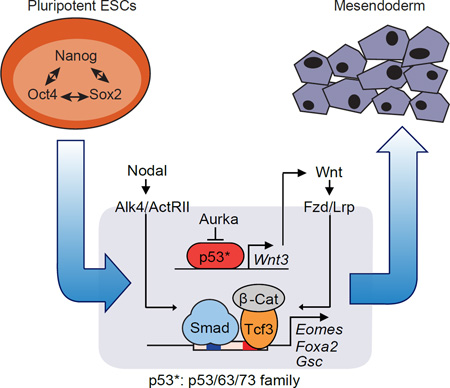
SUMMARY
In this study we outline a regulatory network that involves the p53 tumor suppressor family and the Wnt pathway acting together with the TGF-β pathway in mesendodermal differentiation of mouse and human embryonic stem cells. Knockout of all three members, p53, p63 and p73, shows that the p53 family is essential for mesendoderm specification during exit from pluripotency in embryos and in culture. Wnt3 and its receptor Fzd1 are direct p53 family target genes in this context, and induction of Wnt signaling by p53 is critical for activation of mesendoderm differentiation genes. Globally, Wnt3-activated Tcf3 and nodal-activated Smad2/3 transcription factors depend on each other for co-occupancy of target enhancers associated with key differentiation loci. Our results therefore highlight an unanticipated role for p53 family proteins in a regulatory network that integrates essential Wnt-Tcf and nodal-Smad inputs in a selective and interdependent way to drive mesendodermal differentiation of pluripotent cells.
Graphical abstract
INTRODUCTION
The transcription factor p53 is abundantly expressed in mouse embryo germ layer progenitors and in embryonic stem (ES) cells, suggesting a role in ES cells and early development (Lutzker and Levine, 1996; Schmid et al., 1991). This role is perhaps unrelated to the prominent role of p53 in DNA damage responses in the adult. p53 levels in somatic cells are kept low by complex regulatory mechanisms, rising sharply after DNA damage and other stresses to trigger cell cycle arrest, senescence, or apoptosis (Zilfou and Lowe, 2009). The gene, TP53 (Trp53 in mouse), is the most frequently inactivated tumor suppressor in human cancer (Lane and Levine, 2010; Vousden and Prives, 2009). Despite an extensive understanding of p53 as a tumor suppressor in the adult, its role in ES cells and early embryo development remains unknown.
An involvement of p53 in early development is indicated by the failure of p53-depleted Xenopus embryos to undergo gastrulation (Wallingford et al., 1997). p53 is required for mesendoderm differentiation of mouse ES cells in monolayer culture, though not in ES cell-derived embryoid bodies (EBs) (Shigeta et al., 2013). This effect requires the release of p53 from inhibition by Aurora-A kinase (Aurka) (Lee et al., 2012). p53 inhibits the generation of induced pluripotent stem (iPS) cells (Hong et al., 2009; Kawamura et al., 2009). p53 reactivation in teratocarcinomas can promote cancer cell differentiation (Zhu et al., 2016). Although p53 regulates expression of LIF in the female for embryo implantation (Hu et al., 2007), the role of p53 in the embryo proper remains controversial because Trp53-null mice develop normally, even if they are cancer-prone as adults (Donehower et al., 1992).
Functional redundancy of Trp53 with its other two family members, Trp63 and Trp73, might explain these discrepancies. The transactivating forms of p63 and p73 (TAp63 and TAp73) show structural similarity with p53, bind to the same consensus DNA sequence, and are interchangeable with p53 in certain assays (Dotsch et al., 2010; Flores et al., 2002). However, embryos with homozygous mutations of Trp63 or Trp73 (Mills et al., 1999; Yang et al., 1999; Yang et al., 2000), or double knockouts (KO) of p53 family members develop well beyond gastrulation and germ layer specification, and have only late developmental defects (Flores et al., 2002). No triple KO has been reported.
The mechanism by which p53 family members might control the expression of lineage identity genes in ES cells is also not clear. Germ layer specification genes are directly regulated by various embryo signals, including Wnt and the TGF-β family member nodal. Nodal drives mesendoderm differentiation during gastrulation (Brennan et al., 2001; Zhou et al., 1993). It binds to activin receptors and activates Smad2 and Smad3 transcription factors that directly regulate mesendoderm differentiation genes (Baker and Harland, 1996; Weinstein et al., 1998). Wnt binds to frizzled (Fzd) receptors to activate β-catenin and Tcf transcription factors (Clevers and Nusse, 2012). Wnt cooperates with nodal during mesendoderm induction (Estaras et al., 2015; Funa et al., 2015; Reid et al., 2012), and Tcf factors interact with Smad2/3 in certain contexts (Labbe et al., 2000). p53 was reported to bind Smad proteins for regulation of TGF-β target genes (Cordenonsi et al., 2003), but these observations have not been widely confirmed. Notably, p53 can directly activate the expression of Wnt pathway components in mouse ES cells in response to DNA damaging agents (Lee et al., 2010).
Here we use double and triple KO combinations of p53 family members in mouse and human ES cells to address the role of p53 during the exit from pluripotency as cells undergo differentiation. We show that the p53 family is essential for the activation of mesendoderm differentiation genes in ES cells in vitro and in the embryo. Though these genes lack p53-binding elements, they contain enhancers that are synergistically co-occupied and regulated by nodal-activated Smad2/3 and Wnt-activated Tcf3. We demonstrate that the p53 family enables activation of these genes by stimulating Wnt production as ES cells exit the pluripotent state, thus governing the cooperation of Wnt and nodal for the onset of mesendoderm differentiation.
RESULTS
The p53 Family Redundantly Drives Mesendodermal Differentiation
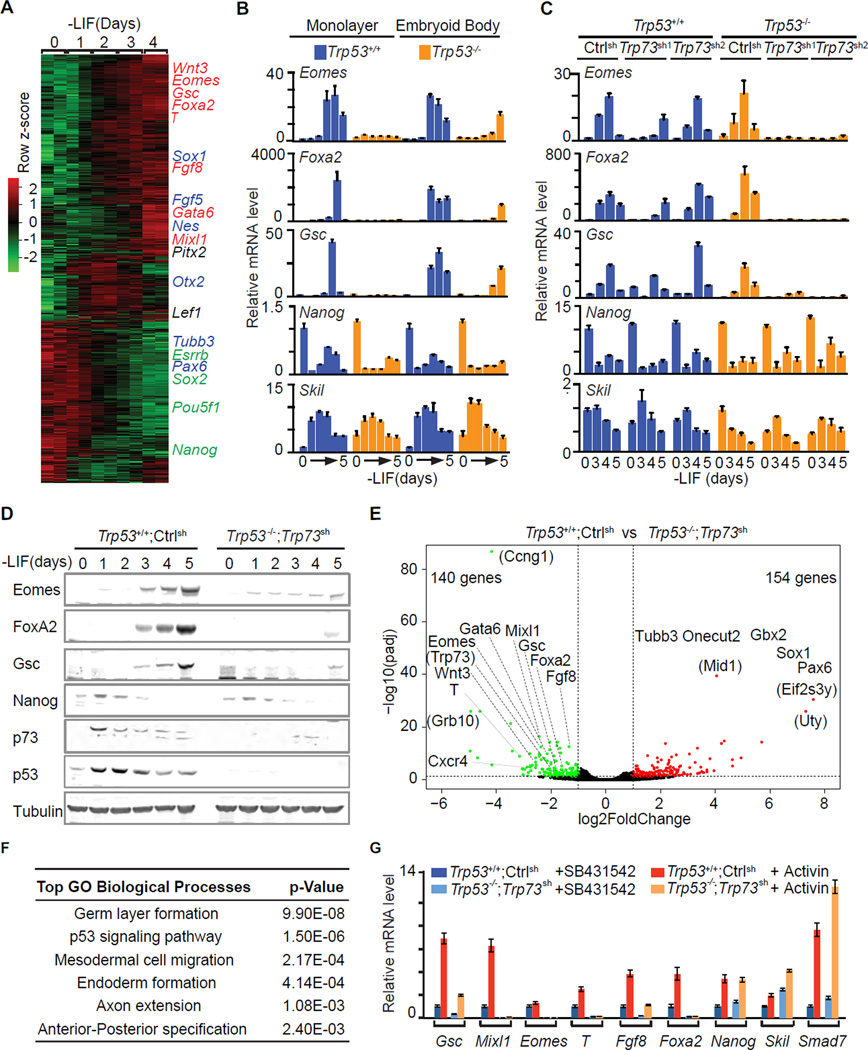
Pluripotent ES cells and early stage EBs formed by these cells under differentiation-permissive conditions [suspension culture in leukemia inhibitory factor (LIF)-free media] recapitulate the signaling and transcriptional events of germ layer specification (Nishikawa et al., 1998; Xi et al., 2011). In LIF-free media, mouse ES cells downregulate the pluripotency genes Nanog first and Sox2 and Pou5f1 (Oct4) thereafter, and gradually induce the expression of mesendoderm marker genes including Eomes, Foxa2, Goosecoid (Gsc), Mixl1, Fgf8, Brachyury (T), and ectoderm marker genes including Sox1, Fgf5, Nestin (Nes) (Figure 1A). Expression of the mesendoderm markers is driven by autocrine nodal through nodal/activin receptor kinases, and can be blocked with the specific kinase inhibitor SB431542 (SB).
Figure 1. The p53 family redundantly drives mesendodermal differentiation.
A. Heatmap presentation of RNA-Seq transcriptomic analysis of mESCs at the indicated times after placement in LIF-free media. Row z scores of genes that were differentially expressed after d0 are included. Representative pluripotency-associated genes (green), early mesendoderm lineage marker genes (red), and early ectoderm marker genes (blue) are highlighted. Two biological replicates at each time point were analyzed.
B. qRT-PCR analysis of mRNA levels of the indicated genes in Trp53+/+ and Trp53−/− cells under EB suspension or ES monolayer differentiation conditions. n=3 for each condition. Error bars represent s.e.m and are used for all the following figures.
C. Levels of the indicated mRNAs during EB differentiation of Trp53+/+ or Trp53−/− cells that were transduced with shRNA vectors targeting control (Luciferase) or two independent shRNA vectors targeting Trp73. Experiment was performed in triplicate and a representative result is presented..
D. Western immunobloting analysis of indicated proteins in control (Trp53+/+;Ctrlsh) and p53/p73-depleted (Trp53−/−;Trp73sh) cells over 5 days under EB differentiation conditions. Tubulin was used as a loading control.
E. Volcano plot of RNA-Seq transcriptome data sets of day-4 EBs derived from control (Trp53+/+;Ctrlsh) and p53/p73-depleted (Trp53−/−;Trp73sh) cells. Green: genes down-regulated in Trp53−/−;Trp73sh (fold change <0.5, p <0.05); red: genes up-regulated in Trp53−/−;Trp73sh (fold change >2, p <0.05). Lineage specification genes for mesendoderm or ectoderm are indicated. Genes in parentheses are not known to have germ layer specification functions. Two biological replicates for each condition were analyzed.
F. Gene Ontology analysis of the genes that were down-regulated in Trp53−/−;Trp73sh cells comparing to Trp53+/+;Ctrlsh cells. The most significantly enriched Biological Processes GO terms with p values are listed.
See also Figure S1.
G. mRNA levels of mesendoderm marker genes (Gsc, Mixl1, T, Eomes, Fgf8 and Foxa2), pluripotency-associated gene (Nanog) and Nodal feedback genes (Skil and Smad7) in d3 EBs that were treated with nodal/activin receptor inhibitor SB431542 (SB) or activin A (AC) for 2 h.
Trp53−/− ES cells failed to express the mesendoderm specification genes Eomes, Foxa2 and Gsc in monolayer culture, but still expressed these genes in EB conditions (Figure 1B) (Shigeta et al., 2013). We investigated if this discrepancy was linked to differences in the expression of functionally redundant p53 family members. Indeed, p53 was expressed both in monolayer and EB conditions, whereas the active form of p73, TAp73, was expressed in EBs but not ES monolayers (Figure S1A). p63 was below the detection limit in both conditions (Figure S1A–B). shRNA-mediated knockdown of Trp73 in Trp53−/− cells (Trp53−/−;Trp73sh cells) eliminated the expression of Eomes, Foxa2 and Gsc in EBs (Figure 1C–D), whereas expression of Nanog and the Smad negative feedback regulator Skil were not perturbed (Figure 1B–D).
RNA-Seq analysis of day 4 (d4) EBs showed reduced expression of 140 genes in Trp53−/−;Trp73sh EBs compared to Trp53+/+;controlsh EBs, whereas 154 genes were upregulated (Figure 1E). Gene Ontology analysis revealed significant down-regulated of gastrulation, mesoderm and endoderm formation genes, including the nodal-regulated mesendoderm genes Eomes, Foxa2, Gsc, Mixl1, Gata6, Cxcr4 and Fgf8 (Figure 1F). Addition of exogenous activin induced the expression of these genes in d3 Trp53+/+;controlsh EBs but not in Trp53−/−;Trp73sh EBs (Figure 1G). Nanog as well as the Smad negative feedback regulators Skil and Smad7 remained responsive to activin in Trp53−/−;Trp73sh EBs (Figure 1G). Notably, the p53/p73-depleted cells adopted a neuroectoderm fate characterized by expression of Pax6, Tubb3, Gbx2, Sox1 and Onecut2 (Figure 1E). This switch is consistent with adoption of an alternative fate upon nodal inhibition (Vallier et al., 2004). These results suggested that p53 and p73 redundantly enable nodal-dependent mesendoderm specification of ES cells.
We investigated this effect in ES cells that were depleted of p53, p63 and p73 by triple KO using CRISPR/Cas9 (Cong et al., 2013; Mali et al., 2013). sgRNAs targeting Trp53, Trp63 and Trp73 genomic loci were transiently transduced with Cas9 into ES cells (Figure S1C). One Trp53+/+;Trp63−/−;Trp73−/− (double knockout, DKO) clone and two Trp53−/−;Trp63−/−;Trp73−/− (triple knockout, TKO) clones were selected and verified by sequencing and western blot analysis (Figure S1D–G). T, Foxa2, Eomes and Gsc expression was detected in p63/p73 DKO EBs but not in TKO EBs, whereas the expression of Nanog and Skil remained unaffected (Figure S1H).
p53/p63/p73 Triple Knockout Leads to Early Defects in Mouse Embryo Development
To extend these observations in vivo, we determined the expression levels of p53 family members by qRT-PCR in mouse embryos ranging from embryonic day E6.5 until E11.5, corresponding to stages of gastrulation and extensive organogenesis. p53 was expressed at E6.5 and thereafter until E10.5 (Figure S2A). Transactivating isoforms of p63 (TAp63α-Υ) and dominant negative isoforms of p63 (ΔNp63α-Υ) were below the qRT-PCR detection limit in E6.5–E7.5 embryos but were highly expressed from E8.5 (Figure S2A). Transactivating isoforms of p73 (TAp73α) and dominant negative isoforms of p73 (ΔNp73α and ζ) were moderately expressed at E6.5, followed by a decrease between E7.5–E8.5 and re-elevation after E9.5 (Figure S2A). Thus, mouse embryos dynamically express all three members of p53 family during gastrulation, raising the possibility of redundant functionality.
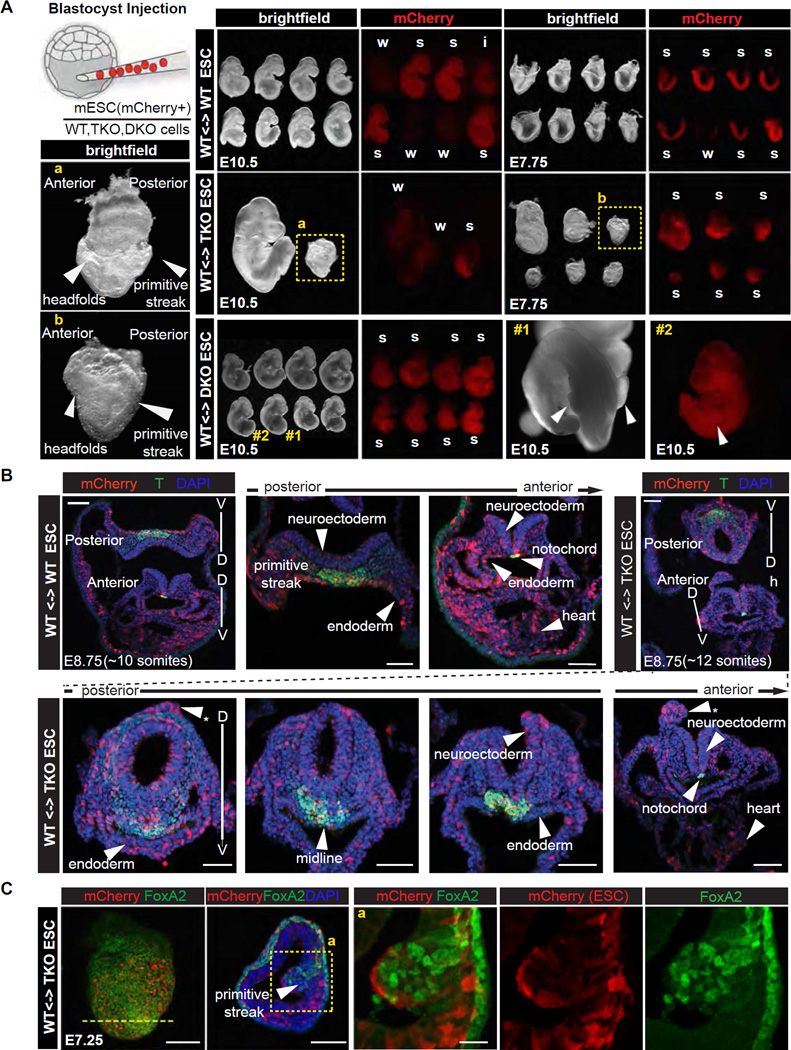
We microinjected wild type (WT), DKO or TKO ES cells labeled with mCherry into WT blastocysts to generate chimeric embryos. Chimeras were dissected at E10.5, corresponding to midgestation, and analyzed for the contribution of mCherry+ cells to major organs (Figure 2A). Regardless of the level of chimerism, which was classified as strong, intermediate or weak according to mCherry fluorescence, all 16 WT control chimeras and another 8 DKO chimeras proceeded to midgestation and exhibited normal morphology (Figure 2A) (Flores et al., 2002). By contrast, 4 chimeras with strong or intermediate TKO cell contribution exhibited profound retardation and resembled E8.0 embryos, whereas weak TKO cell contribution permitted development to slightly later stages (Figure 2A).
Figure 2. Trp53/Trp63/Trp73 triple knockout leads to early defects in mouse embryo development.
A. Scheme of blastocyst injection, brightfield and red fluorescence (mCherry) images of embryo chimeras, comprising control (WT), Trp53/63/73 TKO, and Trp63/73 DKO ES cells, dissected at E10.5 and E7.75. Level of chimerism of each embryo is classified as strong (s), intermediate (i) or weak (s) based on mCherry fluorescence. Panels a and b depict high magnification bright field images of strong TKO chimeric embryos recovered at each stage, but arresting around the same time, highlighting developmental delay and defects in the primitive streak and headfold regions.
B. 3D reconstructions of z-stacks of confocal images of transverse sections of control and TKO ES cell containing embryo chimeras recovered at E8.75 (~10 somite stage wild-type) depicting mCherry (red, ES cell descendants), T expression (green) and nuclear (DAPI, blue) localization. By contrast to control chimeras which exhibited normal morphology, and having uniform and extensive distribution of mCherry cells, TKO ES cell chimeras exhibited morphological defects including an abnormally round neural tube, dorsal neuroectodermal protrusions (asterisks), non-uniform ventral midline, and flattened gut endoderm exhibiting an absence of mCherry cells. Scale bars: 50 µm
C. Confocal microscope images taken of serial sectioned embryo chimeras comprising control and TKO ES cells dissected at E7.25 depicting mCherry (red), FoxA2 expression (green) and nuclear (DAPI, blue) localization. TKO ES cell chimeras exhibit an accumulation of cells at the primitive streak and protrusion into the amniotic cavity (bottom row), which is consistent with a defect at gastrulation. FoxA2 is expressed in WT but not mCherry-positive mutant cells within the expanded primitive streak protrusion. Note that the unaffected FoxA2-positive cell population on the embryo’s surface is visceral endoderm, which is not pluripotent epiblast-derived. Scale bars: whole mount view: 100 µm, transverse section: 50 µm; high magnification view: 20 µm.
See also Figure S2.
Based on the morphology of embryos recovered at E10.5 we estimated that the onset of a defect occurred at around E7.5–E8.5, corresponding to the time of gastrulation when mesoderm and definitive endoderm germ layers are formed. In support of this possibility, at the late headfold to early somite stage (~E7.75), 6 out of 6 strong TKO chimeras exhibited morphological defects symptomatic of defective gastrulation. At E8.5 (6–10 somite stage), 11 out of 18 TKO chimeras exhibited defects in the primitive streak region at the posterior end of the embryo, a kinked neural tube, a paucity of adjacent mesoderm, cardiac defects, as well as defects in the headfold region (Figure S2B).
We performed serial sections to determine whether TKO cells fail to contribute to specific regions of the embryo. Notably, E8.75 TKO chimeras exhibited an abnormal primitive streak and gut endoderm morphology, and a round neural tube rather than a flat neural plate observed in WT chimeras (Figure 2C). Moreover, TKO cells exhibited elevated contribution to ectopic neural protrusions only observed in TKO chimeras, but reduced contribution to gut endoderm, confirming that p53/p63/p73-depletion inhibits endoderm differentiation and favors a neuroectodermal fate (Figure 2B). While, E7.25 TKO chimeras exhibited an accumulation of cells at the primitive streak, which bulged into the amniotic cavity, indicative of a failure in gastrulation (Figure 2B) (Migeotte et al., 2011; Viotti et al., 2014). These results suggested that strong TKO chimeras failed to execute normal gastrulation.
To verify the lineage-specific defects, we performed immunofluorescence analysis of FoxA2, a nascent mesendoderm marker, and T, a primitive streak, mesoderm and midline marker (Wilkinson et al., 1990). FoxA2 expression was interrupted in the midline of E7.75 TKO chimeras (Figure S2C), reminiscent of defects observed in mutants of Lhx1, a downstream target of nodal-Smad signaling (Costello et al., 2015). In E7.25 chimeras mCherry+ TKO cells failed to induce FoxA2 expression whereas surrounding WT cells expressed high levels of FoxA2 (Figure 2C), reinforcing the conclusion that TKO cells were unable to trigger mesendoderm specification. Though T expression was not abolished in TKO cells of E8.75 chimeras, T positive cells were aberrantly organized at the primitive streak and midline structures (Figure 2B). Taken together, our analyses of chimeric embryos strongly suggest that the p53 family plays a critical role in mesendoderm specification.
The p53 Family Selectively Enables Smad Binding to Mesendoderm Genes
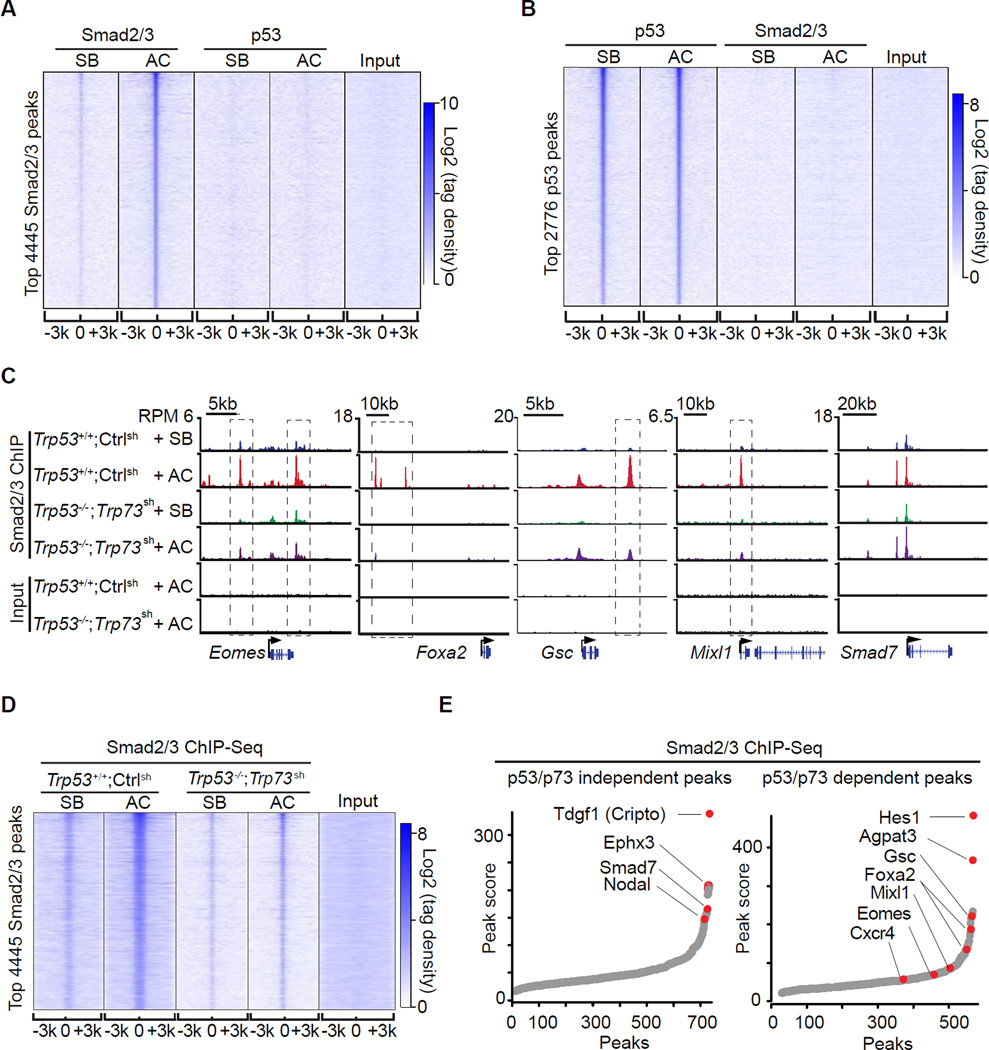
To investigate the mechanistic basis for the role of p53 in the activation of mesendoderm specification genes, we considered the possibility that p53 interacts directly with Smad2/3 on target promoters (Cordenonsi et al., 2003). However, we observed no interaction of p53 family members with Smads 2, 3 or 4 in ES cells or EBs, by immunoblotting or mass spectrometry analysis of Smad-associated proteins (data not shown). We performed p53 and Smad2/3 ChIP-Seq analysis in EBs that were treated with activin or SB431542. Genome-wide, the p53 binding sites were enriched for the p53 consensus binding sequence (Figure S3A), and included the typical p53 target loci Mdm2 and Cdkn1a (Figure S3B). However, the binding patterns of p53 and Smad2/3 did not overlap (Figure 3A–B). There was no significant p53 binding within 100kb centered on the Eomes, Foxa2, Gsc or Mixl1 transcription start sites (TSS) (Figure S3B), regions that contained multiple Smad2/3 binding sites (Figure 3C). Moreover, Smad2/3 binding to chromatin was stimulated by activin whereas p53 binding was not (Figure 3A–B). We concluded that p53 acts as a determinant of nodal action in ES cells without physically contacting Smad target loci.
Figure 3. The p53 family selectively enables Smad binding to mesendoderm genes.
A. Heatmap of ChIP-Seq tag densities for Smad2/3 and p53 within −3kb/+3kb genomic regions surrounding the centers of 4445 high-confidence Smad2/3 binding sites in WT d3 EBs treated with SB or AC. Color key represents log2 tag density distribution.
B. Heatmap of ChIP-Seq tag densities for Smad2/3 and p53 within −3kb/+3kb genomic regions surrounding the centers of 2776 high-confidence p53 binding sites in WT d3 EBs treated with SB or AC. Color key represents log2 tag density distribution.
C. Gene track view for Smad2/3 ChIP-Seq data in the indicated loci, cell lines, and conditions. Gene bodies are schematically represented at the bottom of each track set. Dashed boxes indicate p53/p73-dependent Smad2/3 binding sites.
D. Heatmap of Smad2/3 tag densities within −3kb/+3kb genomic regions surrounding centers of 4445 high-confidence Smad2/3 binding sites, in Trp53+/+;Ctrlsh or Trp53−/−;Trp73sh d3 EBs that were treated for 2 h with SB or AC. Color key represents log2 tag density distribution.
E. Peak score plot for the most significant p53/p73-independent (left) and p53/p73-dependent (right) Smad2/3 binding sites (fold change >2.0 and p <0.01). Genes adjacent to the highest intensity peaks are indicated.
See also Figure S3.
Next we investigated the effect of p53 on the binding of Smad2/3 to target enhancers. To avoid potential clonal variability introduced by CRISPR/Cas9 selection, we utilized Trp53−/−;Trp73sh and Trp53+/+;Ctrlsh EBs. Activin stimulated the binding of Smad2/3 to sites in Eomes, Foxa2, Gsc, Mixl1 and Smad7 in control d3 EBs (Figure 3C). p53/p73-depletion inhibited Smad2/3 binding to a subset of these sites in the differentiation genes but not to sites in Smad7 (Figure 3C). Based on global analysis, Smad2/3 binding peaks can be classified as p53/p73-dependent and p53/p73-independent peaks (Figure 3E). Loci with p53/p73-independent peaks included the nodal feedback regulators Smad7, Tdgf1 (Cripto) and Nodal (Figure 3E), and loci with p53/p73-dependent peaks were enriched for mesendoderm specification genes (Figure 3E and S3C).
The p53 Family Controls Tcf-sensitive nodal Target Genes
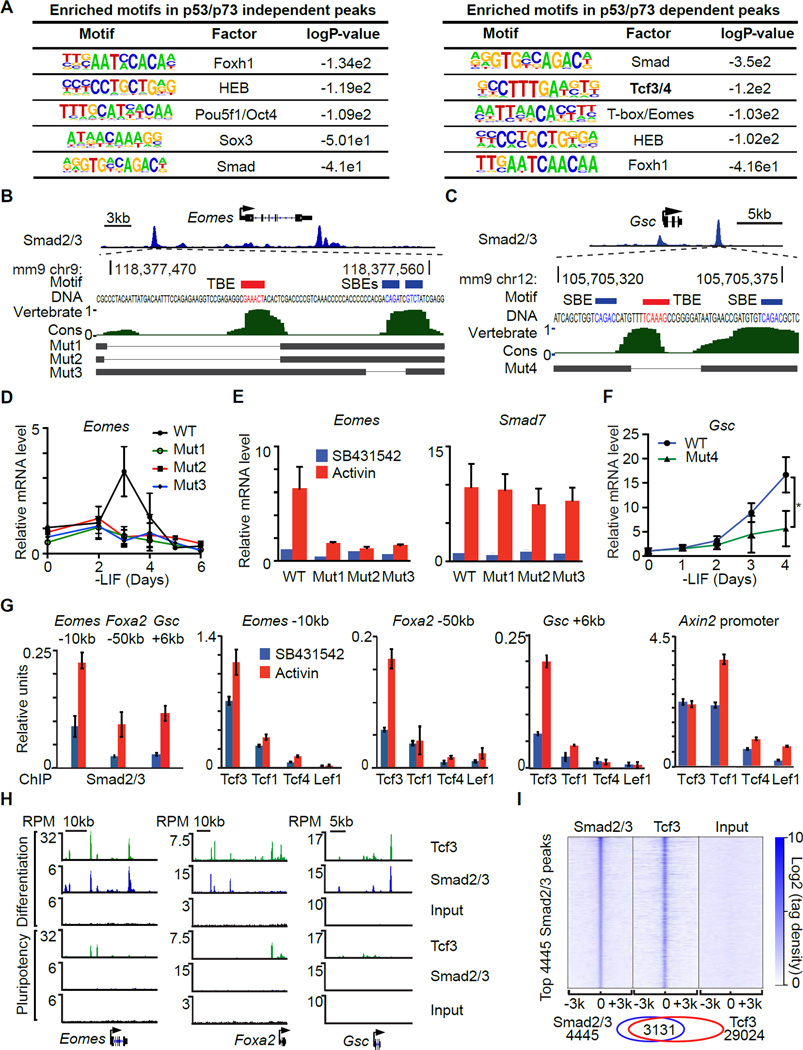
Transcription factor motif analysis revealed that both the p53/p73-dependent and independent Smad binding regions are rich in binding elements for Smad (SBEs) and the Smad partners FoxH1 (Chen et al., 1997; Liu et al., 1997) and HEB (Yoon et al., 2011). Interestingly, a Tcf3/4 binding motif (TBE) was significantly enriched only in p53-dependent Smad2/3 binding sites and not in p53-independent sites (Figure 4A). SBE (CAGAC/T) and TBE (TCAAAG) motifs are located approximately within ~50 bp of each other in the regulatory regions of Eomes, Foxa2, Gsc, Mixl1, Fgf8, and T (Figures 4B–C and S4A–E).
Figure 4. Tcf3 cooperates with Smad2/3 to activate mesendoderm gene enhancers.
A. Enriched known transcription factor binding motifs in p53/p73-independent Smad2/3 binding sites (left) and p53/p73-dependent Smad2/3 binding sites (right).
B. Scheme of CRISPR/Cas9-mediated mutagenesis of the Eomes −10kb distal enhancer region. Gene track view for Smad2/3 ChIP-Seq tag density at Eomes locus at the top panel highlights the targeted region. The DNA sequence of targeted region and the positions of Smad binding elements (CAGAC/T) (SBE, blue) and Tcf binding element (TCAAAG) (TBE, red) are indicated. Middle panel, evolutionary conservation scores in vertebrates. Bottom, black horizontal bars represent the remaining genomic regions in mutant (Mut1–3) clones whereas the thin lines indicate deleted regions.
C. Scheme of the CRISPR/Cas9-mediated mutagenesis of the Gsc +6kb distal enhancer region. Each part is labeled as in B.
D. qRT-PCR analysis of Eomes mRNA expression in d0 to d6 EBs derived from WT or Eomes −10kb enhancer mutant clones. Gene expression level is normalized to d0 WT samples. n=3.
E. qRT-PCR analysis of Eomes and Smad7 expression in AC or SB treated d3 EBs derived from WT or Eomes −10kb enhancer mutant cells. n=3.
F. qRT-PCR analysis of Gsc mRNA expression in WT or Gsc +6kb distal enhancer mutant clone. Gene expression level is normalized to d0 WT samples. n=3. * P<0.05, Mann-Whitney test.
G. Analysis of Smad2/3, Lef1, Tcf1 (Tcf7), Tcf3, and Tcf4 binding (ChIP-qPCR) to the Eomes −10kb, Foxa2 −50kb, Gsc +6kb enhancers, and Axin2 proximal promoter in d3 EBs treated with SB or AC for 2 h.
H. Gene track view of Eomes, Foxa2 and Gsc locus. Tcf3 and Smad2/3 ChIP-Seq were performed in AC-treated d3 EBs (Differentiation), and in d0 non-treated mouse ES cells (Pluripotency). Pre-cleared chromatin prior to primary antibody addition (Input) was used as negative control. Tag densities were normalized to reads per million reads (RPM). The gene structures from RefSeq are schematically represented at the bottom.
I. Heatmap of tag densities of ChIP-Seq data sets from the indicated conditions, within −3kb/+3kb genomic regions surrounding centers of 4445 high-confidence Smad2/3 binding sites. ChIP-Seq data for the indicated proteins and input control are from AC treated d3 EBs. Color key represents log2 tag density distribution.
See also Figure S4.
We used CRISPR/Cas9 to generate focal deletions of neighboring SBEs and TBEs in the Eomes −10kb enhancer and the Gsc +6kb enhancer in ES cells (Figure 4B–C). These deletions caused a significant reduction in Eomes or Gsc induction by endogenous signals or exogenous activin (Figure 4D–F). Smad7 induction by activin remained intact, arguing that the mutant clones were capable of nodal/activin signal transduction. Collectively, these findings suggest that p53 regulates a component that is selectively required for activation of Wnt-responsive nodal target genes.
The Tcf family in mammals includes four members: Lef1, Tcf1 (Tcf7), Tcf3 (Tcf7l1) and Tcf4 (Tcf7l2). ES cells predominantly express Tcf3 and Tcf1 (Figure S4F). Tcf3 is essential for embryo development (Merrill et al., 2004) and ES cell exit from pluripotency (Wray et al., 2011). ChIP analysis showed that Tcf3 is the most highly enriched Tcf family member in the Eomes −10kb enhancer, Foxa2 −50kb enhancer and Gsc +6kb enhancer. Tcf3 binding was further stimulated by activin (Figure 4G). Tcf1 and Tcf3 were highly enriched in the Axin2 promoter, a known Wnt/Tcf target gene (Yan et al., 2001). Moreover, Tcf3 ChIP-Seq analysis showed a high concordance with Smad2/3 binding sites at mesendoderm gene enhancers (Figure 4H) and across the genome (Figure 4I). The binding of Tcf3 and Smad2/3 to these enhancers increased significantly in differentiating EBs compared to pluripotent ES cells (Figure 4H). Collectively these results suggest that Smad2/3 and Tcf3 converge on cis-regulatory elements of p53-dependent mesendoderm specification genes.
Wnt Expression is Rate Limiting for the Onset of Mesendoderm Differentiation
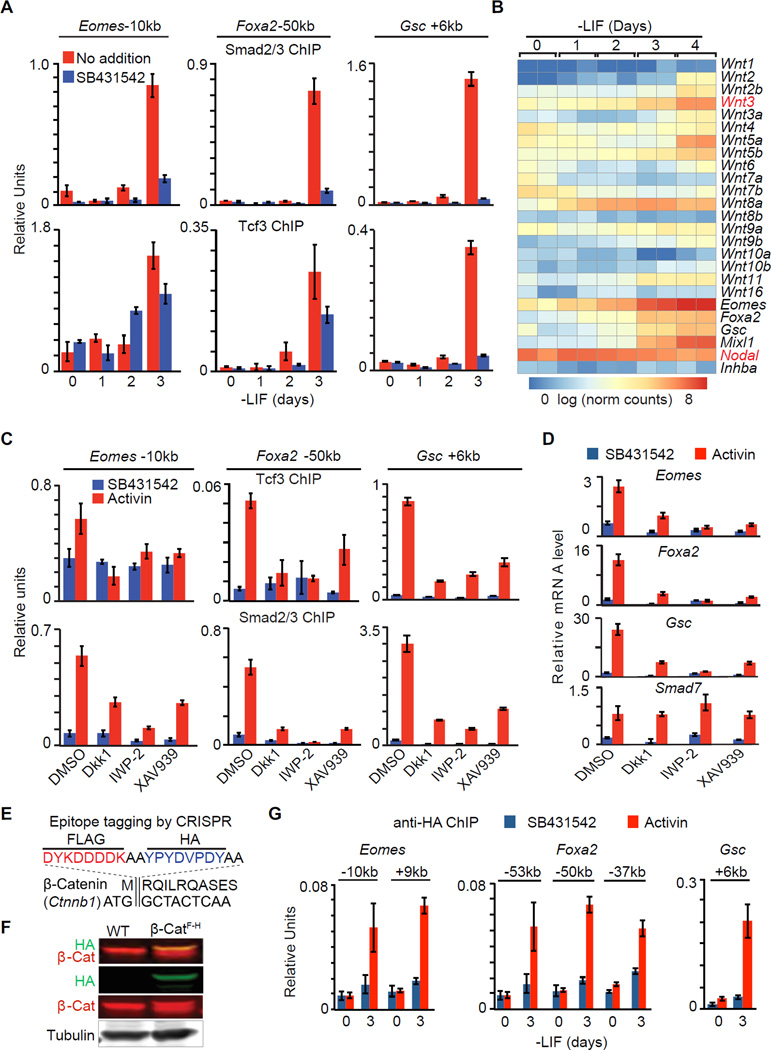
Smad2/3 and Tcf3 binding to the Eomes, Foxa2 or Gsc enhancers in EBs did not occur until d3 (Figure 5A), suggesting the existence of a rate-limiting factor. Nodal was expressed throughout this period (Figure 5B and S5A), and d0 cells, as well as d3, cells responded to activin with formation of Smad2/3-Smad4 and Smad2/3-Trim33 transcriptional complexes (Figure S5B) (Xi et al., 2011). However, Wnt8a was the only Wnt family member expressed before d3 (Figure 5B). Wnt3, an essential factor for mouse primitive streak formation and gastrulation (Liu et al., 1999), was expressed starting on d3, which coincided with the onset of Foxa2, Gsc and Mixl1 induction and peak Eomes expression (Figure 5B, refer to Figure 6G for Wnt3 protein level). Comparable Wnt3 expression kinetics occur during mouse gastrulation (Rivera-Perez and Magnuson, 2005).
Figure 5. Wnt expression is rate limiting for the onset of mesendoderm differentiation.
A. ChIP-qPCR analysis of Smad2/3 and Tcf3 binding to the Eomes −10kb, Foxa2 −50kb and Gsc +6kb enhancers of EBs at the indicated times in LIF-free culture media. SB was added as indicated to inhibit signaling by endogenous nodal.
B. Heatmap of Wnt family mRNA expression levels (RNA-Seq) during d0 to d4 EB differentiation. Scale represents the log2 normalized read counts. Eomes, Foxa2, Gsc, Mixl1, Nodal and Inhba (encoding Activin A) expression levels are also presented for reference. Two biological replicates were analyzed for each condition.
C. ChIP-qPCR analysis of Tcf3 and Smad2/3 binding to the Eomes −10kb, Foxa2 −50kb and Gsc +6kb and enhancers in d3 EBs treated with SB or AC, and with DMSO or Wnt signaling inhibitors, Dkk1, IWP-2 or XAV939 as indicated.
D. Analysis of indicated mRNA levels (qRT-PCR) in d3 EBs that were incubated with SB or AC for 2 h. Wnt signaling inhibitors Dkk1, IWP-2 or XAV939, or DMSO vehicle were added as indicated.
E. Scheme of CRISPR mediated FLAG-HA epitope tagging for Ctnnb1/β-catenin N-terminus.
F. Western immunoblot analysis for β-cateninFLAG-HA (β-CatF-H) cell line that was inserted with FLAG-HA tags at Ctnnb1 N-terminus.
G. Anti-HA ChIP-qPCR analysis at Eomes −10kb and +9kb, Foxa2 −53kb, −50kb and −37kb, and Gsc +6kb enhancers.
See also Figure S5.
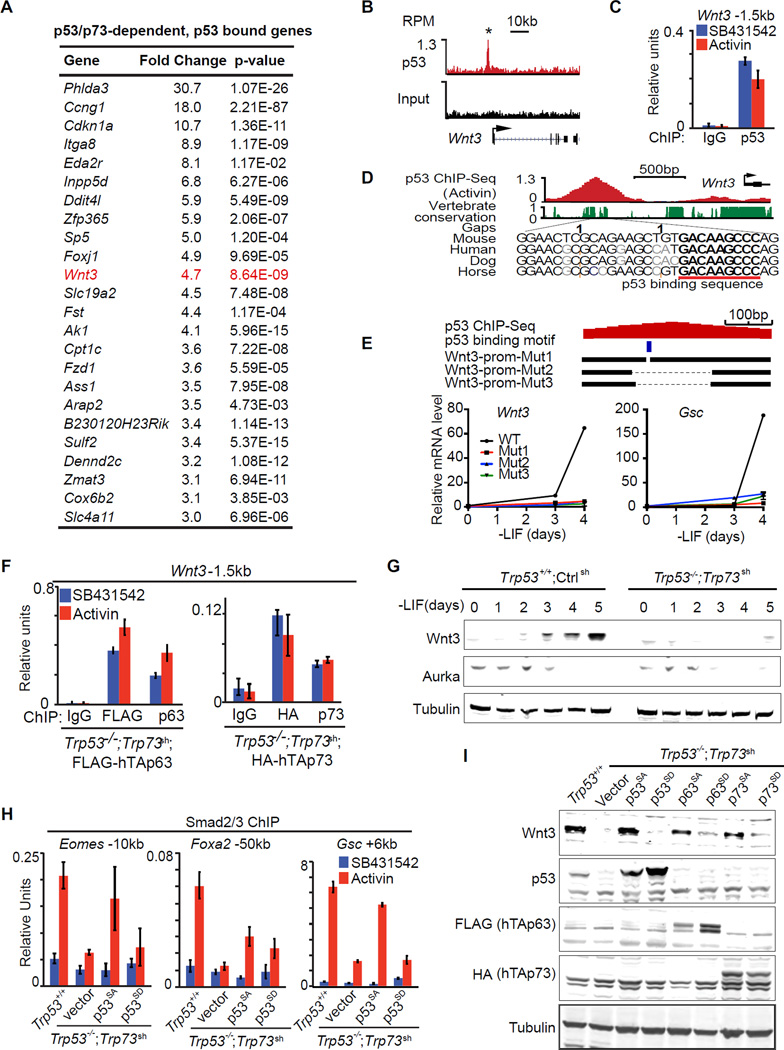
Figure 6. The p53 family controls Wnt3 expression.
A. Table representing the top 24 genes that were differentially expressed (fold change >3, p<0.05, mean normalized read counts > 100) in response to p53/p73-depletion by RNA-Seq, and exhibited significant p53-binding with 50kb from TSS. Genes are ranked by their fold change in RNA-Seq analysis.
B. Gene track view for p53 ChIP-Seq data or input control at the Wnt3 locus. RefSeq gene bodies are schematically represented at the bottom of each track set.
C. Analysis of p53 binding (ChIP-qPCR) to the Wnt3 −1.5kb element in d3 EBs treated with SB or AC for 2 h. ChIP with non-specific IgG was used as negative control.
D. Evolutionary conservation score plot for the p53-binding region near the Wnt3 promoter, and DNA sequence alignment of the peak center in multiple species highlighting the p53 consensus binding sequence.
E. Scheme of CRISPR/Cas9-mediated mutagenesis of the Wnt3 promoter region. Bottom black horizontal bars represent the remaining genomic regions in mutant (Mut1–3) clones whereas the thin lines indicate deleted regions. The p53-binding motif is indicated as a blue box. qRT-PCR analysis of Wnt3 and Gsc upon LIF removal are shown below.
F. ChIP-qPCR analysis of exogenous FLAG-hTAp63 and HA-hTAp73 binding at Wnt3 −1.5kb enhancer region in Trp53−/−;Trp73sh day 4 EB treated with SB or AC for 2 h. Antibodies that recognizes FLAG, HA tags, or specifically human TAp63 and TAp73 were used for ChIP. Non-specific IgG were used as negative control.
G. Western immunoblot analysis of Wnt3 expression in EBs derived from Trp53+/+;Ctrlsh and Trp53−/−;Trp73sh ES cells at indicated time points. Tubulin was used as loading control.
H. Analysis of Smad2/3 binding (ChIP-qPCR) to the Gsc +6kb enhancer in d3 EBs from Trp53+/+;Ctrlsh or Trp53−/−;Trp73sh ES cells transduced with p53SA or p53SD, followed by 2h SB or AC treatment.
I. Western immunoblot analysis of Wnt3 and p53 family protein levels in EBs from control ES cells (Trp53+/+;Ctrlsh), or p53/p73-depleted cells (Trp53−/−;Trp73sh), or p53/p73-depleted cells expressing empty vector or Aurka-resistant (p53SA, p63SA, p73SA) or Aurka phosphorylation mimic (p53SD, p63SD, p73SD) mutant forms of p53 family members. Tubulin was used as loading control.
See also Figure S6.
We used different Wnt pathway inhibitors to assess the role of endogenous Wnt signaling in mesendoderm gene expression. Addition of the Wnt receptor inhibitor Dkk1 (Niida et al., 2004), the Wnt palmitoylation inhibitor IWP-2 (Anastas and Moon, 2013), or the Axin2 stabilizing drug XAV939 (Anastas and Moon, 2013) prevented the binding of Tcf3 to Eomes, Foxa2 and Gsc enhancers (Figure 5C), and the activation of these genes (Figure 5D). Notably, Wnt inhibitors not only diminished the binding of Tcf3 to these enhancers but also that of Smad2/3 (Figure 5C). Conversely, nodal/activin receptor inhibition diminished the binding of Tcf3 as well as Smad2/3 to these sites (Figure 5A). In contrast, Smad7 responded to activin regardless of Wnt inhibition.
To determine whether Tcf3 chromatin binding requires canonical β-catenin signaling in this context, we performed β-catenin ChIP assays. Lacking suitable anti-β-catenin antibodies, we used CRISPR/Cas9 to engineer tandem FLAG and HA epitope tags into the N-terminus of Ctnnb1 (β-catenin) in ES cells (β-CatF-H cells), and validated this modification by sequencing and western blot (Figure 5E–F). Epitope tagging did not significantly alter the differentiation kinetics in β-CatF-H cells (Figure S5C). Anti-HA ChIP confirmed β-catenin binding to Eomes, Foxa2 and Gsc enhancers, and the binding pattern was highly concordant with Tcf3 and Smad2/3 binding (Figure 5G). Collectively, these results suggested that the nodal and Wnt pathways coordinately drive Smad2/3 and Tcf3 to common target enhancers in mesendoderm specification genes, and the expression of autocrine Wnt, as well as the activity of the p53 family, are rate-limiting for this process.
The p53 Family Controls Wnt3 Expression
To search for a link between p53 and Wnt in an unbiased manner, we sorted the 114 genes that are differentially expressed in Trp53−/−;Trp73sh EBs relative to Trp53+/+;controlsh EBs (fold change >3; mean counts >100 in Trp53+/+;controlsh EBs; refer to Figure 1E). Within this group, 24 genes (Figure 6A) showed bound p53 within 50kb from the TSS (refer to Figure 3B). Notably, these 24 genes included Wnt3 and its receptor Fzd1. p53 bound near the TSS in Wnt3 (Figure 6B–C) and at +45kb in Fzd1 (Figure S6A). Both sites contain a p53 consensus binding sequence (Figure 6D and S6A). We generated focal deletions of the p53 binding site in the Wnt3 promoter in ES cells by CRISPR/Cas9 (Figure 6E). These deletions significantly inhibited the expression of Wnt3 itself and the mesendoderm marker Gsc under EB differentiation conditions (Figure 6E), confirming a direct regulation of Wnt3 by p53 binding. ChIP-qPCR analysis of ectopically expressed FLAG-tagged human TAp63 and HA-tagged human TAp73 in Trp53−/−;Trp73sh cells showed that both p63 and p73 can bind to this site (Figure 6F). Importantly, Wnt3 expression after LIF removal was inhibited by p53 depletion in monolayer culture (Figure S6B), and by depletion of p53 and p73 in EBs (Figure S6C and 6G). These results indicate that the p53 family directly controls Wnt3 expression.
p53 activity at this crucial juncture of signal integration is regulated by Aurka (Lee et al., 2012), so we investigated whether p63/73 were regulated in a similar manner. Expression of Aurka in ES cells depends on LIF (Lee et al., 2012). In agreement, Aurka levels in EBs dropped below detection by d3 after LIF removal (Figure 6G). The reported Aurka R(H/Q)S phosphorylation motif in p53 (Ferrari et al., 2005; Lee et al., 2012) is also conserved in p63 and p73 (Figure S6D). We transduced Trp53−/−;Trp73sh cells with vectors encoding p53 proteins with alanine mutations that would prevent phosphorylation of this serine residue (Ser to Ala mutants p53S212A, TAp63S285A and TAp73S235A) or mimic it (Ser to Asp mutants p53S212D, TAp63S285D and TAp73S235D) (Lee et al., 2012). We tested the ability of these mutants to rescue Smad2/3 binding to the Gsc, Foxa2 and Eomes enhancers in Trp53−/−;Trp73sh cells. p53SA but not p53SD rescued the binding (Figure 6H). Moreover, the Ala mutant p53, p63 and p73 but not the Asp mutants rescued the expression of Wnt3 and Gsc (Figure 6I and S6E). These results suggest that p53, p63 and p73 share the ability to trigger Wnt3 expression upon release from inhibition as ES cells exit pluripotency.
Wnt3 Mediates p53 Family Action in Mesendoderm Specification
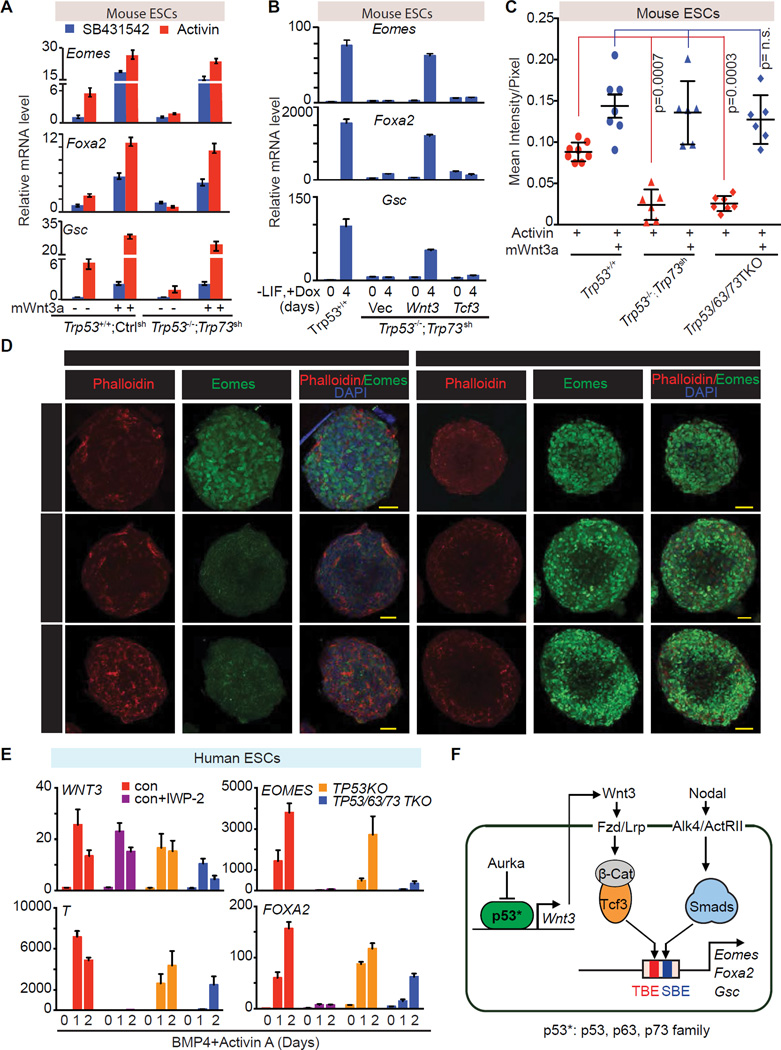
To confirm the specificity of p53 regulation of Wnt signaling, we determined whether enforced Wnt signaling was capable of rescuing mesendoderm marker gene expression in p53 family-deficient ES cells. Addition of recombinant mouse Wnt3a to d3 Trp53−/−;Trp73sh EBs fully rescued the expression of Eomes, Foxa2 and Gsc and the binding of Smad2/3 to the enhancers of these genes in response to activin (Figure 7A and S7A). Wnt3a induction of the feedback regulator Axin2 did not require p53 inputs (Figure S7B). We also engineered WT or Trp53−/−;Trp73sh ES cells with inducible expression vectors encoding mouse Wnt3 or, as a control, Tcf3. Doxycycline-mediated induction of Wnt3, but not that of Tcf3, rescued Eomes, Foxa2, Gsc and Axin2 expression (Figure 7B and S7C).
Figure 7. Wnt3 mediates p53 family action in mesendoderm specification.
A. qRT-PCR analysis of indicated mRNA levels in control ES cells (Trp53+/+;Ctrlsh), or p53/p73-depleted cells (Trp53−/−;Trp73sh), that were treated with or without recombinant mouse Wnt3a (mWnt3a) for 24h, followed by addition of SB or AC for 2 h.
B. qRT-PCR analysis of indicated mRNA levels in p53/p73-depleted cells with doxycycline (Dox)-inducible expression of Wnt3, Tcf3 or empty vector control. Assays were performed at d0 or day 4 of Dox treatment under differentiation conditions. Error bars represent s.e.m.
C. Quantitation of Eomes (green) signal in Panel C. Each dot represents the signal measurement from one EB. P value was calculated using Mann-Whitney test.
D. Immunofluorescence analysis of Eomes (green) in d3 EBs derived from the Trp53+/+;Ctrlsh, Trp53/63/73 TKO, or Trp53−/−;Trp73sh ES cells. Cells were treated with AC for 2h or AC plus mWnt3a for 20h. Scale bars, 50µm. Phalloidin (red) was used to stain F-actin and mark the cell boundaries.
E. qRT-PCR analysis of mRNA levels in control H1 hESC line with or without IWP-2 treatment, and in TP53KO and TP53/63/73 TKO derivative cell lines during BMP4 and AC induced differentiation.
F. Scheme of the p53 family-Wnt-Nodal network driving mesendoderm specification.
See also Figure S7.
Immunofluorescence analysis of EBs showed that Eomes levels and the number of Eomes expressing cells were significantly elevated by activin addition in Trp53+/+;Ctrlsh EBs but not in Trp53−/−;Trp73sh or in TKO EBs. The combined addition of activin and Wnt3a to Trp53−/−;Trp73sh EBs or TKO EBs fully restored Eomes expression and the number of Eomes-expressing cells (Figure 7C–D). The same pattern was observed with FoxA2 expression (Figure S7D), confirming that Wnt3 mediates p53 family action in mouse mesendoderm specification.
WNT signaling also induces primitive streak and mesendoderm differentiation in human ESCs (hESCs) (Jiang et al., 2013; Kurek et al., 2015). The p53 binding sequence in the Wnt3 gene promoter is highly conserved between human and mouse (Figure 6D), raising the possibility that the regulatory mechanism uncovered in mESCs is also operative in developmentally more advanced hESCs. To test this hypothesis we deleted TP53 or all three p53 family members in H1 hESCs by iCRISPR (Gonzalez et al., 2014), as confirmed by DNA sequencing and western blot analysis (Figure S7E–F). TP63 and TP73 were not detectable at the protein level in H1 hESCs, but were deleted to avoid compensatory expression in cells with TP53 deletion (Figure S7G). In the presence of BMP4 and activin, H1 cells rapidly showed expression of WNT3 and the primitive streak marker T at day 1, followed by EOMES and FOXA2 induction (Zhang et al., 2008) (Figure 7E). Expression of the primitive streak genes was abrogated by treatment with WNT Inhibitor IWP-2. Moreover, the TP53/63/73 TKO hESCs, but not the TP53 KO, showed a significant decrease in the expression of WNT3 and primitive streak differentiation genes. Addition of Wnt3a rescued the expression of T and EOMES in TKO hESCs (Figure S7H). These results indicated that the role of p53 family in mesendoderm specification is conserved in mouse and human ESCs.
DISCUSSION
Our results establish the relevance of the p53 family in the differentiation of pluripotent progenitors within the early embryo, as well as pluripotent ESCs in culture, and provide a mechanistic basis for this role. We show that the p53 family governs a regulatory network that integrates Wnt and TGF-β nodal inputs for mesendoderm specification. This network includes two layers of regulation, first, p53 family members that directly control Wnt3 expression, and second, Wnt activated β-catenin/Tcf3 and nodal-activated Smad2/3 that are mutually dependent for binding to, and activation of key mesendoderm identity genes (Figure 7F). The activation of this network is tied to the release of the p53 family from inhibition as ESCs exit from pluripotency (Lee et al., 2012). These findings clarify the role of p53 during early embryogenesis, identify p53 target genes implicated in this process, and highlight the interdependent nature of nodal and Wnt transcriptional mediators in development.
The p53 Family and Mesendoderm Differentiation
Our allelic series of knockouts of p53 family members show that p53, p63 and p73 play a critical role in mesendoderm specification of pluripotent progenitors within the embryo. All three p53 family members can fulfill this role. In line with previous reports (Lin et al., 2005; Lutzker and Levine, 1996; Shigeta et al., 2013), we observed high expression of p53 in early embryo, EBs and ESCs. Expression of p63 and p73 in mouse embryos and EBs varies depending on the culture conditions. Mouse embryos express all p53 family members in a dynamic pattern around the time of gastrulation, providing opportunities for redundant functionality.
The functional overlap of the p53 family members in mesendoderm specification would mask their role in gastrulation in single or double knockout mice. Indeed, Trp53 mutant mice develop normally (Donehower et al., 1992) whereas Trp63 mutant and Trp73 mutant mice have late but not early developmental defects (Mills et al., 1999; Yang et al., 1999; Yang et al., 2000). Evidence for a role of the p53 family in the early embryo rested on transgenic overexpression of ΔNp73, a dominant negative inhibitor of all p53 family members that causes embryonic lethality at gastrulation (Erster et al., 2006; Huttinger-Kirchhof et al., 2006; Yang et al., 2000). We now show that strong TKO chimeras fail to execute normal gastrulation and TKO cells in chimeras do not express mesendodermal differentiation markers.
Compared to naïve mouse ESCs, human ESCs are generally believed to be developmentally more advanced, corresponding to a primed state of pluripotency (Mascetti and Pedersen, 2016). Wnthigh hESCs predominantly form endodermal and cardiac cells, whereas Wntlow hESCs generate primarily neuroectodermal cells (Blauwkamp et al., 2012). We show that TP53/63/73 TKO hESCs are defective in the initiation of primitive streak/mesendoderm differentiation.
p53 Family Links to Wnt
p53 activity in mESCs is kept in check by LIF through Aurka, a constraint that is relieved as Aurka levels drop upon LIF removal (Lee et al., 2012). This regulatory potential seems to be conserved in p63 and p73. The timing of Aurka decline in differentiating EBs coincides with the onset of Wnt3 expression and the induction of mesendoderm differentiation genes. Wnt3 and the Wnt receptor gene Fzd1 are members of a small set of direct p53 family target genes that become activated in differentiating ESCs. p53, 63 and p73 directly bind to a common regulatory element in Wnt3 to drive its expression. Therefore, Wnt3 and Fzd1 are direct p53 transcriptional targets for the control of mesendoderm differentiation.
The ability of p53 to induce the expression of Wnt and Fzd1 in ESCs is not unprecedented. Treatment of pluripotent ESCs with DNA damaging agents can trigger p53-dependent expression of Wnt components (Lee et al., 2010). However, this xenotoxic response was of unknown relevance to the developmental context and, moreover, it was interpreted as a mechanism for p53-dependent inhibition of differentiation. In contrast, our evidence reveals that Wnt expression is low in pluripotent cells, and is significantly elevated by p53/63/73 at the onset of differentiation. Wnt signaling activated under differentiation-permissive conditions cooperates with Nodal signaling and drives mesendoderm specification.
Some of the other p53 target genes associated with ESC differentiation (Ccng1, Cdnk1a, Phlda3; refer to Figure 6A) are also regulated by p53 targets in the context of DNA damage responses in other cell types (el-Deiry et al., 1993; Kawase et al., 2009; Okamoto and Beach, 1994). Overall however, the p53 target gene set in the context of mesendoderm differentiation does not resemble a canonical DNA damage response (Riley et al., 2008). Typical p53-regulated pro-apoptotic genes, for example, are absent. DNA damage caused p53 to repress Nanog in ESCs (Lin et al., 2005), but we observed no change in Nanog expression in p53/73-depleted ESCs. The late developmental phenotypes of Trp63 and Trp73 mutant mice might also involve altered Wnt signaling. For example, the developing limbs in Trp63 mutant embryos lack an apical ectodermal ridge (Mills et al 1999).
Integration of Nodal and Wnt Inputs
The Wnt and TGF-β pathways jointly regulate progenitor identity and differentiation in many contexts (Brennan et al., 2001; Clevers and Nusse, 2012; Massagué, 2012). The present identification of the p53 family as central integrators of Wnt3 and nodal inputs in mesendoderm differentiation provides an expanded view of the synergistic properties of these two pathways, and a previously unrecognized level of interdependency between Tcf and Smad transcription factors.
Nodal target genes in differentiating ESCs fall into two classes depending on the requirement for p53. Genes of the p53-independent class contain Smad2/3 binding enhancers that are not enriched for TCF binding sites or bound by Tcf3. These genes include nodal/Smad pathway feedback regulators, among others. Genes in the p53-dependent class contain Smad2/3 binding enhancers that are co-occupied by Tcf3, and include mesendoderm identity genes. Nodal-activated Smad2/3 and Wnt-activated Tcf3 cooperate in binding to common target enhancers in these genes, driving this crucial developmental transition during gastrulation. Nodal expression and Smad2/3 activation are constitutive in differentiating ESCs, whereas Wnt3 expression for β-catenin/Tcf activation requires the upstream input of the p53 family.
The evidence suggests that the p53 family governs the integration of Wnt and nodal signals for mesendoderm differentiation. Although the present work highlights the importance of this p53-Wnt-Nodal network in mesendoderm differentiation, this process is also regulated by other determinants including histone modifications (Bernstein et al., 2006; Whyte et al., 2012), chromatin topology (Dixon et al., 2015), chromatin remodeling (Alexander et al., 2015) and cell cycle regulators (Pauklin and Vallier, 2013). It will be of interest to discern how these regulatory and epigenetic events converge and interlock to regulate the transition from pluripotency to lineage restricted progenitors during early embryogenesis and possibly also the differentiation of adult stem cells during tissue homeostasis, regeneration and tumorigenesis.
EXPERIMENTAL PROCEDURES
Generation of chimeric embryos
mCherry expressing single ESC colonies were picked and micro-injected 3 days after culture on MEF feeder layers. 10–15 ESCs from each group were injected into E3.5 blastocysts (C57BL/6N Taconic) on a Nikon (Eclipse-Ti) microscope equipped with Narishige micromanipulators. Injected blastocysts were cultured in KSOM /AA (Millipore) at 37°C in an atmosphere of 5% CO2 for 2–3 h until blastocyst cavity expansion and implanted into the uterine horns (10 embryos per horn) of E2.5 pseudopregnant females (CD-1: Charles River) using standard protocols. Chimeric embryos were recovered at E7.25, E7.75, E8.5 and E10.5.
Genome-editing with CRISPR/Cas9
sgRNAs targeting genomic regions of interest were designed using CRISPR Design Tool (http://crispr.mit.edu/) (Hsu et al., 2013) and synthesized by IDT, Inc. Single cells were sorted onto irradiated MEF feeder for increased viability through FACS 72h post-transfection. Mutant clones were first screened through aberrant melting temperature of qPCR products, then verified by PCR, TA-cloning and Sanger sequencing individually. sgRNA target sequences are listed in Supplemental Experimental Procedures. Annealed sgRNA oligos were cloned into pX330-U6-Chimeric_BB-CBh-hSpCas9, pSpCas9(BB)-2A-GFP or pSpCas9n(BB)-2A-GFP Addgene vectors (Ran et al., 2013) and transiently transfected into mouse ES cells with Lipofectamine 3000 (Life Technologies).
H1 hESCs were used for generating TP53/63/73 TKO lines using the iCRISPR method(Gonzalez et al., 2014). Cells were infected with lentivirus-based sgRNAs targeting p53, p63 and p73 (sgRNA sequences are listed in Supplemental Experimental Procedures). Cas9 expression was induced by addition of doxycycline (2 µg/ml) for 5 days. H1 hESCs were grown at single cell density to allow for clonal cell line expansion and isolation. Genotyping and expansion of hESC clones was performed as previously published (Zhu et al., 2014).
Chromatin immunoprecipitation
ChIP was performed as previously described (Xi et al., 2011), mESCs and EBs were collected from cells that were incubated with human recombinant activin A (AC, 50 ng/ml; R&D Systems) for 2 h or SB431542 (SB, 10µM, Tocris) for 2–4 h, as indicated in each experiment. Antibodies and qPCR primers pairs are provided in the Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We thank J. Petrini, S. Lowe, C.J. David, Y-H. Huang and the members of the Massagué lab for insightful discussions, Z. Zhao, R.L. Bowman, L. Studer, for technical advice, J. Huang (National Cancer Institute) for cell lines, L. Dow, E. Flores and I. Lemischka for plasmids. Q.W. was supported by a Postdoctoral Fellowship from NYSTEM. Y.Z. was supported by a Grayer Fellowship. This work was supported by grants R01-CA34610 to J.M., R01-DK084391 to K.H., and P30-CA008748 to MSKCC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental information including 7 figures and detailed experimental procedures is available in the online version of the manuscript.
ACCESSION NUMBERS
All RNA-Seq and ChIP-Seq data were deposited in the Gene Expression Omnibus database under accession number GSE70486.
The authors declare no competing financial interests.
AUTHOR CONTRIBUTIONS
Q.W., Y.Z. and J.M. conceived the project, designed, performed the experimental work and wrote the manuscript. Q.X., J.S. and W.S. assisted with the experiments. Y.Z., Q.W. and C.Z. performed the bioinformatics analysis. S.N., A-K. H. and S.Y.K. performed the embryo analysis. Q.V. L., C-L. S. and D. H. performed human ESC experiments.
REFERENCES
- Alexander JM, Hota SK, He D, Thomas S, Ho L, Pennacchio LA, Bruneau BG. Brg1 modulates enhancer activation in mesoderm lineage commitment. Development. 2015;142:1418–1430. doi: 10.1242/dev.109496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013;13:11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- Baker JC, Harland RM. A novel mesoderm inducer, Madr2, functions in the activin signal transduction pathway. Genes Dev. 1996;10:1880–1889. doi: 10.1101/gad.10.15.1880. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Blauwkamp TA, Nigam S, Ardehali R, Weissman IL, Nusse R. Endogenous Wnt signalling in human embryonic stem cells generates an equilibrium of distinct lineage-specified progenitors. Nat Commun. 2012;3:1070. doi: 10.1038/ncomms2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan J, Lu CC, Norris DP, Rodriguez TA, Beddington RS, Robertson EJ. Nodal signalling in the epiblast patterns the early mouse embryo. Nature. 2001;411:965–969. doi: 10.1038/35082103. [DOI] [PubMed] [Google Scholar]
- Chen X, Weisberg E, Fridmacher V, Watanabe M, Naco G, Whitman M. Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature. 1997;389:85–89. doi: 10.1038/38008. [DOI] [PubMed] [Google Scholar]
- Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordenonsi M, Dupont S, Maretto S, Insinga A, Imbriano C, Piccolo S. Links between tumor suppressors: p53 is required for TGF-beta gene responses by cooperating with Smads. Cell. 2003;113:301–314. doi: 10.1016/s0092-8674(03)00308-8. [DOI] [PubMed] [Google Scholar]
- Costello I, Nowotschin S, Sun X, Mould AW, Hadjantonakis AK, Bikoff EK, Robertson EJ. Lhx1 functions together with Otx2, Foxa2, and Ldb1 to govern anterior mesendoderm, node, and midline development. Genes Dev. 2015;29:2108–2122. doi: 10.1101/gad.268979.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, Ye Z, Kim A, Rajagopal N, Xie W, et al. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518:331–336. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr, Butel JS, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- Dotsch V, Bernassola F, Coutandin D, Candi E, Melino G. p63 and p73, the ancestors of p53. Cold Spring Harb Perspect Biol. 2010;2:a004887. doi: 10.1101/cshperspect.a004887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- Erster S, Palacios G, Rosenquist T, Chang C, Moll UM. Deregulated expression of DeltaNp73alpha causes early embryonic lethality. Cell Death Differ. 2006;13:170–173. doi: 10.1038/sj.cdd.4401743. [DOI] [PubMed] [Google Scholar]
- Estaras C, Benner C, Jones KA. SMADs and YAP compete to control elongation of beta-catenin:LEF-1-recruited RNAPII during hESC differentiation. Mol Cell. 2015;58:780–793. doi: 10.1016/j.molcel.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S, Marin O, Pagano MA, Meggio F, Hess D, El-Shemerly M, Krystyniak A, Pinna LA. Aurora-A site specificity: a study with synthetic peptide substrates. Biochem J. 2005;390:293–302. doi: 10.1042/BJ20050343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores ER, Tsai KY, Crowley D, Sengupta S, Yang A, McKeon F, Jacks T. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature. 2002;416:560–564. doi: 10.1038/416560a. [DOI] [PubMed] [Google Scholar]
- Funa NS, Schachter KA, Lerdrup M, Ekberg J, Hess K, Dietrich N, Honore C, Hansen K, Semb H. beta-Catenin Regulates Primitive Streak Induction through Collaborative Interactions with SMAD2/SMAD3 and OCT4. Cell Stem Cell. 2015 doi: 10.1016/j.stem.2015.03.008. [DOI] [PubMed] [Google Scholar]
- Gonzalez F, Zhu Z, Shi ZD, Lelli K, Verma N, Li QV, Huangfu D. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell. 2014;15:215–226. doi: 10.1016/j.stem.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S. Suppression of induced pluripotent stem cell generation by the p53−p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Feng Z, Teresky AK, Levine AJ. p53 regulates maternal reproduction through LIF. Nature. 2007;450:721–724. doi: 10.1038/nature05993. [DOI] [PubMed] [Google Scholar]
- Huttinger-Kirchhof N, Cam H, Griesmann H, Hofmann L, Beitzinger M, Stiewe T. The p53 family inhibitor DeltaNp73 interferes with multiple developmental programs. Cell Death Differ. 2006;13:174–177. doi: 10.1038/sj.cdd.4401809. [DOI] [PubMed] [Google Scholar]
- Jiang W, Zhang D, Bursac N, Zhang Y. WNT3 is a biomarker capable of predicting the definitive endoderm differentiation potential of hESCs. Stem Cell Reports. 2013;1:46–52. doi: 10.1016/j.stemcr.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Izpisua Belmonte JC. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase T, Ohki R, Shibata T, Tsutsumi S, Kamimura N, Inazawa J, Ohta T, Ichikawa H, Aburatani H, Tashiro F, et al. PH domain-only protein PHLDA3 is a p53-regulated repressor of Akt. Cell. 2009;136:535–550. doi: 10.1016/j.cell.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Kurek D, Neagu A, Tastemel M, Tuysuz N, Lehmann J, van de Werken HJ, Philipsen S, van der Linden R, Maas A, van IWF. Endogenous WNT signals mediate BMP-induced and spontaneous differentiation of epiblast stem cells and human embryonic stem cells. Stem Cell Reports. 2015;4:114–128. doi: 10.1016/j.stemcr.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé E, Letamendia A, Attisano L. Association of Smads with lymphoid enhancer binding factor 1/T cell-specific factor mediates cooperative signaling by the transforming growth factor-beta and wnt pathways. Proc Natl Acad Sci U S A. 2000;97:8358–8363. doi: 10.1073/pnas.150152697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D, Levine A. p53 Research: the past thirty years and the next thirty years. Cold Spring Harbor perspectives in biology. 2010;2:a000893. doi: 10.1101/cshperspect.a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DF, Su J, Ang YS, Carvajal-Vergara X, Mulero-Navarro S, Pereira CF, Gingold J, Wang HL, Zhao R, Sevilla A, et al. Regulation of embryonic and induced pluripotency by aurora kinase-p53 signaling. Cell Stem Cell. 2012;11:179–194. doi: 10.1016/j.stem.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Li M, Michalowski AM, Zhang X, Liao H, Chen L, Xu Y, Wu X, Huang J. A genomewide study identifies the Wnt signaling pathway as a major target of p53 in murine embryonic stem cells. Proc Natl Acad Sci U S A. 2010;107:69–74. doi: 10.1073/pnas.0909734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Chao C, Saito S, Mazur SJ, Murphy ME, Appella E, Xu Y. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol. 2005;7:165–171. doi: 10.1038/ncb1211. [DOI] [PubMed] [Google Scholar]
- Liu F, Pouponnot C, Massagué J. Dual role of the Smad4/DPC4 tumor suppressor in TGFbeta-inducible transcriptional complexes. Genes Dev. 1997;11:3157–3167. doi: 10.1101/gad.11.23.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Wakamiya M, Shea MJ, Albrecht U, Behringer RR, Bradley A. Requirement for Wnt3 in vertebrate axis formation. Nat Genet. 1999;22:361–365. doi: 10.1038/11932. [DOI] [PubMed] [Google Scholar]
- Lutzker SG, Levine AJ. A functionally inactive p53 protein in teratocarcinoma cells is activated by either DNA damage or cellular differentiation. Nat Med. 1996;2:804–810. doi: 10.1038/nm0796-804. [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascetti VL, Pedersen RA. Human-Mouse Chimerism Validates Human Stem Cell Pluripotency. Cell Stem Cell. 2016;18:67–72. doi: 10.1016/j.stem.2015.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J. TGFbeta signalling in context. Nat Rev Mol Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill BJ, Pasolli HA, Polak L, Rendl M, Garcia-Garcia MJ, Anderson KV, Fuchs E. Tcf3: a transcriptional regulator of axis induction in the early embryo. Development. 2004;131:263–274. doi: 10.1242/dev.00935. [DOI] [PubMed] [Google Scholar]
- Migeotte I, Grego-Bessa J, Anderson KV. Rac1 mediates morphogenetic responses to intercellular signals in the gastrulating mouse embryo. Development. 2011;138:3011–3020. doi: 10.1242/dev.059766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- Niida A, Hiroko T, Kasai M, Furukawa Y, Nakamura Y, Suzuki Y, Sugano S, Akiyama T. DKK1, a negative regulator of Wnt signaling, is a target of the beta-catenin/TCF pathway. Oncogene. 2004;23:8520–8526. doi: 10.1038/sj.onc.1207892. [DOI] [PubMed] [Google Scholar]
- Nishikawa SI, Nishikawa S, Hirashima M, Matsuyoshi N, Kodama H. Progressive lineage analysis by cell sorting and culture identifies FLK1+VE-cadherin+ cells at a diverging point of endothelial and hemopoietic lineages. Development. 1998;125:1747–1757. doi: 10.1242/dev.125.9.1747. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Beach D. Cyclin G is a transcriptional target of the p53 tumor suppressor protein. Embo j. 1994;13:4816–4822. doi: 10.1002/j.1460-2075.1994.tb06807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauklin S, Vallier L. The cell-cycle state of stem cells determines cell fate propensity. Cell. 2013;155:135–147. doi: 10.1016/j.cell.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nature protocols. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid CD, Zhang Y, Sheets MD, Kessler DS. Transcriptional integration of Wnt and Nodal pathways in establishment of the Spemann organizer. Dev Biol. 2012;368:231–241. doi: 10.1016/j.ydbio.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- Rivera-Perez JA, Magnuson T. Primitive streak formation in mice is preceded by localized activation of Brachyury and Wnt3. Dev Biol. 2005;288:363–371. doi: 10.1016/j.ydbio.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Schmid P, Lorenz A, Hameister H, Montenarh M. Expression of p53 during mouse embryogenesis. Development. 1991;113:857–865. doi: 10.1242/dev.113.3.857. [DOI] [PubMed] [Google Scholar]
- Shigeta M, Ohtsuka S, Nishikawa-Torikai S, Yamane M, Fujii S, Murakami K, Niwa H. Maintenance of pluripotency in mouse ES cells without Trp53. Scientific reports. 2013;3:2944. doi: 10.1038/srep02944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallier L, Reynolds D, Pedersen RA. Nodal inhibits differentiation of human embryonic stem cells along the neuroectodermal default pathway. Dev Biol. 2004;275:403–421. doi: 10.1016/j.ydbio.2004.08.031. [DOI] [PubMed] [Google Scholar]
- Viotti M, Nowotschin S, Hadjantonakis AK. SOX17 links gut endoderm morphogenesis and germ layer segregation. Nat Cell Biol. 2014;16:1146–1156. doi: 10.1038/ncb3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Wallingford JB, Seufert DW, Virta VC, Vize PD. p53 activity is essential for normal development in Xenopus. Curr Biol. 1997;7:747–757. doi: 10.1016/s0960-9822(06)00333-2. [DOI] [PubMed] [Google Scholar]
- Weinstein M, Yang X, Li C, Xu X, Gotay J, Deng CX. Failure of egg cylinder elongation and mesoderm induction in mouse embryos lacking the tumor suppressor smad2. Proc Natl Acad Sci U S A. 1998;95:9378–9383. doi: 10.1073/pnas.95.16.9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482:221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DG, Bhatt S, Herrmann BG. Expression pattern of the mouse T gene and its role in mesoderm formation. Nature. 1990;343:657–659. doi: 10.1038/343657a0. [DOI] [PubMed] [Google Scholar]
- Wray J, Kalkan T, Gomez-Lopez S, Eckardt D, Cook A, Kemler R, Smith A. Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat Cell Biol. 2011;13:838–845. doi: 10.1038/ncb2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Q, Wang Z, Zaromytidou AI, Zhang XH, Chow-Tsang LF, Liu JX, Kim H, Barlas A, Manova-Todorova K, Kaartinen V, et al. A poised chromatin platform for TGF-beta access to master regulators. Cell. 2011;147:1511–1524. doi: 10.1016/j.cell.2011.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Wiesmann M, Rohan M, Chan V, Jefferson AB, Guo L, Sakamoto D, Caothien RH, Fuller JH, Reinhard C, et al. Elevated expression of axin2 and hnkd mRNA provides evidence that Wnt/beta -catenin signaling is activated in human colon tumors. Proc Natl Acad Sci U S A. 2001;98:14973–14978. doi: 10.1073/pnas.261574498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, Vagner C, Bonnet H, Dikkes P, Sharpe A, et al. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. 2000;404:99–103. doi: 10.1038/35003607. [DOI] [PubMed] [Google Scholar]
- Yoon SJ, Wills AE, Chuong E, Gupta R, Baker JC. HEB and E2A function as SMAD/FOXH1 cofactors. Genes Dev. 2011;25:1654–1661. doi: 10.1101/gad.16800511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Li J, Tan Z, Wang C, Liu T, Chen L, Yong J, Jiang W, Sun X, Du L, et al. Short-term BMP-4 treatment initiates mesoderm induction in human embryonic stem cells. Blood. 2008;111:1933–1941. doi: 10.1182/blood-2007-02-074120. [DOI] [PubMed] [Google Scholar]
- Zhou X, Sasaki H, Lowe L, Hogan BL, Kuehn MR. Nodal is a novel TGF-beta-like gene expressed in the mouse node during gastrulation. Nature. 1993;361:543–547. doi: 10.1038/361543a0. [DOI] [PubMed] [Google Scholar]
- Zhu J, Dou Z, Sammons MA, Levine AJ, Berger SL. Lysine methylation represses p53 activity in teratocarcinoma cancer cells. Proc Natl Acad Sci U S A. 2016 doi: 10.1073/pnas.1610387113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Gonzalez F, Huangfu D. The iCRISPR platform for rapid genome editing in human pluripotent stem cells. Methods Enzymol. 2014;546:215–250. doi: 10.1016/B978-0-12-801185-0.00011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilfou JT, Lowe SW. Tumor suppressive functions of p53. Cold Spring Harbor perspectives in biology. 2009;1:a001883. doi: 10.1101/cshperspect.a001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.