Abstract
Background and aims Brassica napus (AACC, 2n = 38, oilseed rape) is a relatively recent allotetraploid species derived from the putative progenitor diploid species Brassica rapa (AA, 2n = 20) and Brassica oleracea (CC, 2n = 18). To determine the influence of intensive breeding conditions on the evolution of its genome, we analysed structure and copy number of rDNA in 21 cultivars of B. napus, representative of genetic diversity.
Methods We used next-generation sequencing genomic approaches, Southern blot hybridization, expression analysis and fluorescence in situ hybridization (FISH). Subgenome-specific sequences derived from rDNA intergenic spacers (IGS) were used as probes for identification of loci composition on chromosomes.
Key Results Most B. napus cultivars (18/21, 86 %) had more A-genome than C-genome rDNA copies. Three cultivars analysed by FISH (‘Darmor’, ‘Yudal’ and ‘Asparagus kale’) harboured the same number (12 per diploid set) of loci. In B. napus ‘Darmor’, the A-genome-specific rDNA probe hybridized to all 12 rDNA loci (eight on the A-genome and four on the C-genome) while the C-genome-specific probe showed weak signals on the C-genome loci only. Deep sequencing revealed high homogeneity of arrays suggesting that the C-genome genes were largely overwritten by the A-genome variants in B. napus ‘Darmor’. In contrast, B. napus ‘Yudal’ showed a lack of gene conversion evidenced by additive inheritance of progenitor rDNA variants and highly localized hybridization signals of subgenome-specific probes on chromosomes. Brassica napus ‘Asparagus kale’ showed an intermediate pattern to ‘Darmor’ and ‘Yudal’. At the expression level, most cultivars (95 %) exhibited stable A-genome nucleolar dominance while one cultivar (‘Norin 9’) showed co-dominance.
Conclusions The B. napus cultivars differ in the degree and direction of rDNA homogenization. The prevalent direction of gene conversion (towards the A-genome) correlates with the direction of expression dominance indicating that gene activity may be needed for interlocus gene conversion.
Keywords: Brassica napus, allopolyploidy, rDNA, chromosome evolution, gene conversion
INTRODUCTION
Brassica napus (AACC, 2n = 38) has been intensively cultivated since the middle of the 20th century (Baranyk and Fábry, 1999), and is a natural post-Neolithic allotetraploid species that formed approx. 7500 years ago (Chalhoub et al., 2014). Numerous cytogenetic, genetic and genomic studies showed that B. napus formed from crosses between B. oleracea (CC, 2n = 18) and B. rapa (AA, 2n = 20). Its polyphyletic origin was established from chloroplast haplotypes suggesting that B. napus could have been formed at least twice from crosses between different forms of progenitor species (Palmer et al., 1983; Erickson et al., 1983; Allender and King, 2010). Furthermore, cytogenetic studies using genomic in situ hybridization (GISH) (Howell et al., 2008) or GISH-like BAC probes (Leflon et al., 2006) have confirmed that the A and C genomes have largely remained distinct in B. napus and showed disomic inheritance of its chromosomes. However, comparative genetic mapping studies have demonstrated that homeologous recombination occurred between the A and C genomes generating different translocations between the genomes (Parkin et al., 1995; Osborn et al., 2003; Piquemal et al., 2005; Udall et al., 2005; Alix et al., 2008; Chalhoub et al., 2014) indicating that disomic inheritance may be occasionally compromised perhaps as a result of increased transposon element activity in the newly formed allopolyploid nucleus (Sarilar et al., 2013; and reviewed by Tayalé and Parisod, 2013).
The rDNA locus encoding ribosomal 18S, 5·8S and 26S rRNA genes (35S rDNA) has been used in numerous cytogenetic and phylogenetic studies (Poczai and Hyvonen, 2010). It is somewhat paradoxical that, despite conservativity of genes (coding regions), it is one of the most dynamic objects in the genome. This is manifested by sequence variation of intergenic (IGS) (Borisjuk et al., 1997) and internal transcribed (ITS) spacers (Alvarez and Wendel, 2003; Nieto Feliner and Rosselló, 2007), and frequent changes in number and position of loci on chromosomes (Dubcovsky and Dvorak, 1995). In many hybrids and allopolyploids only one parental copy is found while the other is often reduced or lost (Volkov et al., 1999; Kovarik et al., 2005; Doyle et al., 2008; Weiss-Schneeweiss et al., 2012; Mahelka et al., 2013; and reviewed by Volkov et al., 2007; Buggs et al., 2012; Weiss-Schneeweiss et al., 2013). Homogenization may be mediated by different genetic events including locus loss, reduction of copies or replacement with newly amplified ones (Nieto Feliner and Rosselló, 2012). Theoretical models suggest that non-homologous recombination and gene conversion are the mechanisms driving rDNA homogenization processes (Zimmer et al., 1980; Dover, 1982).
The genus Brassica is known to harbour large variability in the number of rDNA loci, ranging between two and five per haploid set (Maluszynska and Heslop-Harrison, 1993; Hasterok et al., 2006; PlantrDNAdatabase – http://www.plantrdnadatabase.com/, Garcia et al., 2012). Brassica rapa, the presumed progenitor species of B. napus, has five loci (two major and three minor) per haploid set while the second presumed genome donor, B. oleracea, harbours two loci (Maluszynska and Heslop-Harrison, 1993). At the cytogenetic level, the cultivars of B. napus show genotypic differences in number, distribution and morphology of rDNA chromosomal loci (Snowdon et al., 1997; Fukui et al., 1998; Hasterok et al., 2001, 2006; Ali et al., 2005; Xiong and Pires, 2011; Amosova et al., 2014). Previous Southern blot hybridization revealed restriction fragments corresponding to both parents indicating Mendelian inheritance of rDNA in B. napus (Bennett and Smith, 1991; Waters and Schaal, 1996). However, several cytogenetic observations suggest that rDNA structural changes occurred in B. napus. First, both natural and synthetic lines show a reduced number of loci compared to the sum of progenitor loci (Maluszynska and Heslop-Harrison, 1993; Snowdon et al., 1997; Kulak et al., 2002; Hasterok et al., 2006). Variability in the number of rDNA loci seems to be limited to minor loci (mostly in the A genome) while major nucleolus organizer regions (NORs) on chromosomes A1, A3, C7 and C8 seem to be intact – for ribosomal RNA genes. We followed the nomenclature allowing attribution of each chromosome to a linkage group in B. rapa (Kim et al., 2009) and B. oleracea (Howell et al., 2002) (www.brassica.info). Second, the rDNA fluorescence in situ hybridization (FISH) signals to C-genome NORs were weaker on the C-genome NORs compared to those in parental B. oleracea (Xiong and Pires, 2011). Third, the C-genome rDNA sites were not totally blocked by the B. oleracea-specific IGS probe (Howell et al., 2008). Finally, chromosome banding showed interpopulation variation in the amount and distribution of heterochromatin adjacent to or overlapping with NORs (Amosova et al., 2014).
In the present study, to shed more light on the evolutionary patterns of rDNA, we carried out a population-level study of gene copies and loci in 21 cultivars of B. napus. We posed the following questions: (1) Are homeologous rRNA genes and loci faithfully inherited in all populations of the allotetraploid? (3) Is the expression status of homeologues additive or biased in different organs according to the structure of the variety? To address these questions, we combined structural and functional analyses. We obtained evidence for gene conversion of thousands of C-genome units that are being replaced by the A-genome type units in cultivar ‘Darmor’. This does not seem to occur in another cultivar ‘Yudal’ where loci and genes remain intact. Variation between cultivars suggests a tentative establishment of bidirectional homogenization amongst the post-Neolithic B. napus allotetraploid.
MATERIALS AND METHODS
Plant material
Oilseed rape cultivars were chosen according to the genetic diversity of the species with mainly ssp. oleifera cultivars but also two accessions of ssp. rapifera (‘Rutabagas 22’ and ‘Rutabaga 95’) and one of ssp. pabularia (‘Asparagus kale’). Fourteen spring B. napus cultivars (‘Asparagus kale’, ‘Brutor’, ‘Loras’, ‘Nachan’, ‘Norin 1’, ‘Norin 6’, ‘Norin 9’, ‘Norin 10’, ‘Oro’, ‘Spok’, ‘Stellar’, ‘Taichung’, ‘Yudal’ and ‘Westar’) and seven winter cultivars (‘Darmor’, ‘Maxol’, ‘Mohican’, ‘Petranova’, ‘Rutabaga 22’, ‘Rutabaga 95’ and ‘Tapidor’) were used for genetic and expression analysis of rDNA (Supplementary Data Table S1). As controls of presumed diploid progenitors, we used Z1, a doubled haploid line of B. rapa provided by AAFC, Canada, and HDEM, a doubled haploid line of B. oleracea provided by BrACySol BRC, Ploudaniel, France. Plants were grown from seeds in a greenhouse. Most seeds were obtained from INRA BrACySol BRC, Ploudaniel, France. Seeds of B. napus ‘Tapidor’ were a gift from the laboratory of Functional Genomics and Proteomics of Plants, CEITEC, Brno, Czech Republic, and were originally obtained from Ian Bancroft’s laboratory, the John Innes Centre (JIC), Norwich, UK.
Southern blot hybridization
Southern blotting followed the protocol described by Koukalova et al. (2010) using rDNA probes, A-genome-specific IGS probe (IGS-A) and C-genome-specific IGS probe (IGS-C) labelled with 32P (DekaPrime kit, Fermentas, Lithuania). The hybridization signals were visualized by Phosphor imaging (Typhoon 9410, GE Healthcare, PA, USA) and signals were quantified using ImageQuant software (GE Healthcare).
Fluorescence in situ hybridization
Preparation of slides and hybridization were carried out according to procedures detailed by Suay et al. (2014). The ribosomal probe used in this study was 35S rDNA (pTa 71 clone) from wheat (Gerlach and Bedbrook, 1979), IGS-A and IGS-C probes described further below and the BAC clone B. oleracea named Bob014O06 (Howell et al., 2002). This BAC clone was used as ‘GISH-like’ to distinguish specifically all C-genome chromosomes in B. napus (Suay et al., 2014). The 35S rDNA and BAC clone were labelled with Alexa-488 dUTP by random priming, the IGS-A with biotin-dUTP (Roche, Mannheim, Germany) using PCR and the IGS-C with biotin-dUTP (Roche) using nick translation (Bionick DNA labelling System, Thermo Fisher Scientific, Waltham, MA, USA). Biotinylated probe was immunodetected by Texas Red avidin DCS (Vector Laboratories, Burlingame, CA, USA) and the signal was amplified with biotinylated anti-avidin D (Vector Laboratories). The chromosomes were mounted and counterstained in Vectashield (Vector Laboratories, Ontario, Canada) containing 2·5 μg mL–1 4′,6-diamidino-2-phenylindole (DAPI). Fluorescence images were captured using a CoolSnap HQ camera (Photometrics, Tucson, AZ, USA) on an Axioplan 2 microscope (Zeiss, Oberkochen, Germany) and analysed using MetaVueTM (Universal Imaging Corporation, Downington, PA, USA).
IGS amplification and subgenome-specific probe generation
Genomic DNA was extracted by a CTAB method from fresh leaves (Saghai-Maroof et al., 1984). IGS regions were amplified by PCR using primers Pr1 (AGACGACTTTAAATACGCGAC) (Garcia and Kovarik, 2013) and a newly designed reverse BrasProm_R primer annealing to promoter (GAGTGCCTACCCCTTATA). Amplification was performed using the following programme: 92 °C for 20s, 62·4 °C for 30 s, 70 °C for 3 min, all for 35 cycles followed by 8 min of extension at 70 °C. The PCR products were separated on an agarose gel, cut and ligated to a pDrive plasmid (Qiagen, Germany). Recombinant clones were amplified and subcloned after restriction. The following restriction enzymes were used: MscI and EcoRI (long IGS variant from ‘Darmor’), MscI and EcoRI (short IGS variant from ‘Darmor’), HincII (‘Asparagus Kale’), MscI and AccI (‘Yudal’). The fragments were ligated to a pDrive vector and sequenced. Sequences were submitted to GenBank under accession numbers KT008109 (‘Darmor’ L), KT008110 (‘Darmor’ S), KT008111 (‘Asparagus kale’) and KT008112 (‘Yudal’). Subrepeats were analysed using the YASS genomic similarity search tool (Noe and Kucherov, 2005) and Tandem Repeats Finder (Benson, 1999). A phylogenetic tree was reconstructed using the Seaview program (Gouy et al., 2010).
The A-genome-specific probe (IGS-A) was prepared by digestion of a plasmid DNA of a B. rapa IGS clone (GenBank: KT008109) with Tru1I. The resulting 1·4-kb Tru1I fragment was cloned and sequenced (pBrapsr2 clone). Its 1436-insert was found to contain the majority of the B. rapa-specific C-subrepeats and part of the B-subrepeats. We checked the specificity by mapping the next-generation sequencing (NGS) reads to the IGS-A sequence. About 1·2 % B. rapa Illumina reads (see further below) were mapped to the insert. There were no longer (>60 bp) regions covered by NGS reads from B. oleracea, and the region between nucleotide positions 400 and 1300 showed essentially no hits. The C-genome-specific probe (IGS-C) was prepared by PCR amplification using newly designed IGS primers Oler_F1 (TGACGGACAGTCCTCGTG) and Oler_F2 (CAGTACACATATCAGCACG). The primers were derived from a B. oleracea IGS repetitive subregion 400–1300 positions downstream from the 26S gene to which essentially only B. oleracea and not B. rapa NGS reads were mapped. In a PCR, B. oleracea DNA template was used at a low concentration (∼1 ng per reaction). The resulting product was purified and labelled without further subcloning.
Expression analysis
The procedures followed those described by Ksiazczyk et al. (2011). Briefly, total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) and contaminating DNA was removed by TURBO™ DNase (Ambion, Austin, TX, USA). Reverse transcription reaction was performed in 40 μL volume and contained: 2 μg RNA, 4 pmol random primers (N9) and 200 U reverse transcriptase (Invitrogen Superscript II RNase H, Paisley, UK), according to conditions recommended by the supplier. Expression of homeologous genes was carried out using a cleaved amplified polymorphism sites (CAPS) assay. The ITS1 region was amplified using 0·5 μL cDNA and primers 18Sfor and 5·8Srev (Kovarik et al., 2005). PCR products were digested with restriction enzyme RsaI in an amplification mixture, separated by electrophoresis and visualized by ethidium bromide staining.
In silico rDNA sequence assembly
Regions (18S, ITS, 26S) of 35S rDNA of B. napus, B. oleracea and B. rapa were assembled into contigs by mapping of Illumina reads to reference 18S–ITS1–5·8S–ITS2–26S sequences obtained from GenBank (AF128100.1, KM538957, KF704394, GQ891874, X51576.1, X13557.1). The 35S rDNA sequence of Arabidopsis thaliana (accession number: X52322) was used as a reference to aid assembly. The following Illumina reads archives were used: B. napus ‘Darmor’ (ERR457740, ERR457754 and ERR457738), B. napus ‘Yudal’ (ERR457757), B. rapa (SRR1296482 and SRR5479996) and B. oleracea (SRR074124). Sequence downloads and basic read manipulations of genomic reads were done with the aid of the Galaxy Server (Goecks et al., 2010). Different regions of 35S rDNA sequences (contigs) were ultimately assembled into one single consensus sequence for each species using CLC Genomics Workbench 6.5.1 (Qiagen, Aarhus, Denmark) via default settings. There were no significant differences in coverage between individual regions and most nucleotides were read >130 times.
Intragenomic variation of rDNA determined from NGS reads
CLC Genomics Workbench 6.5.1 was used to estimate intragenomic variation among 35S rDNA units in B. napus, B. oleracea and B. rapa. Before analysis of single nucleotide polymorphisms (SNPs) all reads with Ns and all reads less than 90 nt in length were removed. In some cases, the number of reads was sampled below 100 million to decrease computing time. Illumina reads used were first mapped to the 35S rDNA consensus sequence of B. oleracea with the following mapping settings: mismatch cost value 2, insertion cost value 3, deletion cost value 3, with both the length fraction value and the similarity fraction value set to 0·8. Variations were then detected via the Probabilistic Variant Detection function tool in CLCbio, using default settings. SNPs were filtered as follows: minimum read coverage 100, Count (the number of countable reads supporting the allele) 10, frequency (the ratio of ‘the number of “countable” reads supporting the allele’ to ‘the number of “countable” reads covering the position of the variant’) ≥20 % (high-frequency SNPs) or 5–20 % (medium-frequency SNPs). Comparative analysis of rDNA variants was carried out using tools with the COMPARE function of CLC Genomics Workbench.
ITS amplicons were obtained by emulsion PCR and sequenced on a Roche 454 GS-FLX platform (EuroFins MWG, Ebersberg, Germany). The reads were mapped to the 256 bp of ITS1 (B. oleracea and B. rapa) and sorted into clusters according to the procedures described by Matyasek et al. (2012). The cut off for scoring SNPs was set to ≥ 10 % reads.
RESULTS
Variability of rDNA homeologue ratios in B. napus cultivars
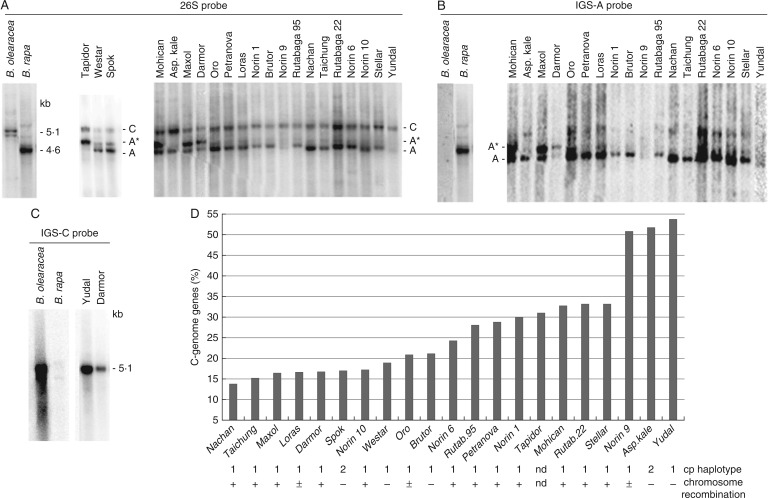
To determine homeologous genes ratios, we carried out Southern blot hybridization of genomic DNA from 21 cultivars of B. napus representing most of the crop diversity, and the presumed diploid progenitor species (Fig. 1). In B. oleracea the 26S probe hybridized to a doublet of the BstNI fragments migrating in the upper part of the gel (Fig. 1A). The large size of the B. oleracea-specific band is consistent with a longer IGS in this species compared to B. rapa (Delseny et al., 1990). In B. rapa, there was a single fast-migrating fragment. All B. napus cultivars inherited major progenitor fragments: the 4·6-kb ‘A’ inherited from B. rapa and the larger 5·1-kb ‘C’ inherited from B. oleracea. A minor B. oleracea fragment was not detected. Four cultivars (‘Darmor’, ‘Maxol’, ‘Mohican’ and ‘Tapidor’) showed a strong ‘A*’ fragment with the size falling between the ‘A’ and ‘C’ bands. The ‘A*’ fragment hybridized strongly with the A-genome-specific IGS-A probe (Fig. 1B) revealing its A-genome origin. The C-genome-specific IGS-C probe hybridized to the upper 5·1-kb ‘C’ band in B. oleracea and B. napus while it did not hybridize with B. rapa and derived A-genome bands of B. napus (Fig. 1C and Supplementary Data Fig. S1). The ratio between the C- and A-genome-specific fragments was determined by quantification of Southern blot bands (Fig. 1A) by counting of radioactivity in ‘A’, ‘A*’ and ‘C’ bands using a Phosphorimager. The proportion of C-genome copies calculated as ‘C’/‘C’ + ‘A’ + ‘A*’ percentages varied from 14 % in B. napus ‘Nachan’ to 54 % in ‘Yudal’ (Fig. 1D). The ‘A’+ ‘A*’/‘C’ band ratio ranged from 0·85 to 6·14 with a median of 3·10. The cultivars analysed in this study were previously genotyped by chloroplast markers (Cifuentes et al., 2010). Two major chloroplast haplotypes are indicated by numbers below the graph (Fig. 1D). There was no apparent relationship to rRNA gene ratios. The frequency of homeologue chromosome pairing in meiosis differed between cultivars (Cifuentes et al., 2010). Cultivars with high A/C ratios seem to belong to a group harbouring a high frequency of homeologue pairing (+ labels below the graph).
Fig. 1.
(A) Southern blot hybridization of genomic DNA from several B. napus cultivars. In the left panel the blot was hybridized with the 26S probe. After stripping the blot was rehybridized with the A-genome-specific IGS probe (B). ‘C’, C-genome bands; ‘A’, A-genome bands; ‘A*’ indicates an IGS family amplified in a subset of B. napus cultivars. (C) Example of a Southern hybridization of the C-genome-specific IGS probe. Note strong hybridization of the probe to ‘Yudal’ DNA. (D) The radioactivity of bands was quantified using a Phosphorimager and homeologue gene number expressed as a proportion of C-genome rDNA to total rDNA. Numbers below indicate two major chloroplast haplotypes identified by Cifuentes et al. (2010). In the same study, the B. napus cultivars were divided into groups with low (–), intermediate (±) and high (+) frequency of homeologue chromosome pairing in meiosis (symbols towards the bottom).
Sequence polymorphisms of rDNA units
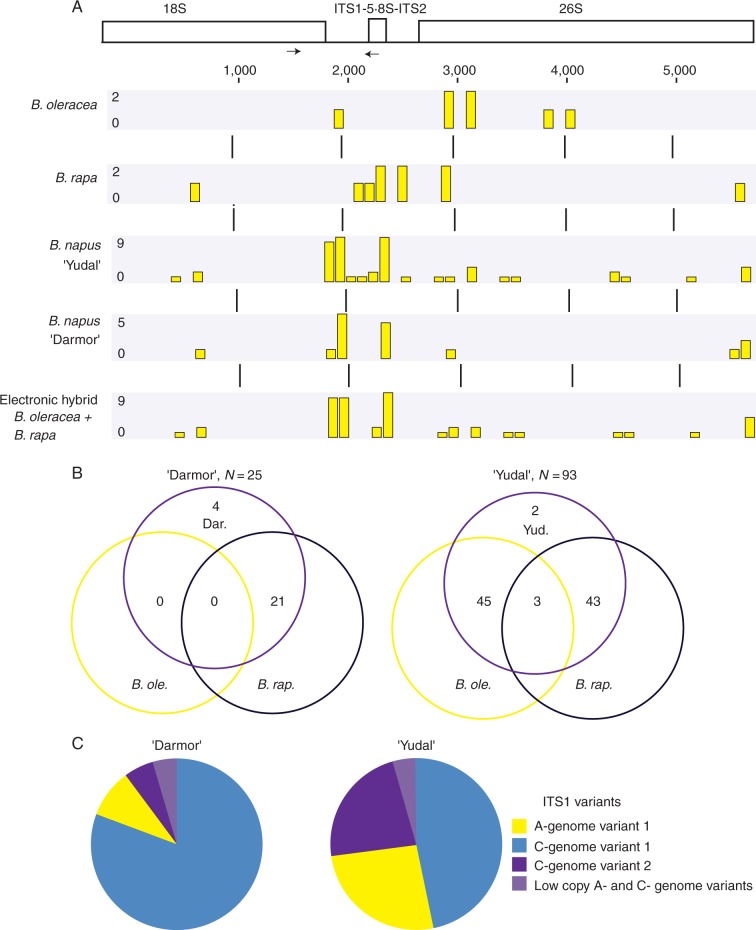
The availability of genomic reads allowed us to reconstruct 18S–5·8S–26S operons in B. napus and the diploid species B. rapa and B. oleracea (Table 1). The reads were mapped to the reference sequence which had been assembled from sequences in GenBank (for accession numbers, see Methods). All three rRNA genes (18S, 5·8S and 26S) were recovered including intervening ITS1 and ITS2 sequences. From the mapped reads, we generated a single rDNA consensus sequence for each diploid species and two cultivars of B. napus (‘Darmor’ and ‘Yudal’). The total length of the recovered 18S–5·8S–26S region was 5803 bp in all samples. Pairwise alignment of consensus sequences revealed 72 polymorphic sites (1·24 % divergence) between B. rapa and B. oleracea units. Brassica rapa had a slightly higher intragenomic variation [ten high- (≥20 %) frequency SNPs per unit] than B. oleracea (seven high-frequency SNPs per unit) which may be related to several minor rDNA loci in the B. rapa genome. Although most variation occurred in the ITS regions, several significant polymorphisms were also found in the 18S and 26S coding regions (Fig. 2A). The consensus sequences constructed from NGS reads of two B. rapa cultivars [sequence archives SRR1296482 and SRR5479996 (susbsp. pekinensis)] were 100 % identical while there were differences in quantitative representation of individual nucleotides at eight polymorphic sites (not shown).
Table 1.
Copy number of rRNA genes in B. napus and progenitor species determined from NGS reads
| Read archive accession | Mapped reads* | Total reads | GP† (%) | rDNA (Mb) | Copies‡ 1C | |
|---|---|---|---|---|---|---|
| B. rapa | SRR1296482 | 1 453 334 | 78 528 998 | 1·86 | 5·80 | 1709 |
| B. oleracea | SRR074124 | 1 437 383 | 146 402 687 | 1·06 | 5·78 | 1162 |
| B. napus ‘Darmor’ | ERR457740, ERR457754, ERR457738 | 838 265 | 60 083 363 | 1·39 | 11·68 | 2832 |
| B. napus ‘Yudal’ | ERR457757 | 780 687 | 72 174 366 | 1·08 | 9·08 | 2201 |
All consensus sequences were 5803 bp long and included the complete 18S–ITS1–5·8S–ITS2–26S subregion.
Genome proportion (GP) is defined as number of reads mapped to rDNA divided by the number of total reads (as a percentage). It would be ∼60 % larger if IGS regions are considered.
From genome proportions copy numbers of rDNA sequences were estimated according to the formula: genome size × genome proportion of rDNA units. The following genome sizes were considered: B. napus, 1182 Mb; B. oleracea, 630 Mb; B. rapa, 530 Mb (Bennett and Leitch, 2012).
Fig. 2.
Intragenomic heterogeneity of rDNA units in progenitor species and two B. napus cultivars determined from whole genomic Illumina reads (A, B) and Roche 454 sequencing of ITS1 amplicons (C). The graphs in A reflect distribution of highly polymorphic sites (>20 % frequency) along the 18S–5·8S–26S region. Each column represents one or more high-frequency SNPs. Coalescence of more SNPs within the 10-bp window is reflected by column height. (B) Venn diagrams showing comparison of rDNA variants between B. napus and its progenitors. Note absence of B. oleracea variants in B. napus ‘Darmor’. (C) Circle chart diagrams showing the distribution of ITS1 variants in the genomes. The scheme of rDNA unit is depicted in A. Arrows indicate positions of primers used in amplicon sequencing and RT-CAPS analysis.
The distribution of high-frequency SNPs in B. napus cultivars is depicted in Fig. 2A. The SNP profiles differed dramatically between ‘Darmor’ and ‘Yudal’ cultivars: B. napus ‘Yudal’ had a complex mutation profile similar to that of the virtual hybrid [constructed from 1 : 1 mixing of rDNA reads from the presumed B. oleracea (SRR074124) and B. rapa (SRR1296482) progenitors]. In contrast, B. napus ‘Darmor’ had fewer SNPs and a relatively smooth mutation profile resembling that of B. rapa. The high-frequency variants were at least three-fold more abundant in B. napus ‘Yudal’ than in B. napus ‘Darmor’ (Fig. 2B). The variants from the presumed progenitors were inherited at comparable ratios in B. napus ‘Yudal’. In contrast, B. napus ‘Darmor’ had 80 % variants inherited from B. rapa, 0 % from B. oleracea and 20 % were unique. Nevertheless, the C-genome variants were found at low copy (<20 % frequency) in B. napus ‘Darmor’ (see further below). Similar results were obtained when analysing promoter regions. For example, the B. oleracea promoter had a C nucleotide at position −7 while B. rapa had A at this position. In ‘Yudal’, 58 % of reads had A (haplotype inherited from B. rapa) and 42 % C (inherited from B. oleracea) at this position. In ‘Darmor’, the ‘A’ and ‘C’ variants accounted for 86 and 14 %, respectively. The promotor region harboured higher (5–10-fold) intragenomic heterogeneity than the coding regions (not shown). Using Roche 454 technology we also sequenced 700-bp PCR amplicons comprising the 18S gene (3′ region) and ITS1 regions. Haplotypic analysis of ‘Darmor’ and ‘Yudal’ amplicons revealed shared identical ITS1 major (≥10 % reads) haplotypes. However, these occurred at markedly different frequencies between the two cultivars (Fig. 2C).
Intragenomic IGS length heterogeneity
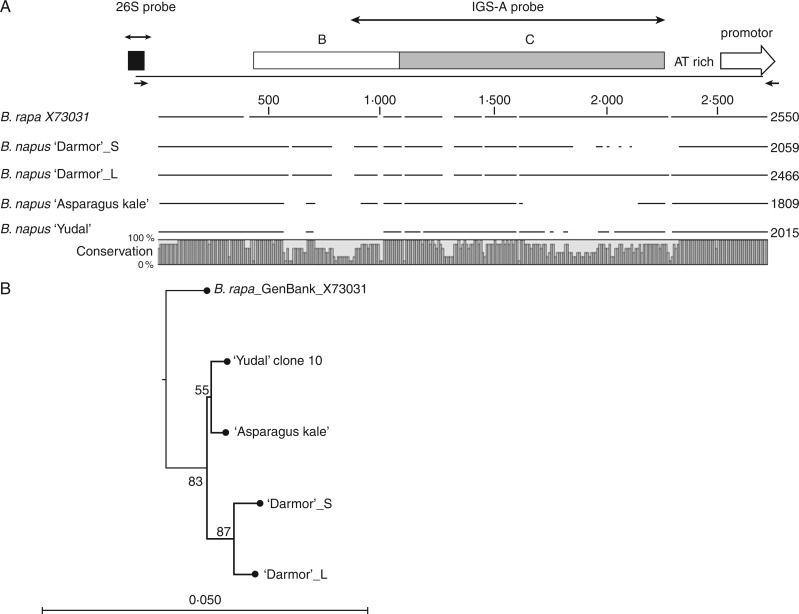
The highly repetitive nature of IGS makes this region refractory to reconstruct from short Illumina reads. We therefore analysed IGS polymorphisms in several clones from B. napus cultivars obtained by cloning of IGS-specific PCR products (see Methods). The organization of aligned A-genome clones is shown in Fig. 3A. The AT-rich region harbouring DNA conformation polymorphisms (Darocha and Bertrand, 1995), core promoter and ∼400 bp downstream of the 26S gene were conserved. In contrast, the long repetitive part was highly variable. In this region multiple indels of variable length were detected, in accordance with variation reported in previous studies (Delseny et al., 1990; Darocha and Bertrand, 1995; Bhatia et al., 1996). In B. napus ‘Darmor’, two IGS variants of different lengths were identified: the long (L) 2059-bp variant differed from the short (S) 2066-bp variant in the organization of the C subrepeats. The B subrepeats (according to the nomenclature of Darocha and Bertrand, 1995) and other parts of IGS were invariant. The C subrepeats located proximally to the promoter formed a higher order structure (HOR) of a basic 421-bp unit (Supplementary Data Fig. S2). There were 2·4 copies of HOR units in the long variant of B. napus ‘Darmor’ and the B. rapa clone (X73031). The short B. napus ‘Darmor’ variant and clones from ‘Asparagus kale’ (1809 bp in length) and ‘Yudal’ (2015 bp in length) had only one copy of 421-bp HOR. The divergence between the first and second copy of HOR was 0·73 %, while the third incomplete copy was less conserved (3·4 %). In the phylogenetic tree (Fig. 3B) both short and long variants of B. napus ‘Darmor’ clustered on the same branch, indicating a common origin. Thus, individual SNPs rather than IGS length polymorphisms exhibit phylogenetic signals in Brassica.
Fig. 3.
(A) Multiple alignment of IGS sequences from B. rapa and B. napus A-genome clones. Note multiple indels occur in each clone. The bottom bar indicates conserved and divergent subregions. ‘B’ and ‘C’ represent blocks of subrepeats. The positions of probes and primers are indicated by double and single arrows, respectively. (B) Unrooted neighbour-joining tree reconstructed from IGS sequences. Note both long (‘Darmor’_L) and short (‘Darmor’_S) clones cluster together. Numbers indicate bootstrap support (1000 replicates). The scale bar indicates the number of substitutions per nucleotide positions (5 base change per 100 nucleotide positions).
rDNA homeologue expression in B. napus cultivars
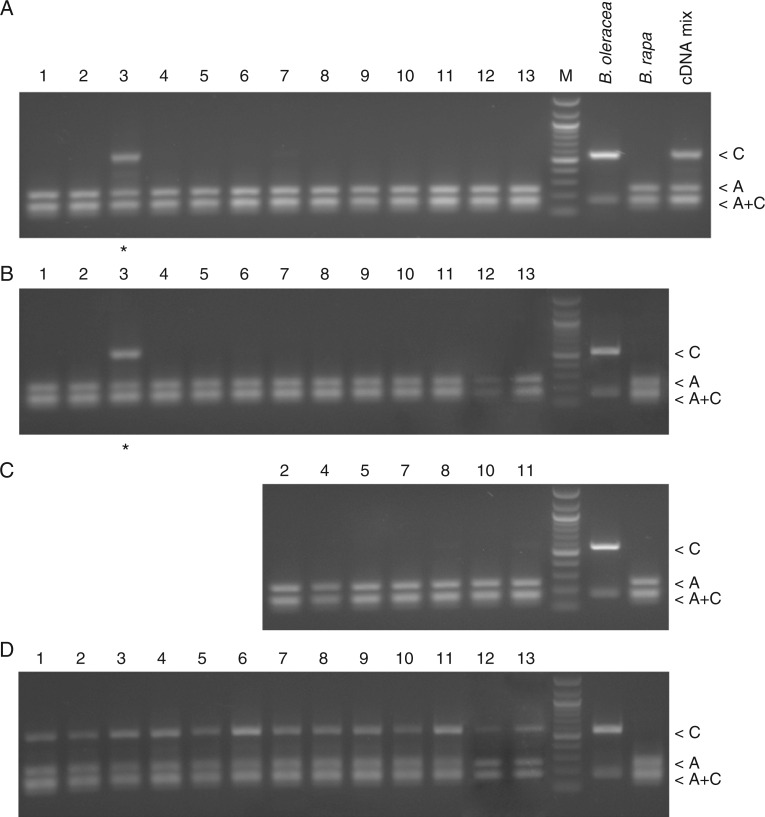
Expression of homeologous genes was analysed in leaf, root and flower bud tissues by the RT-CAPS method (Fig. 4). In both B. oleracea and B. rapa, the RsaI enzyme cleaved RT-PCR products into two fragments. The shortest band was shared between both species while the larger 250-bp (marked as C) and 550-bp (A) bands were specific for B. rapa and B. oleracea progenitors, respectively. In most B. napus cultivars, only the A-genome-specific bands were visualized, indicating expression dominance of A-genome NORs. However, in B. napus ‘Norin 9’ both A- and C-genome bands were amplified at comparable fluorescence intensity, indicating co-dominance in leaf and root. Control amplification of mixed cDNAs from B. rapa and B. oleracea showed equal amplification of both homeologues. Thus, the A-genome NORs are always active while those of the C genome are prevalently silenced.
Fig. 4.
Expression analysis of 35S rDNA. The RT-PCR CAPS was performed using RNA extracted from B. oleracea (HDEM) and B. rapa (Z1) diploids and natural cultivars of B. napus: ‘Darmor’ (1), ‘Taichung’ (2), ‘Norin 9’ (3), ‘Petranova’ (4), ‘Spok’ (5), ‘Asparagus kale’ (6), ‘Norin 6’ (7), ‘Stellar’ (8), ‘Rutabaga’ (9), ‘Loras’ (10), ‘Yudal’ (11), ‘Westar’ (12) and ‘Tapidor’ (13). Gel restriction profiles of ITS1 amplification products obtained from leaf (A), root (B) and flower bud (C) cDNA. Control amplification products from mixed progenitor cDNAs are shown at the right in A. (D) Products from genomic DNA amplification. Asterisks indicate lanes where PCR products corresponding to both homeologues were amplified. The homeologue-specific bands were of ∼250 bp (‘A’, A-genome) and ∼550 bp (‘C’, C-genome) in size. A 100-bp DNA ladder was used as marker (M). The scale bar indicates the number of substitutions per nucleotide positions (5 base change per 100 nucleotide positions).
Genome-specific probes reveal the identity of loci in B. napus chromosomes
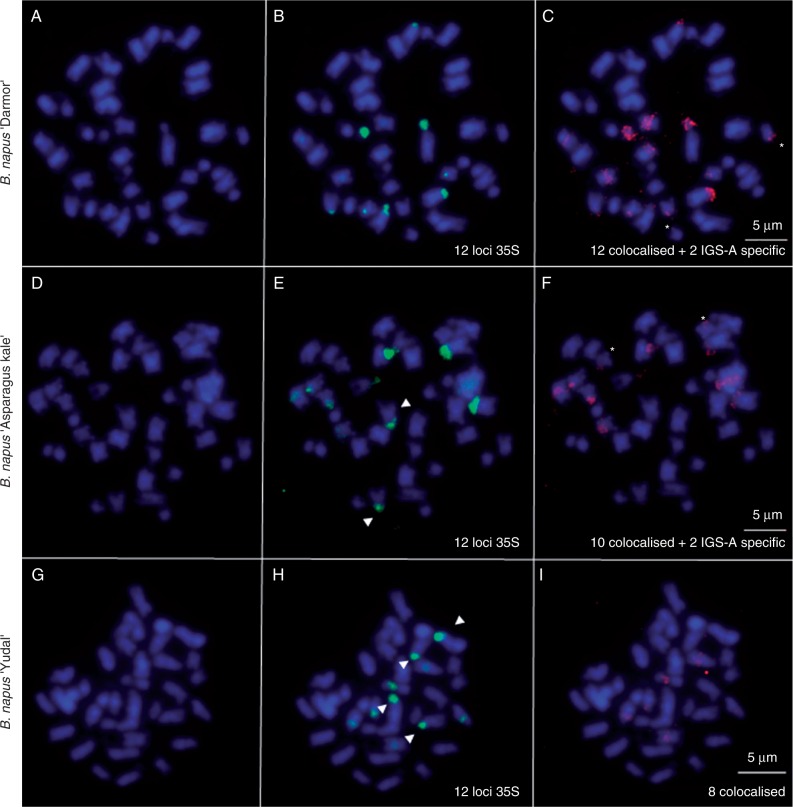
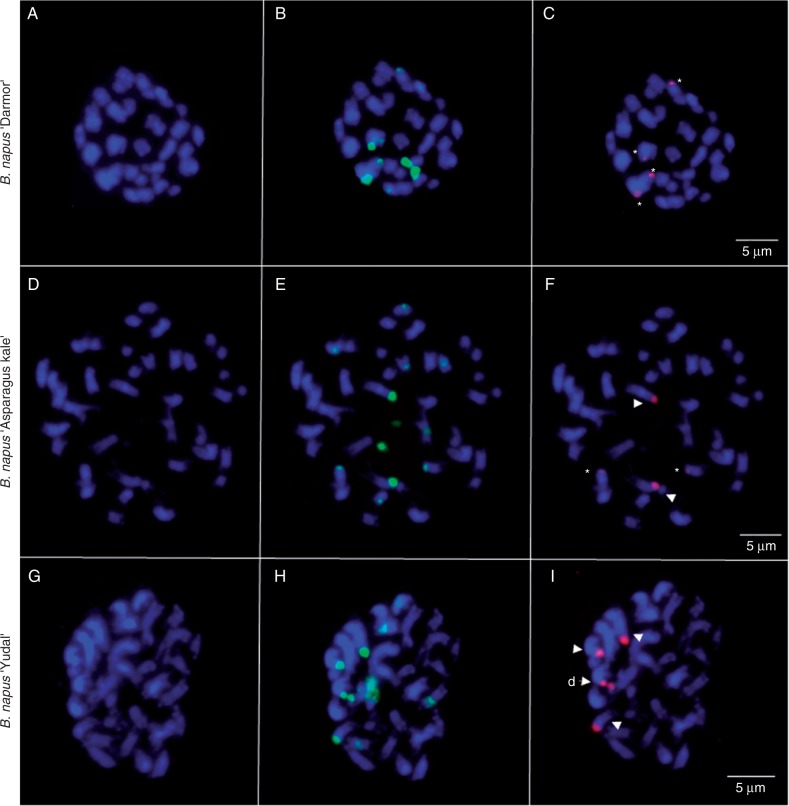
To identify the origin of rDNA loci in B. napus, we isolated specific probes for the A-genome and C-genome NORs. The A-genome-specific (IGS-A) and C-genome-specific rDNA probes (IGS-C) were derived from the variable repetitive part of the IGS region having no or little homology with the analogous region in B. oleracea or B. rapa units, respectively (Fig. 3A). Three B. napus cultivars (‘Darmor’, ‘Yudal’ and ‘Asparagus kale’) together with their putative progenitors were analysed by FISH with the 35S rDNA (green), IGS-A (red) and IGS-C (red) probes (Figs 5 and 6).
Fig. 5.
FISH was carried out using 35S rDNA (green) and IGS-A (red) probes. FISH analyses of somatic metaphase chromosomes of B. napus ‘Darmor’ (A–C), B. napus ‘Aparagus kale’ (D–F) and B. napus ‘Yudal’ (G–I). Arrows indicate 35S-specific signals and the IGS-A additional signals are marked by an asterisk. Chromosomes were counterstained with DAPI (blue). Scale bars = 5 μm.
Fig. 6.
FISH was carried out using 35S rDNA (green) and IGS-C (red) probes. FISH analyses of somatic metaphase chromosomes of B. napus ‘Darmor’ (A–C), B. napus ‘Aparagus kale’ (D–F) and B. napus ‘Yudal’ (G–I). Minor and strong IGS-C signals are marked by asterisks and arrowheads, respectively. A partially decondensed NOR in ‘Yudal’ (I) is indicated by (d-▹). Chromosomes were counterstained with DAPI (blue). Scale bars = 5 μm.
There were ten and four hybridization sites of the 35S probe in B. rapa and B. oleracea, respectively (Supplementary Data Figs. S3 and S4). The IGS-A probe hybridized to B. rapa chromosomes (six signals) but not B. oleracea (Fig. S3). In contrast, the IGS-C probe hybridized to B. oleracea but not B. rapa chromosomes (Fig. S4).
In B. napus ‘Yudal’, the 35S probe hybridized to 12 sites (eight strong and four weak), eight of which co-localized with IGS-A probe (six strong and two weak, Fig. 5I). Four strong 35S sites co-localized with strong IGS-C signals (Fig. 6I).
Brassica napus ‘Darmor’ had also 12 35S sites (six strong, four intermediate and two weak). In this cultivar, the IGS-A probe hybridized to 14 sites (Fig. 5C) out of which 12 co-localized with 35S sites; two sites located outside of rDNA. Two strong and two weak 35S sites co-localized with four weak IGS-C signals (Fig. 6C).
In B. napus ‘Asparagus kale’, the 35S probe hybridized to 12 sites (six strong and six weak). The IGS-A probe hybridized to 12 sites (Fig. 5F), ten of which co-localized with the 35S signals. The IGS-C probe hybridized to two strong and two weak sites (Fig. 6F), all sites overlapping with the green 35S probe signal.
The B. napus cultivars (‘Darmor’, ‘Yudal’ and ‘Asparagus kale’) were also analysed by GISH-like with Bob014O06 (green) and IGS-A (red) probes, which co-localized with the Bob014O06 probe at some sites in ‘ Darmor’ and ‘Asparagus kale’ but not in ‘Yudal’ (Supplementary Data Fig. S5).
DISCUSSION
There has been large diversification of cultivated B. napus round the world since medieval times and especially from the middle of the 20th century (Baranyk and Fábry, 1999). It is therefore interesting to ask how the chromosomes and sequences have evolved under the conditions of intensive breeding. Here, we show that 21 cultivars differ in homeologous rRNA gene ratios, IGS structure and degree of intergenomic homogenization.
Variable rDNA homeologue ratios among B. napus cultivars
In most cultivars of B. napus, the A-genome rDNA units were more abundant than the C units. However, three cultivars (‘Norin 9’, ‘Asparagus kale’ and ‘Yudal’) retained a high number of C-genome genes, indicating that the process may still be in progress, as in some recently formed natural allopolyploid populations of Senecio (Lowe and Abbott, 1996), Tragopogon (Kovarik et al., 2005) and Cardamine (Zozomova-Lihova et al., 2014). No apparent relationship between copy number variation and plastid haplotypes was found, indicating that the putative polyphyletic origin plays little or no role in homogenization direction or that breeding schemes crossing the different cultivars did not allow us to identify the origin. Of note, synthetic lines appear to display more cytogenetic and fewer molecular changes than we observed here (Xiong et al., 2011; Ksiazczyk et al., 2011). For example, Xiong et al. (2011) observed elimination of major NORs that is not encountered in natural cultivars. It is likely that these highly aberrant genotypes may have arisen in early generations of allopolyploids (Szadkowski et al., 2010; Ksiazczyk et al., 2011) while in stabilized species karyotypes with nearly (loss of one of the small loci has been encountered in the A-genome (Hasterok et al., 2006)) additive number of rDNAs seems to be favoured by natural selection.
Gene conversion of rDNA is a variety-specific phenomenon in Brassica
It is striking that several hundred years of intensive cultivation has generated contrasting patterns of rDNA evolution. Both ‘Yudal’ and ‘Darmor’ cultivars of B. napus show similar number of major NORs while their genetic composition (units) is vastly different. In B. napus ‘Yudal’, we observed a high heterogeneity of units that mostly corresponded to additivity of parental contributions. On chromosomes, intactness of parental loci was evidenced by highly localized hybridization signals of the A-genome- (IGS-A) and C-genome- (IGS-C) specific probes. Thus, in ‘Yudal’, concerted evolution has not been operating at rDNA loci until recently. This pattern contrasts markedly with ‘Darmor’ where concerted evolution apparently homogenized most of the genes to the A-genome type. Based on NGS counts, we estimate that about 65 % of C-genome units (∼900 C-genome copies) were eliminated. On chromosomes, all loci were strongly stained with the IGS-A probe while the IGS-C probe hybridized weakly to four loci, indicating these loci carried both A- and C-genome type units. We do not know whether the different unit types are separated or interspersed within the array. The A/C recombinant ITS sequences were not significantly represented among the NGS reads, suggesting that recombinant genes (if present) did not significantly expand in the genome. The IGS family containing rearranged subrepeats was highly amplified in B. napus ‘Darmor’ now comprising most of the rDNA arrays. Also in the coding region about 20 % of highly polymorphic sites appeared to recruit from newly amplified variants. Interestingly, concerted evolution of rDNA in tobacco was associated with rearrangement of IGS (Volkov et al., 1999). Perhaps, interlocus homogenization is preceded by some form of IGS rearrangement and copy number reductions of parental arrays. In B. napus ‘Asparagus kale’, the A-genome IGS probe stained all loci except one. In contrast, the C-genome IGS probe hybridized strongly to only one locus, suggesting that the degree of rDNA homogenization is intermediate to ‘Yudal’ and ‘Darmor’ cultivars. This corroborates another study carried out on a different B. napus cultivar (Xiong and Pires, 2011) that showed reduced rDNA-FISH signals on the C-genome (compared to the B. oleracea progenitor), supporting the hypothesis that the gene richness of C-genome NORs tends to decrease in most natural B. napus. Partial replacement of C-genome genes by A-genome genes potentially explains why B. oleracea IGS was unable to block completely GISH signals on C- genome NORs (Howell et al., 2008).
Dominance of A-genome nucleolar expression
In natural B. napus, epigenetic variability in rRNA expression was much less pronounced than the variability in copy numbers. The majority (95 %) of cultivars showed strong silencing of C-genome genes (A-genome nucleolar dominance) consistent with a previous report (Chen and Pikaard, 1997). The direction of epigenetic silencing in natural B. napus is similar to that observed in synthetic B. napus (Ksiazczyk et al., 2011), indicating that silencing established early in evolution is relatively stable. None of the cultivars showed reverse dominance, i.e. silencing of A-genome NORs. Activation of silent C-genome genes leading to co-dominant phenotype has been reported in certain organs, such as adventious roots (Hasterok and Maluszynska, 2000) or flower organs (Chen and Pikaard, 1997). In our experiments, flowers did not show markedly elevated expression of the C-genome genes and only very faint bands were detected in a minority (15 %) of flower samples after RT-CAPS (data not shown). Nevertheless, we cannot exclude the possibility that certain cell types, not analysed in this study, may specifically express C-genome genes. Partial decondensation of one of the C-genome loci in a ‘strongly’ A-genome-dominant B. napus ‘Yudal’ (Fig. 6I) is consistent with this hypothesis. The co-dominant phenotype in ‘Norin 9’ is intriguing as it occurs irrespective of organ specificity. Perhaps, silencing established at early generations may occasionally be reversed during population divergence.
Why are the A-genome units so invasive?
We observed replacement of C-genome loci by the A-genome units while we did not observe the reverse situation where the C-genome units would be found on the A-genome chromosomes. The question arises as to why the A-genome units are so penetrant and prone to colonize partner chromosomes. There are several hypotheses:
(1) rDNA may follow an overall genomic trend of gene conversion events that appear to be more frequent from the A to C genome than from the C to A genome (Chalhoub et al., 2014). The mechanism of gene conversion remains obscure, but it is believed to involve some form of meiotic recombination (Eickbush and Eickbush, 2007). Indeed, homeologous recombination has been considered as a major source of genetic variability in B. napus (Nicolas et al., 2007; Gaeta and Pires, 2010; Szadkowski et al., 2010, 2011). In support of this, four cultivars (‘Darmor’, ‘Nachan’, ‘Maxol’ and ‘Taichung’) harbouring predominantly the A-genome genes (Fig. 1D) belong to a group showing frequent homeologous recombination (Cifuentes et al., 2010). However, major A- and C-genome NORs occur on non-homeologous chromosomes that pair rarely in meiosis and interlocus recombination is far less frequent than intralocus recombination in Drosophila (Schlotterer and Tautz, 1994). Nevertheless, some form of non-homeologous recombination cannot be excluded due to segmented homology detected between different chromosomes (Parkin et al., 2014). Interlocus homogenization of NORs at non-homeologous chromosomes was also reported in other systems (Kovarik et al., 2004), suggesting the existence of other recombination mechanisms than those acting during meiosis. In Arabidopsis, ectopical rDNA loci physically associate in interphase at a frequency that is higher than random (Pecinka et al., 2004). Hence a translocation of arrays or its part to another position may occur in interphase, perhaps during nucleolar fusions (Kovarik et al., 2008).
(2) Structural features of IGS should be considered. The IGS regions of many plants evolve astonishingly quickly (Saghai-Maroof et al., 1984; Borisjuk et al., 1997; Carvalho et al., 2011; Coutinho et al., 2016). For example, in B. napus progenitor species that diverged less than 3 Mya (Inaba and Nishio, 2002), the rDNAs not only changed the position on chromosomes but also evolved species-specific IGS variants (Bhatia et al., 1996; Delseny et al., 1990; Bennett and Smith 1991). The long (>1·8 kb) region of the A-genome IGS is composed of 21- and 28-bp GC-rich subrepeats which are among the shortest in plants and which form higher order structures (Fig. 3 and Supplementary Data Fig. S2). Given that gene conversion is biased towards GC-rich sequences (Escobar et al., 2011) it may be that novel IGS variants are generated frequently. In accordance with this idea, we identified the A-genome IGS subrepeats as rDNA-independent loci in two B. napus cultivars (‘Darmor’ and ‘Asparagus kale’), undergoing intergenomic homogenization of rDNAs. The solitary IGS loci have been regularly detected in different systems (Stupar et al., 2002; Lim et al., 2004) forming independent satellites of unknown function. We propose that these non-rDNA IGS sites could be a hallmark of recombination events within the NORs possibly arising by integration of extrachromosomal covalent circles (Navratilova et al., 2008).
(3) Epigenetic modification directs homogenization. We previously argued that silent epigenetically modified units may be less vulnerable to homogenization than active units (Kovarik et al., 2008). Heterochromatic marks including histone H3K9 dimethylation and increased cytosine methylation acquired at early generations of synthetic wheat lines lead to their elimination from the tgenome in advanced generations (Guo and Han, 2014). Brassica napus diploid progenitors also differ in number and composition of repeats (Cheung et al., 2009) and epigenetic landscape of their genomes (Braszewska-Zalewska et al., 2010). In synthetic lines of B. napus, we observed immediate silencing of C-genome NORs accompanied by enhanced methylation of promoter regions (Ksiazczyk et al., 2011). Perhaps, long-term silencing of most C-genome units led to their gradual elimination from the genome. In contrast, an open chromatin configuration of intensively transcribed A-genome units may predispose them to homogenization across the genome.
CONCLUSIONS
Patterns of rDNA sequence conversion and provenance of the lost loci are highly idiosyncratic and differ even between allopolyploids of identical parentage, indicating that allopolyploids deriving from the same lower-ploidy-level parental species can follow different evolutionary trajectories (Wendel et al., 1995; Weiss-Schneeweiss et al., 2012). Here, we identified this trend even in relatively recently formed B. napus. We observed shifts in gene ratios and identified intergenomic gene conversion events that partially replaced genes in partner loci. The resulting rDNA homogenization seems to act in a cultivar-specific manner with some preference towards one parental subgenome. This bias is possibly influenced by epigenetic status established early in the allopolyploid nucleus formation. Stabilization of the allopolyploid nucleus is an ongoing process that is accompanied by evolution of distinct rDNA genotypes.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of the following. Table S1: list of Brassica accessions and their analysis. Figure S1: southern blot hybridization of genomic DNA from several B. napus cultivars. Figure S2: structural features of IGS in ‘Darmor’ long and short units. Figure S3: validation of the A-genome probe specificity by Southern hybridization and FISH. Figure S4: validation of the C-genome probe specificity by Southern hybridization (A) and FISH (B). Figure S5: dispersion of A-genome units across the chromosomes analysed by FISH.
Acknowledgements
We thank Dr Zuzana Tesařiková and Květa Dofková (both from Institute of Biophysics ASCR) for the help with DNA extractions. We acknowledge the BrACySol BRC (INRA Ploudaniel, France), which provided most of the seeds of B.napus cultivars as well as the B. rapa and B. oleracea lines used in this study. We thank Dr Ian Bancroft (John Innes Centre, Norwich, UK.) for a gift of B. napus ‘Tapidor’ seeds. The work was funded by the Czech Science Foundation (P501/13/10057S), the French National Research Institute for Agronomy (INRA) and the Czech-French scientific exchange Barrande programme.
Footnotes
These authors contributed equally to the work.
LITERATURE CITED
- Ali HBM, Lysak MA, Schubert I. 2005. Chromosomal localization of rDNA in the Brassicaceae. Genome 48: 341–346. [DOI] [PubMed] [Google Scholar]
- Alix K, Joets J, Ryder CD, et al. 2008. The CACTA transposon Bot1 played a major role in Brassica genome divergence and gene proliferation. The Plant Journal 56: 1030–1044. [DOI] [PubMed] [Google Scholar]
- Allender CJ, King GJ. 2010. Origins of the amphiploid species Brassica napus L. investigated by chloroplast and nuclear molecular markers. BMC Plant Biology 10: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez I, Wendel JW. 2003. Ribosomal ITS sequences and plant phylogenetic inference. Molecular Phylogenetics and Evolution 29: 417–434. [DOI] [PubMed] [Google Scholar]
- Amosova AV, Zemtsova LV, Grushetskaya ZE, et al. 2014. Intraspecific chromosomal and genetic polymorphism in Brassica napus L. detected by cytogenetic and molecular markers. Journal of Genetics 93: 133–143. [DOI] [PubMed] [Google Scholar]
- Baranyk P, Fábry A. 1999. History of the rapeseed (Brassica napus l.) growing and breeding from middle age Europe to Canberra. The regional institute for on-line publishing 4: http://www.regional.org.au/au/gcirc/4/374.htm. [Google Scholar]
- Bennett MD, Leitch I. 2012. Angiosperm DNA C-values database (release 8.0, Dec. 2012). http://data.kew.org/cvalues/.
- Bennett RI, Smith AG. 1991. Use of a genomic clone for ribosomal RNA from Brassica oleracea in RFLP analysis of Brassica species. Plant Molecular Biology 16: 685–688. [DOI] [PubMed] [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Research 27: 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia S, Negi MS, Lakshmikumaran M. 1996. Structural analysis of the rDNA intergenic spacer of Brassica nigra: evolutionary divergence of the spacers of the three diploid Brassica species. Journal of Molecular Evolution 43: 460–468. [DOI] [PubMed] [Google Scholar]
- Borisjuk NV, Davidjuk YM, Kostishin SS, et al. 1997. Structural analysis of rDNA in the genus Nicotiana. Plant Molecular Biology 35: 655–660. [DOI] [PubMed] [Google Scholar]
- Braszewska-Zalewska A, Bernas T, Maluszynska J. 2010. Epigenetic chromatin modifications in Brassica genomes. Genome 53: 203–210. [DOI] [PubMed] [Google Scholar]
- Buggs RJ, Chamala S, Wu W, et al. 2012. Rapid, repeated, and clustered loss of duplicate genes in allopolyploid plant populations of independent origin. Current Biology 22: 248–252. [DOI] [PubMed] [Google Scholar]
- Carvalho A, Guedes-Pinto H, Lima-Brito J. 2011. Intergenic spacer length variants in Old Portuguese bread wheat cultivars. Journal of Genetics 90: 203–208. [DOI] [PubMed] [Google Scholar]
- Chalhoub B., Denoeud F, Liu S, et al. 2014. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345: 1255–1255. [DOI] [PubMed] [Google Scholar]
- Chen ZJ, Pikaard CS. 1997. Transcriptional analysis of nucleolar dominance in polyploid plants: biased expression/silencing of progenitor rRNA genes is developmentally regulated in Brassica. Proceedings of National Academy of Science USA 94: 3442–3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung F, Trick M, Drou N, et al. 2009. Comparative analysis between homoeologous genome segments of Brassica napus and its progenitor species reveals extensive sequence-level divergence. The Plant Cell 21: 1912–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes M, Eber F, Lucas MO, Lode M, Chevre AM, Jenczewski E. 2010. Repeated polyploidy drove different levels of crossover suppression between homoeologous chromosomes in Brassica napus allohaploids. The Plant Cell 22: 2265–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho JP, Carvalho A, Martin A, Ribeiro T, Morais-Cecilio L, Lima-Brito J. 2016. Oak ribosomal DNA: characterization by FISH and polymorphism assessed by IGS PCR–RFLP. Plant Systematics and Evolution 302: 527–544. [Google Scholar]
- Darocha PSCF, Bertrand H. 1995. Structure and comparative-analysis of the rDNA intergenic spacer of Brassica rapa – Implications for the function and evolution of the Cruciferae spacer. European Journal of Biochemistry 229: 550–557. [DOI] [PubMed] [Google Scholar]
- Delseny M, Mcgrath JM, This P, Chevre AM, Quiros CF. 1990. Ribosomal-RNA genes in diploid and amphidiploid Brassica and related species – organization, polymorphism, and evolution. Genome 33: 733–744. [Google Scholar]
- Dover GA. 1982. Molecular drive: a cohesive mode of species evolution. Nature 299: 111–117. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Flagel LE, Paterson AH, et al. 2008. Evolutionary genetics of genome merger and doubling in plants. Annual Review of Genetics 42: 443–461. [DOI] [PubMed] [Google Scholar]
- Dubcovsky J, Dvorak J. 1995. Ribosomal RNA multigene loci – nomads of the Triticeae genomes. Genetics 140: 1367–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickbush TH, Eickbush DG. 2007. Finely orchestrated movements: evolution of the ribosomal RNA genes. Genetics 175: 477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson LR, Straus NA, Beversdorf WD. 1983. Restriction patterns reveal origins of chloroplast genomes in Brassica amphiploids. Theoretical and Applied Genetics 65: 201–206. [DOI] [PubMed] [Google Scholar]
- Escobar JS, Glemin S, Galtier N. 2011. GC-biased gene conversion impacts ribosomal DNA evolution in vertebrates, angiosperms, and other eukaryotes. Molecular Biology and Evolution 28: 2561–2575. [DOI] [PubMed] [Google Scholar]
- Fukui K, Nakayama S, Ohmido N, Yoshiaki H, Yamabe M. 1998. Quantitative karyotyping of three diploid Brassica species by imaging methods and localization of 45s rDNA loci on the identified chromosomes. Theoretical and Applied Genetics 96: 325–330. [DOI] [PubMed] [Google Scholar]
- Gaeta RT, Pires JC. 2010. Homoeologous recombination in allopolyploids: the polyploid ratchet. New Phytologist 186: 18–28. [DOI] [PubMed] [Google Scholar]
- Garcia S, Kovarik A. 2013. Dancing together and separate again: gymnosperms exhibit frequent changes of fundamental 5S and 35S rRNA gene (rDNA) organisation. Heredity (Edinb) 111: 23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia S, Garnatje T, Kovarik A. 2012. Plant rDNA database: ribosomal DNA loci data including other karyological and cytogenetic information in plants. Chromosoma 121: 389–394. [DOI] [PubMed] [Google Scholar]
- Gerlach WL, Bedbrook JR. 1979. Cloning and characterization of ribosomal RNA genes in wheat. Nucleic Acids Research 7: 1869–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J, Team G. 2010. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biology 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular Biology and Evolution 27: 221–224. [DOI] [PubMed] [Google Scholar]
- Guo X, Han F. 2014. Asymmetric epigenetic modification and elimination of rDNA sequences by polyploidization in wheat. The Plant Cell 26: 4311–4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasterok R, Maluszynska J. 2000. Nucleolar dominance does not occur in root tip cells of allotetraploid Brassica species. Genome 43: 574–579. [PubMed] [Google Scholar]
- Hasterok R, Jenkins G, Langdon T, Jones RN, Maluszynska J. 2001. Ribosomal DNA is an effective marker of Brassica chromosomes. Theoretical and Applied Genetics 103: 486–490. [Google Scholar]
- Hasterok R, Wolny E, Hosiawa M, et al. 2006. Comparative analysis of rDNA distribution in chromosomes of various species of Brassicaceae. Annals of Botany 97: 205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell EC, Barker GC, Jones GH, et al. 2002. Integration of the cytogenetic and genetic linkage maps of Brassica oleracea. Genetics 161: 1225–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell EC, Kearsey MJ, Jones GH, King GJ, Armstrong SJ. 2008. A and C genome distinction and chromosome identification in Brassica napus by sequential fluorescence in situ hybridization and genomic in situ hybridization. Genetics 180: 1849–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba R, Nishio T. 2002. Phylogenetic analysis of Brassiceae based on the nucleotide sequences of the S-locus related gene, SLR1. Theoretical and Applied Genetics 105: 1159–1165. [DOI] [PubMed] [Google Scholar]
- Kim H, Choi SR, Bae J, et al. 2009. Sequenced BAC anchored reference genetic map that reconciles the ten individual chromosomes of Brassica rapa. BMC Genomics 10: 432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koukalova B, Moraes AP, Renny-Byfield S, Matyasek R, Leitch AR, Kovarik A. 2010. Fall and rise of satellite repeats in allopolyploids of Nicotiana over c. 5 million years. New Phytologist 186: 148–160. [DOI] [PubMed] [Google Scholar]
- Kovarik A, Matyasek R, Lim KY, et al. 2004. Concerted evolution of 18–5.8–26S rDNA repeats in Nicotiana allotetraploids. Biological Journal of the Linnean Society 82: 615–625. [Google Scholar]
- Kovarik A, Pires JC, Leitch AR, et al. 2005. Rapid concerted evolution of nuclear ribosomal DNA in two Tragopogon allopolyploids of recent and recurrent origin. Genetics 169: 931–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovarik A, Dadejova M, Lim YK, et al. 2008. Evolution of rDNA in Nicotiana allopolyploids: a potential link between rDNA homogenization and epigenetics. Annals of Botany 101: 815–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazczyk T, Kovarik A, Eber F, et al. 2011. Immediate unidirectional epigenetic reprogramming of NORs occurs independently of rDNA rearrangements in synthetic and natural forms of a polyploid species Brassica napus. Chromosoma 120: 557–571. [DOI] [PubMed] [Google Scholar]
- Kulak S, Hasterok R, Maluszynska J. 2002. Karyotyping of Brassica amphidiploids using 5S and 25S rDNA as chromosome markers. Hereditas 136: 144–150. [DOI] [PubMed] [Google Scholar]
- Leflon M, Eber F, Letanneur JC, et al. 2006. Pairing and recombination at meiosis of Brassica rapa (AA) x Brassica napus (AACC) hybrids. Theoretical and Applied Genetics 113: 1467–1480. [DOI] [PubMed] [Google Scholar]
- Lim KY, Skalicka K, Koukalova B, et al. 2004. Dynamic changes in the distribution of a satellite homologous to intergenic 26–18S rDNA spacer in the evolution of Nicotiana. Genetics 166: 1935–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe AJ, Abbott RJ. 1996. Origins of the new allopolyploid species Senecio cambrensis (Asteraceae) and its relationship to the Canary Islands endemic Senecio teneriffae. American Journal of Botany 83: 1365–1372. [Google Scholar]
- Mahelka V, Kopecky D, Baum BR. 2013. Contrasting patterns of evolution of 45S and 5S rDNA families uncover new aspects in the genome constitution of the agronomically important grass Thinopyrum intermedium (Triticeae). Molecular Biology and Evolution 30: 2065–2086. [DOI] [PubMed] [Google Scholar]
- Maluszynska J, Heslop-Harrison JS. 1993. Physical mapping of rDNA loci in Brassica species. Genome 36: 774–781. [DOI] [PubMed] [Google Scholar]
- Matyasek R, Renny-Byfield S, Fulnecek J, et al. 2012. Next generation sequencing analysis reveals a relationship between rDNA unit diversity and locus number in Nicotiana diploids. BMC Genomics 13: 722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navratilova A, Koblizkova A, Macas J. 2008. Survey of extrachromosomal circular DNA derived from plant satellite repeats. BMC Plant Biology 8: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas SD, Le Mignon G, Eber F, et al. 2007. Homeologous recombination plays a major role in chromosome rearrangements that occur during meiosis of Brassica napus haploids. Genetics 175: 487–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto Feliner G, Rosselló JA. 2007. Better the devil you know? Guidelines for insightful utilization of nrDNA ITS in species-level evolutionary studies in plants. Molecular Phylogenetics and Evolution 44: 911–919. [DOI] [PubMed] [Google Scholar]
- Nieto Feliner G, Rosselló JA. 2012. Concerted evolution of multigene families and homeologous recombination In: JF Wendel, ed. Plant genome diversity. Vienna: Springer-Verlag, 171–193. [Google Scholar]
- Noe L, Kucherov G. 2005. YASS: enhancing the sensitivity of DNA similarity search. Nucleic Acids Research 33: W540–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn TC, Butrulle DV, Sharpe AG, et al. 2003. Detection and effects of a homeologous reciprocal transposition in Brassica napus. Genetics 165: 1569–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JD, Shields CR, Cohen DB, Orton TJ. 1983. An unusual mitochondrial-DNA plasmid in the genus Brassica. Nature 301: 725–728. [Google Scholar]
- Parkin IA, Sharpe AG, Keith DJ, Lydiate DJ. 1995. Identification of the A and C genomes of amphidiploid Brassica napus (oilseed rape). Genome 38: 1122–1131. [DOI] [PubMed] [Google Scholar]
- Parkin IA, Koh C, Tang HB. et al. 2014. Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid Brassica oleracea. Genome Biology 15: R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecinka A, Schubert V, Meister A, et al. 2004. Chromosome territory arrangement and homologous pairing in nuclei of Arabidopsis thaliana are predominantly random except for NOR-bearing chromosomes. Chromosoma 113: 258–269. [DOI] [PubMed] [Google Scholar]
- Piquemal J, Cinquin E, Couton F, et al. 2005. Construction of an oilseed rape (Brassica napus L.) genetic map with SSR markers. Theoretical and Applied Genetics 111: 1514–1523. [DOI] [PubMed] [Google Scholar]
- Poczai P, Hyvonen J. 2010. Nuclear ribosomal spacer regions in plant phylogenetics: problems and prospects. Molecular Biology Reports 37: 1897–1912. [DOI] [PubMed] [Google Scholar]
- Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW. 1984. Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proceedings of National Academy of Sciences USA 81: 8014–8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarilar V, Palacios PM, Rousselet A, et al. 2013. Allopolyploidy has a moderate impact on restructuring at three contrasting transposable element insertion sites in resynthesized Brassica napus allotetraploids. New Phytologist 198: 593–604. [DOI] [PubMed] [Google Scholar]
- Schlotterer C, Tautz D. 1994. Chromosomal homogeneity of Drosophila ribosomal DNA arrays suggests intrachromosomal exchanges drive concerted evolution. Current Biology 4: 777–783. [DOI] [PubMed] [Google Scholar]
- Snowdon RJ, Kohler W, Kohler A. 1997. Chromosomal localization and characterization of rDNA loci in the Brassica A and C genomes. Genome 40: 582–587. [DOI] [PubMed] [Google Scholar]
- Stupar RM, Song J, Tek AL, Cheng Z, Dong F, Jiang J. 2002. Highly condensed potato pericentromeric heterochromatin contains rDNA-related tandem repeats. Genetics 162: 1435–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suay L, Zhang DS, Eber F, et al. 2014. Crossover rate between homologous chromosomes and interference are regulated by the addition of specific unpaired chromosomes in Brassica. New Phytologist 201: 645–656. [DOI] [PubMed] [Google Scholar]
- Szadkowski E, Eber F, Huteau V, et al. 2010. The first meiosis of resynthesized Brassica napus, a genome blender. New Phytologist 186: 102–112. [DOI] [PubMed] [Google Scholar]
- Szadkowski E, Eber F, Huteau V, et al. 2011. Polyploid formation pathways have an impact on genetic rearrangements in resynthesized Brassica napus. New Phytologist 191: 884–894. [DOI] [PubMed] [Google Scholar]
- Tayalé A, Parisod C. 2013. Natural pathways to polyploidy in plants and consequences for genome reorganization. Cytogenetic and Genome Research 140: 79–96. [DOI] [PubMed] [Google Scholar]
- Udall JA, Quijada PA, Osborn TC. 2005. Detection of chromosomal rearrangements derived from homeologous recombination in four mapping populations of Brassica napus L. Genetics 169: 967–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov RA, Borisjuk NV, Panchuk II, Schweizer D, Hemleben V. 1999. Elimination and rearrangement of parental rDNA in the allotetraploid Nicotiana tabacum. Molecular Biology and Evolution 16: 311–320. [DOI] [PubMed] [Google Scholar]
- Volkov RA, Komarova NY, Hemleben V. 2007. Ribosomal DNA in plant hybrids: inheritance, rearrangement, expression. Systematics & Biodiversity (NHM London) 5: 261–276. [Google Scholar]
- Waters ER, Schaal BA. 1996. Biased gene conversion is not occurring among rDNA repeats in the Brassica triangle. Genome 39: 150–154. [DOI] [PubMed] [Google Scholar]
- Weiss-Schneeweiss H, Bloch C, Turner B, Villasenor JL, Stuessy TF, Schneeweiss GM. 2012. The promiscuous and the chaste: frequent allopolyploid speciation and its genomic consequences in American daisies (Melampodium Sect. Melampodium; Asteraceae). Evolution 66: 211–228. [DOI] [PubMed] [Google Scholar]
- Weiss-Schneeweiss H, Emadzade K, Jang TS, Schneeweiss GM. 2013. Evolutionary consequences, constraints and potential of polyploidy in plants. Cytogenetic and Genome Research 140: 137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel JF, Schnabel A, Seelanan T. 1995. Bidirectional interlocus concerted evolution following allopolyploid speciation in cotton (Gossypium). Proceedings of National Academy of Sciences USA 92: 280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong ZY, Pires JC. 2011. Karyotype and identification of all homoeologous chromosomes of allopolyploid Brassica napus and its diploid progenitors. Genetics 187: 37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong ZY, Gaeta RT, Pires JC. 2011. Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proceedings of the National Academy USA 108: 7908–7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer EA, Martin SL, Beverley SM, Kan YW, Wilson AC. 1980. Rapid duplication and loss of genes-coding for the alpha-chains of hemoglobin. Proceedings of the National Academy of Sciences USA 77: 2158–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zozomova-Lihova J, Mandakova T, Kovarikova A, et al. 2014. When fathers are instant losers: homogenization of rDNA loci in recently formed Cardamine x schulzii trigenomic allopolyploid. New Phytologist 203: 1096–108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.