑ ileal expression of chemokine KC | TcdA-treated mouse ileal loop | 361 | |
| S. boulardii | Biocodex | No change in total lymphocyte no. or serum antibody levels; cell wall binds complement C3b; ↑ leukocyte chemokinesis; ↑ erythrocyte, total leukocyte, neutrophil, and polynuclear cell numbers | Healthy human PBMCs | 386 | |
| S. cerevisiae | CNCM I-3856 | ↓ IL-6 and IL-8 secretion and CCL20, CXCL2, and CXCL10 expression | ETEC-stimulated porcine epithelial IPEC-1 cells | 174 |
CXCL, C-X-C motif ligand chemokine; DCs, dendritic cells; DSS, dextran sodium sulfate; HLA-DR, human leukocyte antigen, antigen D related; IECs, intestinal epithelial cells; iNOS, inducible nitric oxide synthase; LPS, lipopolysaccharide; MLNs, mesenteric lymph nodes; moDC, monocyte-derived dendritic cell; MPO, myeloperoxidase; NF-κB, nuclear factor kappa light chain enhancer of activated B cells; NO, nitric oxide; PBMCs, peripheral blood mononuclear cells; PMN, polymorphonuclear leukocyte; PPARγ, peroxisome proliferator-activated receptor γ; RORγT, retinoic acid receptor-related orphan receptor gamma T; SCID, severe combined immunodeficiency; sIgA, secretory immunoglobulin A; SOD1, superoxide dismutase 1; TGF-β, transforming growth factor β; TNBS, 2,4,6-trinitrobenzenesulfonic acid; ↑, increase; ↓, decrease.
Effects on neutrophil infiltration and function.
Given the clear association of UC pathology with neutrophil accumulation, the ability of probiotics to regulate the recruitment, function, or apoptosis of neutrophils has the potential to greatly influence the disease course (Table 4). Beneficial effects of probiotics in limiting neutrophil-associated damage may stem from their ability to modulate the production of neutrophil chemotaxins and activators such as IL-17 and IL-8 (311–316).
Probiotics may affect IL-17 levels by modulating the expression of cytokines that promote Th17 responses. Numerous probiotics have been found to downregulate IL-17 production and alleviate colitis, including B. breve (317), B. longum (318), L. acidophilus (319), B. longum subsp. infantis (304), and Streptococcus thermophilus ST28 (320). Additional studies have demonstrated the ability of several probiotic species to reduce the expression of the Th17-promoting cytokines IL-6 and IL-23 (243) in vitro as well as in mouse models of liver fibrosis and GI permeability (188) and colitis (321–323). Further mechanisms of probiotic modulation of Th17 cell differentiation may include the inhibition of the costimulatory molecules CD40 and CD80 on intestinal epithelial cells or the downregulation of the Th17-promoting transcription factors RORγT, STAT3, and NF-κB (243).
Probiotics may also inhibit neutrophil-associated damage through the modulation of IL-8. For example, S. boulardii was shown to produce a soluble factor that can inhibit NF-κB-mediated IL-8 production in IL-1β- and TNF-α-stimulated HT29 cells (314). Another study found that S. boulardii decreased IL-8 expression and neutrophil transmigration during infection of T84 monolayers with Shigella flexneri, possibly by decreasing extracellular signal-regulated kinase (ERK), NF-κB, and c-Jun N-terminal kinase (JNK) signaling (190). Certain probiotics also alter neutrophil function, such as LGG inhibiting the formation of neutrophil extracellular traps formed in response to Staphylococcus aureus and phorbol 12-myristate 13-acetate (PMA) stimulation of in vitro human and murine neutrophils (324). Thus, although more studies are needed to determine the temporal effects of probiotics on neutrophils in colitis models, modulation of neutrophil recruitment and activity is one clear way in which certain probiotics could ameliorate symptoms of UC.
Summary of Probiotic Mechanisms of Action in Ulcerative Colitis and Implications for Future Therapies
UC is characterized by dysbiosis, GI barrier breakdown, and aberrant inflammatory immune responses, as described above (Fig. 1C). Particular microorganisms that are able to ameliorate any of these disease components could be of potential benefit in combination microbial therapy designed for UC.
As the exact species within the microbiota responsible for inciting disease pathology in UC are not well understood, it is possible at present to identify only probiotics that promote a microbiota associated with health and remission as opposed to active UC. It is thus of particular interest that probiotics such as LGG and S. boulardii have been found in some animal models to decrease the levels of Enterobacteriaceae and Porphyromonadaceae, which are increased in active UC (292). The ability of these and other probiotics to speed the restoration of the microbiota following antibiotic treatment and to promote the activity of butyrate-producing bacteria may also help prevent insults that lead to active UC. Further studies are needed to determine the effects of particular probiotics in maintaining or restoring the microbiota specifically in the context of colitis.
Also, as discussed above in the sections on CDI, numerous probiotics are capable of reinforcing epithelial cell barrier function in the context of dysbiosis and inflammation. Strains that are able to prevent TNF-α-induced dysregulation of tight junctions, such as L. plantarum (strain L2) (193) and B. bifidum (strains WU12, WU20, and WU57) (192), may be of particular benefit in cases of UC where levels of this inflammatory cytokine are elevated (237). Reinforcement of barrier integrity may help to reduce immune stimulation and prevent the exacerbation of inflammatory responses to microbial antigens.
It is important to note when considering these studies (Table 4) that most of them relied on in vitro models to evaluate cytokine induction by probiotics. The effect of probiotics on the many other immune cells present in the GI mucosa may lead to strikingly different consequences in vivo than would be predicted from in vitro studies. Furthermore, particular effects of probiotics may depend on the composition of the endogenous microbiota, which is greatly altered in the contexts of UC, antibiotic treatment, and CDI. More in vivo studies are clearly needed to determine the exact effects that probiotic strains will have on cytokine induction in the context of particular diseases such as UC and CDI.
Given that the roles of particular immune cell subsets in UC pathogenesis are still incompletely understood, predicting which probiotics might ameliorate disease symptoms based on their immunological effects becomes a difficult task. Furthermore, particular probiotics may still have beneficial effects in vivo despite inducing what might be considered counterproductive effects on specific immune cell subsets in vitro. E. coli Nissle 1917, for example, has been shown in clinical trials to help mitigate UC (17, 325, 326) yet induces the secretion of the neutrophil chemoattractant IL-8 in vitro (311). Such findings highlight the importance of studying the effects of particular probiotic strains on the epithelium and microbiota as well as on immune cells in vivo, as the relative importance of each mechanism for individual probiotic strains may differ.
Limitations of in vitro and animal studies in the evaluation of probiotic mechanisms of action.
It is worth commenting once more on the fact that most studies evaluating probiotics use in vitro or animal models. While these models are extremely useful in that they enable controlled, easily manipulable systems to test probiotic effects, several caveats should of course be kept in mind when generalizing these results and considering potential clinical applications. There are numerous animal models for both CDI and UC, with each one showing different levels of disease susceptibility and recapitulating different features of human disease (327–329). Only a limited number of models, for example, have been shown to recapitulate reinfection found in a high percentage of human C. difficile infections (330). As might be expected for diseases so dependent on the composition of the microbiota, the animal facilities, supplier, diet, and numerous other factors also influence disease outcome in these models (50). Indeed, recent data demonstrate that animal housing environments profoundly influence immune system development and can skew the nature and distribution of immune cells in laboratory mice (331). Differences in bile salt composition in mice versus humans must also be kept in mind for CDI models (332), although the general effects of primary versus secondary bile salts on C. difficile germination appear to be maintained (113). Clearly, these and numerous other differences must be factored into considerations of the specific identity and doses of probiotics to be tested for human therapies.
THERAPEUTIC USES OF PROBIOTICS
FMT is a promising but as-yet-unrefined therapy for many infectious and autoimmune GI disorders. Both recurrent CDI and UC can be successfully treated with FMT, but the microbial components responsible for benefits are still unknown. Identification of particular strains that confer protection in each case would allow the design of combined microbial therapies to treat disease while eliminating risks associated with the transfer of unknown components of the microbiota from human donors.
The ability to create such tailored therapy will require a thorough understanding of both disease pathogenesis and the in vivo mechanisms of action of particular beneficial microbial strains and combinations of strains. In many cases, an incomplete understanding of the roles played by host and microbial cells will limit the ability to predict effective therapy. Better-tailored microbial therapies for CDI and UC may in fact become possible as further studies continue to clarify the roles played by the microbiota and by host cells, leading to novel targets. Recent data also suggest that the persistence and efficacy of transferred microbial strains will depend on the composition of an individual patient's endogenous microbiota (333). Still, available data regarding the actions of particular beneficial microbial strains, viewed in light of the current understanding of CDI and UC pathogeneses, allow the identification of candidate probiotic strains for further testing (Tables 2 and 4).
Based on the available evidence for probiotic mechanisms of action, it seems unlikely that individual probiotic strains would confer the full repertoire of benefits necessary for protection against disease. Although some trials have shown benefits of S. boulardii and Lactobacillus species for the treatment of CDI and E. coli Nissle 1917 for the treatment of UC, most clinical trials considering single probiotic strains for the treatment of CDI have found no benefit (Table 1). Combinations of strains with complementary actions targeting a variety of factors involved in disease may instead be much more likely to confer protection against disease. General categories of action may include effects on the composition of the microbiota, host epithelial barrier integrity, and immune responses.
Unfortunately, data from in vitro studies suggest that optimal combined therapies may not always be predictable based on studies of individual strains (307). It is possible that some probiotics will have inhibitory effects on other coadministered strains and limit overall efficacy. Thus, although the many studies of individual probiotic strains provide useful information regarding potential mechanisms of action and identify candidates for therapy, further experiments will be necessary to determine if beneficial effects are maintained in combination with other probiotic strains in vivo.
Finally, although this review focuses on CDI and UC as examples, general principles regarding probiotic mechanisms of action can also be applied to similar GI pathogens and other forms of colitis, including CD. Indeed, effects of probiotics on reinforcing the epithelial barrier and limiting immune cell inflammation may be highly beneficial in ameliorating symptoms of many GI diseases. Reinforcement of the GI barrier may help protect the host from increased exposure to either specific toxins, as in the case of CDI or other infections, or components of the microbiota inciting autoimmune inflammation, as in the case of colitis. The ability to direct immune responses against pathogens or to limit aberrant inflammatory responses is an additional clear way in which probiotics could help to ameliorate disease. However, further studies are needed to specifically determine the effects of particular probiotic strains in vivo in the context of each disease.
Given the rising incidences of CDI and UC worldwide, improved therapies for these serious GI conditions are urgently needed. FMT and a few select probiotics already provide some benefit in preventing and treating these diseases. Improving microbial therapies through the use of defined combinations of beneficial strains tailored to each disease holds significant promise for expanding this line of adjuvant therapy and revolutionizing the treatment of these diseases.
ACKNOWLEDGMENTS
This work was supported by the U.S. National Institutes of Health through an NIH Directors New Innovators Award (AI112242) to T.J.L. and a training grant (AI106699) to S.E.A.
We thank Shonna M. McBride for helpful discussions in the creation of the manuscript.
Biographies

Lauren E. Hudson received her bachelor's degree in biology from the College of William and Mary in Virginia. She entered the M.D./Ph.D. program at Emory University in Atlanta, GA, in 2010 and joined the laboratory of Dr. Tracey Lamb. Her dissertation work focused on developing the probiotic yeast Saccharomyces boulardii as a vehicle for vaccine delivery to the gastrointestinal tract. She has recently defended her dissertation and is now continuing her medical education.

Sarah E. Anderson received her B.S. in 2013 from the University of North Carolina (UNC), where she majored in biology and chemistry. She was active in undergraduate research at UNC and also participated in the Undergraduate Research Program at Cold Spring Harbor Laboratory. Following graduation, she was awarded an APHL/CDC Emerging Infectious Diseases Fellowship to conduct research at the Wadsworth Center at the New York State Department of Health. At the Wadsworth Center, she developed strong interests in bacterial genetics and antibiotic resistance, which led her to pursue a Ph.D. in Microbiology and Molecular Genetics at Emory University.

Anita H. Corbett obtained her undergraduate degree in Chemistry at Colgate University in upstate New York. She then attended Vanderbilt University to obtain a Ph.D. in Biochemistry based on her thesis work studying the catalytic mechanism of topoisomerase II. She moved from Nashville, TN, to Boston, MA, to pursue postdoctoral studies at Harvard Medical School examining protein import into the nucleus. In 1997, she was recruited to the Biochemistry Department at the Emory University School of Medicine as a tenure track Assistant Professor. She rose through the ranks and is now a full Professor of Biochemistry and also serves as Codirector of the M.D./Ph.D. Program at Emory University. Her laboratory focuses on the use of Saccharomyces cerevisiae to study evolutionarily conserved processes. However, she has recently branched out in a collaborative effort with Dr. Tracey Lamb to explore the potential to use Saccharomyces boulardii as a probiotic or delivery mechanism.

Tracey J. Lamb was educated in Scotland, obtaining her undergraduate degree at the University of Glasgow and her doctorate at the University of Edinburgh, where her research examined the immune responses mediating resistance and susceptibility to filarial nematodes. In 2010, she was recruited to Emory University to set up a laboratory looking at immunopathogenesis in malaria. As an awardee of the NIH Directors New Innovators Scheme, she began a research project which aims to develop probiotic yeast as a vaccine expression-and-delivery system with a view to developing an inexpensive platform for vaccination that could be implemented in developing countries. In collaboration with Dr. Anita Corbett's laboratory and in research led by Dr. Lauren Hudson, they have developed protocols for the genetic modification of the probiotic yeast Saccharomyces boulardii and demonstrated that S. boulardii is an innocuous vaccine delivery vehicle that will not cause harmful inflammatory responses upon administration.
REFERENCES
- 1.Bakken JS, Borody T, Brandt LJ, Brill JV, Demarco DC, Franzos MA, Kelly C, Khoruts A, Louie T, Martinelli LP, Moore TA, Russell G, Surawicz C. 2011. Treating Clostridium difficile infection with fecal microbiota transplantation. Clin Gastroenterol Hepatol 9:1044–1049. doi: 10.1016/j.cgh.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi HH, Cho Y. 2016. Fecal microbiota transplantation: current applications, effectiveness, and future perspectives. Clin Endosc 49:257–265. doi: 10.5946/ce.2015.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamilton MJ, Weingarden AR, Unno T, Khoruts A, Sadowsky MJ. 2013. High-throughput DNA sequence analysis reveals stable engraftment of gut microbiota following transplantation of previously frozen fecal bacteria. Gut Microbes 4:125–135. doi: 10.4161/gmic.23571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kazerouni A, Burgess J, Burns LJ, Wein LM. 2015. Optimal screening and donor management in a public stool bank. Microbiome 3:75. doi: 10.1186/s40168-015-0140-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drekonja D, Reich J, Gezahegn S, Greer N, Shaukat A, MacDonald R, Rutks I, Wilt TJ. 2015. Fecal microbiota transplantation for Clostridium difficile infection: a systematic review. Ann Intern Med 162:630–638. doi: 10.7326/M14-2693. [DOI] [PubMed] [Google Scholar]
- 6.Schwartz M, Gluck M, Koon S. 2013. Norovirus gastroenteritis after fecal microbiota transplantation for treatment of Clostridium difficile infection despite asymptomatic donors and lack of sick contacts. Am J Gastroenterol 108:1367. doi: 10.1038/ajg.2013.164. [DOI] [PubMed] [Google Scholar]
- 7.Kelly CR, Ihunnah C, Fischer M, Khoruts A, Surawicz C, Afzali A, Aroniadis O, Barto A, Borody T, Giovanelli A, Gordon S, Gluck M, Hohmann EL, Kao D, Kao JY, McQuillen DP, Mellow M, Rank KM, Rao K, Ray A, Schwartz MA, Singh N, Stollman N, Suskind DL, Vindigni SM, Youngster I, Brandt L. 2014. Fecal microbiota transplant for treatment of Clostridium difficile infection in immunocompromised patients. Am J Gastroenterol 109:1065–1071. doi: 10.1038/ajg.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Bella S, Gouliouris T, Petrosillo N. 2015. Fecal microbiota transplantation (FMT) for Clostridium difficile infection: focus on immunocompromised patients. J Infect Chemother 21:230–237. doi: 10.1016/j.jiac.2015.01.011. [DOI] [PubMed] [Google Scholar]
- 9.De Leon LM, Watson JB, Kelly CR. 2013. Transient flare of ulcerative colitis after fecal microbiota transplantation for recurrent Clostridium difficile infection. Clin Gastroenterol Hepatol 11:1036–1038. doi: 10.1016/j.cgh.2013.04.045. [DOI] [PubMed] [Google Scholar]
- 10.Brandt LJ, Aroniadis OC, Mellow M, Kanatzar A, Kelly C, Park T, Stollman N, Rohlke F, Surawicz C. 2012. Long-term follow-up of colonoscopic fecal microbiota transplant for recurrent Clostridium difficile infection. Am J Gastroenterol 107:1079–1087. doi: 10.1038/ajg.2012.60. [DOI] [PubMed] [Google Scholar]
- 11.Alang N, Kelly CR. 2015. Weight gain after fecal microbiota transplantation. Open Forum Infect Dis 2:ofv004. doi: 10.1093/ofid/ofv004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paramsothy S, Borody TJ, Lin E, Finlayson S, Walsh AJ, Samuel D, Van den Bogaerde J, Leong RWL, Connor S, Ng W, Mitchell HM, Kaakoush N, Kamm MA. 2015. Donor recruitment for fecal microbiota transplantation. Inflamm Bowel Dis 21:1600–1606. doi: 10.1097/MIB.0000000000000405. [DOI] [PubMed] [Google Scholar]
- 13.Amirtha T. 2016. Banking on stool despite an uncertain future. Science 352:1261–1262. doi: 10.1126/science.352.6291.1261. [DOI] [PubMed] [Google Scholar]
- 14.Vyas D, Aekka A, Vyas A. 2015. Fecal transplant policy and legislation. World J Gastroenterol 21:6–11. doi: 10.3748/wjg.v21.i1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McFarland L. 2006. Meta-analysis of probiotics for the prevention of antibiotic associated diarrhea and the treatment of Clostridium difficile disease. Am J Gastroenterol 101:812–822. doi: 10.1111/j.1572-0241.2006.00465.x. [DOI] [PubMed] [Google Scholar]
- 16.Ghouri YA, Richards DM, Rahimi EF, Krill JT, Jelinek KA, DuPont AW. 2014. Systematic review of randomized controlled trials of probiotics, prebiotics, and synbiotics in inflammatory bowel disease. Clin Exp Gastroenterol 7:473–487. doi: 10.2147/CEG.S27530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kruis W, Fric P, Pokrotnieks J, Lukás M, Fixa B, Kascák M, Kamm M, Weismueller J, Beglinger C, Stolte M, Wolff C, Schulze J. 2004. Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine. Gut 53:1617–1623. doi: 10.1136/gut.2003.037747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miele E, Pascarella F, Giannetti E, Quaglietta L, Baldassano RN, Staiano A. 2009. Effect of a probiotic preparation (VSL#3) on induction and maintenance of remission in children with ulcerative colitis. Am J Gastroenterol 104:437–443. doi: 10.1038/ajg.2008.118. [DOI] [PubMed] [Google Scholar]
- 19.Claesson MJ, van Sinderen D, O'Toole PW. 2008. Lactobacillus phylogenomics—towards a reclassification of the genus. Int J Syst Evol Microbiol 58:2945–2954. doi: 10.1099/ijs.0.65848-0. [DOI] [PubMed] [Google Scholar]
- 20.MacKenzie D, Defernez M, Dunn W, Brown M, Fuller L, de Herrera S, Günther A, James S, Eagles J, Philo M, Goodacre R, Roberts I. 2008. Relatedness of medically important strains of Saccharomyces cerevisiae as revealed by phylogenetics and metabolomics. Yeast 25:501–512. doi: 10.1002/yea.1601. [DOI] [PubMed] [Google Scholar]
- 21.Brandt K, Barrangou R. 2016. Phylogenetic analysis of the Bifidobacterium genus using glycolysis enzyme sequences. Front Microbiol 7:657. doi: 10.3389/fmicb.2016.00657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Danese S, Fiocchi C. 2011. Ulcerative colitis. N Engl J Med 365:1713–1725. doi: 10.1056/NEJMra1102942. [DOI] [PubMed] [Google Scholar]
- 23.Rupnik M, Wilcox MH, Gerding DN. 2009. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol 7:526–536. doi: 10.1038/nrmicro2164. [DOI] [PubMed] [Google Scholar]
- 24.Moayyedi P, Surette MG, Kim PT, Libertucci J, Wolfe M, Onischi C, Armstrong D, Marshall JK, Kassam Z, Reinisch W, Lee CH. 2015. Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology 149:102–109. doi: 10.1053/j.gastro.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 25.Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, Farley MM, Holzbauer SM, Meek JI, Phipps EC, Wilson LE, Winston LG, Cohen J, Limbago BM, Fridkin SK, Gerding DN, McDonald LC. 2015. Burden of Clostridium difficile infection in the United States. N Engl J Med 372:825–834. doi: 10.1056/NEJMoa1408913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McFarland L, Mulligan M, Kwok R, Stamm W. 1989. Nosocomial acquisition of Clostridium difficile infection. N Engl J Med 320:204–210. doi: 10.1056/NEJM198901263200402. [DOI] [PubMed] [Google Scholar]
- 27.Kim KH, Fekety R, Batts DH, Brown D, Cudmore M, Silva J, Waters D. 1981. Isolation of Clostridium difficile from the environment and contacts of patients with antibiotic-associated colitis. J Infect Dis 143:42–50. doi: 10.1093/infdis/143.1.42. [DOI] [PubMed] [Google Scholar]
- 28.Lawley TD, Clare S, Deakin LJ, Goulding D, Yen JL, Raisen C, Brandt C, Lovell J, Cooke F, Clark TG, Dougan G. 2010. Use of purified Clostridium difficile spores to facilitate evaluation of health care disinfection regimens. Appl Environ Microbiol 76:6895–6900. doi: 10.1128/AEM.00718-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sorg JA, Sonenshein AL. 2008. Bile salts and glycine as cogerminants for Clostridium difficile spores. J Bacteriol 190:2505–2512. doi: 10.1128/JB.01765-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cohen SH, Gerding DN, Johnson S, Kelly CP, Loo VG, McDonald LC, Pepin J, Wilcox MH. 2010. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the Society for Healthcare Epidemiology of America (SHEA) and the Infectious Diseases Society of America (IDSA). Infect Control Hosp Epidemiol 31:431–455. doi: 10.1086/651706. [DOI] [PubMed] [Google Scholar]
- 31.Curry SR, Muto CA, Schlackman JL, Pasculle AW, Shutt KA, Marsh JW, Harrison LH. 2013. Use of multilocus variable number of tandem repeats analysis genotyping to determine the role of asymptomatic carriers in Clostridium difficile transmission. Clin Infect Dis 57:1094–1102. doi: 10.1093/cid/cit475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kyne L, Warny M, Qamar A, Kelly CP. 2000. Asymptomatic carriage of Clostridium difficile and serum levels of IgG antibody against toxin A. N Engl J Med 342:390–397. doi: 10.1056/NEJM200002103420604. [DOI] [PubMed] [Google Scholar]
- 33.Carter GP, Rood JI, Lyras D. 2012. The role of toxin A and toxin B in the virulence of Clostridium difficile. Trends Microbiol 20:21–29. doi: 10.1016/j.tim.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 34.Borriello SP, Wren BW, Hyde S, Seddon S, Sibbons P, Krishna MM, Tabaqchali S, Manek S, Price AB. 1992. Molecular, immunological, and biological characterization of a toxin A-negative, toxin B-positive strain of Clostridium difficile. Infect Immun 60:4192–4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuehne SA, Collery MM, Kelly ML, Cartman ST, Cockayne A, Minton NP. 2014. Importance of toxin A, toxin B, and CDT in virulence of an epidemic Clostridium difficile strain. J Infect Dis 209:83–86. doi: 10.1093/infdis/jit426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drudy D, Fanning S, Kyne L. 2007. Toxin A-negative, toxin B-positive Clostridium difficile. Int J Infect Dis 11:5–10. doi: 10.1016/j.ijid.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 37.LaFrance ME, Farrow M, Chandrasekaran R, Sheng J, Rubin DH, Lacy DB. 2015. Identification of an epithelial cell receptor responsible for Clostridium difficile TcdB-induced cytotoxicity. Proc Natl Acad Sci U S A 112:7073–7078. doi: 10.1073/pnas.1500791112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Na X, Kim H, Moyer MP, Pothoulakis C, LaMont JT. 2008. gp96 is a human colonocyte plasma membrane binding protein for Clostridium difficile toxin A. Infect Immun 76:2862–2871. doi: 10.1128/IAI.00326-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tucker KD, Wilkins TD. 1991. Toxin A of Clostridium difficile binds to the human carbohydrate antigens I, X, and Y. Infect Immun 59:73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan P, Zhang H, Cai C, Zhu S, Zhou Y, Yang X, He R, Li C, Guo S, Li S, Huang T, Perez-Cordon G, Feng H, Wei W. 2015. Chondroitin sulfate proteoglycan 4 functions as the cellular receptor for Clostridium difficile toxin B. Cell Res 25:157–168. doi: 10.1038/cr.2014.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Just I, Selzer J, Wilm M, von Eichel-Streiber C, Mann M, Aktories K. 1995. Glucosylation of Rho proteins by Clostridium difficile toxin B. Nature 375:500–503. doi: 10.1038/375500a0. [DOI] [PubMed] [Google Scholar]
- 42.Just I, Wilm M, Selzer J, Rex G, Von Eichel-Streiber C, Mann M, Aktories K. 1995. The enterotoxin from Clostridium difficile (ToxA) monoglucosylates the Rho proteins. J Biol Chem 270:13932–13936. doi: 10.1074/jbc.270.23.13932. [DOI] [PubMed] [Google Scholar]
- 43.Fiorentini C, Malorni W, Paradisi S, Giuliano M, Mastrantonio P, Donelli G. 1990. Interaction of Clostridium difficile toxin A with cultured cells: cytoskeletal changes and nuclear polarization. Infect Immun 58:2329–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brito GA, Fujji J, Carneiro-Filho BA, Lima AAM, Obrig T, Guerrant RL. 2002. Mechanism of Clostridium difficile toxin A-induced apoptosis in T84 cells. J Infect Dis 186:1438–1447. doi: 10.1086/344729. [DOI] [PubMed] [Google Scholar]
- 45.Gerding DN, Johnson S, Rupnik M, Aktories K. 2014. Clostridium difficile binary toxin CDT: mechanism, epidemiology, and potential clinical importance. Gut Microbes 5:15–27. doi: 10.4161/gmic.26854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Popoff MR, Rubin EJ, Gill DM, Boquet P. 1988. Actin-specific ADP-ribosyltransferase produced by a Clostridium difficile strain. Infect Immun 56:2299–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hecht G, Pothoulakis C, LaMont JT, Madara JL. 1988. Clostridium difficile toxin A perturbs cytoskeletal structure and tight junction permeability of cultured human intestinal epithelial monolayers. J Clin Invest 82:1516–1524. doi: 10.1172/JCI113760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nusrat A, Turner JR, Verkade P, Madara L, Parkos CA. 2001. Clostridium difficile toxins disrupt epithelial barrier function by altering membrane microdomain localization of tight junction proteins. Infect Immun 69:1329–1336. doi: 10.1128/IAI.69.3.1329-1336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kasendra M, Barrile R, Leuzzi R, Soriani M. 2014. Clostridium difficile toxins facilitate bacterial colonization by modulating the fence and gate function of colonic epithelium. J Infect Dis 209:1095–1104. doi: 10.1093/infdis/jit617. [DOI] [PubMed] [Google Scholar]
- 50.Britton RA, Young VB. 2012. Interaction between the intestinal microbiota and host in Clostridium difficile colonization resistance. Trends Microbiol 20:313–319. doi: 10.1016/j.tim.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seekatz A, Rao A, Santhosh K, Young V. 2016. Dynamics of the fecal microbiome in patients with recurrent and nonrecurrent Clostridium difficile infection. Genome Med 8:47. doi: 10.1186/s13073-016-0298-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khanna S, Montassier E, Schmidt B, Patel R, Knights D, Pardi D, Kashyap P. 2016. Gut microbiome predictors of treatment response and recurrence in primary Clostridium difficile infection. Aliment Pharmacol Ther 44:715–727. doi: 10.1111/apt.13750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Freeman J, Wilcox MH. 1999. Antibiotics and Clostridium difficile. Microbes Infect 1:377–384. doi: 10.1016/S1286-4579(99)80054-9. [DOI] [PubMed] [Google Scholar]
- 54.McFarland L. 2008. Update on the changing epidemiology of Clostridium difficile-associated disease. Nat Clin Pract Gastroenterol Hepatol 5:40–48. doi: 10.1038/ncpgasthep1029. [DOI] [PubMed] [Google Scholar]
- 55.McDonald LC, Killgore GE, Thompson A, Owens RC, Kazakova S, Sambol SP, Johnson S, Gerding DN. 2005. An epidemic, toxin gene-variant strain of Clostridium difficile. N Engl J Med 353:2433–2441. doi: 10.1056/NEJMoa051590. [DOI] [PubMed] [Google Scholar]
- 56.Antonopoulos DA, Huse SM, Morrison HG, Schmidt TM, Sogin ML, Young VB. 2009. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun 77:2367–2375. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buffie CG, Jarchum I, Equinda M, Lipuma L, Gobourne A, Viale A, Ubeda C, Xavier J, Pamer EG. 2012. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect Immun 80:62–73. doi: 10.1128/IAI.05496-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gu S, Chen Y, Zhang X, Lu H, Lv T, Shen P, Lv L, Zheng B, Jiang X, Li L. 2016. Identification of key taxa that favor intestinal colonization of Clostridium difficile in an adult Chinese population. Microbes Infect 18:30–38. doi: 10.1016/j.micinf.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 59.Zhang L, Dong D, Jiang C, Li Z, Wang X, Peng Y. 2015. Insight into alteration of gut microbiota in Clostridium difficile infection and asymptomatic C. difficile colonization. Anaerobe 34:1–7. doi: 10.1016/j.anaerobe.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 60.Dethlefsen L, Relman DA. 2011. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A 108:4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lewis BB, Buffie CG, Carter RA, Leiner I, Toussaint NC, Miller LC, Gobourne A, Ling L, Pamer EG. 2015. Loss of microbiota-mediated colonization resistance to Clostridium difficile infection with oral vancomycin compared with metronidazole. J Infect Dis 212:1656–1665. doi: 10.1093/infdis/jiv256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hung YP, Lee JC, Lin HJ, Liu HC, Wu YH, Tsai PJ, Ko WC. 2015. Clinical impact of Clostridium difficile colonization. J Microbiol Immunol Infect 48:241–248. doi: 10.1016/j.jmii.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 63.Lees EA, Miyajima F, Pirmohamed M, Carrol ED. 2016. The role of Clostridium difficile in the paediatric and neonatal gut—a narrative review. Eur J Clin Microbiol Infect Dis 35:1047–1057. doi: 10.1007/s10096-016-2639-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Keller JM, Surawicz CM. 2014. Clostridium difficile infection in the elderly. Clin Geriatr Med 30:79–93. doi: 10.1016/j.cger.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 65.Loo VG, Bourgault A-M, Poirier L, Lamothe F, Michaud S, Turgeon N, Toye B, Beaudoin A, Frost EH, Gilca R, Brassard P, Dendukuri N, Béliveau C, Oughton M, Brukner I, Dascal A. 2011. Host and pathogen factors for Clostridium difficile infection and colonization. N Engl J Med 365:1693–1703. doi: 10.1056/NEJMoa1012413. [DOI] [PubMed] [Google Scholar]
- 66.McDonald LC, Owings M, Jernigan DB. 2006. Clostridium difficile infection in patients discharged from US short-stay hospitals, 1996-2003. Emerg Infect Dis 12:409–415. doi: 10.3201/eid1203.051064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hopkins MJ, Macfarlane GT. 2002. Changes in predominant bacterial populations in human faeces with age and with Clostridium difficile infection. J Med Microbiol 51:448–454. doi: 10.1099/0022-1317-51-5-448. [DOI] [PubMed] [Google Scholar]
- 68.Imhann F, Bonder MJ, Vich Vila A, Fu J, Mujagic Z, Vork L, Tigchelaar EF, Jankipersadsing SA, Cenit MC, Harmsen HJM, Dijkstra G, Franke L, Xavier RJ, Jonkers D, Wijmenga C, Weersma RK, Zhernakova A. 2016. Proton pump inhibitors affect the gut microbiome. Gut 65:740–748. doi: 10.1136/gutjnl-2015-310376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bacon AEI, Fekety R. 1994. Immunoglobulin G directed against toxins A and B of Clostridium difficile in the general population and patients with antibiotic associated diarrhea. Diagn Microbiol Infect Dis 18:205–209. doi: 10.1016/0732-8893(94)90021-3. [DOI] [PubMed] [Google Scholar]
- 70.Viscidi R, Laughon BE, Yolken R, Bo-Linn P, Moench T, Ryder RW, Bartlett JG. 1983. Serum antibody response to toxins A and B of Clostridium difficile. J Infect Dis 148:93–100. doi: 10.1093/infdis/148.1.93. [DOI] [PubMed] [Google Scholar]
- 71.Kelly C, Pothoulakis C, Orellana J, LaMont J. 1992. Human colonic aspirates containing immunoglobulin A antibody to Clostridium difficile toxin A inhibit toxin A-receptor binding. Gastroenterology 102:35–40. doi: 10.1016/0016-5085(92)91781-X. [DOI] [PubMed] [Google Scholar]
- 72.Leung DY, Kelly CP, Boguniewicz M, Pothoulakis C, LaMont JT, Flores A. 1991. Treatment with intravenously administered gamma globulin of chronic relapsing colitis induced by Clostridium difficile toxin. J Pediatr 118:633–637. doi: 10.1016/S0022-3476(05)83393-1. [DOI] [PubMed] [Google Scholar]
- 73.Warny M, Denie C, Delmee M, Lefebvre C. 1995. Gamma globulin administration in relapsing Clostridium difficile-induced pseudomembranous colitis with a defective antibody response to toxin A. Acta Clin Belg 50:36–39. doi: 10.1080/17843286.1995.11718419. [DOI] [PubMed] [Google Scholar]
- 74.Kyne L, Warny M, Qamar A, Kelly CP. 2001. Association between antibody response to toxin A and protection against recurrent Clostridium difficile diarrhoea. Lancet 357:189–193. doi: 10.1016/S0140-6736(00)03592-3. [DOI] [PubMed] [Google Scholar]
- 75.Johnson S, Gerding DN, Janoff EN. 1992. Systemic and mucosal antibody responses to toxin A in patients infected with Clostridium difficile. J Infect Dis 166:1287–1294. doi: 10.1093/infdis/166.6.1287. [DOI] [PubMed] [Google Scholar]
- 76.Warny M, Vaerman JP, Avesani V, Delmée M. 1994. Human antibody response to Clostridium difficile toxin A in relation to clinical course of infection. Infect Immun 62:384–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Salcedo J, Keates S, Pothoulakis C, Warny M, Castagliuolo I, LaMont JT, Kelly CP. 1997. Intravenous immunoglobulin therapy for severe Clostridium difficile colitis. Gut 41:366–370. doi: 10.1136/gut.41.3.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hassett J, Meyers S, McFarland L, Mulligan ME. 1995. Recurrent Clostridium difficile infection in a patient with selective IgG1 deficiency treated with intravenous immune globulin and Saccharomyces boulardii. Clin Infect Dis 20(Suppl 2):S266–S268. doi: 10.1093/clinids/20.Supplement_2.S266. [DOI] [PubMed] [Google Scholar]
- 79.Wilcox M. 2004. Descriptive study of intravenous immunoglobulin for the treatment of recurrent Clostridium difficile diarrhoea. J Antimicrob Chemother 53:882–884. doi: 10.1093/jac/dkh176. [DOI] [PubMed] [Google Scholar]
- 80.Shah N, Shaaban H, Spira R, Slim J, Boghossian J. 2014. Intravenous immunoglobulin in the treatment of severe Clostridium difficile colitis. J Glob Infect Dis 6:82–85. doi: 10.4103/0974-777X.132053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jehangir A, Bennett K, Fareedy SB, Rettew A, Shaikh B, Qureshi A, Jehangir Q, Alweis R. 2015. Recurrent C. difficile in a patient with IgG deficiency. Case Rep Gastrointest Med 2015:356293. doi: 10.1155/2015/356293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McPherson S, Rees CJ, Ellis R, Soo S, Panter SJ. 2006. Intravenous immunoglobulin for the treatment of severe, refractory, and recurrent Clostridium difficile diarrhea. Dis Colon Rectum 49:640–645. doi: 10.1007/s10350-006-0511-8. [DOI] [PubMed] [Google Scholar]
- 83.Juang P, Skledar SJ, Zgheib NK, Paterson DL, Vergis EN, Shannon WD, Ansani NT, Branch RA. 2007. Clinical outcomes of intravenous immune globulin in severe Clostridium difficile-associated diarrhea. Am J Infect Control 35:131–137. doi: 10.1016/j.ajic.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 84.Bagdasarian N, Rao K, Malani PN. 2015. Diagnosis and treatment of Clostridium difficile in adults. JAMA 313:398–408. doi: 10.1001/jama.2014.17103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Louie TJ, Miller MA, Mullane KM, Weiss K, Lentnek A, Golan Y, Gorbach S, Sears P, Shue YK. 2011. Fidaxomicin versus vancomycin for Clostridium difficile infection. N Engl J Med 364:422–431. doi: 10.1056/NEJMoa0910812. [DOI] [PubMed] [Google Scholar]
- 86.Ananthakrishnan AN. 2011. Clostridium difficile infection: epidemiology, risk factors and management. Nat Rev Gastroenterol Hepatol 8:17–26. doi: 10.1038/nrgastro.2010.190. [DOI] [PubMed] [Google Scholar]
- 87.Kelly CP. 2012. Can we identify patients at high risk of recurrent Clostridium difficile infection? Clin Microbiol Infect 18(Suppl 6):21–27. doi: 10.1111/1469-0691.12046. [DOI] [PubMed] [Google Scholar]
- 88.Deakin LJ, Clare S, Fagan RP, Dawson LF, Pickard DJ, West MR, Wren BW, Fairweather NF, Dougan G, Lawley TD. 2012. The Clostridium difficile spo0A gene is a persistence and transmission factor. Infect Immun 80:2704–2711. doi: 10.1128/IAI.00147-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Louie TJ, Byrne B, Emery J, Ward L, Krulicki W, Nguyen D, Wu K, Cannon K. 2015. Differences of the fecal microflora with Clostridium difficile therapies. Clin Infect Dis 60:S91–S97. doi: 10.1093/cid/civ252. [DOI] [PubMed] [Google Scholar]
- 90.Louie TJ, Cannon K, Byrne B, Emery J, Ward L, Eyben M, Krulicki W. 2012. Fidaxomicin preserves the intestinal microbiome during and after treatment of Clostridium difficile infection (CDI) and reduces both toxin reexpression and recurrence of CDI. Clin Infect Dis 55:S132–S142. doi: 10.1093/cid/cis338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rea MC, Dobson A, O'Sullivan O, Crispie F, Fouhy F, Cotter PD, Shanahan F, Kiely B, Hill C, Ross RP. 2011. Effect of broad- and narrow-spectrum antimicrobials on Clostridium difficile and microbial diversity in a model of the distal colon. Proc Natl Acad Sci U S A 108:4639–4644. doi: 10.1073/pnas.1001224107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JF, Tijssen JG, Speelman P, Dijkgraaf MG, Keller JJ. 2013. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med 368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 93.Seekatz AM, Aas J, Gessert CE, Rubin TA, Saman DM, Bakken JS, Young VB. 2014. Recovery of the gut microbiome following fecal microbiota transplantation. mBio 5:e00893-14. doi: 10.1128/mBio.00893-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Khoruts A, Dicksved J, Jansson JK, Sadowsky MJ. 2010. Changes in the composition of the human fecal microbiome after bacteriotherapy for recurrent Clostridium difficile-associated diarrhea. J Clin Gastroenterol 44:354–360. doi: 10.1097/MCG.0b013e3181c87e02. [DOI] [PubMed] [Google Scholar]
- 95.Weingarden A, González A, Vázquez-Baeza Y, Weiss S, Humphry G, Berg-Lyons D, Knights D, Unno T, Bobr A, Kang J, Khoruts A, Knight R, Sadowsky MJ. 2015. Dynamic changes in short- and long-term bacterial composition following fecal microbiota transplantation for recurrent Clostridium difficile infection. Microbiome 3:10. doi: 10.1186/s40168-015-0070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lawley TD, Clare S, Walker AW, Stares MD, Connor TR, Raisen C, Goulding D, Rad R, Schreiber F, Brandt C, Deakin LJ, Pickard DJ, Duncan SH, Flint HJ, Clark TG, Parkhill J, Dougan G. 2012. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog 8:e1002995. doi: 10.1371/journal.ppat.1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Petrof EO, Gloor GB, Vanner SJ, Weese SJ, Carter D, Daigneault MC, Brown EM, Schroeter K, Allen-Vercoe E. 2013. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: “RePOOPulating” the gut. Microbiome 1:3. doi: 10.1186/2049-2618-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Emanuelsson F, Claesson BEB, Ljungström L, Tvede M, Ung K-A. 2014. Faecal microbiota transplantation and bacteriotherapy for recurrent Clostridium difficile infection: a retrospective evaluation of 31 patients. Scand J Infect Dis 46:89–97. doi: 10.3109/00365548.2013.858181. [DOI] [PubMed] [Google Scholar]
- 99.Tvede M, Rask-Madsen J. 1989. Bacteriotherapy for chronic relapsing Clostridium difficile diarrhoea in six patients. Lancet 333:1156–1160. doi: 10.1016/S0140-6736(89)92749-9. [DOI] [PubMed] [Google Scholar]
- 100.Khanna S, Pardi DS, Kelly CR, Kraft CS, Dhere T, Henn MR, Lombardo M, Vulic M, Ohsumi T, Winkler J, Pindar C, McGovern BH, Pomerantz RJ, Aunins JG, Cook DN, Hohmann EL. 2016. A novel microbiome therapeutic increases gut microbial diversity and prevents recurrent Clostridium difficile infection. J Infect Dis 214:173–181. doi: 10.1093/infdis/jiv766. [DOI] [PubMed] [Google Scholar]
- 101.Seres Therapeutics Inc. 2016. Seres Therapeutics announces interim results from SER-109 phase 2 ECOSPOR study in multiply recurrent Clostridium difficile infection. Press release. Seres Therapeutics Inc; Cambridge, MA: http://ir.serestherapeutics.com/phoenix.zhtml?c=254006&p=irol-newsArticle&ID=2190006. [Google Scholar]
- 102.Li M, Liang P, Li Z, Wang Y, Zhang G, Gao H, Wen S, Tang L. 2015. Fecal microbiota transplantation and bacterial consortium transplantation have comparable effects on the re-establishment of mucosal barrier function in mice with intestinal dysbiosis. Front Microbiol 6:692. doi: 10.3389/fmicb.2015.00692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lau CSM, Chamberlain RS. 2016. Probiotics are effective at preventing Clostridium difficile-associated diarrhea: a systematic review and meta-analysis. Int J Gen Med 9:27–37. doi: 10.2147/IJGM.S98280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Johnson S, Maziade PJ, McFarland LV, Trick W, Donskey C, Currie B, Low DE, Goldstein EJC. 2012. Is primary prevention of Clostridium difficile infection possible with specific probiotics? Int J Infect Dis 16:e786–e792. doi: 10.1016/j.ijid.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 105.Dendukuri N, Costa V, McGregor M, Brophy JM. 2005. Probiotic therapy for the prevention and treatment of Clostridium difficile-associated diarrhea: a systematic review. CMAJ 173:167–170. doi: 10.1503/cmaj.050350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Allen SJ, Wareham K, Wang D, Bradley C, Hutchings H, Harris W, Dhar A, Brown H, Foden A, Gravenor MB, Mack D. 2013. Lactobacilli and bifidobacteria in the prevention of antibiotic-associated diarrhoea and Clostridium difficile diarrhoea in older inpatients (PLACIDE): a randomised, double-blind, placebo-controlled, multicentre trial. Lancet 382:1249–1257. doi: 10.1016/S0140-6736(13)61218-0. [DOI] [PubMed] [Google Scholar]
- 107.Dendukuri N, Brophy J. 2007. Inappropriate use of meta-analysis to estimate efficacy of probiotics. Am J Gastroenterol 102:201. doi: 10.1111/j.1572-0241.2007.00916.x. [DOI] [PubMed] [Google Scholar]
- 108.Lewis S. 2007. Response to “Meta-analysis of probiotics for the prevention of antibiotic-associated diarrhea and the treatment of Clostridium difficile disease”. Am J Gastroenterol 2006;101:812–822. Am J Gastroenterol 102:201–202. doi: 10.1111/j.1572-0241.2007.00916.x. [DOI] [PubMed] [Google Scholar]
- 109.Pillai A, Nelson R. 2008. Probiotics for treatment of Clostridium difficile-associated colitis in adults. Cochrane Database Syst Rev 1:CD004611. doi: 10.1002/14651858.CD004611.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mcfarland LV, Surawicz CM, Greenberg RN, Fekety R, Elmer GW, Moyer KA, Melcher SA, Bowen KE, Cox JL, Noorani Z, Harrington G, Rubin M, Greenwald D. 1994. A randomized placebo-controlled trial of Saccharomyces boulardii in combination with standard antibiotics for Clostridium difficile disease. JAMA 271:1913–1918. doi: 10.1001/jama.1994.03510480037031. [DOI] [PubMed] [Google Scholar]
- 111.Reeves AE, Theriot CM, Bergin IL, Huffnagle GB, Schloss PD, Young VB. 2011. The interplay between microbiome dynamics and pathogen dynamics in a murine model of Clostridium difficile infection. Gut Microbes 2:145–158. doi: 10.4161/gmic.2.3.16333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Schubert AM, Sinani H, Schloss PD. 2015. Antibiotic-induced alterations of the murine gut microbiota and subsequent effects on colonization resistance against Clostridium difficile. mBio 6:e00974-15. doi: 10.1128/mBio.00974-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, Littmann E, van den Brink MRM, Jenq RR, Taur Y, Sander C, Cross J, Toussaint NC, Xavier JB, Pamer EG. 2015. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Manges AR, Labbe A, Loo VG, Atherton JK, Behr MA, Masson L, Tellis PA, Brousseau R. 2010. Comparative metagenomic study of alterations to the intestinal microbiota and risk of nosocomial Clostridum difficile-associated disease. J Infect Dis 202:1877–1884. doi: 10.1086/657319. [DOI] [PubMed] [Google Scholar]
- 115.Schubert AM, Rogers MA, Ring C, Mogle J, Petrosino JP, Young VB, Aronoff DM, Schloss PD. 2014. Microbiome data distinguish patients with Clostridium difficile infection and non-C. difficile-associated diarrhea from healthy controls. mBio 5:e01021-14. doi: 10.1128/mBio.01021-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shahinas D, Silverman M, Sittler T, Chiu C, Kim P, Allen-Vercoe E, Weese S, Wong A, Pillai DR. 2012. Toward an understanding of changes in diversity associated with fecal microbiome transplantation based on 16S rRNA gene deep sequencing. mBio 3:e00338-12. doi: 10.1128/mBio.00338-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Song Y, Garg S, Girotra M, Maddox C, von Rosenvinge EC, Dutta A, Dutta S, Fricke WF. 2013. Microbiota dynamics in patients treated with fecal microbiota transplantation for recurrent Clostridium difficile infection. PLoS One 8:e81330. doi: 10.1371/journal.pone.0081330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Antharam VC, Li EC, Ishmael A, Sharma A, Mai V, Rand KH, Wang GP. 2013. Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J Clin Microbiol 51:2884–2892. doi: 10.1128/JCM.00845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dutta SK, Girotra M, Garg S, Dutta A, von Rosenvinge EC, Maddox C, Song Y, Bartlett JG, Vinayek R, Fricke WF. 2014. Efficacy of combined jejunal and colonic fecal microbiota transplantation for recurrent Clostridium difficile infection. Clin Gastroenterol Hepatol 12:1572–1576. doi: 10.1016/j.cgh.2013.12.032. [DOI] [PubMed] [Google Scholar]
- 120.Stecher B. 2013. Finding a sugary foothold: how antibiotics pave the way for enteric pathogens. Cell Host Microbe 14:225–227. doi: 10.1016/j.chom.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 121.Theriot CM, Koenigsknecht MJ, Carlson PEJ, Hatton GE, Nelson AM, Li B, Huffnagle GB, Li J, Young VB. 2014. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun 5:3114. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Antunes LCM, Han J, Ferreira RBR, Lolic P, Borchers CH, Finlay BB. 2011. Effect of antibiotic treatment on the intestinal metabolome. Antimicrob Agents Chemother 55:1494–1503. doi: 10.1128/AAC.01664-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ng KM, Ferreyra J, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, Sonnenburg JL. 2013. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ferreyra JA, Wu KJ, Hryckowian AJ, Bouley DM, Weimer BC, Sonnenburg JL. 2014. Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe 16:770–777. doi: 10.1016/j.chom.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shim JK, Johnson S, Samore MH, Bliss DZ, Gerding DN. 1998. Primary symptomless colonisation by Clostridium difficile and decreased risk of subsequent diarrhoea. Lancet 351:633–636. doi: 10.1016/S0140-6736(97)08062-8. [DOI] [PubMed] [Google Scholar]
- 126.Nagaro KJ, Phillips ST, Cheknis AK, Sambol SP, Zukowski WE, Johnson S, Gerding DN. 2013. Nontoxigenic Clostridium difficile protects hamsters against challenge with historic and epidemic strains of toxigenic BI/NAP1/027 C. difficile. Antimicrob Agents Chemother 57:5266–5270. doi: 10.1128/AAC.00580-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sambol SP, Merrigan MM, Tang JK, Johnson S, Gerding DN. 2002. Colonization for the prevention of Clostridium difficile disease in hamsters. J Infect Dis 186:1781–1789. doi: 10.1086/345676. [DOI] [PubMed] [Google Scholar]
- 128.Villano SA, Seiberling M, Tatarowicz W, Monnot-Chase E, Gerding DN. 2012. Evaluation of an oral suspension of VP20621, spores of nontoxigenic Clostridium difficile strain M3, in healthy subjects. Antimicrob Agents Chemother 56:5224–5229. doi: 10.1128/AAC.00913-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gerding DN, Meyer T, Lee C, Cohen SH, Murthy UK, Poirier A, Van Schooneveld TC, Pardi DS, Ramos A, Barron MA, Chen H, Villano S. 2015. Administration of spores of nontoxigenic Clostridium difficile strain M3 for prevention of recurrent C difficile infection: a randomized clinical trial. JAMA 313:1719–1727. doi: 10.1001/jama.2015.3725. [DOI] [PubMed] [Google Scholar]
- 130.Brouwer MSM, Roberts AP, Hussain H, Williams RJ, Allan E, Mullany P. 2013. Horizontal gene transfer converts non-toxigenic Clostridium difficile strains into toxin producers. Nat Commun 4:2601. doi: 10.1038/ncomms3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ridlon JM. 2005. Bile salt biotransformations by human intestinal bacteria. J Lipid Res 47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 132.Wilson KH. 1983. Efficiency of various bile salt preparations for stimulation of Clostridium difficile spore germination. J Clin Microbiol 18:1017–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Giel JL, Sorg JA, Sonenshein AL, Zhu J. 2010. Metabolism of bile salts in mice influences spore germination in Clostridium difficile. PLoS One 5:e8740. doi: 10.1371/journal.pone.0008740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jump RLP, Polinkovsky A, Hurless K, Sitzlar B, Eckart K, Tomas M, Deshpande A, Nerandzic MM, Donskey CJ. 2014. Metabolomics analysis identifies intestinal microbiota-derived biomarkers of colonization resistance in clindamycin-treated mice. PLoS One 9:e101267. doi: 10.1371/journal.pone.0101267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Allegretti JR, Kearney S, Li N, Bogart E, Bullock K, Gerber GK, Bry L, Clish CB, Alm E, Korzenik JR. 2016. Recurrent Clostridium difficile infection associates with distinct bile acid and microbiome profiles. Aliment Pharmacol Ther 43:1142–1153. doi: 10.1111/apt.13616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Weingarden AR, Chen C, Bobr A, Yao D, Lu Y, Nelson VM, Sadowsky MJ, Khoruts A. 2014. Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. Am J Physiol Gastrointest Liver Physiol 306:G310–G319. doi: 10.1152/ajpgi.00282.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Schoster A, Kokotovic B, Permin A, Pedersen PD, Bello FD, Guardabassi L. 2013. In vitro inhibition of Clostridium difficile and Clostridium perfringens by commercial probiotic strains. Anaerobe 20:36–41. doi: 10.1016/j.anaerobe.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 138.Tejero-Sariñena S, Barlow J, Costabile A, Gibson GR, Rowland I. 2012. In vitro evaluation of the antimicrobial activity of a range of probiotics against pathogens: evidence for the effects of organic acids. Anaerobe 18:530–538. doi: 10.1016/j.anaerobe.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 139.Trejo FM, Minnaard J, Perez PF, De Antoni GL. 2006. Inhibition of Clostridium difficile growth and adhesion to enterocytes by Bifidobacterium supernatants. Anaerobe 12:186–193. doi: 10.1016/j.anaerobe.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 140.Geeraerts S, Ducatelle R, Haesebrouck F, Van Immerseel F. 2015. Bacillus amyloliquefaciens as prophylactic treatment for Clostridium difficile-associated disease in a mouse model. J Gastroenterol Hepatol 30:1275–1280. doi: 10.1111/jgh.12957. [DOI] [PubMed] [Google Scholar]
- 141.Rea MC, Clayton E, O'Connor PM, Shanahan F, Kiely B, Ross RP, Hill C. 2007. Antimicrobial activity of lacticin 3147 against clinical Clostridium difficile strains. J Med Microbiol 56:940–946. doi: 10.1099/jmm.0.47085-0. [DOI] [PubMed] [Google Scholar]
- 142.Ryan MP, Rea MC, Hill C, Ross RP. 1996. An application in cheddar cheese manufacture for a strain of Lactococcus lactis producing a novel broad-spectrum bacteriocin, lacticin 3147. Appl Environ Microbiol 62:612–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Rea MC, Sit CS, Clayton E, O'Connor PM, Whittal RM, Zheng J, Vederas JC, Ross RP, Hill C. 2010. Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc Natl Acad Sci U S A 107:9352–9357. doi: 10.1073/pnas.0913554107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Rea MC, Alemayehu D, Casey PG, O'Connor PM, Lawlor PG, Walsh M, Shanahan F, Kiely B, Ross RP, Hill C. 2014. Bioavailability of the anti-clostridial bacteriocin thuricin CD in gastrointestinal tract. Microbiology 160:439–445. doi: 10.1099/mic.0.068767-0. [DOI] [PubMed] [Google Scholar]
- 145.Castagliuolo I, LaMont JT, Nikulasson ST, Pothoulakis C. 1996. Saccharomyces boulardii protease inhibits Clostridium difficile toxin A effects in the rat ileum. Infect Immun 64:5225–5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Castagliuolo I, Riegler MF, Valenick L, LaMont JT, Pothoulakis C. 1999. Saccharomyces boulardii protease inhibits the effects of Clostridium difficile toxins A and B in human colonic mucosa. Infect Immun 67:302–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Corthier G, Lucas F, Jouvert S, Castex F. 1992. Effect of oral Saccharomyces boulardii treatment on the activity of Clostridium difficile toxins in mouse digestive tract. Toxicon 30:1583–1589. doi: 10.1016/0041-0101(92)90030-9. [DOI] [PubMed] [Google Scholar]
- 148.Qamar A, Aboudola S, Warny M, Michetti P, Pothoulakis C, LaMont JT, Kelly CP. 2001. Saccharomyces boulardii stimulates intestinal immunoglobulin A immune response to Clostridium difficile toxin A in mice. Infect Immun 69:2762–2765. doi: 10.1128/IAI.69.4.2762-2765.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Babcock GJ, Broering TJ, Hernandez HJ, Mandell RB, Donahue K, Boatright N, Stack AM, Lowy I, Graziano R, Molrine D, Ambrosino DM, Thomas WD. 2006. Human monoclonal antibodies directed against toxins A and B prevent Clostridium difficile-induced mortality in hamsters. Infect Immun 74:6339–6347. doi: 10.1128/IAI.00982-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Anosova NG, Cole LE, Li L, Zhang J, Brown AM, Mundle S, Zhang J, Ray S, Ma F, Garrone P, Bertraminelli N, Kleanthous H, Anderson SF. 2015. A combination of three fully human toxin A- and toxin B-specific monoclonal antibodies protects against challenge with highly virulent epidemic strains of Clostridium difficile in the hamster model. Clin Vaccine Immunol 22:711–725. doi: 10.1128/CVI.00763-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lowy I, Molrine DC, Leav BA, Blair BM, Baxter R, Gerding DN, Nichol G, Thomas WDJ, Leney M, Sloan S, Hay CA, Ambrosino DM. 2010. Treatment with monoclonal antibodies against Clostridium difficile toxins. N Engl J Med 362:197–205. doi: 10.1056/NEJMoa0907635. [DOI] [PubMed] [Google Scholar]
- 152.Buts JP, Bernasconi P, Vaerman JP, Dive C. 1990. Stimulation of secretory IgA and secretory component of immunoglobulins in small intestine of rats treated with Saccharomyces boulardii. Dig Dis Sci 35:251–256. doi: 10.1007/BF01536771. [DOI] [PubMed] [Google Scholar]
- 153.Martins FS, Silva AA, Vieira AT, Barbosa FH, Arantes RM, Teixeira MM, Nicoli JR. 2009. Comparative study of Bifidobacterium animalis, Escherichia coli, Lactobacillus casei and Saccharomyces boulardii probiotic properties. Arch Microbiol 191:623–630. doi: 10.1007/s00203-009-0491-x. [DOI] [PubMed] [Google Scholar]
- 154.Rodrigues ACP, Cara DC, Fretez SHGG, Cunha FQ, Vieira EC, Nicoli JR, Vieira LQ. 2000. Saccharomyces boulardii stimulates sIgA production and the phagocytic system of gnotobiotic mice. J Appl Microbiol 89:404–414. doi: 10.1046/j.1365-2672.2000.01128.x. [DOI] [PubMed] [Google Scholar]
- 155.Generoso SV, Viana ML, Santos RG, Arantes RME, Martins FS, Nicoli JR, Machado JAN, Correia MITD, Cardoso VN. 2011. Protection against increased intestinal permeability and bacterial translocation induced by intestinal obstruction in mice treated with viable and heat-killed Saccharomyces boulardii. Eur J Nutr 50:261–269. doi: 10.1007/s00394-010-0134-7. [DOI] [PubMed] [Google Scholar]
- 156.Roller M, Rechkemmer G, Watzl B. 2004. Prebiotic inulin enriched with oligofructose in combination with the probiotics Lactobacillus rhamnosus and Bifidobacterium lactis modulates intestinal immune functions in rats. J Nutr 134:153–156. [DOI] [PubMed] [Google Scholar]
- 157.Sharma R, Kapila R, Dass G, Kapila S. 2014. Improvement in Th1/Th2 immune homeostasis, antioxidative status and resistance to pathogenic E. coli on consumption of probiotic Lactobacillus rhamnosus fermented milk in aging mice. Age 36:9686. doi: 10.1007/s11357-014-9686-4. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 158.Galdeano CM, Perdigo G. 2006. The probiotic bacterium Lactobacillus casei induces activation of the gut mucosal immune system through innate immunity. Clin Vaccine Immunol 13:219–226. doi: 10.1128/CVI.13.2.219-226.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kim YS, Ho SB. 2010. Intestinal goblet cells and mucins in health and disease: recent insights and progress. Curr Gastroenterol Rep 12:319–330. doi: 10.1007/s11894-010-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Johansson M, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. 2008. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A 105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Johansson M, Larsson JMH, Hansson GC. 2011. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci U S A 108:4659–4665. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Engevik M, Yacyshyn MB, Engevik K, Wang J, Darien B, Hassett DJ, Yacyshyn BR, Worrell RT. 2015. Human Clostridium difficile infection: altered mucus production and composition. Am J Physiol Gastrointest Liver Physiol 308:G510–G524. doi: 10.1152/ajpgi.00091.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Eveillard M, Fourel V, Barc MC, Kernéis S, Coconnier MH, Karjalainen T, Bourlioux P, Servin A. 1993. Identification and characterization of adhesive factors of Clostridium difficile involved in adhesion to human colonic enterocyte-like Caco-2 and mucus-secreting HT29 cells in culture. Mol Microbiol 7:371–381. doi: 10.1111/j.1365-2958.1993.tb01129.x. [DOI] [PubMed] [Google Scholar]
- 164.Calabi E, Calabi F, Phillips AD, Fairweather NF, Neil F. 2002. Binding of Clostridium difficile surface layer proteins to gastrointestinal tissues. Infect Immun 70:5770–5778. doi: 10.1128/IAI.70.10.5770-5778.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Krivan HC, Clark GF, Smith DF, Wilkins TD. 1986. Cell surface binding site for Clostridium difficile enterotoxin: evidence for a glycoconjugate containing the sequence Gala1-3Galb1-4GlcNAc. Infect Immun 53:573–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Smith J, Cooke DL, Hyde S, Borriello SP, Long RG. 1997. Clostridium difficile toxin A binding to human intestinal epithelial cells. J Med Microbiol 46:953–958. doi: 10.1099/00222615-46-11-953. [DOI] [PubMed] [Google Scholar]
- 167.Branka JE, Vallette G, Jarry A, Bou-Hana C, Lemarre P, Van PN, Laboisse CL. 1997. Early functional effects of Clostridium difficile toxin A on human colonocytes. Gastroenterology 112:1887–1894. doi: 10.1053/gast.1997.v112.pm9178681. [DOI] [PubMed] [Google Scholar]
- 168.Mattar AF, Teitelbaum DH, Drongowski RA, Yongyi F, Harmon CM, Coran AG. 2002. Probiotics up-regulate MUC-2 mucin gene expression in a Caco-2 cell-culture model. Pediatr Surg Int 18:586–590. doi: 10.1007/s00383-002-0855-7. [DOI] [PubMed] [Google Scholar]
- 169.Mack D, Ahrne S, Hyde L, Wei S, Hollingsworth M. 2003. Extracellular MUC3 mucin secretion follows adherence of Lactobacillus strains to intestinal epithelial cells in vitro. Gut 52:827–833. doi: 10.1136/gut.52.6.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wang L, Cao H, Liu L, Wang B, Walker WA, Acra SA, Yan F. 2014. Activation of epidermal growth factor receptor mediates mucin production stimulated by p40, a Lactobacillus rhamnosus GG-derived protein. J Biol Chem 289:20234–20244. doi: 10.1074/jbc.M114.553800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhang W, Zhu Y, Yang J, Yang G, Zhou D, Wang J. 2015. A selected Lactobacillus rhamnosus strain oromotes EGFR-independent Akt activation in an enterotoxigenic Escherichia coli K88-infected IPEC-J2 cell model. PLoS One 10:e0125717. doi: 10.1371/journal.pone.0125717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Otte J-M, Podolsky DK. 2004. Functional modulation of enterocytes by gram-positive and gram-negative microorganisms. Am J Physiol Gastrointest Liver Physiol 286:G613–G626. doi: 10.1152/ajpgi.00341.2003. [DOI] [PubMed] [Google Scholar]
- 173.Caballero-Franco C, Keller K, Simone C, Chadee K. 2007. The VSL#3 probiotic formula induces mucin gene expression and secretion in colonic epithelial cells. Am J Physiol Gastrointest Liver Physiol 292:G315–G322. [DOI] [PubMed] [Google Scholar]
- 174.Zanello G, Berri M, Dupont J, Sizaret PY, D'Inca R, Salmon H, Meurens F. 2011. Saccharomyces cerevisiae modulates immune gene expressions and inhibits ETEC-mediated ERK1/2 and p38 signaling pathways in intestinal epithelial cells. PLoS One 6:e18573. doi: 10.1371/journal.pone.0018573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Schneider S-M, Girard-Pipau F, Filippi J, Hebuterne X, Moyse D, Hinojosa G-C, Pompei A, Rampal P. 2005. Effects of Saccharomyces boulardii on fecal short-chain fatty acids and microflora in patients on long-term total enteral nutrition. World J Gastroenterol 11:6165–6169. doi: 10.3748/wjg.v11.i39.6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Gaudier E, Rival M, Buisine MP, Robineau I, Hoebler C. 2009. Butyrate enemas upregulate Muc genes expression but decrease adherent mucus thickness in mice colon. Physiol Res 58:111–119. [DOI] [PubMed] [Google Scholar]
- 177.Ouwehand A, Isolauri E, Kirjavainen PV, Salminen SJ. 1999. Adhesion of four Bifidobacterium strains to human intestinal mucus from subjects in different age groups. FEMS Microbiol Lett 172:61–64. doi: 10.1111/j.1574-6968.1999.tb13450.x. [DOI] [PubMed] [Google Scholar]
- 178.Ouwehand A, Isolauri E, Kirjavainen PV, Tölkko S, Salminen SJ. 2000. The mucus binding of Bifidobacterium lactis Bb12 is enhanced in the presence of Lactobacillus GG and Lact. delbrueckii subsp. bulgaricus. Lett Appl Microbiol 30:10–13. doi: 10.1046/j.1472-765x.2000.00590.x. [DOI] [PubMed] [Google Scholar]
- 179.Farquhar MG, Palade GE. 1963. Junctional complexes in various epithelia. J Cell Biol 17:375–412. doi: 10.1083/jcb.17.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Fanning AS, Jameson BJ, Lynne A, Anderson JM, Jesaitis LA, Melvin J. 1998. The tight junction protein ZO-1 establishes a link between the transmembrane protein occludin and the actin cytoskeleton. J Biol Chem 273:29745–29753. doi: 10.1074/jbc.273.45.29745. [DOI] [PubMed] [Google Scholar]
- 181.Madara JL. 1987. Intestinal absorptive cell tight junctions are linked to cytoskeleton. Am J Physiol 253:C171–C175. [DOI] [PubMed] [Google Scholar]
- 182.Perez-Moreno M, Fuchs E. 2006. Catenins: keeping cells from getting their signals crossed. Dev Cell 11:601–612. doi: 10.1016/j.devcel.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Suzuki T, Yoshida S, Hara H. 2008. Physiological concentrations of short-chain fatty acids immediately suppress colonic epithelial permeability. Br J Nutr 100:297–305. doi: 10.1017/S0007114508888733. [DOI] [PubMed] [Google Scholar]
- 184.Peng L, Li Z-R, Green RS, Holzman IR, Lin J. 2009. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. J Nutr 139:1619–1625. doi: 10.3945/jn.109.104638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Sultana R, McBain AJ, O'Neill CA. 2013. Strain-dependent augmentation of tight-junction barrier function in human primary epidermal keratinocytes by Lactobacillus and Bifidobacterium lysates. Appl Environ Microbiol 79:4887–4894. doi: 10.1128/AEM.00982-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Czerucka D, Dahan S, Mograbi B, Rossi B, Rampal P. 2000. Saccharomyces boulardii preserves the barrier function and modulates the signal transduction pathway induced in enteropathogenic Escherichia coli-infected T84 cells. Infect Immun 68:5998–6004. doi: 10.1128/IAI.68.10.5998-6004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Garcia Vilela E, De Lourdes De Abreu Ferrari M, Da Gama Torres HO, Guerra Pinto A, Carneiro Aguirre AC, Paiva Martins F, Andrade Goulart EM, Da Cunha AS. 2008. Influence of Saccharomyces boulardii on the intestinal permeability of patients with Crohn's disease in remission. Scand J Gastroenterol 43:842–848. doi: 10.1080/00365520801943354. [DOI] [PubMed] [Google Scholar]
- 188.Li M, Zhu L, Xie A, Yuan J. 2015. Oral administration of Saccharomyces boulardii ameliorates carbon tetrachloride-induced liver fibrosis in rats via reducing intestinal permeability and modulating gut microbial composition. Inflammation 38:170–179. doi: 10.1007/s10753-014-0019-7. [DOI] [PubMed] [Google Scholar]
- 189.Justino PFC, Melo LFM, Nogueira AF, Costa JVG, Silva LMN, Santos CM, Mendes WO, Costa MR, Franco AX, Lima AA, Ribeiro RA, Souza MHLP, Soares PMG. 2014. Treatment with Saccharomyces boulardii reduces the inflammation and dysfunction of the gastrointestinal tract in 5-fluorouracil-induced intestinal mucositis in mice. Br J Nutr 111:1611–1621. doi: 10.1017/S0007114513004248. [DOI] [PubMed] [Google Scholar]
- 190.Mumy KL, Chen X, Kelly CP, McCormick BA. 2008. Saccharomyces boulardii interferes with Shigella pathogenesis by postinvasion signaling events. Am J Physiol Gastrointest Liver Physiol 294:G599–G609. doi: 10.1152/ajpgi.00391.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Martz SL, McDonald JAK, Sun J, Zhang YG, Gloor GB, Noordhof C, He SM, Gerbaba TK, Blennerhassett M, Hurlbut DJ, Allen-Vercoe E, Claud EC, Petrof EO. 2015. Administration of defined microbiota is protective in a murine Salmonella infection model. Sci Rep 5:16094. doi: 10.1038/srep16094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Hsieh C-Y, Osaka T, Moriyama E, Date Y, Kikuchi J, Tsuneda S. 2015. Strengthening of the intestinal epithelial tight junction by Bifidobacterium bifidum. Physiol Rep 3:e12327. doi: 10.14814/phy2.12327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Wang B, Huang Q, Zhang W, Li N, Li J. 2011. Lactobacillus plantarum prevents bacterial translocation in rats following ischemia and reperfusion injury. Dig Dis Sci 56:3187–3194. doi: 10.1007/s10620-011-1747-2. [DOI] [PubMed] [Google Scholar]
- 194.Madan R, Petri WA. 2012. Immune responses to Clostridium difficile infection. Trends Mol Med 18:658–666. doi: 10.1016/j.molmed.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Riquelme AJ, Calvo MA, Guzmán AM, Depix MS, García P, Pérez C, Arrese M, Labarca JA. 2003. Saccharomyces cerevisiae fungemia after Saccharomyces boulardii treatment in immunocompromised patients. J Clin Gastroenterol 36:41–43. doi: 10.1097/00004836-200301000-00013. [DOI] [PubMed] [Google Scholar]
- 196.Vahabnezhad E, Mochon A, Wozniak L, Ziring D. 2013. Lactobacillus bacteremia associated with probiotic use in a pediatric patient with ulcerative colitis. J Clin Gastroenterol 47:437–439. doi: 10.1097/MCG.0b013e318279abf0. [DOI] [PubMed] [Google Scholar]
- 197.Loftus E. 2004. Clinical epidemiology of inflammatory bowel disease: incidence, prevalence, and environmental influences. Gastroenterology 126:1504–1517. doi: 10.1053/j.gastro.2004.01.063. [DOI] [PubMed] [Google Scholar]
- 198.Kappelman MD, Rifas-Shiman SL, Kleinman K, Ollendorf D, Bousvaros A, Grand RJ, Finkelstein JA. 2007. The prevalence and geographic distribution of Crohn's disease and ulcerative colitis in the United States. Clin Gastroenterol Hepatol 5:1424–1429. doi: 10.1016/j.cgh.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 199.Anderson CA, Boucher G, Lees CW, Franke A, D'Amato M, Taylor KD, Lee JC, Goyette P, Imielinski M, Latiano A, Lagacé C, Scott R, Amininejad L, Bumpstead S, Baidoo L, Baldassano RN, Barclay M, Bayless TM, Brand S, Büning C, Colombel J-F, Denson LA, De Vos M, Dubinsky M, Edwards C, Ellinghaus D, Fehrmann RSN, Floyd JAB, Florin T, Franchimont D, Franke L, Georges M, Glas J, Glazer NL, Guthery SL, Haritunians T, Hayward NK, Hugot J-P, Jobin G, Laukens D, Lawrance I, Lémann M, Levine A, Libioulle C, Louis E, McGovern DP, Milla M, Montgomery GW, Morley KI, Mowat C, et al. 2011. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet 43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Tontini GE, Vecchi M, Pastorelli L, Neurath MF, Neumann H. 2015. Differential diagnosis in inflammatory bowel disease colitis: state of the art and future perspectives. World J Gastroenterol 21:21–46. doi: 10.3748/wjg.v21.i1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Loddenkemper C. 2009. Diagnostic standards in the pathology of inflammatory bowel disease. Dig Dis 27:576–583. doi: 10.1159/000233301. [DOI] [PubMed] [Google Scholar]
- 202.Choi C-HR, Ignjatovic-Wilson A, Askari A, Lee GH, Warusavitarne J, Moorghen M, Thomas-Gibson S, Saunders BP, Rutter MD, Graham TA, Hart AL. 2015. Low-grade dysplasia in ulcerative colitis: risk factors for developing high-grade dysplasia or colorectal cancer. Am J Gastroenterol 110:1461–1471. doi: 10.1038/ajg.2015.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Thomas T, Abrams KA, Robinson RJ, Mayberry JF. 2007. Meta-analysis: cancer risk of low-grade dysplasia in chronic ulcerative colitis. Aliment Pharmacol Ther 25:657–668. doi: 10.1111/j.1365-2036.2007.03241.x. [DOI] [PubMed] [Google Scholar]
- 204.Bernstein CN, Blanchard JF, Rawsthorne P, Yu N. 2001. The prevalence of extraintestinal diseases in inflammatory bowel disease: a population-based study. Am J Gastroenterol 96:1116–1122. doi: 10.1111/j.1572-0241.2001.03756.x. [DOI] [PubMed] [Google Scholar]
- 205.Reif S, Klein I, Lubin F, Farbstein M, Hallak A, Gilat T. 1997. Pre-illness dietary factors in inflammatory bowel disease. Gut 40:754–760. doi: 10.1136/gut.40.6.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Danese S, Sans M, Fiocchi C. 2004. Inflammatory bowel disease: the role of environmental factors. Autoimmun Rev 3:394–400. doi: 10.1016/j.autrev.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 207.Andersson RE, Olaison G, Tysk C, Ekbom A. 2001. Appendectomy and protection against ulcerative colitis. N Engl J Med 344:808–814. doi: 10.1056/NEJM200103153441104. [DOI] [PubMed] [Google Scholar]
- 208.Harries AD, Baird A, Rhodes J. 1982. Non-smoking: a feature of ulcerative colitis. Br Med J 284:706. doi: 10.1136/bmj.284.6317.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Sartor RB. 2006. Mechanisms of disease: pathogenesis of Crohn's disease and ulcerative colitis. Nat Clin Pract Gastroenterol Hepatol 3:390–407. doi: 10.1038/ncpgasthep0528. [DOI] [PubMed] [Google Scholar]
- 210.de Souza HSP, Fiocchi C. 2016. Immunopathogenesis of IBD: current state of the art. Nat Rev Gastroenterol Hepatol 13:13–27. doi: 10.1038/nrgastro.2015.186. [DOI] [PubMed] [Google Scholar]
- 211.Michielan A, D'Incà R. 2015. Intestinal permeability in inflammatory bowel disease: pathogenesis, clinical evaluation, and therapy of leaky gut. Mediators Inflamm 2015:628157. doi: 10.1155/2015/628157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.UK IBD Genetics Consortium, Wellcome Trust Case Control Consortium 2. 2009. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat Genet 41:1330–1334. doi: 10.1038/ng.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Ho G-T, Nimmo ER, Tenesa A, Fennell J, Drummond H, Mowat C, Arnott ID, Satsangi J. 2005. Allelic variations of the multidrug resistance gene determine susceptibility and disease behavior in ulcerative colitis. Gastroenterology 128:288–296. doi: 10.1053/j.gastro.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 214.Schwab M, Schaeffeler E, Marx C, Fromm MF, Kaskas B, Metzler J, Stange E, Herfarth H, Schoelmerich J, Gregor M, Walker S, Cascorbi I, Roots I, Brinkmann U, Zanger UM, Eichelbaum M. 2003. Association between the C3435T MDR1 gene polymorphism and susceptibility for ulcerative colitis. Gastroenterology 124:26–33. doi: 10.1053/gast.2003.50010. [DOI] [PubMed] [Google Scholar]
- 215.Van der Sluis M, De Koning BAE, De Bruijn ACJM, Velcich A, Meijerink JPP, Van Goudoever JB, Buller HA, Dekker J, Van Seuningen I, Renes IB, Einerhand AWC. 2006. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology 131:117–129. doi: 10.1053/j.gastro.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 216.Lennon G, Balfe Á, Early H, Devane LA, Lavelle A, Winter DC, Coffey JC, O'Connell PR. 2014. Influences of the colonic microbiome on the mucous gel layer in ulcerative colitis. Gut Microbes 5:277–285. doi: 10.4161/gmic.28793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Png CW, Linden SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, McGuckin MA, Florin TH. 2010. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol 105:2420–2428. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- 218.Wlodarska M, Thaiss CA, Nowarski R, Henao-Mejia J, Zhang J, Brown EM, Frankel G, Levy M, Katz MN, Philbrick WM, Elinav E, Finlay BB. 2014. NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell 156:1045–1059. doi: 10.1016/j.cell.2014.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, Flavell RA. 2011. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Alipour M, Zaidi D, Valcheva R, Jovel J, Martínez I, Sergi C, Walter J, Mason AL, Wong GK-S, Dieleman LA, Carroll MW, Huynh HQ, Wine E. 2016. Mucosal barrier depletion and loss of bacterial diversity are primary abnormalities in paediatric ulcerative colitis. J Crohns Colitis 10:462–471. doi: 10.1093/ecco-jcc/jjv223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Johansson MEV, Gustafsson JK, Holmén-Larsson J, Jabbar KS, Xia L, Xu H, Ghishan FK, Carvalho FA, Gewirtz AT, Sjövall H, Hansson GC. 2014. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut 63:281–291. doi: 10.1136/gutjnl-2012-303207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Fava F, Danese S. 2011. Intestinal microbiota in inflammatory bowel disease: friend of foe? World J Gastroenterol 17:557–566. doi: 10.3748/wjg.v17.i5.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Ling Z, Liu X, Jia X, Cheng Y, Luo Y, Yuan L, Wang Y, Zhao C, Guo S, Li L, Xu X, Xiang C. 2014. Impacts of infection with different toxigenic Clostridium difficile strains on faecal microbiota in children. Sci Rep 4:7485. doi: 10.1038/srep07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Sartor RB, Mazmanian SK. 2012. Intestinal microbes in inflammatory bowel diseases. Am J Gastroenterol Suppl 1:15–21. doi: 10.1038/ajgsup.2012.4. [DOI] [Google Scholar]
- 225.Giaffer M, Holdsworth C, Duerden B. 1992. Virulence properties of Escherichia coli strains isolated from patients with inflammatory bowel disease. Gut 33:646–650. doi: 10.1136/gut.33.5.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Bullock NR, Booth JCL, Gibson GR. 2004. Comparative composition of bacteria in the human intestinal microflora during remission and active ulcerative colitis. Curr Issues Intest Microbiol 5:59–64. [PubMed] [Google Scholar]
- 227.Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio C-W, Santacruz N, Peterson DA, Stappenbeck TS, Hsieh C-S. 2011. Peripheral education of the immune system by colonic commensal microbiota. Nature 478:250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Round JL, Mazmanian SK. 2009. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 9:313–324. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Sellon RK, Tonkonogy S, Schultz M, Dieleman LA, Grenther W, Balish E, Rennick DM, Sartor RB. 1998. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun 66:5224–5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Dianda L, Hanby A, Wright N, Sebesteny A, Hayday A, Owen MJ. 1997. T cell receptor-alpha beta-deficient mice fail to develop colitis in the absence of a microbial environment. Am J Pathol 150:91–97. [PMC free article] [PubMed] [Google Scholar]
- 231.Hart AL, Al-Hassi HO, Rigby RJ, Bell SJ, Emmanuel A, Knight SC, Kamm MA, Stagg AJ. 2005. Characteristics of intestinal dendritic cells in inflammatory bowel diseases. Gastroenterology 129:50–65. doi: 10.1053/j.gastro.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 232.Selby WS, Janossy G, Bofill M, Jewell DP. 1984. Intestinal lymphocyte subpopulations in inflammatory bowel disease: an analysis by immunohistological and cell isolation techniques. Gut 25:32–40. doi: 10.1136/gut.25.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Zimmerman NP, Vongsa RA, Wendt MK, Dwinell MB. 2008. Chemokines and chemokine receptors in mucosal homeostasis at the intestinal epithelial barrier in inflammatory bowel disease. Inflamm Bowel Dis 14:1000–1011. doi: 10.1002/ibd.20480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Singh UP, Singh NP, Murphy EA, Price RL, Fayad R, Nagarkatti M, Nagarkatti PS. 2016. Chemokine and cytokine levels in inflammatory bowel disease patients. Cytokine 77:44–49. doi: 10.1016/j.cyto.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Wurbel M, McIntire MG, Dwyer P, Fiebiger E. 2011. CCL25/CCR9 interactions regulate large intestinal inflammation in a murine model of acute colitis. PLoS One 6:e16442. doi: 10.1371/journal.pone.0016442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Roda G, Marocchi M, Sartini A, Roda E. 2011. Cytokine networks in ulcerative colitis. Ulcers 2011:391787. doi: 10.1155/2011/391787. [DOI] [Google Scholar]
- 237.Sands BE, Kaplan GG. 2007. The role of TNF-alpha in ulcerative colitis. J Clin Pharmacol 47:930–941. doi: 10.1177/0091270007301623. [DOI] [PubMed] [Google Scholar]
- 238.Jarnerot G, Hertervig E, Friis-Liby I, Blomquist L, Karlen P, Granno C, Vilien M, Strom M, Danielsson A, Verbaan H, Hellstrom PM, Magnuson A, Curman B. 2005. Infliximab as rescue therapy in severe to moderately severe ulcerative colitis: a randomized, placebo-controlled study. Gastroenterology 128:1805–1811. doi: 10.1053/j.gastro.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 239.Fuss IJ, Heller F, Boirivant M, Leon F, Yoshida M, Fichtner-Feigl S, Yang Z, Exley M, Kitani A, Blumberg RS, Mannon P, Strober W. 2004. Nonclassical CD1d-restricted NK T cells that produce IL-13 characterize an atypical Th2 response in ulcerative colitis. J Clin Invest 113:1490–1497. doi: 10.1172/JCI19836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Wang P, Lu Y, Wen Y, Yu D, Ge L, Dong W, Xiang L, Shao J. 2013. IL-16 induces intestinal inflammation via PepT1 upregulation in a pufferfish model: new insights into the molecular mechanism of inflammatory bowel disease. J Immunol 191:1413–1427. doi: 10.4049/jimmunol.1202598. [DOI] [PubMed] [Google Scholar]
- 241.Seegert D, Rosenstiel P, Pfahler H, Pfefferkorn P, Nikolaus S, Schreiber S. 2001. Increased expression of IL-16 in inflammatory bowel disease. Gut 48:326–332. doi: 10.1136/gut.48.3.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Di Sabatino A, Biancheri P, Rovedatti L, MacDonald TT, Corazza GR. 2012. New pathogenic paradigms in inflammatory bowel disease. Inflamm Bowel Dis 18:368–371. doi: 10.1002/ibd.21735. [DOI] [PubMed] [Google Scholar]
- 243.Owaga E, Hsieh R-H, Mugendi B, Masuku S, Shih C-K, Chang J-S. 2015. Th17 cells as potential probiotic therapeutic targets in inflammatory bowel diseases. Int J Mol Sci 16:20841–20858. doi: 10.3390/ijms160920841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Geremia A, Biancheri P, Allan P, Corazza GR, Di Sabatino A. 2014. Innate and adaptive immunity in inflammatory bowel disease. Autoimmun Rev 13:3–10. doi: 10.1016/j.autrev.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 245.Heller F, Florian P, Bojarski C, Richter J, Christ M, Hillenbrand B, Mankertz J, Gitter AH, Bürgel N, Fromm M, Zeitz M, Fuss I, Strober W, Schulzke JD. 2005. Interleukin-13 is the key effector Th2 cytokine in ulcerative colitis that affects epithelial tight junctions, apoptosis, and cell restitution. Gastroenterology 129:550–564. doi: 10.1053/j.gastro.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 246.Zenewicz LA, Antov A, Flavell RA. 2009. CD4 T-cell differentiation and inflammatory bowel disease. Trends Mol Med 15:199–207. doi: 10.1016/j.molmed.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 247.Ito R, Kita M, Shin-Ya M, Kishida T, Urano A, Takada R, Sakagami J, Imanishi J, Iwakura Y, Okanoue T, Yoshikawa T, Kataoka K, Mazda O. 2008. Involvement of IL-17A in the pathogenesis of DSS-induced colitis in mice. Biochem Biophys Res Commun 377:12–16. doi: 10.1016/j.bbrc.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 248.Kobayashi T, Okamoto S, Hisamatsu T, Kamada N, Chinen H, Saito R, Kitazume MT, Nakazawa A, Sugita A, Koganei K, Isobe K, Hibi T. 2008. IL23 differentially regulates the Th1/Th17 balance in ulcerative colitis and Crohn's disease. Gut 57:1682–1689. doi: 10.1136/gut.2007.135053. [DOI] [PubMed] [Google Scholar]
- 249.Sugihara T, Kobori A, Imaeda H, Tsujikawa T, Amagase K, Takeuchi K, Fujiyama Y, Andoh A. 2010. The increased mucosal mRNA expressions of complement C3 and interleukin-17 in inflammatory bowel disease. Clin Exp Immunol 160:386–393. doi: 10.1111/j.1365-2249.2010.04093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Ogawa A, Andoh A, Araki Y, Bamba T, Fujiyama Y. 2004. Neutralization of interleukin-17 aggravates dextran sulfate sodium-induced colitis in mice. Clin Immunol 110:55–62. doi: 10.1016/j.clim.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 251.Zhang Z, Zheng M, Bindas J, Schwarzenberger P, Kolls JK. 2006. Critical role of IL-17 receptor signaling in acute TNBS-induced colitis. Inflamm Bowel Dis 12:382–388. doi: 10.1097/01.MIB.0000218764.06959.91. [DOI] [PubMed] [Google Scholar]
- 252.Fitzpatrick LR, Deml L, Hofmann C, Small JS, Groeppel M, Hamm S, Lemstra S, Leban J, Ammendola A. 2010. 4SC-101, a novel immunosuppressive drug, inhibits IL-17 and attenuates colitis in two murine models of inflammatory bowel disease. Inflamm Bowel Dis 16:1763–1777. doi: 10.1002/ibd.21264. [DOI] [PubMed] [Google Scholar]
- 253.Fitzpatrick LR, Small J, Doblhofer R, Ammendola A. 2012. Vidofludimus inhibits IL-17 and improves hapten-induced colitis in young rats by a unique dual mode of action. J Crohns Colitis 6:S15–S16. doi: 10.1016/S1873-9946(12)60033-6. [DOI] [PubMed] [Google Scholar]
- 254.Geboes K, Riddell R, Ost A, Jensfelt B, Persson T, Löfberg R. 2000. A reproducible grading scale for histological assessment of inflammation in ulcerative colitis. Gut 47:404–409. doi: 10.1136/gut.47.3.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Verspaget HW, Peña A, Weterman IT, Lamers CB. 1988. Diminished neutrophil function in Crohn's disease and ulcerative colitis identified by decreased oxidative metabolism and low superoxide dismutase content. Gut 29:223–228. doi: 10.1136/gut.29.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Hanai H, Takeuchi K, Iida T, Kashiwagi N, Saniabadi AR, Matsushita I, Sato Y, Kasuga N, Nakamura T. 2004. Relationship between fecal calprotectin, intestinal inflammation, and peripheral blood neutrophils in patients with active ulcerative colitis. Dig Dis Sci 49:1438–1443. doi: 10.1023/B:DDAS.0000042243.47279.87. [DOI] [PubMed] [Google Scholar]
- 257.Fournier BM, Parkos CA. 2012. The role of neutrophils during intestinal inflammation. Mucosal Immunol 5:354–366. doi: 10.1038/mi.2012.24. [DOI] [PubMed] [Google Scholar]
- 258.Ferretti S, Bonneau O, Dubois GR, Jones CE, Trifilieff A. 2003. IL-17, produced by lymphocytes and neutrophils, is necessary for lipopolysaccharide-induced airway neutrophilia: IL-15 as a possible trigger. J Immunol 170:2106–2112. doi: 10.4049/jimmunol.170.4.2106. [DOI] [PubMed] [Google Scholar]
- 259.Kolaczkowska E, Kubes P. 2013. Neutrophil recruitment and function in health and inflammation. Nat Rev Immunol 13:159–175. doi: 10.1038/nri3399. [DOI] [PubMed] [Google Scholar]
- 260.Zhang R, Ito S, Nishio N, Cheng Z, Suzuki H, Isobe K. 2011. Up-regulation of Gr1+ CD11b+ population in spleen of dextran sulfate sodium administered mice works to repair colitis. Inflamm Allergy Drug Targets 10:39–46. doi: 10.2174/187152811794352114. [DOI] [PubMed] [Google Scholar]
- 261.Kühl AA, Kakirman H, Janotta M, Dreher S, Cremer P, Pawlowski NN, Loddenkemper C, Heimesaat MM, Grollich K, Zeitz M, Farkas S, Hoffmann JC. 2007. Aggravation of different types of experimental colitis by depletion or adhesion blockade of neutrophils. Gastroenterology 133:1882–1892. doi: 10.1053/j.gastro.2007.08.073. [DOI] [PubMed] [Google Scholar]
- 262.Natsui M, Kawasaki K, Takizawa H, Hayashi SI, Matsuda Y, Sugimura K, Seki K, Narisawa R, Sendo F, Asakura H. 1997. Selective depletion of neutrophils by a monoclonal antibody, RP-3, suppresses dextran sulphate sodium-induced colitis in rats. J Gastroenterol Hepatol 12:801–808. doi: 10.1111/j.1440-1746.1997.tb00375.x. [DOI] [PubMed] [Google Scholar]
- 263.Lee A, Whyte MK, Haslett C. 1993. Inhibition of apoptosis and prolongation of neutrophil functional longevity by inflammatory mediators. J Leukoc Biol 54:283–288. [PubMed] [Google Scholar]
- 264.Cox G. 1996. IL-10 enhances resolution of pulmonary inflammation in vivo by promoting apoptosis of neutrophils. Am J Physiol 271:L566–L571. [DOI] [PubMed] [Google Scholar]
- 265.Salamone G, Giordano M, Trevani A, Gamberale R, Vermeulen M, Schettinni J, Geffner J. 2001. Promotion of neutrophil apoptosis by TNF-alpha. J Immunol 166:3476–3483. doi: 10.4049/jimmunol.166.5.3476. [DOI] [PubMed] [Google Scholar]
- 266.Kucharzik T, Walsh SV, Chen J, Parkos CA, Nusrat A. 2001. Neutrophil transmigration in inflammatory bowel disease is associated with differential expression of epithelial intercellular junction proteins. Am J Pathol 159:2001–2009. doi: 10.1016/S0002-9440(10)63051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Adeyemi E, Hodgson H. 1992. Faecal elastase reflects disease activity in active ulcerative colitis. Scand J Gastroenterol 27:139–142. doi: 10.3109/00365529209165434. [DOI] [PubMed] [Google Scholar]
- 268.Nielsen OH, Munck LK. 2007. Drug insight: aminosalicylates for the treatment of IBD. Nat Clin Pract Gastroenterol Hepatol 4:160–170. doi: 10.1038/ncpgasthep0696. [DOI] [PubMed] [Google Scholar]
- 269.Kornbluth A, Sachar DB, Practice Parameters Committee of the American College of Gastroenterology. 2010. Ulcerative colitis practice guidelines in adults: American College of Gastroenterology Practice Parameters Committee. Am J Gastroenterol 105:501–523. doi: 10.1038/ajg.2009.727. [DOI] [PubMed] [Google Scholar]
- 270.Fraser AG, Orchard TR, Jewell DP. 2002. The efficacy of azathioprine for the treatment of inflammatory bowel disease: a 30 year review. Gut 50:485–489. doi: 10.1136/gut.50.4.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.George J, Present DH, Pou R, Bodian C, Rubin PH. 1996. The long-term outcome of ulcerative colitis treated with 6-mercaptopurine. Am J Gastroenterol 91:1711–1714. [PubMed] [Google Scholar]
- 272.Rutgeerts P, Sandborn WJ, Feagan BG, Reinisch W, Olson A, Johanns J, Travers S, Rachmilewitz D, Hanauer SB, Lichtenstein GR, de Villiers WJS, Present D, Sands BE, Colombel JF. 2005. Infliximab for induction and maintenance therapy for ulcerative colitis. N Engl J Med 353:2462–2476. doi: 10.1056/NEJMoa050516. [DOI] [PubMed] [Google Scholar]
- 273.Reinisch W, Sandborn WJ, Hommes DW, D'Haens G, Hanauer S, Schreiber S, Panaccione R, Fedorak RN, Tighe MB, Huang B, Kampman W, Lazar A, Thakkar R. 2011. Adalimumab for induction of clinical remission in moderately to severely active ulcerative colitis: results of a randomised controlled trial. Gut 60:780–787. doi: 10.1136/gut.2010.221127. [DOI] [PubMed] [Google Scholar]
- 274.Sandborn WJ, Van Assche G, Reinisch W, Colombel J, D'Haens G, Wolf DC, Kron M, Tighe MB, Lazar A, Thakkar RB. 2012. Adalimumab induces and maintains clinical remission in patients with moderate-to-severe ulcerative colitis. Gastroenterology 142:257–265. doi: 10.1053/j.gastro.2011.10.032. [DOI] [PubMed] [Google Scholar]
- 275.Campbell S, Travis S, Jewell D. 2005. Ciclosporin use in acute ulcerative colitis: a long-term experience. Eur J Gastroenterol Hepatol 17:79–84. doi: 10.1097/00042737-200501000-00016. [DOI] [PubMed] [Google Scholar]
- 276.Timmer A, McDonald J, Tsoulis D, Macdonald J. 2012. Azathioprine and 6-mercaptopurine for maintenance of remission in ulcerative colitis. Cochrane Database Syst Rev 9:CD000478. doi: 10.1002/14651858.CD000478.pub3. [DOI] [PubMed] [Google Scholar]
- 277.Cima R. 2010. Timing and indications for colectomy in chronic ulcerative colitis: surgical consideration. Dig Dis 28:501–507. doi: 10.1159/000320409. [DOI] [PubMed] [Google Scholar]
- 278.Navaneethan U, Shen B. 2010. Secondary pouchitis: those with identifiable etiopathogenetic or triggering factors. Am J Gastroenterol 105:51–64. doi: 10.1038/ajg.2009.530. [DOI] [PubMed] [Google Scholar]
- 279.Wu H, Shen B. 2010. Pouchitis and pouch dysfunction. Med Clin North Am 94:75–92. doi: 10.1016/j.mcna.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 280.Angelberger S, Reinisch W, Makristathis A, Lichtenberger C, Dejaco C, Papay P, Novacek G, Trauner M, Loy A, Berry D. 2013. Temporal bacterial community dynamics vary among ulcerative colitis patients after fecal microbiota transplantation. Am J Gastroenterol 108:1620–1630. doi: 10.1038/ajg.2013.257. [DOI] [PubMed] [Google Scholar]
- 281.Rubin DT. 2013. Curbing our enthusiasm for fecal transplantation in ulcerative colitis. Am J Gastroenterol 108:1631–1633. doi: 10.1038/ajg.2013.279. [DOI] [PubMed] [Google Scholar]
- 282.Gupta S, Allen-Vercoe E, Petrof EO. 2016. Fecal microbiota transplantation: in perspective. Therap Adv Gastroenterol 9:229–239. doi: 10.1177/1756283X15607414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Kao D, Hotte N, Gillevet P, Madsen K. 2014. Fecal microbiota transplantation inducing remission in Crohn's colitis and the associated changes in fecal microbial profile. J Clin Gastroenterol 48:625–628. doi: 10.1097/MCG.0000000000000131. [DOI] [PubMed] [Google Scholar]
- 284.Kump PK, Gröchenig H-P, Lackner S, Trajanoski S, Reicht G, Hoffmann KM, Deutschmann A, Wenzl HH, Petritsch W, Krejs GJ, Gorkiewicz G, Högenauer C. 2013. Alteration of intestinal dysbiosis by fecal microbiota transplantation does not induce remission in patients with chronic active ulcerative colitis. Inflamm Bowel Dis 19:2155–2165. doi: 10.1097/MIB.0b013e31829ea325. [DOI] [PubMed] [Google Scholar]
- 285.Rossen NG, Fuentes S, Van Der Spek MJ, Tijssen JG, Hartman JHA, Duflou A, Lowenberg M, Van Den Brink GR, Mathus-Vliegen EMH, De Vos WM, Zoetendal EG, D'Haens GR, Ponsioen CY. 2015. Findings from a randomized controlled trial of fecal transplantation for patients with ulcerative colitis. Gastroenterology 149:110–118. doi: 10.1053/j.gastro.2015.03.045. [DOI] [PubMed] [Google Scholar]
- 286.Colman RJ, Rubin DT. 2014. Fecal microbiota transplantation as therapy for inflammatory bowel disease: a systematic review and meta-analysis. J Crohns Colitis 8:1569–1581. doi: 10.1016/j.crohns.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Tsuda Y, Yoshimatsu Y, Aoki H, Nakamura K, Irie M, Fukuda K, Hosoe N, Takada N, Shirai K, Suzuki Y. 2007. Clinical effectiveness of probiotics therapy (BIO-THREE) in patients with ulcerative colitis refractory to conventional therapy. Scand J Gastroenterol 42:1306–1311. doi: 10.1080/00365520701396091. [DOI] [PubMed] [Google Scholar]
- 288.Ishikawa H, Akedo I, Umesaki Y, Tanaka R, Imaoka A, Otani T. 2003. Randomized controlled trial of the effect of bifidobacteria-fermented milk on ulcerative colitis. J Am Coll Nutr 22:56–63. doi: 10.1080/07315724.2003.10719276. [DOI] [PubMed] [Google Scholar]
- 289.Eloe-Fadrosh EA, Brady A, Crabtree J, Drabek EF, Ma B, Mahurkar A, Ravel J, Haverkamp M, Fiorino AM, Botelho C, Andreyeva I, Hibberd PL, Fraser CM. 2015. Functional dynamics of the gut microbiome in elderly people during probiotic consumption. mBio 6:e00231-15. doi: 10.1128/mBio.00231-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.De Preter V, Raemen H, Cloetens L, Houben E, Rutgeerts P, Verbeke K. 2008. Effect of dietary intervention with different pre- and probiotics on intestinal bacterial enzyme activities. Eur J Clin Nutr 62:225–231. doi: 10.1038/sj.ejcn.1602706. [DOI] [PubMed] [Google Scholar]
- 291.Barc MC, Charrin-Sarnel C, Rochet V, Bourlioux F, Sandré C, Boureau H, Doré J, Collignon A. 2008. Molecular analysis of the digestive microbiota in a gnotobiotic mouse model during antibiotic treatment: influence of Saccharomyces boulardii. Anaerobe 14:229–233. doi: 10.1016/j.anaerobe.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 292.Bajaj JS, Heuman DM, Hylemon PB, Sanyal AJ, Puri P, Sterling RK, Luketic V, Stravitz RT, Siddiqui MS, Fuchs M, Thacker LR, Wade JB, Daita K, Sistrun S, White MB, Noble NA, Thorpe C, Kakiyama G, Pandak WM, Sikaroodi M, Gillevet PM. 2014. Randomised clinical trial: Lactobacillus GG modulates gut microbiome, metabolome and endotoxemia in patients with cirrhosis. Aliment Pharmacol Ther 39:1113–1125. doi: 10.1111/apt.12695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Bull-Otterson L, Feng W, Kirpich I, Wang Y, Qin X, Liu Y, Gobejishvili L, Joshi-Barve S, Ayvaz T, Petrosino J, Kong M, Barker D, McClain C, Barve S. 2013. Metagenomic analyses of alcohol induced pathogenic alterations in the intestinal microbiome and the effect of Lactobacillus rhamnosus GG treatment. PLoS One 8:e53028. doi: 10.1371/journal.pone.0053028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Everard A, Matamoros S, Geurts L, Delzenne NM, Cani PD. 2014. Saccharomyces boulardii administration changes gut microbiota and reduces hepatic steatosis, low-grade inflammation, and fat mass in obese and type 2 diabetic db/db mice. mBio 5:e01011-14. doi: 10.1128/mBio.01011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Al-Sadi R, Boivin M, Ma T. 2009. Mechanism of cytokine modulation of epithelial tight junction barrier. Front Biosci (Landmark Ed) 14:2765–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Lee JS, Tato CM, Joyce-Shaikh B, Gulan F, Cayatte C, Chen Y, Blumenschein WM, Judo M, Ayanoglu G, McClanahan TK, Li X, Cua DJ. 2015. Interleukin-23-independent IL-17 production regulates intestinal epithelial permeability. Immunity 43:727–738. doi: 10.1016/j.immuni.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Roselli M, Finamore A, Nuccitelli S, Carnevali P, Brigidi P, Vitali B, Nobili F, Rami R, Garaguso I, Mengheri E. 2009. Prevention of TNBS-induced colitis by different Lactobacillus and Bifidobacterium strains is associated with an expansion of gamma delta T and regulatory T cells of intestinal intraepithelial lymphocytes. Inflamm Bowel Dis 15:1526–1536. doi: 10.1002/ibd.20961. [DOI] [PubMed] [Google Scholar]
- 298.Mazzon E, Puzzolo D, Caputi AP, Cuzzocrea S. 2002. Role of IL-10 in hepatocyte tight junction alteration in mouse model of experimental colitis. Mol Med 8:353–366. [PMC free article] [PubMed] [Google Scholar]
- 299.Keubler LM, Buettner M, Häger C, Bleich A. 2015. A multihit model: colitis lessons from the interleukin-10-deficient mouse. Inflamm Bowel Dis 21:1967–1975. doi: 10.1097/MIB.0000000000000468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. 1993. Interleukin-10-deficient mice develop chronic enterocolitis. Cell 75:263–274. doi: 10.1016/0092-8674(93)80068-P. [DOI] [PubMed] [Google Scholar]
- 301.Latvala S, Miettinen M, Kekkonen RA, Korpela R, Julkunen I. 2011. Lactobacillus rhamnosus GG and Streptococcus thermophilus induce suppressor of cytokine signalling 3 (SOCS3) gene expression directly and indirectly via interleukin-10 in human primary macrophages. Clin Exp Immunol 165:94–103. doi: 10.1111/j.1365-2249.2011.04408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Hart AL, Lammers K, Brigidi P, Vitali B, Rizzello F, Gionchetti P, Campieri M, Kamm MA, Knight SC, Stagg AJ. 2004. Modulation of human dendritic cell phenotype and function by probiotic bacteria. Gut 53:1602–1609. doi: 10.1136/gut.2003.037325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Gad M, Ravn P, Soborg DA, Lund-Jensen K, Ouwehand AC, Jensen SS. 2011. Regulation of the IL-10/IL-12 axis in human dendritic cells with probiotic bacteria. FEMS Immunol Med Microbiol 63:93–107. doi: 10.1111/j.1574-695X.2011.00835.x. [DOI] [PubMed] [Google Scholar]
- 304.Tanabe S, Kinuta Y, Saito Y. 2008. Bifidobacterium infantis suppresses proinflammatory interleukin-17 production in murine splenocytes and dextran sodium sulfate-induced intestinal inflammation. Int J Mol Med 22:181–185. [PubMed] [Google Scholar]
- 305.Zoumpopoulou G, Foligne B, Christodoulou K, Grangetteb C, Pot B, Tsakalidou E. 2008. Lactobacillus fermentum ACA-DC 179 displays probiotic potential in vitro and protects against trinitrobenzene sulfonic acid (TNBS)-induced colitis and Salmonella infection in murine models. Int J Food Microbiol 121:18–26. doi: 10.1016/j.ijfoodmicro.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 306.Shida K, Nanno M, Nagata S. 2011. Flexible cytokine production by macrophages and T cells in response to probiotic bacteria: a possible mechanism by which probiotics exert multifunctional immune regulatory activities. Gut Microbes 2:109–114. doi: 10.4161/gmic.2.2.15661. [DOI] [PubMed] [Google Scholar]
- 307.Christensen HR, Frøkiaer H, Pestka JJ. 2002. Lactobacilli differentially modulate expression of cytokines and maturation surface markers in murine dendritic cells. J Immunol 168:171–178. doi: 10.4049/jimmunol.168.1.171. [DOI] [PubMed] [Google Scholar]
- 308.Zeuthen LH, Christensen HR. 2006. Lactic acid bacteria inducing a weak interleukin-12 and tumor necrosis factor alpha response in human dendritic cells inhibit strongly stimulating lactic acid bacteria but act synergistically with Gram-negative bacteria. Clin Vaccine Immunol 13:365–375. doi: 10.1128/CVI.13.3.365-375.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Smith IM, Christensen JE, Arneborg N, Jespersen L. 2014. Yeast modulation of human dendritic cell cytokine secretion: an in vitro study. PLoS One 9:e96595. doi: 10.1371/journal.pone.0096595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Thomas S, Przesdzing I, Metzke D, Schmitz J, Radbruch A, Baumgart DC. 2009. Saccharomyces boulardii inhibits lipopolysaccharide-induced activation of human dendritic cells and T cell proliferation. Clin Exp Immunol 156:78–87. doi: 10.1111/j.1365-2249.2009.03878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Lammers KM, Helwig U, Swennen E, Rizzello F, Venturi A, Caramelli E, Kamm MA, Brigidi P, Gionchetti P, Campieri M. 2002. Effect of probiotic strains on interleukin 8 production by HT29/19A cells. Am J Gastroenterol 97:1182–1186. doi: 10.1111/j.1572-0241.2002.05693.x. [DOI] [PubMed] [Google Scholar]
- 312.van der Aa Kühle A, Skovgaard K, Jespersen L. 2005. In vitro screening of probiotic properties of Saccharomyces cerevisiae var. boulardii and food-borne Saccharomyces cerevisiae strains. Int J Food Microbiol 101:29–39. doi: 10.1016/j.ijfoodmicro.2004.10.039. [DOI] [PubMed] [Google Scholar]
- 313.Adouard N, Foligné B, Dewulf J, Bouix M, Picque D, Bonnarme P. 2015. In vitro characterization of the digestive stress response and immunomodulatory properties of microorganisms isolated from smear-ripened cheese. Int J Food Microbiol 197:98–107. doi: 10.1016/j.ijfoodmicro.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 314.Sougioultzis S, Simeonidis S, Bhaskar KR, Chen X, Anton PM, Keates S, Pothoulakis C, Kelly CP. 2006. Saccharomyces boulardii produces a soluble anti-inflammatory factor that inhibits NF-kappaB-mediated IL-8 gene expression. Biochem Biophys Res Commun 343:69–76. doi: 10.1016/j.bbrc.2006.02.080. [DOI] [PubMed] [Google Scholar]
- 315.Schlee M, Harder J, Köten B, Stange EF, Wehkamp J, Fellermann K. 2008. Probiotic lactobacilli and VSL#3 induce enterocyte β-defensin 2. Clin Exp Immunol 151:528–535. doi: 10.1111/j.1365-2249.2007.03587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Imaoka A, Shima T, Kato K, Mizuno S, Uehara T, Matsumoto S, Setoyana H, Hara T, Umesaki Y. 2008. Anti-inflammatory activity of probiotic Bifidobacterium: enhancement of IL-10 production in peripheral blood mononuclear cells from ulcerative colitis patients and inhibition of IL-8 secretion in HT-29 cells. World J Gastroenterol 14:2511–2516. doi: 10.3748/wjg.14.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Ghadimi D, Helwig U, Schrezenmeir J, Heller KJ, de Vrese M. 2012. Epigenetic imprinting by commensal probiotics inhibits the IL-23/IL-17 axis in an in vitro model of the intestinal mucosal immune system. J Leukoc Biol 92:895–911. doi: 10.1189/jlb.0611286. [DOI] [PubMed] [Google Scholar]
- 318.Miyauchi E, Ogita T, Miyamoto J, Kawamoto S, Morita H, Ohno H, Suzuki T, Tanabe S. 2013. Bifidobacterium longum alleviates dextran sulfate sodium-induced colitis by suppressing IL-17A response: involvement of intestinal epithelial costimulatory molecules. PLoS One 8:e79735. doi: 10.1371/journal.pone.0079735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Chen L, Zou Y, Peng J, Lu F, Yin Y, Li F, Yang J. 2015. Lactobacillus acidophilus suppresses colitis-associated activation of the IL-23/Th17 axis. J Immunol Res 2015:909514. doi: 10.1155/2015/909514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Ogita T, Tanii Y, Morita H, Suzuki T, Tanabe S. 2011. Suppression of Th17 response by Streptococcus thermophilus ST28 through induction of IFN-γ. Int J Mol Med 28:817–822. doi: 10.3892/ijmm.2011.755. [DOI] [PubMed] [Google Scholar]
- 321.Mañé J, Lorén V, Pedrosa E, Ojanguren I, Xaus J, Cabré E, Domènech E, Gassull MA. 2009. Lactobacillus fermentum CECT 5716 prevents and reverts intestinal damage on TNBS-induced colitis in mice. Inflamm Bowel Dis 15:1155–1163. doi: 10.1002/ibd.20908. [DOI] [PubMed] [Google Scholar]
- 322.Yoon H, Yoon Y, Kim M, Chung M, Yum D. 2014. A probiotic preparation Duolac-Gold ameliorates dextran sulphate sodium-induced mouse colitis by downregulating the expression of IL-6. Toxicol Res 30:27–32. doi: 10.5487/TR.2014.30.1.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Lee HS, Han SY, Bae EA, Huh CS, Ahn YT, Lee JH, Kim DH. 2008. Lactic acid bacteria inhibit proinflammatory cytokine expression and bacterial glycosaminoglycan degradation activity in dextran sulfate sodium-induced colitic mice. Int Immunopharmacol 8:574–580. doi: 10.1016/j.intimp.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 324.Vong L, Lorentz RJ, Assa A, Glogauer M, Sherman PM. 2014. Probiotic Lactobacillus rhamnosus inhibits the formation of neutrophil extracellular traps. J Immunol 192:1870–1877. doi: 10.4049/jimmunol.1302286. [DOI] [PubMed] [Google Scholar]
- 325.Matthes H, Krummenerl T, Giensch M, Wolff C, Schulze J. 2010. Clinical trial: probiotic treatment of acute distal ulcerative colitis with rectally administered Escherichia coli Nissle 1917 (EcN). BMC Complement Altern Med 10:13. doi: 10.1186/1472-6882-10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Rembacken BJ, Snelling AM, Hawkey PM, Chalmers DM, Axon ATR. 1999. Non-pathogenic Escherichia coli versus mesalazine for the treatment of ulcerative colitis: a randomised trial. Lancet 354:635–639. doi: 10.1016/S0140-6736(98)06343-0. [DOI] [PubMed] [Google Scholar]
- 327.Low D, Nguyen DD, Mizoguchi E. 2013. Animal models of ulcerative colitis and their application in drug research. Drug Des Devel Ther 7:1341–1357. doi: 10.2147/DDDT.S40107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Hutton ML, Mackin KE, Chakravorty A, Lyras D. 2014. Small animal models for the study of Clostridium difficile disease pathogenesis. FEMS Microbiol Lett 352:140–149. doi: 10.1111/1574-6968.12367. [DOI] [PubMed] [Google Scholar]
- 329.Chilton C, Freeman J. 2015. Predictive values of models of Clostridium difficile infection. Infect Dis Clin North Am 29:163–177. doi: 10.1016/j.idc.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 330.Chen X, Katchar K, Goldsmith JD, Nanthakumar N, Cheknis A, Gerding DN, Kelly CP. 2008. A mouse model of Clostridium difficile-associated disease. Gastroenterology 135:1984–1992. doi: 10.1053/j.gastro.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 331.Beura LK, Hamilton SE, Bi K, Schenkel JM, Odumade OA, Casey KA, Thompson EA, Fraser KA, Rosato PC, Filali-Mouhim A, Sekaly RP, Jenkins MK, Vezys V, Haining WN, Jameson SC, Masopust D. 2016. Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature 532:512–516. doi: 10.1038/nature17655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Chiang JY. 2009. Bile acids: regulation of synthesis. J Lipid Res 50:1955–1966. doi: 10.1194/jlr.R900010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Li SS, Zhu A, Benes V, Costea PI, Hercog R, Hildebrand F, Huerta-Cepas J, Nieuwdorp M, Salojärvi J, Voigt AY, Zeller G, Sunagawa S, de Vos WM, Bork P. 2016. Durable coexistence of donor and recipient strains after fecal microbiota transplantation. Science 352:586–589. doi: 10.1126/science.aad8852. [DOI] [PubMed] [Google Scholar]
- 334.Hickson M, D'Souza AL, Muthu N, Rogers TR, Want S, Rajkumar C, Bulpitt CJ. 2007. Use of probiotic Lactobacillus preparation to prevent diarrhoea associated with antibiotics: randomised double blind placebo controlled trial. BMJ 335:80. doi: 10.1136/bmj.39231.599815.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Gao XW, Mubasher M, Fang CY, Reifer C, Miller LE. 2010. Dose-response efficacy of a proprietary probiotic formula of Lactobacillus acidophilus CL1285 and Lactobacillus casei LBC80R for antibiotic-associated diarrhea and Clostridium difficile-associated diarrhea prophylaxis in adult patients. Am J Gastroenterol 105:1636–1641. doi: 10.1038/ajg.2010.11. [DOI] [PubMed] [Google Scholar]
- 336.Thomas MR, Litin SC, Osmon DR, Corr AP, Weaver AL, Lohse CM. 2001. Lack of effect of Lactobacillus GG on antibiotic-associated diarrhea: a randomized, placebo-controlled trial. Mayo Clin Proc 76:883–889. doi: 10.4065/76.9.883. [DOI] [PubMed] [Google Scholar]
- 337.Plummer S, Weaver MA, Harris JC, Dee P, Huter J. 2004. Clostridium difficile pilot study: effects of probiotic supplementation on the incidence of C. difficile diarrhoea. Int Microbiol 7:59–62. [PubMed] [Google Scholar]
- 338.Lawrence SJ, Korzenik JR, Mundy LM. 2005. Probiotics for recurrent Clostridium difficile disease. J Med Microbiol 54:905–906. doi: 10.1099/jmm.0.46096-0. [DOI] [PubMed] [Google Scholar]
- 339.Stein G, Nanim R, Karniel E, Moskowitz I, Zeidman A. 2007. Probiotics as prophylactic agents against antibiotic-associated diarrhea in hospitalized patients. Harefuah 146:520–522, 575. [PubMed] [Google Scholar]
- 340.Beausoleil M, Fortier N, Guénette S, L'Ecuyer A, Savoie M, Franco M, Lachaîne J, Weiss K. 2007. Effect of a fermented milk combining Lactobacillus acidophilus CL1285 and Lactobacillus casei in the prevention of antibiotic-associated diarrhea: a randomized, double-blind, placebo-controlled trial. Can J Gastroenterol 21:732–736. doi: 10.1155/2007/720205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Safdar N, Barigala R, Said A, McKinley L. 2008. Feasibility and tolerability of probiotics for prevention of antibiotic-associated diarrhoea in hospitalized US military veterans. J Clin Pharm Ther 33:663–668. doi: 10.1111/j.1365-2710.2008.00980.x. [DOI] [PubMed] [Google Scholar]
- 342.Wullt M, Hagslätt M-LJ, Odenholt I. 2003. Lactobacillus plantarum 299v for the treatment of recurrent Clostridium difficile-associated diarrhoea: a double-blind, placebo-controlled trial. Scand J Infect Dis 35:365–367. doi: 10.1080/00365540310010985. [DOI] [PubMed] [Google Scholar]
- 343.Sampalis J, Psaradellis E, Rampakakis E. 2010. Efficacy of BIO K+ CL1285 in the reduction of antibiotic-associated diarrhea—a placebo controlled double-blind randomized, multi-center study. Arch Med Sci 6:56–64. doi: 10.5114/aoms.2010.13508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Surawicz CM, McFarland LV, Greenberg RN, Rubin M, Fekety R, Mulligan ME, Garcia RJ, Brandmarker S, Bowen K, Borjal D, Elmer GW. 2000. The search for a better treatment for recurrent Clostridium difficile disease: use of high-dose vancomycin combined with Saccharomyces boulardii. Clin Infect Dis 31:1012–1017. doi: 10.1086/318130. [DOI] [PubMed] [Google Scholar]
- 345.Kotowska M, Albrecht P, Szajewska H. 2005. Saccharomyces boulardii in the prevention of antibiotic-associated diarrhoea in children: a randomized double-blind placebo-controlled trial. Aliment Pharmacol Ther 21:583–590. doi: 10.1111/j.1365-2036.2005.02356.x. [DOI] [PubMed] [Google Scholar]
- 346.Surawicz CM, Elmer GW, Speelman P, McFarland LV, Chinn J, van Belle G. 1989. Prevention of antibiotic-associated diarrhea by Saccharomyces boulardii: a prospective study. Gastroenterology 96:981–988. doi: 10.1016/0016-5085(89)91613-2. [DOI] [PubMed] [Google Scholar]
- 347.Surawicz CM, McFarland L, Elmer G, Chinn J. 1989. Treatment of recurrent Clostridium difficile colitis with vancomycin and Saccharomyces boulardii. Am J Gastroenterol 84:1285–1287. [PubMed] [Google Scholar]
- 348.McFarland LV, Surawicz CM, Greenberg RN, Elmer GW, Moyer KA, Melcher SA, Bowen KE, Cox JL. 1995. Prevention of B-lactam associated diarrhea by Saccharomyces boulardii compared with placebo. Am J Gastroenterol 90:439–448. [PubMed] [Google Scholar]
- 349.Lewis S, Potts L, Barry R. 1998. The lack of therapeutic effect of Saccharomyces boulardii in the prevention of antibiotic-related diarrhoea in elderly patients. J Infect 36:171–174. doi: 10.1016/S0163-4453(98)80008-X. [DOI] [PubMed] [Google Scholar]
- 350.Can M, Beşirbellioglu BA, Avci IY, Beker CM, Pahsa A. 2006. Prophylactic Saccharomyces boulardii in the prevention of antibiotic-associated diarrhea: a prospective study. Med Sci Monit 12:PI19–PI22. [PubMed] [Google Scholar]
- 351.Forsyth CB, Farhadi A, Jakate SM, Tang Y, Shaikh M, Keshavarzian A. 2009. Lactobacillus GG treatment ameliorates alcohol-induced intestinal oxidative stress, gut leakiness, and liver injury in a rat model of alcoholic steatohepatitis. Alcohol 43:163–172. doi: 10.1016/j.alcohol.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Orlando A, Linsalata M, Notarnicola M, Tutino V, Russo F. 2014. Lactobacillus GG restoration of the gliadin induced epithelial barrier disruption: the role of cellular polyamines. BMC Microbiol 14:19. doi: 10.1186/1471-2180-14-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Seth A, Yan F, Polk DB, Rao RK. 2008. Probiotics ameliorate the hydrogen peroxide-induced epithelial barrier disruption by a PKC- and MAP kinase-dependent mechanism. Am J Physiol Gastrointest Liver Physiol 294:G1060–G1069. doi: 10.1152/ajpgi.00202.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Resta-Lenert S, Barrett KE. 2003. Live probiotics protect intestinal epithelial cells from the effects of infection with enteroinvasive Escherichia coli (EIEC). Gut 52:988–997. doi: 10.1136/gut.52.7.988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Yang KM, Jiang ZY, Zheng CT, Wang L, Yang XF. 2014. Effect of Lactobacillus plantarum on diarrhea and intestinal barrier function of young piglets challenged with enterotoxigenic Escherichia coli K88. J Anim Sci 92:1496–1503. doi: 10.2527/jas.2013-6619. [DOI] [PubMed] [Google Scholar]
- 356.Von Bültzingslöwen I, Adlerberth I, Wold A, Dahlén G, Jontell M. 2003. Oral and intestinal microflora in 5-fluorouracil treated rats, translocation to cervical and mesenteric lymph nodes and effects of probiotic bacteria. Oral Microbiol Immunol 18:278–284. doi: 10.1034/j.1399-302X.2003.00075.x. [DOI] [PubMed] [Google Scholar]
- 357.Ewaschuk JB, Diaz H, Meddings L, Diederichs B, Dmytrash A, Backer J, Looijer-van Langen M, Madsen KL. 2008. Secreted bioactive factors from Bifidobacterium infantis enhance epithelial cell barrier function. Am J Physiol Gastrointest Liver Physiol 295:G1025–G1034. doi: 10.1152/ajpgi.90227.2008. [DOI] [PubMed] [Google Scholar]
- 358.Ukena SN, Singh A, Dringenberg U, Engelhardt R, Seidler U, Hansen W, Bleich A, Bruder D, Franzke A, Rogler G, Suerbaum S, Buer J, Gunzer F, Westendorf AM. 2007. Probiotic Escherichia coli Nissle 1917 inhibits leaky gut by enhancing mucosal integrity. PLoS One 2:e1308. doi: 10.1371/journal.pone.0001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Zareie M, Johnson-Henry K, Jury J, Yang P-C, Ngan B-Y, McKay DM, Soderholm JD, Perdue MH, Sherman PM. 2006. Probiotics prevent bacterial translocation and improve intestinal barrier function in rats following chronic psychological stress. Gut 55:1553–1560. doi: 10.1136/gut.2005.080739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Dahan S, Dalmasso G, Imbert V, Peyron J, Rampal P, Czerucka D. 2003. Saccharomyces boulardii interferes with enterohemorrhagic Escherichia coli-induced signaling pathways in T84 cells. Infect Immun 71:766–773. doi: 10.1128/IAI.71.2.766-773.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Chen X, Kokkotou EG, Mustafa N, Bhaskar KR, Sougioultzis S, O'Brien M, Pothoulakis C, Kelly CP. 2006. Saccharomyces boulardii inhibits ERK1/2 mitogen-activated protein kinase activation both in vitro and in vivo and protects against Clostridium difficile toxin A-induced enteritis. J Biol Chem 281:24449–24454. doi: 10.1074/jbc.M605200200. [DOI] [PubMed] [Google Scholar]
- 362.Jahn H-U, Ullrich R, Schneider T, Liehr R-M, Schieferdecker HL, Holst H, Zeitz M. 1996. Immunological and trophical effects of Saccharomyces boulardi on the small intestine in healthy human volunteers. Digestion 57:95–104. [DOI] [PubMed] [Google Scholar]
- 363.Zocco MA, dal Verme LZ, Cremonini F, Piscaglia AC, Nista EC, Lli MC, Novi M, Rigante D, Cazzato IA, Ojetti V, Armuzzi A, Gasbarrini G, Gasbarrini A. 2006. Efficacy of Lactobacillus GG in maintaining remission of ulcerative colitis. Aliment Pharmacol Ther 23:1567–1574. doi: 10.1111/j.1365-2036.2006.02927.x. [DOI] [PubMed] [Google Scholar]
- 364.Furrie E, Macfarlane S, Kennedy A, Cummings JH, Walsh SV, O'Neil DA, Macfarlane GT. 2005. Synbiotic therapy (Bifidobacterium longum/Synergy 1) initiates resolution of inflammation in patients with active ulcerative colitis: a randomised controlled pilot trial. Gut 54:242–249. doi: 10.1136/gut.2004.044834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Kruis W, Schütz E, Fric P, Fixa B, Judmaier G, Stolte M. 1997. Double-blind comparison of an oral Escherichia coli preparation and mesalazine in maintaining remission of ulcerative colitis. Aliment Pharmacol Ther 11:853–858. doi: 10.1046/j.1365-2036.1997.00225.x. [DOI] [PubMed] [Google Scholar]
- 366.Bibiloni R, Fedorak RN, Tannock GW, Madsen KL, Gionchetti P, Campieri M, De Simone C, Sartor RB. 2005. VSL#3 probiotic-mixture induces remission in patients with active ulcerative colitis. Am J Gastroenterol 100:1539–1546. doi: 10.1111/j.1572-0241.2005.41794.x. [DOI] [PubMed] [Google Scholar]
- 367.Tursi A, Brandimarte G, Papa A, Giglio A, Elisei W, Giorgetti GM, Forti G, Morini S, Hassan C, Pistoia MA, Modeo ME, Rodino S, D'Amico T, Sebkova L, Sacca N, Di Giulio E, Luzza F, Imeneo M, Larussa T, Di Rosa S, Annese V, Danese S, Gasbarrini A. 2010. Treatment of relapsing mild-to-moderate ulcerative colitis with the probiotic VSL#3 as adjunctive to a standard pharmaceutical treatment: a double-blind, randomized, placebo-controlled study. Am J Gastroenterol 105:2218–2227. doi: 10.1038/ajg.2010.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Sood A, Midha V, Makharia GK, Ahuja V, Singal D, Goswami P, Tandon RK. 2009. The probiotic preparation, VSL#3 induces remission in patients with mild-to-moderately active ulcerative colitis. Clin Gastroenterol Hepatol 7:1202–1209. doi: 10.1016/j.cgh.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 369.Venturi A, Gionchetti P, Rizzello F, Johansson R, Zucconi E, Brigidi P, Matteuzzi D, Campieri M. 1999. Impact on the composition of the faecal flora by a new probiotic preparation: preliminary data on maintenance treatment of patients with ulcerative colitis. Aliment Pharmacol Ther 13:1103–1108. doi: 10.1046/j.1365-2036.1999.00560.x. [DOI] [PubMed] [Google Scholar]
- 370.Li G, Zeng S, Liao W, Lv N. 2012. The effect of bifid triple viable on immune function of patients with ulcerative colitis. Gastroenterol Res Pract 2012:404752. doi: 10.1155/2012/404752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Guslandi M, Giollo P, Testoni PA. 2003. A pilot trial of Saccharomyces boulardii in ulcerative colitis. Eur J Gastroenterol Hepatol 15:697–698. doi: 10.1097/00042737-200306000-00017. [DOI] [PubMed] [Google Scholar]
- 372.Tamaki H, Nakase H, Inoue S, Kawanami C, Itani T, Ohana M, Kusaka T, Uose S, Hisatsune H, Tojo M, Noda T, Arasawa S, Izuta M, Kubo A, Ogawa C, Matsunaka T, Shibatouge M. 2016. Efficacy of probiotic treatment with Bifidobacterium longum 536 for induction of remission in active ulcerative colitis: a randomized, double-blinded, placebo-controlled multicenter trial. Dig Endosc 28:67–74. doi: 10.1111/den.12553. [DOI] [PubMed] [Google Scholar]
- 373.Tursi A, Brandimarte G, Giorgetti GM, Forti G, Modeo ME, Gigliobianco A. 2004. Low-dose balsalazide plus a high-potency probiotic preparation is more effective than balsalazide alone or mesalazine in the treatment of acute mild-to-moderate ulcerative colitis. Med Sci Monit 10:PI126–PI131. [PubMed] [Google Scholar]
- 374.Ng SC, Plamondon S, Kamm MA, Hart AL, Al-Hassi HO, Guenther T, Stagg AJ, Knight SC. 2010. Immunosuppressive effects via human intestinal dendritic cells of probiotic bacteria and steroids in the treatment of acute ulcerative colitis. Inflamm Bowel Dis 16:1286–1298. doi: 10.1002/ibd.21222. [DOI] [PubMed] [Google Scholar]
- 375.Kato K, Mizuno S, Umesaki Y, Ishii Y, Sugitani M, Imaoka A, Otsuka M, Hasunuma O, Kurihara R, Iwasaki A, Arakawa Y. 2004. Randomized placebo-controlled trial assessing the effect of bifidobacteria-fermented milk on active ulcerative colitis. Aliment Pharmacol Ther 20:1133–1141. doi: 10.1111/j.1365-2036.2004.02268.x. [DOI] [PubMed] [Google Scholar]
- 376.Wildt S, Nordgaard I, Hansen U, Brockmann E, Rumessen JJ. 2011. A randomised double-blind placebo-controlled trial with Lactobacillus acidophilus La-5 and Bifidobacterium animalis subsp. lactis BB-12 for maintenance of remission in ulcerative colitis. J Crohns Colitis 5:115–121. doi: 10.1016/j.crohns.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 377.Peran L, Sierra S, Comalada M, Lara-Villoslada F, Bailón E, Nieto A, Concha A, Olivares M, Zarzuelo A, Xaus J, Gálvez J. 2007. A comparative study of the preventative effects exerted by two probiotics, Lactobacillus reuteri and Lactobacillus fermentum, in the trinitrobenzenesulfonic acid model of rat colitis. Br J Nutr 97:96–103. doi: 10.1017/S0007114507257770. [DOI] [PubMed] [Google Scholar]
- 378.Chauhan R, Vasanthakumari AS, Panwar H, Mallapa RH, Duary RK, Batish VK, Grover S. 2014. Amelioration of colitis in mouse model by exploring antioxidative potentials of an indigenous probiotic strain of Lactobacillus fermentum Lf1. Biomed Res Int 2014:206732. doi: 10.1155/2014/206732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Schreiber O, Petersson J, Phillipson M, Perry M, Roos S, Holm L. 2009. Lactobacillus reuteri prevents colitis by reducing P-selectin-associated leukocyte- and platelet-endothelial cell interactions. Am J Physiol Gastrointest Liver Physiol 296:G534–G542. doi: 10.1152/ajpgi.90470.2008. [DOI] [PubMed] [Google Scholar]
- 380.Zuo L, Yuan K, Yu L, Meng Q, Chung PC, Yang D. 2014. Bifidobacterium infantis attenuates colitis by regulating T cell subset responses. World J Gastroenterol 20:18316–18329. doi: 10.3748/wjg.v20.i48.18316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Lee SK, Kim YW, Chi SG, Joo YS, Kim HJ. 2009. The effect of Saccharomyces boulardii on human colon cells and inflammation in rats with trinitrobenzene sulfonic acid-induced colitis. Dig Dis Sci 54:255–263. doi: 10.1007/s10620-008-0357-0. [DOI] [PubMed] [Google Scholar]
- 382.Dalmasso G, Cottrez F, Imbert V, Lagadec P, Peyron J-F, Rampal P, Czerucka D, Groux H, Foussat A, Brun V. 2006. Saccharomyces boulardii inhibits inflammatory bowel disease by trapping T cells in mesenteric lymph nodes. Gastroenterology 131:1812–1825. doi: 10.1053/j.gastro.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 383.Dalmasso G, Loubat A, Dahan S, Calle G, Rampal P, Czerucka D. 2006. Saccharomyces boulardii prevents TNF-alpha-induced apoptosis in EHEC-infected T84 cells. Res Microbiol 157:456–465. doi: 10.1016/j.resmic.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 384.Girard P, Pansart Y, Gillardin JM. 2005. Inducible nitric oxide synthase involvement in the mechanism of action of Saccharomyces boulardii in castor oil-induced diarrhoea in rats. Nitric Oxide 13:163–169. doi: 10.1016/j.niox.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 385.Soyturk M, Saygili SM, Baskin H, Sagol O, Yilmaz O, Saygili F, Akpinar H. 2012. Effectiveness of Saccharomyces boulardii in a rat model of colitis. World J Gastroenterol 18:6452–6460. doi: 10.3748/wjg.v18.i44.6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Caetano MJ, Paramés M, Babo M, Santos A, Ferreira AB, Freitas A, Coelho MC, Mateus AM. 1986. Immunopharmacological effects of Saccharomyces boulardii in healthy human volunteers. Int J Immunopharmacol 8:245–259. doi: 10.1016/0192-0561(86)90106-2. [DOI] [PubMed] [Google Scholar]