Abstract
Background and Aims Aubrieta is a taxonomically difficult genus from the Brassicaceae family with approximately 20 species centred in Turkey and Greece. Species boundaries and their evolutionary history are poorly understood. Therefore, we analysed bio- and phylogeographic relationships and evaluated morphological variation to study the evolution of this genus.
Methods Phylogenetic analyses of DNA sequence variation of nuclear-encoded loci and plastid DNA were used to unravel phylogeographic patterns. Morphometric analyses were conducted to study species delimitation. DNA sequence-based mismatch distribution and climate-niche analyses were performed to explain various radiations in space and time during the last 2·5 million years.
Key Results Species groups largely show non-overlapping distribution patterns in the eastern Mediterranean and Asia Minor. We recognized 20 species and provide evidence for overlooked species, thereby highlighting taxonomical difficulties but also demonstrating underexplored species diversity. The centre of origin of Aubrieta is probably Turkey, from which various clades expanded independently towards Asia Minor, south to Lebanon and west to Greece and the Balkans during the Pleistocene.
Conclusions Pleistocene climatic fluctuations had a pronounced effect on Aubrieta speciation and radiation during the last 1·1 million years in the Eastern Mediterranean and Asia Minor. In contrast to many other Brassicaceae, speciation processes did not involve excessive formation of polyploids, but displayed formation of diploids with non-overlapping present-day distribution areas. Expansions from the Aubrieta centre of origin and primary centre of species diversity showed adaptation trends towards higher temperature and drier conditions. However, later expansion and diversification of taxa from within the second centre of species diversity in Greece started ∼0·19 Mya and were associated with a general transition of species adaptation towards milder temperatures and less dry conditions.
Keywords: Aubrieta, biodiversity, Brassicaceae, evolutionary history, Greece, multiple radiations, systematics, taxonomy, Turkey
INTRODUCTION
The Brassicaceae (Cruciferae) comprises 51 tribes, 325 genera and 3740 species in the world (Hohmann et al., 2015) and Southwest Asia. Turkey, in particular, is generally considered to be one of the centres of greatest species diversity, and the Irano–Turanian region might possibly be the origin of the family (Franzke et al., 2009, 2011; Karl and Koch, 2013). Indeed, Turkey has at least 606 Brassicaceae species, of which 226 are endemic (Al-Shehbaz et al., 2007; Al-Shehbaz, 2010; Mutlu, 2012), a fact that reflects the tremendous diversity of the family in that particular region. Many of its genera are also highly diversified in the eastern Mediterranean, including Aethionema, Alyssum, Arabis, Aubrieta, Barbarea, Camelina, Conringia, Hesperis, Isatis and Noccaea, to name some that all except Arabis and Aubrieta are still awaiting detailed phylogenetic–systematic analyses.
Aubrieta is a small-sized, taxonomically difficult genus of about 20 species that are primarily centred in Anatolia, with a second centre of diversity in Greece and the Balkan Peninsula (Phitos, 1970; Gustavsson, 1986; Akeroyd and Ball, 1993; Jalas and Suominen, 1994; Appel and Al-Shehbaz, 2002; Al-Shehbaz et al., 2006; Karl and Koch, 2013). The majority of species are montane or (sub)alpine perennials growing on rocks and scree in woodlands or on open slopes. In addition, several species, cultivars and hybrids are widely grown as rock-garden plants, being suitable for border edging and use on dry shady banks, rocks, and walls (Townsend, 1980; Gustavsson, 1986; Akeroyd and Ball, 1993). The genus Aubrieta is the closest relative of Arabis and both are members of tribe Arabideae (Al-Shehbaz, 2012), which underwent radiation during the Pleistocene (Karl and Koch, 2013). Its closest relative and sister taxon is Arabis verna, from which Aubrieta diverged ∼2·7 Mya; it has been shown that the Aubrieta crown group originated in the central-eastern Mediterranean/Anatolian area (Karl and Koch, 2013).
Some 36 binomials in Aubrieta are listed in the International Plant Names Index (IPNI; http://www.uk.ipni.org/), and about a similar number of infraspecific taxa are unaccounted for in that index. This excessive description of taxa reflects the substantial taxonomic difficulties in the genus. The first comprehensive taxonomic study of Aubrieta was presented by Boissier (1867). Seventy years later, Mattfield (1937) presented an updated taxonomic and morphological overview of the genus that was largely influenced by the horticultural relevance of Aubrieta. Reichardt (1959) presented the most valuable and comprehensive morphological and taxonomic work, but unfortunately this has been largely overlooked because it is an unpublished thesis written in German. This contribution summarized the comprehensive knowledge available from the 18th century until the 1950s and added further morphological and biogeographical data, though no key to the species was provided. With the growing body of molecular data on Brassicaceae, the phylogenetic context of Aubrieta has become much clearer during the past 15 years. There has been rapidly accumulating evidence for the placement of Aubrieta in tribe Arabideae (Koch et al., 1999, 2000, 2001, 2007; Warwick et al., 2010), with some of the first evidence that it is nested within the para- and polyphyletic Arabis (Koch et al., 2005; Karl et al., 2012). A series of detailed phylogenetic and phylogeographic studies have unravelled the respective evolution of various clades within the Arabideae (Jordon-Thaden et al., 2010; Koch et al., 2010, 2012a; Karl et al., 2012; Karl and Koch, 2013, 2014) and highlighted the difficulties in identifying reliable morphological characters that distinguish various genera (Al-Shehbaz et al., 2011). The systematics of Arabideae have been challenging because of the extreme convergence in almost all conceivable morphological characters (Koch et al., 2012a). Most characters appear in various combinations in different tribes and evolutionary lineages, and this made Arabis, as delimited prior to molecular studies, the most critical genus of Brassicaceae (Koch et al., 1999, 2000, 2001).
In summary, the Arabideae currently consists of about 508 species (Hohmann et al., 2015) distributed among 17 genera (Koch et al., 2012b; Karl and Koch, 2013; Kiefer et al., 2014). The major Arabideae clades have been identified, and detailed studies have focused on the Arabis alpina clade, the Arabis nordmanniana clade, the Arabis auriculata clade (Arabis s. str.), the Draba clade, the Arabis aucheri clade, the main Arabis clade and the Scapiarabis clade (for a summary see Karl and Koch, 2013). A comprehensive phylogenetic–systematic analysis of Aubrieta is the last step towards unravelling the evolutionary history of the entire Arabideae.
Although the Brassicaceae of Turkey and Greece are fairly well known (Davis, 1965; Davis et al., 1988; Güner et al., 2000; Strid and Tan, 2002; Al-Shehbaz et al., 2007; Mutlu, 2012), new taxa continue to be discovered, especially from areas that have not been adequately explored before. For example, recent fieldwork by one of us (M.A.K.) and collaborative research in various parts of Turkey led to the discovery of the novelty described as Aubrieta ekimii Yüzb., Al-Shehbaz & M.A. Koch (Yüzbaşıoğlu et al., 2015), as well as the re-collection of Aubrieta vulcanica from the putative type locality after more than 100 years since its discovery; the collection of new unassigned accessions is discussed in more detail in this contribution. A compilation of information on the accepted Aubrieta species (e.g. Reichardt, 1959; Phitos, 1970; Gustavsson, 1986; Akeroyd and Ball, 1993; Jalas and Suominen, 1994) indicates that A. columnae, A. deltoidea and A. parviflora are the most widespread species, and the first two represent complexes with several infraspecific taxa (Cullen, 1965; Akeroyd and Ball, 1993; Greuter et al., 1986). Six other species are endemic to Turkey (A. olympica, A. pinardii, A. canescens, A. anamasica, A. ekimii and A. vulcanica), of which the last three are very local endemics (Cullen, 1965; Peşmen and Güner, 1978). Five species are almost endemic to Greece: A. erubescens, A. glabrescens, A. gracilis, A. scyria and A. thessala (Phitos, 1970, 2002). Finally, A. libanotica is restricted to high elevations in Lebanon and adjacent Syria and Turkey (Mouterde, 1986), A. scardica is endemic to the Balkan Peninsula (Albania, Bosnia and Herzogovena, Bulgaria, Croatia, Greece and Macedonia; Akeroyd and Ball, 1993; Phitos, 2002) and A. albanica is endemic to Albania (Meyer, 2011). Two additional taxa were described from Greece: A. intermedia (sometimes considered a synonym of A. deltoidea) and the putative hybrid and local endemic A. × hybrida. Taxonomic treatments of species and subspecies differ among floras, with A. glabrescens and A. scardica treated as subspecies of A. gracilis, and the cultivars are largely combined under the name A. × cultorum, often with A. deltoidea involved.
In the current study, we present a taxonomic–phylogenetic circumscription of the genus Aubrieta combining comprehensive morphometric data with large-scale phylogeographic DNA sequence information from the nuclear and plastid genomes. The ancestral distribution area and subsequent range expansions of Aubrieta are discussed, and we present a spatio-temporal interpretation of this biogeographic data. A key to all species is provided, nomenclatural adjustments are proposed, new chromosome counts are added and comments on putatively new taxa are presented.
MATERIALS AND METHODS
Plant material and taxon sampling
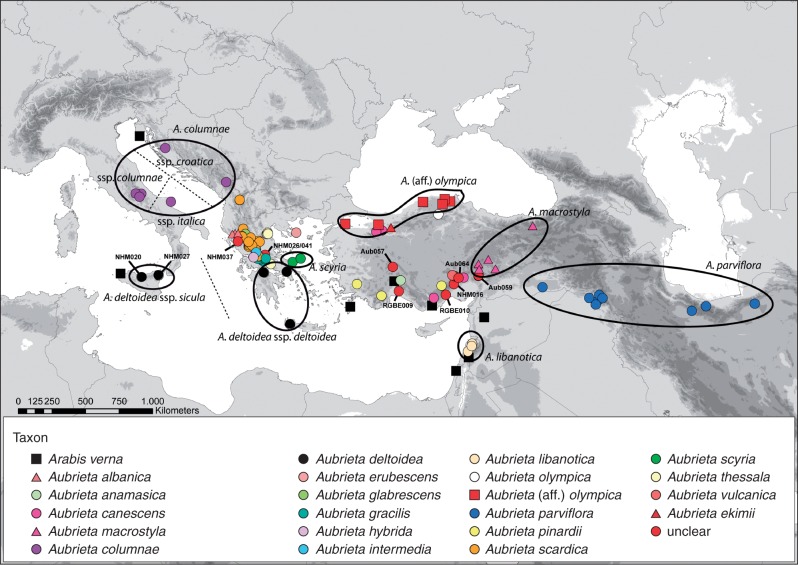
The most important centres of Aubrieta species diversity are Turkey and Greece. Consequently, we focused on material from herbaria housing the majority of important vouchers documenting the floras of Turkey, Greece and the Balkans [Natural History Museum London (BM), Royal Botanic Garden and Herbarium, Edinburgh (E), Lund University Botanical Museum and Herbarium (LD), Jena Botanical Garden and Haussknecht Herbarium (JE) and Berlin Botanical Garden and Herbarium (B)]. The respective collections served as the major source for the circumscription and documentation of the relevant floras. Additional material was added based on our collaboration with Turkish colleagues continuing to collect new and unknown phenotypes (Sırrı Yüzbaşıoğlu, Istanbul; Ali A. Dönmez, Ankara). In addition, our own fieldwork focused on re-collecting species unknown for more than 100 years (e.g. A. vulcanica). In all, we selected 132 accessions of Aubrieta for subsequent morphological and molecular analyses. Ten additional accessions of Arabis verna were selected to cover its entire Mediterranean distribution range and serve as an outgroup for the molecular analyses. The phylogenetic position of annual Arabis verna as sister to Aubrieta was shown earlier (e.g. Karl and Koch, 2013). In total, 142 accessions were analysed, and all voucher details are shown in Supplementary Data Table S1. A distribution map of analysed Aubrieta accession is shown in Fig. 1.
Fig. 1.
Distribution of sampled Aubrieta and Arabis verna accessions in the Mediterranean and Irano–Turanian region. Some accessions are indicated with their code (see Table S1). Aubrieta deltoidea ssp. sicula is newly introduced herein as A. columnae ssp. sicula.
DNA extraction, PCR amplification and sequencing
DNA extractions were carried out using the NucleoSpin Plant II Kit (Macherey-Nagel, Düren, Germany) or according to a slightly modified protocol (Karl et al., 2012) of the CTAB method (Doyle and Doyle, 1987). For phylogenetic inferences we chose the internal transcribed spacer regions (ITSs) 1 and 2 of the nuclear ribosomal (nr) RNA coding region, including the intervening 5·8 S rRNA gene, and three regions from the plastid genome: (1) the trnL-trnF region, including the trnL intron, the second exon of the trnL gene and the trnL-trnF intergenic spacer (hereafter in total named the trnLF region); (2) trnC-ycf6 intergenic spacer; and (3) the trnS-ycf9 intergenic spacer. PCR reactions to amplify the selected regions were performed in a final volume of 25 µL, using 10 µm of each primer, a total of 2·0 mm MgCl2, and 0·5 U of MangoTaq polymerase (Bioline, Luckenwalde, Germany). The following primers and annealing temperatures were used: ITS, forward 5′-GGA AGG AGA AGT CGT AAC AAG G-3′, reverse 5′-GGG TAA TCC CGC CTG ACC TGG-3′ (44 °C); trnL intron, forward 5′-CGA AAT CGG TAG ACG CTA CG-3′, reverse 5′-ATT TGA ACT GGT GAC ACG AG-3′ (52 °C); trnLF intergenic spacer, forward 5′-GGTTCA AGT CCC TCT ATC ATC CC-3′, reverse 5′-TCC TCT GCC AAG AAC CAG ATT TG-3′ (50 °C); trnS-ycf9 intergenic spacer, forward 5′-GAG AGA GAG GGA TTC GAA CC-3′, reverse 5′-CAA AMA CAG CCA ATT GGA AAG C-3′ (56 °C); trnC-ycf6 intergenic spacer, forward 5′-CCA GTT CRA ATC YGG GTG-3′, reverse 5′-GCC CAA GCR AGA CTT ACT ATA TCC AT-3′ (56 °C). The PCR protocols followed Karl et al. (2012). PCR products were purified by centrifugation in a NucleoFast 96 PCR plate (Macherey-Nagel) and directly sequenced using the above primers with respective extensions at the 5′-end (M13 forward 5′-TGT AAA ACG ACG GCC AGT-3′; M13 reverse 5′-CAG GAA ACA GCT ATG ACC-3′). DNA custom sequencing was performed by GATC (Konstanz, Germany) and MWG Eurofins (Ebersberg, Germany).
Alignments and phylogenetic reconstructions
Network analyses were performed with two datasets using the program TCS version 1.21 (Clement et al., 2000). For each marker system (ITS and concatenated plastid trnLF-trnS-ycf9-trnCycf6) a separate analysis was performed. Prior to the analyses, indels more than one base pair in length were coded as one single additional binary character. Gaps were coded (presence/absence) as homologous character states among aligned sequences only if length and position were identical following the ‘simple indel coding’ procedure discussed in detail by Simmons and Ochoterena (2000). Due to insufficient sequencing quality, missing sequence information at the 3′-end and the 5′-end of the DNA alignments was, however, filled with ‘N’s and assigned to the respective haplotypes, according to their remaining sequence. Length variations in polyA, T or G regions were not considered and not coded as characters. Haplotypes of Arabis verna were used as outgroups. After a first run of sequence-type detection, accessions with redundant sequence information were excluded from the final analyses. The connection limit was set to 95 %. We did not show results of tree-building phylogenetic analysis of ITS, because it has already been demonstrated that this marker is poorly resolving within Aubrieta (Karl and Koch, 2013; Yüzbaşıoğlu et al., 2015), and that network analyses as performed here have the best discriminatory power to recognize geographic and phylogenetic/taxonomic patterns (Karl and Koch, 2013, 2014). The concatenated plastid DNA alignment was subjected to phylogenetic analysis using the maximum likelihood criterion and running the programme RAxML (Stamatakis, 2006; Silvestro and Michalak, 2012). The best-fitting nucleotide substitution model was selected using Modeltest version 3.7 (Posada and Crandall, 1998) (GTR + I + Γ). Bootstrapping was performed with 1000 replicates, and Arabis verna was used as outgroup (Karl and Koch, 2013).
Demographic population expansion analyses
In order to test the demographic expansion scenario, mismatch distribution analysis of cpDNA sequence data was performed using a sudden (stepwise) expansion model (Rogers and Harpending, 1992) for the various haplotype groups detected by network analyses at species-group level. Goodness of fit was tested using the sum of squared deviations (SSD) between observed and expected mismatch distributions and Harpending’s raggedness index (HRag; Harpending, 1994). The number of sites where two DNA sequences differ is proportional to the expansion time, making it possible to determine the date of population expansions directly from mismatch distributions (Rogers and Harpending, 1992; Schneider and Excoffier, 1999). For groups with expanding populations, the expansion parameter (τ) was converted to an estimate of time (T, in number of generations) since the start of expansion began using T = τ/2u (Rogers and Harpending, 1992; Rogers, 1995). In this formula, the neutral mutation rate for the entire sequence (haplotype) per generation of u is calculated as u = μkg, where μ is the substitution rate, k is the average sequence length of the DNA region under study, and g is the generation time in years. Generation time g was set to 2 years. Total alignment length was 1757 base pairs. The substitution rate was set to 7·798 × 10−10 mutations site−1 year−1 using the previously defined stem group age of the genus (3·27 Mya, based on cpDNA; Karl and Koch, 2013). A parametric bootstrap approach (Schneider and Excoffier, 1999) with 1000 replicates was used to assess the goodness of fit of the observed mismatch distribution to the sudden expansion model, to test the significance of HRag, and to obtain 95 % confidence intervals (CIs) around τ. Significantly negative D and FS values often indicate population expansion following a severe reduction in population size (Fu, 1997). For the total dataset and each evolutionary lineage, Tajima’s D (Tajima, 1989) and Fu’s FS (Fu, 1997) tests of selective neutrality were conducted. All the above analyses were carried out with Arlequin 3·0 (Excoffier et al., 2005).
Cytogenetic analysis
When seed or living material was available, chromosome numbers were determined using root tips from germinated seeds. Root tips were incubated for 2 h in 2 mm 8-hydroxiquinoline at room temperature and subsequently for 2 h at 4 °C, and transferred to freshly prepared, ice-cold ethanol:acetic acid (3:1). The root tips were stored at − 20 °C for subsequent analysis. Root tips were washed in water, hydrolysed in 5 m HCl for 20 min, washed in water again, mounted on microscope slides, squashed in 45 % acetic acid and fixed at − 80 °C for 30 min. The cover glasses were carefully removed and the slides with fixed root tips were dried and stained with 4 % Giemsa solution (Merck). Additional information on chromosome numbers from various Aubrieta species was obtained from a database by Warwick and Al-Shehbaz (2006) and BrassiBase (Koch et al., 2012b; Kiefer et al., 2014) and recent literature (Yüzbaşıoğlu et al., 2015). In a few cases we also estimated genome sizes following the protocol presented recently (Hohmann et al., 2015) and deposited the data with BrassiBase (Koch et al., 2012b; Kiefer et al., 2014).
Morphometric analysis
Morphological analyses were performed on dried herbarium vouchers deposited at the herbarium at Heidelberg Botanic Garden (HEID) and other herbaria (Table S1). The various measurements were scored by hand using a high-power binocular microscope to measure dimensions on digital live images in high resolution. The removal of some plant organs (e.g. flowers and flower buds) from the original voucher was necessary for appropriate analysis. Here, 100 µL of a pre-washing solution (washing-up liquid, 0·1 %) was added to the removed plant organ, and the organ was further covered with boiling water (total volume 10 mL) for 2 min. The organs were then dissected onto a glass slide, and after removing the water the samples were fixed and mounted on blotting paper and finally dried for 2 d at 40 °C (Koch et al., 2013). In total, 12 characters (continuous, discrete, multistate) were scored (Supplementary Date Table S2) from leaves, flowers and fruit. Initially, these characters were selected from comparative species descriptions and morphological keys to the genus that have been traditionally used to distinguish the different taxa (Cullen, 1965; Gustavsson, 1986; Pollunin, 1987; Hedge, 1968; Phitos, 2002; Ančev and Goranova, 2009; Daşkin and Kaynak, 2011; Meyer, 2011; Yüzbaşıoğlu et al., 2015). The coding of character states did not imply or reflect any evolutionary progression (Möller et al., 2007), and details are listed in Supplementary Data Table S3. From these characters, a final matrix for all individuals (eight quantitative characters, of which two were based on ratios, five were multistate characters and one was a two-state character) was extracted. Investigated individuals of the same species were from different populations, and all measurements of a respective accession refer to one single individual. A principal coordinates analysis (PCoA) was run to analyse the multivariate data using single individuals as operational taxonomic units using MVSP version 3·22 (Kovach Computing Services). PCoA requires that continuous character states are normally distributed, and as our tests demonstrated that no character showed significant skewness, data transformation was unnecessary. The Gower general similarity coefficient was used to generate the similarity/distance matrix (Gower, 1971).
The same dataset was also subjected to cluster analysis to obtain a tree-based visualization of morphological variation. For consistency among analysis we also used the Gower general similarity coefficient and selected the nearest neighbour clustering algorithm implemented in MVSP.
In order to further study the discriminative power of morphological characters and to prepare a genus-wide key to the species, univariate analysis of variance (ANOVA) was run for continuous characters (petal length, style length, length:width fruit ratio and leaf ratio) using SPSS (version 19 for Windows). Based on highly significant P values of < 0·001 of the entire datasets from ANOVA, a posteriori mean separation tests (Hochberg GT2 test; Hochberg, 1974) were conducted for the four characters to illustrate the possibility of using individual characters to distinguish significantly between species pairs. For the a posteriori mean separation tests, we used Hochberg’s GT2 because sample size was not equal among species (Field, 2013). For ANOVA we further had to delete from the analysis any taxon with only one specimen or individual. The analyses were conducted using SPSS (version 19 for Windows).
Bioclimate space evaluation
For this analysis, specific bioclimatic data values (19 bioclimatic variables plus altitude information and data on monthly minimal and maximal temperatures and average precipitation – downloaded at 30 arc-second resolution from http://www.worldclim.org) were extracted for each locality using the program DIVA-GIS (version 7·5·0, http://www.diva-gis.org). Principal component analyses (PCAs) were carried out with the program IBM SPSS Statistics version 19·0·0 using data on these 19 bioclimatic variables and altitude information. In each PCA, a factor analysis was done to reduce the dimensions of the original bioclimatic variables. A correlation matrix was used, and no factor rotation was applied. All principal components with eigenvalues >1 were extracted according to Kaiser’s rule (Zwick and Velicer, 1986).
RESULTS
A plastid DNA phylogenetic network demonstrates strong geographic structure
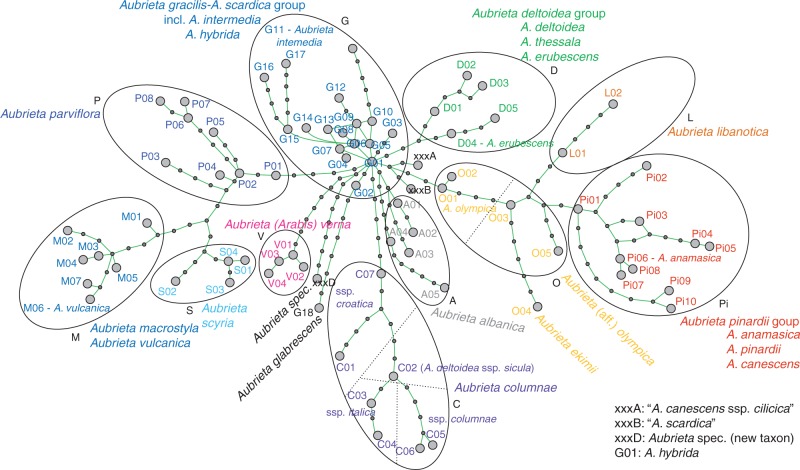
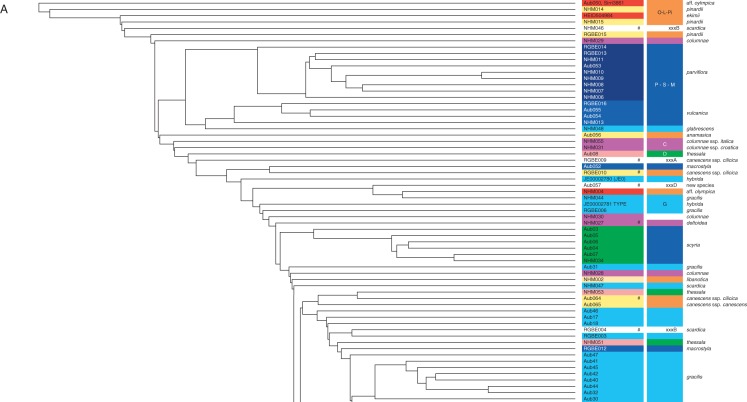
The final plastid DNA alignment consisted of 1756 characters (1735 regular bases, plus 21 positions used for additional gap-coding) and 78 haplotypes. GenBank accession codes are provided in Table S1 and alignments in Supplementary Data Tables S4–S7. The resulting minimum-spanning network from TCS is shown in Fig. 2. This network distinguishes various haplotype groups, named with 11 capital letters.
Fig. 2.
Plastid DNA haplotype network calculated with TCS. Small solid circles without haplotype codes indicate non-sampled or extinct haplotypes.
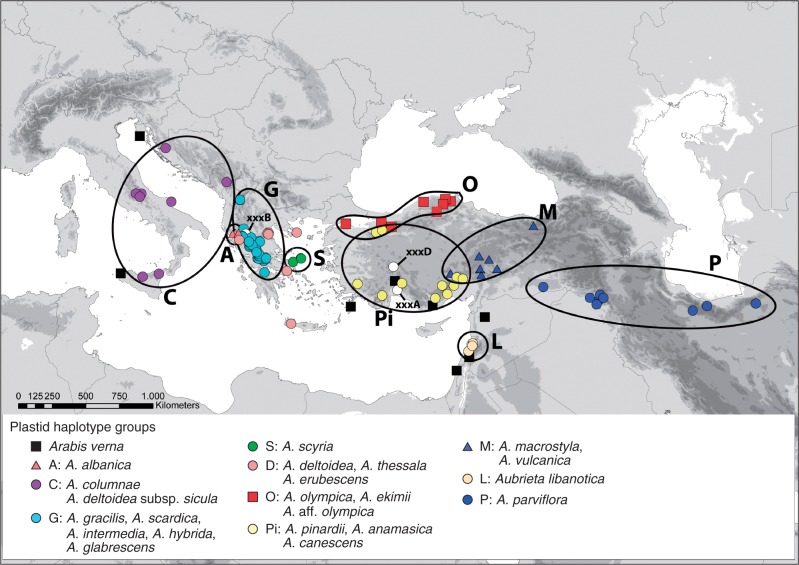
The outgroup, Arabis verna (group V), is connected with the central part of the network, which is fully consistent with phylogenetic analyses (Karl and Koch, 2013). The definition of groups does not follow a strict concept of monophyly (clades) and is influenced by both taxon identity and geographic distribution. Taxon names have been adjusted according to our detailed morphometric analysis (Table S1). Most haplotype groups show a clear biogeographic pattern (Fig. 3) and exhibit largely non-overlapping species distribution areas. Moreover, each of these groups consists of a limited number of species only. The most easterly distributed species is A. parviflora (group P) in East Turkey, Syria, Iraq and Iran. Related to group P is haplotype group M with A. canescens subsp. macrostyla and A. vulcanica from Central Turkey. We will discuss later why A. canescens subsp. macrostyla is best treated at the species level (A. macrostyla), as indicated in Figs 1–3. Further to the south, haplotype group L consists of A. libanotica from Lebanon and Syria. Haplotype group Pi is exclusively found in Turkey, with three species, A. canescens, A. pinardii and A. anamasica. Haplotype group O consists of taxa related to A. olympica in northern Turkey close to the Black Sea.
Fig. 3.
Distribution of plastid haplotypes. Haplotype group definition is in accordance with the TCS network shown in Fig. 2.
This group includes A. ekimii and also unknown taxa as preliminarily described here as A. aff. olympica. In Greece, the highly diverse haplotype group G occurs together with A. gracilis and A. scardica and the largely underexplored A. intermedia, A. hybrida and A. glabrescens. Also distributed in Greece is the haplotype group D. In contrast to haplotype group G, found at higher elevations in inland Greece, haplotypes of group D are distributed closer to the coast at lower elevations. This group consists of A. thessala, A. erubescens and A. deltoidea. Haplotype group S is locally distributed on the Greek island of Skyros and exclusively represents A. scyria. In northwestern Greece towards Albania, haplotype group A is found in A. albanica. Finally, haplotype group C is found on the Balkan Peninsula but also in Italy and further south into Sicily. Haplotype group C consists of A. columnae, and the haplotype network also separates its three subspecies (croatica, italica and columnae). However, in Sicily we also find A. deltoidea subsp. sicula from the same haplotype group, and we might have to assume an ancient hybrid origin between A. columnae subsp. italica and an A. ea representative. Alternatively, this taxon is wrongly assigned and should be better treated as A. columnae subsp. sicula, as discussed below in more detail. Finally, three haplotypes (xxxA, xxxB, and xxxD) belong to accessions not obviously defined morphologically. Haplotype xxxA is found in one accession of A. canescens subsp. cilicica, but other subsp. cilicica carry different haplotypes clearly belonging to A. canescens (haplotype group Pi). Haplotype xxxB is found in A. scardica and A. thessala and is easily associated with haplotype group G and its respective taxa. Finally, one accession was placed in none of the groups, and it was nested in the centre of the network and carried a unique ITS types: haplotype xxxD. This accession was collected by A. Dönmez (March 2001), and it did not match any given species description.
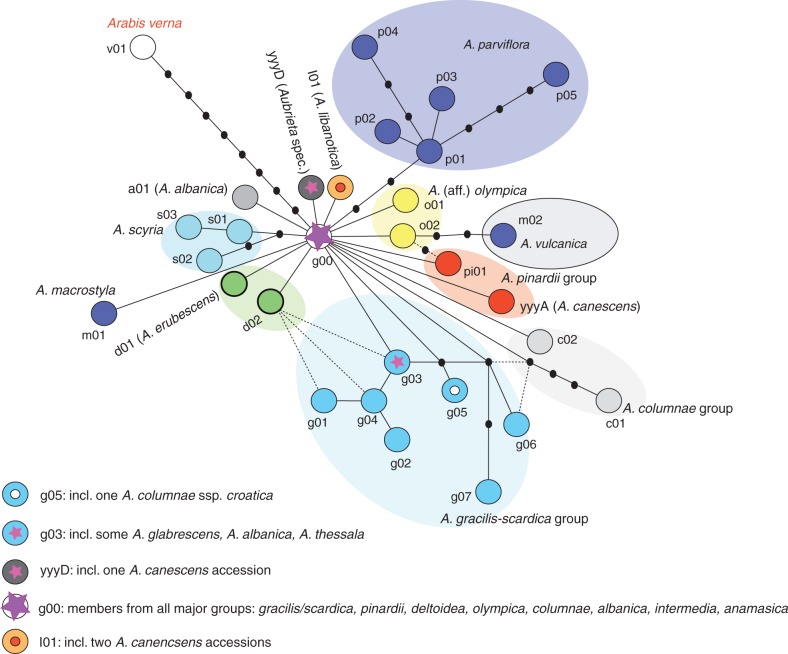
The ITS network does not highly resolve phylogenetic relationships
The final ITS nrDNA alignment consisted of 600 characters (599 nucleotide positions plus one gap-coding character) (Supplementary Data Table S8). GenBank accession codes and ITS types are presented in Table S1. The minimum-spanning network from TCS recognizes 31 ITS haplotypes with only little structure in the whole network (Fig. 4). However, various cpDNA haplotype groups are consistently recognized with ITS data. (1) ITS type m01 (A. macrostyla) corresponds to cpDNA haplotype group M; (2) ITS types p01–05 (A. parviflora) correspond to cpDNA haplotype group P; (3) ITS type s01–s03 (A. scyria) correspond to cpDNA haplotype group S; and (4) ITS type l01 (A. libanotica) corresponds to cpDNA haplotype group L. Other locally distributed taxa, such as A. albanica (ITS type a01), A. erubescens (ITS type d01) and Aubrieta spec. nov. (ITS type yyyD), are also characterized by their own and unique ITS types.
Fig. 4.
TCS network of nuclear-encoded ITS types. Small black circles without haplotype codes indicate non-sampled or extinct haplotypes.
For a few other accessions, ITS data might provide some preliminary hints towards phylogenetic distinctiveness (A. olympica and taxa close to it, A. aff. olympica). In the case of two originally unclear A. canescens accessions, which were identified as A. canescens subsp. cilicica, unusual ITS types (yyyA and yyyD) were found, and the same ITS type (yyyD) was found in the Aubrieta spec. nov. Two other A. canescens subsp. cilicica accessions carry the l01 ITS type found otherwise in A. libanotica. It is probably insignificant in network analysis, but the consistent pattern of unusual distribution of molecular variation in a given taxon (A. canescens subsp. cilicica) may highlight a more complex evolutionary history. The Sicilian A. deltoidea subsp. sicula (carrying an A. columnae cpDNA haplotype) also has different ITS types (c03, g00) compared with most other A. columnae accessions.
Demographic expansion analysis shows several independent radiations
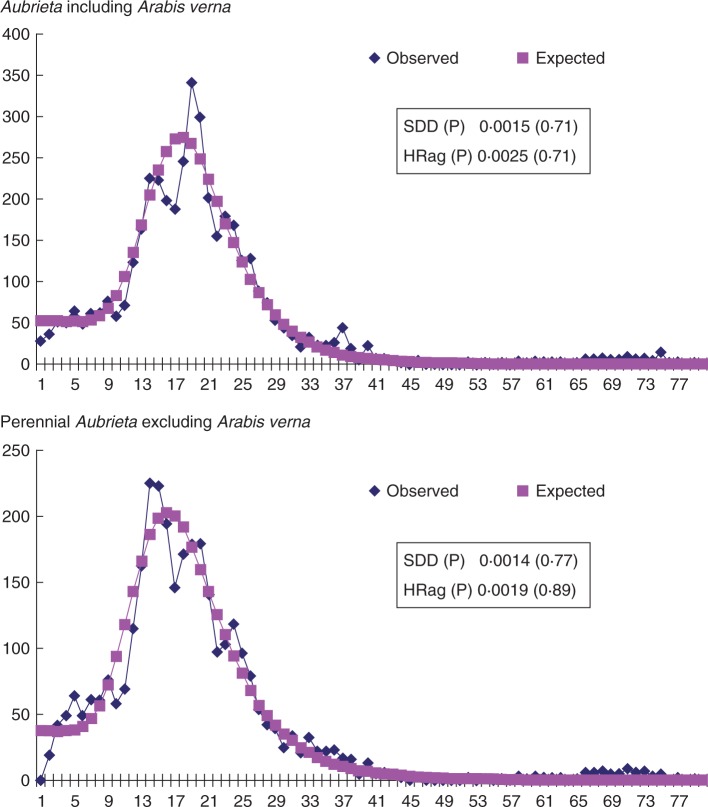
The corrected cpDNA sequence divergence (dA) between the annual Arabis verna and perennial Aubrieta was 0·0051. Using the cpDNA-based divergence time estimated earlier (3·27 Mya; Karl and Koch, 2013), the substitution rate was estimated to be 7·798 ×10−10 mutations site−1 year−1 for the three combined cpDNA regions. Neutrality tests were significantly negative for both Tajima’s D (−2·1199/−2·1805) and Fu’s FS (−24·0245/−24·07625) at the genus level (with/without outgroup, respectively). Mismatch distribution analyses detected unimodal distributions with insignificant SSD (0·0015; P = 0·71/0·0014; P = 0·77) and HRag (0·0025; P = 0·71/0·0019; P = 0·89) statistics at the genus level (with/without Arabis verna) (Fig. 5). Mismatch distribution analysis for the five major cpDNA haplotype groups G, O-L-P, D, C and P-S-M show very similar results (Table 1). For all four haplotype groups, Tajima’s D and Fu’s FS are negative (significant in almost all cases). Unimodal distributions were detected with insignificant SSD and HRag statistics for all cpDNA haplotype groups. Therefore, the evidence for population expansion for each cpDNA haplotype group seems to be relatively strong. The onset of expansion/radiation of the entire genus Aubrieta was estimated to be 1·17 Mya. The P-S-M group started to expand 1·11 Mya, the C-group 0·75 Mya, the O-L-P group 0·53 Mya, the D group 0·41 Mya and the G group 0·19 Mya (Table 1; here the 95 % confidence intervals are also given). Considering Turkey as the centre of origin of the genus (Karl and Koch, 2013), a first expansion (P-S-M) started 1·1 Mya towards the Irano–Turanian region, followed by a second expansion (O-L-Pi) 0·53 Mya further south into Lebanon and Syria in that region. Expansions into Greece (groups D and G), occurred much later, at 0·41 and 0·19 Mya, respectively. Interestingly, the expansion of group C into the Balkan Peninsula and Italy occurred earlier than the Greek expansions (0·75 Mya), but since Tajima’s D and Fu’s FS are not insignificant for group C, this result should be interpreted with caution (Table 1).
Fig. 5.
Mismatch distribution analysis showing unimodal distributions with SSD and HRag statistics at genus level. Mismatch distribution analyses for the five major cpDNA haplotype groups G, O-L-P, D, C and P-S-M show very similar results compared with each other (Table 1).
Table 1.
Summary statistics from the mismatch distribution analyses of plastid DNA sequence variation. Definition of evolutionary lineages follows Figs 2 and 3
| SDD (P value) | HRag (P value) | D (P value) | FS (P value) | τ, mean (95 % confidence interval) | Expansion time (Mya), mean (95 % confidence interval) | |
|---|---|---|---|---|---|---|
| Aubrieta incl. A. verna | 0·0015 (0·70) | 0·0025 (0·71) | −2·1198 (0·00) | −24·02 (0·00) | 15·11 (11·47–26·33) | 1·37 (2·39–1·04) |
| Aubrieta excl. A. verna | 0·0014 (0·77) | 0·0019 (0·89) | −2·1805 (0·00) | −24·07 (0·00) | 12·87 (9·18–25·25) | 1·17 (2·29–0·83) |
| Lineage PSM | 0·0117 (0·38) | 0·0158 (0·64) | −1·6633 (0·03) | −9·57 (0·00) | 12·32 (8·09–15·28) | 1·11 (1·38–0·73) |
| Lineage C | 0·0805 (0·12) | 0·2449 (0·09) | −0·5002 (0·34) | −1·55 (0·08) | 8·43 (4·76–14·16) | 0·75 (1·28–0·43) |
| Lineage OLP | 0·0059 (0·86) | 0·0129 (0·62) | −1·4831 (0·05) | −7·10 (0·00) | 5·88 (2·88–20·88) | 0·53 (1·89–0·26) |
| Lineage D | 0·1185 (0·16) | 0·3000 (0·27) | −2·2098 (0·00) | −1·55 (0·10) | 4·46 (2·19–7·09) | 0·41 (0·64–0·19) |
| Lineage G | 0·0089 (0·86) | 0·0200 (0·57) | −1·9198 (0·01) | −21·27 (0·00) | 2·15 (0·88–8·09) | 0·19 (0·73–0·08) |
Diploids are the prevailing cytotype
Herein we present new chromosome counts for A. canescens, A. olympica, A. macrostyla, A. canescens subsp. cilicica, A. vulcanica and for A. deltoidea subsp. sicula with exclusively diploid accessions (2n = 16). For A. parviflora and A. ekimii, diploid chromosome counts (2n=16) confirmed past results (see the ‘Chromosome number and genome size’ tool in BrassiBase; Koch et al., 2012b; Kiefer et al., 2014). One Arabis verna accession was identified as a tetraploid sample with 32 chromosomes (Table S1). This is also in congruence with published data demonstrating that both ploidy levels (diploid and tetraploid) occur naturally (Kiefer et al., 2014). From voucher label information, we extracted chromosome numbers for tetraploid A. scardica (2n = 32) and diploid A. gracilis (2n = 16). However, published data also provide evidence for tetraploid A. gracilis. All other species for which some chromosome counts do exist are diploids: A. columnae, A. deltoidea, A. erubescens, A. glabrescens and A. scyria. In summary, we can conclude that polyploids are extremely rare and have been found in cpDNA haplotype G from Greece only, and that diploids prevail. However, it should be mentioned that we lack extensive and representative cytogenetic data for most taxa.
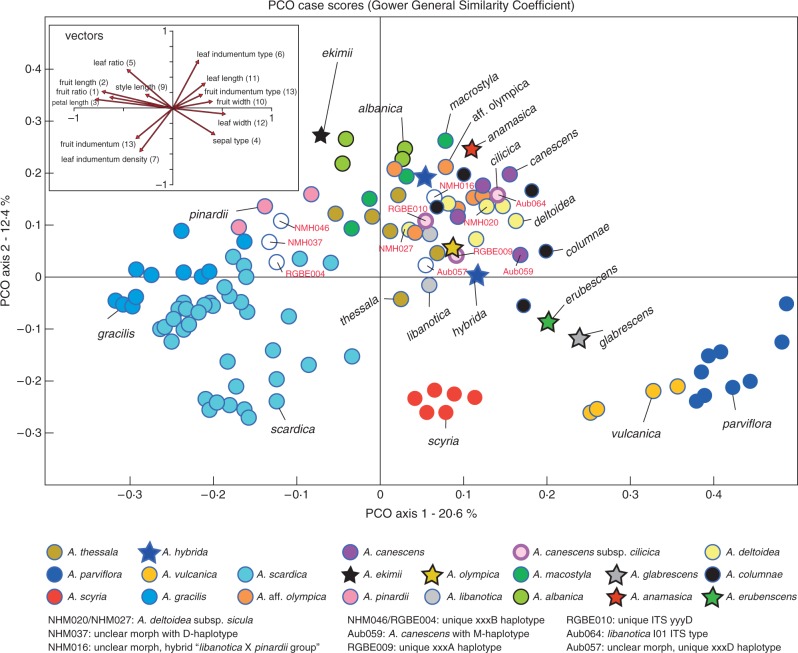
Multivariate morphometric analysis
The results from the morphometric analyses are summarized in Table S2, and a summary is shown with the PCoA in Fig. 6. The first two coordinates explain 33·0 % of the total variation (first, 20·6 %; second, 12·4 %; third, 7·8 %) and a good morphological separation is apparent for many species, such as the Greek A. scardica, A. gracilis and A. scyria, the Albanian A. albanica, the Turkish A. pinardii and A. vulcanica, and the Irano–Turanian A. parviflora. Two endemic taxa from Turkey, A. anamasica and A. ekimii, are also separated from the remaining species. However, other taxa show an overlapping morphological space with various other species close to the centre of the ‘two-dimensional morphospace’ (of the first two coordinates) (Fig. 6). This is particularly true for A. canescens (and subspecies) and A. olympica/A. olympica aff. olympica. A number of accessions showing unexpected genetic composition (morphology versus DNA data) are indicated with their accession codes (Table S1) in Fig. 1, and most of them are also centred within the cloud of PCA data points. Variation along the third axis does not provide any further taxonomic resolution.
Fig. 6.
Results of PCoA of morphological characters. Direction and value of respective eigenvectors are shown in the upper left box. Several accessions are indicated with respective codes, and here some further information on haplotypes is repeated (see text for details; see also Figs 2, 3 and 4).
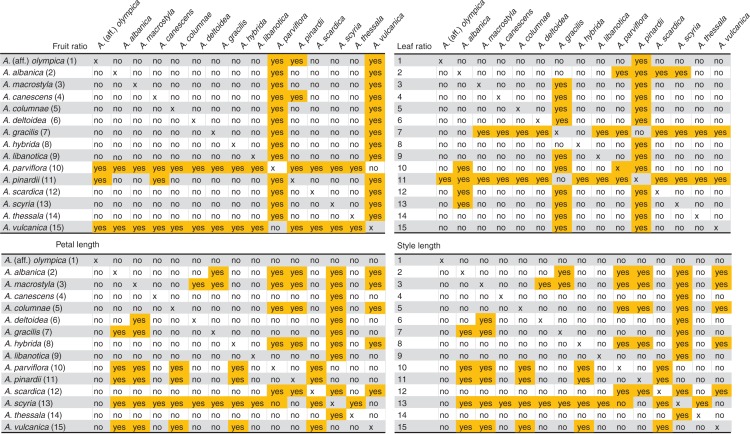
Some of these unexpected results (comparing morphological and genetic data) may best be explained by hybridization and reticulation (Aub059, Aub064, NHM016, NHM037, NHM020, NHM027), others are very unique and very distant genetically (Aub057, NHM046 and RGBE004) and may be best recognized as separate species, and the remaining accessions (RGBE009, RGBE010) may correspond (such as Aub064) with a polyphyletic taxon (A. canescens subsp. cilicica). Analysis of the discriminative power of (quantitative) characters showed similar results (Fig. 7), and species such as A. parviflora, A. vulcanica, A. gracilis, A. pinardii and A. scyria are significantly separated from most other species.
Fig. 7.
Results of the a posteriori mean separation tests (Hochberg GT2 test; Hochberg, 1974) of quantitative morphological characters. Significant differences between species pairs are indicated and highlighted in orange. yes means significant, no means not significant.
Single morphological characters will not be discussed further here, but we have integrated all results into a new morphological key to species and subspecies of Aubrieta, provided at the end of the discussion.
Morphological variation shows limited phylogenetic information
Morphological variation used in phenetic reconstruction has limited power to identify taxonomic groups (Fig. 8). The result is largely congruent with PCoA analysis (Fig. 6). However, only a few species groups are combined into groups congruent with phylogenetically (cpDNA data) defined lineages such as gracilis–scardica and parviflora–vulcanica. The majority of phenotypically based groups are not reflected in plastid types (Fig. 3), evolutionary lineages defined by network and mismatch distribution analyses (Fig. 2) or maximum likelihood analysis (Fig. 8). Accordingly, datasets (cpDNA and morphology) cannot be used in one single combined analysis because of the significant conflict among respective tree topologies.
The bioclimatic space: different preferences
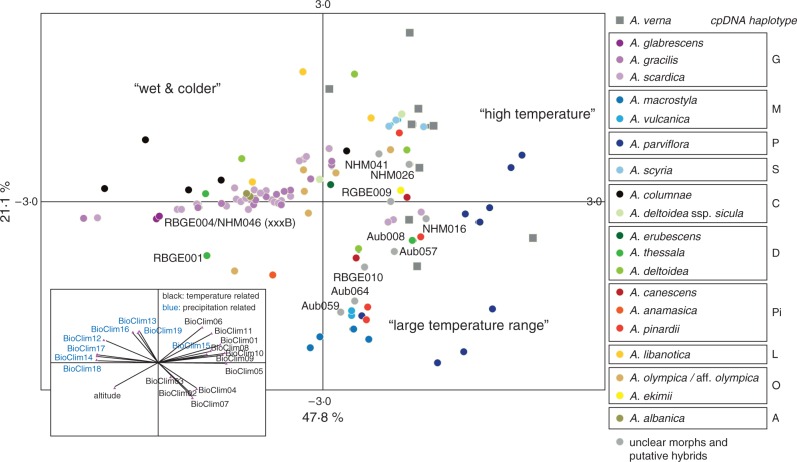
Bioclimatic raw data plus altitudinal information for accessions studied here are provided in Supplementary Data Table S9. The first two components of the PCA explain 68·9 % of the total variation (Fig. 9). For some species, a clearly bioclimatically separated niche is obvious: the Irano–Turanian A. parviflora show association with high temperatures and in parallel with a large range of temperatures. Anatolian and eastern Turkish A. macrostyla and A. vulcanica are also adapted to large temperature ranges. Considering these as the most extreme examples, other species do not exhibit a clear and differentiating pattern. However, from this analysis one major pattern has to be mentioned: the Greek and Balkan species (and plastid DNA haplotype groups A, G, C and D) show bioclimatic niche patterns characterized by increased values for precipitation-based variables and decreased temperature variables. This is in contrast to the other and older species groups from Turkey, Iran, Iraq and Lebanon, which show differentiation towards higher temperatures and much wider temperature ranges in addition to severe draught stress.
Fig. 9.
PCA of geographically defined BioClim data. The respective components are shown in the lower left box. Taxa are indicated with colour codes, and respective haplotype groups are also shown.
DISCUSSION
Phylogenetics and systematics
Aubrieta is monophyletic, and we have not detected any of its taxa placed with any other evolutionary group within the Arabideae, a tribe comprising about 508 species (Karl and Koch, 2013). This finding is not trivial because Arabis, as traditionally delimited, is largely poly- and paraphyletic and has not yet been taxonomically resolved. However, the sister species to Aubrieta is Arabis verna, a species that does not group with any other Arabis member. Its taxonomic position is discussed in a later paragraph. All Aubrieta species are perennials (Fig. 10), and they follow an evolutionary pattern demonstrated earlier to be a more general feature of the Arabideae. It is the sister relationship of one or a few widely distributed annual ‘lowland’ species with a corresponding clade consisting of perennial, montane to alpine species that underwent significant diversification and radiation (Karl and Koch, 2013). Aubrieta diversified into 20 species and several subspecies during the Pleistocene (Figs 1 and 2, Table 1), and most species are geographically well defined, as reflected by the maternally inherited chloroplast DNA sequence variation (Figs 2 and 3). ITS DNA sequence data are not very informative in resolving relationships among Aubrieta, but for several species ITS data are very specific (e.g. A. macrostyla, A. parviflora, A. scyria). In this regard they fully agree with cpDNA data combing these three species into one major haplotype group (P-S-M; Table 1) as the oldest clade in the genus. For other taxa, ITS was unable to resolve species relationships. Instead, either multi single-copy gene datasets or genome-wide fingerprints such as amplified fragment length polymorphisms (AFLPs) or restriction site-associated DNA sequencing (ddRAD/RAD-seq) are needed to provide a highly significant phylogenetic perspective.
Fig. 10.
Diversity of Aubrieta species during high flowering.
Maximum likelihood analysis of plastid DNA data shows a congruent result (Fig. 8) and is largely reflective of phylogenetic network analysis. The comparison of a phylogenetic hypothesis based on DNA sequence data with morphology-based phenetic analysis demonstrates that there is very little congruence of phylogenetic structure. This result reflects the situation in which any morphological character used in ‘Arabis’ and related genera (evolutionary and/or morphologically, such as Aubrieta) is homoplastic and often does even not allow the definition of monophyletic groups on genus level (Koch et al., 1999, 2000, 2010, 2012a; Dobes et al., 2004; Kiefer et al., 2009; Jordon-Thaden et al., 2010; Karl et al., 2012; Karl and Koch, 2013, 2014). However, if individual morphological characters are carefully evaluated, species and subspecies can be determined with sufficient reliability (Fig. 7; a key to the species is provided in Supplementary Data File S1).
Most Aubrieta species are diploid, and tetraploids are rarely found. Upon considering the strong biogeographic structure of maternally inherited cpDNA variation, our data might indicate largely allopatric speciation and limited secondary contact species. Sympatry, hybridization and polyploidy are the prevailing modes of speciation and radiation in the closely related Draba from the same tribe, Arabideae (Jordon-Thaden et al., 2010, 2013). However, some incongruence between cpDNA and nuclear ITS is obvious, and this might indicate past and maybe contemporary gene flow between species. One such example is the Anatolian A. vulcanica which carries a cpDNA haplotype from the A. macrostyla group (Fig. 2) but shows a completely unrelated ITS type (Fig. 4). Aubrieta vulcanica is a very local diploid endemic, and this taxon might be considered as the outcome of local introgression between A. macrostyla and A. canescens. Another example is represented by the various accessions of morphologically defined A. canescens subsp. cilicica (Aub064, RGBE009, RGBE010). It seems that this taxon comprises populations that have a very different evolutionary origin. Furthermore, Aub064 from Turkey shows an A. libanotica cpDNA haplotype, but A. libanotica is geographically and morphologically clearly separated. Consequently, the original Turkish region of A. libanotica (considering the maternal gene pool), might be considered as the source from which A. libanotica migrated into Lebanon. This is further substantiated by accession NHM016, with a morphotype resembling features from both A. libanotica and A. canescens. The A. canescens subsp. cilicica accession RGBE009 carries an unusual cpDNA haplotype (xxxA), which is basal to Turkish haplotypes from groups O, L and Pi. By contrast, RGBE010 carries a cpDNA haplotype (Pi05) that is genetically also different from the other haplotypes. Information on syntypes of the last taxon indicate its geographic location as ‘… in montis Kassan Oghlu ad pagum Gorumse, in cedreto locis rupestribus fissura incolit, 5000…’ collected in 1859 and ‘… Cilicia, Bolkar Daglari, Iter Cilicicum in Tauri alpes Bulgar Dagh. Frequens in alpinis Gusguta dictis ad loca rupestria subhumida …’ collected in 1853 by C.G.T. Kotchy. These regions match our accession RGBE010, which for now is considered as the best ‘true’ A. canescens subsp. cilicica. In summary, this subspecies remains with some uncertainties. It should be mentioned here that A. macrostyla has traditionally been considered as a subspecies of A. canescens, but our molecular data clearly indicate that it should be treated as a distinct species Additional uncertain accessions have been documented from Greece. For example, NHM026 is morphologically close to A. deltoidea (originally labelled as A. deltoidea aff. var. microphylla), and NHM037 and NHM041 are morphologically close to A. deltoidea (originally labelled as A. intermedia). Curiously, NHM037 carries an A. albanica cpDNA haplotype. These accessions might also represent hybrids or introgressed populations, but molecular data do not provide further information. Interestingly, the past taxonomic literature recognized only one naturally occurring hybrid in Aubrieta, namely A. hybrida (Haussknecht, 1893). Haussknecht described this Greek taxon from within a population of A. gracilis. Thus, we might have to assume A. gracilis as the maternal genetic source. He also explicitly mentioned A. intermedia as the other parental species. We tried to test this idea and have analysed the holotype of A. hybrida (accession JE0), and A. gracilis (accession JE05) and A. intermedia (accession JE06) from this particular location. The genetic results are summarized in Table 2, and it is obvious that we are unable to verify Haussknecht’s hybrid hypothesis because A. hybrida does not have an identical haplotype with A. intermedia or with A. gracilis, but instead has its own and unique haplotype G01. Since our ITS failed for A. hybrida, we cannot substantiate that hypothesis any further.
Table 2.
Distribution of ITS and cpDNA types among Aubrieta gracilis, A. intermedia and its putative hybrid A. hybrida. DNA type definition follows Figs 2 and 4
| cpDNA haplotype | ITS type | |
|---|---|---|
| A. gracilis | ||
| Aub30-Aub31 | G06, G10 | g01 |
| RGBE003 | G12 | g03 |
| Aub040-047, NHM044/047 | G6, G9, G14 | g00, g01 |
| JE05 | G16 | g07 |
| A. hybrida | ||
| JE0 | G01 | failed |
| A. intermedia | ||
| JE06 | G11 | g00 |
| NHM037 | A05 | g00 |
| NHM041 | G06 | g00 |
Another remarkable taxon is A. olympica. A comprehensive introduction to the historical–taxonomical problem is given by Reichardt (1959), and some aspects are highlighted and discussed here. The species was described by Boissier in 1867 from Uludağ mountain (Bithynian Olympus) in northwest Turkey (syntypes were collected by G. Clementi in 1850 and E. Boissier in 1862). Additional and geographically fully isolated distribution ranges were listed from Paphlagonia (north towards the Black Sea), Cilicia and Cappodacia (Taurus mountains), and there is complete uncertainty about how such accessions from these different regions belong together. Furthermore, it was also unclear which morphotype represents Boissier’s A. olympica, because the original material includes two different morphotypes – one with glabrous and another with pubescent fruit, as mentioned in the protologue (Boissier, 1867). In addition, further collections from that region by experienced botanists, such as Noë (in 1876) and Pichler (in 1874), argued that some of this material represents A. deltoidea and some represents A. intermedia (Reichardt, 1959). However, the occurrence of A. intermedia is unlikely because it is restricted to Greece. Reichardt (1959) concluded that both glabrous and hairy forms represent a single species. More recently, however, glabrous fruit variants could not be found (Daşkin and Kaynak, 2011). We have solved some of these problems in several ways. First, we described A. ekimii as new from a region close to the Uludağ mountain (Yüzbaşıoğlu et al., 2015). Second, sampling of two populations (SYZB4182 and RGBE007) of an unknown taxon also geographically close to that region confirmed that various species co-occur in the region from which A. olympica was originally described. Aubrieta ekimii and another accession, Aub050, carry A. olympica-related cpDNA haplotypes O04 and O03 respectively. The two unknown taxa SYZB4182 and RGBE007 show haplotype Pi02, which is closely related to the O haplotype group. This result points to the conclusion that A. olympica is a diverse taxon. Third, the distribution of A. olympica morphotypes in Paphlagonia is confirmed in our study, with five accessions, and all carry the O01 or O02 cpDNA haplotype. We were unable to identify any accession from Cilicia and Cappodacia with the O haplotype. Instead, another population (SYZB3843) from northwestern Turkey has been identified to be morphologically close to A. olympica and also to carry a haplotype from the O group (O05). Hence, we might consider A. olympica as a polymorphic species with disjunctly and widely distributed populations in north/northwestern Turkey.
Biogeography, centre of origin and centres of diversity
Our molecular data indicate that Greece and Turkey are the main centres of species and genetic diversity. Mismatch distribution analyses infer Turkey as a centre of origin (from the signal of population expansion) and that clade PSM diversified earliest (∼1·11 Mya; Table 1). Because haplotypes M (A. macrostyla) and P (A. parviflora) define the eastward distribution range, and because A. parviflora shows ancestral haplotypes (compared with the other species from the PSM clade), one might conclude that the Anatolian Diagonal Mountains towards the eastern part of South Anatolia have played an important role during the early evolution of perennial Aubrieta. A similar pattern has been described for Arabis alpina (Karl et al., 2012), which grows sympatrically at many places with different Aubrieta species. The Anatolian mountains have recently been shown to be not only important for the genetic diversification of Arabis alpina (Ansell et al., 2011), but this whole species complex and closely related taxa radiated from that region less than 1·88 Mya (Ansell et al., 2011; Karl et al., 2012). Three main routes were characterized: (1) towards the Irano–Turanian region; (2) along the eastern Mediterranean coastal mountain ranges to Lebanon and further south into the Arabian Peninsula and the East African high mountains; and (3) along the southern Turkish coast to the Balkans and whole of Central Europe, thereby also colonizing various Mediterranean islands. The diversification of Anatolian genetic diversity of the A. alpina complex was calculated at ∼0·7 Mya (Ansell et al., 2011). This value fits well with the second pulse of Aubrieta diversification (C clade), estimated at 0·75 Mya, and it may have started from the South Anatolian region, too. This coincides geographically and temporally with the second oldest diversification, the A. columnae clade, the species of which are distributed in mountain systems close to Mediterranean coasts and also in the westernmost ranges of Aubrieta species in Italy and the Balkans.
According to the divergence time estimates (Table 1), none of the species groups has a postglacial origin. Consequently, all of them must have been affected by past Pleistocene climatic fluctuations, and any evolutionary group must have experienced several environmental changes after radiation and migration. As a result, we can recognize the obvious geographic structure of maternally inherited genetic variation. However, there is also a high correlation between species distribution and Mediterranean refugia, as recently introduced by Médail and Diadema (2009). These authors analysed many different plant species based on published phylogeographic studies from 1993 to 2007, and they identified 52 refugia in the Mediterranean bioclimatic region, of which 19 fall in the eastern part. The patterns of 14 out of our 20 analysed Aubrieta taxa match ten of these refugial regions. In addition, most distribution ranges of Aubrieta species overlap with at least one out of the ten Mediterranean regional hotspots of plant biodiversity (Médail and Quézel, 1997; Véla and Benhouhou, 2007). It should be noted that Médail and Diadema (2009) did not include any Aubrieta species in their study, but Arabis alpina was considered. It was concluded that these refuge areas are determined by cumulative effects (historical and environmental factors) during the Tertiary, rather than solely during the last glacial/postglacial period. It was further argued that these refugia represent climatically ‘stable areas’. If we follow these ideas, then not only are the majority of the older Aubrieta lineages adapted to higher temperatures and draught stress (Fig. 9), but this also reflects the signature of the long evolutionary past rather than contemporary or postglacial differentiation and adaptation. The younger diversification of most Greek taxa during the last 200 000 years is reflected in climate niches generally characterized by less draught and lower temperatures compared with Turkish and Irano–Turanian taxa (Fig. 9). It is noteworthy that Turkish and Greek Aubrieta species diversity is floristically separated by the Rechinger line, and that the onset of species radiation in Greece some 200 Kya might be reflective of this very characteristic and pronounced floristic division (Rechinger, 1943; Strid, 1996). Aubrieta scyria is the only exception because it belongs to a Turkish lineage but is endemic to a Greek island. However, the separation of A. scyria from A. parviflora must be very old (Fig. 2), and this split most likely exceeds 200 Kya. Only a few phytogeographic studies provide detailed time scales for floristic division and radiations separated by the Rechinger line. Bardy et al. (2010) estimated the radiation of Veronica on the southernmost Balkan Peninsula to be within the last 400 000 years, and they argue that it was driven by cooling periods during the Last Glacial Maximum, which correlates with our results on Greek Aubrieta. The example from Aegean Nigella (Bittkau and Comes, 2005) provided only a very rough estimate of divergence ‘less than 1 Mya’. A more recent study on the complex history of the olive tree, Olea europaea, also highlighted this division (Besnard et al., 2013), where the deepest split for any Mediterranean lineage (based on cpDNA haplotype networks) was 284 000 years ago, and was set within 139 000 years for the eastern Mediterranean. Phylogeographic analysis on the laurel tree, Laurus nobilis (Rodríguez-Sánchez et al., 2009), also confirms this divide. However, few, much deeper splits among animals dated within the Pliocene have been reported (reviewed in Bilgin, 2011).
Comments on taxonomy and species delimitation
As indicated above, Arabis is polyphyletic (Karl and Koch, 2013). We closely examined Arabis verna as part of this polyphyletic taxonomic complex because it is sister to perennial Aubrieta, which shows a clear phylogenetic distinctness from any other Arabis s. str. clade. This necessitated a taxonomic reassessment of this annual species, which could be treated either as part of a more broadly defined Aubrieta or as a new monospecific genus. We prefer the second option because of substantial morphological distinctness between Arabis verna and Aubrieta. Arabis verna is an annual or ephemeral versus long-lived perennial (Aubrieta), with auriculate (versus non-auriculate) stem leaves, shortened pedicels and styles both as wide as the fruit (versus non-thickened pedicels and elongated styles much narrower than the fruit), toothless and slender (versus toothed or basally dilated) filaments, narrowly linear siliques (versus broadly linear siliques to subglobose silicles) with uniseriate (versus biseriate) seeds. Hence, Arabis verna will be recognized as a new monotypic genus (Koch and German, unpubl. res.). A further taxonomic change is needed for A. canescens subsp. macrostyla (herein treated as A. macrostyla) because we were unable to confirm the relationship of this taxon to A. canescens. Finally, A. deltoidea subsp. sicula should better be classified within A. columnae s.l., a position indicated molecularly and supported geographically and morphologically. For a few other taxa, future population-based phylogeographic analyses might also provide data to change the present-day taxonomy. For example, our present data on A. thessala are insufficient to make any final conclusion because two accessions, Aub008 and NHM051, show cpDNA haplotypes from different lineages (D05 and G04). Unfortunately, molecular analyses failed for other accessions, and we are unable to adequately circumscribe this species. This agrees with the early concern of Boissieu (1896), who described A. thessala as a new species, and suggested that it might be better to follow a broader concept combining A. gracilis, A. intermedia and A. thessala.
Finally, the unique and morphologically and genetically very distinct Turkish accession Aub057 will be introduced as the new species Aubrieta alshehbazii (Dönmez et al., unpubl. res.; treated herein as Aubrieta sp. nov.).
Taxonomic changes
The following taxonomic changes are presented based on our molecular and morphological analyses. A key to the species integrating our detailed information on morphological characters investigated here is presented in File S1.
Aubrieta macrostyla (Hub.-Mor. & Cullen) M.A. Koch, D.A. German & R. Karl, comb. et stat. nov. ≡ Aubrieta canescens subsp. macrostyla Hub.-Mor. & Cullen, Notes Roy. Bot. Gard. Edinburgh 26: 191. 1965. Type: [Turkey] Prov. Maraş: dist. Göksun, Binboğa Dağ in ravine above Yalak, 1500 m, calc. rocks, 14 July 1952, Davis 19939, Dodds & Çetik (holotype E, isotype K). Chromosome number: 2n = 14 [Bot. Garden Munich, collected 1986 by Mertens et Pasche sub No. 85·133, ex cult., Turkey: Prov. Silvas, 1450mNN, HEID918316].
Aubrieta columnaesubsp. sicula (Strobl) M.A. Koch, D.A. German & R. Karl, comb. nov. ≡ Aubrieta deltoidea var. sicula Strobl, Verh. K. K. Zool.-Bot. Ges. Wien 53: 458. 1903. ≡ Aubrieta deltoidea subsp. sicula (Strobl) Phitos, Candollea 25: 76. 1970. Type: not designated yet. Chromosome number: 2n = 14 (material collected at Rocco Novera sui Peloritani sulla Rocca Salvatesta, Sicily [April 2016, 1180 m asl, 37°59′45″ N 15°.08′47″ E, HEID006219]). edna
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of the following. Table S1: detailed information on accessions and GenBank accession codes. Table S2: original results from morphometric analysis. Table S3: codings of the morphological characters. Tables S4, S5, S6 and S7: alignments of plastids trnLF, trnC-ycf6, trnS-ycf9 and the combined plastid dataset, respectively. Table S8: ITS alignment. Table S9: summary of information on the bioclimatic analysis. File S1: morphological key to the species of Aubrieta.
ACKNOWLEDGEMENTS
We thank the gardener team of the Botanical Garden Heidelberg, particularly Torsten Jakob for continuous support with plant cultivation, and Peter Sack, who helped with the voucher exchanges. We are indebted to Marina Roth, Janka Lampert, Lisa Kretz, Anne-Kathrin Schürholz, Stephen Ansell, Nora Hohmann and Gong Wei for their help in the laboratory and data analyses. We also thank the curators of the herbaria of the Botanical Garden and Botanical Museum Berlin (B), the Natural History Museum, London (BM), Royal Botanic Garden, Edinburgh (E), Botanical Garden and Herbarium Heidelberg (HEID), Botanical Museum Lund (LD) and Jena Botanical Garden and Haussknecht Herbarium (JE) for providing voucher material. Plant or seed material was received from various fellow scientists [Christiane Kiefer (MPIPZ Cologne), Birol Mutlu (Inönü University Malatya), Sırrı Yüzbaşıoğlu (Istanbul University) and Ali A. Dönmez (Haceteppe University)]. We also thank Graham Muir, and in particular Ihsan A. Al-Shehbaz for continuous discussions and helpful comments on the manuscript and the identification key. This work was generously supported by DFG research grants (KO2302 11/1, 12/1 and 13/1 to M.A.K.) and by an EU-SYNTHESIS grant (GB-TAF-849 to M.A.K.).
LITERATURE CITED
- Akeroyd JR, Ball PW. 1993. Aubrieta Adanson In: Tutin TG, Burges NA, Chater AO, et al. , eds. Flora Europaea, 2nd edn Cambridge: Cambridge University Press, 356–358. [Google Scholar]
- Al-Shehbaz IA. 2010. Brassicaceae Burnett In: Flora of North America Editorial Committee, eds. Flora of North America north of Mexico, Vol. 7 New York, 225, 231–234. [Google Scholar]
- Al-Shehbaz IA. 2012. A generic and tribal synopsis of the Brassicaceae (Cruciferae). Taxon 61: 931–954. [Google Scholar]
- Al-Shehbaz IA, Beilstein MA, Kellogg EA. 2006. Systematics and phylogeny of the Brassicaceae (Cruciferae): an overview. Plant Systematics and Evolution 259: 89–120. [Google Scholar]
- Al-Shehbaz IA, Mutlu B, Dönmez AA. 2007. The Brassicaceae (Cruciferae) of Turkey, updated. Turkish Journal of Botany 31: 327–336. [Google Scholar]
- Al-Shehbaz IA, German DA, Karl R, Jordon-Thaden I, Koch MA. 2011. Nomenclatural adjustments in the tribe Arabideae (Brassicaceae). Plant Diversity and Evolution 129: 71–76. [Google Scholar]
- Ančev M, Goranova V. 2009. Aubrieta (Brassicaceae) in the Bulgarian flora. Phytologia Balcanica 15: 43–50. [Google Scholar]
- Ansell SW, Stenoien HK, Grundmann M, et al. 2011. The importance of Anatolian mountains as the cradle of global diversity in Arabis alpina L., a key arctic — alpine species. Annals of Botany 108: 241–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel O, Al-Shehbaz IA. 2002. Cruciferae In: Kubitzki K, Bayer C, eds. The families and genera of vascular plants, Vol. 5 Heidelberg: Springer, 75–174. [Google Scholar]
- Bardy KE, Albach DC, Schneeweiss GM, Fischer MA, Schönswetter P. 2010. Disentangling phylogeography, polyploid evolution and taxonomy of a woodland herb (Veronica chamaedrys group, Plantaginaceae s.l.) in southeastern Europe. Molecular Phylogenetics and Evolution 57: 771–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard G, Khadari B, Navascués M, et al. 2013. The complex history of the olive tree: from Late Quaternary diversification of Mediterranean lineages to primary domestication on the northern Levant. Proceedings of the Royal Society B 280: 20122833 http://dx.doi.org/10.1098/rspb.2012.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilgin R. 2011. Back to the suture: the distribution of intraspecific genetic diversity in and around Anatolia. International Journal of Molecular Sciences 12: 4080–4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittkau C, Comes P. 2005. Evolutionary processes in a continental island system: molecular phylogeny of the Aegean Nigella arvensis alliance (Ranunculaceae) inferred from chloroplast DNA. Molecular Ecology 14: 4065–4083. [DOI] [PubMed] [Google Scholar]
- Boissier E. 1867. Flora orientalis, Vol. 1 Basel: H. Georg. [Google Scholar]
- Boissieu H. 1896. Quelques notes sur la flore d’Orient. Bulletin de la Société Botanique de France 43: 283–291. [Google Scholar]
- Clement M, Posada D, Crandall KA. 2000. TCS: a computer program to estimate gene genealogies. Molecular Ecology 9: 1657–1659. [DOI] [PubMed] [Google Scholar]
- Cullen J. 1965. Aubrieta Adanson In: Davis PH, ed. Flora of Turkey, Vol. 1 Edinburgh: Edinburgh University Press, 444–447. [Google Scholar]
- Daşkin R, Kaynak G. 2011. Conservation status of five endemic species distributed in Northwest Tureky. Phytologia Balcanica 17: 213–219. [Google Scholar]
- Davis PH. 1965. Flora of Turkey and the East Aegean Islands, Vol. 1 Edinburgh: Edinburgh University Press. [Google Scholar]
- Davis PH, Mill RR, Tan K. 1988. Flora of Turkey and the East Aegean Islands (Supplement), Vol. 10 Edinburgh: Edinburgh University Press. [Google Scholar]
- Dobes C, Mitchell-Olds T, Koch M. 2004. Intraspecific diversification in North American Arabis drummondii, A. ×divaricarpa, and A. holboellii (Brassicaceae) inferred from nuclear and chloroplast molecular markers – an integrative approach. American Journal of Botany 91: 2087–2101. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochemical Bulletin 19: 11–15. [Google Scholar]
- Excoffier L, Laval G, Schneider S. 2005. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evolutionary Bioinformatics 1: 47–50. [PMC free article] [PubMed] [Google Scholar]
- Field A. 2013. Discovering statistics using IBM SPSS statistics, 4th edn London: Sage Publications. [Google Scholar]
- Franzke A, German D, Al-Shehbaz IA, Mummenhoff K. 2009. Arabidopsis family ties: molecular phylogeny and age estimates in Brassicaceae. Taxon 58: 425–437. [Google Scholar]
- Franzke A, Lysak MA, Al-Shehbaz IA, Koch MA, Mummenhoff K. 2011. Cabbage family affairs: the evolutionary history of Brassicaceae. Trends in Plant Science 16: 108–116. [DOI] [PubMed] [Google Scholar]
- Fu YX. 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147: 915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gower JC. 1971. A general coefficient of similarity and some of its properties. Biometrics 27: 857‒871. [Google Scholar]
- Greuter W, Burdet HM, Long G. 1986. Med-checklist, Vol. 3 Geneva: Conservatoire et Jardin botaniques de la Ville de Genève, 57‒58. [Google Scholar]
- Güner A, Özhatay N, Ekim T, Başer KHC. 2000. Flora of Turkey and the East Aegean Islands (Supplement 2). Edinburgh: Edinburgh University Press. [Google Scholar]
- Gustavsson LA. 1986. Aubrieta Adanson In: Strid A, ed. Mountain Flora of Greece, Vol. 1 Cambridge: Cambridge University Press, 268‒274. [Google Scholar]
- Harpending RC. 1994. Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Human Biology 66: 591–600. [PubMed] [Google Scholar]
- Haussknecht C. 1893. Symbolae ad floram graecam. Aufzählung der im Sommer 1885 in Griechenland gesammelten Pflanzen. Mittheilungen des Thüringischen Botanischen Vereines 3–4: 96–116. [Google Scholar]
- Hedge I. 1968. Aubrieta Adanson In: Rechinger KH. ed. Flora Iranica, Vol. 1 Graz: Akademische Druck und Verlagsanstalt, 201–210. [Google Scholar]
- Hochberg Y. 1974. Some generalizations of the T-method in simultaneous inference. Journal of Multivariate Analysis 4: 224–234. [Google Scholar]
- Hohmann N, Wolf EM, Lysak M, Koch MA. 2015. A time-calibrated road map of Brassicaceae species radiation and evolutionary history. Plant Cell 27: 2770–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalas J, Suominen J. 1994. Atlas florae Europaeae, Vol. 10. Cruciferae (Sisymbrium to Aubrieta). Helsinki: Committee for Mapping the Flora of Europe and Societas Biologica Fennica Vanamo, 212–215. [Google Scholar]
- Jordon-Thaden I, Hase I, Al-Shehbaz IA, Koch MA. 2010. Molecular phylogeny and systematics of the genus Draba (Brassicaceae) and identification of its most closely related genera. Molecular Phylogenetics and Evolution 55: 524–540. [DOI] [PubMed] [Google Scholar]
- Jordon-Thaden I, Al-Shehbaz IA, Koch MA. 2013. Species richness of a globally distributed, arctic-alpine genus, Draba L. (Brassicaceae). Alpine Botany 123: 97–106. [Google Scholar]
- Karl R, Koch MA. 2013. A world-wide perspective on crucifer speciation and evolution: phylogeny, biogeography and trait evolution in tribe Arabideae. Annals of Botany 112: 983–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl R, Koch MA. 2014. Phylogenetic signatures of adaptation: the Arabis hirsuta species aggregate (Brassicaceae) revisited. Perspectives in Plant Ecology, Evolution and Systematics 16: 247–264. [Google Scholar]
- Karl R, Kiefer C, Ansell S, Koch MA. 2012. Systematics and evolution of arctic-alpine Arabis alpina L. (Brassicaceae) and its closest relatives in the eastern Mediterranean. American Journal of Botany 99: 778–794. [DOI] [PubMed] [Google Scholar]
- Kiefer C, Dobes C, Sharbel T, Koch MA. 2009. Phylogeographic structure of the chloroplast DNA gene pool in North American Boechera – a genus and continental-wide perspective. Molecular Phylogenetics and Evolution 52: 303–311. [DOI] [PubMed] [Google Scholar]
- Kiefer M, Schmickl R, German DA. et al. 2014. BrassiBase: introduction to a novel knowledge database on Brassicaceae evolution. Plant Cell and Physiology 55: e3. [DOI] [PubMed] [Google Scholar]
- Koch M, Bishop J, Mitchell-Olds T. 1999. Molecular systematics and evolution of Arabidopsis and Arabis. Plant Biology 1: 529–537. [Google Scholar]
- Koch M, Haubold B, Mitchell-Olds T. 2000. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis and related genera. Molecular Biology and Evolution 17: 1483–1498. [DOI] [PubMed] [Google Scholar]
- Koch M, Haubold B, Mitchell-Olds T. 2001. Molecular systematics of the Cruciferae: evidence from coding plastome matK and nuclear CHS sequences. American Journal of Botany 88: 534–544. [PubMed] [Google Scholar]
- Koch M, Dobeš C, Matschinger M, Bleeker W, Vogel J, Kiefer M. 2005. Evolution of the plastidic trnF(GAA) gene in Arabidopsis relatives and the Brassicaceae family: monophyletic origin and subsequent diversification of a plastidic pseudogene. Molecular Biology and Evolution 22: 1032–1043. [DOI] [PubMed] [Google Scholar]
- Koch MA, Dobeš C, Kiefer C, Schmickl R, Klimeš L, Lysak MA. 2007. SuperNetwork identifies multiple events of plastid trnF (GAA) pseudogene evolution in the Brassicaceae. Molecular Biology and Evolution 24: 63–73. [DOI] [PubMed] [Google Scholar]
- Koch MA, Karl R, Kiefer C, Al-Shehbaz IA. 2010. Colonizing the American continent – systematics of the genus Arabis in North America (Brassicaceae). American Journal of Botany 97: 1040–1057. [DOI] [PubMed] [Google Scholar]
- Koch MA, Karl R, German DA, Al-Shehbaz IA. 2012a. Systematics, taxonomy and biogeography of three new Asian genera from the Brassicaceae, tribe Arabideae: an ancient distribution circle around the Asian high mountains. Taxon 61: 955–969. [Google Scholar]
- Koch MA, Kiefer M, German DA, et al. 2012b. BrassiBase: tools and biological resources to study characters and traits in the Brassicaceae – version 1.1. Taxon 61: 1001–1009. [Google Scholar]
- Koch MA, Scheriau C, Betzin A, Hohmann N, Sharbel TF. 2013. Evolution of cryptic gene pools in Hypericum perforatum: the influence of reproductive system and gene flow. Annals of Botany 111: 1083–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattfield J. 1937. The species of the genus Aubrieta Adanson. Bulletin of the Alpine Garden Society of Great Britain 7: 157–181, 217–227. [Google Scholar]
- Médail F, Diadema K. 2009. Glacial refugia influence plant diversity patterns in the Mediterranean Basin. Journal of Biogeography 36: 1333–1345. [Google Scholar]
- Médail F, Quézel P. 1997. Hot spot analysis for conservation of plant biodiversity in the Mediterranean Basin. Annals of the Missouri Botanical Garden 84: 112–127. [Google Scholar]
- Meyer FK. 2011. Beiträge zur Flora Albanien. Haussknechtia Beiheft 15: 1–220. [Google Scholar]
- Möller M, Gao LM, Mill RR, Li DZ, Hollingworth ML, Gibby M. 2007. Morphometric analysis of the Taxus wallichiana-complex based on herbarium material. Botanical Journal of the Linnean Society 155: 307–335. [Google Scholar]
- Mouterde P.. 1986. Aubrieta Adans. In: Mouterde P, ed. Nouvelle Flore du Liban et du la Syrie. Vol 2, text. Beyrouth: Dar El-Machreq Éditeurs, 140.
- Mutlu B. 2012. Aubrieta Adans In: Güner A, Aslan S, Ekim T, Vural M, Babaç MT, eds. Türkiye bitkileri listesi (damarlı bitkiler). Istanbul: Nezahat Gökyigit Botanik Bahçesi ve Flora Arastırmaları Dernegi Yayımı, 258–259. [Google Scholar]
- Peşmen H, Güner A. 1978. Four new taxa from Anatolia. Notes from the Royal Botanic Garden, Edinburgh 36: 35. [Google Scholar]
- Phitos D. 1970. Die Gattung Aubrieta in Griechenland. Candollea 25: 69–87. [Google Scholar]
- Phitos D. 2002. Aubrieta Adans In: Strid A, Tan K, eds. Flora Hellenica, Vol. 2 Ruggell: AR Gantner, 192‒195. [Google Scholar]
- Pollunin O. 1987. Flowers of Greece and the Balkans, 2nd edn Oxford: Oxford University Press. [Google Scholar]
- Posada D, Crandall KA. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14: 817–818. [DOI] [PubMed] [Google Scholar]
- Rechinger KH. 1943. Flora Aegea. Flora der Inseln und Halbinseln des ägäischen Meeres Denkschriften, Band 105, Halbband 1. Vienna: Akademie der Wissenschaften Wien, Mathematisch-Naturwissenschaftliche Klasse.
- Reichardt J. 1959. Kritisch-systematische Revision der Gattung Aubrieta Adanson. Diploma thesis, Jena University, Germany.
- Rodríguez-Sánchez F, Guzmán B, Valido A, Vargas P, Arroyo J. 2009. Late Neogene history of the laurel tree (Laurus L., Lauraceae) based on phylogeographical analyses of Mediterranean and Macaronesian populations. Journal of Biogeography 36: 1270–1281. [Google Scholar]
- Rogers A. 1995. Genetic evidence for a Pleistocene population explosion. Evolution 49: 608–618. [DOI] [PubMed] [Google Scholar]
- Rogers AR, Harpending H. 1992. Population growth makes waves in the distribution of pairwise genetic differences. Molecular Biology and Evolution 9: 552–569. [DOI] [PubMed] [Google Scholar]
- Schneider S, Excoffier L. 1999. Estimation of demographic parameters from the distribution of pairwise differences when the mutation rates vary among sites: application to human mitochondrial DNA. Genetics 152: 1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestro D, Michalak I. 2012. raxmlGUI: a graphical front-end for RAxML. Organisms, Diversity and Evolution 12: 335–337. [Google Scholar]
- Simmons MP, Ochoterena H. 2000. Gaps as characters in sequence-based phylogenetic analyses. Systematic Biology 49: 369–381. [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- Strid A. 1996. Phytogeographia Aegaea and the Flora Hellenica Database. Annalen des Naturhistorischen Museums Wien 98B: 279–289. [Google Scholar]
- Strid A, Tan K, eds. 2002. Cruciferae In: Flora Hellenica, Vol. 2 Ruggell: AR Gantner, 116–297. [Google Scholar]
- Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend CC. 1980. Aubrieta Adans In: Townsend CC, Guest E, eds. Flora of Iraq, Vol. 4, Bignoniaceae to Resedaceae. Baghdad: Ministry of Agriculture, 1014–1015. [Google Scholar]
- Véla E Benhouhou S.. 2007. Evaluation d'un nouveau point-chaud de biodiversité végétale dans le bassin méditerranéen (Afrique du nord). Comptes Rendus Biologies 330: 589–605. [DOI] [PubMed] [Google Scholar]
- Warwick SI, Al-Shehbaz IA. 2006. Brassicaceae: chromosome number index and database on CD-Rom. Plant Systematics and Evolution 259: 237–248. [Google Scholar]
- Warwick SI, Mummenhoff K, Sauder C, Koch MA, Al-Shehbaz IA. 2010. Closing the gaps: Phylogenetic relationships in the Brassicaceae based on DNA sequence data of nuclear ribosomal ITS region. Plant Systematics and Evolution 285: 209–232. [Google Scholar]
- Yüzbaşıoğlu S, Koch MA, Al-Shehbaz IA. 2015. Proof of a knowledge database concept. Aubrieta ekimii (Brassicaceae), a new species from NW Anatolia (Turkey): morphological and molecular support. Plant Systematics and Evolution 301: 2043–2055. [Google Scholar]
- Zwick WR, Velicer WF, 1986. Comparison of five rules for determining the number of components to retain. Psychological Bulletin 99: 432–442. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.