Abstract
Primary keratinocytes immortalized by human papillomaviruses (HPVs), along with HPV-induced cervical carcinoma cell lines, are excellent models for investigating neoplastic progression to cancer. By simultaneously visualizing viral DNA and nascent viral transcripts in interphase nuclei, we demonstrated for the first time a selection for a single dominant papillomavirus transcription center or domain (PVTD) independent of integrated viral DNA copy numbers or loci. The PVTD did not associate with several known subnuclear addresses but was almost always perinucleolar. Silent copies of the viral genome were activated by growth in the DNA methylation inhibitor 5-azacytidine. HPV-immortalized keratinocytes supertransduced with HPV oncogenes and selected for marker gene coexpression underwent crisis, and the surviving cells transcribed only the newly introduced genes. Thus, transcriptional selection in response to environmental changes is a dynamic process to achieve optimal gene expression for cell survival. This phenomenon may be critical in clonal selection during carcinogenesis. Examination of HPV-associated cancers supports this hypothesis.
Cellular responses to growth stimuli or stress quickly result in adaptive chromatin remodeling, leading to alterations in gene expression. Modulations made to optimize cell survival can result in the dysregulation of genes governing cell growth and differentiation and in genomic instability (28), such as those associated with neoplastic progression. In cancers in which cellular proto-oncogene amplification has occurred, higher than normal levels of proto-oncoprotein are observed. Whether this is a result of transcription from all amplified gene copies or from a single locus with increased promoter strength or, alternatively, of mRNA stabilization remains to be elucidated. A superb system for studying genetic and transcriptional alternations associated with neoplastic progression can be found in early and late passages of primary keratinocytes immortalized by high-risk human papillomavirus (HPV) genotypes, in HPV-induced cervical cancers, and in cell lines derived therefrom (73). In such cells, substantial changes have occurred in cellular and viral gene expression relative to that in productively infected, benign condylomata and papillomas. In the present study, we used in situ analytic methods to visualize HPV oncogene expression in individual cells of both experimental model and naturally arising neoplasms and observed striking and unexpected changes accompanying progression.
The molecular basis for HPV oncogenesis is predicated on the mechanisms by which these normally benign viruses reproduce themselves. In productive infections, the 8-kb, double-stranded, circular viral DNAs amplify as extrachromosomal nuclear plasmids, and this amplification is restricted to postmitotic differentiated cells of the squamous epithelium. DNA synthesis requires the E2 origin recognition protein and the E1 viral helicase protein, as well as the reactivated host replication machinery. Thus, HPVs encode E6 and E7 oncoproteins that bind to and inactivate the tumor suppressor proteins p53 and the retinoblastoma family of tumor suppressor proteins, respectively, to bypass cell cycle controls (37, 38). Typically, there is little or no viral E6 or E7 oncoprotein gene expression in basal cells due to negative controls exerted on the enhancer-promoter by cellular factors and possibly by the viral E2 protein, which is also a transcription factor (10). As with cellular genes (27), chromatin remodeling by posttranslational modifications of histones has also been implicated in viral gene regulation (15, 72).
In contrast to the noncycling, differentiated keratinocytes comprising the suprabasal strata of benign productive HPV lesions, much or all of the epithelium in high-grade dysplasias and carcinomas consists of basal cell-like proliferating cells in which viral DNA is often integrated and viral E6 and E7 oncogenes are invariably highly expressed (60, 73). These neoplastic changes are probably brought about by inopportune and prolonged expression of the oncogenic HPV E6 and E7 proteins in the normally rather quiescent reserve or stem cells of the epithelia during repeated wounding and healing (10). Consequently, excessive cell proliferation, accumulation of mutations in key host regulatory genes, and chromosome instability occur in these cells (37, 38). Cell lines such as SiHa, CaSki, and HeLa were established from HPV-induced cervical cancers, and they contain integrated HPV DNA from which the continuous expression of the viral oncogenes is critical for the maintenance of cell growth and the transformed phenotypes (3, 14, 33, 41, 48, 50, 67, 70).
Viral DNA integration invariably occurs downstream of the E6/E7 genes, often in the E1 or E2 gene region accompanied by a deletion of downstream sequences, resulting in a loss of feedback control of oncogene expression by the E2 protein. Moreover, transcripts of the E6/E7 genes capture 3′ cellular untranslated sequences and polyadenylation signals (26, 48, 53, 68), and these fusion transcripts are more stable than those derived from extrachromosomal viral DNA. This increase in viral mRNA stability and the resulting elevated levels of viral oncoproteins are considered to be important factors in neoplastic progression (23).
HPV-16 and HPV-18 oncogenes can immortalize primary human keratinocytes in vitro (73). The immortal cells contain multiple integrated HPV DNA, express telomerase and are often hyperdiploid, with a gain or loss of partial or entire chromosomes (36, 54, 57, 59). Early-passage immortal cells are typically not tumorigenic in nude mice, whereas cells at higher passages exhibit more dysplastic histology when grown as organotypic raft cultures, simulating the histopathological characteristics of low-, moderate-, and high-grade dysplasias (21, 58). Eventually, tumorigenic clones may emerge after long-term culturing, presumably due to additional mutations in cellular genes incurred during passage (73). Along with cervical carcinoma cell lines, these immortalized cells serve as experimental models with which to investigate chromosomal changes that occur in patient lesions during the progression to dysplasias and cancers. However, the regulation of viral transcription in these immortalized cells or cancers had not been examined in detail.
In the present study, we used in situ hybridization in conjunction with tyramide-fluorescence enhanced signal deposition (T-FISH) (1, 4, 65) to visualize viral RNA transcription foci in HPV DNA-immortalized human keratinocytes. We also identified chromosomes that harbored active centers in CaSki and SiHa cells by simultaneous detection of nascent viral RNA, integrated viral DNA, or chromosomal territories. Both CaSki and SiHa cells are hyperdiploid. CaSki cells harbor ca. 600 copies of HPV-16 DNA distributed over 11 or more chromosomal sites, mainly in tandem arrays. The complex chromosomal rearrangements and the large number of sites of integration have up till now prevented cytogenetic mapping of the integrated viral DNAs. SiHa cells, in contrast, possess only two copies of HPV DNA, one each in the two copies of chromosome 13 at locus 13q21 (1, 3, 17, 33). Our results demonstrate that, regardless of the copy number of integrated viral DNA or number of distinct integration sites, natural selection favors cells containing only one or at most two transcriptionally active viral DNA center(s), which was preferentially localized to the perinucleolar region. All other integrated viral DNA copies become transcriptionally inactive. This perinucleolar domain, designated the papillomavirus transcriptional domain (PVTD), is distinct from other major nuclear domains commonly studied. Some of the silent copies can be reactivated by growth for one to two cell divisions in medium containing 5-azacytidine, implicating DNA methylation in the silencing. Conversely, the active copy can be silenced under challenge for cell survival. Of particular medical relevance, examination of HPV-associated primary cancers also revealed one nascent viral transcription center per cell. Collectively, these observations indicate that the selection for the fittest cells via transcriptional silencing is a dynamic and reversible process. The implications of this process of transcriptional modulation during oncogenesis are discussed.
MATERIALS AND METHODS
Cell and organotypic cultures.
CaSki and SiHa cells were obtained from the American Type Culture Collection (ATCC; Manassas, Va.) and were cultured in Dulbecco modified Eagle medium with 10% fetal bovine serum at 37°C in 5% CO2. All immortalized keratinocyte cell lines were passaged in keratinocyte-specific SFM (Invitrogen, Carlsbad, Calif.). Raft cultures were prepared as described previously (8, 40).
Nucleic acid hybridization probes and probe labeling.
HPV genomic DNA probes for in situ hybridization were prepared by nick translation of genomic plasmid DNAs in the presence of biotinylated dUTP. The probes were precipitated in the presence of 0.4 μg of Cot-1 DNA, 0.20 μg of yeast tRNA, and 0.6 μg of salmon sperm DNA/μl. The probe cocktail was prepared in Hybridsol VII (Ventana Medical Systems, Tucson, Ariz.) as described previously (65). HPV genomic DNA probes for Southern and Northern blot hybridization were prepared by nick translation in the presence of [α-32P]dCTP (Perkin-Elmer Life Sciences, Boston, Mass.).
In situ detection of nascent viral transcripts, viral DNA, chromosomal DNA, or protein.
Simultaneous detection of nascent viral RNAs and viral DNA or chromosomal domains was performed essentially as described previously (24, 35, 69), with modifications pertaining to the use of tyramide signal amplification (4). Hybridization to nascent RNA transcripts was performed without prior denaturation of nucleic acids. Briefly, cells were permeabilized in CSK solution (19) containing 0.5% Triton X-100 detergent for 3 min at 4°C, fixed with 4% paraformaldehyde-1 mM MgCl2 (pH 7.2) for 10 min, dehydrated in ethanol, and hybridized with probes. After overnight incubation with probes in Hybrizol VII at 37°C, slides were washed extensively and then incubated with a 1:100 dilution of streptavidin-horseradish peroxidase (HRP) conjugate in 4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at 37°C for 1 h. After a washing step, signals were revealed with a tyramide-fluorophor (1:100 dilution in Amplification Diluent; Perkin-Elmer). First-round HRPs were inactivated by 3% hydrogen peroxide in 4× SSC at room temperature prior to further probing.
To detect viral DNA or for interphase chromosome paints, slides were treated with 100 μg of RNase A (Sigma)/ml in 2× SSC at 37°C, dehydrated in ethanol, denatured in 70% formamide-2× SSC (pH 7.0) at 72°C, dehydrated in ethanol, and hybridized with probes. After overnight incubation at 37°C, slides were treated as described for nascent RNA detection. For simultaneous detection of viral DNA and chromosome paints, viral DNA was probed first. Direct fluorescein isothiocyanate (FITC)-conjugated chromosomal paints from Vysis (Downers Grove, Ill.) were detected by using the manufacturer's protocol. Digoxigenin paint probes from Oncor (Gaithersburg, Md.) were detected by using either an anti-digoxigenin (DIG)-FITC or an anti-DIG-Texas red antibody (Roche, Indianapolis, Ind.). Cells were stained with DAPI (4′,6′-diamidino-2-phenylindole; Sigma) at 0.05 mg/ml and mounted in Antifade.
Antibodies used in the present study were: SC35 (Sigma, St. Louis, Mo.); PML (PG-M3), C23 (MS-3; Santa Cruz Biotechnology, Santa Cruz, Calif.), and p220npat (a gift from J. Wade Harper) (31). For C23 antigen detection (Sigma), slides were incubated with a 1:100 dilution of primary antibody in 50% goat serum, 1× phosphate-buffered saline at 37°C, washed, and then incubated again with a 1:100 dilution of a goat anti-mouse-TRITC (tetramethyl rhodamine isothiocyanate) antibody. For the other antigens, Antigen Retrieval (Dako, Carpinteria, Calif.) was performed prior to probing with 1:100 dilution of primary antibodies. After a washing step, a secondary goat anti-rabbit or anti-mouse-HRP antibody was then applied for 1 h at 37°C (Roche). The slides were washed, developed by using a 1:100 dilution of fluorescence-tyramide (Perkin-Elmer), stained with DAPI, and mounted with Antifade.
HPV typing of paraffin-embedded patient biopsy specimens was conducted on DNA recovered from tissue sections by using nested PCR amplification, followed by diagnostic restriction enzyme digestions. After deparaffinization, 4-μm sections were treated with a proteinase K solution (Dako) for 6 min, ethanol dehydrated, and probed for viral RNA and then reprobed for viral DNA with nick-translated probes.
Southern blot analysis, Northern blot analysis, and 3′ RACE.
A total of 7 μg of total cellular DNA from FK16A and FK18B cells at different passages were digested with PstI and separated in a 0.8% agarose gel. Southern blot hybridization was performed with 32P-labeled HPV-16 genomic DNA probes. For Northern blot analysis, total RNA was isolated by using TRIzol reagent (Invitrogen) from cell lines. Total RNA were also isolated from CaSki cells with or without prior treatment with 10 μM 5-azacytidine (Sigma) at 37°C for 40 to 48 h. RNA was digested with RNase-free DNase I before being separated in a 1% formaldehyde gel, transferred to nitrocellulose membranes, and probed. The blots were analyzed with a Molecular Dynamics PhosphorImager. 3′ RACE (rapid amplification of cDNA ends) analysis of HPV RNAs from CaSki cells was performed as described previously (68). Five cDNA clones were randomly selected and sequenced by the UAB Sequencing Core Facility.
Lentiviral vectors and transduction.
An human immunodeficiency virus (HIV)-based vector was used to introduce genes expressing either enhanced green fluorescent protein (GFP)-internal ribosome entry site (IRES)-puro or HPV-18 E6/E7-IRES-puro into the FK18B cell line. The vector was similar to that described (7, 66). The eGFP or E6/E7 coding sequence was inserted 5′ of the IRES, and each was under control of the early cytomegalovirus promoter. Infectious stocks of vectors were prepared by transfection of 293T cells as described previously (39) and used at a multiplicity of infection of 1 or 10 to transduce FK18B cells at passage 109.
Light microscopy.
Slides were viewed with an Olympus AX70 microscope equipped with a Speicher Filter Set (ChromaTechnologies, Brattleboro, Vt.). Photographs were taken by using a Zeiss Axiocam and Zeiss software. Quantification was conducted on at least 200 cells per event.
RESULTS
Detection of HPV DNA by T-FISH.
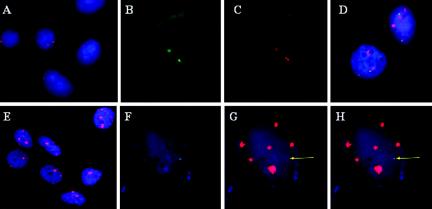
We first determined the sensitivity, specificity and resolution of tyramide-fluorescence in situ hybridization (T-FISH) for viral DNA localization in permeabilized intact interphase cells. Using genomic viral DNA probes, we routinely observed two small signal centers in SiHa cells (Fig. 1A, Cy3, red). In CaSki cells, we detected multiple foci distributed into many distinct big or small centers (Fig. 1E, Cy3, red). We surmise that hybridization signal size and intensity of deposited labeled tyramide reflect viral DNA copy number, with the most intense signals marking the longest viral DNA tandem arrays. Because the signal amplification is an enzyme-driven reaction, a locus with a high-copy-number target generates a diffuse signal that is spatially larger than the actual target size (4).
FIG. 1.
Detection of viral DNA and nascent transcription centers in CaSki and SiHa cervical cancer cell lines. (A and E) Detection of HPV-16 DNA (Cy3; red) in SiHa cells (A) or CaSki cells (E) with nick-translated probes in permeabilized interphase cells. (B and C) Simultaneous in situ detection in a single interphase SiHa cell of nascent HPV-16 RNA (FITC) (B) and HPV-16 DNA (Cy3) (C) by sequential hybridization with a nick-translated genomic probe. (D) Interphase chromosome analysis of SiHa cells for nascent HPV transcripts (FITC) and the chromosome 13 territory (Cy3). (F to H) Simultaneous in situ detection in a single interphase CaSki cell of nascent HPV-16 RNA (FITC) (F) and viral DNA (Cy3) (G) by sequential hybridization with a nick-translated genomic probe. (H) Merged image of panels F and G. Images were captured with a ×40 (A and E) or ×100 (B to D and F to H) objective lens but enlarged to different extent to display several cells (A and E) to show reproducibility or for a single cell for clarity (B to D and F to H). In all panels, nuclear DNA was stained with DAPI (blue).
Coincidence of nascent transcription centers with both HPV DNA loci in SiHa cells.
To determine whether both copies of HPV-16 DNA in SiHa cells are transcriptionally active, we probed for nascent viral RNA. Nuclear and cytoplasmic transcripts that were released from the templates were removed as described in Materials and Methods. Nascent HPV-16 nuclear RNAs were first revealed with a biotinylated nick translated DNA probe (Fig. 1B, FITC, green). After RNase treatment and denaturation, viral DNA was again hybridized with a biotinylated DNA probe, which was then revealed with a different fluor-conjugated tyramide (Fig. 1C, Cy3, red). Two nascent viral RNA transcript foci were visualized, overlapping the viral DNA signals. The cell illustrated in Fig. 1B and C was in late S or G2 phase, since paired signals from one of the two pairs of sister chromatids were clearly visible, whereas the other pair could have been oriented in such a way (above and below one another) that the signals appeared as one. As controls, the nascent RNA foci were RNase sensitive. Using a riboprobe specific for E6 and E7, we demonstrated that both centers are producing E6 and E7 transcripts (data not shown). In another control, viral DNA was not detected when cells were not treated with denaturing conditions (data not shown). Rarely did we observe additional nuclear or cytoplasmic viral RNA. Probing with a nick-translated biotinylated probe specific for HIV contained in a plasmid of similar backbone and size demonstrated no detectable signal (data not shown). Collectively, these results confirm that the signals were indeed nascent transcripts. Finally, we probed for nascent viral RNA and for chromosome 13 with specific paints in interphase cells. Both viral transcription centers (Fig. 1D, green) were observed to associate spatially with chromosome 13 territories (Fig. 1D, red). These results agree with the previous report that SiHa contains two partially deleted, integrated HPV-16 genomes at genetically similar if not identical loci on chromosome 13 (1, 3, 17, 33). We now demonstrate that both copies are transcriptionally active.
One nascent HPV transcription center in CaSki cells.
DNA probes were used to detect transcriptional centers in CaSki cells. Unexpectedly, of the several hundred copies of integrated viral DNA, only a single nuclear RNA hybridization signal was typically observed (Fig. 1F, green). This center consistently colocalized with a low-intensity viral DNA focus (Fig. 1G, red) but not with the much larger tandem arrays of viral DNA. A merged image is shown in Fig. 1H. This nascent RNA center was RNase sensitive and was not observed with HIV probes (data not shown).
Identification of small viral integration centers in the CaSki cells.
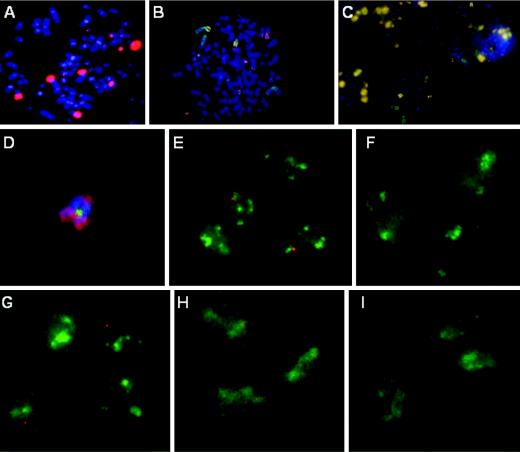
To determine whether the single transcriptionally active, small DNA center in CaSki cells was clonal, or one that differed among individual cells, we proceeded to identify chromosomes that harbor low copy numbers of HPV DNA by using T-FISH on metaphase chromosomes. We found that CaSki cells contained between 75 and 78 chromosomes, and we detected between 12 and 16 distinct integration sites (Fig. 2A, red). On the basis of their fluorescence probe signals, most loci contained multiple copies of viral DNA, and only three chromosomes contained a low copy number of viral DNA.
FIG. 2.
Mapping of the HPV transcription center in CaSki cells. (A) In situ hybridization to detect integrated HPV DNA (Cy3) in metaphase chromosomes (DAPI) in a CaSki cell. (B and C) Localization of HPV-16 DNA in CaSki cells by dual hybridization with a HPV-16 DNA probe (Cy3) and individual chromosome paints (FITC) on metaphase chromosome spreads (DAPI). A high-copy-number of HPV-16 DNA was found on two of the four major chromosomes 2 (green chromosomes with a yellowish arm in panel B). Chromosome 9 harbors no HPV DNA (C). (D) One or a few copies of HPV-16 DNA (FITC) were found on one of three acrocentric chromosomes 14 (Cy3). The paint did not color the entire metaphase chromosome (DAPI). (E to I) Interphase chromosome analysis of CaSki cells for nascent HPV transcription (Cy3) and chromosomal territory domains (FITC) showing chromosome 14 (E), chromosome X (F), chromosome 13 (G), chromosome 6 (H), and chromosome 22 (I). All images were captured with a ×100 objective lens but were enlarged to different extents for clarity.
Sequential hybridization for HPV-16 DNA (in red) and then with individual chromosome paints (in green) on metaphase chromosomes revealed that chromosome 2 or chromosome 2 derivatives (Fig. 2B) contained a high HPV copy number, whereas chromosome 9 (Fig. 2C) and chromosome 13 (data not shown) are devoid of integrated HPV. Low-copy HPV integration was observed for chromosomes 3 and 14 and an unidentified marker chromosome that resembles chromosome 21 or 22 by DAPI banding (data not shown).
Localization of a clonal transcription center to a derivative chromosome 14 in CaSki cells.
To identify the chromosome that harbored the transcriptionally active HPV DNA in CaSki cells, we simultaneously probed for nascent HPV-16 RNA (in red) and for one of several individual chromosome territory domains (in green) in interphase cells. Visual inspection of the spatial localization of viral RNA center revealed a 70% association with chromosome 14 (Fig. 2E) and a statistically insignificant (<15%) association with chromosomes 3 or 21 (data not shown). Because our 3′ RACE analysis suggested a link to chromosome 6 or 22 (see below), we also examined these chromosomal territory domains. Analysis of interphase cells showed a 91% association with chromosome 6 (Fig. 2H) but no significant association with chromosome 22 (Fig. 2I). Indeed, by metaphase chromosome painting, one derivative chromosome 14 was observed to harbor a low copy of HPV integrant (Fig. 2D, in green). The chromosome 14 paint (Fig. 2D, in red) did not hybridize to a small region that was proximal to the HPV DNA but distal to the centromere. Collectively, these results suggest that a clonal selection occurred for a transcriptionally active HPV DNA located on a derivative chromosome 14, which may contain some translocated chromosome 6 materials. However, metaphase paint analysis failed to demonstrate chromosome 6 sequences proximal to the HPV genome (data not shown). Thus, this locus may have a complex chromosomal rearrangement. As controls for specificity, nascent HPV RNA did not associate with large tandem HPV DNA repeats on chromosome 2, nor with chromosomes such as X or 13 (<15% association) that had no integrated HPV (Fig. 2F and G).
Clonal RNA transcription in CaSki cells by 3′ RACE.
To substantiate a clonal selection for the viral transcription center, we performed a 3′ RACE experiment to determine the virus-host RNA junction sequences. Sequence analysis of five independent cDNA clones placed the 3′ viral-cellular junction at HPV-16 nucleotide position 3728 in the overlapping E2/E4 open reading frames. All five cDNAs had the same cellular sequence of 98 or 99 bases, followed by the posttranscriptionally added poly(A), in agreement with previous reports (3, 52, 68). Thus, we are indeed dealing with the same CaSki cells as in previous reports. Interestingly, our cDNAs contained alternative E1∧E4 mRNA splices that differ in the acceptor site at either nucleotide 3357 (two cDNAs) or, one codon later, nucleotide 3360 (three cDNAs). Alternative splicing of this message was not reported previously but was predicted based on comparative sequence analysis (11). BLASTN analysis of the host sequence revealed a homology or identity with sequences that are associated with L2 line elements and a possible match with sequences on chromosomes 6 and 22. The former, but not the latter, is consistent with the chromosome paint analyses. Taken together, these results support the conclusion that a single active HPV center is in direct association with DNA from chromosome 6 located on a derivative chromosome 14.
Selection for a single transcriptional center during passage of HPV-immortalized keratinocytes.
To test whether a single transcription center observed in CaSki cells is a unique case or is a common and reproducible event, we examined four previously well characterized primary human foreskin keratinocyte lines immortalized by either HPV-16 (FK16A, FK16B) or HPV-18 (FK18A, FK18B) (57, 58, 59). The FK16A cell line contains ca. 5 to 10 integrated copies of HPV DNA, whereas FK16B contains more than 100 integrated copies. The FK18A and FK18B lines both contain more than 100 copies of viral DNA, but the two lines might be of clonal origin based on Southern blot hybridization (59). These cell lines went from a normal karyotype to a tetraploid state accompanied by various chromosomal changes. In organotypic cultures, they exhibited progressively more dysplastic phenotypes with increasing passage numbers (58).
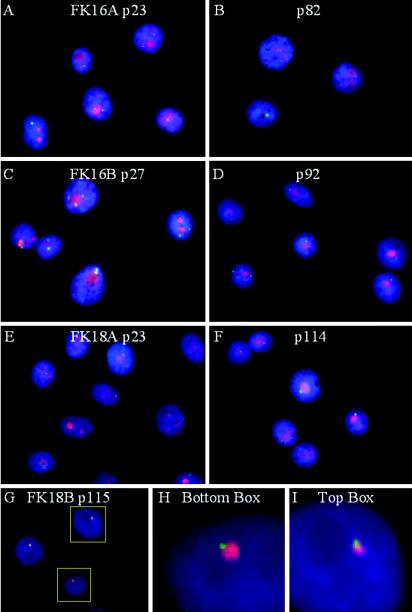
At least 200 interphase cells each of FK16A, FK16B, FK18A, and FK18B at different passages were analyzed for their nascent viral RNA centers. The FK16A cell line demonstrated up to five centers at passage 23, but at passages 64 and 82 there was only a single transcription center (Fig. 3A and B, in green, and data not shown). The FK16B cell line at passage 27 had an average of 3.1 RNA centers per cell, but some cells had as many as 7 centers. At passage 64, the average number of transcription centers per cell had dropped to 2.3. At passage 92, the percentage of cells containing one center increased to 70%, whereas most of the remaining population contained two centers (Fig. 3C and D, in green, and data not shown). Thus, there appears to be a distinct trend toward selection for one transcription center during passage of HPV-16 immortalized cells. In FK18A and FK18B cells, only a single HPV transcription center was observed, even at early passage, and this pattern was maintained at later passages (Fig. 3E to F, in green, and data not shown). Interphase viral DNA analysis of FK18A and FK18B showed that both possessed a single viral DNA integration focus. In these cell lines, the focus of viral RNA transcripts (green) was always positioned at one end of the larger DNA center (red) (Fig. 3G). Enlarged images of two of the three cells in Fig. 3G are shown in Fig. 3H and I. We suggest that that this pattern is consistent with transcription of a DNA template located near or at the 3′ end of the viral DNA array, allowing the use of a downstream host cleavage site and poly(A) signal, as in the cases of the CaSki and SiHa cell lines. Had the entire array been actively transcribed, the RNA signals would have colocalized with the larger DNA signal (see Fig. 7D and E). Collectively, our results strongly suggest that cells with a single transcription center may have a distinct growth advantage over cells with multiple RNA centers.
FIG. 3.
Localization and number of transcriptionally active HPV centers in immortalized human foreskin keratinocytes. (A to F) HPV RNA centers (FITC, green) during passage relative to nucleoli, which was revealed by antibody to nucleolin or C23 (TRITC, red). Nascent viral RNAs were detected by in situ hybridization with DNA probes in FK16A at passages 23 and 82 (A and B), FK16B at passages 27 and 92 (C and D), FK18A at passages 23 and 114 (E and F), or FK18B at passage 115 (G, H, and I). Nuclear DNA was stained with DAPI. (G to I) Localization of HPV RNA center (FITC) relative to HPV DNA in the FK18B. (H and I) Enlargement of two of the three cells in panel G (boxes). All images were acquired with a ×100 objective lens but were enlarged in panels H and I.
FIG. 7.
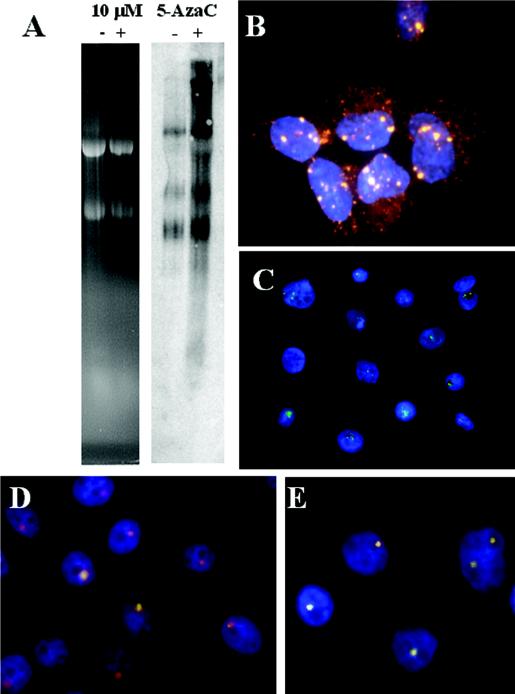
Transcriptional activation of silenced HPV DNA centers by 5-azacytidine. (A) The right panel shows a Northern blot hybridization of HPV-16 RNA in CaSki cells with (lane +) or without (lane −) treatment with 10 μM 5-azacytidine for 48 h. The left panel shows ethidium bromide staining of the 18S and 28S rRNAs of the gel, demonstrating equal loading. (B) In situ detection of nascent HPV-16 RNA (Cy3) in CaSki cells after 5-azacytidine treatment. Because of the high abundance of viral RNAs, some cytoplasmic mRNA was also detected by the probes despite in situ extraction. (C) 5-Azacytidine-treated FK16A cells at passage 82. (D and E) Viral RNA (FITC) and DNA (Cy3) in FK18A cells at passage 113 (D) and in FK18B cells at passage 115 (E) upon treatment with 5-azacytidine. Nuclei were stained with DAPI. Note that nascent RNA (FITC) completely overlaps the DNA signal in some of the cells (white dots). The images in panels B to E were acquired with a ×100 objective lens but are presented in different sizes for clarity and to reveal the reproducible patterns of expression among cells.
Changes in viral RNA expression but not viral DNA integration during passage.
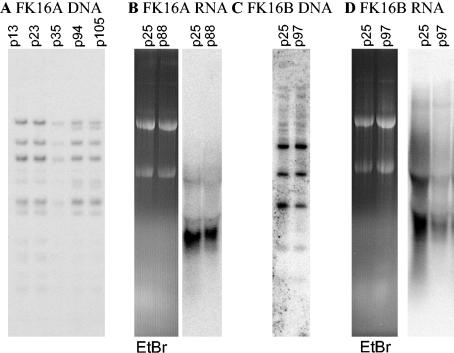
To determine whether a subpopulation of genetically distinct cells was selected during passage leading to the changing transcription, we performed Southern blot hybridization analyses on these cell lines at different passage numbers. In FK16A, we observed two new minor DNA bands in later passages (Fig. 4A), but no changes were observed in FK16B (Fig. 4C) or FK18 cells (59) by Southern blot analyses. Northern blot analysis of total RNA extracted from early and late passages of the FK16A (Fig. 4B) and FK16B (Fig. 4D) were also performed. In the FK16A cell line (Fig. 4B), there was a loss of a high-molecular-weight band in passage 88 relative to passage 25 as the number of centers per cell goes on average from 3 to 1. A more significant change in band pattern and a reduction of intensity were seen in viral RNAs from the late passage FK16B cell line (Fig. 4B), relative to early-passage cells that exhibited up to seven centers per cell. These results suggest epigenetic changes of the integrated viral genes during passage could be responsible for silencing all but one or two transcription centers.
FIG. 4.
Patterns of viral DNA integration and RNA expression in immortalized cell lines at different passages. (A) Southern blot analysis of PstI-digested FK16A DNAs at passages 13, 23, 35, 94, and 105. (B) The right panel shows a Northern blot hybridization of FK16A RNAs at passages 25 and 88, demonstrating a slight decline in expression. The left panel shows ethidium bromide staining of the 18S and 28S rRNA as a loading control. (C) Southern blot analysis of PstI digestion of the FK16B cell line at passages 25 and 97. (D) The right panel shows a Northern blot hybridization of FK16B RNAs at passages 25 and 97, demonstrating a large decrease in viral RNA expression. The left panel shows ethidium bromide staining of the 18S and 28S rRNA as a loading control.
Silencing of the original active center upon selection for newly introduced HPV oncogenes.
To test whether the reduction in transcription centers resulted from an active selection against transcription from multiple centers, FK18B cells were transduced with pseudotyped, recombinant HIV viruses that expressed either HPV-18 E6 and E7 oncogenes or a green fluorescent protein (GFP). The transgene was each placed upstream of an IRES and a puromycin resistance gene (puro). Each expression cassette was driven by the cytomegalovirus immediate-early promoter. Passage 109 of FK18B cells that we showed to harbor a single, established HPV RNA center was infected with either recombinant virus at a multiplicity of infection of 1 or 10 and selected with puromycin. During the drug selection, mock-transduced cells all died, whereas cells infected with the GFP-IRES-puro virus grew vigorously and expressed GFP, a finding consistent with proviral DNA integration (Fig. 5A to C). In contrast, cells transduced with the E6/E7-IRES-puro cassette developed many vacuoles and were slow in growth, and many began to flatten out, which is reminiscent of senescence.
FIG. 5.
Expression of only the newly introduced viral oncogenes in FK18B cells under selection. Lentivirus vector transduction of a GFP-IRES-puro center (A to F) or E6/E7-IRES-puro center (G to I) into FK18B cells at passage 109, followed by puromycin selection. (A) GFP-positive cells (green); (B) DAPI-stained nuclei of the same cells as in panel A; (C) merged image of panels A and B; (D and E) detection of nascent E6-E7 RNA (FITC) (D) and nascent RNA (E) of the puromycin resistance gene (Cy3) in interphase cells with DAPI-stained nuclei; (F) merged image of panels D and E; (G and H) E6-E7 RNA center (FITC) (G) and nascent RNA (H) from the puromycin resistance gene (Cy3); (I) merged image of panels G and H. Images were acquired with a ×10 (A to C) or a ×100 (D to I) objective lens.
On cells that eventually grew out about 5 weeks after the selection, we performed a nascent RNA assay for both puro expression (in red) and for HPV-18 oncogene expression (in green). Analysis of 200 GFP-expressing cells showed all to contain one E6/E7 RNA center, in agreement with previous observation. We found that 5, 82.5, and 11.5% of the cells, respectively, had zero, one, and two puro RNA centers, and a single cell had three centers. None of the puro RNA centers colocalized with the HPV-18 RNA center (Fig. 5D to F), as expected for undirected integration of the provirus. In contrast, in cells transduced with the HPV-18 E6/E7-IRES-puro expression cassette, the HPV-18 RNA center invariably colocalized with the puromycin expression center (Fig. 5G to I). Conversely, no puro center was distinct from the HPV-18 E6-E7 center. Among 200 transduced cells, 5% had no center, whereas 93 and 1.5% had one and two dual-RNA centers, with one cell having three such centers. These observations demonstrate that a switch had occurred in the locus of viral oncogene expression from the original endogenous center to the newly introduced center.
Single transcription centers in human cancer tissues.
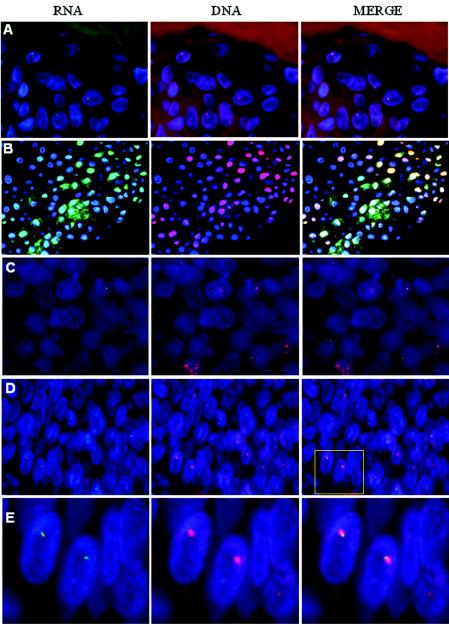
To ascertain the biological relevance of the results obtained from transformed and immortalized cell lines to HPV cancers, we examined viral RNA transcription centers in formalin-fixed, paraffin-embedded tissue biopsies of HPV-associated lesions. To determine the feasibility of using these methods with paraffin-embedded specimens, we first simultaneously detected HPV RNA and DNA in 4-μm sections of comparably fixed and paraffin-embedded organotypic raft cultures of FK16 and FK18 lines. Nascent transcription centers were detected, and the number of bright RNA foci in the nuclei correlated with those present in cells grown in submerged cultures at similar passage numbers. An example of a single DNA and RNA center per cell in the FK18B cells at passage 118 is shown in Fig. 6A. Some nuclei did not contain any RNA center, probably because the relevant portion of the nucleus was sectioned off. In contrast, in productively infected laryngeal papillomas that harbor a large number of extrachromosomal viral DNA, abundant RNA was detected throughout the cells, as previously described (12, 61), masking all nascent transcription centers (Fig. 6B). Thus, a low transcriptional center number is a prerequisite for center detection. These results also suggest a high concentration of nascent viral RNA at the site of transcription relative to low concentrations of completed and dispersed viral transcripts.
FIG. 6.
Simultaneous probing for HPV RNA and DNA in formalin-fixed, paraffin-embedded tissue sections. We used 4-μm sections. Left panels, HPV RNA (FITC, green); middle panels, HPV DNA (Cy3, red); right panels, merged images of RNA and DNA signals. Nuclei were stained with DAPI (blue). (A) Organotypic culture of FK18B passage 118 showing a single small viral RNA and a larger DNA center; (B) HPV-11 laryngeal papilloma; (C and D) representative specimens from 13 cases of cancers induced by HPV-16 or HPV-18 (not shown); (E) enlargement of the boxed area in the right panel of specimen D. Images for A, C, and D were acquired with a ×100 objective lens, and images for B were acquired with a ×40 objective lens.
We then examined representative HPV-containing primary cancers, including nine cases of HPV-16- or HPV-18-positive squamous carcinomas of the cervix or vulva, three cases of HPV-16-containing laryngeal or pharyngeal cancers, and 1 case of HPV-16-positive endocervical carcinoma. Strikingly, all specimens exhibited a single viral RNA center per cell when observable, regardless of the number of DNA foci. Figure 6C demonstrates a typical pattern observed in 11 cases in which only one small RNA center was associated with one of the larger DNA centers. However, we cannot rule out that there might have been a small number of additional centers present at different focal planes or cut off in the thin tissue sections, thus escaping detection. Two remaining cases harbored only a single HPV DNA locus that likely contained a tandem array based on the size of the signal. In both cases, HPV RNA was localized to the edge of the larger DNA signals, possibly signifying a transcriptionally active copy at the virus-host junction, as in the FK18 cells described above. One example is shown in Fig. 6D with an enlargement in Fig. 6E. These results support our hypothesis that there is a selection for a single transcription center during the development of HPV-induced cancers and that, in cases with tandem arrays of integrated viral genomes, only the virus-host junction copy is transcriptionally active.
Activation of transcription from silent viral DNA centers by growth in 5-azacytidine.
To begin to identify mechanisms by which the majority of the integrated HPV DNA copies are transcriptionally inactivated, we treated growing CaSki cells for 40 to 48 h with 10 μM 5-azacytidine, an inhibitor of DNA methylation. Northern blot analysis of RNA revealed an increase in the intensity of the existing messages and the appearance of new transcripts in treated versus untreated cells (Fig. 7A). In situ hybridization revealed a number of large transcription centers in treated cells (Fig. 7B, in red). However, activation was not observed in the majority of cells in these experiments. This incomplete induction might be due to a cell cycle effect, to an incomplete demethylation of the HPV DNA, or to a toxic or lethal effect of the compound (25, 47). Indeed, we observed a decreased cell number in treated cultures compared to the untreated cultures, and cells with activated centers were frequently rounded up. In contrast, treatment of cells with trichostatin A, a potent inhibitor of histone deacetylases, failed to activate the silenced centers (data not shown).
We also tested late passages of HPV-immortalized cell lines that had only one nascent RNA center. In treated FK16A passage 82 cells, multiple centers were observed (Fig. 7C, in green). In treated FK18A passage 113 and FK18B passage 115 cells (Fig. 7D and E), the nascent HPV RNA signals (in green) either colocalized with or were larger in size than the corresponding DNA center (in red), a finding distinctively different from the pattern in untreated cells (Fig. 3G to I). However, as with CaSki cells, some treated cells contained only the original, active 3′ boundary copy (Fig. 7D). These observations suggest that the long-term silencing of HPV DNA centers is mediated at least in part by methylation on cytosine.
Perinucleolar localization of viral transcription.
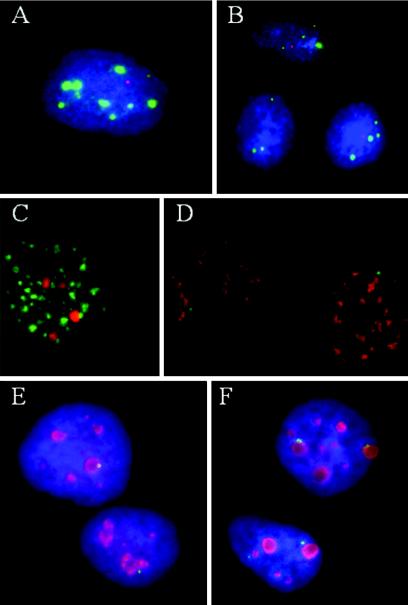
The nucleus is a functionally organized environment, with many gene expression and regulatory processes occurring in distinct subnuclear domains (34, 35, 55). We investigated whether the PVTD is spatially associated with previously described nuclear domains that are known to be involved in transcription regulation, RNA splicing, or cell cycle regulation, such as the nuclear domain 10 (ND10) or PML bodies (revealed by PML) and Cajal bodies (20, 22, 46). Extrachromosomal HPV DNA replication may occur in the neighborhood of ND10 (62), as is the case with a number of other DNA viruses (18). Cajal bodies (20) are nuclear domains implicated in the control of S-phase-associated histone gene transcription mediated by p220npat (31, 71).
Colocalization studies were performed first with CaSki cells by probing for nascent HPV-16 transcripts (in red) and then for PML (Fig. 8A, in green) or for p220npat (Fig. 8B, in green), with or without an antigen retrieval process. We observed no associations. Additional domains involved with gene transcription and RNA metabolism are the interchromatin granule clusters that are visualized as immunofluorescent “speckles” with antibodies to spliceosome assembly factor SC-35 (56). Simultaneous visualization of SC-35 antibody (in green) and HPV-16 DNA in CaSki cells (in red) (Fig. 8C) showed only stochastic colocalization with the larger tandem viral DNA arrays. Although most of the transcripts of oncogenic HPVs spanning the E6 region are spliced, nascent viral RNA (Fig. 8D, in green) did not spatially associate with SC-35 domains (red) (Fig. 8D). This lack of association has also been reported for transcripts of some cellular genes as well (51).
FIG. 8.
Perinucleolar localization of PVTD. With the exception of nucleoli, no association was found between PVTD, as denoted by nascent HPV RNA, and several nuclear domains, as specified. (A to E) CaSki cells; (F) SiHa cells. (A) ND10 domains detected with an antibody to PML in green and viral RNA in red; (B) p220NPAT, a Cajal body-associated protein in green and viral RNA in red; (C) SC35 domain in green and integrated HPV DNA in red; (D) SC35 domain in red and nascent HPV RNA in green; (E and F) antibody to C23 (nucleolin) in red and HPV RNA in green. All images were captured at ×100 magnification and are presented at a similar magnification except panel B, which is shown at a lower magnification.
Localization of nascent viral nuclear RNA (in green) to a perinucleolar region was, however, consistently observed by using an antibody to nucleolin (also called C23) (in red), a marker for nucleoli (30) (Fig. 8E). About 75% of all detectable HPV-16 transcription centers (in green) were perinucleolar (within one to two viral RNA signal diameters from the nucleolus), and another 12% appeared to overlap the nucleoli. Signals that were at first thought to be away from nucleoli were in fact associated with very small C23-positive dots when viewed at high magnification. We then examined SiHa cells for the association with C23, Cajal bodies, and SC35 domains. Only localization to perinucleolar region was consistently noted (Fig. 8F, Table 1, and data not shown). Similarly, nascent viral transcripts in the immortalized cell lines were also perinucleolar regardless of the number of transcription centers in the cells (Fig. 3A to F in red, Table 1). At present, the significance of this perinucleolar PVTD is not understood.
TABLE 1.
Localization of PVTD relative to the nucleolus in HPV-immortalized and -transformed cell linesa
| Cell line | Passage no. | % of nucleolar association
|
||
|---|---|---|---|---|
| Peri | Direct | None | ||
| FK16A | 23 | 87.0 | 7.0 | 6.0 |
| 82 | 86.5 | 10.0 | 3.5 | |
| FK16B | 27 | 78.5 | 8.0 | 13.5 |
| 64 | 78.5 | 14.0 | 7.5 | |
| FK18A | 23 | 84.0 | 5.5 | 10.5 |
| 114 | 78.5 | 10.0 | 11.5 | |
| FK18B | 17 | 88.5 | 4.5 | 7.0 |
| 115 | 87.5 | 6.0 | 6.5 | |
| CaSki | NAb | 75.0 | 12.0 | 13.0 |
| SiHa | NA | 77.5 | 9.5 | 13.0 |
Nascent HPV RNA signals were scored as perinucleolar (Peri) association when they were located within two diameters of the RNA signals from the C23 (nucleolin) signals and as direct association when the two signals overlapped. A total of 200 signals were counted from each culture.
NA, not applicable.
DISCUSSION
Previous studies of the HPV-16 and HPV-18 immortalized and transformed cell lines provide strong support for the notion that these cell lines are faithful experimental models with which to investigate the genetic, epigenetic, and biochemical changes that occur during the progression of dysplastic HPV lesions to cancers. By virtue of the phenotypic inactivation of the tumor suppressor proteins pRB and p53, the oncogenic HPV E7 and E6 initiate a neoplastic process in the anogenital tract epithelium (37, 38, 73). Subsequent selection for and accumulation of mutations in yet-to-be-identified key cellular regulatory genes promotes further progression to cancer. In the present study, by using these model systems along with cancer tissues, we have uncovered an active and parallel epigenetic selection process for cells with certain striking characteristics of viral oncogene transcription during progression.
Using T-FISH to probe for nascent viral RNA simultaneously with integrated viral DNA, metaphase or interphase chromosomes, or nuclear domains with T-IF, we demonstrate here for the first time directly that there is a strong selection for cells containing only a single transcriptionally active viral DNA integrant and that this locus is perinucleolar in the majority of the cells examined, regardless of the number of centers (Fig. 1, 3, 5, and 8). We have named this the PVTD. Additional viral copies do not have associated nascent viral RNA transcripts. The SiHa cell line appears to be an exception, since both copies of HPV-16, one each at genetically identical loci on the pair of chromosomes 13 (1, 3, 17, 33), are transcriptionally active (Fig. 1). We speculate that this double RNA center might even have arisen as a result of positive selection for a chromosome duplication or gene conversion event. Oncoprotein production in their progenitor cells might have been less than optimal and became outcompeted by cells that underwent the duplication during the development of the cancer or the establishment of the cell line. Similarly, cells that no longer express viral oncogenes will not be maintained, as clearly demonstrated by in vitro experimentation (14, 67).
Our data also indicate that, within any one cell line, the single RNA center is clonal rather than stochastically variable among individual cells, implicating a regulated expression. First, our systematic in situ mapping data revealed that the transcriptionally active template in CaSki cells is located at a single-copy or low-copy-number site on a derivative chromosome 14 (Fig. 2), among the dozen loci that together harbor up to 600 copies of viral DNA. These mapping data are supported by 3′ RACE in which five of five cDNAs have the same virus-host junction sequence. Second, in FK18 cells harboring multiple copies of the HPV-18 DNA in a single large tandem array, only the virus-host junction copy appears to be transcribed (Fig. 3). Of greatest relevance, each of 13 primary HPV-associated cancers examined also exhibited one viral transcription center per cell, when observable (Fig. 6).
Our data show that the evolving viral transcription patterns are attributed to epigenetic rather than genetic alterations, in that transcription from the inactive integrants can be reactivated by 5-azacytidine, implicating DNA methylation as at least being partially responsible for silencing transcription (Fig. 7). Reactivation was not achieved by exposing the cells to trichostatin A, an inhibitor of histone deacetylases (data not shown). These results agree with the current view that DNA methylation confers long-term DNA silencing, whereas deacetylases shut off transcription on a short-term basis (13, 43). Our observations are in excellent agreement with a recent report (2) that HPV-16 DNA is targeted for CpG methylation in vivo and that changing methylation patterns were observed with different lesion grades. The data are also consistent with reports that transfected and integrated HPV DNA is heavily methylated and transcriptionally silent (5, 29, 45). However, reactivation was not demonstrated in those studies.
Our findings explain why one predominant viral transcript was identified in cancers and carcinoma cell lines (26, 53, 68) and also why little or no E2 protein is expressed. We speculate that transcription from the internal copies may have contributed relatively little to the expression of viral oncoproteins relative to the junction copy due to low stability of mRNA utilizing the viral poly(A) site (23). Furthermore, there is also a selection against cells that express high levels of E2 which would downregulate the promoter for the viral oncogenes (9, 14, 16, 44, 63, 67). An additional impetus for silencing comes from our recent observation that the HPV E2 protein associates with the mitotic spindles to enable extrachromosomal HPV DNA plasmids to segregate as minichromosomes and to establish persistence (64). However, upon viral DNA integration, E2 might contribute to genomic instability if E2 bound to the viral origin in tandemly integrated HPV DNA creates a viro-centromere, causing chromosome breakage when it is pulled to the opposite centrosome relative to the kinetochore. Mitotic catastrophe associated with the breakage-fusion bridge cycle (32) can be avoided only when the internal tandem viral copies capable of encoding E2 are silenced. Selection for the fittest cells for progression can then begin.
The selection for a single dominant expression locus appears to be a dynamic and plastic process. Various immortalized cell lineages reached this state after different numbers of cell division during passage in vitro, perhaps depending on the number of viral integration sites, the transcriptional activities from these various loci, and the relative mRNA stabilities conferred by the captured 3′ host untranslated regions and poly(A) sequences. The selection may ultimately be predicated on the absolute level of viral oncoprotein expression, as was suggested by our oncogene challenge experiment (Fig. 5). By introducing an extra copy of the HPV oncogenes into a cell line which had already arrived at a single expression center, we presented a striking example in which another round of transcription center selection was attained within a matter of weeks when the cells experienced a new and strong selection pressure for the newly introduced viral oncogenes that are transcriptionally and translationally coupled to a drug resistance gene. The surviving cells that emerged from an apparent “crisis period” turned off the original viral RNA expression center and replaced it with the newly introduced copy, gaining puromycin resistance while maintaining E6/E7 production. Thus, the crisis may have been caused by too much of the viral oncoproteins. Had the higher level of viral oncogene gene expression from both the resident and the newly introduced copies been of no consequence or even advantageous to the cells, we would not have observed the silencing of the original viral RNA center. We presume that the silencing of the original RNA center might be attributed to chromatin remodeling by DNA methylation, as was observed in other cell lines. However, extensive cell death upon exposure to 5-azacytidine prevented us from testing this hypothesis.
Although the selection for cells with more stable transcripts and higher levels of viral oncoproteins in proliferating cells is important at the early stage of viral oncogenesis, in the long run, what seems to be important is an optimal level of oncogene expression from a locus most suitable for the particular environment in which the cells find themselves. Neither too little nor too much of the viral oncoproteins will do. Cells that fail to cycle rapidly are overgrown; those that divide too quickly may not be able to replenish their cellular components and are lost during passage, and others may undergo senescence. Our results with primary HPV cancers are consistent with a clonal selection for the fittest cells, which is in agreement with the notion that cancer is monoclonal in origin (6, 28). We suggest that the ability to detect a single viral RNA center might provide for a novel, highly relevant biomarker of neoplastic progression. Additional experiments will be required to test this hypothesis. Cellular oncogenes are amplified in a variety of cancers, e.g., the N-myc locus in neuroblastomas (49), ErbB2 in breast cancer, and EGRF in other cancers (42). It will be most interesting to determine how many of the amplified copies are actively transcribed. Finally, the ability to conduct multiplex assays with tyramide-enhanced reagents and the gene regulatory concepts described here will have considerable applicability to attempts at cancer control, analysis of diseases, production of transgenic stem cells, and gene replacement therapy.
Acknowledgments
This study was supported by U.S. Public Health Service grants CA36200 to T.R.B. and L.T.C.
We thank Ge Jin, Martha Hayes, Heather Benedict-Hamilton, Brian Streib, and Susie Godbey for technical assistance; Albert Tousson, Julie Decker, and George M. Shaw for support during the initial phase of this project; Mark Bobrow, Kim Fitzpatrick, and Yoshika Sherring from Perkin-Elmer for some of the reagents and advice; and Wade Harper for the p220npat antibody. We also gratefully acknowledge the 1998 Advanced In Situ and Immunohistochemistry Class at Cold Spring Harbor Laboratory for being instrumental to the conduct of this research.
REFERENCES
- 1.Adler, K., K. T. Erickson, and M. Bobrow. 1997. High sensitivity detection of HPV-16 in SiHa and CaSki cells using FISH enhanced by TSA. Histochem. Cell Biol. 108:321-324. [DOI] [PubMed] [Google Scholar]
- 2.Badal, V., L. S. H. Chuang, E. H.-H. Tan, S. Badal, L. L. Villa, C. M. Wheeler, B. F. L. Li, and H.-U. Bernard. 2003. CpG methylation of human papillomavirus type 16 DNA in cervical cancer cell lines and in clinical specimens: genomic hypomethylation correlates with carcinogenic progression. J. Virol. 77:6227-6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, C. C., W. C. Phelps, V. Lindgren, M. J. Braun, M. A. Gonda, and P. M. Howley. 1987. Structural and transcriptional analysis of human papillomavirus 16 sequences in cervical carcinoma cell lines. J. Virol. 61:962-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bobrow, M. N., and P. T. Moen. 2000. Tyramide signal amplification (TSA) systems for the enhancement of ISH signals in cytogenetics, In J. P. Robinson (ed.), Current protocols in cytometry. John Wiley & Sons, Inc., New York, N.Y. [DOI] [PubMed]
- 5.Burnett, T. S., and P. H. Gallimore. 1985. Introduction of cloned human papillomavirus 1a DNA into rat fibroblasts: integration, de novo methylation and absence of cellular morphological transformation. J. Gen. Virol. 66:1063-1072. [DOI] [PubMed] [Google Scholar]
- 6.Cahill, D. P., K. W. Kinzler, B. Vogelstein, and C. Lengauer. 1999. Genetic instability and Darwinian selection in tumours. Trends Cell Biol. 9:M57-M60. [PubMed] [Google Scholar]
- 7.Chen, W., X. Wu, D. N. Levasseur, H. Liu, L. Lai, J. C. Kappes, and T. M. Townes. 2000. Lentiviral vector transduction of hematopoietic stem cells that mediate long-term reconstitution of lethally irradiated mice. Stem Cells 18:352-359. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, S., D.-C. Schmidt-Grimminger, T. Murant, T. R. Broker, and L. T. Chow. 1995. Differentiation-dependent up-regulation of the human papillomavirus E7 gene reactivates cellular DNA replication in suprabasal differentiated keratinocytes. Genes Dev. 9:2335-2349. [DOI] [PubMed] [Google Scholar]
- 9.Chin, M. T., T. R. Broker, and L. T. Chow. 1989. Identification of a novel constitutive enhancer element and an associated binding protein: implications for human papillomavirus type 11 enhancer regulation. J. Virol. 63:2967-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chow, L. T., and T. R. Broker. 1997. Small DNA tumor viruses, In N. Nathanson (ed.), Viral pathogenesis. Lippincott-Raven Publishers, Philadelphia, Pa. p. 267-302.
- 11.Chow, L. T., M. Nasseri, S. M. Wolinsky, and T. R. Broker. 1987. Human papillomavirus types 6 and 11 mRNAs from genital condylomata acuminata. J. Virol. 61:2581-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crum, C. P., G. Nuovo, D. Friedman, and S. J. Silverstein. 1988. Accumulation of RNA homologous to human papillomavirus type 16 open reading frames in genital precancers. J. Virol. 62:84-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curradi, M., A. Izzo, G. Badaracco, and N. Landsberger. 2002. Molecular mechanisms of gene silencing mediated by DNA methylation. Mol. Cell. Biol. 22:3157-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeFilippis, R. A., E. C. Goodwin, L. Wu, and D. DiMaio. 2003. Endogenous human papillomavirus E6 and E7 proteins differentially regulate proliferation, senescence, and apoptosis in HeLa cervical carcinoma cells. J. Virol. 77:1551-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.del Mar Pena, L. M., and L. A. Laimins. 2001. Differentiation-dependent chromatin rearrangement coincides with activation of human papillomavirus type 31 late gene expression. J. Virol. 75:10005-10013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong, G., T. R. Broker, and L. T. Chow. 1994. Human papillomavirus type 11 E2 proteins repress the homologous E6 promoter by interfering with the binding of host transcription factors to adjacent elements. J. Virol. 68:1115-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El Awady, M. K., J. B. Kaplan, S. J. O'Brien, and R. D. Burk. 1987. Molecular analysis of integrated human papillomavirus 16 sequences in the cervical cancer cell line SiHa. Virology 159:389-398. [DOI] [PubMed] [Google Scholar]
- 18.Everett, R. D. 2001. DNA viruses and viral proteins that interact with PML nuclear bodies. Oncogene 20:7266-7273. [DOI] [PubMed] [Google Scholar]
- 19.Fey, E. G., G. Krochmalnic, and S. Penman. 1986. The nonchromatin substructures of the nucleus: the ribonucleoprotein (RNP)-containing and RNP-depleted matrices analyzed by sequential fractionation and resinless section electron microscopy. J. Cell Biol. 102:1654-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gall, J. G. 2000. Cajal bodies: the first 100 years. Annu. Rev. Cell Dev. Biol. 16:273-300. [DOI] [PubMed] [Google Scholar]
- 21.Hurlin, P. J., P. Kaur, P. P. Smith, N. Perez-Reyes, R. A. Blanton, and J. K. McDougall. 1991. Progression of human papillomavirus type 18-immortalized human keratinocytes to a malignant phenotype. Proc. Natl. Acad. Sci. USA 88:570-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen, K., C. Shiels, and P. S. Freemont. 2001. PML protein isoforms and the RBCC/TRIM motif. Oncogene 20:7223-7233. [DOI] [PubMed] [Google Scholar]
- 23.Jeon, S., and P. F. Lambert. 1995. Integration of human papillomavirus type 16 DNA into the human genome leads to increased stability of E6 and E7 mRNAs: implications for cervical carcinogenesis. Proc. Natl. Acad. Sci. USA 92:1654-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson, C. V., R. H. Singer, and J. B. Lawrence. 1991. Fluorescent detection of nuclear RNA and DNA: implications for genome organization. Methods Cell Biol. 35:73-99. [PubMed] [Google Scholar]
- 25.Juttermann, R., E. Li, and R. Jaenisch. 1994. Toxicity of 5-aza-2′-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc. Natl. Acad. Sci. USA 91:11797-11801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klaes, R., S. M. Woener, R. Ridder, N. Wentzensen, M. Düerst, A. Schneider, B. Lotz, P. Melsheimer, and M. von Knebel Doeberitz. 1999. Detection of high-risk cervical intraepithelial neoplasia and cervical cancer by amplification of transcripts derived from integrated papillomavirus oncogenes. Cancer Res. 59:6132-6136. [PubMed] [Google Scholar]
- 27.Kouzarides, T. 2002. Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 12:198-209. [DOI] [PubMed] [Google Scholar]
- 28.Lengauer, C., K. W. Kinzler, and B. Vogelstein. 1998. Genetic instabilities in human cancers. Nature 396:643-649. [DOI] [PubMed] [Google Scholar]
- 29.List, H. J., V. Patzel, U. Zeidler, A. Schopen, G. Ruhl, J. Stollwerk, and G. Klock. 1994. Methylation sensitivity of the enhancer from the human papillomavirus type 16. J. Biol. Chem. 269:11902-11911. [PubMed] [Google Scholar]
- 30.Liu, H. T., and B. Y. Yung. 1999. In vivo interaction of nucleophosmin/B23 and protein C23 during cell cycle progression in HeLa cells. Cancer Lett. 144:45-54. [DOI] [PubMed] [Google Scholar]
- 31.Ma, T., B. A. Van Tine, Y. Wei, M. D. Garrett, D. Nelson, P. D. Adams, J. Wang, J. Qin, L. T. Chow, and J. W. Harper. 2000. Cell cycle-regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 14:2298-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McClintock, B. 1951. Chromosome organization and genic expression. Cold Spring Harbor Symposia Quant. Biol. 16:13-47. [DOI] [PubMed] [Google Scholar]
- 33.Mincheva, A., L. Gissmann, and H. zur Hausen. 1987. Chromosomal integration sites of human papillomavirus DNA in three cervical cancer cell lines mapped by in situ hybridization. Med. Microbiol. Immunol. 176:245-256. [DOI] [PubMed] [Google Scholar]
- 34.Misteli, T. 2001. Protein dynamics: implications for nuclear architecture and gene expression. Science 291:843-847. [DOI] [PubMed] [Google Scholar]
- 35.Moen, P. T., K. P. Smith, and J. B. Lawrence. 1995. Compartmentalization of specific pre-mRNA metabolism: an emerging view. Hum. Mol. Gen. 4:1779-1789. [DOI] [PubMed] [Google Scholar]
- 36.Montgomery, K. D., K. L. Tedford, and J. K. McDougall. 1995. Genetic instability of chromosome 3 in HPV-immortalized and tumorigenic human keratinocytes. Genes Chromosomes Cancer 14:97-105. [DOI] [PubMed] [Google Scholar]
- 37.Münger, K. 2002. The role of human papillomaviruses in human cancers. Front. Biosci. 7:641-649. [DOI] [PubMed] [Google Scholar]
- 38.Münger, K., and P. M. Howley. 2002. Human papillomavirus immortalization and transformation functions. Virus Res. 89:213-228. [DOI] [PubMed] [Google Scholar]
- 39.Naldini, L., U. Blomer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263-267. [DOI] [PubMed] [Google Scholar]
- 40.Parker, J. N., W. Zhao, K. J. Askins, T. R. Broker, and L. T. Chow. 1997. Mutational analyses of differentiation-dependent human papillomavirus type 18 enhancer elements in epithelial raft cultures of neonatal foreskin keratinocytes. Cell Growth Differ. 8:751-762. [PubMed] [Google Scholar]
- 41.Pater, M. M., and A. Pater. 1985. Human papillomavirus types 16 and 18 sequences in carcinoma cell lines of the cervix. Virology 145:313-318. [DOI] [PubMed] [Google Scholar]
- 42.Rajkumar, T., and W. J. Gullick. 1994. The type I growth factor receptors in human breast cancer. Breast Cancer Res. Treat. 29:3-9. [DOI] [PubMed] [Google Scholar]
- 43.Rhee, I., K. E. Bachman, B. H. Park, K. W. Jair, R. W. Yen, K. E. Schuebel, H. Cui, A. P. Feinberg, C. Lengauer, K. W. Kinzler, S. B. Baylin, and B. Vogelstein. 2002. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature 416:552-556. [DOI] [PubMed] [Google Scholar]
- 44.Romanczuk, H., F. Thierry, and P. M. Howley. 1990. Mutational analysis of cis elements involved in E2 modulation of human papillomavirus type 16 P97 and type 18 P105 promoters. J. Virol. 64:2849-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosl, F., A. Arab, B. Klevenz, and H. zur Hausen. 1993. The effect of DNA methylation on gene regulation of human papillomaviruses. J. Gen. Virol. 74:791-801. [DOI] [PubMed] [Google Scholar]
- 46.Salomoni, P., and P. P. Pandolfi. 2002. The role of PML in tumor suppression. Cell 108:165-170. [DOI] [PubMed] [Google Scholar]
- 47.Santi, D. V., C. E. Garrett, and P. J. Barr. 1983. On the mechanism of inhibition of DNA-cytosine methyltransferases by cytosine analogs. Cell 33:9-10. [DOI] [PubMed] [Google Scholar]
- 48.Schneider-Gädicke, A., and E. Schwarz. 1986. Different human cervical carcinoma cell lines show similar transcription patterns of human papillomavirus type 18 early genes. EMBO J. 5:2285-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwab, M. 1993. Amplification of N-myc as a prognostic marker for patients with neuroblastoma. Semin. Cancer Biol. 4:13-18. [PubMed] [Google Scholar]
- 50.Schwarz, E., U. K. Freese, L. Gissmann, W. Mayer, B. Roggenbuck, A. Stremlau, and H. zur Hausen. 1985. Structure and transcription of human papillomavirus sequences in cervical carcinoma cells. Nature 314:111-114. [DOI] [PubMed] [Google Scholar]
- 51.Smith, K. P., P. T. Moen, K. L. Wydner, J. R. Coleman, and J. B. Lawrence. 1999. Processing of endogenous pre-mRNAs in association with SC-35 domains is gene specific. J. Cell Biol. 22:617-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smits, H. L., M. T. Cornelissen, M. F. Jebbink, J. G. van den Tweel, A. P. Struyk, M. Briet, and J. ter Schegget. 1991. Human papillomavirus type 16 transcripts expressed from viral-cellular junctions and full-length viral copies in CaSki cells and in a cervical carcinoma. Virology 182:870-873. [DOI] [PubMed] [Google Scholar]
- 53.Smotkin, D., and F. O. Wettstein. 1986. Transcription of human papillomavirus type 16 early genes in a cervical cancer and a cancer-derived cell line and identification of the E7 protein. Proc. Natl. Acad. Sci. USA 83:4680-4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Solinas-Toldo, S., M. Dürst, and P. Lichter. 1997. Specific chromosomal imbalances in human papillomavirus-transfected cells during progression toward immortality. Proc. Natl. Acad. Sci. USA 94:3854-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spector, D. L. 1993. Macromolecular domains within the cell nucleus. Annu. Rev. Cell Biol. 9:265-315. [DOI] [PubMed] [Google Scholar]
- 56.Spector, D. L. 2001. Nuclear domains. J. Cell Sci. 114:2891-2893. [DOI] [PubMed] [Google Scholar]
- 57.Steenbergen, R. D., M. A. Hermsen, J. M. Walboomers, G. A. Meijer, J. P. Baak, C. J. Meijer, and P. J. Snijders. 1998. Non-random allelic losses at 3p, 11p, and 13q during HPV-mediated immortalization and concomitant loss of terminal differentiation of human keratinocytes. Int. J. Cancer 73:412-417. [DOI] [PubMed] [Google Scholar]
- 58.Steenbergen, R. D., J. N. Parker, S. Isern, P. J. Snijders, J. M. Walboomers, C. J. Meijer, T. R. Broker, and L. T. Chow. 1998. Viral E6-E7 transcription in the basal layer of organotypic cultures without apparent p21cip1 protein precedes immortalization of human papillomavirus type 16- and 18-transfected human keratinocytes. J. Virol. 72:749-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Steenbergen, R. D., J. M. Walboomers, C. J. Meijer, E. M. van der Raaij-Helmer, J. N. Parker, L. T. Chow, T. R. Broker, and P. J. Snijders. 1996. Transition of human papillomavirus type 16 and 18 transfected human foreskin keratinocytes toward immortality: activation of telomerase and allele losses at 3p, 10p, 11q and/or 18q. Oncogene 13:1249-1257. [PubMed] [Google Scholar]
- 60.Stoler, M. H., C. R. Rhodes, A. Whitbeck, S. M. Wolinsky, L. T. Chow, and T. R. Broker. 1992. Human papillomavirus type 16 and 18 gene expression in cervical neoplasias. Hum. Pathol. 23:117-128. [DOI] [PubMed] [Google Scholar]
- 61.Stoler, M. H., S. M. Wolinsky, A. Whitbeck, T. R. Broker, and L. T. Chow. 1989. DifferentiatioN-linked human papillomavirus types 6 and 11 transcription in genital condylomata revealed by in situ hybridization with message-specific RNA probes. Virology 172:331-340. [DOI] [PubMed] [Google Scholar]
- 62.Swindle, C. S., N. Zou, B. A. Van Tine, G. M. Shaw, J. A. Engler, and L. T. Chow. 1999. HPV-11 DNA replication compartments in a transient DNA replication system. J. Virol. 73:1001-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tan, S. H., L. E. Leong, P. A. Walker, and H. U. Bernard. 1994. The human papillomavirus type 16 E2 transcription factor binds with low cooperativity to two flanking sites and represses the E6 promoter through displacement of Sp1 and TFIID. J. Virol. 68:6411-6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Van Tine, B. A., L. D. Dao, S.-Y. Wu, T. M. Sonbuchner, B.-Y. Lin, N. Zou, C.-M. Chiang, T. R. Broker, and L. T. Chow. 2004. Human papillomavirus (HPV) origin-binding protein associates with mitotic spindles to enable viral DNA partitioning. Proc. Natl. Acad. Sci. USA 101:4030-4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Van Tine, B. A., J. F. Knops, A. Butler, P. Deloukas, G. M. Shaw, and P. H. King. 1998. Localization of HuC (ELAVL3) to chromosome 19p13.2 by fluorescence in situ hybridization utilizing a novel tyramide labeling technique. Genomics 53:296-299. [DOI] [PubMed] [Google Scholar]
- 66.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wells, S. I., D. A. Francis, A. Y. Karpova, J. J. Dowhanick, J. D. Benson, and P. M. Howley. 2000. Papillomavirus E2 induces senescence in HPV-positive cells via pRB- and p21CIP-dependent pathways. EMBO J. 19:5762-5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wentzensen, N., R. Ridder, R. Klaes, S. Vinokurova, U. Schaefer, and M. K. Doeberitz. 2002. Characterization of viral-cellular fusion transcripts in a large series of HPV16 and -18 positive anogenital lesions. Oncogene 21:419-426. [DOI] [PubMed] [Google Scholar]
- 69.Xing, Y., C. V. Johnson, P. T. Moen, J. A. McNeil, and J. Lawrence. 1995. J. Nonrandom gene organization: structural arrangements of specific pre-mRNA transcription and splicing with SC-35 domains. J. Cell Biol. 131:1635-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yee, C., I. Krishnan-Hewlett, C. C. Baker, R. Schlegel, and P. M. Howley. 1985. Presence and expression of human papillomavirus sequences in human cervical carcinoma cell. Am. J. Pathol. 119:361-366. [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao, J., B. K. Kennedy, B. D. Lawrence, D. A. Barbie, A. G. Matera, J. A. Fletcher, and E. Harlow. 2000. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 14:2283-2297. [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao, W., F. Noya, W. Y. Chen, T. M. Townes, L. T. Chow, and T. R. Broker. 1999. Trichostatin A up-regulates human papillomavirus type 11 upstream regulatory region-E6 promoter activity in undifferentiated primary human keratinocytes. J. Virol. 73:5026-5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.zur Hausen, H. 1999. Immortalization of human cells and their malignant conversion by high risk human papillomavirus genotypes. Semin. Cancer Biol. l. 9:405-411. [DOI] [PubMed] [Google Scholar]