Abstract
White spot syndrome virus (WSSV) virions were purified from the hemolymph of experimentally infected crayfish Procambarus clarkii, and their proteins were separated by 8 to 18% gradient sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) to give a protein profile. The visible bands were then excised from the gel, and following trypsin digestion of the reduced and alkylated WSSV proteins in the bands, the peptide sequence of each fragment was determined by liquid chromatography-nano-electrospray ionization tandem mass spectrometry (LC-nanoESI-MS/MS) using a quadrupole/time-of-flight mass spectrometer. Comparison of the resulting peptide sequence data against the nonredundant database at the National Center for Biotechnology Information identified 33 WSSV structural genes, 20 of which are reported here for the first time. Since there were six other known WSSV structural proteins that could not be identified from the SDS-PAGE bands, there must therefore be a total of at least 39 (33 + 6) WSSV structural protein genes. Only 61.5% of the WSSV structural genes have a polyadenylation signal, and preliminary analysis by 3′ rapid amplification of cDNA ends suggested that some structural protein genes produced mRNA without a poly(A) tail. Microarray analysis showed that gene expression started at 2, 6, 8, 12, 18, 24, and 36 hpi for 7, 1, 4, 12, 9, 5, and 1 of the genes, respectively. Based on similarities in their time course expression patterns, a clustering algorithm was used to group the WSSV structural genes into four clusters. Genes that putatively had common or similar roles in the viral infection cycle tended to appear in the same cluster.
White spot syndrome virus (WSSV), one of the most devastating viral pathogens of cultured shrimp worldwide, is an enveloped, ellipsoid, large double-stranded DNA virus (15, 34, 49) that also attacks crabs and crayfish as well as many other crustaceans (9, 23, 24, 26). Complete genome sequencing has been performed on three WSSV isolates (5, 6, 47, 56). At ∼300 kbp, the WSSV genome is ∼500 kbp smaller than the giant Mimivirus that infects amoeba (19) and ∼30 kb smaller than the 335,593-bp genome of the Ectocarpus siliculosus virus (EsV-1; family Phycodnaviridae), which is the largest virus genome sequenced to date (43). Studies on the general and molecular characteristics of WSSV (15, 25, 30, 53) and analysis of the complete genome sequence (47, 56) suggest that WSSV does not belong to any known virus family. Recently, the International Committee on Taxonomy of Viruses approved a proposal to erect WSSV as the type species of the genus Whispovirus, family Nimaviridae (www.ncbi.nlm.nih.gov/ICTVdb/Ictv/index.htm). The family name reflects the most notable physical feature of the virus, which is a tail-like projection extending from one end of the WSSV virion.
Because of the large size of the genome and the uniqueness of the proteins that the WSSV open reading frames (ORFs) encode, WSSV has not yet been fully characterized. To date only a few WSSV genes have been studied beyond sequence analysis (13, 14, 17, 20-22, 40-42, 58-60).
In characterizing any virus, its structural proteins are particularly important because these proteins are the first molecules to interact with the host and they therefore play critical roles in cell targeting as well as triggering host defenses. However, it is not always easy to exhaustively identify every structural protein with conventional tools alone. In the case of WSSV, the virions have a complicated sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) protein profile, and so far, SDS-PAGE coupled with Western blotting and/or protein N-terminal sequencing has identified only six structural proteins: VP35, VP28, VP26, VP24, VP19, and VP15 (5, 11, 44, 45). A more comprehensive, global approach is provided by proteomics. In the field of functional genomics, proteomics is defined as the large-scale analysis of the function of genes (37). For this kind of analysis, proteomics combines mass spectrometry with database searches of sequenced genomes. Viral structural proteins are particularly amenable to this approach, since the complete genome sequences of many viruses are known, and viral particles consist of a relatively narrow range of proteins that have constant, stable profiles. Further, for any purified virus, all or nearly all of the proteins that appear in SDS-PAGE are virally encoded. Using this approach, a previous study that combined SDS-PAGE separation with mass spectrometry (14) was able to identify 13 new WSSV structural proteins. However, we hypothesized that many more WSSV structural proteins remained to be discovered, not least because the protein profiles produced by the wide-range gradient SDS-PAGE used in our laboratory consistently contained many more (major) bands. In the present study, we therefore applied proteomic technology not only to rule out the possibility that these bands were the result of contamination, but also to map the complete protein profile and positively identify another 20 previously unknown structural protein genes of WSSV. Because of its completeness and consistency, the WSSV SDS-PAGE protein profile (from crayfish Procambarus clarkii) reported here can also be used as a reference for comparison with the protein profiles of WSSV virions from other hosts. In the present study, all 39 structural protein genes were also subjected to microarray analysis. Temporal gene transcription profiles were generated for each structural protein, and a clustering algorithm was then used to arrange these profiles into four functionally meaningful groups.
MATERIALS AND METHODS
Virus.
The virus used in this study, WSSV Taiwan isolate, was originally isolated from a batch of WSSV-infected Penaeus monodon shrimp collected in Taiwan in 1994 (27, 49). The entire genome sequence of this isolate has been deposited in GenBank under accession no. AF440570.
Proliferation and preparation of intact WSSV virions.
Gill and epithelium tissues from WSSV-infected shrimp (P. monodon; mean weight of 10 g) were homogenized in TNE buffer (50 mM Tris-HCl, 0.1 M NaCl, 1 mM EDTA, pH 7.5) at 0.25 g/ml. After centrifugation at 1,500 × g for 10 min, the supernatant was filtered (0.45-μm-pore-diameter filter) and injected (0.2 ml; 1:10 dilution in TNE) intramuscularly into healthy crayfish P. clarkii between the second and third abdominal segments. Beginning 4 to 6 days later, hemolymph was extracted from the infected crayfish and then centrifuged at 1,500 × g for 10 min. The supernatant was layered on top of a 35% (wt/vol in TNE) sucrose solution and centrifuged at 89,000 × g using a 28 SA rotor in a Hitachi ultracentrifuge (SCP85H2) for 1 h at 4°C. The virus pellet was resuspended with TNE and then subjected to a linear 35 to 65% sucrose gradient centrifugation following the protocols described previously (49). The collected virus band was then mixed with TNE buffer and repelleted at 89,000 × g for 1 h at 4°C. The resulting pellet was again dissolved in TNE. To check for quality and quantity, virus samples were negatively stained with 2% sodium phosphotungstate and examined under a transmission electron microscope (Hitachi).
In-gel digestion for protein identification.
The proteins from purified virions were separated by 8 to 18% gradient SDS-PAGE and stained with Sypro Ruby. Protein bands were manually excised from the gel and cut into pieces. The gel pieces were dehydrated with acetonitrile for 10 min; vacuum dried; rehydrated with 55 mM DTE in 25 mM ammonium bicarbonate, pH 8.5, at 37°C for 1 h; and subsequently alkylated with 100 mM iodoacetamide in 25 mM ammonium bicarbonate, pH 8.5, at room temperature for 1 h. The pieces were then washed twice with 50% acetonitrile in 25 mM ammonium bicarbonate, pH 8.5, for 15 min each time, dehydrated with acetonitrile for 10 min, vacuum dried, and rehydrated with a total of 25 ng of sequencing-grade, modified trypsin (Promega, Madison, Wis.) in 25 mM ammonium bicarbonate, pH 8.5, at 37°C for 16 h. Following digestion, tryptic peptides were extracted twice with 50% acetonitrile containing 5% formic acid for 15 min each time with moderate sonication. The extracted solutions were pooled and evaporated to dryness under vacuum.
LC-nanoESI-MS/MS analysis.
Liquid chromatography-nano-electrospray ionization tandem mass spectrometry (LC-nanoESI-MS/MS) was performed as follows. Selected bands were submitted to an integrated Micromass nano-LC-MS/MS system operated under MassLynx 4.0 control. This system comprised a three-pump Micromass/Waters CapLC system with an autosampler, a stream select module configured for the precolumn plus analytical capillary column, and a Micromass quadrupole/time of flight Ultima API mass spectrometer fitted with nano-LC sprayer. Injected samples were first trapped and desalted isocratically on an LC-Packings PepMap C18 μ-Precolumn cartridge (5 μm, 300-μm inside diameter by 5 mm; Dionex, Sunnyvale, Calif.) for 2 min with 0.1% formic acid delivered by the auxiliary pump at 30 μl/min, after which the peptides were eluted from the precolumn and separated on an analytical C18 capillary column (15 cm by 75 μm inside diameter, packed with 5-μm Zorbax 300 SB C18 particles; Micro-Tech Scientific, Vista, Calif.) connected inline to the mass spectrometer, at 300 nl/min using a 40-min fast gradient of 5 to 80% acetonitrile in 0.1% formic acid.
On-line nanoESI-MS survey scan and data-dependent acquisition of collision-induced dissociation (CID) MS/MS were fully automated and synchronized with the nano-LC runs under the full software control of MassLynx 4.0. Prior to on-line analysis, the nano-LC sprayer and Z-spray source parameters were tuned and optimized with glufibrinopeptide B. Argon was used as the collision gas for CID MS/MS. Calibration was performed using the product ions generated from fragmentation of the doubly charged molecular ion of glufibrinopeptide B at m/z 785.8. For routine protein identification analysis, 1-s survey scans were run over the mass range m/z 400 to 2,000. A maximum of three concurrent MS/MS acquisitions were triggered for 2+, 3+, and 4+ charged precursors detected at an intensity above the predefined threshold (20 counts/s). When the precursor intensity fell below a predefined threshold, or after a maximum of 6 s of acquisition, the system switched back from acquisition mode to survey scan mode.
After data acquisition, the individual MS/MS spectra acquired for each of the precursors within a single LC run were combined, smoothed, deisotoped (fast option, inclusive of simple transformation of multiply charged peaks), and centroided using the Micromass ProteinLynx Global Server (PGS) 2.0 data processing software and output as a single Mascot-searchable peak list file. The peak list files were used to query the nonredundant database at the National Center for Biotechnology Information (NCBInr) using the Mascot program with the following parameters: peptide mass tolerance, 50 ppm; MS/MS ion mass tolerance, 0.25 Da; allowing up to one missed cleavage. Variable modifications considered were methionine oxidation and cysteine carboxyamidomethylation. Both significant hits (as defined by Mascot probability analysis) and hits that exceeded the arbitrarily set acceptance threshold (a peptide ion matching score of more than 20) were regarded as positive identifications.
Homology searches for the WSSV structural proteins.
The amino acid sequences of the WSSV structural proteins were analyzed by BLAST homology search against the NCBInr (1), InterProScan protein domain/functional site search against the protein signatures of the InterPro database (31), and the HMMpfam protein domain families in the Pfam database (3). Putative transmembrane domains in each structural protein sequence were predicted by TMpred using the TMbase database (12). Signal peptide prediction for each structural protein was performed using SignalP V1.1 (35) against the SwissProt database (2), with each search submission consisting of less than 80 amino acids taken from the N terminus of the protein (as deduced from the ORF).
3′ RACE analysis of structural genes.
To check for the existence of a poly(A) tail and to sequence the 3′ untranslated region (UTR) of the structural protein genes, 3′ rapid amplification of cDNA ends (RACE) was performed for 17 structural genes (10 of these genes had the polyadenylation signal and 7 did not). Total RNA was isolated from the WSSV-infected shrimp (P. monodon) 60 h postinfection (hpi), and 3′ RACE was performed with the the oligo(dT)-anchor primer and a primer specific to each gene following the protocol described previously (40).
Structural gene expression analysis by WSSV DNA microarray.
To identify changes in gene expression during the WSSV infection cycle, we constructed a viral microarray containing the 532 WSSV ORFs that consist of 60 amino acids or more. For PCR, specific primers were designed for each ORF, with amplicon sizes from 200 to 600 bp. The PCR products amplified from each ORF were then purified, applied as spots to the glass slide, and used as probes for the DNA microarray. In this array, each ORF was represented by triplicate spots. Actin gene (P. monodon) control spots (six replications) were used to normalize the data across different slides. For the target RNA, healthy subadult P. monodon shrimp (body weight of 15 to 20 g) were experimentally infected with WSSV by the methods described by Liu et al. (22). Total RNA from the gill tissues of the WSSV-infected shrimp was isolated 0, 2, 4, 6, 8, 12, 18, 24, and 36 hpi. Total RNA (20 μg in each reaction) was then reverse transcribed into fluorescently labeled cDNA in the presence of Cy3-dUTP using a CyScribe First-Strand cDNA labeling kit (Amersham). [Please note that in addition to the kit's oligo(T) primer, which is designed to amplify RNA with a poly(A) tail, low-molecular-weight cellular RNAs that were present in the reaction mixture also served as primers for cDNA synthesis, so that RNA without a poly(A) tail could also be reverse transcribed into cDNA; see reference 10.] The Cy3-labeled cDNA was condensed and separated from unincorporated nucleotides with Microcon YM-30 columns (Amicon). The labeled cDNAs (used here as the target) were then subjected to hybridization with all the DNA spots (i.e., the probes) in the WSSV microarray. The microarrays were then scanned with a confocal laser ScanArray 3000 system to measure the fluorescence intensities in each spot, and fluorescent intensities were quantified by Imagen 4.0 (Biodiscovery, Inc.). Because of scan setting and hybridization efficiencies, normalization between arrays was necessary, and the shrimp actin gene was used as the normalization factor. Results are reported as the calibrated expression ratio, which is the ratio of the fluorescence intensity of a WSSV structural gene transcript in infected shrimp gill cells compared to that of the universal control probe (actin gene). A plot of the calibrated expression ratios for each gene over the time course of infection was then used in conjunction with an average linkage hierarchical clustering algorithm, Hierarchical Clustering Explorer 2.0 (39), using the uncentered Pearson correlation coefficient as the similarity metric. After clustering, each gene was allocated its place in a global temporal classification scheme.
RESULTS
SDS-PAGE profile of purified virion structural proteins.
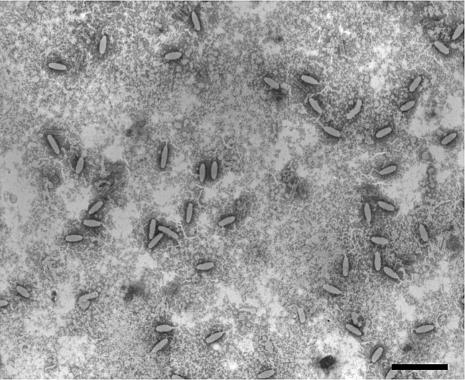
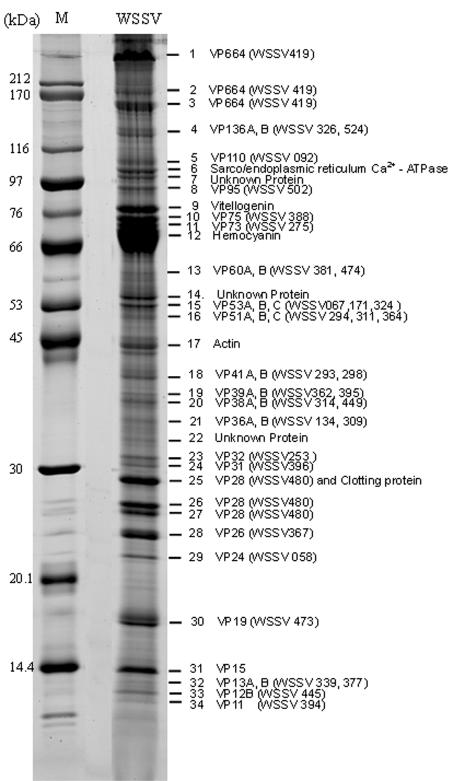
Figure 1 shows that most of the purified WSSV virions were intact. Analysis of the structural proteins of these intact viral particles by 8 to 18% gradient SDS-PAGE revealed the presence of at least 34 protein bands that were consistently visible when stained with Sypro Ruby (Fig. 2).
FIG. 1.
Electron micrograph of intact negatively stained WSSV virions purified from hemolymph of Procambarus clarkii. Scale bar = 500 nm.
FIG. 2.
Sypro Ruby-stained SDS-PAGE of the structural proteins prepared from intact WSSV virions. An 8 to 18% gradient gel was used for electrophoresis. Numbers indicate the excised bands. M, protein molecular mass marker (kilodaltons).
Many of the major bands in Fig. 2 had a greater apparent molecular mass than crayfish hemocyanin (the very-high-intensity band at about 70 kDa). The apparent molecular mass of one of the major bands in our profile (band 1) was much greater than the molecular mass marker of 212 kDa. Although some of the bands were identified as host protein(s), this WSSV virion protein profile is more detailed than any that have been previously published. This was due to our improved protocol for the preparation of large amounts of intact virions and also to our use of a wide-range gradient gel that provided a better separation of the proteins.
Identification of WSSV proteins by mass spectrometry.
After separation of the WSSV virion proteins by gradient SDS-PAGE, 34 bands were excised from the gel. Following trypsin digestion of the reduced and alkylated WSSV proteins, the peptides of each band were sequenced using LC-nanoESI-MS/MS. To identify the proteins, the peptide sequences obtained from the MS/MS data were searched for in the NCBInr database using the Mascot server. Thirty-three of the 34 excised bands consisted mostly of proteins with amino acid sequences that matched WSSV ORFs with coverages from <1 to 44% (Table 1). (Note that even though coverage was sometimes low, the unique sequences of the WSSV ORFs mean that positive identifications could still be made with confidence.) Proteins encoded by WSSV480 and WSSV419 were found in more than one band, while some bands contained more than one protein (Fig. 2; Table 1). Several bands (bands 6, 9, 12, and 17) showed homology to crustacean proteins (sarco/endoplasmic reticulum Ca2+-ATPase, vitellogenin, hemocyanin, and actin, respectively) held in the NCBInr database. Several other minor bands (7, 14, 22) contained proteins that could not be identified.
TABLE 1.
WSSV structural ORFs including partial annotation
| VP no. | Namea | WSSV ORFb | Band no.c | Sequence coverage (%) | ORF size
|
Apparent VP size (mol mass [kDa])e | RGD motiff | TM domainsg | SP sequenceh | Reference | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Amino acid | Mol mass (kDa)d | ||||||||||
| 1 | VP664 | WSSV419 | 1, 2, 3 | 14 | 6077 | 664 | ∼664, 186, 161 | + | This study | ||
| 2 | VP180 | WSSV052 | 1684 | 169 | + | 14 (VP1684) | |||||
| 3 | VP136A | WSSV326 | 4 | 9 | 1219 | 135 | 136 | + | + | This study | |
| 4 | VP136B | WSSV524 | 4 | <1 | 1243 | 138 | 136 | + | This study | ||
| 5 | VP110 | WSSV092 | 5 | 4 | 972 | 108 | 110 | + | + | + | This study |
| 6 | VP95 | WSSV502 | 8 | 10 | 800 | 89.4 | 95 | + | 14 (VP800) | ||
| 7 | VP75 | WSSV388 | 10 | 1 | 786 | 87.6 | 75 | + | This study | ||
| 8 | VP73 | WSSV275 | 11 | 1 | 674 | 76.2 | 73 | + | 14 (VP674) | ||
| 9 | VP60A | WSSV381 | 13 | 11 | 465 | 51.1 | 60 | + | + | This study | |
| 10 | VP60B | WSSV474 | 13 | 2 | 544 | 61.8 | 60 | + | 14 (VP544) | ||
| 11 | VP55 | WSSV051 | 448 | 50.2 | + | 14 (VP448) | |||||
| 12 | VP53A | WSSV067 | 15 | 5 | 1301 | 144 | 53 | + | + | This study | |
| 13 | VP53B | WSSV171 | 15 | 1 | 968 | 108 | 53 | + | This study | ||
| 14 | VP53C | WSSV324 | 15 | 3 | 489 | 56.3 | 53 | + | This study | ||
| 15 | VP51A | WSSV294 | 16 | 6 | 486 | 51.5 | 51 | + | + | This study | |
| 16 | VP51B | WSSV311 | 16 | 19 | 384 | 43.2 | 51 | + | + | 14 (VP384) | |
| 17 | VP51C | WSSV364 | 16 | 21 | 466 | 51.9 | 51 | 14 (VP466) | |||
| 18 | VP41A | WSSV293 | 18 | 16 | 292 | 33.2 | 41 | + | 14 (VP292) | ||
| 19 | VP41B | WSSV298 | 18 | 8 | 300 | 34.4 | 41 | 14 (VP300) | |||
| 20 | VP39A | WSSV362 | 19 | 2 | 419 | 47.5 | 39 | This study | |||
| 21 | VP39B | WSSV395 | 19 | 12 | 283 | 32 | 39 | This study | |||
| 22 | VP38A | WSSV314 | 20 | 8 | 309 | 35.5 | 38 | This study | |||
| 23 | VP38B | WSSV449 | 20 | 4 | 321 | 35.8 | 38 | This study | |||
| 24 | VP36A | WSSV134 | 21 | 3 | 297 | 33.1 | 36 | + | + | This study | |
| 25 | VP36B | WSSV309 | 21 | 4 | 281 | 31.6 | 36 | + | 13, 14 (VP281) | ||
| 26 | VP35 | WSSV019 | 228 | 26.3 | 5, 6 | ||||||
| 27 | VP32 | WSSV253 | 23 | 3 | 278 | 31.4 | 32 | This study | |||
| 28 | VP31 | WSSV396 | 24 | 16 | 261 | 30 | 31 | + | This study | ||
| 29 | VP28 | WSSV480 | 25, 26, 27 | 44 | 204 | 22.1 | 29, 27, 26 | + | + | 46, 47 | |
| 30 | VP26 | WSSV367 | 28 | 38 | 204 | 22.2 | 24 | + | + | 45 | |
| 31 | VP24 | WSSV058 | 29 | 5 | 208 | 23.2 | 22 | + | + | 45 | |
| 32 | VP22 | WSSV359 | 891 | 100 | + | 14 (VP184) | |||||
| 33 | VP19 | WSSV473 | 30 | 9 | 121 | 13.2 | 18 | + | + | 48 | |
| 34 | VP15 | WSSV269 | 31 | 80 | 9.2 | 48 | |||||
| 35 | VP13A | WSSV339 | 32 | 16 | 100 | 11.1 | 13 | + | This study | ||
| 36 | VP13B | WSSV377 | 32 | 11 | 117 | 13.1 | 13 | + | + | This study | |
| 37 | VP12B | WSSV445 | 33 | 19 | 68 | 6.8 | 12 | + | 14 (VP68) | ||
| 38 | VP12A | WSSV065 | 95 | 11 | 14 (VP95) | ||||||
| 39 | VP11 | WSSV394 | 34 | 1 | 433 | 48.2 | 11 | + | + | This study | |
The names used here are based on the apparent molecular mass of the protein on SDS-PAGE. When a protein has previously been referred to by another name, the alternative name is given in parentheses in the reference column. For example, VP180 corresponds to VP1684 in reference 14.
Based on the genome of the Taiwan isolate; GenBank accession no. AF440570.
Band numbers are as shown in Fig. 2.
Predicted molecular mass based on the ORF.
The sizes (molecular mass) of the proteins of the WSSV virion were estimated after separation on SDS-PAGE.
The presence of the putative cell attachment domain is shown by +.
The presence of putative transmembrane (TM) domains is shown by +.
The presence of a putative SP sequence is shown by +.
Characteristics of the WSSV structural genes.
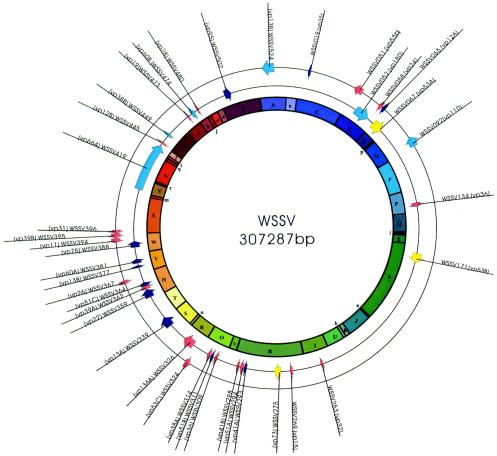
The complete DNA sequence of the WSSV genome of the Taiwan isolate can be assembled into a circular sequence of 307,287 bp (Fig. 3). Of the 532 ORFs that start with an ATG initiation codon and are at least 60 amino acids long, 39 have now been identified as WSSV structural genes (Tables 1 and 2). In some regions of the genome, there are many structural genes, while other regions are quite sparsely populated (Fig. 3). The characterizations and annotations of these structural proteins are presented in Table 2. Altogether, of the 39 structural proteins, 25 include predicted transmembrane domains, 6 have an RGD motif, and 12 have a predicted signal peptide (SP) sequence (Table 1). 82.1% (32 of 39) of the structural genes have a typical TATA box (TATAAA, TATA, or TATAA) in the predicted promoter region. A total of 76.9% (30 of 39) of the structural genes have the initiator (Inr) motif A/TCAC/G/TT, and 83.3% of these putative Inrs are downstream of the TATA box. Twenty-three of the WSSV structural genes also provide a favorable context for efficient eukaryotic translation initiation (PuNNATGPu) (18). A polyadenylation signal sequence (AATAAA), which is found either within the ORF or after the stop codon for only 28.2% of the 532 known WSSV ORFs, was also found in quite a high proportion of the WSSV structural genes (61.5% or 24 of 39; Table 2).
FIG. 3.
Distribution of the 39 WSSV structural protein genes in the WSSV genome. Colors indicate assignment to the corresponding clusters in Fig. 4. The inner circle shows predicted HindIII fragments.
TABLE 2.
5′ and 3′ UTR sequence analysis and microarray transcriptional expression levels of the WSSV structural genes
| VP no. | Name | WSSV ORF | TATA boxa | Inr motifb | Kozak contextc | Poly(A) signald | Transcriptional expression levele |
|---|---|---|---|---|---|---|---|
| 1 | VP664 | WSSV419 | Δ | S | + | L | |
| 2 | VP180 | WSSV052 | Δ | + | S | + | L |
| 3 | VP136A | WSSV326 | Δ | + | S | H | |
| 4 | VP136B | WSSV524 | + | + | W | + | M |
| 5 | VP110 | WSSV092 | + | S | L | ||
| 6 | VP95 | WSSV502 | Δ | + | W | + | L |
| 7 | VP75 | WSSV388 | Δ | S | + | M | |
| 8 | VP73 | WSSV275 | Δ | S | L | ||
| 9 | VP60A | WSSV381 | + | S | + | M | |
| 10 | VP60B | WSSV474 | W | M | |||
| 11 | VP55 | WSSV051 | Δ | + | S | + | L |
| 12 | VP53A | WSSV067 | + | W | + | L | |
| 13 | VP53B | WSSV171 | Δ | S | + | L | |
| 14 | VP53C | WSSV324 | Δ | + | S | H | |
| 15 | VP51A | WSSV294 | Δ | + | W | + | M |
| 16 | VP51B | WSSV311 | + | + | S | + | M |
| 17 | VP51C | WSSV364 | + | + | W | + | H |
| 18 | VP41A | WSSV293 | Δ | + | W | M | |
| 19 | VP41B | WSSV298 | + | S | L | ||
| 20 | VP39A | WSSV362 | Δ | + | W | L | |
| 21 | VP39B | WSSV395 | + | W | L | ||
| 22 | VP38A | WSSV314 | Δ | + | W | + | L |
| 23 | VP38B | WSSV449 | Δ | + | W | L | |
| 24 | VP36A | WSSV134 | + | + | S | + | H |
| 25 | VP36B | WSSV309 | Δ | + | S | + | H |
| 26 | VP35 | WSSV019 | + | + | S | + | M |
| 27 | VP32 | WSSV253 | Δ | + | W | L | |
| 28 | VP31 | WSSV396 | Δ | + | W | L | |
| 29 | VP28 | WSSV480 | Δ | + | S | + | H |
| 30 | VP26 | WSSV367 | + | S | + | H | |
| 31 | VP24 | WSSV058 | Δ | + | W | + | M |
| 32 | VP22 | WSSV359 | + | + | S | M | |
| 33 | VP19 | WSSV473 | + | + | S | + | H |
| 34 | VP15 | WSSV269 | + | + | W | + | H |
| 35 | VP13A | WSSV339 | + | S | M | ||
| 36 | VP13B | WSSV377 | Δ | + | S | M | |
| 37 | VP12B | WSSV445 | + | W | + | M | |
| 38 | VP12A | WSSV065 | + | S | + | H | |
| 39 | VP11 | WSSV394 | Δ | + | S | + | M |
The presence of a TATA box (TATAAA) is indicated by +. The presence of a truncated sequence (TATA or TATAA) is indicated by Δ.
The presence of an Inr motif (A/TCAC/G/TT) upstream of the translation start codon (ATG) is shown by +.
The Kozak context (PuNNATGPu) of the initiation codon is either S (strong) or W (weak).
The presence of a 3′ UTR poly(A) signal (AATAAA) is indicated by +.
Microarray transcriptional expression levels are shown as follows: low (L), baseline mRNA levels increased by a factor of between 3 and 15; medium (M), mRNA increased by a factor of 15 to 60; high (H), increased by a factor of 60 to >300.
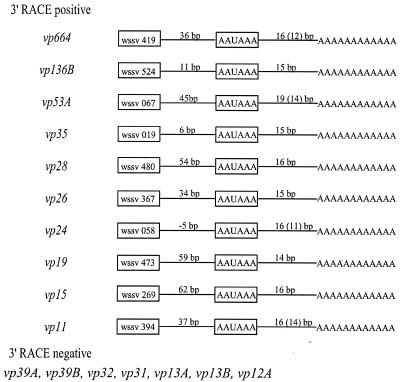
For several structural genes that either had or did not have the poly(A) signal sequence, 3′ RACE was used to confirm empirically the presence or absence of a poly(A) tail. We found that 3′ RACE was unable to amplify the mRNA from any gene that lacked the poly(A) signal sequence. Conversely 3′ RACE successfully amplified every gene that had a polyadenylation signal sequence. For these genes, the polyadenylation started 15 to 19 nucleotides downstream of the poly(A) signal, as shown in Fig. 4. This is within the range of regular poly(A) addition in eukaryotic mRNAs, i.e., 15 to 25 nucleotides downstream of the poly(A) signal (8). The polyadenylation sites for at least five structural protein genes from WSSV-infected P. monodon (vp28, vp26, vp24, vp19, and vp15) are in agreement with the 3′ ends of these genes as determined from RNA isolated from WSSV-infected crayfish (Orconectes limosus) (28).
FIG. 4.
3′ UTR sequencing results based on 3′ RACE of 17 structural protein genes expressed in P. monodon. 3′ RACE-positive genes were those genes whose mRNA was successfully amplified using an oligo(dT) anchor and gene-specific primers. All of these genes had the poly(A) signal sequence. 3′ RACE-negative genes were those whose mRNA could not be amplified. All of these genes lacked the poly(A) signal sequence. Values in parentheses represent the inner end of the poly(A) tail as determined by 3′ RACE, but this does not take into account the fact that the transcript already includes two to five adenosine nucleotides upstream of the actual poly(A) addition site.
When InterProScan and HMMpfam were used to scan the protein sequences of the 39 WSSV structural proteins against the protein domain and functional site databases (InterPro and Pfam, respectively), a putative function was identified for only 12 proteins. These 12 proteins and their inferred functions are listed in Table 3. It is interesting to note that VP60B has a sequence region that is homologous to an adenovirus fiber protein (knob domain). In the adenovirus protein, this domain is involved in specific attachment to the host (55) and is thus a critical factor for virulence and host range.
TABLE 3.
Additional putative functions of 12 of the WSSV structural proteins
| Protein | WSSV ORF | Domain and signature scan result
|
||
|---|---|---|---|---|
| Entry name | Database/accession no. | Inferred function | ||
| VP664 | WSSV419 | Krr1 family | InterPro/IPR007851 | 40S ribosome biogenesis |
| VP180a | WSSV052 | S-Adenosylmethionine synthetase, C-terminal domain | Pfam/PF02773 | ATP binding |
| VP136A | WSSV326 | 3-Octaprenyl-4-hydroxybenzoate carboxy-lyase | InterPro/IPR002830 | Ubiquinone biosynthesis |
| VP73b | WSSV275 | Long hematopoietin receptor, gp130 family 2 | InterPro/IPR003529 | Signaling |
| VP60B | WSSV474 | Adenoviral fiber protein (knob domain) | InterPro/IPR000978 | Virus-specific attachment |
| VP53B | WSSV171 | Ligand-gated ion channel | InterPro/IPR001320 | Glutamate-gated ion channel activity |
| VP51A | WSSV294 | Eggshell protein | InterPro/IPR002952 | Structural component |
| VP38B | WSSV449 | Restriction endonuclease FokI, recognition domain | InterPro/IPR004234 | DNA binding |
| VP36B | WSSV309 | Photosystem II reaction center protein psbp | InterPro/IPR002683 | Photoactivation |
| VP35 | WSSV019 | Calx-beta domain | InterPro/IPR003644 | Domain found in Na/Ca exchangers |
| VP22 | WSSV359 | Transcription factor TFIID | InterPro/IPR000814 | Transcription |
| VP13A | WSSV339 | Photosystem II reaction center M protein (PsbM) | InterPro/IPR007826 | Photoactivation |
Structural gene expression analysis by WSSV DNA microarray.
WSSV structural gene transcription data from the microarrays are presented in Table 4. This table shows the times at which a threefold increase in expression (TF) was seen for each gene with respect to its baseline expression at 0 hpi. TF values provide a reliable indicator of when a particular gene commenced expression. Of the 39 WSSV structural genes, there were 7 genes that commenced expression at 2 hpi, 1 gene that commenced expression at 6 hpi, 4 genes that commenced expression at 8 hpi, 12 genes that commenced expression at 12 hpi, 9 genes that commenced expression at 18 hpi, 5 genes that commenced expression at 24 hpi, and 1 gene that commenced expression at 36 hpi. Table 4 also shows the time of greatest expression for each gene. On these arrays, all of the WSSV structural genes expressed maximally at the late stage, with maximum expression levels either at 24 hpi (41.02%) or at 36 hpi (58.98%).
TABLE 4.
Expression of WSSV structural genes during the WSSV infection cycle
| Gene no. | WSSV ORF | Name | Expression (hpi)
|
Calibrated expression ratio at time (hpi) showna
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TF | Peak | 0 | 2 | 4 | 6 | 8 | 12 | 18 | 24 | 36 | |||
| 1 | 419 | VP664 | 18 | 24 | 0.0144 | 0.0303 | 0.0153 | 0.0343 | 0.0185 | 0.0346 | 0.0579 | 0.1044 | 0.0569 |
| 2 | 52 | VP180 | 24 | 24 | 0.0217 | 0.0243 | 0.0096 | 0.0357 | 0.0446 | 0.0596 | 0.049 | 0.1456 | 0.0971 |
| 3 | 326 | VP136A | 12 | 36 | 0.0097 | 0.0172 | 0.0065 | 0.0183 | 0.0126 | 0.0284 | 0.1453 | 0.6745 | 0.8101 |
| 4 | 524 | VP136B | 12 | 24 | 0.0059 | 0.0158 | 0.002 | 0.0083 | 0.0134 | 0.0178 | 0.0405 | 0.1012 | 0.0535 |
| 5 | 92 | VP110 | 12 | 24 | 0.0089 | 0.0145 | 0.0063 | 0.0161 | 0.0143 | 0.0279 | 0.0158 | 0.0453 | 0.0297 |
| 6 | 502 | VP95 | 18 | 24 | 0.0091 | 0.0059 | 0.0081 | 0.0066 | 0.0083 | 0.0128 | 0.0266 | 0.0606 | 0.0485 |
| 7 | 388 | VP75 | 12 | 24 | 0.0022 | 0.0011 | 0 | 0.0032 | 0.005 | 0.0101 | 0.0936 | 0.1056 | 0.0655 |
| 8 | 275 | VP73 | 2 | 36 | 0.0016 | 0.0047 | 0 | 0.0019 | 0.002 | 0.0031 | 0.0025 | 0.0044 | 0.0075 |
| 9 | 381 | VP60A | 6 | 24 | 0.0038 | 0.0079 | 0.0054 | 0.0145 | 0.0058 | 0.0212 | 0.0321 | 0.0712 | 0.0589 |
| 10 | 474 | VP60B | 8 | 24 | 0.0075 | 0.0189 | 0.0038 | 0.0296 | 0.0219 | 0.0305 | 0.0273 | 0.1235 | 0.0489 |
| 11 | 51 | VP55 | 2 | 36 | 0.0047 | 0.0157 | 0.0077 | 0.0089 | 0.0102 | 0.0207 | 0.0282 | 0.0532 | 0.0711 |
| 12 | 67 | VP53A | 2 | 24 | 0.0076 | 0.0587 | 0.0217 | 0.034 | 0.0392 | 0.0288 | 0.0159 | 0.0713 | 0.0578 |
| 13 | 171 | VP53B | 8 | 36 | 0.0072 | 0.0204 | 0.0068 | 0.0157 | 0.0238 | 0.0147 | 0.0117 | 0.0378 | 0.0419 |
| 14 | 324 | VP53C | 2 | 36 | 0.0005 | 0.0018 | 0.0034 | 0.001 | 0.0044 | 0.0033 | 0.0138 | 0.0195 | 0.034 |
| 15 | 294 | VP51A | 8 | 36 | 0.0011 | 0.0022 | 0.0017 | 0 | 0.0034 | 0.0047 | 0.009 | 0.0134 | 0.0242 |
| 16 | 311 | VP51B | 18 | 24 | 0.0016 | 0.0009 | 0.0003 | 0.0046 | 0.0022 | 0.0027 | 0.0372 | 0.058 | 0.0547 |
| 17 | 364 | VP51C | 2 | 36 | 0 | 0.0052 | 0.0012 | 0.0007 | 0.0037 | 0.0034 | 0.0214 | 0.0334 | 0.0346 |
| 18 | 293 | VP41A | 18 | 24 | 0.002 | 0.0014 | 0.0015 | 0.0002 | 0 | 0.0045 | 0.0146 | 0.0501 | 0.0498 |
| 19 | 298 | VP41B | 24 | 36 | 0.0055 | 0.008 | 0.002 | 0.0053 | 0.0064 | 0.0083 | 0.0087 | 0.0242 | 0.0668 |
| 20 | 362 | VP39A | 24 | 36 | 0.0024 | 0.0001 | 0 | 0.0006 | 0.0032 | 0.0013 | 0.0018 | 0.0105 | 0.0238 |
| 21 | 395 | VP39B | 24 | 36 | 0.0027 | 0.0015 | 0 | 0.0034 | 0.0041 | 0.0046 | 0.0072 | 0.0168 | 0.0299 |
| 22 | 314 | VP38A | 36 | 36 | 0.0172 | 0.001 | 0.002 | 0.0025 | 0.0036 | 0.0024 | 0.0126 | 0.0262 | 0.0688 |
| 23 | 449 | VP38B | 12 | 24 | 0.0256 | 0.044 | 0.0164 | 0.0707 | 0.0421 | 0.094 | 0.095 | 0.2679 | 0.1216 |
| 24 | 134 | VP36A | 18 | 36 | 0.0015 | 0.0013 | 0.0003 | 0.0018 | 0.0027 | 0.0013 | 0.0186 | 0.0556 | 0.0946 |
| 25 | 309 | VP36B | 18 | 36 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0213 | 0.028 | 0.0387 |
| 26 | 19 | VP35 | 12 | 24 | 0.0182 | 0.0164 | 0.0009 | 0.0245 | 0.0397 | 0.0548 | 0.3846 | 0.6298 | 0.385 |
| 27 | 253 | VP32 | 18 | 36 | 0.0027 | 0.0023 | 0.001 | 0.0019 | 0.0054 | 0.0036 | 0.0085 | 0.0295 | 0.0381 |
| 28 | 396 | VP31 | 24 | 36 | 0.0027 | 0.0021 | 0 | 0.0002 | 0 | 0.0008 | 0.007 | 0.0153 | 0.0317 |
| 29 | 480 | VP28 | 12 | 36 | 0.0034 | 0.0017 | 0.0031 | 0.0051 | 0.0049 | 0.0194 | 0.1471 | 0.3699 | 0.7033 |
| 30 | 367 | VP26 | 12 | 36 | 0.0071 | 0.0174 | 0.0058 | 0.0109 | 0.0098 | 0.0308 | 0.1935 | 0.2421 | 0.8689 |
| 31 | 58 | VP24 | 18 | 36 | 0.0079 | 0.0127 | 0.0058 | 0.0085 | 0.0137 | 0.0157 | 0.0338 | 0.062 | 0.2099 |
| 32 | 359 | VP22 | 12 | 24 | 0.0011 | 0.0017 | 0.0021 | 0.0023 | 0.0021 | 0.0043 | 0.0035 | 0.0176 | 0.0168 |
| 33 | 473 | VP19 | 12 | 36 | 0.011 | 0.0201 | 0.0119 | 0.0238 | 0.0288 | 0.0516 | 0.1578 | 0.3933 | 0.6816 |
| 34 | 269 | VP15 | 12 | 36 | 0.0011 | 0.0006 | 0.0021 | 0.002 | 0.0028 | 0.0182 | 0.1348 | 0.2521 | 0.4087 |
| 35 | 339 | VP13A | 2 | 36 | 0.0005 | 0.0068 | 0.0058 | 0.0018 | 0.0054 | 0.0033 | 0.0076 | 0.0141 | 0.0148 |
| 36 | 377 | VP13B | 12 | 24 | 0.0048 | 0.0065 | 0.0043 | 0.0065 | 0.0091 | 0.0228 | 0.0474 | 0.0901 | 0.0681 |
| 37 | 445 | VP12B | 2 | 36 | 0.0147 | 0.0615 | 0.0258 | 0.0227 | 0.0393 | 0.0516 | 0.0982 | 0.1954 | 0.4168 |
| 38 | 65 | VP12A | 8 | 24 | 0.0002 | 0 | 0 | 0.0004 | 0.0015 | 0.0045 | 0.018 | 0.0466 | 0.0394 |
| 39 | 394 | VP11 | 18 | 36 | 0.0032 | 0.0053 | 0.0035 | 0.0016 | 0.004 | 0.005 | 0.0247 | 0.0395 | 0.0806 |
The calibrated ratio is the ratio of the expression of the WSSV structural gene in infected shrimp gill cells compared to a universal control probe (actin). TF is the time (hpi) at which the baseline (0 hpi) expression level increased by at least threefold. Peak expression always occurred at either 24 or 36 hpi.
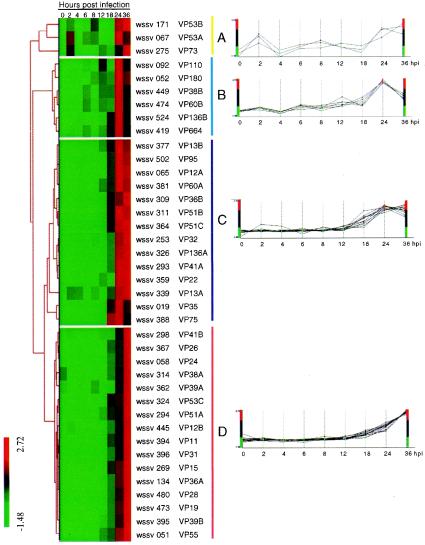
A hierarchical clustering algorithm was used to group the structural genes based on similarities in their patterns of expression (calibrated expression ratios in Table 4) over the time course of infection (Fig. 5). Four major gene expression profiles (A to D) emerged, and Fig. 5 plots the expression profiles of the individual genes within the appropriate cluster. Cluster A genes (VP53A, VP53B, and VP73) began to express very early, at 2 hpi. Surprisingly, the expression profiles of these genes subsequently followed a fluctuating pattern that did not show a steady increase with increasing time postinfection. Genes in clusters B, C, and D had similar transcriptional profiles from 12 hpi, when the genes first started to express, through to 24 hpi. However, the genes in these three clusters could be differentiated by their expression patterns from 24 to 36 hpi. At 36 hpi, cluster B expression levels were down-regulated compared to 24 hpi, cluster C levels were almost unchanged, and cluster D levels increased.
FIG. 5.
Hierarchical clustering of the expression data for WSSV structural genes. Each row in the frame represents a single ORF, and each column indicates the time post-WSSV infection. The expression pattern for each gene was categorized by an average linkage hierarchical clustering program. The color bars on the left indicate the normalized expression levels, with red boxes denoting an expression ratio greater than the mean and green boxes denoting an expression ratio below the mean. Black boxes denote an intermediate level of expression. The four panels (A, B, C, and D) show the expression profiles of the genes in the corresponding clusters.
DISCUSSION
In this study, we used two different WSSV hosts to investigate the WSSV virion structural proteins and temporal gene expression, respectively. Because crayfish are freshwater species that are easy to culture and because they can sustain a heavy WSSV virus load for some time, hemolymph from WSSV-infected crayfish was used as a convenient and stable source of purified virus for SDS-PAGE. The WSSV virion protein profile (Fig. 2) from crayfish P. clarkii is also almost completely the same as that from P. monodon (data not shown). For the microarray temporal expression analysis, however, infected P. monodon shrimp were used as the source of WSSV RNA. P. monodon is very susceptible to the WSSV Taiwan isolate used here, and by using an appropriately managed culture system, the WSSV infection cycle in this shrimp is completed in 36 h (4, 22). As we have argued before (22), in the absence of a suitable cell line, P. monodon is thus a very suitable model for WSSV temporal expression analysis. However, some caution should be exercised in interpreting the temporal expression data here, because in vivo cell cycles are necessarily asynchronous (as opposed to synchronized, which they would usually be in a cell culture model).
Although the virion protein profile of a virus is important, the large size and complexity of the WSSV genome have made such a profile difficult to obtain, so that previous efforts have resulted in only partial, incomplete information (see, e.g., references 14, 32, 33, 45, and 52). In this study, where our purpose was to identify as many structural proteins as possible, by emphasizing completeness at the expense of absolute purity, we were able to use gradient SDS-PAGE separation of intact WSSV virions to produce the most comprehensive profile published to date (Fig. 2). Furthermore, the intactness of the WSSV virions (Fig. 1) ensured that this profile was both consistent (in terms of the molar ratios of the bands) and repeatable (data not shown). After separation and in-gel trypsin digestion, LC-nanoESI-MS/MS techniques identified 33 WSSV structural proteins from 34 bands (Fig. 2, Tables 1 and 2). In addition to 19 WSSV structural proteins that were already known (six of which were not detected here), 20 proteins were identified, bringing the total number of reported WSSV structural proteins to 39. This genomic and proteomic analysis of the structural proteins of WSSV should provide a foundation for further study of the virus-host interaction, the contribution of each WSSV structural protein to virion architecture, and the role of WSSV structural proteins in the completion of the viral infection cycle.
The ORF for WSSV480, which is now known as vp28, encodes a major WSSV envelope protein. This protein, VP28, is widely studied and may be involved in WSSV systemic infection of shrimp (46). In native hosts, VP28 has been reported to occur only in its nonglycosylated form (47), although VP28 is also predicted to have the signal peptide sequence as well as five potential glycosylation sites that have been shown to become glycosylated in insect cells (51). Figure 2 suggests that VP28 has at least three forms with apparent sizes of 25, 26, and 29 kDa, respectively. However, subsequent N-terminal sequencing of these bands showed that while the two lower bands both matched the VP28 gene, the N-terminal sequencing for the 29-kDa band was LHPNLEFQYR?SA??, which is very similar to the crayfish Pacifastacus leniusculus clotting protein LHSNLEYQYRYSGRV (GenBank accession no. 422370). Thus although Huang et al. (14) also reported the existence of the same 29-kDa band, it seems that VP28 must only be a relatively minor component. As for the 25- and 26-kDa bands, in the absence of any C-terminal sequence data, we cannot be sure whether these two proteins have been subjected to posttranslational change or not. We can conclude, however, that since their N-terminal sections are intact (i.e., their signal peptides have not been cleaved), both of these bands represent unglycosylated forms of VP28 protein. Since VP28 is such an important protein, the functional significance of these two forms should be investigated in a future study. The protein encoded by ORF WSSV419 (vp664) also had three forms of different sizes (664, 186, and 161 kDa). However, since the 186- and 161-kDa protein bands were not consistently present in the gels, we tentatively conclude that the two smaller proteins may only be degraded forms of VP664. N-terminal sequencing of these three protein bands was also unsuccessful, although it was not clear whether this was because the N-terminal sections were blocked or because the proteins were too large to be transferred from the gel to the polyvinylidene difluoride membrane. We also note here that the size of VP664 was too large to be estimated by the molecular markers or mass spectrometry. For this protein, since the matched peptide sequences yielded by MS/MS were distributed throughout the entirety of the protein (i.e., from the N terminus to the C terminus), we were able tentatively to predict the molecular mass of the protein in band 1 of Fig. 2 as 664 kDa based on the amino acid sequence deduced from the ORF. We also note here that VP664 appears to be a major capsid protein: in a preliminary immunogold electron microscopy study, we found that the antibody against VP664 bound specifically to the globular subunits of the WSSV nucleocapsid (unpublished data).
The apparent molecular mass of at least two of the proteins was smaller than expected: based on their theoretical molecular masses, the proteins encoded by ORFs WSSV067 and WSSV171 should have sizes of 144 and 108 kDa, respectively, but no bands of these sizes were found. However, the SDS-PAGE band at 53 kDa included two proteins with sequences that matched these two ORFs. It may be that these two proteins were truncated, since the deduced peptide sequences from the MS/MS data corresponded only to their C-terminal amino acid sequences. For this band too, N-terminal sequencing was unsuccessful, and we are now working to prepare sufficient quantities of these proteins for two-dimensional electrophoretic separation.
Bands 6, 9, 12, and 17 (Fig. 2) were identified primarily as host proteins. Bands 9 and 12 were crayfish vitellogenin and hemocyanin, respectively, both of which are present in very high concentrations in crayfish hemolymph (vitellogenin is present only in females) and would thus be quite likely to copurify with the WSSV virions. A recent paper has demonstrated that hemocyanin has antiviral activity (60), which means that the hemocyanin should bind or otherwise interact with viral proteins. This would not only account for the consistent appearance of band 12 in every replication, but also explain the presence of this band in virion samples purified by cesium chloride gradient instead of sucrose (data not shown). This explanation also implies that the appearance of hemocyanin is not simply the result of contamination. In the case of bands 6 and 17, both of which are cytosolic proteins, we follow a suggestion made by Jensen et al. (16) in the context of vaccinia virus and speculate that host actin may play a role in WSSV morphogenesis. Likewise, if host sarco/endoplasmic reticulum Ca2+-ATPase is truly present in the virion, then it might also fulfill some important function for WSSV. We also note that since no WSSV nonstructural proteins (such as DNA polymerase, ribonucleotide reductase subunits RR1 and RR2, thymidine kinase-thymidylate chimeric protein TK-TMK, thymidine synthase TS, dUTPase, etc.) were ever found on the SDS-PAGE gels, the consistent presence of the cytosolic proteins was unlikely to be the result of contamination. Immunogold analysis will be necessary, however, to confirm the presence of these two proteins in the virion.
Based on its apparent molecular mass, band 31 should be VP15 (48). However, although band 31 is a major band that consistently appeared in the SDS-PAGE gels, we were initially unable to identify it by our usual technique of trypsin digestion followed by LC-nanoESI-MS/MS. Since VP15 is rich in arginine, the explanation seemed to be that the trypsin digestion of this protein produced fragments that were too small for LC-nanoESI-MS/MS identification. We therefore used N-terminal sequencing (see reference 54 for the N-terminal sequencing protocols), which yielded the sequence VARSSKTKSRRGSKK and thus confirmed that this band was indeed VP15.
5′ RACE has already been used to show that the transcriptional start sites of some WSSV genes (dnapol, rr1, and rr2) are at ∼26 bp downstream of the TATA box and that they have an A/TCAC/G/TT motif that closely matches the A/C/TCAG/TT Inr motif of arthropods (6, 7). The present paper now shows that 76.9% of the 39 WSSV structural genes also have this Inr motif (A/TCAC/G/TT) located in the predicted promoter region. This is important because RNA polymerase II usually recognizes either the TATA box, the Inr, or the combination of both (29, 36, 57). Since the predicted promoter regions of a high proportion of WSSV structural genes include at least one of these features (82.1% have a TATA box, 76.9% have the Inr motif, and 64.1% have both), and since to date no native WSSV RNA polymerase holoenzyme has been identified, these data suggest that WSSV structural genes (the late genes) may also use the RNA polymerase II of the host for transcription.
In the present paper, we found a relatively high percentage of WSSV structural protein genes that had no readily identified polyadenylation signal. We therefore used 3′ RACE to investigate whether two species of mRNA—i.e., with or without a poly(A) tail—were produced by the WSSV structural genes. We determined that WSSV mRNA probably did not always have a poly(A) tail. However, based on the present results, we cannot rule out the possibility that several genes share a long mRNA with poly(A) and then use the internal ribosomal entrance site to start the translation for each gene. Further study will be needed to clarify this issue.
The presence or absence of a poly(A) tail or the TATA box/Inr is evidently not a good indicator of expression level (cf. the respective columns in Table 2 and the intensity of bands in Fig. 2). However, all the major bands in Fig. 2 (VP664, VP75, VP28, VP26, VP19, and VP15) have both a poly(A) tail and the TATA box/Inr, and except for VP15, all of them also have a strong Kozak context around their translation initiation site.
Some viral proteins, such as human herpesvirus (HHV) viral envelope protein (HHV-8-gB), mediate cell adhesion in an RGD-dependent manner (50). Huang et al. (13) have already reported that VP281 (VP36B in the present paper) contains an RGD motif, and here we show that VP31, VP36A, VP110, VP136A, and VP664 also include this motif. The presence of an RGD motif alone does not guarantee cell adhesion activity because the flanking sequence often influences the protein's conformation. The availability of appropriate integrin receptor molecules on the cell surface also affects the final outcome. It has been reported that for any protein to interact with integrins via the RGD motif, threonine (T) at the fourth position (RGDT) is also critical (38). Since both VP36A and VP31 possess a threonine at the fourth position of the RGD motif, it is possible that these two structural proteins in particular might interact with integrin to generate a cellular state that is more receptive for infection. Further study will be needed to elucidate what role(s) might be mediated by the adhesion of these six RGD structural proteins during viral infection of the target cells.
Although the WSSV genome sequence has been completed for three isolates and is available in the public domain, WSSV protein sequences are unique and show little or no homology to the known proteins from other viruses or organisms (14, 47, 56). In consequence, the putative function of many proteins, including almost all the structural proteins, remains unknown. To date, for the structural proteins, putative additional functions have been assigned only for VP180, VP73, and VP15, which may be a collagen-like protein (20), class I cytokine receptor (14), and DNA-binding protein (58), respectively. The scanning results reported here (Table 3) have now identified 12 new putative functions for WSSV structural proteins. Empirical experiments will be needed to confirm the validity of these predicted protein functions as inferred from the domain/signature scan. Meanwhile, the temporal expression profiling of WSSV structural genes by DNA microarray analysis has shown that genes with common or similar (putative) roles in the viral infection cycle tended to be grouped in the same hierarchical cluster (Fig. 5; Table 4). Thus, of the three structural genes in cluster A, two of them (vp53b and vp73) may play roles in signal transduction pathways. Unusually for structural genes, their expression levels started increasing at the early infection stage (at 2 hpi, cluster A genes showed a 2.8- to ∼7.8-fold increase with respect to 0 hpi), which suggests that these proteins are likely to play a crucial role in the early events of WSSV infection. Genes encoding proteins involved in site-specific endonuclease (vp38b) and biogenesis (vp180 and vp664) were grouped in cluster B with three other genes, while genes encoding proteins involved in transcription (vp22), energy metabolism (vp36B and vp13A), and cofactor for vitamin metabolism (vp136A) were grouped in cluster C. In contrast, in cluster D there were only two structural genes with additional functions: vp51A encodes a protein with a predicted function (eggshell protein; Table 3), while vp15 encodes a protein that functions as a DNA binding protein (58). On the other hand, most of the major WSSV structural genes were assigned to this cluster, including vp28, vp26, vp24, vp19, and vp15. Note that although VP26 was initially thought to be a major capsid protein (45), immunogold electron microscopy with an antibody specific to VP26 has subsequently shown that VP26 is an envelope protein (59).
A comparison of the expression patterns for the different clusters shows that for clusters B and C, expression levels peaked at 24 hpi, whereas peak levels in cluster D were only reached 12 h later, at 36 hpi. Since most of the late structural proteins with additional functionality were grouped in cluster B or C, it is tempting to see a connection here with the sequence of assembly of the WSSV virion (per references 15, 49, and 52 and also based on our own unpublished observations), and hypothesize that the structural proteins related to the first stage of WSSV's morphogenetic assembly (which is to form an empty capsid) should appear in clusters B and C. The fact that the genes coding for a major capsid protein (vp664) and a minor capsid protein (vp35) (5) were assigned to clusters B and C, respectively, is also consistent with this hypothesis. This interpretation can be carried further: in the next stage of virion assembly, the naked nucleocapsid is surrounded by an envelope, and again, the timing of the peak expression levels of the major envelope proteins VP28, VP26, and VP19 (all of which are in cluster D) is consistent with the sequence of morphogenetic assembly. We note that over half (10 of 16) of the structural proteins in cluster D have transmembrane domains, which are often associated with envelope proteins. Moreover, the DNA binding protein gene vp15 is also in cluster D, and since at this stage of morphogenesis the empty capsid now fills with DNA, it may be that VP15 has a function related to DNA packing instead of (or in addition to) the transcription function proposed for this protein by Zhang et al. (58).
Acknowledgments
This investigation was supported financially by the National Science Council grants (NSC90-2311-B-002-059, NSC91-2317-B-002-011, NSC92-2317-B-002-001, and NSC92-2317-B-002-018) and Ministry of Education grant (89-B-FA01-1-4). Proteomic mass spectrometry analyses were performed by the Core Facilities for Proteomics Research located at the Institute of Biological Chemistry, Academia Sinica, supported by a National Science Council grant (91-3112-P-001-009-Y).
We are indebted to Paul Barlow for helpful criticism.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bairoch, A., and B. Boeckmann. 1994. The SWISS-PROT protein sequence data bank: current status. Nucleic Acids Res. 1994:3578-3580. [PMC free article] [PubMed] [Google Scholar]
- 3.Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. Marshall, and E. L. Sonnhammer. 2002. The Pfam Protein Families Database. Nucleic Acids Res. 30:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, P. S., C. F. Lo, Y. C. Wang, and G. H. Kou. 1996. Identification of white spot syndrome associated baculovirus (WSBV) target organs in shrimp, Penaeus monodon, by in situ hybridization. Dis. Aquat. Org. 27:131-139. [Google Scholar]
- 5.Chen, L. L., J. H. Leu, C. J. Huang, C. M. Chou, S. M. Chen, C. H. Wang, C. F. Lo, and G. H. Kou. 2002. Identification of a nucleocapsid protein (VP35) gene of shrimp white spot syndrome virus and characterization of the motif important for targeting VP35 to the nuclei of transfected insect cells. Virology 293:44-53. [DOI] [PubMed] [Google Scholar]
- 6.Chen, L. L., H. C. Wang, C. J. Huang, S. E. Peng, Y. G. Chen, S. J. Lin, W. Y. Chen, C. F. Dai, H. T. Yu, C. H. Wang, C. F. Lo, and G. H. Kou. 2002. Transcriptional analysis of the DNA polymerase gene of shrimp white spot syndrome virus. Virology 301:136-147. [DOI] [PubMed] [Google Scholar]
- 7.Cherbas, L., and P. Cherbas. 1993. The arthropod initiator: the capsite consensus plays an important role in transcription. Insect Biochem. Mol. Biol. 23:81-90. [DOI] [PubMed] [Google Scholar]
- 8.Fitzgerald, M., and T. Shenk. 1981. The sequence 5′-AAUAAA-3′ forms parts of the recognition site for polyadenylation of late SV40 mRNAs. Cell 24:251-260. [DOI] [PubMed] [Google Scholar]
- 9.Flegel, T. W. 1997. Major viral diseases of black tiger prawn (Penaeus monodon) in Thailand. World J. Microbiol. Biotechnol. 13:433-442. [Google Scholar]
- 10.Frech, B., and E. Peterhans. 1994. RT-PCR: ′background priming' during reverse transcription. Nucleic Acids Res. 22:4342-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hameed, A. S. S., M. Anilkumar, M. L. Stephen Raj, and K. Jayaraman. 1998. Studies on the pathogenicity of systemic ectodermal and mesodermal baculovirus and its detection in shrimp by immunological methods. Aquaculture 160:31-45. [Google Scholar]
- 12.Hofmann, K., and W. Stoffel. 1993. Tmbase—a database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler 374:166. [Google Scholar]
- 13.Huang, C., X. Zhang, Q. Lin, X. Xu, and C. Hew. 2002. Characterization of a novel envelope protein (VP281) of shrimp white spot syndrome virus by mass spectrometry. J. Gen. Virol. 83:2385-2392. [DOI] [PubMed] [Google Scholar]
- 14.Huang, C., X. Zhang, Q. Lin, X. Xu, Z. Hu, and C. L. Hew. 2002. Proteomic analysis of shrimp white spot syndrome viral proteins and characterization of a novel envelope protein VP466. Mol. Cell. Proteomics 1:223-231. [DOI] [PubMed] [Google Scholar]
- 15.Inouye, K., S. Miwa, N. Oseko, H. Nakano, and T. Kimura. 1994. Mass mortalities of cultured kuruma shrimp, Penaeus japonicus, in Japan in 1993: electron microscopic evidence of the causative virus. Fish Pathol. 29:149-158. (In Japanese.) [Google Scholar]
- 16.Jensen, O. N., T. Houthaeve, A. Shevchenko, S. Cudmore, T. Ashford, M. Mann, G. Griffiths, and J. Krijnse Locker. 1996. Identification of the major membrane and core proteins of vaccinia virus by two-dimensional electrophoresis. J. Virol. 70:7485-7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khadijah, S., S. Y. Neo, M. S. Hossain, L. D. Miller, S. Mathavan, and J. Kwang. 2003. Identification of white spot syndrome virus latency-related genes in specific-pathogen-free shrimps by use of a microarray. J. Virol. 77:10162-10167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozak, M. 1989. The scanning model for translation: an update. J. Cell Biol. 108:229-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.La Scola, B., S. Audic, C. Robert, L. Jungang, X. de Lamballerie, M. Drancourt, R. Birtles, J. M. Claverie, and D. Raoult. 2003. A giant virus in amoebae. Science 299:2033. [DOI] [PubMed] [Google Scholar]
- 20.Li, Q., Y. Chen, and F. Yang. 2004. Identification of a collagen-like protein gene from white spot syndrome virus. Arch. Virol. 149:215-223. [DOI] [PubMed] [Google Scholar]
- 21.Lin, S. T., Y. S. Chang, H. C. Wang, H. F. Tzeng, T. Z. Chang, J. Y. Lin, C. H. Wang, C. F. Lo, and G. H. Kou. 2002. Ribonucleotide reductase of shrimp white spot syndrome virus (WSSV): expression and enzymatic activity in a baculovirus/insect cell system and WSSV-infected shrimp. Virology 304:282-290. [DOI] [PubMed] [Google Scholar]
- 22.Liu, W. J., H. T. Yu, S. E. Peng, Y. S. Chang, H. W. Pien, C. J. Lin, C. J. Huang, M. F. Tsai, C. J. Huang, C. H. Wang, J. Y. Lin, C. F. Lo, and G. H. Kou. 2001. Cloning, characterization and phylogenetic analysis of a shrimp white spot syndrome virus (WSSV) gene that encodes a protein kinase. Virology 289:362-377. [DOI] [PubMed] [Google Scholar]
- 23.Lo, C. F., J. H. Leu, C. H. Ho, C. H. Chen, S. E. Peng, Y. T. Chen, C. M. Chou, P. Y. Yeh, C. J. Huang, H. Y. Chou, C. H. Wang, and G. H. Kou. 1996. Detection of baculovirus associated with white spot syndrome (WSBV) in penaeid shrimps using polymerase chain reaction. Dis. Aquat. Org. 25:133-141. [Google Scholar]
- 24.Lo, C. F., C. H. Ho, S. E. Peng, C. H. Chen, H. E. Hsu, Y. L. Chiu, C. F. Chang, K. F. Liu, M. S. Su, C. H. Wang, and G. H. Kou. 1996. White spot syndrome associated virus (WSBV) detected in cultured and captured shrimp, crabs and other arthropods. Dis. Aquat. Org. 27:215-225. [Google Scholar]
- 25.Lo, C. F., C. H. Ho, C. H. Chen, K. F. Liu, Y. L. Chiu, P. Y. Yeh, S. E. Peng, H. E. Hsu, H. C. Liu, C. F. Chang, M. S. Su, C. H. Wang, and G. H. Kou. 1997. Detection and tissue tropism of white spot syndrome baculovirus (WSBV) in captured brooders of Penaeus monodon with a special emphasis on reproductive organs. Dis. Aquat. Org. 30:53-72. [Google Scholar]
- 26.Lo, C. F., and G. H. Kou. 1998. Virus-associated white spot syndrome of shrimp in Taiwan: a review. Fish Pathol. 33:365-371. [Google Scholar]
- 27.Lo, C. F., H. C. Hsu, M. F. Tsai, C. H. Ho, S. E. Peng, G. H. Kou, and D. V. Lightner. 1999. Specific genomic DNA fragment analysis of different geographical clinical samples of shrimp white spot syndrome virus. Dis. Aquat. Org. 35:175-185. [Google Scholar]
- 28.Marks, H., M. Mennens, J. M. Vlak, and M. C. van Hulten. 2003. Transcriptional analysis of the white spot syndrome virus major virion protein genes. J. Gen. Virol. 84:1517-1523. [DOI] [PubMed] [Google Scholar]
- 29.Martin, A., G. Ribeiro, M. I. Marques, and J. V. Costa. 1994. Genetic identification and nucleotides sequence of the DNA polymerase gene of African swine fever virus. Nucleic Acids Res. 22:208-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Momoyama, K., M. Hiraoka, H. Nakano, H. Koube, K. Inouye, and N. Oseko. 1994. Mass mortalities of cultured kuruma shrimp, Penaeus japonicus, in Japan in 1993: histopathological study. Fish Pathol. 29:141-148. [Google Scholar]
- 31.Mulder, N. J., R. Apweiler, T. K. Attwood, A. Bairoch, D. Barrell, A. Bateman, D. Binns, M. Biswas, P. Bradley, P. Bork, P. Bucher, R. R. Copley, E. Courcelle, U. Das, R. Durbin, L. Falquet, W. Fleischmann, S. Griffiths-Jones, D. Haft, N. Harte, N. Hulo, D. Kahn, A. Kanapin, M. Krestyaninova, R. Lopez, I. Letunic, D. Lonsdale, V. Silventoinen, S. E. Orchard, M. Pagni, D. Peyruc, C. P. Ponting, J. D. Selengut, F. Servant, C. J. Sigrist, R. Vaughan, and E. M. Zdobnov. 2003. The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res. 31:315-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nadala, E. C. B., Jr., L. M. Tapay, and P. C. Loh. 1998. Characterization of a non-occluded baculovirus-like agent pathogenic to penaeid shrimp. Dis. Aquat. Org. 33:221-229. [DOI] [PubMed] [Google Scholar]
- 33.Nadala, E. C. B., Jr., and C. Loh. 1998. A comparative study of three different isolates of white spot virus. Dis. Aquat. Org. 33:231-234. [DOI] [PubMed] [Google Scholar]
- 34.Nakano, H., H. Koube, S. Umezawa, K. Momoyama, M. Hiraoka, K. Inouye, and N. Oseko. 1994. Mass mortalities of cultured kuruma shrimp, Penaeus japonicus, in Japan in 1993: epizootiological survey and infection trails. Fish Pathol. 29:135-139. [Google Scholar]
- 35.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 36.Nikolov, D. B., and S. K. Burley. 1997. RNA polymerase II transcription initiation: a structural view. Proc. Natl. Acad. Sci. USA 94:15-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pandey, A., and M. Mann. 2000. Proteomics to study genes and genomes. Nature 405:837-846. [DOI] [PubMed] [Google Scholar]
- 38.Plow, E. F., T. A. Haas, L. Zhang, J. Loftus, and J. W. Smith. 2000. Ligand binding to integrins. J. Biol. Chem. 275:21785-21788. [DOI] [PubMed] [Google Scholar]
- 39.Seo, J., and B. Shneiderman. 2002. Understanding hierarchical clustering results by interactive exploration of dendrograms: a case study with genomic microarray data. IEEE Comput. 35:80-86. [Google Scholar]
- 40.Tsai, M. F., C. F. Lo, M. C. W. van Hulten, H. F. Tzeng, C. M. Chou, C. J. Huang, C. H. Wang, J. Y. Lin, J. M. Valk, and G. H. Kou. 2000. Transcriptional analysis of the ribonucleotide reductase genes of shrimp white spot syndrome virus. Virology 277:92-99. [DOI] [PubMed] [Google Scholar]
- 41.Tsai, M. F., H. T. Yu, H. F. Tzeng, J. H. Leu, C. M. Chou, C. J. Huang, C. H. Wang, J. Y. Lin, G. H. Kou, and C. F. Lo. 2000. Identification and characterization of a shrimp white spot syndrome virus (WSSV) gene that encodes a novel chimeric polypeptide of cellular-type thymidine kinase and thymidylate kinase. Virology 277:100-110. [DOI] [PubMed] [Google Scholar]
- 42.Tzeng, H. F., Z. F. Chang, S. E. Peng, C. H. Wang, J. Y. Lin, G. H. Kou, and C. F. Lo. 2002. Chimeric polypeptide of thymidine kinase and thymidylate kinase of shrimp white spot syndrome virus: thymidine kinase activity of the recombinant protein expressed in a baculovirus/insect cell system. Virology 299:248-255. [DOI] [PubMed] [Google Scholar]
- 43.van Etten, J. L., M. V. Graves, D. G. Muller, W. Boland, and N. Delaroque. 2002. Phycodnaviridae—large DNA algal viruses. Arch. Virol. 147:1479-1516. [DOI] [PubMed] [Google Scholar]
- 44.van Hulten, M. C. W., M. F. Tsai, C. A. Schipper, C. F. Lo, G. H. Kou, and J. M. Vlak. 2000. Analysis of a genomic segment of white spot syndrome virus of shrimp containing ribonucleotide reductase genes and repeat regions. J. Gen. Virol. 81:307-316. [DOI] [PubMed] [Google Scholar]
- 45.van Hulten, M. C. W., M. Westenberg, S. D. Goodall, and J. M. Vlak. 2000. Identification of two major virion protein genes of white spot syndrome virus of shrimp. Virology 266:227-236. [DOI] [PubMed] [Google Scholar]
- 46.van Hulten, M. C. W., J. Witteveldt, M. Snippe, and J. M. Vlak. 2001. White spot syndrome virus envelope protein VP28 is involved in the systemic infection of shrimp. Virology 285:228-233. [DOI] [PubMed] [Google Scholar]
- 47.van Hulten, M. C. W., J. Witteveldt, S. Peters, N. Kloosterboer, R. Tarchini, M. Fiers, H. Sandbrink, R. K. Lankhorst, and J. M. Vlak. 2001. The white spot syndrome virus DNA genome sequence. Virology 286:7-22. [DOI] [PubMed] [Google Scholar]
- 48.van Hulten, M. C., M. Reijns, A. M. Vermeesch, F. Zandbergen, and J. M. Vlak. 2002. Identification of VP19 and VP15 of white spot syndrome virus (WSSV) and glycosylation status of the WSSV major structural proteins. J. Gen. Virol. 83:257-265. [DOI] [PubMed] [Google Scholar]
- 49.Wang, C. H., C. F. Lo, J. H. Leu, C. M. Chou, P. Y. Yeh, H. Y. Chou, M. C. Tung, C. F. Chang, M. S. Su, and G. H. Kou. 1995. Purification and genomic analysis of baculovirus associated with white spot syndrome (WSBV) of Panaeus monodon. Dis. Aquat. Org. 23:239-242. [Google Scholar]
- 50.Wang, F.-Z., S. M. Akula, N. Sharma-Walia, L. Zeng, and B. Chandran. 2003. Human herpesvirus 8 envelope glycoprotein B mediates cell adhesion via its RGD sequence. J. Virol. 77:3131-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, H. C., Y. S. Chang, G. H. Kou, and C. F. Lo. White spot syndrome virus: molecular characterization of a major structural protein in a baculovirus expression system and shrimp hemocytes. Mar. Biotechnol., in press.
- 52.Wang, Q., B. T. Poulos, and D. V. Lightner. 2000. Protein analysis of geographic isolates of shrimp white spot syndrome virus. Arch. Virol. 145:263-274. [DOI] [PubMed] [Google Scholar]
- 53.Wongteerasupaya, C., J. E. Vickers, S. Sriurairatana, G. L. Nash, A. Akarajamorn, V. Boosaeng, S. Panyim, A. Tassanakajon, B. Withyachumnarnkul, and T. W. Flegel. 1995. A non-occluded, systemic baculovirus that occurs in the cells of ectodermal and mesodermal origin and causes high mortality in the black tiger prawn Penaeus monodon. Dis. Aquat. Org. 21:69-77. [Google Scholar]
- 54.Wu, C. Y., C. F. Lo, C. J. Huang, H. T. Yu, and C. H. Wang. 2002. The complete genome sequence of Perina nuda picorna-like virus, an insect-infecting RNA virus with a genome organization similar to that of the mammalian picornaviruses. Virology 294:312-323. [DOI] [PubMed] [Google Scholar]
- 55.Xia, D., L. J. Henry, R. D. Gerard, and J. Deisenhofer. 1994. Crystal structure of the receptor-binding domain of adenovirus type 5 fiber protein at 1.7 A resolution. Structure 2:1259-1270. [DOI] [PubMed] [Google Scholar]
- 56.Yang, F., J. He, X. Lin, Q. Li, D. Pan, X. Zhang, and X. Xu. 2001. Complete genome sequence of the shrimp white spot bacilliform virus. J. Virol. 75:11811-11820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Young, R. A. 1991. RNA polymerase II. Annu. Rev. Biochem. 60:689-715. [DOI] [PubMed] [Google Scholar]
- 58.Zhang, X., X. Xu, and C. L. Hew. 2001. The structure and function of a gene encoding a basic peptide from prawn white spot syndrome virus. Virus Res. 79:137-144. [DOI] [PubMed] [Google Scholar]
- 59.Zhang, X., C. Huang, X. Xu, and C. L. Hew. 2002. Transcription and identification of an envelope protein gene (p22) from shrimp white spot syndrome virus. J. Gen. Virol. 83:471-477. [DOI] [PubMed] [Google Scholar]
- 60.Zhang, X., C. Huang, and Q. Qin. 2004. Antiviral properties of hemocyanin isolated from shrimp Penaeus monodon. Antivir. Res. 61:93-99. [DOI] [PubMed] [Google Scholar]