Abstract
Adeno-associated virus type 2 (AAV-2) is a human parvovirus that requires the presence of a helper virus, such as the herpes simplex virus type 1 (HSV-1) to accomplish a complete productive cycle. In the absence of helper virus, AAV-2 can establish a latent infection that is characterized by the absence of expression of viral genes. So far, four HSV-1 early genes, UL5/8/52 (helicase primase complex) and UL29 (single-stranded DNA-binding protein), were defined as sufficient for AAV replication when cells were transfected with a plasmid carrying the wild-type AAV-2 genome. However, none of these viral products was shown to behave as a transcriptional factor able to activate AAV gene expression. Our study provides the first evidence that the immediate-early HSV-1 protein ICP0 can promote rep gene expression in cells latently infected with wild-type AAV-2. This ICP0-mediated effect occurs at the transcriptional level and involves the ubiquitin-proteasome pathway. Furthermore, using deletion mutants, we demonstrate that the localization of ICP0 to ND10 and their disruption is not required for the activation of the rep promoter, whereas binding of ICP0 to the ubiquitin-specific protease HAUSP makes a significant contribution to this effect.
Adeno-associated virus type 2 (AAV-2) is a nonpathogenic human parvovirus that establishes a latent infection in the absence of helper virus (1, 57). During latency, the viral genome persists in a largely repressed episomal or integrated form (14, 54, 63). Infection of latently infected cells with a helper virus such as herpes simplex virus (HSV) or adenovirus (Ad) leads to the reactivation of AAV gene expression, the rescue of the viral genome, and finally to the progression through a productive phase (1, 57).
The 4.7-kb genome of AAV-2 contains two open reading frames (ORFs), rep and cap, flanked by two inverted terminal repeats (ITR) that constitute the cis-acting elements required for DNA replication. Two promoters at map positions 5 and 19 (p5 and p19, respectively) control the expression of the rep ORF leading to the synthesis of Rep78/68 and Rep52/40 proteins, respectively. These proteins are involved in many aspects of the viral life cycle and particularly in the regulation of AAV gene expression. The p40 promoter controls the synthesis of the three proteins (VP1, 2, and 3) that constitute the capsid (1).
During latency, i.e., in the absence of helper virus, the silent state of the AAV promoters, particularly that of the p5 promoter, is generally attributed to repressive activity exerted by cellular factors and the Rep proteins. Indeed, the results of several studies with transient-transfection assays have reported that Rep78 and Rep68 act as site-specific DNA-binding proteins to shut down p5 and p19 transcription (52, 53). The Rep binding site involved in this repressive effect was identified both within the ITR and the p5 promoter (46, 53). In addition, silencing of the p5 promoter was shown to be mediated by its interaction with the cellular transcription factor YY1 bound at position −60 (12, 72). The reactivation from latency that occurs in the presence of a helper virus results in the derepression of the three AAV promoters, particularly that of the p5 promoter, which controls the onset of viral gene expression. Nearly all of the studies on this subject have focused on the helper activities provided by Ad with transient-transfection assays. They have shown that two Ad proteins, E1a and, to a lesser extent, the DNA-binding protein (DBP), are involved in p5 transactivation (11, 12). In particular, the crucial role played by E1a is mediated through its interaction with two cellular proteins, MLTF and YY1 (12, 58, 72). Activation of the p5 promoter leads to the synthesis of Rep78 and Rep68, which in turn act as transactivators of the p19 and p40 promoters while maintaining their repressive effect toward the p5 promoter (44, 67-69).
In contrast, few studies have been conducted on the helper activities provided by HSV. Four early HSV type 1 (HSV-1) gene products, UL29 (DBP) and UL5/8/52 (helicase primase complex) have been identified as essential for AAV replication in cells transfected with a plasmid containing the wild-type (wt) AAV genome (75, 80). In addition, Ward et al. demonstrated that the HSV-1 UL30 gene encoding the viral polymerase was able to initiate AAV DNA synthesis in an in vitro replication assay performed in the absence of cellular extracts and with purified HSV proteins (78). However, none of these HSV factors was shown to be involved in the transactivation or derepression of the AAV promoters, particularly that of the p5 promoter, which constitutes the initial event during AAV replication.
In this study, we focused on the identification of the HSV-1 factors necessary to relieve the repressive state of the AAV p5 promoter. HSV-1 gene expression during lytic infection occurs with a temporal cascade of three groups of genes: immediate-early, early, and late (55). The main viral transactivators required for the expression of the HSV-1 genes are the late protein VP16 and the immediate-early proteins ICP4 and ICP0. VP16, a component of the HSV tegument, activates the expression of the immediate-early genes through a target sequence present in at least one copy on all immediate-early HSV promoters (65). ICP4 is absolutely required for the transactivation of the HSV-1 early and late genes (79). This factor exerts its transactivating activity by binding specifically or nonspecifically to DNA (9, 73). In the absence of ICP4, the HSV-1 life cycle is arrested at the immediate-early phase (16). The ICP0 protein is also an important regulator of the three classes of HSV genes and has been shown to be able to transactivate several other heterologous promoters in transient-transfection assays (28). In the absence of ICP0, the virus is still replication competent but grows poorly and also reactivates from latency at much lower levels than the wt virus in some cell types (7, 56, 71, 74). This multiplicity of infection (MOI)-dependent defect can be overcome in some cell types and by cell cycle status (8, 86). ICP0 does not bind DNA but may act through the interaction with various cellular factors, including cyclin D3, elongation factor EF-1α, transcription factor BMAL1, and ubiquitin-specific protease HAUSP (26, 49-51). Furthermore, several studies reported that ICP0 expresses two E3 ubiquitin ligase activities located in exons 2 and 3 (5, 35, 37, 76). The ubiquitin ligase activity present in exon 2 is associated with a RING finger domain and is responsible for the proteasome-mediated degradation of cellular proteins, including major proteins associated with ND10 nuclear domains such as PML and Sp100, centromeric proteins CENP-A and CENP-C, and the catalytic subunit of DNA-dependent protein kinase (22, 23, 33, 60, 66). The second ubiquitin ligase activity identified in exon 3 is responsible for the degradation of the E2 ubiquitin-conjugating enzyme cdc34 (36, 76). However, none of these substrates were shown to be directly ubiquitinated by ICP0 except, very recently, for p53 (4). The present hypothesis to explain the wide transactivating or derepressing activities of ICP0, as well as its implication in the establishment of lytic replication, is that this protein is able to alter the higher-order structure of chromatin by targeting a repressive factor for degradation (21).
In the present study, we wanted to determine whether ICP0 contributes to AAV reactivation from latency by specifically analyzing its effect on the expression of the rep gene. For these analyses, we used a HeLa cell clone (HA-16) latently infected with wt AAV-2 as a model (17, 77). Our results demonstrated that ICP0 could promote rep gene expression in latently infected HA-16 cells. This ICP0-mediated derepressive effect occurred at the transcriptional level and involved the ubiquitin-proteasome pathway. Furthermore, we demonstrated that the degradation of ND10-associated proteins by ICP0 was not required for the activation of the rep promoter by ICP0, whereas binding to the ubiquitin-specific protease HAUSP makes a significant contribution to this effect.
MATERIALS AND METHODS
Cell and viruses.
wt AAV-2 latently infected human HA-16 derived from HeLa (77), Vero, and U2OS cells were maintained in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% fetal bovine serum (Sigma) and 1% penicillin-streptomycin (5,000 U/ml; Gibco BRL). Ad used in this study were wt Ad type 5 (Ad5) (ATCC VR-5); AdlacZ, encoding the β-galactosidase enzyme; AdTREICP0, containing the ICP0 gene under the control of tetracycline-responsive promoter (TRE) (38); and AdrtTA, encoding for the reverse tetracycline transactivator expressed under the control of the cytomegalovirus promoter (38). In these three recombinant Ad, the transgene was inserted in place of the E1 gene in a ΔE1/ΔE3 adenoviral backbone. All of these viruses were produced and titrated by standard procedures (31) and used at an MOI of 50 on HA-16 cells. wt HSV-1 (F strain), HSV-ΔICP0 (dl1403), and HSV-ΔICP4 (HSV-1-17 Cgal delIE3) were produced on Vero cells or on the ICP4 complementing cell line (62). wt and mutant HSV stocks were titrated on Vero, U2OS, or ICP4 complementing cells by standard procedures (3). Mutated HSV strains were checked for the absence of contaminating wt HSV by a plaque-forming assay performed on Vero cells.
Plasmids.
The pEi11O and p110FXE plasmids contain the wt and RING finger-mutated version of ICP0, respectively. They were obtained by removing the green fluorescent protein gene cloned at the NcoI site of the ICP0 initiation codon from plasmids pEG110 and pEGFXE (kindly provided by P. Lomonte). The p110D9, p110D12, p110D14, and p110E52X plasmids (kindly provided by R. Everett, MRC Virology Unit, Glasgow, United Kingdom) contained a mutated version of the ICP0 gene (18, 19). In all of these plasmids, the ICP0 gene was under the control of its own promoter region. Plasmid pTRE-ICP0 was obtained by cloning the ICP0 gene (NheI/HpaI fragment from pCI110) under the control of the TRE promoter in plasmid pTRE2 at the NheI/EcoRV sites (Clontech).
Antibodies.
The anti-ICP4 (ab6514) and anti-ICP0 (ab6513) mouse monoclonal antibodies were purchased from Abcam (Cambridge, United Kingdom). The anti-ICP0 (r190) rabbit serum was kindly supplied by R. Everett. Anti-Rep 303.9 and anti-Rep76.3 mouse monoclonal antibodies (kindly provided by J. Kleinschmidt, German Cancer Center, Heidelberg, Germany) recognized the four Rep proteins or only unspliced Rep proteins, respectively (81, 82). Anti-α-tubulin (T5168) mouse monoclonal antibody was purchased from Sigma. The anti-PML mouse monoclonal antibody (PG-M3) was purchased from Santa Cruz Biotechnologies.
Western blot analysis.
HA-16 cells were seeded in six-well plates at a density of 106 cells per well and infected the following day with viruses at the MOI indicated in the figure legends. When the proteasome inhibitor was used, the cells were preincubated 2 h prior to infection with 5 μM MG132 (Sigma). All subsequent incubations were performed in the presence of the drug, newly refreshed every 4 h, except during the night period. The cells were then washed in phosphate-buffered saline (PBS) 24 h later and lysed with RIPA buffer (20 mM Tris-HCl [pH 7.4], 50 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40, 0.5% deoxycholate, 0.5% sodium dodecyl sulfate) in the presence of a cocktail of protease inhibitors (Roche). The total cell protein concentration was determined for each sample by a Bradford assay (Bio-Rad). Proteins were loaded on sodium dodecyl sulfate-10% polyacrylamide gels and then transferred to nitrocellulose membranes according to the manufacturer's recommendations (Bio-Rad). The membranes were blocked overnight at 4°C, in PBS-0.1% Tween-5% dried milk, and then incubated for 2 h at room temperature with the appropriate antibody diluted in PBS-0.1% Tween-1% dried milk. Anti-ICP0, anti-ICP4, anti-Rep 303.9, and anti-α-tubulin mouse monoclonal antibodies were used at a 1/4,000, 1/1,600, 1/20, and 1/4,000 dilutions, respectively. After washing, a horseradish peroxidase-conjugated anti-mouse antibody (Amersham Biosciences) was applied at a 1/2,000 dilution for 1 h at room temperature. Finally, after extensive washes in PBS-0.1% Tween, the membranes were soaked in enhanced chemiluminescence reagent (Amersham Biosciences) and exposed to a film. When necessary, the membranes were reprobed with another primary antibody.
Immunofluorescence.
HA-16 cells were seeded onto 12-mm coverslips in a 24-well plate at a density of 105 cells per well. The cells were transfected with plasmid with the Lipofectamine Plus reagent (Gibco-BRL). Twenty four hours later, the cells were washed with PBS, fixed with PBS-4% paraformaldehyde, and permeabilized with PBS-0.2% Triton X-100. The primary antibody was diluted in PBS-1% bovine serum albumin and applied for 1 h at room temperature. The anti-ICP0 rabbit serum r190 was used at a 1/400 dilution, whereas the anti-Rep76.3 mouse monoclonal antibody was used pure. The coverslips were then washed with PBS-1% Tween-20 and incubated for 1 h with the appropriate fluorescein isothiocyanate- or tetramethyl rhodamine isothiocyanate-conjugated anti-mouse antibody or anti-rabbit antibody (Amersham Biosciences) at a 1/400 dilution. To localize the nuclei, fixed cells were stained for 30 min with TO-PRO-3 used at a 1/1,000 dilution (T-3605; Molecular Probes). After extensive washes, the coverslips were mounted onto glass slides with ProLong antifading mounting medium (Molecular Probes). Images were collected on a Leica TCS-SP1 confocal microscope with a 63/1.4× oil immersion lens. The green and red emissions were collected by using two photomultiplier tubes under conditions of no detectable channel overlap. The grayscale digital images were visualized with a 24-bit imaging system including Leica TCS-NT software. The images generated were imported into Adobe Photoshop, version 6.0, pseudocolored, and in some cases, overlapped to produce merged images.
RPA.
Total RNA was isolated from infected HA-16 cells by a guanidium isothiocyanate-based method with the Trizol reagent (Sigma) according to the manufacturer's instructions. RNA was treated with DNase I, resuspended in RNase-free water, and quantified by optical density. The RNase protection assay (RPA) was performed by using an RPA III kit (Ambion) according to the manufacturer′s instructions. The p5 antisense probe was generated by cloning the wt AAV PCR fragment extending from nucletides 255 to 460 into the T7-driven pSP72 transcription vector (Promega). pTRI-β-actin-human plasmid (Ambion) was used to generate an actin antisense control probe of 309 nucleotides. RPAs were performed with 10 μg of total RNA in substantial probe excess. After hybridization, the samples were analyzed by 6% denaturing polyacrylamide sequencing. The gel was dried at 80°C under a vacuum for 2 h and then exposed to a film.
RESULTS
wt HSV-1 but not HSV-1ΔICP0 is able to activate the synthesis of Rep proteins in AAV-2 latently infected cells.
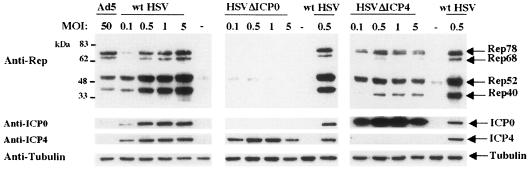
To examine the specific contribution of ICP0 in the reactivation of wt AAV-2 rep gene expression from a latent integrated form, we first compared the ability of different HSV-1 strains to induce Rep protein synthesis upon infection of HA-16, a wt AAV-2 latently infected cell line (77). These cells contain several copies of the AAV-2 genome integrated at the AAV S1 locus in a head-to-tail configuration (17). In these latently infected cells, rep gene expression and AAV production is detected only upon infection with a helper virus. HA-16 cells were infected with either wt HSV-1, HSVΔICP0, or HSVΔICP4 at MOI ranging from 0.1 to 5 PFU/cell. As a control, cells were either left untreated or infected with Ad5. As shown in Fig. 1, uninfected Ha16 cells lack detectable Rep expression, confirming that the AAV genome is highly repressed in these cells (see also Fig. 3). As expected, infection with wt HSV-1 induced the synthesis of the four Rep proteins in an MOI-dependent manner, even at the lowest MOI (Fig. 1, left panel). In contrast, no Rep proteins were detected with an HSV ICP0-null mutant, even at the highest MOI (Fig. 1, middle panel). Eventually, a faint but detectable Rep signal could be observed upon longer exposure or with higher MOI of virus (data not shown). This HSV mutant is impaired for the expression of ICP0 but expresses the other HSV proteins, particularly the ICP4 protein, at detectable levels. As a consequence, this mutant can grow and form plaques, albeit with a reduced efficiency, on Vero and HeLa cells. This result suggested that the presence of ICP0 was required to induce rep gene expression. Accordingly, infection with an HSV ICP4-null mutant, which is impaired for the synthesis of the HSV early and late proteins but overexpresses the ICP0 protein (16), restored the expression of the Rep proteins (Fig. 1, right panel). With this HSV mutant, a maximal level of Rep proteins was observed at an MOI of 0.5. Interestingly, lower amounts of Rep68 and Rep40 were observed with the HSVΔICP4 virus than in wt HSV-1. This observation was also confirmed in further experiments (Fig. 2). Together, these results suggested that the immediate-early ICP0 protein was required to induce the synthesis of the Rep proteins from latently infected cells. To further confirm this observation in a different cell type, we used a clone of human Detroit 6 cells latently infected with wt AAV2 (clone 7374D5), described by Berns et al. (2). Although the 7374D5 cells exhibited a lower AAV-2 genome copy number per cell, we similarly observed Rep synthesis in the presence of ICP0 (data not shown).
FIG. 1.
Effect of wt and mutant HSV-1 strains on the induction of Rep protein synthesis in HA-16 cells. HA-16 cells were infected at the indicated MOIs with either wt HSV-1 (left), HSVΔICP0 (middle), or HSVΔICP4 (right). As positive and negative controls, cells were infected with wt Ad5 at an MOI of 50 or left untreated (−), respectively. At 24 h postinfection, the cells were harvested and protein contents were analyzed by Western blotting with an anti-Rep antibody (303.9) or, on a separate membrane loaded with the same amount of sample, with an anti-α-tubulin antibody (to normalize for protein content). Both membranes were then reprobed with anti-ICP0 and anti-ICP4. ICP0, ICP4, and α-tubulin bands have the expected masses of 110, 175, and 50 kDa, respectively.
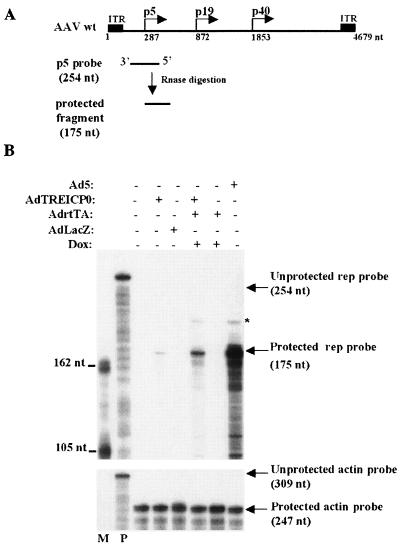
FIG. 3.
Effects of ICP0 on rep RNA accumulation in HA-16 cells. (A) Schematic presentation of the AAV genome and the antisense p5 RNA probe used in this assay. (B) RPA of p5 promoter-initiated transcripts in the presence (+) and absence (−) of ICP0. HA-16 cells were either left untreated (lane 1) or infected with the indicated recombinant Ad at an MOI of 50 (single infection) or 25 (double infection). Where indicated, the cells were additionally treated with 3 μM doxycycline (Dox) to induce ICP0 expression. At 24 h postinfection, RNA was extracted and hybridized either to a p5 antisense rep probe (upper panel) or to a β-actin antisense probe (lower panel). M, molecular size marker; P, unprotected probe. The 175-nucleotide (nt) fragment identified with the rep antisense probe corresponds to transcripts initiated from the consensus transcription initiation site at position 287 of wt AAV-2. The asterisk indicates the size of the protected fragment expected for transcripts initiating from the 5′ ITR.
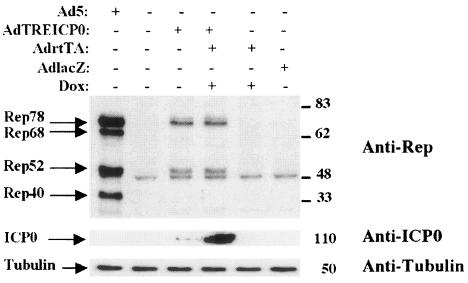
FIG. 2.
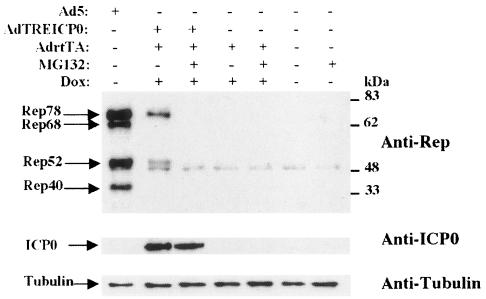
Induction of Rep synthesis with a recombinant Ad coding for ICP0. HA-16 cells were infected with the indicated recombinant Ad at an MOI of 50, except in the case of coinfection, in which the MOI for each virus was 25. Where indicated, the cells were additionally incubated in the presence of 3 μM doxycycline (Dox). As positive and negative controls, the cells were infected with wt Ad5 at an MOI of 50 or left untreated. The cell extracts prepared 24 h later were analyzed by Western blotting with an anti-Rep antibody (303.9). The membrane was then stripped and reprobed with an anti-ICP0 antibody and then with an anti-α-tubulin antibody. +, present; −, absent.
rep gene expression can be induced upon infection with a recombinant Ad encoding ICP0.
To confirm the ability of ICP0 to induce the synthesis of Rep proteins, in the absence of other HSV factors, we next used a recombinant Ad carrying the ICP0 gene under the control of the doxycycline/rtTA-inducible promoter (AdTREICP0) (38). It was of interest to use such a regulatory system to control the expression level of ICP0 because many studies have documented the deleterious effect of this protein on cellular metabolism and growth (43, 59). HA-16 cells were infected with AdTREICP0 either alone or in the presence of AdrtTA and doxycycline, and Rep protein synthesis was evaluated 24 h later by Western blot analysis. Rep protein synthesis was undetectable in uninfected cells or in cells infected with the control adenoviral vectors AdrtTA and AdlacZ (Fig. 2). In contrast, AdTREICP0 alone was able to induce the synthesis of Rep78 and Rep52. A low but detectable level of ICP0 was observed under these conditions, indicating a leakiness of the TRE promoter in the absence of inducers, as previously documented by Halford et al. (38). This observation was also confirmed by immunofluorescence with anti-ICP0 antibodies (data not shown). When HA-16 cells were coinfected with AdTREICP0 and AdrtTA and treated with doxycycline, a net increase in the level of ICP0 was observed (Fig. 2). Interestingly, the level of Rep proteins remained comparable to that observed in the absence of inducers. This result indicated that Rep protein synthesis could be achieved even with very low amounts of ICP0 and was not increased further when more ICP0 protein was produced. Similarly, Hobbs et al. demonstrated that very small amounts of ICP0 produced with a recombinant Ad were sufficient to activate quiescent viral genomes in trans (42). Interestingly, and as already pointed out (Fig. 1), the expression of ICP0 preferentially induced the synthesis of the unspliced Rep proteins Rep78 and Rep52. In this assay, Rep68 and Rep40 could be detected at 48 h postinfection, but their ratio was always greatly reduced compared with the two other Rep proteins (data not shown). This observation is interpreted in Discussion.
Induction of Rep protein synthesis by ICP0 occurs at the transcriptional level.
Earlier studies have documented that ICP0 activates the expression of HSV genes at the level of mRNA synthesis, with no evidence of posttranscriptional effects (48). To analyze this particular point, HA-16 cells were infected with AdTREICP0 in the presence or absence of inducers (AdrtTA and doxycycline) and the levels of rep transcripts were analyzed by RPA. An antisense rep riboprobe spanning the p5 transcription initiation site was used to measure the presence of p5 rep transcripts (Fig. 3A), whereas an actin riboprobe was used to normalize total RNA recovery. As expected, uninfected cells lacked detectable rep transcripts (Fig. 3B), further confirming that the absence of Rep proteins in these cells is due to the silencing of the p5 promoter. Similarly, no rep transcripts were detected when cells were mock infected with AdlacZ or AdrtTA. A protected Rep fragment of the expected size was detected with RNA from cells infected with AdTREICP0 or Ad5 (Fig. 3B). The addition of AdrtTA and doxycycline to cells infected with AdTREICP0 induced a significant increase in the level of rep transcripts. Under this latter condition, as in cells infected with Ad5, a larger protected Rep band was also detected. This signal likely corresponded to transcripts initiated from the 5′ ITR, as previously documented by other investigators (29, 34). Taken together, these data indicated that the activation of rep gene expression by ICP0 occured at the transcriptional level. As discussed below, it is likely that ICP0 exerted this effect indirectly, by affecting chromatin structure or by inactivating a repressive factor, rather than by directly targeting DNA. Strikingly, the increase in rep transcripts in cells coinfected with AdTREICP0 and AdrtTA and treated with doxycycline did not correlate with a similar increase in Rep protein synthesis (compare Fig. 3B and 2). This observation suggested that, in this particular context, the translational step was limiting.
ICP0-induced rep gene expression depends on the presence of the ICP0 RING finger domain and on proteasome activity.
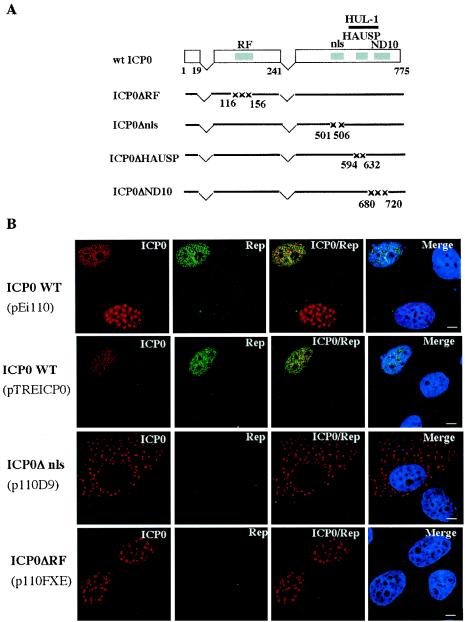
ICP0 is a multifunctional protein that is able to activate a wide variety of genes in transient-transfection assays. Since ICP0 does not bind DNA, it is likely that it functions through interaction with other proteins. Accordingly, several functional domains have been identified within the ICP0 ORF that are responsible for its interaction with cellular proteins or its localization to specific cellular domains (for a review, see reference 21). To determine the role of ICP0 in rep gene expression, we have analyzed HA-16 cells transfected with plasmids encoding wt or mutated ICP0 proteins by immunofluorescence with anti-ICP0 and anti-Rep antibodies (Fig. 4). Upon transfection into HA-16 cells, wt ICP0, expressed from its own promoter or from the TRE promoter, displayed both a punctuate and a more diffuse nuclear pattern. As previously documented by Western blotting (Fig. 3), Rep proteins were detected in cells expressing ICP0. Interestingly, the Rep signal was clearly visible in all cells displaying a more diffuse ICP0 pattern, whereas very low amounts of Rep were observed only in some of the cells displaying a punctate ICP0 pattern. Previous studies performed with infectious HSV have documented that the ICP0 protein, initially present in a punctuate pattern, could present more diffuse staining as infection progressed (24). It is possible that in the context of plasmid transfection, the different patterns assumed by ICP0 reflect a dynamic change that takes place over time or depends upon the amount of protein synthesized. As expected, upon transfection of mutant ICP0Δnls (with a deletion in the nuclear localization signal), the ICP0 protein was found exclusively in the cytoplasm and Rep expression was not observed under these conditions. The following ICP0 mutant tested for its ability to activate rep gene expression, FXE (ICP0ΔRF), had a deletion in the N-terminal RING finger domain that possesses an E3 ubiquitin ligase activity. This domain has been involved in the proteasome-dependent degradation of several cellular proteins (22, 23, 33, 60, 66). In general, the RING finger domain of ICP0 was shown to be required for all biological activities of the protein and for efficient viral growth (19). Consistent with these findings, no Rep synthesis was observed with this mutant (Fig. 4B). These results further confirmed that ICP0 alone was able to induce Rep protein synthesis and suggested that this effect may be dependent upon the proteasome-mediated degradation of some cellular factors. To test this hypothesis, we examined the effect of the proteasome inhibitor MG132 on HA-16 cells expressing the ICP0 protein. Cells infected with AdTREICP0 in the presence of inducers, AdrtTA and doxycycline, were incubated in the presence or absence of MG132 and evaluated for Rep protein synthesis by Western blotting. As shown in Fig. 5, the addition of MG132 strongly inhibited the ability of AdTREICP0 to promote Rep protein synthesis, whereas ICP0 expression was not affected. The same result was observed in the presence of another proteasome inhibitor, lactacystin (data not shown). These results indicated that proteasome activity was required for the induction of rep gene expression by ICP0.
FIG. 4.
The RING finger domain of ICP0 is essential for the induction of rep gene expression. (A) Schematic presentation of the ICP0 constructs used for the immunofluorescence studies shown in panel B and Fig. 6. The two E3 ubiquitin ligase domains of ICP0 are indicated as the RING finger (RF) and the HUL-1 domains. This latter domain is contained between aa 543 and 680 (36). Numbers refer to amino acid positions. (B) HA-16 cells were transfected with the indicated plasmids expressing either the wt or mutated forms of ICP0 (D9 and FXE) and analyzed 24 h later by indirect immunofluorescence with anti-ICP0 rabbit serum and anti-Rep mouse antibody (76.3). The ICP0 signal was detected with a secondary tetramethyl rhodamine isothiocyanate-conjugated anti-rabbit antibody, and the Rep signal was detected with a fluorescein isothiocyanate-conjugated anti-mouse antibody. The last column of panels shows merged images of both labeling schemes, including the staining of the nucleus with TO-PRO-3. Bars, 8 μm. nls, nuclear localization signal.
FIG. 5.
Inhibition of proteasome activity impairs ICP0-induced activation of Rep protein synthesis. HA-16 cells were either uninfected or infected with the indicated recombinant Ad as described in the legend to Fig. 2. Where indicated, the cells were also incubated in the presence of 3 μM doxycycline (Dox) and 5 μM MG132. The synthesis of Rep proteins was analyzed 24 h later by Western blotting. The ICP0 and α-tubulin signals were detected after sequential reprobing of the same membrane. +, present; −, absent.
Role of the ND10 localization domain and of the HAUSP-binding site of ICP0 for the activation of rep gene expression.
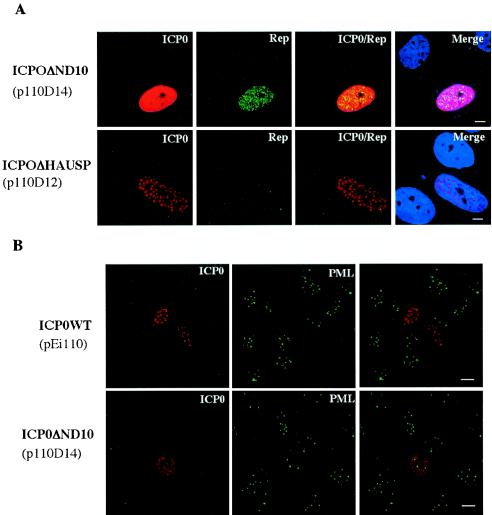
The two other ICP0 mutants analyzed for their ability to activate rep gene expression had deletions in critical functional domains identified for their ability to bind the ubiquitin-specific protease HAUSP (ICP0ΔHAUSP) or to localize to ND10 (ICP0ΔND10) (Fig. 4A). Both mutants were chosen because they were described as critical for the ICP0 transactivating effect. Indeed, the interaction of ICP0 with HAUSP has been described as contributing to its effect on the activation of gene expression and for efficient viral growth (25). We observed that upon expression of this mutant in HA-16 cells, the Rep proteins were synthesized at a nearly undetectable level (Fig. 6A). In contrast, upon transfection of the mutant ICP0ΔND10, a strong Rep signal was clearly observed (Fig. 6A). This ICP0 mutant was initially described as severely impaired for viral gene activation, particularly when acting in synergy with ICP4 (19). It was also unable to localize and disrupt ND10 structures (24). Accordingly, ND10 domains were still detected upon transfection of this mutant into HA-16 cells, whereas they were undetectable in the presence of the wt ICP0 protein (Fig. 6B). Importantly, this last result also indicated that the ND10 domains were not affected by the presence of Rep. As such, the differential effect of these latter two mutants, ICP0ΔHAUSP and ICP0ΔND10, on rep gene activation contrasted with the activity reported previously for other viral promoters (18, 19).
FIG. 6.
(A) Effect of ΔND10 and ΔHAUSP ICP0 mutants on rep gene expression. HA-16 cells were transfected with plasmids p110D14 (ICP0ΔND10) and p110D12 (ICP0ΔHAUSP) and labeled with anti-ICP0 and anti-Rep antibodies as indicated in the legend to Fig. 4B. Bars, 8 μm. (B) Visualization of ND10 domains in HA-16 cells transfected with wt and ΔND10 ICP0 constructs. HA-16 cells transfected with plasmids coding for wt (pEi110) or ΔND10 (p110D14) ICP0 proteins were stained with anti-ICP0 and anti-PML antibodies. The last column of panels shows merged images of both labeling schemes. Bars, 16 μm.
DISCUSSION
Replication and productive infection with AAV-2 requires the presence of a helper virus, typically Ad, HSV-1, or HSV-2 (6, 10). In the absence of helper virus, AAV-2 can establish a latent infection both in vitro and in vivo (14, 32, 39, 41, 54, 63). Many studies have focused on the identification of the helper activities provided by Ad, demonstrating that most of the essential virus-encoded factors were acting to activate AAV gene expression at the transcriptional and posttranscriptional levels (10). In contrast, little information is available about the helper activities provided by HSV-1, particularly concerning the factors involved in AAV gene expression.
The aim of this study was to determine whether the HSV-1 ICP0 protein was able to contribute to AAV-2 reactivation from latency by specifically analyzing the effect of this protein on the transcriptional activity of the p5 rep promoter. Indeed, ICP0 is involved alone or in combination with the immediate-early gene product, ICP4, in the transcriptional activation of the three classes of HSV-1 gene (28). Even more importantly, ICP0 was described as essential for reactivation of HSV from latency both in vitro and in vivo (7, 56). The HA-16 and the 7374D5 cells used for this study are latently infected with wt AAV-2 (2, 77). In this context, as documented in other latently infected cell lines, the AAV genome is silent. Infecting these cells with either Ad or HSV can induce AAV-2 replication and assembly. An essential step during this process is constituted by the reactivation of the AAV-2 promoters, particularly the p5 promoter of the rep gene. This viral promoter controls the synthesis of the two major Rep proteins (Rep78 and Rep68) that constitute the key factors required for the subsequent activation steps leading to rep and cap gene expression (67).
Using this cellular model, we demonstrated that ICP0 is able to activate rep gene expression. This effect was documented with mutated HSV strains, a recombinant Ad encoding ICP0, and by transfection of ICP0 constructs. In addition, we have demonstrated that the effect of ICP0 on the AAV-2 p5 promoter is exerted at the transcriptional level as shown by the detection of p5 mRNA.
ICP0 is considered a key regulator of HSV because of its ability to interact with several cellular proteins and to induce, for some of them, their proteasome-dependent degradation. The N-terminal RING finger domain of ICP0 was shown to possess a ubiquitin E3 ligase activity that is essential to the induction of the degradation of many cellular targets, such as the centromeric proteins CENP-C and CENP-A, the ND10 associated factors PML and Sp100, and the catalytic subunit of DNA-PK (22, 23, 33, 60, 66). Using mutated versions of ICP0, we showed that the activation of rep gene expression by ICP0 required the presence of the RING finger domain and was dependent on proteasome activity. Similarly, the proteasome pathway was also required for reactivation of quiescent HSV-1 in cultured cells (27). In addition, our results indicated that localization of ICP0 to ND10 and their subsequent disruption was not required to activate rep gene expression. In contrast, the ICP0 effect was severely reduced, but not completely eliminated, when using the ICP0 mutant unable to bind the ubiquitin-specific protease HAUSP. The significance of this latter observation is unclear for the moment. Indeed, the functional consequence of the interaction between ICP0 and HAUSP has not yet been elucidated. In addition, previous studies have shown that the ICP0ΔHAUSP mutant can display a different phenotype according to cell type (23). The interpretation of this result is also complicated by the fact that the HAUSP deletion mutant used in this study partially overlaps the region containing the second E3 ubiquitin ligase domain of ICP0, HUL-1 (36). The evaluation of other ICP0 mutants will be performed to further define the precise role of this region in rep gene activation.
All together, these results raise many questions. The first one is how can these findings be reconciled with what is known about the regulation of the AAV-2 p5 promoter? Many studies performed exclusively by transient-transfection assays have documented that the repression of the p5 promoter was due to both the Rep proteins themselves (Rep78 and Rep68) and to cellular factors, notably YY1 (12, 52, 53, 72). As indicated before, the ICP0 protein does not bind DNA directly. As such, a simple hypothesis would be that in HA-16 cells the same viral and cellular factors are involved in silencing the p5 promoter and that ICP0 relieves the repression directly or indirectly through a process involving proteasome-mediated degradation. However, it should be noted that it is presently unknown whether the same factors are indeed binding and repressing the p5 promoter in the context of a latently integrated virus. We are currently performing the analysis of the p5 promoter of HA-16 cells by chromatin immunoprecipitation-PCR to investigate this particular point. An alternative hypothesis is that, as documented for other silenced viral genomes or genes, the integrated AAV genome is embedded in a chromatin structure, preventing the access of transcriptional factors. Indeed, several studies indicate that chromatin conformation can be altered by the posttranslational modifications of histones and DNA, such as acetylation and methylation, that determine whether a specific chromosomal region is transcriptionally active or not (47). In this scenario, ICP0 would act by changing the conformational state of the AAV p5 region, allowing the access of key regulatory transcriptional factors such as YY1 and Rep. The observation that a weak but detectable level of rep gene expression can be observed upon treatment of HA-16 cells with an inhibitor of histone deacetylases, trichostatin A, favors this hypothesis (data not shown). Also, this model fits well with the observation that AAV latency could also be relieved in the absence of helper virus by using cellular stresses or genotoxic agents (83-85).
The second point of discussion concerns the role of ND10 disruption in the AAV life cycle. These nuclear structures, formed by the assembly of several proteins and, notably, PML, are the target of many viruses, such as Ad or HSV, that upon entry into the cell induce their disorganization and dispersal (20). Even if the precise role of ND10 is still unknown, the present hypothesis is that these structures constitute a nuclear depot of proteins that exert an antiviral effect (64, 70). Accordingly, a recent study has shown that ND10 mediates the interferon response to HSV-1 infection (13). On the other hand, overexpression of the major ND10 constituent PML was shown to maintain the ND10 structure intact during HSV infection without interfering with viral growth (61). As such, the question of the role of ND10 disorganization during HSV infection is still a matter of debate. The two major helper viruses for AAV, i.e., Ad and HSV, both encode proteins that target the ND10 structures. As such, it could be expected that disorganization of ND10 induced by the helper virus, either Ad or HSV, is also important for the growth of AAV. In this report, we show that the deletion of the ICP0 domain necessary for the localization of the protein to ND10 and, consequently, their disruption was not required for the reactivation of rep gene expression from a latent silenced form. Interestingly, the same ICP0 mutant was unable to reduce reactivation of quiescent HSV (40). At another level, a recent report by Fraefel et al. has shown that recombinant AAV replication centers could be formed in the absence of ICP0 (30). In addition, the Ad-encoded E4 (ORF 3) product, responsible for ND10 dispersal, has not been identified as a helper factor required for AAV growth, and recombinant AAV vectors can be efficiently produced in the absence of this factor (10, 45). All together, these results strongly suggest that the disruption of ND10 is not required during the AAV life cycle.
The last question concerns the role of ICP0 in the context of the helper activities provided by HSV. So far, four HSV-1 early genes, UL5/8/52 (helicase primase complex) and UL29 (single-stranded DBP), were defined as essential for AAV replication when cells were transfected with a plasmid containing the entire AAV genome (75, 80). However, none of these viral products was shown to behave as a transcriptional activator similar to that described for the E1a protein of Ad (12). A transcriptional activator was not required in these studies, probably because a low but detectable level of Rep protein synthesis can occur upon transfection of cells with a rep gene or a wt AAV2 plasmid even in the absence of helper virus (M. C. Geoffroy and A. Salvetti, unpublished data). In contrast, in our study the integrated rep gene is transcriptionally silent. In this context, our study provides the first evidence that the HSV-1 protein, ICP0, can act as a viral activator of rep gene expression. Interestingly, infection with HSVΔICP0 or AdTREICP0 alone induced the preferential synthesis of the two unspliced Rep proteins compared to the wt virus. This suggests that HSV is able to additionally provide one or more factors involved in splicing and/or the transport of spliced transcripts. Alternatively, it is possible that, when expressed in the absence of any other viral factor, ICP0 inhibits splicing. The observation that at least Rep52 was synthesized upon expression of ICP0 indicated that the effect of ICP0 extended to the p19 promoter. In contrast, no effect on the synthesis of the Cap proteins controlled by the p40 promoter was observed by Western blotting following ICP0 expression (data not shown). This preliminary result suggests that the effect of ICP0 is specific to the rep promoter. As such, it is likely that other HSV factors are required in addition to ICP0 to induce the complete expression of the AAV-2 genome. It will be interesting in the future to evaluate the effect of ICP0 combined with the other HSV helper factors already identified as important for AAV replication.
In conclusion, this study has contributed to the identification of a key HSV-1 helper factor required for the reactivation of AAV-2 rep gene expression from a latent integrated form. We consider these findings to be particularly relevant because both HSV and AAV are able to establish a latent infection and because ICP0 is a protein that has already been characterized as a central element for the switch from latent to lytic HSV infection. Additionally relevant to this point is the observation that AAV infection in humans can occur at the same time as HSV and that AAV, as HSV, has a tropism for neuronal cells (15). As such, it can be envisioned that both viruses establish a latent infection in the same cells and respond to the same viral inductor.
Acknowledgments
We thank Juergen Kleinschmidt for providing the 303.9 and 76.3 antibodies, Priscilla Schaffer for the AdTREICP0 and AdCMVrtTA viruses, Roger Everett for the ICP0-expressing constructs and the anti-ICP0 rabbit serum, and Patrick Lomonte for plasmids pEG110, pEGFXE, and pCI110. We are grateful to Caroline Colombeix for excellent technical assistance during analysis with the confocal microscope. We also thank Roger Everett and Mark Haskins for critically reading the manuscript.
This work was supported by the Association Française contre les Myopathies (AFM), Vaincre les Maladies Lysosomales (VML), Association Nantaise de Thérapie Génique (ANTG), the Fondation pour la Thérapie Génique en Pays de la Loire, and the INSERM.
REFERENCES
- 1.Berns, K. I., and C. Giraud. 1996. Biology of adeno-associated virus. Curr. Top. Microbiol. Immunol. 218:1-23. [DOI] [PubMed] [Google Scholar]
- 2.Berns, K. I., T. C. Pinkerton, G. F. Thomas, and M. D. Hoggan. 1975. Detection of adeno-associated virus (AAV)-specific nucleotide sequences in DNA isolated from latently infected Detroit 6 cells. Virology 68:556-560. [DOI] [PubMed] [Google Scholar]
- 3.Berthomme, H., S. J. Monahan, D. S. Parris, B. Jacquemont, and A. L. Epstein. 1995. Cloning, sequencing, and functional characterization of two subunits of the pseudorabies virus DNA polymerase holoenzyme: evidence for specificity of interaction. J. Virol. 69:2811-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boutell, C., and R. D. Everett. 2003. The herpes simplex virus type 1 (HSV-1) regulatory protein ICP0 interacts with and ubiquitinates p53. J. Biol. Chem. 278:36596-36602. [DOI] [PubMed] [Google Scholar]
- 5.Boutell, C., S. Sadis, and R. D. Everett. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and its isolated RING finger domain act as ubiquitin E3 ligases in vitro. J. Virol. 76:841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buller, R. M., J. E. Janik, E. D. Sebring, and J. A. Rose. 1981. Herpes simplex virus types 1 and 2 completely help adenovirus-associated virus replication. J. Virol. 40:241-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai, W., T. D. Astor, L. M. Liptak, C. Cho, D. Coen, and P. A. Schaffer. 1993. The herpes simplex virus type 1 regulatory protein ICP0 enhances replication during acute infection and reactivation from latency. J. Virol. 67:7501-7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai, W., and P. A. Schaffer. 1991. A cellular function can enhance gene expression and plating efficiency of a mutant defective in the gene for ICP0, a transactivating protein of herpes simplex virus type 1. J. Virol. 65:4078-4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carrozza, M. J., and N. A. DeLuca. 1996. Interaction of the viral activator protein ICP4 with TFIID through TAF250. Mol. Cell. Biol. 16:3085-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carter, B. J. 1990. Adeno-associated virus helper functions, p. 255-282. In P. Tijssen (ed.), Handbook of parvoviruses, vol. 1. CRC Press, Boca Raton, Fla. [Google Scholar]
- 11.Chang, L.-S., and T. Shenk. 1990. The adenovirus DNA-binding protein stimulates the rate of transcription directed by adenovirus and adeno-associated virus promoters. J. Virol. 64:2103-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang, L.-S., Y. Shi, and T. Shenk. 1989. Adeno-associated virus p5 promoter contains an adenovirus E1A-inducible element and a binding site for the major late transcription factor. J. Virol. 63:3479-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chee, A. V., P. Lopez, P. P. Pandolfi, and B. Roizman. 2003. Promyelocytic leukemia protein mediates interferon-based anti-herpes simplex virus 1 effects. J. Virol. 77:7101-7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheung, A. K., M. D. Hoggan, W. W. Hauswirth, and K. I. Berns. 1980. Integration of the adeno-associated virus genome into cellular DNA in latently infected human Detroit 6 cells. J. Virol. 33:739-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davidson, B. L., C. S. Stein, J. A. Heth, I. Martins, R. M. Kotin, T. A. Derksen, J. Zabner, A. Ghodsi, and J. A. Chiorini. 2000. Recombinant adeno-associated virus type 2, 4, and 5 vectors: transduction of variant cell types and regions in the mammalian central nervous system. Proc. Natl. Acad. Sci. USA 97:3428-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeLuca, N. A., A. M. McCarthy, and P. A. Schaffer. 1985. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J. Virol. 56:558-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dutheil, N., F. Shi, T. Dupressoir, and R. M. Linden. 2000. Adeno-associated virus site-specifically integrates into a muscle-specific DNA region. Proc. Natl. Acad. Sci. USA 97:4862-4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Everett, R. D. 1988. Analysis of the functional domains of herpes simplex virus type 1 immediate-early polypeptide Vmw110. J. Mol. Biol. 202:87-96. [DOI] [PubMed] [Google Scholar]
- 19.Everett, R. D. 1989. Construction and characterization of herpes simplex virus type 1 mutants with defined lesions in immediate early gene 1. J. Gen. Virol. 70:1185-1202. [DOI] [PubMed] [Google Scholar]
- 20.Everett, R. D. 2001. DNA viruses and viral proteins that interact with PML nuclear bodies. Oncogene 20:7266-7273. [DOI] [PubMed] [Google Scholar]
- 21.Everett, R. D. 2000. ICP0, a regulator of herpes simplex virus during lytic and latent infection. Bioessays 22:761-770. [DOI] [PubMed] [Google Scholar]
- 22.Everett, R. D., W. C. Earnshaw, J. Findlay, and P. Lomonte. 1999. Specific destruction of kinetochore protein CENP-C and disruption of cell division by herpes simplex virus immediate-early protein Vmw110. EMBO J. 18:1526-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Everett, R. D., P. Freemont, H. Saitoh, M. Dasso, A. Orr, M. Kathoria, and J. Parkinson. 1998. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J. Virol. 72:6581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Everett, R. D., and G. G. Maul. 1994. Maul HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 13:5062-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Everett, R. D., M. Meredith, and A. Orr. 1999. The ability of herpes simplex virus type 1 immediate-early protein Vmw110 to bind to a ubiquitin-specific protease contributes to its roles in the activation of gene expression and stimulation of virus replication. J. Virol. 73:417-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Everett, R. D., M. Meredith, A. Orr, A. Cross, M. Kathoria, and J. Parkinson. 1997. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 16:1519-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Everett, R. D., A. Orr, and C. M. Preston. 1998. A viral activator of gene expression functions via the ubiquitin-proteasome pathway. EMBO J. 17:7161-7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Everett, R. D., C. M. Preston, and N. D. Stow. 1991. Functional and genetic analysis of the role of Vmw110 in herpes simplex virus replication, p. 50-76. In E. K. Wagner (ed.), The control of herpes simplex virus gene expression. CRC Press, Inc., Boca Raton, Fla.
- 29.Flotte, T. R., S. A. Afione, R. Solow, M. L. Drumm, D. Markakis, W. B. Guggino, P. L. Zeitlin, and B. J. Carter. 1993. Expression of the cystic fibrosis transmembrane conductance regulator from a novel adeno-associated virus promoter J. Biol. Chem. 268:3781-3790. [PubMed] [Google Scholar]
- 30.Fraefel, C., A. G. Bittermann, H. Büeler, I. Heid, T. Bächi, and M. Ackermann. 2004. Spatial and temporal organization of adeno-associated virus DNA replication in live cells. J. Virol. 78:389-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graham, F. L., and L. Prevec. 1991. Manipulation of adenovirus vectors, p. 109-128. In E. J. Murray (ed.), Gene transfer and protocol, vol. 7. Humana Press, Inc., Clifton, N.J. [DOI] [PubMed] [Google Scholar]
- 32.Grossman, Z., E. Mendelson, F. Brok-Simoni, F. Mileguir, Y. Leitner, G. Rechavi, and B. Ramot. 1992. Detection of adeno-associated virus type 2 in human peripheral blood cells. J. Gen. Virol. 73:961-966. [DOI] [PubMed] [Google Scholar]
- 33.Gu, H., and B. Roizman. 2003. The degradation of promyelocytic leukemia and Sp100 proteins by herpes simplex virus 1 is mediated by the ubiquitin-conjugating enzyme UbcH5a. Proc. Natl. Acad. Sci. USA 100:8963-8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haberman, R. P., T. J. McCown, and R. J. Samulski. 2000. Novel transcriptional regulatory signals in the adeno-associated virus terminal repeat A/D junction element. J. Virol. 74:8732-8739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hagglund, R., and B. Roizman. 2002. Characterization of the novel E3 ubiquitin ligase encoded in exon 3 of herpes simplex virus-1-infected cell protein 0. Proc. Natl. Acad. Sci. USA 99:7889-7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hagglund, R., and B. Roizman. 2003. Herpes simplex virus 1 mutant in which the ICP0 HUL-1 E3 ubiquitin ligase site is disrupted stabilizes cdc34 but degrades D-type cyclins and exhibits diminished neurotoxicity. J. Virol. 77:13194-13202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hagglund, R., C. Van Sant, P. Lopez, and B. Roizman. 2002. Herpes simplex virus 1-infected cell protein 0 contains two E3 ubiquitin ligase sites specific for different E2 ubiquitin-conjugating enzymes. Proc. Natl. Acad. Sci. USA 99:631-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Halford, W. P., C. D. Kemp, J. A. Isler, D. J. Davido, and P. A. Schaffer. 2001. ICP0, ICP4, or VP16 expressed from adenovirus vectors induces reactivation of latent herpes simplex virus type 1 in primary cultures of latently infected trigeminal ganglion cells. J. Virol. 75:6143-6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Handa, H., and B. J. Carter. 1979. Adeno-associated virus DNA replication complexes in herpes simplex virus or adenovirus-infected cells. J. Biol. Chem. 254:6603-6610. [PubMed] [Google Scholar]
- 40.Harris, R., R. D. Everett, X. Zhu, S. Silverstein, and C. M. Preston. 1989. Herpes simplex virus type 1 immediate-early protein Vmw110 reactivates latent herpes simplex virus type 2 in an in vitro latency system. J. Virol. 63:3513-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hernandez, Y. J., J. Wang, W. G. Kearns, S. Loiler, A. Poirier, and T. R. Flotte. 1999. Latent adeno-associated virus infection elicits humoral but not cell-mediated immune responses in a nonhuman primate model. J. Virol. 73:8549-8558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hobbs, W. E., D. E. Brough, I. Kovesdi, and N. A. DeLuca. 2001. Efficient activation of viral genomes by levels of herpes simplex virus ICP0 insufficient to affect cellular gene expression or cell survival. J. Virol. 75:3391-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hobbs, W. E., and N. A. DeLuca. 1999. Perturbation of cell cycle progression and cellular gene expression as a function of herpes simplex virus ICP0. J. Virol. 73:8245-8255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horer, M., S. Weger, K. Butz, F. Hoppe-Seyler, C. Geisen, and J. A. Kleinschmidt. 1995. Mutational analysis of adeno-associated virus Rep protein-mediated inhibition of heterologous and homologous promoters. J. Virol. 69:5485-5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang, M. M., and P. Hearing. 1989. Adenovirus early region 4 encodes two gene products with redundant effects in lytic infection. J. Virol. 63:2605-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Im, D. S., and N. Muzyczka. 1989. Factors that bind to adeno-associated virus terminal repeats. J. Virol. 63:3095-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jeunwein, T. 2001. Re-SET-ting heterochromatin by histone methyltransferases. Trends Cell Biol. 11:266-273. [DOI] [PubMed] [Google Scholar]
- 48.Jordan, R., and P. A. Schaffer. 1997. Activation of gene expression by herpes simplex virus type 1 ICP0 occurs at the level of mRNA synthesis. J. Virol. 71:6850-6862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kawaguchi, Y., R. Bruni, and B. Roizman. 1997. Interaction of herpes simplex virus 1 alpha regulatory protein ICP0 with elongation factor 1δ: ICP0 affects translational machinery. J. Virol. 71:1019-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kawaguchi, Y., M. Tanaka, A. Yokoymama, G. Matsuda, K. Kato, H. Kagawa, K. Hirai, and B. Roizman. 2001. Herpes simplex virus 1 alpha regulatory protein ICP0 functionally interacts with cellular transcription factor BMAL1. Proc. Natl. Acad. Sci. USA 98:1877-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kawaguchi, Y., C. Van Sant, and B. Roizman. 1997. Herpes simplex virus 1 alpha regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J. Virol. 71:7328-7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kyostio, S. R., R. A. Owens, M. D. Weitzman, B. A. Antoni, N. Chejanovsky, and B. J. Carter. 1994. Analysis of adeno-associated virus (AAV) wild-type and mutant Rep proteins for their abilities to negatively regulate AAV p5 and p19 mRNA levels. J. Virol. 68:2947-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kyostio, S. R., R. S. Wonderling, and R. A. Owens. 1995. Negative regulation of the adeno-associated virus (AAV) P5 promoter involves both the P5 rep binding site and the consensus ATP-binding motif of the AAV Rep68 protein. J. Virol. 69:6787-6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laughlin, C. A., C. B. Cardellichio, and H. C. Coon. 1986. Latent infection of KB cells with adeno-associated virus type 2. J. Virol. 60:515-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lehman, I. R., and P. E. Boehmer. 1999. Replication of herpes simplex virus DNA. J. Biol. Chem. 274:28059-28062. [DOI] [PubMed] [Google Scholar]
- 56.Leib, D. A., D. M. Coen, C. L. Bogard, K. A. Hicks, D. R. Yager, D. M. Knipe, K. L. Tyler, and P. A. Schaffer. 1989. Immediate-early regulatory gene mutants define different stages in the establishment and reactivation of herpes simplex virus latency. J. Virol. 63:759-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leonard, C. J., and K. I. Berns. 1994. Adeno-associated virus type 2: a latent life cycle. Prog. Nucleic Acid Res. Mol. Biol. 48:29-53. [DOI] [PubMed] [Google Scholar]
- 58.Lewis, B. A., G. Tullis, E. Seto, N. Horikoshi, R. Weinmann, and T. Shenk. 1995. Adenovirus E1A proteins interact with the cellular YY1 transcription factor. J. Virol. 69:1628-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lomonte, P., and R. D. Everett. 1999. Herpes simplex virus type 1 immediate-early protein Vmw110 inhibits progression of cells through mitosis and from G1 into S phase of the cell cycle. J. Virol. 73:9456-9467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lomonte, P., K. F. Sullivan, and R. D. Everett. 2001. Degradation of nucleosome-associated centromeric histone H3-like protein CENP-A induced by herpes simplex virus type 1 protein ICP0. J. Biol. Chem. 276:5829-5835. [DOI] [PubMed] [Google Scholar]
- 61.Lopez, P., R. J. Jacob, and B. Roizman. 2002. Overexpression of promyelocytic leukemia protein precludes the dispersal of ND10 structures and has no effect on the accumulation of infectious herpes simplex virus 1 or its proteins. J. Virol. 76:9355-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marconi, P., D. Krisky, T. Oligino, P. L. Poliani, R. Ramakrishnan, W. F. Goins, D. J. Fink, and J. C. Glorioso. 1996. Replication-defective herpes simplex virus vectors for gene transfer in vivo. Proc. Natl. Acad. Sci. USA 93:11319-11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marcus-Sekura, C. J., and B. J. Carter. 1983. Chromatin-like structure of adeno-associated virus DNA in infected cells. J. Virol. 48:79-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Negorev, D., and G. G. Maul. 2001. Cellular proteins localized at and interacting with ND10/PML nuclear bodies/PODs suggest functions of a nuclear depot. Oncogene 20:7234-7242. [DOI] [PubMed] [Google Scholar]
- 65.O'Hare, P. 1993. The virion transactivator of herpes simplex virus. Semin. Virol. 44:751-760. [Google Scholar]
- 66.Parkinson, J., S. P. Lees-Miller, and R. D. Everett. 1999. Herpes simplex virus type 1 immediate-early protein vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J. Virol. 73:650-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pereira, D. J., D. M. McCarty, and N. Muzyczka. 1997. The adeno-associated virus (AAV) Rep protein acts as both a repressor and an activator to regulate AAV transcription during a productive infection. J. Virol. 71:1079-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pereira, D. J., and N. Muzyczka. 1997. The adeno-associated virus type 2 p40 promoter requires a proximal Sp1 interaction and a p19 CArG-like element to facilitate Rep transactivation. J. Virol. 71:4300-4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pereira, D. J., and N. Muzyczka. 1997. The cellular transcription factor SP1 and an unknown cellular protein are required to mediate rep protein activation of the adeno-associated virus p19 promoter. J. Virol. 71:1747-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Regad, T., and M. K. Chelbi-Alix. 2001. Role and fate of PML nuclear bodies in response to interferon and viral infections. Oncogene 20:7274-7286. [DOI] [PubMed] [Google Scholar]
- 71.Sacks, W. R., and P. A. Schaffer. 1987. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J. Virol. 61:829-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shi, Y., E. Seto, L. S. Chang, and T. Shenk. 1991. Transcriptional repression by YY1, a human GLI-Krüppel-related protein, and relief of repression by adenovirus E1A protein. Cell 67:377-388. [DOI] [PubMed] [Google Scholar]
- 73.Smith, C. A., P. Bates, R. Rivera-Gonzales, B. Gu, and N. A. DeLuca. 1993. ICP4 the major transcriptional regulatory protein of herpes simplex virus type 1 forms a tripartite complex with TATA-binding protein and TFIIB. J. Virol. 67:4676-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stow, N. D., and E. C. Stow. 1986. Isolation and characterisation of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate-early polypeptide Vmw110. J. Gen. Virol. 70:695-704. [DOI] [PubMed] [Google Scholar]
- 75.Stracker, T. H., G. D. Cassell, P. Ward, Y. M. Loo, B. van Breukelen, S. D. Carrington-Lawrence, R. K. Hamatake, P. C. van der Vliet, S. K. Weller, T. Melendy, and M. D. Weitzmann. 2004. The Rep protein of adeno-associated virus type 2 interacts with single-stranded DNA-binding proteins that enhance viral replication. J. Virol. 78:441-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Van Sant, C., R. Hagglund, P. Lopez, and B. Roizman. 2001. The infected cell protein 0 of herpes simplex virus 1 dynamically interacts with proteasomes, binds and activates the cdc34 E2 ubiquitin-conjugating enzyme, and possesses in vitro E3 ubiquitin ligase activity. Proc. Natl. Acad. Sci. USA 98:8815-8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Walz, C., and J. R. Schlehofer. 1992. Modification of some biological properties of HeLa cells containing adeno-associated virus DNA integrated into chromosome 17. J. Virol. 66:2990-3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ward, P., M. Falkenberg, P. Elias, M. Weitzman, and R. M. Linden. 2001. Rep-dependent initiation of adeno-associated virus type 2 DNA replication by a herpes simplex virus type 1 replication complex in a reconstituted system. J. Virol. 75:10250-10258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Watson, R. J., and J. B. Clements. 1980. A herpes simplex virus type 1 function continuously required for early and late virus RNA synthesis. Nature 285:329-330. [DOI] [PubMed] [Google Scholar]
- 80.Weindler, F. W., and R. Heilbronn. 1991. A subset of herpes simplex virus replication genes provides helper functions for productive adeno-associated virus replication. J. Virol. 65:2476-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wistuba, A., A. Kern, S. Weger, D. Grimm, and J. A. Kleinschmidt. 1997. Subcellular compartmentalization of adeno-associated virus type 2 assembly. J. Virol. 71:1341-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wistuba, A., S. Weger, A. Kern, and J. A. Kleinschmidt. 1995. Intermediates of adeno-associated virus type 2 assembly: identification of soluble complexes containing Rep and Cap proteins. J. Virol. 69:5311-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yakobson, B., T. A. Hrynko, M. J. Peak, and E. Winocour. 1989. Replication of adeno-associated virus in cells irradiated with UV light at 254 nm. J. Virol. 63:1023-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yakobson, B., T. Koch, and E. Winocour. 1987. Replication of adeno-associated virus in synchronized cells without the addition of a helper virus. J. Virol. 61:972-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yalkinoglu, A. O., R. Heilbronn, A. Burkle, J. R. Schlehofer, and H. zur Huasen. 1988. DNA amplification of adeno-associated virus as a response to cellular genotxic stress. Cancer Res. 48:3123-3129. [PubMed] [Google Scholar]
- 86.Yao, F., and P. A. Schaffer. 1995. An activity specified by the osteosarcoma line U2OS can substitute functionally for ICP0, a major regulatory protein of herpes simplex virus type 1. J. Virol. 69:6249-6258. [DOI] [PMC free article] [PubMed] [Google Scholar]